Abstract
Dysembryoplastic neuroepithelial tumor (DNET) is a benign brain tumor associated with intractable drug-resistant epilepsy. In order to identify underlying genetic alterations and molecular mechanisms, we examined three family members affected by multinodular DNETs as well as 100 sporadic tumors from 96 patients, which had been referred to us as DNETs. We performed whole-exome sequencing on 46 tumors and targeted sequencing for hotspot FGFR1 mutations and BRAF p.V600E was used on the remaining samples. FISH, copy number variation assays and Sanger sequencing were used to validate the findings. By whole exome sequencing of the familial cases, we identified a novel germline FGFR1 mutation, p.R661P. Somatic activating FGFR1 mutations (p.N546K or p.K656E) were observed in the tumor samples and further evidence for functional relevance was obtained by in silico modelling. The FGFR1 p.K656E mutation was confirmed to be in cis with the germline p.R661P variant. In 43 sporadic cases, in which the diagnosis of DNET could be confirmed on central blinded neuropathology review, FGFR1 alterations were also frequent and mainly comprised intragenic tyrosine kinase FGFR1 duplication and multiple mutants in cis (25/43; 58.1%) while BRAF p.V600E alterations were absent (0/43). In contrast, in 53 cases, in which the diagnosis of DNET was not confirmed, FGFR1 alterations were less common (10/53; 19%; p<0.0001) and hotspot BRAF p.V600E (12/53; 22.6%) (p<0.001) prevailed. We observed overexpression of phospho-ERK in FGFR1 p.R661P and p.N546K mutant expressing HEK293 cells as well as FGFR1 mutated tumor samples, supporting enhanced MAP kinase pathway activation under these conditions. In conclusion, constitutional and somatic FGFR1 alterations and MAP kinase pathway activation are key events in the pathogenesis of DNET. These findings point the way towards existing targeted therapies.
Keywords: FGFR signalling, brain development, molecular etiology, whole exome sequencing, epilepsy, targeted therapy
INTRODUCTION
Dysembryoplastic neuroepithelial tumor (DNET) is an epileptogenic histopathologically benign brain tumor (World Health Organisation (WHO) grade I) [6]. These neoplasms, which cause early-onset intractable epilepsy, may be associated with Focal Cortical Dysplasia type IIIb and only rarely progress to malignancy [3]. Their often drug-resistance character leaves surgery as the only effective treatment when seizures develop in the patient.
The histopathological hallmark of DNET is the specific glioneuronal element, i.e. the presence of columns of oligodendroglial-like cells and so-called floating neurons [6]. The concept of “non-specific” DNET proposed by some authors (i.e. tumors showing clinical and imaging similarities with DNET but lacking the specific glioneuronal element) is controversial [6].
The genetic basis of DNETs remains unsettled. Studies have proposed over-activation of the mTOR pathway [3] and NF1 aberrations [2] as possible causes of DNET, and BRAF p.V600E mutations have been described in up to a 30% of DNETs [4,22]. Other molecular alterations reported include a single mutation in the promoter of TERT [18], 3 IDH1 mutations [34] and a single case report of a FGFR1 intragenic duplication of the tyrosine kinase domain [42]. Gains of chromosomes 5 and 7, LOH of 1p/19q and LOH of 10q have been observed [3,29]. Although sporadic DNETs are frequent, multifocal cases and familial forms and are extremely rare, with only two families reported to date [15,32]. The systematic review of existing data highlighted the need of a multifaceted approach to more precisely characterize the molecular nature of DNET. Our approach to understanding DNET was a multicenter, international effort, starting with a three-member family with DNET, followed by a series of 100 sporadic cases submitted to us as DNET.
METHODS
Patients and samples
The study was approved by the Institutional Review Board (IRB) of the Faculty of Medicine of McGill University. Participants were recruited in compliance with the second edition of the Canadian Tri-Council Policy Statement of Ethical Conduct for Research Involving Humans and Eligible Persons or Designates and signed a consent form in accordance with the IRB approvals. Blood from three affected members from the index family and three formalin fixed paraffin embedded (FFPE) blocks (two primary tumors, one from each child plus a recurrence from the daughter) were collected. The sporadic series is composed of a total of 100 cases (29 fresh frozen tumor (FFT) samples and 71 FFPE samples). Samples from 96 persons were recruited under the diagnosis of DNET from the reference centers; age of diagnoses, sex of the patient and location of the tumor was collected with the samples. Recurrence information was also collected when available. Formalin-fixed paraffin-embedded tumor samples from all patients were independently reviewed according to 2007 WHO criteria by three senior neuropathologists (S.A., M.H., W.P). In line with the WHO classification, only tumors containing the specific glioneuronal element were diagnosed as DNET. Because in the current WHO classification, the concept of “non-specific” DNET (i.e. tumors showing clinical and imaging similarities with DNET but lacking the specific glioneuronal element) is controversial, this diagnosis was not made. Three different groups were identified: 1- DNET cases meeting WHO criteria; 2- Non DNET with a differential diagnosis; 3- Unclear cases in which some elements of DNET were present but no glioneuronal element was encountered, or there was disagreement between the reference pathologists and a more definite diagnosis would require an extensive immunohistochemical and or molecular work-up, for which no material was available. (Detailed in Online Resource 1, Supp methods and Online Resource 3, Supp table 1). To be conservative and for statistical purposes, these unclear cases were considered as non-DNETs, from now on we will refer to them as part of the non-DNET cases.
The index family
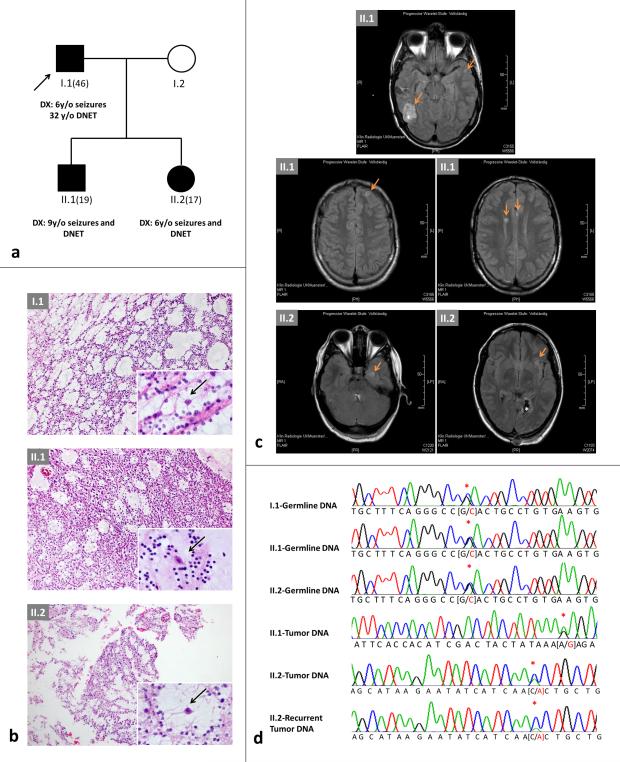
The present study describes a kindred with familial DNETs (Figure 1a-c). In the 46-year old father, focal seizures with eye deviation to the right heralded the diagnosis of a tumor of the left occipital cortex at age six years. After resection, the patient experienced no tumor recurrence and remained seizure-free. His daughter suffered focal seizures with eye deviation at age six years: a non-contrast-enhancing mass in the left occipital cortex was detected (Figure 1b+c). Following tumor excision, she remained seizure-free until age nine, at which time a local recurrence was completely resected. At age 16 years, seizures with visual aura denoted multinodular relapses. After repeat surgery, the patient is seizure-free on lamotrigine. In an update to the original report [15] a DNET in the right temporal cortex was also detected in the proband's son, following onset of focal seizures. Seven years later, evolution to bilateral convulsive seizures led to the finding of multiple lesions in the right posterior horn, the left frontal cortex and both thalami (Figure 1b+c). After surgery of the right posterior lesion, the patient remains seizure-free on lamotrigine (see online resources for further information).
Fig. 1. Family pedigree and imaging, FGFR1 mutations and modelling.
a) Pedigree of the family. Age in brackets = current age; y/o = years old; DNET = dysembryoplastic neuroepithelial tumor. b) H & E of the lesions from the pedigree in a); top: I.1; middle: II.1; bottom: II.2. Note the presence of the specific glioneuronal element (insets, higher magnification) with oligodendroglial-like cells and floating neurons (arrows) in all tumors. c) Magnetic resonance imaging. Fluid attenuated inversion recovery (FLAIR) studies of the brain performed in 2014 in II.2 at age 17 years (left) and in II.1 at age 19 years (right). Note the presence of cortical lesions (arrows) in addition to the previously resected and neuropathologically confirmed DNET (*). d) Chromatograms of germline and somatic mutations identified in the family shown in a); red asterisk denotes single basepair change. p.R661P screening was negative in the germline of I.2
DNA isolation
Germline DNA from members of the index family was isolated from blood using the PAXgene Blood DNA Kit. Tumoral DNA from fresh frozen samples (n = 29) cases was isolated with DNeasy Blood and Tissue Qiagen kits. Tumoral DNA from FFPE sporadic cases (n = 71) and family 1 FFPE samples (n = 3) was isolated with the QIAmp DNA FFPE Tissue kit (Qiagen).
Whole-exome sequencing and mutation detection
In the first instance, WES was performed in the germline DNA from the family members, tumor DNA from the primary tumor and recurrence of II.1 and tumor DNA from II.2. During the validation phase forty-three sporadic tumors from 40 persons were also studied by WES (36 primary tumors and seven recurrences, three of which are primary/recurrence pairs). WES was performed at the McGill University and Génome Québec Innovation Centre (MUGQIC) using Illumina HiSeq 2000. FFT-derived DNA (3 μg) or FFPE-derived DNA (50 ng) of each subject underwent exome capture followed by 100-bp paired-end sequencing. The Agilent SureSelect V4n was used for FFT samples library preparations, and Nextera Rapid-Capture Exome kit was used for FFPE samples, following the manufacturer's protocols. Burrows-Wheeler aligner (BWA) version 0.5.9 was used to align reads to the UCSC hg19 reference genome [20]. Indels were re-aligned using the Genome Analysis Toolkit (GATK) IndelRealigner [23]. GATK was also used to assess capture efficiency and coverage of consensus coding sequence (CCDS) bases [23]. Reads that were marked as PCR duplicates by Picard were excluded from further analysis (http://picard.sourceforge.net/). Variants were called using SAMtools version 1.2 mpileup, and only variants that represented >5% of reads were retained, with a minimum of 5 reads per variant [19]. All variants were then annotated with ANNOVAR, and those most likely to damage the protein (nonsense, canonical splice-site, missense mutations and coding indels) were considered for further analysis [39]. Because matched normal tissues were not available, we applied the following strategies to narrow down the list of variants that could potentially represent somatic mutations: only variants that were not reported in our in-house, non-cancer exome database (with >1,400 exomes), the 1000 Genomes Project database and the National Heart, Lung, and Blood Institute (NHLBI) Grand Opportunity (GO) Exome Sequencing Project (ESP) were retained. The Integrative Genomics Viewer (IGV) was used for the manual examination and visualization of all potential candidate variants [31]. In order to identify genes that carried an excess of mutations in our tumor dataset (indicating their potential functional significance), we used a case–control approach described earlier in Fontebasso et al [10]. Briefly, we compared the frequency of private mutations in each gene in the 40 individual DNET tumor exomes to 1092 control germline exomes from non-cancer patients sequenced at the McGill University and Genome Quebec Innovation Centre. We controlled for false discovery rate using the Benjamini–Hochberg procedure. The most significant genes are listed in Online Resource 4.
Sanger and targeted re-sequencing
FGFR1 (NM_023110) and BRAF p.V600E (NM_004333) mutations were screened for using the Fluidigm access array system and next-generation sequencing for 60 cases, 4 of which were previously study by WES and were used as internal controls. Fluidigm access array system involves an array-based PCR amplification of a specific region of interest (target enrichment). For our purpose, the exons and exon–intron boundaries of FGFR1 (exon 12 and 14) and BRAF (exon 15) were selectively targeted. Parallel amplification of 60 samples was carried out using custom primers designed using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/) and to which CS1 and CS2 tails were added. Samples were barcoded during the targeted enrichment to allow for multiplexed sequencing and amplicons were tagged with adaptor sequences during the PCR amplification reaction. Next-generation sequencing was carried out using the Illumina MiSeq at the MUGQIC. In the samples with potential variants, we then targeting the regions of interest in two independent regular PCR reactions followed by MiSeq sequencing. For mutation detection, we called those variants with allelic-faction larger than 5% after filtering out low base quality (< 30) or mapping quality (< 20) reads, or if the variant was observed in more than one run. Finally, Sanger sequencing was performed to study exons 12 and 14 of FGFR1 and BRAF p.V600E to further validate findings from NGS. Primers and conditions are available upon request.
Structural variation analysis
Tandem duplication (TD) in FGFR1 and FGFR1-TACC1 fusion were screened for by inspecting abnormally mapped read-pairs near previously reported TD region. Since intron 17 and exon 18 of FGFR1, which harbor all of the reported fusion and TD events respectively, are covered at a mean depth of approximately 100X in our exome data, we were confident in our ability to detect those events. At least two pairs of abnormally mapped reads were required to call an event. Then, Pindel version 0.2.5 was used to predict the exact breakpoints in samples with potential TD [40]. Samples with potential FGFR1-TACC1 fusion were first re-aligned using STAR version 2.4.1 [7]. Then, we blatted soft-clipped reads nearby the fusion region, in order to predict the exact breakpoints. Finally, XHMM was used to identify and verify any duplication nearby the FGFR1 region [11].
In 46 samples (3 tumors from the family, 40 sporadic distinct cases and 3 recurrences) we looked for genomic CNV. In brief, ExomeAI [26] detects AI (Allelic Imbalance) events across samples by investigating the B Allele Frequency (BAF) profile of exomes. In order to do this, heterozygous variants (BAF values of 0.05 to 0.95) are first extracted and then segmented to regions of similar values. AI segments are called and then summarized across all samples to detect regions of recurrent AIs. In samples with low quality and/or quantity (e.g. FFPE tumors), AI-based methods produce more reliable calls, as inadequate genome-wide coverage consistency across the tumor and normal sample can lead to false positive copy number calls [26]
Copy number variation (CNV) analysis for Tyrosine Kinase Domain duplication
Tyrosine Kinase Domain duplication and FGFR1 duplication were confirmed by a TaqMan Copy Number quantification assay Hs02882334 (Thermo Fisher). The assay was run in quadruplicates including the 13 (12 distinct patients plus one recurrence) positives cases with known TKD duplications found in the WES phase of this study. The FGFR1 duplicated case was also included. An internal quantification control was used according to the manufacturer's protocol. Results were analyzed with Applied Biosystem Copy Caller software to calculate sample copy number values by relative quantification.
Fluorescence in situ hybridization
Fluorescence in situ hybridization (FISH) analyses were performed on sections from FFPE tissues using analogous protocols to those described by Ventura et al [38]. Breakpoints affecting the FGFR1 locus were investigated with a commercially available XL FGFR1 break apart probe (MetaSystems, Altlussheim, Germany). Evaluation of FISH was conducted according to standard procedures using Zeiss fluorescence microscopes equipped with appropriate filter sets [38]. Digital image acquisition and processing were performed using ISIS digital image analysis system version 5.2.11 (MetaSystems, Altlussheim, Germany).
Cloning confirmation of cis mutations
RNA from double mutant cases II.1, 16, 33, 42 and 66 was isolated using the RecoverAll™ Total Nucleic Acid Isolation Kit for FFPE (Ambion). Reverse transcription was carried out with Superscript III (Invitrogen) followed by amplification of exons 14 and 15 of FGFR1 (primers and conditions are available upon request). Ligation into a TOPO blunt vector (Invitrogen) was performed according manufacturer recommendations and posterior transformation of OneShot TOP10 E.coli competent cells. Bacterial culture was performed overnight in LB agar with kanamycin. A minimum of 25 colonies were cultured overnight; DNA was purified with QIAamp DNA Mini Kit and followed by Sanger sequencing.
Immunohistochemistry
Phospho-ERK expression in the tumor was studied by IHC in FFPE samples. Phospho-ERK antibody CS-4376 (1:200) was optimized in a Ventana machine following recommended protocols. Phospho-ERK expression and localization was analyzed by SA independently to diagnoses and mutational analyses. BRAF-p.V600E evaluation for confirmation of positive cases was optimized in a Ventana machine with the mutation-specific anti-BRAF p.V600E-(VE1) antibody #790-4855 following the manufacturer's protocol.
In silico modelling
To assess the potential effects of the FGFR1 mutations on receptor activity, we constructed models for p.N546K, p.K656E and p.R661P based on structures of FGFR1 in an unliganded state (PDB: 1FGK) and in an activated conformation with bound peptide (PDB: 3GQI) using the program PyMOL (The PyMOL Molecular Graphics System, Version 1.7.4.1 Schrödinger, LLC.).
Expression constructs and lentivirus production
The cDNAs encoding the full-length wild type FGFR1, as well as mutant FGFR1 p.N546K and FGFR1 p.R661P were purchased from OriGene (Rockville, MD, USA). The mutated and wild type FGFR1 cDNAs were amplified and cloned into a pCR8/GW/TOPO TA entry vector and subsequently sub-cloned into a pLenti6.3/TO/V5-DEST Vector (Invitrogen) using the Gateway LR Clonase II Enzyme mix (Invitrogen) according to the manufacturer's protocol. Clones were confirmed by bidirectional Sanger sequencing. HEK293T packaging cells were transfected with pLenti6.3/V5-TOPO plasmids containing wild type and mutants FGFR1, together with ViraPower packaging mix to produce a lentiviral stock. Supernatants were collected at 48, 60 and 72 h after transfection and viral particles were concentrated using PEG-it (System Biosciences). HEK293 cells were transduced with lentiviral particles and stable clones were selected with 10 μg/ml blasticidin.
Cell culture and Phospho-ERK expression measurement
Stable cell lines were culture in presence of blasticidin selection and full growth media (DMEM + 10% FBS) until 12 hours before each experiment when selection was interrupted and each cell line was counted and plated in quadruplicate. After 12 hours in full media cells were washed twice with PBS and grown in serum starvation (0% FBS) during 6 hours. Reactivation was induced by adding fresh full media during 7 minutes. After reactivation, both reactivated and starved cells lines were washed and dislodged by using Strip Cell solution (Wisent Ink) and subsequently fixed with IC fixation buffer eBioscience (Affymetrix) for 20 minutes. Fixation/ permeabilization was then performed in methanol overnight. One million cells were stained with phospho-ERK-PE labelled antibody (#12-9109) according to eBioscience staining intracellular antigens protocol with methanol. Five micrograms of antibody per million cells was used. Samples were analyzed in duplicates by Flow Cytometry in a LSRFortessa machine. Phospho-ERK median expression level was measured in duplicates from 3 independent experiments both under serum starvation and reactivation conditions. Relative mean fluorescence intensity was measured by normalization of mean fluorescence of each mutant against the fluorescence of the WT readout from the same experiment under both starvation and reactivation conditions. To compare among mutants and wild-type overexpressing cell lines a Kruskal-Wallis one-Way ANOVA analysis was performed followed by Student-Newman-Keuls test. To measure the shift of each cell line, basal fluorescence intensity was subtracted of fluorescence intensity after stimulation in each case. An ANOVA one-way analysis followed by a Tukey's multiple comparison test was performed.
Western blotting
Cells were washed with PBS and harvested in RIPA buffer containing protease and phosphatase inhibitors. Lysates were quantified by the Bradford method using BSA as a standard. The proteins were separated on 10% SDS-PAGE and transferred onto polyvinylidene difluoride membranes. Membranes were blocked in 5% BSA for 2h at room temperature and incubated overnight at 4°C with the following primary antibodies: anti-FGFR1 (1:500, ab76464), anti-pFGFR1 (1:750, ab59194), and anti-V5 (1:1500, ab27671). Beta-tubulin (1:2000, ab6046) was used as loading control. Membranes were washed with TBST and then incubated with HRP-conjugated anti-rabbit IgG secondary antibody (1:5000, NA934V) or HRP-conjugated anti-mouse IgG secondary antibody (1:5000; NB7539) for 1h at room temperature and revealed utilizing Amersham ECL detection (Amersham Biosciences).
Senescence assays
Senescence assays were performed as described previously [27].
Statistical analysis for mutation frequency
Exact confidence intervals and tests of association were obtained with Fisher's exact test. Logistic regressions models were fit to estimate the associations between DNET versus non-DNET status and the different types of FGFR mutations.
RESULTS
Discovery whole exome sequencing
WES in all three affected individuals from the index family identified a novel germline variant in FGFR1 (c.1982G>C;p.R661P), localized in the second tyrosine kinase domain of the protein (Figure 1d + Figure 2a). The daughter's primary and recurrent tumors shared a somatic c.1638C>A;p.N546K mutation, whereas the son's tumor harboured a somatic c.1966A>G;p.K656E mutation, both known as “hotspot” mutations in FGFR1 (Figure 1d + Figure 2a). All three variants were confirmed by Sanger sequencing and c.1982G>C;p.R661P was expressed in the lymphocyte RNA of the carriers (Figure 1d and Online Resource 2, Suppl. Figure 1).
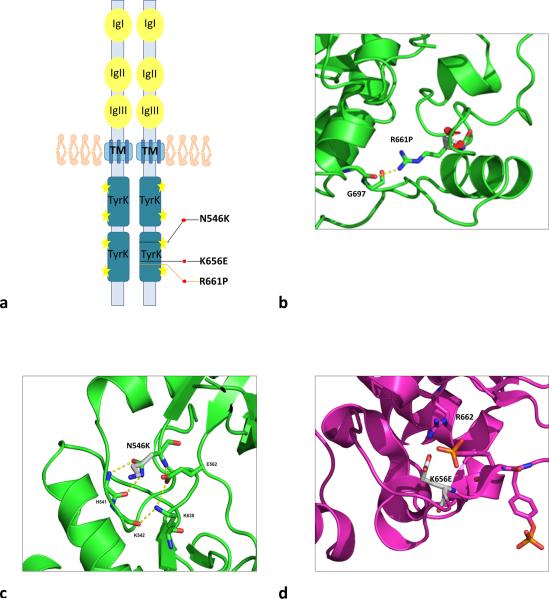
Fig. 2. In silico modelling of FGFR1 mutants found in the index family.
a) Schema of FGFR1 protein and mutations observed in the family. The protein is represented as a dimer. Black lines represent somatic mutations, orange line correspond to germline mutation. Yellow stars represent amino acid sites for phosphorylation of the receptor. Immunoglobulin-like domains (IgI, IgII and IgIII) are the ligand binding domains; TM is the transmembrane domain and TyrK are the tyrosine kinase domains. b) Structural modeling of FGFR1 p.R661P mutation. Ribbon diagram of unphosphorylated FGFR1 structure with the position of p.R661P shown, illustrating potential disruption resulting from the mutation. The R661 residue is located in the C-terminal part of the FGFR1 activation loop. In the inhibited FGFR1K structure, the side chain is extended and forms a hydrogen bond to the carbonyl oxygen of G697. In this position, the side chain blocks the binding site for substrate peptides, representing a mechanism by which it may be auto-inhibited [25]. Substitution of the arginine residue at this position with proline (p.R661P) would prevent the formation of this proposed inhibitory hydrogen bond interaction. c) Cartoon diagrams of the unphosphorylated FGFR1 structure with the position of the mutated residues shown, illustrating potential disruption resulting from the p.N546K mutation. The residue N546 is located in the kinase hinge region (the linker between β5 and αD). Notably, the equivalent residue in FGFR2 was shown to be part of a triad of residues that form a network of hydrogen bonds that dissociate in the phosphorylated structure [5]. The triad was proposed to form a molecule brake that keeps the kinase in an auto-inhibited state. The p.N546K mutation would likely disengage the brake and relax the enzyme towards its active state. d) A cartoon diagram of the activated FGFR1 structure showing how p.K656E might interact with R622 and stabilize the active conformation. K656, on the other hand, maps to the activation loop of FGFR1. In the unphosphorylated structure, the K656 side chain is pointing into the solvent but in the phosphorylated structure, it forms hydrogen bond to one of the phosphorylated tyrosine side chains (Y654), stabilizing the active conformation of the activation loop. However, alterations at the corresponding positions in FGFR2 (p.K659N) and FGFR3 (p.K650E) have been shown to be gain of function mutations. One possibility is that the Asp and Glu side chains of p.K656D/E mutants could be hydrogen bonding to the arginine residue (R622) that is adjacent to the aspartate residue that serves as the catalytic base
In silico modeling
The FGFR1 p.R661P mutation has not been previously described. It localizes within the dimerization region that precedes transphophorilation and activation of the receptor [1]. To assess the potential effects of FGFR1 mutations on receptor activity, we generated homology models for p.N546K, p.K656E and p.R661P based on structures of FGFR1 in an unliganded state (Protein Data Bank (PDB): 1FGK) and in an activated conformation with bound peptide (PDB: 3GQI). Both p.N546K and p.K656E in FGFR1 and their corresponding amino acids in the FGFR2 receptor appear to favor the enzyme in its active state and were previously described as gain-of-function (GOF) mutations [41] (Figure 2a-d). The p.R661 residue is located in the C-terminal part of the activation loop of FGFR1. In the inhibited FGFR1K structure, the side-chain is extended and forms a hydrogen bond to the carbonyl oxygen of p.G697 (Figure 2b). In this position, the side-chain blocks the binding site for substrate peptides and the p.R661P substitution would prevent the formation of this proposed inhibitory hydrogen bond interaction [25].
Central pathology review
To assess the role of mutations in FGF signalling pathway genes in the development of sporadic DNET, we collected 100 tumors from 96 persons (Table 1 and Online Resource 3) previously diagnosed as DNET at various institutions in Canada, Europe and Australia. Three neuropathologists carried out blinded central review of all specimens independently, using the current WHO diagnostic criteria,. A consensus diagnosis of DNET was reached in 43/96 cases (44.8%). The remaining cases (53/96; 55.2%) represented other entities (such as pilocytic astrocytoma) but also cases, in which the diagnosis remained unclear because material was sparse, some elements of DNET (such as nodular growth) were present but no glioneuronal element was encountered or a more definite diagnosis would have required extensive immunohistochemical and molecular work-up, for which no material was available (Online Resource 3).
Table 1.
Characteristics of the 96 cases studied showing age, sex, and location of the tumors and their distributions according to diagnosis and mutational status
| Total | DNET | DNET-like | ||
|---|---|---|---|---|
| Sex | Females | 42 (43.8%) | 19 (44.2%) | 23 (43.4%) |
| Males | 54 (56.2%) | 24 (55.8%) | 30 (56.6%) | |
| Age of diagnoses (years) | Mean Age | 14.6 | 16.7 | 12.9 |
| Mean age in mutated cases | 14.9 | 13.4 | 16.5 | |
| Mean age in cases without mutations | 14.4 | 21.1 | 10.4 | |
| Location | Temporal | 67 (70.5%) | 28 (65.1%) | 39 (75%) |
| Extra temporal | 28 (29.5%) | 15 (34.9%) | 13 (25%) | |
| Unknown | 1 | 0 | 1 | |
Molecular Investigation
Forty-three tumors from 40 persons were studied by WES. The remaining 56 samples were of insufficient quality, necessitating the use of targeted assays informed by earlier WES results. Sanger sequencing and/or immunohistochemistry was used to further confirm mutation-positive cases (Table 2, Figure 3 and Online Resource 2, Suppl. Figure 2). FISH was used to identify chromosomal breakpoints affecting FGFR1; finally all FGFR1 copy number gains were validated by a TaqMan assay. Because of limited quantity or quality for some samples, we could not complete all assays for all samples (Figure 3).
Table 2.
FGFR1 positive cases in the study and their detailed mutational status and phospho-ERK IHC results
| Case | Diagnosis | Location | Age | Sex | Tumor | Mutation | Phospho-ERK IHC |
|---|---|---|---|---|---|---|---|
| 3 | DNET | Midline (septum pellucidum) | 27 | male | Primary tumor | FGFR1 full gene duplication | Nuclear |
| 4 | DNET | R, Temporo-frontal | 21 | male | Primary tumor | p.K656D | Nuclear & cytoplasmic |
| 5 | DNET | R, Temporal | 38 | female | Primary tumor | TKD | Nuclear |
| 6 | DNET | R, Temporo-mesial | 42 | male | Primary tumor | p.K656E | Nuclear |
| 7 | DNET | R, Temporo-parietal | 26 | male | Primary tumor | p.N546K | Nuclear (<5% cytoplasmic) |
| 8 | DNET | R, Temporo-mesial | 43 | female | Recurrence | TKD | Nuclear & cytoplasmic |
| 9 | DNET | R, Parietal | 18 | male | Primary tumor | p.K656E+p.K655V | Nuclear |
| 10 | DNET | R, Parietal | 12 | male | Primary tumor | p.K656E+p.E633A+p.D652G+p.A640A | Nuclear & cytoplasmic |
| 11 | DNET | R, Parietal | 12 | male | Recurrence of case 10 | p.K656E+p.E633A+p.D652G+p.A640A | Nuclear |
| 14 | DNET | L, Temporal | 8 | male | Primary tumor | TKD | Nuclear & cytoplasmic |
| 15 | DNET | R, Occipital | 14 | male | Primary tumor | TKD | Nuclear & cytoplasmic |
| 16 | DNET | L, Frontal | 30 | male | Primary tumor | p.K656D | Nuclear & cytoplasmic |
| 18 | DNET | R, Frontal | 6 | male | Primary tumor | TKD | Nuclear & cytoplasmic |
| 19 | DNET | R, Frontal | 18 | female | Recurrence | FGFR1-TACC1 fusion | Nuclear |
| 20 | DNET | L, Temporal | 1mo | female | Primary tumor | FGFR1 breakpoint | Nuclear |
| 23 | DNET | L, Temporal | 13 | male | Primary tumor | FGFR1 breakpoint | Nuclear |
| 26 | DNET-like | R, Temporal | 31 | male | Primary tumor | p.K656E | Nuclear & cytoplasmic |
| 32 | DNET | L, Temporo-mesial | 12 | female | Primary tumor | p.K656E+ p.K655I | Nuclear & cytoplasmic |
| 33 | DNET | L, Temporo-mesial | 17 | female | Recurrence of case 32 | p.K656E+ p.K655I | Nuclear & cytoplasmic |
| 36 | DNET | L, Temporo-mesial | 5 | male | Primary tumor | p.K656D | Nuclear |
| 38 | DNET | R, Temporal | 13 | male | Recurrence | p.N546K | Nuclear & cytoplasmic |
| 40 | DNET-like | N/A | 38 | female | Primary tumor | p.N546K + FGFR1 breakpoint | N/A, high background |
| 42 | DNET-like~ | L, Frontal opperculum | 20 | female | Primary tumor | p.K656E+ p.D652G | N/A, high background |
| 45 | DNET-like | L, Frontal | 3mo | female | Recurrence | TKD | Nuclear & cytoplasmic |
| 50 | DNET | R, Occipital | 1 | female | Primary tumor | TKD | Nuclear & cytoplasmic |
| 51 | DNET | L, Frontal | 4mo | female | Primary tumor | TKD | Nuclear & cytoplasmic |
| 52 | DNET | R, Temporal | 6mo | female | Primary tumor | TKD | Nuclear & cytoplasmic |
| 59 | DNET | R, Parieto-occipital | 1mo | male | Primary tumor | TKD | Nuclear & cytoplasmic |
| 60 | DNET | R, Parieto-occipital | 14 | male | Recurrence of 59 | TKD | Nuclear & cytoplasmic |
| 61 | DNET | L, Frontal | 1mo | female | Primary tumor | TKD | Nuclear & cytoplasmic |
| 62 | DNET | R, Temporo-parietal | 1mo | female | Primary tumor | TKD | Nuclear & cytoplasmic |
| 66 | DNET | R, Parieto-occipital | 1mo | male | Primary tumor | p.K656D | Nuclear & cytoplasmic |
| 68 | DNET-like | Temporal | 17 | male | Primary tumor | p.K656D | Nuclear & cytoplasmic |
| 87 | DNET-like | Temporal | 31 | female | Primary tumor | p.K656E | Nuclear & cytoplasmic |
| 97 | DNET-like | Temporal | 30 | male | Primary tumor | p.N546K | Nuclear & cytoplasmic |
| 98 | DNET-like^ | L, Medial temporal | 13 | female | Primary tumor | p.N546K | Nuclear |
| 100 | DNET-like | Posterior insula, L temporal lobe | 4 | female | Recurrence | p.N546K+ p.K656N | Nuclear & cytoplasmic |
TKD = tyrosine kinase domain
NB: p.K656D can only be achieved by a double mutation c.1966A>G +c.1968G>C.
Aberrant phospho-ERK IHC expression was considered when the protein was expressed both in nucleous and cytoplasm.
due to low tumor, a final diagnosis was not achieved. So, to be conservative, it was grouped amongst the DNET-like
sample was considered as a possible DNET but, to be conservative, was grouped amongst the DNET-like
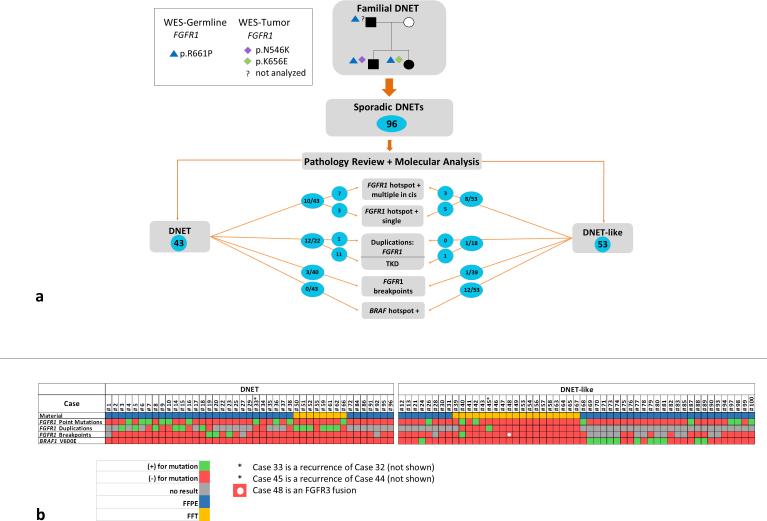
Fig. 3. Summary of results according to pathological diagnosis.
a) Shown here are the key results from the study, divided according to the outcome of the central pathology review. Only the 96 independent samples are included, representing one sample per case. Alterations found in FGFR1 and BRAF are included in the diagram. TKD = tyrosine kinase domain. b) Detailed schema of results per case and analysis performed. Only the 96 independent samples are shown, grouped by final diagnosis of DNET or non-DNET. Each column represents one case. Samples where information for a given test is not available (e.g. TKD not studied due to poor DNA quality) are shown in gray. Green indicates the presence of an alteration, red indicates no alteration. FFPE: formalin-fixed paraffin-embedded tissue; FFT: fresh frozen tissue
Of the 96 distinct samples, 18 (18.7%) had missense mutations in FGFR1 and 12 (12.5%) were BRAF c.1799T>A;p.V600E-positive. Twelve of the forty (30%) that were subjected to WES had an intragenic duplication of the tyrosine kinase domain (Online Resource 2, Suppl. Figure 3), four showed an FGFR1 breakpoint (Online Resource 2, Suppl. Figure 4), one carried a duplication of the entire FGFR1 gene and one carried a FGFR3-TACC3 fusion (Figure 3). We searched for additional candidate genes mutated in the 40 exomes; however, other than FGFR1 we found no statistically significant candidates and no known cancer genes that were mutated in more than 5% of the samples (Online Resource 4). We used ExomeAI [26] to investigate recurrent chromosomal aberrations or Allelic Imbalances across samples (n = 46). A large number of samples (60%) had few or no aberrations (less than 10 Mb AI). The most frequent aberrations were observed on chromosomes 7 and 22 (6/46; 13% for each, Online Resource 2, Suppl. Figure 5). This is consistent with a recent copy number analysis of 64 DNET samples [29], where about 50% of samples had no or few copy number variants and the most frequent aberration was amplification of chromosome 7 (23%) but lower frequency of the loss of chromosome 22 (<10%).
Multiple monoallelic FGFR1 mutants
WES analyses of the individual II. 2 in the family showed that both germline and somatic mutations in the tumor appeared in the same DNA reads, i.e. the mutations are in cis. Among the sporadic FGFR1 mutated cases, 10/18 (55.5%) had multiple FGFR1 mutations. WES analyses revealed that nine out of ten multiple mutants were in cis. We confirmed this by cloning tumor DNA from individual II.2 and cases 16, 33, 42 and 66 (Online Resource 2, Suppl. Figure 6a+b). A review of the pattern of FGFR1 mutated cases unveiled two groups: 8 of 18 were single mutants harbouring a hotspot GOF mutation (p.N546K or p.K656E), while the remaining 10 were double or quadruple mutants. Nine of these multiple mutants consisted of c.1966A>G, coding for p.K656E, plus other variations in cis (Table 2 and Online Resource 2, Suppl. Figure 6b). Multiple mutants could have occurred sequentially or because of a single stochastic event (Online Resource 2, Suppl. Figure 6c-e). The last double mutant harbours the p.N546K and p.K656N mutations and the phase could not be established given that RNA was not available for the study.
Comparison of DNET tumors and other pathologies
Clinical information regarding age of diagnosis, sex and location did not show differences between DNET and the non-DNET group (Table 1). However correlation of molecular and pathology results revealed that 25 of 43 (58.1%) DNETs carried an alteration affecting FGFR1, compared with 10 of 53 (19%) non-DNET tumors. Among 40 samples where point mutations, duplication and breakpoints of FGFR1 were assessed, logistic regression provided significant evidence of more point mutations and more duplications in DNET samples versus non-DNET samples (both p < 0.001). In contrast to previous findings [4,22], no BRAF c.1799T>A, p.V600E mutated samples were diagnosed as DNETs by blinded central review (0/43 vs 12/53, p < 0.001 , Fisher's Exact Test, exact 95% CI, 0 – 0.38, for the odds ratio for mutations in DNETs vs non-DNET samples (Figure 3).
In DNETs, the intragenic tyrosine kinase FGFR1 duplication (Online Resource 2, Suppl. Figure 3) was the most prevalent aberration (11/22, 50%), followed by multiple mutants in cis accounting for 7/43 (16.3%) of DNETs (Figure 3, Table 2 and Online Resource 2, Suppl. Figure 6). Single mutations in FGFR1 were seen in 3/43 cases. Three of forty cases possessed a chromosomal breakpoint affecting the FGFR1 locus; one of which was confirmed to be a FGFR1-TACC1 fusion (Table 2, Online Resource 2, Suppl. Figure 4). One case had a complete FGFR1 gene duplication.
In the non-DNET group, the scenario is more diverse. BRAF p.V600E is the most frequent alteration, present in 12/53 (22.6%) of cases. FGFR1 single mutants (p.N546K or p.K656E) were observed in 5/53 (9.4%) of cases, and 3/53 (5.6%) were FGFR1 double mutants in cis including one carrying the two hotspots amino acids for which phase among mutations could not be identified. Tyrosine kinase duplications were found in 1/18 cases (5.6%), a FGFR1 breakpoint was seen once and another showed a FGFR3-TACC3 fusion.
MAPK-ERK signaling
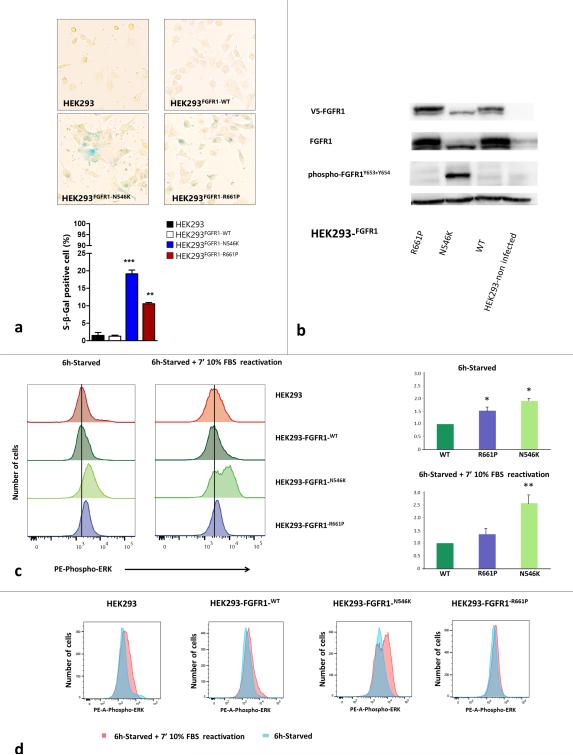
FGFR alterations are known to activate the MAPK-ERK pathway [24,17,16]. Thus we investigated the possible consequences of p.R661P and p.N546K on MAPK-ERK signaling. We transduced HEK293 cell line to stably express FGFR1-WT, FGFR1-R661P mutated and FGFR1-N546K GOF mutations. Increased signaling of the FGFR-RAS-MAPK pathways is known to induce “oncogene”-mediated senescence, which can be assessed using the β-galactosidase marker as a surrogate marker of senescent cells. β-galactosidase staining was present at high levels in p.N546K and p.R661P mutants compared to FGFR1 wild-type overexpressing cells, indicating these mutants induce increased signaling and oncogene-mediated senescence (Figure 4a). Subsequently, immunoblotting of phospho-FGFR1 revealed constitutive phosphorylation of FGFR1 in the p.N546K mutant under normal growth conditions (10% Fetal Bovine Serum, FBS media), confirming the disruption of the inhibitory state predicted by in silico modeling (Figure 4b). To further analyze the effect of FGFR1 mutants on the MAPK-ERK pathway, levels of phospho-ERK were measured under both serum starvation and subsequent serum reactivation. Under starvation conditions, p.N546K showed higher levels of phospho-ERK than cells overexpressing either FGFR1-wild type or p.R661P (both p < 0.05). The latter showed an intermediate level of phospho-ERK (Figures 4c+d and Online Resource 2, Suppl. Figure 7). Serum reactivation increased the levels of phospho-ERK expression after 7 minutes in the p.N546K mutant compared to FGFR1-WT and FGFR1-p.R661P cells (both p < 0.001). The ability of the p.R661P mutant to reactivate was not different from FGFR1-WT overexpressing cells (p = 0.57, Figure 4c+d). Immunohistochemical staining confirmed upregulated phospho-ERK in the nucleus as well as the cytoplasm in pathology sections from 24/35 (69%) of the FGFR1 mutated cases, thus confirming its down-stream repercussions in vivo.
Fig. 4. Functional studies of the p.R661P and p.N546K mutations in FGFR1 Tyrosine Kinase Domain.
a) Senescence marker β-galactosidase staining in p.R661P and p.N546K mutants compared to FGFR1-WT (wild-type) over-expressing cells and non-infected parental cell line HEK293. Graph shows the percentage of β-galactosidase positive cells in HEK293, HEK293-FGFR1-WT, HEK293-FGFR1-N546K and HEK293-FGFR1-R661P. We found a significantly higher number of senescent associated β-galactosidase positive cells (blue cells) in both mutants than in the parental and FGFR-WT overexpressing cell lines (* p < 0.05; ** p < 0.001; *** p < 0.001). Senescence rate was also higher in p.N546K than p.R661P mutants, probably due to a lower level of ERK-signaling in the p.R661P mutant (p < 0.001, one-way ANOVA followed by Tukey's multiple comparison test). b) Immunoblot showing total FGFR1 and phosphorylated FGFR1 across HEK293 parental cell line, FGFR-WT, p.N546K and p.R661P under normal growing conditions (24h growing in 10% FBS media). p.N546K mutant shows a constitutively phosphorylated FGFR1, thus confirming in silico predictions. In contrast, p.R661P mutant shows basal levels of phospho-FGFR1 similar to those observed in FGFR1-WT overexpressing cells. V5-FGFR1 = FGFR1 tagged to V5. c) ERK signalling measurement by assessing phospho-ERK levels in serum deprivation and subsequent reactivation with growth media (10% FBS) for 7 minutes. Left - histograms show phospho-ERK-PE labelled fluorescence in mutants p.R661P, p.N546K and in FGFR1-WT and HEK293 parental cell line. Black vertical line marks median phospho-ERK levels in FGFR1-WT cell line during starvation, highlighting the increased levels in both mutants compare to FGFR1-WT cells. Right - relative fluorescence for p.N546K and p.R661P was calculated from the ratio of fluorescence for each mutant and that of WT from the same experiment, normalizing FGFR1-WT fluorescence to 1.0 under both starvation and stimulation conditions. The results of two technical replicates from three experiments are shown. Error bars shows standard errors in starvation conditions and standard deviations in reactivation conditions. Considering starvation condition as basal level, p.R661P basal levels were higher than FGFR1-WT ( * = p < 0.05 Kruskall Wallis test followed by a Student-Newman-Keuls), differences that were not seen when the stimulation was applied (10% FBS for 7 mins). Under serum deprivation, p.N546K phospho-ERK expression was higher than in both p.R661P and FGFR-WT cells ( both p < 0.05). These differences were accentuated when the cells were reactivated (both p < 0.001). The Kruskall Wallis test followed by a Student-Newman-Keuls method was used in starvation and one-way ANOVA followed by a Student-Newman-Keuls method under reactivation conditions. d) Overlaying of both histograms for phospho-ERK under serum deprivation versus reactivation conditions in each of the cell lines showing the shift in the fluorescence intensity. Only p.N546K shift was significantly increased compare to WT and p.R661P (p < 0.001, one-way ANOVA followed by Tukey's multiple comparison test)
DISCUSSION
FGFR1 abnormalities underpin several developmental disorders of the brain, which can now be extended to include DNETS (Figure 5 and Online Resource 5). In DNETs, the commonest alteration was duplication of the tyrosine kinase domain, followed by two hotspot mutations in cis. Rarely, fusions or breakpoints involving FGFR1 were observed. The final common effect of these alterations appears to be a balanced level of signalling that results in benign rather than malignant tumors. Similar double mutations have been reported in Crouzon syndrome [14] but have been only rarely reported in cancer [17]. The familial germline mutation (c.1982G>C), given the minor functional effect of its resultant protein, p.R661P, (Figure 4c+d) likely acts as an enabling mutation, such that only a single additional mutation in FGFR1 is needed for tumorigenesis. This view is further substantiated by the fact that both children developed multiple DNETs, a finding rarely encountered in sporadic cases. Under this hypothesis, somatic mutations such as p.K656E must arise relatively frequently in normal tissue – a scenario which is supported by recent studies including activating FGFR mutations in the skin [21].
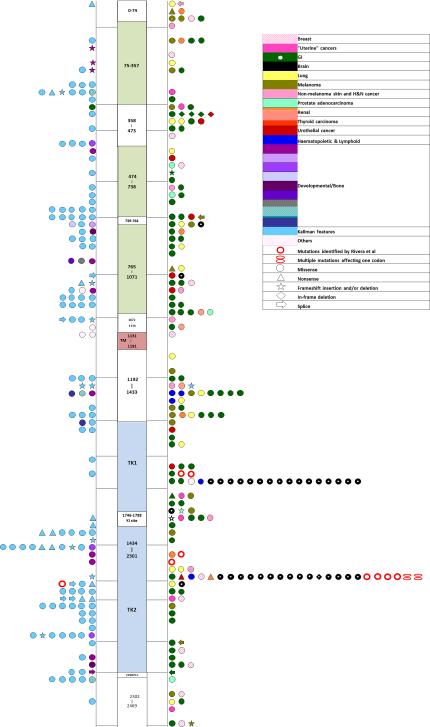
Fig 5. Genotype- phenotype association of previously described mutations in the literature and public databases.
Germline mutations are shown on the left of the figure, somatic mutations to the right. The shapes refer to the type of mutation while color code refers to associated pathology as noted in the color key. In the case of several reports of a single mutation associated to a given pathology, the mutation has been plotted only once. Due to the large number of distinct phenotypic entities, we have grouped them according to the tumor site or described congenital syndrome. Note, brain tumors cluster exclusively within the two hotspots. Online Resource 5 to this figure and lists the specific manifestations found in the literature
FGF signaling plays a ubiquitous role in development, especially in brain development and cell fate [36]. However, diversity and specificity is acquired by combination of different receptors isoforms [13] and ligands reaching a maximum level of complexity depending on cell type, cell context and signalling kinetics. Constitutive activation of the pathway leads to differentiation and senescence, whereas less pronounced signalling will drive proliferation and survival [37].
A wide range of congenital developmental disorders including achondroplasias, Kallmann and Pfeiffer syndromes are associated with inherited mutations leading to FGF receptor dysfunction [28,37]. Our compilation of FGFR1 mutations described in public databases and the literature (Figure 5, Table S4) revealed that FGFR1 somatic mutants that associate with a variety of pathologies (including epithelial malignancies) are fairly evenly distributed along the entire gene. In contrast, mutations associated with brain neoplasms (e.g. pilocytic astrocytomas [17], glioblastomas [33] and rosette forming glioneuronal tumors [12]) cluster exclusively within the GOF hotspots, thus supporting our findings. Furthermore, FGFR mutations result in GOF or loss of function (LOF) depending on the type and localization of the mutation along the gene. The difference between GOF and LOF mutations is key in this genotype-phenotype correlation and the severity of developmental abnormalities caused by defective FGF signalling seems to be dose-dependent [37].
The identification of activating mutations in FGFR1 as the genetic cause of DNETs and the implication of both BRAF and FGFR1 in non-DNET tumors highlights the implication of MAPK/ERK pathway in epileptogenic low-grade glioneuronal tumors. Our study reveals specific FGFR1 alterations as the main molecular driver of DNETs and presents inclusion of molecular data as a complement and support to classical histological categorization. This multifaceted approach responds to the need for a better classification of low-grade glioneuronal tumors, highlighting the existence of molecular differences that could underlie their epileptogenic potential. While our paper was under review, a study focusing on low-grade neuroepithelial tumors was published [30]. This study broadly supports the findings in DNETs reported here.
Our study not only supports the important role of FGFR1 in the etiology of DNET, but also highlights the diagnostic value of molecular findings in difficult or borderline cases. Here, targeted analysis for FGFR1 hotspot mutations may provide a first step to aid the diagnosis of DNET and could be supplemented by analyses for FGFR1 fusion transcripts and FGFR1-TKD. Germline analysis of FGFR1 should be considered in all familial cases, as well as in selected sporadic tumors (e.g. multinodular growth). Importantly, the presence of specific FGFR1 mutations point the way towards existing targeted therapies for children and adults with recurrent or difficult-to-excise tumors [8,9,35,36]. Such therapies could also prove effective in those tumors resistant to conventional anti-seizure treatments.
Several aspects of the clinical management of the patient with a DNET could benefit from these results, from genetic counseling to surveillance and better control of intractable seizures. Ultimately, understanding the precise molecular mechanism by which mutations in the FGF pathway give rise to this specific tumor will provide fundamental knowledge applicable to successful development of new therapies and constitutes an imperative next step in the era of precision medicine [37].
Supplementary Material
Acknowledgements
We particularly thank the family for their unfailing support of this work. We thank Nancy Hamel, Susanne Peetz-Dienhart, Yvonne Crede, Tamiko Nishimura, Christian Young, Leanne de Kock, Leora Witkowski, Greta Gillies, Simon Harvey, Wirginia Maixner, Reina Zühlke-Jenisch and Kate Pope for providing expert technical assistance. We would like to thank Pierre Lepage and Rosalie Fréchette from McGill University and Genome Quebec Innovation Centre for their help with Fluidigm experiments. BR is funded by Fundacion Alfonso Martin Escudero and the FQRNT-Merit scholarship program for foreign students (185460). TG is funded by a CIHR postdoctoral fellowship. PJL is supported by an NHMRC Australia Career Development Fellowship (APP1032364). AMB is the recipient of a Canada Research Chair in Structural Biology, NJ is the recipient of a FRQS Chaire de Recherche, JM is the recipient of a Canada Research Chair in Genomics. Tissues were partly provided from Epilepsy Society Brain and Tissue Bank (UCL, Institute of Neurology; London, United Kingdom supported though the Katy Baggott foundation). This work was partly undertaken at University College London and University College London Hospital, who receive part of their funding from the Department of Health Biomedical Centre scheme.
Funding
This project was supported by the Fonds de Recherche du Quebec-Sante (NJ, JM, WDF); IZKF Münster (Ha3/019/15) and Deutsche Krebshilfe DKH110267 to MH; National Health and Medical Research Council of Australia, the Victorian Government's Operational Infrastructure Support Program and Australian Government NHMRC IRIISS, the Murdoch Childrens Research Institute and the Campbell Edwards Trust to PL and RJL; IMF Münster and Deutsche Krebshilfe (111537) to KK; CIHR grant MOP-114889 to AMB; a National Science Centre grant no. 2014/15/B/NZ4/00744 to MZ; Infrastructure for the research on pediatric tumors is supported by KinderKrebsInitiative Buchholz/Holm-Seppensen to RS and SB; by the Wellcome Trust grant 084730 to SMS.
Footnotes
Conflict of interest: The authors declare no conflict of interest.
Ethical approval: The study was approved by the Institutional Review Board (IRB) of the Faculty of Medicine of McGill University. Participants were recruited in compliance with the second edition of the Canadian Tri-Council Policy Statement of Ethical Conduct for Research Involving Humans and Eligible Persons or Designates and signed a consent form in accordance with the IRB approvals.
Informed consent: Informed consent was obtained from all individual participants included in the study.
References
- 1.Bae JH, Boggon TJ, Tome F, Mandiyan V, Lax I, Schlessinger J. Asymmetric receptor contact is required for tyrosine autophosphorylation of fibroblast growth factor receptor in living cells. Proc Natl Acad Sci U S A. 2010;107:2866–2871. doi: 10.1073/pnas.0914157107. doi:10.1073/pnas.0914157107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barba C, Jacques T, Kahane P, Polster T, Isnard J, Leijten FS, Ozkara C, Tassi L, Giordano F, Castagna M, John A, Oz B, Salon C, Streichenberger N, Cross JH, Guerrini R. Epilepsy surgery in Neurofibromatosis Type 1. Epilepsy Res. 2013;105:384–395. doi: 10.1016/j.eplepsyres.2013.02.021. doi:10.1016/j.eplepsyres.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 3.Blümcke I, Aronica E, Urbach H, Alexopoulos A, Gonzalez-Martinez JA. A neuropathology-based approach to epilepsy surgery in brain tumors and proposal for a new terminology use for long-term epilepsy-associated brain tumors. Acta Neuropathol. 2014;128:39–54. doi: 10.1007/s00401-014-1288-9. doi:10.1007/s00401-014-1288-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chappe C, Padovani L, Scavarda D, Forest F, Nanni-Metellus I, Loundou A, Mercurio S, Fina F, Lena G, Colin C, Figarella-Branger D. Dysembryoplastic neuroepithelial tumors share with pleomorphic xanthoastrocytomas and gangliogliomas BRAF(V600E) mutation and expression. Brain Pathol. 2013;23:574–583. doi: 10.1111/bpa.12048. doi:10.1111/bpa.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H, Ma J, Li W, Eliseenkova AV, Xu C, Neubert TA, Miller WT, Mohammadi M. A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases. Mol Cell. 2007;27:717–730. doi: 10.1016/j.molcel.2007.06.028. doi:10.1016/j.molcel.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daumas-Duport C PT, Hawkins C, Shankar SK. Dysembryoplastic neuroepithelial tumors. In: Louis DNOH, Wiestler OD, Cavanee WK, editors. WHO Classification of Tumours of the Central Nervous System. 4th edn. IARC; Lyon: 2007. pp. 99–102. [Google Scholar]
- 7.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. doi:10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiskus W, Mitsiades N. B-Raf Inhibition in the Clinic: Present and Future. Annu Rev Med. 2016;67:29–43. doi: 10.1146/annurev-med-090514-030732. doi:10.1146/annurev-med-090514-030732. [DOI] [PubMed] [Google Scholar]
- 9.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O'Dwyer PJ, Lee RJ, Grippo JF, Nolop K, Chapman PB. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. doi:10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fontebasso AM, Schwartzentruber J, Khuong-Quang DA, Liu XY, Sturm D, Korshunov A, Jones DT, Witt H, Kool M, Albrecht S, Fleming A, Hadjadj D, Busche S, Lepage P, Montpetit A, Staffa A, Gerges N, Zakrzewska M, Zakrzewski K, Liberski PP, Hauser P, Garami M, Klekner A, Bognar L, Zadeh G, Faury D, Pfister SM, Jabado N, Majewski J. Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol. 2013;125:659–669. doi: 10.1007/s00401-013-1095-8. doi:10.1007/s00401-013-1095-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fromer M, Moran JL, Chambert K, Banks E, Bergen SE, Ruderfer DM, Handsaker RE, McCarroll SA, O'Donovan MC, Owen MJ, Kirov G, Sullivan PF, Hultman CM, Sklar P, Purcell SM. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. American journal of human genetics. 2012;91:597–607. doi: 10.1016/j.ajhg.2012.08.005. doi:10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gessi MM,YA, Hammes J, Goschzik T, Scholz M, Denkhaus D, Waha A, Pietsch T. FGFR1 mutations in Rosette-forming glioneuronal tumors of the fourth ventricle. J Neuropathol Exp Neurol. 2014;73:580–584. doi: 10.1097/NEN.0000000000000080. doi:10.1097/NEN.0000000000000080. [DOI] [PubMed] [Google Scholar]
- 13.Goetz R, Mohammadi M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nat Rev Mol Cell Biol. 2013;14:166–180. doi: 10.1038/nrm3528. doi:10.1038/nrm3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goriely A, McVean GA, van Pelt AM, O'Rourke AW, Wall SA, de Rooij DG, Wilkie AO. Gain-of-function amino acid substitutions drive positive selection of FGFR2 mutations in human spermatogonia. Proc Natl Acad Sci U S A. 2005;102:6051–6056. doi: 10.1073/pnas.0500267102. doi:10.1073/pnas.0500267102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasselblatt M, Kurlemann G, Rickert CH, Debus OM, Brentrup A, Schachenmayr W, Paulus W. Familial occurrence of dysembryoplastic neuroepithelial tumor. Neurology. 2004;62:1020–1021. doi: 10.1212/01.wnl.0000115266.16119.3a. [DOI] [PubMed] [Google Scholar]
- 16.Ho CY, Mobley BC, Gordish-Dressman H, VandenBussche CJ, Mason GE, Bornhorst M, Esbenshade AJ, Tehrani M, Orr BA, LaFrance DR, Devaney JM, Meltzer BW, Hofherr SE, Burger PC, Packer RJ, Rodriguez FJ. A clinicopathologic study of diencephalic pediatric low-grade gliomas with BRAF V600 mutation. Acta Neuropathol. 2015;130:575–585. doi: 10.1007/s00401-015-1467-3. doi:10.1007/s00401-015-1467-3. [DOI] [PubMed] [Google Scholar]
- 17.Jones DT, Hutter B, Jager N, Korshunov A, Kool M, Warnatz HJ, Zichner T, Lambert SR, Ryzhova M, Quang DA, Fontebasso AM, Stutz AM, Hutter S, Zuckermann M, Sturm D, Gronych J, Lasitschka B, Schmidt S, Seker-Cin H, Witt H, Sultan M, Ralser M, Northcott PA, Hovestadt V, Bender S, Pfaff E, Stark S, Faury D, Schwartzentruber J, Majewski J, Weber UD, Zapatka M, Raeder B, Schlesner M, Worth CL, Bartholomae CC, von Kalle C, Imbusch CD, Radomski S, Lawerenz C, van Sluis P, Koster J, Volckmann R, Versteeg R, Lehrach H, Monoranu C, Winkler B, Unterberg A, Herold-Mende C, Milde T, Kulozik AE, Ebinger M, Schuhmann MU, Cho YJ, Pomeroy SL, von Deimling A, Witt O, Taylor MD, Wolf S, Karajannis MA, Eberhart CG, Scheurlen W, Hasselblatt M, Ligon KL, Kieran MW, Korbel JO, Yaspo ML, Brors B, Felsberg J, Reifenberger G, Collins VP, Jabado N, Eils R, Lichter P, Pfister SM, International Cancer Genome Consortium PedBrain Tumor P Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nature genetics. 2013;45:927–932. doi: 10.1038/ng.2682. doi:10.1038/ng.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Jr., Friedman AH, Friedman H, Gallia GL, Giovanella BC, Grollman AP, He TC, He Y, Hruban RH, Jallo GI, Mandahl N, Meeker AK, Mertens F, Netto GJ, Rasheed BA, Riggins GJ, Rosenquist TA, Schiffman M, Shih Ie M, Theodorescu D, Torbenson MS, Velculescu VE, Wang TL, Wentzensen N, Wood LD, Zhang M, McLendon RE, Bigner DD, Kinzler KW, Vogelstein B, Papadopoulos N, Yan H. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. doi:10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. doi:10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. doi:10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, Wedge DC, Fullam A, Alexandrov LB, Tubio JM, Stebbings L, Menzies A, Widaa S, Stratton MR, Jones PH, Campbell PJ. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–886. doi: 10.1126/science.aaa6806. doi:10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marucci G, de Biase D, Visani M, Giulioni M, Martinoni M, Volpi L, Riguzzi P, Rubboli G, Michelucci R, Tallini G. Mutant BRAF in low-grade epilepsy-associated tumors and focal cortical dysplasia. Ann Clin Transl Neurol. 2014;1:130–134. doi: 10.1002/acn3.31. doi:10.1002/acn3.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. doi:10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McWilliams GD, SantaCruz K, Hart B, Clericuzio C. Occurrence of DNET and other brain tumors in Noonan syndrome warrants caution with growth hormone therapy. Am J Med Genet A. 2015;9999:1–7. doi: 10.1002/ajmg.a.37379. doi:10.1002/ajmg.a.37379. [DOI] [PubMed] [Google Scholar]
- 25.Mohammadi M, Schlessinger J, Hubbard SR. Structure of the FGF receptor tyrosine kinase domain reveals a novel autoinhibitory mechanism. Cell. 1996;86:577–587. doi: 10.1016/s0092-8674(00)80131-2. [DOI] [PubMed] [Google Scholar]
- 26.Nadaf J, Majewski J, Fahiminiya S. ExomeAI: detection of recurrent allelic imbalance in tumors using whole-exome sequencing data. Bioinformatics. 2015;31:429–431. doi: 10.1093/bioinformatics/btu665. doi:10.1093/bioinformatics/btu665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neault M, Mallette FA, Vogel G, Michaud-Levesque J, Richard S. Ablation of PRMT6 reveals a role as a negative transcriptional regulator of the p53 tumor suppressor. Nucleic acids research. 2012;40:9513–9521. doi: 10.1093/nar/gks764. doi:10.1093/nar/gks764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Passos-Bueno MR, Wilcox WR, Jabs EW, Sertie AL, Alonso LG, Kitoh H. Clinical spectrum of fibroblast growth factor receptor mutations. Human mutation. 1999;14:115–125. doi: 10.1002/(SICI)1098-1004(1999)14:2<115::AID-HUMU3>3.0.CO;2-2. doi:10.1002/(SICI)1098-1004(1999)14:2<115::AID-HUMU3>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 29.Prabowo AS, van Thuijl HF, Scheinin I, Sie D, van Essen HF, Iyer AM, Spliet WG, Ferrier CH, van Rijen PC, Veersema TJ, Thom M, Schouten-van Meeteren AY, Reijneveld JC, Ylstra B, Wesseling P, Aronica E. Landscape of chromosomal copy number aberrations in gangliogliomas and dysembryoplastic neuroepithelial tumours. Neuropathol Appl Neurobiol. 2015 doi: 10.1111/nan.12235. doi:10.1111/nan.12235. [DOI] [PubMed] [Google Scholar]
- 30.Qaddoumi I, Orisme W, Wen J, Santiago T, Gupta K, Dalton JD, Tang B, Haupfear K, Punchihewa C, Easton J, Mulder H, Boggs K, Shao Y, Rusch M, Becksfort J, Gupta P, Wang S, Lee RP, Brat D, Peter Collins V, Dahiya S, George D, Konomos W, Kurian KM, McFadden K, Serafini LN, Nickols H, Perry A, Shurtleff S, Gajjar A, Boop FA, Klimo PD, Jr., Mardis ER, Wilson RK, Baker SJ, Zhang J, Wu G, Downing JR, Tatevossian RG, Ellison DW. Genetic alterations in uncommon low-grade neuroepithelial tumors: BRAF, FGFR1, and MYB mutations occur at high frequency and align with morphology. Acta Neuropathol. 2016 doi: 10.1007/s00401-016-1539-z. doi:10.1007/s00401-016-1539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nature biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. doi:10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saito TS,K, Yamasaki F, Tominaga A, Kurisu K, Takeshima Y, Hirose T. Familial occurrence of dysembryoplastic neuroepithelial tumor-like neoplasm of the septum pellucidum: case report. Neurosurgery. 2008;63:e370–372. doi: 10.1227/01.NEU.0000320421.82255.63. discussion e372. doi:10.1227/01.NEU.0000320421.82255.63. [DOI] [PubMed] [Google Scholar]
- 33.Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, Liu EM, Reichel J, Porrati P, Pellegatta S, Qiu K, Gao Z, Ceccarelli M, Riccardi R, Brat DJ, Guha A, Aldape K, Golfinos JG, Zagzag D, Mikkelsen T, Finocchiaro G, Lasorella A, Rabadan R, Iavarone A. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–1235. doi: 10.1126/science.1220834. doi:10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thom M, Toma A, An S, Martinian L, Hadjivassiliou G, Ratilal B, Dean A, McEvoy A, Sisodiya SM, Brandner S. One hundred and one dysembryoplastic neuroepithelial tumors: an adult epilepsy series with immunohistochemical, molecular genetic, and clinical correlations and a review of the literature. J Neuropathol Exp Neurol. 2011;70:859–878. doi: 10.1097/NEN.0b013e3182302475. doi:10.1097/NEN.0b013e3182302475. [DOI] [PubMed] [Google Scholar]
- 35.Touat M, Ileana E, Postel-Vinay S, Andre F, Soria JC. Targeting FGFR Signaling in Cancer. Clinical Cancer Research. 2015;21:2684–2694. doi: 10.1158/1078-0432.CCR-14-2329. doi:10.1158/1078-0432.CCR-14-2329. [DOI] [PubMed] [Google Scholar]
- 36.Turner CA, Eren-Kocak E, Inui EG, Watson SJ, Akil H. Dysregulated fibroblast growth factor (FGF) signaling in neurological and psychiatric disorders. Semin Cell Dev Biol. 2015 doi: 10.1016/j.semcdb.2015.10.003. 10.1016/j.semcdb.2015.10.003. doi:10.1016/j.semcdb.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turner NG,R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. doi:10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 38.Ventura RA, Martin-Subero JI, Jones M, McParland J, Gesk S, Mason DY, Siebert R. FISH analysis for the detection of lymphoma-associated chromosomal abnormalities in routine paraffin-embedded tissue. J Mol Diagn. 2006;8:141–151. doi: 10.2353/jmoldx.2006.050083. doi:10.2353/jmoldx.2006.050083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research. 2010;38:e164. doi: 10.1093/nar/gkq603. doi:10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. doi:10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoon K, Nery S, Rutlin ML, Radtke F, Fishell G, Gaiano N. Fibroblast growth factor receptor signaling promotes radial glial identity and interacts with Notch1 signaling in telencephalic progenitors. J Neurosci. 2004;24:9497–9506. doi: 10.1523/JNEUROSCI.0993-04.2004. doi:10.1523/JNEUROSCI.0993-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, Wu G, Miller CP, Tatevossian RG, Dalton JD, Tang B, Orisme W, Punchihewa C, Parker M, Qaddoumi I, Boop FA, Lu C, Kandoth C, Ding L, Lee R, Huether R, Chen X, Hedlund E, Nagahawatte P, Rusch M, Boggs K, Cheng J, Becksfort J, Ma J, Song G, Li Y, Wei L, Wang J, Shurtleff S, Easton J, Zhao D, Fulton RS, Fulton LL, Dooling DJ, Vadodaria B, Mulder HL, Tang C, Ochoa K, Mullighan CG, Gajjar A, Kriwacki R, Sheer D, Gilbertson RJ, Mardis ER, Wilson RK, Downing JR, Baker SJ, Ellison DW, St. Jude Children's Research Hospital-Washington University Pediatric Cancer Genome P Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013;45:602–612. doi: 10.1038/ng.2611. doi:10.1038/ng.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.