Abstract
Hemoglobinopathies, including β-thalassemia and sickle cell disease (SCD), are a heterogeneous group of commonly inherited disorders affecting the function or levels of hemoglobin. Disease phenotype can be severe with substantial morbidity and mortality. Bone marrow transplantation is curative, but limited to those patients with an appropriately matched donor. Genetic therapy, which utilizes a patient’s own cells, is thus an attractive therapeutic option. Numerous therapies are currently in clinical trials or in development, including therapies utilizing gene replacement therapy using lentiviruses and the latest gene editing techniques. In addition, methods are being developed that may be able to expand gene therapies to those with poor access to medical care, potentially significantly decreasing the global burden of disease.
Keywords: gene editing, gene therapy, hemoglobin, hemoglobinopathy, sickle cell anemia, thalassemia
Introduction to hemoglobinopathies
Hemoglobin is a protein found in red blood cells (RBCs) that is critical for the process of oxygen transportation, allowing for oxygen to be transported from the lungs to the tissues. Hemoglobin exists as a heterotetramer, containing two subunits from the α-globin subfamily (ζ, α2, and α1) and two subunits from the β-globin subfamily (ε, Gγ, Aγ, δ, and β) [Forget and Hardison, 2009]. The expression of the different α- and β-globin family subunits are developmentally-regulated processes. Mutated globin proteins or decreased expression of globin proteins (hemoglobinopathies) can lead to poor oxygen transportation and a variety of other damaging physiological outcomes.
Hemoglobinopathies are a large heterogeneous group of inherited disorders of hemoglobin synthesis that can be further subdivided into thalassemia syndromes and structural hemoglobin variants. Thalassemia syndromes result from diminished or absence of either α- or β-globin, leading to α-thalassemia or β-thalassemia, respectively. Structural hemoglobin variants result from mutations within a globin gene, leading to disruption of the normal peptide structure and function.
Hemoglobinopathies can be found throughout the world, but prevalence is increased among certain ethnicities. In particular, sickle cell disease (SCD) is more common among people with sub-Saharan African or Indian ancestry. As a whole, thalassemias are the most common monogenic disorders in the world [Weatherall, 2001]. According to a World Health Organization (WHO) report, approximately 5% of the world’s population carries trait genes for hemoglobin disorders, mainly sickle cell anemia (SCA) and α- or β-thalassemias [WHO, 2011]. Approximately 100,000 Americans have SCA, with 300,000 affected infants born yearly worldwide [Bauer et al. 2012; Yawn et al. 2014]. The global disease burden of hemoglobinopathies is borne enormously by the African continent, where according to the WHO, 5–16% of under-five mortality in some areas of sub-Saharan Africa is attributed to SCA [WHO, 2006]. Additionally, data suggest that children born in Africa with SCA have an early-life mortality of 50–90% [Grosse et al. 2011]. This disproportionate burden on underdeveloped regions is important to remember when considering application of gene therapies for hemoglobinopathies.
Hemoglobinopathies, especially SCA, are prime targets for gene therapy for a variety of reasons. Their high prevalence, significant morbidity and mortality, and the resulting high cost of medical care portends that a curative therapy can greatly improve patient outcomes and significantly reduce associated medical costs. Due to its genetic etiology, genetically-modified hematopoietic stem and progenitor cells (HSPCs) are able to pass on their modified genome to daughter cells, including RBC precursors. Given the self-renewing capacity of HSPCs, a single treatment is potentially curative. With regards to gene editing strategies, many of the mutations resulting in hemoglobinopathies are single point mutations, which typically allow for greater gene correction efficiencies than more complex mutations. Lastly, the hemoglobinopathies have already been successfully treated with hematopoietic stem cell transplant (HSCT), which similar to many gene therapy techniques; requires engraftment of the long-term repopulating hematopoietic stem cells (HSCs). As will be described, gene therapy for hemoglobinopathies can be divided into four general categories (1) gene addition, (2) gene knockdown to improve the β-globinopathy phenotype, (3) globin gene editing, and (4) gene editing of globin regulatory elements.
Thalassemias
α-thalassemia typically results from functional deletion of two or more of the four α-globin genes. Loss of α-globin chains lead to a reduction in the predominant hemoglobin, hemoglobin A (HbA), composed of a α2β2 heterotetramer. Patients with loss of a single α-globin gene (-α/αα) are typically asymptomatic silent carriers. Clinical phenotype of α-thalassemia can range from mild effects on hemoglobin indices to fetal hydrops and intrauterine demise, depending on the number of α-globin genes affected and the specific mutations involved.
Genetic mutations leading to β-thalassemia may be found within the β-globin genes itself, or external to the genes, within the globin locus. Most commonly, the mutations resulting in β-thalassemia are point mutations. Hundreds of different mutations have been described affecting β-globin levels via effects on a wide range of processes, including: transcription, mRNA splicing/processing, RNA stability, translation, and globin peptide stability. The low β-globin content allows the excess α-globin chains to precipitate in erythroid precursors. The α-globin aggregates cause cell membrane damage and lead to early erythroid precursor death. The resultant ineffective erythropoiesis found in patients, if severe, may necessitate frequent blood transfusions. β-globinopathies typically present 6–12 months after birth when γ-globin expression, which is the predominate globin expressed from the β-globin family during fetal life, begins to diminish to residual amounts typically found throughout the remainder of life. Disease severity may range from asymptomatic to severe.
Structural hemoglobinopathies
The most common structural hemoglobinopathy, SCD, is due to a single point mutation in β-globin, an A to T mutation resulting in a Glu6Val substitution. This mutation leads to a mutated β-globin chain, βS, and results in production of hemoglobin S (HbS), composed of two α-globin peptides and two mutated β-globin peptides (α2βS2), instead of the normal HbA (α2β2). Individuals with one copy of βS are typically asymptomatic and referred to as having a sickle cell trait. Any additional pathological mutation in the other β-globin gene results in SCD. An individual exclusively producing HbS (i.e. with two βS genes [βS/βS] or sickle β0 thalassemia [βS/β0]), is referred to as having SCA. Besides SCA, several other structural hemoglobin variants exist (e.g. hemoglobin C, D, and E). All of the structural hemoglobin variants are inherited in an autosomal recessive manner, and compound heterozygotes for any of these variants along with a sickle allele (HbSC, HbSD or HbSE) phenotypically result in SCD. Similarly, inheritance of a β+ thalassemia allele along with a βS allele (HbS-β+ thalassemia) also results in a SCD phenotype, albeit with milder symptomatology.
HbS, is prone to polymerization under reduced oxygen conditions, leading to the cascade of sickling of RBC, RBC hemolysis and aggregation of RBC and adhesion of white blood cells (WBCs) in the microvasculature, and finally vaso-occlusion. This process can lead to a variety of significant adverse sequelae of varying severity (Table 1). The manifestations of SCD are chronic and progressive with significant associated morbidity and mortality. Similar to severe thalassemia, patients with severe SCD often require frequent blood transfusions, and are at risk of transfusion-induced iron overload. Not infrequently, multiple hemoglobinopathies will occur concurrently (e.g. βS and β-thalassemia are often seen together). Concurrent hemoglobinopathies generally lead to a less severe phenotype (i.e. phenotypes of a sickle allele and β+ thalassemia allele or a sickle allele and an HbC allele are typically milder than two sickle alleles, although exceptions exist, such as HbSD).
Table 1.
Common sequelae of sickle cell disease.
| Vaso-occlusive crisis |
| Acute chest syndrome |
| Acute stroke |
| Priapism |
| Hepatobilliary complications |
| Splenic sequestration |
| Acute renal failure |
| Anemia |
| Severe infections (e.g. pneumonia) |
| Chronic obstructive pulmonary disease |
| Asthma |
| Pulmonary hypertension |
| Congestive heart failure |
| Cardiomegaly |
| Arrhythmia |
| Cardiomyopathy |
| Leg ulcers |
| Avascular necrosis |
| Other chronic manifestations |
| Death |
Both structural hemoglobinopathies and the thalassemias can be cured by HSCT [Bernaudin et al. 2007; Lucarelli et al. 2012; Hsieh et al. 2014]. While potentially curative, HSCT is only available to a minority of patients due to the lack of appropriately-matched donors for most patients. For instance, only 15–20% of patients with SCD are able to identify an appropriately-matched donor for possible HSCT. Additionally, HSCT is a rigorous treatment, often with its own morbidity and mortality. Graft versus host disease, toxic effects of conditioning regimens, prolonged and severe cytopenias, susceptibility to infections, and other adverse effects are commonly observed during HSCT. Some of these effects have been mitigated somewhat with the increasing use of nonmyeloablative conditioning regimens.
Principles of gene therapy
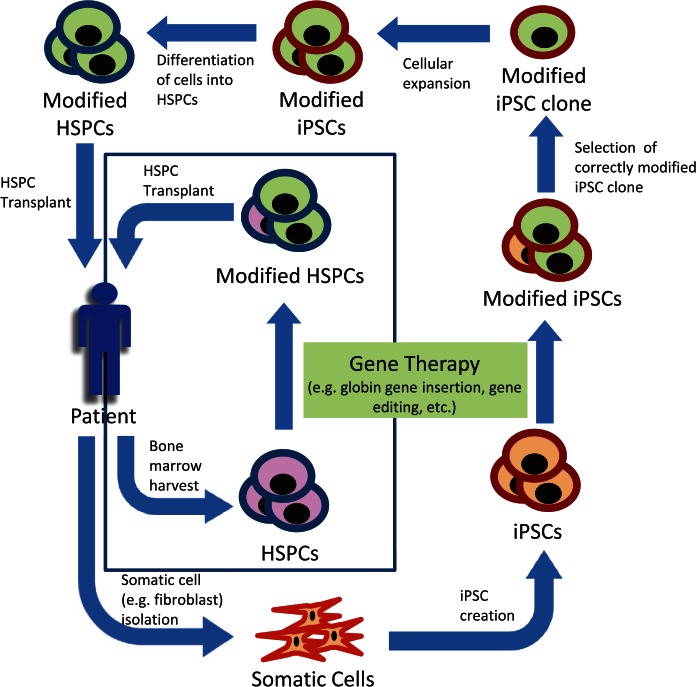
Due to the above considerations, gene therapy utilizing a patient’s autologous stem cells hold great promise, as they would obviate the need to identify matched donors and further mitigate the morbidity and mortality of allogenic HSCT. A general outline of the steps involved in both current and gene therapies in development is shown in Figure 1. Most commonly, gene therapy is accomplished by first harvesting a patient’s HSPCs from either bone marrow, peripheral blood, or umbilical cord blood. Next the cells are exposed to a therapy that introduces designed modifications into the cells genome. Traditional gene therapies currently in clinical trials consist of inserting an additional globin gene (β or γ globin or an anti-sickling β globin) via a lentiviral vector that integrates into the host cell’s genome. The cells are then transplanted back into the patient where the modified cells proliferate and repopulate the hematopoietic compartment. While engraftment of only a small population of corrected HSPCs can result in amelioration of a hemoglobinopathy, highly efficient gene transfer must occur in order to modify a sufficient number of cells able to achieve long-term engraftment. Current gene therapies in clinical trials achieve this by using a lentiviral vector to insert an additional globin gene.
Figure 1.
Typical process of gene therapy for hematopoietic disorders. Hematopoietic stem and progenitor cells (HSPCs), induced pluripotent stem cells (iPSCs). HSPCs can be modified directly, as is the case for currently used therapies. Alternatively, somatic cells, e.g. fibroblasts, can be first reprogrammed into iPSCs before being modified. A correctly modified iPSC clone is then selected and expanded before being differentiated into HSPCs. In either case, the genetically modified HSPCs are then transplanted back into the patient. When HSPCs are modified directly, modification may not occur in every cell.
Future gene therapies may make use of induced pluripotent stem cells (iPSCs) to circumvent the need for high-modification efficiencies. This route involves first isolating somatic cells (e.g. fibroblasts) from a patient. Somatic cells are then reprogrammed into a pluripotent state by expressing certain factors (originally Oct3/4, Sox2, c-Myc, and Klf4) [Takahashi and Yamanaka, 2006]. The iPSCs are then treated to achieve the desired genetic modification. A correctly modified clone is then expanded in vitro. After sufficient expansion, the iPSCs are differentiated into HSPCs. These cells are then transplanted back into the patient. While this strategy would overcome many of the limitations in using HSPCs harvested directly from a patient, it is not yet feasible to use iPSC-derived HSPCs for HSCT, as they currently have only limited repopulation potential.
Gene delivery mechanisms
A variety of gene delivery mechanisms have been devised, but we will focus only on those applicable for treatment of hemoglobinopathies. The applicable delivery mechanisms can be divided into viral and nonviral methods. As mentioned above, traditional gene therapies utilize viral vector delivery systems. Having evolved the ability to transduce mammalian cells over 100 million years ago [Katzourakis et al. 2009] viral vectors are able to deliver engineered genetic material with very high efficiency. Numerous viral systems have been harnessed by researchers for genetic transfer, including gammaretrovirus, lentivirus, foamy virus, adenovirus, and adeno-associated virus (AAV). The most common viral vector used currently is lentivirus. Depending on the nature of the therapy, viral vectors can be chosen based on their ability, or lack of ability, to integrate into the host cells genome. For example, traditional gene therapy, involving insertion of a globin gene, requires genomic integration. Gammaretrovirus, lentivirus, and foamy virus all efficiently integrate into a host cells genome, and are therefore frequently used for this type of therapy. However, many new gene editing techniques benefit from avoiding genomic integration. Constitutive expression of the DNA-cutting elements found in gene editing modalities can lead to increased off-target effects. Therefore, it is often best to express these elements only transiently, which is best achieved using nonintegrating viral vectors. In particular, adenovirus, which does not integrate into a host cells genome, and AAV, which has very low rates of integration, are ideal for this purpose. Additionally, integration-deficient gammaretroviruses and lentiviruses have been engineered, also with low rates of integration.
Traditional gene therapy was first developed in replication-deficient gammaretroviruses. These studies demonstrated the ability to insert globin transgenes into HSPCs, but expression of the globin chain was low and unstable [Dzierzak et al. 1988; Bender et al. 1989; Bodine et al. 1989; Lung et al. 2000]. Subsequently, a variety of modifications were made to the gammaretrovirus vectors, resulting in improved gene expression. Alterations included: inserting portions either the α-locus HS-40 regulatory region or β-globin locus control regions, the main gene regulators of the α- and β-globin gene clusters, respectively [Chang et al. 1992; Plavec et al. 1993; Ren et al. 1996; Nishino et al. 2006]; using the ankyrin gene promoter, promoter of another gene expressed in RBCs, to drive expression of a globin transgene [Sabatino et al. 2000]; or utilizing promoters associated with hereditary persistence of fetal hemoglobin [Katsantoni et al. 2003; Fragkos et al. 2005]. A major problem with early gammaretrovirus vectors that carried the human β-globin gene and β-locus control region (LCR) was that they were significantly unstable with frequent rearrangements [Orkin, 1990]. This instability was overcome through careful optimization of the viral genome, LCR elements, and the β-globin gene (including elimination of an intronic segment and multiple reverse polyadenylation and splicing signals) [Leboulch et al. 1994; Sadelain et al. 1995]. However, despite the above modifications, gene expression levels remained subtherapeutic.
A breakthrough in gene therapy occurred with the advent of lentiviral vector-based vectors [Naldini et al. 1996]. Lentiviral vectors, based on human immunodeficiency virus (HIV)-1, offer several unique advantages compared with those vectors preceding it, including (1) the ability to infect quiescent cells, (2) the ability to package large transgene cassettes, and (3) reduced genotoxicity. As opposed to gammaretroviruses, which integrate preferentially near transcriptional start sites, lentiviruses preferentially integrate within transcribed genes, resulting in lower genotoxicity and insertional oncogenesis. Additionally, safety was increased by the ability to generate vectors efficiently with a self-inactivating (SIN) vector design that deletes the promoters and enhancers from the viral long terminal repeat (LTR) in the provirus. SIN vectors are produced by deleting the viral enhancer/promoter sequences in the U3 region of the 3’ LTR. This deletion is then transferred to the 5’ LTR after reverse transcription and integration into a host cells genome. Without the deleted sequences the LTR is transcriptionally inactive and no viral genes are able to be transcribed, thereby significantly improving the safety profile of the vector, and allowing the vector to be solely driven by the promoter of choice. While first described using gammaretroviral vectors, similar systems were later adapted for use in lentiviral vectors [Yu et al. 1986; Miyoshi et al. 1998; Zufferey et al. 1998]. A total of two seminal studies demonstrated the ability of novel lentiviral vectors to correct relevant mouse models of β-thalassemia and SCD. In the first, a lentivirus carrying DNase I hypersensitive site 2 (HS2), HS3, and HS4 of the LCR and the human β-globin gene was able to ameliorate disease in a β-thalassemia mouse model [May et al. 2000]. In the second, a lentivirus containing a mutant β-globin chain (βA-T87Q) known to have anti-sickling properties and the HS2, 3 and 4 fragments of the LCR was able to ameliorate disease in two mouse models of SCD [Pawliuk et al. 2001].
In addition to viral delivery methods, transposases have been developed that also allow for integration of a transgene into a target cell’s genome. This system consists of a transposase enzyme that is able to cut and paste DNA transposons, such that a transposon supplied on a donor DNA molecule can be pasted into genomic DNA. Transposons consist of a given DNA sequence, flanked by inverted terminal repeats (ITRs). One such transposase is the sleeping beauty transposase system [Geurts et al. 2003; Aronovich et al. 2011; Ivics and Izsvak, 2011]. In this system, a transposon (transgene flanked by ITRs) is inserted into the host cell’s genome at random TA-dinucleotides. An advantage of this system over viral delivery systems is the increased randomness of the integration compared with lentivirus or gammaretrovirus vectors, which have a bias towards integrating into gene rich regions. A version of this system has been used to effectively deliver an anti-sickling globin gene into HSPCs derived from a patient with SCD. The cells derived from the modified HSPCs demonstrated reduced cellular sickling metrics [Sjeklocha et al. 2013].
Significant developments in gene editing/targeting and the advantages of having DNA cutting elements only transiently present has led to an increase in the use of nonviral delivery methods. Common methods include nucleofection, electroporation, and lipofection [Toneguzzo and Keating, 1986; Harrison et al. 1995; Floch et al. 1997; Levetzow et al. 2006; Manzini et al. 2015]. In addition to the ability to deliver DNA, these methods are also capable of delivering RNA and proteins. The ability to deliver proteins is particularly useful for gene editing, as it allows for the delivery of the designer nucleases (discussed below). Nucleofection and electroporation utilize electrical charges to permeabilize cell membranes and deliver the desired agent into the cell. Lipofection utilizes lipid reagents to form liposomes around the agent being delivered. Fusion of the liposomes with the cell membrane results in introduction of the agent into the cellular cytoplasm.
Gene therapy approaches
As mentioned above, current and proposed methods of gene therapy for hemoglobinopathies can be divided into four general categories (1) gene addition, (2) gene knockdown to improve the β-globinopathy phenotype, (3) globin gene editing, and (4) gene editing of globin regulatory elements.
Gene addition
As noted above, traditional gene therapy for hemoglobinopathies involves inserting a globin transgene into a patient’s HSPCs. The globin transgene introduces either an additional copy of an endogenous globin gene, or a globin gene engineered to have disease-ameliorating effects. Engineered globin genes frequently make use of natural polymorphisms that have been shown to reduce the severity of SCD or thalassemia, such as polymorphisms that impair the normal developmental switch from γ-globin to β-globin that occurs during terminal hemoglobin switching [Akinsheye et al. 2011]. Normally the β-globin family members γ- and δ-globin are expressed only at very low levels (<2%) after terminal hemoglobin switching to HbA, and are unable to compensate for hemoglobinopathies [Thein et al. 2009]. However, polymorphisms that impair this switch result in hereditary persistence of fetal hemoglobin (HPFH). In this condition, increased amounts of γ-globin are produced, which is able to form heterotetramers with α-globin (α2γ2) referred to as fetal hemoglobin, or HbF. When HPFH occurs concurrently with β-thalassemia, it reduces the amount of free α-globin chains and its resulting toxicity. RBCs containing HbF are also less prone to βS-induced cell damage and RBC aggregation, resulting in diminished vaso-occlusion [Akinsheye et al. 2011]. Incorporating these polymorphisms into designed globin transgenes thus offers a potential therapeutic advantage. Previous clinical trials using additive globin techniques have proven successful in treating β-thalassemia [Cavazzana-Calvo et al. 2010]. Currently, there are several ongoing clinical trials for SCD and β-thalassemia sponsored by St. Jude Children’s Research Hospital; Cincinnati Children’s Hospital Medical Center; bluebird bio; University of California, Los Angeles; Memorial Sloan Kettering Cancer Center; and IRCCS, San Raffaele, Italy. There are presently no clinical trials for α-thalassemia. The lentiviral vectors used in the above trials have been extensively reviewed previously and summarized briefly below; readers are directed to the following reviews for a more indepth discussion [Villamizar et al. 2001; Dong et al. 2013; Finotti et al. 2015].
Briefly, successful correction of β0/βE thalassemia major has been shown with a β-globin vector carrying a T87Q mutation by Leboulch and coworkers [Bank et al. 2005; Cavazzana-Calvo et al. 2010] (and subsequently licensed by the commercial company, bluebird bio), where all patients received myeloablative chemotherapy conditioning, and following gene therapy achieved a median increase in hemoglobin (contributed by the βT87Q transgene) of 5 g/dl at 6 months, and all patients became transfusion-independent. For β0 thalassemia patients, partial correction occurred in most of the patients with significantly reduced transfusion requirement; one β0/β0 patient attained transfusion independence. Overall, the vector produced approximately 5 g Hb/vector copy. The same vector resulted in correction of the sickle phenotype in at least one of four patients, where high vector copies were achieved (results presented at the American Society of Hematology Meeting, 2015 [Cavazzana-Calvo, 2015]); in the other three patients, modest increases in βT87Q globin was modest. A β-globin lentivirus vector generated by Sadelain and colleagues was used in a β-thalassemia gene therapy trial by Memorial Sloan Kettering with a reduced intensity busulfan conditioning [Sadelain et al. 2010]. The gene marking efficiency was low and therefore, subtherapeutic expression was achieved, with a significant reduction in transfusion in one patient, and a good partial response in a second patient. However, two patients did not show a clinically significant response, of which one was given myeloablative doses of busulfan; and was attributed to low gene-modified HSC. In another gene therapy trial for β-thalassemia being conducted in IRCCS, San Raffaele, Italy by Ferrari and colleagues under the auspices of Glaxo-Smith-Kline using a β-globin lentivirus vector and myeloablative conditioning, one patient has received gene modified cells. Although it is short-term follow up, this patient has shown promising transfusion independence within a month (results presented at the [Cooley’s Anemia Foundation Meeting, 2015]). We have recently opened a gene therapy trial for SCA using a fetal hemoglobin expressing vector and reduced intensity transplant at Cincinnati Children’s Hospital Medical Center and results of this trial are awaited. Another trial for SCA has been opened by Kohn and colleagues [Urbinati et al. 2015; Romero et al. 2013] using an anti-sickling β-globin lentivirus vector designed by Townes and colleagues [Levasseur et al. 2003] and results are awaited. In summary, lentivirus vectors are successfully treating patients with hemoglobinopathies, although optimal hemoglobin levels and reduced chemotherapy conditioning may still be barriers that need to be overcome. Importantly, long-term follow up of patients treated with lentivirus vectors for hemoglobinopathies and other diseases has not shown evidence of genotoxicity thus far.
Gene knockdown to improve the β-globinopathy phenotype
In addition to HPFH, other concurrent genetic variations have inspired potential gene therapies. It has been found that loss of one of the four α-globin genes concurrent with certain β-thalassemia mutations results in a less severe phenotype than β-thalassemia by itself [Kan and Nathan, 1970]. The loss of an α-globin gene leads to less free α-globin, and therefore less toxicity. One way to reduce gene expression is to use RNA interference, whereby defined RNA sequences are able to facilitate mRNA degradation or block protein translation of a specific RNA transcript. Overall, two groups have used RNA interference to decrease α-globin expression in murine primary erythroid cells and a β-thalassemia mouse model [Xie et al. 2007; Voon et al. 2008; Xie et al. 2011]. Knockdown of α-globin resulted in a reduction in reactive oxygen species (ROS) production, improvement in red cell morphology, and RBC counts. While the referenced studies did not perform complete gene knockout, the findings, combined with the patient findings noted above, suggest that deletion of an α-globin may be a plausible approach for treatment of β-thalassemia.
Globin gene editing
Recent advances in the area of gene editing have revolutionized the field of gene therapy. Using designer nucleases, it is now possible to develop therapies that achieve precise genetic modifications (e.g. gene correction). Depending on the delivery system, they are able to avoid nonspecific integration and significantly reduce or completely avoid insertional oncogenesis. Gene editing techniques are not devoid of genotoxicity, as off-target DNA damage can occur at varying rates and nonspecific integration of DNA can occur if a donor sequence is required, the latter at typically low frequencies. The three designer nucleases that are predominantly used are zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats/CRISPR associated protein (CRISPR/Cas).
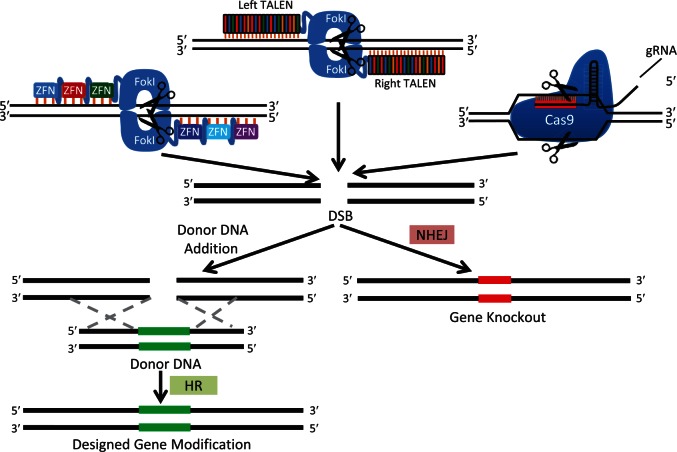
The basic steps involved in the use of designer nucleases to perform site-specific gene editing are depicted in Figure 2. All of the designer nucleases create double strand breaks (DSBs) at precise locations in the genome. Each of the three designer nuclease systems function in a slightly different manner. ZFNs are user-defined zinc finger domains for recognition of specific DNA sequences and a nonspecific FokI catalytic domain for DNA cleavage upon dimerization. By targeting ZFNs to either side of the desired cut site, FokI dimerization occurs, activating its nuclease activity and creating the DSB. TALENs contain multiple DNA-binding domains arranged in tandem (TALE repeats), which are linked to a FokI catalytic domain. Each binding domain contains two defined amino acids that are able to recognize a specific DNA base pair [Moscou and Bogdanove, 2009]. Similar to ZFNs, a pair of TALENs are targeted to either side of the desired cut site, allowing their FokI domains to dimerize and create the DSB with high efficiency [Boch et al. 2009; Miller et al. 2011; Bedell et al. 2012]. The CRISPR/Cas system is composed of a guide-RNA (gRNA) and a Cas protein. The gRNA contains a defined 17–20 nucleotide portion that allows for targeting specific complementary genomic sequences. gRNA first forms a ribonucleoprotein (RNP) complex with a Cas protein and then directs the protein to the correct genomic location. Complementary genomic sequences that are followed by a specific DNA sequence, referred to as a protospacer adjacent motif, are able to be cut by the Cas protein. Several types of both natural and engineered Cas proteins, and a similarly functioning protein, CpfI, have now been described [Ran et al. 2015; Zetsche et al. 2015; Kleinstiver et al. 2016; Slaymaker et al. 2016].
Figure 2.
Three most common gene modification strategies using designer nucleases (ZFN, TALEN, and CRISPR/Cas).
The given nuclease system identifies a specified region of the genome through base pair recognition (orange hash marks) and creates a DSB (scissor). In the absence of a repair donor, NHEJ DNA repair creates an ‘indel’ (red DNA region) resulting in a gene knockout. If a repair donor is supplied, HR may occur, resulting in incorporation of a designed modification (e.g. insertion, deletion, substitution).
Cas, CRISPR associated protein, CRISPR, clustered regularly interspaced short palindromic repeats; DSB, double strand break; gRNA, guide-RNA; HR, homologous recombination; NHEJ, nonhomologous end-joining; TALEN, transcription activator-like effector nuclease; ZFN, zinc-finger nuclease.
All three systems outlined above result in formation of DSBs that are typically repaired by an innate DNA repair process, nonhomologous end joining (NHEJ). The NHEJ repair process is error prone, such that a significant proportion of repair events will result in the addition or subtraction of base pairs at the location of the DSB. This insertion/deletion is referred to as an ‘indel’. Indels can be used to disrupt DNA binding sites and splice sites. Additionally, if an indel occurs within a coding region, a frameshift mutation can result, resulting in formation of a truncated protein, or degradation of the mRNA transcript via nonsense-mediated decay. To achieve a variety of more complicated gene modifications (e.g. gene correction, precise deletions, or insertions) a donor DNA sequence is provided. This allows a cell to make use of another innate DNA repair process, homologous recombination (HR). The donor sequence contains the desired sequence to be introduced into the cell, flanked by homology arms that align with DNA sequences on either side of the region being modified. HR results in seamless and precise repair of the desired locus, replacing the cellular sequence with that in the donor template.
When devising a gene editing modality, multiple factors must be considered, such as choice of endonuclease, donor design, delivery modality, and genomic target of nuclease. Given the variety of considerations, comparisons between different modalities is difficult. Huang and colleagues have reported a side-by-side comparison of ZFNs, TALENs, and CRISPR/Cas and their ability to modify the β-globin locus [Huang et al. 2015]. ZFNs proved to be the least efficient while CRISPR/Cas was the most efficient. Successful use of ZFNs to correct the βS mutation have been reported [Sebastiano et al. 2011; Zou et al. 2011; Hoban et al. 2015]. Most notably, using a ZFN method Hoban and colleagues demonstrated initial significant correction of CD34+ HSPCs isolated from both human umbilical cord blood as well as mobilized peripheral blood, but ultimately poor correction of long-term repopulating HSCs was observed in a xenograft mouse model [Hoban et al. 2015]. Multiple studies have reported on the use of TALENs to correct either the βS or β-thalassemia mutations in iPSCs [Ma et al. 2013; Ramalingam et al. 2014; Sun and Zhao, 2014; Xu et al. 2015]. Instead of correcting β-globin mutations, Voit and colleagues reported the TALEN-facilitated insertion of the entire β-globin cDNA immediately in front of the native β-globin gene [Voit et al. 2014]. In this strategy, the inserted β-globin cDNA is immediately followed by a polyadenylation signal, preventing read-through of the native β-globin gene. Potential advantages of this strategy are its applicability for treatment of any mutation found within the β-globin gene and its ability to express engineered globin genes. Lastly, several studies have demonstrated correction of both β-thalassemia and βS mutations in iPSCs utilizing CRISPR/Cas editing techniques [Xie et al. 2014; Huang et al. 2015; Song et al. 2015].
While the above studies are promising, several obstacles must be overcome for these therapies to become clinically relevant. Most of the above studies necessitate utilizing a selectable marker to select for modified cells. Without the selectable marker, it would be unable to obtain a population of cells with a sufficient proportion of modified cells. This type of selection is feasible when working with iPSCs, which can be maintained in culture without the loss of the self-renewing potential. However, the culture time needed to select cells, and to later remove the marker, would not allow for retention of long-term repopulating HSCs. Editing efficiency of long-term repopulating HSPCs without the use of a selectable marker must be improved in order to achieve therapeutic efficacy. Another obstacle is the need to increase the desired precise HR outcomes over NHEJ. HR and NHEJ are competitive processes, with NHEJ typically being the dominant process with most gene editing systems.
Gene editing of globin regulatory elements
Transcription of β-globin family members is controlled by a large number of transcription factors and other regulatory mechanisms. Researchers have attempted to exploit many of these mechanisms to increase production of fetal hemoglobin. Kruppel-like factor 1 (KLF1) is an erythroid-restricted transcription factor that binds the CACCC box of the β-globin gene in mice and humans and is critical for the expression of the β-globin gene. Manchinu and colleagues were able to increase δ-globin expression by inserting a binding site for the KLF1 transcription factor into the promoter region for δ-globin [Manchinu et al. 2014]. When combined with a mouse model of β-thalassemia, the inserted KLF1 binding site was able to improve the phenotype. Another important transcription factor is BCL11A, which is an important suppressor of γ-globin expression. Guda and colleagues designed an erythroid-specific RNAi technique to knockdown BCL11A, and demonstrated a significant de-repression of γ-globin expression [Guda et al. 2015]. Furthermore, Canver and colleagues were able to knockdown BCL11A in the erythroid lineage by using a CRISPR/Cas technique to delete an erythroid-specific enhancer for BCL11A [Canver et al. 2015]. This resulted in reactivation of fetal hemoglobin. Researchers have also been able to create synthetic transcription factors to influence globin expression. Costa and colleagues increased γ-globin expression by creating an artificial transcription factor composed of a ZFN binding domain recognizing the γ-globin promoter fused to a activation domain (VP64) [Costa et al. 2012].
Conclusion
Effective gene therapy for hemoglobinopathies requires several requirements to be met [Chandrakasan and Malik, 2014]: (1) Therapy must be efficiently transferred into the target cell population. Currently, this requires high-efficiency transfer into HSPCs. However, lower efficiencies would be required for therapies that may one day utilize IPSCs, as this would allow identifying and propagating only corrected clones. (2) Minimal genotoxicity. Therapies that result in insertion of a viral vector may have genotoxic effects if the vector integrates near a putative oncogene. This occurs when the vector either enhances the activity of nearby gene promoters, or functions as a promoter itself. Genotoxicity may lead to formation of clonal dominance and uncontrolled proliferation, often resulting in leukemia. In the case of gene editing strategies, therapies can have varying degrees of off-target effects caused by creation of DSBs at locations other than the desired genomic location. These DSBs can cause genetic mutations, deletions, translocations, and other mutations. (3) Phenotype correction. Therapies must show correction, or at least significant disease amelioration, to be pursued as viable treatment options. There are additional requirements for therapies involving insertion of engineered genes. (4) Consistent, integration site-independent, high-level expression of inserted gene. (5) Erythroid lineage-specific and developmental stage-specific expression of the inserted gene.
Access to genetic therapies
Given the successes of therapies both in clinical trials and in development, gene therapy has the potential to have a profound impact on the treatment of hemoglobinopathies. However, these therapies currently require expensive and sophisticated resources for their implementation, including Good Manufacturing Practice (GMP)-certified manufacturing of viral vectors and hematopoietic stem cell graft products, and facilities capable of performing hematopoietic stem cell transplants. This limitation places these therapies out of reach for most of the patients suffering from hemoglobinopathies, as the vast majority of these patients live in underdeveloped countries and have extremely poor access to even basic medical care, and virtually no access to facilities capable of implementing these treatments. Nevertheless, new techniques are emerging that may increase the availability of these new therapies, especially to those with limited access to medical care.
Gene therapy for hematopoietic disorders has traditionally been achieved by first isolating a patient’s own HSPCs, and genetically altering the cells ex vivo, before transfusing them back to the patient. This method is technically challenging and requires extensive precautions to insure the graft product retains both its sterility and stemness. Direct infusion of a viral vector obviates these considerations. Several studies have demonstrated this methodology, using either gene editing or traditional gene therapy, as a viable alternative to ex vivo manipulation [Carbonaro et al. 2006; Li et al. 2011; Burtner et al. 2014]. Given the high transduction/editing efficiencies often required to achieve a therapeutic effect, ex vivo manipulation of cells may be unavoidable for some therapies.
A new device, the CliniMACS Prodigy (Miltenyi Biotec, Germany), is now available that has the ability to automatically perform cell preparation, enrichment, activation, transduction, and expansion in a closed GMP-certified system [Apel et al. 2013]. This system reduces the technical expertise required by the operator as well as dependency on sophisticated ancillary facilities required for product preparation. Additionally, investigators are developing increasingly less toxic nonmyeloablative transplantation regimens. Less toxicity leads to less transplant complications, which will increase the number and type of facilities capable of performing transplants. Wang and colleagues have provided evidence that STAT5 modulation may increase nonablative stem cell replacement [Wang et al. 2009]. Additionally, Xue and colleagues have demonstrated the potential of a pre-transplantation conditioning using an antibody targeting V-Kit Hardy-Zuckerman 4 Feline Sarcoma Viral Oncogene Homolog (KIT) to facilitate engraftment of transplanted hematopoietic stem cells [Xue et al. 2010]. As new methodologies and technologies advance, there will continue to be a corresponding increase in the access to gene therapy. Hopefully, this will eventually open up these novel therapeutic treatments to populations that have the highest prevalence but poorest healthcare access.
Hemoglobinopathies represent a broad range of inherited diseases leading to reduced hemoglobin levels or improper hemoglobin function. The disorders have significant global prevalence, morbidity, and mortality, but the burden of disease is felt greatest in sub-Saharan Africa, where access to medical care is often limited. For the minority of patients with access to medical care and an available matched donor, HSCT remains a viable curative option. However, gene therapies in clinical trials and earlier in development hold enormous promise for potentially providing a curative therapy to all patients. While promising, significant technical challenges must be overcome for these therapies to reach patients in those remote areas where disease burden is greatest.
Footnotes
Funding: K12 HD000850 – Pediatric Scientist Development Program (K12) 2017-2022; PEDIATRIC PHYSICIAN SCIENTIST PROGRAM AWARD; Pediatric Physician Scientist Program Award; Pediatric Scientist Development Program (PSDP) [K12].
Conflict of interest statement: The authors declare that there is no conflict of interest.
Contributor Information
Michael A. Goodman, Division of Experimental Hematology and Cancer Biology,Division of Allergy and Immunology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA
Punam Malik, Division of Experimental Hematology and Cancer Biology, Cancer and Blood Diseases Institute Cincinnati Children’s Hospital Medical Center, 3333 Burnet Avenue, Cincinnati, OH 45229, USA.
References
- Akinsheye I., Alsultan A., Solovieff N., Ngo D., Baldwin C., Sebastiani P., et al. (2011) Fetal hemoglobin in sickle cell anemia. Blood 118: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel M., Brüning M., Granzin M., Essl M., Stuth J., Blaschke J., et al. (2013) Integrated clinical scale manufacturing system for cellular products derived by magnetic cell separation, centrifugation and cell culture. Chem Ing Tech 85: 103–110. [Google Scholar]
- Aronovich E., McIvor R., Hackett P. (2011) The sleeping beauty transposon system: a nonviral vector for gene therapy. Hum Mol Genet 20: R14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank A., Dorazio R., Leboulch P. (2005) A phase I/II clinical trial of beta-globin gene therapy for beta-thalassemia. Ann NY Acad Sci 1054: 308–316. [DOI] [PubMed] [Google Scholar]
- Bauer D., Kamran S., Orkin S. (2012) Reawakening fetal hemoglobin: prospects for new therapies for the beta-globin disorders. Blood 120: 2945–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell V., Wang Y., Campbell J., Poshusta T., Starker C., Krug R., et al. (2012) In vivo genome editing using a high-efficiency TALEN system. Nature 491: 114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender M., Gelinas R., Miller A. (1989) A majority of mice show long-term expression of a human beta-globin gene after retrovirus transfer into hematopoietic stem cells. Mol Cell Biol 9: 1426–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernaudin F., Socie G., Kuentz M., Chevret S., Duval M., Bertrand Y., et al. (2007) Long-term results of related myeloablative stem-cell transplantation to cure sickle cell disease. Blood 110: 2749–2756. [DOI] [PubMed] [Google Scholar]
- Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., et al. (2009) Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326: 1509–1512. [DOI] [PubMed] [Google Scholar]
- Bodine D., Karlsson S., Papayannopoulou T., Nienhuis A. (1989) Expression of human beta globin genes introduced into primitive murine hematopoietic progenitor cells by retrovirus mediated gene transfer. Prog Clin Biol Res 316B: 219–233. [PubMed] [Google Scholar]
- Burtner C., Beard B., Kennedy D., Wohlfahrt M., Adair J., Trobridge G., et al. (2014) Intravenous injection of a foamy virus vector to correct canine SCID-X1. Blood 123: 3578–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canver M., Smith E., Sher F., Pinello L., Sanjana N., Shalem O., et al. (2015) BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. DOI: 10.1038/nature15521 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonaro D., Jin X., Petersen D., Wang X., Dorey F., Kil K., et al. (2006) In vivo transduction by intravenous injection of a lentiviral vector expressing human ADA into neonatal ADA gene knockout mice: a novel form of enzyme replacement therapy for ADA deficiency. Mol Ther 13: 1110–1120. [DOI] [PubMed] [Google Scholar]
- Cavazzana-Calvo M. (2015). Abstract 0202. Presented at the 57th American Society of Hematology Meeting, 5–8 December 2015, Orlando, FL, USA. [Google Scholar]
- Cavazzana-Calvo M., Payen E., Negre O., Wang G., Hehir K., Fusil F., et al. (2010) Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature 467: 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrakasan S., Malik P. (2014) Gene therapy for hemoglobinopathies: the state of the field and the future. Hematol Oncol Clin of North Am 28: 199–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J., Liu D., Kan Y. (1992) A 36-base-pair core sequence of locus control region enhances retrovirally transferred human beta-globin gene expression. Proc Natl Acad Sci USA 89: 3107–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley’s Anemia Foundation Meeting. (2015) Presented at the Tenth Cooley’s Anemia Symposium Meeting, 18–22 October 2015, Chicago, IL, USA. [Google Scholar]
- Costa F., Fedosyuk H., Neades R., De Los Rios J., Barbas C., Peterson K. (2012) Induction of fetal hemoglobin in vivo mediated by a synthetic gamma-globin zinc finger activator. Anemia DOI: 10.1155/2012/507894 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong A., Rivella S., Breda L. (2013) Gene therapy for hemoglobinopathies: progress and challenges. Transl Res 161: 293–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzierzak E., Papayannopoulou T., Mulligan R. (1988) Lineage-specific expression of a human beta-globin gene in murine bone marrow transplant recipients reconstituted with retrovirus-transduced stem cells. Nature 331: 35–41. [DOI] [PubMed] [Google Scholar]
- Finotti A., Breda L., Lederer C., Bianchi N., Zuccato C., Kleanthous M., et al. (2015) Recent trends in the gene therapy of beta-thalassemia. J Blood Med 6: 69–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floch V., Le Bolc’h G., Audrézet M., Yaouanc J., Clément J., Des Abbayes H., et al. (1997) Cationic phosphonolipids as nonviral vectors for DNA transfection in hematopoietic cell lines and CD34+ cells. Blood Cells Mol Dis 23: 69–87. [DOI] [PubMed] [Google Scholar]
- Forget B., Hardison R. (2009) The normal structure and regulation of human globin gene clusters. In: Steinberg M., Forget B., Higgs D., Weatherall D. (eds), Disorders of Hemoglobin: Genetics, Pathophysiology, and Clinical Management (2nd ed). Cambridge, UK: Cambridge University Press, pp. 46–61. [Google Scholar]
- Fragkos M., Anagnou N., Tubb J., Emery D. (2005) Use of the hereditary persistence of fetal hemoglobin 2 enhancer to increase the expression of oncoretrovirus vectors for human gamma-globin. Gene Ther 12: 1591–1600. [DOI] [PubMed] [Google Scholar]
- Geurts A., Yang Y., Clark K., Liu G., Cui Z., Dupuy A., et al. (2003) Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol Ther 8: 108–117. [DOI] [PubMed] [Google Scholar]
- Grosse S., Odame I., Atrash H., Amendah D., Piel F., Williams T. (2011) Sickle cell disease in Africa: a neglected cause of early childhood mortality. Am J Prev Med 41: S398–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guda S., Brendel C., Renella R., Du P., Bauer D., Canver M., et al. (2015) miRNA-embedded shRNAs for lineage-specific BCL11A knockdown and hemoglobin F induction. Mol Ther 23: 1465–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison G., Wang Y., Tomczak J., Hogan C., Shpall E., Curiel T., et al. (1995) Optimization of gene transfer using cationic lipids in cell lines and primary human CD4+ and CD34+ hematopoietic cells. Biotechniques 19: 816–823. [PubMed] [Google Scholar]
- Hoban M., Cost G., Mendel M., Romero Z., Kaufman M., Joglekar A., et al. (2015) Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood 125: 2597–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh M., Fitzhugh C., Weitzel R., Link M., Coles W., Zhao X., et al. (2014) Nonmyeloablative HLA-matched sibling allogeneic hematopoietic stem cell transplantation for severe sickle cell phenotype. JAMA 312: 48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Wang Y., Yan W., Smith C., Ye Z., Wang J., et al. (2015) Production of gene-corrected adult beta globin protein in human erythrocytes differentiated from patient iPSCs after genome editing of the sickle point mutation. Stem Cells 33: 1470–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z., Izsvak Z. (2011) Nonviral gene delivery with the sleeping beauty transposon system. Hum Gene Ther 22: 1043–1051. [DOI] [PubMed] [Google Scholar]
- Kan Y., Nathan D. (1970) Mild thalassemia: the result of interactions of alpha and beta thalassemia genes. J Clin Invest 49: 635–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsantoni E., Langeveld A., Wai A., Drabek D., Grosveld F., Anagnou N., et al. (2003) Persistent gamma-globin expression in adult transgenic mice is mediated by HPFH-2, HPFH-3, and HPFH-6 breakpoint sequences. Blood 102: 3412–3419. [DOI] [PubMed] [Google Scholar]
- Katzourakis A., Gifford R., Tristem M., Gilbert M., Pybus O. (2009) Macroevolution of complex retroviruses. Science 325: 1512. [DOI] [PubMed] [Google Scholar]
- Kleinstiver B., Pattanayak V., Prew M., Tsai S., Nguyen N., Zheng Z., et al. (2016) High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529: 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leboulch P., Huang G., Humphries R., Oh Y., Eaves C., Tuan D., et al. (1994) Mutagenesis of retroviral vectors transducing human beta-globin gene and beta-globin locus control region derivatives results in stable transmission of an active transcriptional structure. EMBO J 13: 3065–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levasseur D., Ryan T., Pawlik K., Townes T. (2003) Correction of a mouse model of sickle cell disease: lentiviral/antisickling β-globin gene transduction of unmobilized, purified hematopoietic stem cells. Blood 102: 4312–4319. [DOI] [PubMed] [Google Scholar]
- Levetzow G., Spanholtz J., Beckmann J., Fischer J., Kögler G., Wernet P., et al. (2006) Nucleofection, an efficient nonviral method to transfer genes into human hematopoietic stem and progenitor cells. Stem Cells Dev 15: 278–285. [DOI] [PubMed] [Google Scholar]
- Li H., Haurigot V., Doyon Y., Li T., Wong S., Bhagwat A., et al. (2011) In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature 475: 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucarelli G., Isgro A., Sodani P., Gaziev J. (2012) Hematopoietic stem cell transplantation in thalassemia and sickle cell anemia. Cold Spring Harb Perspect Med 2: a011825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung H., Meeus I., Weinberg R., Atweh G. (2000) In vivo silencing of the human gamma-globin gene in murine erythroid cells following retroviral transduction. Blood Cells Mol Dis 26: 613–619. [DOI] [PubMed] [Google Scholar]
- Ma N., Liao B., Zhang H., Wang L., Shan Y., Xue Y., et al. (2013) Transcription activator-like effector nuclease (TALEN)-mediated gene correction in integration-free β-thalassemia induced pluripotent stem cells. J Biol Chem 288: 34671–34679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchinu M., Marongiu M., Poddie D., Casu C., Latini V., Simbula M., et al. (2014) In vivo activation of the human δ-globin gene: the therapeutic potential in β-thalassemic mice. Haematologica 99: 76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzini S., Viiri L., Marttila S., Aalto-Setälä K. (2015) A comparative view on easy to deploy non-integrating methods for patient-specific iPSC production. Stem Cell Rev 11: 900–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May C., Rivella S., Callegari J., Heller G., Gaensler K., Luzzatto L., et al. (2000) Therapeutic haemoglobin synthesis in β-thalassaemic mice expressing lentivirus-encoded human β-globin. Nature 406: 82–86. [DOI] [PubMed] [Google Scholar]
- Miller J., Tan S., Qiao G., Barlow K., Wang J., Xia D., et al. (2011) A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29: 143–148. [DOI] [PubMed] [Google Scholar]
- Miyoshi H., Blomer U., Takahashi M., Gage F., Verma I. (1998) Development of a self-inactivating lentivirus vector. J Virol 72: 8150–8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou M., Bogdanove A. (2009) A simple cipher governs DNA recognition by TAL effectors. Science 326: 1501. [DOI] [PubMed] [Google Scholar]
- Naldini L., Blomer U., Gallay P., Ory D., Mulligan R., Gage F., et al. (1996) In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272: 263–267. [DOI] [PubMed] [Google Scholar]
- Nishino T., Tubb J., Emery D. (2006) Partial correction of murine beta-thalassemia with a gammaretrovirus vector for human gamma-globin. Blood Cells Mol Dis 37: 1–7. [DOI] [PubMed] [Google Scholar]
- Orkin S. (1990) Globin gene regulation and switching: circa 1990. Cell 63: 665–672. [DOI] [PubMed] [Google Scholar]
- Pawliuk R., Westerman K., Fabry M., Payen E., Tighe R., Bouhassira E., et al. (2001) Correction of sickle cell disease in transgenic mouse models by gene therapy. Science 294: 2368–2371. [DOI] [PubMed] [Google Scholar]
- Plavec I., Papayannopoulou T., Maury C., Meyer F. (1993) A human beta-globin gene fused to the human beta-globin locus control region is expressed at high levels in erythroid cells of mice engrafted with retrovirus-transduced hematopoietic stem cells. Blood 81: 1384–1392. [PubMed] [Google Scholar]
- Ramalingam S., Annaluru N., Kandavelou K., Chandrasegaran S. (2014) TALEN-mediated generation and genetic correction of disease-specific human induced pluripotent stem cells. Curr Gene Ther 14: 461–472. [DOI] [PubMed] [Google Scholar]
- Ran F., Cong L., Yan W., Scott D., Gootenberg J., Kriz A., et al. (2015) In vivo genome editing using Staphylococcus aureus CAS9. Nature 520: 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S., Wong B., Li J., Luo X., Wong P., Atweh G. (1996) Production of genetically stable high-titer retroviral vectors that carry a human gamma-globin gene under the control of the alpha-globin locus control region. Blood 87: 2518–2524. [PubMed] [Google Scholar]
- Romero Z., Urbinati F., Geiger S., Cooper A., Wherley J., Kaufman M., et al. (2013) β-globin gene transfer to human bone marrow for sickle cell disease. J Clin Invest. DOI: 10.1172/JCI67930 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatino D., Seidel N., Aviles-Mendoza G., Cline A., Anderson S., Gallagher P., et al. (2000) Long-term expression of gamma-globin mRNA in mouse erythrocytes from retrovirus vectors containing the human gamma-globin gene fused to the ankyrin-1 promoter. Proc Natl Acad Sci USA 97: 13294–13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadelain M., Riviere I., Wang X., Boulad F., Prockop S., Giardina P., et al. (2010) Strategy for a multicenter phase I clinical trial to evaluate globin gene transfer in beta-thalassemia. Ann NY Acad Sci 1202: 52–58. [DOI] [PubMed] [Google Scholar]
- Sadelain M., Wang C., Antoniou M., Grosveld F., Mulligan R. (1995) Generation of a high-titer retroviral vector capable of expressing high levels of the human beta-globin gene. Proc Natl Acad Sci USA 92: 6728–6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiano V., Maeder M., Angstman J., Haddad B., Khayter C., Yeo D., et al. (2011) In situ genetic correction of the sickle cell anemia mutation in human induced pluripotent stem cells using engineered zinc finger nucleases. Stem Cells 29: 1717–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjeklocha L., Wong P., Belcher J., Vercellotti G., Steer C. (2013) Β-globin sleeping beauty transposon reduces red blood cell sickling in a patient-derived CD34+-based in vitro model. PLoS One 8: e80403 DOI: 10.1371/journal.pone.0080403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker I., Gao L., Zetsche B., Scott D., Yan W., Zhang F. (2016) Rationally engineered CAS9 nucleases with improved specificity. Science 351: 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B., Fan Y., He W., Zhu D., Niu X., Wang D., et al. (2015) Improved hematopoietic differentiation efficiency of gene-corrected beta-thalassemia induced pluripotent stem cells by CRISPR/Cas9 system. Stem Cells Dev 24: 1053–1065. [DOI] [PubMed] [Google Scholar]
- Sun N., Zhao H. (2014) Seamless correction of the sickle cell disease mutation of the HBB gene in human induced pluripotent stem cells using TALENs. Biotechnol Bioeng 111: 1048–1053. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676. [DOI] [PubMed] [Google Scholar]
- Thein S., Menzel S., Lathrop M., Garner C. (2009) Control of fetal hemoglobin: new insights emerging from genomics and clinical implications. Hum Mol Genet 18: R216-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toneguzzo F., Keating A. (1986) Stable expression of selectable genes introduced into human hematopoietic stem cells by electric field-mediated DNA transfer. Proc Natl Acad Sci USA 83: 3496–3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbinati F., Hargrove P., Geiger S., Romero Z., Wherley J., Kaufman M., et al. (2015) Potentially therapeutic levels of anti-sickling globin gene expression following lentivirus-mediated gene transfer in sickle cell disease bone marrow CD34+ cells. Exp Hematol 43: 346–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villamizar O., Chambers C., Wilber A. (2001) Gene Therapy for Severe Haemoglobin Disorders. eLS: John Wiley & Sons, Ltd. [Google Scholar]
- Voit R., Hendel A., Pruett-Miller S., Porteus M. (2014) Nuclease-mediated gene editing by homologous recombination of the human globin locus. Nucleic Acids Res 42: 1365–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voon H., Wardan H., Vadolas J. (2008) siRNA-mediated reduction of alpha-globin results in phenotypic improvements in beta-thalassemic cells. Haematologica 93: 1238–1242. [DOI] [PubMed] [Google Scholar]
- Wang Z., Li G., Tse W., Bunting K. (2009) Conditional deletion of STAT5 in adult mouse hematopoietic stem cells causes loss of quiescence and permits efficient nonablative stem cell replacement. Blood 113: 4856–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherall D. (2001) The thalassemias. In: Stamatoyannopoulos G. (ed.), The Molecular Basis of Blood Diseases (3rd ed.), Philadelphia, PA: W.B. Saunders Company. [Google Scholar]
- World Health Organization (2006) Report of a joint WHO–March of Dimes meeting: management of birth defects and haemoglobin disorders. Geneva: World Health Organization. [Google Scholar]
- World Health Organization (2011) Sickle-cell disease and other haemoglobin disorders. WHO fact sheet 308. Geneva: World Health Organization. [Google Scholar]
- Xie F., Ye L., Chang J., Beyer A., Wang J., Muench M., et al. (2014) Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res 24: 1526–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S., Li W., Ren Z., Huang S., Zeng F., Zeng Y. (2011) Correction of beta654-thalassaemia mice using direct intravenous injection of siRNA and antisense RNA vectors. Int J Hematol 93: 301–310. [DOI] [PubMed] [Google Scholar]
- Xie S., Ren Z., Zhang J., Guo X., Wang Q., Wang S., et al. (2007) Restoration of the balanced alpha/beta-globin gene expression in beta654-thalassemia mice using combined RNAi and antisense RNA approach. Hum Mol Genet 16: 2616–2625. [DOI] [PubMed] [Google Scholar]
- Xu P., Tong Y., Liu X., Wang T., Cheng L., Wang B., et al. (2015) Both TALENs and CRISPR/Cas9 directly target the HBB IVS2-654 (c > t) mutation in β-thalassemia-derived iPSCs. Sci Rep 5: 12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X., Pech N., Shelley W., Srour E., Yoder M., Dinauer M. (2010) Antibody targeting KIT as pretransplantation conditioning in immunocompetent mice. Blood 116: 5419–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yawn B., Buchanan G., Afenyi-Annan A., Ballas S., Hassell K., James A., et al. (2014) Management of sickle cell disease: summary of the 2014 evidence-based report by expert panel members. JAMA 312: 1033–1048. [DOI] [PubMed] [Google Scholar]
- Yu S., Von Rüden T., Kantoff P., Garber C., Seiberg M., Rüther U., et al. (1986) Self-inactivating retroviral vectors designed for transfer of whole genes into mammalian cells. Proc Natl Acad Sci USA 83: 3194–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B., Gootenberg J., Abudayyeh O., Slaymaker I., Makarova K., Essletzbichler P., et al. (2015) CPF1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163: 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Mali P., Huang X., Dowey S., Cheng L. (2011) Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease. Blood 118: 4599–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R., Dull T., Mandel R., Bukovsky A., Quiroz D., Naldini L., et al. (1998) Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol 72: 9873–9880. [DOI] [PMC free article] [PubMed] [Google Scholar]