Abstract
Gene body methylation (gbM) is an ancestral and widespread feature in Eukarya, yet its adaptive value and evolutionary implications remain unresolved. The occurrence of gbM within protein-coding sequences is particularly puzzling, because methylation causes cytosine hypermutability and hence is likely to produce deleterious amino acid substitutions. We investigate this enigma using an evolutionarily basal group of Metazoa, the stony corals (order Scleractinia, class Anthozoa, phylum Cnidaria). We show that patterns of coral gbM are similar to other invertebrate species, predicting wide and active transcription and slower sequence evolution. We also find a strong correlation between gbM and codon bias, resulting from systematic replacement of CpG bearing codons. We conclude that gbM has strong effects on codon evolution and speculate that this may influence establishment of optimal codons.
Keywords: DNA methylation, gene body methylation, codon bias, coral, substitution rate, gene expression
Introduction
DNA methylation is an evolutionarily widespread epigenetic modification found in plants, animals, and fungi. It is defined as the covalent addition of a methyl group to the one of the four DNA bases, predominantly on the fifth carbon of cytosines within CG dinucleotides (CpGs), producing 5-methylcytosine (5mC). Unlike other epigenetic modifications, DNA methylation not only alters chromatin structure and transcription, but it also changes the mutation rate of the underlying DNA. This is because 5mC undergoes deamination reactions more readily than normal cytosine (Shen et al. 1994) and deamination produces thymine rather than uracil, which is less likely to be accurately repaired (Zemach and Zilberman 2010). Because of this hypermutability, sequences that are heavily methylated in the germ-line become deficient in CpGs, with corresponding increases in TpGs and CpAs (Sved and Bird 1990). Hence, DNA methylation has evolutionary consequences outside of its direct physiological effects.
Evolutionary effects of 5mC hypermutability are apparent in both vertebrate and invertebrate genomes. In mammals, DNA methylation is ubiquitous, so that nearly all genomic regions show lower than expected frequency of CpGs (Karlin and Mrázek 1996; McGaughey et al. 2014). The exception is regions of elevated CpG content called CG islands that are protected from DNA methylation (Jones 2012). In most invertebrates, DNA methylation is not ubiquitous but patchy, occurring primarily on CpGs within gene bodies (Suzuki et al. 2007; Zemach et al. 2010). This intragenic form of DNA methylation is referred to as gene body methylation (gbM). In invertebrate genomes, gbM occurs preferentially on actively and widely expressed genes, resulting in covariations between genes’ CpG content, function, and expression patterns (Elango et al. 2009; Hunt and Brisson 2010; Zemach et al. 2010, Sarda et al. 2012). Similar patterns of genic methylation are found in plants (Feng et al. 2010; Takuno and Gaut 2013; Tran et al. 2005; Zilberman et al. 2007) and mammals (Baubec et al. 2015).
Despite this widespread phylogenetic occurrence, gbM is by no means universal. In several groups, such as yeast (Saccharomyces cerevisiae), fruit fly (Drosophila melanogaster), worm (Caenorhabditis elegans), and the basal plant (Marchantia polymorpha), genic methylation is extremely scarce or lost altogether (Capuano et al. 2014; Takuno et al. 2016). It has been proposed that the secondary loss of DNA methylation occurs because its mutational costs outweighed its adaptive value (Zemach et al. 2010). Indeed, even within gene bodies, methylation occurs preferentially on exons (Zemach et al. 2010; Wang et al. 2013), where mutations are likely to have the greatest deleterious effect. In humans, genic methylation increases deleterious de novo mutations with paternal age (Francioli et al. 2015). Why, given its apparently nonessential and outright mutagenic nature, has gbM persisted for so long across such a diversity of taxa?
In this study, we investigate the evolutionary consequences of 5mC on invertebrate coding sequences. Using the first direct genome-wide characterization of DNA methylation in a reef-building coral, we confirm previous studies showing that gbM predicts active and stable gene expressive (Elango et al. 2009; Hunt and Brisson 2010; Zemach et al. 2010, Sarda et al. 2012). We also test previous findings that in spite of 5mC hypermutability, gbM predicts slower sequence evolution (Park et al. 2011; Takuno and Gaut 2012). Finally, we examine gbM in the context of synonymous codon usage. Because gbM occurs preferentially on a subset of invertebrate coding genes (Sarda et al. 2012), we hypothesized that its mutagenic effects cause intragenomic variation in codon bias. While methylation is often cited as an explanation for patterns of codon usage (Kanaya et al. 2001; Sterky et al. 2004; Gonzalez-Ibeas et al. 2007; Qin et al. 2013; Duan et al. 2015), direct investigations of this hypothesis have been lacking. As a basal metazoan predicted to have a typical bimodal pattern of gbM (Dixon et al. 2014; Dimond and Roberts 2016), the branching coral Acropora millepora was well suited to address this problem.
Results
Using MBD-Seq to Quantify Gene Body Methylation
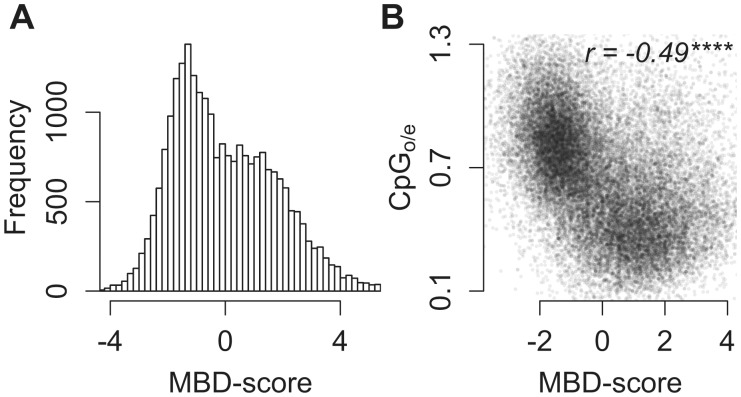
We used Methylation Binding Domain enrichment sequencing (MBD-seq) (Harris et al. 2010) to measure gbM in A. millepora. The strength of methylation for 24,320 coding regions was quantified as the log2-fold difference between captured and flow-through fractions of MBD enrichment preparations. We refer to this log2-fold difference as the MBD-score. Raw read data are publicly available through the NCBI SRA database (SRA accession: SRP074615). Analysis of the distribution of MBD-scores (fig. 1A) showed that it was best described as a mixture of two or more Gaussian components (supplementary fig. S1, Supplementary Material online). MBD-score correlated with CpGo/e, indicating that our measure of gbM overlapped closely with historical patterns of germ-line methylation (fig. 1B). As an MBD-score of 0 indicated equal representation in the captured and flow-through fractions we chose this value to separate strongly and weakly methylated genes. Genes with MBD-scores greater than 0 are referred to as strongly methylated genes, those with scores less than 0 are referred to as weakly methylated.
Fig. 1.
MBD-score is bimodally distributed and correlates with CpGo/e. (A) Distribution of MBD-score (log2-fold difference between enriched and flow-through MBD-seq libraries). Higher values indicate stronger methylation. (B) Scatter plot of MBD-score and CpGo/e. Lower values for CpGo/e are expected with stronger methylation. Asterisks indicate significance based on Spearman’s rank-order correlation test (ns >0.05; * <0.05; ** <0.01; *** <0.001; **** <0.0001).
MBD-Score Is Linked with Gene Function and Expression Patterns
MBD-score was associated with gene function. Analysis of selected GO categories for biological processes revealed that strongly methylated genes tend toward biological functions that are spatially and temporally stable, such as DNA metabolism, ribosome biogenesis, translation, RNA metabolism, and transcription. Weakly methylated genes tended to involve biological functions that are spatially and temporally regulated, such as cell–cell signaling, response to stimulus, signal transduction, cell adhesion, defense response, and development (supplementary fig. S2A, Supplementary Material online). Clustering of KOG categories for higher or lower MBD-scores further supported these results (supplementary fig. S2B, Supplementary Material online).
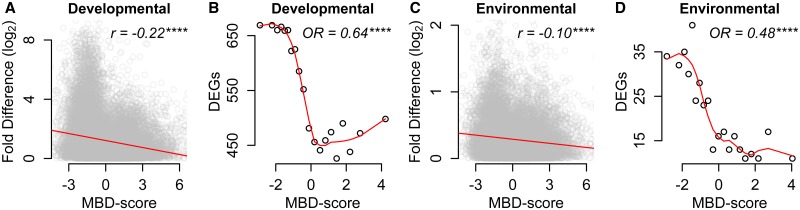
To test if weak gbM is a signature for inducible transcription we correlated MBD-score with RNA-seq data, comparing different developmental stages and environmental conditions. For developmental stage, log2-fold differences in transcript abundance between A. millepora adults and larvae (described in Dixon et al. 2015) were negatively correlated with MBD-score (fig. 2A). Significantly differentially expressed genes (DEGs at FDR <0.01) were 1.4 times more frequent among weakly methylated genes (fig. 2B). A similar trend was found for variation in expression due to environmental conditions (fig. 2C and D). Here, clonal fragments of adult colonies were exposed to two environmentally distinct regimes for 3 months prior to sampling for RNA-seq (Dixon et al. 2014). Differential expression (FDR <0.01) between environmental regimes was 2.2 times more frequent among weakly methylated genes.
Fig. 2.
gbM predicts transcriptional stability across developmental stages and environmental regimes. (A) Scatter plot of MBD-score and transcriptional variation (given as log2-fold differences) between adult colonies and juvenile offspring. Red line shows least squared regression. Asterisks indicate significance based on Spearman’s rho. (B) Distribution of differentially expressed genes (DEGs; FDR <0.01) between juveniles and adults. All genes were divided into 20 quantiles ranked by MBD-score. The number of differentially expressed genes in each quantile was plotted against the median MBD-score for that quantile. Enrichment of DEGs among the weakly methylated genes (MBD-score <0) compared with strongly methylated genes (MBD-score ≥0) is given as the odds ratio (OR) for Fisher’s exact test. Red line shows a smoothed trace of the points. (C, D) The same figures representing transcriptional variation between populations of clonal colony fragments transplanted between distinct habitats described in Dixon et al. (2014). Asterisks indicate significance based on Spearman’s rank-order correlation test (ns >0.05; * <0.05; ** <0.01; *** <0.001; **** <0.0001).
MBD-score also showed weak but significant correlation with transcript abundance (supplementary fig. S3A and B, Supplementary Material online). Highly expressed genes were on average strongly methylated (supplementary fig. S3C and D, Supplementary Material online). The top 5% most strongly methylated genes however showed lower average expression (supplementary fig. S3E, Supplementary Material online). This indicates that while gbM is generally associated with elevated transcription, extreme levels may be inhibitory. This appears to be particularly true for short genes, as the removal of coding sequences shorter than 800 bp mitigated the trend (supplementary fig. S3F, Supplementary Material online).
Phylogeny
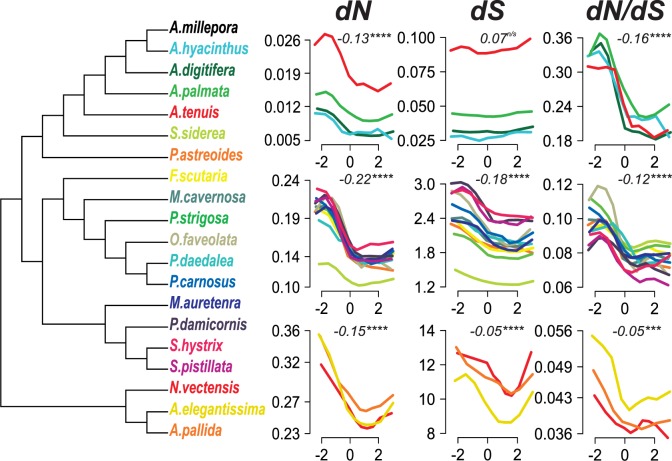
We used a conserved set of 192 coding sequences for phylogenetic construction. These sequences had >75% amino acid identity and 80% representation among the 20 species. Phylogenetic construction was performed using the GTRGAMMA model in RAxML (Stamatakis 2014). All bipartition had 100% bootstrap support based on 1,000 repetitions. All orders, families, and genera formed monophyletic groups (fig. 3). Species from the ‘complex’ and ‘robust’ coral clades (Romano and Palumbi 1996; Kitahara et al. 2010) also formed monophyletic groups. Repetitions of tree building with less conserved sets of orthologs (70%, 60%, 50%, and 40% representation among species) all produced the same topology, but with lower bootstrap values. For the species in which they overlapped, our tree agreed fully with that published by Kitchen et al. (2015).
Fig. 3.
Relationship between MBD-score and substitution rates across the anthozoan phylogeny. All nodes in the phylogeny have 100% bootstrap support based on 1,000 replicates. Line plots trace the mean substitution rates for all genes divided into 10 quantiles ranked by MBD-score. Line color indicates which species A. millepora was compared with to estimate pair-wise substation rates. The top row of line plots shows comparisons within Acropora. The middle row shows corals outside of Acropora. The third row shows comparisons with anemone species. For each panel, the correlation (Spearman’s rho) and statistical significance indicate the median values across all included species. Individual correlations are reported in the Supplementary Material online. Asterisks indicate significance based on Spearman’s rank-order correlation test (ns >0.05; * <0.05; ** <0.01; *** <0.001; **** <0.0001).
Strongly Methylated Genes Evolve Slowly
Pairwise comparisons between orthologs from A. millepora and each other species revealed that strongly methylated genes evolve slowly. The trend was strongest for nonsynonymous substitutions (dN). When orthologs from A. millepora were compared with other Acropora species, mean dN was between 43% and 68% higher for weakly methylated genes than strongly methylated genes (fig. 3; supplementary fig. S4, Supplementary Material online). Pairwise comparisons with all species outside of the Acropora genus produced similar results, with mean dN between 17% and 52% (mean = 36 ± SEM 3%) higher for weakly methylated genes (fig. 3; supplementary fig. S5, Supplementary Material online). Negative correlation between dN and MBD-score was significant for all species comparisons (P ≪ 0.001; Spearman’s Rank test).
The relationship between MBD-score and synonymous substitution rate (dS) was less pronounced than for dN, and varied with evolutionary proximity between species. Comparison of orthologs between A. millepora and other Acropora species showed no relationship (fig. 3). Comparisons with corals outside of Acropora however, showed a significant negative relationship, with an average of 17% higher mean dS for weakly methylated genes (fig. 3; supplementary fig. S6, Supplementary Material online). The correlations with the three anemone species were weaker, although still significant. As most of these comparisons were saturated for synonymous substitutions they should be treated with caution. Analysis of dN/dS values gave similar results to dN for all groups of species (fig. 3).
Strongly Methylated Genes Show Greater Codon Bias
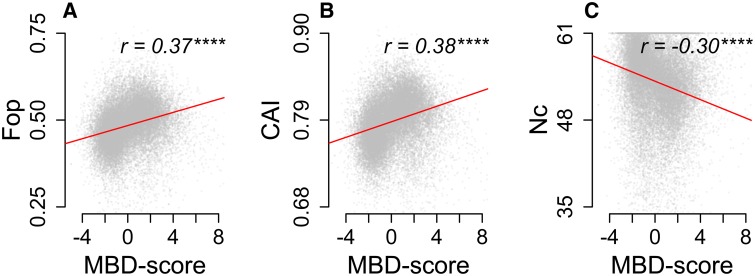
Because DNA methylation alters mutation patterns, we hypothesized that gbM shapes synonymous codon usage in stony corals. Specifically, we predicted that strong gbM produces codon bias via mutational replacement of codons bearing CpG dinucleotides (Kanaya et al. 2001; Qin et al. 2013). To test this we correlated MBD-scores with three distinct indices of codon bias: frequency of optimal codons (Fop) (Ikemura 1981), codon adaptation index (CAI) (Sharp and Li 1987a), and effective number of codons (Nc) (Wright 1990). Fop and CAI each quantify the preference for a set of optimal codons in the coding sequence. Higher values for these metrics indicate stronger codon bias. Nc quantifies nonrandom synonymous codon usage without assuming optimal codons. It is bounded between 1 (indicating complete bias, or use of only 20 codons for the 20 amino acids) and 64 (indicating completely neutral codon usage) (Wright 1990). All three indices correlated significantly with MBD-score (fig. 4). To assess the extent to which codon bias was driven by CpG hypermutability we recalculated CAI estimates using the same relative adaptiveness values (W) (see Methods) for each codon, but excluding the five amino acids coded for by CpG bearing codons (Serine, Proline, Threonine, Alanine, and Arginine). This substantially weakened the correlation from 0.38 (Spearman’s rho; P ≪ 0.0001) to 0.17, although it remained significant (P ≪ 0.0001). In contrast, recalculation of CAI based solely on these five amino acids strengthened the correlation (ρ = 0.42; supplementary fig. S7, Supplementary Material online), indicating that hypermutability of CpGs due to gbM has a strong influence on codon usage.
Fig. 4.
Correlation between MBD-score and indices of codon bias. (A) Fop. (B) CAI. (C) Nc. Red lines trace least squared linear regression. Asterisks indicate significance based on Spearman’s rank-order correlation test (ns >0.05; * <0.05; ** <0.01; *** <0.001; **** <0.0001).
CpG Codons Are under-Represented in Highly Expressed Genes
To further explore the influence of 5mC hypermutability on codon bias, we examined usage of CpG codons in highly expressed genes. As we did not have gene expression data for all species we first examined usage in annotated ribosomal protein genes with the assumption that these genes are highly expressed. For each species, relative synonymous codon usage (RSCU) of CpG codons was depressed in ribosomal protein genes (supplementary fig. S8A, Supplementary Material online). To ensure that this did not result from variation in overall GC content we showed that mean RSCU of CpG codons was significantly lower than that of codons with GC, GG, or CC dinucleotides (t-tests; P for all species <0.01).
For A. millepora, we assessed depression of CpG codons in highly expressed genes using three additional metrics: ΔRSCU (the difference in relative usage between the top 5% and bottom 5% expressed genes), rRSCU (the RSCU calculated for a concatenation of all ribosomal protein genes), and W (the relative adaptiveness of each codon; see Methods). With one exception that had neutral usage, all CpG codons were underrepresented for all three metrics (supplementary fig. S8B–D, Supplementary Material online). Hence, CpG bearing codons are depressed in highly expressed genes.
Underrepresentation of CpG Codons Matches Expectations for 5mC Hypermutability
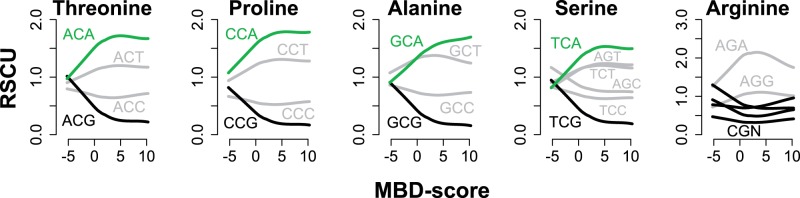
To further illustrate that loss of CpG codons is due to 5mC hypermutability, we examined RSCU for the five amino acids coded for by CpG bearing codons. Four of these (Threonine, Proline, Alanine, and Serine) are coded for by NCG codons, in which the CpG occupies the second and third positions of the codon. For these codons, 5mC > T mutations on the sense strand necessarily produce amino acid changes, which are expected to be rare due to purifying selection. In contrast, 5mC > T substitutions on the antisense strand produce silent substitutions (G > A within the codon) (supplementary fig.S9A, Supplementary Material online). For this reason, we predicted that 5mC hypermutability would increase the usage of NCA codons at the expense of NCG codons. To show this, we plotted RSCU of synonymous codons against MBD-score, illustrating positive relationships for NCA codon usage (Spearman’s rho between 0.156 and 0.196; P ≪ 0.001) and opposing negative relationships for NCG codon usage (fig. 5). Correlations of NCA codon usage with MBD-score were significantly stronger than other non-CpG codons (t-test; P < 0.01), indicating that NCA codons increase preferentially with stronger methylation. Hence for these four amino acids, depression of CpG codons in strongly methylated genes occurs through silent 5mC > T substitutions on the antisense strand. Moreover, all NCA codons were identified as optimal codons (supplementary table S1, Supplementary Material online), and their mean relative adaptiveness (for which the maximum is 1) was 0.99 (supplementary table S2, Supplementary Material online). These data indicate that NCA codons replace NCG codons in strongly methylated genes.
Fig. 5.
Depression of CpG bearing codons occurs via replacement with synonymous NCA codons. Lines show smoothed traces of the relationship between RSCU and MBD-score for the indicated codon. Black lines indicate CpG bearing codons. Green lines indicate NCA codons. Grey lines indicate all other codons. Opposing trends for NCA and NCG codons support the inference that NCA codons replace NCG codons in strongly methylated genes.
The second group of CpG bearing codons is the CGN codons, which code for arginine. These are expected to evolve differently because 5mC > T substitutions on both the sense and antisense stands produce amino acid changes (supplementary fig.S9A, Supplementary Material online). Although the trend is weak (r = −0.06; P < 0.0001), arginine content is negatively correlated with MBD-score (supplementary fig. S9B, Supplementary Material online), suggesting a slight shift in arginine content due to CpG hypermutability.
Summarizing Interrelationships between Gene Characteristics
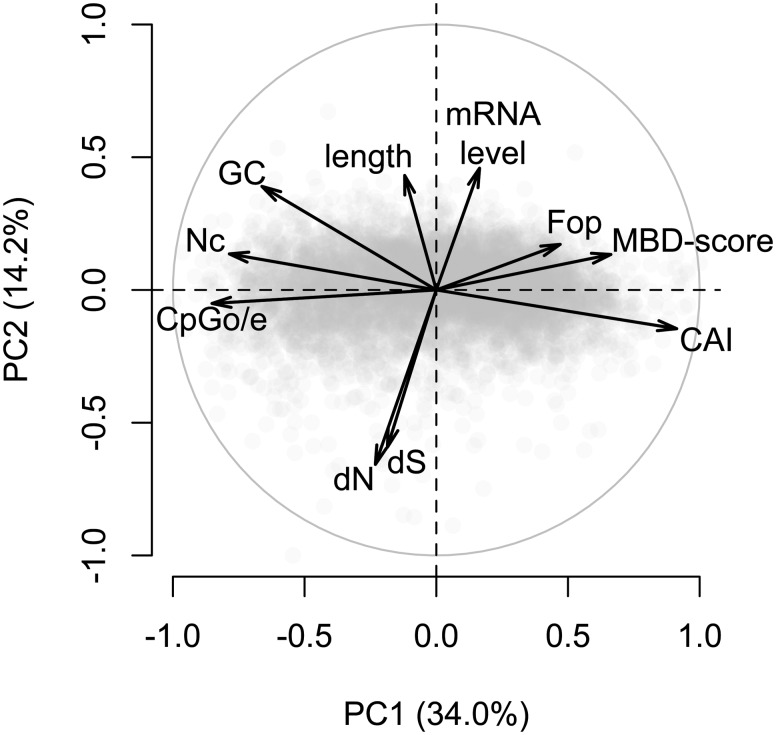
To summarize the relationships between gbM and other gene characteristics we performed principal component analysis (PCA) on all coding regions for which we had MBD-scores and substitution rate estimates. Pair-wise estimates of dN and dS between A. millepora and Siderastrea siderea were used because it was the species outside of the genus Acropora with the greatest number of orthologs. Substitution rates based on other species produced qualitatively similar results. Variation in measures of gbM and codon bias was captured largely by the first principal component (34.0% variance explained) (fig. 6). While the indices of codon bias often correlated most strongly with one another, the strongest alternative predictor for all three was historical germ-line methylation as measured by CpGo/e (supplementary table S3, Supplementary Material online). Variation in transcript abundance, gene length, and substitution rates was captured largely by the second principal component (14.2% variance explained) (fig. 6).
Fig. 6.
PCA of gene features in A. millepora. The first principal component explained 34.0% of variation and correlated primarily with measures of gbM and codon bias. The second principal component explained 14.2% of variation and correlated primarily with gene length, transcript abundance, and substitution rates. Variables included in the analyses are: normalized CpG content (CpGo/e), Nc, GC content of coding regions (GC), nonsynonymous substitution rate (dN), synonymous substitution rate (dS), length of coding region (length), transcript abundance (mRNA level), Fop, log2-fold difference between captured and flow-through fractions of methylation binding domain enrichment libraries (MBD-score), and CAI. Substitution rates are pair-wise estimates between A. millepora and S. siderea.
Discussion
Gene Body Methylation Is a Signature of Broad and Stable Expression
We showed that strongly methylated genes in A. millepora tend to have constitutive and ubiquitous functions and are less likely to be differentially expressed across developmental stages and environmental regimes. These results corroborate earlier findings from diverse taxa including plants (Aceituno et al. 2008; Coleman-Derr and Zilberman 2012; Takuno and Gaut 2012), cnidarians (Sarda et al. 2012; Dimond and Roberts 2016), mollusk (Gavery and Roberts 2010, 2013), arthropods (Elango et al. 2009; Wang et al. 2013), and a basal chordate (Suzuki et al. 2013; Keller et al. 2015). The relationship with differential expression in response to environmental regimes suggests the intriguing possibility that gbM could modulate gene expression plasticity.
We also found a positive correlation between gbM and transcript abundance (supplementary fig. S3, Supplementary Material online), indicating that intermediately methylated genes are highly transcribed, while lowly methylated and extremely strongly methylated genes tend toward lower transcription. These results are similar to previous findings in plants (Zhang et al. 2006; Zilberman et al. 2007; Zemach et al. 2010; Li et al. 2012; Wang et al. 2015), corals (Dimond and Roberts 2016), mollusk (Gavery and Roberts 2013; Wang et al. 2015), and human (Jjingo et al. 2012), indicating that this connection between gbM and expression is evolutionarily ancient and widely conserved.
Gene Body Methylation and Evolutionary Rates
We show that gbM negatively correlates with substitution rates. This finding is consistent with previous results from pants (Takuno and Gaut 2012; Wang et al. 2015) and animals (Park et al. 2011; Sarda et al. 2012; Keller et al. 2015). Still, PCA revealed that while substitution rates are significantly negatively correlated with gbM, they correlate more strongly with transcript abundance—a well-known trend described in bacteria, plants, fungi, and animals (Pál et al. 2001; Subramanian and Kumar 2004; Drummond et al. 2005; Drummond and Wilke 2008; Yang and Gaut 2011). This ubiquitous negative correlation between substitution rate and expression is explained by stronger purifying selection against protein misfolding in highly expressed genes. Because they have a greater cumulative opportunity for errors and misfolding, mutations in highly expressed genes pose greater fitness costs than those in lowly expressed genes (Drummond et al. 2005). Similar logic can be applied to broadly expressed genes, as they are active in a greater number of cells and tissues (Duret and Mouchiroud 2000), must operate in a greater variety of cellular millieus (Hastings 1996), and undergo more translational events at the scale of the entire organism. Whereas nonsynonymous substitutions affect the probability of protein misfolding through direct destabilization, synonymous substitutions most likely exert a similar but weaker effect by lowering translational accuracy (Akashi 1994; Drummond and Wilke 2008). Hence, both dN and dS are expected to be lower in highly and broadly expressed genes. We have shown that in our system, highly expressed genes tend to be strongly methylated (supplementary fig. S3, Supplementary Material online), and strongly methylated genes tend toward broad, constitutive transcription (fig 2; supplementary fig. S2, Supplementary Material online). We conclude that the observed correlation between gbM and substitution rates is most parsimoniously explained by the occurrence of gbM on genes that are under stronger purifying selection because of their expression patterns. A corollary of this conclusion is that purifying selection generally outweighs the effects of 5mC hypermutability. Hence, the paradox that gbM not only causes hypermutability but also correlates with sequence conservation can be explained by the fact that strongly methylated genes tend to undergo strong selection.
Gene Body Methylation Shapes Codon Usage
Codon bias occurs for two reasons. The first is mutational bias, where differences in mutation rates across species and genomic contexts produce nonrandom variation in synonymous codon usage (Plotkin and Kudla 2011). The second mechanism is natural selection, which requires that synonymous mutations affect organismal fitness (Behura and Severson 2013). In our case, mutational processes mediated by gbM appear to be the stronger source of variation. We found that gbM correlates strongly with three separate indices of codon bias (fig. 4). Analysis of RSCU values for NCG codons was consistent with codon bias arising largely through silent 5mC > T substitutions on the antisense stand (fig. 5; supplementary fig. S9A, Supplementary Material online). In other words, gbM causes codon bias by shifting usage of NCG codons to NCA codons.
When assessing whether codon bias is due to selection, researchers examine whether it occurs in genes where translation accuracy and efficiency are most important. Evidence that codon bias is due to selection includes (1) positive correlation with expression level, (2) positive correlation with breadth of expression, and (3) negative correlation with synonymous substitution rate (Sharp and Li 1987b; Duret 2002; Zhang and Li 2004; Plotkin and Kudla 2011; Behura and Severson 2013). As ours and previous studies have shown, gbM covaries with each of these factors. In other words, gbM occurs on the types of genes predicted to undergo strongest selection on codon usage. This fact highlights the need for caution when attributing codon bias to selection, since in our case codon bias results largely from mutation. Here, relationships with dS are of particular interest, because low dS can reflect selection on synonymous codons (Akashi 1994; Drummond and Wilke 2008). In our PCA, dS was nearly orthogonal to measures of gbM and codon bias. This result indicates that if A. millepora harbors codon bias due to selection, it is probably best predicted by expression level, and is dwarfed by mutational effect of gbM.
Although we attribute codon bias largely to mutation, this may still produce a potentially adaptive result—establishing a set of preferred and unpreferred codons in constitutively active genes. Optimal translation dynamics could then be achieved through evolution of tRNA abundances to match these preferred and unpreferred codons, obviating the need for selection of individual codons on a site-by-site basis. To put it another way, selection coefficients for individual synonymous codons will be exceedingly small (Bulmer 1987). In contrast, if a set of preferred codons is mutationally established in constitutively expressed genes, alleles that control the abundance of appropriate tRNAs could have stronger effects more amenable to natural selection. To be clear, we are not proposing that gbM originally evolved for this purpose. However, if its original function was linked with constitutively active expression, as appears to be the case from studies of plants (Takuno and Gaut 2012), invertebrates (Sarda et al. 2012), and mammals (Baubec et al. 2015), then CpG replacement coupled with coevolution of tRNAs provides an efficient means of evolving optimal codons in the genes where they are most beneficial.
An advantage of mutation-driven codon bias is that it could be maintained even in the absence of efficient selection, so it would be particularly beneficial for organisms with relatively small population size or otherwise inefficient selection. If some adaptive value of gbM is indeed related to maintenance of codon usage, it is not surprising that organisms such as yeast, fly, and worm are able to exist without it (Capuano et al. 2014); due to their large population sizes their optimal codon usage can be maintained by selection alone. At a minimum, it seems likely that tRNA pools have evolved in response to methylation-induced codon bias. This hypothesis could be explored through phylogenetic comparison of tRNA abundances between clades that independently lost or retained gbM.
Conclusions and Outlook
Here, we present three primary findings on gbM in stony corals: (1) gbM is most pronounced in genes with broad and stable expression; (2) gbM predicts sequence conservation; and (3) hypermutability due to gbM drives codon bias. Conserved occurrence of gbM on constitutively expressed genes in plants and the basal metazoan examined here indicates an evolutionarily ancient function involving selective pressure for accurate and stable gene expression. One means of improving translation fidelity is the use of optimal codons. Given its capacity to establish preferred and unpreferred codons in actively expressed genes, gbM could potentially influence evolution of optimal codons.
Materials and Methods
For detailed description of methods and materials see supplementary methods, Supplemental Material online. Instructions, scripts, and example output files for computational methods used in this study are available on GitHub (https://github.com/grovesdixon/metaTranscriptomes, last accessed 17 May 2016).
MBD-Seq
To quantify gbM in A. millepora we used methyl-CpG binding domain protein-enriched sequencing (MBD-seq). Enrichment reactions were performed using the MethylCap kit (Diagenode Cat. No. C02020010). Raw reads from the MBD-sequencing libraries were trimmed using cutadapt (Martin 2011) and mapped to coding sequences extracted from the A. millepora reference transcriptome (Moya et al. 2012). MBD-scores were calculated as the log2-fold difference between the MBD-enriched and flow-through libraries using DESeq2 (Love et al. 2014).
Ortholog Comparisons
Transcriptomic data from 17 species of Scleractinia (stony corals) and 3 species of Actiniaria (anemones) were downloaded from the web (supplementary table S4, Supplementary Material online; Schwarz et al. 2008; Sunagawa et al. 2009; Polato et al. 2011; Shinzato et al. 2011; Moya et al. 2012; Kenkel et al. 2013; Traylor-Knowles et al. 2011; Sun et al. 2013; Maor-Landaw et al. 2014; Nordberg et al. 2014; Willette et al. 2014; Kitchen et al. 2015; Davies et al. forthcoming). Coding sequences were extracted based on BlastX (Altschul et al. 1997) alignments to the Nematostella vectensis (Nordberg et al. 2014) and Acropora digitifera (Shinzato et al. 2011) reference proteomes using a custom perl script. Orthologs were identified based on reciprocal best hits (BLASTP) (Altschul et al. 1990) using custom python scripts. Protein alignments were performed using MAFFT (Katoh and Standley 2013) and reverse translation was performed using Pal2Nal (Suyama et al. 2006). Substitution rates were estimated using PAML (Yang 2007). Phylogenetic construction was performed with the rapid bootstrapping algorithm (GTRGAMMA model) with 1,000 iterations using RAxML (Stamatakis 2014). Gene expression datasets were generated using Tag-based RNA-seq (Meyer et al. 2011; Dixon et al. 2014, 2015).
Codon Bias
We tested for relationships between MBD-score and synonymous codon usage using four metrics: RSCU (Sharp et al. 1986), Fop (Ikemura 1981), CAI (Sharp and Li 1987a), and the Nc (Wright 1990). CAI and RSCU were calculated using custom python scripts. Fop and Nc were calculated using CodonW (Peden 1999) (http://codonw.sourceforge.net//culong.html, last accessed 17 May 2016).
Statistical Analyses
Statistical analyses of the relationship between MBD-score and other gene characteristics were performed in R (R Core Team 2015). Significance for correlations was established using Spearman’s rank-order correlation test. Significance tests for differences in counts between the strongly methylated and weakly methylated classes were performed using Fisher’s exact tests (Fisher 1922). PCA was performed using prcomp function in R.
Supplementary Material
Supplementary methods, tables S1–S4, and figures S1–S11 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
Funding for this study was provided by National Science Foundation grant DEB-1054766 to M.V.M. and the grant from the Australian Research Council Center of Excellence for Coral Reef Studies and the Queensland Government to L.K.B.
Supplementary Material
References
- Aceituno FF, Moseyko N, Rhee SY, Gutiérrez RA. 2008. The rules of gene expression in plants: organ identity and gene body methylation are key factors for regulation of gene expression in Arabidopsis thaliana. BMC Genomics 9:438.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H. 1994. Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics 136:927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S, Gish W, Miller W. 1990. Basic Local Alignment Search Tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baubec T, Colombo DF, Wirbelauer C, Schmidt J, Burger L, Krebs AR, Akalin A, Schu D. 2015. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 520:243–247. [DOI] [PubMed] [Google Scholar]
- Behura SK, Severson DW. 2013. Codon usage bias: causative factors, quantification methods and genome-wide patterns: with emphasis on insect genomes. Biol Rev. 88:49–61. [DOI] [PubMed] [Google Scholar]
- Bulmer M. 1987. Coevolution of codon usage and transfer RNA abundance. Nature 325:278–730. [DOI] [PubMed] [Google Scholar]
- Capuano F, Mülleder M, Kok R, Blom HJ, Ralser M. 2014. Cytosine DNA methylation is found in Drosophila melanogaster but absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other yeast species. Anal Chem. 86:3697–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman-Derr D, Zilberman D. 2012. Deposition of histone variant H2A.Z wihtin gene bodies regulates responsive genes. PLoS Genet. 8:10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SW, Marchetti A, Ries JB, Castillo KD. Forthcoming. Thermal and pCO2 stress elicit divergent transcriptomic responses in a resilient coral. Front Mar Sci. FMARS-02-00062. [Google Scholar]
- Dimond JL, Roberts SB. 2016. Germline DNA methylation in reef corals: patterns and potential roles in response to environmental change. Mol Ecol. 25:1895–1904. [DOI] [PubMed] [Google Scholar]
- Dixon GB, Bay LK, Matz MV. 2014. Bimodal signatures of germline methylation are linked with gene expression plasticity in the coral Acropora millepora. BMC Genomics 15:1109.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon GB, Davies SW, Aglyamova GV, Meyer E, Bay LK, Matz MV. 2015. Genomic determinants of coral heat tolerance across latitudes. Science 348:1460–1462. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. 2005. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A. 102:14338–14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. 2008. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134:341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Yi S, Guo X, Wang W. 2015. A comprehensive analysis of codon usage patterns in blunt snout bream (Megalobrama amblycephala) based on RNA-Seq data. Int J Mol Sci. 16:11996–12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L. 2002. Evolution of synonymous codon usage in metazoans. Opin Genet Dev. 12:640–649. [DOI] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. 2000. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol Biol Evol. 17:68–74. [DOI] [PubMed] [Google Scholar]
- Elango N, Hunt BG, Goodisman MAD, Yi SV. 2009. DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera. Proc Natl Acad Sci U S A. 106:11206–11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen P-Y, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. 2010. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A. 107:8689–8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. 1922. On the interpretation of χ2 from contingency tables, and the calculation of P. J R Stat Soc. 85:87–94. [Google Scholar]
- Francioli LC, Polak PP, Koren A, Menelaou A, Chun S, Renkens I, van Duijn CM, Swertz M, Wijmenga C, van Ommen G, et al. 2015. Genome-wide patterns and properties of de novo mutations in humans. Nat Genet. 47:822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavery M, Roberts S. 2010. DNA methylation patterns provide insight into epigenetic regulation in the Pacific oyster (Crassostrea gigas). BMC Genomics 11:483.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavery M, Roberts S. 2013. Predominant intragenic methylation is associated with gene expression characteristics in a bivalve mollusc. Peer J. 1:e215.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Ibeas D, Blanca J, Roig C, González-To M, Picó B, Truniger V, Gómez P, Deleu W, Caño-Delgado A, Arús P, et al. 2007. MELOGEN: an EST database for melon functional genomics. BMC Genomics 8:306.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, et al. 2010. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 28:1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings KE. 1996. Strong evolutionary conservation of broadly expressed protein isoforms in the troponin I gene family and other vertebrate gene families. J Mol Evol. 42:631–640. [DOI] [PubMed] [Google Scholar]
- Hunt B, Brisson J. 2010. Functional conservation of DNA methylation in the pea aphid and the honeybee. Genome Biol Evol. 2:719–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. 1981. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 151:389–409. [DOI] [PubMed] [Google Scholar]
- Jjingo D, Conley AB, Yi SV, Lunyak VV, King I. 2012. On the presence and role of human gene-body DNA methylation. Oncotarget 3:462–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 13:484–492. [DOI] [PubMed] [Google Scholar]
- Kanaya S, Yamada Y, Kinouchi M, Kudo Y, Ikemura T. 2001. Codon usage and tRNA genes in eukaryotes: Correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J Mol Evol. 53:290–298. [DOI] [PubMed] [Google Scholar]
- Karlin S, Mrázek J. 1996. What drives codon choices in human genes? J Mol Biol. 262:459–472. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller TE, Han P, Yi SV. 2015. Evolutionary transition of promoter and gene body DNA methylation across invertebrate-vertebrate boundary. Mol Biol Evol. 33:1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenkel CD, Meyer E, Matz MV. 2013. Gene expression under chronic heat stress in populations of the mustard hill coral (Porites astreoides) from different thermal environments. Mol Ecol. 22:4322–4334. [DOI] [PubMed] [Google Scholar]
- Kitahara MV, Cairns SD, Stolarski J, Blair D, Miller DJ. 2010. A comprehensive phylogenetic analysis of the Scleractinia (Cnidaria, Anthozoa) based on mitochondrial CO1 sequence data. PLoS One 5:e11490.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen SA, Crowder CM, Poole AZ, Weis VM, Meyer E. 2015. De novo assembly and characterization of four anthozoan (phylum Cnidaria) transcriptomes. G3 Genes Genomes Genet. 5:2441–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhu J, Hu F, Ge S, Ye M, Xiang H, Zhang G, Zheng X, Zhang H, Zhang S, et al. 2012. Single-base resolution maps of cultivated and wild rice methylomes and regulatory roles of DNA methylation in plant gene expression. BMC Genomics 13:300.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 15:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maor-Landaw K, Karako-Lampert S, Ben-Asher HW, Goffredo S, Falini G, Dubinsky Z, Levy O. 2014. Gene expression profiles during short-term heat stress in the red sea coral Stylophora pistillata. Glob Chang Biol. 20:3026–3035. [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10–12. [Google Scholar]
- McGaughey DM, Abaan HO, Miller RM, Kropp PA, Brody LC. 2014. Genomics of CpG methylation in developing and developed zebrafish. G3 Genes Genomes Genet. 4:861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E, Aglyamova GV, Matz MV. 2011. Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-Seq procedure. Mol Ecol. 20:3599–3616. [DOI] [PubMed] [Google Scholar]
- Moya A, Huisman L, Ball EE, Hayward DC, Grasso LC, Chua CM, Woo HN, Gattuso J-P, Forêt S, Miller DJ. 2012. Whole transcriptome analysis of the coral Acropora millepora reveals complex responses to CO2-driven acidification during the initiation of calcification. Mol Ecol. 21:2440–2454. [DOI] [PubMed] [Google Scholar]
- Nordberg H, Cantor M, Dusheyko S, Hua S, Poliakov A, Shabalov I, Smirnova T, Grigoriev IV, Dubchak I. 2014. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res. 42:26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pál C, Papp B, Hurst LD. 2001. Highly expressed genes in yeast evolve slowly. Genetics 158:927–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Peng Z, Zeng J, Elango N, Park T, Wheeler D, Werren JH, Yi SV. 2011. Comparative analyses of DNA methylation and sequence evolution using Nasonia genomes. Mol Biol Evol. 28:3345–3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden JF. 1999. Analysis of codon usage [Thesis]. pp. 50–90.
- Plotkin JB, Kudla G. 2011. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet. 12:32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polato NR, Vera JC, Baums IB. 2011. Gene discovery in the threatened elkhorn coral: 454 sequencing of the Acropora palmata transcriptome. PLoS One 6:e28634.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z, Cai Z, Xia G, Wang M. 2013. Synonymous codon usage bias is correlative to intron number and shows disequilibrium among exons in plants. BMC Genomics 14:56.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2015. R: a language and environment for statistical computing.
- Romano SL, Palumbi SR. 1996. Evolution of scleractinian corals inferred from molecular systematics. Science 271:640–642. [Google Scholar]
- Sarda S, Zeng J, Hunt BG, Yi SV. 2012. The evolution of invertebrate gene body methylation. Mol Biol Evol. 29:1907–1916. [DOI] [PubMed] [Google Scholar]
- Schwarz JA, Brokstein PB, Voolstra C, Terry AY, Manohar CF, Miller DJ, Szmant AM, Coffroth MA, Medina M. 2008. Coral life history and symbiosis: functional genomic resources for two reef building Caribbean corals, Acropora palmata and Montastraea faveolata. BMC Genomics 9:97.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li WH. 1987a. The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15:1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li WH. 1987b. The rate of synonymous substitution in enterobacterial genes is inversely related to codon usage bias. Mol Biol Evol. 4:222–230. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Tuohy TMF, Mosurski KR. 1986. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 14:5125–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JC, Rideout WM, Jones PA. 1994. The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res. 22:972–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C, Shoguchi E, Kawashima T, Hamada M, Hisata K, Tanaka M, Fujie M, Fujiwara M, Koyanagi R, Ikuta T, et al. 2011. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476:320–323. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterky F, Bhalerao RR, Unneberg P, Segerman B, Nilsson P, Brunner AM, Charbonnel-Campaa L, Lindvall JJ, Tandre K, Strauss SH, et al. 2004. A Populus EST resource for plant functional genomics. Proc Natl Acad Sci U S A. 101:13951–13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Kumar S. 2004. Gene expression intensity shapes evolutionary rates of the proteins encoded by the vertebrate genome. Genetics 168:373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Chen Q, Lun JCY, Xu J, Qiu JW. 2013. PcarnBase: development of a transcriptomic database for the brain coral Platygyra carnosus. Mar Biotechnol. 15:244–251. [DOI] [PubMed] [Google Scholar]
- Sunagawa S, Wilson EC, Thaler M, Smith ML, Caruso C, Pringle JR, Weis VM, Medina M, Schwarz JA. 2009. Generation and analysis of transcriptomic resources for a model system on the rise: the sea anemone Aiptasia pallida and its dinoflagellate endosymbiont. BMC Genomics 10:258.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34:609–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Kerr ARW, Sousa D, De Bird A. 2007. CpG methylation is targeted to transcription units in an invertebrate genome. Genome Res. 17:625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Yoshinari A, Obara M, Takuno S, Shigenobu S, Sasakura Y, Kerr AR, Webb S, Bird A, Nakayama A. 2013. Identical sets of methylated and nonmethylated genes in Ciona intestinalis sperm and muscle cells. Epigenetics Chromatin. 6:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sved J, Bird A. 1990. The expected equilibrium of the CpG dinucleotide in vertebrate genomes under a mutation model. Proc Natl Acad Sci U S A. 87:4692–4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuno S, Gaut BS. 2012. Body-methylated genes in Arabidopsis thaliana are functionally important and evolve slowly. Mol Biol Evol. 29:219–227. [DOI] [PubMed] [Google Scholar]
- Takuno S, Gaut BS. 2013. Gene body methylation is conserved between plant orthologs and is of evolutionary consequence. Proc Natl Acad Sci U S A. 110:1797–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuno S, Ran J-H, Gaut BS. 2016. Evolutionary patterns of genic DNA methylation vary across land plants. Nat Plants 2:15222.. [DOI] [PubMed] [Google Scholar]
- Tran RK, Henikoff JG, Zilberman D, Ditt RF, Jacobsen SE, Henikoff S. 2005. DNA methylation profiling identifies CG methylation clusters in Arabidopsis genes. Curr Biol. 15:154–159. [DOI] [PubMed] [Google Scholar]
- Traylor-Knowles N, Granger BR, Lubinski TJ, Parikh JR, Garamzegi S, Xia Y, Marto JA, Kaufman L, Finnerty JR. 2011. Production of a reference transcriptome and transcriptomic database (PocilloporaBase) for the cauliflower coral Pocillopora damicornis. BMC Genomics. 12:585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Beyene G, Zhai J, Feng S, Fahlgren N, Taylor NJ, Bart R, Carrington JC, Jacobsen SE, Ausin I. 2015. CG gene body DNA methylation changes and evolution of duplicated genes in cassava. Proc Natl Acad Sci U S A. 112:13729–13734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wheeler D, Avery A, Rago A, Choi J-H, Colbourne JK, Clark AG, Werren JH. 2013. Function and evolution of DNA methylation in Nasonia vitripennis. PLoS Genet. 9:e1003872.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willette DA, Allendorf FW, Barber PH, Barshis DJ, Carpenter KE, Crandall ED, Cresko WA, Fernandez-Silva I, Matz MV, Meyer E, et al. 2014. So, you want to use next-generation sequencing in marine systems? Insight from the Pan-Pacific Advanced Studies Institute. Bull Mar Sci. 90:79–122. [Google Scholar]
- Wright F. 1990. The “effective number of codons” used in a gene. Gene 87:23–29. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang L, Gaut BS. 2011. Factors that contribute to variation in evolutionary rate among Arabidopsis genes. Mol Biol Evol. 28:2359–2369. [DOI] [PubMed] [Google Scholar]
- Zhang L, Li W-H. 2004. Mammalian housekeeping genes evolve more slowly than tissue-specific genes. Mol Biol Evol. 21:236–239. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SWL, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. 2006. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126:1189–1201. [DOI] [PubMed] [Google Scholar]
- Zemach A, Zilberman D. 2010. Evolution of eukaryotic DNA methylation and the pursuit of safer sex. Curr Biol. 20:R780–R785. [DOI] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. 2010. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328:916–919. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. 2007. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 39:61–69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.