Abstract
Epidermal growth factor receptor (EGFR) is a popular therapeutic target for many cancers. EGFR inhibitors have been tested in children with refractory neuroblastoma (NB). Interestingly, partial response or stable disease was observed in a few NB patients. Since EGFR mutations are biomarkers for response to anti-EGFR drugs, primary NB tumors and cell lines were screened for mutations. A novel EGFR extracellular domain deletion mutant, EGFRΔ768, was discovered and the biological and biochemical properties of this mutant were characterized and compared to wild type and EGFRvIII receptors. EGFRΔ768 was found to be constitutively active and localized to the cell surface. Its expression conferred resistance to etoposide and drove proliferation as well as invasion of cancer cells. While EGFRΔ768 had similarity to EGFRvIII, its biological and biochemical properties were distinctly different from both the EGFRvIII and wild type receptors. Even though erlotinib inhibited EGFRΔ768, its effect on the mutant was not as strong as that on wild type EGFR and EGFRvIII. In addition, downstream signaling of EGFRΔ768 was different from that of the wild type receptor. In conclusion, this is the first study to demonstrate that neuroblastoma express not only EGFRvIII, but also a novel EGFR extracellular domain deletion mutant, EGFRΔ768. The EGFRΔ768 also possesses distinct biological and biochemical properties which might have therapeutic implications for neuroblastoma as well as other tumors expressing this novel mutant.
Keywords: EGFR, EGFRvIII, EGFR extracellular mutations, neuroblastoma
Introduction
The tyrosine kinase receptor, EGFR, is well known for its role as an oncogene. Studies over the last 20 years clearly supported that overexpression of EGFR promotes tumorigenesis (1). With such compelling evidence implicating EGFR as a key target for many cancers, the pharmaceutical industry has developed drugs against this important protein. Several EGFR inhibitors are now FDA approved for the treatment of various cancers. Despite the early successes, clinical response to EGFR inhibitors is variable. Molecular predictors for patients’ response to anti-EGFR drugs were only recently discovered. Patients with non-small cell lung cancers that carried somatic mutations in the tyrosine kinase domain of EGFR demonstrated dramatic clinical responses when treated with the EGFR tyrosine kinase inhibitor (TKI), gefitinib (2). These activating kinase mutations conferred exquisite sensitivity to EGFR TKI (3). Other types of EGFR mutations also predict patients’ response to anti-EGFR therapy. For instance, the EGFR extracellular domain deletion mutant, EGFRvIII, is a molecular determinant that predicted glioblastoma response to EGFR TKI. Co-expression of EGFRvIII and PTEN in glioblastoma was significantly associated with a clinical response to EGFR TKI in two independent patient cohorts (4). More recent data suggested that deletion and missense mutations in the EGFR extracellular domain also have therapeutic implications. For instance, an acquired EGFR ectodomain mutation (S492R) conferred resistance to cetuximab by preventing its binding to EGFR (5). An exon 4-deletion variant of EGFR conferred resistance to cisplatin in ovarian cancer cells by upregulating Bcl-2 and downregulating BAD (6). Thus, EGFR mutations are important biomarkers for EGFR-directed therapy response.
The EGFR gene is frequently mutated in many human tumors. EGFRvIII was one of the earliest oncogenic mutations identified in human glioblastoma. It contains the entire exon 1 followed by exon 8 (EGFRΔ801). The splice was in-frame and resulted in the formation of a new codon (GGT) at the splice junction, which translated into a glycine residue. EGFRvIII is constitutively active, does not bind EGFR ligand and is not downregulated upon autophosphorylation, but it has potent pro-oncogenic effects (7). It not only expressed in a high percentage of glioblastoma, but also expressed in other tumor types (8). After the discovery of activating EGFR kinase mutations in non-small cell lung cancer (2), intense focus in the last decade has been on the characterization of these kinase mutants. Now, over 30 kinase mutations have been reported and characterized in primary tumors (9). On the contrary, only a handful of EGFR extracellular domain mutants were identified (10). We recently discovered two novel EGFR extracellular domain deletion mutants in head and neck squamous cell carcinomas, whose expression correlated with advanced disease stage (11). Another EGFR extracellular domain mutant known as the exon 4-deletion variant also displayed enhanced transformation, a higher metastatic potential and a lower sensitivity to cisplatin than wild-type EGFR (6, 12). These findings implied that like the kinase mutations, EGFR extracellular domain mutants might play an important role in tumorigenesis and anti-EGFR therapy response as well.
EGFR inhibitors were in clinical trials for children with refractory solid tumors, including neuroblastoma (NB). The phase I study of erlotinib in children with refractory solid tumors showed that 2/5 NB patients had stable disease for 2 years (13). A 4 year old girl with relapse, refractory NB was treated with a gefitinib containing regimen and remained progression free for 27 months (14). When ten additional refractory NB patients were treated using this regimen, partial response was seen in 3/10 patients (15). These observations suggested that EGFR inhibitors might have a therapeutic effect in a subgroup of NB patients. EGFR expression was readily detectable in neuroblastoma cell lines and primary tumors (16, 17). EGFR inhibitors also have activities against NB cell lines (16, 17). Despite these findings, not all NB patients benefited from EGFR TKI in clinical trials. To understand the reason, we screened for EGFR mutations in 142 primary neuroblastic tumors. Like others (18), we did not find activating kinase mutations in these tumors. Surprisingly, in addition to EGFRvIII, we discovered a novel EGFR extracellular domain deletion mutant, EGFRΔ768, in primary NB. Further characterization of EGFRΔ768 showed that it is constitutively active, responds poorly to erlotinib and is resistant to the chemotherapy, etoposide. It also possesses different biological and biochemical properties than those of activated wild type EGFR and EGFRvIII.
Materials and Methods
Collection of primary neuroblastic tumors
62 Snap frozen primary neuroblastic tumors were provided by the Columbus Biopathology Repository Center (CBRC). These tumors were accrued when excess tissues were available after the initial diagnostic specimen had been processed by the respective Children Oncology Group (COG) institutions. Informed consent for participation in tumor banking and research was obtained at the local institutions and the tumors were sent to the Biopathology Repository Center in Columbus for storage. Pathology reports associated with each tumor were also provided. Next, we made an application to the COG for NB tumor RNA. A total of 80 NB RNA (40 low risk and 40 high risk NB) was approved and sent to our laboratory for testing. The tumor RNA were obtained as part of an ongoing COG neuroblastoma biology study (ANBL00B1) and were characterized by the COG NB reference laboratory. Exempted IRB approval for this study was obtained at our institution.
Cell lines, reagents and antibodies
The following NB cell lines were kindly provided by Dr. Marian Evinger: BE2C, BE2M17, IMR32, SKNSH and SH1N. 3T3 cells were previously obtained and characterized (19, 20). The identity of the NB cell lines were confirmed by STR profiling (Fig. S3). NB cells were grown in 1:1 mixture of Eagle’s minimum essential medium (with nonessential amino acids, 2 mM L-glutamine plus 1 mM sodium pyruvate) and Ham’s nutrient mixture F12 (Gibco, Grand Island, NY), whereas 3T3 cells were maintained in DMEM (Gibco). All media were supplemented with 10% FBS, 1% penicillin/streptomycin (Gibco) and the cells were grown at 37°C in a 5% CO2-humidified incubator. The following antibody was used for immunoprecipitation: cetuximab (ImClone, New York, NY). For western blotting, the following antibodies were used: anti-phosphotyrosine 4G10 (Millipore, Temecula, CA), anti-GFP antibody (Abcam, Cambridge, MA), plasma membrane fraction western blot cocktail (Abcam, Cambridge, MA), EGFR rat antibody (20) and tubulin (Sigma, St. Louis, MO). The fluorescent labeled secondary anti-rabbit, mouse and rat antibodies were purchased (Molecular Probes, Grand Island, NY). The growth factor, EGF, was purchased from Gibco. Geneticin was purchased from Gold Biotechnology (St Louis, MO). Erlotinib (OSI Pharmaceuticals, Northbrook, IL) was reconstituted in sterile water and filtered to yield a final concentration of 2.5 mM. Etoposide (Sigma) was reconstituted in DMSO to yield a stock concentration of 50 mM.
EGFRΔ768 polyclonal rabbit antibody was produced by BioSynthesis, Lewisville, TX. The following splice junction peptide: LEEKKVCQGTCVKKCP was synthesized and purified to greater than 95% in purity. The peptide was then conjugated to the Keyhole Limpet Hemocyanin (KLH) carrier protein using glutaraldehyde crosslinker. Two rabbits (6680 and 6681) were immunized by the KLH-peptide conjugate with 5 times booster injection. The antisera were collected at week 8, 10 and at termination, then, they were loaded to Protein A-Sepharose resin. After washing with phosphate buffered saline (PBS), the bound proteins were eluted with 0.1 mol/L glycine-HCl buffer. The eluted antibodies were adjusted to neutral pH, assessed on 10% SDS-PAGE and shipped to our laboratory for validation. The optimal antibody:protein (in ug) ratio for the best specificity in immunoprecipitation application is 1:250.
Cloning of EGFRΔ768 expression vector and generation of stable clones
An expression vector containing wild type (WT) EGFR with a C-terminal-GFP tag and a neomycin selection cassette was kindly provided by the laboratory of Dr. Dafna Bar-Sagi (21). The EGFRΔ768 and EGFRvIII deletions were subcloned into the WT-EGFR sequence to generate EGFRΔ768 and EGFRvIII expression vectors. We confirmed that no unexpected mutations except the Δ768 and vIII deletions were introduced into the respective subcloned sequences by direct DNA sequencing. Next, 3T3 and BE2C cells were transfected with control GFP, WT-EGFR-GFP, EGFRΔ768-GFP and EGFRvIII-GFP constructs using PolyJet transfection reagent per manufacture protocol (SignaGen, Rockville, MD). Stable 3T3 and BE2C cell clones expressing GFP, WT-EGFR-GFP, EGFRΔ768-GFP and EGFRvIII-GFP were generated by selection with 500 ug/ml geneticin (G418).
Processing of neuroblastic tumors and cell lines for protein, RNA and DNA analyses
Frozen primary neuroblastic tumor was homogenized in 1 ml of Triton X lysis buffer (50mM Tris-Cl, pH = 8, 150mM NaCl and 1% Triton X-100) with protease inhibitor and sodium orthovanadate using the POLYTRON system PT 10–35 GT (Kinematica AG). The homogenates were spun down; pellets discarded and the supernatants were saved for protein analyses. Protein concentrations were determined by Bradford assay. Separate pieces of the frozen tumor were homogenized in 1 ml of Trizol Reagent using the POLYTRON system PT 1200 E (Kinematica AG). Each cell line was also lysed in Trizol reagent. Total RNA was isolated per manufacture recommendation (Life Technologies, Grand Island, NY). RNA concentration and purity were determined using a NanoDrop spectrophotometer. Each tumor was homogenized with a disposable dispersing aggregate to avoid cross contamination between samples. Genomic DNA was isolated from the BE2M17 cell line per manufacture recommendation (Qiagen, Valencia, CA).
RNA/DNA Analyses with PCR and Sequencing
Reverse transcription was carried out using a SuperScript Preamplification Kit (Life Technologies) on 1μg of total RNA aliquots. PCR was performed on cDNA and gDNA using primers as previously described (11) as well as primers to exon 7 sequences upstream of the breakpoint in exon 7 of EGFRΔ768: 5′-CCCGAGGGCAAATACAGC-3′ and to intron 7: 5′-GACAGAGCGGGACAAGGATG-3′ respectively. The PCR end products were resolved by 2% agarose gel electrophoresis and isolated using the Qiagen PCR Extraction Kit. DNA sequencing was performed at the Sequencing Core Facility of the Stony Brook University Medical Center.
Fluorescence Microscopy
The NIH3T3 clonal cells which stably expressing EGFRΔ768-GFP were cultured on a glass coverslip and subsequently fixed with 3.7% formaldehyde. The coverslip was mounted on a glass slide with Immu-Mount (Thermo Scientific, Waltham, MA). Imaging was performed with a Zeiss microscope equipped with 63×/1.4 oil DIC objective and processed with AxioVision Rel4.8 software (Carl Zeiss, Thornwood, NY).
Invasion and XTT assay
Invasion and XTT proliferation assays were performed as previously described (22). For the XTT assay, cells were seeded at 5 × 103 cells/well in a 96 well plate in quintuplicate. The cells were treated the next day with increasing concentration of erlotinib (0.025 – 2.5 uM) or etoposide (0.5 – 2 uM). Activated-XTT reagent was prepared and added to the cells the following day as per protocol. Cell proliferation rates were determined (22). A minimum of three independent experiments were performed. Stable 3T3 clones with close to equivalent WT-EGFR and EGFRΔ768 expression (Fig. S6) as well as clone #5 of BE2C-EGFRΔ768 and clone #2 of BE2C-EGFRvIII (Fig. 5E) were used for the XTT assay respectively.
Plasma Membrane Protein Isolation
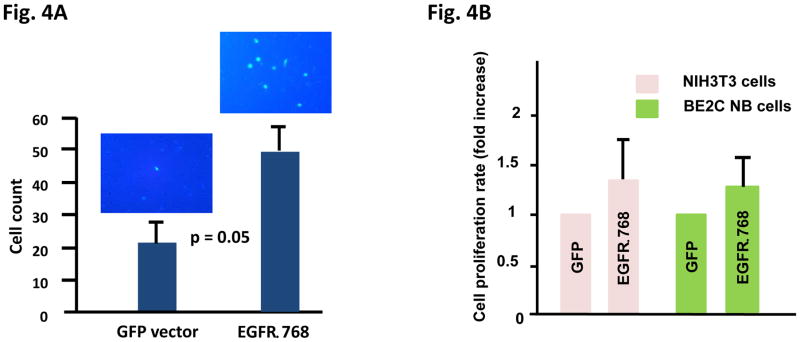
Plasma membrane proteins were isolated from 3T3-EGFRΔ768 cells as per protocol using the plasma membrane protein extraction kit (Abcam, Cambridge, MA). 50 μg of plasma membrane proteins were obtained. 13 ug of nuclear, cytosolic and plasma membrane fractions were analyzed by western blot using the GFP and the plasma membrane fraction cocktail antibodies. The cocktail antibodies contained three monoclonal antibodies, each targeting a specific organelle marker: plasma membrane by anti-sodium/potassium ATPase, cytosol by anti-GAPDH and nucleus by anti-histone H3.
Biochemical analysis
Stable cell clones were serum starved overnight, treated with erlotinib at increasing concentrations (0.025 – 25 uM) or with 50 ug/ml of cetuximab for 30 minutes, then stimulated with 50 ng/ml EGF for 30 minutes before lysing. Cell and tumor lysates were analyzed and quantified for EGFR expression/phosphorylation by IPW as previously described (22). Total cell lysates of the BE2C clones and the transiently transfected IMR32 cells were analyzed for tyrosine phosphorylation by western blots. These lysates were also examined for phosphokinase and receptor tyrosine kinase (RTK) signaling using phospho-kinase and phospho-RTK arrays (R&D Systems). Each biochemical experiment was repeated to confirm the findings.
Statistics Analysis
Statistical analyses were performed using SPSS Statistics 16.0 (SPSS Inc., Chicago, IL). The comparison of EGFR expression (log value) between ΔEGFR+ and ΔEGFR- NB was performed using two samples t-test. EGFR expression was log transformed to normalize the distributions for comparison. Comparison between the different drug treatment groups was performed using ANOVA. Level of statistical significance is 5%. When multiple comparisons were performed, the Bonferroni adjusted P-value was used to determine the level of significance.
Results
Neuroblastoma express a novel EGFR extracellular domain deletion mutant, EGFRΔ768
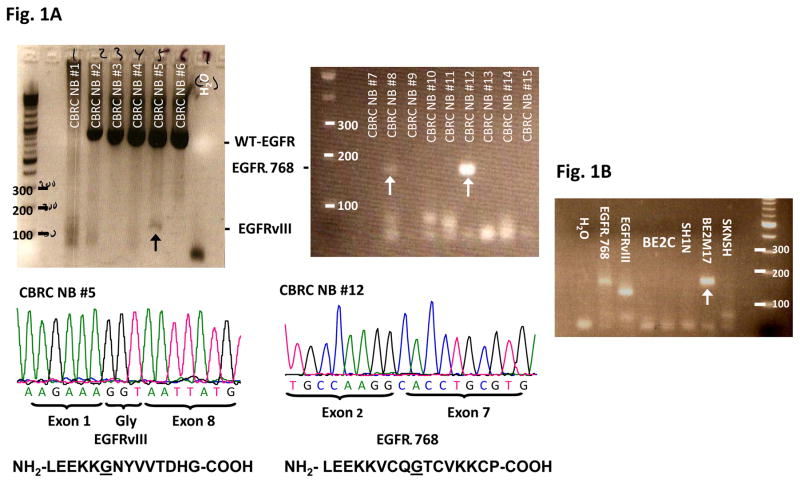
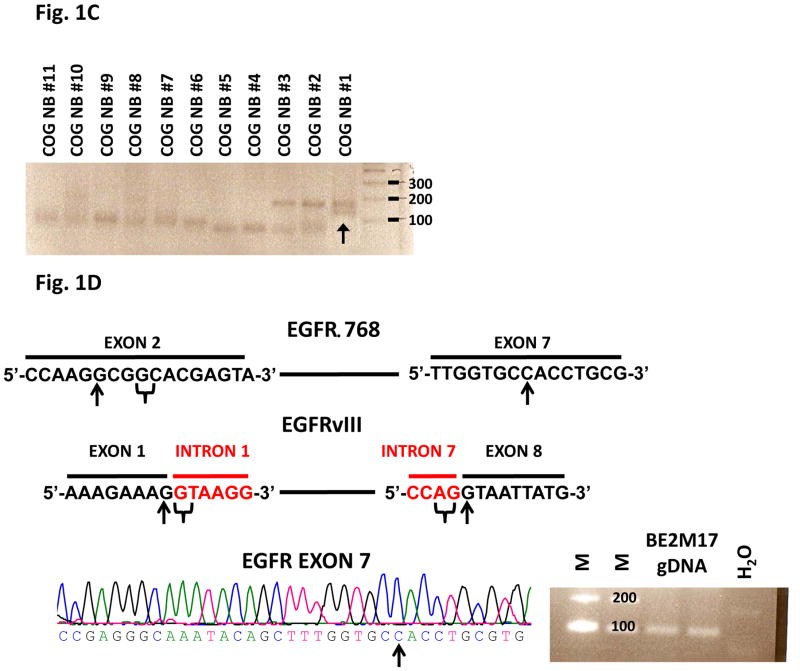
We screened 62 CBRC NB samples for EGFR mutations using RT-PCR and cDNA sequencing. While we did not find activating EGFR kinase mutations in these tumors, we surprisingly discovered that 32% (20/62) of neuroblastic tumors expressed deletion mutations of the EGFR extracellular domain (Fig. 1A & S1). As confirmed by bidirectional sequencing, 14 tumors expressed a novel mutant, EGFRΔ768, whereas 6 expressed EGFRvIII. The EGFRΔ768 mutant has an in-frame deletion from nucleotide 102 of exon 2 to nucleotide 869 of exon 7. The resulting mRNA transcript coded for 11 more amino acids than EGFRvIII. While the EGFRvIII conjoined exon 1 and 8 right at the splice donor and acceptor sites of intron 1 and 7, the 3′ sequences immediately upstream of the breakpoint in exon 7 of EGFRΔ768 do not contain a potential splice acceptor site (i.e. AG), even though there is a potential splice donor site in the vicinity of the 5′ sequences downstream of the breakpoint in exon 2 of EGFRΔ768 (Fig. 1D). Despite the differences in the nucleotide sequences at the spliced junctions of EGFRΔ768 and EGFRvIII, there are similarities in the amino acid sequences between the two mutants (Fig. 1A). First, both mutants have a glycine at the spliced junctions. Second, the N-terminal amino acid sequences immediately upstream of the spliced junctions of both mutants contain the short peptide, LEEKK (Fig. 1A). Next, we screened for EGFRvIII and EGFRΔ768 in a second cohort of NB tumor RNA. Of the 80 NB RNA from the COG, we confirmed that 16% (13/80) expressed the EGFR mutants (Fig. 1C & S2). Interestingly, one of the tumors co-expressed EGFRvIII and EGFRΔ768 (Fig. 1C). We also detected EGFRΔ768 mRNA expression in 1 of 5 NB cell lines (Fig. 1B). Then, we screened the genomic DNA of the EGFRΔ768+ BE2M17 cell line for any sporadic mutation around the exon 7 breakpoint that might create a new splice acceptor site. We confirmed wild type sequences upstream of the exon 7 breakpoint (Fig. 1D). With this finding, we suspected that the EGFRΔ768 is unlikely an alternative spliced variant.
Fig. 1. EGFR extracellular domain mutants in NB.
(A) RT-PCR analysis of EGFR extracellular domain mutations in a representative panel of CBRC NB tumors. Noted that CBRC NB #5 expressed EGFRvIII (black arrow) while CBRC NB #8 and #12 expressed EGFRΔ768 (white arrows). Identities of the bands were confirmed by bidirectional sequencing as shown. The amino acid sequences corresponding to EGFRΔ768 and EGFRvIII were depicted below the nucleotide sequences. The underlined amino acids represented the splice junctions. WT: wild type; Gly: glycine. (B) RT-PCR analysis of EGFR extracellular domain mutations in a panel of NB cell lines. BE2M17 expressed EGFRΔ768 (white arrow). Identity of the band was confirmed by bidirectional sequencing. Gel purified EGFRvIII and EGFRΔ768 cDNA were positive controls. H2O was negative control. (C) RT-PCR analysis of EGFR extracellular domain mutations in a representative panel of COG NB tumors. Noted that COG NB #1 co-expressed EGFRvIII and EGFRΔ768 (black arrow). Identities of the bands were confirmed by bidirectional sequencing. (D) Splice junction analysis of EGFRΔ768 and EGFRvIII. Black arrows pointed to the breakpoints in exon 2/7 of EGFRΔ768 and exon 1/8 of EGFRvIII. The potential/actual splice donor and acceptor sites in the EGFR sequences were bracketed. EGFR exon 7 of BE2M17 genomic DNA was amplified and sequenced as shown in the bottom panel. M: marker; H2O was negative control.
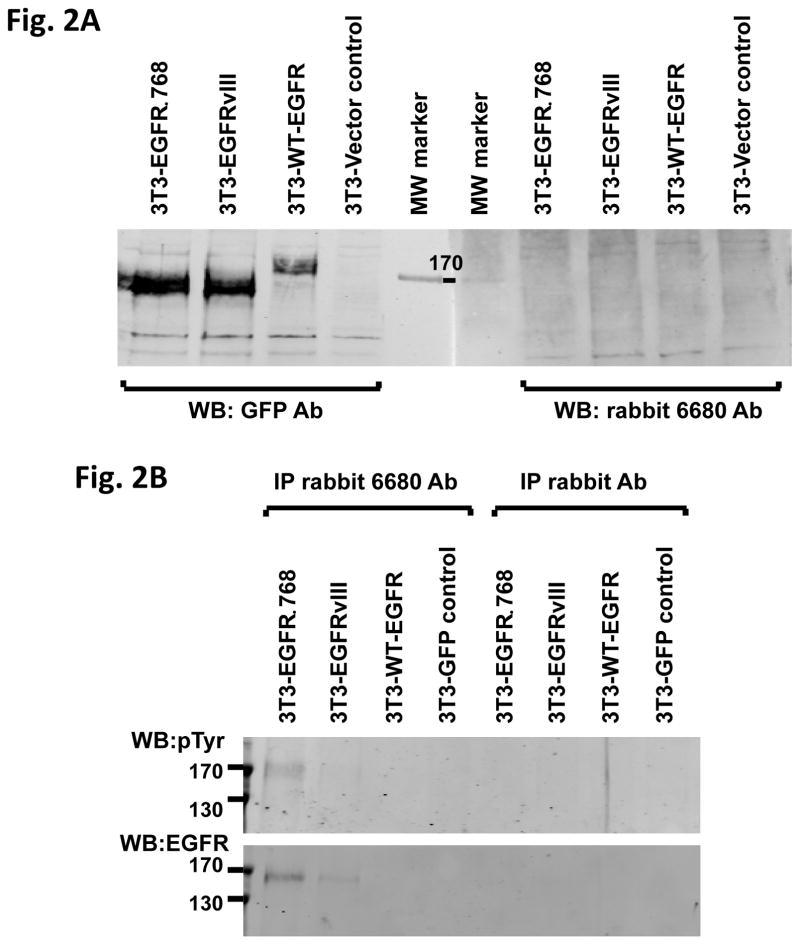
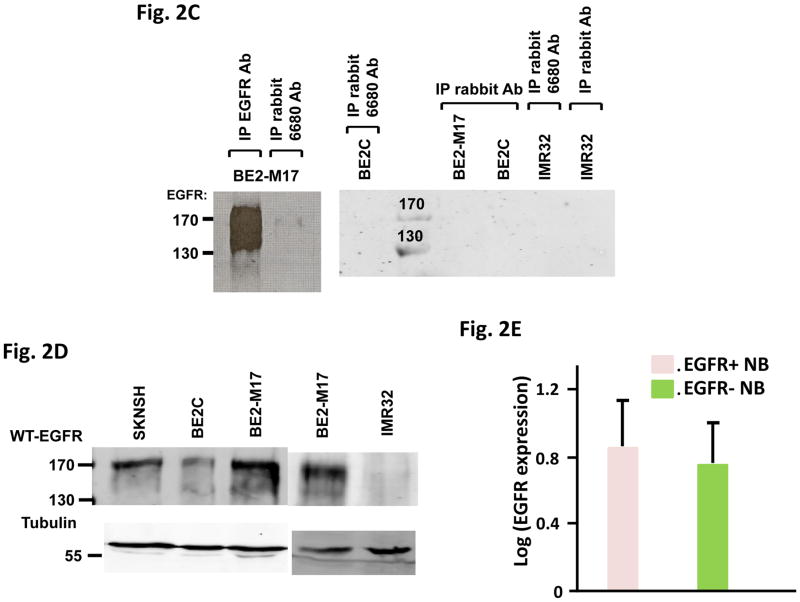
Next, we generated an EGFRΔ768 specific polyclonal antibody. While this antibody did not detect denatured EGFRΔ768 protein on western blot (Fig. 2A), it recognized native EGFRΔ768 and was able to pull down the mutant protein in immunoprecipitation (Fig. 2B). Using this antibody at the optimized condition, we detected the EGFRΔ768 protein expression in the BE2M17 cell lysate (Fig. 2C). Although this antibody binds native EGFRΔ768, its affinity for the endogenous mutant protein is weak. Then, we examined the expression level of WT-EGFR in the NB cell lines. Among the cell lines in our procession, we found that BE2M17, which expressed the EGFRΔ768 mutant, has the highest WT-EGFR level (Fig. 2D). Similarly, the neuroblastic tumors that expressed EGFR mutants (ΔEGFR) have higher WT-EGFR expression than those without the mutations (Fig. 2E). However, the difference in expression level was not statistically significant, implying that there is not a strong correlation between mutants and WT-EGFR expression.
Fig. 2. EGFRΔ768 protein expression in NB.
(A) Western blot analysis of 3T3 cell lysates expressing GFP (vector control), WT-EGFR-GFP, EGFRvIII-GFP and EGFRΔ768-GFP using the purified EGFRΔ768 polyclonal rabbit antibody (6680 Ab) and GFP antibody. MW: molecular weight. (B) Immunoprecipitation followed by western blotting (IPW) analyses of 3T3 cell lysates expressing GFP, WT-EGFR-GFP, EGFRvIII-GFP and EGFRΔ768-GFP using the EGFRΔ768 specific polyclonal rabbit antibody (6680 Ab). The negative controls were IP with rabbit IgG antibody. (C) EGFRΔ768 protein expression in the BE2M17 cell line. BE2M17, BE2C and IMR32 cell lysates were analyzed by IPW using the EGFRΔ768 specific polyclonal rabbit antibody (6680 Ab). The negative controls were IP with rabbit IgG antibody (lane #3, 4 and 6 of the right blot). The positive control was IP with the EGFR antibody, cetuximab (lane #1 of the left blot). (D) WT-EGFR expression in a panel of NB cell lines. (E) Comparison of WT-EGFR expression between ΔEGFR+ (n = 17) and ΔEGFR- (n = 42) CBRC primary NB tumors. ΔEGFR = EGFRvIII + EGFRΔ768. EGFR expression was quantified and compared as previously described (11). 3 of the 20 ΔEGFR+ CBRC NB tumors did not have enough materials for protein analysis. Error bars represent ±2 standard errors.
EGFRΔ768 is biochemically active and localizes to the cell surface
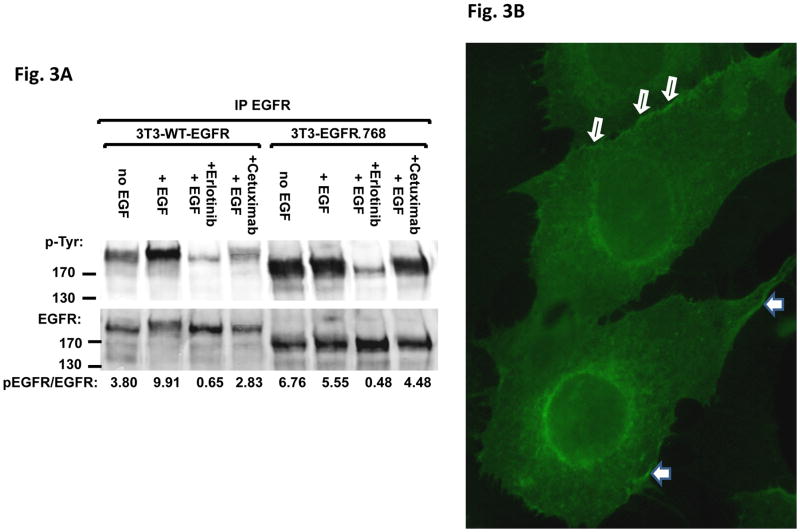
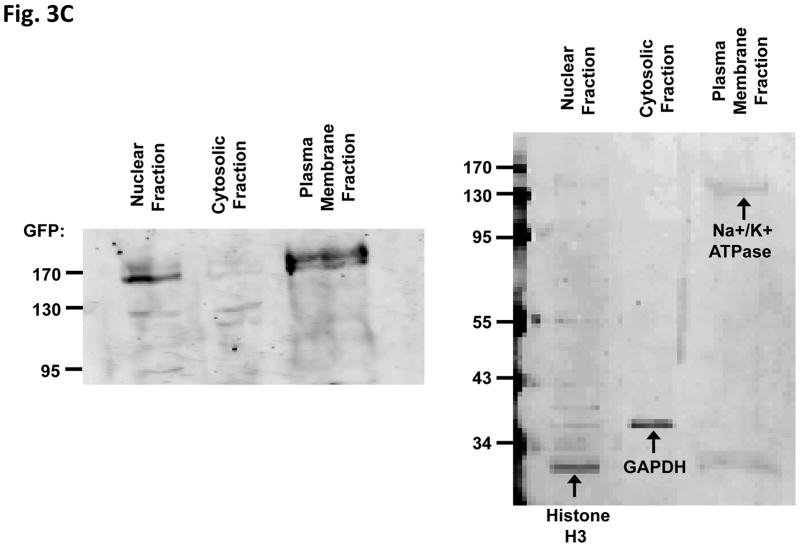
We investigated the biochemical properties of EGFRΔ768. We found that this mutant was constitutively active, but did not respond to EGF stimulation or the EGFR therapeutic antibody, cetuximab. Nevertheless, it responded to high dosage of the EGFR reversible TKI, erlotinib. First, expressing EGFRΔ768 in NIH3T3 cells resulted in a higher phosphorylation signal than expressing WT-EGFR (Fig. 3A, lane 1 and 5). This implied that the mutant is likely capable of self-activation. While EGF stimulation gave a strong phosphorylation signal in WT-EGFR, phosphorylation of EGFRΔ768 did not have an appreciable increase after the addition of EGF (Fig. 3A, lane 2 and 6). This implied that EGFRΔ768 activation is likely ligand independent. While high dose erlotinib inhibited EGFRΔ768, the biochemical effect of cetuximab on EGFRΔ768 was much less than that on the activation of the wild type receptor (Fig. 3A, lane 4 and 8). This is not an unexpected finding for a mutant receptor capable of autophosphorylation. We also made two observations which indirectly supported that some of the EGFRΔ768 receptor localized to the cell surface, which is similar to EGFRvIII (23) and WT-EGFR. First, we performed fluorescence microscopy to observe the expression of EGFRΔ768 in the NIH3T3 clonal cells. We noticed that the protein appeared to be concentrated in membrane ruffles and at the cell to cell contact (Fig. 3B). Furthermore, when we treated the live EGFRΔ768 expressing cells with cetuximab before cell lysis, EGFRΔ768 protein came down when only beads were added to the cell lysate for immunoprecipitation (Fig. 3A). Because cetuximab is too big a molecule to enter the cytosol by itself, this observation implied that cetuximab must have bound to some EGFRΔ768 protein on the cell surface prior to lysing of the cells. Because of an intact domain III in EGFRΔ768, cetuximab was able to bind to the mutant receptor. However, its inhibitory effect on EGFRΔ768 autophosphorylation was not strong (Fig. 3A). To confirm the cell surface expression of EGFRΔ768, we performed cell fractionation experiment on the EGFRΔ768+ 3T3 cells. As shown in Fig. 3C, the EGFRΔ768 protein was detected primarily in the plasma membrane fraction. The relative purity of each fraction was confirmed by the expression of organelle specific proteins. Altogether, these data supported cell surface expression of the EGFRΔ768 receptor.
Fig. 3. Biochemical characterization and localization of EGFRΔ768.
(A) Biochemical characterization of EGFRΔ768. 25 uM of erlotinib and 50 ug/ml of cetuximab were used for the inhibition experiment. No primary antibody was added for IP in the cetuximab treated cell lysates. EGFR phosphorylation/expression was measured as previously described (11). The quantification was depicted as a ratio below the gel. Control IP with IgG antibody were negative (data not shown). (B) Localization of EGFRΔ768-GFP in the NIH3T3 clonal cells. Solid white arrows pointed to GFP concentrated in the membrane ruffles, while hollow white arrows pointed to GFP concentrated at the cell to cell contact. (C) Cell surface expression of EGFRΔ768-GFP in 3T3 cells by cell fractionation and organelle isolation experiment. Noted the strong GFP signal in the plasma membrane fraction (left panel). The identity and relative purity of each fraction was confirmed by the expression of organelle specific markers (right panel).
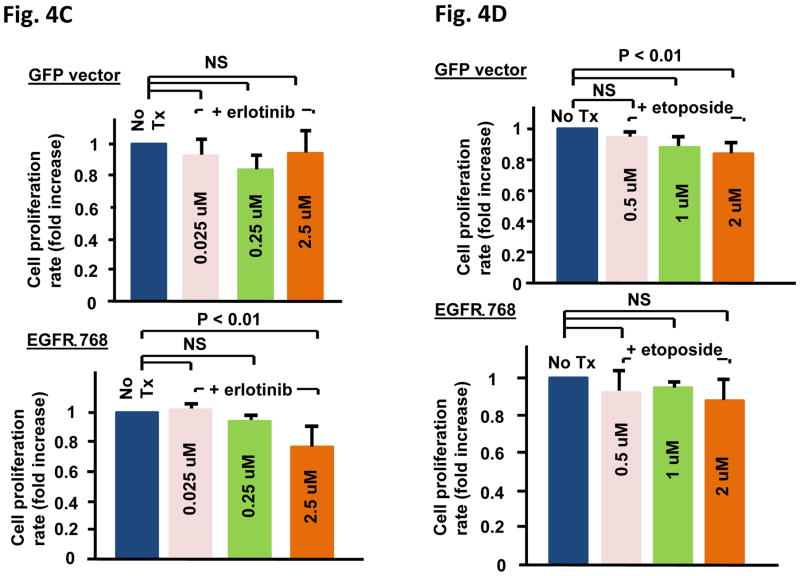
EGFRΔ768 expression increases cellular proliferation and invasion, changes the responsiveness of cancer cells to chemotherapy and EGFR TKI
Next, we investigated the biological effect of EGFRΔ768. We found that EGFRΔ768 expression not only significantly enhanced the invasive potential of BE2C cells (Fig. 4A), but it also augmented the growth potential of both NIH3T3 and BE2C cells (Fig. 4B). Then, we investigated the response of EGFRΔ768+ BE2C cells to erlotinib. While erlotinib did not have a significant inhibitory effect on control BE2C cells, it significantly reduced the proliferation rate of the EGFRΔ768+ BE2C cells at 2.5 uM (Fig. 4C). This result implied that EGFRΔ768 mediates the growth of BE2C cells and that targeting EGFRΔ768 is a plausible therapeutic strategy. Similar results were seen with the NIH3T3 clones (Fig. S4). Finally, we tested the response of the NIH3T3 clones to the chemotherapy, etoposide. While control cells were sensitive to etoposide at concentrations from 1 – 2 uM as expected, EGFRΔ768+ cells did not respond to the chemotherapy at these concentrations (Fig. 4D). These results suggested that EGFRΔ768 expression confers resistance to etoposide.
Fig. 4. Biological characterization of EGFRΔ768.
(A) Cell invasion assay of EGFRΔ768+ BE2C cells and the corresponding control using the modified transwell filter chamber (BD Biosciences) (n = 3). Representative pictures of the stained cells invaded through the Matrigel onto the filters were shown on top of the corresponding bar graphs. Error bars indicate ±2 standard errors. (B) Cell growth analysis of EGFRΔ768+ NIH3T3 (n = 8) and BE2C (n = 9) cells. Cell growth over time was determined using XTT assay (ATCC). The cell proliferation rate of each experiment was calculated as previous described (22). Error bars represent ±2 standard errors. (C) Erlotinib treatment effect on EGFRΔ768+ (bottom panel) and control GFP+ (top panel) BE2C clones (n = 4). Error bars represent ±2 standard errors. Tx: treatment. (D) Etoposide treatment effect on EGFRΔ768+ (bottom panel) and control GFP+ (top panel) 3T3 clones (n = 4 for 0.5 uM, n = 9 for 1 uM and n = 6 for 2 uM). The bonferroni adjusted p value is 0.017. NS: not significant. Error bars represent ±2 standard errors. Tx: treatment.
EGFRΔ768 is biologically and biochemically distinct from WT-EGFR and EGFRvIII
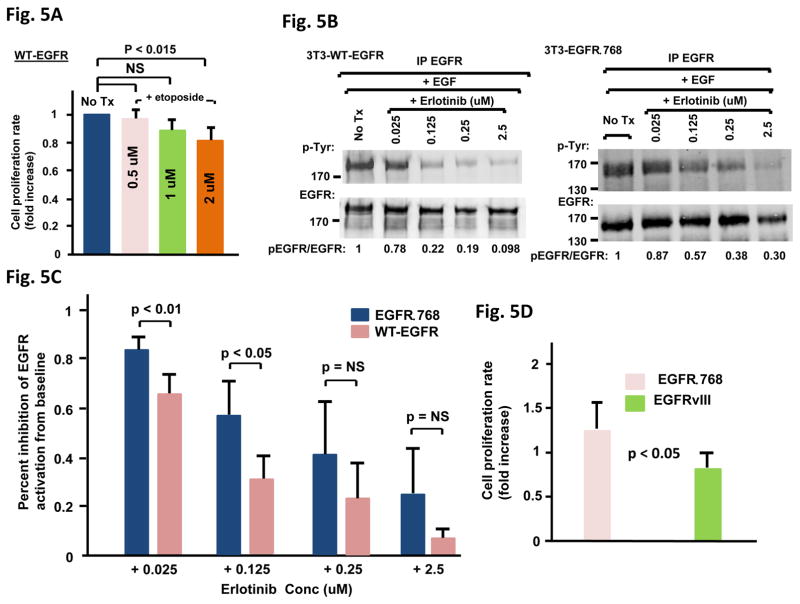
We compared the chemotherapy resistance effect of EGFRΔ768 to that of WT-EGFR. While etoposide significantly reduced the proliferation rate of the WT-EGFR+ 3T3 cells at 2uM (Fig. 5A), EGFRΔ768+ 3T3 cells were resistant to etoposide at that concentration (Fig. 4D). In addition, the inhibitory effect of erlotinib on EGFRΔ768 was not as strong as that on activated WT-EGFR in the range of physiologic concentrations from 0.025 – 2.5 uM (Fig. 5B and C). The biochemical inhibition of EGFRΔ768 by erlotinib was significantly less than that of activated WT-EGFR at the lower concentrations of 0.025 and 0.125 uM (Fig. 5C). These data suggested that EGFRΔ768 has biological and biochemical properties that are different from WT-EGFR. Because of its similarity to EGFRvIII, we also investigated if there are biological differences between the two mutants. We found that the growth rate of EGFRΔ768+ BE2C cells was significantly higher than that of the EGFRvIII+ cells (Fig. 5D). Next, we examined the baseline autophosphorylation status of the two mutants in each stable BE2C clone. Overall, EGFRΔ768 was autophosphorylated at a higher level than EGFRvIII (Fig. 5E). The biochemical response of EGFRvIII to erlotinib was also compared to that of EGFRΔ768. Even though both mutants were similar in their responses to erlotinib at the lowest concentration of 0.025 uM, EGFRΔ768 was significantly less responsive to the biochemical inhibition of erlotinib than EGFRvIII at the higher concentrations from 0.125 – 2.5 uM (Fig. 5F and S5). These results suggested that EGFRΔ768 is biologically and biochemically distinct from EGFRvIII. Taken together, EGFRΔ768 has different biological and biochemical properties from those of WT-EGFR and EGFRvIII.
Fig. 5. Biological and biochemical comparison between EGFRΔ768, WT-EGFR and EGFRvIII.
(A) Etoposide treatment effect on WT-EGFR+ 3T3 clones (n = 4 for 0.5 uM, n = 7 for 1 uM and n = 6 for 2 uM). The bonferroni adjusted p value is 0.017. NS: not significant. Error bars represent ±2 standard errors. Tx: treatment. (B) Biochemical response to erlotinib treatment of WT-EGFR+ and EGFRΔ768+ 3T3 cells. EGFR phosphorylation/expression was analyzed and quantified as previously described (22). Baseline EGFR phosphorylation was normalized to 1 for comparison. Tx: treatment. (C) Comparison of the biochemical response to erlotinib over the range of concentrations from 0.025 – 2.5 uM between the WT-EGFR+ and EGFRΔ768+ 3T3 cells (n = 5 at each concentration). NS: not significant. Error bars represent ±2 standard errors. (D) Comparison of the cell proliferation rate between the EGFRvIII+ (n = 8) and EGFRΔ768+ (n = 9) BE2C clones. Error bars represent ±2 standard errors. (E) Baseline autophosphorylation status of each individual EGFRΔ768+ and EGFRvIII+ BE2C clone. (F) Comparison of the biochemical response to erlotinib over the range of concentrations from 0.025 – 2.5 uM between the EGFRvIII+ and EGFRΔ768+ 3T3 cells (n = 5 at each concentration). NS: not significant. Error bars represent ±2 standard errors.
EGFRΔ768 does not activate the known downstream signaling pathways of WT-EGFR
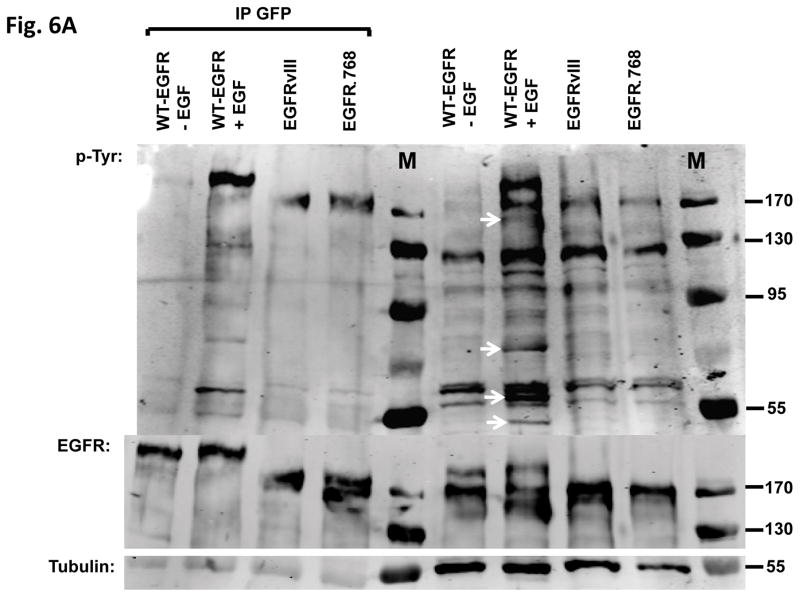
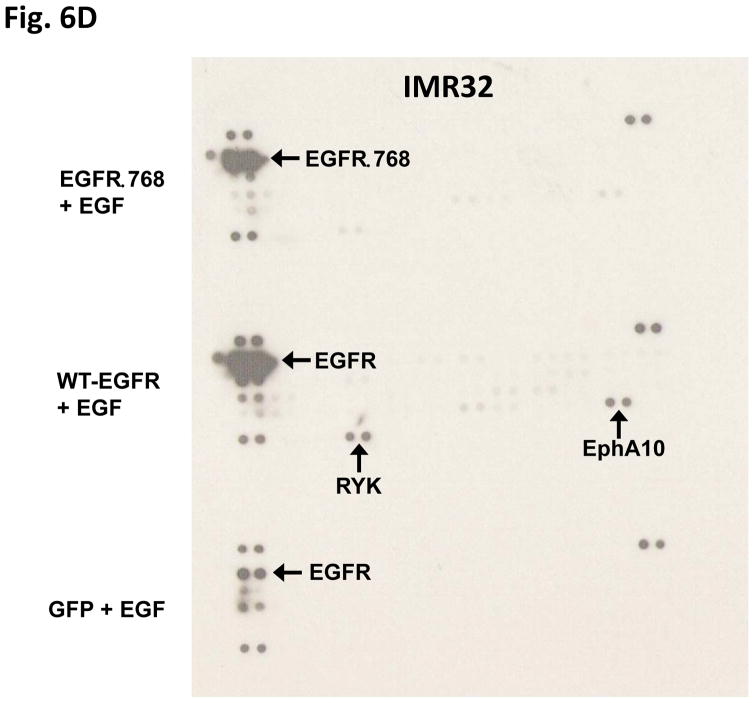
To determine if EGFRΔ768 activates downstream signaling pathways similar to activated WT-EGFR, we compared the tyrosine phosphorylation profiles between activated WT-EGFR and EGFRΔ768 protein lysates. There were several tyrosine phosphorylated bands in the activated WT-EGFR lysates that were not present in the EGFRΔ768 lysates (Fig. 6A). This suggested that EGFRΔ768 activated pathways different from WT-EGFR. To screen for signaling pathways that might be regulated by EGFRΔ768, we probed the NB cell lysates with phosphoarrays. As expected, EGF stimulation of WT-EGFR activated MAPK/Erk and WNK1 in BE2C and IMR32 cells respectively (Fig 6B & C). These are two known downstream intermediaries of WT-EGFR (24–28). On the other hand, EGFRΔ768 expression and phosphorylation did not activate these pathways in these cells (Fig 6B & C). Interestingly, HSP60 signal was not observed in the EGFRΔ768+ IMR32 cell lysate as compared to the GFP and WT-EGFR samples (Fig. 6B). In addition, EGFRΔ768 signaling did not activate receptors such as EphA10 or RYK, which were phosphorylated by activated WT-EGFR in IMR32 cells (Fig. 6D). Overall, these data suggested that downstream signaling of EGFRΔ768 is different from that of activated WT-EGFR. We also performed phosphoarrays on the EGFRvIII sample. Similar to activated WT-EGFR, EGFRvIII expression and phosphorylation in the IMR32 cells activated EphA10 and RYK receptors as well. Interestingly, it also activated the ALK receptor, which was not observed in the activated WT-EGFR and EGFRΔ768 samples (Fig. S7). However, unlike activated WT-EGFR, EGFRvIII did not activate WNK1, but persistent HSP60 signal was observed (Fig. S7). These results hinted that EGFRvIII signaling in neuroblastoma might be different from not only activated WT-EGFR, but also EGFRΔ768.
Fig. 6. Comparison of signaling pathways downstream of EGFRΔ768 and WT-EGFR.
(A) Tyrosine phosphorylation pattern of activated WT-EGFR, EGFRΔ768 and EGFRvIII in the transiently transfected IMR32 cells. White arrows pointed to tyrosine phosphorylated protein bands that were present in the WT-EGFR cell lysate, but not in the EGFRΔ768/vIII lysates. (B) Phosphokinase array analysis of activated WT-EGFR and EGFRΔ768 in the BE2C cells. p53 signaling pathway was activated at baseline in these cells because of a known p53 mutation (48). WT-EGFR activation resulted in phosphorylation of Erk1/2 in these cells. (C) Phosphokinase array analysis of activated WT-EGFR and EGFRΔ768 in the transiently transfected IMR32 cells. WT-EGFR activation resulted in phosphorylation of WNK1 in these cells. Noted that IMR32 cell has baseline HSP60 signal. (D) Phosphoreceptor tyrosine kinase array analysis of activated WT-EGFR and EGFRΔ768 in the transiently transfected IMR32 cells. WT-EGFR activation resulted in phosphorylation of EphA10 and RYK in these cells. Noted the strong EGFRΔ768 phosphorylation signal detected by the array.
Discussion
Neuroblastoma (NB) is the most common extracranial solid tumor of childhood and accounts for more than 7% of childhood malignancies. Despite aggressive therapy, children with high risk neuroblastoma still fare poorly with less than half surviving the disease. Because of the intensified treatment, the survivors also suffer many long term side effects. Therefore, current effort in neuroblastoma research aims to identify driver mutations and potential druggable targets. The NCI sponsored TARGET Project utilized a comprehensive genomic approach to achieve this goal. Using a combination of high throughput sequencing technologies, a few recurrently mutated genes were identified in high-risk neuroblastoma including the oncogene, ALK (29). ALK mutations were first identified as germline mutations in hereditary neuroblastoma (30). Since then, multiple groups have not only found somatically acquired ALK mutations in sporadic neuroblastoma (31–33), but also defined its oncogenic role in animal models (34–36). In fact, ALK inhibitors are currently in clinical trials for neuroblastoma patients (37). Despite these promising discoveries, the mutation rate in neuroblastoma is low and targeted therapies have not yet been established for this disease. Thus, the identification of other potential driver mutations in this tumor may lead to a more effective therapeutic strategy.
To the best of our knowledge, this study is the first to show that neuroblastoma expressed EGFR mutants. We not only discovered the expression of EGFRvIII, but also a novel EGFR extracellular domain mutant, EGFRΔ768. This discovery might have therapeutic implication for neuroblastoma patients. First, we detected the expression of these EGFR mutants in a significant percentage of neuroblastic tumors (~23%). Their prevalence seemed to be higher than that of ALK mutations in neuroblastoma (~ 8%) (38). While most neuroblastoma patients did not respond to EGFR TKI, preselecting those with EGFR mutant expression might improve the response rate. In turn, this might lead to a more rational use of the EGFR inhibitors in clinical trials. Since EGFRvIII is an important oncogenic driver in some glioblastoma (a tumor of the central nervous system) (39), EGFR mutants might also play an important role in the oncogenesis of a peripheral nervous system tumor like neuroblastoma. Second, neuroblastoma response to the EGFR TKI was not dramatic in clinical studies (13, 15). Since our data suggested that the reversible EGFR TKI, erlotinib, might not inhibit EGFRΔ768 as well as it did on activated WT-EGFR or EGFRvIII, it is possible that EGFRΔ768 expressing tumors require a more potent EGFR inhibitor. In addition, it appeared that the EGFR therapeutic antibody, cetuximab, did not block the signaling of the EGFRΔ768 receptor as efficiently as it did on the activation of the wild type receptor. Since EGFRΔ768 activation might not be ligand dependent, the occupation of the ligand binding site in domain III by cetuximab on EGFRΔ768 is unlikely to prevent its activation. Furthermore, since most of domain I is deleted in the EGFRΔ768 (amino acid #33 – 289), it is conceivable that cetuximab might not be able to prevent EGFRΔ768 from dimerization and thus self-activation through the interaction between domain I of the receptor and the Fab region of the cetuximab (40). Together, these results suggested that the EGFR antibodies or the reversible EGFR TKI might not work as well on the EGFRΔ768 mutant as they do on WT-EGFR. In that case, more potent EGFR inhibitors such as the new irreversible inhibitors might be needed to inhibit EGFRΔ768. We are currently investigating the efficacy of the second generation EGFR TKI such as afatinib, dacomitinib and neratinib on EGFRΔ768. We hope that the results from this study will reignite an interest in the role of EGFR inhibitors in neuroblastoma, a disease that is in desperate need of targeted therapy.
The EGFRΔ768/vIII discovery in neuroblastoma was unexpected because prior studies had not identified these mutants. However, the previous study of EGFR mutations in neuroblastoma only screened for mutants in the kinase domain, but not in the extracellular domain (18). These investigators did not find any activating kinase mutations in these tumors and neither did we. Surprisingly, the TARGET project did not detect EGFRΔ768 or EGFRvIII (29). It is possible that the mutations were missed by high throughput sequencings, which is not uncommon with large deletions (41). For examples, both exome and/or transcriptome sequencings did not identify EGFRvIII in head and neck squamous cell carcinoma (HNSCC) or pediatric glioma (42, 43), even though gene specific primers detected its expression in these tumors (11, 44–46). Recent data also suggested that large scale automated sequencing missed EGFRvIII in HNSCC whereas specific assay designed to detect this mutant identify its expression in primary tumors (47). Despite the vast amount of data obtained from high throughput sequencings, the possibility of false negatives exists, which is a limitation of these techniques. Our study illustrated the value of a more comprehensive sequencing approach of a potential oncogene using specifically optimized PCR assay.
Supplementary Material
Implications.
Neuroblastoma expressed a novel EGFR mutant which possesses distinct biological and biochemical properties that might have therapeutic implications.
Acknowledgments
Grant Supports
This work was supported in part by research grants from the American Cancer Society (117718-MRSG-09-172-01-CCE), the National Cancer Institute (1R21CA187554), the Sunrise Fund as well as the Stony Brook Targeted Research Opportunity Grant Program including the Catacosinos Cancer Translational Researcher Award (1088327-54437) and the Clinical Research Award (1096080-37298) (E.L.C.). The Children Oncology Group Biospecimen Bank is supported by the National Cancer Institute (U24CA114766).
Frozen tumor tissues were provided by the Columbus Biopathology Repository Center. Neuroblastoma tumor RNA were kindly provided by the Children Oncology Group. We also liked to acknowledge Mrs. Patti Kelly for her effort in raising the Sunrise Fund to support this research in memory and honor of her loving daughter, Lizzie Kelly.
The abbreviations used are
- EGFR
epidermal growth factor receptor
- FDA
food and drug administration
- TKI
tyrosine kinase inhibitor
- NB
neuroblastoma
- IRB
Institutional Review Board
- FBS
fetal bovine serum
- CBRC
Columbus Biopathology Repository Center
- COG
Children Oncology Group
- NCI
National Cancer Institute
- STR
short tandem repeat
- TARGET
Therapeutically Applicable Research to Generate Effective Treatments
- DMSO
dimethyl sulfoxide
- IPW
immunoprecipitation followed by western blotting
- GFP
green fluorescent protein
- KLH
Keyhole limpet hemocyanin
- PBS
phosphate buffered saline
Footnotes
Conflict of Interest Disclosure Statement
The first and senior authors hold a US patent (#8,962,808) on the method of use of EGFRΔ768.
References
- 1.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37(Suppl 4):S9–15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 2.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 3.Sequist LV, Bell DW, Lynch TJ, Haber DA. Molecular predictors of response to epidermal growth factor receptor antagonists in non-small-cell lung cancer. J Clin Oncol. 2007;25:587–95. doi: 10.1200/JCO.2006.07.3585. [DOI] [PubMed] [Google Scholar]
- 4.Mellinghoff IK, Wang MY, Vivanco I, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–24. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 5.Montagut C, Dalmases A, Bellosillo B, et al. Identification of a mutation in the extracellular domain of the Epidermal Growth Factor Receptor conferring cetuximab resistance in colorectal cancer. Nat Med; 18:221–3. doi: 10.1038/nm.2609. [DOI] [PubMed] [Google Scholar]
- 6.Zhang P, Zhou M, Jiang H, et al. Exon 4 deletion variant of epidermal growth factor receptor enhances invasiveness and cisplatin resistance in epithelial ovarian cancer. Carcinogenesis; 34:2639–46. doi: 10.1093/carcin/bgt216. [DOI] [PubMed] [Google Scholar]
- 7.Pedersen MW, Meltorn M, Damstrup L, Poulsen HS. The type III epidermal growth factor receptor mutation. Biological significance and potential target for anti-cancer therapy. Ann Oncol. 2001;12:745–60. doi: 10.1023/a:1011177318162. [DOI] [PubMed] [Google Scholar]
- 8.Moscatello DK, Holgado-Madruga M, Godwin AK, et al. Frequent expression of a mutant epidermal growth factor receptor in multiple human tumors. Cancer Res. 1995;55:5536–9. [PubMed] [Google Scholar]
- 9.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–81. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 10.Frederick L, Wang XY, Eley G, James CD. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 2000;60:1383–7. [PubMed] [Google Scholar]
- 11.Keller J, Shroyer KR, Batajoo SK, et al. Combination of phosphorylated and truncated EGFR correlates with higher tumor and nodal stage in head and neck cancer. Cancer Invest; 28:1054–62. doi: 10.3109/07357907.2010.512602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Zhou M, Shi B, et al. Identification of an exon 4-deletion variant of epidermal growth factor receptor with increased metastasis-promoting capacity. Neoplasia; 13:461–71. doi: 10.1593/neo.101744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jakacki RI, Hamilton M, Gilbertson RJ, et al. Pediatric phase I and pharmacokinetic study of erlotinib followed by the combination of erlotinib and temozolomide: a Children’s Oncology Group Phase I Consortium Study. J Clin Oncol. 2008;26:4921–7. doi: 10.1200/JCO.2007.15.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donfrancesco A, Jenkner A, De Ioris MA, et al. Prolonged Response to Oral Gefitinib, Cyclophophamide, and Topotecan in Heavily Pretreated Relapsed Stage 4 Neuroblastoma: A Case Report. J Pediatr Hematol Oncol. 2007;29:799–803. doi: 10.1097/MPH.0b013e31815815f6. [DOI] [PubMed] [Google Scholar]
- 15.Donfrancesco A, De Ioris MA, McDowell HP, et al. Gefitinib in combination with oral topotecan and cyclophosphamide in relapsed neuroblastoma: pharmacological rationale and clinical response. Pediatr Blood Cancer; 54:55–61. doi: 10.1002/pbc.22219. [DOI] [PubMed] [Google Scholar]
- 16.Ho R, Minturn JE, Hishiki T, et al. Proliferation of human neuroblastomas mediated by the epidermal growth factor receptor. Cancer Res. 2005;65:9868–75. doi: 10.1158/0008-5472.CAN-04-2426. [DOI] [PubMed] [Google Scholar]
- 17.Richards KN, Zweidler-McKay PA, Van Roy N, et al. Signaling of ERBB receptor tyrosine kinases promotes neuroblastoma growth in vitro and in vivo. Cancer; 116:3233–43. doi: 10.1002/cncr.25073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Izycka-Swieszewska E, Brzeskwiniewicz M, Wozniak A, et al. EGFR, PIK3CA and PTEN gene status and their protein product expression in neuroblastic tumours. Folia Neuropathol; 48:238–45. [PubMed] [Google Scholar]
- 19.Ischenko I, Petrenko O, Gu H, Hayman MJ. Scaffolding protein Gab2 mediates fibroblast transformation by the SEA tyrosine kinase. Oncogene. 2003;22:6311–8. doi: 10.1038/sj.onc.1206742. [DOI] [PubMed] [Google Scholar]
- 20.Agazie YM, Hayman MJ. Molecular mechanism for a role of SHP2 in epidermal growth factor receptor signaling. Mol Cell Biol. 2003;23:7875–86. doi: 10.1128/MCB.23.21.7875-7886.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim HJ, Taylor LJ, Bar-Sagi D. Spatial regulation of EGFR signaling by Sprouty2. Curr Biol. 2007;17:455–61. doi: 10.1016/j.cub.2007.01.059. [DOI] [PubMed] [Google Scholar]
- 22.Keller J, Nimnual AS, Shroyer KR, et al. Ron tyrosine kinase receptor synergises with EGFR to confer adverse features in head and neck squamous cell carcinoma. Br J Cancer; 109:482–92. doi: 10.1038/bjc.2013.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wikstrand CJ, McLendon RE, Friedman AH, Bigner DD. Cell surface localization and density of the tumor-associated variant of the epidermal growth factor receptor, EGFRvIII. Cancer Res. 1997;57:4130–40. [PubMed] [Google Scholar]
- 24.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–54. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 25.Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–16. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 26.Scaltriti M, Baselga J. The epidermal growth factor receptor pathway: a model for targeted therapy. Clin Cancer Res. 2006;12:5268–72. doi: 10.1158/1078-0432.CCR-05-1554. [DOI] [PubMed] [Google Scholar]
- 27.Moniz S, Jordan P. Emerging roles for WNK kinases in cancer. Cell Mol Life Sci; 67:1265–76. doi: 10.1007/s00018-010-0261-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Large MJ, Wetendorf M, Lanz RB, et al. The epidermal growth factor receptor critically regulates endometrial function during early pregnancy. PLoS Genet. 10:e1004451. doi: 10.1371/journal.pgen.1004451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pugh TJ, Morozova O, Attiyeh EF, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet; 45:279–84. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mosse YP, Laudenslager M, Longo L, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–5. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, Takita J, Choi YL, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971–4. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- 32.George RE, Sanda T, Hanna M, et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;455:975–8. doi: 10.1038/nature07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janoueix-Lerosey I, Lequin D, Brugieres L, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455:967–70. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- 34.Heukamp LC, Thor T, Schramm A, et al. Targeted expression of mutated ALK induces neuroblastoma in transgenic mice. Sci Transl Med. 4:141ra91. doi: 10.1126/scitranslmed.3003967. [DOI] [PubMed] [Google Scholar]
- 35.Berry T, Luther W, Bhatnagar N, et al. The ALK(F1174L) mutation potentiates the oncogenic activity of MYCN in neuroblastoma. Cancer Cell; 22:117–30. doi: 10.1016/j.ccr.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cazes A, Lopez-Delisle L, Tsarovina K, et al. Activated Alk triggers prolonged neurogenesis and Ret upregulation providing a therapeutic target in ALK-mutated neuroblastoma. Oncotarget; 5:2688–702. doi: 10.18632/oncotarget.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carpenter EL, Mosse YP. Targeting ALK in neuroblastoma--preclinical and clinical advancements. Nat Rev Clin Oncol; 9:391–9. doi: 10.1038/nrclinonc.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mosse YP. Anaplastic Lymphoma Kinase as a Cancer Target in Pediatric Malignancies. Clin Cancer Res; 22:546–52. doi: 10.1158/1078-0432.CCR-14-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thorne AH, Zanca C, Furnari F. Epidermal growth factor receptor targeting and challenges in glioblastoma. Neuro Oncol. doi: 10.1093/neuonc/nov319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li S, Schmitz KR, Jeffrey PD, Wiltzius JJ, Kussie P, Ferguson KM. Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell. 2005;7:301–11. doi: 10.1016/j.ccr.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 41.Biesecker LG, Green RC. Diagnostic clinical genome and exome sequencing. N Engl J Med; 370:2418–25. doi: 10.1056/NEJMra1312543. [DOI] [PubMed] [Google Scholar]
- 42.Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science; 333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu G, Diaz AK, Paugh BS, et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet; 46:444–50. doi: 10.1038/ng.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sok JC, Coppelli FM, Thomas SM, et al. Mutant epidermal growth factor receptor (EGFRvIII) contributes to head and neck cancer growth and resistance to EGFR targeting. Clin Cancer Res. 2006;12:5064–73. doi: 10.1158/1078-0432.CCR-06-0913. [DOI] [PubMed] [Google Scholar]
- 45.Bax DA, Gaspar N, Little SE, et al. EGFRvIII deletion mutations in pediatric high-grade glioma and response to targeted therapy in pediatric glioma cell lines. Clin Cancer Res. 2009;15:5753–61. doi: 10.1158/1078-0432.CCR-08-3210. [DOI] [PubMed] [Google Scholar]
- 46.Li G, Mitra SS, Monje M, et al. Expression of epidermal growth factor variant III (EGFRvIII) in pediatric diffuse intrinsic pontine gliomas. J Neurooncol; 108:395–402. doi: 10.1007/s11060-012-0842-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wheeler SE, Egloff AM, Wang L, James CD, Hammerman PS, Grandis JR. Challenges in EGFRvIII detection in head and neck squamous cell carcinoma. PLoS One. 10:e0117781. doi: 10.1371/journal.pone.0117781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tweddle DA, Malcolm AJ, Bown N, Pearson AD, Lunec J. Evidence for the development of p53 mutations after cytotoxic therapy in a neuroblastoma cell line. Cancer Res. 2001;61:8–13. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.