Abstract
Leukocyte chemoattractant receptors are members of the G-protein coupled receptor (GPCR) family. Signaling downstream of these receptors directs the localization, positioning and homeostatic trafficking of leukocytes; as well as their recruitment to, and their retention at, inflammatory sites. Ligand induced changes in the molecular conformation of chemoattractant receptors results in the engagement of heterotrimeric G-proteins, which promotes α subunits to undergo GTP/GDP exchange. This results in the functionally release of βγ subunits from the heterotrimers, thereby activating downstream effector molecules, which initiate leukocyte polarization, gradient sensing, and directional migration. Pertussis toxin ADP ribosylates Gαi subunits and prevents chemoattractant receptors from triggering Gαi nucleotide exchange. The use of pertussis toxin revealed the essential importance of Gαi subunit nucleotide exchange for chemoattractant receptor signaling. More recent studies have identified a range of regulatory mechanisms that target these receptors and their associated heterotrimeric G-proteins, thereby helping to control the magnitude, kinetics, and duration of signaling. A failure in these regulatory pathways can lead to impaired receptor signaling and immunopathology. The analysis of mice with targeted deletions of Gαi isoforms as well as some of these G-protein regulatory proteins is providing insights into their roles in chemoattractant receptor signaling.
Keywords: G-protein, RGS proteins, chemokine receptors, sphingosine 1-phosphate, cell trafficking
Graphical Abstract
1. Introduction
Upon ligand binding chemoattractant GPCRs undergoes conformational rearrangements that change their interaction with signal transducer proteins (1–3). The major proximal signal transducers of an activated GPCR are their cognate heterotrimeric G-proteins, although ligand activated GPCRs engage other proteins including G-protein receptor kinases (GRKs) and β-arrestins (4–6). Chemoattractant receptors predominately couple to the Gi family of heterotrimeric G-proteins as their signaling is sensitive to treatment with pertussis toxin, which ADP ribosylates a cysteine residue near the C-termini of αi subunit (7,8). This inhibits Gαi from undergoing GPCR-mediated nucleotide exchange. The GTP/GDP binding status of Gαi controls its interaction with the other members of the trimeric G protein, the Gβγ dimer (2,9). Composed of one of 5 β and one of 12 γ subunits, the Gβγ dimers, once assembled, are inseparable. Receptor activation and Gαi subunit nucleotide exchange causes the Gαi subunit and Gβγ subunits to functionally dissociate. GTP bound Gαi and the freed Gβγ can then engage downstream effector molecules. However, it is the Gβγ effectors that are the most crucial for gradient sensing and directional cell migration (10). The activated Gα subunits remain only transiently GTP bound as they possess an intrinsic GTPase activity, which converts their bound GTP to GDP. This encourages the reassembly of the heterotrimeric G-protein terminating signaling, but also allows the G-protein to engage other activated receptors, thereby initiating another round of signaling. The intrinsic GTPase activity of Gα subunits can be greatly enhanced by GTPase activating proteins termed GAPs. Most Gαi GAPs contain a Regulator of G-protein signaling (RGS) domain (11–13). Other G-protein regulatory proteins include those that have a G-protein regulatory (GPR) domain, also called a GoLoco motif, which acts as a guanine nucleotide dissociation inhibitor (GDI) much like Gβγ (14–16). Both RGS and GPR domain containing proteins impact Gi signaling. This review will focus predominately on the Gαi proteins and their regulators in the context of chemokine and chemoattractant receptor signaling.
2. Gi proteins
The heterotrimeric G proteins are divided into four classes; Gi, Gq, G12/13, and Gs, based on the amino acid sequences of their α subunit (2). The expression levels of the various Gα subunits in murine leukocytes based on mRNA sequencing are shown in Table 1. The “inhibitory class” of heterotrimeric G-proteins were originally named based on the ability of Gi/o proteins to inhibit adenylyl cyclase activity. Based on amino acid sequence homology the Gi family now includes not only Gi/o, but also Gt, Ggust, and Gz. The Gi/o subfamily includes 4 members, three Gi proteins and Go. As Go (Gnao) is poorly expressed in murine leukocytes it will not be further discussed. Of the three Gαi isoforms, murine leukocytes predominately express Gαi2 and Gαi3 with Gnai2 mRNA transcripts 3–10 fold more abundant (16). Gαi1 is nearly undetectable although low levels of Gnai1 mRNA transcripts have been found in mouse TCRγδ cells and Tregs by RNA sequencing (http://www.immgen.org/databrowser/index.html). Each Gαi isoform can theoretically pair with 60 different Gβγ dimers. However, not all of the theoretical dimers are likely to be assembled in vivo and many of the subunits have a rather limited expression profile. For example, murine lymphocytes predominately express mRNA transcripts that encode 2 of the 5 Gβ subunits; Gβ1, Gβ2, and 4 of 12 Gγ subunits; Gγ2, Gγ7, Gγ8, and Gγ10 (16). The signaling specificities of the various Gβγ dimers, in general and for immune cell function specifically, are largely unknown.
Table 1.
Expression of different Gα subunits in various types of leukocytes1
| Cell type | Gnas | Gnai2 | Gnai3 | Gna12 | Gna13 | Gnaq | Gna11 | Gna15 |
|---|---|---|---|---|---|---|---|---|
| B | 12.1 | 13.7 | 2.3 | 0.3 | 3.0 | 0.4 | 0.4 | 0.1 |
| Dendritic | 8.2 | 11.8 | 2.3 | 0.5 | 2.4 | 1.4 | 0.3 | 0.1 |
| Neutrophil | 5.8 | 50.0 | 8.6 | 0.4 | 6.0 | 3.5 | 0.5 | 0.1 |
| Macrophage | 6.4 | 6.9 | 5.7 | 0.3 | 2.2 | 3.8 | 0.5 | 0.3 |
| NK | 16.1 | 13.7 | 3.3 | 0.3 | 2.0 | 0.2 | 0.5 | 0.2 |
| NKT | 6.1 | 3.5 | 2.5 | 0.1 | 3.2 | 0.2 | 0.4 | 0.1 |
| CD4 | 7.7 | 11.8 | 2.7 | 0.2 | 2.8 | 0.1 | 0.4 | 0.1 |
| CD8 | 8.9 | 7.2 | 2.7 | 0.2 | 3.0 | 0.1 | 0.5 | 0.1 |
| Tγδ | 11.0 | 6.9 | 3.6 | 0.3 | 4.1 | 0.2 | 0.3 | 0.1 |
| Treg | 6.0 | 7.1 | 3.2 | 0.3 | 3.3 | 0.3 | 0.4 | 0.1 |
Relative mRNA expression in different cell types. Cells were isolated from 3 male C57BL/6J mouse spleens with the exception of the macrophages, which were isolated from the peritoneal cavity of similar mice. Data are from the Immunological genome project (http://www.immgen.org/databrowser/index.html) and based on RNA sequencing (Illumina) of mRNA from FACS purified cell samples. Very low or undetectable expression of Gnat1, Gnat2, Gnat1, Gnai1, Gnao1, Gnaz, Gnal, and Gna14. Cell type with highest expression of the different Gα subunits is highlighted. Levels of mRNA expression for Gβ and Gγ subunits can be found at the same website. For comparison β-actin expression is approximately 400. Values for each gene (x 103).
While this review is focused on Gαi proteins in GPCR and chemoattractant receptor signaling there is an increasing appreciation that Gαi proteins as well other Gα subunits have functional roles at other intracellular sites. Gαi proteins have been localized in the endoplasmic reticulum, centrosomes, and midbody as well as associated with microtubules (17,18). In model organism and in mammals Gαi proteins have been ascribed functional roles in asymmetric cell divisions. Whether non-canonical Gαi signaling contributes to lymphocyte function remains largely unexplored (16).
2.1. Gαi signaling
The release of GTP-Gαi lowers intracellular cAMP levels by reducing the activity of certain adenylyl cyclase isoforms. While lowering cAMP levels may promote leukocyte chemotaxis, previous studies have not found an essential role for Gαi in chemokine directed cell migration (10). However several recent studies have identified new Gαi2 interacting proteins. One such protein is Homer 3, which acts as a scaffold that spatially organizes actin assembly to support neutrophil polarity and motility downstream of GPCR activation in mice (19). However, in contrast to traditional G-protein effectors Homer 3 binds both the GTP and GDP bound forms of Gαi2. While GTP-Gαi likely has a limited role in chemokine receptor signaling, freed Gβγ subunits are known to activate several signaling pathways needed for gradient sensing and chemotaxis (20–23). An early consequence of the engagement of chemokine receptors is the acquisition of a polarized shape with the cell adopting a leading edge and a trailing uropod. The activation of small GTPases such as Rac, Cdc42, RhoA, and Rap1; many of which are activated by GEFs downstream of Gβγ, are crucial for these early morphological changes and eventually for actin reorganization and cell motility (24). Other direct effector of Gi released Gβγ include PI3Kγ (phosphoinositide 3-kinase γ), which plays a crucial role in inflammatory and allergic processes (25–27) ; phospholipase C β2 and β3 (28,29), which cause increases in intracellular calcium; GRK2, which phosphorylates certain chemokine receptors promoting β-arrestin recruitment (30); and mTor, which has been ascribed a role in chemotaxis (31–33).
2.2. Pertussis toxin
The recognition that pertussis toxin ADP ribosylates the Gαi subunits and inhibits GPCR triggered nucleotide exchange has proven extremely useful for studying GPCRs that signal though Gi (7,8,34–37). While similarly affecting all the Gαi isoforms, the experimental usage of pertussis toxin strongly implicated Gαi signaling in leukocyte trafficking, blood vessel egress, and the organization of lymphoid organ architecture. Ex vivo treatment of human and murine sdneutrophils, monocytes, and lymphocytes with pertussis toxin nearly eliminated in vitro chemotaxis and inhibited chemokine induced firm adhesion. The adoptive transfer of ex vivo treated lymphocytes into mice prevented the transferred lymphocytes from entering the splenic while pulp or entering into lymph nodes (38). Ex vivo treated lymphocytes observed in the high endothelial venules (HEVs) of adoptively transferred mice failed to firmly adhere to high endothelial venules. HEVs are specialized sites in the lymph node microvasculature where blood borne lymphocytes are captured and cross into the lymph node parenchyma. Transgenic expression of pertussis toxin in mice interfered with mature thymocyte reverse transmigration from the thymus medulla into the blood (39). As a consequence mature thymocytes accumulated in the thymus. However, there are some caveats in interpreting results from pertussis toxin experiments (40). Pertussis toxin has an AB5 configuration, one active subunit termed S1 and five binding subunits. The B oligomer binds to glycoconjugate molecules present on most mammalian cell types and the exotoxin enters the cell by endocytosis. As the B subunit oligomer can impact intracellular signaling pathways independent of the enzymatic activity of the S1 subunit care must be taken in not attributing these effects to the inhibition of Gαi signaling. The development of a double mutant PT9K/129G molecule that retains normal cellular binding activity, but lacks enzymatic activity has allowed the discrimination between the ADP-ribosylation activity of the S1 subunit, and signaling initiated by the B subunit oligomer (40). In addition, while Gαi is the major target for pertussis toxin mediated ADP ribosylation, other substrates are also modified. Yet when properly utilized to study GPCR signaling, pertussis toxin sensitivity strongly implicates the involvement of Gαi proteins. Another limitation of pertussis toxin usages is its inability to discriminate between the different pertussis toxin sensitive G-proteins. The development of Gαi isoform specific knock-out mice avoided some of these issues and provided additional insights into the importance of Gi signaling in lymphocytes and other leukocytes.
2.3. Gnai2−/− mice
Despite a compensatory increase in Gαi3 expression Gnai2−/− mice have numerous immune phenotypes (41). These mice develop a Th1-mediated inflammatory colitis reminiscent of human ulcerative colitis, whose penetrance depends upon the genetic background of the mice. A C57BL/6 background confers some resistance to colitis development. C57BL/6 Gnai2−/− mice maintained in ventilated cages at the NIH under do not show evidence of colitis (J. Kehrl, unpublished observation), however a recent report indicates that C57BL/6 kept under conventional conditions do develop colitis (42). Gnai2 deficient mice often lack inguinal and other peripheral lymph nodes; have few, if any, Peyer’s patches; and have an accelerated thymic involution (43–45).
2.3.1 B cells from Gnai2−/− mice
Immune phenotyping of these mice revealed a reduced numbers of splenic marginal zone and T2 transitional B cells; reduced peritoneal B-1a, but increased peritoneal B-1b B cells; and decreased Peyer’s patch B cells (43). Indicating that the B cell phenotypes were lymphocyte intrinsic Gnai2−/− bone marrow reconstituted Rag2−/− mice exhibited a similar set of phenotypes (43). Isolated B cells from the Gnai2−/− mice have diminished responses to chemokines and sphingosine 1-phosphate (44). Consistent with defective chemokine receptor signaling Gnai2−/− B cells adoptively transferred into wild type mice adhered poorly to high endothelial venules (HEVs), entered lymph nodes inefficiently, tended to accumulate around the HEVs, and those cells that gained access to the lymph node moved much less vigorously than wild type B cells (44).
2.3.2 T cells from Gnai2−/− mice
Gnai2−/− mice have thymuses with normal or slightly increased number of thymocytes compared to wild type mice (41). Yet thymic progenitors from them home less efficiently to the thymus than do wild type cells (46). Thymocytes subset analysis (C57BL/6 or 129SvEv prior to colitis) revealed a reduction in double positive (DP) cells with an increase in single positive (SP) cells. Both an increased rate of differentiation of DP to SP cells along with a decreased egress of the SP cells have been proposed to explain the expansion of SP cells in the thymus (47,48). C57BL/6 Gnai2−/− mice maintained at the National Institutes Health have a modest reduction in DP cells, with a significant increase in mature SP cells (unpublished data). CD4 as well as CD8 T cells purified from spleens of these mice responded poorly to chemokines (49). The Gnai2−/− T cells have severe defects in chemokine-induced intracellular calcium mobilization, chemotaxis, and homing, whereas Gnai2+/− T cells exhibit modest defects. Intravital imaging revealed that the Gnai2−/− CD4 T accumulated at the lymph node cortical ridge failing to normally access the T cell zone. These cells also lacked the customary amoeboid-like cell movements of normal CD4 T cells (49).
2.3.2 Neutrophils from Gnai2−/− mice
Gnai2−/− neutrophils also exhibited defective response to chemokines and chemoattractants (50). They arrested poorly on inflamed endothelium and did not accumulate at inflammatory sites. While wild type neutrophils re-inforce a wound site the Gnai2−/− neutrophils did not being excluded from the central neutral cluster (51).
2.3.3 Macrophages from Gnai2−/− mice
Gnai2−/− macrophages exhibited similar defects to those of the Gnai2−/− neutrophils did (52). They were poorly recruited to the peritoneum following thioglycollate-induced peritonitis and to the lung following lipopolysaccharide (LPS)-triggered inflammation. Furthermore, knockdown of Gnai2 mRNA expression decreased both the chemokine induced migration and motility of RAW 264.7 cells, which was rescued by restoring Gαi2 expression (52).
2.4. Gnai3−/− mice
In contrast to the Gnai2−/− mice few immune related phenotypes have been reported in the Gnai3−/− mice although a recent report showed that Gαi3 functions along with Gαi1 in macrophage TLR4 signaling (53). Lymphocytes, neutrophils, and macrophages all express significant levels of Gαi3 yet, chemokine responses are intact in the above cells purified from Gnai3−/− mice (10,52,54,55). There is a report that Gnai3−/− effector T cells actually respond better to certain chemokines than do wild type cells and another that early Gnai3−/− thymocyte progenitors do not populate the thymus as well as wild type progenitors (46,56). The C57BL/6 Gnai3−/− mice maintained at the NIH have normal numbers of thymocytes, slightly reduced numbers of splenocytes, and normal numbers of lymph node cells (unpublished observation). Thymocyte subsets are unperturbed, but splenic and Peyer’s patch B220+ cells are slightly reduced. The report alluded to above (53) showed that TLR4 signaling led to formation of a complex that contained Gαi3, CD14, and Gab1 (growth factor receptor binding 2 (Grb2)-associated binding protein 1), which promoted Akt activation. A deficiency in Gαi3 reduced LPS-induced TLR4 endocytosis and interfered with phosphorylation of interferon regulatory factor 3. Silencing Gnai3/Gnai1 expression in bone marrow-derived macrophages caused a M2-like phenotype with reduced LPS-induced cytokine production. The macrophages used in this study were prepared from mice on a mixed 129 background. Isoform specific immunoblotting indicated that they expressed a significant level of Gαi1. This differs from C57BL/6 macrophages, which express little or no Gnai1 mRNA or Gαi1 protein.
2.5. Gnai2fl/fl mb1-cre mice
Mice lacking Gαi2 expression in the B cell lineage were created by crossing mice carrying a floxed Gnai2 allele to mice expressing the cre recombinase from the mb1 locus (55). These mice delete a portion of the Gnai2 coding sequence at the pre-B cell stage of development and they confirmed the B cell intrinsic origin of most of the B cell phenotypes found in the Gnai2−/− mice. The B cell specific loss of Gαi2 resulted in a severe B cell trafficking problem. Similar to the Gnai2−/− mice marginal zone B cell development and B cell responses to chemokines were both impaired.
2.6. Gnai3−/−Gnai2fl/flmb1-cre mice
Gnai2−/−Gnai3−/− mice are not viable (57); however, conditional deletion of Gnai2 on a Gnai3−/− background is possible. Gnai3−/−Gnai2fl/flmb1-cre mice lack Gαi2 and Gαi3 expression in B cells (55). In these conditional double knock-out mice mucosal sites, splenic marginal zones, and lymph nodes essentially lacked B cells. There was a 50–60% reduction in splenic B cells with few if any marginal zone B cells. The spleen, lymph nodes, and the gastrointestinal tract lacked any organized B cell compartments. Purified splenic B cells were refractory to chemokine stimulation. The mice developed a hyper-IgM like syndrome having an elevated serum IgM level and with reductions in the other serum isotypes. The loss of Gαi2 and Gαi3 in mouse B cells led to a complete failure in their responsiveness to chemoattractants.
3. G-protein regulatory proteins
3.1 Ric-8A
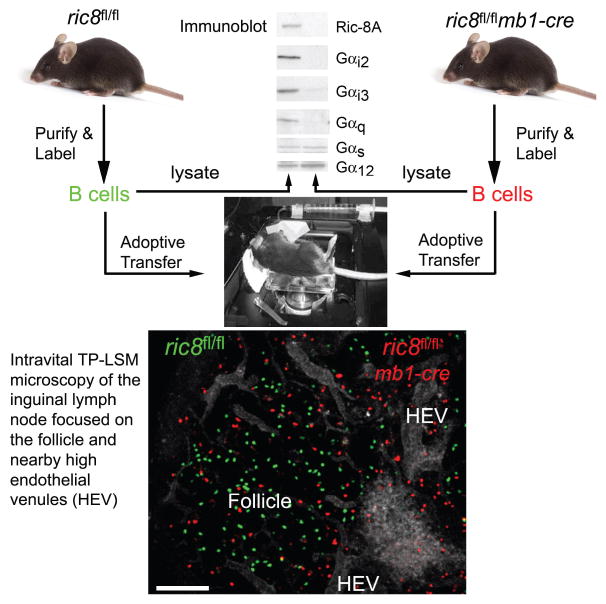
Ric-8 was identified in C. elegans based on its role in asymmetric cell divisions during early development (58–60). A human homologue, Ric-8A, was shown to recruit a signaling complex to the cell cortex that helped orient the mitotic spindle in response to spatial clues (61). Targeting ric8 in mice caused early embryonic lethality, however, derived ric8−/− embryonic cell lines had pleiotropic G protein signaling defects and major losses of Gαi1/2, Gαq, and Gα13 proteins due to rapid protein degradation (62–64). These later observations revealed that Ric-8A functioned as a molecular chaperone to target newly synthesized Gαi, Gαq, and Gα12/13 proteins to cellular membranes. Conditionally targeting ric8 in murine hematopoietic cells using the cre recombinase expressed in either all hematopoietic cells or only in murine B cells also led to severe reductions of Gα proteins with major decreases in Gαi2/i3, Gαq, and Gα13 proteins (65). In the hematopoietic cell specific deletion, the mice had a major reduction of platelets, which likely accounted for their shortened lifespan. The B lymphocyte specific deletion did not impact mouse viability, but led to a severe B cell immune phenotype (65). While bone marrow B cell lymphopoiesis was not, splenic marginal zone B cell development was severely compromised. There was also a marked reduction of B cells numbers in the spleen, lymph nodes, and at other peripheral sites. Not surprisingly B cells from these mice responded poorly to chemoattractants. B cell trafficking and the in situ positioning of B cells was impaired, the lymphoid architecture disorganized, and antibody responses suboptimal. A photograph and schematic of the intravital imaging set-up to analyze the behavior of wild type and Ric-8A deficient B cells is shown (Figure 2). The wild type and Ric-8A deficient B cells were fluorescently labeled and adoptively transferred into a wild type mouse 1 day prior to imaging. The cells were imaged in the inguinal lymph node of an anesthetized mouse. The Ric-8A deficient B cells moved slower and poorly accessed the lymph node follicle when compared to wild type B cells. The major loss of Gαi proteins in the B cells of these mice likely accounts for the majority of the phenotypes. Suggesting that Ric-8A has additional functions in B cells independent of its role as a Gαi chaperone, the number of asymmetric cell divisions detected in vitro in activated B cell and among purified germinal center B cells were reduced.
Fig. 2.
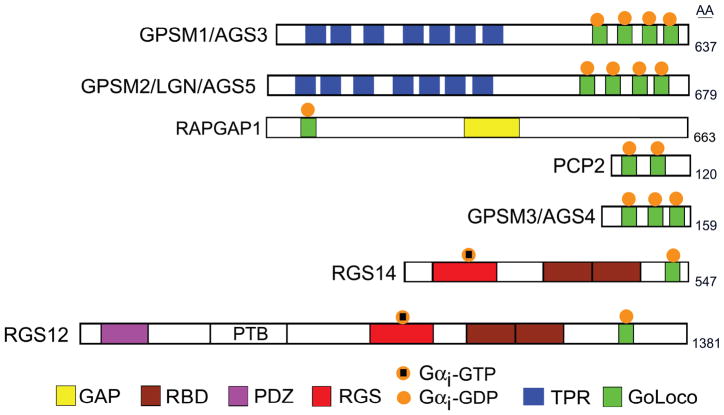
Schematics of the structures of GoLoco motif containing proteins. Proteins that contain GoLoco motifs (GPR domain) are shown. Abbreviations include RBD, Ras Binding Domain; PDZ, Post synaptic density Disc-large Zo-1; RGS, Regulator of G-protein Signaling; GAP, GTPase Activating Protein domain; and TPR, TetratricoPeptide Repeat. RGS domains bind GTP-Gαi and accelerate its intrinsic GTPase activity while the GoLoco motifs bind GDP-Gαi. The number of amino acids of the mouse versions of the indicated proteins is shown on the right.
3.2. AGS proteins
A functional screen in yeast for proteins that activated G-protein signaling in the absence of GPCRs led to the identification of what were termed AGS (Activators of G-protein signaling) proteins (14,66). They were initially divided into three functional groups: group I, guanine nucleotide exchange factors or GEFs of which there was a solitary member AGS1; group II, guanine nucleotide dissociation inhibitors, GDIs, of which there were four members AGS3-6; and group III, Gβγ interactors of which there were 5 members AGS2 & AGS7–10. Some of these AGS proteins are well expressed in immune cells and this review will mention several. Schematics of the domain structure of the known group II AGS proteins are shown (Figure 3). These proteins possess from 1 to 4 GoLoco motifs, which bind GDP, but not GTP bound Gαi much like Gβγ subunits. AGS3-5 (GPSM1-3) are briefly discussed below. A more detailed discussion of these and other AGS proteins can be found in other reviews (14,15,67,68). RGS12 and RGS14 are included in the group II AGS proteins as they each possess a GoLoco motif. However, since they also have an RGS domain they are considered in the section on RGS proteins. Not mention below are PCP2 and RAP1GAP1. PCP2 is expressed exclusively in cerebellar Purkinje cells and retinal bipolar neurons, where it impacts Gi/o signaling (69). The GoLoco motif in Rap1GAP1 is functional and may regulate the GAP activity of the protein (70). Ric-8A mentioned above is a group 1 AGS protein as it can trigger Gαi nucleotide exchange although not when Gα is associated with Gβγ (71). Recent evidence indicates that some group II members when bound to Gαi can sense agonist-induced conformational changes in GPCRs. This would suggest that they have a more complicated relationship with GPCR signaling than originally thought (72).
Fig. 3.
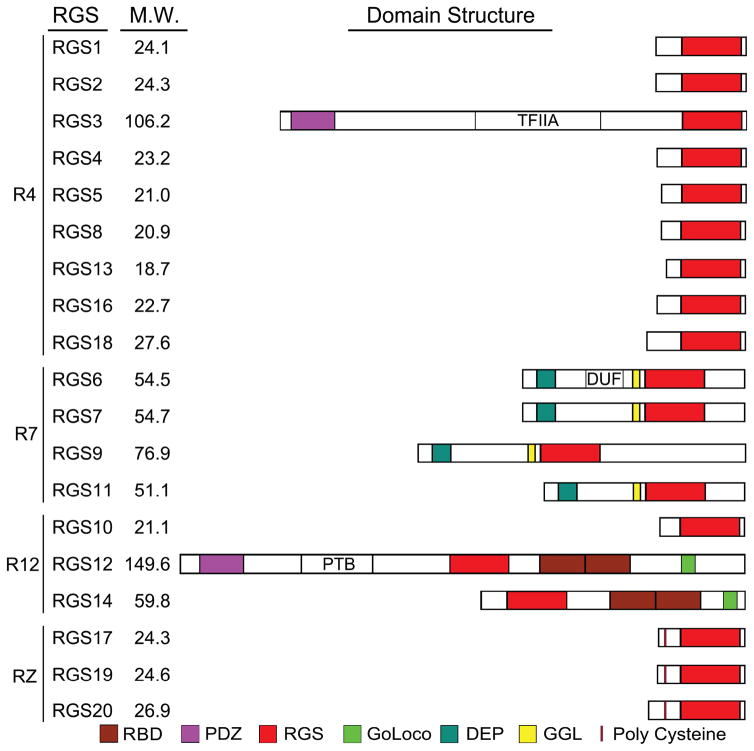
Schematics of the structures of the standard RGS domain containing proteins. Listed are the RGS domain containing proteins that act as GTPase activating proteins for Gαi and/or Gαq. Abbreviations include RBD, Ras Binding Domain; PDZ, Post Synaptic density Disc-large Zo-1; RGS, Regulator of G-protein Signaling; PTB, PhosphoTyrosine Binding domain; DEP; Dishevelled, Egl-10 and Pleckstrin domain, GoLoco (GPR domain); GGL; G protein Gamma-Like domain, DUF; Domain of Unknown Function; and TFIIA, Transcription Factor IIA domain. Predicted molecular weights (M.W.) of the different proteins are shown.
3.2.1. GPSM1/AGS3
The official gene name for AGS3 is Gpsm1, which encodes for a protein with 7 tetratricopeptide (TPR) repeats and 4 GoLoco motifs. The protein is most often referred to as AGS3. As mentioned above the GoLoco motifs can interact with GDP bound Gαi. Multiple TPR motifs (3–16) form a TPR repeat, which acts as a scaffold to mediate protein–protein interactions. Gpsm1 mRNA transcripts are broadly expressed found in most cell type including hematopoietic cells (16). Its expression is elevated in murine macrophages following LPS activation (73). Several studies have implicated AGS3 in chemokine receptor signaling. Gpsm1−/− mouse dendritic cells and lymphocytes exhibit suboptimal responds to chemokines in chemotaxis, calcium mobilization, and effector protein activation assays (74). Conversely, AGS3 may have a positive role in neutrophil chemotaxis. At the neutrophil leading edge GDP-bound Gαi accumulates where it recruits an adaptor molecule Inscuteable (Insc) along with AGS3 and the Par3-aPKC polarity complex (75). Neutrophils lacking Insc poorly stabilize leading edge pseudopods, which can be restored by the addition of wild-type Insc protein, but not by a mutant protein incapable of binding AGS3 (75). In RBL-2H3-CXCR2 cells, a mast cell line overexpressing CXCR2, exposure to a CXCR2 ligand resulted in the rapid formation of a GRK6/AGS3/Gαi2 complex (76). Overexpression of AGS3 in these cells inhibited CXCR2 signaling while knock-down enhanced signaling without affecting receptor internalization (76).
3.2.2. GPSM2/AGS5/LGN
AGS5/GPSM2/LGN also encodes for a protein with 7 TPR and 4 GoLoco motifs. Gpsm2 mRNA transcripts are well expressed in murine cycling pre-B cells, germinal center B cells, and in double positive thymocyte blasts (16). The protein is most commonly referred to as LGN and it has been shown to have an essential roles in asymmetric cell divisions in several different cell types. The binding of Gαi to the C-terminal GoLoco motif releases the auto-inhibited form of LGN allowing the N-terminal TPR motifs to interact with mInsc and NuMA, 2 proteins involved in cell division (77,78). Whether LGN has a role in asymmetric cell divisions in lymphocytes is unknown.
3.2.3. GPSM3/AGS4
GPSM3/AGS4 encodes for a protein that has 3 GoLoco motifs, but it lacks the TPR motifs. Gpsm3 mRNA has a selective tissue distribution being well expressed in immune system cells with its highest level of expression in neutrophils (16). Besides interacting with GDP-Gαi via its GPR motif, AGS4 also interacts with Gβ independent of any interaction with Gα or Gγ. The interaction with Gβ may act to stabilize to Gβ prior to its binding of Gγ. Genome-wide association studies have shown that single nucleotide polymorphisms in GPSM3 are associated with chronic inflammatory diseases (16).
3.3. RGS proteins
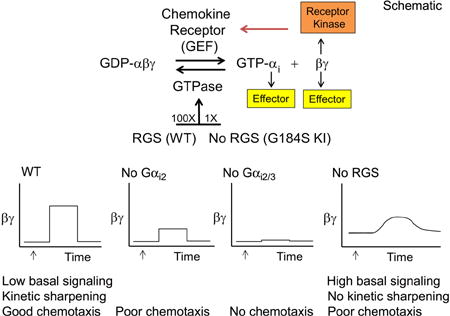
RGS proteins are a diverse family of proteins that are defined by the presence of a Regulator of G-protein Signaling (RGS) domain (11,13,79). The mammalian RGS family has more than 30 members, if those proteins with domains that exhibit weak homology to the RGS domain are included. The overall domain structure of the classical RGS proteins is shown (Figure 3). RGS family members can act as GAPs for Gαi and/or Gαq and several can attenuate Gs or G12/13 signaling. Most RGS domains bind with high affinity to Gαi subunits undergoing GTP hydrolysis. This interaction accelerates the intrinsic GTPase activity of the Gα subunit as much as a 100 fold. By shortening the duration that Gα is GTP bound, RGS proteins reduce the duration that GTP-Gα can interact with effectors. Since GDP-Gα can rebind freed Gβγ RGS proteins also limit Gβγ signaling. As such RGS proteins set a threshold for Gα-protein activation and sharpen the decay of signaling responses. Initially, RGS proteins were predicted to decrease steady-state GPCR agonist sensitivity, but this was not found. Explaining this observation several studies showed that RGS proteins not only increased the rate of G-protein deactivation, but also the rate of G-protein activation. The enhanced onset of signaling was sufficient to maintain agonist sensitivity and response amplitude. Several models have been proposed to account for this. An attractive model is termed “kinetic scaffolding” or “spatial focusing” (80,81). This model proposes that G-protein activation becomes saturated near spatially constrained GPCRs. This leads to GTP hydrolysis, rather than GDP release, becoming the rate limiting step in the G-protein cycle. Available RGS proteins by accelerating Gα GTP hydrolysis would promote heterotrimers re-assembly and provide additional heterotrimeric G-proteins for GPCR-activation. In the absence of RGS proteins, slow diffusion and collision events can not provide the needed heterotrimeric G-proteins for rapid and sustained activation of the signaling pathway. A prediction of this model is that loss of all Gαi/RGS protein interactions would lead to a substantial disruption of Gαi-mediated GPCR signaling (see section 3.3.11). The graphical abstract for this review illustrates the impact of the loss of Gαi proteins and RGS/Gαi protein interactions on the output of an idealized GPCR signaling pathway. While many RGS proteins predominately consist of an RGS domain, other RGS proteins contain multifunctional motifs and domains. These help mediate cross talk between GPCR-dependent and -independent signaling pathways. Furthermore, cell/tissue-specific expression patterns of many of the RGS proteins and their ability to interact with other signaling molecules helps restrict their signaling specificity. Many RGS proteins are expressed in human and murine immune cells (see Table 2, murine results shown) and there is considerable evidence that they influence immune cell function. The R4 RGS proteins are particularly well expressed and many of them impact chemokine receptor signaling by acting as GAPs for Gαi proteins (16). The R4 family includes RGS1-5, RGS8, RGS13, RGS16, RGS18, and RGS21 (82). In humans and mice, 9 of the 10 genes encoding the R4 RGS proteins are located in groups of two or more genes on chromosome 1. The sole exception is RGS3, which is located on chromosome 9 in humans and chromosome 4 in mice. Interestingly, the R4 RGS genes are closely linked to an MHC paralogon on chromosome 1 (83). The RGS proteins known to have roles in G-protein signaling in leukocytes are discussed below and table 3 outlines the impact of RGS protein gene targeting in mice.
Table 2.
Expression of different RGS protein in various types of leukocytes1
| Cell type | Rgs1 | Rgs2 | Rgs3 | Rgs10 | Rgs12 | Rgs14 | Rgs16 | Rgs18 | Rgs19 |
|---|---|---|---|---|---|---|---|---|---|
| B | 0.1 | 1.0 | 0.1 | 0.2 | 0.1 | 0.7 | 0.0 | 0.1 | 1.3 |
| Dendritic | 6.9 | 14.4 | 0.5 | 0.9 | 3.0 | 0.3 | 0.0 | 0.3 | 0.8 |
| Neutrophil | 0.1 | 28.2 | 1.0 | 0.1 | 0.0 | 3.4 | 0.0 | 0.7 | 3.1 |
| Macrophage | 0.1 | 3.7 | 0.3 | 2.2 | 0.0 | 0.3 | 0.0 | 1.2 | 1.0 |
| NK | 1.5 | 2.1 | 1.8 | 0.0 | 0.1 | 0.5 | 0.1 | 0.0 | 0.6 |
| NKT | 1.9 | 2.1 | 3.7 | 0.2 | 0.1 | 0.4 | 0.0 | 0.0 | 0.3 |
| CD4 | 0.4 | 0.3 | 0.7 | 1.8 | 0.1 | 1.0 | 0.1 | 0.0 | 0.6 |
| CD8 | 0.1 | 0.2 | 0.9 | 1.8 | 0.0 | 0.6 | 0.0 | 0.0 | 0.6 |
| Tγδ | 0.4 | 0.5 | 1.7 | 0.7 | 0.3 | 0.5 | 0.1 | 0.0 | 0.4 |
| Treg | 2.3 | 0.7 | 0.8 | 2.6 | 0.1 | 0.7 | 0.5 | 0.0 | 1.1 |
Shown are the relative mRNA expression levels in different cell types. Cells were isolated from 3 male C57BL/6J mouse spleens with the exception of the macrophages, which were isolated from the peritoneal cavity of similar mice. Data are from Immunological genome project (http://www.immgen.org/databrowser/index.html) and based on RNA sequencing of mRNA from FACS purified cell samples. Cell type with highest expression of individual RGS protein mRNA expression is highlighted. Little or no expression for Rgs4, Rgs5, Rgs6, Rgs7, Rrgs8, Rgs13, Rgs17, Rgs20, or Rgs22. Non-detectible expression levels of Rgs9 and Rgs11 were found in the different cell subsets with the exception of CD4 and CD8 T cells, where low amounts were found (0.1). β-actin mRNA level is approximately 400. Values for each gene (x 103).
Table 3.
Impact of targeting Gαi regulatory genes expressed in murine leukocytes
| Gene | Sites of expression1 | Immune phenotypes2 |
|---|---|---|
| Rgs1 | DC, NK, NKT, Treg, Tγδ, GC B, Ba, MC | Altered antibody responses, B and gut T cell trafficking defects, colitis, enhanced macrophage accumulation in atherosclerotic plaques. |
| Rgs2 | GN, DC, MF, NK, Ba, MC | Decreased platelet mediated hemostasis, decreased T cell proliferation, impaired antiviral immunity. |
| Rgs3 | NKT, NK, GN, Tγδ, Treg | Defective T cell trafficking, altered cytokine production, enhanced T cell infiltration into the lung in an asthma model. |
| Rgs10 | MF, Treg, T, DC, B, MC | Enhanced LPS induced inflammatory cytokine production by microglia, decreased M2 macrophage priming |
| Rgs12 | DC, O | Increased bone mass with decreased osteoclast numbers. |
| Rgs13 | GC B, MC | Increased extrafollicular antibody response, large germinal centers, decreased autoantibody production in autoimmune mice, increased mast cell chemotaxis. |
| Rgs14 | GN, T, Treg, B | Normal T and B cell compartments3 |
| Rgs16 | Treg, T | Altered T cell trafficking and cytokine production in allergic lung inflammation. |
| Rgs19 | MF, B, DC, T, Ba, MC | Altered antibody responses, B cell trafficking defects, expanded B cell compartment4 |
| Gpsm1 | Broadly in leukocytes, highest in NK | Reduced lymphocyte and dendritic cell chemotaxis. Impact on NK function unknown. |
| Gpsm2 | GC B, double positive thymocytes | No reported phenotypes in leukocytes. |
| Gpsm3 | Broadly in leukocytes, highest in GN | Myeloid cells had reduced migration to chemokines and enhanced apoptosis in vitro. No reported phenotypes in GN. |
| Ric8 | Broadly in leukocytes, highest GN | B cell specific deletion resulted in a severe reduction in Gαi, Gαq, and Gα13; and a severe B cell immunodeficiency. |
Based on mRNA expression, abbreviations as follows DC – dendritic cell, NK - natural killer cell, NKT – natural killer T cell, Treg- regulatory T cell, Tγδ-gamma delta T cell, GC B–germinal center B cell, MF- macrophage, GN – granulocyte, Ba – basophil, MC – mast cell, and O – osteoclast.
Immune phenotypes for mice lacking the indicated genes. References are included in the text.
Mice lacking Rgs14 have normal T and B cell compartments and normal antibodies responses (J. Kehrl, unpublished observation).
Mice lacking Rgs19 expression exhibit altered antibody responses, B cell trafficking defects, and an expanded B cell compartment in the spleen (J. Kehrl, unpublished observation).
3. 3.1. RGS1
RGS1 is an R4 RGS protein. It and RGS2 were the founding members of the RGS protein family (84,85). Alterations in the RGS1 gene are associated with celiac disease, multiple sclerosis, type I diabetes, and various types of human cancer (68,86). RGS1 is highly expressed in regulatory T cells, intraepithelial lymphocytes, activated B cells, NK-T cells, basophils, eosinophils, mast cells, dendritic cells, and activated macrophages (16). Expression is much higher in T cells from human gut versus peripheral blood, and that this can be exaggerated in intestinal inflammation (87). RGS1 expression is upregulated in many cell types by exposure to hypoxia or interferons (68,88). Reduced RGS1 levels have been shown to enhance T cell, B cell, dendritic, and macrophage responsiveness to chemokines (87,89,90). The roles of RGS1 in mast cell, eosinophil, and basophil biology are largely unknown, but the particularly high levels of expression of RGS1 in these cell types suggests that is functionally important for their biology.
3.3.2. RGS2
RGS2 is an R4 RGS protein and its coding region is located in close proximity to Rgs1 and Rgs13 on chromosome 1. In immune system cell types Rgs2 mRNA expression is very similar to that of Rgs1 mRNA with a couple of exceptions. Rgs2 is highly expressed in neutrophils, while Rgs1 is not, and in contrast to Rgs1, Rgs2 expression is not interferon inducible (16). Despite the similar expression pattern to Rgs1, Rgs2 has not been linked to diseases of the immune system, but rather to cardiovascular and central nervous system dysfunction (91,92). However, analysis of mice lacking Rgs2 expression did reveal a role for RGS2 in T cell proliferation and anti-viral immune responses (93). RGS2 differs from other RGS proteins as its GAP activity is largely restricted to Gαq, showing little activity toward Gαi. RGS2 also interacts with Gαs and adenylyl cyclase (AC), suppressing Gαs signaling pathways independently of its GAP activity (94–96). Thus, RGS2 is unlikely to impact chemokine receptor signaling, but to influence signaling through Gq- and Gs-linked receptors.
3.3.3. RGS3
With the exception of RGS3 the other R4 RGS protein family members are predominately an RGS domain with an n-terminal and c-terminal extension. There are several RGS3 splice variants that encode for proteins with additional domains. One variant termed PDZ-RGS3 has N terminal PDZ domain, which can bind type B ephrins, an ATP/GTP-binding site, and a proline-rich region of unknown function (97,98). A loss of PDZ-RGS3 in mice caused an early cell cycle exit and precocious differentiation of neural progenitor cells located in the developing cerebral cortex, a phenotype similar to that observed in the ephrin-B1 knockout mice. This phenotype was linked to dysregulated CXCR4 signaling (99). The original human RGS3 cDNA lacked the coding region for the PDZ domain and the predicted protein was termed RGS3L to distinguish it from a smaller RGS3 protein designated RGS3S (84). Both RGS3L and RGS3S are expressed in immune cells while PDZ-RGS3 is more prominent in neurons. Among immune cell types murine NK, NK-T, T cells, and neutrophils best express RGS3 mRNA transcripts (16). In a mouse model of asthma RGS3 affected the recruitment of inflammatory T cells and modulation of RGS3 levels affected the chemotaxis of a human T cell line (100).
3.3.4. RGS10
Among the RGS proteins RGS10 is particularly well expressed in cells of the immune system (101). Based on homology of its RGS domain to those of other RGS proteins it is considered a member of the R12 subfamily. High levels of expression are present in murine dendritic cells, mast cells, macrophages, and CD4 T cells. Although found at lower levels in the B lymphocyte compartment, it is relatively enriched in germinal center and marginal zone B cells (16). In the central nervous system it is well expressed in microglial cells. There are two isoforms of murine RGS10 termed RGS10L (181 amino acids) and RGS10S (167 amino acids), although the former predominates in mouse leukocytes. While several RGS proteins use an amphipathic helix or cysteine string to target cellular membranes, RGS10 lacks these structures and relies on the palmitoylation of an n-terminal cysteine to target membranes (102). Loss of RGS10 in mice results in dysregulated microglial cell cytokine production with a pronounced increase in LPS induced pro-inflammatory cytokine production (103). Similarly, Rgs10−/− macrophages produce higher levels of pro-inflammatory cytokines in response to LPS treatment and exerted higher cytotoxicity towards neuroblastoma cells. The deficient macrophages exhibited a blunted M2 phenotype upon IL-4 priming (104). The role of RGS10 in lymphocytes has received relatively little attention. A gain of function/loss of function study using a human T cell line revealed that RGS10 inhibited chemokine receptor Gαi-dependent T cell adhesion mediated by the integrins α4β1 and αLβ2 (105). RGS10 regulated chemokine induced adhesion by limiting the activation of the Vav1-Rac1 pathway. RGS10 also limited T cell chemotaxis and inhibited chemokine induced activation of the small GTPase cdc42 (105). In depth studies of B and T cell function in RGS10 knock-out mice have not been reported.
3.3.5 RGS12
RGS12 is the largest protein of the RGS protein family and a member of the R12 subfamily along with RGS10 and RGS14 (see below). Full length RGS12 possesses a PDZ/PTB domain, post synaptic density disc-large zo-1 (PDZ) and phosphotyrosine binding (PTB) along with an RGS domain, 2 Ras binding domains (RBD), and a GoLoco motif. Alternative splicing creates multiple different isoforms of the RGS12 protein (106–108). In immune cell Rgs12 mRNA expression is most prominent in dendritic cells with lesser amounts in macrophages (16). It is also strongly expressed in osteoclasts. In a recent study Rgs12fl/fl Mx1-Cre transgenic mice had the expression of Rgs12 deleted at postnatal day 10 in interferon-responsive cells. This resulted in growth retardation, increased bone mass, and reduced numbers of osteoclasts in the mice (109). Similar results were found in Rgs12fl/fl Cd11b-cre mice, which would predominately delete Rgs12 expression in neutrophils and macrophages (110). No assessment of dendritic cell function was performed in these mice. The N-terminal PDZ domain of RGS12 is known to interact with the C-terminus of CXCR2, however the functional importance of this interaction remains unknown (107). In muscle cells Gβγ signaling can lead to a PI 3-kinase-γ- and c-Src- dependent tyrosine phosphorylation of Gαi. This results in the recruitment of RGS12 to Gαi and a reduction in its activity (111). Whether such a mechanism is operant in dendritic cells or macrophages has not been reported.
3.3.6. RGS13
RGS13 is a R4 RGS protein and it is one of the smallest of the RGS family (112,113). The human and mouse proteins are composed of 159 and 158 amino acids, respectively. Although largely composed on an RGS domain, RGS13 also interacts with and inhibits the activity of phosphatidylinositol-3-kinase (PI3K) in mast cells and phosphorylated cyclic AMP response element binding protein (pCREB) in B cells (114,115). RGS13 has one of the most restricted patterns of expression among the RGS proteins. In immune system cells Rgs13 mRNA transcripts are largely confined to murine germinal center B cells and mast cells although they are also present in thymic medullary epithelial cells (16). Analysis of the B cell compartment in Rgs13 deficient mice (C57BL/6 background) revealed that RGS13 constrains extra-follicular plasma cell generation, germinal center sizes, and germinal center B cell numbers (116). In an autoimmune strain of mice, BXD2, Rgs13 is highly expressed in germinal center B cells, but also in T follicular helper (TFH) cells (117). This contrasts with C57BL/6 mice where TFH cells do not express detectable Rgs13 mRNA. Surprisingly many of the B cell abnormalities noted in BXD2 mice were partially rescued by loss of Rgs13 (117). While manipulating Rgs13 expression in human B cell lines affected responses to chemokines, surprisingly, the loss of Rgs13 expression did not materially affect murine germinal center B cell chemotaxis towards CXCL12 or CXCL13 (116). The reason for this is unclear although it may reflect the difficulties in assessing germinal center B cell chemokine responses. Stimulation of B cell adrenergic receptors results in the translocation of RGS13 to the nucleus, where it inhibits pCREB-mediated transcription (115). Germinal center B cells from Rgs13 deficient mice expressed increased levels of several pCREB target genes (116,117).
3.3.7. RGS14
RGS14 is a member of the R12 subfamily of RGS proteins and a selective GAP for Gαi/o. Mouse RGS14 is a 547 amino acid proteins that has a N-terminal RGS domain, two Ras binding domains, and a GoLoco motif. RGS14 can simultaneously bind GDP-Gαi via it GoLoco motif and act as a GAP for GTP bound Gαi (108,118–120). RGS14 can interacts with the monomeric G proteins Rap1, Rap2, and H-Ras (121). In the immune system Rgs14 expression is abundant in neutrophils, but also found in most immune system cell types (16). In the brain Rgs14 is expressed in neurons in the hippocampus and olfactory cortex, where it suppresses synaptic plasticity. Mice lacking Rgs14 perform better than do wild-type mice in hippocampus-dependent tasks (91). The impact of the loss of Rgs14 on the adaptive or innate immune system has not been reported.
3.3.8. RGS16
RGS16 is a member of the R4 family of RGS proteins. A cDNA that encodes RGS16 was initially cloned from the retina and subsequent studies demonstrated widespread tissue expression (82,122). It has a demonstrated role in the circadian regulation of intracellular G-protein signaling, which controls rhythmicity in the suprachiasmatic nucleus (123). Within cells of the immune system Rgs16 mRNA is found at low levels in NK cells, CD4 T cells and platelets with higher levels in regulatory T cells (16). Several studies have suggested a role for RGS16 in allergic airway disease. T cell-specific Rgs16 transgenic mice had reduced T cell trafficking to the lung following allergic airway while the lungs of allergen exposed Rgs16 deficient mice had more TH2 cells than similarly challenged WT mice (124,125). Rgs16 mRNA transcripts were strongly upregulated in human monocyte derived DCs treated with LPS and IL-10. Gain and loss of function experiments using a human monocyte cell line showed that RGS16 overexpression reduces the expression of pro-inflammatory cytokines while reduction of RGS16 had the opposite effect (126).
3.3.9. RGS18
RGS18 is also an R4 RGS protein that is abundantly expressed in platelets and, to a lesser extent in megakaryocytes, macrophages, osteoclasts, neutrophils, and bone marrow progenitors (16,127–130). RGS18 inhibits both Gi- and Gq-mediated signaling. RGS18 has been implicated in the control of osteoclastogenesis mediated by RANKL by modulating signaling through the proton sensing GPCR OGR1 (131). Several recent studies have documented a role for RGS18 in thrombopoiesis and platelet function (132–136). Following exposure to thrombin or thromboxane A2 RGS18 is phosphorylated on serine 49 and serine 218, which promotes its binding to 14-3-3 proteins leading to an inhibition of RGS18 GAP activity. Rgs18 deficient mice are mildly thrombocytopenic due to a decrease in platelet production, but are also hypercoagulable due to increased platelet sensitivity to platelet activators. These studies suggest that RGS18 modulates both megakaryocyte differentiation and platelet hemostatic function.
3.3.10. RGS19
RGS19 is a member of the RZ subfamily of RGS proteins (11,137). It is broadly expressed although at the highest levels in murine hematopoietic cells. Among immune cell types it is most prevalent in neutrophils, eosinophils, basophils, and mast cells. It is expressed at lower levels in lymphocytes, dendritic cells, natural killer cells, and macrophages (16). It has been implicated in cancer; described to suppress Wnt-dependent signaling and Ras-induced cell proliferation; and to be a downstream target of Notch signaling and involved in the activation of Akt (138–140). Like other RZ family members RGS19 has an amino-terminal cysteine string motif, an RGS domain, and a short carboxyl-terminal (141,142). Its GAP activity exhibits some specificity as it preferentially acts on Gαi3 and Gαo showing little activity towards Gαi2. It has also been shown to function as a GDI for Gαi3 and Gαo even though it lacks a GoLoco motif (143). Thereby, RGS19 can retain GDP bound Gαi3 following GTP hydrolysis. This would tend to suppress Gαi3 activity, but potentiate Gβγ signaling. The results of targeting Rgs19 in mice have not been reported, however knocking down Rgs19 expression during mouse development suggested that Rgs19 functions to limit the expression of Wnt-responsive genes, which are needed for proper midline fusion of the mouse palate (144).
3.3.11 RGS insensitive Gai2
Because many cell types can express multiple RGS proteins the loss of an individual RGS protein may result in a minimal or no phenotype. Mapping the site of interaction of RGS proteins with Gαi proteins provided a partial solution to the problem of compensation by other family members (145). A single mutation in Gαi proteins was found to render them insensitive to RGS proteins as it blocked the interaction between the two proteins. This mutation does not affect Gαi binding to receptors, Gβγ, or effectors; nor does it affect Gαi expression (146). Mice with such a mutation in the Gnai2 locus have been made (147). This mutat ion results in a serine at posit ion 184 of the protein instead of the usual glycine (Gαi2 G184S). The mutant Gαi protein is expressed at levels similar to that of the wild type protein. Study of these mice has revealed abnormal cardiac function and central nervous system dysfunction (148). Mirroring the Rgs18 deficient mice, the mice carrying the Gαi2 G184S mutation have enhanced platelet aggregation and increased platelet accumulation following vascular injury (149). Neutrophils with the Gαi2 G184S mutation accumulated in the bone marrow and mobilized poorly to inflammatory sites. They displayed an enhanced sensitivity to background signals, altered kinetics of chemoattractant receptor signaling, and inappropriate CXCR2 downregulation. As neutrophils prominently express several RGS proteins, this phenotype has not been phenocopied by the loss of an individual RGS protein. The analyses of the G184S Gαi2 neutrophils support a role for RGS proteins in setting a threshold for Gαi2 activation, which helps to coordinate desensitization mechanisms (150). B cells from these same mice also had defective chemokine receptor signaling. The Gαi2 G184S B cells had an elevated basal intracellular calcium level; enhanced non-specific migration, but poor chemokine (CXCL12, CXCL13, and CCL19) induced chemotaxis. Analysis of the intracellular calcium response and the activation of downstream effectors in the Gαi2 G184S B cells indicated that both the onset of signaling and the termination of signaling were disrupted by the loss of Gαi2/RGS protein interactions (151). As discussed earlier the loss of RGS proteins can affect both the activation and de-activation of GPCR signaling pathways (80). In vivo the Gαi2 G184S B cells homed poorly to lymph nodes and exhibited improper in situ positioning, findings consistent with impaired chemokine receptor signaling. Thus, the RGS proteins in B cells act in a similar fashion to those in neutrophils helping to coordinate chemokine receptor Gαi2 signaling and desensitization mechanisms (151). Surprisingly, the loss of Gαi2/RGS protein interactions did not similarly affect signaling though another GPCR expressed in B cells, the sphingosine 1-phosphate receptor 1 (S1PR1) (151). The S1P/S1PR1 axis facilitates the egress of thymocytes from the thymus into the blood, and of B and T cells from lymph nodes into the efferent lymph, the latter a requirement for lymphocyte recirculation (152). In contrast to impaired chemokine receptor signaling, exposure of Gαi2 G184S B cells to S1P elicited higher intracellular calcium responses and enhanced in vitro chemotaxis (151). Furthermore, there was no obvious defect in B cell egress from lymph nodes. This was perplexing as S1PR1 also signals by activating Gαi. Perhaps explaining the discrepancy, S1PR1 preferred to pre-couple to Gαi3 versus Gαi2 (>10-fold difference), while the chemokine receptor CXCR4 had no such preference. This conclusion was based on an in vitro BRET (Bioluminescence Resonance Energy Transfer) assay using transfected proteins (151). Assuming that other B cell chemokine receptors also have no isoform bias, the higher expression of Gαi2 versus Gαi3 (5–10 fold difference) would lead to chemokine receptors being predominately pre-coupled to the mutant Gαi2 protein, while S1PR1 would be pre-coupled more to the wild type Gαi3, which would be subject to normal regulation by RGS proteins. Thus, signaling through S1PR1 via Gαi3 would proceed normally, while the limited activation of Gαi2 G184S might potentiate the signaling output. Further studies are needed to better understand the G-protein selectivity of the S1PR1 receptor and to better understand the role of RGS proteins in regulating Gαi2 versus Gαi3 signaling. Finally, macrophages carrying the Gαi2 G184S mutation had a decrease in LPS-induced cytokine production when compared to wild type macrophages (153). This indicates that the loss of Gαi2/RGS protein interactions in macrophages impacts TLR4 signaling and that RGS proteins dampen cytokine production. Whether this is a direct or an indirect effect of Gαi2 on TLR4 signaling remains controversial.
4. Conclusions
Many of the GPCRs expressed by leukocytes function as chemoattractant receptors. The signals emanating from these receptors recruit leukocytes to inflammatory sites; organize the positioning of leukocytes in immune organs, help maintain the overall architecture of immune organs; coordinate the movements of leukocytes through tissues; and facilitate the trafficking of leukocytes into and out of lymph nodes, the bone marrow, skin, and the gut associated lymphoid tissues (GALT). The signaling output of GPCRs depends on the level of GPCR expression and the levels of heterotrimeric G proteins and their regulators. For chemoattractant receptors Gαi is crucial. Lymphocyte chemoattractant signaling is highly dependent upon Gαi2, but absolutely dependent upon Gαi2 and Gαi3. In their absence murine B and T cells can no longer respond to these chemoattractant signals as other heterotrimeric G-proteins cannot substitute for their loss. Likely, Gαi2/3 are similarly important in chemoattractant receptor signaling in other leukocytes, but this needs confirmation. This review focused on some of the regulatory mechanisms that impact Gαi. Those that directly impact the receptor such as the G-protein regulatory kinases and β-arrestins were not covered, although they have considerable importance for chemoattractant receptor intracellular trafficking and receptor desensitization. The RGS proteins known to affect Gαi signaling in leukocytes were reviewed. The RGS domain-containing proteins appeared early in eukaryote evolution, they expanded in conjunction with the Gα subunits, and they have served as a primary mechanism to regulate the sensing of environmental signals by GPCRs. The considerable redundancy of RGS protein expression remains confounding although the analysis of the Gαi2 G184S mice has provided important insights into the overall importance of RGS proteins expressed in neutrophils, platelets, and lymphocytes. Based on some of the studies reviewed here and others, RGS proteins have multiple roles in GPCR signaling. They set a threshold for Gαi activation, function to limit Gαi signaling, sharpen Gαi signaling output, coordinate with the β-arrestin system to control receptor desensitization, and is some instance serve as multi domain intracellular platforms for interactions with GDP-Gαi, GTP-Gαi, Gβγ, GPCRs, and other proteins.
Fig. 1.
Intravital imaging of wild type and Ric-8A deficient B cells in the inguinal lymph node of a live mouse. Wild type (green) and Ric-8A deficient (red) B cells were adoptively transferred into a wild type mouse 24 hours before imaging. The mouse was anesthetized, a small incision made over the inguinal lymph node, and images were collected with a multiphoton microscope. The blood vessels including the high endothelial venules were outlined by injection of a fluorescent intravascular dye. A snapshot from the imaging is shown. Of not the Ric-8A deficient B cells localized near the HEVs and entered poorly into the lymph node follicle, a process that depends upon CXCR5 signaling. The expression of different Gα subunits in the wild type and Ric-8A deficient B cells is shown by immunoblotting cell lysates with specific antibodies.
Acknowledgments
The author would like to thank the members of his laboratory over the years that contributed to some of the studies described in this review and to thank Dr. Anthony Fauci for his long standing support. The intramural program of the National Institutes of Allergy and Infectious Diseases supported some of the research described in this review.
Footnotes
Conflict of interest
There are no conflict of interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Duc NM, Kim HR, Chung KY. Structural mechanism of G protein activation by G protein-coupled receptor. Eur J Pharmacol. 2015;763:214–222. doi: 10.1016/j.ejphar.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 2.Wettschureck N, Offermanns S. Mammalian G proteins and their cell type specific functions. Physiol Rev. 2005;85:1159–1204. doi: 10.1152/physrev.00003.2005. [DOI] [PubMed] [Google Scholar]
- 3.Zhang D, Zhao Q, Wu B. Structural Studies of G Protein-Coupled Receptors. Mol Cells. 2015;38:836–842. doi: 10.14348/molcells.2015.0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walther C, Ferguson SS. Arrestins: role in the desensitization, sequestration, and vesicular trafficking of G protein-coupled receptors. Prog Mol Biol Transl Sci. 2013;118:93–113. doi: 10.1016/B978-0-12-394440-5.00004-8. [DOI] [PubMed] [Google Scholar]
- 5.Shukla AK. Biasing GPCR signaling from inside. Sci Signal. 2014;7:pe3. doi: 10.1126/scisignal.2005021. [DOI] [PubMed] [Google Scholar]
- 6.Zhou L, Bohn LM. Functional selectivity of GPCR signaling in animals. Curr Opin Cell Biol. 2014;27:102–108. doi: 10.1016/j.ceb.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dillon SB, Verghese MW, Snyderman R. Signal transduction in cells following binding of chemoattractants to membrane receptors. Virchows Arch B Cell Pathol Incl Mol Pathol. 1988;55:65–80. doi: 10.1007/BF02896561. [DOI] [PubMed] [Google Scholar]
- 8.Cyster JG, Goodnow CC. Pertussis toxin inhibits migration of B and T lymphocytes into splenic white pulp cords. J Exp Med. 1995;182:581–586. doi: 10.1084/jem.182.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaziro Y, Itoh H, Kozasa T, Nakafuku M, Satoh T. Structure and function of signal-transducing GTP-binding proteins. Annu Rev Biochem. 1991;60:349–400. doi: 10.1146/annurev.bi.60.070191.002025. [DOI] [PubMed] [Google Scholar]
- 10.Neptune ER, Iiri T, Bourne HR. Galphai is not required for chemotaxis mediated by Gi-coupled receptors. J Biol Chem. 1999;274:2824–2828. doi: 10.1074/jbc.274.5.2824. [DOI] [PubMed] [Google Scholar]
- 11.Woodard GE, Jardin I, Berna-Erro A, Salido GM, Rosado JA. Regulators of G-protein-signaling proteins: negative modulators of G-protein-coupled receptor signaling. Int Rev Cell Mol Biol. 2015;317:97–183. doi: 10.1016/bs.ircmb.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 12.Kehrl JH. Heterotrimeric G protein signaling: roles in immune function and fine-tuning by RGS proteins. Immunity. 1998;8:1–10. doi: 10.1016/s1074-7613(00)80453-7. [DOI] [PubMed] [Google Scholar]
- 13.Kach J, Sethakorn N, Dulin NO. A finer tuning of G-protein signaling through regulated control of RGS proteins. Am J Physiol Heart Circ Physiol. 2012;303:H19–35. doi: 10.1152/ajpheart.00764.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cismowski MJ, Takesono A, Bernard ML, Duzic E, Lanier SM. Receptor-independent activators of heterotrimeric G-proteins. Life Sci. 2001;68:2301–2308. doi: 10.1016/s0024-3205(01)01019-0. [DOI] [PubMed] [Google Scholar]
- 15.Willard FS, Kimple RJ, Siderovski DP. Return of the GDI: the GoLoco motif in cell division. Annu Rev Biochem. 2004;73:925–951. doi: 10.1146/annurev.biochem.73.011303.073756. [DOI] [PubMed] [Google Scholar]
- 16.Boularan C, Kehrl JH. Implications of non-canonical G-protein signaling for the immune system. Cell Signal. 2014;26:1269–1282. doi: 10.1016/j.cellsig.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hewavitharana T, Wedegaertner PB. Non-canonical signaling and localizations of heterotrimeric G proteins. Cell Signal. 2012;24:25–34. doi: 10.1016/j.cellsig.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho H, Kehrl JH. Localization of Gi alpha proteins in the centrosomes and at the midbody: implication for their role in cell division. J Cell Biol. 2007;178:245–255. doi: 10.1083/jcb.200604114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu J, Pipathsouk A, Keizer-Gunnink A, Fusetti F, Alkema W, Liu S, Altschuler S, Wu L, Kortholt A, Weiner OD. Homer3 regulates the establishment of neutrophil polarity. Mol Biol Cell. 2015;26:1629–1639. doi: 10.1091/mbc.E14-07-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamp ME, Liu Y, Kortholt A. Function and Regulation of Heterotrimeric G Proteins during Chemotaxis. IntJ Mol Sci. 2015:17. doi: 10.3390/ijms17010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Surve CR, Lehmann D, Smrcka AV. A chemical biology approach demonstrates G protein betagamma subunits are sufficient to mediate directional neutrophil chemotaxis. J Biol Chem. 2014;289:17791–17801. doi: 10.1074/jbc.M114.576827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan J, Jin T. Signaling network from GPCR to the actin cytoskeleton during chemotaxis. Bioarchitecture. 2012;2:15–18. doi: 10.4161/bioa.19740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerr JS, Jacques RO, Moyano Cardaba C, Tse T, Sexton D, Mueller A. Differential regulation of chemotaxis: role of Gbetagamma in chemokine receptor-induced cell migration. Cell Signal. 2013;25:729–735. doi: 10.1016/j.cellsig.2012.12.015. [DOI] [PubMed] [Google Scholar]
- 24.Gambardella L, Vermeren S. Molecular players in neutrophil chemotaxis--focus on PI3K and small GTPases. J Leukoc Biol. 2013;94:603–612. doi: 10.1189/jlb.1112564. [DOI] [PubMed] [Google Scholar]
- 25.Sasaki T, Irie-Sasaki J, Jones RG, Oliveira-dos-Santos AJ, Stanford WL, Bolon B, Wakeham A, Itie A, Bouchard D, Kozieradzki I, Joza N, Mak TW, Ohashi PS, Suzuki A, Penninger JM. Function of PI3Kgamma in thymocyte development, T cell activation, and neutrophil migration. Science. 2000;287:1040–1046. doi: 10.1126/science.287.5455.1040. [DOI] [PubMed] [Google Scholar]
- 26.Li Z, Jiang H, Xie W, Zhang Z, Smrcka AV, Wu D. Roles of PLC-beta2 and -beta3 and PI3Kgamma in chemoattractant-mediated signal transduction. Science. 2000;287:1046–1049. doi: 10.1126/science.287.5455.1046. [DOI] [PubMed] [Google Scholar]
- 27.Hirsch E, Katanaev VL, Garlanda C, Azzolino O, Pirola L, Silengo L, Sozzani S, Mantovani A, Altruda F, Wymann MP. Central role for G protein-coupled phosphoinositide 3-kinase gamma in inflammation. Science. 2000;287:1049–1053. doi: 10.1126/science.287.5455.1049. [DOI] [PubMed] [Google Scholar]
- 28.Bach TL, Chen QM, Kerr WT, Wang Y, Lian L, Choi JK, Wu D, Kazanietz MG, Koretzky GA, Zigmond S, Abrams CS. Phospholipase cbeta is critical for T cell chemotaxis. J Immunol. 2007;179:2223–2227. doi: 10.4049/jimmunol.179.4.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu X, Gera N, Li H, Yun M, Zhang L, Wang Y, Wang QJ, Jin T. GPCR-mediated PLCbetagamma/PKCbeta/PKD signaling pathway regulates the cofilin phosphatase slingshot 2 in neutrophil chemotaxis. Mol Biol Cell. 2015;26:874–886. doi: 10.1091/mbc.E14-05-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnon TI, Xu Y, Lo C, Pham T, An J, Coughlin S, Dorn GW, Cyster JG. GRK2-dependent S1PR1 desensitization is required for lymphocytes to overcome their attraction to blood. Science. 2011;333:1898–1903. doi: 10.1126/science.1208248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dillenburg-Pilla P, Patel V, Mikelis CM, Zarate-Blades CR, Doci CL, Amornphimoltham P, Wang Z, Martin D, Leelahavanichkul K, Dorsam RT, Masedunskas A, Weigert R, Molinolo AA, Gutkind JS. SDF-1/CXCL12 induces directional cell migration and spontaneous metastasis via a CXCR4/Galphai/mTORC1 axis. FASEB J. 2015 doi: 10.1096/fj.14-260083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He Y, Li D, Cook SL, Yoon MS, Kapoor A, Rao CV, Kenis PJ, Chen J, Wang F. Mammalian target of rapamycin and Rictor control neutrophil chemotaxis by regulating Rac/Cdc42 activity and the actin cytoskeleton. Mol Biol Cell. 2013;24:3369–3380. doi: 10.1091/mbc.E13-07-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasaki AT, Firtel RA. Regulation of chemotaxis by the orchestrated activation of Ras, PI3K, and TOR. Eur J Cell Biol. 2006;85:873–895. doi: 10.1016/j.ejcb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Broxmeyer HE, Youn BS, Kim C, Hangoc G, Cooper S, Mantel C. Chemokine regulation of hematopoiesis and the involvement of pertussis toxin-sensitive G alpha i proteins. Ann N Y Acad Sci. 2001;938:117–127. doi: 10.1111/j.1749-6632.2001.tb03580.x. discussion 127–118. [DOI] [PubMed] [Google Scholar]
- 35.Spangrude GJ, Sacchi F, Hill HR, Van Epps DE, Daynes RA. Inhibition of lymphocyte and neutrophil chemotaxis by pertussis toxin. J Immunol. 1985;135:4135–4143. [PubMed] [Google Scholar]
- 36.Braaten BA, Spangrude GJ, Daynes RA. Molecular mechanisms of lymphocyte extravasation. II. Studies of in vitro lymphocyte adherence to high endothelial venules. J Immunol. 1984;133:117–122. [PubMed] [Google Scholar]
- 37.Spangrude GJ, Braaten BA, Daynes RA. Molecular mechanisms of lymphocyte extravasation. I. Studies of two selective inhibitors of lymphocyte recirculation. J Immunol. 1984;132:354–362. [PubMed] [Google Scholar]
- 38.Kehrl JH, Hwang IY, Park C. Chemoattract receptor signaling and its role in lymphocyte motility and trafficking. Curr Top Microbiol Immunol. 2009;334:107–127. doi: 10.1007/978-3-540-93864-4_5. [DOI] [PubMed] [Google Scholar]
- 39.Chaffin KE, Perlmutter RM. A pertussis toxin-sensitive process controls thymocyte emigration. Eur J Immunol. 1991;21:2565–2573. doi: 10.1002/eji.1830211038. [DOI] [PubMed] [Google Scholar]
- 40.Carbonetti NH, Artamonova GV, Andreasen C, Bushar N. Pertussis toxin and adenylate cyclase toxin provide a one-two punch for establishment of Bordetella pertussis infection of the respiratory tract. Infect Immun. 2005;73:2698–2703. doi: 10.1128/IAI.73.5.2698-2703.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudolph U, Finegold MJ, Rich SS, Harriman GR, Srinivasan Y, Brabet P, Boulay G, Bradley A, Birnbaumer L. Ulcerative colitis and adenocarcinoma of the colon in G alpha i2-deficient mice. Nat Genet. 1995;10:143–150. doi: 10.1038/ng0695-143. [DOI] [PubMed] [Google Scholar]
- 42.Wiege K, Ali SR, Gewecke B, Novakovic A, Konrad FM, Pexa K, Beer-Hammer S, Reutershan J, Piekorz RP, Schmidt RE, Nurnberg B, Gessner JE. Galphai2 is the essential Galphai protein in immune complex-induced lung disease. J Immunol. 2013;190:324–333. doi: 10.4049/jimmunol.1201398. [DOI] [PubMed] [Google Scholar]
- 43.Dalwadi H, Wei B, Schrage M, Spicher K, Su TT, Birnbaumer L, Rawlings DJ, Braun J. B cell developmental requirement for the G alpha i2 gene. J Immunol. 2003;170:1707–1715. doi: 10.4049/jimmunol.170.4.1707. [DOI] [PubMed] [Google Scholar]
- 44.Han SB, Moratz C, Huang NN, Kelsall B, Cho H, Shi CS, Schwartz O, Kehrl JH. Rgs1 and Gnai2 regulate the entrance of B lymphocytes into lymph nodes and B cell motility within lymph node follicles. Immunity. 2005;22:343–354. doi: 10.1016/j.immuni.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 45.Ohman L, Franzen L, Rudolph U, Birnbaumer L, Hornquist EH. Regression of Peyer’s patches in G alpha i2 deficient mice prior to colitis is associated with reduced expression of Bcl-2 and increased apoptosis. Gut. 2002;51:392–397. doi: 10.1136/gut.51.3.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin Y, Wu MX. Requirement of Galphai in thymic homing and early T cell development. Mol Immunol. 2008;45:3401–3410. doi: 10.1016/j.molimm.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Finegold MJ, Jin Y, Wu MX. Accelerated transition from the double-positive to single-positive thymocytes in G alpha i2-deficient mice. Int Immunol. 2005;17:233–243. doi: 10.1093/intimm/dxh204. [DOI] [PubMed] [Google Scholar]
- 48.Elgbratt K, Jansson A, Hultgren-Hornquist E. A quantitative study of the mechanisms behind thymic atrophy in Galphai2-deficient mice during colitis development. PLoS One. 2012;7:e36726. doi: 10.1371/journal.pone.0036726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hwang IY, Park C, Kehrl JH. Impaired trafficking of Gnai2+/− and Gnai2−/− T lymphocytes: implications for T cell movement within lymph nodes. J Immunol. 2007;179:439–448. doi: 10.4049/jimmunol.179.1.439. [DOI] [PubMed] [Google Scholar]
- 50.Zarbock A, Deem TL, Burcin TL, Ley K. Galphai2 is required for chemokine-induced neutrophil arrest. Blood. 2007;110:3773–3779. doi: 10.1182/blood-2007-06-094565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lammermann T, Afonso PV, Angermann BR, Wang JM, Kastenmuller W, Parent CA, Germain RN. Neutrophil swarms require LTB4 and integrins at sites of cell death in vivo. Nature. 2013;498:371–375. doi: 10.1038/nature12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiege K, Le DD, Syed SN, Ali SR, Novakovic A, Beer-Hammer S, Piekorz RP, Schmidt RE, Nurnberg B, Gessner JE. Defective macrophage migration in Galphai2- but not Galphai3-deficient mice. J Immunol. 2012;189:980–987. doi: 10.4049/jimmunol.1200891. [DOI] [PubMed] [Google Scholar]
- 53.Li X, Wang D, Chen Z, Lu E, Wang Z, Duan J, Tian W, Wang Y, You L, Zou Y, Cheng Y, Zhu Q, Wan X, Xi T, Birnbaumer L, Yang Y. Galphai1 and Galphai3 regulate macrophage polarization by forming a complex containing CD14 and Gab1. Proc Natl Acad Sci U S A. 2015;112:4731–4736. doi: 10.1073/pnas.1503779112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hwang IY, Park C, Harrision KA, Huang NN, Kehrl JH. Variations in Gnai2 and Rgs1 expression affect chemokine receptor signaling and the organization of secondary lymphoid organs. Genes Immun. 2010;11:384–396. doi: 10.1038/gene.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hwang IY, Park C, Luong T, Harrison KA, Birnbaumer L, Kehrl JH. The loss of Gnai2 and Gnai3 in B cells eliminates B lymphocyte compartments and leads to a hyper-IgM like syndrome. PLoS One. 2013;8:e72596. doi: 10.1371/journal.pone.0072596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson BD, Jin Y, Wu KH, Colvin RA, Luster AD, Birnbaumer L, Wu MX. Inhibition of G alpha i2 activation by G alpha i3 in CXCR3-mediated signaling. J Biol Chem. 2007;282:9547–9555. doi: 10.1074/jbc.M610931200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gohla A, Klement K, Piekorz RP, Pexa K, vom Dahl S, Spicher K, Dreval V, Haussinger D, Birnbaumer L, Nurnberg B. An obligatory requirement for the heterotrimeric G protein Gi3 in the antiautophagic action of insulin in the liver. Proc Natl Acad Sci U S A. 2007;104:3003–3008. doi: 10.1073/pnas.0611434104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.David NB, Martin CA, Segalen M, Rosenfeld F, Schweisguth F, Bellaiche Y. Drosophila Ric-8 regulates Galphai cortical localization to promote Galphai-dependent planar orientation of the mitotic spindle during asymmetric cell division. Nat Cell Biol. 2005;7:1083–1090. doi: 10.1038/ncb1319. [DOI] [PubMed] [Google Scholar]
- 59.Miller KG, Emerson MD, McManus JR, Rand JB. RIC-8 (Synembryn): a novel conserved protein that is required for G(q)alpha signaling in the C. elegans nervous system. Neuron. 2000;27:289–299. doi: 10.1016/s0896-6273(00)00037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tall GG, Gilman AG. Resistance to inhibitors of cholinesterase 8A catalyzes release of Galphai-GTP and nuclear mitotic apparatus protein (NuMA) from NuMA/LGN/Galphai-GDP complexes. Proc Natl Acad Sci U S A. 2005;102:16584–16589. doi: 10.1073/pnas.0508306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woodard GE, Huang NN, Cho H, Miki T, Tall GG, Kehrl JH. Ric-8A and Gi alpha recruit LGN, NuMA, and dynein to the cell cortex to help orient the mitotic spindle. Mol Cell Biol. 2010;30:3519–3530. doi: 10.1128/MCB.00394-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tonissoo T, Lulla S, Meier R, Saare M, Ruisu K, Pooga M, Karis A. Nucleotide exchange factor RIC-8 is indispensable in mammalian early development. Dev Dyn. 2010;239:3404–3415. doi: 10.1002/dvdy.22480. [DOI] [PubMed] [Google Scholar]
- 63.Gabay M, Pinter ME, Wright FA, Chan P, Murphy AJ, Valenzuela DM, Yancopoulos GD, Tall GG. Ric-8 proteins are molecular chaperones that direct nascent G protein alpha subunit membrane association. Sci Signal. 2011;4:ra79. doi: 10.1126/scisignal.2002223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chan P, Thomas CJ, Sprang SR, Tall GG. Molecular chaperoning function of Ric-8 is to fold nascent heterotrimeric G protein alpha subunits. Proc Natl Acad Sci U S A. 2013;110:3794–3799. doi: 10.1073/pnas.1220943110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boularan C, Hwang IY, Kamenyeva O, Park C, Harrison K, Huang Z, Kehrl JH. B Lymphocyte-Specific Loss of Ric-8A Results in a Galpha Protein Deficit and Severe Humoral Immunodeficiency. J Immunol. 2015;195:2090–2102. doi: 10.4049/jimmunol.1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takesono A, Cismowski MJ, Ribas C, Bernard M, Chung P, Hazard S, 3rd, Duzic E, Lanier SM. Receptor-independent activators of heterotrimeric G-protein signaling pathways. J Biol Chem. 1999;274:33202–33205. doi: 10.1074/jbc.274.47.33202. [DOI] [PubMed] [Google Scholar]
- 67.Cismowski MJ. Non-receptor activators of heterotrimeric G-protein signaling (AGS proteins) Semin Cell Dev Biol. 2006;17:334–344. doi: 10.1016/j.semcdb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 68.Billard MJ, Gall BJ, Richards KL, Siderovski DP, Tarrant TK. G protein signaling modulator-3: a leukocyte regulator of inflammation in health and disease. Am J Clin Exp Immunol. 2014;3:97–106. [PMC free article] [PubMed] [Google Scholar]
- 69.Walton JC, Schilling K, Nelson RJ, Oberdick J. Sex-dependent behavioral functions of the Purkinje cell-specific Galphai/o binding protein, Pcp2(L7) Cerebellum. 2012;11:982–1001. doi: 10.1007/s12311-012-0368-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Willard FS, Low AB, McCudden CR, Siderovski DP. Differential G-alpha interaction capacities of the GoLoco motifs in Rap GTPase activating proteins. Cell Signal. 2007;19:428–438. doi: 10.1016/j.cellsig.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 71.Thomas CJ, Tall GG, Adhikari A, Sprang SR. Ric-8A catalyzes guanine nucleotide exchange on G alphai1 bound to the GPR/GoLoco exchange inhibitor AGS3. J Biol Chem. 2008;283:23150–23160. doi: 10.1074/jbc.M802422200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Robichaux WG, 3rd, Oner SS, Lanier SM, Blumer JB. Direct Coupling of a Seven-Transmembrane-Span Receptor to a Galphai G-Protein Regulatory Motif Complex. Mol Pharmacol. 2015;88:231–237. doi: 10.1124/mol.115.097741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vural A, Al-Khodor S, Cheung GY, Shi CS, Srinivasan L, McQuiston TJ, Hwang IY, Yeh AJ, Blumer JB, Briken V, Williamson PR, Otto M, Fraser ID, Kehrl JH. Activator of G-Protein Signaling 3-lnduced Lysosomal Biogenesis Limits Macrophage Intracellular Bacterial Infection. J Immunol. 2016;196:846–856. doi: 10.4049/jimmunol.1501595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Branham-O’Connor M, Robichaux WG, 3rd, Zhang XK, Cho H, Kehrl JH, Lanier SM, Blumer JB. Defective chemokine signal integration in leukocytes lacking activator of G protein signaling 3 (AGS3) J Biol Chem. 2014;289:10738–10747. doi: 10.1074/jbc.M113.515031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kamakura S, Nomura M, Hayase J, Iwakiri Y, Nishikimi A, Takayanagi R, Fukui Y, Sumimoto H. The cell polarity protein mlnsc regulates neutrophil chemotaxis via a noncanonical G protein signaling pathway. Dev Cell. 2013;26:292–302. doi: 10.1016/j.devcel.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 76.Singh V, Raghuwanshi SK, Smith N, Rivers EJ, Richardson RM. G Protein-coupled receptor kinase-6 interacts with activator of G protein signaling-3 to regulate CXCR2-mediated cellular functions. J Immunol. 2014;192:2186–2194. doi: 10.4049/jimmunol.1301875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Du Q, Macara IG. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell. 2004;119:503–516. doi: 10.1016/j.cell.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 78.Yuzawa S, Kamakura S, Iwakiri Y, Hayase J, Sumimoto H. Structural basis for interaction between the conserved cell polarity proteins Inscuteable and Leu-Gly-Asn repeat-enriched protein (LGN) Proc Natl Acad Sci U S A. 2011;108:19210–19215. doi: 10.1073/pnas.1110951108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cho H, Kehrl JH. Regulation of immune function by G protein-coupled receptors, trimeric G proteins, and RGS proteins. Prog Mol Biol Transl Sci. 2009;86:249–298. doi: 10.1016/S1877-1173(09)86009-2. [DOI] [PubMed] [Google Scholar]
- 80.Lambert NA, Johnston CA, Cappell SD, Kuravi S, Kimple AJ, Willard FS, Siderovski DP. Regulators of G-protein signaling accelerate GPCR signaling kinetics and govern sensitivity solely by accelerating GTPase activity. Proc Natl Acad Sci U S A. 2010;107:7066–7071. doi: 10.1073/pnas.0912934107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhong H, Wade SM, Woolf PJ, Linderman JJ, Traynor JR, Neubig RR. A spatial focusing model for G protein signals. Regulator of G protein signaling (RGS) protien-mediated kinetic scaffolding. J Biol Chem. 2003;278:7278–7284. doi: 10.1074/jbc.M208819200. [DOI] [PubMed] [Google Scholar]
- 82.Xie Z, Chan EC, Druey KM. R4 Regulator of G Protein Signaling (RGS) Proteins in Inflammation and Immunity. AAPS J. 2015 doi: 10.1208/s12248-015-9847-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Suurvali J, Robert J, Boudinot P, Ruutel Boudinot S. R4 regulators of G protein signaling (RGS) identify an ancient MHC-linked synteny group. Immunogenetics. 2013;65:145–156. doi: 10.1007/s00251-012-0661-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Druey KM, Blumer KJ, Kang VH, Kehrl JH. Inhibition of G-protein-mediated MAP kinase activation by a new mammalian gene family. Nature. 1996;379:742–746. doi: 10.1038/379742a0. [DOI] [PubMed] [Google Scholar]
- 85.Koelle MR, Horvitz HR. EGL-10 regulates G protein signaling in the C. elegans nervous system and shares a conserved domain with many mammalian proteins. Cell. 1996;84:115–125. doi: 10.1016/s0092-8674(00)80998-8. [DOI] [PubMed] [Google Scholar]
- 86.Guo H, Fortune MD, Burren OS, Schofield E, Todd JA, Wallace C. Integration of disease association and eQTL data using a Bayesian colocalisation approach highlights six candidate causal genes in immune-mediated diseases. Hum Mol Genet. 2015;24:3305–3313. doi: 10.1093/hmg/ddv077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gibbons DL, Abeler-Dorner L, Raine T, Hwang IY, Jandke A, Wencker M, Deban L, Rudd CE, Irving PM, Kehrl JH, Hayday AC. Cutting Edge: Regulator of G protein signaling-1 selectively regulates gut T cell trafficking and colitic potential. J Immunol. 2011;187:2067–2071. doi: 10.4049/jimmunol.1100833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tran T, Paz P, Velichko S, Cifrese J, Belur P, Yamaguchi KD, Ku K, Mirshahpanah P, Reder AT, Croze E. Interferonbeta-1b Induces the Expression of RGS1 a Negative Regulator of G-Protein Signaling. Int J Cell Biol. 2010;2010:529376. doi: 10.1155/2010/529376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moratz C, Hayman JR, Gu H, Kehrl JH. Abnormal B-cell responses to chemokines, disturbed plasma cell localization, and distorted immune tissue architecture in Rgs1−/− mice. Mol Cell Biol. 2004;24:5767–5775. doi: 10.1128/MCB.24.13.5767-5775.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Patel J, McNeill E, Douglas G, Hale AB, de Bono J, Lee R, Iqbal AJ, Regan-Komito D, Stylianou E, Greaves DR, Channon KM. RGS1 regulates myeloid cell accumulation in atherosclerosis and aortic aneurysm rupture through altered chemokine signalling. Nat Commun. 2015;6:6614. doi: 10.1038/ncomms7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gerber KJ, Squires KE, Hepler JR. Roles for Regulator of G Protein Signaling Proteins in Synaptic Signaling and Plasticity. Mol Pharmacol. 2016;89:273–286. doi: 10.1124/mol.115.102210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Patane S. Regulator of G-protein signaling 2 (RGS2) in cardiology and oncology. Int J Cardiol. 2015;179:63–65. doi: 10.1016/j.ijcard.2014.10.088. [DOI] [PubMed] [Google Scholar]
- 93.Oliveira-Dos-Santos AJ, Matsumoto G, Snow BE, Bai D, Houston FP, Whishaw IQ, Mariathasan S, Sasaki T, Wakeham A, Ohashi PS, Roder JC, Barnes CA, Siderovski DP, Penninger JM. Regulation of T cell activation, anxiety, and male aggression by RGS2. Proc Natl Acad Sci U S A. 2000;97:12272–12277. doi: 10.1073/pnas.220414397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Salim S, Sinnarajah S, Kehrl JH, Dessauer CW. Identification of RGS2 and type V adenylyl cyclase interaction sites. J Biol Chem. 2003;278:15842–15849. doi: 10.1074/jbc.M210663200. [DOI] [PubMed] [Google Scholar]
- 95.Kehrl JH, Sinnarajah S. RGS2: a multifunctional regulator of G-protein signaling. Int J Biochem Cell Biol. 2002;34:432–438. doi: 10.1016/s1357-2725(01)00141-8. [DOI] [PubMed] [Google Scholar]
- 96.Sinnarajah S, Dessauer CW, Srikumar D, Chen J, Yuen J, Yilma S, Dennis JC, Morrison EE, Vodyanoy V, Kehrl JH. RGS2 regulates signal transduction in olfactory neurons by attenuating activation of adenylyl cyclase III. Nature. 2001;409:1051–1055. doi: 10.1038/35059104. [DOI] [PubMed] [Google Scholar]
- 97.Lu Q, Sun EE, Klein RS, Flanagan JG. Ephrin-B reverse signaling is mediated by a novel PDZ-RGS protein and selectively inhibits G protein-coupled chemoattraction. Cell. 2001;105:69–79. doi: 10.1016/s0092-8674(01)00297-5. [DOI] [PubMed] [Google Scholar]
- 98.Kehrl JH, Srikumar D, Harrison K, Wilson GL, Shi CS. Additional 5′ exons in the RGS3 locus generate multiple mRNA transcripts, one of which accounts for the origin of human PDZ-RGS3. Genomics. 2002;79:860–868. doi: 10.1006/geno.2002.6773. [DOI] [PubMed] [Google Scholar]
- 99.Qiu R, Wang J, Tsark W, Lu Q. Essential role of PDZ-RGS3 in the maintenance of neural progenitor cells. Stem Cells. 2010;28:1602–1610. doi: 10.1002/stem.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Williams JW, Yau D, Sethakorn N, Kach J, Reed EB, Moore TV, Cannon J, Jin X, Xing H, Muslin AJ, Sperling AI, Dulin NO. RGS3 controls T lymphocyte migration in a model of Th2-mediated airway inflammation. Am J Physiol Lung Cell Mol Physiol. 2013;305:L693–701. doi: 10.1152/ajplung.00214.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee JK, Tansey MG. Physiology of RGS10 in Neurons and Immune Cells. Prog Mol Biol Transl Sci. 2015;133:153–167. doi: 10.1016/bs.pmbts.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 102.Tu Y, Popov S, Slaughter C, Ross EM. Palmitoylation of a conserved cysteine in the regulator of G protein signaling (RGS) domain modulates the GTPase-activating activity of RGS4 and RGS10. J Biol Chem. 1999;274:38260–38267. doi: 10.1074/jbc.274.53.38260. [DOI] [PubMed] [Google Scholar]
- 103.Lee JK, McCoy MK, Harms AS, Ruhn KA, Gold SJ, Tansey MG. Regulator of G-protein signaling 10 promotes dopaminergic neuron survival via regulation of the microglial inflammatory response. J Neurosci. 2008;28:8517–8528. doi: 10.1523/JNEUROSCI.1806-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee JK, Chung J, Kannarkat GT, Tansey MG. Critical role of regulator G-protein signaling 10 (RGS10) in modulating macrophage M1/M2 activation. PLoS One. 2013;8:e81785. doi: 10.1371/journal.pone.0081785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Garcia-Bernal D, Dios-Esponera A, Sotillo-Mallo E, Garcia-Verdugo R, Arellano-Sanchez N, Teixido J. RGS10 restricts upregulation by chemokines of T cell adhesion mediated by alpha4beta1 and alphalLbeta2 integrins. J Immunol. 2011;187:1264–1272. doi: 10.4049/jimmunol.1002960. [DOI] [PubMed] [Google Scholar]
- 106.Ponting CP. Raf-like Ras/Rap-binding domains in RGS12- and still-life-like signalling proteins. J Mol Med (Berl) 1999;77:695–698. doi: 10.1007/s001099900054. [DOI] [PubMed] [Google Scholar]
- 107.Snow BE, Hall RA, Krumins AM, Brothers GM, Bouchard D, Brothers CA, Chung S, Mangion J, Gilman AG, Lefkowitz RJ, Siderovski DP. GTPase activating specificity of RGS12 and binding specificity of an alternatively spliced PDZ (PSD-95/Dlg/ZO-1) domain. J Biol Chem. 1998;273:17749–17755. doi: 10.1074/jbc.273.28.17749. [DOI] [PubMed] [Google Scholar]
- 108.Snow BE, Antonio L, Suggs S, Gutstein HB, Siderovski DP. Molecular cloning and expression analysis of rat Rgs12 and Rgs14. Biochem Biophys Res Commun. 1997;233:770–777. doi: 10.1006/bbrc.1997.6537. [DOI] [PubMed] [Google Scholar]
- 109.Yang S, Li YP, Liu T, He X, Yuan X, Li C, Cao J, Kim Y. Mx1-cre mediated Rgs12 conditional knockout mice exhibit increased bone mass phenotype. Genesis. 2013;51:201–209. doi: 10.1002/dvg.22373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yuan X, Cao J, Liu T, Li YP, Scannapieco F, He X, Oursler MJ, Zhang X, Vacher J, Li C, Olson D, Yang S. Regulators of G protein signaling 12 promotes osteoclastogenesis in bone remodeling and pathological bone loss. Cell Death Differ. 2015;22:2046–2057. doi: 10.1038/cdd.2015.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huang J, Nalli AD, Mahavadi S, Kumar DP, Murthy KS. Inhibition of Galphai activity by Gbetagamma is mediated by PI 3-kinase-gamma- and cSrc-dependent tyrosine phosphorylation of Galphai and recruitment of RGS12. Am J Physiol Gastrointest Liver Physiol. 2014;306:G802–810. doi: 10.1152/ajpgi.00440.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shi GX, Harrison K, Wilson GL, Moratz C, Kehrl JH. RGS13 regulates germinal center B lymphocytes responsiveness to CXC chemokine ligand (CXCL)12 and CXCL13. J Immunol. 2002;169:2507–2515. doi: 10.4049/jimmunol.169.5.2507. [DOI] [PubMed] [Google Scholar]
- 113.Johnson EN, Druey KM. Functional characterization of the G protein regulator RGS13. J Biol Chem. 2002;277:16768–16774. doi: 10.1074/jbc.M200751200. [DOI] [PubMed] [Google Scholar]
- 114.Bansal G, Xie Z, Rao S, Nocka KH, Druey KM. Suppression of immunoglobulin E-mediated allergic responses by regulator of G protein signaling 13. Nat Immunol. 2008;9:73–80. doi: 10.1038/ni1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xie Z, Geiger TR, Johnson EN, Nyborg JK, Druey KM. RGS13 acts as a nuclear repressor of CREB. Mol Cell. 2008;31:660–670. doi: 10.1016/j.molcel.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hwang IY, Hwang KS, Park C, Harrison KA, Kehrl JH. Rgs13 constrains early B cell responses and limits germinal center sizes. PLoS One. 2013;8:e60139. doi: 10.1371/journal.pone.0060139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang JH, New JS, Xie S, Yang P, Wu Q, Li J, Luo B, Ding Y, Druey KM, Hsu HC, Mountz JD. Extension of the germinal center stage of B cell development promotes autoantibodies in BXD2 mice. Arthritis Rheum. 2013;65:2703–2712. doi: 10.1002/art.38059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Traver S, Bidot C, Spassky N, Baltauss T, De Tand MF, Thomas JL, Zalc B, Janoueix-Lerosey I, Gunzburg JD. RGS14 is a novel Rap effector that preferentially regulates the GTPase activity of galphao. Biochem J. 2000;350(Pt 1):19–29. [PMC free article] [PubMed] [Google Scholar]
- 119.Cho H, Kozasa T, Takekoshi K, De Gunzburg J, Kehrl JH. RGS14, a GTPase-activating protein for Gialpha, attenuates Gialpha- and G13alpha-mediated signaling pathways. Mol Pharmacol. 2000;58:569–576. doi: 10.1124/mol.58.3.569. [DOI] [PubMed] [Google Scholar]
- 120.Hepler JR, Cladman W, Ramineni S, Hollinger S, Chidiac P. Novel activity of RGS14 on Goalpha and Gialpha nucleotide binding and hydrolysis distinct from its RGS domain and GDI activity. Biochemistry. 2005;44:5495–5502. doi: 10.1021/bi048359d. [DOI] [PubMed] [Google Scholar]
- 121.Zhao P, Nunn C, Ramineni S, Hepler JR, Chidiac P. The Ras-binding domain region of RGS14 regulates its functional interactions with heterotrimeric G proteins. J Cell Biochem. 2013;114:1414–1423. doi: 10.1002/jcb.24483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Snow BE, Antonio L, Suggs S, Siderovski DP. Cloning of a retinally abundant regulator of G-protein signaling (RGS-r/RGS16): genomic structure and chromosomal localization of the human gene. Gene. 1998;206:247–253. doi: 10.1016/s0378-1119(97)00593-3. [DOI] [PubMed] [Google Scholar]
- 123.Mieda M, Ono D, Hasegawa E, Okamoto H, Honma K, Honma S, Sakurai T. Cellular clocks in AVP neurons of the SCN are critical for interneuronal coupling regulating circadian behavior rhythm. Neuron. 2015;85:1103–1116. doi: 10.1016/j.neuron.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 124.Lippert E, Yowe DL, Gonzalo JA, Justice JP, Webster JM, Fedyk ER, Hodge M, Miller C, Gutierrez-Ramos JC, Borrego F, Keane-Myers A, Druey KM. Role of regulator of G protein signaling 16 in inflammation-induced T lymphocyte migration and activation. J Immunol. 2003;171:1542–1555. doi: 10.4049/jimmunol.171.3.1542. [DOI] [PubMed] [Google Scholar]
- 125.Shankar SP, Wilson MS, DiVietro JA, Mentink-Kane MM, Xie Z, Wynn TA, Druey KM. RGS16 attenuates pulmonary Th2/Th17 inflammatory responses. J Immunol. 2012;188:6347–6356. doi: 10.4049/jimmunol.1103781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Suurvali J, Pahtma M, Saar R, Paalme V, Nutt A, Tiivel T, Saaremae M, Fitting C, Cavaillon JM, Ruutel Boudinot S. RGS16 restricts the pro-inflammatory response of monocytes. Scand J Immunol. 2015;81:23–30. doi: 10.1111/sji.12250. [DOI] [PubMed] [Google Scholar]
- 127.Gagnon AW, Murray DL, Leadley RJ. Cloning and characterization of a novel regulator of G protein signalling in human platelets. Cell Signal. 2002;14:595–606. doi: 10.1016/s0898-6568(02)00012-8. [DOI] [PubMed] [Google Scholar]
- 128.Yowe D, Weich N, Prabhudas M, Poisson L, Errada P, Kapeller R, Yu K, Faron L, Shen M, Cleary J, Wilkie TM, Gutierrez-Ramos C, Hodge MR. RGS18 is a myeloerythroid lineage-specific regulator of G-protein-signalling molecule highly expressed in megakaryocytes. Biochem J. 2001;359:109–118. doi: 10.1042/0264-6021:3590109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Park IK, Klug CA, Li K, Jerabek L, Li L, Nanamori M, Neubig RR, Hood L, Weissman IL, Clarke MF. Molecular cloning and characterization of a novel regulator of G-protein signaling from mouse hematopoietic stem cells. J Biol Chem. 2001;276:915–923. doi: 10.1074/jbc.M005947200. [DOI] [PubMed] [Google Scholar]
- 130.Nagata Y, Oda M, Nakata H, Shozaki Y, Kozasa T, Todokoro K. A novel regulator of G-protein signaling bearing GAP activity for Galphai and Galphaq in megakaryocytes. Blood. 2001;97:3051–3060. doi: 10.1182/blood.v97.10.3051. [DOI] [PubMed] [Google Scholar]
- 131.Iwai K, Koike M, Ohshima S, Miyatake K, Uchiyama Y, Saeki Y, Ishii M. RGS18 acts as a negative regulator of osteoclastogenesis by modulating the acid-sensing OGR1/NFAT signaling pathway. J Bone Miner Res. 2007;22:1612–1620. doi: 10.1359/jbmr.070612. [DOI] [PubMed] [Google Scholar]
- 132.Louwette S, Van Geet C, Freson K. Regulators of G protein signaling: role in hematopoiesis, megakaryopoiesis and platelet function. J Thromb Haemost. 2012;10:2215–2222. doi: 10.1111/j.1538-7836.2012.04903.x. [DOI] [PubMed] [Google Scholar]
- 133.Louwette S, Labarque V, Wittevrongel C, Thys C, Metz J, Gijsbers R, Debyser Z, Arnout J, Van Geet C, Freson K. Regulator of G-protein signaling 18 controls megakaryopoiesis and the cilia-mediated vertebrate mechanosensory system. FASEB J. 2012;26:2125–2136. doi: 10.1096/fj.11-198739. [DOI] [PubMed] [Google Scholar]
- 134.Gegenbauer K, Elia G, Blanco-Fernandez A, Smolenski A. Regulator of G-protein signaling 18 integrates activating and inhibitory signaling in platelets. Blood. 2012;119:3799–3807. doi: 10.1182/blood-2011-11-390369. [DOI] [PubMed] [Google Scholar]
- 135.Ma P, Ou K, Sinnamon AJ, Jiang H, Siderovski DP, Brass LF. Modulating platelet reactivity through control of RGS18 availability. Blood. 2015;126:2611–2620. doi: 10.1182/blood-2015-04-640037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Alshbool FZ, Karim ZA, Vemana HP, Conlon C, Lin OA, Khasawneh FT. The regulator of G-protein signaling 18 regulates platelet aggregation, hemostasis and thrombosis. Biochem Biophys Res Commun. 2015;462:378–382. doi: 10.1016/j.bbrc.2015.04.143. [DOI] [PubMed] [Google Scholar]
- 137.Nunn C, Mao H, Chidiac P, Albert PR. RGS17/RGSZ2 and the RZ/A family of regulators of G-protein signaling. Semin Cell Dev Biol. 2006;17:390–399. doi: 10.1016/j.semcdb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 138.Wang Y, Tong Y, Tso PH, Wong YH. Regulator of G protein signaling 19 suppresses Ras-induced neoplastic transformation and tumorigenesis. Cancer Lett. 2013;339:33–41. doi: 10.1016/j.canlet.2013.07.025. [DOI] [PubMed] [Google Scholar]
- 139.Tso PH, Wang Y, Yung LY, Tong Y, Lee MM, Wong YH. RGS19 inhibits Ras signaling through Nm23H1/2-mediated phosphorylation of the kinase suppressor of Ras. Cell Signal. 2013;25:1064–1074. doi: 10.1016/j.cellsig.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 140.Sangphech N, Osborne BA, Palaga T. Notch signaling regulates the phosphorylation of Akt and survival of lipopolysaccharide-activated macrophages via regulator of G protein signaling 19 (RGS19) Immunobiology. 2014;219:653–660. doi: 10.1016/j.imbio.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.De Vries L, Mousli M, Wurmser A, Farquhar MG. GAIP, a protein that specifically interacts with the trimeric G protein G alpha i3, is a member of a protein family with a highly conserved core domain. Proc Natl Acad Sci USA. 1995;92:11916–11920. doi: 10.1073/pnas.92.25.11916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.De Vries L, Elenko E, Hubler L, Jones TL, Farquhar MG. GAIP is membrane-anchored by palmitoylation and interacts with the activated (GTP-bound) form of G alpha i subunits. Proc Natl Acad Sci U S A. 1996;93:15203–15208. doi: 10.1073/pnas.93.26.15203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lin C, Koval A, Tishchenko S, Gabdulkhakov A, Tin U, Solis GP, Katanaev VL. Double suppression of the Galpha protein activity by RGS proteins. Mol Cell. 2014;53:663–671. doi: 10.1016/j.molcel.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 144.Sohn WJ, Ji YR, Kim HS, Gwon GJ, Chae YM, An CH, Park HD, Jung HS, Ryoo ZY, Lee S, Kim JY. Rgs19 regulates mouse palatal fusion by modulating cell proliferation and apoptosis in the MEE. Mech Dev. 2012;129:244–254. doi: 10.1016/j.mod.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 145.Lan KL, Sarvazyan NA, Taussig R, Mackenzie RG, DiBello PR, Dohlman HG, Neubig RR. A point mutation in Galphao and Galphai1 blocks interaction with regulator of G protein signaling proteins. J Biol Chem. 1998;273:12794–12797. doi: 10.1074/jbc.273.21.12794. [DOI] [PubMed] [Google Scholar]
- 146.Fu Y, Zhong H, Nanamori M, Mortensen RM, Huang X, Lan K, Neubig RR. RGS-insensitive G-protein mutations to study the role of endogenous RGS proteins. Methods Enzymol. 2004;389:229–243. doi: 10.1016/S0076-6879(04)89014-1. [DOI] [PubMed] [Google Scholar]
- 147.Huang X, Fu Y, Charbeneau RA, Saunders TL, Taylor DK, Hankenson KD, Russell MW, D’Alecy LG, Neubig RR. Pleiotropic phenotype of a genomic knock-in of an RGS-insensitive G184S Gnai2 allele. Mol Cell Biol. 2006;26:6870–6879. doi: 10.1128/MCB.00314-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Neubig RR. RGS-insensitive G Proteins as In Vivo Probes of RGS Function. Prog Mol Biol Transl Sci. 2015;133:13–30. doi: 10.1016/bs.pmbts.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 149.Signarvic RS, Cierniewska A, Stalker TJ, Fong KP, Chatterjee MS, Hess PR, Ma P, Diamond SL, Neubig RR, Brass LF. RGS/Gi2alpha interactions modulate platelet accumulation and thrombus formation at sites of vascular injury. Blood. 2010;116:6092–6100. doi: 10.1182/blood-2010-05-283846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cho H, Kamenyeva O, Yung S, Gao JL, Hwang IY, Park C, Murphy PM, Neubig RR, Kehrl JH. The loss of RGS protein-Galpha(i2) interactions results in markedly impaired mouse neutrophil trafficking to inflammatory sites. Mol Cell Biol. 2012;32:4561–4571. doi: 10.1128/MCB.00651-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hwang IY, Park C, Harrison K, Boularan C, Gales C, Kehrl JH. An essential role for RGS protein/Galphai2 interactions in B lymphocyte-directed cell migration and trafficking. J Immunol. 2015;194:2128–2139. doi: 10.4049/jimmunol.1401952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cyster JG, Schwab SR. Sphingosine-1-phosphate and lymphocyte egress from lymphoid organs. Annu Rev Immunol. 2012;30:69–94. doi: 10.1146/annurev-immunol-020711-075011. [DOI] [PubMed] [Google Scholar]
- 153.Li P, Neubig RR, Zingarelli B, Borg K, Halushka PV, Cook JA, Fan H. Toll-like receptor-induced inflammatory cytokines are suppressed by gain of function or overexpression of Galpha(i2) protein. Inflammation. 2012;35:1611–1617. doi: 10.1007/s10753-012-9476-z. [DOI] [PMC free article] [PubMed] [Google Scholar]