Abstract
Melanoma is one of the most lethal forms of skin cancer due to its early metastatic spread. The variant form of CD44 (CD44v), a cell surface glycoprotein, is highly expressed on metastatic melanoma. The mechanisms of regulation of CD44 alternative splicing in melanoma and its pathogenic contributions are so far poorly understood. Here, we investigated the expression level of CD44 in a large set of melanocytic lesions at different stages. We found that the expression of CD44v8-10 and a splicing factor, U2AF2, is significantly increased during melanoma progression, while CD82/KAI1, a tetraspanin family of tumor suppressor, is reduced in metastatic melanoma. CD44v8-10 and U2AF2 expressions which are negatively correlated with CD82 levels are dramatically elevated in primary melanoma compared with dysplastic nevi and further increased in metastatic melanoma. We also showed that patients with higher CD44v8-10 and U2AF2 expression levels tended to have shorter survival. By using both in vivo and in vitro assays, we demonstrated that CD82 inhibits the production of CD44v8-10 on melanoma. Mechanistically, U2AF2 is a downstream target of CD82 and in malignant melanoma facilitates CD44v8-10 alternative splicing. U2AF2-mediated CD44 isoform switch is required for melanoma migration in vitro and lung and liver metastasis in vivo. Notably, overexpression of CD82 suppresses U2AF2 activity by inducing U2AF2 ubiquitination. In addition, our data suggested that enhancement of melanoma migration by U2AF2-dependent CD44v8-10 splicing is mediated by Src/FAK/RhoA activation and formation of stress fibers as well as CD44-E-selectin binding reinforcement. These findings uncovered a hitherto unappreciated function of CD82 in severing the linkage between U2AF2-mediated CD44 alternative splicing and cancer aggressiveness, with potential prognostic and therapeutic implications in melanoma.
Keywords: CD82, U2AF2, CD44v, alternative splicing
Introduction
Melanoma is one of the most deadly and malignant tumors with high propensity to metastasize to distant organs 1, 2. Previously, it was shown that metastatic melanoma cells possess aberrantly high levels of CD44v on their surface that can be associated with E-selectin on endothelium to mediate cell adhesion in flow and induce endothelial junction breakdown 3-5. CD44 is a glycoprotein displaying size heterogeneity due to alternative splicing and differential post-translational glycosylations6. Standard CD44 (CD44s) is generated by splicing out all variable exons, while variant forms of CD44 (CD44v) are created by selectively including multiple variant exons (exon 6-14). The isoform switching from CD44s to CD44v in response to certain stimuli confers metastatic potentials on tumors. The process of CD44v pre-mRNA splicing is composed of spliceosome complex assembly favoring inclusion of variant exons7. It has been shown that CD44 variable exon splicing is mediated by Epithelial splicing regulatory protein (ESRP) in immortalized epithelial cells, breast cancer and head & neck carcinoma cells 8-10. ESRP1 and ESRP2 were shown to regulate splicing of v8-v10 and v6-v10 exons of CD44, maintaining cell epithelial phenotypes11. Other splicing factors, like Tra2β and SRm160, also mediate CD44 alternative splicing in gastric epithelial cells and Hela cells 12, 13. It was recently reported that primary melanoma with tendency for brain metastasis expresses high levels of CD44v6. Splicing factors, ESRP1, ESRP2, PTBP1 and U2 snRNP auxiliary factor (U2AF2), correlate with CD44v6 expression in primary melanoma, and ESRP1 knockdown in melanoma cells resected from lymph node metastases decreases CD44v6 expression. Despite all these discoveries, very little is known about the functional roles of splicing factors in melanoma metastasizing to other organs, especially lung and liver, and the exact mechanisms of CD44 splicing in melanoma.
The KAI1/CD82, firstly identified as a tumor suppressor in prostate cancer, is a member of the tetraspanin superfamily with a wide spectrum of biological functions14. Studies demonstrated that CD82 limits tumor progression and metastasis in a number of ways. CD82 can inhibit cell motility by reorganizing specific tetraspanin-enriched web, called TERM15. TERM facilitates CD82 association with various receptors and cytoplasmic signaling molecules, modulating lateral cross-talk in the plasma membrane16. CD82 can be associated with EGFR and regulate its ubiquitylation, thereby inhibiting communications between heparin sulfate proteoglycans and ligand-bound EGFR17. The modulatory effect of CD82 is also dependent on trafficking of CD82 in endocytic organelles like late endosomes and lysosomes17, 18. Noteworthy, CD82 participates in regulating angiogenesis by disturbing the interaction among CD44, ganglioside and lipid rafts, leading to enhanced clathrin-independent endocytosis of CD4419. Nevertheless, it is unknown whether CD82 regulates melanoma migration by altering the structure and function of CD44. In the current study, we demonstrated that ectopic expression of CD82 in metastatic melanoma reduces tumor migration and spontaneous metastasis by decreasing the ratio of CD44v to CD44s. In addition, CD82 inhibits CD44 alternative splicing by inducing U2AF2 ubiquitination.
Results
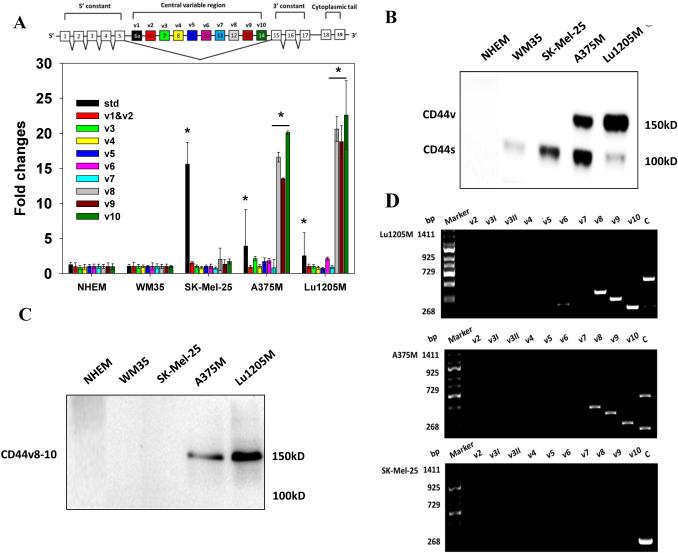
CD44v8-10 is upregulated during melanoma progression and correlated with poor prognosis
CD44 variant isoforms are upregulated in cancer stem cell-like cells and advanced cancer, playing roles in tumor adhesion, hematogenous metastasis, drug resistance and tumorigenesis 6, 20-23. To identify the CD44 isoform present on malignant melanoma, real-time PCR was employed to measure variable exon selections in Lu1205M, A375M 2, SK-Mel-25 and WM35 cells with different metastatic potentials (Table 1) as well as Normal Human Epidermal Melanocytes (NHEMs). Lu1205M cells were derived from Lu1205 cells 4 and underwent extensive aggressiveness selection. Therefore, Lu1205M cells exhibited high malignant and metastatic properties with elevated potentials for colonizing lungs and livers (Table 1). Real-time PCR revealed that SK-Mel-25 cells expressed appreciable amounts of CD44s but low levels of variable exons, while exons 12-14 (v8-v10) were selectively expressed on A375M and Lu1205M cells (Fig. 1A). The performance of each primer set was evaluated with CD44v2-10-expressing keratinocyte HaCaT cells. All primer sets efficiently amplified target exons in HaCaT mRNA (Supplementary Fig.1A). PCR results were validated with Western blotting (2C5 antibody) which demonstrated that NHEM and WM35 don’t express any CD44 molecules, SK-Mel-25 only expresses CD44s yielding an intense 100 kD band, A375M expresses the 100 kD CD44s and a 150 kD protein, and high levels of the 150 kD protein were also detectable on Lu1205M cells (Fig. 1B). PCR results motivated us to surmise that the 150 kD protein is CD44v8-10. To verify the status of the protein, we generated a CD44v8-10 epitope-specific antibody (Supplementary Fig. 2A-B). This antibody specifically recognized the 150 kD protein without any cross-reactivity with other proteins on Lu1205M and A375M cells, suggesting that the protein is CD44v8-10 (Fig. 1C). The lack of reactivity of VFF-18 towards any epitopes in SK-Mel-25, A375M and Lu1205M samples implies that melanoma cells don’t express detectable amounts of CD44v6 protein (Supplementary Fig. 3A). A more precise determination of the exon composition of variant isoforms was achieved by the run-off analysis which verified the continuous alignment of v8, v9 and v10 exons up to CD44v8-10 (729 bp) in the transcripts from Lu1205M and A375M cells, while the predominant isoform on SK-Mel-25 cells was CD44s (268 bp) (Fig. 1D). As a positive control, exon-specific primers used in the run-off analysis detected CD44v2-10, CD44v6-10 and CD44v8-10 which are the major CD44v isoforms being expressed by HaCaT (Supplementary Fig. 1B).
Table 1.
Characterization of invasiveness and aggressiveness of melanoma cell lines
| Cell line | Potentials | |||||
|---|---|---|---|---|---|---|
| Invasiona | chemotaxisb | Adhesionc | ||||
| ICAM-1 | CD44 | α4β1
(VLA-4) |
α v β 3 | |||
| Lu1205Md | ++++ | +++ | +++ | ++++ | ++ | +++ |
| Lu1205 | +++ | ++ | +++ | ++ | + | ++ |
| A375M | ++ | ++ | ++ | +++ | ++ | + |
| SK-Mel-25 | + | + | ++ | + | n/a | + |
| WM35 | − | − | + | − | − | − |
Cell invasiveness was measure with transwell assay in a 24-well plate (Corning). 8 μm pore transwell membrane was coated with 1 mg/ml of Matrigel (BD Biosciences).The lower chamber was filled with 20% FBS as chemoattractant. Cells were allowed to migrate for 24 hr.
Cell chemotaxis was determined with a 48 well Boyden chamber with IV collage as chemoattractant. Number of migrated cells were stained with crystal violet and visualized with microscopy.
Cell adhesion was measured in fibronectin-coated 3.5 cm dishes. Cell adhesive molecules were examined with flow cytometry with relevant antibodies.
Cytogenetics analysis revealed Lu1205M cells have hypertriploid range with many unbalanced rearrangements. DNA flow cytometry results show an aneuploidy peak with a value of 1.5 D.I., corresponding to triploid range in Lu1205M. The presence of T1799A B-Raf mutation was detected with PCR.
Figure 1.
CD44v8-10 is highly expressed on metastatic melanoma cell lines. (A) The expressions of central variable regions of CD44 and standard (std) CD44 transcript in NHEM, WM35, SK-Mel-25, A375M and Lu1205M were measured by quantitative real-time PCR with respective primer sets using GAPDH as an endogenous control. Values were expressed as fold changes (mean±SEM, n=3) relative to respective levels in WM35 cells. *p<0.05 compared with WM35. The structure of CD44 pre-mRNA is shown in the insert. (B) Western blotting analysis of the presence of CD44 isoforms on a variety of melanoma cell lines and NHEM with 2C5 antibody. CD44v:150 kD; CD44s: 100 kD. (C) Verification of the status of 150 kD CD44 on melanoma cell lines with CD44v8-10-secific antibody. (D) Run-off analysis of CD44 isoform expression on Lu1205M, A375M and SK-Mel-25. Exon-specific PCRs were performed using 5’ primers for the variant exons (v2-v10) and constant 5’ primer C13, respectively, in combination with 3’ primer C2A. PCR products were resolved in agarose gel. Numbers in the left indicate marker sizes in bp. Representative results are shown from three independent experiments.
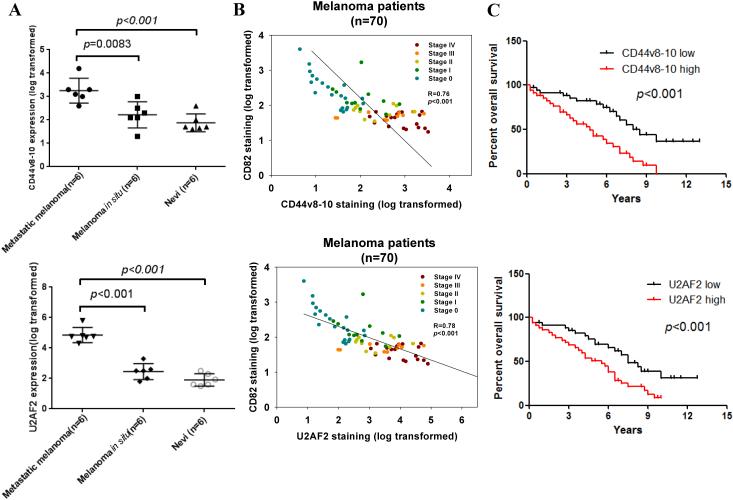
To evaluate the clinical implication of molecular alteration of CD44, we analyzed melanoma patient tissue samples (Table 2). We included melanoma in situ which is designated as TNM stage 0 (TisN0M0) into our analysis. In 70 melanoma patients, CD44v8-10 staining was not correlated with age and gender, while it was significantly correlated with tumor differentiation status and TNM stages. Strong CD44v8-10 expression was found in 20% and 40% cases with stage I and II, while 73% and 80% of stage III and IV melanoma had strong CD44v8-10 expression. CD44v8-10 staining intensity increased in primary melanoma compared with dysplastic nevi and further rose in metastatic melanomas (p<0.001) (Fig. 2A). Nevi, primary melanoma and metastatic melanoma showed a weak expression of CD44v6 (Supplementary Fig. 3B). No significant difference in staining intensity of CD44v6 was found in nevi compared to primary and metastatic melanoma (p>0.05). In analysis of Kaplan-Meier survival curves, we found that elevated CD44v8-10 was significantly correlated with a poorer overall survival of all melanoma patients (Fig. 2C). These data suggest that elevated CD44v8-10 may have pathological roles in melanoma metastasis.
Table 2.
CD44v8-10 expression levels and clinicopathologic characteristics of melanoma Patients
| Total | CD44v8-10 expression* | p-value | ||||
|---|---|---|---|---|---|---|
|
|
||||||
| − | + | ++ | +++ | |||
| Gender | 0.7107a | |||||
| Male | 38 | 2 | 8 | 12 | 16 | |
| Female | 32 | 0 | 7 | 11 | 14 | |
| Age | 0.9789a | |||||
| ≤60 | 31 | 1 | 6 | 11 | 13 | |
| >60 | 39 | 1 | 9 | 12 | 17 | |
| Differentiation | <0.001b | |||||
| High | 19 | 1 | 13 | 3 | 2 | |
| Moderate | 26 | 1 | 1 | 13 | 11 | |
| Poor | 25 | 0 | 1 | 7 | 17 | |
| TNM stage | <0.001b | |||||
| 0 | 19 | 2 | 12 | 5 | 0 | |
| I | 10 | 0 | 3 | 5 | 2 | |
| II | 10 | 0 | 0 | 6 | 4 | |
| III | 11 | 0 | 0 | 3 | 8 | |
| IV | 20 | 0 | 0 | 4 | 16 | |
P value when expression levels were compared using Mann-Whitney test
P value when expression levels were compared using Kruskal-Wallis test
TNM, tumor-node-metastasis staging classification system. From National Comprehensive Cancer Network (NCCN) guidelines.
CD44 expression was scored according to both staining intensity and percentage of CD44v8-10-posititve cells which were evaluated in a blinded manner by 3 independent observers.
CD44v8-10 expression was scored as − (negative), +(weak), ++(moderate) and +++(strong).
Figure 2.
CD44v8-10 expression is positively correlated with U2AF2 expression and negatively correlated with CD82 level in metastatic melanoma tissues. (A) Analysis of expressions of CD44v8-10 and U2AF2 with IHC in nevi from 6 non-melanoma patients and 12 samples of melanoma in situ and metastatic melanoma from melanoma patients. Protein levels were quantified in FFPE tissue samples. Mann-Whitney U test was used. (B) The correlation between CD82 relative expression levels (log transformed) and CD44v8-10 relative expression levels (log transformed) in 70 FFPE melanoma tissue samples from patients with clinical TNM stage 0-IV. Pearson’s R test was used. R- and p-values are calculated as indicated. (C) Association between CD44v8-10 and U2AF2 expression and survival of melanoma patients (n=70). The melanoma patients were stratified according to CD44v8-10 and U2AF2 expression levels into high and low groups. Patients expressing high CD44v8-10 and U2AF2 levels displayed significantly shorter survival (log-rank test).
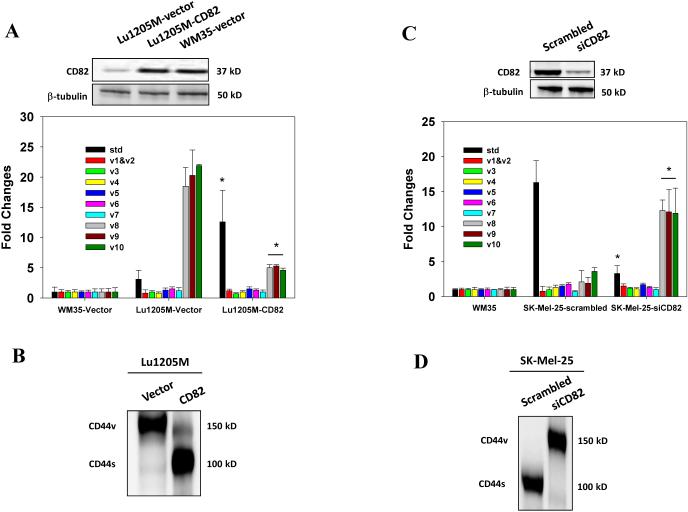
CD82 prevents CD44s/CD44v8-10 isoform switch in melanoma
Previously, it was reported that endothelial CD82 inhibited the cell surface presence of CD4419. Furthermore, downregulation of CD82 is common in advanced human cancer24. Therefore, we examined the effect of modulation of CD82 expression on CD44 isoform expression in melanoma. Compared with WM35, Lu1205M expresses low levels of CD82 (Fig. 3A). When CD82 was overexpressed in Lu1205M, CD44v8-10 was reduced and CD44s upregulated at both mRNA and protein levels (Fig. 3A-B and Supplementary Fig. 4A-B). Conversely, knockdown of endogenous CD82 with siRNA in SK-Mel-25 resulted in a remarkable increase in CD44v8-10 and a concomitant reduction of CD44s expression (Fig. 3C-D). These results suggest that CD82 functions as an important regulator to suppress the isoform switch from CD44s to CD44v.
Figure 3.
Regulation of alternative splicing of CD44 pre-mRNA by CD82. (A) After transfection of WM35 or Lu1205M with a plasmid encoding CD82 or vector alone, amounts of CD44 standard and variable exons (v1-v10) in the central viable region were measured with real-time PCR. Values were expressed as fold changes (mean±SEM, n=3) relative to respective levels in WM35-vector cells. *p<0.05 compared with Lu1205M-vector. CD82 transfection efficiency was assessed by Western blotting. (B) The effect of CD82 overexpression on CD44 isoform switches in Lu1205M was measured by Western blotting. (C) The amounts of standard CD44 standard and variable exons (v1-v10) in SK-Mel-25 cells which received scrambled or CD82 siRNA were measured with quantitative real-time PCR. Values were expressed as fold changes (mean±SEM, n=3) relative to respective levels in WM35 cells. *p<0.05 compared with SK-Mel-25-siCD82. CD82 knockdown efficiency was assessed by Western blotting. (D) The effect of CD82 knockdown on CD44 isoform switches in SK-Mel-25 cells was measured by Western blotting. Representative results are shown from three independent experiments.
Then, we determined the association between CD82 and CD44v8-10 expression in clinical samples. CD82 level tended to drop as the TNM staging advanced and high abundance of CD82 correlated with a better relapse-free survival (Fig. 2B). In addition, CD82 expression was negatively correlated with levels of CD44v8-10 as detected by CD44v8-10 antibody and commercial CD44v7/8, CD44v9 and CD44v10 antibodies (p<0.001, R=0.76) (Fig. 2B and Supplementary Fig. 5).
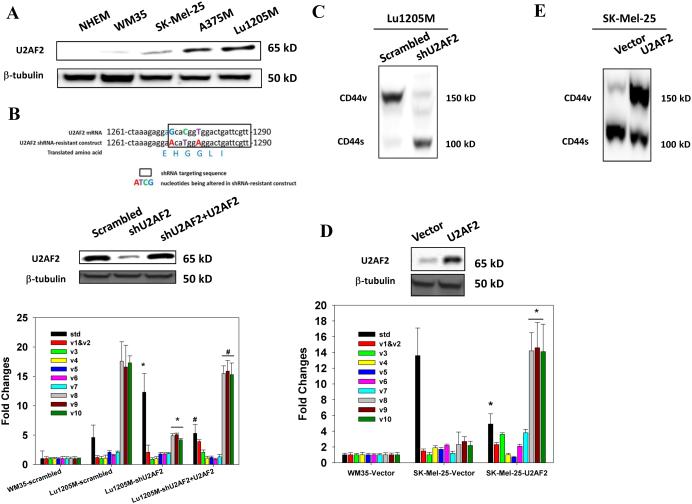
Splicing factor U2AF2 promotes CD44 alternative splicing in metastatic melanoma
To investigate whether CD82 suppresses CD44v8-10 expression through regulation of splicing, we silenced a set of splicing factors, Tra2β, SRp20, ESRP1, YB-1, SRm160, Sam68 and U2AF2, which have been shown to mediate CD44v splicing 10, 12, 13, 25, 26. Only knockdown of U2AF2 dramatically reduced CD44v8-10 expression (Supplementary Fig. 6). Of five cell lines, U2AF2 expression was associated with metastatic potentials with the highest level found on Lu1205M (Fig. 4A). U2AF2 knockdown with shRNA significantly increased CD44s and attenuated CD44v8-10 expressions (Fig. 4B-C, Supplementary Fig. 4C-D). Then, we generated a shRNA-resistant U2AF2 construct which contains three mutated nucleotides in shRNA seeding region without changing amino acid sequence (Fig. 4B upper panel)27. Overexpression of shRNA-resistant U2AF2 in U2AF2-silenced Lu1205M rescued CD44v8-10 expression, suggesting that U2AF2 is the splicing factor mediating CD44s/CD44v isoform switch (Fig. 4B middle and lower panels). To further evaluate the role of U2AF2 in CD44 alternative splicing, U2AF2 construct was transfected into SK-Mel-25 cells which expressed low levels of this splicing factor. Overexpression of U2AF2 triggered CD44v8-10 expression at both mRNA and protein levels (Fig. 4D-E).
Figure 4.
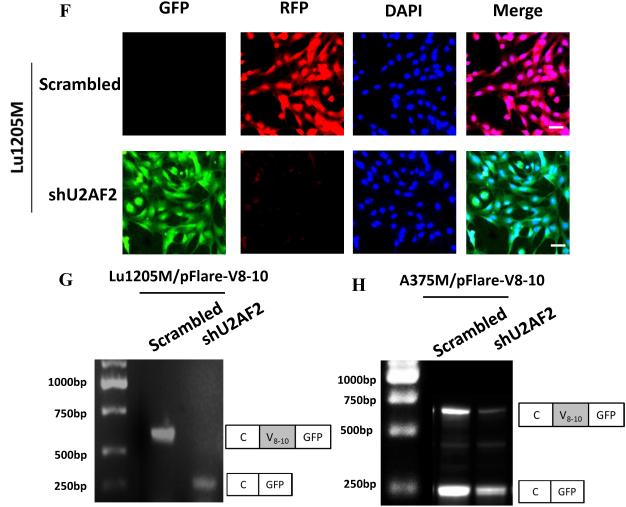
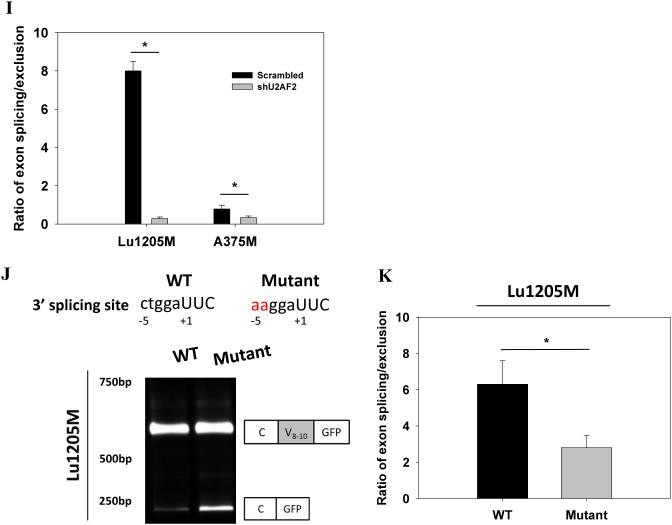
U2AF2 promotes exon v8-10 splicing of CD44 pre-mRNA. (A) Western blotting analysis of the presence of U2AF2 on a variety of metastatic melanoma cell lines and NHEM. (B) Top panel, Design and sequence of shRNA-resistant U2AF2 construct are shown. Middle panel, U2AF2 expression in Lu1205M transfected with different constructs was assessed by Western blotting. Bottom panel, The amounts of CD44 standard and variable exons (v1-v10) in WM35 or Lu1205M cells which received scrambled shRNA, shU2AF2 or shU2AF2 plus shRNA-resistant U2AF2 were measured with real-time PCR. Values were expressed as fold changes (mean±SEM, n=3) relative to respective levels in WM35-scrambled cells. *p<0.05 compared with Lu1205M-scrambled. #p<0.05 compared with Lu1205M-shU2AF2. (C) shU2AF2 transfection suppressed CD44v8-10 expression and increased CD44s expression as assessed by Western blotting. (D) The amounts of CD44 standard and variable exons (v1-v10) in WM35 or SK-Mel-25 cells which received vector or U2AF2 were measured with quantitative real-time PCR. Values were expressed as fold changes (mean±SEM, n=3) relative to respective levels in WM35-vector cells. *p<0.05 compared with SK-Mel-25-vector. The U2AF2 expression in SK-Mel-25 cells transfected with different constructs was assessed by Western blotting. (E) The effect of U2AF2 overexpression on CD44 isoform switches in SK-Mel-25 cells was measured by western blotting. (F) Fluorescence microscopy analysis of Lu1205M cells expressing pFlare-V8-10 minigene. Lu1205M/pFlare-V8-10 cells were transfected with either scrambled shRNA or shU2AF2. Bar=50 μm. (G-I) Real-time-PCR analysis of V8-10 exon splicing of CD44 from pFlare-V8-10-expressing stable Lu1205M and A375M cell lines which were transfected with scrambled or U2AF2 shRNA. *p<0.05 compared with scrambled. The V8-10 included product is C-V8-10-GFP; the V8-10 skipped product is C-GFP. (J-K) U2AF2 mediates CD44v8-10 splicing by binding to 3’ polypyrimidine tract flanking V8 exon. Sequences of 3’-splicing site in wild type (WT) and mutant minigenes are shown in the upper panel. Exonic and intronic regions are indicated in uppercase and lowercase, respectively. Alternative splicing events were monitored by real-time PCR. (K) The ratio of the amount of exon V8-10 spliced variant (splicing) to that of V8-10 skipped variant (exclusion) is shown among samples. Data is represented as mean±SEM from three independent assays. *p<0.05 compared with WT.
To gain more insight into the molecular basis for the function of U2AF2 in regulating CD44 splicing, we utilized a minigene with pFlare-RFP/GFP reporter system28 where start condon for GFP is split by inserting V8-10 exon and its flanking introns. This insertion results in GFP expression when V8-10 exon is skipped and RFP synthesis when it is included. Our results showed that RFP was dominantly expressed, suggesting that CD44v8-10 is spliced in control Lu1205M (Fig.4F). In contrast, knockdown of U2AF2 in Lu1205M attenuated RFP signal and increased GFP expression. Knockdown of U2AF2 in Lu1205M and A375M promoted exon skipping and significantly reduced the ratio of V8-10 exon splicing/exclusion (Fig. 4G-I). U2AF2 can bind to weak polypyrimidine tract in 3’-splicing site to facilitate exon splicing26. To find whether 3’-splicing site of V8-10 contains U2AF2 response element, we introduced mutations in 3’ polypyrimidine tract. Point mutations from CT to AA at −4 and −5 positions sharply comprised the stimulative effect of U2AF2 on V8-10 splicing (Fig. 4J-K). These results imply that by targeting 3’ polypyrimidine tract, U2AF2 enforces CD44 v8-10 splicing in melanoma.
U2AF2 participates in CD82-mediated CD44 alternative splicing
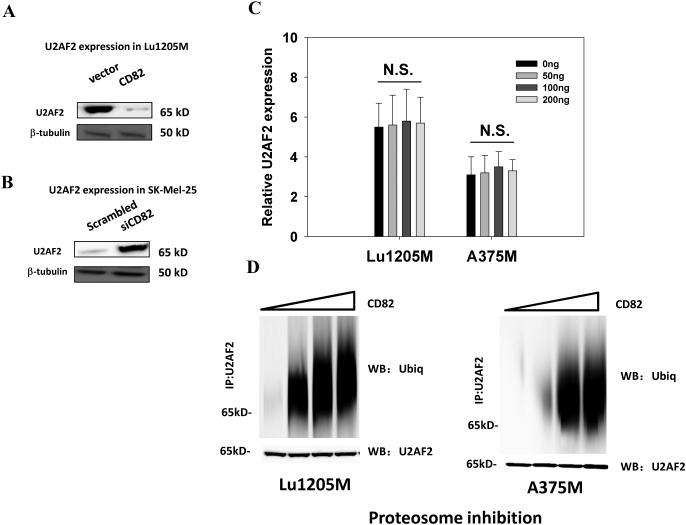
To demonstrate that U2AF2 is under control of CD82 to mediate CD44 splicing, U2AF2 expression was examined in Lu1205M following CD82 overexpression. Ectopic expression of CD82 in Lu1205M diminished U2AF2 expression (Fig. 5A). On the other hand, knockdown of CD82 by siRNA in SK-Mel-25 resulted in an increased expression of U2AF2 (Fig. 5B). Surprisingly, although CD82 knockdown in Lu1205M and A375M induced a dramatic change of protein levels of U2AF2, mRNA levels did not correlate with this change (Fig. 5C). Therefore, CD82 may modulate U2AF2 activities at posttranslational level. Accordingly, we tested the hypothesis that CD82 reduces U2AF2 protein stability by inducing its ubiquitination. Our results showed that CD82 increased U2AF2 ubiquitination in a dose-dependent manner in both Lu1205M and A375M (Fig. 5D).
Figure 5.
CD82 suppresses U2AF2 activity by mediating U2AF2 ubiquitination. (A-B) The effects of CD82 overexpression in Lu1205M and CD82 knockdown in SK-Mel-25 on U2AF2 expression were measured by Western blotting. (C) Ectopic expression of CD82 has no effect on U2AF2 mRNA levels as measured by real-time PCR using GAPDH as an endogenous control. Lu1205M and A375M cells were transfected with 0, 50, 100 and 200 ng CD82 before mRNA was extracted. Data is represented as mean±SEM from three independent assays. N.S., not significant. (D) Ectopic expression of CD82 enhances U2AF2 ubiquitination. Lu1205M and A375M cells were transfected with 0, 50, 100 and 200 ng CD82 and treated with 10 μM MG132 for 6 hr before harvesting. U2AF2 ubiquitination was detected by immunoprecipitation with U2AF2 antibody and immunoblotting with anti-ubiquitin antibody (ubiq). The results of one of three independent experiments are shown.
As with CD44v8-10, enhanced expression of U2AF2 was observed during clinical melanoma progression (Fig. 2A). U2AF2 level was drastically increased in primary melanoma compared with dysplastic nevi (P <0.001) and further upregulated in metastatic melanoma (P<0.001). In addition, decreased U2AF2 staining was strongly associated with a better patient survival (Fig. 2C). The 5-year survival rate dropped from 75% in patients with low U2AF2 expression to 48% in those with positive U2AF2 expression. There was a significant negative correlation between CD82 and U2AF2 levels in melanoma samples (Fig. 2B). Altogether, these results suggest that in metastatic melanoma, downregulation of CD82 expression elevated U2AF2 expression, which increased the ratio of CD44v8-10 to CD44s.
CD82 inhibits melanoma motility by interfering U2AF2-mediated CD44v8-10 splicing
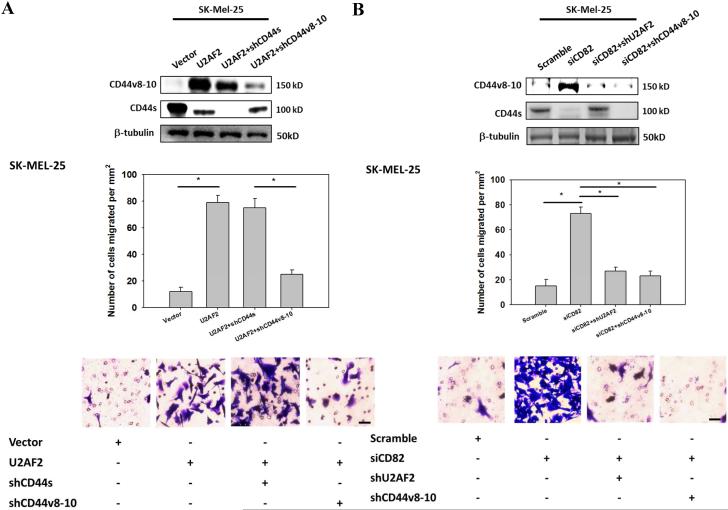
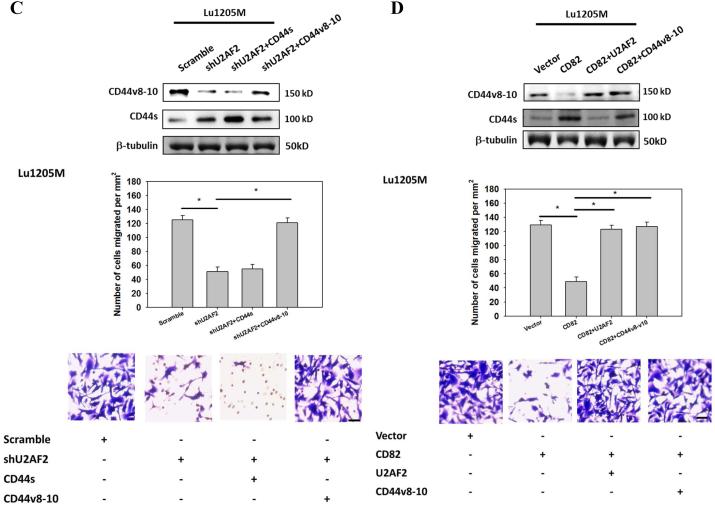
Since CD44v isoform expression in cancer may contribute to tumor adhesion, migration and metastasis, we next assessed the roles of CD82 in CD44v8-10-dependent cell motility6, 29. Overexpression of U2AF2 significantly enhanced SK-Mel-25 migration (Fig. 6A). However, the pro-migratory effect of U2AF2 can be attenuated by CD44v8-10 shRNA but not by CD44s shRNA. When endogenous CD82 in SK-MEL-25 were inhibited, cell migration was enhanced by 5-fold (Fig. 6B). Knockdown of U2AF2 or CD44v8-10 markedly reduced the migration potentials of SK-Mel-25/siCD82 cells. Either knockdown of U2AF2 or overexpression of CD82 in Lu1205M suppressed Lu1205M motility (Fig. 6C and D). Restoration of CD44v8-10 but not CD44s rescued the suppressive effect of shU2AF2 on melanoma migration (Fig. 6C). Similarly, migratory potentials of Lu1205M/CD82 were increased by reconstitution of U2AF2 or CD44v8-10 (Fig. 6D). A lack of involvement of CD44v6 in melanoma migration was evident as knockdown of CD44v6 did not affect Lu1205M migration (Supplementary Fig. 3C). These results suggest that CD82 suppresses melanoma motility through regulating U2AF2 expression and CD44s/CD44v8-10 isoform switching.
Figure 6.
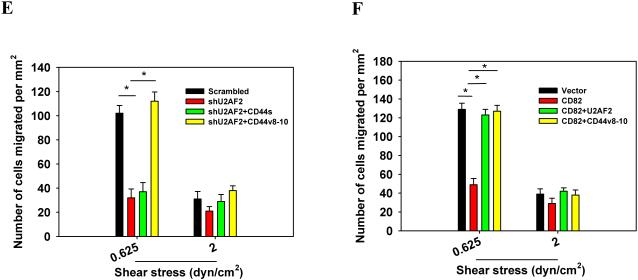
CD82 suppresses melanoma invasiveness and migration by modulating U2AF2-dependent CD44 isoform switching. (A) SK-Mel-25 cells that were transfected with vector or U2AF2 were cotransfected with shCD44s or shCD44v8-10. (B) SK-Mel-25 cells that were transfected with scrambled or CD82 siRNA were cotransfected with shU2AF2 or shCD44v8-10. (C, E) Lu1205M cells that were transfected with scrambled or U2AF2 shRNA were cotransfected with CD44s or CD44v8-10. (D, F) Lu1205M cells that were transfected with vector or CD82 were cotransfected with U2AF2 or CD44v8-10. (A-D) SK-Mel-25 or Lu1205M cells were tested for static migration abilities in transwell system. HUVEC monolayer was seeded on transwell insert membrane. The downward side of the membrane was stained with crystal violet and migrated cells were counted using an inverted microscope. Image magnification: 200X. Values are shown as mean±SEM from 12 fields from each of three independent experiments. *p<0.05. Bar=50 μm. (E-F) Lu1205M migration potentials under flow conditions were assessed with flow migration assays which were carried out for 4 hr at shear stresses of 0.625 and 2 dyn/cm2. The migrated cells were stained and counted. Values are shown as mean±SEM from 12 fields from each of three independent experiments. *p<0.05.
Flow regulated cancer migration plays important roles in tumor metastasis. To evaluate the roles of CD82-regulated CD44 splicing on melanoma migration under hydrodynamic conditions, we utilized a flow migration assay1, 30. At a shear stress of 0.625 dyn/cm2 but not 2 dyn/cm2, knockdown of endogenous U2AF2 or overexpression of CD82 reduced the transendothelial migration of Lu1205M (Fig. 6E and F). Conversely, restoration of CD44v8-10 but not CD44s abrogated the U2AF2 knockdown-dependent reduction in melanoma extravasation (Fig. 6E). The suppressive effect of CD82 overexpression on melanoma migration in flow was rescued by transfection of U2AF2 or CD44v8-10 (Fig. 6F). Therefore, CD44v8-10 may contribute to melanoma migration under low shear conditions.
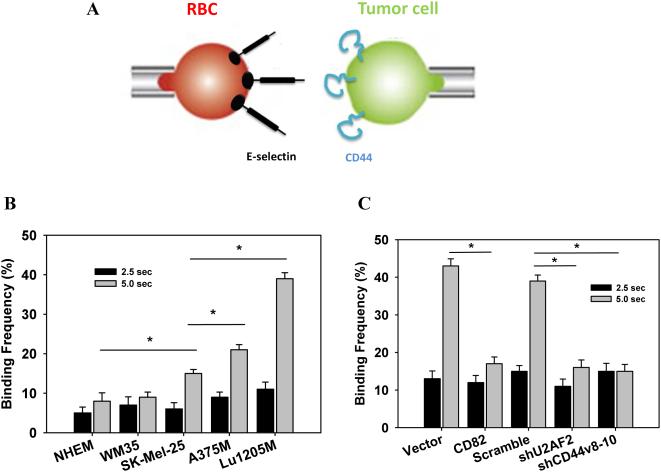
Cell tethering mediated by reversible interactions between selectins and sialyl-LewisX-decorated receptors on tumor surface is the first step for tumor metastasis31, 32. To determine the effect of CD44 isoform switching on tumor adhesion, we assessed binding frequency between E-selectin and melanoma with a micropipette aspiration assay (Fig. 7A). Upon 5-sec contacts, CD44s-expressing SK-Mel-25 adhered more frequently to E-selectin compared with WM35 or NHEM (p<0.05) (Fig. 7B). CD44v8-10-expressing A375M or Lu1205M had higher affinity for E-selectin than SK-Mel-25 (p<0.05). Overexpression of CD82 or knockdown of U2AF2 or CD44v8-10 sharply diminished the adhesion probability of Lu1205M (Fig. 7C). Interestingly, the difference of adhesion probability among different cell lines was not observed for short contact duration (2.5 sec) (Fig. 7B-C).
Figure 7.
U2AF2-mediated CD44 isoform switching increases the affinity of tumor CD44 for E-selectin. Binding affinity of melanoma cells for E-selectin was analyzed by the micropipette assay.(A) Schematic representation of the dual micropipette setup. A CD44-expressing tumor cell and an E-selectin-coated RBC were held by 2 opposing micropipettes and allowed to contact for different time durations. Binding events were recorded by analyzing the deflection of RBC membrane. (B) Adhesion frequency of melanoma to E-selectin upon receptor/ligand contact for 5 sec was increased progressively with increasing metastatic potentials. (C) Lu1205M cells were transfected with vector, CD82, scrambled shRNA, shU2AF2 or shCD44v8-10 and allowed to contact with E-selectin coated RBC for 2.5 and 5 sec before binding frequency was measured. Values are shown as mean±SEM from 6 independent experiments. *p<0.05.
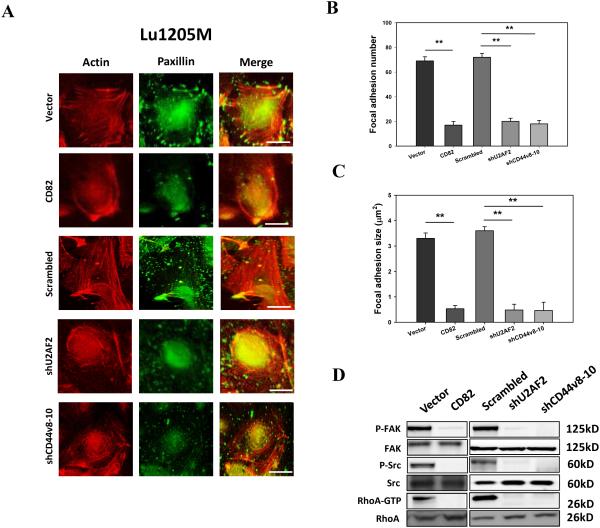
In migrating Lu1205M, robust stress fiber structures formed with thick actin filaments traversing the cell bodies (Fig. 8A). In sharp contrast, ectopic expression of CD82 disrupted the stress fiber structure with only thin filaments visible in the cell peripheries. Likewise, silencing U2AF2 or CD44v8-10 abolished actin polymerization. By staining paxillin, a scaffold adaptor protein localized in focal adhesion, it was demonstrated that control cells had bright punctate focal adhesion associated with the ends of thick stress fibers. But overexpression of CD82 or knockdown of U2AF2 or CD44v8-10 reduced the size and number of focal adhesions where paxillin spots disengaged with actin (Fig. 8A-C). Then, the activation status of migratory machinery, including focal adhesion kinase (FAK), Src, and RhoA, was assessed. CD82 overexpression or knockdown of U2AF2 or CD44v8-10 suppressed the phosphorylation of FAK and Src and GTP-coupling of RhoA without affecting total FAK, Src and RhoA levels (Fig. 8D).
Figure 8.
CD82 suppresses melanoma motile machinery by inhibiting U2AF2-mediated CD44v8-10 generation. (A) Lu1205M cells were transfected with vector, CD82, scrambled shRNA, shU2AF2 or shCD44v8-10 before being stained with rhodamine-phalloidin and paxillin antibody (Geen=paxillin, Red=F-actin). Bar=10μm. The average number (B) and size (μm2) (C) of paxillin-containing focal adhesions in transfected Lu1205M were quantified using ImageJ software. 12 cells were analyzed per condition in each experiment. Data were expressed as mean±SEM of three independent experiments. **p < 0.01 compared with control. (D) Lu1205M cells were transfected with vector, CD82, scrambled shRNA, shU2AF2 or shCD44v8-10 before being lysed and subjected to Western blotting analysis of phosphorylation of FAK and Src and GTP-coupling of RhoA. Total FAK, Src, and RhoA were used as loading controls.
CD82 targets U2AF2-mediated CD44v8-10 splicing to inhibit melanoma metastasis in vivo
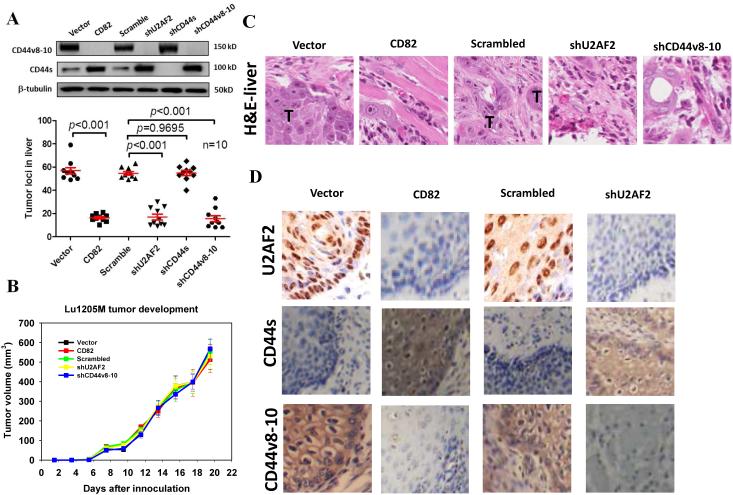
To assess whether modulation of CD82, U2AF2, CD44s and CD44v8-10 expression could affect melanoma metastasis in vivo, we used two metastatic models: xenograft liver metastasis and experimental lung metastasis. Compared with control groups, xenograft tumors derived from CD82-overexpressing, U2AF2-slienced and CD44v8-10-silenced Lu1205M metastasized less efficiently to the liver 20 days after subcutaneous implantation (Fig. 9A). CD82 overexpression, U2AF2 knockdown or CD44v8-10 knockdown reduced the number of metastatic lesions by 80%. On the contrary, CD44s knockdown did not significantly reduce Lu1205M liver metastasis compared with control. To determine whether melanoma tumorigenesis was affected, size of primary tumors was measured on alternative days up to 19.5 days. Overexpression of CD82 or knockdown of U2AF2 or CD44v8-10 did not alter the rate at which Lu1205M tumors grew in comparison to control (Fig. 9B). Thus, CD82-regulated CD44v8-10 splicing had no effect on melanoma tumorigenesis. H&E staining verified tumor nodule quantification results for involvement of CD82, U2AF2 and CD44v8-10 in melanoma liver metastasis (Fig. 9C). In tumor migrating fronts, CD44v8-10 was localized in cell membrane, while U2AF2 exhibited a nucleus and cytoplasma staining pattern (Fig. 9D). CD44s was absent from tumor migrating fronts. CD82 overexpression or U2AF2 knockdown reduced the robust staining for CD44v8-10 and U2AF2, whereas CD44s levels were increased (Fig. 9D). As for experimental lung metastasis, Lu1205M-LUC/CD82 cells migrated less efficiently than control cells (Supplementary Fig. 7A and B). Further, overexpression of U2AF2 or CD44v8-10 rescued tumor lung metastasis. Notably, inoculation of Lu1205M-LUC co-transfected with CD82 and CD44v8-10 resulted in even more lung seedings than Lu1205M-LUC/vector melanoma.
Figure 9.
CD82 regulates melanoma experimental metastasis in vivo by suppressing U2AF2-mediated CD44 isoform switching. (A) Quantification of tumor loci in liver 20 days post implantation of vector, CD82, scrambled shRNA, shU2AF2, shCD44s and shCD44v8-10 transfected-Lu1205M cells into nude mice. Ten mice were injected under each condition. The livers in each group were collected and sectioned. 6 sections through the center of the liver were examined under dissecting microscopy for the presence of metastatic tumor loci. Western blotting analysis was conducted to determine the levels of CD44v8-10 and CD44s in lysates from implanted tumors. (B) Quantification of primary tumor’s tumorigenic potentials. Data are mean±SEM. n=10. (C) Representative images of H&E staining of liver metastases of vector, CD82, scrambled shRNA, shU2AF2 and shCD44v8-10-transfected Lu1205M cells in mice. T: metastatic tumor. (D) IHC staining of U2AF2, CD44s and CD44v8-10 in invasive fronts of primary tumor.
Western blotting analysis demonstrated lower expressions of CD44v8-10 and U2AF2 in CD82-expressing lung metastases in comparison with control (Supplementary Fig. 7C). Reconstitution of U2AF2 expression in CD82-expressing melanomas rescued the suppressed CD44v8-10 expression. Restoring CD44v8-10 in melanoma did not alter the expression levels of U2AF2 and CD44s, indicating that CD44v8-10 is located in the downstream of U2AF2. Our findings suggest that CD82 suppresses melanoma metastasis in vivo by inhibiting U2AF2-dependent CD44v8-10 splicing.
Discussion
Our study revealed that CD44v8-10 is not only a prognostic marker but also a functional receptor for melanoma metastasis. In agreement with earlier studies, our results showed that elevated level of CD44v8-10 is associated with tumor metastatic potentials11. It was reported that CD44v8-10 splicing promotes tumorigenesis by driving stem cell-like properties25, 33. In the current study, our findings provided novel evidence for the role of CD82 in regulating pre-mRNA splicing in melanoma and proposed a mechanism by which CD82 suppresses melanoma metastasis by modulating U2AF2-dependent CD44v8-10 splicing (Fig. 10). This hypothesis is supported by: 1) overexpression of CD82 in highly metastatic melanoma reduces the ratio of CD44v8-10 to CD44s; 2) CD82 regulates U2AF2 protein stability by facilitating its ubiquitination; 3) overexpression of CD82 in melanoma impedes tumor migration under static and flow conditions, which can be rescued by restoration of U2AF2 or CD44v8-10 expression; 4) the suppressive effect of CD82 on tumor migration is dependent on altered affinity of CD44 for selectin, inactivation of Src/FAK/RhoA, and disassembly of stress fibers and focal adhesions.
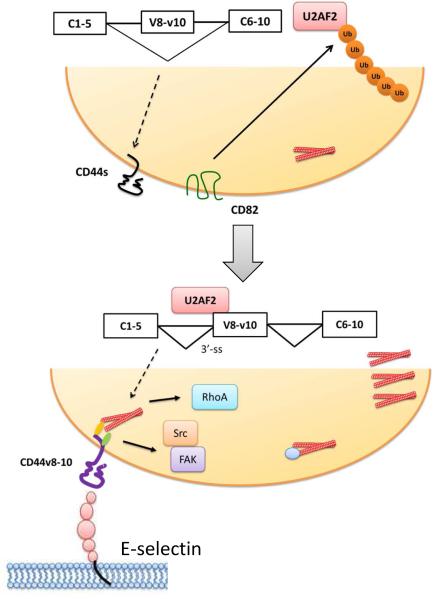
Figure 10.
Model of inhibition of U2AF2-dependent CD44 alternative splicing by CD82 in metastatic melanoma. Upper panel, in non-metastatic melanoma, expression of CD82 induces ubiquitination and degradation of U2AF2, which leads to exon V8-10 skipping of CD44 pre-mRNA. Lower panel, loss of CD82 expression results in stabilization and binding of U2AF2 to 3’-splicing site of the intron adjacent to exon V8. This process facilitates exon V8-10 splicing, enhances the binding affinity of CD44 to E-selectin and promotes stress fiber formation and Src/FAK/RhoA activation, leading to increased motility and metastatic potential of melanoma.
A dramatic decrease in CD82 expression is one of the characteristics of late-stage human cancers14. The function of CD82 in impeding cancer motility and invasion may be related to modulating the activity of receptor tyrosine kinases, inhibiting p130CAS-CrkII coupling, and suppressing integrin-dependent crosstalk with c-Met and Src 34, 35. Strikingly, a previous study showed that upon association with Duffy antigen receptor group (DARC), CD82 limits melanoma invasion by disrupting IL-8-mediated VE-cadherin disassembly36. It was also shown that CD82 reduces EGFR ubiquitination and alters the kinetics of the recruitment of receptors to early endosomes17. The relationship between CD82 and CD44 was explored by a recent study showing that CD82 reduces endothelial migration by suppressing lipid raft formation and enhancing endocytosis of CD4419. It is unlikely that CD82 directly regulates spliceosome, since it lacks enzymatic motifs in its cytoplasmic tails14. Instead, CD82 may be associated with integrins in tetraspanin web to regulate integrin signaling. In agreement with this, the role of integrins in gene splicing has been documented in that α3β1 integrin induces COX-2 alternative splicing in breast cancer37.
Disruption of exon recognition and misregulation of alternative splicing are a common cause of human disease. In the current study, we showed that CD44v8-10 splicing was U2AF2-dependent, as the minigene system reported a significant reduction of V8-10 splicing following U2AF2 knockdown. The essential pre-mRNA splicing factor, U2AF2, is a non-snRNP protein required for binding U2 snRNP to pre-mRNA38. Recruitment of U2AF2 to 3’-splicing site is a primary determinant for exon selection and the initiation of prespliceosome complex formation38. It was shown that U2AF2 expression in melanoma tissue samples is correlated with CD44v6/s ratio 39. Although ESRP1 has been reported in multiple epithelial tumors to promote CD44v8-10 splicing, in mesenchymal-type melanoma cells, like Lu1205M, ESRP1 was barely detectable (data not shown). Thus, ESRP1 may be dispensable for CD44 splicing in late-stage melanoma. It was reported that YB-1 may assist exon inclusion of CD44v by enhancing the recruitment of U2AF2 to weak polypyrimidine tracts26. Therefore, it is of interest to investigate whether other splice factors participate in U2AF2-induced CD44v8-10 splicing in the future studies.
Posttranslational modifications, including phosphorylation and ubiquitination, are implicated in modulation of activities of a number of splicing factors, allowing fine-tuning of the splicing reactions40. SF2/ASF protein can be phosphorylated by Cdc2 kinase in a cell-cycle-dependent manner to regulate spliceosome function41. Serine/threonine kinase, Plk1 interacts with splicing factor, UAP56, triggering its phosphorylation, ubiquitination and degradation through proteasomes42. A more recent report showed that dopamine and cyclic adenosine monophosphate-regulated phosphoprotein (DARPP-32) interacts and stabilizes SRp20, thereby increasing its splicing activity and CD44v8-10 expression25. In the presence of DARPP-32, SRp20 ubiquitination is inhibited, highlighting the importance of ubiquitination in controlling splicing factor activities. In the current study, CD82 may utilize some indirect pathways to activate ubiquitin ligases which induce ubiquitination and degradation of U2AF2 14.
Tumor extravasation in blood stream requires cell rolling and tethering mediated by Sialyl-LewisX-decorated receptors followed by integrin-mediated stable adhesion22, 43. It was widely accepted that CD44v rather than CD44s binds to selectin in flow to promote colon and breast cancer rolling and tethering44. In agreement with these studies, our micropipette aspiration assay revealed that CD44v8-10 on metastatic melanoma cells has higher affinity for E-selectin than CD44s. Due to the presence of variant regions, CD44v has greater molecular length and more glycosaminoglycan modifications which facilitate bond formation with selectins compared with CD44s6. Therefore, the pro-migratory effect of U2AF2-dependent CD44 splicing may be attributed to diverse affinities of CD44v8-10 and CD44s for E-selectin. Since CD44v has a higher affinity for E-selectin than CD44s, a isoform switching from CD44s to CD44v8-10 can serve as an effective receptor for circulating melanoma to be captured by endothelium under physiological flow conditions32. On the other hand, by tethering to selectins, CD44v8-10 may also function as a signaling platform mediating mechanosensor docking to activate integrin which further enhances melanoma migration45. This is supported by the observation that CD44v8-10 is important for melanoma motility by promoting stress fiber formation and focal adhesion assembly.
In conclusion, we demonstrated that CD82 controls melanoma metastasis by suppressing U2AF2-mediated CD44 alternative splicing. The CD82-U2AF2 axis may be critical for regulating CD44v8-10 splicing that promotes melanoma metastasis. Our work provides rationales for further exploration of these molecules as therapeutic targets to treat melanoma metastasis.
Materials and Methods
Cell culture
NHEMs (obtained from PromoCell) were cultured in Melanocyte Growth Medium. Lu1205M and A375M, highly metastatic variants of melanoma cell lines, Lu1205 and A375 (ATCC), were derived by in vivo injecting Lu1205 and A375 cells into nude mice and, 25 days later, establishing cell lines from tumors, which had formed in the lungs. Cells were maintained in Dulbecco's modified Eagle's medium (GIBCO) supplemented with 10% FBS and 100 U/ml of penicillin-streptomycin. SK-Mel-25 (ATCC) and WM35 melanoma cells (provided by Dr. Meenhard Herlyn, Wistar Institute, Philadelphia, PA) were maintained in Roswell Memorial Park Institute (RPMI) supplemented with 10% FBS and 100 U/ml of penicillin-streptomycin. All cells were maintained in a humidified incubator at 37°C and 5% CO2.
Human tissue samples
Tissue collection and analysis in this study were approved by the Ethics Committee of Third Military Medical University of China, and written informed consent was obtained from all participants. Formalin-fixed paraffin-embedded tissue (FFPE) sections of 6 dysplastic melanocytic nevi, 19 primary melanomas and 51 metastatic melanomas samples were collected. All clinicopathological data are made available for all melanoma cases. The 70 melanoma patients consist of 32 females and 38 males, aging 18 to 83years (median, 53years). Cases were selected from original hematoxylin and eosin (H&E) stainings. The differences of variables, like age and sex, among TNM stages are statistically insignificant.
Xenograft tumor model
All the procedures involving animals were reviewed and approved by Pennsylvania State University Institutional Animal Care and Use Committee. For orthotopic metastasis assay, five-week-old athymic nu/nu male Balb/c mice (Herlan, Indianapolis, IN) were randomized in different groups and implanted subcutaneously in the right flank with Lu1205M cells (1×106 cells/100 μl PBS/Matrigel). 10 successful tumor xenografts were utilized in each group, according to statistical analysis of sample size. Evaluation of tumor size and metastatic tumor loci was conducted by two independent investigators who were blinded to the study groups. The tumor size was measured by caliper. After 20 days, there were no significant differences of primary tumor weight among groups (data not shown). To examine the tumor metastasis, the livers in each group were collected and submitted in 4% neutral buffered formalin. 6 sections through the center of the liver were examined under dissecting microscopy for presence of metastases and the number of metastases per section was determined. In selected cases, the tumor tissues were fixed in formalin and embedded in paraffin for immunohistochemistry and routine H&E staining.
Immunohistochemistry (IHC)
Polyclonal antibodies against CD44v8-10 molecule were generated and purified as described previously46, 47. Briefly, New Zealand White female rabbits were immunized with a GST-CD44v8-10 fusion protein, and the resulting anti-CD44v8-10 antibody was immunoaffinity purified with a GST-CD44v8-10 affinity column. FFPE from patient and mouse samples were incubated with commercial antibodies, anti-CD44v7/8 (VFF-17, Serotec, Inc., Raleigh, NC), CD44v9 (11.24, IgG 1), CD44v10 (Millipore, Billerica, MA), U2AF2 (Abcam, Eugene, OR) or CD82 (Novus Biologicals, Littleton, CO) or immunopurified anti-CD44v8-10. Then, the samples were incubated with horseradish peroxidase (HRP) conjugated-secondary antibodies (Abcam, Eugene, OR). The target proteins were detected with 3,3′diaminobenzidine (DAB). Nuclei counterstained with hematoxylin. The gray values of staining were quantified with ImageJ.
cDNA transfection
cDNA clones of CD44s, CD44v8-10, hemagglutinin-tagged CD82 and GST-tagged U2AF2 (Origene, Rockville, MD)were inserted into the expression vector pcDNA3.1(Invitrogen). 200 ng CD44s, CD44v8-10, U2AF2, CD82 or pcDNA3.1 vector control constructs were transfected into cells with Mirus TransIT (Mirus Bio LLC.) and then subjected to G418 selection for 2–3 weeks.
Silencing CD82, U2AF2, CD44s and CD44v8-10 with siRNA or shRNA
siRNA targeting CD82 and si/shRNA transfection were described previously36. siRNA oligonucleotides targeting U2AF2, CD44s or CD44v8-10 were synthesized (Life technologies, Carlsbad, CA) and ligated into pSUPER.neo.gfp expression vector (oligoengine, Inc, Seattle, WA). The following oligonucleotides were used (underlined, sense and antisense sequences; bold, restriction enzyme sites; light italics, polIII termination signals; bold italics, loop with linker): CD44v8-10, 5’-GATCCCCGGAAGAAGATAAAGACCATTTCAAGAGAATGGTCTTTATCTTCTTCCTTTTTC-3’; CD44s, 5’-GATCCCCCCTGCTACCAGAGACCAAGTTCAAGAGACTTGGTCTCTGGTAGCAGGTTTTTC-3’; U2AF2, 5’-GATCCCCGCACGGTGGACGATTCGTTTCAAGAGAACGAATCGTCCACCGTGCTTTTTC-3’. A tested scrambled sequence (Life technologies) was used as a negative control.
Construction of pFlare-v8-10 minigene
A reporter construct was generated using standard cloning techniques. The CD44 genomic DNA, including CD44v8-10 exon and flanking introns (500 bp each) was PCR amplified from the Human Genomic DNA Library (Promega, Madison, WI, USA). The PCR product was cloned into MluI and BamHI enzyme sites in pFlare9A-Dup34 vector28.
Real-time PCR and run-off analysis
Real-time PCR assay and primer sets for standard and variant exons were described previously3,13. Run-off analysis was performed as described48, 49.
Western blotting and immunoprecipitation
Western blotting analysis of protein expression was described previously5. CD44 (clone 2C5), CD82 (Life Technologies), U2AF2 (Abcam, Eugene, OR), FAK (Abcam, Eugene, OR), pY397 FAK (Abcam, Eugene, OR), Src (Santa Cruz, Dallas, TX), pY416 Src (Cell Signaling Technology, Danvers, MA), and RhoA (Abcam, Eugene, OR) were detected with corresponding primary monoclonal antibodies (1:1000 diluted in blocking buffer). For examining the effect of CD82 overexpression on U2AF2 ubiquitination, melanoma cells were treated with 10 μM MG132 for 6 hr before U2AF2 was immunoprecipitated, resolved in SDS-PAGE, transferred to a nitrocellulose membrane and developed with anti-ubiquitin P4D1 (1:1,000, Santa Cruz) and anti-U2AF2 antibodies.
Immunofluorescence
Detailed procedure was described previously50.
Transwell migration
Transmigration was assessed by the ability of cells to migrate through a porous (8μm) polycarbonate membrane of a transwell device (Corning, NY) towards collagen IV. Migrated cells at the bottom of the filter were fixed with 4% formaldehyde and stained with 0.5% crystal violet.
Micropipette binding assay
A micropipette frequency assay to measure the binding affinity of tumor CD44 isoforms for E-selectin was described previously3, 51, 52.
Flow migration assay
The assay was described previously1, 36. Briefly, a monolayer of HUVECs was grown on polycarbonate filters (8 μm pore size; NeuroProbe, Gaithersburg, MD). The center 12 wells of bottom plate were filled with collagen IV. Then, melanoma cells were drawn across HUVEC monolayer with a peristaltic pump at a shear flow (0.625 or 2 dyn/cm2) for 4 hr. Migrated cells were counted using an inverted microscope (IX71, Olympus) with the NIH ImageJ software.
Statistical analysis
Data were obtained from at least three independent experiments and are expressed as means±SEM. Statistical significance of differences was determined by Mann-Whitney U test, Student’s t-test or analysis of variance (ANOVA). Tukey’s test was used for post hoc analysis for ANOVA. To assess the normal distribution and equal variance of the data, Kolmogorov-Smirnov and Levene tests were employed. The correlation between molecule expression and patient survival was analyzed with Kaplan-Meier survival curve and log-rank test. Probability values of p<0.05 and p<0.01 were chosen as statistical significance.
Supplementary Material
Acknowledgements
We thank Go J. Yoshida (Tokyo medical and dental University) for helpful discussion. This study was funded by Fundamental Research Funds for the Central Universities (XDJK2014C176 to P.Z., XDJK2012B010 to H.F.Z., XDJK2015C154 to Q.R. and XDJK2015C155 to S.F.), the Start-up Foundation of Southwest University (SWU114017 to P.Z.), National Natural Science Foundation of China (NSFC) (NSFC-81402393 to P.Z., NSFC-81572678 to P.Z., NSFC-81272433 to G.T.L., NSFC-81472732 to G.T.L. and NSFC-81073084 to H.F.Z.) and National Science Foundation grants (CBET-0729091 to C.D.).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- 1.Huh SJ, Liang S, Sharma A, Dong C, Robertson GP. Transiently entrapped circulating tumor cells interact with neutrophils to facilitate lung metastasis development. Cancer Res. 2010;70:6071–6082. doi: 10.1158/0008-5472.CAN-09-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma A, Tran MA, Liang S, Sharma AK, Amin S, Smith CD, et al. Targeting mitogen-activated protein kinase/extracellular signal-regulated kinase kinase in the mutant (V600E) B-Raf signaling cascade effectively inhibits melanoma lung metastases. Cancer Res. 2006;66:8200–8209. doi: 10.1158/0008-5472.CAN-06-0809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang P, Goodrich C, Fu C, Dong C. Melanoma upregulates ICAM-1 expression on endothelial cells through engagement of tumor CD44 with endothelial E-selectin and activation of a PKCalpha-p38-SP-1 pathway. FASEB J. 2014;28:4591–4609. doi: 10.1096/fj.11-202747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang P, Ozdemir T, Chung CY, Robertson GP, Dong C. Sequential binding of alphaVbeta3 and ICAM-1 determines fibrin-mediated melanoma capture and stable adhesion to CD11b/CD18 on neutrophils. J Immunol. 2011;186:242–254. doi: 10.4049/jimmunol.1000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang P, Fu C, Bai H, Song E, Song Y. CD44 variant, but not standard CD44 isoforms, mediate disassembly of endothelial VE-cadherin junction on metastatic melanoma cells. FEBS Lett. 2014;588:4573–4582. doi: 10.1016/j.febslet.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 6.Zoller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11:254–267. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
- 7.Prochazka L, Tesarik R, Turanek J. Regulation of alternative splicing of CD44 in cancer. Cell Signal. 2014;26:2234–2239. doi: 10.1016/j.cellsig.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 8.Ishii H, Saitoh M, Sakamoto K, Kondo T, Katoh R, Tanaka S, et al. Epithelial splicing regulatory proteins 1 (ESRP1) and 2 (ESRP2) suppress cancer cell motility via different mechanisms. J Biol Chem. 2014;289:27386–27399. doi: 10.1074/jbc.M114.589432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reinke LM, Xu Y, Cheng C. Snail represses the splicing regulator epithelial splicing regulatory protein 1 to promote epithelial-mesenchymal transition. J Biol Chem. 2012;287:36435–36442. doi: 10.1074/jbc.M112.397125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, et al. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest. 2011;121:1064–1074. doi: 10.1172/JCI44540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yae T, Tsuchihashi K, Ishimoto T, Motohara T, Yoshikawa M, Yoshida GJ, et al. Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat Commun. 2012;3:883. doi: 10.1038/ncomms1892. [DOI] [PubMed] [Google Scholar]
- 12.Cheng C, Sharp PA. Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol Cell Biol. 2006;26:362–370. doi: 10.1128/MCB.26.1.362-370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeo K, Kawai T, Nishida K, Masuda K, Teshima-Kondo S, Tanahashi T, et al. Oxidative stress-induced alternative splicing of transformer 2beta (SFRS10) and CD44 pre-mRNAs in gastric epithelial cells. Am J Physiol Cell Physiol. 2009;297:C330–338. doi: 10.1152/ajpcell.00009.2009. [DOI] [PubMed] [Google Scholar]
- 14.Tsai YC, Weissman AM. Dissecting the diverse functions of the metastasis suppressor CD82/KAI1. FEBS Lett. 2011;585:3166–3173. doi: 10.1016/j.febslet.2011.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charrin S, le Naour F, Silvie O, Milhiet PE, Boucheix C, Rubinstein E. Lateral organization of membrane proteins: tetraspanins spin their web. Biochem J. 2009;420:133–154. doi: 10.1042/BJ20082422. [DOI] [PubMed] [Google Scholar]
- 16.Berditchevski F, Odintsova E. Tetraspanins as regulators of protein trafficking. Traffic. 2007;8:89–96. doi: 10.1111/j.1600-0854.2006.00515.x. [DOI] [PubMed] [Google Scholar]
- 17.Odintsova E, van Niel G, Conjeaud H, Raposo G, Iwamoto R, Mekada E, et al. Metastasis suppressor tetraspanin CD82/KAI1 regulates ubiquitylation of epidermal growth factor receptor. J Biol Chem. 2013;288:26323–26334. doi: 10.1074/jbc.M112.439380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Odintsova E, Sugiura T, Berditchevski F. Attenuation of EGF receptor signaling by a metastasis suppressor, the tetraspanin CD82/KAI-1. Curr Biol. 2000;10:1009–1012. doi: 10.1016/s0960-9822(00)00652-7. [DOI] [PubMed] [Google Scholar]
- 19.Wei Q, Zhang F, Richardson MM, Roy NH, Rodgers W, Liu Y, et al. CD82 restrains pathological angiogenesis by altering lipid raft clustering and CD44 trafficking in endothelial cells. Circulation. 2014;130:1493–1504. doi: 10.1161/CIRCULATIONAHA.114.011096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobs PP, Sackstein R. CD44 and HCELL: Preventing hematogenous metastasis at step 1. Febs Letters. 2011;585:3148–3158. doi: 10.1016/j.febslet.2011.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagano O, Okazaki S, Saya H. Redox regulation in stem-like cancer cells by CD44 variant isoforms. Oncogene. 2013;32:5191–5198. doi: 10.1038/onc.2012.638. [DOI] [PubMed] [Google Scholar]
- 22.Konstantopoulos K, Thomas SN. Cancer cells in transit: the vascular interactions of tumor cells. Annu Rev Biomed Eng. 2009;11:177–202. doi: 10.1146/annurev-bioeng-061008-124949. [DOI] [PubMed] [Google Scholar]
- 23.Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 24.Kim JH, Kim B, Cai L, Choi HJ, Ohgi KA, Tran C, et al. Transcriptional regulation of a metastasis suppressor gene by Tip60 and beta-catenin complexes. Nature. 2005;434:921–926. doi: 10.1038/nature03452. [DOI] [PubMed] [Google Scholar]
- 25.Zhu S, Chen Z, Katsha A, Hong J, Belkhiri A, El-Rifai W. Regulation of CD44E by DARPP-32-dependent activation of SRp20 splicing factor in gastric tumorigenesis. Oncogene. 2015 doi: 10.1038/onc.2015.250. e-pub ahead of print 29 Jun 2015; doi: 10.1038/onc.2015.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei WJ, Mu SR, Heiner M, Fu X, Cao LJ, Gong XF, et al. YB-1 binds to CAUC motifs and stimulates exon inclusion by enhancing the recruitment of U2AF to weak polypyrimidine tracts. Nucleic Acids Res. 2012;40:8622–8636. doi: 10.1093/nar/gks579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, et al. Widespread siRNA "off-target" transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoilov P, Lin CH, Damoiseaux R, Nikolic J, Black DL. A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. Proc Natl Acad Sci U S A. 2008;105:11218–11223. doi: 10.1073/pnas.0801661105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanley WD, Burdick MM, Konstantopoulos K, Sackstein R. CD44 on LS174T colon carcinoma cells possesses E-selectin ligand activity. Cancer Res. 2005;65:5812–5817. doi: 10.1158/0008-5472.CAN-04-4557. [DOI] [PubMed] [Google Scholar]
- 30.Liang S, Sharma A, Peng HH, Robertson G, Dong C. Targeting mutant (V600E) B-Raf in melanoma interrupts immunoediting of leukocyte functions and melanoma extravasation. Cancer Res. 2007;67:5814–5820. doi: 10.1158/0008-5472.CAN-06-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McEver RP, Zhu C. Rolling cell adhesion. Annu Rev Cell Dev Biol. 2010;26:363–396. doi: 10.1146/annurev.cellbio.042308.113238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wirtz D, Konstantopoulos K, Searson PC. The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nat Rev Cancer. 2011;11:512–522. doi: 10.1038/nrc3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng Y, Wodzenski D, Gao D, Shiraishi T, Terada N, Li Y, et al. Stress-response protein RBM3 attenuates the stem-like properties of prostate cancer cells by interfering with CD44 variant splicing. Cancer Res. 2013;73:4123–4133. doi: 10.1158/0008-5472.CAN-12-1343. [DOI] [PubMed] [Google Scholar]
- 34.Zhou B, Liu L, Reddivari M, Zhang XA. The palmitoylation of metastasis suppressor KAI1/CD82 is important for its motility- and invasiveness-inhibitory activity. Cancer Res. 2004;64:7455–7463. doi: 10.1158/0008-5472.CAN-04-1574. [DOI] [PubMed] [Google Scholar]
- 35.Sridhar SC, Miranti CK. Tetraspanin KAI1/CD82 suppresses invasion by inhibiting integrin-dependent crosstalk with c-Met receptor and Src kinases. Oncogene. 2006;25:2367–2378. doi: 10.1038/sj.onc.1209269. [DOI] [PubMed] [Google Scholar]
- 36.Khanna P, Chung CY, Neves RI, Robertson GP, Dong C. CD82/KAI expression prevents IL-8-mediated endothelial gap formation in late-stage melanomas. Oncogene. 2014;33:2898–2908. doi: 10.1038/onc.2013.249. [DOI] [PubMed] [Google Scholar]
- 37.Subbaram S, Lyons SP, Svenson KB, Hammond SL, McCabe LG, Chittur SV, et al. Integrin alpha3beta1 controls mRNA splicing that determines Cox-2 mRNA stability in breast cancer cells. J Cell Sci. 2014;127:1179–1189. doi: 10.1242/jcs.131227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mackereth CD, Madl T, Bonnal S, Simon B, Zanier K, Gasch A, et al. Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature. 2011;475:408–411. doi: 10.1038/nature10171. [DOI] [PubMed] [Google Scholar]
- 39.Marzese DM, Liu M, Huynh JL, Hirose H, Donovan NC, Huynh KT, et al. Brain metastasis is predetermined in early stages of cutaneous melanoma by CD44v6 expression through epigenetic regulation of the spliceosome. Pigment Cell Melanoma Res. 2015;28:82–93. doi: 10.1111/pcmr.12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Bruderer S, Rafi Z, Xue J, Milburn PJ, Kramer A, et al. Phosphorylation of splicing factor SF1 on Ser20 by cGMP-dependent protein kinase regulates spliceosome assembly. EMBO J. 1999;18:4549–4559. doi: 10.1093/emboj/18.16.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okamoto Y, Onogi H, Honda R, Yasuda H, Wakabayashi T, Nimura Y, et al. cdc2 kinase-mediated phosphorylation of splicing factor SF2/ASF. Biochem Biophys Res Commun. 1998;249:872–878. doi: 10.1006/bbrc.1998.9247. [DOI] [PubMed] [Google Scholar]
- 42.Xiong F, Lin Y, Han Z, Shi G, Tian L, Wu X, et al. Plk1-mediated phosphorylation of UAP56 regulates the stability of UAP56. Mol Biol Rep. 2012;39:1935–1942. doi: 10.1007/s11033-011-0940-x. [DOI] [PubMed] [Google Scholar]
- 43.Shirure VS, Liu T, Delgadillo LF, Cuckler CM, Tees DF, Benencia F, et al. CD44 variant isoforms expressed by breast cancer cells are functional E-selectin ligands under flow conditions. Am J Physiol Cell Physiol. 2015;308:C68–78. doi: 10.1152/ajpcell.00094.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanley WD, Napier SL, Burdick MM, Schnaar RL, Sackstein R, Konstantopoulos K. Variant isoforms of CD44 are P- and L-selectin ligands on colon carcinoma cells. FASEB J. 2006;20:337–339. doi: 10.1096/fj.05-4574fje. [DOI] [PubMed] [Google Scholar]
- 45.Yago T, Shao BJ, Miner JJ, Yao LB, Klopocki AG, Maeda K, et al. E-selectin engages PSGL-1 and CD44 through a common signaling pathway to induce integrin alpha(L)beta(2)-mediated slow leukocyte rolling. Blood. 2010;116:485–494. doi: 10.1182/blood-2009-12-259556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi K, Stamenkovic I, Cutler M, Saya H, Tanabe KK. CD44 hyaluronate binding influences growth kinetics and tumorigenicity of human colon carcinomas. Oncogene. 1995;11:2223–2232. [PubMed] [Google Scholar]
- 47.Okamoto I, Morisaki T, Sasaki J, Miyake H, Matsumoto M, Suga M, et al. Molecular detection of cancer cells by competitive reverse transcription-polymerase chain reaction analysis of specific CD44 variant RNAs. J Natl Cancer Inst. 1998;90:307–315. doi: 10.1093/jnci/90.4.307. [DOI] [PubMed] [Google Scholar]
- 48.Konig H, Moll J, Ponta H, Herrlich P. Trans-acting factors regulate the expression of CD44 splice variants. EMBO J. 1996;15:4030–4039. [PMC free article] [PubMed] [Google Scholar]
- 49.van Weering DH, Baas PD, Bos JL. A PCR-based method for the analysis of human CD44 splice products. PCR Methods Appl. 1993;3:100–106. doi: 10.1101/gr.3.2.100. [DOI] [PubMed] [Google Scholar]
- 50.Zhang P, Feng S, Bai H, Zeng P, Chen F, Wu C, et al. Polychlorinated biphenyl quinone induces endothelial barrier dysregulation by setting the cross talk between VE-cadherin, focal adhesion, and MAPK signaling. Am J Physiol Heart Circ Physiol. 2015;308:H1205–1214. doi: 10.1152/ajpheart.00005.2015. [DOI] [PubMed] [Google Scholar]
- 51.Chesla SE, Selvaraj P, Zhu C. Measuring two-dimensional receptor-ligand binding kinetics by micropipette. Biophys J. 1998;75:1553–1572. doi: 10.1016/S0006-3495(98)74074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu C, Tong C, Wang M, Gao Y, Zhang Y, Lu S, et al. Determining beta2-integrin and intercellular adhesion molecule 1 binding kinetics in tumor cell adhesion to leukocytes and endothelial cells by a gas-driven micropipette assay. J Biol Chem. 2011;286:34777–34787. doi: 10.1074/jbc.M111.281642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.