Abstract
Recently we identified that PREX1 overexpression is critical for metastatic but not tumorigenic growth in a mouse model of NRAS-driven melanoma. In addition, a PREX1 gene signature correlated with and was dependent on ERK mitogen-activated protein kinase (MAPK) activation in human melanoma cell lines. In the current study, the underlying mechanism of PREX1 overexpression in human melanoma was assessed. PREX1 protein levels were increased in melanoma tumor tissues and cell lines compared with benign nevi and normal melanocytes, respectively. Suppression of PREX1 by siRNA impaired invasion but not proliferation in vitro. PREX1-dependent invasion was attributable to PREX1-mediated activation of the small GTPase RAC1 but not the related small GTPase CDC42. Pharmacologic inhibition of ERK signaling reduced PREX1 gene transcription and additionally regulated PREX1 protein stability. This ERK-dependent upregulation of PREX1 in melanoma, due to both increased gene transcription and protein stability, contrasts with the mechanisms identified in breast and prostate cancers, where PREX1 overexpression was driven by gene amplification and HDAC-mediated gene transcription, respectively. Thus, although PREX1 expression is aberrantly upregulated and regulates RAC1 activity and invasion in these three different tumor types, the mechanisms of its upregulation are distinct and context-dependent.
Keywords: PREX1, RhoGEF, Rac-GEF, RAC1, ERK, melanoma
INTRODUCTION
Driver roles in cancer have been identified for several members of the Dbl family of Rho guanine nucleotide exchange factors (RhoGEFs), most prominently ECT2, TIAM1, VAV1/2/3 and PREX1/2 (1,2). Increased expression and activation of these RhoGEFs result in enhanced activity of their Rho family small GTPase substrates in a context-dependent manner. For example, we recently identified overexpression of ECT2 protein in ovarian cancer, mediated by gene amplification, that resulted in activation of RHOA in the cytosol and RAC1 in the nucleus (3). The critical importance of RAC1 in cancer cell migration and invasion (4) has further focused attention on the mechanisms regulating activators of RAC1, such as the Dbl family of RhoGEFs.
The highly related Dbl RhoGEFs PREX1 and PREX2 (56% overall amino acid identity), which are GEFs for RAC1 and other Rho family small GTPases such as CDC42 (5), have been implicated as cancer drivers in several tumor types. The first cancer-driving role for PREX1 was described in prostate cancer (6), where limited analyses of tumor tissue revealed elevated levels of PREX1 protein. PREX1 was also elevated in metastatic but not primary prostate tumor cell lines. Suppression of PREX1 by RNA interference in PC-3 human prostate cancer cells decreased the levels of activated RAC and impaired tumor cell migration and invasion in vitro. Conversely, ectopic expression of PREX1 stimulated RAC activation, and promoted metastatic but not primary tumor growth of CWR22Rv1 prostate tumor cells. A follow-up study identified a histone deacetylase (HDAC)-mediated increase in PREX1 gene transcription as a basis for the increased levels of PREX1 protein in prostate cancer (7).
PREX1 overexpression was also identified in estrogen receptor-positive luminal and HER2-positive breast cancers (8-10). PREX1 protein was detected in ~60% of breast tumors but not in normal breast tissue. PREX1 is located in a chromosomal region frequently amplified in breast cancers and PREX1 gene amplification was detected in breast cancer cell lines, supporting gene amplification as a mechanism for PREX1 protein overexpression in these tumor types. Silencing of PREX1 expression by RNA interference reduced HER2-stimulated activation of RAC1, and impaired tumor cell motility and invasion in vitro and tumorigenic growth in vivo (8-10).
A role in cancer for the related RhoGEF PREX2 has also been identified, but not by overexpression. Instead, missense mutations in PREX2 have been identified in 25% of malignant melanomas (11). Although no clear mutational hotspots have been seen in melanomas, experimental studies support a gain-of-function consequence of these mutations (11,12). PREX2 missense mutations have also been found in 38% of cutaneous squamous cell carcinomas (13) and 17% of stomach adenocarcinomas (14). To date, a similar level of activating missense mutations in PREX1 in cancer has not been reported. However, the occurrence of activating missense mutations (e.g., P29S) in the PREX1 substrate, RAC1, in ~11% of melanomas (15,16) is also consistent with a driver role for overexpressed PREX1 in this disease.
Our previous studies revealed overexpression of PREX1 protein in melanoma cell lines and tumor tissue (17). Further, in a mouse model of melanoma, we determined that Prex1-deficient mice were impaired in forming tumor metastases but not primary tumors. Here, extending our mouse model studies, we demonstrate that PREX1 protein is increased in human melanoma tumor tissue and that PREX1 is required for human melanoma cell invasion but not proliferation.
In a separate earlier study, we also identified PREX1 as a gene upregulated by the ERK mitogen-activated protein kinase (MAPK) in melanoma (18). The RAF-MEK-ERK protein kinase cascade is aberrantly activated in up to 80% of melanomas through BRAF or NRAS mutation, and serves as a critical therapeutic target in this disease (19-22). These observations supported the possibility that PREX1 protein overexpression in melanoma is driven by ERK activation. Additionally, since PREX1 has a demonstrated driver role in other cancers, PREX1 overexpression may be a key driver of ERK-dependent melanoma growth. In the present study, we determined that PREX1 protein overexpression is blocked by pharmacologic inhibitors of RAF-MEK-ERK signaling, and that ERK regulates not only PREX1 gene transcription but also PREX1 protein stability. Thus, there are significant cancer type-distinct mechanisms that drive PREX1 overexpression in cancer.
MATERIALS AND METHODS
Human melanoma tissue and immunohistochemistry (IHC)
Following institutional review board approval, primary and metastatic melanomas were retrieved from a series of patients treated at UNC Healthcare. Immunohistochemical staining was performed in the UNC Department of Dermatology Dermatopathology Laboratory as we have recently described (23). Briefly, freshly cut 4-μm thick sections of formalin-fixed and paraffin-embedded melanoma tissue blocks were stained using the fully automated Leica Bond III system. Sections were pretreated using an onboard heat-induced epitope retrieval in EDTA buffer. Following incubation with PREX1 antibody (6F12; provided by Marcus Thelen, IRB, Switzerland), chromogenic detection was performed using the Leica Refined Red polymer detection system (Leica Microsystems). Some sections were also counterstained with hematoxylin and eosin (H&E). PREX1 antibody staining intensity was scored in a blinded manner by a pathologist (AJ Finn) as high, medium, low or none.
Tissue culture
Cutaneous melanoma, breast cancer and prostate cancer cell lines were obtained from ATCC. Cells were maintained in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), and were not cultured for longer than 6 months after receipt from cell banks.
siRNA transfection, proliferation, and invasion assays
For siRNA knockdown, A375, WM2664, SK-MEL-119 and Mel224 cells were plated in 6-well plates. Cells were transfected with 10 nM siRNA against PREX1 (Thermo Fisher s33364, s33365, s33366; PREX1 #1, #2, #3, respectively), RAC1 (Thermo Fisher s11711, s11712, s11713; RAC1 #1, #2, #3, respectively) or mismatch control (Dharmacon #D-001210-05), using Lipofectamine RNAimax (Life Technologies). Cells were serum-starved overnight for 18 h and then seeded for invasion assays after 48 h of siRNA knockdown. For the proliferation assay, cells were seeded at 2-3 × 103 cells/well in 96-well plates and allowed to grow for 72 h before incubation for 3 h in 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT). MTT was solubilized in DMSO and the absorbance was read at A570. For the Boyden chamber invasion assay, 1-3 × 104 cells were seeded into the upper chamber of Matrigel-coated invasion chambers in duplicate (Corning BioCoat) and allowed to invade towards 20% FBS in DMEM for 24 h. Invasion chambers were fixed and stained using a Diff-Quik staining kit (GE). Invasion chambers were imaged using a 10x objective lens on a Nikon TS100 microscope at 5 fields per insert, and images were analyzed to calculate invaded cells per field using ImageJ software. For the collagen spheroid invasion assay, we slightly modified a published protocol (24,25). Briefly, 5-10 × 103 cells were seeded in ultra-low attachment round-bottomed 96-well plates (Corning) for 96 h, a sufficient time for the cells to organize into spheroids. Spheroids were then transferred to 48-well plates and embedded in collagen (1 mg/ml rat-tail collagen, BD). Spheroids were imaged at 0 h and after 72 h of invasion using a 5x objective on a Nikon TS100 microscope. Total spheroid area was calculated as fold-change in area of 72 h outgrowth versus 0 h spheroid area, using ImageJ.
Pulldown assay to detect GTPase activity
Levels of active, GTP-bound RAC1 and CDC42 were assessed by an affinity pulldown assay as we described previously (26). Briefly, after 48 h of siRNA-mediated PREX1 knockdown, whole cell lysates were exposed to GST-PAK-PBD, which contains the binding domain of the shared RAC1/CDC42 effector PAK1. After resolving pulldown samples on 15% SDS-PAGE gels and western blotting for RAC1 (clone 23A9, BD) and CDC42 (BD), levels of each GTP-bound GTPase were normalized to both total protein and the vinculin loading control (Sigma) by densitometry analysis performed in ImageJ.
Drug treatment and western blotting
BRAF-mutant A375 and WM2664 cells were treated with BRAF inhibitor vemurafenib (Selleckchem) or ERK inhibitor SCH772984 (Merck, kindly provided by Ahmed Samatar), and NRAS-mutant SK-MEL-119 and Mel224 cells were treated with MEK inhibitor trametinib (Selleckchem) or SCH772984 for 24 and 48 h before samples were collected in RIPA lysis buffer. For PREX1 protein stability experiments, cells were co-treated with cycloheximide (50 μg/ml) and SCH772984 for a 24 h timecourse before samples were collected in RIPA. Whole cell lysates were resolved on 10% SDS-PAGE gels and western blotting was performed using antibodies to phospho-ERK1/2 (Thr202/Tyr204), total ERK1/2, phospho-RSK (Ser308), phospho-RSK (Thr359/Ser363), total RSK1/2/3, and c-myc (MYC) (Cell Signaling); β-actin and vinculin (Sigma), and PREX1 (6F12) (27). IRDye800-conjugated anti-mouse and anti-rabbit secondary antibodies were from Rockland Immunochemicals.
Quantitative PCR
Total RNA was isolated using an RNeasy kit (Qiagen) and reverse transcription was performed using the High Capacity RNA-to-cDNA kit (Thermo Fisher). Real time quantitative Taqman PCR was performed on the QuantStudio 6 Flex (Thermo Fisher) with FAM/MGB labeled probes against PREX1 (Hs00368207_m1, Hs_001031512, Thermo Fisher) and endogenous control VIC/TAMRA labeled β-actin (Thermo Fisher).
Statistical analysis
Data were analyzed using GraphPad Prism 6 software and statistical analyses were performed as indicated in the Figure Legends.
RESULTS
PREX1 overexpression is correlated with elevated ERK activation
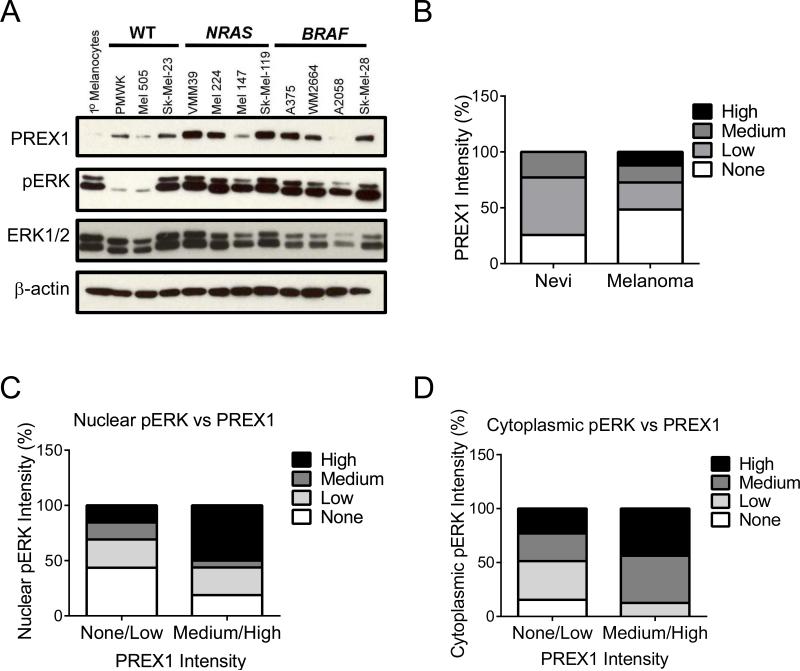
We recently identified overexpression of PREX1 protein in melanomas, determined that Prex1 deficiency impaired mouse melanoblast migration in vivo, and demonstrated that Prex1 expression is required for metastasis in an Nras-mutant genetically engineered mouse model of cutaneous melanoma (17). Since our previous gene array analyses identified PREX1 as an ERK activation-dependent gene (18), here we assessed a relationship among BRAF and NRAS mutation status, ERK activation and PREX1 protein overexpression in human melanoma. We first investigated the expression of PREX1 in a panel of human melanoma cell lines that did or did not harbor BRAF or NRAS mutations. The majority of BRAF- (3 of 4) or NRAS- (3 of 4) mutant cell lines exhibited substantially higher PREX1 protein expression when compared with normal melanocytes or with BRAF/NRAS wild type cell lines (Figure 1A). Generally, the level of activated, phosphorylated ERK (pERK) correlated with the level of PREX1. In our analyses, normal melanocytes may exhibit low pERK levels (18), or they may also display low PREX1 protein levels even in the presence of high pERK levels, as shown here.
Figure 1. PREX1 protein levels are elevated in melanoma patient tumor tissues and cell lines, along with phospho-ERK.
(A) Western blot analysis of PREX1 protein, phospho-ERK (pERK) and total ERK1/2 in a panel of WT, BRAF- or NRAS-mutant human melanoma tumor cell lines. (B-D) Human tissue samples of benign melanocytic nevi and malignant skin cutaneous melanoma were subjected to IHC for PREX1 and pERK. Shown are (B) the distribution of PREX1 expression in nevi versus melanoma samples as measured by IHC; n=35 and 33, respectively. Samples were first binned according to no, low, medium or high staining intensity for each protein, and then the distribution was graphed to show the relationship between PREX1 and the percent of samples that stained positive for (C) nuclear pERK or (D) cytoplasmic pERK.
We next utilized immunohistochemistry (IHC) to evaluate PREX1 protein expression and pERK levels in melanoma patient tissues. We first compared PREX1 expression in benign melanocytic nevi (n=35) and human melanoma tumors (n=33) (Figure 1B). A range of expression was seen in nevi, with ~75% expressing low-to-medium levels of PREX1. Since BRAF and NRAS mutations are found in a high percentage of nevi (28,29), it is not surprising to find PREX1 in nevi as well as in melanoma tissue. However, high level PREX1 expression was detected only in melanomas (~10%, Figure 1B).
ERK can phosphorylate numerous substrates present in both the nucleus and the cytoplasm (30,31), few of which have been firmly linked to specific outcomes of ERK-mediated signaling. Melanoma responses to pharmacological inhibitors of the RAF-MEK-ERK pathway (e.g., clinically, to BRAF inhibition (32) and preclinically, to inhibitors of ERK dimerization (33,34)) correlated with suppression of cytoplasmic pERK. We therefore evaluated the distribution of pERK in our human melanoma tissues, and found that levels of pERK were correlated with those of PREX1 both in the nucleus (Figure 1C) and in the cytoplasm (Figure 1D). Both nuclear and cytoplasmic ERK activity may contribute to increased expression of PREX1.
PREX1 regulates invasion in a complex manner in both BRAF- and NRAS-mutant melanoma cell lines
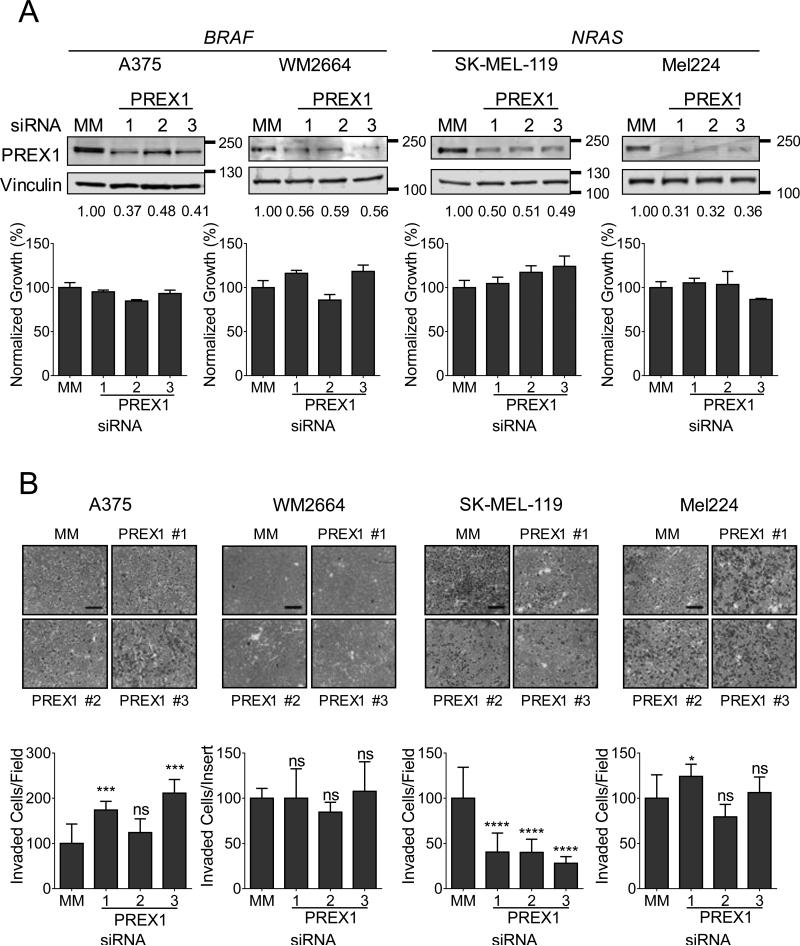
An unexpected observation in our studies of PREX1 function in a mouse model of melanoma was that Prex1 deficiency greatly impaired metastatic but not tumorigenic growth. This result contrasts with studies evaluating the role of PREX1 overexpression in human breast cancer cells, where stable shRNA-mediated suppression of PREX1 reduced their tumorigenic growth (8-10). We therefore compared the effect of PREX1 suppression in human melanoma cell lines on both proliferation and invasion in vitro.
To evaluate the role of PREX1 overexpression in cell growth, we first used three independent siRNAs to knock down PREX1 in two BRAF-mutant (A375 and WM2664) and two NRAS-mutant cell lines (SK-MEL-119 and Mel224) (Figure 2A, upper panels). We found that transient (72 h) suppression of PREX1 did not significantly reduce their proliferation in vitro (Figure 2A, lower panels), consistent with the lack of effect of Prex1 deficiency on the growth of primary melanomas in mice. Next, we evaluated the role of PREX1 in invasion by analysis of invasion through Matrigel towards serum as a chemoattractant. We observed a surprisingly heterogeneous response to PREX1 knockdown that was independent of BRAF or NRAS mutational status. For example, the NRAS-mutant line SK-MEL-119 exhibited a 60-70% decrease in invasion upon knockdown of PREX1 (p<0.0001, Figure 2B) whereas the BRAF-mutant cell line A375 conversely exhibited a ~2-fold increase (p<0.001). In contrast, the already very low degree of directed invasion of the BRAF-mutant line WM2664 was unaffected by PREX1 knockdown. Similarly, the invasive NRAS-mutant cell line Mel224 was largely unaffected, despite efficient knockdown of PREX1. These data demonstrate that PREX1 plays a complex and variable role in directed invasion towards an attractant.
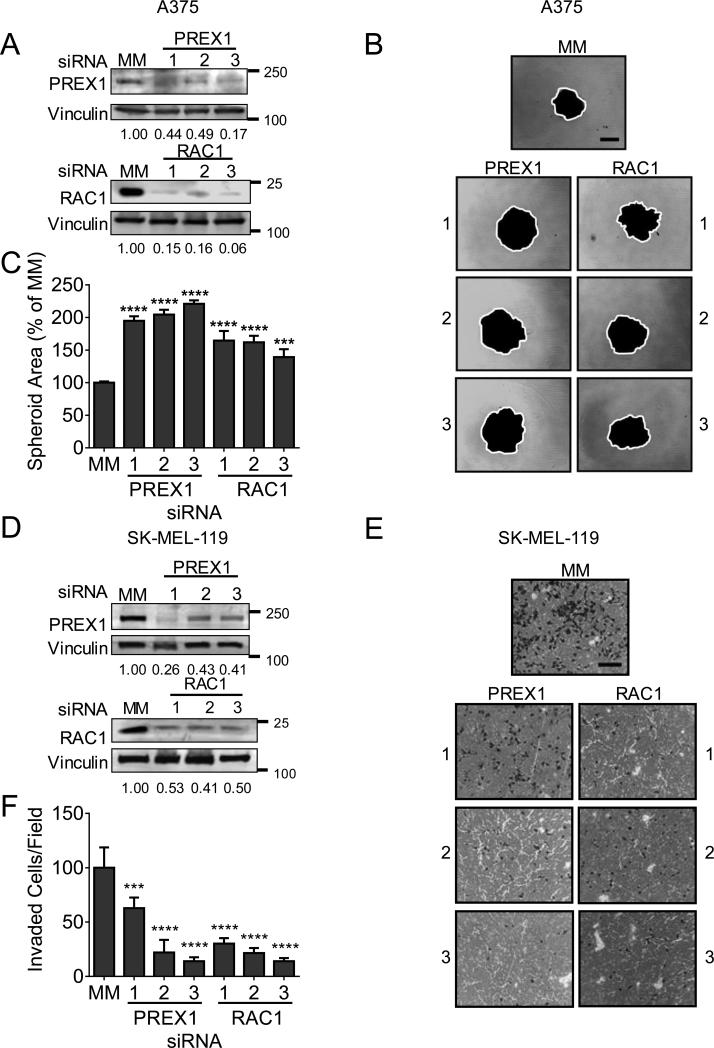
Figure 2. PREX1 regulates spheroid formation and invasion, but not proliferation, of BRAF- and NRAS-mutant melanoma cells in a context-dependent manner.
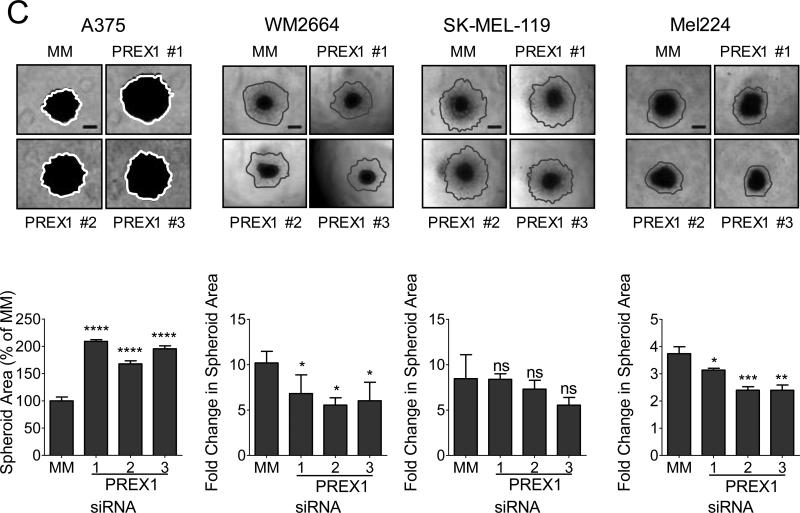
(A) BRAF-mutant A375 and WM2664 and NRAS-mutant SK-MEL-119 and Mel224 cells were transfected with siRNA against PREX1 or a mismatch control (MM) for 48 h, and knockdown was confirmed by western blot (upper panels). Apparent molecular weights are indicated to the right of each panel; vinculin served as a loading control. Fold changes in protein expression compared to MM control are shown in numbers below each blot. Effects of PREX1 knockdown on growth in monolayer culture were determined by MTT assay at 72 hr (lower panels). (B) To determine the effects of PREX1 knockdown on invasion, cells were seeded in the upper chamber of a Matrigel-coated Boyden chamber, and allowed to invade towards serum for 24 h, then stained and imaged. ImageJ was used to quantitate invaded cells per field for 5 fields per insert in duplicate inserts (A375, SK-MEL-119, Mel224) or invaded cells over both inserts (WM2664). (C) For spheroid collagen invasion assays, spheroids were allowed to form for 4 days. Total spheroid area was normalized to that of mismatch control-treated cells; impaired spheroid formation is indicated by increased area of the flattened spheroid. WM2664, SK-MEL-119, and Mel224 spheroids were embedded in a collagen matrix and imaged (day 0) and the extent of cell outgrowth/invasion was imaged 3 days later (thick gray lines). Fold change in area from day 3 to day 0 was calculated in ImageJ. Data are represented as mean ± SD and statistical significance was evaluated by Student's t-test, where *: p<0.05, **: p<0.01, ***: p<0.001, ****: p<0.0001. Scale bar represents 250 μm for invasion assays and 500 μm for spheroids. Experiments shown are representative of two (WM2664, SK-MEL-119) or three (A375, SK-MEL-119) independent experiments.
Next, we investigated the role of PREX1 in a three-dimensional spheroid formation and collagen invasion assay, which mimics the in vivo tumor environment of human skin (35). Figure 2C illustrates both spheroid formation and the subsequent invasion of cells from the spheroid into the surrounding collagen matrix. In three of the four cell lines, knockdown of PREX1 impaired spheroid invasion into collagen, either trending (SK-MEL-119) or significantly so (Mel224, WM2664). In contrast, A375 spheroids were defective in formation and did not invade the surrounding collagen matrix. The nearly doubled total spheroid area of PREX1-knockdown A375 cells compared to mismatch control cells observed after 4 days in culture was caused by a flattening of the three-dimensional spheroid structure and not by increased invasion or by increased proliferation; no change in proliferation occurred upon loss of PREX1 (Fig. 2A).
Collectively, our results suggest a complex and context-dependent role for PREX1 in driving both directed invasion and three-dimensional spheroid collagen outgrowth of human melanomas, and one that is not dependent on BRAF or NRAS mutational status.
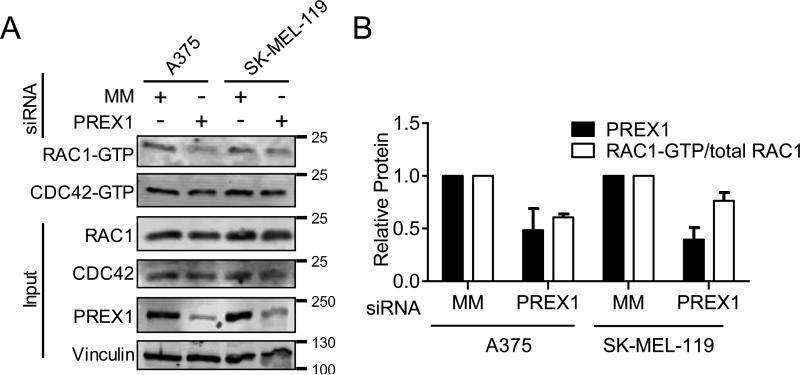
PREX1 regulates active, GTP-bound RAC1 but not CDC42 in melanoma cells
Although PREX1 is considered a RAC-selective GEF (36), PREX1 is also active on CDC42 (27). We therefore investigated which Rho family small GTPases are activated downstream of PREX1 in human melanoma cells. We found that knockdown of PREX1 decreased the levels of activated RAC1, as measured by RAC1-GTP pulldown, in both BRAF-mutant A375 and NRAS-mutant SK-MEL-119 cells (Figure 3). Despite the continued presence of other RacGEFs capable of inducing nucleotide exchange on RAC1, even incomplete loss of PREX1 was sufficient to cause a substantial decrease in RAC1-GTP (Figure 3). This effect was selective for RAC1, as the levels of activated CDC42 did not decrease (Figure 3). These results support a role for PREX1 in regulating RAC1 activity and subsequent RAC1-driven invasive behavior of melanomas.
Figure 3. PREX1 regulates active RAC1-GTP, but not active CDC42-GTP, in melanoma cells.
A375 and SK-MEL-119 cells were transfected with pooled siRNAs #1-3 against PREX1 or MM control for 48 h, then starved overnight (18 h). RAC1-GTP and CDC42-GTP were measured by GST-PAK-PBD pulldown (A). Apparent molecular weights are indicated to the right of each panel; vinculin served as a loading control. Quantification (mean ± SD) using ImageJ (B) is representative of two independent experiments.
We next asked if the loss of RAC1 was sufficient to phenocopy the impairment of invasion that we observed upon loss of PREX1. We found that knockdown of either PREX1 or RAC1 with three independent siRNAs for each (Figure 4A) was sufficient to substantially impair spheroid formation of A375 cells, as demonstrated by an increase in the flattened spheroid area (Figure 4B,C). The degree of impairment upon knockdown of RAC1 was highly significant (p<0.0001, Figure 4C) and comparable to the degree of impairment observed upon knockdown of PREX1 (p<0.0001, Figure 4C). Next, we observed that loss of RAC1 (Figure 4D) was sufficient to prevent the vast majority of SK-MEL-119 cell invasion in the Boyden chamber assay (Figure 4E). This decrease in invasion was similar to the decrease seen upon PREX1 knockdown (p<0.0001, Figure 4F). The ability of RAC1 to phenocopy PREX1 in impairing both spheroid formation and directed invasion supports the idea that RAC1 is the most critical Rho family small GTPase downstream of PREX1 in regulating invasive melanoma behavior. Of note, other Rho family small GTPases such as RND3 have also been shown to be regulated by the RAF-MEK-ERK pathway and to contribute to melanoma invasion and spheroid outgrowth (37,38). However, as a Rho-like rather than a Rac-like GTPase, RND3 is unlikely to be a target of PREX1 in this context (39).
Figure 4. RAC1 phenocopies PREX1 in regulating spheroid formation in A375 and invasion in SK-MEL-119.
A375 and SK-MEL-119 cells were transfected with siRNA against PREX1, RAC1, or MM control for 48 h before seeding into invasion chambers. Knockdown was confirmed by western blot in A375 (panel A) and SK-MEL-119 (panel D). Apparent molecular weights are on the right of each panel; vinculin was a loading control. Total spheroid area of A375 cells was quantified after 4 days using ImageJ (white line); increased flattened spheroid area indicates impaired spheroid formation (panels B,C). SK-MEL-119 cells were starved overnight and seeded in a Boyden chamber assay with 20% serum as a chemoattractant and allowed to invade for 24h (10x magnification) (E). Stained inserts were quantified for invaded cells/field, 10 fields per condition, using ImageJ (F). Data are represented as mean ± SD. Student's t-test, where *: p<0.05, **: p<0.01, ***: p<0.001, ****: p<0.0001). Scale bar represents 250 μm for invasion assays and 500 μm for spheroids.
PREX1 protein levels are positively regulated by ERK activity in melanoma
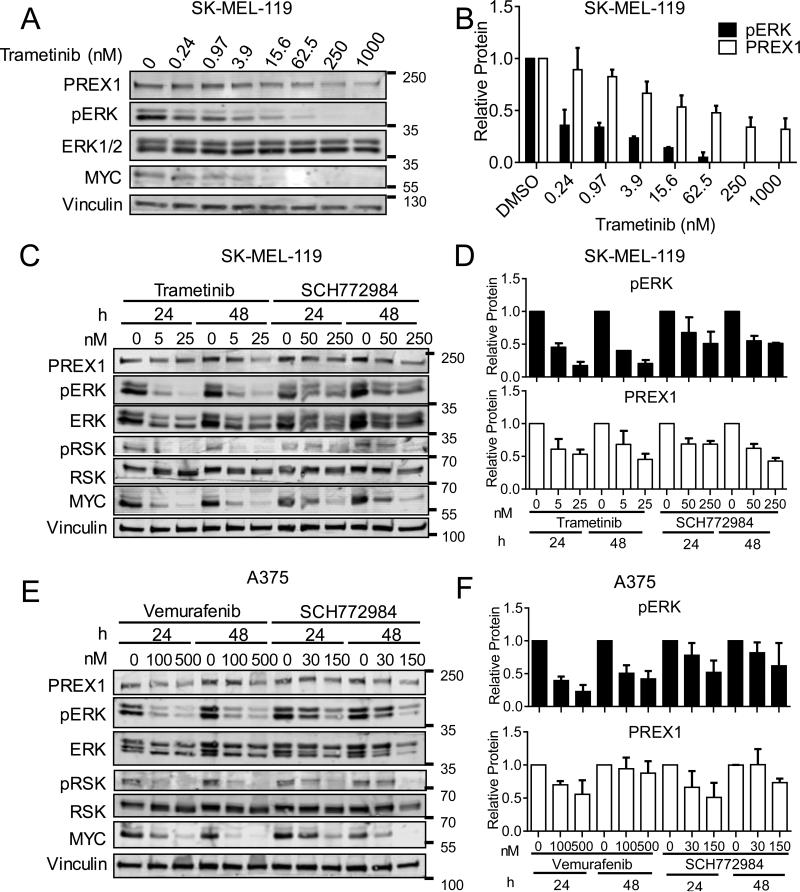
Our evaluation of human melanoma cell lines and tumor tissue found a correlation between phosphorylated ERK and PREX1 protein overexpression. To directly address whether ERK activation is required for PREX1 overexpression, we evaluated whether pharmacologic inhibition of RAF-MEK-ERK signaling would reduce PREX1 protein levels in BRAF- and NRAS-mutant melanoma. We first treated NRAS-mutant SK-MEL-119 cells with increasing concentrations of the MEK inhibitor trametinib. The more effective the inhibition of MEK, as measured by decreasing levels of phosphorylated and activated ERK (pERK) and total MYC (an ERK substrate; ERK phosphorylation blocks degradation), the greater the decrease in PREX1 protein (Figures 5A,B), suggesting a direct correlation between ERK activity and PREX1 protein levels. We next wanted to determine whether this dose-dependent decrease in PREX1 protein would also occur when the ERK MAPK cascade was inhibited at different nodes, and whether such an effect is time-dependent. We therefore treated SK-MEL-119 cells with two different concentrations (1 × EC50 and 5 × EC50 for growth) of either trametinib or the ERK inhibitor SCH772984, for either 24 or 48 h (Figures 5C,D). PREX1 protein levels tracked closely with the level of pERK at each concentration of inhibitor, and this was sustained for 48 h. Next, we investigated whether PREX1 protein was similarly regulated downstream of the ERK-MAPK cascade in BRAF-mutant melanoma cells. We treated A375 cells similarly but with the BRAF inhibitor vemurafenib or SCH772984 for 24 or 48 h (Figures 5E,F). Similarly to SK-MEL-119 cells, levels of PREX1 in A375 cells tracked closely with the levels of pERK and demonstrated a time-dependent effect. In both SK-MEL-119 and A375 cells, phosphorylation of the ERK substrate RSK (pRSK) and total MYC served as effective markers to demonstrate inhibition of ERK, as we have observed in other settings (40). These results indicate that ERK MAPK activity is an important contributor to the total amount of PREX1 protein in melanoma cells.
Figure 5. PREX1 protein levels are regulated by the ERK kinase cascade.
NRAS-mutant SK-MEL-119 cells were first treated with the indicated concentrations of MEKi trametinib for 48 h, and lysates immunoblotted for PREX1, pERK and MYC (A; quantified in B). SK-MEL-119 cells were next treated with trametinib or the ERK inhibitor SCH772984 for 24 or 48 h, and lysates probed for pERK and PREX1 (C; quantified in D), and for pRSK and total MYC to monitor ERK pathway inhibition (C). Similarly, BRAF-mutant A375 cells were treated with the BRAF inhibitor vemurafenib or with SCH772984 for 24 or 48 h and lysates probed as above (E; quantified in F). Quantification is of n=3 experiments for SK-MEL-119 and n=4 for A375. Data are represented as mean ± SD.
PREX1 levels are regulated by ERK both transcriptionally and post-transcriptionally in melanoma
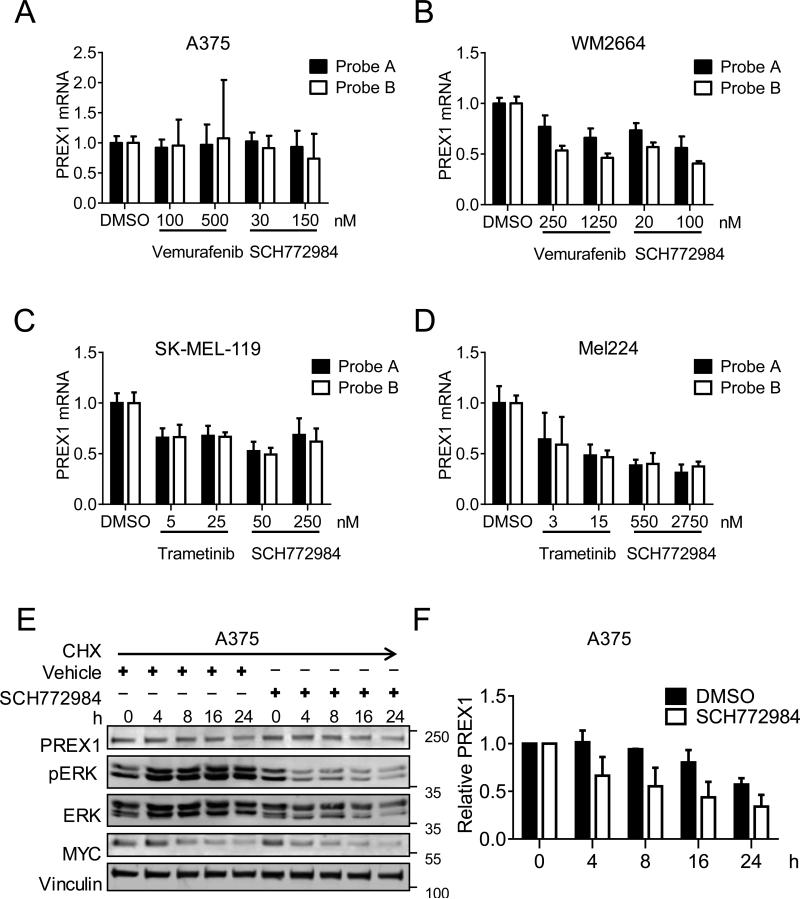
To determine if the loss of PREX1 protein upon blockade of the ERK MAPK cascade was due to loss of PREX1 mRNA, we treated two BRAF-mutant and two NRAS-mutant melanoma cell lines with ERK MAPK cascade inhibitors. BRAF-mutant A375 and WM2664 cells were treated for 24 h with vemurafenib or SCH772984 as above. Taqman quantitative PCR analysis revealed that PREX1 mRNA, measured by two independent probes, did not change upon inhibition of BRAF or ERK in A375 cells (Figure 6A) but decreased dose-dependently in WM2664 cells upon inhibition of either BRAF or ERK (Figure 6B). NRAS-mutant cells were treated with trametinib or SCH772984 as above. We observed that PREX1 mRNA also decreased upon ERK inhibition in both SK-MEL-119 (Figure 6C) and Mel224 cells (Figure 6D). Additional melanoma lines also exhibited reduced PREX1 mRNA levels when treated with inhibitors of the ERK MAPK cascade, including BRAF-mutant SK-MEL-28 and NRAS-mutant SK-MEL-147 cells (Figures S3A,B). Thus, in the majority of melanoma cell lines, ERK MAPK regulates PREX1 protein levels both transcriptionally and post-transcriptionally.
Figure 6. PREX1 levels are regulated by ERK both transcriptionally and post-transcriptionally.
BRAF-mutant A375 and WM2664 cells were treated with vemurafenib or SCH772984 for 24 h and PREX1 mRNA levels were measured by Taqman qPCR using two independent probes (A,B). NRAS-mutant SK-MEL-119 and Mel224 cells were treated with trametinib or SCH772984 for 24 h, and PREX1 mRNA measured as above (C,D). Taqman analyses indicate compiled results of n=2 experiments for WM2664 and Mel224, and n=3 experiments for A375 and SK-MEL-119. To test posttranscriptional regulation, A375 cells were treated with vehicle or SCH772984 in the presence of 50 μg/ml cycloheximide, and lysates were probed by western blot for PREX1, pERK and MYC (E). Quantification of PREX1 levels using ImageJ (F) is representative of n=2 independent experiments. Data are represented as mean ± SD.
That ERK MAPK activity altered PREX1 protein levels in A375 melanoma cells without changes at the transcriptional level suggested a post-transcriptional mechanism for ERK MAPK-mediated regulation of PREX1 protein in these cells. To investigate this possibility, we treated A375 cells with vehicle or SCH772984 in the presence of the protein synthesis inhibitor cycloheximide at various time points over 24 h (Figures 6E,F). Inhibition of ERK led to greater loss of PREX1 protein in the presence of cycloheximide compared to vehicle-treated cells. Similar results were obtained upon treatment of SK-MEL-119 cells with trametinib in the presence of cycloheximide (Figures S3C,D). These results indicate that ERK can regulate protein stability as well as transcription of PREX1 in melanoma cell lines.
Finally, we tested the possibility that PREX1 is not only an ERK target but also an ERK activator. Since it has been demonstrated that PREX1 can regulate MEK-ERK signaling through RAC1 in breast cancer (8,41), we also examined whether PREX1 can regulate ERK1/2 phosphorylation in our melanoma lines. We found that knockdown of PREX1 did not alter ERK1/2 phosphorylation in the BRAF-mutant cell lines, A375 and WM2664, or the NRAS-mutant cell lines, SK-MEL-119 and Mel224 (Figures S4A,B and S4C,D, respectively).
To determine the generality of ERK regulation of PREX1 expression in no-nmelanoma tumor types, we also tested whether they held true in breast and prostate cancer cell lines. PREX1 has been shown to be overexpressed in these tumor types, but the role of ERK in its expression has not been explored. Unlike our observations in melanoma cells, we observed that inhibition of the ERK MAPK cascade in T47D and MCF7 breast cancer cells did not reduce PREX1 protein (Figures S5A,C) or mRNA (Figures S5B,D). Conversely, in PC-3 prostate cancer cells, inhibition of the ERK MAPK cascade reduced PREX1 mRNA levels (Figure S5F) but had minimal effects on PREX1 protein (Figure S5E). These results support distinct mechanisms of regulating PREX1 expression in melanoma through ERK1/2 that do not apply in breast or prostate cancer, cancers in which BRAF and RAS mutation frequencies are low. Overall, our results support that both ERK regulation of PREX1 abundance and PREX1 regulation of ERK phosphorylation are context-dependent, and may differ between breast and prostate cancers and cutaneous melanoma.
DISCUSSION
We determined that the ERK MAPK cascade plays an important role in driving PREX1 protein overexpression in both BRAF- and NRAS-mutant melanomas. In contrast, ERK is not the key driver for PREX1 overexpression in prostate (6,7) or in breast carcinomas (8-10,42), where its abundance is associated with HDAC-dependent (7) PREX1 gene transcription and with HDAC- and methylation-dependent PREX1 gene transcription (42) and gene amplification (8,10,42), respectively. Thus, there are striking cancer-type differences in mechanisms driving PREX1 overexpression. In support of this idea, PREX1 and PREX2 display distinct expression and mutation patterns in breast cancer, prostate cancer, and melanoma (Figure S1), and PREX1 in particular is differentially amplified in breast cancer, prostate cancer and cutaneous melanoma (Figure S2). Our findings also suggest that loss of PREX1-RAC1 signaling may contribute to the clinical response of patients with BRAF-mutant melanomas to BRAF and MEK inhibitors.
The effectiveness of these inhibitors provides compelling evidence that aberrant ERK signaling is a major driver of melanoma growth. Despite this clear driver role, the ERK targets important for melanoma growth remain poorly characterized. The ERK1/2 kinases can phosphorylate more than 200 known substrates (30,31) and serve as master regulators of numerous transcription factors (43), both directly and indirectly, through both transcriptional and post-translational mechanisms (40,43-45) We have determined that ERK regulates PREX1 expression levels in part via protein stability, a mechanism also not observed in other cancers. ERK regulation of PREX1 protein stability presents a previously unknown mechanism of maintaining PREX1 protein expression and may explain the basis for the relatively high PREX1 expression in malignant melanomas where the ERK MAPK cascade is upregulated.
Finally, the PREX1-related RhoGEF PREX2 is activated by missense mutations in 25% of metastatic melanomas, especially by truncating mutations mutations (11,12), and mutational activation of the PREX1/2 target RAC1 has been observed in ~11% of melanomas (15,16). The rarity of PREX1 truncating mutations and lack of apparent hotspots among the few missense mutations argue that this is not a significant mechanism of PREX1 activation in melanoma. Instead, our determination that PREX1 is overexpressed and regulated at multiple levels in response to the ERK MAPK cascade characterizes a third mechanism for driving aberrant RAC1 signaling in melanoma. Our findings that loss of RAC1 phenocopies loss of PREX1 with respect to invasive behavior regardless of BRAF or NRAS mutation status supports the importance of the PREX1-RAC1 relationship as a promoter of melanoma cell invasion. Interestingly, downregulation of the RacGEF TIAM1 by mutant BRAF was shown to enhance invasion of human melanoma cells (46). Although PREX1 was not examined in that particular study, PREX1 has consistently demonstrated a positive role in invasion (6,17,47), whereas TIAM1 can be either a positive or a negative regulator of this process (1,46,48-50). Thus, the relative input from different upstream activators of RAC1 can have a profound influence on melanoma invasion.
In summary, we have demonstrated that the ERK MAPK cascade mediates overexpression of PREX1 in melanoma at multiple levels, and by mechanisms that are distinct from those identified previously in other cancer types. Our results contribute to a better understanding of how RacGEFs are modulated in distinct cancer contexts.
Supplementary Material
Acknowledgments
Financial Support: CJD and ADC are supported by grants from the NIH (CA42978, CA179193, CA175747, CA199235) and the Pancreatic Cancer Action Network-AACR. CJD is also supported by the Lustgarten Pancreatic Cancer Foundation. MBR has fellowship support from the NIH (NRSA predoctoral fellowship, F31 CA192829).
Footnotes
Conflicts of Interest: The authors declare no conflict of interest.
Implications:
This study identifies a ERK-dependent mechanism that drives PREX1 upregulation and subsequent RAC1-dependent invasion in BRAF- and NRAS-mutant melanoma.
AUTHORS’ CONTRIBUTIONS
MB Ryan, CJ Der and AD Cox conceived and designed the study. MB Ryan performed all experiments except those shown in Figures 1A and 1B. KH Pedone performed the experiment shown in Figure 1A. AJ Finn performed the experiment shown in Figure 1B. NE Thomas provided melanoma tumor samples. MB Ryan, CJ Der and AD Cox wrote the manuscript, and all authors reviewed it.
REFERENCES
- 1.Cook DR, Rossman KL, Der CJ. Rho guanine nucleotide exchange factors: regulators of Rho GTPase activity in development and disease. Oncogene. 2014;33(31):4021–35. doi: 10.1038/onc.2013.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10(12):842–57. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huff LP, Decristo MJ, Trembath D, Kuan PF, Yim M, Liu J, et al. The Role of Ect2 Nuclear RhoGEF Activity in Ovarian Cancer Cell Transformation. Genes Cancer. 2013;4(11-12):460–75. doi: 10.1177/1947601913514851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Symons M, Segall JE. Rac and Rho driving tumor invasion: who's at the wheel? Genome Biol. 2009;10(3):213. doi: 10.1186/gb-2009-10-3-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Welch HC. Regulation and function of P-Rex family Rac-GEFs. Small GTPases. 2015;6(2):49–70. doi: 10.4161/21541248.2014.973770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin J, Xie Y, Wang B, Hoshino M, Wolff DW, Zhao J, et al. Upregulation of PIP3-dependent Rac exchanger 1 (P-Rex1) promotes prostate cancer metastasis. Oncogene. 2009;28(16):1853–63. doi: 10.1038/onc.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong CY, Wuriyanghan H, Xie Y, Lin MF, Abel PW, Tu Y. Epigenetic regulation of phosphatidylinositol 3,4,5-triphosphate-dependent Rac exchanger 1 gene expression in prostate cancer cells. J Biol Chem. 2011;286(29):25813–22. doi: 10.1074/jbc.M110.211292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dillon LM, Bean JR, Yang W, Shee K, Symonds LK, Balko JM, et al. P-REX1 creates a positive feedback loop to activate growth factor receptor, PI3K/AKT and MEK/ERK signaling in breast cancer. Oncogene. 2015;34(30):3968–76. doi: 10.1038/onc.2014.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montero JC, Seoane S, Ocana A, Pandiella A. P-Rex1 participates in Neuregulin-ErbB signal transduction and its expression correlates with patient outcome in breast cancer. Oncogene. 2011;30(9):1059–71. doi: 10.1038/onc.2010.489. [DOI] [PubMed] [Google Scholar]
- 10.Sosa MS, Lopez-Haber C, Yang C, Wang H, Lemmon MA, Busillo JM, et al. Identification of the Rac-GEF P-Rex1 as an essential mediator of ErbB signaling in breast cancer. Mol Cell. 2010;40(6):877–92. doi: 10.1016/j.molcel.2010.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485(7399):502–6. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lissanu Deribe Y, Shi Y, Rai K, Nezi L, Amin SB, Wu CC, et al. Truncating PREX2 mutations activate its GEF activity and alter gene expression regulation in NRAS-mutant melanoma. Proc Natl Acad Sci U S A. 2016;113(9):E1296–305. doi: 10.1073/pnas.1513801113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li YY, Hanna GJ, Laga AC, Haddad RI, Lorch JH, Hammerman PS. Genomic analysis of metastatic cutaneous squamous cell carcinoma. Clin Cancer Res. 2015;21(6):1447–56. doi: 10.1158/1078-0432.CCR-14-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–9. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44(9):1006–14. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindsay CR, Lawn S, Campbell AD, Faller WJ, Rambow F, Mort RL, et al. PRex1 is required for efficient melanoblast migration and melanoma metastasis. Nat Commun. 2011;2:555. doi: 10.1038/ncomms1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shields JM, Thomas NE, Cregger M, Berger AJ, Leslie M, Torrice C, et al. Lack of extracellular signal-regulated kinase mitogen-activated protein kinase signaling shows a new type of melanoma. Cancer Res. 2007;67(4):1502–12. doi: 10.1158/0008-5472.CAN-06-3311. [DOI] [PubMed] [Google Scholar]
- 19.Goetz EM, Ghandi M, Treacy DJ, Wagle N, Garraway LA. ERK mutations confer resistance to mitogen-activated protein kinase pathway inhibitors. Cancer Res. 2014;74(23):7079–89. doi: 10.1158/0008-5472.CAN-14-2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwong LN, Boland GM, Frederick DT, Helms TL, Akid AT, Miller JP, et al. Co-clinical assessment identifies patterns of BRAF inhibitor resistance in melanoma. J Clin Invest. 2015;125(4):1459–70. doi: 10.1172/JCI78954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryan MB, Der CJ, Wang-Gillam A, Cox AD. Targeting RAS-mutant cancers: is ERK the key? Trends Cancer. 2015;1(3):183–98. doi: 10.1016/j.trecan.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sullivan R, LoRusso P, Boerner S, Dummer R. Achievements and challenges of molecular targeted therapy in melanoma. Am Soc Clin Oncol Educ Book. 2015:177–86. doi: 10.14694/EdBook_AM.2015.35.177. [DOI] [PubMed] [Google Scholar]
- 23.Pearlstein MV, Zedek DC, Ollila DW, Treece A, Gulley ML, Groben PA, et al. Validation of the VE1 immunostain for the BRAF V600E mutation in melanoma. J Cutan Pathol. 2014;41(9):724–32. doi: 10.1111/cup.12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan KT, Asokan SB, King SJ, Bo T, Dubose ES, Liu W, et al. LKB1 loss in melanoma disrupts directional migration toward extracellular matrix cues. J Cell Biol. 2014;207(2):299–315. doi: 10.1083/jcb.201404067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vultur A, Villanueva J, Krepler C, Rajan G, Chen Q, Xiao M, et al. MEK inhibition affects STAT3 signaling and invasion in human melanoma cell lines. Oncogene. 2014;33(14):1850–61. doi: 10.1038/onc.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiordalisi JJ, Keller PJ, Cox AD. PRL tyrosine phosphatases regulate rho family GTPases to promote invasion and motility. Cancer Res. 2006;66(6):3153–61. doi: 10.1158/0008-5472.CAN-05-3116. [DOI] [PubMed] [Google Scholar]
- 27.Welch HC, Condliffe AM, Milne LJ, Ferguson GJ, Hill K, Webb LM, et al. P-Rex1 regulates neutrophil function. Curr Biol. 2005;15(20):1867–73. doi: 10.1016/j.cub.2005.09.050. [DOI] [PubMed] [Google Scholar]
- 28.Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33(1):19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 29.Poynter JN, Elder JT, Fullen DR, Nair RP, Soengas MS, Johnson TM, et al. BRAF and NRAS mutations in melanoma and melanocytic nevi. Melanoma Res. 2006;16(4):267–73. doi: 10.1097/01.cmr.0000222600.73179.f3. [DOI] [PubMed] [Google Scholar]
- 30.Yoon S, Seger R. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors. 2006;24(1):21–44. doi: 10.1080/02699050500284218. [DOI] [PubMed] [Google Scholar]
- 31.Roskoski R., Jr. ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol Res. 2012;66(2):105–43. doi: 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 32.Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467(7315):596–9. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casar B, Pinto A, Crespo P. Essential role of ERK dimers in the activation of cytoplasmic but not nuclear substrates by ERK-scaffold complexes. Mol Cell. 2008;31(5):708–21. doi: 10.1016/j.molcel.2008.07.024. [DOI] [PubMed] [Google Scholar]
- 34.Herrero A, Pinto A, Colon-Bolea P, Casar B, Jones M, Agudo-Ibanez L, et al. Small Molecule Inhibition of ERK Dimerization Prevents Tumorigenesis by RASERK Pathway Oncogenes. Cancer Cell. 2015;28(2):170–82. doi: 10.1016/j.ccell.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Smalley KS, Haass NK, Brafford PA, Lioni M, Flaherty KT, Herlyn M. Multiple signaling pathways must be targeted to overcome drug resistance in cell lines derived from melanoma metastases. Mol Cancer Ther. 2006;5(5):1136–44. doi: 10.1158/1535-7163.MCT-06-0084. [DOI] [PubMed] [Google Scholar]
- 36.Welch HC, Coadwell WJ, Ellson CD, Ferguson GJ, Andrews SR, Erdjument-Bromage H, et al. P-Rex1, a PtdIns(3,4,5)P3- and Gbetagamma-regulated guanine-nucleotide exchange factor for Rac. Cell. 2002;108(6):809–21. doi: 10.1016/s0092-8674(02)00663-3. [DOI] [PubMed] [Google Scholar]
- 37.Klein RM, Spofford LS, Abel EV, Ortiz A, Aplin AE. B-RAF regulation of Rnd3 participates in actin cytoskeletal and focal adhesion organization. Mol Biol Cell. 2008;19(2):498–508. doi: 10.1091/mbc.E07-09-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klein RM, Aplin AE. Rnd3 regulation of the actin cytoskeleton promotes melanoma migration and invasive outgrowth in three dimensions. Cancer Res. 2009;69(6):2224–33. doi: 10.1158/0008-5472.CAN-08-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Damoulakis G, Gambardella L, Rossman KL, Lawson CD, Anderson KE, Fukui Y, et al. P-Rex1 directly activates RhoG to regulate GPCR-driven Rac signalling and actin polarity in neutrophils. J Cell Sci. 2014;127(Pt 11):2589–600. doi: 10.1242/jcs.153049. [DOI] [PubMed] [Google Scholar]
- 40.Hayes TK, Neel NF, Hu C, Gautam P, Chenard M, Long B, et al. Long-Term ERK Inhibition in KRAS-Mutant Pancreatic Cancer Is Associated with MYC Degradation and Senescence-like Growth Suppression. Cancer Cell. 2016;29(1):75–89. doi: 10.1016/j.ccell.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ebi H, Costa C, Faber AC, Nishtala M, Kotani H, Juric D, et al. PI3K regulates MEK/ERK signaling in breast cancer via the Rac-GEF, P-Rex1. Proc Natl Acad Sci U S A. 2013;110(52):21124–9. doi: 10.1073/pnas.1314124110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barrio-Real L, Benedetti LG, Engel N, Tu Y, Cho S, Sukumar S, et al. Subtype-specific overexpression of the Rac-GEF P-REX1 in breast cancer is associated with promoter hypomethylation. Breast Cancer Res. 2014;16(5):441. doi: 10.1186/s13058-014-0441-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morrison DK. MAP kinase pathways. Cold Spring Harb Perspect Biol. 2012;4(11) doi: 10.1101/cshperspect.a011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14(19):2501–14. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitmarsh AJ. Regulation of gene transcription by mitogen-activated protein kinase signaling pathways. Biochim Biophys Acta. 2007;1773(8):1285–98. doi: 10.1016/j.bbamcr.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 46.Monaghan-Benson E, Burridge K. Mutant B-RAF regulates a Rac-dependent cadherin switch in melanoma. Oncogene. 2013;32(40):4836–44. doi: 10.1038/onc.2012.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Campbell AD, Lawn S, McGarry LC, Welch HC, Ozanne BW, Norman JC. PRex1 cooperates with PDGFRbeta to drive cellular migration in 3D microenvironments. PLoS One. 2013;8(1):e53982. doi: 10.1371/journal.pone.0053982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minard ME, Herynk MH, Collard JG, Gallick GE. The guanine nucleotide exchange factor Tiam1 increases colon carcinoma growth at metastatic sites in an orthotopic nude mouse model. Oncogene. 2005;24(15):2568–73. doi: 10.1038/sj.onc.1208503. [DOI] [PubMed] [Google Scholar]
- 49.Uhlenbrock K, Eberth A, Herbrand U, Daryab N, Stege P, Meier F, et al. The RacGEF Tiam1 inhibits migration and invasion of metastatic melanoma via a novel adhesive mechanism. J Cell Sci. 2004;117(Pt 20):4863–71. doi: 10.1242/jcs.01367. [DOI] [PubMed] [Google Scholar]
- 50.Xu K, Tian X, Oh SY, Movassaghi M, Naber SP, Kuperwasser C, et al. The fibroblast Tiam1-osteopontin pathway modulates breast cancer invasion and metastasis. Breast Cancer Res. 2016;18(1):14. doi: 10.1186/s13058-016-0674-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.