Abstract
Salmonellae invasion and intracellular replication within host cells results in a range of diseases including gastroenteritis, bacteraemia, enteric fever and focal infections. In recent years, considerable progress has been made in our understanding of the molecular mechanisms by which Salmonellae alter host cell physiology through the delivery of effector proteins with specific activities, and through the modulation of defence and stress response pathways in the host. In this Review, we summarize our current knowledge of the complex interplay of bacterial and host factors that lead to inflammation, disease and in most cases, control of Salmonellae infection, particularly Salmonella enterica serovar Typhimurium, by its animal hosts. We also highlight gaps in our knowledge of the contributions of Salmonellae and the host to disease pathogenesis and suggest future avenues for further study.
ToC
In this Review, Miller and colleagues discuss the arsenal of effector proteins that salmonellae use to manipulate their animal hosts, in addition to the host response to these infections. The authors also discuss the challenges ahead for unravelling the mechanistic details of effector function.
Salmonellae are motile, Gram-negative bacteria that cause enteric diseases in a wide range of animals. The species Salmonella enterica is comprised of over 2,500 serovars, on the basis of flagellar and lipopolysaccharide antigens, and includes both typhoidal and non-typhoidal Salmonella (NTS) strains. Salmonella enterica serovar Typhi and Salmonella enterica serovar Paratyphi (the typhoidal serovars) are human-restricted pathogens that cause the systemic disease enteric (typhoid) fever, which is characterized by fever and abdominal pain. The NTS, Salmonella enterica serovar Enteritidis and Salmonella enterica serovar Typhimurium (S. Typhimurium), are broad host range pathogens that cause acute self-limiting gastroenteritis in humans, cattle, swine and poultry, but can cause bacteraemia and systemic infection in immunosuppressed hosts, in very young and older individuals and occasionally in healthy adult humans and animals1. The majority of studies discussed herein have used S. Typhimurium.
Salmonellae are usually acquired by oral ingestion of contaminated food or water and survive gastric acidity to gain access to the intestinal epithelium. NTS strains elicit inflammatory changes in the intestinal epithelium, including the infiltration of neutrophils and fluid into the intestinal lumen, resulting in inflammatory diarrhea2. The inflammatory reaction is essential for the release of factors (such as tetrathionate) that can be used as nutritional sources by NTS, which provides the pathogen with a growth advantage over the intestinal microbiota3, 4. Salmonellae can be engulfed by intestinal luminal phagocytes to facilitate invasion of the mucosal barrier; however, the bacterium usually induces its own uptake by epithelial cells and traverses the epithelial barrier through microfold cells (M cells) that overlay the intestinal lymphoid tissue or Peyer’s patches5.
Salmonellae can invade and survive inside a variety of mammalian cells types including macrophages, but are rapidly cleared by neutrophils6, 7. The intracellular lifestyle of salmonellae within epithelial cells and macrophages facilitates the avoidance of neutrophil-mediated killing and is essential for pathogenesis. Intracellular survival requires the bacteria to recognize and resist components of the innate immune system, including cationic antimicrobial peptides and the acidic pH of the phagocytic vacuole. The recognition of host innate immunity by salmonellae results in transcriptional activation of genes important for the remodelling of the bacterial cell surface to promote intracellular survival8. Sensing of the intracellular environment and subsequent bacterial membrane remodelling is dependent on regulatory proteins, including the two-component systems PhoP-PhoQ, OmpR-EnvZ, PmrA-PmrB, RcsB-RcsC, and Cya-Crp8, 9. S. Typhimurium pathogenesis is also highly dependent on two distinct type III secretion systems (T3SSs) encoded on Salmonella pathogenicity islands (SPI) 1 and 2, which function to transport effector proteins to the host cell cytoplasm. These effectors target host cell processes to promote S. Typhimurium invasion and intracellular survival (Supplementary Table S1). The two T3SSs are encoded at distinct locations on the chromosome and analysis of the genome suggests that they were acquired by horizontal gene transfer10. The need for two distinct T3SSs is probably linked to the differential use of these secretion systems under different environmental conditions, as other transcriptional and post-transcriptional regulatory mechanisms could presumably be used to deliver a different set of effectors through a single apparatus.
Following contact with host cells, the S. Typhimurium SPI-1 T3SS transports proteins across the plasma membrane to enable bacterial invasion, and results in the induction of intestinal inflammatory responses. By contrast, the SPI-2 T3SS transports proteins that are important for intracellular survival and vacuolar movement across the membrane of the Salmonella-containing vacuole (SCV)9. Many effectors translocated by the SPI-1 T3SS persist within host cells after bacterial internalization and may also be important for intracellular survival within the SCV11–16. In contrast to NTS, S. Typhi does not elicit a pronounced inflammatory response within the host gastrointestinal tract, and a number of factors that may contribute to such differences are discussed in Box 1.
Box 1. Salmonella enterica serovar Typhi.
Most of our understanding of salmonellae-host interactions is based on studies using S. Typhimurium infection of cultured cells and the inbred mouse model. Although the findings of such studies are often generalized to all serovars, much less is definitively known about the interactions between human host cells and the causative agent of typhoid fever, S. Typhi. The ability of S. Typhi to cause a human-restricted systemic disease is through the acquisition of novel genes and loss of others through gene loss or inactivation. For example, the S. Typhi-specific typhoid toxin is a tripartite exotoxin that causes DNA damage and subsequent cell-cycle arrest. This is mediated by the active subunit of cytolethal distending toxin, CdtB, which associates with PltA and PltB (which are homologues of pertussis toxin), and the tripartite complex is delivered to the extracellular milieu, thereby intoxicating neighboring cells195, 196. Secretion of the tripartite toxin is dependent on the muramidase TtsA, which is predicted to bind to peptidoglycan in the cell wall and may mediate secretion of typhoid toxin through a secretion mechanism that is thought to have evolved from phage endolysins197. However, the contribution of the toxin to virulence and host specificity is poorly understood, although its binding to specific carbohydrates may have a role in its localization in host cells. In addition, the absence of the effector GtgE (which cleaves and inactivates Rab29) enables Rab29-dependent vesicular export of typhoid toxin, which contributes to efficient trafficking of the toxin to its targets198, 199. S. Typhi also expresses a capsular polysaccharide that reduces intestinal inflammatory responses200, 201. This, in combination with the inability to produce very long O-antigen chains as part of LPS and the differential regulation of the SPI-1 T3SS, results in a reduced inflammatory response to S. Typhi compared with S. Typhimurium202.
Genome reduction contributes to the ability of S. Typhi to cause human-restricted systemic disease. Several virulence factors that are present in serovars that cause broad host range gastroenteritis are pseudogenes in S. Typhi and S. Paratyphi203. As mentioned above, GtgE is a cysteine protease secreted by NTS that cleaves specific Rab GTPases in their regulatory switch regions and this prevents their recruitment to the SCV; however, this protease is absent in S. Typhi. Heterologous expression of GtgE in S. Typhi promotes intracellular replication in human macrophages and survival in non-permissive mouse macrophages198, 204. Furthermore, the effectors SseJ and SopD2 are absent in S. Typhi, and their loss may contribute to the ability of S. Typhi to cause systemic, rather than gastrointestinal, disease203, 205.
Between 3–5% of individuals that become acutely infected with S. Typhi become chronic carriers and actively shed bacteria206. S. Typhi can access the gall bladder during acute infection, where it can reside in biofilms for decades and be sheltered from killing by conventional antibiotic treatment76. This finding, together with the development of an appropriate animal model for the carrier state, will greatly aid investigations that aim to eradicate the human reservoir. S. Typhi does not naturally replicate in mice, and considerable research into the development of an animal model has led to a number of models for infection which can give us important insights into this pathogen. These models include humanized mouse models that develop lethal S. Typhi infections, albeit the pathological features do not fully reflect human infection189–191. Ultimately, human genetics has an important role in susceptibility to infectious diseases, and to truly understand S. Typhi interaction with humans there must be a shift from the reliance upon cultured cells or mouse cell lines that have genetically similar backgrounds. Perhaps the future lies in more advanced human intestinal and immune cell models (such as the epithelial cell culture system developed to produce a panel of cell lines from individuals which exhibited differential adherence phenotypes for pathogenic E. coli) to understand individual responses to S. Typhi188, 207.
Together, the effector proteins that are delivered into the host cell by the two T3SSs promotes entry, survival and replication of bacteria within host tissues. Salmonellae induce actin cytoskeleton ruffling and macropinocytosis to promote bacterial uptake into non-phagocytic cells and the SPI-1 T3SS effectors reverse these changes after entry. Phagocytosis by macrophages also involves macropinocytosis, which occurs by both SPI1-dependent or SPI1-independent mechanisms. Macropinocytic uptake of salmonellae occurs rapidly, and bacteria initially reside in a spacious phagosomal compartment17, which undergoes a maturation process to form a unique compartment known as the SCV18. Salmonellae modify the lipid and protein content of the SCV and induce morphological changes, including cytosolic membrane associated actin polymerization and endosomal tubulation of the vacuolar membrane using the SPI-2 T3SS effectors9. Although some of the host factors manipulated by salmonellae effectors that are released from the SCV are known, many of the eukaryotic targets have yet to be discovered and the downstream activities of the majority of these effectors is still elusive. Most of the effectors are specifically localized to the SCV, whereas others are localized to the apical surface of polarized epithelial cells. Furthermore, effectors can have multiple mammalian binding partners, and their enzymatic activities may require these binding partners or specific post-translocation modifications, which complicates the identification of their specific functions. Understanding the molecular basis of the complex interplay between effectors delivered across the SCV membrane and host cells should lead to a greater understanding of the overall strategy that salmonellae use to achieve a successful intracellular lifestyle. Although many questions remain unanswered, despite intense study of these model organisms, in this Review, we summarize what is currently known about the complex interplay between the arsenal of effectors that Salmonella secretes and the pathways altered in host cells, including cytoskeletal changes, inflammatory responses, membrane modifications, vacuolar trafficking and autophagy.
Invasion of non-phagocytic epithelia
Salmonellae effectors translocated by the SPI-1 T3SS are important for bacterial invasion of non-phagocytic epithelia (Figure 1). The effector protein SopB is an inositol phosphatase that affects a number of cellular pathways during infection, including membrane ruffling, initiation of M-cell development and inhibition of SCV fusion with lysosomes13, 19–21. Bacterial invasion is mediated in part by SopB activation of Rho kinase-dependent actin rearrangements at the host cell membrane22–24. SopB also recruits AnxA2 which functions as a platform for actin rearrangements25. On the SCV, SopB can generate Phosphatidylinositol 3-phosphate (PI(3)P) by recruiting Rab5 and the PI3 kinase, Vps3426. SopB decreases levels of acidic lipids on the SCV, which may alter Rab family GTPase trafficking and antagonize SCV fusion with lysosomes21. SopB also influences the ability of the bacterium to transform follicular associated epithelial cells into M-cells through activation of Wnt/β-catenin signalling20; induction of Wnt signalling leads to activation of the receptor-activator of NF-κB ligand (RANKL) and its receptor RANK, which are sufficient to initiate M-cell development and can lead to promotion of salmonellae invasion20. The distinct activities of SopB are spatiotemporally controlled in vivo through mono-ubiquitination of distinct residues at specific times during the infection, which results in the persistence of SopB on the SCV but it is removed from the plasma membrane following entry16, 27.
Figure 1. Host pathways manipulated by Salmonella during epithelial cell invasion.
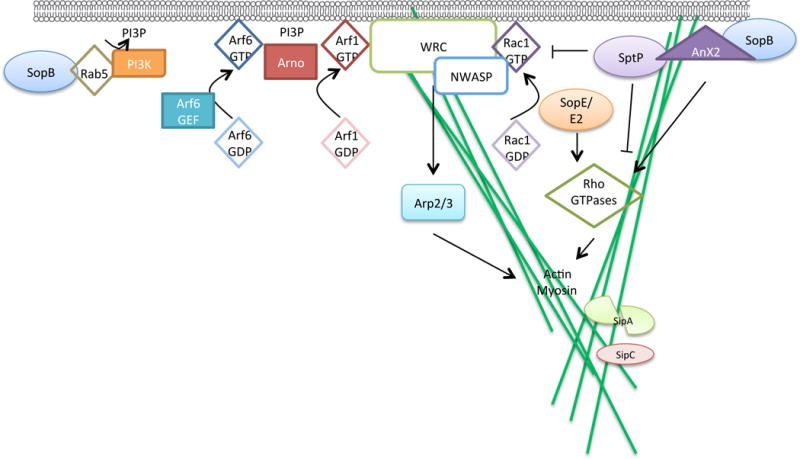
Following contact with host cells, Salmonella spp. translocate effectors via the Salmonella pathogenicity island 1 (SPI-1) type III secretion system (T3SS) to mediate invasion. On the SCV and potentially at the plasma membrane, SopB recruits Rab5 and the PI3K Vps34 to generate PI(3)P. SopB can also recruit AnxA2 to the membrane, which functions as a platform for actin rearrangements, and SopB also activates Rho-kinase dependent actin rearrangements. SopE and SopE2 activate host Rho GTPases to promote bacterial internalization by actin reorganization. Activation of Rac1 (conversion from Rac1-GDP to Rac1-GTP) by SopE in particular promotes the recruitment of factors to the host cell membrane (such as WAVE regulatory complex (WRC) and N-WASP), which promotes actin rearrangements at the host cell membrane, proximal to extracellular Salmonella spp.. The host Arf1 GEF, ARNO, is activated by binding to Arf6 and is recruited to the plasma membrane by PI(3)P, which is possibly generated by SopB. Activation of Arf1 promotes WRC-dependent actin polymerization and bacterial uptake. WRC and N-WASP activate actin related proteins (Arp2/3) which results in localized actin polymerization to promote bacterial uptake into non-phagocytic cells. The effectors SipA and SipC promote bacterial invasion through their actin bundling functions. SipA may need to be cleaved into two domains (potentially by caspase-3) for activation. SptP promotes restoration of epithelial cell architecture after bacterial entry by reversing the activation of Rho GTPases.
Reorganization of the actin cytoskeleton is also triggered following activation of the small Rho GTPases Rac1 and Cdc42 by the guanine nucleotide exchange factor (GEF) activity of the SPI-1 effectors SopE and SopE228–32. Salmonellae effectors trigger localized actin polymerization by influencing recruitment and activation of the nucleation promoting factors (WAVE and N-WASP)33, 34. The host Arf1 GEF, ARNO, is recruited by PI(3)P, possibly owing to its generation by SopB at the plasma membrane34. ARNO (which is activated by the binding active Arf6, Arf6-GTP) activates the GTPase Arf1, which then promotes WAVE regulatory complex (WRC)-dependent membrane ruffling and bacterial invasion34, 35. The GEF activity of SopE triggers recruitment of WRC and N-WASP, leading to activation of actin related protein (Arp2/3) at the cell membrane, resulting in localized actin polymerization and membrane ruffling which promotes bacterial uptake into phagosomes inside the non-phagocytic epithelial cell33, 34.
Bacterial internalization is also promoted by SipA and SipC, which bind directly to actin at the site of insertion of the T3SS translocon, of which SipC is a component (Figure 1). SipA inhibits actin depolymerization and increases actin bundling at the site of bacterial entry into epithelial cells36, 37, and SipC bundles and nucleates actin on insertion of the translocon to promote invasion38, 39. SipC may also interact with intermediate filament proteins40, 41. After bacterial entry into host cells, the architecture of the cytoskeleton is restored. The GTPase activating protein (GAP) SptP reverses the activation of Rac1 and Cdc42 by SopE, SopE2 and SopB, to restore the architecture of epithelial cells42, and SopE is inactivated by proteasomal degradation43.
SPI-1 mediated inflammation
Localized inflammation of the intestinal tract is essential for providing a growth advantage for luminal NTS salmonellae, and enhances bacterial transmission through multiple mechanisms4. Many of the SPI-1 effectors (including SopE, SopE2, SopB, SipA, SipC and SopA) contribute to intestinal inflammation44–46 by stimulating production of the proinflammatory cytokine IL-8 through the mitogen-activated protein kinase (MAPK) and NF-κB pathways, destabilizing tight junctions and stimulating neutrophil transepithelial migration into the intestinal lumen (Figure 2)1, 47, 48. Host Toll-like receptors (TLRs) are activated following the sensing of salmonellae components (for example, lipopolysaccharide (LPS) which promotes macrophage activation and increased killing by the phagosomal compartment, as well as transcriptional activation of inflammatory caspase genes49.
Figure 2. Salmonella-induced inflammation promotes pathogen transmission.
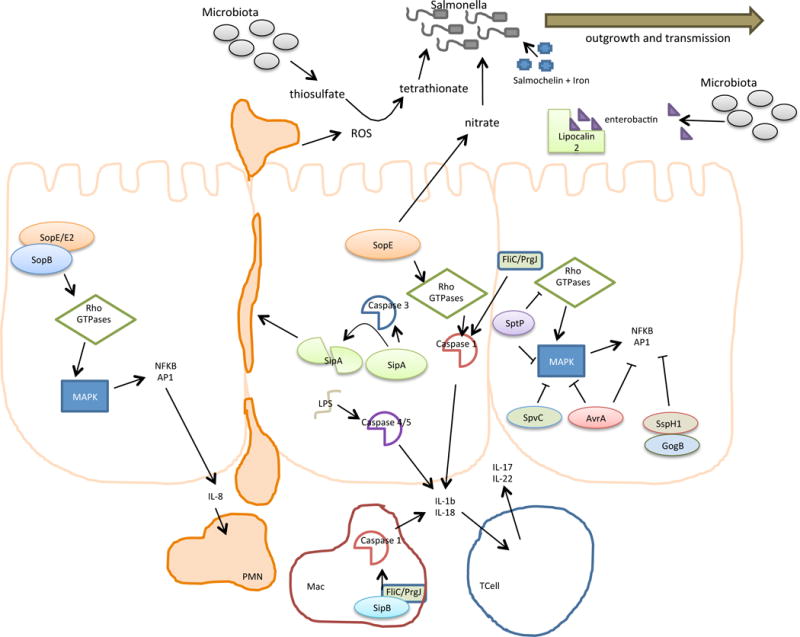
Localized inflammation in the intestinal tract is important for promoting transmission of salmonellae. Activation of Rho GTPases by the effectors SopE, SopE2 and SopB induces MAPK pathways, thereby activating NFκB and AP1, which stimulates production of the proinflammatory cytokine IL-8 to promote transepithelial migration of neutrophils into the intestinal lumen. SipA activates Caspase-3 and may be subsequently cleaved by this protease;; cleavage of SipA also promotes transepithelial migration of neutrophils. Caspase-1 is activated by FliC (a flagellin protein), PrgJ (a rod protein of the T3SS apparatus) and possibly by the effector SipB in macrophages, which may stimulate the release of IL-1β and IL-18. SopE activation of Rho GTPases can also activate Caspase-1 in epithelial cells. Intracellular LPS can activate Caspases4 and 5, resulting in the release of mature IL-1β and IL-18, which promotes the synthesis of IL-17 and IL-22 by T-cells and amplifies inflammation in the intestinal mucosa. SopE also activates the production of nitrate by host cells, which can be used by luminal Salmonella spp. for respiration. Pathogen invasion also induces the release of reactive oxygen species (ROS) and lipocalin-2. ROS converts the respiratory by-product thiosulfate (generated by the microbiota) into tetrathionate, which can be used by Salmonella spp. (but not the microbiota) for respiration. Lipocalin-2 sequesters the iron siderophore enterochelin from the microbiota, but the Salmonella spp. siderophore salmochelin escapes sequestration. Together, these changes promote outgrowth and transmission of Salmonella spp. that reside in the intestinal lumen. The MAPK inflammatory responses are dampened by the activities of SptP, SpvC, AvrA, SspH1 and GogB, but the mechanistic basis of this is not fully understood.
Inflammation in response to Salmonellae is also induced by activation of inflammatory caspases 1 and 11 in mice and caspases 1, 4, and 5 in human cells50. Cytoplasmic delivery of the flagellar FliC filament protein and the PrgJ rod protein by the T3SS-1 apparatus activates caspase-1 in macrophages51, 52 (Figure 2). Caspase-1 activation triggers release of the proinflammatory cytokines IL-18 and IL-1β by macrophages, that in turn, promote the release of IL-17 and IL-22 by T cells, thereby amplifying inflammation in the intestinal mucosa53. The SPI-1 translocon protein SipB is inserted in the plasma membrane and may migrate to the mitochondria, damages to plasma and mitochondrial membranes might also contribute to caspase-1 activation.54. In addition to the role of SopE in invasion of non-phagocytic cells, its GEF activity for the Rho GTPases Rac and Cdc42 can activate caspase-1 in stromal cells, which elicits gut inflammation during infection, probably as a result of the reorganization of actin in response to bacterial-mediated endocytosis55. Caspase 11 in mice and caspases 4 and 5 in humans may have a role in intestinal inflammation and restriction of NTS through their activation by cytoplasmic LPS, which may be generated by destabilization of the SCV in epithelial cells or another unknown transport mechanism50, 56. SipA may also have a direct role in inflammation, independent of its role in bacterial invasion, by promoting PMN migration across the intestinal epithelium57 and by activating caspase-3 in host cells58.
Competition with the intestinal microbiota
Salmonellae have several mechanisms to compete with the gut microbiota. For example, intestinal inflammation results in luminal accumulation of lipocalin-2, a host protein that sequesters the bacterial siderophore enterochelin, which is required by the microbiota for the acquisition of iron. The salmonellae siderophore salmochelin is not sequestered by lipocalin-2, which provides the pathogen with a growth advantage compared to other enteric bacteria59 (Figure 2). Salmonellae invasion of cells results in the release of reactive oxygen species into the lumen, which reacts with thiosulfate, a respiratory by-product generated by the microbiota and the intestinal lumen, to generate tetrathionate. Tetrathionate is used by Salmonella spp. but not the microbiota, as a terminal electron acceptor to support anaerobic or microaerophillic growth3. Furthermore, Salmonella spp. induce SopE-dependent production of nitrate by the host, which can be used by anaerobic nitrate respiration to enhance growth of Salmonella spp. and other members of the Enterobacteriaceae60. These and other changes allow Salmonella spp. to outgrow the intestinal microbiota, providing them with the opportunity for more effective transmission owing to increased proliferation in the intestinal lumen.
Dampening of inflammation after entry
After entry into host cells, salmonellae reverse actin cytoskeleton rearrangements (which are required for entry) and dampen inflammation using other effectors. The tyrosine phosphatase SptP reverses MAPK-mediated inflammation and IL-8 secretion61, 62, and it persists during the late stages of infection to dephosphorylate the AAA+ ATPase valosin-containing protein, which is important for intracellular replication and endosomal tubule formation15. Both the SPI-1 effector AvrA and the SPI-1/2 E3 ubiquitin ligase effector SspH1, contribute to downregulation of Salmonella-induced IL-8 production by epithelial cells. SspH1 binds to and ubiquitinates PKN1, which inhibits NF-κB activity63–65. The acetyltransferase activity of AvrA can suppress the inflammation that is triggered by apoptosis of macrophages and enhance bacterial intracellular survival66. Several other effectors target the MAPK and NF-κB pathways to down-modulate inflammation. These include AvrA67, 68, the phosphothreonine lyase SpvC, which increases Salmonella spp. dissemination in mice69, and the anti-inflammatory effector GogB which blocks ubiquitination and degradation of IκB, leading to inhibition of NF-κB proinflammatory signalling70. In addition, the SPI-1/2 effector SpvC reduces inflammation by inhibiting the production of IL-8 and TNF-α71. Another SPI-1/2 effector, SteA elicits changes in the expression of several genes in HeLa cells, including the activation of genes that regulate extracellular matrix organization, cell proliferation and serine/threonine kinase signaling pathways; and the repression of genes involved in regulating immune processes, cell death, adhesion and migration72, which suggests that this effector can also modulate inflammation in the host. The effectors SseF, SifA, SspH2, SlrP, PipB2 and SseI can inhibit the migration of dendritic cells73, and several of these effectors (SspH2, SlrP, PipB2, SifA, SopD2 and SseFG) also inhibit antigen presentation by dendritic cells and inhibit T-cell proliferation74. Collectively, the results of these studies suggest that both the innate and adaptive immune response of the host is activated and inhibited during salmonellae infection.
Survival and replication within the SCV
For many years, it has been recognized that intracellular salmonellae can exist in both replicative and non-replicative states75. In recent years, increased attention has been devoted to the study of the non-replicating population because they display a reversible, antibiotic-tolerant phenotype and persist following antibiotic treatment. These so-called “persisters” or drug-tolerant phenotypic variants of salmonellae seem to arise stochastically and are now more amenable to study owing to the development of new technologies (Box 2). In addition to antibiotic tolerance, this phenotypic diversity of individual cells during infection is an interesting area for further exploration, particularly in the case of S. Typhi, which causes a relapsing infection with a chronic phase that might be dominated by non-replicating bacteria that are intracellular or in biofilms associated with gall stones and bladder stones76. The molecular basis of phenotypic heterogeneity among individual bacteria and SCV variability is an area that requires further study to determine whether specific populations of bacteria may have different functions in the infection process.
Box 2. Salmonella persister cells.
Phenotypic heterogeneity of Salmonella spp. populations within endocytic vacuoles was originally documented in the early 1990’s and indicated that salmonellae populations could divided into two subpopulations: those that rapidly replicate and those that do not replicate but are still viable75. These non-replicating variants are recalcitrant to killing by antibiotics (termed antibiotic tolerance) and have been termed persisters. The subject of bacterial persistence is an area of renewed interest owing to the increasing number of multi-drug resistant bacteria combined with the dwindling pipeline of novel antibiotics. Persisters arise stochastically owing to the inherent heterogeneity of bacterial populations208, and their formation can also be induced following exposure to stresses such as sub-inhibitory concentrations of antibiotics209. Phenotypic heterogeneity in isogenic bacterial populations has been observed frequently, but the mechanisms behind this phenomenon have been difficult to dissect with traditional tools that employ bulk methodologies. Bacterial signalling using second messengers, like cyclic di-GMP, enables a rapid response to changing environmental conditions and is one way in which phenotypic variation can be achieved. A FRET-based biosensor has been used to measure the variation in the expression of cyclic di-GMP in individual cells, which has been key to our understanding of how differences in the levels of this factor between cells correlate with phenotypic diversity210. The fluorescence dilution technique has also been used to detect persisters by monitoring the growth rate of single cells in vivo and was recently been applied to examine the emergence of S. Typhimurium persisters194. Internalization of S. Typhimurium by macrophages was found to rapidly induce the persistence phenotype as a result of vacuolar acidification and nutritional deprivation. The emergence of S. Typhimurium persisters in macrophages was also found to be dependent on the Lon protease which degrade the antitoxins of 14 putative toxin-antitoxin modules, resulting in the release of toxin activity and subsequent growth arrest194. As persisters can revert back to growth in new environmental conditions, they may have a role in re-occurring infections. The methods described above and other developing technologies to follow and/or isolate individual cells from a population will greatly enhance our ability to understand phenotypic diversity within isogenic populations.
Despite the challenges associated with studying intracellular bacteria and SCV diversity because of the large number of transient interactions and modifications that take place, several principles about the SCV within epithelial cells are known. Beginning immediately after entry into epithelial cells, the SCV undergoes several vacuolar modifications that distinguish it morphologically from typical phagosomal compartments. Although defined functions for the large repertoire of salmonellae SPI-2 effectors remain to be established, these proteins are required for many of the SCV vacuolar modifications, and are involved in its microtubule-based vacuolar movement in the host cell (Figure 3). Our current knowledge regarding the identity, targets and functions of the SPI-1 and SPI-2 effector proteins is detailed in Supplementary Table S1. Maturation of the SCV involves transient interactions with early endosomes, which result in the recruitment of early endosomal markers, including the transferrin receptor, early endosomal antigen-1 and the small GTPase Rab577, 78. These markers are subsequently replaced with late endosomal markers and often lysosomal markers including the lysosome-associated membrane proteins (LAMPs), the GTPase Rab7, RILP, vacuolar ATPase and cholesterol77, 79–81. Other lysosomal markers including mannose-6-phosphate receptor (MPR) and lysobisphosphatidic acid can be absent from the mature SCV in some cell types (mostly HeLa cells)80, although bacteria can survive within the SCV in macrophages despite direct fusion of the SCV with the lysosomal compartment82. The observed variation in SCV surface markers in different experimental systems is probably a reflection of whether the SPI-1 T3SS is used for entry (in macrophages), the degree of caspase-1 activation and cell toxicity, and diversity in the secretion of SPI-2 effectors and bacterial gene expression within the SCV. Despite this complexity and the lack of uniformity in experimental systems, it is likely that modifications of the SCV surface by SPI-2 effectors contribute to intracellular survival.
Figure 3. Salmonella manipulation of host membranes.
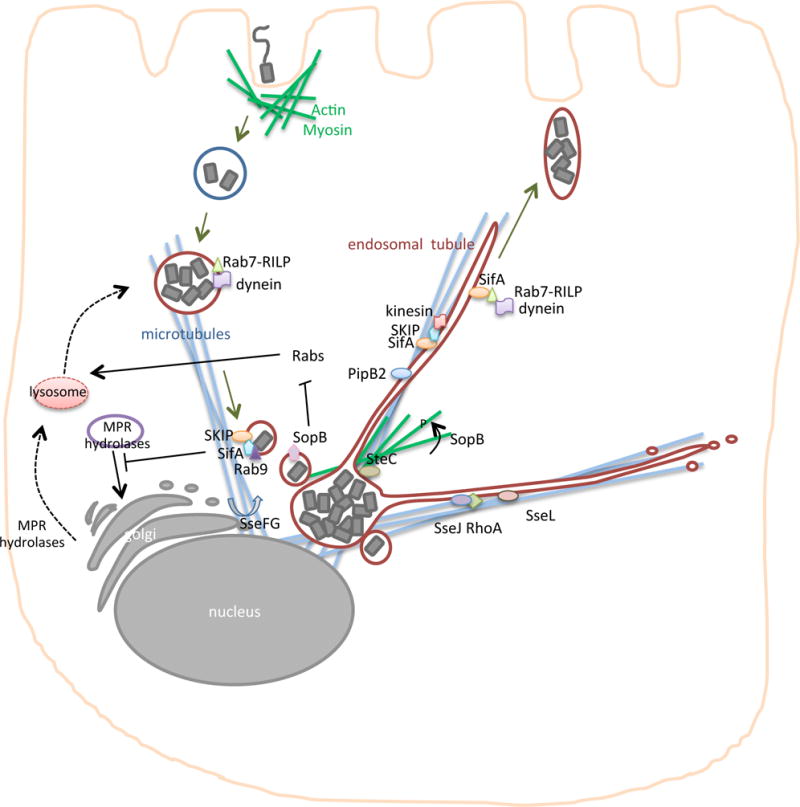
After invasion, Salmonella reside in a vacuolar compartment that undergoes various surface modifications and alterations that distinguish it morphologically, forming the Salmonella-containing vacuole (SCV). In some cell types (such as epithelial cells), the SCV acquires markers of late endosomal compartments including LAMP-1 (purple), Rab7, vacuolar ATPase and cholesterol. Rab7 interacts with Rab7-interacting lysosomal protein (RILP) and the microtubular motor protein, dynein, and this complex is important for centripetal movement of the SCV in the cell early in infection. Salmonella pathogenicity island 2 (SPI-2) effectors (shown in deep red) are released across the SCV membrane into the host cell cytosol by the T3SS. These effectors distribute to different locations in the cell and are required for SCV vacuolar modifications and microtubular-based movement, including promoting the dynamic formation of endosomal tubules (ETs). Binding of SseJ to RhoA results in direct modification of the lipid content of the SCV and ETs (producing cholesterol esters), whereas SseL counteracts this activity by decreasing lipid droplet accumulation in cells. SseF and SseG promote microtubule bundling and the aggregation of endosomal vesicles;; in addition, recruit Golgi-derived vesicles to the SCV. SteC induces actin rearrangements around the SCV and can alter the position of the SCV within the cell, and SopB induces phosphorylation and activation of myosin II indirectly. SopB phosphatase activity reduces the membrane charge of the SCV by preventing the accumulation of Rabs, which are important for promoting SCV fusion to lysosomes. SifA binds to the eukaryotic SifA and kinesin interacting protein (SKIP), which activates the microtubular motor protein kinesin to mediate the extension of ETs. PipB2 is involved in recruiting kinesin to the SCV and ETs,and SifA-SKIP is suggested to bind Rab7 and Rab9. SifA binding to Rab7 may disrupt interactions between Rab7-RILP and dynein, which would result in increased peripheral movement of ETs. SifA binding to Rab9 blocks retrograde transport of mannose 6 phosphate receptor (MPR) hydrolyases and MPR to the Golgi, which blocks lysosomal fusion with the SCV. Salmonella spp. manipulate host membranes to direct the SCV towards the nucleus and then promotes SCV movement towards the cell periphery where bacteria are presumably released into the intestinal lumen to infect neighbouring cells.
Further evidence that a major function of the SPI-2 T3SS is modification of the endosomal membrane is that many SPI-2 effector proteins localize to the SCV, including SifA and SseJ, which are the the best characterized. The main function of SseJ on the SCV is activation of its glycerophospholipid:cholesterol acyltransferase activity (by binding to RhoA) to directly modify the composition of cholesterol and phospholipids in these membranes, ultimately increasing the accumulation of cholesterol esters within lipid droplets in the cytoplasm83–85 (Figure 3). The lipid changes on the SCV have the potential to dramatically alter the repertoire of proteins that associate with the SCV; for example, SifA is recruited and functions as a link to connect the SCV to the microtubular network86. The effector SseL, which is a deubiquitinase, may prevent lipid droplet accumulation in cells, potentially antagonizing SseJ activity and preventing lipid composition on the SCV87. Many other effectors that localize to the SCV may alter protein or lipid content, but their specific activities are currently unknown. As detailed in Supplementary Table S1, proteins such as SspH2 (a ubiquitin ligase with unknown targets) localize to the SCV in non-polarized cells and the apical surface of polarized epithelial cells, and may function in the removal of host proteins through ubiquitination. The specific enzymatic activities of the wide array of SPI-2 effector proteins may manipulate small GTPases, remove proteins from the SCV by post-translational modification, inhibit the trafficking of proteins through endocytic recycling pathways or deubiquitination, among other complex activities, all of which result in a vacuole with unique protein and lipid content.
SPI-2 mediated movement of the SCV
The SCV traffics via the microtubule network to the perinuclear region of non-polarized host cells (such as HeLa) in a manner that is dependent on SPI-2 effectors (Figure 3)88. Salmonellae induce sorting nexin tubule formation early after invasion, which contributes to the recruitment of Rab7 and LAMP-189, 90. Rab7-interacting lysosomal protein (RILP) links active Rab7 to the motor protein dynein91, a complex that is important for the centripetal movement of the SCV at earlier stages of infection92, 93. At later stages of infection, the SCV is displaced to the cell periphery where it could be important for bacterial transfer between epithelial cells94. The SPI-2 effector SifA links the SCV to the microtubular network through its interaction with the host SifA and kinesin-interacting protein (SKIP), which is relocated from the microtubule organizing centre to the SCV95. Other SPI-2 effectors, including PipB2 and SseF, bind to the SCV and microtubular motor proteins and may have a role in specific intracellular movement96, 97, suggesting that a major function of SPI-2 could be to move the SCV within the host cell cytoplasm. It is interesting to speculate that this movement along the microtubular network may be directional when bacteria are within polarized epithelial cells that have a specific microtubular orientation.
Further evidence that SPI-2 effectors function in the manipulation of the microtubular network comes from work with S. Typhimurium, which has shown that the bacterium induces the formation of dynamic LAMP-1 positive microtubule-dependent endosomal tubule (ET) extensions, which are historically termed Salmonella-induced filaments (Sifs)98–100. The term Sif is somewhat confusing as traditional eukaryotic filaments are actin based fibres, whereas Sifs are formed via actin-independent, but microtubular-dependent, alteration of endosomal membranes. The SPI-2 effectors, SifA, PipB2, SseF, SseG, SseJ, SpvB, SteA and SopD2 all contribute to ET dynamics and SCV movement100–106. Recruitment of the microtubular motor protein kinesin to the SCV results in ET extension, which is dependent on PipB296 and the host Arf GTPase Arl8B107. Following its recruitment, kinesin is activated by the eukaryotic factor SKIP, which is complexed to SifA (Figure 3)86. SifA also associates with the SCV and ETs through its carboxyl-terminus CAAX motif (C is cysteine residue, AA are two aliphatic residues, and X represents any C-terminal amino acid), which is post-translationally lipidated108, 109. The SifA-SKIP complex links the SCV to the microtubular network and initiates the fission of periphery directed transport vesicles, triggering ET formation along microtubules86, 110 (Figure 3). SifA may recruit Rab7 to displace the Rab7-RILP-dynein motor complex and thereby increase the peripheral movement of ETs92. In HeLa cells, SifA-SKIP sequesters Rab9 and blocks Rab9-dependent retrograde trafficking of mannose-6-phosphate receptors, which decreases lysosomal function and promotes bacterial survival111. Salmonellae induce tubulation of membranes that are positive for secretory carrier membrane protein 3 (SCAMP3)112, a protein normally found in the trans-Golgi network, which suggests that the tubular network surrounding Salmonella spp. is derived from both the Golgi and endosomes. Interestingly, there is evidence that the host secretory pathway communicates with the SCV, since S. Typhimurium disrupts exocytosis suggesting that secretory traffic may be directed to the SCV as part of the mechanism of this unique compartment’s longevity within cells113. Although no physiological function has been attributed to membrane tubulation, this process is correlated with the ability of S. Typhimurium to cause disease as mutation of effectors (such as PipB2, SopD, SopD2, SseJ, SifA) required for ET formation results in virulence attenuation in inbred mice96, 102, 114, 115. The hypotheses that ETs may be used to collect membrane and nutrients important for salmonellae survival, to dilute lysosomal enzymes or to promote the stability of the SCV membrane have yet to be verified experimentally. Endosomal tubulation could be a result of effector inhibition of fission, which typically occurs to recycle endosomes back to the plasma membrane and hence, the inhibition of this step might maintain membranes and proteins (that are normally recycled) as components of the SCV.
Overlapping functions of effectors
Irrespective of the mechanisms involved, SPI-2 and its effectors promote replication of S. Typhimurium in bone marrow-derived macrophages from mice and virulence in inbred mice116. In the systemic mouse model, strains lacking individual SPI-2 effectors have attenuated virulence, although these single deletions do not lead to the more pronounced attenuation that accompanies inactivation of the SPI-2 T3SS, suggesting that effectors have overlapping and partially redundant functions. Much attention has been placed on the S. Typhimurium mouse model of infection, in which the importance of individual effectors is measured on the basis of virulence phenotypes. However, the acquisition of effectors rarely has a neutral effect on fitness. and their expression alone suggests that the effector has a pivotal role in pathogenesis, and the seemingly partially redundant functions may reflect their differential importance in different animal hosts. Another important and often overlooked variable in experiments using strains that lack specific effectors, is the possible increased delivery of other effectors that may mask, enhance or otherwise affect virulence phenotypes. These issues can only be circumvented by testing the effects of point mutations of crucial functional residues of the effector, but these studies are only feasible after function and/or binding partners have been identified. This is best illustrated by our understanding of the virulence defect of the SifA deletion mutant. Strains that lack SifA have the most pronounced virulence defects117, and SseJ and SopD2 promote release of bacteria into the cytosol in this mutant106, 118. The presence of cytosolic salmonellae results in rapid bacterial clearance in macrophages owing to the induction of host stress response pathways that are not normally activated in response to intravacuolar bacteria. Vacuolar integrity is however not the only variable that is responsible for the virulence defect of sifA-null bacteria, as the virulence defect is SopD2-dependent but not SseJ-dependent106, 118. Other SPI-2 effectors that confer virulence defects when deleted individually are SpvB, SseJ, SseF, SseG, SseI, SopD2, SteA and PipB297, 105, 114, 115, 117, 119–122; and other than SseJ, in which a site-directed mutation that confers a loss in activity has been generated, it is important to interpret these results as preliminary since they could reflect an increased delivery of other effectors that may contribute to attenuated virulence.
Host modification of effectors
Inside host cells, salmonellae effectors undergo modifications and participate in interactions similar to host proteins, which enables them to perform their functions by influencing their localization and activity. Lipidation of some effectors has been documented, which probably affects their membrane localization. For example, SifA undergoes prenylation108 and SspH2 and SseI are S-palmitoylated123. SopB27, SptP43 and SopA124 undergo ubiquitination, and phosphorylation of AvrA68 is important for activity. In addition, caspase-3 can cleave SipA into two domains, but this occurs extracellularly and not inside cells where SipA functions, therefore cleavage by Caspase-3 may not be required for SipA function58. In the case of SseJ, its enzymatic activity is induced by the host factor RhoA84 and its localization to endosomal membranes is dependent on its association with RhoA84. Co-expression of SseJ and SifA or SseJ and RhoA induces the formation of ETs in HeLa cells, supporting the idea of cooperativity between these proteins to induce tubulation95. The carboxy termini of SifA and SifB have a fold similar to GEFs of Rho GTPases although neither effector has known GEF activity95. SifA binds GDP-bound RhoA95, 125 and promotes the activity of a yeast RhoA homologue (Rho1) on peroxisomes126, suggesting that it may regulate SseJ enzymatic activity by activating RhoA95, 125. Despite having a structural fold similar to GEFs, SifA lacks conserved catalytic residues important for GEF activity and they differ from those in other bacterial GEFs127, which supports the idea that SifA functions as a scaffold for the assembly of other proteins on GTPases; however, a biochemical link between SifA and SseJ remains to be defined.
Induction of host stress response pathways
Activation of autophagy
Autophagy is a stress-induced metabolic adaptation that is triggered in eukaryotic cells in response to sublethal stress, and is characterized by lysosomal degradation of cytosolic components, including bacteria. Its role in the clearance of salmonellae131 is unknown because Salmonella spp. largely (and potentially exclusively) reside within the SCV. However, in tissue culture models and inbred mice, a small percentage of NTS escape the vacuole and hyper-replicate, leading to the extrusion of the infected cell132. Data using inbred mice models suggest that host cell extrusion may be physiologically relevant in restricting intraepithelial proliferation in the intestinal mucosa {Sellin, 2014 #302}, although since it occurs for only a minority of cells, its overall importance in the infectious process is unknown. Cytosolic detection may be most relevant to induction of innate and acquired immune responses that are related to autophagy and the inflammasome. Deficiencies in autophagy related genes restrict the replication of intracellular Salmonella spp. in the unicellular model organism Dictostyleum discoideum and in the multicellular model organisms Caenorhabditis elegans and Drosophila melanogaster133. In the mouse model, autophagy in intestinal epithelial cells protects against dissemination of invasive bacteria134. Polymorphisms in the autophagy related gene ATG16L1, which is associated with susceptibility to Crohn’s disease in humans, correlate with reduced clearance of salmonellae in cells and animal models, owing to decreased induction of autophagy and increased IL-1β secretion135, 136. However, these polymorphisms also influence granule formation in Paneth cells in an autophagy-independent manner137, suggesting that some effects on bacterial clearance may not be a consequence of altered autophagy alone. Irrespective of whether NTS escape to the cytoplasm138 through alteration of the SCV by SPI-2 effectors, it is plausible that autophagy-mediated targeting of the SCV for destruction, and its interactions with other host innate immune defence pathways, has an effect on host clearance of NTS.
Cellular models have shown that damaged SCVs and cytosolic bacteria can be targeted for autophagic degradation (Figure 4) as they recruit autophagosomes through ubiquitin-LC3adaptor proteins, such as p62 (sequestosome SQSTM1) and nuclear dot protein 52 (NDP52)139. Phosphorylation of the adaptor optineurin (OPTN) by protein kinase TANK binding kinase 1 (TBK1) can also promote autophagy of ubiquitin-coated cytosolic salmonellae140. Ubiquitination results in the recruitment of additional components of the autophagy machinery, including the Atg16L1 complex, the ULK1 complex and Atg9L1 around cytosolic salmonellae139, 141–143. Furthermore, SCV membrane damage promotes the accumulation of galectins 3 and 8, which recruit NDP52 to initiate autophagy144. The lipid second messenger diacylglycerol (DAG) is also observed on damaged SCVs, independent of ubiquitin accumulation145. DAG activation can also activate SQSTM1 and protein kinase Cδ to induce the production of reactive oxygen species via NOX2 complex assembly145. These host factors contribute to autophagic clearance of cytosolic bacteria and damaged SCVs, but salmonellae use counteractive mechanisms to subvert this process. The reactivation and recruitment of mammalian target of rapamycin (mTOR) to SCVs, which occurs following the acute and transient amino acid starvation response that is induced on infection, inhibits the induction of autophagy and possibly provides an escape mechanism for the bacteria146. It has been suggested that in macrophages, an unknown SPI-2 effector is involved in the recruitment of focal adhesion kinase to the SCV, which can then activate mTOR through Akt147. Furthermore, it is likely that autophagic targeting of damaged or altered SCVs influences antigen presentation, as well as adaptive and autoimmunity, and a deeper understanding of this process should expand our knowledge of chronic human diseases.
Figure 4. Autophagy and inflammasome activation in response to Salmonella spp. infection.
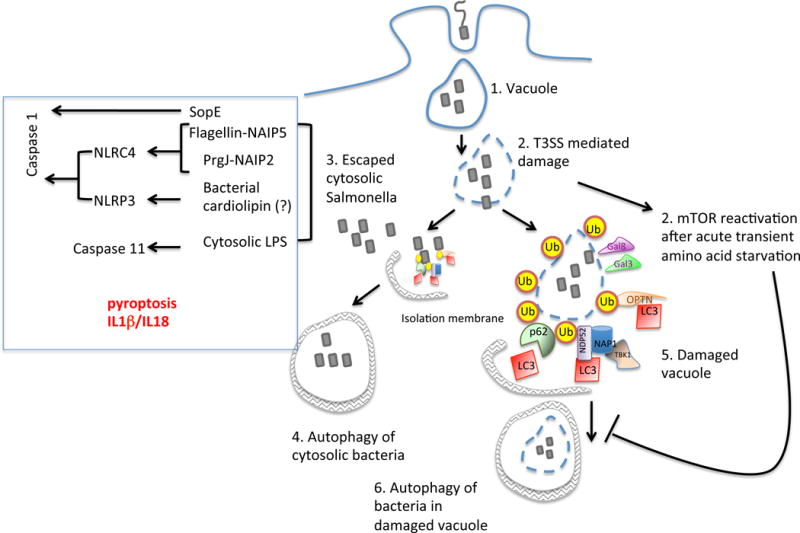
Most intracellular S. Typhimurium reside in SCVs but in some cellular models of infection, some bacteria escape into the host cytosol. Both cytosolic bacteria (shown in step 3) and damaged SCVs (shown in step 5) can be ubiquitinated, and are therefore recognized by the bridging adaptors such as NDP52, OPTN or p62, which leads to the recruitment of LC3 and autophagic clearance. In parallel, Salmonella spp. mediate an acute and transient amino acid starvation response, which reactivates mTOR, thereby inhibiting autophagy. The physiologic significance of this on bacterial clearance remains unclear, since most Salmonella spp. remain inside the SCV. Detection of specific bacterial components (such as flagellin, the T3SS rod protein PrgJ and LPS) in the host cytosol can trigger activation of inflammasome-mediated cell death via caspase-1 and -11 (shown in step 7). SPI-1 mediated detection of PrgJ and flagellin by NAIP2 and NAIP5 respectively in mice trigger NLRC4 mediated Caspase-1 activation, while citrate and potentially bacterial cardiolipin can trigger Caspase-1 activation via NLRP3 inflammasome. The detection of cytosolic LPS in the host can trigger Caspase-11 activation.
Inflammasome activation
In addition to autophagy, inflammasome activation can occur following the detection of salmonellae. Inflammasomes are molecular platforms that respond to pathogens and tissue damage by promoting cell death (in a process known as pyroptosis) and pro-inflammatory signalling148. Their importance for innate immune defence against salmonellae is evidenced by mice that lack key components (such as caspases-1 and 11, IL-1β or IL-18), which succumb earlier to infection and have higher bacterial loads149, 150. Caspase-1 activation by salmonellae can occur via the activation of either the NLRC4 or NLRP3 inflammasome, both of which are individually dispensable in vivo, indicating that they have redundant roles early in infection151, 152. Coordinated activation of both NLRs sustains the release of IL1β, which is important for progression of the infection in mice153. The Apoptosis Speck like CARD domain containing protein (ASC) associates with the inflammasome and is crucial for salmonellae-induced IL-1β secretion152. The physiological significance of ASC containing foci is unknown, as the CARD domain of NLRC4 can interact directly with the CARD domain of Caspase-1, which suggests that ASC may not be required as a linker. However, it has been proposed that ASC might have a role in signal amplification function and activation of NLRP3 and NLRC4 requires ASC in vitro152, 154, 155.
The NLRP3 inflammasome can be activated by citrate, which is secreted by S. Typhimurium mutants that lack the TCA cycle enzyme aconitase156. Aconitase-deficient salmonellae show reduced acute systemic virulence in mice after oral administration and are defective in their ability to persist. Although mitochondrial cardiolipin is a direct ligand for NLRP3157, it is unknown whether cardiolipin in bacterial membranes, (which may be altered by dysregulation of the TCA cycle), can also activate NLRP3. The cardiolipin content of the salmonellae outer membrane increases in a PhoPQ-dependent manner, indicating that it might increase in host cells where PhoPQ is activated, and could be involved in inflammasome activation158. Caspase-1 activation by NLRC4 depends largely on the expression of flagellin159–163 and the presence of cytosolic flagellin is sufficient to activate NLRC4, whereas mutation of three conserved carboxy-terminal hydrophobic amino acids in flagellin abolishes recognition160. As mentioned above, in addition to flagellin, the inner rod proteins of the salmonellae T3SSs are capable of activating NLRC4. These rod proteins adopt an α-helical secondary structure and may oligomerize to form a protein translocation channel154, 166. Cytosolic recognition of flagellin and inner rod proteins is mediated by the NLR family Apoptosis Inhibitory Proteins (NAIP)167, 168. Mouse NAIP5 directly recognizes flagellin and mouse NAIP2 recognizes the SPI-1 T3SS rod protein PrgJ, but not its SPI-2 homologue SsaI167. The ligand specificity of NAIPs is mediated by an internal region that encompasses several NBD-associated α-helical domains169, and ligand binding is required for NAIP oligomerization with NLRC4. Unlike the multiple NAIP genes in mice, the human NAIP gene locus consists of only a single full-length orthologue and recognizes the salmonellae SPI-1 T3SS needle protein PrgI170. Recognition of the T3SS components may be more important than recognition of flagellin since the T3SSs have a stronger association with bacterial virulence. Copy number variation in NAIP has been reported for geographically-segregated human populations and could contribute to the variable responses of individuals to infection171, 172. The redundancy between NAIP5 and NAIP2, as well as NLRC4 and NLRP3 in the detection of salmonellae may enable the host to activate the Caspase-1 pathway despite changes in the expression of pathogen ligands that these complexes recognize, e.g. detection may be dependent on NLRP3 when flagellin expression is down-regulated during the systemic phase of infection173.
Detection of cytosolic salmonellae or direct recognition of cytoplasmic LPS173 can also induce inflammasome-mediated cell death through Caspase-11174, 175, which is non-essential for the activation of Caspase-1 and may represent a form of inflammasome activation that occurs later in the infection50. It is unclear whether Caspase-11 interacts with the Caspase 1 inflammasome components NLRC4 and ASC175, 176, but in the absence of Caspase-1, Caspase-11-mediated cell death seems to be exploited by salmonellae to cause inflammation in the host. Mice lacking Caspase-1 are more susceptible to infection with S. Typhimurium than mice lacking both Caspase-1 and Caspase-1156, 150. Although activation of the inflammasome pathway may be beneficial in defence against cytosolic bacteria 50, aberrant hyperactivation, possibly when LPS concentrations reach a high level in the cytosol (known as endotoxic shock), is likely to be detrimental177.
Interactions between stress response pathways
Recent studies are now beginning to unravel the integral coupling of the inflammasome and autophagy pathways. It is possible that hyperactivation of inflammasomes occurs when autophagy fails to clear the pathogen, given that autophagy maintains cellular homeostasis and tightly controls inflammasome activation in the host. The sensing of pathogens by TLRs, which is important for the upregulation of Salmonellae SPI-2 transcription and formation of the replicative compartment, the SCV, in macrophages49, promotes autophagosome formation178–180. Ectopic expression of the effector SipA activates Nod1 and Nod2 in epithelial cells181, which also induces autophagosome formation182. In turn, autophagy regulates the transcription, processing and secretion of cytokines (IL1β, IL1α and IL18) that are triggered by the inflammasome183, and autophagosomes can sequester and degrade inflammasome components184. Moreover, the inhibition of mitophagy185, 186 can also trigger inflammasome activation. The exquisite control exerted by autophagy on inflammasome components is apparent by its ability to both activate and inhibit Caspase-11; Caspase-11 activation requires the lysis of pathogen containing vacuoles by IRGs187, but interestingly, autophagy also counteracts Caspase-11 activation, possibly by sequestering bacteria that escape the vacuole and preventing LPS release into the cytosol. Autophagic clearance of pathogen components in the cytosol may temper overt inflammasome activation, and the timing of all of these events in the host may determine the ultimate outcome.
Conclusions and future prospects
Considerable progress has been made in our understanding of how salmonellae intricately adapt host signalling and vesicle trafficking pathways to make the usually inhospitable intracellular environment a replication niche. The discrete functions of many of the numerous salmonellae effector proteins are not fully understood, and it is likely that many of their functions may only be elucidated when their activities are studied in the context of other effectors and in a spatiotemporal context within the host. The pace at which the activities of effector proteins are elucidated is likely to only increase through a more systematic investigation of their functions. Furthermore, much of the work to date has used non-polarized, immortalized cell lines, which greatly confounds our ability to understand the importance of directional movement within intestinal polarized epithelial cells, as well as the interplay between salmonellae and cell death pathways. Recent advances in the culturing and differentiation of primary epithelial cells from both mice and humans should add to our knowledge of the specific and diverse responses of these cell types to salmonellae188. It is most likely that not all of the effectors function in all cell types, which must be investigated for substantial progress towards understanding the important niches of salmonellae in the host to be made. It is possible that the seemingly redundant functions of effector proteins are important for bacterial survival or replication in different cell types or in different hosts, particularly for those serovars that have a broad host range (such as S. Typhimurium). This would be consistent with data that shows that host-restricted serovars (such as S. Typhi) have lost a number of effector proteins that are potentially important for causing gastrointestinal disease in a diverse range of hosts, but are possibly detrimental to causing systemic disease in specific host species. Only a limited number of unique virulence factors have been identified in S. Typhi suggesting that loss-of-function mutations may have a pivotal role in restricting host range and promoting systemic disease. It is interesting to speculate that many of the S. Typhimurium effectors that S. Typhi lacks are most important within polarized gastrointestinal epithelial cells as S. Typhi does not typically reside in this niche in human hosts, except during chronic colonization of the gallbladder. Research on S. Typhi has obviously lagged behind our understanding of NTS, but the availability of humanized mouse models, as well as advances in studying the organism in human cells, should teach us more about the pathogenesis of this human-specific pathogen189–191. The increase in the number of whole genome sequences of S. Typhimurium strains and other non-typhoidal and typhoidal strains that cause systemic or gastrointestinal disease should help to tease apart the effector proteins that are involved in determining host range and possible causing different disease manifestations. Similarly, advances in human diversity in inflammasome and autophagy pathways should provide important information about human susceptibility to salmonellae infection.
Substantial progress has been made in understanding how NTS can outcompete the microbiota to cause disease and disseminate, with induction of inflammation being an important trigger for release of host factors that promote proliferation. Salmonellae infection, which has reduced substantially owing to modern hygiene, may have provided a selective pressure for predisposition to increased inflammatory responses during times of increasing population density. Thus, future studies that examine host-salmonellae interactions in the context of different human populations will expand our understanding of the role of both the host and the bacterium in pathogenesis. Finally, considerable interest in phenotypic variation of individual salmonellae cells within host cells and tissues has led to renewed interest in the observation that different populations of bacteria have variable growth rates in vivo192–194. It will be interesting to examine these diverse populations and the expression profiles of important virulence factors that modify host cell responses, as this should help to elucidate interactions between bacteria in the host in the context of distinct subpopulations. Such studies are expected to improve our understanding of whether bacteria have specialized functions to replicate and resist host defences in specific mammalian niches.
Supplementary Material
Online summary.
Salmonella spp. deliver proteins into host cells to promote replication and survival.
Effector proteins translocated by the Salmonella pathogenicity island 1 Type III secretion system (T3SS) are important for bacterial invasion into non-phagocytic cells.
Induction of inflammation enhances extracellular growth of salmonellae and enables them to outcompete the gut microbiota
Effector proteins delivered by the Salmonella pathogenicity island-2 T3SS modify the Salmonella containing vacuole, associated endosomal membranes and proteins, all of which promotes intracellular replication.
Interplay between the host response pathways of autophagy and pyroptosis is involved in the detection of intracellular Salmonella spp.
Distinct functions for many of the salmonellae effector proteins are not fully understood, and it is likely that many of their functions will only be elucidated when their activities are studied in the context of other effectors and are considered in a spatiotemporal context within the host.
Glossary
- Autophagy
A catabolic cellular process in which cytoplasmic contents, such as damaged organelles and proteins, are targeted for lysosomal degradation
- Pyroptosis
An inflammatory, programmed cell death pathway that is triggered by activation of caspases 1 and 11 (in mice) and caspases 4/5 and 11 (in humans).
- Peyers Patches
Organized lymphoid regions of the small intestine monolayer that function in immune surveillance and the generation of a localized immune response
- Pathogenicity Island
A large horizontally acquired region of genomic DNA that often encodes virulence factors
- Type III Secretion System (T3SS)
A needle-like apparatus that is assembled by pathogenic bacteria for the delivery of bacterial effectors directly into host cells
- Pathogen recognition receptors (PRRs)
Proteins of the innate immune system that recognize pathogen-associated molecular patterns and initiate an innate immune cascade that facilitates pathogen clearance by triggering cytokine and chemokine expression
- Toll-like receptor (TLR)
Membrane bound receptors of the innate immune system that recognize specific pathogen associated molecular patterns
- Nod-like receptor (NLR)
Intracellular receptors of the innate immune system that recognize pathogen associated molecular patterns
- Endocytosis
The process by which extracellular particles are engulfed by eukaryotic cells and enclosed in a vesicle
- Jnk Pathway
The JNK signalling pathway responds to stress to regulate apoptosis, cell proliferation and immune responses
- Stress Fibre
Cytoskeletal structures comprised of bundles of actin filaments and are important for cell motility and endocytosis
- Microfold cells
Specialized epithelial cells that phagocytose molecules in the intestinal lumen for transepithelial transport to enable immunological sampling of antigens
- Tight junctions
The adhesive contact comprised of protein complexes that function to limit the movement of molecules and ions through the space between cells in a monolayer
- Transcytosis
The process of vesicular transport of molecules across a cell
- Caspase
Family of cysteine proteases that have an essential role in cell death pathways
- Rho GTPase
A family of GTPases that are important molecular switches for regulating actin dynamics
- Siderophore
Small molecules that have high affinity for iron and are secreted by bacteria to scavenge iron
- Reactive Oxygen Species (ROS)
Chemically reactive species containing oxygen, which are produced as a by-product of aerobic respiration and function in combating microbial infections
- Endotoxic shock
Severe inflammatory reaction induced by high levels of endotoxin in the bloodstream
- Macropinocytosis
A non-selective form of endocytosis that involves membrane ruffles
- Microtubular network
The dynamic network of tubulin polymers that comprise an important component of the cell cytoskeleton. This network is utilized for intracellular transport, including movement of secretory vesicles and organelles, including the Salmonella containing vacuole
- Sorting nexin
A family of proteins that are characterized by the presence of a phox-homology domain that functions by binding phosphatidylinositol-3-monophosphate. Sorting nexins associate with the endocytic network and are involved in endocytosis, endosomal sorting and endosomal signalling
- Peroxisome
Organelles that degrade long-chain fatty acids in eukaryotes
- Prenylation
The attachment of farnesyl or geranyl-geranyl groups to C-terminal cysteine residues that are present in specific prenylation motifs of proteins, which promotes membrane association or protein-protein interactions
- Palmitoylation
The reversible covalent posttranslational attachment of lipids (usually palmitate) to proteins on cysteine residues
- Mitophagy
The process of targeting mitochondria for autophagic degradation
- Stromal cells
Connective tissue cells that include enterocytes, tissue fibroblasts, and vascular endothelial cells
- Periphery directed transport vesicles
Vesicles, such as those from the SCV, that move towards the periphery of cells along microtubules
- Peroxisome
Eukaryotic organelles involved in the catabolism of fatty acids
Biographies
Doris LaRock received her Ph.D. in the lab of Sam Miller at the University of Washington, Seattle, WA, where she studied the function of S. Typhimurium T3SS effectors. She is currently a postdoctoral research associate in the lab of Scott Lesley at the Joint Center for Structural Genomics at The Scripps Research Institute, San Diego, CA. Her current research is focused on the structure and function of proteins that are secreted by gut microbiota.
Anu Chaudhary received her Ph.D. in the lab of Glenn D. Prestwich, and then studied cell signalling with Joan S. Brugge and Jonathan A. Cooper. After a stint at Wyeth Research and Los Alamos National labs, she now studies human genetic susceptibility to autophagy and inflammasome activation pathways with Sam Miller in the Department of Microbiology at the University of Washington.
Samuel I. Miller is Professor of Medicine, Microbiology and Genome Sciences at the University of Washington. He received his M.D. from Baylor School of Medicine and his medical training at MGH and Harvard Medical School. His group focuses on defining the molecular basis of bacterial pathogenesis and interactions with eukaryotic cells, with an emphasis on innate immune responses to Gram-negative bacteria.
References
- 1.Pegues DA, Ohl ME, Miller SI. In: Salmonella species, including Salmonella typhi. Mandell GL, Bennett JE, Dolin R, editors. Elsevier/Churchill Livingstone; New York: 2005. [Google Scholar]
- 2.Harris JC, Dupont HL, Hornick RB. Fecal leukocytes in diarrheal illness. Ann Intern Med. 1972;76:697–703. doi: 10.7326/0003-4819-76-5-697. [DOI] [PubMed] [Google Scholar]
- 3.Winter SE, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. Reference 3 provided the first molecular evidence that intestinal inflammation promotes pathogen over microbiota growth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stecher B, et al. Salmonella enterica Serovar Typhimurium Exploits Inflammation to Compete with the Intestinal Microbiota. PLoS Biol. 2007;5:e244. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones BD, Ghori N, Falkow S. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer’s patches. J Exp Med. 1994;180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez-Moya M, de Pedro MA, Schwarz H, Garcia-del Portillo F. Inhibition of Salmonella intracellular proliferation by non-phagocytic eucaryotic cells. Res Microbiol. 1998;149:309–318. doi: 10.1016/s0923-2508(98)80436-1. [DOI] [PubMed] [Google Scholar]
- 7.Niedergang F, Sirard JC, Blanc CT, Kraehenbuhl JP. Entry and survival of Salmonella typhimurium in dendritic cells and presentation of recombinant antigens do not require macrophage-specific virulence factors. Proceedings of the National Academy of Sciences. 2000;97:14650–14655. doi: 10.1073/pnas.97.26.14650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalebroux ZD, Miller SI. Salmonellae PhoPQ regulation of the outer membrane to resist innate immunity. Current Opinion in Microbiology. 2014;17:106–113. doi: 10.1016/j.mib.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nat Rev Micro. 2008;6:53–66. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- 10.McClelland M, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 11.Steele-Mortimer O, et al. The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cellular Microbiology. 2002;4:43–54. doi: 10.1046/j.1462-5822.2002.00170.x. [DOI] [PubMed] [Google Scholar]
- 12.Giacomodonato MN, et al. SipA, SopA, SopB, SopD and SopE2 effector proteins of Salmonella enterica serovar Typhimurium are synthesized at late stages of infection in mice. Microbiology. 2007;153:1221–1228. doi: 10.1099/mic.0.2006/002758-0. [DOI] [PubMed] [Google Scholar]
- 13.Hernandez LD, Hueffer K, Wenk MR, Galán JE. Salmonella modulates vesicular traffic by altering phosphoinositide metabolism. Science. 2004;304:1805–1807. doi: 10.1126/science.1098188. [DOI] [PubMed] [Google Scholar]
- 14.Brawn LC, Hayward RD, Koronakis V. Salmonella SPI1 Effector SipA Persists after Entry and Cooperates with a SPI2 Effector to Regulate Phagosome Maturation and Intracellular Replication. Cell Host & Microbe. 2007;1:63–75. doi: 10.1016/j.chom.2007.02.001. Reference 14 gave the first evidence that SPI1 effectors can persist to influence the SCV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Humphreys D, Hume PJ, Koronakis V. The Salmonella Effector SptP Dephosphorylates Host AAA+ ATPase VCP to Promote Development of its Intracellular Replicative Niche. Cell Host & Microbe. 2009;5:225–233. doi: 10.1016/j.chom.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel JC, Hueffer K, Lam TT, Galan JE. Diversification of a Salmonella Virulence Protein Function by Ubiquitin-Dependent Differential Localization. Cell. 2009;137:283–294. doi: 10.1016/j.cell.2009.01.056. Reference 16 demonstrated that SopB multiple functions are controlled by ubiquitination. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alpuche-Aranda CM, Racoosin EL, Swanson JA, Miller SI. Salmonella stimulate macrophage macropinocytosis and persist within spacious phagosomes. J Exp Med. 1994;179:601–608. doi: 10.1084/jem.179.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh YK, et al. Rapid and complete fusion of macrophage lysosomes with phagosomes containing Salmonella typhimurium. Infection and Immunity. 1996;64:3877–3883. doi: 10.1128/iai.64.9.3877-3883.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Norris FA, Wilson MP, Wallis TS, Galyov EE, Majerus PW. SopB, a protein required for virulence of Salmonella dublin, is an inositol phosphate phosphatase. Proceedings of the National Academy of Sciences. 1998;95:14057–14059. doi: 10.1073/pnas.95.24.14057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tahoun A, et al. Salmonella Transforms Follicle-Associated Epithelial Cells into M Cells to Promote Intestinal Invasion. Cell Host & Microbe. 2012;12:645–656. doi: 10.1016/j.chom.2012.10.009. Reference 20 shows that SopB can induce transformation of epithelial cells into M cells. [DOI] [PubMed] [Google Scholar]
- 21.Bakowski MA, et al. The Phosphoinositide Phosphatase SopB Manipulates Membrane Surface Charge and Trafficking of the Salmonella-Containing Vacuole. Cell Host & Microbe. 2010;7:453–462. doi: 10.1016/j.chom.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 22.Mirold S, et al. Salmonella Host Cell Invasion Emerged by Acquisition of a Mosaic of Separate Genetic Elements, IncludingSalmonella Pathogenicity Island 1 (SPI1), SPI5, and sopE2. Journal of Bacteriology. 2001;183:2348–2358. doi: 10.1128/JB.183.7.2348-2358.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou D, Chen LM, Hernandez L, Shears SB, Galán JE. A Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host cell actin cytoskeleton rearrangements and bacterial internalization. Molecular Microbiology. 2001;39:248–260. doi: 10.1046/j.1365-2958.2001.02230.x. [DOI] [PubMed] [Google Scholar]
- 24.Hanisch J, et al. Activation of a RhoA/Myosin II-Dependent but Arp2/3 Complex-Independent Pathway Facilitates Salmonella Invasion. Cell Host & Microbe. 2011;9:273–285. doi: 10.1016/j.chom.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 25.Jolly C, Winfree S, Hansen B, Steele-Mortimer O. The Annexin A2/p11 complex is required for efficient invasion of Salmonella Typhimurium in epithelial cells. Cellular Microbiology. 2014;16:64–77. doi: 10.1111/cmi.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mallo GV, et al. SopB promotes phosphatidylinositol 3-phosphate formation on Salmonella vacuoles by recruiting Rab5 and Vps34. The Journal of Cell Biology. 2008;182:741–752. doi: 10.1083/jcb.200804131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knodler LA, Winfree S, Drecktrah D, Ireland R, Steele-Mortimer O. Ubiquitination of the bacterial inositol phosphatase, SopB, regulates its biological activity at the plasma membrane. Cellular Microbiology. 2009;11:1652–1670. doi: 10.1111/j.1462-5822.2009.01356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stender S, et al. Identification of SopE2 from Salmonella typhimurium, a conserved guanine nucleotide exchange factor for Cdc42 of the host cell. Molecular Microbiology. 2000;36:1206–1221. doi: 10.1046/j.1365-2958.2000.01933.x. [DOI] [PubMed] [Google Scholar]
- 29.Friebel A, et al. SopE and SopE2 from Salmonella typhimurium Activate Different Sets of RhoGTPases of the Host Cell. Journal of Biological Chemistry. 2001;276:34035–34040. doi: 10.1074/jbc.M100609200. [DOI] [PubMed] [Google Scholar]
- 30.Hardt WD, Chen LM, Schuebel KE, Bustelo XR, Galán JE. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell. 1998;93:815–826. doi: 10.1016/s0092-8674(00)81442-7. [DOI] [PubMed] [Google Scholar]
- 31.Wood MW, Rosqvist R, Mullan PB, Edwards MH, Galyov EE. SopE, a secreted protein of Salmonelladublin, is translocated into the target eukaryotic cell via a sip‐ dependent mechanism and promotes bacterial entry. Molecular microbiology. 1996;22:327–338. doi: 10.1046/j.1365-2958.1996.00116.x. [DOI] [PubMed] [Google Scholar]
- 32.Bakshi CS, et al. Identification of SopE2, a SalmonellaSecreted Protein Which Is Highly Homologous to SopE and Involved in Bacterial Invasion of Epithelial Cells. Journal of Bacteriology. 2000;182:2341–2344. doi: 10.1128/jb.182.8.2341-2344.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lebensohn AM, Kirschner MW. Activation of the WAVE Complex by Coincident Signals Controls Actin Assembly. Molecular Cell. 2009;36:512–524. doi: 10.1016/j.molcel.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Humphreys D, Davidson A, Hume PJ, Koronakis V. Salmonella Virulence Effector SopE and Host GEF ARNO Cooperate to Recruit and Activate WAVE to Trigger Bacterial Invasion. Cell Host & Microbe. 2012;11:129–139. doi: 10.1016/j.chom.2012.01.006. Reference 34 demonstrate the molecular mechanisms involved in Salmonella induced cell invasion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humphreys D, Davidson AC, Hume PJ, Makin LE, Koronakis V. Arf6 coordinates actin assembly through the WAVE complex, a mechanism usurped by Salmonella to invade host cells. Proceedings of the National Academy of Sciences. 2013;110:16880–16885. doi: 10.1073/pnas.1311680110. Reference 35 demonstrate the molecular mechanisms involved in Salmonella induced cell invasion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou D, Mooseker MS, Galán JE. Role of the S. typhimurium actin-binding protein SipA in bacterial internalization. Science. 1999;283:2092–2095. doi: 10.1126/science.283.5410.2092. [DOI] [PubMed] [Google Scholar]
- 37.Zhou D, Mooseker MS, Galán JE. An invasion-associated Salmonella protein modulates the actin-bundling activity of plastin. Proceedings of the National Academy of Sciences. 1999;96:10176–10181. doi: 10.1073/pnas.96.18.10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayward RD, Koronakis V. Direct nucleation and bundling of actin by the SipC protein of invasive Salmonella. EMBO J. 1999;18:4926–4934. doi: 10.1093/emboj/18.18.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Myeni SK, Zhou D. The C Terminus of SipC Binds and Bundles F-actin to Promote Salmonella Invasion. Journal of Biological Chemistry. 2010;285:13357–13363. doi: 10.1074/jbc.M109.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scherer CA, Cooper E, Miller SI. The Salmonella type III secretion translocon protein SspC is inserted into the epithelial cell plasma membrane upon infection. Molecular Microbiology. 2000;37:1133–1145. doi: 10.1046/j.1365-2958.2000.02066.x. [DOI] [PubMed] [Google Scholar]
- 41.Carlson SA, Omary MB, Jones BD. Identification of cytokeratins as accessory mediators of Salmonella entry into eukaryotic cells. Life Sciences. 2002;70:1415–1426. doi: 10.1016/s0024-3205(01)01512-0. [DOI] [PubMed] [Google Scholar]
- 42.Fu Y, Galán JE. A Salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature. 1999;401:293–297. doi: 10.1038/45829. [DOI] [PubMed] [Google Scholar]
- 43.Kubori T, Galán JE. Temporal Regulation of Salmonella Virulence Effector Function by Proteasome-Dependent Protein Degradation. Cell. 2003;115:333–342. doi: 10.1016/s0092-8674(03)00849-3. [DOI] [PubMed] [Google Scholar]
- 44.Zhang S, et al. The Salmonella enterica serotype typhimurium effector proteins SipA, SopA, SopB, SopD, and SopE2 act in concert to induce diarrhea in calves. Infection and Immunity. 2002;70:3843–3855. doi: 10.1128/IAI.70.7.3843-3855.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hapfelmeier S, et al. Role of the Salmonella Pathogenicity Island 1 Effector Proteins SipA, SopB, SopE, and SopE2 in Salmonella enterica Subspecies 1 Serovar Typhimurium Colitis in Streptomycin-Pretreated Mice. Infection and Immunity. 2004;72:795–809. doi: 10.1128/IAI.72.2.795-809.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhavsar AP, et al. The Salmonella Type III Effector SspH2 Specifically Exploits the NLR Co-chaperone Activity of SGT1 to Subvert Immunity. PLoS Pathog. 2013;9:e1003518. doi: 10.1371/journal.ppat.1003518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCormick BA, Miller SI, Carnes D, Madara JL. Transepithelial signaling to neutrophils by salmonellae: a novel virulence mechanism for gastroenteritis. Infection and Immunity. 1995;63:2302–2309. doi: 10.1128/iai.63.6.2302-2309.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCormick BA, et al. Surface attachment of Salmonella typhimurium to intestinal epithelia imprints the subepithelial matrix with gradients chemotactic for neutrophils. The Journal of Cell Biology. 1995;131:1599–1608. doi: 10.1083/jcb.131.6.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arpaia N, et al. TLR signaling is required for Salmonella typhimurium virulence. Cell. 2011;144:675–88. doi: 10.1016/j.cell.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knodler Leigh A, et al. Noncanonical Inflammasome Activation of Caspase-4/Caspase-11 Mediates Epithelial Defenses against Enteric Bacterial Pathogens. Cell Host & Microbe. 2014;16:249–256. doi: 10.1016/j.chom.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miao EA, et al. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proceedings of the National Academy of Sciences. 2010;107:3076–3080. doi: 10.1073/pnas.0913087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miao EA, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nat Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 53.Godinez I, et al. T Cells Help To Amplify Inflammatory Responses Induced by Salmonella enterica Serotype Typhimurium in the Intestinal Mucosa. Infection and Immunity. 2008;76:2008–2017. doi: 10.1128/IAI.01691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hersh D, et al. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proceedings of the National Academy of Sciences. 1999;96:2396–2401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muller AJ, et al. The S. Typhimurium Effector SopE Induces Caspase-1 Activation in Stromal Cells to Initiate Gut Inflammation. Cell Host & Microbe. 2009;6:125–136. doi: 10.1016/j.chom.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 56.Shi J, et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014 doi: 10.1038/nature13683. advance online publication. Reference 56 identifies caspases 4/5/11 as receptors for intracellular LPS. [DOI] [PubMed] [Google Scholar]
- 57.Lee CA, et al. A secreted Salmonella protein induces a proinflammatory response in epithelial cells, which promotes neutrophil migration. Proceedings of the National Academy of Sciences. 2000;97:12283–12288. doi: 10.1073/pnas.97.22.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Srikanth CV, et al. Salmonella Pathogenesis and Processing of Secreted Effectors by Caspase-3. Science. 2010;330:390–393. doi: 10.1126/science.1194598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raffatellu M, et al. Lipocalin-2 Resistance Confers an Advantage to Salmonella enterica Serotype Typhimurium for Growth and Survival in the Inflamed Intestine. Cell Host & Microbe. 2009;5:476–486. doi: 10.1016/j.chom.2009.03.011. Reference 59 demonstrates that Salmonella upregulation of inflammation promotes release of antimicrobial factors that function against the microbiota. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lopez CA, et al. Phage-Mediated Acquisition of a Type III Secreted Effector Protein Boosts Growth of Salmonella by Nitrate Respiration. mBio. 2012;3 doi: 10.1128/mBio.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin SL, Le TX, Cowen DS. SptP, a Salmonella typhimurium type III-secreted protein, inhibits the mitogen-activated protein kinase pathway by inhibiting Raf activation. Cellular Microbiology. 2003;5:267–275. doi: 10.1046/j.1462-5822.2003.t01-1-00274.x. [DOI] [PubMed] [Google Scholar]
- 62.Murli S, Watson RO, Galán JE. Role of tyrosine kinases and the tyrosine phosphatase SptP in the interaction of Salmonella with host cells. Cellular Microbiology. 2001;3:795–810. doi: 10.1046/j.1462-5822.2001.00158.x. [DOI] [PubMed] [Google Scholar]
- 63.Haraga A, Miller SI. A Salmonella enterica Serovar Typhimurium Translocated Leucine-Rich Repeat Effector Protein Inhibits NF-KB-Dependent Gene Expression. Infection and Immunity. 2003;71:4052–4058. doi: 10.1128/IAI.71.7.4052-4058.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haraga A, Miller SI. A Salmonella type III secretion effector interacts with the mammalian serine/threonine protein kinase PKN1. Cellular Microbiology. 2006;8:837–846. doi: 10.1111/j.1462-5822.2005.00670.x. [DOI] [PubMed] [Google Scholar]
- 65.Keszei AF, et al. Structure of an SspH1-PKN1 complex reveals the basis for host substrate recognition and mechanism of activation for a bacterial E3 ubiquitin ligase. Molecular and cellular biology. 2014;34:362–373. doi: 10.1128/MCB.01360-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu H, Jones RM, Neish AS. The Salmonella effector AvrA mediates bacterial intracellular survival during infection in vivo. Cellular Microbiology. 2012;14:28–39. doi: 10.1111/j.1462-5822.2011.01694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jones RM, et al. Salmonella AvrA Coordinates Suppression of Host Immune and Apoptotic Defenses via JNK Pathway Blockade. Cell Host & Microbe. 2008;3:233–244. doi: 10.1016/j.chom.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 68.Du F, Galán JE. Selective Inhibition of Type III Secretion Activated Signaling by the Salmonella Effector AvrA. PLoS Pathog. 2009;5:e1000595. doi: 10.1371/journal.ppat.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haneda T, et al. Salmonella type III effector SpvC, a phosphothreonine lyase, contributes to reduction in inflammatory response during intestinal phase of infection. Cellular Microbiology. 2012;14:485–499. doi: 10.1111/j.1462-5822.2011.01733.x. [DOI] [PubMed] [Google Scholar]
- 70.Pilar AV, Reid-Yu SA, Cooper CA, Mulder DT, Coombes BK. GogB is an anti-inflammatory effector that limits tissue damage during Salmonella infection through interaction with human FBXO22 and Skp1. PLoS Pathog. 2012;8:e1002773. doi: 10.1371/journal.ppat.1002773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mazurkiewicz P, et al. SpvC is a Salmonella effector with phosphothreonine lyase activity on host mitogen-activated protein kinases. Molecular Microbiology. 2008;67:1371–1383. doi: 10.1111/j.1365-2958.2008.06134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cardenal-Muñoz E, Gutiérrez G, Ramos-Morales F. Global impact of Salmonella type III secretion effector SteA on host cells. Biochemical and Biophysical Research Communications. doi: 10.1016/j.bbrc.2014.05.056. [DOI] [PubMed] [Google Scholar]
- 73.McLaughlin LM, et al. A microfluidic-based genetic screen to identify microbial virulence factors that inhibit dendritic cell migration. Integrative Biology. 2014;6:438–449. doi: 10.1039/c3ib40177d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Halici S, Zenk SF, Jantsch J, Hensel M. Functional Analysis of the Salmonella Pathogenicity Island 2-Mediated Inhibition of Antigen Presentation in Dendritic Cells. Infection and Immunity. 2008;76:4924–4933. doi: 10.1128/IAI.00531-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abshire KZ, Neidhardt FC. Growth rate paradox of Salmonella typhimurium within host macrophages. Journal of Bacteriology. 1993;175:3744–3748. doi: 10.1128/jb.175.12.3744-3748.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Crawford RW, et al. Gallstones play a significant role in Salmonella spp. gallbladder colonization and carriage. Proceedings of the National Academy of Sciences. 2010;107:4353–4358. doi: 10.1073/pnas.1000862107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Steele-Mortimer O, Méresse S, Gorvel JP, Toh BH, Finlay BB. Biogenesis of Salmonella typhimurium-containing vacuoles in epithelial cells involves interactions with the early endocytic pathway. Cellular Microbiology. 1999;1:33–49. doi: 10.1046/j.1462-5822.1999.00003.x. [DOI] [PubMed] [Google Scholar]
- 78.Smith AC, Cirulis JT, Casanova JE, Scidmore MA, Brumell JH. Interaction of the Salmonella-containing Vacuole with the Endocytic Recycling System. Journal of Biological Chemistry. 2005;280:24634–24641. doi: 10.1074/jbc.M500358200. [DOI] [PubMed] [Google Scholar]
- 79.Brumell JH, Tang P, Mills SD, Finlay BB. Characterization of Salmonella-induced filaments (Sifs) reveals a delayed interaction between Salmonella-containing vacuoles and late endocytic compartments. Traffic. 2001;2:643–653. doi: 10.1034/j.1600-0854.2001.20907.x. [DOI] [PubMed] [Google Scholar]
- 80.Garcia-del Portillo F, Finlay BB. Targeting of Salmonella typhimurium to vesicles containing lysosomal membrane glycoproteins bypasses compartments with mannose 6-phosphate receptors. The Journal of Cell Biology. 1995;129:81–97. doi: 10.1083/jcb.129.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Méresse S, Steele-Mortimer O, Finlay BB, Gorvel JP. The rab7 GTPase controls the maturation of Salmonella typhimurium-containing vacuoles in HeLa cells. EMBO J. 1999;18:4394–4403. doi: 10.1093/emboj/18.16.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eswarappa SM, Negi VD, Chakraborty S, Chandrasekhar Sagar BK, Chakravortty D. Division of the Salmonella-Containing Vacuole and Depletion of Acidic Lysosomes in Salmonella-Infected Host Cells Are Novel Strategies of Salmonella enterica To Avoid Lysosomes. Infection and Immunity. 2010;78:68–79. doi: 10.1128/IAI.00668-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Christen M, et al. Activation of a Bacterial Virulence Protein by the GTPase RhoA. Science Signaling. 2009;2:ra71. doi: 10.1126/scisignal.2000430. Reference 83 shows the first bacterial effector activated by a mammalian GTPase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.LaRock DL, Brzovic PS, Levin I, Blanc MP, Miller SI. A Salmonella typhimurium-translocated Glycerophospholipid:Cholesterol Acyltransferase Promotes Virulence by Binding to the RhoA Protein Switch Regions. Journal of Biological Chemistry. 2012;287:29654–29663. doi: 10.1074/jbc.M112.363598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nawabi P, Catron DM, Haldar K. Esterification of cholesterol by a type III secretion effector during intracellular Salmonella infection. Molecular Microbiology. 2008;68:173–185. doi: 10.1111/j.1365-2958.2008.06142.x. [DOI] [PubMed] [Google Scholar]
- 86.Boucrot E, Henry T, Borg JP, Gorvel JP, Méresse S. The Intracellular Fate of Salmonella Depends on the Recruitment of Kinesin. Science. 2005;308:1174–1178. doi: 10.1126/science.1110225. [DOI] [PubMed] [Google Scholar]
- 87.Arena ET, et al. The Deubiquitinase Activity of the Salmonella Pathogenicity Island 2 Effector, SseL, Prevents Accumulation of Cellular Lipid Droplets. Infection and Immunity. 2011;79:4392–4400. doi: 10.1128/IAI.05478-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ramsden AE, Mota LJ, Munter S, Shorte SL, Holden DW. The SPI-2 type III secretion system restricts motility of Salmonella-containing vacuoles. Cellular Microbiology. 2007;9:2517–2529. doi: 10.1111/j.1462-5822.2007.00977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Braun V, et al. Sorting nexin 3 (SNX3) is a component of a tubular endosomal network induced by Salmonella and involved in maturation of the Salmonella‐containing vacuole. Cellular microbiology. 2010;12:1352–1367. doi: 10.1111/j.1462-5822.2010.01476.x. [DOI] [PubMed] [Google Scholar]
- 90.Bujny MV, et al. Sorting nexin-1 defines an early phase of Salmonella-containing vacuole-remodeling during Salmonella infection. Journal of cell science. 2008;121:2027–2036. doi: 10.1242/jcs.018432. [DOI] [PubMed] [Google Scholar]
- 91.Cantalupo G, Alifano P, Roberti V, Bruni CB, Bucci C. Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 2001;20:683–693. doi: 10.1093/emboj/20.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Harrison RE, et al. Salmonella Impairs RILP Recruitment to Rab7 during Maturation of Invasion Vacuoles. Molecular Biology of the Cell. 2004;15:3146–3154. doi: 10.1091/mbc.E04-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guignot J, et al. Microtubule motors control membrane dynamics of Salmonella-containing vacuoles. Journal of Cell Science. 2004;117:1033–1045. doi: 10.1242/jcs.00949. [DOI] [PubMed] [Google Scholar]
- 94.Szeto J, Namolovan A, Osborne SE, Coombes BK, Brumell JH. Salmonella-Containing Vacuoles Display Centrifugal Movement Associated with Cell-to-Cell Transfer in Epithelial Cells. Infection and Immunity. 2009;77:996–1007. doi: 10.1128/IAI.01275-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ohlson MB, et al. Structure and Function of Salmonella SifA Indicate that Its Interactions with SKIP, SseJ, and RhoA Family GTPases Induce Endosomal Tubulation. Cell Host & Microbe. 2008;4:434–446. doi: 10.1016/j.chom.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Henry T, et al. The Salmonella effector protein PipB2 is a linker for kinesin-1. Proceedings of the National Academy of Sciences. 2006;103:13497–13502. doi: 10.1073/pnas.0605443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Deiwick J, et al. The Translocated Salmonella Effector Proteins SseF and SseG Interact and Are Required To Establish an Intracellular Replication Niche. Infection and Immunity. 2006;74:6965–6972. doi: 10.1128/IAI.00648-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Garcia-del Portillo F, Zwick MB, Leung KY, Finlay BB. Salmonella induces the formation of filamentous structures containing lysosomal membrane glycoproteins in epithelial cells. Proceedings of the National Academy of Sciences. 1993;90:10544–10548. doi: 10.1073/pnas.90.22.10544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stein M, Leung K, Zwick M, Garcia-del Portillo F, Finlay B. Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Molecular Microbiology. 1996;20:151–164. doi: 10.1111/j.1365-2958.1996.tb02497.x. [DOI] [PubMed] [Google Scholar]
- 100.Birmingham CL, Jiang X, Ohlson MB, Miller SI, Brumell JH. Salmonella-Induced Filament Formation Is a Dynamic Phenotype Induced by Rapidly Replicating Salmonella enterica Serovar Typhimurium in Epithelial Cells. Infection and Immunity. 2005;73:1204–1208. doi: 10.1128/IAI.73.2.1204-1208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Domingues L, Holden DW, Mota LJ. The Salmonella effector SteA contributes to the control of membrane dynamics of Salmonella-containing vacuoles. Infection and Immunity. 2014 doi: 10.1128/IAI.01385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Brumell J, Goosney D, Finlay B. SifA, a Type III Secreted Effector of Salmonella typhimurium, Directs Salmonella-Induced Filament (Sif) Formation Along Microtubules. Traffic. 2002;3:407–415. doi: 10.1034/j.1600-0854.2002.30604.x. [DOI] [PubMed] [Google Scholar]
- 103.Knodler LA, Steele-Mortimer O. The Salmonella Effector PipB2 Affects Late Endosome/Lysosome Distribution to Mediate Sif Extension. Molecular Biology of the Cell. 2005;16:4108–4123. doi: 10.1091/mbc.E05-04-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Guy RL, Gonias LA, Stein MA. Aggregation of host endosomes by Salmonella requires SPI2 translocation of SseFG and involves SpvR and the fms–aroE intragenic region. Molecular Microbiology. 2000;37:1417–1435. doi: 10.1046/j.1365-2958.2000.02092.x. [DOI] [PubMed] [Google Scholar]
- 105.Ruíz-Albert J, et al. Complementary activities of SseJ and SifA regulate dynamics of the Salmonella typhimurium vacuolar membrane. Molecular Microbiology. 2002;44:645–661. doi: 10.1046/j.1365-2958.2002.02912.x. [DOI] [PubMed] [Google Scholar]
- 106.Schroeder N, et al. The Virulence Protein SopD2 Regulates Membrane Dynamics of Salmonella Containing Vacuoles. PLoS Pathog. 2010;6:e1001002. doi: 10.1371/journal.ppat.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kaniuk NA, et al. Salmonella exploits Arl8B-directed kinesin activity to promote endosome tubulation and cell-to-cell transfer. Cellular Microbiology. 2011;13:1812–1823. doi: 10.1111/j.1462-5822.2011.01663.x. [DOI] [PubMed] [Google Scholar]
- 108.Boucrot E, Beuzón CR, Holden DW, Gorvel JP, Méresse S. Salmonella typhimurium SifA Effector Protein Requires Its Membrane-anchoring C-terminal Hexapeptide for Its Biological Function. Journal of Biological Chemistry. 2003;278:14196–14202. doi: 10.1074/jbc.M207901200. [DOI] [PubMed] [Google Scholar]
- 109.Reinicke AT, et al. A Salmonella typhimurium Effector Protein SifA Is Modified by Host Cell Prenylation and S-Acylation Machinery. Journal of Biological Chemistry. 2005;280:14620–14627. doi: 10.1074/jbc.M500076200. [DOI] [PubMed] [Google Scholar]
- 110.Dumont A, et al. SKIP, the Host Target of the Salmonella Virulence Factor SifA, Promotes Kinesin-1-Dependent Vacuolar Membrane Exchanges. Traffic. 2010;11:899–911. doi: 10.1111/j.1600-0854.2010.01069.x. [DOI] [PubMed] [Google Scholar]
- 111.McGourty K, et al. Salmonella Inhibits Retrograde Trafficking of Mannose-6-Phosphate Receptors and Lysosome Function. Science. 2012;338:963–967. doi: 10.1126/science.1227037. Reference 111 demonstrates that SifA inhibits lysosome-SCV fusion by binding Rab9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mota LJ, Ramsden AE, Liu M, Castle JD, Holden DW. SCAMP3 is a component of the Salmonella-induced tubular network and reveals an interaction between bacterial effectors and post-Golgi trafficking. Cellular Microbiology. 2009;11:1236–1253. doi: 10.1111/j.1462-5822.2009.01329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Perrett CA, Zhou D. Salmonella type III effector SopB modulates host cell exocytosis. Emerg Microbes Infect. 2013;2:e32. doi: 10.1038/emi.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jiang X, et al. The related effector proteins SopD and SopD2 from Salmonella enterica serovar Typhimurium contribute to virulence during systemic infection of mice. Molecular Microbiology. 2004;54:1186–1198. doi: 10.1111/j.1365-2958.2004.04344.x. [DOI] [PubMed] [Google Scholar]
- 115.Freeman JA, Ohl ME, Miller SI. The Salmonella enterica Serovar Typhimurium Translocated Effectors SseJ and SifB Are Targeted to the Salmonella-Containing Vacuole. Infection and Immunity. 2003;71:418–427. doi: 10.1128/IAI.71.1.418-427.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Figueira R, Watson KG, Holden DW, Helaine S. Identification of Salmonella pathogenicity island-2 type III secretion system effectors involved in intramacrophage replication of S. enterica serovar Typhimurium: implications for rational vaccine design. MBio. 2013;4:e00065–13. doi: 10.1128/mBio.00065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Beuzón CR, et al. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 2000;19:3235–3249. doi: 10.1093/emboj/19.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ohlson MB, Fluhr K, Birmingham CL, Brumell JH, Miller SI. SseJ Deacylase Activity by Salmonella enterica Serovar Typhimurium Promotes Virulence in Mice. Infection and Immunity. 2005;73:6249–6259. doi: 10.1128/IAI.73.10.6249-6259.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McLaughlin LM, et al. The Salmonella SPI2 Effector SseI Mediates Long-Term Systemic Infection by Modulating Host Cell Migration. PLoS Pathog. 2009;5:e1000671. doi: 10.1371/journal.ppat.1000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Knodler L, et al. Salmonella type III effectors PipB and PipB2 are targeted to detergent-resistant microdomains on internal host cell membranes. Molecular Microbiology. 2003;49:685–704. doi: 10.1046/j.1365-2958.2003.03598.x. [DOI] [PubMed] [Google Scholar]
- 121.Browne SH, Hasegawa P, Okamoto S, Fierer J, Guiney DG. Identification of Salmonella SPI-2 secretion system components required for SpvB-mediated cytotoxicity in macrophages and virulence in mice. FEMS Immunology & Medical Microbiology. 2008;52:194–201. doi: 10.1111/j.1574-695X.2007.00364.x. [DOI] [PubMed] [Google Scholar]
- 122.Geddes K, Worley M, Niemann G, Heffron F. Identification of New Secreted Effectors in Salmonella enterica Serovar Typhimurium. Infection and Immunity. 2005;73:6260–6271. doi: 10.1128/IAI.73.10.6260-6271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hicks SW, Charron G, Hang HC, Galán JE. Subcellular Targeting of Salmonella Virulence Proteins by Host-Mediated S-Palmitoylation. Cell host & microbe. 2011;10:9–20. doi: 10.1016/j.chom.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang Y, Higashide W, Dai S, Sherman DM, Zhou D. Recognition and ubiquitination of Salmonella type III effector SopA by a ubiquitin E3 ligase, HsRMA1. Journal of Biological Chemistry. 2005;280:38682–38688. doi: 10.1074/jbc.M506309200. [DOI] [PubMed] [Google Scholar]
- 125.Arbeloa A, et al. EspM2 is a RhoA guanine nucleotide exchange factor. Cellular Microbiology. 2010;12:654–664. doi: 10.1111/j.1462-5822.2009.01423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Vinh DB, Ko DC, Rachubinski RA, Aitchison JD, Miller SI. Expression of the Salmonella spp. virulence factor SifA in yeast alters Rho1 activity on peroxisomes. Molecular biology of the cell. 2010;21:3567–3577. doi: 10.1091/mbc.E10-06-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Huang Z, et al. Structural insights into host GTPase isoform selection by a family of bacterial GEF mimics. Nat Struct Mol Biol. 2009;16:853–860. doi: 10.1038/nsmb.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Martinon F, Chen X, Lee AH, Glimcher LH. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010;11:411–8. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Oslowski Christine M, et al. Thioredoxin-Interacting Protein Mediates ER Stress-Induced β Cell Death through Initiation of the Inflammasome. Cell Metabolism. 2012;16:265–273. doi: 10.1016/j.cmet.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 131.Jia K, et al. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc Natl Acad Sci U S A. 2009;106:14564–9. doi: 10.1073/pnas.0813319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Knodler LA, et al. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc Natl Acad Sci U S A. 2010;107:17733–8. doi: 10.1073/pnas.1006098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Manzanillo PS, et al. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013;501:512–6. doi: 10.1038/nature12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Benjamin JL, Sumpter R, Jr, Levine B, Hooper LV. Intestinal epithelial autophagy is essential for host defense against invasive bacteria. Cell Host Microbe. 2013;13:723–34. doi: 10.1016/j.chom.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lassen KG, et al. Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proceedings of the National Academy of Sciences. 2014;111:7741–7746. doi: 10.1073/pnas.1407001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. Impaired autophagy of an intracellular pathogen induced by a Crohn’s disease associated ATG16L1 variant. PLoS One. 2008;3:e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cadwell K, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–63. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Knodler LA, Nair V, Steele-Mortimer O. Quantitative assessment of cytosolic Salmonella in epithelial cells. PLoS One. 2014;9:e84681. doi: 10.1371/journal.pone.0084681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cemma M, Kim PK, Brumell JH. The ubiquitin-binding adaptor proteins p62/SQSTM1 and NDP52 are recruited independently to bacteria-associated microdomains to target Salmonella to the autophagy pathway. Autophagy. 2011;7:341–5. doi: 10.4161/auto.7.3.14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wild P, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. Reference 140 identifies a phosphorylation-dependent regulation of autophagy receptor OPTN. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fujita N, et al. Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J Cell Biol. 2013;203:115–28. doi: 10.1083/jcb.201304188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ponpuak M, et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity. 2010;32:329–41. doi: 10.1016/j.immuni.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Randow F. How cells deploy ubiquitin and autophagy to defend their cytosol from bacterial invasion. Autophagy. 2011;7:304–9. doi: 10.4161/auto.7.3.14539. [DOI] [PubMed] [Google Scholar]
- 144.Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–8. doi: 10.1038/nature10744. Reference 144 shows galectin 8 can detect damaged vesicles in the cytosol to trigger host autophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shahnazari S, et al. A diacylglycerol-dependent signaling pathway contributes to regulation of antibacterial autophagy. Cell Host Microbe. 2010;8:137–46. doi: 10.1016/j.chom.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tattoli I, et al. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe. 2012;11:563–75. doi: 10.1016/j.chom.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 147.Owen KA, Meyer CB, Bouton AH, Casanova JE. Activation of Focal Adhesion Kinase by Salmonella Suppresses Autophagy via an Akt/mTOR Signaling Pathway and Promotes Bacterial Survival in Macrophages. PLoS Pathog. 2014;10:e1004159. doi: 10.1371/journal.ppat.1004159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Angosto D, et al. Evolution of inflammasome functions in vertebrates: Inflammasome and caspase-1 trigger fish macrophage cell death but are dispensable for the processing of IL-1beta. Innate Immun. 2012;18:815–24. doi: 10.1177/1753425912441956. [DOI] [PubMed] [Google Scholar]
- 149.Raupach B, Peuschel SK, Monack DM, Zychlinsky A. Caspase-1-mediated activation of interleukin-1beta (IL-1beta) and IL-18 contributes to innate immune defenses against Salmonella enterica serovar Typhimurium infection. Infect Immun. 2006;74:4922–6. doi: 10.1128/IAI.00417-06. References 149 show that inflammasome activation is important in Salmonella pathogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Broz P, et al. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature. 2012;490:288–91. doi: 10.1038/nature11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lara-Tejero M, et al. Role of the caspase-1 inflammasome in Salmonella typhimurium pathogenesis. J Exp Med. 2006;203:1407–12. doi: 10.1084/jem.20060206. References 151 show that inflammasome activation is important in Salmonella pathogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Broz P, et al. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J Exp Med. 2010;207:1745–55. doi: 10.1084/jem.20100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Man SM, et al. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc Natl Acad Sci U S A. 2014;111:7403–8. doi: 10.1073/pnas.1402911111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Miao EA, Warren SE. Innate immune detection of bacterial virulence factors via the NLRC4 inflammasome. J Clin Immunol. 2010;30:502–6. doi: 10.1007/s10875-010-9386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Proell M, Gerlic M, Mace PD, Reed JC, Riedl SJ. The CARD plays a critical role in ASC foci formation and inflammasome signalling. Biochem J. 2013;449:613–21. doi: 10.1042/BJ20121198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wynosky-Dolfi MA, et al. Oxidative metabolism enables Salmonella evasion of the NLRP3 inflammasome. J Exp Med. 2014 doi: 10.1084/jem.20130627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Iyer SS, et al. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity. 2013;39:311–23. doi: 10.1016/j.immuni.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Dalebroux ZD, Matamouros S, Whittington D, Bishop RE, Miller SI. PhoPQ regulates acidic glycerophospholipid content of the Salmonella Typhimurium outer membrane. Proceedings of the National Academy of Sciences. 2014 doi: 10.1073/pnas.1316901111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Franchi L, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol. 2006;7:576–82. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 160.Lightfield KL, et al. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat Immunol. 2008;9:1171–8. doi: 10.1038/ni.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Miao EA, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. 2006;7:569–75. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 162.Mariathasan S, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–8. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 163.Franchi L, et al. NLRC4-driven production of IL-1beta discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat Immunol. 2012;13:449–56. doi: 10.1038/ni.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Andersen-Nissen E, et al. Evasion of Toll-like receptor 5 by flagellated bacteria. Proc Natl Acad Sci U S A. 2005;102:9247–52. doi: 10.1073/pnas.0502040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yoon SI, et al. Structural basis of TLR5-flagellin recognition and signaling. Science. 2012;335:859–64. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhong D, et al. The Salmonella type III secretion system inner rod protein PrgJ is partially folded. J Biol Chem. 2012;287:25303–11. doi: 10.1074/jbc.M112.381574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhao Y, et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 168.Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 2011;477:592–5. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tenthorey JL, Kofoed EM, Daugherty MD, Malik HS, Vance RE. Molecular Basis for Specific Recognition of Bacterial Ligands by NAIP/NLRC4 Inflammasomes. Molecular cell. 2014;54:17–29. doi: 10.1016/j.molcel.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yang J, Zhao Y, Shi J, Shao F. Innate immune recognition of bacterial type III secretion needle protein by the NAIP inflammasome in human and mouse. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1306376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Romanish MT, Nakamura H, Lai CB, Wang Y, Mager DL. A novel protein isoform of the multicopy human NAIP gene derives from intragenic Alu SINE promoters. PLoS One. 2009;4:e5761. doi: 10.1371/journal.pone.0005761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Boniotto M, et al. Population variation in NAIP functional copy number confers increased cell death upon Legionella pneumophila infection. Hum Immunol. 2012;73:196–200. doi: 10.1016/j.humimm.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 173.Cummings LA, Wilkerson WD, Bergsbaken T, Cookson BT. In vivo, fliC expression by Salmonella enterica serovar Typhimurium is heterogeneous, regulated by ClpX, and anatomically restricted. Mol Microbiol. 2006;61:795–809. doi: 10.1111/j.1365-2958.2006.05271.x. [DOI] [PubMed] [Google Scholar]
- 174.Kayagaki N, et al. Noncanonical Inflammasome Activation by Intracellular LPS Independent of TLR4. Science. 2013 doi: 10.1126/science.1240248. References 174 show that caspase 11 is activated by intracellular LPS and protects against cytosolic bacteria. [DOI] [PubMed] [Google Scholar]
- 175.Aachoui Y, et al. Caspase-11 protects against bacteria that escape the vacuole. Science. 2013;339:975–8. doi: 10.1126/science.1230751. References 175 show that caspase 11 is activated by intracellular LPS and protects against cytosolic bacteria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Akhter A, et al. Caspase-11 promotes the fusion of phagosomes harboring pathogenic bacteria with lysosomes by modulating actin polymerization. Immunity. 2012;37:35–47. doi: 10.1016/j.immuni.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. Cytoplasmic LPS activates caspase-11: implications in TLR4 independent endotoxic shock. Science. 2013;341:1250–1253. doi: 10.1126/science.1240988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Into T, Inomata M, Takayama E, Takigawa T. Autophagy in regulation of Toll-like receptor signaling. Cell Signal. 2012;24:1150–62. doi: 10.1016/j.cellsig.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 179.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13:722–37. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J Biol Chem. 2008;283:33175–82. doi: 10.1074/jbc.M804478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Keestra AM, et al. A Salmonella virulence factor activates the NOD1/NOD2 signaling pathway. MBio. 2011;2 doi: 10.1128/mBio.00266-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Travassos LH, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 183.Saitoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–8. doi: 10.1038/nature07383. References 183 show crosstalk between autophagy and inflammasome pathways. [DOI] [PubMed] [Google Scholar]
- 184.Shi CS, et al. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012;13:255–63. doi: 10.1038/ni.2215. References 184 show crosstalk between autophagy and inflammasome pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–5. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 186.Nakahira K, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–30. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Meunier E, et al. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature. 2014;509:366–70. doi: 10.1038/nature13157. [DOI] [PubMed] [Google Scholar]
- 188.VanDussen KL, et al. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut. 2014 doi: 10.1136/gutjnl-2013-306651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Mathur R, et al. A Mouse Model of Salmonella Typhi Infection. Cell. 2012;151:590–602. doi: 10.1016/j.cell.2012.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Song J, et al. A Mouse Model for the Human Pathogen Salmonella Typhi. Cell Host & Microbe. 2010;8:369–376. doi: 10.1016/j.chom.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Libby SJ, et al. Humanized nonobese diabetic-scid IL2rγnull mice are susceptible to lethal Salmonella Typhi infection. Proceedings of the National Academy of Sciences. 2010;107:15589–15594. doi: 10.1073/pnas.1005566107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Núñez-Hernández C, Alonso A, Pucciarelli MG, Casadesús J, García-del Portillo F. Dormant Intracellular Salmonella enterica Serovar Typhimurium Discriminates among Salmonella Pathogenicity Island 2 Effectors To Persist inside Fibroblasts. Infection and Immunity. 2014;82:221–232. doi: 10.1128/IAI.01304-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Arnoldini M, et al. Bistable Expression of Virulence Genes in Salmonella Leads to the Formation of an Antibiotic-Tolerant Subpopulation. PLoS Biol. 2014;12:e1001928. doi: 10.1371/journal.pbio.1001928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Helaine S, et al. Internalization of Salmonella by Macrophages Induces Formation of Nonreplicating Persisters. Science. 2014;343:204–208. doi: 10.1126/science.1244705. Reference 194 uses a fluorescence dilution technique to study single cell persisters. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Spano S, Ugalde JE, Galan JE. Delivery of a Salmonella Typhi Exotoxin from a Host Intracellular Compartment. Cell Host & Microbe. 2008;3:30–38. doi: 10.1016/j.chom.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 196.Haghjoo E, Galan JE. Salmonella typhi encodes a functional cytolethal distending toxin that is delivered into host cells by a bacterial-internalization pathway. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4614–4619. doi: 10.1073/pnas.0400932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hodak H, Galan JE. A Salmonella Typhi homologue of bacteriophage muramidases controls typhoid toxin secretion. EMBO Rep. 2013;14:95–102. doi: 10.1038/embor.2012.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Spano S, Galan JE. A Rab32-Dependent Pathway Contributes to Salmonella Typhi Host Restriction. Science. 2012;338:960–963. doi: 10.1126/science.1229224. Reference 198 demonstrates that loss of a T3S effector contributes to S. Typhi host restriction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Song J, Gao X, Galán JE. Structure and function of the Salmonella Typhi chimaeric A2B5 typhoid toxin. Nature. 2013;499:350–354. doi: 10.1038/nature12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Winter SE, Raffatellu M, Wilson RP, Rüssmann H, Bäumler AJ. The Salmonella enterica serotype Typhi regulator TviA reduces interleukin‐8 production in intestinal epithelial cells by repressing flagellin secretion. Cellular microbiology. 2008;10:247–261. doi: 10.1111/j.1462-5822.2007.01037.x. [DOI] [PubMed] [Google Scholar]
- 201.Haneda T, et al. The capsule-encoding viaB locus reduces intestinal inflammation by a Salmonella pathogenicity island 1-independent mechanism. Infection and immunity. 2009;77:2932–2942. doi: 10.1128/IAI.00172-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Crawford RW, et al. Loss of Very-Long O-Antigen Chains Optimizes Capsule-Mediated Immune Evasion by Salmonella enterica Serovar Typhi. mBio. 2013;4:e00232–13. doi: 10.1128/mBio.00232-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.McClelland M, et al. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat Genet. 2004;36:1268–1274. doi: 10.1038/ng1470. [DOI] [PubMed] [Google Scholar]
- 204.Spano S, Liu X, Galan JE. Proteolytic targeting of Rab29 by an effector protein distinguishes the intracellular compartments of human-adapted and broad-host Salmonella. Proceedings of the National Academy of Sciences. 2011;108:18418–18423. doi: 10.1073/pnas.1111959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Trombert AN, Berrocal L, Fuentes J, Mora GS. Typhimurium sseJ gene decreases the S. Typhi cytotoxicity toward cultured epithelial cells. BMC Microbiology. 2010;10:312. doi: 10.1186/1471-2180-10-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Levine MM, Black RE, Lanata C. Precise Estimation of the Numbers of Chronic Carriers of Salmonella typhi in Santiago, Chile, an Endemic Area. Journal of Infectious Diseases. 1982;146:724–726. doi: 10.1093/infdis/146.6.724. [DOI] [PubMed] [Google Scholar]
- 207.Miyoshi H, Stappenbeck TS. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protocols. 2013;8:2471–2482. doi: 10.1038/nprot.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial Persistence as a Phenotypic Switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 209.Dörr T, Vulić M, Lewis K. Ciprofloxacin Causes Persister Formation by Inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010;8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Christen M, et al. Asymmetrical Distribution of the Second Messenger c-di-GMP upon Bacterial Cell Division. Science. 2010;328:1295–1297. doi: 10.1126/science.1188658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


