Abstract
Despite the increasing prevalence of the nosocomial pathogen Acinetobacter baumannii, little is known about which genomic components contribute to clinical presentation of this important pathogen. Most whole-genome comparisons of A. baumannii have focused on specific genomic regions associated with phenotypes in a limited number of genomes. In this work, we describe the results of a whole-genome comparative analysis of 254 surveillance isolates of Acinetobacter species, 203 of which were A. baumannii, isolated from perianal swabs and sputum samples collected as part of an infection control active surveillance program at the University of Maryland Medical Center. The collection of surveillance isolates includes both carbapenem-susceptible and -resistant isolates. Based on the whole-genome phylogeny, the A. baumannii isolates collected belong to two major phylogenomic lineages. Results from multilocus sequence typing indicated that one of the major phylogenetic groups of A. baumannii was comprised solely of strains from the international clonal lineage 2. The genomic content of the A. baumannii isolates was examined using large-scale BLAST score ratio analysis to identify genes that are associated with carbapenem-susceptible and -resistant isolates, as well as genes potentially associated with the source of isolation. This analysis revealed a number of genes that were exclusive or at greater frequency in each of these classifications. This study is the most comprehensive genomic comparison of Acinetobacter isolates from a surveillance study to date and provides important information that will contribute to our understanding of the success of A. baumannii as a human pathogen.
INTRODUCTION
The Acinetobacter genus is comprised of 34 named species, many of which are found ubiquitously in the environment (1). The most clinically relevant species are members of what is known as the Acinetobacter calcoaceticus-baumannii (Acb) complex, which include A. baumannii, A. nosocomialis, A. pittii, and A. calcoaceticus (2). The organisms of the Acb complex are difficult to distinguish from each other using traditional culturing and molecular diagnostic methods employed in the current clinical microbiology laboratories (3). A. baumannii is the most frequently isolated Acinetobacter species in the health care setting (4), partly due to its ability to survive on abiotic surfaces for long periods of time (5), as well as its ability to rapidly develop antibiotic resistance (6). It is an important emerging nosocomial pathogen associated with common infections, multidrug resistance, and a high transmission rate within hospitals all over the world (7).
Prior genomic studies of A. baumannii have mainly focused on multidrug-resistant strains and have identified key determinants associated with resistance (8–14). A genomic comparison of six clinical isolates of A. baumannii concluded that despite extensive genome-wide similarity, the resistance gene repertoire was highly variable, even in the genomically closely related isolates (10). In addition, another comparison of multidrug-resistant strains isolated from diverse body sites concluded that there was no gene content exclusivity based on site of isolation (11). Limited whole-genome studies have provided insight into A. baumannii and identified a common core set of genes and a highly variable accessory genome that includes many of the acquired antibiotic resistance genes (14, 15).
One of the main characteristics that accounts for the ability of A. baumannii to persist in the clinical setting is its ability to acquire the DNA for numerous antimicrobial resistance mechanisms (16). The bacterium has been demonstrated to acquire resistance by either mutation of existing DNA or acquisition of mobile elements, such as plasmids (17), transposons (18), and resistance islands, that carry the genes responsible for antimicrobial resistance (19–21).
The group of β-lactam antibiotics known as the carbapenems is commonly used to treat A. baumannii infections (22). However, A. baumannii can acquire resistance to carbapenems by acquiring carbapenem-hydrolyzing class D β-lactamase genes such as OXA-23 (23). Another OXA gene, OXA-51, is chromosomal and believed to be native to most, if not all, A. baumannii isolates (24). Although it is typically a weak carbapenemase, acquiring mutations or movement from the chromosome to a plasmid, thereby effectively increasing the copy number, has been reported to increase activity in some isolates (25, 26). Since the carbapenems are one of the last lines of defense in terms of antibiotics against this pathogen, efforts to elucidate factors that contribute to the acquisition of resistance will be important to further our understanding of how to treat infections and develop new therapies, and genomic studies such as the present one will prove important to further this understanding.
Multilocus sequence typing (MLST) is a common molecular method used to distinguish different clonal lineages of A. baumannii as well as outbreaks in a health care setting (27–30). There are two major MLST schemes used for typing of A. baumannii strains, Pasteur and Oxford (31, 32). Numerous clonal groups of A. baumannii species have been identified by MLST, and these clonal groups are often associated with specific geographic locations (33, 34). International clonal lineages I, II, and III are the most frequently observed clonal lineages isolated in the hospital setting (33, 35–37). While International clonal lineages I and II are identified worldwide, lineage III appears to be more prevalent in Europe (38).
Although MLST has been a relatively effective typing scheme for this pathogen, whole-genome sequencing has provided additional insight into the potential virulence and evolution of this important human pathogen (39). Previous whole-genome comparisons of Acinetobacter, particularly A. baumannii, have included a number of isolate collections (9, 11, 40). A total of 1,136 genomes are publically available in GenBank at the time of the writing of the manuscript, and although there are many genomes in GenBank, not all have been examined in the comparative studies listed above. Most of the strains genomically characterized to date have been from outbreaks, from cases of hospital infections, or from retrospective collections (8, 10). In a comparison of multidrug-resistant A. baumannii isolates from diverse human body sites, Sahl et al. did not identify genomic regions that were exclusive to any particular phenotype or site of isolation; however, this comparison only included a limited number of genomes (11). While these analyses have provided important insight into the genomic characteristics, resistance mechanisms, and outbreak potential of this pathogen, inclusion of strains from surveillance samples, such as the ones examined in the present study, may allow for a more meaningful and unbiased comparison of associated genetic features.
In this study, we sequenced the genomes of 254 Acinetobacter species isolates and characterized these genomes using whole-genome phylogenetic analysis (41), MLST (31, 32), and large-scale BLAST score ratio analysis (LS-BSR) (42). The genomic content within these isolates, in addition to 61 sequenced reference genomes, demonstrates tremendous diversity in the Acinetobacter species. However, we have identified a limited number of genetic features that may further our understanding of the success of this pathogen in the clinical setting.
MATERIALS AND METHODS
Strain selection.
Isolates were obtained from a collection of patient surveillance samples collected as part of a cohort study at the University of Maryland Medical Center (UMMC). This surveillance cohort included a total of 2,220 Acinetobacter isolates from 985 patients treated in UMMC medical and surgical intensive care units, with isolation dates ranging from 2005 to 2012 (43, 44). Isolates were obtained which provide samples from a broad range of years, subjects, and human body isolation locations. Swabs were cultured on selective media for Acinetobacter species, and carbapenem susceptibility was tested using the Kirby-Bauer disk diffusion method (45). A total of 96 sputum and 158 perirectal isolates were selected for sequencing. Additionally, 61 publicly available reference Acinetobacter strains were included in these genomic analyses (see Table S1 in the supplemental material).
DNA isolation and sequencing.
For genomic DNA isolation, strains were grown overnight at 37°C with shaking at 250 rpm in Luria-Bertani broth, and the Sigma GenElute genomic kit (Sigma-Aldrich) was used to isolate DNA. Genomic DNA was extracted with standard methods (46) and sequenced on the Illumina HiSeq 2000 platform. The resulting 100-bp reads were assembled with the MaSurRCA Assembler (47) at the University of Maryland Institute for Genome Sciences, Genome Resource Center. The resulting assembly metrics of the entire genomic content and corresponding accession numbers are shown in Table S1 in the supplemental material.
Phylogenetic analysis.
The whole-genome phylogeny was inferred using the In Silico Genotyper (ISG), an open-source pipeline (41). In brief, 315 assembled genomes, including the 254 newly sequenced genomes, were aligned, and single-nucleotide polymorphisms (SNP) were called using the fully sequenced A. baumannii isolate TYTH-1 as the reference (48). The program FigTree v1.4.0 was used for visualization (http://tree.bio.ed.ac.uk/software/figtree/).
LS-BSR.
Large-scale BLAST score ratio (LS-BSR) analysis was performed as described previously (42). Briefly, the LS-BSR pipeline includes the program Prodigal (49), which was used to predict all coding sequences from query genomes. Coding sequences were then dereplicated using USEARCH (50) and clustered, and one representative sequence from the cluster, known as a centroid, was used to generate the BSR values. One could consider the centroid sequences as the complete collection of nonredundant coding sequences in this collection of genomes. All unique coding sequences were then translated with BioPython, and a reference bit score was calculated based on a TBLASTN alignment. Each query sequence was aligned against each genome and a query bit score was generated. Dividing the query bit score by the reference bit score then generated the final BSR values. Centroids considered exclusive to a group had a BSR value of ≥0.8 in genomes in the target group and ≤0.4 in all genomes in the nontarget group. Core centroids all had BSR values of ≥0.8 in all genomes analyzed. Centroids considered unique in one genome had a BSR value of ≥0.8 in a single genome. The matrix of BSR values was visualized using Multiple Experiment Viewer (MeV) (51), and the Interactive Tree of Life project (52) was used to generate a heatmap for the comparison of the virulence genes.
In order to determine centroids present in greater frequency among phylogenetic or clinical groups, centroids within groups were counted and a matrix of these values was generated. The R program was used to examine the values using Fisher's exact test (53). The output included P values for each of the centroid frequencies within the specific groupings. A P value threshold of ≤0.001 was considered significant for all comparisons.
MLST analysis.
In silico multilocus sequence type (MLST) profiles were assigned using the sequences for MLST markers for the Oxford typing scheme (gltA, recA, cpn60, gyrB, gdhB, rpoD, and gpi) (31) and Pasteur typing scheme (gltA, recA, cpn60, fusA, pyrG, rpoB, and rplB) (32). Sequences were downloaded from the pubMLST database (http://pubmlst.org/abaumannii) and then used to extract allele sequences from each of the 203 A. baumannii genomes using the BLASTN tool. The extracted sequences were then uploaded to the pubMLST database to assign both existing and novel sequence types.
Accession numbers.
Sequence data for the 254 Acinetobacter species isolates have been deposited in GenBank, and the accession numbers are listed in Table S1 in the supplemental material.
RESULTS AND DISCUSSION
Genome statistics.
The 254 Acinetobacter species isolates were sequenced, assembled, and deposited in GenBank. Summary details of descriptive statistics for isolates by year are listed in Table 1. The genome assembly statistics are provided in Table S1 in the supplemental material. A total of 203 A. baumannii isolates were selected for further analysis to include the greatest period of time between isolates (2006 to 2012) for carbapenem-resistant and -susceptible isolates, as well as isolates collected from both the perirectal and sputum surveillance sources. The average genome size for all A. baumannii genomes was 4.09 Mb (range, 3.70 Mb to 4.85 Mb), and the average GC content was 39.1% (range, 38.8% to 39.4%), which is common for this species.
TABLE 1.
Summary of all Acinetobacter isolates newly sequenced in this study by year
| Yr | Total no. of isolates | % of total isolates | No. (%) of isolates |
No. (%) of samples |
||
|---|---|---|---|---|---|---|
| Carbapenem resistant | Carbapenem sensitive | Sputum (%) | Perirectal (%) | |||
| Before 2009 | 42 | 17 | 31 (74) | 11 (26) | 2 (5) | 40 (95) |
| After 2009 | 212 | 83 | 99 (47) | 110 (52) | 94 (44) | 118 (56) |
There was no bias of genome size based on location of isolation or resistance phenotype observed (data not shown). There was no statistical difference in average genome size for carbapenem-resistant isolates, 4.11 Mb (3.84 to 4.85 Mb), compared to susceptible isolates, 4.05 Mb (3.68 to 4.70 Mb), using a P value cutoff of <0.01. In addition, the %GC was similar for resistant and susceptible isolates, 39.1% and 39.0%, respectively. Likewise, a comparison of the genome size for isolates collected from sputum versus by perirectal swab, 4.07 Mb (3.68 to 4.51 Mb) and 4.10 Mb (3.70 to 4.85 Mb), respectively, showed no statistical difference (P < 0.01).
Phylogenetic analysis of Acinetobacter strains.
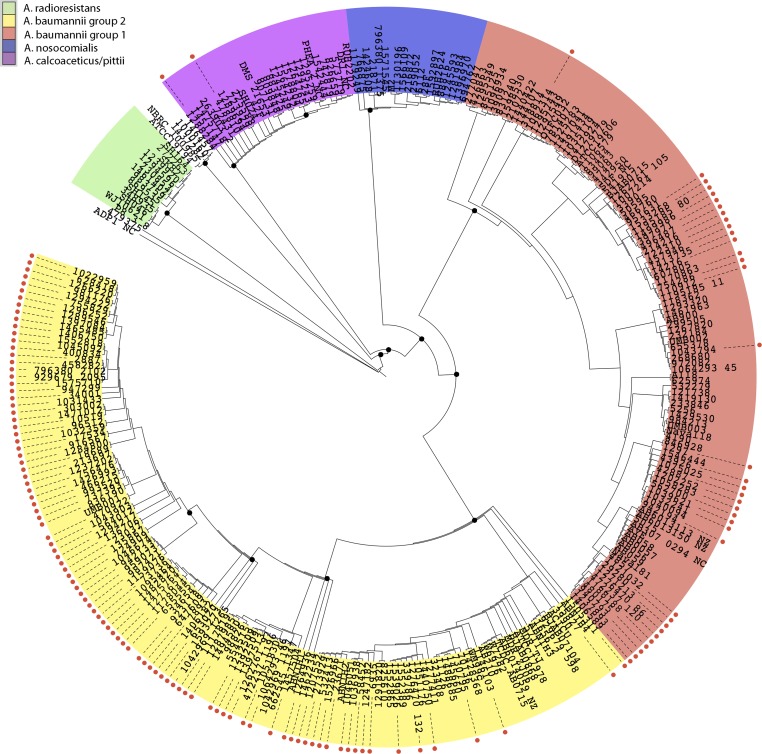
The whole-genome phylogeny was inferred using the 254 Acinetobacter species sequenced in this project, as well as 61 additional reference Acinetobacter species genomes that were publically available in GenBank (see Table S1 in the supplemental material). When the genomes were compared using ISG (41), a total of 22,788 informative SNPs were identified. Based on the genome clustering with characterized reference genomes, 242/254 (95.3%) of the isolates selected for genomic analysis were part of the Acinetobacter calcoaceticus-baumannii (Acb) complex (Fig. 1). Twelve of the genomes in this analysis clustered with the reference genome for A. radioresistens, which is known to colonize human skin but is not considered a clinically relevant member of the Acb complex (54). The majority of the newly sequenced isolates, 79.9% (203/254), could be further classified as A. baumannii based on the phylogenetic similarity to reference isolates, and 16.2% (39/242) of isolates identified as species inside the Acb complex but not A. baumannii.
FIG 1.
Phylogenomic analysis of Acinetobacter genomes. Phylogenetic analysis of 254 newly sequenced Acinetobacter genomes and 61 genomes that had been previously sequenced. Genomes were aligned and SNPs were called using the program MUMmer. GATK Unified Genotyper was used to call and determine ambiguity of SNPs and to generate a VCF file. RAxML 7.2.8 was used to infer the phylogenetic tree with 100 bootstrap replicates. The phylogeny was rooted with the A. baumannii TYTH-1 complete genome.
The inferred phylogeny shown in Fig. 1 identifies two major phylogenomic groups of A. baumannii (Fig. 1, yellow and salmon groups). These two phylogenomic groups are akin to the epidemic or global clones that have been previously discussed in the literature (12, 14), with our phylogenomic group 1 being similar to the clonal complex 1. The distribution of isolates based on carbapenem susceptibility reveals that a greater proportion, 80% (92/115), of the isolates present in the A. baumannii phylogenomic group 2 are carbapenem resistant, whereas only 41% (36/88) of the isolates in phylogenomic group 1 are resistant. This was found to be statistically significant (P < 0.001) with a t test.
When the isolates were examined based on the isolation source and resistance phenotype, 72% (56/78) of the sputum isolates were resistant to carbapenems, and 60% (75/125) of the perirectal isolates were resistant. This difference was not statistically significant (P > 0.001). Examination of the isolation source within the two A. baumannii phylogenetic groups revealed a greater proportion of perirectal isolates in phylogenetic group 1. A total of 33% (29/88) of isolates in phylogenetic group 1 and 44% (51/115) in phylogenetic group 2 were isolated from sputum, whereas 67% (59/88) of the isolates from phylogenetic group 1 and 56% (64/115) from phylogenetic group 2 were isolated from perirectal swabs. These differences also were not found to be statistically significant (P > 0.001).
Overall, the newly sequenced isolates in this study are overrepresented by perirectal carbapenem-resistant isolates that are members of A. baumannii phylogroup 2 compared to other large-scale A. baumannii comparative genomics studies (13–15, 39, 55).
MLST.
The results of the MLST profiling of the genomes included in these analyses using two different MLST schemes (Pasteur and Oxford) are presented in Table S1 in the supplemental material. MLST sequence types (ST) were determined and the results overlaid onto the whole-genome phylogeny (see Fig. S1a and b in the supplemental material). The results indicate that the Pasteur (32) and Oxford (31) schemes differ remarkably in terms of distribution of similar STs compared to the phylogenetic tree inferred based on the whole-genome analysis. The STs assigned by the Pasteur scheme are more consistent with the results of the whole-genome phylogeny (see Fig. S1a in the supplemental material), with most strains in A. baumannii phylogenomic group 2 assigned to Pasteur ST 2. In contrast, the results from the Oxford MLST schema were less congruent with the whole-genome phylogeny and there was a greater number of STs represented and a significant number of strains with novel STs, suggesting that the allele sequences for this MLST scheme were more variable among the isolates examined. A similar finding of the lack of congruence with pulsed-field gel electrophoresis and MLST typing was also previously noted (56).
LS-BSR analysis.
The genome contents of the A. baumannii isolates were analyzed using LS-BSR (46). Generation of the LS-BSR matrix resulted in the reduction of 959,329 potential coding sequences present in the newly sequenced A. baumannii genomes to a nonredundant set of 13,585 centroids (see Data File S1 in the supplemental material). The average number of centroids present at a BSR value of ≥0.8 in each strain was 4,675 (range, 4,154 to 5,312). The analysis identified 1,507 centroids that were conserved (BSR, ≥0.8) among all 203 A. baumannii isolates (Table 2). In contrast, 3,237 centroids were present in only a single genome, indicating that there is an average of ∼16 unique centroids per A. baumannii genome sequenced in this study. This value is similar to those of other comparable A. baumannii genomic studies (15). A previous genomic analysis by Sahl et al. on a limited number of genomes identified a single gene, annotated as a hypothetical protein (GenBank accession number ACJ58112), that was conserved in >99% of the A. baumannii isolates via PCR assay (11). Using LS-BSR in silico analysis, we were able to identify the presence of this predicted marker in 96% (195/203) of the A. baumannii isolates in this study and 2% (1/51) of the non-A. baumannii isolates. These results suggest that this genetic region is a molecular marker for the identification of A. baumannii isolates.
TABLE 2.
LS-BSR analysis grouping summary
| Species | No. (%) of: |
|||
|---|---|---|---|---|
| Isolates | Centroids | Conserved centroidsa | Unique centroidsb | |
| A. baumannii only | 203 | 13,586 | 1,507 (11) | 3,237 |
| A. baumannii and A. nosocomialis | 220 | 16,676 | 1,695 (10) | 3,497 |
| A. baumannii/nosocomialis/calcoaceticus (Acb complex) | 242 | 21,485 | 2,140 (10) | 4,250 |
| A. baumannii/nosocomialis/radioresistens | 229 | 26,485 | 1,907 (7) | 7,216 |
| A. baumannii/calcoaceticus/radioresistens | 237 | 32,560 | 2,346 (7) | 8,209 |
| A. baumannii/radioresistens | 212 | 23,369 | 1,928 (8) | 7,010 |
| A. baumannii/nosocomialis/calcoaceticus/radioresistens | 254 | 35,242 | 2,297 (7) | 8,283 |
Defined as BSR value of ≥0.8 in all genomes.
Centroids present in only a single genome.
The LS-BSR analysis also allowed identification of centroids that were unique to each of the major phylogenomic groups of the inferred phylogeny in Fig. 1. LS-BSR was used to compare all A. baumannii and non-baumannii phylogroups as identified through the whole-genome phylogenetic analysis. Using the strict criteria of selected centroids identified being present in all isolates of one phylogenomic group but in none of the isolates from any other phylogenomic group, unique centroids were found for each major phylogroup except A. baumannii group 1 (see Table S2 in the supplemental material). While the majority of these predicted genes are currently annotated as hypothetical, they do provide potential molecular targets for identification of these closely related species. Interestingly, no centroids were found to be present exclusively in all A. baumannii isolates and lacking in other species isolates. Further analysis revealed two centroids that were unique to all isolates in A. baumannii phylogenetic group 2 compared to A. baumannii phylogenomic group 1, as well as all other non-baumannii isolates. These centroids, 993569 and 204780, were annotated as a hypothetical protein and an enzyme in the cupin superfamily. Conversely, there were no centroids found to be exclusive to all isolates of A. baumannii phylogenetic group 1 compared to group 2. This finding highlights the diversity of the genomes in this species.
Diversity based on date of isolation.
Isolation dates for the surveillance samples analyzed in this study ranged from 2006 to 2012 (see Table S1 in the supplemental material). In order to determine if there was emergence or selection of specific genes during this time period, a comparison was completed of samples before 2009 versus those obtained after 2009. The majority of the samples, 81% (164/203), were isolated after 2009, compared to 19% (39/203) obtained before 2009. There were 140 centroids found at higher frequency in the pre-2009 isolates (P ≤ 0.001) (see Table S3a in the supplemental material). A large proportion (∼40%) of these centroids were annotated as hypothetical with no assigned function. The remaining 60% of these centroids encode a wide diversity of proteins, as indicated by their assigned annotation (see Table S3a).
There were 68 centroids found to be present at greater frequency (P ≤ 0.001) in isolates collected after 2009 (see Table S3b in the supplemental material); 33 of these were found to be present exclusively in post-2009 isolates but were not present in all isolates of that group. These exclusive centroids encoded phage integrases, proteases, RHS repeat-associated proteins, an MreB/MBL family protein, membrane proteins, pyocin activator proteins, and numerous hypothetical proteins. In addition, there were 35 centroids that were found at greater frequency post-2009 but were not found exclusively in these genomes. These encoded many phage/plasmid-like proteins and hypotheticals, as well as a hydrolase and a phosphatase (see Table S3b).
It is interesting that a majority of centroids (54/68) found at greater frequency in the post-2009 isolates were found exclusively in A. baumannii phylogroup 1. This suggests that phylogroup 2 is more clonal and the genomes of these isolates are more stable and resistant to acquisition of new genetic elements. Conversely, members of A. baumannii phylogroup 1 are more diverse, with more dynamic genomic content that is vulnerable to loss and acquisition of new genetic elements.
LS-BSR analysis by location of isolation.
A comparison of the genomes of A. baumannii isolates from all sputum samples versus all perirectally obtained samples identified that there were no centroids found in all genomes from one isolation location and not in the other. In addition, there were no centroids that were exclusive to either group. However, using Fisher's exact test we were able to identify centroids that were found more frequently in the genomes from one isolation source relative to the frequency in the genomes of the comparison group. There were 42 centroids more common in the sputum isolates and 4 centroids more common in the perirectal isolate genomes (see Table S4a and b in the supplemental material). The sputum-specific genes include 15 genes with no functional annotation and a number of genes associated with flagellar assembly and motility, as well as multiple regulatory proteins, suggesting that motility and regulation of A. baumannii genes are critical in the survival or transmission in sputum (see Table S4a). Interestingly, it has been described that A. baumannii lacks motility via flagella (57); however, our annotation suggests that there are some genes annotated via our pipeline that have enough homology to be associated with flagellar assembly and motility genes; however, as previously described, no isolate contained a complete flagellar operon. Whereas the four genes that are specific to the perirectal isolates include lipases, an HPP family protein, and a regulatory protein, with currently unknown roles in survival or tropism for this environment, it is possible that these features facilitate colonization, but these isolates may not all be continuous colonizers of the gastrointestinal tract (i.e., they are passing through but are not established). These centroids provide some insight into the potential success of the isolates at each body site but also confirm previous studies that suggest that A. baumannii isolates are generalists in terms of site of isolation (see Table S4a and b). Overall, these molecular targets provide a limited gene set with which to start functional characterization of these phenotypes or isolates with a tropism for these body sites.
LS-BSR analysis by resistance status.
When the genomes of the carbapenem-resistant A. baumannii isolates were compared to those of the carbapenem-susceptible A. baumannii isolates using LS-BSR, eight centroids were identified to be unique and present only in the resistant isolates (see Table S5a and b in the supplemental material). Interestingly, none of these centroids matched with 100% identity to previously identified A. baumannii genes currently in GenBank by BLAST search, suggesting that these genes have been acquired relatively recently by A. baumannii. Two of the centroids matched with 99% nucleotide identity to a gene in an Acinetobacter capsule biosynthesis gene cluster, wzx (locus tag AN415_03886) (58). Conversely, there were 319 centroids that were found only in the genomes of carbapenem-susceptible A. baumannii isolates compared to carbapenem-resistant A. baumannii isolates (see Table S5b). These centroids include genes involved in type IV secretion systems and CRISPR systems (59), as well as several transcriptional regulatory genes. In addition, 141 (44%) hypothetical genes, one transcriptional regulator, and many phage-related genes are included in this list.
The difference in the number of genes that are preferentially in each of these groups is striking and suggests that the resistant isolates are lacking novel genes due to selective processes, or that the susceptible isolates represent a more diverse population which could be considered generalist, thus requiring a greater diversity of genes for increased survival in multiple environments. For example, the identification of the CRISPR genes in the susceptible isolates suggests the loss of these genes was an important step in acquiring genetic elements necessary for carbapenem resistance. CRISPR-cas systems provide bacteria a type of immunity against foreign genetic elements such as plasmids and bacteriophage; therefore, the loss of these genes may allow genetic elements to be acquired more readily (59).
LS-BSR analysis by resistance status and location of isolation.
When the additional criteria of location of isolation and resistance status are combined, there is a further focusing on genes that may play a role in virulence in the specific body location. Comparison of genomes from carbapenem-susceptible A. baumannii isolates from the perirectum compared to resistant A. baumannii isolates from the perirectum identified 135 centroids unique to the perirectal isolates that were susceptible (see Table S6a in the supplemental material). These centroids include genes for CRISPR-cas systems as well as phage-related genes. In contrast, only seven centroids were identified to be exclusively in the perirectal resistant A. baumannii isolates compared to susceptible A. baumannii isolates from the perirectum (see Table S6b). All seven of these centroids were also found to be unique to the resistant isolates irrespective of site of isolation. The only centroid identified as unique among resistant isolates which was not found to be unique to the perirectal resistant group was a centroid that encodes a type IV pilin structural subunit (centroid 365901).
Comparisons of the genomes from A. baumannii carbapenem-susceptible sputum isolates to A. baumannii carbapenem-resistant sputum isolates revealed 31 centroids unique to the susceptible isolates (see Table S6c in the supplemental material). Many of these coding sequences were annotated as hypothetical proteins; however, several centroids encoded Rhs element Vgr family domain proteins which have been shown to be involved in type VI secretion system (T6SS) functions (60). A T6SS has been previously characterized in some isolates of A. baumannii (61). In contrast, the comparison of genomes from carbapenem-resistant sputum isolates to carbapenem-susceptible sputum isolates revealed only a single centroid unique to sputum-resistant isolates, which encoded the carbapenemase OXA-23 gene (centroid 805287) (62).
Prevalence of the OXA genes in A. baumannii.
The OXA-51 gene is known to be encoded on the chromosome and believed to be present in most, if not all, A. baumannii isolates (24). In the current study, 200 of 203 isolates contain this version of the gene (see Table S14 in the supplemental material). Additionally, the oxacillinase gene, blaOXA-23, which is known to be largely restricted to the Acinetobacter genus, has been found worldwide in the chromosome or plasmids of A. baumannii isolates (16). The presence of this gene was investigated, and it was found to be present in only 61 of the 203 newly sequenced A. baumannii genomes (see Table S14 in the supplemental material). However, a greater proportion of isolates in A. baumannii phylogenomic group 2 contained this gene than A. baumannii phylogenomic group 1, 43% (50/115) versus 10% (9/88), respectively. Interestingly, 40% of the isolates that were phenotypically resistant to imipenem did not contain this gene or any other OXA-like gene, which is commonly identified in resistant isolates. Likewise, we identified isolates that were phenotypically susceptible to imipenem but contained the blaOXA-23 gene, suggesting that there are other factors involved in the phenotype of antibiotic sensitivity and resistance in A. baumannii. Other OXA genes were also examined for prevalence in this data set, but only a limited number, 6 or 7 isolates, encoded OXA-24/40-like or OXA-143-like genes, respectively. Further study will be required to characterize the integration of these OXA-like genes as well as other factors in the development of resistance.
Prevalence of previously identified virulence-associated genes in A. baumannii genomes.
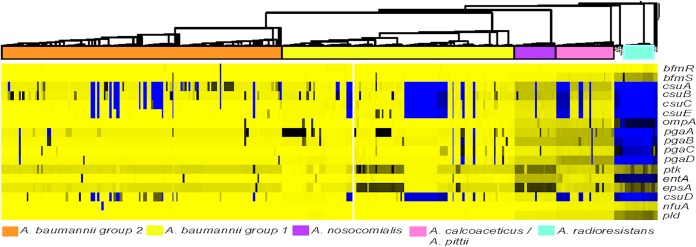
Numerous genes have been identified as contributing to the virulence of A. baumannii. The presence of these genes was determined in the genomes of the newly sequenced A. baumannii isolates (Fig. 2). The list of virulence genes analyzed can be found in Table 3. These include genes for a two-component regulatory system, an usher-chaperone system, outer membrane proteins, genes involved in polysaccharide production, a protein tyrosine kinase, genes involved in iron transport and siderophore biosynthesis, and a phospholipase protein (57, 63–69). These virulence-associated genes appear to be highly conserved in all of the A. baumannii genomes analyzed, with the exception of the genes of the CsuA/BABCDE chaperone-usher complex, which is important for the assembly and production of pili involved in surface adhesion. Overall, the previously identified A. baumannii virulence factors are highly conserved in this collection of sequenced surveillance isolates.
FIG 2.
Heat map of known virulence-associated factors in A. baumannii. The amino acid sequences for genes previously identified as being associated with virulent phenotypes were obtained from the National Center for Biotechnology database (Table 3). LS-BSR analysis to detect if these genes were present in the newly sequenced A. baumannii isolates was performed using TBLASTN. The BSR values were generated as described previously. The heat map generated was visualized with phylogeny using Tree of Life v3.0 software.
TABLE 3.
Virulence genes analyzed
| Gene | Protein ID | Function | Reference |
|---|---|---|---|
| bfmR | AAX40744.1 | Two-component system | 58 |
| bfmS | AAX40745.1 | Two-component system | 58 |
| csuA | AAP43036.1 | Usher-chaperone system | 59 |
| csuB | AAP43037.1 | Usher-chaperone system | 59 |
| csuC | AAP43038.1 | Usher-chaperone system | 59 |
| csuD | WP_044557399.1 | Usher-chaperone system | 59 |
| csuE | AAP43040.1 | Usher-chaperone system | 59 |
| ompA | AHW42434.1 | Outer membrane protein | 60 |
| pgaA | ACV90433.1 | PNAG synthesis | 61 |
| pgaB | ACV90434.1 | PNAG synthesis | 61 |
| pgaC | ACV90435.1 | PNAG synthesis | 61 |
| pgaD | ACV90436.1 | PNAG synthesis | 61 |
| ptk | WP_001075368 | Protein tyrosine kinase | 62 |
| entA | WP_001257961.1 | Siderophore biosynthesis | 63 |
| epsA | WP_000872534.1 | Polysaccharide export outer membrane protein | 62 |
| nfuA | ABO11411.2 | Iron transport protein | 64 |
| Phospholipase D | ABO13297.2 | Phospholipase | 65 |
Conclusion.
The goal of this study was to characterize the genomic diversity of A. baumannii surveillance isolates from the University of Maryland Medical Center and compare it to known collections of sequenced A. baumannii. Two major phylogenomic groups within A. baumannii were identified, termed A. baumannii phylogroup 1 and A. baumannii phylogroup 2 (Fig. 1). Interestingly, both phylogenomic groups contained carbapenem-resistant isolates; however, A. baumannii phylogroup 2 contains a greater proportion of carbapenem-resistant isolates (80%; 92/115). The presence of the beta-lactamase gene blaOXA-23, which has been correlated with carbapenem resistance, did not always correlate with the observed resistance phenotype previously described (23). Whole-genome comparisons identified two centroids that were in all genomes of A. baumannii group 2 and were not present in any genomes from A. baumannii group 1, which will require additional functional studies.
Using the LS-BSR analysis, genomic regions were identified that are unique to specific phylogenetic groups, sites of isolation, or resistance phenotypes. The LS-BSR analysis identified 1,507 conserved centroids among the 203 A. baumannii isolates analyzed (Table 4). This value is within the range of previous estimates, which have put the number of core genes between 1,455 and 2,830 genes (9, 13–15, 55, 70–72). This information provides possible targets for future diagnostic tests to distinguish members of the Acb complex from each other and other Acinetobacter species. Although we did not find a single centroid that was present in all A. baumannii isolates compared to other Acinetobacter species groups, sequences were identified that were present in all isolates of other phylogenetic groups, such as hydrolases unique to A. calcoaceticus and several hypothetical proteins in A. nosocomialis. These sequences, in addition to the conserved sequence previously identified (11) as being present in the majority of A. baumannii isolates in a prior study, may provide a useful combination of tests to positively distinguish A. baumannii from other species of the Acb complex using multiplex PCR. In addition to the diagnostics aspects of these types of data sets, we have also narrowed the number of targets to be functionally examined that may play a role in the development of antimicrobial resistance or tropism to the lung. These functional studies are beyond the scope of the current study, but it provides a framework for future functional studies.
TABLE 4.
Summary of results from Fisher's exact analysis of LS-BSR
| Group | No. of centroids |
|
|---|---|---|
| Present in higher frequencya | Exclusive to groupb | |
| Resistant | 1,246 | 8 |
| Sensitive | 1,038 | 319 |
| Perirectal | 4 | 0 |
| Sputum | 42 | 0 |
| Perirectal resistantc | 904 | 7 |
| Sputum resistantd | 327 | 1 |
| Perirectal sensitive | 556 | 135 |
| Sputum sensitive | 315 | 31 |
Based on a P value of ≤0.001.
Number of centroids found exclusively in the group but not necessarily in every isolate of that group.
Perirectal resistant genomes were compared to perirectal sensitive genomes.
Sputum resistant genomes were compared to sputum sensitive genomes.
Overall, this study demonstrates that the A. baumannii genome is more diverse than previously anticipated and is lacking an extensive common core that is associated with resistance to carbapenem antibiotics, suggesting that there are other unidentified factors responsible for this phenotype.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported in part by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract number HHSN272200900009C, NIH grant numbers K24AI079040 and R01HS02229103, and startup funds from the state of Maryland. L.W. was supported by the Defense Threat Reduction Agency and the Department of Defense SMART Scholarship Program.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00477-16.
REFERENCES
- 1.Jung J, Park W. 2015. Acinetobacter species as model microorganisms in environmental microbiology: current state and perspectives. Appl Microbiol Biotechnol 99:2533–2548. doi: 10.1007/s00253-015-6439-y. [DOI] [PubMed] [Google Scholar]
- 2.Nemec A, Krizova L, Maixnerova M, van der Reijden TJ, Deschaght P, Passet V, Vaneechoutte M, Brisse S, Dijkshoorn L. 2011. Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU). Res Microbiol 162:393–404. doi: 10.1016/j.resmic.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Gerner-Smidt P, Tjernberg I, Ursing J. 1991. Reliability of phenotypic tests for identification of Acinetobacter species. J Clin Microbiol 29:277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wisplinghoff H, Paulus T, Lugenheim M, Stefanik D, Higgins PG, Edmond MB, Wenzel RP, Seifert H. 2012. Nosocomial bloodstream infections due to Acinetobacter baumannii, Acinetobacter pittii and Acinetobacter nosocomialis in the United States. J Infect 64:282–290. doi: 10.1016/j.jinf.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Jawad A, Seifert H, Snelling AM, Heritage J, Hawkey PM. 1998. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J Clin Microbiol 36:1938–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergogne-Berezin E, Towner KJ. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev 9:148–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antunes LC, Visca P, Towner KJ. 2014. Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis 71:292–301. doi: 10.1111/2049-632X.12125. [DOI] [PubMed] [Google Scholar]
- 8.Fournier PE, Vallenet D, Barbe V, Audic S, Ogata H, Poirel L, Richet H, Robert C, Mangenot S, Abergel C, Nordmann P, Weissenbach J, Raoult D, Claverie JM. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet 2:e7. doi: 10.1371/journal.pgen.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, Jamison JJ, MacDonald IJ, Martin KM, Russo T, Campagnari AA, Hujer AM, Bonomo RA, Gill SR. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J Bacteriol 190:8053–8064. doi: 10.1128/JB.00834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adams MD, Chan ER, Molyneaux ND, Bonomo RA. 2010. Genomewide analysis of divergence of antibiotic resistance determinants in closely related isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 54:3569–3577. doi: 10.1128/AAC.00057-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahl JW, Johnson JK, Harris AD, Phillippy AM, Hsiao WW, Thom KA, Rasko DA. 2011. Genomic comparison of multi-drug resistant invasive and colonizing Acinetobacter baumannii isolated from diverse human body sites reveals genomic plasticity. BMC Genomics 12:291. doi: 10.1186/1471-2164-12-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan SY, Chua SL, Liu Y, Hoiby N, Andersen LP, Givskov M, Song Z, Yang L. 2013. Comparative genomic analysis of rapid evolution of an extreme-drug-resistant Acinetobacter baumannii clone. Genome Biol Evol 5:807–818. doi: 10.1093/gbe/evt047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu F, Zhu Y, Yi Y, Lu N, Zhu B, Hu Y. 2014. Comparative genomic analysis of Acinetobacter baumannii clinical isolates reveals extensive genomic variation and diverse antibiotic resistance determinants. BMC Genomics 15:1163. doi: 10.1186/1471-2164-15-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Liu F, Zhang Y, Wang X, Zhao C, Chen H, Zhang F, Zhu B, Hu Y, Wang H. 2015. Evolution of carbapenem-resistant Acinetobacter baumannii revealed through whole-genome sequencing and comparative genomic analysis. Antimicrob Agents Chemother 59:1168–1176. doi: 10.1128/AAC.04609-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan AP, Sutton G, DePew J, Krishnakumar R, Choi Y, Huang XZ, Beck E, Harkins DM, Kim M, Lesho EP, Nikolich MP, Fouts DE. 2015. A novel method of consensus pan-chromosome assembly and large-scale comparative analysis reveal the highly flexible pan-genome of Acinetobacter baumannii. Genome Biol 16:143. doi: 10.1186/s13059-015-0701-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roca I, Espinal P, Vila-Farres X, Vila J. 2012. The Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan-drug-resistant menace. Front Microbiol 3:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamidian M, Holt KE, Pickard D, Dougan G, Hall RM. 2014. A GC1 Acinetobacter baumannii isolate carrying AbaR3 and the aminoglycoside resistance transposon TnaphA6 in a conjugative plasmid. J Antimicrob Chemother 69:955–958. doi: 10.1093/jac/dkt454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Post V, White PA, Hall RM. 2010. Evolution of AbaR-type genomic resistance islands in multiply antibiotic-resistant Acinetobacter baumannii. J Antimicrob Chemother 65:1162–1170. doi: 10.1093/jac/dkq095. [DOI] [PubMed] [Google Scholar]
- 19.Shaikh F, Spence RP, Levi K, Ou HY, Deng Z, Towner KJ, Rajakumar K. 2009. ATPase genes of diverse multidrug-resistant Acinetobacter baumannii isolates frequently harbour integrated DNA. J Antimicrob Chemother 63:260–264. [DOI] [PubMed] [Google Scholar]
- 20.Ramirez MS, Vilacoba E, Stietz MS, Merkier AK, Jeric P, Limansky AS, Marquez C, Bello H, Catalano M, Centron D. 2013. Spreading of AbaR-type genomic islands in multidrug resistance Acinetobacter baumannii strains belonging to different clonal complexes. Curr Microbiol 67:9–14. doi: 10.1007/s00284-013-0326-5. [DOI] [PubMed] [Google Scholar]
- 21.Post V, Hall RM. 2009. AbaR5, a large multiple-antibiotic resistance region found in Acinetobacter baumannii. Antimicrob Agents Chemother 53:2667–2671. doi: 10.1128/AAC.01407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livermore DM. 2002. The impact of carbapenemases on antimicrobial development and therapy. Curr Opin Investig Drugs 3:218–224. [PubMed] [Google Scholar]
- 23.Poirel L, Nordmann P. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect 12:826–836. doi: 10.1111/j.1469-0691.2006.01456.x. [DOI] [PubMed] [Google Scholar]
- 24.Brown S, Young HK, Amyes SG. 2005. Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin Microbiol Infect 11:15–23. doi: 10.1111/j.1469-0691.2004.01016.x. [DOI] [PubMed] [Google Scholar]
- 25.Smith CA, Antunes NT, Stewart NK, Frase H, Toth M, Kantardjieff KA, Vakulenko S. 2015. Structural basis for enhancement of carbapenemase activity in the OXA-51 family of class D beta-lactamases. ACS Chem Biol 10:1791–1796. doi: 10.1021/acschembio.5b00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell JM, Leonard DA. 2014. Common clinical substitutions enhance the carbapenemase activity of OXA-51-like class D beta-lactamases from Acinetobacter spp. Antimicrob Agents Chemother 58:7015–7016. doi: 10.1128/AAC.03651-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zarrilli R, Di Popolo A, Bagattini M, Giannouli M, Martino D, Barchitta M, Quattrocchi A, Iula VD, de Luca C, Scarcella A, Triassi M, Agodi A. 2012. Clonal spread and patient risk factors for acquisition of extensively drug-resistant Acinetobacter baumannii in a neonatal intensive care unit in Italy. J Hosp Infect 82:260–265. doi: 10.1016/j.jhin.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Qiao F, Yu R, Gao Y, Zong Z. 2013. Clonal diversity of Acinetobacter baumannii clinical isolates revealed by a snapshot study. BMC Microbiol 13:234. doi: 10.1186/1471-2180-13-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stietz MS, Ramirez MS, Vilacoba E, Merkier AK, Limansky AS, Centron D, Catalano M. 2013. Acinetobacter baumannii extensively drug resistant lineages in Buenos Aires hospitals differ from the international clones I-III. Infect Genet Evol 14:294–301. doi: 10.1016/j.meegid.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 30.Wisplinghoff H, Hippler C, Bartual SG, Haefs C, Stefanik D, Higgins PG, Seifert H. 2008. Molecular epidemiology of clinical Acinetobacter baumannii and Acinetobacter genomic species 13TU isolates using a multilocus sequencing typing scheme. Clin Microbiol Infect 14:708–715. doi: 10.1111/j.1469-0691.2008.02010.x. [DOI] [PubMed] [Google Scholar]
- 31.Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodriguez-Valera F. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol 43:4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. 2010. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. doi: 10.1371/journal.pone.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gogou V, Pournaras S, Giannouli M, Voulgari E, Piperaki ET, Zarrilli R, Tsakris A. 2011. Evolution of multidrug-resistant Acinetobacter baumannii clonal lineages: a 10 year study in Greece (2000-09). J Antimicrob Chemother 66:2767–2772. doi: 10.1093/jac/dkr390. [DOI] [PubMed] [Google Scholar]
- 34.Villalon P, Valdezate S, Medina-Pascual MJ, Rubio V, Vindel A, Saez-Nieto JA. 2011. Clonal diversity of nosocomial epidemic Acinetobacter baumannii strains isolated in Spain. J Clin Microbiol 49:875–882. doi: 10.1128/JCM.01026-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villalon P, Valdezate S, Cabezas T, Ortega M, Garrido N, Vindel A, Medina-Pascual MJ, Saez-Nieto JA. 2015. Endemic and epidemic Acinetobacter baumannii clones: a twelve-year study in a tertiary care hospital. BMC Microbiol 15:47. doi: 10.1186/s12866-015-0383-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams-Haduch JM, Onuoha EO, Bogdanovich T, Tian GB, Marschall J, Urban CM, Spellberg BJ, Rhee D, Halstead DC, Pasculle AW, Doi Y. 2011. Molecular epidemiology of carbapenem-nonsusceptible Acinetobacter baumannii in the United States. J Clin Microbiol 49:3849–3854. doi: 10.1128/JCM.00619-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D'Arezzo S, Capone A, Petrosillo N, Visca P, Grab Ballardini M, Bartolini S, Bordi E, Di Stefano A, Galie M, Minniti R, Meledandri M, Pacciani L, Parisi G, Prignano G, Santini C, Valmarin M, Venditti M, Ziantoni S. 2009. Epidemic multidrug-resistant Acinetobacter baumannii related to European clonal types I and II in Rome (Italy). Clin Microbiol Infect 15:347–357. doi: 10.1111/j.1469-0691.2009.02668.x. [DOI] [PubMed] [Google Scholar]
- 38.Zarrilli R, Pournaras S, Giannouli M, Tsakris A. 2013. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents 41:11–19. doi: 10.1016/j.ijantimicag.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 39.Sahl JW, Gillece JD, Schupp JM, Waddell VG, Driebe EM, Engelthaler DM, Keim P. 2013. Evolution of a pathogen: a comparative genomics analysis identifies a genetic pathway to pathogenesis in Acinetobacter. PLoS One 8:e54287. doi: 10.1371/journal.pone.0054287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ou HY, Kuang SN, He X, Molgora BM, Ewing PJ, Deng Z, Osby M, Chen W, Xu HH. 2015. Complete genome sequence of hypervirulent and outbreak-associated Acinetobacter baumannii strain LAC-4: epidemiology, resistance genetic determinants and potential virulence factors. Sci Rep 5:8643. doi: 10.1038/srep08643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sahl JW, Beckstrom-Sternberg SM, Babic-Sternberg J, Gillece JD, Hepp CM, Auerbach RK, Tembe W, Wagner DM, Keim PS, Pearson T. The In Silico Genotyper (ISG): an open-source pipeline to rapidly identify and annotate nucleotide variants for comparative genomics applications. bioRxiv doi: 10.1101/015578. [DOI] [Google Scholar]
- 42.Sahl JW, Caporaso JG, Rasko DA, Keim P. 2014. The large-scale blast score ratio (LS-BSR) pipeline: a method to rapidly compare genetic content between bacterial genomes. PeerJ 2:e332. doi: 10.7717/peerj.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harris AD, Johnson JK, Thom KA, Morgan DJ, McGregor JC, Ajao AO, Moore AC, Comer AC, Furuno JP. 2011. Risk factors for development of intestinal colonization with imipenem-resistant Pseudomonas aeruginosa in the intensive care unit setting. Infect Control Hosp Epidemiol 32:719–722. doi: 10.1086/660763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson JK, Smith G, Lee MS, Venezia RA, Stine OC, Nataro JP, Hsiao W, Harris AD. 2009. The role of patient-to-patient transmission in the acquisition of imipenem-resistant Pseudomonas aeruginosa colonization in the intensive care unit. J Infect Dis 200:900–905. doi: 10.1086/605408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing: eighteenth informational supplement M100-S18. CLSI, Wayne, PA. [Google Scholar]
- 46.Sahl JW, Lloyd AL, Redman JC, Cebula TA, Wood DP, Mobley HL, Rasko DA. 2011. Genomic characterization of asymptomatic Escherichia coli isolated from the neobladder. Microbiology 157:1088–1102. doi: 10.1099/mic.0.043018-0. [DOI] [PubMed] [Google Scholar]
- 47.Zimin AV, Marcais G, Puiu D, Roberts M, Salzberg SL, Yorke JA. 2013. The MaSuRCA genome assembler. Bioinformatics 29:2669–2677. doi: 10.1093/bioinformatics/btt476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liou ML, Liu CC, Lu CW, Hsieh MF, Chang KC, Kuo HY, Lee CC, Chang CT, Yang CY, Tang CY. 2012. Genome sequence of Acinetobacter baumannii TYTH-1. J Bacteriol 194:6974. doi: 10.1128/JB.01860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 51.Eisen MB, Spellman PT, Brown PO, Botstein D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kunin V, Raes J, Harris JK, Spear JR, Walker JJ, Ivanova N, von Mering C, Bebout BM, Pace NR, Bork P, Hugenholtz P. 2008. Millimeter-scale genetic gradients and community-level molecular convergence in a hypersaline microbial mat. Mol Syst Biol 4:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 54.Seifert H, Dijkshoorn L, Gerner-Smidt P, Pelzer N, Tjernberg I, Vaneechoutte M. 1997. Distribution of Acinetobacter species on human skin: comparison of phenotypic and genotypic identification methods. J Clin Microbiol 35:2819–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Touchon M, Cury J, Yoon EJ, Krizova L, Cerqueira GC, Murphy C, Feldgarden M, Wortman J, Clermont D, Lambert T, Grillot-Courvalin C, Nemec A, Courvalin P, Rocha EP. 2014. The genomic diversification of the whole Acinetobacter genus: origins, mechanisms, and consequences. Genome Biol Evol 6:2866–2882. doi: 10.1093/gbe/evu225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hamouda A, Evans BA, Towner KJ, Amyes SG. 2010. Characterization of epidemiologically unrelated Acinetobacter baumannii isolates from four continents by use of multilocus sequence typing, pulsed-field gel electrophoresis, and sequence-based typing of bla(OXA-51-like) genes. J Clin Microbiol 48:2476–2483. doi: 10.1128/JCM.02431-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomaras AP, Dorsey CW, Edelmann RE, Actis LA. 2003. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149:3473–3484. doi: 10.1099/mic.0.26541-0. [DOI] [PubMed] [Google Scholar]
- 58.Hu D, Liu B, Dijkshoorn L, Wang L, Reeves PR. 2013. Diversity in the major polysaccharide antigen of Acinetobacter baumannii assessed by DNA sequencing, and development of a molecular serotyping scheme. PLoS One 8:e70329. doi: 10.1371/journal.pone.0070329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karah N, Samuelsen O, Zarrilli R, Sahl JW, Wai SN, Uhlin BE. 2015. CRISPR-cas subtype I-Fb in Acinetobacter baumannii: evolution and utilization for strain subtyping. PLoS One 10:e0118205. doi: 10.1371/journal.pone.0118205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koskiniemi S, Lamoureux JG, Nikolakakis KC, t'Kint de Roodenbeke C, Kaplan MD, Low DA, Hayes CS. 2013. Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci U S A 110:7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weber BS, Miyata ST, Iwashkiw JA, Mortensen BL, Skaar EP, Pukatzki S, Feldman MF. 2013. Genomic and functional analysis of the type VI secretion system in Acinetobacter. PLoS One 8:e55142. doi: 10.1371/journal.pone.0055142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Donald HM, Scaife W, Amyes SG, Young HK. 2000. Sequence analysis of ARI-1, a novel OXA beta-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob Agents Chemother 44:196–199. doi: 10.1128/AAC.44.1.196-199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gaddy JA, Tomaras AP, Actis LA. 2009. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect Immun 77:3150–3160. doi: 10.1128/IAI.00096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Choi AH, Slamti L, Avci FY, Pier GB, Maira-Litran T. 2009. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-beta-1-6-N-acetylglucosamine, which is critical for biofilm formation. J Bacteriol 191:5953–5963. doi: 10.1128/JB.00647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tomaras AP, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA. 2008. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology 154:3398–3409. doi: 10.1099/mic.0.2008/019471-0. [DOI] [PubMed] [Google Scholar]
- 66.Russo TA, Luke NR, Beanan JM, Olson R, Sauberan SL, MacDonald U, Schultz LW, Umland TC, Campagnari AA. 2010. The K1 capsular polysaccharide of Acinetobacter baumannii strain 307-0294 is a major virulence factor. Infect Immun 78:3993–4000. doi: 10.1128/IAI.00366-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jacobs AC, Hood I, Boyd KL, Olson PD, Morrison JM, Carson S, Sayood K, Iwen PC, Skaar EP, Dunman PM. 2010. Inactivation of phospholipase D diminishes Acinetobacter baumannii pathogenesis. Infect Immun 78:1952–1962. doi: 10.1128/IAI.00889-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Penwell WF, Arivett BA, Actis LA. 2012. The Acinetobacter baumannii entA gene located outside the acinetobactin cluster is critical for siderophore production, iron acquisition and virulence. PLoS One 7:e36493. doi: 10.1371/journal.pone.0036493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zimbler DL, Park TM, Arivett BA, Penwell WF, Greer SM, Woodruff TM, Tierney DL, Actis LA. 2012. Stress response and virulence functions of the Acinetobacter baumannii NfuA Fe-S scaffold protein. J Bacteriol 194:2884–2893. doi: 10.1128/JB.00213-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Imperi F, Antunes LC, Blom J, Villa L, Iacono M, Visca P, Carattoli A. 2011. The genomics of Acinetobacter baumannii: insights into genome plasticity, antimicrobial resistance and pathogenicity. IUBMB life 63:1068–1074. doi: 10.1002/iub.531. [DOI] [PubMed] [Google Scholar]
- 71.Peleg AY, de Breij A, Adams MD, Cerqueira GM, Mocali S, Galardini M, Nibbering PH, Earl AM, Ward DV, Paterson DL, Seifert H, Dijkshoorn L. 2012. The success of acinetobacter species; genetic, metabolic and virulence attributes. PLoS One 7:e46984. doi: 10.1371/journal.pone.0046984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eijkelkamp BA, Stroeher UH, Hassan KA, Paulsen IT, Brown MH. 2014. Comparative analysis of surface-exposed virulence factors of Acinetobacter baumannii. BMC Genomics 15:1020. doi: 10.1186/1471-2164-15-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.