Abstract
We characterized two epidemiologically similar Acinetobacter baumannii clusters from two separate intensive care units (ICU) using core genome multilocus sequence typing. Clonal spread was confirmed in ICU-1 (12 of 14 isolates shared genotypes); in ICU-2, all genotypes (13 isolates) were diverse, thus excluding transmissions and enabling adequate infection control measures.
TEXT
Acinetobacter baumannii is often reported as a cause of hospital-acquired infections (1, 2) and is associated with respiratory infections, bacteremia, meningitis, and wound infections (1, 3, 4). A. baumannii is transmitted via direct or indirect contact, and its ability to survive for months on inanimate surfaces hampers infection control measures (5). The emergence of multidrug-resistant (MDR) A. baumannii (6, 7) and the frequent association with nosocomial outbreaks (8–10) make MDR A. baumannii a pathogen of serious concern (11, 12).
If bacterial pathogens occur in clusters, epidemiological investigations are triggered and frequently complemented with molecular typing methods to confirm whether or not clusters are due to the simultaneous occurrence of identical bacterial genotypes. Currently, pulsed-field gel electrophoresis and other methods (6, 13–15) are increasingly being replaced by whole-genome sequence (WGS)-based methods, which provide the highest possible discriminatory power (16). WGS-based approaches rely either on the characterization of single nucleotide polymorphisms (SNP) (16, 17) or on gene-by-gene allelic profiling of core genome genes, called core genome multilocus sequence typing (cgMLST) (18–20).
As a published cgMLST scheme for A. baumannii is not yet available, here we investigated the ability of an ad hoc scheme to differentiate two epidemiologically determined clusters of A. baumannii that occurred in two unrelated intensive care units (ICU) and compared our ad hoc cgMLST scheme with SNP typing.
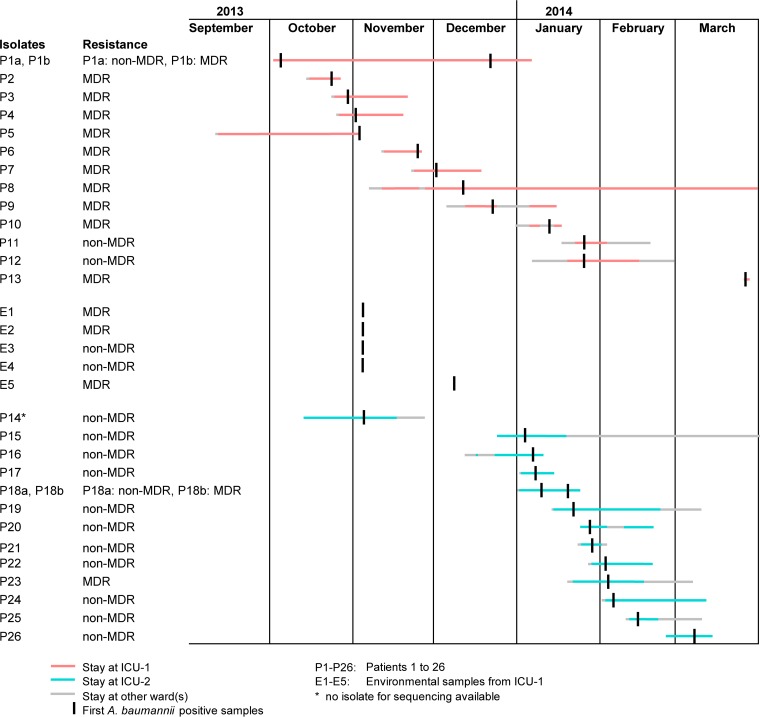
During routine surveillance efforts, which comprise a daily review of patients' charts and microbiological data, two clusters were detected in two ICUs at the University Hospital Muenster, Muenster, Germany, a 1,457-bed tertiary care hospital, between October 2013 and March 2014. In ICU-1 (cluster I), in addition to intensified disinfection measures to eliminate potential environmental reservoirs, extra training of staff members regarding compliance with hand hygiene measures and contact precautions, and isolation of patients in single rooms, a weekly microbiological screening of patients was established. Moreover, two series of environmental samplings (5 November and 12 December 2013) were initiated after the first putative transmission event was recognized at the end of October 2013. In total, five environmental (isolates E1 to E5) and 14 samples from 13 A. baumannii-positive patients (P1 to P13; P1 exhibited a switch from non-MDR [P1a] to MDR [P1b]) were detected (isolates P1b, P10, and P13 from the skin, P12 from a central intravenous catheter, and all others from respiratory samples). The majority of these were rated as MDR A. baumannii, P1b to P10 exhibiting resistance to piperacillin, 3rd/4th generation cephalosporins, and fluoroquinolones but susceptibility to most carbapenems (see the supplemental methods). Only three isolates (P1a, P11, and P12) were susceptible, i.e., non-MDR, against the majority of antibiotics tested (Fig. 1; Table 1). Cluster II from ICU-2 encompassed 12 patients and was noticed in January 2014. In contrast to cluster I, all but two A. baumannii isolates were non-MDR. Epidemiological investigations also included one further patient who stayed in ICU-2 in October/November 2013 (P14); however, the isolate of this patient was not available for typing. Since one patient (P18) showed a change from non-MDR A. baumannii (P18a) to MDR A. baumannii (P18b), overall 13 isolates from cluster II were sequenced. In total, 32 A. baumannii isolates were subjected to WGS on a MiSeq platform (Illumina Inc., San Diego, CA, USA), which is described in detail in the supplemental methods, and the resulting reads were quality trimmed and de novo assembled as recently described (21). Using SeqSphere+ software version 2.0 beta (Ridom GmbH, Muenster, Germany), all coding regions (CDS) were extracted and compared in a gene-by-gene approach (cgMLST) using A. baumannii strain ATCC 17978 (GenBank accession number CP000521.1) as the reference sequence. The clonal relationship was displayed in a minimum spanning tree that was generated using the same software (see supplemental methods).
FIG 1.
Linelist of all 26 patients and 5 environmental samples positive for A. baumannii for the two clusters in ICU-1 and ICU-2 during September 2013 and March 2014. P, patient; E, environment; MDR, multidrug-resistant phenotype (for details, see Table 1).
TABLE 1.
Antibiotic susceptibility patterns of the A. baumannii isolates investigateda
| Antibiotic substance | Result for isolate(s): |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1a | P1b, P2, P9, E1, E2, E5 | P3 | P4-P6, P8, P10 | P7 | P11, P16 | P12, P22, P25, P26 | P13 | E3 | E4 | P14 | P15 | P17, P18a | P18b | P19 | P20 | P21 | P23 | P24 | |
| Piperacillin | S | R | R | R | R | S | I | I | NT | NT | R | R | R | R | R | I | I | I | I |
| Piperacillin/tazobactam | S | R | R | R | R | S | S | S | S | S | S | S | S | R | S | S | S | S | S |
| Ceftazidime | R | R | R | R | R | S | S | S | I | I | I | S | S | R | S | S | S | I | I |
| Cefepime | NT | NT | R | R | R | S | S | S | NT | NT | I | NT | S | R | R | R | S | I | I |
| Imipenem | S | S | I | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| Meropenem | S | S | I | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| Gentamicin | S | R | R | R | R | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| Ciprofloxacin | S | R | R | R | R | S | S | R | I | S | S | S | S | I | S | S | S | R | S |
| Tigecycline | S | NT | I | I | R | S | S | NT | NT | NT | S | S | S | I | S | S | S | NT | S |
| Trimethoprim-sulfamethoxazole | S | R | R | R | R | S | S | S | S | S | S | S | S | S | S | S | R | S | I |
| Overall rated as | Non-MDR | MDR | MDR | MDR | MDR | Non-MDR | Non-MDR | MDR | Non-MDR | Non-MDR | Non-MDR | Non-MDR | Non-MDR | MDR | Non-MDR | Non-MDR | Non-MDR | MDR | Non-MDR |
Antibiotic susceptibility was determined using a Vitek II system (bioMérieux, Marcy-l'Étoile, France); testing was performed in accordance with EUCAST guidelines (versions 3.1 to 5.0). S, susceptible; I, intermediate; R, resistant; NT, not tested.
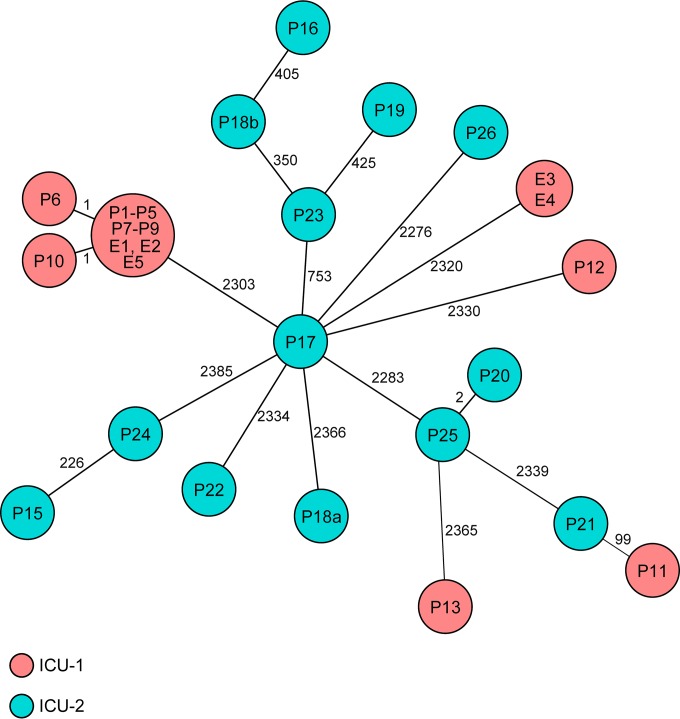
After WGS of the 32 isolates, the 3,319 A. baumannii genes of the ad hoc typing scheme were queried in all genomes and further analyzed (see Table S1 and the supplemental data set in the supplemental material). Of the 3,319 genes, 2,592 to 2,876 genes were present (mean 2,682 genes). In cluster I, 12 of 19 isolates (P1 to P5, P7 to P9, E1, E2, and E5) were identical, and two isolates showed only a single allelic variation (P6 and P10) indicating clonal spread; the remaining five isolates were only distantly related, exhibiting >2,300 differing genes (Fig. 2). In contrast, in cluster II nearly all isolates were genotypically distantly related (≥226 differing genes). Only two isolates (P20 and P25) differed in only two genes (Fig. 2). SNP-based typing of the 32 isolates corroborated both our findings based on cgMLST and the results of a recent study by Fitzpatrick et al. (16), where they determined a maximum distance of two SNPs as a threshold among isolates belonging to an outbreak: of the 19 cluster I isolates, 14 were identical (the 12 isolates identical by cgMLST and the 2 isolates differing only in one allele), the remaining isolates were only distantly related (see Fig. S1 in the supplemental material). A similar situation was detected among cluster II isolates. To investigate whether our cgMLST approach also results in results similar to the data of Fitzpatrick et al., we constructed a minimum spanning tree of the outbreak isolates. Indeed, cgMLST also resulted in a similar clustering of isolates with a maximum difference of 12 alleles in a pairwise comparison (see Fig. S2 in the supplemental material).
FIG 2.
Minimum spanning tree of 32 A. baumannii isolates detected for cluster I (ICU-1) and cluster II (ICU-2) based on the allelic profiles of up to 3,319 target genes (see Tables S1 and S2 and the supplemental data set in the supplemental material) that were present in the isolates with the “pairwise ignoring missing values” option turned on in SeqSphere+ software during comparison. Each circle represents an allelic profile, i.e., the genotype, and is named by the isolate(s). The numbers on the connecting lines give the number of differing alleles. The size of the circle is proportional to the number of isolates with an identical genotype and the color of the circles represent the different wards.
Our results shown here enabled us to concentrate infection control measures on patients and the environment of the affected ward and indicated that the detected cluster I was localized rather than a hospital-wide problem with transmissions on several wards. This is further underlined by the fact that from discharge of the last patient of cluster I until the day of writing this paper no further A. baumannii isolate with the cluster I genotype has been found among the 14 MDR A. baumannii isolates detected up to mid-2016.
When the first isolates of cluster II were detected, the WGS workflow for A. baumannii typing had already been established. This enabled an immediate response with respect to infection control procedures, avoiding extensive measures that would have been withdrawn after typing data were available. Since, starting from the first A. baumannii of cluster II determined, the isolates exhibited a genotype different from those of both the cluster I isolates and among the cluster II isolates, no additional infection control procedures were implemented to prevent nosocomial spread; only one single putative transmission (patients P20 and P25) of non-MDR A. baumannii within cluster II was recognized. Despite efforts to elucidate the origin of cluster II, a final explanation for this temporary increase in A. baumannii detections was not determined. Moreover, we could not finally explain the detection of two genotypically different isolates of P18. The most likely explanation is that at the time of the first strain's isolation, P18 already carried more than one A. baumannii variants that were unrecognized. Furthermore, the two environmental isolates, E3 and E4, warrant some comments. We assume that E3 and E4 originated from a previous patient in this ward as A. baumannii in general is not an environmental bacterium but is able to survive for a long time in the environment.
Our approach clearly demonstrates the technical evolution in our laboratory of the two clusters, enabling prospective WGS typing to rapidly confirm or refute clonal spread of the pathogens as recently described by Roach et al. (22). Spurred on by these results, we have been constantly monitoring the molecular epidemiology of MDR A. baumannii using WGS. The cgMLST approach allows immediate comparisons of newly determined genotypes with historical data, enabling continuous surveillance, in contrast to SNP-based approaches that have to be recalculated once the data set changes. From a technical perspective, two challenges remain: first, a universal cgMLST nomenclature, i.e., a published cgMLST scheme, should be available to ensure interlaboratory comparability of data analogous to classical MLST results (23); and second, a threshold for genomic similarity should be defined to facilitate detection of transmissions as suggested by Salipante et al. (24). For some species, e.g., Listeria monocytogenes and Enterococcus faecium, these challenges have already been resolved (19, 25). Finally, future studies should address whether such WGS-based surveillance for infection control purposes is cost-effective: whereas costs for sequencing reagents are easily calculated (in this study approximately 120€ per isolate), determination of the financial impact on patient care remains challenging.
Accession number.
All raw reads generated in this study were submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena/) under the study accession number PRJEB7302.
Supplementary Material
ACKNOWLEDGMENTS
The skillful technical assistance of T. Böking, U. Keckevoet, and I. Höfig is greatly appreciated. Furthermore, we thank F. Schaumburg for confirmatory susceptibility testing and E. J. Bernhard for critical reading of the manuscript.
We declare no conflicts of interest.
Funding Statement
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00721-16.
REFERENCES
- 1.Munoz-Price LS, Weinstein RA. 2008. Acinetobacter infection. N Engl J Med 358:1271–1281. doi: 10.1056/NEJMra070741. [DOI] [PubMed] [Google Scholar]
- 2.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 3.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 4.Bergogne-Bérézin E, Towner KJ. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev 9:148–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jawad A, Seifert H, Snelling AM, Heritage J, Hawkey PM. 1998. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J Clin Microbiol 36:1938–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. 2010. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. doi: 10.1371/journal.pone.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doi Y, Murray GL, Peleg AY. 2015. Acinetobacter baumannii: evolution of antimicrobial resistance-treatment options. Semin Respir Crit Care Med 36:85–98. doi: 10.1055/s-0034-1398388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baang JH, Axelrod P, Decker BK, Hujer AM, Dash G, Truant AR, Bonomo RA, Fekete T. 2012. Longitudinal epidemiology of multidrug-resistant (MDR) Acinetobacter species in a tertiary care hospital. Am J Infect Control 40:134–137. doi: 10.1016/j.ajic.2011.04.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turton JF, Gabriel SN, Valderrey C, Kaufmann ME, Pitt TL. 2007. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin Microbiol Infect 13:807–815. doi: 10.1111/j.1469-0691.2007.01759.x. [DOI] [PubMed] [Google Scholar]
- 10.Munoz-Price LS, Zembower T, Penugonda S, Schreckenberger P, Lavin MA, Welbel S, Vais D, Baig M, Mohapatra S, Quinn JP, Weinstein RA. 2010. Clinical outcomes of carbapenem-resistant Acinetobacter baumannii bloodstream infections: study of a 2-state monoclonal outbreak. Infect Control Hosp Epidemiol 31:1057–1062. doi: 10.1086/656247. [DOI] [PubMed] [Google Scholar]
- 11.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Health Organization. 2014. Antimicrobial resistance: global report on surveillance. World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf. [Google Scholar]
- 13.Higgins PG, Dammhayn C, Hackel M, Seifert H. 2010. Global spread of carbapenem-resistant Acinetobacter baumannii. J Antimicrob Chemother 65:233–238. doi: 10.1093/jac/dkp428. [DOI] [PubMed] [Google Scholar]
- 14.Seifert H, Dolzani L, Bressan R, van der Reijden T, van Strijen B, Stefanik D, Heersma H, Dijkshoorn L. 2005. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J Clin Microbiol 43:4328–4335. doi: 10.1128/JCM.43.9.4328-4335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seifert H, Dijkshoorn L, Gerner-Smidt P, Pelzer N, Tjernberg I, Vaneechoutte M. 1997. Distribution of Acinetobacter species on human skin: comparison of phenotypic and genotypic identification methods. J Clin Microbiol 35:2819–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fitzpatrick MA, Ozer EA, Hauser AR. 2016. Utility of whole-genome sequencing in characterizing Acinetobacter epidemiology and analyzing hospital outbreaks. J Clin Microbiol 54:593–612. doi: 10.1128/JCM.01818-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis T, Loman NJ, Bingle L, Jumaa P, Weinstock GM, Mortiboy D, Pallen MJ. 2010. High-throughput whole-genome sequencing to dissect the epidemiology of Acinetobacter baumannii isolates from a hospital outbreak. J Hosp Infect 75:37–41. doi: 10.1016/j.jhin.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 18.Maiden MC, Jansen van Rensburg MJ, Bray JE, Earle SG, Ford SA, Jolley KA, McCarthy ND. 2013. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol 11:728–736. doi: 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruppitsch W, Pietzka A, Prior K, Bletz S, Fernandez HL, Allerberger F, Harmsen D, Mellmann A. 2015. Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Listeria monocytogenes. J Clin Microbiol 53:2869–2876. doi: 10.1128/JCM.01193-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, Rico A, Prior K, Szczepanowski R, Ji Y, Zhang W, McLaughlin SF, Henkhaus JK, Leopold B, Bielaszewska M, Prager R, Brzoska PM, Moore RL, Guenther S, Rothberg JM, Karch H. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751. doi: 10.1371/journal.pone.0022751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bletz S, Mellmann A, Rothgänger J, Harmsen D. 2015. Ensuring backwards compatibility: traditional genotyping efforts in the era of whole genome sequencing. Clin Microbiol Infect 21:347.e341–347.e344. doi: 10.1016/j.cmi.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Roach DJ, Burton JN, Lee C, Stackhouse B, Butler-Wu SM, Cookson BT, Shendure J, Salipante SJ. 2015. A year of infection in the intensive care unit: prospective whole genome sequencing of bacterial clinical isolates reveals cryptic transmissions and novel microbiota. PLoS Genet 11:e1005413. doi: 10.1371/journal.pgen.1005413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salipante SJ, SenGupta DJ, Cummings LA, Land TA, Hoogestraat DR, Cookson BT. 2015. Application of whole-genome sequencing for bacterial strain typing in molecular epidemiology. J Clin Microbiol 53:1072–1079. doi: 10.1128/JCM.03385-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Been M, Pinholt M, Top J, Bletz S, Mellmann A, van Schaik W, Brouwer E, Rogers M, Kraat Y, Bonten M, Corander J, Westh H, Harmsen D, Willems RJ. 2015. Core genome multilocus sequence typing scheme for high-resolution typing of Enterococcus faecium. J Clin Microbiol 53:3788–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.