ABSTRACT
Sphingomonas sp. strain Ndbn-20 degrades and utilizes the herbicide dicamba as its sole carbon and energy source. In the present study, a tetrahydrofolate (THF)-dependent dicamba methyltransferase gene, dmt, was cloned from the strain, and three other genes, metF, dhc, and purU, which are involved in THF metabolism, were found to be located downstream of dmt. A transcriptional study revealed that the four genes constituted one transcriptional unit that was constitutively transcribed. Lysates of cells grown with glucose or dicamba exhibited almost the same activities, which further suggested that the dmt gene is constitutively expressed in the strain. Dmt shared 46% and 45% identities with the methyltransferases DesA and LigM from Sphingomonas paucimobilis SYK-6, respectively. The purified Dmt catalyzed the transfer of methyl from dicamba to THF to form the herbicidally inactive metabolite 3,6-dichlorosalicylic acid (DCSA) and 5-methyl-THF. The activity of Dmt was inhibited by 5-methyl-THF but not by DCSA. The introduction of a codon-optimized dmt gene into Arabidopsis thaliana enhanced resistance against dicamba. In conclusion, this study identified a THF-dependent dicamba methyltransferase, Dmt, with potential applications for the genetic engineering of dicamba-resistant crops.
IMPORTANCE Dicamba is a very important herbicide that is widely used to control more than 200 types of broadleaf weeds and is a suitable target herbicide for the engineering of herbicide-resistant transgenic crops. A study of the mechanism of dicamba metabolism by soil microorganisms will benefit studies of its dissipation, transformation, and migration in the environment. This study identified a THF-dependent methyltransferase, Dmt, capable of catalyzing dicamba demethylation in Sphingomonas sp. Ndbn-20, and a preliminary study of its enzymatic characteristics was performed. Introduction of a codon-optimized dmt gene into Arabidopsis thaliana enhanced resistance against dicamba, suggesting that the dmt gene has potential applications for the genetic engineering of herbicide-resistant crops.
INTRODUCTION
Dicamba (3,6-dichloro-2-methoxybenzoic acid) is a type of synthetic auxin herbicide that has been widely used to control more than 200 kinds of broadleaf weeds. Because of its low cost, high efficiency, broad weed control spectrum, easy degradation in the environment, and low mammalian toxicity (1–4), dicamba is considered a suitable target herbicide for the engineering of herbicide-resistant transgenic crops (5). Recently, the biotechnology company Monsanto successfully developed dicamba- and glyphosate (or bialaphos)-resistant soybean and cotton by introducing a dicamba monooxygenase (DMO) gene and a glyphosate resistance gene (encoding CP4 EPSPS) (or bialaphos resistance gene bar) (5). These genetically modified (GM) crops received regulatory approval from the U.S. Department of Agriculture (USDA) in August 2014 (http://www.aphis.usda.gov/newsroom/2014/08/pdf/brs_eis.pdf) and have been commercially available since 2015. The increasing usage of dicamba has raised concerns regarding its dissipation and microbial degradation mechanism in the environment. Furthermore, the dicamba degradation or detoxification gene has potential applications in the construction of herbicide-resistant transgenic crops.
Dicamba was chemically stable but did not persist in soil and water, suggesting that its dissipation in the environment was biologically mediated (6, 7, 8, 9). Several dicamba-utilizing strains have been isolated, including the aerobic strains Stenotrophomonas (formerly Pseudomonas) maltophilia DI-6 (6), Sphingomonas sp. RW5 (10), Sphingobium sp. Ndbn-10, and Sphingomonas sp. Ndbn-20 (11) and the anaerobic strain Moorella thermoacetica (7). The first step in the degradation of dicamba in these strains was demethylation to generate the herbicidally inactive metabolite 3,6-dichlorosalicylic acid (DCSA) (6, 9). To date, two types of dicamba demethylation systems have been documented. In S. maltophilia DI-6, a DMO was responsible for the demethylation of dicamba. The DMO belonged to the Rieske nonheme iron oxygenase (RHO) system (12) and consisted of three components: a glutathione reductase (GR)-type reductase (DdmA), a [2Fe-2S] ferredoxin (DdmB), and an oxygenase (DdmC). In the reaction catalyzed by the DMO, the electron-transport chain (ETC, consisting of DdmB and DdmA) transferred reducing equivalents from NAD(P)H to the oxygenase component DdmC, which then oxidized dicamba to DCSA and formaldehyde (13, 14, 15). In the anaerobic bacterium M. thermoacetica, a tetrahydrofolate (THF)-dependent methyltransferase system, Mtv, was responsible for the dicamba demethylation. Mtv also consisted of three components: methyltransferase I (MtvB), corrinoid protein (MtvC) and methyltransferase II (MtvA) (16, 17). In the reaction catalyzed by Mtv, the methyl moiety of dicamba was first transferred by methyltransferase I to the corrinoid protein and then further transferred by methyltransferase II to THF to form 5-methyl-THF. The metabolite DCSA was hydroxylated to 3,6-dichlorogentisate by an unknown monooxygenase in S. maltophilia DI-6 (18), and in Sphingomonas sp. RW5, 3,6-dichlorogentisate was further transformed to 3,6-dichloromaleylpyruvate through ortho cleavage by a gentisate 1,2-dioxygenase (GtdA) (10).
Previously, we isolated two dicamba-utilizing sphingomonads, Sphingobium sp. Ndbn-10 and Sphingomonas sp. Ndbn-20, from activated sludge and compost samples, respectively (11). The degradation of dicamba in both sphingomonads is initiated by demethylation but occurs through different mechanisms. In Sphingobium sp. Ndbn-10, demethylation is catalyzed by a NADH-dependent dicamba monooxygenase, which shares 99% identity with the DMO of S. maltophilia DI-6. However, in Sphingomonas sp. Ndbn-20, dicamba demethylation requires THF but not NAD(P)H as a cofactor, and no dicamba:hydroxocobalamin methyl transfer activity (MtvB) has been detected in cell lysates (11), indicating that dicamba demethylation in the strain occurs via an unknown THF-dependent methyltransferase that differs from the DMO or Mtv. To date, only two THF-dependent methoxylated aromatics methyltransferases, DesA (19) and LigM (20), have been reported. Both DesA and LigM are involved in the demethylation of lignin-derived aromatics in Sphingomonas paucimobilis SYK-6. DesA catalyzes the demethylation of vanillate and syringate, and LigM catalyzes the demethylation of vanillate and 3-methoxygallate. However, the enzymatic characteristics of DesA and LigM have not been studied.
In this study, a THF-dependent methyltransferase, Dmt, capable of catalyzing dicamba demethylation was identified in strain Ndbn-20. The transfer of a codon-optimized dmt gene into Arabidopsis thaliana resulted in resistance against dicamba, indicating that the dmt gene is a good candidate for the genetic engineering of dicamba-resistant crops.
MATERIALS AND METHODS
Chemicals and media.
Dicamba (99.3% purity), THF (≥65% purity), 5-methyl-THF (97% purity), anisate (96% purity), syringate (95% purity), and vanillate (97% purity) were purchased from Sigma-Aldrich (Shanghai, China). DSCA (98% purity) was obtained from Qingdao Chemical Reagent Co., Ltd. (Qingdao, China). Chromatographic-grade methanol and acetonitrile and analytical-grade acetic acid were purchased from Shanghai Chemical Reagent Co., Ltd. (Shanghai, China). Luria-Bertani (LB) broth and LB agar were purchased from Difco Laboratories (Detroit, MI, USA). MS medium for plant culture was purchased from Beijing Solarbio Science & Technology Co., Ltd. (Beijing, China). The minimal salts medium (MSM) consisted of the following components: 1.3 g K2HPO4, 0.86 g KH2PO4, 0.66 g (NH4)2SO4, 0.097 g MgSO4, 0.025 g MnSO4·H2O, 0.005 g FeSO4·7H2O, and 0.0013 g CaSO4·6H2O per liter water (pH 7.0).
Strains, vectors, and culture conditions.
The bacterial strains and vectors used in this study are listed in Table 1, and the primers are listed in Table 2. Strain Ndbn-20 was grown at 30°C in LB or MSM containing 4.5 mM dicamba. Sphingomonas wittichii DC-6 was grown at 30°C in LB, unless otherwise indicated. Escherichia coli strains and Agrobacterium tumefaciens GV3101 were grown at 37°C in LB broth supplemented with the appropriate antibiotics, as indicated in Table 1.
TABLE 1.
Strains and vectors used in this study
| Strain or vector | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| Sphingomonas sp. Ndbn-20 (= CCTCC M 2014550) | Dicamba utilizer; Strr (50 μg/ml) | 11 |
| Sphingomonas wittichii DC-6 (= KACC 16600) | Could not degrade dicamba; Strr (50 μg/ml) | 33 |
| Agrobacterium tumefaciens GV3101 | Strain for transformation of Arabidopsis thaliana; Genr (50 μg/ml) and Rfpr (10 μg/ml) | Novagen |
| Escherichia coli BL21(DE3) | Host strain for expressing vector | TaKaRa |
| Escherichia coli DH5α | Host strain for cloning vector | TaKaRa |
| Escherichia coli HB101 (pRK600) | Conjugation helper strain; Cmr (50 μg/ml) | This lab |
| Vectors | ||
| pBBR1MCS-5 | Broad-host-range cloning vector; Genr (50 μg/ml) | 27 |
| pBBRdmt02c | pBBR1MCS-5 derivative carrying methyltransferase gene cluster from scaffold 02; Genr (50 μg/ml) | This study |
| pBBRdmt02 | pBBR1MCS-5 derivative carrying methyltransferase gene from scaffold 02; Genr (50 μg/ml) | This study |
| pBBRdmt66c | pBBR1MCS-5 derivative carrying methyltransferase gene cluster from scaffold 66; Genr (50 μg/ml) | This study |
| pBBRdmt66 | pBBR1MCS-5 derivative carrying methyltransferase gene from scaffold 66; Genr (50 μg/ml) | This study |
| pET-29a(+) | Expression vector; Kanr (100 μg/ml) | Novagen |
| pETdmt02 | pET 29a(+) derivative carrying methyltransferase gene from scaffold 02; Kanr (100 μg/ml) | This study |
| pETdmt66 | pET 29a(+) derivative carrying methyltransferase gene from scaffold 66; Kanr (100 μg/ml) | This study |
| pGEM-T | Clone vector; Ampr (100 μg/ml) | Promega |
| pDBN01-T | pGEM-T derivative carrying dmt gene; Ampr (100 μg/ml) | This study |
| pDBNBC-01 | Binary expression vector derived from pCAMBIA2301; AtUbi10, AtCTP2, tCaMV35S, prCaMV35S, PATb; Kanr (100 μg/ml) | Lab stock |
| pDBN100879 | pDBNBC-01 derivative carrying dmt gene; Kanr (100 μg/ml) | This study |
Str, streptomycin; Gen, gentamicin; RFP, rifapentine; Cm, chloramphenicol; Kan, kanamycin; Amp, ampicillin.
AtUbi10, promoter of Arabidopsis thaliana polyubiquitin10 gene; AtCTP2, chloroplast transit peptide coding sequence of Arabidopsis thaliana EPSP synthase gene; tCaMV35S, terminator of cauliflower mosaic virus 35S gene; prCaMV35S, promoter of cauliflower mosaic virus 35S gene; PAT, marker gene of genetically modified crops.
TABLE 2.
Primers used in this study
| Primer | DNA sequence (5′ to 3′)a | Purpose |
|---|---|---|
| pBBRdmt02-F | GGGGTACCCATGTTGTCGGGGTGAAGGAAAGTG (KpnI) | Amplification of a fragment containing the methyltransferase gene from scaffold 02 for functional study in strain DC-6 |
| pBBRdmt02-R | GCTCTAGATGTCCTGGGCCGCAAGGGCGAGTT (XbaI) | |
| pBBRdmt02c-F | CGGGGTACCCAATGGCGGCCGCCCTGATGCTC (KpnI) | Amplification of a fragment containing the methyltransferase gene cluster from scaffold 02 for functional study in strain DC-6 |
| pBBRdmt02c-R | CCGGAATTCTGGCTTCGGCATCGCCACCGAATGC (EcoR I) | |
| pBBRdmt66-F | CGGAATTCGGGTCAGGGCGCGGCCGCTAGG (EcoR I) | Amplification of a fragment containing the methyltransferase gene from scaffold 66 for functional study in strain DC-6 |
| pBBRdmt66-R | GCTCTAGAGCACGGCGAGAATAACCCCGAAGCA (XbaI) | |
| pBBRdmt66c-F | CTAGTCTAGACGTTGCAGCTCATCAACACCCACGA (XbaI) | Amplification of a fragment containing the methyltransferase gene cluster from scaffold 66 for functional study in strain DC-6 |
| pBBRdmt66c-R | CCCAAGCTTTCAAGTCCGCCATCAGGAGCGAA (HindIII) | |
| pET-dmt02-F | GGAATTCCATATGAAGAGCCCATTGATGAAATATCGG (NdeI) | Amplification of the methyltransferase gene from scaffold 02 for expression in Escherichia coli |
| pET-dmt02-R | CCGCTCGAGTCAGTCTTTCTTGATGACCTTCTCG (XhoI) | |
| pET-dmt66-F | GGAATTCCATATGGTGCGGTCGGTTCAGGA (NdeI) | Amplification of the methyltransferase gene from scaffold 66 for expression in Escherichia coli |
| pET-dmt66-R | CCGCTCGAGGAGCGTCGCGCGGACCCGGC(XhoI) | |
| RTdmt-F | GCCTTTCATTCCTGCCGAGTTCA | Amplification of a 525-bp fragment of dmt by RT-PCR |
| RTdmt-R | ATGCCATGCCGCAGGATGTGAACGC | |
| RTmetF-F | CGGCGCTCATCGTCACGCAATT | Amplification of a 301-bp fragment of metF by RT-PCR |
| RTmetF-R | AAAGGGTAGAAGTGCAGGCGGACAT | |
| RTpurU-F | TTGACCATTGCCTGGGTGACCTGCT | Amplification of a 540-bp fragment of purU by RT-PCR |
| RTpurU-R | TTCAGGAAGACCCGGCCCTCCAGAT | |
| RTdhc-F | CATAAAGCCTGCGAAGGACGTGGA | Amplification of a 166-bp fragment of dhc by RT-PCR |
| RTdhc-R | TCGACTTTCCGATCACCACAGCG | |
| RTdmt-metF-F | TTTACACCAGCGACGCAATGAA | Amplification of a 1,167-bp fragment of dmt-metF-spanning region |
| RTdmt-metF-R | CTGGCAATCAACTGCATGCTATCGG | |
| RTmetF-purU-F | GTCCGCATCCGTGCTGAGCAAATAT | Amplification of a 604-bp fragment of metF-purU-spanning region |
| RTmetF-purU-R | GAACGGGACGCCACGGAGCGATGAG | |
| RTpurU-dhc-F | ATCGCCCAGGATACCGAGGTCGT | Amplification of a 663-bp fragment of purU-dhc-spanning region |
| RTpurU-dhc-R | TCGACTTTCCGATCACCACAGCG | |
| RT-dmt-F | GCCTTTCATTCCTGCCGAGTTCA | Amplification of a 161-bp fragment of dmt by RT-qPCR |
| RT-dmt-R | TGGAGAAGGTGGACGGCGACAGATA | |
| RT-metF-F | GTCCGCATCCGTGCTGAGCAAATAT | Amplification of a 138-bp fragment of metF by RT-qPCR |
| RT-metF-R | AAAGGGTAGAAGTGCAGGCGGACAT | |
| RT-purU-F | TTGACCATTGCCTGGGTGACCTGCT | Amplification of a 128-bp fragment of purU by RT-qPCR |
| RT-purU-R | GAACGGGACGCCACGGAGCGATGAG | |
| RT-dhc-F | CATAAAGCCTGCGAAGGACGTGGA | Amplification of a 167-bp fragment of dhc by RT-qPCR |
| RT-dhc-R | TCGACTTTCCGATCACCACAGCG | |
| RT-16S-F | GGCGACGATCCATAGCTGGTCTGAG | Amplification of a 141-bp fragment of 16S rRNA by RT-qPCR |
| RT-16S-R | TTCATCACTCACGCGGCATTGCTG |
Restriction sites are underlined.
DNA and amino acid sequence analysis.
DNA manipulation was performed as described by Sambrook and Russell (21). The draft genome of strain Ndbn-20 was acquired by Shanghai Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China), using an Illumina HiSeq2000 system (22). De novo gene prediction was performed through Glimmer software (version 3.0) (http://cbcb.umd.edu/software/glimmer) (23, 24). Functional annotation was accomplished by BLAST analysis of protein sequences in the non-redundant protein (NR), KEGG (25), Swiss-Prot, and COG (26) databases. Analyses of DNA and amino acid sequences were performed using OMIGA 2.0 software. DNA and amino acid sequence identity searches were conducted using the BLASTN and BLASTP tools (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The promoter and transcription terminators were predicted by the online tools Neural Network Promoter Prediction (http://fruitfly.org:9005/seq_tools/promoter.html) and ARNold (http://rna.igmors.u-psud.fr/toolbox/arnold/index.php), respectively.
Functional studies of the putative dicamba methyltransferase gene.
The fragments containing each putative dicamba methyltransferase gene or gene cluster (including the methyltransferase gene, metF, dhc, and purU) were amplified by PCR from the genome of strain Ndbn-20 using PrimeSTAR HS DNA polymerase (TaKaRa Biotechnology Co. Ltd., Dalian, China). The primers were designed to include approximately 500 bp upstream of each methyltransferase gene to ensure inclusion of the promoter. The fragment was digested and ligated into the corresponding site of broad-host-range vector pBBR1MCS-5 (27). The resulting recombinant vector was transformed into E. coli DH5α. The inserted fragment was verified by PCR and sequenced. The recombinant vector was then introduced into strain DC-6 by triparental mating with pRK600 as the helper. The ability of the recombinant to degrade dicamba was determined through a whole-cell biotransformation test in MSM, as described by Liu et al. (28). The dicamba concentration in the cell suspension was 1.8 mM. The disappearance of dicamba was monitored by high-performance liquid chromatography (HPLC), as described below.
Expression of the putative dicamba methyltransferase genes and purification of the products.
The methyltransferase genes in scaffold 02 and scaffold 66 were amplified from the genomic DNA of strain Ndbn-20. The PCR products were digested and ligated into the corresponding sites of pET-29a(+) to generate pETdmt02 and pETdmt66. The two recombinant vectors were then transformed into E. coli BL21(DE3). The two genes were induced, and the C-terminal His6-tagged proteins were purified using the method described by Wang et al. (29). The molecular weights were determined by SDS-PAGE and Coomassie blue staining, and the protein concentrations were quantified by the Bradford method using bovine serum albumin as the standard (30, 31).
Preparation of cell lysate of strain Ndbn-20.
The strain was cultured in MSM with 4.5 mM dicamba or 4.5 mM glucose as the carbon source. The cells were harvested at the mid-exponential phase, washed twice with 100 mM Tris HCl buffer (pH 8.0), and resuspended in 15 ml of the same buffer. Cell lysates were obtained by sonication according to the method described by Guo et al. (32).
Determination of methyltransferase activity.
The methyltransferase activity of the cell lysate and the purified Dmt toward dicamba, syringate, vanillate, or anisate was determined in a 300-μl mixture containing 100 mM Tris HCl buffer (pH 8.0), 2.0 mM THF, 1.0 mM each substrate, and 0.1 μg of Dmt. The mixture was incubated for 5 min at 30°C under anaerobic conditions according to the method described by Abe et al. (20), and then the reaction was terminated by boiling at 100°C for 1 min. The conversion of substrates was analyzed by HPLC as described below. One unit of methyltransferase activity was defined as the amount of enzyme that catalyzed the conversion of 1 nmol of substrate per min. The temperature and pH ranges of the enzyme were determined according to the methods described by Chen et al. (33). The optimal reaction temperature was determined under standard conditions at pH 8.0 (100 mM Tris HCl buffer) and different temperatures (20 to 45°C). For the optimal pH test, Dmt was incubated at 30°C with three different buffering systems: 100 mM phosphate buffer (pH 5.0 to 6.5), 100 mM Tris HCl buffer (pH 6.2 to 8.5), and 100 mM glycine-NaOH buffer (pH 8.2 to 10.0). Each value reported was the average standard deviation from three independent experiments. To identify the products, the enzymatic mixture was incubated for 10 min, and the products were then determined by HPLC and tandem mass spectrometry as described below.
HPLC and tandem mass spectrometry analysis.
The samples were freeze-dried, dissolved in isovolumetric solvents, and then filtered through a 0.22-μm Millipore membrane. A separation column (internal diameter, 4.6 mm; length, 250 mm) filled with Kromasil was used for the HPLC analysis. For dicamba, DCSA, syringate, vanillate, and anisate determination, the samples were dissolved in isovolumetric methanol, the isocratic mobile phase was a mixture of ultrapure water (58.4%), acetonitrile (31.7%), methanol (7.5%), and acetic acid (2.4%), and the detection wavelengths were 275 nm for dicamba, syringate, and vanillate, 319 nm for DCSA, and 252 nm for anisate. The flow rate was 1.0 ml/min, and the injection volume was 20 μl. THF and its derivative were determined according to the method described in previous studies (34, 35, 36). Briefly, the samples were dissolved in isovolumetric 0.1 M KH2PO4 buffer (pH 6.8) (with 1% ascorbic acid and 0.1% β-mercaptoethanol), the isocratic mobile phase was a mixture of 0.05 M KH2PO4 buffer (pH 3.0) (90%) and acetonitrile (10%), and the detection wavelength was 298 nm. The products were identified by tandem mass spectrometry (Agilent G6410B) according to the method described by Dong et al. (37), the metabolites were separated and confirmed by standard mass spectrometry, ionized by electrospray with a positive polarity, and scanned in the normal mass range from 200 m/z (mass to charge ratio) to 600 m/z. The capillary temperature was 350°C, and the source voltage was set at 4 kV. The sheath gas was set at 35 arbitrary units. Characteristic fragment ions were detected using second-order mass spectrometry.
RNA isolation and transcription analysis.
Strain Ndbn-20 was grown in MSM supplemented with 4.5 mM glucose or 4.5 mM dicamba, and the cultures were incubated at 30°C and 150 rpm on a rotary shaker. The cells were harvested at the mid-exponential growth phase by centrifugation. Total RNA was extracted with an RNA isolation kit (TaKaRa) and treated with 1.5 U of RQ1 RNase-free DNase (Promega, Madison, WI) according to the manufacturer's instructions. Reverse transcription (RT)-PCR was performed with a PrimeScript RT reagent kit (TaKaRa). Then, 1 μl of 1:5 diluted cDNA samples was used as the template for quantitative real-time PCR in a RealPlex2 system (Eppendorf, Germany) with 10 μl SYBR Premix Ex Taq II (TaKaRa) and the primer pairs as shown in Table 2 in a total volume of 20 μl, according to the procedure described by Wang et al. (38). The 16S rRNA gene was used as an internal control, and all of the samples were analyzed in triplicate.
Construction of transgenic Arabidopsis thaliana carrying a codon-optimized dmt gene and evaluation of its tolerance to dicamba.
The codons of the dmt gene were optimized using the GenScript OptimumGene Codon Optimization system in terms of codon usage bias and GC content to ensure that the gene would be well expressed in plants. The codon-optimized dmt was synthesized by GenScript Co., Ltd. (Nanjing, China) and was ligated into the clone vector pGEM-T to generate pDBN01-T. The codon-optimized dmt was cleaved from pDBN01-T by digestion with SpeI and KasI and ligated into the corresponding site of the pDBNBC-01 vector to construct pDBN10879 (see Fig. S1 in the supplemental material), and pDBN100879 was then introduced into Agrobacterium tumefaciens GV3101 by the liquid nitrogen method (39). The Arabidopsis thaliana ecotype Columbia was transfected with Agrobacterium tumefaciens GV3101 cells bearing pDBN100879 by the floral dip method (40). The seeds of the transgenic plants were screened on MS medium containing 10 mg/liter dicamba, and the resistant seedlings (T1 generation transgenic plants) were transferred to soil. For the dicamba treatments, 28-day-old plants were sprayed with dicamba at a level of 560 g/ha (the normal field application rate) using an Amway spray bottle.
Accession number(s).
The GenBank accession numbers of the draft genome of strain Ndbn-20, scaffold 02, scaffold 66, and the codon-optimized dmt gene are LGVA01000000, KT372210, KT372212, and KX013225, respectively.
RESULTS AND DISCUSSION
Screening of the THF-dependent dicamba methyltransferase gene.
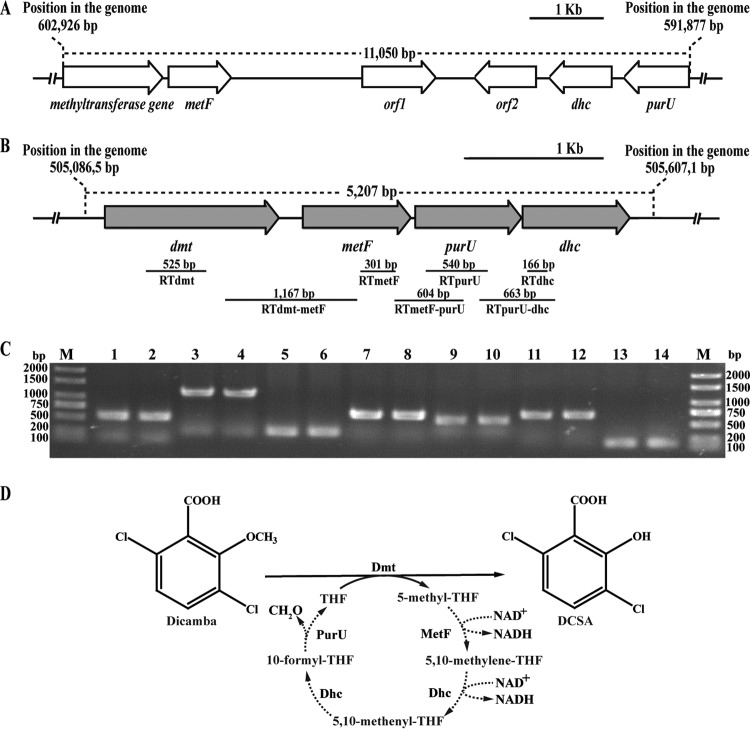
The draft genome of strain Ndbn-20 was resolved into 127 contigs consisting of 5.35 Mb, and 4,911 genes were predicted. The THF-dependent methyltransferases DesA and LigM were used for BLASTP searches of the genome. This screening resulted in the identification of two putative genes located in scaffold 02 and scaffold 66 (Fig. 1 and Table 3). The methyltransferase gene in scaffold 02 shared 43% and 42% identities at the amino acid level with LigM and DesA, respectively, whereas the methyltransferase gene in scaffold 66 shared 46% and 45% identities with LigM and DesA, respectively. An open reading frame (ORF) analysis also revealed that three other genes, which exhibited similarities with metF (encoding 5,10-methylene-THF reductase), dhc (encoding the bifunctional enzyme 5,10-methylene-THF dehydrogenase/5,10-methenyl-THF cyclohydrolase), and purU (encoding formyl-THF deformylase), were located downstream of each methyltransferase gene. The three genes have been reported to be involved in THF metabolism (34, 35, 36). This analysis suggested that the methyltransferase gene and its adjacent genes in each scaffold may be organized in a single cluster and involved in the demethylation of dicamba. Furthermore, a putative dioxygenase gene that exhibited 99% identity to gtdA of Sphingomonas sp. RW5 was retrieved from the genome but was not located in the immediate vicinity of the dmt gene. gtdA is responsible for the cleavage of 3,6-dichlorogentisate, the hydroxylation production of DCSA (10). The presence of gtdA in the genome implied that DCSA may be further degraded via the gentisate pathway in the strain.
FIG 1.
(A, B) Organization of the methyltransferase gene cluster in scaffold 02 (A) and scaffold 66 (B). The gray arrows indicate the sizes and transcriptional direction of each gene in scaffold 66. The locations of the primer sets RTdmt, RTdmt-metF, RTmetF, RTmetF-purU, RTpurU, RTpurU-dhc, and RTdhc and the DNA fragments amplified by RT-PCR are indicated below. (C) Transcription analysis of dmt, metF, purU, and dhc in scaffold 66 by RT-PCR. Total RNAs of strain Ndbn-20 grown with glucose or dicamba were prepared for RT-PCR. Lane M, molecular markers; lanes 2, 4, 6, 8, 10, 12, and 14 (template from cells grown with dicamba), products obtained from the reactions of the RTdmt, RTdmt-metF, RTmetF, RTmetF-purU, RTpurU, RTpurU-dhc, and RTdhc primer sets, respectively, with the RT products. Lanes 1, 3, 5, 7, 9, 11, and 13, corresponding negative controls. (D) Proposed dicamba O-demethylation system linked to the THF-mediated C1 metabolism in strain Ndbn-20. The reactions indicated by a dotted line have not been confirmed by experiments. MetF, 5,10-methylene-THF reductase; Dhc, the bifunctional enzyme 5,10-methylene-THF dehydrogenase/5,10-methenyl-THF cyclohydrolase; PurU, formyl-THF deformylase.
TABLE 3.
Deduced function of each ORF within the fragments containing the methyltransferase genes in scaffold 02 and scaffold 66
| Gene name | Position, product size (amino acids) | Homologous protein and source | % identity |
|---|---|---|---|
| Fragment in scaffold 02 | |||
| Methyltransferase gene | 37–1407, 443 | LigM (vanillate/3-O-methylgallate O-demethylase) (BAK65949), Sphingomonas paucimobilis SYK-6 | 43 |
| metF | 1474–2343, 290 | MetF (5,10-methylenetetrahydrofolate reductase) (AEG51610), Sphingobium chlorophenolicum L-1 | 81 |
| orf1 | 6629–7612, 328 | RNase Z (A1KL99.1), Mycobacterium bovis BCG strain Pasteur 1173P2 | 30 |
| orf2 | 8145–9017, 291 | Hydroquinone 1,2-dioxygenase (WP_007686007), Sphingomonas sp. MM-1 | 51 |
| dhc | 9942–9097, 268 | Dhc (5,10-methylene-tetrahydrofolate dehydrogenase) (WP_013849824), Sphingobium chlorophenolicum | 79 |
| purU | 11003–10116, 296 | PurU (formyltetrahydrofolate deformylase) (WP_037457233), Sphingobium chlorophenolicum | 86 |
| Fragment in scaffold 66 | |||
| dmt | 1881–3278, 466 | LigM (vanillate/3-O-methylgallate O-demethylase) (BAD61059), Sphingomonas paucimobilis SYK-6 | 46 |
| metF | 3467–4333, 289 | MetF (5,10-methylenetetrahydrofolate reductase) (EQB14749), Sphingobium quisquiliarum P25 | 66 |
| purU | 4370–5224, 285 | PurU (formyltetrahydrofolate deformylase) (EQB14750), Sphingobium quisquiliarum P25 | 66 |
| dhc | 5227–6087, 287 | Dhc (methenyltetrahydrofolate cyclohydrolase) (EXS71521), Sphingobium sp. Ant17 | 74 |
Functional studies of the two putative methyltransferases.
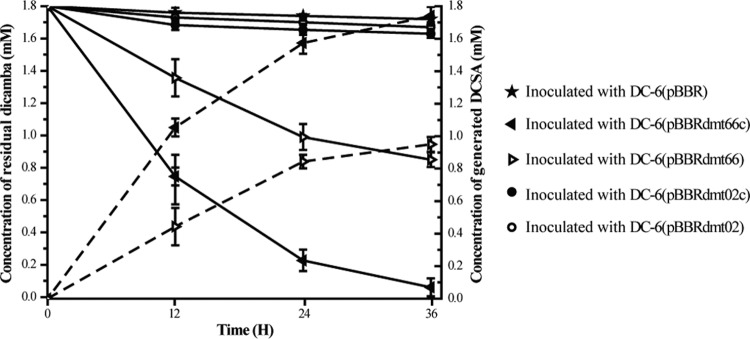
Recombinant vectors containing the putative methyltransferase genes or gene clusters were introduced into Sphingomonas wittichii DC-6, which lacks the ability to degrade dicamba. Whole-cell transformation experiments showed that recombinants DC-6pBBRdmt02 (containing the methyltransferase gene from scaffold 02) and DC-6pBBRdmt02c (containing the methyltransferase gene cluster from scaffold 02) did not degrade dicamba, whereas DC-6pBBRdmt66 (containing the methyltransferase gene from scaffold 66) and DC-6pBBRdmt66c (containing the methyltransferase gene cluster from scaffold 66) acquired the abilities to convert dicamba to DCSA, and the amount of generated DCSA was equal to the amount of the disappeared dicamba (Fig. 2). These results suggested that the methyltransferase gene from scaffold 66 was involved in the demethylation of dicamba. It is interesting that the conversion efficiency of DC-6pBBRdmt66 was much slower than that of DC-6pBBRdmt66c. A possible reason is that DC-6pBBRdmt66 did not include metF, dhc, and purU, which are involved in THF metabolism, and that the absence of these genes affected the recycling of THF.
FIG 2.
Time course of dicamba conversion by Sphingomonas sp. DC-6(pBBR) (containing the pBBR1MCS-5 vector), DC-6(pBBRdmt66c) (containing the dmt gene cluster from scaffold 66), DC-6(pBBRdmt66) (containing the dmt gene from scaffold 66), DC-6(pBBRdmt02c) (containing the methyltransferase gene cluster from scaffold 02), and DC-6(pBBRdmt02) (containing the methyltransferase gene from scaffold 02). The solid and dotted lines indicate the residual dicamba and generated DCSA, respectively. The data were derived from three independent measurements, and the error bars indicate standard deviations.
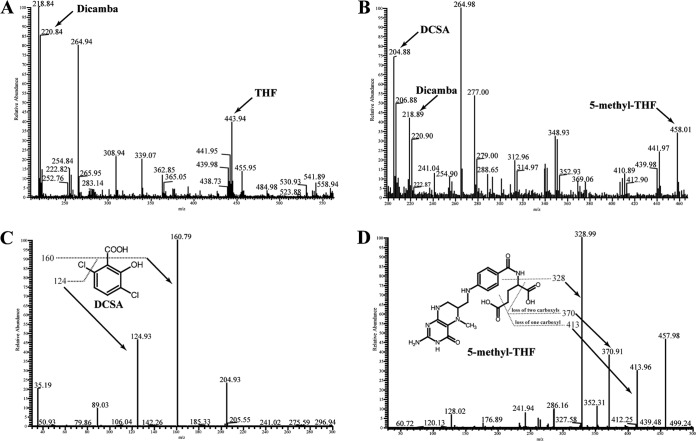
The two putative methyltransferase genes were further cloned into pET-29a(+) and expressed in E. coli BL21(DE3). The recombinant proteins were purified by Ni affinity chromatography. The molecular masses of the two denatured methyltransferases were determined to be approximately 50 kDa (see Fig. S2 in the supplemental material), which is similar to the values calculated from the deduced amino acid sequences (51.3 kDa and 52.5 kDa, respectively). The dicamba demethylation activities of the two purified methyltransferases were determined. HPLC and mass spectrometry analysis showed that in the control without enzyme (Fig. 3A; see also Fig. S3A and B in the supplemental material) or the reaction with methyltransferase from scaffold 02 added (data not shown), dicamba and THF were not transformed. Whereas in the reaction with methyltransferase from scaffold 66 added, concentrations of dicamba and THF were reduced, and two metabolites were produced (Fig. 3B; see also Fig. S3C and D in the supplemental material). Metabolite 1 had a retention time of 7.86 min, which was equal to that of the authentic DCSA standard. A mass spectrometry analysis showed a prominent protonated molecular ion at m/z = 205 (M + H)+ and two fragment ion peaks at m/z = 161 (loss of a -COOH group) and m/z = 125 (loss of a -COOH and a -Cl) (Fig. 3C); thus, metabolite 1 was identified as DCSA. Metabolite 2 had a retention time of 11.23 min, which was equal to that of the authentic 5-methyl-THF standard. A mass spectrometry analysis showed a prominent protonated molecular ion at m/z = 458 (M + H)+ and characteristic fragment ion peaks at m/z = 414 (loss of a -COOH), m/z = 371 (loss of two -COOH groups), and m/z = 329 (loss of a -COOH-(CH2)3-COOH) (Fig. 3D); as a result, metabolite 2 was identified as 5-methyl-THF. In the subsequent study, the methyltransferase gene in scaffold 66 was designated dmt (dicamba methyltransferase). Dmt catalyzed the transfer of a methyl group from dicamba to THF, producing DCSA and 5-methyl-THF.
FIG 3.
Tandem mass spectrometry (MS) analysis of the products generated during dicamba conversion by Dmt. (A and B) First-order MS spectrum of the extracts obtained from reaction mixtures without or with Dmt, respectively. (C) The second-order MS analysis of the m/z 205 [M + H]+, which corresponded to DCSA. (D) Second-order MS analysis of the m/z 458 [M + H]+, which corresponded to 5-methyl-THF.
ORF analysis and transcriptional study of the dmt gene cluster.
As determined through an online sequence analysis, a promoter (−240 bp to −195 bp) and a transcriptional start site (TSS) (−200 bp) were located upstream of the initiator codon ATG of the dmt gene (see Fig. S4 in the supplemental material), and a terminator was located downstream from dhc. The above analysis suggested the possibility that the four continuous genes constituted one transcriptional unit. RT-PCR was performed with RNA derived from strain Ndbn-20 grown with glucose or dicamba. The four genes, dmt, metF, purU, and dhc, were proven to form part of one transcriptional operon and were transcribed in cells grown with glucose and dicamba (Fig. 1C). The quantitative real-time PCR (qPCR) results showed no obvious differences in the transcription levels of the four genes between the two conditions (see Fig. S5 in the supplemental material). Furthermore, cell lysates of strain Ndbn-20 grown with glucose or dicamba presented almost the same methyltransferase activities (0.51 ± 0.04 nmol/min/mg and 0.49 ± 0.16 nmol/min/mg, respectively). These results confirmed that the dmt gene cluster was constitutively expressed in the strain.
Characteristics of Dmt.
Dicamba methyltransferase activity was detected from 4 to 45°C and at pH values ranging from 5.0 to 10.0, with the highest activity at 30°C and pH 8.0 (see Fig. S6 in the supplemental material). THF was necessary for this activity, whereas the addition of NAD(P)H, ATP, or menadione had no obvious effect on the activity of this enzyme. The incubation of the purified Dmt at pH 8.0 and 30°C for 5 min resulted in specific activity of 114 ± 5 nmol/min/mg toward dicamba. However, Dmt did not catalyze the demethylation of some other methoxy group-containing aromatic compounds, such as syringate, vanillate, and anisate.
Dmt was inhibited by its catalytic product 5-methyl-THF rather than DCSA.
It is interesting that in the time course of the enzyme activity determination (1.0 mM dicamba, 2.0 mM THF, and 0.1 μg purified Dmt were added), we found that the detected velocity of Dmt gradually slowed over time (240 min) (Fig. 4). During the first 10 min of the incubation, 0.21 mM dicamba was converted, whereas 0.09 mM dicamba was consumed during the following 50 min, and only 0.02 mM dicamba was converted in the subsequent 180 min.
FIG 4.
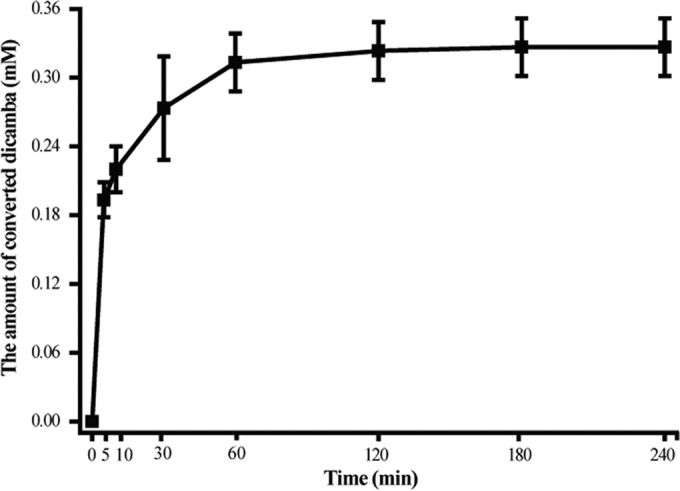
Time course of dicamba conversion by purified Dmt. The reaction mixtures contained 10 mM Tris HCl (pH 8.0), 0.1 μg of purified Dmt66, 1.0 mM dicamba, and 2.0 mM THF in a final volume of 300 μl. Each value shows the average and standard deviation (error bar) from at least three measurements.
There are several possible reasons for this phenomenon. First, Dmt might have easily lost its activity during the experimental period. However, this possibility was ruled out because we found that purified Dmt retained more than 90% of its original activity after preincubation at 30°C for 240 min. The second possibility is that Dmt might be inhibited by the products DCSA or 5-methyl-THF. To confirm this, different amounts (0, 0.1, 0.3, 0.5, 0.7, and 1.0 mM) of DCSA or 5-methyl-THF were subsequently added to the above-mentioned reaction mixture, and the amounts of converted dicamba over a 120-min period were determined. As expected, the addition of 5-methyl-THF decreased the amounts of converted dicamba in an approximately dose-dependent manner (Fig. 5). In contrast, no such inhibition was found with the addition of different amounts of DCSA to the reaction mixture because 0.3 mM converted dicamba was always detected under all conditions. These results clearly suggested that the product 5-methyl-THF but not DCSA (at least in our experimental conditions) was a negative inhibitor of Dmt.
FIG 5.
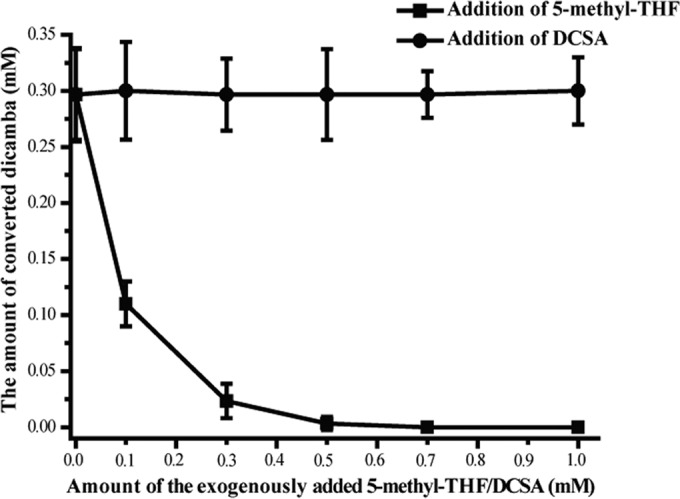
Effect of exogenously added DCSA or 5-methyl-THF on the conversion of dicamba. The reaction mixtures contained 10 mM Tris HCl (pH 8.0), 0.1 μg of purified Dmt, 1.0 mM dicamba, 2.0 mM THF, and various concentrations (0, 0.1, 0.3, 0.5, 0.7, and 1.0 mM) of 5-methyl-THF or DCSA in a final volume of 300 μl and were incubated for 120 min at 30°C.
Arabidopsis thaliana carrying a codon-optimized dmt gene exhibited obvious resistance to dicamba.
The cultivation of herbicide-tolerant transgenic crops is an effective way to control weeds and reduce agriculture costs. Glyphosate-resistant crops, which include a glyphosate resistance gene (encoding CP4 EPSPS), have been commercialized and widely planted (41). However, the continued use of glyphosate-resistant crops has resulted in the worldwide occurrence of glyphosate-resistant weeds (42). Dicamba can effectively kill glyphosate-resistant weeds and is thus considered an ideal complement to glyphosate (5). Therefore, dicamba detoxification genes have potential applications in the genetic engineering of herbicide-resistant plants. In this study, the codons of the dmt gene were optimized to improve its expression efficiency in plants (see Fig. S7 in the supplemental material), and the dicamba detoxification effect of the codon-optimized dmt gene was investigated in Arabidopsis thaliana, which is quite sensitive to treatment with dicamba at a level of 560 g/ha. A total of 195 independently derived T1 generation Arabidopsis thaliana plants carrying the codon-optimized dmt gene were acquired, and 29 of them were randomly selected to investigate their resistance to dicamba. Twenty-one days after treatment with dicamba, all of the nontransgenic plants died, whereas the transgenic plants carrying the codon-optimized dmt gene exhibited minimal damage. Among the 29 plants investigated, 17 plants grew up to set seeds, 8 plants stayed alive but could not bolt, and only 4 plants died (Fig. 6 and Table 4). These results indicated that the dmt gene conferred obvious dicamba resistance to Arabidopsis thaliana and is thus a potential candidate for the engineering of dicamba-resistant transgenic crops.
FIG 6.

Effects of dicamba treatment on nontransgenic Arabidopsis thaliana plants and plants carrying a codon-optimized dmt gene (21 days after spraying with dicamba at a level of 560 g/ha). (A) Untreated-nontransgenic plant. (B to E) Four independently derived T1 generation plants carrying a codon-optimized dmt gene and treated with dicamba. (F) Nontransgenic plant treated with dicamba.
TABLE 4.
State of T1 generation Arabidopsis thaliana plants carrying an optimized dmt gene and nontransgenic plants 21 days after treatment with dicamba at a level of 560 g/ha
| Genotype | No. of plants: |
|||
|---|---|---|---|---|
| Bolting and seed bearing | Alive but nonbolting | Dead | Total | |
| T1 generation plants carrying an optimized dmt gene | 17 | 8 | 4 | 29 |
| Nontransgenic plants | 0 | 0 | 27 | 27 |
As a conclusion, a one-component and THF-dependent dicamba methyltransferase Dmt was identified in Sphingomonas sp. Ndbn-20. The demethylation activity of Dmt was inhibited the by-product 5-methyl-THF but not that of DCSA. Arabidopsis thaliana introduced with the codon-optimized dmt gene acquired obvious resistance against dicamba, indicating that the dmt gene is a good candidate for the genetic engineering of herbicide-resistant crops.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (31570105), the Program for New Century Excellent Talents in University (NCET-13-0861), and Genetically Modified Organisms Breeding Major Projects of China (grant 2016ZX08011003-008).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01201-16.
REFERENCES
- 1.Stevens JT, Sumner DD. 1991. Handbook of pesticide toxicology, p 1317−1408. Academic Press, New York, NY. [Google Scholar]
- 2.Hazardous Substance Databank. 1995. US National Library of Medicine, Bethesda, MD. [Google Scholar]
- 3.Environmental Protection Agency. 1998. Health advisory summary: Dicamba. Office of Drinking Water, US Environmental Protection Agency, Washington, DC. [Google Scholar]
- 4.Tomlin CDS. 2006. The pesticide manual: a world compendium, 14th ed British Crop Protection Council, Farnham, Surrey, United Kingdom. [Google Scholar]
- 5.Behrens M, Mutlu N, Chakraborty S, Dumitru R, Jiang WZ, LaVallee BJ, Herman PL, Clemente TE, Weeks DP. 2007. Dicamba resistance: enlarging and preserving biotechnology-based weed management strategies. Science 316:1185–1188. doi: 10.1126/science.1141596. [DOI] [PubMed] [Google Scholar]
- 6.Krueger JP, Butz RG, Atallah YH, Cork DJ. 1989. Isolation and identification of microorganisms for the degradation of dicamba. J Agric Food Chem 37:534–538. doi: 10.1021/jf00086a057. [DOI] [Google Scholar]
- 7.Taraban RH, Berry DF, Berry DA, Walker HL. 1993. Degradation of dicamba by an anaerobic consortium enriched from wetland soil. Appl Environ Microbiol 59:2332–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu JD, Cheng SP, Gu JG. 2001. Degradation of the herbicide dicamba under strictly anaerobic conditions. J Environ Sci China 22:111–113. [PubMed] [Google Scholar]
- 9.Milligan PW, Häggblom MM. 1999. Biodegradation and biotransformationof dicamba under different reducing conditions. Environ Sci Technol 33:1224–1229. doi: 10.1021/es981117e. [DOI] [Google Scholar]
- 10.Werwath J, Arfmann HA, Pieper DH, Timmis KN, Wittich RM. 1998. Biochemical and genetic characterization of a gentisate 1,2-dioxygenase from Sphingomonas sp. strain RW5. J Bacteriol 180:4171–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao L, Jia XJ, Zhao JD, Cao Q, Xie XT, Yu LL, He J, Tao Q. 2015. Degradation of the herbicide dicamba by two sphingomonads via different demethylation mechanisms. Int Biodeterior Biodegr 104:324–332. doi: 10.1016/j.ibiod.2015.06.016. [DOI] [Google Scholar]
- 12.Kweon O, Kim SJ, Baek S, Chae JC, Adjei MD, Baek DH, Kim YC, Cerniglia CE. 2008. A new classification system for bacterial Rieske nonheme iron aromatic ring-hydroxylating oxygenases. BMC Biochem 9:11. doi: 10.1186/1471-2091-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herman PL, Behrens M, Chakraborty S, Chrastil BM, Barycki J, Weeks DP. 2005. A three-component dicamba O-demethylase from Pseudomonas maltophilia, strain DI-6. J Biol Chem 280:24759–24767. doi: 10.1074/jbc.M500597200. [DOI] [PubMed] [Google Scholar]
- 14.Dumitru R, Jiang WZ, Weeks DP, Wilson MA. 2009. Crystal structure of dicamba monooxygenase: a Rieske nonheme oxygenase that catalyzes oxidative demethylation. J Mol Biol 392:498–510. doi: 10.1016/j.jmb.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang XZ, Li B, Herman PL, Weeks DP. 1997. A three-component enzyme system catalyzes the O-demethylation of the herbicide dicamba in Pseudomonas maltophilia DI-6. Appl Environ Microbiol 63:1623–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naidu DSW, Ragsdale SW. 2001. Characterization of a three-component vanillate O-demethylase from Moorella thermoacetica. J Bacteriol 183:3276–3281. doi: 10.1128/JB.183.11.3276-3281.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meßmer M, Reinhardt S, Wohlfarth G, Diekert G. 1996. Studies on methyl chloride dehalogenase and O-demethylase in cell extracts of the homoacetogen strain MC based on a newly developed coupled enzyme assay. Arch Microbiol 165:18–25. doi: 10.1007/s002030050291. [DOI] [Google Scholar]
- 18.Cork DJ, Krueger JP. 1991. Microbial transformations of herbicides and pesticides. Adv Appl Microbiol 36:1−66. doi: 10.1016/S0065-2164(08)70450-7. [DOI] [PubMed] [Google Scholar]
- 19.Masai E, Sasaki M, Minakawa Y, Abe T, Sonoki T, Katayama Y, Fukuda M. 2004. A novel tetrahydrofolate-dependent O-demethylase gene is essential for growth of Sphingomonas paucimobilis SYK-6 with syringate. J Bacteriol 186:2757–2765. doi: 10.1128/JB.186.9.2757-2765.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abe T, Masai E, Miyauchi K, Katayama Y, Fukuda M. 2005. A tetrahydrofolate-dependent O-demethylase, LigM, is crucial for catabolism of vanillate and syringate in Sphingomonas paucimobilis SYK-6. J Bacteriol 187:2030–2037. doi: 10.1128/JB.187.6.2030-2037.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 22.Ansorge WJ. 2009. Next-generation DNA sequencing techniques. New Biotechnol 25:195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, Li S, Yang H, Wang J, Wang J. 2010. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delcher AL, Bratke KA, Powers EC, Salzberg SL. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. 2012. KEGG for integration and interpretation of large-scale molecular datasets. Nucleic Acids Res 40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 28.Liu H, Wang SJ, Zhang JJ, Dai H, Tang H, Zhou NY. 2011. Patchwork assembly of nag-like nitroarene dioxygenase genes and the 3-chlorocatechol degradation cluster for evolution of the 2-chloronitrobenzene catabolism pathway in Pseudomonas stutzeri ZWLR2-1. Appl Environ Microbiol 77:4547–4552. doi: 10.1128/AEM.02543-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang BZ, Guo P, Hang BJ, Li L, He J, Li SP. 2009. Cloning of a novel pyrethroid-hydrolyzing carboxylesterase gene from Sphingobium sp. strain JZ-1 and characterization of the gene product. Appl Environ Microbiol 75:5496–5500. doi: 10.1128/AEM.01298-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 31.Wang F, Zhou J, Li ZK, Dong WL, Hou Y, Huang Y, Cui ZL. 2015. Involvement of the cytochrome P450 system EthBAD in the N-deethoxymethylation of acetochlor by Rhodococcus sp. strain T3-1. Appl Environ Microbiol 81:2182–2188. doi: 10.1128/AEM.03764-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo P, Wang BZ, Hang BJ, Li L, Ali SW, He J, Li SP. 2009. Pyrethroid-degrading Sphingobium sp. JZ-2 and the purification and characterization of a novel pyrethroid hydrolase. Int Biodeterior Biodegr 63:1107–1112. [Google Scholar]
- 33.Chen Q, Wang CH, Deng SK, Wu YD, Li Y, Yao L, Jiang JD, Yan X, He J, Li SP. 2014. A novel three-component Rieske non-heme iron oxygenase (RHO) system catalyzing the N-dealkylation of chloroacetanilide herbicides in sphingomonads DC-6 and DC-2. Appl Environ Microbiol 80:5078–5085. doi: 10.1128/AEM.00659-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pantùčková P, Křivánková L. 2010. Analysis of 5-methyltetrahydrofolate in human blood, serum and urine by on-line coupling of capillary isotachophoresis and zone electrophoresis. Electrophoresis 31:3391–3399. doi: 10.1002/elps.201000193. [DOI] [PubMed] [Google Scholar]
- 35.Shin HC, Shimoda M, Kokute E, Takahashi Y. 1993. Identification of 10-formyltetrahydrofolate, tetrahydrofolate and 5-methyltetrahydrofolate as major reduced folate derivates in rat bile. J Chromatogr 620:39–46. doi: 10.1016/0378-4347(93)80049-A. [DOI] [PubMed] [Google Scholar]
- 36.Etienne MC, Speziale N, Milano G. 1993. HPLC of folinic acid diastereoisomers and 5-methyltetrahydrofolate in plasma. Clin Chem 39:82–86. [PubMed] [Google Scholar]
- 37.Dong WL, Wang F, Huang F, Wang YC, Zhou J, Ye XF, Li ZK, Hou Y, Huang Y, Ma JF, Jiang M, Cui ZL. 2016. Metabolic pathway involved in 6-chloro-2-benzoxazolinone degradation by Pigmentiphaga sp. strain DL-8 and identification of the novel metal-dependent hydrolase CbaA. Appl Environ Microbiol 82:4169−4179. doi: 10.1128/AEM.00532-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang CH, Chen Q, Wang R, Shi C, Yan X, He J, Hong Q, Li SP. 2014. A novel angular dioxygenase gene cluster encoding 3-phenoxybenzoate 1,2-dioxygenase in Sphingobium wenxiniae JZ-1. Appl Environ Microbiol 80:3811–3818. doi: 10.1128/AEM.00208-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen H, Nelson RS, Sherwood JL. 1994. Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 16:664–670. [PubMed] [Google Scholar]
- 40.Zhang XR, Henriques R, Lin SS, Niu QW, Chua NH. 2006. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1:641–646. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- 41.Cerdeira AL, Duke SO. 2010. Effects of glyphosate-resistant crop cultivation on soil and water quality. GM Crops 1:16–24. doi: 10.4161/gmcr.1.1.9404. [DOI] [PubMed] [Google Scholar]
- 42.Powles SB. 2008. Evolved glyphosate-resistant weeds around the world: lessons to be learnt. Pest Manag Sci 64:360–365. doi: 10.1002/ps.1525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.