Abstract
Acetylation is an important, reversible post-translational modification affecting histone and non-histone proteins with critical roles in gene transcription, DNA replication, DNA repair, and cell cycle progression. Key regulatory enzymes include histone deacetylase (HDACs) and histone acetyltransferases (HATs). Overexpressed HDACs have been identified in many human cancers, resulting in repressed chromatin states that interfere with vital tumor suppressor functions. Inhibition of HDAC activity has been pursued as a mechanism for re-activating repressed genes in cancers, with some HDAC inhibitors showing promise in the clinical setting. Dietary compounds and their metabolites also have been shown to modulate HDAC activity or expression. Out of this body of research, attention increasingly has shifted towards non-histone targets of HDACs and HATs, such as transcriptions factors, hormone receptors, DNA repair proteins, and cytoskeletal components. These aspects are covered in present review, along with the possible clinic significance. Where such data are available, examples are cited from the literature of studies with short chain fatty acids, polyphenols, isoflavones, indoles, organosulfur compounds, organoselenium compounds, sesquiterpene lactones, isoflavones, and various miscellaneous agents. By virtue of their effects on both histone and non-histone proteins, dietary chemopreventive agents modulate the cellular acetylome in ways that are only now becoming apparent. A better understanding of the molecular mechanisms will likely enhance the potential to more effectively combat diseases harboring altered epigenetic landscapes and dysregulated protein signaling.
Dietary chemopreventive agents modulate the cellular acetylome by affecting both histone and non-histone proteins, which will likely enhance their potential to more effectively combat diseases harboring altered epigenetic landscapes.
Keywords: Acetylation, diet, epigenetics, phytochemicals, HDAC, non-histone
Graphical abstract

1. INTRODUCTION
1.1 HDAC inhibition and histone acetylation
The genome is organized into arrays of nucleosomes composed of histones and DNA. The nucleosome is a fundamental and highly dynamic unit consisting of chromatin structures involved in key cellular functions such as cell cycle progression, mitosis, DNA replication, DNA damage recognition, DNA repair, heterochromatin silencing, and gene transcription. During such processes, histones are post-translationally modified in various ways, including methylation, ubiquitination, phosphorylation, and acetylation1. Acetylation on histones neutralizes their positive charge, decreasing the interactions with negatively charged DNA and associated regulatory proteins. This open or “euchromatin” conformation allows genes to be actively transcribed2. In contrast, deacetylation of histones results in condensation, or “heterochromatin”, leading to gene repression.
Histone acetylation is regulated in large measure by the opposing actions of histone acetyltransferases (HATs), which add acetyl groups, and histone deacetylases (HDACs), which remove the acetyl groups3. Acetylation and other post-translation modifications on N-terminal tails of histones influence the state of gene expression by providing a binding platform for various proteins, such as transcriptional factors, chromatin remodeling/modifying components, and co-activators/repressors4. Thus, manipulation of histone acetylation through HDAC inhibition has been proposed as a mechanism for derepressing genes dysregulated in chronic conditions such as cancer, neurodegenerative diseases, psychological disorders, and lung pathologies.
At present, 18 HDACs have been identified in humans that fall into four classes: class I HDACs (HDAC1, HDAC2, HDAC3 and HDAC8) are localized mainly in the nucleus, class II HDACs (HDAC4, HDAC5, HDAC6, HDAC7, HDAC9 and HDAC10) are restricted to certain tissues and can be present in both nuclear and cytoplasmic compartments, class III HDACs are NAD-dependent sirtuins (SIRT1 to SIRT7), and HDAC11, the only class IV HDAC, which shares conserved residues with both class I and II HDACs5. Non-sirtuin HDACs are zinc-dependent enzymes.
The online website clinicaltrials.gov lists numerous human trials with HDAC inhibitors, either as monotherapies or combination treatments. Vorinostat and Romidepsin are the best-studied HDAC inhibitors, approved by the US Food and Drug Administration (FDA) for the treatment of advanced cutaneous T-cell lymphoma. However, the list of HDAC inhibitors is growing rapidly, and comprises compounds from unrelated chemical classes. Despite the different HDAC isoform specificity, these agents typically exhibit similar toxicity and resistance profiles6. This could be related to the fact that they share a common mechanistic theme, that is, inhibition of zinc-dependent functions, thus affecting all but the NAD-dependent sirtuins as “pan-HDAC inhibitors”.
For these reasons, the pursuit of novel and improved HDAC inhibitors has shifted towards dietary factors, isolated phytochemicals, and novel metabolites with wider safety margins or improved HDAC selectivity7, 8. Mechanisms being pursued include direct binding to the catalytic site, as competitive HDAC inhibitors, or interactions with allosteric sites critical for protein-protein interactions (e.g., in HDAC/co-repressor complexes). Other dietary compounds affect HDAC expression and stability indirectly, for example, via kinase-mediated mechanisms and ubiquitylation pathways (Fig. 1). Such mechanisms often involve cross-talk among post-translational modifications, including acetylation changes on non-histone proteins.
Fig 1. Mechanisms of HDAC inhibition by dietary compounds.

Dietary compounds inhibit HDAC activity and/or expression via direct or indirect mechanisms. (A) Hydroxamic acids interact with the zinc atom in the catalytic site via a bidentate ligand. In dietary compounds such as butyrate, the corresponding interaction likely involves a carboxyl moiety221; whereas in garlic organosulfur compounds, it is the sulfhydryl group of AM222. (B) Allosteric site binding, as predicted for the SFN-NAC metabolite in the inositol tetraphosphate (IP4) pocket that lies between HDAC3 and its corepressor partner SMRT94. (C) Dissociation of co-repressor complexes due to SFN-mediated CK2 kinase recruitment and HDAC3/SMRT phosphorylation172. (D) Ubiquitylation leading to HDAC3 protein turnover, triggered in cancer cells by dietary indoles104.
1.2 Acetylation of non-histone proteins
Many non-histone proteins (transcription factors, regulators of DNA repair, recombination and replication, chaperones, viral proteins, and others) are subject to acetylation. Indeed, a recent investigation used high-resolution mass spectrometry to identify 3,600 separate acetylation sites in 1,750 human proteins, implicating lysine acetylation in the regulation of nearly all nuclear functions and many cytoplasmic processes9, 10. It is noteworthy that the non-histone targets are involved in cellular processes such as chromatin remodeling, cell cycle regulation, apoptosis, autophagy, and actin nucleation, which are dysregulated in cancer etiology10.
We have summarized some of the well-known non-histone protein targets for which the acetylation status is regulated by opposing actions of HATs and HDACs (Table 1). Unlike acetylation on histones, the acetylation of non-histone targets is associated with either activation or inactivation of protein function11. These non-histone proteins regulate many key biological processes, including cell proliferation, cell survival, and apoptosis, with the acetylation status influencing protein stability, enzymatic activity, sub-cellular localization, protein–DNA interactions, and protein–protein partnering11 (Fig 2).
Table 1.
Non-histone targets of HATs and HDACs
| Non-histone | HAT | HDAC |
|---|---|---|
| p53 | p300/CBP46, 47 | HDAC148, SIRT149 |
| YY1 | p300/CBP50 | HDAC251 |
| STAT3 | p300/CBP17 | HDAC317 |
| HMG | CBP, PCAF52 | NA |
| c-Myc | PCAF/GCN5, TIP6053 | NA |
| AR | p300/CBP, TIP6054 | HDAC155 |
| ER | p30056 | NA |
| GATA | CBP (GATA1)57, p300 and PCAF (GATA2)58 | HDAC3(GATA2)59 |
| EKLF | p300/CBP60 | NA |
| MyoD | PCAF, CBP, and p30061, 62 | HDAC163 |
| E2F/Rb | PCAF, CBP, and p30064 | HDAC165 |
| NF-kB | p300/CBP66 | HDAC366, SIRT167 |
| HIF1α | ARD168 | HDAC469 |
| Smad7 | p30070 | |
| α-Tubulin | NA | HDAC625, SIRT223 |
| Importin-α | CBP, p30071 | NA |
| Ku70 | PCAF, CBP72 | SIRT172 |
| Hsp90 | NA | HDAC673 |
| E1A | p300, PCAF74 | NA |
| CtIP | GCN594 | HDAC394, SIRT675 |
| β-catenin | PCAF76, p30077 | SIRT178, HDAC679 |
| Survivin | NA | HDAC680 |
| FoxO1 | CBP/p30081 | SIRT182 |
| FoxO3a | CBP/p30082 | SIRT182, SIRT283, SIRT384 |
| PGC1- α | GCN585 | SIRT174, 86 |
| FXR | p30087 | SIRT188 |
NA: Not available
Fig 2. Dietary deacetylase inhibitors and (de)acetylation targets.
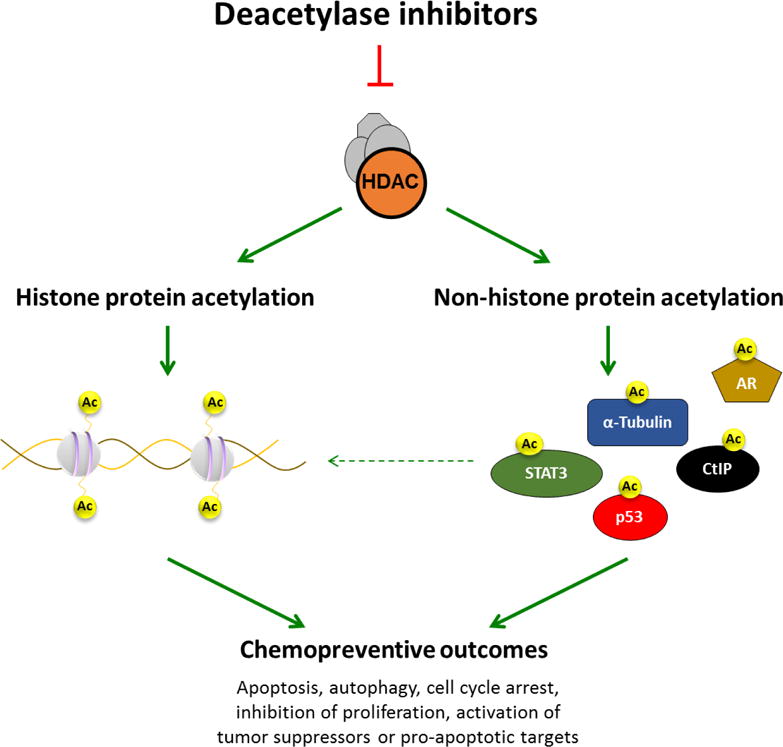
Deacetylase inhibitors induce histone hyperacetylation leading to an open chromatin conformation that correlates with gene activation. Acetylation of non-histone proteins modulates their function, stability, cellular localization, and/or protein–protein interactions. Transcription factors such as STAT3 and p53 are directly acetylated by HDAC inhibitors, which can positively or negatively alter their capacity to bind DNA and modulate gene expression. Through the combined effects on histone and non-histone protein acetylation, deacetylase inhibitors exert chemopreventive outcomes ranging from inhibition of cell growth, cell cycle arrest, apoptosis, autophagy, or necrosis.
The well-studied tumor suppressor p53 is subject to acetylation by p300/CBP at the C-terminal DNA binding domain12. As a result, the activity of p53 is altered during the DNA damage response12. Acetylation of p53 by p300/CBP is opposed by HDAC1, and subsequent steps can involve MDM2-mediated ubiquitination, leading to p53 turnover13, 14. Thus, HATs and HDACs balance the acetylation status of p53 and fine-tune its stability and half-life. In contrast to the actions of HDAC1, deacetylation by the class III HDAC SIRT1 decreases the transcriptional activity of p53, and activates p21 in cell cycle regulation and DNA repair process15.
Another well-known transcription factor regulated by SIRT1 is signal transducer and activator of transcription 3 (STAT3), which has key roles in cytokine signaling16. Like phosphorylation, acetylation status influences dimerization, nuclear-cytoplasmic localization, and the transcriptional activity of STAT family members17. On histone tails, a phospho-acetyl switch has been identified18, 19, for which parallels might exist in regulating STAT3 and other non-histone proteins.
Nuclear receptors also represent important non-histone targets for acetylation and deacetylation. Acetylation of the androgen receptor (AR) by p300/CBP and TIP60 at multiple lysines in the DNA binding domain enhances hormone-dependent transactivation, whereas deacetylation of AR by HDAC1 represses its function20. Further, ubiquitination by MDM2 occurs upon removal of acetylation on AR14. Interestingly, in chronic obstructive pulmonary disease (COPD) and other lung pathologies, it is loss of HDAC2, not HDAC1, that is implicated in excessive GR signaling pathways7, 21, 22
In addition to transcription factors and nuclear receptors, the cytoskeletal protein α-tubulin is a well-recognized non-histone target for acetylation. Deacetylation of α-tubulin by HDAC6 and SIRT2 induces cell motility and enhances microtubule depolymerization23–25. HDAC8 also associates with β-actin and, via changes in its acetylation status, regulates smooth muscle contractility26.
In the following paragraphs, we further discuss protein acetylation in the context of dietary agents and their modulatory effects (Table 2), highlighting key mechanistic questions or gaps in the literature.
Table 2.
Histone and non-histone target of dietary HDAC inhibitors
| Class of Compound | Phytochemical | Chemical structure | HAT/HDAC | Histone | Non-histone | Outcome |
|---|---|---|---|---|---|---|
| Short-chain fatty acid | Sodium butyrate |

|
HDAC1 and HDAC334 | H3, H434 | NA | Inhibition of tumor growth, Apoptosis, Cell cycle arrest |
| Isothiocyanates | Sulforaphane |
|
HDAC1, 2, 3, 4 HDAC7, HDAC689–92 |
H3, H489–92 | α-Tubulin93, CtIP*94 | Inhibition of tumor growth, Apoptosis, Cell cycle arrest |
| Allyl-ITC |
|
HDAC95 | H3, H495 | NA | Inhibition of tumor growth, Apoptosis, Cell cycle arrest | |
| BITC |
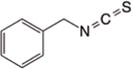
|
HDAC1 and HDAC396 | H396 | FoxO197 | Inhibition of tumor growth, Apoptosis, Cell cycle arrest, Autophagy | |
| PHITC |
|
HDAC and p30098, HDAC1 and HDAC299 | H3, H499–101 | NA | Apoptosis | |
| PEITC |
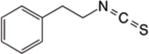
|
NA | H3102 | α-Tubulin103 | Attenuating cell proliferation, Apoptosis | |
| Indol-3-carbinol | I3C |
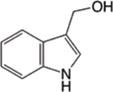
|
HDACs104 HDAC1, HDAC2105, HDAC3106, 107 |
H3, H4104 | NA | Cell cycle arrest, T cell activation |
| Sesquiterpene lactone | Parthenolide |

|
HDAC1108 | H3108 | NA | Inhibition of tumorigenesis, Apoptosis |
| Allium compounds | Diallyl disulfide |
|
N-acetyltransferase109 HDACs |
H3, H4110–113 | NA | Adipocyte differentiation, Apoptosis |
| Allyl mercaptan |
|
HDACs114 | H3, H495, 114–117 | NA | Inhibition of tumor growth, Apoptosis, Cell cycle arrest | |
| Selenium | MSA |
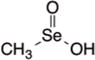
|
HDAC2 and GCN5118 | H3119 H3K18 and H3K9118 | α-Tubulin119 | Inhibition of tumor growth, Apoptosis, Cell cycle arrest |
| Selenite |
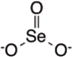
|
HDAC120 | H3K9120 | NA | Inhibition of tumor growth, Apoptosis, Cell cycle arrest | |
| MSP |

|
HDAC1 and HDAC8121 | H3K9/14, H4121,26 | NA | Apoptosis | |
| Polyphenols | EGCG |

|
HDAC1, HDAC2, and HDAC3122,123 HAT124 | H3K9/14 H4K5,12,16,H3K 9/18122,125,126 |
p53127, 128 FoxO1129 AR*130 Smad2/3*131 |
Inhibition of tumor growth, Apoptosis, Cell cycle arrest |
| Curcumin |

|
HDAC1, HDAC2132 HDAC3 HDAC4133 HDAC6134 HDAC8135 P300 |
H3, H4135,136 H3K18ac, H4K16ac137 | p53132,138 | Inhibition of tumor growth, Apoptosis, Cell cycle arrest | |
| Resveratrol |
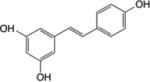
|
SIRT1 HDACs139, SIRT2140 | H3, H4139 | p53141, FXR88, PGC1- α142, FOXO3a143 | Inhibition of tumor growth and cell proliferation, Apoptosis, Cell cycle arrest | |
| Chrysin |

|
HDAC8144 | H3acK14, H4acK12, H4acK16144 | NA | Cell cycle arrest, Inhibition of proliferation | |
| Ellagic acid |
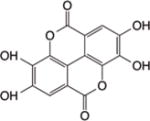
|
HDAC6145, HDAC9146 | NA | NA | Inhibition of proliferation, Apoptosis, Cell cycle arrest | |
| Isoflavones | Quercetin |

|
SIRT6147,148 | H3K56ac148 | NA | Inhibition of a β-catenin pathway, Inhibition of tumor growth, Apoptosis, Cell cycle arrest |
| Apigenin |
|
HDAC1, HDAC3149 | H3, H4149 | NA | p53 activation, Generation of ROS, apoptosis, Inhibition of tumor invasion, Cell cycle arrest | |
| Genistein |
|
HAT1150, HDAC1151 SIRT1152 | H3K9150, 152, H3153, H4154 | NA | Apoptosis, Inhibition of cell proliferation and growth | |
| Luteolin |
|
CBP/p300155, SIRT6147 | H3, H4156 | NA | Apoptosis, Inhibition of invasiveness | |
| Royal jelly | (E)-10-hydroxy-2-decenoic acid (10HDA) |
|
HDAC3157 | H3, H4158 | NA | Apoptosis, Cell cycle arrest |
| Vitamin | Nicotinamide |
|
SIRT1159 | H3160 | p53161, 162, BCL6163, BAZ2a, NPM1, MOF, WRN, Ku70 and Ku80160 | Cell cycle arrest, Apoptosis |
| Tyrosine derived compound | Psammaplin A |

|
HDAC1, HDAC6164, SIRT165 | H3, H4164 | Tubulin164 | Apoptosis, Inhibition of proliferation, Cell cycle arrest |
| Methyl xanthine alkaloid | Caffeine |
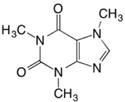
|
Hdac1 and Hdac2166 | H3K9 and H3K14166 | NA | Fetal development |
CtIP* is pulled down with lysine antibody in the presence of SFN. AR* and Smad2/3* are deacetylated in the presence of EGCG in human lung cancer cells.
NA: Not available
2.0 PROTEIN ACETYLATION AND DIETARY CHEMOPREVENTIVE AGENTS
2.1 Short Chain Fatty Acids
Short chain fatty acids (SCFAs) are the end product of fermentation of dietary fiber and they exhibit anticancer effects such as inhibition of proliferation, apoptosis induction, or cell cycle arrest27–29. Such effects are thought to be mediated via the inhibition of HDAC activity. In many studies, SCFA treatment resulted in hyper acetylation of histone H3 and/or H4 and the activation of tumor suppressor and oxidative stress response genes in cancer cells27, 30. Sodium butyrate (IC50: 0.3 mM) was reported over three decades ago to induce histone acetylation in cultured leukemia cells31, 32. However, it was not formally identified as an HDAC inhibitor until quite recently. In vitro, butyrate acts as a competitive HDAC inhibitor, with an apparent Ki of 46 μM when tested in whole cell lysates of human MCF-7 breast cancer cells, compared with an apparent Ki of 1 nM for trichostatin A (TSA) under the same conditions33. Interestingly, butyrate and pyruvate have been reported as selective inhibitors of HDAC1 and HDAC3, whereas lactate, a precursor in the pathway of carbohydrate metabolism, had no such effect on HDAC activity34, 35. These results are surprising since butyrate is usually thought of as a class I HDAC inhibitor (if not a pan HDAC inhibitor) that has both histone and non-histone targets, for example HMG protein in colon cancer cells36, 37, 38. Tributyrin and valproic acid (IC50: 39μM~2mM) are other small molecule HDAC inhibitors in the same category39, 40, 41. Tributyrin shares p21WAF1 as a common gene target for de-repression in cancer cells41. Similar to butyrate, tributyrin has also been shown to acetylate p5342, and valproic acid has been shown to acetylate Ku70 in medulloblastoma cells43. More recently, it was reported that the ketone body d-β-hydroxybutyrate (βOHB) (IC50: 2~5mM) is an endogenous and specific inhibitor of class I HDACs. Treatment of cells with βOHB increased global histone acetylation and activated oxidative stress genes such as FOXO3A and MT2 through inhibition of HDAC1 and HDAC244.
2.2 Selenium
Selenium is an essential trace element found in the soil, and is also bio-accumulated as organic forms in foods such as Brazil nuts, cruciferous vegetables, and allium vegetables. Anticancer effects of selenium compounds have been reported in various cancer cell lines such as colon, prostate, and ovary45. In addition, metabolites of organoselenium compounds have been implicated in regulating gene expression via epigenetic mechanisms. For example, methylseleninic acid (MSA) inhibited the growth of esophageal squamous cell carcinoma cells118 and diffuse large B-cell lymphoma cell lines119, and this was associated with decreased HDAC activity. In the former study, MSA acetylated both histone and non-histone proteins, such as α-tubulin, and this required conversion to its presumed metabolites methylselenol and dimethylselenide118. Recently, we reported that a novel seleno-α-keto acid, methylselenopyruvate (MSP) (IC50: 20μM), triggered global histone acetylation in human colon cancer cells and in several other cancer cell lines. Apoptosis was activated via direct transcriptional changes on the gene promoter for Bcl-2-modifying factor (BMF)26. Based on our data, HDAC8 de-recruitment rather than STAT3 acetylation likely served as the primary driver of BMF transcriptional activity in response to MSP26.
2.3 Isothiocyanates
Brassica or cruciferous vegetables are a rich source of glucosinolates. The hydrolysis of these glucosinolates by the plant enzyme myrosinase generates biologically active isothiocyanates (ITC)167 and indoles168. Many studies have shown that ITCs exert anti-cancer effects through inhibition of HDAC activity and histone hyperacetylation, as reported for sulforaphane (SFN)169, 170, allyl isothiocyanate (allyl-ITC)95, benzyl isothiocyanate (BITC)96, phenylhexyl isothiocyanate (PHITC)98, phenethyl isothiocyanate (PEITC)102, and other longer-chain isothiocyanates171.
BITC decreased HDAC activity and the expression levels of HDAC1 and HDAC3 proteins in Capan-2 and BxPC-3 pancreatic cancer cells, but not in normal cells96. HDAC inhibition by BITC resulted in histone hyperacetylation on the p21WAF1 gene, induction of p21waf1 protein expression, and cell cycle arrest. Recently, Xiao et al. reported that BITC induced autophagy by increasing acetylation of FoxO1, a non-histone target, in breast cancer cells97.
In silico studies have indicated that metabolites of dietary ITCs have structural features similar to pharmacological HDAC inhibitors, allowing them to dock into the HDAC catalytic site169, 171 or bind to allosteric sites on HDAC complexes94. In addition to histone targets, ITCs have been reported to alter the acetylation status of non-histone proteins. SFN decreased HDAC3 and/or HDAC6 protein expression in colon and prostate cancer cells, leading to hyperacetylation of histones and α-tubulin172,93, 173. Others have examined such relationships in keratinocytes89 and lymphocytes174. In the latter study, SFN increased histone acetylation and attenuated lymphocyte HDAC activity to control levels174.
We reported that SFN and related longer-chain ITCs induced DNA damage in colon cancer cells, but not in normal colonic epithelial cells94. DNA damage was associated with acetylation and turnover of C-terminal binding protein interacting protein (CtIP), a key component of DNA repair pathways involving homologous recombination and DNA repair94. Data from in vivo studies revealed that targeted magnetic microspheres containing SFN inhibited tumor growth and reduced histone deacetylase levels in C57BL/6 mice175. In the prostate cancer (TRAMP) model, SFN also attenuated the expression levels of HDAC1, HDAC4, and HDAC791.
PEITC is another promising chemopreventive agent, with efficacy in preclinical models against various types of cancer176. PEITC has been found to affect both promoter demethylation and the chromatin states in prostate cancer cells, re-activating genes such as glutathione S-transferase P1 (GSTP1)177, 178. Recently, Chang et al. reported that PEITC mediated α-tubulin acetylation in breast cancer cells, although they did not formally test the hypothesis that PEITC might selectively target HDAC6 or its α-tubulin deacetylase activity103.
2.4 Indole-3-carbinol and 3,3′-diindolylmethane
Cruciferous vegetables contain glucosinolates such as glucobrassicin, the precursor of indole-3-carbinol (I3C) (IC50: 0.1–20uM). I3C and its acid condensation products, such as 3,3′-diindolylmethane (DIM) (IC50: 0.1–20uM), have been examined extensively for their cancer chemoprotective properties179. Bhatnagar et al. reported that DIM inhibited the expression of HDAC1, HDAC2 and HDAC3 in colon cancer cells leading to inhibition of Survivin mRNA and protein expression106. Li et al. demonstrated that DIM-induced HDAC depletion involved proteasome-mediated HDAC protein turnover104. Although the authors found negligible increases in the acetylation of histones associated with selected gene promoters, a reduction in the levels of repressive HDACs bound to the p21WAF1 and p27 promoters coincided with cell cycle arrest104. Beaver et al. recently reported that DIM significantly inhibited HDAC activity and reduced HDAC2 expression in both androgen sensitive and insensitive prostate cancer cells, and this was correlated with increased expression of p21WAF1105. In mice, Staphylococcal enterotoxin B (SEB)-induced activation of T cells was attenuated by I3C and DIM treatment, and this was associated with the inhibition of class I HDACs107.
2.5 Parthenolide
Parthenolide (PN) is a sesquiterpene lactone of the germacranolide class isolated from Tanacetum parthenium and found in many flowers and fruits. In addition to its other actions, PN treatment was associated with depleting HDAC1 protein without affecting other class I/II HDACs108. Recently, PN was shown to inhibit tumorigenesis due to the activation of p21WAF1 and repression of CYCLIN D1, with evidence for alteration in local histone H3 acetylation180, 108, but perhaps involving non-histone targets of HDAC1.
2.6 Allium compounds
Garlic, onions, shallots and other members of the allium family contain an interesting mix of water-soluble and fat-soluble organosulfur compounds, some of which have been implicated as chemopreventive agents45, 181. Allyl derivatives from garlic were among the first compounds described to impact histone acetylation status in cancer cells. Allyl mercaptan (AM), diallyl disulfide (DADS), S-allylcysteine (SAC), S-allylmercaptocysteine (SAMC) and allicin increased histone acetylation in various human cancer cells95, 115–117, implicating HDACs as possible targets. AM was the most effective HDAC inhibitor among several garlic-derived organosulfur compounds tested in vitro114. Based on the IC50 values, intracellular metabolism to small molecular thiols, such as AM, likely accounts for most of the HDAC inhibition associated with garlic organosulfur compounds. At relatively low concentrations, AM caused histone H3 hyperacetylation in human colon cancer cells, and facilitated Sp3 and p53 binding on the p21WAF1 promoter114.
Several reports suggest that DADS can induce histone acetylation, as shown in leukemia111, colon113, breast112 and prostate182 cancer cells. DADS treatment also triggered global histone hyperacetylation during adipocyte differentiation110. Further, adipocyte differentiation-related genes such as LPL, FAS, SREBP1c, aP2 and PPAR-γ were increased, while PREF-1, a preadipocyte marker gene, was decreased by DADS. These findings suggested that DADS affects adipocyte differentiation through histone acetylation at an early phase110. In vivo studies with DADS revealed increased histone acetylation in rat colonocytes183. Although metabolites of DADS are thought to act as HDAC inhibitors in various cancer cell lines, one group focused on DADS and its ability to decrease the activity and expression of N-acetyltransferase (NAT) in human esophagus epidermoid carcinoma CE 81T/VGH cells109.
Whereas global and local histone acetylation is induced under such conditions, little is known about the specific HDACs affected. Molecular docking and enzyme kinetics studies supported AM-HDAC8 interactions114, but other HDACs remain to be examined. If, indeed, the final common metabolite generated by such organosulfur compounds is AM, the small size and highly reactive sulfhydryl group would predict ready access and strong interactions with the active site zinc atom in multiple HDACs. This lack of HDAC specificity might offer no greater benefit over currently used pan-HDAC inhibitors undergoing clinical trials.
2.7 Polyphenols
2.7.1 Epigallocatechin-3-gallate
Polyphenols occur naturally in many foods and beverages, including green tea, curry spices, grapes, soy, and berries. (−)-Epigallocatechin-3-gallate (EGCG) is the most abundant polyphenol in green tea. Although it was first identified as an “antioxidant” in vitro184, the possible relevance of this “antioxidant” activity to anticancer properties of EGCG is far from established in vivo. EGCG was reported to inhibit enzymes involved in DNA methylation, and was subsequently identified as a histone modifier122, 125, 126. EGCG inhibited HDAC activity and increased histone acetylation in prostate122, skin125, and breast cancer cells126. Pandey et al. demonstrated that EGCG reduced mRNA expression of HDAC1, HDAC2, and HDAC3, leading to re-expression of GSTP1 in prostate cancer cells122. In human oral squamous carcinoma cells, EGCG decreased SIRT3 activity and expression185.
Li et al. showed that EGCG reactivated the estrogen receptor (ERα) in breast cancer cells, due to decreased binding of a transcription repressor complex containing HDAC1 (Rb/p130-E2F4/5-HDAC1-SUV39H1-DNMT1)126. On the contrary, Choi et al. identified EGCG as a HAT inhibitor that suppressed transcription factor p65 (RelA) acetylation, thereby inhibiting nuclear factor kappa B (NFκB), interleukin 6 (IL6), and other downstream target genes124.
In addition to the HAT and HDAC modulatory activities, EGCG also inhibited polycomb group (PcG) proteins, such as Ezh2123. Reduced Ezh2 and HDAC1 levels at the TIMP-3 gene were associated with a decrease in the histone repressive mark H3K27me3, and an increase in the activation mark H3K9/18ac123. EGCG also was shown to prevent hydrogen peroxide-induced senescence in human dermal fibroblasts by the suppression of p53 acetylation, indicating that p53 is a non-histone target of EGCG127. In human prostate cancer cells, EGCG inhibited class I HDACs to activate p53 transcriptional activity, via increased acetylation on p53128. Recently, Liu et al. reported that EGCG is involved in acetylating FoxO1 in high-glucose treated H9c2 cardiomyoblasts129. Acetylated FoxO1 was associated with increased reactive oxygen species (ROS) and autophagy pathways in cardiomyoblasts. Another non-histone target for EGCG is the androgen receptor (AR), which is regulated by acetylation in prostate cancer cells130. EGCG also has been reported to act via HAT inhibition131. In human lung cancer cells, EGCG treatment resulted in the deacetylation of Smad2 and Smad3, which inhibited TGF-β1-mediated epithelial-mesenchymal transition131.
2.7.2 Curcumin
Curcumin, a component of turmeric (Curcuma longa), has reported antioxidant, anti-inflammatory, and chemopreventive properties, although the underlying molecular mechanisms have yet to be fully elucidated. In clinical studies, curcumin has been tested for its chemosensitizing potential. Like EGCG, curcumin has been evaluated as a p300 (HAT) inhibitor. Kang et al. observed that curcumin treatment resulted in histone hypoacetylation in brain cancer cells, and induced differentiation and cell death in neurons, but not astrocytes186. This finding suggested that curcumin exerts selective HAT inhibitory activity in neural cancer stem cells. In another study, curcumin reduced DNA repair activity by inhibiting HATs and ATR187. The authors showed that curcumin specifically suppressed homologous recombination by reducing the expression of the BRCA1 gene, via altered histone acetylation at the BRCA1 promoter187. However, in other studies, curcumin was reported as an HDAC inhibitor, and increased histone acetylation137 on gene promoters such as suppressor of cytokine signaling (SOCS) genes SOCS1 and SOCS3135 and BAX, a proapoptotic gene136. Curcumin inhibited HDAC4 in medulloblastoma cells133 and HDAC6 in leukemia (K-562 and HL-60) cell lines134.
Curcumin also has been shown to affect non-histone protein acetylation. In cervical cancer cells, curcumin inhibited HDAC1 and HDAC2 activity and increased acetylation of p53132. Similar results were found in prostate cancer cells138, and p53 acetylation was found to regulate its stability, transcriptional activity, and subcellular localization138.
2.7.3 Resveratrol
Resveratrol is a polyphenolic phytoestrogen found in certain types of grapes and red wine. It has attracted attention due to reports on increased longevity, and the treatment of age-related diseases such as diabetes, neurodegeneration, and cancer. Different modes of action have been proposed for resveratrol, including the activation of sirtuins188. Resveratrol has also been shown to inhibit sirtuins, e.g. SIRT2 in glioblastoma stem cells140. Some reports have shown that resveratrol can also inhibit other HDACs. Indeed, Venturelli et al. demonstrated that resveratrol inhibited all classes of HDACs in a dose-dependent manner139. Using in silico molecular docking, they showed that resveratrol fits into the active site of HDAC2, HDAC4, HDAC7, and HDAC8, and inhibits their deacetylase activity139. Others reported that resveratrol increases histone and non-histone protein acetylation status, for example, associated with p53141, the nuclear bile acid receptor FXR88, and PGC1α142, despite the fact that SIRT1 deacetylase activity may be enhanced under the same conditions. In another study, resveratrol increased acetylation on p53 and FOXO3a proteins in Hodgkin lymphoma-derived L-428 cells143. Similar to resveratrol, piceatannol, another polyphenol found in red wine, is known to be a SIRT1 activator189–192. Taken together, the results suggest that resveratrol may have differential effects on histone and non-histone protein acetylation, depending on the interplay between different HDACs.
2.7.4 Chrysin
Chrysin is a naturally-occurring flavone belonging to a group of polyphenolic compounds found in mushrooms, olive oil, tea, red wine, and passion fruit flowers, with antitumor effects in various cancer cells193. Pal-Bhadra et al. showed that chrysin inhibited HDAC2 and HDAC8 activity in melanoma (A375) cells leading to histone acetylation and p21WAF1 activation, via the recruitment of STAT1, STAT3, and STAT5 proteins to the corresponding gene promoter144. Further studies are warranted on the apparent selective inhibition by chrysin of HDAC2 and HDAC8.
2.7.5 Ellagic acid
Ellagic acid is a naturally-occurring polyphenolic acid abundant in fruits and nuts, such as pomegranate, berries, and walnuts. Chemopreventive effects of ellagic acid have been observed against various cancers, including oral cancer, where it was recently shown to block the VEGF signaling pathway in a hamster model of oral cancer145. The anti-angiogenic activity of ellagic acid can be mediated, in part, by blocking hypoxia and VEGF/VEGFR2 signaling pathways, through the suppression of HDAC6 and HIF-1α145.
2.8 Isoflavones
2.8.1 Quercetin
Quercetin is a flavonoid naturally occurring in fruits, vegetables, leaves, and grains. Based on pharmacophore modeling, quercetin was predicted to bind to the SIRT6 catalytic pocket147. Kokkonen et al. demonstrated that quercetin is a potent inhibitor of SIRT6, inducing histone acetylation (H3K56ac) in cultured cells148. Interestingly, quercetin recruited p300 HAT to the promoter region of the cyclooxygenase-2 (COX-2) gene, an important regulator of inflammation and tumorigenesis, thereby enhancing COX-2 expression in melanoma cells194.
2.8.2 Apigenin
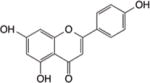
Apigenin (4′,5,7,-trihydroxyflavone) is a plant flavone abundant in grapefruit, parsley, and chamomile. Anticancer effects of apigenin involve mechanisms associated with cell cycle arrest and apoptosis195. Activation of p53 and the generation of ROS were observed in prostate cancer cells treated with apigenin196. Interestingly, Pandey et al. recently reported that apigenin inhibited the activity and expression of HDAC1 and HDAC3, leading to the acetylation of histones on the p21WAF1 gene, and apoptosis induction in human prostate cancer cells149. Apigenin also is known to suppress DNA methylation levels by decreasing mRNA and protein levels of DNMT proteins in skin cancer cells197.
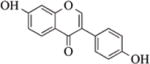
2.8.3 Genistein
Genistein is abundantly found in soybeans and soy products and has been shown to prevent and inhibit breast cancer by activating tumor suppressors genes such as p21, p16 and ERα198. Genistein has been reported to have HDAC inhibitory properties in cancer cells151. It has been shown to inhibit HDAC1 in colon cancer cells151, HDAC6 and SIRT1 in prostate cancer cells199,152, and cause histone methylation and acetylation changes in breast cancer cell lines200.
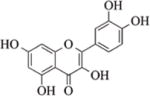
2.8.4 Luteolin
Luteolin is a flavone abundant in plants including fruits, vegetables, and medicinal herbs. Recently, Attoub et al. have shown that the anti-cancer and anti-metastatic properties of luteolin are associated with changes in histone acetylation156. A computational modeling study has shown that luteolin can bind to SIRT6 protein147.
2.9 Other dietary HDAC inhibitors
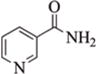
Queen bee phenotypes such as fertility and larvae size are determined at an early stage by differential nutrition with Royal jelly secreted from the nurse bees201. Recently, Spannhoff et al. demonstrated that Royal jelly has HDAC inhibitory activity associated with (E)-10-hydroxy-2-decenoic acid (10HDA), a major component of Royal jelly158. Structural similarities between 10HDA and Vorinostat might explain the increased global histone acetylation levels and re-activation of the FAS gene in NIH3T3 cells158.
Nicotinamide occurs in meat, fish, nuts, and mushrooms, as well as to a lesser extent in some vegetables. Nicotinamide is believed to be a pan-sirtuin inhibitor, and has demonstrated sirtuin inhibitory activity in gastric cancer202, lung cancer203, and breast cancer cells204. TIP5, the largest subunit of NoRC chromatin-remodeling complex, known to silence ribosomal RNA (rRNA) genes by establishing a heterochromatic structure can be acetylated by nicotinamide through inhibition of SIRT1205. Acetylation regulates the interaction of NoRC with promoter-associated RNA (pRNA), which in turn affects heterochromatin formation, nucleosome positioning and rDNA silencing205.
Psammaplin A present in margin sponge inhibits class I HDACs and induces cell cycle arrest and apoptosis206, 207. Baud et al. demonstrated that Psammaplin A is a highly potent HDAC1 inhibitor in vitro (IC50: 0.9 nM) and can induce acetylation on histones and α-tubulin164.
Caffeine is a central nervous system (CNS) stimulant of the methylxanthine class found in the seeds, nuts, or leaves of a number of plants, the most well-known source being the coffee plant. In pregnant rats, 120mg/kg/d caffeine reduced expression of a key transcription factor steroidogenic factor-1 (SF-1) in the fetal adrenal. It was found that caffeine treatment increased the mRNA expression of DNA methyltransferase (Dnmt) 1, Dnmt3a, Hdac1, and Hdac2. This correlated with increased methylation and decreased histone acetylation on the SF-1 promoter166.
2.10 Clinical Significance
In clinicalTrials.gov, there are several studies testing HDAC inhibitors and acetylation changes in cancer and other diseases. However, few trials address the role of dietary fat, fiber, cruciferous vegetable intake, or phytochemicals such as resveratrol and SFN on epigenetic endpoints. Resveratrol has been evaluated for its effect on sirtuins in cardiovascular, diabetes and Alzheimer’s disease. Sirtuin activity has also been tested in high-fat diet feeding studies in men where high fat has been reported to decrease NAD(+) and sirtuin activity in muscle biopsies, resulting in hyperacetylation of peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α)208.
The FACT study proposes to test the relationship between dietary fiber intake and colon cell turnover, to identify new molecular targets based on global protein acetylation analysis209. There are also several ongoing studies testing nicotinamide in Friedreich’s ataxia, polycystic kidney disease, lung cancer and Alzheimer’s disease.
Several studies on broccoli and/or SFN supplementation that evaluate HDAC inhibition have been reported in clinicalTrials.gov. One study aims at identifying the distribution of SFN and its metabolites and evaluates HDAC inhibition following broccoli sprout supplementation in subjects at risk for prostate cancer. The study also proposes to investigate the effects on DNA methylation status and proliferation markers in a pre-biopsy setting (Portland VA Medical Center, OR). Another study testing broccoli sprout extract supplements for treating ductal carcinoma in situ and/or atypical ductal hyperplasia has been completed. Isothiocyanates were detected in urine samples and changes in proliferation (Ki-67) and HDAC activity have been reported in peripheral blood mononuclear cells (PBMCs) of subjects who consumed the supplement as compared to baseline values (OHSU Knight Cancer Institute, OR).
Moreover, a single intake of 68 g broccoli sprouts in human subjects can inhibited HDAC activity in PBMC 3 h and 6 h following consumption170. More recent studies have confirmed that fresh broccoli sprouts are far more superior in lowering HDAC activity as compared to a broccoli sprout extract (BSE)210 or a myrosinase inactivated broccoli supplement211,212. However, mature broccoli (200 g) intake has been reported to be ineffective in lowering HDAC activity213. The lowering of HDAC activity by broccoli supplements in humans has been recently reported to activate p21 and long terminal repeats (LTRs)214. In addition, consuming BSE or a diet high on cruciferous vegetables reduced HDAC3 expression in circulating PBMCs and colon biopsies (unpublished data). These differential responses likely relate to the concentrations of bioactive ITC metabolites achieved in the circulation or target tissues.
3.0 CONCLUSIONS
HDAC inhibitors continue to be pursued as a promising class of anticancer agents across a broad spectrum of hematologic and solid neoplasms. These inhibitors are associated with diverse phenotypic outcomes, including suppression of cell proliferation, induction of differentiation, and activation of autophagy/apoptosis. In human clinical trials, however, off-target effects and resistance mechanisms have been observed with currently approved pan-HDAC inhibitors. HDAC-selective agents will likely improve therapeutic outcomes, while clarifying the molecular targets, especially the cadre of non-histone proteins affected by changes in acetylation status.
This review article addresses natural compounds, predominantly found in diet that hold great promise in terms of cancer chemoprevention and treatment. These compounds have both advantages and drawbacks as compared to anti-cancer drugs. Dietary phytochemicals tend to be relatively weak HDAC ligands unlike non-dietary compounds such as TSA, although intracellular metabolism can generate more potent HDAC inhibitor intermediates and provide a means of selective toxicity. In this regard, some dietary compounds can be viewed as “prodrugs” with the potential to generate HDAC inhibitor(s) at the appropriate time and place, taking advantage of dysregulated metabolic pathways in cancer cells.
Dietary compounds also work in synergy with pharmacological agents. The combination of anticancer agents and phytochemicals holds great promise as a therapeutic strategy for the future. However, rather than the random, trial-and-error approaches used historically, a rational development of appropriate combinations ideally should be based on mechanistic considerations. For example, green tea polyphenols acting on DNMTs combined with the HDAC inhibition of SFN might account for augmented ER receptor signaling and the inactivation of co-repressor complexes on the ER gene promoter215. Curcumin mechanisms impacting HATs and HDACs might cooperate with drugs such as valproic acid136 or hydroxamic acids216 to activate p38 MAPK-mediated signaling pathways and arrest the growth of human leukemia cells. Similar cooperativity might explain the synergy observed for TSA combined with HDAC inhibitors in Royal jelly158. Recently, the combination of EGCG and sodium butyrate was shown to efficiently induce Survivin expression in colon cancer cells217. Further, the combination of selenium compounds and green tea extract showed improved anticancer activity in vitro and in vivo218. These reports likely represent an incomplete view of all such studies in the literature, but they provide illustrative examples for future work on combination treatments through rational design.
Protein acetylation is a reversible post-translational modification that is tightly regulated in various cellular contexts. Acetylomics studies10, 219, 220 have identified many differentially acetylated proteins in addition to those summarized in Table 1. However, the role of specific HATs and/or HDACs is far from clear, and the functional significance of acetylation on these proteins requires further investigation. As we begin to define the different sub-categories of non-histone proteins regulated by acetylation, it is becoming apparent that the number of cellular targets implicated is much larger than initially anticipated. Isoform-specific HDAC inhibitors and/or knockdown of selected HDACs, coupled with genome-wide approaches, will likely reveal many new mechanistic targets for therapeutic intervention. These findings will be applied to specific cancer sub-types, as well as in other chronic conditions characterized by altered protein acetylation states and dysregulated gene expression.
Acknowledgments
Work cited from the authors’ laboratories was supported by NIH grants CA090890, CA122959, CA90176, CA111842, P30 ES00210, P30 ES023512, and a Chancellor’s Research Initiative from Texas A&M University.
Abbreviations
- HDACs
Histone deacetylase
- HATs
Histone acetyltransferases
- SIRT
Sirtuins
- FDA
US Food and Drug Administration
- STAT3
Signal transducer and activator of transcription 3
- AR
Androgen receptor
- COPD
Chronic obstructive pulmonary disease
- SCFAs
Short chain fatty acids
- TSA
Trichostatin A
- βOHB
β-hydroxybutyrate
- High-mobility-group
HMG
- Estrogen Receptor
ER
- GATA
Globin transcription factor
- EKLF
Erythroid Kruppel like fator
- YY1
Ying Yang 1
- Rb
Retinoblastoma protein
- HIF 1α
Hypoxia inducible factor
- CtIP
C-terminal binding protein interacting protein
- Hsp90
Heat Shock Protein 90
- FOXO
Forkhead box O
- PGC-1α
Peroxisome proliferator-activated receptor-γ coactivator-1α
- GCN5
General control nonderepressible 5
- CBP
CREB-binding protein
- PCAF
P300/CBP-associated factor
- TIP60
Tat interacting protein 60
- FXR
Farnesoid X receptor
- MSA
Methylseleninic acid
- SFN
Sulforaphane
- MSP
Methylselenopyruvate
- ITCs
Isothiocyanates
- allyl-ITC
Allyl isothiocyanate
- BITC
Benzyl isothiocyanate
- PHITC
Phenylhexyl isothiocyanate
- PEITC
Phenethyl isothiocyanate
- TRAMP
Tyrosine Rich Acidic Matrix Protein
- GSTP1
Glutathione S-transferase gene
- I3C
Indole-3-carbinol
- DIM
3,3′-diindolylmethane
- SEB
Staphylococcal enterotoxin B
- PN
Parthenolide
- AM
Allyl mercaptan
- DADS
Diallyl disulfide
- SAC
S-allylcysteine
- SAMC
S-allylmercaptocysteine
- NAT
N-acetyltransferase
- EGCG
Epigallocatechin-3-gallate
- NFκB
Nuclear factor kappa B
- IL6
Interleukin 6
- PcG
Polycomb group
- ROS
Reactive oxygen species
- SOCS
Suppressor of cytokine signaling
- COX-2
Cyclooxygenase-2
- pRNA
Promoter-associated RNA
- rRNA
Ribosomal RNA
- PBMC
Peripheral blood mononuclear cells
- BSE
Broccoli sprout extract
- LTRs
Long terminal repeats
- BMF
Bcl2 modifying factor
- SF-1
Steroidogenic factor-1
- Dnmt
DNA methyltransferase
- PBMCs
Peripheral blood mononuclear cells
Footnotes
Competing interests
The author(s) confirm no conflicts of interest associated with this review article.
References
- 1.Voss TC, Hager GL. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet. 2014;15(2):69–81. doi: 10.1038/nrg3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128(4):707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 3.Peserico A, Simone C. Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J Biomed Biotechnol. 2011;2011:371832. doi: 10.1155/2011/371832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12(1):7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 5.Marks PA, Xu WS. Histone deacetylase inhibitors: Potential in cancer therapy. J Cell Biochem. 2009;107(4):600–608. doi: 10.1002/jcb.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Subramanian Srividya, B SE, Wright John J, Espinoza-Delgado Igor, Piekarz RL. Clinical Toxicities of Histone Deacetylase Inhibitors. Pharmaceuticals. 2010;3:2751–2767. doi: 10.3390/ph3092751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajendran P, Ho E, Williams DE, Dashwood RH. Dietary phytochemicals, HDAC inhibition, and DNA damage/repair defects in cancer cells. Clin Epigenetics. 2011;3(1):4. doi: 10.1186/1868-7083-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajendran P, Williams DE, Ho E, Dashwood RH. Metabolism as a key to histone deacetylase inhibition. Crit Rev Biochem Mol Biol. 2011;46(3):181–199. doi: 10.3109/10409238.2011.557713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson ES, Kornbluth S. Life, death, and the metabolically controlled protein acetylome. Curr Opin Cell Biol. 2012;24(6):876–880. doi: 10.1016/j.ceb.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 11.Singh BN, Zhang G, Hwa YL, Li J, Dowdy SC, Jiang SW. Nonhistone protein acetylation as cancer therapy targets. Expert Rev Anticancer Ther. 2010;10(6):935–954. doi: 10.1586/era.10.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12(18):2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito A, Lai CH, Zhao X, Saito S, Hamilton MH, Appella E, Yao TP. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 2001;20(6):1331–1340. doi: 10.1093/emboj/20.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito A, Kawaguchi Y, Lai CH, Kovacs JJ, Higashimoto Y, Appella E, Yao TP. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 2002;21(22):6236–6245. doi: 10.1093/emboj/cdf616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107(2):137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 16.Nie Y, Erion DM, Yuan Z, Dietrich M, Shulman GI, Horvath TL, Gao Q. STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat Cell Biol. 2009;11(4):492–500. doi: 10.1038/ncb1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307(5707):269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 18.Sawicka A, Seiser C. Histone H3 phosphorylation - a versatile chromatin modification for different occasions. Biochimie. 2012;94(11):2193–2201. doi: 10.1016/j.biochi.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawicka A, Seiser C. Sensing core histone phosphorylation - a matter of perfect timing. Biochim Biophys Acta. 2014;1839(8):711–718. doi: 10.1016/j.bbagrm.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu M, Wang C, Reutens AT, Wang J, Angeletti RH, Siconolfi-Baez L, Ogryzko V, Avantaggiati ML, Pestell RG. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J Biol Chem. 2000;275(27):20853–20860. doi: 10.1074/jbc.M000660200. [DOI] [PubMed] [Google Scholar]
- 21.Delage B, Dashwood RH. Dietary manipulation of histone structure and function. Annu Rev Nutr. 2008;28:347–366. doi: 10.1146/annurev.nutr.28.061807.155354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barnes PJ. Role of HDAC2 in the pathophysiology of COPD. Annu Rev Physiol. 2009;71:451–464. doi: 10.1146/annurev.physiol.010908.163257. [DOI] [PubMed] [Google Scholar]
- 23.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11(2):437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 24.Matsuyama A, Shimazu T, Sumida Y, Saito A, Yoshimatsu Y, Seigneurin-Berny D, Osada H, Komatsu Y, Nishino N, Khochbin S, Horinouchi S, Yoshida M. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 2002;21(24):6820–6831. doi: 10.1093/emboj/cdf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417(6887):455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 26.Kang Y, Nian H, Rajendran P, Kim E, Dashwood WM, Pinto JT, Boardman LA, Thibodeau SN, Limburg PJ, Lohr CV, Bisson WH, Williams DE, Ho E, Dashwood RH. HDAC8 and STAT3 repress BMF gene activity in colon cancer cells. Cell Death Dis. 2014;5:e1476. doi: 10.1038/cddis.2014.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gui CY, Ngo L, Xu WS, Richon VM, Marks PA. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc Natl Acad Sci U S A. 2004;101(5):1241–1246. doi: 10.1073/pnas.0307708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin H, Lee YS, Lee YC. Sodium butyrate-induced DAPK-mediated apoptosis in human gastric cancer cells. Oncol Rep. 2012;27(4):1111–1115. doi: 10.3892/or.2011.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edmond V, Brambilla C, Brambilla E, Gazzeri S, Eymin B. SRSF2 is required for sodium butyrate-mediated p21(WAF1) induction and premature senescence in human lung carcinoma cell lines. Cell Cycle. 2011;10(12):1968–1977. doi: 10.4161/cc.10.12.15825. [DOI] [PubMed] [Google Scholar]
- 30.Qiu J, Gao Z, Shima H. Growth of human prostate cancer cells is significantly suppressed in vitro with sodium butyrate through apoptosis. Oncol Rep. 2012;27(1):160–167. doi: 10.3892/or.2011.1470. [DOI] [PubMed] [Google Scholar]
- 31.Berni Canani R, Di Costanzo M, Leone L. The epigenetic effects of butyrate: potential therapeutic implications for clinical practice. Clin Epigenetics. 2012;4(1):4. doi: 10.1186/1868-7083-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donohoe DR, Collins LB, Wali A, Bigler R, Sun W, Bultman SJ. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol Cell. 2012;48(4):612–626. doi: 10.1016/j.molcel.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sekhavat A, Sun JM, Davie JR. Competitive inhibition of histone deacetylase activity by trichostatin A and butyrate. Biochem Cell Biol. 2007;85(6):751–758. doi: 10.1139/o07-145. [DOI] [PubMed] [Google Scholar]
- 34.Thangaraju M, Carswell KN, Prasad PD, Ganapathy V. Colon cancer cells maintain low levels of pyruvate to avoid cell death caused by inhibition of HDAC1/HDAC3. Biochem J. 2009;417(1):379–389. doi: 10.1042/BJ20081132. [DOI] [PubMed] [Google Scholar]
- 35.Thangaraju M, Gopal E, Martin PM, Ananth S, Smith SB, Prasad PD, Sterneck E, Ganapathy V. SLC5A8 triggers tumor cell apoptosis through pyruvate-dependent inhibition of histone deacetylases. Cancer Res. 2006;66(24):11560–11564. doi: 10.1158/0008-5472.CAN-06-1950. [DOI] [PubMed] [Google Scholar]
- 36.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schauber J, Iffland K, Frisch S, Kudlich T, Schmausser B, Eck M, Menzel T, Gostner A, Luhrs H, Scheppach W. Histone-deacetylase inhibitors induce the cathelicidin LL-37 in gastrointestinal cells. Mol Immunol. 2004;41(9):847–854. doi: 10.1016/j.molimm.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 38.Ugrinova I, Pashev IG, Pasheva EA. Post-synthetic acetylation of HMGB1 protein modulates its interactions with supercoiled DNA. Mol Biol Rep. 2009;36(6):1399–1404. doi: 10.1007/s11033-008-9327-z. [DOI] [PubMed] [Google Scholar]
- 39.Licciardi PV, Ververis K, Karagiannis TC. Histone deacetylase inhibition and dietary short-chain Fatty acids. ISRN Allergy. 2011;2011:869647. doi: 10.5402/2011/869647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heidor R, Furtado KS, Ortega JF, de Oliveira TF, Tavares PE, Vieira A, Miranda ML, Purgatto E, Moreno FS. The chemopreventive activity of the histone deacetylase inhibitor tributyrin in colon carcinogenesis involves the induction of apoptosis and reduction of DNA damage. Toxicol Appl Pharmacol. 2014;276(2):129–135. doi: 10.1016/j.taap.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 41.Rocchi P, Tonelli R, Camerin C, Purgato S, Fronza R, Bianucci F, Guerra F, Pession A, Ferreri AM. p21Waf1/Cip1 is a common target induced by short-chain fatty acid HDAC inhibitors (valproic acid, tributyrin and sodium butyrate) in neuroblastoma cells. Oncol Rep. 2005;13(6):1139–1144. [PubMed] [Google Scholar]
- 42.de Conti A, Tryndyak V, Koturbash I, Heidor R, Kuroiwa-Trzmielina J, Ong TP, Beland FA, Moreno FS, Pogribny IP. The chemopreventive activity of the butyric acid prodrug tributyrin in experimental rat hepatocarcinogenesis is associated with p53 acetylation and activation of the p53 apoptotic signaling pathway. Carcinogenesis. 2013;34(8):1900–1906. doi: 10.1093/carcin/bgt124. [DOI] [PubMed] [Google Scholar]
- 43.Hacker S, Karl S, Mader I, Cristofanon S, Schweitzer T, Krauss J, Rutkowski S, Debatin KM, Fulda S. Histone deacetylase inhibitors prime medulloblastoma cells for chemotherapy-induced apoptosis by enhancing p53-dependent Bax activation. Oncogene. 2011;30(19):2275–2281. doi: 10.1038/onc.2010.599. [DOI] [PubMed] [Google Scholar]
- 44.Shimazu T, Hirschey MD, Newman J, He W, Shirakawa K, Le Moan N, Grueter CA, Lim H, Saunders LR, Stevens RD, Newgard CB, Farese RV, Jr, de Cabo R, Ulrich S, Akassoglou K, Verdin E. Suppression of oxidative stress by beta-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science. 2013;339(6116):211–214. doi: 10.1126/science.1227166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nian H, Delage B, Ho E, Dashwood RH. Modulation of histone deacetylase activity by dietary isothiocyanates and allyl sulfides: studies with sulforaphane and garlic organosulfur compounds. Environ Mol Mutagen. 2009;50(3):213–221. doi: 10.1002/em.20454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Espinosa JM, Emerson BM. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol Cell. 2001;8(1):57–69. doi: 10.1016/s1097-2765(01)00283-0. [DOI] [PubMed] [Google Scholar]
- 47.Luo J, Li M, Tang Y, Laszkowska M, Roeder RG, Gu W. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci U S A. 2004;101(8):2259–2264. doi: 10.1073/pnas.0308762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408(6810):377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 49.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107(2):149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 50.Yao YL, Yang WM, Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol Cell Biol. 2001;21(17):5979–5991. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Glenn DJ, Wang F, Chen S, Nishimoto M, Gardner DG. Endothelin-stimulated human B-type natriuretic peptide gene expression is mediated by Yin Yang 1 in association with histone deacetylase 2. Hypertension. 2009;53(3):549–555. doi: 10.1161/HYPERTENSIONAHA.108.125088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Munshi N, Agalioti T, Lomvardas S, Merika M, Chen G, Thanos D. Coordination of a transcriptional switch by HMGI(Y) acetylation. Science. 2001;293(5532):1133–1136. doi: 10.1126/science.293.5532.1133. [DOI] [PubMed] [Google Scholar]
- 53.Patel JH, Du Y, Ard PG, Phillips C, Carella B, Chen CJ, Rakowski C, Chatterjee C, Lieberman PM, Lane WS, Blobel GA, McMahon SB. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol Cell Biol. 2004;24(24):10826–10834. doi: 10.1128/MCB.24.24.10826-10834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gaughan L, Logan IR, Cook S, Neal DE, Robson CN. Tip60 and histone deacetylase 1 regulate androgen receptor activity through changes to the acetylation status of the receptor. J Biol Chem. 2002;277(29):25904–25913. doi: 10.1074/jbc.M203423200. [DOI] [PubMed] [Google Scholar]
- 55.Fu M, Wang C, Wang J, Zhang X, Sakamaki T, Yeung YG, Chang C, Hopp T, Fuqua SA, Jaffray E, Hay RT, Palvimo JJ, Janne OA, Pestell RG. Androgen receptor acetylation governs trans activation and MEKK1-induced apoptosis without affecting in vitro sumoylation and trans-repression function. Mol Cell Biol. 2002;22(10):3373–3388. doi: 10.1128/MCB.22.10.3373-3388.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang C, Fu M, Angeletti RH, Siconolfi-Baez L, Reutens AT, Albanese C, Lisanti MP, Katzenellenbogen BS, Kato S, Hopp T, Fuqua SA, Lopez GN, Kushner PJ, Pestell RG. Direct acetylation of the estrogen receptor alpha hinge region by p300 regulates transactivation and hormone sensitivity. J Biol Chem. 2001;276(21):18375–18383. doi: 10.1074/jbc.M100800200. [DOI] [PubMed] [Google Scholar]
- 57.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396(6711):594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 58.Hayakawa F, Towatari M, Ozawa Y, Tomita A, Privalsky ML, Saito H. Functional regulation of GATA-2 by acetylation. J Leukoc Biol. 2004;75(3):529–540. doi: 10.1189/jlb.0603389. [DOI] [PubMed] [Google Scholar]
- 59.Ozawa Y, Towatari M, Tsuzuki S, Hayakawa F, Maeda T, Miyata Y, Tanimoto M, Saito H. Histone deacetylase 3 associates with and represses the transcription factor GATA-2. Blood. 2001;98(7):2116–2123. doi: 10.1182/blood.v98.7.2116. [DOI] [PubMed] [Google Scholar]
- 60.Zhang W, Kadam S, Emerson BM, Bieker JJ. Site-specific acetylation by p300 or CREB binding protein regulates erythroid Kruppel-like factor transcriptional activity via its interaction with the SWI-SNF complex. Mol Cell Biol. 2001;21(7):2413–2422. doi: 10.1128/MCB.21.7.2413-2422.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Polesskaya A, Duquet A, Naguibneva I, Weise C, Vervisch A, Bengal E, Hucho F, Robin P, Harel-Bellan A. CREB-binding protein/p300 activates MyoD by acetylation. J Biol Chem. 2000;275(44):34359–34364. doi: 10.1074/jbc.M003815200. [DOI] [PubMed] [Google Scholar]
- 62.Sartorelli V, Puri PL, Hamamori Y, Ogryzko V, Chung G, Nakatani Y, Wang JY, Kedes L. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol Cell. 1999;4(5):725–734. doi: 10.1016/s1097-2765(00)80383-4. [DOI] [PubMed] [Google Scholar]
- 63.Mal A, Sturniolo M, Schiltz RL, Ghosh MK, Harter ML. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J. 2001;20(7):1739–1753. doi: 10.1093/emboj/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martinez-Balbas MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19(4):662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marzio G, Wagener C, Gutierrez MI, Cartwright P, Helin K, Giacca M. E2F family members are differentially regulated by reversible acetylation. J Biol Chem. 2000;275(15):10887–10892. doi: 10.1074/jbc.275.15.10887. [DOI] [PubMed] [Google Scholar]
- 66.Chen L, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293(5535):1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 67.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23(12):2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jeong JW, Bae MK, Ahn MY, Kim SH, Sohn TK, Bae MH, Yoo MA, Song EJ, Lee KJ, Kim KW. Regulation and destabilization of HIF-1alpha by ARD1-mediated acetylation. Cell. 2002;111(5):709–720. doi: 10.1016/s0092-8674(02)01085-1. [DOI] [PubMed] [Google Scholar]
- 69.Geng H, Harvey CT, Pittsenbarger J, Liu Q, Beer TM, Xue C, Qian DZ. HDAC4 protein regulates HIF1alpha protein lysine acetylation and cancer cell response to hypoxia. J Biol Chem. 2011;286(44):38095–38102. doi: 10.1074/jbc.M111.257055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gronroos E, Hellman U, Heldin CH, Ericsson J. Control of Smad7 stability by competition between acetylation and ubiquitination. Mol Cell. 2002;10(3):483–493. doi: 10.1016/s1097-2765(02)00639-1. [DOI] [PubMed] [Google Scholar]
- 71.Bannister AJ, Miska EA, Gorlich D, Kouzarides T. Acetylation of importin-alpha nuclear import factors by CBP/p300. Curr Biol. 2000;10(8):467–470. doi: 10.1016/s0960-9822(00)00445-0. [DOI] [PubMed] [Google Scholar]
- 72.Cohen HY, Lavu S, Bitterman KJ, Hekking B, Imahiyerobo TA, Miller C, Frye R, Ploegh H, Kessler BM, Sinclair DA. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol Cell. 2004;13(5):627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 73.Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18(5):601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 74.Zhang Q, Yao H, Vo N, Goodman RH. Acetylation of adenovirus E1A regulates binding of the transcriptional corepressor CtBP. Proc Natl Acad Sci U S A. 2000;97(26):14323–14328. doi: 10.1073/pnas.011283598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329(5997):1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Ge X, Jin Q, Zhang F, Yan T, Zhai Q. PCAF acetylates {beta}-catenin and improves its stability. Mol Biol Cell. 2009;20(1):419–427. doi: 10.1091/mbc.E08-08-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wolf D, Rodova M, Miska EA, Calvet JP, Kouzarides T. Acetylation of beta-catenin by CREB-binding protein (CBP) J Biol Chem. 2002;277(28):25562–25567. doi: 10.1074/jbc.M201196200. [DOI] [PubMed] [Google Scholar]
- 78.Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, Bhimavarapu A, Luikenhuis S, de Cabo R, Fuchs C, Hahn WC, Guarente LP, Sinclair DA. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS One. 2008;3(4):e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Y, Zhang X, Polakiewicz RD, Yao TP, Comb MJ. HDAC6 is required for epidermal growth factor-induced beta-catenin nuclear localization. J Biol Chem. 2008;283(19):12686–12690. doi: 10.1074/jbc.C700185200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Riolo MT, Cooper ZA, Holloway MP, Cheng Y, Bianchi C, Yakirevich E, Ma L, Chin YE, Altura RA. Histone deacetylase 6 (HDAC6) deacetylates survivin for its nuclear export in breast cancer. J Biol Chem. 2012;287(14):10885–10893. doi: 10.1074/jbc.M111.308791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, Miyagishi M, Nakajima T, Fukamizu A. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci U S A. 2004;101(27):10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116(4):551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 83.Wang F, Nguyen M, Qin FX, Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell. 2007;6(4):505–514. doi: 10.1111/j.1474-9726.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- 84.Jacobs KM, Pennington JD, Bisht KS, Aykin-Burns N, Kim HS, Mishra M, Sun L, Nguyen P, Ahn BH, Leclerc J, Deng CX, Spitz DR, Gius D. SIRT3 interacts with the daf-16 homolog FOXO3a in the mitochondria, as well as increases FOXO3a dependent gene expression. Int J Biol Sci. 2008;4(5):291–299. doi: 10.7150/ijbs.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. 2006;3(6):429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 86.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J Biol Chem. 2005;280(16):16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 87.Fang S, Tsang S, Jones R, Ponugoti B, Yoon H, Wu SY, Chiang CM, Willson TM, Kemper JK. The p300 acetylase is critical for ligand-activated farnesoid X receptor (FXR) induction of SHP. J Biol Chem. 2008;283(50):35086–35095. doi: 10.1074/jbc.M803531200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kemper JK, Xiao Z, Ponugoti B, Miao J, Fang S, Kanamaluru D, Tsang S, Wu SY, Chiang CM, Veenstra TD. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metab. 2009;10(5):392–404. doi: 10.1016/j.cmet.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dickinson SE, Rusche JJ, Bec SL, Horn DJ, Janda J, Rim SH, Smith CL, Bowden GT. The effect of sulforaphane on histone deacetylase activity in keratinocytes: Differences between in vitro and in vivo analyses. Mol Carcinog. 2014 doi: 10.1002/mc.22224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Su ZY, Zhang C, Lee JH, Shu L, Wu TY, Khor TO, Conney AH, Lu YP, Kong AN. Requirement and epigenetics reprogramming of Nrf2 in suppression of tumor promoter TPA-induced mouse skin cell transformation by sulforaphane. Cancer Prev Res (Phila) 2014;7(3):319–329. doi: 10.1158/1940-6207.CAPR-13-0313-T. [DOI] [PubMed] [Google Scholar]
- 91.Zhang C, Su ZY, Khor TO, Shu L, Kong AN. Sulforaphane enhances Nrf2 expression in prostate cancer TRAMP C1 cells through epigenetic regulation. Biochem Pharmacol. 2013;85(9):1398–1404. doi: 10.1016/j.bcp.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fan H, Zhang R, Tesfaye D, Tholen E, Looft C, Holker M, Schellander K, Cinar MU. Sulforaphane causes a major epigenetic repression of myostatin in porcine satellite cells. Epigenetics. 2012;7(12):1379–1390. doi: 10.4161/epi.22609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Clarke JD, Hsu A, Yu Z, Dashwood RH, Ho E. Differential effects of sulforaphane on histone deacetylases, cell cycle arrest and apoptosis in normal prostate cells versus hyperplastic and cancerous prostate cells. Mol Nutr Food Res. 2011;55(7):999–1009. doi: 10.1002/mnfr.201000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rajendran P, Kidane AI, Yu TW, Dashwood WM, Bisson WH, Lohr CV, Ho E, Williams DE, Dashwood RH. HDAC turnover, CtIP acetylation and dysregulated DNA damage signaling in colon cancer cells treated with sulforaphane and related dietary isothiocyanates. Epigenetics. 2013;8(6):612–623. doi: 10.4161/epi.24710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lea MA, Randolph VM, Lee JE, desBordes C. Induction of histone acetylation in mouse erythroleukemia cells by some organosulfur compounds including allyl isothiocyanate. Int J Cancer. 2001;92(6):784–789. doi: 10.1002/ijc.1277. [DOI] [PubMed] [Google Scholar]
- 96.Batra S, Sahu RP, Kandala PK, Srivastava SK. Benzyl isothiocyanate-mediated inhibition of histone deacetylase leads to NF-kappaB turnoff in human pancreatic carcinoma cells. Mol Cancer Ther. 2010;9(6):1596–1608. doi: 10.1158/1535-7163.MCT-09-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xiao D, Bommareddy A, Kim SH, Sehrawat A, Hahm ER, Singh SV. Benzyl isothiocyanate causes FoxO1-mediated autophagic death in human breast cancer cells. PLoS One. 2012;7(3):e32597. doi: 10.1371/journal.pone.0032597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ma X, Fang Y, Beklemisheva A, Dai W, Feng J, Ahmed T, Liu D, Chiao JW. Phenylhexyl isothiocyanate inhibits histone deacetylases and remodels chromatins to induce growth arrest in human leukemia cells. Int J Oncol. 2006;28(5):1287–1293. [PubMed] [Google Scholar]
- 99.Beklemisheva AA, Fang Y, Feng J, Ma X, Dai W, Chiao JW. Epigenetic mechanism of growth inhibition induced by phenylhexyl isothiocyanate in prostate cancer cells. Anticancer Res. 2006;26(2A):1225–1230. [PubMed] [Google Scholar]
- 100.Lu Q, Lin X, Feng J, Zhao X, Gallagher R, Lee MY, Chiao JW, Liu D. Phenylhexyl isothiocyanate has dual function as histone deacetylase inhibitor and hypomethylating agent and can inhibit myeloma cell growth by targeting critical pathways. J Hematol Oncol. 2008;1:6. doi: 10.1186/1756-8722-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xiao L, Huang Y, Zhen R, Chiao JW, Liu D, Ma X. Deficient histone acetylation in acute leukemia and the correction by an isothiocyanate. Acta Haematol. 2010;123(2):71–76. doi: 10.1159/000264628. [DOI] [PubMed] [Google Scholar]
- 102.Wang LG, Liu XM, Fang Y, Dai W, Chiao FB, Puccio GM, Feng J, Liu D, Chiao JW. De-repression of the p21 promoter in prostate cancer cells by an isothiocyanate via inhibition of HDACs and c-Myc. Int J Oncol. 2008;33(2):375–380. [PubMed] [Google Scholar]
- 103.Cang S, Ma Y, Chiao JW, Liu D. Phenethyl isothiocyanate and paclitaxel synergistically enhanced apoptosis and alpha-tubulin hyperacetylation in breast cancer cells. Exp Hematol Oncol. 2014;3(1):5. doi: 10.1186/2162-3619-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li Y, Li X, Guo B. Chemopreventive agent 3,3′-diindolylmethane selectively induces proteasomal degradation of class I histone deacetylases. Cancer Res. 2010;70(2):646–654. doi: 10.1158/0008-5472.CAN-09-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Beaver LM, Yu TW, Sokolowski EI, Williams DE, Dashwood RH, Ho E. 3,3′-Diindolylmethane, but not indole-3-carbinol, inhibits histone deacetylase activity in prostate cancer cells. Toxicol Appl Pharmacol. 2012;263(3):345–351. doi: 10.1016/j.taap.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bhatnagar N, Li X, Chen Y, Zhou X, Garrett SH, Guo B. 3,3′-diindolylmethane enhances the efficacy of butyrate in colon cancer prevention through down-regulation of survivin. Cancer Prev Res (Phila) 2009;2(6):581–589. doi: 10.1158/1940-6207.CAPR-08-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Busbee PB, Nagarkatti M, Nagarkatti PS. Natural indoles, indole-3-carbinol and 3,3′-diindolymethane, inhibit T cell activation by staphylococcal enterotoxin B through epigenetic regulation involving HDAC expression. Toxicol Appl Pharmacol. 2014;274(1):7–16. doi: 10.1016/j.taap.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gopal YN, Arora TS, Van Dyke MW. Parthenolide specifically depletes histone deacetylase 1 protein and induces cell death through ataxia telangiectasia mutated. Chem Biol. 2007;14(7):813–823. doi: 10.1016/j.chembiol.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 109.Yu FS, Yu CS, Lin JP, Chen SC, Lai WW, Chung JG. Diallyl disulfide inhibits N-acetyltransferase activity and gene expression in human esophagus epidermoid carcinoma CE 81T/VGH cells. Food Chem Toxicol. 2005;43(7):1029–1036. doi: 10.1016/j.fct.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 110.Lee JH, Kim KA, Kwon KB, Kim EK, Lee YR, Song MY, Koo JH, Ka SO, Park JW, Park BH. Diallyl disulfide accelerates adipogenesis in 3T3-L1 cells. Int J Mol Med. 2007;20(1):59–64. [PubMed] [Google Scholar]
- 111.Zhao J, Huang WG, He J, Tan H, Liao QJ, Su Q. Diallyl disulfide suppresses growth of HL-60 cell through increasing histone acetylation and p21WAF1 expression in vivo and in vitro. Acta Pharmacol Sin. 2006;27(11):1459–1466. doi: 10.1111/j.1745-7254.2006.00433.x. [DOI] [PubMed] [Google Scholar]
- 112.Altonsy MO, Habib TN, Andrews SC. Diallyl disulfide-induced apoptosis in a breast-cancer cell line (MCF-7) may be caused by inhibition of histone deacetylation. Nutr Cancer. 2012;64(8):1251–1260. doi: 10.1080/01635581.2012.721156. [DOI] [PubMed] [Google Scholar]
- 113.Druesne-Pecollo N, Pagniez A, Thomas M, Cherbuy C, Duee PH, Martel P, Chaumontet C. Diallyl disulfide increases CDKN1A promoter-associated histone acetylation in human colon tumor cell lines. J Agric Food Chem. 2006;54(20):7503–7507. doi: 10.1021/jf061369w. [DOI] [PubMed] [Google Scholar]
- 114.Nian H, Delage B, Pinto JT, Dashwood RH. Allyl mercaptan, a garlic-derived organosulfur compound, inhibits histone deacetylase and enhances Sp3 binding on the P21WAF1 promoter. Carcinogenesis. 2008;29(9):1816–1824. doi: 10.1093/carcin/bgn165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lea MA, Randolph VM, Patel M. Increased acetylation of histones induced by diallyl disulfide and structurally related molecules. Int J Oncol. 1999;15(2):347–352. doi: 10.3892/ijo.15.2.347. [DOI] [PubMed] [Google Scholar]
- 116.Lea MA, Randolph VM. Induction of histone acetylation in rat liver and hepatoma by organosulfur compounds including diallyl disulfide. Anticancer Res. 2001;21(4A):2841–2845. [PubMed] [Google Scholar]
- 117.Lea MA, Rasheed M, Randolph VM, Khan F, Shareef A, desBordes C. Induction of histone acetylation and inhibition of growth of mouse erythroleukemia cells by S-allylmercaptocysteine. Nutr Cancer. 2002;43(1):90–102. doi: 10.1207/S15327914NC431_11. [DOI] [PubMed] [Google Scholar]
- 118.Hu C, Liu M, Zhang W, Xu Q, Ma K, Chen L, Wang Z, He S, Zhu H, Xu N. Upregulation of KLF4 by methylseleninic acid in human esophageal squamous cell carcinoma cells: Modification of histone H3 acetylation through HAT/HDAC interplay. Mol Carcinog. 2014 doi: 10.1002/mc.22174. [DOI] [PubMed] [Google Scholar]
- 119.Kassam S, Goenaga-Infante H, Maharaj L, Hiley CT, Juliger S, Joel SP. Methylseleninic acid inhibits HDAC activity in diffuse large B-cell lymphoma cell lines. Cancer Chemother Pharmacol. 2011;68(3):815–821. doi: 10.1007/s00280-011-1649-1. [DOI] [PubMed] [Google Scholar]
- 120.Xiang N, Zhao R, Song G, Zhong W. Selenite reactivates silenced genes by modifying DNA methylation and histones in prostate cancer cells. Carcinogenesis. 2008;29(11):2175–2181. doi: 10.1093/carcin/bgn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nian H, Bisson WH, Dashwood WM, Pinto JT, Dashwood RH. Alpha-keto acid metabolites of organoselenium compounds inhibit histone deacetylase activity in human colon cancer cells. Carcinogenesis. 2009;30(8):1416–1423. doi: 10.1093/carcin/bgp147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pandey M, Shukla S, Gupta S. Promoter demethylation and chromatin remodeling by green tea polyphenols leads to re-expression of GSTP1 in human prostate cancer cells. Int J Cancer. 2010;126(11):2520–2533. doi: 10.1002/ijc.24988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Deb G, Thakur VS, Limaye AM, Gupta S. Epigenetic induction of tissue inhibitor of matrix metalloproteinase-3 by green tea polyphenols in breast cancer cells. Mol Carcinog. 2014 doi: 10.1002/mc.22121. [DOI] [PubMed] [Google Scholar]
- 124.Choi KC, Jung MG, Lee YH, Yoon JC, Kwon SH, Kang HB, Kim MJ, Cha JH, Kim YJ, Jun WJ, Lee JM, Yoon HG. Epigallocatechin-3-gallate, a histone acetyltransferase inhibitor, inhibits EBV-induced B lymphocyte transformation via suppression of RelA acetylation. Cancer Res. 2009;69(2):583–592. doi: 10.1158/0008-5472.CAN-08-2442. [DOI] [PubMed] [Google Scholar]
- 125.Nandakumar V, Vaid M, Katiyar SK. (−)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p16INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis. 2011;32(4):537–544. doi: 10.1093/carcin/bgq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li Y, Yuan YY, Meeran SM, Tollefsbol TO. Synergistic epigenetic reactivation of estrogen receptor-alpha (ERalpha) by combined green tea polyphenol and histone deacetylase inhibitor in ERalpha-negative breast cancer cells. Mol Cancer. 2010;9:274. doi: 10.1186/1476-4598-9-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Han DW, Lee MH, Kim B, Lee JJ, Hyon SH, Park JC. Preventive effects of epigallocatechin-3-O-gallate against replicative senescence associated with p53 acetylation in human dermal fibroblasts. Oxid Med Cell Longev. 2012;2012:850684. doi: 10.1155/2012/850684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Thakur VS, Gupta K, Gupta S. Green tea polyphenols increase p53 transcriptional activity and acetylation by suppressing class I histone deacetylases. Int J Oncol. 2012;41(1):353–361. doi: 10.3892/ijo.2012.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu J, Tang Y, Feng Z, Hou C, Wang H, Yan J, Liu J, Shen W, Zang W, Liu J, Long J. Acetylated FoxO1 mediates high-glucose induced autophagy in H9c2 cardiomyoblasts: regulation by a polyphenol -(−)-epigallocatechin-3-gallate. Metabolism. 2014;63(10):1314–1323. doi: 10.1016/j.metabol.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 130.Lee YH, Kwak J, Choi HK, Choi KC, Kim S, Lee J, Jun W, Park HJ, Yoon HG. EGCG suppresses prostate cancer cell growth modulating acetylation of androgen receptor by anti-histone acetyltransferase activity. Int J Mol Med. 2012;30(1):69–74. doi: 10.3892/ijmm.2012.966. [DOI] [PubMed] [Google Scholar]
- 131.Ko H, So Y, Jeon H, Jeong MH, Choi HK, Ryu SH, Lee SW, Yoon HG, Choi KC. TGF-beta1-induced epithelial-mesenchymal transition and acetylation of Smad2 and Smad3 are negatively regulated by EGCG in human A549 lung cancer cells. Cancer Lett. 2013;335(1):205–213. doi: 10.1016/j.canlet.2013.02.018. [DOI] [PubMed] [Google Scholar]
- 132.Chakraborty S, Das K, Saha S, Mazumdar M, Manna A, Chakraborty S, Mukherjee S, Khan P, Adhikary A, Mohanty S, Chattopadhyay S, Biswas SC, Sa G, Das T. Nuclear Matrix Protein SMAR1 Represses c-Fos-mediated HPV18 E6 Transcription through Alteration of Chromatin Histone Deacetylation. J Biol Chem. 2014;289(42):29074–29085. doi: 10.1074/jbc.M114.564872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lee SJ, Krauthauser C, Maduskuie V, Fawcett PT, Olson JM, Rajasekaran SA. Curcumin-induced HDAC inhibition and attenuation of medulloblastoma growth in vitro and in vivo. BMC Cancer. 2011;11:144. doi: 10.1186/1471-2407-11-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sarkar R, Mukherjee A, Mukherjee S, Biswas R, Biswas J, Roy M. Curcumin augments the efficacy of antitumor drugs used in leukemia by modulation of heat shock proteins via HDAC6. J Environ Pathol Toxicol Oncol. 2014;33(3):247–263. doi: 10.1615/jenvironpatholtoxicoloncol.2014010913. [DOI] [PubMed] [Google Scholar]
- 135.Chen CQ, Yu K, Yan QX, Xing CY, Chen Y, Yan Z, Shi YF, Zhao KW, Gao SM. Pure curcumin increases the expression of SOCS1 and SOCS3 in myeloproliferative neoplasms through suppressing class I histone deacetylases. Carcinogenesis. 2013;34(7):1442–1449. doi: 10.1093/carcin/bgt070. [DOI] [PubMed] [Google Scholar]
- 136.Chen J, Wang G, Wang L, Kang J, Wang J. Curcumin p38-dependently enhances the anticancer activity of valproic acid in human leukemia cells. Eur J Pharm Sci. 2010;41(2):210–218. doi: 10.1016/j.ejps.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 137.Collins HM, Abdelghany MK, Messmer M, Yue B, Deeves SE, Kindle KB, Mantelingu K, Aslam A, Winkler GS, Kundu TK, Heery DM. Differential effects of garcinol and curcumin on histone and p53 modifications in tumour cells. BMC Cancer. 2013;13:37. doi: 10.1186/1471-2407-13-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shankar S, Srivastava RK. Involvement of Bcl-2 family members, phosphatidylinositol 3′-kinase/AKT and mitochondrial p53 in curcumin (diferulolylmethane)-induced apoptosis in prostate cancer. Int J Oncol. 2007;30(4):905–918. [PubMed] [Google Scholar]
- 139.Venturelli S, Berger A, Bocker A, Busch C, Weiland T, Noor S, Leischner C, Schleicher S, Mayer M, Weiss TS, Bischoff SC, Lauer UM, Bitzer M. Resveratrol as a pan-HDAC inhibitor alters the acetylation status of histone [corrected] proteins in human-derived hepatoblastoma cells. PLoS One. 2013;8(8):e73097. doi: 10.1371/journal.pone.0073097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sayd S, Thirant C, El-Habr EA, Lipecka J, Dubois LG, Bogeas A, Tahiri-Jouti N, Chneiweiss H, Junier MP. Sirtuin-2 activity is required for glioma stem cell proliferation arrest but not necrosis induced by resveratrol. Stem Cell Rev. 2014;10(1):103–113. doi: 10.1007/s12015-013-9465-0. [DOI] [PubMed] [Google Scholar]
- 141.Kai L, Samuel SK, Levenson AS. Resveratrol enhances p53 acetylation and apoptosis in prostate cancer by inhibiting MTA1/NuRD complex. Int J Cancer. 2010;126(7):1538–1548. doi: 10.1002/ijc.24928. [DOI] [PubMed] [Google Scholar]
- 142.Higashida K, Kim SH, Jung SR, Asaka M, Holloszy JO, Han DH. Effects of resveratrol and SIRT1 on PGC-1alpha activity and mitochondrial biogenesis: a reevaluation. PLoS Biol. 2013;11(7):e1001603. doi: 10.1371/journal.pbio.1001603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Frazzi R, Valli R, Tamagnini I, Casali B, Latruffe N, Merli F. Resveratrol-mediated apoptosis of hodgkin lymphoma cells involves SIRT1 inhibition and FOXO3a hyperacetylation. Int J Cancer. 2013;132(5):1013–1021. doi: 10.1002/ijc.27748. [DOI] [PubMed] [Google Scholar]
- 144.Pal-Bhadra M, Ramaiah MJ, Reddy TL, Krishnan A, Pushpavalli SN, Babu KS, Tiwari AK, Rao JM, Yadav JS, Bhadra U. Plant HDAC inhibitor chrysin arrest cell growth and induce p21WAF1 by altering chromatin of STAT response element in A375 cells. BMC Cancer. 2012;12:180. doi: 10.1186/1471-2407-12-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kowshik J, Giri H, Kishore TK, Kesavan R, Vankudavath RN, Reddy GB, Dixit M, Nagini S. Ellagic Acid Inhibits VEGF/VEGFR2, PI3K/Akt and MAPK Signaling Cascades in the Hamster Cheek Pouch Carcinogenesis Model. Anticancer Agents Med Chem. 2014;14(9):1249–1260. doi: 10.2174/1871520614666140723114217. [DOI] [PubMed] [Google Scholar]
- 146.Kang I, Okla M, Chung S. Ellagic acid inhibits adipocyte differentiation through coactivator-associated arginine methyltransferase 1-mediated chromatin modification. J Nutr Biochem. 2014;25(9):946–953. doi: 10.1016/j.jnutbio.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 147.Ravichandran S, Singh N, Donnelly D, Migliore M, Johnson P, Fishwick C, Luke BT, Martin B, Maudsley S, Fugmann SD, Moaddel R. Pharmacophore model of the quercetin binding site of the SIRT6 protein. J Mol Graph Model. 2014;49:38–46. doi: 10.1016/j.jmgm.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kokkonen P, Rahnasto-Rilla M, Mellini P, Jarho E, Lahtela-Kakkonen M, Kokkola T. Studying SIRT6 regulation using H3K56 based substrate and small molecules. Eur J Pharm Sci. 2014;63:71–76. doi: 10.1016/j.ejps.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 149.Pandey M, Kaur P, Shukla S, Abbas A, Fu P, Gupta S. Plant flavone apigenin inhibits HDAC and remodels chromatin to induce growth arrest and apoptosis in human prostate cancer cells: in vitro and in vivo study. Mol Carcinog. 2012;51(12):952–962. doi: 10.1002/mc.20866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Phillip CJ, Giardina CK, Bilir B, Cutler DJ, Lai YH, Kucuk O, Moreno CS. Genistein cooperates with the histone deacetylase inhibitor vorinostat to induce cell death in prostate cancer cells. BMC Cancer. 2012;12:145. doi: 10.1186/1471-2407-12-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Groh IA, Chen C, Luske C, Cartus AT, Esselen M. Plant polyphenols and oxidative metabolites of the herbal alkenylbenzene methyleugenol suppress histone deacetylase activity in human colon carcinoma cells. J Nutr Metab. 2013;2013:821082. doi: 10.1155/2013/821082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kikuno N, Shiina H, Urakami S, Kawamoto K, Hirata H, Tanaka Y, Majid S, Igawa M, Dahiya R. Genistein mediated histone acetylation and demethylation activates tumor suppressor genes in prostate cancer cells. Int J Cancer. 2008;123(3):552–560. doi: 10.1002/ijc.23590. [DOI] [PubMed] [Google Scholar]
- 153.Wang H, Li Q, Chen H. Genistein affects histone modifications on Dickkopf-related protein 1 (DKK1) gene in SW480 human colon cancer cell line. PLoS One. 2012;7(7):e40955. doi: 10.1371/journal.pone.0040955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Majid S, Dar AA, Shahryari V, Hirata H, Ahmad A, Saini S, Tanaka Y, Dahiya AV, Dahiya R. Genistein reverses hypermethylation and induces active histone modifications in tumor suppressor gene B-Cell translocation gene 3 in prostate cancer. Cancer. 2010;116(1):66–76. doi: 10.1002/cncr.24662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kim HJ, Lee W, Yun JM. Luteolin inhibits hyperglycemia-induced proinflammatory cytokine production and its epigenetic mechanism in human monocytes. Phytother Res. 2014;28(9):1383–1391. doi: 10.1002/ptr.5141. [DOI] [PubMed] [Google Scholar]
- 156.Attoub S, Hassan AH, Vanhoecke B, Iratni R, Takahashi T, Gaben AM, Bracke M, Awad S, John A, Kamalboor HA, Al Sultan MA, Arafat K, Gespach C, Petroianu G. Inhibition of cell survival, invasion, tumor growth and histone deacetylase activity by the dietary flavonoid luteolin in human epithelioid cancer cells. Eur J Pharmacol. 2011;651(1–3):18–25. doi: 10.1016/j.ejphar.2010.10.063. [DOI] [PubMed] [Google Scholar]
- 157.Wang Wen Xiang, T LQ, Huang Qiang, Wu Xiao Bo, Zeng Zhi Jiang. Effects of 10-Hydroxy-2-decenoic acid on the development of honey bee (Apis mellifera) larvae. International bee research association. 2014;53(1):171–176. [Google Scholar]
- 158.Spannhoff A, Kim YK, Raynal NJ, Gharibyan V, Su MB, Zhou YY, Li J, Castellano S, Sbardella G, Issa JP, Bedford MT. Histone deacetylase inhibitor activity in royal jelly might facilitate caste switching in bees. EMBO Rep. 2011;12(3):238–243. doi: 10.1038/embor.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang T, Cui H, Ma N, Jiang Y. Nicotinamide-mediated inhibition of SIRT1 deacetylase is associated with the viability of cancer cells exposed to antitumor agents and apoptosis. Oncol Lett. 2013;6(2):600–604. doi: 10.3892/ol.2013.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Scholz C, Weinert BT, Wagner SA, Beli P, Miyake Y, Qi J, Jensen LJ, Streicher W, McCarthy AR, Westwood NJ, Lain S, Cox J, Matthias P, Mann M, Bradner JE, Choudhary C. Acetylation site specificities of lysine deacetylase inhibitors in human cells. Nat Biotechnol. 2015;33(4):415–423. doi: 10.1038/nbt.3130. [DOI] [PubMed] [Google Scholar]
- 161.Puca R, Nardinocchi L, Sacchi A, Rechavi G, Givol D, D’Orazi G. HIPK2 modulates p53 activity towards pro-apoptotic transcription. Mol Cancer. 2009;8:85. doi: 10.1186/1476-4598-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Solomon JM, Pasupuleti R, Xu L, McDonagh T, Curtis R, DiStefano PS, Huber LJ. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol Cell Biol. 2006;26(1):28–38. doi: 10.1128/MCB.26.1.28-38.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bereshchenko OR, Gu W, Dalla-Favera R. Acetylation inactivates the transcriptional repressor BCL6. Nat Genet. 2002;32(4):606–613. doi: 10.1038/ng1018. [DOI] [PubMed] [Google Scholar]
- 164.Baud MG, Leiser T, Haus P, Samlal S, Wong AC, Wood RJ, Petrucci V, Gunaratnam M, Hughes SM, Buluwela L, Turlais F, Neidle S, Meyer-Almes FJ, White AJ, Fuchter MJ. Defining the mechanism of action and enzymatic selectivity of psammaplin A against its epigenetic targets. J Med Chem. 2012;55(4):1731–1750. doi: 10.1021/jm2016182. [DOI] [PubMed] [Google Scholar]
- 165.Pereira R, Benedetti R, Perez-Rodriguez S, Nebbioso A, Garcia-Rodriguez J, Carafa V, Stuhldreier M, Conte M, Rodriguez-Barrios F, Stunnenberg HG, Gronemeyer H, Altucci L, de Lera AR. Indole-derived psammaplin A analogues as epigenetic modulators with multiple inhibitory activities. J Med Chem. 2012;55(22):9467–9491. doi: 10.1021/jm300618u. [DOI] [PubMed] [Google Scholar]
- 166.Ping J, Wang JF, Liu L, Yan YE, Liu F, Lei YY, Wang H. Prenatal caffeine ingestion induces aberrant DNA methylation and histone acetylation of steroidogenic factor 1 and inhibits fetal adrenal steroidogenesis. Toxicology. 2014;321:53–61. doi: 10.1016/j.tox.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 167.Stan SD, Singh SV, Whitcomb DC, Brand RE. Phenethyl isothiocyanate inhibits proliferation and induces apoptosis in pancreatic cancer cells in vitro and in a MIAPaca2 xenograft animal model. Nutr Cancer. 2014;66(4):747–755. doi: 10.1080/01635581.2013.795979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hayes JD, Kelleher MO, Eggleston IM. The cancer chemopreventive actions of phytochemicals derived from glucosinolates. Eur J Nutr. 2008;47(Suppl 2):73–88. doi: 10.1007/s00394-008-2009-8. [DOI] [PubMed] [Google Scholar]
- 169.Myzak MC, Karplus PA, Chung FL, Dashwood RH. A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer Res. 2004;64(16):5767–5774. doi: 10.1158/0008-5472.CAN-04-1326. [DOI] [PubMed] [Google Scholar]
- 170.Myzak MC, Tong P, Dashwood WM, Dashwood RH, Ho E. Sulforaphane retards the growth of human PC-3 xenografts and inhibits HDAC activity in human subjects. Exp Biol Med (Maywood) 2007;232(2):227–234. [PMC free article] [PubMed] [Google Scholar]
- 171.Dashwood RH, Myzak MC, Ho E. Dietary HDAC inhibitors: time to rethink weak ligands in cancer chemoprevention? Carcinogenesis. 2006;27(2):344–349. doi: 10.1093/carcin/bgi253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rajendran P, Delage B, Dashwood WM, Yu TW, Wuth B, Williams DE, Ho E, Dashwood RH. Histone deacetylase turnover and recovery in sulforaphane-treated colon cancer cells: competing actions of 14-3-3 and Pin1 in HDAC3/SMRT corepressor complex dissociation/reassembly. Mol Cancer. 2011;10:68. doi: 10.1186/1476-4598-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gibbs A, Schwartzman J, Deng V, Alumkal J. Sulforaphane destabilizes the androgen receptor in prostate cancer cells by inactivating histone deacetylase 6. Proc Natl Acad Sci U S A. 2009;106(39):16663–16668. doi: 10.1073/pnas.0908908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Katoch O, Kumar A, Adhikari JS, Dwarakanath BS, Agrawala PK. Sulforaphane mitigates genotoxicity induced by radiation and anticancer drugs in human lymphocytes. Mutat Res. 2013;758(1–2):29–34. doi: 10.1016/j.mrgentox.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 175.Enriquez GG, Rizvi SA, D’Souza MJ, Do DP. Formulation and evaluation of drug-loaded targeted magnetic microspheres for cancer therapy. Int J Nanomedicine. 2013;8:1393–1402. doi: 10.2147/IJN.S43479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Gupta P, W S, Kim S, HSrivastava SK. Phenethyl isothiocyanate: A comprehensive review of anti-cancer mechanisms. Biochim Biophys Acta. 2014;1846(2):405–424. doi: 10.1016/j.bbcan.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Xue C, Pasolli HA, Piscopo I, Gros DJ, Liu C, Chen Y, Chiao JW. Mitochondrial structure alteration in human prostate cancer cells upon initial interaction with a chemopreventive agent phenethyl isothiocyanate. Cancer Cell Int. 2014;14(1):30. doi: 10.1186/1475-2867-14-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wang LG, Beklemisheva A, Liu XM, Ferrari AC, Feng J, Chiao JW. Dual action on promoter demethylation and chromatin by an isothiocyanate restored GSTP1 silenced in prostate cancer. Mol Carcinog. 2007;46(1):24–31. doi: 10.1002/mc.20258. [DOI] [PubMed] [Google Scholar]
- 179.Ahmad A, Sakr WA, Rahman KM. Anticancer properties of indole compounds: mechanism of apoptosis induction and role in chemotherapy. Curr Drug Targets. 2010;11(6):652–666. doi: 10.2174/138945010791170923. [DOI] [PubMed] [Google Scholar]
- 180.Ghantous A, Saikali M, Rau T, Gali-Muhtasib H, Schneider-Stock R, Darwiche N. Inhibition of tumor promotion by parthenolide: epigenetic modulation of p21. Cancer Prev Res (Phila) 2012;5(11):1298–1309. doi: 10.1158/1940-6207.CAPR-12-0230. [DOI] [PubMed] [Google Scholar]
- 181.Iciek M, Kwiecien I, Wlodek L. Biological properties of garlic and garlic-derived organosulfur compounds. Environ Mol Mutagen. 2009;50(3):247–265. doi: 10.1002/em.20474. [DOI] [PubMed] [Google Scholar]
- 182.Arunkumar A, Vijayababu MR, Gunadharini N, Krishnamoorthy G, Arunakaran J. Induction of apoptosis and histone hyperacetylation by diallyl disulfide in prostate cancer cell line PC-3. Cancer Lett. 2007;251(1):59–67. doi: 10.1016/j.canlet.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 183.Druesne-Pecollo N, Chaumontet C, Pagniez A, Vaugelade P, Bruneau A, Thomas M, Cherbuy C, Duee PH, Martel P. In vivo treatment by diallyl disulfide increases histone acetylation in rat colonocytes. Biochem Biophys Res Commun. 2007;354(1):140–147. doi: 10.1016/j.bbrc.2006.12.158. [DOI] [PubMed] [Google Scholar]
- 184.Katiyar S, Elmets CA, Katiyar SK. Green tea and skin cancer: photoimmunology, angiogenesis and DNA repair. J Nutr Biochem. 2007;18(5):287–296. doi: 10.1016/j.jnutbio.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 185.Tao L, Park JY, Lambert JD. Differential prooxidative effects of the green tea polyphenol, (−)-epigallocatechin-3-gallate, in normal and oral cancer cells are related to differences in sirtuin 3 signaling. Mol Nutr Food Res. 2015;59(2):203–211. doi: 10.1002/mnfr.201400485. [DOI] [PubMed] [Google Scholar]
- 186.Kang SK, Cha SH, Jeon HG. Curcumin-induced histone hypoacetylation enhances caspase-3-dependent glioma cell death and neurogenesis of neural progenitor cells. Stem Cells Dev. 2006;15(2):165–174. doi: 10.1089/scd.2006.15.165. [DOI] [PubMed] [Google Scholar]
- 187.Ogiwara H, Ui A, Shiotani B, Zou L, Yasui A, Kohno T. Curcumin suppresses multiple DNA damage response pathways and has potency as a sensitizer to PARP inhibitor. Carcinogenesis. 2013;34(11):2486–2497. doi: 10.1093/carcin/bgt240. [DOI] [PubMed] [Google Scholar]
- 188.Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430(7000):686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 189.Pietrocola F, Marino G, Lissa D, Vacchelli E, Malik SA, Niso-Santano M, Zamzami N, Galluzzi L, Maiuri MC, Kroemer G. Pro-autophagic polyphenols reduce the acetylation of cytoplasmic proteins. Cell Cycle. 2012;11(20):3851–3860. doi: 10.4161/cc.22027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hong KS, Park JI, Kim MJ, Kim HB, Lee JW, Dao TT, Oh WK, Kang CD, Kim SH. Involvement of SIRT1 in hypoxic down-regulation of c-Myc and beta-catenin and hypoxic preconditioning effect of polyphenols. Toxicol Appl Pharmacol. 2012;259(2):210–218. doi: 10.1016/j.taap.2011.12.025. [DOI] [PubMed] [Google Scholar]
- 191.Frombaum M, Therond P, Djelidi R, Beaudeux JL, Bonnefont-Rousselot D, Borderie D. Piceatannol is more effective than resveratrol in restoring endothelial cell dimethylarginine dimethylaminohydrolase expression and activity after high-glucose oxidative stress. Free Radic Res. 2011;45(3):293–302. doi: 10.3109/10715762.2010.527337. [DOI] [PubMed] [Google Scholar]
- 192.Kawakami S, Kinoshita Y, Maruki-Uchida H, Yanae K, Sai M, Ito T. Piceatannol and its metabolite, isorhapontigenin, induce SIRT1 expression in THP-1 human monocytic cell line. Nutrients. 2014;6(11):4794–4804. doi: 10.3390/nu6114794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Chang H, Mi M, Ling W, Zhu J, Zhang Q, Wei N, Zhou Y, Tang Y, Yuan J. Structurally related cytotoxic effects of flavonoids on human cancer cells in vitro. Arch Pharm Res. 2008;31(9):1137–1144. doi: 10.1007/s12272-001-1280-8. [DOI] [PubMed] [Google Scholar]
- 194.Xiao X, Shi D, Liu L, Wang J, Xie X, Kang T, Deng W. Quercetin suppresses cyclooxygenase-2 expression and angiogenesis through inactivation of P300 signaling. PLoS One. 2011;6(8):e22934. doi: 10.1371/journal.pone.0022934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Shukla S, Gupta S. Apigenin-induced cell cycle arrest is mediated by modulation of MAPK, PI3K-Akt, and loss of cyclin D1 associated retinoblastoma dephosphorylation in human prostate cancer cells. Cell Cycle. 2007;6(9):1102–1114. doi: 10.4161/cc.6.9.4146. [DOI] [PubMed] [Google Scholar]
- 196.Shukla S, Gupta S. Apigenin-induced prostate cancer cell death is initiated by reactive oxygen species and p53 activation. Free Radic Biol Med. 2008;44(10):1833–1845. doi: 10.1016/j.freeradbiomed.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Paredes-Gonzalez X, Fuentes F, Su ZY, Kong AN. Apigenin reactivates Nrf2 anti-oxidative stress signaling in mouse skin epidermal JB6 P + cells through epigenetics modifications. AAPS J. 2014;16(4):727–735. doi: 10.1208/s12248-014-9613-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Li Y, Meeran SM, Patel SN, Chen H, Hardy TM, Tollefsbol TO. Epigenetic reactivation of estrogen receptor-alpha (ERalpha) by genistein enhances hormonal therapy sensitivity in ERalpha-negative breast cancer. Mol Cancer. 2013;12:9. doi: 10.1186/1476-4598-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Basak S, Pookot D, Noonan EJ, Dahiya R. Genistein down-regulates androgen receptor by modulating HDAC6-Hsp90 chaperone function. Mol Cancer Ther. 2008;7(10):3195–3202. doi: 10.1158/1535-7163.MCT-08-0617. [DOI] [PubMed] [Google Scholar]
- 200.Dagdemir A, Durif J, Ngollo M, Bignon YJ, Bernard-Gallon D. Histone lysine trimethylation or acetylation can be modulated by phytoestrogen, estrogen or anti-HDAC in breast cancer cell lines. Epigenomics. 2013;5(1):51–63. doi: 10.2217/epi.12.74. [DOI] [PubMed] [Google Scholar]
- 201.Page RE, Jr, Peng CY. Aging and development in social insects with emphasis on the honey bee, Apis mellifera L. Exp Gerontol. 2001;36(4–6):695–711. doi: 10.1016/s0531-5565(00)00236-9. [DOI] [PubMed] [Google Scholar]
- 202.Lu J, Zhang L, Chen X, Lu Q, Yang Y, Liu J, Ma X. SIRT1 counteracted the activation of STAT3 and NF-kappaB to repress the gastric cancer growth. Int J Clin Exp Med. 2014;7(12):5050–5058. [PMC free article] [PubMed] [Google Scholar]
- 203.Hassan MK, Watari H, Salah-Eldin AE, Sultan AS, Mohamed Z, Fujioka Y, Ohba Y, Sakuragi N. Histone deacetylase inhibitors sensitize lung cancer cells to hyperthermia: involvement of Ku70/SirT-1 in thermo-protection. PLoS One. 2014;9(4):e94213. doi: 10.1371/journal.pone.0094213. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 204.Jafary H, Ahmadian S, Soleimani M. The enhanced apoptosis and antiproliferative response to combined treatment with valproate and nicotinamide in MCF-7 breast cancer cells. Tumour Biol. 2014;35(3):2701–2710. doi: 10.1007/s13277-013-1356-0. [DOI] [PubMed] [Google Scholar]
- 205.Zhou Y, Schmitz KM, Mayer C, Yuan X, Akhtar A, Grummt I. Reversible acetylation of the chromatin remodelling complex NoRC is required for non-coding RNA-dependent silencing. Nat Cell Biol. 2009;11(8):1010–1016. doi: 10.1038/ncb1914. [DOI] [PubMed] [Google Scholar]
- 206.Kim DH, Shin J, Kwon HJ. Psammaplin A is a natural prodrug that inhibits class I histone deacetylase. Exp Mol Med. 2007;39(1):47–55. doi: 10.1038/emm.2007.6. [DOI] [PubMed] [Google Scholar]
- 207.Ahn MY, Jung JH, Na YJ, Kim HS. A natural histone deacetylase inhibitor, Psammaplin A, induces cell cycle arrest and apoptosis in human endometrial cancer cells. Gynecol Oncol. 2008;108(1):27–33. doi: 10.1016/j.ygyno.2007.08.098. [DOI] [PubMed] [Google Scholar]
- 208.Seyssel K, Alligier M, Meugnier E, Chanseaume E, Loizon E, Canto C, Disse E, Lambert-Porcheron S, Brozek J, Blond E, Rieusset J, Morio B, Laville M, Vidal H. Regulation of energy metabolism and mitochondrial function in skeletal muscle during lipid overfeeding in healthy men. J Clin Endocrinol Metab. 2014;99(7):E1254–1262. doi: 10.1210/jc.2013-4379. [DOI] [PubMed] [Google Scholar]
- 209.Corfe BM, Williams EA, Bury JP, Riley SA, Croucher LJ, Lai DY, Evans CA. A study protocol to investigate the relationship between dietary fibre intake and fermentation, colon cell turnover, global protein acetylation and early carcinogenesis: the FACT study. BMC Cancer. 2009;9:332. doi: 10.1186/1471-2407-9-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Atwell LL, Hsu A, Wong CP, Stevens JF, Bella D, Yu TW, Pereira CB, Lohr CV, Christensen JM, Dashwood RH, Williams DE, Shannon J, Ho E. Absorption and chemopreventive targets of sulforaphane in humans following consumption of broccoli sprouts or a myrosinase-treated broccoli sprout extract. Mol Nutr Food Res. 2015;59(3):424–433. doi: 10.1002/mnfr.201400674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Clarke JD, Riedl K, Bella D, Schwartz SJ, Stevens JF, Ho E. Comparison of isothiocyanate metabolite levels and histone deacetylase activity in human subjects consuming broccoli sprouts or broccoli supplement. J Agric Food Chem. 2011;59(20):10955–10963. doi: 10.1021/jf202887c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Clarke JD, Hsu A, Riedl K, Bella D, Schwartz SJ, Stevens JF, Ho E. Bioavailability and inter-conversion of sulforaphane and erucin in human subjects consuming broccoli sprouts or broccoli supplement in a cross-over study design. Pharmacol Res. 2011;64(5):456–463. doi: 10.1016/j.phrs.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Hariharan R, Babu JM, R P, Pillai MR. Mutational analysis of Smad7 in human cervical cancer. Oncol Rep. 2009;21(4):1001–1004. doi: 10.3892/or_00000315. [DOI] [PubMed] [Google Scholar]
- 214.Baier SR, Zbasnik R, Schlegel V, Zempleni J. Off-target effects of sulforaphane include the derepression of long terminal repeats through histone acetylation events. J Nutr Biochem. 2014;25(6):665–668. doi: 10.1016/j.jnutbio.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Meeran SM, Patel SN, Li Y, Shukla S, Tollefsbol TO. Bioactive dietary supplements reactivate ER expression in ER-negative breast cancer cells by active chromatin modifications. PLoS One. 2012;7(5):e37748. doi: 10.1371/journal.pone.0037748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Chen J, Bai H, Wang C, Kang J. Trichostatin A improves the anticancer activity of low concentrations of curcumin in human leukemia cells. Pharmazie. 2006;61(8):710–716. [PubMed] [Google Scholar]
- 217.Saldanha SN, Kala R, Tollefsbol TO. Molecular mechanisms for inhibition of colon cancer cells by combined epigenetic-modulating epigallocatechin gallate and sodium butyrate. Exp Cell Res. 2014;324(1):40–53. doi: 10.1016/j.yexcr.2014.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Hu Y, McIntosh GH, Le Leu RK, Nyskohus LS, Woodman RJ, Young GP. Combination of selenium and green tea improves the efficacy of chemoprevention in a rat colorectal cancer model by modulating genetic and epigenetic biomarkers. PLoS One. 2013;8(5):e64362. doi: 10.1371/journal.pone.0064362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Rauh D, Fischer F, Gertz M, Lakshminarasimhan M, Bergbrede T, Aladini F, Kambach C, Becker CF, Zerweck J, Schutkowski M, Steegborn C. An acetylome peptide microarray reveals specificities and deacetylation substrates for all human sirtuin isoforms. Nat Commun. 2013;4:2327. doi: 10.1038/ncomms3327. [DOI] [PubMed] [Google Scholar]
- 220.Xing S, Poirier Y. The protein acetylome and the regulation of metabolism. Trends Plant Sci. 2012;17(7):423–430. doi: 10.1016/j.tplants.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 221.Myzak MC, Ho E, Dashwood RH. Dietary agents as histone deacetylase inhibitors. Mol Carcinog. 2006;45(6):443–446. doi: 10.1002/mc.20224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Whittaker M, Floyd CD, Brown P, Gearing AJ. Design and therapeutic application of matrix metalloproteinase inhibitors. Chem Rev. 1999;99(9):2735–2776. doi: 10.1021/cr9804543. [DOI] [PubMed] [Google Scholar]


