Using superresolution and quantitative fluorescence microscopy, Das et al. have revealed that iron-transferrin–containing endosomes directly interact with mitochondria, facilitating iron transfer in epithelial cells. Their findings further enrich the repertoire of organelle–organelle direct interactions to accomplish a functional significance.
Abstract
Transient “kiss and run” interactions between endosomes containing iron-bound transferrin (Tf) and mitochondria have been shown to facilitate direct iron transfer in erythroid cells. In this study, we used superresolution three-dimensional (3D) direct stochastic optical reconstruction microscopy to show that Tf-containing endosomes directly interact with mitochondria in epithelial cells. We used live-cell time-lapse fluorescence microscopy, followed by 3D rendering, object tracking, and a distance transformation algorithm, to track Tf-endosomes and characterize the dynamics of their interactions with mitochondria. Quenching of iron sensor RDA-labeled mitochondria confirmed functional iron transfer by an interacting Tf-endosome. The motility of Tf-endosomes is significantly reduced upon interaction with mitochondria. To further assess the functional role of iron in the ability of Tf-endosomes to interact with mitochondria, we blocked endosomal iron release by using a Tf K206E/K534A mutant. Blocking intraendosomal iron release led to significantly increased motility of Tf-endosomes and increased duration of endosome–mitochondria interactions. Thus, intraendosomal iron regulates the kinetics of the interactions between Tf-containing endosomes and mitochondria in epithelial cells.
Introduction
The endocytic basis of organelle biogenesis, maturity, and function is well documented (Sigismund et al., 2012; Bissig and Gruenberg, 2013; Antonescu et al., 2014; Bökel and Brand, 2014; Di Fiore and von Zastrow, 2014; Eaton and Martin-Belmonte, 2014; Gautreau et al., 2014). However, the reciprocal modulation of endosomal dynamics by their direct interaction with organelles such as lysosomes, ER, and mitochondria is only beginning to be dissected (Duclos et al., 2003; Sheftel et al., 2007; Friedman et al., 2013; Rowland et al., 2014). Endosomal fission is mediated by ER contact sites around the endosome (Rowland et al., 2014). Late endosomal maturity is facilitated by its sustained contact with the ER, further highlighting the importance of endosome–organelle interactions on the regulation of endocytic trafficking (Friedman et al., 2013).
Metabolite shuttle has been proposed to be at the basis of several organelle–organelle interactions via specific membrane contacts (Calore et al., 2010; McBride, 2015; Murley et al., 2015). Cellular iron uptake by transferrin (Tf) endocytosis occurs in all cell types and has been well characterized (van Bockxmeer and Morgan, 1977; Bleil and Bretscher, 1982; Ciechanover et al., 1983; Morgan and Baker, 1988; Aisen et al., 2001). Iron release from the Tf–Tf receptor (TfR) complex into the endosomal lumen occurs early in the endocytic pathway due to the establishment of an acidification gradient (Dautry-Varsat et al., 1983; Harding et al., 1983; Dautry-Varsat, 1986; Aisen, 2004).
The mitochondrial matrix is the site of vital iron utilization processes (Ponka, 1997). However, the iron transport itinerary from the Tf-endosomal lumen to the mitochondrial matrix remained elusive until recently, when Tf-endosomes were shown to undergo a transient “kiss and run” interaction with mitochondria, facilitating iron transfer in erythroid cells (Sheftel et al., 2007; Richardson et al., 2010). Although this finding successfully addressed the limitations of the cytosolic labile iron pool theory (Greenberg and Wintrobe, 1946; Jacobs, 1977; Ponka, 1997; Mikhael et al., 2010; Chepelev and Willmore, 2011) by establishing direct endosome–mitochondria interactions, detailed characterization of these interactions has not been achieved. Moreover, there is a lack of evidence supporting the endosome–mitochondria kiss and run hypothesis in nonerythroid cells.
In this study, we provide visual evidence of direct endosome–mitochondria interactions at superresolution using 3D direct stochastic optical reconstruction microscopy (STORM [dSTORM]). Using diffraction-limited live-cell time-lapse fluorescence microscopy coupled with semiautomated and unbiased quantitation, we have confirmed Tf-endosome to mitochondrial iron transfer by using a mitochondria-specific iron sensor dye called rhodamine B-[(2,2-bipyridin-4-yl) aminocarbonyl] benzyl ester (RDA; Rauen et al., 2007). We have characterized the Tf-endosomal kiss and run interactions with mitochondria based on alterations in endosomal motility. Furthermore, we have evaluated the functional role of iron release in endosome–mitochondria interactions using human Tf (hTf) mutants that do not release iron (lock-hTf), wherein an amino acid point mutation on each N and C lobe (K206E/K534A) of the recombinant hTf prevents Fe(III) release upon endosomal acidification (Halbrooks et al., 2003). The semiautomated and unbiased image analysis followed throughout this study has enabled us to characterize and compare the dynamics of individual hTf- and lock-hTf–endosomes interacting with mitochondria. Lock-hTf–endosomes that interact with mitochondria traffic at significantly higher speeds than the wild-type hTf-endosomes. Interestingly, the lock-hTf–endosomes also interact with mitochondria for longer durations. Our results suggest that iron-mediated alterations in cargo conformation or intraluminal milieu can significantly impact Tf-endosome–mitochondria interactions.
Results
3D dSTORM imaging reveals interactions between Tf-endosomes and mitochondria
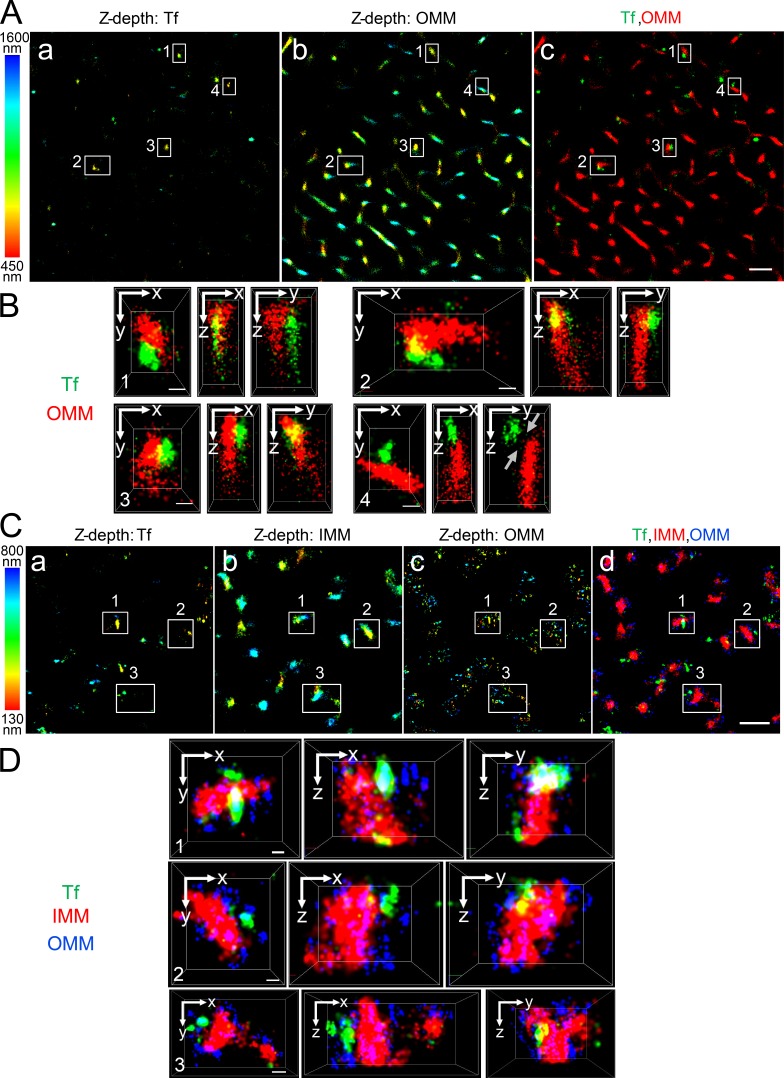
In developing erythroid cells, the demonstration of iron transfer from the Tf-endosome to the mitochondria has led to the kiss and run hypothesis (Sheftel et al., 2007; Richardson et al., 2010). Such intracellular organelle–organelle interactions, occurring at nanometer range, can be convincingly demonstrated using STORM, which is based on high-accuracy localization of photoswitchable fluorophores providing single molecule detection and improved lateral resolution up to nanometer range (Rust et al., 2006). STORM imaging has helped study clathrin-mediated endocytosis (Jones et al., 2011; Xie et al., 2016) and spatial resolution of mitochondrial outer and inner membrane organization (Huang et al., 2008a; Shim et al., 2012; Jakobs and Wurm, 2014; Klotzsch et al., 2015). To explore the interactions between Tf-endosomes and mitochondria at early time points of the Tf endocytic pathway in nonerythroid cells, MDCK transfected with rabbit polymeric lgA receptor and hTf receptor (MDCK-PTR) epithelial cells were incubated with Alexa Fluor (AF) 647–Tf for 2 min at 37°C and chased with fresh imaging medium for another 2 min (see Materials and methods; Maxfield and McGraw, 2004). Cells were then fixed and subjected to immunofluorescence using antibodies against Tom20 and OxPhos to label the outer mitochondrial membrane (OMM; Fig. 1, A–D) and the inner mitochondrial membrane (IMM; Fig. 1, C and D), respectively. 3D dSTORM imaging of the double- (Fig. 1, A and B) and triple-labeled (Fig. 1, C and D) immunostained cells was performed by introducing a cylindrical lens in the light path before the detector and comparing the x- and y-width profiles of the emission point spread function of individual fluorophores against a predetermined calibration curve (Huang et al., 2008b). This enabled us to distinguish the z positions of fluorophores situated 50 nm apart from each other within images with a total z depth of ∼1,150 nm (Fig. 1 A, a and b) and ∼670 nm (Fig. 1 C, a–c). Images were collected at the cell periphery, where the majority of the Tf-endosomes were found to be located close to mitochondria (Fig. 1, A c and C d), suggesting a complex interconnected spatial distribution of the two organelles. Different regions of interest (ROIs; Fig. 1 A, ROIs 1–4; and Fig. 1 C, ROIs 1–3) were enlarged to reveal distinct interactions between Tf-endosomes and mitochondria in all three dimensions (Fig. 1, B and D; and Videos 1 and 2). In sharp contrast, a Tf-endosome, despite being located fairly proximal to a mitochondrion, is clearly not interacting with the OMM (Fig. 1 B and Video 1, ROI 4). However, because of the close proximity between the two organelles, this Tf-endosome may be in the process of initiating or have already completed an interaction event with the mitochondrion. Using 3D dSTORM imaging, we were able to spatially resolve the OMM and IMM (Fig. 1, C and D) that are separated by an intermembrane distance varying between ∼14 nm and ∼100 nm at the region of cristae (Reichert and Neupert, 2002; Kopek et al., 2012; Gold et al., 2014). The discontinuous appearance of Tom20 immunostaining is in agreement with the clustered distribution of Tom complexes on the OMM revealed by nanoscale imaging (Schmidt et al., 2008; Wurm et al., 2011). In contrast, the OxPhos immunostaining representing the IMM appears continuous and dense because of its abundance and uniform distribution (Betzig et al., 2006; van de Linde et al., 2008). Furthermore, STORM imaging confirms the expected size difference between Tf-endosomes and mitochondria, and hence, several Tf-endosomes can be detected in close proximity to a mitochondrion (Fig. 1 D). Thus, for the first time, using superresolution 3D dSTORM imaging, we have shown that Tf-endosomes directly interact with mitochondria in epithelial cells.
Figure 1.
Tf-endosome–mitochondrion interactions using 3D dSTORM. MDCK-PTR cells pulsed with AF647-Tf for 2 min at 37°C, chased for 2 min, and fixed, and OMM and/or IMM were immunolabeled with antibodies against Tom20 and OxPhos, respectively. (A) Representative double-labeled 3D dSTORM image of the z position information of AF647-Tf–endosomes (a) and AF568-labeled anti-OMM (b), color coded according to a 450–1,600-nm range; merged (c) to show Tf (green) and OMM (red). Bar, 1 µm. (B) Magnified x–y, x–z, and y–z cross sections of ROIs 1–4 (white boxes in A). ROIs 1–3 depict AF647-Tf–endosomes directly interacting with AF568-labeled anti-OMM in all three dimensions, whereas ROI 4 shows a noninteraction (gray arrows) despite being in close proximity. See Video 1. Bars, 100 nm. (C) Representative triple-labeled 3D dSTORM image of the z position information of AF568-Tf–endosomes (a), AF647-labeled anti-OMM (b), and Atto488-labeled anti-IMM (c), color coded according to a 130–800-nm range; merged (d) to show Tf (green), IMM (red), and OMM (blue). Bar, 1 µm. (D) Magnified 3D cross sections of ROIs 1–3 (white boxes in C) show direct interactions between AF568-Tf–endosomes and AF647-labeled anti-OMM, which distinctly envelopes Atto488-labeled anti-IMM. See Video 2. Bars, 100 nm.
Quantitative evaluation of endosome–mitochondria interactions
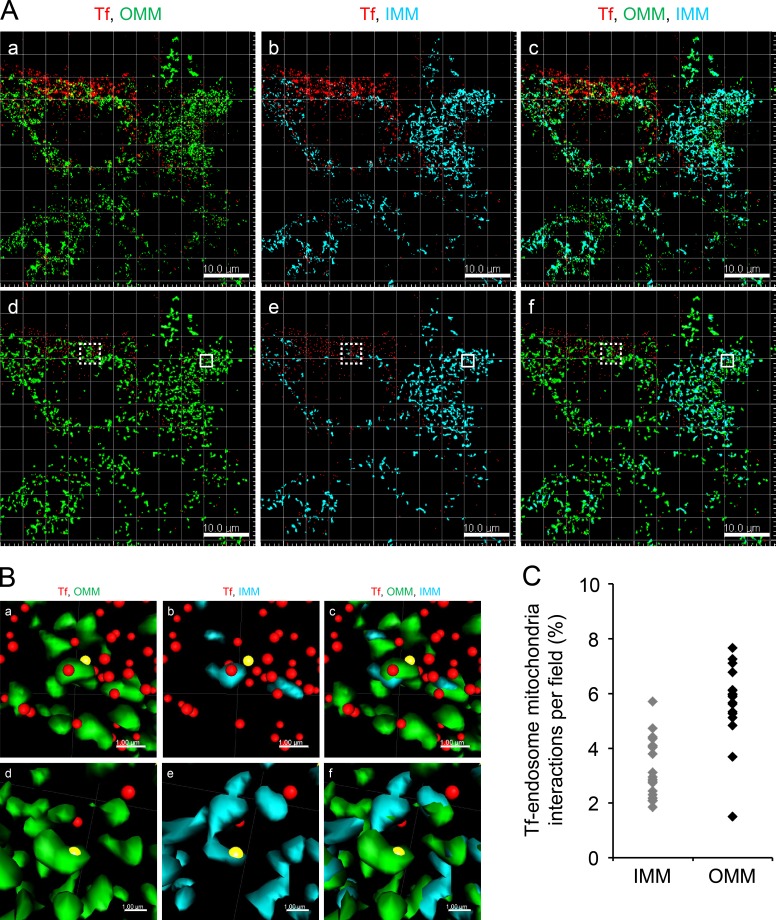
To investigate the prevalence of endosome–mitochondria interactions within the early endocytic pathway, we established a criterion to quantitatively measure interactions in an unbiased and semiautomated manner. Previously, quantitative analyses of cell-to-cell (Malide et al., 2012; McKee et al., 2013) or organelle–organelle interactions (Whalen et al., 2012; Bouvet et al., 2013; Wang et al., 2015) were performed using the distance transformation (DT) algorithm. DT is a representation of the digital image in terms of pixels with assigned values based on their respective distances from the boundary of a specific object. Therefore, 3D rendering of the object in question is a prerequisite for carrying out the DT operation. Here, we used the DT algorithm, built in the image analysis software Imaris, for quantitative determination of the distance of Tf-endosomes from the boundary of 3D-rendered mitochondrial surfaces, which are considered to be the anchor organelle, as they are less dynamic than endosomes. The DT criterion was evaluated for its consistency and reliability on immunofluorescent fixed cells (Fig. 2) before it was used in the analysis of time-lapse videos to study the dynamics of endosome–mitochondria interactions in live cells (see Figs. 3, 4, and 5).
Figure 2.
DT criterion to determine Tf-endosome–mitochondria interactions. (A, a–c) Representative confocal z stack image of MDCK-PTR cells pulsed with AF647-Tf (red) for 2 min, chased, fixed, and immunolabeled with AF555 anti-OMM (green) and AF488 anti-IMM (cyan). (d–f) Corresponding Imaris-based 3D-rendered images depicting OMM and IMM by surface and early Tf-endosomes by spots. Bars, 10 µm. (B, a–c) Magnified ROIs (dotted boxes in A, d–f) where the Tf-endosome under consideration (yellow spots) was in close proximity to the OMM (a) and subsequently validated by DT = 0. Upon deselecting the OMM and selecting the IMM surface render, the Tf-endosome showed failed interaction with the IMM of the same mitochondrion (b), also confirmed by a DT > 0 value with respect to the IMM surface; merged image (c). (d–f) Magnified ROIs (solid boxes in A, d–f) where another Tf-endosome (yellow spots) was identified to be in close proximity to both OMM (d) and IMM (e). Bars, 1 µm. The brightness and contrast of images have been enhanced equally across each panel to aid visualization. (C) Percentage of Tf-endosomes detected in close proximity to OMM or IMM per z stack of an image field (four to seven cells per field).
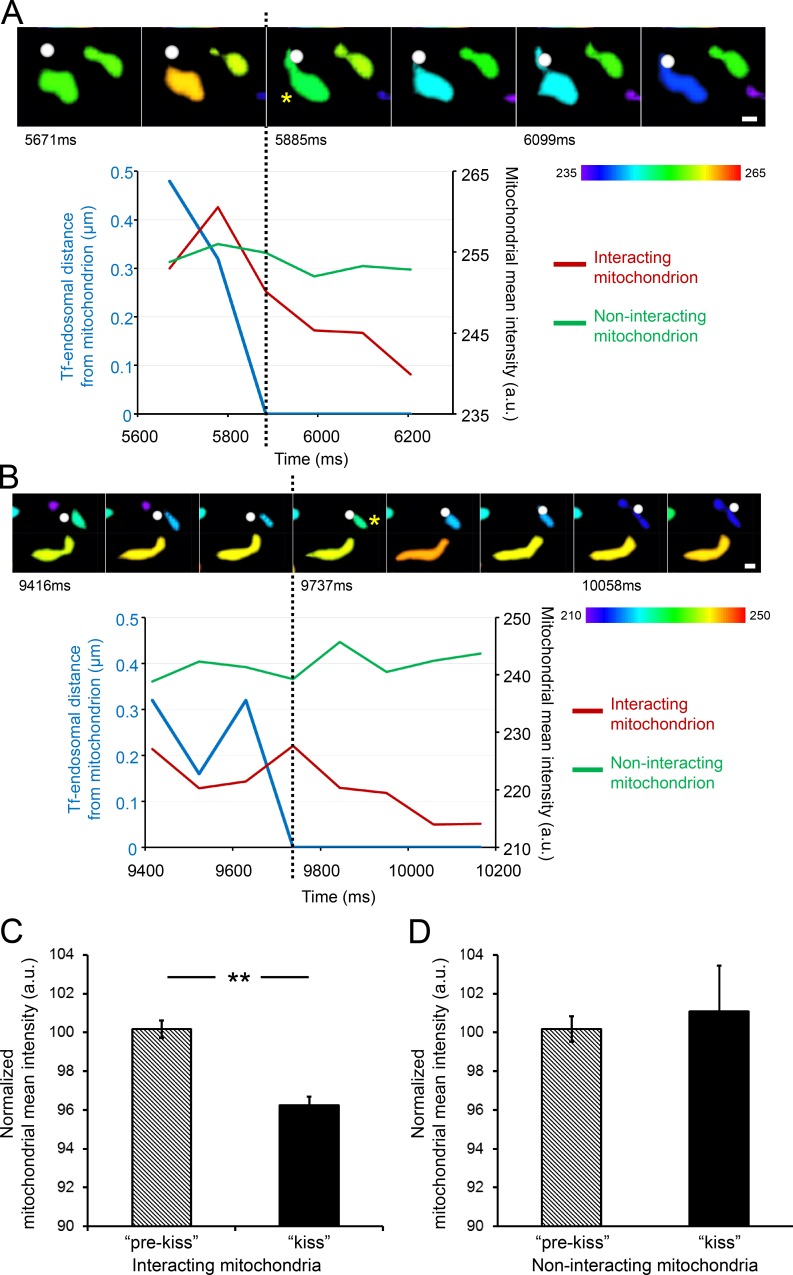
Figure 3.
Tf-endosome–mitochondria interactions facilitate functional iron transfer. MDCK-PTR cells with iron sensor RDA-labeled mitochondria were pulsed with AF647-Tf for 2 min at 37°C and chased, and time-lapse images of 15-s duration (107-ms interval) were acquired at 5 min after the Tf pulse to capture the early time points of Tf-endosomal trafficking. (A and B) Videomicrographs of distinct kiss interactions between 3D-rendered Tf-endosomes (white spots) and surface-rendered mitochondria (asterisks), proceeding from left to right at 107-ms intervals. The rendered mitochondrial color, indicated by the color bars, is based on the mitochondria’s mean fluorescence intensity. See Videos 3 and 4. Graphs represent Tf-endosomal interaction in terms of its distance from the mitochondrion (blue lines) and corresponding change in mean intensity of interacting (red lines) and noninteracting mitochondria (green lines). The dotted lines mark the beginning of the kiss interaction, after which the interacting mitochondrial mean intensity gradually quenches, whereas that of the noninteracting mitochondria remains unchanged. Bars, 0.5 µm. (C) Normalized mean intensity of interacting mitochondria (n = 12) is significantly reduced after the kiss interaction. (D) Mean intensity of noninteracting mitochondria (n = 31) remains unchanged over time. Error bars: 95% confidence interval; **, P < 0.001, Student’s t test. a.u., arbitrary units.
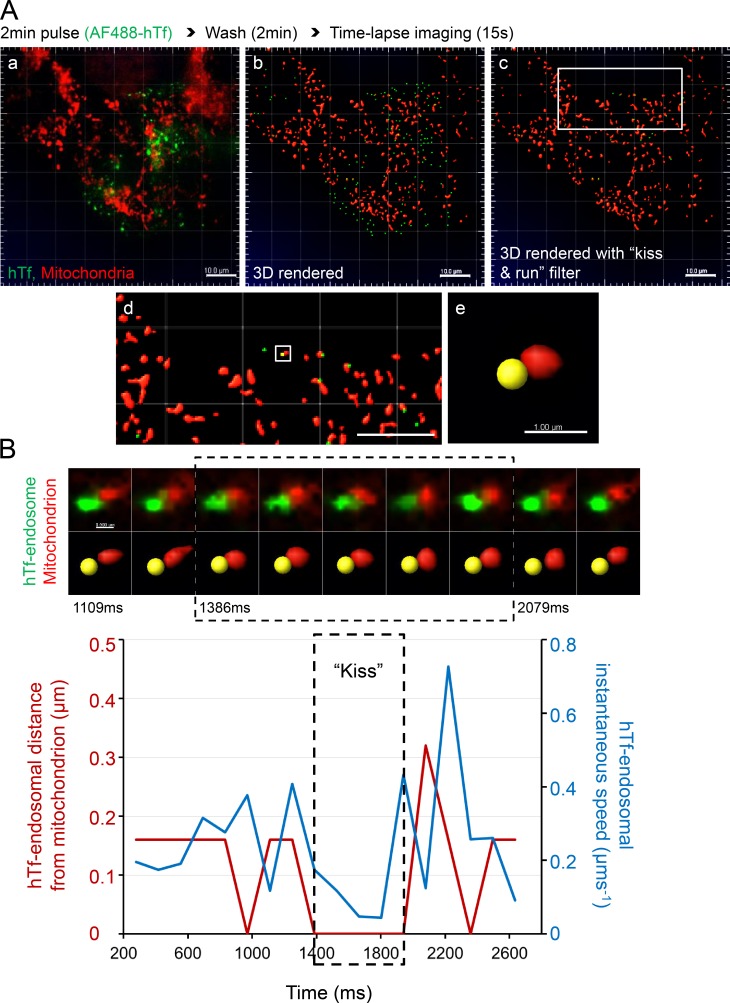
Figure 4.
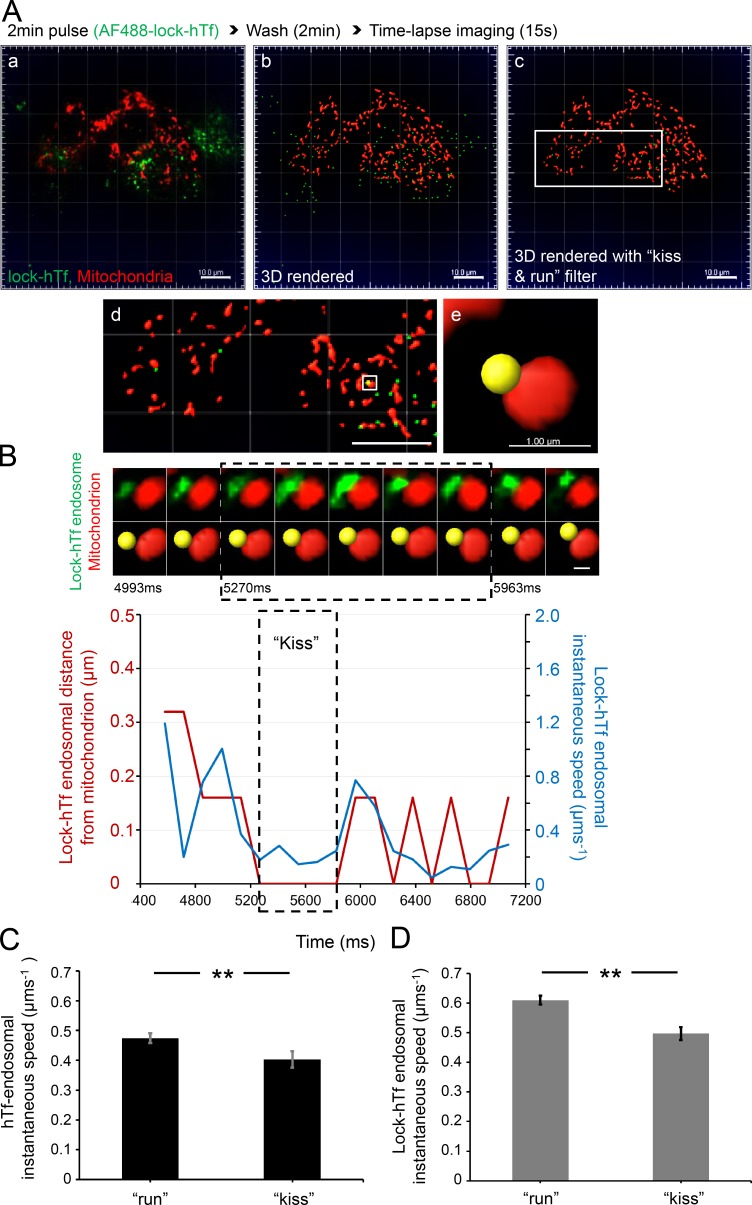
Tf-endosomal motility is reduced upon interaction with mitochondria. MitoTracker orange–labeled MDCK-PTR cells were pulsed with AF488-hTf for 2 min at 37°C and chased, and time-lapse images of 15-s duration (139-ms interval) were acquired at 5 min after the hTf pulse. (A) Double-labeled fluorescence image from a representative time lapse (a); corresponding 3D render where AF488 hTf-endosomes are green spots and mitochondria are red surfaces (b); 3D-rendered image subjected to a DT 0–based kiss and run filter selecting only those Tf-endosomes that are in close proximity to mitochondria (c; see Video 5); magnified ROI (d; white box in c); magnified ROI (white box in d) where the selected hTf-endosome (yellow spot) is in close proximity to a mitochondrion (e). Bars: (a–d) 10.0 µm; (e) 1.0 µm. (B) Videomicrograph (top row, fluorescence; bottom row, 3D rendered) of the kiss and run interaction between the selected hTf-endosome and mitochondrion (A e) proceeds from left to right at a 139-ms frame interval. Graph represents the hTf-endosomal instantaneous speed (blue line) with respect to its corresponding DT-based distance from the mitochondrion (red line) over time. Dashed box in videomicrograph and graph indicates the kiss phase. hTf-endosomal instantaneous speed is markedly reduced when it is in close proximity to the mitochondrion, denoted by the overlapping of the red line with the x axis. See Videos 6 and 7, which show two additional hTf-endosomal kiss and run events. Bar, 0.5 µm.
Figure 5.
Tf-endosome–mitochondrial interaction does not depend on intraendosomal iron release. (A) Double-labeled fluorescence image from a representative time-lapse video of MitoTracker orange–labeled MDCK-PTR cells pulsed with AF488 lock-hTf (a); corresponding 3D rendering of AF488 lock-hTf–endosomes and mitochondria (b); the 3D-rendered image selecting kiss and run undergoing lock-hTf–endosomes (c; see Video 8); magnified ROI (d; white box in c); magnified ROI (white box in d) where the selected lock-hTf–endosome (yellow spot) is in close proximity to a mitochondrion (e). Bars: (a–d) 10.0 µm; (e) 1.0 µm. (B) Videomicrograph of the kiss and run interaction between the selected lock-hTf–endosome and mitochondrion (A e). Graph shows a definite decrease in lock-hTf–endosomal instantaneous speed (blue line) during the kiss phase (dashed boxes), defined by DT = 0 (red line). See Video 9. Video 10 shows two additional lock-hTf–endosomal kiss and run events with mitochondria displaying a similar trend of decreased endosomal instantaneous speed upon interaction. Bar, 0.5 µm. (C and D) Mean hTf-endosomal (C: 311 kiss and run events, 131 endosomes, and 6 cells) and lock-hTf–endosomal (D: 603 kiss and run events, 356 endosomes, and 10 cells) instantaneous speeds were significantly higher during run noninteractions than during their kiss phases. Error bars: 95% confidence interval; **, P < 0.001, Student’s t test.
To test the performance and sensitivity of the DT algorithm, confocal z stacks of MDCK-PTR cells pulse labeled with AF647-Tf and immunolabeled with anti-OMM and anti-IMM were acquired and deconvolved using Huygens Professional package v.15.05 (Scientific Volume Imaging BV; Fig. 2 A, a–c). 3D rendering of OMM and IMM (Fig. 2 A, d–f) was performed using the “surface” feature in Imaris (see Materials and methods). After 3D rendering, the DT operation was performed individually for OMM and IMM surfaces to generate respective pixel maps. Pixels occupying the boundaries of the surfaces were assigned a DT value of 0, and the values of subsequent pixels progressively increased with their distance away from the pixel with DT = 0 (x–y pixel dimension = 0.14 µm and z = 0.497 µm). Finally, the Tf-endosomes were 3D rendered as “spots” (Fig. 2 A, d-f). A Tf-endosome spot lying on a pixel with DT = 0 with respect to an OMM or IMM surface therefore implied that it occupied the same pixel as the boundary of the corresponding mitochondrial surface. This was considered a potential interaction between the Tf-endosome and the mitochondria by virtue of being in the closest proximity of detection (i.e., within a pixel). Consequently, a Tf-endosome with DT > 0 indicated its distance in pixels from the nearest mitochondrial surface and hence was an obvious noninteraction. Typical Tf-endosomes with DT = 0 (Fig. 2 B, a–f, yellow spots, to distinguish them from other Tf-endosomes denoted by red spots) with respect to either an OMM (Fig. 2 B, a and d) or IMM (Fig. 2 B, b and e) were manually confirmed to be in close proximity to a mitochondrion by zooming into the ROIs in the 3D-rendered confocal z stacks (Fig. 2 A, d–f; dotted and solid boxes are enlarged in Fig. 2 B, a–c and d–f, respectively) using the Imaris 3D visualization interface. Even though the Tf-endosome (Fig. 2 B a, yellow spot) that scored a DT of 0 with respect to the OMM is found to be in its close proximity, it clearly failed to show contact with the IMM of the same mitochondrion (Fig. 2 B b), which was concordant with its DT being >0 with reference to the IMM surface. In another instance (Fig. 2 B, d–f), a particular Tf-endosome that scored a DT of 0 with both OMM and IMM surfaces was found to be in close proximity to both mitochondrial markers (Fig. 2 B, d and e), suggesting a potential interaction. Thus, by manually inspecting Tf-endosomes showing DT = 0 and confirming their proximity to mitochondria, we established the reliability of the DT algorithm to quantitatively determine Tf-endosome–mitochondrion interactions in our endocytic pulse-chase assay. In Fig. 2 C, each data point on the scatter plot denotes an image field containing the percentage of Tf-endosomes interacting with IMM or OMM as observed in the z stack (n = 21 and 17 different fields of confocal z stacks for IMM and OMM surfaces, respectively). Using the DT analysis, 5–8% of all detected Tf-endosomes were in close proximity to OMM, whereas only 2–5% of them were proximal to IMM.
Direct interaction between Tf-endosomes and mitochondria facilitates iron transfer
Although 3D dSTORM imaging has revealed direct interactions between Tf-endosomes and mitochondria, whether these interactions mediate iron transfer still remains unknown. To address the functional significance of this interaction, we used an iron-quenching reporter dye, RDA, which specifically labels the mitochondria without affecting their dynamics or normal functioning (Petrat et al., 2002; Rauen et al., 2007). RDA fluoresces upon excitation with a red laser, and its emission signal undergoes rapid quenching in the presence of iron, thus behaving as an adequate iron sensor.
MDCK-PTR cells were incubated with RDA to label mitochondria and then subjected to a 2-min pulse with AF647-Tf and a 2-min chase at 37°C (see Materials and methods). Time-lapse images of 15-s duration were immediately acquired within 5–6 min of Tf pulse. Imaging was performed using a total internal reflection fluorescence (TIRF) microscope with oblique laser illumination to excite fluorophores at ∼100–200 nm from the surface of the coverglass. 3D rendering of RDA-labeled mitochondria as surfaces and AF647-Tf endosomes as spots, based on true localization and fluorescence intensity in raw images, was performed as above (Fig. 2). Typical endosome–mitochondria interaction events, identified based on the DT = 0 criterion, are depicted in the time series videomicrographs (Fig. 3, A and B; and Videos 3 and 4), where the Tf-endosomes are denoted by white spots and the mitochondria surfaces are color coded according to their mean fluorescence intensity. Corresponding graphs in Fig. 3 (A and B) coupling the Tf-endosomal distance from the interacting mitochondrion (blue lines) and the mean mitochondrial intensity (red lines) demonstrate quenching of the interacting mitochondrial emission (asterisks) after the kiss event (dotted black lines). In contrast, the intensity of a noninteracting mitochondrion (Fig. 3, A and B, green lines) in the same field does not decrease significantly, discounting the effect of photobleaching. The mean intensity of mitochondria interacting with Tf-endosomes decreased significantly after kiss (Fig. 3 C; n = 12, interacting mitochondria), whereas the intensity of noninteracting mitochondria remained unchanged (Fig. 3 D; n = 31, noninteracting mitochondria collected in images showing endosome–mitochondria kiss events). Hence, the decrease in fluorescence intensity of RDA-labeled mitochondria after interacting with a Tf-endosome should be solely attributed to quenching mediated by functional iron transfer.
Tf-endosomal motility is reduced upon its interaction with a mitochondrion
Because Tf-endosomes are highly mobile at 37°C during the early endocytic pathway when compared with larger-sized and less dynamic mitochondria, we performed live-cell time-lapse imaging to investigate the effect of endosome–mitochondria interactions on Tf-endosomal motility. Based on the rationale that the speed of an object is reduced upon interaction with another object of greater mass, we predicted that the Tf-endosomal instantaneous speed would undergo a significant reduction upon interaction with a mitochondrion. To test this, we labeled mitochondria in MDCK-PTR cells with MitoTracker orange and performed AF488-hTf pulse-chase assay and imaging exactly as before (Fig. 4 A a and Video 5). 3D spot rendering of AF488-hTf–endosomes and surface rendering of MitoTracker orange–labeled mitochondria was performed (Fig. 4 A b and Video 5). Although all hTf-endosomes were 3D rendered as spots, tracks of motion were generated for only those Tf-endosomes with a DT of 0 with respect to mitochondrial surfaces (kiss and run filter; Fig. 4 A c and Video 5) using the Imaris built-in spot-tracking features. The endosome-tracking algorithm was validated by selecting an hTf-endosome (Fig. 4 A, d and e, yellow spot) interacting with a mitochondrion and manually inspecting its motion frame by frame along the fluorescence as well as in the 3D-rendered time-lapse videos. The time series videomicrograph depicting this particular kiss and run interaction in fluorescence (Fig. 4 B, top row) and corresponding 3D-rendered images (Fig. 4 B, bottom row) proceeds from left to right at the same rate as the time-lapse acquisition (i.e., 139-ms interval; Fig. 4 B). To characterize the hTf-endosomal motility upon interaction with the mitochondrion, the instantaneous speed of the hTf-endosome (Fig. 4 B, blue line) was plotted along with its corresponding distance from the mitochondrion (Fig. 4 B, red line) versus time. The hTf-endosomal instantaneous speed was observed to decrease as it interacted with the mitochondrion (Fig 4 B, red line denoting DT reaching 0), remaining low during the entire period of interaction (kiss phase of 0.7 s), and increased once again as the endosome moved away from the mitochondrion (Video 6, event 1). The trend of hTf-endosomal instantaneous speed decreasing upon interaction with a mitochondrion and increasing as it departs was consistent in other kiss and run events (Video 7, events 2 and 3). However, we also detected instances in which the hTf-endosomes displayed an increased burst of instantaneous speed during the kiss phase (Video 7, event 3). Such hTf-endosomal instantaneous speed increments during the kiss phase could be attributed to the relatively less mobile nature of the mitochondria itself, resulting in the concomitant movement of both organelles. In summary, the motility of early endosomes is significantly affected by their interaction with mitochondria in epithelial cells.
Iron release from Tf is not required for the interaction of endosomes with mitochondria
Although we confirmed Tf-endosomal interactions with mitochondria at nanometer-range resolution as well as an interaction-mediated functional iron transfer, an obvious question that remained unanswered was whether the intraendosomal iron release from the Tf–TfR complex was essential for the kiss and run interaction. To address this, we used a recombinant mutant Tf (lock-hTf) with an amino acid point mutation in each of its two lobes (K206E/K534A), thus preventing iron release upon endosomal acidification (Halbrooks et al., 2003). Mitochondrial labeling, Tf-endocytic pulse-chase assay, and live-cell time-lapse imaging were performed exactly as before, except this time with AF488–lock-hTf (Fig. 5 A a and Video 8). Upon 3D rendering (Fig. 5 A, b and c; and Video 8), a typical lock-hTf–endosome undergoing kiss and run interaction with a mitochondrion was detected based on DT = 0 and its Imaris-assigned identity number (Fig. 4 A, d and e). The time series videomicrograph depicting the interaction (Fig. 5 B) shows that the lock-hTf–endosomal instantaneous speed also decreases upon interaction with the mitochondrion (Fig. 5 B, blue line), remaining low during the entire kiss phase (Fig. 5 B, dashed box, 0.7 s), and increases as it departs (Video 9). The mitochondrial interaction-dependent reduction of lock-hTf–endosomal instantaneous speed was consistent in other detected interactions as well (Video 10).
We then quantitated the instantaneous speed values during all time points of run (DT > 0) versus kiss (DT = 0) for all detected hTf- and lock-hTf–endosomes interacting with mitochondria. For the unmutated hTf-endosomes (311 kiss and run events, 131 endosomes, and 6 cells), we found a statistically significant difference (P < 0.001) between the mean endosomal instantaneous speed during kiss interactions and noninteractions (Fig. 5 C, run). Similarly, in the case of lock-hTf–endosomes (603 kiss and run events, 356 endosomes, and 10 cells), the mean instantaneous speed during run was significantly higher (P < 0.001) when compared with that during kiss (Fig. 5 D). These results suggest that the intraluminal iron release from the Tf–TfR complex is not necessary for Tf-endosome–mitochondria interactions to occur. Moreover, Tf-endosomal motility is reduced upon interaction with mitochondria independent of the presence or absence of luminal iron.
Lock-hTf–endosomes that interact with mitochondria show increased motility
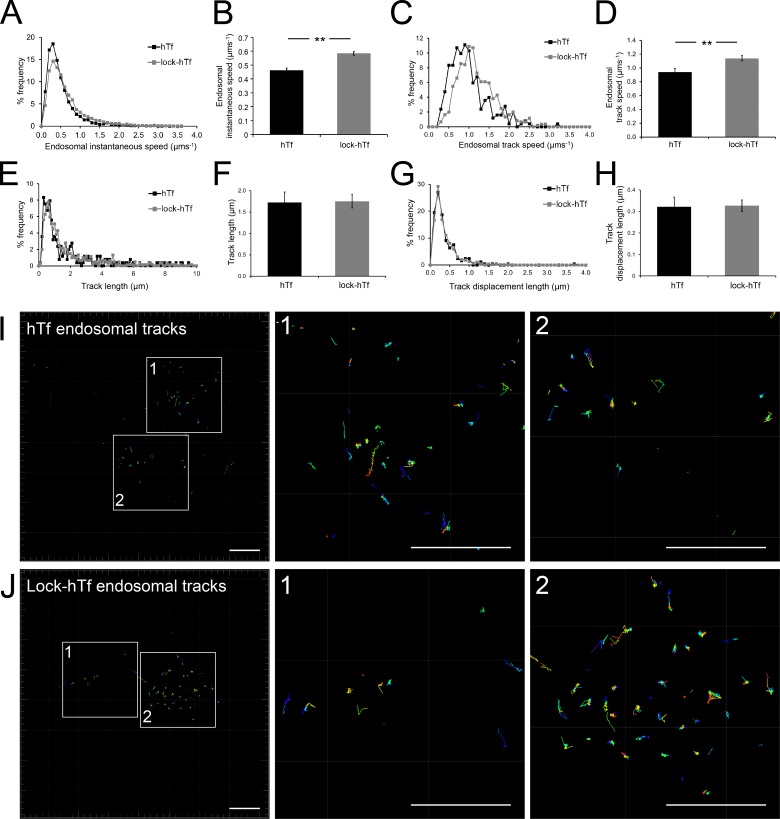
Blocking iron release from Tf prevents iron-mediated conformational changes in the Tf–TfR complex (Eckenroth et al., 2011), thus allowing us to test whether an altered cargo conformation affects the motility of Tf-endosomes that interact with mitochondria. Surprisingly, lock-hTf–endosomes interacting with mitochondria were detected to have higher instantaneous speeds during both kiss and run phases compared with the unmutated hTf-endosomes (Fig. 5, C and D). To test our hypothesis of perturbed endosomal cargo affecting the motility of interacting Tf-endosomes, we compared the percentage of frequency distribution of overall hTf- and lock-hTf–endosomal instantaneous speeds, combining those during kiss and run phases. Although the majority of the Tf-endosomes in both cases (hTf and lock-hTf) was found to have an instantaneous speed in the range of 0.2–0.5 µm/s, we noticed that a higher percentage of the lock-hTf–endosomes trafficked at increased instantaneous speeds (>0.5 µm/s) compared with the hTf-endosomes (Fig. 6 A). The mean instantaneous speed of interacting lock-hTf–endosomes at 0.59 µm/s was found to be significantly higher (P < 0.001) than that of the hTf-endosomes at 0.47 µm/s (Fig. 6 B). We then obtained the track speed of each interacting endosome by dividing its track length by its total track time in the time-lapse video. The interacting lock-hTf–endosomes were found to traffic at higher track speeds compared with the interacting hTf-endosomes (Fig. 6 C). The mean track speed of all interacting lock-hTf–endosomes (n = 356) was found to be significantly higher (P < 0.001) than that of the hTf-endosomes (n = 131; Fig. 6 D). The increased motility of interacting lock-hTf compared with hTf-endosomes is strengthened by the absence of any significant differences observed among their total track lengths (Fig. 6, E and F) as well as track displacement lengths (shortest linear distance between the first and last points of the track; Fig. 6 H). Interacting endosomal tracks in representative hTf (Fig. 6 I, enlarged ROIs 1 and 2) and lock-hTf (Fig. 6 J, enlarged ROIs 1 and 2) time-lapses show similar distribution of lengths. These results imply that iron-mediated Tf–TfR conformational changes impact the motility of endosomes that interact with mitochondria.
Figure 6.
Lock-hTf–endosomes interacting with mitochondria have increased speeds. (A) Percent frequency distribution of overall instantaneous speeds (during kiss and run phases combined) of interacting hTf-endosomes versus lock-hTf–endosomes indicates that lock-hTf–endosomes have higher motility. (B) The mean instantaneous speed of lock-hTf–endosomes was significantly higher than that of hTf-endosomes. (C) Percent frequency distribution of endosomal track speeds (total length of track divided by time taken to form the track) of interacting hTf-endosomes versus lock-hTf–endosomes also indicate higher speeds for the lock-hTf–endosomes. (D) The mean track speed of the lock-hTf–endosomes was significantly higher than that of the hTf-endosomes. (E and F) The percent frequency distribution of track lengths for hTf- and lock-hTf–endosomes was similar (E), and the difference between their mean track lengths (F) was not statistically significant. (G and H) The percent frequency distribution of track displacement lengths (the shortest linear distance between the starting and ending point of a track) for hTf- and lock-hTf–endosomes was also similar (G), and the difference between their mean values (H) was not statistically significant. Error bars: 95% confidence interval; **, P < 0.001, Student’s t test. (I) Interacting hTf-endosomal tracks magnified (ROIs 1 and 2) from a representative time lapse. (J) Interacting lock-hTf–endosomal tracks magnified (ROIs 1 and 2) from a representative time lapse. Bars, 10 µm.
Blocked iron release from endosomal Tf lengthens endosomal interaction with mitochondria
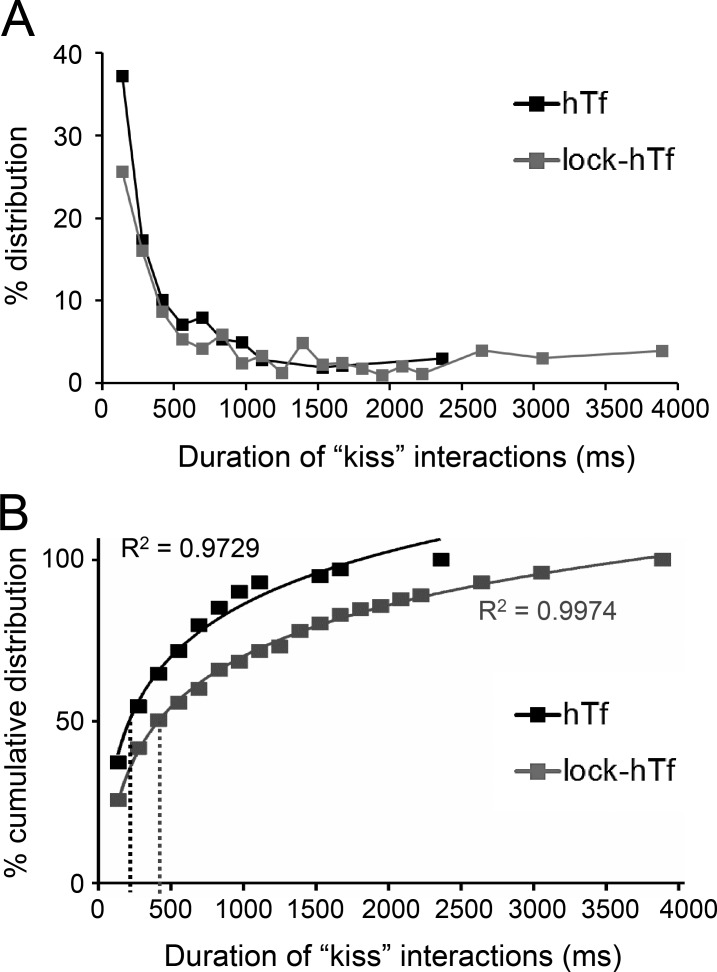
To further address the impact of perturbed endosomal cargo on the dynamics of endosome–mitochondria interactions, we compared the duration of kiss interactions between all detected hTf- and lock-hTf–endosomes with mitochondria. The duration (milliseconds) of each kiss phase between each individual hTf- and lock-hTf–endosome with mitochondria was calculated from the known interval of acquired time-lapse (139 ms) and the number of consecutive image frames that were detected proximal to a mitochondrion (cross-verified by DT = 0). Only completely detected kiss and run events defined by one or more frames of kiss flanked before and after by at least one frame each of noninteraction (DT > 0) with mitochondria were considered for analysis. Although the majority of the detected endosome–mitochondria interactions by both the hTf- and lock-hTf–endosomes were observed to last for durations of <0.5 s, an increased number of longer interactions was observed in the case of the lock-hTf–endosomes (Fig. 7 A). To resolve this further, we plotted the percent cumulative distribution of the duration of interactions and fitted the data points using a logarithmic equation trend line (Fig. 7 B). We found that the half-life of duration of kiss events by hTf-endosomes (Fig. 7 B, black dotted line) derived from the trend line equation y = 23.544ln(x) − 76.472 is 216 ms, whereas the half-life of duration of kiss events by lock-hTf–endosomes (Fig. 7 B, gray dotted line) derived from the trend line equation y = 22.877ln(x) − 87.928 is 416 ms. The substantial increase in the half-life of lock-hTf–endosomal kiss duration suggests that blocking intraluminal iron release from the Tf–TfR complex and the subsequent changes in the Tf–TfR conformation prolong Tf-endosome–mitochondria kiss interactions.
Figure 7.
Lock-hTf–endosomes interact longer with mitochondria. The duration of each individual hTf- or lock-hTf–endosome–mitochondrion kiss phase was calculated by multiplying the number of consecutive frames in which they were seen to be interacting (DT = 0) by the known interval (139 ms) of time-lapse acquisition. (A) Percent distribution of the duration of kiss events between hTf-endosomes (n = 311 events) and lock-hTf–endosomes (n = 603 events) with mitochondria. (B) Percent cumulative distribution of duration of kiss events with a logarithmic equation trend line to fit data points. The half-life duration of kiss events by hTf-endosomes (black dotted line) is 216 ms (y = 23.544ln(x) − 76.472), whereas for the lock-hTf–endosomes (gray dotted line), it is 416 ms (y = 22.877ln(x) − 87.928).
Discussion
In erythroid cells, direct transient interactions between Tf-endosomes and mitochondria have been suggested as an alternative mechanism of facilitating iron transfer by bypassing the oxygen-rich cytosol (Sheftel et al., 2007; Richardson et al., 2010). However, a key unanswered question was whether such Tf-endosome–mitochondria kiss and run interactions also occurred in nonerythroid tissues with a basal level of iron homeostasis. Elucidating the iron logistics in nonerythroid cells is physiologically important, considering that faulty sequestration-related iron overload is a hallmark of Friedreich’s ataxia, Alzheimer’s disease, Parkinson’s disease, and multiple sclerosis (Zecca et al., 2004; Lehmann et al., 2006; Stankiewicz et al., 2007; Williams et al., 2011; Ward et al., 2014).
A direct interaction between the endosomal membrane and the OMM occurs at the nanometer range, requiring a superresolution microscopy approach for detection. For the first time, we have used superresolution 3D dSTORM to image interactions between fluorescently labeled Tf-endosomes and the OMM in epithelial cells. Based on the well-characterized Tf-endosomal pathway kinetics and subcellular localization (Maxfield and McGraw, 2004; Mesaki et al., 2011), we used a short pulse-chase Tf-internalization protocol and imaged an area at the periphery of the cell, away from the perinuclear region, to capture a complex pattern of early Tf-endosomes proximal to mitochondria. Upon magnification of several individual interactions, Tf-endosomal association with mitochondria was detected at both the lateral and axial dimensions. We further confirmed that these Tf-endosome–mitochondria interactions are functionally relevant for iron delivery to mitochondria. Two lines of evidence support this observation. One is the quenching of RDA upon Tf-endosome–mitochondria interaction, and the other relies on a well-characterized mutant hTf that fails to release iron upon endosomal acidification (Halbrooks et al., 2003; Mayle et al., 2012; Chiu et al., 2014).
To further characterize Tf-endosome–mitochondria interactions, we tested two main hypotheses. First, we assessed the effect of endosome–mitochondria interactions on the dynamics of endocytic trafficking, using endosomal instantaneous speed as an indicator of endosomal motility. We observed a significant decrease in the motility of Tf-endosomes upon interaction with mitochondria. The Tf-endosomal motility remained low during the entire kiss period and increased when the endosome moved away. Despite the inherent cellular heterogeneity of endosomal dynamics, this trend of a significantly reduced Tf-endosomal motility during interaction with mitochondria persisted in all detected kiss and run events in multiple cells. A similar trend of decreased endosomal motility has been observed when late endosomes interact with the ER (Raiborg et al., 2015). Moreover, axonal cargos, such as lysosomes, have been shown to slow down or pause transiently within the proximity of stationary organelles, such as mitochondria (Che et al., 2016). Thus, we have quantitatively demonstrated that the motility of a mobile organelle, such as an endosome, is a crucial determinant in defining its interactions with other organelles, especially in diffraction-limited images.
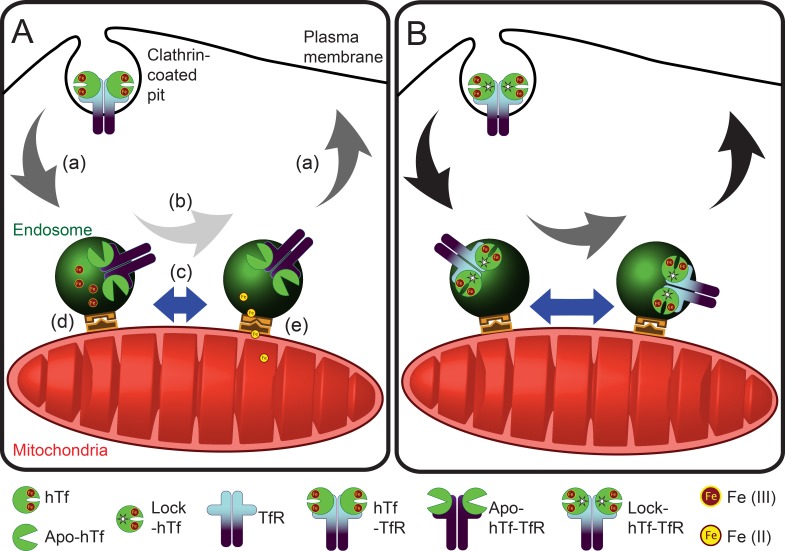
Second, we inquired whether alterations in the endosomal cargo, particularly in the iron release ability of Tf–TfR complexes by using lock-hTf, affected endosome–mitochondria interactions. Our quantitative analysis of the duration of kiss events revealed an increased number of longer interactions by the mutant hTf-endosomes with mitochondria. The overall interaction can be perceived to comprise of at least three discrete events: proximity-dependent interaction, functional accomplishment of iron transfer, and the Tf-endosome’s departure from mitochondria. Our results suggest that these docking and release mechanisms may be disrupted when iron release is blocked (Fig. 8). Putative candidates for this docking step are the divalent metal transporter 1 (DMT-1) that was found both in the endosomal membrane as well as in the OMM and the voltage-dependent anion-selective channel (VDAC1) that was shown to colocalize with DMT-1 (Wolff et al., 2014).
Figure 8.
Model for endosome–mitochondria interactions in nonerythroid cells. (A and B) Comparing the behavior of unmutated hTf- (A) and lock-hTf–endosomal interaction (B) with mitochondria suggests that blocking iron release from Tf–TfR complexes affects the ability of endosomes to interact with mitochondria because of an iron-mediated cargo and/or endosomal milieu alterations. (a) Arrows represent endosomal speed during run; black arrows in lock-hTf indicate higher speeds than dark gray arrows in the case of hTf-endosomes. (b) Arrows represent endosomal speed during kiss; light gray arrow in hTf-endosome indicates slower speed than the dark gray arrow in the case of lock-hTf–endosomes. (c) Length of blue bars with double arrowheads indicates duration of kiss events, which is longer in the case of lock-hTf–endosomes. (d and e) Schematic representations of putative docking complexes involved in iron transfer to mitochondria.
We noticed that blocking intraluminal iron release led to increased motility of the Tf-endosomes that interacted with mitochondria. The lock-hTf–endosomes are strikingly different from hTf-endosomes because of the absence of free iron in the endosomal milieu. It has also been shown that conformation of the Tf–TfR complex undergoes significant changes as a result of iron binding and release (Eckenroth et al., 2011). Thus, it is logical to infer that altered endosomal cargo and milieu have direct bearings on the endosomal motility and the duration of its interaction with mitochondria (Fig. 8). Motility plays an important role in early endosomal function and behavior by modulating its maturation (Mesaki et al., 2011; Padilla-Parra et al., 2012; Friedman et al., 2013), receptor-mediated signaling (Saxena et al., 2005; Lanahan et al., 2010; Leo et al., 2014), and cytoskeletal interaction (Zajac et al., 2013; Schafer et al., 2014). Early endosomes show a complex motility behavior that has been associated with their cargo content (Lakadamyali et al., 2006). Furthermore, early endosomal motility also plays a crucial role in regulating the intracellular movement of other organelles such as the ER, lipid droplets, and peroxisomes (Guimaraes et al., 2015). Several lines of evidence suggest the existence of two subpopulations of early endosomes based on their motility being differently regulated by actin or microtubule cytoskeletons (Padilla-Parra et al., 2012; Zajac et al., 2013; Schafer et al., 2014). An interesting hypothesis is that an intricate cross talk occurs between the endosomal cargo and milieu and the cytoplasmic molecular motors and adapters responsible for the regulation of endocytic trafficking. Hence, the most likely explanation for an increased motility of lock-hTf–endosomes is the difference in the structural conformation of the cytoplasmic domain of the Tf–TfR complex in the presence or absence of bound iron, which would affect the way it communicates with the molecular motors and/or the adapter proteins governing endosomal motility.
In summary, our compelling visual evidence of direct interaction between Tf-endosomes and mitochondria in nonerythroid cells, for the first time at nanometer resolution, strongly supports the endosome–mitochondria kiss and run hypothesis (Sheftel et al., 2007; Richardson et al., 2010). Furthermore, our results suggest that alterations in the endosomal cargo and milieu modulate the dynamics of endosome–mitochondria interactions (Fig. 8).
Materials and methods
Cell culture
MDCK-PTR cells (MDCK cells stably expressing human TfR; Odorizzi et al., 1996) were cultured in DMEM supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (Atlanta Biologicals) and penicillin/streptomycin at 37°C and 5% CO2 according to standard tissue culture protocols. For imaging, cells were seeded at a density of 105 cells/dish in No. 1.5 coverglass-bottom (0.16–0.19 mm) 35-mm Petri dishes coated with poly-d-lysine (MatTek Corporation). Cell culture medium and additives were purchased from Gibco. MDCK-PTR cells on MatTek dishes were incubated at 37°C and 5% CO2 for ∼20–24 h, after which the medium was replaced with a freshly prepared imaging medium comprised of phenol red–free and serum-free DMEM supplemented with 4 mM l-glutamine and 20 mM Hepes, pH 7.4 (Sigma-Aldrich), for 1 h before Tf pulse-chase assay and live imaging or fixing for immunofluorescence.
Live-cell mitochondrial staining and Tf-endocytic pulse-chase assay
Live mitochondrial staining was achieved by adding 100 nM iron sensor dye RDA (a gift from U. Rauen, Institute of Physiological Chemistry, University Hospital Essen, Essen, Germany) or 100 nM membrane potential-dependent dye MitoTracker orange CM-H2TMRos (Molecular Probes) to the last 15 min of the 1-h preincubation step with imaging media. Then, 50 µg/ml Tf conjugated to AF647 (Invitrogen) or hTf or lock-hTf conjugated to AF488 (Invitrogen) was added to the cells for a 2-min pulse at 37°C. After this, the cells were briefly washed and chased with imaging medium for another 2 min. Starting at time point 5–6 min after Tf pulse, time-lapse videos of 15-s duration were acquired. This entire assay was performed with the dish with cells placed on the oil immersion objective lens inside the temperature-controlled live-cell incubation chamber of the TIRF microscope (ZEISS) to avoid delay in the acquisition of early time points of Tf-endocytic trafficking.
Immunofluorescence staining
For immunofluorescence staining, after the 1-h preincubation step with imaging media, cells were pulsed with 50 µg/ml fluorescently labeled Tf for strictly 2 min at 37°C, briefly washed, and chased with imaging medium. The cells were then fixed at 5–6 min after the Tf pulse using chilled 4% (vol/vol) paraformaldehyde in PBS for 10 min at room temperature. Upon fixation, cells were permeabilized with 0.1% Triton X-100 (vol/vol; Sigma-Aldrich) in PBS for 15 min and blocked in PBS containing 5% fish gelatin, 1% BSA (wt/vol), and 0.05% Triton X-100 for 2 h at room temperature. This was followed by incubation with anti–human ATP Synthase Subunit IF1 (IMM marker) mouse monoclonal antibody (1:300 in wash buffer containing 1% fish gelatin, 1% BSA, and 0.5% Triton X-100 in PBS; A21355; Molecular Probes) for 6 h at 4°C. Cells were then washed four times for 10 min each with wash buffer at room temperature. This was followed by another incubation with anti–human Tom20 (OMM marker) rabbit polyclonal antibody (1:250 in wash buffer; sc-11415; Santa Cruz Biotechnology, Inc.), and washing steps were performed as described previously. The cells were then incubated with a mixture containing AF488-linked goat anti–mouse F(ab’)2 (1:250; A-11017; Thermo Fisher Scientific) or Atto488 (38371-1KT; Sigma-Aldrich)-linked goat anti–mouse antibody (1:250; 31182; Thermo Fisher Scientific) and AF555-linked goat anti–rabbit F(ab’)2 (1:250; A-21430; Thermo Fisher Scientific) or AF568-linked donkey anti–rabbit antibody (1:250; A10042; Thermo Fisher Scientific) for 2 h at room temperature. The secondary antibodies were aspirated, and cells were washed four times for 10 min each with wash buffer at room temperature. Finally, a postfixation step was performed by incubating the cells with 4% (vol/vol) paraformaldehyde in PBS for 10 min at room temperature, washing them three times, and storing them in PBS at 4°C until microscopic imaging.
Confocal imaging
Immunostained cells were imaged with a laser-scanning microscope system (LSM 510META-NLO; ZEISS) equipped with an air-cooled argon laser at 488 nm, helium/neon lasers at 543 and 633 nm, and a plan-Apochromat 63×/1.4 oil differential interference contrast objective (ZEISS). Acquisition was run by a Zen 2010 microscope and imaging software (ZEISS) using optimal settings to avoid saturation. Images were collected for each channel sequentially at 512 × 512 pixels, 16-bit depth, and 2× magnification. Photomultiplier tube gain and black-level settings, laser power, and pinhole values were set at identical levels and remained unchanged for all image acquisitions. Voxel dimension was 0.14 µm (x–y) and 0.497 µm (z).
3D dSTORM imaging and data processing
Immunostained cells were incubated with STORM-imaging buffer, which included 7 µl GLOX (made by dissolving 14 mg glucose oxidase in 50 µl catalase [17 mg/ml] + 200 µl Tris-buffered saline [10 mM Tris, pH 8.0, + 50 mM NaCl]), 7 µl 2-mercaptoethanol, and 690 µl Tris-buffered saline (50 mM Tris, pH 8.0, + 10 mM NaCl + 10% glucose) during the entire imaging session. All ingredients were purchased from Sigma-Aldrich. dSTORM imaging was performed using an inverted microscope system (Ti-E Perfect Focus; Nikon) equipped with a monolithic laser combiner (MLC400; Agilent Technologies) with solid-state lasers of wavelengths 405 nm, 488 nm, and 561 nm at 50 mW (maximum fiber output power) and a 647-nm laser at 125 mW. After locating a suitable field at the periphery of the cell, a diffraction-limited TIRF image was acquired for reference, followed by a STORM acquisition. Depending on whether we were imaging double- or triple-labeled cells, appropriate lasers starting from the longer wavelength were sequentially fired at 100% power to excite all possible fluorophore molecules and photoswitch them into a nonemitting dark state. The emitted wavelengths from AF647, AF568, and Atto488 fluorophores were then sequentially acquired by the plan-Apochromat 100×/1.49 TIRF objective (Nikon), filtered by an emission filter set (Nikon TIRF Cube consisting of a TRF89902-EM filter set; Chroma Technology Corp.), and detected on an electron-multiplying charge-coupled device camera (Ixon DU-897; Andor Technology). The image acquisition was performed at 110 Hz. A series of 50,000 frames for double-labeled and 30,000 frames for triple-labeled samples (256 × 256 pixels) were captured for each channel. The image acquisition, lateral drift correction, and data processing were done using NIS Elements software with STORM package (version 4.30 build 1053; Nikon). Before sample imaging, the alignment between three colors was done using fiducial multicolor (blue, green, orange, and dark red) fluorescent microspheres of 100-nm diameter (T-7279; Thermo Fisher Scientific) plated on a No.1.5 glass-bottomed MatTek dish and allowed to air dry. For 3D dSTORM image acquisition, a cylindrical lens was introduced to the emission path in front of the detector. The cylindrical lens was calibrated by imaging the fiducial multicolor microspheres through a depth of 1,600 nm in the z axis (from −800 to +800 nm) in 10-nm increments with the help of a Piezo-Z stage (Nikon), and the calibration file was saved as a reference file.
Live-cell fluorescence imaging
Live-cell imaging was performed using an Axiovision software–managed laser TIRF microscope (ZEISS). A MatTek dish with live cells was placed on a holder enclosed by a temperature-controlled incubator system mounted on the microscope and maintained at 37°C throughout the course of Tf pulse-chase assay and imaging. The sample was sequentially excited by appropriate lasers using oblique laser illumination conditions, whereby the evanescent wave field effectively excited fluorophores located within ∼100–200 nm of the surface of the coverglass, thus eliminating the majority of out-of-focus fluorescence. Emitted fluorescence was collected by a plan-Apochromat 100×/1.6 TIRF objective (ZEISS) and filtered by standard emission filter sets before being detected on an electron-multiplying charge-coupled device camera (Photometrics). Definite focus was not used to acquire the images (512 × 512 pixels) because significant out-of-focus drift was not observed in the 15-s duration of time-lapse acquisition. Pixel dimension was 0.16 µm (x–y). For the RDA-labeled mitochondrial quenching assay, a frame rate of >9 Hz (107-ms interval) was obtained using the fast acquisition mode with sequential dual laser excitations at 639 nm and 561 nm. For the hTf and lock-hTf pulse-chase assay to determine endosomal motility, a frame rate of >7 Hz (139-ms interval) was obtained using sequential excitations at 561 nm and 488 nm.
Live-cell time-lapse analysis: 3D rendering, object tracking, and mitochondrial mean intensity quantitation
The fluorescence time-lapse videos were 3D rendered using Imaris version 7.6.5 (Bitplane Inc.) to analyze and derive the quantitative values determining Tf-endosome–mitochondrion interactions (DT), Tf-endosomal speeds, and mitochondrial mean intensity (for RDA sensor assay). We generated 3D surface rendering of the mitochondria and spot rendering of the Tf-endosomes based on the intensity of their fluorescent signals in every image of the time-lapse videos. To determine endosomal interaction (kiss) or not (run) with the nearest mitochondrion over the time-lapse, we obtained the distance of the 3D-rendered Tf-endosome spots from the boundary of the interacting mitochondrial surface using a Matlab-based DT algorithm in Imaris software. Every 3D render, be it Tf-endosome spots or mitochondrial surfaces, was assigned a unique ID number, which enabled us to identify it from the pool. The Tf-endosome spots that occupied the same pixels as the boundary of a mitochondrial surface were assigned a DT value of 0, depicting a kiss event. The DT values for each spot progressed in increments of 0.16 µm (true x–y pixel dimension) away from the closest mitochondrial surface depending on the number of pixels away they were located. These individual spots in consecutive images of the time-lapse were connected, based on an optimum maximum permissible distance value, to form tracks describing the paths of motion of Tf-endosomes along the time-lapse video. Quality control of the data was performed by manually validating randomly selected individual Tf-endosomal tracks for consistency and comparing their Imaris-derived DT values against image-by-image visual inspections of kiss interactions or noninteractions. A benchmark to define a complete Tf-endosome–mitochondrion kiss and run event was set to include only those endosomal tracks that were observed for a minimum length of three images, including at least one image of kiss flanked by at least one image of run before and after. Applying a track length filter, we eliminated the Tf-endosomes with tracks shorter than three consecutive images and considered only those Tf-endosomal tracks that were at least three images or more in length for all analysis shown in this study. For the RDA-labeled mitochondrial intensity–quenching assay, the interacting mitochondrion was identified based on its 3D surface rendered ID, and its image-by-image mean intensity values were compared against the corresponding image-by-image Tf-endosomal DT values.
Statistical methods
All graphical plots, histograms, and statistical analyses were generated in Excel (Microsoft) except for Fig. 7 B, which was generated using Prism software (GraphPad Software). Error bars represent a 95% confidence interval. Significances were tested using two-tailed unpaired t tests. Differences were considered significant at p-values <0.001.
Online supplemental material
Video 1 shows two-color 3D dSTORM (Fig. 1 B) and Video 2 shows three-color 3D dSTORM (Fig. 1 D) images with rotated viewing for better visualization of distinct Tf-endosome–mitochondria direct interactions. Video 3 (Fig. 3 A) and Video 4 (Fig. 3 B) depict two distinct events of an iron sensor–labeled mitochondrial quenching upon interaction with a Tf-endosome denoting an iron transfer. Video 5 comprises a live-cell time-lapse of fluorescently labeled mitochondria and hTf-endosomes in MDCK-PTR cells (Fig. 4 A a), the corresponding 3D-rendered video (Fig. 4 A b) followed by a 3D rendering filtered to include only the interacting hTf-endosomes (Fig. 4 A c). Video 6 is a composite of fluorescence and corresponding 3D-rendered time-lapse of a particular kiss and run event between an hTf-endosome and mitochondrion along with a graphical demonstration of the interaction-dependent modulation of the hTf-endosomal motility indicated by its instantaneous speed. Video 7 shows two additional distinct hTf-endosome–mitochondria interactions. Video 8 comprises a live-cell time-lapse of fluorescently labeled mitochondria and lock-hTf–endosomes in MDCK-PTR cells (Fig. 5 A a) and the corresponding 3D-rendered video (Fig. 5 A b) followed by a 3D rendering filtered to include only the interacting lock-hTf–endosomes (Fig. 5 A c). Video 9 depicts the kiss and run interaction between the lock-hTf–endosome and mitochondrion shown in Fig. 5 B. Video 10 shows two additional distinct lock-hTf–endosome–mitochondria interactions. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201602069/DC1.
Supplementary Material
Acknowledgments
We thank Matthew J. Gastinger for his assistance in 3D rendering and object tracking using Imaris. We also thank Sean Christie for his assistance in 3D dSTORM image acquisition and data analysis. We thank all members of the Barroso laboratory for their stimulating discussion. We are grateful to Dr. Elizabeth Sztul for her critical reading of the manuscript.
This work was partially supported by funding from the Namm’65 research award from 2011 to 2016.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AF
- Alexa Fluor
- dSTORM
- direct STORM
- DT
- distance transformation
- hTf
- human Tf
- IMM
- inner mitochondrial membrane
- OMM
- outer mitochondrial membrane
- ROI
- region of interest
- STORM
- stochastic optical reconstruction microscopy
- Tf
- transferrin
- TfR
- Tf receptor
- TIRF
- total internal reflection fluorescence
References
- Aisen P. 2004. Transferrin receptor 1. Int. J. Biochem. Cell Biol. 36:2137–2143. 10.1016/j.biocel.2004.02.007 [DOI] [PubMed] [Google Scholar]
- Aisen P., Enns C., and Wessling-Resnick M.. 2001. Chemistry and biology of eukaryotic iron metabolism. Int. J. Biochem. Cell Biol. 33:940–959. 10.1016/S1357-2725(01)00063-2 [DOI] [PubMed] [Google Scholar]
- Antonescu C.N., McGraw T.E., and Klip A.. 2014. Reciprocal regulation of endocytosis and metabolism. Cold Spring Harb. Perspect. Biol. 6:a016964 10.1101/cshperspect.a016964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E., Patterson G.H., Sougrat R., Lindwasser O.W., Olenych S., Bonifacino J.S., Davidson M.W., Lippincott-Schwartz J., and Hess H.F.. 2006. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 313:1642–1645. 10.1126/science.1127344 [DOI] [PubMed] [Google Scholar]
- Bissig C., and Gruenberg J.. 2013. Lipid sorting and multivesicular endosome biogenesis. Cold Spring Harb. Perspect. Biol. 5 10.1101/cshperspect.a016816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleil J.D., and Bretscher M.S.. 1982. Transferrin receptor and its recycling in HeLa cells. EMBO J. 1:351–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bökel C., and Brand M.. 2014. Endocytosis and signaling during development. Cold Spring Harb. Perspect. Biol. 6 10.1101/cshperspect.a017020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet S., Golinelli-Cohen M.P., Contremoulins V., and Jackson C.L.. 2013. Targeting of the Arf-GEF GBF1 to lipid droplets and Golgi membranes. J. Cell Sci. 126:4794–4805. 10.1242/jcs.134254 [DOI] [PubMed] [Google Scholar]
- Calore F., Genisset C., Casellato A., Rossato M., Codolo G., Esposti M.D., Scorrano L., and de Bernard M.. 2010. Endosome-mitochondria juxtaposition during apoptosis induced by H. pylori VacA. Cell Death Differ. 17:1707–1716. 10.1038/cdd.2010.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che D.L., Chowdary P.D., and Cui B.. 2016. A close look at axonal transport: Cargos slow down when crossing stationary organelles. Neurosci. Lett. 610:110–116. 10.1016/j.neulet.2015.10.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepelev N.L., and Willmore W.G.. 2011. Regulation of iron pathways in response to hypoxia. Free Radic. Biol. Med. 50:645–666. 10.1016/j.freeradbiomed.2010.12.023 [DOI] [PubMed] [Google Scholar]
- Chiu R.Y., Tsuji T., Wang S.J., Wang J., Liu C.T., and Kamei D.T.. 2014. Improving the systemic drug delivery efficacy of nanoparticles using a transferrin variant for targeting. J. Control. Release. 180:33–41. 10.1016/j.jconrel.2014.01.027 [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Schwartz A.L., and Lodish H.F.. 1983. Sorting and recycling of cell surface receptors and endocytosed ligands: the asialoglycoprotein and transferrin receptors. J. Cell. Biochem. 23:107–130. 10.1002/jcb.240230111 [DOI] [PubMed] [Google Scholar]
- Dautry-Varsat A. 1986. Receptor-mediated endocytosis: the intracellular journey of transferrin and its receptor. Biochimie. 68:375–381. 10.1016/S0300-9084(86)80004-9 [DOI] [PubMed] [Google Scholar]
- Dautry-Varsat A., Ciechanover A., and Lodish H.F.. 1983. pH and the recycling of transferrin during receptor-mediated endocytosis. Proc. Natl. Acad. Sci. USA. 80:2258–2262. 10.1073/pnas.80.8.2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore P.P., and von Zastrow M.. 2014. Endocytosis, signaling, and beyond. Cold Spring Harb. Perspect. Biol. 6 10.1101/cshperspect.a016865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclos S., Corsini R., and Desjardins M.. 2003. Remodeling of endosomes during lysosome biogenesis involves ‘kiss and run’ fusion events regulated by rab5. J. Cell Sci. 116:907–918. 10.1242/jcs.00259 [DOI] [PubMed] [Google Scholar]
- Eaton S., and Martin-Belmonte F.. 2014. Cargo sorting in the endocytic pathway: a key regulator of cell polarity and tissue dynamics. Cold Spring Harb. Perspect. Biol. 6:a016899 10.1101/cshperspect.a016899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckenroth B.E., Steere A.N., Chasteen N.D., Everse S.J., and Mason A.B.. 2011. How the binding of human transferrin primes the transferrin receptor potentiating iron release at endosomal pH. Proc. Natl. Acad. Sci. USA. 108:13089–13094. 10.1073/pnas.1105786108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J.R., Dibenedetto J.R., West M., Rowland A.A., and Voeltz G.K.. 2013. Endoplasmic reticulum-endosome contact increases as endosomes traffic and mature. Mol. Biol. Cell. 24:1030–1040. 10.1091/mbc.E12-10-0733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautreau A., Oguievetskaia K., and Ungermann C.. 2014. Function and regulation of the endosomal fusion and fission machineries. Cold Spring Harb. Perspect. Biol. 6 10.1101/cshperspect.a016832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold V.A., Ieva R., Walter A., Pfanner N., van der Laan M., and Kühlbrandt W.. 2014. Visualizing active membrane protein complexes by electron cryotomography. Nat. Commun. 5:4129 10.1038/ncomms5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg G.R., and Wintrobe M.M.. 1946. A labile iron pool. J. Biol. Chem. 165:397. [PubMed] [Google Scholar]
- Guimaraes S.C., Schuster M., Bielska E., Dagdas G., Kilaru S., Meadows B.R., Schrader M., and Steinberg G.. 2015. Peroxisomes, lipid droplets, and endoplasmic reticulum “hitchhike” on motile early endosomes. J. Cell Biol. 211:945–954. 10.1083/jcb.201505086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbrooks P.J., He Q.Y., Briggs S.K., Everse S.J., Smith V.C., MacGillivray R.T., and Mason A.B.. 2003. Investigation of the mechanism of iron release from the C-lobe of human serum transferrin: mutational analysis of the role of a pH sensitive triad. Biochemistry. 42:3701–3707. 10.1021/bi027071q [DOI] [PubMed] [Google Scholar]
- Harding C., Heuser J., and Stahl P.. 1983. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 97:329–339. 10.1083/jcb.97.2.329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Jones S.A., Brandenburg B., and Zhuang X.. 2008a Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat. Methods. 5:1047–1052. 10.1038/nmeth.1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Wang W., Bates M., and Zhuang X.. 2008b Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 319:810–813. 10.1126/science.1153529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs A. 1977. Low molecular weight intracellular iron transport compounds. Blood. 50:433–439. [PubMed] [Google Scholar]
- Jakobs S., and Wurm C.A.. 2014. Super-resolution microscopy of mitochondria. Curr. Opin. Chem. Biol. 20:9–15. 10.1016/j.cbpa.2014.03.019 [DOI] [PubMed] [Google Scholar]
- Jones S.A., Shim S.H., He J., and Zhuang X.. 2011. Fast, three-dimensional super-resolution imaging of live cells. Nat. Methods. 8:499–505. 10.1038/nmeth.1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotzsch E., Smorodchenko A., Löfler L., Moldzio R., Parkinson E., Schütz G.J., and Pohl E.E.. 2015. Superresolution microscopy reveals spatial separation of UCP4 and F0F1-ATP synthase in neuronal mitochondria. Proc. Natl. Acad. Sci. USA. 112:130–135. 10.1073/pnas.1415261112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopek B.G., Shtengel G., Xu C.S., Clayton D.A., and Hess H.F.. 2012. Correlative 3D superresolution fluorescence and electron microscopy reveal the relationship of mitochondrial nucleoids to membranes. Proc. Natl. Acad. Sci. USA. 109:6136–6141. 10.1073/pnas.1121558109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakadamyali M., Rust M.J., and Zhuang X.. 2006. Ligands for clathrin-mediated endocytosis are differentially sorted into distinct populations of early endosomes. Cell. 124:997–1009. 10.1016/j.cell.2005.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanahan A.A., Hermans K., Claes F., Kerley-Hamilton J.S., Zhuang Z.W., Giordano F.J., Carmeliet P., and Simons M.. 2010. VEGF receptor 2 endocytic trafficking regulates arterial morphogenesis. Dev. Cell. 18:713–724. 10.1016/j.devcel.2010.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann D.J., Worwood M., Ellis R., Wimhurst V.L., Merryweather-Clarke A.T., Warden D.R., Smith A.D., and Robson K.J.. 2006. Iron genes, iron load and risk of Alzheimer’s disease. J. Med. Genet. 43:e52 10.1136/jmg.2006.040519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leo M.D., Bannister J.P., Narayanan D., Nair A., Grubbs J.E., Gabrick K.S., Boop F.A., and Jaggar J.H.. 2014. Dynamic regulation of β1 subunit trafficking controls vascular contractility. Proc. Natl. Acad. Sci. USA. 111:2361–2366. (published erratum appears in Proc. Natl. Acad. Sci. USA. 111:7879) 10.1073/pnas.1317527111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malide D., Métais J.Y., and Dunbar C.E.. 2012. Dynamic clonal analysis of murine hematopoietic stem and progenitor cells marked by 5 fluorescent proteins using confocal and multiphoton microscopy. Blood. 120:e105–e116. 10.1182/blood-2012-06-440636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield F.R., and McGraw T.E.. 2004. Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5:121–132. 10.1038/nrm1315 [DOI] [PubMed] [Google Scholar]
- Mayle K.M., Le A.M., and Kamei D.T.. 2012. The intracellular trafficking pathway of transferrin. Biochim. Biophys. Acta. 1820:264–281. 10.1016/j.bbagen.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride H.M. 2015. Open questions: seeking a holistic approach for mitochondrial research. BMC Biol. 13:8 10.1186/s12915-015-0120-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee A.S., Burchill M.A., Munks M.W., Jin L., Kappler J.W., Friedman R.S., Jacobelli J., and Marrack P.. 2013. Host DNA released in response to aluminum adjuvant enhances MHC class II-mediated antigen presentation and prolongs CD4 T-cell interactions with dendritic cells. Proc. Natl. Acad. Sci. USA. 110:E1122–E1131. 10.1073/pnas.1300392110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesaki K., Tanabe K., Obayashi M., Oe N., and Takei K.. 2011. Fission of tubular endosomes triggers endosomal acidification and movement. PLoS One. 6:e19764 10.1371/journal.pone.0019764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhael M., Sheftel A.D., and Ponka P.. 2010. Ferritin does not donate its iron for haem synthesis in macrophages. Biochem. J. 429:463–471. 10.1042/BJ20100346 [DOI] [PubMed] [Google Scholar]
- Morgan E.H., and Baker E.. 1988. Role of transferrin receptors and endocytosis in iron uptake by hepatic and erythroid cells. Ann. NY Acad. Sci. 526:65–82. 10.1111/j.1749-6632.1988.tb55493.x [DOI] [PubMed] [Google Scholar]
- Murley A., Sarsam R.D., Toulmay A., Yamada J., Prinz W.A., and Nunnari J.. 2015. Ltc1 is an ER-localized sterol transporter and a component of ER-mitochondria and ER-vacuole contacts. J. Cell Biol. 209:539–548. 10.1083/jcb.201502033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G., Pearse A., Domingo D., Trowbridge I.S., and Hopkins C.R.. 1996. Apical and basolateral endosomes of MDCK cells are interconnected and contain a polarized sorting mechanism. J. Cell Biol. 135:139–152. 10.1083/jcb.135.1.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla-Parra S., Matos P.M., Kondo N., Marin M., Santos N.C., and Melikyan G.B.. 2012. Quantitative imaging of endosome acidification and single retrovirus fusion with distinct pools of early endosomes. Proc. Natl. Acad. Sci. USA. 109:17627–17632. 10.1073/pnas.1211714109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrat F., Weisheit D., Lensen M., de Groot H., Sustmann R., and Rauen U.. 2002. Selective determination of mitochondrial chelatable iron in viable cells with a new fluorescent sensor. Biochem. J. 362:137–147. 10.1042/bj3620137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponka P. 1997. Tissue-specific regulation of iron metabolism and heme synthesis: distinct control mechanisms in erythroid cells. Blood. 89:1–25. [PubMed] [Google Scholar]
- Raiborg C., Wenzel E.M., Pedersen N.M., Olsvik H., Schink K.O., Schultz S.W., Vietri M., Nisi V., Bucci C., Brech A., et al. 2015. Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature. 520:234–238. 10.1038/nature14359 [DOI] [PubMed] [Google Scholar]
- Rauen U., Springer A., Weisheit D., Petrat F., Korth H.G., de Groot H., and Sustmann R.. 2007. Assessment of chelatable mitochondrial iron by using mitochondrion-selective fluorescent iron indicators with different iron-binding affinities. ChemBioChem. 8:341–352. 10.1002/cbic.200600311 [DOI] [PubMed] [Google Scholar]
- Reichert A.S., and Neupert W.. 2002. Contact sites between the outer and inner membrane of mitochondria-role in protein transport. Biochim. Biophys. Acta. 1592:41–49. 10.1016/S0167-4889(02)00263-X [DOI] [PubMed] [Google Scholar]
- Richardson D.R., Lane D.J., Becker E.M., Huang M.L., Whitnall M., Suryo Rahmanto Y., Sheftel A.D., and Ponka P.. 2010. Mitochondrial iron trafficking and the integration of iron metabolism between the mitochondrion and cytosol. Proc. Natl. Acad. Sci. USA. 107:10775–10782. 10.1073/pnas.0912925107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland A.A., Chitwood P.J., Phillips M.J., and Voeltz G.K.. 2014. ER contact sites define the position and timing of endosome fission. Cell. 159:1027–1041. 10.1016/j.cell.2014.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust M.J., Bates M., and Zhuang X.. 2006. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods. 3:793–796. 10.1038/nmeth929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena S., Bucci C., Weis J., and Kruttgen A.. 2005. The small GTPase Rab7 controls the endosomal trafficking and neuritogenic signaling of the nerve growth factor receptor TrkA. J. Neurosci. 25:10930–10940. 10.1523/JNEUROSCI.2029-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer J.C., Baetz N.W., Lapierre L.A., McRae R.E., Roland J.T., and Goldenring J.R.. 2014. Rab11-FIP2 interaction with MYO5B regulates movement of Rab11a-containing recycling vesicles. Traffic. 15:292–308. 10.1111/tra.12146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R., Wurm C.A., Jakobs S., Engelhardt J., Egner A., and Hell S.W.. 2008. Spherical nanosized focal spot unravels the interior of cells. Nat. Methods. 5:539–544. 10.1038/nmeth.1214 [DOI] [PubMed] [Google Scholar]
- Sheftel A.D., Zhang A.S., Brown C., Shirihai O.S., and Ponka P.. 2007. Direct interorganellar transfer of iron from endosome to mitochondrion. Blood. 110:125–132. 10.1182/blood-2007-01-068148 [DOI] [PubMed] [Google Scholar]
- Shim S.H., Xia C., Zhong G., Babcock H.P., Vaughan J.C., Huang B., Wang X., Xu C., Bi G.Q., and Zhuang X.. 2012. Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. Proc. Natl. Acad. Sci. USA. 109:13978–13983. 10.1073/pnas.1201882109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigismund S., Confalonieri S., Ciliberto A., Polo S., Scita G., and Di Fiore P.P.. 2012. Endocytosis and signaling: cell logistics shape the eukaryotic cell plan. Physiol. Rev. 92:273–366. 10.1152/physrev.00005.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz J., Panter S.S., Neema M., Arora A., Batt C.E., and Bakshi R.. 2007. Iron in chronic brain disorders: imaging and neurotherapeutic implications. Neurotherapeutics. 4:371–386. 10.1016/j.nurt.2007.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bockxmeer F.M., and Morgan E.H.. 1977. Identification of transferrin receptors in reticulocytes. Biochim. Biophys. Acta. 468:437–450. 10.1016/0005-2736(77)90293-0 [DOI] [PubMed] [Google Scholar]
- van de Linde S., Sauer M., and Heilemann M.. 2008. Subdiffraction-resolution fluorescence imaging of proteins in the mitochondrial inner membrane with photoswitchable fluorophores. J. Struct. Biol. 164:250–254. 10.1016/j.jsb.2008.08.002 [DOI] [PubMed] [Google Scholar]
- Wang C., Wang Y., Li Y., Bodemann B., Zhao T., Ma X., Huang G., Hu Z., DeBerardinis R.J., White M.A., and Gao J.. 2015. A nanobuffer reporter library for fine-scale imaging and perturbation of endocytic organelles. Nat. Commun. 6:8524 10.1038/ncomms9524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward R.J., Zucca F.A., Duyn J.H., Crichton R.R., and Zecca L.. 2014. The role of iron in brain ageing and neurodegenerative disorders. Lancet Neurol. 13:1045–1060. 10.1016/S1474-4422(14)70117-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen K., Reitzel A.M., and Hamdoun A.. 2012. Actin polymerization controls the activation of multidrug efflux at fertilization by translocation and fine-scale positioning of ABCB1 on microvilli. Mol. Biol. Cell. 23:3663–3672. 10.1091/mbc.E12-06-0438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R., Rohr A.M., Wang W.T., Choi I.Y., Lee P., Berman N.E., Lynch S.G., and LeVine S.M.. 2011. Iron deposition is independent of cellular inflammation in a cerebral model of multiple sclerosis. BMC Neurosci. 12:59 10.1186/1471-2202-12-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff N.A., Ghio A.J., Garrick L.M., Garrick M.D., Zhao L., Fenton R.A., and Thévenod F.. 2014. Evidence for mitochondrial localization of divalent metal transporter 1 (DMT1). FASEB J. 28:2134–2145. 10.1096/fj.13-240564 [DOI] [PubMed] [Google Scholar]
- Wurm C.A., Neumann D., Lauterbach M.A., Harke B., Egner A., Hell S.W., and Jakobs S.. 2011. Nanoscale distribution of mitochondrial import receptor Tom20 is adjusted to cellular conditions and exhibits an inner-cellular gradient. Proc. Natl. Acad. Sci. USA. 108:13546–13551. 10.1073/pnas.1107553108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S., Bahl K., Reinecke J.B., Hammond G.R., Naslavsky N., and Caplan S.. 2016. The endocytic recycling compartment maintains cargo segregation acquired upon exit from the sorting endosome. Mol. Biol. Cell. 27:108–126. 10.1091/mbc.E15-07-0514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajac A.L., Goldman Y.E., Holzbaur E.L., and Ostap E.M.. 2013. Local cytoskeletal and organelle interactions impact molecular-motor-driven early endosomal trafficking. Curr. Biol. 23:1173–1180. 10.1016/j.cub.2013.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecca L., Youdim M.B., Riederer P., Connor J.R., and Crichton R.R.. 2004. Iron, brain ageing and neurodegenerative disorders. Nat. Rev. Neurosci. 5:863–873. 10.1038/nrn1537 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.