Abstract
Studies attempting to characterize the membrane translocation of antimicrobial and cell-penetrating peptides are frequently limited by the resolution of conventional light microscopy. This study shows that spheroplasts provide a valuable approach to overcome these limits. Spheroplasts produce less ambiguous images and allow for more systematic analyses of localization. Data collected with spheroplasts are consistent with studies using normal bacterial cells and imply that a particular peptide may not always follow the same mechanism of action.
TEXT
Antimicrobial peptides (AMPs) represent a promising alternative to conventional therapeutics in the face of concerns about the rise of antibiotic-resistant bacteria in clinical settings (1). Traditionally, AMPs were believed to kill bacteria through membrane disruption. While many AMPs do induce membrane permeabilization, researchers have identified increasing numbers of peptides that function by translocating into bacterial cells and targeting intracellular components (2). Thus, it has become increasingly important for researchers to reliably determine whether AMPs are able to effectively translocate into bacterial cells (3). Many researchers have turned to confocal microscopy in order to assess cell entry (4–11). However, bacterial cells are so small that effective imaging is limited by the resolution of conventional light microscopes. For example, in order to distinguish whether any observed signal from peptides arises from inside the cell versus on the cell membrane, researchers ideally should examine individual focal plane images throughout cells. However, if signal on the membrane is sufficiently strong it can “contaminate” slices ostensibly taken “inside” the cell, as we have observed in measurements of the membrane-localized dye di-8-ANEPPS (Fig. 1).
FIG 1.

Confocal images from a z-stack taken of an E. coli cell incubated with the fluorescent membrane labeling dye di-8-ANEPPS. z positions are given relative to the middle image.
In order to overcome these resolution limits, we have employed bacterial spheroplasts (12–14). Spheroplasts are produced by culturing bacteria in the presence of an antibiotic, such as cephalexin, that prevents division while still allowing cells to grow. The resulting elongated bacterial “snakes” are then exposed to lysozyme, which digests the outer cell wall and produces spherical spheroplasts that are typically 2 to 5 μm in diameter (see Fig. S1 in the supplemental material). Perhaps even more important than larger size, the spherical shape allows one to obtain consistent slices regardless of how a spheroplast is oriented during imaging.
In order to test the validity of using spheroplasts to assess peptide translocation, we have measured the cellular localization of four previously characterized peptides (Table 1). To this end, we exposed Escherichia coli spheroplasts to peptides with an N-terminally conjugated fluorescein isothiocyanate (FITC) label for imaging; detailed methods for spheroplast preparation and peptide incubation are provided in the supplemental material. As one set of positive and negative controls, we chose buforin II (BF2), arguably the best-studied membrane-translocating AMP (15), and BF2 with a P11A mutation that dramatically decreases the peptide's ability to enter cells and lipid vesicles (6, 16). As an additional nontranslocating control, we employed magainin 2, a prototypical AMP that acts at the cell membrane (16). As in previous studies, BF2 and magainin peptides included F10W and F5W variations, respectively, which allow for straightforward quantification without significantly altering the peptide activity or mechanism. We also considered HipC, a cell-penetrating peptide without antibacterial activity that was previously observed to enter E. coli (5).
TABLE 1.
Sequences of peptides used in the study and percentages of imaged spheroplasts showing translocation and membrane localization for each peptidea
| Peptide | Sequence | No. of spheroplasts | % translocating | % membrane localized |
|---|---|---|---|---|
| BF2 | TRSSRAGLQWPVGRVHRLLRK | 67 | 63 | 37 |
| P11A BF2 | TRSSRAGLQWAVGRVHRLLRK | 101 | 26 | 74 |
| Magainin 2 | GIGKWLHSAKKFGKAFVGEIMNS | 60 | 18 | 82 |
| HipC | GNYAHRVGAGAPVWL | 67 | 97 | 3 |
Data for each peptide were collected from at least two independently prepared batches of spheroplasts, characterized over a total of at least five separate imaging sessions for each peptide; data for each spheroplast batch are given in Tables S1 to S4 in the supplemental material.
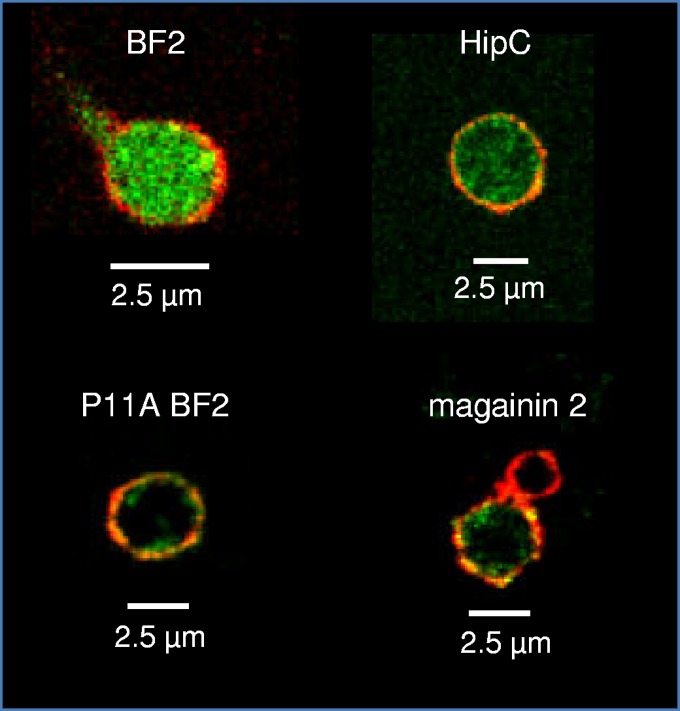
All four control peptides showed the same behavior in spheroplasts as when studied with normal E. coli cells (Fig. 2). Both BF2 and HipC clearly showed entry into the majority of spheroplasts, while P11A BF2 and magainin typically colocalized with membrane dye. For all samples, we found that the use of a membrane dye made it significantly easier to visually distinguish membrane localization from cytosol entry, and no samples showed membrane dye signal contamination on image slices taken from the inside of spheroplasts, regardless of dye intensity.
FIG 2.
Confocal images of representative E. coli spheroplasts incubated with FITC-labeled peptides (BF2, P11A BF2, HipC, or magainin 2) and di-8-ANEPPS. Images from the middle of a z-stack of each spheroplast were chosen, and the merged fluorescence of FITC (green) and di-8-ANEPPS (red) is shown.
In addition to providing improved confocal images, working with spheroplasts also allows us to obtain appreciably more individual images than possible when working with normal cells. While the smaller samples of images possible with bacterial cells can allow one to demonstrate qualitative trends, the difficulty of obtaining sufficiently high-quality images makes it infeasible to perform more systematic analyses of entry data. However, with spheroplasts we can consider the percentage of images showing translocation or membrane localization, providing more systematic data (Table 1). Again, these percentages support the previously observed trends for membrane entry (5, 6, 16), with BF2 and HipC entering significantly more spheroplasts than P11A BF2 and magainin. Interestingly, none of these peptides exclusively exhibit membrane localization or membrane translocation behavior. It is possible that spheroplast behavior differs from bacterial cells or that the observed heterogeneity was related to the exact time of imaging; for example, perhaps all spheroplasts would show entry with BF2 if allowed to incubate for a longer time. However, our observation could also be consistent with the idea that a given AMP may not always follow a single, exclusive mechanism. In fact, there is some evidence for this in previous studies, such as measurements showing that the “translocating” BF2 peptide does induce low levels of membrane permeabilization (17) and that the P11A mutation in BF2 reduces but does not eliminate translocation into lipid vesicles (16). It will be interesting for future studies on spheroplasts, bacterial cells, and other model systems to further evaluate this possibility. It is also worth noting that it is impossible to know for certain whether a particular spheroplast is alive, in the process of dying, or dead in our images, based on the time frame between peptide incubation and mounting and focusing a slide. While this limitation also occurs for studies with normal bacteria, the optical advantages of spheroplasts may make studies looking at the time frame of AMP effects on cells more feasible.
In summary, bacterial spheroplasts provide a promising approach for the effective visualization of AMP interactions with bacterial cells. Clearly, there are differences between “normal” bacterial cells and spheroplasts, in particular the lack of the outer cell wall. Researchers will need to take care to ensure that the lack of cell wall does not affect the results observed in spheroplast experiments for peptides. For example, the cell wall may have a “sieving” effect with some larger peptides that would be lost in spheroplasts, requiring additional controls comparing spheroplasts and “normal” cells in other assays (18). However, even with these caveats we believe that spheroplasts provide an excellent model system compared to other alternatives to overcome size and shape limitations, such as giant unilamellar vesicles (19–21), as spheroplasts preserve a physiological bacterial membrane composition and are viable if returned to growth conditions (13, 22). Moreover, although spheroplasts have generally been produced from E. coli, protocols can be adjusted to make them from strains of other species (23). Thus, we believe that the use of bacterial spheroplasts can be a useful addition to the toolbox of researchers characterizing AMPs and other membrane-active agents, such as cell-penetrating peptides.
Supplementary Material
Funding Statement
Donald E. Elmore is a Henry Dreyfus Teacher-Scholar (Camille and Henry Dreyfus Foundation, Inc.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01008-16.
REFERENCES
- 1.Wang G, Mishra B, Lau K, Lushnikova T, Golla R, Wang X. 2015. Antimicrobial peptides in 2014. Pharmaceuticals 8:123–150. doi: 10.3390/ph8010123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hale JD, Hancock RE. 2007. Alternative mechanisms of action of cationic antimicrobial peptides on bacteria. Expert Rev Anti Infect Ther 5:951–959. doi: 10.1586/14787210.5.6.951. [DOI] [PubMed] [Google Scholar]
- 3.Henriques ST, Melo MN, Castanho MA. 2007. How to address CPP and AMP translocation? Methods to detect and quantify peptide internalization in vitro and in vivo (review). Mol Membr Biol 24:173–184. doi: 10.1080/09687860601102476. [DOI] [PubMed] [Google Scholar]
- 4.Libardo MD, Cervantes JL, Salazar JC, Angeles-Boza AM. 2014. Improved bioactivity of antimicrobial peptides by addition of amino-terminal copper and nickel (ATCUN) binding motifs. ChemMedChem 9:1892–1901. doi: 10.1002/cmdc.201402033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bustillo ME, Fischer AL, LaBouyer MA, Klaips JA, Webb AC, Elmore DE. 2014. Modular analysis of hipposin, a histone-derived antimicrobial peptide consisting of membrane translocating and membrane permeabilizing fragments. Biochim Biophys Acta 1838:2228–2233. doi: 10.1016/j.bbamem.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park CB, Yi KS, Matsuzaki K, Kim MS, Kim SC. 2000. Structure-activity analysis of buforin II, a histone H2A-derived antimicrobial peptide: the proline hinge is responsible for the cell-penetrating ability of buforin II. Proc Natl Acad Sci U S A 97:8245–8250. doi: 10.1073/pnas.150518097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anunthawan T, de la Fuente-Nunez C, Hancock RE, Klaynongsruang S. 2015. Cationic amphipathic peptides KT2 and RT2 are taken up into bacterial cells and kill planktonic and biofilm bacteria. Biochim Biophys Acta 1848:1352–1358. doi: 10.1016/j.bbamem.2015.02.021. [DOI] [PubMed] [Google Scholar]
- 8.Hayden RM, Goldberg GK, Ferguson BM, Schoeneck MW, Libardo MD, Mayeux SE, Shrestha A, Bogardus KA, Hammer J, Pryshchep S, Lehman HK, McCormick ML, Blazyk J, Angeles-Boza AM, Fu R, Cotten ML. 2015. Complementary effects of host defense peptides piscidin 1 and piscidin 3 on DNA and lipid membranes: biophysical insights into contrasting biological activities. J Phys Chem B 119:15235–15246. doi: 10.1021/acs.jpcb.5b09685. [DOI] [PubMed] [Google Scholar]
- 9.Shin A, Lee E, Jeon D, Park YG, Bang JK, Park YS, Shin SY, Kim Y. 2015. Peptoid-substituted hybrid antimicrobial peptide derived from papiliocin and magainin 2 with enhanced bacterial selectivity and anti-inflammatory activity. Biochemistry 54:3921–3931. doi: 10.1021/acs.biochem.5b00392. [DOI] [PubMed] [Google Scholar]
- 10.Koo YS, Kim JM, Park IY, Yu BJ, Jang SA, Kim KS, Park CB, Cho JH, Kim SC. 2008. Structure-activity relations of parasin I, a histone H2A-derived antimicrobial peptide. Peptides 29:1102–1108. doi: 10.1016/j.peptides.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 11.Pavia KE, Spinella SA, Elmore DE. 2012. Novel histone-derived antimicrobial peptides use different antimicrobial mechanisms. Biochim Biophys Acta 1818:869–876. doi: 10.1016/j.bbamem.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinac B, Rohde PR, Cranfield CG, Nomura T. 2013. Patch clamp electrophysiology for the study of bacterial ion channels in giant spheroplasts of E. coli. Methods Mol Biol 966:367–380. doi: 10.1007/978-1-62703-245-2_23. [DOI] [PubMed] [Google Scholar]
- 13.Sun Y, Sun TL, Huang HW. 2014. Physical properties of Escherichia coli spheroplast membranes. Biophys J 107:2082–2090. doi: 10.1016/j.bpj.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinac B, Buechner M, Delcour AH, Adler J, Kung C. 1987. Pressure-sensitive ion channel in Escherichia coli. Proc Natl Acad Sci U S A 84:2297–2301. doi: 10.1073/pnas.84.8.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho JH, Sung BH, Kim SC. 2009. Buforins: histone H2A-derived antimicrobial peptides from toad stomach. Biochim Biophys Acta 1788:1564–1569. doi: 10.1016/j.bbamem.2008.10.025. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi S, Takeshima K, Park CB, Kim SC, Matsuzaki K. 2000. Interactions of the novel antimicrobial peptide buforin 2 with lipid bilayers: proline as a translocation promoting factor. Biochemistry 39:8648–8654. doi: 10.1021/bi0004549. [DOI] [PubMed] [Google Scholar]
- 17.Cutrona KJ, Kaufman BA, Figueroa DM, Elmore DE. 2015. Role of arginine and lysine in the antimicrobial mechanism of histone-derived antimicrobial peptides. FEBS Lett 589:3915–3920. doi: 10.1016/j.febslet.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Decad GM, Nikaido H. 1976. Outer membrane of gram-negative bacteria. XII. Molecular-sieving function of cell wall. J Bacteriol 128:325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Islam MZ, Ariyama H, Alam JM, Yamazaki M. 2014. Entry of cell-penetrating peptide transportan 10 into a single vesicle by translocating across lipid membrane and its induced pores. Biochemistry 53:386–396. doi: 10.1021/bi401406p. [DOI] [PubMed] [Google Scholar]
- 20.Ambroggio EE, Separovic F, Bowie JH, Fidelio GD, Bagatolli LA. 2005. Direct visualization of membrane leakage induced by the antibiotic peptides: maculatin, citropin, and aurein. Biophys J 89:1874–1881. doi: 10.1529/biophysj.105.066589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wheaten SA, Ablan FD, Spaller BL, Trieu JM, Almeida PF. 2013. Translocation of cationic amphipathic peptides across the membranes of pure phospholipid giant vesicles. J Am Chem Soc 135:16517–16525. doi: 10.1021/ja407451c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruthe H-J, Adler J. 1985. Fusion of bacterial spheroplasts by electric fields. Biochim Biophys Acta 819:105–113. doi: 10.1016/0005-2736(85)90200-7. [DOI] [PubMed] [Google Scholar]
- 23.Rowe I, Elahi M, Huq A, Sukharev S. 2013. The mechanoelectrical response of the cytoplasmic membrane of Vibrio cholerae. J Gen Physiol 142:75–85. doi: 10.1085/jgp.201310985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.