Abstract Abstract
Accurate stock assessments for each of the dominant species of sand lances in the northeast Atlantic Ocean and adjacent areas are not available due to the lack of a reliable identification procedure; therefore, appropriate measures of fisheries management or conservation of sand lances cannot be implemented. In this study, detailed morphological and molecular features are assessed to discriminate between four species of sand lances belonging to the genera Ammodytes and Hyperoplus.
Morphological characters described by earlier authors as useful for identification of the genera are confirmed, and two additional distinguishing characters are added. A combination of the following morphological characters is recommended to distinguish between the genera Hyperoplus and Ammodytes: the protrusibility of the premaxillae, the presence of hooked ends of the prevomer, the number of dermal plicae, and the pectoral-fin length as a percentage of the standard length. The discriminant function analysis revealed that morphometric data are not very useful to distinguish the species of each of the two genera. The following meristic characters improve the separation of Hyperoplus lanceolatus from Hyperoplus immaculatus: the number of lower arch gill rakers, total number of gill rakers, numbers of caudal vertebrae and total vertebrae, and numbers of dorsal-fin and anal-fin rays. It is confirmed that Ammodytes tobianus differs from Ammodytes marinus by its belly scales that are organised in tight chevrons, scales which are present over the musculature at the base of the caudal fin, as well as by the lower numbers of dermal plicae, dorsal-fin rays, and total vertebrae.
In contrast to the morphological data, mitochondrial COI sequences (DNA barcodes) failed to separate unambiguously the four investigated species. Ammodytes tobianus and Hyperoplus lanceolatus showed an overlap between intraspecific and interspecific K2P genetic distances and cannot be reliably distinguished using the common DNA barcoding approach. Ammodytes marinus and Hyperoplus immaculatus exhibited gaps between intraspecific and interspecific K2P distances of 2.73 and 3.34% respectively, indicating that their DNA barcodes can be used for species identification. As an alternative, short nuclear Rhodopsin sequences were analysed and one diagnostic character was found for each of the species Ammodytes marinus, Hyperoplus lanceolatus, and Hyperoplus immaculatus. Ammodytes tobianus can be characterised by the lack of species-specific mutations when compared to the other three species. In contrast to COI, the short nuclear sequences represent a useful alternative for rapid species identification whenever an examination of morphological characters is not available.
Keywords: Ammodytes, COI, DNA barcoding, Hyperoplus, meristic characters, mitochondrial DNA, morphology, morphometrics, northeast Atlantic, nuclear gene, Rhodopsin, Sand lances, species identification
Introduction
Sand lances of the family Ammodytidae are small fishes that live primarily in marine and adjacent brackish waters with sandy substrates of the northern hemisphere, where they are able to quickly dive into the substrate to escape predation (Randall and Ida 2014, Orr et al. 2015). These fishes are characterised by elongated and subcylindrical bodies and possess relatively low elongated dorsal and anal fins without spines, which are separated from the forked caudal fin (e.g. Reay 1986). The number of principal caudal rays is reduced and there is no pelvic fin in most species (e.g. Ida et al. 1994). Sand lances have an increased number of vertebrae in which the number of pre-caudal vertebrae is higher than the number of caudal vertebrae. The lower jaws project beyond the upper jaws. Small and unobtrusive scales are present (e.g. Reay 1986) and the body is often covered in oblique skinfolds (so-called plicae).
The family Ammodytidae comprises 31 species in seven genera (e.g. Randall and Ida 2014, Orr et al. 2015) of which the two genera Ammodytes and Hyperoplus are distributed circumboreally (Ida et al. 1994). Five species of sand lances belonging to three genera occur in northeast Atlantic waters (Sparholt 2015). This includes the Common sand eel Ammodytes tobianus Linnaeus, 1758 and the Lesser sand eel Ammodytes marinus Raitt, 1934, currently recognised together with four further species in the genus Ammodytes (Orr et al. 2015). Additionally, both species of the genus Hyperoplus, Corbin´s sand eel Hyperoplus immaculatus (Corbin, 1950) and the Greater sand eel Hyperoplus lanceolatus (Le Sauvage, 1824), can be found in the eastern north Atlantic area (Reay 1986), as well as Gymnammodytes semisquamatus (Jourdain, 1879). The latter can morphologically be distinguished from the species mentioned above by having a branched lateral line, a body not covered in oblique plicae (Cameron 1959), and scales that are loosely scattered and restricted to the posterior third of the body (Reay 1986), whereas the genera Hyperoplus and Ammodytes exhibit plicae along the body and an unbranched lateral line.
In identification keys these two genera are often distinguished by showing clear protrusible premaxillae and no vomerine teeth (Ammodytes) or no clear protrusible premaxillae and a pair of vomerine teeth (Hyperoplus, e.g. Reay 1986). Hyperoplus lanceolatus can be separated from Hyperoplus immaculatus by the occurrence of a conspicuous dark spot on either side of the snout below the anterior nostril. This spot is lacking in Hyperoplus immaculatus. Ammodytes tobianus is generally distinguished from Ammodytes marinus by its characteristic belly scales that are organised in tight chevrons and having scales present over the musculature at the base of the caudal fin, whereas these features are not present in Ammodytes marinus (Reay 1986).
However, the distinguishing features mentioned above are not easy to observe for the untrained eye when comparative material of different species is not available. Furthermore, an accurate species identification, especially of juvenile individuals, is difficult and even sub-adult and adult sand lances are difficult to identify (Sparholt 2015), if identification procedure is restricted to the few morphological characters mentioned above. In this context, Naevdal and Thorkildsen (2002) mentioned the difficulties regarding morphological separation of some of the five species of sand lances found in the northeast Atlantic and suggested a method for successful species identification on the basis of allozyme variation. DNA restriction fragment patterns have also been proposed to distinguish between some of the Atlantic sand eel species (Mitchell et al. 1998) as an alternative to morphological characters.
The difficult identification of sand lance species contributed to the current situation that accurate stock assessments are not available separately for each of the species in the North Sea and adjacent areas (see Sparholt 2015). However, sand lances here are subject to large-scale, industrial exploitation for fish meal and oil production and are also a major prey for many predators such as piscivorous fish, birds, and mammals (e.g. Reay 1986). It is known that exploitation of sand lances affects the food availability for these predators and that the abundance of sand lances is sensitive to recruitment variation (Sparholt 2015). Sand lances are divided into seven stock components for stock assessments in the North Sea based on the most abundant species Ammodytes marinus. With this approach, the stock situation of the single species cannot be evaluated, as it does not consider that sand lances represent a mix of different species. Clearly, another drawback is that an evaluation of the conservation status of the single species of sand lances is not possible (Thiel et al. 2013).
Molecular-based identification methods of fish species have been developed over the last decades (for an overview see Teletchea 2009). In this context, DNA barcoding constitutes the most popular and effective technique by using partial (COI) sequences for a standardised and routine identification of specimens to species level (Hebert et al. 2003). For a successful application of DNA barcoding as a tool for specimen identification, reliable sequence reference libraries such as the Barcode of Life Database (BOLD, Ratnasingham and Hebert 2007) were developed. Newly generated DNA barcodes can be uploaded and analysed together with data already available on BOLD in order to provide taxonomic identification. Additionally, barcode sequences were automatically analysed on BOLD and a (BIN) is assigned according to the calculated sequence clusters (Ratnasingham and Hebert 2013). Taxonomic conflicts apparently occur if sequences assigned to the same species name can be found within different BIN clusters.
For fish, the species discrimination success of DNA barcoding was demonstrated in many studies including freshwater as well as marine faunas from many regions all over the world (e.g. Ward et al. 2005, Hubert et al. 2008, Ward et al. 2008a, Steinke et al. 2009, April et al. 2011, Mabragaña et al. 2011, Costa et al. 2012, Zhang and Hanner 2012, Keskin and Atar 2013, McCusker et al. 2013, Geiger et al. 2014, Knebelsberger et al. 2014, Knebelsberger and Thiel 2014, Knebelsberger et al. 2015). DNA barcodes have also been successfully used to identify fish larvae (Pegg et al. 2006, Victor et al. 2009, Hubert et al. 2010, Kim et al. 2010), and fins (Holmes et al. 2009), and can provide evidence for cryptic diversity (Hubert et al. 2010, Ward et al. 2008b, Zemlak et al. 2009, Puckridge et al. 2013, Geiger et al. 2014, Knebelsberger et al. 2015).
For the North Sea and adjacent areas, two DNA barcoding studies revealed successful differentiation of all investigated species (Knebelsberger et al. 2014, Knebelsberger and Thiel 2014). Altogether, 105 species belonging to 88 genera were analysed. Most of the genera were represented by only one species. As an exception, the genus Pomatoschistus was represented by five closely related species.
One of these studies already provided DNA barcodes for the two sand lance species Ammodytes marinus and Hyperoplus immaculatus and demonstrated a clear separation of these two species (Knebelsberger et al. 2014). A former study from continental Portugal Atlantic waters included DNA barcodes for Hyperoplus lanceolatus but other species of sand lances were missing (Costa et al. 2012). Studies including congeneric species of the genus Ammodytes revealed inconsistencies between morphological and DNA barcode-based identification: for two species from the northwest Atlantic Ocean, namely Ammodytes americanus and Ammodytes dubius, barcoding fails to separate these species, which may be caused by inadequate taxonomy (McCusker et al. 2013). Inadequate taxonomy may also concern the two species Ammodytes personatus and Ammodytes hexapterus from the north Pacific (Turanov and Kartavtsev 2014). In both cases the taxonomic status of the species is questionable and may require comprehensive taxonomic revision. In order to examine the application of DNA barcoding for the identification of sand lances from the North Sea area, all closely related species from this region must be included. This concerns Ammodytes marinus and Hyperoplus immaculatus as well as Ammodytes tobianus and Hyperoplus lanceolatus. For the latter two species reliable COI data from the North Sea are still missing.
This paper presents the first comprehensive study combining morphological and molecular methods for the discrimination of four species of sand lances belonging to the genera Ammodytes and Hyperoplus occurring in the northeast Atlantic Ocean and adjacent waters. The suitability of two morphological types of parameters (meristic characters and morphometric measurements) and two genetic approaches (mitochondrial COI (DNA barcoding region) and partial nuclear Rhodopsin DNA sequences) for accurate species identification is examined. A detailed and accurate species identification matrix is presented, based on the integration of morphological and molecular traits.
Materials and methods
Material
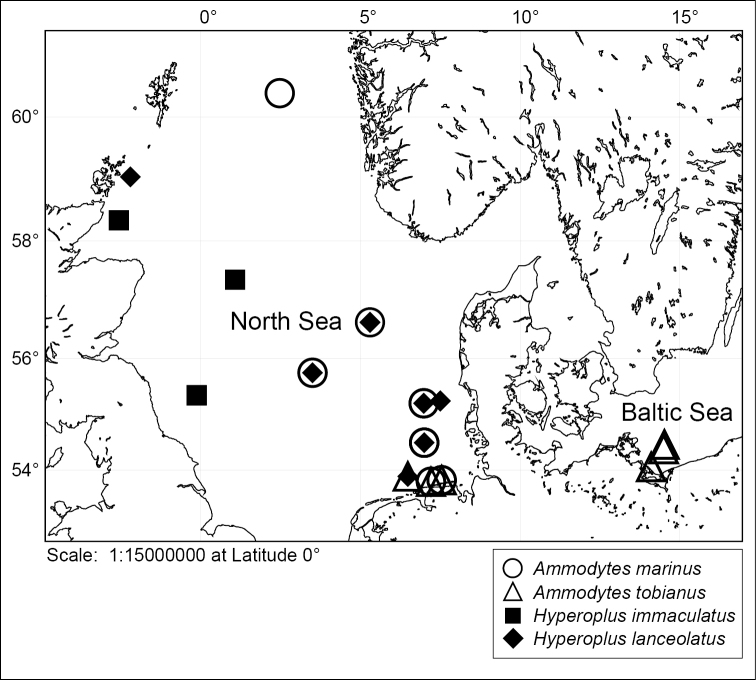
In this study 85 specimens representing two species of genus Ammodytes and two species of genus Hyperoplus were sampled from the North and the Baltic Seas (Suppl. material 1 and 2, Figure 1). For the molecular analysis 70 samples were collected from the North Sea during several cruises conducted by the Thünen Institute of Sea Fisheries (Hamburg, Germany) and the research vessel of the Senckenberg Institute (Wilhelmshaven, Germany). Tissue samples were taken from each of the 70 specimens and preserved in 96% ethanol for molecular analysis at Senckenberg’s German Center for Marine Biodiversity Research (DZMB, Wilhelmshaven, Germany). Specimens were preserved in 70% ethanol. The remaining 15 individuals belonging to the species Ammodytes tobianus were used for morphological analyses only and collected from the Baltic Sea during three different cruises conducted by the German Oceangraphic Museum (Stralsund, Germany). Immediately after catch, specimens were preserved in 4% formaldehyde solution. All 85 voucher specimens were databased and morphologically investigated at the (ZMH, Hamburg, Germany). Finally, the material was stored for future reference in the ZMH fish collection. All COI sequences and related metadata belonging to the 70 voucher specimens from the North Sea are available on the Barcode of Life Data System (www.barcodinglife.org; Ratnasingham and Hebert 2007). DNA barcodes of eight specimens of Hyperoplus immaculatus and 22 specimens of Ammodytes marinus were obtained from the BOLD project (BNSFI) (Knebelsberger et al. 2014). Newly generated barcodes belonging to five specimens of Ammodytes marinus, six of Ammodytes tobianus, and 29 of Hyperoplus lanceolatus were uploaded to the BOLD project (BNSSE). In addition to COI, nuclear Rhodopsin DNA sequences were generated from all 70 North Sea specimens (Suppl. material 1). For comparison, published Rhodopsin data was downloaded from GenBank (Ammodytes tobianus: AY141306; Hyperoplus lanceolatus: EU492010 and EU492011).
Figure 1.
Sampling sites of sand lance species of the genera Ammodytes and Hyperoplus.
Morphological analyses
Meristic parameters (Table 1) were analysed at the left-hand side of the specimens and supplemented with right-side counts when the left side was damaged. Counts of dorsal, ventral and principal caudal-fin rays as well as of vertebrae were taken from radiographs (Figure 2) made by an X-ray imaging system (Faxitron LX-60). The first caudal vertebra was defined as the first centrum with a long haemal spine, and the centrum fused to the hypural plate was counted as the last vertebra. Counts of dorsal-fin rays were made using the method of Nizinski et al. (1990). Counting dorsal-fin rays began with the first visible ray and excluded the one or two anterior rayless pterygiophores. However, these counts included the last two rays that were each supported by a pterygiophore. Counts of anal-fin rays included all rays visible from the outside. Gill rakers were counted on the lower and upper arch separately. Gill rakers of the lower arch included the raker at the junction between upper and lower parts of the arch. Dermal plicae included those anterior and posterior to the lateral-line pores.
Table 1.
Data of estimated morphological characters of four species of sand lances of the genera Ammodytes and Hyperoplus. If possible, each meristic character and morphometric measurement is presented with its range (before the semicolon), mean value with standard deviation and the number of specimens analysed (in brackets). Morphometric measurements are given as proportion of SL.
| Species | Ammodytes marinus | Ammodytes tobianus | Hyperoplus immaculatus | Hyperoplus lanceolatus |
|---|---|---|---|---|
| Meristic characters | ||||
| (DP) | 140–150; 143.1 ± 2.6 (24) | 123–135; 128.5 ± 3.0 (21) | 179–196; 187.8 ± 5.4 (8) | 169–194; 182.6 ± 6.4 (29) |
| (DR) | 56–62; 59.1 ± 1.4 (27) | 51–55; 52.8 ± 1.1 (21) | 59–61; 60.1 ± 0.6 (8) | 54–57; 55.9 ± 0.8 (29) |
| (AR) | 28–31; 29.5 ± 0.8 (27) | 25–30; 27.8 ± 1.3 (21) | 31–32; 31.8 ± 0.5 (8) | 28–30; 29.1 ± 0.8 (29) |
| (PR) | 12–15; 13.6 ± 0.6 (26) | 12–13; 13.0 ± 0.2 (21) | 13–15; 14.1 ± 0.6 (8) | 13–14; 13.5 ± 0.5 (29) |
| (CR) | 15 ± 0 (27) | 15 ± 0 (21) | 15 ± 0 (8) | 15 ± 0 (29) |
| (UR) | 5–6; 5.0 ± 0.2 (24) | 5 ± 0 (21) | 5 ± 0 (8) | 5 ± 0 (29) |
| (LR) | 18–20; 18.6 ± 0.8 (24) | 20 ± 0 (21) | 23–25; 24.3 ± 0.7 (8) | 20–22; 20.7 ± 0.8 (29) |
| (GR) | 23–26; 23.6 ± 0.9 (24) | 25 ± 0 (21) | 28–30; 29.1 ± 0.6 (8) | 25–27; 25.7 ± 0.8 (29) |
| (PV) | 42–44; 42.9 ± 0.7 (27) | 36–41; 38.1 ± 1.4 (21) | 42–43; 42.9 ± 0.4 (8) | 38–42; 40.3 ± 0.8 (29) |
| (CV) | 26–28; 26.7 ± 0.5 (27) | 24–26; 25.1 ± 0.6 (21) | 29–30; 29.5 ± 0.5 (8) | 25–28; 26.5 ± 0.7 (29) |
| (TV) | 68–71; 69.6 ± 0.9 (27) | 60–66; 63.2 ± 1.4 (21) | 72–73; 72.4 ± 0.5 (8) | 65–68; 66.8 ± 0.8 (29) |
| (MDAS) | yes (27) | yes (21) | yes (8) | yes (29) |
| (BCOP) | yes (27) | yes (21) | yes (8) | yes (29) |
| (PCP) | yes (27) | yes (21) | no (8) | no (29) |
| (VTP) | no (27) | no (21) | yes (8) | yes (29) |
| (DSSS) | no (27) | no (21) | no (8) | yes (29) |
| (BSTC) | no (27) | yes (21) | yes (8) | not clearly detectable (29) |
| (SBCF) | no (27) | yes (21) | yes (8) | yes (29) |
| (SADF) | no (27) | yes (21) | yes (8) | yes (29) |
| Morphometric measurements | ||||
| (SL, mm) | 61.2–193; 136.7 ± 29.7 (27) | 121.5–146.4; 134.1 ± 7.7 (21) | 220.4–270.1; 251.4 ± 15.0 (8) | 165.0–291.0; 219.3 ± 32.7 (29) |
| Percentage standard length | ||||
| (BDD) | 5.9–11.1; 9.1 ± 1.3 (26) | 8.3–11.9; 9.5 ± 0.8 (21) | 5.8–7.9; 6.8 ± 0.9 (8) | 5.6–8.8; 7.0 ± 0.9 (29) |
| (BDA) | 4.9–10.1; 8.1 ± 1.2 (27) | 7.3–10.4; 9.4 ± 0.8 (20) | 6.6–8.2; 7.5 ± 0.5 (8) | 5.2–8.0; 6.7 ± 0.6 (28) |
| (BWD) | 4.1–6.4; 5.3 ± 0.7 (26) | 4.0–7.6; 6.0 ± 0.9 (21) | 5.6–8.2; 6.5 ± 0.8 (8) | 4.3–7.2; 5.4 ± 0.8 (29) |
| (HL) | 17.6–22.6; 20.0 ± 1.1 (26) | 18.4–21.4; 19.6 ± 0.8 (21) | 18.3–20.2; 19.4 ± 0.7 (8) | 20.2–23.0; 21.7 ± 0.6 (29) |
| (SNL) | 5.1–6.1; 5.7 ± 0.3 (26) | 5.1–6.0; 5.4 ± 0.3 (21) | 6.0–6.3; 6.1 ± 0.1 (8) | 5.0–8.1; 7.2 ± 0.5 (29) |
| (OD) | 2.7–4.4; 3.2 ± 0.5 (26) | 2.6–3.5; 3.0 ± 0.3 (21) | 1.9–2.5; 2.3 ± 0.2 (8) | 2.0–4.5; 2.5 ± 0.5 (29) |
| (IW) | 2.0–3.5; 2.4 ± 0.3 (27) | 2.1–2.9; 2.4 ± 0.2 (21) | 2.8–3.6; 3.2 ± 0.3 (8) | 2.7–3.7; 3.3 ± 0.3 (29) |
| (UJL) | 4.6–10.3; 6.7 ± 1.0 (26) | 5.4–7.0; 6.4 ± 0.4 (21) | 4.7–6.0; 5.3 ± 0.4 (8) | 6.5–8.1; 7.1 ± 0.3 (29) |
| (CPD) | 2.2 -2.9; 2.6 ± 0.2 (26) | 2.8–3.2; 3.0 ± 0.1 (21) | 2.5–2.9; 2.7 ± 0.1 (8) | 2.3–2.8; 2.5 ± 0.1 (29) |
| (CPL) | 3.2–5.6; 4.1 ± 0.5 (27) | 3.5–5.4; 4.3 ± 0.5 (21) | 4.4–6.4; 4.8 ± 0.7 (8) | 3.9–5.7; 4.9 ± 0.4 (29) |
| (PPL) | 16.6–21.7; 18.6 ± 1.0 (25) | 14.8–20.4; 18.6 ± 1.1 (21) | 17.9–18.7; 18.4 ± 0.3 (8) | 19.5–22.1; 20.8 ± 0.7 (29) |
| (PDL) | 23.5–28.1; 25.3 ± 1.0 (26) | 24.0–27.3; 25.4 ± 1.0 (21) | 26.1–27.6; 26.6 ± 0.5 (8) | 25.7–30.4; 27.9 ± 1.0 (29) |
| (PAL) | 63.4–66.8; 64.9 ± 1.1 (27) | 60.3–67.0; 63.7 ± 1.9 (21) | 60.2–64.1; 62.8 ± 1.4 (8) | 45.4–70.6; 64.9 ± 4.1 (29) |
| (PFL) | 8.6–11.9; 9.9 ± 0.7 (25) | 9.4–11.4; 10.4 ± 0.5 (21) | 7.2–8.2; 7.7 ± 0.4 (8) | 6.7–8.7; 7.7 ± 0.6 (28) |
| (DFBL) | 66.6–72.0; 69.8 ± 1.4 (27) | 66.9–70.8; 69.2 ± 1.1 (21) | 68.1–69.8; 69.1 ± 0.7 (8) | 65.5–69.2; 67.4 ± 1.1 (29) |
| (AFBL) | 28.9–32.4; 30.5 ± 1.0 (27) | 26.7–33.4; 31.6 ± 1.9 (21) | 30.5–33.8; 32.4 ± 1.2 (8) | 28.4–32.1; 29.8 ± 1.0 (29) |
| (CFL) | 8.3–11.5; 10.1 ± 0.7 (26) | 9.0–12.0; 10.3 ± 0.7 (21) | 8.5–9.7; 8.9 ± 0.4 (8) | 7.6–10.3; 8.9 ± 0.7 (28) |
| (DFH) | 3.6–6.0; 4.7 ± 0.7 (27) | 3.9–6.1; 4.8 ± 0.6 (21) | 3.4–4.6; 4.1 ± 0.4 (8) | 2.7–5.7; 3.9 ± 0.6 (29) |
| (AFH) | 2.6–6.2; 4.7 ± 0.8 (27) | 3.4–5.8; 4.8 ± 0.5 (21) | 2.5–4.6; 3.9 ± 0.7 (8) | 2.7–5.6; 4.1 ± 0.6 (29) |
Figure 2.
Radiograph of Common sand eel Ammodytes tobianus (Linnaeus, 1758) indicating the meristic characters evaluated from X-ray pictures. Depicted specimen: ZMH 26098-3, standard length 128.1 mm.
Morphometric measurements (Table 2) were taken by vernier calipers to one tenth of a millimetre. Measurements were done following Hubbs and Lagler´s (1958) method, with the following changes: (SL) was measured from the front of the upper lip in the median plane to the midbase of the caudal fin (end of hypural plate). The front of the upper lip was used as the anterior point of all other horizontal measurements. (HL) was measured from the front of the upper lip to the posterior end of the opercular membrane. Body depth was measured twice, as the (BDD) and as the (BDA). Body width was measured as the maximum (BWD). (OD) is the maximum fleshy diameter. (IW) is the least fleshy width. (CPD) is the smallest depth, and (CPL) the horizontal distance between verticals at the rear base of the anal fin and the caudal-fin base. (DFH) and (AFH) were measured from their tips to the body contour. (CFL) was taken horizontally from the caudal-fin base to a vertical point at the tip of the longest ray. The values obtained were standardised by multiplying them by 100 and dividing them by the standard length.
Table 2.
Standardised coefficients of the first three discriminant functions (DF1, DF2, DF3) separating the four species of Ammodytes and Hyperoplus based on meristic characters. In bold, characters with the greatest weight in DF1 and DF2.
| Meristic characters | DF1 | DF2 | DF3 |
|---|---|---|---|
| DP | 0.884 | -0.320 | -0.444 |
| DR | 0.056 | 0.664 | -0.096 |
| AR | 0.112 | -0.155 | 0.159 |
| PR | 0.015 | 0.079 | -0.172 |
| UR | -0.428 | -0.135 | -0.289 |
| LR | 0.432 | -0.140 | 0.891 |
| PV | -0.017 | 0.523 | -0.061 |
| CV | 0.426 | 0.516 | 0.471 |
| Percentage of explained variance | 71.438 | 20.317 | 8.245 |
| Eigenvalue | 40.392 | 11.488 | 4.662 |
| Cumulative variance in % | 71.438 | 91.755 | 100.00 |
Statistical treatment of morphological data
All morphological data were statistically processed, involving ranges, means, and standard deviations. Morphological data of all specimens that had a complete suite of meristic and morphometric character data were used to conduct two multiple (DFA) to determine if the four species of sand lances could be differentiated based on meristic and/or morphometric parameters using XLSTAT (version 2013.0.04, Addinsoft), a statistical analysis add-in for Microsoft Excel®. DFA was used to demonstrate the degree of separation in multivariate space defined by the main patterns of morphological variation among species which is described via the discriminant functions. It also shows which character contributes more to the differentiation. The standardised discriminant function coefficients represent the contributions of every variable to the discriminatory power of the function. Hence, the larger the standardised coefficient, the larger the weight of the variable in the function. Both discriminant analyses were conducted for 76 individuals (22 Ammodytes marinus, 20 Ammodytes tobianus, 8 Hyperoplus immaculatus, 26 Hyperoplus lanceolatus). Morphological variables without any variation (e.g. (CR)), variables, where other variables are included (e.g. (TV)) and qualitative variables (e.g. (PCP)) were excluded from the DFA procedures. The first DFA was performed for the following eight quantitative meristic characters: dermal plicae, dorsal-fin rays, anal-fin rays, pectoral-fin rays, upper arch gill rakers, lower arch gill rakers, precaudal vertebrae, and caudal vertebrae. The second DFA was conducted for the following 19 morphometric parameters: body depth at dorsal-fin origin, body depth at anal-fin origin, body width at dorsal-fin origin, head length, snout length, orbit diameter, interorbital width, upper jaw length, caudal peduncle depth, caudal peduncle length, prepectoral length, predorsal length, preanal length, pectoral-fin length, dorsal-fin base length, anal-fin base length, caudal-fin length, dorsal-fin height, and anal-fin height.
DNA isolation, PCR amplification, and sequencing
Genomic DNA was extracted at the DZMB using the Qiagen DNeasy Blood and Tissue Kit for single columns as described by Knebelsberger and Stöger (2012). A 652-bp fragment of the (mt) COI gene was amplified for 38 samples using a M13 tailed primer cocktail (C_FishF1t1-C_FishR1t1) including FishF2_t1 (5’-TGTAAAACGACGGCCAGTCGACTAATCATAAAGATATCGGCAC), FishR2_t1 (5’-CAGGAAACAGCTATGACACTTCAGGGTGACCGAAGAATCAGAA), VF2_t1 (5’-TGTAAAACGACGGCCAGTCAACCAACCACAAAGACATTGGCAC) and FR1d_t1 (5’-CAGGAAACAGCTATGACACCTCAGGGTGTCCGAARAAYCARAA) (Ivanova et al. 2007). As a second marker, a 464 bp fragment of the (nc) Rhodpsin gene was amplified for all 70 samples using the forward primer Rh545 (5’-GCAAGCCCATCAGCAACTTCCG) and the reverse primer Rh1039r (5’-TGCTTGTTCATGCAGATGTAGA) developed by Chen et al. (2003). Each PCR reaction mixture contained 1 µl DNA template, 2.25 µl of 10X reaction buffer (including MgCl2), 0.5 µl dNTPs (2mM each), 0.5 µl of each primer (10 pmol/µl), 0.5 µl Taq polymerase (5 U/µl; Qiagen) and molecular grade water for a total volume of 25µl. Thermal cycling was performed with an initial denaturation for 2 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at the annealing temperature of 50°C, 60 s at 72°C with a final extension of 10 min at 72°C. All PCR products were checked by a 1% agarose gel. Amplicons were purified by incubating 10 µl of PCR products with 0.5 µl of Exonuclease I (20 U/µl) and 2 µl of Alkaline Phosphatase (1 U/µl) for 15 min at 37°C followed by 20 min at 75°C. Purified amplicons were sequenced by Macrogen Europe (Amsterdam, Netherlands).
Sequence alignment and data analyses
Forward and reverse sequences of COI and Rhodpsin were assembled and edited using Geneious (version 7.1.9. http://www.geneious.com). Consensus sequences were submitted to GenBank (for accession numbers see Suppl. material 1). Variance in sequence length, base composition, number of invariable sites and the presence of stop codons were analysed using Geneious. The nc and mt sequences were aligned independently using MUSCLE (Edgar 2004) with default settings as implemented in MEGA version 6.06 (Tamura et al. 2013). Primer sequences were cut from the alignment. Rhodopsin gene sequence alignment was checked by eye for species specific diagnostic characters. For COI, (K2P) distances were calculated in MEGA, as K2P is used as standard model for barcoding analyses and enables direct comparison with other studies. (NJ) topology (Saitou and Nei 1987) was built in MEGA using the “pairwise deletion” option for the treatment of gaps and missing data, in order to retain all sites initially, excluding them as necessary. Node support for the NJ topology was evaluated by a non-parametric bootstrap analysis (Felsenstein 1985) with 10,000 replicates. In order to quantify the distinctness between species at the barcode locus, genetic distances were used to calculate the difference between the maximum intraspecific genetic distance and the minimum distance to the nearest neighbor (barcode gap). For the calculation of genetic distances at genus and family level, we used BOLDs “Distance Summary” tool by choosing K2P distance model and MUSCLE (Edgar 2004) alignment algorithm.
On BOLD, DNA barcodes were automatically assigned to (OTUs), generated through (RESL) analyses (Ratnasingham and Hebert 2013). Finally, a unique alphanumeric code is assigned to each of the OTUs, constituting the so called (BIN). It has been shown that BINs are highly congruent with existing species assignments (Ratnasingham and Hebert 2013). Here, the ‘BIN Discordance Report’ analysis tool was applied to analyse our dataset together with public sequences on BOLD, and to get hints on cryptic diversity (species) or to identify cases of haplotype sharing between species.
Furthermore, BOLD’s “Diagnostic Characters” sequence analysis tool was applied to the COI dataset choosing MUSCLE (Edgar 2004) alignment algorithm. Sequences were grouped by species names in order to categorise consensus bases by their diagnostic potential.
Results
General results of morphological analysis
Meristic characters and morphometric measurements of the four examined species of sand lances are given in Table 1. The number of individuals per analysed character ranged from 24 to 27 for Ammodytes marinus, from 20 to 21 for Ammodytes tobianus, from 28 to 29 for Hyperoplus lanceolatus, and was eight individuals for Hyperoplus immaculatus (Table 1).
The data of the present study confirmed that the two genera of Ammodytes and Hyperoplus can be distinguished by qualitative meristic characters, i.e. by having a (PCP) and no (VTP) (Ammodytes) or no clear protrusible premaxillae and two vomerine teeth (Hyperoplus) (Table 1). Hyperoplus can also be separated from Ammodytes by its significantly higher number of (DP). It is also possible to distinguish Hyperoplus from Ammodytes by its obviously lower (PFL), and to a somewhat lesser significance, also by its greater mean (SNL), since no sexual dimorphism has been reported for the last two characters in both genera.
Hyperoplus lanceolatus can be separated from Hyperoplus immaculatus by the presence of a conspicuous (DSSS) which is lacking in Hyperoplus immaculatus (Table 1). Furthermore, Hyperoplus lanceolatus differs from Hyperoplus immaculatus by its lower numbers of total and lower arch gill rakers (GR, LR), total and caudal vertebrae (TV, CV), as well as dorsal and anal-fin rays (DR, AR).
Ammodytes tobianus can be distinguished from Ammodytes marinus by having (BSTC) and having (SBCF) and in the midline anterior to dorsal fin (SADF), whereas these characters are not present in Ammodytes marinus (Table 1). Ammodytes tobianus differs from Ammodytes marinus also by its lower numbers of (DP), (DR), and precaudal and total vertebrae numbers (PV, TV).
Discriminant Function Analysis with meristic characters
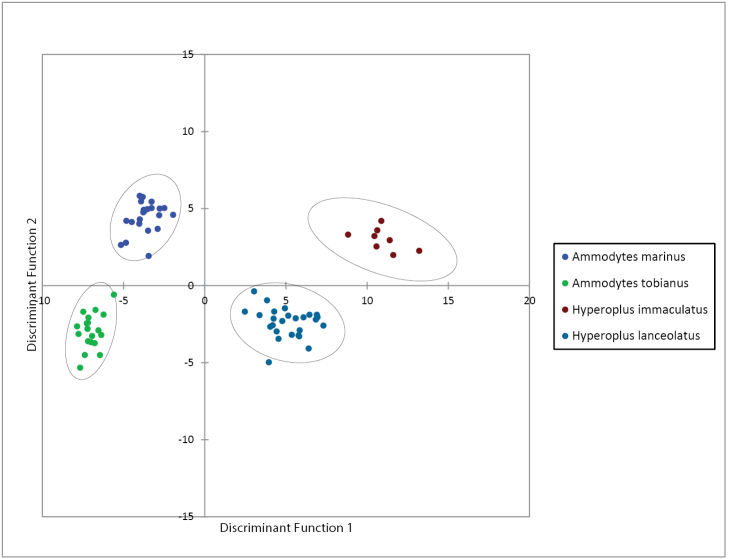
DFA based on meristic characters provided three significant functions (Box-Test with χ2 = 790.916 and p<0.0001; Wilks´ lambda = 0.0003 and p<0.0001). These three functions explain 100% of the total variation in the data. The first two functions explain 91,755% of the total variation in the data (Table 2), which is sufficient for the further detailed analysis. The third discriminant function explains 8.245% of total variation.
Individual specimens are projected onto the first two discriminant functions in Figure 3. Because all four species were clearly separated in the discriminant space defined by the first two functions, the third function was not used. The first discriminant function explains 71.438% of total variation (Table 2). It mainly separates Ammodytes tobianus and Hyperoplus immaculatus and to a lesser extent the species pairs of Ammodytes tobianus and Hyperoplus lanceolatus, Ammodytes marinus and Hyperoplus immaculatus, Ammodytes marinus and Hyperoplus lanceolatus as well as Hyperoplus lanceolatus and Hyperoplus immaculatus (Figure 3). Ammodytes tobianus and Ammodytes marinus cannot be so clearly separated by the first discriminant function.
Figure 3.
Plot of all analysed Ammodytes and Hyperoplus specimens onto the first and second discriminant functions based on a set of eight meristic characters. Circles include 95% of specimens in each species.
From the standardised coefficients (Table 2), the two characters that have the greatest influence on the first discriminant function (characters most discriminatory) are the (DP) and (LR) (Table 2). In general Hyperoplus immaculatus and Hyperoplus lanceolatus have a much higher number of dermal plicae than Ammodytes tobianus and Ammodytes marinus (Table 1). The numbers of DP of both species of Hyperoplus are overlapping, whereas the Ammodytes species have different numbers of DP. The number of (LR) is higher in Hyperoplus immaculatus in comparison with the other three species, which have overlapping numbers of LR.
The second discriminant function accounts for 20.317% of total variation. Ammodytes tobianus and Ammodytes marinus are clearly and the species pairs of Ammodytes tobianus and Hyperoplus immaculatus, Ammodytes marinus and Hyperoplus lanceolatus as well as Hyperoplus immaculatus and Hyperoplus lanceolatus are to a lesser extent discriminated by this function. Ammodytes tobianus and Hyperoplus lanceolatus and Ammodytes marinus and Hyperoplus immaculatus cannot be clearly separated by the second discriminant function. The contrasts between the numbers of (DR) and the numbers of (PV) of the species are mainly responsible for this discrimination. DR is lowest in Ammodytes tobianus and highest in Hyperoplus immaculatus (Table 1). PV is lowest in Ammodytes tobianus and highest in Ammodytes marinus and Hyperoplus immaculatus.
Discriminant Function Analysis with morphometric measurements
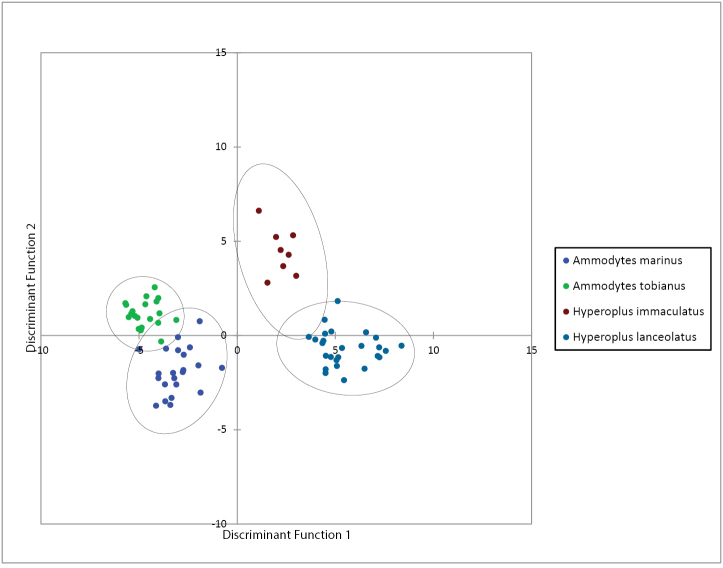
Three significant DFA functions were estimated based on morphometric measurements (Box-Test with χ2 = 944.979 and p < 0.0001; Wilks´ lambda = 0.003 and p < 0.0001). Together these functions explain 100% of the total variation in the data. The first two functions explain 93.144% of the total variation in the data (Table 3), which is sufficient for the further detailed analysis. The third discriminant function explains 6.856% of total variation.
Table 3.
Standardised coefficients of the first three discriminant functions (DF1, DF2, DF3) separating the four species of Ammodytes and Hyperoplus based on morphometric measurements. In bold, characters with the greatest weight in DF1 and DF2.
| Morphometric measurements | DF1 | DF2 | DF3 |
|---|---|---|---|
| BDD | -0.135 | -0.184 | 0.085 |
| BDA | -0.178 | -0.038 | -0.208 |
| BWD | -0.054 | 0.317 | 0.222 |
| HL | 0.290 | -0.084 | -0.603 |
| SNL | 0.380 | -0.040 | -0.045 |
| OD | -0.231 | 0.078 | 0.497 |
| IW | 0.198 | 0.164 | 0.237 |
| UJL | -0.034 | -0.868 | -0.260 |
| CPD | -0.228 | 0.542 | -0.473 |
| CPL | 0.112 | 0.418 | -0.147 |
| PPL | 0.204 | 0.101 | -0.076 |
| PDL | 0.140 | 0.251 | 0.143 |
| PAL | -0.020 | 0.008 | 0.103 |
| PFL | -0.612 | -0.280 | -0.528 |
| DFBL | -0.060 | 0.104 | 0.551 |
| AFBL | 0.098 | 0.429 | -0.208 |
| CFL | -0.148 | 0.520 | 0.390 |
| DFH | -0.078 | -0.125 | 0.223 |
| AFH | 0.136 | -0.333 | -0.259 |
| Percentage of explained variance | 78.576 | 14.568 | 6.856 |
| Eigenvalue | 20.555 | 3.811 | 1.794 |
| Cumulative variance in % | 78.576 | 93.144 | 100.00 |
Figure 4 presents the individual specimens projected onto the first two discriminant functions. Because all four species were clearly separated in the discriminant space defined by the first two functions, the third function was not used. The first discriminant function explains 78.576% of total variation (Table 3). It mainly separates Ammodytes tobianus and Hyperoplus lanceolatus and to a lesser extent the species pairs of Ammodytes tobianus and Hyperoplus immaculatus, Ammodytes marinus and Hyperoplus lanceolatus as well as Ammodytes marinus and Hyperoplus immaculatus (Figure 4). The species pairs of Ammodytes marinus and Ammodytes tobianus as well as of Hyperoplus immaculatus and Hyperoplus lanceolatus cannot be separated by the first discriminant function.
Figure 4.
Plot of all analysed Ammodytes and Hyperoplus specimens onto the first and second discriminant functions based on a set of 19 morphometric characters. Circles include 95% of specimens in each species.
The two measurement characters that have the greatest weight on the first discriminant function are (PFL) and the (SNL) (Table 3). Both species of the genus Ammodytes have a greater PFL than both species of the genus Hyperoplus (Table 1). In contrast, both Hyperoplus species have a greater SNL than both Ammodytes species. PFL and SNL are relatively similar for the species of the same genera.
The second discriminant function accounts for 14.568% of total variation. Especially the species within the genera Ammodytes and Hyperoplus, namely Ammodytes tobianus and Ammodytes marinus as well as Hyperoplus immaculatus and Hyperoplus lanceolatus are separated by this function (Figure 4).
(UJL) and (CPD) are the two measurements, for which no sexual dimorphism is known, and that have the greatest weight on the second discriminant function (Table 3).
Mt DNA barcoding
Mitochondrial DNA barcodes were obtained for 70 specimens belonging to four species of the family Ammodytidae investigated in this study (Suppl. material 1). The DNA sequences did not show any ambiguous base calls (Ns) or stop codons, and no insertions or deletions were found within the sequence alignment. Sequence length ranged from 619 to 652 bp (mean and standard deviation: 650.5 ± 5.7 bp). The average base composition was 22.8% adenine (A), 29.7% cytosine (C), 18.3% guanine (G) and 29.3% thymine (T); GC content was 48%. The sequence alignment showed 588 identical sites.
The NJ analysis of the K2P distances revealed well supported monophyletic clusters for the species Ammodytes marinus and Hyperoplus immaculatus with bootstrap values of 97 and 100, respectively (Figure 5). In contrast, Ammodytes tobianus and Hyperoplus lanceolatus sequences were grouped together in one monophyletic cluster with a bootstrap support of 100. Within this cluster the sequences of Ammodytes tobianus were grouped together without bootstrap support indicating that there is no sharing of haplotypes between these two species. The analysis of the K2P genetic distances revealed an overlap between intraspecific (range: 0.0-0.77%; mean and standard deviation: 0.22 ± 0.17%) and interspecific distances (0.15-7.27%; 4.73 ± 1.7%). The overlap was caused by the two species Ammodytes tobianus and Hyperoplus lanceolatus: in Ammodytes tobianus, the minimum distance to the nearest neighbour species was even lower than the maximum intraspecific distance, whereas both values were equal in Hyperoplus lanceolatus (Table 4). In contrast, the species Ammodytes marinus and Hyperoplus immaculatus exhibited barcode gaps of 2.73% and 3.34% respectively, which indicates an undoubtedly separation from the other species. At genus and family level, the genetic distances between species of the same genus varied between 4.46-7.09% and the distances between species belonging to different ranged from 0.15-7.27%.
Figure 5.
NJ dendrogram based on K2P pairwise genetic distances. Values at nodes indicate the result of the bootstrap test (10.000 pseudo replicates). Only values ≥ 50 are shown. For Ammodytes tobianus (grey box) and Hyperoplus lanceolatus all analysed individuals are shown. In case of Ammodytes marinus and Hyperoplus immaculatus the number of specimens is given in brackets.
Table 4.
Minimum and maximum intraspecific genetic K2P distances (%) for each species including mean and standard deviation. The barcoding gap indicates the difference between the maximum intraspecific and the minimum interspecific (nearest neighbour) genetic distance. Additionally K2P genetic distances are given in brackets, if they differ from p-distances.
| Species | Specimens | Mean Distance | SD* | Minimum Distance | Maximum Distance | Nearest Neighbor | Distance to Nearest Neighbor | Barcoding Gap |
|---|---|---|---|---|---|---|---|---|
| Ammodytes marinus | 27 | 0.24 | 0.19 | 0.00 | 0.77 | Hyperoplus immaculatus | 3.50 | 2.73 |
| Ammodytes tobianus | 6 | 0.15 | 0.01 | 0.00 | 0.15 | Hyperoplus lanceolatus | 0.15 | no gap |
| Hyperoplus lanceolatus | 29 | 0.22 | 0.15 | 0.00 | 0.62 | Ammodytes tobianus | 0.15 | no gap |
| Hyperoplus immaculatus | 8 | 0.07 | 0.08 | 0.00 | 0.16 | Ammodytes marinus | 3.50 | 3.34 |
BIN report
The BIN discordance report tool on BOLD assigned three different BIN numbers to the 70 COI haplotypes. The BIN BOLD:ACF3320 was found to be “concordant” and exclusively comprised 32 specimens of the species Ammodytes marinus, of which five individuals were not provided by this study. The “discordant” BIN BOLD:AAC5676 comprised 57 specimens, 14 identified as Ammodytes tobianus and 43 as Hyperoplus lanceolatus. From the former species eight specimens and from the latter 14 specimens were not provided by our study but also support the findings of this study. The third BIN BOLD:AAJ2299 was also specified as discordant and comprised ten specimens, eight (in our study) identified as Hyperoplus immaculatus and two identified as Ammodytes marinus. The two Ammodytes marinus entries may represent cases of misidentification as 32 Ammodytes marinus individuals were grouped together in BIN BOLD:ACF3320.
Diagnostic characters
The analysis revealed four diagnostic characters for the species Ammodytes marinus and 16 for Hyperoplus immaculatus (results not shown). The two species Ammodytes tobianus and Hyperoplus lanceolatus did not show any diagnostic characters on species level. Consequently, only two of the four investigated species can be identified using diagnostic characters on the basis of COI barcode sequences.
Nc DNA analysis
The nc Rhodopsin sequence alignment showed a length of 464 bp after primer trimming. The complete fragment could be amplified and sequenced for all 70 specimens used for the mt DNA barcode analysis. The number of variable sites was very low and the alignment could be easily evaluated by eye. One diagnostic character was found for each of the species Ammodytes marinus (Table 5; site 460: C instead of A), Hyperoplus lanceolatus (site 82: T instead of C), and Hyperoplus immaculatus (site 433: A instead of G). Ammodytes tobianus showed no species specific mutation but could be characterised by a combination of all tree variable sites (Table 5, underlined bases). The Rhodopsin sequences from GenBank were compared with our sequences; the two Hyperoplus lanceolatus sequences (GenBank accessions: EU492010, EU492011) showed concordant results. In the case of the Ammodytes tobianus sequence (GenBank accession: AY141306) no data was available for site 460 but the two other sites were in agreement with our results.
Table 5.
Variable sites identified for the nc Rhodopsin gene fragment sequence alignment. Bases in bold indicate species specific diagnostic characters. The three underlined bases are distinctive for Ammodytes tobianus.
| Nucleotide position | ||||
|---|---|---|---|---|
| Species | Specimens | 82 | 433 | 460 |
| Ammodytes marinus | 27 | C | G | C |
| Ammodytes tobianus | 7* | C | G | A |
| Hyperoplus lanceolatus | 30** | T | G | A |
| Hyperoplus immaculatus | 8 | C | A | A |
*one /**two sequences downloaded from GenBank.
Discussion
Identification of genera and species using morphological characters
The primary objective of this study was to contribute to robust genera- and species-level identifications, combining morphological and molecular methods, of four closely related species of sand lances of the genera Ammodytes and Hyperoplus occurring in the northeast Atlantic Ocean and adjacent waters.
The detailed morphological analyses confirmed findings described by other authors (e.g. Duncker and Mohr 1939, Reay 1986): the genus Ammodytes can be distinguished by two morphological characters from the genus Hyperoplus. Ammodytes has clear protrusible premaxillae and no vomerine teeth. In contrast, Hyperoplus has no clear protrusible premaxillae and a pair of vomerine teeth. It should be noted here that Kayser (1961) found out that Hyperoplus has no real vomerine teeth, but anterior hooked ends of the prevomer instead.
Subsequently, Ida et al. (1994) pointed out that the tip of the prevomer in Ammodytes is straight, not protruded from the roof of the mouth, whereas in Hyperoplus the tip of the prevomer curved downwards, protruding from the roof of the mouth. According to Wiecaszek et al. (2007), the genus Ammodytes also has a longer lower jaw when compared to the length of pectoral-fin, while this relationship is reversed in Hyperoplus.
This study adds three more characters helpful in distinguishing between both genera of sand lances based on the four species considered. Firstly, the number of dermal plicae is significantly higher in Hyperoplus compared to Ammodytes. Secondly, Hyperoplus has a lower pectoral-fin length in relation to (SL) than Ammodytes. Goltberg (1910) also reported a lower value of pectoral-fin length expressed as a proportion of head length for Hyperoplus lanceolatus than for Ammodytes tobianus. Thirdly, Hyperoplus has a larger mean snout length in % SL than Ammodytes. However, the last mentioned character is less recommended for practical taxonomical assignments, since its ranges overlap between the genera to a relatively large extent. Therefore, a combination of the following four characters remains, which seems to be useful to distinguish between the genera Hyperoplus and Ammodytes: protrusibility of premaxillae, presence of the hooked ends of prevomer, number of dermal plicae, and pectoral-fin length in % SL.
As indicated by the results of discriminant function analysis, morphometric measurements seem not to be characters of the first choice to distinguish the two species of each of the two genera, since they could not be discriminated by the first discriminant function.
According to the results presented here, six meristic characters (the number of lower arch gill rakers, the total number of gill rakers, the number of caudal vertebrae, the number of total vertebrae, and the number of dorsal-fin and anal-fin rays) are more useful than morphometric measurements to distinguish between Hyperoplus immaculatus and Hyperoplus lanceolatus. The use of these additional characters would support and refine the current methods to separate Hyperoplus lanceolatus from Hyperoplus immaculatus. Searching only for the occurrence of a conspicuous dark spot on either side of snout below anterior nostril could be unsuccessful in the case of preserved specimens.
In the case of Ammodytes tobianus and Ammodytes marinus, these results support the information on useful distinguishing characters between both species reported for instance by Reay (1986): Ammodytes tobianus differs from Ammodytes marinus by its belly scales that are organised in tight chevrons, scales which are present over musculature at base of caudal fin, as well as by lower numbers of dermal plicae, dorsal-fin rays and vertebrae. It should be mentioned that our analyses included also Ammodytes tobianus from the Baltic Sea, for which no meristic or morphometric data had been published, except for the number of vertebrae and pectoral-fin length (Wiecaszek et al. 2007).
Discrimination of genera and species based on molecular data
The successful discrimination of the two sand lance species Ammodytes marinus and Hyperoplus immaculatus by DNA barcoding was already demonstrated by Knebelsberger et al. (2014) and could be confirmed by the present study. An additional three specimens of Ammodytes marinus were added to the dataset and the NJ analysis revealed well-supported monophyletic species clusters for Ammodytes marinus and Hyperoplus immaculatus, indicating an unambiguous separation of these two species. Successful species discrimination can also be demonstrated by the presence of gaps between intra- and interspecific genetic distances (Meyer and Paulay 2005), which were in case of Ammodytes marinus and Hyperoplus immaculatus 2.73 and 3.34 respectively. The BIN analysis performed on BOLD revealed two separate species BINs: one concordant BIN exclusively contained sequences which were taxonomically annotated as Ammodytes marinus, and a second discordant BIN contained all specimens of Hyperoplus lanceolatus and two further entries referring to as Ammodytes marinus. These two individuals were provided by other sources and may represent cases of misidentification, as all other Ammodytes marinus entries appeared in the concordant BIN.
Surprisingly, the two species Ammodytes tobianus and Hyperoplus lanceolatus belonging to different genera cannot be clearly separated on the basis of genetic distances, as the lowest distance (K2P) between these two species was only 0.15% and within species variation was found to be 0.15 and 0.62% respectively. In the NJ dendrogram both species appeared together in a well supported clade and were also found within the same BIN cluster when analysed together with data on BOLD. However, Ammodytes tobianus and Hyperoplus lanceolatus do not show haplotype sharing, as Ammodytes tobianus sequences appeared together in a separate cluster. The two species may therefore be separated by applying tree-based approaches like GMYC or model-based ones like ABGD.
In contrast to the barcoding results, both genera of Ammodytes and Hyperoplus can undoubtedly be separated by morphological character traits as discussed above. DNA barcoding failure between closely related congeneric species is usually more common than between species belonging to different genera (e.g. McCusker et al. 2013, Knebelsberger et al. 2015). For congeneric species of the genus Ammodytes inconsistencies between morphological data and DNA barcodes have already been demonstrated. For instance Ammodytes americanus DeKay, 1842 and Ammodytes dubius Reinhardt, 1837 from the northwest Atlantic Ocean could not be separated by DNA barcoding, possibly caused by inadequate taxonomy (McCusker et al. 2013), which may also concern the two species Ammodytes personatus Girard, 1856 and Ammodytes hexapterus Pallas, 1814 from the north Pacific (Turanov and Kartavtsev 2014).
In the present work, inadequate taxonomy, erroneous species designation or identification error can be excluded as possible explanation for DNA barcoding failure in unambiguously separating Ammodytes tobianus from Hyperoplus lanceolatus. In addition, true biological phenomena such as the occurrence of hybridisation or incomplete lineage sorting seem to be unlikely, as no interspecific haplotype sharing was found. In cases where mitochondrial COI sequences fail to distinguish between species, the application of nuclear DNA markers may be tested alternatively. In fish, the nuclear Rhodopsin gene has already been proposed as supplementary marker in order to identify species (Sevilla et al. 2007). However, most studies demonstrated reduced species discrimination success using nuclear Rhodopsin sequences compared to COI barcodes (Hanner et al. 2011, Collins et al. 2012, Behrens-Chapuis et al. 2015). In our study, the analysis of a short nuclear Rhodpsin gene fragment revealed diagnostic nucleotides for the species Ammodytes marinus, Hyperoplus lanceolatus and Hyperoplus immaculatus. The species Ammodytes tobianus can be characterised by the lack of species specific mutations compared to the other three species. Consequently, all four species of sand lances can be identified using the diagnostic character approach in combination with nuclear Rhodopsin sequences. In contrast to that, COI provided diagnostic characters only for the two species Ammodytes marinus and Hyperoplus immaculatus. Ammodytes tobianus and Hyperoplus lanceolatus cannot be characterised by this approach.
Our study clearly demonstrated that nuclear Rhodopsin constitutes a preferable alternative marker to discriminate successfully between the four investigated species of sand lances.
Finally, it should be pointed out that the present results are not meant to provide a phylogenetic reconstruction with regard to the genera Ammodytes and Hyperoplus, since the latter requires a more detailed study of more species of both genera, as well as other members of the group. However, accurate identification of these sand lance species is the basis to assess the status of their stocks and to implement appropriate measures of fisheries management or conservation, and as such, the aim of successfully identifying the NE Atlantic species has been accomplished.
Conclusion
With this study a robust genus- and species-level discrimination of the four most abundant and closely related species of sand lances of the genera Ammodytes and Hyperoplus in the NE Atlantic Ocean and adjacent waters has been provided. It is expected that these results will facilitate the accurate identification of Ammodytes marinus, Ammodytes tobianus, Hyperoplus immaculatus, and Hyperoplus lanceolatus combining morphological and molecular methods.
Acknowledgements
We thank the Thünen Institute of Sea Fisheries for supporting sampling regimes. Many thanks go also to Irina Eidus, Elena Hauten, Renate Thiel, and Laura Wichmann for their helpful support during this study. The molecular work was funded by the Federal Ministry of Education and Research (Grant No. 03F0499A) and the Land Niedersachsen. A part of the Ammodytes tobianus specimen was sampled in the framework of a study which was assigned by the German Federal Agency for Nature Conservation and sponsored by the Federal Ministry for the Environment, Nature Conservation and Nuclear Safety under grant number 80385220.
Citation
Thiel R, Knebelsberger T (2016) How reliably can northeast Atlantic sand lances of the genera Ammodytes and Hyperoplus be distinguished? A comparative application of morphological and molecular methods. ZooKeys 617: 139–164. doi: 10.3897/zookeys.617.8866
Supplementary materials
Table S1
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Ralf Thiel, Thomas Knebelsberger
Data type: specimen data
Explanation note: Supplementary metadata for specimens used for both morphological and genetic analyses; Museum and Sample IDs are specimen identifiers, BOLD Process IDs are unique codes automatically generated for each record on BOLD, GenBank Accession NOs represent sequence identifiers.
Table S2
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Ralf Thiel, Thomas Knebelsberger
Data type: specimen data
Explanation note: Museum IDs and collection data for specimens of Ammodytes tobianus used for morphological analyses only.
References
- April J, Mayden RL, Hanner RH, Bernatchez L. (2011) Genetic calibration of species diversity among North America’s freshwater fishes. Proceedings of the National Academy of Sciences of the United States of America 108: 10602–10607. doi: 10.1073/pnas.1016437108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens-Chapuis S, Herder F, Geiger MF, Esmaeili HR, Hamidan NA, Özuluğ M, Šanda R. (2015) Adding nuclear rhodopsin data where mitochondrial COI indicates discrepancies–can this marker help to explain conflicts in cyprinids? DNA Barcodes 3(1): 187–199. doi: 10.1515/dna-2015-0020 [Google Scholar]
- Collins RA, Armstrong KF, Meier R, Yi Y, Brown SD, Cruickshank RH, et al. (2012) Barcoding and border biosecurity: identifying cyprinid fishes in the aquarium trade. PLoS ONE 7(1): . doi: 10.1371/journal.pone.0028381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron J. (1959) The larval and post-larval stages of Gymnammodytes semisquamatus (Jourdain). Journal of the Marine Biological Association of the UK 38: 17–25. doi: 10.1017/S002531540001554X [Google Scholar]
- Costa FO, Landi M, Martins R, Costa MH, Costa ME, Carneiro M, Alves MJ, Steinke D, Carvalho GR. (2012) A ranking system for reference libraries of DNA barcodes: application to marine fish species from Portugal. PLoS ONE 7: . doi: 10.1371/journal.pone.0035858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger MF, Herder F, Monaghan MT, Almada V, Barbieri R, Bariche M, Berrebi P, Bohlen J, Casal-Lopez M, Delmastro GB, Denys GPJ, Dettai A, Doadrio I, Kalogianni E, Kärst H, Kottelat M, Kovačić M, Laporte M, Lorenzoni M, Marčić Z, Özuluğ M, Perdices A, Perea S, Persat H, Porcelotti S, Puzzi C, Robalo J, Šanda R, Schneider M, Šlechtová V, Stumboudi M, Walter S, Freyhof J. (2014) Spatial Heterogeneity in the Mediterranean Biodiversity Hotspot Affects Barcoding Accuracy of its Freshwater Fishes. Molecular Ecology Resources, online in advance of print. doi: 10.1111/1755-0998.12257 [DOI] [PubMed]
- Goltberg G. (1910) Ammodytes - arterna vid Finlands kuster. Acta Societatis pro Fauna et Flora Fennica 33: 1–39. [Google Scholar]
- Duncker G, Mohr E. (1939) Revision der Ammodytidae. Mitteilungen Zoologisches Museum Berlin 24(1): 8–31. [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. doi: 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1985) Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791. doi: 10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- Hanner R, Floyd R, Bernard A, Collette BB, Shivji M. (2011) DNA barcoding of billfishes. Mitochondrial dna 22(sup1): 27–36. doi: 10.3109/19401736.2011.596833 [DOI] [PubMed] [Google Scholar]
- Hebert PD, Cywinska A, Ball SL. (2003) Biological identifications through DNA barcodes. Proceedings of the Royal Society of London B: Biological Sciences 270(1512): 313–321. doi: 10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbs CL, Lagler KF. (1958) Fishes of the Great Lakes Region, revised edition. University of Michigan Press, Michigan, 213 pp. [Google Scholar]
- Hubert N, Hanner R, Holm E, Mandrak NE, Taylor E, Burridge M, Watkinson D, Dumont P, Curry A, Bentzen P, Zhang J, April J, Bernatchez L. (2008) Identifying Canadian Freshwater Fishes through DNA Barcodes. PLoS ONE 3: . doi: 10.1371/journal.pone.0002490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert N, Delrieu-Trottin E, Irisson J-O, Meyer C, Planes S. (2010) Identifying coral reef fish larvae through DNA barcoding: a test case with the families Acanthuridae and Holocentridae. Molecular Phylogenetics and Evolution 55: 1195–203. doi: 10.1016/j.ympev.2010.02.023 [DOI] [PubMed] [Google Scholar]
- Holmes BH, Steinke D, Ward RD. (2009) Identification of shark and ray fins using DNA barcoding. Fisheries Research 95: 280–88. doi: 10.1016/j.fishres.2008.09.036 [Google Scholar]
- Ida H, Sirimontaporn P, Monkolprasit S. (1994) Comparative morphology of the fishes of the family Ammodytidae, with a description of two new genera and two new species. Zoological Studies 33(4): 251–277. [Google Scholar]
- Kayser JL. (1961) Vergleichende Untersuchung über Vorstreckmechanismen der Oberkiefer bei Fischen. Zoologische Beiträge 7: 321–445. [Google Scholar]
- Keskin E, Atar HH. (2013) DNA barcoding commercially important fish species of Turkey. Molecular Ecology Resources 13: 788–97. doi: 10.1111/1755-0998.12120 [DOI] [PubMed] [Google Scholar]
- Kim JK, Watson W, Hyde J, Lo N, Kim JY, Kim S, Kim YS. (2010) Molecular identification of Ammodytes (PISCES, Ammodytidae) larvae, with ontogenetic evidence on separating populations. Genes & Genomics 32(5): 437–445. doi: 10.1007/s13258-010-0017-6 [Google Scholar]
- Knebelsberger T, Stöger I. (2012) DNA extraction, preservation, and amplification. In: Kress WJ, Erickson DL. (Eds) DNA barcodes: Methods and protocols, methods in molecular biology, vol. 858, Springer Science+Business Media, LLC; 2012, 311–338. doi: 10.1007/978-1-61779-591-6_14 [DOI] [PubMed] [Google Scholar]
- Knebelsberger T, Landi M, Neumann H, Kloppmann M, Sell AF, Campbell PD, Laakmann S, Raupach MJ, Carvalho GR, Costa FO. (2014) A reliable DNA barcode reference library for the identification of the North European shelf fish fauna. Molecular Ecology Resources 14: 1060–1071. doi: 10.1111/1755-0998.12238 [DOI] [PubMed] [Google Scholar]
- Knebelsberger T, Thiel R. (2014) Identification of gobies (Teleostei: Perciformes: Gobiidae) from the North and Baltic Seas combining morphological analysis and DNA barcoding. Zoological Journal of the Linnean Society 172: 831–845. doi: 10.1111/zoj.12189 [Google Scholar]
- Knebelsberger T, Dunz AR, Neumann D, Geiger MF. (2015) Molecular diversity of Germany’s freshwater fishes and lampreys assessed by DNA barcoding. Molecular ecology resources 15(3): 562–572. doi: 10.1111/1755-0998.12322 [DOI] [PubMed] [Google Scholar]
- Mabragaña E, Díaz de Astarloa JM, Hanner R, Zhang J, González Castro M. (2011) DNA barcoding identifies Argentine fishes from marine and brackish waters. PLoS ONE 6: . doi: 10.1371/journal.pone.0028655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer CP, Paulay G. (2005) DNA barcoding: error rates based on comprehensive sampling. PLoS Biology 3: . doi: 10.1371/journal.pbio.0030422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker MR, Denti D, Van Guelpen L, Kenchington E, Bentzen P. (2013) Barcoding Atlantic Canada’s commonly encountered marine fishes. Molecular Ecology Resources 13: 177–188. doi: 10.1111/1755-0998.12043 [DOI] [PubMed] [Google Scholar]
- Mitchell A, McCarthy E, Verspoor E. (1998) Discrimination of the North Atlantic lesser sandeels Ammodytes marinus, A. tobianus, A. dubius and Gymnammodytes semisquamatus by mitochondrial DNA restriction fragment patterns. Fisheries research 36(1): 61–65. doi: 10.1016/S0165-7836(98)00081-2 [Google Scholar]
- Naevdal G, Thorkildsen S. (2002) Genetic studies on species composition and population structure of sand eels (Genera: Ammodytes, Hyperoplus and Gymnammodytes) in Norwegian waters. Journal of Applied Ichthyology 18: 124–126. doi: 10.1046/j.1439-0426.2002.00310.x [Google Scholar]
- Nizinski MS, Collett BB, Washington BB. (1990) Separation of two species of sand lances, Ammodytes americanus and A. dubius, in the western north Atlantic. Fishery Bulletin, U.S. 88: 241–255. [Google Scholar]
- Orr JW, Wildes S, Kai Y, Raring N, Nakabo T, Katugin O, Guyon J. (2015) Systematics of North Pacific sand lances of the genus Ammodytes based on molecular and morphological evidence, with the description of a new species from Japan. Fishery Bulletin 113(2): 129–156. doi: 10.7755/FB.113.2.3 [Google Scholar]
- Pegg GG, Sinclair B, Briskey L, Aspden WJ. (2006) MtDNA barcode identification of fish larvae in the southern Great Barrier Reef, Australia. Scientia Marina 70: 7–12. doi: 10.3989/scimar.2006.70s27 [Google Scholar]
- Puckridge M, Andreakis N, Appleyard S, Ward RD. (2013) Cryptic diversity in flathead fishes (Scorpaeniformes: Platycephalidae) across the Indo-West Pacific uncovered by DNA barcoding. Molecular Ecology Resources 13: 32–42. doi: 10.1111/1755-0998.12022 [DOI] [PubMed] [Google Scholar]
- Randall JE, Ida H. (2014) Three new species of sand lances (Perciformes: Ammodytidae) from the southwest Indian Ocean. Journal of the Ocean Science Foundation 12: 1–11. [Google Scholar]
- Ratnasingham S, Hebert PD. (2007) BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Molecular ecology notes 7(3): 355–364. doi: 10.1111/j.1471-8286.2007.01678.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnasingham S, Hebert PD. (2013) A DNA-based registry for all animal species: The Barcode Index Number (BIN) System. PLoS ONE 8(7): . doi: 10.1371/journal.pone.0066213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reay PJ. (1986) Ammodytidae. In: Whitehead PJP, Beauchot ML, Hureau JC, Nielsen J, Tortonese E. (Eds) Fishes of the Northeastern Atlantic and Mediterranean. Unesco, Paris, 945–950. [Google Scholar]
- Sparholt H. (2015) 63. Sandeels (Ammodytidae). In: Heessen H, Daan N, Ellis JR. (Eds) Fish atlas of the Celtic Sea, North Sea, and Baltic Sea: Based on international research-vessel surveys. Wageningen Academic Publishers, Wageningen, 377–381. [Google Scholar]
- Saitou N, Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4: 406–25. [DOI] [PubMed] [Google Scholar]
- Sevilla RG, Diez A, Noren M, Mouchel O, Jerome M, Verrez-Bagnis V, et al. (2007) Primers and polymerase chain reaction conditions for DNA barcoding teleost fish based on the mitochondrial cytochrome b and nuclear rhodopsin genes. Molecular Ecology Notes 7: 730–734. doi: 10.1111/j.1471-8286.2007.01863.x [Google Scholar]
- Steinke D, Zemlak TS, Boutillier J, Hebert PDN. (2009) DNA barcoding of Pacific Canada’s fishes. Marine Biology 156: 2641–47. doi: 10.1007/s00227-009-1284-0 [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution 30: 2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teletchea F. (2009) Molecular identification methods of fish species: reassessment and possible applications. Reviews in Fish Biology and Fisheries 19(3): 265–293. doi: 10.1007/s11160-009-9107-4 [Google Scholar]
- Thiel R, Winkler HM, Böttcher U, Dänhardt A, Fricke R, George M, Kloppmann M, Schaarschmidt T, Ubl C, Vorberg R. (2013) Rote Liste und Gesamtartenliste der etablierten Fische und Neunaugen (Elasmobranchii, Actinopterygii & Petromyzontida) der marinen Gewässer Deutschlands. In: Becker N, Haupt H, Hofbauer N, Ludwig G, Nehring S. (Eds) Rote Liste gefährdeter Tiere, Pflanzen und Pilze Deutschlands, Band 2: Meeresorganismen. Naturschutz und Biologische Vielfalt; 70(2): 11–76. [Google Scholar]
- Turanov SV, Kartavtsev YP. (2014) The taxonomic composition and distribution of sand lances from the genus Ammodytes (Perciformes: Ammodytidae) in the North Pacific. Russian Journal of Marine Biology 40(6): 447–454. doi: 10.1134/S1063074014060212 [Google Scholar]
- Victor BC, Hanner R, Shivji M, Hyde J, Caldow C. (2009) Identification of the larval and juvenile stages of the Cubera Snapper, Lutjanus cyanopterus, using DNA barcoding. Zootaxa 2215: 24–36. [Google Scholar]
- Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. (2005) DNA barcoding Australia’s fish species. Philosophical transactions of the Royal Society of London, Series B 360: 1847–1857. doi: 10.1098/rstb.2005.1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RD, Bronwyn HH, William TW, Last PR. (2008a) DNA barcoding Australasian chondrichthyans: results and potential uses in conservation. Marine and Freshwater Research 59: 57–71. doi: 10.1071/MF07148 [Google Scholar]
- Ward RD, Costa F, Holmes B, Steinke D. (2008b) DNA barcoding of shared fish species from the North Atlantic and Australasia: minimal divergence for most taxa, but Zeus faber and Lepidopus caudatus each probably constitute two species. Aquatic Biology 3: 71–8. doi: 10.3354/ab00068 [Google Scholar]
- Wiecaszek B, Krzykawski S, Antoszek A. (2007) Meristic and morphometric characters of small sandeel, Ammodytes tobianus L. (Actinopterygii: Ammodytidae), from the Gulf of Gdansk, Baltic Sea. Acta Ichthyologica et Piscatoria 37(1): 37–45. doi: 10.3750/AIP2007.37.1.06 [Google Scholar]
- Zemlak TS, Ward RD, Connell AD, Holmes BH, Hebert PDN. (2009) DNA barcoding reveals overlooked marine fishes. Molecular Ecology Resources 9: 237–242. doi: 10.1111/j.1755-0998.2009.02649.x [DOI] [PubMed] [Google Scholar]
- Zhang J, Hanner R. (2012) Molecular approach to the identification of fish in the South China Sea. PLoS ONE 7: . doi: 10.1371/journal.pone.0030621 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Ralf Thiel, Thomas Knebelsberger
Data type: specimen data
Explanation note: Supplementary metadata for specimens used for both morphological and genetic analyses; Museum and Sample IDs are specimen identifiers, BOLD Process IDs are unique codes automatically generated for each record on BOLD, GenBank Accession NOs represent sequence identifiers.
Table S2
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Ralf Thiel, Thomas Knebelsberger
Data type: specimen data
Explanation note: Museum IDs and collection data for specimens of Ammodytes tobianus used for morphological analyses only.