Abstract
Transcription of nonprotein-coding DNA is widespread in eukaryotes and plays important regulatory roles for many genes, including genes that are misregulated in cancer cells. Its pervasiveness presents the potential for a wealth of diverse regulatory roles for noncoding transcription. We previously showed that the act of transcribing noncoding DNA (ncDNA) across the promoter of the protein-coding SER3 gene in Saccharomyces cerevisiae positions nucleosomes over the upstream activating sequences, leading to strong repression of SER3 transcription. To explore the possibility of other regulatory roles for ncDNA transcription, we selected six candidate S. cerevisiae genes that express ncRNAs over their promoters and analyzed the regulation of one of these genes, ECM3, in detail. Because noncoding transcription can lead to changes in the local chromatin landscape that impinge on the expression of nearby coding genes, we surveyed the effects of various chromatin regulators on the expression of ECM3. These analyses identified roles for the Paf1 complex in positively regulating ECM3 transcription through methylation of histone H3 at lysine 4 (K4) and for Paf1 in controlling the pattern of intergenic transcription at this locus. By deleting a putative promoter for the noncoding transcription unit that lies upstream of ECM3, we provide evidence for a positive correlation between intergenic transcription and ECM3 expression. Our results are consistent with a model in which cotranscriptional methylation of histone H3 K4, mediated by the Paf1 complex and noncoding transcription, leads to activation of ECM3 transcription.
Keywords: noncoding RNA, histone methylation, RNA polymerase II, cryptic unstable transcripts, Paf1 complex
Widespread pervasiveness of transcription is a common feature of eukaryotic genomes. The importance of pervasive nonprotein-coding transcription is highlighted by the results of the human genome project, which revealed that over 80% of the human genome displays biochemical activities associated with transcription in at least one cell type, even though only about 1% of the transcribed regions contain protein-coding exons (Venter et al. 2001; Borel et al. 2008; ENCODE Project Consortium 2012). Noncoding transcripts carry out a diverse array of regulatory functions. For example, microRNAs (miRNAs) associate with Argonaute proteins to regulate gene expression at a posttranscriptional level or by directing chromatin modifications (Bartel 2004). Several long noncoding RNAs (lncRNAs), such as Xist and HOTAIR, two important developmental regulators, associate with and direct the Polycomb repressive complex, PRC2, to specific genetic loci. PRC2 is then able to alter the local chromatin state and lead to the regulation of gene expression (Rinn et al. 2007; Lee 2009; Tsai et al. 2010; Simon and Kingston 2013). PCGEM1, a lncRNA associated with prostate cancer, directly binds the transcription factor c-Myc, activates transcription of c-Myc target genes, and regulates several metabolic pathways including nucleotide and lipid biosynthetic pathways and the tricarboxylic acid cycle (Hung et al. 2014).
In these examples, the noncoding RNA (ncRNA) molecule itself plays a regulatory role. The act of transcribing ncDNA can also alter the local chromatin environment and the regulation of neighboring genes. Transcription can alter the occupancy and positions of nucleosomes, posttranslational histone modifications, and potentially higher order chromatin structures. Our previous studies in Saccharomyces cerevisiae revealed an interesting role for the act of transcribing ncDNA, SRG1, at the promoter of a protein-coding gene, SER3, which represses transcription of SER3 (Martens et al. 2004, 2005). The mechanism by which SRG1 transcription represses SER3 requires histone chaperones, Spt6 and the FACT complex, that travel with RNA polymerase II (Pol II) during transcription elongation. During transcription of SRG1, these histone chaperones place nucleosomes over the upstream regulatory sequences for the SER3 gene, creating a barrier that prevents the transcriptional machinery from accessing the SER3 promoter (Hainer et al. 2011).
Several other cases of gene regulation by ncDNA transcription have been reported. In Schizosaccharomyces pombe, the fbp1+ gene is activated by stepwise displacement of nucleosomes over the fbp1+ promoter, which is mediated by induction of a series of long noncoding transcripts across the promoter (Hirota et al. 2008). In maturing B cells, noncoding transcription at the IgL loci is required to evict histone H2A/H2B dimers to allow recombination factors to access the DNA for V(D)J recombination (Bevington and Boyes 2013). This example demonstrates that the regulatory potential for noncoding transcription is not limited to transcription and may be extended to all DNA-templated processes. In these examples, transcription of ncDNA exerts its regulatory effect by altering nucleosome occupancy.
At some loci, noncoding transcription has been shown to alter chromatin structure through posttranslational histone modifications. Transcribed loci display a characteristic set of posttranslational histone modifications, including acetylation of histones over promoter regions, trimethylation of H3 lysine 4 (H3 K4me3) at 5′ ends of transcription units, monoubiquitylation of H2B (at K123 in S. cerevisiae or K120 in humans) and methylation of H3 K79 throughout the bodies of transcription units, and methylation of H3 K36 at the 3′ ends of transcription units (reviewed in Smolle and Workman 2013). The histone modification states of nonprotein-coding transcribed regions can influence the expression of nearby protein-coding genes. For example, trimethylation of H3 K4 due to transcription of ncDNA at the GAL1-10 locus is required for histone deacetylation, which leads to repression of GAL1 and GAL10 in the presence of glucose (Houseley et al. 2008; Pinskaya et al. 2009). Two other S. cerevisiae genes, DCI1 and DUR3, are regulated by noncoding transcription across their promoters through a mechanism in which methylation of H3 K4 and subsequent Set3-mediated deacetylation of histones leads to gene repression (Kim et al. 2012). Transcriptional interference, as has been observed at the yeast IME4 gene, presents an alternative mode of regulation by intergenic transcription that does not require alteration of chromatin but can be explained by the collision of traveling Pol II complexes (Prescott and Proudfoot 2002; Hongay et al. 2006; Gelfand et al. 2011; Hobson et al. 2012).
In this study, we sought to expand our knowledge of gene regulatory mechanisms by focusing on genes that harbor noncoding transcription units in their promoters and exploring the importance of both chromatin regulatory proteins and the ncDNA in the regulation of these genes. To begin, we identified candidate genes that might be regulated by noncoding transcription and selected one of these genes, ECM3, for detailed study. Our analyses of ECM3 expression revealed an integral role for the Paf1 complex, H3 K4 methylation, and two histone acetyltransferases in the positive regulation of ECM3. In addition, our findings indicate that transcription of the intergenic ncDNA at ECM3 correlates with expression of the gene, potentially through establishment of a permissive chromatin structure.
Materials and Methods
S. cerevisiae strains and media
S. cerevisiae strains used in this study are listed in Supplemental Material, Table S1. The strains used to perform the anchor away experiment are W303 derivatives purchased from Euroscarf or generously provided by Patrick Cramer (Schulz et al. 2013). For anchor away experiments, cells were grown at 30° in YPD medium (1% yeast extract, 2% peptone, and 2% dextrose) until cultures reached a density of 2 × 107 cells per ml. Rapamycin was then added to cultures for 1 hr at a final concentration of 1 μg/ml from a stock of 1 mg/ml rapamycin suspended in ethanol. All other strains used in this study are derived from a GAL2+ S288C isolate using standard genetic crosses and transformations (Winston et al. 1995). The EUC1 promoter deletions were made by two-step integration of an HA-URA3-HA cassette that was PCR-amplified from the plasmid pMPY-3XHA (Schneider et al. 1995). This resulted in strains where a portion of the EUC1 promoter has been replaced with a DNA sequence encoding one copy of the 3XHA tag, which is serving as spacer DNA. Cells were grown at 30° in YPD medium until cultures reached a density of 1–2 × 107 cells per ml for isolation of either RNA or chromatin for use in northern blotting, primer extension, and chromatin immunoprecipitation (ChIP) analyses.
Northern blot analysis
Northern blot analyses were performed using 20 μg total RNA samples resolved in gels containing 2% agarose, 6.5% formaldehyde, and 1 × MOPS as previously described (Ausubel 1987). Double-stranded probes were generated by random-primed labeling and single-stranded probes were generated by asymmetric PCR with α-32P-dATP (Rio 2011). Probe templates were amplified from genomic DNA to contain the following sequences relative to the +1 start codon of the protein-coding gene at each locus: EUC1 (−541 to −100), ECM3 (+545 to +976), ARO2-CUT (−494 to −49), ARO8-CUT (−429 to −28), CLN3-CUT (−671 to −321), FET4-CUT (−479 to −114), KNH1-CUT (−496 to −235), and SCR1 (−182 to +284). SCR1 RNA levels serve as an internal loading control. The oligonucleotides used to generate probe templates are listed in Table S2. Images were generated by phosphorimaging and quantified using ImageJ software. For each experiment, the data from at least three biological replicates were averaged.
Primer extension analysis
Primer extension assays were performed as previously described using 20 μg total RNA samples (Ausubel 1987). Sequencing reactions were performed with Sequenase following the manufacturer’s guidelines (Affymetrix USB) using a purified PCR product as a template. Oligonucleotides were gel-purified and end-labeled with γ-32P-ATP and T4 polynucleotide kinase using standard protocols (Ausubel 1987). The EUC1 and ECM3 transcription start sites were mapped by primer extension using two different oligonucleotides for each major start site listed in Table S2.
ChIP
Chromatin was isolated and sheared by sonication using a Misonix 3000 sonicator as previously described (Shirra et al. 2005). Immunoprecipitations were performed by incubating sheared chromatin with 5 μl antisera to histone H3 (Tomson et al. 2011) or 2.5 μl of antibody to H3 K4me3 (Active Motif, catalog number 39159) at 4° overnight, followed by precipitation using Protein A sepharose beads (GE Healthcare) for 2 hr at 4°. All ChIP results were determined by quantitative PCR amplification of immunoprecipitated DNA compared to input DNA. Real-time PCR reactions were performed using SYBR green reagents (Fermentas) and a Step One Plus instrument (Applied Biosystems). The oligonucleotides used for qPCR amplification are listed in Table S2. Data were analyzed using the Pfaffl relative quantitation method (Pfaffl 2001). H3K4me3 occupancy values were normalized to total H3 occupancy values.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Identification of genes with promoter-localized cryptic unstable transcripts (CUTs)
With the goal of identifying chromatin-mediated gene regulatory mechanisms associated with ncDNA transcription, we selected candidate genes that might be regulated by transcription across their promoter regions. Our selection criteria for candidate genes were intentionally simple to minimize the introduction of bias. The only criterion that we required of a candidate gene was the presence of a noncoding transcript over the promoter of a protein-coding gene in a tandem orientation, resembling the structure of the SRG1-SER3 locus. Several types of noncoding transcripts have been characterized in S. cerevisiae. We focused our studies on cryptic unstable transcripts (CUTs), which are short transcripts that are terminated by the Nrd1-Nab3-Sen1 termination pathway, polyadenylated by the TRAMP complex, and degraded by the nuclear exosome (Arndt and Reines 2015; Porrua and Libri 2015). Enabling their detection, degradation of CUTs can be disrupted by deletion of TRF4, which encodes a subunit of the TRAMP complex, or RRP6, which encodes a catalytic subunit of the nuclear exosome (Wyers et al. 2005; Arigo et al. 2006; Davis and Ares 2006). Although we anticipate that regulation by intergenic transcription is not limited to the nature of the intergenic transcript, we limited this study to CUTs, as they are rapidly degraded. Due to the instability of CUTs in wild-type cells, we reasoned that regulatory events associated with CUT expression would likely be due to the act of transcription rather than the ncRNA molecule itself. We selected genes from microarray expression data, which mapped CUTs genome-wide (Neil et al. 2009), and confirmed that Pol II occupancy was detected across the promoters of these genes in genome-wide ChIP studies (Steinmetz et al. 2006). The genes we selected for initial characterization were ARO2, ARO8, CLN3, ECM3, FET4, and KNH1.
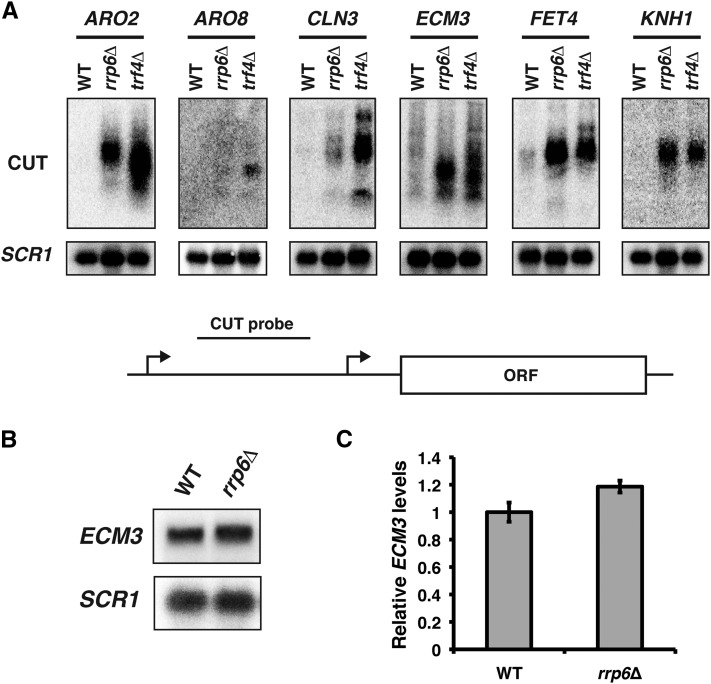
To confirm the microarray expression data in our strain background, we performed northern analyses to detect CUT expression over the candidate gene promoters. Using wild-type strains and strains lacking either Rrp6 or Trf4, we were able to detect CUTs over the promoter regions of all six candidate genes (Figure 1A).
Figure 1.
Confirmation of CUT expression at candidate gene promoters. (A) Northern analysis was performed using RNA isolated from a wild-type strain (FY4) or strains where CUTs are stabilized, either rrp6Δ (YJ744) or trf4Δ (KY1975) mutants. Probes were designed to detect transcripts produced upstream of the neighboring protein-coding gene as diagrammed below. SCR1 serves as a loading control. (B) Representative northern analysis of ECM3 mRNA levels in a wild-type strain (YJ1125) compared to an rrp6Δ (YJ1126) strain. SCR1 serves as a loading control. (C) Quantitation of ECM3 transcript levels in (B) relative to wild-type levels from three biological replicates. Error bars represent the SEM. CUT, cryptic unstable transcript; mRNA, messenger RNA; ORF, open reading frame; WT, wild-type.
We focused our attention on the ECM3 locus for mechanistic characterization. ECM3 is a nonessential protein-coding gene that was first identified in a screen for sensitivity to the cell wall stressor, calcofluor white (Lussier et al. 1997). For this reason, it is thought that ECM3 might be involved in cell wall maintenance. We detected two major transcription start sites for ECM3 (Figure S1), which were also detected by TIF-Seq analysis (Pelechano et al. 2013) and which overlap with the 3′ end of the upstream CUT as observed in microarray expression data (Neil et al. 2009). This presented an opportunity to observe isoform-specific regulation. We refer to the CUT across the ECM3 promoter as the ECM3 upstream CUT (EUC1). Upon deletion of the RRP6 gene, we did not detect a significant change in ECM3 expression, suggesting that ECM3 expression is not regulated by stability of the EUC1 transcript itself (Figure 1, B and C).
The Paf1 complex and methylation of H3 K4 positively regulate ECM3 expression
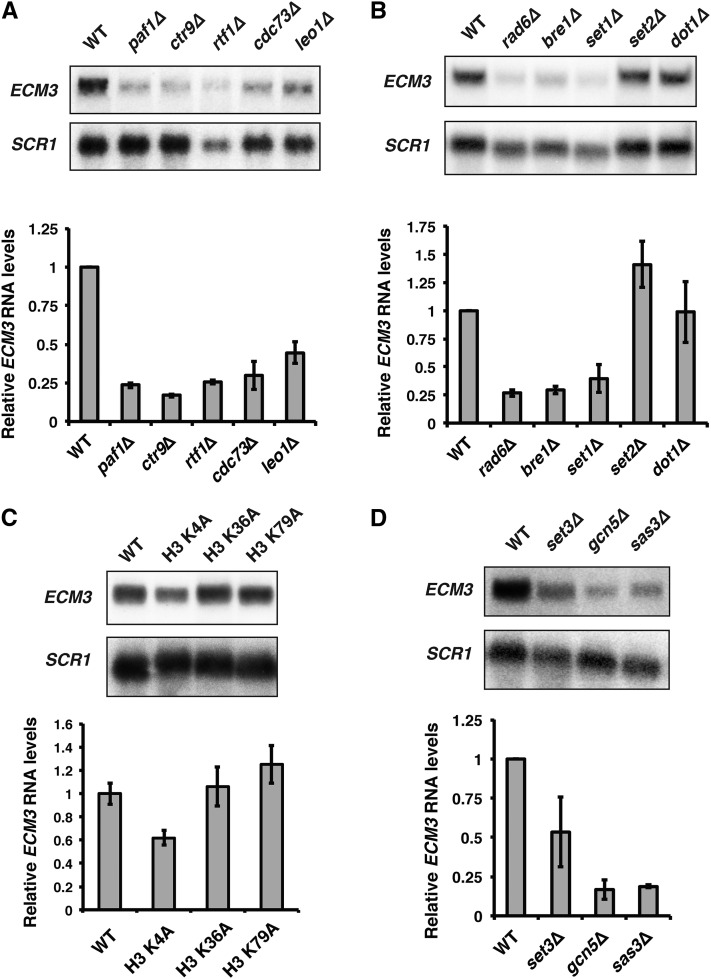
Because noncoding transcription can alter the local chromatin environment in a way that impinges upon protein-coding gene regulation (Fu 2014; Rinn and Guttman 2014; Yang et al. 2014), we surveyed various chromatin regulators for their effects on ECM3 expression. Through these analyses, we determined that the Paf1 complex positively regulates ECM3 expression, as deletion of any one of the five members of the Paf1 complex results in reduced ECM3 transcript levels (Figure 2A).
Figure 2.
The Paf1 complex and methylation of H3 K4 positively regulate ECM3 expression. (A) Representative northern blot analysis of ECM3 transcript levels in a wild-type strain (FY4) or strains lacking one of the five subunits of the Paf1 complex, either paf1∆ (YJ807), ctr9∆ (KY2170), rtf1∆ (YJ788), cdc73∆ (KY2171), or leo1∆ (KY1805). (B) Representative northern blot analysis of ECM3 transcript levels in a wild-type strain (FY4) and strains where the genes encoding histone modifiers that work in concert with the Paf1 complex have been deleted (rad6∆, KY1712; bre1∆, KY1713; set1∆, KY2720; set2∆, KY2723; and dot1∆, KY2725). (C) Representative northern blot analysis of ECM3 transcript levels in a wild-type control strain, lacking one copy of the genes for H3 and H4 (JDY86), and derivatives of JDY86 in which the only copy of the H3-H4 genes encodes the indicated amino acid substitution in H3. (D) Representative northern blot analysis of ECM3 transcript levels in strains lacking a subunit of the Set3 HDAC complex (set3∆, KY2782), the SAGA HAT complex (gcn5∆, KY1743), or the NuA3 HAT complex (sas3∆, ECY394) compared to wild-type levels (YJ1125). Quantitation below shows the average ECM3 mRNA levels relative to WT (set to 1) from at least three biological replicates. Error bars represent the SEM. SCR1 serves as a loading control. mRNA, messenger RNA; WT, wild-type.
The Paf1 complex associates with Pol II during transcription elongation and plays a crucial role in establishing patterns of histone modifications across transcribed loci (reviewed in Tomson and Arndt 2013). Therefore, to investigate the mechanism by which the Paf1 complex stimulates ECM3 transcription, we analyzed ECM3 mRNA levels in strains that lack histone modifiers known to depend on the Paf1 complex for function. Our results indicate that ubiquitylation of H2B K123 and methylation of H3 K4 are required to activate ECM3 as the enzymes that perform these modifications, the ubiquitin conjugase Rad6 and ubiquitin-protein ligase Bre1 for H2B K123 ubiquitylation and the H3 K4 methyltransferase Set1, are required for normal ECM3 expression (Figure 2B). Other histone modifications dependent on the Paf1 complex, H3 K79 di- and trimethylation and H3 K36 trimethylation, do not appear to play a role in stimulating ECM3 transcription. Mutations that delete DOT1 or SET2, which encode the H3 K79 and H3 K36 methyltransferases, respectively, did not reduce ECM3 mRNA levels (Figure 2B). Targeted mutation of H3 K4 to an unmodifiable residue, H3 K4A, reduced ECM3 transcript levels, consistent with the idea that Set1 stimulates ECM3 expression by methylating H3 K4 (Figure 2C). The magnitude of the decrease in ECM3 mRNA levels was greater for the set1∆ mutant than for the H3 K4A mutant. In addition to methylating H3 K4, it is possible that Set1 targets a nonhistone substrate that contributes to ECM3 expression. Alternatively, because the H3 K4A mutant has only a single copy of the H3 and H4 genes, reduced H3-H4 dosage may partially rescue the H3 K4A effect on ECM3 transcription. A previous study has shown that histone dosage impacts expression of a gene regulated by noncoding transcription as deletion of a single copy of the H3 and H4 genes, HHT1 and HHF1, results in SER3 derepression (Hainer and Martens 2011). Furthermore, histone dosage affects the expression of many genes in S. cerevisiae (Wyrick et al. 1999). Collectively, these data indicate that H2B K123 ubiquitylation by Rad6-Bre1, and subsequent H3 K4 methylation by Set1, positively regulate ECM3 expression.
Methylation of H3 K4 may serve as a signal to downstream activators of ECM3 expression. To test this, we explored the effects of complexes that recognize various H3 K4 methylation states on ECM3 expression. The recruitment of SAGA, NuA3, and NuA4 histone acetyltransferase complexes and the Set3 histone deacetylase complex is stimulated by methylated H3 K4 in S. cerevisiae (Martin et al. 2006; Ginsburg et al. 2009, 2014; Kim and Buratowski 2009; Bian et al. 2011). We evaluated the contributions of Set3, SAGA, and NuA3 to ECM3 regulation by northern analysis using strains lacking a single member of each complex. Consistent with previous studies showing that the Set3 histone deacetylase complex represses transcription (Kim et al. 2012), we did not observe a strong or consistent positive effect of SET3 on ECM3 expression (Figure 2D). The NuA3 and SAGA complexes have various effects on chromatin and have been generally associated with transcriptional activation. Deletion of the genes encoding Gcn5 and Sas3, the catalytic subunits of SAGA and NuA3, respectively, caused a dramatic reduction in ECM3 expression (Figure 2D). We note that ECM3 transcript levels are lower in gcn5Δ and sas3Δ mutants than in the set1Δ mutant. SAGA and NuA3 are both large, multisubunit complexes that associate with chromatin through multiple interactions. It is possible that the loss of H3 K4 methylation results in a partial loss of SAGA and NuA3 activity at ECM3 and that some function may be retained through other interactions.
Transcription of the EUC1 ncDNA does not require H3 K4 methylation
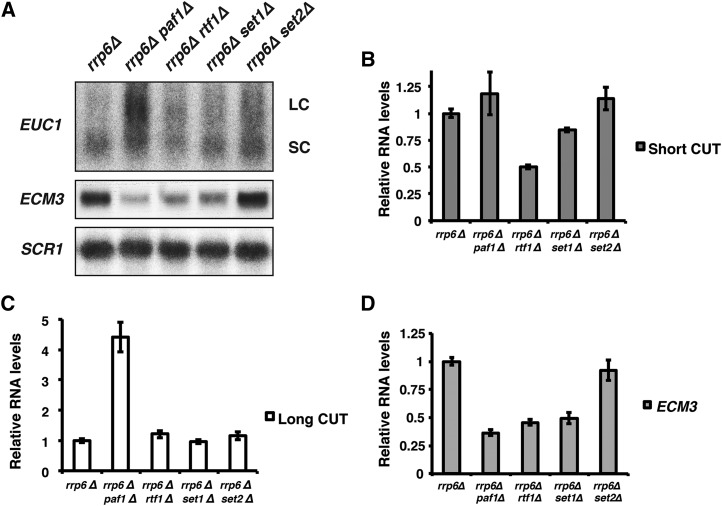
Expression of many genes is influenced by the regulation of neighboring transcription units. To determine if methylation of H3 K4 could be regulating ECM3 indirectly by acting farther upstream to regulate transcription of EUC1, we analyzed EUC1 RNA levels in strains lacking H3 K4 methylation. Northern analysis of EUC1 transcription in an rrp6∆ background showed only a slight reduction in EUC1 transcript levels in the absence of H3 K4 methylation (rrp6∆ set1∆ strain), although ECM3 mRNA levels remained low (Figure 3, A and B–D). In the rtf1∆ rrp6∆ double mutant, EUC1 levels were lower than those detected in rrp6∆ or set1∆ rrp6∆ strains (Figure 3, A and B–D), consistent with the observation that Rtf1 has functions in addition to regulating H3 K4 methylation (Warner et al. 2007). In the absence of Paf1, levels of the EUC1 isoform present in rrp6∆ cells, termed EUC1 SC for short CUT, were not significantly altered (Figure 3, A and B–D). However, a larger, more abundant EUC1 isoform, which we refer to as EUC1 LC for long CUT, was enriched in the paf1∆ rrp6∆ strain (Figure 3, A and C). Thus, although the set1∆, paf1∆, and rtf1∆ mutations all reduce ECM3 expression (Figure 3, A and D), they do not affect the pattern of EUC1 transcripts in the same way. Instead, the correlation with ECM3 expression is most clearly related to the loss of H3 K4 methylation. Fitting with this idea, a set2∆ rrp6∆ double mutant, which lacks H3 K36 methylation, does not show a change in EUC1 or ECM3 expression compared to the rrp6∆ control strain (Figure 3). These data do not exclude a role for EUC1 in ECM3 regulation; however, ECM3 transcript levels remain low in the absence of H3 K4 methylation even though an EUC1 transcript is produced in the rrp6∆ set1∆ double mutant strain. Thus, if EUC1 transcription is involved in ECM3 regulation, methylation of H3 K4 may work downstream of this effect. These results suggest that H3 K4 methylation does not indirectly regulate ECM3 expression by acting farther upstream to modulate the levels of EUC1 transcription.
Figure 3.
Effect of the Paf1 complex and histone methyltransferases on EUC1 transcription. (A) Northern blot analysis of RNA isolated from strains lacking RRP6 to stabilize CUTs and also lacking subunits of the Paf1 complex (paf1∆ rrp6∆, KY2727 and rtf1∆ rrp6∆, YJ1143) or histone methyltransferases (set1∆ rrp6∆, YJ1140 and set2∆ rrp6∆, YJ1146). The rrp6∆ control strain was YJ746. (B–D) Quantitation shows average transcript levels relative to those observed in the rrp6∆ strain, which were set to 1. Averaged results from three biological replicates for the EUC1 short isoform (SC, panel B), the EUC1 long isoform (LC, panel C), and the ECM3 ORF transcript (panel D) are shown. Error bars represent the SEM. SCR1 serves as a loading control. CUT, cryptic unstable transcript; mRNA, messenger RNA; ORF, open reading frame; WT, wild-type.
Loss of EUC1 transcription correlates with a reduction in ECM3 transcription
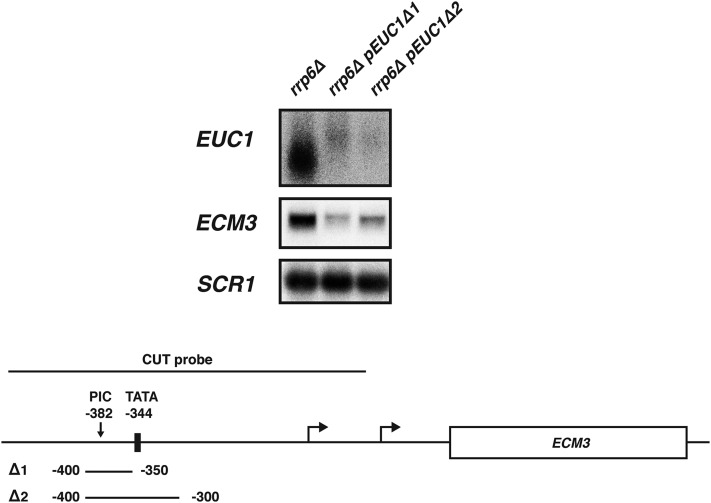
As the Paf1 complex associates with actively transcribing polymerases and H3 K4 methylation patterns are established during transcription, one possible model is that transcription of EUC1 functions to place this modification over the ECM3 promoter to positively regulate ECM3 expression. To investigate a possible role for EUC1 transcription in regulating ECM3 expression, we devised a strategy to disrupt the EUC1 promoter and eliminate CUT transcription. To identify putative EUC1 promoter elements, we performed a sequence alignment of the intergenic region 5′ of ECM3 in four related yeast species (S. cerevisiae, S. mikatae, S. bayanus, and S. paradoxus) (Figure S2). This analysis identified a conserved putative TATA sequence 344 nucleotides upstream of the ECM3 start codon. In ChIP-exo analyses, Rhee and Pugh (2012) identified a site of preinitiation complex (PIC) assembly 382 nucleotides upstream of the ECM3 start codon. Guided by this information, we generated promoter deletion mutations at the endogenous ECM3 locus using a two-step integration method, which replaced the putative EUC1 promoter sequence with an unrelated DNA sequence (177 bp encoding the 3XHA tag and linker DNA). Deletion 1 (pEUC1∆1) replaced bases −400–−350 and deletion 2 (pEUC1∆2) replaced bases −400–−300 relative to the ECM3 start codon. These deletions eliminate the detected site of PIC assembly and pEUC1∆2 also eliminates the putative TATA sequence (Rhee and Pugh 2012). Both deletions greatly reduce EUC1 transcript levels (Figure 4). A very small amount of a slightly larger transcript is detected with the EUC1 probe in RNA isolated from these promoter deletion strains. Based on the location of the northern probes, the nature of the promoter deletion mutations, and the size of the RNA product, this transcript may initiate downstream of the major EUC1 start sites and extend partially into ECM3. Because the amount of this transcript in pEUC1∆2 strains is very small, we continued our analyses with this mutation. Interestingly, both deletions also lower ECM3 mRNA levels (Figure 4). These results indicate that EUC1 transcription is positively correlated with ECM3 transcription.
Figure 4.
Deletion of putative EUC1 promoter sequences 5′ of ECM3 results in reduced ECM3 expression. Representative northern blot analysis of RNA isolated from strains carrying an RRP6 deletion and containing either a WT ECM3 locus (YJ1126) or the indicated EUC1 promoter deletion mutations. Promoter deletion mutations were introduced at the endogenous ECM3 locus and replaced either 50 bp (pEUC1∆1; region −400 to −350 deleted; YJ1128) or 100 bp (pEUC1∆2; region −400 to −300 deleted; YJ1131) upstream of the +1 start codon of ECM3, as diagrammed below. The locations of a PIC identified by Rhee and Pugh (2012) and a putative TATA sequence are indicated on the diagram below. SCR1 serves as a loading control. CUT, cryptic unstable transcript; PIC, preinitiation complex; WT, wild-type.
Loss of EUC1 transcription is associated with a reduction in H3 K4me3 levels at the ECM3 promoter
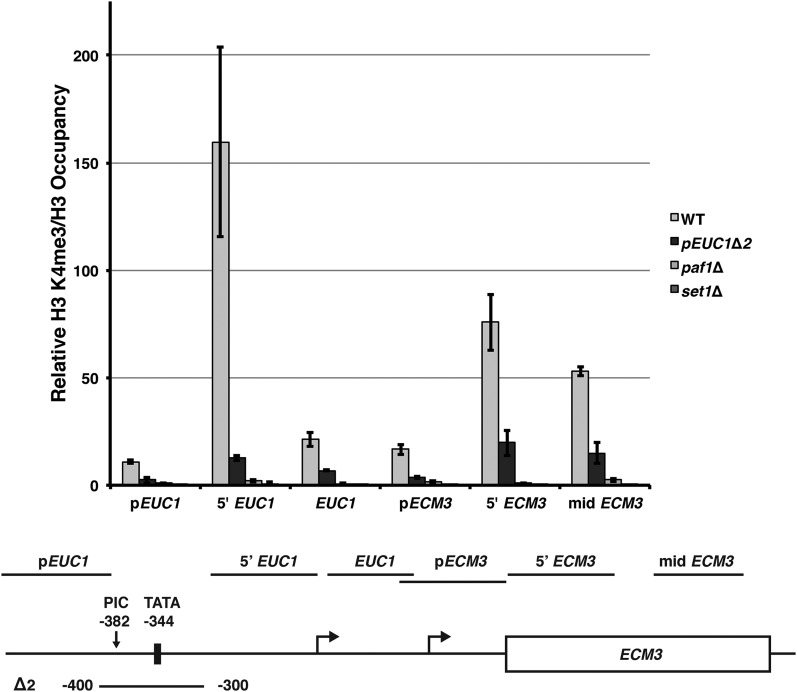
To further investigate the connections between H3 K4 methylation and ECM3 transcription, we analyzed the local chromatin landscape of the EUC1-ECM3 locus by ChIP analysis. In particular, we analyzed the levels of H3 K4 trimethylation (me3) in the presence and absence of EUC1 transcription as well as in paf1∆ and set1∆ mutants, which served as controls for loss of the modification. As expected, H3 K4me3 levels were nearly undetectable in the paf1∆ and set1∆ mutants (Figure 5). In wild-type cells, two peaks of H3 K4me3 were observed, one over the 5′ end of EUC1 and one over the 5′ end of ECM3. Interestingly, deletion of the putative EUC1 promoter in the pEUC1∆2 mutant reduced H3 K4me3 levels at both of these locations (Figure 5), consistent with the possibility that EUC1 transcription positively regulates ECM3 transcription by promoting H3 K4me3.
Figure 5.
EUC1 promoter deletion reduces H3K4me3 levels across the ECM3 locus. ChIP analysis of H3 K4me3 levels at the ECM3 locus. Immunoprecipitations were performed in biological triplicate using chromatin isolated from WT (FY4, FY5, and YJ1125), pEUC1∆2 (YJ1133, YJ1134, and YJ1135), paf1∆ (YJ807, YJ809, and KY1701), and set1∆ (KY1755, KY1715, and KY2722) strains. Enrichment of H3 K4me3 relative to input DNA was measured by qPCR and normalized to H3 occupancy. Error bars represent the SEM of three biological replicates. The relative locations of qPCR primers are indicated on the diagram below (the mid ECM3 primer set is not shown to scale). ChIP, chromatin immunoprecipitation; PIC, preinitiation complex; qPCR, quantitative polymerase chain reaction; WT, wild-type.
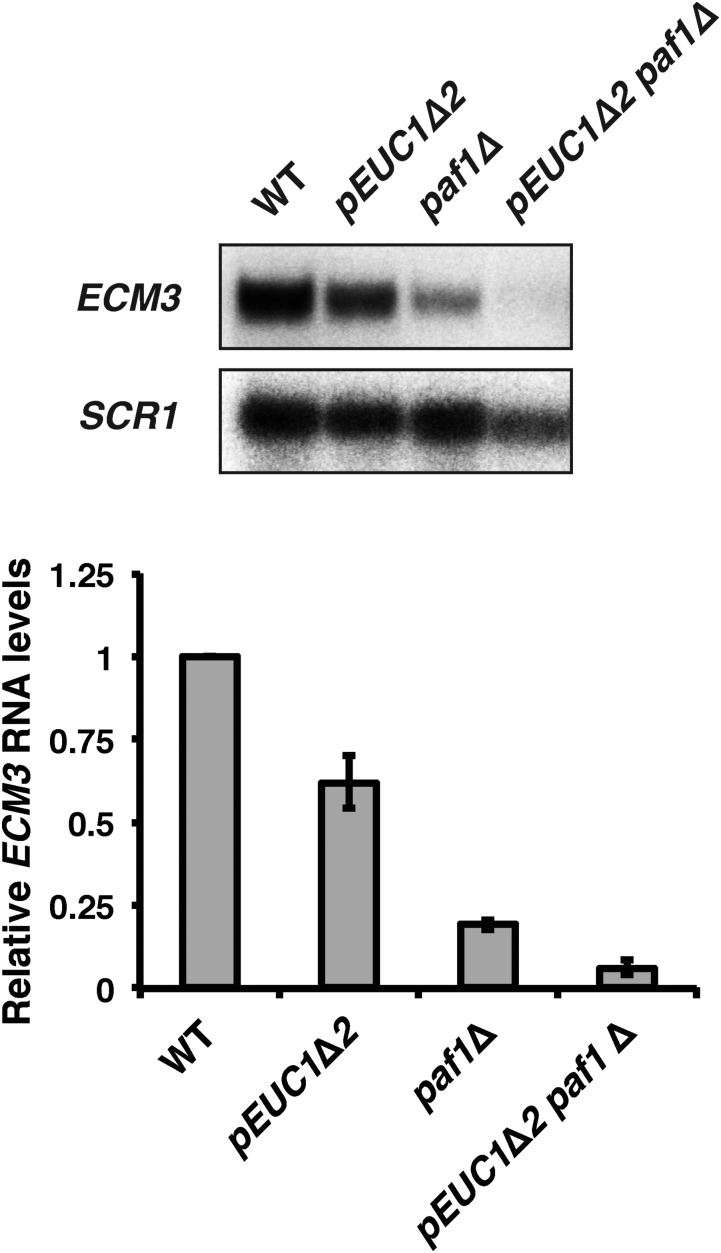
The Paf1 complex and EUC1 impact ECM3 transcription through independent pathways
To determine if Paf1 regulates ECM3 transcription in a manner that requires synthesis of the upstream CUT, we analyzed ECM3 transcript levels in pEUC1∆2 paf1∆ double mutants. In combination, deletion of the putative EUC1 promoter and deletion of PAF1 caused a greater defect in ECM3 expression than either mutation alone (Figure 6). Although the interpretation of these data are complicated somewhat by the fact that the pEUC1∆2 mutation is not equivalent to an EUC1 null allele, as it does not completely eliminate EUC1 transcription (Figure 4), the results are consistent with the Paf1 complex and EUC1 transcription having separable roles that contribute to ECM3 expression. Our data suggest that one shared role of EUC1 and the Paf1 complex is promoting methylation of H3 K4 (Figure 5), but each of these factors may have other contributions to ECM3 regulation that remain to be identified.
Figure 6.
The Paf1 complex and EUC1 impact ECM3 expression through independent pathways. Representative northern blot analysis of ECM3 transcript levels in a pEUC1∆2 strain (YJ1135), a paf1∆ strain (KY1701), and a pEUC1∆2 paf1∆ double mutant strain (YJ1138) compared to a WT strain (YJ1125). Bar graphs show the average ECM3 mRNA levels relative to WT strains (set to 1) from three biological replicates. Error bars represent the SEM. SCR1 serves as a loading control. WT, wild-type.
Defective termination of EUC1 is not sufficient to repress ECM3 expression
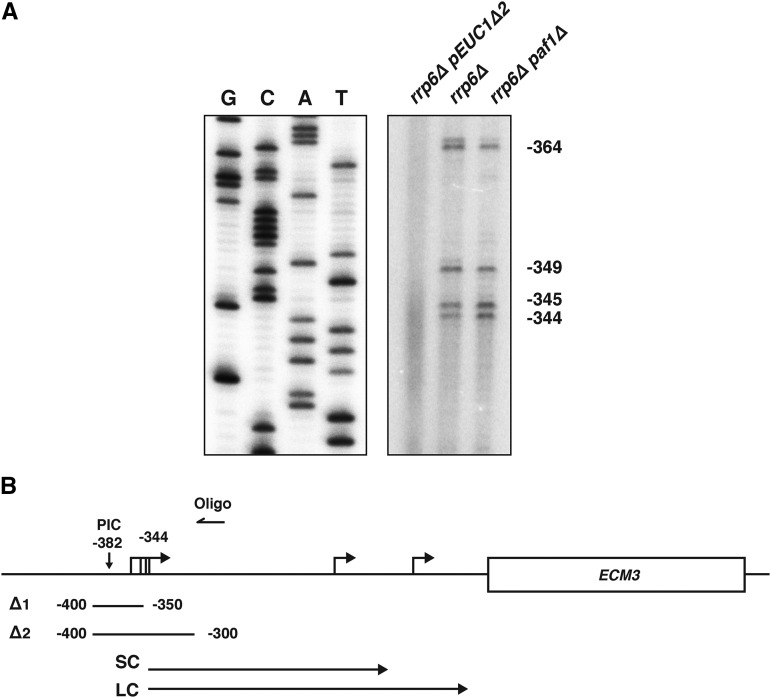
We next investigated whether the long isoform of EUC1 (LC), which is enriched in paf1∆ strains (Figure 3), plays a role in repressing ECM3 transcription. Northern analysis using strand-specific probes to detect sense transcripts showed that the LC isoform, like the SC isoform, is transcribed from the sense strand relative to the ECM3 ORF (Figure S3). Neither the SC nor the LC isoform is detected with an antisense strand-specific northern probe. In addition, the LC transcript is not detected in paf1∆ strains carrying a wild-type RRP6 allele, indicating that it is also unstable (Figure S3). Because Paf1 is required for proper termination by the Nrd1-Nab3-Sen1 pathway (Sheldon et al. 2005; Tomson et al. 2011, 2013), we hypothesized that the longer EUC1 isoform might be a read-through product of the smaller isoform that terminates farther downstream. In support of this, primer extension analysis revealed that paf1∆ rrp6∆ strains use the same EUC1 transcription start sites as an rrp6∆ control strain, which are located between 344 and 364 nucleotides upstream of the ECM3 start codon (Figure 7).
Figure 7.
Evidence that the short and long CUT isoforms of EUC1 initiate from the same transcription start sites. (A) Primer extension analysis of the 5′ ends of EUC1 transcripts produced in strains that express the EUC1 SC transcript (rrp6∆, YJ746) or both the EUC1 SC and LC transcripts (paf1∆ rrp6∆, KY2729). The pEUC1∆2 mutant (YJ1130) was used as a negative control as this strain displays severely reduced EUC1 transcription. A DNA sequencing ladder is shown on the left. (B) A schematic diagram of the ECM3 locus with the positions of the upstream EUC1 CUTs and the pEUC1∆1 and pEUC1∆2 mutations indicated. For simplicity, the EUC1 SC and LC isoforms are diagrammed as initiating at a single start site to reflect that the closely positioned start sites detected in (A) do not appear as distinct isoforms by northern blot analysis. CUT, cryptic unstable transcript; LC, long CUT; SC, short CUT; PIC, preinitiation complex.
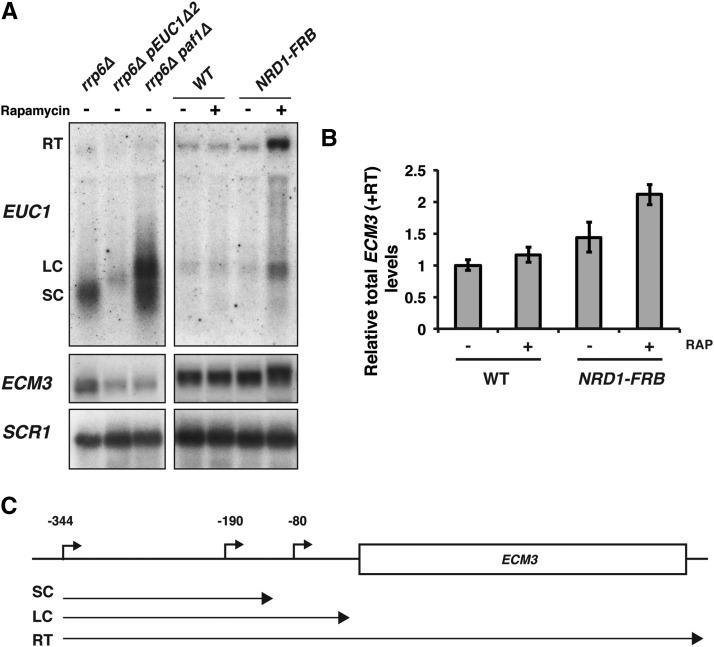
We also analyzed the potential effect of alternative EUC1 termination by depletion of Nrd1 from the nucleus by the anchor-away method (Haruki et al. 2008). Upon depletion of Nrd1 from the nucleus by addition of rapamycin to a NRD1-FRB strain, a prominent EUC1 transcript, which comigrated with the LC isoform observed in the paf1∆ background, was observed. This result is consistent with the idea that the LC isoform detected in paf1∆ rrp6∆ strains arises from transcriptional read-through of a CUT terminator. In the Nrd1-depleted strain, an additional transcript was detected by the EUC1 probe and this transcript corresponds to the size of an EUC1 transcript reading through (RT) the entire ECM3 ORF (Figure 8A). The relative locations of these isoforms based on northern probe hybridization and relative size are diagrammed in Figure 8C. The RT isoform overlaps with the ECM3 mRNA bands in the northern blot in Figure 8A and is included in quantitation of ECM3 levels. Upon addition of rapamycin, there is no significant change in ECM3 transcript levels in the control strain used for the anchor away technique (labeled WT in Figure 8). There is also no significant fold change in ECM3 transcript levels in the NRD1-FRB strain after addition of rapamycin (Figure 8B). This result indicates that a defect in Nrd1-dependent termination of the EUC1 CUT does not significantly change the overall level of ECM3 transcripts. Although we have not ruled out a role for the longer EUC1 isoform in fine-tuning ECM3 expression, these data indicate that transcription of the long EUC1 isoform alone is not sufficient to regulate ECM3 expression under these conditions. Therefore, of the two described functions of the Paf1 complex that we have explored, stimulation of H3 K4 methylation is important for ECM3 transcription, while regulation of EUC1 termination plays little if any role in the expression of ECM3. As indicated by our genetic data (Figure 6), we anticipate that the Paf1 complex has multiple functions that contribute to ECM3 regulation that will be interesting to explore.
Figure 8.
Disruption of CUT termination produces the long EUC1 isoform. (A) Representative northern blot analysis comparing EUC1 and ECM3 transcript patterns in rrp6∆ (YJ746), rrp6∆ pEUC1∆2 (YJ1131), and paf1∆ rrp6∆ (KY2729) strains to those of a strain in which Nrd1 (OKA292) has been depleted from the nucleus by the anchor away method. An untagged anchor away strain was used as a control (OKA279). SCR1 serves as a loading control. The following transcripts were detected with the EUC1 probe: RT, LC, and SC. Lanes 1–3 and 4–7 were taken from the same exposure of the same blot. Intervening lanes were removed for clarity. (B) Quantitation of ECM3 levels detected in (A) from three replicates. (+) and (−) indicate the presence or absence of Rap. Quantitation includes signal from the RT isoform and the values are relative to the WT control (−) Rap. Error bars represent the SEM. (C) Diagram showing the relative positions of EUC1 isoforms at the ECM3 locus. CUT, cryptic unstable transcript; LC, long CUT; Rap, rapamycin; RT, read through transcript; SC, short CUT; WT, wild-type.
Discussion
Prompted by previous mechanistic work that revealed a repressive effect of noncoding transcription on expression of the yeast SER3 gene, we sought to identify additional cases in which transcription of intergenic DNA upstream of a protein-coding gene impacts the expression of that gene. We focused on the ECM3 locus, where a promoter-associated CUT, EUC1, is synthesized in the sense direction relative to the ORF. We investigated the role of transcription-associated chromatin alterations in the regulation of ECM3, as many genes regulated by intergenic transcription employ mechanisms that alter the local chromatin environment. A survey of transcription-associated chromatin regulators uncovered an integral role for the Paf1 complex in the positive regulation of ECM3 expression. Each of the five subunits of the Paf1 complex is necessary for proper ECM3 expression. Our data suggest that the role of the Paf1 complex in promoting H3 K4 methylation is required for proper ECM3 expression. Loss of Set1, the histone methyltransferase for H3 K4, or the ubiquitin conjugase and ligase enzymes that catalyze H2B K123 monoubiquitylation, the prerequisite modification for H3 K4 di- and trimethylation, results in reduced ECM3 expression. Additionally, we provide evidence that the role of H3 K4 methylation may be to serve as a signal for HAT activity at the ECM3 promoter. Loss of catalytic subunits of the SAGA and NuA3 HAT complexes, which both recognize methylated states of H3 K4, results in a dramatic reduction of ECM3 expression. In addition to the stimulatory role of this network of chromatin modifiers, ECM3 transcription appears to be positively correlated with noncoding transcription across its promoter. Differences in the levels and isoforms of the EUC1 noncoding transcripts do not strongly affect ECM3 transcription. Instead, the common link between factors stimulating ECM3 expression appears to be their role in promoting H3 K4 methylation. Consistent with this idea, a EUC1 promoter deletion reduced H3 K4me3 levels across both the EUC1 and ECM3 transcription units.
One explanation for our observations is that transcription of EUC1 positively regulates ECM3 expression by promoting the methylation of H3 K4, which may lead to downstream histone acetylation at the ECM3 promoter. A key result in support of this model is that deletion of a 50 bp sequence just upstream of the EUC1 transcription start sites severely impaired both EUC1 and ECM3 expression. Although we cannot completely rule out the possibility that this deletion (pEUC1∆1) may remove a transcription factor binding site that could directly activate the ECM3 promoter, several pieces of data indicate that this sequence most likely contains key regulatory elements of the EUC1 promoter. First, our ChIP data show two distinct peaks of H3 K4me3 for the EUC1 and ECM3 transcripts, suggesting that these promoters are distinct. Second, preliminary data in environmental conditions where we have observed slight changes in ECM3 expression show a corresponding change in expression of the short EUC1 isoform. Although these effects are subtle, they suggest that the correlation between EUC1 and ECM3 expression is not limited to a context where the EUC1 promoter has been deleted. Third, we report multiple lines of evidence for the methylation of H3 K4 as a positive regulatory event for ECM3 expression. As this histone modification is coupled to transcriptional activity, it is likely that transcription of EUC1 plays a role in placing this mark across the ECM3 promoter. Though we are not aware of any transcription factors that both bind to the sequence deleted in pEUC1∆1 and upregulate ECM3 transcription in a manner not dependent on EUC1, we cannot rule out their existence. Our attempts to prevent EUC1 transcription through strategies other than generating the pEUC1∆1 and pEUC1∆2 mutations were unsuccessful. Introduction of an exogenous terminator sequence within EUC1 had nonspecific effects on ECM3 transcription start site selection, and targeted mutation of the predicted TATA element failed to eliminate EUC1 transcription. This is consistent with previous reports detecting PIC assembly further upstream of the putative TATA sequence and indicates that this TATA element is not likely to be the predominant site of PIC assembly for the EUC1 transcript. The overall AT-richness of this region may allow for transcription initiation at multiple sites that may not be abolished by targeted mutations.
While both appear to be important for establishing the H3 K4 methylation pattern across the ECM3 promoter, EUC1 transcription and the Paf1 complex also have separable functions in facilitating ECM3 expression. A double mutant lacking both EUC1 transcription and PAF1 is more severely compromised for ECM3 expression than either single mutant strain. This indicates that the Paf1 complex stimulates ECM3 transcription through a mechanism in addition to its role in promoting H3 K4 methylation. This is further evidenced by the Leo1 subunit of the Paf1 complex having a positive role in ECM3 regulation, despite the fact that Leo1 is not required for H3 K4me3 (Ng et al. 2003). Although termination of the EUC1 transcript is altered in a paf1∆ strain, our results of Nrd1 depletion do not support a major role for alternative termination of EUC1 in regulating levels of ECM3 expression. However, it is possible that there are transient isoform-specific effects or that these isoforms have different roles under biologically relevant conditions. An intriguing feature of the locus is that the EUC1 and ECM3 transcripts overlap in a region of DNA occupied by a nucleosome (Mavrich et al. 2008). It is interesting to speculate that the position and modification state of this nucleosome could be a major determinant in promoting or preventing transcription of ECM3. This nucleosome could also be a determinant in which isoforms of EUC1 and ECM3 are expressed. It would be interesting to relate the position of this nucleosome with where termination of EUC1 and initiation of ECM3 occur.
Our finding of a positive role for H3 K4 methylation at a gene that lies downstream of a noncoding transcription unit is interesting, as others have shown this modification to be repressive to initiation of transcription at the GAL1-10, DCI1, and DUR3 loci (Houseley et al. 2008; Pinskaya et al. 2009; Kim et al. 2012). In these cases, noncoding transcription places H3 K4 methylation across the promoters of these genes and results in the recruitment of HDACs, such as Set3. Our data indicate that the functionally important readers of the H3 K4 methyl marks at ECM3 are the HAT complexes SAGA and NuA3, which would explain the opposite effect of H3 K4 methylation on ECM3 expression compared to GAL1-10, DCI1, and DUR3. Differential recruitment of readers could depend on whether the histones at the promoters of these genes display predominantly tri- or dimethylation of H3 K4 or other differences in the local chromatin environment. The observation that one histone modification associated with noncoding transcription can have opposite effects on the regulation of a neighboring gene adds to the increasing diversity of mechanisms by which noncoding transcription can regulate gene expression. This points to a potential plethora of regulatory mechanisms imparted by noncoding transcription that will be important to study at individual genes.
Supplementary Material
Acknowledgments
We thank Fred Winston for critical review of this manuscript and the members of the Martens and Arndt labs for helpful discussions and experimental guidance, especially Margaret Shirra and Christine Cucinotta. We also thank Justin Pruneski, Mary Braun, Kostadin Petrov, and Elia Crisucci for strain construction. Patrick Cramer kindly provided the NRD1 anchor away strains. We are grateful to Craig Kaplan and Lars Steinmetz for helpful discussions in developing this work. This work was funded by grants from the National Institutes of Health (GM080470 to J.M. and GM052593 to K.A.) and the University of Pittsburgh.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.033118/-/DC1
Communicating editor: A. M. Dudley
Literature Cited
- Arigo J. T., Eyler D. E., Carroll K. L., Corden J. L., 2006. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell 23: 841–851. [DOI] [PubMed] [Google Scholar]
- Arndt K. M., Reines D., 2015. Termination of Transcription of Short Noncoding RNAs by RNA Polymerase II. Annu. Rev. Biochem. 84: 381–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F. M., 1987 Current protocols in molecular biology. Greene/Wiley, New York. [Google Scholar]
- Bartel D. P., 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Bevington S., Boyes J., 2013. Transcription-coupled eviction of histones H2A/H2B governs V(D)J recombination. EMBO J. 32: 1381–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian C., Xu C., Ruan J., Lee K. K., Burke T. L., et al. , 2011. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J. 30: 2829–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borel C., Gagnebin M., Gehrig C., Kriventseva E. V., Zdobnov E. M., et al. , 2008. Mapping of small RNAs in the human ENCODE regions. Am. J. Hum. Genet. 82: 971–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C. A., Ares M., Jr, 2006. Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 103: 3262–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium , 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. D., 2014. Non-coding RNA: a new frontier in regulatory biology. Natl. Sci. Rev. 1: 190–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand B., Mead J., Bruning A., Apostolopoulos N., Tadigotla V., et al. , 2011. Regulated antisense transcription controls expression of cell-type-specific genes in yeast. Mol. Cell. Biol. 31: 1701–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg D. S., Govind C. K., Hinnebusch A. G., 2009. NuA4 lysine acetyltransferase Esa1 is targeted to coding regions and stimulates transcription elongation with Gcn5. Mol. Cell. Biol. 29: 6473–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg D. S., Anlembom T. E., Wang J., Patel S. R., Li B., et al. , 2014. NuA4 links methylation of histone H3 lysines 4 and 36 to acetylation of histones H4 and H3. J. Biol. Chem. 289: 32656–32670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer S. J., Martens J. A., 2011. Identification of histone mutants that are defective for transcription-coupled nucleosome occupancy. Mol. Cell. Biol. 31: 3557–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer S. J., Pruneski J. A., Mitchell R. D., Monteverde R. M., Martens J. A., 2011. Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev. 25: 29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki H., Nishikawa J., Laemmli U. K., 2008. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell 31: 925–932. [DOI] [PubMed] [Google Scholar]
- Hirota K., Miyoshi T., Kugou K., Hoffman C. S., Shibata T., et al. , 2008. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature 456: 130–134. [DOI] [PubMed] [Google Scholar]
- Hobson D. J., Wei W., Steinmetz L. M., Svejstrup J. Q., 2012. RNA polymerase II collision interrupts convergent transcription. Mol. Cell 48: 365–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongay C. F., Grisafi P. L., Galitski T., Fink G. R., 2006. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell 127: 735–745. [DOI] [PubMed] [Google Scholar]
- Houseley J., Rubbi L., Grunstein M., Tollervey D., Vogelauer M., 2008. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol. Cell 32: 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung C. L., Wang L. Y., Yu Y. L., Chen H. W., Srivastava S., et al. , 2014. A long noncoding RNA connects c-Myc to tumor metabolism. Proc. Natl. Acad. Sci. USA 111: 18697–18702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T., Buratowski S., 2009. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell 137: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T., Xu Z., Clauder-Munster S., Steinmetz L. M., Buratowski S., 2012. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell 150: 1158–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. T., 2009. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 23: 1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussier M., White A. M., Sheraton J., di Paolo T., Treadwell J., et al. , 1997. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 147: 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens J. A., Laprade L., Winston F., 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429: 571–574. [DOI] [PubMed] [Google Scholar]
- Martens J. A., Wu P. Y., Winston F., 2005. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 19: 2695–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. G., Grimes D. E., Baetz K., Howe L., 2006. Methylation of histone H3 mediates the association of the NuA3 histone acetyltransferase with chromatin. Mol. Cell. Biol. 26: 3018–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich T. N., Ioshikhes I. P., Venters B. J., Jiang C., Tomsho L. P., et al. , 2008. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 18: 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil H., Malabat C., d’Aubenton-Carafa Y., Xu Z., Steinmetz L. M., et al. , 2009. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457: 1038–1042. [DOI] [PubMed] [Google Scholar]
- Ng H. H., Robert F., Young R. A., Struhl K., 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11: 709–719. [DOI] [PubMed] [Google Scholar]
- Pelechano V., Wei W., Steinmetz L. M., 2013. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature 497: 127–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinskaya M., Gourvennec S., Morillon A., 2009. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J. 28: 1697–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrua O., Libri D., 2015. Transcription termination and the control of the transcriptome: why, where and how to stop. Nat. Rev. Mol. Cell Biol. 16: 190–202. [DOI] [PubMed] [Google Scholar]
- Prescott E. M., Proudfoot N. J., 2002. Transcriptional collision between convergent genes in budding yeast. Proc. Natl. Acad. Sci. USA 99: 8796–8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee H. S., Pugh B. F., 2012. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483: 295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J., Guttman M., 2014. RNA Function. RNA and dynamic nuclear organization. Science 345: 1240–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J. L., Kertesz M., Wang J. K., Squazzo S. L., Xu X., et al. , 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio D. C., 2011. RNA: a laboratory manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Schneider B. L., Seufert W., Steiner B., Yang Q. H., Futcher A. B., 1995. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast 11: 1265–1274. [DOI] [PubMed] [Google Scholar]
- Schulz D., Schwalb B., Kiesel A., Baejen C., Torkler P., et al. , 2013. Transcriptome surveillance by selective termination of noncoding RNA synthesis. Cell 155: 1075–1087. [DOI] [PubMed] [Google Scholar]
- Sheldon K. E., Mauger D. M., Arndt K. M., 2005. A Requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol. Cell 20: 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirra M. K., Rogers S. E., Alexander D. E., Arndt K. M., 2005. The Snf1 protein kinase and Sit4 protein phosphatase have opposing functions in regulating TATA-binding protein association with the Saccharomyces cerevisiae INO1 promoter. Genetics 169: 1957–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J. A., Kingston R. E., 2013. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 49: 808–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolle M., Workman J. L., 2013. Transcription-associated histone modifications and cryptic transcription. Biochim. Biophys. Acta 1829: 84–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz E. J., Warren C. L., Kuehner J. N., Panbehi B., Ansari A. Z., et al. , 2006. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol. Cell 24: 735–746. [DOI] [PubMed] [Google Scholar]
- Tomson B. N., Arndt K. M., 2013. The many roles of the conserved eukaryotic Paf1 complex in regulating transcription, histone modifications, and disease states. Biochim. Biophys. Acta 1829: 116–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomson B. N., Davis C. P., Warner M. H., Arndt K. M., 2011. Identification of a role for histone H2B ubiquitylation in noncoding RNA 3′-end formation through mutational analysis of Rtf1 in Saccharomyces cerevisiae. Genetics 188: 273–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomson B. N., Crisucci E. M., Heisler L. E., Gebbia M., Nislow C., et al. , 2013. Effects of the Paf1 complex and histone modifications on snoRNA 3′-end formation reveal broad and locus-specific regulation. Mol. Cell. Biol. 33: 170–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M. C., Manor O., Wan Y., Mosammaparast N., Wang J. K., et al. , 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter J. C., Adams M. D., Myers E. W., Li P. W., Mural R. J., et al. , 2001. The sequence of the human genome. Science 291: 1304–1351. [DOI] [PubMed] [Google Scholar]
- Warner M. H., Roinick K. L., Arndt K. M., 2007. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol. Cell. Biol. 27: 6103–6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Dollard C., Ricupero-Hovasse S. L., 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11: 53–55. [DOI] [PubMed] [Google Scholar]
- Wyers F., Rougemaille M., Badis G., Rousselle J. C., Dufour M. E., et al. , 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737. [DOI] [PubMed] [Google Scholar]
- Wyrick J. J., Holstege F. C., Jennings E. G., Causton H. C., Shore D., et al. , 1999. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402: 418–421. [DOI] [PubMed] [Google Scholar]
- Yang L., Froberg J. E., Lee J. T., 2014. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem. Sci. 39: 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.