Abstract
Oxidative stress has a critical role in the pathogenesis of vitiligo. However, the specific molecular mechanism involved in oxidative stress-induced melanocyte death is not well characterized. Given the powerful role of microRNAs (miRNAs) in the regulation of cell survival as well as the fact that the generation of miRNAs can be affected by oxidative stress, we hypothesized that miRNAs may participate in vitiligo pathogenesis by modulating the expression of vital genes in melanocytes. In the present study, we initially found that miR-25 was increased in both serum and lesion samples from vitiligo patients, and its serum level was correlated with the activity of vitiligo. Moreover, restoration of miR-25 promoted the H2O2-induced melanocyte destruction and led to the dysfunction of melanocytes. Further experiments proved that MITF, a master regulator in melanocyte survival and function, accounted for the miR-25-caused damaging impact on melanocytes. Notably, other than the direct role on melanocytes, we observed that miR-25 inhibited the production and secretion of SCF and bFGF from keratinocytes, thus impairing their paracrine protective effect on the survival of melanocytes under oxidative stress. At last, we verified that oxidative stress could induce the overexpression of miR-25 in both melanocytes and keratinocytes possibly by demethylating the promoter region of miR-25. Taken together, our study demonstrates that oxidative stress-induced overexpression of miR-25 in vitiligo has a crucial role in promoting the degeneration of melanocytes by not only suppressing MITF in melanocytes but also impairing the paracrine protective effect of keratinocytes. Therefore, it is worthy to investigate the possibility of miR-25 as a potential drug target for anti-oxidative therapy in vitiligo.
Vitiligo is a disfiguring dermatosis with an incidence rate of approximately 0.5–1.0% in the populations worldwide.1 Characterized by patchy depigmentation of the skin, the disease can affect the patients' self-image, or even cause depression, thus substantially decreasing life quality among vitiligo patients.2 Although several etiological theories, including genetic predisposition,3, 4, 5 autoimmunity,6, 7 melanocytorrhagy8, 9 and toxic metabolites10 have been proposed to participate in the pathogenesis of vitiligo, the exact mechanism of melanocyte degeneration in depigmented lesions still remains unclear.
The generation of oxidative stress has long been demonstrated to have a crucial role in the onset and progression of vitiligo.11, 12 Owing to the pro-oxidant state generated during melanin synthesis, melanocytes are particularly vulnerable to oxidative stress.13 In vitiligo, accumulation of toxic intermediates such as 6- and 7-BH4 and catecholamine,14, 15 concomitant with reduced levels and activity of catalase and several other antioxidant enzymes16, 17, 18 have been demonstrated in patients' epidermis. Because of these intracellular metabolic disorder and compromised intrinsic antioxidant defenses, hydrogen peroxide (H2O2) and other reactive oxygen species (ROS) accumulate in melanocytes,19 which causes DNA damage and the peroxidation of lipid and protein,20, 21, 22, 23, 24 thus disrupting the homeostasis of melanocytes. However, the specific molecular mechanism involved in oxidative stress-induced melanocyte death in vitiligo is not fully clarified, which impedes the discovery of novel drug target for the antioxidant therapy in vitiligo.
MicroRNAs (miRNAs) are a class of noncoding RNAs that suppress gene expression by targeting messenger RNAs for translational repression or, less frequently, degradation.25, 26 It is known that miRNAs can regulate the growth, differentiation and even death of cells, and the aberrant expression of miRNAs has been found to be involved in the pathogenesis of many diseases.27 Recently, several studies have found that the expression profile of miRNAs could be regulated by oxidative stress and mediate the pathogenic effect of ROS in some diseases and animal models, which deepens the understanding of redox biology and implicates new therapeutic strategies for oxidative stress-related diseases.28, 29, 30 Given this crosstalk between redox signaling and miRNAs, we hypothesized that enhanced oxidative stress in vitiligo could induce the aberrant expression of miRNAs, which promotes the degeneration of melanocytes.
To test our hypothesis, a microarray analysis was initially conducted to identify alterations in the miRNAome of vitiligo patients. We discovered that miR-25, one of the most highly elevated miRNAs in vitiligo serum, had a significant correlation with the disease activity and also increased in vitiligo lesion. Subsequent experiments revealed the regulatory effect of miR-25 on the destruction of melanocytes under oxidative stress and the dysfunction of melanocytes, which implicated an important role of miR-25 in the pathogenesis of vitiligo. Meanwhile, the production of miR-25 and the mechanism involved were also investigated in the present study.
Results
Characteristic alterations in the vitiligo miRNAome
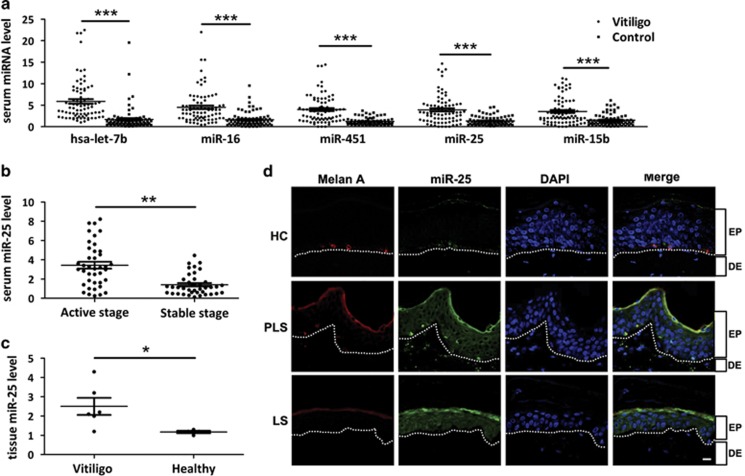
In view of the possibility to use serum miRNAs as disease biomarkers,31 we performed a miRNA microarray study using serum samples from vitiligo patients (n=50) and healthy controls (n=50). The assay identified 48 aberrantly expressed miRNAs (>twofold changes, P<0.01), among which 24 miRNAs (e.g., let-7b) were upregulated, whereas another 24 (e.g., miR-1914) were downregulated (Supplementary Table S1). We examined the five highest increased miRNAs (let-7b, miR-16, miR-451, miR-25 and miR-15b) in serum samples from vitiligo patients (n=80) and healthy controls (n=80) using real-time quantitative reverse transcription-PCR (qRT-PCR). Consistent with the microarray data, all the five miRNAs are significantly (P<0.001) upregulated in vitiligo (Figure 1a).
Figure 1.
Characteristic alteration in the miRNAome of vitiligo serum and skin specimens. (a) The five miRNAs were analyzed using qRT-PCR in the serum of 80 healthy individuals and 80 patients with vitiligo. ***P<0.001. (b) Serum miR-25 level of patients at active stage (n=40) versus stable stage (n=40) of vitiligo is measured by qRT-PCR. **P<0.01. (c) miR-25 expression was analyzed in healthy skin (n=4) and vitiligo lesion (n=6) using qRT-PCR. *P<0.05. (d) FISH revealed endogenous miR-25 expression (green) in vitiligo lesional skin (LS), perilesional skin (PLS) and healthy control (HC). Melanocytes in the epidermis were demonstrated by immune staining with antibody to Melan-A (red). Nuclei were counterstained with DAPI (blue). Data are representative of three biological replicates. Scale bar=50 μm. EP, epidermis; DE, dermis
Among the five highest increased miRNAs, only miR-25 showed higher level in the serum samples from patients in active stage (n=40) than those in stable stage (n=40, P<0.01), while the other four miRNAs showed no difference (Figure 1b and Supplementary Figure S1). We then tested miR-25 level in vitiligo lesion (n=6) and healthy controls (n=4). In accordance with its serum level, miR-25 was significantly (P<0.05) elevated in vitiligo lesion (Figure 1c).
To clarify the location of increased miR-25 in vitiligo lesion, we collected lesional and perilesional skin from vitiligo patients as well as normal skin from healthy individuals, and detected miR-25 by fluorescence in situ hybridization (FISH). As a result, miR-25 is expressed primarily in melanocytes (marked by Melan-A) at the basal layer in healthy epidermis, while the whole epidermis layer of lesional and perilesional skin, most of which are obviously keratinocytes, exhibited much enhanced miR-25 expression. Notably, melanocytes in perilesional skin showed intensely upregulated expression of miR-25 possibly because of some etiological factors like oxidative stress that already existed at the edge of vitiligo lesion (Figure 1d).
MiR-25 promotes the destruction of melanocytes under oxidative stress
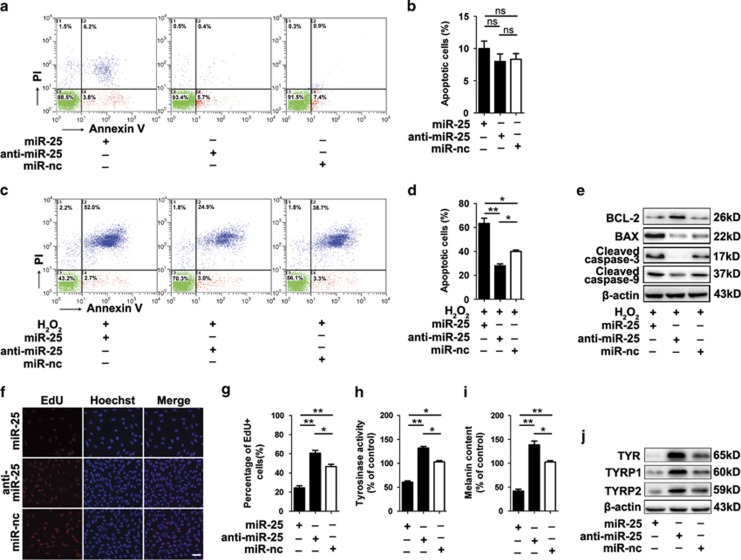
To investigate whether increased miR-25 is responsible for melanocyte loss in vitiligo, normal human melanocytes were transfected with miR-25 mimics, miR-25 inhibitor and negative control and examined by annexin V-FITC/PI staining. However, there was no significant distinction among their apoptosis rates (Figures 2a and b). As oxidative stress is a driven factor for melanocyte loss in vitiligo, we treated those cells with H2O2 (a main form of oxidative stress in vitiligo) and found that the percentage of apoptotic cells was decreased with miR-25 knockdown (28.7 versus 41%), whereas overexpressed miR-25 elevated apoptosis level (65 versus 41%) (Figures 2c and d). We further found that overexpressed miR-25 reduced the level of anti-apoptotic B-cell lymphoma 2 (Bcl-2) but upregulated some pro-apoptotic proteins, including B-cell lymphoma 2-associated X protein (Bax), the active cleavages of caspase-3 and caspase-9, whereas anti-miR-25 treatment presented with the opposite effect (Figure 2e). These results indicated that miR-25 could facilitate melanocyte apoptosis under oxidative stress.
Figure 2.
MiR-25 promotes melanocyte apoptosis under oxidative stress and leads to the dysfunction of melanocytes. (a–d) Normal human melanocytes were pre-transfected by miR-25 mimics, miR-25 inhibitor and negative control (miR-nc) for 48 h. The apoptosis level of melanocytes with or without the treatment of 400 μM H2O2 for 24 h was examined by annexin V-FITC/PI staining. Bar graphs represent the mean values of flow cytometry data (B corresponding to A, D corresponding to C, n=3). *P<0.05, **P<0.01. (e) The level of apoptosis-related proteins was detected by western blotting, and the results for the protein collected at 24 h after H2O2 treatment are shown. Data are representative of three independently performed experiments. β-Actin was detected as loading control. (f, g) Detection of EdU add-in cells (red) at 48 h after transfection. Nuclei were counterstained with Hoechst (blue). Scale bar=50 μm. Bar graphs represent the mean values of five wells that accepted the same treatment. *P<0.05, **P<0.01. (h) Tyrosinase activity and (i) melanin content in melanocytes with different transfections were measured using an ELISA reader. Mean±S.D. is shown. *P<0.05, **P<0.01. (j) The level of TYR, TYRP1and TYRP2 in melanocytes with different transfections was detected by western blotting. Data are representative of three independently performed experiments. β-Actin was detected as loading control
Moreover, EdU cell proliferation assay was adopted in our study, and the results showed that the EdU incorporation rate (percentage of cells that undergo cell division) was reduced (P<0.01) in the cells overexpressing miR-25, whereas anti-miR-25 treatment increased the number of EdU-positive cells (P<0.05) (Figures 2f and g).
MiR-25 leads to the dysfunction of melanocytes
The melanin synthesis in melanocytes has been reported to be impaired in vitiligo.12 In our study, we found that in melanocytes, overexpressed miR-25 inhibited the activity of tyrosinase (TYR) that is the rate-limiting enzyme in melanogenesis (Figure 2h) and significantly decreased the melanin content (Figure 2i). Along with TYR, tyrosinase-related protein 1 (TYRP1) and tyrosinase-related protein 2 (TYRP2) are also crucial genes in melanin biosynthesis. Using western blotting, we observed the reduction of TYR, TYRP1 and TYRP2 protein level in melanocytes transfected with miR-25 mimics (Figure 2j). These results indicated that miR-25 suppresses the expression of TYR, TYRP1 and TYRP2, and decreases the activity of tyrosinase, thereby inhibiting melanin biosynthesis.
We further investigated whether miR-25 affected the distribution of melanosomes by detecting gp100 (a marker for melanosomes). As shown in Supplementary Figure S2, melanosomes were mainly located in the periphery and the dendrites of melanocytes, whereas in cells transfected with miR-25 mimics, we observed a perinuclear aggregation of melanosomes.
MITF are required for miR-25-induced destruction and dysfunction of melanocytes
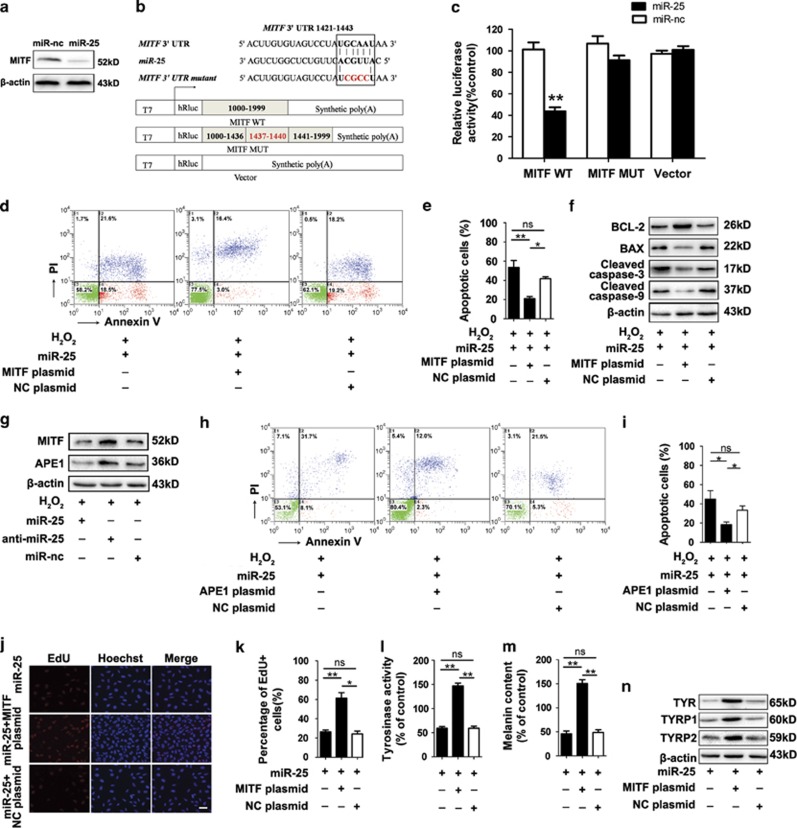
Three bioinformatics tools including miRanda, PICTAR5 and Targetscan were used to screen the potential targets of miR-25 that may explain its function in melanocytes (Supplementary Figure S3), and microphthalmia-associated transcription factor (MITF) was selected out for its key role in melanocyte survival and function. We tested MITF level in melanocytes transfected with miR-25 mimics and found that overexpressed miR-25 reduced the expression of MITF at protein level but not at mRNA level (Figure 3a and Supplementary Figure S4a).
Figure 3.
MITF is required for miR-25-induced destruction and dysfunction of melanocytes. (a) The level of MITF in melanocytes transfected by miR-25 mimics or negative control (miR-nc) is detected by western blotting. β-Actin was detected as loading control. Data are representative of three independently performed experiments. (b) The sequences of human miR-25 and its complementary sequence in mRNA 3′UTR of MITF WT and MITF mutant are shown on the top, in which the letters in red represent the four nucleotides we mutated. The structure of the reporter plasmids named MITF WT and MITF MUT are shown below. (c) Normal human melanocytes were co-transfected by MITF WT, MITF MUT or the control vector with miR-25 mimics or negative control for 48 h. Firefly luciferase levels were then detected. Data are expressed as the ratio of luciferase activity detected in cells transfected by miR-25 mimics to that in negative control-treated cells. **P<0.01. (d, e) Normal human melanocytes were transfected by miR-25 mimics with MITF plasmids or control plasmids (NC-plasmids). The apoptosis level of melanocytes after exposure to 400 μM H2O2 for 24 h was examined by annexin V-FITC/PI staining. Bar graphs represent the mean values of flow cytometry data (n=3). *P<0.05, **P<0.01. (f) The level of apoptosis-related proteins was detected by western blotting and the results for the protein collected at 24 h after H2O2 treatment are shown. Data are representative of three independently performed experiments. β-Actin was detected as loading control. (g) The level of MITF and APE1 was detected by western blotting and the results for the protein collected at 24 h after H2O2 treatment are shown. Data are representative of three independently performed experiments. β-Actin was detected as loading control. (h, i) Normal human melanocytes were transfected by miR-25 mimics with APE1 plasmids or control plasmids (NC-plasmids). The apoptosis level of melanocytes after exposure to 400 μM H2O2 for 24 h was examined by annexin V-FITC/PI staining. Bar graphs represent the mean values of flow cytometry data (n=3). *P<0.05. (j, k) Detection of EdU add-in cells (red) at 48 h after transfection. Nuclei were counterstained with Hoechst (blue). Scale bar=50 μm. Bar graphs represent the mean values of five wells that accepted the same treatment. *P<0.05, **P<0.01. (l) Tyrosinase activity and (m) melanin content in melanocytes following transfection was measured using an ELISA reader. Mean±S.D. is shown. **P<0.01. (n) The level of TYR, TYRP1 and TYRP2 in melanocytes with different transfections was detected by western blotting. Data are representative of three independently performed experiments. β-Actin was detected as loading control
We performed luciferase reporter assays to verify whether miR-25 repressed MITF expression by targeting the predicted sequences in the 3′ untranslated regions (3′UTR) of MITF mRNA. To this end, part of the 3′UTR (1000 nucleotides) with a putative binding site for miR-25 in the middle was inserted into the reporter plasmids and named MITF WT. Furthermore, we mutated four nucleotides within the seed-matching sequences in plasmids named MITF MUT (Figure 3b). All the reporter plasmids were co-transfected into melanocytes with miR-25 mimics or negative control, and luciferase activity was analyzed 48 h after transfection. In the cells transfected with MITF WT, miR-25 reduced the luciferase activity (P<0.01). However, miR-25 mimics failed to influence the reporter activity of MITF MUT (Figure 3c). These results indicated that miR-25 directly regulates MITF expression by binding to the predicted target sites in the 3′UTR of its mRNAs.
The inhibition of miR-25 on MITF expression encouraged us to investigate whether the decreased MITF was required for the effect of miR-25 in melanocytes. To test this, melanocytes overexpressing miR-25 were co-transfected with 3′UTR-deleted MITF plasmids or control plasmids (transfection efficiency shown in Supplementary Figure S5), and flow cytometry was used to evaluate cell apoptosis following H2O2 treatment. The re-introduction of MITF decreased the percentage of apoptotic cells (P<0.05) (Figures 3d and e), and did invert the downregulation of anti-apoptotic BCL2 and the upregulation of pro-apoptotic Bax, cleaved caspase 3 and caspase 9 induced by miR-25 in melanocytes (Figure 3f).
According to previous cytometry analysis, the overexpression of miR-25 alone does not affect melanocyte apoptosis (Figures 2a and b), thus we speculated that its suppression on MITF promoted ROS-induced melanocyte apoptosis by inhibiting the antioxidant response. As a transcriptional target for MITF,32 apurinic/apyrimidinic endonuclease (APE1) can promote intracellular antioxidant defenses by enhancing the DNA-binding activities of stress-inducible transcription factors, including p53,33 hypoxia-inducible factor-1 (HIF-1),34 activator protein-1 (AP-1)35 and antioxidant factors like nuclear factor-erythroid-related factor 2 (NRF2).36 Besides, using siRNA to deplete APE1 could promote the apoptosis of cells under oxidative stress.37 On the basis of these findings, we conducted western blotting analysis and found that miR-25 simultaneously downregulated the expression of both MITF and APE1 with or without the stimulation of H2O2 treatment (Figure 3g and Supplementary Figure S6). We then found that the transfection of APE1 plasmids into melanoctyes overexpressing miR-25 (transfection efficiency shown in Supplementary Figure S7) significantly reduced the miR-25-promoted apoptosis rate of melanocytes under oxidative stress (P<0.05) (Figures 3h and i). Moreover, we detected the mRNA level of heme oxygenase 1 (HO-1), superoxide dismutase 1 (SOD1), catalase (CAT), glutathione peroxidase 1 (GPX1) and NAD(P)H dehydrogenase, quinone 1 (NQO1), five antioxidant genes downstream of NRF2,38 in melanocytes transfected by miR-25 mimics, miR-25 inhibitor or negative control. The results showed that the mRNA levels of HO-1 and GPX1 were suppressed by miR-25 in melanocytes with or without the stimulation of H2O2 (Supplementary Figure S8). Taken together, these findings suggested that miR-25 could suppress the antioxidant ability of melanocytes through inhibiting the MITF-APE1 pathway, thus making melanocytes more susceptible to oxidative stress-induced apoptosis.
We went on to investigate whether miR-25 regulated melanocyte proliferation and function through its inhibition on MITF. The re-introduction of MITF promoted the proliferation of melanocytes transfected with miR-25 mimics (Figures 3j and k), and successfully upregulated the activity of tyrosinase and the melanin content in melanocytes compared with control (Figures 3l and m). Further western blot assay showed the restoring expression of TYR, TYRP1 and TYRP2 after re-introduction of MITF in melanocytes overexpressing miR-25 (Figure 3n). In addition, transfection with MITF plasmids in melanocytes overexpressing miR-25 facilitated the re-location of melanosomes in the periphery (Supplementary Figure S2).
MiR-25 inhibits the production and secretion of growth factors in keratinocytes
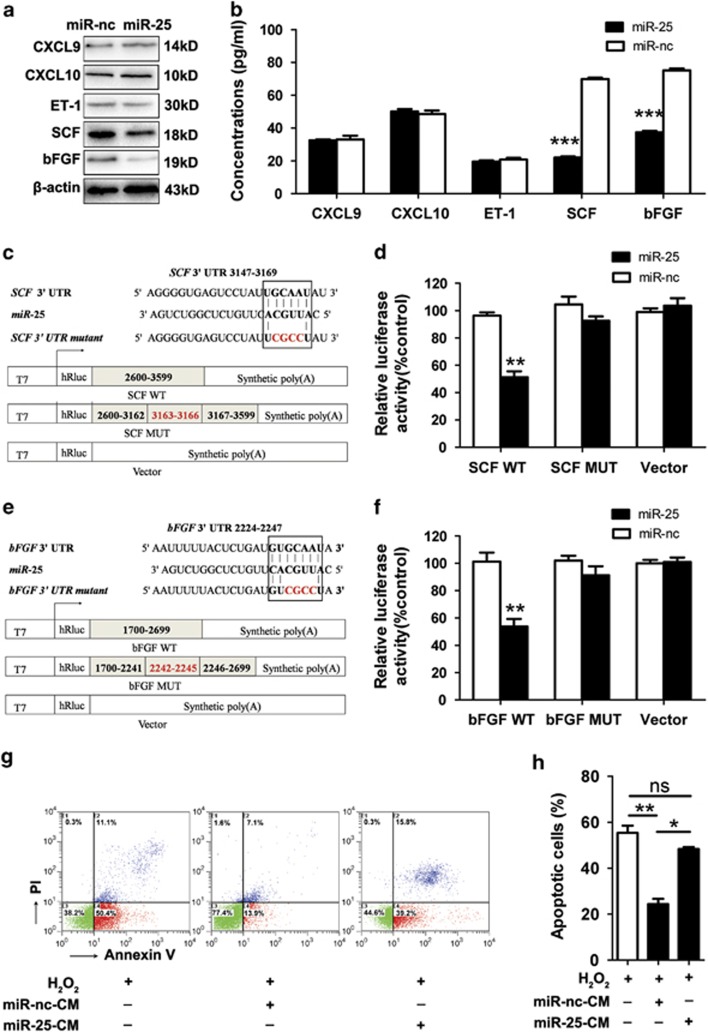
Studies have found that keratinocytes regulate the proliferation and survival of melanocytes through paracrine mechanism. To investigate the role of keratinocytes overexpressing miR-25 in vitiligo, we treated normal human keratinocytes with miR-25 mimics and tested both the expression and the secretion level of some cytokines and growth factors that have been reported to relate with vitiligo development.39, 40 Decreased level of of stem cell factor (SCF) and basic fibroblast growth factor (bFGF) were observed in keratinocytes treated with miR-25 mimics. However, the level of endothelin-1 (ET-1), CXCL9 or CXCL10 seemed to be independent on miR-25 in keratinocytes (Figures 4a and b).
Figure 4.
MiR-25 downregulates the expression and secretion of SCF and bFGF from human keratinocytes. (a) The protein level of CXCL9, CXCL10, ET-1, SCF and bFGF was detected by western blotting. Data are representative of three independently performed experiments. β-Actin was used as the internal standard. (b) The culture medium of normal human keratinocytes transfected with miR-25 mimics or negative control (miR-nc) was analyzed by ELISA to determine the secretion level of CXCL9, CXCL10, ET-1, SCF and bFGF. Mean±S.D. is shown. ***P<0.001. (c, e) The sequences of human miR-25 and its complementary sequence in mRNA 3′UTR of SCF WT, SCF mutant, bFGF WT and bFGF mutant are shown, in which the letters in red represent the four nucleotides we mutated in each gene. The structure of the reporter plasmids named SCF WT, SCF MUT, bFGF WT and bFGF MUT are shown below. (d, f) SCF WT, SCF MUT, bFGF WT, bFGF MUT, and the empty vectors were individually transfected into normal human keratinocytes along with miR-25 mimics or negative control. Forty-eight hours after transfection, firefly luciferase levels were detected. Data are expressed as the ratio of luciferase activity detected in cells transfected by miR-25 mimics to that in negative control-treated cells. **P<0.01. (g, h) Determination of 400 μM H2O2-induced apoptosis for melanocytes treated with serum-free media supplemented by culture medium (CM) that was derived from keratinocytes pre-transfected by miR-25 mimics or negative control. Bar graphs represent the mean values of flow cytometry data (n=3). *P<0.05, **P<0.01
According to bioinformatics tools, SCF and bFGF are putative target genes of miR-25. Although their protein levels were suppressed by miR-25 as previously described, their mRNA levels were not affected by miR-25 in keratinocytes (Supplementary Figures S4b and S4c), which suggested that miR-25 regulated SCF and bFGF at the posttranscriptional level. We then performed luciferase reporter assays to verify whether miR-25 represses the expression of the two genes by targeting the predicted sequences in their 3′UTR. Reporter plasmids named SCF WT, SCF MUT, bFGF WT and bFGF MUT were constructed in the same way as described before (Figures 4c and e), and co-transfected with miR-25 mimics or negative control into normal human keratinocytes. It turned out that miR-25 reduced the luciferase activity in cells transfected with SCF WT or bFGF WT (P<0.01), but did not influence the reporter activity of SCF MUT and bFGF MUT (Figures 4d and f). Therefore, miR-25 suppresses the level of SCF and bFGF by binding to the predicted target sites in the 3′UTR of their mRNAs.
We further treated melanocytes with serum-free media supplemented by culture medium derived from keratinocytes to investigate whether miR-25 influenced the paracrine regulation of keratinocytes on melanocytes. We found that culture medium from miR-25-overexpressing keratinocytes failed to protect melanocytes from H2O2-induced apoptosis, whereas in the control group, the protective effect was obvious (Figures 4g and h). Collectively, these results suggested that miR-25 may interfere with the production and the secretion of SCF or bFGF from keratinocytes, thus facilitating melanocyte loss under oxidative stress.
Oxidative stress induces the overexpression of miR-25 in melanocytes and keratinocytes by promoting DNA demethylation
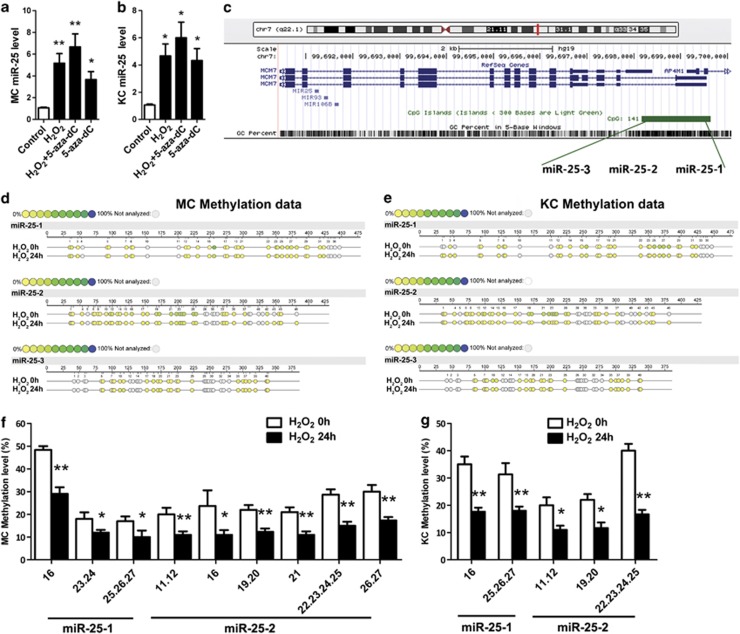
As oxidative stress has been demonstrated to affect miRNA expression, we speculated that the increased miR-25 in both melanocytes and keratinocytes that we observed in previous FISH analysis might result from enhanced oxidative stress in vitiligo. Our qRT-PCR analysis showed that miR-25 was increased in both melanocytes (P<0.01) and keratinocytes (P<0.05) following H2O2 stimulation (Figures 5a and b). Previous studies have reported that oxidative stress can regulate gene expression by promoting DNA demethylation.41 We thus adopted the DNA demethylating agent 5-aza-2′-deoxycytidine (5-aza-dC) to test whether such an epigenetic regulation was involved in miR-25 expression. 5-Aza-dC upregulated the level of miR-25 (P<0.05) (Figures 5a and b), and the maximum increase occurred when the cells were treated with H2O2 and 5-aza-dC simultaneously (P<0.01 for melanocytes and P<0.05 for keratinocytes).
Figure 5.
Oxidative stress induces the overexpression of miR-25 and demethylates the promoter region of miR-25 in melanocytes and keratinocytes. (a, b) MiR-25 expression was analyzed in normal human melanocytes (a) and keratinocytes (b) exposed to H2O2 (400 μM for melanocytes and 1000 μM for keratinocytes), 5-aza-dC (5 mM) or both for 24 h and control groups using qRT-PCR. *P<0.05, **P<0.01. (c) Schematic diagram of human miR-25 promoter. MiR-25 is located within intron 13 of the minichromosome maintenance protein 7 (MCM7) gene (the red line) on chromosome 7q22.1. The location of promoter region of miR-25 are indicated by green bar. The distal, central and proximal DNA fragments of the promoter sequences were separated and labeled as miR-25-1, miR-25-2 and miR-25-3, respectively. (d–g) The methylation levels of CpG sites in miR-25 promoter regions from H2O2 groups (melanocytes exposed to 400 μM H2O2 for 24 h and keratinocytes exposed to 1000 μM H2O2 for 24 h) and control groups were detected and compared. The results of quantitative methylation analysis are shown in a color scale: yellow (0% methylation), green (50% methylation) and dark blue (100% methylation). The white circles represent the missing data at a given CpG site. The below bar graphs represent the mean methylation levels of CpG sites, in which only those had a significant decrease of methylation level after the treatment of H2O2 are shown. The data are expressed as mean±S.D. *P<0.05, **P<0.01. MC, melanocyte; KC, keratinocyte
We went on to investigate whether H2O2 demethylated the promoter region of miR-25. Genomic DNA was extracted from melanocytes and keratinocytes with or without H2O2 treatment and subjected to bisulfite sequencing analysis. The distal, central and proximal DNA fragments of the promoter sequences of miR-25 were separated and labeled as miR-25-1, miR-25-2 and miR-25-3 (Figure 5c), with 46, 37 and 41 CpG sites involved, respectively (Supplementary Figure S9). The methylation levels varied at different CpG sites (Figures 5d and e). For melanocytes, the highest methylation level (50%) occurred at the sixteenth CpG site of miR-25-1 (Figure 5f), whereas in keratinocytes the highest methylation level (43%) occurred at the twenty-second to twenty-fifth CpG sites of miR-25-2 (Figure 5g). After H2O2 stimulation, the methylation level of multiple CpG sites in miR-25-1 and miR-25-2 was reduced in both melanocytes and keratinocytes, though there was no change in miR-25-3 (Figures 5f and g).
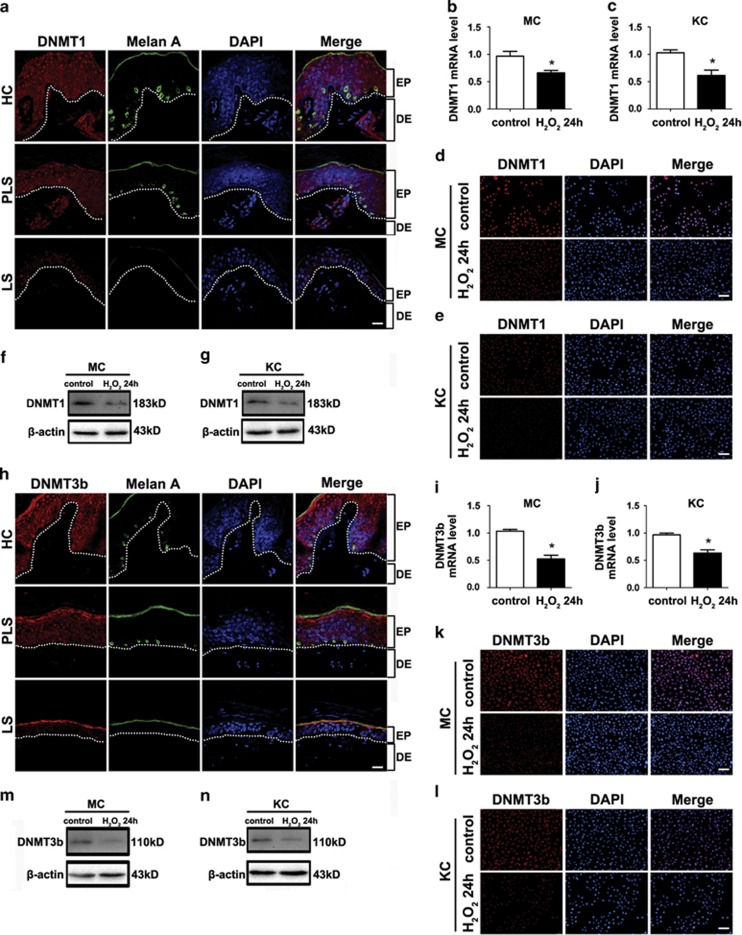
It is reported that in Alzheimer's disease, oxidative stress causes DNA demethylation by decreasing the expression of DNA methyltransferases (DNMTs) possibly through the activation of c-Jun amino terminal kinase (JNK) signaling pathway.41 To investigate the level of DNMTs in vitiligo, immunofluorescence assay was performed using the three kinds of skin specimens as previously described. Although there was no distinction in DNMT3a expression among the three groups (Supplementary Figure S10), DNMT1 and DNMT3b were both positive in the epidermis of healthy skin but absent in lesional and perilesional epidermis (Figures 6a and h), which is opposite of the expression pattern of miR-25 in vitiligo. Given this in vivo phenomenon, the expression of DNMT1 and DNMT3b in melanocytes and keratinocytes under oxidative stress was further evaluated. Both DNMT1 and DNMT3b were downregulated in cells following H2O2 treatment at both mRNA level (P<0.05, Figures 6b, c, i and j) and protein level (Figures 6d and g and k-n). In addition, H2O2 increased the expression of phosphorylated JNK in both melanocytes and keratinocytes, and the adoption of the JNK inhibitor elevated the expression of both DNMT1 and DNMT3b in H2O2-treated melanocytes and keratinocytes (Supplementary Figure S11). Collectively, these results suggested that ROS could reduce the expression of DNMTs by promoting the phosphorylation of JNK in melanocytes and keratinocytes.
Figure 6.
DNMT1 and DNMT3b are downregulated in vitiligo lesion and can be suppressed by oxidative stress in vitro. (a) and (h) The expression and location of DNMT1 and DNMT3b (red) were visualized by immunofluorescence in vitiligo lesional skin (LS), perilesional skin (PLS) and healthy control (HC). Melanocytes in the epidermis were demonstrated by immune staining with antibody to Melan-A (green). Nuclei were counterstained with DAPI (blue). Data are representative of three biological replicates. Scale bar=50 mm. EP, epidermis; DE, dermis (b, c) and (i, j) DNMT1 and DNMT3b mRNAs were analyzed in normal human melanocytes exposed to 400 μM H2O2, normal human keratinocytes and exposed to 1000 μM H2O2 and control groups using qRT-PCR. *P<0.05. (d, e) and (k, l) DNMT1 and DNMT3b expression (each in red) was shown in normal human melanocytes exposed to 400 μM H2O2, normal human keratinocytes exposed to 1000 μM H2O2 and control groups using immunofluorescence. Nuclei were counterstained with DAPI (blue). Data are representative of three biological replicates. MC, melanocyte; KC, keratinocyte. Scale bar=50 mm. (f, g) and (m, n) The protein level of DNMT1 and DNMT3b was analyzed in normal human melanocytes exposed to 400 μM H2O2, normal human keratinocytes and exposed to 1000 μM H2O2 and control groups using western blotting. Data are representative of three independently performed experiments. β-Actin was detected as loading control
Discussion
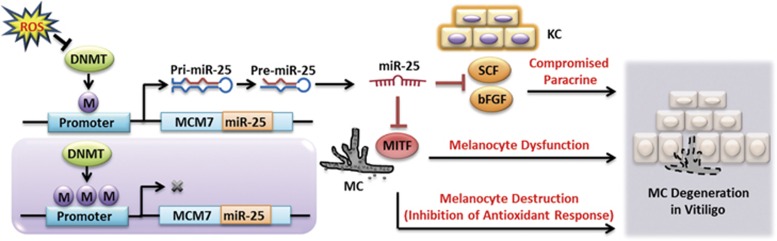
We found increased miR-25 in serum and lesion samples from vitiligo patients. Further experiments showed that miR-25 facilitated H2O2-induced apoptosis by impairing the antioxidant response and led to the dysfunction of melanocytes owing to its suppression on MITF. Noteworthily, miR-25 could inhibit the production and secretion of SCF and bFGF from keratinocytes and impair their paracrine protective effect on the survival of melanocytes under oxidative stress. At last, we verified that oxidative stress induced miR-25 generation in both melanocytes and keratinocytes by demethylating its promoter region. Our findings indicate that oxidative stress-induced overexpression of miR-25 has a crucial role in mediating the degeneration of melanocytes in vitiligo (Figure 7).
Figure 7.
ROS-induced miR-25 promotes the degeneration of melanocytes in vitiligo. ROS suppress the expression of DNMTs, thus demethylating the promoter region of miR-25. MiR-25 is increased following demethylation and suppresses the level of MITF in melanocytes, which leads to the dysfunction of melanocytes and facilitates the oxidative stress-induced apoptosis by inhibiting the intracellular antioxidant response in melanocytes. Moreover, miR-25 suppresses the expression of SCF and bFGF in keratinocytes, thus impairing their paracrine protective effect on melanocytes. Collectively, oxidative stress-induced overexpression of miR-25 promotes the degeneration of melanocytes in vitiligo. MC, melanocyte; KC, keratinocyte
Our microarray analysis implicated the overexpression of miR-25 in vitiligo serum, which is consistent with a report by Shi et al.42 Importantly, miR-25 was closely related with the activity of vitiligo and upregulated in the whole lesional epidermis. Although the melanocytes in lesional skin had been disappeared, we observed an intensively positive expression of miR-25 in perilesional skin that was harvested at the edge of vitiligo lesion. We speculated that this phenomenon resulted from oxidative stress in vitiligo. Further in vitro assays confirmed the inductive effect of oxidative stress on miR-25 production in both melanocytes and keratinocytes.
The destruction of melanocytes is a key event in the development of vitiligo, the underlying mechanism of which is still puzzling. Studies have confirmed melanocyte apoptosis in vitiligo by observing nuclear shrinkage, vacuolization and detachment.43 We proved that miR-25 could promote H2O2-induced melanocyte apoptosis by suppressing the expression of MITF, a master regulator of melanocyte survival and function.44, 45, 46, 47 Subsequent experiments demonstrated that APE1, a transcription target of MITF important for the antioxidant response, could be inhibited by miR-25, and its re-introduction could counteract the facilitating effect of miR-25 on ROS-induced melanocyte apoptosis. We further detected the downregulation of HO-1 and GPX1, two antioxidant genes, in melanocytes overexpressing miR-25. These findings suggested that miR-25 could suppress the antioxidant response through inhibiting MITF-APE1 pathway, thus making melanocytes more susceptible to oxidative stress-induced destruction.
As poorly functioning melanocytes have been isolated from lesional vitiligo skin and cultured in vitro,48, 49, 50, 51 the deterioration of melanocyte function may be an early and crucial event in vitiligo. We found that miR-25 downregulated the expression of TYR, TYRP1 and TYRP2, as well as tyrosinase activity in melanocytes, thus impairing melanin synthesis. Rescue experiments further verified that the regulation depended on the suppression of MITF, which is consistent with previous findings that MITF is a transcription activator regulating those melanogenesis-related genes.52, 53, 54 Furthermore, the process of melanosome transportation could be disturbed by miR-25. In that case, miR-25 is possibly involved in the entire pathological development of vitiligo, from the initial dysfunction to the final disappearance of melanocytes.
The homeostasis of melanocytes is influenced by a complex paracrine network consisting of growth factors secreted by keratinocytes. The deficiency of this paracrine network may cause an adverse microenvironment, which can lead to the loss of melanocytes in vitiligo. We found that miR-25 impaired the production and secretion of SCF and bFGF from keratinocytes, both of which have been reported to promote melanocyte growth and show lower expression in vitiligo lesions.55 Consistent with previous findings, the culture medium of miR-25-knockin keratinocytes failed to conduct the protective effect on H2O2-treated melanocytes. Given the above, we believe that increased miR-25 in keratinocytes also contributes to melanocyte destruction in vitiligo.
It is reported that oxidative stress could inhibit the expression of DNMTs through JNK signaling pathway, thus inducing DNA demethylation and contributing to the development of Alzheimer's disease.41 In our study, oxidative stress successfully caused the demethylation of CpG sites in the promoter region of miR-25, and demethylation reagent did elevate miR-25 level in melanocytes and keratinocytes. In addition, we found that H2O2 reduced the expression of DNMTs by promoting JNK phosphorylation in both melanocytes and keratinocytes. It is worth mentioning that except for miR-25, decreased DNMTs may break the balance between DNA methylation and demethylation and cause aberrant expression of many other genes that may have a role in the development of vitiligo.
In conclusion, miR-25 is an important miRNA that promotes oxidative stress-induced melanocyte degeneration by not only suppressing MITF in melanocytes but also impairing the paracrine protective effect of keratinocytes. To our knowledge, this is the first study to describe the role of miRNAs in the pathogenesis of vitiligo, and our findings, to some extent, deepen the mechanism of melanocyte degeneration caused by oxidative stress in vitiligo. Further studies using appropriate animal models of vitiligo are still needed to fully clarify the pathogenic role of miR-25 in vivo and evaluate its potential to be a therapeutic target in vitiligo.
Materials and methods
Clinical assessment and patient materials
Serum samples for microarray and qRT-PCR analysis were collected from 130 patients with vitiligo and 130 healthy subjects. Skin specimens for qRT-PCR analysis were taken from the abdomen of six vitiligo patients and four healthy volunteers. Skin tissues for FISH and immunofluorescence were later taken from another three vitiligo patients and three healthy donors. The patients were diagnosed according to the criteria of the Vitiligo Area Severity Index (VASI) and the Vitiligo European Task Force (VETF).56 All the vitiligo cases were diagnosed by clinical examination and Wood's lamp. All of the patients enrolled in our study had no other autoimmune or systemic diseases, and none of the patients received systemic treatment, including glucocorticoids, immunomodulating agents and phototherapy, within 1 month prior to the sample collection period. Segmental vitiligo was excluded from the present study. Disease stability is assessed using the combined criteria of a clinical scoring system (VASI and VETF), patient self-reporting and serial digital imaging of specific lesions over 12 months.1 All of the patients and control subjects were pair-matched in age and gender, and informed consent was obtained from all the patients and control subjects. The research protocol was designed and executed according to the principles of the Declaration of Helsinki and was approved by the ethics review board of Fourth Military Medical University.
Microarray analysis
MiRNA microarrays were performed using Agilent's miRNA Microarray System (Agilent, Santa Clara, CA, USA) according to the manufacturer's instructions. Briefly, total RNA was isolated from serum sample using the one-step protocol of the mir Vana miRNA Isolation Kit (Ambion, Austin, TX, USA). For each sample and replicate, 100 ng of total RNA were fluorescently labeled with Cyanine 3-pCp and hybridized to oligonucleotide arrays for 20 h at 55 °C. After washing the microarrays in an ozone-depleted room, the array slides were scanned and analyzed using an Agilent DNA microarray scanner. Microarray images were processed using Feature Extraction Software 9.5.3 (Agilent). Processed data were imported into Microsoft Office Excel 2010 for further analysis. The Wilcoxon Rank Sum test was used to compare the difference in serum miRNAs between vitiligo patients and healthy controls. P-values were adjusted for multiple testing using the Benjamini and Hochberg method. Only miRNAs with false discovery rate<0.01 were considered significant. MiRNAs were considered differentially regulated if at least twofold upregulation or downregulation was observed. Hierarchical clustering was performed using TIGR Multi Experiment Viewer Version 4.0.
MiRNA isolation and real-time qRT-PCR
MiRNA isolation from serum and skin samples were performed using the miRNeasy Serum/Plasma Kit and Micro Kit (Qiagen, Valencia, CA, USA) following the manufacturer's protocol, respectively. MiRNeasy Serum/Plasma Spike-In Control (C. elegans miR-39) was added to serum as controls. Serum and skin cDNA were synthesized from miRNA using miRNA cDNA First Strand Synthesis and miRNA SYBR qRT-PCR Kit (Qiagen). QRT-PCR was performed with a BIO-RAD Multicolor Real-time PCR Detection System (iQTM5; Bio-Rad, Hercules, CA, USA) using primers and templates that were mixed with SYBR Premix. The primers of cel-miR-39, hsa-let-7b, hsa-miR-16, hsa-miR-451, hsa-miR-25 and hsa-miR-15b were purchased from Qiagen. DNA was amplified for 40 cycles of denaturation for 20 s at 95 °C and annealed for 60 s at 60 °C. Threshold cycle (CT) for each miRNA was determined using the iQ5-standard Edition Optical System ver 2.1 (Bio-Rad). Relative quantification was performed according to the ΔΔCT method, and results were expressed in the linear form using the formula 2-ΔΔCT. Serum transcript levels of hsa-miR-25 were normalized to those of cel-miR-39, while skin transcript levels of hsa-miR-25 were normalized to those of RUNU6.
Fluorescence in situ hybridization
FISH was performed on formalin-fixed paraffin-embedded sections (10 μm in thickness) of skin biopsy specimens from three vitiligo patients and three healthy individuals. After dewaxation, sections were treated with proteinase K (2 μg/ml) at 37 °C for 15 min, washed and prehybridized for 30 min at 37 °C. Hybridization with FAM-labeled locked nucleic acid-based probes specific for mir-25 (Exiqon, Vedbaek, Denmark) was performed overnight at 60 °C. Slides were then washed at 60 °C. Co-staining for melanocytes was performed by incubation with Melan-A (mouse monoclonal anti-Melan A antibody, 1 : 100, Abcam Inc., Cambridge, MA, USA) for 1 h at 36 °C, followed by 1 h incubation with appropriate Alexa Fluor 640 secondary antibodies (Life Technologies, Carlsbad, CA, USA). DAPI (Dako, Glostrup, Denmark) was used as a counterstain.
Cell culture and treatment
Normal human melanocytes that were isolated from human foreskin specimens obtained during circumcision surgery were cultured in Medium 254 (Gibco, Grand Island, NY, USA) supplemented with Human Melanocyte Growth Supplement (Gibco) at 37 °C in the presence of 5% CO2. The second- or third-passage keratinocytes were used in all experiments. Normal human keratinocytes were extracted from plastic surgery skin obtained from healthy individuals with cells cultured in the serum-free keratinocyte growth medium (Gibco). The CM of normal human keratinocytes was collected at 48 h after transfection and stored at −80 °C until use. Oxidative stress in normal human melanocytes was induced by treatment of 400 μM H2O2 (Sigma, St. Louis, MO, USA) for 24 h. Oxidative stress in normal human keratinocytes was induced by treatment of 1000 μM H2O2 (Sigma) for 24 h. Demethylation is induced by treatment of 5 mM 5-aza-dC (Sigma) for 24 h. JNK inhibition was induced by pretreatment on the cells for 1 h of JNKi (SP600125) (Selleck Chemicals, Houston, TX, USA) stimulation (10 mmol/l) before the H2O2 treatment.
Oligonucleotide transfection into cells
Hsa-miR-25 mimics, hsa-miR-25 inhibitor and negative control were double-stranded RNA oligonucleotides that were purchased from Invitrogen (Carlsbad, CA, USA). They were transfected into normal human melanocytes or normal human keratinocytes using Lipofectamine 3000 (Invitrogen) according to the manufacturer's instructions.
Detection of apoptosis
To quantify apoptotic death, normal human melanocytes were plated in six-well plates at a density of 2–3 × 105 cells per well overnight. At 48 h after transfection, the cells were directly collected or switched into serum-free, antibiotic-free DMEM (with or without the mixture of keratinocyte CM with an 1 : 1 ratio) and exposed to 400 μM H2O2 (Sigma) for 24 h before collection. Apoptotic cells were identified using the Annexin V-PE Apoptosis Detection Kit (MaiBio, Shanghai, China) following the manufacturer's instructions. The number of apoptotic cells was quantified by flow cytometry (Beckman Coulter, Miami, FL, USA), and the analysis was carried out using the Expo32 software (Beckman Coulter). Each sample was repeated in triplicate.
Western blotting
After treatment, cells were washed two times with ice-cold phosphate-buffered saline (PBS) and then lysed with cell lysis solution (DSL, Webster, TX, USA). Proteins were extracted from cells and quantified using BCA Protein Assay kit (Pierce, Rockford, IL, USA). Equal amounts of protein were separated by 10% SDS-PAGE (Bio-Rad) and were then transferred from the gel to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). Page Ruler Plus Prestained Protein Ladder (Fermentas, Hanover, MD, USA) was used to confirm protein electrophoresis and transferring. After blocking in a solution of 5% non-fat dry milk diluted in Tris-buffered saline, the membranes were washed and then incubated with primary antibodies (rabbit polyclonal anti-MITF, 1 : 1000, Abcam; rabbit monoclonal anti-Bcl-2, 1 : 500, Abcam; rabbit monoclonal to Bax, 1 : 2000, Abcam; rabbit monoclonal anti-Cleaved Caspase-3,1 : 1000, Cell Signaling Technology (Danvers, MA, USA); rabbit polyclonal anti-Cleaved Caspase-9,1 : 1000, Cell Signaling Technology; mouse monoclonal to Tyrosinase, 1 : 250, Abcam; rabbit monoclonal to TYRP1, 1 : 10000, Abcam; rabbit polyclonal anti-TYRP2, 1 : 500, Abcam; mouse monoclonal anti-APE1, 1 : 2000, Abcam; rabbit monoclonal anti-SCF, 1 : 10000, Abcam; mouse monoclonal anti-Endothelin 1, 1 : 500, Abcam; rabbit monoclonal anti-bFGF, 1 : 1000, Cell Signaling Technology; rabbit polyclonal anti-CXCL10, 1 : 1000, Abcam; rabbit polyclonal anti-CXCL9, 1 : 5000, Abcam; rabbit polyclonal anti-DNMT1 antibody, 1 : 1000, Abcam; rabbit polyclonal anti-DNMT3b, 1 : 500, Abcam; rabbit monoclonal anti-Bax, 1 : 2000, Abcam; rabbit monoclonal anti-phospho-JNK, 1 : 1000, Cell Signaling Technology; rabbit monoclonal anti-β-actin, 1 : 5000, Abcam) overnight at 4 °C. After being washed, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (Goat Anti-Mouse IgG, 1 : 5000; Goat Anti-Rabbit IgG, 1 : 10000) for 2 h at room temperature. Bound antibodies were detected using the ECL western blotting detection system (Millipore).
5-Ethynyl-2′-deoxyuridine assay
Forty-eight hours after transfection, EdU (Cell Light EdU DNA imaging Kit, RiboBio, Guangzhou, China) was added at a 10 mM final concentration to the transfected cells 24 h before harvesting. The cells were then stained according to the following protocol: discard the EdU medium mixture, add 4% paraformaldehyde to fix cells at room temperature for 30 min, wash with glycine (2 mg/ml) for 5 min in a shaker, add 0.2% Triton X-100 for 10 min, wash with PBS for two times, add click reaction buffer (Tris-HCl, pH 8.5, 100 mM; CuSO4, 1 mM; Apollo 550 fluorescent azide, 100 mM; ascorbic acid, 100 mM) for 30 min while protecting from light, wash with 0.5% Triton X-100 for three times, stain with Hoechst (5 mg/ml) for 30 min at room temperature, wash with 0.5% Triton X-100 for five times and, finally, add 150 ml PBS. Images were taken and analyzed using High Content Imaging Pathway 855 (BD, Franklin Lakes, NJ, USA). EdU-positive cells were calculated by (EdU add-in cells/Hoechst stained cells) × 100%.
Tyrosinase activity and measurement of melanin content
Tyrosinase activity was determined as described by Tomita et al.57 with slight modification. Normal human melanocytes were plated at a density of 2.5 × 103 cells/well in 96-well plates. After incubation with test substances for 48 h, cells were washed with ice-cold PBS and lysed with phosphate buffer (pH 6.8) containing 1% Triton-X/PBS (90 μl/well) and then frozen at −80 °C for 30 min. After thawing and mixing, 10 μl of 1% L-DOPA was added to each well. Following incubation at 37 °C for 2 h, the absorbance was measured at 492 nm using an ELISA reader (Thermo Fisher Scientific Inc., Waltham, MA USA). Melanin content was measured according to the method of Tsuboi et al.58 with slight modifications. Briefly, cells were treated with test substances for 2 days, and cell pellets containing a known number of cells (5 × 105) were dissolved in 0.5 ml of 1 M NaOH at 100 °C for 30 min and centrifuged for 20 min at 16 000 × g. Optical densities of supernatants were measured at 410 nm using an ELISA reader (Thermo Fisher Scientific Inc.).
Detection of melanosome transport
Immunocytochemistry was used to detect the melanosome transport. Normal human melanocytes were cultured on glass coverslips and accepted transfection. Forty-eight hours later, sections were washed and fixed in 4% paraformaldehyde for 10 min. Subsequently, the sections were incubated with 0.5% BSA for 30 min. To detect the melanosome marker gp100, cells were incubated with a polyclonal mouse anti-human gp100 antibody (1 : 200, Abcam) overnight at 4 °C. After that, the cells were incubated for 1 h with a rabbit anti-mouse secondary antibody (Dako). Next, DAPI was used at a 1 : 1000 dilution for detection of cell nuclei, and coverslips were mounted using glycerol (Dako).
Enzyme-linked immunosorbent assay
CXCL9, CXCL10, ET-1, SCF and bFGF were assayed using the commercially available assay kits (Westang Biotechnology, Shanghai, China) according to the manufacturer's instructions. The absorbance (A450) was measured with a plate reader (MRP-2100, Syntron, Carlsbad, CA, USA). Detection sensitivity of this ELISA system was at least 15 pg/ml. All samples were diluted to 1 : 50 before the assay.
Bioinformatics analysis for target genes of miRNAs
Multiple target gene prediction software, including miRanda (http://www.mocrorna.org), TargetScan (http://www.targetscan.org) and PicTar (http://pictar.mdc-berlin.de/) were used to predict potential targets of miR-25.
RNA isolation and qRT-PCR
Total RNA was isolated using Trizol reagent (Invitrogen), and then reverse-transcribed to single-strand cDNA using reverse transcription reagents according to the manufacturer's instructions. QRT-PCR experiments were performed using the IQ SYBR Green Supermix (Bio-Rad) and BIO-RAD MJ Mini Opticon Real-Time PCR System. The resulting amplification and melt curves were analyzed to ensure the identity of the specific PCR product. Threshold cycle values were used to calculate the fold change in the transcript levels by using the 2-ΔΔCT method. The relative mRNA expression levels were normalized to the actin gene. The primer sequences are listed in Supplementary Table S2.
Plasmids construction for luciferase reporter assay
The full-length human MITF 3′UTR comprises approximately 3089 bp (NCBI reference number NM_000248), with the miR-25 interaction site corresponding to nt 1436–1442. The full-length human SCF 3′UTR comprises approximately 4426 bp (NCBI reference number NM_003994), with the miR-25 interaction site corresponding to nt 3162–3168. The full-length human bFGF 3′UTR comprises approximately 5839 bp (NCBI reference number NM_002006), with the miR-25 interaction site corresponding to nt 2239–2246. The 1000–1999 nt region of the MITF 3′UTR, the 2600–3599 nt region of the SCF 3′UTR and the 1700–2699 nt region of the bFGF 3′UTR were amplified by PCR and inserted into the psiCHECK-2 Vector (Promega Corporation, Madison, WI, USA) downstream of the stop codon of the luciferase gene, between NotI and XhoI restriction sites, to generate the MITF WT plasmid, SCT WT and bFGF WT. Nucleotides from nt 1437 to 1440 within the seed-matching sequences in MITF WT plasmid plasmids, nucleotides from nt 3163 to 3166 within the seed-matching sequences in SCT WT plasmid plasmids and nucleotides from nt 2242 to 2245 within the seed-matching sequences in bFGF WT plasmid plasmids were mutated to generated plasmids named MITF MUT, SCF WT and bFGF WT, respectively.
Luciferase reporter assay
Transfections of melanocytes were performed using Lipofectamine 3000 (Invitrogen) according to the manufacturer's recommendations. All transfections were performed in triplicate. Luciferase vectors (100 ng) (MITF WT, MITF MUT, SCF WT, SCF MUT, bFGF WT, bFGF MUT and the empty vectors) were individually transfected into normal human melanocytes or normal human keratinocytes along with 100 nM hsa-miR-25 mimics (Invitrogen) or 100 nM negative control (Invitrogen). Forty-eight hours after transfection, luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega Corporation). For each transfection, the luciferase activity of three replicates was averaged.
Plasmid construction and transfection for rescue experiments
Human 3′UTR-deleted MITF mRNA and Human APE1 mRNA was amplified by RT-PCR and cloned into the GV141 vector (Genechem, Shanghai, China) between the EcoRI and BamHI restriction sites, respectively. The vectors were sequenced to confirm the insert direction and sequences.
DNA methylation analysis
Genomic DNA was extracted from the cultured cells by using Wizard HSV Genomic DNA Purification System (Promega Corporation) according to the manufacturer's instructions. The integrity and purity of DNA were spectrophotometrically examined according to its A260/A280 absorption. A total of 200 ng genomic DNA from each sample was bisulfite-treated with the Methylamp DNA Modification Kit (Epigentek, Brooklyn, NY, USA). The quality of bisulfite conversion was evaluated using the PCR products without methyl groups as the control. Sequenom Mass ARRAY platform (CapitalBio, Beijing, China), which was composed of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and combined with RNA base-specific cleavage was used to analyze miR-25 methylation quantitatively (Gen-Bank Accession Number: NM_053304.1). PCR primers were designed using Methprimer (http://www.urogene.org/methprimer/). For each reverse primer, an additional T7 promoter tag for in vivo transcription was added, whereas a 10-m tag on the forward primer was used to adjust melting temperature differences. Supplementary Table S3 lists all the primers used in this study. The methylation ratios were generated by Epityper software version 1.0 (Sequenom, San Diego, CA, USA). The generated data were put into the EPI 3.1 Database (EpiData Association, Odense, Denmark) and analyzed with SPSS 11.5 software (SPSS Inc, Chicago, IL, USA). Independent t-tests were conducted to evaluate the difference between the experimental and control groups. P<0.05 was considered statistically significant.
Immunofluorescence
Paraffin-embedded tissue sections were deparaffinized and rehydrated with graded ethanol dilutions. After antigen retrieval in sodium citrate buffer (10 mM, pH 6.0), immunofluorescence staining was performed by incubating the paraffin sections with a primary antibody (rabbit polyclonal anti-DNMT1 antibody, 1 : 1000, Abcam; rabbit polyclonal anti-DNMT3a antibody, 1 : 50, Abcam; rabbit polyclonal anti-DNMT3b antibody, 1 : 50, Abcam). Co-staining for melanocytes was performed by incubation with Melan-A (mouse monoclonal anti-Melan A antibody, 1 : 100, Abcam) overnight at 4 °C, followed by 1 h incubation with appropriate secondary antibodies (goat anti-rabbit Alexa Fluor 640 Dye 1:5000 and goat anti-mouse Alexa Fluor 488 Dye, 1 : 5000, Life Technologies). DAPI (Dako) was used as a counterstain. Tissue sections were analyzed by confocal laser scanning microscopy (LSM510; Carl Zeiss AB, Jena, Germany).
Melanocytes and keratinocytes for immunofluorescence staining were first seeded on coverslips in a 24-well plate and allowed to adhere before staining. Cells were washed with PBS, fixed with 4% paraformaldehyde for 10–20 min, and permeabilized with 10% saponin for 5 min followed by acetone treatment at −20 °C for 10 min. After washing the slides with PBS, cells were blocked with normal goat sera at room temperature for 1 h. Cells were incubated with primary antibody diluted in the blocking solution at 4 °C overnight. The primary antibodies were as follows: rabbit polyclonal anti-DNMT1 antibody, 1 : 1000 (Abcam); rabbit polyclonal anti-DNMT3b antibody, 1 : 50 (Abcam). Cells were washed with PBS and incubated with secondary antibodies (goat anti-rabbit Alexa Fluor 640 Dye, 1 : 5000, Life Technologies) at room temperature for 1 h. Nuclei were stained with DAPI (Dako) and subsequently analyzed by confocal laser scanning microscopy (LSM 510).
Statistical analysis
Each experiment was performed at least for three times, and statistical analyses of the data were performed using unpaired, two-tailed Student's t-tests built into GraphPad Prism (GraphPad Software 3.0; San Diego, CA, USA). All the data are expressed as the mean±S.D. P-values of<0.05 were considered statistically significant.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (no. 81172749, no. 81472863, no. 30972528 and no. 81130032).
Glossary
- miRNAs
microRNAs
- H2O2
hydrogen peroxide
- ROS
reactive oxygen species
- qRT-PCR
real-time quantitative reverse transcription-PCR
- FISH
fluorescence in situ hybridization
- MITF
microphthalmia-associated transcription factor
- Bcl-2
B cell lymphoma 2
- Bax
B cell lymphoma 2-associated X protein
- TYR
tyrosinase
- TYRP1
tyrosinase-related protein 1
- TYRP2
tyrosinase-related protein 2
- 3′UTR
3′ untranslated regions
- APE1
apurinic/apyrimidinic endonuclease
- HO-1
heme oxygenase 1 (HO-1)
- GPX1
glutathione peroxidase 1
- SCF
stem cell factor
- bFGF
basic fibroblast growth factor
- ET-1
endothelin-1
- DNMTs
DNA methyltransferases
- 5-aza-dC
5-aza-2′-deoxycytidine
- JNK
c-Jun amino terminal kinase
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by G Melino
Supplementary Material
References
- Ezzedine K, Lim HW, Suzuki T, Katayama I, Hamzavi I, Lan CC et al. Revised classification/nomenclature of vitiligo and related issues: the Vitiligo Global Issues Consensus Conference. Pigment Cell Melanoma Res 2012; 25: E1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongenae K, Van Geel N, De Schepper S, Naeyaert JM. Effect of vitiligo on self-reported health-related quality of life. Br J Dermatol 2005; 152: 1165–1172. [DOI] [PubMed] [Google Scholar]
- Jin Y, Birlea SA, Fain PR, Ferrara TM, Ben S, Riccardi SL et al. Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat Genet 2012; 44: 676–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan C, Ren YQ, Xiang LH, Sun LD, Xu AE, Gao XH et al. Genome-wide association study for vitiligo identifies susceptibility loci at 6q27 and the MHC. Nat Genet 2010; 42: 614–618. [DOI] [PubMed] [Google Scholar]
- Jin Y, Birlea SA, Fain PR, Gowan K, Riccardi SL, Holland PJ et al. Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo. N Engl J Med 2010; 362: 1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval-Cruz M, Garcia-Carrasco M, Sanchez-Porras R, Mendoza-Pinto C, Jimenez-Hernandez M, Munguia-Realpozo P et al. Immunopathogenesis of vitiligo. Autoimmun Rev 2011; 10: 762–765. [DOI] [PubMed] [Google Scholar]
- Gey A, Diallo A, Seneschal J, Leaute-Labreze C, Boralevi F, Jouary T et al. Autoimmune thyroid disease in vitiligo: multivariate analysis indicates intricate pathomechanisms. Br J Dermatol 2013; 168: 756–761. [DOI] [PubMed] [Google Scholar]
- Gauthier Y, Cario Andre M, Taieb A. A critical appraisal of vitiligo etiologic theories. Is melanocyte loss a melanocytorrhagy? Pigment Cell Res 2003; 16: 322–332. [DOI] [PubMed] [Google Scholar]
- Gauthier Y, Cario-Andre M, Lepreux S, Pain C, Taieb A. Melanocyte detachment after skin friction in non lesional skin of patients with generalized vitiligo. Br J Dermatol 2003; 148: 95–101. [DOI] [PubMed] [Google Scholar]
- Schallreuter KU, Wood JM, Ziegler I, Lemke KR, Pittelkow MR, Lindsey NJ et al. Defective tetrahydrobiopterin and catecholamine biosynthesis in the depigmentation disorder vitiligo. Biochim Biophys Acta 1994; 1226: 181–192. [DOI] [PubMed] [Google Scholar]
- Maresca V, Roccella M, Roccella F, Camera E, Del Porto G, Passi S et al. Increased sensitivity to peroxidative agents as a possible pathogenic factor of melanocyte damage in vitiligo. J Invest Dermatol 1997; 109: 310–313. [DOI] [PubMed] [Google Scholar]
- Jimbow K, Chen H, Park JS, Thomas PD. Increased sensitivity of melanocytes to oxidative stress and abnormal expression of tyrosinase-related protein in vitiligo. Br J Dermatol 2001; 144: 55–65. [DOI] [PubMed] [Google Scholar]
- Koga S, Nakano M, Tero-Kubota S. Generation of superoxide during the enzymatic action of tyrosinase. Arch Biochem Biophys 1992; 292: 570–575. [DOI] [PubMed] [Google Scholar]
- Cucchi ML, Frattini P, Santagostino G, Orecchia G. Higher plasma catecholamine and metabolite levels in the early phase of nonsegmental vitiligo. Pigment Cell Res 2000; 13: 28–32. [DOI] [PubMed] [Google Scholar]
- Schallreuter KU, Moore J, Wood JM, Beazley WD, Peters EM, Marles LK et al. Epidermal H(2)O(2) accumulation alters tetrahydrobiopterin (6BH4) recycling in vitiligo: identification of a general mechanism in regulation of all 6BH4-dependent processes? J Invest Dermatol 2001; 116: 167–174. [DOI] [PubMed] [Google Scholar]
- Schallreuter KU, Wood JM, Berger J. Low catalase levels in the epidermis of patients with vitiligo. J Invest Dermatol 1991; 97: 1081–1085. [DOI] [PubMed] [Google Scholar]
- Schallreuter KU, Rubsam K, Gibbons NC, Maitland DJ, Chavan B, Zothner C et al. Methionine sulfoxide reductases A and B are deactivated by hydrogen peroxide (H2O2) in the epidermis of patients with vitiligo. J Invest Dermatol 2008; 128: 808–815. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Li CY, Li K, Wang T, Zhang B, Gao TW. Decreased methionine sulphoxide reductase A expression renders melanocytes more sensitive to oxidative stress: a possible cause for melanocyte loss in vitiligo. Br J Dermatol 2009; 161: 504–509. [DOI] [PubMed] [Google Scholar]
- Schallreuter KU, Moore J, Wood JM, Beazley WD, Gaze DC, Tobin DJ et al. In vivo and in vitro evidence for hydrogen peroxide (H2O2) accumulation in the epidermis of patients with vitiligo and its successful removal by a UVB-activated pseudocatalase. J Investig Dermatol Symp Proc 1999; 4: 91–96. [DOI] [PubMed] [Google Scholar]
- Khan R, Satyam A, Gupta S, Sharma VK, Sharma A. Circulatory levels of antioxidants and lipid peroxidation in Indian patients with generalized and localized vitiligo. Arch Dermatol Res 2009; 301: 731–737. [DOI] [PubMed] [Google Scholar]
- Koca R, Armutcu F, Altinyazar HC, Gurel A. Oxidant-antioxidant enzymes and lipid peroxidation in generalized vitiligo. Clin Exp Dermatol 2004; 29: 406–409. [DOI] [PubMed] [Google Scholar]
- Giovannelli L, Bellandi S, Pitozzi V, Fabbri P, Dolara P, Moretti S. Increased oxidative DNA damage in mononuclear leukocytes in vitiligo. Mutat Res 2004; 556: 101–106. [DOI] [PubMed] [Google Scholar]
- Schallreuter KU, Elwary SM, Gibbons NC, Rokos H, Wood JM. Activation/deactivation of acetylcholinesterase by H2O2: more evidence for oxidative stress in vitiligo. Biochem Biophys Res Commun 2004; 315: 502–508. [DOI] [PubMed] [Google Scholar]
- Schallreuter KU, Gibbons NC, Zothner C, Abou Elloof MM, Wood JM. Hydrogen peroxide-mediated oxidative stress disrupts calcium binding on calmodulin: more evidence for oxidative stress in vitiligo. Biochem Biophys Res Commun 2007; 360: 70–75. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003; 425: 415–419. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Chang TC, Mendell JT. microRNAs in vertebrate physiology and human disease. Annu Rev Genomics Hum Genet 2007; 8: 215–239. [DOI] [PubMed] [Google Scholar]
- Magenta A, Cencioni C, Fasanaro P, Zaccagnini G, Greco S, Sarra-Ferraris G et al. miR-200c is upregulated by oxidative stress and induces endothelial cell apoptosis and senescence via ZEB1 inhibition. Cell Death Differ 2011; 18: 1628–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Liu X, Cheng Y, Yang J, Huo Y, Zhang C. Involvement of MicroRNAs in hydrogen peroxide-mediated gene regulation and cellular injury response in vascular smooth muscle cells. J Biol Chem 2009; 284: 7903–7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Zhang Q, Niu J, Lu K, Xie B, Cui D et al. Screening of microRNAs associated with Alzheimer's disease using oxidative stress cell model and different strains of senescence accelerated mice. J Neurol Sci 2014; 338: 57–64. [DOI] [PubMed] [Google Scholar]
- Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids—the mix of hormones and biomarkers. Nat Rev Clin Oncol 2011; 8: 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Fu Y, Meyskens FL Jr. MiTF regulates cellular response to reactive oxygen species through transcriptional regulation of APE-1/Ref-1. J Invest Dermatol 2009; 129: 422–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiddon C, Moorthy NC, Prives C. Ref-1 regulates the transactivation and pro-apoptotic functions of p53 in vivo. EMBO J 1999; 18: 5609–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LE, Arany Z, Livingston DM, Bunn HF. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J Biol Chem 1996; 271: 32253–32259. [DOI] [PubMed] [Google Scholar]
- Xanthoudakis S, Curran T. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J 1992; 11: 653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki K, Mackenzie EL, Hailemariam K, Sakamoto K, Tsuji Y. Hemin-mediated regulation of an antioxidant-responsive element of the human ferritin H gene and role of Ref-1 during erythroid differentiation of K562 cells. Mol Cell Biol 2006; 26: 2845–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung H, Demple B. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol Cell 2005; 17: 463–470. [DOI] [PubMed] [Google Scholar]
- Kaspar JW, Niture SK, Jaiswal AK. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic Biol Med 2009; 47: 1304–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AY. Role of keratinocytes in the development of vitiligo. Ann Dermatol 2012; 24: 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashighi M, Agarwal P, Richmond JM, Harris TH, Dresser K, Su MW et al. CXCL10 is critical for the progression and maintenance of depigmentation in a mouse model of vitiligo. Sci Transl Med 2014; 6: 223ra23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Sun J, Li S, Wu X, Li L. Oxidative stress induces DNA demethylation and histone acetylation in SH-SY5Y cells: potential epigenetic mechanisms in gene transcription in Abeta production. Neurobiol Aging 2013; 34: 1069–1079. [DOI] [PubMed] [Google Scholar]
- Shi YL, Weiland M, Li J, Hamzavi I, Henderson M, Huggins RH et al. MicroRNA expression profiling identifies potential serum biomarkers for non-segmental vitiligo. Pigment Cell Melanoma Res 2013; 26: 418–421. [DOI] [PubMed] [Google Scholar]
- Boissy RE, Liu YY, Medrano EE, Nordlund JJ. Structural aberration of the rough endoplasmic reticulum and melanosome compartmentalization in long-term cultures of melanocytes from vitiligo patients. J Invest Dermatol 1991; 97: 395–404. [DOI] [PubMed] [Google Scholar]
- Vachtenheim J, Borovansky J. ‘Transcription physiology' of pigment formation in melanocytes: central role of MITF. Exp Dermatol 2010; 19: 617–627. [DOI] [PubMed] [Google Scholar]
- Cheli Y, Ohanna M, Ballotti R, Bertolotto C. Fifteen-year quest for microphthalmia-associated transcription factor target genes. Pigment Cell Melanoma Res 2010; 23: 27–40. [DOI] [PubMed] [Google Scholar]
- Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med 2006; 12: 406–414. [DOI] [PubMed] [Google Scholar]
- Bentley NJ, Eisen T, Goding CR. Melanocyte-specific expression of the human tyrosinase promoter: activation by the microphthalmia gene product and role of the initiator. Mol Cell Biol 1994; 14: 7996–8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Poole IC, van den Wijngaard RM, Westerhof W, Dutrieux RP, Das PK. Presence or absence of melanocytes in vitiligo lesions: an immunohistochemical investigation. J Invest Dermatol 1993; 100: 816–822. [DOI] [PubMed] [Google Scholar]
- Tobin DJ, Swanson NN, Pittelkow MR, Peters EM, Schallreuter KU. Melanocytes are not absent in lesional skin of long duration vitiligo. J Pathol 2000; 191: 407–416. [DOI] [PubMed] [Google Scholar]
- Kim YC, Kim YJ, Kang HY, Sohn S, Lee ES. Histopathologic features in vitiligo. Am J Dermatopathol 2008; 30: 112–116. [DOI] [PubMed] [Google Scholar]
- Gottschalk GM, Kidson SH. Molecular analysis of vitiligo lesions reveals sporadic melanocyte survival. Int J Dermatol 2007; 46: 268–272. [DOI] [PubMed] [Google Scholar]
- Yasumoto K, Yokoyama K, Shibata K, Tomita Y, Shibahara S. Microphthalmia-associated transcription factor as a regulator for melanocyte-specific transcription of the human tyrosinase gene. Mol Cell Biol 1994; 14: 8058–8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotto C, Busca R, Abbe P, Bille K, Aberdam E, Ortonne JP et al. Different cis-acting elements are involved in the regulation of TRP1 and TRP2 promoter activities by cyclic AMP: pivotal role of M boxes (GTCATGTGCT) and of microphthalmia. Mol Cell Biol 1998; 18: 694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumoto K, Yokoyama K, Takahashi K, Tomita Y, Shibahara S. Functional analysis of microphthalmia-associated transcription factor in pigment cell-specific transcription of the human tyrosinase family genes. J Biol Chem 1997; 272: 503–509. [DOI] [PubMed] [Google Scholar]
- Hirobe T. Role of keratinocyte-derived factors involved in regulating the proliferation and differentiation of mammalian epidermal melanocytes. Pigment Cell Res 2005; 18: 2–12. [DOI] [PubMed] [Google Scholar]
- Kawakami T, Hashimoto T. Disease severity indexes and treatment evaluation criteria in vitiligo. Dermatol Res Pract 2011; 2011: 750342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita Y, Maeda K, Tagami H. Melanocyte-stimulating properties of arachidonic acid metabolites: possible role in postinflammatory pigmentation. Pigment Cell Res 1992; 5: 357–361. [DOI] [PubMed] [Google Scholar]
- Tsuboi T, Kondoh H, Hiratsuka J, Mishima Y. Enhanced melanogenesis induced by tyrosinase gene-transfer increases boron-uptake and killing effect of boron neutron capture therapy for amelanotic melanoma. Pigment Cell Res 1998; 11: 275–282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.