A previously uncharacterized WD40 domain–containing protein named TSSC1 is shown to interact with the GARP and EARP tethering complexes, promoting retrograde transport of Shiga toxin from endosomes to the TGN, as well as recycling internalized transferrin from endosomes to the plasma membrane.
Abstract
Endosomes function as a hub for multiple protein-sorting events, including retrograde transport to the trans-Golgi network (TGN) and recycling to the plasma membrane. These processes are mediated by tubular-vesicular carriers that bud from early endosomes and fuse with a corresponding acceptor compartment. Two tethering complexes named GARP (composed of ANG2, VPS52, VPS53, and VPS54 subunits) and EARP (composed of ANG2, VPS52, VPS53, and Syndetin subunits) were previously shown to participate in SNARE-dependent fusion of endosome-derived carriers with the TGN and recycling endosomes, respectively. Little is known, however, about other proteins that function with GARP and EARP in these processes. Here we identify a protein named TSSC1 as a specific interactor of both GARP and EARP and as a novel component of the endosomal retrieval machinery. TSSC1 is a predicted WD40/β-propeller protein that coisolates with both GARP and EARP in affinity purification, immunoprecipitation, and gel filtration analyses. Confocal fluorescence microscopy shows colocalization of TSSC1 with both GARP and EARP. Silencing of TSSC1 impairs transport of internalized Shiga toxin B subunit to the TGN, as well as recycling of internalized transferrin to the plasma membrane. Fluorescence recovery after photobleaching shows that TSSC1 is required for efficient recruitment of GARP to the TGN. These studies thus demonstrate that TSSC1 plays a critical role in endosomal retrieval pathways as a regulator of both GARP and EARP function.
INTRODUCTION
Early endosomes (EEs) are a major sorting station in the endomembrane system of eukaryotic cells where various transport pathways intersect (Maxfield and McGraw, 2004; Bonifacino and Rojas, 2006; Jovic et al., 2010; Figure 1). EEs receive cargoes by endocytosis from the plasma membrane and the extracellular space, as well as by biosynthetic transport from the trans-Golgi network (TGN). Some cargoes are subsequently delivered from EEs to late endosomes and lysosomes, whereas others are retrieved to the plasma membrane or the TGN. For example, the transferrin receptor (TfR) carrying iron-loaded holo-transferrin (Tf) is delivered from the plasma membrane to EEs by clathrin-dependent endocytosis. After dissociation of iron in the acidic pH of EEs, TfR bound to the resulting apo-Tf returns to the plasma membrane via various recycling endosomal compartments (Figure 1). In contrast, mannose-6-phosphate receptors (MPRs) carrying lysosomal hydrolase precursors are delivered to EEs from the TGN. The acidic pH of EEs also causes release of the lysosomal hydrolase precursors, after which the enzymes are delivered to lysosomes and the MPRs are returned by retrograde transport to the TGN (Figure 1). Other cargoes entering EEs do not recycle to their compartments of origin but traffic along a single route that they transiently share with the recycling cargoes. A prominent example is the bacterial Shiga toxin, which binds to surface globotriaosylceramide (Gb3) and undergoes internalization into the EE, followed by retrograde transport to the TGN (Johannes and Romer, 2010; Figure 1).
FIGURE 1:
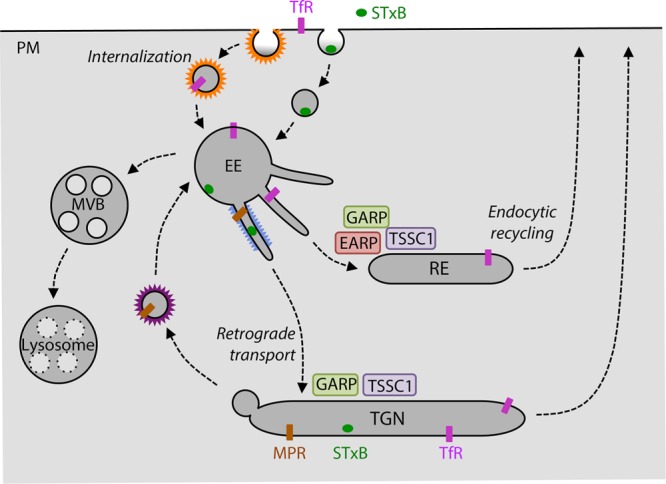
Schematic representation of endosomal retrieval pathways. Trafficking routes of the transferrin receptor (TfR), mannose 6-phosphate receptor (MPR), and Shiga toxin B subunit (STxB) through various compartments of the endomembrane system. The sites of action of the GARP and EARP complexes are indicated, as is the deduced functional localization of TSSC1. MVB, multivesicular body; PM, plasma membrane; RE, recycling endosome; TGN, trans-Golgi network.
Whereas other intercompartmental transport steps involve small, coated vesicles, endosomal retrieval is largely mediated by tubular-vesicular carriers that bud from EEs and fuse with either recycling endosomes (REs) or the TGN (Gallon and Cullen, 2015; Hierro et al., 2015). Both budding and fusion of the carriers depend on complex molecular machineries that are recruited from the cytosol onto membranes. A core component of the endosomal budding machinery is the retromer complex, which comprises a VPS26-VPS29-VPS35 heterotrimer and various combinations of sorting nexin (SNX) proteins, including a SNX1/2-SNX5/6 heterodimer, SNX3, and/or SNX27 (Gallon and Cullen, 2015; Hierro et al., 2015). Various other components, such as membrane phosphatidylinositol 3-phosphate (PtdIns3P; Burda et al., 2002), the small GTPase Rab7 (Rojas et al., 2008; Seaman et al., 2009), the Rab-GAP TBC1D5 (Seaman et al., 2009), the WASH complex (Gomez and Billadeau, 2009), and dynactin (Hong et al., 2009; Wassmer et al., 2009), interact with retromer to mediate different steps in the process of tubule formation and translocation. Fusion of the tubules with acceptor RE or TGN compartments is mediated by specific sets of soluble N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE) proteins, which have been best characterized for the TGN (Mallard et al., 2002). Two multisubunit tethering complexes—endosome-associated recycling protein (EARP; composed of ANG2, VPS52, VPS53, and Syndetin subunits; Gillingham et al., 2014; Schindler et al., 2015) and Golgi-associated retrograde protein (GARP; composed of ANG2, VPS52, VPS53, and VPS54 subunits; Conibear et al., 2003; Liewen et al., 2005; Fridmann-Sirkis et al., 2006; Perez-Victoria et al., 2008, 2010; Perez-Victoria and Bonifacino, 2009)—are believed to promote SNARE complex assembly required for fusion with REs and the TGN, respectively. Whereas EARP does not mediate retrograde transport to the TGN, GARP to some extent also participates in endocytic recycling (Schindler et al., 2015). There is evidence in Drosophila that EARP interacts with Rab4 (Gillingham et al., 2014), whereas studies in yeast and mammalian cells suggest that GARP interacts with the small GTPases Rab6 and Arl1 (Panic et al., 2003; Liewen et al., 2005). However, it is not known whether these interactions contribute the recruitment of the complexes to their corresponding compartments, particularly in mammals. Moreover, in contrast to the abundance of retromer interactors, no other proteins have been shown to interact with EARP and GARP in the performance of their functions as tethering factors.
To identify proteins that participate with EARP and GARP in endosomal retrieval pathways, we performed affinity purification using EARP/GARP subunits as baits, followed by mass spectrometry. This approach allowed us to identify a protein named tumor-suppressing subchromosomal transferable fragment candidate gene 1 (TSSC1) as novel interactor of both EARP and GARP. TSSC1 is a phylogenetically conserved and ubiquitously expressed protein predicted to comprise a WD40/β-propeller domain. The association of TSSC1 with both EARP and GARP was confirmed by coimmunoprecipitation and gel filtration analyses. In addition, confocal fluorescence microscopy showed colocalization of TSSC1 with both EARP and GARP. Silencing of TSSC1 in human cells impaired the transport of internalized Shiga toxin B subunit (STxB) to the TGN and of internalized TfR back to the plasma membrane, consistent with its involvement in both retrograde transport and endocytic recycling pathways. Fluorescence recovery after photobleaching (FRAP) analysis showed that TSSC1 is required for efficient recruitment of GARP to membranes. These findings thus identify TSSC1 as a novel component of the endosomal retrieval machinery that cooperates with both GARP and EARP in the processes of retrograde transport and endocytic recycling, respectively.
RESULTS
Affinity purification and mass spectrometry identify TSSC1 as a novel interactor of EARP and GARP
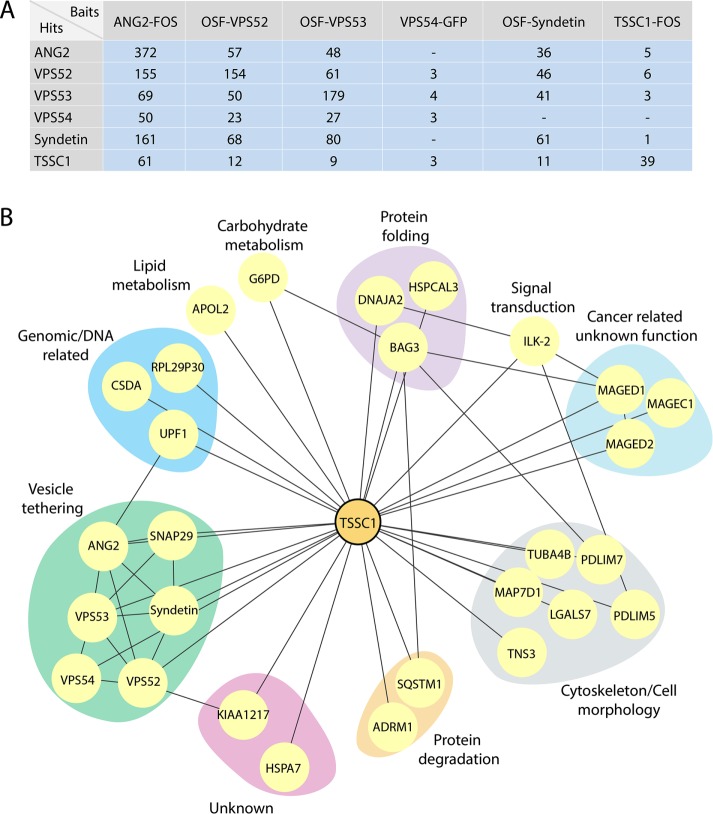
To identify proteins that interact with EARP and GARP, we performed affinity purification followed by mass spectrometry using detergent extracts from H4 human neuroglioma cells stably expressing each of the subunits of these complexes appended with a C-terminal FLAG/One-STrEP (FOS), an N-terminal One-STrEP/FLAG (OSF), or a green fluorescent protein (GFP) tag (Figure 2A; a complete set of results is shown in Supplemental Table S1). To remove frequent contaminants in this kind of analysis, the raw data were filtered against the Contaminant Repository for Affinity Purification (CRAPome) database (Mellacheruvu et al., 2013). Only proteins with an average spectral count of <0.25 hit per experiment in this database were considered for further analysis. We also set a threshold of six unique peptides in initial FOS and OSF data sets to restrict our analysis to the more abundant and reliable interactors. The resulting set of proteins showed that ANG2, VPS52, and VPS53 (shared subunits of both EARP and GARP) copurified with each other, as well as with VPS54 (unique subunit of GARP) and Syndetin (unique subunit of EARP; Figure 2A). Syndetin copurified with ANG2, VPS52, and VPS53 but not with VPS54 (Figure 2A). VPS54 copurified with VPS52 and VPS53 but not with ANG2 and Syndetin (Figure 2A). All of these results are in agreement with the known subunit composition of GARP and EARP (Schindler et al., 2015), except for the absence of ANG2 in the VPS54 isolates, which we attribute to the low efficiency of the purification using GFP as a tag (notice low peptide number in Figure 2A). Of interest, after the GARP/EARP subunits, the next most abundant interactor for all five subunits was a previously uncharacterized protein named TSSC1 (Figure 2A). Reciprocal affinity purification-mass spectrometry using TSSC1-FOS as bait coisolated ANG2, VPS52, VPS53, and Syndetin but not VPS54 (Figure 2A). The absence of VPS54 from this purification was also likely due to the low peptide number for the EARP/GARP subunits, since VPS54-GFP did pull down TSSC1 (Figure 2A), and subsequent analyses showed coimmunoprecipitation of VPS54 with TSSC1 (see later discussion). By combining the interactions found in our study with those in the BioGRID database (Stark et al., 2006) and previous high-throughput protein interaction screens (Hein et al., 2015; Huttlin et al., 2015), we generated a TSSC1 interaction network that highlighted a relationship of this protein with the GARP/EARP subunits and the SNARE protein SNAP29 (Figure 2B). These interactions are conserved in human cell lines derived from neuroglioma, embryonic kidney, and cervix adenocarcinoma, as well as in rat brain tissue (Supplemental Figure S1). The clustering of TSSC1 with proteins involved in tethering and fusion suggested that it could play a role in these processes.
FIGURE 2:
Affinity purification and mass spectrometric analysis of GARP and EARP interactors. (A) Table showing the proteins (Hits) coisolated by affinity purification from H4 cells expressing the FOS-, OSF-, or GFP-tagged proteins indicated on top (Baits). Total numbers of peptides found in mass spectrometry. ANG2 and Syndetin are also designated VPS51 and VPS50, respectively, by the HUGO Gene Nomenclature Committee. (B) Network analysis showing the interactions between the hits from our screens filtered by the CRAPome (www.crapome.org) and interactions mapped using the BioGRID database (thebiogrid.org). Proteins were grouped according to their Gene Ontology classification.
Bioinformatic analysis of TSSC1
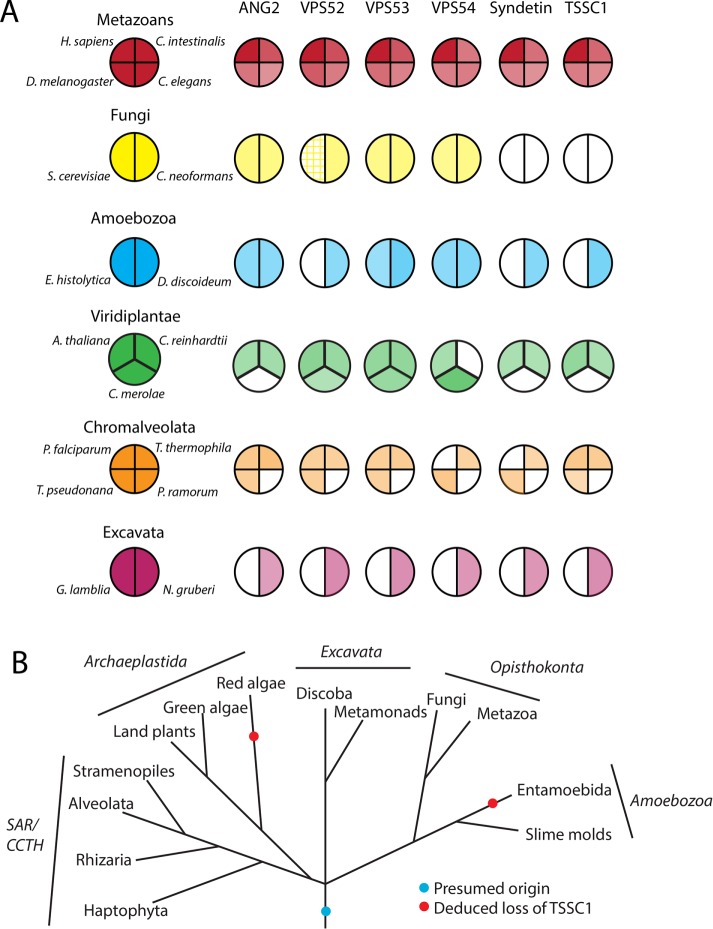
TSSC1 was previously identified as the product of a gene encoded in the subchromosomal 11p15.5 tumor-suppressor gene region (Hu et al., 1997; Scelfo et al., 1998) and regulated by the transcription factor Runx2 (Wang et al., 2013). The human gene encodes a 387–amino acid protein with a predicted molecular mass of 43.6 kDa. The Phyre2 server (Kelley and Sternberg, 2009) predicts with 100% confidence that the protein consists of a single WD40/β-propeller domain. A phylogenetic analysis by reciprocal homology searches with the Homo sapiens TSSC1 protein sequence using the BLASTP algorithm (Altschul et al., 1990) and subsequent searches with multi-iteration PSI-BLAST indicated that TSSC1 is conserved in all eukaryotic supergroups (classified according to Barlow et al., 2014) and has a shared phylogeny with members of the GARP/EARP complexes (Figure 3, A and B; Koumandou et al., 2007). Exceptions to this conservation are Phytophthora ramorum and Giardia lamblia, in which there is an apparent loss of GARP, EARP, and TSSC1, and Cyanidioschyzon merolae and Entamoeba histolytica, which show a partial loss of both complexes accompanied by loss of TSSC1 (Figure 3, A and B). The conservation of TSSC1 in most eukaryotes is suggestive of an essential physiological function, and the general trend of coevolution with GARP/EARP supports a functional relationship.
FIGURE 3:
Phylogenetic analysis of GARP and EARP subunits and TSSC1. (A) Coulson plot (Field et al., 2013) showing the existence of ANG2, VPS52, VPS53, VPS54, Syndetin, and TSSC1 orthologues in a variety of eukaryotes chosen for their diversity as in Hirst et al. (2011). Color intensity represents the degree of identity based on a BLASTP alignment with the human homologue. Hatched shading indicates a known homologue that shares little primary sequence homology. (B) Deduced evolutionary history of TSSC1 (based on Walker et al., 2011).
Biochemical analyses confirm the interaction of TSSC1 with both EARP and GARP
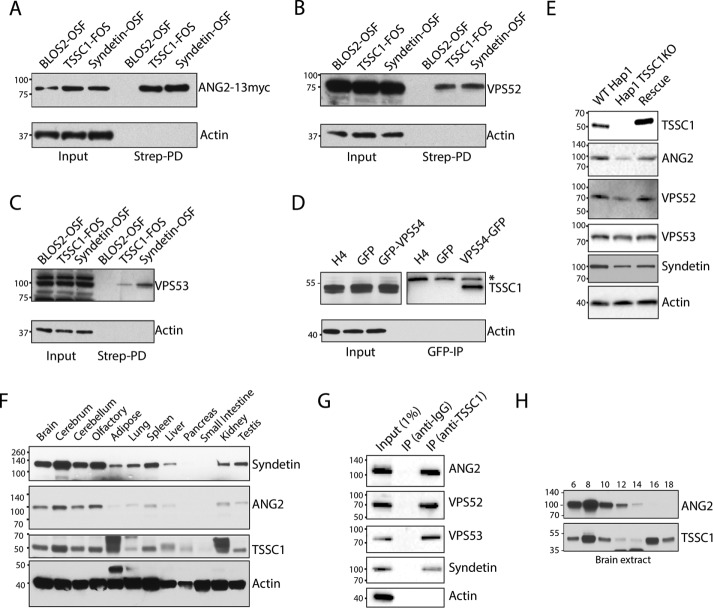
We next performed additional biochemical analyses to confirm the specificity of the interaction of TSSC1 with GARP and EARP. StrepTactin pull down from H4 cells expressing TSSC1-FOS followed by immunoblotting with antibodies to GARP/EARP subunits showed coisolation of TSSC1 with transgenic ANG2-13myc and endogenous VPS52 and VPS53 (Figure 4, A–C). These interactions were robust and specific, as demonstrated by the coisolation of these same proteins with Syndetin-OSF (positive control) but not with the BLOS2 subunit of the BLOC-1 (Starcevic and Dell’Angelica, 2004) and BORC (Pu et al., 2015) complexes tagged with OSF (negative control; Figure 4, A–C). Because we do not have an antibody that recognizes endogenous VPS54, we performed a GFP-trap pull down from H4 cells stably expressing GFP (negative control) or VPS54-GFP. Immunoblot analysis with antibody to endogenous TSSC1 detected this protein in the VPS54-GFP but not GFP or untransfected cell pull downs (Figure 4D). To validate the specificity of the antibody to TSSC1, we performed immunoblot analysis of extracts from three lines of human HAP1 cells: wild type (WT), TSSC1 clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 knockout (KO), and TSSC1-KO rescued by stable expression of TSSC1-FOS (Figure 4E). We observed a TSSC1 band in WT and TSSC1-rescued cells but not in TSSC1-KO cells, thus demonstrating the specificity of the antibody (Figure 4E).
FIGURE 4:
Biochemical analysis of TSSC1. (A–C) Extracts from H4 cells stably expressing BLOS2-OSF (negative control), Syndetin-OSF (positive control), or TSSC1-FOS were analyzed by pull down with StrepTactin beads, followed by SDS–PAGE and immunoblotting for coexpressed ANG2-13myc and endogenous VPS52, VPS53, and actin (negative control). (D) Extracts from untransfected and stably expressing GFP or VPS54-GFP cells were analyzed by pull down with a nanobody to GFP conjugated to magnetic beads, followed by SDS–PAGE and immunoblotting for endogenous TSSC1 and actin (negative control). *Nonspecific band. (E) WT and TSSC1-KO HAP1 cells, and TSSC1-KO HAP1 cells rescued by stable expression of TSSC1-FOS, were analyzed by SDS–PAGE and immunoblotting with antibodies to TSSC1, ANG2, VPS52, VPS53, Syndetin, or actin (negative control). (F) SDS–PAGE and immunoblot analysis of the expression of Syndetin, ANG2, TSSC1, and actin in multiple rat tissues. (G) A rat brain extract was subjected to immunoprecipitation with antibodies to immunoglobulin G (negative control) or TSSC1 followed by SDS–PAGE and immunoblot analysis of the precipitates with antibodies to ANG2, VPS52, VPS53, Syndetin, and actin. A 1% fraction of the input extract was run alongside for comparison. (H) Gel filtration of rat brain extract on a Superose 6 column and analysis of the fractions indicated on top by SDS–PAGE and immunoblotting with antibodies to ANG2 and TSSC1. In all the immunoblots, the positions of molecular mass markers (in kilodaltons) are indicated on the left.
Immunoblot analysis of multiple rat tissues showed that TSSC1 is expressed ubiquitously, albeit with higher levels in the brain (Figure 4F), as previously shown for GARP and EARP (Schindler et al., 2015). Coimmunoprecipitation from a rat brain cytosolic extract using antibody to TSSC1 confirmed the association of this protein with endogenous ANG2, VPS52, VPS53, and Syndetin (Figure 4G). Finally, gel filtration of rat brain extract on Superose 6 revealed that a fraction of TSSC1 coeluted with ANG2 (a subunit of both GARP and EARP), whereas another fraction eluted with a smaller hydrodynamic radius, likely corresponding to free TSSC1 (Figure 4H).
Taken together, all of the foregoing biochemical experiments coincided in demonstrating that TSSC1 is a specific interactor of both GARP and EARP.
Colocalization of TSSC1 with GARP and EARP
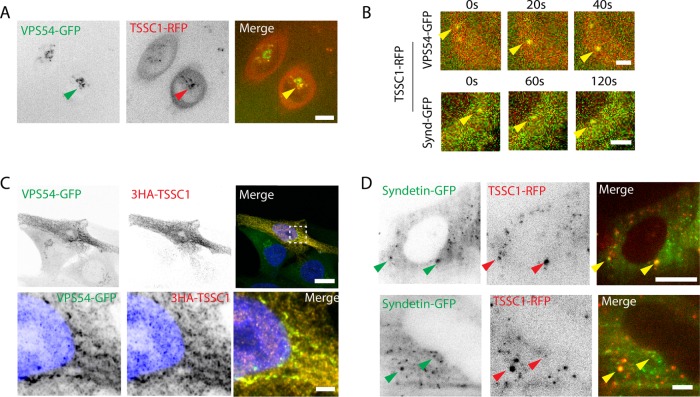
To try to understand the function of TSSC1, we next examined the subcellular localization of this protein. Attempts to localize TSSC1 in various cell lines using the antibody to the endogenous protein yielded only background staining. Similarly, TSSC1-FOS or TSSC1-GFP expressed by transfection into HeLa cells showed only cytosolic staining (unpublished data). However, transient expression of TSSC1-RFP in H4 cells stably expressing VPS54-GFP and live-cell imaging using a spinning-disk confocal microscope revealed partial colocalization of these proteins to a juxtanuclear structure characteristic of the TGN (Figure 5A). Punctae exhibiting colocalization of TSSC1 with VPS54 were comotile (Figure 5B). Further analyses using a higher-resolution Airyscan confocal microscope allowed unequivocal demonstration of the colocalization of 3-hemagglutinin (HA)-TSSC1 with VPS54-GFP to TGN structures in fixed H4 cells (Figure 5C). Colocalization of TSSC1 with Syndetin was more difficult to demonstrate using these methodologies because of the disperse distribution of Syndetin (Schindler et al., 2015). However, we succeeded in observing colocalization of transiently expressed TSSC1–red fluorescent protein (RFP) and Syndetin-GFP in H4 cells by live-cell total internal reflection fluorescence (TIRF) microscopy, which allows low-background visualization of cytoplasmic particles within an ∼200-nm evanescent field (Figure 5D). We concluded that, at least in some settings, TSSC1 colocalizes with both GARP and EARP.
FIGURE 5:
Colocalization of TSSC1 with VPS54. (A) Live H4 cells expressing VPS54-GFP stably and TSSC1-RFP transiently were imaged by spinning-disk confocal microscopy. Eleven z-stacks of five slices each were imaged with a 20-s interval between each stack. The z-stacks were individually compiled into a time series of maximum intensity projections, which were then combined into an average intensity projection representing the whole time series. Arrowheads point to colocalization at the TGN. Bar, 10 μm. (B) Three frames from the time series in A showing VPS54-GFP/TSSC1-RFP and Syndetin-GFP/TSSC1-RFP comotility. (C) Airyscan imaging of 3HA-TSSC1 transiently expressed in the VPS54-GFP stable H4 cell line. Cells were stained with primary antibody to the HA epitope, Atto488-conjugated GFP nanobody (GFP booster), and DAPI (for nuclear staining) Scale bars, 10 μm (top), 2 μm (bottom). (D) TIRF microscopy of Syndetin-GFP and TSSC1-RFP. Scale bars, 10 μm (top), 2 μm (bottom). Arrowheads point to colocalizing organelles. Top row, low magnification, and bottom row, high magnification, of different cells.
TSSC1 is required for retrograde transport of STxB and recycling of internalized Tf
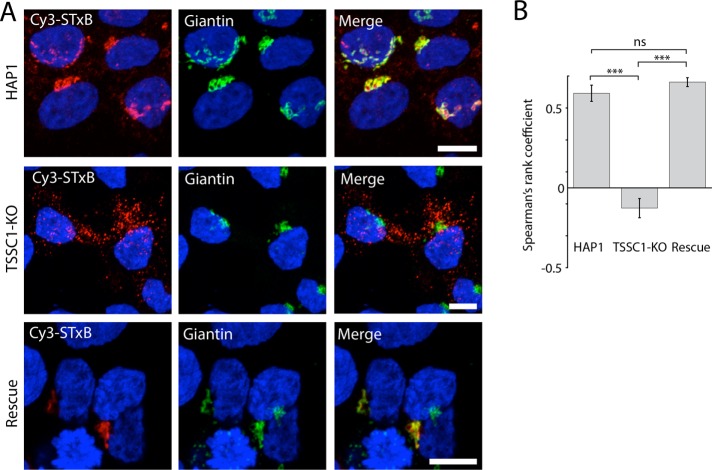
The interaction and colocalization of TSSC1 with the VPS54 subunit of GARP prompted us to test the involvement of TSSC1 in retrograde transport from endosomes to the TGN. To this end, we examined the localization of the Cy3-tagged Shiga toxin B subunit (Cy3-STxB) after 15 min of internalization and 1 h of chase in WT, TSSC1-KO, and TSSC1-rescued HAP1 cells. Cells were subsequently immunostained for the Golgi marker giantin and analyzed by confocal fluorescence microscopy. We observed that Cy3-STxB became concentrated in the Golgi region labeled for giantin in the WT and TSSC1-rescued cells but remained in a population of disperse vesicular structures in the TSSC1-KO cells (Figure 6A). Quantification using Spearman’s rank correlation analysis of multiple cells from several experiments showed the difference in colocalization between the WT and TSSC1-KO cells to be highly significant and rescuable with transfected TSSC1 (Figure 6B).
FIGURE 6:
TSSC1 KO impairs STxB retrograde transport. (A) Live WT and TSSC1-KO HAP1 cells, and TSSC1-KO HAP1 cells rescued by stable expression of TSSC1-FOS, were incubated with Cy3-STxB for 1 h, fixed, stained for the Golgi marker giantin, and examined by confocal fluorescence microscopy. Scale bar, 10 μm. (B) Spearman’s rank coefficient quantification of data set in A, expressed as mean; error bars, SEM. Minimum of nine cells. ***p ≤ 0.001.
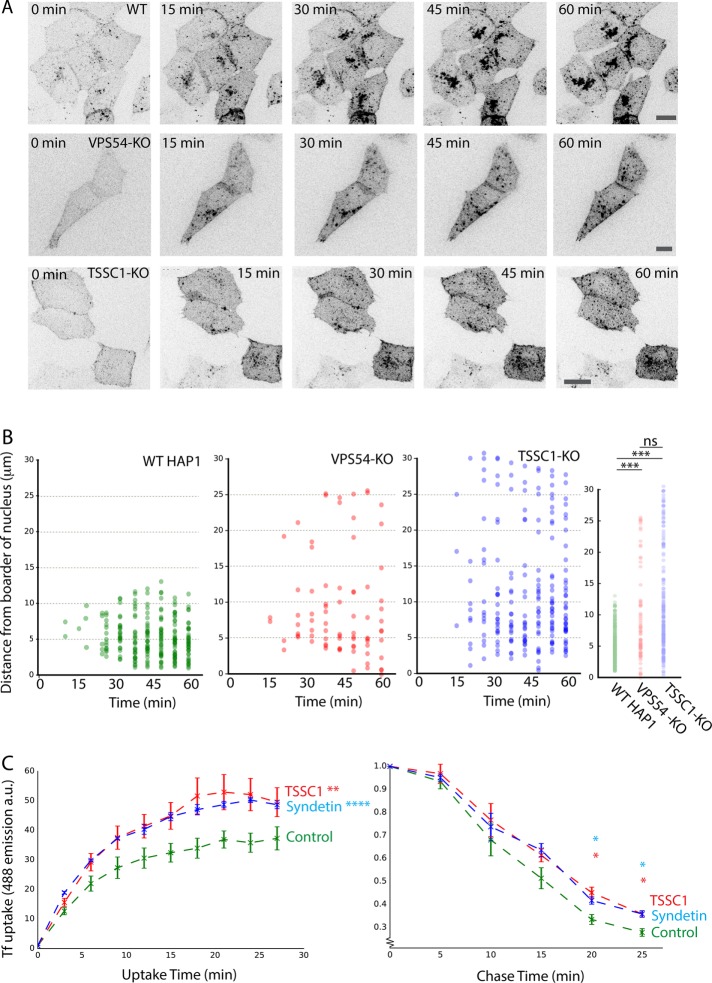
More detailed live-cell imaging analyses of Cy3-STxB uptake in WT cells showed that a sizable fraction of the toxin subunit reached the Golgi region as early as in 15 min; this fraction became much larger over the next 45 min of imaging (Figure 7A and Supplemental Video S1), consistent with previous work on the kinetics of STxB retrograde transport to the Golgi complex (Popoff et al., 2007). Quantification of Cy3-STxB distribution across all time points using three-dimensional (3D) reconstructions showed a mean distance of 5.2 μm from the center of the nucleus (Figure 7B). In contrast, in TSSC1-KO and VPS54-KO cells, Cy3-STxB accumulated in vesicular structures scattered throughout the cytoplasm (Figure 7A and Supplemental Videos S2 and S3), with mean distances of 11.4 and 9.1 μm, respectively (Figure 7B). Cy3-STxB–containing vesicles in TSSC1-KO cells did not colocalize with early and recycling endosomal markers (EEA1 and TfR, respectively), consistent with them being intermediates in transport from endosomes to the TGN (Supplemental Figure S2; Perez-Victoria et al., 2008). These experiments indicated that TSSC1 plays a role in retrograde transport of STxB from endosomes to the TGN.
FIGURE 7:
(A) Live-cell imaging of Cy3-STxB uptake in WT, VPS54-KO, and TSSC1-KO HAP1 cells. Cells were incubated for 1 h in the continuous presence of Cy3-STxB and imaged over the course of 1½ h by spinning-disk confocal microscopy. At each time point, a z-stack of the whole cell was taken and averaged into a maximum intensity projection. Scale bar, 10 μm. See Supplemental Videos S1–S3 for complete time course. (B) Distance quantification of data set from A. Each point plotted represents an individual detectable punctate structure, plotted showing its distance from the center of the nucleus. Left, combination of at least 70 data points from multiple cells over time; far right, combination of all data, demonstrating the relative proximity of the STxB punctae to the nucleus in the WT cells. Significance was calculated using Student’s t test. (C) FACS analysis of Alexa 488–Tf uptake into control HeLa cells or HeLa cells treated with siRNAs to TSSC1 or Syndetin. The plot represents the geometric mean of Alexa 488–Tf intensity of the population (∼30,000 cells) as a function of time. Error is the SEM from multiple independent experiments. Left, continuous exposure to Alexa 488–Tf (significance calculated as two-way analysis of variance); right, chase experiment after 30-min uptake (significance calculated using Student’s t test at selected time points). ns: p > 0.05, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
To determine whether TSSC1 also participates in endocytic recycling, we examined the TfR-mediated uptake of Alexa 488–labeled Tf in HeLa cells treated with no small interfering RNA (siRNA; control) or siRNAs to TSSC1 or Syndetin, as previously described (Schindler et al., 2015). Cells were incubated with Alexa 488–Tf for 0–27 min, uptake was measured by fluorescence-activated cell sorting (FACS) at 3-min intervals, and the geometric mean of the population was calculated at each time point. We observed that knockdown (KD) of Syndetin or TSSC1 increased the accumulation of Alexa 488–Tf relative to control cells (Figure 7C). To determine whether this increased accumulation was due to reduced recycling, control, Syndetin KD, or TSSC1 KD, cells were incubated for 30 min with Alexa-488-Tf, washed, and chased for different times in regular culture medium. We observed that Syndetin- or TSSC1-KD cells retained more Tf compared with the WT cells (about double the amount at 20–25 min; Figure 7C). Taken together, these observations indicated that TSSC1 also plays a role in endocytic recycling of Tf.
TSSC1 promotes GARP recruitment to the TGN
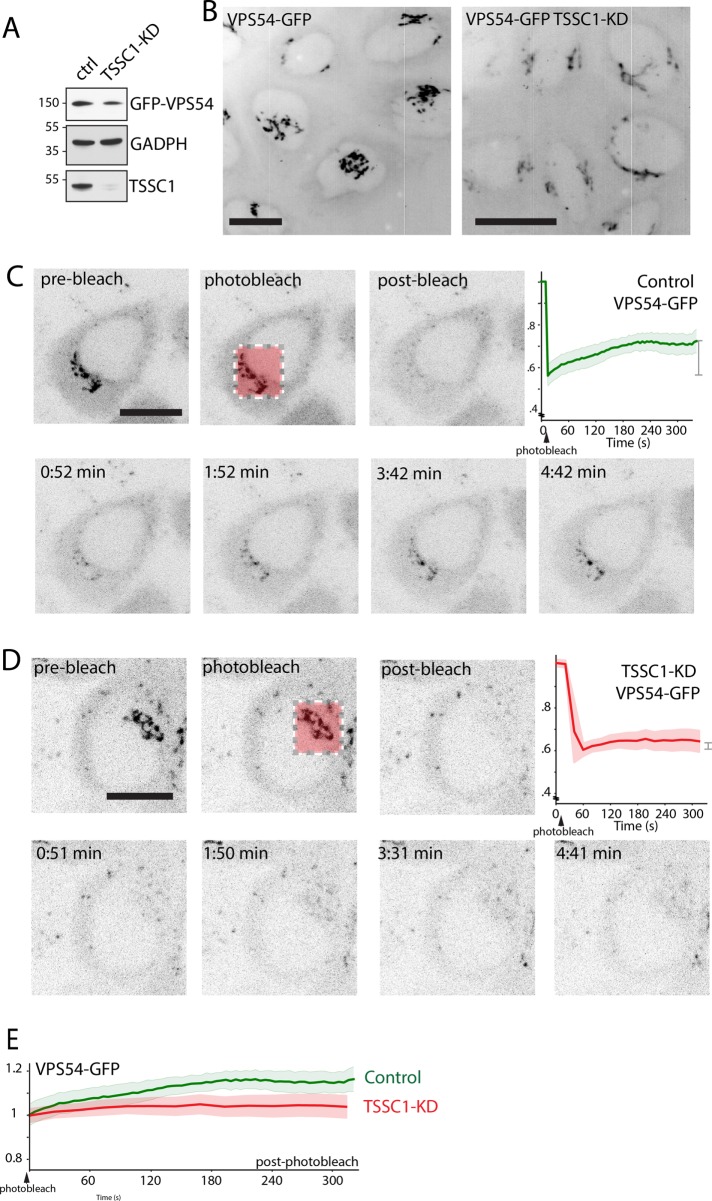
To gain insight into the mechanism by which TSSC1 functions in retrograde transport, we examined the effect of TSSC1 KD on the localization and dynamics of VPS54-GFP stably expressed in H4 cells (Figure 8). We observed that TSSC1-KD cells (Figure 8A) exhibited less intense VPS54-GFP fluorescence at the TGN (Figure 8B). FRAP analysis of the TGN in control VPS54-GFP–expressing cells showed recovery of ∼38% of the original fluorescence after 180 s, revealing that VPS54, and thus the GARP complex, exchanges rapidly between the TGN and the cytosol (Figure 8, C and E). In contrast, FRAP of TSSC1-KD cells revealed a much slower recovery of VPS54-GFP at the TGN (Figure 8, D and E), indicating that TSSC1 contributes to GARP recruitment to the TGN.
FIGURE 8:
TSSC1 regulates the recruitment of VPS54-GFP to the TGN. (A) SDS–PAGE and immunoblot analysis of VPS54-GFP in control and TSSC1-KD H4 cells. The positions of molecular mass markers (in kilodaltons) are indicated on the left. (B) Live-cell imaging of VPS54-GFP in a series of sequential z-stacks. The z-stacks were combined into a maximum intensity projection, and then the whole time series was combined into a maximum intensity projection, resulting in a four-dimensional projection. Scale bar. 10 μm. (C, D) FRAP analysis of VPS54-GFP at the TGN of control and TSSC1-KD H4 cells. Scale bar, 10 μm. The graphs represent the fluorescence intensity as a function of time for each cell line. (E) Graph comparing the recovery of VPS54-GFP fluorescence in control (green) and TSSC1-KD cells (red) with postbleaching intensity normalized to 1.
DISCUSSION
In contrast to the growing number of proteins that are being found to cooperate with retromer in the formation and translocation of tubular-vesicular carriers (Gallon and Cullen, 2015; Hierro et al., 2015), there is a dearth of knowledge on proteins that participate with EARP and GARP in the process of SNARE-mediated fusion of the carriers with REs and the TGN, respectively. We addressed this deficiency by performing affinity purification and mass-spectrometric analysis of proteins that interact with EARP/GARP subunits. Our experiments resulted in the identification of the previously uncharacterized TSSC1 protein as a specific interactor of both EARP and GARP (Figure 1). TSSC1 also appeared as an interactor of EARP and GARP subunits in previous high-throughput interaction screens of human proteins (Hein et al., 2015; Huttlin et al., 2015), and the Drosophila TSSC1 and EARP subunit orthologues coisolated with Rab4 in a comprehensive screen for Rab-family interactors (Gillingham et al., 2014). The specificity and functional significance of these interactions, however, were not validated in these previous studies. Our experiments now demonstrate that TSSC1 is a bona fide interactor of EARP and GARP.
Live-cell imaging and Airyscan microscopy revealed that TSSC1 partially colocalizes with VPS54, and by extension with GARP, at the TGN (Figure 5, A–C). Moreover, silencing of TSSC1 or VPS54 similarly impaired retrograde transport of STxB from endosomes to the TGN (Figures 6 and 7). Previous high-throughput RNA interference (RNAi; Bassik et al., 2013) and CRISPR interference (Gilbert et al., 2014) screens also identified a requirement of TSSC1 and/or GARP subunits for the lethal effects of other protein toxins, such as ricin, cholera toxin, and diphtheria toxin. Taken together, these findings indicate that TSSC1 and GARP are generally required for retrograde transport of various protein toxins to the TGN. TIRF microscopy also revealed colocalization of TSSC1 with Syndetin (Figure 5D), and KD of these proteins similarly increased intracellular retention of internalized Tf (Figure 7C). These experiments are consistent with TSSC1 playing roles in both retrograde transport to the TGN and recycling to the plasma membrane in conjunction with GARP and EARP, respectively.
The exact molecular function of TSSC1 remains to be elucidated. A fraction of TSSC1 coelutes with the ANG2 subunit of EARP/GARP on gel filtration of rat brain extracts (Figure 3I). It is not clear, however, whether this result means that TSSC1 is an additional subunit of EARP/GARP or a transient interactor. FRAP analysis shows that TSSC1 promotes the association of GARP with the TGN (Figure 8), suggesting that it may function as a membrane-docking factor for the complex. TSSC1 is thus the first protein shown to contribute to GARP recruitment to the TGN.
Analysis of the TSSC1 interactome (Figure 2B) revealed the noteworthy presence of another protein that copurified with TSSC1 and ANG2: the SNARE protein SNAP29. This protein is a Q-SNARE that functions in multiple trafficking pathways, including recycling to the cell surface and retrograde transport to the TGN (Steegmaier et al., 1998; Xu et al., 2004; Sato et al., 2011), possibly through interactions with the RE/TGN Q-SNAREs Syntaxin 6 and 16 (Wong et al., 1999; Xu et al., 2014). SNAP29 also interacts with EHD1 (Rotem-Yehudar et al., 2001; Xu et al., 2004), another protein involved in endocytic recycling (Lin et al., 2001; Caplan et al., 2002). TSSC1 could aid the interaction of EARP/GARP with SNAP29, thereby contributing to the recruitment of these complexes to membranes and/or the assembly of fusogenic trans-SNARE complexes.
TSSC1 is predicted to consist of a single WD40 domain. These domains have a β-propeller structure and serve as platforms for protein–protein interactions (Stirnimann et al., 2010). Of note, other WD40-domain proteins have been implicated in endocytic recycling (Shi et al., 2011) or membrane fusion events involving SNAREs and multisubunit tethering complexes related to EARP/GARP, such as the exocyst and NRZ complexes (Fujita et al., 1998; Lehman et al., 1999; Zhang et al., 2005; Civril et al., 2010). For example, the yeast WD40-domain proteins Ere1 and Ere2 function as cargo-specific adaptors for recycling of the arginine transporter Can1 to the cell surface through the retromer pathway (Shi et al., 2011). Members of the WD40-domain lethal giant larvae family, on the other hand, interact with the exocyst complex and SNAREs to promote fusion of exocytic vesicles with the plasma membrane (Fujita et al., 1998; Lehman et al., 1999; Zhang et al., 2005). Finally, the WD40-domain protein NAG assembles with ZW10 and RINT1 to form the NRZ complex, a tethering factor that functions in transport between the Golgi complex and the endoplasmic reticulum (Civril et al., 2010). Although the exact molecular functions of TSSC1 and the aforementioned WD40-domain proteins remain to be elucidated, the involvement of WD40-domain proteins in tethering and fusion is emerging as a common theme in the mechanisms of vesicle trafficking.
In sum, our experiments identify TSSC1 as a novel component of the molecular machinery that mediates cargo recycling to the plasma membrane and retrograde transport to the TGN through interactions with EARP and GARP, respectively. Our findings also show that TSSC1 and EARP/GARP participate in a network of interactions with SNARE proteins, consistent with a role of all of these proteins in tethering and fusion of endosome-derived carriers to the corresponding acceptor compartments. This role is likely critical for cell and organismal viability, as demonstrated by the lethality of null mutations in TSSC1 and GARP subunits in some settings (Schmitt-John et al., 2005; Blomen et al., 2015). Furthermore, failure of these mechanisms likely underlies the pathogenesis of hereditary disorders such as progressive cerebello-cerebral atrophy type 2 (Feinstein et al., 2014) and cerebral dysgenesis, neuropathy, ichthyosis, and keratoderma syndrome (Sprecher et al., 2005), which are caused by mutations in VPS54 and SNAP29, respectively.
MATERIALS AND METHODS
Antibodies and other reagents
The following antibodies were used for immunoblotting (sources and working dilutions are indicated in parentheses): horseradish peroxidase (HRP)–conjugated mouse monoclonal antibody to myc (1:200; 9E10; Santa Cruz Biotechnology, Santa Cruz, CA), rabbit polyclonal antibody to VPS52 (1:1000; Perez-Victoria et al., 2008), rabbit polyclonal antibody to ANG2 (1:1000; Perez-Victoria et al., 2010; HPA039650; Atlas Antibodies, Stockholm, Sweden), mouse monoclonal antibody to Syndetin (1:1000; 2D11, H00055610-M01; Abnova, Taipei City, Taiwan), rabbit polyclonal antibody to VPS53 (1:200; HPA024446; Atlas Antibodies), mouse monoclonal antibody to actin (1:1000; 612656; BD Biosciences, Franklin Lakes, NJ), rabbit polyclonal antibody to TSSC1 (1:1000 for immunoblotting, 1:300 for immunoprecipitation; ab96595; Abcam, Cambridge, United Kingdom), and HRP-conjugated mouse monoclonal antibody to GFP (MACS Miltenyi Biotec, Bergisch Gladbach, Germany; 130-091-833) (1:2000). The following antibodies were used for immunofluorescence microscopy: rabbit polyclonal antibodies to giantin (Covance, Princeton, NJ; PRB-114C; 1:3000) and HA epitope (ThermoFisher Scientific, Waltham, MA; OPA1-10980; 1:1000). To increase GFP signal in fixed cells, GFP-booster from ChromoTek (Planegg-Martinsried, Germany) was used (GFP-Booster_Atto488; 1:200). Cy3-STxB was a kind gift from Ludger Johannes (Curie Institute, Paris, France). Alexa-488-Tf was from ThermoFisher Scientific (T13342). Alexa-conjugated secondary antibodies used for immunofluorescence at 1:1000 dilution were from Invitrogen (Carlsbad, CA).
Recombinant plasmids
Most plasmids used have been previously published (Perez-Victoria et al., 2008; Perez-Victoria and Bonifacino, 2009; Schindler et al., 2015). To generate TSSC1-FOS, TSSC1 was amplified using 5′-CCGGAATTCGCCACCATGGAGGACGATGCACCAGTG-3′ and 5′-CGGGGTACCTAGCAGGATGTGGTACTTCAGGG-3′ primers and subloned into pcDNA-FOS. To generate TSSC1-RFP, TSSC1 was subcloned using Gibson assembly (New England Biolabs, Ipswich, MA) into EcoR1 (New England Biolabs)-linearized RFP-N1 using 5′-GGACTCAGATCTCGAGCTCAAGCTTCGAATGGAGGACGATGCACCAGTG-3′ and 3′-CCCTGAAGTACCACATCCTGCTACTGCAGTCGACGGTACCGCGGGCCCGGGATCCACCG-5′. To generate 3HA-TSSC1, 5′-CCGGAATTCATGGAGGACGATGCACCAGTG-3′ and 5′-ACGCGTCGACTCATAGCAGGATGTGGTACTTCAG-3′ were used to amplify and subclone TSSC1 into a pCIneo-HA vector backbone.
RNAi
Transfection with siRNAs was performed using Lipofectamine 2000 (ThermoFisher Scientific). Cells were plated at 60% confluency in a 24-well plate and adhered overnight. Cells were then transfected with siRNA using Lipofectamine 2000 according to the manufacturer’s instructions, incubated overnight, split into three wells of a 24-well plate, and adhered overnight again before being transfected with overnight incubation. Cells were finally split again onto either coverslips or Lab-Tek chambers (for imaging; 155380; ThermoFisher Scientific) or into a 96-well plate (for Tf uptake followed by FACS). Proteins targeted by siRNAs used were TSSC1 (ON-TARGETplus TSSC1 siRNA; J-011410-05-0005; Dharmacon, Lafayette, CO), VPS54 (Perez-Victoria et al., 2008; Schindler et al., 2015; UCACGAUGUUUGCAGUUAAUU; J-021174-07 Dharmacon), and Syndetin (Schindler et al., 2015; 5′ AGAACAGAUGUACGGUUAA-3′).
Tandem affinity purification and mass spectrometry
Affinity purifications and pull downs using FOS- or OSF-tagged constructs were performed as described by Schindler et al. (2015), with analysis by liquid chromatography–mass spectrometry done at the Taplin Mass Spectrometry Facility (Harvard Medical School, Boston, MA). For VPS54-GFP mass spectrometry, nanobody to GFP coupled to magnetic beads (ChromoTek) was used on VPS54-GFP and GFP-expressing H4 cell lines, with lysate generated from a 15-cm plate at 90% confluence. ChromoTek lysis buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40, 0.02% Thimerosal) supplemented with cOmplete EDTA-free Protease Inhibitor tablets obtained from Roche (Penzberg, Germany; 11873580001) was used. Samples were eluted from the beads with 50 μl of 0.2 M glycine, pH 2.5, and neutralized with 5 μl of 1 M Tris base, pH 10.4. Samples were precipitated using trichloroacetic acid and air-dried. Mass spectrometric analysis was done at the National Institute of Neurological Disorders and Stroke Protein/Peptide Sequencing Facility using an Orbitrap Elite instrument.
Generation of cointeraction network
To generate the interaction network, mass spectrometry data from TSSC1-FOS, ANG2-FOS, VPS52-OSF, VPS53-OSF, Syndetin-OSF, and VPS54-GFP were imported into Python 2.7 (www.python.org). Negative controls were subtracted. To remove commonly occurring negative results that were not in our negative controls, we filtered data sets using the CRAPome API (www.crapome.org; Mellacheruvu et al., 2013) by generating a threshold based on average spectral count by hits per experiment in the CRAPome negative control database and filtering to a threshold set to 0.25. Filtered hits were combined with data from previously published findings using the BioGRID API (http://thebiogrid.org; Stark et al., 2006) to find cointeractors between all hits and identify interacting complexes. These were then mapped as a network using the NetworkX package (https://networkx.github.io) with each line representing an interaction (either novel or published). Protein ontology searches were manually performed and the hits grouped and annotated using Adobe Illustrator (Adobe Systems, San Jose, CA).
Phylogenetic analysis
To generate the Coulson plot and phylogenetic tree, BLASTP searches were performed with the appropriate human reference sequence (Uniprot IDs TSSC1 Q53HC9, Syndetin Q96JG6, VPS54 Q9P1Q0, VPS53 Q5VIR6, VPS52 Q8N1B4, and ANG2 Q9UID3) against the relevant organisms. The hit with the highest homology was then reciprocally used to perform a BLASTP search against the human proteome. If the search resulted in the original human protein with the highest identity, this was considered a positive match and the match plotted. If no hit could be found by this method, additional approaches were taken to identify more distant homologues, using either the human protein or a more closely related homologue (e.g., for Viridiplantae, the Arabidopsis thaliana homologue). These were used for a multi-iteration PSI-BLAST search on either the specific organism or the supergroup. Iterations were repeated until convergence, and any hits were probed for validity using a reverse-BLASTP or reverse-PSI-BLAST against the human proteome. For the Coulson plot, the intensity was plotted by the intensity based on a BLASTP alignment. To generate the cladogram of TSSC1, a similar reciprocal BLASTP search was performed against the specified organisms.
Cell lines, tissue extraction, and gel filtration
H4 stable cells lines for VPS54-GFP and GFP were generated by transfection with the previously described VPS54-GFP construct (Perez-Victoria et al., 2008) and a soluble GFP construct using Lipofectamine 2000. After 2 d, G418 (10131-035; Life Technologies, Carlsbad, CA) was added to the cells at a concentration of 1 mg/ml, and the cells were split into a 96-well plate at a concentration of 1 cell per well. Wells were visually inspected for GFP expression in single colonies using a Zeiss (Carl Zeiss, Oberkochen, Germany) epifluorescence microscope and selected for confirmation by immunoblot analysis for GFP expression. WT, TSSC1-KO, and VPS54-KO HAP1 cell lines were from Haplogen Genomics (Vienna, Austria; HAP1_TSSC1_1451-00, and HAP1_VPS54_12303-0). Mouse tissue extraction for immunoblot analysis was done as described by Schindler et al. (2015). For gel filtration analysis, rat brain tissue from Sprague Dawley rats (embryonic day 18) was extracted in lysis buffer (0.25 M sucrose, 50 mM Tris-HCl, pH 7.4, 50 mM NaCl, and 0.5% Triton supplemented with EDTA-free protease inhibitor tablets from Roche [11873580001]) by manual dicing using a razor blade on a glass Petri dish followed by disruption by 20 strokes with a Dounce homogenizer and incubation of homogenates on ice for 30 min with thorough pipetting every 10 min and centrifugation in 1.5-ml tubes for 20 min at a top speed and 4°C in a bench-top microcentrifuge to remove cell debris. Gel filtration was done on a Superose 6 10/300 GL column (GE Bioscience, Pittsburgh, PA) as described by Schindler et al. (2015).
Immunoprecipitations
For immunoprecipitation of VPS54-GFP, anti-GFP nanobody-conjugated beads (ChromoTek) were used as per manufacturer’s instructions. Cell lysate was generated from the VPS54-GFP stable H4 cell line described here. Cells were grown in a 100-mm cell culture plate. For immunoprecipitation of endogenous TSSC1, cortical rat brain tissue from Sprague Dawley rats (embryonic day 18) was lysed in lysis buffer (0.25 M sucrose, 50 mM Tris, pH 7.4, 50 mM NaCl, and 0.5% Triton supplemented with EDTA-free protease inhibitor tablets from Roche [11873580001]) by manual dicing, disruption, and centrifugation as before. Lysate was precleared at 4°C for 2 h with protein A–Sepharose beads (GE Healthcare, Pittsburgh, PA), and simultaneously antibody was prebound to 40 μl of phosphate-buffered saline (PBS)–washed beads in PBS. After incubation, both lysate beads and antibody-bound beads were pelleted in a microcentrifuge for 2 s at 16,000 × g (maximum speed; 4°C), and the supernatant from the lysate was added to the antibody-bound beads. In addition, 100 μl of the lysate was set aside to be used as input. Beads were incubated with antibody overnight at 4°C. After incubation, beads were washed three times in wash buffer (50 mM Tris, pH 7.4, 50 mM NaCl) and once in PBS before being analyzed by SDS–PAGE and immunoblotting.
Immunofluorescence staining
Cells were transfected with either Lipofectamine 2000 (Thermo Fisher Scientific) or FuGENE HD (Promega, Madison, WI) on glass coverslips (EF15973A; Daigger), and after allowing gene expression for 1 d, cells were washed twice in PBS(CaMg) (PBS supplemented with calcium and magnesium ions), fixed for 30 min in 4% paraformaldehyde in PBS at room temperature, washed with PBS(CaMg), and permeabilized in 0.2% Triton X-100 for 5 min at room temperature. Cells were blocked in PBS(CaMg) and 3% bovine serum albumin (BSA) for 2 h at room temperature. Primary and secondary antibodies were used at the foregoing concentrations in the blocking buffer, incubating at 37°C for 30 min with four washes in PBS(CaMg) between each step. Cells were mounted on glass slides with Fluoromount containing 4′,6-diamidino-2-phenylindole (DAPI) to visualize the nucleus (17989-97; EMS Immuno Mount; Electron Microscopy Sciences, Hatfield, PA).
Microscopy
Live-cell imaging was performed on a Nikon Eclipse Ti Microscope System (Nikon Instruments, Melville, NY) at 37°C and 5% CO2. Images were captured using an electron-multiplying charge-coupled device camera (Photometrics Evolve 512; Photometrics, Tucson, AZ) and a Plan Apo VC 60× H objective (numerical aperture [NA] 1.40). Diffraction-limited fixed-cell imaging was performed with a Zeiss 780 (Carl Zeiss). Superresolution imaging was performed using a Zeiss 880 Airyscan microscope and a 63×/1.40 NA Plan-Apochromat Oil DIC M27 objective lens (Carl Zeiss) and processed at setting 6. Image processing was performed in ImageJ (http://imagej.nih.gov/ij/), including cropping and average/maximum intensity projections. Photobleaching experiments were performed at 37°C and 5% CO2 on a Nikon Eclipse Ti Microscope System equipped with a Galvo miniscanner to allow photobleaching and subsequent spinning-disk confocal imaging. TIRF images were acquired with an Apo TIRF 100× objective.
Microscopy quantification
Quantification of Spearman’s r values were performed in ImageJ using the PSC colocalization plug-in (French et al., 2008). Organelle distance mapping was performed using the Imaris imaging platform (Bitplane, Belfast, United Kingdom). A pseudonucleus was added to the observable nuclear center, with a diameter of 10 μm. Shiga toxin–containing organelles in z-stacked time series were mapped using an estimated xy-diameter of 1 μm and threshold of 600. Distance of at least 75 detectable organelles was measured. Using a distance transformation from the pseudonucleus, the 3D distance of each detectable object was calculated. Data were processed and plotted in Python 2.7.
Uptake and FACS analysis
Cy3-conjugated Shiga toxin B subunit (Cy3-STxB) uptake was performed as previously described (Perez-Victoria et al., 2008). In brief, cells were incubated for 30 min in HAP1 uptake medium (Iscove’s modified Dulbecco’s medium [IMDM; ThermoFisher Scientific] –FBS + 1% BSA). Cy3-STxB was added at 0.5 μg/ml and incubated for 5 min. For live-cell imaging, to avoid disturbing the cells and thus allow continuous imaging of STxB uptake, STxB was not removed from the medium but supplemented with a fivefold excess of complete IMDM prewarmed to 37°C. For analysis of Cy3-STxB uptake in fixed cells, cells were incubated with 0.5 μg/ml Cy3-STxB for 15 min and washed with prewarmed 37°C complete IMDM (+10% FBS) before being incubated for 1 h with prewarmed complete IMDM. After 1 h, cells were washed in PBS(CaMg) twice before being fixed with paraformaldehyde as described. Antibody staining was as described. The Tf uptake assays were performed in 96-well plates. Knockdown and WT cells were plated, and 30 min before the experiment, the medium was exchanged for 37°C prewarmed uptake medium (DMEM –FPB +1% BSA) and further supplemented with 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) to buffer pH changes during the experiment. In a 37°C water bath, Alexa 488–conjugated Tf was added in 3-min time steps to the plated cells with a multichannel pipette. After the completed time course (in which the first cells had been incubated for 27 min with the fluorescent Tf), the medium was removed from the plates and the cells washed twice in ice-cold PBS on ice. Cells were then fixed with 4% paraformaldehyde for 10 min on ice. To remove cells from the plate for FACS, cells were incubated for 1 h with 10 mM EDTA and manually agitated. Suspended cells were analyzed using an LSRFortessa (BD Biosciences) with HTS plate reader module, and data were analyzed using FlowJo software (Ashland, OR; www.flowjo.com). For the Tf recycling assay, KD and WT cells were plated onto six-well plates (one plate per time point) in DMEM, which, before the experiment, was exchanged with 37°C prewarmed uptake medium (DMEM – FPB +1% BSA), further supplemented with 25 mM HEPES to buffer pH changes during the experiment. Alexa 488–conjugated Tf was incubated for 30 min with the cells, after which it was removed and chased for the indicated times with complete DMEM supplemented with HEPES. The cells were analyzed as described using FACS.
Supplementary Material
Acknowledgments
We thank Yan Li at the National Institute of Neurological Disorders and Stroke Protein/Peptide Sequencing Facility for the mass-spectrometric analysis of VPS54-GFP isolates, Jing Pu for mouse tissues used in immunoblotting, Eric Balzer (Nikon Instruments) for assistance with photobleaching experiments, Matthew Gastinger (Imaris, Bitplane) for help with quantification of data, and Ludger Johannes for his generous gift of Cy3-STxB. This work was funded by the Intramural Program of the National Institute of Child Health and Human Development, National Institutes of Health (ZIA HD001607).
Abbreviations used:
- API
application program interface
- BLAST
basic local alignment search tool
- EARP
endosome-associated recycling protein complex
- EE
early endosomes
- FACS
fluorescence-activated cell sorting
- FOS
FLAG/One-STrEP epitope tag
- FRAP
fluorescence recovery after photobleaching
- GARP
Golgi-associated retrograde protein complex
- GFP
green fluorescent protein
- HA
3-hemagglutinin epitope tag
- KD
knockdown
- KO
knockout
- MPR
mannose-6-phosphate receptor
- MVB
multivesicular body
- PM
plasma membrane
- RE
recycling endosome
- RFP
red fluorescent protein
- siRNA
small interfering RNA
- SNARE
soluble N-ethylmaleimide–sensitive factor attachment protein receptor
- STxB
Shiga toxin B subunit
- TfR
transferrin receptor
- TGN
trans-Golgi network
- TIRF
total internal reflection fluorescence
- TSSC1
tumor-suppressing subchromosomal transferable fragment candidate gene 1
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-04-0209) on July 20, 2016.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Barlow LD, Dacks JB, Wideman JG. From all to (nearly) none: tracing adaptin evolution in fungi. Cell Logist. 2014;4:e28114. doi: 10.4161/cl.28114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassik MC, Kampmann M, Lebbink RJ, Wang S, Hein MY, Poser I, Weibezahn J, Horlbeck MA, Chen S, Mann M, et al. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013;152:909–922. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomen VA, Majek P, Jae LT, Bigenzahn JW, Nieuwenhuis J, Staring J, Sacco R, van Diemen FR, Olk N, Stukalov A, et al. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350:1092–1096. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Rojas R. Retrograde transport from endosomes to the trans-Golgi network. Nat Rev Mol Cell Biol. 2006;7:568–579. doi: 10.1038/nrm1985. [DOI] [PubMed] [Google Scholar]
- Burda P, Padilla SM, Sarkar S, Emr SD. Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J Cell Sci. 2002;115:3889–3900. doi: 10.1242/jcs.00090. [DOI] [PubMed] [Google Scholar]
- Caplan S, Naslavsky N, Hartnell LM, Lodge R, Polishchuk RS, Donaldson JG, Bonifacino JS. A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. EMBO J. 2002;21:2557–2567. doi: 10.1093/emboj/21.11.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civril F, Wehenkel A, Giorgi FM, Santaguida S, Di Fonzo A, Grigorean G, Ciccarelli FD, Musacchio A. Structural analysis of the RZZ complex reveals common ancestry with multisubunit vesicle tethering machinery. Structure. 2010;18:616–626. doi: 10.1016/j.str.2010.02.014. [DOI] [PubMed] [Google Scholar]
- Conibear E, Cleck JN, Stevens TH. Vps51p mediates the association of the GARP (Vps52/53/54) complex with the late Golgi t-SNARE Tlg1p. Mol Biol Cell. 2003;14:1610–1623. doi: 10.1091/mbc.E02-10-0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein M, Flusser H, Lerman-Sagie T, Ben-Zeev B, Lev D, Agamy O, Cohen I, Kadir R, Sivan S, Leshinsky-Silver E, et al. VPS53 mutations cause progressive cerebello-cerebral atrophy type 2 (PCCA2) J Med Genet. 2014;51:303–308. doi: 10.1136/jmedgenet-2013-101823. [DOI] [PubMed] [Google Scholar]
- Field HI, Coulson RM, Field MC. An automated graphics tool for comparative genomics: the Coulson plot generator. BMC Bioinformatics. 2013;14:141. doi: 10.1186/1471-2105-14-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French AP, Mills S, Swarup R, Bennett MJ, Pridmore TP. Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat Protoc. 2008;3:619–628. doi: 10.1038/nprot.2008.31. [DOI] [PubMed] [Google Scholar]
- Fridmann-Sirkis Y, Kent HM, Lewis MJ, Evans PR, Pelham HR. Structural analysis of the interaction between the SNARE Tlg1 and Vps51. Traffic. 2006;7:182–190. doi: 10.1111/j.1600-0854.2005.00374.x. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Shirataki H, Sakisaka T, Asakura T, Ohya T, Kotani H, Yokoyama S, Nishioka H, Matsuura Y, Mizoguchi A, et al. Tomosyn: a syntaxin-1-binding protein that forms a novel complex in the neurotransmitter release process. Neuron. 1998;20:905–915. doi: 10.1016/s0896-6273(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Gallon M, Cullen PJ. Retromer and sorting nexins in endosomal sorting. Biochem Soc Trans. 2015;43:33–47. doi: 10.1042/BST20140290. [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Sinka R, Torres IL, Lilley KS, Munro S. Toward a comprehensive map of the effectors of rab GTPases. Dev Cell. 2014;31:358-373. doi: 10.1016/j.devcel.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev Cell. 2009;17:699–711. doi: 10.1016/j.devcel.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- Hierro A, Gershlick DC, Rojas AL, Bonifacino JS. Formation of tubulovesicular carriers from endosomes and their fusion to the trans-Golgi network. Int Rev Cell Mol Biol. 2015;318:159–202. doi: 10.1016/bs.ircmb.2015.05.005. [DOI] [PubMed] [Google Scholar]
- Hirst J, Barlow LD, Francisco GC, Sahlender DA, Seaman MN, Dacks JB, Robinson MS. The fifth adaptor protein complex. PLoS Biol. 2011;9:e1001170. doi: 10.1371/journal.pbio.1001170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Yang Y, Zhang C, Niu Y, Li K, Zhao X, Liu JJ. The retromer component SNX6 interacts with dynactin p150(Glued) and mediates endosome-to-TGN transport. Cell Res. 2009;19:1334–1349. doi: 10.1038/cr.2009.130. [DOI] [PubMed] [Google Scholar]
- Hu RJ, Lee MP, Connors TD, Johnson LA, Burn TC, Su K, Landes GM, Feinberg AP. A 2.5-Mb transcript map of a tumor-suppressing subchromosomal transferable fragment from 11p15.5, and isolation and sequence analysis of three novel genes. Genomics. 1997;46:9–17. doi: 10.1006/geno.1997.4981. [DOI] [PubMed] [Google Scholar]
- Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, et al. The BioPlex network: a systematic exploration of the human interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes L, Romer W. Shiga toxins–from cell biology to biomedical applications. Nat Rev Microbiol. 2010;8:105–116. doi: 10.1038/nrmicro2279. [DOI] [PubMed] [Google Scholar]
- Jovic M, Sharma M, Rahajeng J, Caplan S. The early endosome: a busy sorting station for proteins at the crossroads. Histol Histopathol. 2010;25:99–112. doi: 10.14670/hh-25.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Koumandou VL, Dacks JB, Coulson RM, Field MC. Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol Biol. 2007;7:29. doi: 10.1186/1471-2148-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman K, Rossi G, Adamo JE, Brennwald P. Yeast homologues of tomosyn and lethal giant larvae function in exocytosis and are associated with the plasma membrane SNARE, Sec9. J Cell Biol. 1999;146:125–140. doi: 10.1083/jcb.146.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liewen H, Meinhold-Heerlein I, Oliveira V, Schwarzenbacher R, Luo G, Wadle A, Jung M, Pfreundschuh M, Stenner-Liewen F. Characterization of the human GARP (Golgi associated retrograde protein) complex. Exp Cell Res. 2005;306:24–34. doi: 10.1016/j.yexcr.2005.01.022. [DOI] [PubMed] [Google Scholar]
- Lin SX, Grant B, Hirsh D, Maxfield FR. Rme-1 regulates the distribution and function of the endocytic recycling compartment in mammalian cells. Nat Cell Biol. 2001;3:567–572. doi: 10.1038/35078543. [DOI] [PubMed] [Google Scholar]
- Mallard F, Tang BL, Galli T, Tenza D, Saint-Pol A, Yue X, Antony C, Hong W, Goud B, Johannes L. Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J Cell Biol. 2002;156:653–664. doi: 10.1083/jcb.200110081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield FR, McGraw TE. Endocytic recycling. Nat Rev Mol Cell Biol. 2004;5:121–132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panic B, Whyte JR, Munro S. The ARF-like GTPases Arl1p and Arl3p act in a pathway that interacts with vesicle-tethering factors at the Golgi apparatus. Curr Biol. 2003;13:405–410. doi: 10.1016/s0960-9822(03)00091-5. [DOI] [PubMed] [Google Scholar]
- Perez-Victoria FJ, Bonifacino JS. Dual roles of the mammalian GARP complex in tethering and SNARE complex assembly at the trans-golgi network. Mol Cell Biol. 2009;29:5251–5263. doi: 10.1128/MCB.00495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Victoria FJ, Mardones GA, Bonifacino JS. Requirement of the human GARP complex for mannose 6-phosphate-receptor-dependent sorting of cathepsin D to lysosomes. Mol Biol Cell. 2008;19:2350–2362. doi: 10.1091/mbc.E07-11-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Victoria FJ, Schindler C, Magadan JG, Mardones GA, Delevoye C, Romao M, Raposo G, Bonifacino JS. Ang2/Fat-free is a conserved subunit of the Golgi-associated retrograde protein (GARP) complex. Mol Biol Cell. 2010;21:3386–3395. doi: 10.1091/mbc.E10-05-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popoff V, Mardones GA, Tenza D, Rojas R, Lamaze C, Bonifacino JS, Raposo G, Johannes L. The retromer complex and clathrin define an early endosomal retrograde exit site. J Cell Sci. 2007;120:2022–2031. doi: 10.1242/jcs.003020. [DOI] [PubMed] [Google Scholar]
- Pu J, Schindler C, Jia R, Jarnik M, Backlund P, Bonifacino JS. BORC, a multiprotein complex that regulates lysosome positioning. Dev Cell. 2015;33:176–188. doi: 10.1016/j.devcel.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas R, van Vlijmen T, Mardones GA, Prabhu Y, Rojas AL, Mohammed S, Heck AJ, Raposo G, van der Sluijs P, Bonifacino JS. Regulation of retromer recruitment to endosomes by sequential action of Rab5 and Rab7. J Cell Biol. 2008;183:513–526. doi: 10.1083/jcb.200804048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem-Yehudar R, Galperin E, Horowitz M. Association of insulin-like growth factor 1 receptor with EHD1 and SNAP29. J Biol Chem. 2001;276:33054–33060. doi: 10.1074/jbc.M009913200. [DOI] [PubMed] [Google Scholar]
- Sato M, Saegusa K, Sato K, Hara T, Harada A, Sato K. Caenorhabditis elegans SNAP-29 is required for organellar integrity of the endomembrane system and general exocytosis in intestinal epithelial cells. Mol Biol Cell. 2011;22:2579–2587. doi: 10.1091/mbc.E11-04-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scelfo R, Sabbioni S, Barbanti-Brodano G, Negrini M. Subchromosomal assignment1 of the TSSC1 gene to human chromosome band 11p15.5 near the HBB gene cluster. Cytogenet Cell Genet. 1998;83:52–53. doi: 10.1159/000015123. [DOI] [PubMed] [Google Scholar]
- Schindler C, Chen Y, Pu J, Guo X, Bonifacino JS. EARP, a multisubunit tethering complex involved in endocytic recycling. 2015;17:639–650. doi: 10.1038/ncb3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt-John T, Drepper C, Mussmann A, Hahn P, Kuhlmann M, Thiel C, Hafner M, Lengeling A, Heimann P, Jones JM, et al. Mutation of Vps54 causes motor neuron disease and defective spermiogenesis in the wobbler mouse. Nat Genet. 2005;37:1213–1215. doi: 10.1038/ng1661. [DOI] [PubMed] [Google Scholar]
- Seaman MN, Harbour ME, Tattersall D, Read E, Bright N. Membrane recruitment of the cargo-selective retromer subcomplex is catalysed by the small GTPase Rab7 and inhibited by the Rab-GAP TBC1D5. J Cell Sci. 2009;122:2371–2382. doi: 10.1242/jcs.048686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Stefan CJ, Rue SM, Teis D, Emr SD. Two novel WD40 domain-containing proteins, Ere1 and Ere2, function in the retromer-mediated endosomal recycling pathway. Mol Biol Cell. 2011;22:4093–4107. doi: 10.1091/mbc.E11-05-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprecher E, Ishida-Yamamoto A, Mizrahi-Koren M, Rapaport D, Goldsher D, Indelman M, Topaz O, Chefetz I, Keren H, O’Brien TJ, et al. A mutation in SNAP29, coding for a SNARE protein involved in intracellular trafficking, causes a novel neurocutaneous syndrome characterized by cerebral dysgenesis, neuropathy, ichthyosis, and palmoplantar keratoderma. Am J Hum Genet. 2005;77:242–251. doi: 10.1086/432556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starcevic M, Dell’Angelica EC. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1) J Biol Chem. 2004;279:28393–28401. doi: 10.1074/jbc.M402513200. [DOI] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steegmaier M, Yang B, Yoo JS, Huang B, Shen M, Yu S, Luo Y, Scheller RH. Three novel proteins of the syntaxin/SNAP-25 family. J Biol Chem. 1998;273:34171–34179. doi: 10.1074/jbc.273.51.34171. [DOI] [PubMed] [Google Scholar]
- Stirnimann CU, Petsalaki E, Russell RB, Muller CW. WD40 proteins propel cellular networks. Trends Biochem Sci. 2010;35:565–574. doi: 10.1016/j.tibs.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Walker G, Dorrell RG, Schlacht A, Dacks JB. Eukaryotic systematics: a user’s guide for cell biologists and parasitologists. Parasitology. 2011;138:1638–1663. doi: 10.1017/S0031182010001708. [DOI] [PubMed] [Google Scholar]
- Wang DC, Wang HF, Yuan ZN. Runx2 induces bone osteolysis by transcriptional suppression of TSSC1. Biochem Biophys Res Commun. 2013;438:635–639. doi: 10.1016/j.bbrc.2013.07.131. [DOI] [PubMed] [Google Scholar]
- Wassmer T, Attar N, Harterink M, van Weering JR, Traer CJ, Oakley J, Goud B, Stephens DJ, Verkade P, Korswagen HC, et al. The retromer coat complex coordinates endosomal sorting and dynein-mediated transport, with carrier recognition by the trans-Golgi network. Dev Cell. 2009;17:110–122. doi: 10.1016/j.devcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SH, Xu Y, Zhang T, Griffiths G, Lowe SL, Subramaniam VN, Seow KT, Hong W. GS32, a novel Golgi SNARE of 32 kDa, interacts preferentially with syntaxin 6. Mol Biol Cell. 1999;10:119–134. doi: 10.1091/mbc.10.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Mohtashami M, Stewart B, Boulianne G, Trimble WS. Drosophila SNAP-29 is an essential SNARE that binds multiple proteins involved in membrane traffic. PLoS One. 2014;9:e91471. doi: 10.1371/journal.pone.0091471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Shi H, Wei S, Wong SH, Hong W. Mutually exclusive interactions of EHD1 with GS32 and syndapin II. Mol Membr Biol. 2004;21:269–277. doi: 10.1080/09687680410001716871. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang P, Gangar A, Zhang J, Brennwald P, TerBush D, Guo W. Lethal giant larvae proteins interact with the exocyst complex and are involved in polarized exocytosis. J Cell Biol. 2005;170:273–283. doi: 10.1083/jcb.200502055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.