Study of Drosophila oogenesis reveals that the nuclear localization of actin is controlled by both development and Fascin. Fascin regulates both endogenous nuclear actin and ectopic nuclear actin rod formation by controlling Cofilin.
Abstract
Drosophila oogenesis provides a developmental system with which to study nuclear actin. During Stages 5–9, nuclear actin levels are high in the oocyte and exhibit variation within the nurse cells. Cofilin and Profilin, which regulate the nuclear import and export of actin, also localize to the nuclei. Expression of GFP-tagged Actin results in nuclear actin rod formation. These findings indicate that nuclear actin must be tightly regulated during oogenesis. One factor mediating this regulation is Fascin. Overexpression of Fascin enhances nuclear GFP-Actin rod formation, and Fascin colocalizes with the rods. Loss of Fascin reduces, whereas overexpression of Fascin increases, the frequency of nurse cells with high levels of nuclear actin, but neither alters the overall nuclear level of actin within the ovary. These data suggest that Fascin regulates the ability of specific cells to accumulate nuclear actin. Evidence indicates that Fascin positively regulates nuclear actin through Cofilin. Loss of Fascin results in decreased nuclear Cofilin. In addition, Fascin and Cofilin genetically interact, as double heterozygotes exhibit a reduction in the number of nurse cells with high nuclear actin levels. These findings are likely applicable beyond Drosophila follicle development, as the localization and functions of Fascin and the mechanisms regulating nuclear actin are widely conserved.
INTRODUCTION
Actin was first reported to be in the nucleus >40 yr ago (Lane, 1969). Initially, this finding was not widely accepted, as the level of actin in the nucleus was very low compared with that in the cytoplasm, there were concerns about the purity of isolated nuclei, nuclear filamentous actin (F-actin) could not be visualized, and functions for nuclear actin were unknown (for reviews, see Vartiainen, 2008; Viita and Vartiainen, 2016). These concerns have been addressed by recent studies, which have firmly established that actin is inside the nucleus. Such studies have identified antibodies that recognize nuclear actin (Gonsior et al., 1999; Schoenenberger et al., 2005) and elucidated the mechanisms regulating the nuclear localization of actin (Wada et al., 1998; Stuven et al., 2003; Munsie et al., 2012). Furthermore, functional studies have implicated actin in regulating transcription, chromatin remodeling, nuclear organization/structure, and DNA damage repair (Vartiainen, 2008; Visa and Percipalle, 2010; Grosse and Vartiainen, 2013; Percipalle, 2013; Belin et al., 2015; Viita and Vartiainen, 2016).
Recent studies have provided insight into the structure of nuclear actin. Fluorescence recovery after photobleaching (FRAP) and fluorescence correlation spectroscopy studies of green fluorescent protein (GFP)–tagged Actin indicate there are two pools of nuclear actin (McDonald et al., 2006). One pool has a slower turnover rate and is believed to be polymeric actin. The other pool of actin turns over faster and could be monomeric or polymeric actin that is rapidly associating and dissociating with protein complexes in the nucleus. Belin et al. (2013) generated nuclear actin probes to label both monomeric and polymeric actin. They found that monomeric nuclear actin is punctate and localizes to nuclear speckles and sites of RNA processing. Submicrometer-length polymeric actin does not localize to chromatin and likely generates a viscoelastic structure within the nucleus (Belin et al., 2013). Recent studies from the same group implicate submicrometer actin filaments in DNA damage repair and nuclear oxidation (Belin et al., 2015). Although these studies have significantly advanced our understanding of nuclear actin, much remains to be learned about its structure, including what factors, such as actin binding proteins, regulate its architecture.
Certain circumstances can result in nuclear actin filament formation (for reviews, see Grosse and Vartiainen, 2013; Hendzel, 2014). Stressors, including heat shock, treatment with dimethyl sulfoxide, and ATP depletion, can induce intranuclear actin filaments termed actin rods (Fukui and Katsumaru, 1979; Osborn and Weber, 1980; Iida et al., 1986; Nishida et al., 1987; Vartiainen et al., 2007; Munsie et al., 2012). Although nuclear actin rods have primarily been studied in cultured cell systems, they have also been observed in vivo. Nuclear actin rods are observed in Dictyostelium spores and disappear as germination occurs (Sameshima et al., 2001). Similar rods are seen in patients with actin myopathies (Goebel and Warlo, 2001; Domazetovska et al., 2007a, b). Expression of Lamin mutants in Drosophila larval muscles also results in nuclear actin rods (Dialynas et al., 2010). In addition, actin rod formation is linked to neurodegenerative diseases, including Alzheimer’s, Huntington’s, and Parkinson’s diseases (Minamide et al., 2000; Maloney et al., 2005; Lim et al., 2007; Munsie et al., 2011). These and other studies have led to the idea that actin rods form under conditions of cellular stress and function as a prosurvival mechanism. However, failure to remove the rods is toxic to cells (Maloney and Bamburg, 2007). Alterations in the actin cytoskeleton or the nuclear import/export of actin also result in nuclear actin rods (Pendleton et al., 2003; Stuven et al., 2003; Bohnsack et al., 2006; Dopie et al., 2012; Munsie et al., 2012; Sen et al., 2015). In addition, nuclear actin rods are observed in cultured cells in response to extracellular signaling induced by cell spreading (Plessner et al., 2015). As discussed earlier, DNA damage induces submicrometer nuclear actin filament formation, and these filaments play a critical role in repair (Belin et al., 2015). Together these findings suggest that nuclear actin filaments and rods have physiological functions and may occur in other in vivo contexts.
The structure and composition of nuclear actin rods remain unclear. Initially, nuclear actin rods were not believed to label with phalloidin. Such findings resulted in the idea that these rods are structurally distinct from cytoplasmic F-actin. Indeed, some nuclear actin rods have been shown to be composed of actin that is fully bound along its length by Cofilin (Nishida et al., 1987; Munsie et al., 2012); such Cofilin binding precludes phalloidin staining. Recent studies indicate that some nuclear actin rods are phalloidin positive (Rohn et al., 2011; Belin et al., 2015; Dopie et al., 2015; Plessner et al., 2015; Sen et al., 2015). It is important to note that imaging conditions necessary to visualize the phalloidin-stained nuclear actin rods require that the cytoplasmic phalloidin signal be significantly overexposed. Thus it is unclear whether there are different types of nuclear actin rods—those that are phalloidin versus Cofilin positive—or the differential staining of the structures simply reflects nuclear actin rod dynamics.
Here we present our novel finding that Drosophila oogenesis, or follicle development, is a model for studying the structure and regulation of nuclear actin. The Drosophila ovary is composed of ∼15 ovarioles—chains of sequentially maturing egg chambers or follicles. Each follicle consists of ∼1000 somatic cells termed follicle cells and 16 germline cells, including 15 nurse or support cells and one oocyte. Oogenesis is divided into 14 morphological stages, from the germarium to Stage 14 (S14). Here we primarily focus on S5–9 of development. During these stages, the follicle cells transition from being mitotic (S1–6) to endocycling (S7–9), the nurse cells transition from polytene (S1–5) to polyploid (S6 and after) and are endocycling, the follicle is growing rapidly in size, and oocyte polarity is established (reviewed in Theurkauf et al., 1992; Spradling, 1993; Dobens and Raftery, 2000; Claycomb and Orr-Weaver, 2005). We find by both subcellular fractionation and immunofluorescence studies that actin is in the nucleus during Drosophila follicle development. Specifically, during S5–9, varying levels of nuclear actin are observed in the nurse cell nuclei, and high levels are seen within the germinal vesicle (oocyte nucleus). Germline expression of GFP-tagged Actin results in nuclear actin rod formation in a percentage of the nurse cells and the germinal vesicle during these same stages. These rods are both Cofilin and phalloidin positive. We also find that manipulation of the actin binding protein Fascin, which we recently found localizes to the nucleus and nuclear periphery in addition to the cytoplasm (Groen et al., 2015; Jayo et al., 2016), alters both endogenous nuclear actin and nuclear GFP-Actin rod formation. Our data suggest that Fascin modulates nuclear actin by regulating Cofilin. Thus Drosophila oogenesis provides an in vivo, multicellular system with which to uncover new means of regulating nuclear actin.
RESULTS
Developmental regulation of nuclear actin
Previously we found that germline expression of the actin-labeling reagents GFP-Utrophin and Lifeact-GFP resulted in nuclear actin rods in the nurse cells and/or germinal vesicles during S5–9 of Drosophila oogenesis (Spracklen et al., 2014). Because these reagents can stabilize endogenous actin structures, the stage-specific formation of nuclear actin rods suggests that nuclear actin might normally play an important role during this period of follicle development.
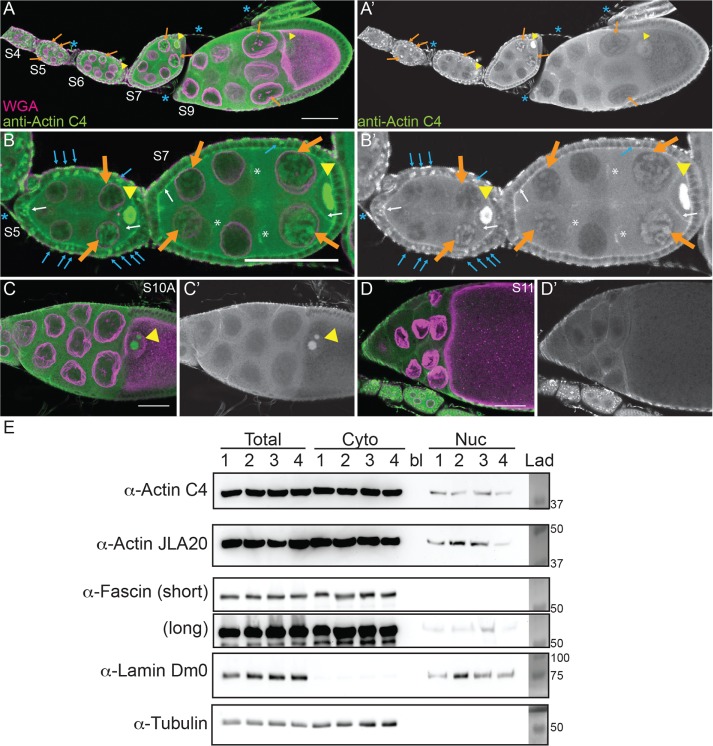
To address this possibility, we used a broad-specificity actin antibody that has been used to examine nuclear actin—anti–actin C4. This actin antibody recognizes a highly evolutionarily conserved region in actin and labels all types of vertebrate actin and actin in lower eukaryotes, including Dictyostelium and slime mold (Lessard, 1988). The actin C4 antibody has been widely used to examine nuclear actin, including during oocyte development (Parfenov et al., 1995), in Cajal bodies in multiple cell types (Gedge et al., 2005; Lenart et al., 2005; Maslova and Krasikova, 2012), in association with RNA Pol II (Hofmann et al., 2004), and during cellular senescence (Spencer et al., 2011). Immunofluorescence images reveal that actin is indeed found in nuclei during early oogenesis (Supplemental Movie S1). Nurse cells during S5–9 exhibit varying levels of nuclear actin (Figure 1, A–B′). Some nurse cells within a follicle have nuclear actin that exhibits a structured or blobby appearance, and other nurse cells within the same follicle exhibit a nuclear actin haze (Figure 1, A–B′; orange arrows indicate structured nuclear actin; see quantification in Figure 8E later in this article and Supplemental Table 1B). As follicle development proceeds (S10 and later), only unstructured or hazy nuclear actin is observed in the nurse cells (Figure 1, C–D′). Nuclear actin is also observed in a subset of the follicle cells during early oogenesis (germarium-S7), with more follicle cells being labeled in the germarium-S5 and decreasing to only a few cells during S7 (Figure 1, B and B′, blue arrows, and unpublished data). In addition, throughout oogenesis, the germinal vesicle exhibits a very high level of nuclear actin (Figure 1, A–C′, yellow arrowheads, and unpublished data). The actin C4 antibody also labels some F-actin structures, including the muscle sheath (blue asterisk in Figure 1, A–B′), follicle cell basal cortical actin and oocyte cortical actin (white arrows in Figure 1, B and B′), the ring canals connecting the nurse cells (white asterisk in Figure 1, B and B′), and in S10A and later stages, the nurse cell cortical actin (Figure 1, C–D′, and unpublished data). Supporting our immunofluorescence studies, subcellular fractionation analysis of whole ovaries indicates that a low level of actin is found in the nuclear fraction using both the actin C4 antibody and another actin antibody (Figure 1E).
FIGURE 1:
Nuclear accumulation of actin is developmentally regulated. (A–D′) Maximum projections of two to four confocal slices of wild-type follicles. (A–D) Merged images of nuclear envelope (wheat germ agglutinin, WGA) in magenta and anti–actin C4 staining in green. (A′–D′) Anti–actin C4, white. Scale bars, 50 μm. The nurse cells during S5–9 exhibit varying levels of nuclear actin (A–B′), with some exhibiting structured actin (orange arrows) and others exhibiting a low haze of nuclear actin (unmarked). The germinal vesicles have very high levels of nuclear actin (A–C′, yellow arrowheads). In addition, actin is observed in the nuclei of a subset of the follicle cells during early oogenesis (B and B′, blue arrows). The actin C4 antibody also labels some F-actin structures, including the basal cortical actin of the follicle cells and the oocyte cortical actin during early oogenesis (B and B′, white arrows), the nurse cell cortical actin later in follicle development (>S10A; C–D′), ring canals (B and B′, white asterisks), and the muscle sheath (A–B′, blue asterisk). (E) Representative Western blots of subcellular fractionation with four independent samples, labeled 1–4, of whole ovary lysates from wild-type flies (total lysate, cytoplasmic fraction, nuclear fraction) blotted for actin (actin C4 and JLA20), Fascin (two exposures), Lamin Dm0 (nuclear marker), and α-Tubulin (cytoplasmic marker). bl, blank lane; Lad, ladder with molecular weight markers labeled. Actin and Fascin are found in the nuclear fraction of wild-type ovary lysates.
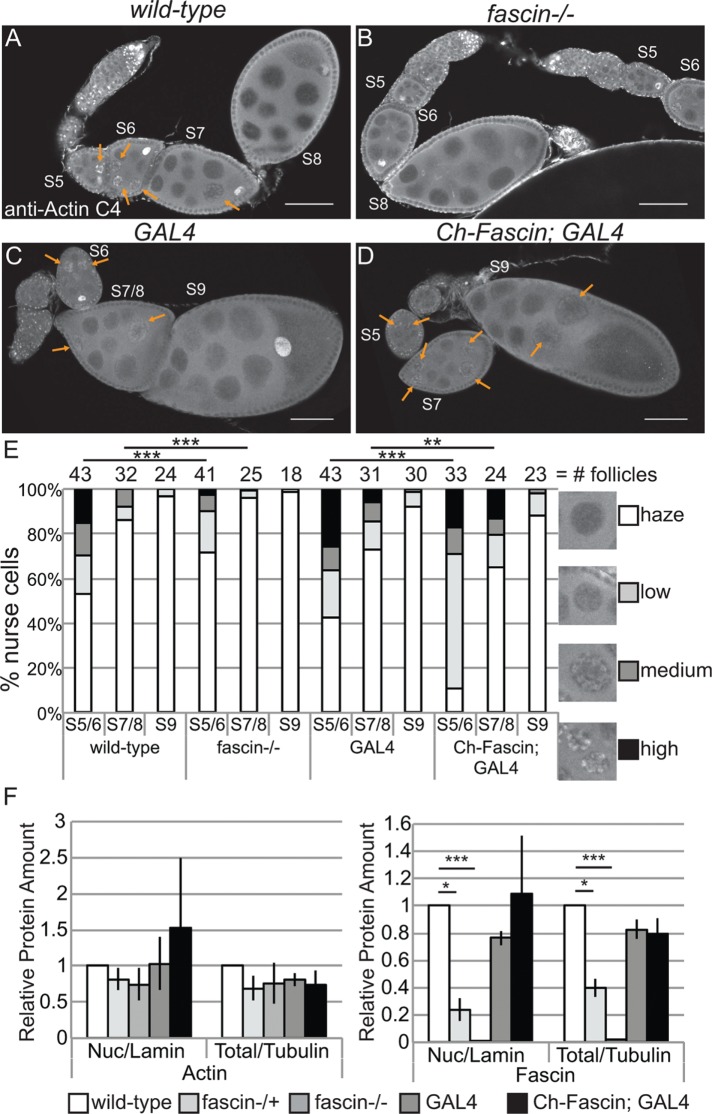
FIGURE 8:
Manipulating Fascin alters endogenous nuclear actin. (A–D) Maximum projections of two to four confocal slices of follicles of the indicated genotypes stained for endogenous nuclear actin (anti–actin C4). Scale bars, 50 μm. (E) Chart quantifying the percentage of nurse cells in S5–6, S7–8, and S9 of the indicated genotypes exhibiting unstructured or structured nuclear actin; the number of follicles (n) examined is indicated across the top of the chart. Each follicle within a confocal stack was scored, in a genotypically blinded manner, for the percentage of nurse cells exhibiting unstructured nuclear actin haze or low, medium, or high levels of structured nuclear actin. Loss of Fascin (fascinsn28/sn28) results in a low uniform haze of nuclear actin and a lack of structure compared with wild-type follicles (B vs. A, quantified in E; ***p < 0.001, Pearson’s chi-squared test), whereas overexpression of Fascin appears to increase the number of nurse cells exhibiting high levels of nuclear actin (orange arrows) compared with GAL4 alone (D vs. C, quantified in E; ***p < 0.001 and **p < 0.01, Pearson’s chi-squared test). (F) Charts quantifying the relative amount of nuclear protein (endogenous actin or endogenous Fascin) to nuclear Lamin and total protein (endogenous actin or endogenous Fascin) to total Tubulin in the indicated genotypes from Western blots of three subcellular fractionation experiments. Protein amount was normalized to wild-type. Error bars, SE. *p < 0.05 and ***p < 0.001, unpaired t test with Welch’s correction. Neither loss nor overexpression of Fascin significantly alters nuclear actin at the whole ovary level compared with their respective controls (fascin-/- to wild-type, p = 0.4; Ch-Fascin to GAL4, p = 0.7, unpaired t test with Welch’s correction).
Given that the anti–actin C4 nuclear labeling pattern is unique, we wanted to verify the specificity of the antibody. By immunoblotting, the actin C4 antibody recognizes a single band the size of actin, just like another actin antibody (JLA20; Supplemental Figure S1A). Although this finding indicates that the antibody recognizes Drosophila actin, it is possible that the antibody also recognizes something nonspecifically by immunofluorescence. To address this possibility, we used a number of approaches. First, we attempted to use other actin antibodies—2G2 (Gonsior et al., 1999) and 1C7 (Schoenenberger et al., 2005)—that have been used to examine nuclear actin in other systems. In our hands, neither of these antibodies, using multiple fixation conditions, labels nuclear structures within Drosophila follicles or recognizes Drosophila actin by immunoblot (Supplemental Figure S1, B–D, and unpublished data). This lack of labeling is not unexpected, as the antigen for 2G2 is a nonsequential region of actin found in the profilin–actin complex from rabbit skeletal muscle (Gonsior et al., 1999), whereas the antigen for 1C7 is a chemically cross-linked actin dimer, also from rabbit skeletal muscle, that is structurally similar to lower-dimer actin (Schoenenberger et al., 2005). Thus the conformation of Drosophila nuclear actin is likely sufficiently divergent to prevent labeling with these antibodies. Furthermore, these two nuclear actin antibodies label different nuclear actin structures within the same vertebrate cells (Schoenenberger et al., 2005; Asumda and Chase, 2012). These findings suggest that nuclear actin likely exists in numerous conformations or structures that are only labeled by specific reagents (Jockusch et al., 2006).
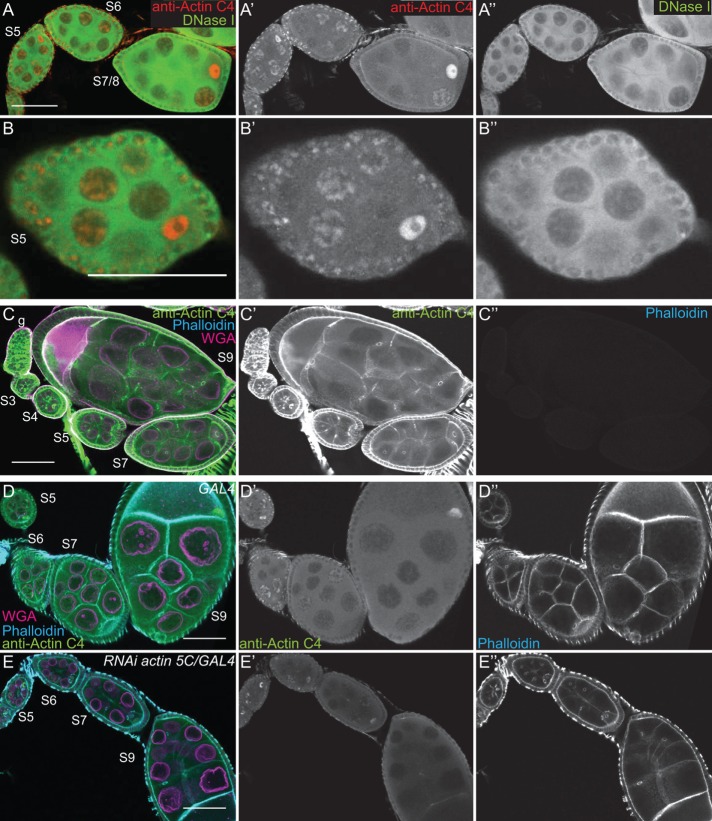
In addition to these antibodies, we used DNase I, which labels monomeric or G-actin (Hitchcock, 1980), to examine nuclear actin. We found that DNase I uniformly labels a blobby structure within the nurse cell nuclei that strikingly resembles the structure labeled by the actin C4 antibody (Figure 2, A′′ and B′′, and unpublished data). Indeed, costaining showed that DNase I and actin C4 colocalize (Figure 2, A–B′′, and Supplemental Movie S2). Of note, DNase I did not label the actin within the germinal vesicles (Figure 2, A–B′′). Because the actin C4 antibody can label both monomeric and F-actin (Lessard, 1988) whereas DNase I labels only monomeric actin, the actin C4 labeling within the nurse cell nuclei and the germinal vesicles may reflect the polymerization state of nuclear actin.
FIGURE 2:
The actin C4 antibody recognizes Drosophila actin. (A–E′′) Maximum projections of two to four confocal slices of early stage wild-type follicles; images C–C′′ are of methanol-fixed follicles. (A, B) Merged images: anti–actin C4, red; DNase I, green. (A′, B′) Anti–actin C4, white. (A′′, B′′) DNase I, white. (C–E) Merged images: anti–actin C4, green; phalloidin, cyan; WGA, magenta. (C′–E′) Anti–actin C4, white. (C′′–E′′) Phalloidin, white. Scale bars, 50 μm. DNase I uniformly labels a blobby structure within the nurse cell nuclei (A, B, A′′, B′′), and the actin C4 antibody labels the same structure within a subset of the nuclei (A, B, A′, B′). Of note, DNase I does not label the germinal vesicle (A–B′′). On methanol fixation, the actin C4 antibody no longer labels the structured nuclear actin but instead labels F-actin structures within the follicles (C, C′); such fixation prevents phalloidin staining (C′′). RNAi-mediated knockdown of actin 5C within the germline results in decreased structured nuclear actin (anti–actin C4) and phalloidin staining of the cortical actin within the nurse cells, whereas both the antibody and phalloidin still label the muscle sheath and phalloidin strongly marks the cortical actin within the follicle cells (E–E′′ vs. D–D′′).
To verify that the actin C4 antibody can label F-actin, we used methanol fixation to make the actin within filaments more accessible to the antibody. The actin C4 antibody no longer labeled the structured nuclear actin but instead labeled all canonical F-actin structures (Figure 2, C and C′). As expected, such fixation prevents phalloidin from labeling F-actin (Figure 2C′′).
Because actin 5C is the most abundantly expressed actin gene during oogenesis (ModENCODE [www.modencode.org/]; Tootle et al., 2011), we altered its levels to further test the specificity of the actin C4 antibody. RNA interference (RNAi)–mediated germline knockdown of actin 5C results in a reduction in nuclear anti–actin C4 labeling (Figure 2, E–E′′ vs. D–D′′). Knockdown was evident by both immunoblotting (Supplemental Figure S1E) and reduced phalloidin staining within the germline, but normal phalloidin labeling remained in the somatic cells and the muscle sheath (Figure 2, E′′ vs. D′′). Conversely, as discussed in detail later, overexpression of GFP-Actin 5C results in nuclear actin rod formation. These rods label with the actin C4 antibody, and rod formation results in the loss of the structured nuclear actin observed with the actin C4 antibody (see later discussion of Figure 6, A–B′′). Thus both the level of labeling and the structures recognized by the antibody are affected by altering the level of actin within the cells.
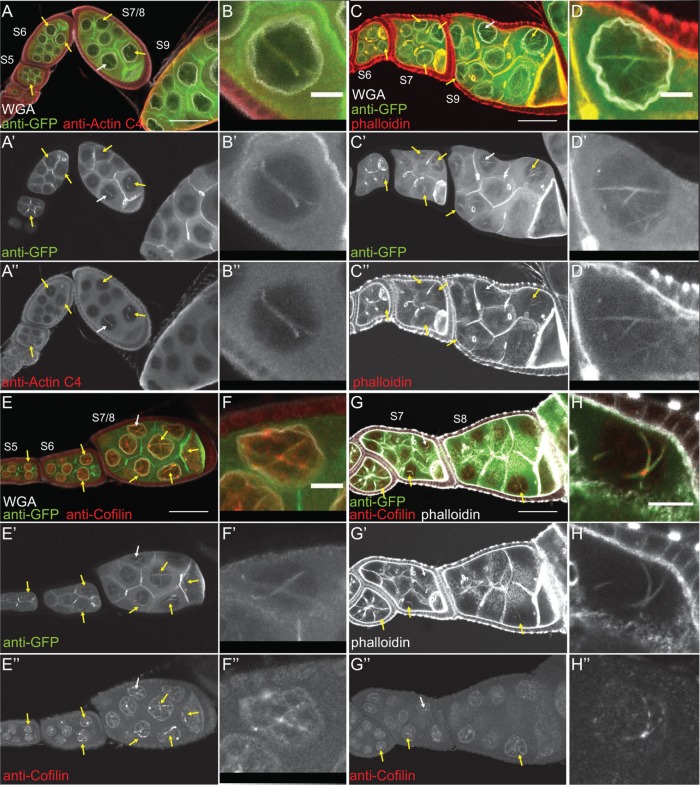
FIGURE 6:
Nuclear GFP-Actin rods contain Cofilin and label with phalloidin. (A–H′′) Maximum projections of two to four confocal slices of follicles from GFP-Actin 5C; matGAL4 females stained with the indicated antibodies and reagents. (A–F) Merged images: WGA, white; anti-GFP, green; other antibody or stain, red (A, B, anti–actin C4; C, D, phalloidin; E, F, anti-Cofilin). (G–H) Merged images: anti-GFP, green; phalloidin, white; anti-Cofilin, red. (B–B′′, D–D′′, F–F′′, H–H′′) Zoomed-in image of the actin rods pointed out by the white arrows in A–A′′, C–C′′, E–E′′, and G–G′′, respectively. Scale bars, 50 μm (A, C, E, G), and 10 μm (B, D, F, H). Nuclear GFP-Actin rods (yellow arrows) label with antibodies to nuclear actin (A–B′′) and Cofilin (E–F′′) and are weakly labeled by phalloidin (C–D′′). Costaining for both Cofilin and phalloidin reveals that the GFP-Actin rods exhibit regions that label with phalloidin, and in the weak phalloidin regions, the rods contain Cofilin (G–H′′).
Together the foregoing data support that the actin C4 antibody recognizes Drosophila actin, both in the cytoplasm and within the nucleus. Throughout the rest of this article, the nuclear actin C4 antibody labeling will be referred to as endogenous nuclear actin. However, it is important to note that this antibody may only recognize a subset, that is, a particular structure and/or modification state, of nuclear actin.
Cofilin and Profilin localize to the nucleus
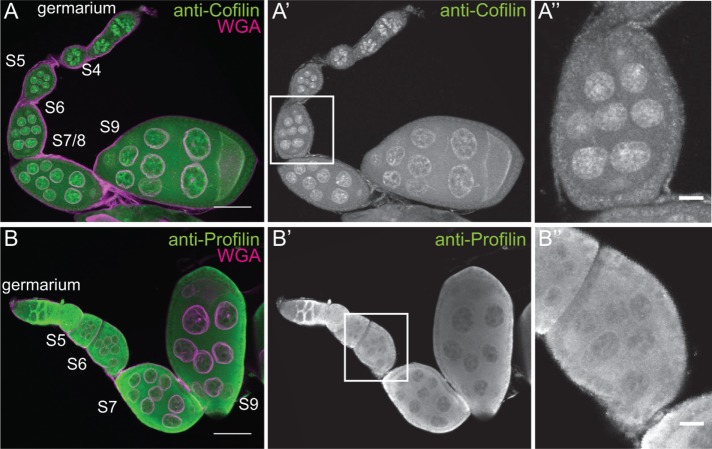
We interpret the varying levels of endogenous nuclear actin observed within the nurse cells during S5–9 as an indication that nuclear actin is dynamically regulated during this period of development. The localization of actin to the nucleus is regulated by Cofilin (Drosophila Twinstar) and Profilin (Drosophila Chickadee) in other systems (Wada et al., 1998; Pendleton et al., 2003; Stuven et al., 2003; Dopie et al., 2012). Immunofluorescence images reveal that both Cofilin and Profilin are found in the nurse cell nuclei during the same stages of follicle development as endogenous nuclear actin (Figure 3, A–B′′). Therefore the factors needed to regulate nuclear actin levels are present during the appropriate developmental time. Subcellular fractionation analysis of Profilin and Cofilin is not currently possible, as Profilin fails to be retained in the nuclei during fractionation (Groen et al., 2015), and the Cofilin antibody works poorly for immunoblotting (unpublished data).
FIGURE 3:
Cofilin and Profilin are present in the nurse cell nuclei. (A–B′′) Maximum projections of two to four confocal slices of wild-type early stage follicles. (A, B) Merged images of nuclear envelope (WGA) in magenta and antibody staining in green. (A–A′′) Anti-Cofilin. (B–B′′) Anti-Profilin. (A′′, B′′) Zoomed-in images of the regions boxed in A′ and B′, respectively. Scale bars, 50 μm (A, B), 10 μm (A′′, B′′). The levels of nuclear Cofilin (A–A′′) and Profilin (B–B′′) appear fairly constant during early oogenesis.
Germline expression of GFP-Actin induces stage-specific nuclear actin rod formation
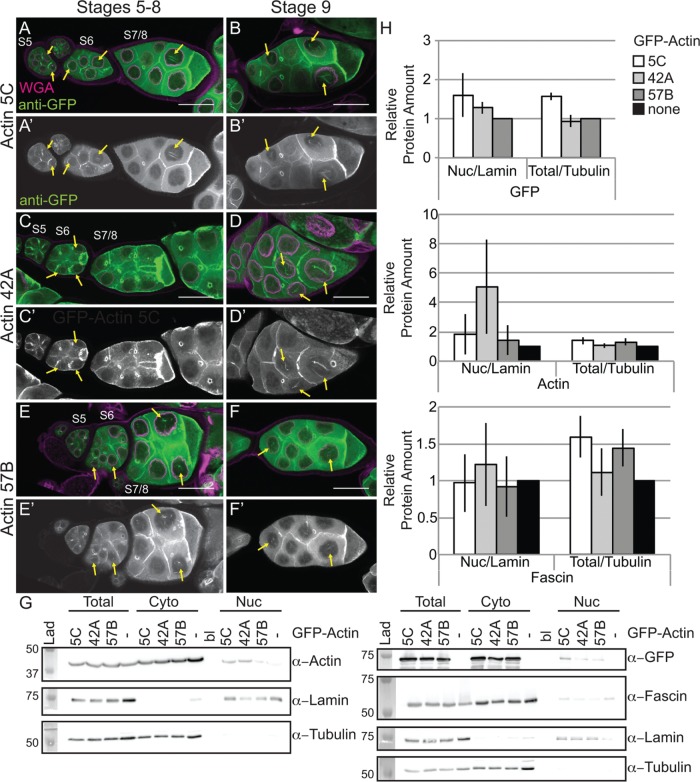
Our prior work revealed that while germline expression of GFP-Utrophin or Lifeact-GFP using the UAS/GAL4 system (Rorth, 1998) induces nuclear actin rods, these tools also cause severe cytoskeletal defects (Spracklen et al., 2014). Another group demonstrated that N-terminally tagged GFP-Actin could be expressed within the germline using the same UAS/GAL4 system without causing major defects (Roper et al., 2005). Thus we assessed whether GFP-Actin could be used to examine nuclear actin. There are six actins in Drosophila; two are strongly expressed in the ovary (5C and 42A), one appears to be weakly expressed (57B), and three are not expressed (79B, 87E, and 88F; ModENCODE; Tootle et al., 2011). In our initial studies on the six actins, we found that strong germline expression (matGAL4) of only the GFP-Actins normally expressed during oogenesis (5C, 42A, and 57B) resulted in nuclear actin rod formation in the nurse cell nuclei and the germinal vesicle during S5–9 (Figure 4, A–F′, and unpublished data). It is worth noting that both weak (nanosGAL4) and strong (matGAL4) germline expression of any of the six GFP-Actins results in severely reduced female fertility or sterility, which does not appear to be due to cytoskeletal defects (Supplemental Figure 2C and unpublished data).
FIGURE 4:
Germline expression of GFP-Actin induces nuclear actin rods. (A–F′) Maximum projections of two to four confocal slices of early follicles expressing the indicated GFP-Actin in the germline (matGAL4). (A–F) Merged images: WGA, magenta; anti-GFP, green. (A′–F′) Anti-GFP, white. Scale bars, 50 μm. Expression of GFP-Actin in the germline results in nuclear actin rod formation (A–F′, yellow arrows). (G) Representative Western blots of subcellular fractionation samples (total lysate, cytoplasmic fraction, nuclear fraction) from the indicated GFP-Actin or GAL4 only (–) blotted for GFP (GFP-Actin), actin (JLA20), Fascin, Lamin Dm0 (nuclear marker), and α-Tubulin (cytoplasmic marker). bl, blank lane; Lad, ladder. (H) Charts quantifying the relative amount of nuclear protein (actin, Fascin, or GFP-Actin) to nuclear Lamin and total protein (actin, Fascin, or GFP-Actin) to total Tubulin from Western blots of three subcellular fractionation experiments. For actin and Fascin values, protein amount was normalized to GAL4 only; GFP protein amount was normalized to GFP-Actin 57B. Error bars, SE. Nuclear and total levels of actin, Fascin, and GFP are not significantly different between GFP-Actin 5C, 42A, or 57B (p > 0.05, ordinary one-way analysis of variance).
Both the developmental stage and the GFP-Actin being expressed appeared to affect nuclear actin rod formation. To characterize these differences, we scored the frequency and number of nurse cells exhibiting nuclear actin rods and the length of the rods during S5–6, S7–8, and S9 from confocal stacks labeled for GFP and the nuclear envelope (wheat germ agglutinin [WGA]). Follicles were scored as having 0, ≤25, 25–75, or ≥75% of the nurse cells exhibiting actin rods. Rod length was scored as short (≤1/4 diameter of the nucleus), medium (∼1/2 diameter of the nucleus), or long (≥1 diameter of the nucleus). We found that the frequency of nuclear actin rod formation is generally higher in the earlier stages (S5–8) and decreases with development (S9) for GFP-Actin 5C (p < 0.01, Fisher’s exact test) and trends similarly with GFP-Actin 42A (p = 0.14; Supplemental Figure S2A′ and Supplemental Table S1D). Of note, the early stages are where we also observe higher endogenous nuclear actin levels (Figure 1, A–B′′; see later quantification in Figure 8E and statistical analysis in Supplemental Table S1B; p < 0.0001, Fisher’s exact test). GFP-Actin 5C and 42A expression strongly induces nuclear actin rods, and the rods that form tend to be long, whereas GFP-Actin 57B expression results in a low frequency of short nuclear actin rods (Supplemental Figure S2, A–B′; statistical analysis in Supplemental Table S1D). Similar results were observed using a different germline GAL4 driver (oskGAL4; unpublished data). These phenotypic differences are not due to fixation conditions, as live imaging revealed similar differences in nuclear actin rod length (Supplemental Figure S3). The differences in actin rod formation also do not appear to be due to the expression level of the transgenes or the level of nuclear GFP-Actin or endogenous nuclear actin (Figure 4, G and H, and unpublished data). This finding suggests the different GFP-Actins have distinct abilities to form nuclear actin rods, similar to the prior finding that the GFP-Actins also differentially incorporate into cytoplasmic actin structures during oogenesis (Roper et al., 2005). All subsequent experiments use GFP-Actin 5C.
Nuclear GFP-Actin rod dynamics and composition
We next wanted to assess how rapidly the nuclear GFP-Actin rods form and disassemble. Our initial live-imaging experiments suggested that the rods are stable, as no apparent changes in rod length or number were observed over multiple minutes (Supplemental Movie S3, ∼8 min). To assess whether actin treadmilling occurred on the nuclear GFP-Actin rods, we performed FRAP analysis. A small segment in the middle of a rod was photobleached, and recovery was assessed; if treadmilling were occurring, the bleached area should move along the rod, and fluorescence should recover in the area that was bleached. We found that there was no recovery over 3 min (Figure 5 and Supplemental Movie S4). Thus, whereas nuclear actin levels vary during development, with decreased numbers of nurse cells with high levels of nuclear actin (Figure 1, A–B′; quantified later in Figure 8E and statistical analysis in Supplemental Table S1B) and GFP-Actin rod formation (Supplemental Figure S2, A-B′; statistical analysis in Supplemental Table S1D) in the later stages, when rods are present, they are fairly static.
FIGURE 5:
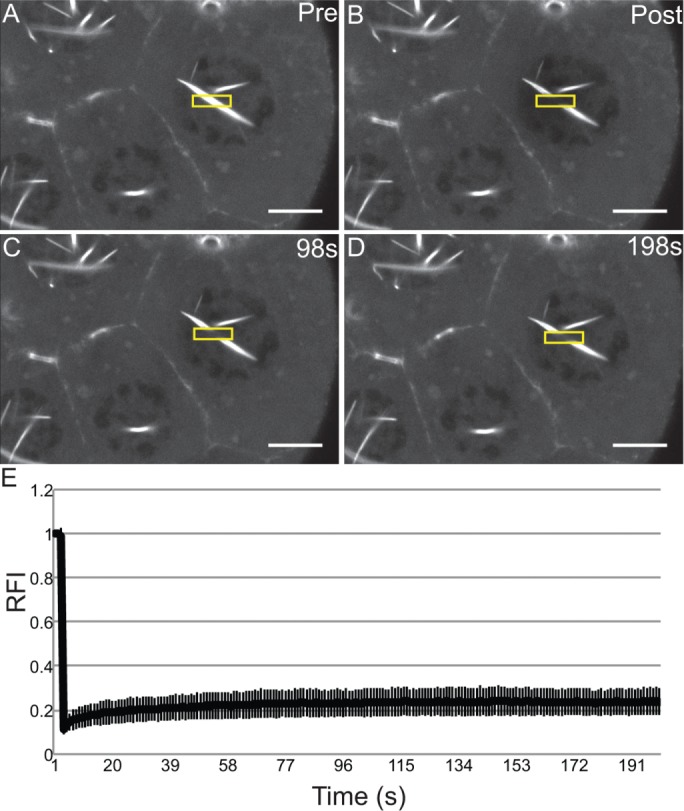
Nuclear GFP-Actin rods are stable. (A–D) Representative example of FRAP experiments. Single-slice confocal images of GFP-Actin 5C (UAS GFP-Actin 5C; mat3Gal4) during FRAP time course showing prebleach, postbleach, middle, and endpoint. Scale bar, 10 μm. (E) Average FRAP recovery curve. Relative fluorescence intensity over time for nine different nuclei from five follicles. Error bars, SD. The nuclear GFP-Actin rods are stable, as the bleached region fails to recover.
Because the composition of nuclear actin rods appears to vary across systems, we sought to determine what type of actin rods were forming due to GFP-Actin expression. First, we validated that the GFP rods we observed were indeed actin rods. Using the actin C4 antibody, we found that the nuclear rods colabel for both actin and GFP (Figure 6, A–B′′). We next asked whether the nuclear actin rods label with phalloidin and/or Cofilin. Our images showed that the nuclear actin rods labeled weakly with phalloidin (Figure 6, C–D′′) and exhibited nonuniform Cofilin colocalization (Figure 6, E–F′′). Costaining revealed that the rods labeled with both markers, and, as expected, Cofilin appeared strongest in the areas of weaker phalloidin staining (Figure 6, G–H′′).
Fascin regulates nuclear actin rod formation and length
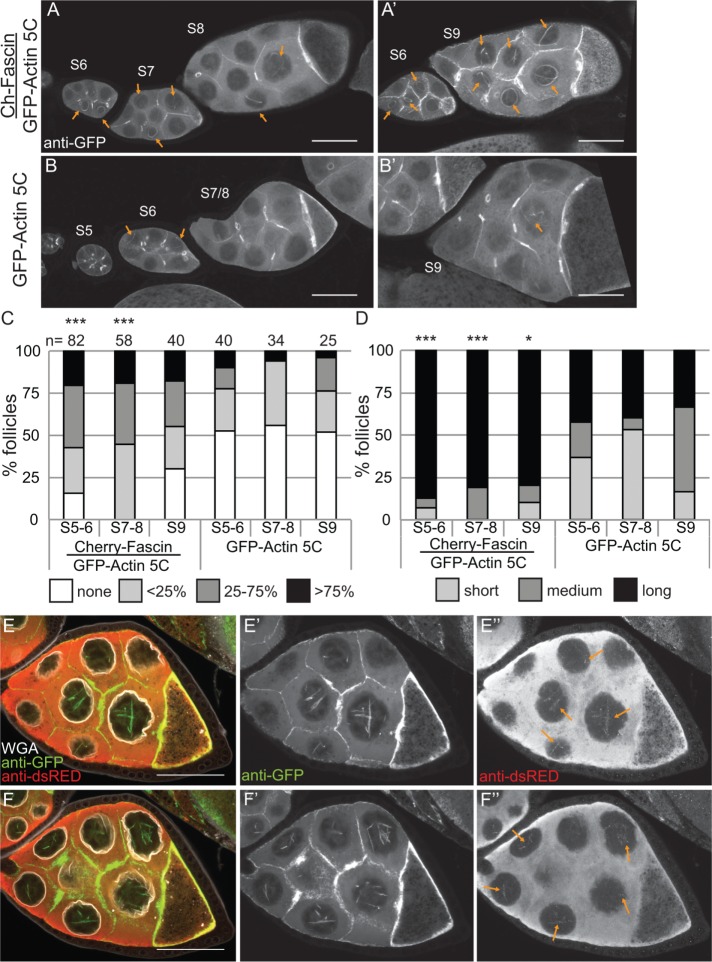
We recently found that the actin bundling protein Fascin localizes to the nucleus during Drosophila oogenesis (Figure 1E; Groen et al., 2015). Because the best-characterized function of Fascin is to bind to and alter the structure of actin, we assessed how changing the level of Fascin expression alters nuclear GFP-Actin rod formation. Images revealed that coexpression of mCherry-Fascin and GFP-Actin 5C in the germline results in increased rod formation compared with follicles from siblings expressing only GFP-Actin (Figure 7, A and A′ vs. B and B′). To further characterize the change in nuclear GFP-Actin rod formation, we quantified the frequency and length of the rods as described earlier. We find that expression of Fascin significantly increases rod formation in S5–6 (p < 0.001; Pearson’s chi-squared test) and S7–8 (p < 0.0001), and rods are longer in all stages examined (S5–6, p < 0.001; S7–8, p < 0.0001; and S9, p < 0.05; Figure 7, C and D; statistical analysis in Supplemental Table S1A).
FIGURE 7:
Overexpression of Fascin enhances GFP-Actin rod formation. (A–B′) Maximum projections of two to four confocal slices of S5–9 follicles stained for GFP-Actin (anti-GFP). (A, A′). UAS mCherry-Fascin/GFP-Actin 5C; oskGAL4/+. (B, B′) GFP-Actin 5C/+; oskGAL4/+. Orange arrows indicate nuclear GFP-Actin rods. Scale bars, 50 μm. (C, D) Charts of the percentage of follicles of the indicated stages and transgenes expressed using oskGAL4 from 5- to 8-d-old females that exhibit particular frequencies and lengths of nuclear GFP-Actin rods; n values for both graphs are indicated across the top of the graph in C. Each follicle within a confocal stack was scored, in a genotypically blinded manner, for the percentage of nurse cells exhibiting nuclear actin rods (C; 0, ≤25, 25–75, or ≥75%) and the length of those rods (D; short, ≤1/4 of the nuclear diameter; medium, ∼1/2 diameter; or long, ≥1 diameter). Overexpression of Fascin enhances both the frequency and length of the nuclear actin rods in S5–6 and S7–8 but only the length during S9 compared with GFP-Actin alone during the same stages (***p < 0.001 and *p < 0.05, Pearson’s chi-squared test). (E–F′′) Maximum projections of two to four confocal slices of two different planes of a S8 UAS mCherry-Fascin/GFP-Actin 5C; oskGAL4/+ follicle. (E, F) Merged images: WGA, white; anti-GFP, green; anti-dsRed, red. (E′, F′) Anti-GFP, white. (E′′, F′′) Anti-dsRed, white. Scale bars, 50 μm. mCherry-Fascin labels nuclear GFP-Actin rods (orange arrows in E′′ and F′′).
Overexpression of Fascin could increase GFP-Actin rod formation by a number of mechanisms. Fascin may alter the total level of nuclear actin, thereby increasing the formation of nuclear actin rods. Subcellular fractionation studies reveal that the level of nuclear GFP-Actin, endogenous actin, and endogenous Fascin are not increased by Fascin overexpression (Supplemental Figure S4). Another means by which overexpression of Fascin could increase nuclear actin rod formation and length is by binding to and stabilizing actin rods within the nucleus. Supporting this idea, we find that mCherry-Fascin labeled GFP-Actin rods (Figure 7, E–F′′, and Supplemental Movie S5).
Fascin regulates endogenous nuclear actin
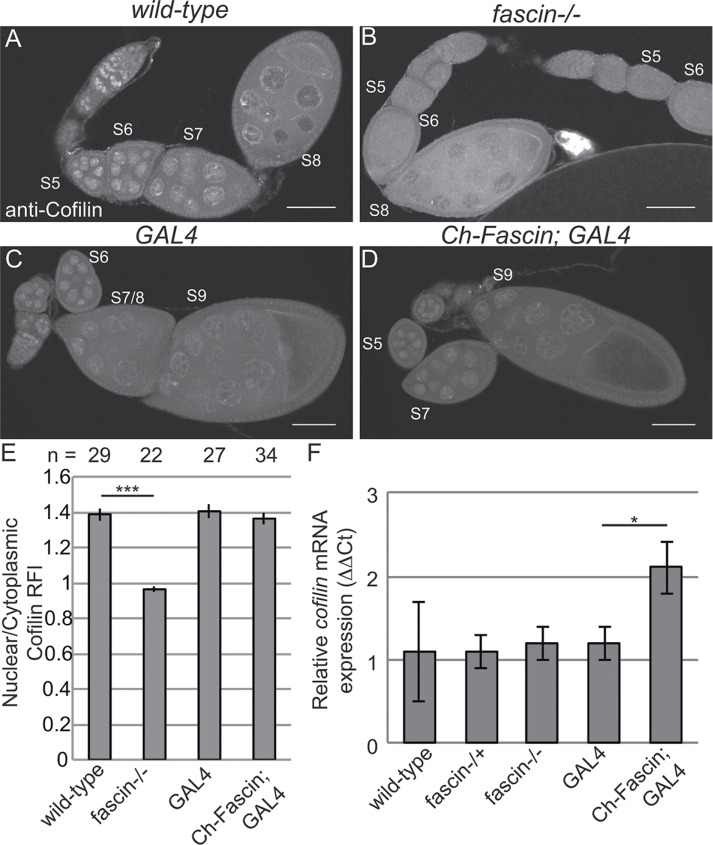
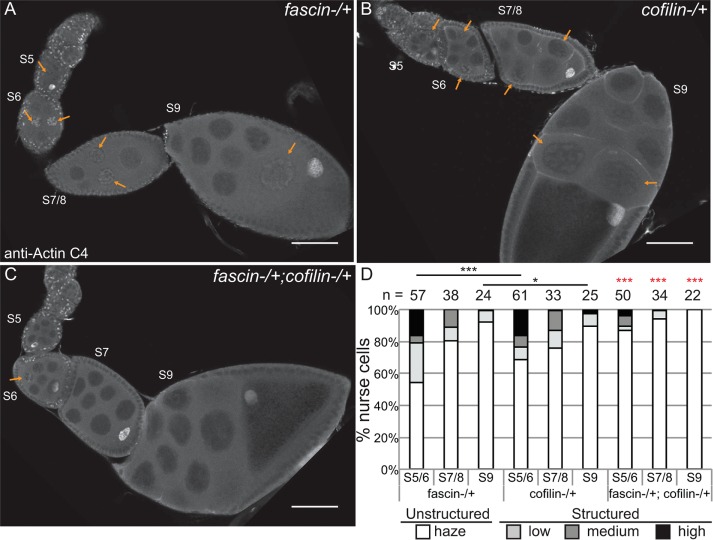
Although the foregoing studies showed that Fascin regulates GFP-Actin rod formation, it remains to be determined whether Fascin regulates endogenous nuclear actin. Immunofluorescence images reveal that loss of Fascin results in a more uniform, low level of nuclear actin in the nurse cells (Figure 8, B vs. A), whereas overexpression of Fascin results in an increase in the number of nurse cells exhibiting higher levels of endogenous, structured nuclear actin during S5–9 (Figure 8, D vs. C). To further characterize these differences, we scored the percentage of nurse cells exhibiting unstructured nuclear actin (haze) and low, medium, or high levels of structured nuclear actin (Figure 8E; statistical analysis in Supplemental Table S1B). This quantification reveals that structured nuclear actin is present at the highest frequency in S5–6 (∼47% total) and decreases as follicle development progresses (13.5% in S7/8 and 3.3% in S9; p < 0.0001, Fisher’s exact test). Loss of Fascin reduces the number of nurse cells exhibiting structured nuclear actin, with 28.6% of S5–6 (p < 0.0001; Pearson’s chi-squared test), 4.3% of S7–8 (p < 0.0001), and 1.1% of S9 (nonsignificant [n.s.]) nurse cells having structured nuclear actin. GAL4 expression alone, compared with wild type, increases the frequency of nurse cells with structured nuclear actin (statistical analysis in Supplemental Table S1B): 57.4% in S5–6 (p < 0.0001; Pearson’s chi-squared test), 27.3% in S7–8 (p < 0.001), and 8% in S9 (n.s.). Expression of mCherry-Fascin further increases the frequency of nurse cells with structured nuclear actin compared with GAL4 expression alone, with 88.9% in S5–6 (p < 0.001, Pearson’s chi-squared test), 35% in S7–8 (p < 0.01), and 11.6% in S9 (n.s.).
The Fascin-dependent changes in endogenous nuclear actin could be due to globally altered nuclear actin levels. To assess this possibility, we used subcellular fractionation (Supplemental Figure S5). We found that reduction or loss of Fascin did not alter nuclear actin levels compared with wild type at the whole ovary level; similarly, overexpression of Fascin did not affect total nuclear actin levels compared with the GAL4 control (Figure 8F, p > 0.05, and Supplemental Figure S5). Endogenous nuclear Fascin levels, as expected, were reduced by the loss of Fascin. Whereas overexpression of Fascin did not affect endogenous nuclear Fascin levels (Figure 8F), total nuclear Fascin levels increased due to mCherry-Fascin localizing to the nucleus (Supplemental Figure S5, red box, and unpublished data). These data indicate that Fascin regulates the accumulation or retention of actin within a subset of nuclei.
Fascin modulates Cofilin to regulate nuclear actin
One means by which Fascin may regulate endogenous nuclear actin is by altering the localization of the factors regulating the nuclear import and export of actin—Cofilin and Profilin, respectively. Immunofluorescence analyses reveal that, qualitatively, the localization of Profilin is normal when Fascin levels are manipulated (Supplemental Figure 6, B–C′ vs. A–A′). To quantify Profilin localization, we generated fluorescence intensity plots from the cytoplasm into the nucleus of nurse cells from S7/8 follicles and used the peak intensity for WGA to define the nuclear boundary. This quantification revealed that the nuclear-to-cytoplasmic ratios of Profilin are unaffected by loss or overexpression of Fascin (Supplemental Figure S6D). Surprisingly, total Profilin levels are reduced by the loss of Fascin (Supplemental Figure S6E). Decreased Profilin levels would be predicted to result in increased nuclear actin due to reduced export of actin from the nucleus. Because loss of Fascin results in decreased endogenous nuclear actin (Figure 8, B and E), it seems unlikely that the reduced Profilin level contributes to the nuclear actin phenotype. The reduction in Profilin may play a role in the late stage cytoplasmic actin defects observed in fascin mutant follicles (Cant et al., 1994). Alternatively, the reduced Profilin level may simply reflect altered follicle composition of the ovaries in the fascin mutant; indeed, the mutant ovaries exhibit a reduction in the number of late stage follicles. Thus we interpret these data to suggest that the main mechanism by which Fascin alters nuclear actin accumulation is not due to Fascin regulating Profilin. However, Fascin does affect Cofilin. Although nuclear Cofilin levels do not appear to be altered by overexpression of Fascin (Figure 9, D vs. C), loss of Fascin qualitatively decreased nuclear Cofilin (Figure 9, B vs. A). Quantification of nuclear-to-cytoplasmic ratios of Cofilin by fluorescence intensity plots reveals that the wild-type ratio is 1.38 ± 0.035 (SE), and this ratio is reduced to 0.962 ± 0.013 in fascin mutants (Figure 9E, p < 0.0001). Total cofilin transcript levels, as assessed by quantitative reverse transcriptase PCR, are unaffected by the loss of Fascin but increase when Fascin is overexpressed (Figure 9F). Together these data suggest that Fascin regulates Cofilin by an unknown mechanism to modulate nuclear actin levels.
FIGURE 9:
Loss of Fascin reduces nuclear Cofilin levels. (A–D) Maximum projections of two to four confocal slices of follicles of the indicated genotypes stained for Cofilin; note that these are the same follicles shown in Figure 8. Scale bars, 50 μm. (E) Chart depicting the average nuclear-to-cytoplasmic relative fluorescence intensity of Cofilin in nurse cells from S7–8 follicles; the number of nurse cells (n) examined is indicated across the top of the chart. Briefly, the fluorescence intensity along a line traversing from the cytoplasm into the nucleus from two to three nurse cells from a minimum of seven different follicles from each genotype, in a genotypically blinded manner, was analyzed as described in Materials and Methods. Loss of Fascin significantly reduced the nuclear level of Cofilin, whereas overexpression of Fascin did not alter the distribution of Cofilin. Error bars, SE. ***p < 0.001, unpaired t test with Welch’s correction. (F) Chart depicting the normalized mRNA expression of cofilin from whole ovary preparations. Error bars, SE. Reduction of Fascin expression does not alter cofilin expression relative to wild-type. Overexpression of Fascin results in a significant increase in cofilin expression relative to its GAL4 control (Ch-Fascin to GAL4, p < 0.05, unpaired t test with Welch’s correction).
Supporting this idea, we find that Fascin and Cofilin dominantly interact. Heterozygosity for a null mutation in fascin has little effect on the frequency of nurse cells exhibiting structured nuclear actin (Figure 10, A and quantified in D; statistical analysis in Supplemental Table S1C), and heterozygosity for cofilin (Drosophila twinstar) slightly reduces the number of nurse cells with structured nuclear actin during S5–6 (p < 0.001, Fisher’s exact test) and S9 (p < 0.05; Figure 10, B and quantified in D). However, follicles from flies that are heterozygous for mutations in both fascin and cofilin exhibit a significant loss of structured nuclear actin compared with either heterozygote alone in S5–9 (Figure 10, C and quantified in D; p < 0.001, Fisher’s exact test; statistical analysis in Supplemental Table S1C). This loss of nurse cells with structured nuclear actin is not due to a global decrease in nuclear actin levels, as subcellular fractionation analysis of whole ovaries indicates that total nuclear actin levels are similar between wild-type, fascin−/+, cofilin−/+, and fascin−/+;cofilin−/+ (Supplemental Figure S7). These data support a model in which Fascin regulates Cofilin to control nuclear actin, determining when and which nuclei accumulate higher levels of structured nuclear actin.
FIGURE 10:
Fascin genetically interacts with Cofilin to regulate nuclear actin. (A–C) Maximum projections of two to four confocal slices of follicles stained for nuclear actin (anti–actin C4). (A) fascinsn28/ +, (B) cofilintsr1/+, and (C) fascinsn28/ +; cofilintsr1/+. Orange arrows indicate structured nuclear actin within the nurse cells. Scale bars, 50 μm. (D) Chart quantifying the percentage of nurse cells in S5–6, S7–8, and S9 of the indicated genotypes exhibiting unstructured or structured nuclear actin; the number of follicles (n) examined is indicated across the top of the chart. Each follicle within a confocal stack was scored, in a genotypically blinded manner, for the percentage of nurse cells exhibiting unstructured nuclear actin haze or low, medium or high levels of structured nuclear actin. Heterozygosity for a null allele of fascin has minimal effect on the frequency of nurse cells with structured nuclear actin levels, whereas heterozygosity for cofilin (Drosophila twinstar, tsr) exhibits reduced structured nuclear actin in S5/6 and S9 compared with fascin−/+ (A and B, quantified in D). Double heterozygotes for mutations in fascin and cofilin result in a striking reduction in the number of nurse cells with structured nuclear actin compared with either heterozygote alone (C vs. A and B; quantified in D); red asterisks indicate significant difference from both individual heterozygotes at the same stages. ***p < 0.001 and *p < 0.05; Fisher’s exact test.
DISCUSSION
Nuclear actin is found at varying levels within the nurse cells, is present in a subset of early follicle cells, and is highly enriched in the germinal vesicle during S5–9 of Drosophila oogenesis. We used the broad-specificity actin C4 antibody (Lessard, 1988) to examine endogenous nuclear actin. Analysis of this antibody supports that it recognizes actin, because altering actin expression affects its labeling pattern, when it labels nuclear structures it colocalizes with nuclear DNase I, which marks monomeric actin, and finally, methanol fixation relocalizes its labeling to F-actin structures. However, not all nuclear actin is recognized by the antibody. Indeed, DNase I uniformly labels a blobby structure within the nurse cell nuclei, whereas the actin C4 antibody labels only a subset of these nuclei. Furthermore, in many studies on nuclear actin, different antibodies to actin routinely label distinct nuclear structures (Schoenenberger et al., 2005; Jockusch et al., 2006; Asumda and Chase, 2012). Thus the actin C4 antibody likely labels a subset of nuclear actin. It remains to be determined what the structure of this actin is and whether other pools of nuclear actin exist in these cells.
During the same stages in which we observe endogenous nuclear actin, ectopic germline expression of GFP-Actin exhibits nuclear actin rod formation. Similarly, expression of the actin-labeling reagents GFP-Utrophin and Lifeact-GFP results in nuclear actin structures during these stages (Spracklen et al., 2014). These structures likely form due to increased nuclear actin levels resulting from the higher expression of actin in the case of GFP-Actin and stabilization of nuclear actin in the cases of GFP-Utrophin and Lifeact-GFP. These findings suggest that nuclear actin levels and structures are tightly regulated during development.
Although the physiological relevance of nuclear actin rods remains debated, cell culture–based screens assessing nuclear actin rods have identified novel regulators of nuclear actin (Rohn et al., 2011). Here we used nuclear GFP-Actin rod formation to uncover the role of the actin bundling protein Fascin in modulating nuclear actin in an in vivo, developmental context. Overexpression of Fascin enhances both the length and frequency of formation of nuclear GFP-Actin rods. This finding is not specific to nuclear GFP-Actin, as overexpression of Fascin increases the frequency of nurse cells with high levels of endogenous, structured nuclear actin, whereas loss of Fascin results in a uniform, low level of nuclear actin. Because the total level of nuclear actin within the ovary is not affected by genetically manipulating Fascin, these findings suggest that Fascin regulates cell-specific accumulation of nuclear actin. Thus, by screening for modifiers of nuclear GFP-Actin rod formation during Drosophila oogenesis, regulators of endogenous nuclear actin can be identified.
Multiple lines of evidence indicate that Fascin regulates Cofilin to control nuclear actin. Loss of Fascin results in decreased nuclear Cofilin, whereas total Cofilin levels appear unchanged. In addition, Fascin and Cofilin genetically interact. Follicles from flies that are heterozygous for mutations in both fascin and cofilin exhibit a striking reduction in the number of nurse cells with high levels of structured nuclear actin compared with either heterozygote alone. These data lead to a model in which Fascin promotes the nuclear localization of Cofilin, and therefore actin, in a cell-specific manner within the Drosophila germline. Given that we recently showed that Fascin localizes to the nucleus and nuclear periphery in addition to the cytoplasm (Groen et al., 2015; Jayo et al., 2016), Fascin could act at any of these cellular locations to modulate Cofilin and nuclear actin.
Cofilin has both cytoplasmic and nuclear roles in regulating nuclear actin. In the cytoplasm, Cofilin binds to and severs F-actin (Lappalainen and Drubin, 1997); this activity alters the cellular levels of F- versus G-actin. Because increased G-actin levels lead to nuclear actin accumulation (Pendleton et al., 2003), cytoplasmic Cofilin regulates the pool of actin available for transport to the nucleus. Of interest, Cofilin severing activity is enhanced on Fascin-bundled actin filaments (Breitsprecher et al., 2011). Thus loss of Fascin’s bundling activity is predicted to decrease Cofilin activity and G-actin levels, reducing nuclear actin accumulation. Cofilin also directly binds monomeric actin and facilitates its import into the nucleus (Pendleton et al., 2003; Dopie et al., 2012). In the nucleus, Cofilin can stabilize nuclear actin structures by forming Cofilin–actin rods (Munsie et al., 2012). Because Fascin localizes to the nucleus, nuclear Fascin may prevent nuclear export of Cofilin, thereby stabilizing nuclear actin structures.
Phosphorylation inhibits both Cofilin’s actin severing (Agnew et al., 1995) and nuclear import functions (Dopie et al., 2012). Two kinases phosphorylate Cofilin: LIM kinase (LIMK) and Tes kinase (Drosophila Cdi). Both kinases are expressed during Drosophila oogenesis (ModENCODE). Of note, Fascin binds to LIMK in other systems and relocates it to sites of actin bundle formation (Jayo et al., 2012). Thus cytoplasmic Fascin may regulate LIMK to control the phosphorylation of Cofilin and thereby regulate nuclear actin. Fascin may also regulate the expression or activity of Ssh (Drosophila Slingshot), the phosphatase for Cofilin. Of interest, a recent genetic screen for modifiers of nuclear actin rod formation uncovered a number of proteins that regulate Cofilin (Dopie et al., 2015). It will be interesting to examine whether Fascin affects any of these newly identified Cofilin regulators to modulate nuclear actin.
Fascin also regulates the structure of nuclear actin. Overexpression of Fascin results in curved and often circular nuclear actin rods. Loss of Fascin also alters nuclear actin structure, as there is a reduction in the number of nuclei exhibiting structured, endogenous nuclear actin. Because mCherry-Fascin localizes to nuclear GFP-Actin rods, Fascin modulation of nuclear actin structure could be direct. It is possible that Fascin also indirectly regulates nuclear actin structure. Although we favor the model in which nuclear Fascin controls nuclear actin structure, Fascin at any cellular location may modulate the complement of actin binding proteins within the nucleus to regulate nuclear actin architecture.
Our finding that nuclear actin is tightly controlled during Drosophila oogenesis strongly suggests that nuclear actin plays important but unknown functions. Supporting this idea, germline expression of any of the six GFP-Actins results in female sterility without any striking defects in cytoplasmic F-actin structures or follicle morphogenesis. One potential role of nuclear actin may be to modulate DNA replication within the nurse cells. During S5–9, the nurse cells undergo asynchronous endocycles (Lilly and Spradling, 1996). Nuclear actin may serve a similar role in the follicle cells, as patches of these cells within a given follicle also exhibit varying levels of nuclear actin. Alternatively, nuclear actin may regulate transcription. The nurse cells actively transcribe components that will ultimately be provided to the oocyte to mediate embryonic development. Of interest, strong germline expression of GAL4 increases the frequency of nurse cells with structured nuclear actin; this increase in nuclear actin may be due to increased transcription within the cells or strong expression of GAL4 causing cellular stress. Another potential role of nuclear actin is structural, maintaining nuclear shape in response to mechanical forces. The nurse cell nuclei undergo dramatic shape changes during S5–9. During S5, the nuclei are round, but as development progresses, the nuclei become increasingly invaginated (Cummings and King, 1970). Of note, endogenous nuclear actin and GFP-Actin rod formation are inversely correlated with nuclear envelope invagination in the nurse cells. The high level of nuclear actin within the germinal vesicle may also play a structural role, as it is in a very large cytoplasm and likely has to resist substantial pressure. Thus a nuclear actin meshwork similar to that in Xenopus oocytes (Bohnsack et al., 2006) may maintain germinal vesicle shape and integrity in Drosophila. Nuclear actin may also serve as a storage facility for monomeric actin. During S5–9, the nurse cells rapidly grow in size and need a large pool of G-actin for cortical actin growth. However, given that the endogenous, structured nuclear actin substantially decreases in S9 compared with S5–8 and is rarely detected in S10A, when the nurse cells are still increasing in size, it is unlikely that this is a major role of structured nuclear actin during follicle development. Further studies are needed to uncover which of these potential actions or other novel functions nuclear actin plays during Drosophila oogenesis.
Our finding that Fascin regulates Cofilin to modulate nuclear actin is likely relevant beyond Drosophila oogenesis. Indeed, Fascin’s diverse cellular locations and activities are conserved from invertebrates to humans (Jayo and Parsons, 2010; Jayo et al., 2016; Groen et al., 2015). Furthermore, the majority of the new nuclear actin regulators discovered in screens using cultured Drosophila cells also modulate nuclear actin in mammalian cells (Rohn et al., 2011; Dopie et al., 2015). Thus our study provides the foundation to explore the in vivo, physiologically important regulation and function of nuclear actin.
MATERIALS AND METHODS
Fly stocks
Fly stocks were maintained on cornmeal/agar/yeast food at 21°C, except where noted. Before immunofluorescence or Western blot analysis, flies were fed wet yeast paste daily for 3–7 d. Unless otherwise noted, yw was used as the wild-type control. The following stocks were obtained from the Bloomington Drosophila Stock Center (Bloomington, IN): UASp RNAi actin 5C (TRiP.HMS02487), UASp GFP-Actin 5C, UASp GFP-Actin 42A, UASp-GFP-Actin 57B, UASp GFP-Actin 79B, UASp GFP-Actin 87E, UASp GFP-Actin 88F, snx2, tsr1, tsrN121, tsrk05633, and matαGal4 (third chromosome). The sn28, UASp mCherry-Fascin, and nanosGAL4 lines were a generous gift from Jennifer Zanet (Centre de Biologie du Développement, Université de Toulouse, Toulouse, France; Zanet et al., 2012), and the oskarGal4 lines (second and third chromosomes) were a generous gift from Anne Ephrussi (European Molecular Biology Laboratory, Heidelberg, Germany; Telley et al., 2012). Expression of UASp RNAi actin 5C was achieved by crossing to matαGal4, maintaining fly crosses at 21 or 25°C, and maintaining progeny at 29°C. Expression of UASp GFP-Actin and/or UASp Cherry-Fascin was achieved by crossing to matαGal4 or oskarGal4 flies, maintaining fly crosses at 21°C, and maintaining progeny at 25°C.
Fertility assays
Three females (∼4 d old, fed wet yeast every day before mating) of the indicated genotypes were allowed to mate with two or three wild-type (yw) males for 2 d. Matings were performed in triplicate for each genotype. Fresh wet yeast was provided daily. The flies were then transferred to a fresh vial, provided wet yeast, and allowed to lay eggs for 24 h. The adults were removed after 24 h, and the resulting adult progeny were counted ∼18 d later. The number of progeny per female was determined for each vial, and the average and SD of the three independent vials per genotype was calculated. Fertility assays were performed at 25°C. Statistical analysis was performed using Prism (GraphPad Software, La Jolla, CA).
Immunofluorescence
Whole-mount Drosophila ovary samples were dissected into Grace’s insect medium (Lonza, Walkersville, MD) and fixed for 10 min at room temperature in 4% paraformaldehyde in Grace’s insect medium, except for the instance of methanol fixation, which was done for 10 min at 4°C in −20°C methanol. Briefly, samples were blocked by washing in Triton antibody wash (1× phosphate-buffered saline [PBS], 0.1% Triton X-100, and 0.1% bovine serum albumin) six times for 10 min each at room temperature. Primary antibodies were incubated overnight at 4°C, except for actin C4, 2G2, and 1C7, and Cofilin which were incubated for a minimum of 20 h at 4°C. The following additional antibodies and concentrations were used: rabbit anti-GFP 1:2000 (preabsorbed on yw ovaries at 1:20 and used at 1:100; Torrey Pines Biolabs, Secaucus, NJ); goat anti-GFP 1:2000 (preabsorbed on yw ovaries at 1:20 and used at 1:100; Fitzgerald, Acton, MA); rabbit anti-DsRed 1:500 (Clontech, Mountain View, CA); mouse anti–actin C4 1:50 (EMB Millipore, Billerica, MA); mouse anti–actin 2G2 1:25 (EMB Millipore); mouse anti-1C7 1:25 (BS Antibody Facility, Braunschweig, Germany); rabbit anti-cofilin AB3 1:100 (SAB Signalway, College Park, MD); and mouse anti-Profilin undiluted (obtained from the Developmental Studies Hybridoma Bank [DSHB] developed under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biology, University of Iowa, Iowa City, IA; chi 1J, Cooley, L. [ Verheyen and Cooley, 1994]). After six washes in Triton antibody wash (10 min each), secondary antibodies were incubated overnight at 4°C or for ∼4 h at room temperature. The following secondary antibodies were used at 1:250–1:500: AF488::goat anti-mouse, AF568::goat anti-mouse (immunoglobulin M–specific secondary was used for actin 2G2 staining), AF633::goat anti-mouse, AF488::goat anti-rabbit, AF568::goat anti-rabbit, AF488::donkey anti-goat, and AF568::donkey anti-rabbit (Life Technologies, Grand Island, NY). Alexa Fluor 647–, rhodamine- or Alexa Fluor 488–conjugated phalloidin (Life Technologies) was included with secondary antibodies at a concentration of 1:100–1:250. Alexa Fluor 555– or Alexa Fluor 647–conjugated WGA (Life Technologies) was included with the secondary antibody at a concentration of 1:500. After six washes in Triton antibody wash (10 min each), 4′,6-diamidino-2-phenylindole (5 mg/ml) staining was performed at a concentration of 1:5000 in 1× PBS for 10 min at room temperature. Ovaries were mounted in 1 mg/ml phenylenediamine in 50% glycerol, pH 9 (Platt and Michael, 1983). All experiments were performed a minimum of three independent times.
Image acquisition and processing
Microscope images of fixed Drosophila follicles were obtained using LAS AF SPE Core software on a Leica TCS SPE mounted on a Leica DM2500 using an ACS APO 20×/0.60 IMM CORR -/D or an ACS APO 63×/1.30 Oil CS 0.17/E objective (Leica Microsystems, Buffalo Grove, IL) Maximum projections (two to five confocal slices), merged images, rotation, and cropping were performed using ImageJ software (Abramoff et al., 2004). To aid in visualization, all panels were brightened by 30% in Photoshop (Adobe, San Jose, CA).
Live imaging
Whole ovaries were dissected from flies fed wet yeast paste for 4–6 d and maintained at 25°C. Ovaries were dissected in Stage 9 medium (Prasad et al., 2007): Schneider’s medium (Life Technologies), 0.6× penicillin/streptomycin (Life Technologies), 0.2 mg/ml insulin (Sigma-Aldrich, St. Louis, MO), and 15% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA). Ovarioles and individual early stage follicles (S3–9) were hand dissected and either placed in a drop of medium or embedded in 1.25% low-melt agarose (IBI Scientific, Peosta, IA) made with Stage 9 media on a coverslip-bottom dish (MatTek, Ashland, MA; Groen and Tootle, 2015). Live imaging of GFP-Actin was performed using Zen software on a Zeiss 700 LSM mounted on an Axio Observer.Z1 using a LD C-APO 40×/1.1 W/0 objective (Carl Zeiss Microscopy, Thornwood, NY). For FRAP experiments, GFP-Actin was photobleached using 100% laser power of the 488-nm laser for 50 iterations. From 100 to 200 images were obtained in a time series (three prebleach and the rest postbleach) with no delay between images. FRAP recovery curve analysis was performed using the FRAP Calculator Package plug-in in ImageJ (Abramoff et al., 2004). FRAP recovery curves of multiple nuclei were averaged and analyzed using Excel (Microsoft, Redmond, WA).
Quantification of nuclear actin rod frequency and length
Quantification of nuclear actin rods was performed on confocal image stacks of follicles stained with anti-GFP and WGA. Genotypically deidentified images were analyzed using ImageJ; as necessary, brightness and contrast were adjusted to score all of the actin rods present. Data were collected for S5–6, S7–8, and S9. Follicle staging was assigned based on morphology and size. For each follicle, the percentage of nurse cells exhibiting nuclear actin rods was assessed and binned into four categories: none, ≤25, 25–75, or ≥75%. The rods were then scored for length: short (≤1/4 diameter of nucleus), medium (∼1/2 diameter of nucleus), or long (≥1 diameter). Data were analyzed using Excel. Statistical analysis was performed using R (www.r-project.org).
Quantification of endogenous nuclear actin from immunofluorescence
Quantification of endogenous nuclear actin was performed on confocal image stacks of follicles stained with anti-actin C4 and WGA. Genotypically deidentified images were analyzed using ImageJ; as necessary, brightness and contrast were adjusted to score all of the structured nuclear actin present. Data were collected for S5–6, S7–8, and S9. For each follicle, the number of nurse cells exhibiting unstructured (haze) or structured nuclear actin, binned into three categories, low, medium, and high levels, was scored. Data were analyzed using Excel. Statistical analysis was performed using R.
Quantification of nuclear/cytoplasmic levels from immunofluorescence
Quantification of the nuclear-to-cytoplasmic ratios of Profilin and Cofilin was performed on confocal image stacks of follicles stained with anti-Profilin or anti-Cofilin, respectively, and WGA. Fluorescence intensity plots of nurse cell cytoplasm and nuclei were generated from a single slice of deidentified 20× confocal images using ImageJ software (Abramoff et al., 2004). Briefly, a line segment was drawn from the cytoplasm into the nucleus, and the plot profile function was used to generate a fluorescence intensity plot for each desired channel. The raw data files generated by these plot profiles were analyzed in Excel, with each plot line normalized to the peak value within that plot, creating intensity plots where the maximum observed fluorescence of a given line is represented by a value of 1.0 relative fluorescence intensity (RFI). The nuclear boundary was marked by the highest WGA value. All RFI averaging was performed in Excel. The cytoplasm was defined as all of the plot points to the left of the nuclear boundary. The nucleus was defined as the plot points at and to the right of the peak WGA value. Statistical analysis was performed using R.
Quantitative reverse transcriptase PCR
Whole ovaries were dissected from adult flies provided wet yeast paste for 3–5 d in room temperature Grace’s insect medium and homogenized in TRIzol (Thermo Scientific, Waltham, MA). Total RNA was extracted following manufacturer’s protocol, and samples were treated overnight with RNase-free DNase (Roche, Indianapolis, IN). After quantification, 400 ng of RNA was subjected to first-strand cDNA synthesis using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Quantitative real-time PCR analysis was preformed using iQ SYBR Green Supermix (Bio-Rad) following the manufacturer’s protocol on the Bio-Rad CFX96 Real-Time system. Primers for qPCR experiments were designed to span exon–exon junctions against twinstar (forward, 5′-GTGAAAGAAGGCGGAAGGTTAA-3′; reverse, 5′-CACAGTTACACCAGAAGCCATT-3′) and reference genes succinate dehydrogenase, subunit A (forward, 5′-CAAGGTTGTCGATAGGTCG-3′; reverse, 5′-CTCACAATAGTCATCTGGGC-3′) and cyclophillin-33 (forward, 5′-TGATACCCGAGTTTATGTGTC-3′; reverse, 5′-GGCCATTGAAAGAGTTCCA-3′).
Each primer was ∼100% efficient when examined against a serial dilution of cDNA and produced a single product observed through endpoint melt-curve analysis. The fold change in twinstar expression was determined using the 2-ΔΔCt method of quantification against two reference genes. Each sample was assessed in triplicate from three independent experiments. Data were analyzed and statistical significance determined against respective control using Excel.
Subcellular fractionation
Subcellular fractionation was performed using methods modified from Guilluy et al. (2011). In short, whole ovaries from adult females that had been provided wet yeast paste for 3–5 d were dissected in room temperature Grace’s insect medium. Ovaries were lysed in 1.5-ml microcentrifuge tubes on ice in 100 μl of hypotonic buffer (10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, 20 μg/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride) with gentle grinding with a plastic pestle. A rough nuclear pellet was separated from the cytoplasmic fraction by centrifugation at 300 × g for 10 min at 4°C (Centrifuge 5415R; Eppendorf, Hauppauge, NY). The nuclear pellet was further clarified using a 25% iodixanol gradient (OptiPrep 60% iodixanol solution [Sigma-Aldrich] diluted with 0.25 M sucrose, 25 mM KCl, 5 mM MgCl2, 20 mM Tris-Cl, pH 7.8). Specifically, the nuclear pellet was resuspended in iodixanol and centrifuged at 10,000 × g for 10 min at 4°C. Iodixanol supernatant was removed and the step repeated one time before resuspending the nuclear pellet in 50 mM Tris-HCl, pH 7.6, 500 mM NaCl, 1% Triton X-100, 0.1% SDS, and 10 mM MgCl2. Equivalent volumes of protein lysates for total cell lysate, cytoplasmic fraction, and nuclear fraction were analyzed by standard SDS–PAGE/Western blot analysis (see later description) using the following primary antibodies: mouse anti-Fascin 1:50–1:100 (sn7c, Cooley, L., DSHB), rabbit anti-GFP (Torre Pines) 1:10000; mouse anti-Lamin Dm0 (ADL195, Fisher, P.A., DSHB) 1:200; mouse anti-actin (JLA20, Lim, J.J-C, DSHB) 1:100–1:500; mouse anti–actin C4 (Signalway) 1:5000; and mouse anti-tubulin (Sigma-Aldrich) 1:5000. Nuclear Fascin, actin, and GFP-Actin densitometry was normalized to nuclear Lamin, and total levels were normalized to total α-Tubulin. Densitometry values were made relative to respective experimental controls. Data were analyzed using Excel and Prism. A minimum of three independent experiments were performed for each subcellular fractionation experiment.
Western blotting
Western blots were performed using standard methods with the primary antibodies referred to in the Subcellular fractionation section and the following antibodies: mouse anti-Profilin (chi 1J, Cooley, L., DSHB) 1:10; mouse anti–actin 2G2 (Millipore) 1:1000, and mouse anti–actin 1C7 (BS Antibody Facility) 1:500. All blots had 0.1% Tween 20 added to the primary antibody in 5% milk diluted in 1× Tris-buffered saline. The following secondary antibodies were used: Peroxidase-AffiniPure Goat Anti-Rabbit IgG (H + L) (1:5000) or Peroxidase-AffiniPure Goat Anti-Mouse IgG (H + L) (1:5000; Jackson ImmunoResearch Laboratories, West Grove, PA). Blots were developed with SuperSignal West Pico or Femto Chemiluminescent Substrate (Thermo Scientific, Waltham, MA) and imaged using ChemiDoc-It Imaging System and VisionWorksLS software (UVP, Upland, CA). Bands were quantified using the gel analyzer function of ImageJ (Abramoff et al., 2004). Densitometry values were made relative to respective experimental controls. Data were analyzed using Excel and Prism. A minimum of three independent experiments were performed for each Western blotting experiment.
Supplementary Material
Acknowledgments
We thank the Lin and Frank labs and Martine Dunnwald for helpful discussions and members of the Tootle lab for helpful discussions and careful review of the manuscript. Information Technology Services–Research Services, University of Iowa, provided data storage support. This project was supported by National Science Foundation Grant MCB-1158527, National Institutes of Health Grant R01GM116885, and a Carver Trust Medical Research Initiative Grant. D.J.K. is partially supported by National Institute of Neurological Disorders and Stroke Grant T32NS045549.
Abbreviations used:
- F-actin
filamentous actin
- FRAP
fluorescence recovery after photobleaching
- Sx
Stage x of oogenesis
- WGA
wheat germ agglutinin.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-09-0634) on August 17, 2016.
REFERENCES
- Abramoff M, Magalhaes P, Ram S. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Agnew BJ, Minamide LS, Bamburg JR. Reactivation of phosphorylated actin depolymerizing factor and identification of the regulatory site. J Biol Chem. 1995;270:17582–17587. doi: 10.1074/jbc.270.29.17582. [DOI] [PubMed] [Google Scholar]
- Asumda FZ, Chase PB. Nuclear cardiac troponin and tropomyosin are expressed early in cardiac differentiation of rat mesenchymal stem cells. Differentiation. 2012;83:106–115. doi: 10.1016/j.diff.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Belin BJ, Cimini BA, Blackburn EH, Mullins RD. Visualization of actin filaments and monomers in somatic cell nuclei. Mol Biol Cell. 2013;24:982–994. doi: 10.1091/mbc.E12-09-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin BJ, Lee T, Mullins RD. DNA damage induces nuclear actin filament assembly by Formin -2 and Spire-(1/2) that promotes efficient DNA repair. [corrected] Elife. 2015;4:e07735. doi: 10.7554/eLife.07735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Stuven T, Kuhn C, Cordes VC, Gorlich D. A selective block of nuclear actin export stabilizes the giant nuclei of Xenopus oocytes. Nat Cell Biol. 2006;8:257–263. doi: 10.1038/ncb1357. [DOI] [PubMed] [Google Scholar]
- Breitsprecher D, Koestler SA, Chizhov I, Nemethova M, Mueller J, Goode BL, Small JV, Rottner K, Faix J. Cofilin cooperates with fascin to disassemble filopodial actin filaments. J Cell Sci. 2011;124:3305–3318. doi: 10.1242/jcs.086934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cant K, Knowles BA, Mooseker MS, Cooley L. Drosophila singed, a fascin homolog, is required for actin bundle formation during oogenesis and bristle extension. J Cell Biol. 1994;125:369–380. doi: 10.1083/jcb.125.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb JM, Orr-Weaver TL. Developmental gene amplification: insights into DNA replication and gene expression. Trends Genet. 2005;21:149–162. doi: 10.1016/j.tig.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Cummings MR, King RC. Ultrastructural changes in nurse and follicle cells during late stages of oogenesis in Drosophila melanogaster. Z Zellforch Microsk Anat. 1970;110:1–8. doi: 10.1007/BF00343981. [DOI] [PubMed] [Google Scholar]
- Dialynas G, Speese S, Budnik V, Geyer PK, Wallrath LL. The role of Drosophila Lamin C in muscle function and gene expression. Development. 2010;137:3067–3077. doi: 10.1242/dev.048231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobens LL, Raftery LA. Integration of epithelial patterning and morphogenesis in Drosophila ovarian follicle cells. Dev Dyn. 2000;218:80–93. doi: 10.1002/(SICI)1097-0177(200005)218:1<80::AID-DVDY7>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Domazetovska A, Ilkovski B, Cooper ST, Ghoddusi M, Hardeman EC, Minamide LS, Gunning PW, Bamburg JR, North KN. Mechanisms underlying intranuclear rod formation. Brain. 2007a;130:3275–3284. doi: 10.1093/brain/awm247. [DOI] [PubMed] [Google Scholar]
- Domazetovska A, Ilkovski B, Kumar V, Valova VA, Vandebrouck A, Hutchinson DO, Robinson PJ, Cooper ST, Sparrow JC, Peckham M, et al. Intranuclear rod myopathy: molecular pathogenesis and mechanisms of weakness. Ann Neurol. 2007b;62:597–608. doi: 10.1002/ana.21200. [DOI] [PubMed] [Google Scholar]
- Dopie J, Rajakyla EK, Joensuu MS, Huet G, Ferrantelli E, Xie T, Jaalinoja H, Jokitalo E, Vartiainen MK. Genome-wide RNAi screen for nuclear actin reveals a network of cofilin regulators. J Cell Sci. 2015;128:2388-2400. doi: 10.1242/jcs.169441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopie J, Skarp KP, Rajakyla EK, Tanhuanpaa K, Vartiainen MK. Active maintenance of nuclear actin by importin 9 supports transcription. Proc Natl Acad Sci USA. 2012;109:E544–E552. doi: 10.1073/pnas.1118880109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y, Katsumaru H. Nuclear actin bundles in Amoeba, Dictyostelium and human HeLa cells induced by dimethyl sulfoxide. Exp Cell Res. 1979;120:451–455. doi: 10.1016/0014-4827(79)90412-9. [DOI] [PubMed] [Google Scholar]
- Gedge LJ, Morrison EE, Blair GE, Walker JH. Nuclear actin is partially associated with Cajal bodies in human cells in culture and relocates to the nuclear periphery after infection of cells by adenovirus 5. Exp Cell Res. 2005;303:229-239. doi: 10.1016/j.yexcr.2004.06.034. [DOI] [PubMed] [Google Scholar]
- Goebel HH, Warlo IA. Surplus protein myopathies. Neuromuscul Disord. 2001;11:3–6. doi: 10.1016/s0960-8966(00)00165-6. [DOI] [PubMed] [Google Scholar]
- Gonsior SM, Platz S, Buchmeier S, Scheer U, Jockusch BM, Hinssen H. Conformational difference between nuclear and cytoplasmic actin as detected by a monoclonal antibody. J Cell Sci. 1999;112:797–809. doi: 10.1242/jcs.112.6.797. [DOI] [PubMed] [Google Scholar]
- Groen CM, Jayo A, Parsons M, Tootle TL. Prostaglandins regulate nuclear localization of Fascin and its function in nucleolar architecture. Mol Biol Cell. 2015;26:1901–1917. doi: 10.1091/mbc.E14-09-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen CM, Tootle TL. Visualization of actin cytoskeletal dynamics in fixed and live Drosophila egg chambers. Methods Mol Biol. 2015;1328:113–124. doi: 10.1007/978-1-4939-2851-4_8. [DOI] [PubMed] [Google Scholar]
- Grosse R, Vartiainen MK. To be or not to be assembled: progressing into nuclear actin filaments. Nat Rev Mol Cell Biol. 2013;14:693–697. doi: 10.1038/nrm3681. [DOI] [PubMed] [Google Scholar]
- Guilluy C, Dubash AD, Garcia-Mata R. Analysis of RhoA and Rho GEF activity in whole cells and the cell nucleus. Nat Protoc. 2011;6:2050–2060. doi: 10.1038/nprot.2011.411. [DOI] [PubMed] [Google Scholar]
- Hendzel MJ. The F-act’s of nuclear actin. Curr Opin Cell Biol. 2014;28:84–89. doi: 10.1016/j.ceb.2014.04.003. [DOI] [PubMed] [Google Scholar]
- Hitchcock SE. Actin deoxyroboncuclease I interaction. Depolymerization and nucleotide exchange. J Biol Chem. 1980;255:5668–5673. [PubMed] [Google Scholar]
- Hofmann WA, Stojiljkovic L, Fuchsova B, Vargas GM, Mavrommatis E, Philimonenko V, Kysela K, Goodrich JA, Lessard JL, Hope TJ, et al. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat Cell Biol. 2004;6:1094–1101. doi: 10.1038/ncb1182. [DOI] [PubMed] [Google Scholar]
- Iida K, Iida H, Yahara I. Heat shock induction of intranuclear actin rods in cultured mammalian cells. Exp Cell Res. 1986;165:207–215. doi: 10.1016/0014-4827(86)90545-8. [DOI] [PubMed] [Google Scholar]
- Jayo A, Malboubi M, Antoku S, Chang W, Ortiz-Zapater E, Groen C, Pfisterer K, Tootle T, Charras G, Gundersen GG, Parsons M. Fascin regulates nuclear movement and deformation in migrating cells. Dev Cell. 2016;38:371–383. doi: 10.1016/j.devcel.2016.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayo A, Parsons M. Fascin: a key regulator of cytoskeletal dynamics. Int J Biochem Cell Biol. 2010;42:1614–1617. doi: 10.1016/j.biocel.2010.06.019. [DOI] [PubMed] [Google Scholar]
- Jayo A, Parsons M, Adams JC. A novel Rho-dependent pathway that drives interaction of fascin-1 with p-Lin-11/Isl-1/Mec-3 kinase (LIMK) 1/2 to promote fascin-1/actin binding and filopodia stability. BMC Biol. 2012;10:72. doi: 10.1186/1741-7007-10-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jockusch BM, Schoenenberger CA, Stetefeld J, Aebi U. Tracking down the different forms of nuclear actin. Trends Cell Biol. 2006;16:391–396. doi: 10.1016/j.tcb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Lane NJ. Intranuclear fibrillar bodies in actinomycin D-treated oocytes. J Cell Biol. 1969;40:286–291. doi: 10.1083/jcb.40.1.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen P, Drubin DG. Cofilin promotes rapid actin filament turnover in vivo. Nature. 1997;388:78–82. doi: 10.1038/40418. [DOI] [PubMed] [Google Scholar]
- Lenart P, Bacher CP, Daigle N, Hand AR, Eils R, Terasaki M, Ellenberg J. A contractile nuclear actin network drives chromosome congression in oocytes. Nature. 2005;436:812–818. doi: 10.1038/nature03810. [DOI] [PubMed] [Google Scholar]
- Lessard JL. Two monoclonal antibodies to actin: one muscle selective and one generally reactive. Cell Motil Cytoskeleton. 1988;10:349–362. doi: 10.1002/cm.970100302. [DOI] [PubMed] [Google Scholar]
- Lilly MA, Spradling AC. The Drosophila endocycle is controlled by Cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev. 1996;10:2514–2526. doi: 10.1101/gad.10.19.2514. [DOI] [PubMed] [Google Scholar]
- Lim MK, Kawamura T, Ohsawa Y, Ohtsubo M, Asakawa S, Takayanagi A, Shimizu N. Parkin interacts with LIM Kinase 1 and reduces its cofilin-phosphorylation activity via ubiquitination. Exp Cell Res. 2007;313:2858–2874. doi: 10.1016/j.yexcr.2007.04.016. [DOI] [PubMed] [Google Scholar]
- Maloney MT, Bamburg JR. Cofilin-mediated neurodegeneration in Alzheimer’s disease and other amyloidopathies. Mol Neurobiol. 2007;35:21–44. doi: 10.1007/BF02700622. [DOI] [PubMed] [Google Scholar]
- Maloney MT, Minamide LS, Kinley AW, Boyle JA, Bamburg JR. Beta-secretase-cleaved amyloid precursor protein accumulates at actin inclusions induced in neurons by stress or amyloid beta: a feedforward mechanism for Alzheimer’s disease. J Neurosci. 2005;25:11313–11321. doi: 10.1523/JNEUROSCI.3711-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslova A, Krasikova A. Nuclear actin depolymerization in transcriptionally active avian and amphibian oocytes leads to collapse of intranuclear structures. Nucleus. 2012;3:300–311. doi: 10.4161/nucl.20393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Carrero G, Andrin C, de Vries G, Hendzel MJ. Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J Cell Biol. 2006;172:541–552. doi: 10.1083/jcb.200507101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamide LS, Striegl AM, Boyle JA, Meberg PJ, Bamburg JR. Neurodegenerative stimuli induce persistent ADF/cofilin-actin rods that disrupt distal neurite function. Nat Cell Biol. 2000;2:628–636. doi: 10.1038/35023579. [DOI] [PubMed] [Google Scholar]
- Munsie L, Caron N, Atwal RS, Marsden I, Wild EJ, Bamburg JR, Tabrizi SJ, Truant R. Mutant huntingtin causes defective actin remodeling during stress: defining a new role for transglutaminase 2 in neurodegenerative disease. Hum Mol Genet. 2011;20:1937–1951. doi: 10.1093/hmg/ddr075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munsie LN, Desmond CR, Truant R. Cofilin nuclear-cytoplasmic shuttling affects cofilin-actin rod formation during stress. J Cell Sci. 2012;125:3977–3988. doi: 10.1242/jcs.097667. [DOI] [PubMed] [Google Scholar]
- Nishida E, Iida K, Yonezawa N, Koyasu S, Yahara I, Sakai H. Cofilin is a component of intranuclear and cytoplasmic actin rods induced in cultured cells. Proc Natl Acad Sci USA. 1987;84:5262–5266. doi: 10.1073/pnas.84.15.5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn M, Weber K. Dimethylsulfoxide and the ionophore A23187 affect the arrangement of actin and induce nuclear actin paracrystals in PtK2 cells. Exp Cell Res. 1980;129:103–114. doi: 10.1016/0014-4827(80)90335-3. [DOI] [PubMed] [Google Scholar]
- Parfenov VN, Davis DS, Pochukalina GN, Sample CE, Bugaeva EA, Murti KG. Nuclear actin filaments and their topological changes in frog oocytes. Exp Cell Res. 1995;217:385–394. doi: 10.1006/excr.1995.1101. [DOI] [PubMed] [Google Scholar]
- Pendleton A, Pope B, Weeds A, Koffer A. Latrunculin B or ATP depletion induces cofilin-dependent translocation of actin into nuclei of mast cells. J Biol Chem. 2003;278:14394–14400. doi: 10.1074/jbc.M206393200. [DOI] [PubMed] [Google Scholar]
- Percipalle P. Co-transcriptional nuclear actin dynamics. Nucleus. 2013;4:43–52. doi: 10.4161/nucl.22798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt JL, Michael AF. Retardation of fading and enhancement of intensity of immunofluorescence by p-phenylenediamine. J Histochem Cytochem. 1983;31:840–842. doi: 10.1177/31.6.6341464. [DOI] [PubMed] [Google Scholar]
- Plessner M, Melak M, Chinchilla P, Baarlink C, Grosse R. Nuclear F-actin formation and reorganization upon cell spreading. J Biol Chem. 2015;290:11209–11216. doi: 10.1074/jbc.M114.627166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad M, Jang AC, Starz-Gaiano M, Melani M, Montell DJ. A protocol for culturing Drosophila melanogaster stage 9 egg chambers for live imaging. Nat Protoc. 2007;2:2467–2473. doi: 10.1038/nprot.2007.363. [DOI] [PubMed] [Google Scholar]
- Rohn JL, Sims D, Liu T, Fedorova M, Schock F, Dopie J, Vartiainen MK, Kiger AA, Perrimon N, Baum B. Comparative RNAi screening identifies a conserved core metazoan actinome by phenotype. J Cell Biol. 2011;194:789–805. doi: 10.1083/jcb.201103168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper K, Mao Y, Brown NH. Contribution of sequence variation in Drosophila actins to their incorporation into actin-based structures in vivo. J Cell Sci. 2005;118:3937–3948. doi: 10.1242/jcs.02517. [DOI] [PubMed] [Google Scholar]
- Rorth P. Gal4 in the Drosophila female germline. Mech Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- Sameshima M, Kishi Y, Osumi M, Minamikawa-Tachino R, Mahadeo D, Cotter DA. The formation of actin rods composed of actin tubules in Dictyostelium discoideum spores. J Struct Biol. 2001;136:7–19. doi: 10.1006/jsbi.2001.4424. [DOI] [PubMed] [Google Scholar]
- Schoenenberger CA, Buchmeier S, Boerries M, Sutterlin R, Aebi U, Jockusch BM. Conformation-specific antibodies reveal distinct actin structures in the nucleus and the cytoplasm. J Struct Biol. 2005;152:157–168. doi: 10.1016/j.jsb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Sen B, Xie Z, Uzer G, Thompson WR, Styner M, Wu X, Rubin J. Intranuclear actin regulates osteogenesis. Stem Cells. 2015;33:3065–3076. doi: 10.1002/stem.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer VA, Costes S, Inman JL, Xu R, Chen J, Hendzel MJ, Bissell MJ. Depletion of nuclear actin is a key mediator of quiescence in epithelial cells. J Cell Sci. 2011;124:123–132. doi: 10.1242/jcs.073197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spracklen AJ, Fagan TN, Lovander KE, Tootle TL. The pros and cons of common actin labeling tools for visualizing actin dynamics during Drosophila oogenesis. Dev Biol. 2014;393:209–226. doi: 10.1016/j.ydbio.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC. Developmental genetics of oogenesis. In: Martinez-Arias B, editor. The Development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 1–70. [Google Scholar]
- Stuven T, Hartmann E, Gorlich D. Exportin 6: a novel nuclear export receptor that is specific for profilin.actin complexes. EMBO J. 2003;22:5928–5940. doi: 10.1093/emboj/cdg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telley IA, Gaspar I, Ephrussi A, Surrey T. Aster migration determines the length scale of nuclear separation in the Drosophila syncytial embryo. J Cell Biol. 2012;197:887–895. doi: 10.1083/jcb.201204019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf WE, Smiley S, Wong ML, Alberts BM. Reorganization of the cytoskeleton during Drosophila oogenesis: implications for axis specification and intercellular transport. Development. 1992;115:923–936. doi: 10.1242/dev.115.4.923. [DOI] [PubMed] [Google Scholar]
- Tootle TL, Williams D, Hubb A, Frederick R, Spradling A. Drosophila eggshell production: identification of new genes and coordination by Pxt. PLoS One. 2011;6:e19943. doi: 10.1371/journal.pone.0019943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen MK. Nuclear actin dynamics–from form to function. FEBS Lett. 2008;582:2033–2040. doi: 10.1016/j.febslet.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Vartiainen MK, Guettler S, Larijani B, Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 2007;316:1749–1752. doi: 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- Verheyen EM, Cooley L. Profilin mutations disrupt multiple actin-dependent processes during Drosophila development. Development. 1994;120:717–728. doi: 10.1242/dev.120.4.717. [DOI] [PubMed] [Google Scholar]
- Viita T, Vartiainen MK. From cytoskeleton to gene expression: actin in the nucleus. Handb Exp Pharmacol. 2016. doi: 10.1007/164_2016_27. [DOI] [PubMed]
- Visa N, Percipalle P. Nuclear functions of actin. Cold Spring Harb Perspect Biol. 2010;2:a000620. doi: 10.1101/cshperspect.a000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada A, Fukuda M, Mishima M, Nishida E. Nuclear export of actin: a novel mechanism regulating the subcellular localization of a major cytoskeletal protein. EMBO J. 1998;17:1635–1641. doi: 10.1093/emboj/17.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanet J, Jayo A, Plaza S, Millard T, Parsons M, Stramer B. Fascin promotes filopodia formation independent of its role in actin bundling. J Cell Biol. 2012;197:477–486. doi: 10.1083/jcb.201110135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.