Abstract
Wilson disease (WD) is an autosomal recessive disorder that is caused by the toxic accumulation of copper (Cu) in the liver. The ATP7B gene, which is mutated in WD, encodes a multitransmembrane domain adenosine triphosphatase that traffics from the trans‐Golgi network to the canalicular area of hepatocytes, where it facilitates excretion of excess Cu into the bile. Several ATP7B mutations, including H1069Q and R778L that are two of the most frequent variants, result in protein products, which, although still functional, remain in the endoplasmic reticulum. Thus, they fail to reach Cu excretion sites, resulting in the toxic buildup of Cu in the liver of WD patients. Therefore, correcting the location of these mutants by leading them to the appropriate functional sites in the cell should restore Cu excretion and would be beneficial to help large cohorts of WD patients. However, molecular targets for correction of endoplasmic reticulum‐retained ATP7B mutants remain elusive. Here, we show that expression of the most frequent ATP7B mutant, H1069Q, activates p38 and c‐Jun N‐terminal kinase signaling pathways, which favor the rapid degradation of the mutant. Suppression of these pathways with RNA interference or specific chemical inhibitors results in the substantial rescue of ATP7BH1069Q (as well as that of several other WD‐causing mutants) from the endoplasmic reticulum to the trans‐Golgi network compartment, in recovery of its Cu‐dependent trafficking, and in reduction of intracellular Cu levels. Conclusion: Our findings indicate p38 and c‐Jun N‐terminal kinase as intriguing targets for correction of WD‐causing mutants and, hence, as potential candidates, which could be evaluated for the development of novel therapeutic strategies to combat WD. (Hepatology 2016;63:1842‐1859)
Abbreviations
- BCS
bathocuproine disulfonate
- CFTR
cystic fibrosis transmembrane conductance regulator
- CS3
copper sensor 3
- EM
electron microscopy
- ER
endoplasmic reticulum
- ERAD
ER‐associated protein degradation
- ERES
ER export site
- ERK
extracellular signal‐regulated kinase
- GFP
green fluorescent protein
- GO
gene ontology
- ICP‐MS
inductively coupled plasma mass spectrometry
- JNK
c‐Jun N‐terminal kinase
- MAPK
mitogen‐activated protein kinase
- MS
mass spectrometry
- PM
plasma membrane
- ROS
reactive oxygen species
- TGN
trans‐Golgi network
- WD
Wilson disease
The liver is essential for the maintenance of copper (Cu) homeostasis as it plays a central role in the excretion of this essential, yet toxic metal. This is highlighted by Wilson disease (WD), an autosomal recessive disorder in which biliary excretion of Cu is severely impaired, causing the toxic accumulation of the metal in the liver.1, 2
The ATP7B gene (defective in WD) encodes a Cu‐transporting P‐type adenosine triphosphatase that pumps cytosolic Cu across cellular membranes, using the energy derived from adenosine triphosphate hydrolysis (Fig. 1A). Increased Cu levels prompt ATP7B to traffic from the Golgi to compartments that are involved in Cu excretion.3, 4 WD‐associated mutations affect the intracellular trafficking of ATP7B to the canalicular area of hepatocytes and/or the protein's ability to transfer Cu across the membrane.3, 4 This results in the failure of hepatocytes to remove excess Cu into the bile and, thus, leads to the accumulation of the metal, which causes cell death and Cu accumulation in extrahepatic tissues. Therefore, clinical features of WD often include hepatic abnormalities, neurological defects, and psychiatric symptoms. When left untreated, liver failure may result in death.1, 2
Figure 1.
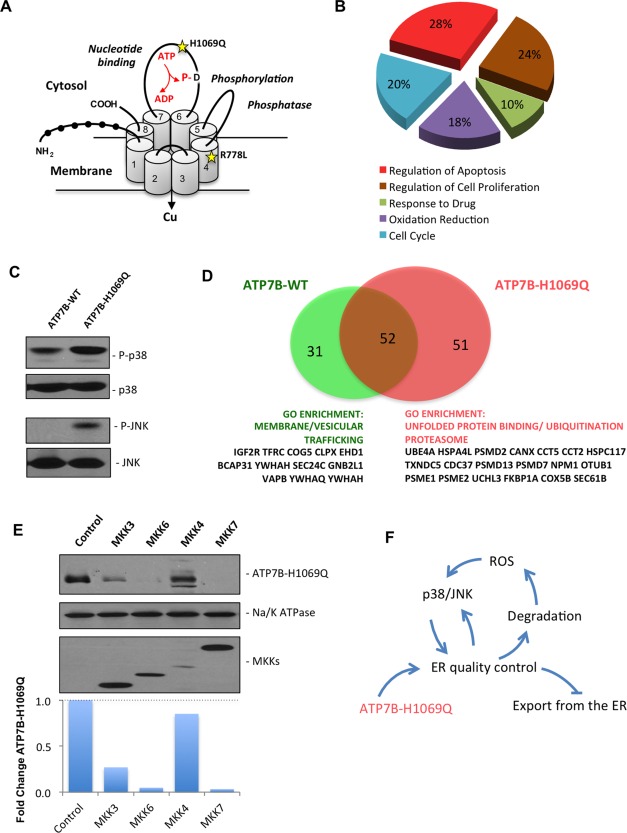
Expression of the ATP7BH1069Q mutant is associated with activation of p38 and JNK signaling pathways. (A) Schematic structure of ATP7B. Black circles show N‐terminal metal binding domains. Numbers indicate transmembrane helices. The domains which regulate adenosine triphosphatase activity are indicated in italic with D residue for catalytic phosphorylation. Yellow stars indicate the position of the most frequent WD‐causing mutations, H1069Q and R778L. (B) HepG2 cells were infected with Ad‐ATP7BWT‐GFP or Ad‐ATP7BH1069Q‐GFP and prepared for microarray analysis (see Materials and Methods). Genes that were differently expressed in cells expressing ATP7BH1069Q were analyzed for GO enrichment. The pie diagram shows the GO categories that were enriched among the altered genes in ATP7BH1069Q‐expressing cells, as opposed to cells expressing ATP7BWT (see also Supporting Table S1). Genes involved in the regulation of apoptosis constituted the largest group of genes whose expression was altered by the ATP7BH1069Q mutant. (C) HepG2 cells were infected with Ad‐ATP7BWT‐GFP or Ad‐ATP7BH1069Q‐GFP and analyzed with western blot. Phosphorylated forms of p38 or JNK increased in cells expressing the ATP7BH1069Q mutant, while overall amounts of p38 or JNK remained similar in wild type‐expressing and mutant‐expressing cells. (D) Putative interactors of ATP7BWT and ATP7BH1069Q were identified using a proteomics approach (see Materials and Methods). The diagram shows the number of interactors that were specific for ATP7BWT or for ATP7BH1069Q, as well as the number of common interactors. GO analysis revealed ATP7BWT interactors to be enriched in proteins belonging to membrane trafficking categories, while mutant‐specific interactors were enriched in proteins involved in ER‐associated protein quality control and degradation. (E) HepG2 cells expressing ATP7BH1069Q were transfected with activators of p38 (MKK3 and MKK6) or JNK (MKK4 and MKK7). Western blot (see also quantification graph) revealed a decrease in ATP7BH1069Q levels in cells expressing p38 or JNK activators. Na/K‐adenosine triphosphatase was used as input control. The modest decrease in ATP7BH1069Q in cells transfected with MKK4 is due to lower overexpression of MKK4 in comparison to other MKKs. (F) The schematic drawing shows a vicious circle that is generated by expression of the ATP7BH1069Q mutant, which leads to activation of ER quality control and degradation of ATP7BH1069Q. As a consequence of ATP7BH1069Q loss, ROS increase and stimulate p38 and JNK signaling, which triggers further retention and degradation of the mutant in the ER, preventing it from being exported to the secretory pathway. Abbreviations: ADP, adenosine diphosphate; ATP, adenosine triphosphate; P‐, phosphorylated.
WD treatment may be successfully approached with zinc (Zn) salts and Cu‐chelating agents. However, these treatments do have serious toxicities.2, 5 Moreover, about one‐third of WD patients do not respond efficiently either to Zn or to Cu chelators.6 All considered, developing novel WD treatment strategies has become an important goal. When approaching therapeutic solutions, properties of WD‐causing mutants should be carefully considered. The most frequent ATP7B mutations (Fig. 1A), H1069Q (40%‐75% in the white patient population) and R778L (10%‐40% of Asian patients), result in ATP7B proteins with significant residual activities,7, 8, 9 which, however, are strongly retained in the endoplasmic reticulum (ER).10 Notably, many other WD‐causing ATP7B mutants with substantial Cu‐translocating activity undergo complete or partial arrest in the ER.11 Thus, although potentially able to transport Cu, these ATP7B mutants cannot reach the Cu excretion sites to remove excess Cu from hepatocytes. ER retention of such ATP7B mutants occurs due to their misfolding10, 11 and increased aggregation12 and, hence, their failure to fulfill the requirements of the ER quality control machinery. As a result, the cellular proteostatic network recognizes ATP7B mutants as defective and directs them toward the ER‐associated protein degradation (ERAD) pathway.9 Therefore, identifying molecular targets for the recovery of partially or fully active ATP7B mutants from the ER to the appropriate functional compartment(s) would be beneficial for the majority of WD patients.
Here, we demonstrate, using both systems biology and classical approaches, that the degradation of the most frequent ATP7BH1069Q mutant is under the control of the stress kinases p38 and c‐Jun N‐terminal kinase (JNK). Suppressing both p38 and JNK resulted in the efficient correction of the mutant, thus allowing it to be transported from the ER to the trans‐Golgi network (TGN), and supported its Cu‐induced trafficking to the post‐Golgi vesicles and canalicular surface of polarized hepatic cells. As a consequence, treatments with p38 or JNK inhibitors reduced Cu accumulation in cells expressing ATP7BH1069Q and attenuated degradation of the mutant due to its improved sorting from the ER into the secretory pathway. Our findings suggest that p38 and JNK signaling pathways may serve as attractive targets for the correction of WD‐causing mutants and could be explored for the development of therapeutic approaches that counteract WD.
Materials and Methods
Antibodies, Reagents, and DNAs
Antibodies against phospho‐p38, phospho‐JNK, phospho‐extracellular signal‐regulated kinase (ERK), p38, JNK, and ERK were from Cell Signaling Technology (Beverly, MA). The DNAs of ATP7A and its mutants were already reported.13 DNAs of flag‐tagged MKK3, MKK4, MKK6, and MKK7 were obtained from Addgene (Cambridge MA). To obtain the R778L, D765N, L776V, A874V, and L1083F mutants of ATP7B, the phosphorylated enhanced green fluorescent protein (GFP) C1‐ATP7B construct was used as a template and site‐directed mutagenesis was performed according to the manufacturer's instructions using the QuickChange kit (Stratagene, La Jolla, CA). Other reagents, antibodies, DNAs, and viruses have been reported.14
Cell Culture
HeLa and HepG2 cells and primary mouse hepatocytes were grown in Dulbecco's modified Eagle's medium supplemented with fetal calf serum 10% (decomplemented for HepG2 cells), l‐glutamine, and penicillin/streptomycin.
Immunoprecipitation and Mass Spectrometric Analysis
To compare interactomes of ATP7BH1069Q‐GFP or ATP7BWT‐GFP, the tagged proteins were immunoprecipitated from cell lysates with the anti‐GFP antibody. Pull‐downs were fractionated by sodium dodecyl sulfate‐polyacrylamide gel electrophoresis, and the protein bands were digested with trypsin. Peptide mixtures were analyzed by nano‐chromatography tandem mass spectrometry (MS/MS) on a CHIP MS Ion Trap XCT Ultra (Agilent Technologies, Palo Alto, CA). Peptide analysis was performed using data‐dependent acquisition of one MS scan (mass range 400‐2000 m/z) followed by MS/MS scans of the three most abundant ions in each MS scan. Raw data from nano liquid chromatography‐MS/MS analyses were employed to query a nonredundant protein database using in‐house MASCOT software (Matrix Science, Boston, MA).
Microarray Analysis
To identify the effects of ATP7BH1069Q expression on the transcriptome, total RNA was extracted from HepG2 cells (expressing either ATP7BH1069Q‐GFP or ATP7BWT‐GFP) and hybridized on Affymetrix GeneChips. All of the raw microarray data were formatted for the Gene Expression Omnibus to comply with Minimum Information About a Microarray Experiment and Microarray Gene Expression Database group standards (Gene Expression Omnibus number GSE51818). Microarray analyses were carried out with R, a free software environment. Genes that passed all filtering criteria were entered into the DAVID Gene Functional Annotation Tool. The gene ontology (GO) options GOTERM_BP_ALL and GOTERM_MF_ALL were selected, and a functional annotation chart was generated. A maximum P value of 0.05 was chosen to select only the significant categories.
Statistical Analysis
Data were expressed as mean values ± standard deviation. The Student t test was used to compare differences between two groups by GraphPad Prism 6 software (GraphPad Prism Inc., San Diego, CA). Statistically significant differences between them are indicated by the following: *P < 0.05, **P < 0.01, and ***P < 0.001.
Other Methods
Adenoviral and vesicular stomatitis viral infection, DNA transfection, RNA interference, quantitative polymerase chain reaction, fluorescent microscopy, immuno‐electron microscopy (EM), inductively coupled plasma mass spectrometry (ICP‐MS), and western blot analysis were done as reported.14 For details see the Supporting Information.
Results
Expression of the ATP7BH1069Q Mutant Induces Activation of p38 and JNK
We used unbiased approaches to understand which molecular pathway could be targeted to correct mutants causing WD. We first sought to determine how ATP7BH1069Q mutant expression affects the transcriptome of hepatic cells. Comparison of microarrays from ATP7BH1069Q‐expressing and ATP7BWT‐expressing cells revealed 626 differently expressed genes (GSE51818). GO analysis of these hits indicated enrichment in genes that regulate cell death and response to stress (Fig. 1B; Supporting Table S1). Several of these genes (SQSTM1, TXNIP, IL6R, FOSL1, MAP2K6, and NET1) have been reported to operate in mitogen‐activated protein kinase (MAPK) signaling pathways mediated by two proapoptotic stress kinases, p38 and JNK.15 This might be linked to WD pathogenesis because reactive oxygen species (ROS) accumulating in WD3 can stimulate the activity of p38 and JNK.15, 16, 17, 18 Indeed, several ROS‐sensitive genes (SOD2, MSRA, TP53I3) were up‐regulated in ATP7BH1069Q‐expressing cells. This prompted us to investigate whether the activity of these kinases rises upon ATP7BH1069Q expression. Western blot analysis revealed that phosphorylated p38 and JNK increased significantly in cells expressing the mutant (Fig. 1C), indicating that both kinases were activated. In the meantime the MAPK kinase/ERK pathway of the MAPK signaling cascade was not activated as the levels of phosphorylated ERK remained unaltered in mutant‐expressing cells (Supporting Fig. S1A).
We also compared the interactomes of ATP7BWT and ATP7BH1069Q. Proteomics analysis revealed 31 ATP7BWT‐specific and 51 ATP7BH1069Q‐specific interactors (Supporting Tables S2 and S3). GO analysis indicated that specific binding partners of ATP7BWT were enriched in “intracellular trafficking” proteins (Fig. 1D). In contrast, the ATP7BH1069Q interactome contained numerous proteins belonging to the “unfolded protein binding,” “proteasome,” and “ubiquitination” GO categories (Fig. 1D). These differences are in line with an enhanced association of the mutant with the components of the ER‐associated quality control machinery, resulting in ER retention of the mutant.9, 10 Notably, specific MAPK pathways, which include p38 and JNK, have recently been shown to affect proteostasis of proteins such as cystic fibrosis transmembrane conductance regulator (CFTR)19 and to regulate the expression of ER quality control/degradation genes.15 We thus examined whether activation of p38 and JNK might lead to the accelerated degradation of ATP7BH1069Q. Indeed, overexpression of the upstream activators of p38 (MKK3 and MKK6) or JNK (MKK4 and MKK7) reduced ATP7BH1069Q levels, indicating increased mutant degradation (Fig. 1E).
On the whole, the above observations indicate that induction of p38 and JNK may be triggered by ATP7BH1069Q expression. This activation, in turn, may promote ER retention and degradation of the mutant, an increase in ROS levels (due to Cu accumulation), and further perturbation of ER proteostasis, generating a vicious circle (Fig. 1F). In this context, p38 and JNK represent regulatory centerpieces and potential druggable targets to prevent ER retention of the mutant and, hence, to rescue its localization and function.
Suppression of p38 and JNK Rescues Appropriate Localization and Trafficking of the ATP7BH1069Q Mutant
To test whether deactivating the p38 and JNK pathways facilitates ATP7BH1069Q export from the ER, we suppressed the activity of p38 and JNK first by using specific chemical inhibitors, SB202190 (SB90) and SP600125 (SP125), and then by RNA interference. We initially evaluated the impact of p38/JNK‐specific inhibitors and small interfering RNAs in HeLa cells, given that silencing of the genes of interest can be easily achieved in this cell type. In cells treated with the Cu chelator bathocuproine disulfonate (BCS), ATP7BWT was mainly detected over the Golgi membranes, while ATP7BH1069Q remained in the ER (Fig. 2A, upper row). Neither p38 and JNK inhibitor (added for 24 hours) affected ATP7BWT (Supporting Fig. S2A), but both reduced ATP7BH1069Q within the ER and improved the delivery of the mutant to the Golgi (Fig. 2A[upper row],B). The correction of ATP7BH1069Q localization was remarkably effective even at quite low concentrations (Supporting Fig. S2B). Interestingly, the combination of p38 and JNK inhibitors did not further increase mutant relocation from the ER to the Golgi (Supporting Fig. S3), suggesting that the two inhibitors target similar molecular pathways for correction of ATP7BH1069Q. In control experiments the ERK inhibitor failed to correct the mutant from the ER to the Golgi (Supporting Fig. S1B). This indicates that only p38 and JNK branches of the MAPK signaling cascade are involved in mutant retention in the ER. Importantly, acute suppression of p38 and JNK did not impact ER export of ATP7BH1069Q (Supporting Fig. S4). Therefore, inhibitors apparently act through the transcriptional regulation of quality control and/or trafficking genes rather than by reducing the direct phosphorylation of the mutant or components of its trafficking machinery.
Figure 2.
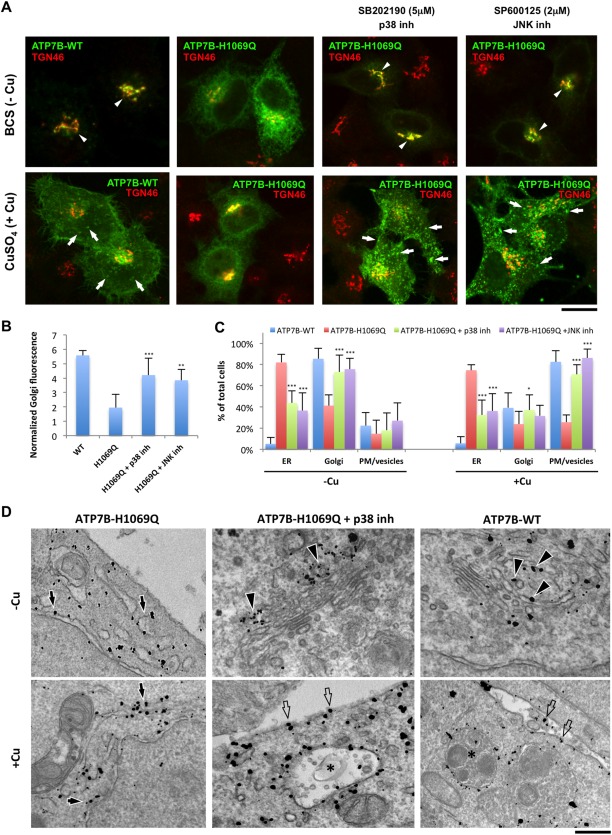
Inhibitors of p38 and JNK correct localization and trafficking of ATP7BH1069Q mutant. (A) HeLa cells were infected with Ad‐ATP7BWT‐GFP or Ad‐ATP7BH1069Q‐GFP, incubated overnight with 200 μM BCS, and fixed or incubated for an additional 2 hours with 100 μM CuSO4. In response to Cu, ATP7BWT (left column) traffics from the Golgi (arrowheads in upper panel) to the PM and vesicle (arrows in lower panel), while ATP7BH1069Q (second column) is retained within the ER under both low‐Cu and high‐Cu conditions. The p38 inhibitor SB202190 (5 µM) or the JNK inhibitor SP600125 (2 µM) was added to the cells 24 hours before fixation (as indicated in the corresponding panels). Fixed cells were further labeled for TGN46 and visualized under a confocal microscope. Both p38 and JNK inhibitors corrected ATP7BH1069Q from the ER to the Golgi (arrowheads in upper panels) under low‐Cu conditions and to the PM and vesicles (arrows in lower panels) in high‐Cu conditions. (B) Cells were treated with BCS and drugs as shown in (A). Fluorescence of ATP7B signal in the Golgi (average ± standard deviation, n = 40 cells) was quantified and normalized to total cell fluorescence. Both inhibitors caused an increase of ATP7BH1069Q signal in the Golgi region. (C) Cells were treated as in (A). The percentage of cells (average ± standard deviation, n = 10 fields) with an ATP7B signal in the ER, Golgi, or PM/vesicles was calculated. Both p38 and JNK inhibitors reduced the percentage of cells exhibiting ATP7BH1069Q in the ER and increased the number of cells in which ATP7B was corrected to the Golgi under low‐Cu conditions and to the PM and vesicles upon Cu stimulation. (D) Cells were treated as in (A) and prepared for immuno‐EM (see Materials and Methods). Arrows indicate ATP7BH1069Q in the ER. Arrowheads show ATP7BWT or ATP7BH1069Q in the Golgi. Empty arrows indicate ATP7BWT or ATP7BH1069Q signal at the PM, while asterisks label ATP7B‐positive multivesicular body‐like vesicles. Scale bars = 5 μm (A) and 260 nm (D). Abbreviations: inh, inhibitor; WT, wild type.
Next, we investigated whether the inhibitors rescue Cu‐responsive trafficking of ATP7BH1069Q. CuSO4 addition induced redistribution of ATP7BWT from the TGN to the plasma membrane (PM) and peripheral vesicular structures but failed to induce ATP7BH1069Q relocation from the ER (Fig. 2A, lower row). Instead, when corrected by p38/JNK antagonists, ATP7BH1069Q efficiently moved from the Golgi to the PM and vesicles in response to elevated Cu (Fig. 2A,C).
The above results were confirmed by biochemistry and EM. Western blot exhibited reduced amounts of ATP7BH1069Q in the PM subcellular fractions in comparison to ATP7BWT, while both p38 and JNK inhibitors significantly increased quantities of the mutant delivered to the cell surface (Supporting Fig. S5). In turn, EM revealed ATP7BH1069Q mainly over the ER membrane profiles, while cells incubated with p38 antagonists contained the mutant in TGN membranes (Fig. 2D). In response to CuSO4 treatment inhibitor‐treated cells delivered ATP7BH1069Q from the TGN to the cell surface and endosome/lysosome‐like “vesicles” (Fig. 2D), which normally receive ATP7BWT upon Cu overload.14 Thus, it turns out that, once corrected to the Golgi by p38/JNK inhibitors, the mutant reacts to increasing Cu by trafficking to Cu excretion sites, just as ATP7BWT does.
To further verify the above conclusions, we silenced different isoforms of p38 (MAPK11‐MAPK14) and JNK (MAPK8‐MAPK10) in cells expressing ATP7BH1069Q (Fig. 3A). Both reduction in ER retention and recovery of Golgi and vesicle targeting of the mutant were detected after depletion of MAPK8, MAPK11, and MAPK14 (Fig. 3B,C). In addition, we sought to suppress MAP3K11, the upstream activator of both p38 and JNK,20 to evaluate whether this treatment improved ATP7BH1069Q recovery from the ER. MAP3K11 has been reported to effectively correct Δ508‐CFTR.19 Indeed, depletion of MAP3K11 resulted in strong reduction of ATP7BH1069Q in the ER (Fig. 3B,C).
Figure 3.
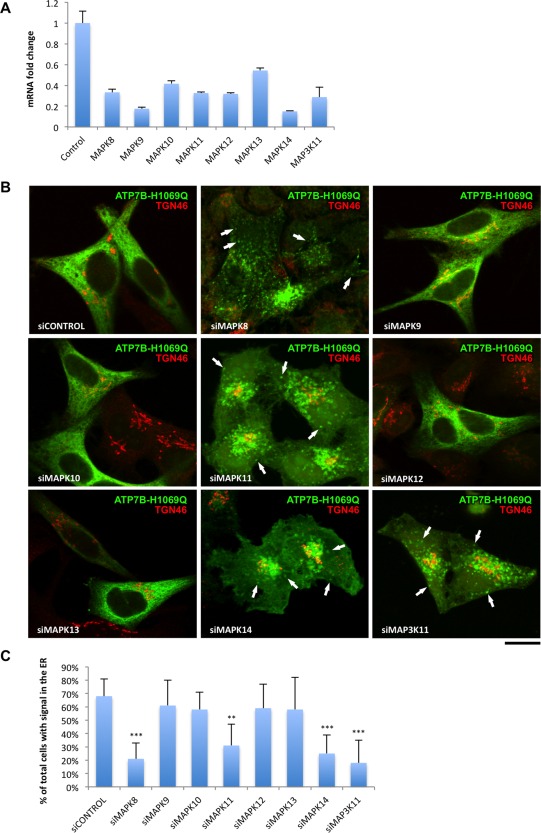
Silencing of several p38 and JNK isoforms corrects localization and trafficking of the ATP7BH1069Q mutant. (A) HeLa cells were incubated with small interfering RNA, which targets specific genes (indicated in graph) belonging to the p38 and JNK pathways. The efficiency of silencing for each gene was evaluated by quantitative reverse‐transcription polymerase chain reaction and expressed as fold‐change (average ± standard deviation, n = 3 experiments) after normalization to messenger RNA levels of the same gene in control cells (treated with scramble small interfering RNA). (B) HeLa cells were silenced for different p38 and JNK isoforms as in (A), then infected with Ad‐ATP7BH1069Q‐GFP and incubated for 2 hours with 100 μM CuSO4. Fixed cells were then labeled for TGN46 and visualized under a confocal microscope. Silencing of MAPK8, MAPK11, MAPK14, or MAP3K11 showed the rescue of ATP7BH1069Q from the ER and its movement to the post‐Golgi vesicles (arrows) and PM. (C) Cells were treated as in (B). The percentage of cells (average ± standard deviation, n = 10 fields) with ATP7BH1069Q signal in the ER was calculated. RNA interference of MAPK8, MAPK11, MAPK14, and MAP3K11 reduced the percentage of cells exhibiting ATP7BH1069Q in the ER. Scale bar = 4.7 μm (B). Abbreviations: mRNA, messenger RNA; si, small interfering.
We next investigated whether inhibitors of p38 or JNK overcome the ER retention of other ATP7B mutants with residual activity.7, 11 Correction was achieved for D765N, L776V, and R778L mutants but not for A874V and L1083F variants (Fig. 4). Meanwhile, p38 and JNK antagonists failed to correct ATP7A mutants with post‐Golgi trafficking defects (Fig. 5), indicating that inhibitors act mainly at the ER level. In summary, our observations suggest that suppressing p38 or JNK allows various ATP7B mutants to escape ER retention in a manner that depends on the type of mutation.
Figure 4.
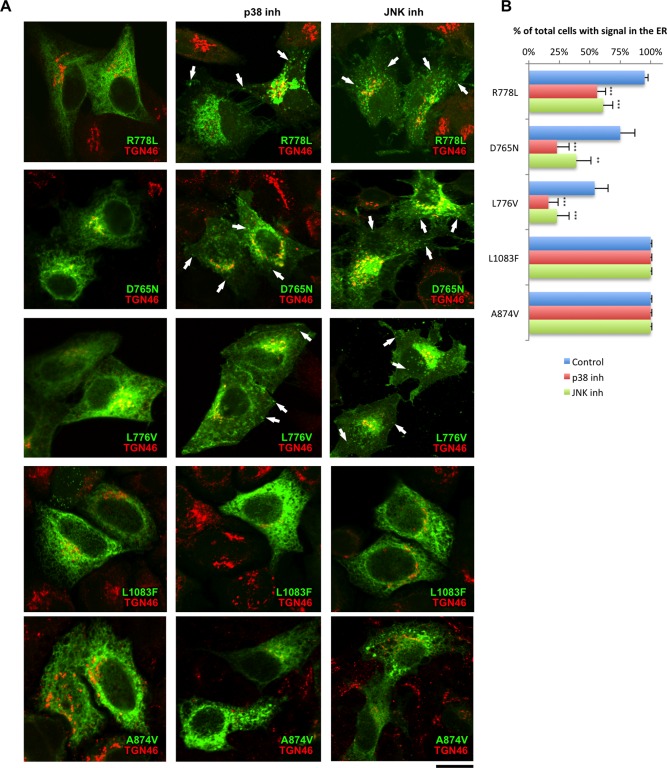
Impact of p38 and JNK inhibitors on localization of ER‐retained ATP7B mutants. (A) HeLa cells were transfected with different ATP7B mutants (indicated in each panel), then treated for 24 hours with either p38 or JNK inhibitor and exposed to 100 μM CuSO4 for 2 hours. Cells were then fixed, labeled for TGN46, and visualized under a confocal microscope. All mutants exhibited clear ER patterns in cells that were not treated with inhibitors. Incubation with either p38 or JNK inhibitor allowed R778L, D765N, and L776V mutants of ATP7B to be transported to the Golgi, post‐Golgi vesicles (arrows), and PM. In contrast, inhibitors did not recover either L1083F or A874V mutants of ATP7B from the ER (two bottom rows). (B) Cells were treated as in (A). The percentage of cells (average ± standard deviation, n = 10 fields) with ATP7B signal in the ER was calculated for each mutant. p38 and JNK inhibitors reduced the percentage of cells that exhibited R778L, D765N, and L776V in the ER. Scale bar = 5 μm (A). Abbreviation: inh, inhibitor.
Figure 5.
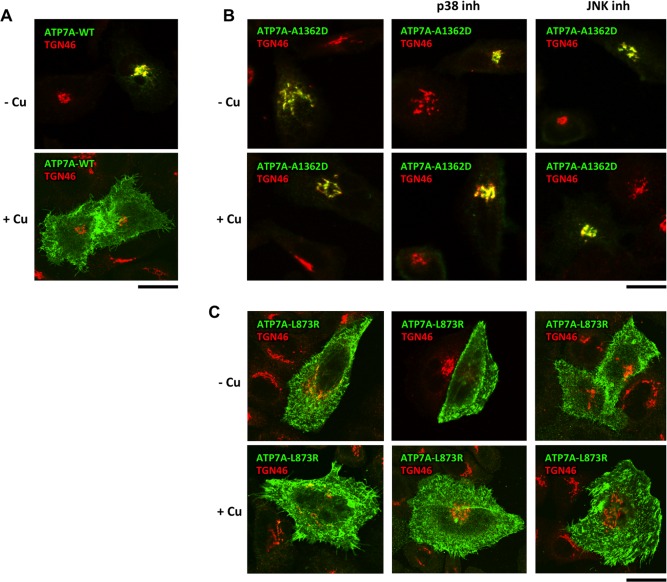
p38 and JNK inhibitors do not rescue post‐Golgi trafficking of ATP7A mutants. HeLa cells were transfected with either ATP7AWT (A), ATP7AA1362D (B), or ATP7AL873R (C) mutant. Then, cells were exposed to a 200 μM treatment with BCS (‐Cu) and fixed directly or incubated with 100 μM CuSO4 for an additional 2 hours (+Cu). p38 or JNK inhibitors were added (as indicated in B and C) for 24 hours prior to fixation. Fixed cells were labeled for TGN46 and visualized under a confocal microscope. (A) ATP7AWT moved from the Golgi (top panel) to the PM (bottom panel) after Cu increase. (B) ATP7BA1362D mutant remained associated with TGN46 under both low‐Cu and high‐Cu conditions (left column). Neither p38 nor JNK inhibitors allowed the mutant to be transported from the Golgi to the PM in response to increasing Cu (middle and right columns). (C) The ATP7AL873R mutant remained distributed over the PM, independently of the Cu levels (left column). Incubation with p38 or JNK inhibitors did not correct the mutant from the PM to the Golgi (middle and right columns), even when Cu levels were low (top row). Scale bar = 4.8 μm (A‐C). Abbreviation: inh, inhibitor.
p38 and JNK Inhibitors Rescue ATP7BH1069Q Trafficking and Targeting in Hepatic Cells
As a further step, we tested whether suppressing p38 and JNK is efficient at rescuing the mutant in WD‐relevant cellular systems such as HepG2 cells and primary hepatocytes.21 In HepG2 cells, ATP7BWT moved from the Golgi to the vesicles and cell membrane in response to Cu (Fig. 6A), while most of the ATP7BH1069Q was arrested in the ER, regardless of changes in Cu concentrations (Fig. 6A). However, incubation with either p38 inhibitor or JNK inhibitor allowed significant amounts of ATP7BH1069Q to reach the Golgi and to be transported toward the PM and associated vesicles in response to Cu overload (Fig. 6A,B). Moreover, the ATP7BH1069Q mutant was targeted to the appropriate canalicular surface domain in polarized HepG2 cells (Supporting Fig. S6). Importantly, primary hepatocytes exhibited a similar response to p38 inhibitor and JNK inhibitor, which rescued ATP7BH1069Q from the ER and allowed it to traffic toward Golgi and post‐Golgi Cu excretion sites (Fig. 6C,D). Therefore, hepatic cells tend to deliver the ATP7B mutant to the right compartments upon suppression of p38 and JNK signaling pathways.
Figure 6.
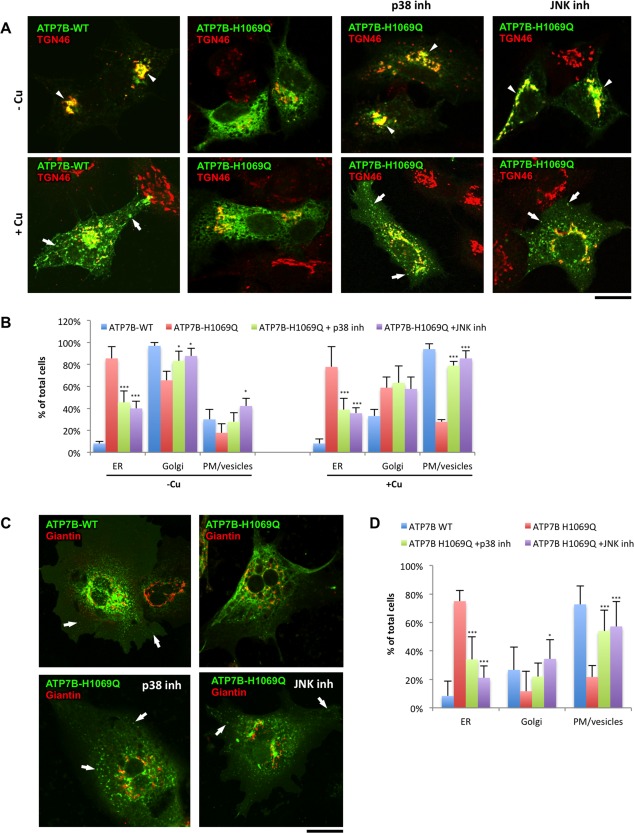
Suppression of p38 and JNK corrects localization and trafficking of ATP7BH1069Q mutant in hepatic cells. (A) HepG2 cells were infected with Ad‐ATP7BWT‐GFP or Ad‐ATP7BH1069Q‐GFP, incubated overnight with 200 μM BCS, and fixed or incubated for an additional 2 hours with 100 μM CuSO4. In response to Cu, ATP7BWT (left column) traffics from the Golgi (arrowheads in upper panels) to the PM and vesicles (arrows in lower panels), while ATP7BH1069Q (second column) is retained in the ER under both low‐Cu and high‐Cu conditions. p38 or JNK inhibitors were added to the cells 24 hours before fixation (as indicated in the corresponding panels). Fixed cells were then labeled for TGN46 and visualized under a confocal microscope. Both p38 and JNK inhibitors corrected ATP7BH1069Q from the ER to the Golgi (arrowheads in upper panels) under low‐Cu conditions and to the PM and vesicles (arrows in lower panels) upon Cu exposure. (B) Cells were treated as in (A). The percentage of cells (average ± standard deviation, n = 10 fields) with ATP7B signal in the ER, Golgi, or PM/vesicles was calculated. Both p38 and JNK inhibitors reduced the percentage of cells exhibiting ATP7BH1069Q in the ER independently from the Cu levels and increased the number of cells in which ATP7B was corrected to the Golgi under low‐Cu conditions and to the PM and vesicles upon Cu stimulation. (C) Primary hepatocytes isolated from mouse liver cells were infected with Ad‐ATP7BWT‐GFP or Ad‐ATP7BH1069Q‐GFP and then incubated with 100 μM CuSO4 for 2 hours. p38 or JNK inhibitors were added to the cells 24 hours before fixation (as indicated in the corresponding panels). Fixed cells were then labeled for giantin and visualized under a confocal microscope. Both p38 and JNK inhibitors corrected ATP7BH1069Q from the ER to the PM and vesicles (arrows). (D) Primary hepatocytes were treated as in (C). The percentage of cells (average ± standard deviation, n = 10 fields) with ATP7B signal in the ER, Golgi, or PM/vesicles was calculated. Both p38 and JNK inhibitors reduced the percentage of cells exhibiting ATP7BH1069Q in the ER and increased the number of cells in which ATP7B was corrected to the PM and vesicles upon Cu stimulation. Scale bars = 5 μm (A) and 5.3 μm (C). Abbreviation: inh, inhibitor.
Correction of the ATP7BH1069Q Mutant With p38 or JNK Inhibitors Facilitates Cu Excretion
We reasoned that correcting ATP7BH1069Q from the ER to the appropriate Cu excretion sites should reduce the total amount of intracellular Cu. To test this hypothesis we used ICP‐MS. ICP‐MS measurements revealed higher concentrations of the metal in cells expressing ATP7BH1069Q, while treatment with p38 or JNK inhibitor significantly reduced intracellular Cu levels (Fig. 7A).
Figure 7.
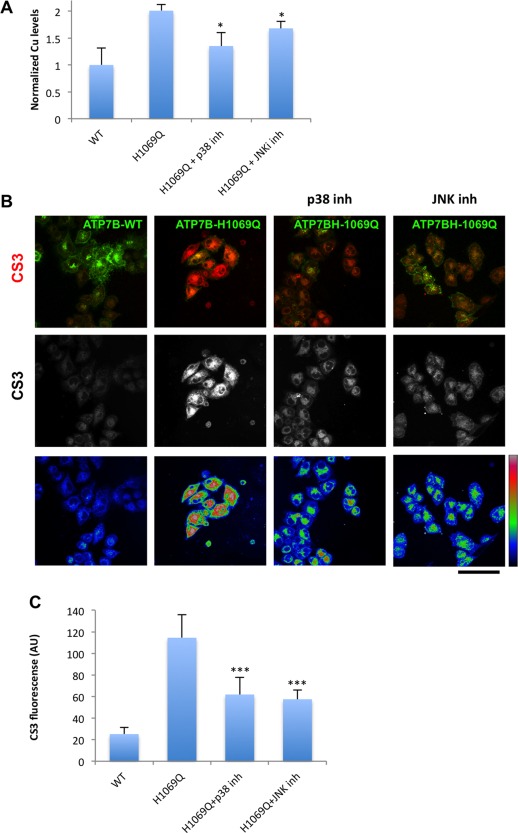
Inhibitors of p38 and JNK reduce Cu levels in cells expressing ATP7BH1069Q mutant. (A) HepG2 cells were infected with Ad‐ATP7BWT‐GFP or Ad‐ATP7BH1069Q‐GFP and incubated with p38 or JNK inhibitor. CuSO4 at 100 μM concentration was added to the cells for the last 2 hours of incubation with the inhibitors. The cells were examined by ICP‐MS (see Materials and Methods), which revealed an increase in normalized intracellular Cu levels (average ± standard deviation, n = 3 experiments) in ATP7BH1069Q‐expressing cells. Both p38 and JNK inhibitors reduced Cu levels in ATP7BH1069Q‐expressing cells. (B) HepG2 cells were infected with Ad‐ATP7BWT‐GFP or Ad‐ATP7BH1069Q‐GFP, incubated for 2 hours with 100 μM CuSO4, and loaded with CS3 before fixation. p38 or JNK inhibitors were added to the cells 24 hours before fixation (as indicated in the corresponding panels). Confocal microscopic images show CS3 fluorescence in red (top row), in white (middle row), and in false color scale (bottom row). Cells expressing ATP7BH1069Q exhibited higher CS3 signals than cells expressing ATP7BWT, while both p38 and JNK inhibitors decreased CS3 fluorescence in ATP7BH1069Q‐expressing cells. (C) Quantification revealed an increase in CS3 fluorescence (average ± standard deviation, n = 10 fields) in cells expressing ATP7BH1069Q and a decrease of CS3 signal upon incubation with either p38 or JNK inhibitor. Scale bar = 14 μm (C). Abbreviations: AU, arbitrary units; inh, inhibitor; WT, wild type.
To further support these findings, we used copper sensor 3 (CS3), which becomes fluorescent in the presence of bioavailable Cu.22 We found low CS3 signal in cells expressing ATP7BWT, whereas CS3 fluorescence was higher in mutant‐expressing cells (Fig. 7B). p38 and JNK inhibitors significantly decreased CS3 signal in cells expressing ATP7BH1069Q (Fig. 7B,C). More importantly, reduction in CS3 fluorescence (and hence in Cu levels) correlated with the amount of ATP7BH1069Q expressed in inhibitor‐treated cells (Supporting Fig. S7). Collectively, the above findings indicate that correcting the mutant to the appropriate compartments with p38/JNK antagonists allows the cells to eliminate excess Cu.
p38 and JNK Inhibitors Reduce Degradation of ATP7BH1069Q by Improving Mutant Sorting Into the Secretory Pathway
In the ER the failure of misfolded protein to pass the quality control check directs such a protein to degradation.23 Therefore, we analyzed whether p38 or JNK inhibitors counteract ATP7BH1069Q degradation. We found that the ATP7BH1069Q protein product increased in response to p38 and MAPK inhibition (Fig. 8A), while the levels of its messenger RNA remained the same in treated cells (not shown). This indicated that suppressing p38 and JNK attenuates degradation of the mutant. To support this conclusion we treated cells with cycloheximide and found that p38 and JNK inhibitors significantly slowed down the kinetics of mutant turnover (Fig. 8B,C). Thus, our results suggest that suppression of the p38 and JNK pathways allows ATP7BH1069Q to circumvent ERAD.
Figure 8.
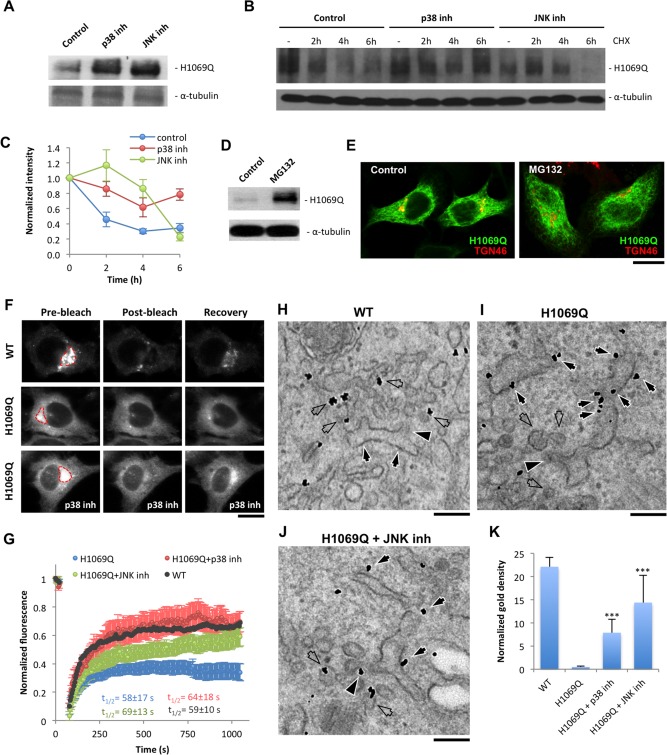
p38 and JNK inhibitors reduce degradation of ATP7BH1069Q mutant and improve its sorting into the secretory pathway. (A) HepG2 cells were infected with Ad‐ATP7BH1069Q and treated with p38 or JNK inhibitor. Western blot revealed an increase in the amount of ATP7BH1069Q after suppression of p38 or JNK. (B,C) HepG2 cells expressing ATP7BH1069Q were treated with p38 and JNK inhibitors and then exposed to 100 μM cycloheximide for different time intervals. Western blot (B) and quantification of ATP7BH1069Q bands (C) showed a decrease of ATP7BH1069Q signals (means ± standard deviation, n = 3 experiments) in control cells exposed to cycloheximide, while p38 and JNK inhibitors attenuated the decay of ATP7BH1069Q levels. (D,E) HepG2 cells expressing ATP7BH1069Q were treated with 20 μM of the proteasome inhibitor MG132. Although western blot indicated an increase in ATP7BH1069Q protein levels (D), the localization of the mutant (E) was similar in control and MG132‐treated cells. (F,G) HeLa cells expressing either ATP7BWT‐GFP or ATP7BH1069Q‐GFP were observed in vivo. p38 or JNK inhibitor was added to some ATP7BH1069Q‐GFP‐expressing cells 24 hours before the experiment. The GFP signal within the Golgi area (outlined in red) was photo‐bleached (see Materials and Methods), and the kinetics of its recovery in the bleached region was analyzed. Both time‐lapse images (F) and a kinetics plot (G) revealed more efficient recovery of ATP7BWT‐GFP in comparison to the mutant. Incubation with p38 or JNK inhibitor led to recovery of higher ATP7BH1069Q‐GFP levels (means ± standard deviation, n = 6 cells) in the bleached Golgi area (F,G), although the half time (t 1/2) of the recovery process remained similar in control and treated cells (G). (H‐K) HepG2 cells expressing ATP7BWT‐GFP or ATP7BH1069Q‐GFP were fixed and prepared for immuno‐EM with anti‐GFP antibody. Some of the ATP7BH1069Q‐GFP‐expressing cells were incubated with p38 or JNK inhibitor. ER (arrows), buds (arrowheads), and ERES vesicular/tubular profiles (empty arrows) are indicated in (H‐J). (K) An increase in normalized density of gold particles associated with ATP7BH1069Q‐GFP (means ± standard deviation, n = 20 ERES) at the ERES structures in inhibitor‐treated cells. Scale bars = 6.5 μm (E), 5 μm (F), and 180 nm (H‐J). Abbreviations: CHX, cycloheximide; inh, inhibitor; WT, wild type.
How do p38 and JNK antagonists suppress ATP7BH1069Q degradation? The first possibility is that they directly inhibit the ERAD components that comprise the proteasome.23 Being protected from ERAD, aberrant ATP7B molecules would reside in the ER longer and, therefore, may have a higher chance to be incorporated into carriers that transport them from the ER to the Golgi. To test whether this is the case, we treated ATP7BH1069Q‐expressing cells with the proteasome inhibitor MG132. MG132 increased ATP7BH1069Q levels (Fig. 8D) but failed to overcome mutant retention in the ER (Fig. 8E). This indicates that inhibitor‐mediated recovery of ATP7BH1069Q from the ER does not involve ERAD components.
Therefore, ATP7BH1069Q may be protected from degradation as a result of accelerated trafficking from the ER. To verify this we first observed whether p38 and JNK inhibitors accelerated overall trafficking rates in the secretory pathway. This was not the case as the trafficking kinetics of the generic cargo protein VSVG was not altered in SB90‐treated or SP125‐treated cells (Supporting Fig. S8).
We further hypothesized that the suppression of p38 and JNK might specifically accelerate trafficking rates of the ATP7BH1069Q mutant from the ER to the Golgi. To test this hypothesis we used selective photo‐bleaching, which allowed us to analyze protein exchange between different compartments in live cells.24 We bleached ATP7BWT‐GFP or ATP7BH1069Q‐GFP in the Golgi region and observed how the material from the ER recovered GFP signal in the bleached area (Fig. 8F). Recovery of ATP7BH1069Q‐GFP fluorescence in the Golgi reached the plateau with the same dynamics (t 1/2 ≈ 1 min) in both control and inhibitor‐treated cells (Fig. 8G). However, the amount of ATP7BH1069Q‐GFP that reappeared within the Golgi over the same time interval was markedly higher in cells incubated with p38 or JNK inhibitor (Fig. 8F,G). These observations indicate that suppressing p38 and JNK does not shorten the time needed for a given ATP7BH1069Q molecule to be transported from the ER to the Golgi. Instead, p38 and JNK antagonists allow a higher number of ATP7BH1069Q molecules to be sorted from the ER and transported to the Golgi during this interval.
To further support this point we evaluated the distribution of ATP7BH1069Q at the ER export sites (ERES) using immuno‐EM. ERES contain coated buds (Fig. 8H‐J, arrowheads) emerging from the ER cisternae and associated vesicular‐tubular membranes (Fig. 8H‐J, empty arrows) that carry newly synthetized proteins to the Golgi. Normally, membrane proteins directed into the secretory pathway (like ATP7BWT) undergo enrichment in the ERES membranes (Fig. 8H, empty arrows) over regular ER cisternae.25 Such concentration in ERES was not detected for ATP7BH1069Q. Indeed, ATP7BH1069Q‐associated gold particles decorated mainly ER cisternae (Fig. 8I, arrows) and were scarce in the coated buds (Fig. 8I, arrowhead) and neighboring vesicles (Fig. 8I, empty arrows). Incubation with either p38 or JNK inhibitor increased the density of ATP7BH1069Q in the ERES buds (Fig. 8J, arrowhead) and vesicular profiles (Fig. 8J, empty arrows) and reduced ATP7BH1069Q signal in ER cisternae (Fig. 8J, arrows; quantification in Fig. 8H). This trend indicates that inhibiting p38 and JNK facilitates sorting of ATP7BH1069Q from the ER into nascent Golgi‐directed transport intermediates.
Discussion
The main finding in this study is that p38 and JNK play an important role in WD by promoting retention and degradation of the ATP7BH1069Q mutant in the ER. Thus, suppression of p38 and JNK allows ATP7BH1069Q to reach the post‐Golgi vesicles and the apical surface in hepatocytes, from where it can contribute to the removal of excess Cu from the cell. As a consequence, treatment with p38 or JNK inhibitor reduces Cu accumulation in cells expressing ATP7BH1069Q. Thus, p38 and JNK represent attractive targets for correction of the ATP7B mutant localization and function and could be considered for the development of novel therapeutic strategies to counteract WD.
A large proportion of WD‐causing ATP7B mutations result in a protein product that is still able to pump Cu but remains retained within the ER and undergoes accelerated degradation. Therefore, a therapeutic strategy that can target the proteostasis of these ATP7B mutants to reduce their degradation and to improve their folding and export would be beneficial for the large cohort of WD patients.
In recent years, several attempts have been made in this direction, with the use of correctors such as curcumin and phenylbutyrate.9 The rationale for using these drugs is that they are effective at rescuing misfolded CFTR or lysosomal enzymes,26, 27 which result in cystic fibrosis and lysosomal storage diseases, respectively. These drugs were shown to reduce the degradation of several ER‐retained mutants of ATP7B.9 Whether and how these correctors rescue the appropriate localization of these ATP7B mutants and, therefore, recover the ability of these aberrant ATP7B proteins to eliminate Cu remains to be demonstrated.
Several proteins have also been reported to modulate the degradation of the ATP7B mutants and/or their retention in the ER. These include COMMD1,28 clusterin,29 and CRYAB.12 However, these proteins do not belong to the “druggable” category; thus, their potential for the development of drugs to cure WD remains questionable. Instead, both p38 and JNK represent bona fide druggable targets, and their inhibitors show high efficiency at rescuing several of these pathogenic ATP7B variants. Their ability to recover ATP7BH1069Q trafficking in primary hepatocytes indicates potential efficacy in the hepatic tissue. This property of p38 and JNK inhibitors is really important for therapy because the number of active compounds which are effective in immortalized cell lines frequently fail to provide the same beneficial impact in primary cells or animals.30 Unfortunately an animal model that recapitulates ATP7BH1069Q mutation remains to be developed. Therefore, evaluation of the inhibitor efficiency in vivo represents a further challenging and important goal.
Of note, the therapeutic potential of some p38 and JNK antagonists has been explored for the treatment of different disorders. Some p38 and JNK inhibitors have entered phase 2 clinical studies for other disorders (ClinicalTrials.gov identifiers NCT00316771, NCT00303563, NCT00383188, NCT00620685, NCT00395577, NCT00205478, NCT00390845, NCT00570752, NCT01630252).31 Thus, in theory the passage of such p38 and JNK inhibitors into clinical use for WD might be rapid as their toxicity level has been reported to be relatively low32 and may be further decreased through specific delivery using ligands of the asialoglycoprotein receptor.33
Which molecular mechanisms operate behind the correction of the ATP7BH1069Q mutant? As we have demonstrated, inhibition of p38 and JNK manifests two main phenomena: a reduction in ATP7BH1069Q degradation and an improved ATP7BH1069Q delivery to the Golgi apparatus and to the post‐Golgi compartments. It turns out that a reduction in the degradation of ATP7BH1069Q occurs as a consequence of increasingly efficient sorting of ATP7BH1069Q into the secretory pathway. Indeed, photo‐bleaching experiments and immuno‐EM indicate that p38 and JNK inhibitors facilitate ATP7B mutant sorting from the ER into the secretory pathway at the level of the ER exit sites, while overall membrane trafficking rates remain unchanged.
Therefore, corrections of ATP7BH1069Q from the ER into the secretory route probably arise through modulation of the proteostatic network, which improves the ability of ATP7BH1069Q to be captured by the export machinery in inhibitor‐treated cells. ATP7BH1069Q interactome exhibits a significant enrichment in components of ER quality control and ERAD. Thus, an excessive amount of quality control proteins that bind the mutant may prevent its transition from the ER to the secretory pathway. Such a chaperone‐mediated ER trap was reported for the CFTR mutant.34 The inhibitors of p38 and JNK might reduce the amounts of quality control proteins that bind the ATP7B mutant and in this way allow ATP7BH1069Q to escape into the secretory pathway.
Alternatively, inhibition of p38 and JNK may result in the up‐regulation of some chaperones that are negatively regulated by these stress kinases. Thus, up‐regulation of such chaperones could improve mutant folding and stability in cells treated with p38 or JNK antagonists. We have shown that the CRYAB chaperone significantly facilitates ATP7BH1069Q exit from the ER.12 Its activity and expression appear to be regulated by p3835 and, therefore, might also be modulated by p38 inhibition. It remains to be investigated whether p38 antagonists can induce CRYAB up‐regulation in hepatocytes that express ATP7B mutants.
Although p38 and JNK inhibitors were effective at correcting several ER‐retained ATP7B variants, they did not rescue a few other ATP7B mutants from the ER. Interestingly, mutations in the same domain (like ATP7BH1069Q and ATP7BL1083F) are differently sensitive to p38 and JNK inhibitors. This indicates that p38‐dependent and JNK‐dependent machineries have a certain level of specificity and that inhibiting them does not seem to threaten the global quality‐control processes at the level of the ER. On the other hand, p38 and JNK inhibitors also rescue some, but not all, ER‐retained mutants of other membrane proteins, such as CFTR and P‐glycoprotein.19 Therefore, these p38 and JNK inhibitors might modulate specific mechanisms involved in the recognition of common (or similar) protein‐folding defects.
In conclusion, p38 and JNK signaling pathways may serve as attractive targets to counteract WD by rescuing ER‐retained ATP7B mutants. It remains to be established to what extent suppression of these pathways would be able to prevent toxic Cu accumulation in model animals or patients carrying corresponding ATP7B mutations. On the other hand, the impact of p38 and JNK antagonists on the key components of proteostatic networks requires further detailed investigation. This may expand the use of kinase inhibitors to correction of other ER‐retained pathogenic mutants and, hence, to open new avenues for therapeutic strategies to combat disorders that are caused by protein misfolding.
Author names in bold designate shared co‐first authorship.
Supporting information
Additional Supporting Information may be found at onlinelibrary.wiley.com/doi/10.1002/hep.28398/suppinfo.
Supporting Information
Acknowledgment
We thank those who provided us with antibodies, reagents, and cells. Christopher J. Chang and Jefferson Chan for help with CS3; Diego di Bernardo, Fabiana Ciciriello, and the Telethon Institute of Genetics and Medicine (TIGEM) Bioinformatics Core for analysis of proteomics and microarray results; TIGEM Advanced Microscopy and Imaging Core for microscopy support; and TIGEM Vector Core for production of adenoviruses.
Potential conflict of interest: Nothing to report.
Supported by grants from Telethon (TGM11CB4, to R.S.P., TGM11CB5, to A.L.), AIRC (to R.S.P. and A.L.), and MIUR (FaReBio, PON01‐00117, PON01‐00862, PON03‐00025, EPIGEN, and Invecchiamento, to A.L.).
REFERENCES
- 1. Gitlin JD. Wilson disease. Gastroenterology 2003;125:1868‐1877. [DOI] [PubMed] [Google Scholar]
- 2. Ferenci P. Review article: diagnosis and current therapy of Wilson's disease. Alimentary Pharmacol Ther 2004;19:157‐165. [DOI] [PubMed] [Google Scholar]
- 3. Lutsenko S, Barnes NL, Bartee MY, Dmitriev OY. Function and regulation of human copper‐transporting ATPases. Physiol Rev 2007;87:1011‐1046. [DOI] [PubMed] [Google Scholar]
- 4. La Fontaine S, Mercer JF. Trafficking of the copper‐ATPases, ATP7A and ATP7B: role in copper homeostasis. Arch Biochem Biophys 2007;463:149‐167. [DOI] [PubMed] [Google Scholar]
- 5. Roberts EA, Schilsky ML. Diagnosis and treatment of Wilson disease: an update. Hepatology 2008;47:2089‐2111. [DOI] [PubMed] [Google Scholar]
- 6. Iorio R, D'Ambrosi M, Marcellini M, Barbera C, Maggiore G, Zancan L, et al. Serum transaminases in children with Wilson's disease. J Pediatr Gastroenterol Nutr 2004;39:331‐336. [DOI] [PubMed] [Google Scholar]
- 7. Forbes JR, Cox DW. Functional characterization of missense mutations in ATP7B: Wilson disease mutation or normal variant? Am J Hum Genet 1998;63:1663‐1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iida M, Terada K, Sambongi Y, Wakabayashi T, Miura N, Koyama K, et al. Analysis of functional domains of Wilson disease protein (ATP7B) in Saccharomyces cerevisiae . FEBS Lett 1998;428:281‐285. [DOI] [PubMed] [Google Scholar]
- 9. van den Berghe PV, Stapelbroek JM, Krieger E, de Bie P, van de Graaf SF, de Groot RE, et al. Reduced expression of ATP7B affected by Wilson disease‐causing mutations is rescued by pharmacological folding chaperones 4‐phenylbutyrate and curcumin. Hepatology 2009;50:1783‐1795. [DOI] [PubMed] [Google Scholar]
- 10. Payne AS, Kelly EJ, Gitlin JD. Functional expression of the Wilson disease protein reveals mislocalization and impaired copper‐dependent trafficking of the common H1069Q mutation. Proc Natl Acad Sci USA 1998;95:10854‐10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huster D, Kuhne A, Bhattacharjee A, Raines L, Jantsch V, Noe J, et al. Diverse functional properties of Wilson disease ATP7B variants. Gastroenterology 2012;142:947‐956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. D'Agostino M, Lemma V, Chesi G, Stornaiuolo M, Cannata Serio M, D'Ambrosio C, et al. The cytosolic chaperone alpha‐crystallin B rescues folding and compartmentalization of misfolded multispan transmembrane proteins. J Cell Sci 2013;126:4160‐4172. [DOI] [PubMed] [Google Scholar]
- 13. Vonk WI, de Bie P, Wichers CG, van den Berghe PV, van der Plaats R, Berger R, et al. The copper‐transporting capacity of ATP7A mutants associated with Menkes disease is ameliorated by COMMD1 as a result of improved protein expression. Cell Mol Life Sci 2012;69:149‐163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Polishchuk EV, Concilli M, Iacobacci S, Chesi G, Pastore N, Piccolo P, et al. Wilson disease protein ATP7B utilizes lysosomal exocytosis to maintain copper homeostasis. Dev Cell 2014;29:686‐700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Darling NJ, Cook SJ. The role of MAPK signalling pathways in the response to endoplasmic reticulum stress. Biochim Biophys Acta 2014;1843:2150‐2163. [DOI] [PubMed] [Google Scholar]
- 16. Nawaz M, Manzl C, Lacher V, Krumschnabel G. Copper‐induced stimulation of extracellular signal‐regulated kinase in trout hepatocytes: the role of reactive oxygen species, Ca2+, and cell energetics and the impact of extracellular signal‐regulated kinase signaling on apoptosis and necrosis. Toxicol Sci 2006;92:464‐475. [DOI] [PubMed] [Google Scholar]
- 17. Chen SH, Lin JK, Liu SH, Liang YC, Lin‐Shiau SY. Apoptosis of cultured astrocytes induced by the copper and neocuproine complex through oxidative stress and JNK activation. Toxicol Sci 2008;102:138‐149. [DOI] [PubMed] [Google Scholar]
- 18. Rupesh KR, Priya AM, Sundarakrishnan B, Venkatesan R, Lakshmi BS, Jayachandran S. 2,2′‐Bipyridyl based copper complexes down regulate expression of pro‐inflammatory cytokines and suppress MAPKs in mitogen induced peripheral blood mononuclear cells. Eur J Med Chem 2010;45:2141‐2146. [DOI] [PubMed] [Google Scholar]
- 19. Hegde R, Parashuraman S, Iorio F, Capuani F, Ciciriello F, Carissimo A, et al. Unravelling druggable signalling networks that control F508del‐CFTR proteostasis. Elife. December 23, 2015;4. pii: e10365. doi: 10.7554/eLife.10365. [Correction added March 7, 2016, after original online publication: reference was updated to include more recent publication information.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brancho D, Ventura JJ, Jaeschke A, Doran B, Flavell RA, Davis RJ. Role of MLK3 in the regulation of mitogen‐activated protein kinase signaling cascades. Mol Cell Biol 2005;25:3670‐3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cater MA, La Fontaine S, Shield K, Deal Y, Mercer JF. ATP7B mediates vesicular sequestration of copper: insight into biliary copper excretion. Gastroenterology 2006;130:493‐506. [DOI] [PubMed] [Google Scholar]
- 22. Dodani SC, Domaille DW, Nam CI, Miller EW, Finney LA, Vogt S, et al. Calcium‐dependent copper redistributions in neuronal cells revealed by a fluorescent copper sensor and X‐ray fluorescence microscopy. Proc Natl Acad Sci USA 2011;108:5980‐5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum‐associated degradation. Nat Rev Mol Cell Biol 2008;9:944‐957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Polishchuk EV, Di Pentima A, Luini A, Polishchuk RS. Mechanism of constitutive export from the Golgi: bulk flow via the formation, protrusion, and en bloc cleavage of large trans‐Golgi network tubular domains. Mol Biol Cell 2003;14:4470‐4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bannykh SI, Rowe T, Balch WE. The organization of endoplasmic reticulum export complexes. J Cell Biol 1996;135:19‐35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Singh OV, Pollard HB, Zeitlin PL. Chemical rescue of deltaF508‐CFTR mimics genetic repair in cystic fibrosis bronchial epithelial cells. Mol Cell Proteomics 2008;7:1099‐1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mu TW, Ong DS, Wang YJ, Balch WE, Yates JR 3rd, Segatori L, et al. Chemical and biological approaches synergize to ameliorate protein‐folding diseases. Cell 2008;134:769‐781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Bie P, van de Sluis B, Burstein E, van de Berghe PV, Muller P, Berger R, et al. Distinct Wilson's disease mutations in ATP7B are associated with enhanced binding to COMMD1 and reduced stability of ATP7B. Gastroenterology 2007;133:1316‐1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Materia S, Cater MA, Klomp LW, Mercer JF, La Fontaine S. Clusterin (apolipoprotein J), a molecular chaperone that facilitates degradation of the copper‐ATPases ATP7A and ATP7B. J Biol Chem 2011;286:10073‐10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Trzcinska‐Daneluti AM, Nguyen L, Jiang C, Fladd C, Uehling D, Prakesch M, et al. Use of kinase inhibitors to correct DeltaF508‐CFTR function. Mol Cell Proteomics 2012;11:745‐757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Damjanov N, Kauffman RS, Spencer‐Green GT. Efficacy, pharmacodynamics, and safety of VX‐702, a novel p38 MAPK inhibitor, in rheumatoid arthritis: results of two randomized, double‐blind, placebo‐controlled clinical studies. Arthritis Rheum 2009;60:1232‐1241. [DOI] [PubMed] [Google Scholar]
- 32. Kumar S, Boehm J, Lee JC. p38 MAP kinases: key signalling molecules as therapeutic targets for inflammatory diseases. Nat Rev Drug Discov 2003;2:717‐726. [DOI] [PubMed] [Google Scholar]
- 33. Pujol AM, Cuillel M, Renaudet O, Lebrun C, Charbonnier P, Cassio D, et al. Hepatocyte targeting and intracellular copper chelation by a thiol‐containing glycocyclopeptide. J Am Chem Soc 2011;133:286‐296. [DOI] [PubMed] [Google Scholar]
- 34. Wang X, Venable J, LaPointe P, Hutt DM, Koulov AV, Coppinger J, et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell 2006;127:803‐815. [DOI] [PubMed] [Google Scholar]
- 35. Clements RT, Feng J, Cordeiro B, Bianchi C, Sellke FW. p38 MAPK‐dependent small HSP27 and alphaB‐crystallin phosphorylation in regulation of myocardial function following cardioplegic arrest. Am J Physiol Heart Circ Physiol 2011;300:H1669‐H1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found at onlinelibrary.wiley.com/doi/10.1002/hep.28398/suppinfo.
Supporting Information


