Abstract
A key quantity in the analysis of structured populations is the parameter K, which describes the number of subpopulations that make up the total population. Inference of K ideally proceeds via the model evidence, which is equivalent to the likelihood of the model. However, the evidence in favor of a particular value of K cannot usually be computed exactly, and instead programs such as Structure make use of heuristic estimators to approximate this quantity. We show—using simulated data sets small enough that the true evidence can be computed exactly—that these heuristics often fail to estimate the true evidence and that this can lead to incorrect conclusions about K. Our proposed solution is to use thermodynamic integration (TI) to estimate the model evidence. After outlining the TI methodology we demonstrate the effectiveness of this approach, using a range of simulated data sets. We find that TI can be used to obtain estimates of the model evidence that are more accurate and precise than those based on heuristics. Furthermore, estimates of K based on these values are found to be more reliable than those based on a suite of model comparison statistics. Finally, we test our solution in a reanalysis of a white-footed mouse data set. The TI methodology is implemented for models both with and without admixture in the software MavericK1.0.
Keywords: population structure, K, model evidence, thermodynamic integration, model comparison
THE detection and characterization of population structure is one of the cornerstones of modern population genetics. Ever since Wright (1949) and his contemporaries (Malécot 1948) it has been recognized that genetic samples obtained from a large population may be better understood as a series of draws from multiple partially isolated subpopulations or demes. While traditional methods (such as those based on the fixation index, ) assume that the allocation of individuals to demes is known a priori, many modern programs such as Structure (Pritchard et al. 2000; Falush et al. 2003a, 2007; Hubisz et al. 2009) take a different approach, attempting to infer the group allocation from the observed data. What makes this possible is the simple genetic mixture modeling framework used by these programs, together with the efficiency of Markov chain Monte Carlo (MCMC) methods for sampling from this broad class of models.
However, even within the flexible framework of Bayesian mixture models, the number of demes (denoted K) is difficult to ascertain. While the allocation of individuals to demes is a parameter within a particular model, the value of K is fixed for a given mixture model, and so the problem of estimating K involves a comparison between models. One of the most common ways of comparing between models in a Bayesian setting is through the model evidence, defined as the probability of the observed data under the model (equivalently the likelihood of the model). This quantity can be estimated for a range of K, and the model with the highest evidence value can then become the focus of our analysis. However, there are two potential issues with this approach. The first one is philosophical and revolves around the idea that there is a single true value of K that we can estimate from the data. In reality populations are rarely divided into discrete subpopulations, and so the idea of a single true value of K does not strictly apply. This does not mean that K cannot be a useful quantity, but it is better viewed as a flexible parameter that describes just one point on a continuously varying scale of population structure. This flexible interpretation of K has been advocated by a number of previous authors (Raj et al. 2014; Jombart and Collins 2015), including the authors of the Structure program (Pritchard et al. 2010).
The second issue is purely statistical—computing the model evidence in complex, multidimensional models is not straightforward. For this reason it is common to resort to heuristic estimators of the true evidence. These heuristics tend to have some direct mathematical connection to the model evidence, but also make certain simplifying assumptions in their derivation. For example, in the original article on which Structure is based, Pritchard et al. (2000) comment on the difficulties in obtaining the model evidence directly and instead opt for an ad hoc procedure in which a heuristic (denoted here) is used as an approximation of The derivation of this statistic rests on certain simplifying assumptions, and the authors are careful to emphasize that these assumptions are “dubious.”
Here we focus on the latter problem: reliable estimation of the model evidence. Rather than resorting to heuristics, what we want is a direct way of estimating the model evidence that is both accurate and straightforward to implement. As noted by Gelman and Meng (1998), such a method already exists and has been known in the physical sciences for some time. This method—referred to in the statistical literature as thermodynamic integration (TI)—uses the output of several closely related MCMC chains to obtain a direct estimate of the evidence. Crucially, this is not just another heuristic. Rather, it is a true statistical estimator that can be evaluated to an arbitrary degree of precision by simply increasing the number of MCMC iterations used in the calculation. The TI methodology was introduced into population genetics by Lartillot and Philippe (2006) and has since been applied to a range of problems in phylogenetics and coalescent theory, including comparing models of demographics (Baele et al. 2012), migration (Beerli and Palczewski 2010), relaxed molecular clocks (Lepage et al. 2007), and sequence evolution (Blanquart and Lartillot 2006).
In the remainder of this article we demonstrate the effectiveness of TI as a method for estimating K in simple genetic mixture models. For small data sets we find that the TI estimator is several orders of magnitude more accurate and precise than the estimator for the same computational effort. We also explore the ability of different statistics to correctly estimate K for larger data sets, finding that TI outperforms Evanno’s (Evanno et al. 2005), the Akaike information criterion (AIC), the Bayesian information criterion (BIC), and the deviance information criterion (DIC). Finally we reanalyze data from an earlier study on the genetic structure of white-footed mouse populations in New York City (Munshi-South and Kharchenko 2010b). All of the methods described here are made available through the program MavericK (www.bobverity.com/MavericK).
Materials and Methods
Evidence and Bayes factors
In a Bayesian setting the problem of deciding between competing models can be addressed using Bayes’ rule. The posterior probability of the model given the observed data can be written
| (1) |
The quantity —the probability of the observed data given just the model —is defined as the model evidence.
The ratio of the evidence between competing models, known as the Bayes factor, can be used to measure the strength of evidence in favor of one model over another. Bayes factors can be used on their own, or they can be combined with priors on the different models to arrive at the posterior odds:
| (2) |
A large Bayes factor in (2) provides evidence in favor of model over model whereas a small Bayes factor provides evidence in favor of model over model A useful scale for interpreting Bayes factors can be found in Kass and Raftery (1995); however, it is important to note that this scale is meaningful only if priors are chosen appropriately (see Discussion).
The problem of estimating the number of demes in a structured population can be understood in this light: If we let denote a genetic mixture model in which K demes are assumed, then the problem of estimating K becomes one of comparing between different models. Ideally we want to solve this problem using the exact model evidence, Unfortunately, however, calculating the model evidence in complex, multidimensional models is not straightforward, as most of the time we cannot write down the probability of the data under the model without also conditioning on certain known parameters, denoted Obtaining the evidence from the likelihood requires that we integrate over a prior on
| (3) |
It is this integration step that makes calculating the model evidence difficult in practice. In genetic mixture models might represent the allele frequencies in all K demes, perhaps alongside some additional admixture parameters, making the integral in (3) extremely high dimensional (a 100-dimensional integral would not be uncommon). For this reason it makes practical sense to turn to numerical methods or heuristic approximations.
Estimating and approximating the evidence
Perhaps the simplest way of estimating the model evidence is through the harmonic mean estimator, (Newton and Raftery 1994),
| (4) |
where for denotes a series of draws from the posterior distribution of Part of the appeal of this estimator is its simplicity—it is straightforward to calculate from the output of a single MCMC run. As an example, the program Structurama (Huelsenbeck and Andolfatto 2007; Huelsenbeck et al. 2011), which contains within it a version of the basic Structure model, has an option for using to estimate the model evidence (we note that this is not the primary purpose of Structurama, which also implements a Dirichlet process model). However, in spite of its intuitive appeal, the harmonic mean estimator has been widely criticized due to its instability; has been found to be very sensitive to the choice of prior, often being dominated by the reciprocal of a few small values (Neal 1994; Raftery et al. 2006).
To avoid some of the problems inherent in the harmonic mean estimator, the approach taken by Pritchard et al. (2000) was to define the heuristic estimator (our notation) as
| (5) |
where and are simple statistics that can be calculated from the posterior draws (see Supplemental Material, File S1 for a more detailed derivation of this and other statistics). The key assumption that underpins this heuristic is that the posterior deviance is approximately normally distributed, which may or may not be true in practice. is usually evaluated for a range of K, and the smallest (corresponding to the largest evidence) is used as an indication of the most likely model. Alternatively, these values can be transformed out of log space to provide direct estimates of the evidence that, once normalized, can be used to approximate the full posterior distribution of K:
| (6) |
This procedure is rarely carried out in practice, despite being recommended in the Structure software documentation (Pritchard et al. 2010).
Thermodynamic integration
The TI estimator differs fundamentally from in the sense that it is not a heuristic estimator—it makes no simplifying assumptions about the distribution of the likelihood. It also differs from in that it is well behaved, having finite and quantifiable variance. The approach centers around the “power posterior” (Friel and Pettitt 2008), defined as follows:
| (7) |
This is nothing more than the ordinary posterior distribution of but with the likelihood raised to the power β [the value is a normalizing constant that ensures the distribution integrates to 1]. In the same way that we can design an MCMC algorithm to draw from the posterior distribution of we can design a similar algorithm to draw from the power posterior distribution. Details of the MCMC steps are given in the Appendix for models both with and without admixture. The resulting draws from the power posterior are written where the superscript β indicates the power used when generating the draws. The TI methodology then proceeds in two simple steps. First, we calculate the mean log-likelihood of the power posterior draws:
| (8) |
[It is important to note that the notation refers to values drawn from the power posterior with power β; it does not indicate that the values of (or these likelihoods) are raised to the power β]. This step is repeated for a range of values for spanning the interval Second, we calculate the area under the curve made by the values using a suitable numerical integration scheme, such as the trapezoidal rule:
| (9) |
The value is the TI estimator of the model evidence (see File S1 for a more detailed derivation). It can be seen that is straightforward to calculate, although it does require us to run multiple MCMC chains to obtain a single estimate of the evidence, making it computationally intensive. On the other hand, the method has greater precision than some alternatives that can be calculated faster. In our comparisons this trade-off was taken into account by using the same number of MCMC iterations for all methods.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article and are available upon request.
Results
Comparison against the exact model evidence
Our first objective was to measure the accuracy and precision of different estimators of the model evidence against the exact value, obtained by brute force (see Appendix). The difficulty in calculating the exact model evidence meant that this was possible only for very small simulated data sets of diploid individuals at loci, generated from the same without-admixture model implemented in the program Structure2.3.4. A total of 1000 simulated data sets were produced, with K ranging from 1 to 10 (100 simulations each) and with for each of alleles (see Table A1 for a list of parameters). Each data set was then analyzed using the program MavericK1.0. This program is written in C++ and was designed specifically to carry out TI for structured populations via the algorithms described in the Appendix. In addition, MavericK1.0 implements certain features that lead to efficient and reliable exploration of the posterior, including solving the label switching problem via the method of Stephens (2000) (see File S2 for further details of the main algorithm). The output of MavericK1.0 includes values of and the TI estimator Calculation of was compared extensively against Structure2.3.4 to ensure agreement. For the TI estimator the number of “rungs” used (the value of r) was set to 50, while for and the analysis was repeated 50 times to obtain a global mean and standard error over replicates, thereby ensuring that the same computational effort was expended for all methods. A total of 10,000 samples were obtained from the posterior distribution in each MCMC analysis, with a burn-in of 1000 iterations.
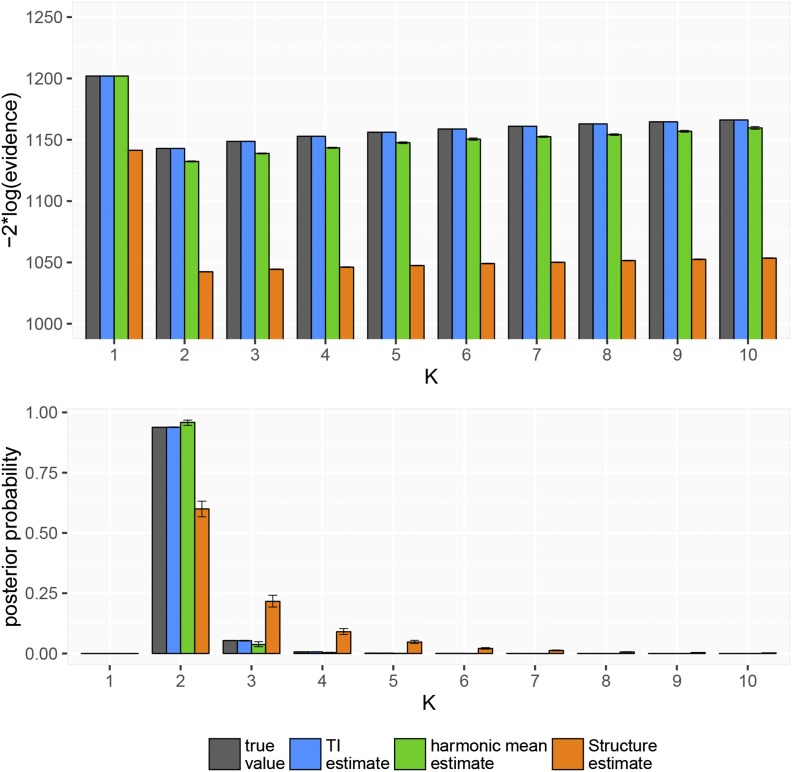
Figure 1 shows the results of one such analysis, in which the true number of demes was
Figure 1.
True and estimated values of the model evidence in log space and in linear space. Error bars give 95% confidence intervals around estimates.
It can be seen that both and are negatively biased in this example, leading to estimates of that are smaller than the true value. Any bias that is constant over K goes away after transforming to a linear scale and normalizing; however, and particularly still give poor estimates of the true posterior distribution.
The accuracy and precision of the different estimators was evaluated across all 1000 simulated data sets in the form of the mean signed difference (MSD) and the mean absolute difference (MAD). The MSD measures the average difference between the true and estimated values and hence can be considered a measure of bias, while the MAD measures the average absolute difference and hence is influenced by both the bias and the precision of the estimator (small values represent estimates that are both accurate and precise). Results are given in Table 1, broken down by the value of K used in the inference step (a more detailed breakdown can be found in Table S1).
Table 1. Accuracy of estimation methods compared with the exact model evidence.
| −2 log(evidence) | Normalized evidence | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MSD | MAD | MSD | MAD | |||||||||
| K | ||||||||||||
| 1 | 0.00e + 00 | 0.00 | 28.72 | 0.00e + 00 | 0.00 | 28.72 | 7.03e-06 | −6.50e-03 | −2.76e-02 | 4.59e-05 | 6.50e-03 | 2.88e-02 |
| 2 | −1.54e-03 | 2.52 | 42.65 | 6.99e-03 | 2.52 | 42.65 | 6.67e-07 | 7.76e-03 | 4.93e-02 | 2.37e-04 | 1.41e-02 | 6.49e-02 |
| 3 | −1.95e-03 | 3.51 | 46.65 | 7.71e-03 | 3.51 | 46.65 | −6.00e-05 | 3.19e-02 | 8.96e-02 | 5.19e-04 | 3.96e-02 | 1.37e-01 |
| 4 | −1.96e-03 | 3.60 | 46.74 | 7.17e-03 | 3.60 | 46.74 | −8.42e-08 | 4.07e-02 | 1.37e-01 | 6.25e-04 | 5.54e-02 | 2.27e-01 |
| 5 | −1.61e-03 | 3.37 | 45.59 | 6.70e-03 | 3.37 | 45.59 | −9.85e-06 | 3.23e-02 | 1.37e-02 | 6.09e-04 | 5.16e-02 | 1.27e-01 |
| 6 | −1.39e-03 | 3.10 | 44.72 | 6.73e-03 | 3.10 | 44.72 | 1.59e-05 | 1.42e-02 | −5.46e-02 | 6.62e-04 | 4.08e-02 | 8.33e-02 |
| 7 | −1.47e-03 | 2.85 | 44.78 | 6.47e-03 | 2.85 | 44.78 | −7.72e-06 | −6.09e-03 | −6.99e-02 | 6.67e-04 | 3.17e-02 | 7.97e-02 |
| 8 | −1.18e-03 | 2.61 | 45.11 | 5.94e-03 | 2.61 | 45.11 | 2.01e-05 | −2.56e-02 | −6.74e-02 | 6.28e-04 | 3.17e-02 | 8.03e-02 |
| 9 | −1.21e-03 | 2.43 | 45.53 | 5.99e-03 | 2.43 | 45.53 | 4.17e-05 | −4.09e-02 | −5.11e-02 | 6.22e-04 | 4.23e-02 | 7.94e-02 |
| 10 | −1.44e-03 | 2.26 | 45.90 | 5.77e-03 | 2.26 | 45.90 | −2.17e-05 | −5.30e-02 | −2.08e-02 | 5.79e-04 | 5.31e-02 | 9.44e-02 |
| Mean | −1.38e-03 | 2.63 | 43.64 | 5.95e-03 | 2.63 | 43.64 | −1.41e-06 | −5.23e-04 | −2.12e-04 | 5.19e-04 | 3.67e-02 | 1.00e-01 |
Shown are mean signed difference (MSD) and mean absolute difference (MAD) of various estimation methods compared with the exact value, obtained by brute force. Formulas for and are given in Equations 9, 4, and 5, respectively. Values are shown in log space (columns 2–7) and linear space after exponentiating and normalizing to sum to 1 (columns 8–13). Values of K here denote the value used in the inference step, with each row being an average over 1000 simulations (a more detailed breakdown can be found in Table S1).
It can be seen that the average MAD of the estimator after normalizing is ∼0.1, while the MAD of the estimator is for the same computational effort. The harmonic mean estimator is intermediate between these values, differing from the true evidence by ∼0.04 on average. Based on these results we would expect estimates of the posterior distribution of K made using or to be qualitatively different from the true posterior distribution.
Accuracy for larger data sets
Although the results in Table 1 are suggestive of a weakness in heuristic estimators, we are limited here to looking at small data sets in which the exact model evidence can be calculated by brute force. It is plausible based on these results that the bias in and could be amplified in small data sets due to a lack of information and would cease to be a problem if more data were available. Here we therefore use larger simulated data sets to address the question of whether the TI method produces improvements that would be of practical importance. Although we cannot calculate the true evidence by brute force here, the advantage of using simulated data sets is that we can generate observations from the exact model used in the inference step and for a known value of K. We can then measure the proportion of times that the true value of K is correctly identified. As well as comparing the estimators and in which the smallest value of the estimator indicates the most likely model, we also compared Evanno’s (Evanno et al. 2005), in which the largest value indicates the point of maximum curvature of and the AIC, BIC, and DIC statistics, in which the smallest value indicates the best-fitting model. Values of the DIC were calculated using the method of Spiegelhalter et al. (2002) () as well as the method of Gelman et al. (2004) (). To ensure that our results were not driven by a lack of information, larger data sets of diploid individuals at 20, and 50 loci were generated from the same without-admixture model used above. As before, 1000 simulated data sets were produced with K ranging from 1 to 10 (100 simulations each). MavericK1.0 was run under the without-admixture model with 1000 burn-in iterations and 10,000 sampling iterations. For the TI estimator 50 rungs were used, and for and the analysis was repeated 50 times.
Table 2 gives the proportion of times that the correct value of K was identified by each of the methods. It can be seen that the TI-based method of choosing K provided reliable results across all simulated data sets. Estimates of K based on were less reliable, although still reasonable when the number of loci was large, whereas estimates based on were generally not reliable and particularly poor when the number of loci was small. This appears to be due to the well-documented tendency of to continually increase with larger values of K (Pritchard et al. 2010), also giving the false impression that is highly accurate when in this example. Evanno’s mitigated this to some extent, but still did not provide consistently reliable results (note that cannot be calculated on the smallest or largest K in any analysis as a consequence of how it is derived). Of the model comparison statistics the AIC was the most consistently reliable, providing accurate estimates across a range of simulations.
Table 2. Percentage times K correctly identified.
| K | AIC | BIC | ||||||
|---|---|---|---|---|---|---|---|---|
| 10 loci | ||||||||
| 1 | 100.0 | 76.0 | 0.0 | — | 88.0 | 100.0 | 0.0 | 100.0 |
| 2 | 100.0 | 83.0 | 0.0 | 100.0 | 99.0 | 100.0 | 0.0 | 98.0 |
| 3 | 100.0 | 83.0 | 0.0 | 76.0 | 100.0 | 100.0 | 0.0 | 94.0 |
| 4 | 100.0 | 77.0 | 0.0 | 67.0 | 95.0 | 99.0 | 0.0 | 89.0 |
| 5 | 100.0 | 71.0 | 0.0 | 58.0 | 98.0 | 92.0 | 0.0 | 90.0 |
| 6 | 100.0 | 72.0 | 1.0 | 45.0 | 96.0 | 75.0 | 0.0 | 90.0 |
| 7 | 100.0 | 65.0 | 3.0 | 43.0 | 96.0 | 35.0 | 0.0 | 94.0 |
| 8 | 100.0 | 46.0 | 6.0 | 42.0 | 93.0 | 7.0 | 0.0 | 84.0 |
| 9 | 100.0 | 57.0 | 17.0 | 14.0 | 96.0 | 1.0 | 0.0 | 94.0 |
| 10 | 100.0 | 100.0 | 100.0 | — | 96.0 | 0.0 | 100.0 | 99.0 |
| Mean | 100.0 | 73.0 | 12.7 | 55.6 | 95.7 | 60.9 | 10.0 | 93.2 |
| 20 loci | ||||||||
| 1 | 100.0 | 100.0 | 0.0 | — | 95.0 | 100.0 | 0.0 | 89.0 |
| 2 | 100.0 | 100.0 | 2.0 | 100.0 | 100.0 | 100.0 | 0.0 | 86.0 |
| 3 | 100.0 | 100.0 | 64.0 | 95.0 | 100.0 | 100.0 | 10.0 | 92.0 |
| 4 | 100.0 | 99.0 | 85.0 | 93.0 | 100.0 | 100.0 | 44.0 | 97.0 |
| 5 | 100.0 | 98.0 | 90.0 | 97.0 | 100.0 | 100.0 | 70.0 | 100.0 |
| 6 | 100.0 | 92.0 | 88.0 | 94.0 | 100.0 | 100.0 | 78.0 | 100.0 |
| 7 | 100.0 | 94.0 | 85.0 | 93.0 | 100.0 | 94.0 | 84.0 | 99.0 |
| 8 | 100.0 | 91.0 | 87.0 | 97.0 | 100.0 | 73.0 | 78.0 | 100.0 |
| 9 | 100.0 | 87.0 | 82.0 | 88.0 | 100.0 | 26.0 | 70.0 | 97.0 |
| 10 | 100.0 | 100.0 | 100.0 | — | 100.0 | 3.0 | 100.0 | 100.0 |
| Mean | 100.0 | 96.1 | 68.3 | 94.6 | 99.5 | 79.6 | 53.4 | 96.0 |
| 50 loci | ||||||||
| 1 | 100.0 | 100.0 | 45.0 | — | 100.0 | 100.0 | 17.0 | 25.0 |
| 2 | 100.0 | 99.0 | 21.0 | 100.0 | 100.0 | 100.0 | 100.0 | 19.0 |
| 3 | 100.0 | 90.0 | 30.0 | 99.0 | 100.0 | 100.0 | 100.0 | 17.0 |
| 4 | 100.0 | 97.0 | 32.0 | 100.0 | 100.0 | 100.0 | 100.0 | 23.0 |
| 5 | 100.0 | 98.0 | 28.0 | 100.0 | 100.0 | 100.0 | 100.0 | 20.0 |
| 6 | 100.0 | 97.0 | 42.0 | 100.0 | 100.0 | 100.0 | 100.0 | 27.0 |
| 7 | 100.0 | 98.0 | 47.0 | 100.0 | 100.0 | 100.0 | 100.0 | 30.0 |
| 8 | 100.0 | 95.0 | 58.0 | 99.0 | 100.0 | 95.0 | 100.0 | 47.0 |
| 9 | 100.0 | 96.0 | 63.0 | 99.0 | 100.0 | 83.0 | 100.0 | 57.0 |
| 10 | 100.0 | 100.0 | 99.0 | — | 100.0 | 45.0 | 100.0 | 100.0 |
| Mean | 100.0 | 97.0 | 46.5 | 99.6 | 100.0 | 92.3 | 91.7 | 36.5 |
Shown is the percentage times K is correctly identified by each method, broken down by the value of K used when generating the data. Formulas for and are given in Equations 9, 4, and 5, respectively, while formulas for AIC, BIC, and are given in File S1 Equations 40, 43, 46, and 47, respectively. The formula for can be found in Evanno et al. (2005).
Returning to the question of whether the inaccuracy in and in Table 1 was driven by a lack of information, it can be seen from Table 2 that the quantity of data certainly plays a role. However, the fact that TI provides reliable estimates across the range of simulations indicates that there is sufficient signal in the data to detect the value of K even in relatively small data sets. Thus, the increased precision of the TI approach is of practical as well as theoretical importance.
Reanalysis of white-footed mouse data
Our main reason for focusing on simulated data sets above is for the purposes of comparing different statistical methods under very controlled circumstances. By simulating data from the exact model used in the inference step we can tease apart the issue of whether inaccuracies are due to statistical problems or simply a lack of model fit to the data (the latter being ruled out). However, ultimately our interest lies in real-world analyses of population structure. Here the parameter K has a less literal meaning and should be seen as a convenient way of summarizing the structure in the available data, rather than as an exact description of the number of demes.
To test MavericK1.0 in a realistic setting we reanalyzed data from a study by Munshi-South and Kharchenko (2010b), made available through the Dryad digital repository (Munshi-South and Kharchenko 2010a). The data consist of diploid genotypes at 18 putatively neutral microsatellite loci in 312 white-footed mice (Peromyscus leucopus), sampled from 15 distinct locations in and around New York City (see the original article for details). White-footed mice are known to be urban adaptors, and so the original study investigated the effects of urbanization and habitat fragmentation on the mouse population, concluding that there has been pervasive genetic differentiation and the emergence of strong population structure. The authors carried out a range of statistical tests, including but not limited to an analysis with Structure2.3 under the admixture model with correlated allele frequencies and with α inferred as part of the MCMC. They explored values of K from 1 to 20 (repeating each analysis 10 times), finding that the mean peaked at while Evanno’s had peaks at and although generally the distribution of this statistic was complex (see figure 2 in Munshi-South and Kharchenko 2010b).
We carried out a similar analysis in MavericK1.0, using TI to estimate the evidence for K as well as using and We used the same admixture model as in the original study, in which α is inferred as part of the MCMC; however, the correlated allele frequencies model is not implemented in MavericK1.0 and so we assumed a model of independent allele frequencies. For this reason our results are not directly comparable with those of the original study, although our assumptions are broadly similar. We explored K from 1 to 20. When carrying out TI we used rungs, and for the other estimation methods we took the mean and standard error over 21 replicates. For each MCMC analysis we ran 10 chains, each with 10,000 burn-in iterations and 50,000 sampling iterations, before trimming and merging chains to obtain 500,000 sampling iterations (we found that this gave better results than running one long chain).
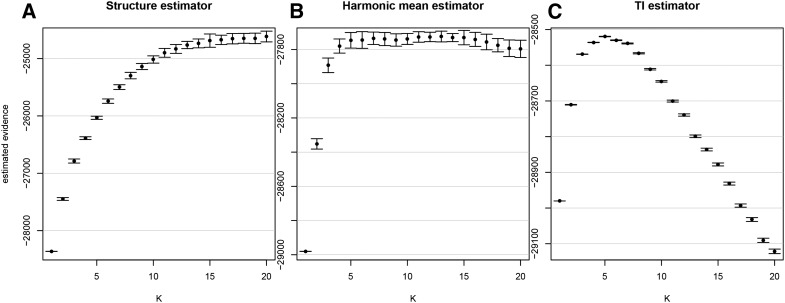
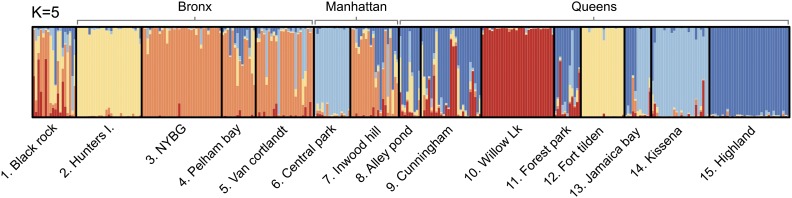
The results of this analysis are shown in Figure 2. It can be seen that increases smoothly with K, in a trend similar to that found by Munshi-South and Kharchenko (2010b), the difference being that we find no peak at The harmonic mean estimator increases rapidly until but at this point saturates and cannot distinguish between higher values of K. In contrast to both of these statistics, the TI estimator has a strong peak at with narrow confidence intervals. Based on the arguments presented above we conclude that this is the most accurate curve for the model evidence, and so has the strongest support under this model. The posterior allocation plot for is shown in Figure 3 (plots for all values of K can be found in Figure S1). Comparing this with figure 3 in Munshi-South and Kharchenko (2010b), we see some striking similarities—for example, the strong population differentiation in the Hunters Island and Willow Lake (a.k.a. Flushing Meadows) samples and the greater uncertainty in samples from the Black Rock Forest location. However, we also group together several populations that were previously found to be distinct, including locations 3, 4, 5, and 7 (all from the Bronx) and locations 8, 9, 11, and 15 (all from central Queens). The fact that we found evidence for fewer distinct populations than the original study may be due to our use of an uncorrelated allele frequencies model, although the geographical proximity of these regions gives us some confidence that this clustering is biologically plausible. Moreover, the striking difference between Figure 2A and Figure 2C demonstrates that different estimation methods can lead to quantitatively different conclusions even conditional on the same underlying model.
Figure 2.
Estimates of the model evidence for obtained using (A) the Structure estimator (B) the harmonic mean estimator and (C) the TI estimator For A and B, solid points give the mean over 21 replicates and error bars give 95% confidence intervals calculated from the variance over replicates. For C the TI estimation procedure results in a single point estimate of the evidence and an estimate of the 95% confidence interval without the need to average over replicates.
Figure 3.
Posterior assignment of all 312 individuals into clusters. Site names correspond to locations in and around New York City, and major landmasses are also given (the Black Rock Forest site is not within any of the five New York City boroughs). Further details of sampling sites can be found in Munshi-South and Kharchenko (2010b).
Discussion
Model-based clustering methods have proved extremely useful within population genetics. The probabilistic allocation of individuals to demes employed by programs such as Structure has made it possible to tease apart population subdivision within a wide range of organisms, including humans (Rosenberg et al. 2002; Li et al. 2008; Tishkoff et al. 2009), human pathogens (Falush et al. 2003b), plants (Garris et al. 2005), and animals (Parker et al. 2004). However, these posterior assignments are always produced conditional on the known value of K. Choosing an appropriate value of K is statistically much more challenging than estimating population assignments, as it involves a comparison between models rather than simple parameter estimation within a given model. Thermodynamic integration offers a way to do this, providing estimates of the evidence for K that are both accurate and precise. Our reanalysis of the white-footed mouse data demonstrates that this is of practical as well as theoretical importance, with the potential to lead to quantitatively different conclusions about the data.
The main disadvantage of TI is the computational cost. Multiple MCMC chains are needed, each drawn from a different version of the power posterior, to compute a single estimate of the model evidence. If the number of rungs is too low, then the trapezoidal rule step in (9) will not capture the shape of the underlying curve that it is approximating, leading to bias in the estimator. We must also be careful to take account of autocorrelation in the samples. This is dealt with automatically in MavericK1.0 through the use of effective sample size (ESS) calculations (see File S1 for details), which result in estimates of the model evidence that are accurate even in the presence of autocorrelation. However, it is still the case that high levels of autocorrelation require us to obtain a large number of posterior draws, and so we cannot ignore autocorrelation completely. This is a particular problem for the admixture model with α free to vary, where the much higher dimensionality of the model (compared with the without-admixture case) tends to result in poor MCMC mixing.
For this reason, TI may be suitable only for small- to medium-sized data sets of the sort analyzed here, at least for the time being. The use of TI for large SNP data sets—for example, data from the Human Genome Diversity Project (HGDP) analyzed by Li et al. (2008)—is therefore not practically possible at this stage without devoting significant computational resources to the problem. Good results will tend to be obtained when applied to data sets on the order of hundreds of individuals and tens to hundreds of loci, depending on the parameter set used. Fortunately, the accuracy of some heuristic estimators and traditional model comparison statistics appears to improve for larger data sets, and so it may be possible to sidestep this issue. It is also worth noting that, when genetic markers are sufficiently dense, loci can no longer be considered independent, alternative approaches such as chromosome painting may be more appropriate (Lawson et al. 2012).
An important consequence of working with the model evidence is that we must be careful in our choice of priors. In ordinary parameter estimation it is common practice to use relatively uninformative priors—the logic being that the model should be free to be driven by the data and not by our prior assumptions. However, when calculating the evidence (as in Equation 3), the thinness of the prior has an effect that is not diminished by adding more data. This can result in models being unduly punished if the observed data are extremely unlikely a priori. For example, our use of independent Dirichlet priors on the allele frequencies in all populations can be considered a fairly thin prior, as no combination of allele frequencies is any more likely than any other a priori. This will tend to result in conservative estimates of K, as there is a large cost (in evidence terms) of adding more populations unless they can justify their existence by a commensurate increase in the likelihood. Alternative model formulations, such as the correlated allele frequencies model of Falush et al. (2003a), may therefore be better at detecting subtle signals of population subdivision. This model is likely to feature in later versions of MavericK.
Finally, it is important to keep in mind that when thinking about population structure, we should not place too much emphasis on any single value of K. The simple models used by programs such as Structure and MavericK are highly idealized cartoons of real life, and so we cannot expect the results of model-based inference to be a perfect reflection of true population structure (see discussion in Waples and Gaggiotti 2006). Thus, while TI can help ensure that our results are statistically valid conditional on a particular evolutionary model, it can do nothing to ensure that the evolutionary model is appropriate for the data. Similarly—in spite of the results in Table 2—we do not advocate using the model evidence (estimated by TI or any other method) as a way of choosing the single “best” value of K. The chief advantage of the evidence in this context is that it can be used to obtain the complete posterior distribution of K, which is far more informative than any single point estimate. For example, by averaging over the distribution of K, weighted by the evidence, we can obtain estimates of parameters of biological interest (such as the admixture parameter α) without conditioning on a single population structure. Although one value of K may be most likely a posteriori, in general a range of values will be plausible, and we should entertain all of these possibilities when drawing conclusions.
The MavericK program and documentation can be downloaded from www.bobverity.com/MavericK.
Acknowledgments
We are grateful to Jason Munshi-South and Katerina Kharchenko for making the data from their 2010 white-footed mouse analysis publicly available, to James Borrell for patiently testing early versions of the program, and to three anonymous reviewers whose suggestions substantially improved this article.
Appendix
MCMC Under the Without-Admixture Model
To carry out the TI estimation approach we need to be able to draw from the power posterior distribution. This is straightforward in the case of genetic mixtures and requires nothing more than a simple extension of existing MCMC algorithms. In the following we strive to bring our notation in line with previous studies wherever possible, but the complexities of certain likelihood functions also motivate us to define some new notation (see Table A1). It is worth noting, for example, that we will write individual genotypes in simple list form (as in Pritchard et al. 2000), using the notation for the lth locus of the ith individual, but also in allelic partition form (as in Huelsenbeck and Andolfatto 2007), using the notation For example, a diploid individual homozygous for the third allele at a particular locus can be written or equivalently where there are five possible alleles to choose from in this example. Conditioning on the model is also implicit throughout this section.
In the basic algorithm described by Pritchard et al. (2000) there are two free parameters to keep track of—the allocation of individuals to demes, denoted here, and the allele frequencies in all K demes, denoted Under the assumptions of Hardy–Weinberg and linkage equilibrium it is possible to write the probability of the observed data given the known values of these free parameters, Combining this likelihood with a prior on the allele frequencies at each locus, we can derive the conditional posterior distribution of the allele frequencies given the known group allocation, Alternatively, multiplying by an equal prior on the allocation of individuals to demes, we can derive the conditional posterior distribution of the group allocation given the known allele frequencies, Algorithm 1 of Pritchard et al. (2000) works by alternately sampling from each of these conditional distributions, resulting (after sufficient burn-in) in a series of draws from the full posterior distribution. More often than not we are interested in the posterior allocation, in which case the posterior allele frequencies can simply be ignored.
However, as stated in the original derivation of Rannala and Mountain (1997) and reiterated by later authors (Corander et al. 2003; Pella and Masuda 2006; Huelsenbeck and Andolfatto 2007), it is possible to integrate over the allele frequencies analytically, thereby greatly reducing the dimensionality of the problem. The new likelihood, conditional only on the group allocation, can be written
| (A1) |
(see Table A1 for parameter definitions). This expression is extremely useful to us, as it means the likelihood can be calculated without having to take into account an explicit representation of the unknown allele frequencies—our uncertainty in the allele frequencies has already been integrated out of the problem.
Rather than using (A1) directly, later authors including Corander et al. (2003), Pella and Masuda (2006), and Huelsenbeck and Andolfatto (2007) used this analytical solution to define an efficient MCMC algorithm. Dividing the probability of the data by the probability of the data with the ith observation removed, denoted we obtain the conditional probability of observation i given all others. Using the fact that we obtain
| (A2) |
Computing (A2) for all k and normalizing, we obtain the conditional posterior probability that individual i belongs to deme k:
| (A3) |
By repeatedly drawing new group allocations for all individuals from (A3), we obtain a series of draws from the posterior distribution without ever needing to invoke the unknown allele frequencies. Thus, the two-step algorithm of Pritchard et al. (2000) can be reduced to the more efficient one-step algorithm of Corander et al. (2003).
The reason these results are pertinent to our problem is that we can make use of the same gains in efficiency when designing an MCMC algorithm for the purposes of TI. In fact, the only difference when carrying out TI is that the likelihood in (A1) should be raised to the power β, allowing us to draw from the power posterior. On making this change we find that the conditional posterior distribution in (A2) should also be raised to the power β [this follows from the fact that (A2) can be derived as a ratio of two ordinary likelihoods]. Thus, we arrive at a new expression for the probability of individual i being assigned to group k:
| (A4) |
By repeatedly sampling new group allocations for all individuals from (A4), we obtain a series of allocation vectors drawn from the power posterior (note that when we are essentially drawing from the prior). The likelihood of each vector can then be computed using (A1), at which point we have everything we need to calculate as in (8). Carrying out this entire procedure for a range of values we obtain a series of points that can be used to calculate the TI estimator as in (9). The complete TI algorithm for the model without admixture can be defined as follows:
Algorithm 1 (without admixture)
-
For r distinct values of spanning the interval
Perform MCMC by repeatedly drawing from (A4) for all This results (after discarding burn-in) in t draws from the power posterior group allocation.
Calculate the likelihood of each group allocation, using (A1).
Calculate as the average log-likelihood, as in (8). If calculating the variance of the estimator, calculate using the formula in File S1, taking care to use an appropriate value of the ESS.
Use the values to calculate in a suitable numerical integration scheme, for example using the trapezoidal rule as in (9).
MCMC Under the Admixture Model
The model with admixture described by Pritchard et al. (2000) is slightly complicated by the fact that each gene copy is free to originate from a different deme. However, we can still apply the same basic logic described above to arrive at a simple one-step algorithm for sampling from the power posterior. First, we note that the probability of the data conditional on the known group allocation is identical in this model to the probability in the without-admixture model and is given by (A1). This is true because we make the same assumption that gene copies are drawn independently from demes, and we apply the same Dirichlet priors on allele frequencies, meaning the final likelihood does not change. The difference in the admixture model is that the group allocation takes place at the level of the gene copy, rather than at the level of the individual, and so the values are no longer restricted to being the same for all This is reflected in the values used to keep track of the gene copies allocated to a particular deme, which are now free to contain only a partial contribution of the genome of each individual.
Following the same approach as for the without-admixture model, we can obtain the conditional probability of gene copy by dividing through the probability of the complete data by the probability of the data with this element removed [denoted ]. Most of the terms in the resulting expression cancel out, leading to the following simple result:
| (A5) |
As before, this likelihood should be combined with the prior probability of assignment to each deme. If the admixture proportions for individual i are given by the vector then, under the assumptions of the model described by Pritchard et al. (2000), the number of gene copies in this individual that are allocated to each deme can be considered a multinomial draw from Integrating over a prior on these frequencies, we obtain
| (A6) |
We can use this expression to write down the prior probability of gene copy a at locus l in individual i being allocated to deme k, conditional on the allocations of all other gene copies:
| (A7) |
Bringing together the prior with the likelihood raised to the power β, we obtain the following expression for the power posterior probability of an individual gene copy being allocated to deme k:
| (A8) |
By repeatedly sampling new allocations for all gene copies at all loci within all individuals (i.e., all ), we obtain a series of draws from the power posterior group allocation under the admixture model. Again, this algorithm is made more efficient by the fact that the unknown allele frequencies in all populations and the unknown admixture proportions in all individuals have been integrated out of the problem at an early stage.
A common extension to the basic admixture model is to leave α as a free parameter, updating it as part of the MCMC. This can be accommodated within the TI framework by using a simple Metropolis–Hastings step. If is a new value of α, drawn from some suitable proposal distribution then the acceptance probability under Metropolis–Hastings is given by
| (A9) |
Note that the core probability that drives this expression is the prior probability of the allocation which is given in (A6). The actual probability of the data—i.e., the expression that is raised to the power β in the power posterior calculation—does not feature here. Thus, we can use the same Metropolis–Hastings step to update α irrespective of the value of β.
The complete TI algorithm for the model with admixture can be defined as follows:
Algorithm 2 (with admixture)
-
For r distinct values of spanning the interval
Perform MCMC by repeatedly drawing from (A8) for all gene copies at all loci in all individuals (all ). If α is a free parameter, then update this value using a Metropolis–Hastings step, as in (A9). This results (after discarding burn-in) in t draws from the power posterior group allocation.
Calculate the likelihood of each group allocation, using (A1).
Calculate as the average log-likelihood, as in (8). If calculating the variance of the estimator, calculate using the formula in File S1, taking care to use an appropriate value of the ESS.
Use all the values to calculate in a suitable numerical integration scheme, for example using the trapezoidal rule as in (9).
Finally, we note that the expressions derived in this section can be used to obtain the exact model evidence by brute force in restricted settings. For example, focusing on the model without admixture, we could sum over the likelihood of all possible group allocations to obtain the true model evidence,
| (A10) |
where is given by (A1), and for this model for all group allocations. Although this is possible in theory, the sheer number of allocations that are required to sum over makes this method impractical in all but the simplest situations. Even if we exploit redundancies in the labeling of different allocations, we are still restricted to values of n and K not much >10. This method is therefore only really useful as a way of checking the accuracy of other estimation methods.
Table A1. Definitions of parameters used in this study.
| Parameter | Description |
|---|---|
| α | Dirichlet parameter on admixture proportions |
| β | Power used in power posterior calculation |
| Evaluation of a parameter while excluding information for individual i (c could be any of the parameters listed above) | |
| Evaluation of a parameter while excluding information for gene copy a at locus l in individual i (c could be any of the parameters listed here) | |
| No. of unique alleles observed at locus l | |
| K | No. of populations |
| L | No. of loci |
| Dirichlet parameter for frequency of allele j at locus l [where ] | |
| Sum of the Dirichlet parameters for locus l [i.e., ] | |
| n | No. of individuals sampled |
| Allele frequencies in all populations at all loci | |
| Admixture proportions in individual i | |
| Allelic partition of alleles in individual i at locus l. For example, the genotype can be written in allelic partition form, where in this example | |
| No. of copies of allele j at locus l in individual i [where ] | |
| No. of copies of any allele at locus l in individual i [i.e., ] | |
| Denotes the gamma function. | |
| Partition of gene copies to populations in all individuals | |
| Partition of gene copies to populations in individual i | |
| No. of gene copies in individual i assigned to population k | |
| No. of gene copies in individual i assigned to any population [i.e., ] | |
| Genetic information for all individuals | |
| Genetic information for individual i | |
| Genetic information for individual i at locus l | |
| Allelic type of the gene copy in individual i at locus l [where ] | |
| Allelic partition at all loci of all gene copies assigned to population k | |
| Allelic partition at locus l of all gene copies assigned to population k | |
| No. of copies of allele j at locus l assigned to population k | |
| No. of copies of any allele at locus l assigned to population k [i.e., ] | |
| Assignment of all gene copies in all individuals | |
| Assignment of gene copy a at locus l in individual i to a population [where ] | |
| Assignment of individual i to a population. When referring to it is implied that is identical for all meaning all gene copies within this individual are assigned together |
Footnotes
Communicating editor: N. A. Rosenberg
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.180992/-/DC1.
Literature Cited
- Baele G., Lemey P., Bedford T., Rambaut A., Suchard M. A., et al. , 2012. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol. Biol. Evol. 29: 2157–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli P., Palczewski M., 2010. Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics 185: 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanquart S., Lartillot N., 2006. A Bayesian compound stochastic process for modeling nonstationary and nonhomogeneous sequence evolution. Mol. Biol. Evol. 23: 2058–2071. [DOI] [PubMed] [Google Scholar]
- Corander J., Waldmann P., Sillanpää M. J., 2003. Bayesian analysis of genetic differentiation between populations. Genetics 163: 367–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G., Regnaut S., Goudet J., 2005. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- Falush D., Stephens M., Pritchard J. K., 2003a Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush, D., T. Wirth, B. Linz, J. K. Pritchard, M. Stephens et al., 2003b Traces of human migrations in helicobacter pylori populations. Science 299: 1582–1585. [DOI] [PubMed]
- Falush D., Stephens M., Pritchard J. K., 2007. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol. Ecol. Notes 7: 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friel N., Pettitt A. N., 2008. Marginal likelihood estimation via power posteriors. J. R. Stat. Soc. Ser. B Stat. Methodol. 70: 589–607. [Google Scholar]
- Garris A. J., Tai T. H., Coburn J., Kresovich S., McCouch S., 2005. Genetic structure and diversity in Oryza sativa l. Genetics 169: 1631–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman A., Meng X.-L., 1998. Simulating normalizing constants: from importance sampling to bridge sampling to path sampling. Stat. Sci. 13: 163–185. [Google Scholar]
- Gelman A., Carlin J. B., Stern H. S., Rubin D. B., 2004. Bayesian Data Analysis, 2nd edition. Chapman & Hall/CRC, Boca Raton, Florida. [Google Scholar]
- Hubisz M. J., Falush D., Stephens M., Pritchard J. K., 2009. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 9: 1322–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J. P., Andolfatto P., 2007. Inference of population structure under a Dirichlet process model. Genetics 175: 1787–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J. P., Andolfatto P., Huelsenbeck E. T., 2011. Structurama: Bayesian inference of population structure. Evol. Bioinform. Online 7: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart, T., and C. Collins, 2015 A tutorial for discriminant analysis of principal components (dapc) using adegenet 2.0.0. Available at: http://adegenet.r-forge.r-project.org/files/tutorial-dapc.pdf.
- Kass R. E., Raftery A. E., 1995. Bayes factors. J. Am. Stat. Assoc. 90: 773–795. [Google Scholar]
- Lartillot N., Philippe H., 2006. Computing Bayes factors using thermodynamic integration. Syst. Biol. 55: 195–207. [DOI] [PubMed] [Google Scholar]
- Lawson D. J., Hellenthal G., Myers S., Falush D., 2012. Inference of population structure using dense haplotype data. PLoS Genet. 8: e1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage T., Bryant D., Philippe H., Lartillot N., 2007. A general comparison of relaxed molecular clock models. Mol. Biol. Evol. 24: 2669–2680. [DOI] [PubMed] [Google Scholar]
- Li, J. Z., D. M. Absher, H. Tang, A. M. Southwick, A. M. Casto et al., 2008 Worldwide human relationships inferred from genome-wide patterns of variation. Science 319: 1100–1104. [DOI] [PubMed]
- Malécot, G., 1948 Mathématiques de l’Hérédité. Masson & Cie, Paris.
- Munshi-South, J., and K. Kharchenko, 2010a Data from Rapid, pervasive genetic differentiation of urban white-footed mouse (peromyscus leucopus) populations in New York City. Dryad digital repository. Available at http//dx..org/10.5061/dryad.1893.10.5061/dryad.1893 [DOI] [PubMed]
- Munshi-South J., Kharchenko K., 2010b Rapid, pervasive genetic differentiation of urban white-footed mouse (peromyscus leucopus) populations in New York City. Mol. Ecol. 19: 4242–4254. [DOI] [PubMed] [Google Scholar]
- Neal R. M., 1994. Contribution to the discussion of “Approximate Bayesian inference with the weighted likelihood bootstrap” by Michael A. Newton and Adrian E. Raftery. J. R. Stat. Soc. B 56: 41–42. [Google Scholar]
- Newton M. A., Raftery A. E., 1994. Approximate Bayesian inference with the weighted likelihood bootstrap. J. R. Stat. Soc. B 56: 3–48. [Google Scholar]
- Parker, H. G., L. V. Kim, N. B. Sutter, S. Carlson, T. D. Lorentzen et al, 2004 Genetic structure of the purebred domestic dog. Science 304: 1160–1164. [DOI] [PubMed]
- Pella J., Masuda M., 2006. The Gibbs and split merge sampler for population mixture analysis from genetic data with incomplete baselines. Can. J. Fish. Aquat. Sci. 63: 576–596. [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K., X. Wen, and D. Falush, 2010 Documentation for structure software: version 2.3. Available at: http://pritchardlab.stanford.edu/structure_software/release_versions/v2.3.4/structure_doc.pdf.
- Raftery, A. E., M. A. Newton, J. M. Satagopan, and P. N. Krivitsky, 2006 Estimating the integrated likelihood via posterior simulation using the harmonic mean identity. Bayes. Stat. 8: 1–45.
- Raj A., Stephens M., Pritchard J. K., 2014. Faststructure: variational inference of population structure in large SNP data sets. Genetics 197: 573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannala B., Mountain J. L., 1997. Detecting immigration by using multilocus genotypes. Proc. Natl. Acad. Sci. USA 94: 9197–9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg, N. A., J. K. Pritchard, J. L. Weber, H. M. Cann, K. K. Kidd et al., 2002 Genetic structure of human populations. Science 298: 2381–2385. [DOI] [PubMed]
- Spiegelhalter D. J., Best N. G., Carlin B. P., Van Der Linde A., 2002. Bayesian measures of model complexity and fit. J. R. Stat. Soc. Ser. B Stat. Methodol. 64: 583–639. [Google Scholar]
- Stephens M., 2000. Dealing with label switching in mixture models. J. R. Stat. Soc. Ser. B Stat. Methodol. 62: 795–809. [Google Scholar]
- Tishkoff S. A., Reed F. A., Friedlaender F. R., Ehret C., Ranciaro A., et al. , 2009. The genetic structure and history of Africans and African Americans. Science 324: 1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples R. S., Gaggiotti O., 2006. Invited review: What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol. Ecol. 15: 1419–1439. [DOI] [PubMed] [Google Scholar]
- Wright S., 1949. The genetical structure of populations. Ann. Eugen. 15: 323–354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article and are available upon request.