Abstract
Information from various public and private data sources of extremely large sample sizes are now increasingly available for research purposes. Statistical methods are needed for utilizing information from such big data sources while analyzing data from individual studies that may collect more detailed information required for addressing specific hypotheses of interest. In this article, we consider the problem of building regression models based on individual-level data from an “internal” study while utilizing summary-level information, such as information on parameters for reduced models, from an “external” big data source. We identify a set of very general constraints that link internal and external models. These constraints are used to develop a framework for semiparametric maximum likelihood inference that allows the distribution of covariates to be estimated using either the internal sample or an external reference sample. We develop extensions for handling complex stratified sampling designs, such as case-control sampling, for the internal study. Asymptotic theory and variance estimators are developed for each case. We use simulation studies and a real data application to assess the performance of the proposed methods in contrast to the generalized regression (GR) calibration methodology that is popular in the sample survey literature.
Keywords: Case-control study, Empirical likelihood, Generalized regression estimator, Misspecified model, Profile-likelihood
1 INTRODUCTION
Population based biomedical science is now going through a paradigm shift as extremely large data sets are becoming increasingly available for research purposes. Sources of such big data include, but are not limited to, population based census data, disease registries, health care databases and various consortia of individual studies. The power of such large data sets lies in their sample size. They, however, often do not contain detailed information at the level of individual analytic studies, which may be much smaller in size, but have been designed to answer specific hypotheses of interest. There is a growing need for a statistical framework for combining information from data sets that are large but have relatively crude information with that available from studies that are small but contain more detailed information on each subject. Methods that can work with summary level information, as opposed to individual level data, are particularly appealing due to practical reasons such as data sharing, storage and computing, as well as for ethical reasons, such as maintenance of the privacy of the study subjects and protection of the future research interests of data generating institutions and investigators.
In this article, we consider the problem of building regression models using individual level information from an analytic study while incorporating summary level information from an external large data source. Our goal is to work within a semiparametric framework that allows the distribution of all covariates to remain completely unspecified, so that analysis results are not sensitive to modeling assumptions. One class of methods that could potentially be used for this purpose is to use calibration techniques that are popular in sample-survey theory. Chen and Chen (2000), for example, studied the extension of the generalized regression (GR) method for developing regression models using data from double sampling or two-phase designs in cases where the internal study is a sub-sample of the external one. There exists a rich literature on how to form optimal calibration equations for improving efficiency of parameter estimates within various classes of unbiased estimators (Deville and Sarndal 1992; Robins, Rotnitzki and Zhao 1994; Wu and Sitter 2001; Wu 2003; Lumley, Shaw and Dai, 2011). Unlike GR, however, the application of many of these more optimal methods in the current setting requires access to individual level data from the external study.
The methodology for “model-based” maximum likelihood estimation has also been studied previously in some special cases of this problem, where it can be assumed that the covariate information available in the external data source can be summarized into discrete strata. In particular, in the setting of two-phase studies where it can be assumed that the internal study is a sub-sample of the external study, a number of researchers have proposed semiparametric maximum likelihood (SPML) methods for various types of regression models, while accounting for complex sampling designs (Scott and Wild 1997; Breslow and Holubkov 1997; Lawless, Wild and Kalbflesich 1999). Most recently, Qin et al. (2015) studied the problem of fitting a logistic regression model to case-control data utilizing information on stratum-specific disease probability rates from external sources. The assumption that only discretized information is available from the external source is a major limitation of these methods. In practice, the external data set may often include combinations of many variables and summarizing this information into strata can be subjective and inefficient.
In this article, we develop a general SPML estimation methodology, where we assume that the external information is summarized, not by a discrete set of strata defined by the study variables, but by a finite set of parameters obtained from fitting a model to the external data. We identify very general equations imposed by the external model, regard-less of whether the model is correctly specified or not, and use these equations to develop constrained maximum-likelihood estimation methodology. The broad framework allows arbitrary types of covariates and arbitrary types of regression models, including non-nested models for the internal and external data and complex sampling designs. For inference, we consider an empirical likelihood estimation technique, as well as a synthetic maximum likelihood approach that allows incorporating externally available estimates of the covariate distribution. We evaluate the performance of these maximum likelihood methods together with the GR-type calibration estimator in a wide variety of settings of practical interest. Finally, we illustrate an application of the method for developing an updated model for predicting risk of breast cancer using multiple data sources.
2 METHODS
2.1 Models and Notation
Let Y be an outcome of interest and X be a set of covariates. We assume a model for the predictive distribution gθ(y|x) has been built based on an external big data set. In general, we will assume that we only have access to the model parameters, θ, but not necessarily to the individual level data from the “external” study based on which the original model was built. We assume that data on Y, X and a new set of covariates Z are available to us from an “internal” study for building a model of the form fβ(y|x, z). Throughout, we will refer to fβ(y|x, z) and gθ(y|x) as the “full” and “reduced” models, respectively. We assume fβ(y|x, z) is correctly specified, but the external model gθ(Y|X) need not be. In practice, although all models are going to be wrong to some extent, investigators will control the specification fβ(Y|X, Z) and can carry out suitable model diagnostics. Let F(X, Z) denote the distribution function of all risk factors for the underlying population, which, for the time being is assumed to be the same for the “external” and “internal” studies. This assumption, however, will be inspected more closely later.
2.2 The Key Constraints Relating Model Parameters of Full and Reduced Models
Let U(Y|X; θ) = ∂log{gθ(Y|X)}/∂θ be the score function associated with the reduced model, and θ the parameter in the reduced model. The population parameter value θ* of θ underlying the external reduced model satisfies the equation
| (1) |
where pr(y, x) = pr(y|x)pr(x) is the true underlying joint distribution of (Y, X). When the model gθ(y|x) is misspecified, gθ(y|x) ≠ pr(y|x), but (1) still holds true under mild conditions (e.g. White 1882). Under the assumption that fβ(Y|X, Z) is correctly specified, we can write
with β0 the true value of β. Thus the constraint imposed by equation (1) can be re-written, after changing some ordering of integrals, as
| (2) |
The equation essentially converts the external information to a set of constraints, which we utilize in our analysis of internal data to improve efficiency of parameter estimates and generalizability of models. The dimension of the constraints is the same as the number of parameters by which the external model has been summarized.
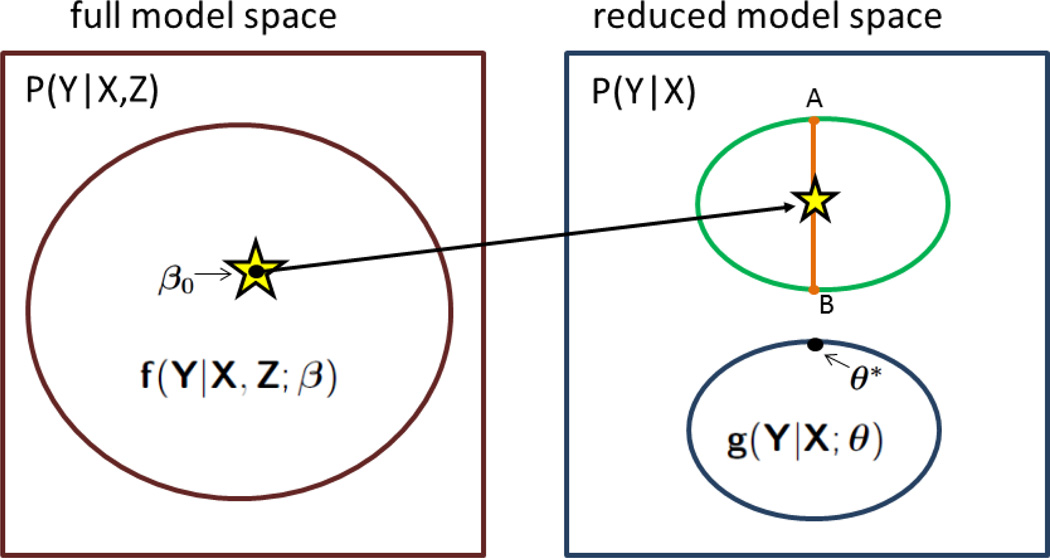
Figure 1 provides a geometric perspective for how the external reduced model provides information for building the full model based on the internal study. The true probability distribution for P0(Y|X, Z) (shown in the left panel), which is assumed to belong to the class generated by a parametric family fβ(Y|X, Z), induces a true value for P0(Y|X) = ∫ fβ0(Y|X, z)dF(z|X)dz (shown in the right panel). The reduced model space under consideration, gθ(Y|X), may not contain P0(Y|X). Nevertheless, a value of θ = θ* that solves the score equation E{U(Y|X, θ)} = 0 has a valid interpretation in that it minimizes the Kullback-Leibler distance (Huber 1967; White 1982) between the fixed P0(Y|X) and the model space of gθ(Y|X). Thus, from a reverse perspective, it is intuitive that if the value of θ* is given, then the search for β0 could be constrained to a space so that Pθ*(Y|X) remains the minimizer between the model space of gθ(Y|X) and any fixed point in the induced model space of Pβ(Y|X) = ∫ fβ(Y|X, z)dF(z|X)dz. In the figure, the constrained space for the induced model is represented by the chord .
Figure 1.
Geometric interpretation of the problem. The true distribution function P0(Y|X) for Y given (X, Z) across all values of β is depicted in the left panel, with the true value of β, i.e., β0 shown. In the right top panel, we see the true distribution function P0(Y|X) for Y given X across all values of β with β0 shown. The assumed external data model is given in the bottom right panel across all values of θ, with the solution θ* to (1) being shown as the minimizer of the Kullback-Leibler distance between the fixed P0(Y|X) and the model space of gθ(Y|X).
2.3 Semiparametric Maximum Likelihood
One class of methodology we consider is SPML methodology that allows the distribution F(X, Z) to be completely unspecified. Importantly, two-phase design maximum likelihood methodology requires X to be discrete as the number of constraints increases with the number of distinct levels of (Y, X). In contrast, here Y and X can be arbitrary in nature and yet the number of constraints, defined by the dimension of the reduced model parameter θ, remains finite.
2.3.1 Maximum-Likelihood Under Simple Random Sampling for the Internal Studies
Suppose we have data on (Yi, Xi, Zi) for i = 1, …, N randomly selected subjects in the internal study. The likelihood is given by
Our goal is to maximize log{Lβ,F} with respect to β and F(·) while maintaining the constraint given by (2). We assume that θ* is given to us externally, that is, θ is fixed at θ*. Thus, from now on, for simplicity we use θ to denote θ*.
Define
| (3) |
We propose to maximize lλ = log(Lβ,F) + λT ∫ uβ(X, Z; θ)dF(X, Z), where λ is a vector of Lagrange multipliers with the same dimension as θ. Assuming that nonparametric maximum likelihood estimation (NPMLE) of F(X, Z) has masses only at the unique observed data points in the internal study, F(·, ·) can be characterized by the corresponding masses (δ1, …, δm), where m denotes the number of unique values of (Xi, Zi), i = 1 …, N. Using standard empirical likelihood (or profile likelihood) computation steps (see e.g. Qin and Lawless 1994; Scott and Wild 1997), we can show that the values of β and λ that satisfy the constrained maximum likelihood equations also satisfy the score-equations associated with a “pseudo-loglikelihood” given by
| (4) |
2.3.2 Maximum Likelihood Under a Case-Control Sampling Design for the Internal Study
Even if we only have a case-control or retrospective sample from the internal study, as long as we have the external reduced model for the underlying population, we can estimate all of the parameters of the full model fβ(Y|X, Z) using this constrained maximum-likelihood approach. A special case is when the external information is simply the disease prevalence in the underlying population; it is well know that such information can be used to augment the case-control sample to estimate all of the parameters of a logistic or other binary disease risk models (see e.g., Scott and Wild 1992).
Suppose Y is binary and let p1 = pr(Y = 1) = 1 − p0 = ∫ fβ(Y = 1|x, z)dF(x, z) denote the underlying marginal disease probability in the population, for arbitrary values of β. The likelihood for the internal case-control study is given by
| (5) |
where N1 and N0 denote the numbers of cases and controls sampled. The goal is to maximize . Again, assuming that the NPMLE of F(X, Z) has masses only within the unique observed data points of (Xi, Zi), i = 1, …, N1 + N0, in Appendix we show that the constrained maximum likelihood estimation problem is equivalent to solving the score equation associated with the pseudo-loglikelihood
| (6) |
The pseudo-loglikelihood in the case-control sampling setting requires introduction of one additional nuisance parameter μ1 = N1/p1, with μ0 = N0/p0 defined by μ1. In principle, this theory could be generalized to account for more complex stratified sampling designs for the internal study, assuming the external model is rich enough that it allows identification of all the parameters of the full logistic model even if certain parameters are not identifiable from the internal study.
The problem stated above has a strong connection with methodology in two-phase design semiparametric maximum likelihood estimation as developed by Scott and Wild (1997) and Breslow and Holubkov (1997). In fact, if the data (Y, X) from the external study can be summarized into a frequency table defined by fixed sets of strata, this problem essentially can be studied using previous theory with some modification for the fact that the internal study may not be a subset of the external study. The categorization of phase-I covariate data (X) is needed in these and other previously developed semiparametric methods, as all of them intrinsically rely on functionals that are smoothed over Z but not X. In contrast, in our maximum likelihood theory, the constraints are represented by functionals that are smoothed with respect to both X and Z. As a result, we avoid the “curse of dimensionality” problem that is typically faced in semiparametric estimation theory for covariate missing data and measurement error problems; see Roeder, Carroll, and Lindsay (1996) for a discussion on the latter topic.
2.3.3 Numerical Computation and Asymptotic Theory
For ease of exposition, when discussing computation and asymptotic theory, we transform the parameter μ1 to α defined in Appendix S.2 of the Supplementary Material for the case-control design. Further, we absorb the parameter α into β in the case-control design, so that the notation for certain functions such as uβ(X, Z; θ) can be unified under both the simple random and case-control sampling designs.
As mentioned above and detailed in Appendix, the pseudo-loglikelihood given in (4) or (6) is in fact the Lagrange function for the constrained loglikelihood obtained by Lagrange multipliers and profiling out the infinite dimensional parameter F(·, ·). By the theory of Lagrange multipliers (Chiang and Wainwright, 1984), the proposed semiparametric constrained maximum likelihood estimator for β0, denoted by β̂, is then obtained by directly solving for the stationary point, indeed the saddle point, over the expanded parameter space η = (βT, λT)T for the pseudo-loglikelihood function. We solve the resulting stationary equation to obtain β̂ by the usual Newton-Raphson method, which is quite stable and performs efficiently in our numerical studies when the initial value of λ is set to 0. In the simulations and data analysis performed in this work, all results converged within 10 iterations of the Newton-Raphson algorithm employed. Numerical optimizations were performed using PROC IML of SAS (version 9.3). Formulae for the score and Hessian for both simple random sampling and for case-control studies are given in Appendix S.2 of the Supplementary Material.
Let η̂ = (β̂T, λ̂T)T be the stationary point of the pseudo-loglikelihood function given in (4) or (6); namely, η̂ is the solution to the score equation or when the internal study is under simple random or case-control sampling. Explicit expressions for the score function, the first derivative of the pseudo-loglikelihood with respect to η = (βT, λT)T, are given in Appendix S.2 in the Supplementary Material. The following result confirms that the constrained maximum likelihood estimator for β can be obtained by solving the score equation. The proof is detailed in Appendix S.5 of the Supplementary Material.
Lemma 1
Under regularity conditions for model fβ(y|x, z) and conditions (i)–(iv) given in Appendix S.4 of the Supplementary Material, the pseudo-loglikelihood function or is maximized at β = β̂ with probability one, and η̂ = (β̂, λ̂) is the solution to or .
The following proposition establishes the asymptotic normality of the constrained maximum likelihood estimator proposed.
Proposition 1
Let η̂ = (β̂T, λ̂T)T be the solution to or , and with β0 the true value of β, 0 denoting a ℓ-vector of zeros and ℓ the dimension of λ. Under regularity conditions for fβ(y|x, z) and conditions (i)–(v) in Appendix S.4 of the Supplementary Material, as N → ∞, N1/2(η̂ − η0) converges in distribution to a zero-mean normal distribution with covariance matrix given by
| (7) |
where B = E{iββ(Y, X, Z)}, C = E{cβ(X, Z; θ)}, and , and where iββ(Y, X, Z) and cβ(X, Z; θ) are defined in (S.4) and (S.2), respectively, in Appendix S.2 in the Supplementary Material.
The proof is in Appendix S.5 of the Supplementary Material. A simple consistent estimator for the covariance matrix (7) is obtained by using the corresponding sample means for the expected quantities in the expression. In Section S.5 of the Supplementary Material, we show how to modify Proposition 1 when there is uncertainty about θ when it is estimated from a finite external study.
From Proposition 1 we see two interesting facts. First, the asymptotic variance of β̂ is (B + CL−1CT)−1 = B−1 − B−1C(L + CTB−1C)−1CTB−1, and hence the constrained maximum likelihood estimator is asymptotically more efficient than the estimator based only on internal sample data, whose asymptotic variance is B−1. Second, β̂ and λ̂ are asymptotically uncorrelated, a phenomenon also shared by other empirical likelihood methods such as that of Qin and Lawless (1994).
2.4 Synthetic Maximum Likelihood
So far we have assumed that the underlying populations for the internal and external studies are identical, which may be violated in practice. In particular, as we will demonstrate through simulation studies, various types of calibrations methods, either maximum likelihood or not, can lead to substantial bias in parameter estimates if the distributions of the underlying risk factors are different between the internal and external populations. In this section, we consider the situation when an external reference sample may be available for unbiased estimation of the covariate distribution for the external population.
Let F†(X, Z) denote the underlying distribution for the external population, and consider the setting where it differs from the distribution F(X, Z) in the internal population. We continue to assume that the regression model fβ(Y|X, Z) correctly holds for both of the populations and that the underlying true parameters β0 are the same. We assume data are available from the external reference sample in the form , j = 1, …, Nr, where Nr is the size of the external reference sample. When the internal study sample is obtained under the simple random sampling design, the synthetic constrained loglikelihood is defined as , with F̃† the empirical distribution of (X†, Z†) in the external reference sample, and the synthetic constrained maximum likelihood (SCML) estimator (β̃, λ̃) for (β, λ) can be obtained by solving the estimating equations and , completely ignoring F(·, ·) because it factors out from the likelihood of the internal study.
When the internal sample represents a case-control study, however, the synthetic constrained loglikelihood is defined as
from which F(·, ·) cannot be factored out. As before, we consider NPMLE estimation of F(X, Z) allowing it to have masses at each of the unique observed data points of (Xi, Zi) (i = 1 …, N1 + N0) in the internal study. In this setting, following Prentice and Pyke (1979) we can show that the SCML estimate of β can be obtained by maximization of a pseudo-loglikelihood of the form
where
with
| (8) |
and α = log (μ1/μ0). Here, corresponds to the standard “prospective likelihood” for case-control data that is known to produce equivalent inference for β as the retrospective likelihood (Prentice and Pyke 1979; Scott and Wild 1997). In general, a likelihood of the form (8) may not be able to identify all of the parameters of the original model without additional information. In particular, for the logistic model, the intercept parameters become completely confounded by the nuisance parameters α and cannot be estimated from case-control data alone. However, in the setting considered here, the additional constraint (2) defined by the external model allows estimation of all of the parameters of the full model even when the distribution of the risk-factors may differ in the two underlying populations.
2.4.1 Computation and Asymptotic Theory
As in the procedure considered in Section 2.3, we use the Newton-Raphson method to solve the stationary equations for the synthetic constrained loglikelihood or , depending on whether the internal study is based on simple random or case-control sampling. Formulae for the score and Hessian for simple random sampling and for case-control sampling are given in Appendix S.3 of the Supplementary Material.
As mentioned previously, to simplify exposition, in the case-control setting we absorb the nuisance parameter α into β, and let η = (βT, λT)T. Denote by q and ℓ the dimensions of β and λ. The following lemma shows that β̃ obtained from or indeed maximizes the log-likelihood function log(Lβ,F) or with probability tending to one under the constraint ∫ uβ(X, Z; θ)dF̃†(X, Z) = 0.
Lemma 2
Suppose that q > ℓ, and Nr/N → κ > 0. Under regularity conditions for the model fβ(y|x, z) and conditions (i)–(iv) given in Appendix S.4 of the Supplementary Material, the loglikelihood functions log(Lβ,F) or is maximized at β = β̃ with probability approaching one under the constraint ∫ uβ(X, Z; θ)dF̃†(X, Z) = 0, where β̃ is in the interior of a neighborhood of true parameter value β0, and η̃ = (β̃T, λ̃T)T is the solution to or .
We also provide asymptotic distribution theory for the SCML estimator η̃ = (β̃T, λ̃T)T. The proofs of these theoretical results are given in Appendix S.5 of Supplementary Material.
Proposition 2
Recall that Nr/N → κ > 0. with 0 denoting a ℓ-vector of zeros and ℓ the dimension of λ. Under the assumptions in Lemma 2 and condition (v) in Appendix S.4 of the Supplementary Material, the estimator η̃ = (β̃T, λ̃T)T satisfying or is asymptotically normal as N → ∞, such that N1/2(η̃ − η0) converges in distribution to a zero-mean normal distribution with covariance matrix
| (9) |
where with E† denoting expectation over the external covariate distribution F†(X, Z), G = CTB−1C, B = E{iββ(Y, X, Z)}, C = E†{cβ(X, Z; θ)}, , and iββ(Y, X, Z) and cβ(X, Z; θ) defined in (S.4) and (S.2), respectively, in Appendix S.2 of the Supplementary Material.
The variance-covariance matrix (9) for η̃ can be readily estimated by replacing the component quantities in the expression with their sample analogies. In Section S.5 of the Supplementary Material, we show how to modify Proposition 2 when there is uncertainty in the parameter estimates of the external study.
From Propositions 1 and 2, we see differences between (β̂, λ̂) and (β̃, λ̃) in their asymptotic theory. First, unlike β̂, the SCML estimator β̃ is not guaranteed to be more efficient than the internal-sample only estimator. However, β̃ is usually more efficient than the latter estimator, especially when κ is large, i.e., the size of the reference sample for estimating F(·, ·) is sufficiently large relative to that of the internal sample. In particular, in the special case of κ → ∞, the matrix B−1CG−1CTB−1 is positive definite and hence β̃ is always more efficient than that of the internal-sample only estimator. Second, unlike (β̂, λ̂), (β̃ λ̃) are correlated asymptotically, although the correlation vanishes as κ → ∞.
3 SIMULATION STUDIES
We conduct simulation studies to evaluate the performance of the proposed methods in a wide variety of settings of practical interest. We consider developing models for a binary outcome Y using logistic and non-logistic link functions. In all simulations, it is assumed (X, Z) is bivariate normal with zero marginal means, unit marginal variances and a correlation of 0.3.
In other numerical studies (not shown here), the conclusions from simulation studies did not change qualitatively if we used an alternative to the bivariate normal distribution for simulating (X, Z).
We study three different settings, in two of which we assume that the full model of interest has the form
where h−1 denotes the inverse link function corresponding to a logistic or a probit model. In one of these settings, we assume that the external model is under-specified but involves both covariates with the form
and in the second setting we assume that the external model is missing the covariate Z altogether and has the form
In the third setting, we consider a measurement error problem where it is assumed that Z is the true covariate of interest and X is a surrogate of Z in the sense that Y is independent of X given Z. In this setting, we assume that the full and reduced models of interest are
respectively.
In each setting, we simulate data under the correct full model given a set of parameter values and then obtain the values of external parameters by fitting the reduced model, which by definition is incorrectly specified, to a very large data set. In the under-specified and missing covariate scenarios, the parameter values of the true model (β0, βX, βZ, βXZ) = (−1.6, 0.4, 0.4, 0.2); in the measurement error scenario, they are (β0, βZ) = (−1.6, 0.4). In all models, the parameter specifications lead to a population disease prevalence around 20%. For simulating case-control samples for the internal study, in each simulation, we first generate a random sample and then select fixed and equal numbers of cases and controls. In both the simple random and case-control sampling settings, the size of the internal sample is N = 1000. The external data are generated with a very large sample size and fixed throughout the simulations.
As a benchmark for comparison, in each simulation, in addition to the constrained maximum likelihood estimator proposed, we obtain the internal-sample-only estimate β̂I, and implement a generalized regression (GR) estimator, popular in the survey literature and developed for regression inference with double or two-phase sampling by Chen and Chen (2000). Specifically, the estimator takes the form
| (10) |
In (10), θ̃E and θ̃I are estimates for θ using the external and internal samples, respectively, while
where sβ(Y, X, Z) = ∂log fβ(Y|X, Z)/∂β, and where EI denotes the sample expectation based on the internal study. In the current implementation of such a calibration method, unlike in the original proposal, we disregard the uncertainty associated with θ̂E, since in the current setting such uncertainty is assumed to be negligible compared with the uncertainty in the internal sample. As shown in Chen and Chen (2000), the asymptotic covariance matrix of is given as
| (11) |
which accounts for the uncertainty of θ̂I, and can be estimated by replacing each component quantity with its sample analogue.
Further, since the method of Chen and Chen (2000) was originally developed for simple random sampling designs only, when implementing their method for logistic regression analysis of a case-control design, we make an ad-hoc modification to GR estimator which we denote as mGR. Instead of applying the calibration formula (10) to the full set of regression parameters β, we apply it only to the subset of β excluding the intercept parameter. Such a modification is based on the rationale that, according to Prentice and Pyke (1979), for fβ(·) following a logistic regression model, the prospective maximum likelihood estimator provides valid and efficient estimates of all the parameters of the model except the intercept.
Tables 1–3 display simulation results with N = 1000. We also examine the cases with N = 400, which lead to similar conclusions and are relegated to Tables S.1–S.4 in the Supplementary Material. From Tables 1–3 we conclude that the constrained maximum likelihood estimator β̂CML is always more efficient than the internal-sample-only estimator β̂I. For the under-specified and missing covariate scenarios, substantial efficiency gains are observed for regression parameters corresponding to the covariates that are also included in the reduced model fitted with external data, i.e., for (β0, βX, βZ) and (β0, βX), respectively. In the measurement error setting, the efficiency gain is observed for the main covariate of interest (Z) where the reduced model only includes an error-prone surrogate (X). These observations hold under both the simple random and case-control sampling designs. Under the simple random sampling design, the estimator β̂GR performs similarly to or slightly worse than our estimator β̂CML in the under-specified and missing covariate settings. However, in the measurement error setting, β̂GR is far less efficient than β̂CML. Under the case-control sampling design, the mGR estimator incurred substantial bias in the under-specified setting, but not in the other two settings. From these simulation results and those conducted in a smaller sample-size setting (N = 400), we can conclude that the asymptotic distribution theory provided in Proposition 1 for β̂CML performs quite well, as seen by the generally close agreement between the estimated (ESE) and simulation standard errors (SE), and between the nominal and simulation coverage probabilities of the Wald-type confidence intervals based on asymptotic normality. Tables S.5–S.7 in the Supplementary Material show the performance of the different estimators in the setting of probit models under the random-sampling design. The results are generally similar to those for the logistic model in Tables 1–3.
Table 1.
Simulation results for the under-specification setting, in which the full model of interest has the form h−1 {pr(D = 1)} = β0 + βXX + βZZ + βXZXZ, where h−1 denotes the inverse link function corresponding to a logistic model, and where the external model is under-specified but involves both covariates with the form h−1 {pr(D = 1)} = θ0 + θXX + θZ. Results multiplied by 103 are presented, and the coverage probabilities (CP) are reported as percents.
| Int | β0 GR/ mGR |
CML | Int | βX GR/ mGR |
CML | Int | βZ GR/ mGR |
CML | Int | βXZ GR/ mGR |
CML | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| simple random; N = 1000 | ||||||||||||
| Bias | −8.94 | −0.79 | −1.12 | 2.42 | 2.16 | 2.46 | 1.29 | 1.29 | 1.65 | 1.39 | 1.30 | 1.87 |
| SE | 91.4 | 24.4 | 24.4 | 96.8 | 20.1 | 19.9 | 94.3 | 20.2 | 19.8 | 89.4 | 89.6 | 89.3 |
| ESE | 91.8 | 22.9 | 23.5 | 92.3 | 19.8 | 19.7 | 92.4 | 19.9 | 19.8 | 85.8 | 85.3 | 86.9 |
| MSE | 8.42 | 0.59 | 0.59 | 9.38 | 0.41 | 0.40 | 8.89 | 0.41 | 0.39 | 7.98 | 8.02 | 7.97 |
| CP | 95.4 | 88.6 | 90.8 | 94.3 | 94.6 | 95.2 | 94.6 | 94.4 | 95.5 | 93.6 | 93.4 | 93.8 |
| case-control; N = 1000 | ||||||||||||
| Bias | - | - | 1.70 | 2.40 | 28.4 | 2.23 | 5.06 | 27.6 | 1.73 | −1.51 | −1.20 | −2.30 |
| SE | - | - | 17.4 | 75.7 | 11.3 | 16.7 | 72.2 | 11.4 | 16.6 | 72.9 | 72.8 | 71.7 |
| ESE | - | - | 16.6 | 73.3 | 11.6 | 16.5 | 73.1 | 11.6 | 16.5 | 71.4 | 71.3 | 70.1 |
| MSE | - | - | 0.30 | 5.73 | 0.93 | 0.28 | 5.24 | 0.89 | 0.28 | 5.31 | 5.29 | 5.15 |
| CP | - | - | 90.7 | 94.2 | 31.4 | 94.5 | 95.4 | 33.1 | 94.8 | 94.7 | 94.6 | 93.7 |
Int: internal-data only method
GR: generalized regression, mGR: modified GR for case-control sampling
CML: constrained maximum likelihood
ESE: estimated standard error
MSE: mean squared error
CP: coverage probability of a 95% confidence interval interval
Table 3.
Simulation results for the measurement error setting in which the full and reduced models are h−1 {pr(D = 1)} = β0 + βZZ and h−1 {pr(D = 1)} = θ0 + θXX, respectively, and where h−1 denotes the inverse link function corresponding to a logistic model. Results presented are multiplied by 103, and the coverage probability (CP) is in percents.
| Int | β0 GR/mGR |
CML | Int | βZ GR/mGR |
CML | |
|---|---|---|---|---|---|---|
| simple random; N = 1000 | ||||||
| Bias | −2.12 | −3.73 | 0.20 | 0.80 | 1.23 | 1.13 |
| SE | 87.7 | 25.1 | 15.1 | 89.6 | 84.7 | 40.1 |
| ESE | 87.1 | 23.9 | 15.2 | 86.3 | 82.5 | 38.7 |
| MSE | 7.69 | 0.64 | 0.23 | 8.02 | 7.17 | 1.61 |
| CP | 95.9 | 92.6 | 94.1 | 94.2 | 94.0 | 94.1 |
| case-control; N = 1000 | ||||||
| Bias | - | - | 0.99 | 2.85 | 2.91 | 1.74 |
| SE | - | - | 12.8 | 66.0 | 62.5 | 37.6 |
| ESE | - | - | 12.9 | 66.6 | 63.8 | 36.3 |
| MSE | - | - | 0.16 | 4.36 | 3.63 | 1.42 |
| CP | - | - | 95.6 | 95.7 | 96.1 | 94.6 |
Int: internal-data only method
GR: generalized regression, mGR: modified GR for case-control sampling
CML: constrained maximum likelihood
ESE: estimated standard error
MSE: mean squared error
CP: coverage probability of a 95% confidence interval interval
We next investigate the properties of the different estimators when the distribution of (X, Z) differs between the internal and external populations. All of the steps are identical as before except that we assume corr(X, Z) is 0.1 in the external population. In this scenario, we implement the SCML method assuming a reference sample for the external population is available to estimate the underlying covariate distribution. We assume that the sample size for the reference sample is the same as that of the internal study, and in each simulation the random reference sample for (X, Z) is drawn from the distribution that is the same as that for the external population. We obtain the value of the external parameter by fitting a reduced model to a very large data set simulated using the external covariate distribution. Table 4 shows the results under the missing covariate setting for logistic regression with simple random and case-control sampling.
Table 4.
Simulation results for the missing covariate setting as in the caption for Table 2, but when the covariate distributions are different between the internal and external populations. Results multiplied by 103 are presented, and the coverage probabilities (CP) are reported as percents.
| Int | β0 CML |
SCML | Int | βX CML |
SCML | Int | βZ CML |
SCML | Int | βXZ CML |
SCML | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| simple random; N = Nr = 1000 | ||||||||||||
| Bias | −8.61 | −32.4 | −1.67 | 4.33 | −85.2 | 2.63 | 1.10 | 0.30 | −0.69 | 1.38 | 4.41 | −0.04 |
| SE | 91.9 | 32.2 | 27.6 | 96.8 | 38.4 | 30.7 | 97.9 | 97.9 | 96.8 | 89.3 | 89.5 | 88.0 |
| ESE | 91.8 | 33.5 | 26.2 | 92.5 | 39.2 | 30.1 | 92.5 | 92.5 | 91.0 | 85.9 | 85.2 | 85.6 |
| MSE | 8.51 | 2.09 | 0.76 | 9.39 | 8.72 | 0.95 | 9.58 | 9.58 | 9.36 | 7,97 | 8.03 | 7.74 |
| CP | 95.3 | 90.6 | 92.0 | 93.5 | 42.3 | 93.0 | 93.2 | 93.3 | 93.4 | 93.5 | 93.6 | 93.9 |
| case-control; N = Nr = 1000 | ||||||||||||
| Bias | - | −27.0 | −1.13 | 0.76 | −89.0 | 0.58 | 2.74 | 4.15 | 1.94 | 4.03 | −1.90 | 2.30 |
| SE | - | 23.6 | 23.1 | 71.4 | 27.1 | 26.8 | 74.1 | 74.1 | 73.6 | 73.7 | 73.5 | 72.7 |
| ESE | - | 23.2 | 23.1 | 73.3 | 27.9 | 27.4 | 73.3 | 72.7 | 72.5 | 71.6 | 69.8 | 71.2 |
| MSE | - | 1.29 | 0.53 | 5.09 | 8.65 | 0.72 | 5.50 | 5.51 | 5.42 | 5.45 | 5.40 | 5.29 |
| CP | - | 82.0 | 94.2 | 95.0 | 7.9 | 93.7 | 94.3 | 94.0 | 94.1 | 93.6 | 93.5 | 94.3 |
Int: internal-data only method
CML: constrained maximum likelihood method
SCML: synthetic constrained maximum likelihood method
ESE: estimated standard error
MSE: mean squared error
CP: coverage probability of a 95% confidence interval
Since results from the regression calibration β̂GR and the constrained maximum likelihood β̂CML estimates are quite similar in this setting, results for the former are omitted in Table 4. As can be seen in Table 4, β̂, as well as β̂GR, is subject to remarkable bias for parameters corresponding to the covariates included in the reduced model. This is expected since β̂, as well as β̂GR, is derived under the assumption of complete homogeneity between the internal and external studies, and incorporation of the inconsistent external information can result in biased parameter estimates. On the other hand, we see from Table 4 that the SCML estimate β̃ performs quite well under both the simple random and case-control sampling: it is virtually unbiased, and more efficient than the estimate β̂I based on the internal sample only. The efficiency gain of β̃ over β̂I is particularly large for parameters corresponding to the covariates included in the reduced model. The standard error estimators and confidence intervals based on Proposition 2 for the synthetic constrained maximum likelihood estimation perform quite well in the settings considered.
4 DATA APPLICATION: BREAST CANCER RISK MODELING
We illustrate an application of our methodology by re-analysis of data from the Breast Cancer Detection and Demonstration Project (BCDDP) used by Chen et al (2006) for building a model for breast cancer risk prediction. The Breast Cancer Risk Assessment Tool (BCRAT), sometimes known as the Gail Model, is a widely used model for predicting the risk of breast cancer based on a handful of standard risk factors, including age at menarche (agemen), age at first live birth (ageflb), weight, number of first-degree relatives with breast cancer (numrel) and number of previous biopsies (nbiops). Chen et al (2006) developed an updated model, known as BCRAT2, to include mammographic density (MD), the areal proportion of breast tissue that is radiographically dense, that is known to be a strong risk factor for breast cancer. They used data available from 1217 cases and 1610 controls within the BCDDP study on whom data were available on these standard risk factors as well as MD. To increase efficiency, however, they used two-phase design methodology, utilizing data on standard risk factors that were available on a larger sample of subjects, including about 2808 cases and 3119 controls within the BCDDP study. Details of the BCDDP study design, case-control sample selections and two-phase design methodology used can be found in their previous publication (Chen et al. 2006; 2008).
Within the last decade, epidemiologic studies of cancers and many other chronic diseases have gone through a major transition due to the formation of various consortia that allow for the powerful analysis of common factors across many studies based on a very large set of samples. For example, two major consortia have been formed to study breast cancer: one based on cohort studies, e.g., the BPC3 study of Canzian et al, 2010, and others based on case-control studies (Breast Cancer Association Consortium, 2006) have been formed. These studies have led to extremely powerful investigation of various types of hypotheses, such as genetic association, based on tens of thousands of cases and controls depending on the types of risk factors that are being analyzed. Many of the studies participating in these consortia have standard risk factors available, although sometimes in a crude form. However, mammographic density (MD), which is much harder to evaluate, is rarely available in these studies.
To illustrate the utility of the proposed methodology, we develop a model for predicting breast cancer risk using data available on the full set of risk factors from the 1217 cases and 1610 controls of the “internal” BCDDP study and calibrating to the parameters from a standard risk factor model built from 12802 cases and 14296 controls from the “external” BPC3 consortium. The BPC3 standard risk factor model did not include mammographic density or the number of biopsies (nbiops); it included family history (famhist) as yes/no instead of the actual number of affected relatives, and recorded weight in tertiles as opposed to as a continuous variable. We used only summary level information from the BPC3 study, namely the estimates of the log-odds-ratio parameters and their standard errors. Our goal is to build a model similar to BCRAT2, i.e. a model with the standard risk factors as coded in BCRAT, and mammographic density.
Table 5 presents the results for model fit based on CML- and GR-based calibration approaches, together with the standard logistic regression analysis of the BCDDP data alone. In the model, the variable agemen is trichotomized according to age at menarche ≥ 14, 12–13, or < 12, and the two dummy variables for the latter two categories are denoted as agemen1 and agemen2; the variable ageflb is categorized into four groups according to age at first live birth < 20, 20–24, 25–29, and ≥ 30, and the latter three groups are denoted as ageflb1, ageflb2, and ageflb3. For both CML and GR methods, the standard errors were adjusted to account for uncertainty in the parameter estimates of the external model (see Appendix S.5 in Supplementary Material as well as in the proofs of Propositions 1 and 2). As expected, both CML and GR methods led to much smaller standard errors for the parameter estimates associated with covariates included in the external model than the analysis of the BCDDP data alone. For a number of these factors, including number of first-degree relatives with breast cancer and age at menarche, the use of the external model seems to change point estimates of model coefficients to a degree that cannot be explained by uncertainty alone. Closer inspection of the estimates of the parameters of the reduced model from the internal and external studies, also shown in Table 5, indicates that breast cancer associations for these two factors were different between the two studies to a degree that may be indicative of true population differences. Since BPC3 represents a consortium of cohort studies underlying a broader population than that underlying the BCDDP study, a risk model that is built based on the calibrated estimates could potentially be more broadly applicable. However, future validation studies would be needed to verify such an assertion.
Table 5.
Analysis results of BCDDP data. The variables in the full model include: number of first-degree relatives with breast cancer (numrel), age at menarche (two dummy variables agemen1–agemen2 according to ≥ 14, 12–13, or < 12 years), age at first live birth (three dummy variables ageflb1–ageflb3 according to < 20, 20–24, 25–29, and ≥ 30 years), weight (in kg), number of previous biopsies (nbiops), and mammographic density (MD). The variables in the reduced model include: family history (famhist, binary according to yes/no), age at menarche (two dummy variables agemen1–agemen2 according to ≥ 14, 12–13, or < 12 years), age at first live birth (three dummy variables ageflb1–ageflb3 according to < 20, 20–24, 25–29, and ≥ 30 years), and weight (two dummy variables weight1–weight2 according to < 62.6, 62.6–73.1, and ≥ 73.1 kg)
| Full model† | Reduced model† | |||||
|---|---|---|---|---|---|---|
| variable | Internal data Est. (SE) |
mGR Est. (SE) |
CML Est. (SE) |
variable | Internal data Est. (SE) |
External data Est. (SE) |
| numrel | 0.648 (0.090) | 0.371 (0.038) | 0.297 (0.033) | famhist | 0.716 (0.101) | 0.354 (0.030) |
| agemen1 | 0.083 (0.091) | 0.074 (0.034) | 0.077 (0.035) | agemen1 | 0.059 (0.090) | 0.052 (0.030) |
| agemen2 | 0.468 (0.124) | 0.185 (0.041) | 0.167 (0.042) | agemen2 | 0.387 (0.120) | 0.081 (0.031) |
| ageflb1 | −0.018 (0.146) | −0.109 (0.054) | −0.117 (0.057) | ageflb1 | 0.046 (0.142) | −0.053 (0.048) |
| ageflb2 | 0.086 (0.144) | 0.003 (0.052) | −0.005 (0.055) | ageflb2 | 0.261 (0.139) | 0.171 (0.043) |
| ageflb3 | 0.251 (0.173) | 0.173 (0.067) | 0.163 (0.070) | ageflb3 | 0.449 (0.167) | 0.358 (0.057) |
| weight | 0.020 (0.004) | 0.022 (0.003) | 0.024 (0.002) | weight1 | 0.061 (0.088) | 0.106 (0.031) |
| weight2 | 0.135 (0.106) | 0.214 (0.031) | ||||
| nbiops | 0.180 (0.070) | 0.178 (0.069) | 0.165 (0.073) | |||
| MD | 0.430 (0.044) | 0.428 (0.045) | 0.441 (0.047) | |||
: adjusted for 5-year age strata
mGR: modified GR for case-control sampling
CML: constrained likelihood method
Est.: estimated coefficient
SE: estimated standard error
Of the two calibration estimators, both CML and GR method produced comparable point estimates, but CML produced noticeably smaller standard errors for number of first-degree relatives with breast cancer and weight, two variables that were included in cruder forms in the external model. These results are consistent with higher efficiency of CML over GR in the simulation setting of measurement error. For the number of previous biopsies and mammographic density, two factors which were not included in the external model, both CML and GR produced results similar to the standard analysis of the BCDDP data alone. In Table S.8 of the Supplementary Material, we present results for CML and GR proceeding as though the external model parameters came from a data set that is so large that uncertainty can be ignored. In this case, as expected, we observe that the efficiency of both CML and GR further increases relative to BCDDP-only analysis. Moreover, the relative efficiency of CML over GR increases for the parameters associated with weight and number of first-degree relatives with breast cancer.
5 DISCUSSION
We have proposed alternative maximum likelihood methods for utilizing information from external big data sets while building refined regression models based on an individual analytic study. External information, when properly used, can increase both efficiency of underlying parameter estimates and the generalizability of the overall models to broader populations. In recognition of the potential of external data, survey methodologists have long used various types of “design-based” or “model-assisted” calibration techniques for estimating target parameters of interest without relying on a full probability model for the data. In this report, we provide a framework for a very general model-based, yet semiparametric, maximum likelihood inferential framework that requires only summary-level information from external sources.
Our simulation studies and data analysis show that the constrained maximum likelihood (CML) and synthetic constrained maximum likelihood (SCML) methods can achieve major efficiency gains over generalized regression (GR) type calibration estimators for covariates in a model that are measured with a poorer instrument in the external study. On the other hand, for covariates that are measured the same way in the external and internal studies, the efficiency of these two methods was similar. It is, however, noteworthy that the modified GR estimator we implemented for the case-control study is not a proper model-free calibration estimator in the sense survey methodologists use. The method is only applicable for logistic models and is likely to be more efficient than a proper design-based GR estimator when the model is correct. Future research is merited to explore the theoretical properties of such modified GR estimators and their connection with ML estimators.
Our simulation studies show that model calibration using external information has important caveats as well. In particular, if the risk factor distribution differs between the underlying populations for the internal and external studies, any type of calibration method, model-based or not, can produce severe bias in estimates of the underlying regression parameters, even when the regression relationship fβ(Y|X, Z) is exactly the same in the two populations. The assumption of complete exchangeability of populations that is required in calibration methods is more likely to be violated in the kind of applications we envision than in survey-sampling or two-phase design applications, where by design there is a common underlying population. Ideally, in this setting, model calibration should be performed with respect to a risk factor distribution that is representative of the external population. Our synthetic conditional maximum likelihood (SCML) method allows this by importing covariate distributions from an external reference sample. Even when such a sample is not available, the SCML method can be used to perform sensitivity analysis under various hypothetical or simulated covariate distributions that may be considered realistic for the external population. It is further important to note that likelihood-based methods require the assumption that the full model fβ(Y|X, Z) is correctly specified for both the internal and the external populations. Although the internal study can be used for performing model diagnostics for the underlying population, the assumption is not testable for the external population because of lack of information on Z and inaccessibility of individual level data.
The proposed methods may have applications in other areas, such as bench-marking small area estimates to match larger area estimates (Mugglin and Carlin, 1998; Bell, et al., 2013; Zhang, et al, 2014), analyzing randomized clinical trial data so that they are generalizable to larger populations (Greenhouse, et al., 2008; Frangakis, 2009; Stuart, et al., 2011; Pearl and Bareinboim, 2014; Hartman, et al., 2015) and standardization and control of confounding for observational studies (Keiding and Clayton, 2014). In general, we foresee that model synthesis using disparate types of data sources will be increasingly important for biomedical research in the future. The key constraints we identify to relate models of varying size, i.e. equation (2), could be useful for model synthesis in more general settings than we have considered here. More research is needed to extend the framework for developing models incorporating information from studies that may have collected different, possibly overlapping, sets of covariates. In this setting, each study or a combination of studies that collect similar covariates, can provide information on a particular type of reduced model. Future research is also merited to explore methods that can incorporate these constraints in a “softer” fashion to account for sources of uncertainty of the external models and their parameters.
Supplementary Material
Table 2.
Simulation results for the missing covariate setting, in which the full model of interest has the form h−1 {pr(D = 1)} = β0 + βXX + βZZ + βXZXZ, where h−1 denotes the inverse link function corresponding to a logistic model, and where the external model is h−1 {pr(D = 1)} = θ0 + θXX. Results multiplied by 103 are presented, and the coverage probabilities (CP) are reported as percents.
| Int | β0 GR/ mGR |
CML | Int | βX GR/ mGR |
CML | Int | βZ GR/ mGR |
CML | Int | βXZ GR/ mGR |
CML | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| simple random; N = 1000 | ||||||||||||
| Bias | −8.94 | 2.67 | 2.84 | 2.42 | 3.30 | 3.37 | 1.29 | 1.50 | 0.95 | 1.33 | 1.27 | 2.42 |
| SE | 91.4 | 32.5 | 32.4 | 96.8 | 39.0 | 38.9 | 94.3 | 94.4 | 94.3 | 89.4 | 89.4 | 89.5 |
| ESE | 91.8 | 32.1 | 32.3 | 92.3 | 38.8 | 38.9 | 92.4 | 92.3 | 92.5 | 85.8 | 85.6 | 86.9 |
| MSE | 8.42 | 1.06 | 1.06 | 9.38 | 1.53 | 1.53 | 8.89 | 8.91 | 8.89 | 7.98 | 7.99 | 8.01 |
| CP | 95.4 | 94.7 | 95.3 | 94.3 | 93.4 | 94.0 | 94.6 | 94.5 | 95.1 | 93.6 | 93.7 | 93.8 |
| case-control; N = 1000 | ||||||||||||
| Bias | - | - | 2.59 | 2.40 | 14.8 | 0.88 | 5.06 | 5.01 | 5.11 | −1.51 | −1.53 | −1.57 |
| SE | - | - | 22.7 | 75.7 | 25.1 | 26.8 | 72.2 | 72.3 | 72.2 | 72.9 | 72.9 | 72.8 |
| ESE | - | - | 22.8 | 73.3 | 26.1 | 27.9 | 73.1 | 73.2 | 73.2 | 71.4 | 71.4 | 71.6 |
| MSE | - | - | 0.52 | 5.73 | 0.85 | 0.72 | 5.24 | 5.24 | 5.24 | 5.31 | 5.31 | 5.30 |
| CP | - | - | 94.7 | 94.2 | 91.3 | 96.2 | 95.4 | 95.6 | 95.4 | 94.7 | 94.4 | 94.5 |
Int: internal-data only method
GR: generalized regression, mGR: modified GR for case-control sampling
CML: constrained maximum likelihood
ESE: estimated standard error
MSE: mean squared error
CP: coverage probability of a 95% confidence interval interval
Acknowledgments
Chatterjee’s and Maas’s research were supported by the intramural program of the US National Cancer Institute. Chen’s research was supported by Ministry of Science and Technology of Taiwan (NSC101-2118-M-001-002-MY3). Carroll’s research was supported by a grant from the National Cancer Institute (U01-CA057030). The authors are grateful to the editor and two referees for their very helpful comments.
Appendix: Derivation of the Pseudo-Loglikelihood
We first derive the pseudo-loglikelihood (4) under the simple random sampling design. The semiparametric likelihood is to be maximized under the constraint ∫ uβ(X, Z; θ)dF(X, Z) = 0, where uβ(X, Z; θ) is defined in (3), and F is treated nonparametrically by assigning masses (δ1, …, δm) to the unique values of the data (Xi, Zi) (i = 1, …, N), with m the number of unique data points and . Let nj be the number of (Xi, Zi) equal to the jth distinct pair value. For notational convenience, write fβ,i = fβ(Yi, Xi, Zi), uβ,i = uβ(Xi, Zi; θ), and Fi = F(Xi, Zi) (i = 1, …, N). Applying the Lagrange multiplier method, we solve the stationary point to
where λ and ϕ are Lagrange multipliers. By differentiating with respect to λ and ϕ, we obtain the stationary equation: and , as desired. Further, by differentiating with respect to δj, we obtain the stationary equation
which, by multiplying δj and summing over j on both sides leads to ϕ = −N since and . Hence
Plugging this δj into log(Lβ,F) results in a profile likelihood that is equivalent to (4), with λ rescaled by a factor of N.
Now consider the semiparametric constrained maximum likelihood under the case-control sampling design of the internal study. In this case we want to maximize the case-control loglikelihood given in (5) subject to the constraint ∫ uβ(X, Z; θ)dF(X, Z) = 0. Define for y = 0, 1, and . By the characterization of F as above and the Lagrange multiplier method, we solve the stationary equation for
Again, by differentiating with respect to λ and ϕ, we obtain the stationary equations: and . The stationary equation by differentiating δj leads to
Multiplying by δj, and summing over j on both sides leads to ϕ = 0 given and . Hence
where μy = Ny/py (y = 0, 1) and μ0 in fact links to μ1 since p0 = 1 − p1. Plugging δj into then gives the profile likelihood equivalent to (6).
Contributor Information
Nilanjan Chatterjee, Email: chattern@mail.nih.gov, National Cancer Institute, Rockville MD 20852, U.S.A.
Yi-Hau Chen, Email: yhchen@stat.sinica.edu.tw, Institute of Statistical Science, Academia Sinica, Taipei 11529, Taiwan.
Paige Maas, Email: paige.maas@mail.nih.gov, National Cancer Institute, Rockville MD 20852, U.S.A.
Raymond J. Carroll, Email: carroll@stat.tamu.edu, Department of Statistics, Texas A&M University, College Station TX 77843–3143, U.S.A., and Department of Mathematics and Statistics, University of Technology Sydney, PO Box 123, Broadway, NSW, 2007.
REFERENCES
- Bell WR, Datta GS, Ghosh M. Benchmarking Small Area Estimators. Biometrika. 2013;100:189–202. [Google Scholar]
- Breast Cancer Association Consortium. Commonly Studied Single-Nucleotide Polymorphisms and Breast Cancer: Results From the Breast Cancer Association Consortium. Journal of the National Cancer Institute. 2006;98:1382–1396. doi: 10.1093/jnci/djj374. [DOI] [PubMed] [Google Scholar]
- Breslow NE, Holubkov R. Maximum Likelihood Estimation of Logistic Regression Parameters Under Two-Phase, Outcome-Dependent Sampling. Journal of the Royal Statistical Society, Series B. 1997;59:447–461. [Google Scholar]
- Canzian F, Cox D, Setiawan V, Wendy, Stram D, Ziegler R, Dossus L, Beckmann L, Blanch H, Barricarte A, Berg C, Bingham S, Buring J, Buys S, Calle E, Chanock S, Clavel-Chapelon F, DeLancey J, Diver W, Dorronsoro M, Haiman C, Hallmans G, Hankinson S, Hunter D, Hsing A, Isaacs C, Khaw K, Kolonel L, Kraft P, Le Marchand L, Lund E, Overvad K, Panico S, Peeters P, Pollak M, Thun M, Tjnneland A, Trichopoulos D, Tumino R, Yeager M, Hoover R, Riboli E, Thomas G, Henderson B, Kaaks R, Feigelson H. Comprehensive Analysis of Common Genetic Variation in 61 Genes Related to Steroid Hormone and Insulin-Like Growth Factor-I Metabolism and Breast Cancer Risk in the NCI Breast and Prostate Cancer Cohort Consortium. Human Molecular Genetics. 2010;19:3873–3884. doi: 10.1093/hmg/ddq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Pee D, Ayyagari R, Graubard B, Schairer C, Byrne C, Benichou J, Gail MH. Projecting Absolute Invasive Breast Cancer Risk in White Women With a Model That Includes Mammographie Density. Journal of the National Cancer Institute. 2006;98:1215–1226. doi: 10.1093/jnci/djj332. [DOI] [PubMed] [Google Scholar]
- Chen J, Ayyagari R, Chatterjee N, Pee DY, Schairer C, Byrne C, Benichou J, Gail MH. Breast Cancer Relative Hazard Estimates from Case-Control and Cohort Designs with Missing Data on Mammographic Density. Journal of the American Statistical Association. 2008;103:976–988. [Google Scholar]
- Chen Y-H, Chen H. A Unified Approach to Regression Analysis under Double Sampling Design. Journal of the Royal Statistical Society, Series B. 2000;62:449–460. [Google Scholar]
- Chiang AC, Wainwright K. Fundamental Methods of Mathematical Economics. 3rd. New York: McGraw-Hill; 1984. [Google Scholar]
- Deville JC, Sarndal CE. Calibration Estimators in Survey Ssampling. Journal of the American Statistics Association. 1992;87:376–382. [Google Scholar]
- Frangakis C. The Calibration of Treatment from Clinical Trials to Target Populations. Clinical Trials. 2009;6:136–140. doi: 10.1177/1740774509103868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhouse JB, Kaizar EE, Kelleher K, Seltman H, Gardner W. Generalizing from Clinical Trial Data: a Case Study: the Risk of Suicidality Among Pediatric Antidepressant Users. Statistics in Medicine. 2008;27:1801–1813. doi: 10.1002/sim.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman E, Grieve R, Ramsahai R, Sekhon J. From SATT to PATT: Combining Experimental with Observational Studies to Estimate Population Treatment Effects. Journal of the Royal Statistical Society, Series B. 2015;178:757–778. [Google Scholar]
- Huber P. Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability. Vol. 1. Berkeley: University of California Press; 1967. The Behavior of the Maximum Likelihood Estimates under Nonstandard Conditions; pp. 221–233. [Google Scholar]
- Keiding N, Clayton D. Standardization and Control for Confounding in Observational Studies: A Historical Perspective. Statistical Science. 2014;29:529–558. [Google Scholar]
- Lawless JF, Wild CJ, Kalbfleisch JD. Semiparametric Methods for Response-Selective and Missing Data Problems in Regression. Journal of the Royal Statistical Society, Series B. 1999;61:413–438. [Google Scholar]
- Lumley T, Shaw PA, Dai JY. Connections Between Survey Calibration Estimators and Semiparametric Models for Incomplete Data. International Statistitical Review. 2011;79:200–220. doi: 10.1111/j.1751-5823.2011.00138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugglin A, Carlin B. Hierarchical Modeling in Geographic Information Systems: Population Interpolation over Incompatible Zones. Journal of Agricultural, Biological, and Environmental Statistics. 1998;3:111–130. [Google Scholar]
- Pearl J, Bareinboim E. External Validity: From do-Calculus to Transportability Across Populations. Statistical Science. 2014;29:579–595. [Google Scholar]
- Prentice RL, Pyke R. Logistic Disease Incidence Models and Case-Control Studies. Biometrika. 1979;66:403–412. [Google Scholar]
- Qin J, Lawless J. Empirical Likelihood and General Estimating Equations. Annals of Statistics. 1994;22:300–325. [Google Scholar]
- Qin J, Zhang H, Li P, Albanes D, Yu K. Using Covariate-Specific Disease Prevalence Information to Increase the Power of Case-Control Studies. Biometrika. 2015;102:169–180. [Google Scholar]
- Robins JM, Rotnitzky A, Zhao LP. Estimation of Regression Coefficients When Some Regressors Are Not Always Observed. Journal of the American Statistical Association. 1994;89:846–866. [Google Scholar]
- Roeder K, Carroll RJ, Lindsay BG. A Nonparametric Maximum Likelihood Approach to Case-control Studies with Errors in Covariables. Journal of the American Statistical Association. 1996;91:722–732. [Google Scholar]
- Scott AJ, Wild CJ. Fitting Regression Models to Case-Control Data by Maximum Likelihood. Biometrika. 1997;84:57–71. [Google Scholar]
- Stuart EA, Cole SR, Bradshaw CP, Leaf PJ. The Use of Propensity Scores to Assess the Generalizability of Results From Randomized Trials. Journal of the Royal Statistical Society, Series A. 2011;174:369–386. doi: 10.1111/j.1467-985X.2010.00673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White H. Maximum Likelihood Estimation of Misspecified Models. Econometrica. 1982;50:1–26. [Google Scholar]
- Wu C. Optimal Calibration Estimators in Survey Sampling. Biometrika. 2003;90:937–951. [Google Scholar]
- Wu C, Sitter RR. A Model-Calibration Approach to Using Complete Auxiliary Information from Survey Data. Journal of the American Statistical Association. 2001;96:185–193. [Google Scholar]
- Zhang X, Holt JB, Lu H, Wheaton AG, Ford ES, Greenlund KJ, Croft JB. Multilevel Regression and Poststratification for Small-Area Estimation of Population Health Outcomes: A Case Study of Chronic Obstructive Pulmonary Disease Prevalence Using the Behavioral Risk Factor Surveillance System. American Journal of Epidemiology. 2014;179:1025–1033. doi: 10.1093/aje/kwu018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.