Abstract
Purpose
There is a need for improved subclassification of urothelial carcinoma (UC) at diagnosis. A major aim of this study was to search for novel genomic subgroups.
Experimental design
We assessed 160 tumors for genome-wide copy number alterations and mutation in genes implicated in UC. These comprised all tumor grades and stages and included 49 high-grade stage T1 (T1G3) tumors.
Results
Our findings point to the existence of genomic subclasses of the “gold-standard” grade/stage groups. The T1G3 tumors separated into 3 major subgroups that differed with respect to the type and number of copy number events and to FGFR3 and TP53 mutation status. We also identified novel regions of copy number alteration, uncovered relationships between molecular events, and elucidated relationships between molecular events and clinico-pathologic features. FGFR3 mutant tumors were more chromosomally stable than their wild-type counterparts and a mutually exclusive relationship between FGFR3 mutation and overrepresentation of 8q was observed in non-muscle-invasive tumors. In muscle-invasive (MI) tumors, metastasis was positively associated with losses of regions on 10q (including PTEN), 16q and 22q, and gains on 10p, 11q, 12p, 19p, and 19q. Concomitant copy number alterations positively associated with TP53 mutation in MI tumors were losses on 16p, 2q, 4q, 11p, 10q, 13q, 14q, 16q, and 19p, and gains on 1p, 8q, 10q, and 12q. Significant complexity was revealed in events affecting chromosome 9.
Conclusions
These findings may lead to improved biologic understanding and the development of prognostic biomarkers. Novel regions of copy number alteration may reveal potential therapeutic targets.
Introduction
Urothelial carcinoma (UC) of the bladder is the 6th most common cancer in the United States and the United Kingdom (>70,000 and >10,000 new cases per annum, respectively). The majority of tumors (70%) are low-grade (grade 1/2), low-stage (Ta) lesions not penetrating the basement membrane, and these infrequently progress to muscle invasion (10%–15%). However, tumor recurrence is a major feature and the need for repeated cystoscopic monitoring places a great burden on both patients and healthcare resources. Muscle-invasive (MI) disease (Stages T2–T4) represents 20% of cases. Of these, approximately 50% develop metastases and 5-year survival is less than 50%. Stage T1 tumors that have penetrated the basement membrane but not invaded muscle represent 10% to 20% of cases. A significant proportion of these have potential to become invasive, with progression rates of up to 50% (1). High-grade T1 tumors (T1G3) in particular exhibit highly divergent behavior and their clinical management is challenging.
Prediction of risk of recurrence, invasion, and metastasis in individual patients with UC remain major goals. Histopathologic characteristics have limited ability to predict outcome for individual patients. Although some molecular features of UC show association with clinical phenotype, none are currently suitable for application as prognostic biomarkers. Frequent genomic alterations include copy number loss and/or LOH involving chromosome 9 in UC of all stages and grades, FGFR3 and PIK3CA mutation in low-grade Ta tumors (2) and TP53 mutation and inactivation of the retinoblastoma (RB) pathway in MI tumors (3, 4). High-throughput methodologies at the mRNA, microRNA, protein, epigenetic, and copy number levels have reported associations with clinico-pathologic characteristics. These include mRNA and microRNA expression signatures associated with distinct clinico-pathologic features (5, 6) and a range of aberrant methylation or histone modifications (e.g., 7, 8). Although some expression signatures have been validated in independent sample series (9), the use of such markers in the clinic remains unproven.
DNA copy number differences underlie a significant proportion of differences in gene expression detected between cancer subtypes and studies in other tumor types have described prognostic subtypes based on DNA copy number alterations. In addition, DNA represents a more stable template for marker assessment than RNA. The use of DNA copy number profiles to define molecular subtypes of UC, and the relationships between copy number, mutational events, and clinico-pathologic data have not been fully assessed. Genome-wide analysis of copy number aberrations and LOH using array-based comparative genomic hybridisation (CGH) or single-nucleotide polymorphism array profiling have revealed regions of alteration (10–15). In general, fewer copy number alterations are found in low-stage and low-grade tumors, and more complex patterns in MI tumors. Several regions of high-level amplification have been identified (14, 16) and integration of copy number and expression data has identified candidate genes (14). A few studies have searched for concomitant copy number events (14, 17), or have combined assessment of copy number alterations with mutation status of FGFR3 or TP53 and found differences in chromosomal stability relating to mutation (11, 18). Copy number alterations have been associated with stage, grade, recurrence, carcinoma in situ (CIS), and outcome (12, 13, 19, 20), but few studies have conducted in-depth correlations of findings with clinico-pathologic information, and where this has been done, mutation status was not assessed (21), full copy number information was not included in the analysis (22), or tumors of all stages and grades together were subdivided on the basis of gene expression profiles before copy number analysis (23).
We predict that further genomic subdivision of UC remains to be revealed, and that this may be achieved through integrated analysis of genomic events in large panels of tumors. Here, we assessed 160 bladder tumors, including the largest panel of T1G3 tumors analyzed to date (n = 49), for both genome-wide copy number alterations and mutation of 8 key genes implicated in UC. We identified concomitant molecular events and assessed the relationships of molecular alterations to clinico-pathologic data. Potential subclasses of the “gold-standard” grade/stage groups of UC defined by DNA copy number and mutation status have been identified.
Materials and Methods
Patient samples and clinical information
Study approval was granted by the Leeds-East Research Ethics Committee and informed consent obtained from all patients. Cold cup biopsies were snap-frozen and stored in liquid nitrogen and the remainder of the sample embedded in paraffin for diagnostic assessment (24, 25). The sample set consisted of 3 TaG1, 39 TaG2, 16 TaG3, 11 T1G2, 49 T1G3, and 42 ≥T2 (all grades) tumors (Supplementary Table S1). Median follow-up time was 6 years.
Nucleic acid isolation
DNA was extracted from frozen sections containing at least 70% tumor cells using the QIAamp DNA Mini Kit (Qiagen). DNA was extracted from venous blood using a Nucleon DNA extraction kit (Nucleon Biosciences) or by salt precipitation. Total RNA was extracted using the Pico-Pure RNA Isolation Kit (Nikon UK Limited).
Array CGH
Two types of 1-Mb resolution CGH array were used; Centre for Microarray Resources (CMR; University of Cambridge, Cambridge, UK) and Sanger (26). Coverage on the 2 arrays was essentially the same (details available on request). Hybridization was as described (16). Reference samples were paired normal DNA from blood. In 54 cases blood DNA was not available at the time of CGH analysis and an unmatched lymphoblastoid cell line was used. BlueFuse software (BlueGnome) was used to define spots, subtract background, and calculate normalized fluorescence intensities. Breakpoints and regions of gain and loss were detected using the BlueFuse software aCGH-smooth algorithm (27) with calling thresholds for determining copy number gain and loss set to ±0.15 log2 ratio (test/reference) for CMR arrays or ±0.2 for Sanger arrays. These thresholds were determined by observation of normal samples and samples with known changes. Alterations were confirmed visually by 2 observers. Amplifications and homozygous deletions (HD) were defined where the log2 ratio was ≥1.2 and ≥1.2, respectively. Physical positions of clones are according to hg18/NCBI Build 36 (March 2006). Fraction of genome altered (FGA) was defined as the percentage of clones reporting significantly altered copy number.
Mutation analysis
High-resolution melting (HRM) curve analysis was used to screen for mutations in FGFR3, PIK3CA, HRAS, KRAS, and NRAS (28). Fluorescent single-stranded conformational polymorphism analysis or HRM analysis was used to screen TSC1 (28) and TP53. AKT1 mutations were detected as described (29). GRIN2A was analyzed using HRM analysis or direct sequencing. Potential mutations were confirmed by sequencing tumor and matched blood samples to establish somatic mutation status. Primer sequences and protocols are available for TP53 and GRIN2A on request.
Loss of heterozygosity analysis
Microsatellite markers in the CDKN2A region (D9S1748, D9S1749), TSC1 region (D9S1830, D9S1199, D9S149, D9S66), and the PTEN region (D10S1765, D10S215, D10S541) were used. Forward primers were fluorescently labeled, products run on an ABI 3130 sequencer (Applied Biosystems), and analyzed using GeneMapper v3.7 software (Applied Biosystems). LOH was scored as a reduction of at least 60% signal from 1 allele.
Immunohistochemistry
Sections were stained for FGFR3 (1:200; B9; Santa Cruz), PTEN (1:100; 9559; Cell Signaling Technology), p53 (1:100; 18-01; Novacastra), and USP7 (1:100; IHC-00018; Bethyl Laboratories Inc.). All runs included a no primary antibody control.
Statistics and cluster analysis
Smoothed log2 ratio values were used for frequency plot visualization and statistical comparisons of frequency data within the Nexus Copy Number Professional 3.1 Software (BioDiscovery). Using the Comparisons function, a Fishers Exact test (P ≤ 0.05; differential threshold 25%) was conducted to determine significant differences between sample subgroups. Hierarchical clustering, Kruskal–Wallis tests, Pearson correlations, and Kaplan–Meier survival analysis were conducted using the Partek Genomics Suite 6.5 (Partek Inc.). To conduct cluster analysis, each individual BAC (Bacterial Artificial Chromosome) array clone was assigned a copy number class (0 = no copy number aberration; 1 = gain; −1 = loss; 2 = high-level gain; −2 = high-level loss) for each sample. One-way unsupervised hierarchical clustering was conducted using Euclidean distance and the Ward method of linkage. The Kruskal–Wallis test was used to test for significant differences in FGA between tumor subgroups and bootstrap analysis was applied to test the robustness of the data set. χ2 tests were conducted to test associations with stage and grade. Pearson correlations were carried out between DNA copy number (0 = no copy number alteration; 1 = gain; 2 = high-level amplification) and gene expression levels for 8q22.2-q22.3 candidate genes as determined by quantitative real-time PCR (qRT-PCR).
cDNA synthesis and qRT-PCR
cDNA was synthesized using Superscript II (Invitrogen) according to the manufacturer’s instructions. Candidate gene expression was assessed by RT-PCR using TaqMan assays (Applied Biosystems) for YWHAZ (Hs01122445_g1), ANKRD46 (Hs01569215_m1), SNX31 (Hs00381645_m1), ZNF706 (Hs00706404_s1), NACAP1 (Hs04233493_sH), GRHL2 (Hs00227745_m1), and GRIN2A (Hs00168219_m1). Levels of expression were normalized to SDHA (Hs00417200_m1) and measured relative to a pool of uncultured normal human urothelial cells isolated from human ureters obtained at nephrectomy (30).
Results
Relationship of genome-wide copy number alterations to tumor stage and grade
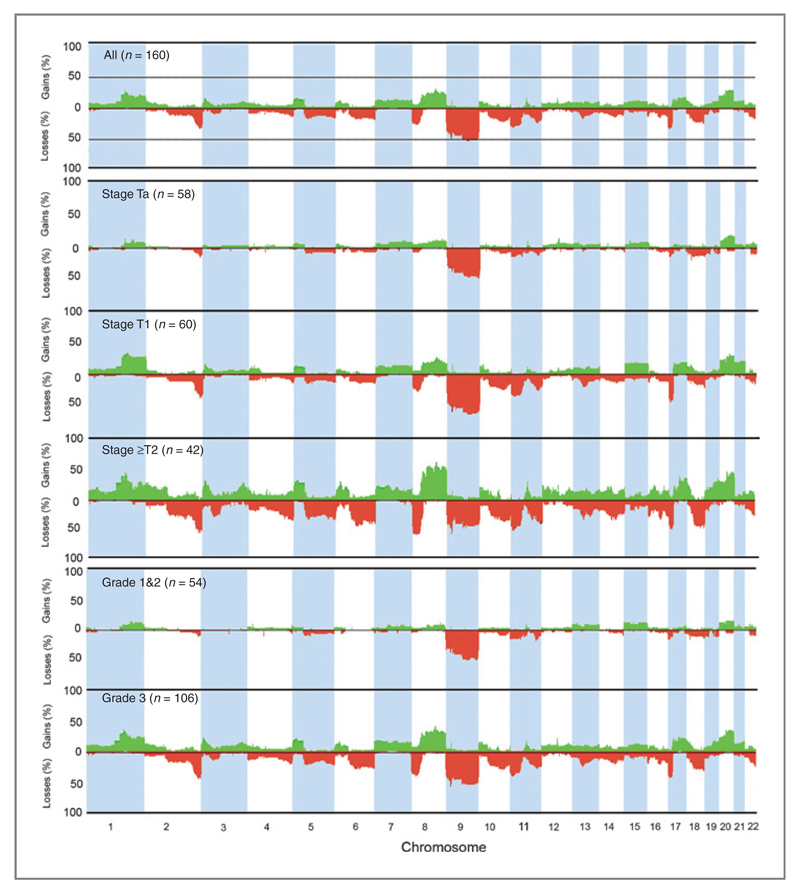
1Mb-resolution array-CGH was used to assess copy number alterations in 160 UC samples. Tumor grade and stage currently form a major component in assessment of risk of progression in non-MI bladder cancer (31). Thus, we first constructed frequency plots of DNA copy number alterations according to stage and grade (Fig. 1). We next examined whether the proportions of different types of alteration (i.e., those involving whole chromosomes, chromosome arms, or smaller regions) differed between stages and grades. No significant differences were observed but the frequencies of such alterations increased with stage and grade (Table 1).
Figure 1.
Genome-wide frequency plots of copy number alterations identified in 160 bladder tumors. Regions of chromosomal copy number imbalance were identified using the aCGH-smooth algorithm within BlueFuse. The smoothed log2 ratio values generated from these analyses were used as input values for the Nexus software package. The x-axis corresponds to chromosomes 1 to 22 and the y-axis corresponds to the percentage of gains and losses. Copy number gains are shown in green and losses in red. Frequencies of copy number alterations for all tumors, and according to stage and grade are shown.
Table 1. Recurrent genomic alterations by chromosome arm.
| Stage/grade | Losses (frequency %) | Gains (frequency %) |
|---|---|---|
| TaG1/G2 (n = 42) | 9p (28%), 9q (48%), 11p (17%), 11q (21%), 17p (19%), 18q (19%), 19p (19%), 19q (19%) |
|
| TaG3 (n = 16) | 2q (31%), 4p (19%), 6q (25%), 8p (25%), 9p (62%), 9q (69%), 10q (25%), 11p (37%), 11q (25%), 12q (19%), 13q (19%), 14q (19%), 18p (19%), 18q (31%) |
1q (31%), 4q (31%), 7p (25%), 7q (19%), 8p (25%), 8q (31%), 9p (25%), 10q (19%), 11q (19%), 12q (37%), 20p (31%), 20q (31%) |
| T1G2 (n = 11) | 2q (27%), 4q (18%), 8p (18%), 9p (82%), 9q (73%), 11p (27%), 11q (45%), 14q (18%), 17p (18%), 22q (18%) |
1q (27%), 6p (18%), 8q (18%), 10q (18%), 13q (18%), 15q (18%) |
| T1G3 (n = 49) | 2q (47%), 4q (27%), 5q (25%), 6q (22%), 8p (33%), 8q (16%), 9p (65%), 9q (53%), 10q (35%), 11p (39%), 11q (29%), 12q (16%), 13q (35%), 14q (20%), 16p (22%), 17p (55%), 18p (16%), 18q (27%), 22q (27%) |
1q (41%), 3p (20%), 4p (16%), 5p (18%), 7p (18%), 7q (16%), 8p (18%), 8q (29%), 9p (22%), 10p (16%), 12q (20%), 13q (16%), 15q (20%), 16q (20%), 17q (27%), 20p (27%), 20q (39%), 21q (25%) |
| ≥T2 all grades (n = 42) | 2q (59%), 3p (36%), 3q (24%), 4p (33%), 4q (38%), 5q (43%), 6p (29%), 6q (52%), 8p (57%), 9p (59%), 9q (50%), 10p (21%), 10q (40%), 11p (50%), 11q (43%), 12q (26%), 13q (43%), 14q (40%), 15q (21%), 16p (38%), 16q (38%), 17p (50%), 18q (50%), 19p (24%), 19q (19%), 22q (31%) |
1p (33%), 1q (59%), 2p (38%), 2q (26%), 3p (38%), 3q (33%), 4p (26%), 4q (29%), 5p (36%), 5q (19%), 6p (36%), 7p (26%), 7q (29%), 8p (43%), 8q (69%), 9p (31%), 10p (29%), 10q (31%), 11q (31%), 12p (29%), 12q (33%), 13q (33%), 14q (31%), 15q (21%), 16p (38%), 16q (21%), 17p (48%), 18p (29%), 19p (24%), 19q (40%), 20p (33%), 20q (59%), 21q (19%), 22q (24%) |
NOTE: Only those alterations that occurred at frequencies ≥15% are shown.
Molecular alterations involving chromosome 9
Deletion of all or part of chromosome 9 is common in UC and critical regions have been identified at 9p21.3 (CDKN2A, CDKN2B), 9q22 (PTCH), 9q33 (DBC1), and 9q34 (TSC1). HD at 9p21.3 was detected in 16 tumors and in most cases included CDKN2A. In 6 samples, deletion did not include CDKN2A. Two had deletion of a more distal region containing 15 genes (IFN1α cluster, IFNE, KLHL9, miR-31) and 4 had deletion proximal to CDKN2A containing ELAVL2 only. Four other HD were detected, each in a single tumor; 9q32-q33 (DBC1, ASTN, and part of TLR2); 9p24.2-p24.3 (DOCK8-SMARCA2), 9p22.2-p22.3 (SNAPC3-BNC2), and 9q21.33 (DAPK1).
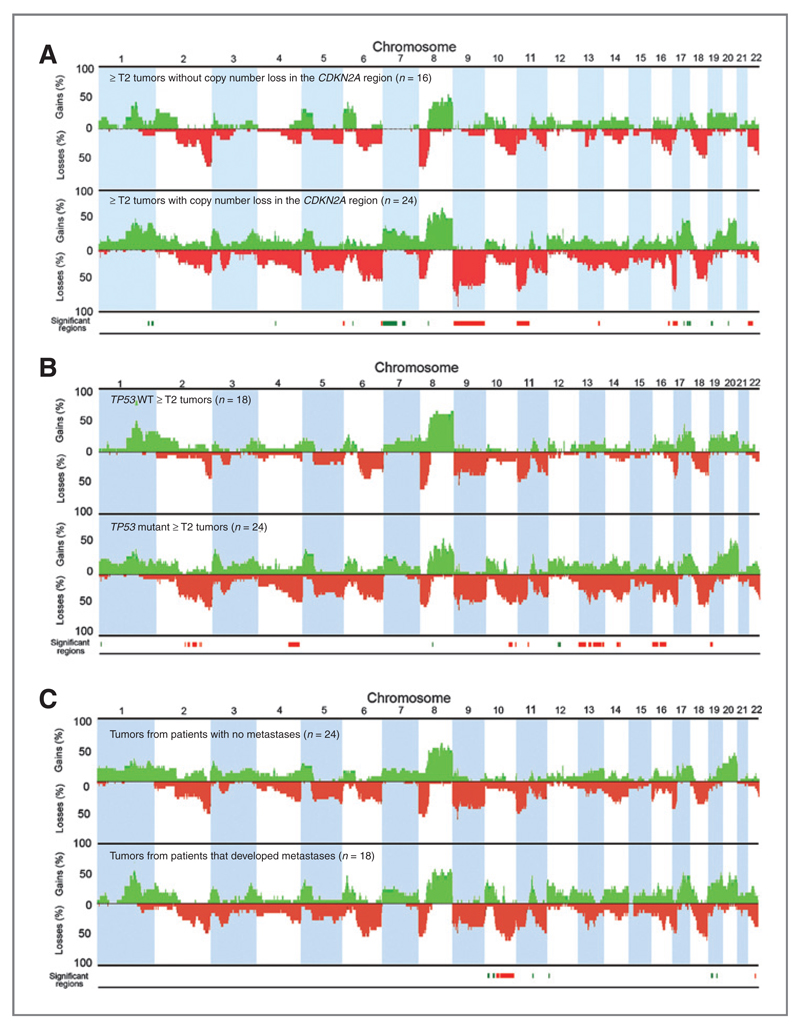
9p21.3 deletions were associated with high stage (P = 0.01) and grade (P = 0.02). In ≥T2 tumors, deletions on 6p, 6q, 9p, 9q, 11p, 13q, and 17p, and gains on 1q, 4q, 7p, 7q, 8p, 17q, and 20q were positively associated with copy number loss in the CDKN2A region (Fig. 2A). In other tumor groups, the same chromosome regions were associated with CDKN2A deletion, but the association did not reach significance. TSC1 LOH (9q34) was associated with grade (P = 0.005) but not with stage. No associations between TSC1 copy number loss and stage, grade or other copy number alterations were observed.
Figure 2.
CDKN2A copy number loss, TP53 mutation status, metastasis and genome-wide comparisons of copy number alterations in ≥T2 tumors. Genome-wide frequency plots of copy number alterations in (A) tumors with (n = 24) and without (n = 16) loss of copy number in the CDKN2A region, (B) TP53 wild type (WT; n = 18) and mutant (n = 24) tumors, and (C) tumors from patients that did (n = 18) or did not (n = 24) develop metastases. Significant regions (0.05 P value cutoff) of copy number gain (green) and loss (red) positively associated with copy number loss in the CDKN2A region, TP53 mutation or metastasis are highlighted at the bottom of each figure. Two tumors with increased copy number in the CDKN2A region were not included in the analyses.
A region of high-level amplification at 9p24.3 (FOXD4-DMRT2) was present in one tumor and a larger region of amplification containing 38 genes from SMARCA2 to PTPRD (9p23-p24.3) in another. Two discrete regions of gain were detected at 9p13.3-p21 (TMEM215-C9orf25) in 2 tumors and 9q21.12 (C9orf135-TRPM3) in one tumor.
Other regions of HD and high-level amplification
Fifty-one HD, including those on chromosome 9 were detected in 29 tumors (Table 2). These included 1p34.1 (PTCH2), 2q36.1-q36.3 (CUL3), 11p11.2 (DDB2), 18p11.22-p11.21 (FAM38B), and 19q12 (TSHZ3). One hundred and seventy-one high-level amplifications were detected in 54 tumors (3/42 TaG2, 5/16 TaG3, 20/49 T1G3, 1/11 T1G2, 25/42 ≥T2; Table 2). The most frequent (n = 11) was on 11q13.2-q13.3 (containing CCND1). Other regions were 3p25.3-p25.1, 8q22.2-q22.3, and 12q15 (containing RAF1, YWHAZ, and MDM2, respectively). Concomitant amplification of YWHAZ and a paralogous gene YWHAQ (14-3-3-theta, 2p25), which has been reported in UC (14), was not observed here, but concomitant gain or amplification was observed for 3p25.3-p25.1 (RAF1) and 8q22.2-q22.3 in 21 tumors. Amplification of both regions was detected in 3 samples (2 T1G3, 1≥T2G3), gain of 8q22.2-q22.3 in 6 samples with 3p25.3-p25.1 amplification (1 TaG2, 1 T1G3, 4 T2G3), gain of 3p25.3-p25.1 in 4 samples with 8q22.2-q22.3 amplification (2 T1G3, 2≥T2G3), and gain of both regions in 8 samples (2 TaG3, 2 T1G3, 4 ≥T2G3).
Table 2. Recurrent regions of HD and high-level amplification.
| Chromosome | Cytoband | Positiona (Mb) | Number of genes | Candidate genesb | Number of tumorsc | Loss or gain frequencyd (n = 160) |
|---|---|---|---|---|---|---|
| HD | ||||||
| 1 | 1p34.1 | 44.61–45.35 | 12 | PTCH2 | 2 | 4% |
| 2 | 2q36.1-q36.3 | 224.01–227.04 | 9 |
SGC2, AP1S3, WDFY1, MRPL44, SERPINE2, FAM124B, CUL3, DOCK10, KIAA1486 |
2 | 28.6% |
| 9 | 9p21.3 | 21.15–21.73 | 14 |
IFNA1α cluster, IFNE, KLHL9, miR-31 |
13 | 49.7% |
| 9 | 9p21.3 | 21.73–22.15 | 4 |
MTAP, CDKN2A, CDKN2B, ANRIL |
16 | 50.9% |
| 9 | 9p21.3 | 22.47–24.09 | 1 | ELAVL2 | 4 | 38.5% |
| 11 | 11p11.2 | 45.04–47.29 | 29 | DDB2, CRY2 | 2 | 24.8% |
| 18 | 18p11.22-p11.21 | 10.58–11.06 | 1 | FAM38B | 2 | 11.8% |
| 19 | 19q12 | 35.76–36.89 | 1 | TSHZ3 | 3 | 6.2% |
| Amplification | ||||||
| 1 | 1q23.2-q23.3 | 158.15–161.21 | 72 | TAGLN2 | 9 | 28% |
| 3 | 3p25.3- p25.1 | 10.78–13.47 | 17 | RAF1, PPARG | 11 | 16% |
| 6 | 6p22.3 | 20.01–22.58 | 5 |
MBOAT1, E2F3, CDKAL1, SOX4, PRL |
8 | 11.8% |
| 8 | 8p12- p11.22 | 37.37–39.84 | 27 | FGFR1, TACC1, RCP | 5 | 16.1% |
| 8 | 8q22.2-q22.3 | 101.43–103.80 | 11 | YWHAZ | 9 | 30.4% |
| 11 | 11p15.5 | 0.26–1.58 | 46 | HRAS | 3 | 6.2% |
| 11 | 11q13.2-q13.3 | 68.70–69.30 | 5 |
MYEOV, CCND1, ORAOV1, FGF19, FGF4 |
14 | 14.9% |
| 11 | 11q14.1 | 78.20–79.64 | 1 | ODZ4 | 3 | 8.1% |
| 12 | 12q15 | 66.93–68.44 | 14 | MDM2 | 9 | 14.9% |
| 12 | 12q24.21-q24.22 | 114.12–115.49 | 1 | MED13L | 3 | 10.6% |
| 17 | 17q12-q21.2 | 34.62–35.68 | 36 | ERBB2 | 3 | 13.0% |
| 20 | 20q12-q13.2 | 40.79–43.89 | 52 | YWHAB | 7 | 28.6% |
| 20 | 20q13.32-q13.33 | 57.73–59.89 | 7 |
PHACTR3, SYCP2, PPP1R3D, CDH26, C20orf197, miR-646, CDH4 |
3 | 27.9% |
Physical position is according to hg18/NCBI build 36.
All candidate genes are listed for regions where the total number of genes is ≤10. For regions where the total number of genes >10, candidate genes listed were selected based on known or putative cancer-associated function.
HDs were classified as those regions where the normalized log2 ratio was ≥–1.2. HDs detected in 2 or more tumors are listed. Amplifications were classified as those regions where the normalized log2 ratio was ≥1.2. Amplifications detected in 3 or more tumors are listed.
The frequency of loss or gain across candidate regions was estimated as the percentage of samples with log2 ratio ≥–0.15 for CMR arrays or ≥–0.2 for Sanger arrays.
Mutation status
Mutation status of FGFR3, TP53, PIK3CA, HRAS, KRAS, NRAS, AKT1, and TSC1 was determined (Supplementary Table S1). FGFR3 mutation (n = 77) was associated with low stage (P < 0.0001) and grade (P < 0.0001) and TP53 mutation (n = 44) with high stage (P < 0.0001) and grade (P < 0.0001). Eleven tumors had both FGFR3 and TP53 mutation. Forty-one PIK3CA, 22 TSC1, 7 HRAS, 7 KRAS, 1 NRAS, and 5 AKT1 mutations were detected and had no association with stage or grade.
FGA
FGA was defined as the percentage of clones reporting significantly altered copy number and provides a measure of chromosomal instability. FGA groups A (<1%), B (1%– <10%), C (10%–<30%), and D (≥30%) were defined (Supplementary Table S1). Median FGA was higher in tumors of higher grade and stage (Supplementary Fig. S1). Some tumors exhibited few or no copy number alterations. Thirty-five had <5% FGA and 19 of these had <1% FGA. The majority of these chromosomally stable tumors were of low stage and grade (TaG1 n = 3; TaG2 n = 21; TaG3 n = 4; T1G2 n = 2; T1G3 n = 3; T3G3 n = 2). Fifty-eight mutations were detected in this subgroup (26 FGFR3; 2 HRAS; 3 KRAS; 20 PIK3CA; 2 AKT1; 5 TP53) with at least one mutation in each tumor.
Relationships between FGA, mutation status, copy number alterations, and clinico-pathologic data
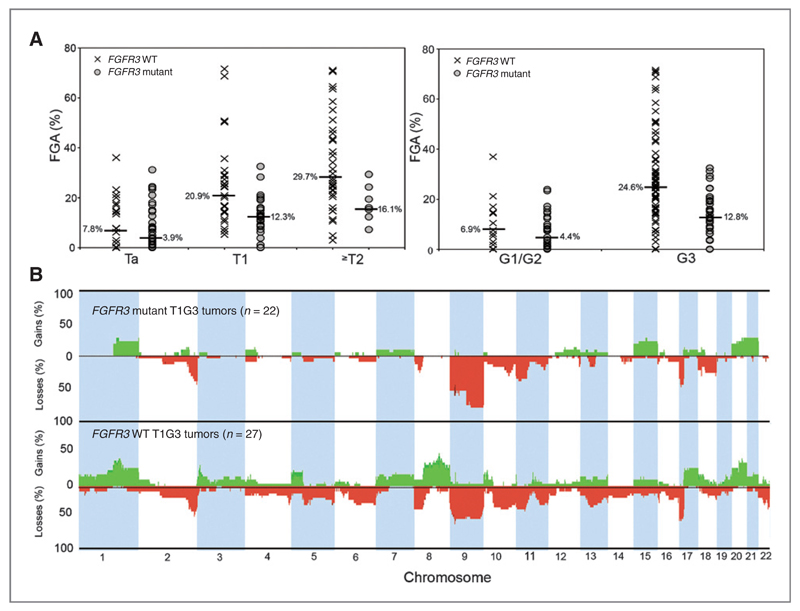
Lower median FGA was found in T1 (P = 0.005), T2 (P = 0.02), and grade 3 (P = 0.00002) tumors with FGFR3 mutation (Fig. 3A; Supplementary Table S1). Interestingly, FGFR3 wild-type Ta tumors commonly showed gain of the long arm of chromosome 8 (P < 0.0001). This inverse relationship was also found in FGFR3 mutant (n = 22) and wild-type (n = 27) T1G3 tumors (P < 0.0001; Fig. 3B). In most cases, all of 8q was gained but 4 tumors showed focal amplification (Supplementary Fig. S2A). The minimal region (8q22.2-q22.3) contained 13 genes (ANKRD46-KLF10). A previous study at tiling path resolution identified a core region extending more than 1.8Mb from POLR2K to NCALD and revealed strongest correlation between YWHAZ copy number and expression (14). Information from this and the present study defines a minimal region (ANKRD46-NCALD) (Supplementary Fig. S2B). We measured expression of YWHAZ, and 5 other genes (ANKRD46, SNX31, ZNF706, NACAP1, and GRHL2) in 46 T1G3 tumors, including those with amplification. YWHAZ showed the best correlation (r = 0.61554; P = 0.000005; Supplementary Fig. S2C).
Figure 3.
FGFR3 mutation status, FGA, and genome-wide comparisons of copy number alterations. A, FGA (%) values and FGFR3 mutation status according to stage and grade. B, genome-wide frequency plots of copy number alterations in FGFR3 wild type (WT; n = 27) and mutant T1G3 tumors (n = 22).
PIK3CA and FGFR3 mutation are commonly found together in UC (2). Compatible with this, we found that as for FGFR3 mutation, PIK3CA mutation was associated with a lower FGA in Ta (P = 0.003), T1 (P = 0.02), G2 (P = 0.01), and G3 (P = 0.005) tumors. No relationships between TSC1 or RAS gene mutation, FGA, and/or copy number events were observed.
Higher FGA was associated with TP53 mutation in T1, T2, and grade 3 tumors (P = 0.006, 0.02, and 0.0003, respectively; Supplementary Fig. S1). However, some TP53 mutant tumors had low FGA. A comparison of specific copy number alterations in TP53 mutant (n = 24) and wild-type (n = 18) ≥T2 samples revealed significant differences (Fig. 2B). Deletions on 2q, 4q, 11p, 10q, 13q, 14q, 16p, 16q, and 19p and gains on 1p, 8q, 10q, and 12q were positively associated with TP53 mutation. A region extending from 16p13.2 to 16p13.3 (6,846,409-10,432,694bp) had the best P value, and contains 10 genes (A2BP1-ATF7IP2). The same association of 16p loss with TP53 mutation was not detected in T1G3 tumors.
Two genes from 16p13.2-p13.3 (USP7 and GRIN2A) were studied further. The deubiquitinating enzyme USP7 (HAUSP) was considered a good p53-related candidate, and GRIN2A (NMDAR2A) was recently proposed as a tumor suppressor gene in melanoma following detection of frequent mutations (32). USP7 expression examined by immunohistochemistry (IHC) did not correlate with 16p loss in the TP53 mutant subgroup. Mutation scanning of 27 ≥T2 tumors including 9 with TP53 mutation and loss of 16p13.2-p13.3 revealed one missense mutation (p. R181S). As GRIN2A has been reported as a target for methylation (33), we measured GRIN2A mRNA expression in 26 stage ≥T2 tumors. GRIN2A expression was low in normal human urothelium and the majority of tumors had comparable or lower expression. Expression did not correlate with TP53 mutation status or 16p loss in ≥T2 tumors and a similar pattern of expression was observed in 14 stage Ta tumors (data not shown).
FGA was not associated with metastasis or overall survival in ≥T2 tumors (P > 0.05), but higher FGA was positively associated with progression in T1G3 tumors (P = 0.03). The presence of high-level amplification(s) did not correlate with outcome. In ≥T2 tumors, deletions on 10q, 16q, and 22q, and gains on 10p, 11q, 12p, 19p, and 19q were independently associated with metastasis (Fig. 2C). PTEN is located on 10q23.3 and its inactivation is common in advanced bladder cancer and associated with poor outcome. Eleven of 14 tumors with copy number loss in the PTEN region were in the metastatic group. Kaplan–Meier analysis (Supplementary Fig. S3) showed that time to metastasis was shorter for tumors with PTEN deletion (P = 0.00003; log-rank test). As TP53 mutation and loss of RB1 are frequent events in muscle invasive UC, we conducted similar analyses. TP53 mutation had no effect either alone or in combination with PTEN copy number loss. RB1 copy number loss had a nonsignificant (P = 0.053; log-rank test) trend for association with shorter time to metastasis when assessed alone, and significant association (P = 0.0002; log-rank test) in combination with PTEN loss (Supplementary Fig. S3). We also examined PTEN protein expression by IHC (Supplementary Fig. S4). However, it was PTEN copy number that showed the strongest association with metastasis. Supplementary Fig. S4 summarizes the relationships between clinical outcome data and alterations involving selected components of the RB/p53/PI3-kinase-related pathways and common regions of amplification.
Tumor subgroups based on genomic profile
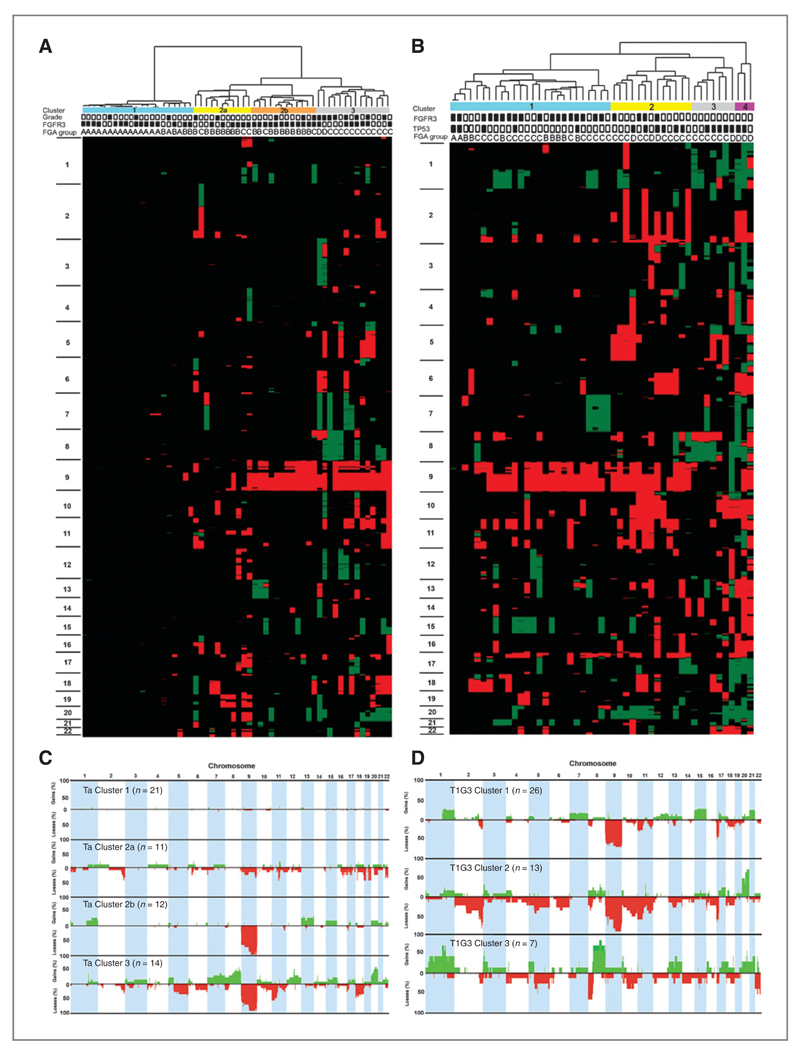
A key aim was to search for novel genomic subgroups to provide improved subclassification at diagnosis. One-way unsupervised hierarchical cluster analysis on copy number data separated tumors primarily but not exclusively according to stage and grade into 3 main clusters (Supplementary Fig. S5A). We then used the same approach to assess heterogeneity within tumor subgroups (Ta, ≥T2, T1G3) in relation to FGA and mutation data.
The 58 Ta tumors (TaG1/2 = 42; TaG3 = 16) formed 4 main clusters (Fig. 4A and C). FGFR3 mutation was frequent and evenly distributed. Only 2 TP53 mutations were detected (Cluster 1 and Cluster 2a). Cluster 1 contained mostly chromosomally stable tumors. Cluster 3 contained mainly grade 3 tumors (n = 8) with high levels of chromosomal instability. Gain of 8q was a feature of this cluster and was mutually exclusive with FGFR3 mutation. The remaining TaG3 tumors were spread across all other clusters. No association was found between local disease recurrence or multifocality and cluster assignment.
Figure 4.
Unsupervised hierarchical cluster analysis of aCGH data and genome-wide frequency plots of copy number alterations in individual clusters from stage Ta and T1G3 tumors. Each tumor was scored for copy number gains and losses and these were assigned a copy number class (2 high-level gain, 1 gain, 0 no change, −1 loss, −2 high-level loss). Copy number class data was used in 1-way hierarchical cluster analysis of (A) 58 stage Ta and (B) 49 T1G3 tumors. Each column of the heat map represents 1 sample and each row represents the genomic position of individual clones on the array. Green, copy number gain; red, copy number loss. Chromosome number is shown on the left-hand side of the heat map. Four main clusters of Ta tumors were identified and these are indicated by the color bars at the top of panel (A): Cluster 1, blue; Cluster 2a, yellow; Cluster 2b, orange; Cluster 3, gray.
The grade of each tumor is shown at the top of the figure (black box, G3; white box, G1/G2) along with FGFR3 mutation status (back box, mutant; white box, wild type) and FGA group (A–D). Four clusters of T1G3 tumors were identified and these are indicated by the color bars at the top of panel (B): Cluster 1, blue; Cluster 2, yellow; Cluster 3, gray; Cluster 4, purple. The TP53 and FGFR3 mutation status of each tumor is also shown at the top of the figure (black box, mutant; white box, wild type) along with FGA group (A–D). C and D, frequency plots of copy number events for individual Ta clusters and T1G3 clusters with copy number gains shown in green and losses in red. T1G3 Cluster 4 consisted of only 3 samples therefore frequency data is not presented.
Three main clusters were identified in ≥T2 samples (Supplementary Fig. S5B and S5C). Cluster 3 samples were all FGA group D and displayed losses of 3p, 5q, and 13q (including the RB1 region), and gains of 4p, 4q, and 5p and frequent TP53 mutation (83.3%). Metastasis occurred in 6 tumors from each cluster and overall survival did not differ significantly between clusters.
Patients with T1G3 tumors are particularly difficult to manage as the risk of progression to muscle-invasion is high and prognostic biomarkers are lacking. Such tumors have been underrepresented in the majority of previous genomic studies. TP53 and FGFR3 mutations were independently distributed in this subset (FGFR3, 45%; TP53, 37%), with the combined mutation distribution being FGFR3wt/TP53wt (33%), FGFR3mut/TP53wt (31%), FGFR3wt/TP53mut (22%), and FGFR3mut/TP53mut (14%). One-way unsupervised hierarchical cluster analysis revealed 3 primary clusters and a 4th smaller cluster consisting of 3 highly chromosomally unstable samples (Fig. 4B). We compared copy number alterations and TP53/FGFR3 mutation frequency in these clusters to those in Ta and ≥T2 tumors (Supplementary Table S2; Fig. 4D). FGFR3 and p53 protein expression was examined in 38 of the 49 T1G3 tumors (data not shown). As in Ta tumors, Cluster 1 samples had frequent FGFR3 mutation (with increased FGFR3 protein expression) and low median FGA (12%). Clusters 3 and 4 resembled ≥T2 Cluster 3, with frequent TP53 mutation, gain of 8q and high median FGA (Cluster 3, 25%; Cluster 4, 69%). Three tumors in Cluster 3 and 2 in Cluster 4 showed p53 overexpression, associated with TP53 mutation. No samples in Cluster 3 were FGFR3 mutant, and there was infrequent chromosome 9 loss. Cluster 2 tumors had 24% median FGA, many were TP53 mutant, only 4 were FGFR3 mutant and gain of 8q was less frequent than in Cluster 3. On the basis of FGA and FGFR3/TP53 mutation frequency, Cluster 2 more closely resembled ≥T2 Clusters 1 and 2.
Four of 5 T1G3 tumors that metastasized during the course of the study were in Cluster 2. Three other patients with initial T1G3 tumors showed progression to stage T2 or more. Two of these were from Cluster 2 and 1 from Cluster 1. Clinico-pathologic features were examined, including size, recurrence, multifocality, and the presence of CIS (where recorded) but were not defining features of the clusters or of samples that progressed or metastasized. One or more high-level amplifications were detected in 19%, 54%, 71%, and 100% of tumors from Clusters 1, 2, 3, and 4, respectively, and were detected in 5 of 8 tumors that progressed or metastasized. A copy number signature for Cluster 2 could not be defined as many copy number alterations were shared between clusters. However, copy number frequency plots of the 3 primary clusters showed an overall prevalence of losses in this cluster (Fig. 4D).
For each tumor group (Ta, T1G3, ≥T2), we determined the average numbers of clones reporting gain or loss, and the numbers of gain and loss events for tumors within individual clusters (Supplementary Table S3; Supplementary Fig. S6). Of the 3 primary T1G3 clusters, Cluster 2 showed the highest clone loss:gain ratio (3.9:1) and this prevalence of losses was also evident when separate events were assessed. Ta Cluster 2a also showed a high clone loss: gain ratio (2.7:1). Several other clusters showed an excess of losses, in most cases due to loss of all or most of chromosome 9.
In samples that metastasized, we examined copy number alterations and mutation in detail. All 4 tumors from T1G3 Cluster 2 had loss of distal 2q, chromosome 9, and 13q including RB1. Three also had loss of 10q and the other had TSC1 mutation. In addition, 3 were TP53 mutant and 1 had MDM2 amplification. The Cluster 1 sample that metastasized had HD of PTEN, MDM2 amplification and loss of RB1.
Discussion
The clinical and pathologic heterogeneity of bladder cancer presents a challenge in disease management. It is likely that combined molecular and histopathologic classification will provide optimal tools for informed clinical decision making. Here we have identified novel regions of copy number alteration, elucidated relationships between molecular alterations and clinico-pathologic features, uncovered relationships between molecular events, and revealed novel genomic subgroups.
Previous studies have identified 2 groups of UC on the basis of chromosomal alterations (3, 4). Here, we confirmed the distinction between low-grade Ta tumors, with low complexity of chromosomal changes, frequent FGFR3 mutation and infrequent TP53 mutation, and MI tumors, with more complex chromosomal changes, infrequent FGFR3 mutation, and frequent TP53 mutation. Hierarchical cluster analysis of copy number events in the entire series generated 3 main clusters primarily according to stage and grade. We then assessed the complexity of chromosomal changes within these diagnostic histopathologic groups to identify additional features that improve biologic understanding or have clinical utility.
One cluster of Ta tumors had strikingly few copy number alterations (<1% FGA). These were characterized by point mutations, including 74% in FGFR3. Almost certainly, further heritable events remain to be identified in this group. Some Ta tumors showed considerable complexity of chromosomal changes. Subcluster (2a) was characterized by no chromosome 9 loss, a common event in other clusters, and these showed more widespread changes than Cluster 2b and represent an interesting group for further analysis. Cluster 3 contained the majority of grade 3 tumors, with high FGA and similar alterations to T2 tumors. T2 tumors also comprised 3 clusters, all with high chromosomal complexity. It was reported that MI tumors with higher FGA had poorer prognosis (12), but this was not clear here. Although we identified no association between cluster assignment and disease recurrence in Ta or metastasis and survival in T2 groups, numbers in the final clusters are relatively small. Analysis of much larger numbers is needed to assess this and to define subgroup signatures.
T1G3 tumors represent an intermediate and heterogeneous group. This is a clinically challenging group, in which the decision to give conservative local treatment with close surveillance, or radical cystectomy, is currently made in the absence of validated prognostic biomarkers. The 49 tumors analyzed here represent the largest series analyzed for mutation and copy number alterations. Previously a large series of T1G3 tumors was assessed for FGFR3 and TP53 mutation. Unlike Ta tumors in which these mutations are virtually mutually exclusive, mutations were independently distributed (34). We confirmed this independent distribution, although here the overall frequency of FGFR3 mutation was higher and TP53 frequency lower. This indicates the existence of distinct T1G3 subgroups.
In support of this, we defined 3 major T1G3 clusters and a 4th smaller cluster that differed with respect to both copy number events and FGFR3 and TP53 mutation status. Cluster 1 had frequent FGFR3 mutation (69.2%), with few alterations apart from losses of 17p and chromosome 9, and gains of 1q, 7, and 15. Cluster 3 were FGFR3 wild type and mostly TP53 mutant (71%), with more complex chromosomal changes but strikingly low frequency of chromosome 9 loss. Cluster 4 comprised 3 highly chromosomally unstable samples. Cluster 2 tumors contained fewer alterations than Cluster 3 but showed the highest rate of stage progression/metastasis. Interestingly, these had a prevalence of copy number losses rather than gains. A recent conventional CGH study of 67 Ta and T1 UC, including 21 T1G3 tumors, defined 3 groups with even distribution of the T1G3 cases, and found that those with more losses had worst outcome (15). Similarly, an analysis of lung cancer showed that copy number loss was associated with reduced survival (35). Such profiles may reflect inactivation of multiple tumor suppressor genes. Because of small numbers of patients with adverse outcome, we cannot define a progression classifier, but some combinations of genomic events were apparent in samples that progressed, including p53 pathway alterations (TP53 mutation or MDM2 amplification), RB1 loss, and losses of 2q, 9q, and 10q (PTEN).
The distinct characteristics of these T1G3 subgroups and the independent distribution of events shared by low-grade Ta or MI UC may indicate either different developmental pathways for one or more of these T1G3 groups, or an origin in one or other of the 2 major pathogenesis pathways, e.g., as CIS or a low-grade Ta tumor. Deep resequencing or CGH analysis of single cells or small regions of these tumors may reveal heterogeneity that will allow pathogenesis pathways to be inferred.
Events affecting chromosome 9 showed significant complexity. Candidate tumor suppressor genes at 9p21 (CDKN2A, CDKN2B), 9q22 (PTCH), 9q33 (DBC1), and 9q34 (TSC1) have been identified previously. We found 9p21.3 HD, which did not include CDKN2A, implicating another target in this region. Candidate genes include IFN-α genes, INFE, KLHL9, and miR-31. Homozygous codeletion of miR-31 and CDKN2A has been described (36) and reduced expression is associated with invasion (37). Our finding of HD of miR-31 alone indicates that it may be an independent target. ELAVL2 is the only gene in the HD region proximal to CDKN2A. ELAVL2 (HuB; HelN1) is a member of the Hu family of RNA-binding proteins implicated in post-transcriptional regulation and differentiation. Several putative mRNA targets of Hu proteins have been identified including the cyclin-dependent kinase inhibitor p21 (38). HD of ELAVL2 has been reported in other cancers (39, 40) and mutation in glioblastoma (40). We also detected HD at 9p24.2-p24.3 and 9p22.2-p22.3. HD at 9p24 and 9p23 were detected previously in UC (14). We also found HD of DAPK1, hypermethylation of which has been reported in UC previously (41). Whole genome sequencing should clarify the status of all chromosome 9 genes. Indeed, exome sequencing of 9 invasive UC, detected mutations in several genes on chromosome 9 including DOCK8, which showed HD here (42).
We detected amplification at 9p24.3 and 9p23-p24.3 and regions of gain at 9p13.3-p21 and 9q21.12. Amplifications at 9p13.3 and 9p22.3-p24.3 are reported in carcinomas of tongue and larynx (43) and in glioma, with GLIS3 implicated as a 9p23 candidate gene (44). Amplification of 9p21.3-pter in seminomas is associated with overexpression of DNMT1 (45). Our observations suggest that copy number increases on chromosome 9 should be further elucidated in UC.
The observed FGFR3 mutation distribution is compatible with previous studies suggesting that this is a key event in non-invasive tumors with good prognosis (46) that are chromosomally stable (18). An inverse relationship between FGFR3 mutation status and gains of chromosome 6, 8q, and 11q in Ta tumors was reported (18) and gains of 8q11.23-q24.21 in FGFR3 wild-type tumors (47). Here, FGFR3 mutation and overrepresentation of 8q were mutually exclusive in all non-MI tumors and in T1G3 tumors. Several tumors had high-level amplification at 8q22.2-q22.3, which has been associated with more aggressive UC phenotype (e.g., 12, 14, 17). On the basis of current and published data (14) YWHAZ (14-3-3-zeta) is a good candidate gene and potential therapeutic target. Compatible with this, amplification and overexpression contributes to chemotherapy resistance and recurrence of breast cancer (48). The 14-3-3 protein family regulate many pathways in normal and cancer cells by interacting with key target proteins. Indeed, the binding and sequestration of proapoptotic proteins by YWHAZ has been implicated in the development of castration-resistant prostate cancer (49).
We found a subgroup of ≥T2 tumors with TP53 mutation and concomitant 16p13.2-p13.3 deletion. 16p LOH and deletion in UC is associated with high grade and progression (10). Although USP7 and GRIN2A appeared good 16p candidates, we found no evidence for their inactivation. However, as several deletions involved larger regions or the whole of 16p, more than one candidate region cannot be discounted. For example, TSC2 is outside the minimal region and mutations were recently detected in UC (50). However, it should be noted that here, copy number losses of 16p that included TSC2 were not mutually exclusive with alterations involving TSC1.
In MI tumors, we found association between metastasis and 10q loss, including PTEN. Previous studies have reported loss of PTEN in invasive UC (28) and conditional knockout mouse models show that Pten loss with p53 or Lkb1 loss leads to invasive and metastatic UC (51, 52). We found no association between LKB1 deletion and metastasis and when both PTEN deletion and TP53 mutation were examined, PTEN copy number loss alone best predicted metastasis.
Our findings indicate that the “gold-standard” grade/stage groups of UC are heterogeneous and contain genetically distinct subgroups. Further studies of larger numbers of tumors will be required to confirm these genomic signatures. This may improve our understanding of UC pathogenesis, provide prognostic biomarkers and reveal novel therapeutic targets.
Supplementary Material
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Translational Relevance.
Heterogeneity in clinical behavior exhibited by bladder tumors of similar grade and stage indicates the existence of molecularly distinct subgroups. Understanding of this is required for rational approaches to clinical management. Here we used a genomic approach to tumor subclassification. Our findings point to the existence of multiple subclasses, independent of the “gold-standard” grade/stage groups. The study included the largest series of T1G3 tumors analyzed for copy number events and mutations to date and identified a subgroup that contained the majority that showed disease progression. Further development of this classification may allow such tumors to be stratified for conservative treatment or cystectomy. We identified novel regions of copy number alteration, relationships between molecular events, and associations with clinico-pathologic features. Some novel regions of copy number alteration may contain potential therapeutic targets. Overall, our findings add information to conventional grading and staging and may allow development of robust prognostic biomarkers.
Acknowledgments
The authors are extremely grateful to Joanne Brown for tissue collection and processing and to Filomena Esteves for immunohistochemistry.
Grant Support
This work was supported by grants from Cancer Research UK (C6228/A5433; C6228/A12512; C37059/A11941).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Disclosure of Potential Confiicts of Interest
No potential conflicts of interest were disclosed.
Authors' Contributions
Conception and design: C.D. Hurst, F. Platt, M.A. Knowles
Development of methodology: C.D. Hurst, F. Platt
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): C.D. Hurst, F. Platt, C.F. Taylor
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): C.D. Hurst, F. Platt, C.F. Taylor, M.A. Knowles
Writing, review, and/or revision of the manuscript: C.D. Hurst, F. Platt, C.F. Taylor, M.A. Knowles
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): C.D. Hurst, F. Platt, C.F. Taylor
Study supervision: M.A. Knowles
References
- 1.Montie JE, Clark PE, Eisenberger MA, El-Galley R, Greenberg RE, Herr HW, et al. Bladder cancer. J Natl Compr Canc Netw. 2009;7:8–39. doi: 10.6004/jnccn.2009.0002. [DOI] [PubMed] [Google Scholar]
- 2.López-Knowles E, Hernández S, Malats N, Kogevinas M, Lloreta J, Carrato A, et al. PIK3CA mutations are an early genetic alteration associated with FGFR3 mutations in superficial papillary bladder tumors. Cancer Res. 2006;66:7401–4. doi: 10.1158/0008-5472.CAN-06-1182. [DOI] [PubMed] [Google Scholar]
- 3.Wu XR. Urothelial tumorigenesis: a tale of divergent pathways. Nat Rev Cancer. 2005;5:713–25. doi: 10.1038/nrc1697. [DOI] [PubMed] [Google Scholar]
- 4.Knowles MA. Molecular subtypes of bladder cancer: Jekyll and Hyde or chalk and cheese? Carcinogenesis. 2006;27:361–73. doi: 10.1093/carcin/bgi310. [DOI] [PubMed] [Google Scholar]
- 5.Catto JW, Alcaraz A, Bjartell AS, De Vere White R, Evans CP, Fussel S, et al. MicroRNA in prostate, bladder, and kidney cancer: a systematic review. Eur Urol. 2011;59:671–81. doi: 10.1016/j.eururo.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 6.Sjodahl G, Lauss M, Lovgren K, Chebil G, Gudjonsson S, Veerla S, et al. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res. 2012;18:3377–86. doi: 10.1158/1078-0432.CCR-12-0077-T. [DOI] [PubMed] [Google Scholar]
- 7.Wolff EM, Chihara Y, Pan F, Weisenberger DJ, Siegmund KD, Sugano K, et al. Unique DNA methylation patterns distinguish noninvasive and invasive urothelial cancers and establish an epigenetic field defect in premalignant tissue. Cancer Res. 2010;70:8169–78. doi: 10.1158/0008-5472.CAN-10-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reinert T, Modin C, Castano FM, Lamy P, Wojdacz TK, Hansen LL, et al. Comprehensive genome methylation analysis in bladder cancer: identification and validation of novel methylated genes and application of these as urinary tumor markers. Clin Cancer Res. 2011;17:5582–92. doi: 10.1158/1078-0432.CCR-10-2659. [DOI] [PubMed] [Google Scholar]
- 9.Dyrskjot L, Zieger K, Real FX, Malats N, Carrato A, Hurst C, et al. Gene expression signatures predict outcome in non-muscle-invasive bladder carcinoma: a multicenter validation study. Clin Cancer Res. 2007;13:3545–51. doi: 10.1158/1078-0432.CCR-06-2940. [DOI] [PubMed] [Google Scholar]
- 10.Hoque MO, Lee CC, Cairns P, Schoenberg M, Sidransky D. Genome-wide genetic characterization of bladder cancer: a comparison of high-density single-nucleotide polymorphism arrays and PCR-based microsatellite analysis. Cancer Res. 2003;63:2216–22. [PubMed] [Google Scholar]
- 11.Primdahl H, Wikman FP, von der Maase H, Zhou XG, Wolf H, Orntoft TF. Allelic imbalances in human bladder cancer: genome-wide detection with high-density single-nucleotide polymorphism arrays. J Natl Cancer Inst. 2002;94:216–23. doi: 10.1093/jnci/94.3.216. [DOI] [PubMed] [Google Scholar]
- 12.Blaveri E, Brewer JL, Roydasgupta R, Fridlyand J, DeVries S, Koppie T, et al. Bladder cancer stage and outcome by array-based comparative genomic hybridization. Clin Cancer Res. 2005;11:7012. doi: 10.1158/1078-0432.CCR-05-0177. [DOI] [PubMed] [Google Scholar]
- 13.Koed K, Wiuf C, Christensen LL, Wikman FP, Zieger K, Moller K, et al. High-density single nucleotide polymorphism array defines novel stage and location-dependent allelic imbalances in human bladder tumors. Cancer Res. 2005;65:34–45. [PubMed] [Google Scholar]
- 14.Heidenblad M, Lindgren D, Jonson T, Liedberg F, Veerla S, Chebil G, et al. Tiling resolution array CGH and high density expression profiling of urothelial carcinomas delineate genomic amplicons and candidate target genes specific for advanced tumors. BMC Med Genomics. 2008;1:3. doi: 10.1186/1755-8794-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prat E, del Rey J, Ponsa I, Nadal M, Camps J, Plaja A, et al. Comparative genomic hybridization analysis reveals new different subgroups in early-stage bladder tumors. Urology. 2010;75:347–55. doi: 10.1016/j.urology.2009.04.080. [DOI] [PubMed] [Google Scholar]
- 16.Hurst CD, Fiegler H, Carr P, Williams S, Carter NP, Knowles MA. High-resolution analysis of genomic copy number alterations in bladder cancer by microarray-based comparative genomic hybridization. Oncogene. 2004;23:2250–63. doi: 10.1038/sj.onc.1207260. [DOI] [PubMed] [Google Scholar]
- 17.Veltman JA, Fridlyand J, Pejavar S, Olshen AB, Korkola JE, DeVries S, et al. Array-based comparative genomic hybridization for genome-wide screening of DNA copy number in bladder tumors. Cancer Res. 2003;63:2872–80. [PubMed] [Google Scholar]
- 18.Junker K, van Oers JMM, Zwarthoff EC, Kania I, Schubert J, Hartmann A. Fibroblast growth factor receptor 3 mutations in bladder tumors correlate with low frequency of chromosome alterations. Neoplasia. 2008;10:1–7. doi: 10.1593/neo.07178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zieger K, Marcussen N, Borre M, Ørntoft TF, Dyrskjøt L. Consistent genomic alterations in carcinoma in situ of the urinary bladder confirm the presence of two major pathways in bladder cancer development. Int J Cancer. 2009;125:2095–103. doi: 10.1002/ijc.24619. [DOI] [PubMed] [Google Scholar]
- 20.Nord H, Segersten U, Sandgren J, Wester K, Busch C, Menzel U, et al. Focal amplifications are associated with high grade and recurrences in stage Ta bladder carcinoma. Int J Cancer. 2010;126:1390–402. doi: 10.1002/ijc.24954. [DOI] [PubMed] [Google Scholar]
- 21.Nishiyama N, Arai E, Nagashio R, Fujimoto H, Hosoda F, Shibata T, et al. Copy number alterations in urothelial carcinomas: their clinicopathological significance and correlation with DNA methylation alterations. Carcinogenesis. 2011;32:462–9. doi: 10.1093/carcin/bgq274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindgren D, Frigyesi A, Gudjonsson S, Sjodahl G, Hallden C, Chebil G, et al. Combined gene expression and genomic profiling define two intrinsic molecular subtypes of urothelial carcinoma and gene signatures for molecular grading and outcome. Cancer Res. 2010;70:3463–72. doi: 10.1158/0008-5472.CAN-09-4213. [DOI] [PubMed] [Google Scholar]
- 23.Lindgren D, Sjodahl G, Lauss M, Staaf J, Chebil G, Lovgren K, et al. Integrated genomic and gene expression profiling identifies two major genomic circuits in urothelial carcinoma. PLoS One. 2012;7:e38863. doi: 10.1371/journal.pone.0038863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.WHO. Histological typing of urinary bladder tumours. Int Histol Class Tumours. 1973;10 [Google Scholar]
- 25.UICC. TNM classification of malignant tumors, bladder. 3rd ed. Geneva: Union Internationale Contre le Cancer; 1978. pp. 113–7. [Google Scholar]
- 26.Fiegler H, Carr P, Douglas EJ, Burford DC, Hunt S, Smith J, et al. DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer. 2003;36:361–74. doi: 10.1002/gcc.10155. [DOI] [PubMed] [Google Scholar]
- 27.Jong K, Marchiori E, Meijer G, Vaart AV, Ylstra B. Breakpoint identification and smoothing of array comparative genomic hybridization data. Bioinformatics. 2004;20:3636–7. doi: 10.1093/bioinformatics/bth355. [DOI] [PubMed] [Google Scholar]
- 28.Platt FM, Hurst CD, Taylor CF, Gregory WM, Harnden P, Knowles MA. Spectrum of phosphatidylinositol 3-kinase pathway gene alterations in bladder cancer. Clin Cancer Res. 2009;15:6008–17. doi: 10.1158/1078-0432.CCR-09-0898. [DOI] [PubMed] [Google Scholar]
- 29.Askham JM, Platt F, Chambers PA, Snowden H, Taylor CF, Knowles MA. AKT1 mutations in bladder cancer: identification of a novel oncogenic mutation that can co-operate with E17K. Oncogene. 2010;29:150–5. doi: 10.1038/onc.2009.315. [DOI] [PubMed] [Google Scholar]
- 30.Southgate J, Hutton KA, Thomas DF, Trejdosiewicz LK. Normal human urothelial cells in vitro: proliferation and induction of stratification. Lab Invest. 1994;71:583–94. [PubMed] [Google Scholar]
- 31.Sylvester RJ, van der Meijden AP, Oosterlinck W, Witjes JA, Bouffioux C, Denis L, et al. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: a combined analysis of 2596 patients from seven EORTC trials. Eur Urol. 2006;49:466–5. doi: 10.1016/j.eururo.2005.12.031. discussion 475–7. [DOI] [PubMed] [Google Scholar]
- 32.Wei X, Walia V, Lin JC, Teer JK, Prickett TD, Gartner J, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43:442–6. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim MS, Lebron C, Nagpal JK, Chae YK, Chang X, Huang Y, et al. Methylation of the DFNA5 increases risk of lymph node metastasis in human breast cancer. Biochem Biophys Res Commun. 2008;370:38–43. doi: 10.1016/j.bbrc.2008.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hernandez S, Lopez-Knowles E, Lloreta J, Kogevinas M, Jaramillo R, Amoros A, et al. FGFR3 and Tp53 mutations in T1G3 transitional bladder carcinomas: independent distribution and lack of association with prognosis. Clin Cancer Res. 2005;11:5444. doi: 10.1158/1078-0432.CCR-05-0122. [DOI] [PubMed] [Google Scholar]
- 35.Belvedere O, Berri S, Chalkley R, Conway C, Barbone F, Pisa F, et al. A computational index derived from whole-genome copy number analysis is a novel tool for prognosis in early stage lung squamous cell carcinoma. Genomics. 2012;99:18–24. doi: 10.1016/j.ygeno.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 36.Veerla S, Lindgren D, Kvist A, Frigyesi A, Staaf J, Persson H, et al. MiRNA expression in urothelial carcinomas: important roles of miR-10a, miR-222, miR-125b, miR-7 and miR-452 for tumor stage and metastasis, and frequent homozygous losses of miR-31. Int J Cancer. 2009;124:2236–42. doi: 10.1002/ijc.24183. [DOI] [PubMed] [Google Scholar]
- 37.Wszolek MF, Rieger-Christ KM, Kenney PA, Gould JJ, Silva Neto B, Lavoie AK, et al. A MicroRNA expression profile defining the invasive bladder tumor phenotype. Urol Oncol. 2011;29:794–801. e1. doi: 10.1016/j.urolonc.2009.08.024. [DOI] [PubMed] [Google Scholar]
- 38.Yano M, Okano HJ, Okano H. Involvement of Hu and heterogeneous nuclear ribonucleoprotein K in neuronal differentiation through p21 mRNA post-transcriptional regulation. J Biol Chem. 2005;280:12690–9. doi: 10.1074/jbc.M411119200. [DOI] [PubMed] [Google Scholar]
- 39.Cairns P, Okami K, King P, Bonacum J, Ahrendt S, Wu L, et al. Genomic organization and mutation analysis of Hel-N1 in lung cancers with chromosome 9p21 deletions. Cancer Res. 1997;57:5356–9. [PubMed] [Google Scholar]
- 40.Wong KK, Tsang YT, Chang YM, Su J, Di Francesco AM, Meco D, et al. Genome-wide allelic imbalance analysis of pediatric gliomas by single nucleotide polymorphic allele array. Cancer Res. 2006;66:11172–8. doi: 10.1158/0008-5472.CAN-06-2438. [DOI] [PubMed] [Google Scholar]
- 41.Tada Y, Wada M, Taguchi K, Mochida Y, Kinugawa N, Tsuneyoshi M, et al. The association of death-associated protein kinase hypermethylation with early recurrence in superficial bladder cancers. Cancer Res. 2002;62:4048–53. [PubMed] [Google Scholar]
- 42.Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–8. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jarvinen AK, Autio R, Kilpinen S, Saarela M, Leivo I, Grenman R, et al. High-resolution copy number and gene expression microarray analyses of head and neck squamous cell carcinoma cell lines of tongue and larynx. Genes Chromosomes Cancer. 2008;47:500–9. doi: 10.1002/gcc.20551. [DOI] [PubMed] [Google Scholar]
- 44.Cooper LA, Gutman DA, Long Q, Johnson BA, Cholleti SR, Kurc T, et al. The proneural molecular signature is enriched in oligodendrogliomas and predicts improved survival among diffuse gliomas. PLoS One. 2010;5:e12548. doi: 10.1371/journal.pone.0012548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Looijenga LH, Hersmus R, Gillis AJ, Pfundt R, Stoop HJ, van Gurp RJ, et al. Genomic and expression profiling of human spermatocytic seminomas: primary spermatocyte as tumorigenic precursor and DMRT1 as candidate chromosome 9 gene. Cancer Res. 2006;66:290–302. doi: 10.1158/0008-5472.CAN-05-2936. [DOI] [PubMed] [Google Scholar]
- 46.Billerey C, Chopin D, Aubriot-Lorton MH, Ricol D, Gil Diez de Medina S, Van Rhijn B, et al. Frequent FGFR3 mutations in papillary non-invasive bladder (pTa) tumors. Am J Pathol. 2001;158:1955–9. doi: 10.1016/S0002-9440(10)64665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zieger K, Dyrskjot L, Wiuf C, Jensen JL, Andersen CL, Jensen KM, et al. Role of activating fibroblast growth factor receptor 3 mutations in the development of bladder tumors. Clin Cancer Res. 2005;11:7709–19. doi: 10.1158/1078-0432.CCR-05-1130. [DOI] [PubMed] [Google Scholar]
- 48.Li Y, Zou L, Li Q, Haibe-Kains B, Tian R, Desmedt C, et al. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat Med. 2010;16:214–8. doi: 10.1038/nm.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zoubeidi A, Zardan A, Wiedmann RM, Locke J, Beraldi E, Fazli L, et al. Hsp27 promotes insulin-like growth factor-I survival signaling in prostate cancer via p90Rsk-dependent phosphorylation and inactivation of BAD. Cancer Res. 2010;70:2307–17. doi: 10.1158/0008-5472.CAN-09-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sjodahl G, Lauss M, Gudjonsson S, Liedberg F, Hallden C, Chebil G, et al. A systematic study of gene mutations in urothelial carcinoma; inactivating mutations in TSC2 and PIK3R1. PLoS One. 2011;6:e18583. doi: 10.1371/journal.pone.0018583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Puzio-Kuter AM, Castillo-Martin M, Kinkade CW, Wang X, Shen TH, Matos T, et al. Inactivation of p53 and Pten promotes invasive bladder cancer. Genes Dev. 2009;23:675–80. doi: 10.1101/gad.1772909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shorning BY, Griffiths D, Clarke AR. Lkb1 and Pten synergise to suppress mTOR-mediated tumorigenesis and epithelial-mesenchymal transition in the mouse bladder. PLoS One. 2011;6:e16209. doi: 10.1371/journal.pone.0016209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.