Abstract
Large-scale transcription start site (TSS) profiling produces a high-resolution, quantitative picture of transcription initiation and core promoter locations within a genome. However, application of TSS profiling to date has largely been restricted to a small set of prominent model systems. We sought to characterize the cis-regulatory landscape of the water flea Daphnia pulex, an emerging model arthropod that reproduces both asexually (via parthenogenesis) and sexually (via meiosis). We performed Cap Analysis of Gene Expression (CAGE) with RNA isolated from D. pulex within three developmental states: sexual females, asexual females, and males. Identified TSSs were utilized to generate a “Daphnia Promoter Atlas,” i.e., a catalog of active promoters across the surveyed states. Analysis of the distribution of promoters revealed evidence for widespread alternative promoter usage in D. pulex, in addition to a prominent fraction of compactly-arranged promoters in divergent orientations. We carried out de novo motif discovery using CAGE-defined TSSs and identified eight candidate core promoter motifs; this collection includes canonical promoter elements (e.g., TATA and Initiator) in addition to others lacking obvious orthologs. A comparison of promoter activities found evidence for considerable state-specific differential gene expression between states. Our work represents the first global definition of transcription initiation and promoter architecture in crustaceans. The Daphnia Promoter Atlas presented here provides a valuable resource for comparative study of cis-regulatory regions in metazoans, as well as for investigations into the circuitries that underpin meiosis and parthenogenesis.
Keywords: CAGE, cis-regulatory regions, Daphnia, meiosis, parthenogenesis, promoter architecture, transcription initiation
ALL biological processes, including development, differentiation, and maintenance of homeostasis, rely upon precise, coordinate regulation of gene expression. A key early step in gene expression is transcription initiation at the core promoter, a short genomic region containing the transcription start site (TSS) (Kadonaga 2012). During initiation, sequences within core promoters recruit general transcription factors (GTFs), which is followed by binding of RNA polymerase II (RNAPII) and formation of the preinitiation complex (PIC) (Cosma 2002). Identifying the locations and composition of promoters is fundamental for understanding the basis for gene expression regulation. Recent work (Frith et al. 2008; Hoskins et al. 2011) demonstrates that core promoters are more structurally diverse than previously appreciated. This diversity is thought to reflect large numbers of developmental programs and regulatory strategies (Lenhard et al. 2012), but the precise rules and mechanisms underlying promoter function remain unclear.
Genome-scale TSS profiling has identified promoters in a number of metazoans [Lenhard et al. 2012; FANTOM Consortium and the RIKEN PMI and CLST (DGT) 2014]. Cap Analysis of Gene Expression (CAGE) (Kodzius et al. 2006; Kurosawa et al. 2011), the most prominent TSS profiling method, identifies core promoter positions at high resolution. This approach revealed that most genes do not possess a single TSS, but instead exhibit sets of closely spaced TSSs that will be referred to as Transcription start regions (TSRs) in the following. While the largest number of TSS profiling studies have been performed in mammalian (i.e., human and mouse) systems [Djebali et al. 2008; FANTOM Consortium and the RIKEN PMI and CLST (DGT) 2014], CAGE has also been performed in nonmammalian metazoans, including fruit fly (Hoskins et al. 2011), nematode (Nepal et al. 2013), and zebrafish (Haberle et al. 2014). Overall, these studies indicate that the majority of core promoters in metazoan genomes lack TATA elements (Lenhard et al. 2012), an unanticipated finding given previously established models of transcription initiation. At least two major promoter classes are evident. In human and mouse, the largest class is known as CpG island promoters (CPI) (Saxonov et al. 2006; Lenhard et al. 2012). These promoters are located near CpG islands and are generally of high GC-content and depleted for TATA elements. Sequences in the other major promoter class, called “low-CpG,” exhibit low GC-content and are enriched for TATA boxes. This latter class of promoter is consistent with conventional models of promoter structure, such as TATA-dependent transcription initiation (Kadonaga 2012).
Characterization of CAGE-defined promoters in a wider taxonomic context uncovered two distinct patterns of TSS distributions within a given promoter (Carninci et al. 2006; Hoskins et al. 2011; Lenhard et al. 2012). “Peaked” promoters exhibit CAGE signal from a narrow genomic region surrounding a single prominent TSS, whereas “broad” promoters instead feature multiple TSSs distributed across a wide (30 bp and longer) genomic region (Kadonaga 2012; Lenhard et al. 2012). These TSS distribution patterns appear to coincide with the aforementioned (mammalian) classes of promoter architecture: peaked promoters are highly associated with the low-CpG promoter class, whereas broad TSS distributions tend to be found at high-CpG promoters. Peaked and broad promoters also regulate separate functional gene classes: genes with peaked promoters tend to be developmentally regulated or tissue-specific, while genes with broad promoters tend to be housekeeping genes exhibiting constitutive expression (Lenhard et al. 2012). Recent work using CAGE from a variety of mammalian cell types unexpectedly detected widespread enrichment of TSSs at enhancers (Andersson et al. 2014). The new class of RNA defined by this work, enhancer RNAs (eRNAs), are short, transient, RNAPII-derived transcripts generated at active enhancer regions (Kim et al. 2010). While enhancers appeared to be distinguishable from promoters on the basis of transcript stability and bidirectionality (Andersson et al. 2014), subsequent reports suggest that enhancers and promoters possess common properties, including motif composition and activity (Arner et al. 2015).
Despite recent progress, considerable gaps remain in the understanding of promoter architecture across metazoan diversity. To date, high-resolution TSS profiling has been reported in just two arthropod species, both closely-related drosophilids: Drosophila melanogaster (Hoskins et al. 2011) and D. pseudoobscura (Chen et al. 2014). Promoter profiling in a broader set of taxa is necessary to establish robust comparative genomic analyses of cis-regulatory regions in metazoa. To address this need, we performed TSS profiling using CAGE in the water flea Daphnia pulex. A freshwater microcrustacean with a cosmopolitan distribution, D. pulex is notable for its ability to reproduce both sexually and asexually, high levels of heterozygosity, and relatively large effective population sizes (Ne) compared to other broadly dispersed arthropods (Haag et al. 2009; Tucker et al. 2013). D. pulex serves as a key model system throughout the biological sciences, from ecosystem ecology to molecular genetics. By mapping TSSs for D. pulex from active promoters within three developmental states—sexual females, asexual females, and adult (sexual) males—we sought to characterize the architecture of core promoters in D. pulex and also explore meiosis- and sex-specific gene regulatory programs. We successfully identified TSSs at high resolution across the entire genome, defining promoters for all genes expressed under the experimental conditions. We then performed computational de novo motif discovery using this set of mapped TSSs, obtaining consensus sequences of canonical core promoter elements, including TATA and Initiator (Inr). The quantitative tag counts from the CAGE datasets allowed us to identify differentially-expressed genes within each of the three states surveyed, including those regulated in a sex-specific manner. The resultant D. pulex promoter atlas extends our knowledge of metazoan cis-regulation into Crustacea, a taxonomic expansion that will also serve as a public resource for functional and comparative genomics.
Materials and Methods
Focal genotype and maintenance of individuals
The D. pulex genotype used in this work was isolated from Portland Arch Pond (Warren County, IN; geographic coordinates: 40.2096°, −87.3294°) and is identified as PA13-42 (hereafter PA42). The PA42 clone originates from a well-characterized natural population (Lynch et al. 1989). D. pulex individuals from the PA42 clone are cyclical parthenogens, meaning that they are capable of reproducing both asexually through eggs that develop directly or sexually through diapausing eggs. All individuals used in this study were the result of asexual reproduction. Females were maintained in 3 liter containers containing COMBO media (Kilham et al. 1998) (diluted 1:1 with water) at 20° and fed Scenedesmus at ∼100,000 cells/ml. New offspring were removed and placed in new containers daily. Asexual females, preephipial (sexual) females, and males were isolated from culture on separate occasions using strainers of differential sizes and visual identification under a dissecting microscope. Males can be visually distinguished from females based on the criteria of enlarged atennules and flattened ventral carapace margin. The current reproductive mode of females can be determined by phenotyptic differences in yolk-filled ovaries: females currently reproducing asexually have more “bulbous” ovaries that tend to be more green in color, while females currently reproducing sexually have blackish yolks of reduced size and a smoother external margin.
RNA isolation and quantification
Whole D. pulex individuals (∼50–75) were collected from fresh cultures from each of the three aforementioned states. Collections were homogenized manually using a small pestle in microcentrifuge tubes containing lysis buffer. Isolation of total RNA was performed using solid phase extraction (Bioline). Samples were snap-frozen in liquid nitrogen and stored at −80°. RNA samples were quantified and evaluated for quality and using the Bioanalyzer 2100 (Agilent Technologies).
CAGE library preparation and sequencing
A multiplexed CAGE library was constructed as described (Takahashi et al. 2012a) from 5 μg total RNA sample using the nAnT-iCAGE protocol (Murata et al. 2014) (K. K. DNAForm, Yokohama, Japan). Briefly, total RNA was reverse transcribed using a random “N6 plus base 3” primer (TCTNNNNNN), using Superscript III reverse transcriptase (Thermo Fisher). Following oxidation (with sodium peroxide) and biotinylation of the m7G cap structures, first-strand-complete messenger RNA: complementary DNA (mRNA:cDNA) hybrids were bound with streptavidin beads, pulled down with a magnet, and released. This was followed by ligation of the 5′ linker, which includes the three nucleotide (nt) barcode (e.g., iCAGE_01 N6 5′-CGACGCTCTTCCGATCTACCNNNNNN-3′) followed by 3′ linker ligation. Finally, second-strand synthesis was performed using the nAnT-iCAGE second primer (5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTT-3′), creating the final double-stranded DNA product. For a more detailed protocol, please see the following (Murata et al. 2014).
Quantitative RT-PCR evaluation of CAGE libraries
Prior to sequencing, relative mRNA:rRNA (ribosomal RNA) ratios were measured for each CAGE library using quantitative RT-PCR (qRT-PCR) with SYBR Green I (Life Technologies). The control gene GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was selected, (Forward Primer: 5′-ACCACTGTCCATGCCATCACT-3′, Reverse Primer: 5′-CACGCCACAACTTTCCAGAA-3′) and was measured against 18S ribosomal mRNA (Forward Primer: 5′-CCGGCGACGTATCTTTCAA-3′, Reverse Primer: 5′-CACGCCACAACTTTCCAGAA-3′). Biological replicates of each of the three states were reflected in the final CAGE library (n = 3 for both female groups, n = 2 for males). Finally, the completed CAGE library was sequenced using Illumina HiSeq 2000 (single-end, 50-bp reads) at the University of California, Berkeley Genome Sequencing Laboratory (Berkeley, CA).
CAGE processing, alignment, and rRNA filtering
All CAGE-adapted sequence reads (1.82 × 108) were demultiplexed (http://hannonlab.cshl.edu/fastx_toolkit/index.html), creating eight separate fastq files corresponding to the original CAGE libraries. All CAGE-adapted sequences (47 bp) from each library were aligned separately using bwa (Li and Durbin 2009) to the D. pulex assembly v1.1 [Joint Genome Institute (JGI)] (Colbourne et al. 2011). Prior to downstream analysis, CAGE alignments (in BAM format) were subjected to a filtering step (rRNAdust; http://fantom.gsc.riken.jp/5/sstar/Protocols:rRNAdust) to remove rRNA sequences (28S, 18S, and 5S). The SAM flags of identified rRNA reads in the alignment were changed to “unmapped.” Overall, 1.22 × 108 CAGE reads (67.0% of the total) mapped successfully (Table 1), and these were utilized in subsequent analyses. Evaluations of CAGE alignments and pooling of multiple libraries was performed using Samtools (Li et al. 2009). The distribution of CAGE tags within the D. pulex genome was determined using BEDtools (Quinlan 2014). Nonoverlapping genomic intervals were created using BEDtools from the JGI’s Frozen Gene Catalog annotation (“FrozenGeneCatalog20110204.gff3”) located at http://genome.jgi.doe.gov/Dappu1/Dappu1.download.html.
Table 1. Summary of CAGE libraries in this study.
| Number | Library name | Number of sequenced CAGE tags | Number of mapped CAGE reads |
|---|---|---|---|
| 1 | Asexual females-1 | 28,803,508 | 18,601,744 |
| 2 | Asexual females-2 | 16,701,216 | 10,839,287 |
| 3 | Asexual females-3 | 29,786,273 | 20,754,759 |
| 4 | Sexual females-1 | 24,076,420 | 15,861,581 |
| 5 | Sexual females-2 | 24,567,545 | 15,163,393 |
| 6 | Sexual females-3 | 15,115,501 | 9,621,093 |
| 7 | Males-1 | 18,512,317 | 12,412,516 |
| 8 | Males-2 | 24,655,373 | 16,960,704 |
| Total | – | 182,218,153 | 120,215,077 |
The value at the end of each library name refers to the biological replicate number. CAGE, Cap Analysis of Gene Expression.
Analysis of mapped CAGE tags
TSRs were defined from mapped CAGE tags using the CAGEr package (Haberle et al. 2015) in R Bioconductor (Huber et al. 2015). Aligned reads from each library were normalized by fitting to a power law distribution as described [FANTOM Consortium and the RIKEN PMI and CLST (DGT) 2014]. The 5′ coordinate (CAGE adapter-adjacent) of each aligned read was designated as a CTSS (CAGE-detected TSS), and the CAGE tag abundance at each genomic position was quantified in tags per million (tpm). CTSSs with CAGE tag support above 2 tpm (significant CTSSs; sCTSSs) were clustered into TSRs using the distclu algorithm in CAGEr, which merges sCTSSs below a maximum distance of 20 bp apart. Correlation of sCTSS abundance across biological replicates showed extremely high within-sample concordance (R2 > 0.97). TSR width was defined as the length of the genomic segment occupied by sCTSSs within a TSR. Where specified, we calculated TSR width using the interquantile range using the “quantilePositions” function in CAGEr (Haberle et al. 2015). We selected the interquantile range between the 10th and 90th percentile of all CAGE signals within a TSR, where the nth percentile refers to the genomic position where n% of the CAGE signal is 5′ of the entirety of the CAGE signal within a TSR (Haberle et al. 2015).
Promoter definitions:
TSRs were reported for a given condition if the evidence from all replicates were in agreement (n = 3 for sexual females and asexual females, n = 2 for males). Consensus promoters are the genomic coordinates of promoters found in all CAGE datasets and were calculated using interquantile widths (10th–90th) using CAGEr (Haberle et al. 2015). CAGE definitions are illustrated in Figure 1.
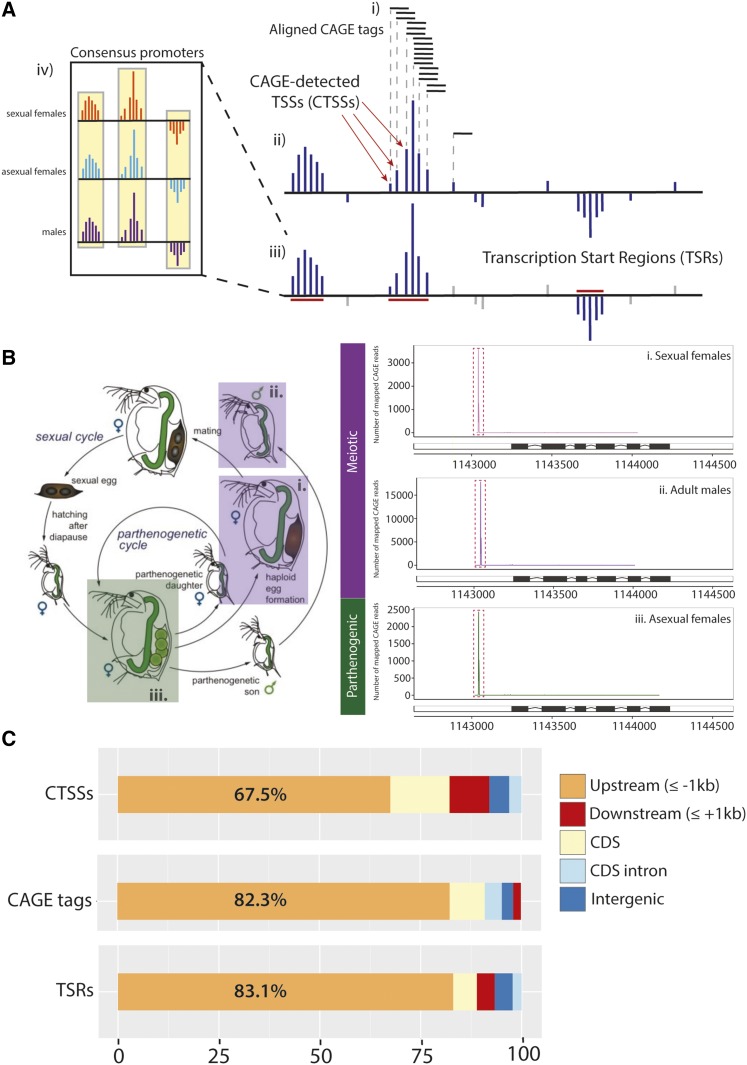
Figure 1.
TSS profiling in D. pulex using CAGE. (A) Schematic of CAGE annotations. (i) Individual sequenced CAGE tags (represented by short, horizontal black lines) are aligned to the genome in a strand-specific manner, (ii) defining distinct CTSSs (represented by dark blue vertical lines). (iii) CTSSs with CAGE tag support above 2 tpm are spatially clustered into TSRs (indicated with red lines). CTSSs (gray vertical lines) below the 2 tpm threshold are ignored during this clustering step and are not included in the eventual TSRs. (iv) TSRs with evidence across three states are classified as consensus promoters. (B) A summary of the developmental stages surveyed in this study. We sequenced CAGE-adapted cDNA libraries originating in (i) sexual females, (ii) males, and (iii) asexual females. The life cycle of D. pulex is summarized (left panel), showing the parthenogenic (ameiotic) and sexual (meiotic) cycles. A representative visualization of CAGE tag densities for a single promoter region across the three states is presented at right. The illustration of the Daphnia life cycle in the left panel is adapted from an illustration by D. B. Vizoso (Freiburg University) in (Ebert 2005), and is used with permission. (C) Proportions of CAGE annotations by genomic location. Locations of all aligned CAGE tags, CTSSs, and TSRs by genome segment are shown, including 1 kb upstream of the CDS (orange), 1 kb downstream of CDS (red), within the CDS (light yellow), CDS introns (light blue), and within intergenic (i.e., exclusive to the other categories) regions (dark blue). CAGE, Cap Analysis of Gene Expression; cDNA, complementary DNA; CDS, coding sequence; CTSS, CAGE-detected TSS; tpm, tags per million; TSR, transcription start region; TSS, transcription start site.
Classification of promoter shape:
We measured promoter shape by calculating the diversity of CTSSs within a given TSR or consensus promoter. To do this, we applied the Shape Index (SI) as described (Hoskins et al. 2011), which is itself based on the Shannon entropy (Shannon 1948). The SI is calculated as follows using TSSs within a given promoter:
| (1) |
where p is the probability of CTSS position i being observed among all L CTSS positions within the TSR (or consensus promoter). TSRs that contain a single unique CTSS position will have a SI equal to 2, while the SI value becomes more negative as the number of distinct CTSSs within the TSR increases.
TSRs and consensus promoters were labeled as either broad (SI < −2), peaked (SI > 1.5), or unclassified (all others) according to their associated SI values.
Test for bimodality of TSR shapes:
We tested the calculated shape value (in units of SI, as described above) of all consensus promoters for bimodality. The distribution of shape values was evaluated using the Expectation-Maximization (EM) algorithm implemented in the Mixtools package (Benaglia et al. 2009) in R. The results support a two-component mixture within the distribution. Fitted Gaussian densities of the two components (shaded in coral and blue, respectively) were plotted against the overall distribution of calculated consensus promoter shapes (Figure 2A, inset).
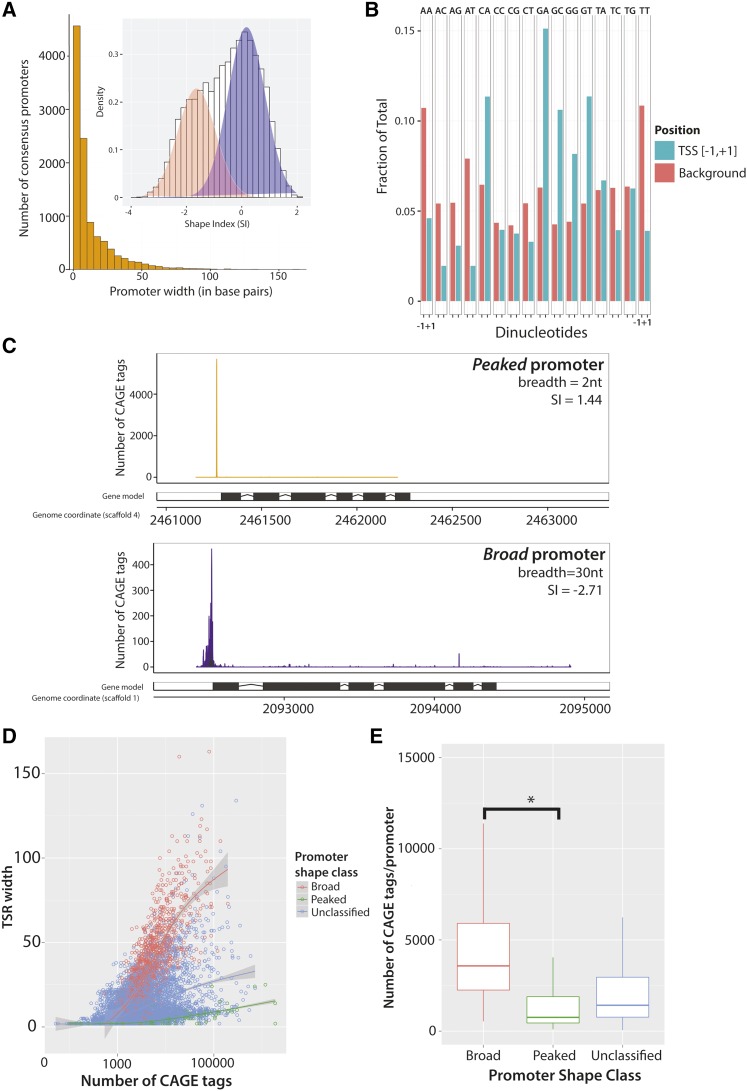
Figure 2.
Distributions of consensus promoter width and shape in the D. pulex promoter atlas. (A) A histogram representing the distribution of calculated consensus promoter (n = 10,580) widths is shown in orange (outer figure). Each bin width represents 5 bp. Inset: Consensus promoter shapes have a bimodal distribution. A histogram representing the shapes (measured with the SI) of all consensus promoters (n = 10,580) is shown in white, with each bin indicating 0.1 of a SI. The densities of broad (coral) and peaked (purple) consensus promoter shapes were fitted from the overall distribution of SI values (see Materials and Methods). (B) Distinct dinucleotide preferences at transcription initiation sites in D. pulex. The dinucleotide frequencies at CTSSs ([−1,+1]; aqua) are compared to background (coral). CTSSs show a twofold or greater preference for the dinucleotides CA, GA, GC, and GT, and are similarly depleted for AA, AT, and TT. (C) Representative examples of canonical CAGE tag distribution patterns observed in D. pulex consensus promoters. Peaked consensus promoters (above) exhibit narrow CAGE tag distributions, whereas broad consensus promoters (below) are typified by more a dispersed distribution of CAGE tags. (D) Consensus promoter activity correlates with shape more strongly than width. Consensus promoter activity (measured according to total number of CAGE tags) is plotted against TSR width in base pairs bp. Peaked (SI > 1), broad (SI < −1.5) and unclassified (all other) consensus promoters are identified by green, red, and blue circles, respectively. (E) Broad consensus promoters have greater activity than peaked consensus promoters. A significantly greater number of CAGE tags are observed in broad consensus promoters relative to peaked consensus promoters (* P < 0.0005; Tukey’s HSD). Box-and-whisker plots representing the distributions of the consensus promoter activity in three shape classes (broad, red; peaked, green; and unclassified, blue) are shown. CAGE, Cap Analysis of Gene Expression; CTSS, CAGE-detected TSS; HSD, honest significant difference; SI, Shape Index; TSR, transcription start region.
Dinucleotide preference at initiation sites
Dinucleotide frequencies were calculated using bedtools nuc (Quinlan 2014) from 2 bp intervals (position: [−1,+1]) created from (i) pooled CTSSs and (ii) randomly sampled background intervals derived from the D. pulex genome. A statistical test of the observed dinucleotide preferences was performed by repeating this procedure iteratively for all consecutive dinucleotides within the [−1,−100] window (the control) relative to +1, and evaluating the resulting dinucleotide frequencies observed for each. Dinucleotide frequencies within the window [−1,+1] relative to CTSSs were considered significant if they fell in the top or bottom five (0.05) of all control observations. We did not test dinucleotide frequencies downstream of +1 in our test to avoid the potential confounding effects of codon bias.
Promoter orientation
The relationship between adjacent CAGE-identified promoters was obtained using BEDtools (Quinlan 2014). We identified for the nearest adjacent, nonoverlapping consensus promoter interval on either strand (bedtools closest -id -io -D ref), as well as the distance between them in base pairs. To retrieve the data relating to promoters of different orientations, we subdivided the dataset to retrieve promoters in tandem, divergent, and convergent orientations, as illustrated in Figure 3A. Overlap of divergent promoters with annotated coding genes was measured using the GenomicRanges package in R Bioconductor (Huber et al. 2015). The distribution of distances between promoters was plotted using ggplot in R. Genome browser-like plots of individual alignments were created using the ggbio package (Yin et al. 2012) in R. A slightly expanded (n=11,399) collection of consensus promoters was used for the promoter orientation comparisons. Due to the large number of short scaffolds in the D. pulex TCO assembly, not all consensus promoters exist in a promoter pair.
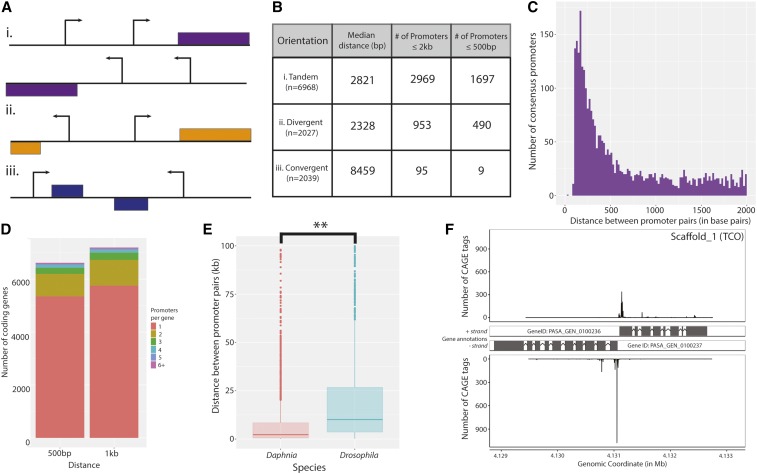
Figure 3.
Compact promoter orientations in D. pulex. (A) Illustration of possible configurations of promoter pairs. Promoters can be found in (i) tandem, (ii) divergent, and (iii) convergent orientations. (B) Distance measures between promoter pairs for each of the orientations in (A) are shown. The median distance (in bp) of promoter pairs in (i) tandem, (ii) divergent, and (iii) convergent orientations are listed, in addition to the number of consensus promoter pairs within 2 kb and 500 bp. (C) A histogram of the distribution of distances between tandem promoter pairs is shown, which reveals a large number of closely-spaced consensus promoters. Only those promoter pairs within 2 kb are shown. (D) Putative alternative promoter usage in D. pulex. Two barplots representing annotated coding genes by number of associated consensus promoters (left, within 500 bp; right, within 1 kb of the annotated translation start site) are shown. (E) Divergent promoters in D. pulex are more closely-spaced than those in D. melanogaster. Boxplots representing the overall distances between convergent promoters in D. pulex (coral) and D. melanogaster (cyan) are shown. (** P < 0.001 Welch two sample t-test) (F) A representative example of closely-spaced divergent promoter pairs in D. pulex. Aligned CAGE reads from the selected coordinates (4,129,000–4,133,122 on scaffold 1) are shown, with the CAGE tag abundance (in number of CAGE tags) presented on the y-axis. CAGE, Cap Analysis of Gene Expression.
Consensus promoter annotations for D. melanogaster were generated using the CAGE sequence data from Hoskins et al. (2011). Sequence reads from mixed larval samples were retrieved from the NCBI Sequence Read Archive (SRA) (Accession: SRR035178) and aligned to the Dm3 assembly (http://hgdownload.cse.ucsc.edu/downloads.html#fruitfly) as described previously. TSS mapping, abundance, and promoter identification were completed using the same analysis pipeline described for D. pulex.
De novo motif discovery
Daphnia core promoter motifs were discovered using hypergeometric enrichment in Homer (Heinz et al. 2010). This procedure was performed as follows: first, CAGE peaks (using the peak-finding algorithm of Homer) from pooled (i.e., in all three states) alignments were detected using annotatePeaks.pl to create a peak interval file. Next, we retrieved motifs that were enriched within 100 bp sequences ([−50,+50]) surrounding the CAGE peaks relative to background (findMotifsGenome.pl). We searched for motifs of 6, 8, 10, and 12 bp, reflecting the typical size range of cis-regulatory motifs.
Statistical validation of predicted de novo motifs:
Promoter motifs were determined using a multi-step workflow, as follows. First, we performed 10-fold cross-validation. The CAGE peak position file was divided into 10 folds (subsamples) of equal size. For each round of validation, one of the folds was labeled as the test set, and the other nine were identified as the training set. This process was iterated 10 times, such that each fold served as the test set exactly once. De novo motif prediction was performed on each of the 10 training sets using Homer, as described above. We evaluated motifs within all 10 training sets by measuring the consistency with which a motif is found within a training set. For example, if a given motif is found only in a handful of the 10 training sets, it is unlikely to be a bona fide core promoter motif. We retrieved all motifs predicted by Homer (homerMotifs.all.motifs) in each training set, and selected the top 25 according to the log P-value of enrichment, provided the P-value of each was below a cutoff of 1e−10. Predicted motifs from each of the 10 training sets were grouped and clustered according to their pairwise distance (Pearson correlation coefficient) using the Tomtom module (Gupta et al. 2007) of the MEME Suite package (Bailey et al. 2015). To group identical motifs within the training set, we generated a graph with the python module “NetworkX” (Schult and Swart 2008) from the significant hits between motifs from the Tomtom output, with each pairwise match between motifs becoming an undirected edge. We identified connected components containing eight or more nodes, and selected all motifs associated with these. Eight groups met this criterion; these were used to build corresponding eight motif sets. Finally, position weight matrices (PWMs) from each motif set were iteratively merged to create a single consensus PWM, generating eight motifs overall. These consensus PWMs were designated Daphnia (core) promoter motifs (Dpm). Motif logos were generated for each Dpm PWM using the motif2images command from MEME Suite (Bailey et al. 2015). The similarity of each member of the Dpm motif set to core promoter elements in D. melanogaster was determined by sequence alignment STAMP (Mahony and Benos 2007) against the JASPAR database (Portales-Casamar et al. 2009). The E-value of the best alignment was recorded for every Dpm motif. The enrichment P-value of a representative PWM from the motif set was selected to reflect each Dpm motif in Figure 4. Code used to perform the de novo motif discovery workflow described here is found in the MoVRs (Motif set Reduction and Validation) software package hosted on GitHub: https://github.com/BrendelGroup/MoVRs.
Figure 4.
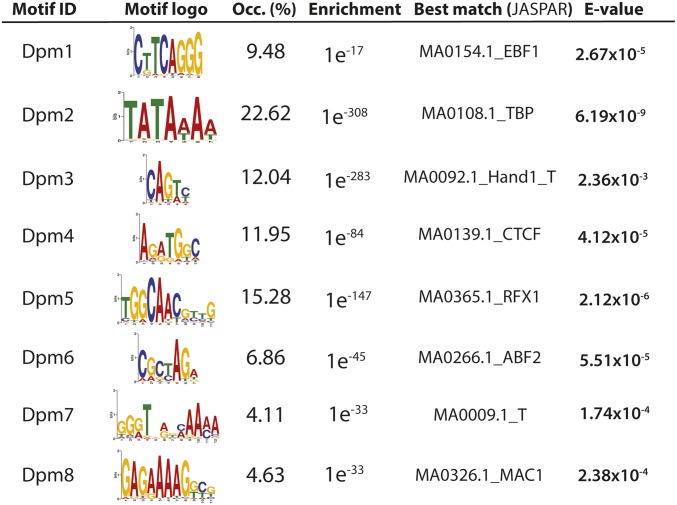
De novo discovery of core promoter elements in D. pulex. The D. pulex core promoter motifs identified in this study are listed. For each identified motif (n = 8), we show a logo representing the PWM of each motif, its frequency relative to regions surrounding major CAGE peaks ([−200,+50]) (see Materials and Methods), observed motif enrichment E-value, and the E-value of the most similar motif within the JASPAR database (Portales-Casamar et al. 2009). The motif enrichment E-value represents the probability that a motif of equal length would be discovered in an equivalent number of randomly-derived sequences with the same underlying nucleotide frequencies with equal or lower likelihood. CAGE, Cap Analysis of Gene Expression; Dpm, Daphnia (core) promoter motif; Occ., occurrence; PWM, position weight matrix.
Analysis of differential activity of consensus promoters
Differential expression of promoters was performed using defined consensus promoters (n = 10,580) along with their normalized expression values (in tags per million; tpm) observed in each condition. We utilized the most recent version of the limma package in R (Ritchie et al. 2015) to identify differentially-active (DA) promoters across all three conditions. Limma, which implements a linear modeling algorithm, also incorporates voom (variance modeling at the observational level), a method that estimates the mean–variance relationship in a counts-based fashion (Law et al. 2014).
Analysis of mean–variance and linear model:
Genomic coordinates and activity values (in tpm) for all consensus promoters within a library were used to create an ExpressionSet object (Lawrence and Morgan 2014) in R. Biological replicates from a given stage were labeled and used to construct a “contrasts matrix” to initiate comparisons between stages (e.g., males and sexual females). Analysis of mean variance (voom) was performed for every consensus promoter containing more than 25 tags (CTSSs) on aggregate across all CAGE samples. The log-ratios from the previous step were fitted to a linear model (lmFit; Ritchie et al. 2015), followed by a “contrasts fit” using the aforementioned contrasts matrix, which calculates the standard error for each contrast, or between-stage comparison. An empirical Bayes method (eBayes) was applied to the model fits from the previous step, generating moderated t- and F-statistics, respectively, and a log-odds differential expression value for each consensus promoter. A decide test (decideTest) was then performed on this set of t-statistics, where consensus promoters with P-values < 0.01 (after Benjamini–Hochberg FDR correction) were deemed to be significantly DA. DA promoters from each comparison were retrieved for subsequent analysis.
Heatmaps:
The normalized expression levels (in all CAGE libraries) of promoters classified as DA were extracted and plotted as a hierarchically-clustered heatmaps in R using the gplots package (Warnes et al. 2015).
Analysis of functional enrichment
Gene ontology:
Consensus promoters were assigned to annotated genes (the “Frozen Gene Catalog”) using their genomic coordinates. The complete gene ontology (GO) dataset for D. pulex (http://genome.jgi.doe.gov/cgi-bin/ToGo?species=Dappu1) was downloaded and GO terms were associated with the gene annotation. We applied the Fisher’s Exact Test in the topGO package (Alexa and Rahnenfuhrer 2010) in R, asking which GO terms were overrepresented among genes shown to have differentially-regulated promoters (see previous section). Enrichment analysis was performed separately using terms from the GO categories “Molecular Function” and “Biological Process,” respectively. GO Terms with P-values < 0.01 were classified as “significantly enriched”
Pathway analysis:
We extracted the KEGG (Kyoto Encyclopedia of Genes and Genomes; http://www.genome.jp/kegg/) pathway identifier, using the same promoter-to-gene-annotation dataset described for the GO analysis. Using the set of terms for DA consensus promoters, we performed a test for statistical enrichment of KEGG pathways using a Python script (C. Jackson personal communications). KEGG terms with P-values < 0.01 were considered significantly enriched.
Data availability
Data from this manuscript were deposited in the NCBI Gene Expression Omnibus (Edgar et al. 2002) (http://www.ncbi.nlm.nih.gov/geo) and are accessible through GEO Series accession number GSE80141 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE80141) with the BioProject identifier of PRJNA318020. Sequence data were deposited in the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) and have been assigned the accession number SRP073105.
Results
Profiling capped 5′ mRNA ends characterizes the global landscape of transcription initiation
Interrogation of 5′-ends of capped RNAs identifies the locations and patterns of transcription initiation within a genome. Through biochemical capture of these 5′ transcript ends (see Materials and Methods), CAGE ultimately generates short, strand-specific sequences (CAGE tags), the 5′-ends of which correspond to the first base of the associated mRNA. Sequenced CAGE tags (47 bp in length) were aligned to the genome (Figure 1A, panel i). The coordinate corresponding to the 5′ aligned base of each aligned read is defined as a CAGE-detected TSS (CTSS; Figure 1A, panel ii). Multiple CAGE tags mapping to identical CTSS coordinates provide a quantitative measure of the abundance of mRNA ends that originated from that position. Individual CTSSs supported by sufficient numbers of CAGE tags (sCTSSs; see Materials and Methods) occurring in close proximity in the genome were clustered to yield TSRs that correspond to genomic intervals that coincide with transcriptionally active promoters (Figure 1A, panel iii). Finally, when CAGE data from multiple conditions or tissues were compared, we define TSRs that agree (i.e., overlap) in all cases as “consensus promoters” (Figure 1A, panel iv).
A promoter atlas in Daphnia pulex
D. pulex can reproduce asexually, through ameiotically-produced eggs that develop directly, and sexually, through diapausing eggs (Ebert 2005). We generated CAGE datasets from three distinct adult ‘states’ of D. pulex (Figure 1B; leftmost panel): males, parthenogenetic females (hereafter asexual females), and preephippial females (hereafter sexual females). These states were chosen to potentially identify distinct genes and gene networks associated with meiosis, parthenogenesis, and sex-specificity. We sequenced eight libraries, generating 1.82 × 108 CAGE reads overall (Table 1), of which 1.22 × 108 (67.0%) mapped successfully to the current version (JGIv1.1) of the D. pulex assembly (Colbourne et al. 2011). After normalization (see Materials and Methods), biological replicates for each state were highly correlated (Pearson coefficient > 0.97; Supplemental Material, Figure S1 and Figure S2). We then applied a computational analysis pipeline to identify CTSSs, TSRs, and consensus promoters (Figure 1A) from CAGE reads across each of the three states (See Materials and Methods) (File S1).
We evaluated our CAGE definitions in their entirety by considering their locations within the D. pulex genome. Among CTSSs (n = 2,332,582) pooled across all states, we observe that a sizable fraction (67.5%) were located within 1 kb of a coding sequence (CDS), while 9.88% were present in the first 1 kb downstream of a stop codon (Figure 1C), an observation also reported in D. melanogaster (Hoskins et al. 2011). When CAGE tags are considered individually (rather than unique CTSSs alone), we report a substantially larger percentage (82.3%) located within the first 1 kb upstream of the translation start site of coding genes, while only a small fraction (1.95%) were located downstream of annotated CDSs (Figure 1C). From this, we conclude that CTSSs supported by many CAGE reads are more likely to be positioned upstream of coding genes than those supported by fewer reads.
Similar numbers of TSRs (between 11,289 and 11,558) are identified within the three individual states, totaling 12,662 unique TSRs overall (Table 2). The majority of identified promoters (83.1%) were positioned within the first 1 kb upstream of coding genes, indicating general but incomplete agreement with the current D. pulex gene annotation (Figure 1C). This work represents a comprehensive, sex-specific promoter atlas in adult D. pulex, the first of its kind in crustaceans.
Table 2. Summary of CAGE evidence generated in this study.
| Sample name | Number of mapped reads | TSRs (unique) | Consensus promoters |
|---|---|---|---|
| Asexual females | 50,195,790 | 11,496 (316) | – |
| Sexual females | 40,646,067 | 11,289 (231) | – |
| Males | 29,373,220 | 11,558 (557) | – |
| Total | 120,215,077 | 12,662 | 10,580 |
CAGE, Cap Analysis of Gene Expression; TSRs; transcription start regions.
Promoter shape, base composition, and expression class:
The property of the distribution of TSSs is known to be a key descriptor of the structure and composition of the underlying promoter in metazoans (Rach et al. 2009; Hoskins et al. 2011). We evaluated CAGE tag distributions at consensus promoters (n = 10,580) using two criteria. The first is width, which is defined as the length of the genomic segment occupied by all CTSSs within a TSR or consensus promoter. We observe an ample range of widths (2–163 bp), including a small number (1104; 10.4%) of TSRs with widths > 30 bp (Figure 2A). Overall, we observe a median width of 5 bp, and a mean width of 12 bp for all consensus promoters. We applied a second metric, promoter shape, which measures the stability of the CAGE tag distribution at a TSR. For example, a TSR with a sharp distribution of CAGE tags surrounding a single major CTSS would be considered peaked, whereas a TSR with numerous distinct CTSSs supported by roughly equivalent numbers of CAGE tags would be broad. We applied the Hoskins SI (Hoskins et al. 2011) to measure shape across all consensus promoters. We also observed a wide range of consensus promoter shapes (Figure 2A, inset); the observed median and mean SI values were −0.42 and −0.54, respectively.
Two distinct promoter classes have been proposed in mouse, human, and Drosophila, defined according to the shape of empirical (generally CAGE-based) 5′-end distributions (Carninci et al. 2006; Hoskins et al. 2011; Kadonaga 2012). We reasoned that if two distinct classes of promoter exist in D. pulex, then the shapes we observe should be bimodally-distributed. We fit the distribution of consensus promoter shapes using an EM algorithm (see Materials and Methods), and see strong support for a two-component mixture model (Figure 2A, inset), consistent with broad and peaked consensus promoter shapes. This result provides evidence for the existence of two classes of promoter in D. pulex and is consistent with previous findings. We classified consensus promoters into categories according to SI, peaked (n = 738), broad (n = 1318), or unclassified (see Materials and Methods). Examples of peaked and broad consensus promoters found within our CAGE dataset are shown in Figure 2C. We then asked if promoter activity (the abundance of CAGE tags associated with a consensus promoter) varied by promoter shape class (see Materials and Methods). We find that broad TSRs have significantly higher activities overall (P < 0.0003710) than peaked and unclassified TSRs (Figure 2, D and E). However, we do not observe a similar relationship between activity and promoter width (data not shown). This suggests that, in D. pulex, shape is more reflective of promoter properties than width.
Dinucleotide preferences of D. pulex TSSs:
Global studies of transcription initiation across metazoan diversity identified distinct dinucleotide compositions at the TSS (Frith et al. 2008; Nepal et al. 2013). We investigated dinucleotide preferences in D. pulex, measuring the dinucleotide frequencies present within the [−1,+1] interval relative to CTSSs. We observe a strong preference for CA, GA, GC, GG, and GT relative to background (P < 0.01; see Materials and Methods) and considerable depletion for AT-rich dinucleotides AA, AT, and TT (P < 0.02, 0.01, and 0.01, respectively; Figure 2B).
Promoter orientation in D. pulex
We considered the arrangement of consensus promoters within the genome. As with genes, promoters can assume one of three relative orientations: (i) tandem (also known as head-to-tail), (ii) divergent (also known as head-to-head), and (iii) convergent, which are illustrated in Figure 3A. Evaluating the interpromoter distance, we observe an unexpectedly large number of closely-spaced consensus promoters. We find 1697 (24.4% of all tandemly-arranged pairs) tandemly-arranged consensus promoters within 500 bp, and nearly 3000 (n = 2969; 42.6% of all such pairs) within 2 kb (Figures 3, B and C). By comparison, compact convergent consensus promoter arrangement is rare; we observe only nine consensus promoter pairs within 500 bp (Figure 3B).
Alternative promoter usage:
Reasoning that consensus promoters in proximal, tandem arrangement could represent cases of alternative promoter usage, we examined this set further. We evaluated the number of tandemly-arranged consensus promoters associated with annotated coding genes, inquiring as to the frequency of putative alternative promoter usage. While most genes are associated with a single consensus promoter (n = 5347), we observe that a prominent fraction (n = 1266; 19.1%) of genes are associated with at least two consensus promoters (Figure 3D). Interestingly, a small number (n = 188; 2.84%) of genes are associated with four. This evidence suggests that alternative promoter usage is not infrequent in D. pulex.
Divergent promoters:
We next considered those consensus promoters that assume divergent orientations (Figure 3A; n = 2027 pairs). Nearly one-quarter (n = 490; 24.2%) of divergent consensus promoter pairs are located within 500 bp, and 953 (40.9% value) within 1 kb (Figures 3B). Intrigued by the large proportion of divergently-arranged consensus promoters, we asked whether both members of a given promoter pair were associated with annotated coding genes. A majority of divergent promoter pairs were found to each associate with coding genes: 554 (58.1%) of pairs within 1 kb, and 386 (40.5%) of those within 500 bp. For perspective, we compared the distances between divergent promoter pairs to those observed in D. melanogaster. Divergent promoters in D. pulex are much more compact than their counterparts in Drosophila, exhibiting median distances of 2328 and 11,694 bp, respectively (Figure 3E). In addition, the fraction of divergent promoters that exhibit a compact (within 500 bp) orientation is ∼10-fold higher in D. pulex (490 of 1549; 31.6%) than those in D. melanogaster (43 of 1318; 3.26%). We then investigated the relationship of the activities of divergent promoter pairs, investigating the possibility that they are coregulated. We find no correlation in promoter activity between closely-spaced (≤ 500 bp) divergent promoters (data not shown), suggesting that, overall, closely-spaced divergent promoters in D. pulex are not coregulated under the conditions surveyed here. A representative example of a closely-spaced divergent promoter pair (geneIDs: PASA_GEN_0100236 and PASA_GEN_0100237) is presented in Figure 3F.
De novo discovery of consensus promoter elements in D. pulex
Core promoter elements and their motif consensus sequences have been identified in D. melanogaster (Ohler et al. 2002; Down et al. 2007; Kadonaga 2012), mammals [Carninci et al. 2006; FANTOM Consortium and the RIKEN PMI and CLST (DGT) 2014], and other metazoan model organisms: worm (Caenorhabditis elegans) (Saito et al. 2013) and zebrafish (Danio rerio) (Nepal et al. 2013; Haberle et al. 2014).
Cis-regulatory motifs of any kind in D. pulex are unknown, so we sought to identify core promoter elements using the CAGE data generated in this study. To accomplish this, we performed de novo motif discovery using CAGE evidence (see Materials and Methods), applying sequence windows corresponding to core promoters ([−50,+50]). This procedure revealed a set of eight core promoter elements in D. pulex (Figure 4). To evaluate their similarity to known core promoter elements, we performed sequence alignment of each PWM against two motif sets: the complete JASPAR database (Portales-Casamar et al. 2009) and a curated list of 14 nonredundant core promoter motifs in D. melanogaster. We find two motifs within our set with strong sequence identity to the most well-characterized metazoan core promoter elements. The motif Dpm2, which has the consensus TATAWAA, has significant identity to the TBP-binding motif consensus in JASPAR (MA0108.1_TBP, E-value = 6.19 × 10−9) in addition to the TATA element of D. melanogaster (E-value = 7.49 × 10−10). The TATA-like Dpm2 was observed in 9.48% of promoters. The motif Dpm3, with the consensus NCAGTY, has significant sequence similarity to the Initiator (Inr) element (consensus TCAKTY) (E-value = 6.10 × 10−6) of D. melanogaster and is found at 12.0% of promoters.
In addition to TATA and Inr, we report a variety of motifs within our set of D. pulex core promoter elements (Figure 4). Dpm5 (consensus TGGCAACNYYG), exhibits significant similarity (E-value = 5.76 × 10−8) to the “Ohler8” motif in D. melanogaster (Ohler et al. 2002). All of the remaining motifs match significantly with at least one motif in the JASPAR database. Among these, three motifs exhibit similarity to well-characterized transcription factor binding sites (TFBSs): Dpm4 (consensus ARATGGC) matches the CTCF motif in JASPAR (MA0139.1_CTCF) (E-value = 5.51 × 10−5); Dpm6 (CGCTAGA) matches the ABF transcription factor binding site consensus (MA0266.1_ABF2) (E-value = 5.51 × 10−6) (Portales-Casamar et al. 2009), and the motif Dpm5 (consensus CARCGTTGCC) exhibits a significant match to the TFBS consensus of RFX1 (MA0365.1) (E-value = 2.12 × 106).
Motif cooccurrence at promoters
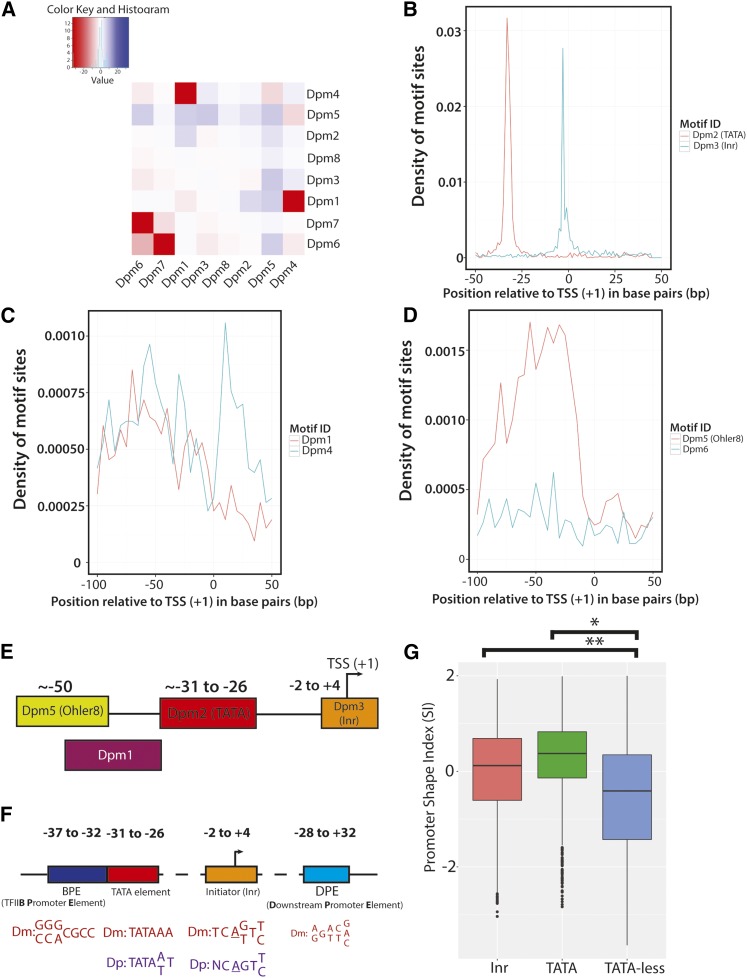
After completing de novo discovery of core promoter elements in D. pulex (Figure 4), we sought to characterize the overall motif composition of promoters within the Daphnia Promoter Atlas. We used the consensus sequences of each of the eight motifs in the Daphnia promoter set and searched within a sequence window of [−200,+50] surrounding the midpoint of all annotated promoters. Using this information, we constructed a cooccurrence matrix for all identified promoter motifs, asking as to the overall coincidence of motifs within promoter regions. Several patterns of motif cooccurrence are observed among the Dpm motifs (Figure 5A). We find that TATA (Dpm2)-containing promoters are not enriched for other identified Daphnia motifs and are depleted for Dpm4 and Dpm5. Inr (Dpm3) promoters are enriched for Dpm6 and have fewer Dpm4 and Dpm5 motifs than expected. Dpm4, while strongly enriched for Dpm1, also exhibits significant enrichment for Dpm5 and Dpm6. We observe strong cooccurrence between Dpm6 and Dpm7. Three motifs, Dpm1, Dpm6, and Dpm7, have greater than expected frequencies of cooccurrence. Of note, none of the other core promoter elements are coenriched with (Dpm2), and two (Dpm4 and Dpm5) are depleted. This line of evidence suggests that TATA-containing promoters do not frequently act in combination with the other identified elements.
Figure 5.
The cooccurrence and distribution of identified D. pulex core promoter motifs within promoter regions. (A) Heatmap of cooccurrence frequencies among identified D. pulex motifs. The log of each P-value is plotted within the heatmap. The frequency distributions of Dpm2 and Dpm3 (B), Dpm1 and Dpm4 (C), and Dpm5 and Dpm6 (D) relative to identified promoters (TSRs) are shown (the distributions of Dpm7 and Dpm8 are not shown). (E) Current model of core promoter composition in D. pulex derived from the evidence in this study. A cartoon illustration of the Daphnia core promoter motifs that exhibit strong positional distributions are shown, with their approximate locations relative to the TSS (+1). (F) Model representing the positions and consensus sequences of canonical core promoter elements between D. pulex and D. melanogaster. The four major core promoter elements in D. melanogaster are displayed, along with their typical positions relative to the TSS (+1). The consensus sequence of each element, if present, is shown for D. melanogaster (Dm; red) and D. pulex (Dp; purple). Note that an individual core promoter may have none, all, or some of the elements listed in the illustration. Graphic adapted from (Butler and Kadonaga 2002). (G) Comparison of promoter shape between TATA- and Inr-containing promoters and those lacking TATA. The box-and-whisker plots representing the distributions of calculated SI values for consensus promoters with Inr (coral), TATA (green), and those lacking TATA (blue) are shown. Inr- (**) and TATA-containing (*) consensus promoters possess a significantly more peaked shape (P < 0.001) than TATA-less promoters. Dpm, Daphnia (core) promoter motif; Inr, Initiator; SI, Shape Index; TSR, transcription start region; TSS, transcription start site.
Positional enrichment of identified D. pulex core promoter elements
Many characterized core promoter elements are known to occur at specific locations relative to the TSS (+1). To determine the spatial characteristics of each of the D. pulex motifs, we evaluated their positional distributions relative to CTSSs and found that four of the eight Dpm motifs exhibit positional enrichment. We observe strong positional enrichment of Dpm2 (TATA-like) and Dpm3 (Inr-like) relative to D. pulex promoters (Figure 5B), with peaks at −30 and +1, respectively, consistent with the positions of TATA and Inr within other metazoans (Kadonaga 2012). Dpm1 displays a modest peak at ∼+50, while Dpm5 is enriched between −50 and −40 (Figure 5, B and C). Dpm4 shows an irregular distribution within promoters, with two distinct peaks near −50 and +10 (Figure 5D). We do not observe a positional enrichment for motifs Dpm6, Dpm7, and Dpm8 (Figure 5D and data not shown). Taken together, this positional information allows us to construct an initial working model of the known core promoter elements in D. pulex (Figure 5E), and draw a comparison between canonical core promoter elements in D. pulex and D. melanogaster (Figure 5F).
Patterns of transcription initiation are known to relate to underlying promoter architecture in Drosophila (Rach et al. 2009; Hoskins et al. 2011) and mammals (Kadonaga 2012), so we asked whether possession of the two major core promoter elements Inr and TATA (Dpm2 and Dpm3, respectively) is associated with TSR shape in D. pulex. Using the SI (as previously described) to measure the focus and dispersion of CTSSs within a promoter, we find that both Inr- and TATA-containing consensus promoters are significantly more peaked overall (P < 0.001) than TATA-less promoters (Figure 5G), consistent with our expectations and the evidence in other metazoan model organisms including D. melanogaster (Rach et al. 2009; Hoskins et al. 2011).
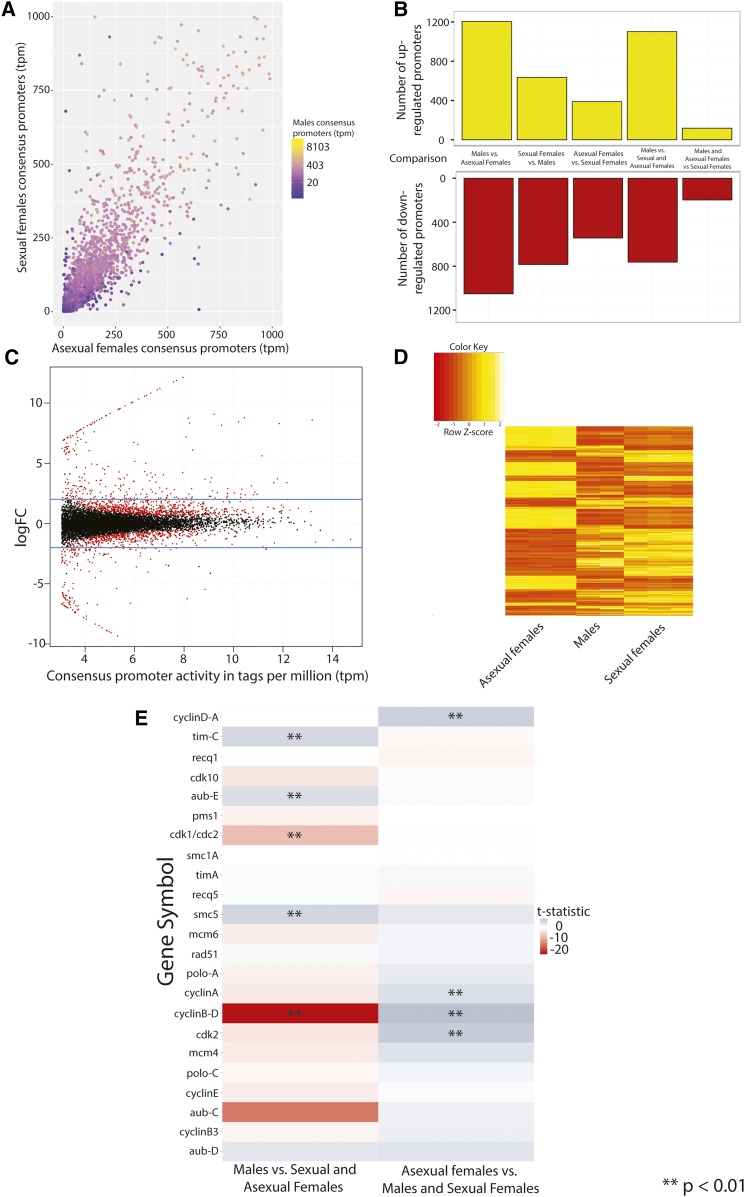
Differential activity of D. pulex promoters
The abundance of CAGE tags that map to a putative promoter region provides quantitative measurement of the extent of transcription initiation at that site; this is capable of estimating expression of the associated genes (Balwierz et al. 2009; Murata et al. 2014). In this way, we sought to identify promoters with differential activity, differentially-expressed genes across the three states surveyed by our CAGE experiment. We used our defined set of consensus promoters (Table 2; n = 10,580) and compared the normalized quantities of CAGE reads within a given state. Consensus promoter activity (i.e., the abundance of CAGE tags present at a consensus promoter in a given state) was measured using the number of mapped CAGE tags within the promoter and were represented in units of tpm. An illustration of tag abundance within consensus promoters across the three states surveyed in this study is presented in Figure 6A. We carried out differential expression analysis across all libraries using limma (Ritchie et al. 2015), applying the mean–variance relationship of log-tpm (see Materials and Methods). During our analysis, we compared promoter activities between each state separately (e.g., sexual females vs. asexual females, etc.) in addition to the following comparisons: males vs. both females, sexual vs. asexual females, comprising five comparisons in total. We observe that an average of 1359 consensus promoters have differential activity within each comparison: an average of 690 promoters exhibited significantly increased activity and 669 promoters had significantly decreased activity (Figure 6B). We observe the greatest number of DA promoters (n = 1206, upregulated; n = 1052, downregulated) in the comparison between males and asexual females. Consensus promoters with differential activity exhibit a complex topology of enrichment patterns across all three states; representative comparisons for asexual females are shown in Figure 6, C and D. Heatmaps of DA promoters from other comparisons are presented in Figure S3.
Figure 6.
Analysis of differential activity of D. pulex consensus promoters. (A) Representation of consensus promoter activities among the states surveyed in this study. A scatterplot of consensus promoter activity (in tpm) within all three states measured within our study is shown, with the value for asexual females (x-axis) plotted against sexual females (y-axis). Corresponding promoter activity values for males are represented according to a color gradient in log-scale. A small number of consensus promoters (n = 145) that lie outside the area of the plot are not shown. (B) Barplot of DA promoters between pairs of the states surveyed. (C) MA plot of consensus promoter activity within asexual compared to sexual females. MA activity of consensus promoters (x-axis) is plotted against the log FC of the ratio of the activity of consensus promoters between asexual females and sexual females (y-axis). DA consensus promoters (P < 0.01 are represented by red dots; all others are colored in black. Top and bottom blue lines on the plot indicate the log(FC) of 2 and −2, respectively. (D) Heatmap of the activities of DA (P < 0.01) consensus promoters between asexual females and sexual females. (E) Heatmap grid of relative activity of consensus promoters of D. pulex meiosis genes within two selected comparisons: males vs. females and asexual females vs. sexuals. Cells are shaded according to the calculated t-statistic of a given comparison. Instances of significant differential activity (P < 0.01) are labeled with two asterisks (**). DA, differentially-active; fold-change, FC; MA, mean–average; tpm, tags per million.
Genes associated with DA promoters are enriched for endocrine and environmental response functions:
We investigated the set of DA promoters between each state, asking if the members of each respective gene set were enriched for common functions. We carried this out using the Gene Ontology (GO), using GO terms associated with the gene adjacent to each DA consensus promoter. We observe significantly enriched GO categories for every comparison (data not shown). Results for the inferred differentially-expressed genes between asexual and sexual females are summarized in Figure S5. Among asexual females, enriched categories among upregulated genes include “nitrogen compound metabolic process” (GO:0006807; P < 1.2 × 10−7). In sexual females (Figure S5), we observe enrichment of several GO categories, including “hormone activity” (GO:0003735; P < 0.014) and “organic cyclic compound metabolic process” (GO:1901360; P < 2.9 × 10−6).
Differential upregulation of promoters of meiosis genes in asexual (parthenogenetic) females:
We then asked whether there was evidence for specific pathway enrichment within genes associated with DA promoters. Among genes upregulated in asexual females (vs. sexual females) (Figure 6B), we detect enrichment of pathways associated with cell cycle progression and oocyte meiosis (Figure S6), including cell cycle (04110; P < 1.57 × 10−5), p53 signaling pathways (04115; P < 3.80 × 10−3), and oocyte meiosis (04114; 6.88 × 10−3). Upon inspection of the genes associated with these terms, we observe substantial overlaps with annotated meiotic genes in D. pulex. Of the inferred differentially-expressed genes associated with the cell cycle KEGG pathway (Kanehisa et al. 2016) (Figure S6), five out of nine (Cdc20, CycA, CycB, CycE, and Cdk2; 55.6%) are functionally designated as “meiotic” by at least one study (Schurko et al. 2009). Additionally, three of seven upregulated genes within the “oocyte meiosis” category (Cdc20, Cdk2, and CycE) are annotated in meiosis within D. pulex (Schurko et al. 2009), with two others [Plk1 (Pahlavan et al. 2000) and AurA (Crane et al. 2004)] being directly implicated in meiosis in other model systems. Given their positions within gene networks, upregulation of these genes would be expected to have a negative regulatory impact on meiotic progression overall. Relative activities of the detected promoters of meiosis genes from two comparisons: males vs. females and asexual females vs. sexuals (i.e., males and sexual females), are shown (Figure 6E).
We investigated the set of inferred upregulated genes in the (facultatively) asexual females within our study, asking about the extent of the concordance between the differentially-upregulated genes and scaffolds known to be physically linked to obligate asexuality (Tucker et al. 2013). Considering the genomic locations of differentially-upregulated genes, we unexpectedly find that a fraction (four of 15 genes) are located on scaffolds linked to “asexual” chromosomes. This list includes Cdk2 (scaffold_77/ChrVIII), Tim-C (scaffold_76/ChrVIII), Plk1-C (scaffold_9/ChrIX), and HDAC (scaffold_13/ChrIX). We also note that two of the 15 genes, CycE (scaffold_163) and β-TrCP (scaffold_169), are located on short scaffolds that were not previously tested (Tucker et al. 2013).
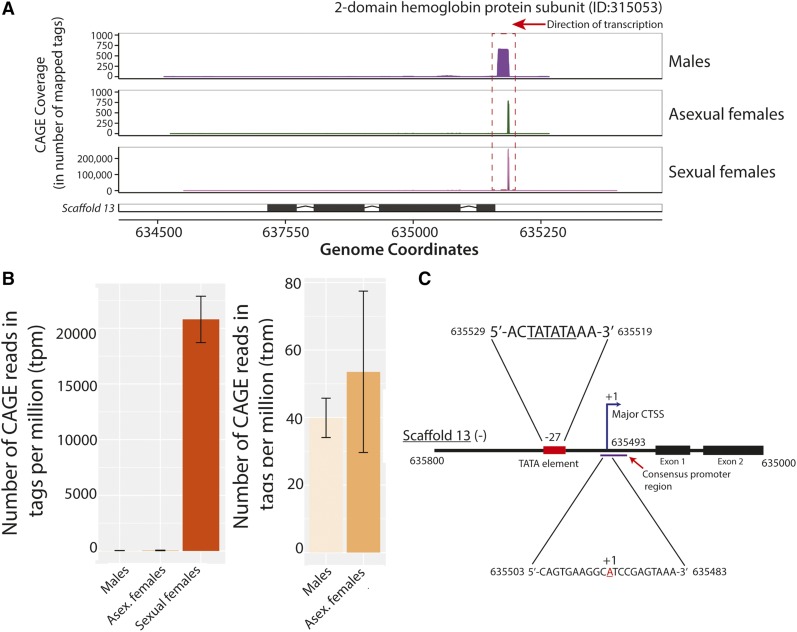
Dramatic, sex-specific differential expression of a hemoglobin gene
In evaluating the differential activity consensus promoter data (Figure 6A), we note several genes that are dramatically upregulated in a single condition. Among these is the 2-domain hemoglobin protein subunit (ID: 315053) gene on scaffold 13. We observe ∼400-fold more CAGE tags at the promoter of this gene within sexual females than the other two states (males and asexual females) (Figure 7, A and B), indicating considerable apparent state-specific upregulation of hemoglobin. The striking abundance of CAGE tags at the consensus promoter in sexual females (20,791 tpm) represents just over 2% of all sequenced CAGE tags within that state. An illustration of the core and proximal promoter region of the gene is shown in Figure 7C, including the consensus promoter region and major CTSS identified by this study. The core promoter contains a TATA box (5′-TATATA-3′) at −27. We looked in the proximal promoter region for the juvenoid response element (JRE; 5′-CTGGTTA-3′) identical to the one reported in D. magna (Gorr et al. 2006), but did not find one. An additional example of sex-specific expression is shown in Figure S4, where upregulation of the consensus promoter for the gene encoding the egg protein vitellogenin among asexual females is presented.
Figure 7.
Extreme upregulation observed at the putative promoter of a hemoglobin gene in D. pulex sexual females. (A). Mapped CAGE tags from each of the three surveyed states to an annotated hemoglobin gene (ID:315053) on scaffold 13 are shown. The frequency of CAGE tags observed at each genomic coordinate (x-axis) are indicated by the y-axes of each plot. Note that larger y-axis scales are applied for the sexual females plot due to the dramatically higher number of mapped CAGE tags observed at the same locus. (B) Consensus promoter activity (in tpm) at the same genomic locus as (A) across all three states is presented in the left panel; in the right panel only the values for males and asexual females are shown to provide perspective. The standard error of the mean of all replicates is shown for each individual plot. (C) Schematic illustration of the core and proximal promoter region of the hemoglobin gene (ID:315053). The major CTSS (+1) is identified by the blue arrow, and the TATA consensus sequence is represented by the red rectangle. The purple line represents the consensus promoter region identified by CAGE. The genomic coordinates for the sequences (all on scaffold 13) are shown in black. Note that the sequence for the negative strand is shown; the illustration was flipped to improve legibility. The drawing was not made to scale. CAGE, Cap Analysis of Gene Expression; CTSS, CAGE-detected TSS; tpm, tags per million.
Discussion
In this study, we performed CAGE (Kodzius et al. 2006; Takahashi et al. 2012b) to map 5′-mRNA ends and identify active promoters within the ubiquitous aquatic microcrustacean D. pulex, providing a taxonomic extension to the picture of metazoan promoter architecture. We report an average of 11,448 TSRs across the three conditions, 12,662 unique TSRs, and 10,580 consensus promoters. This D. pulex promoter atlas provides the first comprehensive collection of cis-regulatory elements within Crustacea.
We measured the occurrence of our CAGE-derived annotations with sites within the D. pulex genome, finding that they are generally located in positions consistent with promoter regions. The observation of CTSSs downstream of coding regions is consistent with the findings in D. melanogaster, where 17% of CAGE peaks were detected within annotated 3′UTR regions (Hoskins et al. 2011). The possible functions of CTSSs observed in CDSs and downstream of coding genes are challenging to interpret; they could represent the biochemical background of CAGE (Hoskins et al. 2011) or could alternatively represent bona fide RNA Pol II-derived transcripts. The latter case would suggest conflict with existing gene annotations, which can be resolved as more transcriptome analysis is performed in D. pulex. Approximately 82% of total aligned CAGE tags map upstream of annotated protein-coding genes (Figure 1C), a similar figure to that reported in Drosophila embryos (86%) (Hoskins et al. 2011). The overall incidence of TSRs upstream of coding genes (83%) mirrors that of CAGE tags (82.3%), suggesting that most TSRs in our dataset are positioned in locations consistent with the promoters of coding genes. The collection of TSRs (17%) located elsewhere is likely to contain a number of bona fide promoters.
The total number of unique TSRs defined here, 12,662, is close to the total of 12,454 promoters reported in D. melanogaster (Hoskins et al. 2011). This result may indicate a greater similarity in the number of protein-coding genes between D. pulex and D. melanogaster than is presently predicted by the present genome annotation. The predicted gene count for D. pulex (30,907) (Colbourne et al. 2011) is considerably larger than the ∼14,000 (13,918) protein-coding genes in the most recent annotation of D. melanogaster (Matthews et al. 2015). The high depth of sampling and variety of stages measured in this study would be expected to reveal a similar ratio of active TSRs to annotated genes to what was observed in D. melanogaster (Hoskins et al. 2011). However, given the limited functional genomic evidence in D. pulex currently available, we cannot unequivocally conclude how many of the TSRs we report are, in fact, “true” promoters, beyond evaluating their relationship to the current gene annotation. As it currently stands, this reality may lend greater weight to those TSRs that are found upstream of annotated coding genes. Further functional genomic [e.g., RNA sequencing (RNA-seq)] analysis will be helpful to reconcile these existing discrepancies. We propose that the promoter atlas presented here be utilized to form an important component of an improved gene annotation in D. pulex.
We explored the properties of the consensus promoters within our D. pulex promoter atlas. Overall, the distribution of consensus promoter widths observed are consistent with those determined in D. melanogaster using CAGE (Figure 2A) (Hoskins et al. 2011; Chen et al. 2014). A proportion of the consensus promoter widths are long, including 1104 (10.4%) with widths longer than 30 bp (Figure 2A). This value is also similar to the amount observed (10.8%) in D. melanogaster (Hoskins et al. 2011). Promoters with similarly long widths have also been observed in human, mouse (Carninci et al. 2006), and, more recently, C. elegans (Saito et al. 2013). The distribution of consensus promoter shapes (Figure 2A, inset) indicates that both broad and peaked transcription initiation patterns are observed at D. pulex promoters. The observation that shape distribution is bimodal (Figure 2A, inset) agrees with previous models of promoter classes (Rach et al. 2009; Kadonaga 2012) and provides rationale for the classification of promoters according to shape. We found that broad promoters exhibited higher activity than did peaked promoters (Figure 2E), but we did not observe the same relationship between width and activity (data not shown). This suggests that shape is a more faithful representation of CTSS distribution and TSR properties than breadth alone. Our finding that broad promoters have higher promoter activity agrees with the available evidence in other species. In D. melanogaster, promoter width was positively associated with CAGE tag count (the equivalent to “activity” as defined here) (Hoskins et al. 2011). In D. melanogaster and elsewhere, broad promoters are associated with higher expression and genes with constitutive expression (Lenhard et al. 2012). While we did not directly address the relationship between promoter class and gene function in this study, such a comparison will be possible using these data, particularly as the functional annotation (e.g., the GO) of D. pulex genes improves.
We further examined closely-spaced consensus promoters in tandem orientation, revealing widespread alternative promoter usage in D. pulex (Figure 3, A–C). Slightly fewer than one-fifth (19%) of all consensus promoter-associated genes were found to have multiple (i.e., two or more) alternative promoters. This study is the first genome-scale observation of alternative promoter usage among crustaceans; this is largely consistent with what is known elsewhere in metazoans. In D. melanogaster, the most closely related species with available data, Batut et al. (2012) reported that 40% of developmentally-expressed genes in D. melanogaster exhibit multiple promoters. The differences between the samples measured in both studies (i.e., adult vs. both larval and adult tissues), complicates determinations of the precise extent of alternative promoter usage in D. pulex relative to D. melanogaster. Alternative promoter usage has been reported in other metazoans, and has been most extensively characterized in mammalian systems: namely human (Kawaji et al. 2006; Kimura et al. 2006) and mouse (Kawaji et al. 2006). Several theories have been promulgated to explain the high incidence of alternative promoters. Kimura et al. (2006) propose that alternative promoters lead to alternative first exon usage, which contributes to proteome diversity. Using the panel of CAGE data in human cell lines generated by FANTOM5 [FANTOM Consortium and the RIKEN PMI and CLST (DGT) 2014], Sardar et al. (2014) presented evidence in support of this hypothesis, demonstrating that alternative promoter usage causes putative inclusion or exclusion of entire domains in the amino termini of proteins. While this hypothesis is enticing, its drawback is the relative lack of transcript connectivity evidence; in nearly all circumstances each CAGE peak is informatically connected to a downstream gene body, thus, the identity of the actual transcript that is produced must be inferred in all cases. However, a limited number of other studies provide evidence of transcript connectivity, albeit on a smaller number of datasets. Using a tool to integrate CAGE peaks and RNA-seq data from Drosophila, Boley et al. (2014) detected links between putative alternative promoters and bona fide transcripts, which supports with previous reports of distal TSS connectivity in fruit fly (Manak et al. 2006; Batut et al. 2012). Future study of alternative promoter usage in D. pulex would benefit from separate analysis of individual tissue types. The TSS profiling method employed by Batut et al. (2012), RAMPAGE (Batut and Gingeras 2013), is well-positioned to address alternative promoter usage in this and other species because it provides paired-end sequence information along with the 5′-end of the transcript.
The current study relied on whole individuals, making it impossible to determine how many of the alternative promoters we report are tissue-specific or active in a broad array of tissue types. This study detected a large proportion of densely-arrayed promoter pairs in divergent configurations in D. pulex. The extent of this compactness is striking; almost one-third (31.6%) of divergent promoter pairs had short (i.e., within 500 bp) upstream distances, a 10-fold greater proportion compared to those found in D. melanogaster. In addition, most of these promoter pairs are associated with gene annotations, providing evidence that these promoters each regulate expressed coding genes. The relationship between gene (and therefore promoter) organization and function is an ongoing subject of interest; the former varies widely within eukaryotes (Hurst et al. 2004). Genes with head-to-head (i.e., divergent) upstream gene orientations are dramatically expanded in D. melanogaster and mammals, where they outnumber genes with head-to-tail upstream orientations 10:1 (Woo and Li 2011). Studying a panel of six diverse eukaryotes, Woo and Li (2011) found that (compact) head-to-head genes exhibit lower expression variability than their counterparts in a head-to-tail orientation. Supported by evidence from budding yeast, the authors posit that this is partly due to more stably positioned −1 and +1 nucleosomes in these genes, which would disfavor expression divergence among the gene pairs (Woo and Li 2011). The evidence we present for compact, divergent promoters in D. pulex is consistent with previous findings about the organization of its genome: the D. pulex genome has small intergenic distances relative to other metazoans, featuring densely-arranged clusters of gene families, of which some are proposed to represent tandem duplicates (Colbourne et al. 2011). While we did not evaluate which of the compact, divergent (or head-to-head) promoters we report are members of tandem gene clusters, these promoter pairs, and the evolutionary fates of the genes they regulate, provide an appealing avenue for future study. The evidence presented here suggests that D. pulex may provide a useful model for investigating spatial constraint on cis-regulatory sequence size and genome architecture in metazoans.
We observe a preference for specific dinucleotides (CA, GA, GC, GG, and GT) at CTSSs (Figure 2B). These results are partly in line with what is known elsewhere; the CA dinucleotide is located at [−1,+1] in Inr-containing promoters (Kadonaga 2012), and purines (A and G) are enriched at the TSS in metazoans, where studied (Fitzgerald et al. 2006; Sandelin et al. 2007; Nepal et al. 2013). However, three of the four overrepresented dinucleotides (GA, GC, and GG) have guanines at −1, which is observed less commonly in metazoans. D. melanogaster, the most closely related species for which CAGE data are available (Hoskins et al. 2011; Chen et al. 2014), is enriched for YR at [−1,+1]; no enrichment of dinucleotides with G at −1 is reported. In human, where core promoters tend to be GC-rich (Fitzgerald et al. 2004, 2006), YR, but no GN dinucleotides, are enriched at initiation sites (Sandelin et al. 2007; Frith et al. 2008).
Overall, some of the initiation dinucleotide preferences we observe in D. pulex appear to be distinct to those of other metazoans that have been similarly surveyed. Some similarities are evident; the CA dinucleotide at ∼12% of CTSSs, which is identical to the canonical YR code at initiation sites, and coincides with the sequence of Inr at the TSS [−1,+1] position (Butler and Kadonaga 2002). In contrast, the other four statistically-enriched dinucleotides reported here are observed less frequently at promoter initiation sites in other metazoans. In light of these differences, it is important to note that the analysis dinucleotide frequencies at CTSSs carried out here was performed with CTSS data irrespective of annotated genomic position. As such, the dinucleotide frequencies include contributions from all CTSSs, including those that were not associated with promoters (Figure 1C). As the D. pulex becomes more well-defined, an evaluation of dinucleotide frequencies from CTSSs according to their genomic position may help to clarify this. As an example, Nepal et al. (2013) performed a dinucleotide analysis using genomic position in zebrafish, and report considerable dinucleotide frequency differences between promoter and intergenic CTSSs, including above-average frequencies of two GN dinucleotides: GG and GC. We exclude the trivial explanation, 5′ guanine addition bias sometimes observed in CAGE studies (Carninci et al. 2006), for the observed GN enrichment because these were corrected for by our analysis pipeline (see Materials and Methods).
Our de novo discovery revealed eight distinct enriched motifs that we call the D. pulex core promoter set (Dpm1-Dpm7; Figure 4). Of the eight D. pulex core promoter elements, three have significant sequence identity to a core promoter element in D. melanogaster. We find correspondence to major metazoan core promoter elements: Dpm2, with the consensus TATAWAA, displays similarity to the TATA element in Drosophila (TATAAA), and the consensus of the putative Inr motif Dpm3 (NCAGT) has significant identity to the Inr motif of fruit fly, which is NCAKTY (Ohler et al. 2002) (Figure 5F). The putative TATA Dpm2 and Inr Dpm3 are enriched between −30 and +1 (Figure 5B), respectively, consistent with their positions elsewhere within metazoans (Juven-Gershon and Kadonaga 2010). This almost certainly suggests that we have identified the TATA and Inr motifs in D. pulex. The motif Dpm5 (TGGCAAC), observed at 15.3% of promoters, bears significant identity to the Ohler8 motif (–YGGCARC–) in D. melanogaster (Ohler et al. 2002). Dpm5 is enriched at ∼+50 (Figure 5D); the D. melanogaster Ohler8 motif has an equivalent, but more modest, peak at the same position (Down et al. 2007). The cis-regulatory role of Ohler8 is unknown, but it has been validated separately on several occasions since its initial discovery (Fitzgerald et al. 2006; Hoskins et al. 2011). In our study, the Ohler8-like Dpm5 motif was observed in a smaller fraction of promoters than observed in D. melanogaster (15.3% vs. 23.2%) (Ohler et al. 2002).
The remainder of the Daphnia promoter motif set is less well-characterized. The five other motifs within our D. pulex core promoter set, Dpm1, Dpm6, Dpm7, and Dpm8 (Figure 4), lack similarity to any member of the core promoter list in D. melanogaster. Two of these exhibit a degree of positional enrichment relative to the TSS. Dpm1 is enriched broadly between ∼−40 and −75. Dpm4 exhibits a sharp positional enrichment at −10, and a second, wider distribution surrounding −50. No positional enrichment was observed among Dpm6, Dpm7, and Dpm8 (Figure 5D and data not shown), suggesting that they may lack location preferences within core promoter regions.
The core promoter motif discovery described in this study is the first comprehensive glimpse into the cis-regulatory repertoire of D. pulex, and indeed for any crustacean. We observe strong cognates to core promoter elements in more well-studied metazoan genomes, including D. melanogaster. Collectively, these data support an initial model for the composition of the D. pulex core promoter (Figure 5E). Comparisons between our D. pulex core promoter model and the established model in D. melanogaster highlight the similarity of the reported TATA and Inr elements between the two species, but also underscores the apparent absence of two canonical fly core promoter elements (BRE and DPE) (Butler and Kadonaga 2002) in our set of core promoters (Figure 5F). A finely-tuned motif discovery approach that selects only specific promoter classes (e.g., only Inr-containing promoters) is necessary as it would be more suited for discovery of BRE and DPE, which are less abundant than TATA and Inr.
In total, three of eight Dpm motifs identified by our study lack obvious homologs in Drosophila. While we cannot propose precise functions for these putative core promoter elements, the overall positional enrichment and motif cooccurrence data (Figure 5, A–D) suggests that core promoters in D. pulex may group into TATA and TATA-less categories. In D. melanogaster, promoters that contain TATA, Inr, and a small number of other elements [including Pause Button (Hendrix et al. 2008), which we did not detect in our set] are very likely to exhibit a peaked shape (Hoskins et al. 2011). By contrast, broad promoters are depleted for TATA and Inr (Rach et al. 2009; Hoskins et al. 2011); in mammals, they are associated with CpG Islands (Lenhard et al. 2012). Our finding that TATA and Inr-containing promoters have a more peaked shape than TATA-less promoters (Figure 5G) is consistent with this model. A complete characterization of the relationship between core promoter motif composition (especially TATA and Inr) and TSR shape and expression will require further analysis of the evidence generated in this study.
D. pulex is an important model in which to study the maintenance of sexual and asexual reproduction (Hebert 1981; Tucker et al. 2013). We analyzed the genes associated with differentially-active promoters observed between asexual females and sexual females (Figure 6, C and D) and both sexuals (sexual females and adult males; Figure S3). Our observation of strong enrichment cell cycle pathways (KEGG IDs: 04110 and 04115) among genes upregulated in asexual females (Figure S6) was unexpected. Upon closer inspection, we find strong overlap between genes in these categories and those belonging to two enriched meiosis-related pathways [Progesterone-mediated oocyte maturation (04914) and Oocyte meiosis (04114)]; a number have been annotated as meiotic in D. pulex (Schurko et al. 2009). The observation of upregulated meiosis genes in asexual females (Figure 6E) was surprising, but is consistent with what is known about the functions of some of the genes in question. The most compelling of these examples is Cdc20 (ID:326123; NCBI_GNO_7600067), which is more than twofold upregulated (169.4–76.2 tpm) in asexual females. In mammals, Cdc20 acts with the APC to trigger progression through prophase during Meiosis I (Homer et al. 2009). Increased expression of Cdc20 would be expected to hasten the exit from Meiosis I-like cell-division. Cdc20 misexpression is known to disrupt Meiosis I; mice hypomorphic for Cdc20 were shown to be infertile (or nearly so) due to chromosomal lagging and misalignment during Meiosis I (Jin et al. 2010).
Although we lack comparable sources of expression data in Daphnia, the apparent increase in Cdc20 expression we observe here in parthenogenic individuals is consistent with current model of parthenogenic oogenesis in D. pulex, which is known to consist of abortive Meiosis I followed by a normal, Meiosis II-like division (Hiruta et al. 2010). The apparent differential regulation of meiotic and cell cycle genes observed here may provide a glimpse of the transcriptional changes that accompany parthenogenesis in D. pulex. However, it must be emphasized that additional molecular and cytological work will be required to appropriately address this possibility.
Finally, the identity and genomic position of several genes upregulated in asexual females on scaffolds associated with the evolution of asexuality (Figure 6E) is worth noting. Among these are Cdc20 (scaffold 76/ChrVIII) and HDAC (scaffold 13/ChrIX), two genes that were recently shown to be strongly upregulated in cyclic parthenogenesis (relative to obligate parthenogenesis) in bdelloid rotifers (Hanson et al. 2013).
Taken together, our large-scale analysis of transcription initiation in the microcrustacean D. pulex provides the first glimpse of cis-regulation and core promoter architecture in Crustacea. We find that D. pulex exhibits similar features of promoter architectures relative to fly and mammals, including peaked promoters associated with TATA and Inr and constitutively-expressed broad promoters. We also detect major constituents of Daphnia’s core promoter that lack an obvious ortholog in fly, suggesting some degree of novelty within the core promoter of D. pulex. It is intended that the data presented here, including the Daphnia Promoter Atlas, serve as a resource for future investigations within D. pulex, and comparative genomic analysis across metazoan diversity. We anticipate that, using this resource, comparisons between D. pulex and the fruit fly and fellow arthropod D. melanogaster, which are ∼600 MY diverged (Hedges et al. 2006), will be of particular utility.
Acknowledgments
We are grateful to Peter Cherbas, Nathan Keith, Dave Tang, and Sen Xu for critical comments to the manuscript, and to Teresa Crease for assistance with aspects of the methodology. We thank Kim Young for her work culturing Daphnia collections. We thank Xiangyu Yao for contributing to our motif discovery workflow. This work was supported by a grant-in-aid to M.L. from the National Institutes of Health (NIH) (identifier: 1R01GM101672-01A1). We used genomic resources from the Vincent J. Coates Genomics Sequencing Laboratory at the University of California, Berkeley, which was supported by NIH S10 Instrumentation grants S10RR029668 and S10RR027303. Research computing resources were provided in part by the National Center for Genome Analysis Support (NCGAS), supported by NSF Division of Biological Infrastructure (DBI) (grant 1458641).
Author contributions: R.T.R. coconceived the idea, performed the computational analyses and experimental work, and wrote the paper. K.S. performed experimental and culturing work, and contributed to the paper. V.P.B. developed the idea, contributed to, and edited the paper. M.L. conceived the idea, contributed to, and edited the paper. The authors declare that they have no competing interests.
Footnotes
Communicating editor: L. M. Steinmetz
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.193334/-/DC1.
These authors contributed equally to this work.
Literature Cited
- Alexa, A., and J. Rahnenfuhrer, 2010 topGO: Enrichment analysis for Gene Ontology, R package version 2.20. 0. Available at: http://bioconductor.org/packages/release/bioc/html/topGO.html. Accessed: June 24, 2015.
- Andersson R., Refsing Andersen P., Valen E., Core L. J., Bornholdt J., et al. , 2014. Nuclear stability and transcriptional directionality separate functionally distinct RNA species. Nat. Commun. 5: 5336. [DOI] [PubMed] [Google Scholar]
- Arner E., Daub C. O., Vitting-Seerup K., Andersson R., Lilje B., et al. FANTOM consortium , 2015. Gene regulation. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science 347: 1010–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L., Johnson J., Grant C. E., Noble W. S., 2015. The MEME suite. Nucleic Acids Res. 43: W39–W49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balwierz P. J., Carninci P., Daub C. O., Kawai J., Hayashizaki Y., et al. , 2009. Methods for analyzing deep sequencing expression data: constructing the human and mouse promoterome with deepCAGE data. Genome Biol. 10: R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batut P. J., Gingeras T. R., 2013. RAMPAGE: Promoter Activity Profiling by Paired-End Sequencing of 5’-complete cDNAs. Curr. Protoc. Mol. Biol. 104: Unit 25B.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batut P. J., Dobin A., Plessy C., Carninci P., Gingeras T. R., 2012. High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression. Genome Res. 23: 169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benaglia T., Chauveau D., Hunter D., 2009. mixtools: an R package for analyzing finite mixture models. J. Stat. Softw. 32: 1–29. [Google Scholar]
- Boley N., Stoiber M. H., Booth B. W., Wan K. H., Hoskins R. A., et al. , 2014. Genome-guided transcript assembly by integrative analysis of RNA sequence data. Nat. Biotechnol. 32: 341–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J. E. F., Kadonaga J. T., 2002. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 16: 2583–2592. [DOI] [PubMed] [Google Scholar]
- Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., et al. , 2006. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 38: 626–635. [DOI] [PubMed] [Google Scholar]
- Chen Z.-X., Sturgill D., Qu J., Jiang H., Park S., et al. , 2014. Comparative validation of the D. melanogaster modENCODE transcriptome annotation. Genome Res. 24: 1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbourne J. K., Pfrender M. E., Gilbert D., Thomas W. K., Tucker A., et al. , 2011. The ecoresponsive genome of Daphnia pulex. Science 331: 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma M. P., 2002. Ordered recruitment: gene-specific mechanism of transcription activation. Mol. Cell 10: 227–236. [DOI] [PubMed] [Google Scholar]
- Crane R., Gadea B., Littlepage L., Wu H., Ruderman J. V., 2004. Aurora A, meiosis and mitosis. Biol. Cell 96: 215–229. [DOI] [PubMed] [Google Scholar]
- Djebali S., Kapranov P., Foissac S., Lagarde J., Reymond A., et al. , 2008. Efficient targeted transcript discovery via array-based normalization of RACE libraries. Nat. Methods 5: 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down T. A., Bergman C. M., Su J., Hubbard T. J. P., 2007. Large-scale discovery of promoter motifs in Drosophila melanogaster. PLOS Comput. Biol. 3: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert, D., 2005 Ecology, Epidemiology, and Evolution of Parasitism in Daphnia. Available at: http://www.ncbi.nlm.nih.gov/books/NBK2036/. Accessed: April 12, 2016.
- Edgar R., Domrachev M., Lash A. E., 2002. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FANTOM Consortium and the RIKEN PMI and CLST (DGT) , 2014. A promoter-level mammalian expression atlas. Nature 507: 462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald P. C., Shlyakhtenko A., Mir A. A., Vinson C., 2004. Clustering of DNA sequences in human promoters. Genome Res. 14: 1562–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald P. C., Sturgill D., Shyakhtenko A., Oliver B., Vinson C., 2006. Comparative genomics of Drosophila and human core promoters. Genome Biol. 7: R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith M. C., Valen E., Krogh A., Hayashizaki Y., Carninci P., et al. , 2008. A code for transcription initiation in mammalian genomes. Genome Res. 18: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorr T. A., Rider C. V., Wang H. Y., Olmstead A. W., LeBlanc G. A., 2006. A candidate juvenoid hormone receptor cis-element in the Daphnia magna hb2 hemoglobin gene promoter. Mol. Cell. Endocrinol. 247: 91–102. [DOI] [PubMed] [Google Scholar]
- Gupta S., Stamatoyannopoulos J. A., Bailey T. L., Noble W. S., 2007. Quantifying similarity between motifs. Genome Biol. 8: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag C. R., McTaggart S. J., Didier A., Little T. J., Charlesworth D., 2009. Nucleotide polymorphism and within-gene recombination in Daphnia magna and D. pulex, two cyclical parthenogens. Genetics 182: 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V., Li N., Hadzhiev Y., Plessy C., Previti C., et al. , 2014. Two independent transcription initiation codes overlap on vertebrate core promoters. Nature 507: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V., Forrest A. R., Hayashizaki Y., Carninci P., Lenhard B., 2015. CAGEr: precise tss data retrieval and high-resolution promoterome mining for integrative analyses. Nucleic Acids Res. 43: e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson S. J., Stelzer C.-P., Welch D. B. M., Logsdon J. M., 2013. Comparative transcriptome analysis of obligately asexual and cyclically sexual rotifers reveals genes with putative functions in sexual reproduction, dormancy, and asexual egg production. BMC Genomics 14: 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P. D. N., 1981. Obligate asexuality in Daphnia. Am. Nat. 117: 784–789. [Google Scholar]
- Hedges S. B., Dudley J., Kumar S., 2006. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22: 2971–2972. [DOI] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., et al. , 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix D. A., Hong J.-W., Zeitlinger J., Rokhsar D. S., Levine M. S., 2008. Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 105: 7762–7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiruta C., Nishida C., Tochinai S., 2010. Abortive meiosis in the oogenesis of parthenogenetic Daphnia pulex. Chromosome Res. 18: 833–840. [DOI] [PubMed] [Google Scholar]
- Homer H., Gui L., Carroll J., 2009. A spindle assembly checkpoint protein functions in prophase I arrest and prometaphase progression. Science 326: 991–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins R. A., Hoskins R. A., Landolin J. M., Landolin J. M., Brown J. B., et al. , 2011. Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res. 21: 182–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W., Carey V. J., Gentleman R., Anders S., Carlson M., et al. , 2015. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12: 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst L. D., Pál C., Lercher M. J., 2004. The evolutionary dynamics of eukaryotic gene order. Nat. Rev. Genet. 5: 299–310. [DOI] [PubMed] [Google Scholar]
- Jin F., Hamada M., Malureanu L., Jeganathan K. B., Zhou W., et al. , 2010. Cdc20 Is Critical for meiosis I and fertility of female mice. PLoS Genet. 6: e1001147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T., Kadonaga J. T., 2010. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 339: 225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J. T., 2012. Perspectives on the RNA polymerase II core promoter. Wiley Interdiscip. Rev. Dev. Biol. 1: 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M., 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44: D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H., Frith M. C., Katayama S., Sandelin A., Kai C., et al. , 2006. Dynamic usage of transcription start sites within core promoters. Genome Biol. 7: R118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilham S. S., Kreeger D. A., Lynn S. G., Goulden C. E., Herrera L., 1998. COMBO: a defined freshwater culture medium for algae and zooplankton. Hydrobiologia 377: 147–159. [Google Scholar]
- Kim T.-K., Hemberg M., Gray J. M., Costa A. M., Bear D. M., et al. , 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., Wakamatsu A., Suzuki Y., Ota T., Nishikawa T., et al. , 2006. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 16: 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodzius R., Kojima M., Nishiyori H., Nakamura M., Fukuda S., et al. , 2006. CAGE: cap analysis of gene expression. Nat. Methods 3: 211–222. [DOI] [PubMed] [Google Scholar]
- Kurosawa J., Nishiyori H., Hayashizaki Y., 2011. Deep cap analysis of gene expression. Methods Mol. Biol. 687: 147–163. [DOI] [PubMed] [Google Scholar]
- Law C. W., Chen Y., Shi W., Smyth G. K., 2014. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15: R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M., Morgan M., 2014. Scalable genomics with R and bioconductor. Stat. Sci. 29: 214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard B., Sandelin A., Carninci P., 2012. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat. Rev. Genet. 13: 233–245. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. 1000 Genome Project Data Processing Subgroup , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Spitze K., Crease T., 1989. The distribution of life-history variation in the Daphnia pulex complex. Evolution 43: 1724–1736. [DOI] [PubMed] [Google Scholar]
- Mahony S., Benos P. V., 2007. STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 35: W253–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manak J. R., Dike S., Sementchenko V., Kapranov P., Biemar F., et al. , 2006. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat. Genet. 38: 1151–1158. [DOI] [PubMed] [Google Scholar]
- Matthews B. B., dos Santos G., Crosby M. A., Emmert D. B., St Pierre S. E., et al. , 2015. Gene model annotations for Drosophila melanogaster: impact of high-throughput data. G3 (Bethesda) 5: 1721–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata M., Nishiyori-Sueki H., Kojima-Ishiyama M., Carninci P., Hayashizaki Y., et al. , 2014. Detecting expressed genes using CAGE, pp. 67–85 in Transcription Factor Regulatory Networks. Springer, New York. [DOI] [PubMed] [Google Scholar]
- Nepal C., Hadzhiev Y., Previti C., Haberle V., Li N., et al. , 2013. Dynamic regulation of the transcription initiation landscape at single nucleotide resolution during vertebrate embryogenesis. Genome Res. 23: 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler, U., G.-C. Liao, H. Niemann, and G. M. Rubin, 2002 Computational analysis of core promoters in the Drosophila genome. Genome Biol. 3: 0087.1–0087.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahlavan G., Polanski Z., Kalab P., Golsteyn R., Nigg E. A., et al. , 2000. Characterization of polo-like kinase 1 during meiotic maturation of the mouse oocyte. Dev. Biol. 220: 392–400. [DOI] [PubMed] [Google Scholar]
- Portales-Casamar E., Thongjuea S., Kwon A. T., Arenillas D., Zhao X., et al. , 2009. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 38: D105–D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., 2014. BEDTools: the swiss-army tool for genome feature analysis. Curr. Protoc. Bioinformatics 47: 11.12.1–11.12.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rach E. A., Yuan H.-Y., Majoros W. H., Tomancak P., Ohler U., 2009. Motif composition, conservation and condition-specificity of single and alternative transcription start sites in the Drosophila genome. Genome Biol. 10: R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., et al. , 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T. L., Hashimoto S., Gu S. G., Morton J. J., Stadler M., et al. , 2013. The transcription start site landscape of C. elegans. Genome Res. 23: 1348–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A., Carninci P., Lenhard B., Ponjavic J., Hayashizaki Y., et al. , 2007. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat. Rev. Genet. 8: 424–436. [DOI] [PubMed] [Google Scholar]
- Sardar A. J., Oates M. E., Fang H., Forrest A. R., Kawaji H., et al. , 2014. The evolution of human cells in terms of protein innovation. Mol. Biol. Evol. 31: 1364–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxonov S., Berg P., Brutlag D. L., 2006. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. USA 103: 1412–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schult, D. A., and P. Swart, 2008 Exploring network structure, dynamics, and function using NetworkX, pp. 11–16 in 7th Python in Science Conferences (SciPy), edited by G. Varoquaux, T. Vaught, and J. Millman. Pasadena, CA. [Google Scholar]
- Schurko A. M., Logsdon J. M., Eads B. D., 2009. Meiosis genes in Daphnia pulex and the role of parthenogenesis in genome evolution. BMC Evol. Biol. 9: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon C. E., 1948. A mathematical theory of communication. Bell Syst. Tech. J. 27: 379–423, 623–656. [Google Scholar]
- Takahashi H., Kato S., Murata M., Carninci P., 2012a CAGE (cap analysis of gene expression): a protocol for the detection of promoter and transcriptional networks. Methods Mol. Biol. 786: 181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Lassmann T., Murata M., Carninci P., 2012b 5′ end–centered expression profiling using cap-analysis gene expression and next-generation sequencing. Nat. Protoc. 7: 542–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker A. E., Ackerman M. S., Eads B. D., Xu S., Lynch M., 2013. Population-genomic insights into the evolutionary origin and fate of obligately asexual Daphnia pulex. Proc. Natl. Acad. Sci. USA 110: 15740–15745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes, G. R., B. Bolker, L. Bonebakker, R. Gentleman, W. H. A. Liaw, et al., 2015 gplots: Various R Programming Tools for Plotting Data. R package version 2.17.0. Available at: https://CRAN.R-project.org/package=gplots. Accessed: February 1, 2016.
- Woo Y. H., Li W.-H., 2011. Gene clustering pattern, promoter architecture, and gene expression stability in eukaryotic genomes. Proc. Natl. Acad. Sci. USA 108: 3306–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin T., Cook D., Lawrence M., 2012. Ggbio: an r package for extending the grammar of graphics for genomic data. Genome Biol. 13: R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data from this manuscript were deposited in the NCBI Gene Expression Omnibus (Edgar et al. 2002) (http://www.ncbi.nlm.nih.gov/geo) and are accessible through GEO Series accession number GSE80141 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE80141) with the BioProject identifier of PRJNA318020. Sequence data were deposited in the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) and have been assigned the accession number SRP073105.