Abstract
Ten strains representing four lineages of the Pseudomonas fluorescens group (P. chlororaphis, P. corrugata, P. koreensis, and P. fluorescens subgroups) were evaluated for toxicity to the tobacco hornworm Manduca sexta and the common fruit fly Drosophila melanogaster. The three strains within the P. chlororaphis subgroup exhibited both oral and injectable toxicity to the lepidopteran M. sexta. All three strains have the gene cluster encoding the FitD insect toxin and a ΔfitD mutant of P. protegens strain Pf-5 exhibited diminished oral toxicity compared to the wildtype strain. Only one of the three strains, P. protegens Pf-5, exhibited substantial levels of oral toxicity against the dipteran D. melanogaster. Three strains in the P. fluorescens subgroup, which lack fitD, consistently showed significant levels of injectable toxicity against M. sexta. In contrast, the oral toxicity of these strains against D. melanogaster was variable between experiments, with only one strain, Pseudomonas sp. BG33R, causing significant levels of mortality in repeated experiments. Toxin complex (Tc) gene clusters, which encode insecticidal properties in Photorhabdus luminescens, were identified in the genomes of seven of the ten strains evaluated in this study. Within those seven genomes, six types of Tc gene clusters were identified, distinguished by gene content, organization and genomic location, but no correlation was observed between the presence of Tc genes and insect toxicity of the evaluated strains. Our results demonstrate that members of the P. fluorescens group have the capacity to kill insects by both FitD-dependent and independent mechanisms.
Introduction
Pseudomonas is a diverse genus of γ-Proteobacteria known for its ubiquity in the natural world, capacity to utilize a striking variety of organic compounds as energy sources, and production of a remarkable array of exoenzymes, toxins, and secondary metabolites. The genus currently comprises at least 144 species [1] exhibiting varied lifestyles in a wide range of environments, including soil, water, plant surfaces and animals. Within the genus, the Pseudomonas fluorescens group is particularly heterogeneous, encompassing bacteria that have been classified into many subgroups and more than 60 named species, including P. protegens, P. chlororaphis, P. brassicacearum, and P. fluorescens itself [2]. Many plant-associated bacteria in these species have the capacity to suppress diseases caused by a spectrum of bacterial, fungal, and oomycete pathogens [3]. These beneficial bacteria are important components of the soil and plant microbiome contributing to plant health [4,5] and some strains have been used commercially for biological control of plant disease [6,7]. Certain strains within the P. fluorescens group exhibit insecticidal activity [8,9,10,11,12,13,14], opening the potential to use these bacteria for management of diseases and insect pests of plants.
Two types of insect toxin genes have been identified in strains of the P. fluorescens group and both types are similar to genes first described in entomopathogenic bacteria in the genera Xenorhabdus and Photorhabdus. A gene encoding FitD (fluorescens insecticidal toxin) is present in the genomes of P. protegens [11] and the related species P. chlororaphis [15,16]. FitD is similar to the Mcf (“Makes caterpillars floppy”) toxin of Photorhabdus luminescens, which exhibits injectable toxicity towards insects via apoptosis [17,18,19]. When injected into larvae of the tobacco hornworm Manduca sexta or the wax moth Galleria mellonella, fitD-containing strains of P. protegens are lethal [11]. Furthermore, FitD is a major determinant of the oral toxicity of P. protegens CHA0 and P. chlororaphis PCL1391 to three agriculturally-important lepidopteran insect pests, the African cotton leafworm Spodoptera littoralis, the tobacco budworm Heliothis virescens, and the diamondback moth Plutella xylostella [13]. In addition to fitD, some strains in the P. fluorescens group [16,20] have genes similar to those encoding Toxin complex (Tc) proteins, which are best characterized in the entomopathogen Photorhabdus luminescens strain W14 [21]. Tc toxins have three components (A, B, and C), each of which have conserved domains (Fig 1) [20]. All three components are necessary for full toxicity [22] with the A component serving as the primary toxin and the B and C components thought to be potentiators enhancing toxicity of the protein complex [21,23]. Tc genes are present in the genomes of many bacteria, including species without known associations with insects. Within the genus Pseudomonas, Tc gene clusters have been described in P. syringae pv. syringae B728a and P. fluorescens Pf0-1 [20] and a tccC gene has been associated with insect toxicity of Pseudomonas taiwanensis [14].
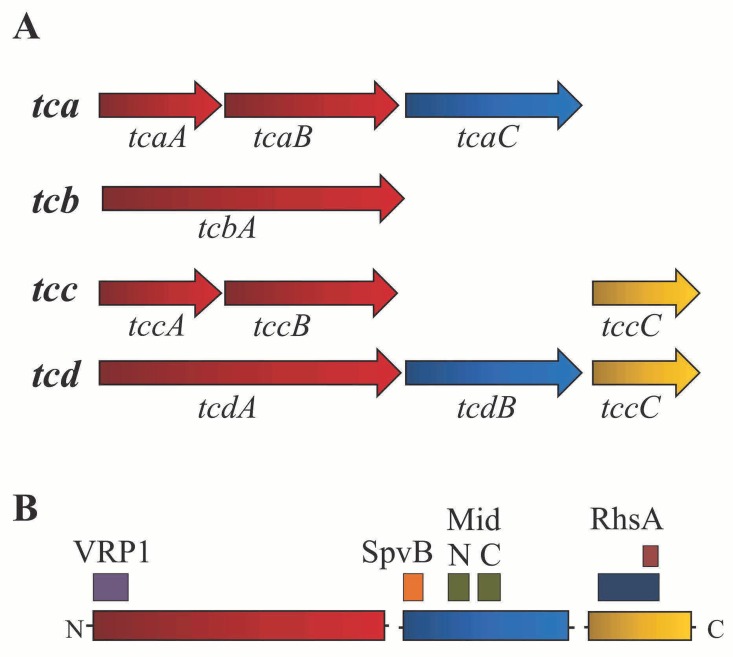
Fig 1. Tc gene clusters in Photorhabdus luminescens W14.
(A) The four Tc gene clusters (tca, tcb, tcc and tcd) present in the genome of P. luminescens W14. Genes are colored according to the Tc component encoded: red (Component A), blue (Component B) and yellow (Component C). Component A can be encoded by two genes or by a single, large gene. (B) Conserved domains present in each Tc component (A, B and C). The A component (TcaA/TcdA) has a conserved VRP1 domain near the N-terminus. The B component (TcaC/TcdB) has the SpvB domain near the N-terminus and the MidN and MidC domains near the center. The C component (TccC) has an RhsA domain with an Rhs repeat-associated core domain.
The aims of this study were to evaluate ten strains of the P. fluorescens group for insect toxicity and to characterize the putative Tc genes (tcaA, tcaB, tcaC, tcdA, tcdB, tccC) in the genomes of these ten strains. The strains tested were isolated from soil, root or leaf surfaces and known for their biological control of plant diseases [16,24,25]. They represent four subgroups (P. chlororaphis, P. corrugata, P. koreensis, and P. fluorescens) of the P. fluorescens group (Fig 2). This study demonstrated that the P. fluorescens group includes strains that are toxic to insects representing two orders (Diptera and Lepidoptera) and that the known insect toxin FitD is responsible for some, but not all, of the insect toxicity of these bacteria.
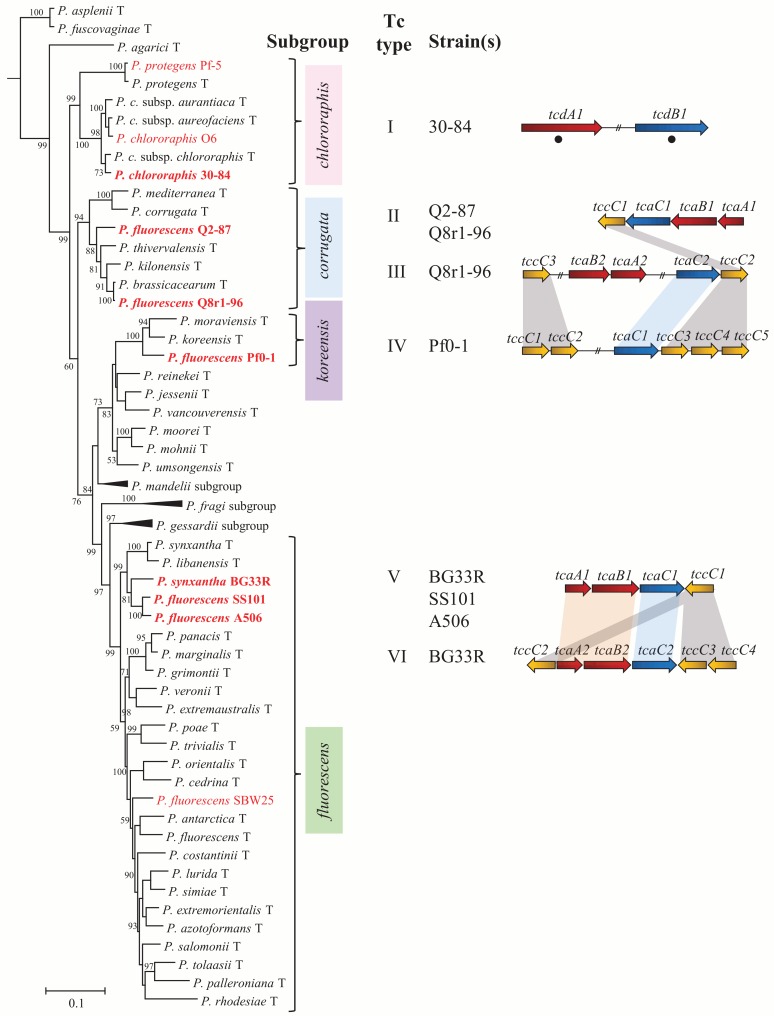
Fig 2. Seven of the ten strains of Pseudomonas spp. evaluated in this study have Tc clusters.
The ten strains evaluated in this study fall into four subgroups within the P. fluorescens group, as shown in a phylogenetic tree based on concatenated alignments of gyrB, rpoB, rpoD and 16S rRNA of the type strains within the P. fluorescens group. The ten strains examined in this study are shown in red font and subgroups containing these strains are labelled to the right (pink, chlororaphis subgroup; blue, corrugata subgroup; purple, koreensis subgroup; green, fluorescens subgroup). The tree is artificially rooted on the type strain of P. aeruginosa. Subgroups lacking any of the ten strains are collapsed and labeled. Bootstrap support less than 50% is not shown. Branch lengths indicate the number of nucleotide substitutions per site. Strains evaluated in this study that contain a Tc cluster are shown in bold, red font and are also listed to the right of the tree. Tc cluster Types I-VI are distinguished from one another by gene organization and genome location. Genes are colored according to the Tc component encoded: red (component A), blue (component B) and yellow (component C). Type I: 30–84 (Pchl3084_2947 and Pchl3084_2950); Type II: Q2-87 (PflQ2_0667–0670) and Q8r1-96 (PflQ8_0736–0739); Type III: Q8r1-96 (PflQ8_4696, PflQ8_4570–4571 and PflQ8_4580–4581); Type IV: Pf0-1 (Pfl01_0947–0948 and Pfl01_4453–4456); Type V: A506 (PflA506_3065–3068), SS101 (PflSS101_2971–2974) and BG33R (PseBG33_3189–3192); Type VI: BG33R (PseBG33_3799–3804). Numbers following gene names distinguish genes within a single genome that encode the same Tc component. Black circles denote genes located on genomic islands. Among genomes, homologous genes, defined by genomic location and phylogenetic relationships, are connected with shading of the same color.
Results
Phylogenetic analysis of ten strains of Pseudomonas spp. evaluated for insect toxicity
The ten strains evaluated in this study, which were the subjects of an earlier comparative genomic analysis [16], were placed in four subgroups of P. fluorescens based on multilocus sequence analysis (MLSA; Fig 2). Previously, the ten strains evaluated in this study were assigned to three sub-clades of the P. fluorescens group based on an MLSA of strains of Pseudomonas spp. with fully-sequenced genomes [16]. By including the type species within the P. fluorescens group in the MLSA shown in Fig 2, the ten strains were placed in four established subgroups of the P. fluorescens group. The three strains previously assigned to sub-clade 3 (P. chlororaphis 30–84, P. chlororaphis O6, and P. protegens Pf-5), are in the P. chlororaphis subgroup. Two strains previously assigned to sub-clade 2, P. brassicacearum Q8r1-96 and Pseudomonas sp. Q2-87, are in the P. corrugata subgroup. Pseudomonas sp. Pf0-1, which was assigned previously to sub-clade 2 [16], is in the P. koreensis subgroup. The four strains previously assigned to sub-clade 3 (SBW25, A506, SS101, and BG33R) are in the P. fluorescens subgroup (Fig 2). The results of our current MLSA, which was based on concatenated sequences of the housekeeping genes gyrB, rpoD, rpoB, and 16S rRNA (Fig 2), are congruent with those published recently by other researchers [1,2]. The ten strains of this study were included in the study of Garrido-Sanz et al. [2] and were assigned to the same subgroups in both analyses. Here, the previously defined sub-clades [16] were related to subgroups defined by type species within the P. fluorescens group.
Characterization of Tc genes
A bioinformatic survey of the ten strains used in this study identified Tc genes in seven of the ten genomes. A total of 38 Tc genes were identified from a BLASTN search using characterized Tc genes of P. luminescens W14 as queries. Predicted amino acid sequences for each gene have conserved domains characteristic of the A, B, or C components of Tc proteins present in entomopathogenic bacteria (S1–S3 Tables). Amino acid sequences of candidate tcaA genes from five genomes and the much larger tcdA of strain 30–84 have a conserved domain called VRP1 (S1 Table). The VRP1 domain corresponds to SpvA, the product of a plasmid-borne gene associated with virulence of Salmonella spp. [26]. The VRP1 domain is also present in known TccA, TcaA, TcbA TcdA proteins of entomopathogenic bacteria such as P. luminescens strain W14 [27]. Amino acid sequences of candidate tcaC/tcdB genes from seven genomes also have three conserved domains called SpvB, MidN and MidC (S2 Table). These domains are found in TcaC/TcdB sequences of insect-associated bacteria [27]. The TccC sequences identified in the Pseudomonas strains of this study contain the RhsA and Rhs repeat-associated core domains (S3 Table), which are commonly found in secreted bacterial toxins [28]. The presence of these conserved domains in candidate genes provides further evidence for their identity as tcaA/tcdA, tcaC/tcdB and tccC genes.
The organization of Tc genes varies among the seven genomes, falling into six types (I-VI) (Fig 2) distinguished by gene content and organization. Type I, which contains genes encoding putative A and B components, is present in the genome of P. chlororaphis 30–84. These Tc genes are approximately twice as large as any other Tc gene found in the P. fluorescens group but are similar in size to tcdA and tcdB of P. luminescens (S1 and S2 Tables) [27]. The Type I Tc gene cluster of strain 30–84 does not include a gene encoding a C component, which is considered a ‘potentiator’ and necessary for full toxicity of the Tc complex of P. luminescens and P. taiwanensis [14,21,27]. The Type II Tc cluster, which is present in strains Q8r1-96 and Q2-87, is composed of four contiguous genes (tcaA1, tcaB1, tcaC1 and tccC1) located at the same site in both genomes (Fig 2). Type III, which is present in strain Q8r1-96 only, is composed of genes for all three Tc components located at three sites distributed over 169 kb of the genome. Although these genes are not contiguous in the genome of Q8r1-96, they could be functional if gene expression is coordinated. The Type IV Tc gene cluster, which is present in strain Pf0-1, lacks an A component but has a single B component and five genes encoding putative C components in two clusters distal to one another in the genome. By analogy to the Tc cluster in P. luminescens where the A component encodes the primary toxin [21,23], the Type IV cluster may not be functional. Type V, which is present in strains A506, SS101 and BG33R, is composed of four contiguous genes (tcaA1, tcaB1, tcaC1 and tccC1) located at the same site in all three genomes (Fig 2). Type VI is a single cluster in BG33R containing contiguous genes: tcaA2, tcaB2, tcaC2 and three copies of tccC. Two strains, Q8r1-96 and BG33R, have two Tc gene clusters of different types.
Phylogenetic analyses were performed using the amino acid sequences of each Tc component present in the seven genomes of the P. fluorescens group. Trees were created using characterized Tc peptide sequences from P. luminescens W14 and BLASTP hits for each P. fluorescens group sequence with greater than 75% query coverage and 50% identity. Phylogenetic relationships support the placement of Tc clusters into the six types described above, with components of the same Tc type from different strains grouping within the same clades (S1–S3 Figs). Genes from different Tc types that fall together within well-supported clades of the phylogenetic trees are shown with shading in Fig 2. The most striking similarities are within the Type V and Type VI clusters, as sequences from these clusters group together in each of the three trees representing the three components of Tc clusters. The peptide sequences of the B and C components of type III and Type IV clusters are also closely related phylogenetically (S2 and S3 Figs) and genes encoding these components are located in identical regions of Q8r1-96 and Pf0-1 genomes. It seems likely that genes encoding the B and C components of these Tc types were inherited from a common ancestor prior to divergence of the clusters, with duplication of genes for the C components in Pf0-1 and either acquisition of the A component genes in strain Q8r1-96 or loss of the A component genes in strain Pf0-1.
Five of the six types of Tc gene clusters (Types II through VI) are in conserved regions of the genome shared by all ten strains of the P. fluorescens group evaluated in this study. In contrast, the Type I cluster of P. chlororaphis 30–84 is in a genomic island, unique to P. chlororaphis 30–84, that has characteristics of a phage, including the presence of genes encoding a phage integrase, a transposase and cointegrate resolution proteins. Sequence bias in this region was found using the Alien Hunter program that searches for regions of genomic plasticity [29]. Furthermore, the G+C content of tcdA1 (55.4%) and tcdB1 (56.3%) differ significantly (P< 0.001) from the genomic average (62.9%) of strain 30–84, as determined from a chi-square analysis. In our phylogenetic analysis, TcdA1 (Pchl3084_2947) and TcdB1 (Pchl3084_2950) of P. chlororaphis strain 30–84 are the only peptide sequences to have closely related hits outside of the Pseudomonas genus. TcdA1 is closely related to the products of uncharacterized genes in the soil bacterium Mesorhizobium alhagi and TcdB1 (Pchl3084_2950) is closely related to a gene product in Marinomonas posidonica (S1 and S2 Figs). Based on proximity to genes conferring mobilization functions, gene sequence bias and high identity to gene products of other bacterial taxa, we conclude that the Type I Tc gene cluster was likely acquired through horizontal gene transfer (HGT).
Strains in the P. fluorescens group differ in injectable toxicity to M. sexta
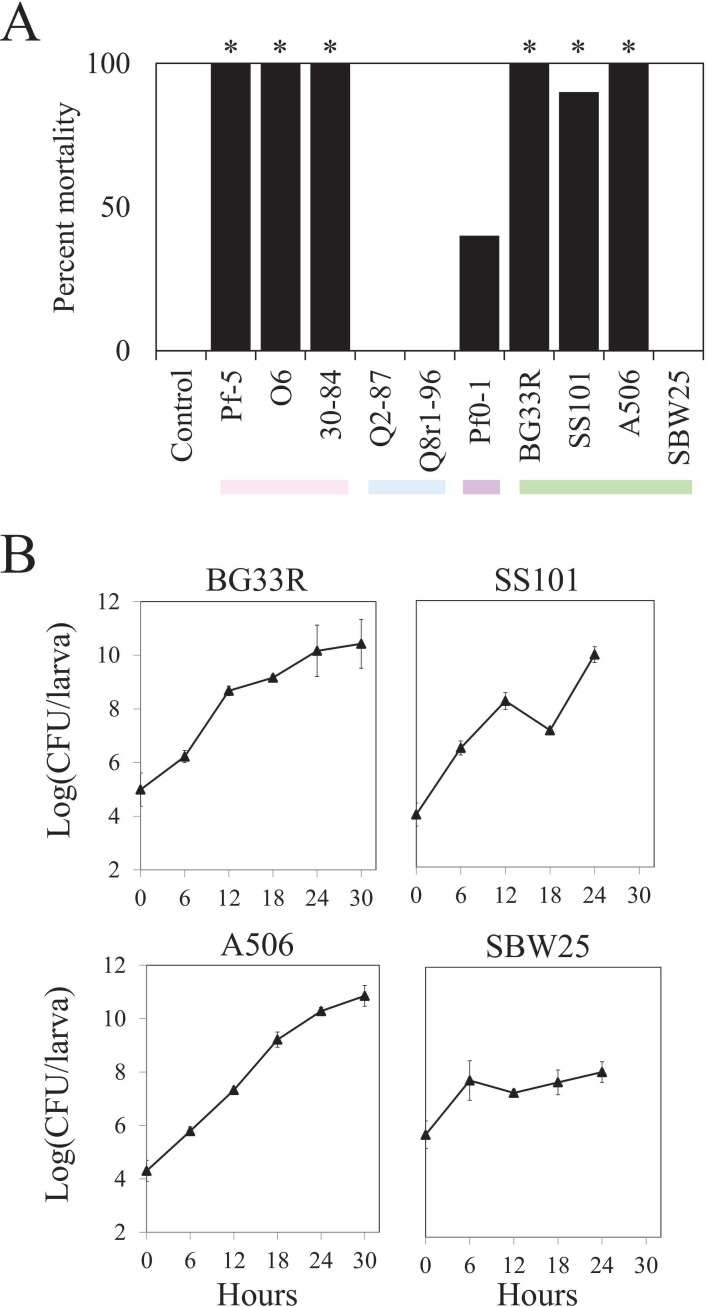
Of the ten strains of P. fluorescens evaluated in this study, six consistently killed M. sexta when injected at ca. 5 log (CFU/larva) into fifth instar larvae and three strains (Q2-87, Q8r1-96, and SBW25) exhibited no significant toxicity (Fig 3, S4 Fig). One strain, Pf0-1, caused a significant increase in mortality in one experiment (Fig 3), but did not do so in a second experiment (S4 Fig). The greatest levels of hornworm mortality were caused by strains in the P. chlororaphis subgroup and three of the four strains in the P. fluorescens subgroup (Fig 3, S4 Fig). At 72 h following injection, the six strains caused similar levels of mortality, but three strains in the P. chlororaphis subgroup killed the insects within 24 h following injection whereas mortality was slower for insects injected with strains in the P. fluorescens subgroup (S4 Fig). All three strains (Pf-5, O6 and 30–84) in the P. chlororaphis subgroup have the fitD gene, which encodes the FitD toxin. Because FitD is known to be lethal to M. sexta and a primary determinant of the injectable insect toxicity phenotype of Pf-5 [11], it is likely that FitD is largely responsible for the injectable toxicity of P. chlororaphis strains 30–84 and O6 as well. The four strains in the P. fluorescens subgroup differed in toxicity, with strains A506, BG33R, and SS101 causing significant mortality and strain SBW25 causing no mortality when injected at ca. 5 log (CFU/larva) (Fig 3A). When inoculated at ten-fold higher inoculum densities (ca. 6 log [CFU/larva]), however, all of the strains caused significant mortality in at least one of the two experiments in which they were evaluated (S5 Fig).
Fig 3. Injectable toxicity of ten strains of Pseudomonas spp. and colonization of larvae of M. sexta by the strains post-injection.
(A) Mortality of M. sexta was assessed 72 h following injection with ca. 5 log (CFU/larva) of the designated strain. Values are from one of three experiments (S4 Fig), each evaluating ten replicate larvae per treatment. Asterisks represent significant differences from the water control (P<0.05, d.f. = 1, χ2 test). Colors below the graph denote the subgroup of the strain tested for insect toxicity: pink, P. chlororaphis; blue, P. corrugata; purple, P. koreensis; green, P. fluorescens. (B) Rifampicin-resistant derivatives of strains BG33R, SS101, A506 and SBW25 were injected into ultimate instar larvae of M. sexta at ca. 5 log (CFU/larva). Internal population sizes of each strain were estimated from ten replicates of surface-sterilized larvae over time. Bacterial population sizes were log transformed and the means and standard errors are shown. No rifampicin-resistant Pseudomonas spp. were re-isolated from control larvae injected with sterile water.
The differential lethalities of the four strains in the P. fluorescens subgroup may be due to their different capacities to colonize the larvae of M. sexta. To explore this possibility, the growth of each strain in larvae of M. sexta was assessed over time. Ultimate instar M. sexta were injected with ca. 5 log (CFU/larva) and sampled every six hours (Fig 3B). The populations of all strains increased over time, and three strains (A506, BG33R, and SS101) reached population sizes of or exceeding 10 log (CFU/larva) at 24 h or 30 h after injection. In contrast to the other strains, SBW25 established lower population sizes in the larvae, reaching only ca. 8 log (CFU/larva) at 24 h, the last time point sampled. SBW25 did not kill larvae of M. sexta in this experiment whereas the other three strains caused significant levels of mortality (Fig 3A), which suggests a correlation between the toxicity and larval colonization phenotypes of the strains. It is likely that a strain that colonizes larvae would have a greater ability to cause mortality but conversely, it is also possible that a dying animal provides a habitat more conducive to colonization than a healthy animal. The two possibilities were not distinguished in this study.
A role for FitD in oral toxicity of P. protegens Pf-5 to M. sexta
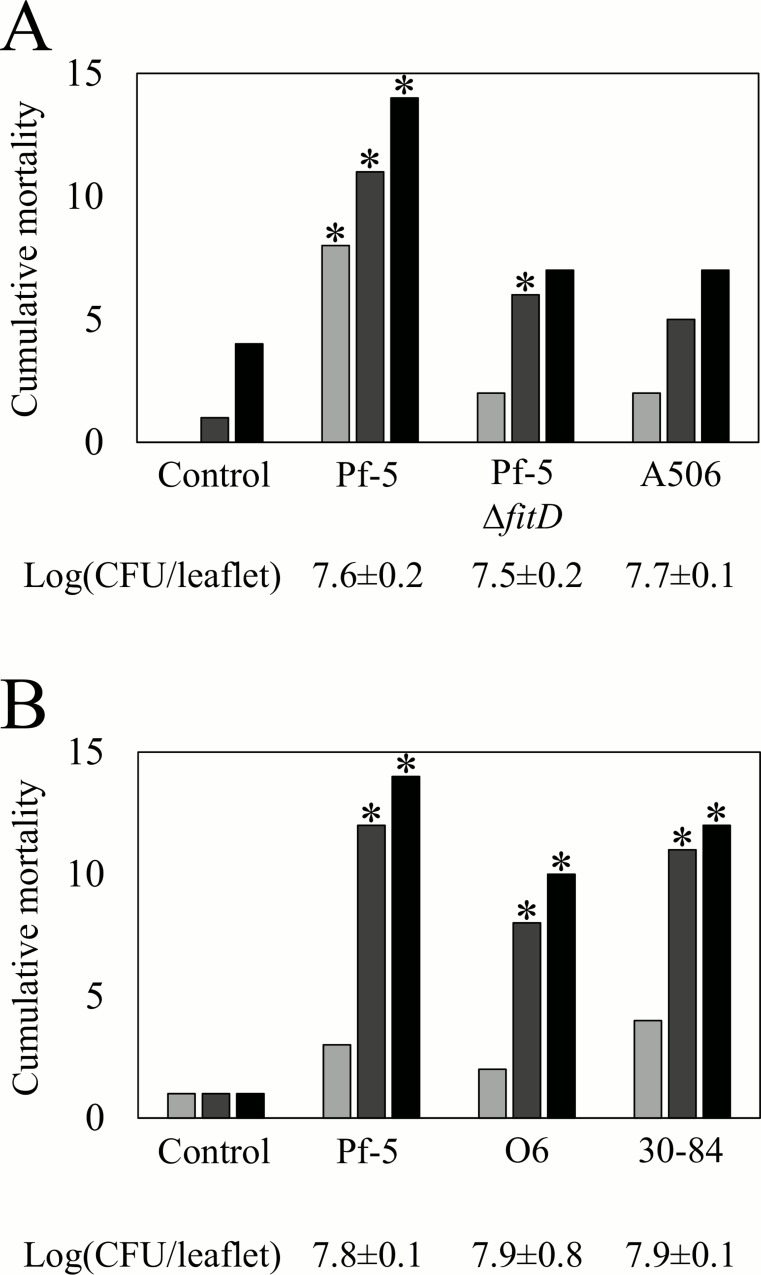
Due to the established role of FitD in injectable toxicity of Pf-5 to M. sexta [11], we developed an assay to assess the role of FitD in oral toxicity to the insect. Larvae of M. sexta were placed on tomato leaves that had been previously inoculated with one of the ten strains of Pseudomonas spp. As the larvae fed on the tomato leaves, they ingested cells of the inoculated bacterial strains, which were present at population sizes averaging 7.7 log (CFU/leaflet). Larvae feeding on Pf-5-inoculated leaves had a significantly higher level of mortality than non-inoculated controls in the seven experiments of this study (Fig 4 and S6 Fig). We attribute the mortality caused by Pf-5 to the FitD toxin, as mortality of the larvae on leaves colonized by the ΔfitD mutant did not differ significantly (P<0.05) from those on non-inoculated leaves (Fig 4A and S6 Fig). Like Pf-5, the two strains of P. chlororaphis (30–84 and O6), which also have fitD [16], caused significant oral toxicity to M. sexta (Fig 4B and S6 Fig). In contrast, P. fluorescens A506, a member of the P. fluorescens subgroup that lacks fitD, did not show oral toxicity to M. sexta (Fig 4A and S6 Fig). Similarly, we tested the other six strains of Pseudomonas spp. that lack fitD, each in a single experiment, and only BG33R caused significant levels of oral toxicity to M. sexta (S6 Fig).
Fig 4. Oral toxicity of the FitD-containing P. chlororaphis subgroup to M. sexta.
Cumulative mortality of M. sexta was assessed by counting the number of dead larvae at 2 d (), 4 d () and 6 d (■) after larvae were placed on tomato leaves supporting epiphytic populations of the specified bacterial strain. Bars show the cumulative mortality of larvae on leaves previously inoculated with (A) P. protegens Pf-5, a ΔfitD mutant of Pf-5, and P. fluorescens A506, a member of the P. fluorescens subgroup that lacks fitD; and (B) three strains in the P. chlororaphis subgroup, which possess fitD. Control larvae were placed on leaves that had not been inoculated with bacteria. Fifteen replicate larvae were evaluated per treatment in each of two experiments that yielded similar results; data presented are from a single experiment. Asterisks represent significant differences from the water control (P<0.05, d.f. = 1, χ2 test). The epiphytic population size of each strain on tomato leaflets, determined at the time that larvae were placed on the leaves, is shown below each graph.
Strains in the P. fluorescens group differ in oral toxicity to D. melanogaster
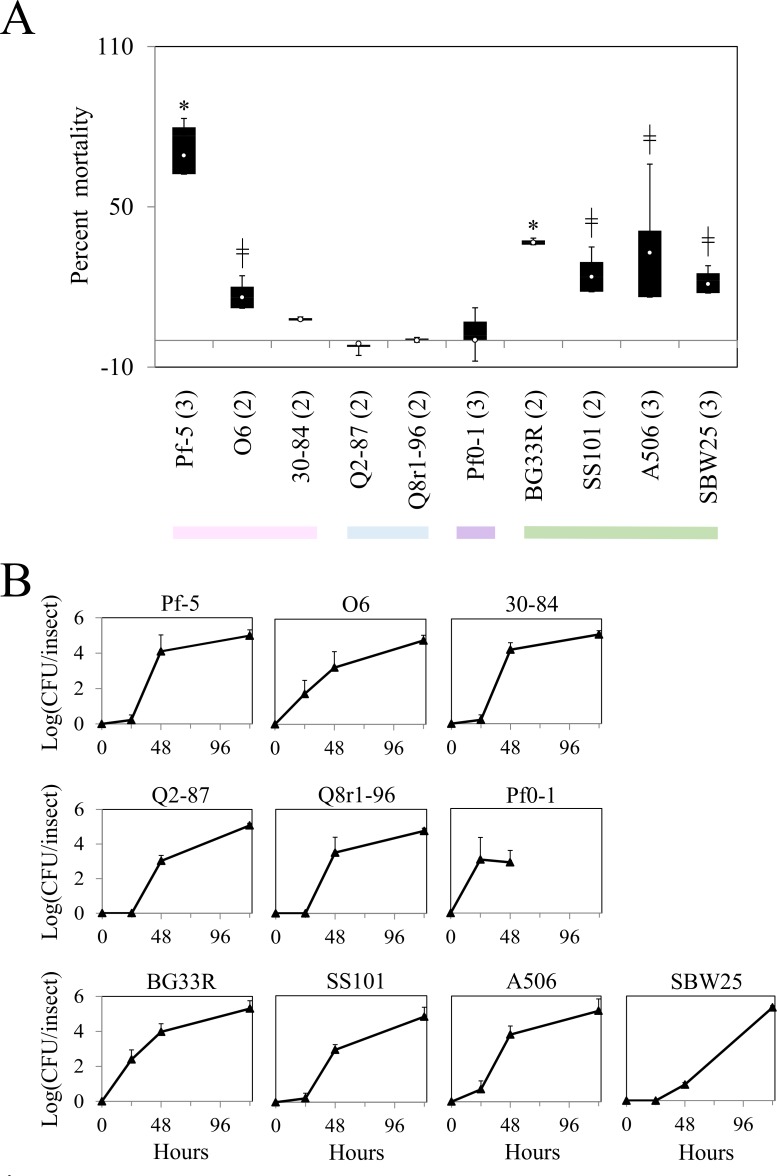
The ten strains of Pseudomonas spp. were evaluated for oral toxicity to D. melanogaster using a previously-developed non-invasive assay [10]. Averaging the results from three replicated experiments, 13% of the larvae died in the control treatment, in which larvae of D. melanogaster were fed with water-treated, killed yeast. In contrast, an average of 69% of the larvae fed P. protegens Pf-5-inoculated yeast died (Fig 5, S4 Table), which corresponds well with the previously-reported oral toxicity of Pf-5 to D. melanogaster [10]. The two other strains in the P. chlororaphis subgroup were evaluated in two experiments. Strain 30–84 caused no significant levels of mortality in either experiment whereas strain O6 caused a small but significant level of mortality in one experiment (Fig 5, S4 Table). In an earlier study, we reported that strain SBW25, a member of the P. fluorescens subgroup caused mortality of D. melanogaster [10]. In the present study, all four strains in the P. fluorescens subgroup caused significant mortality in at least one of the two to three experiments in which they were evaluated (Fig 5, S4 Table). Mortality caused by three of these strains (SS101, SBW25, and A506) varied among experiments, whereas BG33R caused significant mortality in both experiments in which it was evaluated. Neither strain in the P. corrugata subgroup (Q2-87 and Q8r1-96) nor Pf0-1 caused mortality of D. melanogaster in this assay (Fig 5, S4 Table). Populations of all ten strains increased inside larvae of D. melanogaster over the course of the experiment (Fig 5B), so it appears that all strains had the capacity to grow within the insect regardless of the magnitude of their influences on mortality.
Fig 5. Oral toxicity of strains of Pseudomonas spp. to D. melanogaster.
(A). The mortality of D. melanogaster was assessed 12 d after larvae were fed with yeast grains inoculated with ca. 7 log (CFU/plate). For each treatment, the percent mortality was calculated from counts of the number of adults per larvae in each replicate corrected for average larval to adult mortality in the control using the Schneider-Orelli formula. The white circle in each box shows the mean value from two or three experiments. The number of experiments evaluated for each strain is shown in parentheses. Boxes are bound at the top by the third quartile and at the bottom by the first quartile, with the whiskers representing the minimum and maximum values. An asterisk (*) denotes treatments that differed significantly from the control in all experiments and the double-cross symbol (╫) denotes treatments that differed significantly from the control in one experiment (P<0.05, d.f. = 2, χ2 test) (see S4 Table for data from individual experiments). The horizontal line indicates zero mortality. Colors denote the subgroup of the strain tested for insect toxicity: pink, P. chlororaphis; blue, P. corrugata; purple, P. koreensis; green, P. fluorescens. (B) The internal population size of each strain was estimated from surface-sterilized larvae at 24 h, 48 h, and 120 h. The population size of Pf0-1 was assessed only at 24 and 48 h. Bacterial population sizes were log transformed and the means of three replicate larvae are shown as triangular symbols. Error bars denoting standard errors are sometimes obscured by the symbols.
When fed yeast inoculated with any of the four strains in the P. fluorescens subgroup, larvae consistently developed a systemic melanization of the hemolymph during third instar stage prior to larval death (Fig 6B & 6C compared to 6A), as described earlier for strain SBW25 [10]. The activation of the prophenoloxidase cascade leading to the production of melanin is an important component of Drosophila innate immunity that is normally tightly regulated to prevent damage to the host due to overproduction of quinones and excessive melanization [30,31,32]. This regulation appears to be relaxed when the insect ingests bacteria in the P. fluorescens subgroup. In addition, other larvae died as pupae because certain phases of normal pupariation did not occur or were less successful. In some cases, the animals failed to undergo head involution and their larval mouth hooks extended through the pupal case (Fig 6E compared to 6D). In some animals, head involution occurred but the mouth hooks were not extruded successfully leading to a failure to seal the anterior pupal case and subsequent death as pharate adults (Fig 6F). As these animals developed, the head and eyes were smaller than in pupae that had formed normally and most failed to emerge as adults (Fig 6F). Larvae that did not develop the systemic melanization appeared to pupate at normal rates, indicating that ingestion of strains in the P. fluorescens subgroup did not cause developmental delay as found with sub-lethal doses of Pf-5 (S7 Fig).
Fig 6. Larval and pupal phenotypes of D. melanogaster fed with strains of Pseudomonas spp.
(A) Ventral view of a third instar control larva showing normal clear hemolymph and internal organs. (B) Side view of a dead SBW25-fed third instar larva showing systemic melanization of the hemolymph. (C) Dorsal view of a dead BG33R-fed third instar larva with complete melanization of the hemolymph. (D) Ventral view of a mid-stage control pupa showing normal extrusion of the mouthparts (arrow) and normal size of pupal eyes (brackets). (E) Dorsal view of a dead SS101-fed pre-pupa with extended mouthparts (arrow) and no head involution. (F) Dorsal view of a dead A506-fed pharate adult with extruded mouthparts caught within the pupal case (arrow). The head and eyes (brackets) are smaller and more recessed than in normal pupae.
Ingestion of P. protegens Pf-5 is known to cause a delay in the development of D. melanogaster [10]. Accordingly, larvae that fed on Pf-5 pupated later than the control in this study (S7 Fig). Of the other nine strains evaluated in this study, none caused notable delays in metamorphosis (S7 Fig), which is consistent with an earlier report that strains Pf0-1 and SBW25 do not cause developmental delay [10]. Our results suggest that developmental delay of D. melanogaster is associated with ingestion of the most toxic strain, Pf-5, but is not a common outcome of ingesting bacteria in the P. fluorescens group.
Discussion
The results of this study demonstrated that strains representing two lineages of the large and diverse P. fluorescens group exhibited toxicity against the lepidopteran M. sexta and the dipteran D. melanogaster. Three strains in the P. chlororaphis subgroup showed both injectable and oral toxicity to larvae of the tobacco hornworm, M. sexta, and one strain, P. protegens Pf-5, was also toxic to larvae of D. melanogaster when ingested. Three of four strains in the P. fluorescens subgroup were toxic when injected into M. sexta, and one strain consistently caused mortality of D. melanogaster when ingested. These results build upon a growing body of literature highlighting the insect toxicities of bacteria in the P. fluorescens group [12], including a recent study by Flury et al. [33] evaluating 26 strains of Pseudomonas spp. for toxicity to three lepidopteran species. Different insects and Pseudomonas strains were evaluated in the present study and in the study of Flury et al. [33] but the results of both studies show that strains in the P. chlororaphis subgroup and certain strains in the P. fluorescens subgroup exhibited insect toxicity whereas strains in the P. corrugata and P. koreensis subgroups did not exhibit consistent toxicity to insects. It remains to be seen if these taxonomic distinctions in insect toxicity hold up as more strains in the corrugata and koreensis subgroups are evaluated in the future.
Genes predicted to encode two distinct insect toxins (FitD/Mcf and Tc) known to be active in Photorhabdus spp. and other bacterial genera [34,35,36,37,38] are present in eight of the ten genomes of Pseudomonas spp. evaluated in this study. The three strains in the P. chlororaphis subgroup have the fit cluster, which is highly conserved among the strains in that subgroup that have been sequenced to date [15,16,33]. Seven of the ten strains have Tc gene clusters, identified from the presence of conserved domains and sequence similarity to regions encoding components of Tc toxins in Photorhabdus spp. and other entomopathogenic bacteria. Only P. chlororaphis 30–84 has both fitD and a Tc gene cluster, and bioinformatic analysis of the Tc gene cluster in strain 30–84 revealed characteristics of HGT. Here, we categorized the Tc gene clusters into six types based on their organization, phylogenies, and locations in the genome. For the most part, strains in a given subgroup shared Tc gene clusters of a single type, suggesting an ancestral inheritance of these clusters. Two of the ten strains evaluated in this study have two Tc clusters of different types in their genomes. Taken together, our analysis indicates that Tc gene clusters are inherited through a complex process involving HGT as well as vertical transmission through defined lineages of Pseudomonas.
A central objective of this study was to relate insect toxicity to the inventory of insect toxin genes in the genomes of diverse strains within the P. fluorescens group. The ten strains evaluated in this study have fully sequenced genomes that have been evaluated for the presence of genes for the production of exoenzymes, toxins, and metabolites that participate in the interactions of Pseudomonas spp. with insects, plants, and microorganisms [16]. Our results support a role for the FitD toxin in both injectable and oral toxicity to larvae of the lepidopteran M. sexta. All three strains (Pf-5, O6, and 30–84) that have a fit cluster caused injectable and oral toxicity to M. sexta, and a ΔfitD mutant of Pf-5 exhibited reduced oral toxicity (this study) and injectable toxicity to this insect [11,13]. These results support the known role of FitD in toxicity towards lepidopteran insects [11,13] and extend that role to oral toxicity against another insect pest, M. sexta. Only one of the seven strains lacking a fit cluster exhibited a significant level of oral toxicity to M. sexta in the single replicated experiment in which it was tested (S6 Fig). Therefore, fitD was the only factor identified in this study that contributed to oral toxicity of these ten strains against M. sexta. In contrast to oral toxicity, three strains lacking the fit cluster caused significant levels of injectable toxicity to M. sexta. These results provide convincing evidence that fitD, while important, is not the sole determinant of lepidopteran insect toxicity in the P. fluorescens group, as suggested earlier [11,13]. Identifying genes other than fitD that contribute to insect toxicity in these bacteria is a worthy goal for future study. Towards that end, this study identified strains within the P. fluorescens group that consistently cause injectable toxicity to M. sexta that could be explored in the future.
Tc gene clusters are among the many candidate genes that could contribute to injectable toxicity but we observed no correlation between the presence of Tc gene clusters and insect toxicity of the ten strains investigated here. We recognize, however, that different types of Tc gene clusters could vary in toxicity. For example, strains Q2-87 and Q8r1-96, which exhibited no insect toxicity in any of the three assays evaluated, have Type II and III Tc gene clusters whereas strains BG33R, A506 and SS101, which showed significant levels of injectable toxicity against M. sexta, have Type V Tc gene clusters. Strain BG33R, which also has a Type VI Tc gene cluster, consistently caused oral toxicity to D. melanogaster. The possibility that the Type V or VI Tc gene clusters contribute to insect toxicity is another worthy objective for future research. A tccC gene in P. taiwanesis is the only component of a Tc gene cluster that has been demonstrated to contribute to insect toxicity of Pseudomonas spp. [39]. Additional candidate insect toxicity genes may be identified from comparative genomic analysis [16] of strains A506, BG33R and SS101 versus strains that exhibited no toxicity to M. sexta in this study.
A different set of strains exhibited oral toxicity to the dipteran D. melanogaster versus the lepidopteran M. sexta, suggesting that distinct mechanisms of toxicity are operating in these insect hosts. Whereas fitD was the primary determinant of oral toxicity against M. sexta, as discussed above, this was not the case for toxicity against D. melanogaster. All three strains in the P. chlororaphis group have the fit cluster [16], but only Pf-5 consistently caused significant mortality of D. melanogaster in our assays. These results are consistent with those from a recent study demonstrating that FitD is not required for oral toxicity of P. protegens Pf-5 to D. melanogaster [40]. Instead, analogs of rhizoxin, orfamide A, and chitinase are the primary determinants of oral toxicity of Pf-5 against D. melanogaster. Rhizoxin is a 16-member macrolide that binds to β-tubulin, thereby interfering with microtubule dynamics during mitosis, and is toxic to many eukaryotes [41]. Orfamide A is a cyclic lipopeptide with surfactant properties that aids in bacterial motility across surfaces and solubilization of certain substrates [42], inhibition of some oomycetes and fungi [42,43,44], and toxicity to the green peach aphid [45]. Chitinases can degrade the peritrophic membrane, a chitin-based matrix in the insect mid-gut that functions in protection against mechanical and chemical damage and serves as a barrier to infection by pathogens [46]. Of the ten strains evaluated in this study, only Pf-5 produces orfamide A or rhizoxin analogs, but several strains that exhibited some level of toxicity to D. melanogaster have genes for the production of cyclic lipopeptides, chitinase, as well as other secondary metabolites that have not yet been evaluated for their potential roles in insect toxicity [16].
A striking observation of this study was the induction of melanization in adults that escaped mortality when fed any of the four strains in the P. fluorescens subgroup. Melanization is a conspicuous immune response that results in the production of quinones that are toxic to microorganisms [32]. It is intriguing that the strains in the P. fluorescens subgroup cause a pronounced melanization response in larvae and adults, but the animals that survive infection by P. protegens Pf-5, the most lethal strain of this study, do not exhibit this response. The mechanisms responsible for inducing this immune response are unknown but worthy of future study.
The results of this study highlight the specificity of the insect-bacterial interaction, as different strains caused mortality on the two insect hosts and when inoculated by feeding versus injection. Furthermore, evaluation of fitD mutants in this and a companion study [40] show that even a specific bacterial strain, such as P. protegens Pf-5, employs different lethality mechanisms on different insect hosts. Clearly, there are distinct mechanisms by which different strains of Pseudomonas spp. kill insects, and this study identified systems that could be explored to identify novel mechanisms of insect toxicity.
Materials and Methods
Generation of Multi-Locus Sequence Analysis (MLSA) phylogenetic tree
MAFFT v. 7.245 [47] was used to generate alignments for gyrB, rpoD, rpoB, and 16S rRNA from the type strains of the P. fluorescens group (S5 Table) and from the ten strains of Pseudomonas sp. listed in Table 1. Accession numbers for all sequences are provided in S5 Table. Alignments of gappy columns were trimmed using Gblocks [48]. The concatenated nucleotide sequences from each organism were used as input for a partitioned maximum-likelihood phylogenetic analysis using RAxML v. 8.1.21 [49], using GTR+GAMMA as the substitution model for each partition. Trees were generated according to the guidelines provided in the RAxML v. 8 user’s manual, with 100 individual maximum-likelihood searches performed and 450 bootstrap replicates were completed using the extended majority rule-based bootstrapping criterion [49].
Table 1. Strains of the Pseudomonas fluorescens group evaluated in this study.
| Strain | Site where strain was isolated | Description | Source |
|---|---|---|---|
| P. chlororaphis subgroup: | |||
| P. chlororaphis 30–84 | Wheat rhizosphere, Washington, USA | Suppresses take-all of wheat. RifR, [50] | L.S. Pierson II, Texas A&M, College Station, TX USA |
| P. chlororaphis O6 | Soil, Utah, USA | Suppresses several plant diseases [51] | A. Anderson, Utah State University, Logan Utah, USA |
| P. protegens Pf-5 | Soil, Texas, USA | Suppresses seedling emergence diseases [24,52]. Also called JL4585. | C. Howell, USDA-ARS, College Station, TX, USA |
| P. protegens Pf-5 ΔfitD | Mutant of Pf-5 with a deletion in fitD [40] | ||
| P. corrugata subgroup: | |||
| P. brassicacearum Q8r1-96 | Wheat rhizosphere, Washington, USA | Suppresses take-all of wheat [53] | D. Weller, USDA-ARS, Pullman WA, USA |
| Q2-87 | Wheat rhizosphere, Washington, USA | Suppresses take-all of wheat [54] | D. Weller, USDA-ARS, Pullman WA, USA |
| P. koreensis subgroup: | |||
| Pf0-1 | Soil, Massachusetts, USA | Soil bacterium [55] | M. Silby, University of Massachusetts, Princeton, MA, USA |
| P. fluorescens subgroup: | |||
| SBW25 | Sugar beet phyllosphere, Oxfordshire, UK | Phyllosphere bacterium [55] | G. Preston, Oxford University, UK |
| SBW25-RifR | Spontaneous mutant of SBW25 selected for resistance to rifampicin, RifR | J. M. Raaijmakers, Netherlands Institute of Ecology, Wageningen, The Netherlands | |
| A506 | Pear phyllosphere, California, USA | Suppresses fire blight of pear and apple; frost injury, fruit russet. RifR [7,56] | S. E. Lindow, University of California, Berkeley, CA, USA |
| SS101 | Wheat rhizosphere, The Netherlands | Suppresses diseases caused by Pythium spp. and Phytophthora spp. RifR [57, 58] | J. M. Raaijmakers, Netherlands Institute of Ecology, Wageningen, The Netherlands |
| BG33R | Peach rhizosphere, South Carolina, USA | Suppresses the plant-parasitic nematode Mesocriconema xenoplax [59] | D. Kluepfel, USDA-ARS, Davis, CA, USA |
| BG33R-RifR | Spontaneous mutant of BG33R selected for resistance to rifampicin, RifR. | This study | |
Abbreviation: RifR, resistant to rifampicin (100 μg/ml).
Bioinformatic analysis of Tc clusters in genomes of Pseudomonas spp.
Using the nucleotide sequences of well-characterized Tc genes from P. luminescens W14 as queries, a BLASTN search was performed against the ten genomes of P. fluorescens to identify Tc genes. Conserved domains were identified by searching the Pfam database [60] and the conserved domain database of the National Center for Biotechnology Information (NCBI) [61]. Multiple sequence alignments of the Tc peptide sequences were executed using the MAFFT option in MegAlign Pro (DNAStar). Three unrooted phylogenetic trees were created using BIONJ with each putatively annotated amino acid sequence for each Tc component: A (TcaA, TcaB, and TcdA), B (TcaC and TcdB) and C (TccC). The G+C contents of tcdA and tcdB were determined using the MBCF Oligo Calculator (http://mbcf.dfci.harvard.edu/docs/oligocalc.html) and normalized to gene size. Significant differences in percent G+C from the genomic average were identified by chi-square analysis using a two-tailed P-value.
Assessing injectable toxicity of Pseudomonas spp. to M. sexta
Injectable toxicities of ten strains of Pseudomonas spp. (Table 1) to M. sexta were assessed as described previously [11]. Briefly, bacteria were grown in 5 ml of King’s medium B (KMB) broth [62] for 24 h at 27°C. Cells were collected by centrifugation, washed, resuspended in sterile water at OD600 = 0.01 (ca. 7 log[CFU/ml]), and diluted to obtain cell densities specified in the Results. Dilutions of the cell suspensions were spread on KMB to determine titers of the inoculum injected into larvae of M. sexta. For each treatment, ten fifth instar larvae were injected between the second and third abdominal segments with 10 μl of water or bacterial suspension. Larvae were then placed in individual containers, maintained in an incubator at 16:8 h (L:D) and 27°C, and assessed for mortality over time. Each bacterial strain was evaluated in at least two replicated experiments. A chi-squared analysis was performed for each experiment, in which the negative control (sterile water) was compared to each treatment individually.
To monitor bacterial colonization of M. sexta, larvae were inoculated with ca. 5 log (CFU/larva) rifampicin-resistant mutants of Pseudomonas spp. and incubated as described above. Larvae with a healthy appearance were sampled immediately after inoculation and at 6, 12, 18, 24, and 30 h after injection. Each larva was surface-disinfested for 30 s in 70% ethanol, rinsed with sterile water, and then homogenized for 30 s in 10 ml of sterile distilled H2O using a Tekmar SDT Tissumizer (Cincinnati, OH, USA). Serial dilutions were prepared from the resulting homogenate and plated onto KMB containing rifampicin (100 μg/ml) and cycloheximide (50 μg/ml) to select for the injected strains. Plates were incubated for 24 h at 27°C prior to enumeration of bacterial colonies. The experiment was done twice with similar results and a representative experiment is presented.
Assessing oral toxicity of Pseudomonas spp. to M. sexta
Oral toxicity of Pseudomonas spp. to M. sexta was assessed on excised leaves of tomato (cv. Patio). Excised leaves were dipped in an aqueous bacterial suspension prepared from overnight cultures grown in KMB broth; cells were collected by centrifugation and suspended in sterile water to an OD600 of 0.5, which corresponds to approximately 9 log (CFU/ml). Leaves were placed in plastic containers lined with moistened paper towels. Containers were placed in a lighted growth chamber at 27°C to allow epiphytic bacterial populations to develop on leaf surfaces. After the 24 h incubation period, three leaflets were removed from each treatment, placed in separate tubes containing 10 mM potassium phosphate buffer (pH 7.0), and vortexed. The number of CFU of bacterial strains was assessed by serial dilution of leaflet washings on KMB. For each treatment, fifteen larvae, ranging in size from 18 to 30 mg, were placed on leaves in separate containers (i.e., one larvae per container; 15 containers/treatment). Containers were placed in a growth chamber maintained at 27°C with 17:7 h (L:D), 40–50% relative humidity). Larval mortality was assessed at 2, 4, and 6 d after their placement on leaves. One to five of the fifteen larvae feeding on water-treated leaves died in each of the seven experiments, which was probably due in part to injuries caused by handling the larvae as they were placed on the leaves. Due to logistical constraints, only four treatments could be assessed in each experiment. A chi-squared analysis was performed to compare each bacterial treatment to the water treatment within each experiment.
Assessing oral toxicity of Pseudomonas spp. to D. melanogaster
Oral toxicity of Pseudomonas spp. to D. melanogaster was assessed using a non-invasive assay described previously [10]. Briefly, adult flies were transferred to Petri plates containing Apple Agar (http://cshprotocols.cshlp.org/content/2011/9/pdb.recO65672.short) prepared without Nipagin and supplemented by placing killed yeast grains (20 mg) on the solidified agar surface. Plates were incubated for four hours at 25°C to allow egg lay. From these plates, thirty eggs were transferred aseptically to the surface of non-nutritive agar (2% wt/vol agar in water) with 2 to 3 mg killed yeast grains distributed on the agar surface in a 35mm Petri plate. On Day 1, the number of first instar larvae was determined by counting the number of empty egg cases. On Day 2, 200 μl of a yeast suspension was added to the middle of the plate to serve as a food source for second instar larvae. The yeast suspension was prepared by dissolving 0.2 g yeast in 1.2 ml of sterile water (control) or a bacterial suspension (7.4 ± 0.6 log[CFU/ml]), prepared as described above. The plates were then incubated at 22°C and, starting at Day 4, larvae were fed with 100 μl of a yeast suspension (0.2 mg yeast/1.2 ml sterile water) at 48-h intervals as long as live larvae were observed in the dish. Numbers of pupae and adults were counted daily. Each bacterial strain was tested in at least two experiments having three replications per strain (S4 Table). For each experiment, percent mortality for each treatment was calculated from the number of adults on Day 12 per the number of larvae on Day 1 and averaged over the replicate plates. Mean percent mortality values for each experiment were corrected for the mortality in the control yeast-only plates using the Schneider-Orelli formula (% corrected adult mortality = [(% adult mortality of treated larvae—% adult mortality of control larvae)/(100—% adult mortality of control larvae)] [63]. A chi-squared analysis of the corrected mortality data was used to detect significant differences of treatments from the control in each experiment.
Population sizes of the bacterial strains internal to larvae or pupae of D. melanogaster were estimated from surface-sterilized larvae. Three larvae were placed in a drop of a freshly-made sterilizing solution (2.5 ml bleach and 45 μl 0.01% Triton-X in 10 ml water) for 60 seconds and then transferred serially through four drops of sterile water and placed individually into a 1.5 ml microcentrifuge tube. Each insect was then homogenized in 50 μl of sterile distilled water, serially diluted, and the dilutions were spread on KMB. Plates were incubated overnight at 27°C prior to counting colonies that exhibited the green-blue fluorescence characteristic of the inoculated strains.
Larval and pupal morphologies were documented through images captured by a digital camera mounted on a dissecting scope with images adjusted only for contrast and brightness in Photoshop (Adobe Systems Inc., San Jose, CA, USA) as needed.
Supporting Information
The A-component tree contains seven TcaA sequences, seven TcaB sequences, and one TcdA sequence from the ten strains within the P. fluorescens group examined in this study (shown in red font) as well as BLASTP hits with greater than 75% query coverage and 50% identity to one or more of these sequences. The tree also includes all of the characterized A-component peptide sequences (TcaA, TcbA, TcdA) from P. luminescens W14 (shown in blue font). Phylogenetic relationships support the placement of the Tc clusters into the six types (Types I to VI), with components of the same Tc type from different strains grouping within the same clades. Boxes show the Tc type and are colored to denote the subgroup of strains shown in red font: pink, chlororophis; blue, corrugata; green, fluorescens, as depicted in Fig 2.
(PDF)
The B component tree contains eight TcaC sequences and one TcdB sequence from the ten strains within the P. fluorescens group examined in this study (shown in red font) as well as BLASTP hits with greater than 75% query coverage and 50% identity to one or more of these sequences. The tree also includes all of the characterized B-component peptide sequences (TcaC and TcdB) from P. luminescens W14 (shown in blue font). All TcaC sequences grouped together with homologs from other Pseudomonas sp. in a single large clade, whereas the Type I TcdC sequence fell outside of this clade and grouped with the only BLASTP hit outside of the Pseudomonas genus. Phylogenetic relationships support the placement of the Tc clusters into the six types (Types I to VI), with components of the same Tc type from different strains grouping together. Boxes show the Tc type and are colored to denote the subgroup of strains shown in red font: pink, chlororophis; blue, corrugata; purple, koreensis; green, fluorescens, as depicted in Fig 2.
(PDF)
The C-component tree contains 15 TccC sequences from the ten strains within the P. fluorescens group examined in this study (shown in red font) as well as BLASTP hits with greater than 75% query coverage and 50% identity to one or more of these sequences. The tree also includes all characterized C-component peptide sequences (TccC) from P. luminescens W14 (shown in blue font). The six types of Tc clusters (I to VI) fall into distinct clades, but both the Type II and Type III TccC sequences are dispersed in the tree. Boxes show the Tc type and are colored to denote the subgroup of strains shown in red font: pink, chlororaphis; blue, corrugata; purple, koreensis; green, fluorescens, as depicted in Fig 2.
(PDF)
Lethality of strains in the P. fluorescens group to fifth instar of the tobacco hornworm, M. sexta. Mortality was assessed at 24h (), 48h (), or 72h (■) following injection with ca. 5 log(CFU per larva) or water, as a control. Three experiments (A, B and C), each evaluating ten replicate larvae per treatment, are presented. The 72 h observations for experiment A are shown in Fig 3. Asterisks represent a significant difference in mortality between a treatment and the control (P<0.05, d.f. = 1, χ2 test).
(TIF)
Lethality of high cell densities of strains in the P. fluorescens group to fifth instar of the tobacco hornworm, M. sexta. Mortality was assessed at 24h (), 48h (), or 72h (■) following injection with ca. 6 log(CFU per larva) or water, as a control. Two experiments (A and B), each evaluating ten replicate larvae per treatment, are presented. Asterisks represent a significant difference in mortality between a treatment and the control (P<0.05, d.f. = 1, χ2 test).
(TIF)
Cumulative mortality of M. sexta was assessed by counting the number of dead larvae at 2 d (), 4 d () and 6 d (■) after larvae were placed on tomato leaves supporting epiphytic populations of the specified bacterial strain. Strain Pf0-1 gacA+ (also called LK194) is a derivative of strain Pf0-1 with a chromosomal insertion of gacA [64]. Controls were larvae on leaves that were not inoculated with bacteria. (A-E) Each panel shows the results from an individual experiment, with fifteen replicate larvae evaluated per treatment in each experiment. Values that differ significantly from the control at the designated time are shown with an asterisk (P<0.05) or a diamond (P<0.10) (d.f. = 1, χ2 test). The epiphytic population size of each strain on tomato leaflets, determined at the time that larvae were placed on the leaves, is shown below each graph.
(TIF)
Developmental time course of D. melanogaster after ingestion of strains in the A) P. chlororaphis, B) P. koreensis or P. corrugata, or C) P. fluorescens subgroups. Second instar larvae were fed with a yeast suspension having no bacteria (black) or amended with bacterial strains. Initial population sizes of bacterial strains [log (CFU/plate)] are shown to the right of each panel. The percentage of larvae that pupated, counted as prepupae and/or pupae were determined over time. The percentage of larvae that emerged as adults is shown at the 288 hpi time point. Values represent the mean and standard errors from three replicates per treatment, with each replicate evaluating the larvae and adults that developed from 30 eggs. A ΔgacA mutant of Pf-5 (JL4975) [65] was included as a negative control, as it was shown previously to lack toxicity to D. melanogaster [10].
(TIF)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
Acknowledgments
We gratefully acknowledge Ashley Bixenstein, Kise Bond, Max Kohen, Molly Albrecht, Kelly Donahue, and Amanda Lake for assisting with the experiments described herein. We thank Anne Anderson, Charles Howell, Dan Kleupfel, Steve Lindow, Gail Preston, Leland S. Pierson II, Jos Raaijmakers, Mark Silby, and David Weller for providing bacterial strains used in this study.
Data Availability
All relevant data are within the paper and supporting information files.
Funding Statement
LIR was supported by a Provost’s Distinguished Graduate Fellowship from Oregon State University. This work was supported by Agriculture and Food Research Initiative Competitive Grant 2008-35600-18770 and 2011-67019-30192 from the United States Department of Agriculture National Institute of Food and Agriculture.
References
- 1.Gomila M, Pena A, Mulet M, Lalucat J, Garcia-Valdes E (2015) Phylogenomics and sytematics in Pseudomonas. Front Microbiol 6: 214 10.3389/fmicb.2015.00214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garrido-Sanz D, Meier-Kolthoff JP, Göker M, Martín M, Rivilla R, et al. (2016) Genomic and genetic diversity within the Pseudomonas fluorescens complex. PLoS ONE 11: e0150183 10.1371/journal.pone.0150183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Défago G, Haas D (1990) Pseudomonads as antagonists of soilborne plant pathogens: modes of action and genetic analysis In: Bollag J-M, Stotzky G, editors. Soil Biochemistry. New York: Marcel Dekker, Inc; pp. 249–291. [Google Scholar]
- 4.Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, et al. (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332: 1097–1100. 10.1126/science.1203980 [DOI] [PubMed] [Google Scholar]
- 5.Weller DM, Raaijmakers JM, McSpadden Gardener BB, Thomashow LS (2002) Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu Rev Phytopathol 40: 309–348. [DOI] [PubMed] [Google Scholar]
- 6.Höfte M, Altier N (2010) Fluorescent pseudomonads as biocontrol agents for sustainable agricultural systems. Res Microbiol 161: 464–471. 10.1016/j.resmic.2010.04.007 [DOI] [PubMed] [Google Scholar]
- 7.Stockwell VO, Stack JP (2007) Using Pseudomonas spp. for integrated biological control. Phytopathology 97: 244–249. 10.1094/PHYTO-97-2-0244 [DOI] [PubMed] [Google Scholar]
- 8.Commare RR, Nandakumar R, Kandan A, Suresh S, Bharathi M, et al. (2002) Pseudomonas fluorescens based bio-formulation for the management of sheath blight disease and leaffolder insect in rice. Crop Protect 21: 671–677. [Google Scholar]
- 9.Otsu Y, Matsuda Y, Mori H, Ueki H, Nakajima T, et al. (2004) Stable phylloplane colonization by entomopathogenic bacterium Pseudomonas fluorescens KPM-018P and biological control of phytophagous ladybird beetles Epilachna vigintioctopunctata (Coleoptera: Coccinellidae). Biocontrol Sci Technol 14: 427–439. [Google Scholar]
- 10.Olcott MH, Henkels MD, Rosen KL, Walker FL, Sneh B, et al. (2010) Lethality and developmental delay in Drosophila melanogaster larvae after ingestion of selected Pseudomonas fluorescens strains. PLoS ONE 5: e12504 10.1371/journal.pone.0012504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Péchy-Tarr M, Bruck D, Maurhofer M, Fischer E, Vogne C, et al. (2008) Molecular analysis of a novel gene cluster encoding an insect toxin in plant-associated strains of Pseudomonas fluorescens. Environ Microbiol 10: 2368–2386. 10.1111/j.1462-2920.2008.01662.x [DOI] [PubMed] [Google Scholar]
- 12.Kupferschmied P, Maurhofer M, Keel C (2013) Promise for plant pest control: root-associated pseudomonads with insecticidal activities. Front Plant Sci 4: 287 10.3389/fpls.2013.00287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruffner B, Péchy-Tarr M, Ryffel F, Hoegger P, Obrist C, et al. (2013) Oral insecticidal activity of plant-associated pseudomonads. Environ Microbiol 15: 751–763. 10.1111/j.1462-2920.2012.02884.x [DOI] [PubMed] [Google Scholar]
- 14.Chen W-J, Hsieh F-C, Hsu F-C, Tasy Y-F, Liu J-R, et al. (2014) Characterization of an insecticidal toxin and pathogenicity of Pseudomonas taiwanensis against insects. PLoS Path 10: e1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruffner B, Péchy-Tarr M, Höfte M, Bloemberg G, Grunder J, et al. (2015) Evolutionary patchwork of an insecticidal toxin shared between plant-associated pseudomonads and the insect pathogens Photorhabdus and Xenorhabdus. BMC Genomics 16: 609 10.1186/s12864-015-1763-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loper JE, Hassan KA, Mavrodi DV, Davis EW II, Lim CK, et al. (2012) Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet 8: e1002784 10.1371/journal.pgen.1002784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daborn PJ, Waterfield N, Silva CP, Au CPY, Sharma S, et al. (2002) A single Photorhabdus gene, makes caterpillars floppy (mcf), allows Escherichia coli to persist within and kill insects. Proc Natl Acad Sci USA 99: 10742–10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dowling AJ, Daborn PJ, Waterfield NR, Wang P, Streuli CH, et al. (2004) The insecticidal toxin Makes caterpillars floppy (Mcf) promotes apoptosis in mammalian cells. Cell Microbiol 6: 345–353. [DOI] [PubMed] [Google Scholar]
- 19.Waterfield NR, Daborn PJ, Dowling AJ, Yang GW, Hares M, et al. (2003) The insecticidal toxin makes caterpillars floppy 2 (Mcf2) shows similarity to HrmA, an avirulence protein from a plant pathogen. FEMS Microbiol Lett 229: 265–270. [DOI] [PubMed] [Google Scholar]
- 20.Ffrench-Constant RH, Dowling A, Waterfield NR (2007) Insecticidal toxins from Photorhabdus bacteria and their potential use in agriculture. Toxicon 49: 436–451. [DOI] [PubMed] [Google Scholar]
- 21.Ffrench-Constant RH, Waterfield NR (2006) An ABC guide to the bacterial toxin complexes. Adv Appl Microbiol 58: 169–183. [PubMed] [Google Scholar]
- 22.Ffrench-Constant RH, Bowen DJ (2000) Novel insecticidal toxins from nematode-symbiotic bacteria. Cell Mol Life Sci 57: 828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waterfield NR, Hares M, Yang G, Dowling AJ, Ffrench-Constant RH (2005) Potentiation and cellular phenotypes of the insecticidal toxin complexes of Photorhabdus bacteria. Cell Microbiol 7: 373–382. [DOI] [PubMed] [Google Scholar]
- 24.Paulsen IT, Press CM, Ravel J, Kobayashi DY, Myers GSA, et al. (2005) Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat Biotechnol 23: 873–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silby MW, Cerdeño-Tarraga AM, Vernikos GS, Giddens SR, Jackson RW, et al. (2009) Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol 10: R51 10.1186/gb-2009-10-5-r51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guiney DG, Fierer J (2011) The role of the spv genes in Salmonella pathogenesis. Front Microbiol 2: 129 10.3389/fmicb.2011.00129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waterfield NR, Bowen DJ, Fetherston JD, Perry RD, ffrench-Constant RH (2001) The tc genes of Photorhabdus: a growing family. Trends Microbiol 9: 185–191. [DOI] [PubMed] [Google Scholar]
- 28.Joo Lee P, Ahn J-Y, Kim Y-H, Wook Kim S, Kim J-Y, et al. (2004) Cloning and heterologous expression of a novel insecticidal gene (tccC1) from Xenorhabdus nematophilus strain. Biochem Biophys Res Commun 319: 1110–1116. [DOI] [PubMed] [Google Scholar]
- 29.Vernikos GS, Parkhill J (2006) Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics 22: 2196–2203. [DOI] [PubMed] [Google Scholar]
- 30.Bidla G, Hauling T, Dushay MS, Theopold U (2009) Activation of insect phenoloxidase after injury: endogenous versus foreign elicitors. J Innate Immun 1: 301–308. 10.1159/000168009 [DOI] [PubMed] [Google Scholar]
- 31.Kan H, Kim C-H, Kwon H-M, Park J-W, Roh K-B, et al. (2008) Molecular control of phenoloxidase-induced melanin synthesis in an insect. J Biol Chem 283: 25316–25323. 10.1074/jbc.M804364200 [DOI] [PubMed] [Google Scholar]
- 32.Tang H (2009) Regulation and function of the melanization reaction in Drosophila. Fly (Austin) 3: 105–111. [DOI] [PubMed] [Google Scholar]
- 33.Flury P, Aellen N, Ruffner B, Péchy-Tarr M, Fataar S, et al. (2016) Insect pathogenicity in plant-beneficial pseudomonads: phylogenetic distribution and comparative genomics. ISME J. 10.1038/ismej.2016.5 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang G, Dowling AJ, Gerike U, Ffrench-Constant RH, Waterfield NR (2006) Photorhabdus virulence cassettes confer injectable insecticidal activity against the wax moth. J Bacteriol 188: 2254–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuchs T, Bresolin G, Marcinowski L, Schachtner J, Scherer S (2008) Insecticidal genes of Yersinia spp.: taxonomical distribution, contribution to toxicity towards Manduca sexta and Galleria mellonella, and evolution. BMC Microbiol 8: 214 10.1186/1471-2180-8-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hinchliffe SJ, Hares MC, Dowling AJ, Ffrench-Constant RH (2010) Insecticidal toxins from the Photorhabdus and Xenorhabdus bacteria. Open Tox J 3: 101–118. [Google Scholar]
- 37.Hurst MRH, Jones SA, Binglin T, Harper LA, Jackson TA, et al. (2011) The main virulence determinant of Yersinia entomophaga MH96 is a broad-host-range toxin complex active against insects. J Bacteriol 193: 1966–1980. 10.1128/JB.01044-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shrestha YK, Jang EK, Yu YS, Kwon M, Shin JH, et al. (2011) Oral toxicity of symbiotic bacteria Photorhabdus spp. against immature stages of insects. J Asia-Pacif Entomol 14: 127–130. [Google Scholar]
- 39.Liu J-R, Lin Y-D, Chang S-T, Zeng Y-F, Wang S-L (2010) Molecular cloning and characterization of an insecticidal toxin from Pseudomonas taiwanensis. J Agric Food Chem 58: 12343–12349. 10.1021/jf103604r [DOI] [PubMed] [Google Scholar]
- 40.Loper JE, Henkels MD, Rangel LI, Olcott MH, Rosen KL, et al. (2016) Rhizoxin analogs, orfamide A, and chitinase production contribute to the toxicity of Pseudomonas protegens strain Pf-5 to Drosophila melanogaster. Environ Microbiol. 10.1111/1462-2920.13369 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 41.Gupta S, Bhattacharyya B (2003) Antimicrotubular drugs binding to vinca domain of tubulin. Mol Cell Biochem 253: 41–47 [DOI] [PubMed] [Google Scholar]
- 42.Gross H, Stockwell VO, Henkels MD, Nowak-Thompson B, Loper JE, et al. (2007) The genomisotopic approach: a systematic method to isolate products of orphan biosynthetic gene clusters. Chem Biol 14: 53–63. [DOI] [PubMed] [Google Scholar]
- 43.Olorunleke FE, Hua GKH, Kieu NP, Ma Z, Höfte M (2015) Interplay between orfamides, sessilins and phenazines in the control of Rhizoctonia diseases by Pseudomonas sp. CMR12a. Environ Microbiol Rep 7: 774–781. 10.1111/1758-2229.12310 [DOI] [PubMed] [Google Scholar]
- 44.Ma Z, Geudens N, Kieu NP, Sinnaeve D, Ongena M, et al. (2016) Biosynthesis, chemical structure and structure-activity relationship of orfamide lipopeptides produced by Pseudomonas protegens and related species. Front Microbiol 7: 382 10.3389/fmicb.2016.00382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jang JY, Yang SY, Kim YC, Lee CW, Park MS, et al. (2013) Identification of Orfamide A as an insecticidal metabolite produced by Pseudomonas protegens F6. J Agric Food Chem 61: 6786–6791. 10.1021/jf401218w [DOI] [PubMed] [Google Scholar]
- 46.Lehane MJ (1997) Peritrophic matrix structure and function. Annu Rev Entomol 42: 525–550. [DOI] [PubMed] [Google Scholar]
- 47.Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540–552. [DOI] [PubMed] [Google Scholar]
- 49.Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomashow LS, Weller DM, Bonsall RF, Pierson LS (1990) Production of the antibiotic phenazine-1-carboxylic acid by fluorescent Pseudomonas species in the rhizosphere of wheat. Appl Environ Microbiol 56: 908–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim MS, Kim YC, Cho BH (2004) Gene expression analysis in cucumber leaves primed by root colonization with Pseudomonas chlororaphis O6 upon challenge-inoculation with Corynespora cassiicola. Plant Biol 6: 105–108. [DOI] [PubMed] [Google Scholar]
- 52.Howell CR, Stipanovic RD (1979) Control of Rhizoctonia solani on cotton seedlings with Pseudomonas fluorescens and with an antibiotic produced by the bacterium. Phytopathology 69: 480–482. [Google Scholar]
- 53.Raaijmakers JM, Weller DM (1998) Natural plant protection by 2,4-diacetylphloroglucinol: producing Pseudomonas spp. in take-all decline soils. Mol Plant Microbe Interact 11: 144–152. [Google Scholar]
- 54.Vincent MN, Harrison LA, Brackin JM, Kovacevich PA, Mukerji P, et al. (1991) Genetic analysis of the antifungal activity of a soilborne Pseudomonas aureofaciens strain. Appl Environ Microbiol 57: 2928–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silby MW, Cerdeno-Tarraga AM, Vernikos GS, Giddens SR, Jackson RW, et al. (2009) Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol 10: R51 10.1186/gb-2009-10-5-r51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson M, Lindow SE (1993) Interactions between the biological control agent Pseudomonas fluorescens A506 and Erwinia amylovora in pear blossoms. Phytopathology 83: 117–123. [Google Scholar]
- 57.Mazzola M, Zhao X, Cohen MF, Raaijmakers JM (2007) Cyclic lipopeptide surfactant production by Pseudomonas fluorescens SS101 is not required for suppression of complex Pythium spp. populations. Phytopathology 97: S72–S72. [DOI] [PubMed] [Google Scholar]
- 58.Tran H, Ficke A, Asiimwe T, Hofte M, Raaijmakers JM (2007) Role of the cyclic lipopeptide massetolide A in biological control of Phytophthora infestans and in colonization of tomato plants by Pseudomonas fluorescens. New Phytol 175: 731–742. [DOI] [PubMed] [Google Scholar]
- 59.Kluepfel DA, Mcinnis TM, Zehr EI (1993) Involvement of root-colonizing bacteria in peach orchard soils suppressive of the nematode Criconemella xenoplax. Phytopathology 83: 1240–1245. [Google Scholar]
- 60.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, et al. (2012) The Pfam protein families database. Nucleic Acids Res 40: D290–D301. 10.1093/nar/gkr1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, et al. (2015) CDD: NCBI's conserved domain database. Nucleic Acids Res 43: D222–D226. 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 44: 301–307. [PubMed] [Google Scholar]
- 63.Püntener W (1981) Manual for Field Trials in Plant Protection. Basel, Switzerland: Ciba-Geigy Limited; 68. [Google Scholar]
- 64.Seaton SC, Silby MW, Levy SB (2013) Pleiotropic effects of GacA on Pseudomonas fluorescens Pf0-1 in vitro and in soil. Appl Environ Microbiol 79: 5405–5410. 10.1128/AEM.00819-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Henkels MD, Kidarsa TA, Shaffer BT, Goebel NC, Burlinson P, et al. (2014) Pseudomonas protegens Pf-5 causes discoloration and pitting of mushroom caps due to the production of antifungal metabolites. Mol Plant Microbe Interact 27: 733–746. 10.1094/MPMI-10-13-0311-R [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The A-component tree contains seven TcaA sequences, seven TcaB sequences, and one TcdA sequence from the ten strains within the P. fluorescens group examined in this study (shown in red font) as well as BLASTP hits with greater than 75% query coverage and 50% identity to one or more of these sequences. The tree also includes all of the characterized A-component peptide sequences (TcaA, TcbA, TcdA) from P. luminescens W14 (shown in blue font). Phylogenetic relationships support the placement of the Tc clusters into the six types (Types I to VI), with components of the same Tc type from different strains grouping within the same clades. Boxes show the Tc type and are colored to denote the subgroup of strains shown in red font: pink, chlororophis; blue, corrugata; green, fluorescens, as depicted in Fig 2.
(PDF)
The B component tree contains eight TcaC sequences and one TcdB sequence from the ten strains within the P. fluorescens group examined in this study (shown in red font) as well as BLASTP hits with greater than 75% query coverage and 50% identity to one or more of these sequences. The tree also includes all of the characterized B-component peptide sequences (TcaC and TcdB) from P. luminescens W14 (shown in blue font). All TcaC sequences grouped together with homologs from other Pseudomonas sp. in a single large clade, whereas the Type I TcdC sequence fell outside of this clade and grouped with the only BLASTP hit outside of the Pseudomonas genus. Phylogenetic relationships support the placement of the Tc clusters into the six types (Types I to VI), with components of the same Tc type from different strains grouping together. Boxes show the Tc type and are colored to denote the subgroup of strains shown in red font: pink, chlororophis; blue, corrugata; purple, koreensis; green, fluorescens, as depicted in Fig 2.
(PDF)
The C-component tree contains 15 TccC sequences from the ten strains within the P. fluorescens group examined in this study (shown in red font) as well as BLASTP hits with greater than 75% query coverage and 50% identity to one or more of these sequences. The tree also includes all characterized C-component peptide sequences (TccC) from P. luminescens W14 (shown in blue font). The six types of Tc clusters (I to VI) fall into distinct clades, but both the Type II and Type III TccC sequences are dispersed in the tree. Boxes show the Tc type and are colored to denote the subgroup of strains shown in red font: pink, chlororaphis; blue, corrugata; purple, koreensis; green, fluorescens, as depicted in Fig 2.
(PDF)
Lethality of strains in the P. fluorescens group to fifth instar of the tobacco hornworm, M. sexta. Mortality was assessed at 24h (), 48h (), or 72h (■) following injection with ca. 5 log(CFU per larva) or water, as a control. Three experiments (A, B and C), each evaluating ten replicate larvae per treatment, are presented. The 72 h observations for experiment A are shown in Fig 3. Asterisks represent a significant difference in mortality between a treatment and the control (P<0.05, d.f. = 1, χ2 test).
(TIF)
Lethality of high cell densities of strains in the P. fluorescens group to fifth instar of the tobacco hornworm, M. sexta. Mortality was assessed at 24h (), 48h (), or 72h (■) following injection with ca. 6 log(CFU per larva) or water, as a control. Two experiments (A and B), each evaluating ten replicate larvae per treatment, are presented. Asterisks represent a significant difference in mortality between a treatment and the control (P<0.05, d.f. = 1, χ2 test).
(TIF)
Cumulative mortality of M. sexta was assessed by counting the number of dead larvae at 2 d (), 4 d () and 6 d (■) after larvae were placed on tomato leaves supporting epiphytic populations of the specified bacterial strain. Strain Pf0-1 gacA+ (also called LK194) is a derivative of strain Pf0-1 with a chromosomal insertion of gacA [64]. Controls were larvae on leaves that were not inoculated with bacteria. (A-E) Each panel shows the results from an individual experiment, with fifteen replicate larvae evaluated per treatment in each experiment. Values that differ significantly from the control at the designated time are shown with an asterisk (P<0.05) or a diamond (P<0.10) (d.f. = 1, χ2 test). The epiphytic population size of each strain on tomato leaflets, determined at the time that larvae were placed on the leaves, is shown below each graph.
(TIF)
Developmental time course of D. melanogaster after ingestion of strains in the A) P. chlororaphis, B) P. koreensis or P. corrugata, or C) P. fluorescens subgroups. Second instar larvae were fed with a yeast suspension having no bacteria (black) or amended with bacterial strains. Initial population sizes of bacterial strains [log (CFU/plate)] are shown to the right of each panel. The percentage of larvae that pupated, counted as prepupae and/or pupae were determined over time. The percentage of larvae that emerged as adults is shown at the 288 hpi time point. Values represent the mean and standard errors from three replicates per treatment, with each replicate evaluating the larvae and adults that developed from 30 eggs. A ΔgacA mutant of Pf-5 (JL4975) [65] was included as a negative control, as it was shown previously to lack toxicity to D. melanogaster [10].
(TIF)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and supporting information files.