A novel, interacting protein with acetyl-CoA carboxylase resembles a known subunit but functions as a dominant-negative regulator of this multienzyme complex.
Abstract
Acetyl-CoA carboxylase (ACCase) catalyzes the committed step of de novo fatty acid biosynthesis. In prokaryotes, green algae, and most plants, this enzyme is a heteromeric complex requiring four different subunits for activity. The plant complex is recalcitrant to conventional purification schemes and hence the structure and composition of the full assembly have been unclear. In vivo coimmunoprecipitation using subunit-specific antibodies identified a novel family of proteins in Arabidopsis thaliana annotated as biotin/lipoyl attachment domain containing (BADC) proteins. Results from yeast two-hybrid and coexpression in Escherichia coli confirmed that all three BADC isoforms interact with the two biotin carboxyl carrier protein (BCCP) isoforms of Arabidopsis ACCase. These proteins resemble BCCP subunits but are not biotinylated due to a mutated biotinylation motif. We demonstrate that BADC proteins significantly inhibit ACCase activity in both E. coli and Arabidopsis. Targeted gene silencing of BADC isoform 1 in Arabidopsis significantly increased seed oil content when normalized to either mass or individual seed. We conclude the BADC proteins are ancestral BCCPs that gained a new function as negative regulators of ACCase after initial loss of the biotinylation motif. A functional model is proposed.
INTRODUCTION
Plant oils are an important renewable source of hydrocarbons for food, energy, and industrial feedstocks (Thelen and Ohlrogge, 2002a). Acyl chains stored as triacylglycerol are produced by the de novo fatty acid biosynthesis (FAS) pathway localized to plastids. The committed step of de novo FAS is catalyzed by acetyl-CoA carboxylase, which carboxylates acetyl-CoA to form malonyl-CoA in a two-step reaction requiring ATP, bicarbonate, and biotin cofactor (for recent review, see Salie and Thelen, 2016). Two forms of acetyl-CoA carboxylase are present in nature: a heteromeric (hetACCase) and a homomeric form (homACCase). In prokaryotes, and in plastids of dicots and nongraminaceous monocots, hetACCase is a complex requiring four distinct subunits: biotin carboxylase (BC), biotin carboxyl carrier protein (BCCP), and α- and β-carboxyltransferase (CT) (Guchhait et al., 1974; Konishi and Sasaki, 1994). In contrast, graminaceous monocots possess a homACCase form for plastidial de novo FAS, wherein the catalytic components are fused in tandem as a single polypeptide. A homACCase has also been identified in the plastids of dicot species (Schulte et al., 1997), but its metabolic role is unclear. Structural models for the plant hetACCase are based on the Escherichia coli homolog. The E. coli hetACCase is composed of two enzymatic subcomplexes: an α/β-CT heterotetramer and a BC/BCCP heterooctamer (Cronan and Waldrop, 2002; Thelen and Ohlrogge, 2002b; Sasaki and Nagano, 2004; Baud and Lepiniec, 2009). The components of each subcomplex form stable associations, while the two subcomplexes themselves show a relatively weak interaction with one another. This property has contributed to the difficulties in biochemical and structural characterization of hetACCase from plants.
Regulation of hetACCase activity involves multiple mechanisms. HetACCase is activated by light, during which its activity increases severalfold in the plastid. This activation occurs within minutes and is reportedly due to the increase in stromal pH from 7.0 to 8.0 (Hunter and Ohlrogge, 1998) and thioredoxin-dependent reduction of a disulfide bond in CT (Sasaki et al., 1997; Kozaki et al., 2001). HetACCase activity is also suppressed by feedback inhibition through increased levels of acyl-acyl carrier protein (Davis and Cronan, 2001; Andre et al., 2012). Recently, an interaction between hetACCase and a 2-oxoglutarate binding protein PII was shown to reduce hetACCase activity (Feria Bourrellier et al., 2010). Recombinant PII protein was shown to pull down BCCP from chloroplast lysates in the absence of 2-oxoglutarate, likely through a direct interaction with the biotin cofactor. Addition of 2-oxoglutarate abrogates the interaction. Lastly, two types of BCCPs were identified in plants and have been shown to be under different transcriptional control (Thelen et al., 2001). The BCCP1 isoform is constitutively expressed, while BCCP2 is predominantly expressed in the seed under the control of the WRINKLED1 transcription factor (Thelen et al., 2001; Ruuska et al., 2002; Baud et al., 2009). Of these two isoforms, BCCP2 was shown to be more active in vitro (Thelen et al., 2001) but nonessential to plant growth (Li et al., 2011).
Other properties of the plant hetACCase suggest additional, noncatalytic components may be involved in its function or regulation: (1) the observed size of the plant hetACCase complex is larger than the calculated mass of the known subunits (Reverdatto et al., 1999; Olinares et al., 2010); (2) the α- and β-CT subunits contain large domains of 200 to 300 residues that are not required for catalytic activity, are not present in prokaryotic homologs, and have no known function (Kozaki et al., 2001; Cronan and Waldrop, 2002); (3) the CT subcomplex associates with the plastid inner envelope through an unknown, non-ionic interaction (Thelen and Ohlrogge, 2002b); and 4) the α-CT subunit is phosphorylated on multiple sites but its functional role is unclear (Nakagami et al., 2010; Meyer et al., 2012). Collectively, these observations suggest that hetACCase may possess additional factors that facilitate the various forms of regulation. To discover novel interactors with hetACCase, we employed coimmunoprecipitation coupled with liquid chromatography-tandem mass spectrometry (LC-MS/MS). The results presented here show that a family of genes annotated to encode biotin attachment domain-containing (BADC) proteins interacts with and inhibits hetACCase activity in both E. coli and Arabidopsis thaliana.
RESULTS
Two Novel Proteins, BADC1 and BADC2, Coimmunoprecipitate with hetACCase
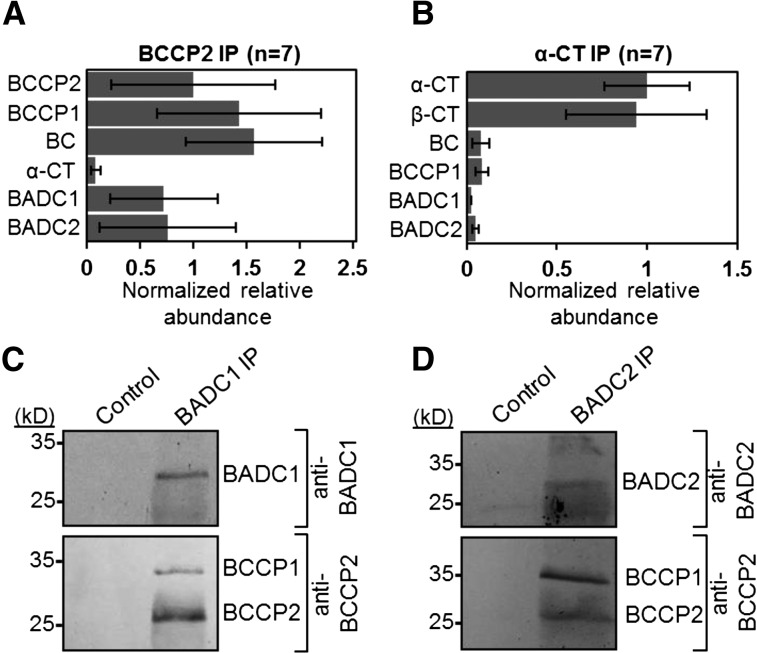
To discover unknown protein interactors with the hetACCase, quantitative coimmunoprecipitation (co-IP) analyses were performed. Clarified chloroplast lysates from 14-d-old Arabidopsis seedlings were incubated with Protein A Sepharose beads coated with polyclonal antibodies against either BCCP2 or α-CT. Control precipitations were performed using uncoated beads. Precipitated proteins were identified by LC-MS/MS analysis of trypsin-digested peptides. From seven biological replicates of α-CT co-IPs, all four catalytic subunits of the hetACCase complex were identified. Likewise, all catalytic subunits, except β-CT, were identified from co-IPs with antibodies against BCCP2 (Figures 1A and 1B). As expected, the BC/BCCP and α/β-CT subcomplexes were more abundant in the BCCP2 and α-CT co-IPs, respectively. Additionally, two proteins of unknown function containing a presumptive biotin attachment domain-containing region (TAIR), hereafter termed BADC1 and BADC2, were identified from both sets of co-IPs. The BADC1 protein was present in seven and one replicates of the BCCP2 and α-CT co-IPs, respectively, while BADC2 was present in six and two replicates of the BCCP2 and α-CT co-IPs, respectively. For comparison, BCCP1 was present in seven and three replicates of the BCCP2 and α-CT co-IPs, respectively, while BC was present in seven and five replicates of the BCCP2 and α-CT co-IPs, respectively. The normalized, relative abundance of BADC proteins was more commensurate with BC and BCCP abundance than α- and β-CT from both co-IP analyses. In addition, the BADC proteins were the most abundant non-hetACCase proteins in the BCCP2 co-IPs, implicating them as putative interacting proteins. A summary of the mass spectrometry data used to calculate the normalized relative abundance can be found in Supplemental Figure 1. Reciprocal co-IPs using antibodies specific to BADC1 and BADC2 precipitated both BCCP isoforms (Figures 1C and 1D). Thus, BADC1 and BADC2 appear to preferentially interact with the BC/BCCP component of hetACCase.
Figure 1.
Coimmunoprecipitation of ACCase and BADC Proteins from Arabidopsis Seedlings.
(A) and (B) Proteins were precipitated from Arabidopsis crude chloroplast lysate using antibodies specific for ACCase subunits BCCP2 (A) or α-CT (B) and identified by LC-MS/MS. Control precipitations were performed with uncoated Protein A Sepharose beads. For both sets of experiments, n = 7. Normalized relative abundance averages signify the average number of peptide spectral matches divided by the protein molecular weight and normalized to BCCP2 (A) or α-CT (B). Error bars represent se of normalized relative abundance among replicates in which the protein was identified.
(C) and (D) Protein blot analysis of reciprocal co-IPs from the same Arabidopsis lysate showed that the BCCP subunits of ACCase coprecipitated with BADC1 (C) and BADC2 (D). Blots are representative of three biological replicates. Each biological replicate consisted of a chloroplast lysate isolated from at least 5 g harvested leaf tissue.
Yeast Two-Hybrid and Heterologous Coexpression Confirm Direct Interactions between BADC and BCCP Isoforms from Arabidopsis
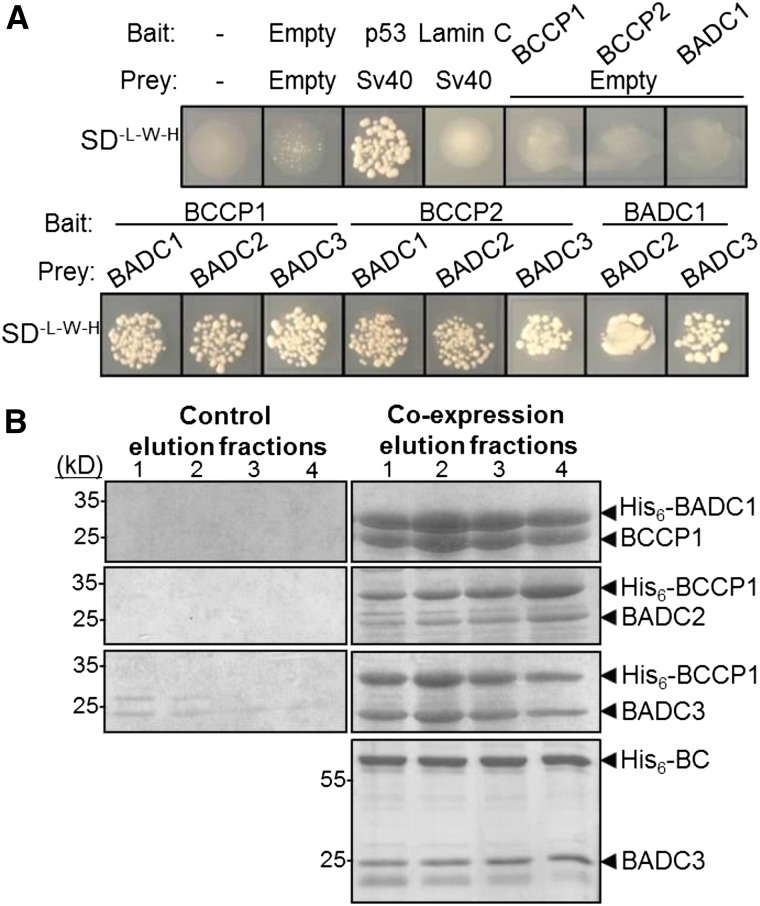
To confirm the co-IP results suggesting interaction with BC/BCCP, targeted yeast two-hybrid analysis was employed. In addition to the two experimentally identified BADCs, we also tested a third, putative BADC isoform, termed BADC3 (AT3G15690), identified by BLAST interrogation of the Arabidopsis genome. This protein shares 61% amino acid sequence identity with BADC2, suggesting similar function. Yeast two-hybrid analyses showed each of the three BADC proteins interacted with each BCCP isoform (Figure 2A). Additionally, the BADC isoforms are capable of interacting with each other.
Figure 2.
BADC Proteins Directly Interact with BCCP Subunits of ACCase.
(A) Strain AH109 yeast was transformed with bait and prey constructs containing the genes shown and plated on medium lacking Trp, Leu, and His. Negative controls showed minimal or no growth. Sv40 and p53 were used as a positive control. Lamin C was used as a negative control. Results shown are representative of three biological replicates. Each biological replicate was performed by transforming a separate culture of AH109 yeast.
(B) Coomassie-stained gels showing the elution fractions of Ni2+-NTA-purified protein from E. coli. At right, a native protein was coexpressed with a His6-tagged protein. At left, the native protein was expressed alone. Protein identities were confirmed by LC-MS/MS.
To further verify the interaction between BADC and BCCP, Arabidopsis BCCP1 was coexpressed with each of the three Arabidopsis BADC proteins in E. coli. In these experiments, either the BADC or BCCP1 protein was expressed with a His6 tag, while the other contained no affinity tag. When the His6-tagged protein was purified by Ni2+-NTA affinity chromatography, the respective “untagged” protein was present in the same elution fractions (Figure 2B). As a control, it was verified that untagged proteins were unable to bind to the affinity column. This approach confirmed the interaction between BADC and BCCP isoforms. In addition, BADC3 was observed to copurify with His6-BC using the same method (Figure 2B), suggesting that BADCs can bind to BC directly.
Biotin Is Not Required for BADC-BCCP Interaction
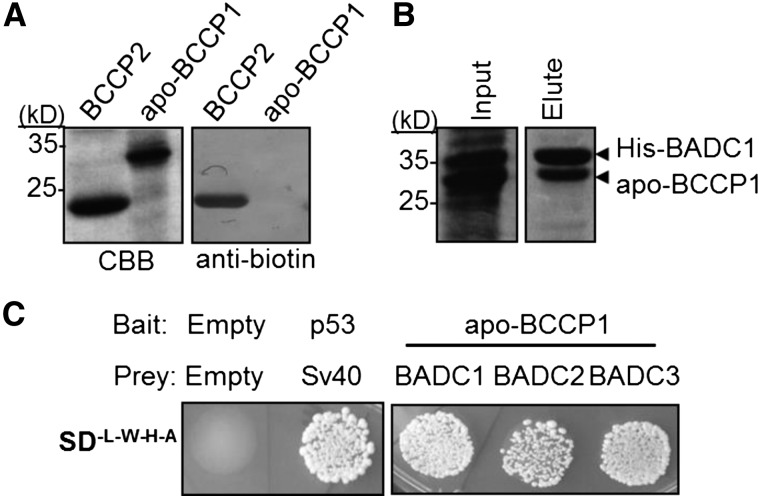
To determine if the BCCP-BADC interaction involves the biotin cofactor, as previously reported for the PII interaction with hetACCase (Gerhardt et al., 2015), the biotinyl Lys-245 residue on BCCP1 was mutated to Arg by site-directed mutagenesis. This mutation prevents biotinylation of BCCP1 (Figure 3A). Using this ”apo-BCCP1,” we repeated yeast two-hybrid and coexpression analysis with BADCs. All BADC isoforms were shown to interact with apo-BCCP1 (Figures 3B and 3C).
Figure 3.
Biotinylation Is Not Required for BADC-BCCP Interaction.
(A) Purified recombinant His6-tagged BCCP2 or apo-BCCP1 was resolved by SDS-PAGE and stained with Coomassie blue or blotted with antibiotin antibody.
(B) Coomassie-stained gels showing the elution fractions of Ni2+-NTA-purified protein from E. coli. Input shows that both proteins are expressed. Apo-BCCP1 was strongly present in the elution fraction with His6-BADC1. Protein identities were confirmed by LC-MS/MS.
(C) Strain AH109 yeast was transformed with bait and prey constructs containing the genes shown and plated on medium lacking Trp, Leu, His, and adenine. Empty vector negative control showed no growth. Sv40 and p53 were used as a positive control. Results shown are representative of three biological replicates, i.e., three separate transformation events.
Recombinant At-BADC1 and At-BADC3 Form Homodimers through a Disulfide Bond
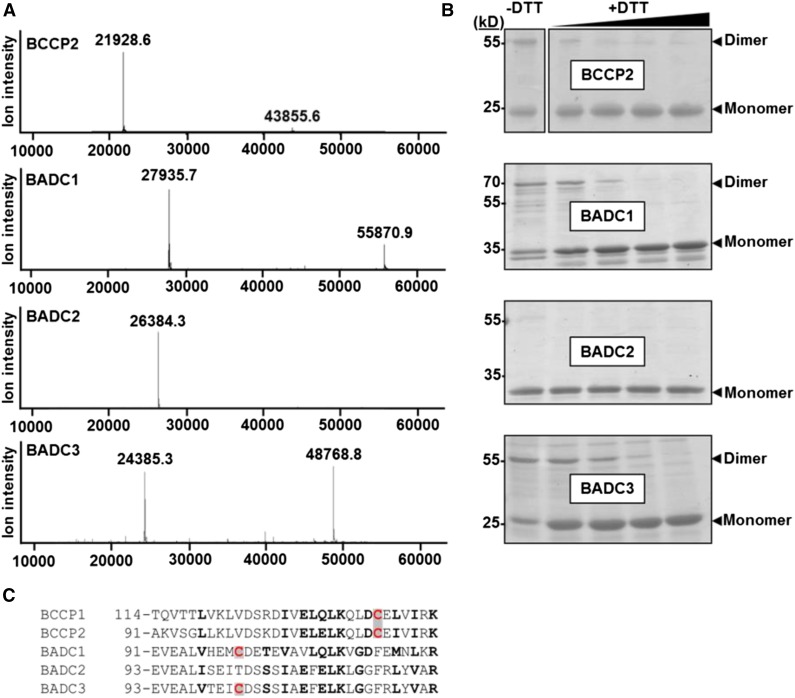
Previous analysis of E. coli BCCP suggested that this subunit forms homodimers when incorporated into the BC/BCCP subcomplex in vivo (Cronan, 2001). Through intact mass analysis of purified recombinant BCCP2, we observed that plant BCCP can also form homodimers (Figure 4A). In addition, analysis of recombinant BADCs showed that BADC1 and BADC3, but not BADC2, can form homodimers (Figure 4A). The observed monomer mass for each BADC was in agreement with the predicted mass, indicating that these proteins are unmodified. During nonreducing SDS-PAGE, purified recombinant BCCP2, BADC1, and BADC3 show both a monomer and dimer band, while BADC2 shows only a monomer band (Figure 4B). Addition of the reducing agent DTT led to the disappearance of the homodimer band, suggesting a disulfide bond is involved in homodimer formation of BADCs and plant BCCPs. Disulfide bonds are also used to link the α- and β-CT subunits of the CT subcomplex (Kozaki et al., 2001).
Figure 4.
Recombinant BADC Proteins Form Homodimers through Disulfide Bonds.
(A) Spectra show the intact mass spectrometry analysis of purified recombinant BCCP2, BADC1, BADC2, and BADC3. All proteins showed a monomer and homodimer peak except for BADC2.
(B) Purified recombinant proteins were incubated in 3× SDS sample buffer containing 0, 1, 2.5, 5, or 10 mM DTT, resolved by SDS-PAGE, and Coomassie-stained. Monomer and homodimer bands are marked. Protein identities were confirmed by mass spectrometry. Gels are representative of three replicates. Each replicate consisted of incubation of recombinant protein with DTT.
(C) Partial alignment of protein sequences from Arabidopsis. Conserved and similar residues are in bold; Cys residues are highlighted in red.
The presumptive mature primary sequences of the BADC proteins in Arabidopsis contain only two or three Cys residues (Supplemental Figure 2). Of these, only one Cys residue is conserved in BADC1 and BADC3, but not BADC2 (Figure 4C). Therefore, this Cys residue is the most likely candidate for disulfide bond formation. Similarly, the mature BCCP subunits only contain two Cys residues, one of which is only 15 residues downstream of the candidate Cys residue in BADCs (Figure 4C), implicating this residue in disulfide bond formation.
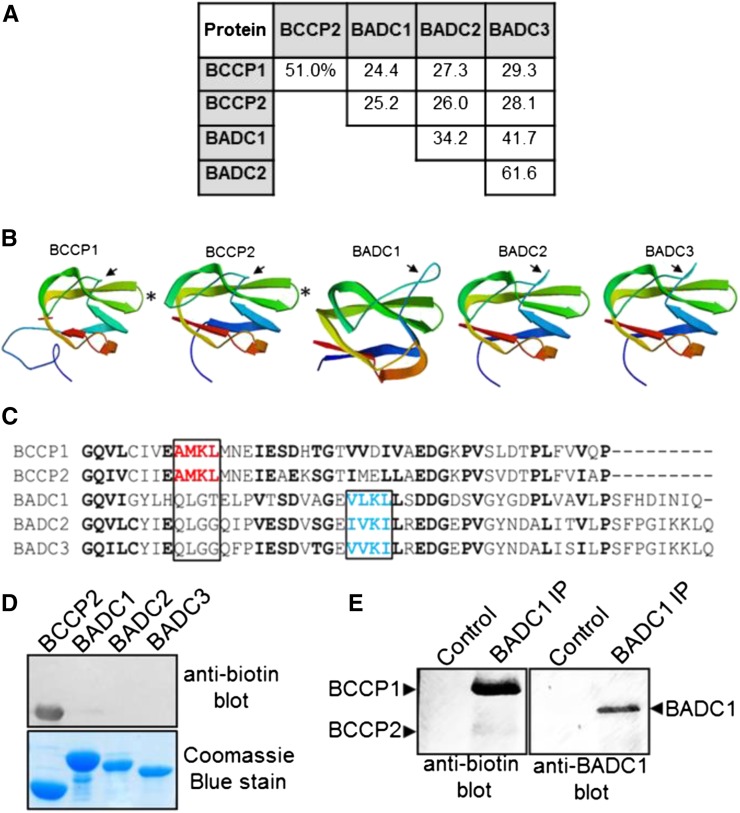
BADCs Resemble BCCPs but Are Not Biotinylated
The ability to form homodimers is a shared characteristic between BADCs and BCCPs. Interestingly, the three BADC isoforms share many other characteristics with the two BCCP isoforms from Arabidopsis: (1) these proteins contain a canonical plastid target peptide and are plastid localized based upon co-IP results here using purified chloroplasts and proteomic analysis of purified chloroplasts (Olinares et al., 2010); (2) the BADC isoforms share 24 to 29% amino acid sequence identity with the BCCP isoforms (Figure 5A); (3) structural predictions of the BADC and BCCP proteins show similar β-sheet secondary structure with a characteristic “thumb motif” as previously observed for the E. coli BCCP (Cronan, 2001) (Figure 5B). Despite these similarities, BADCs lack the canonical biotinylation motif (Figure 5C). The tetrapeptide (Ala/Val)-Met-Lys-(Met/Leu) biotinylation motif is usually located 34 or 35 residues from the C terminus (Tissot et al., 1996; Alban et al., 2000). However, the BADC proteins do possess a conserved Lys residue in a similar (Val/Ile)-(Leu/Val)-Lys-(Leu/Ile) motif located near the C terminus, suggesting the possibility of a noncanonical biotinylation motif. To test this possibility, we purified BADC proteins expressed in E. coli and the native BADC1 protein from Arabidopsis seedlings and probed for biotinylation using a biotin-specific antibody (Figures 5D and 5E). The results of these experiments confirmed the BADC proteins are not biotinylated in vivo, unlike the BCCP controls.
Figure 5.
The BADC Proteins Resemble BCCP Isoforms but Are Not Biotinylated.
(A) Protein sequences from Arabidopsis were aligned and percent amino acid identity was calculated using ClustalW.
(B) Structures of each protein were generated using Swiss-Model. The solved structure for E. coli BCCP (PDB 4HR7) was the closest matching solved structure to all five proteins and was used as a template. Protein sequences lacking the predicted transit peptide residues were used as the input. The N and C termini are colored blue and red, respectively. Arrows indicate thumb domain. Asterisks indicate location of the biotinyl Lys residue on the BCCPs.
(C) Alignment of the C termini of the Arabidopsis BCCP and BADC proteins. Conserved residues are in bold. Highlighted in red is the canonical biotinylation motif containing the modified Lys in BCCP. Highlighted in blue is the chemically similar motif found in the BADC proteins.
(D) Protein blot analysis of recombinant Arabidopsis proteins using a biotin-specific antibody.
(E) Protein blot analysis of immunoprecipitated in vivo BADC1 from Arabidopsis seedlings. Blotting precipitate with BADC1-specific antibody showed the presence BADC1 in the sample, while blotting with biotin-specific antibody showed no recognition of BADC1.
Identification of BADC Orthologs and Co-Occurrence Analysis Suggest That BADCs First Appeared in Red Algae
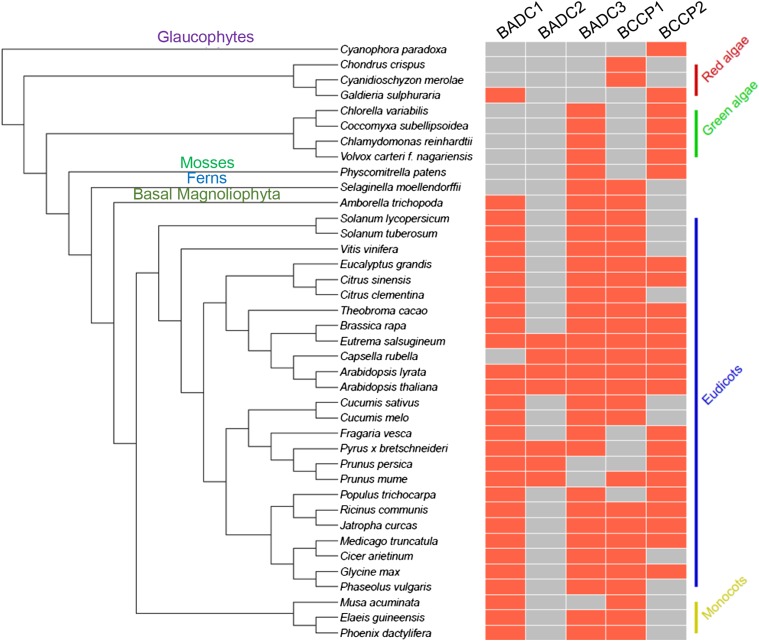
The evidence of a direct BADC-BCCP interaction suggests that BADC function is linked to hetACCase. Thus, orthologs to Arabidopsis BADCs (At-BADC) would be expected to reside only in organisms that contain hetACCase, not the homomeric form that predominates in eukaryotes. To search for the presence of At-BADC orthologous proteins, the primary sequence of each At-BADC was used to search against all complete plant, animal, and prokaryotic genomes available on the KEGG Sequence Similarity database (Kanehisa et al., 2016). Putative orthologs were confirmed by reciprocal BLAST searches against the Arabidopsis proteome. Orthologs to At-BADC were found only in algae and plants. All At-BADC orthologs lacked the conserved biotinyl Lys found in BCCPs. In total, 59, 14, and 60 orthologous proteins were identified for At-BADC1, At-BADC2, and At-BADC3, respectively, across 73 different species of land plants and algae (Supplemental Data Set 1). Full-length BADC orthologs were used to generate a maximum-likelihood phylogenetic tree (Supplemental Figure 3 and Supplemental File 1). It was apparent that all species that harbor a putative At-BADC ortholog also contain the heteromeric form of ACCase. No orthologs were detected in organisms that contain only the homACCase. Additionally, no At-BADC orthologs were detected in prokaryotes, which also contain a hetACCase. The presence of orthologs in algae but not prokaryotes suggests that BADCs first appeared in algae.
To determine if BADCs arose from a previously functional BCCP in algae, co-occurrence analysis was performed. Figure 6 shows the species containing At-BCCP and/or At-BADC orthologs that were used in this analysis. With the exception of two red algae and the freshwater glaucophyte alga Cyanophora paradoxa, all species contained At-BCCP and At-BADC orthologs. In red algae, only one putative At-BADC1 ortholog (GenBank ID: XP_005708748.1) was identified in the species Galdieria sulphuraria. This protein shares the same number of identical (31) and similar (46) amino acid residues with both At-BADC1 and At-BADC2, as well as 30 identical and 44 similar amino acid residues with At-BADC3. However, the BLAST search attributed the highest score to At-BADC1. In addition, two putative BCCP proteins were identified in the red algae species Chondrus crispus and Cyanidioschyzon merolae that lack the biotinylation motif but share higher sequence similarity to At-BCCP2 than At-BADCs (GenBank IDs XP_005715802.1 and XP_005535248.1, respectively), suggesting that BADCs originated from a BCCP gene duplication and loss-of-function mutation in red algae. From this observation, we can infer not only that BADCs and BCCPs are related, but also that the branch point between these proteins occurred in red algae, particularly since the more primitive glaucophytes contain no At-BADC orthologs.
Figure 6.
Co-Occurrence of BADC and BCCP Proteins during Evolution of the Plant Kingdom.
A schematic illustrating the species included in the co-occurrence analysis. At left, an unrooted taxonomic tree depicting the relation of each species is shown. Species names are in black, clades are in various colors. At right, boxes indicate whether the given species contains an ortholog to At-BADC or At-BCCP (red, yes; gray, no).
BADC Expression Reduces hetACCase Activity in a Temperature-Sensitive E. coli Mutant
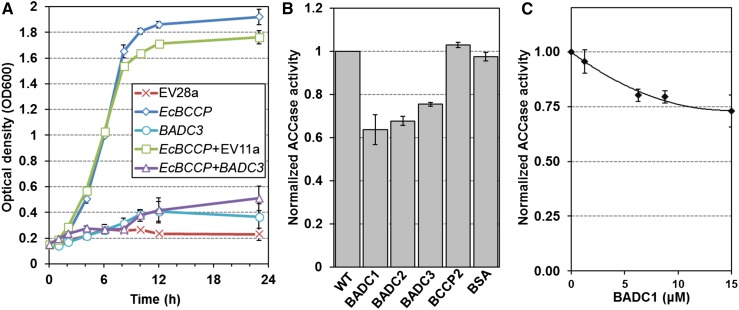
Due to their similarity with BCCPs but lack of a conserved biotinylation motif, it was hypothesized that BADCs are negative regulators of hetACCase activity. As E. coli contains hetACCase but lacks BADC orthologs, this system was appropriate to test this theory in vivo. Growth assays in E. coli accb strain L8 were performed to evaluate the potential effect of the BADC proteins on hetACCase activity. This strain has been studied in detail (Li and Cronan, 1992; Chapman-Smith et al., 1999; Cronan, 2001), and it is known that cell growth at 37°C is directly proportional to hetACCase activity when lacking an exogenous source of fatty acids. This phenotype is due to temperature-sensitive (Ts) mutations in the BCCP gene (accB) that prevents de novo FAS. Experiments were performed in minimal medium containing only glucose and glycerol as carbon sources. To complement the Ts phenotype, the native E. coli BCCP (EcBCCP) gene was cloned into L8 cells in a T7 polymerase expression system. Although this strain does not contain a T7 polymerase for optimal expression, a basal level of expression was observed by protein gel blot analysis (Supplemental Figure 4A). Expression of EcBCCP rescued cell growth at 37°C in medium lacking fatty acids, while empty vector controls showed minimal growth (Figure 7A). In the same way, the Arabidopsis BADC3 gene was cloned into L8 cells and was unable to complement the Ts phenotype. However, coexpression of BADC3 with EcBCCP reduced the complementing effect of EcBCCP expression by ∼72% after 23 h growth. Affinity pull-down assays with tagged BADC3 confirmed the inhibition was mediated by interaction with EcBCCP (Supplemental Figure 4B). Control experiments showed that BADC3 was not toxic to wild-type cells (Supplemental Figure 5). This result demonstrates that the BADC proteins affect the ability of EcBCCP to form active ACCase complexes.
Figure 7.
BADCs Reduce ACCase Activity in E. coli and Arabidopsis.
(A) Growth curves showing the optical density of L8 E. coli cells over time. Cultures were grown in liquid culture at 37°C. Transformed cells contained the following vectors: empty pET28a (EV28a), pET28a containing the E. coli BCCP gene (EcBCCP), empty pET11a (EV11a), and/or pET11a containing the Arabidopsis BADC3 gene (BADC3). BADC3 expression alone showed no statistical difference from EV control except at T = 10 h, while coexpression of BADC3 with EcBCCP showed an approximate 72% reduction in growth compared with EcBCCP+EV11a at T = 24 h. Results shown are representative of three separate experiments. Error bars represent sd.
(B) Protein extracted from 10-d-old Arabidopsis siliques was assayed for ACCase activity by incorporation of radiolabeled sodium bicarbonate into acid-stable products. Assays were performed in the absence (WT) or presence of 10 μM recombinant BADC1, BADC2, BADC3, BCCP2, or BSA. Specific activities were calculated for each assay and then normalized to wild-type control. Specific activities of controls ranged from 1.5 to 2.5 nmol/mg/min. Four biological replicates were performed for each trial. Each replicate consisted of three light-adapted whole siliques. Error bars denote se.
(C) ACCase activity was monitored in 20-d-old leaf extracts with increasing concentrations of recombinant BADC1; 0 μM controls were normalized to 1. Specific activity values for controls ranged from 0.84 to 2.00 nmol/min/mg. Shown is average of four biological replicates. Each biological replicate contained four light-adapted leaves. Error bars denote se.
Recombinant BADC Inhibits Plant hetACCase Activity
To test if the BADCs can also inhibit plant hetACCase, enzyme activity assays were performed on 10-d-old Arabidopsis silique extracts. The activity of hetACCase was monitored in vitro by measuring the incorporation of H14CO3 into acid-stable products. Assays were performed in the presence of 10 μM purified recombinant BADC1, BADC2, BADC3, BCCP2, or BSA (Supplemental Figure 6) and compared with buffer control (wild type). A high, nonphysiological concentration was chosen for this assay given the short incubation time and to determine if the BADC isoforms differed in their ability to affect ACCase activity. The average of four biological replicates showed that all three BADCs inhibited hetACCase activity by 25 to 37%, while BCCP2 and BSA showed no effect at the same concentration (Figure 7B). These results, in addition to the E. coli expression results (Figure 7A), confirm that the BADCs negatively affect hetACCase activity. Titration of BADC levels revealed a Ki of 4.3 µM (Figure 7C). The high Ki value is likely due to the brief 5-min incubation period, which is necessary for optimal in vitro activity but likely insufficient to reach complex equilibrium.
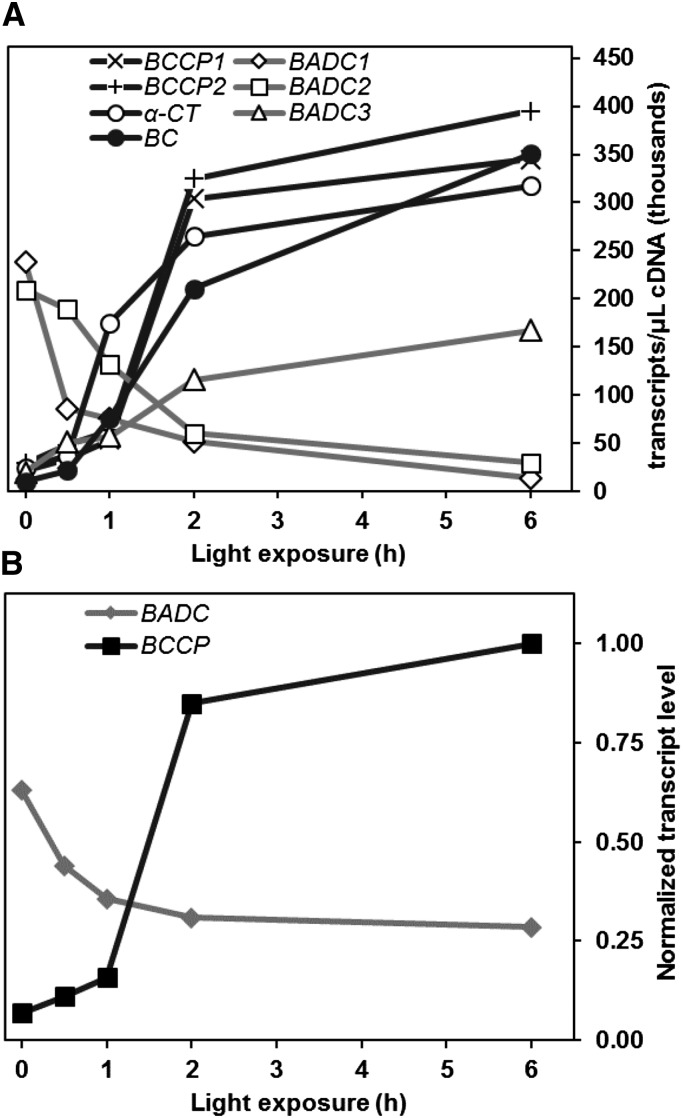
Expression Profiles of BADC and hetACCase Subunits Respond Differently to Light
As mentioned previously, hetACCase activity is enhanced upon light exposure in photosynthetic cells. Although biochemical changes to the stromal environment are a major reason for this catalytic enhancement, transcriptional regulation could also be important. To characterize the effect of light on gene expression, absolute transcript levels of the BADCs and nucleus-encoded hetACCase subunits were monitored in 10-d-old Arabidopsis siliques. Whole siliques were used for analysis, as over 90% of total BADC and hetACCase gene expression was observed to be localized to the seed tissue (Supplemental Figure 7). Siliques were harvested after dark adaption or time-dependent exposure to light. Quantitative RT-PCR analysis of RNA extracts from these samples showed that gene expression for each nucleus-encoded catalytic subunit to hetACCase increased significantly in response to light. After 6 h, expression of BCCP1, BCCP2, and α-CT increased ∼15-fold, while BC expression increased 35-fold (Figure 8A). In contrast, BADC1 and BADC2 expression was reduced approximately 10-fold, while BADC3 expression increased 8-fold. Despite the conflicting changes in BADC isoforms, the total BADC transcript level was reduced by half after 6 h light exposure (Figure 8B). The total BADC:BCCP transcript ratio was ∼9:1 prior to light exposure and then shifted to almost 1:4 after 6 h light exposure (Figure 8B), suggesting that BADC protein levels are greater than BCCP protein levels in the dark, and vice versa in the light.
Figure 8.
Light-Dependent Changes in Gene Expression of BADC and hetACCase in Arabidopsis Siliques.
(A) Graph shows the absolute expression level of the given genes obtained by qPCR. Ten-day-old Arabidopsis siliques were collected after various amounts of light exposure. RNA extracted from these tissues was used to create cDNA for this analysis. Average values of four biological replicates are shown, with each replicate containing 10 siliques. Three technical replicates were performed and averaged for each biological replicate. se was ∼5 to 10% for all data points.
(B) Graph depicts the shift in BADC and BCCP total transcript level in response to light. The sum of transcript levels from BCCPs and BADCs in (A) for each time point were normalized to the sum of BCCP transcripts at 6 h light exposure. At T = 0, the ratio of BADC:BCCP transcript is 9:1. At T = 6, the ratio shifts to 1:4.
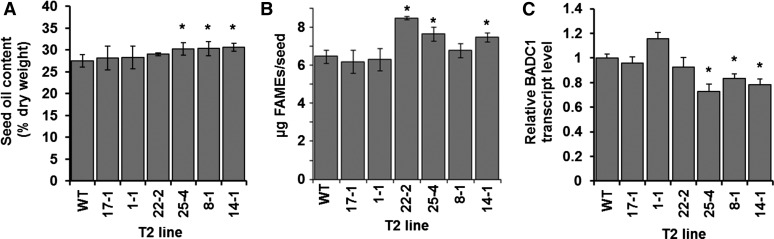
Seed-Specific RNAi Silencing of BADC1 Increases Seed Oil Content in Arabidopsis
Biochemical analysis showed that BADCs can function as negative regulators of hetACCase. Therefore, reduction of BADC expression in planta might increase hetACCase activity. Since ACCase activity has previously been shown to correlate with seed oil content in transgenic lines altered in ACCase function (Roesler et al., 1997; Thelen and Ohlrogge, 2002c), RNAi silencing of BADC1 in Arabidopsis using a seed-specific promoter was performed. Seed oil content analysis showed a significant increase in three of six independent T2 seeds, as normalized to either seed mass or individual seed (Figures 9A and 9B). Additionally, RT-PCR analysis of whole silique tissue showed a significant reduction in BADC1 transcript level of ∼22% on average in the three lines containing significantly higher seed oil (Figure 9C). Fractional silencing is partly due to the use of whole silique tissue instead of isolated seed for RT-PCR analysis and the fact these seed are segregating for the transgene. These results agree with the data shown in Figure 7, demonstrating that BADC proteins are negative regulators of hetACCase.
Figure 9.
Seed-Specific RNAi Silencing of BADC1 Increases Seed Oil Content in Arabidopsis.
(A) Bar graph shows total seed oil content in wild-type and basta-resistant T2 Arabidopsis lines containing a construct that silences BADC1 expression in the seed. Each bar represents the average of four plants. Error bars denote sd.
(B) Oil content per seed. Graph shows the average amount of fatty acid methyl esters identified per seed of four biological replicates, in micrograms.
(C) RT-PCR analysis of BADC1 RNAi silencing lines. BADC1 transcript level was quantified relative to actin transcript level and normalized to the wild type. RNA used for analysis was extracted from four biological replicates of 10-d-old siliques. Error bars denote se. In all graphs, statistical significance was determined by Student’s t test (*P < 0.05).
DISCUSSION
The BADC proteins described here are annotated as proteins with unknown function. When mentioned in previous reports, these proteins were labeled as BCCP-like due to their sequence similarity to BCCP. The BADCs were observed to coelute with Arabidopsis hetACCase during size-exclusion chromatography (Olinares et al., 2010) and were also detected in pull-down experiments with the 2-oxoglutarate binding protein PII along with both BCCP isoforms (Feria Bourrellier et al., 2010). In both cases, a direct interaction between ACCase catalytic subunits and the BADCs was not established, and a functional role was neither determined nor proposed. Here, we present evidence that the BADCs not only directly interact with hetACCase, but also function as negative regulators of this multienzyme complex. Thus, we have termed them BADCs to avoid confusion with BCCPs. At least four lines of evidence demonstrate these proteins are not functional BCCPs: (1) the BADCs are not biotinylated (Figure 5E); (2) BADC protein expression cannot complement the Ts BCCP in L8 E. coli (Figure 7A); (3) addition of recombinant BADCs to Arabidopsis silique extracts inhibits hetACCase activity while biotinylated BCCP2 has no effect (Figure 7B); and (4), seed-specific silencing of BADC protein expression resulted in increased seed oil content (Figure 9). Therefore, we conclude that the BADC proteins function as negative regulators of hetACCase activity.
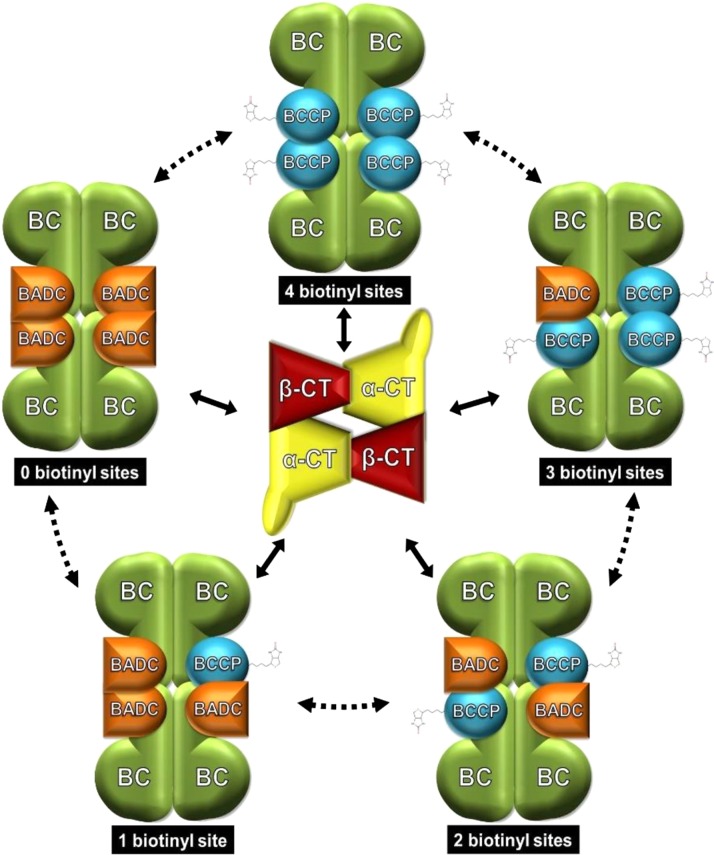
In vitro, BADC1 shows concentration-dependent inhibition of hetACCase (Figure 7C). Taken alone, this result does not differentiate between competitive or uncompetitive inhibition. However, given the similarity of BADC and BCCP with respect to primary amino acid sequence (Figure 5A), predicted structure (Figure 5B), and dimerization capability (Figure 4), the parsimonious interpretation of these data is that BADC proteins inhibit hetACCase activity by competing with BCCP for binding to other hetACCase subunits. The crystal structure of the E. coli BC/BCCP subcomplex consists of two dimers of BC held together by four BCCPs, thus having a BCCP:BC ratio of 1:1 (Broussard et al., 2013). In contrast, proteomics (Ishihama et al., 2008), in vivo labeling (Choi-Rhee and Cronan, 2003), and ribosome profiling studies (Li et al., 2014) have observed a BCCP:BC ratio of 2:1. Our co-IP experiments do not distinguish between these possibilities in plants. In either case, the subcomplex theoretically allows for the incorporation of up to four BADC proteins and enables BADC proteins to exert concentration-dependent inhibition on hetACCase. Assuming a BCCP:BC ratio of 1:1, we propose a model where multiple BC/BCCP/BADC subcomplexes can be formed (Figure 10). Coupled with the observation that BADC proteins can interact with one another (Figure 2A), it appears that minimally five different BC/BCCP/BADC subcomplexes are possible, wherein the steady state level of each subcomplex is dependent on the concentration of the BADC and BCCP subunits. With two BCCP and three BADC isoforms in Arabidopsis, the combinatorial possibilities increase exponentially. Such a mechanism would enable fine control of hetACCase function through variable BCCP and BADC expression. Although we propose BADCs act competitively, alternative modes of inhibition cannot be ruled out without further kinetic analysis.
Figure 10.
Model of BADC Competitive Inhibition of hetACCase.
Schematic illustrating the proposed mechanism of BADC inhibition. The BC/BCCP subcomplex design was made based on the crystal structure in E. coli (Broussard et al., 2013), consisting of two dimers of BC and four BCCP proteins. The BADC proteins compete with BCCP for binding to BC. Binding of BADC prevents binding of the essential BCCP subunit. The pool of BC/BCCP and BC/BCCP/BADC subcomplexes then compete for interaction with the CT subcomplex (design based on crystal structure in E. coli; Bilder et al., 2006), leading to variable reductions in ACCase activity. While a transient association of the two ACCase half reactions is known, it is unclear whether BADC can displace BCCP from an assembled BC/BCCP subcomplex, hence the dashed arrows.
Since the BADCs resemble BCCP but are not biotinylated, a BC/BCCP/BADC subcomplex could be considered pseudo-poisoned. Natural examples of pseudo-poison complex regulation are infrequent, though at least one bioengineered example has been reported. Overexpressing BCCP2 in Arabidopsis seed markedly reduced both hetACCase activity and seed oil content due to increased levels of apo-BCCP (Thelen and Ohlrogge, 2002c; Chen et al., 2009). Pseudo-poisoned hetACCase complexes were formed, reducing carbon flux through de novo FAS. This mechanism appears to be similar for BADC proteins, whereby they act as natural, apo-BCCP mimics and, thus, hetACCase agonists. We can conceive of at least one advantage this form of metabolic regulation provides for de novo FAS. This proposed regulatory model would simplify the genetic control necessary for hetACCase activity, as the cell would require strict control over only the BADC or BCCP genes rather than the entire complement of plastid and nuclear hetACCase genes. Subtle expression variations in either BADC or BCCP isoforms, all residing in the nuclear genome, could determine hetACCase activity analogous to a molecular rheostat.
Such a model implies that BCCP and BADC proteins are under differential regulation and are potentially inducible. We show evidence of differential gene regulation in Arabidopsis siliques (Figure 8). After exposing dark-adapted siliques to 6 h of light, total BADC transcript levels decreased approximately 2-fold, while total BCCP transcript levels increased ∼15-fold (Figure 8A). Therefore, during dark adaption, the total BADC transcript level is approximately 9-fold higher than the total BCCP transcript level (Figure 8B). After 6 h of light exposure, the total BCCP transcript load becomes 3-fold higher than the total BADC transcript load. If the BADC and BCCP protein levels change in accordance with transcript levels even to a small degree, the competitive model suggests hetACCase activity will be higher in light conditions and lower in dark conditions. As mentioned previously, hetACCase activity is known to increase in response to light. Therefore, the competitive model of BADC regulation is consistent with previous reports on the light activation of hetACCase.
The literature also gives evidence of the inducible nature of BADC and BCCP gene expression. First, transcriptomic analysis of the Arabidopsis wrinkled1 mutant of WRI1, a transcription factor that regulates ∼18 enzymes involved in central metabolism, showed differential expression for only the BCCP2, BADC2, and BADC3 subunits of hetACCase (Ruuska et al., 2002). Second, overexpression of WRI1 significantly increased expression of only the BCCP2 subunit of hetACCase (Baud et al., 2007; Fukuda et al., 2013); according to the model, we appear to finally understand how WRI1 upregulates ACCase by targeting only the BCCP2 subunit of this multienzyme complex. Third, global transcriptome and proteomic analysis of BCCP2 overexpression lines in Arabidopsis showed that targeted upregulation of BCCP2 produced a concomitant increase in BADC2 transcript and protein levels; quantitative immunoblotting showed no changes to any other hetACCase subunit (Chen et al., 2009). Thus, it appears that expression of BADC and BCCP subunits are under different transcriptional control than other hetACCase subunits.
Regulation of hetACCase by BADCs must now be considered among the other regulatory mechanisms for this multienzyme complex. As mentioned previously, these mechanisms include feedback inhibition, PII-mediated inhibition, light, and redox chemistry. However, regulation of hetACCase by the BADC family of proteins is unusual in that transcriptional control of these genes can directly affect hetACCase activity. Purified, recombinant BADC, lacking any posttranslational modifications, by itself was sufficient to inhibit hetACCase in vivo (Figure 7A). By extension, targeted downregulation of one BADC gene was also sufficient to enhance ACCase function and accumulation of triacylglyerol in the seed (Figure 9). From these observations, it is tempting to speculate what effect the reduced expression of all three BADC genes might have on seed oil content or plant function in general. Given their efficacy, it is possible this could produce unintended, deleterious effects, particularly if silencing is not targeted to cell types capable of storing excess fatty acids originating from the plastid, i.e., maturing, nondividing embryonic cells. This is presently being explored by systematic tandem RNAi and by investigating the collection of T-DNA knockout lines available for Arabidopsis.
In conclusion, our results implicate the BADC proteins as negative regulators of hetACCase in diverse autotrophs. Due to the essential role of hetACCase in de novo FAS and the identification of BADC orthologs in almost every major oil crop and green algae species currently in use to produce biofuels (Supplemental Data Set 1), further study could provide new mechanisms for engineering increased oil content in these species.
METHODS
Plant Materials and Growth Conditions
Wild-type Arabidopsis thaliana (ecotype-Columbia-0) were grown in a growth chamber with 12 h day (23°C, 50% humidity, 50 μmol m−2 s−1, white light, fluorescent bulbs) and 12 h night (20°C, 50% humidity) conditions. For co-IP experiments, 8.5 × 8.5-cm pots were filled with moist soil (Sunshine Mix #6; Sun Gro Horticulture), covered with a screen (1 mm2 pore size), and coated with seeds. For hetACCase activity experiments, two plants were grown in opposite corners of 8.5 × 8.5-cm pots.
Co-IP of hetACCase from Arabidopsis Seedlings
Crude chloroplasts were isolated from 14-d-old Arabidopsis seedlings after 1 h light exposure. Fresh leaves were homogenized in ice-cold grinding buffer (50 mM HEPES-KOH, pH 8.0, 330 mM sorbitol, 1.5 mM MnCl2, 2 mM MgCl2, 2 mM EDTA, and 0.1% [w/v] BSA) using a Waring blender. Homogenate was filtered through two layers of Miracloth and centrifuged at 2600g at 4°C for 5 min. Chloroplasts were lysed for 30 min in ice-cold lysis buffer (50 mM HEPES-KOH, pH 8.0, 10% [v/v] glycerol, and 0.5% [v/v] Triton X-100). Lysates were homogenized 10 times in a Dounce homogenizer on ice and then centrifuged at 30,000g for 20 min at 4°C. Then, 1 mL of the 30,000g supernatant was added to 25 μL Protein A Sepharose beads either uncoated (control) or coated with 2 μL antibody specific for α-CT, BCCP2, BADC1, or BADC2. Co-IPs were performed at 4°C for 3 h with gentle mixing. The beads were washed twice before precipitated protein was eluted with 30 μL 6× SDS sample buffer (350 mM Tris-HCl, pH 6.8, 350 mM SDS, 30% [v/v] glycerol, 100 mM DTT, and 2.5 mM bromophenol blue) and heating at 65°C for 5 min. Eluted proteins were resolved on 10% SDS-PAGE gels for protein gel blot and mass spectrometry analysis.
Mass Spectrometry Sample Preparation and Analysis
Precipitated proteins from co-IPs were resolved by 10% SDS-PAGE and stained with colloidal Coomassie Brilliant Blue G 250. Each lane was separated into 0.5-cm segments and subsequently diced into ∼1-mm3 gel pieces. Gel pieces were digested with trypsin and peptides were extracted as described previously (Shevchenko et al., 2006). Tryptic peptides were lyophilized and stored at −20°C until analysis by LC-MS/MS. Lyophilized peptides were prepared for mass spectrometry analysis as described previously (Swatek et al., 2011). Samples were analyzed on a LTQ Orbitrap XL ETD (Thermo Fisher Scientific) with the same settings as described by Swatek et al. (2014), except that peptides were eluted using a 30-min acetonitrile gradient (5 to 43% acetonitrile), the top eight masses from the precursor scan were selected for data-dependent acquisition, and precursor ions were fragmented using collision-induced dissociation. Dynamic exclusion was enabled with the following settings: repeat count, 3; repeat duration, 30 s; exclusion list, 50; and exclusion list duration, 30 s.
Database Searching
Acquired spectra were searched against the TAIR10 protein database (70,773 entries, downloaded on 06/11/2012), concatenated to a randomized TAIR10 database as a decoy. Raw data files were searched using SEQUEST and filtered to <1% false discovery rate using Proteome Discoverer 1.3 (Thermo Fisher Scientific) and the following filters: Max 2 missed cleavages, fixed Cys carbamidomethylation, variable Met oxidation, 10 ppm peptide mass deviation, and two peptide minimum. Proteins identified from SEQUEST searches were compared against uncoated Sepharose bead controls treated in an identical manner to the hetACCase subunit co-IPs. Only proteins identified in the hetACCase subunit co-IPs were considered putative interacting clients. All other proteins were excluded. The hetACCase subunits were never identified in controls.
Yeast Two-Hybrid Construct Design and Interaction Assay
The open reading frame of genes of interest were inserted into bait and prey vectors PGBKT7 and pGADT7 (Clontech) using the restriction enzymes NdeI and BamHI. Primers were designed to exclude the N-terminal target peptide from the coding region, as predicted by TargetP (Nielsen et al., 1997) (Supplemental Figure 8). Genes were amplified from cDNA clones obtained from the ABRC. Amplicons were first inserted into Zero Blunt TOPO vector (Life Technologies) and checked for errors by DNA sequencing. Error-free amplicons were then subcloned into either pGBKT7 or pGADT7 vector and used for yeast transformation.
Targeted yeast two-hybrid assays were performed using an adaptation of the lithium acetate method (Gietz and Schiestl, 2007). Strain AH109 yeast were transformed with 100 ng of bait and prey vector. Pelleted transformed cells were resuspended in 300 μL sterile water. Aliquots of 100 μL cell suspension were plated on synthetic dropout (SD) medium lacking leucine, tryptophan, and histidine. Plates were incubated at 30°C for 4 d and then imaged. Images shown are representative of at least three replicate experiments.
Recombinant Protein Expression and Purification
The open reading frames of BCCP1, BCCP2, BADC1, BADC2, and BADC3 were amplified via PCR from cDNA clones U15855, U85042, U12534, C00214, and U15571, respectively (ABRC). The primer pairs for these amplifications were the same as those used in the yeast two-hybrid construct formation (Supplemental Figure 8). The amplified open reading frame of all five genes was cloned into either the expression vector pET28a or pET11a, producing an N-terminal His-tagged fusion protein or an untagged recombinant protein, respectively. All constructs were sequence confirmed via DNA sequencing and then transformed into Escherichia coli strain BL21 (B2685; Sigma-Aldrich). Recombinant protein was expressed and purified from transformed BL21 cells as described (Swatek et al., 2011). For coexpression experiments, ∼200 ng of each plasmid was used to transform BL21 cells.
Immunoblotting
Proteins resolved by SDS-PAGE were transferred to PVDF membrane and stained with the appropriate primary antibody (1:5000 dilution in PBS-T) overnight at 4°C for protein gel blot analysis. Antibodies used in this study were derived from rabbits immunized with recombinant Pisum sativum α-CT (Thelen and Ohlrogge, 2002b), recombinant Arabidopsis BCCP2 (Thelen and Ohlrogge, 2002c), recombinant Arabidopsis BADC2 short peptide (Chen et al., 2009), or recombinant Arabidopsis BADC1 (this study). Blots were probed in secondary antibody for 1 h at room temperature and developed as described previously (Thelen and Ohlrogge, 2002b). Goat anti-rabbit IgG secondary antibody conjugated to alkaline phosphatase was obtained from Sigma-Aldrich (cat. A3687, lot 123K6037).
Structural Predictions of BADC Proteins
Predicted models of At-BCCP and At-BADC proteins were constructed using SWISS-MODEL (Biasini et al., 2014). The primary sequence of each protein without the transit peptide was used for the model. Transit peptide sequences were predicted using TargetP (Nielsen et al., 1997) (Supplemental Figure 8). Models were constructed from a template, and the E. coli BCCP protein structure (PDB 4HR7) was found to be the closest matching template for all five proteins. Default parameters were used in generating the model.
Co-Occurrence Analysis
Orthologs to At-BADC proteins were identified as explained in the Supplemental Methods. The primary sequences of confirmed BCCP and BADC orthologs from all species present in the maximum-likelihood tree were used for co-occurrence analysis. In addition, BCCP and BADC orthologs from Cyanophora paradoxa and three red algae species with whole genome sequences were included to trace the origin of BADC proteins. The ortholog assignment was confirmed by reciprocal BLAST searches. The species tree was generated using phyloT (http://phylot.biobyte.de) based on NCBI Taxonomy and the Tree of Life project (http://tolweb.org/tree/). The protein co-occurrence pattern was drawn using ggtree (http://bioconducter.org/packages/release/bioc/html/ggtree.html).
Growth Assays in L8 Strain E. coli Cells
Full details can be found in the Supplemental Methods. In brief, the temperature-sensitive (Ts) L8 strain E. coli was obtained from the Coli Genetic Stock Center (Yale, New Haven, CT) and transformed with the vectors in the text using the heat shock method. Transformants were selected by antibiotic resistance and confirmed by PCR. Prior to the growth experiment, cultures were grown overnight in LB medium at 30°C. Overnight cultures were centrifuged at 3000g and resuspended in 5 mL sterile deionized water. Cultures were centrifuged again and resuspended in M63 minimal medium to make OD600 = 3.75. Then, 200 μL cell suspension was added to 7 mL M63 medium plus antibiotics in 15-mL sterile culture tubes. Cultures contained Kan, and Amp if necessary, at 50 μg/mL each as well as 1 μM isopropyl β-d-1-thiogalactopyranoside at T = 0.
RNAi Silencing of BADC1 in Arabidopsis
Inverted repeats targeting At-BADC1 were inserted into the pMU103 vector (Flores et al., 2008). The repeats coded for bases 774 to 1034 of the cDNA sequence (accession AT3G56130.1). Primers used to amplify the sequence were 5′-GTGTTAGTCACATCTCCCGCAGT-3′ and 5′-GATGTTGATGTCGTGGAAAGATGGC-3′. Details of construct design can be found in the Supplemental Methods. A sequence confirmed construct was transformed into Arabidopsis ecotype Col-0 using the floral dip method (Clough and Bent, 1998). Basta herbicide screening was used to identify independent lines. Expression of the RNAi cassette was driven by the glycinin promoter (Cho et al., 1989; Sims and Goldberg, 1989; Fatihi et al., 2013). For monitoring seed oil content, T2 plants from each independent line were grown to maturity alongside wild-type plants. Dry seed was harvested for analysis.
Fatty Acid Methyl Ester Analysis
Seed oil was derivatized as described previously (Li et al., 2006). Heptadecanoic acid was used as an internal standard. Fatty acid methyl esters were analyzed by a Hewlett Packard 6890 gas chromatograph as described previously (Chen and Thelen, 2013). For the wild type and each independent line, 5 mg seed from 13 and 4 plants, respectively, was analyzed. Seeds were dried over desiccant for 1 week prior to analysis.
RT-PCR and qPCR Analysis
RNA for RT-PCR and qPCR analysis was extracted from 10-d-old siliques using the RNeasy Plant Mini Kit (Qiagen). cDNA was synthesized from 500 ng RNA of four biological replicates. One biological replicate consisted of 10 whole siliques collected at the same time and pooled. Three technical replicates were performed for each biological replicate and averaged to obtain expression values for each biological replicate. Primers used in analysis were: BADC1 sense, 5′-GCTCCTAGCCCATCTCAAGC-3′; BADC1 antisense, 5′-TCCAGATGCCTCCAAAGCAG-3′; Actin 8 sense, 5′-CCAGATCTTCATCGTCGTGGT-3′; Actin 8 antisense, 5′-ATCCAGCCTTAACCATTCCAGT-3′. qPCR assays were performed on an ABI 7500 system (Applied Biosystems). Reaction volumes were 20 μL and contained SYBR Green PCR Master Mix (Applied Biosystems). Control reactions contained no template and were performed in triplicate. Amplicon identity was confirmed through melting curve analysis. For qPCR analysis, absolute transcript quantities were calculated using a standard curve of serially diluted amplicons of known concentrations.
ACCase Activity Assays
HetACCase activity was directly monitored in 10-d-old siliques or 21-d-old leaves through the incorporation of 14CO2 into acid-stable products as described (Thelen and Ohlrogge, 2002c). Details can be found in the Supplemental Methods. Arabidopsis wild-type Col-0 10-d-old siliques were harvested after 6 h of light exposure, while leaves were grown under constant light conditions and harvested near midday. In each trial, four biological replicates of three siliques or four leaves were assayed. Tissue was pulverized in homogenization buffer (20 mM TES, pH 7.5, 10% glycerol, 5 mM EDTA, 2 mM DTT, 2 mM benzamidine, 2 mM PMSF, and 1% Triton X-100), centrifuged at 10,000g for 15 s, and assayed within 5 min of harvest to minimize loss of hetACCase activity. Assays were performed in the presence of 10 μM haloxyfop to eliminate homomeric ACCase activity. Enzyme activity values for minus acetyl-CoA controls were subtracted from +acetyl-CoA trials to determine the true hetACCase activity levels. Purified recombinant protein was added to assay tube prior to addition of tissue lysate. The Ki value for BADC1 was calculated using the equation fit to the data y = 0.0012x2-0.036x+0.9979, where y equals the normalized ACCase activity at half-maximal inhibition and x equals inhibitor concentration. Maximum inhibition was constrained to the 15 mM BADC1 trial (0.73 normalized ACCase activity).
Accession Numbers
Sequence data from this article can be found in the TAIR database libraries (arabidopsis.org) under the following accession numbers: α-CT, AT2G38040; β-CT, ATCG00500; BC, AT5G35360; BCCP1, AT5G16390; BCCP2, AT5G15530; BADC1, AT3G56130; BADC2, AT1G52670; and BADC3, AT3G15690.
Supplemental Data
Supplemental Figure 1. Mass spectrometry analysis of hetACCase coimmunoprecipitations.
Supplemental Figure 2. Multiple sequence alignment of BADC and BCCP primary sequences.
Supplemental Figure 3. A maximum-likelihood phylogenetic tree of BADC full-length proteins.
Supplemental Figure 4. Recombinant protein expression in L8 E. coli cells.
Supplemental Figure 5. E. coli growth during BADC3 expression.
Supplemental Figure 6. Purity of recombinant protein purifications for ACCase activity assays.
Supplemental Figure 7. hetACCase and BADC gene expression in whole silique, seed, and silique pod/septa.
Supplemental Figure 8. Primers used in cloning experiments.
Supplemental Data Set 1. Complete list of identified BADC orthologs.
Supplemental File 1. Alignments used for phylogenetic tree construction.
Supplementary Material
Acknowledgments
We thank John Ohlrogge for the α-CT and BCCP antibody, Melissa Mitchum for the yeast strain and two-hybrid vectors, Brian Mooney and Beverly DaGue for the intact mass analysis, and Shannon King for aiding in the study of BADC dimerization. This work was supported by National Science Foundation Grant PGRP IOS-1339385 and National Institutes of Health Grant T32 GM008396.
AUTHOR CONTRIBUTIONS
M.J.S. designed and performed the experiments and wrote the manuscript. N.Z. and D.X. performed the evolutionary analysis. V.L. performed the qPCR and RT-PCR analysis. J.J.T. conceived the idea for the project and wrote the manuscript with M.J.S.
Glossary
- FAS
fatty acid biosynthesis
- BC
biotin carboxylase
- BCCP
biotin carboxyl carrier protein
- CT
carboxyltransferase
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- co-IP
coimmunoprecipitation
Footnotes
Articles can be viewed without a subscription.
References
- Alban C., Job D., Douce R. (2000). Biotin metabolism in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51: 17–47. [DOI] [PubMed] [Google Scholar]
- Andre C., Haslam R.P., Shanklin J. (2012). Feedback regulation of plastidic acetyl-CoA carboxylase by 18:1-acyl carrier protein in Brassica napus. Proc. Natl. Acad. Sci. USA 109: 10107–10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S., Lepiniec L. (2009). Regulation of de novo fatty acid synthesis in maturing oilseeds of Arabidopsis. Plant Physiol. Biochem. 47: 448–455. [DOI] [PubMed] [Google Scholar]
- Baud S., Wuillème S., To A., Rochat C., Lepiniec L. (2009). Role of WRINKLED1 in the transcriptional regulation of glycolytic and fatty acid biosynthetic genes in Arabidopsis. Plant J. 60: 933–947. [DOI] [PubMed] [Google Scholar]
- Baud S., Mendoza M.S., To A., Harscoët E., Lepiniec L., Dubreucq B. (2007). WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 50: 825–838. [DOI] [PubMed] [Google Scholar]
- Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., Schwede T. (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42: W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilder P., Lightle S., Bainbridge G., Ohren J., Finzel B., Sun F., Holley S., Al-Kassim L., Spessard C., Melnick M., Newcomer M., Waldrop G.L. (2006). The structure of the carboxyltransferase component of acetyl-coA carboxylase reveals a zinc-binding motif unique to the bacterial enzyme. Biochemistry 45: 1712–1722. [DOI] [PubMed] [Google Scholar]
- Broussard T.C., Kobe M.J., Pakhomova S., Neau D.B., Price A.E., Champion T.S., Waldrop G.L. (2013). The three-dimensional structure of the biotin carboxylase-biotin carboxyl carrier protein complex of E. coli acetyl-CoA carboxylase. Structure 21: 650–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman-Smith A., Morris T.W., Wallace J.C., Cronan J.E. Jr. (1999). Molecular recognition in a post-translational modification of exceptional specificity. Mutants of the biotinylated domain of acetyl-CoA carboxylase defective in recognition by biotin protein ligase. J. Biol. Chem. 274: 1449–1457. [DOI] [PubMed] [Google Scholar]
- Chen M., Thelen J.J. (2013). ACYL-LIPID DESATURASE2 is required for chilling and freezing tolerance in Arabidopsis. Plant Cell 25: 1430–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Mooney B.P., Hajduch M., Joshi T., Zhou M., Xu D., Thelen J.J. (2009). System analysis of an Arabidopsis mutant altered in de novo fatty acid synthesis reveals diverse changes in seed composition and metabolism. Plant Physiol. 150: 27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho T.J., Davies C.S., Nielsen N.C. (1989). Inheritance and organization of glycinin genes in soybean. Plant Cell 1: 329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi-Rhee E., Cronan J.E. (2003). The biotin carboxylase-biotin carboxyl carrier protein complex of Escherichia coli acetyl-CoA carboxylase. J. Biol. Chem. 278: 30806–30812. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cronan J.E., Jr. (2001). The biotinyl domain of Escherichia coli acetyl-CoA carboxylase. Evidence that the “thumb” structure id essential and that the domain functions as a dimer. J. Biol. Chem. 276: 37355–37364. [DOI] [PubMed] [Google Scholar]
- Cronan J.E. Jr., Waldrop G.L. (2002). Multi-subunit acetyl-CoA carboxylases. Prog. Lipid Res. 41: 407–435. [DOI] [PubMed] [Google Scholar]
- Davis M.S., Cronan J.E. Jr (2001). Inhibition of Escherichia coli acetyl coenzyme A carboxylase by acyl-acyl carrier protein. J. Bacteriol. 183: 1499–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatihi A., Zbierzak A.M., Dörmann P. (2013). Alterations in seed development gene expression affect size and oil content of Arabidopsis seeds. Plant Physiol. 163: 973–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feria Bourrellier A.B., Valot B., Guillot A., Ambard-Bretteville F., Vidal J., Hodges M. (2010). Chloroplast acetyl-CoA carboxylase activity is 2-oxoglutarate-regulated by interaction of PII with the biotin carboxyl carrier subunit. Proc. Natl. Acad. Sci. USA 107: 502–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores T., Karpova O., Su X., Zeng P., Bilyeu K., Sleper D.A., Nguyen H.T., Zhang Z.J. (2008). Silencing of GmFAD3 gene by siRNA leads to low alpha-linolenic acids (18:3) of fad3-mutant phenotype in soybean [Glycine max (Merr.)]. Transgenic Res. 17: 839–850. [DOI] [PubMed] [Google Scholar]
- Fukuda N., Ikawa Y., Aoyagi T., Kozaki A. (2013). Expression of the genes coding for plastidic acetyl-CoA carboxylase subunits is regulated by a location-sensitive transcription factor binding site. Plant Mol. Biol. 82: 473–483. [DOI] [PubMed] [Google Scholar]
- Gerhardt E.C., Rodrigues T.E., Müller-Santos M., Pedrosa F.O., Souza E.M., Forchhammer K., Huergo L.F. (2015). The bacterial signal transduction protein GlnB regulates the committed step in fatty acid biosynthesis by acting as a dissociable regulatory subunit of acetyl-CoA carboxylase. Mol. Microbiol. 95: 1025–1035. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl R.H. (2007). Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2: 35–37. [DOI] [PubMed] [Google Scholar]
- Guchhait R.B., Polakis S.E., Dimroth P., Stoll E., Moss J., Lane M.D. (1974). Acetyl coenzyme A carboxylase system of Escherichia coli. Purification and properties of the biotin carboxylase, carboxyltransferase, and carboxyl carrier protein components. J. Biol. Chem. 249: 6633–6645. [PubMed] [Google Scholar]
- Hunter S.C., Ohlrogge J.B. (1998). Regulation of spinach chloroplast acetyl-CoA carboxylase. Arch. Biochem. Biophys. 359: 170–178. [DOI] [PubMed] [Google Scholar]
- Ishihama Y., Schmidt T., Rappsilber J., Mann M., Hartl F.U., Kerner M.J., Frishman D. (2008). Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics 9: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. (2016). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44 (D1): D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T., Sasaki Y. (1994). Compartmentalization of two forms of acetyl-CoA carboxylase in plants and the origin of their tolerance toward herbicides. Proc. Natl. Acad. Sci. USA 91: 3598–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozaki A., Mayumi K., Sasaki Y. (2001). Thiol-disulfide exchange between nuclear-encoded and chloroplast-encoded subunits of pea acetyl-CoA carboxylase. J. Biol. Chem. 276: 39919–39925. [DOI] [PubMed] [Google Scholar]
- Li G.W., Burkhardt D., Gross C., Weissman J.S. (2014). Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157: 624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.J., Cronan J.E. Jr (1992). The gene encoding the biotin carboxylase subunit of Escherichia coli acetyl-CoA carboxylase. J. Biol. Chem. 267: 855–863. [PubMed] [Google Scholar]
- Li X., Ilarslan H., Brachova L., Qian H.R., Li L., Che P., Wurtele E.S., Nikolau B.J. (2011). Reverse-genetic analysis of the two biotin-containing subunit genes of the heteromeric acetyl-coenzyme A carboxylase in Arabidopsis indicates a unidirectional functional redundancy. Plant Physiol. 155: 293–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Beisson F., Pollard M., Ohlrogge J. (2006). Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915. [DOI] [PubMed] [Google Scholar]
- Meyer L.J., Gao J., Xu D., Thelen J.J. (2012). Phosphoproteomic analysis of seed maturation in Arabidopsis, rapeseed, and soybean. Plant Physiol. 159: 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H., Sugiyama N., Mochida K., Daudi A., Yoshida Y., Toyoda T., Tomita M., Ishihama Y., Shirasu K. (2010). Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 153: 1161–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H., Engelbrecht J., Brunak S., von Heijne G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10: 1–6. [DOI] [PubMed] [Google Scholar]
- Olinares P.D., Ponnala L., van Wijk K.J. (2010). Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering. Mol. Cell. Proteomics 9: 1594–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverdatto S., Beilinson V., Nielsen N.C. (1999). A multisubunit acetyl coenzyme A carboxylase from soybean. Plant Physiol. 119: 961–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesler K., Shintani D., Savage L., Boddupalli S., Ohlrogge J. (1997). Targeting of the Arabidopsis homomeric acetyl-coenzyme A carboxylase to plastids of rapeseeds. Plant Physiol. 113: 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuska S.A., Girke T., Benning C., Ohlrogge J.B. (2002). Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14: 1191–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salie M.J., Thelen J.J. (2016). Regulation and structure of the heteromeric acetyl-CoA carboxylase. Biochim. Biophys. Acta 1861: 1207–1213. [DOI] [PubMed] [Google Scholar]
- Sasaki Y., Nagano Y. (2004). Plant acetyl-CoA carboxylase: structure, biosynthesis, regulation, and gene manipulation for plant breeding. Biosci. Biotechnol. Biochem. 68: 1175–1184. [DOI] [PubMed] [Google Scholar]
- Sasaki Y., Kozaki A., Hatano M. (1997). Link between light and fatty acid synthesis: thioredoxin-linked reductive activation of plastidic acetyl-CoA carboxylase. Proc. Natl. Acad. Sci. USA 94: 11096–11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte W., Töpfer R., Stracke R., Schell J., Martini N. (1997). Multi-functional acetyl-CoA carboxylase from Brassica napus is encoded by a multi-gene family: indication for plastidic localization of at least one isoform. Proc. Natl. Acad. Sci. USA 94: 3465–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havlis J., Olsen J.V., Mann M. (2006). In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1: 2856–2860. [DOI] [PubMed] [Google Scholar]
- Sims T.L., Goldberg R.B. (1989). The glycinin Gy1 gene from soybean. Nucleic Acids Res. 17: 4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swatek K.N., Graham K., Agrawal G.K., Thelen J.J. (2011). The 14-3-3 isoforms chi and epsilon differentially bind client proteins from developing Arabidopsis seed. J. Proteome Res. 10: 4076–4087. [DOI] [PubMed] [Google Scholar]
- Swatek K.N., Wilson R.S., Ahsan N., Tritz R.L., Thelen J.J. (2014). Multisite phosphorylation of 14-3-3 proteins by calcium-dependent protein kinases. Biochem. J. 459: 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen J.J., Ohlrogge J.B. (2002a). Metabolic engineering of fatty acid biosynthesis in plants. Metab. Eng. 4: 12–21. [DOI] [PubMed] [Google Scholar]
- Thelen J.J., Ohlrogge J.B. (2002b). The multisubunit acetyl-CoA carboxylase is strongly associated with the chloroplast envelope through non-ionic interactions to the carboxyltransferase subunits. Arch. Biochem. Biophys. 400: 245–257. [DOI] [PubMed] [Google Scholar]
- Thelen J.J., Ohlrogge J.B. (2002c). Both antisense and sense expression of biotin carboxyl carrier protein isoform 2 inactivates the plastid acetyl-coenzyme A carboxylase in Arabidopsis thaliana. Plant J. 32: 419–431. [DOI] [PubMed] [Google Scholar]
- Thelen J.J., Mekhedov S., Ohlrogge J.B. (2001). Brassicaceae express multiple isoforms of biotin carboxyl carrier protein in a tissue-specific manner. Plant Physiol. 125: 2016–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissot G., Job D., Douce R., Alban C. (1996). Protein biotinylation in higher plants: characterization of biotin holocarboxylase synthetase activity from pea (Pisum sativum) leaves. Biochem. J. 314: 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.