Abstract
Proper gene expression is essential for the survival of every cell. Once thought to be a passive transporter of genetic information, RNA has recently emerged as a key player in nearly every pathway in the cell. A full description of its structure is critical to understanding RNA function. Decades of research have focused on utilizing chemical tools to interrogate the structures of RNAs, with recent focus shifting to performing experiments inside living cells. This Review will detail the design and utility of chemical reagents used in RNA structure probing. We also outline how these reagents have been used to gain a deeper understanding of RNA structure in vivo. We review the recent merger of chemical probing with deep sequencing. Finally, we outline some of the hurdles that remain in fully characterizing the structure of RNA inside living cells, and how chemical biology can uniquely tackle such challenges.
The control of gene expression lies at the heart of fundamental biological processes. A comprehensive understanding of gene expression programs would thus provide continued insight into normal biological processes and the ways that their alteration results in disease. Although much is known about DNA sequences and transcription factors that regulate RNA expression1,2, control over gene expression is also driven by RNA functions that extend beyond serving as a template for protein expression.
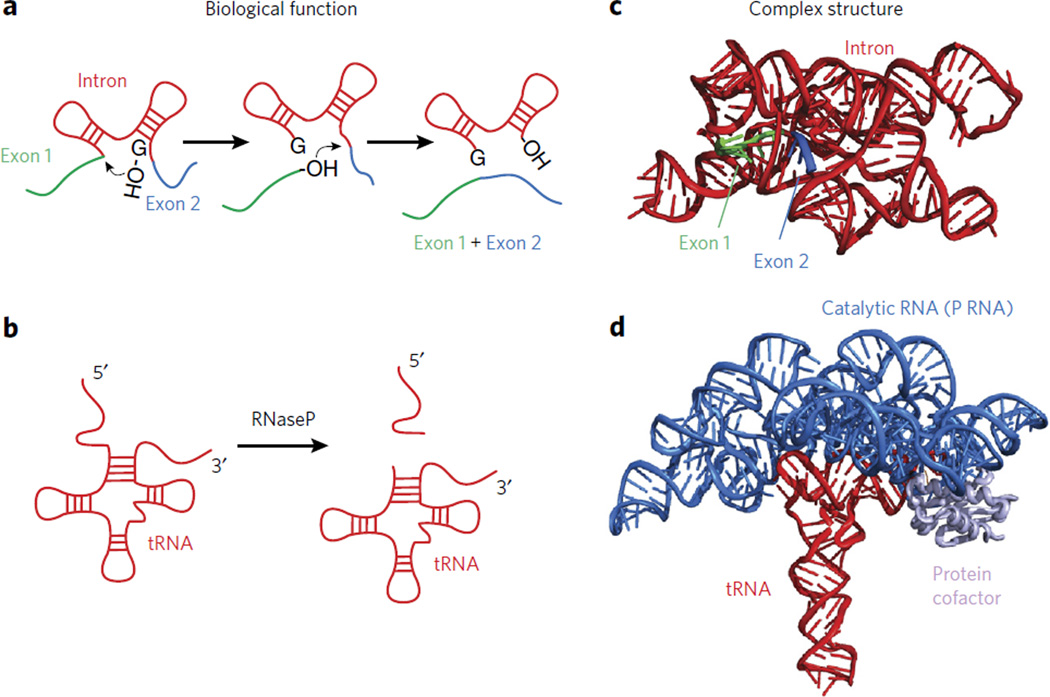
Early research focused on characterizing RNA function primarily centered on messenger RNA (mRNA), transfer RNA (tRNA) and ribosomal RNA (rRNA). Although these RNAs play critical roles for the survival of every cell, the discovery of catalytic RNA in the 1980s3,4 propelled the race to identify RNA molecules that perform more exotic functions. The discovery of catalytic self-splicing introns3, which precisely remove the intron and link exons together in the correct order (Fig. 1a), and the RNA component of ribonuclease (RNase) P4 (Fig. 1b), the enzyme that matures tRNA precursors, first highlighted the catalytic roles of RNA. The peptidyltransferase active site of the ribosome has also been shown to be composed of RNA5,6. Several species of bacteria utilize small molecule–binding RNAs, called ‘riboswitches’, to control the expression of genes involved in metabolic pathways7. Long noncoding RNAs have been shown to target chromatin-remodeling complexes to establish or reinforce an epigenetic state8,9. As these examples illustrate, the range of functions performed by RNA continues to expand.
Figure 1. The functional and structural complexity of RNA.
(a) A cartoon depicting the function of group I introns. (b) A cartoon depicting the function of RNase P in trimming tRNA. (c) A crystal structure of the group I intron (PDB 3BO2). (d) A crystal structure of the catalytic component of RNase P in complex with a tRNA substrate (PDB 3Q1R).
The base-pairing properties of RNA endows it with the capacity to form extensive intramolecular and intermolecular structures, which are known to influence practically every step of gene expression1. Unique RNA structure elements provide scaffolds for splicing and for the binding of other RNA sequences and proteins10–12. RNA structural motifs are necessary for RNA subcellular localization13 and are critical in the regulation of transcript half-life and decay14. Many RNA structure elements have been discovered to play critical roles in the onset of cancer metastasis and neurological disorders14–16. Despite the growing list of functional RNA motifs, only a relatively small number have had their three-dimensional structures studied at high resolution.
The crystal structure of the Group I intron first revealed the principles employed by RNA intron-exon splicing (Fig. 1c). X-ray crystallographic analysis of the catalytic component of RNase P revealed the way that an RNA active site orients the tRNA substrate for trimming (Fig. 1d). Several structures of metabolite-binding riboswitches have revealed how RNA can fold into highly complex structures to sense small molecules7. High-resolution structures definitively showed the ribosome is a ribozyme6. NMR analysis has been extremely useful for dissecting the mechanisms of viral RNAs, telomerase RNA and riboswitches17–19. These investigations, and many others, highlight the power of applying three-dimensional structural approaches to RNA biology. Despite such progress, however, many structures of individual RNAs remain to be solved, and this is especially true within the native environment of the cell.
With few exceptions, the majority of the aforementioned investigations of RNA have been performed on well-studied smaller RNAs. The size of many RNAs inside the cell, which can be greater than 1 kb in length, renders most of them intractable to three-dimensional analysis by NMR and X-ray crystallography. Researchers have instead turned to chemical probing, which has been used to characterize RNA secondary structure, RNA-protein interactions, and even more dynamic processes such as RNA folding20. Importantly, many of the reagents used in these experiments are small enough to traverse the cell membrane and thus can probe RNA structure inside the cellular environment.
In this Review, we will detail the use and the evolution of chemical tools used to interrogate RNA structure and discuss the use and resurgence of chemical probing applied to RNA inside living cells. We will highlight how the merger between RNA structure probing and deep sequencing has allowed a systems-level view of RNA structure and revealed some general principles of RNA regulation in gene expression pathways. Finally, we will comment on some of the challenges that lie ahead in interrogating RNA structure inside cells and outline how the field of chemical biology is poised to make a great impact in this area.
Chemical methods to probe RNA structure in vitro
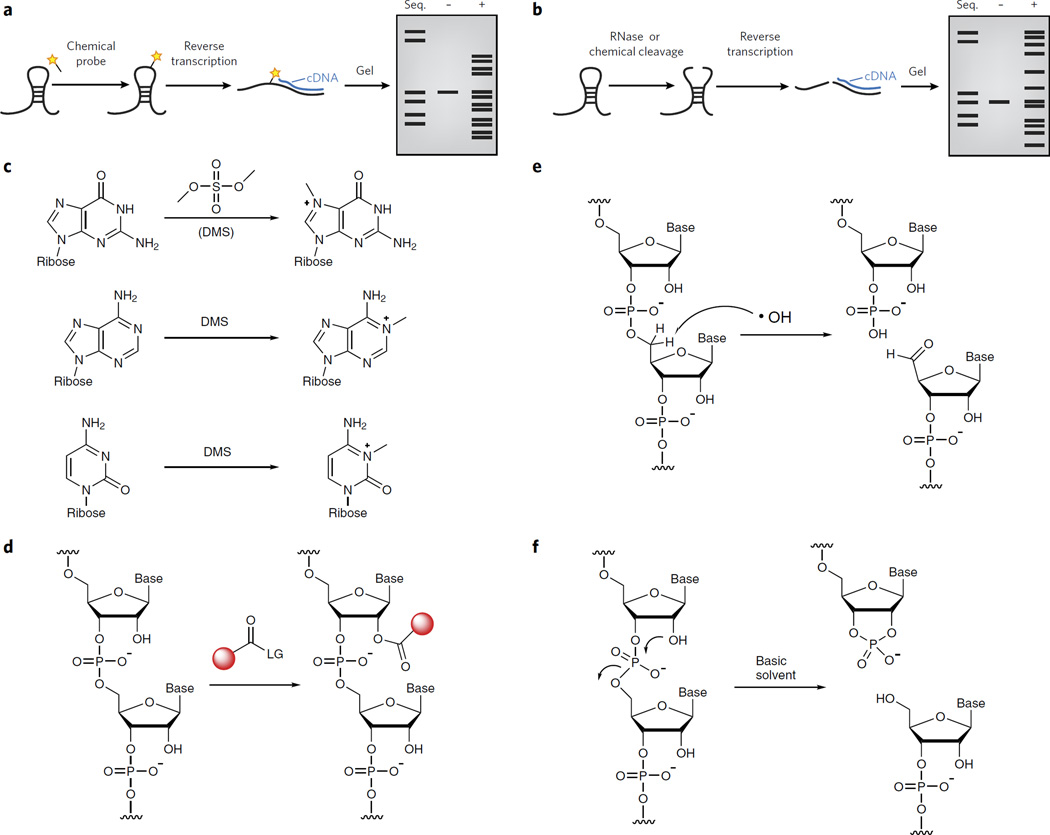
Chemical probing methods for characterizing RNA structural elements rely on chemical approaches that either introduce a chemical adduct (Fig. 2a) or induce strand scission (Fig. 2b) in the target RNA. These probing events are then detected in several ways. Sites of chemical adducts, which generally block polymerase extension, are detected by termination of reverse transcription (Fig. 2a). Sites of strand scission are identified either by direct labeling of the 5′ end of the RNA (with 32P) or by reverse transcription (Fig. 2b). Denaturing gel analysis, followed by mapping of cDNA sites back to the primary sequence, is used to measure the chemical reactivities at each position of the sequence20,21. Secondary structure prediction algorithms can be dramatically improved by integrating such data derived from probing experiments (Box 1)22,23. We will discuss the type of reactions that can be used to arrive at structure reactivities in detail below.
Figure 2. Chemical methods to probe RNA structure.
(a) A depiction of a typical chemical probing experiment. The RNA is first folded and then treated with a reagent that reacts covalently. The site of reaction is read out by reverse transcription and synthesis of cDNA that can be resolved by denaturing gel electrophoresis. (b) A depiction of a typical RNA structure probing experiment in which RNA cleavage is used to probe structure. The RNA is first folded and then treated with a reagent that cleaves the RNA backbone. The site of cleavage is read out by reverse transcription and synthesis of cDNA that can be resolved by denaturing gel electrophoresis. (c) Dimethylsulfate can alkylate the N7 of guanosine, the N1 of adenosine and the N3 of cytidine. (d) RNA SHAPE analysis measures the propensity of the 2′-OH to become activated as a nucleophile and undergo an acylation reaction. (e) Hydroxyl radical probing at the C5′ position leads to strand cleavage, resulting in the formation of a 3′-phosphate and 5′-aldehyde. (f) In-line probing results in a 2′,3′-cyclic phosphate and a 5′-hydroxyl.
Box 1. Structure probing in RNA structure predictions.
Chemical probing experiments provide data sets for building models of RNA structure. However, these methods do not present the complete picture and can lead to hypothetical structural models. Incorporating experimental restraints from chemical probing experiments as ‘pseudo-energies’ can improve the accuracy of structure prediction programs such as RNAstructure85, UNAfold86 and RNAfold87. For instance, the secondary structure model of 16S rRNA was improved to 97% accuracy, from 49.7%, as a result of restraints on the model introduced from SHAPE data88. A recent excellent review outlines each of these programs and their limitations in predicting RNA secondary structure89.
The major challenge facing all RNA structure prediction methods is that of parameter estimation90. That is, the accuracy of predictions made by free energy minimization is limited by the quality of the free energy parameters in the underlying model. It is still unclear whether the structural estimations gathered by studying RNAs with a significant amount of structural stability (such as ribosomal RNA, ribozymes and riboswitches) can be extrapolated to other types of RNAs within cells (such as mRNAs). The recent observation that mRNA structures are quite dynamic within the cell63,64 further suggests that the estimates gathered by studying highly structured RNAs may not apply.
Methods to model RNA structure on entire transcriptomes are currently being pursued as well. One program, SeqFold91, transforms experimental RNA structural data into a structure preference profile (SPP) and uses it to select stable RNA structure candidates representing the structural ensemble. Under a high-dimensional classification framework, SeqFold efficiently matches a given SPP to the most likely cluster of structures sampled from the Boltzmann-weighted ensemble. Another program, StructureFold, directly integrates probing data to restrain RNA secondary structure prediction via the RNAstructure and ViennaRNA package algorithms92. Overall, the merger of RNA structure probing with prediction algorithms is sure to increase the accuracy of RNA structure predictions and models.
Understanding how primary RNA sequence variation leads to alterations in the structural ensemble of RNA is another active area of study and will increase in importance as more genomic data, some with disease relevance, identifies functional RNA mutations that occur outside of coding regions. A recent study toward this goal has taken publicly available RNA structure probing data and benchmarked several RNA structure prediction algorithms93. Prediction algorithms designed to identify changes in RNA structure due to mutations performed the best among the 11 algorithms tested. More traditional prediction programs (RNAfold and RNAstructure) performed better when base-pairing probabilities, rather than free energy calculations, were implemented. However, overall algorithmic performance was low, suggesting the need for continued improvement. Nevertheless, these results demonstrate that structural rearrangements are possible as a result of genetic variation and these should be taken into consideration when dissecting mutational data in genomic data sets.
The first chemical probing reagents were reactive alkylating agents that form chemical adducts with RNA. Dimethylsufate (DMS) was first used in the 1980s as a reagent to probe single-stranded RNA24; it alkylates the N7 position of guanosine, the N1 position of adenosine and the N3 position of cytidine (Fig. 2c). Since then additional reagents have been developed to probe the Watson-Crick face of guanosine. Kethoxal reacts with the N1 and C2 exocyclic amine of guanosine residues21. The silyl derivative N,N-(dimethylamino) dimethylchlorosilane has also been used to probe the solvent accessibility of guanosines in complex RNA structures25. Carbodiimides react primarily with N3 of uridine and N1 of guanine, modifying two sites responsible for hydrogen bonding on the bases26. Each of these reagents reacts with specific functional groups on unique bases, but other structural probing tools have been developed to examine other RNA functional groups.
Selective hydroxyl acylation analyzed by primer extension (SHAPE) is perhaps the most widely employed method of utilizing chemical modification to probe RNA. SHAPE takes advantage of the observations that electrophilic reagents can acylate the 2′-hydroxyl group of RNA nucleotides (Fig. 2d) and that the reactivity of these 2′-hydroxyl groups is different depending on local RNA structure: groups at flexible positions sample conformations that transiently enhance their nucleophilicity and thus are more highly modified27,28. SHAPE chemistry has been used to understand the structure of the entire HIV RNA genome29, to characterize RNA-protein interactions30 and to characterize RNA folding and structural transitions27. Because SHAPE chemistry is less selective than base-specific chemical probing methods, it allows the user to interrogate all nucleotide positions within an RNA molecule simultaneously and provides direct measurements of RNA backbone flexibility.
The second major class of chemical probing techniques is those that induce strand scission at specific sites. The dominant tool for site-specific RNA cleavage has been RNase enzymes. RNase enzymes cut at their binding sites, resulting in the formation of a 2′,3′-cyclic phosphate and a 5′-hydroxyl, and can be used to probe specific structure elements in RNA (Fig. 2b). RNase S1 recognizes single-stranded domains in RNA, whereas RNase V1 cuts double-stranded regions31, and RNase TI recognizes single-stranded RNA sequences and cuts at a guanosine residues32. RNase probing is limited by the footprints of the enzymes. Instead, researchers have turned to small molecules for higher-resolution probing experiments.
Chemical methods are also widely used to create RNA strand breaks. For example, hydroxyl radical probing is used to probe the solvent accessibility of the RNA backbone. In one mechanism, hydroxyl radicals abstract hydrogens from the C5′ position of the backbone, eventually leading to strand scission, with 3′-phosphate and 5′-aldehyde products (Fig. 2e)33. Alterations in cut sites reveal changes in solvent accessibility rather than Watson-Crick pairing. The rate of backbone cleavage is fast enough (milliseconds) to be used to infer RNA folding, RNA-protein interaction and solvent effects through careful time-course experiments34,35.
‘In-line probing’ is a methodology that takes advantage of the intrinsic chemical reactivity of the 2′-hydroxyl group to induce phosphate backbone cleavage under slightly basic conditions. The optimal reaction geometry for intramolecular RNA backbone scission requires that the nucleophilic 2′-hydroxyl must be positioned directly opposite (at 180 degrees to or ‘in line’ with) the departing 5′-phosphate group (Fig. 2f)36. Residues at flexible RNA nucleotides have a higher likelihood of adopting an in-line conformation and are therefore more likely to undergo cleavage at that site. In-line probing has been widely used to elucidate changes in RNA flexibility that occur due to the binding of small molecules and for the structural interrogation of metabolite-binding riboswitch RNAs36. One drawback to in-line probing is that the typical experiment may need to be performed for up to 40 h to induce enough ‘spontaneous’ cleavage. This prevents its use for analyzing RNA folding or RNA structure transitions that occur on faster time scales.
Chemical probing has provided a powerful method for analyzing RNA secondary structure in vitro. The small molecule–based methods developed initially for use in the test tube are now finding use in living cells and are helping to elucidate the role that RNA structure plays in cellular RNA regulation.
Progress on in vivo chemical probing of RNA structure
The structural characteristics of RNA in vivo are likely more complex than those in vitro. RNA structure is easily influenced by the rate of transcription37–40, local solution conditions41, the binding of small molecules42 and interactions with RNA-binding proteins12. These observations hint that the physical state of RNA within the cell is very important for its function, but little is known of how intracellular RNA structure can contribute to the specificity of such events. A key bottleneck is the ability to ‘observe’ RNA structure in living cells. Probing RNA structure inside cells will provide a more realistic understanding of the physical nature of RNA in living systems.
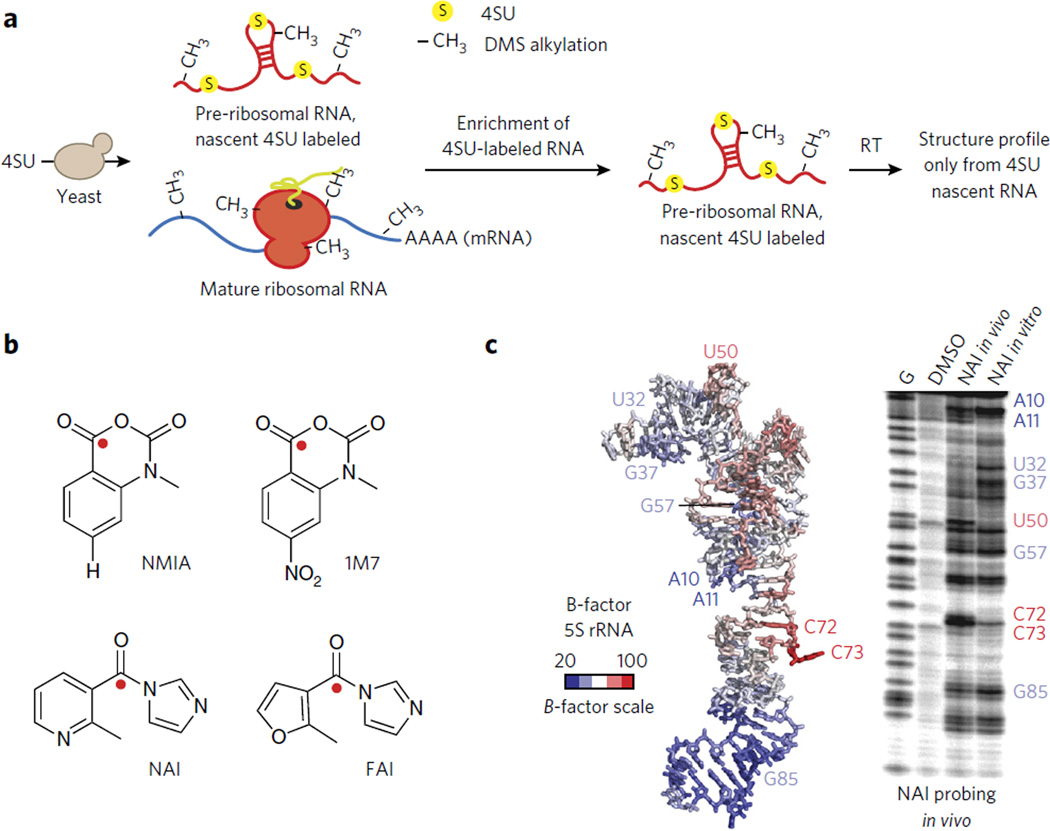
Dimethylsufate RNA alkylation has been the most widely used method of analyzing RNA structure inside cells. The small size of DMS makes it cell permeant. Further, DMS reaction times can be limited to just a few minutes, thereby capturing a somewhat short window of time for structure probing. DMS probing has been used to elucidate RNA-protein interactions43, study RNA enzyme function in living cells44 and even probe RNA structural rearrangements caused by RNA remodeling proteins45. The strength of DMS probing was recently highlighted when it was used to investigate the structure of ribosomal RNA during pre-ribosome maturation46. Ribosomes undergo many structural rearrangements during maturation that are critical to the final ribosome structure. Probing experiments do not select for nascent RNA structures, and thus early structural intermediates are notoriously difficult to probe. 4-thiouracil (4-thioU) RNA labeling46, in which newly transcribed RNAs are selectively marked with 4-thioU (see Fig. 3a below for a more detailed discussion of this approach), was used to overcome this challenge. In vivo structure probing by DMS modification of newly synthesized (4-thioU-labeled) 20S pre-rRNA was evaluated over time and revealed a remarkably flexible structure throughout ribosomal subunit biogenesis, with little stable RNA-protein interaction observed. DMS probing at different time points revealed structural changes that are associated with small nucleolar RNA (snoRNA) binding, a critical event in ribosome maturation. Finally, the analyses indicated that many parts of ribosomal RNA structure mature early, but additional protection appears subsequently, presumably reflecting protein binding. This paper represents just one important demonstration of the power of DMS probing to enhance mechanistic understanding of RNA structure and function in cells, particularly throughout the lifetime of RNA folding and maturation.
Figure 3. Methods to measure RNA structure inside living cells.
(a) A schematic representation of using 4-thioU (4SU) RNA labeling to enrich for newly transcribed RNAs. DMS chemical probing was merged with 4SU labeling to study the structure of pre-mature rRNA in vivo (see ref. 46). In this method DMS chemical probing is performed in vivo and only the RNA structure pattern for premature ribosomal RNA is obtained. Both the pre-folded and mature RNA are present. However, as a result of 4SU enrichment, only the pre-mature rRNA is probed. (b) A chemical schematic of RNA SHAPE reagents. The site of 2′-OH attack is represented as a red sphere. (c) Demonstration of NAI structure probing of 5S rRNA in living cells. A denaturing gel is shown at right and the B-factors of the 5S rRNA from a corresponding crystal structure at left. Images in c are reproduced from ref. 49 with permission from Nature Publishing Group.
As discussed above, SHAPE has emerged as a key method for chemical probing of RNA. Using SHAPE in vivo would dramatically increase our understanding of how RNA structure is manipulated by the intracellular environment by means of trans-acting proteins and RNAs. The 1M7 SHAPE reagent (Fig. 3b) has been used to probe the structure of ribosomal RNA47,48 and a riboswitch RNA41 in vivo. Further exploration of reagent design has extended the ability of SHAPE to be used on weakly expressed RNAs, such as noncoding RNAs and even mRNAs.
Recent advances in SHAPE electrophile design have extended the SHAPE tool kit. NAI and FAI are two new SHAPE reagents that have an acyl imidazole scaffold (Fig. 3b)49. Both reagents have much higher solubility (>200 mM) and extended half-lives (~30 min) as compared to earlier SHAPE reagents. Initial analysis of NAI reactivity suggested chemical probing patterns similar to those of other SHAPE reagents. SHAPE probing of 5S rRNA, comparing in vitro and in vivo data, identified regions of the RNA that are in contact with the RNA-binding protein L5 and 16S rRNA. Comparison of SHAPE probing and a recent crystal structure of the ribosome demonstrated that NAI is highly reactive, with residues that have high B-factors (thermal flexibility, Fig. 3c). Importantly, NAI was also used to read out the secondary structure of the 5S rRNA across five different species and those of less abundant nuclear RNAs in mammalian cells. Additional studies have used NAI when comparing RNA structures inside and outside cells, further underscoring its usefulness50,51. Although the approach is powerful, the long incubation times (minutes) of SHAPE and DMS reagents render them incapable of measuring RNA dynamics and folding in cells.
Hydroxyl radical cleavage is perhaps the most promising method for measuring RNA dynamics in vivo52. Recent extension of this method has permitted the analysis of RNA solvent accessibility53 in living cells54. Synchrotron-generated X-rays were used to create hydroxyl radicals within cells, which permitted the analysis of RNA folding and observation of RNA-protein complexes in vivo. In a typical experiment, a 100-ms exposure to the high flux of a synchrotron X-ray beam is sufficient to probe the folding and dynamics of intracellular RNAs55. In vivo hydroxyl radical probing has revealed the critical interplay between ribosomal RNA and associated proteins56 and the use of RNA chaperones57 that are necessary for ribosome assembly. Hydroxyl radical probing has been solidified as a powerful method to analyze RNA structure folding inside living cells.
Systems-based approaches to RNA structure in cells
Most analyses of RNA structure have come from examining a few well-studied RNAs one at a time. Recent efforts have been focused on combining the precision of chemical probing experiments with the power of transcriptomics to gain a holistic understanding of RNA structure, even on understudied messenger and other less abundant noncoding RNAs.
RNA molecules act in a concerted manner to control biological pathways. Groups of RNAs are localized to the same cellular destination through consensus RNA structure elements58. RNA-binding proteins can bind to many classes of RNAs, all based on conserved primary and secondary structure elements59. RNA structures can control the decay of gene sets to control their abundance14. Therefore, moving beyond single-transcript analysis to analyzing groups of RNAs may reveal how RNA structure contributes to cell biology on a systems level.
Coupling RNA structure measurements to deep sequencing provides two key pieces of information. First, sequencing of the reverse transcriptase–generated cDNA (as outlined in Fig. 2a) catalogs RNA modification sites across the entire transcriptome for each experimental condition. Second, precise structural features can be inferred, in the same manner as with denaturing gel electrophoresis, by quantifying the abundance of each cDNA molecule from the deep sequencing data. There are a series of recent articles detailing genome-wide methods of probing RNA structure60,61. How these efforts have increased our understanding of RNA structure inside the cell and how that controls post-transcriptional pathways are discussed below.
SHAPE-seq was the first method used to probe the structures of many RNAs in parallel in vitro62. In SHAPE-seq, a pool of in vitro–transcribed RNAs is subjected to modification by conventional SHAPE reagents followed by deep sequencing of the reverse transcription products (cDNAs). SHAPE-seq was initially used to simultaneously measure the structures of hundreds of RNA molecules, even revealing structural changes resulting from mutations62. This was a landmark study, as it demonstrated the sequencing methods could be used to not only measure RNA structure but also discern between alternative RNA structures that are the result of single-nucleotide differences. Overall, this early effort set the stage for transcriptome-wide measurements of RNA structure.
DMS was the first chemical successfully applied to transcriptome-wide structure probing inside living cells63. This study revealed that mRNAs associated with stress responses tend to have more single-strandedness, longer maximal loop length and higher free energy per nucleotide. Such features may allow these RNAs to undergo conformational changes in response to environmental conditions63. Consensus RNA structure elements shared among similar classes of RNAs may be an underappreciated means to control gene expression.
In a parallel study also using DMS, it was revealed that mRNA structures seem to sample many different conformations; however, highly important RNA structures, such as those involved in localization, have similar profiles inside and outside the cell64. ATP-dependent RNA remodeling enzymes (helicases) were shown to be responsible for the structure probing variation. Their analysis hints at the possibility that important, conserved structure elements must have a high degree of thermodynamic stability, whereas other portions of the RNA sequence have a much greater ensemble of alternate structures. These studies highlight important examples of the information that in vivo RNA structural measurements can reveal about basic RNA biology.
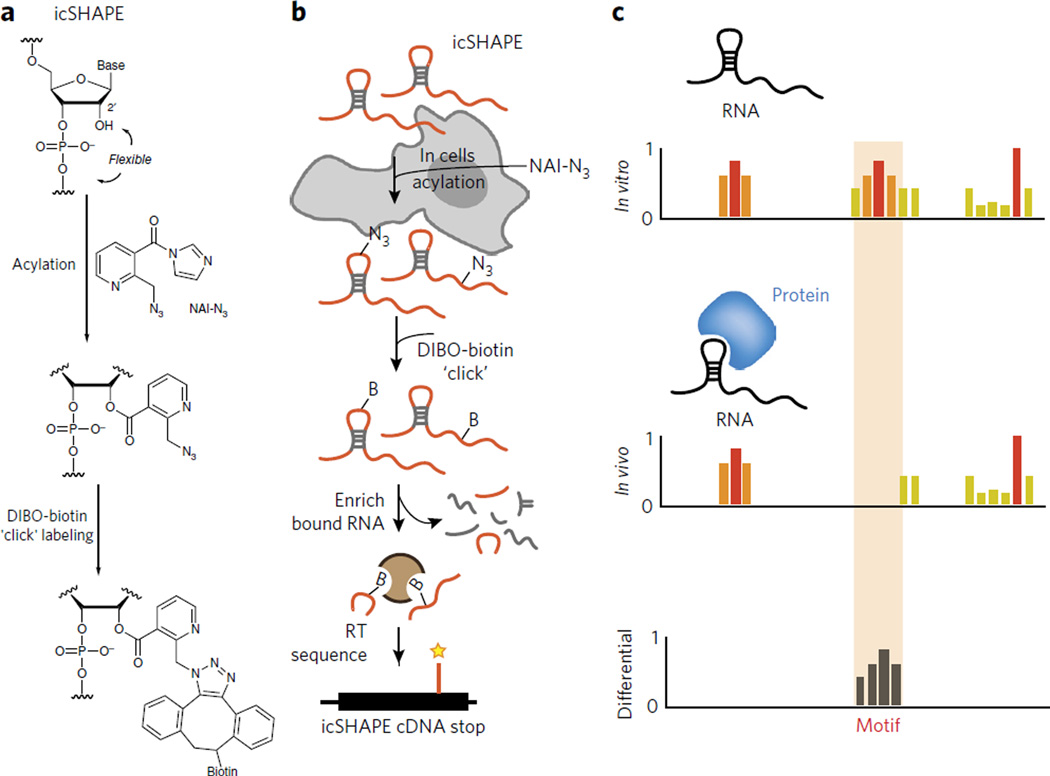
In sequencing experiments, the sampling of low-abundance RNAs can be problematic, and the low signal-to-noise ratio from chemical probing experiments presents an even greater challenge. Designing and synthesizing chemical tools that have dual functionalities—RNA structure probing and modified RNA-enriching properties—to select for chemically modified RNAs could overcome such shortcomings. Recently developed SHAPE reagents are amenable to transcriptome-wide measurements of RNA structure in vivo65. Building off the previously designed in vivo SHAPE probe49, a dual-functioning SHAPE electrophile was produced with the addition of an azide to the nicotinic acid ring (at position 2), yielding NAI-N3 (Fig. 4a). Incorporation of the azido moiety allowed covalent attachment of biotin to the SHAPE reagent and RNA enrichment using copper-free ‘click’ chemistry (Fig. 4a). This method, termed in vivo click-selective 2′-hydroxyl acylation and profiling experiment (icSHAPE), affords the user greater signal-to-noise ratio and higher specificity over traditional chemical structure probing methods by enabling the selective purification of SHAPE-probed RNA (Fig. 4b). Comparing structure probing profiles inside and outside the cell can reveal sites of structural differences afforded by the intracellular environment (Fig. 4c). For instance, implementation of icSHAPE in vitro and in vivo recapitulated known global structural features of mammalian transcriptomes, while also reading out RNA-protein interactions and RNA methylation for the first time.
Figure 4. icSHAPE is a novel chemical probing method that permits transcriptome-wide interrogation of RNA structure.
(a) Chemical scheme for the preparation of acylated RNA, which can be purified by biotin-streptavidin purification. DIBO, dibenzocyclooctyne. (b) Schematic of icSHAPE modification and purification steps to generate a sequencing library. (c) Schematic of subtractive RNA structure probing, which can be used to study RNA-protein interactions to identify the role of RNA structure in regulating post-transcriptional interactions65.
icSHAPE has also been used to identify protein-binding sites and RNA modification sites on cellular RNAs. The former was accomplished for RBFOX and HuR, two proteins important for regulating RNA splicing and stability, respectively. In addition to protein-binding sites, icSHAPE also was able to identify sites of RNA methylation: m6A chemical modifications were identified with a >90% true positive rate65. These structural studies revealed, for the first time, that m6A modification sites are base-paired, which is contradictory to the single-stranded structure proposed66. The structural motifs probed by icSHAPE were also later corroborated using focused biochemistry and traditional RNA structure footprinting67. Overall, these studies demonstrate the power and importance of chemical enrichment strategies, such as icSHAPE, for studying RNA structure in vivo.
The progress from the initial studies probing RNA structure with DMS to the present day has been impressive. The field now has a strong toolset with which to go after the structure of nearly any RNA. Nevertheless, transcriptome-wide studies in living cells are just recently being tackled, and there are still many challenges to overcome. We discuss below some of the key remaining obstacles that need to be addressed to further expand our understanding of RNA structure inside living cells and how chemical biology is poised to help the field move forward.
Future challenges and the role of chemical biology
A first major limitation to all methods of probing RNA inside cells is that the probing pattern obtained is from an average of structures. This problem is especially important to address when considering co-transcriptional folding. The structure of RNA as it is being transcribed is likely to be different from the fully folded structure. In classic in vitro experiments, RNA is denatured and annealed, such that it is the final folded molecule that is probed. Whereas small RNAs fold on the microsecond to millisecond time scale in vitro, large RNA molecules may require minutes to hours to reach the functional state68. However, the time scales of RNA structure formation in vivo may be much faster. This is likely to be the case when taking into account the rate of transcription, which can be 10–20 nt·s−1 for human RNA polymerase II or 20–80 nt·s−1 for bacterial RNA polymerases69.
The majority of our knowledge of co-transcriptional folding has been garnered from extensively studied model systems. RNA structure formation of the Group I intron was demonstrated to be much faster in vivo, where folding is limited by the disruption of prematurely formed non-native structures, as compared with fully synthesized and denatured RNA in vitro70,71. Studies using the hairpin ribozyme in Saccharomyces cerevisiae demonstrated that the primary sequence direction and order can influence the formation of functional structures, consistent with a sequential folding model in which the outcome is determined by the structure that forms first during transcription72. Thus, determining how RNA structures are being formed co-transcriptionally could dramatically increase our understanding of how RNA structures fold and reach their final functional state.
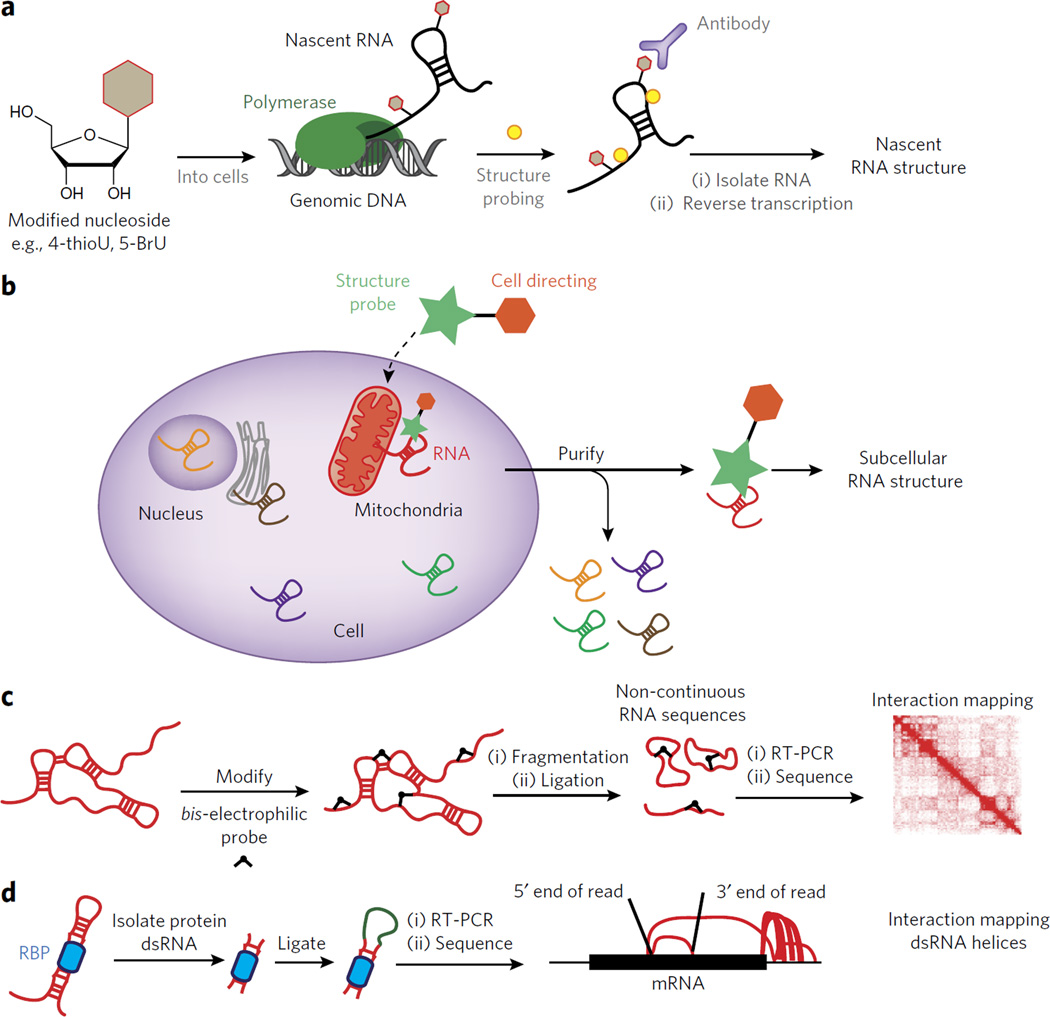
Studying co-transcriptional folding on a transcriptome scale is a grand challenge, and novel methods for isolating RNA as it is being transcribed present an opportunity for chemical biology. Nascent RNAs can be isolated using chemically modified nucleoside analogs that are added into RNA by endogenous metabolic pathways. GRO-seq utilizes a bromouridine (BrU) analog, which can be incorporated into nascent RNA and purified with an anti-BrU antibody73. Similarly, 4-thiouracil (4-thioU) pulse labeling can be used to purify nascent transcripts by enrichment. Such an approach was used to probe the structure of early ribosome assembly intermediates in living S. cerevisiae cells46. Any RNA or RNA–protein complex is potentially amenable to such an approach, as 4-thioU incorporation occurs in all RNA classes. Marrying affinity purification of nascent transcripts with DMS and SHAPE probing transcriptome-wide could prove powerful for elucidating co-transcriptional RNA structure formation inside living cells (depicted in Fig. 5a).
Figure 5. Outstanding challenges for understanding RNA structure inside living cells.
(a) Outline of an experiment for interrogating RNA structure formation during transcription. In such a case a modified nucleoside can be introduced into the cell and then used to enrich for co-transcriptionally probed RNA structure. (b) A schematic for the design of a dual-functioning chemical probe to measure RNA structure within unique subcellular compartments. (c) A depiction of how a chemical probe can be used to identify three-dimensional contacts within folded RNAs. The results are mapped to an interaction map, depicting the spatial relationship between two points in the RNA sequence. (d) A schematic for the recently developed method known as hiCLIP84. In hiCLIP non-contiguous reads are mapped to genes and represented by rainbow maps to connect primary sequence points through space.
RNA localization is another facet of RNA biology driven by RNA structure. In S. cerevisiae spatial control over protein expression during cell division is driven by mRNA localization74. Fluorescence in situ hybridization (FISH) revealed that 71% of the 3,370 genes expressed in the developing Drosophila melanogaster embryo are differentially localized75. As such, evaluating RNA structure in different cellular compartments may lead to a better understanding of how structure motifs contribute to spatial organization.
The structure of RNAs in subcellular compartments can be obtained by chemical probing before lysis, followed by cellular fractionation. However, fractionation methods are notoriously dirty, with many complexes forming and dissolving upon cellular lysis76,77. Because transcripts can exist in different parts of the cell at different times, chemical probing data ends up being a mixture of what are likely many structural states at different spatial points. An alternative would be to probe the RNA in different cellular compartments in such a way that only the RNAs within that compartment are subjected to chemical treatment (Fig. 5b).
There are many different cell-directing functional groups that could be co-opted to localize modification reagents so as to enrich subcellular reactivity. For example, the benzimidazole dyes (such as Hoechst 33258) are exclusively localized in the nucleus78. Triphenylphosphonium salts have been shown to localize to the mitochondrial matrix as a result of the negative potential formed during oxidative phosphorylation79. Triphenylphosphonium RNA structure reagents could be used to understand the mitochondrial transcriptome and the unique mitochondrial ribosome. A myristoyl lipopeptide derived from the N terminus of Src-family proteins is membrane permeant and can localize to the inside surface of the plasma membrane80. The literature provides numerous examples of self-localizing chemical moieties, which provide a strong starting point for the development of localized reagents that would permit RNA structure analysis.
RNA structure reagents have been primarily designed to measure secondary structure. However, three-dimensional folds, mediated by long-range base-pairing or other structural interactions, are important for regulating RNA function. The development of methods to restrain RNA structure prediction programs to account for long-distance or three-dimensional contacts should be a primary focus of future efforts. One such method recently developed with this goal in mind is crosslinking, ligation and sequencing of hybrids (CLASH)81. In CLASH, irradiation at 254 nm causes the formation of RNA-RNA crosslinks. Sequencing CLASH crosslinks can identify RNA-RNA crosslinks that can be separated by great lengths of primary sequence. Hydroxyl radical probing has also been merged with discrete molecular dynamics (DMD) simulations of RNA to generate three-dimensional structural ensembles consistent with experimental radical footprinting measurements35. These methods are starting to bring structural probing into three dimensions, but there is still much work to be done in this area.
Designing chemical reagents that can mark two points in space on the same RNA molecule would be ideal for developing a three-dimensional profile of the RNA. If a reagent was designed that could ligate two functional domains of the same RNA together (Fig. 5c), deep sequencing could identify them. A hint for analyzing such data comes from analogous methods that have been developed for three-dimensional modeling of the genome. For example, hi-C captures three-dimensional contacts in the genome through formaldehyde proximity crosslinking82,83. A two-dimensional heatmap could be constructed to identify sites of RNA structure interaction that are close in three-dimensional space (Fig. 5c). Developing such a method would go a long way toward allowing more accurate representations of RNA structures, and this may be feasible given the potential for small molecules to traverse the cell membrane and probe RNA structure in living cells.
All the suggested challenges in this section have been limited to interrogating RNA structure on its own. However, RNA is rarely without a protein partner in the cell, and thus understanding of the RNA-protein interface and how structure controls such interactions is critical to a holistic view of RNA structure. The description of how structure controls protein binding, on a global scale, is still severely limited. RNA hybrid and iCLIP (hiCLIP)84 permits the detection of RNA-RNA intermolecular duplexes that are bound by a protein, which are then identified using high-throughput sequencing (Fig. 5d). When hiCLIP was applied to the RNA-binding protein Staufen 1, double-stranded regions of RNA were identified through sequencing of noncontiguous insertions, which were mapped back to the primary sequence. These interactions included the hybridization of regions in the 3′-untranslated region, just downstream of the stop codon, with regions further downstream near the poly(A) site or upstream near the start codon. The invention of hiCLIP suggests that merging biochemical methods for the isolation of RNA structure segments with deep sequencing can reveal a genome-wide view of the regulatory role that RNA structure elements play in protein recognition and cell biology.
These challenges represent just a fraction of what needs to be accomplished in order to better understand RNA structure and function inside the cell. At the heart of each of these hurdles is the need to develop novel chemical tools or biochemical methods that can measure RNA structure in ways that are currently not possible. Future developments are sure to open the door to further exploration of RNA form and function.
Conclusions
Although once regarded as a passive genetic transporter, RNA has emerged as a key player in nearly every biological pathway and processes. At the heart of RNA’s ability to perform so many functions is its inherent ability to fold into complex structures that control its role inside the cell. Methods that utilize chemical modification reagents to analyze RNA structure have emerged as the main approaches for dissecting RNA structure formation, analyzing structural interactions with proteins and other trans-acting factors, and gaining a physical perspective on RNA function. However, performing such assays on cellular RNA has proven to be difficult, partly because of a lack of chemical methods that work robustly in the cellular environment. Recent progress in the design of novel reagents and protocols has begun to permit RNA structural analysis inside cells. Further, the merger of chemical and genomic technologies has opened the door to gaining a holistic understanding of how RNA structure elements regulate gene expression and cell biology. Despite such progress, there remains much work to be done. Analyzing RNA structure as a transcript is being transcribed should reveal the early stages of RNA folding and the eventual final form. Deciphering the structures of RNAs within unique cellular compartments will reveal the physical nature of genetic organization in the cell. Finally, obtaining a three-dimensional perspective on RNA structure will unravel how motifs are organized and controlled to guide transcripts throughout their lifetime. With the intersection of chemistry and biology leading the way toward novel methods of structure probing, the future looks very bright.
Acknowledgments
We thank members of the Spitale laboratory and R. Flynn for helpful discussions. We also thank professors S. Woodson and A. Laederach for critical reading of the manuscript. The University of California, Irvine and the US National Institutes of Health Director’s New Innovator Award (grant 1DP2GM119164-01) support RNA research in the Spitale lab.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Xie X, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badis G, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kruger K, et al. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 4.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 5.Noller HF, Hoffarth V, Zimniak L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science. 1992;256:1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- 6.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 7.Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McHugh CA, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stefl R, Skrisovska L, Allain FH. RNA sequence- and shape-dependent recognition by proteins in the ribonucleoprotein particle. EMBO Rep. 2005;6:33–38. doi: 10.1038/sj.embor.7400325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kedde M, et al. A Pumilio-induced RNA structure switch in p27–3′ UTR controls miR-221 and miR-222 accessibility. Nat. Cell Biol. 2010;12:1014–1020. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- 13.Chartrand P, Meng XH, Singer RH, Long RM. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr. Biol. 1999;9:333–336. doi: 10.1016/s0960-9822(99)80144-4. [DOI] [PubMed] [Google Scholar]
- 14.Goodarzi H, et al. Metastasis-suppressor transcript destabilization through TARBP2 binding of mRNA hairpins. Nature. 2014;513:256–260. doi: 10.1038/nature13466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sobczak K, de Mezer M, Michlewski G, Krol J, Krzyzosiak WJ. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 2003;31:5469–5482. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan R, Sharma S, Xia Q, Garber K, Jin P. Towards understanding RNA-mediated neurological disorders. J. Genet. Genomics. 2014;41:473–484. doi: 10.1016/j.jgg.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Houck-Loomis B, et al. An equilibrium-dependent retroviral mRNA switch regulates translational recoding. Nature. 2011;480:561–564. doi: 10.1038/nature10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang M, Eichhorn CD, Feigon J. Structural determinants for ligand capture by a class II preQ1 riboswitch. Proc. Natl. Acad. Sci. USA. 2014;111:E663–E671. doi: 10.1073/pnas.1400126111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang J, et al. The architecture of Tetrahymena telomerase holoenzyme. Nature. 2013;496:187–192. doi: 10.1038/nature12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weeks KM. Advances in RNA structure analysis by chemical probing. Curr. Opin. Struct. Biol. 2010;20:295–304. doi: 10.1016/j.sbi.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ziehler WA, Engelke DR. Probing RNA structure with chemical reagents and enzymes. Curr. Protoc. Nucleic Acid Chem. 2001;Ch. 6:6.1.1–6.1.21. doi: 10.1002/0471142700.nc0601s00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Low JT, Weeks KM. SHAPE-directed RNA secondary structure prediction. Methods. 2010;52:150–158. doi: 10.1016/j.ymeth.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathews DH, et al. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lempereur L, et al. Conformation of yeast 18S rRNA. Direct chemical probing of the 5′ domain in ribosomal subunits and in deproteinized RNA by reverse transcriptase mapping of dimethyl sulfate-accessible. Nucleic Acids Res. 1985;13:8339–8357. doi: 10.1093/nar/13.23.8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mortimer SA, Johnson JS, Weeks KM. Quantitative analysis of RNA solvent accessibility by N-silylation of guanosine. Biochemistry. 2009;48:2109–2114. doi: 10.1021/bi801939g. [DOI] [PubMed] [Google Scholar]
- 26.Burgstaller P, Kochoyan M, Famulok M. Structural probing and damage selection of citrulline- and arginine-specific RNA aptamers identify base positions required for binding. Nucleic Acids Res. 1995;23:4769–4776. doi: 10.1093/nar/23.23.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilkinson KA, Merino EJ, Weeks KM. RNA SHAPE chemistry reveals nonhierarchical interactions dominate equilibrium structural transitions in tRNAAsp transcripts. J. Am. Chem. Soc. 2005;127:4659–4667. doi: 10.1021/ja0436749. [DOI] [PubMed] [Google Scholar]
- 28.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 29.Watts JM, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGinnis JL, Duncan CD, Weeks KM. High-throughput SHAPE and hydroxyl radical analysis of RNA structure and ribonucleoprotein assembly. Methods Enzymol. 2009;468:67–89. doi: 10.1016/S0076-6879(09)68004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan Y, Qu K, Ouyang Z, Chang HY. Genome-wide mapping of RNA structure using nuclease digestion and high-throughput sequencing. Nat. Protoc. 2013;8:849–869. doi: 10.1038/nprot.2013.045. [DOI] [PubMed] [Google Scholar]
- 32.Peng Y, Soper TJ, Ouyang Z, Woodson SA. RNase footprinting of protein binding sites on an mRNA target of small RNAs. Methods Mol. Biol. 2012;905:213–224. doi: 10.1007/978-1-61779-949-5_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ingle S, Azad RN, Jain SS, Tullius TD. Chemical probing of RNA with the hydroxyl radical at single-atom resolution. Nucleic Acids Res. 2014;42:12758–12767. doi: 10.1093/nar/gku934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costa M, Monachello D. Probing RNA folding by hydroxyl radical footprinting. Methods Mol. Biol. 2014;1086:119–142. doi: 10.1007/978-1-62703-667-2_7. [DOI] [PubMed] [Google Scholar]
- 35.Ding F, Lavender CA, Weeks KM, Dokholyan NV. Three-dimensional RNA structure refinement by hydroxyl radical probing. Nat. Methods. 2012;9:603–608. doi: 10.1038/nmeth.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods Mol. Biol. 2008;419:53–67. doi: 10.1007/978-1-59745-033-1_4. [DOI] [PubMed] [Google Scholar]
- 37.Heilman-Miller SL, Woodson SA. Effect of transcription on folding of the Tetrahymena ribozyme. RNA. 2003;9:722–733. doi: 10.1261/rna.5200903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perdrizet GA, II, Artsimovitch I, Furman R, Sosnick TR, Pan T. Transcriptional pausing coordinates folding of the aptamer domain and the expression platform of a riboswitch. Proc. Natl. Acad. Sci. USA. 2012;109:3323–3328. doi: 10.1073/pnas.1113086109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahen EM, Watson PY, Cottrell JW, Fedor MJ. mRNA secondary structures fold sequentially but exchange rapidly in vivo. PLoS Biol. 2010;8:e1000307. doi: 10.1371/journal.pbio.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan T, Artsimovitch I, Fang XW, Landick R, Sosnick TR. Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc. Natl. Acad. Sci. USA. 1999;96:9545–9550. doi: 10.1073/pnas.96.17.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tyrrell J, McGinnis JL, Weeks KM, Pielak GJ. The cellular environment stabilizes adenine riboswitch RNA structure. Biochemistry. 2013;52:8777–8785. doi: 10.1021/bi401207q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frieda KL, Block SM. Direct observation of cotranscriptional folding in an adenine riboswitch. Science. 2012;338:397–400. doi: 10.1126/science.1225722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tijerina P, Mohr S, Russell R. DMS footprinting of structured RNAs and RNA-protein complexes. Nat. Protoc. 2007;2:2608–2623. doi: 10.1038/nprot.2007.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waldsich C, Masquida B, Westhof E, Schroeder R. Monitoring intermediate folding states of the td group I intron in vivo. EMBO J. 2002;21:5281–5291. doi: 10.1093/emboj/cdf504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waldsich C, Grossberger R, Schroeder R. RNA chaperone StpA loosens interactions of the tertiary structure in the td group I intron in vivo. Genes Dev. 2002;16:2300–2312. doi: 10.1101/gad.231302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swiatkowska A, et al. Kinetic analysis of pre-ribosome structure in vivo. RNA. 2012;18:2187–2200. doi: 10.1261/rna.034751.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McGinnis JL, Weeks KM. Ribosome RNA assembly intermediates visualized in living cells. Biochemistry. 2014;53:3237–3247. doi: 10.1021/bi500198b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGinnis JL, et al. In-cell SHAPE reveals that free 30S ribosome subunits are in the inactive state. Proc. Natl. Acad. Sci. USA. 2015;112:2425–2430. doi: 10.1073/pnas.1411514112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spitale RC, et al. RNA SHAPE analysis in living cells. Nat. Chem. Biol. 2013;9:18–20. doi: 10.1038/nchembio.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwok CK, Ding Y, Tang Y, Assmann SM, Bevilacqua PC. Determination of in vivo RNA structure in low-abundance transcripts. Nat. Commun. 2013;4:2971. doi: 10.1038/ncomms3971. [DOI] [PubMed] [Google Scholar]
- 51.Hector RD, et al. Snapshots of pre-rRNA structural flexibility reveal eukaryotic 40S assembly dynamics at nucleotide resolution. Nucleic Acids Res. 2014;42:12138–12154. doi: 10.1093/nar/gku815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Latham JA, Cech TR. Defining the inside and outside of a catalytic RNA molecule. Science. 1989;245:276–282. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- 53.Sclavi B, Woodson S, Sullivan M, Chance MR, Brenowitz M. Time-resolved synchrotron X-ray “footprinting”, a new approach to the study of nucleic acid structure and function: application to protein-DNA interactions and RNA folding. J. Mol. Biol. 1997;266:144–159. doi: 10.1006/jmbi.1996.0775. [DOI] [PubMed] [Google Scholar]
- 54.Sclavi B, Sullivan M, Chance MR, Brenowitz M, Woodson SA. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- 55.Adilakshmi T, Soper SF, Woodson SA. Structural analysis of RNA in living cells by in vivo synchrotron X-ray footprinting. Methods Enzymol. 2009;468:239–258. doi: 10.1016/S0076-6879(09)68012-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim H, et al. Protein-guided RNA dynamics during early ribosome assembly. Nature. 2014;506:334–338. doi: 10.1038/nature13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clatterbuck Soper SF, Dator RP, Limbach PA, Woodson SA. In vivo X-ray footprinting of pre-30S ribosomes reveals chaperone-dependent remodeling of late assembly intermediates. Mol. Cell. 2013;52:506–516. doi: 10.1016/j.molcel.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blower MD. Molecular insights into intracellular RNA localization. Int. Rev. Cell. Mol. Biol. 2013;302:1–39. doi: 10.1016/B978-0-12-407699-0.00001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mortimer SA, Kidwell MA, Doudna JA. Insights into RNA structure and function from genome-wide studies. Nat. Rev. Genet. 2014;15:469–479. doi: 10.1038/nrg3681. [DOI] [PubMed] [Google Scholar]
- 61.Kwok CK, Tang Y, Assmann SM, Bevilacqua PC. The RNA structurome: transcriptome-wide structure probing with next-generation sequencing. Trends Biochem. Sci. 2015;40:221–232. doi: 10.1016/j.tibs.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 62.Lucks JB, et al. Multiplexed RNA structure characterization with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Proc. Natl. Acad. Sci. USA. 2011;108:11063–11068. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ding Y, et al. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 64.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spitale RC, et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519:486–490. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwartz S, et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell. 2013;155:1409–1421. doi: 10.1016/j.cell.2013.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu N, et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zarrinkar PP, Wang J, Williamson JR. Slow folding kinetics of RNase P RNA. RNA. 1996;2:564–573. [PMC free article] [PubMed] [Google Scholar]
- 69.Zemora G, Waldsich C. RNA folding in living cells. RNA Biol. 2010;7:634–641. doi: 10.4161/rna.7.6.13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wong TN, Pan T. RNA folding during transcription: protocols and studies. Methods Enzymol. 2009;468:167–193. doi: 10.1016/S0076-6879(09)68009-5. [DOI] [PubMed] [Google Scholar]
- 71.Pan T, Sosnick T. RNA folding during transcription. Annu. Rev. Biophys. Biomol. Struct. 2006;35:161–175. doi: 10.1146/annurev.biophys.35.040405.102053. [DOI] [PubMed] [Google Scholar]
- 72.Mahen EM, Harger JW, Calderon EM, Fedor MJ. Kinetics and thermodynamics make different contributions to RNA folding in vitro and in yeast. Mol. Cell. 2005;19:27–37. doi: 10.1016/j.molcel.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 73.Danko CG, et al. Identification of active transcriptional regulatory elements from GRO-seq data. Nat. Methods. 2015;12:433–438. doi: 10.1038/nmeth.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beach DL, Bloom K. ASH1 mRNA localization in three acts. Mol. Biol. Cell. 2001;12:2567–2577. doi: 10.1091/mbc.12.9.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lécuyer E, et al. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 76.Riley KJ, Yario TA, Steitz JA. Association of Argonaute proteins and microRNAs can occur after cell lysis. RNA. 2012;18:1581–1585. doi: 10.1261/rna.034934.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mayerle M, Woodson SA. Specific contacts between protein S4 and ribosomal RNA are required at multiple stages of ribosome assembly. RNA. 2013;19:574–585. doi: 10.1261/rna.037028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Panja S, Schu DJ, Woodson SA. Conserved arginines on the rim of Hfq catalyze base pair formation and exchange. Nucleic Acids Res. 2013;41:7536–7546. doi: 10.1093/nar/gkt521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lucas CH, Calvez M, Babu R, Brown A. Altered subcellular localization of the NeuN/Rbfox3 RNA splicing factor in HIV-associated neurocognitive disorders (HAND) Neurosci. Lett. 2014;558:97–102. doi: 10.1016/j.neulet.2013.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kudla G, Granneman S, Hahn D, Beggs JD, Tollervey D. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc. Natl. Acad. Sci. USA. 2011;108:10010–10015. doi: 10.1073/pnas.1017386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Belton JM, et al. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods. 2012;58:268–276. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yaffe E, Tanay A. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat. Genet. 2011;43:1059–1065. doi: 10.1038/ng.947. [DOI] [PubMed] [Google Scholar]
- 84.Sugimoto Y, et al. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature. 2015;519:491–494. doi: 10.1038/nature14280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mathews DH. RNA secondary structure analysis using RNAstructure. Curr. Protoc. Bioinformatics. 2014;46:12.6.1–12.6.25. doi: 10.1002/0471250953.bi1206s46. [DOI] [PubMed] [Google Scholar]
- 86.Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol. Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- 87.Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Washietl S, et al. Computational analysis of noncoding RNAs. Wiley Interdiscip Rev. RNA. 2012;3:759–778. doi: 10.1002/wrna.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Andronescu M, Condon A, Hoos HH, Mathews DH, Murphy KP. Computational approaches for RNA energy parameter estimation. RNA. 2010;16:2304–2318. doi: 10.1261/rna.1950510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ouyang Z, Snyder MP, Chang HY. SeqFold: genome-scale reconstruction of RNA secondary structure integrating high-throughput sequencing data. Genome Res. 2013;23:377–387. doi: 10.1101/gr.138545.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tang Y, et al. StructureFold: genome-wide RNA secondary structure mapping and reconstruction in vivo. Bioinformatics. 2015;31:2668–2675. doi: 10.1093/bioinformatics/btv213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Corley M, Solem A, Qu K, Chang HY, Laederach A. Detecting riboSNitches with RNA folding algorithms: a genome-wide benchmark. Nucleic Acids Res. 2015;43:1859–1868. doi: 10.1093/nar/gkv010. [DOI] [PMC free article] [PubMed] [Google Scholar]