Abstract
Chromosomal rearrangements can shape the structure of genetic variation in the genome directly through alteration of genes at breakpoints or indirectly by holding combinations of genetic variants together due to reduced recombination. The third chromosome of Drosophila pseudoobscura is a model system to test hypotheses about how rearrangements are established in populations because its third chromosome is polymorphic for >30 gene arrangements that were generated by a series of overlapping inversion mutations. Circumstantial evidence has suggested that these gene arrangements are selected. Despite the expected homogenizing effects of extensive gene flow, the frequencies of arrangements form gradients or clines in nature, which have been stable since the system was first described >80 years ago. Furthermore, multiple arrangements exist at appreciable frequencies across several ecological niches providing the opportunity for heterokaryotypes to form. In this study, we tested whether genes are differentially expressed among chromosome arrangements in first instar larvae, adult females and males. In addition, we asked whether transcriptional patterns in heterokaryotypes are dominant, semidominant, overdominant, or underdominant. We find evidence for a significant abundance of differentially expressed genes across the inverted regions of the third chromosome, including an enrichment of genes involved in sensory perception for males. We find the majority of loci show additivity in heterokaryotypes. Our results suggest that multiple genes have expression differences among arrangements that were either captured by the original inversion mutation or accumulated after it reached polymorphic frequencies, providing a potential source of genetic variation for selection to act upon. These data suggest that the inversions are favored because of their indirect effect of recombination suppression that has held different combinations of differentially expressed genes together in the various gene arrangement backgrounds.
Keywords: gene expression, inversions, expression inheritance, adaptation
MODIFIERS reducing rates of recombination are likely to play an important role in evolution when a species is adapting to a heterogenous environment (Otto and Barton 2001). When a species encounters a heterogeneous environment, positive modifiers of recombination may initially be favored to generate many different gene combinations as a way to best adapt to the novel habitat. The problem then becomes how to preserve these adaptive combinations once good solutions are found. Negative modifiers of recombination will be favored if they capture adaptive combinations of genes in linkage disequilibrium that provide higher fitness in the new niche (Feldman 1972; Charlesworth and Charlesworth 1973; Charlesworth 1974). Inversion mutations are a type of negative recombination modifier that could allow an organism to hold associations between beneficial combinations of genes together.
Inversions are a class of structural mutation where the order of genes within a chromosomal segment is reversed. Not only do chromosomal inversions modify gene order, but they suppress recombination in heterokaryotypes (Dobzhansky and Epling 1944; Levine and Levine 1954; Levine 1956), and have been shown to change levels of expression of genes that flank the breakpoints (Puig et al. 2004). Since their initial discovery nearly a century ago in Drosophila melanogaster, inversions have been implicated in such evolutionary processes as adaptation, disease susceptibility, sex chromosome evolution, and speciation (Sturtevant 1921; Hartl 1975; Lahn and Page 1999; Noor et al. 2001; Stefansson et al. 2005; Kirkpatrick and Barton 2006; Evans et al. 2007). Numerous fixed inversion differences have been detected among homologous chromosomes from different species, including members of the Drosophila genus and between humans and chimpanzees (Carson and Yoon 1982; Wasserman 1982; Feuk et al. 2005; Bhutkar et al. 2008; Kirkpatrick 2010). Furthermore, chromosomal inversions are also polymorphic within species, including humans (Sperlich and Pfriem 1986; Andolfatto et al. 2001; Bansal et al. 2007). Despite their ubiquity in nature, the evolutionary factors influencing the origin, spread, and maintenance of inversions in populations are not well understood.
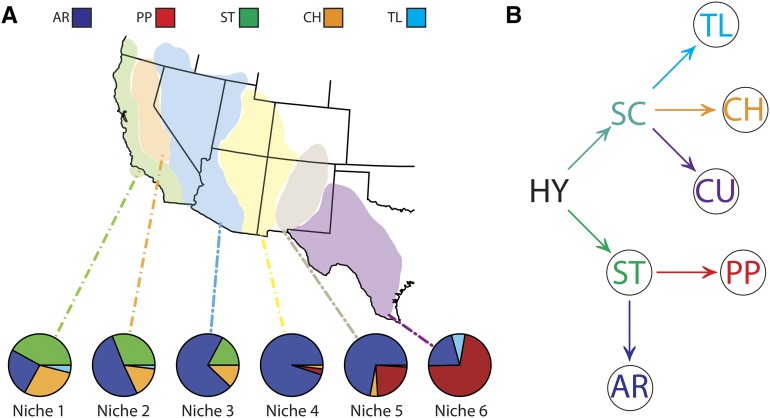
The third chromosome (Muller element C) (Schaeffer et al. 2008) of D. pseudoobscura offers an ideal system to test hypotheses regarding the evolutionary forces responsible for the establishment and maintenance of inversions in natural populations. This chromosome is polymorphic for >30 naturally occurring gene arrangements that were generated by a series of overlapping inversion mutations (Dobzhansky and Queal 1938; Dobzhansky and Sturtevant 1938; Dobzhansky 1944). Circumstantial evidence has suggested that inversions are adaptive. The relative frequencies of arrangements in D. pseudoobscura populations form a stable east-to-west cline across the American Southwest (Dobzhansky 1944; Anderson et al. 1991; Schaeffer and Miller 1992) where the frequency shifts coincide with major climatic and geographic differences that fall into one of six ecological niches (Lobeck 1950; Schaeffer 2008). Niches are found to be either polymorphic or monomorphic for gene arrangements (Figure 1). For example in niche 1, three gene arrangements dominate the westernmost region of California. In niche 4, however, one chromosome type is near fixation in the Four Corners region of the Colorado Plateau. In the eastern niche 6, two arrangements are frequent in western Texas. The cline has been stable since the 1940s despite sufficient gene flow to homogenize frequencies among populations (Anderson et al. 1991; Schaeffer and Miller 1992). Previous studies have provided evidence of extensive migration and low genetic differentiation across the genome, with the exception of the inverted regions (Riley et al. 1989; Kovacevic and Schaeffer 2000; Schaeffer et al. 2003). Furthermore, population structure is not observed on Muller C within arrangements (Schaeffer et al. 2003). Additional evidence for selection includes seasonal cycling of arrangements, altitudinal gradients of inversion frequencies, and laboratory demonstration of nontrivial equilibria in population cage experiments (Dobzhansky 1948a,b). It is therefore hypothesized that selection has acted on these inversion mutations, but it is not clear if selection acted to establish the different arrangements in populations and what numbers of genes might be involved (Schaeffer et al. 2003).
Figure 1.
The D. pseudoobscura third chromosome inversion polymorphism system. (A) The frequency distribution of D. pseudoobscura arrangements within six ecological niches of the American Southwest. (B) The phylogeny of chromosome arrangements inferred from cytogenetic and molecular data. Circled arrangements indicate those used in this study. AR, Arrowhead; CH, Chiricahua; CU, Cuernavaca; HY, hypothetical; PP, Pikes Peak; SC, Santa Cruz; ST, standard; TL, Tree Line.
Several mechanisms have been proposed for how inversion mutations are established in populations (Kirkpatrick and Barton,2006). Inversions could generate selectable variation at the inversion breakpoints either because genes are disrupted or expression of genes at the boundary of the breaks could be altered, i.e., direct or position effects (Sperlich and Pfriem 1986; Puig et al. 2004). Meiotic drive of duplication and deficiency recombinant chromosomes has also been suggested as a direct effect of inversions (Novitski 1951). Inversions could be beneficial in their roles as negative modifiers of recombination because they capture: (1) sets of genes free of recessive lethal mutations (Nei et al. 1967; Ohta 1968), (2) sets of epistatically interacting genes (Dobzhansky 1950), or (3) sets of genes involved in local adaptation (Kirkpatrick and Barton 2006). An inversion could be established if it hitchhikes with a selectively sweeping gene (Smith and Haigh 1974). Finally, inversions may not be selected at all and increase in frequency via random genetic drift (Lande 1984). These potential evolutionary mechanisms are not mutually exclusive.
If selection plays a role in adaptation of the inversions, then amino acid variation in proteins, differential gene expression, or variation in nonprotein coding genes are the likely types of genetic diversity it has acted upon. This study will test whether strains that carry different inversions show evidence of differential gene expression. Differences in levels of gene transcription can contribute to phenotypic variability within a species (Fay et al. 2004; Gilad et al. 2006; Choi and Kim 2007; Hoffman and Goodisman 2007). For example, several recent studies in the Drosophila genus have associated adaptive phenotypes and traits with changes in gene expression (Chen et al. 2015; Zhao et al. 2015). It is possible selection has acted on adaptive transcriptional variation at multiple loci, which was captured by the inversions and differences in gene expression that contribute to phenotypic and fitness variability between arrangements.
Populations of D. pseudoobscura show evidence of panmixia and the formation of homokaryotypes and heterokaryotypes occur at frequencies expected under Hardy–Weinberg (Dobzhansky 1944; Schaeffer 2008). Because multiple arrangements are found in most niches, heterokaryotypes can form at appreciable frequencies. Several models have been proposed to explain this stable equilibrium (Anderson et al. 1991; Schaeffer et al. 2003) and formation of heterokaryotypes, including selection for coadapted gene complexes, local adaptation with associative overdominance against recessive deleterious mutations, and epistatic selection across heterogeneous environments (Dobzhansky and Levene 1951; Kirkpatrick and Barton 2006; Schaeffer 2008). If overdominant selection targets transcriptional variation, differences in gene expression between the heterokaryotype and either parental homokaryotic arrangement background are expected. Selection for heterokaryotypes could occur through epistatic selection if the expression levels of genes have higher fitness when they are heterozygous and reduced fitness when recombination shuffles transcriptional variation among arrangements and breaks apart long-range associations (Otto and Barton 2001; Schaeffer 2008).
To investigate potential mechanisms that maintain frequencies of third chromosome gene arrangements in D. pseudoobscura, we used next-generation sequencing technologies to quantify gene expression in third chromosome homo- and heterokaryotypes. RNA sequencing (RNA-seq) was performed on homokaryotypes of six of the most common third chromosome arrangements: Arrowhead (AR), Chiricahua (CH), Pikes Peak (PP), Cuernavaca (CU), Standard (ST) and Tree Line (TL). RNA-seq was similarly performed for commonly occurring heterokaryotypes of arrangements in the six southwestern niches that D. pseudoobscura occupies: AR/ST, AR/CH, ST/CH, AR/PP, and AR/AR (control). Differential gene expression between karyotypes was tested across three different life stages: adult male, adult female, and first instar larvae. First instar larvae were included under the hypothesis that they may face greater challenges to viability than other life stages (Schaeffer 2008). While adults can move freely across large distances, first instar larvae are essentially confined to the immediate location where they hatch and may have to survive under random environmental conditions. Selection might therefore be expected to act very strongly on such traits as the ability to find food, avoid desiccation, and grow quickly. Furthermore, we characterize the mode of inheritance of gene expression (dominant, overdominant, underdominant, or additive) for the heterokaryotic crosses to investigate the mechanisms maintaining their formation. Our results demonstrate that chromosomal rearrangements have captured sets of genes that differ in their expression levels and suggest that gene expression is a potential target for selection on gene arrangements.
Materials and Methods
Fly strains
Isofemale lines were set up from collections of D. pseudoobscura from the following locations (collector): Mount St. Helena, CA (W. W. Anderson, University of Georgia, Athens, GA); James Reserve, CA (W. W. Anderson); Santa Cruz Island, CA (L. Matzkin, University of Alabama, Huntsville, AL); Kaibab National Forest, AZ (S. W. Schaeffer); Davis Mountains, TX (S. W. Schaeffer); and San Pablo Etla, Oaxaca, Mexico (T. A. Markow, University of California, San Diego). Crosses with the Lobe or Blade balancer strains were used to create homozygous strains for the third chromosome (Dobzhansky and Queal 1938). The Lobe balancer carries the dominant marker Lobe eyes, which is on a Santa Cruz background and was used to make non-Santa Cruz chromosomes homozygous. The Blade balancer carries the Blade wings, which is on an Arrowhead arrangement and was used to make non-Arrowhead chromosomes homozygous. After balancer crosses were completed, the gene arrangement of each isochromosomal strain was diagnosed with preparations of the larval salivary gland polytene chromosomes (Painter 1934). Santa Cruz strains from Lobe crosses and Arrowhead strains from Blade crosses were discarded. The third chromosome (Muller C) gene arrangements carried by these isochromosomal strains were either Arrowhead (AR), Chiricahua (CH), Pikes Peak (PP), Cuernavaca (CU), Standard (ST), or Tree Line (TL) (Table 1; Powell 1992). Chromosome arms XL, XR, 2, 4, and 5 (Muller A, A/D, B, E, and F) are a mixed background from the balancer strain and the wild-type chromosomes from the original isofemale lines. All D. pseudoobscura strains used in this study are available at the Drosophila Species Stock Center (San Diego).
Table 1. Drosophila pseudoobscura strains isochromosomal for the third chromosome.
| Arrangement | Strain ID | DSSC | Sampling location | Balancer chromosome |
|---|---|---|---|---|
| Arrowhead | AR DM1005 | 14011-0121.224 | Davis Mountains, TX | Lobe |
| Arrowhead | AR KB945 | 14011-0121.237 | Kaibab Natl. Forest, AZ | Lobe |
| Arrowhead | AR MSH126 | 14011-0121.230 | Mount St. Helena, CA | Lobe |
| Chiricahua | CH JR272 | 14011-0121.258 | James Reserve, CA | Blade |
| Chiricahua | CH KB888 | 14011-0121.253 | Kaibab Natl. Forest, AZ | Lobe |
| Chiricahua | CH MSH202 | 14011-0121.254 | Mount St. Helena, CA | Blade |
| Cuernavaca | CU SPE123 1.2 | 14011-0121.271 | San Pablo Etla, Mexico | Blade |
| Cuernavaca | CU SPE123 4.1 | 14011-0121.272 | San Pablo Etla, Mexico | Blade |
| Cuernavaca | CU SPE123 5.2 | 14011-0121.273 | San Pablo Etla, Mexico | Blade |
| PikesPeak | PP DM1049 | 14011-0121.241 | Davis Mountains, TX | Blade |
| PikesPeak | PP DM1054 | 14011-0121.242 | Davis Mountains, TX | Lobe |
| PikesPeak | PP DM1065 | 14011-0121.243 | Davis Mountains, TX | Lobe |
| Standard | ST JR138 | 14011-0121.251 | James Reserve, CA | Lobe |
| Standard | ST MSH177 | 14011-0121.246 | Mount St. Helena, CA | Blade |
| Standard | ST MSH49 | Mount St. Helena, CA | Lobe | |
| TreeLine | TL SCI 12.2 | 14011-0121.265 | Santa Cruz Island, CA | Blade |
| TreeLine | TL SPE123 7.1 | 14011-0121.269 | San Pablo Etla, Mexico | Blade |
| TreeLine | TL SPE123 8.1 | 14011-0121.270 | San Pablo Etla, Mexico | Blade |
Each strain is descended from a single individual collected from the wild. Wild-caught flies were crossed and backcrossed with laboratory strains carrying nonrecombining balancer third chromosomes to create lineages isochromosomal for the third chromosome. All strains are available at the Drosophila Species Stock Center (DSSC) (San Diego).
RNA collection from homokaryotic flies
We isolated total RNA from three biological replicates of AR, CH, CU, PP, ST, and TL isochromosomal fly strains (Table 1). Three life stages were collected for each strain: first instar larvae of indeterminate sex, 4-day-old virgin adult females, and 4-day-old virgin adult males. Flies and larvae were snap frozen in liquid nitrogen immediately after collection and stored at −80° prior to RNA extraction. Frozen tissues of the same life stage and strain were pooled for RNA extraction, with an average of 668, 20, and 22 individuals pooled per extraction for the larvae, females, and males, respectively. RNA was purified with RNeasy spin columns (QIAGEN, Valencia, CA) following manufacturer’s instructions and immediately snap frozen in liquid nitrogen and stored at −80° before RNA-seq.
First instar larvae were harvested from grape agar (Genesee Scientific, San Diego, CA) plates where adult females had laid eggs. Adult males and females were acclimated to grape–agar media within egg laying chambers (Genesee Scientific) for 5 days. Adult flies were then transferred to fresh grape–agar in 4-hr time periods. After adult flies were removed, agar plates were examined under a compound microscope to confirm that eggs had been laid. Larvae were collected after 50 hr by washing the surface of the grape–agar dishes through a fine-mesh sieve using deionized water. After washing, the sieve contained a mixture of larvae and unhatched eggs. Larvae were separated from eggs.
Adult flies were harvested from bottles containing standard cornmeal–agar–molasses food media with yeast. Newly emerging adults were collected after clearing bottles 12–16 hr the night before. Flies were separated by sex and placed on fresh media for 4 days prior to RNA isolation. The bottles used to store these virgin female flies were retained for 2 weeks after fly collection. If larvae were observed in the bottles after this period, flies collected from that bottle were discarded.
RNA collection from heterokaryotic flies
Gene expression was assayed in AR/ST, AR/CH, ST/AR, and AR/PP inversion heterozygotes. For each cross, RNA was collected individually from three technical replicates. These heterokaryotypes were chosen because they were expected to have frequencies >15% in samples collected in 1940 ± 2 years in at least one of the six different niches (see figure 1 in Schaeffer 2008). Recent collections have shown an increase of the TL arrangement (Anderson et al. 1991), but we did not include any TL heterozygotes at this time. Reciprocal crosses were performed to generate AR/ST, AR/CH, ST/CH, and AR/PP heterokaryotypes. In addition, two unrelated Arrowhead chromosomes were crossed to generate an AR/AR control (Table 2). The rearing and collection methods for homokaryotypes were identical to those used for the homokaryotic flies, except bottles to collect adult heterokaryotypes were cleared after 20 days to ensure that there would be no contamination from homokaryotypic parents. First instar larvae of indeterminate sex, 4-day-old virgin adult females, and 4-day-old adult male inversion heterozygotes were used to prepare RNA for RNA-seq analysis.
Table 2. Reciprocal crosses between third chromosome homokaryotypic D. pseudoobscura strains to generate heterokaryotypes.
| ID | Male homokaryotype | Female homokaryotype |
|---|---|---|
| Cross 1A | AR KB945 | ST MSH49 |
| Cross 1B | ST MSH49 | AR KB945 |
| Cross 2A | AR KB945 | CH MSH202 |
| Cross 2B | CH MSH202 | AR KB945 |
| Cross 3A | ST MSH49 | CH MSH202 |
| Cross 3B | CH MSH202 | ST MSH49 |
| Cross 4A | AR KB945 | PP DM1054 |
| Cross 4B | PP DM1054 | AR KB945 |
| Cross 5A | AR KB945 | AR DM1005 |
| Cross 5B | AR DM1005 | AR KB945 |
Each cross was replicated three times for RNA-seq.
RNA was purified with RNeasy spin columns (QIAGEN) following manufacturer’s instructions, and quantified with a NanoDrop 200× UV-Vis Spectrometer (Thermo Scientific). As genomic imprinting has been detected in Drosophila (Lloyd et al. 1999) equimolar quantities of RNA for each reciprocal cross were combined for RNA sequencing.
RNA-seq
Illumina RNA-seq (Wang et al. 2009) was performed following standard protocols by the Baylor College of Medicine Human Genome Sequencing Center (Houston) on an Illumina HiSequation 2000 sequencing platform. Briefly, poly-A+ messenger RNA (mRNA) was extracted from 1 μg total RNA using Oligo(dT)25 Dynabeads (Life Technologies, cat. no. 61002) followed by fragmentation of the mRNA by heat at 94° for 3 min [for samples with RNA Integrity Number (RNI) = 3–6] or 4 min (for samples with RIN of ≥6.0). First-strand complementary DNA (cDNA) was synthesized using the Superscript III reverse transcriptase (Life Technologies, cat. no. 18080-044) and purified using Agencourt RNAClean XP beads (Beckman Coulter, cat. no. A63987). During second-strand cDNA synthesis, deoxynucleoside triphosphate (dNTP) mix containing deoxyuridine triphosphate was used to introduce strand specificity. For Illumina paired-end library construction, the resultant cDNA was processed through end repair and A-tailing, ligated with Illumina PE adapters, and then digested with 10 units of uracil–DNA glycosylase (New England Biolabs, Ipswich, MA; cat. no. M0280L). Amplification of the libraries was performed for 13 PCR cycles using the Phusion High-Fidelity PCR Master Mix (New England Biolabs, cat. no. M0531L); 6-bp molecular barcodes were also incorporated during this PCR amplification. These libraries were then purified with Agencourt AMPure XP beads after each enzymatic reaction, and after quantification using the Agilent Bioanalyzer 2100 DNA Chip 7500 (cat. no. 5067-1506), libraries were pooled in equimolar amounts for sequencing. Sequencing was performed on Illumina HiSeq2000s, generating 100-bp paired-end reads.
Read mapping and statistical tests for differential expression
Paired-end reads were mapped to the D. pseudoobscura reference genome (FlyBase version 3.03) using the subjunc aligner (version 1.4.6) under default parameters (Liao et al. 2013). Because subjunc soft clips the ends of reads with low-mapping quality (MAPQ) scores, read ends were not trimmed prior to alignment as recommended in the software guidelines. The number of reads mapping to each annotated exon in the reference genome in each resulting BAM file were counted using featureCounts (version 1.4.6), and the counts summarized independently for the different life stages (Liao et al. 2014). Genes that did not have at least 10 reads mapping in at least three samples were filtered from downstream analyses.
Between-lane upper-quartile normalization was performed on the filtered read counts using the R package RUVSeq (Risso et al. 2014). Such normalization controls for variation between sequencing lanes and RNA samples, which would otherwise confound genuine signals of differential gene expression between gene arrangements. Normalization was performed separately for each life stage, with homokaryotype and heterokaryotypes of the same life stage analyzed together. The RUVs method implemented in RUVSeq performs a factor analysis on a set of negative control genes for which no differential expression is expected between samples where the covariates of interest (i.e., chromosome arrangement) are constant to remove unwanted variation from the data. We used a set of housekeeping genes previously identified by Brown and Bachtrog (2014) (provided by E. Brown, personal communication; Supplemental Material, Table S1) as our negative controls. Because the factor analysis is performed for the set of negative controls between control samples, it does not exclude those genes from being detected as differentially expressed between other test samples in the full data set. Furthermore, RUVs has been demonstrated to be robust to the selection of the set of negative control genes (Peixoto et al. 2015). For our control samples, we used the AR/AR heterokaryotype control cross-replicates and the AR homokaryotypes since transcriptional variation captured by the inversion is expected to be constant between them. Before the normalization step, we first validated this expectation by testing for differential expression between the AR homokaryotypes and AR/AR control cross. With a false discovery rate (FDR) of 0.05 to control for multiple testing (Benjamini and Hochberg 1995), five genes in larvae, zero genes in females, and one gene in males are differentially expressed, none of which are located on Muller C. Using a more stringent FDR cutoff of 0.01, no genes are detected as differentially expressed. Because the two AR homokaryotypes come from different populations, these results also seem to reject a strict interpretation of Dobzhansky’s (1950) coadaptation hypothesis. It is, however, possible one of the AR chromosomes could be a recent migrant.
Differential expression was analyzed using the R package edgeR (version 3.10.2) (Robinson et al. 2010) on the normalized read counts. The design matrix for the negative binomial generalized linear model (GLM) was constructed using the first factor of unwanted variation (k = 1) and the covariates of interest, here the chromosome arrangements. Briefly, the GLM takes the form of
where Y is the matrix containing the read counts for each gene, W is the matrix containing the factors of “unwanted variation,” X is the matrix containing the covariates of interest, and O is a matrix of offsets estimated through upper-quartile normalization. α and β correspond to the parameters for the factors of unwanted variation and covariates of interest (i.e., “treatment effect,” here the chromosome arrangement), respectively.
The homokaryotype dataset included three biological replicates for each arrangement (Table 1). To test for differential expression among the homokaryotypes, a one-way ANOVA-like analysis was performed using the quasi-likelihood F-test implemented in edgeR to provide a more robust control rate for error and variability due to the relatively small sample size of the replicates (Lund et al. 2012). We applied a stringent FDR cutoff of 0.01 because the null hypothesis of no differential gene expression across all arrangements can be rejected by differences in one or more groups of six arrangements (A full list of genes is available in Table S2, Table S3, Table S4, Table S5, Table S6, and Table S7). Therefore, there are multiple ways in which the null hypothesis can be rejected for a given gene. We provide a full list of genes detected in the ANOVA-like analysis at a FDR cutoff of 0.05 in Table S8, Table S9, and Table S10; however, we only consider genes with a FDR of <0.01 as significant here. Similar tests were performed in edgeR to detect differential expression between heterokaryotypes and homokaryotype arrangement backgrounds. Here, for each pairwise comparison, we relaxed our FDR cutoff to 0.05 for considering genes as significant because the null hypothesis can only be rejected when differences exist between the two groups tested.
Analysis of genetic variation
From the aligned reads, we called variants using FreeBayes version 0.9.21 (Garrison and Marth 2012) under default parameters, with the exception of setting the ploidy option to 1 for X chromosome mapping scaffolds in males. We filtered out segregating sites with a genotype quality phred score of <30. To examine the structure of genetic variation in the expression data, we performed a principal component analysis (PCA) from the called genotypes in homokaryotypes using the R function prcomp. Only sites that had coverage for all individuals were considered to mitigate the effects of expression level on the analysis.
Classification of expression inheritance in heterokaryotypes
We classified the inheritance patterns of genes identified as differentially expressed with pairwise comparisons of the two homokaryotypes and the corresponding heterokaryotypic cross. Genes were classified as either dominant, underdominant, overdominant, or additive. Following convention (Gibson et al. 2004; McManus et al. 2010; Bougas et al. 2013; Schaefke et al. 2013), here dominant, overdominant, and underdominant refer to the mode of inheritance, not selection or fitness. These categories of expression patterns were defined similarly to previous studies (McManus et al. 2010; Bell et al. 2013). Genes that exhibited significant expression differences between homokaryotypes but had intermediate expression levels in the heterokaryotypes state were classified as additive. If expression was significantly greater in the heterokaryotype than either homokaryotype, genes were considered overdominant. Contrarily, if expression was significantly reduced in the heterokaryotype relative to either homokaryotype, the gene was classified as underdominant. If expression was significantly different between the heterokaryotype and only one of the two homokaryotypes, it was classified as dominant. Genes showing evidence of dominant or additive inheritance were further classified by which homokaryotype arrangement had the greater relative level of expression.
Data availability
Fly strains are available upon request. Raw sequencing reads are available from the National Center for Biotechnology Information as a BioProject (PRJNA326536).
Results
Transcriptome sequencing and mapping
An RNA-seq approach was used to quantify gene expression for six D. pseudoobscura third chromosome homokaryotypes (AR, CU, CH, PP, ST, and TL) and four heterokaryotypes (AR/ST, AR/CH, AR/PP, and ST/CH) in three different life forms: first instar larvae, adult females, and adult males. In addition, an AR/AR heterozygote cross between two different Arrowhead chromosomes from two different localities were sequenced for each life stage as a control. In total, ∼2.3 billion read pairs were generated from larvae, 1.9 billion read pairs from females, and 2.4 billion read pairs from males, resulting in a total of >6 billion paired-end Illumina sequences and between 7 million and 127 million read pairs per individual library.
An average of 64.1, 71.6, and 70.6% of reads successfully mapped to annotated features on the D. pseudoobscura reference genome in larvae, females, and males, respectively. After filtering out genes that did not meet the minimum criteria of at least 10 reads being mapped in at least three individuals, 13,819 genes in larvae, 13,362 in females, and 15,438 genes in males were retained for downstream analyses.
Differential expression among third chromosome inversion homokaryotypes
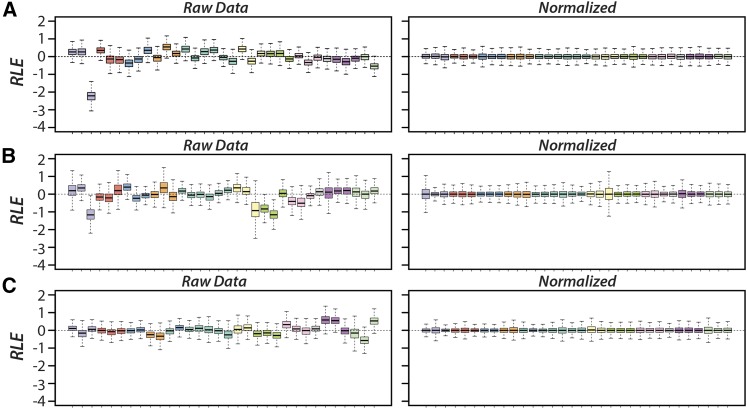
A one-way ANOVA-like analysis was performed to test for differential expression among the six third chromosome arrangement homokaryotypes. Following read normalization in RUVSeq (Figure 2), a preliminary PCA and relative log-expression profiling performed in edgeR identified strain CH_MSH202 in larvae and TL_SCI12.2 in males as potential outliers. For each case, the single outlier strains accounted for >10% of the total variation in the entire data set and had significantly higher variance in the relative log-expression profile. These samples were therefore excluded in subsequent differential expression tests.
Figure 2.
Removal of unwanted variation from the RNA-seq data for males (A), females (B), and larvae (C). The boxplots on the left depict the relative log expression (RLE) profiles of each individual for the raw, unnormalized read counts. After removing unwanted variation through normalization between lanes, using 4390 housekeeping genes and between AR replicates, the RLE distributions are centered around zero and similar to one another as expected.
Significant differences in expression between gene arrangements were detected for 81 genes in larvae, 101 genes in females, and 335 genes in males across the genome (for a full list of differentially expressed genes, see Table S4, Table S5, and Table S6). Of these genes, 23 exhibit differential expression in both males and females and 5 are differentially expressed across all three life stages. The 5 genes differentially expressed across all life stages are located within inverted segments of the third chromosome and have no annotated biological functions. A PCA based on the called genotypes in the RNA-seq reads reveals structure associated with the arrangements for transcripts on Muller C (Figure S1A). However, the PCA provides strong evidence that the structure of genetic variation on non-Muller C chromosomes results from the balancer strain used (Figure S1B). Furthermore, it is possible that sections of uncontrolled wild-type chromosomes are present in the lines as a result of our isogenic crossing scheme, which may lead to cis-acting effects on gene expression on non-Muller C chromosomes. Although these differentially expressed genes could be the result of trans-acting elements associated with genetic variation on chromosome 3, there are possible alternative sources of expression differences. As a result, we do not consider genes located on non-Muller C chromosomes for all subsequent analyses. Of the differentially expressed genes, 45 for larvae, 45 for females, and 182 for males are located on chromosome 3.
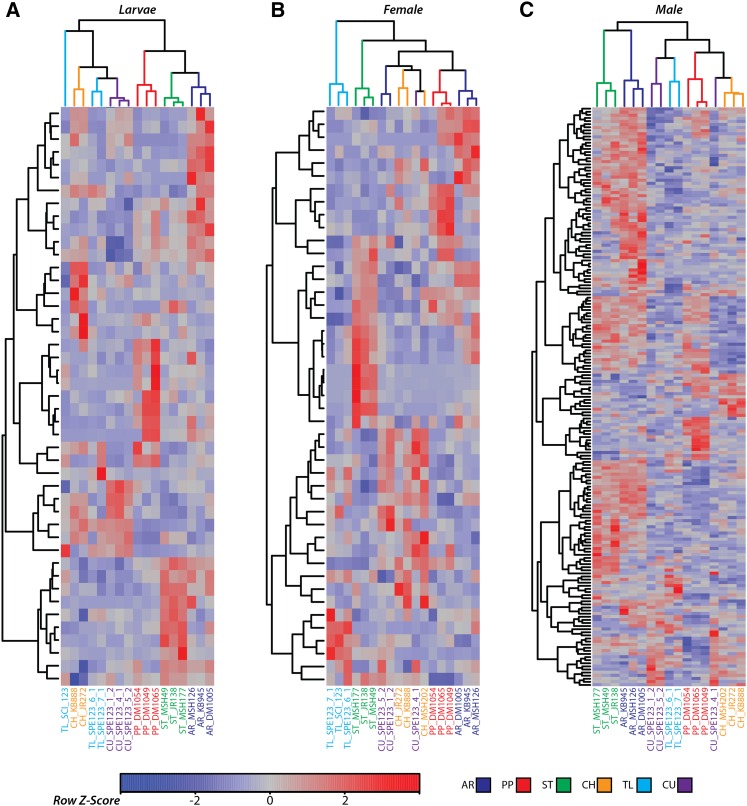
Gene expression differences among the six gene arrangements suggest that the transcriptional variants are in linkage disequilibrium. We investigated the associations of gene expression variations with individual homokaryotypes using unsupervised hierarchical clustering methods. For each life stage, a matrix was constructed from the log2 transformed reads per kilobase per million mapped reads (RPKM) (Mortazavi et al. 2008) obtained from the read counts and lengths of significantly differentially expressed genes. The unsupervised hierarchical clustering (Figure 3) is based on Pearson’s correlation coefficients for each biological replicate (column) and for each gene (row) in the RPKM matrix. In nearly every case, the biological replicates for each chromosomal arrangement cluster together and the relationship among arrangements tends to recover the accepted molecular and cytogenetic phylogeny (Dobzhansky and Queal 1938; Wallace et al. 2011). For males, the phylogenetic relationship is generally recovered in the unsupervised clustering, with the exception of CH falling on a node closest to PP. Additionally, a single CU individual groups in between the PP and CH nodes. In females and larvae there are a small number of discrepancies where a single replicate is placed on an internal node leading to a different cluster of arrangement replicates. For larvae, all AR, CH, CU, PP, and ST individuals form monophyletic clusters. A single TL individual falls outside the CH, CU, and TL clusters, which are all derived from the SC arrangement. For females, ST, TL, AR, and PP all form monophyletic clusters. However, two CH and CU individuals cluster together. The recovery of the cytological phylogeny from gene expression data suggests nonrandom association or linkage disequilibrium of transcriptional variation with gene arrangement type.
Figure 3.
Differentially expressed genes depicted as a heat map for larvae (A), females (B), and males (C). The genes (rows) and individuals (columns) have been ordered based on unsupervised hierarchical clustering. The color of each individual represents the arrangement. Rows are colored according to the deviation from the average level of expression across all individuals.
To test for an enrichment of any particular biological functions within each set of differentially expressed genes, we performed a gene ontology (GO) analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) software under default parameters (Huang et al. 2008). We note that not all D. pseudoobscura genes are annotated and DAVID may contain incomplete published pathway interactions, so the reported results may be an underrepresentation of the true biological enrichment (Wadi et al. 2016). In males, genes involved in sensory perception are significantly overrepresented after correcting for multiple testing [q < 0.05; Benjamini–Hochberg (BH) corrected; Table 3]. Specifically, genes associated with sensory perception of taste (q < 3.4 × 10−3) and olfaction (q < 0.048) are significantly enriched. Although differential expression was detected in females and larvae for genes involved in sensory perception, detoxification and cuticle formation, no significant overrepresentation of any gene function was detected for these groups.
Table 3. Significantly enriched GO terms among differentially expressed genes on chromosome 3 in males.
| GO ID | GO term | Count | P-value (BH corrected) |
|---|---|---|---|
| GO:0007606 | Sensory perception of chemical stimulus | 9 | 1.07 × 10−4 |
| GO:0050890 | Cognition | 9 | 1.14 × 10−4 |
| GO:0007166 | Cell surface receptor | 10 | 2.24 × 10−3 |
| GO:0050909 | Sensory perception of taste | 5 | 3.41 × 10−3 |
| GO:0004984 | Olfaction | 6 | 4.80 × 10−2 |
Position effects of inversion mutations on gene expression
We tested for significant differential expression of genes located at the boundaries of inversion breakpoints as an indicator of direct physical position effects (Puig et al. 2004) The approximate locations of the inversion breakpoints have previously been inferred from the concordance of the D. pseudoobscura reference sequence with the polytene chromosome map (Richards et al. 2005; Schaeffer et al. 2008, Z. L. Fuller, G. D. Haynes, S. Richards, and S. W. Schaeffer, unpublished data).
Of the genes immediately adjacent to breakpoints, only GA22082 shows evidence of significantly elevated levels of transcription (FDR < 0.01) for larvae in ST relative to the PP (log2 fold change: 1.086). This gene is adjacent to the chromosome region containing the proximal hypothetical (HY)-to-ST breakpoint (pHYST) and the distal ST-to-PP breakpoint (dSTPP). The pHYST and dSTPP breakpoints are thought to have occurred independently in the same location in different inversion mutation events (Z. L. Fuller, G. D. Haynes, S. Richard, and S. W. Schaeffer, unpublished data). The ortholog of GA22082 in D. melanogaster (CG9864) has previously been shown to be a member of the major facilitator superfamily and has been identified in phosphate response pathways (Bergwitz et al. 2012). However, no regulatory regions or enhancer sites are known in the current D. pseudoobscura or D. melanogaster genome annotations. Significant differential gene expression was not detected in any other gene adjacent to the approximate location of inversion breakpoints. With the possible exception of the pHYST/dSTPP breakpoint in larvae, we conclude that inversions can alter gene expression near breakpoints but position effects do not appear to be a general mechanism for generating genetic variation for selection to act (Sperlich and Pfriem 1986). Our data do not preclude the possibility that gene expression was altered near the inversion breakpoints when the mutation first occurred, yet returned to normal levels in the population through stabilizing selection.
Differential expression between heterokaryotype and homokaryotype arrangements
For each of the Muller C heterokaryotypes, we tested for differential gene expression between the heterokaryotype and both parental homokaryotype backgrounds (for a full list of genes, see Table S11, Table S12, and Table S13). In each life stage, the greatest number of differentially expressed genes is found between the two homokaryotic arrangements (Table 4). The largest number of differentially expressed genes detected in any of the heterokaryotype–homokaryotype comparisons is found between ST/CH and CH in females with five. Interestingly, differential expression was not detected between ST/CH and the other homokaryotype background (ST) in either males, females, or larvae. There is a significant enrichment (q < 0.012; BH corrected) of seven-transmembrane receptors (7-TMRs) among genes differentially expressed between AR/PP and AR in males. Only one of these 7-TMR genes (GA15816) is also differentially expressed between ST/CH and CH in females. Significant enrichment for biological categories or GO terms was not found for any of the other comparisons between heterokaryotype and homokaryotype arrangement backgrounds.
Table 4. Gene expression differences between parental third chromosome homokaryotypes and crossed heterokaryotypes for genes located on the third chromosome.
| AR/ST | AR/CH | ST/CH | AR/PP | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Test | P1-C | P2-C | P1–P2 | P1-C | P2-C | P1–P2 | P1-C | P2-C | P1–P2 | P1-C | P2-C | P1–P2 |
| Male | 6 | 1 | 25 | 3 | 1 | 94 | 0 | 0 | 22 | 9 | 0 | 63 |
| Female | 1 | 0 | 5 | 0 | 0 | 30 | 0 | 5 | 11 | 0 | 0 | 1 |
| Larvae | 0 | 1 | 1 | 1 | 1 | 17 | 0 | 0 | 30 | 0 | 0 | 10 |
P1 and P2 represent either of the homokaryotype arrangements. C indicates the heterokaryotype.
We performed a one-way ANOVA-like analysis to test for differential expression among the four arrangement heterokaryotypes in each life stage. Significant differential expression was detected for 1 gene in larvae, 0 genes in females, and 17 genes in males. No biological categories or GO terms are significantly enriched for any of the comparisons. Furthermore, unsupervised hierarchical clustering of expression levels could not cluster all biological replicates for a heterokaryotype arrangement together in any case. Therefore, we conclude either that gene expression patterns are similar among the heterokaryotypes included in our study or that we did not have sufficient power to detect major differences. Under both conclusions, it is clear that differences in patterns of gene expression are greater and more pronounced between homozygous chromosome arrangements than between mixed, heterokaryotic backgrounds.
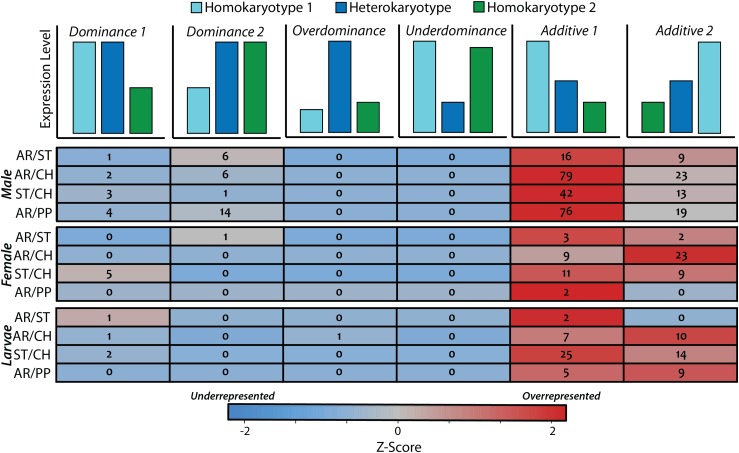
The inheritance of gene expression in heterokaryotypes
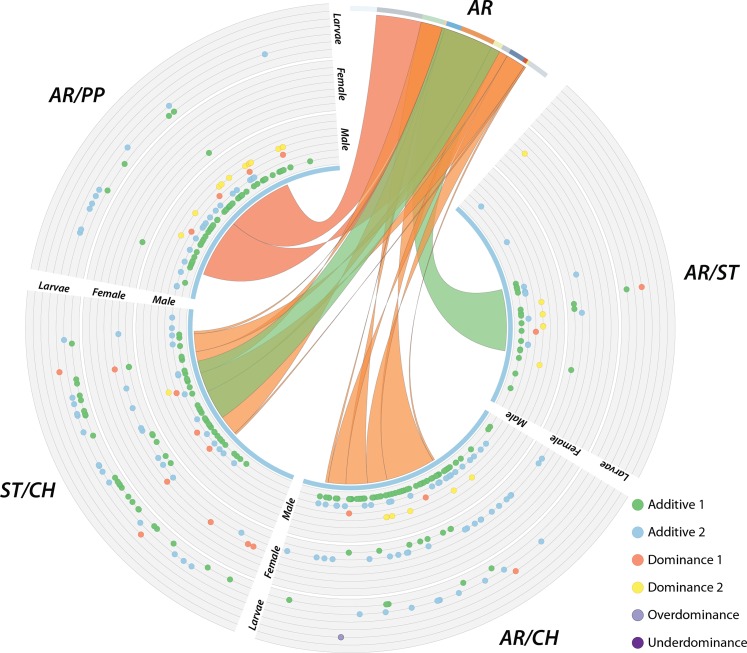
The count of genes displaying additive, dominant, overdominant, and underdominant inheritance across the genome among heterokaryotic individuals is summarized in Figure 4 (for a full list of genes, see Table S14, Table S15, and Table S16). Although dominant, overdominant, and underdominant are terms widely used to describe selection or fitness, here they specifically refer to the mode of expression inheritance. We find that genes exhibiting an additive mode of inheritance are overrepresented for males, females, and larvae (P < 0.0001, χ2). This abundance of additive inheritance explains the overall lower number of differentially expressed genes detected between heterokaryotypes and homokaryotypes than between the two homokaryotypes. Genes displaying a pattern of underdominant inheritance, where the heterokaryotype is significantly under expressed relative to both homokaryotypes, was not observed in any test. Furthermore, there is a significantly greater proportion of genes displaying an additive mode of inheritance located within nonhomosequential regions in the heterokaryotype (Figure 5).
Figure 4.
The inheritance of expression in heterokaryotypes for genes located on the third chromosome. The cartoons at the top represent the level of expression in the homokaryotypes and heterokaryotype for a hypothetical gene of each class. Rows are colored by the over- or under-abundance of a class.
Figure 5.
The location of third chromosome genes classified according to their mode of inheritance. The outermost track depicts larvae, the middle females, and the inner males. The ribbons connecting the AR chromosome to each heterokaryotype represents how the regions map relative to the reference sequence gene order.
For all genes showing evidence for an additive mode of inheritance, analysis using DAVID software under default parameters (Huang et al. 2008) reported a significant enrichment of GO terms associated with the following categories: sensory perception of chemical stimulus (q < 6.9 × 10−3), odorant binding (q < 7.6 × 10−3), and glutathione S-transferase activity (q < 1.7 × 10−2). Among genes showing evidence for nonadditive effects in the heterokaryotype, analysis in DAVID found a significant enrichment for 7-TMRs associated with chemoreception (q < 1.6 × 10−3). All 7-TMR genes displayed a dominant mode of expression inheritance.
Overall, the largest number of genes classified as dominant in heterokaryotypes show expression levels similar to the ST arrangement. ST is ancestral to AR and PP and is believed to be derived from the presumed ancestral D. pseudoobscura arrangement, hypothetical (HY) (Dobzhansky and Epling 1944; Wallace et al. 2011). Furthermore, ST dominance occurs at a significantly greater frequency than would be expected if it could be inherited from either arrangement with equal probability (P < 0.001, χ2). No other chromosome arrangement is significantly overrepresented in dominant expression inheritance. This may provide evidence for recessive deleterious mutations in AR and PP associated with transcription in these genes.
Discussion
Gene expression as a potential target of selection
To investigate the role that chromosome inversions may play in evolutionary processes, we used RNA-seq to ask whether gene expression differs among gene arrangement backgrounds of D pseudoobscura. Changes in gene expression can profoundly affect an organism’s physiology, behavior, and fitness, and can therefore provide targets for natural selection to act upon (Gilad et al. 2006). By directly testing for differential expression, we identify potential candidate gene targets for natural selection that would not otherwise be readily apparent from genomic sequence analysis.
The existence of stable clinal frequencies of D. pseudoobscura third chromosomal arrangements in the face of extensive gene flow suggests that natural selection played a role in both the initial spread of inversion mutations and in maintaining their subsequent frequencies within wild populations (Schaeffer and Miller 1992; Schaeffer et al. 2003). Inversion mutations may directly alter gene expression by disrupting regulatory regions or modifying the relationship between a gene and its regulatory elements. Inversions can also “capture” gene expression variants at multiple loci and maintain their associations by inhibiting recombination. Here, we have examined differential expression in third chromosome homo- and heterokaryotypes to investigate transcriptional differences that may serve as potential targets of selection that contribute to the stable inversion polymorphism system.
On Muller C, we detected significant expression differences in 45 genes for larvae, 45 genes for females, and 181 genes in males. We found one case for gene expression directly affected at the breakpoints of inversion mutations. The gene GA22082, which is adjacent to the reused breakpoint (distal ST to PP/proximal HY to ST) showed elevated expression levels in ST relative to PP in larvae, suggesting that the breakpoint reusage may have played a role in altering the regulation of the gene. Other than this single case, none of the other breakpoints altered gene expression of their adjacent genes, indicating that position effects are not a prominent mechanism in generating potential genetic diversity for selection to act upon. Instead, we find a significant overrepresentation of multiple differentially expressed genes located within inverted segments of the third chromosome relative to the noninverted segments (P < 0.0001 for all life stages). This suggests that if selection is acting to maintain transcriptional differences between arrangements, then it has acted through the indirect effect of suppressed recombination maintaining associations of multiple differentially expressed genes within the inversions.
Suppressed recombination maintains differential expression differences
Previous studies of the inverted segments of the D. pseudobscura third chromosome have demonstrated both suppressed recombination and elevated genetic differentiation between gene arrangements, relative to the noninverted regions (Wallace et al. 2013; Fuller et al. 2014). Unsupervised hierarchical clustering of gene expression data recapitulates the accepted gene arrangement phylogeny, supporting the hypothesis that multiple transcriptional variants are in linkage disequilibrium with the different inversion types (Charlesworth and Charlesworth 1973). Suppressed recombination allows for a selective advantage of an inversion that captures two or more alleles contributing to local adaptation, regardless of epistatic interactions, because it prevents the exchange of maladaptive alleles between two different locally adapted chromosomes while maintaining a beneficial combination of alleles from escaping an adaptive gene arrangement (Kirkpatrick and Barton 2006; Kirkpatrick 2010). Over time, new mutations coupled with natural selection may increase the frequency of additional locally adaptive alleles within the inverted chromosome segment. Under this scenario, it follows that locally adaptive alleles that influence gene expression could lead to significant transcriptional differences between chromosomes. Consistent with selection acting on the indirect effects of suppressed recombination, the proportion of genes exhibiting differential expression between arrangement homokaryotypes for all life stages is significantly greater within inverted segments on the third chromosome than in segments where recombination can proceed freely.
GO analysis gives an indication of which biological functions may be targets of natural selection by identifying biological pathways that are “enriched” with differentially expressed genes. Male homokaryotypes showed by far the greatest enrichment among their differentially expressed genes. On Muller C, enrichment was detected for differentially expressed genes involved with sensory perception, in particular chemoreception (q < 1.1 × 10−4) and olfaction (q < 0.042). In females and larvae, we do not find evidence for any overrepresented functional gene categories, although a number of odorant binding genes exhibit differential expression in both these life stages.
The overrepresentation of differentially expressed chemoreception genes among males, along with differential expression of chemoreceptor genes in larvae and females, indicates that chemoreception is a putative target of selection. Chemoreception is essential in insect feeding, reproduction, and predator avoidance, and changes to chemoreception genes can underlie adaptation to environmental shifts (Sánchez-Gracia et al. 2009). Furthermore, sensory and chemoreception genes are differentially expressed between high and low latitude populations of D. melanogaster, consistent with a role of selection maintaining transcriptional differences across heterogeneous environments (Zhao et al. 2015). The chemical environment of D. pseudoobscura no doubt differs greatly between regions, habitats, and seasons, providing ample opportunity for selection to correspondingly favor differing expression of chemosensory genes. The boundaries of the six niches D. pseudoobscura occupies are defined by major climatic and physiogeographic shifts (Lobeck 1950; Schaeffer 2008). Specific expression patterns of genes involved in odorant and taste reception may confer fitness advantages in particular habitats, and the effects of suppressed recombination mediated by the inversions may hold together these locally adaptive sets of genes.
Another class of genes that show evidence of differential expression are involved in the limonene and pinene degradation pathway. This includes two cytochrome P450 genes (GA10189 and GA17561 in males) that degrade α-pinene. D. pseudoobscura cooccurs with ponderosa pine in the western United States (Dobzhansky and Epling 1944; Smith 1977). Levels of α-pinene in ponderosa pine vary clinally with lower levels in California and higher levels in Arizona. These data provide an intriguing link into the ecology of D. pseudoobscura. Despite >80 years of research on the genetics of D. pseudoobscura, the definitive breeding sites for this species have yet to be discovered (Carson 1951). Although the DAVID analysis does not show enrichment of these genes, our results warrant further study of aspects of ponderosa pine biology to uncover the natural breeding sites for D. pseudoobscura. It is interesting to note that 14 of the 28 genes involved in the limonene and pinene degradation pathway are located on chromosome three, raising the question of whether genes controlling this complex trait are nonrandomly arranged in the genome.
Additive inheritance in heterokaryotypes as a consequence of local adaptation in homokaryotypes
Early studies invoked balancing selection and overdominance to explain the stable equilbria of frequencies of D. pseudoobscura arrangements in nature and the observations from population cage experiments, through a model of coadapted gene complexes (Dobzhansky and Levene 1951; Wallace 1968). We find no evidence for significant differential expression between individuals with AR chromosomes derived from different populations (AR/AR control cross) and AR homokaryotypes from any of the populations tested in this study. This result seems to reject the strict interpretation of the coadaptation hypothesis (Dobzhansky 1950) where homosequential chromosomes from different populations had lower fitness than heterokaryotypes with chromosomes from the same population. The expression data here does not support the conclusion that homosequential AR chromosomes from different populations carry dissimilar transcriptional information. Associative overdominance has also been hypothesized to play a role in the maintenance chromosomal inversion systems (Haldane 1957; Kojima 1967; Nei et al. 1967; Ohta 1971). Similarly, Kirkpatrick and Barton (2006) explained this apparent case of balancing selection in chromosomal inversion systems with a model of local adaptation where each arrangement harbors recessive deleterious mutations leading to associative overdominance. Here, selection acts against deleterious recessives and prevents any chromosome from completely fixing within a population. However, the AR arrangement exists at near fixation in the Four Corners region of the American Southwest and strong evidence for the accumulation of recessive deleterious mutations on other arrangements has yet to be found. Meanwhile, a model of selection across heterogeneous environments does not require the frequencies of arrangements to be maintained through global overdominance, but predicts that heterokaryotypes are the result of the average heterosis over variable microhabitats. This is similar in theory to a form of balancing selection termed marginal overdominance, where the mean fitness of the heterozygote is greater than either homozygote in a temporal environmentally heterogeneous environment (Wills 1975; Bergland et al. 2014). Numerical solutions of migration–selection recursions under this model result in a mixture of directional, overdominant, and underdominant selection acting on different arrangements to produce the observed frequencies of chromosomes in particular niches (Schaeffer 2008).
Our results are consistent with a hypothesis of selection for local adaptation in homokaryotypes, yet the absence of significant differential expression in heterokaryotype comparisons would appear to reject the notion of selection acting directly to maintain transcriptional differences in arrangement heterozygotes. Instead, we find prevalent additive inheritance of gene expression in heterokaryotypes where the level of transcription is intermediate to either homozygous arrangement. If local environments where selection is acting are imagined as optimal fitness gradients instead of abrupt shifts in microhabitats, it is possible to envision a scenario where intermediate levels of transcription are actually favored. This would simultaneously support the local adaptation models of Kirkpatrick and Barton (2006) and the appearance of multiple modes of selection acting to maintain heterokaryotype frequencies. For a particular gene, the optimal level of expression may be low in one local environment and gradually increase across the ecotone to where high expression is favored in a different local environment. If high or low gene expression is controlled by differences in cis-regulatory elements, individuals heterozygous for alleles associated with either extreme would have an intermediate level of transcription (Lemos et al. 2008). Here, selection would appear as directional or underdominant at either end of this fitness gradient and overdominant near the center where the optimal level of expression is intermediate. If heterozygotes are favored in transitional habitats and two or more loci are involved in the local adaptation of homokaryotypes, recombination would break apart these sets of beneficial alleles in the absence of a suppression mechanism, such as chromosomal inversions. Hence, this process may also explain how inversions are initially established across environmental clines.
We find evidence of cis-regulatory differences on Muller C, prevalent additive inheritance of gene expression in heterokaryotypes and the maintenance of transcriptional differences between arrangements, which would support such a model. In this specific case, the level of gene expression is related to fitness and is the target of selection; however, it is possible that other intermediate phenotypes may behave in a similar fashion across environmental clines. Indeed, this phenomenon is termed “bounded hybrid superiority” when describing hybrid zone structure between closely related species and is thought to occur in a variety of systems, including birds and plants (Moore 1977; Arnold 1996; Good et al. 2000; Abbott and Brennan 2014). A recent study found extensive additive effects and intermediate expression in hybrids for transcripts that are differentially expressed between two populations of Pacific oyster that differ substantially in water temperature (Sussarellu et al. 2015). The authors conclude this additivity underlies the invasiveness of hybrids and allows them to persist in environments different from the locally adapted parent populations. Although much more theoretical work is needed to fully formalize this model, and specifically for the case of chromosomal inversions, our results provide empirical support for local adaptation along with additive genetic inheritance in the maintenance of arrangement frequencies in D. pseudoobscura. Future work may also be directed at examining specific environmental interactions on gene expression and the relationship between fitness and the environment to further explore this model. The identity of differentially expressed genes provides valuable clues about the molecular basis for fitness differences.
In this study, we provide evidence that significant transcriptional variation exists among chromosome arrangements. Furthermore, our results support a model of local adaptation across a spatially heterogeneous environment. Although our findings are consistent with selection acting on the indirect effects of chromosomal inversions, we are limited in identifying the specific loci that show evidence of selection at this time. Additional work is needed to determine the loci at the genomic level that show evidence of selection. Furthermore, future functional experiments will be required to identify the molecular basis of adaptive variation at the phenotypic level.
Acknowledgments
We thank Monte E. Turner of the University of Akron who pointed us to the literature on α-pinene. We also thank the staff at the Baylor College of Medicine Human Genome Sequencing Center for their contributions and two anonymous reviewers who suggested that we examine the SNP background of non-Muller C chromosomes, which revealed a balancer effect. This work was supported by National Institutes of Health (NIH) grant R01-GM098478 to S.W.S. and the Penn State University NIH-funded Computation, Bioinformatics, and Statistics Predoctoral Training Program to Z.L.F.
Footnotes
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.191429/-/DC1.
Communicating editor: D. J. Begun
Literature Cited
- Abbott, R. J., and A. C. Brennan, 2014 Altitudinal gradients, plant hybrid zones and evolutionary novelty. Philos. Trans. R. Soc. B Biol. Sci. 369: 1648. Available at: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4071520/. [DOI] [PMC free article] [PubMed]
- Anderson W. W., Arnold J., Baldwin D. G., Beckenbach A. T., Brown C. J., et al. , 1991. Four decades of inversion polymorphism in Drosophila pseudoobscura. Proc. Natl. Acad. Sci. USA 88: 10367–10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P., Depaulis F., Navarro A., 2001. Inversion polymorphisms and nucleotide variability in Drosophila. Genet. Res. 77: 1–8. [DOI] [PubMed] [Google Scholar]
- Arnold M., 1996. Natural Hybridization and Evolution, Oxford University Press, New York. [Google Scholar]
- Bansal V., Bashir A., Bafna V., 2007. Evidence for large inversion polymorphisms in the human genome from HapMap data. Genome Res. 17: 219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. D. M., Kane N. C., Rieseberg L. H., Adams K. L., 2013. RNA-Seq analysis of allele-specific expression, hybrid effects, and regulatory divergence in hybrids compared with their parents from natural populations. Genome Biol. Evol. 5: 1309–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57: 289–300. [Google Scholar]
- Bergland A. O., Behrman E. L., O’Brien K. R., Schmidt P. S., Petrov D. A., 2014. Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila. PLoS Genet. 10: e1004775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergwitz C., Rasmussen M. D., DeRobertis C., Wee M. J., Sinha S., et al. , 2012. Roles of major facilitator superfamily transporters in phosphate response in Drosophila. PLoS One 7: e31730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutkar A., Schaeffer S. W., Russo S. M., Xu M., Smith T. F., et al. , 2008. Chromosomal rearrangement inferred from comparisons of 12 Drosophila genomes. Genetics 179: 1657–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougas B., Normandeau E., Audet C., Bernatchez L., 2013. Linking transcriptomic and genomic variation to growth in brook charr hybrids (Salvelinus fontinalis, Mitchill). Heredity 110: 492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E. J., Bachtrog D., 2014. The chromatin landscape of Drosophila: comparisons between species, sexes, and chromosomes. Genome Res. 24: 1125–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson H. L., 1951. Breeding sites of Drosophila pseudoobscura and Drosophila persimilis in the transition zone of the Sierra Nevada. Evolution 5: 91–96. [Google Scholar]
- Carson H., Yoon S. J., 1982. Genetics and evolution of Hawaiian Drosophila, pp. 297–344 in The Genetics and Biology of Drosophila, Academic Press, New York. [Google Scholar]
- Charlesworth B., 1974. Inversion polymorphism in a two-locus genetic system. Genet. Res. 23: 259–280. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., 1973. Selection of new inversions in multi-locus genetic systems. Genet. Res. 21: 167–183. [Google Scholar]
- Chen J., Nolte V., Schlötterer C., 2015. Temperature stress mediates decanalization and dominance of gene expression in Drosophila melanogaster. PLoS Genet. 11: e1004883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. K., Kim S. C., 2007. Environmental effects on gene expression phenotype have regional biases in the human genome. Genetics 175: 1607–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T., 1944 Chromosomal races in Drosophila pseudoobscura and Drosophila persimilis. Pub 554:47–144, Carnegie Institute, Washington, DC.
- Dobzhansky T., 1948a Genetics of natural populations. Xvi. Altitudinal and seasonal changes produced by natural selection in certain populations of Drosophila pseudoobscura and Drosophila persimilis. Genetics 33: 158–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T., 1948b Genetics of natural populations. Xviii. Experiments on chromosomes of Drosophila pseudoobscura from different geographic regions. Genetics 33: 588–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T., 1950. Genetics of natural populations. Xix. Origin of heterosis through natural selection in populations of Drosophila pseudoobscura. Genetics 35: 288–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T., Queal M. L., 1938. Genetics of natural populations. II. Genic variation in populations of Drosophila pseudoobscura inhabiting isolated mountain ranges. Genetics 23: 463–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T., Sturtevant A. H., 1938. Inversions in the chromosomes of Drosophila pseudoobscura. Genetics 23: 28–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. G. and C. C. Epling 1944 Contributions to the genetics, taxonomy, and ecology of Drosophila pseudoobscura and its relatives. Carnegie Institute, Washington, DC.
- Dobzhansky T., Levene H., 1951. Development of heterosis through natural selection in experimental populations of Drosophila pseudoobscura. Am. Nat. 85: 247–264. [Google Scholar]
- Evans A. L., Mena P. A., McAllister B. F., 2007. Positive selection near an inversion breakpoint on the Neo-X chromosome of Drosophila americana. Genetics 177: 1303–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay J. C., McCullough H. L., Sniegowski P. D., Eisen M. B., 2004. Population genetic variation in gene expression is associated with phenotypic variation in Saccharomyces cerevisiae. Genome Biol. 5: R26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M. W., 1972. Selection for linkage modification. I. Random mating populations. Theor. Popul. Biol. 3: 324–346. [DOI] [PubMed] [Google Scholar]
- Feuk L., MacDonald J. R., Tang T., Carson A. R., Li M., et al. , 2005. Discovery of human inversion polymorphisms by comparative analysis of human and chimpanzee DNA sequence assemblies. PLoS Genet. 1: e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller Z.L., G. DHaynes, DZhu, MBatterton, HChao et al, 2014. Evidence for stabilizing selection on codon usage in chromosomal rearrangements of Drosophila pseudoobscura. G3 (Bethesda) 4: 2433–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison, E., and G. Marth, 2012 Haplotype-based variant detection from short-read sequencing. ArXiv:1207.3907 Q-Bio. Available at: http://arxiv.org/abs/1207.3907.
- Gibson G., Riley-Berger R., Harshman L., Kopp A., Vacha S., et al. , 2004. Extensive sex-specific nonadditivity of gene expression in Drosophila melanogaster. Genetics 167: 1791–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y., Oshlack A., Rifkin S. A., 2006. Natural selection on gene expression. Trends Genet. TIG 22: 456–461. [DOI] [PubMed] [Google Scholar]
- Good T. P., Ellis J. C., Annett C. A., Pierotti R., 2000. Bounded hybrid superiority in an avian hybrid zone: effects of mate, diet, and habitat choice. Evolution 54: 1774–1783. [DOI] [PubMed] [Google Scholar]
- Haldane J. B. S., 1957. The conditions for coadaptation in polymorphism for inversions. J. Genet. 55: 218–225. [Google Scholar]
- Hartl D. L., 1975. Genetic dissection of segregation distortion II. Mechanism of suppression of distortion by certain inversions. Genetics 80: 539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman E. A., Goodisman M. A. D., 2007. Gene expression and the evolution of phenotypic diversity in social wasps. BMC Biol. 5: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A., 2008. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M., 2010. How and why chromosome inversions evolve. PLoS Biol. 8: pii: e1000501. Available at: http://www.ncbi.nlm.nih.gov/pubmed/20927412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M., Barton N., 2006. Chromosome inversions, local adaptation and speciation. Genetics 173: 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K.-I., 1967. Likelihood of establishing newly induced inversion chromosomes in small populations. Cienc. Cult. 19: 67–77. [Google Scholar]
- Kovacevic M., Schaeffer S. W., 2000. Molecular population genetics of X-linked genes in Drosophila pseudoobscura. Genetics 156: 155–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn B. T., Page D. C., 1999. Four evolutionary strata on the human X chromosome. Science 286: 964–967. [DOI] [PubMed] [Google Scholar]
- Lande R., 1984. The expected fixation rate of chromosomal inversions. Evolution 38: 743–752. [DOI] [PubMed] [Google Scholar]
- Lemos B., Araripe L. O., Fontanillas P., Hartl D. L., 2008. Dominance and the evolutionary accumulation of cis- and trans-effects on gene expression. Proc. Natl. Acad. Sci. USA 105: 14471–14476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine R. P., 1956. Crossing over and inversions in coadapted systems. Am. Nat. 90: 41–45. [Google Scholar]
- Levine R. P., Levine E. E., 1954. The genotypic control of crossing over in Drosophila pseudoobscura. Genetics 39: 677–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K., Shi W., 2013. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41: e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K., Shi W., 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. [DOI] [PubMed] [Google Scholar]
- Lloyd V. K., Sinclair D. A., Grigliatti T. A., 1999. Genomic imprinting and position-effect variegation in Drosophila melanogaster. Genetics 151: 1503–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobeck A. K., 1950. Physiographic Diagram of North America, Geographical Press, Columbia University, NY. [Google Scholar]
- Lund S. P., Nettleton D., McCarthy D. J., Smyth G. K., 2012. Detecting differential expression in RNA-sequence data using quasi-likelihood with shrunken dispersion estimates. Stat. Appl. Genet. Mol. Biol. 11 Available at: http://www.degruyter.com/view/j/sagmb.2012.11.issue-5/1544–6115.1826/1544–6115.1826.xml. [DOI] [PubMed] [Google Scholar]
- McManus C. J., Coolon J. D., Duff M. O., Eipper-Mains J., Graveley B. R., et al. , 2010. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res. 20: 816–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore W. S., 1977. An evaluation of narrow hybrid zones in vertebrates. Q. Rev. Biol. 52: 263–277. [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B., 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Nei M., Kojima K.-I., Schaffer H. E., 1967. Frequency changes of new inversions in populations under mutation-selection equilibria. Genetics 57: 741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor M. A. F., Grams K. L., Bertucci L. A., Reiland J., 2001. Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. USA 98: 12084–12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitski E., 1951. Non-random disjunction in Drosophila. Genetics 36: 267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T., 1971. Associative overdominance caused by linked deleterious mutations. Genet. Res. 18: 227–286. [PubMed] [Google Scholar]
- Ohta T. and K. I. Kojima, 1968. Survival probabilities f new inversions in large populations. Biometrics 24: 501–516. [PubMed] [Google Scholar]
- Otto S. P., Barton N. H., 2001. Selection for recombination in small populations. Evolution 55: 1921–1931. [DOI] [PubMed] [Google Scholar]
- Painter T. S., 1934. A new method for the study of chromosome aberrations and the plotting of chromosome maps in Drosophila melanogaster. Genetics 19: 175–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixoto L., Risso D., Poplawski S. G., Wimmer M. E., Speed T. P., et al. , 2015. How data analysis affects power, reproducibility and biological insight of RNA-seq studies in complex datasets. Nucleic Acids Res. 43: 7664–7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J. R., 1992. Inversion polymorphisms in Drosophila pseudoobscura and Drosophila persimilis, pp. 73–126 in Drosophila Inversion Polymorphism, CRC Press, Ann Arbor, MI. [Google Scholar]
- Puig M., Cáceres M., Ruiz A., 2004. Silencing of a gene adjacent to the breakpoint of a widespread Drosophila inversion by a transposon-induced antisense RNA. Proc. Natl. Acad. Sci. USA 101: 9013–9018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S., Liu Y., Bettencourt B. R., Hradecky P., Letovsky S., et al. , 2005. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res. 15: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M. A., Hallas M. E., Lewontin R. C., 1989. Distinguishing the forces controlling genetic variation at the Xdh locus in Drosophila pseudoobscura. Genetics 123: 359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D., Ngai J., Speed T. P., Dudoit S., 2014. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 32: 896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K., 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Gracia A., Vieira F. G., Rozas J., 2009. Molecular evolution of the major chemosensory gene families in insects. Heredity 103: 208–216. [DOI] [PubMed] [Google Scholar]
- Schaeffer S. W., 2008. Selection in heterogeneous environments maintains the gene arrangement polymorphism of Drosophila pseudoobscura. Evolution 62: 3082–3099. [DOI] [PubMed] [Google Scholar]
- Schaeffer S. W., Miller E. L., 1992. Estimates of gene flow in Drosophila pseudoobscura determined from nucleotide sequence analysis of the alcohol dehydrogenase region. Genetics 132: 471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer S. W., Goetting-Minesky M. P., Kovacevic M., Peoples J. R., Graybill J. L., et al. , 2003. Evolutionary genomics of inversions in Drosophila pseudoobscura: Evidence for epistasis. Proc. Natl. Acad. Sci. USA 100: 8319–8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer S. W., Bhutkar A., McAllister B. F., Matsuda M., Matzkin L. M., et al. , 2008. Polytene chromosomal maps of 11 Drosophila species: the order of genomic scaffolds inferred from genetic and physical maps. Genetics 179: 1601–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefke B., Emerson J. J., Wang T.-Y., Lu M.-Y. J., Hsieh L.-C., et al. , 2013. Inheritance of gene expression level and selective constraints on trans- and cis-regulatory changes in yeast. Mol. Biol. Evol. 30: 2121–2133. [DOI] [PubMed] [Google Scholar]
- Smith J. M., Haigh J., 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35. [PubMed] [Google Scholar]
- Smith, R. H., 1977 Monoterpenes of Ponderosa Pine Xylem Resin in Western United States. Department of Agriculture, Economic Research Service, Washington, DC. Available at: https://ideas.repec.org/p/ags/uerstb/158103.html.
- Sperlich, D., and P. Pfriem, 1986 Chromosomal polymorphism in natural and experimental populations. In: The Genetics and Biology of Drosophila, 3rd Ed. pp. 257–309. Academic Press, London. [Google Scholar]
- Stefansson H., Helgason A., Thorleifsson G., Steinthorsdottir V., Masson G., et al. , 2005. A common inversion under selection in Europeans. Nat. Genet. 37: 129–137. [DOI] [PubMed] [Google Scholar]
- Sturtevant A. H., 1921. A case of rearrangement of genes in Drosophila. Proc. Natl. Acad. Sci. USA 7: 235–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussarellu R., Huvet A., Lapègue S., Quillen V., Lelong C., et al. , 2015. Additive transcriptomic variation associated with reproductive traits suggest local adaptation in a recently settled population of the Pacific oyster, Crassostrea gigas. BMC Genomics 16: 808 Available at: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4613751/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadi L., Meyer M., Weiser J., Stein L. D., Reimand J., 2016. Impact of knowledge accumulation on pathway enrichment analysis. bioRxiv, 049288 DOI: http://dx.doi.org/10.1101/049288. [Google Scholar]
- Wallace A. G., Detweiler D., Schaeffer S. W., 2011. Evolutionary history of the third chromosome gene arrangements of Drosophila pseudoobscura inferred from inversion breakpoints. Mol. Biol. Evol. 28: 2219–2229. [DOI] [PubMed] [Google Scholar]
- Wallace A. G, D. Detweiler, S. W. Schaeffer, 2013 Molecular population genetics of inversion breakpoint regions in Drosophila pseudoobscura. G3 (Bethesda) 3: 1151–1163. [DOI] [PMC free article] [PubMed]
- Wallace B., 1968. Topics in Population Genetics, W. W. Norton, New York. [Google Scholar]
- Wang Z., Gerstein M., Snyder M., 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman M., 1982. Evolution of the repleta group, pp. 61–139 in The Genetics and Biology of Drosophila, Vol. 3b. Academic Press, New York. [Google Scholar]
- Wills C., 1975. Marginal overdominance in Drosophila. Genetics 81: 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Wit J., Svetec N., Begun D. J., 2015. Parallel gene expression differences between low and high latitude populations of Drosophila melanogaster and D. simulans. PLoS Genet. 11: e1005184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Fly strains are available upon request. Raw sequencing reads are available from the National Center for Biotechnology Information as a BioProject (PRJNA326536).