Abstract
The global regulator CodY controls the expression of dozens of metabolism and virulence genes in the opportunistic pathogen Staphylococcus aureus in response to the availability of isoleucine, leucine and valine (ILV), and GTP. Using RNA-Seq transcriptional profiling and partial activity variants, we reveal that S. aureus CodY activity grades metabolic and virulence gene expression as a function of ILV availability, mediating metabolic reorganization and controlling virulence factor production in vitro. Strains lacking CodY regulatory activity produce a PIA-dependent biofilm, but development is restricted under conditions that confer partial CodY activity. CodY regulates the expression of thermonuclease (nuc) via the Sae two-component system, revealing cascading virulence regulation and factor production as CodY activity is reduced. Proteins that mediate the host-pathogen interaction and subvert the immune response are shut off at intermediate levels of CodY activity, while genes coding for enzymes and proteins that extract nutrients from tissue, that kill host cells, and that synthesize amino acids are among the last genes to be de-repressed. We conclude that S. aureus uses CodY to limit host damage to only the most severe starvation conditions, providing insight into one potential mechanism by which S. aureus transitions from a commensal bacterium to an invasive pathogen.
Keywords: CodY, Staphylococcus aureus, virulence, regulation, branched-chain amino acids
Graphical abstract
Introduction
Staphylococcus aureus is a ubiquitous, Gram-positive commensal bacterium that asymptomatically colonizes the anterior nare epithelium of 20% of the human population stably and 30% of the population intermittently (Lowy, 1998, Casewell & Hill, 1986, Noble et al., 1967). However, under permissive conditions, S. aureus converts to an opportunistic pathogen and is the leading cause of skin and soft tissue infections, pneumonia, endocarditis, peritonitis, osteomyelitis, bacteremia, and sepsis (Salgado-Pabón et al., 2013, Lowy, 1998, Kumar et al., 2015, Gillaspy et al., 1995a, Bubeck Wardenburg et al., 2007). In addition to healthcare-associated infections, community-acquired infections are increasing in frequency and pose a major health threat to otherwise healthy individuals (Herold et al., 1998, Naimi et al., 2003). The augmented capacity of community-acquired strains to cause disease has been attributed to the acquisition of genes that encode Panton-Valentine Leukocidin (PVL) and genes that provide for wider metabolic versatility during survival on host skin (Joshi et al., 2011, Vandenesch et al., 2003). Indeed, S. aureus is a successful pathogen largely due to its metabolic versatility and its ability to subvert the immune response.
S. aureus virulence genes encode a vast arsenal of surface-associated and secreted factors that are deployed in stages, allowing the bacterium to survive and replicate in the host (Cheng et al., 2009). These genes can be separated into two major categories: one containing genes that mediate colonization; the other containing genes that mediate tissue damage and cellular toxicity (Novick, 2003). Regulation of virulence gene expression is complex and is tightly controlled by a plethora of Two-Component Systems (TCSs), global regulators and small RNAs that sense a variety of signals including growth phase and population density. Notable regulators include SaeRS, repressor of toxins Rot, the alternative sigma factor B (SigB), the Sar family of proteins and RNAIII (Cheung et al., 2008, Giraudo et al., 1999, McNamara et al., 2000, Recsei et al., 1986, Wu et al., 1996). Because survival and growth in the host requires the scavenging of essential nutrients (Weinberg, 1975), nutrient availability and virulence are inextricably linked.
The global transcriptional regulator CodY directly or indirectly controls the expression of over 200 genes in S. aureus (Majerczyk et al., 2010, Pohl et al., 2009). CodY acts mainly as a repressor of metabolic genes and virulence genes, though CodY stimulates a small number of genes as well. CodY is activated as a DNA-binding protein by two known classes of effectors: the branched-chain amino acids isoleucine, leucine and valine (ILV) and GTP, effectively linking the nutritional status of the cell with the production of virulence factors (Majerczyk et al., 2010). Although the site of GTP binding has remained elusive, isoleucine and valine have been shown crystallographically to bind to the N-terminal GAF domain of the Bacillus subtilis CodY protein (Levdikov et al., 2006). The interaction with ILV triggers a substantial conformational change that facilitates binding to a site-specific DNA sequence first recognized in Lactococcus lactis and revised specifically for S. aureus that is essential for full regulation of its targets (den Hengst et al., 2005, Levdikov et al., 2009, Majerczyk et al., 2010). CodY regulates many of the virulence genes indirectly by regulating the agr locus and the levels of RNAIII by an as yet unknown mechanism. Recently, CodY was shown to be capable of overriding the quorum sensing control of agr-mediated gene expression, emphasizing that nutrient availability is a major factor in adjusting virulence gene expression (Roux et al., 2014).
Changes in medium composition, growth rate, and physiological state present significant challenges when measuring expression of gene targets in vitro under nutrient depletion. For one, factors other than the regulator of interest can alter gene expression. We previously reported that genes within the CodY regulon in B. subtilis are controlled in a hierarchical manner using a method that exploits the known structure of CodY (Brinsmade et al., 2014). Herein, we report that in S. aureus, strains producing CodY proteins with single-amino acid substitutions in the ILV binding pocket retain varying amounts of residual regulatory activity and mimic situations of decreasing ILV availability where the fraction of active CodY is sequentially reduced; such graded expression is likely reflective of the true mechanism of regulation during the host-pathogen interaction. Using RNA-Seq, we found clear transcriptional trends to prioritize amino acid metabolism pathways upon onset of nutrient limitation. Targeted metabolite profiling supported the transcriptional response, revealing alterations in steady-state abundance of several amino acids and their precursors. As the simulated nutrient limitation became more severe, we observed de-repression of genes whose products mediate biofilm development, destruction of host tissue, immune response evasion and dissemination. In doing so, we present, to our knowledge, the first such analysis for a global regulator in a pathogen, providing a foundation for determining the strategy by which S. aureus integrates metabolism and virulence functions relevant to infection.
Results
CodY target gene expression level changes as nutrients are depleted in vitro
The DNA-binding activity of S. aureus CodY is modulated by ILV and GTP (Majerczyk et al., 2010). Thus, CodY-dependent genes should exhibit changes in expression level if these nutrients are depleted. To test this hypothesis, we isolated RNA from the USA200 methicillin-sensitive nosocomial osteomyelitis isolate UAMS-1 (Gillaspy et al., 1995a) during exponential growth in chemically defined medium (CDM) (Hussain et al., 1991) with decreasing ILV concentrations and performed quantitative, real-time RT-PCR (qRT-PCR) analysis on two gene targets that are repressed by CodY: ilvD (coding for dihydroxy acid dehydratase) and hld (coding for delta toxin) (Fink, 1993, Janzon et al., 1989, Majerczyk et al., 2010, Pohl et al., 2009). We used UAMS-1 for the bulk of our studies because the CodY effect is well-characterized in this genetic background and a reference genome was recently published (Sassi et al., 2015). In CDM medium containing 1,500 μM ILV (a concentration that maximizes potential CodY activity), ilvD and hld were de-repressed 147- and 59-fold in the codY mutant relative to the UAMS-1 wild-type strain, respectively (Table 1). Because the expression levels of ilvD and hld differ greatly in magnitude, we assigned an expression value of 0% for each gene in UAMS-1 (WT; CodY activity is maximal) and 100% for each gene in the codY null mutant (no CodY activity) during growth with 1,500 μM ILV. We then calculated expression relative to these levels for each gene during growth in CDM with concentrations of ILV spanning three orders of magnitude. ilvD expression in UAMS-1 increased to a level similar to that observed in the codY null mutant when ILV was reduced to 15 μM (Table 1). In contrast, hld expression increased only moderately, achieving an expression level at 1.5 μM ILV that is ~10% of that measured in the codY null mutant (Table 1).
Table 1.
Transcript abundance of CodY-repressed targets during ILV titration.
| ilvD | hld | |||||
|---|---|---|---|---|---|---|
| Relevant genotype |
[ILV] (μM) |
doubling time (min) |
Transcript1 | % expression2 |
Transcript | % expression |
| WT3 | 1,500 | 56 (3) | 0.03 (0.001) | 0 | 8.8 (1.9) | 0 |
| 150 | 53 (5) | 0.05 (0.006) | 0.5 | 11.5 (3.4) | 0.5 | |
| 15 | 120 (21) | 7.60 (1.40) | 173 | 81.9 (3.9) | 14 | |
| 1.5 | 99 (9) | 5.20 (0.90) | 118 | 60.1 (7.4) | 10 | |
| codY | 1,500 | 66 (2) | 4.40 (0.37) | 100 | 518.9 (159.1) | 100 |
Transcript abundance normalized to polC abundance during growth in CDM containing the indicated concentration of ILV. Data are presented as the means (SEM) of at least two independent experiments.
% expression calculated using the equation: ((WT[ILV]-WT1500)/(codY1500/WT1500)) × 100.
UAMS-1
During exponential growth, intracellular ILV and GTP pools are expected to be relatively high, maximizing CodY activity. However, entrance into post-exponential and stationary phases is expected to coincide with nutrient depletion, a condition monitored by CodY. To determine the extent to which CodY activity is retained under these conditions, we constructed a nuc-gfp transcriptional fusion (nuc is repressed by CodY (Majerczyk et al., 2010)) and followed the production of GFP throughout the growth cycle. During exponential growth in tryptic soy broth (TSB; a rich, complex medium), nuc-gfp remained repressed in UAMS-1 and cells exhibited relatively low fluorescence (Fig 1, 0-2 h, grey circles). As the culture exited exponential phase, we measured an increase in fluorescence; the pattern of fluorescence was essentially identical to that observed in the codY null mutant, though the magnitude was higher. In fact, fluorescence increased nearly continuously from 0-3.5 h as expected given CodY’s role in overriding the population density signal (Roux et al., 2014); at 3.5 h nuc-gfp expression was ~12-fold higher relative to that measured in UAMS-1 at the same time point. Taken together, our results show that CodY target gene expression can be modulated by severe ILV limitation in defined medium, but mild nutrient depletion during post-exponential growth in TSB does not lead to significant loss of presumably potent CodY-mediated repression of nuc-gfp. Moreover, some gene targets appear to be more sensitive to perturbations in CodY activity than others.
Fig. 1. Post-exponential nutrient limitation does not relieve strong CodY-mediated nuc repression.
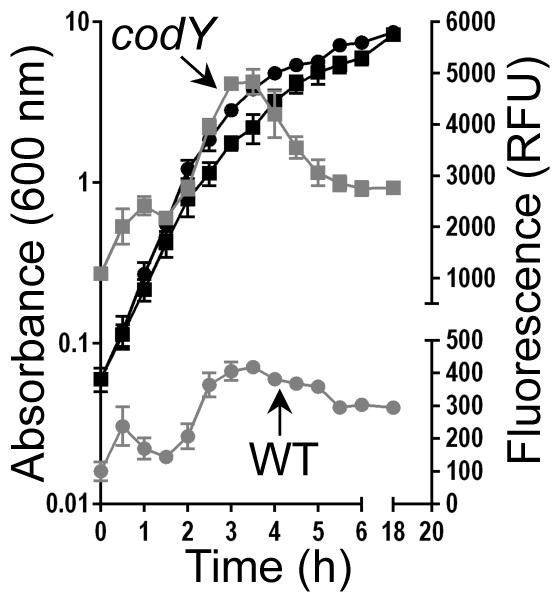
S. aureus UAMS-1 and the codY null mutant carrying the nuc-gfp reporter were grown in TSB medium (black curves). At the indicated times, samples were withdrawn, washed and resuspended in PBS buffer and GFP fluorescence was measured, normalizing fluorescence to cell growth (Relative Fluorescence Units [RFUs]; grey curves). The data are presented as mean ± SEM from two independent experiments. Where error bars appear absent, they are obscured by the curve symbol. UAMS-1, circles; codY null mutant, squares.
Staphylococcus aureus uses CodY to regulate metabolism and virulence genes sequentially
Although one can determine the threshold level of CodY activity required to turn on and off various genes using the methods described above, these experimental designs remain uncontrolled for physiological state, changes in growth rate, and factors that alter expression when the medium composition is changed. The analysis of Bacillus subtilis strains producing variant CodY proteins with varying amounts of residual DNA-binding activity revealed graded gene expression (Brinsmade et al., 2014) and avoids such experimental design problems. Using the crystal structure of B. subtilis CodY as a guide (80% overall similarity to S. aureus CodY; 76% similar for residues 1-155 containing the GAF domain in which the ILV-binding pocket is located), we targeted a conserved arginine residue at position 61, substituting a variety of amino acids that alter the biochemical properties of this key ligand-interacting residue (Levdikov et al., 2006). Alleles coding for these proteins were constructed using allelic exchange at the native codY locus in UAMS-1 to preserve the native regulation and copy number. Using qRT-PCR, we empirically and indirectly screened the activity of the resulting variant proteins by measuring transcript abundance for select CodY-regulated genes during steady-state exponential growth in TSB. As in B. subtilis, Arg61 proved to be a critical residue: CodYR61K, CodYR61H and CodYR61E proteins retain varying amounts of residual activity as judged by the loss of regulation of known repressed and stimulated gene targets. Relatively low regulatory activity was measured for an additional variant substituting aspartate for glycine at position 129 (CodYG129D) near the ILV binding pocket (Fig. S1). Cellular abundance and mobility of our CodY variants remained virtually unchanged compared to the wild-type UAMS-1 protein as judged by immunoblot analysis, suggesting that the CodY variants are stable and changes in gene expression are the result of changes in CodY specific activity (Fig S2). Collectively, our strains producing either wild-type CodY or a mutant version of CodY reflect the fraction of active CodY at varying levels of nutrient depletion; the codY null mutant behaves as though ILV are depleted.
To systematically profile CodY regulation of gene expression, we generated RNA-Seq data from UAMS-1 and the CodY mutant strains described above during exponential growth in TSB (see Experimental Procedures). Under these conditions, ILV and GTP pools are highest, CodY activity is maximized and cells are at physiological steady state, allowing for a robust, genome-wide measurement of the response to decreasing nutrients likely experienced in vivo without confounding variables associated with medium composition or growth rate changes.
We first compared RNA-Seq data from UAMS-1 and its codY null mutant to generate a list of genes under CodY control and to determine the extent to which each gene is maximally regulated. We identified 353 genes negatively regulated (i.e., repressed) by CodY and 29 genes positively regulated (i.e., stimulated) by CodY (fold change > 2, adjusted p-value <0.01). The RNA-Seq data compare favorably to our independent validation by qRT-PCR (Table 2). There was significant overlap between the list of CodY targets revealed by our analysis and those identified previously (Majerczyk et al., 2010, Pohl et al., 2009). Genes overexpressed in the null mutant (i.e., repressed by CodY) include direct and indirect targets of CodY and were the most strongly regulated; the ilv-leu operon that codes for enzymes and proteins for ILV synthesis showed the highest derepression (~200- to ~800-fold) (Dataset S1). Genes under-expressed in UAMS-1 (i.e., under positive CodY control) were at most ~11-fold regulated (Dataset S2). Comparison with previous microarray experiments yielded some exceptions. For example, we failed to detect significant upregulation of rot, autolysin SAR2040 or the putative transcriptional regulator SA2585 (Majerczyk et al., 2010, Pohl et al., 2009). Strain differences, cultivation conditions, the high false discovery rate of microarrays and the signal-to-noise ratio in both RNA-Seq and microarray approaches when trying to observe relatively small differences in transcript abundance may account for these discrepancies.
Table 2.
RNA-Seq Data Validation
| qRT-PCR | RNA-Seq | ||||
|---|---|---|---|---|---|
| transcript abundance | % expression1 |
RPKMO value | % expression1 |
||
| WT | 1 (0)2 | 0 | 4 (0) | 0 | |
| R61K | 6 (1) | 2 | 51 (6) | 3 | |
| ilvD | R61H | 68 (7) | 28 | 652 (77) | 46 |
| R61E | 132 (7) | 56 | 1176 (40) | 82 | |
| G129D | 216 (35) | 91 | 1429 (15) | 100 | |
| Δ codY | 236 (32) | 100 | 1427 (5) | 100 | |
| WT | 18 (1) | 0 | 108 (13) | 0 | |
| R61K | 59 (13) | 3 | 402 (129) | 5 | |
| hld | R61H | 81 (6) | 5 | 554 (104) | 8 |
| R61E | 293 (4) | 20 | 1912 (286) | 31 | |
| G129D | 894 (321) | 65 | 5479 (2562) | 93 | |
| Δ codY | 1364 (44) | 100 | 5887 (1438) | 100 | |
| WT | 6 (1) | 0 | 107 (1) | 0 | |
| R61K | 8 (2) | 1 | 115 (9) | 0 | |
| nuc | R61H | 16 (1) | 3 | 196 (3) | 4 |
| R61E | 54 (2) | 17 | 722 (2) | 28 | |
| G129D | 137 (5) | 45 | 1666 (234) | 70 | |
| Δ codY | 297 (40) | 100 | 2337 (116) | 100 | |
| WT | 7 (0) | 0 | 84 (0) | 0 | |
| R61K | 43 (2) | 33 | 469 (6) | 54 | |
| brnQ2 | R61H | 60 (7) | 48 | 743 (24) | 93 |
| R61E | 80 (3) | 66 | 770 (20) | 97 | |
| G129D | 95 (2) | 79 | 818 (21) | 103 | |
| Δ codY | 117 (25) | 100 | 794 (22) | 100 | |
| WT | 22 (7) | 100 | 271 (45) | 100 | |
| R61K | 30 (5) | 141 | 318 (26) | 119 | |
| fnbA | R61H | 22 (2) | 101 | 242 (24) | 88 |
| R61E | 6 (1) | 15 | 75 (6) | 20 | |
| G129D | 4 (1) | 8 | 42 (3) | 7 | |
| Δ codY | 3 (1) | 0 | 25 (0) | 0 | |
Set to 100% in the strain where the expression is maximal (UAMS-1 [WT] for stimulated genes, AcodY for repressed genes using the equation ((mutant-WT)/(codY-WT)) × 100. Intermediate values are calculated on this scale.
Data are presented as the mean and (standard error) of ≥ two biological replicates.
Using a fixed concentration of active CodY, Majerczyk et al. identified differential enrichment of chromosomal regions containing putative CodY sequence motifs with varying degrees of degeneracy relative to a consensus sequence, suggesting that the S. aureus CodY protein transiently occupies its many sites based the active fraction of CodY to generate a graded response to nutrient availability. If true, we reasoned that we should be able to identify groups of genes whose expression patterns were similar. To that end, we applied a k-means cluster-based approach (Tavazoie et al., 1999). We first assigned an expression value of 100% for repressed genes in the codY null mutant; positively regulated genes were set to 100% in UAMS-1. We then calculated expression relative to these levels for each gene across the set of strains producing CodY proteins with partial regulatory activity. To identify the optimum number of clusters, we examined the distribution of genes in as few as two and as many as 10 clusters each for repressed and stimulated genes. We chose six clusters (four and two, respectively) because the responses to changes in CodY activity were clearly differentiated. As in B. subtilis, we observed variable sensitivity of genes to changes in CodY activity (Dataset S3, Fig. S3). For example, Clusters 5 and 6 contained the positively regulated genes; on average, genes most sensitive to changes in CodY activity (cluster 5) showed <20% expression in the CodYR61K-producing strain. Cluster 6 contained genes whose average expression level dropped below 20% in the R61E mutant. Negatively regulated genes fell into four clusters. Those genes most sensitive to changes in CodY activity (i.e., genes in cluster 1) achieved, on average, complete de-repression in the R61K mutant, whereas those genes least sensitive to changes in CodY activity (i.e., genes in cluster 4) showed >50% expression in the G129D mutant. Genes crossed the 50% threshold in the R61H and R61E mutants for clusters 2 and 3, respectively. In general, metabolic and virulence genes were dispersed throughout each cluster (Dataset S3, Fig. S3B, compare grey vs. black lines) and genes either co-transcribed or sharing a biological function fell into the same cluster (Dataset S3). For example, the majority of the purine biosynthesis genes (i.e., purCSQLFMHD) fell into cluster 2; purEK fell into cluster 3. Genes for heme-based iron acquisition (isdCDE), genes of the agr RNAII transcript (agrBDCA) and the majority of the ILV biosynthetic genes (ilvDBHC, leuABCD, ilvA) all fell into cluster 3; hld (delta toxin; agr RNAIII transcript), the unlinked ILV biosynthetic gene ilvE, and the capsule biogenesis genes (cap5ABCDEFGHIJK) all fell into cluster 4. Moreover, positively-regulated enterotoxin-like protein genes (i.e., eto1-6) and genes of the accessory Sec locus (i.e., asp1-3 and secY) fell into cluster 5.
Genes for amino acid uptake are sensitive to changes in CodY activity
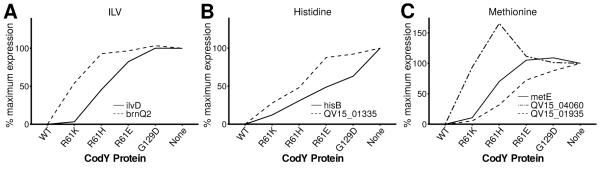
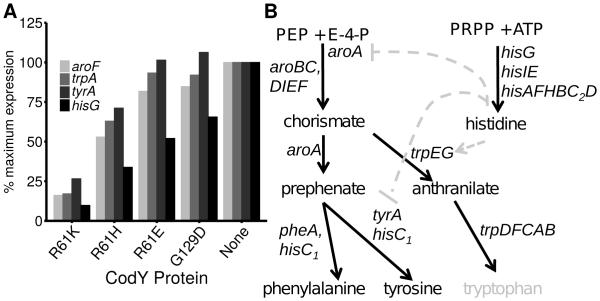
Analysis of the k-means clustering revealed that nutrient uptake and biosynthesis are separated along the ILV gradient. We focused our analysis on transport systems shown or believed to salvage available branched-chain amino acids (i.e., ILV; brnQ1, brnQ2; QV15_00670, QV15_01265), methionine (metNIQ2; QV15_04055-04065) and histidine (QV15_01335), as well as genes that direct the synthesis of these amino acids (ilv-leu; metICHE, mdh; hisZGDC2HAFIE) because they have been previously characterized (Burke & Pattee, 1972, Kaiser et al., 2015, Majerczyk et al., 2010, Rodionov et al., 2004, Schoenfelder et al., 2013). We consistently observed that ILV, histidine and methionine uptake genes were relatively sensitive to changes in CodY activity compared to their cognate biosynthesis genes, falling into clusters 2, 2 and 1, respectively (Dataset S3, Fig. 2). A notable exception is a duplicate set of putative methionine uptake genes (metNIQ1; QV15_01935-01945) that fall into cluster 3 that are relatively resistant to changes in CodY activity. In contrast, biosynthetic genes for ILV, histidine and methionine fell into clusters 3 and 4, 3 and 2, respectively. Interestingly, genes encoding enzymes and proteins for the synthesis of aromatic amino acids (trp, aro, tyr), lysine (lys, dap, asd) and methionine are more sensitive to changes in CodY activity than those genes for ILV and histidine synthesis and fall into cluster 2 (Dataset S3, Fig 3A).
Fig. 2. S. aureus prioritizes amino acid uptake in response to reductions in CodY activity.
All data here and in subsequent figures are presented as the means of at least two independent experiments. RNA-Seq data are shown; % maximal expression relative to the codY null mutant (SRB372, None) is plotted across strains producing varying amounts of CodY regulatory activity: CodYWT, SRB337; CodYR61K, SRB506; CodYR61H, SRB623; CodYR61E, SRB577; CodYG129D, SRB493. In each panel, genes for nutrient transport are more sensitive to changes in CodY activity and reach 100% prior to genes for nutrient biosynthesis. A. CodY-repressed genes coding for the uptake (brnQ2, QV15_01265) and synthesis (ilvD) of ILV. B. CodY-repressed genes coding for the uptake (proposed histidine transporter, QV15_01335) and synthesis (hisB) of histidine. C. CodY-repressed genes coding for the uptake (metNIQ1 [QV15_01935-01945], metNIQ2 [QV15_04055-04065]) and synthesis (metE) of methionine.
Fig. 3. S. aureus represses aromatic amino acid synthesis less stringently than histidine biosynthesis.
A. Representative CodY-repressed genes coding for the synthesis of aromatic amino acids and histidine are shown; RNA-Seq data in CodY variant strains are presented as a percentage of the value obtained in the codY mutant strain (SRB372, none), set to 100% expression. R61K, SRB506; R61H, SRB623; R61E, SRB577; G129D, SRB493. Genes for aromatic amino acid biosynthesis include aroF (chorismate synthase), trpA (tryptophan synthase), and tyrA (prephenate dehydrogenase). hisG encodes ATP phosphoribosyltransferase (catalytic subunit) for histidine biosynthesis. WT, SRB337 was set at 0% and is not shown. B. Phenylalanine and tyrosine are derived from the common intermediate prephenate. UAMS-1 is likely a tryptophan auxotroph (grey) due to a frameshift in trpD. Histidine is synthesized from phosphoribosyl pyrophosphate (PRPP) and ATP and, at least in B. subtilis, influences aromatic amino acid synthesis transcriptionally (aroA and tyrA) and allosterically (TrpEG), and is indicated by the grey dashed arrows. Arrows, positive effect; blunt arrow, negative effect.
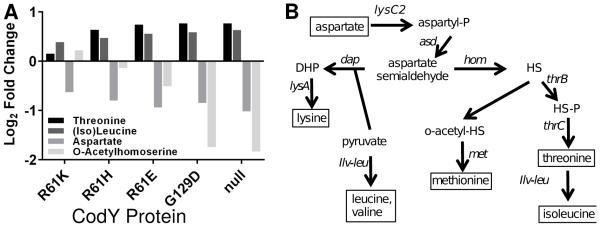
To determine the extent to which these changes in CodY-dependent gene expression alter the steady state levels of these amino acids and their intermediates, we quantified steady-state intracellular abundances using targeted tandem liquid chromatography-TOF MS (LC-MS) analysis of cell lysates during exponential phase in TSB. Samples were collected within one generation of those collected for gene expression analysis. Consistent with our RNA-Seq results, we observed an increase in intracellular threonine in the codY null mutant relative to UAMS-1 (Fig. 4A; p < 0.05). Isoleucine and leucine, though not resolved in our mobile phase, were also significantly increased. Aspartate and acetyl homoserine (methionine biosynthetic intermediate) pools were reduced in the codY null mutant relative to UAMS-1, reflecting their use in biosynthesis (Fig. 4; p < 0.05). As CodY activity was reduced, we observed small, but dose-dependent changes in these metabolite abundances across our collection of strains (Fig. 4A). We also measured significant increases in intracellular serine pools as well as significant decreases in proline and pyrrole carboxylate as CodY activity decreased. A full list of compounds analyzed and the statistical significance of their changes is available in the Supporting Information (Dataset S4).
Fig. 4. Metabolite profiling reveals increases in steady state pools of amino acids and biosynthetic intermediates as cognate biosynthetic genes are de-repressed.
A. S. aureus strains were cultivated exponentially in TSB medium and sampled at OD600~0.5-0.6. Changes in relative intracellular abundance of selected amino acids and biosynthetic intermediates are shown and plotted on a log2 scale relative to UAMS-1. Mean intracellular metabolite abundances are >1.5-fold altered in the codY null mutant relative to UAMS-1 as judged by a Mann-Whitney U test (p < 0.05) and across all six strains as judged by Cuzick’s test for trend (p < 0.01) (Cuzick, 1985). Complete data and statistical analysis can be found in Dataset S4. B. Lysine, methionine and threonine are derived from aspartate and are collectively known as the aspartate family of amino acids. Pyruvate serves as a precursor for both lysine and ILV synthesis. The pathway bifurcates at aspartate semialdehyde. Differential expression in the codY null mutant suggests increased flux through this pathway. o-acetyl-HS, o-acetyl homoserine; HS, homoserine; HS-P, homoserine-phosphate.
Reductions in CodY activity result in increases in static biofilm development
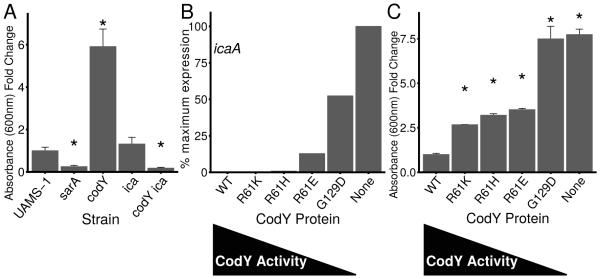
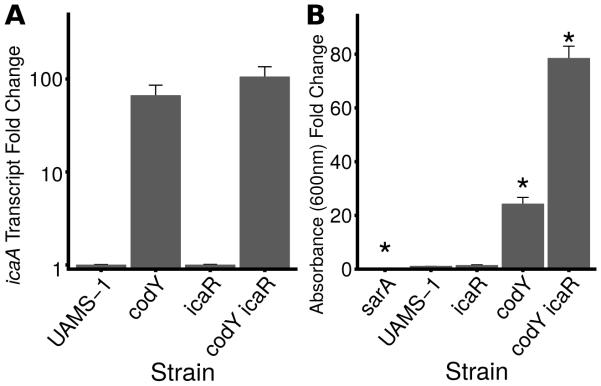
To determine whether sequential regulation of CodY gene targets leads to detectable changes in protein production and behavior, we examined two well characterized in vitro phenotypes – static biofilm formation and the production of thermonuclease, whose activity is required for the development of biofilms under flow conditions (Kiedrowski et al., 2011, Moormeier et al., 2014). CodY-deficient strains of UAMS-1 have been shown to form thicker biofilms relative to UAMS-1; this has been attributed to the production of PIA due to de-repression of the ica locus (Atwood et al., 2015, Majerczyk et al., 2010, Majerczyk et al., 2008). Our RNA-Seq data and static biofilm assays confirm the entire ica locus (icaADBC; except the divergently-transcribed regulator icaR) is over-expressed 37- to 225-fold in the codY null mutant (Dataset S1) and the codY null mutant forms significantly more biofilm relative to the wild-type parent strain (Fig. 5A, compare UAMS-1 vs. codY). We used a sarA mutant to confirm we had removed the majority of non-adherent cells because of its known defect in biofilm development (Beenken et al., 2003). Interestingly, we saw no statistical difference relative to UAMS-1 in static biofilm development when we deleted the ica locus in otherwise wild-type cells. However, CodY-deficient cells formed relatively little static biofilm when also lacking an intact ica locus, providing the most convincing genetic evidence to date for a PIA-dependent biofilm when CodY is inactive in UAMS-1 (Fig. 5A, compare UAMS-1 vs. ica and ica vs. ica codY).
Fig. 5. CodY suppresses PIA-dependent biofilm formation.
Strains were grown exponentially for several generations prior to analysis. A. Static biofilm development in UAMS-1 strains was measured 24 h following inoculation to tissue culture-treated microtiter plates. Adherent cells were detected by measuring crystal violet retained as absorbance at 600 nm. Data are relative to UAMS-1 (fold change=1). UAMS1, SRB337; UAMS-929, sarA::kan (negative control, known to be defective in static biofilm formation); codY::erm, SRB372; Δica::tet, AR11; codY::erm Δica::tet, AR12. Asterisks denote statistically significant comparisons to the codY null mutant (p < 0.05) using the Games-Howell test (Games et al., 1979). B. icaA expression data (representative of ica locus expression) were extracted from the RNA-Seq data. Values in CodY variant strains are presented as a percentage of the value obtained in the codY mutant strain, set to 100% expression. C. Static biofilm development in tissue culture-treated microtiter plates indicates that threshold CodY activities promote stepwise increases in static biofilm formation. Adherent cells were detected after 24 h by measuring crystal violet retained as increase in absorbance at 600 nm. Data are relative to UAMS-1 (fold change =1). Asterisks denote statistically significant comparisons to the codY null mutant (p < 0.05) using the Games-Howell test (Games et al., 1979).
ica gene expression is regulated by the icaR transcriptional repressor (Jefferson et al., 2004). To determine whether CodY regulates icaADBC gene expression through IcaR, we constructed icaR, codY, and icaR codY double mutants and measured transcript abundance of icaA using qRT-PCR. In UAMS-1 and the icaR single mutant, icaA transcript abundance was extremely low and indistinguishable from our no-reverse transcriptase control (Fig 6A). icaA was > 60-fold overexpressed in the codY null mutant; in the icaR codY double mutant, icaA transcript was essentially unchanged (Fig 6A). Whereas UAMS-1 and the icaR null mutant both formed relatively little static biofilm (crystal violet absorbance < 1), the codY icaR double mutant formed ~3-fold more biofilm than the codY single mutant (Fig 6B). From these experiments, we conclude that CodY is epistatic to IcaR.
Fig 6. CodY is epistatic to IcaR.
Strains were grown exponentially for several generations prior to analysis. In both panels, data are relative to UAMS-1 (fold change=1). A. qRT-PCR measurement of icaA transcript abundance during exponential growth suggests that CodY overrides loss of IcaR-mediated repression of the ica locus. The transcript abundance of icaA in UAMS-1 and in the icaR null mutant was below the limit of detection (0.08 copies of icaA relative to rpoC). B. Static biofilm development of derivatives of UAMS-1 was measured 24 h following inoculation to tissue culture-treated microtiter plates. Adherent cells were detected by measuring crystal violet retained as absorbance at 600 nm. Asterisks denote statistically significant comparisons to the codY null mutant (p < 0.05) using the Games-Howell test (Games et al., 1979). UAMS-1, SRB337; icaR::φΝΣ, SRB813; codY:: φΝΣ[erm::tet(M)], SRB814; icaR::φΝΣ codY::φΝΣ[erm::tet(M)], SRB817. sarA::kan (UAMS-929) was used as a negative control because of its known role in static biofilm development.
The icaADBC genes fall into cluster 4, indicating that expression of these genes is relatively robust to changes in CodY activity (Dataset S3, Fig. 5B). This suggested that the thickest PIA biofilms would be produced when CodY activity is eliminated. To answer this question, we measured the ability of our strains producing variable CodY activity to form a biofilm and found that the CodYR61K, CodYR61H and CodYR61E variant-producing strains show a two- to three-fold increase in biofilm formation, whereas biofilm formation was maximal in both the CodYG129D variant-producing strain and the CodY-deficient strain (Fig. 6C). Taken together, our results indicate UAMS-1 produces an ica- and PIA-dependent biofilm when CodY is inactive, and that although ica expression is turned on under conditions that mimic relatively severe ILV depletion, small changes in ica expression appear to trigger biofilm formation.
CodY controls secreted nuclease production by grading saePQRS expression
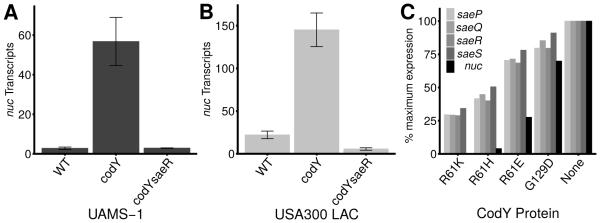
nuc, coding for thermonuclease, is directly controlled by the SaePQRS two-component system (TCS) and is a CodY-regulated virulence factor (Majerczyk et al., 2010, Olson et al., 2013). The nuc locus is ~22-fold de-repressed in the codY null mutant relative to UAMS-1 (Dataset S1), but regulation of nuc by CodY is likely to be indirect, as no obvious CodY motif was detected in or near the nuc promoter region (Majerczyk et al., 2010). We did, however, measure an approximate three- to eight-fold increase in saePQRS expression in the codY null mutant (Dataset S1). In order to determine whether CodY regulates nuc expression via the SaeRS TCS, we performed an epistasis experiment and measured nuc transcript abundance in exponentially growing S. aureus cells. Consistent with our RNA-Seq data, we saw a ~50-fold increase in nuc expression when we knocked out codY (Fig. 7A). In a codY saeR double mutant, expression of nuc was ablated, indicating that SaeR is required for nuc overexpression in a codY null mutant (Fig. 7A, compare codY vs. codY saeR). To determine whether the effect of CodY is specific to UAMS-1, we repeated the experiment in a community-acquired MRSA isolate (LAC, USA300 lineage) (Voyich et al., 2005) and found that again, overexpression of nuc in a CodY mutant requires an intact sae locus (Fig. 7B).
Fig. 7. CodY regulates thermonuclease expression via the Sae TCS.
Overexpression of nuc in a codY mutant requires saeR. Strains were grown exponentially and nuc transcript abundance measured using qRT-PCR. Data are presented as the mean ± standard error of at least two independent experiments. A. UAMS-1: WT, SRB337; codY::erm, SRB372; codY::erm saeR::φΝΣ[erm::tet(M)], SRB759. B. USA300 LAC: WT, SRB687; codY, SRB746; codY saeR::φΝΣ[erm::tet(M)], SRB761. C. Loss of CodY-mediated repression at saePQRS leads to subsequent loss of nuc repression. CodY-repressed gene targets coding for the Sae TCS (saePQRS) and thermonuclease (nuc) are shown. Values in CodY variant strains are presented as a percentage of the value obtained in the codY mutant strain, set to 100% expression.
Interestingly, saePQR (cluster 3) and saeS (cluster 2) respond to moderate changes in CodY activity, whereas nuc (cluster 4) is de-repressed only when CodY activity is substantially reduced (Fig. 7C). To determine whether the increases in nuc expression in the codY mutant strains lead to increased secretion of thermonuclease enzyme, we used a FRET-based assay to quantify nuclease present in culture supernatants (Kiedrowski et al., 2011). During exponential growth in TSB, we detected 1.5 units (U) ml−1 OD600−1of thermonuclease activity in UAMS-1 culture supernatants. In contrast, the codY null mutant accumulated 14.3 U ml−1 OD600−1 of thermonuclease activity in culture supernatants (Fig. 8A, compare UAMS-1 versus codY). This activity is approximately 50% of the activity measured in sarA culture supernatants known to overproduce thermonuclease (Kiedrowski et al., 2011); as expected, culture supernatants of a strain lacking nuc showed no secreted nuclease activity (Fig. 8B). We found coinciding trends when comparing nuclease activity, nuc transcript numbers, and nuc RNA-Seq counts, indicating that rising levels of saePQRS and nuc transcripts lead to increases in secreted nuclease as CodY activity is reduced (Figs. 7, 8; Dataset S1, Table 2). Taken together, these results show that CodY represses the expression and production of thermonuclease during rapid, exponential growth, and this graded regulation is dependent on the Sae TCS.
Fig. 8. CodY controls the secretion of thermonuclease in exponentially growing cells.
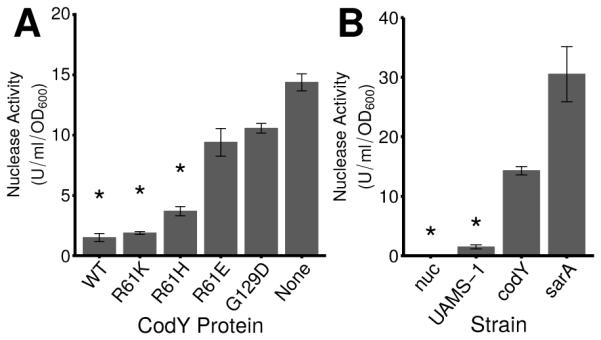
A. Secreted thermonuclease activity in exponentially growing UAMS-1 cell culture supernatants was quantified using a FRET-based assay in strains producing varying amounts of residual CodY activity as described in Experimental Procedures. UAMS-1 (WT), SRB337; codY null mutant (None), SRB372; and the CodY variants R61K (SRB506), R61H (SRB508), R61E (SRB577) and G129D (SRB493). B. Secreted nuclease activity in a CodY-deficient strain relative to a SarA-deficient strain during exponential growth in TSB. nuc, UAMS-1471; UAMS-1, SRB337; codY, SRB372; sarA, UAMS-929. SarA has been shown previously to suppress Nuc enzyme levels (Kiedrowski et al., 2011) and was used as a positive control. Asterisks (*) denote statistical significance by Games-Howell pairwise test (p < 0.05) when compared to the codY null mutant. The data are presented as the mean ± the standard error of at least 2 independent experiments.
Discussion
Gram-positive bacteria like S. aureus are metabolically versatile, can reside in and on human hosts as commensal bacteria, and as opportunistic pathogens, can cause life-threatening infections. A complex network of regulatory factors adjusts gene expression and physiology to ensure survival and promote multiplication in diverse environments. To that end, transcription factors that bind key intracellular metabolites can regulate many genes simultaneously, mediating, at least in part, the shift between commensal and pathogenic lifestyles. Despite observations that the expression of the most important virulence genes often correlates with the exhaustion of available nutrients (Somerville & Proctor, 2009), there is limited knowledge of how the signaling of nutrient status and the resulting physiological responses are coordinated. While the targets of CodY are known in S. aureus, virtually nothing is known about the threshold levels of CodY activity required to regulate each CodY-controlled gene. The ILV binding pocket in B. subtilis has been characterized (Villapakkam et al., 2009) and is highly similar to that of S. aureus. Using available structural data from the B. subtilis CodY protein, we sought to modulate the activity of S. aureus CodY by constructing a collection of strains that produce CodY proteins with varying amounts of residual regulatory activity. This approach allows us to simulate an ILV limitation while growing all cells under identical conditions where the only variable is CodY activity. We report the first linkage of turning on and off metabolism and virulence gene expression to increasing nutrient depletion, revealing non-binary modulation of a regulon built to mediate adaptation to dynamic environments.
CodY directs the metabolism of key nutrients sequentially along an ILV gradient
Our data suggest that S. aureus uses CodY to prioritize nutrient uptake and biosynthesis during growth in a complex mixture of carbon and nitrogen sources and generally, uptake genes are more sensitive to changes in CodY activity than their cognate biosynthetic pathways. Such a strategy makes physiological sense and allows the bacterium to scavenge available nutrients before diverting carbon, nitrogen and energy to their biosynthesis. Interestingly, while one set of uptake genes was de-repressed with a relatively small change in CodY activity, another putative set of methionine uptake genes, metNIQ1 (QV15_01935-01945) fell into in cluster 3 and is turned on at a lower level of CodY activity required to turn on methionine biosynthesis (Fig. 2C). Although these uptake systems have not been biochemically characterized, we speculate that they may differ in affinity as is the case for ILV uptake proteins (Kaiser et al., 2015), and our data potentially reveal a strategy in which a low-affinity system may be rapidly turned on to import relatively abundant methionine. As amino acids including methionine become increasingly scarce, a high-affinity uptake system may be induced along with the genes for methionine synthesis.
We found that not all synthesis pathways are regulated at the same level of residual CodY activity. First, aromatic amino acid biosynthetic genes are turned on at a lower threshold CodY activity compared to other amino acid biosynthetic genes (cluster 2). Histidine (synthesized via his; cluster 3) is known to act post-transcriptionally to cross-regulate aromatic amino acid synthesis in B. subtilis (Fig. 3B) (Kane & Jensen, 1970, Nester, 1968). Thus, S. aureus may prioritize aromatic amino acid synthesis to avoid negative cross-pathway regulation by rising histidine levels to maintain a steady pool of aromatic amino acids for protein production as well as for folic acid and ubiquinone synthesis. Second, although lysine, methionine, threonine and, by extension, isoleucine, are all synthesized from aspartate and are collectively known as the aspartate family of amino acids (Fig. 4B) (Paulus, 1993), we consistently saw lysine and methionine biosynthetic genes turned on with relatively minor reductions in CodY activity (cluster 2) compared to threonine and ILV biosynthetic genes (cluster 3). Because S. aureus uses lysine to synthesize peptidoglycan (Mengin-Lecreulx et al., 1999, Schleifer & Kandler, 1972), it is possible that S. aureus prioritizes lysine synthesis to support both peptidoglycan synthesis and protein production even when pools of amino acids are only slightly reduced. Because methionine is limited in physiological fluids and undetectable in nasal secretions at the site of commensal carriage (Basavanna et al., 2013, Krismer et al., 2014), the positions of the met biosynthetic genes within the overall CodY expression hierarchy may reflect the adaption of S. aureus to the human host and the need to synthesize this amino acid.
As gene annotations improve and biochemical data regarding biosynthetic and degradative pathways become increasingly available, our metabolite data can be further mined for physiological significance. For example, the genes responsible for conversion of threonine to serine via glycine (SAR1326 and glyA [SAR2201]) were not differentially expressed between the wild type and codY null mutant strains. However, serine was > 6-fold more abundant in the codY null mutant compared to wild type cells (p < 0.05). It has been suggested that serine can be synthesized from 3-phosphoglycerate (Lee et al., 2009b). serA (SAR1801; coding for 3-phosphoglycerate dehydrogenase) is overexpressed 80-fold in the codY null mutant, but genes encoding the phosphoserine transaminase and phosphatase appear to be absent in UAMS-1. At this time we cannot rule out the possibility for increased serine transport or synthesis via an uncharacterized pathway.
We note here that our data reflect steady state abundances and only suggest altered fluxes through a particular pathway; in the future, use of isotopic labeling can provide information regarding carbon and nitrogen flow. Though not significant, we detected a trend toward increasing homocysteine and decreasing S-adenosylmethionine (SAM) as CodY activity decreased; our methods did not allow for reliable detection of redox-labile cysteine. These observations are consistent with the upregulation of genes for methionine synthesis via aspartate (Fig 4). Cysteine synthesis can proceed via a dedicated pathway and, in some bacteria including B. subtilis, the reverse transsulfuration pathway whereby methionine is converted to cysteine via homocysteine and the SAM recycling pathway (Rodionov et al., 2004). Based on our data, we have no evidence for altered regulation of the reverse transsulfuration pathway.
CodY links biofilm development to nutrient availability
The effect of a codY null mutation on biofilm formation in UAMS-1 was documented previously; CodY-deficient cells behave as though nutrients are completely exhausted. Our genetic analysis of ica operon regulation suggests that CodY is epistatic to IcaR. That is, during exponential growth, there is no effect of disrupting icaR (Fig. 6). This is likely because CodY is active and represses transcription by binding to a site internal to icaB (Majerczyk et al., 2010); however, we were unable to measure a quantitative difference between icaB transcript abundance upstream or downstream of the binding site (data not shown). Interestingly, though there is a trend toward an increase in expression, we see no significant up-regulation of ica when we measure icaA transcript abundance in the icaR codY double mutant relative to the codY single mutant. It is possible that this increase is responsible for the 3-fold increase in static biofilm production. Alternatively, there may be additional transcriptional or post-transcriptional regulation. For example, the 3’ untranslated region (3’-UTR) of icaR contains an element that base pairs with the Shine-Delgarno region of the icaR messenger RNA, regulating icaR translation and IcaR abundance (Ruiz de los Mozos et al., 2013). The environmental signals or factors that control this regulation are unknown but may include nutrient availability and/or CodY. This could also explain the mild increase in biofilm formed in the strains producing the R61K and R61H variants of CodY (Fig 5C) despite no apparent change in ica expression. The exact mechanism for CodY-mediated repression of ica is unclear, but given the position of the CodY binding site, may involve a roadblock mechanism (Belitsky & Sonenshein, 2011). This is under investigation in our lab.
We found that the ica deletion mutant shows no detectable biofilm phenotype after 24 h (Fig. 6), suggesting that the mild biofilm produced by UAMS-1 under static biofilm conditions is PIA-independent. However, PIA accumulates during exponential growth and, after the transition to post-exponential and stationary phases, carbon is directed to capsule synthesis, which shares the same precursors (Sadykov et al., 2010, Majerczyk et al., 2008). This is also in good agreement with the fact that CodY represses capsule biosynthesis genes during exponential phase ((Majerczyk et al., 2010), Dataset S1). Thus, we are hesitant to say that UAMS-1 biofilms are completely PIA-independent. Alternatively, our ILV titration and nutrient exhaustion experiments are consistent with the notion that nutrients must be severely depleted before genes within clusters 3 and 4 are de-repressed (Table 1, Fig. 1). icaADBC are among those genes relatively insensitive to changes in CodY activity (cluster 4). Thus, CodY activity may remain sufficiently high to repress the ica locus in the UAMS-1 wild type strain and prevent the production of a biofilm. Deletion of the ica locus in a codY null mutant eliminates the hyper-biofilm phenotype observed in the codY null mutant and in fact these cells fail to produce biofilms to the extent measured in UAMS-1 (Fig. 6). This phenotype is reminiscent of fnbAB strains of USA300 LAC that fail to produce appreciable amounts of PIA and fail to form significant biofilm (Atwood et al., 2015, McCourt et al., 2014). Thus, in strains other than UAMS-1, cell attachment and biofilm development are likely synchronized with nutrient availability.
In UAMS-1, codY null mutants produce PIA-dependent biofilms but the biofilm yields measured in our variant strains were stepwise in nature and did not follow a smooth increase in the mutants that might have been predicted by the ica gene expression patterns alone (Figs. 6B, C). We propose that UAMS-1 produces PIA-dependent biofilms as CodY activity drops. However, at a critical threshold level of CodY activity, biofilm development is restricted (Fig. 6C). We postulate this is due to CodY-dependent, sequential de-repression of genes whose products have been shown to be important for remodeling the S. aureus biofilms (Kiedrowski et al., 2011, Moormeier et al., 2014, Mootz et al., 2013). For example, CodY controls the expression of the protease genes sspA, sspB, sspC and aur, and thermonuclease nuc. These genes are 3- to 44-fold regulated by CodY and are relatively insensitive to changes in CodY activity (Datasets S1, S3; clusters 3 and 4). As nutrients continue to be depleted, CodY is further inactivated and PIA production may override the effect of nucleases and proteases. Interestingly, it has been proposed that biofilm towers are important for surviving environmental stress, and gene expression in microfluidic devices has revealed biofilm towers contain metabolically distinct microniches (Moormeier et al., 2013). Because nutrients likely diffuse poorly into biofilms and because CodY controls metabolic and virulence gene expression, it is tempting to speculate that CodY is responsible, at least in part, for distinct patterns of gene expression and subsequent regions of cells with specific roles. An examination of biofilm development under biologically relevant flow where nutrients are replenished over time is underway and will enable us to more clearly define CodY’s role in sculpting staphylococcal biofilms.
CodY instigates cascading virulence factor regulation
In S. aureus, CodY has been shown to control the expression of virulence genes directly by binding to sites containing a CodY motif and indirectly by controlling the expression of agr (Majerczyk et al., 2010, Majerczyk et al., 2008, Pohl et al., 2009). Additionally, Majerczyk et al. identified saeRS to be regulated by CodY (Majerczyk et al., 2010). However, the dynamics of this regulation have remained unexplored. We document the first example of cascading regulation of virulence genes by CodY in S. aureus by demonstrating CodY grades sae expression under conditions that mimic ILV depletion to induce nuc expression. The positions of saePQRS (clusters 2 and 3) and nuc (cluster 4) within the CodY expression hierarchy reveal that the threshold for saePQRS induction is lower than for nuc regulation. That is, saePQRS expression precedes nuc expression (Fig. 7C). The sae P1 promoter is activated by agr, and repressed by Rot and the alternative sigma factor SigB, each of which is effected by CodY (Geisinger et al., 2006, Li & Cheung, 2008, Majerczyk et al., 2010, Novick & Jiang, 2003, Pohl et al., 2009), suggesting regulation by CodY is indirect. Interestingly, it has been proposed that the ratio of activated SaeR (SaeR~P) relative to SaeR rather than absolute abundance of SaeR dictates target gene transcription. coa and hla, regulated by CodY and targets of SaeR, showed no increase in expression during exponential and post-exponential phases when saeR was placed under the control of an inducible promoter (Mainiero et al., 2010). Under these conditions, CodY is active and likely masks expression of these targets as SaeR~P increases. We propose that nutrient availability, monitored by CodY, ensures precise dosing and timing of Sae-dependent gene expression. This is in agreement with the notion put forth by Novick and Jiang whereby the sae locus collects converging environmental and population density signals to adjust gene expression (Novick & Jiang, 2003). Precisely how CodY controls the expression and activity of the Sae TCS is under investigation in our laboratory.
Staphylococcus aureus uses graded CodY activity to modulate metabolism and virulence gene expression in vitro
In this report, we used strains producing variable CodY activity to pinpoint the threshold activities required to regulate each metabolic and virulence gene, offering snapshots of transcript abundance at discrete points along the CodY activity continuum. Changes in transcript abundance of selected CodY targets in these strains mirror changes seen when wild-type cells are grown in defined medium with varying concentrations of exogenous ILV. For example, ilvD (cluster 3) is de-repressed at a lower CodY activity threshold than hld (cluster 4); when grown in medium containing 15 μM ILV, ilvD was completely de-repressed while hld exhibited only ~10% of the expression observed in the codY null mutant. These experiments should be interpreted with caution, as medium- and growth-rate dependent effects can confound the analysis. Indeed, at low ILV concentrations, the doubling time of S. aureus strains increases ~2-fold (Table 1). As the intracellular pools of ILV and guanine nucleotides drop during nutrient depletion, the active fraction of CodY decreases. If one considers nutrient sufficiency as the moment prior to limitation, our use of partial CodY activity variants can serve as a surrogate to our initial time course experiment (Fig 1). Indeed, based on our results, CodY gene targets can be arranged into temporal classes with clear “early” and “late” gene expression (Fig. S3, Fig. 9, Dataset S3). Such a classification is consistent with the sensitivity of selective gene targets to changes in CodY activity (Fig. S1). For example, S. aureus generally turns on nutrient uptake before devoting carbon, nitrogen and energy to biosynthesis, whose genes are turned on relatively late when nutrients are nearly depleted. Genes coding for proteins and enzymes for utilizing other carbon and nitrogen sources are turned on and off at various points along the ILV gradient. For instance, transporters for oligopeptides (QV15_04450-04470), p-aminobenzoyl-glutamate (QV15_12780), and galactitol (QV15_00960) are turned on early. pycA (QV15_05170), sucA (QV15_06885) and QV15_00370, coding for pyruvate carboxylase, 2-oxoglutarate dehydrogenase and acetoin reductase - all of which control the distribution of pyruvate and 2-oxoglutarate - are also turned on early. In contrast, a transporter for trehalose (treP, QV15_02000) and ldh (QV15_13410; coding for lactate dehydrogenase) are turned on late, suggesting that trehalose and lactate are utilized after other nutrients are exhausted. QV15_04760, coding for an Fe3+ transport protein and QV15_11350-11355, coding for the anabolic acetolactate decarboxylase and synthase alsDS are positively regulated by CodY and are turned off as nutrients are depleted.
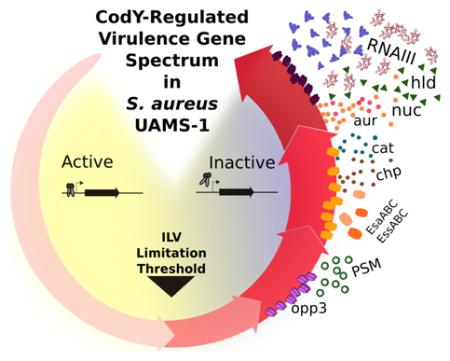
Fig. 9. Working model for how a spectrum of CodY activities generates a developmental program based on the availability of ILV and GTP.
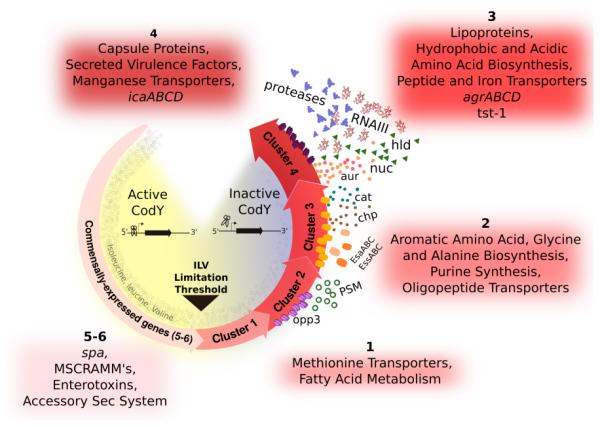
During periods of nutrient sufficiency, CodY is active and promotes colonization or persistence on or within the host. Under conditions of increasing nutrient depletion, CodY turns off these commensal lifestyle genes and sequentially turns on (by loss of repression) genes required for transporting preformed compounds, de novo amino acid synthesis, and virulence factors to subvert host defenses and damage host cells and tissues. Salvaging ILV and purines from the host adjusts CodY-dependent gene expression appropriate for that microenvironment.
Most revealing in this experiment is the placement of many of the well-studied virulence genes within the CodY-dependent gene expression hierarchy in S. aureus, linking the availability of nutrients to the ability to cause disease (Fig. 9). In our working model, during periods of relative nutrient abundance, positively regulated genes coding for microbial surface components recognizing adhesive matrix molecule (MSCRAMM) proteins like fnbA, spa, clfB and sdrC are expressed as are genes encoding 8 of the 29 enterotoxin-like proteins located within the same genomic context as the egc cluster (Thomas et al., 2006). These genes are among the first to be shut off as ILV and GTP are depleted. Although the enterotoxic properties have not been confirmed, one possibility is that S. aureus may attempt to prevent eliciting an immune response as a survival strategy once nutrients become limiting. MSCRAMM proteins are shut off upon nutrient limitation to potentially limit persistence and promote dissemination. As nutrients continue to be depleted, sae as well as genes coding for a type VII secretion system (esa) and chp are turned on. agr and genes coding for enzymes and proteins that extract nutrients from tissue (e.g., lipases, proteases, hyaluronidase) and those that elicit cellular toxicity (e.g., hld) are among the last genes to be de-repressed, suggesting that S. aureus limits the most severe damage to the host until nutrients are nearly depleted.
Hosts often sequester essential nutrients that challenge bacteria to adjust gene expression to nutrient limiting environments (Weinberg, 1975). Moreover, reactive oxygen species serve as an additional host defense and can damage essential iron-sulfur clusters in IlvD and LeuD (Jang & Imlay, 2007, Brown et al., 1995), potentially rendering S. aureus an ILV auxotroph during infection. Human plasma and ocular fluids contain millimolar concentrations of ILV, but anterior nares contain 10-fold lower concentrations of leucine and valine and lack detectable isoleucine (Durham, 1970, Nasset et al., 1979, Krismer et al., 2014, Nakatsukasa et al., 2011). Moreover, staphylococcal abscesses have been proposed to be limiting in guanine nucleotides (Valentino et al., 2014). Thus, S. aureus likely encounters varying levels of ILV and guanine nucleotides in host tissue during infection, potentially resulting in variable CodY activity and CodY-dependent gene expression in various host niches. A recent study showed that community-acquired S. aureus (CA-MRSA) strains lacking CodY are hypervirulent in two mouse models of infection, demonstrating that CodY regulates virulence gene expression in vivo (Montgomery et al., 2012). However, the extent to which CodY is active in these infection sites is unknown. Indeed, CodY may serve as an important regulator whose graded activity can mediate the switch between commensal and pathogenic lifestyles. We are actively pursuing these questions in our laboratory.
Experimental Procedures
Strains and growth conditions
Bacterial strains used in this study are listed in Tables 3 and S1. Lennox (L) medium lacking glucose (Lennox, 1955) was used to cultivate Escherichia coli strains; tryptic soy broth (TSB) or chemically defined medium (CDM) (Hussain et al., 1991) was used to cultivate S. aureus strains. When necessary, media were solidified with agar at 1.5% [w/v]. Media were supplemented with antibiotics at the following concentrations to maintain selection: ampicillin (Ap), 50 μg ml−1; chloramphenicol (Cm), 10 μg ml−1; erythromycin (Em), 5 μg ml−1; tetracycline (Tc), 1.5 μg ml−1. 5-bromo-4-chloro-3-indolyl-beta-D-galacto-pyranoside (X-gal) was included at a concentration of 80 μg ml−1 to detect BgaB activity on solid media. Experiments were performed in 125 ml or 250 ml DeLong shake flasks with either 25 ml or 50 ml culture volumes, respectively (5:1 flask-to-volume ratio). Experiments were initiated by inoculating overnight cultures incubated in TSB at 37°C to an optical density at 600 nm (OD600) of 0.05. Flasks were incubated at 37°C in an Innovo orbital shaking water bath (New Brunswick) with vigorous shaking (280 RPM) until OD600 ~1.0. In order to dilute out the effect on transcription of residual quorum-sensing signals accumulated during overnight growth (e.g., RNAIII), cultures were re-diluted into fresh medium to OD600 = 0.05, maintaining the flask-to-volume ratio. Growth was monitored as increase in absorbance at 600 nm using a Beckman DU350 UV-visible spectrophotometer (Beckman Coulter).
Table 3.
S. aureus strains used.
| Strain | Description or genotype | Source or referencea |
|---|---|---|
| AR2 | Newman Δica::tet | GB Pier via AL Sonenshein (Kropec et al., 2005) |
| AR11 | SRB337 Δica::tet | |
| AR12 | SRB337 Δica::tet ΔcodY::erm | |
| NE1132 | JE2 icaR::ϕN∑ | (Fey et al., 2013) |
| NE1555 | JE2 codY::ϕN∑ | (Fey et al., 2013) |
| NE1622 | JE2 saeR::ϕN∑ | (Fey et al., 2013) |
| RN4220 | Restriction-deficient, highly transformable | (Kreiswirth et al., 1983) |
| MS-1 | ΔcodY::erm | (Majerczyk et al., 2008) |
| SRB337 | USA200 MSSA UAMS-1 | (Gillaspy et al., 1995b) |
| SRB372 | SRB337 ΔcodY::erm | |
| SRB373 | SRB337 ΔsucD2::cat194 | |
| SRB487 | SRB337 ΔilvE1 codY489 CodYG129D | |
| SRB493 | SRB337 codY489 CodYG129D | |
| SRB506 | SRB337 codY57 CodYR61K | |
| SRB577 | SRB337 codY66 CodYR61E | |
| SRB623 | SRB337 codY58 CodYR61H | |
| SRB687 | USA300 CA-MRSA EmS (LAC*)(AH1263) | A.R. Horswill |
| SRB742 | JE2 saeR::ϕN∑[erm::tet(M)] | |
| SRB746 | SRB687 ΔcodY::erm | |
| SRB759 | SRB337 ΔcodY::erm saeR::ϕN∑[erm::tet(M)] | |
| SRB761 | SRB687 ΔcodY::erm saeR::ϕN∑[erm::tet(M)] | |
| SRB769 | SRB337/pMRSI-nuc (nuc-gfp) | |
| SRB770 | SRB372/pMRS1-nuc (nuc-gfp) | |
| SRB798 | JE2 codY::ϕN∑ [erm::tet(M)] | |
| SRB813 | SRB337icaR::ϕN∑ | |
| SRB814 | SRB337 codY::ϕN∑[erm::tet(M)] | |
| SRB817 | SRB337 icaR::ϕN∑ codY::ϕN∑[erm::tet(M)] | |
| UAMS-929 | UAMS-1 sarA::kan | (Blevins et al., 2002) |
| UAMS-1471 | UAMS-1 Δnuc | (Tsang et a1., 2008) |
Unless otherwise noted, strains were constructed during the course of this study.
Molecular biology and genetic techniques
Oligonucleotides used in this study were synthesized by Integrated DNA Technologies (Coralville, IA). Non-radioactive Sanger sequencing was performed by GeneWiz (Germantown, MD). Plasmids were introduced into S. aureus strain RN4220 by electroporation as previously described (Schenk & Laddaga, 1992). Plasmids and marked mutations were mobilized between S. aureus strains by φ11-mediated transduction (Novick, 1991).
Strain and plasmid construction
Fragments for generating mutant alleles were synthesized using overlap PCR (Vallejo et al., 2008) using oligonucleotides in Table S2. Mutant strains producing variant CodY proteins or deleted for ilvE or sucD were constructed by allelic exchange using pMAD and confirmed by sequencing (Arnaud et al., 2004). When possible, mutants were reconstructed in UAMS-1 by transduction. The mutant producing CodYG129D (encoded by the codY489 allele) was isolated as a spontaneous mutant during the construction of an ilvE null mutant. We moved the codY489 allele into UAMS-1 by cotransduction with a linked sucD+ allele into a sucD::cat194 mutant, selecting for growth on chemically defined medium (Lee et al., 2009a) lacking glucose as the carbon source.
codY and saeR mutant construction
S. aureus JE2 containing transposon insertions in the codY gene and the saeR gene were obtained from the Nebraska Transposon Mutant Library (NTML, strains NE1555 and NE1622, respectively) (Fey et al., 2013). The pTET plasmid from the NTML genetic toolkit was used to replace the erythromycin resistance cassette with a tetracycline-resistant cassette as previously described (Bose et al., 2013). These mutations were then moved into UAMS-1 by transduction.
Generation of the nuc-sGFP reporter fusion
To generate the nuc-sGFP fusion, we used a double reporter shuttle vector designated as pMRSI (Fig. S4). The pMRSI plasmid, a derivative of pDM4 (Moormeier et al., 2013), was constructed by PCR using primers KPPE-TIR-f and NNSB-TIR*-r to replace divergently transcribed cidABC and lrgAB promoter regions of the pDM4 with a multiple cloning site (MCS). Then, the PCR fragment was treated with the restriction endonuclease DpnI (to hydrolyze the pDM4 template DNA), self-ligated and transformed into the E. coli strain DH5α. The replacement of the cidABC and lrgAB promoter regions with the MCS and integrity of the TIRs and sDsRed and sGFP genes in the pMRSI plasmid were confirmed by DNA sequencing using primers sDsRed-S-r, sGFP-S-r, pCN51-S1-f, and pCN51-S-r.
Primers used for construction of the nuc-sGFP reporter fusion were generated based on the sequence of S. aureus strain MRSA252 (NC_002952.2). A 382-nucleotide (nt) PCR fragment containing the promoter region, ribosome binding site (RBS) and translation initiating codon (ATG) of the S. aureus UAMS-1 nuc (SAR0847) gene was amplified using primers SphI-Pnuc-f and Pnuc1-r1. Following digestion with the restriction endonuclease SphI the PCR product was used to replace the SphI-SmaI DNA fragment of the plasmid pMRSI containing sDsRed gene, MCS and translation initiating codon (ATG) of the sGFP gene. The resulting recombinant plasmid designated as pMRSI-nuc was transformed into strain RN4220 by electroporation and then introduced into UAMS-1 and UAMS-codY strains by phage Φ11-mediated transduction.
RNA sampling and preparation
Samples (4 ml) of exponentially-growing cultures were rapidly withdrawn from each flask, mixed by vortexing with an equal volume of 100% ethanol-acetone quench solution (1:1 [v/v], pre-chilled to −20°C) and immediately frozen on dry ice and stored at −80°C. Nucleic acids were extracted from cell pellets after washing twice with 0.5 ml TE buffer (10 mM Tris-Cl [pH 8], 1 mM EDTA) by mechanical disruption using a Precellys 24 homogenizer (Bertin Technologies) in 4 ml Trizol (Invitrogen) with three 30 sec pulses at 6,800 RPM with 1 min incubations on wet ice between pulses. RNAs were isolated from clarified cell lysates using the Direct-Zol kit (Zymo Research Corporation) as per the manufacturer’s recommendations. Genomic DNA was reduced to mathematically insignificant levels from samples using the Turbo DNA-free DNase treatment and removal kit (Ambion) according to the manufacturer’s “rigorous-treatment” instructions. Total RNA preparations were quantified using a computer-controlled NanoDrop ND-1000 spectrophotometer (Thermo Scientific) running quantification software version 3.1.0.
Quantitative, real-time RT-PCR
250 ng of total RNA from each sample was used as template to synthesize cDNA using the Protoscript first strand cDNA synthesis kit (NEB) and random primers as per the manufacturer’s instructions. Parallel reactions lacking reverse transcriptase were used to control for any remaining residual DNA contamination. Quantitative PCR (qPCR) was performed using a C1000 thermal cycler fitted with a CFX96 detection module (Bio-Rad) and SsoAdvanced Universal SYBR Green Supermix (Bio-Rad). Oligonucleotides for qPCR (Table S2) were used at a concentration of 400 nM. The reaction mixture was incubated at 98°C for 2 min followed by thermal cycling between 98°C and 60°C. No-template and no-RT controls were run on each plate for each assay, and specificity of each amplification product was verified using dissociation curve analysis. Standard curves were generated from serial dilutions of chromosomal DNA spanning at least 6 orders of magnitude. All reactions proceeded within 90% to 110% efficiency, and data points lay within linear regression with correlation coefficients (r2 values) >98%. Data are presented as copies of target transcript relative to copies of rpoC or polC transcript because expression was constant between strains and replicates.
Transcriptional profiling with RNA-Seq and analysis
RNA-Seq was performed on two independent biological replicates for each strain. An 8.6-μg sample of nucleic acid was depleted of DNA using the Turbo DNA-free kit (Ambion). RNA integrity was assessed using a 2100 Bioanalyzer and the RNA 6000 Pico kit (Agilent); samples exhibiting RNA Integrity Numbers > 8 were used for downstream applications. DNA-depleted RNAs (negative PCR amplification of rpoC after 25 cycles using oSRB239 and oSRB240) were then depleted of rRNA using the Ribo-Zero magnetic kit for Gram-positive bacteria (Illumina). When necessary, RNAs were concentrated using the RNA Clean & Concentrator-5 kit (Zymo Research Corporation). Samples containing < 3% 16s and 23s rRNA were used for library preparation. Libraries were constructed using the TruSeq Stranded mRNA Library Prep Kit (Illumina), and sequenced in a single, high-throughput flow cell lane with TruSeq V3 chemistry using an Illumina (HiSeq 2500) at the Tufts University Genomics Core Facility. Single-ended reads were filtered to retain reads ~ 50bp, which were then cleaned and aligned to the S. aureus UAMS-1 genome (GI:727808807) using BWA (Li & Durbin, 2009). Because the published UAMS-1 annotation is incomplete, we first annotated the UAMS-1 genome with a custom BLAST+ pipeline, obtaining the best match from the USA300_TCH1516 annotation for protein sequences. If that failed, we obtained the best match from the MRSA252 genome. Genes previously listed as hypothetical or missing from these two genomes were manually searched against the NCBI nr database to find probable homologs. Additional annotations for the USA300 and MRSA252 strains were obtained from KEGG (Kanehisa & Goto, 2000, Kanehisa et al., 2014). These sequencing data have been deposited in the Sequence Read Archive of the National Center for Biotechnology (accession number xxxxx).
Using a counts table generated with HTSeq (Anders et al., 2015) containing mapped fragments per gene, all subsequent analyses were performed in R. DESeq2 (Love et al., 2014) was used to determine differential expression and normalization. Normalized counts from DESeq2 were converted to Reads Per Kilobase per Million reads mapped to Open Reading Frames (ORFs), or RPKMOs, and then converted to percentages of maximal expression (i.e., expression in the CodY null mutant for negatively regulated genes and expression in the WT for positively regulated genes). K-means clustering of the least and most active CodY variant strains was used to group similarly expressed genes. Virulence genes were manually annotated by referencing previous literature (Cheng et al., 2011, Majerczyk et al., 2010, Majerczyk et al., 2008, Kobayashi et al., 2011, Rogasch et al., 2006).
Metabolite sampling and analysis
Cells for metabolite analysis were grown in TSB as described above. At OD600 ~0.5, a 13 ml sample of cells was collected via vacuum filtration on 0.2 μm nitrocellulose filters (Millipore) and washed with 5 ml ice cold PBS. Filters were quenched in Acetonitrile/Methanol/H2O (40:40:20) supplemented with 0.1 M formic acid on a bed of dry ice. Cells were mechanically disrupted with 0.1 mm zirconia beads in Precellys 24 homogenizer (Bertin Technologies) for four, 30 sec pulses at 6,000 rpm with a 2 min cooling period on dry ice in between pulses. Lysates were clarified by a 15 min centrifugation at 18,213 × g at −11° C. Residual peptide concentration was used to normalize for biomass in these samples and was quantified using the Micro BCA Protein Assay Kit (Pierce) according to manufacturer’s instructions. Tandem liquid chromatography coupled to Time-Of-Flight mass spectrometry (LC-MS) was performed as described previously (Brinsmade et al., 2014). Experiments were performed independently at least three times.
LC-MS data processing and analysis
To normalize samples by biomass, raw ion counts obtained from LC-MS analysis were divided by the residual peptide concentration of each sample. To correct for batch effects between samples, the average ion count for all metabolites within a sample was calculated. The average ion count for each sample was divided by the average ion count for the WT-1 sample to generate a ratio. The biomass-normalized ion counts for all metabolites within a sample were each divided by this ratio to yield a corrected ion count. For the comparison of WT S. aureus (SRB337) with the codY null mutant (SRB372), a Mann-Whitney U test was used to determine significant differences in intracellular abundance. To compare the significance of differences across all six strains, we used Cuzick’s nonparametric test for trend (Cuzick, 1985). Statistical analyses were performed in either Microsoft Excel or StataSE 14 (StataCorp).
Measuring nuc expression using a nuc-gfp transcriptional reporter
Strains were cultivated as described above. At the indicated time point, samples were removed, washed twice with phosphate buffered saline (PBS) and re-suspended in PBS to remove background fluorescence from TSB medium. Fluorescence was measured using a computer-controlled Tecan Infinite F200 Pro plate reader (equipped with 485 nm and 535 nm excitation and emission filters, respectively) and i-control software (Ver. 1.11). GFP signal acquisition settings were kept constant: gain, 35%; flash number, 25; integration time, 20 μs; lag time, 0 μs; settle time, 0 ms. Relative fluorescence units (RFUs) were calculated by subtracting the fluorescence from SRB337 (UAMS-1, lacks GFP reporter) and dividing by OD600 to correct for cell density.
Secreted Nuclease activity assays
Secreted nuclease activity was quantified essentially as described previously employing a Fluorescence Resonance Energy Transfer (FRET) assay (Kiedrowski et al., 2011). In short, cells were grown as described above; supernatants were sterilized using 0.22 μm Spin X filters (Corning) and mixed with the FRET substrate: a single-stranded 15-mer oligonucleotide with the 5’ end modified with a Cy3 fluorophore and the 3’ end modified with BHQ2 quencher (Table S2). Fluorescence from cleaved substrate was measured using a computer-controlled Tecan Infinite F200 Pro plate reader (equipped with 535 (25) nm and 590 (20) nm excitation and emission filters, respectively) and Magellan software (Ver. 7.1). Nuclease activity was related to a standard activity curve generated with purified micrococcal nuclease enzyme (Worthington Biochemicals).
Static biofilm assays
Static biofilm development was assayed in tissue-culture treated microtiter plates as previously described (Loughran et al., 2014), except that the well bottom geometry was round rather than flat. Briefly, 200μl of freshly prepared biofilm medium (BFM; TSB supplemented with 0.5 % [w/v] glucose and 3 % [w/v] NaCl) was distributed to wells of the microtiter plate. S. aureus strains were grown as described above, inoculating BFM to an initial optical density of 0.01 in a total volume of 200 μl. Plates were incubated statically for 24 hours at 37°C. All proceeding manipulations from this point were performed at room temperature. Medium and non-adherent bacteria were removed from the microtiter plate by gentle inversion; fluid was completely removed by tapping against absorbent paper. The wells were then washed four times with 200 μl of Phosphate Buffered Saline (PBS; Ambion) to remove non-adherent cells. The PBS was also removed by gentle blotting against absorbent paper. Washed biofilms were fixed with 200 μl 100% [v/v] ethanol for 20 min followed by staining with 200 μl 0.1% [w/v] crystal violet (in 12 % [v/v] ethanol) for 8 min. The crystal violet was decanted and the wells were again washed four times each with 200 μl of PBS. Plates were inverted and dried for 10 min, at which time crystal violet was eluted into 200 μl of 95% ethanol for 10 min. Absorbance (600 nm) of each sample well was measured using an Infinite F200 Pro plate reader (Tecan), controlled by the Magellan software. When necessary, samples were diluted to remain within the linear range of the plate reader.
Immunoblot analysis
Immunoblotting was performed as described (Brinsmade et al., 2014) except that we incubated samples with lysostaphin (2 mg ml−1) prior to sonication and rabbit anti-CodY antibody was applied at 1:3,000 dilution.
Statistical Significance
Unless otherwise specified, statistical significance for experimental data was determined using Games-Howell test (Games et al., 1979) chosen for its robustness across data types for pairwise comparisons. Unless noted elsewhere, experimental data reported are the result of at least two independent replicates.
Supplementary Material
Acknowledgements
We thank Jeffrey Cavanaugh for advice on the nuclease activity assay, and Alex Horswill and Mark Smeltzer for the gift of strains. We also thank Abraham “Linc” Sonenshein and Boris Belitsky for helpful advice, as well as Agnès Roux, Deepak Sharma, Kevin Mlynek, Kevin Craig and Nivedita Sengupta for technical assistance as well as Jennifer Fox and Matt Hamilton for advice on statistical approaches.
Funding Information
This work was funded in part by an NIH Pathway to Independence Award (grant GM099893) and faculty startup funds to SRB, as well as a Research Project Grant (grant GM042219) prior to work performed at Georgetown University. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
References
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud M, Chastanet A, Débarbouillé M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 2004;70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood DN, Loughran AJ, Courtney AP, Anthony AC, Meeker DG, Spencer HJ, Gupta RK, Lee CY, Beenken KE, Smeltzer M. Comparative impact of diverse regulatory loci on Staphylococcus aureus biofilm formation. Microbiologyopen. 2015;4:436–451. doi: 10.1002/mbo3.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basavanna S, Chimalapati S, Maqbool A, Rubbo B, Yuste J, Wilson RJ, Hosie A, Ogunniyi AD, Paton JC, Thomas G, Brown JS. The effects of methionine acquisition and synthesis on Streptococcus pneumoniae growth and virulence. PLoS One. 2013;8:e49638. doi: 10.1371/journal.pone.0049638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenken KE, Blevins JS, Smeltzer M. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect Immun. 2003;71:4206–4211. doi: 10.1128/IAI.71.7.4206-4211.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky BR, Sonenshein AL. Roadblock repression of transcription by Bacillus subtilis CodY. J Mol Biol. 2011;411:729–743. doi: 10.1016/j.jmb.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins JS, Beenken KE, Elasri MO, Hurlburt BK, Smeltzer MS. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect immun. 2002;70:470–480. doi: 10.1128/IAI.70.2.470-480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose JL, Fey PD, Bayles KW. Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl Environ Microbiol. 2013;79:2218–2224. doi: 10.1128/AEM.00136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinsmade S, Alexander E, Livny J, Stettner A, Segrè D, Rhee K, Sonenshein A. Hierarchical expression of genes controlled by the Bacillus subtilis global regulatory protein CodY. Proc Natl Acad Sci U S A. 2014;111:8227–8232. doi: 10.1073/pnas.1321308111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown OR, Smyk-Randall E, Draczynska-Lusiak B, Fee JA. Dihydroxy-acid dehydratase, a [4Fe-4S] cluster-containing enzyme in Escherichia coli: effects of intracellular superoxide dismutase on its inactivation by oxidant stress. Arch Biochem Biophys. 1995;319:10–22. doi: 10.1006/abbi.1995.1262. [DOI] [PubMed] [Google Scholar]
- Bubeck Wardenburg J, Patel R, Schneewind O. Surface proteins and exotoxins are required for the pathogenesis of Staphylococcus aureus pneumonia. Infect Immun. 2007;75:1040–1044. doi: 10.1128/IAI.01313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke ME, Pattee PA. Histidine biosynthetic pathway in Staphylococcus aureus. Can J Microbiol. 1972;18:569–576. doi: 10.1139/m72-090. [DOI] [PubMed] [Google Scholar]
- Casewell M, Hill R. The carrier state: methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 1986;18(Suppl A):1–12. doi: 10.1093/jac/18.supplement_a.1. [DOI] [PubMed] [Google Scholar]
- Cheng A, DeDent A, Schneewind O, Missiakas D. A play in four acts: Staphylococcus aureus abscess formation. Trends Microbiol. 2011;19:225–232. doi: 10.1016/j.tim.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Kim H, Burts M, Krausz T, Schneewind O, Missiakas D. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J. 2009;23:3393–3404. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A, Nishina K, Trotonda M, Tamber S. The SarA protein family of Staphylococcus aureus. Int J Biochem Cell Biol. 2008;40:355–361. doi: 10.1016/j.biocel.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzick J. A Wilcoxon-type test for trend. Stat Med. 1985;4:87–90. doi: 10.1002/sim.4780040112. [DOI] [PubMed] [Google Scholar]
- den Hengst CD, van Hijum SAFT, Geurts JMW, Nauta A, Kok J, Kuipers OP. The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J Biol Chem. 2005;280:34332–34342. doi: 10.1074/jbc.M502349200. [DOI] [PubMed] [Google Scholar]
- Durham D. Distribution of free amino acids in human intraocular fluids. Trans Am Ophthalmol Soc. 1970;68:462–500. [PMC free article] [PubMed] [Google Scholar]
- Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio. 2013;4:e00537–00512. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink PS. Biosynthesis of the branched-chain amino acids. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and Other Gram-Positive Bacteria. American Society for Microbiology; Washington, DC: 1993. pp. 307–317. [Google Scholar]
- Games PA, Keselman HJ, Clinch JJ. Tests for homogeneity of variance in factorial designs. Psychol Bull. 1979;86:978–984. [Google Scholar]
- Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol Microbiol. 2006;61:1038–1048. doi: 10.1111/j.1365-2958.2006.05292.x. [DOI] [PubMed] [Google Scholar]
- Gillaspy A, Hickmon S, Skinner R, Thomas J, Nelson C, Smeltzer M. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect Immun. 1995a;63:3373–3380. doi: 10.1128/iai.63.9.3373-3380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillaspy AF, Hickmon SG, Skinner RA, Thomas JR, Nelson CL, Smeltzer MS. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 1995b;63:3373–3380. doi: 10.1128/iai.63.9.3373-3380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudo A, Calzolari A, Cataldi A, Bogni C, Nagel R. The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol Lett. 1999;177:15–22. doi: 10.1111/j.1574-6968.1999.tb13707.x. [DOI] [PubMed] [Google Scholar]
- Herold B, Immergluck L, Maranan M, Lauderdale D, Gaskin R, Boyle-Vavra S, Leitch C, Daum R. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA. 1998;279:593–598. doi: 10.1001/jama.279.8.593. [DOI] [PubMed] [Google Scholar]
- Hussain M, Hastings JG, White PJ. A chemically defined medium for slime production by coagulase-negative staphylococci. J Med Microbiol. 1991;34:143–147. doi: 10.1099/00222615-34-3-143. [DOI] [PubMed] [Google Scholar]
- Jang S, Imlay JA. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J. Biol. Chem. 2007;282:929–937. doi: 10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzon L, Löfdahl S, Arvidson S. Identification and nucleotide sequence of the delta-lysin gene, hld, adjacent to the accessory gene regulator (agr) of Staphylococcus aureus. Mol Gen Genet. 1989;219:480–485. doi: 10.1007/BF00259623. [DOI] [PubMed] [Google Scholar]
- Jefferson KK, Pier DB, Goldmann DA, Pier GB. The Teicoplanin-Associated Locus Regulator (TcaR) and the Intercellular Adhesin Locus Regulator (IcaR) Are Transcriptional Inhibitors of the ica Locus in Staphylococcus aureus. J Bacteriol. 2004;186:2449–2456. doi: 10.1128/JB.186.8.2449-2456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi G, Spontak J, Klapper D, Richardson A. Arginine catabolic mobile element encoded speG abrogates the unique hypersensitivity of Staphylococcus aureus to exogenous polyamines. Mol Microbiol. 2011;82:9–20. doi: 10.1111/j.1365-2958.2011.07809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser J, Omer S, Sheldon J, Welch I, Heinrichs D. Role of BrnQ1 and BrnQ2 in branched-chain amino acid transport and virulence in Staphylococcus aureus. Infect Immun. 2015;83:1019–1029. doi: 10.1128/IAI.02542-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane JF, Jensen RA. Metabolic interlock. The influence of histidine on tryptophan biosynthesis in Bacillus subtilis. J Biol Chem. 1970;245:2384–2390. [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiedrowski M, Kavanaugh J, Malone C, Mootz J, Voyich J, Smeltzer M, Bayles K. Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus. PLoS One. 2011;6:e26714. doi: 10.1371/journal.pone.0026714. H. AR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi SD, Malachowa N, Whitney AR, Braughton KR, Gardner DJ, Long D, Bubeck Wardenburg J, Schneewind O, Otto M, Deleo FR. Comparative analysis of USA300 virulence determinants in a rabbit model of skin and soft tissue infection. J Infect DIs. 2011;204:937–941. doi: 10.1093/infdis/jir441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiswirth BN, Löfdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- Krismer B, Liebeke M, Janek D, Nega M, Rautenberg M, Hornig G, Unger C, Weidenmaier C, Lalk M, Peschel A. Nutrient limitation governs Staphylococcus aureus metabolism and niche adaptation in the human nose. PLoS Pathog. 2014;10:e1003862. doi: 10.1371/journal.ppat.1003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropec A, Maira-Litran T, Jefferson KK, Grout M, Cramton SE, Götz F, Goldmann DA, Pier GB. Poly-N-acetylglucosamine production in Staphylococcus aureus is essential for virulence in murine models of systemic infection. Infect Immun. 2005;73:6868–6876. doi: 10.1128/IAI.73.10.6868-6876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, David M, Boyle-Vavra S, Sieth J, Daum R. High Staphylococcus aureus colonization prevalence among patients with skin and soft tissue infections and controls in an urban emergency department. J Clin Microbiol. 2015;53:810–815. doi: 10.1128/JCM.03221-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D-S, Burd H, Liu J, Almaas E, Wiest O, Barabási A-L, Oltavi ZN, Kapatral V. Comparative genome-scale metabolic reconstruction and flux balance analysis of multiple Staphylococcus aureus genomes identify novel antimicrobial drug targets. J Bacteriol. 2009a;191:4015–4024. doi: 10.1128/JB.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DS, Burd H, Liu J, Almaas E, Wiest O, Barabási AL, Oltvai ZN, Kapatral V. Comparative genome-scale metabolic reconstruction and flux balance analysis of multiple Staphylococcus aureus genomes identify novel antimicrobial drug targets. J Bacteriol. 2009b;191:4015–4024. doi: 10.1128/JB.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennox ES. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955;1:190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Levdikov VM, Blagova E, Colledge VL, Lebedev AA, Williamson DC, Sonenshein AL, Wilkinson AJ. Structural rearrangement accompanying ligand binding in the GAF domain of CodY from Bacillus subtilis. J Mol Biol. 2009;390:1007–1018. doi: 10.1016/j.jmb.2009.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levdikov VM, Blagova E, Joseph P, Sonenshein AL, Wilkinson AJ. The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in Gram-positive bacteria. J Biol Chem. 2006;281:11366–11373. doi: 10.1074/jbc.M513015200. [DOI] [PubMed] [Google Scholar]
- Li D, Cheung A. Repression of hla by rot is dependent on sae in Staphylococcus aureus. Infect Immun. 2008;76:1068–1075. doi: 10.1128/IAI.01069-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughran AJ, Atwood DN, Anthony AC, Harik NS, Spencer HJ, Beenken KE, Smeltzer MS. Impact of individual extracellular proteases on Staphylococcus aureus biofilm formation in diverse clinical isolates and their isogenic sarA mutants. Microbiologyopen. 2014;3:897–909. doi: 10.1002/mbo3.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy F. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- Mainiero M, Goerke C, Geiger T, Gonser C, Herbert S, Wolz C. Differential target gene activation by the Staphylococcus aureus two-component system saeRS. J Bacteriol. 2010;192:613–623. doi: 10.1128/JB.01242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerczyk CD, Dunman PM, Luong TT, Lee CY, Sadykov MR, Somerville GA, Bodi K, Sonenshein AL. Direct targets of CodY in Staphylococcus aureus. J Bacteriol. 2010;192:2861–2877. doi: 10.1128/JB.00220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerczyk CD, Sadykov MR, Luong TT, Lee C, Somerville GA, Sonenshein AL. Staphylococcus aureus CodY negatively regulates virulence gene expression. J Bacteriol. 2008;190:2257–2265. doi: 10.1128/JB.01545-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCourt J, O'Halloran DP, McCarthy H, O'Gara JP, Geoghegan JA. Fibronectin-binding proteins are required for biofilm formation by community-associated methicillin-resistant Staphylococcus aureus strain LAC. FEMS Microbiol Lett. 2014;353:157–164. doi: 10.1111/1574-6968.12424. [DOI] [PubMed] [Google Scholar]
- McNamara PJ, Milligan-Monroe KC, Khalili S, Proctor RA. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J Bacteriol. 2000;182:3197–3203. doi: 10.1128/jb.182.11.3197-3203.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengin-Lecreulx D, Falla T, Blanot D, van Heijenoort J, Adams DJ, Chopra I. Expression of the Staphylococcus aureus UDP-N-acetylmuramoyl-L-alanyl-D-glutamate:L-lysine ligase in Escherichia coli and effects on peptidoglycan biosynthesis and cell growth. J Bacteriol. 1999;181:5909–5914. doi: 10.1128/jb.181.19.5909-5914.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery CP, Boyle-Vavra S, Roux A, Ebine K, Sonenshein AL, Daum RS. CodY deletion enhances in vivo virulence of community-associated methicillin-resistant Staphylococcus aureus USA300. Infect Immun. 2012 doi: 10.1128/IAI.06172-11. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moormeier D, Endres J, Mann E, Sadykov M, Horswill A, Rice K, Fey P, Bayles K. Use of microfluidic technology to analyze gene expression during Staphylococcus aureus biofilm formation reveals distinct physiological niches. Appl Environ Microbiol. 2013;79:3413–3424. doi: 10.1128/AEM.00395-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moormeier DE, Bose JL, Horswill AR, Bayles KW. Temporal and stochastic control of Staphylococcus aureus biofilm development. MBio. 2014;5:e01341–01314. doi: 10.1128/mBio.01341-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootz JM, Malone CL, Shaw LN, Horswill AR. Staphopains modulate Staphylococcus aureus biofilm integrity. Infect Immun. 2013;81:3227–3238. doi: 10.1128/IAI.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naimi T, LeDell K, Como-Sabetti K, Borchardt S, Boxrud D, Etienne J, Johnson S, Vandenesch F, Fridkin S, O'Boyle C, Danila R, Lynfield R. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA. 2003;290:2976–2984. doi: 10.1001/jama.290.22.2976. [DOI] [PubMed] [Google Scholar]
- Nakatsukasa M, Sotozono C, Shimbo K, Ono N, Miyano H, Okano A, Hamuro J, Kinoshita S. Amino Acid profiles in human tear fluids analyzed by high-performance liquid chromatography and electrospray ionization tandem mass spectrometry. Am J Ophthalmol. 2011;151:799–808. doi: 10.1016/j.ajo.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Nasset ES, Heald FP, Calloway DH, Margen S, Schneeman P. Amino acids in human blood plasma after single meals of meat, oil, sucrose and whiskey. J Nutr. 1979;109:621–630. doi: 10.1093/jn/109.4.621. [DOI] [PubMed] [Google Scholar]
- Nester EW. Cross pathway regulation: effect of histidine on the synthesis and activity of enzymes of aromatic acid biosynthesis in Bacillus subtilis. J Bacteriol. 1968;96:1649–1657. doi: 10.1128/jb.96.5.1649-1657.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble W, Valkenburg H, Wolters C. Carriage of Staphylococcus aureus in random samples of a normal population. J Hyg (Lond) 1967;65:567–573. doi: 10.1017/s002217240004609x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- Novick R, Jiang D. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology. 2003;149:2709–2717. doi: 10.1099/mic.0.26575-0. [DOI] [PubMed] [Google Scholar]
- Novick RP. Genetic systems in staphylococci. Methods Enzymol. 1991;204 doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- Olson M, Nygaard T, Ackermann L, Watkins R, Zurek O, Pallister K, Griffith S, Kiedrowski M, Flack C, Kavanaugh J, Kreiswirth B, Horswill A, Voyich J. Staphylococcus aureus nuclease is an SaeRS-dependent virulence factor. Infect Immun. 2013;81:1316–1324. doi: 10.1128/IAI.01242-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulus H. Biosynthesis of the aspartate family of amino acids. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and Other Gram-Positive Bacteria. American Society for Microbiology; Washington, DC: 1993. pp. 237–267. [Google Scholar]
- Pohl K, Francois P, Stenz L, Schlink F, Geiger T, Herbert S, Goerke C, Schrenzel J, Wolz C. CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J Bacteriol. 2009;191:2953–2963. doi: 10.1128/JB.01492-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recsei P, Kreiswirth B, O'Reilly M, Schlievert P, Gruss A, Novick R. Regulation of exoprotein gene expression in Staphylococcus aureus by agr. Mol Gen Genet. 1986;202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the methionine metabolism in Gram-positive bacteria: a variety of regulatory systems. Nucleic Acids Res. 2004;32:3340–3353. doi: 10.1093/nar/gkh659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogasch K, Ruhmling V, Pane-Farre J, Hoper D, Weinberg C, Fuchs S, Schmudde M, Broker BM, Wolz C, Hecker M, Engelmann S. Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J Bacteriol. 2006;188:7742–7758. doi: 10.1128/JB.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux A, Todd D, Velázquez J, Cech N, Sonenshein A. CodY-mediated regulation of the Staphylococcus aureus Agr system integrates nutritional and population density signals. J Bacteriol. 2014;196:1184–1196. doi: 10.1128/JB.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz de los Mozos I, Vergara-Irigaray M, Segura V, Villanueva M, Bitarte N, Saramago M, Domingues S, Arraiano CM, Fechter P, Romby P, Valle J, Solano C, Lasa I, Toledo-Arana A. Base pairing interaction between 5'- and 3'-UTRs controls icaR mRNA translation in Staphylococcus aureus. PLos Genet. 2013;9:e1004001. doi: 10.1371/journal.pgen.1004001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadykov MR, Mattes TA, Luong TT, Zhu Y, Day SR, Sifri CD, Lee CY, Somerville GA. Tricarboxylic acid cycle-dependent synthesis of Staphylococcus aureus Type 5 and 8 capsular polysaccharides. J Bacteriol. 2010;192:1459–1462. doi: 10.1128/JB.01377-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado-Pabón W, Breshears L, Spaulding A, Merriman J, Stach C, Horswill A, Peterson M, Schlievert P. Superantigens are critical for Staphylococcus aureus Infective endocarditis, sepsis, and acute kidney injury. MBio. 2013;4:e00494–00513. doi: 10.1128/mBio.00494-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassi M, Sharma D, Brinsmade S, Felden B, Augagneur Y. Genome Sequence of the Clinical Isolate Staphylococcus aureus subsp. aureus Strain UAMS-1. Genome Announc. 2015;3:e01584–01514. doi: 10.1128/genomeA.01584-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk S, Laddaga RA. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett. 1992;73:133–138. doi: 10.1016/0378-1097(92)90596-g. [DOI] [PubMed] [Google Scholar]
- Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder SM, Marincola G, Geiger T, Goerke C, Wolz C, Ziebuhr W. Methionine biosynthesis in Staphylococcus aureus is tightly controlled by a hierarchical network involving an initiator tRNA-specific T-box riboswitch. PLoS Pathog. 2013;9:e1003606. doi: 10.1371/journal.ppat.1003606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville GA, Proctor RA. At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol Mol Biol Rev. 2009;73:233–248. doi: 10.1128/MMBR.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM. Systematic determination of genetic network architecture. Nat Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- Thomas DY, Jarraud S, Lemercier B, Cozon G, Echasserieau K, Etienne J, Gougeon ML, Lina G, Vandenesch F. Staphylococcal enterotoxin-like toxins U2 and V, two new staphylococcal superantigens arising from recombination within the enterotoxin gene cluster. Infect Immun. 2006;74:4724–4734. doi: 10.1128/IAI.00132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang LH, Cassat JE, Shaw LN, Beenken KE, Smeltzer MS. Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PLoS One. 2008;3:e3361. doi: 10.1371/journal.pone.0003361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentino M, Foulston L, Sadaka A, Kos V, Villet R, Santa Maria JJ, Lazinski D, Camilli A, Walker S, Hooper D, Gilmore M. Genes contributing to Staphylococcus aureus fitness in abscess- and infection-related ecologies. MBio. 2014;5:e01729–01714. doi: 10.1128/mBio.01729-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo AN, Pogulis RJ, Pease LR. PCR mutagenesis by overlap extension and gene SOE. CSH Protoc. 2008 doi: 10.1101/pdb.prot4861. 2008:pdb.prot4861. [DOI] [PubMed] [Google Scholar]
- Vandenesch F, Naimi T, Enright M, Lina G, Nimmo G, Heffernan H, Liassine N, Bes M, Greenland T, Reverdy M, Etienne J. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg Infect Dis. 2003;9:978–984. doi: 10.3201/eid0908.030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villapakkam AC, Handke LD, Belitsky BR, Levdikov VM, Wilkinson AJ, Sonenshein AL. Genetic and biochemical analysis of the interaction of Bacillus subtilis CodY with branched-chain amino acids. J Bacteriol. 2009;191:6865–6876. doi: 10.1128/JB.00818-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voyich J, Braughton K, Sturdevant D, Whitney A, Saïd-Salim B, Porcella S, Long R, Dorward D, Gardner D, Kreiswirth B, Musser J, DeLeo F. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J Immunol. 2005;175:3907–3919. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- Weinberg E. Nutritional immunity. Host's attempt to withold iron from microbial invaders. JAMA. 1975;231:39–41. doi: 10.1001/jama.231.1.39. [DOI] [PubMed] [Google Scholar]
- Wu S, de Lencastre H, Tomasz A. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J Bacteriol. 1996;178:6036–6042. doi: 10.1128/jb.178.20.6036-6042.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.