Abstract
Friend Leukemia Virus Induced erythroleukemia-1 (Fli-1), an ETS transcription factor, was isolated a quarter century ago through a retrovirus mutagenesis screen. Fli-1 has since been recognized to play critical roles in normal development and homeostasis. For example, it transcriptionally regulates genes that drive normal hematopoiesis and vasculogenesis. Indeed, Fli-1 is one of 10 key regulators of hematopoietic stem/progenitor cell maintenance and differentiation. Aberrant expression of Fli-1 also underlies a number of virally induced leukemias, including Friend virus-induced erythroleukemia and various types of human cancers, and it is the target of chromosomal translocations in childhood Ewing’s sarcoma. Abnormal expression of Fli-1 is important in the aetiology of auto-immune diseases such as Systemic Lupus Erythematosus (SLE) and Systemic Sclerosis (SSc). These studies establish Fli-1 as a strong candidate for drug development. Despite difficulties in targeting transcription factors, recent studies identified small molecule inhibitors for Fli-1. Here we review past and ongoing research on Fli-1 with emphasis on its mechanistic function in autoimmune disease and malignant transformation. The significance of identifying Fli-1 inhibitors and their clinical applications for treatment of disease and cancer with deregulated Fli-1 expression are discussed.
Keywords: ETS genes, Fli-1, Hematopoiesis, Vasculogenesis, Cancer, Autoimmunity
Introduction
Fli-1 was first identified in 1990 by Ben-David Y. et al., as a common site for proviral integration in Friend Murine Leukemia Virus (F-MuLV)-Induced erythroleukemia.1 Induction of Fli-1 expression as a result of proviral integration in the vicinity of this gene was shown to be responsible for the development of erythroleukemia.2 This locus was also identified as a preferred proviral integration site in Cas-Br-E-induced leukemias.3 A year after its discovery, Fli-1 was found to be a target of translocation in a majority (85%) of Ewing Sarcoma, a paediatric cancer of bones. The EWS-Fli-1 translocation (t[11;22]) generates a fusion protein, EWS-Fli-1, with strong transforming activity.4 The chromosomal translocation creates a fusion of the 5′ trans-activation domain of EWS with the 3′ Ets domain of Fli-1. The role of ESW-Fli-1 in Ewing’s sarcoma has been extensively evaluated.5–8 In addition, Fli-1 overexpression was detected in various types of cancer and disease. Here we review the effect of Fli-1 on hematopoiesis, vasculogenesis and cancer as well as certain diseases associated with abnormal expression of this transcription factors (TF).
Fli-1 is a member of the ETS gene family of TFs.9 This gene family shares a DNA binding domain, the ETS domain, responsible for sequence-specific DNA recognition on target promoters. In vertebrates, there are 29 different ETS genes expressed in distinct tissues. Based on sequence homology of the ETS domain, this gene family is divided into 13 groups: ETS, ER71, GABP, PEA3, ERG, ERF, ELK, DETS4, ELF, ESE, TEL, YAN and SPI.9 The ERG group comprises Fli-1 and 3 other genes ERG, ERV and ETV.9
I. Fli-1 in hematopoietic Stem/progenitor maintenance and proliferation
Recent studies have identified TF networks that control hematopoietic stem cell self-renewal and differentiation into various mature blood lineages.10 A genome-wide computational analysis of complex binding patterns identified 10 TFs: SCL/TAL1, LYL1, LMO2, GATA2, RUNX1, MEIS1, PU.1, ERG, FLI-1 and GFI1B, which are important for hematopoietic stem/progenitor cells maintenance and differentiation.11 Three of these genes (PU.1, ERG and FLI-1) are ETS members. Among these, ERG is most closely related to FLI-1. While this study identified protein-protein interactions among SCL, LYL1, LMO2, GATA2, RUNX1, ERG and FLI-1, direct interaction leading to stable DNA binding was only observed between four (GATA2, RUNX1, SCL and ERG). Despite these results, Fli-1 and ERG, which recognize a similar DNA binding motif,12,13 were shown to be required for hematopoietic stem cell development and megakaryocytic lineage commitment.14–16 Indeed, using single and double knock-out mice for Fli-1 and Erg, this study demonstrated that disruption of both ETS genes in the hematopoietic linage significantly reduces the number of HSC/progenitors and mature megakaryocytes.16 As fli-1 and erg were derived through gene duplication of an ancestral gene,2 overlapping functions of these genes may reflect the enormous demand in a living organism to maintain HSC and produce distinct mature blood cells.
II. Role of Fli-1 in hematopoiesis and development of various blood cell lineages
a. Erythroid development
Fli-1 is expressed at high levels in various hematopoietic progenitors/mature cells and endothelial cells, and at lower levels in lung, heart and ovaries.2,17–19 In erythroblasts, Fli-1 expression is downregulated during (Epo)-induced differentiation of erythroblasts to mature erythroid cells.20 Downregulation of Fli-1 triggers erythroid progenitors to undergo differentiation and generate erythroid cells. Indeed, exogenous Fli-1 expression in erythroid progenitors (through transfection or viral infection) blocks differentiation and promotes uncontrolled cell proliferation.21 Deregulated/overexpressed Fli-1 in erythroid progenitors alters a cascade of events that switches Epo-induced differentiation to Epo-induced proliferation by activating Ras signaling.21 Importance of Fli-1 in erythroid differentiation was further established in vivo using chimeric and knock-out mice models of Fli-1 as described below.14,22,23
b. T-cell development
Expression of Fli-1 and several other ETS members is elevated in various stages of T-cell development.24 In T-cells, the fli-1 promoter is tightly regulated by several members of the ETS gene family. Its expression is upregulated by Ets1, Ets2, Fli-1 and Elf1 alone or in combination with GATA factors, but inhibited by Tel.25 The role of Fli-1 in T-cell development was demonstrated in Fli-1 knock-out mice, in which an N-terminal region was deleted by gene targeting (designated Fli-1ΔNT). These mutant mice are viable but exhibit thymic hypo-cellularity. This defect was not associated with a specific subpopulation of thymocytes or apoptosis, implicating Fli-1 in prethymic T-cell progenitors.19 This initial Fli-1 mutant mouse strain expressed a truncated Fli-1 protein owing to an internal translation-initiation site and alternative splicing around the neo cassette used for gene targeting. Two subsequent Fli-1 null mice succumbed to embryonic lethality at day 11.5–12.5, precluding analysis of T cell development.23,26 Interestingly, transgenic mice overexpressing Fli-1 in hematopoietic progenitors exhibit delayed double-negative (DN) to double-positive (DP) transition in vitro, and inhibition of CD4 differentiation and enhanced CD8 development both in vitro and in vivo.27 This eventually leads to a pre-T cell lymphoblastic leukaemia/lymphoma. Increased NOTCH1 expression was detected in these Fli-1 transgenic T cells and, accordingly, activating Notch1 mutations were later identified in all tumors.27
c. B-cell development
Fli-1 Transgenic overexpression of Fli-1 in various mouse tissues, with highest levels in thymus and spleen, induces high incidence of a progressive immunological renal disease and ultimately renal failure. The presence of hypergammaglobulinemia, splenomegaly, B-cell hyperplasia, accumulation of abnormal CD3+/B220+-T lymphoid cells and CD5+/B220+-B cells in peripheral lymphoid tissues as well as detection of various auto-antibodies in sera of these transgenic mice, implicate Fli-1 in B-cell proliferation and survival.28 A role of Fli-1 in B-cells was also observed in a recently generated Fli-1 knock-out mice engineered to lack the CTA domain (Fli-1ΔCTA), resulting in expression of mutant mRNA and protein.29,30 Fli-1ΔCTA homozygous mice are viable but exhibit partial perinatal lethality with reduced platelet numbers. These mutant mice have significantly fewer splenic follicular B cells and more transitional and marginal zone B cells relative to wild-type mice.30 Expression of genes implicated in B cell development including Igα, Pax-5, E2A and Egr-1 is reduced, while Id1 and Id2 expression increases.30 In addition, naive B-cells from Fli-1ΔCTA mice show reduced responsiveness to mitogens.31
d. Megakaryocyte development
Fli-1 is expressed at high levels in megakaryocytic progenitors and further induced during differentiation.32 A dominant phenotype of Fli-1ΔCTA mice is a significant reduction in the number of mature megakaryocytes and thrombocytopenia.29 As noted, a complete knock-out of Fli-1 results in embryonic lethality. This is associated with loss of vascular integrity leading to bleeding within the vascular plexus of the cerebral meninges and specific downregulation of Tek/Tie-2, the receptor for angiopoietin-1.26 These mice also exhibit a defect in megakaryopoiesis, a phenotype similar to Jacobsen or Paris-Trousseau syndrome, a relatively rare congenital disorder in which fli-1 is commonly deleted. Clinical abnormalities of this disease include growth and mental retardation, cardiac defects, dysmorphogenesis of the digits and face, pancytopenia and thrombocytopenia.33–35 Fli-1 in collaboration with GATA-1 was shown to regulate expression of several megakaryocytic specific genes including c-mpl, gpIIb, gpIV, gpIX, PF4, NF-E2, MafG, HOxa10 and Rab27B, further highlighting the importance of Fli-1 in megakaryopoiesis.29,36 Further evidence for a role of Fli-1 in megakaryopoiesis has recently been demonstrated using conditional knockout mice.14
A clinical relevance of Fli-1 in megakaryopoiesis was uncovered by Stockley et al., who detected mutations in Fli-1 and Runx1 in 6 families with excessive bleeding and defects in platelet development. The discovery of inactivating mutation in these TFs affecting megakaryocytopoeisis suggests a common genetic aetiology for defective platelet dense granule secretion and mild thrombocytopenia.37
e. Development of other blood cells
In addition to megakaryocytes, Fli-1 is involved in development of other myeloid derived cells. Fli-1ΔCTA mice exhibit significant reduction in the number of mature monocytes, marcrophages and dendritic cells.38 In addition, analysis of Fli-1−/−:+/+ chimeric mice, generated through morula-stage aggregation, revealed reduced neutrophillic granulocytes and monocytes as well as increased number of natural killer (NK) cells.22
III. The role of Fli-1 in vasculogenesis and angiogenesis
1. Vasculogenesis
Vascular endothelial cells (EC) form the luminal layer of blood vessels. A role of ETS genes in vasculogenesis and angiogenesis was initially suggested from studies in Xenopus laevis. Transgenic overexpression of Xenopous Fli-1 (Xi-fli-1) during early embryogenesis led to anomalies in head and heart development and delayed erythroid differentiation.39,40 Analysis of fli-1 mutant Zebrafish also revealed defects in the circulating system, indicating early evolutionary role for this TF in development of blood and vascular systems.41 Based on these observations, Lawson et al., generated transgenic zebrafish in which enhanced green florescence protein (eGFP) is placed under control of the fli-1 promoter. The fli-1/egfp transgenic zebrafish expressed eGFP in all blood vessels throughout embryogenesis, providing a unique model system to continuously monitor Fli-1 expression during vertebrate embryonic vasculature development in vivo.42
In a related study, Pham et al., discovered high expression of fli-1 and three other ets genes, erg, ets1 and etsrp, in the zebrafish vascular system.43 To uncover their roles in vasculogenesis, these genes were knocked out individually or in combination. Individual fli-1, erg, ets or etsrp knock-out mice showed partial inhibition of endothelial differentiation, whereas loss of all 4 genes blocked endothelial differentiation43, hence revealing extensive redundancy among these ets genes in endothelial cells. In another direction, genome-wide analysis of ETS genes in zebrafish revealed that erg, fli-1 and etsrp act cooperatively and are required for angiogenesis, possibly through direct regulation of the endothelial cell junction protein VE-cadherin.44 Xenopus and zebrafish embryos lacking fli-1 exhibit a blockade in hemangioblast development. In addition, Fli-1 induces expression of key hemangioblast genes including Scl/Tal1, Lmo2, Gata2, Etsrp and Flk1.45 These intriguing findings establish Fli-1 at the apex of a transcriptional network that regulates blood and endothelial development.
In a recent and related study, Li et al., demonstrated that Angiogenic factor with G patch and FHA domains 1 (Aggf1) acts upstream of Fli-1 in zebrafish. Knockdown of Aggf1 in Zebrafish resulted in a dramatic reduction in expression of the hemangioblast markers Fli-1, Etsrp, lmo2 and Scl, This result indicates that aggf1 is involved in differentiation of both hematopoietic and endothelial lineages, and that aggf1 acts upstream of fli-1 to specify hemangioblasts.46 Overall, these results corroborate previous observations on the presence of DNA-binding recognition sites for Fli-1 and other ETS factors on promoters of master developmental regulators of blood and endothelial cells.41,43,47–50
In mouse and human, ECs express high levels of Fli-1 and several other ETS genes including ERG, ETS1 and ELF-1.51 In human umbilical vein endothelial cells (HUVEC), expression of these ets genes is significantly induced after stimulation with Vascular Endothelial Growth Factor (VEGF).51 While Fli-1 overexpression in xenopus showed an endothelial-specific phenotype, such effect was not observed in Fli-1 transgenic mice. These mice however developed a B-cell phenotype associated with severe renal and auto-immune disease.28 The absence of a xenopus-like EC effect in these mice may be due to promoter selective activity, resulting in insufficient expression of Fli-1. As noted, Fli-1 null mice die at gestation day 11.5–12.5 due to massive cerebral hemorrhage and loss of vessel integrity.23,26 Whether this cerebral hemorrhage is the result of EC dysfunction remains to be determined.
The ETS genes ETV2, FLI-1 and ERG1 specify the differentiation of pluripotent stem cells into induced vascular endothelial cells (iVECs).44,52,53 EC specification by these ETS proteins requires cooperation with other TFs including SCL/TAL1, GATA2 and LMO2.52 These iVECs are unstable and drift toward nonvascular cell fate. In a recent study, human midgestation c-Kit(−) lineage-committed amniotic cells (ACs) were successfully reprogrammed into vascular endothelial cells (rAC-VECs) without transitioning through a pluripotent state. Transient ETV2 expression in ACs generated immature rAC-VECs, whereas co-expression with FLI-1/ERG1 endowed rAC-VECs with a vascular repertoire and morphology that matched mature ECs. Brief inhibition by transforming growth factorβ (TGFβ) functionalized VEGFR2 signaling, and increased specification of ACs into rAC-VECs.54 These results demonstrate that short-term ETV2 expression and TGFβ inhibition with constitutive ERG1/FLI-1 co-expression reprogram mature ACs into durable rAC-VECs that may be used to treat diverse vascular diseases.
2. Angiogenesis
During angiogenesis, new blood vessels arise from existing ones by budding out of endothelial cell capillaries. Fli-1 regulates this process by controlling expression of key angiogenic genes. For example, expression of Fli-1 as well as ERG and ELF-1 in ECs activates Endoglin during angiogenesis.47 Endoglin is upregulated in ECs to control cellular response to TGFβ. Fli-1/ERG in cooperation with GATA2 also regulates VE-Statin/egf17,55 which is expressed in embryonic and adult ECs and controls the recruitment and proliferation of smooth muscle cells and pericytes, two key events in the stabilization of newly formed capillaries during angiogenesis.56 In ECs, Fli-1 is directly regulated by ETS-1 implicating both ETS genes in endothelial cell fate.57
IV. Role of Fli-1 in immunity and auto-immune diseases
A. Systemic Lupus Erythematosus (SLE)
As mentioned above, high levels of Fli-1 expression is detected in B-cells, pre- and resting T-cells as well as in several other types of immune cells.24,27,30,58,59 Initial observations implicating Fli-1 in immunity and immune disorders came from analysis of transgenic mice overexpressing Fli-1. Transgenic expression of Fli-1 via the h2k promoter induced B-cell proliferation and auto-immunity with similar clinical symptoms observed in patients with SLE.28 This SLE-like phenotype was associated with an increased antibody production, proteinuria, renal pathology and mortality. In accordance, breeding of heterozygous mice carrying a Fli-1 null mutation into lupus MRL/lpr mice led to significantly reduced autoimmune disease.60 Likewise, breeding Fli-1+/− heterozygous mice with NZM2410 mice, another lupus murine model, also significantly increased survival, which was associated with reduced levels of auto-antibodies including anti-dsDNA and anti-glomerular basement antigen.61 The Fli-1+/−:NZM2410 mice also exhibited a reduced development of kidney disease (glomerulonephritis) associated with decreased monocyte chemo-attractant protein-1 (MCP-1) expression in endothelial cells in the kidney.62 In a related study, transplantation of bone marrow (BM) cells from Fli-1+/−:MRL/lpr mice into MRL/lpr recipients reduced the symptoms of autoimmunity such as auto-antibodies, proteinuria and renal disease and prolonged survival compared to control mice.63 These results demonstrate that Fli-1 over-expression in hematopoietic cells underlies autoimmunity in lupus.
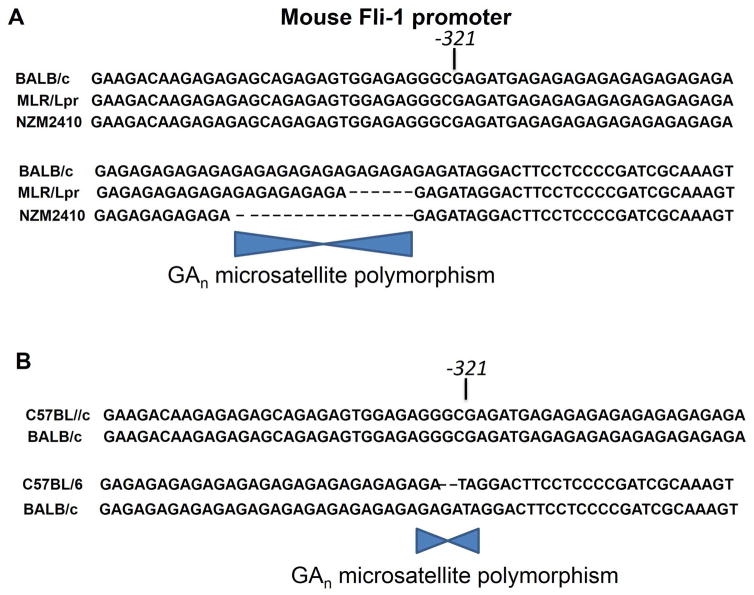
Consistent with these mouse studies, Fli-1 is expressed in peripheral blood mononucleated cells (PBMCs) and its over-expression correlates with severity of disease in Lupus patients.58 Interestingly, a polymorphic microsatellite consisting of GA repeats is shorter in MRL/lpr B and NZM2410 mice than in parental mice. Promoter analysis revealed that activity of the mouse fli-1 promoter inversely correlates with length of the GA repeats (Figure 2A).64 In addition, clinical data and genomic DNA analysis of SLE patients revealed that the human microsatellite exhibits a similar inverse correlation between GA repeats and promoter activity. The specific polymorphic microsatellite in the human fli-1 promoter was significantly more prevalent in SLE patients without nephritis and tended to be more prevalent in SLE patients with serenities.65 Together, these data suggest that a shorter microsatellite repeat in the fli-1 promoter may increase gene expression and contribute to the pathogenesis of lupus and other disease. Interestingly, Fli-1 has also been identified as the leishmania major susceptibility locus lmr2.66 In contrast to lupus with a longer GAn repeat deletion, a shorter GAn repeat in the fli-1 promoter was found in the resistant strain (C57Bl/6) versus susceptible strain (BALB/c) (Figure 2B). This one GA repeat deletion results in a lower Fli-1 expression and may be a major contributor to cutaneous leishmaniasis.
Figure 2.
(A) The fli-1 promoter contains a GAn microsatellite that is polymorphic in lupus prone mice. Shown is a sequence comparison of fli-1 promoter regions containing the GAn microsatellite from three mouse strains. The GAn microsatellite sequence, located in the 5′ end of exon 1 and begins at bp -321, is marked. Transcription of the Fli-1 gene negatively correlates with size of the GAn repeats (adopted from Ref. 64). (B) Effect of the GAn microsatellite repeats in the fli-1 promoter on susceptibility to Leishmenia. The location of GA polymorphism between C57BL/6 and BALB/c are marked (Adopted from Ref 66).
b. Systemic Sclerosis or Scleroderma (SSc)
SSc is a complex autoimmune connective tissue disease characterized by vasculopathy, activation of the immune system and wide-spread organ fibrosis.67 Although the onset of fibrosis in SSc typically correlates with production of auto-antibodies, whether the latter contributes to disease pathogenesis or simply serves as a marker of disease remains controversial. Moreover, the mechanism for auto-antibodies induction is largely unknown. A recent study suggests that epigenetic downregulation of Fli-1 in fibroblasts of SSC patients plays a pivotal role in the pathogenesis of disease.68 In accordance, targeted disruption of Fli-1 in fibroblasts or endothelial cells reproduced the histopathologic features of fibrosis and vasculopathy seen in SSc, respectively.69,70 Since Fli-1 regulates genes involved in vessel maturation and stabilization, reduced levels of Fli-1 in SSc vasculature could trigger unstable development of vessels that are prone to regression, as observed in the human disease.
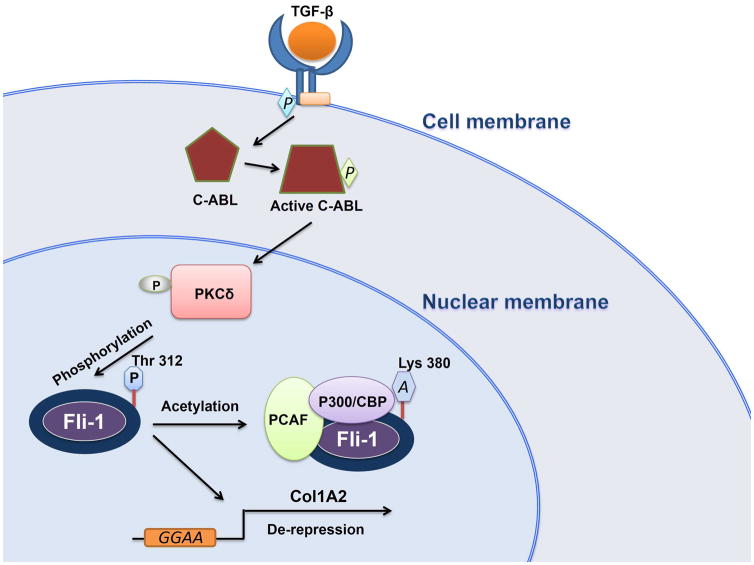
Interestingly, Fli-1 is phosphorylated on threonine 312 at high levels in SSc fibroblasts, and this correlates with induction of PKCδ nuclear localization by TGF-β and its downstream effector c-Abl, suggesting that a c-Abl/PKCδ/phospho-Fli-1 pathway is constitutively activated in these cells.71 Phosphorylation of Fli-1 facilitates its binding to the p300/CREB-binding protein-associated factor (PCAF), resulting in acetylation of lysine 312 (Fig. 3).71–73 Fli-1 acetylation in human fibroblasts decreases its stability and DNA binding, leading to de-repression of the alpha2(I) collagen (COL1A2) promoter and higher collagen production. Thus, as proposed by the authors, blocking the TGFβ/c-Abl/PKCδ/phospho-Fli-1 pathway may be an attractive approach for SSc therapy.71,74
Figure 3.
A model for SSc induction by Fli-1. In fibroblasts, the phosphorylation of C-Abl by TGF-β induces nuclear translocation of PKCδ. In the nucleus, PKCδ phosphorylates Fli-1 at threonine 312, which initiates acetylation of Fli-1 by PCAF/P300/CBP, resulting in dissociation of Fli-1 from the Col1A2 promoter and transcriptional de-repression. Induction of collagen expression through Fli-1 phosphorylation/acetylation is proposed as a mechanism for SSC.
The cardiotonic steroid hormone marinobufagenin (MBG), a digitalis-like substance in mam mals, is implicated in pathogenesis of experimental uremic cardiomyopathy, characterized by progressive cardiac fibrosis. A negative correlation between Fli-1 level and MBG has been observed in various cell lines and knock-out models of Fli-1.75–77 Moreover, a causal relationship between MBG-induced PKCδ modification results in phosphorylation and decreased nuclear Fli-1 levels, and increased collagen production.75 Interestingly, in contrast to TGF-β-mediated Fli-1 modification in SSc (Figure 3), MBG mediated PKCδ phosphorylation resulted in downregulation of Fli-1 in fibroblasts. Since MBG phosphorylates PKCδ through a signal cascade involving PLC,75 it is possible that this glycoside initiates Fli-1 modification in a different amino acid position. Future studies may determine the position of the MBG-mediated Fli-1 phosphoryation that induces its degradation. Overall, these findings point to the PKCδ/phospho-Fli-1 pathway as a potential therapeutic target for uremic cardiomyopathy and other conditions associated with excessive fibrosis.
V. Role of Fli-1 in malignant transformation
As noted, Fli-1 was first identified as a proto-oncogene activated by proviral integration in F-MuLV-induced erythroleukemias and later in Cas-Br-E-induced Non-T/B-cell and 10A1 stem cell like-induced leukemias.1–3,78 The human homologue of Fli-1 was also identified during this period.4,79,80 Its oncogenic role was functionally demonstrated in studies in which over-expression of Fli-1 in erythroblasts resulted in inhibition of differentiation.20,21,81–83 Overexpression of Fli-1 in erythroblasts also resulted in continuous activation of the Epo-Epo-R signal transduction pathway and uncontrolled proliferation.20,21,84 However, transgenic mice over-expressing Fli-1 under control of the h2k promoter developed B-cell proliferation and lupus-like symptom, not erythroleukemia.28 Lack of leukemia in these mice may be due to incorrect cell-of-origin of Fli-1 activation via the h2k promoter, to insufficient expression of the transgene or to genetic background/modifiers that suppress tumorigenesis. Notably, cre-mediated inducible expression of EWS-Fli-1 fusion protein under control of the ubiquitous promoter rosa26 led to erythroid/myeloid leukemia in mice.85
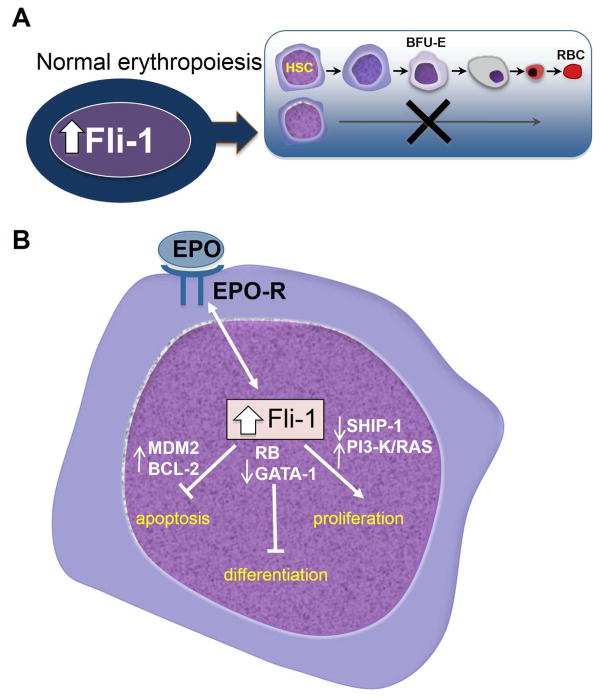
An important insight into the mechanism by which Fli-1 affects erythroid transformation was gained through identification of its target genes. Fli-1 acts in different contexts as a transcriptional activator or suppressor to regulate genes involved in cell proliferation, survival or differentiation (Figure 1).86–92 Thus, transcriptional downregulation of gata-1 and Retinoblastoma (rb) inhibits erythroid differentiation,20,86 and up-regulation of bcl-2 and mdm2 blocks apoptosis.20,87,88 Direct upregulation of MDM2 by Fli-1 destabilizes the anti-apoptotic protein p53, which plays a critical role in the initial transformation of erythroblasts by Friend virus.89 Downregulation of p53 by transcriptional upregulation of MDM2 through Fli-1 in erythroblasts also accelerates tumor progression by inducing genomic instability.89 Similarly, inhibition of p53 by the EWS-Fli-1 fusion protein through direct protein-protein interaction and/or activation of NOTCH signaling accelerates sarcoma progression despite retention of the p53 gene.90,91 Fli-1 overexpression in erythroblasts also increases tyrosine phosphorylation of the p85 subunit of PI3-Kinase and phosphorylation of Shc/Ras pathway, two critical regulators of cell survival and proliferation.21 At lower Fli-1 expression, Epo-EpoR activation in erythroid cells results in differentiation. This observation suggests that high and low levels of Fli-1 expression may induce a different phosphorylation of Epo-EPOR, leading to either erythroid differentiation or proliferation, respectively.21 Moreover, Fli-1 negatively regulates phosphatidyl-inositol polyphosphate 5-phosphatase (ship1) gene expression, leading to a higher phosphorylation of AKT/PKB by PI3K and erythroid proliferation.92 While the mechanism responsible for RAS activation has not been established, this is likely mediated by the Epo-Epo-R signal transduction pathway, which is constitutively activated during F-MuLV-induced erythroleukemia transformation.21 In addition to the above downstream effectors of EPO-EPOR signaling pathway, Fli-1 also positively regulates ribosomal gene expression and ribosome biogenesis, which may play roles in erythroleukemogenesis.93
Figure 1.
(A) Fli-1 overexpression in erythroblasts induces erythroleukemias by inhibiting differentiation. (B) Fli-1 over-expression in erythroblasts results in a switch from Epo-induced differentiation to proliferation. Fli-1 regulates this process through its target genes, which affect apoptosis, differentiation and proliferation. HSC: Hematopoietic Stem Cells; BFU-E: Burst Forming Units-Erythroid; RBC: Red Blood Cells.
Importantly, high Fli-1 expression is observed in a subset of human erythroleukemia, mostly in the absence of genomic amplification.94 In addition, shRNA-mediated knock-down of Fli-1 in murine and human erythroleukemic cells suppresses cell proliferation, induces differentiation and accelerates apoptosis associated with decreased expression of its target genes including gata-1 and Bcl-2.95 A summary of target genes regulated by Fli-1 is shown in Table I.
Table I.
A list of Fli-1 target genes, suppression/activation effect on these genes, their target cells and mechanistic function with appropriate references.
| Fli-1 target gene | Effect | Target cells | Function | Ref |
|---|---|---|---|---|
| MDM2 | Activation | Erythroid | Cell cycle, apoptosis, genomic instability, cancer | 88 |
| BCL-2 | Activation | Erythroid | Apoptosis, cancer | 87 |
| GATA-1 | Suppression | Erythroid | Cell cycle, cancer | 86 |
| RB | Suppression | Erythroid | Cell cycle, apoptosis, cancer | 20 |
| SHIP-1 | Suppression | Erythroid | Phosphatase, signal transduction, cancer | 92 |
| Several ribosomal genes | Activation | Erythroid | Ribosomal biogenesis, erythroid transformation | 93 |
| NOTCH1 | Activation | T-cells | Signal transduction, cancer | 27 |
| Fli-1 | Activation | T-cells | Transcriptional regulation, hematopoiesis, cancer, vasculogenesis | 25 |
| SCL/TAL1 | Activation | Hematopoietic cells | Hematopoiesis | 49,50 |
| c-mpl, gpIIb, gpIV, gpIX, PF4, NF-E2/p45, MafG, HOxa10 and Rab27B | Activation | Megakaryocytes | Megakaryopoiesis | 29,36 |
| MCP-1 | Activation | Endothelial cells | Chemo-attractant factor | 62 |
| Tek/Tie-2 | Activation | Endothelial cells | Angiogenesis, vasculogenesis | 26 |
| VE-cadherin | Activation | Endothelial cells | Cell junction protein interaction, angiogenesis | 44 |
| VE-Statin/egf17 | Activation | Endothelial cells | Controls the recruitment and proliferation of smooth muscle cells and pericytes | 56 |
| VEGF-A | Activation | Endothelial cells | Angiogenesis, vasculogenesis | 109 |
| COL1A2 | Suppression | Fibroblasts | Involved in construction of type I collagen | 72,73 |
In addition to erythroleukemia, deregulated Fli-1 can induce other hematological malignancies. For examples, retroviral transduction of Fli-1 into murine T-cell progenitors disrupts normal development and induces pre-T-cell lymphoblastic lymphoma.27 Transgenic expression of Fli-1 results in a lupus like disease associated with a significant increase in B-cell proliferation and autoimmunity.28 Abnormal Fli-1 expression is also associated with progression of acute myeloid leukemia (AML).96 In this study, both high and low expression of Fli-1 was associated with AML progression. In support of these observations, a recent study identified fli-1 gene amplification within a 11q23–25 amplicon in several cases of AML and diffuse large B-cell lymphomas.97 In leukemia, the ETS protein Tel, but not the translocated protein Tel-AML, binds and inhibits Fli-1 transcriptional activity.98,99 It appears that Tel-translocation contributes to the de-repression of Fli-1 activity by abrogating the normally inhibiting function of the untranslocated Tel allele. Since Tel translocation occurs widely in myeloid/lymphoid tumors, Fli-1 could act as a major driver of various hematological tumors. Moreover, Fli-1 and its closely related ERG protein facilitate binding of the fusion protein AML1-ETO to its cognate DNA, thus highlighting the dual importance of these ETS proteins in leukemias harboring t(8;21) translocation.100
In addition to hematological malignancies, high Fli-1 expression promotes the development of divergent types of solid tumors. The best example is the above mentioned EWS-FLI-1 translocation (t[11;22]) in Ewing sarcoma and certain neuroectodermal neoplasms.4,101 Furthermore, while Fli-1 expression is normally limited to very few lineages including hematopoietic, vascularogenesis and nervous system,24,27,30,47,48,55–59,69–74 its expression is readily detected by immunostaining in various benign and malignant tumors. Analysis of 4323 tumours revealed that FLI-1 is expressed at high percentage in 46/62 Ewing’s sarcoma/primitive neuroectodermal tumors (EWS/PNETs), 2/3 olfactory neuroblastomas, 7/102 small cell carcinomas of the lung, 10/34 Merkel cell carcinomas (MCCs), 19/132 non-Hodgkin’s lymphomas, 9/29 medullar carcinomas of the breast, 2/3 desmoplastic small round cell tumors (DSRCTs), and 53/74 benign and malignant vascular tumours.102
FLI-1 is highly expressed in several triple-negative breast cancer cell lines (MDA-MB231, MDA-MB436, BT-549 and HCC1395). Moreover, over-expression of FLI-1 in the luminal breast cancer cell line, MCF7, which expresses a negligible level of this TF, suppressed apoptosis upon serum depletion. Inhibition of apoptosis was associated with upregulation of Bcl-2, a direct target of Fli-1.103 Similar to breast cancer, FLI-1 expression is also detected in several melanoma cell lines and in formalin-fixed/paraffin-embedded tissue sections from 97 melanomas including 69 cases of primary and 28 metastatic melanomas by immunohistochemistry. Higher FLI-1 expression was detected in metastatic than in primary melanoma tumors, and this was positively correlated with high Ki67 index and ulceration of the primary tumor.104 Fli-1 has been found fused to the solute carrier family 45 member 3 (SLC45A3) gene in a small subset of prostate cancer.105 Together, these studies demonstrate high expression as well as translocations involving Fli-1 in a wide range of hematopoietic and solid tumors. Additional analysis is needed to determine whether these malignancies are driven by FLI-1 and whether tumors are addicted to continuous expression of this ETS factor.
VI. Role of Fli-1 in tumor angiogenesis
Angiogenesis is required for proliferating tumor cells to receive nutrients and metabolites. Tumor cells regulate angiogenesis through secretion of factors that stimulate neo-vascularisation. As described above, endothelial cells express high levels of Fli-1.51 This has prompted the use Fli-1 as a marker for tumor angiogenesis. High Fli-1 expression is also detected in the majority of benign and malignant vascular tumours.106–108 The EWS-Fli-1 fusion protein directly activates vascular endothelial growth factor-A (VEGF-A), leading to increased angiogenesis and malignant progression.109 High levels of VEGF were also detected in the tumor microenvironment of Fli-1 overexpressing erythroleukemias and shown to play a critical role in tumor initiation.110
VII. Therapeutic aspects of Fli-1
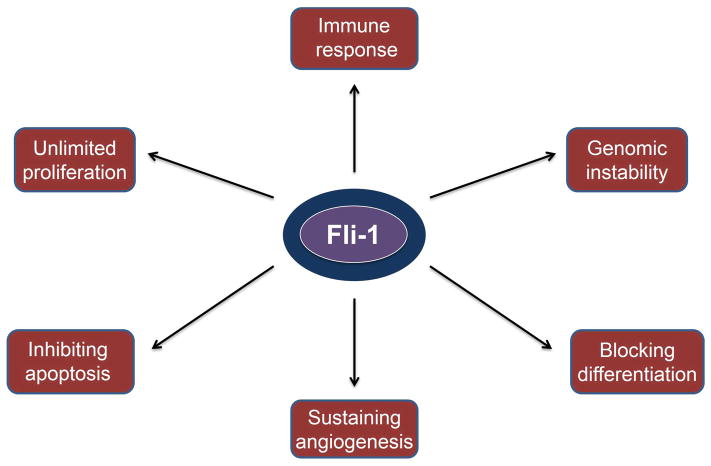
As described above, Fli-1 regulates genes and pathways associated with hallmarks of cancer initiation and progression including sustained proliferation, angiogenesis, genomic instability, inhibition of apoptosis and differentiation; its abnormal expression or translocation induces diverse tumor types (Figure 4). In addition, FLI-1 controls the fate of hematopoietic stem cells and progenitors and is directly involved in several auto-immune diseases. Some immune cells with over-expressed Fli-1 (unpublished results) have been found to act as microenvironmental niche to support expansion of leukemic cells in vivo and in vitro.110 Therefore, Fli-1 represents an attractive therapeutic target. Toward this end, several inhibitors of DNA- or RNA-binding activity of the EWS-Fli-1 fusion protein were developed.111–114 The effectiveness of these compounds for the treatment of Ewing’s sarcoma and other malignancies with a EWS-Fli-1 translocation is yet to be determined in randomized clinical trials. In a recent study, additional inhibitors of EWS-Fli-1 have been identified from libraries of small molecules/compounds with known biological activities. In addition to anti-EWS-Fli-1 activity, these drugs possessed strong anti-Fli-1 activity, resulting in suppression of proliferation of human and murine erythroleukemia cell lines at low drug concentrations (nM - low μM range). Some of these Fli-1 inhibitors could suppress progression of F-MuLV-induced erythroleukemias in vivo, in which leukemic cells acquire activated Fli-1 through retroviral insertional mutagenesis.115 Many of these Fli-1 inhibitors have known biological activities and are presently used to treat of cancers. Two of these drugs, etoposide and dactinomycin, are topoisomerase inhibitors used to treat diverged cancer types including Ewing’s sarcoma.116–118 These chemotherapeutic agents likely kill tumor cells both by inducing DNA damage and by suppressing Fli-1 expression. About 30% of anti-Fli-1 compounds were from cardiac glycoside family of drugs (similar to MBG, see section II.b), which downregulates Fli-1 expression.115 Interestingly, one of the compounds (Calcimycin), a calcium ionophore, inhibits Fli-1 DNA-binding by inducing a Fli-1 modification that is associated with downregulation of PKCδ, an interesting observation that warrants further investigation.
Figure 4.
Aberrant Fli-1 expression enhances hallmarks of malignant transformation and immune response.
A recent study has identified microRNA-145 as a potent inhibitor of both Fli-1 and EWS-Fli-1 protein expression.119,120 Development of a delivery system to introduce mir-145 into cancer cells over-expressing this TF may therefore be therapeutic. As described above, Fli-1 plays a critical role in survival and proliferation of angiogenic cells. Thus, Fli-1 inhibitors may not only inhibit proliferation of Fli-1 over-expressing tumors, but also cut off their blood supply. Table II lists anti-Fli-1 compounds/molecules identified to date with their respected mechanism.
Table II.
List of small molecules/compounds with anti-Fli-1/EWS-Fli-1 activity and their mechanism of action.
| Name | Function | Fli-1 inhibition | EWS/Fli-1 inhibition | Mechanism of Fli-1 inhibition | Ref |
|---|---|---|---|---|---|
| Calcimycin (A23187) | Calcium Ionophore | Yes | Unknown | Inhibition of Fli-1 phosphorylation | 115 |
| Peruvosid | Cardiac glycoside (Na+, K− ATPase) | Yes | Unknown | Transcriptional downregulation of Fli-1 | 115 |
| Camptothecin | Topoisomerase inhibitor | Yes | Unknown | Post-transcriptional downregulation of Fli-1 | 115 |
| Etoposide | Chemotherapeutic drug | Yes | Yes | Topoisomerase II inhibition resulting Fli-1 downregulation | 115 |
| dactinomycin | Chemotherapeutic drug | Yes | Yes | topoisomerase II inhibition resulting Fli-1 downregulation | 115 |
| doxorubicin | Chemotherapeutic drug | Yes | Yes | Unkonwn | 111 |
| cycloheximide | Chemotherapeutic drug | Yes | Unknown | Unkonwn | 115 |
| midostaurin (PKC412) | Kinase inhibitor | Unknown | Yes | Induction of apoptosis by downregulating ESW-Fli-1 target genes | 111 |
| Ecteinascidin 743 (ET-743) | Marine derived anti-tumor agent | Unknown | Yes | Downregulation of ESW-Fli-1 target genes | 112 |
| YK-4–279 | RNA helicase A (RHA) inhibitor | Unknown | Yes | Inhibits function by blocking RHA binding to EWS-FLI-1 | 113 |
| Mithramycin | A tricyclic pentaglycosidic antibiotic with anti-cancer activity | Unknown | Yes | Downregulation of ESW-Fli-1 target genes | 114 |
| Ciprofloxacin | Fluoroquinolone antibiotic with anti-cancer activity | Yes | Unknown | Transcriptional downregulation of Fli-1 | 121 |
We expect that useful Fli-1 inhibitors (or agonists) to be effective only if they specifically target this TF but not other related ETS members. Thus, the challenge in identifying Fli-1 modulators is compounded by the general difficulty of identifying antagonists for a transcription factor and the need for specificity. For example, inhibitors that interfere with Fli-1 binding to DNA elements on promoters may simultaneously inhibit other family members and therefore may have inadvertent adverse effects. Thus, putative inhibitors should be analyzed side-by-side with specific shRNA/RNAi for Fli-1 to determine if they phenocopy the effect of knocking down this TF.
In addition to cancer, Fli-1 inhibitors may also be beneficial in the treatment of autoimmune diseases such as SLE, which express high levels of FLI-1. Fli-1 also plays a critical role in maintenance of hematopoietic stem/progenitors and their differentiation to various mature blood cells. Indeed, a recent study has demonstrated that conditional Fli-1 loss in several myeloid lineages affects commitment decisions leading to a drastic decrease in mature megakaryocytes, proliferation arrest and inhibition of terminal erythrocytic differentiation.14 It is therefore possible that syndromes such as red cell anemia and thrombocytopenia can also be treated with Fli-1 inhibiting compounds. Overall, generation of potent anti-Fli-1 inhibitors with good pharmacokinetic properties may revolutionize treatment of multiple diseases and cancers overexpressing Fli-1.
While these diseases may benefit from Fli-1 inhibitors, others such as SSc, which are characterized by low Fli-1 expression, may be ameliorated by treatment with small molecules capable of increasing/activating endogenous Fli-1. Indeed, a recent study has identified the fluoroquinolone antibiotic ciproflovacin as an antifibrotic compound in SSc. Ciproflovacin was shown to affect SSc through downregulation of Dnmt1 and upregulation of Fli-1.121 Thus, both agonists and antagonists of Fli-1 may provide therapeutic benefits for the treatment of immune-diseases and cancers, respectively.
Summary
In the past two and half decades, data from over 1000 publications establish Fli-1 as a key regulator of normal development and malignant transformation. These studies reveal an essential role of Fli-1 in stem cell maintenance and differentiation, hematopoiesis, vasculogenesis and angiogenesis. Abnormal expression of Fli-1 was shown to drive various diseases, hematological malignancies and solid tumors. Thus, Fli-1 is emerging as a new and exciting therapeutic target. The development of small molecules targeting different aspects of Fli-1 expression or activity could potentially impact treatment of auto-immune diseases and cancers driven by abnormal expression of this ETS factor.
Acknowledgments
This work was supported by Canadian Institute of Health Research to YBD (MOP-110952), Canadian Breast Cancer Foundation to EZ, the Science and Technology Department of Guizhou Province innovation and project grants (6012, 4001) to YBD.
Footnotes
Conflict of interest: These authors declare no conflict of interest.
References
- 1.Ben-David Y, Giddens EB, Bernstein A. Identification and mapping of a common proviral integration site Fli-1 in erythroleukemia cells induced by Friend murine leukemia virus. Proc Natl Acad Sci U S A. 1990;87:1332–1336. doi: 10.1073/pnas.87.4.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-David Y, Giddens EB, Letwin K, Bernstein A. Erythroleukemia induction by Friend murine leukemia virus: insertional activation of a new member of the ets gene family, Fli-1, closely linked to c-ets-1. Genes Dev. 1991;5:908–918. doi: 10.1101/gad.5.6.908. [DOI] [PubMed] [Google Scholar]
- 3.Bergeron D, Poliquin L, Houde J, Barbeau B, Rassart E. Analysis of proviruses integrated in Fli-1 and Evi-1 regions in Cas-Br-E MuLV-induced non-T-, non-B-cell leukemias. Virology. 1992;191:661–669. doi: 10.1016/0042-6822(92)90241-g. [DOI] [PubMed] [Google Scholar]
- 4.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 5.Ordóñez JL, Osuna D, Herrero D, de Alava E, Madoz-Gúrpide J, Ordóñez JL, et al. Advances in Ewing’s sarcoma research: where are we now and what lies ahead? Cancer Res. 2009;69:7140–7150. doi: 10.1158/0008-5472.CAN-08-4041. [DOI] [PubMed] [Google Scholar]
- 6.Leavey PJ, Collier AB. Ewing sarcoma: prognostic criteria, outcomes and future treatment. Expert Rev Anticancer Ther. 2008;8:617–24. doi: 10.1586/14737140.8.4.617. [DOI] [PubMed] [Google Scholar]
- 7.Riggi N, Stamenkovic I. The Biology of Ewing sarcoma. Cancer Lett. 2007;254:1–10. doi: 10.1016/j.canlet.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 8.Owen LA, Lessnick SL. Identification of target genes in their native cellular context: an analysis of EWS/FLI in Ewing’s sarcoma. Cell Cycle. 2006;5:2049–2053. doi: 10.4161/cc.5.18.3213. [DOI] [PubMed] [Google Scholar]
- 9.Laudet V, Hänni C, Stéhelin D, Duterque-Coquillaud M. Molecular phylogeny of the ETS gene family. Oncogene. 1999;18:1351–1359. doi: 10.1038/sj.onc.1202444. [DOI] [PubMed] [Google Scholar]
- 10.Schütte J, Moignard V, Göttgens B. Establishing the stem cell state: insights from regulatory network analysis of blood stem cell development. Wiley Interdiscip Rev Syst Biol Med. 2012 May-Jun;4:285–295. doi: 10.1002/wsbm.1163. [DOI] [PubMed] [Google Scholar]
- 11.Wilson NK, Foster SD, Wang X, Knezevic K, Schütte J, Kaimakis P, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 12.Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camões MJ, Paulo P, Ribeiro FR, Barros-Silva JD, Almeida M, Costa VL, et al. Potential downstream target genes of aberrant ETS transcription factors are differentially affected in Ewing’s sarcoma and prostate carcinoma. PLoS One. 2012;7:e49819. doi: 10.1371/journal.pone.0049819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Starck J, Weiss-Gayet M, Gonnet C, Guyot B, Vicat JM, Morlé F. Inducible Fli-1 gene deletion in adult mice modifies several myeloid lineage commitment decisions and accelerates proliferation arrest and terminal erythrocytic differentiation. Blood. 2010;116:4795–805. doi: 10.1182/blood-2010-02-270405. [DOI] [PubMed] [Google Scholar]
- 15.Carmichael CL, Metcalf D, Henley KJ, Kruse EA, Di Rago L, Mifsud S, et al. Hematopoietic overexpression of the transcription factor Erg induces lymphoid and erythro-megakaryocytic leukemia. Proc Natl Acad Sci U S A. 2012;109:15437–15442. doi: 10.1073/pnas.1213454109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kruse EA, Loughran SJ, Baldwin TM, Josefsson EC, Ellis S, Watson DK, et al. Dual requirement for the ETS transcription factors Fli-1 and Erg in hematopoietic stem cells and the megakaryocyte lineage. Proc Natl Acad Sci U S A. 2009;106:13814–13819. doi: 10.1073/pnas.0906556106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pusztaszeri MP, Seelentag W, Bosman FT. Immunohistochemical expression of endothelial markers CD31, CD34, von Willebrand factor, and Fli-1 in normal human tissues. J Histochem Cytochem. 2006;54:385–395. doi: 10.1369/jhc.4A6514.2005. [DOI] [PubMed] [Google Scholar]
- 18.Hewett PW, Nishi K, Daft EL, Clifford MJ. Selective expression of erg isoforms in human endothelial cells. Int J Biochem Cell Biol. 2001;33:347–355. doi: 10.1016/s1357-2725(01)00022-x. [DOI] [PubMed] [Google Scholar]
- 19.Mélet F, Motro B, Rossi DJ, Zhang L, Bernstein A. Generation of a novel Fli-1 protein by gene targeting leads to a defect in thymus development and a delay in Friend virus-induced erythroleukemia. Mol Cell Biol. 1996;16:2708–2718. doi: 10.1128/mcb.16.6.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamir A, Howard J, Higgins RR, Li YJ, Berger L, Zacksenhaus E, et al. Fli-1, an Ets-related transcription factor, regulates erythropoietin-induced erythroid proliferation and differentiation: evidence for direct transcriptional repression of the Rb gene during differentiation. Mol Cell Biol. 1999;19:4452–4464. doi: 10.1128/mcb.19.6.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zochodne B, Truong AH, Stetler K, Higgins RR, Howard J, Dumont D, et al. Epo regulates erythroid proliferation and differentiation through distinct signaling pathways: implication for erythropoiesis and Friend virus-induced erythroleukemia. Oncogene. 2000;19:2296–2304. doi: 10.1038/sj.onc.1203590. [DOI] [PubMed] [Google Scholar]
- 22.Masuya M, Moussa O, Abe T, Deguchi T, Higuchi T, Ebihara Y, et al. Dysregulation of granulocyte, erythrocyte, and NK cell lineages in Fli-1 gene-targeted mice. Blood. 2005;105:95–102. doi: 10.1182/blood-2003-12-4345. [DOI] [PubMed] [Google Scholar]
- 23.Spyropoulos DD, Pharr PN, Lavenburg KR, Jackers P, Papas TS, Ogawa M, et al. Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol Cell Biol. 2000;20:5643–5652. doi: 10.1128/mcb.20.15.5643-5652.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson MK, Hernandez-Hoyos G, Diamond RA, Rothenberg EV. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development. 1999;126:3131–3148. doi: 10.1242/dev.126.14.3131. [DOI] [PubMed] [Google Scholar]
- 25.Svenson JL, Chike-Harris K, Amria MY, Nowling TK. The mouse and human Fli1 genes are similarly regulated by Ets factors in T cells. Genes Immun. 2010;11:161–172. doi: 10.1038/gene.2009.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hart A, Melet F, Grossfeld P, Chien K, Jones C, Tunnacliffe A, et al. Fli-1 is required for murine vascular and megakaryocytic development and is hemizygously deleted in patients with thrombocytopenia. Immunity. 2000;13:167–177. doi: 10.1016/s1074-7613(00)00017-0. [DOI] [PubMed] [Google Scholar]
- 27.Smeets MF, Chan AC, Dagger S, Bradley CK, Wei A, Izon DJ. Fli-1 overexpression in hematopoietic progenitors deregulates T cell development and induces pre-T cell lymphoblastic leukaemia/lymphoma. PLoS One. 2013;8:e62346. doi: 10.1371/journal.pone.0062346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Eddy A, Teng YT, Fritzler M, Kluppel M, Melet F, et al. An immunological renal disease in transgenic mice that overexpress Fli-1, a member of the ets family of transcription factor genes. Mol Cell Biol. 1995;15:6961–6970. doi: 10.1128/mcb.15.12.6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moussa O, LaRue AC, Abangan RS, Jr, Williams CR, Zhang XK, Masuya M, et al. Thrombocytopenia in mice lacking the carboxy-terminal regulatory domain of the Ets transcription factor Fli1. Mol Cell Biol. 2010;30:5194–5206. doi: 10.1128/MCB.01112-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang XK, Moussa O, LaRue A, Bradshaw S, Molano I, Spyropoulos DD, et al. The transcription factor Fli-1 modulates marginal zone and follicular B cell development in mice. J Immunol. 2008;181:1644–1654. doi: 10.4049/jimmunol.181.3.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bradshaw S, Zheng WJ, Tsoi LC, Gilkeson G, Zhang XK. A role for Fli-1 in B cell proliferation: implications for SLE pathogenesis. Clin Immunol. 2008;129:19–30. doi: 10.1016/j.clim.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okada Y, Nobori H, Shimizu M, Watanabe M, Yonekura M, Nakai T, et al. Multiple ETS family proteins regulate PF4 gene expression by binding to the same ETS binding site. PLoS One. 2011;6:e24837. doi: 10.1371/journal.pone.0024837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krishnamurti L, Neglia JP, Nagarajan R, Berry SA, Lohr J, Hirsch B, et al. Paris-Trousseau syndrome platelets in a child with Jacobsen’s syndrome. Am J Hematol. 2001;66:295–299. doi: 10.1002/ajh.1061. [DOI] [PubMed] [Google Scholar]
- 34.Favier R, Jondeau K, Boutard P, Grossfeld P, Reinert P, Jones C, et al. Paris-Trousseau syndrome: clinical, hematological, molecular data of ten new cases. Thromb Haemost. 2003;90:893–897. doi: 10.1160/TH03-02-0120. [DOI] [PubMed] [Google Scholar]
- 35.Wenger SL, Grossfeld PD, Siu BL, Coad JE, Keller FG, Hummel M. Molecular characterization of an 11q interstitial deletion in a patient with the clinical features of Jacobsen syndrome. Am J Med Genet A. 2006;140:704–708. doi: 10.1002/ajmg.a.31146. [DOI] [PubMed] [Google Scholar]
- 36.Gosiengfiao Y, Horvat R, Thompson A. Transcription factors GATA-1 and Fli-1 regulate human HOXA10 expression in megakaryocytic cells. DNA Cell Biol. 2007;26:577–587. doi: 10.1089/dna.2007.0575. [DOI] [PubMed] [Google Scholar]
- 37.Stockley J, Morgan NV, Bem D, Lowe GC, Lordkipanidzé M, Dawood B, et al. Enrichment of FLI1 and RUNX1 mutations in families with excessive bleeding and platelet dense granule secretion defects. Blood. 2013;122:4090–4093. doi: 10.1182/blood-2013-06-506873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki E, Williams S, Sato S, Gilkeson G, Watson DK, Zhang XK. The transcription factor Fli-1 regulates monocyte, macrophage and dendritic cell development in mice. Immunology. 2013;139:318–327. doi: 10.1111/imm.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Remy P, Sénan F, Meyer D, Mager AM, Hindelang C. Overexpression of the Xenopus Xl-fli gene during early embryogenesis leads to anomalies in head and heart development and erythroid differentiation. Int J Dev Biol. 1996;40:577–589. [PubMed] [Google Scholar]
- 40.Meyer D, Wolff CM, Stiegler P, Sénan F, Befort N, Befort JJ, Remy P. Xl-fli, the Xenopus homologue of the fli-1 gene, is expressed during embryogenesis in a restricted pattern evocative of neural crest cell distribution. Mech Dev. 1993;44:109–121. doi: 10.1016/0925-4773(93)90061-2. [DOI] [PubMed] [Google Scholar]
- 41.Brown LA, Rodaway AR, Schilling TF, Jowett T, Ingham PW, Patient RK, Sharrocks AD. Insights into early vasculogenesis revealed by expression of the ETS-domain transcription factor Fli-1 in wild-type and mutant zebrafish embryos. Mech Dev. 2000;90:237–252. doi: 10.1016/s0925-4773(99)00256-7. [DOI] [PubMed] [Google Scholar]
- 42.Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- 43.Pham VN, Lawson ND, Mugford JW, Dye L, Castranova D, Lo B, Weinstein BM. Combinatorial function of ETS transcription factors in the developing vasculature. Dev Biol. 2007;303:772–783. doi: 10.1016/j.ydbio.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu F, Patient R. Genome-wide analysis of the zebrafish ETS family identifies three genes required for hemangioblast differentiation or angiogenesis. Circ Res. 2008;103:1147–1154. doi: 10.1161/CIRCRESAHA.108.179713. [DOI] [PubMed] [Google Scholar]
- 45.Liu F, Walmsley M, Rodaway A, Patient R. Fli1 acts at the top of the transcriptional network driving blood and endothelial development. Curr Biol. 2008;18:1234–1240. doi: 10.1016/j.cub.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 46.Li L, Chen D, Li J, Wang X, Wang N, Xu C, Wang QK. Aggf1 acts at the top of the genetic regulatory hierarchy in specification of hemangioblasts in zebrafish. Blood. 2013 Nov 25; doi: 10.1182/blood-2013-07-514612. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pimanda JE, Chan WY, Donaldson IJ, Bowen M, Green AR, Göttgens B. Endoglin expression in the endothelium is regulated by Fli-1, Erg, and Elf-1 acting on the promoter and a -8-kb enhancer. Blood. 2006;107:4737–4745. doi: 10.1182/blood-2005-12-4929. [DOI] [PubMed] [Google Scholar]
- 48.Landry JR, Kinston S, Knezevic K, Donaldson IJ, Green AR, Göttgens B. Fli1, Elf1, and Ets1 regulate the proximal promoter of the LMO2 gene in endothelial cells. Blood. 2005;106:2680–2687. doi: 10.1182/blood-2004-12-4755. [DOI] [PubMed] [Google Scholar]
- 49.Göttgens B, Broccardo C, Sanchez MJ, Deveaux S, Murphy G, Göthert JR, et al. The scl +18/19 stem cell enhancer is not required for hematopoiesis: identification of a 5′ bifunctional hematopoietic-endothelial enhancer bound by Fli-1 and Elf-1. Mol Cell Biol. 2004;24:1870–1883. doi: 10.1128/MCB.24.5.1870-1883.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Göttgens B, Nastos A, Kinston S, Piltz S, Delabesse EC, Stanley M, et al. Establishing the transcriptional programme for blood: the SCL stem cell enhancer is regulated by a multiprotein complex containing Ets and GATA factors. EMBO J. 2002;21:3039–3050. doi: 10.1093/emboj/cdf286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heo SH, Choi YJ, Ryoo HM, Cho JY. Expression profiling of ETS and MMP factors in VEGF-activated endothelial cells: role of MMP-10 in VEGF-induced angiogenesis. J Cell Physiol. 2010;224:734–742. doi: 10.1002/jcp.22175. [DOI] [PubMed] [Google Scholar]
- 52.De Val S, Black BL. Transcriptional control of endothelial cell development. Dev Cell. 2009;16:180–195. doi: 10.1016/j.devcel.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McLaughlin F, Ludbrook VJ, Cox J, von Carlowitz I, Brown S, Randi AM. Combined genomic and antisense analysis reveals that the transcription factor Erg is implicated in endothelial cell differentiation. blood. 2001;98:3332–3339. doi: 10.1182/blood.v98.12.3332. [DOI] [PubMed] [Google Scholar]
- 54.Ginsberg M, James D, Ding BS, Nolan D, Geng F, Butler JM, et al. Efficient direct reprogramming of mature amniotic cells into endothelial cells by ETS factors and TGFβ suppression. Cell. 2012;151:559–575. doi: 10.1016/j.cell.2012.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Le Bras A, Samson C, Trentini M, Caetano B, Lelievre E, Mattot V, et al. VE-statin/egfl7 expression in endothelial cells is regulated by a distal enhancer and a proximal promoter under the direct control of Erg and GATA-2. PloS One. 2010;5:e12156. doi: 10.1371/journal.pone.0012156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soncin F, Mattot V, Lionneton F, Spruyt N, Lepretre F, Begue A, et al. VE-statin, an endothelial repressor of smooth muscle cell migration. EMBO J. 2003;22:5700–5711. doi: 10.1093/emboj/cdg549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lelièvre E, Lionneton F, Mattot V, Spruyt N, Soncin F. Ets-1 regulates fli-1 expression in endothelial cells. Identification of ETS binding sites in the fli-1 gene promoter. J Biol Chem. 2002;277:25143–25151. doi: 10.1074/jbc.M201628200. [DOI] [PubMed] [Google Scholar]
- 58.Georgiou P, Maroulakou I, Green J, Dantis P, Romanospica V, Kottaridis S, et al. Expression of ets family of genes in systemic lupus erythematosus and Sjogren’s syndrome. Int J Oncol. 1996;9:9–18. [PubMed] [Google Scholar]
- 59.Suzuki E, Williams S, Sato S, Gilkeson G, Watson DK, Zhang XK. The transcription factor Fli-1 regulates monocyte, macrophage and dendritic cell development in mice. Immunology. 2013;139:318–327. doi: 10.1111/imm.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang XK, Gallant S, Molano I, Moussa OM, Ruiz P, Spyropoulos DD, et al. Decreased expression of the Ets family transcription factor Fli-1 markedly prolongs survival and significantly reduces renal disease in MRL/lpr mice. J Immunol. 2004;173:6481–6489. doi: 10.4049/jimmunol.173.10.6481. [DOI] [PubMed] [Google Scholar]
- 61.Mathenia J, Reyes-Cortes E, Williams S, Molano I, Ruiz P, Watson DK, et al. Impact of Fli-1 transcription factor on autoantibody and lupus nephritis in NZM2410 mice. Clin Exp Immunol. 2010;162:362–371. doi: 10.1111/j.1365-2249.2010.04245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suzuki E, Karam E, Williams S, Watson DK, Gilkeson G, Zhang XK. Fli-1 transcription factor affects glomerulonephritis development by regulating expression of monocyte chemoattractant protein-1 in endothelial cells in the kidney. Clin Immunol. 2012;145:201–208. doi: 10.1016/j.clim.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Molano I, Mathenia J, Ruiz P, Gilkeson GS, Zhang XK. Decreased expression of Fli-1 in bone marrow-derived haematopoietic cells significantly affects disease development in Murphy Roths Large/lymphoproliferation (MRL/lpr) mice. Clin Exp Immunol. 2010;160:275–282. doi: 10.1111/j.1365-2249.2009.04080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nowling TK, Fulton JD, Chike-Harris K, Gilkeson GS. Ets factors and a newly identified polymorphism regulate Fli1 promoter activity in lymphocytes. Mol Immunol. 2008;45:1–12. doi: 10.1016/j.molimm.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morris EE, Amria MY, Kistner-Griffin E, Svenson JL, Kamen DL, Gilkeson GS, et al. A GA microsatellite in the Fli1 promoter modulates gene expression and is associated with systemic lupus erythematosus patients without nephritis. Arthritis Res Ther. 2010;12:R212. doi: 10.1186/ar3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sakthianandeswaren A, Curtis JM, Elso C, Kumar B, Baldwin TM, Lopaticki S, et al. Fine mapping of Leishmania major susceptibility Locus lmr2 and evidence of a role for Fli1 in disease and wound healing. Infect Immun. 2010;78:2734–2744. doi: 10.1128/IAI.00126-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tani C, Bellando Randone S, Guiducci S, Della Rossa A. Systemic sclerosis: a critical digest of the recent literature. Clin Exp Rheumatol. 2013;31:172–179. [PubMed] [Google Scholar]
- 68.Wang Y, Fan PS, Kahaleh B. Association between enhanced type I collagen expression and epigenetic repression of the FLI1 gene in scleroderma fibroblasts. Arthritis Rheum. 2006;54:2271–2279. doi: 10.1002/art.21948. [DOI] [PubMed] [Google Scholar]
- 69.Asano Y, Bujor AM, Trojanowska M. The impact of Fli1 deficiency on the pathogenesis of systemic sclerosis. J Dermatol Sci. 2010;59:153–162. doi: 10.1016/j.jdermsci.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Asano Y, Stawski L, Hant F, Highland K, Silver R, Szalai G, et al. Endothelial Fli1 deficiency impairs vascular homeostasis: a role in scleroderma vasculopathy. Am J Pathol. 2010;176:1983–1998. doi: 10.2353/ajpath.2010.090593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bujor AM, Asano Y, Haines P, Lafyatis R, Trojanowska M. The c-Abl tyrosine kinase controls protein kinase Cd-induced Fli-1 phosphorylation in human dermal fibroblasts. Arthritis Rheum. 2011;63:1729–1737. doi: 10.1002/art.30284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Asano Y, Trojanowska M. Phosphorylation of Fli1 at threonine 312 by protein kinase C delta promotes its interaction with p300/CREB-binding protein-associated factor and subsequent acetylation in response to transforming growth factor beta. Mol Cell Biol. 2009;29:1882–1894. doi: 10.1128/MCB.01320-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Asano Y, Czuwara J, Trojanowska M. Transforming growth factor-beta regulates DNA binding activity of transcription factor Fli1 by p300/CREB-binding protein-associated factor-dependent acetylation. J Biol Chem. 2007;282:34672–34683. doi: 10.1074/jbc.M703907200. [DOI] [PubMed] [Google Scholar]
- 74.Noda S, Asano Y, Akamata K, Aozasa N, Taniguchi T, Takahashi T, et al. Constitutive activation of c-Abl/protein kinase C-d/Fli1 pathway in dermal fibroblasts derived from patients with localized scleroderma. Br J Dermatol. 2012;167:1098–1105. doi: 10.1111/j.1365-2133.2012.11055.x. [DOI] [PubMed] [Google Scholar]
- 75.Elkareh J, Periyasamy SM, Shidyak A, Vetteth S, Schroeder J, Raju V, et al. Marinobufagenin induces increases in procollagen expression in a process involving protein kinase C and Fli-1: implications for uremic cardiomyopathy. Am J Physiol Renal Physiol. 2009;296:F1219–F1226. doi: 10.1152/ajprenal.90710.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haller ST, Kennedy DJ, Shidyak A, Budny GV, Malhotra D, Fedorova OV, et al. Monoclonal antibody against marinobufagenin reverses cardiac fibrosis in rats with chronic renal failure. Am J Hypertens. 2012;25:690–696. doi: 10.1038/ajh.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nikitina ER, Mikhailov AV, Nikandrova ES, Frolova EV, Fadeev AV, Shman VV, et al. In preeclampsia endogenous cardiotonic steroids induce vascular fibrosis and impair relaxation of umbilical arteries. J Hypertens. 2011;29:769–776. doi: 10.1097/HJH.0b013e32834436a7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ott DE, Keller J, Rein A. 10A1 MuLV induces a murine leukemia that expresses hematopoietic stem cell markers by a mechanism that includes fli-1 integration. Virology. 1994;205:563–568. doi: 10.1006/viro.1994.1680. [DOI] [PubMed] [Google Scholar]
- 79.Prassad DD, Rao VN, Reddy ES. Structure and expression of human Fli-1 gene. Cancer Res. 1992;52:5833–5837. [PubMed] [Google Scholar]
- 80.Watson DK, Smyth FE, Thompson DM, Cheng JQ, Testa JR, Papas TS, Seth A. The ERGB/Fli-1 gene: isolation and characterization of a new member of the family of human ETS transcription factors. Cell Growth Differ. 1992;3:705–713. [PubMed] [Google Scholar]
- 81.Ano S, Pereira R, Pironin M, Lesault I, Milley C, Lebigot I, et al. Erythroblast transformation by FLI-1 depends upon its specific DNA binding and transcriptional activation properties. J Biol Chem. 2004;279:2993–3002. doi: 10.1074/jbc.M303816200. [DOI] [PubMed] [Google Scholar]
- 82.Athanasiou M, Mavrothalassitis G, Sun-Hoffman L, Blair DG. FLI-1 is a suppressor of erythroid differentiation in human hematopoietic cells. Leukemia. 2000;14:439–445. doi: 10.1038/sj.leu.2401689. [DOI] [PubMed] [Google Scholar]
- 83.Pereira R, Quang CT, Lesault I, Dolznig H, Beug H, Ghysdael J. FLI-1 inhibits differentiation and induces proliferation of primary erythroblasts. Oncogene. 1999;18:1597–1608. doi: 10.1038/sj.onc.1202534. [DOI] [PubMed] [Google Scholar]
- 84.Lee CR, Cervi D, Truong AH, Li YJ, Sarkar A, Ben-David Y. Friend virus-induced erythroleukemias: a unique and well-defined mouse model for the development of leukemia. Anticancer Res. 2003;23:2159–2166. [PubMed] [Google Scholar]
- 85.Torchia EC, Boyd K, Rehg JE, Qu C, Baker SJ. EWS/FLI-1 induces rapid onset of myeloid/erythroid leukemia in mice. Mol Cell Biol. 2007;27:7918–7934. doi: 10.1128/MCB.00099-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Athanasiou M, Mavrothalassitis G, Sun-Hoffman L, Blair DG. FLI-1 is a suppressor of erythroid differentiation in human hematopoietic cells. Leukemia. 2000;14:439–445. doi: 10.1038/sj.leu.2401689. [DOI] [PubMed] [Google Scholar]
- 87.Lesault I, Quang CT, Frampton J, Ghysdael J. Direct regulation of BCL-2 by FLI-1 is involved in the survival of FLI-1-transformed erythroblasts. EMBO J. 2002;21:694–703. doi: 10.1093/emboj/21.4.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Truong AH, Cervi D, Lee J, Ben-David Y. Direct transcriptional regulation of MDM2 by Fli-1. Oncogene. 2005;24:962–969. doi: 10.1038/sj.onc.1208323. [DOI] [PubMed] [Google Scholar]
- 89.Wong KS, Li YJ, Howard J, Ben-David Y. Loss of p53 in F-MuLV induced-erythroleukemias accelerates the acquisition of mutational events that confers immortality and growth factor independence. Oncogene. 1999;18:5525–5534. doi: 10.1038/sj.onc.1202938. [DOI] [PubMed] [Google Scholar]
- 90.Li Y, Tanaka K, Fan X, Nakatani F, Li X, Nakamura T, Takasaki M, et al. Inhibition of the transcriptional function of p53 by EWS-Fli1 chimeric protein in Ewing Family Tumors. Cancer Lett. 2010;294:57–65. doi: 10.1016/j.canlet.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 91.Ban J, Bennani-Baiti IM, Kauer M, Schaefer KL, Poremba C, Jug G, et al. EWS-FLI1 suppresses NOTCH-activated p53 in Ewing’s sarcoma. Cancer Res. 2008;68:7100–7109. doi: 10.1158/0008-5472.CAN-07-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lakhanpal GK, Vecchiarelli-Federico LM, Li YJ, Cui JW, Bailey ML, Spaner DE, et al. The inositol phosphatase SHIP-1 is negatively regulated by Fli-1 and its loss accelerates leukemogenesis. Blood. 2010;116:428–436. doi: 10.1182/blood-2009-10-250217. [DOI] [PubMed] [Google Scholar]
- 93.Juban G, Giraud G, Guyot B, Belin S, Diaz JJ, Starck J, et al. Spi-1 and Fli-1 directly activate common target genes involved in ribosome biogenesis in Friend erythroleukemic cells. Mol Cell Biol. 2009;29:2852–2864. doi: 10.1128/MCB.01435-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Klemsz MJ, Maki RA, Papayannopoulou T, Moore J, Hromas R. Characterization of the ets oncogene family member, fli-1. J Biol Chem. 1993;268:5769–5773. [PubMed] [Google Scholar]
- 95.Cui JW, Vecchiarelli-Federico LM, Li YJ, Wang GJ, Ben-David Y. Continuous Fli-1 expression plays an essential role in the proliferation and survival of F-MuLV-induced erythroleukemia and human erythroleukemia. Leukemia. 2009;23:1311–1319. doi: 10.1038/leu.2009.20. [DOI] [PubMed] [Google Scholar]
- 96.Kornblau SM, Qiu YH, Zhang N, Singh N, Faderl S, Ferrajoli A, et al. Abnormal expression of FLI1 protein is an adverse prognostic factor in acute myeloid leukemia. Blood. 2011;118:5604–5612. doi: 10.1182/blood-2011-04-348052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tyybäkinoja A, Saarinen-Pihkala U, Elonen E, Knuutila S. Amplified, lost, and fused genes in 11q23–25 amplicon in acute myeloid leukemia, an array-CGH study. Genes Chromosomes Cancer. 2006;45:257–264. doi: 10.1002/gcc.20288. [DOI] [PubMed] [Google Scholar]
- 98.Kwiatkowski BA, Zielinska-Kwiatkowska AG, Bauer TR, Jr, Hickstein DD. The ETS family member Tel antagonizes the Fli-1 phenotype in hematopoietic cells. Blood Cells Mol Dis. 2000;26:84–90. doi: 10.1006/bcmd.2000.0282. [DOI] [PubMed] [Google Scholar]
- 99.Kwiatkowski BA, Bastian LS, Bauer TR, Jr, Tsai S, Zielinska-Kwiatkowska AG, Hickstein DD. The ets family member Tel binds to the Fli-1 oncoprotein and inhibits its transcriptional activity. J Biol Chem. 1998;273:17525–17530. doi: 10.1074/jbc.273.28.17525. [DOI] [PubMed] [Google Scholar]
- 100.Martens JH, Mandoli A, Simmer F, Wierenga BJ, Saeed S, Singh AA, et al. ERG and FLI1 binding sites demarcate targets for aberrant epigenetic regulation by AML1-ETO in acute myeloid leukemia. Blood. 2012;120:4038–4048. doi: 10.1182/blood-2012-05-429050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Johnson AD, Pambuccian SE, Andrade RS, Dolan MM, Aslan DL. Ewing sarcoma and primitive neuroectodermal tumor of the esophagus: report of a case and review of literature. Int J Surg Pathol. 2010;18:388–393. doi: 10.1177/1066896908316903. [DOI] [PubMed] [Google Scholar]
- 102.Mhawech-Fauceglia P, Herrmann FR, Bshara W, Odunsi K, Terracciano L, Sauter G, et al. Friend leukaemia integration-1 expression in malignant and benign tumours: a multiple tumour tissue microarray analysis using polyclonal antibody. J Clin Pathol. 2007;60:694–700. doi: 10.1136/jcp.2006.039230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sakurai T, Kondoh N, Arai M, Hamada J, Yamada T, Kihara-Negishi F, et al. Functional roles of Fli-1, a member of the Ets family of transcription factors, in human breast malignancy. Cancer Sci. 2007;98:1775–1784. doi: 10.1111/j.1349-7006.2007.00598.x. [DOI] [PubMed] [Google Scholar]
- 104.Torlakovic EE, Slipicevic A, Flørenes VA, Chibbar R, DeCoteau JF, Bilalovic N. Fli-1 expression in malignant melanoma. Histol Histopathol. 2008;23:1309–1314. doi: 10.14670/HH-23.1309. [DOI] [PubMed] [Google Scholar]
- 105.Paula P, Barros-Silva JD, Ribeiro FR, Ramalho-Carvalho J, Jerónimo C, Henrique R, et al. FLI1 is a novel ETS transcription factor involved in gene fusions in prostate cancer. Genes Chromosomes Cancer. 2012;51:240–249. doi: 10.1002/gcc.20948. [DOI] [PubMed] [Google Scholar]
- 106.Cuda J, Mirzamani N, Kantipudi R, Robbins J, Welsch MJ, Sundram UN. Diagnostic utility of Fli-1 and D2–40 in distinguishing atypical fibroxanthoma from angiosarcoma. Am J Dermatopathol. 2013;35:316–318. doi: 10.1097/DAD.0b013e318266b197. [DOI] [PubMed] [Google Scholar]
- 107.Brown JG, Folpe AL, Rao P, Lazar AJ, Paner GP, Gupta R, et al. Primary vascular tumors and tumor-like lesions of the kidney: a clinicopathologic analysis of 25 cases. Am J Surg Pathol. 2010;34:942–949. doi: 10.1097/PAS.0b013e3181e4f32a. [DOI] [PubMed] [Google Scholar]
- 108.Gill R, O’Donnell RJ, Horvai A. Utility of immunohistochemistry for endothelial markers in distinguishing epithelioid hemangioendothelioma from carcinoma metastatic to bone. Arch Pathol Lab Med. 2009;133:967–972. doi: 10.5858/133.6.967. [DOI] [PubMed] [Google Scholar]
- 109.Nagano A, Ohno T, Shimizu K, Hara A, Yamamoto T, Kawai G, et al. EWS/Fli-1 chimeric fusion gene upregulates vascular endothelial growth factor-A. Int J Cancer. 2010;126:2790–2798. doi: 10.1002/ijc.24781. [DOI] [PubMed] [Google Scholar]
- 110.Shaked Y, Cervi D, Neuman M, Chen L, Klement G, Michaud CR, et al. The splenic microenvironment is a source of proangiogenesis/inflammatory mediators accelerating the expansion of murine erythroleukemic cells. Blood. 2005;105:4500–4507. doi: 10.1182/blood-2004-08-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Boro A, Prêtre K, Rechfeld F, Thalhammer V, Oesch S, Wachtel M, et al. Small-molecule screen identifies modulators of EWS/FLI1 target gene expression and cell survival in Ewing’s sarcoma. Int J Cancer. 2012;131:2153–2164. doi: 10.1002/ijc.27472. [DOI] [PubMed] [Google Scholar]
- 112.Grohar PJ, Griffin LB, Yeung C, Chen QR, Pommier Y, Khanna C, et al. Ecteinascidin 743 interferes with the activity of EWS-FLI1 in Ewing sarcoma cells. Neoplasia. 2011;13:145–153. doi: 10.1593/neo.101202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Erkizan HV, Kong Y, Merchant M, Schlottmann S, Barber-Rotenberg JS, Yuan L, et al. A small molecule blocking oncogenic protein EWS-FLI1 interaction with RNA helicase A inhibits growth of Ewing’s sarcoma. Nat Med. 2009;15:750–756. doi: 10.1038/nm.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Grohar PJ, Woldemichael GM, Griffin LB, Mendoza A, Chen QR, Yeung C, et al. Identification of an inhibitor of the EWS-FLI1 oncogenic transcription factor by high-throughput screening. J Natl Cancer Inst. 2011;103:962–978. doi: 10.1093/jnci/djr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li YJ, Zhao X, Vecchiarelli-Federico LM, Li Y, Datti A, Cheng Y, et al. Drug-mediated inhibition of Fli-1 for the treatment of leukemia. Blood Cancer J. 2012;2:e54. doi: 10.1038/bcj.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bhatia S, Krailo MD, Chen Z, Burden L, Askin FB, Dickman PS, et al. Therapy-related myelodysplasia and acute myeloid leukemia after Ewing sarcoma and primitive neuroectodermal tumor of bone: A report from the Children’s Oncology Group. Blood. 2007;109:46–51. doi: 10.1182/blood-2006-01-023101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ladenstein R, Pötschger U, Le Deley MC, Whelan J, Paulussen M, Oberlin O, et al. Primary disseminated multifocal Ewing sarcoma: results of the Euro-EWING 99 trial. J Clin Oncol. 2010;28:3284–3291. doi: 10.1200/JCO.2009.22.9864. [DOI] [PubMed] [Google Scholar]
- 118.Miser JS, Krailo MD, Tarbell NJ, Link MP, Fryer CJ, Pritchard DJ, et al. Treatment of metastatic Ewing’s sarcoma or primitive neuroectodermal tumor of bone: evaluation of combination ifosfamide and etoposide--a Children’s Cancer Group and Pediatric Oncology Group study. J Clin Oncol. 2004;22:2873–2876. doi: 10.1200/JCO.2004.01.041. [DOI] [PubMed] [Google Scholar]
- 119.Zhang J, Guo H, Zhang H, Wang H, Qian G, Fan X, Hoffman AR, Hu JF, Ge S. Putative tumor suppressor miR-145 inhibits colon cancer cell growth by targeting oncogene Friend leukemia virus integration 1 gene. Cancer. 2011;117:86–95. doi: 10.1002/cncr.25522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ban J, Jug G, Mestdagh P, Schwentner R, Kauer M, Aryee DN, et al. Hsa-mir-145 is the top EWS-FLI1-repressed microRNA involved in a positive feedback loop in Ewing’s sarcoma. Oncogene. 2011;30:2173–2180. doi: 10.1038/onc.2010.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bujor AM, Haines P, Padilla C, Christmann RB, Junie M, Sampaio-Barros PD, et al. Ciprofloxacin has antifibrotic effects in scleroderma fibroblasts via downregulation of Dnmt1 and upregulation of Fli1. Int J Mol Med. 2012;30:1473–1480. doi: 10.3892/ijmm.2012.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]