Abstract
Introduction
Outbreaks of the zoonotic disease tularemia occurred in north-east Bulgaria in the 1960s. Then came 30 years of epidemiological silence until new outbreaks occurred in west Bulgaria in the 1990s. To investigate how bacterial strains of Francisella tularensis causing tularemia in wildlife and humans in the 1960s and the 1990s were related, we explored their genetic diversity.
Material and methods
Ten F. tularensis genomes from the 1960s (n=3) and the 1990s (n=7) were sequenced, assigned to canonical single-nucleotide polymorphism (canSNP) clades, and compared to reference genomes. We developed four new canSNP polymerase chain reaction (PCR) assays based on the genome sequence information.
Results and discussion
The genetic analysis showed that the outbreaks in the 1960s as well as in the 1990s involved multiple clones and new genetic diversity. The smallest genetic difference found between any of the Bulgarian strains was five SNPs between the strains L2 and 81 isolated 43 years apart, indicating that F. tularensis may persist locally over long time periods without causing outbreaks. The existence of genetically highly similar strain-pairs isolated the same year in the same area from different hosts supports a hypothesis of local expansion of clones during outbreaks. Close relationship (two SNPs) was found between one strain isolated 1961 in northeast Bulgaria and one strain isolated 5 years before in USSR. Historical data coinciding with the actual time point describe the introduction of water rats from USSR into the Bulgarian outbreak area, which may explain the close genetic relationship and the origin of the outbreak.
Conclusion
Genome analysis of strains from two outbreaks in the 1960s and the 1990s provided valuable information on the genetic diversity and persistence of F. tularensis in Bulgaria.
Keywords: Francisella, genome, sequencing, Illumina, outbreak, phylogeography, subtyping, canSNP, assay, zoonosis
The zoonosis tularemia often presents as recurrent seasonal disease outbreaks in geographically restricted areas and is reported to be emerging in Europe (1, 2). Disease in humans is associated with exposure to Francisella tularensis (F. tularensis)-infected arthropods, animals, food, water, fomites, or aerosols.
Tularemia is one of the diseases that historically adhere to the ‘natural nidality of disease’ concept formulated in 1939 by the Russian parasitologist Yevgeny Pavlovsky (3). The nidality concept bears many resemblances with the modern One Health concept adopted by the west in the first decade of the 2000s (4). In essence, the ‘natural nidality of disease’ concept postulates that diseases occur naturally in wildlife in certain places (nidus) over time and may be transmitted from wildlife to humans by arthropod vectors in regions with certain landscape features (3).
The advent of modern high-resolution typing methods of pathogens now allows for further investigating the natural nidality concept. Whole genome sequence comparisons and canonical single-nucleotide polymorphism (canSNP) genotyping have provided insights into the genetic diversity of F. tularensis subsp. holarctica (5, 6). Because of the genetic homogeneity of F. tularensis strains, whole genome sequencing is the most appropriate typing method. Previous work has shown the utility of genome sequencing for assigning strains into robust genetic groups, and that canSNP assays subsequently can be used for identifying these groups (5–7).
In 1961–1969, a prolonged tularemia outbreak occurred in the Srebarna lake reserve in Bulgaria, where an epizootic among muskrats was followed by human tularemia cases (Supplementary file 1). Then, a three-decade period of epidemiological silence followed. It was not until November 1997 that a new tularemia focus appeared and new F. tularensis strains were isolated in Bulgaria. Recent genomic epidemiological studies of tularemia in Sweden and Norway have revealed strains that are almost 100% identical at the genome level despite being isolated many years apart and/or separated by several hundreds of km (8, 9). In light of these findings, we asked if the clone/clones that caused the recent epidemics in Bulgaria were the same as those that occurred in the early 1960s. We analyzed 10 genome sequences from both outbreaks to investigate this and to place Bulgarian F. tularensis isolates in a global phylogenetic context.
Materials and methods
Tularemia outbreaks
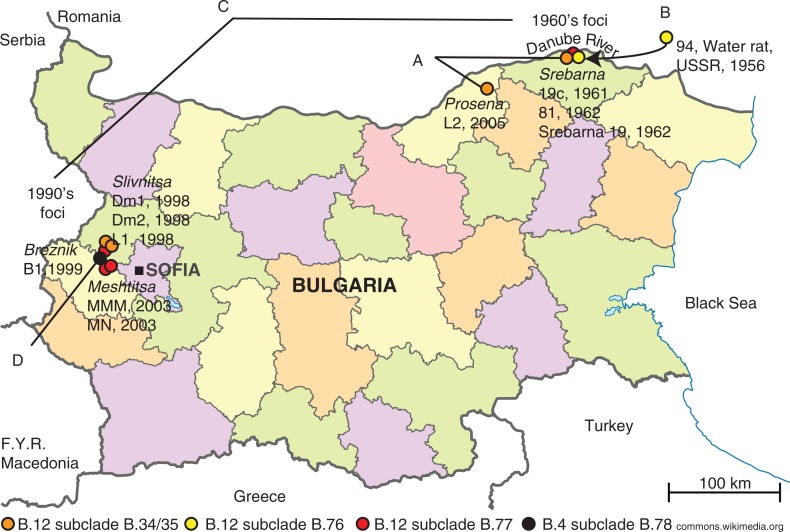
The old 1960s tularemia focus was in the Srebarna lake nature reserve and its surroundings (Fig. 1 and Supplementary file 1). The focus covered an area of about 7 km2 where the marsh lake abuts with the Danube River. The new 1990s focus covered an area of 1,000–1,500 km2 situated on 500–800 m altitude adjacent to mountains and the capital of Sofia (Fig. 1 and Supplementary file 1).
Fig. 1.
The old and new tularemia foci in Bulgaria. A. The genetic distance between strains L2 and 81 (orange dots) comprised five SNPs in 33 years. B. The genetic distance between strain 94 from USSR and strain 19c from Srebarna (yellow dots) comprised two SNPs in 5 years. C. There was an accumulation of >20 SNPs between the Srebarna 19 from the old focus and L1, MN, and MMM in the new focus (red dots). D. The first Bulgarian genome in the B.4 basal clade was represented by strain B1 isolated in Breznik in 1999 (black dot).
Tularemia outbreak strains
Three strains from the 1960s outbreak in Srebarna lake reserve in northeast Bulgaria and seven strains from the new 1990s focus in west Bulgaria were genome sequenced (Table 1 and Fig. 1). In addition, two strains recovered in the old focus in the 1960s were canSNP typed. The 1960s strains had been lyophilized after their isolation and kept at room temperature until culturing for genome sequencing in 2011. The isolation and storage of the seven strains from the 1990s outbreaks have been previously described (10). One archive strain isolated 1956 in USSR was sequenced for comparison purposes. For a more detailed epidemiology description, see Supplementary file 1 (10–16).
Table 1.
Francisella tularensis strains investigated in the study
| Original ID | Collection ID | Whole genome | Source | Year of isolation | Place of isolation | Fig. 3 Basal clade (subclade) |
|---|---|---|---|---|---|---|
| L1a,b | FDC179 | Yes | Hare | 1998 | Slivnitsa | B.12 (B.77) |
| Dm1a | FDC180 | Yes | Tick, Dermacentor marginatus | 1998 | Slivnitsa | B.12 (B.34/35)c |
| Dm2a | FDC181 | Yes | Tick, D. marginatus | 1998 | Slivnitsa | B.12 (B.34/35)c |
| B1a | FDC182 | Yes | Tick, Ixodes ricinus, collected from goat | 1999 | Breznik | B.4 (B.78) |
| MMMa | FDC183 | Yes | Common vole | 2003 | Meshtitsa | B.12 (B.77) |
| MNa | FDC184 | Yes | Human | 2003 | Meshtitsa | B.12 (B.77) |
| L2a,b | FDC185 | Yes | Hare | 2005 | Prosena | B.12 (B.34/35)c |
| Srebarna 19a | FDC186 | Yes | Muskrat | 1962 | Srebarna | B.12 (B.77) |
| Srebarna 5a | No | 1962 | Srebarna | B.12 (B.23)d | ||
| Srebarna 56a | No | 1962 | Srebarna | B.12 (B.23)d | ||
| 19ce | FSC930 | Yes | Muskrat | 1961 | Srebarnaf | B.12 (B.76) |
| 81e | FSC937 | Yes | Water | 1962 | Srebarnaf | B.12 (B.34/B.35) |
| 94 or N94 | FSC931 | Yes | Water rat | 1956 | USSR | B.12 (B.76) |
The strains are from the strain collection of the Military Medical Academy (MMA), Sofia, Bulgaria. The strains Srebarna 5, Srebarna 19, and Srebarna 56 were recovered by an MMA team (Gotev/Kupenov) and originate from the Bulgarian Type Culture Collection (BTCC) in Sofia
Note that the strain L1 is not identical to a strain also named L1 in the reference (16)
B.34/35 should be read as that the strain showed the derived SNP state for B.34 and the ancestral state for B.35
No further subtyping with B.77 was performed on these two strains due to insufficient amount of DNA (Fig. 3)
The strains 19c, 81, and 94 are part of a strain collection maintained at the Military Institute of Hygiene and Epidemiology (MIHE) in Warsaw, Poland, and originate from a strain collection at the former Institute of Marine Medicine in Gdansk, Poland (21). Strains 19c and 94 originated from the Medical Academy, Moscow, USSR and strain 81 originated from Centre of Epidemiology and Microbiology in Prague, former Czechoslovakia
The original documentation reads Bulgaria (21). No other outbreaks than the Srebarna outbreak nor other areas of strains isolation are described in the literature, institutional records, or known by personal communication to our knowledge (Marinov, personal communication).
DNA preparation
The strains Srebarna 5, Srebarna 19, Srebarna 56, L1, L2, Dm1, Dm2, B1, MMM, and MN were cultured as previously described at the Military Medical Academy (MMA) in Sofia, Bulgaria (10). Lysis of the bacteria at MMA was achieved by suspending the samples in 1.0 ml 0.9% NaCl and heat-treated at 65°C for 2 h.
The strains 19c, 81, and 94 were re-cultured at Military Institute of Hygiene and Epidemiology (MIHE) in Pulawy, Poland, and sent to the Swedish Defence Research Agency (FOI) in Umeå. Prior to DNA preparation, strains 19c, 81, and 94 were re-cultured at FOI on GC II agar with 1% hemoglobin and 1% IsoVitaleX at 37°C (17)
DNA from L1, L2, Dm1, B1, MN, 19c, 81, and 94 was prepared at FOI using phenol-chloroform extraction, as previously described (18), while DNA from Dm2, MMM, Srebarna 5, Srebarna 19, and Srebarna 56 was prepared using the EZ1 robot preparation method, as previously described (19). DNA concentrations were measured using the NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and the Qubit (Life Technologies, Carlsbad, CA, USA). The amount of DNA prepared from Srebarna 5 and Srebarna 56 stock samples was insufficient for DNA sequencing and therefore only genotyped with the canSNP markers B.12 and B.23.
Genome sequencing and phylogenetic analysis
Genome sequencing of isolates was performed at SciLifeLab, Uppsala, Sweden, using the Illumina HiSeq 2000 platform with TruSeq® DNA Library Prep Kits (strain 91 with TruSeq® Nano DNA HT sample prep kit) according to standard protocols to produce 100 bp pair-end data. The resulting sequence data were assembled using ABySS versions 1.3.2 and 1.3.3 using standard parameters (Table 2). Multiple genome alignments were performed using progressive Mauve version 2.3.1. Nucleotide distances and phylogeny were inferred using Mega 5.2 (complete deletion option and the number of differences method, maximum parsimony). The sequence data are available from the NCBI under BioProject accession PRJNA285143.
Table 2.
Genome data for 10 Bulgarian strains and one archive strain sequenced in this study
| Original ID | Total number of bases (>1,000 bp) | Total number of contigs (>1,000 bp) | N50a | Number of reads/pairs | Coverage | Sample accessionb | Sample providerc |
|---|---|---|---|---|---|---|---|
| L1 | 1,886,108 | 95 | 28469 | 26,517,196 | 2812 | SAMN03773843 | MMA |
| Dm1 | 1,883,038 | 91 | 28802 | 25,066,628 | 2662 | SAMN03773844 | MMA |
| Dm2 | 1,881,120 | 93 | 28426 | 8,579,883 | 912 | SAMN03773845 | MMA |
| B1 | 1,897,672 | 93 | 28339 | 22,520,234 | 2373 | SAMN03773846 | MMA |
| MMM | 1,875,643 | 95 | 28022 | 8,821,904 | 941 | SAMN03773847 | MMA |
| MN | 1,884,503 | 94 | 28467 | 34,247,320 | 3635 | SAMN03773848 | MMA |
| L2 | 1,889,172 | 93 | 28557 | 28,616,993 | 3030 | SAMN03773849 | MMA |
| Srebarna 19 | 1,881,962 | 94 | 28470 | 6,532,837 | 694 | SAMN03773850 | MMA |
| 19c | 1,847,256 | 97 | 28076 | 18,541,564 | 2007 | SAMN03774233 | MIHE |
| 81 | 1,832,473 | 93 | 29527 | 24,573,197 | 2682 | SAMN03774238 | MIHE |
| 94 or N94 | 1,818,365 | 97 | 27934 | 7,428,933 | 817 | SAMN03774234 | MIHE |
N50 is the length of the smallest contig in the set that contains the fewest (largest) contigs whose combined length represents at least 50% of the assembly
NCBI BioProject PRJNA285143
MMA=Military Medical Academy, Sofia, Bulgaria; MIHE=Military Institute of Hygiene and Epidemiology, Pulawy, Poland.
Typing and design of four new SNP assays for clades that include Bulgarian isolates
In silico typing of genomes was performed using canSNPer (7). The strains were SNP genotyped with an SYBR® Green-based PCR assay adopted from the protocol developed by Svensson et al. (6). Changes were made from the original protocol to simplify the procedure by reducing the number of components and to increase the discriminatory power of the assays. The PCR mix used by Svensson et al. (6) was exchanged for SYBR® Green PCR mix (Applied Biosystems, Life Technologies Corporation, Carlsbad, CA, USA) that contains all the necessary components except the primers and template. SNP-typing was performed in two separate reactions, differing by the two allele specific forward primers, specifically designed for each marker. Each 10 µl reaction contained 1×SYBR® Green PCR mix (Applied Biosystems, Life Technologies Corporation, Carlsbad, CA, USA), 300 nM of each forward and reverse primer, and 1 ng template. The PCR was initiated by an activation and denaturation step for 10 min at 95°C, followed by 35 cycles of 15 s at 95°C and 60 s at 55°C, and plate reading. Finally, product dissociation (melt analysis) was performed at 65–95°C at 0.2°C increments. The SNP state was determined by comparing the Cq values between the product generated by the ancestor and derived primers, respectively (Supplementary file 2).
The primers and markers used in these assays were as far as possible adopted from previous publications (5, 6, 20) (Table 3). However, intentional mismatches at the penultimate or antepenultimate nucleotide (already present in the primers published by Vogler et al. (5) and Gyuranecz et al. (20)) were introduced in the primers published by Svensson et al. (6).
Table 3.
Four new assays B.76−B.79 for Bulgarian clades. Existing assays using modified primer sequences from previous publications
| canSNP | SNP state | Position SCHU S4a | Primerb | Primer sequence 5’–3’ |
|---|---|---|---|---|
| First order of discrimination | ||||
| B.16c | T | 608,245 | D | gcgggcagggcggcATGCTAGCAAATTACCATCAAAAcT |
| G | A | ATGCTAGCAAATTACCATCAAAAtG | ||
| C | AACTCTTCTCGCCATCAACTTCTAT | |||
| B.4d | T | 823,672 | D | cggggcggggcggggGACTTTTGGGGTTTTTGCTGgT |
| A | A | GACTTTTGGGGTTTTTGCTGgA | ||
| C | GCGATCACCAAAACTACCYATAACCACACTG | |||
| B.6d | A | 713,647 | D | ggggcggggcggggTGTTATTGAATGTAAGGAAAATGATTCcA |
| G | A | tttttttttttttttttttTGTTATTGAATGTAAGGAAAATGATTCaG | ||
| C | GAAATAGTAAGTCCAAACGCATGAAATA | |||
| B.19c | A | 1,373,999 | D | gcgggcTTGCTACTGATGGTTTAAgTA |
| C | A | gcgggcagggcggcTTGCTACTGATGGTTTAAaTC | ||
| C | CAATACGTCACTTATGCAGTGAT | |||
| Second order of discrimination | ||||
| B.10d | G | 387,537 | D | cggggcggggcggggGCCATCACTAGTAAATACCACATTAAaC |
| A | A | TGCCATCACTAGTAAATACCACATTAAgT | ||
| C | TTGTAATATTAGCTMGAAAAGTAGATGAA | |||
| B.11d | A | 1,282,029 | D | cggggcggggcggggTGTGATCAAGGGAAATGCTCAgA |
| G | A | TGTGATCAAGGGAAATGCTCAtG | ||
| C | TATTCTGTTATATTGTTGCGCATAGGCATAC | |||
| B.20c | G | 1,396,082 | D | gcgggcagggcggcTCTGATGAAGAATATCTTACtG |
| A | 1,789,417 | A | gcgggcTCTGATGAAGAATATCTTACcA | |
| C | ATTATGGCAAAACTATACCTT | |||
| B.23c | T | 253,120 | D | gcgggcTTACTACAAATTCGCCTCTgAT |
| G | A | gcgggcagggcggcTTACTACAAATTCGCCTCTtAG | ||
| C | AGCAAAAGAGCTTACTAAACAATTTGA | |||
| Third order of discrimination | ||||
| B.34e | A | 766,614 | D | ggggcggggcggggcTAGCGAGCATTATTTGCTGGgTT |
| G | A | GTAGCGAGCATTATTTGCTGGtTC | ||
| C | ATAAAACTATAAATTTACATAAAATGAAAACTTCTC | |||
| B.76 | T | 1,509,966 | D | ccccgccccgccccgATATCCGAGCATATTCCTAGaT |
| C | A | ATATCCGAGCATATTCCTAGtC | ||
| C | ATGGAGGTAGAGAGTATCTTATAG | |||
| B.77 | A | 1,802,833 | D | cggggcggggcggggTGTTGAAAAGCCAAAAGTTcT |
| G | A | ATGTTGAAAAGCCAAAAGTTtC | ||
| C | GTAAATGCTGCTGCCATC | |||
| B.78 | C | 1,634,094 | Df | cggggcggggcggggcAGTGTGCGTGGTTTCTTgCG |
| T | A | GGAAGTGTGCGTGGTTTCTTtCA | ||
| C | AAAGGTACAGGGAATAATACATCAT | |||
| B.79 | A | 1,661,376 | D | cggggcggggcggggACCTGGGATTCTTGGcGT |
| G | A | gcgggcATACCTGGGATTCTTGGtGC | ||
| C | ggggcggggGAATGGCTGCATATAATATGATAT | |||
SCHU S4 has GenBank accession no. AJ749949.2
D=Derived SNP state primer, A=Ancestral SNP state primer, C=Common primer
Svensson et al. (6). A real-time PCR array for hierarchical identification of Francisella isolates. B.19 was used for identifying branch B.12 (Fig. 2 and Fig. 3)
Vogler et al. (5). Phylogeography of Francisella tularensis: Global expansion of a highly fit clone
Gyuranecz et al. (20) Phylogeography of Francisella tularensis subsp. holarctica, Europe
Primer generates a product with double melt peaks.
Results and discussion
Following a three-decade epidemiological silence, an outbreak of tularemia occurred in 1997 in west Bulgaria, where tularemia had never been registered previously, 300 km from the old outbreak area in northeast Bulgaria (Fig. 1). In the following decade, there were also some sporadic cases close to the old focus. By genome sequencing of 10 strains from Bulgaria, three from the old outbreaks in 1960s and seven strains from the new outbreaks in 1990s, we were able to assign the strains to genetic groups using canSNPs and compare them with reference genomes (Table 1). The two strains Srebarna 5 and Srebarna 56 were subtyped by canSNP typing assays only and were assigned to the genetic subclade B.12 (B.23).
Genetic diversity and persistence of tularemia in Bulgaria
The genetic analysis of the 12 strains showed that the 1960s as well as the 1990s outbreaks of tularemia in Bulgaria involved multiple clones of F. tularensis with five and six members of the two B.12 subclades B.20 and B.23, respectively, and one member of the basal clade B.4 isolated in 1999 (Table 1 and Fig. 2). The relatively large genetic distances between the clones isolated in the old and new foci (Fig. 2 and Fig. 3) argue against the fact that the same clone caused the epidemics in the new focus. The old and new foci are geographically and ecologically separated (Fig. 1). They are divided by the Balkan Mountain, a long and high mountain chain that limits natural movement of vectors, rodents, and hares that could transfer F. tularensis. Thus, neither common ecological factors nor close genetic kinships suggest that there was a transfer of F. tularensis from the old focus to the new focus.
Fig. 2.
Phylogeny. Whole genome phylogeny of F. tularensis subsp. holarctica based on SNPs in an alignment of nine published genomes (strain names are indicated) showing the placement of the Bulgarian strains in a global phylogenetic framework. Bulgarian outbreak strains and the non-outbreak strain 94 sequenced in this study are indicated in bold text. B.4, B.6, B.12, and B.16 indicate the four major canSNP clades of the subspecies. B.20 and B.23 indicate two B.12 subclades that included Bulgarian strains The tree was mid-point rooted in FSC022. The total length of the root branch was 3,317 base differences.
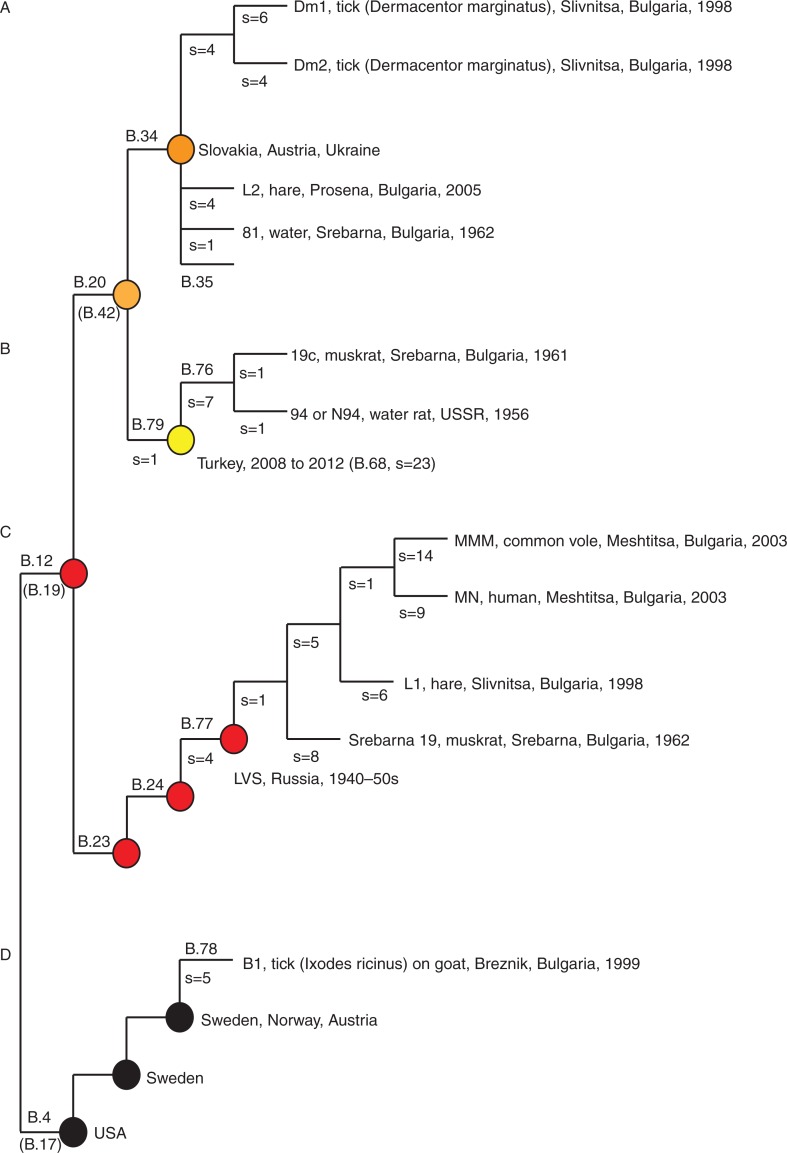
Fig. 3.
The new Bulgarian genomes placed in the phylogenetic framework of canSNPs. Four assays B.76 − B.79 were developed in this study to target subclades, where strains from Bulgaria were members. B.4, B.12, B.23, B.24, B.34, and B.35 have been previously published (5, 6, 20). Alternative canSNPs (B.17, B.19, and B.42) are indicated in gray below the first published canSNP in indicated branch (6, 19). s=the numbers of canSNPs on the indicated branch.
A local persistence of F. tularensis is suggested in the old focus by the close kinship of strain 81 and strain L2. Strain 81 was isolated in 1962 in Srebarna (Fig. 3A, subclade B.34/35) and the L2 strain was isolated in close proximity to Srebarna in 2005 (Fig. 3A, subclade B.34/35), and the genomes of the strains differed by only five SNPs. This finding may suggest that F. tularensis was preserved in the Srebarna area over 43 years and thus would lend support to Pavlovsky's inclusion of tularemia as one of the diseases that adhere to the nidality concept (3). However, the close genetic relationships of strains from Bulgaria with strains from Hungary, Sweden, the former Soviet Union, and Turkey caution for making firm conclusions on persistence (see Fig. 2). There was similarly a close genetic relationship but little epidemiological connection between the Bulgarian strain B1 and isolates from Sweden, Norway, and Austria (see basal clade B.4, Fig. 3D).
The 19c strain isolated in 1961 in the old focus and member of the subclade B.79 showed a very close relationship to the reference strain 94 isolated from a water rat in 1956 in USSR, with only one unique SNP each ( Fig. 3B). This corroborates previous findings on the possible long-distance movement of F. tularensis clones (8, 9). In this case, a USSR origin is also supported by historical data on movement of 24 muskrats (Ondatra zibethica) from the USSR into the Srebarna reserve in 1956 for the purpose of hunting and production of fur (11). Five years later, in 1961, the muskrat population had reached approximately 20,000 individuals and an epizootic of tularemia occurred. The same year, F. tularensis strain 19c was isolated from a dead muskrat in the reserve (11). Potentially, a parental clone to strain 19c and the USSR strain 94 of 1956 may have been transferred along with muskrats into the reserve (13).
Assays for Bulgarian specific clades
The genetic analysis showed that the 1960s as well as the 1990s outbreaks involved new genetic diversity. The new diversity was characterized by four new canSNPs denoted B.76, B.77, B.78, and B.79 in the basal clades B.12 (B.76, B.77, and B.79) and B.4 (B.78) (Fig. 3). We developed canSNP assays for these new genetic branches of F. tularensis for use in future epidemiological investigations in Bulgaria (Table 3). The canSNP system is a generic taxonomic system for detailed intra-subspecies division that has been developed for F. tularensis (5, 6) and other strictly clonal bacteria such as Bacillus anthracis (22), Yersinia pestis (23), and Coxiella burnetii (24). The system is based on identification of selected representative (canonical) SNPs for each branch in a phylogenetic tree making it possible to use canSNP assays for rapidly placing samples into the taxonomic system, for example, during a suspected outbreak.
Conclusions
The existence of strain-pairs isolated in the same year and same area (Dm1 and Dm2; MMM and MN), and different hosts (MMM and MN), supports an expansion of individual F. tularensis clones in local areas during outbreaks among wildlife and humans. The data were inconclusive to infer a common origin of F. tularensis strains that caused the Bulgarian tularemia outbreaks in the 1960s and the 1990s. The finding of very few SNP differences (n=5) across the whole genome in two strains isolated in the Srebarna region in 1961 and 2005, respectively, indicates local persistence and/or slow natural mutation rate in this bacterial species. A modified low-cost canSNP detection system was developed that can be used in future outbreak investigations to place F. tularensis strains into the global phylogenetic context. Taken together, this study provides information on the genetic diversity of F. tularensis in Bulgaria and suggests that muskrat implantation from USSR contributed to this diversity.
Supplementary Material
Acknowledgements
The authors thank Malin Granberg for DNA preparations and Adrian Lärkeryd and Caroline Öhrman for genome assembly work. KrM and MN provided strains and metadata. KM carried out the bioinformatics and genetic analyses and drafted the manuscript. EK carried out the assays. KrM, KM, AJ, and MF conceived of the study, participated in its design and coordination, and helped draft the manuscript. All authors read and approved the final manuscript. This work was supported by the Swedish Ministry of Defence (FOI project no. A4040) and the Swedish Ministry of Foreign Affairs (FOI project no. A4952). The authors acknowledge the support of the National Genomics Infrastructure (NGI)/ Uppsala Genome Center. The work performed at NGI/Uppsala Genome Center was funded by RFI/VR and Science for Life Laboratory, Sweden.
Footnotes
These authors contributed equally to this work.
Conflict of interest and funding
The authors have not received any funding or benefits from industry or elsewhere to conduct this study.
References
- 1.Maurin M, Gyuranecz M. Tularaemia: clinical aspects in Europe. Lancet Infect Dis. 2016;16:113–24. doi: 10.1016/S1473-3099(15)00355-2. [DOI] [PubMed] [Google Scholar]
- 2.Hestvik G, Warns-Petit E, Smith LA, Fox NJ, Uhlhorn H, Artois M, et al. The status of tularemia in Europe in a one-health context: a review. Epidemiol Infect. 2014;143:2137–60. doi: 10.1017/S0950268814002398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pavlovsky EN. The natural nidality of transmissible diseases. In: Levine ND, editor. Natural nidality of transmissible diseases. Urbana, IL: University of Illinois Press (Urbana, III. 61803); 1966. pp. 93–129. [Google Scholar]
- 4.World Organization for Animal Health (OIE) Contributing to one world, one health: a strategic framework for reducing risks of infectious diseases at the animal-human-ecosystems interface. Paris: World Organization for Animal Health; 2008. [Google Scholar]
- 5.Vogler AJ, Birdsell D, Price LB, Bowers JR, Beckstrom-Sternberg SM, Auerbach RK, et al. Phylogeography of Francisella tularensis: global expansion of a highly fit clone. J Bacteriol. 2009;191:2474–84. doi: 10.1128/JB.01786-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Svensson K, Granberg M, Karlsson L, Neubauerova V, Forsman M, Johansson A. A real-time PCR array for hierarchical identification of Francisella isolates. PLoS One. 2009;4:e8360. doi: 10.1371/journal.pone.0008360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lärkeryd A, Myrtennäs K, Karlsson E, Dwibedi CK, Forsman M, Larsson P, et al. CanSNPer: a hierarchical genotype classifier of clonal pathogens. Bioinformatics. 2014;30:1762–4. doi: 10.1093/bioinformatics/btu113. [DOI] [PubMed] [Google Scholar]
- 8.Johansson A, Lärkeryd A, Widerström M, Mörtberg S, Myrtennäs K, Öhrman C, et al. An outbreak of respiratory tularemia caused by diverse clones of Francisella tularensis . Clin Infect Dis. 2014:59: 1546–53. doi: 10.1093/cid/ciu621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Afset JE, Larssen KW, Bergh K, Lärkeryd A, Sjödin A, Johansson A, et al. Phylogeographical pattern of Francisella tularensis in a nationwide outbreak of tularaemia in Norway, 2011. Euro Surveill. 2015;20:21125. [PubMed] [Google Scholar]
- 10.Marinov KT, Georgieva ED, Ivanov IN, Kantardjiev TV. Characterization and genotyping of strains of Francisella tularensis isolated in Bulgaria. J Med Microbiol. 2009;58:82–5. doi: 10.1099/jmm.0.003426-0. [DOI] [PubMed] [Google Scholar]
- 11.Gotev N. Epizooty of tularemia among ondatras in the Reserve Srebarna Lake. Voenno-Medicinsko Delo (Military Medcine) 1962;3:70–6. [Google Scholar]
- 12.Dinev T, Zlatanov Z. Bacteriological study of a natural focus of tularemia in the Lake Reserve of Srebrna, the Silistra region, Bulgaria. J Hyg Epidemiol Microbiol Immunol. 1972;16:341–5. [PubMed] [Google Scholar]
- 13.Kupenov N, Gotev N, Symnaliev M, Tomov A, Khristov L, Baev V, et al. A natural tularemia focus in Bulgaria. Zh Mikrobiol Epidemiol Immunobiol. 1964;41:124–31. [PubMed] [Google Scholar]
- 14.Christova I, Velinov T, Kantardjiev T, Galev A. Tularaemia outbreak in Bulgaria. Scand J Infect Dis. 2004;36:785–9. doi: 10.1080/00365540410021199. [DOI] [PubMed] [Google Scholar]
- 15.Mladenov K, Marinov K, Tsvetkova E, Kasovski V. Microbiological researches on the focus of tularemia in West Bulgaria. Probl Infect Parasit Dis. 2003;31:17–18. [Google Scholar]
- 16.Kantardjiev T, Ivanov I, Velinov T, Padeshki P, Popov B, Nenova R, et al. Tularemia outbreak, Bulgaria, 1997–2005. Emerg Infect Dis. 2006;12:678–80. doi: 10.3201/eid1204.050709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.World Health Organization. WHO guidelines on tularaemia: epidemic and pandemic alert and response. Geneva: World Health Organization; 2007. [Google Scholar]
- 18.Larsson P, Elfsmark D, Svensson K, Wikström P, Forsman M, Brettin T, et al. Molecular evolutionary consequences of niche restriction in Francisella tularensis, a facultative intracellular pathogen. PLoS Pathog. 2009;5:e1000472. doi: 10.1371/journal.ppat.1000472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karlsson E, Svensson K, Lindgren P, Byström M, Sjödin A, Forsman M, et al. The phylogeographic pattern of Francisella tularensis in Sweden indicates a Scandinavian origin of Eurosiberian tularaemia. Environ Microbiol. 2013;15:634–45. doi: 10.1111/1462-2920.12052. [DOI] [PubMed] [Google Scholar]
- 20.Gyuranecz M, Birdsell DN, Splettstoesser W, Seibold E, Beckstrom-sternberg SM, Makrai L, et al. Phylogeography of Francisella tularensis subsp. holarctica, Europe. Emerg Infect Dis. 2012;18:290–3. doi: 10.3201/eid1802.111305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dominowska C. Bulletin of the Institute of Marine Medicine in Gdansk. Cambridge: Cambridge University Press; 1967. Properties and typing of Francisella tularensis; pp. 18: 131–156. [Google Scholar]
- 22. Van Ert MN, Easterday WR, Huynh LY, Okinaka RT, Hugh-Jones ME, Ravel J, et al. Global genetic population structure of Bacillus anthracis. PLoS One. 2007;2:e461. doi: 10.1371/journal.pone.0000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morelli G, Song Y, Mazzoni CJ, Eppinger M, Roumagnac P, Wagner DM, et al. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat Genet. 2010;42:1140–3. doi: 10.1038/ng.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlsson E, Macellaro A, Byström M, Forsman M, Frangoulidis D, Janse I, et al. Eight new genomes and synthetic controls increase the accessibility of rapid melt-MAMA SNP typing of Coxiella burnetii. PLoS One. 2014;9:e85417. doi: 10.1371/journal.pone.0085417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.