Abstract
After being a matter of hot debate for years, the presence of lipid membranes in the last common ancestor of extant organisms (i.e., the cenancestor) now begins to be generally accepted. By contrast, cenancestral cell walls have attracted less attention, probably owing to the large diversity of cell walls that exist in the three domains of life. Many prokaryotic cell walls, however, are synthesized using glycosylation pathways with similar polyisoprenol lipid carriers and topology (i.e., orientation across the cell membranes). Here, we provide the first systematic phylogenomic report on the polyisoprenol biosynthesis pathways in the three domains of life. This study shows that, whereas the last steps of the polyisoprenol biosynthesis are unique to the respective domain of life of which they are characteristic, the enzymes required for basic unsaturated polyisoprenol synthesis can be traced back to the respective last common ancestor of each of the three domains of life. As a result, regardless of the topology of the tree of life that may be considered, the most parsimonious hypothesis is that these enzymes were inherited in modern lineages from the cenancestor. This observation supports the presence of an enzymatic mechanism to synthesize unsaturated polyisoprenols in the cenancestor and, since these molecules are notorious lipid carriers in glycosylation pathways involved in the synthesis of a wide diversity of prokaryotic cell walls, it provides the first indirect evidence of the existence of a hypothetical unknown cell wall synthesis mechanism in the cenancestor.
Keywords: Polyisoprenol, Lipid carrier, Cenancestor, LUCA, Cell walls, Glycosylation, Archaea, Bacteria, Eukaryotes
Introduction
Cells from the three domains of life (archaea, bacteria and eukaryotes) are bound by cell membranes of which the main lipid components are the phospholipids. At first, the strong chemical and biosynthetic dissimilarities that exist between the archaeal phospholipids and those from bacteria and eukaryotes triggered a hot debate about the existence of lipid membranes in the last common ancestor of extant organisms, namely the cenancestor or LUCA (Koga et al., 1998; Wächtershäuser, 2003; Martin & Russell, 2003; Peretó, López-García & Moreira, 2004). Later, new phylogenomic analyses inferred the presence of metabolic pathways responsible for the synthesis of most phospholipid components in the cenancestor (Peretó, López-García & Moreira, 2004; Lombard & Moreira, 2011; Lombard, López-García & Moreira, 2012a; Lombard, López-García & Moreira, 2012c). Therefore, the debates have progressively shifted from arguing the existence of membranes to discuss their composition and properties (Lane & Martin, 2012; Lombard, López-García & Moreira, 2012b; Sojo, Pomiankowski & Lane, 2014). Contrary to cell membranes, the early evolution of cell walls has been scarcely studied. Cell walls indeed provide essential structural support and external interactions in modern organisms (Albers & Meyer, 2011), but their extreme diversity makes the study of their early evolution very challenging.
Despite the stunning diversity that exists among prokaryotic cell envelopes, the synthesis of many of their main components (e.g., N-or O-glycosylated S-layer proteins, peptidoglycan, O-antigen LPS, teichoic acids, exopolysaccharides) relies on comparable glycosylation pathways (Hartley & Imperiali, 2012; Lombard, 2016). These pathways are all located in the cell membranes, are mediated by similar lipid carriers and have the same orientation across the membrane. Their respective mechanisms can be described in three major steps: (1) synthesis of an oligomer on a lipid carrier in the cytoplasmic side of the cell membranes; (2) “flipping” of the oligomer-linked lipid carrier to the opposite side of the membrane; and (3) oligomer transfer from the lipid carrier to the acceptor molecule (e.g., protein or external polymer). The eukaryotic protein N-glycosylation also meets these criteria, although it takes place in the ER membranes instead of the plasma membranes. The proteins involved in these glycosylation pathways belong to a small number of protein superfamilies, some of which could be traced back to the cenancestor (Lombard, 2016). The large diversity of the glycosylation pathways makes it difficult to untangle the specific evolutionary relationships that exist among them but, despite their differences, the prokaryotic cell wall synthesis mechanisms have at least one element in common: they all use polyisoprenol phosphate lipid carriers (Hartley & Imperiali, 2012).
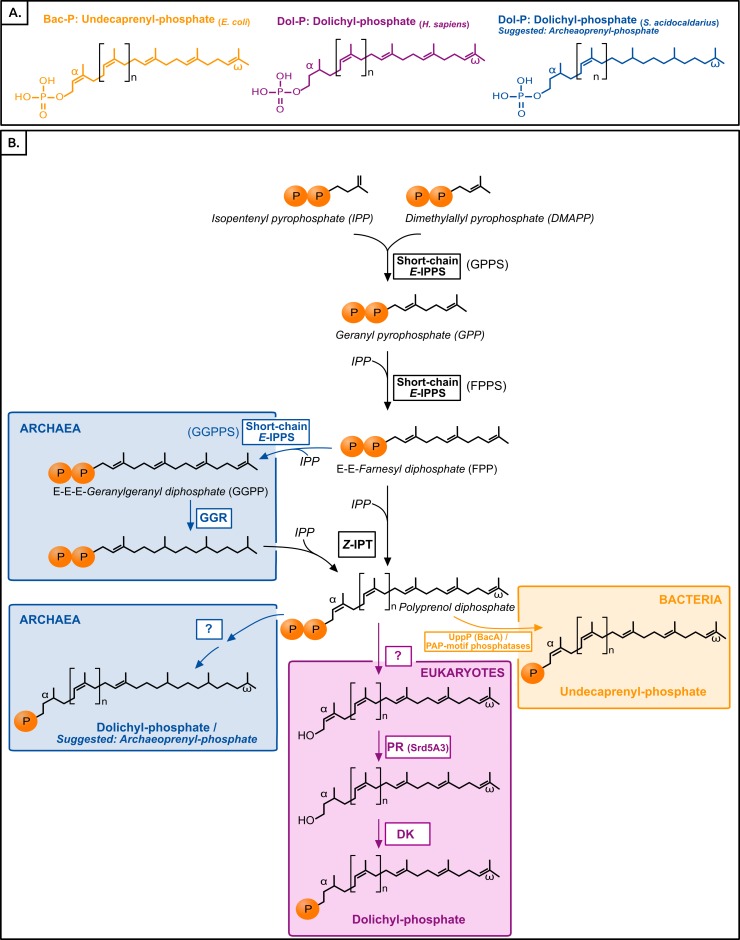
Polyisoprenol phosphates are long unsaturated chains with a varying number of isoprene units and an α-terminal phosphate group (Fig. 1). The eukaryotic polyisoprenol phosphate is called dolichol-phosphate (Dol-P) and it is characterized by the saturation of its α-unit. Archaeal lipid carriers are α- and ω-polysaturated and also called Dol-P. Bacterial polyisoprenols are fully unsaturated and their names vary according to their length (Meyer & Albers, 2013), so they will be referred to as bactoprenol phosphate (Bac-P) for simplicity.
Figure 1. (A) Typical polyisoprenol lipid carriers in the three domains of life. (B) Polyisoprenol-phosphate biosynthesis pathways in the three domains of life.
Steps outside boxes are common to all pathways, whereas steps in boxes are characteristic of only one domain of life. Unknown steps are labeled with a question mark. GPPS, geranyl diphosphate synthase; FPPS, farnesyl diphosphate synthase; GGPPS, geranylgeranyl diphosphate synthase; GGR, geranylgeranylreductase; IPT, isopentenyl transferase; UppP, undecaprenyl pyrophosphate phosphatase; PR, polyprenol reductase; DK, dolichol kinase.
Similar to other isoprenoids, the polyisoprenols are initially synthesized through the sequential condensation of isopentenyl pyrophosphate (IPP) and allylic prenyl diphosphates, the first of which is dimethylallyl pyrophosphate (DMAPP, Fig. 1). The first 3–4 condensations in polyisoprenol biosynthesis require regular E-prenyltransferases (IPPS) but, contrary to other isoprenoids, later elongations involve Z-type links (Ogura & Koyama, 1998; Schenk, Fernandez & Waechter, 2001; Guan et al., 2011). The rest of the polyisoprenol phosphate synthesis pathways differs in each domain of life (Fig. 1). In bacteria, polyprenol diphosphate must be dephosphorylated into Bac-P. Most (75%) of this activity is catalyzed by an undecaprenyl pyrophosphate phosphatase (UppP/BacA) (El Ghachi et al., 2004; Chang et al., 2014; Manat et al., 2015), and complemented by the members of a promiscuous family of PAP-motif phosphatases (Stukey & Carman, 1997; El Ghachi et al., 2005; Touzé, Blanot & Mengin-Lecreulx, 2008). The involvement in this dephosphorylation step of other components, such as a supplementary cytoplasmic phosphatase or a polyprenol diphosphate translocation mechanism, has been suggested (see below), but their identity remains unknown. In archaea, the ω-terminal and internal saturated units are reduced by the geranylgeranyl reductases (GGR, Fig. 1) either before the elongation of FPP/GGPP into Dol-P or later on (Sato et al., 2008; Guan et al., 2011; Naparstek, Guan & Eichler, 2012). The rest of the archaeal Dol-P biosynthesis pathway remains unknown. Finally, in eukaryotes, polyprenol diphosphate is dephosphorylated, α-reduced (Cantagrel et al., 2010) and rephosphorylated (Cantagrel & Lefeber, 2011, Fig. 1). Here, a phylogenomic analysis of the proteins involved in the polyisoprenol phosphate biosynthesis pathways was performed in the three domains of life. This analysis provides promising insights into the unexplored question of the early evolution of cell walls.
Material and Methods
The original sequence seeds were selected from KEGG (http://www.genome.jp/kegg/) and verified in the literature. BLASTp (Altschul et al., 1990) were carried out against a selection of representative genomes (Fig. S1) or the non-redundant (nr) dataset on GenBank (http://www.ncbi.nlm.nih.gov/Genbank), depending on results. When homologues were difficult to detect, more distant homologues were searched for using PSI-BLAST (Altschul et al., 1997) or HMM profiles and the hmmsearch tool implemented in the HMMER v3.1b2 webserver (http://hmmer.org/download.html and Finn et al., 2015). The alignments were carried out with MUSCLE v3.8.31 (Edgar, 2004) and trimmed with the program NET of the MUST package (Philippe, 1993). The preliminary phylogenies were reconstructed using FastTree v.2.1.7 (Price, Dehal & Arkin, 2010). Representative sequences were selected from these phylogenies to carry out more accurate analyses, and realigned using MUSCLE on the GUIDANCE server (http://guidance.tau.ac.il/ver2/, Penn et al., 2010). The final alignments are provided in Fig. S2 and were trimmed based on the statistical scores provided by GUIDANCE. The final phylogenies were constructed using MrBayes 3.2.6 (Ronquist et al., 2012) (LG substitution model (Le & Gascuel, 2008); 4 Γ categories; 4 chains of 1,000,000 generations; tree sampling every 100 generations; 25% generations discarded as “burn in”) and RaxML-HPC2 8.1.24 (Stamatakis, 2014) (LG +Γ model (Le, Dang & Gascuel, 2012); 4 rate categories; 100 bootstrap replicates) implemented in the CIPRES Science Gateway (Miller, Pfeiffer & Schwartz, 2010). Bayesian and maximum likelihood phylogenies gave comparable results.
Results and Discussion
Despite being widespread in archaea, bacteria and eukaryotes, IPP and DMAPP are synthesized by respective pathways in each domain of life (Lombard & Moreira, 2011; Dellas et al., 2013; Vranová, Coman & Gruissem, 2013). A previous study on the evolution of these pathways concluded that the cenancestor had a primitive mevalonate pathway that could have synthesized IPP and DMAPP in this organism, and that each domain of life adopted its specific biosynthesis pathway later on (Lombard & Moreira, 2011). The E-type prenyl condensations are carried out by E-prenyltransferases (IPPS). The evolution of these proteins has been described in prokaryotes (Lombard, López-García & Moreira, 2012a) and the analysis is now extended to eukaryotes (see Fig. S3 for details). In summary, these analyses are not conclusive on the specific relationships that may exist among IPPS from the three domains of life, but they support the presence of IPPS homologues in the respective common ancestor of each domain of life. As a result, their presence can also be inferred in the cenancestor independently of the topology of the tree of life that may be favored.
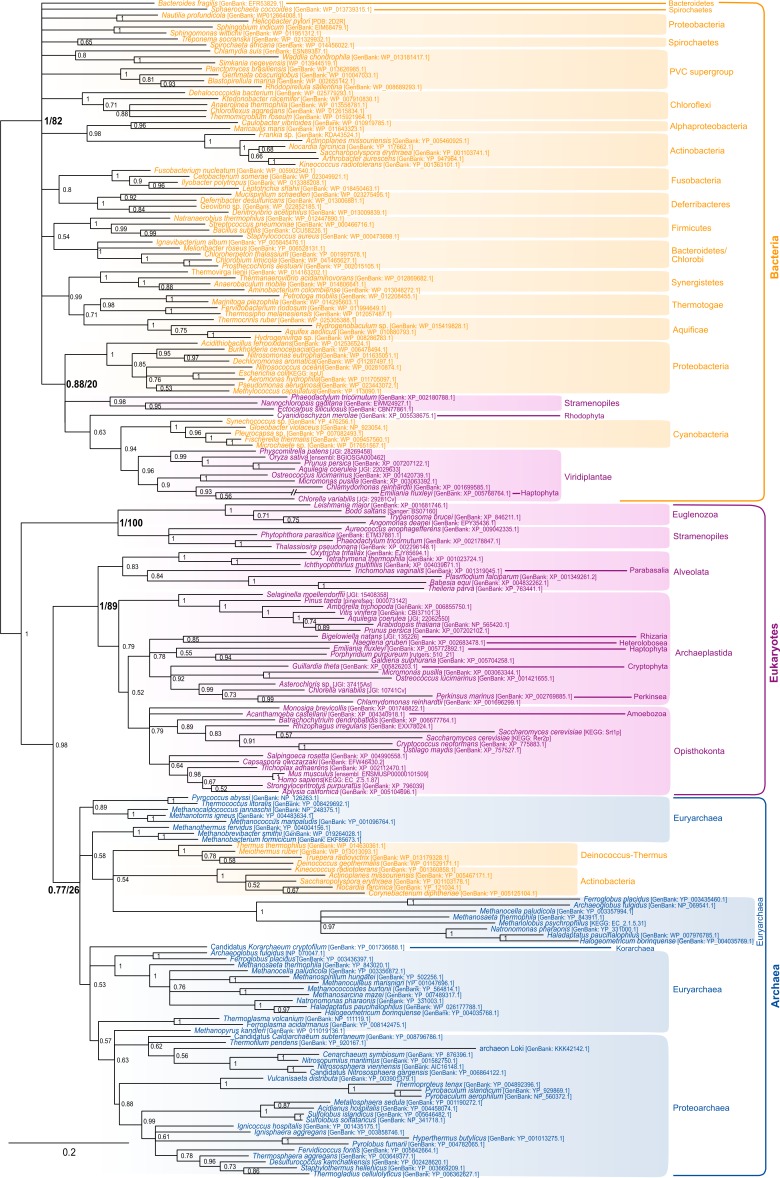
The most characteristic enzyme of the polyisoprenol phosphate biosynthesis pathway is the Z-isopentenyltransferase (Z-IPTase, Fig. 1) (Shimizu, Koyama & Ogura, 1998). The phylogeny of the Z-IPTases (Fig. 2) shows archaeal and bacterial monophyletic clades (Bayesian posterior probability (BPP) = 0.77 and BPP = 1, respectively) and several groups of eukaryotic sequences. The internal relationships within the archaeal clade are poorly resolved, which is not unexpected for a single-gene tree. The euryarchaea are paraphyleticand contain a group made up of distant paralogues and bacterial sequences probably acquired through horizontal gene transfer (HGT). Nevertheless, Z-IPTase homologues are widespread among archaea and the monophyly of the archaeal and proteoarchaeal (TACK superphylum, Petitjean et al., 2015) sequences support the presence of this gene in the Last Archaeal Common Ancestor. The deep phylogenetic relationships within the bacterial clade are also unresolved. Yet, the wide diversity of bacteria in which Z-IPTase homologues may be detected, together with the fact that most bacterial sequences group according to their taxonomic classification supports the ancestral presence of this gene in bacteria. Some polyphyletic sequences from plastid-bearing eukaryotes branch within a group of cyanobacteria and proteobacteria (BPP = 0.88) and likely have plastidial ancestors. The rest of the eukaryotic sequences are paraphyletic. The largest eukaryotic clade (BPP = 1) is made up of diverse sequences that cluster according to major eukaryotic taxa (e.g., opisthokonts, streptophytes, alveolates). Some internal branching is suggestive of inter-eukaryotic HGT, such as in organisms known to have had secondary plastid endosymbioses, which branch among archaeplastida. A smaller cluster of divergent euglenozoan and stramenopile sequences (BPP = 1) forms a sister group to the large eukaryotic/archaeal clade. Some of euglenozoa are known to synthesize shorter Dol-P molecules than other eukaryotes, so it is possible that the phylogenetic position of these sequences reflects a reconstruction artifact due to their high divergence (Schenk, Fernandez & Waechter, 2001). Ultimately, if we accept that the few paraphylies and polyphylies in this tree result from reconstruction artifacts due to the extreme sequence divergence of Z-IPTases across the three domains of life, this (unrooted) phylogeny supports the presence of Z-IPTases in the respective last common ancestors of each domain of life and, regardless of the topology of tree of life that may be invoked, in the cenancestor.
Figure 2. Bayesian phylogeny of the Z-IPTase homologues.
The tree is unrooted and was reconstructed using 197 representative sequences and 202 conserved sites. Multifurcations correspond to branches with Bayesian posterior probabilities <0.5, whereas numbers at nodes indicate Bayesian posterior probabilities higher than 0.5. The bootstrap values from the maximum likelihood analyses have been reported on basal and major nodes. Colors of leaves represent the affiliation of sequences to their respective domain of life: archaea (blue), bacteria (orange) and eukaryotes (purple).
The following steps in the polyisoprenol phosphate biosynthesis pathways are specific to each domain of life (Fig. 1). In bacteria, the polyprenol diphosphate is dephosphorylated into Bac-P by UppP (BacA) or a family of periplasmic PAP-motif phosphatases. UppP and the PAP-motif phosphatases are not evolutionary related to each other and, until recently, it was thought that they were respectively responsible for the cytoplasmic de novo synthesis or the periplasmic recycling of polyprenol diphosphate (Bickford & Nick, 2013; Manat et al., 2014). The recent discovery that they all function in the periplasmic side of the membrane implies that the de novo polyprenol diphosphate dephosphorylation requires either the activity of an unknown cytoplasmic phosphatase, or the translocation to the periplasmic side of the membrane through an unknown mechanism (Chang et al., 2014; Manat et al., 2015; Teo & Roper, 2015). Although some candidates have been suggested to mediate the polyprenol (di)phosphate translocation (Sanyal & Menon, 2010; Chang et al., 2014; Manat et al., 2015), these hypotheses require biochemical verification. Thus, only the two known phosphatase families will be studied here.
UppP homologues were detected in archaea and a wide diversity of bacteria but not in eukaryotes, even when HMM searches for distantly related sequences were carried out (see ‘Methods’). Since relatively little is known about the last steps in archaeal polyisoprenol biosynthesis, the possible participation of these archaeal homologues in this pathway seems worth testing experimentally. The phylogeny of the UppP homologues is discussed in more detail in Fig. S4 but, in summary, the respective monophyly of archaeal and bacterial UppP homologues (BPP = 0.96) suggests that this gene could have been present in the respective ancestors of both archaea and bacteria and, therefore, in the cenancestor. Importantly, this suggests that the cenancestor could have had a minimum set of proteins representative of all the enzymes currently known to be required to synthesize not only polyisoprenol chains but also functional lipid carriers (Bac-P). Regarding the PAP-motif phosphatases, several representatives of the family have been biochemically characterized (Fernández & Rush, 2001; El Ghachi et al., 2005; Manabe et al., 2009) and their homologues are widespread in the three domains of life, but their phylogeny is poorly resolved and dominated by the presence of paralogues and xenologues (see Fig. S5 for details). As a result, it is not possible at this stage to discriminate between a cenancestral or more recent origin of the PAP-motif phosphatases.
In archaea, the ω-terminal and internal units are reduced by the GGRs (Fig. 1). A recent phylogenomic analysis of the GGRs suggested (1) the ancestral presence of this gene in archaea, followed by a complicated history of duplications and HGTs; (2) an unclear origin for these genes in bacteria; and (3) a likely plastidial origin of these genes in eukaryotes (Lombard, López-García & Moreira, 2012a); thus, the presence of GGRs in the cenancestor is not supported. The rest of the archaeal Dol-P biosynthesis pathway remains uncharacterized, so particular attention was drawn to the possible archaeal homologues of the enzymes in the eukaryotic Dol-P synthesis pathway.
The specific steps in the eukaryotic pathway consist in the dephosphorylation, α-reduction and rephosphorylation of the polyprenol diphosphate (Fig. 1). The eukaryotic polyprenol diphosphate phosphatase is unknown (Cantagrel & Lefeber, 2011; Bickford & Nick, 2013). Sequences similar to the polyisoprenol α-unit reductases (PR, Fig. 1) are widespread in eukaryotes but they are absent in archaea and rare in bacteria (these are likely xenologues). This implies that archaea use an alternative way to saturate the α-unit of their Dol-P. The PR phylogeny (see Fig. S6 for details) suggests that this protein is an early eukaryotic innovation that was developed before the Last Eukaryotic Common Ancestor and duplicated later in several eukaryotic lineages. Finally, homologues of the dolichol kinase (DK) were easily detected among a large diversity of eukaryotes, but only in a few prokaryotes. PSI-BLAST searches (Altschul et al., 1997) revealed a distant evolutionary relationship between the DKs and the CDP-diacylglycerol synthases (CdsA) involved in phospholipid synthesis (Sparrow & Raetz, 1985). The evolution of both functions was studied (Fig. S7) but the quality of the sequence alignment was very poor, the resulting phylogenies are unreliable and so are the conclusions that can be taken from them. This analysis, however, questions the existence of DK functional orthologues in most prokaryotes, especially in Euryarchaea. The lack of homologues of the last steps of eukaryotic Dol-P synthesis in most archaeal genomes raises the question of how these organisms finish the synthesis of their polyisoprenol phosphate lipid carriers.
Conclusions
The initial metabolic steps of the polyisoprenol biosynthesis pathways (from IPP/DMAPP synthesis to polyprenol diphosphate) are widespread in the three domains of life, and their presence can be inferred in at least the respective ancestor of each prokaryotic domain of life (Fig. 3). The later steps differ in each domain. Bacterial UppP (BacA) may be ancestral to both bacteria and archaea, but the evolution of the PAP-motif phosphatases is less clear. PR, and probably DK as well, are eukaryotic innovations. Archaea use their ancestral GGR, they do not have PR homologues and most of them lack DK homologues. Although neither the archaeal nor the eukaryotic pathways are fully described yet, the available information suggests that these domains have different means of synthesizing their polyisoprenol phosphates. This is congruent with the fact that, despite having the same name, eukaryotic Dol-P and archaeal Dol-P are chemically different from each other (Fig. 1). In order to avoid any confusion, we propose the archaeal polysaturated polyisoprenols (Guan et al., 2010; Guan et al., 2011; Chang et al., 2015) to be called “archaeoprenols” from now on.
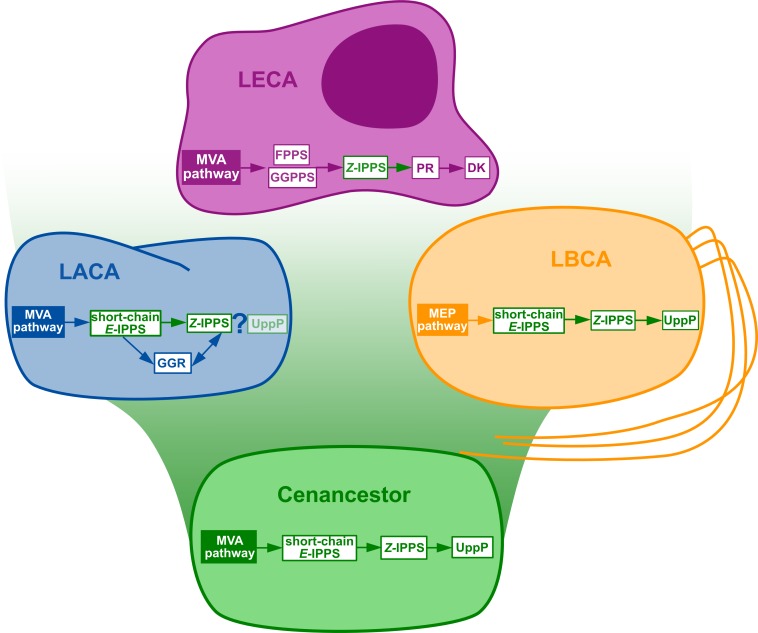
Figure 3. Summary of the phylogenomic results of the enzymes involved in the polyisoprenol biosynthesis pathways.
Only known genes are presented, so the eukaryotic and archaeal pathways are not complete. MVA, mevalonate; MEP, Methylerythritol phosphate; LACA, Last Archaeal Common Ancestor; LBCA, Last Bacterial Common Ancestor; LECA, Last Eukaryotic Common Ancestor. Refer to Fig. 1 for other abbreviations.
Polyprenol diphosphate biosynthesis is ancestral to each domain of life and, therefore, the most parsimonious hypothesis to explain its distribution is that it was inherited from the cenancestor. Although the mechanism that provides this molecule to the periplasmic phosphatases remains unknown (Chang et al., 2014; Manat et al., 2015), the possibility that UppP homologues may also be ancestral to both prokaryotic domains similarly suggests that the cenancestor could even have been able to synthesize some sort of Bac-P. Polyisoprenols may carry out pleiotropic functions (Cantagrel & Lefeber, 2011; Hartley & Imperiali, 2012), but their most notorious role is as lipid carriers in a number of glycosylation pathways (Lombard, 2016). In prokaryotes, these glycosylations are systematically related to the synthesis of cell wall components, so it is reasonable to postulate that hypothetical cenancestral polyisoprenols may have played the role of lipid carriers in early glycosylation mechanisms, possibly related to cell wall synthesis. Yet, this hypothesis will need to be updated when more information about the polyisoprenol biosynthesis pathways will be available.
Although the presence of a polyisoprenol phosphate biosynthesis pathway in the cenancestor is a promising piece of evidence to support the existence of cell walls in this organism, the large diversity of microbial cell walls remains an obstacle to risk a hypothesis on the specific nature of this cenancestral structure. Interestingly, the presence of at least two promising glycosyltransferase superfamilies (HPT and MurG) has been inferred in the cenancestor (Lombard, 2016). The proteins in these families are responsible for the transfer of soluble monosaccharides to polyisoprenol phosphate lipid carriers in bacterial peptidoglycan synthesis (Mohammadi et al., 2007) and archaeal S-layer protein N-glycosylation (Meyer & Albers, 2013). If we sum up the results presented here and those about glycosyltransferases (Lombard, 2016), it is tempting to say that the cenancestor could have been able to use its HPT and MurG homologues to synthesize an oligosaccharide on a Bac-P lipid carrier. The question that remains open is the final acceptor of that hypothetical cenancestral oligosaccharide, as that is the element that would define the nature of the putative cenancestral cell wall. One way to tackle this question would be studying the transferases that convey the glycans from the lipid carrier to various donors. These transferases were phylogenomically analyzed in (Lombard, 2016), but none of them revealed a widespread function that would provide an obvious candidate to make up a particular type of cell wall in the cenancestor. For instance, S-layers have been put forward as possible first cell wall components (Albers & Meyer, 2011; Fagan & Fairweather, 2014). The protein N-glycosylation oligosaccharyltransferases and O-mannosylation transferases responsible for S-layer protein glycosylation are indeed probably ancestral to eukaryotes and archaea, but their ancestral presence in bacteria is uncertain (Lombard, 2016). Therefore, the presence of glycosylated S-layers proteins in the cenancestor remains inconclusive.
It has been argued that the strong conservation of polyisoprenol phosphates as lipid carriers may be functional: they may play important regulatory roles on glycan mechanisms, their biochemistry may be particularly adapted to glycan translocation across phospholipid bilayers or they can be beneficial to the formation of large protein complexes with which they frequently interact (Hartley & Imperiali, 2012). Contrary to these possible strong pressures to maintain polyisoprenol phosphates as lipid carriers, the difficulty to determine the final acceptor of the hypothetical cenancestral oligosaccharide may be explained by the likely strong selective pressures that acted on the exposed cell wall components to diversify. For instance, one specific type of cell wall may have actually existed in the cenancestor, but the strong selective pressures exerted on the cell wall glycocalyx may have resulted in the independent replacement of the original cell wall in many lineages. Another possibility is that the cenancestral cell wall glycosylation was promiscuous and that specific cell wall types only evolved later in modern lineages, following the same selective pressures. We are at a time of dynamic characterization of prokaryotic envelopes and comparative studies, so new evidence may soon shed light on more mechanistic similarities among cell wall syntheses in the three domains of life and complement our knowledge of the cenancestral envelopes.
Supplemental Information
Acknowledgments
I thank U Gophna for the stimulating conversations that crystallized in the development of this project and D Moreira and TA Richards for their advice at later steps. I also thank D Moreira, TA Richards, C Petitjean, F Savory, R Bamford, V Attah, five reviewers from previous submissions and two reviewers from this submission for their comments on the manuscript.
Funding Statement
This work was supported by the National Science Foundation through the National Evolutionary Synthesis Center (grant number NSF #EF-0905606). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Jonathan Lombard conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, wrote the paper, prepared figures and/or tables, reviewed drafts of the paper.
Data Availability
The following information was supplied regarding data availability:
The raw data has been supplied as Supplementary File.
References
- Albers & Meyer (2011).Albers S-V, Meyer BH. The archaeal cell envelope. Nature Reviews Microbiology. 2011;9:414–426. doi: 10.1038/nrmicro2576. [DOI] [PubMed] [Google Scholar]
- Altschul et al. (1990).Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul et al. (1997).Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickford & Nick (2013).Bickford JS, Nick HS. Conservation of the PTEN catalytic motif in the bacterial undecaprenyl pyrophosphate phosphatase, BacA/UppP. Microbiology. 2013;159:2444–2455. doi: 10.1099/mic.0.070474-0. [DOI] [PubMed] [Google Scholar]
- Cantagrel & Lefeber (2011).Cantagrel V, Lefeber DJ. From glycosylation disorders to dolichol biosynthesis defects: a new class of metabolic diseases. Journal of Inherited Metabolic Disease. 2011;34:859–867. doi: 10.1007/s10545-011-9301-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantagrel et al. (2010).Cantagrel V, Lefeber DJ, Ng BG, Guan Z, Silhavy JL, Bielas SL, Lehle L, Hombauer H, Adamowicz M, Swiezewska E, De Brouwer AP, Blümel P, Sykut-Cegielska J, Houliston S, Swistun D, Ali BR, Dobyns WB, Babovic-Vuksanovic D, Van Bokhoven H. SRD5A3 is required for converting polyprenol to dolichol and is mutated in a congenital glycosylation disorder. Cell. 2010;142:203–217. doi: 10.1016/j.cell.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang et al. (2014).Chang H-Y, Chou C-C, Hsu M-F, Wang AHJ. Proposed carrier lipid-binding site of undecaprenyl pyrophosphate phosphatase from Escherichia coli. The Journal of Biological Chemistry. 2014;289:18719–18735. doi: 10.1074/jbc.M114.575076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang et al. (2015).Chang MM, Imperiali B, Eichler J, Guan Z. N-Linked glycans are assembled on highly reduced dolichol phosphate carriers in the hyperthermophilic archaea Pyrococcus furiosus. PLoS ONE. 2015;10:e2626. doi: 10.1371/journal.pone.0130482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellas et al. (2013).Dellas N, Thomas ST, Manning G, Noel JP. Discovery of a metabolic alternative to the classical mevalonate pathway. ELife. 2013;2:e2626. doi: 10.7554/eLife.00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar (2004).Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan & Fairweather (2014).Fagan RP, Fairweather NF. Biogenesis and functions of bacterial S-layers. Nature Reviews Microbiology. 2014;12:211–222. doi: 10.1038/nrmicro3213. [DOI] [PubMed] [Google Scholar]
- Fernández & Rush (2001).Fernández F, Rush JS, Toke DA, Han G-S, Quinn JE, Carman GM, Choi J-Y, Voelker DR, Aebi M, Waechter CJ. The CWH8 gene encodes a dolichyl pyrophosphate phosphatase with a luminally oriented active site in the endoplasmic reticulum of Saccharomyces cerevisiae. Journal of Biological Chemistry. 2001;276:41455–41464. doi: 10.1074/jbc.M105544200. [DOI] [PubMed] [Google Scholar]
- Finn et al. (2015).Finn RD, Clements J, Arndt W, Miller BL, Wheeler TJ, Schreiber F, Bateman A, Eddy SR. HMMER web server: 2015 update. Nucleic Acids Research. 2015;43:W30–W38. doi: 10.1093/nar/gkv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Ghachi et al. (2004).El Ghachi M, Bouhss A, Blanot D, Mengin-Lecreulx D. The bacA gene of Escherichia coli encodes an undecaprenyl pyrophosphate phosphatase activity. Journal of Biological Chemistry. 2004;279:30106–30113. doi: 10.1074/jbc.M401701200. [DOI] [PubMed] [Google Scholar]
- El Ghachi et al. (2005).El Ghachi M, Derbise A, Bouhss A, Mengin-Lecreulx D. Identification of multiple genes encoding membrane proteins with undecaprenyl pyrophosphate phosphatase (UppP) activity in Escherichia coli. Journal of Biological Chemistry. 2005;280:18689–18695. doi: 10.1074/jbc.M412277200. [DOI] [PubMed] [Google Scholar]
- Guan et al. (2011).Guan Z, Meyer BH, Albers S-V, Eichler J. The thermoacidophilic archaeon Sulfolobus acidocaldarius contains an unsually short, highly reduced dolichyl phosphate. Biochimica et Biophysica Acta. 2011;1811:607–616. doi: 10.1016/j.bbalip.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan et al. (2010).Guan Z, Naparstek S, Kaminski L, Konrad Z, Eichler J. Distinct glycan-charged phosphodolichol carriers are required for the assembly of the pentasaccharide N-linked to the Haloferax volcaniiS-layer glycoprotein. Molecular Microbiology. 2010;78:1294–1303. doi: 10.1111/j.1365-2958.2010.07405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley & Imperiali (2012).Hartley M, Imperiali B. At the membrane frontier: a prospectus on the remarkable evolutionary conservation of polyprenols and polyprenyl-phosphates. Archives of Biochemistry and Biophysics. 2012;517:83–97. doi: 10.1016/j.abb.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga et al. (1998).Koga Y, Kyuragi T, Nishihara M, Sone N. Did archaeal and bacterial cells arise independently from noncellular precursors? A hypothesis stating that the advent of membrane phospholipid with enantiomeric glycerophosphate backbones caused the separation of the two lines of descent. Journal of Molecular Evolution. 1998;46:54–63. doi: 10.1007/PL00006283. [DOI] [PubMed] [Google Scholar]
- Lane & Martin (2012).Lane N, Martin WF. The origin of membrane bioenergetics. Cell. 2012;151:1406–1416. doi: 10.1016/j.cell.2012.11.050. [DOI] [PubMed] [Google Scholar]
- Le, Dang & Gascuel (2012).Le SQ, Dang CC, Gascuel O. Modeling protein evolution with several amino acid replacement matrices depending on site rates. Molecular Biology and Evolution. 2012;29:2921–2936. doi: 10.1093/molbev/mss112. [DOI] [PubMed] [Google Scholar]
- Le & Gascuel (2008).Le SQ, Gascuel O. An improved general amino acid replacement matrix. Molecular Biology and Evolution. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- Lombard (2016).Lombard J. The multiple evolutionary origins of the eukaryotic N-glycosylation pathway. Biology Direct. 2016;11:36. doi: 10.1186/s13062-016-0137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard, López-García & Moreira (2012a).Lombard J, López-García P, Moreira D. Phylogenomic investigation of phospholipid synthesis in archaea. Archaea. 2012a;2012:630910. doi: 10.1155/2012/630910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard, López-García & Moreira (2012b).Lombard J, López-García P, Moreira D. The early evolution of lipid membranes and the three domains of life. Nature Reviews Microbiology. 2012b;10:507–515. doi: 10.1038/nrmicro2815. [DOI] [PubMed] [Google Scholar]
- Lombard, López-García & Moreira (2012c).Lombard J, López-García P, Moreira D. An ACP-independent fatty acid synthesis pathway in archaea: implications for the origin of phospholipids. Molecular Biology and Evolution. 2012c;29:3261–3265. doi: 10.1093/molbev/mss160. [DOI] [PubMed] [Google Scholar]
- Lombard & Moreira (2011).Lombard J, Moreira D. Origins and early evolution of the mevalonate pathway of isoprenoid biosynthesis in the three domains of life. Molecular Biology and Evolution. 2011;28:87–99. doi: 10.1093/molbev/msq177. [DOI] [PubMed] [Google Scholar]
- Manabe et al. (2009).Manabe F, Itoh YH, Shoun H, Wakagi T. Membrane-bound acid pyrophosphatase from Sulfolobus tokodaii, a thermoacidophilic archaeon: heterologous expression of the gene and characterization of the product. Extremophiles. 2009;13:859–865. doi: 10.1007/s00792-009-0273-z. [DOI] [PubMed] [Google Scholar]
- Manat et al. (2015).Manat G, El Ghachi M, Auger R, Baouche K, Olatunji S, Kerff F, Touzé T, Mengin-Lecreulx D, Bouhss A. Membrane topology and biochemical characterization of the Escherichia coli BacA undecaprenyl-pyrophosphate phosphatase. PLoS ONE. 2015;10:e2626. doi: 10.1371/journal.pone.0142870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manat et al. (2014).Manat G, Roure S, Auger R, Bouhss A, Barreteau H, Mengin-Lecreulx D, Touzé T. Deciphering the metabolism of undecaprenyl-phosphate: the bacterial cell-wall unit carrier at the membrane frontier. Microbial Drug Resistance. 2014;20:199–214. doi: 10.1089/mdr.2014.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin & Russell (2003).Martin W, Russell MJ. On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2003;358:59–83. doi: 10.1098/rstb.2002.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer & Albers (2013).Meyer BH, Albers S-V. Hot and sweet: protein glycosylation in Crenarchaeota. Biochemical Society Transactions. 2013;41:384–392. doi: 10.1042/BST20120296. [DOI] [PubMed] [Google Scholar]
- Miller, Pfeiffer & Schwartz (2010).Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE); 2010. pp. 1–8. [Google Scholar]
- Mohammadi et al. (2007).Mohammadi T, Karczmarek A, Crouvoisier M, Bouhss A, Mengin-Lecreulx D, Den Blaauwen T. The essential peptidoglycan glycosyltransferase MurG forms a complex with proteins involved in lateral envelope growth as well as with proteins involved in cell division in Escherichia coli. Molecular Microbiology. 2007;65:1106–1121. doi: 10.1111/j.1365-2958.2007.05851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naparstek, Guan & Eichler (2012).Naparstek S, Guan Z, Eichler J. A predicted geranylgeranyl reductase reduces the ω-position isoprene of dolichol phosphate in the halophilic archaeon, Haloferax volcanii. Biochimica et Biophysica Acta. 2012;1821:923–933. doi: 10.1016/j.bbalip.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura & Koyama (1998).Ogura K, Koyama T. Enzymatic aspects of isoprenoid chain elongation. Chemical Reviews. 1998;98:1263–1276. doi: 10.1021/cr9600464. [DOI] [PubMed] [Google Scholar]
- Penn et al. (2010).Penn O, Privman E, Ashkenazy H, Landan G, Graur D, Pupko T. GUIDANC : a web server for assessing alignment confidence scores. Nucleic Acids Research. 2010;38:W23–W28. doi: 10.1093/nar/gkq443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretó, López-García & Moreira (2004).Peretó J, López-García P, Moreira D. Ancestral lipid biosynthesis and early membrane evolution. Trends in Biochemical Sciences. 2004;29:469–477. doi: 10.1016/j.tibs.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Petitjean et al. (2015).Petitjean C, Deschamps P, López-García P, Moreira D. Rooting the domain archaea by phylogenomic analysis supports the foundation of the new kingdom Proteoarchaeota. Genome Biology and Evolution. 2015;7:191–204. doi: 10.1093/gbe/evu274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe (1993).Philippe H. MUST, a computer package of management utilities for sequences and trees. Nucleic Acids Research. 1993;21:5264–5272. doi: 10.1093/nar/21.22.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, Dehal & Arkin (2010).Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e2626. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist et al. (2012).Ronquist F, Teslenko M, Van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal & Menon (2010).Sanyal S, Menon AK. Stereoselective transbilayer translocation of mannosyl phosphoryl dolichol by an endoplasmic reticulum flippase. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11289–11294. doi: 10.1073/pnas.1002408107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato et al. (2008).Sato S, Murakami M, Yoshimura T, Hemmi H. Specific partial reduction of geranylgeranyl diphosphate by an enzyme from the thermoacidophilic archaeon Sulfolobus acidocaldarius yields a reactive prenyl donor, not a dead-end product. Journal of Bacteriology. 2008;190:3923–3929. doi: 10.1128/JB.00082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk, Fernandez & Waechter (2001).Schenk B, Fernandez F, Waechter CJ. The ins(ide) and outs(ide) of dolichyl phosphate biosynthesis and recycling in the endoplasmic reticulum. Glycobiology. 2001;11:61R–79R. doi: 10.1093/glycob/11.5.61R. [DOI] [PubMed] [Google Scholar]
- Shimizu, Koyama & Ogura (1998).Shimizu N, Koyama T, Ogura K. Molecular cloning, expression and purification of undecaprenyl diphosphate synthase. Journal of Biological Chemistry. 1998;273:19476–19481. doi: 10.1074/jbc.273.31.19476. [DOI] [PubMed] [Google Scholar]
- Sojo, Pomiankowski & Lane (2014).Sojo V, Pomiankowski A, Lane N. A bioenergetic basis for membrane divergence in archaea and bacteria. PLoS Biology. 2014;12:e2626. doi: 10.1371/journal.pbio.1001926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow & Raetz (1985).Sparrow CP, Raetz CR. Purification and properties of the membrane-bound CDP-diglyceride synthetase from Escherichia coli. Journal of Biological Chemistry. 1985;260:12084–12091. [PubMed] [Google Scholar]
- Stamatakis (2014).Stamatakis A. RAxMLversion 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukey & Carman (1997).Stukey J, Carman GM. Identification of a novel phosphatase sequence motif. Protein Science. 1997;6:469–472. doi: 10.1002/pro.5560060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo & Roper (2015).Teo A.CK, Roper DI. Core teps of membrane-bound peptidoglycan biosynthesis: Recent advances, insight and opportunities. Antibiotics. 2015;4:495–520. doi: 10.3390/antibiotics4040495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touzé, Blanot & Mengin-Lecreulx (2008).Touzé T, Blanot D, Mengin-Lecreulx D. Substrate specificity and membrane topology of Escherichia coli PgpB, an undecaprenyl pyrophosphate phosphatase. Journal of Biological Chemistry. 2008;283:16573–16583. doi: 10.1074/jbc.M800394200. [DOI] [PubMed] [Google Scholar]
- Vranová, Coman & Gruissem (2013).Vranová E, Coman D, Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annual Review of Plant Biology. 2013;64:665–700. doi: 10.1146/annurev-arplant-050312-120116. [DOI] [PubMed] [Google Scholar]
- Wächtershäuser (2003).Wächtershäuser G. From pre-cells to Eukarya—a tale of two lipids. Molecular Microbiology. 2003;47:13–22. doi: 10.1046/j.1365-2958.2003.03267.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
The raw data has been supplied as Supplementary File.