SUMMARY
Epigenetic changes are present in all human cancers and are now known to cooperate with genetic alterations to drive the cancer phenotype. These changes involve DNA methylation, histone modifiers and readers, chromatin remodelers, microRNAs, and other components of chromatin. Cancer genetics and epigenetics are inextricably linked in generating the malignant phenotype; epigenetic changes can cause mutations in genes, and, conversely, mutations are frequently observed in genes that modify the epigenome. Epigenetic therapies, in which the goal is to reverse these changes, are now one standard of care for a preleukemic disorder and form of lymphoma. The application of epigenetic therapies in the treatment of solid tumors is also emerging as a viable therapeutic route.
Epigenetic changes (e.g., DNA hypomethylation) are present in all human cancers and help to drive tumorigenesis. Drugs that reverse such changes are emerging as effective cancer therapies.
OVERVIEW
Cancer is caused by the somatically heritable deregulation of genes that control the processes governing when cells divide, die, and move from one part of the body to another. During carcinogenesis, genes can become activated in such a way that enhances division or prevents cell death (oncogene). Alternatively, genes can become inactivated so that they are no longer available to apply the brakes to these processes (tumor-suppressor gene). It is the interplay between these two classes of genes that results in the formation of cancer.
Tumor-suppressor genes (TSGs) can become inactivated by at least three pathways: (1) through mutations, in which their functions become disabled; (2) a gene can be completely lost and thus not be available to work appropriately (loss of heterozygosity); and (3) a gene can be switched off in a somatically heritable fashion by epigenetic changes, rather than by mutation of the DNA sequence. Epigenetic silencing can occur by deregulation of the epigenetic machinery at several different levels; it may involve inappropriate methylation of cytosine (C) residues in CpG sequence motifs that reside within control regions governing gene expression. Also, changes to histone posttranslational modifications (PTMs) or aberrations in the way histone-modifying enzymes function may occur. A change in a protein’s ability to read histone marks, and hence bind to chromatin, or alterations in the way nucleosome-remodeling or histone exchange complexes function can result. Finally, changes in regulatory microRNA (miRNA) expression patterns have been noted.
This article focuses predominantly on how cancer is affected by this third pathway (i.e., epigenetic mechanisms). The basic molecular mechanisms responsible for maintaining the silenced state are quite well understood, as outlined in this collection. Consequently, we also know that epigenetic silencing has profound implications for cancer prevention, detection, and therapies. We now have drugs approved by the U.S. Food and Drug Administration (FDA) that are used to reverse epigenetic changes and restore gene activity to cancer cells. Also, because changes in DNA methylation can be detected with a high degree of sensitivity, many strategies are able to detect cancer early by finding changes in DNA methylation. The translational opportunities for epigenetics in human cancer research, detection, prevention, and treatment are, therefore, quite extraordinary.
1. THE BIOLOGICAL BASIS OF CANCER
Cancer is ultimately a disease of gene expression in which the complex networks governing homeostasis in multicellular organisms become deranged, allowing cells to grow without reference to the needs of the organism as a whole. Great advancements have been made in delineating the subset of cellular control pathways subject to derangement in human cancer (Table 1). The realization that distinct sets of cellular control pathways are affected and heritably disabled in almost all cancers is a key concept that has advanced the field (Hanahan and Weinberg 2011). Historically, research has focused on the genetic basis of cancer, particularly, in terms of how mutational activation of oncogenes or inactivation of tumor-suppressor genes (TSGs) underpins these above pathway changes. However, since the 1990s, a growing research endeavor has centered on the recognition that heritable changes, regulated by epigenetic alterations, may also be critical for the evolution of all human cancer types (Baylin and Jones 2011).
Table 1.
Examples of key cellular pathways disrupted in human cancers by genetic and epigenetic mechanisms
| Pathway | Example of genetic alteration | Example of epigenetic alteration |
|---|---|---|
| Self sufficiency in growth signals | Mutations in Ras gene | Methylation of RASSFIA gene |
| Insensitivity to antigrowth signals | Mutation in TGF-β receptors | Down-regulation of TGF-β receptors |
| Tissue invasion and metastasis | Mutation in E-cadherin gene | Methylation of E-cadherin promoter |
| Limitless replicative potential | Mutation in p16 and pRb genes | Silencing of p16 or pRb genes by promoter methylation |
| Sustained angiogenesis | Silencing of thrombospondin-1 | |
| Evading apoptosis | Mutation in p53 | Methylation of DAPK, ASC/TMS1, and HIC1 |
| DNA repair capacity | Mutations in MLH1, MSH2 | Methylation of GST Pi, O6-MGMT, MLH1 |
| Monitoring genomic stability | Mutations in Chfr | Methylation of Chfr |
| Protein ubiquination functions regulating mitotic control genes | Mutations in Chfr | Methylation of Chfr |
TGF-β, transforming growth factor β; DAPK, death-associated protein kinase.
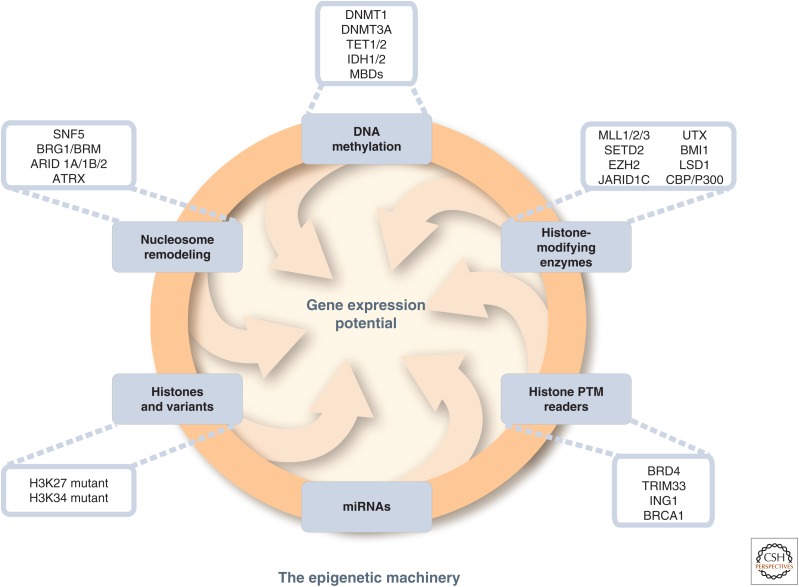
Epigenetic alterations can be observed as abnormal patterns of DNA methylation, disrupted patterns of histone posttranslational modifications (PTMs), and changes in chromatin composition and/or organization. Changes in the epigenome largely occur through disrupting the epigenetic machinery, and Figure 1 illustrates the different elements of the epigenetic machinery that are now known to be perturbed in cancer. These epigenomic changes not only are associated with altered patterns of expression for otherwise wild-type genes, but, in some cases, may also be causal to their changed expression state. The recognition of an epigenetic component in tumorigenesis, or the existence of a cancer “epigenome,” has led to new opportunities for the understanding, detection, treatment, and prevention of cancer.
Figure 1.
Genetic mutations of epigenetic modifiers in cancer. The drawing shows the input of epigenetic processes in specifying gene expression patterns. Recent whole-exome sequencing studies show that mutations in various classes of epigenetic modifiers are frequently observed in many types of cancers, further highlighting the cross talk between genetics and epigenetics. Examples of some, but not all, of these mutations are illustrated here and listed in Table 2. The mutations of epigenetic modifiers potentially cause genome-wide epigenetic alterations in cancer, but, save for isocitrate dehydrogenase (IDH) mutations as discussed in the text, these have yet to be shown on a genome-wide scale. Understanding the relationship of genetic and epigenetic changes in cancer will offer novel insights for cancer therapies. MBDs, methylcytosine-binding proteins; PTM, posttranslational modification. (Adapted from You and Jones 2012.)
Signaling gene (oncogene) mutations in many human cancers are often dominant and drive the formation of cancers. An example would be ras, which when mutated, enhances the activity of the gene product to stimulate growth. Genetic mutations or epigenetic silencing of TSGs, on the other hand, are often recessive, requiring disruptive events in both allelic copies of a gene for the full expression of the transformed phenotype. The idea that both copies of a TSG have to be incapacitated in a malignant cell line was proposed by Knudson (2001) in his “two- or multiple-hit” hypothesis and has found wide acceptance. It is now realized that three classes of “hits” can participate in different combinations to cause a complete loss of activity of TSGs. Direct mutations in the coding sequence may occur, loss of parts or entire copies of genes, or epigenetic silencing, can cooperate with each other to result in the disablement of key control genes. Another growing concept discussed in this article is that there is an intense cooperation between genetic and epigenetic abnormalities to drive the initiation and progression of cancer (Fig. 1) (Baylin and Jones 2011; You and Jones 2012; Garraway and Lander 2013; Shen and Laird 2013). Most recently, excitement has centered on the realization that most cancers actually harbor frequent mutations in genes that encode for components of the epigenetic machinery, potentially resulting in abnormalities in the epigenome, which may affect gene expression patterns and genomic stability (Baylin and Jones 2011; You and Jones 2012; Garraway and Lander 2013; Shen and Laird 2013). Some of the growing list of genes frequently mutated in cancer, encoding proteins central to establishing normal control of chromatin and DNA methylation patterns, are illustrated in Figure 1 and more exhaustively, listed in Table 2 or the Appendices of Audia and Campbell (2014) (Baylin and Jones 2011; You and Jones 2012; Garraway and Lander 2013; Shen and Laird 2013). Although most of the consequences of these mutations remain to be elucidated, this concept is critical not only for understanding the biology of cancer, but also for implications regarding cancer therapy. Conversely, epigenetic silencing or activation of genes may predispose cells to further mutations (e.g., the epigenetic silencing of the key MLH1 DNA repair protein leads to new mutations because of a lack of efficient DNA repair). Other articles in this collection provide details concerning our understanding of how the various epigenetic processes contribute to regulating the genome and can become deregulated in cancer.
Table 2.
Mutations in selected epigenetic modifiers in human cancers
| Process | Gene | Function | Tumor type | Alteration |
|---|---|---|---|---|
| DNA methylation | DNMT1 | DNA methyltransferase | Colorectal, Non–small cell lung, pancreatic, gastric, breast cancer |
Mutation (Kanai et al. 2003) Overexpression (Wu et al. 2007) |
| DNMT3A | DNA methyltransferase | MDS; AML | Mutation (Ley et al. 2010; Yamashita et al. 2010; Yan et al. 2011) | |
| DNMT3B | DNA methyltransferase | ICF syndrome, SNPs in breast and lung adenoma |
Mutation (Wijmenga et al. 2000) Mutation (Shen et al. 2002) |
|
| MBD1/2 | Methyl-binding protein | Lung and breast cancer | Mutation (Sansom et al. 2007) | |
| TET1 | 5′-Methylcytosine hydroxylase | AML | Chromosome translocation (De Carvalho et al. 2010; Wu and Zhang 2010) | |
| TET2 | 5′-Methylcytosine hydroxylase | MDS, myeloid malignancies, gliomas | Mutation/silencing (Araki et al. 2009) | |
| IDH1/2 | Isocitrate dehydrogenase | Glioma, AML | Mutation (Figueroa et al. 2010; Lu et al. 2012; Turcan et al. 2012) | |
| AID | 5′-Cytidine deaminase | CML | Aberrant expression (De Carvalho et al. 2010) | |
| MLL1/2/3 | Histone methyltransferase H3K4 | Bladder TCC, hematopoietic, non-Hodgkin lymphoma, B-cell lymphoma, prostate (primary) | Translocation, mutation, aberrant expression (Gui et al. 2011; Morin et al. 2011) | |
| Histone modification enzymes | EZH2 | Histone methyltransferase H3K27 | Breast, prostate, bladder, colon, pancreas, liver, gastric, uterine tumors, melanoma, lymphoma, myeloma, and Ewing’s sarcoma | Mutation, aberrant expression (Chase and Cross 2011; Tsang and Cheng 2011) |
| BMI-1 | PRC1 subunit | Ovarian, mantle cell lymphomas, and Merkel cell carcinomas | Overexpression (Jiang and Song 2009; Lukacs et al. 2010) | |
| G9a | Histone methyltransferase H3K9 | HCC, cervical, uterine, ovarian, and breast cancer | Aberrant expression (Varier and Timmers 2011) | |
| PRMT1/5 | Protein arginine methyltransferase | Breast/gastric | Aberrant expression (Miremadi et al. 2007) | |
| LSD1 | Histone demethyltransferase H3K4/H3K9 | Prostate | Mutation (Rotili and Mai 2011) | |
| UTX (KDM6A) | Histone demethyltransferase H3K27 | Bladder, breast, kidney, lung, pancreas, esophagus, colon, uterus, brain, hematological malignancies | Mutation (Rotili and Mai 2011) | |
| JARID1B/C (KDM5C) | Histone demethyltransferase H3K4/H3K9 | Testicular and breast, RCCC | Overexpression (Rotili and Mai 2011) | |
| EP300 (P300/KAT3B) | Histone acetyltransferase | Breast, colorectal, pancreatic cancer | Mutation (Miremadi et al. 2007) | |
| CREBBP (CBP/KAT3A) | Histone acetyltransferase | Gastric and colorectal, epithelial, ovarian, lung, esophageal cancer | Mutation, overexpression (Miremadi et al. 2007) | |
| PCAF | Histone acetyltransferase | Epithelial | Mutation (Miremadi et al. 2007) | |
| HDAC2 | Histone deacetyltransferase | Colonic, gastric, endometrial cancer | Mutation (Ropero et al. 2006) | |
| SIRT1, HDAC5/7A | Histone deacetyltransferase | Breast, colorectal, prostate cancer | Mutation, aberrant expression (Miremadi et al. 2007) | |
| Chromatin-remodeling enzymes | SNF5 (SMARCB1, INI1) | BAF subunit | Kidney malignant rhabdoid tumors, atypical rhabdoid/teratoid tumors (extrarenal), epithelioid sarcomas, small cell hepatoblastomas, extraskeletal myxoid chondrosarcomas, and undifferentiated sarcomas | Mutation, silencing, loss of expression (Wilson and Roberts 2011) |
| BRG1 (SMARCA4) | ATPase of BAF | Lung, rhabdoid, medulloblastoma | Mutation, low expression (Wilson and Roberts 2011) | |
| BRM (SMARCA2) | ATPase of BAF | Prostate, basal cell carcinoma | Mutation, low expression (Sun et al. 2007; de Zwaan and Haass 2010) | |
| ARID1A (BAF250A) | BAF subunit | Ovarian clear cell carcinomas, 30% of endometrioid carcinomas, endometrial carcinomas | Mutation, genomic rearrangement, low expression (Jones et al. 2010; Guan et al. 2011) | |
| ARID2 (BAF200) | PBAF subunit | Primary pancreatic adenocarcinomas | Mutation (Li et al. 2011) | |
| BRD7 | PBAF subunit | Bladder TCC | Mutation (Drost et al. 2010) | |
| PBRM1 (BAF180) | PBAF subunit | Breast tumors | Mutation (Varela et al. 2011) | |
| SRCAP | ATPase of SWR1 | Prostate | Aberrant expression (Balakrishnan et al. 2007) | |
| P400/Tip60 | ATPase of SWR1, acetylase of SWR1 | Colon, lymphomas, head and neck, breast | Mutation, aberrant expression (Mattera et al. 2009) | |
| CHD4/5 | ATPase of NuRD | Colorectal and gastric cancer, ovarian, prostate, neuroblastoma, hematopoietic | Mutation (Bagchi et al. 2007; Kim et al. 2011; Wang et al. 2011) | |
| CHD7 | ATP-dependent helicase | Gastric and colorectal | Mutation (Wessels et al. 2010) |
Adapted from You and Jones 2012.
MDS, myelodysplastic syndromes; AML, acute myeloid leukemia; ICF, immunodeficiency, centromere instability, and facial anomalies; SNPs, single-nucleotide polymorphisms; TCC, transitional cell carcinoma; HCC, hepatocellular carcinoma; RCCC, renal clear cell carcinoma; TET, ten-eleven translocation; NuRD, nucleosome remodeling and deacetylation.
2. THE IMPORTANCE OF CHROMATIN TO CANCER
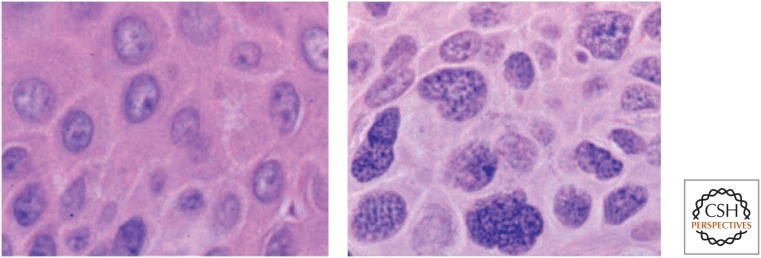
Despite the major advances in understanding the key molecular lesions in cellular control pathways that contribute to cancer, it is true that microscopic examination of nuclear structure by a pathologist remains a gold standard in cancer diagnosis. The human eye can accurately discern changes in nuclear architecture, which largely involve the state of chromatin configuration, and definitively diagnose the cancer phenotype in a single cell. Foremost in the cues used by pathologists are the size of the nucleus, nuclear outline, a condensed nuclear membrane, prominent nucleoli, dense “hyperchromatic” chromatin, and a high nuclear/cytoplasmic ratio. These structural features, visible under a microscope (Fig. 2), likely correlate with profound alterations in chromatin structure and function, with resultant changes in gene expression states and/or chromosome stability. Linking changes observable at a microscopic level with the molecular marks discussed throughout this collection remains one of the great challenges in cancer research. In this article, we review epigenetic marks that are abnormally distributed in cancer cells, typified by changes in DNA cytosine methylation at CpG dinucleotides, changes in histone modifications, nucleosomal composition (i.e., the incorporation of histone variants), and nucleosome positioning.
Figure 2.
Chromatin structural changes in cancer cells. These two photomicrographs were taken from a patient with a squamous cell carcinoma of the skin. The left panel shows normal epidermal cells within one millimeter of the contiguous tumor shown at the same magnification on the right. The chromatin, which stains purple as a result of its affinity to hematoxylin, appears much more coarse and granular in the cancer cells than in normal epidermis. Such changes in the staining characteristics of chromatin are used by pathologists as diagnostic criteria for cancer.
Understanding what the pathologist’s visible cellular phenotype means will require researchers to link it to the relationship between nuclear organization, chromatin structure, molecular marks, and genome function. This is an exciting new domain of research only touched on in this article, but it is likely to yield important contributions to our understanding of cancer initiation and progression, thanks to the continued advances in technologies, such as chromosome conformation capture (see Dekker and Misteli 2014), epigenome-wide mapping studies, massive parallel sequencing, genome tethering techniques, and advanced fluorescence microscopy modeling (Bernstein et al. 2010; Cancer Genome Atlas Research Network 2013b; Garraway and Lander 2013; Reddy and Feinberg 2013).
One of the most recent exciting developments in the understanding of normal and cancer epigenomes comes from the results of whole-exon sequencing, whole-genome sequencing, genome-wide DNA methylation and chromatin analyses, and RNA expression approaches, which all supersede previous genome-wide analyses (Bernstein et al. 2010; Jones 2012; Cancer Genome Atlas Research Network 2013b; Garraway and Lander 2013; Reddy and Feinberg 2013). We, therefore, now recognize that epigenetic control involves not only canonical coding genes, but also noncoding RNA (ncRNA), microRNAs (miRNAs), and other regions that provide important genome regulatory function (Bernstein et al. 2010; Jones 2012; Cancer Genome Atlas Research Network 2013b; Garraway and Lander 2013; Reddy and Feinberg 2013). Thousands of solid and liquid tumors have been analyzed, showing, as introduced above, that there is an unexpected plethora of mutations in genes that control the function of the epigenome (Fig. 1; Table 2) (Baylin and Jones 2011; Dawson et al. 2011; You and Jones 2012; Garraway and Lander 2013; Shen and Laird 2013; Timp and Feinberg 2013; Audia and Campbell 2014). Importantly, many of these mutations occur at high enough frequencies to justify their roles as “driver” mutations in the cancers—that is, the results clearly show that disruption of the epigenome by mutations may lead to the initiation and/or progression of cancer. A major challenge, however, is to understand their precise contribution to cancer-specific alterations in chromatin and DNA methylation, and the exact consequences of these mutations in the key steps of tumorigenesis. It is important to remember that epigenetic changes in cancer may arise independently of mutations in chromatin-modifying factors; the epigenome is also subject to damage and heritable alterations induced by environmental or physiological events inherent to cancer risk states and steps during cancer progression (O’Hagan et al. 2008, 2011; Zheng et al. 2012), as will be discussed.
3. THE ROLE OF DNA METHYLATION IN CANCER
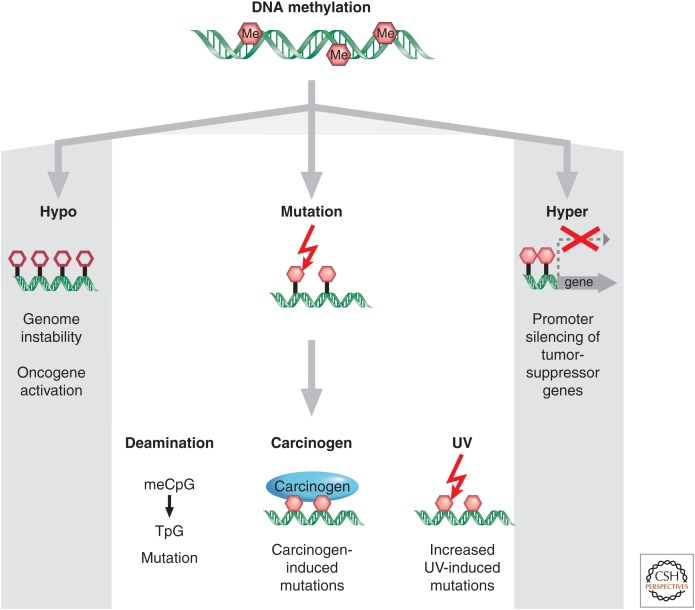
The initial discovery that the cytosine base in DNA can be methylated to become 5-methylcytosine (5mC), sometimes referred to as the 5th base, soon led to the proposal that alterations in DNA methylation may contribute to oncogenesis (Table 3). Over the last 40 years, there have been many studies showing that alterations in the 5mC distribution patterns can distinguish cancer cells from normal cells. At least three major routes have been identified by which CpG methylation can contribute to the oncogenic phenotype. The first is by general hypomethylation of the cancer genome. Second, focal hypermethylation at TSG promoters may occur. Third, direct mutagenesis of 5mC-containing sequences by deamination, UV irradiation, or exposure to other carcinogens is possible (Fig. 3) (Jones and Laird 1999; Jones and Baylin 2002; Herman and Baylin 2003; Baylin and Jones 2011). It is significant that all three of these alterations generally occur simultaneously to contribute to cancer, suggesting that altered homeostasis of epigenetic mechanisms is central to the evolution of human cancer.
Table 3.
Time line for elucidating the role of DNA methylation in cancer
| Observation | Reference |
|---|---|
| Hypothesis of “methylases as oncogenic agents” | Srinivasan and Borek 1964 |
| Decreased levels of 5-methylcytosine in animal tumors | Lapeyre and Becker 1979 |
| 5-Azacytidine and 5-aza-2′-deoxycytidine inhibit methylation and activate genes | Jones and Taylor 1980 |
| Decreased genomic and gene-specific methylation in human tumors | Ehrlich et al. 1982; Feinberg and Vogelstein 1983; Flatau et al. 1984 |
| Inhibitors of DNA methylation alter tumorigenic phenotype | Frost et al. 1984 |
| Methylation of a CpG island in cancer | Baylin et al. 1987 |
| Hot spots for p53 mutations are methylated CpG sites | Rideout et al. 1990 |
| Allele-specific methylation of the retinoblastoma TSG | Sakai et al. 1991 |
| Loss of imprinting in cancer | Rainier et al. 1993 |
| Hypermethylation of CpG islands is associated with aging | Issa et al. 1994 |
| Mice with decreased methylation develop fewer tumors | Laird et al. 1995 |
| Coupling DNA methylation and HDAC inhibitors leads to rapid isolation of TSGs | Suzuki et al. 2002; Yamashita et al. 2002 |
| DNA repair gene (MLH1) is methylated in somatic cells Hypomethylation contributes to cancer |
Gazzoli et al. 2002 Gaudet et al. 2003 |
| 5-Azacytidine is FDA approved for treatment of myelodysplastic syndrome Discovery of the 5-hydroxymethyl-cytosine base and the TET1/2/3 enzymes that catalyze this conversion |
Kaminskas et al. 2005 Kriaucionis and Heintz 2009; Tahiliani et al. 2009 |
Adapted from You and Jones 2012.
HDAC, histone deacetylase; FDA, [U.S.] Food and Drug Administration; TSG, tumor-suppressor gene; TET, ten-eleven translocation.
Figure 3.
Epigenetic alterations involving DNA methylation can lead to cancer by various mechanisms. Loss of DNA cytosine methylation (white hexagons) illustrated in the hypo column results in genome instability. Focal hypermethylation (pink hexagons) at gene promoters shown in the hyper column causes heritable silencing and, therefore, inactivation of tumor suppressors and other genes. Additionally, methylated CpG sites (pink hexagons) are prone to mutation: They are hot spots for C to T transition mutations caused by spontaneous hydrolytic deamination; or methylation of CpG sites can increase the binding of some chemical carcinogens to DNA; and it increases the rate of UV-induced mutations.
3.1. DNA Hypomethylation in Cancer
The most prominent and earliest recognized change in DNA methylation patterns in cancer cells was regional decreases in this modification (Feinberg and Vogelstein 1983; Ehrlich and Lacey 2013), now recognized by genome-wide analyses as a global DNA hypomethylation (Hansen et al. 2011; Berman et al. 2012; Bert et al. 2013). Although all of the ramifications of these losses still need definition, DNA demethylation potentially contributes to genomic instability and increases in aneuploidy (Ehrlich and Lacey 2013), which are both classic hallmarks of cancer. Indeed, deletion or reduction of the maintenance DNA methyltransferase, Dnmt1, results in increased mutation rates, aneuploidies, and tumor induction, a clear indication that DNA hypomethylation plays an active role in increasing chromosomal fragility (Chen et al. 1998; Narayan et al. 1998; Gaudet et al. 2003; Ehrlich and Lacey 2013). Loss of DNA methylation may be accompanied by the activation of transcription, allowing transcription of repeats, transposable elements (TEs), and oncogenes (Jones and Baylin 2007; Ehrlich and Lacey 2013; Hur et al. 2014). Activation of repeats may predispose the genome of a cell to recombination, as corroborated by the increased frequency of chromosomal recombination at certain genomic regions (hot spots) or may express nearby proto-oncogenes (Wolffe 2001; Jones and Baylin 2007; Ehrlich and Lacey 2013; Hur et al. 2014). Indeed, the activation of TEs is another potential source of mutations during the transposition process.
We know that most of the CpGs in the genome, apart from CpG-rich regions, are 80% methylated. In cancer, the average CpG methylation levels are 40%–60%. Advances in mapping technologies are allowing researchers to map the patterns more precisely. Such studies have revealed that DNA hypomethylation can be concentrated in blocks of 28 kb–10 Mb, covering about one-third of the genome (Hansen et al. 2011; Berman et al. 2012; Hon et al. 2012; Bert et al. 2013). The exact mechanisms by which DNA methylation is lost from the cancer epigenome and how functional consequences occur are not yet fully understood; however, we are beginning to be able to dissect these mechanisms. For example, a leading possibility is that many regions of DNA hypomethylation could be integrally tied to broad shifts in chromatin organization, typical in cancer (discussed further in Sec. 6). The broad epigenomic changes, in turn, could, in some instances, result from mutations in chromatin regulators that affect DNA methylation homeostasis, such that the active or passive process of removing DNA methylation is promoted. This could occur, for example, as discussed below and in other articles, by the deregulated activation of ten-eleven translocation (TET) family members or the partial loss of function of the DNA methyltransferase (DNMT) proteins.
3.2. DNA Hypermethylation in Cancer
A well-chronicled DNA methylation change in cancer is abnormal hypermethylation of CpG islands in the 5′ regions of cancer-related genes (i.e., hypermethylation, Fig. 3). This change can be integrally associated with transcriptional silencing, providing an alternative mechanism to mutation for the inactivation of genes with tumor-suppressor function (Jones and Baylin 2007; Baylin and Jones 2011; Shen and Laird 2013). In this regard, 60% of all gene promoters have CpG islands, most of which are not DNA methylated at any time in normal development or in adult cell renewal systems (Jones and Baylin 2007; Baylin and Jones 2011; Shen and Laird 2013). This lack of methylation is fundamental to the more open chromatin states, and active, or ready to be activated, expression status of these genes (Jones and Baylin 2007; Baylin and Jones 2011; Shen and Laird 2013). The fact that methylated CpG island promoters are so prevalent in cancers (∼5%–10% of CGI genes) and are known to directly contribute to carcinogenesis has led to new possibilities in the area of epigenetic therapy—that is, where epigenetic changes are targeted for therapeutic reversal, as discussed further in Section 9 (Egger et al. 2004; Spannhoff et al. 2009; Kelly et al. 2010; Bernt et al. 2011; Daigle et al. 2011; Dawson et al. 2012; Azad et al. 2013).
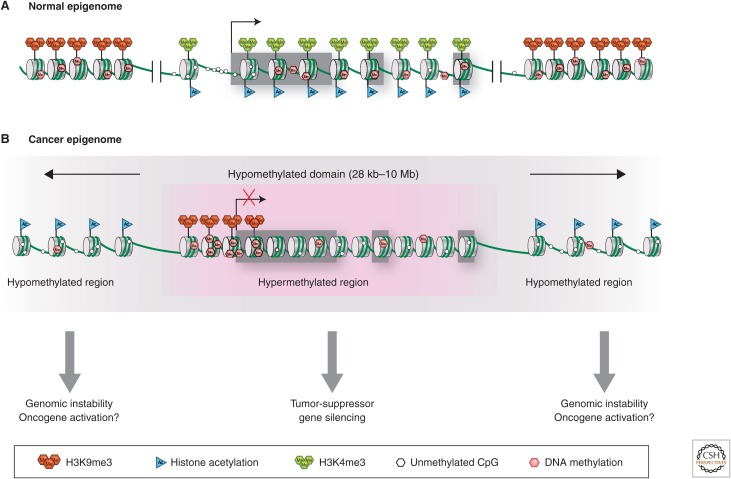
It should be noted that 5mC commonly occurs in the gene body of active genes and functional ramifications in this region may often be opposite to presence of this modification in promoters (Jones 2012; Kulis et al. 2012; Shen and Laird 2013). Thus, rather than being associated with repression of transcription, gene body DNA methylation may facilitate transcriptional elongation and enhance gene expression (Fig. 4) (Jones 2012; Kulis et al. 2012; Shen and Laird 2013). Interestingly, DNMT3A somatic mutations that occur in certain patients with acute myeloid leukemia (AML) may predispose them to a loss of gene body DNA methylation (Cancer Genome Atlas Research Network 2013a), the causal consequences of which are currently unclear.
Figure 4.
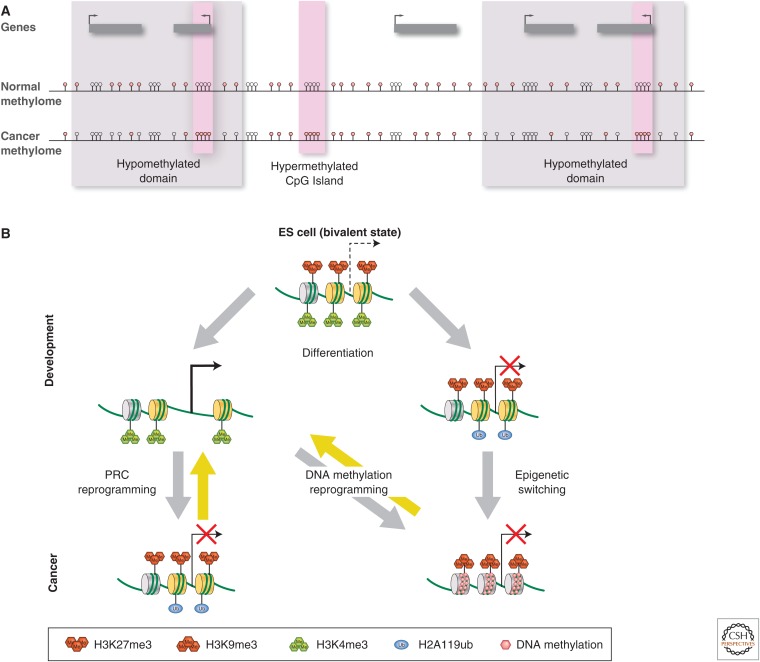
Chromatin structural changes in cancer cells. (A) In a typical cell, a CpG-island-containing active gene can be recognized by virtue of a nucleosome-depleted promoter, absence of promoter DNA methylation, but marked by H3K4me3 surrounding the promoter and histone acetylation along the locus. Gene body CpG methylation often can be observed. Nongenic regions flanking an active gene are frequently marked by repressive epigenetic marks, such as H3K9me3 and 5mC. (B) The cancer epigenome is characterized by simultaneous global losses in DNA methylation (gray shading), interspersed with silenced genes that have abnormal gains of DNA methylation and repressive histone modifications in CpG island promoter regions. These silenced genes may be hypomethylated in their gene body, similar to surrounding chromatin. The hypomethylated regions can have an abnormally open nucleosome configuration and acetylated histone lysines. Conversely, abnormal DNA hypermethylation in promoter CpG islands of silenced genes is associated with nucleosomes positioned over the transcription start sites.
Our mechanistic understanding of how DNA methylation homeostasis may be disturbed in cancer is continually being enriched by discoveries challenging two key assumptions in epigenetics and cancer: All mammalian DNA methylation is confined to CpG sequences and it is a very stable mark. The first assumption was challenged when DNA methylation at CpHpG sequences was documented in human embryonic stem (ES) cells (Lister et al. 2009). The significance of this remains to be determined and it has not been well documented in cancers. The second assumption was challenged following proof that methylated cytosines can be actively demethylated; this has been extremely significant in both the epigenetic and cancer fields (described in Kriaucionis and Tahiliani 2014; elaborated in Sec. 3 of Li and Zhang 2014). DNA demethylation was first discovered through the identification of oxidative derivatives of 5mC, including 5-hydroxymethylcytosine (Kriaucionis and Heintz 2009; Tahiliani et al. 2009), 5-formylcytosine, and 5-carboxylcytosine. Simultaneously, the TET1, -2, and -3 proteins (ten-eleven translocations) were shown to catalyze these oxidative steps (Wu and Zhang 2011a,b), suggesting that these are some of the effectors of active and/or passive DNA demethylation pathways (see Fig. 6 of Li and Zhang 2014).
Figure 6.
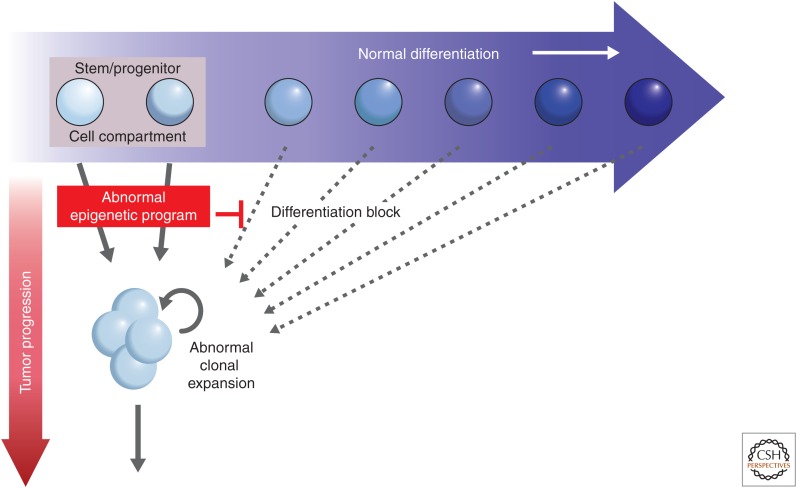
Epigenetic gene-silencing events and tumorigenesis. The earliest steps in tumorigenesis are depicted as abnormal clonal expansion, which evolves during the stress of cell renewal. This is caused by factors, such as aging and chronic injury from, for example, inflammation. These cell clones are those at risk of subsequent genetic and epigenetic events that could drive tumor progression. Abnormal epigenetic events, such as the aberrant gene silencing focused on in this article, could be the earliest heritable causes, in many instances, for a potential role in inducing the abnormal clonal expansion from within stem/progenitor cell compartments in a renewing adult cell system. The gene silencing is triggered by chromatin modifications that repress transcription, and the DNA hypermethylation of this chromatin serves as the tight lock to stabilize the heritable silencing. The gene silencing, in turn, disrupts normal homeostasis, preventing stem and progenitor cells from moving properly along the differentiation pathway for a given epithelial cell system (blue arrow) and channels them into the abnormal clonal expansion (red arrow).
The suggestion that mutations in the TET enzymes may be associated with a DNA hypermethylation phenotype in cancer (Figueroa et al. 2010) is still being debated (Cancer Genome Atlas Research Network 2013a). However, TET-mediated DNA demethylation has been linked to altered cellular metabolism and cancer through mutations in the upstream isocitrate dehydrogenase enzymes, IDH1 and IDH2. These enzymes normally produce α-ketoglutarate, an essential cofactor for the TET hydroxylases (elaborated in Sec. 5.2) (Lu et al. 2012, 2013; Shen and Laird 2013; Venneti et al. 2013). Mutations in IDH1/2, however, lead to a marked increase in the formation of an abnormal metabolite, 2-hydroxy-glutarate, formed from α-ketoglutarate (see Fig. 6 of Berger and Sassone-Corsi 2014). In this scenario, an increased frequency of DNA hypermethylation can be observed, as seen with leukemias and brain tumors (Noushmehr et al. 2010; Turcan et al. 2012; Shen and Laird 2013). The fact that TET and IDH mutations in cancer are mutually exclusive underscores the need for constant demethylation in ensuring the correct level of cellular 5mC (Williams et al. 2011). Importantly, IDH mutations in the hematopoietic system (Sasaki et al. 2012a) appear to drive tumorigenesis because it blocks a cell’s response to differentiation cues and, hence, skews lineage choice (Borodovsky et al. 2013; Turcan et al. 2013). Importantly, the experimental drug reversal of abnormal DNA methylation patterns associated with IDH mutations appears to restore an element of cellular differentiation responses, showing therapeutic promise for treating these types of cancers (Borodovsky et al. 2013; Turcan et al. 2013).
3.3. Mutation of 5mCs
A third mechanism, which we have known of for some time, by which methylation of cytosine residues (5mC) contributes disproportionately to cancer is its propensity of cytosines to be mutated in this sequence context (Fig. 3). Thus, when looking at the human germline, CpG sites typically methylated in the soma constitute more than a third of all transition mutations. Early examples of such mutations were documented in the cancer-causing p53 gene (Rideout et al. 1990). More surprising is the observation that this mechanism also operates in somatic tissues, contributing significantly to the formation of inactivating mutations in many TSGs. This occurs because methylation of the 5 position of the cytosine ring increases the rate of hydrolytic deamination of the base in double-stranded DNA. The deamination product of 5mC, however, is thymine rather than uracil, as is the case for cytosine (Fig. 3). DNA repair mechanisms are subsequently less efficient at repairing deamination-induced mismatches in DNA. For example, >50% of all of the p53 mutations, which are acquired in sporadic colorectal cancers, occur at sites of cytosine methylation (Greenblatt et al. 1994). Thus, the modification of DNA by the DNMTs substantially increases the risk of getting cancer by this endogenous mechanism.
Methylation of cytosine residues have also been shown to favor the formation of carcinogenic adducts between DNA and carcinogens, such as benzo(a)pyrene in cigarette smoke (Fig. 3). In this case, methylation of the cytosine residue increases the formation of carcinogenic adducts between an adjacent guanine residue and benzo(a)pyrene diol epoxide, resulting in increased mutations at CpG sites in the lungs of cigarette smokers (Greenblatt et al. 1994; Pfeifer et al. 2000).
Interestingly, DNA methylation can also alter the rate of mutations in the p53 gene in sunlight-exposed skin (Greenblatt et al. 1994; Pfeifer et al. 2000). This is because the methyl group changes the absorption spectrum for cytosine into the range of incident sunlight, increasing the formation of pyrimidine dimers in the DNA of skin cells exposed to sunlight. In summary, the 5mC modification of DNA not only increases spontaneous mutagenesis, but can influence the way DNA interacts with carcinogens and UV light (Pfeifer et al. 2000).
4. HYPERMETHYLATED GENE PROMOTERS IN CANCER
A main focus of this article, described in this section, is the characterization and role of DNA methylation in cancer and, in particular, its effect on TSGs. We are beginning to see how this intersects with other modes of epigenetic regulation, discussed further in Section 6.
4.1. The Genes Involved
The most well-understood mechanism by which DNA methylation contributes to cancer is through association with the focal hypermethylation of promoters at TSGs. This clearly is a significant pathway by which genes that would normally suppress cancer development are heritably silenced (Jones and Baylin 2002, 2007; Herman and Baylin 2003; Baylin and Jones 2011; Shen and Laird 2013). Usually, DNA hypermethylation occurs at CpG-rich regions or CpG islands that are located in and around the transcriptional start site of abnormally silenced genes in cancer (Fig. 4). Typically, 5%–10% of these CpG island promoters are DNA methylated in cancer (Baylin and Jones 2011). It is important to recognize that cytosine methylation in CpG islands is usually restricted to the vicinity of the gene start site position often spanning the transcription start site, but also occurring in the island at proximal upstream or downstream positions; this same DNA modification occurring within bodies of genes generally has either no correlation to transcription status or, as discussed earlier, can actually accompany increased gene expression, possibly through facilitating the transcriptional elongation process (Jones 2012; Kulis et al. 2012; Shen and Laird 2013).
The list of cancer-related genes affected by transcription disruption through DNA hypermethylation continues to grow and involves genes found at all chromosome locations. In an individual tumor, hundreds of genes can be disrupted by promoter hypermethylation and this mechanism holds true for virtually every type of cancer (Jones and Baylin 2002, 2007; Baylin and Jones 2011; Hammerman et al. 2012; Cancer Genome Atlas Research Network 2013a; Shen and Laird 2013). Indeed, as more deep analyses of DNA methylation are being performed in multiple tumor types, the frequency of this epigenetic change appears to be outnumbering gene mutations in human tumors (Jones and Baylin 2002, 2007; Baylin and Jones 2011; Hammerman et al. 2012; Cancer Genome Atlas Research Network 2013a; Shen and Laird 2013), promoter regions occur in genes involved in virtually every signaling pathway altered in tumorigenesis. Involvement of such a large number of genes has created one of the most important conundrums for the cancer epigenetics field: Why would so many genes be involved in cancer and which silencing events are truly important for the process of tumorigenesis? Clearly, experimentally, it is difficult to test whether each gene is critical for tumor initiation and progression by loss-of-function analyses. However, as well reviewed and mentioned below, some of the genes involved are clearly driver TSGs (Esteller 2007; Jones and Baylin 2007; Baylin and Jones 2011; Shen and Laird 2013). Moreover, just as analyzing signaling pathway participation has been important for understanding the myriad of genetic alterations in cancer, categorizing DNA hypermethylated genes in this manner has great potential to facilitate our understanding of their significance in the process of tumorigenesis (Jones and Baylin 2007; Baylin and Jones 2011; Shen and Laird 2013).
A first group of DNA hypermethylated genes found in cancers constitutes those in which loss of function clearly has a “driver function” for all stages of cancer evolution (Jones and Baylin 2007; Baylin and Jones 2011; Shen and Laird 2013). Typically, true cancer driver mutations involve a relatively limited group of genes. The first examples of epigenetically silenced genes to be characterized were instrumental in defining gene silencing by promoter hypermethylation as an important mechanism for loss of TSG function in cancer (Table 4). The genes were easily recognized as classic TSGs, known to cause inherited forms of cancer when mutated in the germline of families (Jones and Laird 1999; Jones and Baylin 2002, 2007; Esteller 2008; Shen and Laird 2013). They were also often mutated in sporadic forms of cancers and, notably, were also frequently hypermethylated on one or both alleles in such tumors (Jones and Laird 1999; Jones and Baylin 2002; Herman and Baylin 2003). Also, for these genes, it was noted that promoter hypermethylation sometimes constituted the “second hit” in Knudson’s hypothesis, that is, the first hit constituted a germline mutation in familial tumors, whereas the second hit arose from the loss of function through DNA methylation of the second copy of the gene (Grady et al. 2000; Esteller et al. 2001a). In some instances, 5-azacytidine (5-aza-CR) treatment in cultured tumor cells induced the reactivation of these genes, restoring the key TSG function lost during tumor progression. This was shown for the mismatch repair gene, MLH1, which is typically silenced in colon cancer cells (Herman et al. 1998).
Table 4.
Discovery classes of hypermethylated genes
| Class of hypermethylated gene | Examples |
|---|---|
| Known TSGa |
VHL E-cadherin P16Ink4a MLH1 APC Stk4 Rb |
| Candidate TSG |
FHIT Rassf1a O6-MGMT Gst-Pi GATA4/5 DAP-kinase |
| Gene discovered through random screens for hypermethylated genes |
HIC-1 SFRP1, -2, -4, -5 BMP-3 SLC5A8 SSI1 |
a A classic tumor-suppressor gene (TSG) is known to be mutated in the germline of families with hereditary cancer syndromes.
A second group of epigenetically silenced genes are those previously identified as candidate TSGs by virtue of their function, but were not found to have an appreciable frequency of mutational inactivation, which would indicate that they are true driver mutations (Table 4). Despite the paucity of known cancer-associated mutations in this category of genes, they often reside in chromosome regions frequently suffering deletions in cancers. Examples include RasFF1a and FHIT, located on chromosome arm 3p, frequently deleted in lung and other types of tumors (Dammann et al. 2000; Burbee et al. 2001). Other candidate TSGs fit into this category because they are known to encode proteins, which subserve functions critical for the prevention of tumor progression, such as the proapoptotic gene, DAP-kinase (Katzenellenbogen et al. 1999), families of genes that antagonize WNT signaling (Suzuki et al. 2004; Jones and Baylin 2007; Zhang et al. 2008; Baylin and Jones 2011; Shen and Laird 2013). Yet, others qualify because it is now recognized that promoter CpG island hypermethylation can silence noncoding miRNA genes, which are necessary for modulating signaling networks (Saito and Jones 2006; Saito et al. 2006; Chaffer et al. 2013; Tam and Weinberg 2013; Nickel and Stadler 2014; Sun et al. 2014). These genes present an important challenge for the field of cancer epigenetics because, although they are often hypermethylated in tumors, many of them are not frequently mutated, making it difficult to be sure that they actually contribute to tumorigenesis. Section 4.3 describes the strategies being used to determine whether these are truly TSGs.
The third and largest group of genes (Table 4) continues to be populated as more and more genome-wide screens randomly identify aberrant DNA hypermethylation, involving coding and noncoding regions (Baylin and Jones 2011; Shen and Laird 2013; Taberlay et al. 2014). As compared with genes in the first two groups, it is a challenge to place these genes into a functional context for cancer progression because their precise roles are not yet obvious.
A very important relationship exists between a large number of genes hypermethylated in cancer and their tendency to evolve promoter CpG island methylation with age (Issa 2014; Maegawa et al. 2014). This has been very well shown for genes in the colon in which this increasing DNA methylation virtually parallels the age-related risk for colon cancer (Issa et al. 1994; Toyota et al. 1999; Issa 2014; Maegawa et al. 2014). This relationship has now been well documented for other cancers as well and appears to relate to such increases with age that occur not only in humans, but across mammalian species also (Maegawa et al. 2014). The mechanisms for these changes need further dissection, but clearly, this epigenetic change is closely linked to risk for human cancer.
4.2. Technology Used to Identify DNA Methylation Patterns
Class II hypermethylated genes, shown in Table 4, are categorized on the presumption that any abnormal DNA methylation is potentially a causal mechanism in the loss of TSG function, especially when genetic mutations are lacking, yet expression of the gene is low or absent in tumor versus normal tissue. These characteristics provide the basis for a candidate gene approach. The robust application of global mapping assays of genome-wide DNA methylation patterns is now a mandatory approach to identify new hypermethylated genes involved in cancer (i.e., class III in Table 4). Importantly, these technologies also can now place the promoter changes under discussion here into context for their importance, as compared with other regions in which there are DNA methylation changes in tumorigenesis (Bernstein et al. 2010; Cancer Genome Atlas Research Network 2012a; Taberlay et al. 2014). These newer platforms for genome-wide hybridization and/or next-generation sequencing yield comprehensive genome coverage of the DNA methylation landscape. These assays are being used in consortial efforts to map DNA methylation in both normal and disease cells, and illustrate the power to rapidly identify large numbers of hypermethylated genes and other cancer DNA methylation abnormalities (Cancer Genome Atlas Research Network 2012b, 2013a; ENCODE 2012; Shen and Laird 2013).
Many mapping studies to date have used a high-throughput method, which is very cost-effective, when there is a need to broadly screen DNA methylation in many human samples. Termed the Illumina Infinium 450K microarray platform, the assay involves bisulfite treatment of genomic DNA and subsequent hybridization to approximately 450,000 candidate CpG sites throughout the genome. Bisulfite treatment distinguishes methylated from unmethylated cytosines by virtue of the fact that cytosines are converted to uracil, whereas 5mC is resistant to this modification. The Infinium 450K platform queries sites not only at gene promoters, but also at other candidate sequences, including enhancer and ncRNA promoter regions. However, the coverage, although wide, is often not deep in a given sequence region and valuable as a first screening tool, to be followed by deeper probing of selected samples from among those being studied (Dedeurwaerder et al. 2011). This platform is currently used by the Cancer Genome Atlas project for matching DNA methylation abnormalities to genome-wide screens for gene mutations, copy number alterations, translocations, expression changes, and their integration for delineating signaling pathway abnormalities in cancer (Cancer Genome Atlas Research Network 2012b, 2013a; ENCODE 2012; Shen and Laird 2013). A prime goal is to outline cancer-specific abnormalities that suggest new therapy targets for development and biomarker strategies for cancer detection and prognostic predictions. These studies have already produced large lists of newly defined genes with epigenetic abnormalities for brain, colon, lung, breast, and other (Cancer Genome Atlas Research Network 2012b, 2013a; ENCODE 2012; Shen and Laird 2013).
Other integrative studies, such as the ENCODE project and the Epigenome Roadmap projects (Cancer Genome Atlas Research Network 2012b, 2013a; ENCODE 2012), are also increasing our understanding of the role of DNA methylation and chromatin abnormalities in cancer and, specifically, the hypermethylated genes under discussion. The more costly approaches involve methods, such as capturing differentially DNA methylated sequences by either methylcytosine antibodies, antibodies recognizing methylcytosine-binding proteins or their binding domains or sequences generated by methylation-sensitive restriction enzymes, and then identifying these via next-generation sequencing (Harris et al. 2010; Aryee et al. 2013). Even more extensive information is also being compiled by direct sequencing of virtually all candidate CpG sites following bisulfite treatment of DNA (Lister et al. 2009; Lister et al. 2011; Berman et al. 2012). All of these approaches are providing a detailed view of DNA methylation patterns inherent to normal development in normal mature tissues and primary and cultured tumor samples.
The high-throughput DNA methylation detection approaches can be combined with data obtained by treatment of cultured cells with demethylating agents, such as 5-aza-CR or 5-aza-2′-deoxycytidine (5-aza-CdR). RNA from before and after drug treatment is hybridized to gene microarrays, or subject to RNA-sequencing (RNA-seq) analysis, to detect drug-induced up-regulated genes (Suzuki et al. 2002; Yamashita et al. 2002; Schuebel et al. 2007). It must be recognized, however, that the very low expression levels of many of the induced genes before and after drug treatment challenge the sensitivity of gene expression platforms and reduce the efficiency of these approaches (Suzuki et al. 2002; Schuebel et al. 2007). Use of quantitative RNA-seq assays may provide a more dynamic range of gene expression changes, which enhances the utility of combining induced gene expression with genome-wide DNA methylation assays.
4.3. Determining the Functional Importance of Genes Hypermethylated in Cancer
The large number of genes with hypermethylated DNA at their promoters in cancer presents a formidable research challenge for understanding the functional scope of these changes. Frequent promoter hypermethylation in a given gene does not, in and of itself, guarantee that the silenced gene has a functional significance in cancer, as is often the case for genetic mutations. This is especially the case when the hypermethylated gene is not a known tumor suppressor and there is no evidence that the gene is frequently mutated in cancers. Thus, it is obligatory that the gene in question is studied in such a way as to determine the significance of loss of function, in terms of both the processes controlled by the encoded protein and the implications for tumor progression. In fact, sorting out the driver versus passenger roles for this class of genes is one of the biggest research challenges in cancer epigenetics.
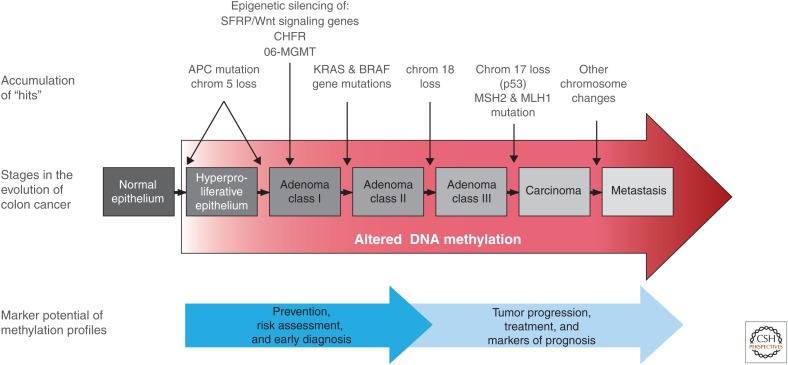
Several initial steps are useful, but do not absolutely confirm the importance of a given gene in cancer (summarized in Table 5). First, of course, is the precise documentation of its cancer-specific hypermethylation profile, including its position in the gene promoter and consequences on the expression state of the gene. This might include assessing the ability of the gene to undergo reexpression following drug-induced promoter demethylation. Second, the incidence of hypermethylation and gene silencing must be well established in primary, as well as cultured, tumor samples. Third, it is often useful to know at what point the silencing of the gene occurs in tumor progression, as exemplified in Figure 5 for colon cancer.
Table 5.
Steps in documenting the importance of a hypermethylated gene for tumorigenesis
| 1. Document CpG island promoter methylation and correlate with transcriptional silencing of the gene and ability to reverse the silencing with demethylating drugs in culture. |
| 2. Document correlation of promoter hypermethylation with specificity for this change in tumor cells (cell culture and primary tumors) versus normal cell counterparts and incidence for the hypermethylation change in primary tumors. |
| 3. Document the position of the hypermethylation change for tumor progression of given cancer types. |
| 4. Document the potential significance for the gene silencing in tumorigenesis through gene reinsertion studies in culture and effects on soft agar cloning, growth of tumor cells in nude mouse explants, etc. |
| 5. Establish function of the protein encoded by the silenced gene, through either known characteristics of the gene or testing for activity of recognized protein motifs in culture systems, etc. |
| 6. Document tumor-suppressor activity and functions of the gene for cell renewal, etc., especially for totally unknown genes, through mouse knockout studies. |
Figure 5.
The position of abnormal DNA methylation in tumor progression. This is depicted in the classic model (Kinzler and Vogelstein 1997) for genetic alterations during the evolution of colon cancer. Altered DNA methylation is shown to occur from very early on in tumorigenesis (red arrow), as discussed in the text, during the conversion of normal to hyperplastic epithelium, accruing during the progression from noninvasive to invasive and, ultimately, metastatic tumors. This places it in a strategic position for channeling stem cells into abnormal clonal expansion (illustrated in Fig. 6) by cooperating with key genetic alterations. These epigenetic abnormalities also have connotations for cancer treatment and markers of prognosis.
Confirming a gene is a bona fide TSG requires studies that assess its contribution to tumorigenicity following loss of function. The function of the encoded protein is important and can be established through knowledge about the type of protein, aspects of the protein structure, and/or relationships to gene families and signaling pathways. In an age when many known genes have been subjected to genetic knockout studies, the phenotypes produced and attendant biology can be informative in pointing to the potential contribution of gene silencing in tumorigenesis. Candidate TSGs can be assayed for their tumorigenic potential following gene knockout by assessing in cultured cells the effects of their loss on (1) soft agar cloning (to detect any capacity for malignant transformation) and (2) tumorigenicity of the cells when grown as heterotransplants in immunocompromised mice and (3) assessing the cellular properties, such as the induction of apoptosis following gene reinsertion. Ultimately, however, additional transgenic knockout approaches may be needed to establish the role of a gene as a tumor suppressor and to understand the functions of the encoded protein in development, adult cell renewal, etc. Mouse knockout studies documenting the function of the transcription factor and developmental gene, HIC-1, provide an example of how this gene was experimentally validated as a TSG (Chen et al. 2003, 2004). It was initially identified through screens of genomic regions that have undergone loss of heterozygosity in cancer cells (Wales et al. 1995). Clearly, discovering genes that are epigenetically silenced in cancer is of great value, yet, the major scope of work lies ahead in definitively trying to show where loss of function for the gene is important in cancer.
5. THE IMPORTANCE OF EPIGENETIC GENE SILENCING IN EARLY TUMOR PROGRESSION
In the classic view of cancer evolution, as articulated by Vogelstein and colleagues (Kinzler and Vogelstein 1997; Vogelstein et al. 2013), a series of genetic changes drives progression from early premalignant stages through the appearance of invasive cancer to the onset of metastatic disease (Fig. 5), although this progression does not necessarily occur in the same exact linear order from tumor to tumor. We now know that epigenetic changes are occurring throughout this course of events, and this includes the early appearance of widespread loss of normal DNA methylation and more focal gains in gene promoters, discussed in Section 4 (Fig. 4). Other features of the epigenome can also be deregulated, including the altered occurrence and distribution of histone marks, and this may be caused by mutations in components of the epigenetic machinery. Thus, there is the potential for interaction between epigenetic and genetic events to drive progressive cellular abnormalities throughout the entire course of neoplastic progression (Fig. 1). Two epigenetic processes, loss of gene imprinting or LOI (as discussed in Zoghbi and Beaudet 2014) and epigenetic gene silencing, are extremely important mechanisms contributing to the very early stages of cancer development.
5.1. Loss of Imprinting
Loss of imprinting (LOI) and epigenetic gene silencing are the most studied processes involving epigenetic aberrations that affect tumor evolution. LOI is a process in which the silenced allele of an imprinted gene becomes activated during tumorigenesis. This results in biallelic expression of the gene, generating excess gene product (Rainier et al. 1993). The most studied example of LOI occurs at the IGF2 gene in tumors, such as colon cancer (Kaneda and Feinberg 2005). This occurs because hypermethylation of a regulatory element upstream of the neighboring imprinted H19 gene removes its insulator function (illustrated in Fig. 8 of Barlow and Bartolomei 2014). This insulator, which normally prevents the IGF2 gene from being activated through interaction with its distal enhancer, allows IGF2 to become expressed on the maternal copy of chromosome 11p in some cancers (Kaneda and Feinberg 2005). The resultant biallelic IGF2 expression leads to excess production of the growth-promoting IGF2 protein. Experimental evidence suggests that this could play a role in the very early progression steps of colon cancer (Kaneda and Feinberg 2005; Sakatani et al. 2005). In fact, studies in mouse models suggest that LOI events alone may be sufficient to initiate the tumorigenesis process (Holm et al. 2005).
Figure 8.
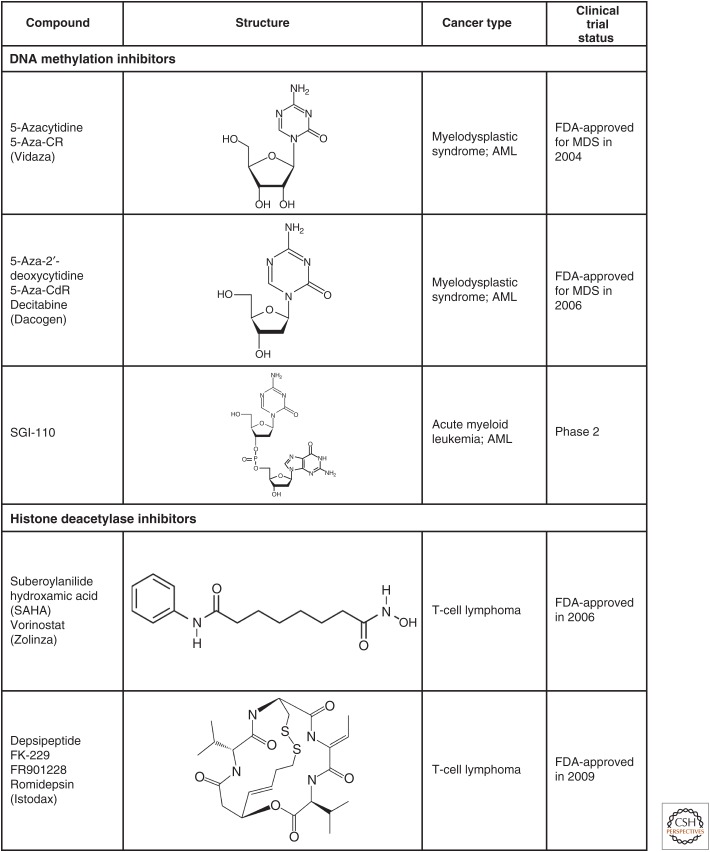
Structures of selected epigenetic drugs. Three nucleoside analogs are known that can inhibit DNA methylation after incorporation into DNA. 5-aza-CR (Vidaza) and 5-aza-CdR (decitabine) have been FDA approved for the treatment of the preleukemic disorder, myelodysplasia. Two HDAC inhibitors are also FDA approved for cutaneous T-cell lymphoma and several others are in clinical trials. Drugs targeting other epigenetic processes are in earlier stages of clinical development (see also Figs. 5 and 6 of Audia and Campbell 2014).
5.2. IDH Mutations Leading to Epigenetic Deregulation in Oncogenesis
Another compelling story illustrating how epigenetic regulation is central in cancer initiation and progression involves IDH1 and IDH2 oncogenic mutations in brain, colon, and hematologic cancers (Figueroa et al. 2010; Noushmehr et al. 2010; Prensner and Chinnaiyan 2011; Turcan et al. 2012; Cancer Genome Atlas Research Network 2013b; Losman and Kaelin 2013). The IDH1/2 alterations appear to alter the DNA and histone demethylation pathways, resulting in imbalances in histone methylation levels, such as increases in H3K36, H3K9 methylation (Lu et al. 2012; Lu et al. 2013; Venneti et al. 2013). There is also an associated increase in the frequency of promoter region CpG island DNA hypermethylation, which resembles the well-characterized CpG island methylator phenotype (CIMP) in colon and other cancers (Figueroa et al. 2010; Noushmehr et al. 2010; Turcan et al. 2012). The genes heavily involved are those with a history of an embryonic state chromatin pattern and often involved in the regulation of development (discussed further in Sec. 6.3).
Delineating the precise causes for these above chromatin and DNA methylation changes is an ongoing area of investigation. The leading data-driven hypothesis is that the changes result from the accumulation of 2-hydroxy-glutarate from α-ketoglutarate in cancer cells. This abnormal 2-hydroxy-glutarate metabolite, which increases to mm levels in cells with the IDH mutations, constituting a biomarker in and of itself, competes with the necessary α-ketoglutarate metabolite needed by the TET and lysine (K) demethylase (KDM) enzymes, which regulate either chromatin demethylase function or levels of DNA methylation. Interestingly, other Krebs cycle control genes, when mutated in certain tumor types, can also result in decreased α-ketoglutarate levels and similar chromatin and DNA methylation abnormalities (Xiao et al. 2012; Mason and Hornick 2013). Experiments have specifically shown that this leads to the buildup of repressive histone marks in gene promoter regions and, subsequently, DNA hypermethylation in what may constitute a molecular progression (Lu et al. 2012, 2013; Venneti et al. 2013). Mouse models for IDH1 or IDH2 mutagenesis suggest that these mutations are implicated in early tumor progression events (Sasaki et al. 2012b). Engineering the mutations into mice or cells, in vitro, appears to trap stem/progenitor cells in states of abnormal self-renewal and/or diminishes their capacity for lineage commitment and differentiation, as illustrated in Figure 6 (Lu et al. 2012; Turcan et al. 2012; Borodovsky et al. 2013). Then, inducing DNA demethylation can partially restore the capacity of cells with the mutations to respond to differentiation cues (Borodovsky et al. 2013; Turcan et al. 2013).
5.3. Early Event IDH or H3 Mutations Drive Oncogenesis
Beyond the IDH1/2 example in cancer, as discussed earlier, there are an increasing number of common mutations being found in genes coding for proteins that establish and maintain appropriate chromatin configurations (i.e., the normal epigenome [Figs. 1 and 4]). In fact, recent studies looking at the pathways that are activated during typical early cancer cell insults implicates the epigenetic machinery, and this is beginning to explain why epigenetic alterations are a common event in the early stages of cancer and even in precancerous changes preceding frank malignancy. Intriguingly, the timing of key mutations and cell compartments in which they occur may actually dictate and/or accompany the evolution of tumor subtypes. This may involve either a prominent presence of DNA methylation abnormalities or chromatin changes, both of which can play a major driver role.
A dramatic example of contrasting epigenetic patterns in cancer is evident when comparing IDH mutations in a pediatric subtype of brain tumor versus a histone mutation occurring at a key PTM site in another subtype of brain tumor. The IDH mutations are associated with CIMP and confined to low-grade gliomas arising in proneural progenitor cells in younger patients with better survival rates than those with advanced gliomas. These tumors arise in glial cell progenitors (Parsons et al. 2008; Noushmehr et al. 2010). In contrast, in the other subtype, mutations in H3K27 have been recently described and these tumors do not have CIMP. Although these H3K27 mutations are present in only one of the many H3 alleles, they apparently exert a dominant negative effect, which blunts all activity of the EZH2 enzyme catalyzing H3K27 methylation. The result is a dramatic loss of H3K27me3 (Chan et al. 2013; Lewis et al. 2013; Shen and Laird 2013), which probably leads to the activation of many genes that can drive tumorigenesis in a particular progenitor cell within a cell compartment.
5.4. Known Examples of TSG Epigenetic Silencing in Oncogenesis
Evidence for the involvement of specific genes in cancer progression continues to build. p16, for example, is a classic TSG that can be mutated or epigenetically silenced in human cancers. In lung cancer, the epigenetic silencing of p16ink4a (listed in Table 1) occurs very early in populations of premalignant cells before tumor formation (Swafford et al. 1997). In breast cancer, small populations of hyperplastic epithelial cells are also prone to p16ink4a epigenetic silencing (Holst et al. 2003). In fact, in cell culture (on plastic), normal human mammary epithelial cells require this type of p16 silencing as a prerequisite for the very early steps toward cell transformation (Kiyono et al. 1998; Romanov et al. 2001). This loss of gene function through epigenetic means accompanies a failure of subsets of the mammary cells to reach a mortality checkpoint, allowing these cells to then develop progressive chromosomal abnormalities and the reexpression of telomerase as they continue to proliferate. Furthermore, it also involves the expansion of stem cells, as observed in p16 mouse knockout models (Janzen et al. 2006).
A second example concerns the mismatch repair gene MLH1. This gene is typically mutated in the germline of families predisposed to a type of colon cancer; this form displays multiple genetic alterations and the “microsatellite” instability phenotype (Fishel et al. 1993; Liu et al. 1995). However, 10%–15% of patients with this tumor phenotype have nonfamilial colon cancer, in which the MLH1 gene is epigenetically silenced rather than genetically mutated (Herman et al. 1998; Veigl et al. 1998). It follows that its loss of function in a DNA repair capacity could lead to multiple genetic alterations and microsatellite instability. Indeed, in cell culture, reexpression of epigenetically silenced MLH1 produces a functional protein, which restores a considerable portion of the DNA damage mismatch repair function (Herman et al. 1998). This illustrates the clear link between genetics and epigenetics for these types of colon cancers in which MLH1 is epigenetically silenced. However, we do not have a full understanding of all the mechanisms involved yet; for instance, it is intriguing that virtually all of these colon tumors have the CpG island hypermethylator phenotype (Toyota et al. 1999; Weisenberger et al. 2006; Hinoue et al. 2012) and mutations of the B-RAF oncogene (Weisenberger et al. 2006; Shen and Laird 2013). Recent work by Hitchins et al. (2011) has interestingly shown that a single nucleotide variant in the promoter region of the MLH1 gene, which results in reduced expression of the allele, predisposes it to becoming methylated. Such reduced transcription may bias these alleles to evolve DNA methylation at promoters, which may deepen the silencing and make the gene more difficult to transcribe. It is most important, however, to pursue the underlying mechanisms leading to these outcomes.
Another example of a gene that is subject to early and important epigenetic changes is Chfr, a checkpoint-regulating gene that also controls genomic integrity, chromosomal stability, and ploidy (Table 1) (Sanbhnani and Yeong 2012). This gene is infrequently mutated in tumors, although it is often epigenetically silenced in lung and other cancers and, importantly, silenced early in the progression of colon cancer (Fig. 5) (Mizuno et al. 2002). Mouse knockout studies have revealed a tumor-suppressor role for this gene based on its function as an E3 ubiquitin ligase that regulates Aurora A, a control gene for mitosis (Yu et al. 2005). Consequently, embryonic cells from the mice display chromosomal instability and a predisposition to transformation.
5.5. Defining Epigenetic Silencing of TSGs as Drivers or Passengers of Oncogenesis
Many of the hypermethylated genes in cancer can only be defined as candidate TSGs and often only have a history of epigenetic change, but no genetic mutations. Research, as highlighted in Section 4.3, is needed to determine whether these genes are silenced early, which would represent a key event in early tumor progression. For example, the DNA repair gene, O6-MGMT, is silenced early in premalignant stages of colon cancer progression (Fig. 5) (Esteller et al. 2001b) and this loss of function can predispose cells to persistent alkylation damage at guanosines, resulting in G to A point mutations. Indeed, silencing of this gene occurs in premalignant colon polyps before the appearance of a high rate of p53 and RAS gene mutations in later colon tumor progression phases (Esteller et al. 2001b; Wolf et al. 2001). Similarly, the GST-Pi gene is epigenetically silenced via promoter hypermethylation in virtually all premalignant lesions that are predisposing to prostate cancer, putting cells at risk of oxidative damage at adenine (Lee et al. 1994).
The random screening approaches used to identify DNA hypermethylated genes in cancer has uncovered a particularly intriguing scenario in the progression of colon cancer: Epigenetic loss of function seems to occur in a number of components of the Wnt signaling family of genes, as discovered through a microarray approach (Suzuki et al. 2002). Silencing of genes required for regulating signal transduction may, thus, allow the abnormal activation of the WNT developmental pathway, driving early cancer progression (Suzuki et al. 2004; Jones and Baylin 2007; Zhang et al. 2008; Baylin and Jones 2011). For instance, frequent mutations (genetic and epigenetic) in another member of the Wnt pathway, the APC tumor suppressor, are also known to be universally involved with the initiation and progression of this disease and, hence, can be considered as driver mutations for this type of cancer. Other components of the Wnt signaling pathway were later implicated in colon tumorigenesis and gene silencing, such as the family of secreted frizzled-related protein genes (SFRPs) (Suzuki et al. 2004) and the transcription factor SOX17 (Zhang et al. 2008). The silencing of SFRPs relieves repression of the pathway at the level of membrane and cytoplasmic events. Loss of SOX17, which normally antagonizes the β-catenin transcription factor, relieves repression of this nuclear step that normally blocks Wnt ligand signal transduction (Finch et al. 1997; Zorn et al. 1999; Zhang et al. 2008), resulting in the up-regulation of downstream cellular β-catenin transcription factor levels. These silencing events occur in very early lesions predisposing to colon cancer, sometimes before common mutations in downstream Wnt pathway proteins (Suzuki et al. 2004; Zhang et al. 2008). Thus, early activation of the Wnt pathway by epigenetic events promotes the early expansion of cells. Persistence of both epigenetic and genetic alterations seems to complement one another in further driving progression of the disease (Suzuki et al. 2004).
The HIC-1 (hypermethylated-in-cancer 1) gene, which encodes a zinc finger transcriptional repressor, provides a final example of how a putative TSG, when its expression is epigenetically altered, can be cancer driving. HIC-1 was discovered by a random screening looking for hypermethylated CpG islands in a hot spot for chromosomal loss in cancer cells (Wales et al. 1995). HIC-1, although not mutated, was epigenetically silenced early on in cancer progression and, through mouse knockout modeling, proven to be a tumor suppressor (Chen et al. 2003). It complements p53 mutations (Chen et al. 2003) leading to up-regulation of SIRT1 (Chen et al. 2005), which contributes to enhanced stem/progenitor cell growth (Howitz et al. 2003; Nemoto et al. 2004; Kuzmichev et al. 2005). In pediatric medulloblastoma tumors, Hic1 silencing was shown to exert cancer-driving function by depressing the Atoh1 transcription factor required for neuronal cell growth (Briggs et al. 2008).
A key issue for understanding the processes leading to altered DNA methylation and chromatin patterns in early tumorigenesis is elucidating the causative factors that may trigger them. In this regard, some of the environmental factors that induce cellular stress responses, as highlighted in Figure 37 of Allis et al. (2014), appear critical. These exposure scenarios are linked to multiple disease states, including cancer. Recent experimental evidence, for instance, has directly linked exposure to cellular stress and the reversion of key cell population to a stem/progenitor state for survival, with the recruitment of protein silencing complexes involving PcG, histone deactylases (HDACs), and DNMTs, to CpG-rich gene promoters, and consequent gene silencing (O’Hagan et al. 2011). The molecular progression to DNA methylation is often then triggered at vulnerable low-expressed genes (see O’Hagan et al. 2011). Examples of stress stimuli, often observed in the cancer risk state of chronic inflammation and injury, are increases in reactive oxygen species (ROS) or DNA double-strand breaks (O’Hagan et al. 2008, 2011). The genes subject to permanent chromatin and DNA methylation changes at promoters after such insults may be those for which loss of function sets the stage for cell survival (Hahn et al. 2008; O’Hagan et al. 2011). Such cells are then poised for clonal expansion as stem/progenitor types and will be predisposed to later genetic and epigenetic events that drive tumor progression (Fig. 6) (Easwaran et al. 2014).
All of the data discussed above support the hypothesis outlined in Figure 5, which suggests that some of the earliest heritable changes in the evolution of tumors are epigenetic ones, particularly the transcriptional silencing of genes maintained by promoter DNA methylation. Although the precise effects of mutations in epigenetic regulators on cellular phenotypes and the epigenome of the cancer cell are not fully understood, these genetic alterations have highlighted how important epigenetic changes are in tumor initiation and progression. The key challenges now, as outlined in Table 6 and discussed more fully in Section 7, are to understand the molecular dynamics of epigenomic changes causal to cancer progression. This, in turn, will feed into discovering molecular strategies aimed at the prevention and early intervention of cancer, as well as providing more markers for improved diagnosis and prognosis of cancers.
Table 6.
Major research challenges for understanding the molecular events mediating epigenetic gene silencing in cancer
| Questions to be addressed | Research required |
|---|---|
| The cancer methylome | Elucidate links between simultaneous losses and gains of DNA methylation in the same cancer cells. |
| Chromatin boundaries | Determine the molecular nature of boundaries, and how they change during tumorigenesis, that separate areas of transcriptionally active zones encompassing gene promoters from the transcriptionally repressive areas that surround them and which may prevent the repressive chromatin from spreading through the active zone. Among the candidate mechanisms are roles that may be played by key histone modifications, insulator proteins, chromatin-remodeling proteins, etc. |
| Hierarchy of epigenetic events leading to gene silencing | What is the order of events for the evolution of gene silencing in cancer with respect to histone modifications, DNA methylation, etc.? Which comes first and what are the key protein complexes that target the processes (DNA methylating enzymes, histone deacetylating and methylation enzymes, cytosine methyl-binding proteins, polycomb silencing complexes, etc.) that determine the events? |
| Targeting and composition of DNA methylation machinery | Which specific DNA methylating enzymes are required for initiating and/or maintaining the most stable gene silencing and what protein complexes contain them, including their interaction with key histone posttranslational modifications? |
| Composition of the epigenetic machinery in maintaining silencing | Once established, what are all of the components of chromatin and DNA methylation machinery, and the hierarchy of their involvement, required to maintain the gene silencing and how are they reversible? |
6. THE MOLECULAR ANATOMY OF EPIGENETICALLY SILENCED CANCER GENES
Knowing which genes are silenced in neoplastic cells is important for understanding what contributes to the initiation and maintenance of cancer. Silenced loci also serve as excellent models for understanding how gene silencing is initiated and maintained, and how the mammalian genome is packaged to facilitate regions of transcription and repression. An understanding of chromatin function, which is a major emphasis of many of the articles in this collection, is facilitating our understanding of what may trigger aberrant gene silencing in cancer and how the components of this silencing maintain the attendant transcriptional repression. Also, they are unveiling how (onco)genes and regions can be transcriptionally derepressed and what bearing that has on cancer development.
6.1. Chromatin Characteristics of Active and Repressive Genomic Regions
This article has concentrated on DNA methylation in cancer, as well as associated chromatin changes, which may occur in association with or without altered DNA methylation. In particular, we have described the finding and role of aberrant DNA methylation in the gene silencing of TSGs and our understanding of factors involved in DNA methylation homeostasis (i.e., DNMTs, TET enzymes, and IDH1/2). The fundamental defect in cancers with regard to these abnormalities, particularly in proximal gene promoters, appears to be a disruption of chromatin borders that normally separate transcriptionally repressive from active chromatin. In this regard, several laboratories have highlighted how, in cancer cells, chromatin configuration found at hypermethylated CpG islands near the promoters of aberrantly silenced genes differs from when these genes are basally expressed in normal settings (Kelly et al. 2012; Yang et al. 2012). The promoter CpG islands of active genes in normal (or cancer) cells are characterized by a zone of open chromatin, a lack of DNA methylation, nucleosome depletion (detected by hypersensitive sites), and histone PTMs, which are typical for active genes (Fig. 4A) (Kelly et al. 2012; Yang et al. 2012). Active covalent histone marks at gene promoters, which typically become altered along with abnormal DNA methylation in cancer, include acetylation of H3 at lysines 9 and 14 (H3K9ac and H3K14ac) and methylation of H3K4 (Nguyen et al. 2001; Fahrner et al. 2002; McGarvey et al. 2008; Baylin and Jones 2011; Shen and Laird 2013). In addition, the histone variant H2A.Z is present in the nucleosomes flanking the start sites, and its presence is strongly anticorrelated with DNA methylation (Zilberman et al. 2008; Yang et al. 2012).
Beyond the 5′ and 3′ borders of active genes, there appears to be a stark transition in chromatin structure with characteristics of transcriptionally repressed genomic regions (Fig. 4A). Historically, chromatin characterization has been restricted to analyses of relatively short DNA stretches that are biologically relevant. Such studies, using normal cells, revealed that just upstream of promoter CpG islands, the less frequent CpG sites are mostly methylated (Berman et al. 2005; Hansen et al. 2011). These sites were found to recruit methylcytosine-binding proteins (MBDs) and their partners (e.g., HDACs) (illustrated in Fig. 9 of Li and Zhang 2014) and are accessible to enzymes that catalyze repressive histone methylation marks, particularly H3K9me2, accompanied by deacetylation of key histone residues (Nguyen et al. 2001; Fahrner et al. 2002; Kondo et al. 2003; McGarvey et al. 2008; Baylin and Jones 2011; Shen and Laird 2013).
6.2. The Mistargeting of Epigenetic Machinery Recruited by Oncogenic Translocation Products
Several examples showing that chromatin-modifying activities play a role in human cancer have been known for some time (Wolffe 2001). For example, the use of HDACs is altered by chromosomal translocations in AML and acute promyelocytic leukemia (PML) (Di Croce et al. 2002). Histone acetylation is associated with open transcriptionally active chromatin regions (see Marmorstein and Zhou 2014; Pirrotta 2014; Seto and Yoshida 2014). In PML, the PML gene is fused to the retinoic acid receptor (RAR). The PML part of the fusion receptor recruits HDAC and DNA methylation activity and causes a state of transcriptional silencing at RAR target loci (Di Croce et al. 2002). This ultimately participates in a cellular differentiation block (Di Croce et al. 2002). In AML, the DNA-binding domain of the transcription factor AML-1 is fused to a protein called ETO, which, similar to PML, interacts with a HDAC. The mistargeted HDAC contributes to aberrant gene repression, blocking cellular differentiation and ultimately leading to leukemia (Amann et al. 2001).
Another translocation occurring in infants with a highly aggressive form of acute leukemia involves the mixed lineage leukemia (MLL) gene, which codes for a histone K methyltransferase (KMT). The MLL gene product normally catalyzes the formation of the histone H3K4me3 active mark, which helps to repel the de novo DNA methylation machinery (Popovic and Licht 2012). The MLL translocation, however, inactivates the enzyme, thus losing the ability to generate the active histone mark. This fusion gene product can then associate with DNA hypermethylation at some promoters, which may contribute to the disease phenotype (Stumpel et al. 2009). These are just three examples of the direct involvement of chromatin-modifying factors contributing to the oncogenic phenotype.
6.3. The Makeup and Distribution of Typical Epigenomic Alterations in Cancer
More recently, in-depth analyses of CpG methylation across genomes are providing an exciting and enriched look at chromatin transitions at CpG-island-containing promoters, which are prone to abnormal DNA methylation in cancer. These studies suggest that for both normal and cancer cells, there are important configurations across defined, megabase regions of most chromosomes (∼100 kb–10 Mb). In normal ES cell and differentiated cell types, the majority of these megabase domains are not CpG rich, although where they occur, these CpGs are heavily methylated, but in a mosaic fashion, across different tissue types, which have been termed partially methylated domains, that is, ∼80% methylated (Fig. 7A) (Hansen et al. 2011; Berman et al. 2012; Bert et al. 2013; Shen and Laird 2013). In cancer, substantial loss of normal DNA methylation is found throughout these regions, creating hypomethylated domains, with only ∼40%–60% of CpGs methylated, as documented in colon and other cancers (Hansen et al. 2011; Berman et al. 2012; Bert et al. 2013; Shen and Laird 2013). This creates megabase “islands” of reduction, located frequently throughout the genome, commonly termed “hypomethylated blocks” or “domains.” Other epigenomic mapping approaches have termed broadly similar regions in cancer as “large organized chromatin K” domains, corresponding to regions rich in histone lysine methylation, such as H3K9 (Wen et al. 2009; Hansen et al. 2011; Hon et al. 2012). A key question that needs deciphering is whether these broadly defined regions are configured in a repressive chromatin environment (e.g., H3K9me3) or in a more open chromatin environment, as depicted in Figure 4B. Data indicate that both exist in cancer, the significance and consequence of which are actively being investigated (Berman et al. 2012; Hon et al. 2012; Brennan et al. 2013; Reddy and Feinberg 2013; Timp and Feinberg 2013).
Figure 7.
Reprogramming of DNA methylation patterns and abnormal modes of gene silencing in cancer. (A) Common DNA methylome changes observable in cancer versus a normal somatic cell are illustrated. This is shown in the context of large hypomethylated blocks (gray shading) of the genome seen in cancer interspersed with focal hypermethylation of promoter region CpG-island-containing genes (pink shading). In normal cells, background DNA methylation is high (pink shaded hexagons) with the exception of CpG islands (densely packed white shaded hexagons). In the cancer methylome, overall genome DNA methylation declines, particularly in the hypomethylated blocks, whereas CpG island promoter genes frequently become methylated (pink shading), most of which are located in the hypomethylated blocks. (B) The currently suggested routes to abnormally silenced CpG-island-containing genes in cancer are shown. Genes that are active in cells throughout development and adult cell renewal initially have active promoter chromatin, which is characterized by the presence of the bivalent histone modification pattern consisting of H3K4me, the repressive H3K27me3 mark, and a lack of DNA methylation. Genes that become transcriptionally active lose much of their Polycomb-mediated repressive H3K27 methylation, whereas those that become silenced (indicated by a red X) can do so by the loss of H3K4 methylation and acquisition of, or increases in, Polycomb-mediated repressive chromatin (PRC) mark and H2A119 ubiquitination. During tumor progression, active genes may become silenced through either the aberrant PRC-mediated reprogramming (bottom left) or DNA methylation and H3K9me marks (bottom right). Some normally silent genes may change the way in which they are transcriptionally repressed from H3K27-methylation-type repression to H3K9-methylation-based silencing and/or DNA hypermethylation (epigenetic switching). The reverse yellow arrows indicate the potential for epigenetic abnormalities in cancer to be corrected by epigenetic therapies. Representative of such therapies are DNMT inhibitors, HDAC inhibitors, KMT inhibitors, and others, as discussed in this and other articles. These inhibitors can all potentially promote gene activation by producing losses of DNA methylation, or deacetylating lysines, or alleviating silencing mediated by histone methylation PTMs, such as H3K27 methylation. (A, Adapted from Reddington et al. 2014; B, adapted from Sharma et al. 2010.)
Of great interest for the focus of this article, and what may be most functionally significant about hypomethylated blocks in cancer, is the occurrence of opposite, focal gains in promoter CpG island, or gains in DNA methylation for genes embedded within these regions (Berman et al. 2012). Although there is some disagreement about the exact positioning of this methylation, it seems to be within the CpG islands of gene promoters that reside within hypomethylated domains. These promoter islands are virtually always protected from methylation in normal cells, even when they reside in partially methylated domains best characterized in differentiated cells (Berman et al. 2012). Thus, these large domains may harbor a much higher than expected percentage of genes that are vulnerable to abnormal CpG island DNA hypermethylation (Ohm et al. 2007; Schlesinger et al. 2007; Widschwendter et al. 2007; Berman et al. 2012). Thus, hypomethylated blocks consist of juxtaposed regions of losses and more focal CpG island gains of DNA methylation in cancer (Fig. 7A).
Several laboratories have now identified that hypermethylated genes are heavily biased to Polycomb repressive complex 2 (PRC2)-regulated, H3K27me3-marked genes, in ES and adult stem cells (Fig. 7B) (Ohm et al. 2007; Schlesinger et al. 2007; Widschwendter et al. 2007). Interestingly, the above partially DNA methylated or hypomethylated domains in which many of these genes reside broadly correspond to late replicating and lamin-associated domains at the nuclear periphery, generally associated with repressive chromatin domains and PcG-marked, often bivalent, genes in ES cells (Peric-Hupkes and van Steensel 2010; Peric-Hupkes et al. 2010; Berman et al. 2012). This PcG-mediated transcriptional repression in a stem-cell setting is most often, in the context of bivalent chromatin (i.e., dually marked with H3K27me3 and H3K4me3), thought to mediate a low, poised transcription state for genes important for cell commitment and/or that must not be highly expressed to preserve states of stem cell self-renewal (Bernstein et al. 2006; Chi and Bernstein 2009). Importantly, these bivalently marked promoters are virtually never associated with the presence of DNA methylation in normal cells at any stage of development (Fig. 7B) (Bernstein et al. 2006; Chi and Bernstein 2009; Baylin and Jones 2011; Shen and Laird 2013). A working model envisions a molecular progression during tumorigenesis during which, in the abnormally expanding adult stem or progenitor cell compartments depicted in Figure 6, the bivalent and/or PcG-repressed chromatin at CpG island promoters is replaced with more stable silencing states associated with DNA methylation and H3K9 methylation (Ohm et al. 2007), or, for some genes, they can remain in an abnormal state of PRC reprogramming (Baylin and Jones 2011; Easwaran et al. 2012). Also, what has been observed for these long-range chromatin domains are instances in which genes can be abnormally activated (Bert et al. 2013). The mechanisms appear to involve either focal losses of DNA methylation in genes with low-density CpG islands or a switch to alternative transcription start sites, because the canonical sites harbor focal gains in CpG island methylation (Bert et al. 2013).
What is essential for extending the concept of chromatin transitions at bivalently marked genes during tumorigenesis is to unravel the molecular mechanisms underlying this progression. Scenarios can be hypothesized in which abnormal retention of PcG complexes may initially occur and, then, DNA methylation subsequently ensues (i.e., epigenetic switching, Fig. 7B). Once DNA methylation evolves, the PcG complex and accompanying H3K27me3 histone may be completely or quantitatively replaced (Gal-Yam et al. 2008; McGarvey et al. 2008; Bartke et al. 2010). Experimental data corroborating this show that methylated DNA, when in a nucleosomal context, is resistant to the presence of PcG complexes and, hence, the imposition of the silencing H3K27me marks (Schlesinger et al. 2007; Widschwendter et al. 2007; Gal-Yam et al. 2008; Bartke et al. 2010). A scenario in which active bivalent genes are PRC reprogrammed (Fig. 7B) could be explained by surrounding hypomethylation allowing PRC2 access, which could then extend repression to neighboring active genes (reviewed in Reddington et al. 2014). Continued research is needed to understand the interplay between different repressive mechanisms.
6.4. Chromatin Boundaries
We also need to understand that although, in normal cells, CpG islands at promoters of resident genes have a narrow band of protection from the surrounding DNA methylation (O’Hagan et al. 2011; Berman et al. 2012), why does the molecular maintenance of chromatin and DNA methylation boundaries “break down” during tumor progression (Fig. 4B)? One idea is that factors, such as insulator proteins (e.g., CTCF), which separate transcriptionally repressive and active chromatin states, may be altered (Taberlay et al. 2014). Also, the chromatin-modifying machinery may be altered and cause shifts in chromatin boundaries and configuration (O’Hagan et al. 2011). Cancer risk states, such as chronic inflammation and DNA damage, can participate in inducing such shifts (Hahn et al. 2008; O’Hagan et al. 2011). Within these hypomethylated domains, there could also be alterations in function and/or targeting of the recently identified TET proteins, which normally contribute to maintaining promoter CpG islands free of DNA methylation (Williams et al. 2011). All of these possibilities create rich substrates for the next era of defining normal and cancer epigenomes.
6.5. Involvement of the DNA Methylation Machinery in Tumorigenesis
The question of how DNMTs are targeted and then establish and maintain abnormal patterns of DNA methylation in cancer cells needs continued study, most especially the complexes through which these enzymes act cooperatively to target gene promoters and modify DNA methylation patterns. For DNMT1, the protein UHFR1 and proteins associated with it seem to facilitate the targeting of this protein to DNA replication and other sites (Bostick et al. 2007; Nishiyama et al. 2013). Although less is known about the targeting DNMT3A and -B, specific types of DNA configuration (e.g., DNA-RNA triplex structures) may exert targeting effects for DNMT3B (Schmitz et al. 2010). Very importantly, past and present studies indicate that transcriptional repression complexes, which include histone-modifying enzymes, such as methyltransferases (KMTs) and demethylases (KDMs) are key for the recruitment of, or being recruited by, DNMTs, as elaborated in Figure 7 and Section 2.2.2 of Almouzni and Cedar (2014) (Di Croce et al. 2002; Fuks et al. 2003; Brenner et al. 2005; O’Hagan et al. 2011). Indeed, some studies suggest that deregulation of the chromatin machinery precedes DNA methylation changes (Bachman et al. 2003; O’Hagan et al. 2011; Sproul et al. 2011, 2012). As noted above, the changes discussed in key cancer risk states, such as chronic inflammation and the buildup of ROS, appear capable of rapidly triggering the assembly of DNMTs with HDAC and MBD protein partners and recruiting them to promoter CpG islands (O’Hagan et al. 2011). In these events, there is a rapid tightening of DNMT1 and SIRT1 (an HDAC) to chromatin. The DNMT1 step seems to be upstream in this process, highlighting the multitasking potential for this protein, in addition to it catalyzing DNA methylation (O’Hagan et al. 2011).
Genetic disruption studies of DNMTs in cultured colon cancer cells indicate that the maintenance of most DNA methylation, including at hypermethylated promoters and its attendant gene silencing, requires both DNMT1 and DNMT3b (Rhee et al. 2000, 2002). Studies in other cancer cell types have produced more variable results (Leu et al. 2003; Jones and Liang 2009). Whatever the mechanism of molecular progression to hypermethylated bivalent genes in cancer, bear in mind that mammalian DNMTs appear to have complex functions, which include not only catalytic DNMT activity at the carboxy-terminal regions, but also direct transcriptional repression activities at their amino-terminal domains (Robertson et al. 2000; Rountree et al. 2000; Fuks et al. 2001; Clements et al. 2012). Thus, a role for DNMTs potentially has many facets in transcriptional silencing, from initiation to maintenance, and is not necessarily restricted to steps involving DNA methylation (see also Cheng 2014; Patel 2014).
7. SUMMARY OF MAJOR RESEARCH ISSUES FOR UNDERSTANDING EPIGENETIC GENE SILENCING IN CANCER
Despite progress in understanding the molecular events that can drive the epigenetic abnormalities typifying the cancer epigenome, Table 6 summarizes some of the most important questions that remain to be resolved through future research. First, molecular events determining the simultaneous appearance of overall DNA hypomethylation and more localized promoter DNA hypermethylation must continue to be elucidated. These juxtaposed states suggest a broad mistargeting of chromatin states in cancer cells. We particularly need to relate DNA methylation patterns in cancer with other chromatin marks, such as H3K9 methylation, histone acetylation, and H3K27 methylation. We also need more research relating how all these epigenetic features are organized in a three-dimensional fashion in normal versus cancer cells, and whether nuclear architecture is a regulating factor in the deregulation that occurs in cells during transformation. What we learn about how the changing cancer epigenome contributes to cancer etiology should prove equally illuminating in understanding how mammalian cells normally package their genomes for proper patterns of gene expression and the maintenance of chromosome integrity.
A second important question will be to identify the determinants and function of chromatin boundaries. This will obviously need to be performed in the context of how DNA methylation patterns around individual gene promoters relate to the general chromatin configuration of other surrounding regions, such as gene enhancers, bodies, and insulators; this will need to be worked out in both normal and abnormal states of transcription. A third consideration is addressing the evolution of chromatin states across the various regulatory regions of cancer relevant loci during the course of tumorigenesis and comparing them to normal developmental scenarios. Fourth, key components of this must be dissected, particularly during specific phases of tumor initiation and progression. This should include assessing the molecular interactions determining the constitution and targeting of DNMTs and other silencing complexes, such as Polycomb repressive complexes (described in Grossniklaus and Paro 2014) and how this relates to gene expression, PTM signatures, and ncRNAs. Also, determining what are truly the causal epigenetic mechanisms that cause TSG silencing must be resolved. Finally, once abnormal heritable gene silencing is established in cancer, what is the precise hierarchy of molecular steps that maintain it? This latter question is not only a key basic question, but also central to the translational implications discussed in Section 8 for using epigenetic abnormalities as cancer biomarkers and, in Section 9, for reversing abnormal gene silencing as a cancer prevention or therapy strategy.
8. DNA METHYLATION ABNORMALITIES AS BIOMARKERS FOR CANCER DETECTION AND MONITORING CANCER PROGNOSIS
The pervasive nature of epigenetic abnormalities being characterized at all stages of cancer development constitutes an ever-increasing pool of potential biomarkers, which can be developed for predicting cancer risk states, the early detection of cancer, and for use as prognostic indicators. Methods that can sensitively detect changes in DNA methylation and chromatin have already been developed and more are being pursued not only for use on tumor and other tissue biopsies, but also those that can be applied to body fluids for noninvasive detection methods.
Focal, promoter region DNA hypermethylation of CpG islands, which is so common in cancer, is currently the most well-studied and developed biomarker. A number of very sensitive polymerase chain reaction (PCR)-based assays have been developed to be used in combination with sodium bisulfite pretreated DNA to detect levels of DNA methylation (Herman et al. 1996; Laird 2003). PCR approaches, such as methylation-specific PCR, now being used quantitatively, and new nano-assay approaches, in which primers are designed to amplify only methylated regions, are very sensitive (Bailey et al. 2010). Other methods to detect methylated DNA include techniques based on real-time PCR, such as “MethyLight” (Campan et al. 2009), in which a fluorescent probe can only bind to methylated DNA. These techniques can detect one methylated allele in a background of about 1000–50,000 alleles, depending on the particular assay design and specific needs of application. Thus, these approaches are applicable to a mixture of cells or even various biological fluids, such as plasma, urine, or sputum (Laird 2003).
Cancer detection by identification of altered cytosine methylation is quite robust because of the inherent stability of DNA compared with RNA or proteins. Also, because altered methylation patterns are often cancer-specific, these approaches may be able to distinguish one type of cancer from another. There are now a host of studies providing “proof of principle” for the use of promoter DNA hypermethylated sequences as an extremely sensitive strategy for predicting cancer risk and/or detection. For example, synchronous detection of abnormal promoter DNA methylation in tumor and chest lymph nodes, which were deemed microscopically free, shows promise for predicting the rapid recurrence of early stage lung cancer (Brock et al. 2008). Similarly, sensitive detection of such abnormalities in DNA from stool may offer a test for predicting the presence of colon tumors (Hong and Ahuja 2013; Imperiale et al. 2014). The detection of both CpG island DNA methylation and specific mutations is even more promising for detecting colon polyps and/or cancer by assaying stool blood DNA (Hong and Ahuja 2013; Imperiale et al. 2014) and this approach is moving toward clinical practice. The clinical value of this approach is being tested in larger studies in which the current hypotheses can be fully validated over the next few years. Likewise, detection of DNA hypermethylated genes in prostate needle biopsies is now being used clinically to augment histological detection of prostate cancer (Van Neste et al. 2012).
Several approaches for using CpG island hypermethylation to predict cancer patients’ response to therapies are very promising. Examples include the detection of this change in the promoter of the O6MGMT gene to predict the response to alkylating agents as the main treatment approach in gliomas (Esteller et al. 2000; Hegi et al. 2005). Use of this methylation marker is now becoming standard practice in the management of patients with gliomas. Tumors in which O6MGMT is silenced in association with DNA methylation changes are more sensitive to alkylating therapy because the repair gene is not available to remove guanosine adducts from the genome (Esteller et al. 2000; Hegi et al. 2005). Another recent promising example includes the promoter methylation of SMAD1 to predict resistance to the chemotherapeutic agent, doxorubicin, in patients with diffuse large B-cell lymphoma (DBCL) (Clozel et al. 2013). The silencing of this gene, when reversed by low doses of DNMT inhibitors, appears to be key in mediating the reversal of this chemoresistance (Clozel et al. 2013). Early findings in a phase I clinical study of patients with DBCL suggests that low doses of azacitidine can prime for increased responses to chemotherapy (Clozel et al. 2013).
9. EPIGENETIC THERAPY
The heritable inactivation of cancer-related genes by altered DNA methylation and chromatin modification has led to the realization that silenced chromatin may represent a viable target for cancer therapy. Thus, a new treatment approach called “epigenetic therapy” has been developed in which drugs that can modify chromatin or DNA methylation patterns are used alone or in combination to affect therapeutic outcomes (Egger et al. 2004; Kelly et al. 2010; Dawson and Kouzarides 2012; Azad et al. 2013; Ahuja et al. 2014).
9.1. DNMT Inhibitors
Powerful mechanism-based inhibitors of DNA cytosine methylation represent the most advanced epigenetic therapeutics currently available for cancer treatment. The nucleoside analogs, 5-aza-CR (Vidaza) and 5-aza-CdR (Dacogen or Decitibine), have been in clinical trials for many years. More recently, a new prodrug-like agent for 5-aza-CdR, called SGI-110, has entered the scene showing promise (Fig. 8) (Chuang et al. 2005; Yoo et al. 2007). These drugs, or the prodrug derivative, are incorporated into the DNA of replicating cells after they have been metabolized to the appropriate deoxynucleoside triphosphate or cleaved by phosphodiesterase in the case of SGI-110 (Chuang et al. 2005; Yoo et al. 2007). Once incorporated into DNA, they interact with all three known DNMTs to form covalent intermediates, which ultimately inhibit DNA methylation in subsequent rounds of DNA synthesis. The mechanism of action of these compounds for blocking the catalytic site of DNMTs is quite well understood and they have been used for some time to reactivate silenced genes in tissue culture or xenograft models (Santi et al. 1984; Ghoshal et al. 2005; Kelly et al. 2010; Tsai and Baylin 2011; Azad et al. 2013). However, it is often overlooked that the above DNA-demethylating agents not only induce the above catalytic block, but also cause degradation of the DNMTs (Ahuja et al. 2014). This latter action is quickly triggered, even by low doses of the drugs, when used in vivo (Tsai and Baylin 2011).This protein loss is potentially very important for the DNMT inhibitors to achieve reexpression of key cancer genes because, experimentally, all three biologically active DNMTs can exert transcriptional repression independent of catalyzing DNA methylation (Fuks et al. 2000; Robertson et al. 2000; Rountree et al. 2000; Bachman et al. 2001; Clements et al. 2012). These latter events are related to the potential scaffolding properties of these proteins with respect to binding key mediators of gene silencing, such as HDAC1 and HDAC2 (Fuks et al. 2000; Robertson et al. 2000; Rountree et al. 2000; Bachman et al. 2001; Clements et al. 2012). Thus, loss of DNMTs as proteins cannot be overlooked as a key event linked to any therapeutic efficacy of the drugs discussed above.
Initially, when used at what we now know in retrospect to be very high doses, the DNMT inhibitors were too toxic to patients to gain any traction for the treatment of cancer. Later, however, as the doses were profoundly lowered, these agents have now found application in the treatment of certain hematological malignancies, particularly myelodysplastic syndrome, which is a preleukemic condition occurring mainly in elderly patients (Lubbert 2000; Wijermans et al. 2000; Silverman et al. 2002; Issa et al. 2004). Clinical responses for patients with this disorder, and with leukemias that may have progressed from the preleukemic stage, are becoming increasingly dramatic. Accordingly, drugs with the clinical names Vidaza and Dacogen, for 5-aza-CR and 5-aza-CdR, respectively, have now been approved by the U.S. FDA for the treatment of patients with these disorders (Fig. 8). Although Vidaza and decitabine have been shown to be clinically efficacious, it has been more difficult to establish with clarity whether the targets of drug action are methylated gene promoters. Preliminary experiments suggested that the p15 TSG became demethylated following decitabine treatment (Daskalakis et al. 2002); however, it remains to be shown whether the drugs act by inducing gene expression or some other mechanism, such as triggering an immune response to the tumor. Based on preclinical studies looking at the above responses, and applying the approaches to solid tumor models, it seems likely that at very low nanomolar doses, both Vidaza and Dacogen can “reprogram” cancer cells and cause antitumor responses, which are most likely caused by the specific targeting of DNMTs rather than producing other less off-target effects (Tsai et al. 2012).
Using these concepts, DNA-demethylating agents, poised for new therapeutic applications, may assume a major role in cancer therapy. Newer versions of DNA-demethylating drugs are being developed with these concepts in mind. For example, as noted earlier, SGI-110, which is a dinucleotide prodrug of 5-aza-CdR, is also an inhibitor of DNMTs after cleavage by phosphodiesterases. Also, it has a longer half-life in patients because it is not deaminated by plasma cytidine deaminase, which causes the rapid inactivation of the 5-azanucleosides (Chabot et al. 1983; Qin et al. 2011). To date, effective inhibitors that do not require incorporation into DNA have not been developed, but these might be more desirable in the clinic because they might have fewer side effects. Numerous approaches to synthesize and/or discover such drugs are now ongoing.
9.2. HDAC Inhibitors
Another key set of proteins being targeted for cancer therapy are the HDACs (Dawson and Kouzarides 2012; Bose et al. 2014; West and Johnstone 2014). This large family of enzymes removes acetylation marks from histone tails (as well as other nonhistone proteins), typically acting in the context of larger protein complexes, sometimes associated with DNA methylation, to establish repressive chromatin environments (the topic of Seto and Yoshida 2014). Inhibitors of HDACs (HDACis) have a general transcriptional activating effect and their therapeutic use in cancer treatment has been presumed to be largely through the activation of abnormally silenced TSGs, although this eminently remains to be proven. Two of these inhibitors, suberoylanilide hydroxamic acid (SAHA or Vorinostat) and depsipeptide (Romidepsin), which are more specific inhibitors of HDACs (Fig. 8B), have now been approved by the FDA for the treatment of cutaneous T-cell lymphoma. However, the molecular mechanisms responsible for the unusual sensitivity of this tumor type to these drugs are still unclear. A significantly larger number of drugs are known to cause substantial inhibition of HDACs (see our Fig. 8 and Sec. 7 of Seto and Yoshida 2014 for more detail). Some of these, such as 4-phenylbutyrate or valproic acid (VPA), have been in clinical use to treat other conditions for some time (Marks et al. 2001; Richon and O’Brien 2002), whereas newer ones are now in clinical trials. HDACis, used alone, however, have had little success, especially in solid tumors (Azad et al. 2013; Ahuja et al. 2014). Interestingly, preclinical studies have recently suggested that these drugs may be able to reprogram cancer cells in a way that reverses treatment resistance or sensitizes cancers to conventional chemotherapy and newer targeted therapies (Sharma et al. 2010). Pursuant to these concepts, clinical data in patients with advanced non–small cell lung carcinoma (NSCLC) and breast cancer are accruing to corroborate this hypothesis. For example, a newer HDACi called entinostat, combined with the epidermal growth factor receptor inhibitor erlotinib, showed a significant overall survival benefit in patients with recurrent advanced NSCLC (Witta et al. 2012). Also, vorinostat therapy increased response rates significantly in combination with carboplatin and paclitaxel as a front line treatment of patients with metastatic NSCLC, and may extend overall survival (Ramalingam et al. 2010). Moreover, entinostat significantly increased survival of patients with advanced breast cancer in combination with an aromatase inhibitor (Yardley et al. 2013).
9.3. Epigenetic Drug Development
The clinical successes with current epigenetic drugs have led to a big increase in interest from the pharmaceutical industry in developing compounds that will target epigenetic abnormalities in cancers (Kelly et al. 2010; Dawson et al. 2011; Arrowsmith et al. 2012). The challenges and strategies being adopted by the research and industry sector are discussed in Section 3 of Audia and Campbell (2014). Examples of epigenetic drugs in development include a potent small-molecule DOT1L inhibitor, which can selectively kill MLL cells (Daigle et al. 2011). Inhibitors of BRD4 represent another class of small-molecule epigenetic therapeutics that has been developed to interfere with their capacity for reading histone acetyllysine marks (Filippakopoulos et al. 2010; Nicodeme et al. 2010; also reviewed in Qi 2014; Schaefer 2014). The BRD4 protein constitutes part of the machinery that activates transcription and, in particular, may be key for multiple gene activation events controlled by the pervasive oncogene, c-MYC (illustrated in Fig. 1 of Qi 2014) (Filippakopoulos et al. 2010; Delmore et al. 2011; Zuber et al. 2011; Dawson and Kouzarides 2012). The BRD4 inhibitors appear very effective in preclinical studies for the treatment of MLL-fusion leukemias (Dawson et al. 2011) and might be a therapeutic strategy for countering c-MYC overactivity (Delmore et al. 2011).
9.4. Combination Epigenetic Therapy
One of the major concepts emerging from all of the above clinical trial and drug development activities is that of combination epigenetic therapy. This is currently being tested in the clinic for the older drugs, targeting DNA demethylation and inhibiting HDACs. This will surely emerge for the newer drugs either in combination with these above drugs or other novel combinatorial strategies. In terms of the older drugs, the approach has been to exploit preclinical data showing that blocking HDAC activity subsequent to inhibiting DNA methylation can additively lead to the reexpression of DNA hypermethylated genes (Cameron et al. 1999; Suzuki et al. 2002; Cai et al. 2014). This concept exploits the fact that interactions between HDAC-mediated histone deacetylation (especially via HDAC1 and -2), collaborate with DNA methylation for the silencing of these genes (Cameron et al. 1999; Suzuki et al. 2002; Cai et al. 2014), as discussed in Section 6. This treatment paradigm has been applied in the clinical treatment of hematologic malignancies. The first study used Vidaza and the older HDACi, sodium phenyl butyrate, on patients with myelodysplastic syndrome and AML (Gore et al. 2006). This was well tolerated, and clinical responses were frequent, with five of 14 patients achieving complete or partial response. Another pilot study resulted in three out of 10 patients with myelodysplastic syndrome or AML developing a partial response (Maslak et al. 2006). Investigators at M.D. Anderson Cancer Center administered decitabine and VPA, and 12 of 54 patients achieved complete remissions (Garcia-Manero 2008). Subsequently, a study of Vidaza and VPA also suggests increased efficacy in high-risk myelodysplastic syndrome (Voso et al. 2009).
Controversy over the efficacy of DNA-demethylating agents used in combination with HDAC is in myelodysplastic syndrome/AML has arisen in subsequent studies. Thus, the U.S. Leukemia Intergroup undertook a study with a randomized phase using entinostat (HDACi) in combination with Vidaza (DNMTi). The combination did not show increased efficacy and suggested even less efficacy (Prebet et al. 2014). The reasons for the mixed results are not clear, but the approach still bears promise for myelodysplastic syndrome/AML. However, it remains to be established whether combination therapies are more effective than single-agent demethylating therapies, how to best use such agents together, and what molecular mechanisms account for any efficacies seen.
Much less has been performed to test the efficacy of combination therapy in solid tumors. A recent study on a lung cancer model in mice has shown promise that DNA methylation inhibitors (e.g., azacytidine) and HDAC inhibitors (e.g., entinostat) might have strong synergistic, antitumor effects (Belinsky et al. 2011). Closely related to this, recently completed clinical trials in 65 patients with advanced lung cancer, the deadliest of all human cancers, show promise that these approaches can, in a small subset of patients, induce robust, durable responses (Juergens et al. 2011). Moreover, in these same trials, there is an early indication that the epigenetic therapy may lead to sensitization to subsequent therapies in many more (Juergens et al. 2011). These latter include not only standard chemotherapies, but also, intriguingly, an exciting new immunotherapy (Brahmer et al. 2012; Topalian et al. 2012), which targets breaking lymphocyte immune tolerance to render these cells immune competent (Wrangle et al. 2013). This last possibility is backed in the laboratory by work suggesting that, in lung cancer cells and other solid tumor types, DNA-demethylating agents up-regulate a very complex, immune attraction effect with pathways harboring hundreds of genes (Wrangle et al. 2013; Li et al. 2014). Both the possibilities for sensitizing patients with advanced lung cancer to chemotherapy and immunotherapy are now being tested in larger trials that are under way. It is worthwhile noting that others are reporting that DNA-demethylating agents can sensitize patients with advanced ovarian cancer to subsequent chemotherapy (Matei et al. 2012), as well as the aforementioned beneficial effects of HDACi in sensitizing patients to chemotherapy.
Combinations of newer agents targeting additional steps in chromatin assembly are just starting to be explored at the preclinical level for cancer therapy paradigms. Examples include the finding that synergistic antitumor activity is achieved using a BRD4 inhibitor and an HDACi, and when using a LSD1 inhibitor and HDACi in the treatment of human AML cells (Fiskus et al. 2014a, 2014b). The concept underlying the first therapeutic strategy relies on combinatorially activating the histone acetylation pathway via HDAC is to reexpress abnormally repressed TSGs, whereas the BET (double bromodomain proteins) inhibitor interferes with myc oncogene-activated genes. The second strategy also activates histone acetylation while boosting the H3K4me3 activating mark to combinatorially target and activate abnormally repressed genes.
In summary, the concept of epigenetic therapy for cancer has a rationale and expanding basis in theory, and clinical efficacies are emerging, which suggest great promise. However, much needs to be performed at a mechanistic and clinical level to realize this promise, especially for the common human cancers. One problem that is broadly discussed is the lack of specificity of some of the older agents being used, like the DNA-demethylating agents. However, most steps in epigenetic regulation control many genes and pathways in normal and cancer cells (Jones and Baylin 2007; Baylin and Jones 2011; Jones 2012). This is the very nature of epigenetic control of a cell program. In cancer, the epigenome is widely altered and drugs that can broadly “reprogram” such cells and blunt many tumor pathways may be the most valuable (Jones and Baylin 2007; Baylin and Jones 2011; Dawson et al. 2012; Azad et al. 2013; Ahuja et al. 2014). These arguments are not to say that targeting individual genes abnormally regulated in cancer would not be a highly desirable goal for personalizing cancer therapy. A second problem is always the possibility of the inadvertent reactivation of normal genes as the result of therapy. This, however, in terms of contribution to therapy related toxicities has not been documented. Nor has increased tumorigencitiy associated with this possibility been noted in scenarios such as MD/AML, in which the older epigenetic therapy drugs have been used the longest. Thus, the cancer epigenome as a target for cancer therapies remains a vital possibility and one for which exciting advances are anticipated in the coming years.
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, Danny Reinberg, and Monika Lachner
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
*Reference is also in this subject collection.
- Ahuja N, Easwaran H, Baylin SB. 2014. Harnessing the potential of epigenetic therapy to target solid tumors. J Clin Invest 124: 56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Allis CD, Jenuwein T, Reinberg D. 2014. Overview and concepts. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018739. [DOI] [Google Scholar]
- *.Almouzni G, Cedar H. 2014. Maintenance of epigenetic information. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann JM, Nip J, Strom DK, Lutterbach B, Harada H, Lenny N, Downing JR, Meyers S, Hiebert SW. 2001. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol Cell Biol 21: 6470–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki S, Doi H, Sano Y, Tanaka S, Miyake Y. 2009. Preparation and CO(2) adsorption properties of aminopropyl-functionalized mesoporous silica microspheres. J Colloid Interface Sci 339: 382–389. [DOI] [PubMed] [Google Scholar]
- Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. 2012. Epigenetic protein families: A new frontier for drug discovery. Nat Rev Drug Discov 11: 384–400. [DOI] [PubMed] [Google Scholar]
- Aryee MJ, Liu W, Engelmann JC, Nuhn P, Gurel M, Haffner MC, Esopi D, Irizarry RA, Getzenberg RH, Nelson WG, et al. 2013. DNA methylation alterations exhibit intraindividual stability and interindividual heterogeneity in prostate cancer metastases. Sci Transl Med 5: 169ra110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Audia JE, Campbell RM. 2014. Histone modifications and cancer. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad N, Zahnow CA, Rudin CM, Baylin SB. 2013. The future of epigenetic therapy in solid tumours—Lessons from the past. Nat Rev Clin Oncol 10: 256–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman KE, Rountree MR, Baylin SB. 2001. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J Biol Chem 276: 32282–32287. [DOI] [PubMed] [Google Scholar]
- Bachman KE, Park BH, Rhee I, Rajagopalan H, Herman JG, Baylin SB, Kinzler KW, Vogelstein B. 2003. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Cancer Cell 3: 89–95. [DOI] [PubMed] [Google Scholar]
- Bagchi A, Papazoglu C, Wu Y, Capurso D, Brodt M, Francis D, Bredel M, Vogel H, Mills AA. 2007. CHD5 is a tumor suppressor at human 1p36. Cell 128: 459–475. [DOI] [PubMed] [Google Scholar]
- Bailey VJ, Zhang Y, Keeley BP, Yin C, Pelosky KL, Brock M, Baylin SB, Herman JG, Wang TH. 2010. Single-tube analysis of DNA methylation with silica superparamagnetic beads. Clin Chem 56: 1022–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan A, Bleeker FE, Lamba S, Rodolfo M, Daniotti M, Scarpa A, van Tilborg AA, Leenstra S, Zanon C, Bardelli A. 2007. Novel somatic and germline mutations in cancer candidate genes in glioblastoma, melanoma, and pancreatic carcinoma. Cancer Res 67: 3545–3550. [DOI] [PubMed] [Google Scholar]
- *.Barlow DP, Bartolomei MS. 2014. Genomic imprinting in mammals. Cold Spring Harb Perspect Biol 6: a018382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. 2010. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 143: 470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. 2011. A decade of exploring the cancer epigenome—Biological and translational implications. Nat Rev Cancer 11: 726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Fearon ER, Vogelstein B, de Bustros A, Sharkis SJ, Burke PJ, Staal SP, Nelkin BD. 1987. Hypermethylation of the 5′ region of the calcitonin gene is a property of human lymphoid and acute myeloid malignancies. Blood 70: 412–417. [PubMed] [Google Scholar]
- Belinsky SA, Grimes MJ, Picchi MA, Mitchell HD, Stidley CA, Tesfaigzi Y, Channell MM, Liu Y, Casero RA Jr, Baylin SB, et al. 2011. Combination therapy with Vidaza and entinostat suppresses tumor growth and reprograms the epigenome in an orthotopic lung cancer model. Cancer Res 71: 454–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Berger SL, Sassone-Corsi P. 2014. Metabolic signaling to chromatin. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H, Zhang J, Crawford YG, Gauthier ML, Fordyce CA, McDermott KM, Sigaroudinia M, Kozakiewicz K, Tlsty TD. 2005. Genetic and epigenetic changes in mammary epithelial cells identify a subpopulation of cells involved in early carcinogenesis. Cold Spring Harb Symp Quant Biol 70: 317–327. [DOI] [PubMed] [Google Scholar]
- Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CP, van Dijk CM, Tollenaar RA, et al. 2012. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet 44: 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. 2010. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol 28: 1045–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, et al. 2011. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 20: 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bert SA, Robinson MD, Strbenac D, Statham AL, Song JZ, Hulf T, Sutherland RL, Coolen MW, Stirzaker C, Clark SJ. 2013. Regional activation of the cancer genome by long-range epigenetic remodeling. Cancer Cell 23: 9–22. [DOI] [PubMed] [Google Scholar]
- Borodovsky A, Salmasi V, Turcan S, Fabius AW, Baia GS, Eberhart CG, Weingart JD, Gallia GL, Baylin SB, Chan TA, et al. 2013. 5-azacytidine reduces methylation, promotes differentiation and induces tumor regression in a patient-derived IDH1 mutant glioma xenograft. Oncotarget 4: 1737–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose P, Dai Y, Grant S. 2014. Histone deacetylase inhibitor (HDACI) mechanisms of action: Emerging insights. Pharmacol Ther pii: S0163-7258: 00092–00098. 10.1016/j.pharmthera.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. 2007. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317: 1760–1764. [DOI] [PubMed] [Google Scholar]
- Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al. 2012. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 366: 2455–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al. 2013. The somatic genomic landscape of glioblastoma. Cell 155: 462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner C, Deplus R, Didelot C, Loriot A, Vire E, De Smet C, Gutierrez A, Danovi D, Bernard D, Boon T, et al. 2005. Myc represses transcription through recruitment of DNA methyltransferase corepressor. Embo J 24: 336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs KJ, Corcoran-Schwartz IM, Zhang W, Harcke T, Devereux WL, Baylin SB, Eberhart CG, Watkins DN. 2008. Cooperation between the Hic1 and Ptch1 tumor suppressors in medulloblastoma. Genes Dev 22: 770–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock MV, Hooker CM, Ota-Machida E, Han Y, Guo M, Ames S, Glockner S, Piantadosi S, Gabrielson E, Pridham G, et al. 2008. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med 358: 1118–1128. [DOI] [PubMed] [Google Scholar]
- Burbee D, Forgacs E, Zochbauer-Muller S. 2001. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst 93: 691–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Geutjes EJ, de Lint K, Roepman P, Bruurs L, Yu LR, Wang W, van Blijswijk J, Mohammad H, de Rink I, et al. 2014. The NuRD complex cooperates with DNMTs to maintain silencing of key colorectal tumor suppressor genes. Oncogene 33: 2157–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. 1999. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet 21: 103–107. [DOI] [PubMed] [Google Scholar]
- Campan M, Weisenberger DJ, Trinh B, Laird PW. 2009. MethyLight. Methods Mol Biol 507: 325–337. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. 2012a. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. 2012b. Comprehensive genome characterization of squamous cell lung cancers. Nature 489: 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. 2013a. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499: 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. 2013b. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368: 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot GG, Bouchard J, Momparler RL. 1983. Kinetics of deamination of 5-aza-2′-deoxycytidine and cytosine arabinoside by human liver cytidine deaminase and its inhibition by 3-deazauridine, thymidine or uracil arabinoside. Biochem Pharmacol 32: 1327–1328. [DOI] [PubMed] [Google Scholar]
- Chaffer CL, Marjanovic ND, Lee T, Bell G, Kleer CG, Reinhardt F, D’Alessio AC, Young RA, Weinberg RA. 2013. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell 154: 61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K-M, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, Gupta N, Mueller S, James CD, Jenkins R, et al. 2013. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev 27: 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase A, Cross NC. 2011. Aberrations of EZH2 in cancer. Clin Cancer Res 17: 2613–2618. [DOI] [PubMed] [Google Scholar]
- Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R. 1998. DNA hypomethylation leads to elevated mutation rates. Nature 395: 89–93. [DOI] [PubMed] [Google Scholar]
- Chen WY, Zeng X, Carter MG, Morrell CN, Chiu Yen RW, Esteller M, Watkins DN, Herman JG, Mankowski JL, Baylin SB. 2003. Heterozygous disruption of Hic1 predisposes mice to a gender-dependent spectrum of malignant tumors. Nat Genet 33: 197–202. [DOI] [PubMed] [Google Scholar]
- Chen W, Cooper TK, Zahnow CA, Overholtzer M, Zhao Z, Ladanyi M, Karp JE, Gokgoz N, Wunder JS, Andrulis IL, et al. 2004. Epigenetic and genetic loss of Hic1 function accentuates the role of p53 in tumorigenesis. Cancer Cell 6: 387–398. [DOI] [PubMed] [Google Scholar]
- Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. 2005. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell 123: 437–448. [DOI] [PubMed] [Google Scholar]
- *.Cheng X. 2014. Structural and functional coordination of DNA and histone methylation. Cold Spring Harb Perspect Biol 6: a018747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi AS, Bernstein BE. 2009. Developmental biology. Pluripotent chromatin state. Science 323: 220–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JC, Yoo CB, Kwan JM, Li TW, Liang G, Yang AS, Jones PA. 2005. Comparison of biological effects of non-nucleoside DNA methylation inhibitors versus 5-aza-2′-deoxycytidine. Mol Cancer Ther 4: 1515–1520. [DOI] [PubMed] [Google Scholar]
- Clements EG, Mohammad HP, Leadem BR, Easwaran H, Cai Y, Van Neste L, Baylin SB. 2012. DNMT1 modulates gene expression without its catalytic activity partially through its interactions with histone-modifying enzymes. Nucleic Acids Res 40: 4334–4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clozel T, Yang S, Elstrom RL, Tam W, Martin P, Kormaksson M, Banerjee S, Vasanthakumar A, Culjkovic B, Scott DW, et al. 2013. Mechanism-based epigenetic chemosensitization therapy of diffuse large B-cell lymphoma. Cancer Discov 3: 1002–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, Johnston LD, Scott MP, Smith JJ, Xiao Y, et al. 2011. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 20: 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann R, Li C, Yoon J, Chin P, Bates S, Pfeifer G. 2000. Epigenetic inactivation of a RAS association domain family protein from the lung tumor suppressor locus 3p21.3. Nat Genet 25: 315–319. [DOI] [PubMed] [Google Scholar]
- Daskalakis M, Nguyen TT, Nguyen C, Guldberg P, Kohler G, Wijermans P, Jones PA, Lubbert M. 2002. Demethylation of a hypermethylated P15/INK4B gene in patients with myelodysplastic syndrome by 5-Aza-2′-deoxycytidine (decitabine) treatment. Blood 100: 2957–2964. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T. 2012. Cancer epigenetics: From mechanism to therapy. Cell 150: 12–27. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. 2011. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478: 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T, Huntly BJ. 2012. Targeting epigenetic readers in cancer. N Engl J Med 367: 647–657. [DOI] [PubMed] [Google Scholar]
- De Carvalho DD, You JS, Jones PA. 2010. DNA methylation and cellular reprogramming. Trends Cell Biology 20: 609–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zwaan SE, Haass NK. 2010. Genetics of basal cell carcinoma. Australasian J Dermatol 51: 81–92; quiz 93–84. [DOI] [PubMed] [Google Scholar]
- Dedeurwaerder S, Defrance M, Calonne E, Denis H, Sotiriou C, Fuks F. 2011. Evaluation of the Infinium Methylation 450K technology. Epigenomics 3: 771–784. [DOI] [PubMed] [Google Scholar]
- *.Dekker J, Misteli T. 2014. Long-range chromatin interactions. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. 2011. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146: 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Croce L, Raker VA, Corsaro M, Fazi F, Fanelli M, Faretta M, Fuks F, Lo Coco F, Kouzarides T, Nervi C, et al. 2002. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science 295: 1079–1082. [DOI] [PubMed] [Google Scholar]
- Drost J, Mantovani F, Tocco F, Elkon R, Comel A, Holstege H, Kerkhoven R, Jonkers J, Voorhoeve PM, Agami R, et al. 2010. BRD7 is a candidate tumour suppressor gene required for p53 function. Nat Cell Biol 12: 380–389. [DOI] [PubMed] [Google Scholar]
- Easwaran H, Johnstone SE, Van Neste L, Ohm J, Mosbruger T, Wang Q, Aryee MJ, Joyce P, Ahuja N, Weisenberger D, et al. 2012. A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome Res 22: 837–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easwaran H, Tsai HC, Baylin SB. 2014. Cancer epigenetics: Tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol Cell 54: 716–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger G, Liang G, Aparicio A, Jones PA. 2004. Epigenetics in human disease and prospects for epigenetic therapy. Nature 429: 457–463. [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Lacey M. 2013. DNA hypomethylation and hemimethylation in cancer. Adv Exp Med Biol 754: 31–56. [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, Gehrke C. 1982. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucl Acids Res 10: 2709–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE 2012. Decoding ENCODE. Nat Chem Biol 8: 871. [DOI] [PubMed] [Google Scholar]
- Esteller M. 2007. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet 8: 286–298. [DOI] [PubMed] [Google Scholar]
- Esteller M. 2008. Epigenetics in cancer. N Engl J Med 358: 1148–1159. [DOI] [PubMed] [Google Scholar]
- Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG. 2000. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 343: 1350–1354. [DOI] [PubMed] [Google Scholar]
- Esteller M, Fraga MF, Guo M, Garcia-Foncillas J, Hedenfalk I, Godwin AK, Trojan J, Vaurs-Barriere C, Bignon YJ, Ramus S, et al. 2001a. DNA methylation patterns in hereditary human cancers mimic sporadic tumorigenesis. Hum Mol Genet 10: 3001–3007. [DOI] [PubMed] [Google Scholar]
- Esteller M, Risques RA, Toyota M, Capella G, Moreno V, Peinado MA, Baylin SB, Herman JG. 2001b. Promoter hypermethylation of the DNA repair gene O(6)-methylguanine-DNA methyltransferase is associated with the presence of G:C to A:T transition mutations in p53 in human colorectal tumorigenesis. Cancer Res 61: 4689–4692. [PubMed] [Google Scholar]
- Fahrner JA, Eguchi S, Herman JG, Baylin SB. 2002. Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer Res 62: 7213–7218. [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. 1983. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301: 89–92. [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. 2010. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. 2010. Selective inhibition of BET bromodomains. Nature 468: 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch PW, He X, Kelley MJ, Uren A, Schaudies RP, Popescu NC, Rudikoff S, Aaronson SA, Varmus HE, Rubin JS. 1997. Purification and molecular cloning of a secreted, Frizzled-related antagonist of Wnt action. Proc Natl Acad Sci 94: 6770–6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R. 1993. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 75: 1027–1038. [DOI] [PubMed] [Google Scholar]
- Fiskus W, Sharma S, Qi J, Valenta JA, Schaub LJ, Shah B, Peth K, Portier BP, Rodriguez M, Devaraj SG, et al. 2014a. Highly active combination of BRD4 antagonist and histone deacetylase inhibitor against human acute myelogenous leukemia cells. Mol Cancer Ther 13: 1142–1154. [DOI] [PubMed] [Google Scholar]
- Fiskus W, Sharma S, Shah B, Portier BP, Devaraj SG, Liu K, Iyer SP, Bearss D, Bhalla KN. 2014b. Highly effective combination of LSD1 (KDM1A) antagonist and pan-histone deacetylase inhibitor against human AML cells. Leukemia. 10.1038/leu.2014.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatau E, Gonzales FA, Michalowsky LA, Jones PA. 1984. DNA methylation in 5-aza-2′-deoxycytidine-resistant variants of C3H 10T1/2 C18 cells. Mol Cell Biol 4: 2098–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost JK, Ball WC Jr, Levin ML, Tockman MS, Baker RR, Carter D, Eggleston JC, Erozan YS, Gupta PK, Khouri NF, et al. 1984. Early lung cancer detection: Results of the initial (prevalence) radiologic and cytologic screening in the Johns Hopkins study. Am Rev Respir Dis 130: 549–554. [DOI] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. 2000. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet 24: 88–91. [DOI] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T. 2001. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. Embo J 20: 2536–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Deplus R, Kouzarides T. 2003. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res 31: 2305–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Yam EN, Egger G, Iniguez L, Holster H, Einarsson S, Zhang X, Lin JC, Liang G, Jones PA, Tanay A. 2008. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci 105: 12979–12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Manero G. 2008. Demethylating agents in myeloid malignancies. Curr Opin Oncol 20: 705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Lander ES. 2013. Lessons from the cancer genome. Cell 153: 17–37. [DOI] [PubMed] [Google Scholar]
- Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. 2003. Induction of tumors in mice by genomic hypomethylation. Science 300: 489–492. [DOI] [PubMed] [Google Scholar]
- Gazzoli I, Loda M, Garber J, Syngal S, Kolodner RD. 2002. A hereditary nonpolyposis colorectal carcinoma case associated with hypermethylation of the MLH1 gene in normal tissue and loss of heterozygosity of the unmethylated allele in the resulting microsatellite instability-high tumor. Cancer Res 62: 3925–3928. [PubMed] [Google Scholar]
- Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, Jacob ST. 2005. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol 25: 4727–4741. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gore SD, Baylin S, Sugar E, Carraway H, Miller CB, Carducci M, Grever M, Galm O, Dauses T, Karp JE, et al. 2006. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res 66: 6361–6369. [DOI] [PubMed] [Google Scholar]
- Grady WM, Willis J, Guilford PJ, Dunbier AK, Toro TT, Lynch H, Wiesner G, Ferguson K, Eng C, Park JG, et al. 2000. Methylation of the CDH1 promoter as the second genetic hit in hereditary diffuse gastric cancer. Nat Genet 26: 16–17. [DOI] [PubMed] [Google Scholar]
- Greenblatt MS, Bennett WP, Hollstein M, Harris CC. 1994. Mutations in the p53 tumor suppressor gene: Clues to cancer etiology and molecular pathogenesis. Cancer Res 54: 4855–4878. [PubMed] [Google Scholar]
- *.Grossniklaus U, Paro R. 2014. Transcriptional silencing by Polycomb-group proteins. Cold Spring Harb Perspect Biol 6: a019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan B, Mao TL, Panuganti PK, Kuhn E, Kurman RJ, Maeda D, Chen E, Jeng YM, Wang TL, Shih Ie M. 2011. Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am J Surg Pathol 35: 625–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, Wu R, Chen C, Li X, Zhou L, et al. 2011. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet 43: 875–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Hahn T, Lee DH, Esworthy RS, Kim BW, Riggs AD, Chu FF, Pfeifer GP. 2008. Methylation of polycomb target genes in intestinal cancer is mediated by inflammation. Cancer Res 68: 10280–10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerman PS, Hayes DN, Wilkerson MD, Schultz N, Bose R, Chu A, Collisson EA, Cope L, Creighton CJ, Getz G, et al. 2012. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489: 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: The next generation. Cell 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, et al. 2011. Increased methylation variation in epigenetic domains across cancer types. Nat Genet 43: 768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, et al. 2010. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol 28: 1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. 2005. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 352: 997–1003. [DOI] [PubMed] [Google Scholar]
- Herman JG, Baylin SB. 2003. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 349: 2042–2054. [DOI] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. 1996. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci 93: 9821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, et al. 1998. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci 95: 6870–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoue T, Weisenberger DJ, Lange CP, Shen H, Byun HM, Van Den Berg D, Malik S, Pan F, Noushmehr H, van Dijk CM, et al. 2012. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res 22: 271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchins MP, Rapkins RW, Kwok CT, Srivastava S, Wong JJ, Khachigian LM, Polly P, Goldblatt J, Ward RL. 2011. Dominantly inherited constitutional epigenetic silencing of MLH1 in a cancer-affected family is linked to a single nucleotide variant within the 5′UTR. Cancer Cell 20: 200–213. [DOI] [PubMed] [Google Scholar]
- Holm TM, Jackson-Grusby L, Brambrink T, Yamada Y, Rideout WM 3rd, Jaenisch R. 2005. Global loss of imprinting leads to widespread tumorigenesis in adult mice. Cancer Cell 8: 275–285. [DOI] [PubMed] [Google Scholar]
- Holst CR, Nuovo GJ, Esteller M, Chew K, Baylin SB, Herman JG, Tlsty TD. 2003. Methylation of p16(INK4a) promoters occurs in vivo in histologically normal human mammary epithelia. Cancer Res 63: 1596–1601. [PubMed] [Google Scholar]
- Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, Valsesia A, Ye Z, Kuan S, Edsall LE, et al. 2012. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res 22: 246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L, Ahuja N. 2013. DNA methylation biomarkers of stool and blood for early detection of colon cancer. Genet Test Mol Biomarkers 17: 401–406. [DOI] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, et al. 2003. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425: 191–196. [DOI] [PubMed] [Google Scholar]
- Hur K, Cejas P, Feliu J, Moreno-Rubio J, Burgos E, Boland CR, Goel A. 2014. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut 63: 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, Ahlquist DA, Berger BM. 2014. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med 370: 1287–1297. [DOI] [PubMed] [Google Scholar]
- Issa JP. 2014. Aging and epigenetic drift: A vicious cycle. J Clin Invest 124: 24–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. 1994. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet 7: 536–540. [DOI] [PubMed] [Google Scholar]
- Issa JP, Garcia-Manero G, Giles FJ, Mannari R, Thomas D, Faderl S, Bayar E, Lyons J, Rosenfeld CS, Cortes J, et al. 2004. Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2′-deoxycytidine (decitabine) in hematopoietic malignancies. Blood 103: 1635–1640. [DOI] [PubMed] [Google Scholar]
- Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, Cheng T, Depinho RA, Sharpless NE, Scadden DT. 2006. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16(INK4a). Nature 443: 421–426. [DOI] [PubMed] [Google Scholar]
- Jiang L, Li J, Song L. 2009. Bmi-1, stem cells and cancer. Acta Biochim Biophys Sinica 41: 527–534. [DOI] [PubMed] [Google Scholar]
- Jones PA. 2012. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat Rev Genet 13: 484–492. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. 2002. The fundamental role of epigenetic events in cancer. Nat Rev Genet 3: 415–428. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. 2007. The epigenomics of cancer. Cell 128: 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Laird PW. 1999. Cancer epigenetics comes of age. Nat Genet 21: 163–167. [DOI] [PubMed] [Google Scholar]
- Jones PA, Liang G. 2009. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet 10: 805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Taylor SM. 1980. Cellular differentiation, cytidine analogs and DNA methylation. Cell 20: 85–93. [DOI] [PubMed] [Google Scholar]
- Jones S, Wang TL, Shih Ie M, Mao TL, Nakayama K, Roden R, Glas R, Slamon D, Diaz LA Jr, Vogelstein B, et al. 2010. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 330: 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, Coleman B, Sebree R, Rodgers K, Hooker CM, Franco N, et al. 2011. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov 1: 598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminskas E, Farrell A, Abraham S, Baird A, Hsieh LS, Lee SL, Leighton JK, Patel H, Rahman A, Sridhara R, et al. 2005. Approval summary: Azacitidine for treatment of myelodysplastic syndrome subtypes. Clin Cancer Res 11: 3604–3608. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Ushijima S, Nakanishi Y, Sakamoto M, Hirohashi S. 2003. Mutation of the DNA methyltransferase (DNMT) 1 gene in human colorectal cancers. Cancer Lett 192: 75–82. [DOI] [PubMed] [Google Scholar]
- Kaneda A, Feinberg AP. 2005. Loss of imprinting of IGF2: A common epigenetic modifier of intestinal tumor risk. Cancer Res 65: 11236–11240. [DOI] [PubMed] [Google Scholar]
- Katzenellenbogen RA, Baylin SB, Herman JG. 1999. Hypermethylation of the DAP-kinase CpG island is a common alteration in B-cell malignancies. Blood 93: 4347–4353. [PubMed] [Google Scholar]
- Kelly TK, De Carvalho DD, Jones PA. 2010. Epigenetic modifications as therapeutic targets. Nat Biotechnol 28: 1069–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TK, Liu Y, Lay FD, Liang G, Berman BP, Jones PA. 2012. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res 22: 2497–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Chung NG, Kang MR, Yoo NJ, Lee SH. 2011. Genetic and expressional alterations of CHD genes in gastric and colorectal cancers. Histopathology 58: 660–668. [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. 1997. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature 386: 761–763. [DOI] [PubMed] [Google Scholar]
- Kiyono T, Foster SA, Koop JI, McDougall JK, Galloway DA, Klingelhutz AJ. 1998. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 396: 84–88. [DOI] [PubMed] [Google Scholar]
- Knudson AG. 2001. Two genetic hits (more or less) to cancer. Nat Rev Cancer 1: 157–162. [DOI] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Issa JP. 2003. Critical role of histone methylation in tumor suppressor gene silencing in colorectal cancer. Mol Cell Biol 23: 206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. 2009. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324: 929–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Kriaucionis S, Tahiliani M. 2014. Expanding the epigenetic landscape: Novel modificatons of cytosine in genomic DNA. Cold Spring Harb Perspect Biol 6: a018630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulis M, Heath S, Bibikova M, Queiros AC, Navarro A, Clot G, Martinez-Trillos A, Castellano G, Brun-Heath I, Pinyol M, et al. 2012. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet 44: 1236–1242. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, et al. 2005. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci 102: 1859–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird PW. 2003. The power and the promise of DNA methylation markers. Nat Rev Cancer 3: 253–266. [DOI] [PubMed] [Google Scholar]
- Laird PW, Jackson-Grusby L, Fazeli A, Dickinson SL, Jung WE, Li E, Weinberg RA, Jaenisch R. 1995. Suppression of intestinal neoplasia by DNA hypomethylation. Cell 81: 197–205. [DOI] [PubMed] [Google Scholar]
- Lapeyre JN, Becker FF. 1979. 5-Methylcytosine content of nuclear DNA during chemical hepatocarcinogenesis and in carcinomas which result. Biochem Biophys Res Commun 87: 698–705. [DOI] [PubMed] [Google Scholar]
- Lee WH, Morton RA, Epstein JI, Brooks JD, Campbell PA, Bova GS, Hsieh WS, Isaacs WB, Nelson WG. 1994. Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc Natl Acad Sci 91: 11733–11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu YW, Rahmatpanah F, Shi H, Wei SH, Liu JC, Yan PS, Huang TH. 2003. Double RNA interference of DNMT3b and DNMT1 enhances DNA demethylation and gene reactivation. Cancer Res 63: 6110–6115. [PubMed] [Google Scholar]
- Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. 2013. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340: 857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et al. 2010. DNMT3A mutations in acute myeloid leukemia. N Engl J Med 363: 2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Chiappinelli KB, Guzzetta AA, Easwaran H, Yen RW, Vatapalli R, Topper MJ, Luo J, Connolly RM, Azad NS, et al. 2014. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget 5: 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Li E, Zhang Y. 2014. DNA methylation in mammals. Cold Spring Harb Perspect Biol 6: a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhao H, Zhang X, Wood LD, Anders RA, Choti MA, Pawlik TM, Daniel HD, Kannangai R, Offerhaus GJ, et al. 2011. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet 43: 828–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S, et al. 2011. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471: 68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Nicolaides NC, Markowitz S, Willson JK, Parsons RE, Jen J, Papadopolous N, Peltomaki P, de la Chapelle A, Hamilton SR, et al. 1995. Mismatch repair gene defects in sporadic colorectal cancers with microsatellite instability. Nat Genet 9: 48–55. [DOI] [PubMed] [Google Scholar]
- Losman JA, Kaelin WG Jr, 2013. What a difference a hydroxyl makes: Mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev 27: 836–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. 2012. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483: 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Venneti S, Akalin A, Fang F, Ward PS, Dematteo RG, Intlekofer AM, Chen C, Ye J, Hameed M, et al. 2013. Induction of sarcomas by mutant IDH2. Genes Dev 27: 1986–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubbert M. 2000. DNA methylation inhibitors in the treatment of leukemias, myelodysplastic syndromes and hemoglobinopathies: Clinical results and possible mechanisms of action. Curr Top Microbiol Immunol 249: 135–164. [DOI] [PubMed] [Google Scholar]
- Lukacs RU, Memarzadeh S, Wu H, Witte ON. 2010. Bmi-1 is a crucial regulator of prostate stem cell self-renewal and malignant transformation. Cell Stem Cell 7: 682–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maegawa S, Gough SM, Watanabe-Okochi N, Lu Y, Zhang N, Castoro RJ, Estecio MR, Jelinek J, Liang S, Kitamura T, et al. 2014. Age-related epigenetic drift in the pathogenesis of MDS and AML. Genome Res 24: 580–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. 2001. Histone deacetylases and cancer: Causes and therapies. Nat Rev Cancer 1: 194–202. [DOI] [PubMed] [Google Scholar]
- *.Marmorstein R, Zhou M-M. 2014. Writers and readers of histone acetylation: Structure, mechanism, and inhibition. Cold Spring Harb Perspect Biol 6: a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslak P, Chanel S, Camacho LH, Soignet S, Pandolfi PP, Guernah I, Warrell R, Nimer S. 2006. Pilot study of combination transcriptional modulation therapy with sodium phenylbutyrate and 5-azacytidine in patients with acute myeloid leukemia or myelodysplastic syndrome. Leukemia 20: 212–217. [DOI] [PubMed] [Google Scholar]
- Mason EF, Hornick JL. 2013. Succinate dehydrogenase deficiency is associated with decreased 5-hydroxymethylcytosine production in gastrointestinal stromal tumors: Implications for mechanisms of tumorigenesis. Mod Pathol 26: 1492–1497. [DOI] [PubMed] [Google Scholar]
- Matei D, Fang F, Shen C, Schilder J, Arnold A, Zeng Y, Berry WA, Huang T, Nephew KP. 2012. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res 72: 2197–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattera L, Escaffit F, Pillaire MJ, Selves J, Tyteca S, Hoffmann JS, Gourraud PA, Chevillard-Briet M, Cazaux C, Trouche D. 2009. The p400/Tip60 ratio is critical for colorectal cancer cell proliferation through DNA damage response pathways. Oncogene 28: 1506–1517. [DOI] [PubMed] [Google Scholar]
- McGarvey KM, Van Neste L, Cope L, Ohm JE, Herman JG, Van Criekinge W, Schuebel KE, Baylin SB. 2008. Defining a chromatin pattern that characterizes DNA-hypermethylated genes in colon cancer cells. Cancer Res 68: 5753–5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miremadi A, Oestergaard MZ, Pharoah PD, Caldas C. 2007. Cancer genetics of epigenetic genes. Hum Mol Genet 16: R28–R49. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Osada H, Konishi H, Tatematsu Y, Yatabe Y, Mitsudomi T, Fujii Y, Takahashi T. 2002. Aberrant hypermethylation of the CHFR prophase checkpoint gene in human lung cancers. Oncogene 21: 2328–2333. [DOI] [PubMed] [Google Scholar]
- Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, et al. 2011. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476: 298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan A, Ji W, Zhang XY, Marrogi A, Graff JR, Baylin SB, Ehrlich M. 1998. Hypomethylation of pericentromeric DNA in breast adenocarcinomas. Int J Cancer 77: 833–838. [DOI] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. 2004. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science 306: 2105–2108. [DOI] [PubMed] [Google Scholar]
- Nguyen CT, Gonzales FA, Jones PA. 2001. Altered chromatin structure associated with methylation-induced gene silencing in cancer cells: Correlation of accessibility, methylation, MeCP2 binding and acetylation. Nucleic Acids Res 29: 4598–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel A, Stadler SC. 2014. Role of epigenetic mechanisms in epithelial-to-mesenchymal transition of breast cancer cells. Transl Res pii: S1931-524400125–X 10.1016/j.trsl.2014.04.001. [DOI] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, et al. 2010. Suppression of inflammation by a synthetic histone mimic. Nature 468: 1119–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Yamaguchi L, Sharif J, Johmura Y, Kawamura T, Nakanishi K, Shimamura S, Arita K, Kodama T, Ishikawa F, et al. 2013. Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature 502: 249–253. [DOI] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. 2010. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17: 510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hagan HM, Mohammad HP, Baylin SB. 2008. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet 4: e1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, Clements EG, Cai Y, Van Neste L, Easwaran H, et al. 2011. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG islands. Cancer Cell 20: 606–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. 2007. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet 39: 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. 2008. An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Patel DJ. 2014. A structural perspective on readout of epigenetic histone and DNA methylation marks. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, van Steensel B. 2010. Role of the nuclear lamina in genome organization and gene expression. Cold Spring Harb Symp Quant Biol 75: 517–524. [DOI] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. 2010. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 38: 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer GP, Tang M, Denissenko MF. 2000. Mutation hotspots and DNA methylation. Curr Top Microbiol Immunol 249: 1–19. [DOI] [PubMed] [Google Scholar]
- *.Pirrotta V. 2014. The necessity of chromatin: A view in perspective. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic R, Licht JD. 2012. Emerging epigenetic targets and therapies in cancer medicine. Cancer Discov 2: 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prebet T, Sun Z, Figueroa ME, Ketterling R, Melnick A, Greenberg PL, Herman J, Juckett M, Smith MR, Malick L, et al. 2014. Prolonged administration of azacitidine with or without entinostat for myelodysplastic syndrome and acute myeloid leukemia with myelodysplasia-related changes: Results of the US Leukemia intergroup trial E1905. J Clin Oncol 32: 1242–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner JR, Chinnaiyan AM. 2011. The emergence of lncRNAs in cancer biology. Cancer Discov 1: 391–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Qi J. 2014. Bromodomain and extraterminal domain inhibitors (BETi) for cancer therapy: Chemical modulation of chromatin structure. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin T, Castoro R, El Ahdab S, Jelinek J, Wang X, Si J, Shu J, He R, Zhang N, Chung W, et al. 2011. Mechanisms of resistance to decitabine in the myelodysplastic syndrome. PLoS One 6: e23372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainier S, Dobry CJ, Feinberg AP. 1995. Loss of imprinting in hepatoblastoma. Cancer research 55: 1836–1838. [PubMed] [Google Scholar]
- Rainier S, Johnson LA, Dobry CJ, Ping AJ, Grundy PE, Feinberg AP. 1993. Relaxation of imprinted genes in human cancer. Nature 362: 747–749. [DOI] [PubMed] [Google Scholar]
- Ramalingam SS, Maitland ML, Frankel P, Argiris AE, Koczywas M, Gitlitz B, Thomas S, Espinoza-Delgado I, Vokes EE, Gandara DR, et al. 2010. Carboplatin and Paclitaxel in combination with either vorinostat or placebo for first-line therapy of advanced non-small-cell lung cancer. J Clin Oncol 28: 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddington JP, Sproul D, Meehan RR. 2014. DNA methylation reprogramming in cancer: Does it act by re-configuring the binding landscape of Polycomb repressive complexes? Bioessays 36: 134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KL, Feinberg AP. 2013. Higher order chromatin organization in cancer. Semin Cancer Biol 23: 109–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee I, Jair KW, Yen RW, Lengauer C, Herman JG, Kinzler KW, Vogelstein B, Baylin SB, Schuebel KE. 2000. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature 404: 1003–1007. [DOI] [PubMed] [Google Scholar]
- Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, Kinzler KW, et al. 2002. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416: 552–556. [DOI] [PubMed] [Google Scholar]
- Richon VM, O’Brien JP. 2002. Histone deacetylase inhibitors: A new class of potential therapeutic agents for cancer treatment. Clin Cancer Res 8: 662–664. [PubMed] [Google Scholar]
- Rideout WM 3rd, Coetzee GA, Olumi AF, Jones PA. 1990. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science 249: 1288–1290. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. 2000. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet 25: 338–342. [DOI] [PubMed] [Google Scholar]
- Romanov SR, Kozakiewicz BK, Holst CR, Stampfer MR, Haupt LM, Tlsty TD. 2001. Normal human mammary epithelial cells spontaneously escape senescence and acquire genomic changes. Nature 409: 633–637. [DOI] [PubMed] [Google Scholar]
- Ropero S, Fraga MF, Ballestar E, Hamelin R, Yamamoto H, Boix-Chornet M, Caballero R, Alaminos M, Setien F, Paz MF, et al. 2006. A truncating mutation of HDAC2 in human cancers confers resistance to histone deacetylase inhibition. Nat Genet 38: 566–569. [DOI] [PubMed] [Google Scholar]
- Rotili D, Mai A. 2011. Targeting histone demethylases: A new avenue for the fight against cancer. Genes Cancer 2: 663–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rountree MR, Bachman KE, Baylin SB. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet 25: 269–277. [DOI] [PubMed] [Google Scholar]
- Saito Y, Jones PA. 2006. Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle 5: 2220–2222. [DOI] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. 2006. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 9: 435–443. [DOI] [PubMed] [Google Scholar]
- Sakai T, Toguchida J, Ohtani N, Yandell DW, Rapaport JM, Dryja TP. 1991. Allele-specific hypermethylation of the retinoblastoma tumor-suppressor gene. Am J Hum Genet 48: 880–888. [PMC free article] [PubMed] [Google Scholar]
- Sakatani T, Kaneda A, Iacobuzio-Donahue CA, Carter MG, de Boom Witzel S, Okano H, Ko MS, Ohlsson R, Longo DL, Feinberg AP. 2005. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science 307: 1976–1978. [DOI] [PubMed] [Google Scholar]
- Sanbhnani S, Yeong FM. 2012. CHFR: A key checkpoint component implicated in a wide range of cancers. Cell Mol Life Sci 69: 1669–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom OJ, Maddison K, Clarke AR. 2007. Mechanisms of disease: Methyl-binding domain proteins as potential therapeutic targets in cancer. Nat Clin Practice Oncol 4: 305–315. [DOI] [PubMed] [Google Scholar]
- Santi DV, Norment A, Garrett CE. 1984. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci 81: 6993–6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Knobbe CB, Itsumi M, Elia AJ, Harris IS, Chio II, Cairns RA, McCracken S, Wakeham A, Haight J, et al. 2012a. D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev 26: 2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brustle A, Harris IS, Holmes R, Wakeham A, Haight J, et al. 2012b. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature 488: 656–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Schaefer U. 2014. Pharmacological inhibition of bromodomain-containing proteins in inflammation. Cold Spring Harb Perspect Biol 6: a018671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. 2007. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 39: 232–236. [DOI] [PubMed] [Google Scholar]
- Schmitz KM, Mayer C, Postepska A, Grummt I. 2010. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 24: 2264–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuebel KE, Chen W, Cope L, Glockner SC, Suzuki H, Yi JM, Chan TA, Van Neste L, Van Criekinge W, van den Bosch S, et al. 2007. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet 3: 1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Seto E, Yoshida M. 2014. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb Perspect Biol 6: a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. 2010. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Laird PW. 2013. Interplay between the cancer genome and epigenome. Cell 153: 38–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen XM, Ohno K, Fukudome T, Tsujino A, Brengman JM, De Vivo DC, Packer RJ, Engel AG. 2002. Congenital myasthenic syndrome caused by low-expressor fast-channel AChR delta subunit mutation. Neurology 59: 1881–1888. [DOI] [PubMed] [Google Scholar]
- Silverman LR, Demakos EP, Peterson BL, Kornblith AB, Holland JC, Odchimar-Reissig R, Stone RM, Nelson D, Powell BL, DeCastro CM, et al. 2002. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: A study of the cancer and leukemia group B. J Clin Oncol 20: 2429–2440. [DOI] [PubMed] [Google Scholar]
- Spannhoff A, Hauser AT, Heinke R, Sippl W, Jung M. 2009. The emerging therapeutic potential of histone methyltransferase and demethylase inhibitors. ChemMedChem 4: 1568–1582. [DOI] [PubMed] [Google Scholar]
- Sproul D, Nestor C, Culley J, Dickson JH, Dixon JM, Harrison DJ, Meehan RR, Sims AH, Ramsahoye BH. 2011. Transcriptionally repressed genes become aberrantly methylated and distinguish tumors of different lineages in breast cancer. Proc Natl Acad Sci 108: 4364–4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproul D, Kitchen RR, Nestor CE, Dixon JM, Sims AH, Harrison DJ, Ramsahoye BH, Meehan RR. 2012. Tissue of origin determines cancer-associated CpG island promoter hypermethylation patterns. Genome Biol 13: R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan PR, Borek E. 1964. Enzymatic alteration of nucleic acid structure. Science 145: 548–553. [DOI] [PubMed] [Google Scholar]
- Stumpel DJ, Schneider P, van Roon EH, Boer JM, de Lorenzo P, Valsecchi MG, de Menezes RX, Pieters R, Stam RW. 2009. Specific promoter methylation identifies different subgroups of MLL-rearranged infant acute lymphoblastic leukemia, influences clinical outcome, and provides therapeutic options. Blood 114: 5490–5498. [DOI] [PubMed] [Google Scholar]
- Sun A, Tawfik O, Gayed B, Thrasher JB, Hoestje S, Li C, Li B. 2007. Aberrant expression of SWI/SNF catalytic subunits BRG1/BRM is associated with tumor development and increased invasiveness in prostate cancers. Prostate 67: 203–213. [DOI] [PubMed] [Google Scholar]
- Sun J, Song Y, Wang Z, Wang G, Gao P, Chen X, Gao Z, Xu H. 2014. Clinical significance of promoter region hypermethylation of microRNA-148a in gastrointestinal cancers. Onco Targets Ther 7: 853–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Gabrielson E, Chen W, Anbazhagan R, van Engeland M, Weijenberg MP, Herman JG, Baylin SB. 2002. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet 31: 141–149. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van Engeland M, et al. 2004. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet 36: 417–422. [DOI] [PubMed] [Google Scholar]
- Swafford DS, Middleton SK, Palmisano WA, Nikula KJ, Tesfaigzi J, Baylin SB, Herman JG, Belinsky SA. 1997. Frequent aberrant methylation of p16INK4a in primary rat lung tumors. Mol Cell Biol 17: 1366–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlay PC, Statham AL, Kelly TK, Clark SJ, Jones PA. 2014. Reconfiguration of nucleosome depleted regions at distal regulatory elements accompanies DNA methylation of enhancers and insulators in cancer. Genome Res pii: gr.163485.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam WL, Weinberg RA. 2013. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med 19: 1438–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timp W, Feinberg AP. 2013. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat Rev Cancer 13: 497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. 2012. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 366: 2443–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. 1999. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci 96: 8681–8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HC, Baylin SB. 2011. Cancer epigenetics: Linking basic biology to clinical medicine. Cell Res 21: 502–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HC, Li H, Van Neste L, Cai Y, Robert C, Rassool FV, Shin JJ, Harbom KM, Beaty R, Pappou E, et al. 2012. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell 21: 430–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang DP, Cheng AS. 2011. Epigenetic regulation of signaling pathways in cancer: Role of the histone methyltransferase EZH2. J Gastroenterol Hepatol 26: 19–27. [DOI] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. 2012. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483: 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S, Fabius AW, Borodovsky A, Pedraza A, Brennan C, Huse J, Viale A, Riggins GJ, Chan TA. 2013. Efficient induction of differentiation and growth inhibition in IDH1 mutant glioma cells by the DNMT Inhibitor Decitabine. Oncotarget 10: 1729–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Neste L, Bigley J, Toll A, Otto G, Clark J, Delree P, Van Criekinge W, Epstein JI. 2012. A tissue biopsy-based epigenetic multiplex PCR assay for prostate cancer detection. BMC Urol 12: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, et al. 2011. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varier RA, Timmers HT. 2011. Histone lysine methylation and demethylation pathways in cancer. Biochim Biophys Acta 1815: 75–89. [DOI] [PubMed] [Google Scholar]
- Veigl ML, Kasturi L, Olechnowicz J, Ma A, Lutterbaugh JD, Periyasamy S, Li GM, Drummond J, Modrich PL, Sedwick WD, et al. 1998. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc Natl Acad Sci 95: 8698–8702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venneti S, Felicella MM, Coyne T, Phillips JJ, Gorovets D, Huse JT, Kofler J, Lu C, Tihan T, Sullivan LM, et al. 2013. Histone 3 lysine 9 trimethylation is differentially associated with isocitrate dehydrogenase mutations in oligodendrogliomas and high-grade astrocytomas. J Neuropathol Exp Neurol 72: 298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. 2013. Cancer genome landscapes. Science 339: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voso MT, Santini V, Finelli C, Musto P, Pogliani E, Angelucci E, Fioritoni G, Alimena G, Maurillo L, Cortelezzi A, et al. 2009. Valproic acid at therapeutic plasma levels may increase 5-azacytidine efficacy in higher risk myelodysplastic syndromes. Clin Cancer Res 15: 5002–5007. [DOI] [PubMed] [Google Scholar]
- Wales MM, Biel MA, el Deiry W, Nelkin BD, Issa JP, Cavenee WK, Kuerbitz SJ, Baylin SB. 1995. p53 activates expression of HIC-1, a new candidate tumour suppressor gene on 17p13.3. Nat Med 1: 570–577. [DOI] [PubMed] [Google Scholar]
- Wang J, Chen H, Fu S, Xu ZM, Sun KL, Fu WN. 2011. The involvement of CHD5 hypermethylation in laryngeal squamous cell carcinoma. Oral Oncol 47: 601–608. [DOI] [PubMed] [Google Scholar]
- Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, et al. 2006. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 38: 787–793. [DOI] [PubMed] [Google Scholar]
- Wen K, Azevedo MS, Gonzalez A, Zhang W, Saif LJ, Li G, Yousef A, Yuan L. 2009. Toll-like receptor and innate cytokine responses induced by lactobacilli colonization and human rotavirus infection in gnotobiotic pigs. Vet Immunol Immunopathol 127: 304–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels K, Bohnhorst B, Luhmer I, Morlot S, Bohring A, Jonasson J, Epplen JT, Gadzicki D, Glaser S, Gohring G, et al. 2010. Novel CHD7 mutations contributing to the mutation spectrum in patients with CHARGE syndrome. Eur J Med Genet 53: 280–285. [DOI] [PubMed] [Google Scholar]
- West AC, Johnstone RW. 2014. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest 124: 30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. 2007. Epigenetic stem cell signature in cancer. Nat Genet 39: 157–158. [DOI] [PubMed] [Google Scholar]
- Wijermans P, Lubbert M, Verhoef G, Bosly A, Ravoet C, Andre M, Ferrant A. 2000. Low-dose 5-aza-2′-deoxycytidine, a DNA hypomethylating agent, for the treatment of high-risk myelodysplastic syndrome: A multicenter phase II study in elderly patients. J Clin Oncol 18: 956–962. [DOI] [PubMed] [Google Scholar]
- Wijmenga C, Hansen RS, Gimelli G, Bjorck EJ, Davies EG, Valentine D, Belohradsky BH, van Dongen JJ, Smeets DF, van den Heuvel LP, et al. 2000. Genetic variation in ICF syndrome: Evidence for genetic heterogeneity. Human Mutat 16: 509–517. [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. 2011. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473: 343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Roberts CW. 2011. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 11: 481–492. [DOI] [PubMed] [Google Scholar]
- Witta SE, Jotte RM, Konduri K, Neubauer MA, Spira AI, Ruxer RL, Varella-Garcia M, Bunn PA Jr, Hirsch FR. 2012. Randomized phase II trial of erlotinib with and without entinostat in patients with advanced non-small-cell lung cancer who progressed on prior chemotherapy. J Clin Oncol 30: 2248–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf P, Hu YC, Doffek K, Sidransky D, Ahrendt SA. 2001. O(6)-Methylguanine-DNA methyltransferase promoter hypermethylation shifts the p53 mutational spectrum in non-small cell lung cancer. Cancer Res 61: 8113–8117. [PubMed] [Google Scholar]
- Wolffe AP. 2001. Chromatin remodeling: Why it is important in cancer. Oncogene 20: 2988–2990. [DOI] [PubMed] [Google Scholar]
- Wrangle J, Wang W, Koch A, Easwaran H, Mohammad HP, Vendetti F, Vancriekinge W, Demeyer T, Du Z, Parsana P, et al. 2013. Alterations of immune response of non-small cell lung cancer with azacytidine. Oncotarget 4: 2067–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. 2010. Active DNA demethylation: Many roads lead to Rome. Nav Rev Mol Cell Biol 11: 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. 2011a. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev 25: 2436–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. 2011b. Tet1 and 5-hydroxymethylation: A genome-wide view in mouse embryonic stem cells. Cell Cycle 10: 2428–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Strawn E, Basir Z, Halverson G, Guo SW. 2007. Aberrant expression of deoxyribonucleic acid and methyltransferases DNMT1, DNMT3A, and DNMY3B in women with endometriosis. Fertil Steril 87: 24–32. [DOI] [PubMed] [Google Scholar]
- Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, Liu L, Liu Y, Yang C, Xu Y, et al. 2012. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev 26: 1326–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita K, Upadhyay S, Osada M, Hoque MO, Xiao Y, Mori M, Sato F, Meltzer SJ, Sidransky D. 2002. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell 2: 485–495. [DOI] [PubMed] [Google Scholar]
- Yamashita Y, Yuan J, Suetake I, Suzuki H, Ishikawa Y, Choi YL, Ueno T, Soda M, Hamada T, Haruta H, et al. 2010. Array-based genomic resequencing of human leukemia. Oncogene 29: 3723–3731. [DOI] [PubMed] [Google Scholar]
- Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, Shi JY, Zhu YM, Tang L, Zhang XW, et al. 2011. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet 43: 309–315. [DOI] [PubMed] [Google Scholar]
- Yang X, Noushmehr H, Han H, Andreu-Vieyra C, Liang G, Jones PA. 2012. Gene reactivation by 5-aza-2′-deoxycytidine-induced demethylation requires SRCAP-mediated H2A.Z insertion to establish nucleosome depleted regions. PLoS Genet 8: e1002604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yardley DA, Ismail-Khan RR, Melichar B, Lichinitser M, Munster PN, Klein PM, Cruickshank S, Miller KD, Lee MJ, Trepel JB. 2013. Randomized phase II, double-blind, placebo-controlled study of exemestane with or without entinostat in postmenopausal women with locally recurrent or metastatic estrogen receptor-positive breast cancer progressing on treatment with a nonsteroidal aromatase inhibitor. J Clin Oncol 31: 2128–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CB, Jeong S, Egger G, Liang G, Phiasivongsa P, Tang C, Redkar S, Jones PA. 2007. Delivery of 5-aza-2′-deoxycytidine to cells using oligodeoxynucleotides. Cancer Res 67: 6400–6408. [DOI] [PubMed] [Google Scholar]
- You JS, Jones PA. 2012. Cancer genetics and epigenetics: Two sides of the same coin? Cancer Cell 22: 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Minter-Dykhouse K, Malureanu L, Zhao WM, Zhang D, Merkle CJ, Ward IM, Saya H, Fang G, van Deursen J, et al. 2005. Chfr is required for tumor suppression and Aurora A regulation. Nat Genet 37: 401–406. [DOI] [PubMed] [Google Scholar]
- Zhang W, Glockner SC, Guo M, Machida EO, Wang DH, Easwaran H, Van Neste L, Herman JG, Schuebel KE, Watkins DN, et al. 2008. Epigenetic inactivation of the canonical Wnt antagonist SRY-box containing gene 17 in colorectal cancer. Cancer Res 68: 2764–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Dai H, Zhou M, Li X, Liu C, Guo Z, Wu X, Wu J, Wang C, Zhong J, et al. 2012. Polyploid cells rewire DNA damage response networks to overcome replication stress-induced barriers for tumour progression. Nat Commun 3: 815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Zoghbi HY, Beaudet AL. 2014. Epigenetics and human disease. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. 2008. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456: 125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn AM, Barish GD, Williams BO, Lavender P, Klymkowsky MW, Varmus HE. 1999. Regulation of Wnt signaling by Sox proteins: XSox17 α/β and XSox3 physically interact with β-catenin. Mol Cell 4: 487–498. [DOI] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. 2011. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478: 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]