SUMMARY
Helicobacter pylori colonizes the human stomach and is a potential cause of peptic ulceration or gastric adenocarcinoma. H. pylori secretes a pore-forming toxin known as vacuolating cytotoxin A (VacA). The 88 kDa secreted VacA protein, composed of an N-terminal p33 domain and a C-terminal p55 domain, assembles into water-soluble oligomers. The structural organization of membrane-bound VacA has not been characterized in any detail and the role(s) of specific VacA domains in membrane binding and insertion are unclear. We show that membrane-bound VacA organizes into hexameric oligomers. Comparison of the two-dimensional averages of membrane-bound and soluble VacA hexamers generated using single particle electron microscopy reveals a structural difference in the central region of the oligomers (corresponding to the p33 domain), suggesting that membrane association triggers a structural change in the p33 domain. Analyses of the isolated p55 domain and VacA variants demonstrate that while the p55 domain can bind membranes, the p33 domain is required for membrane insertion. Surprisingly, neither VacA oligomerization nor the presence of putative transmembrane GXXXG repeats in the p33 domain is required for membrane insertion. These findings provide new insights into the process by which VacA binds and inserts into the lipid bilayer to form membrane channels.
Keywords: Gastric cancer, electron microscopy, pore-forming toxins
Graphical Abstract
INTRODUCTION
Helicobacter pylori is a Gram-negative bacterium that colonizes the human stomach and causes gastric inflammation, leading to the development of peptic ulceration and gastric cancer in a subset of infected individuals (Atherton and Blaser, 2009; Cover and Blaser, 2009; Marshall and Warren, 1984; Suerbaum and Michetti, 2002). H. pylori infects about 50% of humans (more than 3 billion people), with a prevalence as high as 90% in some developing nations (Amieva and El-Omar, 2008; Atherton, 2006; Nomura et al., 1991; Parsonnet et al., 1991; Suerbaum and Michetti, 2002). Gastric cancer is the third leading cause of cancer-related deaths worldwide (de Martel et al., 2012; Fuchs and Mayer, 1995). H. pylori-associated gastric adenocarcinoma is a rare example of a human cancer attributed to a specific bacterial infection, a connection that led the World Health Organization to classify H. pylori as a type 1 carcinogen (Anonymous, 1994).
One of the major virulence factors secreted by H. pylori is the pore-forming exotoxin VacA (vacuolating cytotoxin A), named for its ability to induce the formation of large vacuoles in eukaryotic cells (Cover and Blanke, 2005). VacA causes multiple cellular effects that include cell vacuolation (Cover and Blanke, 2005), depolarization of membrane potential (Szabo et al., 1999), mitochondrial dysfunction (Domanska et al., 2010; Galmiche et al., 2000; Jain et al., 2011; Willhite and Blanke, 2004), autophagy (Terebiznik et al., 2009), cell death (Calore et al., 2010; Jain et al., 2011; Radin et al., 2011), activation of mitogen-activated protein kinases (Nakayama et al., 2004), and inhibition of T cell activities (Gebert et al., 2003; Sundrud et al., 2004). Most VacA-induced cellular alterations require oligomerization of the toxin (Genisset et al., 2006; Ivie et al., 2008; Vinion-Dubiel et al., 1999), insertion into the lipid bilayer to form a membrane channel (Czajkowsky et al., 1999; Iwamoto et al., 1999; Szabo et al., 1999; Tombola et al., 1999a), and internalization into the cell (Calore et al., 2010; Gauthier et al., 2007; Willhite and Blanke, 2004). The molecular mechanisms underlying each of these steps and the order in which they occur are not understood.
VacA does not exhibit sequence relatedness to any other known bacterial toxins. Secreted as an 88 kDa protein, VacA contains an N-terminal p33 domain and a C-terminal p55 domain. Three polymorphic regions have been identified (Gangwer et al., 2010): an N-terminal region (s1 or s2) (Atherton et al., 1995), an intermediate region (i1 or i2) located within the p33 domain (Rhead et al., 2007), and a p55 mid-region (m1 or m2) (Atherton et al., 1995; Pagliaccia et al., 1998). Sequence variations at the N-terminus (s1 or s2) and within the p55 mid-region (m1 or m2) are associated with differences in the ability of VacA to form pores or differences in the binding of VacA to cells, respectively (Atherton et al., 1995; Ji et al., 2000; Letley and Atherton, 2000; McClain et al., 2001b; Pagliaccia et al., 1998; Sewald et al., 2008; Wang et al., 2001). Sequence variation in the p33 intermediate-region and p55 mid-region may modulate differences in cellular tropism (Gonzalez-Rivera et al., 2012; Ji et al., 2000; Pagliaccia et al., 1998; Rhead et al., 2007; Wang et al., 2001). The vacA genotype of a strain is strongly associated with the risk of peptic ulceration and gastric cancer, and the various forms of VacA differ in activity in vitro. Specifically, humans infected with H. pylori strains containing vacA s1/i1/m1 alleles have an increased risk of developing gastric cancer compared to individuals who are infected with H. pylori strains containing vacA s2/i2/m2 alleles (Atherton et al., 1995; Cover, 2016; Figueiredo et al., 2002; Memon et al., 2014; Rhead et al., 2007).
VacA forms anion-selective channels in planar lipid bilayers (Czajkowsky et al., 1999; Iwamoto et al., 1999; Szabo et al., 1999; Tombola et al., 1999a; Tombola et al., 1999b), an activity that correlates with the predicted formation of VacA channels in the plasma membrane, endosomal membranes, and mitochondrial membranes of host cells (Campello et al., 2002; Debellis et al., 2001; Galmiche et al., 2000; Papini et al., 1998a; Papini et al., 1998b; Szabo et al., 1999; Willhite and Blanke, 2004; Willhite et al., 2003). Single channel analyses have provided evidence predicting that VacA pores are composed of six subunits (Iwamoto et al., 1999) and that they mimic the action of endogenous anion-selective channels (Czajkowsky et al., 2005; Domanska et al., 2010; Tombola et al., 1999b). Deep-etch electron microscopy (EM) and atomic force microscopy (AFM) have been utilized to visualize VacA bound to mica-supported lipid bilayers, but these studies were not able to conclusively determine either the type(s) of oligomers bound to the membrane or the conformation of the central pore-forming region of VacA in this lipid environment (Adrian et al., 2002; Czajkowsky et al., 1999; Geisse et al., 2004). Thus, very little is known about structural features of membrane-bound VacA.
The amino-terminus of VacA (within the p33 domain) is predicted to be highly hydrophobic (McClain et al., 2003; Vinion-Dubiel et al., 1999). A VacA mutant protein lacking a portion of this region (VacA Δ6–27) does not form pores in planar lipid bilayers and lacks cell-vacuolating activity (McClain et al., 2001b; Vinion-Dubiel et al., 1999), which indicates that sequences within the N-terminal hydrophobic region of the VacA p33 domain are required for the formation of membrane channels. The N-terminal portion of the p33 domain contains three GXXXG sequences, a motif often involved in facilitating oligomeric interactions between transmembrane (TM) helices (Russ and Engelman, 2000; Teese and Langosch, 2015). A computationally generated structural model of the predicted VacA TM domain sequence suggests that the N-terminal GXXXG motifs pack together to form an anion-channel (Kim et al., 2004). Studies using genetically modified VacA show that the N-terminal 32 hydrophobic residues of p33, especially the proline residue at position 9 (P9) and the G14XXXG18 motif, are required for channel activity and cell-vacuolating activity (McClain et al., 2001a; McClain et al., 2003). These studies also showed that the GXXXG motifs facilitate dimerization in the context of membrane based on experiments using a TOXCAT system (McClain et al., 2001a; McClain et al., 2003), a method developed to study TM helix-helix associations (Russ and Engelman, 1999, 2000).
In solution, VacA forms hexamers, heptamers, dodecamers and tetradecamers, with the majority of soluble oligomers (>90%) organizing into double layered structures (dodecamers and tetradecamers) (Adrian et al., 2002; Chambers et al., 2013; Lanzavecchia et al., 1998; Lupetti et al., 1996). Although there is no atomic resolution structure of the 88 kDa VacA holotoxin, there is a 2.4 Å three-dimensional (3D) structure of the p55 domain (Gangwer et al., 2007; Sewald et al., 2008), and 3D structures of “snowflake”-like soluble VacA oligomers have been determined using negative stain EM (Chambers et al., 2013; El-Bez et al., 2005). Our structural analysis showed that while soluble hexamers and heptamers form chiral structures with the “tips” of the snow-flakes all curving in the same direction, the dodecamers and tetradecamers are achiral (Chambers et al., 2013). VacA p55 domains have been localized to the peripheral arms of oligomers, and p33 is localized to the central region of the oligomers (Chambers et al., 2013; El-Bez et al., 2005; Gangwer et al., 2007).
In order to form a functional anion-specific channel, VacA must oligomerize and insert into the lipid bilayer (Adrian et al., 2002; Czajkowsky et al., 1999; Geisse et al., 2004; Tombola et al., 1999a). However, the mechanisms by which these steps occur are not fully understood. In the current study, we show that VacA oligomerizes on membranes under buffer conditions that do not support oligomerization in solution, and that membrane-bound VacA is also membrane-inserted. Studies of individual VacA domains show that while p55 can interact with membranes, p33 is required for insertion into the lipid bilayer. Importantly, we show that VacA organizes into single layered oligomers on membranes, and show that the p33 central region of VacA hexamers has a different structural organization when comparing membrane-associated hexamers to soluble hexamers. These are the first in-depth studies of VacA oligomer conformation in the context of membranes and provide important new insights into the process by which VacA inserts into membranes.
RESULTS
VacA s1/i1/m1 oligomerizes as chiral hexamers on membranes
As an initial step towards characterizing the structural organization of membrane-bound VacA, we first confirmed VacA’s ability to bind vesicles using lipid compositions previously used in planar lipid bilayer experiments that characterized VacA channel activity (Czajkowsky et al., 1999; Iwamoto et al., 1999; Tombola et al., 1999a; Vinion-Dubiel et al., 1999). For these assays, 0.8 µM of VacA s1/i1/m1 was acid activated to disassemble soluble oligomers into monomers (Cover et al., 1997; Molinari et al., 1998) and then mixed with liposomes composed of DOPC, ePC/DOPS/chol (55/15/30 mol%), DOPC/DOPS/eSM (45/20/35 mol%), or DOPC/DOPS/eSM/chol (35/20/35/10 mol%) before being pelleted by centrifugation. BSA at 1.1 µM was used as a negative control to measure non-specific membrane binding. Each binding experiment was done in triplicate and samples were analyzed by Coomassie staining, quantifying VacA or BSA in the supernatant (unbound) and the vesicle pellet (bound) (Fig. S1). The results show that VacA s1/i1/m1 bound to all lipid compositions tested (Fig. 1A). Statistically significant differences in VacA binding to specific lipid compositions were detected, but the differences in magnitude were small and probably not biologically meaningful (Fig. 1A). The negative control BSA did not bind the liposomes (Fig. 1B).
Figure 1. VacA s1/i1/m1 organizes on membranes as hexamers.
(A–B) Large unilamellar vesicles (LUVs) with various lipid compositions were incubated with 0.8 µM VacA s1/i1/m1 or 1.1 µM BSA, and the proportion of protein bound to the vesicles was determined. p < 0.05, *; p <0.01, **. Results represent the mean + SEM based on three independent binding assays. PC: 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC); ePC: L-α-phosphatidylcholine from egg; PS: 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS); SM: sphingomyelin from egg; chol: cholesterol. (C) Representative negative stain image of VacA s1/i1/m1 bound to ePC/DOPS/chol (55/15/30 mol%) LUVs. Scale bar, 50 nm. (D) Representative class averages of membrane-bound VacA s1/i1/m1 hexamers. Number of particles in each average shown in bottom left. *, marks the average used for comparison between soluble and membrane bound VacA s1/i1/m1 hexamers in Figure 2. False colored average shows the location of the p55 (red) and p33 (blue) domains. Side length of panels, 42 nm.
AFM, deep-etch EM, and 2D crystallization techniques have been used to visualize VacA s1/i1/m1 associated with membranes (Adrian et al., 2002; Czajkowsky et al., 1999; Geisse et al., 2004; Vinion-Dubiel et al., 1999). However, the limited resolution of the images in these studies made it difficult to determine whether the membrane-bound oligomers were single or double layer oligomers and impossible to visualize the conformation of the central pore-forming region of membrane-bound VacA. To define the structural organization of VacA s1/i1/m1 associated with lipid bilayers, we used negative stain EM to image VacA bound to ePC/DOPS/chol (55/15/30 mol%) and heart lipid LUVs (Figs. 1C and data not shown), two lipid compositions that support formation of anion-selective VacA pores (Czajkowsky et al., 1999; Vinion-Dubiel et al., 1999). From this analysis, we found that VacA can oligomerize on membranes in an acidic buffer condition (i.e. pH of 3) that does not support oligomerization in solution (Cover et al., 1997; Molinari et al., 1998), suggesting that monomeric VacA can bind to lipid and then oligomerize. From our EM images we collected datasets of greater than 2,000 particles for both ePC/DOPS/chol- and heart lipid-bound VacA s1/i1/m1. The datasets were classified into 2D class averages using multi-reference alignment (Figs. S2A–D). Analyses of the resulting classes showed that VacA s1/i1/m1 binds to membranes of either composition as an oligomer with six chiral “arms” (Figs. 1D and S2A–D). While a small number of oligomers with seven chiral arms were observed, there were not enough in the dataset to form a stable class average. Oligomers with achiral arms were not detected. Previous studies showed that oligomers with six or seven chiral arms correspond to hexamers or heptamers, whereas achiral oligomers correspond to dodecamers or tetradecamers (Adrian et al., 2002; Chambers et al., 2013; Cover et al., 1997; El-Bez et al., 2005). From these data we conclude that VacA s1/i1/m1 assembles on membranes predominantly as hexamers.
Membrane-bound VacA hexamers are structurally distinct from soluble oligomers and are inserted into the lipid bilayer
To determine whether there are structural differences between soluble and membrane-bound VacA hexamers, we compared 2D class averages of the two types of oligomers using radial density distribution plots, previously used to visualize differences between pre-pore and pore-formed Perfringolysin O (Dang et al., 2005), and difference maps. Radial density distribution plots were generated from normalized 2D class averages of soluble and membrane-bound VacA s1/i1/m1 hexamers (Fig. 2A,B and S2E). The plots were overlaid to compare the density across the radius of the soluble and membrane-bound hexamers (Fig. 2C). Additionally, a difference map was generated by subtracting the average of the membrane-bound VacA hexamer from the average of the soluble VacA hexamer (Fig. S3A–C). Both analyses showed that there is less density in the central, p33, region of membrane-bound s1/i1/m1 hexamers compared to soluble hexamers, suggesting that the p33 pore-forming domain undergoes a structural change when associated with membrane. To determine whether this difference is membrane composition-specific, we compared the radial density distribution plots and a difference map of soluble VacA s1/i1/m1 and VacA s1/i1/m1 bound to heart lipid membranes and obtained results similar to what was observed for VacA hexamers bound to ePC/DOPS/chol (Fig. S3D–G), indicating that the gross structural change observed in the central region of membrane-bound VacA s1/i1/m1 hexamers is not dependent on membrane composition.
Figure 2. The p33 domain of VacA s1/i1/m1 changes structure when bound to membranes.
(A–D) Comparison of soluble and membrane-bound VacA s1/i1/m1 hexamers. (A) Negative stain 2D average of soluble VacA s1/i1/m1 hexamers; 243 particles in class. (B) Negative stain 2D average of VacA s1/i1/m1 bound to ePC/DOPS/chol (55/15/30 mol%) LUVs; 917 particles in class. Same average shown in Figure 1D. Side length of averages, 42 nm. (C) Overlapping radial density distribution plots of normalized 2D averages of the soluble and membrane-bound s1/i1/m1 hexamers shown in panels A and B. Solid line = soluble hexamer; dotted line = membrane-bound hexamer. Yellow bar highlights the region of maximum difference. (D) Diagram showing the use of sodium carbonate stripped membranes to address whether membrane-bound proteins are peripheral or integral. Membranes treated with sodium carbonate are loaded at the bottom of an alkaline sucrose gradient (the highest density). After centrifugation, proteins peripherally associated with membranes remain in the highest density fraction, while membrane-inserted proteins float with the membranes into the lower density fractions. (E–F) Alkaline sucrose gradient analysis of Cdc15 F-BAR-E30K/E152K (E) and VacA s1/i1/m1 (F) without (−) or with (+) LUVs. Cdc15 F-BAR-E30K/E152K was bound to LUVs composed of DOPC/DOPS (80/20 mol%), and VacA s1/i1/m1 was bound to LUVs composed of ePC/DOPS/chol (55/15/30 mol%) before being treated with sodium carbonate and centrifuged in an alkaline sucrose gradient. Fractions were collected from top (low density) to bottom (high density) and analyzed by SDS-PAGE and Coomassie staining.
One explanation for decreased central region density of membrane-bound oligomers could be that the p33 domains inserted into the lipid bilayer, where they would not be visible by negative stain EM analysis; however, it is also possible that binding to membrane causes a loss of structural organization in the p33 domain which would also lead to a loss of density in the 2D average. Since the negative stain 2D averages were not sufficient to definitively differentiate among these models, we used alkaline carbonate sucrose gradient centrifugation to test whether the membrane-bound VacA hexamers were also membrane-inserted (Fig. 2D). This treatment separates integral membrane proteins from peripheral membrane proteins (Fossati et al., 2014). In this assay, the VacA bound LUVs were generated as described above and then treated with sodium carbonate before being loaded at the bottom (highest density fraction) of an alkaline sucrose gradient. Peripheral membrane-associated proteins are stripped from the LUVs under alkaline conditions and pellet in the high density sucrose fractions during centrifugation, while membrane-inserted proteins remain bound to the membranes and float into the lower density sucrose fractions (Fig. 2D). To verify this assay’s sensitivity for peripheral membrane binding proteins, we tested a mutant F-BAR domain from Schizosaccharomyces pombe Cdc15 (E30K/E152K), which binds but does not insert into membranes (McDonald et al., 2015). Cdc15 F-BAR-E30K/E152K was mixed with DOPC/DOPS (80/20 mol%) and acid activated VacA s1/i1/m1 was mixed with ePC/DOPS/chol (55/15/30 mol%) liposomes. Gradient fractions were collected from the top (low density) to the bottom (high density) and visualized by Coomassie staining on a SDS-PAGE gel. Cdc15 F-BAR-E30K/E152K sedimented in the highest density fraction, both in the absence and presence of liposomes (Fig. 2E), as expected for a peripheral membrane binding protein. VacA s1/i1/m1 remained in the highest density fraction when no liposomes were present, but floated into the lower density fractions in the presence of liposomes (Fig. 2F). The inability to strip VacA from membranes under alkaline conditions indicates that membrane-bound VacA s1/i1/m1 hexamers are membrane-inserted. Based on these results and combined with our analysis of the 2D averages of soluble and membrane-bound s1/i1/m1 hexamers (Fig. 2C and S3F,G), we conclude that the structural differences seen between soluble and membrane-bound VacA hexamers arise from insertion of the p33 domain into the membrane, a step required for pore formation.
Oligomerization is not required for insertion into the lipid bilayer
Next we investigated whether VacA Δ346–347, a mutant that cannot oligomerize in solution (Ivie et al., 2008), could oligomerize and insert into the lipid bilayer when bound to membrane. VacA Δ346–347 lacks two residues in the p55 domain (Fig. 3A), and consequently cannot form soluble oligomers even under neutral conditions, as determined by gel filtration (Ivie et al., 2008) and negative stain EM (Fig. 3B). Importantly, 2D averages of VacA s1/i1/m1 at pH 3 (a condition that causes soluble oligomers to disassemble into monomers) and VacA Δ346–347 at pH 7 were very similar, indicating that the two amino acid deletion in VacA Δ346–347 does not alter the overall structure of p88 monomers (Fig. 3C–E). Using liposome co-pelleting assays, we confirmed that VacA Δ346–347 binds to the same liposome compositions as those bound by VacA s1/i1/m1 (Fig. 3F and Fig. S4A–D); however, VacA Δ346–347 binds at reduced levels compared to wild-type VacA. We visualized VacA Δ346–347 bound to liposomes composed of ePC/DOPS/chol (55/15/30 mol%) using negative stain EM. Although VacA Δ346–347 bound to the LUVs (Fig. 3F), it remained monomeric (Fig. 3G). Thus, VacA residues 346 and 347 are essential for VacA oligomerization, both in solution and in the presence of membrane. In addition, the reduced binding of VacA Δ346–347 compared to wild-type VacA suggests that oligomerization may contribute to VacA-membrane interaction.
Figure 3. VacA Δ346–347, a non-oligomerizing mutant, can insert into the lipid bilayer.
(A) Schematic of VacA Δ346–347 mutant. p33 = blue; p55 = red; i1 = light blue; m1= black, location of Δ346–347 = yellow. (B) Representative negative stain image of soluble VacA Δ346–347. Scale bar, 50 nm. (C-D) 2D average of negatively stained acidified VacA s1/i1/m1 monomers (C) and VacA Δ346–347 at neutral pH (D). Side length of panels, 22.4 nm. Number of particles in each average shown in bottom right corner. (E) Cartoon of p88 to scale with averages in C and D. p55 (PDB: 2QV3(Gangwer et al., 2007)) in red, p33 in blue. (F) LUVs with various lipid compositions were incubated with 0.8 µM VacA Δ346–347 and the proportion of VacA Δ346–347 bound to the vesicles was determined. p <0.01, **; p < 0.001, ***. Results represent the mean + SEM based on three independent binding assays. PC: 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC); ePC: L-α-phosphatidylcholine from egg; PS: 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS); SM: sphingomyelin from egg; chol: cholesterol. (G) Representative negative stain image of VacA Δ346–347 bound to ePC/DOPS/chol (55/15/30 mol%) LUVs. Scale bar, 50 nm. (H) Alkaline sucrose gradient analysis of VacA Δ346–347 without (−) or with (+) LUVs. VacA Δ346–347 alone and bound to ePC/DOPS/chol (55/15/30 mol%) LUVs were treated with sodium carbonate and centrifuged in an alkaline sucrose gradient. Fractions were collected from top (low density) to bottom (high density) and analyzed by SDS-PAGE and Coomassie staining.
We next tested whether VacA Δ346–347 acts like a peripheral or integral membrane protein using the sodium carbonate extraction vesicle flotation assay. In the absence of liposomes, VacA Δ346–347 pelleted in the highest density gradient fractions. However, when combined with vesicles, VacA Δ346–347 floated into the low density gradient fractions (Fig. 3H), indicating the monomeric toxin had inserted into the bilayer. This finding shows that oligomerization is not required for VacA to transition from a soluble protein to a membrane-inserted protein.
Role of VacA p55 and p33 domains in membrane binding and insertion
With the unexpected finding that VacA can insert into membranes without first oligomerizing, we next investigated whether specific VacA domain(s) are required for membrane binding and insertion. First we tested whether the p55 domain alone can bind to membrane. p55 (amino acids 312–821) was expressed and purified as described (Gangwer et al., 2007) and used in liposome co-pelleting experiments. Similar to full length VacA s1/i1/m1, p55 bound to all the lipid compositions tested (Fig. 4A and S4E–H). Using negative stain EM analysis, the membrane-bound p55 was not visible (data not shown), consistent with the expected failure of membrane-bound p55 to form large oligomeric structures. To determine whether p55 binds as a peripheral membrane binding protein or inserts into the membrane, p55-bound vesicles were treated with sodium carbonate and subjected to centrifugation in an alkaline sucrose gradient. p55 sedimented in the highest density fraction, both in the absence and presence of liposomes (Fig. 4B), indicating that it is a peripheral membrane binding protein that does not insert into the membrane. These results provide direct biochemical evidence that while p55 binds membranes, the p33 domain is required for membrane insertion of VacA.
Figure 4. The roles of VacA domains in membrane insertion and binding.
(A) LUVs with various lipid compositions were incubated with 1.3 µM p55 and the proportion of p55 bound to the vesicles was then determined. p <0.05, *. Results represent the mean + SEM based on three independent binding assays. PC: 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC); ePC: L-α-phosphatidylcholine from egg; PS: 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS); SM: sphingomyelin from egg; chol: cholesterol. (B) Alkaline sucrose gradient analysis of p55 without (−) or with (+) LUVs. p55 alone or p55 bound to ePC/DOPS/chol (55/15/30 mol%) LUVs were treated with sodium carbonate and centrifuged in an alkaline sucrose gradient. Fractions were collected from top (low density) to bottom (high density) and analyzed by SDS-PAGE and Coomassie staining. (C) LUVs with various lipid compositions were incubated with 0.9 µM VacA Δ6–27 and the proportion of VacA Δ6–27 bound to the vesicles was determined. Graph represents quantification of three independent binding assays. (D) Alkaline sucrose gradient of VacA Δ6–27 without (−) or with (+) LUVs. VacA Δ6–27 alone or bound to LUVs composed of ePC/DOPS/chol (55/15/30 mol%) were treated with sodium carbonate and centrifuged in an alkaline sucrose gradient. Fractions were collected from top (low density) to bottom (high density) and analyzed by SDS-PAGE and Coomassie staining. (E) Representative negative stain image of VacA Δ6–27 bound to ePC/DOPS/chol (55/15/30 mol%) LUVs. Scale bar, 50 nm. (F) Representative class averages of membrane-bound VacA Δ6–27 hexamers and heptamers. Number of particles in each average shown in bottom left. Side length of panels, 42 nm.
VacA’s p33 domain contains three GXXXG motifs, is essential for anion-selective channel formation (McClain et al., 2003), and has an altered structure when bound to membrane as compared to soluble hexamers (Fig. 2A–C), suggesting that it binds and inserts into membrane. Since recombinantly expressed p33 is insoluble and only folds correctly in the presence of p55 (Gonzalez-Rivera et al., 2010), it was not possible to use isolated p33 in our assays. Therefore, to test the contribution of the N-terminus and the GXXXG repeats in membrane insertion, we examined the ability of VacA Δ6–27, a mutant lacking all three GXXXG repeats (Vinion-Dubiel et al., 1999), to bind membrane, insert into the lipid bilayer, and oligomerize. The GXXXG repeats are required for channel activity (McClain et al., 2003) and have been computationally modeled to form an α-helix that traverses the membrane and participates in toxin oligomerization (Kim et al., 2004). The 3D structures of soluble VacA Δ6–27 oligomers lack an organized central density (Chambers et al., 2013). VacA Δ6–27 bound to all lipid compositions tested (Figs. 4C and S4I–L). To determine whether the three GXXXG motifs are required for membrane insertion, VacA Δ6–27 was mixed with liposomes and subjected to sodium carbonate extraction followed by centrifugation in an alkaline sucrose gradient. VacA Δ6–27 remained in the highest density fraction in the absence of liposomes, but surprisingly, this mutant floated with membranes into the low density fractions, indicating that it had inserted into the membrane (Fig. 4D). Therefore, while the GXXXG motifs are required for active pore formation (McClain et al., 2003), our analysis suggests that these repeats are not the only region of the p33 domain that inserts into the membrane. To explore whether the GXXXG repeats are required for VacA oligomerization when bound to membranes, we used negative stain EM to visualize membrane-bound VacA Δ6–27. Unlike membrane-bound VacA Δ346–347, which remains monomeric (Fig. 3G), membrane-bound VacA Δ6–27 oligomerized into chiral hexamers (87%) and heptamers (13%) (Figs. 4E,F and S5A–D). Thus, the putative transmembrane N-terminal GXXXG repeats are not required for either VacA membrane insertion or oligomerization.
Finally, we investigated the interactions of type s2 forms of VacA with membranes. In comparison to type s1 forms of VacA, type s2 VacA variants contain a 12-residue hydrophilic N-terminal extension (Atherton et al., 1995). The s2 variants of VacA form ion channels less efficiently than s1 variants, and do not vacuolate cells (Atherton et al., 1995; Letley and Atherton, 2000; Letley et al., 2003; McClain et al., 2001b). The N-terminal extension could potentially ablate VacA function by disrupting toxin oligomerization on membranes, blocking insertion into the lipid bilayer, and/or altering pore conformation. While 3D structural analysis of soluble VacA s2/i1/m1 oligomers found no significant differences between soluble s1/i1/m1 and s2/i1/m1 dodecamers and tetradecamers (Chambers et al., 2013), the effect of this extension on VacA interactions with membranes has not been tested. To investigate this topic, we first confirmed that VacA s2/i1/m1 bound to all membrane compositions previously assayed (Fig. 5A and S4M–P). To determine whether the s2 VacA variant can insert into the lipid bilayer, VacA s2/i1/m1 bound vesicles were treated with sodium carbonate and centrifuged in an alkaline sucrose gradient. VacA s2/i1/m1 sedimented in the highest density fraction in the absence of liposomes, but when added to vesicles it floated with the membranes into the low density fractions (Fig. 5B). Therefore, the 12-amino acid extension does not inhibit membrane insertion. Visualizing VacA s2/i1/m1 bound to LUVs using single particle negative stain EM showed that the s2 variant oligomerizes into chiral hexamers (80%), heptamers (20%) (Fig. 5C,D and S5E–H), and under certain lipid compositions, even octamers (Fig. S5H). In contrast, heptamers and octamers were infrequently observed under these conditions in experiments with s1/i1/m1 VacA (Fig. 1 and S2). Thus, the addition or the deletion of residues at the N-terminus of VacA alters the types of membrane-bound oligomers that are able to form.
Figure 5. The N-terminal extension on VacA s2/i1/m1 does not alter membrane binding or membrane insertion, but does influence the types of oligomers that form.
(A) LUVs with various lipid compositions were incubated with 0.8 µM VacA s2/i1/m1 and the proportion of VacA s2/i1/m1 bound to the vesicles was determined. p <0.01, **. Results represent the mean + SEM based on three independent binding assays. PC: 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC); ePC: L-α-phosphatidylcholine from egg; PS: 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS); SM: sphingomyelin from egg; chol: cholesterol. (B) Alkaline sucrose gradient of VacA s2/i1/m1 without (-) or with (+) LUVs. VacA s2/i1/m1 alone or bound to LUVs composed of ePC/DOPS/chol (55/15/30 mol%) were treated with sodium carbonate and centrifuged in an alkaline sucrose gradient. Fractions were collected from top (low density) to bottom (high density) and analyzed by SDS-PAGE and Coomassie staining. (C) Representative negative stain image of VacA s2/i1/m1 bound to ePC/DOPS/chol (55/15/30 mol%) LUVs. Scale bar, 50 nm. (D) Representative class averages of membrane-bound VacA s2/i1/m1 hexamers and heptamers. Number of particles in each average shown in bottom left. Side length of panels, 42 nm.
DISCUSSION
Our studies provide the first detailed structural characterization of membrane-bound VacA and address the role(s) of specific regions of VacA in membrane binding, oligomerization, and membrane-insertion. We show that VacA oligomerizes into hexamers when bound to membrane and that this oligomerization occurs under pH conditions that do not support oligomerization of soluble VacA. Comparison of soluble and membrane-bound VacA hexamers reveals a prominent structural difference in the central region of the oligomers. Combined with our biochemical results indicating that membrane-bound VacA hexamers are also membrane-inserted, we conclude that this structural difference results from insertion of the p33 domains into the membrane to form a hexameric pore. Importantly, membrane insertion is not dependent on toxin oligomerization.
Under neutral buffer conditions in solution, we previously showed that VacA organizes into a heterogeneous combination of hexamers, heptamers, dodecamers, and tetradecamers, with ~90% of soluble VacA found as achiral, double layer dodecamers and tetradecamers (Chambers et al., 2013). In contrast, we find that membrane-bound VacA s1/i1/m1 organizes mainly into chiral hexamers, with no dodecamers or tetradecamers present. The oligomerization into membrane-bound VacA hexamers is not dependent on membrane composition, since hexamers formed on membranes composed of either DOPC or heart lipids. Additionally, membrane-bound VacA hexamers formed independent of whether lipid raft components, such as cholesterol and sphingomyelin, were present. Our negative stain EM analysis concurs with the results of single channel and computational modeling studies that predicted VacA pores would be hexamers composed of six subunits (Iwamoto et al., 1999; Kim et al., 2004). The double layer oligomers found in solution are presumed to result from hydrophobic interactions between p88 molecules that would normally be made with membranes. Altogether, these data indicate that in the presence of membranes, VacA monomers organize into stable hexamers and these hexamers form membrane-inserted pores.
It has long been appreciated that maximum toxin activity in vitro requires acid activation of purified VacA before it is added to cells (Cover et al., 1997; de Bernard et al., 1995; McClain et al., 2000; Molinari et al., 1998; Yahiro et al., 1999). The need for acid activation has been attributed to changes in VacA structure, most notably the disassembly of VacA oligomers in an acidic environment, but it has also been proposed that low pH changes the conformation of p88 so that it can penetrate the lipid bilayer more efficiently (de Bernard et al., 1995; McClain et al., 2000; Molinari et al., 1998; Pagliaccia et al., 2000). While there are many precedents for toxins undergoing conformational changes upon exposure to low pH environments (Boquet et al., 1976; Choe et al., 1992; Leka et al., 2014; O'Keefe et al., 1992; Pruitt et al., 2010), our results for VacA show that exposure to low pH does not lead to a gross structural rearrangement. Indeed, 2D class averages of VacA s1/i1/m1 monomers at pH 3 and VacA Δ346–347 at pH 7 are indistinguishable. Additionally, VacA Δ346–347 still inserts into the lipid bilayer at a neutral pH, showing that acid activation is not required for membrane insertion. From these results we conclude that the main importance of acid activating VacA is to simply dissociate the dodecamers and tetradecamers that make up ~90% of the sample at a neutral pH. The released p88 monomers can then oligomerize on membranes (perhaps via hydrophobic regions that were inaccessible in the double layer oligomers), or alternatively, the monomers may bind specific cell surface receptors with increased affinity compared to that of oligomerized toxin (Yahiro et al., 1999). Once bound to the membrane, we predict that regions of p33 undergo structural rearrangement(s) that allow VacA to insert into the bilayer and oligomerize.
Multiple lines of evidence suggest that the p33 domain forms the VacA anion-selective channel, and both the p33 and p55 domains are involved in VacA binding to host cells and oligomerization (Cover and Blanke, 2005; Domanska et al., 2010; Gonzalez-Rivera et al., 2010; McClain et al., 2003; Torres et al., 2005). Difficulties in purifying properly folded recombinant p33 have made it a challenge to examine the individual contributions of the p33 and p55 domains to membrane binding, membrane insertion, oligomerization, and pore formation. We found that while recombinant p55 binds membranes, it does not oligomerize on membranes or insert into the lipid bilayer. In previous studies using the TOXCAT system, a method developed to study TM helix-helix associations (Russ and Engelman, 1999, 2000), the N-terminal hydrophobic region of VacA (amino acids 1–32) was found to insert into the membrane and drive dimerization (McClain et al., 2001a; McClain et al., 2003). Conversely, in the current study we found that VacA Δ6–27, a mutant lacking all three N-terminal GXXXG repeats, was still able to insert into the lipid bilayer. This finding suggests that there are previously unrecognized regions of p33 that are important for facilitating the transition of VacA from a soluble toxin into an active ion channel; since these regions presumably insert into membranes, they could potentially be important structural components of the VacA ion channel.
It is currently not clear which regions in addition to the hydrophobic N-terminus can insert into the membrane. Analysis of the p33 domain hydrophobicity plot does not predict any obvious TM domains in addition to the previously recognized GXXXG motifs (McClain et al., 2003; Vinion-Dubiel et al., 1999). However, previous studies using VacA deletion mutants to map regions in p33 that are required for oligomerization in solution (Genisset et al., 2006; Vinion-Dubiel et al., 1999) provided evidence that multiple regions of p33 other than the N-terminus contribute to oligomerization. Additionally, one study, using a combination of proteolytic digestion and mass spectrometry analysis, reported that VacA residues 40–66, 111–169, 205–266, 548-–574 and 723–767 were protected from proteolysis because of their interaction with the membrane (Wang et al., 2000). Two of these protected peptides (548–574 and 723–767) are in p55, a domain that we show does not insert into the membrane. The other three peptides (40–66, 111–169 and 205–266) are located in the p33 domain, making them possible candidates for membrane insertion. Membrane-protected peptides spanning residues 1–32 were not detected, leading the authors to suggest that the N-terminus of VacA regulates channel formation through an unknown mechanism that does not involve forming the actual channel (Wang et al., 2000). While this conclusion is consistent with their mass spectrometry results, it does not fit with other studies showing that the N-terminus of VacA is required for insertion of the protein into membranes and formation of an anion channel (Kim et al., 2004; McClain et al., 2001b; McClain et al., 2003; Vinion-Dubiel et al., 1999). High-resolution structural analysis of membrane-bound VacA will be needed to elucidate the membrane topology of VacA in the context of the lipid bilayer and conclusively define which regions of p33 are essential and sufficient for pore formation.
While the current results show that VacA Δ6–27 can still insert into membranes, several lines of evidence indicate that the first 32 amino acids of the p33 domain are essential for the formation of a functional ion channel. Deletion of amino acids Δ6–27 abrogates the capacity of VacA to form pores in planar lipid bilayers and its capacity to cause vacuolation of cells (McClain et al., 2001b; Vinion-Dubiel et al., 1999), and mutant proteins containing specific point mutations near or in the third G14XXXG18 motif lack channel activity and do not cause cellular vacuolation (McClain et al., 2001b; McClain et al., 2003). Additionally, VacA s2/i1/m1, a mutant that contains a 12-amino acid N-terminal extension, has reduced pore activity and does not vacuolate cells (McClain et al., 2001b). Collectively, these studies show that the p33 N-terminal region is required for pore formation and is sensitive to structural alterations. While the first 32 amino acids are not required for oligomerization in solution or on membrane (Chambers et al., 2013; El-Bez et al., 2005; Vinion-Dubiel et al., 1999), modifying the VacA N-terminus, either by adding or deleting amino acids, alters the ratio of hexamer and heptamer formation on membranes. Thus, the first 32 amino acids of VacA play at least an ancillary role in organizing oligomer formation of membrane-bound VacA. This conclusion is consistent with previous studies showing that point mutations made in the N-terminus of VacA reduce TM dimerization (McClain et al., 2001a; McClain et al., 2003), as well as the prediction made by computational modeling that the GXXXG motifs make numerous intramolecular contacts required for the proper organization of a hexameric ion channel (Kim et al., 2004).
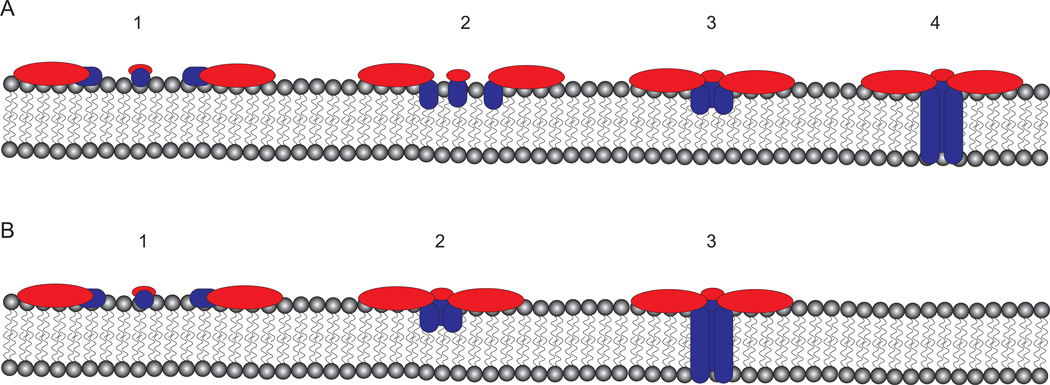
In summary, we have shown that membrane-bound VacA assembles into hexameric structures, that there is a distinct structural difference in the central region of soluble hexamers compared to membrane-bound hexamers, and that insertion of VacA into the lipid membrane is not dependent on oligomerization. Our findings predict that VacA pore formation requires multiple regions of VacA in addition to the N-terminal GXXXG repeats. We propose a model where monomeric VacA binds to lipids and the p33 domain inserts into the lipid bilayer to oligomerize and form a pore (Fig. 6). The insertion of p33 into the membrane leads to a clear structural difference between soluble and membrane-bound VacA hexamers. Since oligomerization is not required for p33 to insert into the membrane, we propose a model in which 1) VacA monomers bind to membrane, 2) the p33 domain inserts into the lipid-bilayer, and 3) the pore is formed (Fig. 6). However, our data do not resolve questions about the temporal order of membrane-insertion, oligomerization, and pore formation. Thus, multiple models are possible, including one where VacA membrane insertion and pore formation occur concomitantly (Fig. 6). Continuing biophysical and structural analyses of membrane-bound VacA s1/i1/m1 and VacA mutants will be essential for dissecting the interrelated steps of oligomerization, insertion, and pore formation, and elucidating the molecular organization of the VacA anion channel.
Figure 6. Models of VacA pore formation in membranes.
Monomeric VacA binds to membrane where regions of p33 insert into the lipid bilayer, either before (A) or concomitant (B) with p88 oligomerization into a hexamer forming a functional pore. Further biophysical studies are needed to determine the temporal order of membrane insertion, oligomerization, and pore formation. p55 depicted in red, while p33 is shown in blue.
EXPERIMENTAL PROCEDURES
Purification of H. pylori VacA, p55 recombinant VacA, and Schizosaccharomyces pombe Cdc15 F-BAR
H. pylori strain 60190 expressing VacA s1/i1/m1 and mutant strains expressing VacA s2/i1/m1 or VacA Δ6–27 have been previously described (McClain et al., 2001b; Vinion-Dubiel et al., 1999). Strains were grown in Brucella broth supplemented with cholesterol (Jimenez-Soto et al., 2012) and VacA was isolated as previously described (Cover et al., 1997; Vinion-Dubiel et al., 1999). To purify a non-oligomerizing form of VacA, an H. pylori strain expressing VacA Δ346–347 (Willhite and Blanke, 2004; Ye et al., 1999) was modified so that it produced VacA containing an internal strep tag inserted at amino acid 312. VacA Δ346–347-Str312 was then purified using Strep-Tactin resin (IBA, Goettingen, Germany). The VacA p55 domain (residues 312–821) was expressed in Escherichia coli and purified as described (Gangwer et al., 2007). Recombinant S. pombe Cdc15 F-BAR-E30K/E152K was expressed and purified as described (McDonald et al., 2015). Recombinant proteins were visualized using SDS-PAGE and Coomassie staining.
Liposome co-pelleting assays
Large unilamellar vesicles (LUVs) were generated as described (Itoh et al., 2005), using the following lipid mixtures: 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC); L-α-phosphatidylcholine from egg (ePC), 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS), and ovine cholesterol (55/15/30 % of total moles (mol%)); DOPC, DOPS, and egg sphingomyelin (eSM) (45/20/35 mol%); DOPC, DOPS, eSM, and cholesterol (35/20/35/10 mol%). All lipids were purchased from Avanti Polar Lipids (Alabaster, AL). Briefly, lipids in chloroform (CHCl3) were mixed at the desired ratios and evaporated under a nitrogen stream, further dried in a vacuum overnight, and rehydrated in 10 mM HEPES pH 7.2 and 100 mM potassium chloride (KCl) to a final concentration of 1 mg/mL lipid. Lipid mixtures were vortexed, exposed to three cycles of freeze thawing using a dry ice/ethanol bath and 37°C water bath, and then extruded using an Avanti extruder with an 800 nm filter. Before being mixed with the LUVs, VacA (s1/i1/m1, s2/i1/m1, and Δ6–27) was acid activated by lowering the pH to 3.0 resulting in disassembly of oligomers as previously described (Cover et al., 1997). VacA Δ346–347 was not acid activated since this mutant protein is defective in oligomerization and does not require acid-induced disassembly (Ivie et al., 2008). Acid activated VacA s1/i1/m1, s2/i1/m1, Δ6–27, Δ346–347, p55, and bovine serum albumin (BSA) were mixed at increasing concentrations (µM) with 100 µg LUVs and incubated for 15 minutes at 25°C. Samples were spun down using a Beckman Coulter (Pasadena, CA) TL-100 ultracentrifuge and TLA-100 rotor at 156,425 × g for 45 minutes at 25°C. The supernatant (unbound sample) was removed and the liposome pellet (bound sample) suspended in 10 mM HEPES pH 7.2 and 100 mM KCl. Samples were run on 4–12% Bis-Tris SDS-PAGE gels (Invitrogen, Carlsbad CA) and stained with Coomassie. Gels were digitized using a CanonScan 8800F digital scanner and quantified using ImageJ (Schneider et al., 2012). Each liposome co-pelleting assay was repeated three times in independent experiments. The percent of VacA bound to membrane was determined by averaging intensity measurements of bands corresponding to membrane-bound and unbound VacA. Results represent the mean + standard error of the mean (SEM) based on three independent experiments. Graphs and statistics were generated using GraphPad Prism (version 5.0a).
VacA insertion assay
VacA s1/i1/m1, s2/i1/m1, Δ6–27, Δ346–347, and p55 were incubated with ePC/DOPS/chol (55/15/30 mol%) at a lipid-to-protein ratio (LPR) of 50:1 (w/w) for 15 minutes at room temperature. Cdc15 F-BAR-E30K/E152K was mixed with DOPC/DOPS (80/20 mol%) LUVs at a LPR of 5:1 (w/w) and incubated for 15 minutes at room temperature. Buffer only controls were prepared for all protein-lipid samples. Protein-bound liposome samples were alkalinized with an equal volume of 0.2 M sodium carbonate (Na2CO3) for 30 minutes on ice to compete off peripherally bound protein. This treatment strips peripheral, but not integral, bound proteins from the membrane (Fossati et al., 2014). A final concentration of 1.2 M sucrose in 0.1 M Na2CO3 was then added. A discontinuous sucrose gradient was prepared with alkalinized protein-liposomes at the bottom, followed by layers of 1 M, 0.5 M, 0.25 M, 0.15 M, and 0 M sucrose in 0.1 M Na2CO3. The gradients were spun at 108,759 × g for 16 hours at 4°C to allow flotation of liposomes into lower density fractions (Beckman Coulter, Pasadena, CA). Fractions of 700 µL were collected and precipitated with TCA. Samples were hydrated with 10 mM HEPES pH 7.2, 100 mM KCl and 2× SDS sample loading dye. Samples were run on a 10% Bis-Tris SDS-PAGE gel (Life Technologies) and Coomassie stained.
Sample preparation for EM analysis
For negative stain single particle EM of membrane-bound VacA, LUVs composed of ePC/DOPS/chol (55/15/30 mol%) and heart lipid extract in 10 mM HEPES pH 7.2, 100 mM KCl were prepared as described above. VacA s1/i1/m1, s2/i1/m1, and Δ6–27 were acid activated by lowering the pH to 3.0 (Cover et al., 1997), mixed with LUVs (either ePC/DOPS/chol or heart lipids), and incubated for 15 minutes at 37°C. A LPR of 50:1 (w/w) was used for each VacA sample. For analyzing membrane-bound VacA Δ346–347, LUVs composed of ePC/DOPS/chol (55/15/30 mol%) or DOPC/DOPS/eSM/chol (35/20/35/10 mol%) in 10 mM HEPES pH 7.2, 100 mM KCl at LPRs of 100:1 (w/w) and 50:1 (w/w), were prepared as described above. VacA Δ346–347 was not acid activated prior to mixing with LUVs, since this mutant protein is defective in oligomerization and does not require acid-induced disassembly (Ivie et al., 2008). Grids containing soluble VacA s1/i1/m1, s2/i1/m1, Δ346–347, and Δ6–27 were made as described (Chambers et al., 2013). To visualize soluble monomeric VacA s1/i1/m1, the pH of the sample was lowered to 3.0 before making grids.
Electron Microscopy (EM)
For negative stain EM grids, samples were adsorbed to a glow discharged 200-mesh copper grid covered with carbon-coated collodion film (EMS, Hatfield, PA). Grids were washed in two drops of water and stained with two drops of uranyl formate (0.75%) (EMS, Hatfield, PA) as described (Ohi et al., 2004). Samples were visualized on a Tecnai TF20 electron microscope (FEI, Hillsboro, OR) equipped with a field emission gun at an acceleration voltage of 200 kV under low-dose conditions. Images were taken at a magnification of 62,000× at a defocus value of −1.5 µm and recorded on a 4k × 4k CCD Ultrascan camera (Gatan, Pleasanton, CA). Images were converted to mixed raster content (mrc) format and binned by two, resulting in final images with a pixel size of 3.5 Å/pixel. Images of VacA Δ346–347-LUVs were taken on a FEI Morgagni (FEI, Hillsboro, OR) equipped with a tungsten filament at an acceleration voltage of 100kV and a magnification of 28,000×.
Classification and difference mapping
Particles were selected interactively using BOXER in the program EMAN (Ludtke, 2010) and windowed into 120 × 120 pixel images (42 nm × 42 nm). Boxed particles were rotationally and translationally aligned and subjected to ten rounds of reference free alignment using the program SPIDER (Shaikh et al., 2008). Datasets used include: 5,763 particles for soluble VacA s1/i1/m1; 2,207 particles for VacA s1/i1/m1-ePC/DOPS/chol LUVs; 3,800 particles for VacA s1/i1/m1-heart lipid LUVs; 2,142 particles for VacA s2/i1/m1-ePC/DOPS/chol LUVs; 2,919 particles for s2/i1/m1-heart lipid LUVs; 4,186 particles for VacA Δ6–27; 2,429 particles for VacA Δ6–27-ePC/DOPS/chol LUVs; and 2,111 particles for VacA Δ6–27-heart lipid LUVs. Four or five representative averages for each VacA variant were chosen and particles were subjected to reference based alignment. 2D averages of soluble and membrane-bound oligomers were compared by generating and then overlaying radial density distribution plots of normalized 2D averages as described (Dang et al., 2005). Averages were normalized and values for the radial density distribution plots and difference maps were calculated using the program SPIDER (Shaikh et al., 2008).
Supplementary Material
SIGNIFICANCE.
Helicobacter pylori colonizes the stomach and increases the risk of gastroduodenal disease in infected individuals. H. pylori VacA, a secreted toxin, forms channels in cell membranes and causes alterations in host cells. The vacA genotype of a strain correlates with the risk of H. pylori-associated gastric cancer and the various forms of VacA have different activities in vitro. We show that upon binding to membrane, monomeric VacA oligomerizes into membrane-inserted hexamers. There is a visible structural difference in the pore-forming domain of VacA when comparing soluble and membrane-bound hexamers. These results provide important new insights into mechanisms of VacA oligomerization and membrane insertion.
Acknowledgments
This work was supported by NIH F31AI112324 to T.M.P.; T32GM08320 to N.J.F.; T32AI007821 to C.G.; AHA fellowship 15PRE21780003 to N.A.M.; AI039657, CA116087, AI118932 and the Department of Veterans Affairs to T.L.C.; RO1AI039657, AI118932 and the Vanderbilt Center for Structural Biology to M.D.O. We thank members of the Cover, Gould, and Ohi labs and Dr. Borden Lacy for helpful scientific discussions.
Footnotes
All authors confirm that there are no conflicts of interest.
REFERENCES
- Adrian M, Cover TL, Dubochet J, Heuser JE. Multiple oligomeric states of the Helicobacter pylori vacuolating toxin demonstrated by cryo-electron microscopy. Journal of molecular biology. 2002;318:121–133. doi: 10.1016/S0022-2836(02)00047-5. [DOI] [PubMed] [Google Scholar]
- Amieva MR, El-Omar EM. Host-bacterial interactions in Helicobacter pylori infection. Gastroenterology. 2008;134:306–323. doi: 10.1053/j.gastro.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Anonymous. International Agency for Research on Cancer. France: Lyons; 1994. Schistosomes, liver flukes and Helicobacter pylori IARC monographs on the evaluation of carcinogenic risks to humans. [PMC free article] [PubMed] [Google Scholar]
- Atherton JC. The pathogenesis of Helicobacter pylori-induced gastro-duodenal diseases. Annual review of pathology. 2006;1:63–96. doi: 10.1146/annurev.pathol.1.110304.100125. [DOI] [PubMed] [Google Scholar]
- Atherton JC, Blaser MJ. Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. The Journal of clinical investigation. 2009;119:2475–2487. doi: 10.1172/JCI38605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton JC, Cao P, Peek RM, Jr, Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori Association of specific vacA types with cytotoxin production and peptic ulceration. The Journal of biological chemistry. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- Boquet P, Silverman MS, Pappenheimer AM, Jr, Vernon WB. Binding of triton X-100 to diphtheria toxin, crossreacting material 45, and their fragments. Proceedings of the National Academy of Sciences of the United States of America. 1976;73:4449–4453. doi: 10.1073/pnas.73.12.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calore F, Genisset C, Casellato A, Rossato M, Codolo G, Esposti MD, Scorrano L, de Bernard M. Endosome-mitochondria juxtaposition during apoptosis induced by H. pylori VacA. Cell death and differentiation. 2010;17:1707–1716. doi: 10.1038/cdd.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campello S, Tombola F, Cabrini G, Zoratti M. The vacuolating toxin of Helicobacter pylori mimicks the CFTR-mediated chloride conductance. FEBS letters. 2002;532:237–240. doi: 10.1016/s0014-5793(02)03656-6. [DOI] [PubMed] [Google Scholar]
- Chambers MG, Pyburn TM, Gonzalez-Rivera C, Collier SE, Eli I, Yip CK, Takizawa Y, Lacy DB, Cover TL, Ohi MD. Structural analysis of the oligomeric states of Helicobacter pylori VacA toxin. Journal of molecular biology. 2013;425:524–535. doi: 10.1016/j.jmb.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Bennett MJ, Fujii G, Curmi PM, Kantardjieff KA, Collier RJ, Eisenberg D. The crystal structure of diphtheria toxin. Nature. 1992;357:216–222. doi: 10.1038/357216a0. [DOI] [PubMed] [Google Scholar]
- Cover TL. Helicobacter pylori Diversity and Gastric Cancer Risk. mBio. 2016;7 doi: 10.1128/mBio.01869-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover TL, Blanke SR. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nature reviews. 2005;3:320–332. doi: 10.1038/nrmicro1095. [DOI] [PubMed] [Google Scholar]
- Cover TL, Blaser MJ. Helicobacter pylori in health and disease. Gastroenterology. 2009;136:1863–1873. doi: 10.1053/j.gastro.2009.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover TL, Hanson PI, Heuser JE. Acid-induced dissociation of VacA, the Helicobacter pylori vacuolating cytotoxin, reveals its pattern of assembly. The Journal of cell biology. 1997;138:759–769. doi: 10.1083/jcb.138.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czajkowsky DM, Iwamoto H, Cover TL, Shao Z. The vacuolating toxin from Helicobacter pylori forms hexameric pores in lipid bilayers at low pH. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:2001–2006. doi: 10.1073/pnas.96.5.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czajkowsky DM, Iwamoto H, Szabo G, Cover TL, Shao Z. Mimicry of a host anion channel by a Helicobacter pylori pore-forming toxin. Biophysical journal. 2005;89:3093–3101. doi: 10.1529/biophysj.105.066746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang TX, Hotze EM, Rouiller I, Tweten RK, Wilson-Kubalek EM. Prepore to pore transition of a cholesterol-dependent cytolysin visualized by electron microscopy. Journal of structural biology. 2005;150:100–108. doi: 10.1016/j.jsb.2005.02.003. [DOI] [PubMed] [Google Scholar]
- de Bernard M, Papini E, de Filippis V, Gottardi E, Telford J, Manetti R, Fontana A, Rappuoli R, Montecucco C. Low pH activates the vacuolating toxin of Helicobacter pylori, which becomes acid and pepsin resistant. The Journal of biological chemistry. 1995;270:23937–23940. doi: 10.1074/jbc.270.41.23937. [DOI] [PubMed] [Google Scholar]
- de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, Plummer M. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. The Lancet. Oncology. 2012;13:607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- Debellis L, Papini E, Caroppo R, Montecucco C, Curci S. Helicobacter pylori cytotoxin VacA increases alkaline secretion in gastric epithelial cells. American journal of physiology. Gastrointestinal and liver physiology. 2001;281:G1440–G1448. doi: 10.1152/ajpgi.2001.281.6.G1440. [DOI] [PubMed] [Google Scholar]
- Domanska G, Motz C, Meinecke M, Harsman A, Papatheodorou P, Reljic B, Dian-Lothrop EA, Galmiche A, Kepp O, Becker L, et al. Helicobacter pylori VacA toxin/subunit p34: targeting of an anion channel to the inner mitochondrial membrane. PLoS pathogens. 2010;6:e1000878. doi: 10.1371/journal.ppat.1000878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Bez C, Adrian M, Dubochet J, Cover TL. High resolution structural analysis of Helicobacter pylori VacA toxin oligomers by cryo-negative staining electron microscopy. Journal of structural biology. 2005;151:215–228. doi: 10.1016/j.jsb.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Figueiredo C, Machado JC, Pharoah P, Seruca R, Sousa S, Carvalho R, Capelinha AF, Quint W, Caldas C, van Doorn LJ, et al. Helicobacter pylori and interleukin 1 genotyping: an opportunity to identify high-risk individuals for gastric carcinoma. Journal of the National Cancer Institute. 2002;94:1680–1687. doi: 10.1093/jnci/94.22.1680. [DOI] [PubMed] [Google Scholar]
- Fossati M, Goud B, Borgese N, Manneville JB. An investigation of the effect of membrane curvature on transmembrane-domain dependent protein sorting in lipid bilayers. Cellular logistics. 2014;4:e29087. doi: 10.4161/cl.29087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs CS, Mayer RJ. Gastric carcinoma. The New England journal of medicine. 1995;333:32–41. doi: 10.1056/NEJM199507063330107. [DOI] [PubMed] [Google Scholar]
- Galmiche A, Rassow J, Doye A, Cagnol S, Chambard JC, Contamin S, de Thillot V, Just I, Ricci V, Solcia E, et al. The N-terminal 34 kDa fragment of Helicobacter pylori vacuolating cytotoxin targets mitochondria and induces cytochrome c release. The EMBO journal. 2000;19:6361–6370. doi: 10.1093/emboj/19.23.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangwer KA, Mushrush DJ, Stauff DL, Spiller B, McClain MS, Cover TL, Lacy DB. Crystal structure of the Helicobacter pylori vacuolating toxin p55 domain. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16293–16298. doi: 10.1073/pnas.0707447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangwer KA, Shaffer CL, Suerbaum S, Lacy DB, Cover TL, Bordenstein SR. Molecular evolution of the Helicobacter pylori vacuolating toxin gene vacA. Journal of bacteriology. 2010;192:6126–6135. doi: 10.1128/JB.01081-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier NC, Monzo P, Gonzalez T, Doye A, Oldani A, Gounon P, Ricci V, Cormont M, Boquet P. Early endosomes associated with dynamic F-actin structures are required for late trafficking of H. pylori VacA toxin. The Journal of cell biology. 2007;177:343–354. doi: 10.1083/jcb.200609061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert B, Fischer W, Weiss E, Hoffmann R, Haas R. Helicobacter pylori vacuolating cytotoxin inhibits T lymphocyte activation. Science (New York, N.Y. 2003;301:1099–1102. doi: 10.1126/science.1086871. [DOI] [PubMed] [Google Scholar]
- Geisse NA, Cover TL, Henderson RM, Edwardson JM. Targeting of Helicobacter pylori vacuolating toxin to lipid raft membrane domains analysed by atomic force microscopy. The Biochemical journal. 2004;381:911–917. doi: 10.1042/BJ20031719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genisset C, Galeotti CL, Lupetti P, Mercati D, Skibinski DA, Barone S, Battistutta R, de Bernard M, Telford JL. A Helicobacter pylori vacuolating toxin mutant that fails to oligomerize has a dominant negative phenotype. Infection and immunity. 2006;74:1786–1794. doi: 10.1128/IAI.74.3.1786-1794.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rivera C, Algood HM, Radin JN, McClain MS, Cover TL. The Intermediate Region of Helicobacter pylori VacA Is a Determinant of Toxin Potency in a Jurkat T Cell Assay. Infection and immunity. 2012;80:2578–2588. doi: 10.1128/IAI.00052-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rivera C, Gangwer KA, McClain MS, Eli IM, Chambers MG, Ohi MD, Lacy DB, Cover TL. Reconstitution of Helicobacter pylori VacA toxin from purified components. Biochemistry. 2010;49:5743–5752. doi: 10.1021/bi100618g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Developmental cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Ivie SE, McClain MS, Torres VJ, Algood HM, Lacy DB, Yang R, Blanke SR, Cover TL. Helicobacter pylori VacA subdomain required for intracellular toxin activity and assembly of functional oligomeric complexes. Infection and immunity. 2008;76:2843–2851. doi: 10.1128/IAI.01664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto H, Czajkowsky DM, Cover TL, Szabo G, Shao Z. VacA from Helicobacter pylori: a hexameric chloride channel. FEBS letters. 1999;450:101–104. doi: 10.1016/s0014-5793(99)00474-3. [DOI] [PubMed] [Google Scholar]
- Jain P, Luo ZQ, Blanke SR. Helicobacter pylori vacuolating cytotoxin A (VacA) engages the mitochondrial fission machinery to induce host cell death. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16032–16037. doi: 10.1073/pnas.1105175108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Fernandez T, Burroni D, Pagliaccia C, Atherton JC, Reyrat JM, Rappuoli R, Telford JL. Cell specificity of Helicobacter pylori cytotoxin is determined by a short region in the polymorphic midregion. Infection and immunity. 2000;68:3754–3757. doi: 10.1128/iai.68.6.3754-3757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Soto LF, Rohrer S, Jain U, Ertl C, Sewald X, Haas R. Effects of cholesterol on Helicobacter pylori growth and virulence properties in vitro. Helicobacter. 2012;17:133–139. doi: 10.1111/j.1523-5378.2011.00926.x. [DOI] [PubMed] [Google Scholar]
- Kim S, Chamberlain AK, Bowie JU. Membrane channel structure of Helicobacter pylori vacuolating toxin: role of multiple GXXXG motifs in cylindrical channels. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:5988–5991. doi: 10.1073/pnas.0308694101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzavecchia S, Bellon PL, Lupetti P, Dallai R, Rappuoli R, Telford JL. Three-dimensional reconstruction of metal replicas of the Helicobacter pylori vacuolating cytotoxin. Journal of structural biology. 1998;121:9–18. doi: 10.1006/jsbi.1997.3941. [DOI] [PubMed] [Google Scholar]
- Leka O, Vallese F, Pirazzini M, Berto P, Montecucco C, Zanotti G. Diphtheria toxin conformational switching at acidic pH. The FEBS journal. 2014;281:2115–2122. doi: 10.1111/febs.12783. [DOI] [PubMed] [Google Scholar]
- Letley DP, Atherton JC. Natural diversity in the N terminus of the mature vacuolating cytotoxin of Helicobacter pylori determines cytotoxin activity. Journal of bacteriology. 2000;182:3278–3280. doi: 10.1128/jb.182.11.3278-3280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letley DP, Rhead JL, Twells RJ, Dove B, Atherton JC. Determinants of non-toxicity in the gastric pathogen Helicobacter pylori. The Journal of biological chemistry. 2003;278:26734–26741. doi: 10.1074/jbc.M304071200. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ. 3-D structures of macromolecules using single-particle analysis in EMAN. Methods in molecular biology (Clifton, N.J. 2010;673:157–173. doi: 10.1007/978-1-60761-842-3_9. [DOI] [PubMed] [Google Scholar]
- Lupetti P, Heuser JE, Manetti R, Massari P, Lanzavecchia S, Bellon PL, Dallai R, Rappuoli R, Telford JL. Oligomeric and subunit structure of the Helicobacter pylori vacuolating cytotoxin. The Journal of cell biology. 1996;133:801–807. doi: 10.1083/jcb.133.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- McClain MS, Cao P, Cover TL. Amino-terminal hydrophobic region of Helicobacter pylori vacuolating cytotoxin (VacA) mediates transmembrane protein dimerization. Infection and immunity. 2001a;69:1181–1184. doi: 10.1128/IAI.69.2.1181-1184.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain MS, Cao P, Iwamoto H, Vinion-Dubiel AD, Szabo G, Shao Z, Cover TL. A 12-amino-acid segment, present in type s2 but not type s1 Helicobacter pylori VacA proteins, abolishes cytotoxin activity and alters membrane channel formation. Journal of bacteriology. 2001b;183:6499–6508. doi: 10.1128/JB.183.22.6499-6508.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain MS, Iwamoto H, Cao P, Vinion-Dubiel AD, Li Y, Szabo G, Shao Z, Cover TL. Essential role of a GXXXG motif for membrane channel formation by Helicobacter pylori vacuolating toxin. The Journal of biological chemistry. 2003;278:12101–12108. doi: 10.1074/jbc.M212595200. [DOI] [PubMed] [Google Scholar]
- McClain MS, Schraw W, Ricci V, Boquet P, Cover TL. Acid activation of Helicobacter pylori vacuolating cytotoxin (VacA) results in toxin internalization by eukaryotic cells. Molecular microbiology. 2000;37:433–442. doi: 10.1046/j.1365-2958.2000.02013.x. [DOI] [PubMed] [Google Scholar]
- McDonald NA, Vander Kooi CW, Ohi MD, Gould KL. Oligomerization but Not Membrane Bending Underlies the Function of Certain F-BAR Proteins in Cell Motility and Cytokinesis. Developmental cell. 2015;35:725–736. doi: 10.1016/j.devcel.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memon AA, Hussein NR, Miendje Deyi VY, Burette A, Atherton JC. Vacuolating cytotoxin genotypes are strong markers of gastric cancer and duodenal ulcer-associated Helicobacter pylori strains: a matched case-control study. Journal of clinical microbiology. 2014;52:2984–2989. doi: 10.1128/JCM.00551-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari M, Galli C, de Bernard M, Norais N, Ruysschaert JM, Rappuoli R, Montecucco C. The acid activation of Helicobacter pylori toxin VacA: structural and membrane binding studies. Biochemical and biophysical research communications. 1998;248:334–340. doi: 10.1006/bbrc.1998.8808. [DOI] [PubMed] [Google Scholar]
- Nakayama M, Kimura M, Wada A, Yahiro K, Ogushi K, Niidome T, Fujikawa A, Shirasaka D, Aoyama N, Kurazono H, et al. Helicobacter pylori VacA activates the p38/activating transcription factor 2-mediated signal pathway in AZ-521 cells. The Journal of biological chemistry. 2004;279:7024–7028. doi: 10.1074/jbc.M308898200. [DOI] [PubMed] [Google Scholar]
- Nomura A, Stemmermann GN, Chyou PH, Kato I, Perez-Perez GI, Blaser MJ. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. The New England journal of medicine. 1991;325:1132–1136. doi: 10.1056/NEJM199110173251604. [DOI] [PubMed] [Google Scholar]
- O'Keefe DO, Cabiaux V, Choe S, Eisenberg D, Collier RJ. pH-dependent insertion of proteins into membranes: B-chain mutation of diphtheria toxin that inhibits membrane translocation, Glu-349----Lys. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:6202–6206. doi: 10.1073/pnas.89.13.6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliaccia C, de Bernard M, Lupetti P, Ji X, Burroni D, Cover TL, Papini E, Rappuoli R, Telford JL, Reyrat JM. The m2 form of the Helicobacter pylori cytotoxin has cell type-specific vacuolating activity. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:10212–10217. doi: 10.1073/pnas.95.17.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliaccia C, Wang XM, Tardy F, Telford JL, Ruysschaert JM, Cabiaux V. Structure and interaction of VacA of Helicobacter pylori with a lipid membrane. European journal of biochemistry / FEBS. 2000;267:104–109. doi: 10.1046/j.1432-1327.2000.00970.x. [DOI] [PubMed] [Google Scholar]
- Papini E, Satin B, de Bernard M, Molinari M, Arico B, Galli C, Telford JR, Rappuoli R, Montecucco C. Action site and cellular effects of cytotoxin VacA produced by Helicobacter pylori. Folia microbiologica. 1998a;43:279–284. doi: 10.1007/BF02818613. [DOI] [PubMed] [Google Scholar]
- Papini E, Satin B, Norais N, de Bernard M, Telford JL, Rappuoli R, Montecucco C. Selective increase of the permeability of polarized epithelial cell monolayers by Helicobacter pylori vacuolating toxin. The Journal of clinical investigation. 1998b;102:813–820. doi: 10.1172/JCI2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, Sibley RK. Helicobacter pylori infection and the risk of gastric carcinoma. The New England journal of medicine. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- Pruitt RN, Chambers MG, Ng KK, Ohi MD, Lacy DB. Structural organization of the functional domains of Clostridium difficile toxins A and B. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13467–13472. doi: 10.1073/pnas.1002199107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radin JN, Gonzalez-Rivera C, Ivie SE, McClain MS, Cover TL. Helicobacter pylori VacA induces programmed necrosis in gastric epithelial cells. Infection and immunity. 2011;79:2535–2543. doi: 10.1128/IAI.01370-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhead JL, Letley DP, Mohammadi M, Hussein N, Mohagheghi MA, Eshagh Hosseini M, Atherton JC. A new Helicobacter pylori vacuolating cytotoxin determinant, the intermediate region, is associated with gastric cancer. Gastroenterology. 2007;133:926–936. doi: 10.1053/j.gastro.2007.06.056. [DOI] [PubMed] [Google Scholar]
- Russ WP, Engelman DM. TOXCAT: a measure of transmembrane helix association in a biological membrane. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:863–868. doi: 10.1073/pnas.96.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ WP, Engelman DM. The GxxxG motif: a framework for transmembrane helix-helix association. Journal of molecular biology. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewald X, Fischer W, Haas R. Sticky socks: Helicobacter pylori VacA takes shape. Trends in microbiology. 2008;16:89–92. doi: 10.1016/j.tim.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, Frank J. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nature protocols. 2008;3:1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suerbaum S, Michetti P. Helicobacter pylori infection. The New England journal of medicine. 2002;347:1175–1186. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- Sundrud MS, Torres VJ, Unutmaz D, Cover TL. Inhibition of primary human T cell proliferation by Helicobacter pylori vacuolating toxin (VacA) is independent of VacA effects on IL-2 secretion. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7727–7732. doi: 10.1073/pnas.0401528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo I, Brutsche S, Tombola F, Moschioni M, Satin B, Telford JL, Rappuoli R, Montecucco C, Papini E, Zoratti M. Formation of anion-selective channels in the cell plasma membrane by the toxin VacA of Helicobacter pylori is required for its biological activity. The EMBO journal. 1999;18:5517–5527. doi: 10.1093/emboj/18.20.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teese MG, Langosch D. Role of GxxxG Motifs in Transmembrane Domain Interactions. Biochemistry. 2015;54:5125–5135. doi: 10.1021/acs.biochem.5b00495. [DOI] [PubMed] [Google Scholar]
- Terebiznik MR, Raju D, Vazquez CL, Torbricki K, Kulkarni R, Blanke SR, Yoshimori T, Colombo MI, Jones NL. Effect of Helicobacter pylori's vacuolating cytotoxin on the autophagy pathway in gastric epithelial cells. Autophagy. 2009;5:370–379. doi: 10.4161/auto.5.3.7663. [DOI] [PubMed] [Google Scholar]
- Tombola F, Carlesso C, Szabo I, de Bernard M, Reyrat JM, Telford JL, Rappuoli R, Montecucco C, Papini E, Zoratti M. Helicobacter pylori vacuolating toxin forms anion-selective channels in planar lipid bilayers: possible implications for the mechanism of cellular vacuolation. Biophysical journal. 1999a;76:1401–1409. doi: 10.1016/S0006-3495(99)77301-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombola F, Oregna F, Brutsche S, Szabo I, Del Giudice G, Rappuoli R, Montecucco C, Papini E, Zoratti M. Inhibition of the vacuolating and anion channel activities of the VacA toxin of Helicobacter pylori. FEBS letters. 1999b;460:221–225. doi: 10.1016/s0014-5793(99)01348-4. [DOI] [PubMed] [Google Scholar]
- Torres VJ, Ivie SE, McClain MS, Cover TL. Functional properties of the p33 and p55 domains of the Helicobacter pylori vacuolating cytotoxin. The Journal of biological chemistry. 2005;280:21107–21114. doi: 10.1074/jbc.M501042200. [DOI] [PubMed] [Google Scholar]
- Vinion-Dubiel AD, McClain MS, Czajkowsky DM, Iwamoto H, Ye D, Cao P, Schraw W, Szabo G, Blanke SR, Shao Z, et al. A dominant negative mutant of Helicobacter pylori vacuolating toxin (VacA) inhibits VacA-induced cell vacuolation. The Journal of biological chemistry. 1999;274:37736–37742. doi: 10.1074/jbc.274.53.37736. [DOI] [PubMed] [Google Scholar]
- Wang WC, Wang HJ, Kuo CH. Two distinctive cell binding patterns by vacuolating toxin fused with glutathione S-transferase: one high-affinity m1-specific binding and the other lower-affinity binding for variant m forms. Biochemistry. 2001;40:11887–11896. doi: 10.1021/bi010065u. [DOI] [PubMed] [Google Scholar]
- Wang X, Wattiez R, Paggliacia C, Telford JL, Ruysschaert J, Cabiaux V. Membrane topology of VacA cytotoxin from H. pylori. FEBS letters. 2000;481:96–100. doi: 10.1016/s0014-5793(00)01978-5. [DOI] [PubMed] [Google Scholar]
- Willhite DC, Blanke SR. Helicobacter pylori vacuolating cytotoxin enters cells, localizes to the mitochondria, and induces mitochondrial membrane permeability changes correlated to toxin channel activity. Cellular microbiology. 2004;6:143–154. doi: 10.1046/j.1462-5822.2003.00347.x. [DOI] [PubMed] [Google Scholar]
- Willhite DC, Cover TL, Blanke SR. Cellular vacuolation and mitochondrial cytochrome c release are independent outcomes of Helicobacter pylori vacuolating cytotoxin activity that are each dependent on membrane channel formation. The Journal of biological chemistry. 2003;278:48204–48209. doi: 10.1074/jbc.M304131200. [DOI] [PubMed] [Google Scholar]
- Yahiro K, Niidome T, Kimura M, Hatakeyama T, Aoyagi H, Kurazono H, Imagawa K, Wada A, Moss J, Hirayama T. Activation of Helicobacter pylori VacA toxin by alkaline or acid conditions increases its binding to a 250-kDa receptor protein-tyrosine phosphatase beta. The Journal of biological chemistry. 1999;274:36693–36699. doi: 10.1074/jbc.274.51.36693. [DOI] [PubMed] [Google Scholar]
- Ye D, Willhite DC, Blanke SR. Identification of the minimal intracellular vacuolating domain of the Helicobacter pylori vacuolating toxin. The Journal of biological chemistry. 1999;274:9277–9282. doi: 10.1074/jbc.274.14.9277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.