Abstract
Transcriptional noise is a necessary consequence of the molecular events that drive gene expression in prokaryotes. However, some environmental microorganisms that inhabit polluted sites, for example, the m-xylene degrading soil bacterium Pseudomonas putida mt-2 seem to have co-opted evolutionarily such a noise for deploying a metabolic diversification strategy that allows a cautious exploration of new chemical landscapes. We have examined this phenomenon under the light of deterministic and stochastic models for activation of the main promoter of the master m-xylene responsive promoter of the system (Pu) by its cognate transcriptional factor (XylR). These analyses consider the role of co-factors for Pu activation and determinants of xylR mRNA translation. The model traces the onset and eventual disappearance of the bimodal distribution of Pu activity along time to the growth-phase dependent abundance of XylR itself, that is, very low in exponentially growing cells and high in stationary. This tenet was validated by examining the behaviour of a Pu-GFP fusion in a P. putida strain in which xylR expression was engineered under the control of an IPTG-inducible system. This work shows how a relatively simple regulatory scenario (for example, growth-phase dependent expression of a limiting transcription factor) originates a regime of phenotypic diversity likely to be advantageous in competitive environmental settings.
Introduction
Noise in gene expression, or stochastic fluctuations of the gene products (mRNAs and proteins) around their average values, is an unavoidable source of randomness and cell heterogeneity in isogenic cell populations, both in prokaryotes (Taniguchi et al., 2010) and eukaryotes (Newman et al., 2006). Although extrinsic noise (fluctuations in cellular components affecting many genes) is an important source of cell-to-cell variability in eukaryotes (Newman et al., 2006; Volfson et al., 2006), most of the variability observed when gene products are in low copies is attributable to the stochastic character of the biochemical reactions (intrinsic noise; Taniguchi et al., 2010). Especially in bacteria, many mRNA, transcription factors or post-transcriptional regulators such as small non-coding RNAs are present at a few copies per cell, inducing large fluctuations in protein abundance. But noise is not always a nuisance that organisms must overcome to deploy robust functions. Stochasticity may also have a beneficial role by promoting phenotypic diversity (Raser and O'Shea, 2005; Raj and van Oudenaarden, 2008) and cellular decision-making (Balazsi et al., 2011). For instance, different microbial populations exploit phenotypic variation to survive to adverse changes in environmental conditions (Balaban et al., 2004; Zimmermann et al., 2015), a strategy known as bet-hedging (Ackermann, 2013, 2015; Grimbergen et al., 2015). In one common scenario, different phenotypes coexist within an isogenic population in a given environment. These subpopulations are usually sustained by the presence of simultaneous stable expression states of specific genes, with molecular noise driving transitions between them (Maamar et al., 2007; Acar et al., 2008), a mechanism known as stochastic switching. This mechanism can favour adaptation to unpredictable environmental changes (Kussell and Leibler, 2005; Acar et al., 2008). In other cases, an initially homogeneous population shows phenotypic diversification after a nutritional or physicochemical shift. This situation, known as responsive diversification (Grimbergen et al., 2015), has been recently observed as an adaptation strategy to changes in metabolic conditions (Kotte et al., 2014; New et al., 2014; Venturelli et al., 2015). In this scenario, there is a transition from a single gene expression state to two different stable states driven by a change in substrate. In all these cases, the presence of positive feedback is required to generate and maintain the different expression states (Acar et al., 2005; Maamar and Dubnau, 2005; Suel et al., 2006; Veening et al., 2008; Kotte et al., 2014; Gallie et al., 2015; Venturelli et al., 2015), while molecular noise may enable phenotype switching. The existence of two simultaneous expression states without positive feedback is a much less-frequent phenomenon, and to the best of our knowledge has never been observed in this context. We will use the term 'bimodality' to refer to the coexistence of two different states, which does not necessarily implies stability of both states (bistability). Bimodality without feedback, that is, with a purely stochastic origin has been predicted theoretically (Kaern et al., 2005; Ochab-Marcinek and Tabaka, 2010). Also, it has been reported in a few designed experimental set-ups due to transcription bursts (Blake et al., 2006), noise in transcription factors coupled to slow promoter kinetics (Nevozhay et al., 2009) or external noise (Dublanche et al., 2006). But whether or not it happens as a natural mechanism of phenotypic diversification—metabolic or otherwise remains unknown thus far.
We have recently examined the expression dynamics of catabolic promoters involved in biodegradation of m-xylene of the soil bacterium Pseudomonas putida mt-2 (Silva-Rocha and de Lorenzo, 2012). The metabolic and regulatory circuit borne by plasmid pWW0 (that is, the so-called TOL network) is responsible for the stepwise degradation of the pathway substrate through the action of two operons (upper and lower, Figure 1). Single-cell analysis by flow cytometry of fluorescent reporters from the different promoters revealed a striking variability in cell-to-cell expression levels. In particular, both the Pu promoter of the upper pathway and the Ps promoter that drives expression of xylS (Figure 1) showed strong bimodal responses after exposure to m-xylene. This is intriguing for various reasons: Pu is triggered by activation of XylR, itself the product of the Pr promoter that showed a unimodal distribution of fluorescence levels (Silva-Rocha and de Lorenzo, 2012). Moreover, there are no positive-feedback interactions between this gene and other regulatory elements of the network. Second, this bimodal expression regime was observed when cells actively growing on a different C source (for example, succinate) were exposed to TOL inducers. The same induction experiments conducted with cells arrested in stationary phase revealed only unimodal responses. Cells can thus utilize their internal state to modulate population heterogeneity when confronted with different nutrient conditions, as a kind of metabolic versification program.
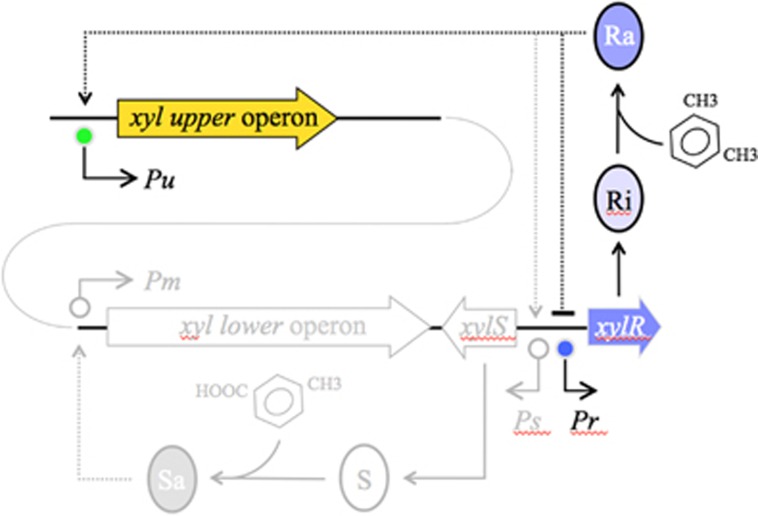
Figure 1.
Organization of the TOL network of the TOL plasmid pWW0 of P. putida mt-2. The TOL pathway encoded by plasmid pWW0 consists of two different operons: the upper operon, the products of which transform m-xylene into 3-methylbenzoate, and the lower operon that produces enzymes for further metabolism of this compound into TCA cycle intermediates. XylR and XylS are the transcriptional regulators that control expression of either operon. The master regulatory gene xylR is encoded in a location adjacent to the end of the meta operon and is expressed from the Pr promoter. XylR is produced in an inactive form (Ri) that, in the presence of the pathway substrate (m-xylene) changes to an active form (Ra). Ra then activates both Pu and Ps, triggering expression of the upper pathway and XylS, respectively. At the same time, Ra acts as repressor of its transcription, thereby decreasing its own expression. The part of the network under study is highlighted in colour, the rest is faded. Operons and regulatory elements not to scale.
In this work, we use a mathematical stochastic model of the upper TOL subnetwork combined with single-cell measurements of Pu-GFP activity to elucidate the mechanism underlying such behaviour. We show below that the strongly variable (bimodal) responses of this catabolic promoter after induction in exponential phase are caused by the few molecules of XylR present in this phase, which is kept at low levels by post-transcriptional repression of the xylR mRNA. This originates large fluctuations in this transcription factor, which is required to activate the TOL network response, inducing long-lasting random episodes where the Pu promoter remains inactive. This mechanism of phenotypic diversification adds to the repertoire of genetic, regulatory and biochemical devices through which bacteria cope with nutritionally changing environments (Kotte et al., 2014; Ackermann, 2015; Gallie et al., 2015).
Materials and methods
Strains, plasmids and growth conditions
Escherichia coli (E. coli) CC118λpir (de Lorenzo and Timmis, 1994) was used as the host for the plasmid constructs. E. coli HB101 (pRK600) was employed as conjugative helper in four-parental matings, as previously described (Choi et al., 2005; de Las Heras and de Lorenzo, 2012). All E. coli strains were grown at 37 °C in Luria Bertani (LB) medium supplemented when required with gentamicin (Gm, 10 μg ml−1), kanamycin (Km, 50 μg ml−1), cloramphenicol (Cm, 30 μg ml−1), streptomycin (Sm, 50 μg ml−1) and ampicillin (Ap, 50 μg ml−1). P. putida Mmt-2-Pu (Silva-Rocha and de Lorenzo, 2012) was used as the reference for single-cell analysis of Pu expression in native conditions. This strain derives from the natural P. putida mt-2 isolate bearing the TOL plasmid pWW0, but it has been engineered to bear in its chromosome a transcriptional Pu-GFP fusion (Silva-Rocha and de Lorenzo, 2012). P. putida Mmt-2-Pu thus keeps all the regulatory components of the TOL plasmid in its native configuration and connectivity. A second reporter strain that transcribes xylR in response to adding isopropyl β-d-1-thiogalactopyranoside (IPTG) to the medium was constructed as follows. First, a NotI DNA segment bearing an expression construct lacIq/Ptrc → xylR was excised from plasmid pVTR-XylR plasmid (Perez-Martin and de Lorenzo, 1996) and cloned in the corresponding site of the mini-Tn7-Gm delivery vector pTn7-M (Zoebel et al., 2015), resulting in plasmid pIB. The thereby assembled mini-Tn7 [L-Gm lacIq/Ptrc → xylR-R] transposon was then delivered to the chromosome of strain P. putida [Pu-GFP] (Garmendia et al., 2008) as explained in (de Lorenzo et al., 1990; Martínez-García et al., 2011; de Las Heras et al., 2012). This target strain is a plasmid-less P. putida KT2440 specimen bearing a chromosomal Pu-GFP fusion inserted with a mini-Tn5 transposon vector (de Lorenzo et al., 1990; Martínez-García et al., 2011; de Las Heras et al., 2012). To verify the correct insertion of mini-Tn7 [L-Gm lacIq/Ptrc → xylR-R] into the att locus of P. putida [Pu-GFP] we selected some GmR clones amplified diagnostic segments by PCR using primer combinations 5-Pput-glmS UP (5′AGTCAGAGTTACGGAATTGTAGG3′) with 3-Tn7L (5′ATTAGCTTACGACGCTACACCC3′) and 5-PpuglmS DOWN (5′TTACGTGGCCG TGCTAAAGGG3′) with 3-Tn7R (5′CACAGCATA ACTGGACTGATTTC3′) and the products of amplification having a size of 400 and 200bp, respectively (Schweizer, 2001; Choi et al., 2005; Choi and Schweizer, 2006). One of the clones with the correct banding patterns was then named P. putida KT-IB1 and employed as required. These strains were grown at 30 °C in M9 minimal medium (Sambrook et al., 1989) supplemented with 2 mM MgSO4 and 25 mM of either citrate or succinate as sole carbon source and proper antibiotics when required. IPTG was added to the media at the concentrations indicated in each case. When required, the XylR/Pu regulatory node of the strain under inspection was induced by exposing the corresponding cultures to saturating vapours of m-xylene.
Single-cell analysis by flow cytometry
Single-cell experiments were performed with a Gallios (Beckman Coulter, Danvers, MA, USA) flow cytometer. To this end, GFP was excited at 488 nm, and the fluorescence signal was recovered with a 525(40) BP filter. For the assays of Pu-GFP under native regulation conditions, P. putida Mmt-2-Pu were pre-grown overnight in M9-succinate medium, regrown in the same conditions until early exponential phase and exposed to saturating m-xylene vapours for the next samples taken and processed explained in detail in (Fraile et al., 2001; Silva-Rocha and de Lorenzo, 2011). In the case of P. putida KT-IB1, overnight-grown cells were also diluted in fresh M9-succinate medium, added with different concentrations of IPTG and incubated for 4 h at 30 °C. After this pre-incubation (cells reaching approximately mid-exponential phase, at OD600 of 0.3–0.4), bacteria were exposed to m-xylene as before. In either case, cultures were then incubated with aeration at 30 °C, and each hour after induction an aliquot of each sample was stored on ice until analysis. For every aliquot, 20000 events were analysed. The data processing was performed using FCS express 4 software and MATLAB (The Mathworks, Inc., Natick, MA, USA) for 3D graphics.
Mathematical modelling
Stochastic simulations were carried out based on the interactions and biochemical reactions sketched in Figure 2b, using Gillespie's algorithm (Gillespie, 1977) and custom scripts programmed in FORTRAN. A complete description of the mathematical model adopted in this work can be found in the Supplementary Information.
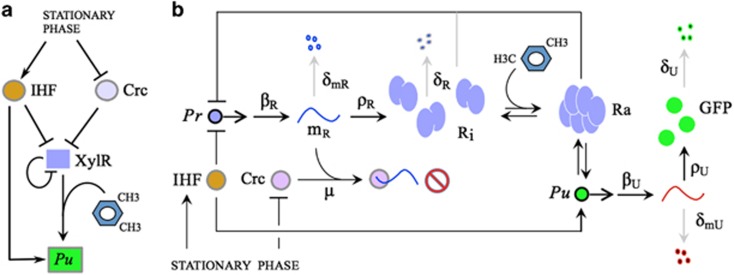
Figure 2.
The Pr/XylR/Pu node and formalization of regulatory interactions. (a) Basic interactions between the Pu promoter, its cognate regulator XylR and the global regulators IHF and Crc whose levels are modulated by growth state. (b) Biochemical reactions involved in the upper route of the TOL pathway. We take into account transcription from the Pr and Pu promoters (with rates βR and βU, respectively), translation of the corresponding mRNAs (with rates ρR and ρU), and degradation of the molecular species: mRNAs (with degradation rates δmR and δmU) and proteins (with rates δR and δU). Translational repression of xylR mRNA (mR) by Crc is described by the effective association rate constant μ. We also consider, sketched as double arrows, the activation (mediated by m-xylene) and inactivation reactions of XylR, as well as the binding/unbinding reactions of active XylR (Ra) to the Pu promoter. A detailed description of the model is provided in Supplementary Information, and the values of the reaction rates specified in Table 1.
Results and discussion
Components and known parameters of the TOL upper network
The upper route of the TOL catabolic pathway is encoded in an operon expressed by the Pu promoter (Figure 1). Pu activity is triggered by the presence of m-xylene once this aromatic substrate binds the transcriptional factor called XylR. The Pr promoter initiates transcription of xylR mRNA at two different sites. Once loaded with m-xylene, an inactive dimer of the XylR protein (Ri) binds ATP and oligomerizes, likely as a hexamer (Valls and de Lorenzo, 2003), undergoing conformational changes that lead to the active form of the regulator (Ra). In addition to Ra, Pu transcription necessitates the action of the host factor IHF, which binds to Pu promoter as a heterodimer favouring DNA bending (Valls et al., 2002). Ra and IHF are both needed for Pu activity, acting as an AND logic gate (Silva-Rocha and de Lorenzo, 2011). Intriguingly, IHF has also the effect of downregulating Pr activity (Figure 2a) forming an unusual type 4 incoherent feed-forward loop with the Pr/Pu system (Silva-Rocha and de Lorenzo, 2011). XylR levels are further negatively regulated at the transcriptional level by Ri and Ra, which are able to bind the Pr promoter self-repressing its activity (Marques et al., 1998). Finally, production of XylR in vivo is checked by a complex post-transcriptional mechanism (Moreno et al., 2010) in which the so-called catabolic repressor control protein (Crc) acts as an operative translational co-repressor along with the default RNA-binding factor Hfq (Milojevic et al., 2013; Moreno et al., 2015).
Interestingly, the levels of the global regulators IHF and Crc are subject to growth control: on one hand, IHF concentration increases approximately sevenfold (~2000 monomers to ~14 000 monomers) from exponential to stationary phase (Valls et al., 2002). On the other hand, Crc is abundant in exponential phase but drops about three- to fourfold in stationary phase (Ruiz-Manzano et al., 2005). Therefore, exponential growth in rich media lowers the IHF level and increases Crc, implying more xylR mRNA but little translation. The stationary phase leads to the opposite situation: less xylR mRNA but very efficient translation. Under culture conditions where the effect of Crc is maximal (for example, LB or amino-acid-rich media), XylR protein levels are expected to increase in stationary phase, as it is the case (Fraile et al., 2001). But, even in growth or culture conditions where both effects compensate and average levels of XylR or of the ensuing Pu product are similar (Silva-Rocha and de Lorenzo, 2011, 2012), these two different regulatory mechanisms could induce differences in variability around mean product levels, affecting individual cell heterogeneity in a clonal population. Indeed, the amount of variability or noise, quantified as the coefficient of variation (s.d. over the mean) of protein levels in a population distribution, has been shown to be dependent on the regulatory mechanism (Levine et al., 2007; Mehta et al., 2008; Guantes et al., 2012). In the following, we translate all these interactions, sketched in Figure 2a, into a mathematical stochastic model to elucidate the origins of the cell-to-cell variability observed in the TOL network.
A mathematical model to study the stochasticity of the TOL network
To account for all the possible sources of intrinsic variability in Pu activity (experimentally measured by flow cytometry) while keeping the system as simple as possible, we explicitly describe the dynamics and detailed biochemical reactions of Pr and Pu products, as well as XylR activation by m-xylene and Pu activation by Ra (Figure 2b). The regulators IHF and Crc are considered as factual effectors whose levels change between exponential and stationary phase conditions, modulating the rate constants of the interactions involved. Specifically, we take into account the stochastic dynamics of seven variables: xylR mRNA (mR), inactive XylR (Ri), active XylR (Ra), Pu inactive promoter (Puoff), Pu activated by both Ra and IHF (Puon), Pu-GFP mRNA (mU) and GFP protein (GFP), which is the experimental readout. The biochemical reactions considered, sketched in Figure 2b, together with the corresponding parameter values are detailed in Supplementary Information and Table 1. Parameters are estimated from previous experimental observations or manually adjusted following the procedure in Parameter fitting section (Supplementary Information). We notice that parameters are tuned in exponential phase conditions to reproduce experimentally observed average values, induction time and GFP distributions, but in stationary phase conditions only IHF/Crc levels are varied.
Table 1. Model parameters: definitions and estimated values.
| Parameter | Meaning | Value | Reference |
|---|---|---|---|
| βR | Basal transcription rate of xylR-mRNA | 240 molecules per houra | (Vogel and Jensen, 1994) (and adjusted) |
| 80 molecules per hourb | (Proshkin et al., 2010) | ||
| fR, fR3 | Fold changes for negative autoregulation by XylR | 3 | (Fraile et al., 2001) |
| fIHF | Fold change for Pr repression by IHF | 5 | (Silva-Rocha and de Lorenzo, 2011) |
| θR,θR3 | Thresholds for negative autoregulation by XylR | 15 molecules | Adjusted to levels of XylR in (Fraile et al., 2001) |
| θIHF | Threshold for Pr repression by IHF | 4000 molecules | Adjusted to levels of IHF in (Valls et al., 2002) |
| μ | Crc-xylR mRNA association | 1 per molecule per hour | (Levine et al., 2007). |
| mxyl/θm | m-Xylene above threshold after induction | 10 | Saturating conditions |
| fm | Fold change in Ra due to m-xylene activation | 0.018 | Adjusted |
| kR | Basal Ra formation in non-inducing conditions | 10−5 per molecule2 per hour | Adjusted to give negligible Ra |
| KRoff | Ra dissociation | 10 per hour | Adjusted |
| θu | Threshold for IHF activation of Pu promoter | 4000 molecules | Adjusted to levels of IHF in (Valls et al., 2002) |
| fu | Fold change in Pu activity by IHF | 0.1 | (Valls et al., 2002) |
| ku | Basal Pu activation in the absence of IHF | 0.08 per molecule per hour | Adjusted from induction time of GFP distributions |
| Kuoff | Dissociation of Ra from Pu promoter | 0.5 per hour | Adjusted |
| βu | Transcription rate from the active Pu promoter | 200 molecules per houra | (Vogel and Jensen, 1994) and adjusted |
| 60 molecules per hourb | (Proshkin et al., 2010) | ||
| ρR | Ri translation rate | 20 per houra | (Proshkin et al., 2010) and adjusted |
| 7 per hourb | |||
| ρu | Pu-GFP translation rate | 20 per houra | (Proshkin et al., 2010) |
| 7 per hourb | |||
| δmR,δmu | xylR/Pu-GFP mRNAs degradation rates | 10 per houra | (Bernstein et al., 2002) |
| 5 per hourb | |||
| δR,δGFP | Ri, Pu-GFP degradation rates | 1 per houra | Stable proteins |
| 0.5 per hourb | |||
| IHF | IHF levels | 2000 moleculesa | (Valls et al., 2002) |
| 14 000 moleculesb | |||
| Crc | Crc levels | 200 moleculesa | (Ruiz-Manzano et al., 2005) and adjusted |
| 50 moleculesb | |||
| γR | Basal transcription from lacI-Ptrc promoter | 2400 molecules per hour | (de Lorenzo et al., 1993; Silva-Rocha and de Lorenzo, 2011) |
| a | Ratio of total LacI and lacI repression threshold | 10 | Arbitrary value >>1 (Ozbudak et al., 2004) |
| fL | Fold change in lacI-Ptrc-xylR expression due to LacI repression | 3.8 | Adjusted (see Materials and methods) |
| θIPTG | Threshold for LacI inactivation by IPTG | 22.6 | Adjusted (see Materials and methods) |
Exponential phase.
Stationary phase. Except stated otherwise, the parameters were taken/adjusted in exponential phase conditions and the same values used in stationary phase.
Growth state modulates variability in TOL pathway activation after induction
To investigate how the cell growth state affects activation of the upper TOL pathway and heterogeneity in the response to induction by aromatic compounds, we used a fusion of Pu to GFP placed in the chromosome of P. putida Mmt-2-Pu (Materials and methods). Cells were grown in liquid M9 media containing succinate as the sole carbon source, induced with saturating vapours of m-xylene and samples collected at different time points after induction to measure fluorescence of the cell culture by flow cytometry. Two different experimental situations were analysed: in one of them, growing cells were incubated to mid-exponential phase and then exposed to m-xylene. In the other, cells were forced to remain in the stationary phase (that is, perfectly viable but without any chance of growing) and subsequently collected, exposed to m-xylene and analysed by flow cytometry. In all the samples investigated, we observed a behaviour similar to the one shown in Figures 3a and c: clear bimodal responses in exponential phase, where a fraction of the population remained inactive at any one time after induction by m-xylene (with a slow leakage to the induced state, Figure 3a), and unimodal responses in stationary phase indicating that the whole population is induced at comparable rates (Figure 3c).
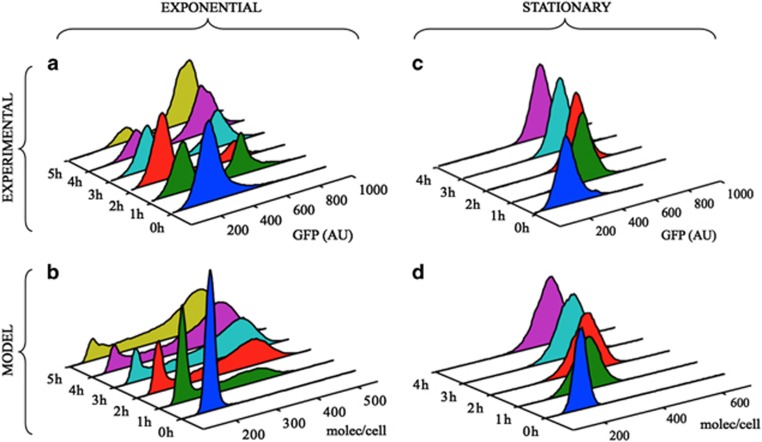
Figure 3.
Single-cell measurements of Pu-GFP activity and stochastic model simulations. (a) Cells grown overnight were diluted in fresh M9-succinate media, incubated to the mid-exponential phase and then exposed to saturating vapours of m-xylene. The fluorescence distributions were calculated from flow cytometry measurements at time intervals of 1 h. (b) Distribution of number of GFP molecules per cell as obtained from 20 000 stochastic trajectories using the model parameters in exponential phase (see Supplementary Information and Table 1). (c) Fluorescence distribution of Pu-GFP levels in P. putida Mmt-2-Pu cells arrested in stationary phase (viable but without any growth), at different time points after exposure to m-xylene vapours. (d) Distribution of number of GFP molecules per cell from ensembles of trajectories generated by the stochastic model in stationary phase conditions.
To mimic the experimental situation, we run ensembles of independent 10 000–20 000 stochastic trajectories simulating the induced conditions in both growth phases (see Mathematical model in Supplementary Information), recorded the numbers of GFP molecules for each independent trajectory at the experimental time points and calculated the corresponding distributions at each time. Most model parameters were taken from previous experimental data or adjusted within known physiological ranges to match experimental observations (Supplementary Information and Table 1). The main free parameters were the rates of activation/deactivation of both Ra and the Pu promoter that we fitted to the experimental induction curves in exponential phase (Supplementary Figure S1 and Parameter fitting in Supplementary Information). These parameters reproduced the observed bimodal behaviour in exponential phase (Figure 3b). The main differences in stationary phase arise from the different levels of the two global regulatory factors IHF and Crc (Figure 2a). For the simulations in stationary phase, we thus kept the same activation/deactivation rates of XylR and Pu used in exponential phase, but varied the amount of IHF and Crc, treated as intracellular ‘inducers' in our mathematical model, according to the experimental observations: approximately fourfold decrease in Crc (Ruiz-Manzano et al., 2005) and approximately sevenfold increase in IHF (Valls et al., 2002). By just altering the levels of these two factors, we recover the unimodal distributions observed in stationary phase, Figure 3d. This supports the idea that P. putida heterogeneous responses are altogether determined by growth phase modulation of these global factors.
Bimodality is originated by low-frequency activation events
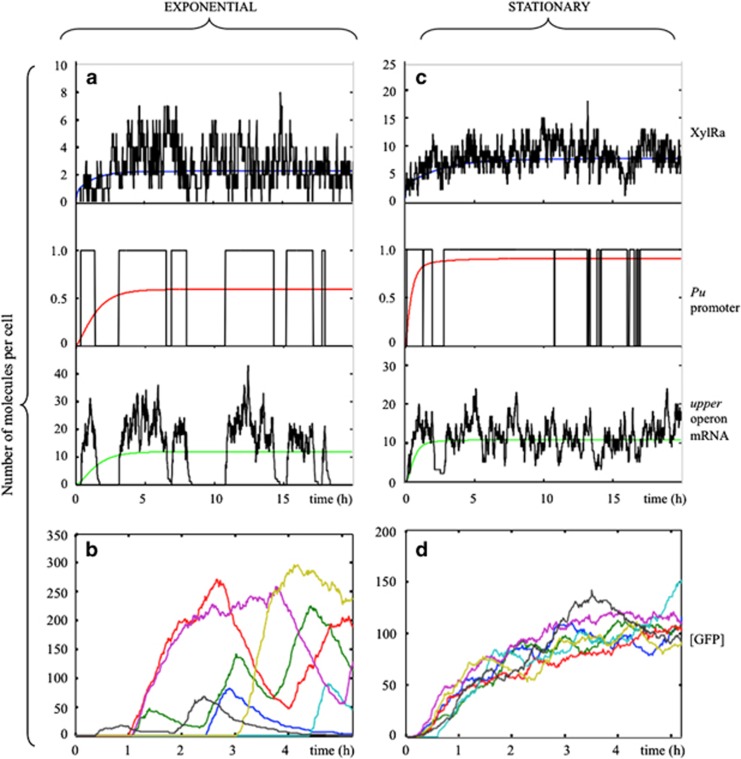
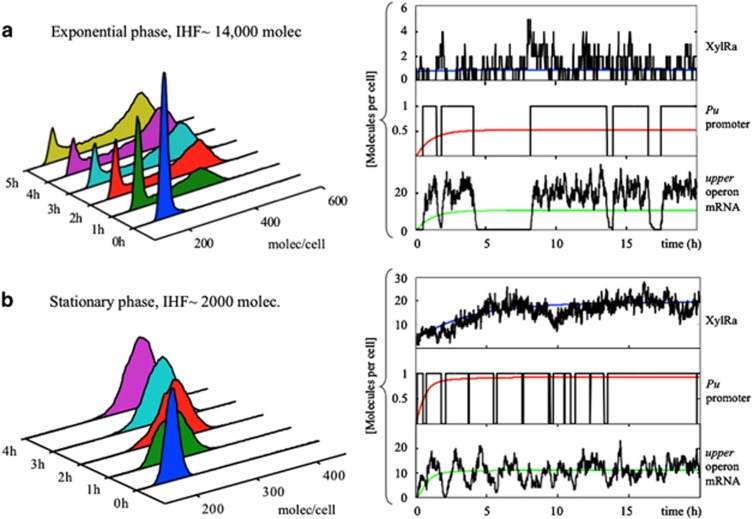
As the absence of positive feedback rules out bistability (coexistence of two stable expression states) in the TOL network, the bimodality observed in the Pu-GFP distributions must be exclusively due to the stochastic dynamics of Pu activation. Previous experimental work in yeast (Blake et al., 2006; To and Maheshri, 2010) and bacteria (So et al., 2011) has demonstrated that transcription bursts can originate bimodal single-cell distributions without bistability. This bimodality is caused by slow promoter fluctuations between active and inactive states, and promoters with multiple binding sites/states for activation can provide such infrequent events (Blake et al., 2006; To and Maheshri, 2010). To elucidate if this is the case in the system under study, we examined the time course of the different species involved in the network. In Figures 4a and c we show typical stochastic (in black) and the corresponding deterministic trajectories (coloured lines) in both growth phases. Particularly illustrative is the activation/inactivation dynamics of the Pu promoter (Figures 4a and c; mid panel). In exponential phase, activation events are less frequent and separated by long episodes of inactivation (the deterministic result, red line, gives the fraction of time the promoter is in the active state, ~50% of the time). In stationary phase promoter activation is more frequent and inactivation episodes much shorter. This has the consequence that, despite the amount of Pu mRNA on average is the same in exponential and stationary phases (green lines in Figures 4a and c, low panels), in exponential phase Pu transcription is produced in large, infrequent bursts, while Pu mRNA noise is attenuated in stationary phase. In Figures 4b and d we plot the time trace of individual GFP trajectories in both conditions, showing that GFP production can be highly delayed in exponential phase, and even decay to low levels once activated, with the result that a fraction of the cells will be inactive even at long times after induction.
Figure 4.
Deterministic and stochastic dynamics of TOL network components in different growth phases. Differences in Pu promoter activation, Pu mRNA variability and GFP induction in exponential (a, b) and stationary (c, d) phases. (a) Time evolution of active XylR (top panel), active Pu promoter (intermediate panel) and Pu mRNA (low panel). Black line is the stochastic simulation and coloured lines the deterministic results. (b) Seven stochastic trajectories mimicking GFP induction in different cells during the time course of experiments. (c, d) The same variables and GFP trajectories in stationary phase conditions.
Post-transcriptional repression is responsible for noise-induced bimodality
The activation of the Pu promoter requires both the binding of XylR hexamers (Ra) and IHF. We first notice that, as observed experimentally (Fraile et al., 2001), total XylR levels increase around fivefold in stationary phase. This is reflected in the amount of Ra present in the different growth phases, see the time traces of Ra in Figures 4a and c (upper panels). Due to the small number of XylR molecules in exponential phase (~15), Ra is at very low copies (~0–6) with large relative fluctuations. This could be reflected in infrequent activation of the Pu promoter. The amount of XylR is negatively controlled by Crc and IHF (Figure 2a), but IHF repression produces a modest decrease (approximately twofold) in both phases (Silva-Rocha and de Lorenzo, 2011), indicating that most of the differences observed in XylR abundance must come from the post-transcriptional repression exerted by Crc.
On the other hand, IHF is necessary for Pu activity, so that IHF level differences in both phases could be modulating Pu dynamics and hence GFP variability. To test whether direct activation (due to IHF) or indirect (through Crc control of XylR amount) originates the different behaviours observed, we performed two types of stochastic simulations: in one of them, IHF levels were increased in exponential phase and decreased in stationary phase, contrary to the real situation, to see how growth state differences in IHF abundance affect Pu activation. The results of Figure 5a show that in spite of increasing IHF amount approximately sevenfold in exponential phase, the population distributions are very similar to the experimental situation with low IHF levels. Although one could reason in principle that high IHF levels would favour Pu expression by increasing the activation frequency, we observe that this frequency is indeed very similar to that observed at low IHF levels (red line in right panel of Figure 5a, compared with Figure 4a). We recall that both IHF and Ra are required to activate Pu transcription. As Ra is still present at extremely low copies (~0–6 copies per cell, blue line in the right panel of Figure 5a), it acts as a limiting factor precluding Pu activation despite the high levels of IHF. Moreover, as IHF also represses Pr (Figures 1 and 2a), XylR production is strongly shut off by the combined action of high Crc and high IHF levels, counteracting the positive effect of IHF on Pu. In Figure 5b, we show with numerical simulations the effect of decreasing IHF in stationary phase. Stochastic distributions remain unimodal although wider than in the normal situation. As IHF is not a limiting factor, reducing IHF only increases deactivation frequency (mid panel in Figure 5b, right), that is, Pu undergoes frequent deactivation events but of short duration, as the high amount of Ra present (upper panel in Figure 5b, right), compensates eventual dissociations of IHF from the Pu promoter. The basic effect of these fast inactivation events is to increase the size of transcriptional bursts (lower panel in Figure 5b, right), and hence the noise, but is not enough to create bimodality, which requires the presence of slow promoter fluctuations (To and Maheshri, 2010).
Figure 5.
Effect of IHF levels in response variability. (a) Simulation of increasing intracellular IHF levels in exponential phase to the concentration present in stationary conditions (~14 000 molecules). Distributions remain bimodal (left panel) and Pu promoter dynamics exhibits slow fluctuations (right panel). (b) Simulation of decreasing IHF levels in exponential phase to those of exponential conditions (~2000 molecules). The population distributions of GFP are still unimodal (left panel) although noise increases, due to larger fluctuations in Pu mRNA (right panel).
The outcome of these numerical experiments hints to the presence of low copy numbers of XylR as the main reason for the bimodal responses observed in exponential phase. XylR would act as a limiting factor for Pu activation, inducing slow fluctuations in promoter dynamics. In support of this idea, we carried out numerical simulations in exponential phase using low Crc levels (comparable to those in stationary phase), thus releasing post-transcriptional repression of XylR. Our results (Supplementary Figure S2) show a noticeable rise in Ra levels, yielding unimodal population distributions of Pu-GFP activity with low variability.
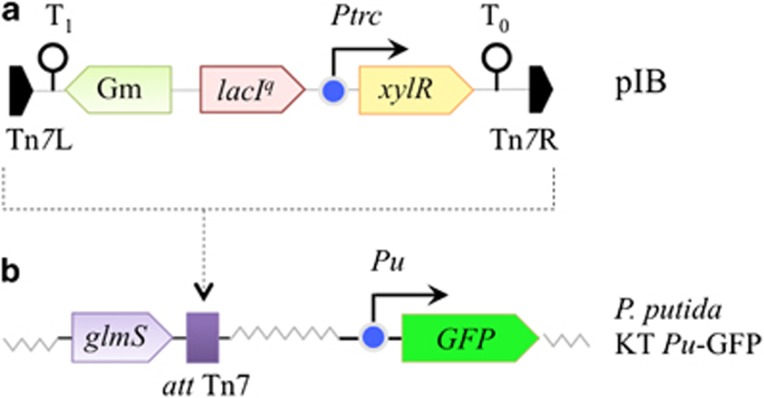
Overproduction of XylR defeats Pu bimodality in vivo
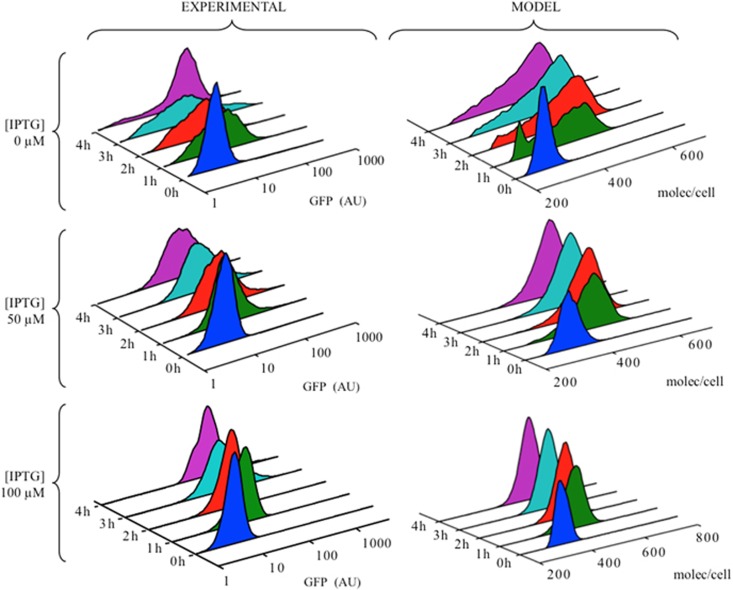
The key tenet of the model above is that phenotypic divergence of Pu activity can ultimately be traced to the very low levels of XylR during exponential growth. The logical prediction of this scenario is that artificially increasing such levels should overcome the observed bimodality. One strategy to this end could be the use of a crc mutant of P. putida to allow efficient translation of XylR during exponential phase. However, as Crc is a global regulator affecting many cellular processes (Moreno et al., 2009; Linares et al., 2010), we adopted instead an experimental protocol to increase XylR expression in a controllable way with minimum interference with cell metabolism and state. To this end, we engineered a mini-Tn7 transposon carrying an expression cassette LacIq-Ptrc → xylR (Perez-Martin and de Lorenzo, 1996) and inserted it into the chromosome of P. putida KT2440 strain bearing in its chromosome a transcriptional Pu-GFP fusion (Figure 6 and Materials and methods). To control XylR production, different concentrations of IPTG—the LacIq-Ptrc inducer—were added to the medium during cell growth, while m-xylene was added in exponential phase to activate XylR. The mathematical model was modified accordingly to describe IPTG induction of XylR by the synthetic LacIq-Ptrc promoter (Supplementary Information). Pu activity was monitored by flow cytometry as previously and the fluorescence distributions compared with numerical simulations (Figure 7). We noticed that, in the absence of the inducer IPTG responses were very heterogeneous with a large range of Pu-GFP levels (Figure 7 top left). However, the two distinct subpopulations observed in the wild-type scenario were not separated well. This is probably due to the fact that the basal activity of the Ptrc promoter is ⩾2–3-fold higher than the activity of the native Pr promoter (de Lorenzo et al., 1993; Silva-Rocha and de Lorenzo, 2011). Taking into account this fact in the mathematical model (Supplementary Information), we see that except for a small fraction of cells still uninduced at t=1 h after m-xylene addition, the numerical distributions reproduce well these blurred and largely heterogeneous responses. Note that except for XylR production, the rest of reactions and parameters in the model are the same as those used for the wild-type conditions in exponential phase (Figures 3 and 4). Addition of IPTG, and subsequent augmented expression of XylR, reduced heterogeneity and yielded clear unimodal distributions, with faster induction times and less variability as the inducer concentration increases (Figure 7, middle and bottom panels). Taken together, these results accredit the role of XylR as a limiting factor controlling the variability in the P. putida response to aromatic carbon sources, a quality that is encoded in the growth status of cells mostly through changes in post-transcriptional repression mediated by the global regulator Crc.
Figure 6.
Genetic strategy to conditionally overexpress XylR in P. putida upon IPTG addition. (a) Organization of the mini-Tn7 carrying xylR under control of lacIq/Ptrc system and assembled in vector pTn7-M to yield the mini-Tn7 [L-Gm lacIq/Ptrc → xylR-R] transposon delivery plasmid pIB. (b) The business part of the mini tranposon is then inserted into the attTn7 site (close to the glmS gene) of recipient strain P. putida KT [Pu-GFP], which bears a chromosomally determined transcriptional Pu-GFP fusion (see Materials and methods). In the resulting strain (P. putida KT-IB1), XylR production depends on the concentration of IPTG added to the medium, while m-xylene is added to activate XylR for triggering Pu promoter activity.
Figure 7.
Overexpression of XylR reduces cellular heterogeneity in exponential phase. Experimental (left) and numerical (right) distribution of GFP outputs in exponential phase conditions in Pu-GFP engineered strain P. putida KT-IB1, which is inserted with the lacIq/Ptrc → xylR genetic construct sketched in Figure 6. Even without inducer ([IPTG]=0), the basal activity of the Ptrc promoter is ~2–3-fold than that of the native Pr promoter, blurring its bimodality but still producing very heterogeneous responses. Addition of IPTG reduces variability in exponential phase and yields only unimodal distributions.
Conclusion
To the best of our knowledge, this report provides for the first time a mechanistic basis for typical population phenomena involving phenotypic diversification in environmental bacteria, specifically the control of metabolic response variability linked to the cellular growth state. Changes in gene expression and growth rate are intimately coupled processes following nutrient changes (Scott et al., 2010; Scott and Hwa, 2011). This coupling has been shown to underlie phenotypic variability through bistability in bacteria evolving antibiotic resistance (Deris et al., 2013) as well as in stem cell differentiation (Kueh et al., 2013). The bistable behaviour observed in these examples and others (Sureka et al., 2008; Tan et al., 2009; Ghosh et al., 2011) is of deterministic type, due to a growth mediated positive feedback in the expression of certain genes that stabilizes particular expression states. Here we inspect a different effect mediated by the growth status of the cell: noise-induced bimodality in a bacterial metabolic response. As growth state has a global impact on gene expression and cellular components, it is natural to expect that changes in global factors modulate gene expression noise in a cell-wide manner (extrinsic noise; Guido et al., 2007). In this work, we demonstrate that variable phenotypic responses can be also modulated by growth coupling to the expression of specific genes (intrinsic noise). This modulation is achieved by the alternative actions of two global regulators (Crc and IHF), the levels of which are linked to the growth status of the cell. On one hand, the opposing effects of Crc and IHF act as a homoeostatic mechanism during cell growth balancing the activity of the Pu promoter, whose levels are maintained on average along the growth curve. On the other hand, the different regulatory mechanisms and strengths of IHF and Crc on the Pu activator, XylR, produce vast differences in heterogeneity around these average Pu levels. Strong post-transcriptional repression of the xylR mRNA mediated by Crc during exponential phase keeps active XylR at very low copy numbers, causing slow activation dynamics of the Pu promoter and bimodal responses after m-xylene induction. The release of post-transcriptional inhibition due to diminished Crc levels during stationary phase drastically reduces heterogeneity in this growth state. Our study highlights the importance of cell physiology and internal composition and its impact on phenotypic variability. Also, the model presented here emphasizes the need to understand regulatory complexity under the light of population behaviour (Silva-Rocha et al., 2013) and community function (Ackermann, 2013; Kotte et al., 2014; Gallie et al., 2015; Zimmermann et al., 2015) rather than just individual benefit.
Acknowledgments
We thank Angel Goñi for critical reading of the manuscript. This work was supported by the CAMBIOS Program of the Spanish Ministry of Economy and Competitiveness, the ARISYS, EVOPROG and EMPOWERPUTIDA Contracts of the EU, The ERANET-IB and the PROMT Project of the CAM. RG acknowledges financial support from the Spanish Ministry of Economy and Competitiveness (grant BFU2013-45918-R) and the AIRBIOTA project of the CAM.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Acar M, Becskei A, van Oudenaarden A. (2005). Enhancement of cellular memory by reducing stochastic transitions. Nature 435: 228–232. [DOI] [PubMed] [Google Scholar]
- Acar M, Mettetal JT, van Oudenaarden A. (2008). Stochastic switching as a survival strategy in fluctuating environments. Nat Genet 40: 471–475. [DOI] [PubMed] [Google Scholar]
- Ackermann M. (2013). Microbial individuality in the natural environment. ISME J 7: 465–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann M. (2015). A functional perspective on phenotypic heterogeneity in microorganisms. Nat Rev Microbiol 13: 497–508. [DOI] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. (2004). Bacterial persistence as a phenotypic switch. Science 305: 1622–1625. [DOI] [PubMed] [Google Scholar]
- Balazsi G, van Oudenaarden A, Collins JJ. (2011). Cellular decision making and biological noise: from microbes to mammals. Cell 144: 910–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein JA, Khodursky AB, Lin PH, Lin-Chao S, Cohen SN. (2002). Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc Natl Acad Sci USA 99: 9697–9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake WJ, Balazsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y et al. (2006). Phenotypic consequences of promoter-mediated transcriptional noise. Mol Cell 24: 853–865. [DOI] [PubMed] [Google Scholar]
- Choi KH, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR et al. (2005). A Tn7-based broad-range bacterial cloning and expression system. Nat Methods 2: 443–448. [DOI] [PubMed] [Google Scholar]
- Choi KH, Schweizer HP. (2006). mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc 1: 153–161. [DOI] [PubMed] [Google Scholar]
- de Las Heras A, de Lorenzo V. (2012). Engineering whole-cell biosensors with no antibiotic markers for monitoring aromatic compounds in the environment. Methods Mol Biol 834: 261–281. [DOI] [PubMed] [Google Scholar]
- de Las Heras A, Fraile S, de Lorenzo V. (2012). Increasing signal specificity of the TOL network of Pseudomonas putida mt-2 by rewiring the connectivity of the master regulator XylR. PLoS Genet 8: e1002963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V, Eltis L, Kessler B, Timmis KN. (1993). Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123: 17–24. [DOI] [PubMed] [Google Scholar]
- de Lorenzo V, Herrero M, Jakubzik U, Timmis KN. (1990). Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in Gram-negative eubacteria. J Bacteriol 172: 6568–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V, Timmis KN. (1994). Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol 235: 386–405. [DOI] [PubMed] [Google Scholar]
- Deris JB, Kim M, Zhang Z, Okano H, Hermsen R, Groisman A et al. (2013). The innate growth bistability and fitness landscapes of antibiotic-resistant bacteria. Science 342: 1237435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dublanche Y, Michalodimitrakis K, Kümmerer N, Foglierini M, Serrano L. (2006). Noise in transcription negative feedback loops: simulation and experimental analysis. Mol Syst Biol 2: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraile S, Roncal F, Fernandez LA, de Lorenzo V. (2001). Monitoring intracellular levels of XylR in Pseudomonas putida with a single-chain antibody specific for aromatic-responsive enhancer-binding proteins. J Bacteriol 183: 5571–5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie J, Libby E, Bertels F, Remigi P, Jendresen CB, Ferguson GC et al. (2015). Bistability in a metabolic network underpins the de novo evolution of colony switching in Pseudomonas fluorescens. PLoS Biol 13: e1002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmendia J, de las Heras A, Galvão TC, de Lorenzo V. (2008). Tracing explosives in soil with transcriptional regulators of Pseudomonas putida evolved for responding to nitrotoluenes. Microb Biotechnol 1: 236–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Sureka K, Ghosh B, Bose I, Basu J, Kundu M. (2011). Phenotypic heterogeneity in mycobacterial stringent response. BMC Syst Biol 5: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie DT. (1977). Exact stochastic simulation of coupled chemical reactions. J Phys Chem 81: 2340–2361. [Google Scholar]
- Grimbergen AJ, Siebring J, Solopova A, Kuipers OP. (2015). Microbial bet-hedging: the power of being different. Curr Opin Microbiol 25: 67–72. [DOI] [PubMed] [Google Scholar]
- Guantes R, Cayrol B, Busi F, Arluison V. (2012). Positive regulatory dynamics by a small noncoding RNA: speeding up responses under temperature stress. Mol Biosyst 8: 1707–1715. [DOI] [PubMed] [Google Scholar]
- Guido NJ, Lee P, Wang X, Elston TC, Collins JJ. (2007). A pathway and genetic factors contributing to elevated gene expression noise in stationary phase. Biophys J 93: L55–L57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaern M, Elston TC, Blake WJ, Collins JJ. (2005). Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet 6: 451–464. [DOI] [PubMed] [Google Scholar]
- Kotte O, Volkmer B, Radzikowski JL, Heinemann M. (2014). Phenotypic bistability in Escherichia coli's central carbon metabolism. Mol Sys Biol 10: 736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueh HY, Champhekar A, Nutt SL, Elowitz MB, Rothenberg EV. (2013). Positive feedback between PU.1 and the cell cycle controls myeloid differentiation. Science 341: 670–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussell E, Leibler S. (2005). Phenotypic diversity, population growth, and information in fluctuating environments. Science 309: 2075–2078. [DOI] [PubMed] [Google Scholar]
- Levine E, Zhang Z, Kuhlman T, Hwa T. (2007). Quantitative characteristics of gene regulation by small RNA. PLoS Biol 5: e229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares JF, Moreno R, Fajardo A, Martinez-Solano L, Escalante R, Rojo F et al. (2010). The global regulator Crc modulates metabolism, susceptibility to antibiotics and virulence in Pseudomonas aeruginosa. Environ Microbiol 12: 3196–3212. [DOI] [PubMed] [Google Scholar]
- Maamar H, Dubnau D. (2005). Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol Microbiol 56: 615–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar H, Raj A, Dubnau D. (2007). Noise in gene expression determines cell fate in Bacillus subtilis. Science 317: 526–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques S, Gallegos MT, Manzanera M, Holtel A, Timmis KN, Ramos JL. (1998). Activation and repression of transcription at the double tandem divergent promoters for the xylR and xylS genes of the TOL plasmid of Pseudomonas putida. J Bacteriol 180: 2889–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García E, Calles B, Arévalo-Rodríguez M, de Lorenzo V. (2011). pBAM1: an all-synthetic genetic tool for analysis and construction of complex bacterial phenotypes. BMC Microbiol 11: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P, Goyal S, Wingreen NS. (2008). A quantitative comparison of sRNA-based and protein-based gene regulation. Mol Sys Biol 4: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milojevic T, Grishkovskaya I, Sonnleitner E, Djinovic-Carugo K, Blasi U. (2013). The Pseudomonas aeruginosa catabolite repression control protein Crc is devoid of RNA binding activity. PLoS One 8: e64609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno R, Fonseca P, Rojo F. (2010). The Crc global regulator inhibits the Pseudomonas putida pWW0 toluene/xylene assimilation pathway by repressing the translation of regulatory and structural genes. J Biol Chem 285: 24412–24419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno R, Hernandez-Arranz S, La Rosa R, Yuste L, Madhushani A, Shingler V et al. (2015). The Crc and Hfq proteins of Pseudomonas putida cooperate in catabolite repression and formation of ribonucleic acid complexes with specific target motifs. Environ Microbiol 17: 105–118. [DOI] [PubMed] [Google Scholar]
- Moreno R, Martinez-Gomariz M, Yuste L, Gil C, Rojo F. (2009). The Pseudomonas putida Crc global regulator controls the hierarchical assimilation of amino acids in a complete medium: evidence from proteomic and genomic analyses. Proteomics 9: 2910–2928. [DOI] [PubMed] [Google Scholar]
- Nevozhay D, Adams RM, Murphy KF, Josic K, Balázsi G. (2009). Negative autoregulation linearizes the dose-response and suppresses the heterogeneity of gene expression. Proc Natl Acad Sci USA 106: 5123–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New AM, Cerulus B, Govers SK, Perez-Samper G, Zhu B, Boogmans S et al. (2014). Different levels of catabolite repression optimize growth in stable and variable environments. PLoS Biol 12: e1001764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL et al. (2006). Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441: 840–846. [DOI] [PubMed] [Google Scholar]
- Ochab-Marcinek A, Tabaka M. (2010). Bimodal gene expression in noncooperative regulatory systems. Proc Natl Acad Sci USA 107: 22096–22101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Lim HN, Shraiman BI, Van Oudenaarden A. (2004). Multistability in the lactose utilization network of Escherichia coli. Nature 427: 737–740. [DOI] [PubMed] [Google Scholar]
- Perez-Martin J, de Lorenzo V. (1996). VTR expression cassettes for engineering conditional phenotypes in Pseudomonas: activity of the Pu promoter of the TOL plasmid under limiting concentrations of the XylR activator protein. Gene 172: 81–86. [DOI] [PubMed] [Google Scholar]
- Proshkin S, Rahmouni AR, Mironov A, Nudler E. (2010). Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328: 504–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A. (2008). Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135: 216–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK. (2005). Noise in gene expression: origins, consequences, and control. Science 309: 2010–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Manzano A, Yuste L, Rojo F. (2005). Levels and activity of the Pseudomonas putida global regulatory protein Crc vary according to growth conditions. J Bacteriol 187: 3678–3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor: New York, USA. [Google Scholar]
- Schweizer HP. (2001). Vectors to express foreign genes and techniques to monitor gene expression in Pseudomonads. Curr Opin Biotechnol 12: 439–445. [DOI] [PubMed] [Google Scholar]
- Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T. (2010). Interdependence of cell growth and gene expression: origins and consequences. Science 330: 1099–1102. [DOI] [PubMed] [Google Scholar]
- Scott M, Hwa T. (2011). Bacterial growth laws and their applications. Curr Opin Biotechnol 22: 559–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Rocha R, de Lorenzo V. (2011). A composite feed-forward loop I4-FFL involving IHF and Crc stabilizes expression of the XylR regulator of Pseudomonas putida mt-2 from growth phase perturbations. Mol Biosyst 7: 2982–2990. [DOI] [PubMed] [Google Scholar]
- Silva-Rocha R, de Lorenzo V. (2012). Stochasticity of TOL plasmid catabolic promoters sets a bimodal expression regime in Pseudomonas putida mt-2 exposed to m-xylene. Mol Microbiol 86: 199–211. [DOI] [PubMed] [Google Scholar]
- Silva-Rocha R, Perez-Pantoja D, de Lorenzo V. (2013). Decoding the genetic networks of environmental bacteria: regulatory moonlighting of the TOL system of Pseudomonas putida mt-2. ISME J 7: 229–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So L-h, Ghosh A, Zong C, Sepulveda LA, Segev R, Golding I. (2011). General properties of transcriptional time series in Escherichia coli. Nat Genet 43: 554–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. (2006). An excitable gene regulatory circuit induces transient cellular differentiation. Nature 440: 545–550. [DOI] [PubMed] [Google Scholar]
- Sureka K, Ghosh B, Dasgupta A, Basu J, Kundu M, Bose I. (2008). Positive feedback and noise activate the stringent response regulator rel in mycobacteria. PLoS One 3: e1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C, Marguet P, You L. (2009). Emergent bistability by a growth-modulating positive feedback circuit. Nat Chem Biol 5: 842–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J et al. (2010). Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329: 533–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To TL, Maheshri N. (2010). Noise can induce bimodality in positive transcriptional feedback loops without bistability. Science 327: 1142–1145. [DOI] [PubMed] [Google Scholar]
- Valls M, Buckle M, de Lorenzo V. (2002). In vivo UV laser footprinting of the Pseudomonas putida sigma 54 Pu promoter reveals that integration host factor couples transcriptional activity to growth phase. J Biol Chem 277: 2169–2175. [DOI] [PubMed] [Google Scholar]
- Valls M, de Lorenzo V. (2003). Transient XylR binding to the UAS of the Pseudomonas putida sigma 54 promoter Pu revealed with high intensity UV footprinting in vivo. Nucleic Acids Res 31: 6926–6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening JW, Stewart EJ, Berngruber TW, Taddei F, Kuipers OP, Hamoen LW. (2008). Bet-hedging and epigenetic inheritance in bacterial cell development. Proc Natl Acad Sci USA 105: 4393–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturelli OS, Zuleta I, Murray RM, El-Samad H. (2015). Population diversification in a yeast metabolic program promotes anticipation of environmental shifts. PLoS Biol 13: e1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel U, Jensen KF. (1994). The RNA chain elongation rate in Escherichia coli depends on the growth rate. J Bacteriol 176: 2807–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volfson D, Marciniak J, Blake WJ, Ostroff N, Tsimring LS, Hasty J. (2006). Origins of extrinsic variability in eukaryotic gene expression. Nature 439: 861–864. [DOI] [PubMed] [Google Scholar]
- Zimmermann M, Escrig S, Hubschmann T, Kirf MK, Brand A, Inglis RF et al. (2015). Phenotypic heterogeneity in metabolic traits among single cells of a rare bacterial species in its natural environment quantified with a combination of flow cell sorting and NanoSIMS. Front Microbiol 6: 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zobel S, Benedetti I, Eisenbach L, de Lorenzo V, Wierckx N, Blank LM. (2015). Tn7-Based device for calibrated heterologous gene expression in Pseudomonas putida. ACS Synth Biol; e-pub ahead of print 20 July 2015. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.