Abstract
Innate and adaptive immunity depends critically on host recognition of pathogen-associated molecules. Toll-like receptors (TLRs) are key mediators of pathogen surveillance at the cell or phagocytic vacuole surface. However, mechanisms underlying recognition of pathogens in other cellular compartments remain unclear, and responses elicited by cytosolic challenge are poorly characterized. We therefore used mouse cDNA microarrays to investigate gene expression triggered by infection of bone marrow-derived macrophages with cytosol- and vacuole-localized Listeria monocytogenes (Lm), a model cytosolic pathogen. The resulting gene expression program included two basic categories of induced genes: an “early/persistent” cluster consistent with NF-κB-dependent responses downstream of TLRs, and a subsequent “late response” cluster largely composed of IFN-responsive genes (IRGs). The early/persistent cluster was observed upon infection with WT, heat-killed, or mutant Lm lacking listeriolysin O, the pore-forming hemolysin that promotes escape from phagocytic vacuoles. However, the IRG cluster depended on entry of WT Lm into the cytosol. Infection with listeriolysin O-expressing, cytosolic Bacillus subtilis (Bs) strikingly recapitulated the expression profile associated with WT Lm, including IRG induction. IRG up-regulation was associated with MyD88-independent induction of IFN-β transcription and activity. Whereas Staphylococcus aureus (Sa) lipoteichoic acid treatment confirmed that many late-response genes could also be stimulated through TLRs, our study identified a cytosol-specific transcriptional program independent of TLR signaling through MyD88. Further characterization of cytosolic surveillance pathway(s) and their points of convergence with TLR- and IFN-dependent pathways will enhance our understanding of the means by which mammals detect and respond to pathogens.
A central problem in cellular immunity is how immune effector cells integrate multiple signals, of microbial and host origin, to induce an appropriate immune response. Rapid detection of pathogens is crucial for the induction of innate and adaptive immune responses. The primary detection mechanism is mediated by plasma membrane-bound pathogen recognition receptors (PRRs), such as Toll-like receptors (TLRs), which allow self-nonself discrimination by selective engagement of pathogen-associated molecules (1). TLRs interact with various microbe-associated molecules, and subsequent activation induces up-regulation of costimulatory molecule expression, production of antimicrobial effector molecules, and secretion of proinflammatory cytokines and chemokines. These responses are mediated largely through NF-κB-dependent pathways (1). TLR signaling occurs at the cell surface or from within phagosomes and requires the MyD88 adaptor molecule for full activation (2, 3). However, little is known about innate immune signaling initiated within the cytosol. An emerging family of intracellular PRRs, called NOD-LRRs (nucleotide-binding oligomerization domain–leucine-rich repeats) may enable the innate immune system to sense intracellular pathogens. NOD-LRRs are involved in regulation of apoptosis and inflammation and have been linked to chronic inflammatory disorders (4). Although little is known about their ligands, NOD-LRRs may mediate immune responses to cytosolic pathogens. Indeed, cytosol-triggered NF-κB activation and cytokine gene transcription have been described, supporting the possibility of a TLR-independent, cytosolic surveillance pathway (5). Because bacterial pathogens have evolved to exploit different cellular niches, activation of the innate immune system from either the cell surface, or from within the vacuole or cytosol, may allow the host to recognize and respond to microbes that occupy distinct cellular compartments.
A Gram-positive intracellular pathogen, Listeria monocytogenes (Lm) is an excellent model organism for probing the innate response to cytosolic pathogens. Upon phagocytosis, Lm is initially enclosed within host cell vacuoles. Subsequent secretion by Lm of the hemolysin listeriolysin O (LLO) (encoded by hly) promotes vacuolar escape and cytosol entry. Cytosolic growth and cell-to-cell spread allow dissemination of Lm while maintaining its intracellular niche and avoiding the adaptive response. Several well characterized mutants cannot progress through the stages of the Lm life cycle and are thus constrained to distinct cellular compartments. LLO-deficient Lm (Δhly) cannot escape from host vacuoles, enter the cytosol, replicate, or spread from cell to cell (6). Because the Δhly mutant is vacuole-confined, it is a valuable tool for distinguishing between surface (TLR)- and cytosol-dependent gene expression programs.
Microarrays are a powerful tool for analysis of gene expression programs associated with diverse immunological processes, including the host–pathogen interaction (7). Both core and organism-specific transcriptional responses to various microbes have been described for several host cell types (8–11). Like other cell types, macrophages undergo a generalized response to a broad range of pathogen-associated molecules, including up-regulation of proinflammatory genes, receptors, signaling molecules, and transcription factors (12). Whereas TLRs play a critical role in this response, some pathogen-specific gene expression programs indicate the possible involvement of alternative signaling pathways (8, 12). However, little is known about the transcriptional response resulting from cytosolic localization of intracellular pathogens. We therefore characterized the gene expression program triggered in response to infection of primary bone marrow-derived macrophages (BMDM) with WT Lm, vacuole-restricted Lm mutants, and related nonpathogenic bacteria to identify genes induced by cytosolic surveillance. We found that cytosolic pathogen surveillance induces a transcriptional program encompassing many genes regulated by IFN-β. Several of these genes are also induced by Gram-positive Staphylococcus aureus (Sa) lipoteichoic acid (LTA) and a high multiplicity of infection (moi) of vacuole-restricted Δhly Lm, implying that both TLR and cytosolic surveillance pathways converge with Type I IFN pathways. We also describe a distinctly cytosol-specific gene expression program. Further characterization of signaling events that activate cytosol-mediated induction of both IFN-responsive gene (IRG) and non-IRG genes will help elucidate mechanisms by which hosts recognize and respond to cytosolic pathogens.
Materials and Methods
Bacterial Strains and Growth. Congenic Lm 10403S (WT) and DP-L2161 (Δhly) were inoculated into liquid brain–heart infusion and incubated overnight at 30°C without shaking. Before infection, Lm were normalized to 2 × 109 colony-forming units (cfu)/ml, washed twice, and resuspended in PBS. Congenic asporogenous (spoIIE::Tn917ΩHU181) Bacillus subtilis (Bs) DP-L980 (SPβ::pAG58-ble1-hly; LLO+) and DP-L1066 (SPβ::pAG58-ble1; LLO–) 37°C mid-log cultures were normalized to 2 × 108 cfu/ml, washed twice, and resuspended in PBS (13).
Cell Culture, Infections, and Liposomes. BMDM were isolated from 4- to 6-week-old female MyD88–/– (gift of R. Medzhitov, Yale University School of Medicine, New Haven, CT) or C57BL/6 (The Jackson Laboratory) mice (14). Sixteen hours prior to infection, 3 × 107 BMDM per condition per time point were plated in L929 cell conditioned medium and incubated overnight at 37°Cin5%CO2. BMDM were infected with WT Lm at an moi of ≈6:1, resulting in ≈99% infected with 1–5 Lm per cell. Δhly Lm were added at mois of ≈6:1, or ≈300:1. At 0.5 hours postinfection (hpi), plates were washed three times with 37°C DMEM, and fresh media were added. At 1 hpi, gentamycin was added to give a final concentration of 50 μg/ml. Mock infections were conducted in parallel. mRNA was prepared with FastTrack 2.0 kits (Invitrogen). For Bs infections, isopropyl β-d-thiogalactoside (IPTG) was added to BMDM to give a final concentration of 1 mM, and normalized cultures were added to an moi of ≈6:1. At 0.5 hpi, plates were washed three times with DMEM, and fresh IPTG– media were added. Subsequent steps were as above. Heat-killed (HK) Lm were added to BMDM at an moi of ≈6:1. Additionally, HK Lm were added to BMDM at an moi of ≈6:1 and not washed off. Filtered Sa LTA (Sigma) was used at a final concentration of 10 μg/ml. Subsequent steps were as described for WT Lm. phosphatidylethanolamine/cholesterylhemisuccinate (PE/CHEMS) liposomes packaged with recombinant LLO (15) were added to BMDM at a final concentration of 100 ng/ml LLO. At 1 hpi, BMDM were washed three times with 37°C DMEM with subsequent steps as before. Except for the Sa LTA time course, all treatments were independently replicated two to three times.
Microarrays. Fluorescently labeled cDNAs were hybridized as follows: Bs+LLO vs. Bs–LLO, LLO+liposomes vs. LLO–liposomes, and all other conditions vs. mock infections. Data were normalized and have been archived at http://microarray-pubs.stanford.edu/Lm (16). The significance analysis of microarrays (SAM) algorithm (17) was used with two-class unpaired designs to identify genes differentially expressed relative to their aggregate T0 expression. Δ values giving approximately two false significant genes per treatment per time point were chosen, and a 3-fold threshold was superimposed. Sa LTA treatment genes were selected on the basis of a 3-fold threshold alone. We removed spots with signal <2-fold above background in both channels and/or a regression correlation <0.6. Significance at any one time point was sufficient for inclusion in a master gene list, which was used to retrieve the LLO ± liposome treatment data. Best-effort averages of replicates were calculated. Genes with <75% good data were excluded, and remaining data were clustered and visualized (18).
RT-PCR and Protein Assays. First-strand cDNA was produced by using 1 μg of total RNA and Applied Biosystems RT-PCR reagents. Quantitative PCR (q-PCR) was performed by using the GeneAmp 5700 Sequence Detection System using SYBR Green PCR reagents (Applied Biosystems). Primer sequences are available on request. Culture supernatant proteins were analyzed by using IFN-α (Pierce) and IFN-γ (R & D Systems) ELISA kits. IFN-type I bioassays were as described (19).
Results and Discussion
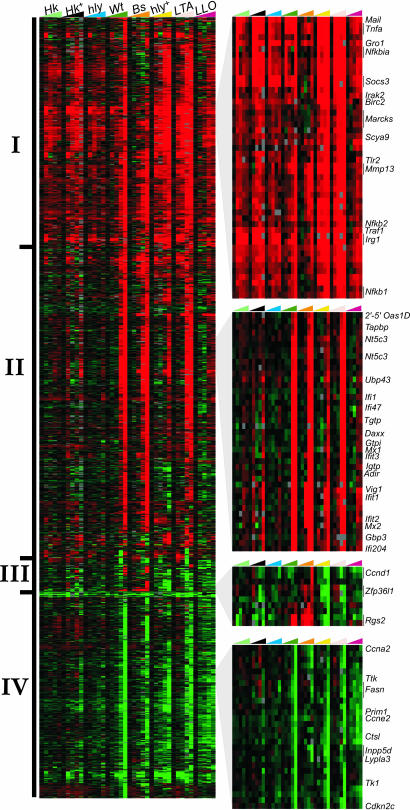
Cytosolic Lm Induce a Biphasic Transcriptional Response Whereas Vacuolar Lm Primarily Induce an Early TLR/NF-κB-Mediated Response. Macrophages are crucial mediators of innate immunity and have been used extensively as a model for host–pathogen interactions. Macrophages are also important sites of growth and dissemination of intracellular pathogens. We therefore used mouse cDNA microarrays to analyze the gene expression profiles of BMDM infected with cytosolic or vacuole-restricted Lm. To elucidate the transcriptional responses attributable to cytoplasmic localization of Lm, we first compared the response of BMDM to WT Lm, which enter the host cytosol, with that elicited by an equivalent initial moi of vacuole-restricted mutant Lm (Δhly) (Fig. 1, WT, Δhly). Consistent with previous studies, we found that the host–pathogen interaction resulted in significant expression level changes for a large number of host genes (Fig. 1; 1,445 vary >3-fold) (12, 9). The transcriptional response to WT Lm could be broadly divided into an “early” activation phase by 1–2 hpi (Fig. 1, group I) and a “late” phase by 4–8 hpi (Fig. 1, groups II and IV). However, this biphasic response was unlike the pattern observed previously in response to WT Lm infection of human Caco-2 epithelial-like cells, which primarily manifest an early-and-persistent response (9). This difference may reflect host or cell-type specificity or could be attributable to the postinduction repression of Ifnb expression noted in human cells (5). In our system, Δhly Lm also induced the early response, but the late cluster was unique to WT Lm. Because TLRs would presumably have a comparable ability to signal the presence of WT and Δhly Lm, we hypothesized that the early response was attributable to the initial cell surface–Lm interaction. To determine whether the early response could also be elicited by Lm-associated molecules, we exposed BMDM transiently (0.5 h; Fig. 1, HK) or persistently (8 h; Fig. 1, HK+) to vacuole-restricted, HK Lm. We found that, like the Δhly mutant, HK and HK+ Lm induced only the early cluster. Thus, the early response did not depend upon any interaction with live Lm but was instead a generalized response presumably mediated by surface/vacuole-bound PRRs.
Fig. 1.
Macrophage activation program elicited by intracytosolic or vacuolar bacteria and bacterial components. Shown is an Eisen plot of 1,445 genes selected as described in Materials and Methods. Columns represent BMDM treatments at 0, 1, 2, 4 and 8 hpi; rows colorimetrically represent expression ratios of individual genes. Cy5-labeled treatments were compared with matched Cy3-labeled uninfected or untreated controls. Intensity of red or green shading is proportional to, respectively, gene induction or repression relative to the uninfected or control. Zoom boxes represent clusters described in the text. HK, heat-killed Lm, 0.5-h treatment; HK+, persistent 8-h treatment; Hly, Δhly Lm (moi = 1); WT, WT Lm; Bs, B. subtilis ± LLO; Hly+, Δhly Lm (moi = 50); LTA, S. aureus LTA; LLO, liposomes ± LLO.
The early response induced by vacuole-restricted Lm was dominated by two basic patterns: a strong, transient response beginning by 1 hpi and diminishing by 4–8 hpi and an early/persistent response, lasting the duration of the time course. In neither case was cytosolic localization required for gene induction; Δhly and HK Lm induced as efficiently as WT Lm. Most genes in this subset seemed to be components of the general response described for both bacteria-associated molecules and for Lm in the context of the Caco2 and THP1 cell lines (8, 9, 11). The early cluster was strongly enriched for genes with regulatory or effector roles in innate immunity, including receptors, signal transducers, proinflammatory cytokines, and chemokines. Indeed, many of these genes are direct members of TLR/NF-κB signaling pathways or are induced by their stimulation. Induced NF-κB pathway members included NF-κB precursors p105 and p49/p100, and the NF-κB inhibitor IκB. Among the induced cytokine genes were IL-1β and tumor necrosis factor (TNF)-α, both hallmarks of NF-κB activation. Consistent with the role of macrophages in effector cell recruitment, we observed induction of large numbers of chemokines, predominantly from the C–C ligand family. Induced innate immunity receptor genes included Tlr2, Cd14, and the n-formyl peptide receptor, Fpr1. Whereas the majority fell into the early/persistent category, it is interesting to note that the early/transient category included the gene encoding immediate early response 3 (IER3), which counters the proapoptotic properties of TNF-α. Thus, both WT and vacuole-bound Lm induce an early TLR/NF-κB-associated response whereas cytosolic localization of WT Lm was required to induce the late response.
The Presence of Cytosolic Lm Stimulates a Distinct Late Expression Cluster. Unlike the universal early/persistent cluster, the late cluster (Fig. 1, group II) was observed upon infection with WT, but not Δhly or HK Lm. The late cluster coincided with times (4–8 hpi) at which Lm localized to the cytoplasm. At later time points, BMDM infected with WT Lm had a significantly higher bacterial load than those infected with an equivalent initial moi of Δhly Lm (Fig. 5, which is published as supporting information on the PNAS web site). To determine whether differences between WT and Δhly gene expression was due to the disparity in bacterial numbers, we infected BMDM with Δhly Lm at an initial moi 50-fold higher than previously used (hly+, 300:1). This increased moi resulted in a bacterial load of Δhly Lm equal to or exceeding the number observed in WT infections at 8 hpi (median 40 Lm per cell; Fig. 6, which is published as supporting information on the PNAS web site). As expected, this “overinfection” led to powerful stimulation of genes associated with the cell surface receptor-mediated early/persistent cluster (Fig. 1, hly+). Also, whereas some late cluster genes previously uninduced by Δhly and HK Lm infection did then exhibit activation upon Δhly+ infection, the majority remained uninduced. Thus, some overlap exists between Lm-mediated surface/vacuolar and cytosolic signaling, but cytoplasmic localization of Lm was necessary for up- or down-regulation of most late cluster genes.
Alteration of Cell Cycle Regulation and Nucleic Acid Metabolism Is the Dominant Theme of the Late Repressed Cluster. Unlike the early/persistent cluster, which was strongly enriched for activated genes, the late response cluster featured both activated and repressed genes (Fig. 1, groups II and IV, respectively). The repressed cluster included many genes involved in cell cycle regulation, DNA replication, and nucleic acid metabolism, consistent with the role of IFNs as physiological growth inhibitors affecting the cell cycle. Significantly, the gene encoding the oncogenic Myc transcription factor is repressed. Myc induction promotes cell proliferation by activation of growth-promoting genes such as ornithine decarboxylase and Cdc25a, both of which are group IV members (20, 21). Myc repression has been previously associated with growth inhibition by type I IFNs. The repression of cellular growth-related genes attendant to WT Lm infection may serve to reserve energy or resources for sustaining the immune response or to inhibit further bacterial growth and/or spread. Growth inhibition instigated by intracytosolic Lm may be attributable to IFN induction.
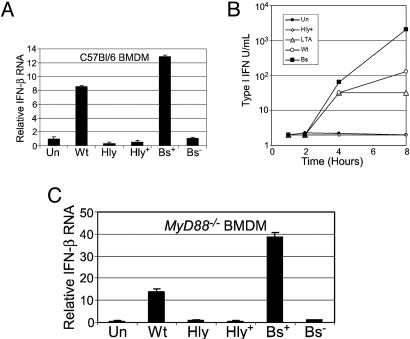
The Late Cytoplasmic Cluster Is Dominated by IFN-Regulated Genes. A striking feature of the late activation cluster (Fig. 1, group II) is the predominance of IRGs. Both Type I (α/β) and Type II (γ) IFNs play significant roles in the host response to numerous pathogens, predominantly by mediating induction of genes crucial to development of innate and adaptive immunity (22, 23). In our system, ELISA analysis of culture supernatants showed that neither IFN-α nor IFN-γ production mediated the observed IRG induction (data not shown). Historically, it was thought that IFN-β was produced primarily in response to viral pathogens. However, we and others have demonstrated that IFN-β production can also be elicited by bacterial pathogens and pathogen-associated molecules (5, 19, 24). Cytosolic Lm infection resulted in significant IFN-β transcription and secretion, but even a high initial dose of Δhly Lm did not (Fig. 3).
Fig. 3.
Cytosolic but not vacuolar bacteria significantly induce IFN-β mRNA and protein expression. (A) Mouse BMDM treated with cytosolic bacteria [WT, WT Lm; Bs+, Bs plus LLO] or vacuolar bacteria [hly, Δhly Lm (moi = 6), hly+, Δhly Lm (moi = 300)]. Shown is total RNA from 6 hpi, Ifnb RNA levels estimated by quantitative PCR, mean of three experiments ± SD. (B) Type I IFN bioassay, supernatants from BMDM treated with bacteria or bacterial products, legend as in Fig. 1 except Bs = B. subtilis-expressing LLO. Shown are the means of two separate bioassays of samples pooled from three experiments ± SEM. Data shown for 4 and 8 hpi; all samples before 4 hpi had titers ≤4. (C) Total RNA from MyD88–/– BMDM at 6 hpi. Ifnb levels were estimated by quantitative PCR. Values represent the means of three experiments ± SD. In all panels, Un = untreated control.
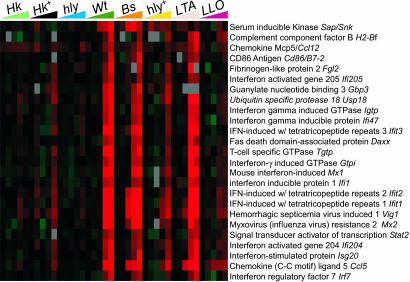
The significance of IFN-β production in the context of bacterial infections is unclear although type I IFNs may sensitize macrophages to Lm-induced cell death (25). Interestingly, the late IRG cluster included genes encoding anti-viral factors, such as myxovirus resistance proteins and oligoadenylate 2′-5′ synthetase, and genes involved in MHC class I antigen presentation, such as Tapasin and proteasome component Lmp2 (Fig. 2). Whereas the precise functions of many IRGs are unknown, some IRGs, such as Ccl5/Rantes, encode chemokines promoting recruitment of inflammatory cells. Thus, cytosolic Lm-induced IFN-β and subsequent IRG activation may serve to enhance antigen processing and presentation, as well as to recruit and activate inflammatory cells, just as in viral infections.
Fig. 2.
The late expression cluster is dominated by IFN-dependent genes. Both cytosolic and LTA-induced signaling induce IFN-regulated gene expression; shown are the 25 most highly expressed (WT, 8 h) group II genes previously shown to exhibit IFN dependence. Abbreviations are as in Fig. 1.
Extensive activation of IFN signaling genes was noted, including the signal transducers and activators of transcription Stat1, Stat2, and Stat3, and the Jak2 kinase required for later IFN-γ signaling. Notably, the late cluster includes genes encoding IL-12 and IL-18, which synergistically promote secretion of IFN-γ by NK and T cells, and direct the immune response toward a protective Th1 response. IFN regulatory factors (IRF) Irf1 and Irf7, which encode factors that bind IFN-stimulated response elements in the promoter regions of IRGs, were also induced. IRF7 is associated with transcription of IFN-α genes, and, despite the absence of IFN-α production during our time course, this observation suggests that α-IFNs may nevertheless be produced later. The extent to which the IFN response depends on IFN-β secretion or type I IFN receptor binding is unclear; future studies will investigate whether IRFs such as IRF3 may directly stimulate transcription of IRGs in response to Lm.
Finally, IRG induction could contribute to clinical manifestations of listeriosis; we note the induction of Fgl2, encoding fibrinogen-like protein 2, an IFN-regulated protease that cleaves prothrombin. FGL2-mediated thrombin deposition in the vascular epithelium of veins and arteries nourishing the placenta causes fibrin deposition, and complement and leukocyte activation leading to termination of placental blood flow. Fgl2 induction may thus contribute to Lm-induced spontaneous abortion (26).
IRG Induction Is Recapitulated by Cytoplasmic Localization of Bs, Indicating a Generalized Rather than Lm-Specific Response. The dependence of the IRG cluster on cytosolic localization of Lm led us to question whether the IFN response was specific to Lm infection. We therefore tested whether cytoplasmic localization of a related nonpathogenic bacteria would recapitulate the WT Lm transcriptional response. We infected BMDM with a strain of Bs engineered to secrete LLO (27). Bs expressing LLO (Bs+LLO) escape the phagocytic vacuole and enter the cytosol but persist rather than proliferate (Fig. 7, which is published as supporting information on the PNAS web site). Strikingly, we observed that infection of BMDM with Bs+LLO, but not a non-LLO-producing control (Bs–LLO), closely mimicked the response induced by WT Lm (Fig. 1, Bs). This response included significant induction of both Ifnb transcription and IFN-β secretion upon infection by Bs+LLO, but not by Bs–LLO (Fig. 3B). Except for a small set of genes induced only by Bs+LLO (Fig. 1, group III), the overwhelming majority of group II genes exhibited similar expression patterns under both infection regimes. However, because the Lm virulence factor LLO was used in both instances to mediate cytosolic localization, we examined the transcriptional effects of LLO alone.
LLO has been shown to trigger several host-signaling pathways, but many of these experiments directly exposed cell cultures to purified LLO (e.g., 28). Exogenous LLO treatment damages the cell membrane and differs greatly from the way that host cells would be exposed to LLO during Lm infection. To imitate a more physiological exposure to LLO, we used an acid-labile liposome system to deliver purified LLO directly to the cytosol (Fig. 8, which is published as supporting information on the PNAS web site). We compared relative BMDM transcription occurring upon treatment with LLO+ and LLO– liposomes and found that LLO modestly activated many genes of the early/persistent cluster. However, we found significant induction by 1 hpi of Tnfa, consistent with earlier studies showing LLO-induced tumor necrosis factor (TNF)-α production (28). This result was also consistent with the somewhat stronger stimulation of the early cluster observed in WT relative to HK or Δhly Lm infections. Significantly, LLO+ liposomes had little effect on the majority of genes in the late cluster (Fig. 1, LLO). Thus, LLO played no significant role in triggering the cytosolic surveillance pathway but was able to induce NF-κB-regulated genes, possibly through crosstalk with pathway(s) downstream of TLRs. We therefore conclude that late IRG cluster induction principally required general recognition of cytosolic bacteria rather than depending on some specific feature of Lm pathogenesis.
The IRG Cluster Is Induced by Sa LTA, Implying Convergence of TLR, IFN, and Cytosolic Surveillance Pathways. Although we observed little evidence for IRG activation by noncytosolic bacteria, others report that extracellular stimulation of TLR3 and TLR4 by double-stranded RNA (dsRNA) and lipopolysaccharide (LPS), respectively, can lead to the production of IFN-β and activation of IRGs (29, 30). Lm presents neither LPS nor dsRNA, and the late IRG cluster remained uninduced by high numbers of Δhly Lm. Given that TLR2 has not been implicated in IRF3/IFN-β pathways, and if TLR2 is the primary mediator of noncytosolic Lm detection, one would predict little or no induction of the IRG cluster by Lm-associated molecules alone.
We wished to treat BMDM with a strong TLR stimulus to both maximize identification of TLR-induced IRGs and to further distinguish cytosol-specific genes. We therefore exposed BMDM to a high dose (10 μg/ml) of Sa LTA, a Gram-positive TLR ligand. Despite our inability to efficiently stimulate the late IRG cluster with a high moi of Δhly Lm, we were surprised to find that the majority of the genes in that cluster were induced strongly by this purely extracellular stimulus, and with kinetics similar to those observed upon infection with cytosolic bacteria (Fig. 1, LTA, group II). Although some group II genes were induced to a lesser degree by Sa LTA treatment than by WT Lm or Bs+LLO infection, IRG expression was similar across these conditions, indicating that both cytosolic and surface/vacuolar stimulation could lead to IRG induction.
Whether Sa LTA is a TLR2 or TLR4 ligand is controversial, and endotoxin contamination of commercial LTA preparations may explain this confusion (31). Using Tlr4-deficient C3H/HeJ BMDM, we observed that Ifnb induction by our Sa LTA preparation was entirely TLR4-dependent (data not shown). Thus, the gene induction observed upon Sa LTA treatment may have been attributable to contaminating LPS signaling through a TLR4-mediated pathway not normally triggered by Lm, leading to induction of the IRG cluster. Alternatively, Sa LTA may naturally signal through TLR4. Regardless, these results point to links among TLR, IFN, and cytosolic signaling pathways. Indeed, evidence for convergence of these pathways has been described. All TLRs share a common adaptor protein, MyD88, which is required for early and complete activation of NF-κB/mitogen-activated protein kinase (MAPK) signaling (1). MyD88 is not absolutely required for signaling through all TLRs, and several alternate adaptors have been identified that interact with specific TLRs and transmit signals independently of MyD88. For example, the TRIF adaptor mediates MyD88-independent, TLR3-mediated induction of IFN-β and IRGs by double-stranded RNA, by means of IRF3 activation (32). TRIF also confers MyD88-independent, TLR4-mediated induction of IFN-β and IRG expression by LPS, also by means of IRF3 activation (33). In contrast to TRIF, the TRAM adaptor mediates a MyD88-independent pathway specific to TLR4 (34).
Using MyD88–/– BMDM, we found MyD88-independent Ifnb induction by WT Lm and by Bs+LLO, but not by vacuole-restricted bacteria (Fig. 3C). This result corroborates research showing that Ifnb induction upon WT Lm infection is not TLR4-dependent (5). We conclude that Ifnb induction by cytosolic Lm and Bs is a general response independent of TLR signaling through MyD88, and may thus be triggered by an alternate, cytosolic surveillance pathway. Furthermore, we interpret the IRG induction by Sa LTA as evidence that both pathways are active in BMDM and converge on common regulatory factors that induce Ifnb expression. It will be of major interest to determine at what point(s) the cytosolic surveillance pathway may intersect with TLR-mediated pathways leading to IFN-β production; IRF3 is one likely candidate (25).
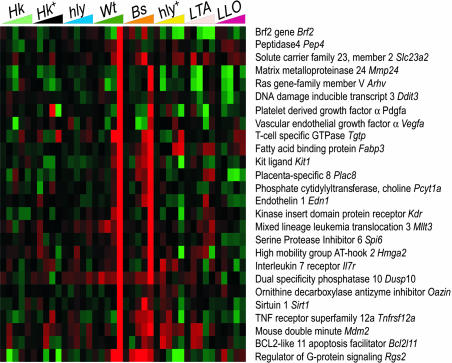
Cytosolic Lm and Bs Induce Cytosol-Specific Genes. Despite evidence for convergence of TLR, IFN, and cytosolic signaling pathways in IRG induction, we also observed a distinct, cytosol-specific induction of genes by WT Lm and Bs+LLO. Twenty-six genes were highly induced at 8 hpi by WT Lm and Bs+LLO, but not by Sa LTA or Δhly+ (Fig. 4). These genes represent a diversity of transcription and growth factors, mediators of cell–cell interaction, migration, nutrient acquisition, and vascularization, but the functional significance and transcriptional regulation of this subset are unknown. Because of its unusual expression pattern and inhibitory role in signal transduction, Rgs2 is of special interest. Although WT Lm and Bs+LLO significantly induce Rgs2, Sa LTA treatment and Δhly overinfection both repress this gene (Fig. 4, LTA and hly+). Rgs2 encodes regulator of G protein signaling 2, which belongs to a class of proteins that negatively regulate heterotrimeric G proteins by increasing their GTPase activity. RGS2 specifically inhibits Gαq-mediated signaling, and Rgs2–/– mice have impaired anti-viral immunity as a consequence of T cell hyporesponsiveness (35). Whereas the identity of the G protein-coupled receptor(s) (GPCRs) modulated by RGS2 upon Lm infection is unknown, we note that some chemokine receptors are Gαq-coupled (36). Other Gaq-associated GPCRs involved in inflammation include the angiotensin-2R, α-1-adrenergicR, vasopressin-1R, P2Y1R, G2AR, and several eicosanoid receptors. Thus, a distinct cytosol-specific program of gene expression, induced only by cytosolic bacteria, may modulate the host inflammatory response. It will be interesting to determine which signaling pathways are influenced by the expression of this class of genes, and to determine how these genes function to regulate the innate response to cytosolic pathogens.
Fig. 4.
Intracytosolic Lm and Bs induce cytosol-specific genes. Abbreviations are as in Fig. 1. Twenty-six genes representing the cytosol-specific pathway were selected based on the following criteria: >3-fold induction for WT Lm at 8 hpi, <3-fold induction by Sa LTA at 4 and 8 hpi, and a >3-fold difference between 8 hpi WT Lm and 8 hpi LTA expression levels.
NOD-LRRs Are Candidate PRRs for Cytosolic Gram-Positives. Whereas TLR-mediated pathogen surveillance at the cell surface is well established, our data contribute to a body of evidence that this is not the exclusive mode of microbe detection. TLR-mediated pathways are, in principle, ill-suited to the prolonged surveillance of intracellular pathogens, such as Lm, that gain access to the host cytoplasm and spread directly from cell to cell without further engaging extracellular receptors. Whereas the mechanism of cytosolic signaling in response to Lm or Bs+LLO infection is unknown, the NOD-LRRs are possible receptor candidates. This family of putative PRRs includes ≈25 members that share homology with plant disease-resistance proteins, several of which are cytosolic molecules inducing localized cell death upon pathogen infection (4).
The role of specific mammalian NODs is unclear, but three (Nod1, Nod2, and Nalp3) have been implicated in immunodeficiency or inflammatory disorders such as Crohn's disease (4). Both NOD1 and NOD2 are activated by specific peptidoglycan derivatives, and NOD1 has been directly associated with NF-κB activation in response to the Gram-negative cytosolic pathogen Shigella flexneri (37). Stimulation of NOD2 activates proinflammatory and costimulatory molecule induction by monocytes and dendritic cells, suggesting a TLR-like role in linking innate and adaptive immunity (38). Furthermore, synergism of NOD ligands with LPS indicates that NODs and TLRs may cooperate in immune responses against bacteria (39).
Whether NOD-LRRs play a direct role in the immune response to cytosolic Lm is undetermined. The NOD2 ligand, muramyl dipeptide (MDP) can potentially be generated from Lm peptidoglycan upon cleavage by the Lm autolysins p60 and NamA (40). It is unknown whether Lm MDP is a NOD2 ligand and if so, whether it would be immunostimulatory or immunomodulatory. However, any NOD-LRR ligand in our system is not specific to naturally intracellular pathogens because cytosolic Bs efficiently induced cytosol-specific gene induction.
Although we are unaware of evidence for NOD-LRRs contributing to IRG expression, NOD-LRR-signaling pathways could, like TLR pathways, intersect with IFN pathways, potentially explaining the MyD88-independent induction of Ifnb by cytosolic but not vacuole-bound bacteria. TLR and NOD-LRR pathways share common signaling intermediates and transcriptional outputs and must thus converge at specific points in their respective signaling cascades. For example, the receptor-interacting protein 2 (RIP2) kinase, identified as immediately downstream of NOD1, is required for full signaling through TLR2, –3, and –4 (41). Rip2–/– mice are impaired in their ability to defend against Lm infection (42).
In summary, we have identified both distinct and overlapping TLR-, IFN- and cytosol-mediated transcriptional programs induced in primary macrophages upon infection with cytosolic and vacuolar bacteria. Identification of genes induced by cytosol-specific signaling supports a growing body of evidence pointing to the existence of a TLR-independent cytosolic surveillance system. We also identified a subset of IRGs that could be induced both by TLR and cytosolic signaling, further supporting evidence of overlap between TLR, IFN, and cytosolic pathways. It will be of great interest to determine whether cytosolic Lm infection results directly in NOD-LRR-mediated signaling, and at which point, if any, NOD-LRR pathways overlap with IFN-associated pathways. Although the significance of crosstalk among the TLR, IFN, and NOD-LRR pathways in the innate immune response is unclear, it will be important to elucidate the collective roles these pathways play in the host response to cytosolic pathogens.
Supplementary Material
Acknowledgments
We thank A. Alizadeh, V. Auerbuch, L. Cheng, A. McCaffrey, D. Relman, and D. Raulet for comments. This work was supported by Defense Advanced Research Planning Agency Grant N65236-99-1-5428 (to P.O.B.) and National Institutes of Health Grants R01 A127655 and R37 AI029619 (to D.A.P.); R01 CA77097 (to P.O.B.); and F31 AI50250-01 (to R.L.M.). P.O.B. is a Howard Hughes Medical Institute investigator.
Abbreviations: PRR, pathogen recognition receptor; TLR, Toll-like receptor; NOD-LRR, nucleotide-binding oligomerization domain–leucine-rich repeat; LLO, listeriolysin O; BMDM, bone marrow-derived macrophage; LTA, lipoteichoic acid; moi, multiplicity of infection; HK, heat-killed; IRG, IFN-responsive gene; IRF, IFN regulatory factor; LPS, lipopolysaccharide; Lm, Listeria monocytogenes; Sa, Staphylococcus aureus; Bs, Bacillus subtilis; hpi, hours postinfection.
References
- 1.Medzhitov, R. (2001) Nat. Rev. Immunol. 1, 135–145. [DOI] [PubMed] [Google Scholar]
- 2.Underhill, D. M. & Ozinsky, A. (2002) Curr. Opin. Immunol. 14, 103–110. [DOI] [PubMed] [Google Scholar]
- 3.Janssens, S. & Beyaert, R. (2002) Trends Biochem. Sci. 27, 474–482. [DOI] [PubMed] [Google Scholar]
- 4.Inohara, N. & Nunez, G. (2003) Nat. Rev. Immunol. 3, 371–382. [DOI] [PubMed] [Google Scholar]
- 5.O'Riordan, M., Yi, C. H., Gonzales, R., Lee, K.-D. & Portnoy, D. A. (2002) Proc. Natl. Acad. Sci. USA 99, 13861–13866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Portnoy, D. A., Auerbuch, V. & Glomski, I. J. (2002) J. Cell Biol. 158, 409–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Staudt, L. M. & Brown, P. O. (2000) Annu. Rev. Immunol. 18, 829–859. [DOI] [PubMed] [Google Scholar]
- 8.Boldrick, J. C., Alizadeh, A. A., Diehn, M., Dudoit, S., Liu, C. L., Belcher, C. E., Botstein, D., Staudt, L. M., Brown, P. O. & Relman, D. A. (2002) Proc. Natl. Acad. Sci. USA 99, 972–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baldwin, D. N., Vanchinathan, V., Brown, P. O. & Theriot, J. A. (2002) Genome Biol. 4, R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang, Q., Liu, D., Majewski, P., Schulte, L. C., Korn, J. M., Young, R. A., Lander, E. S. & Hacohen, N. (2001) Science 294, 870–875. [DOI] [PubMed] [Google Scholar]
- 11.Cohen, P., Bouaboula, M., Bellis, M., Baron, V., Jbilo, O., Poinot-Chazel, C., Galiegue, S., Hadibi, E. H. & Casellas, P. (2000) J. Biol. Chem. 275, 11181–11190. [DOI] [PubMed] [Google Scholar]
- 12.Nau, G. J., Richmond, J. F., Schlesinger, A., Jennings, E. G., Lander, E. S. & Young, R. A. (2002) Proc. Natl. Acad. Sci. USA 99, 1503–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Portnoy, D. A., Tweten, R. K., Kehoe, M. & Bielecki, J. (1992) Infect. Immun. 60, 2710–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Portnoy, D. A., Jacks, P. S. & Hinrichs, D. J. (1988) J. Exp. Med. 167, 1459–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, K.-D., Oh, Y.-K., Portnoy, D. A. & Swanson, J. A. (1996) J. Biol. Chem. 271, 7249–7252. [PubMed] [Google Scholar]
- 16.Gollub, J., Ball, C. A., Binkley, G., Demeter, J., Finkelstein, D. B., Hebert, J. M., Hernandez-Boussard, T., Jin, H., Kaloper, M., Matese, J. C., et al. (2003) Nucleic Acids Res. 31, 94–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tusher, V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Havell, E. A. (1986) J. Infect. Dis. 153, 960–969. [DOI] [PubMed] [Google Scholar]
- 20.Wagner, A. J., Meyers, C., Laimins, L. A. & Hay, N. (1993) Cell Growth Differ. 4, 879–883. [PubMed] [Google Scholar]
- 21.Galaktionov, K., Chen, X. & Beach, D. (1996) Nature 382, 511–517. [DOI] [PubMed] [Google Scholar]
- 22.LeBon, A. & Tough, D. F. (2002) Curr. Opin. Immunol. 14, 432–436. [DOI] [PubMed] [Google Scholar]
- 23.Shtrichman, R. & Samuel, C. E. (2001) Curr. Opin. Microbiol. 4, 251–259. [DOI] [PubMed] [Google Scholar]
- 24.Herzog, P. J., O'Neill, L. A. & Hamilton, J. A. (2003) Trends Immunol. 24, 534–539. [DOI] [PubMed] [Google Scholar]
- 25.Stockinger, S., Materna, T., Stoiber, D., Bayr, L., Steinborn, R., Kolbe, T., Unger, H., Chakaborty, T., Levy, D., Müller, M., et al. (2002) J. Immunol. 169, 6522–6529. [DOI] [PubMed] [Google Scholar]
- 26.Knackstedt, M. K., Zenclussen, A. C., Hertwig, K., Hagen, E., Dudenhausen, J. W., Clark, D. A. & Arck, P. C. (2003) Am. J. Reprod. Immunol. 49, 210–220. [DOI] [PubMed] [Google Scholar]
- 27.Bielecki, J., Youngman, P., Connelly, P. & Portnoy, D. A. (1990) Nature 345, 175–176. [DOI] [PubMed] [Google Scholar]
- 28.Kayal, S., Lilienbaum, A., Join-Lambert, O., Li, X., Israel, A. & Berche, P. (2002) Mol. Microbiol. 44, 1407–1419. [DOI] [PubMed] [Google Scholar]
- 29.Alexopoulou, L., Holt, A. C., Medzhitov, R. & Flavell, R. A. (2001) Nature 413, 732–738. [DOI] [PubMed] [Google Scholar]
- 30.Toshchakov, V., Jones, B. W., Perera, P., Thomas, K., Cody, M. J., Zhang, S., Williams, B. R. G., Major, J., Hamilton, T. A., Fenton, M. J., et al. (2002) Nat. Immunol. 3, 392–398. [DOI] [PubMed] [Google Scholar]
- 31.Gao, J. J., Xue, Q., Zuvanich, E. G., Haghi, K. R. & Morrison, D. C. (2001) Infect. Immunol. 69, 751–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamamoto, M., Sato, S., Mori, K., Hoshino K., Takeuchi O., Takeda, K. & Akira, S. (2002) J. Immunol. 169, 6668–6672. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto, M., Sato, S., Hemmi, H., Hoshino K., Kaisho, T., Sanjo, H., Takeuchi O., Sugiyama, M., Okabe, M., Takeda, K., et al. (2003) Science 301, 640–643. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto, M., Sato, S., Hemmi, H., Uematsu, S., Hoshino K., Kaisho, T., Takeuchi O., Takeda, K. & Akira, S. (2003) Nat. Immunol. 4, 1144–1150. [DOI] [PubMed] [Google Scholar]
- 35.Oliveira-Dos-Santos, A. J., Matsumoto, G., Snow, B. E., Bai, D., Houston, F. P., Whishaw, I. Q., Mariathasan, S., Sasaki, T., Wakeham, A., Ohashi, P. S., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 12272–12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arai, H. & Charo, I. F. (1996) J. Biol. Chem. 271, 21814–21819. [DOI] [PubMed] [Google Scholar]
- 37.Girardin, S. E., Tournebize, R., Mavris, M., Page, A. L., Li, X., Stark, G. R., Bertin, J., DiStefano, P. S., Yaniv, M., Sansonetti, P. J., et al. (2001) EMBO Rep. 2, 736–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Girardin, S. E., Travassos, L. H., Herve, M., Blanot, D., Boneca, I. G., Philpott, D. J., Sansonetti, P. J. & Mengin-Lecreulx, D. (2003) J. Biol. Chem. 278, 41702–41708. [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi, K., Eynon, E. E. & Flavell, R. A. (2003) Nat. Immunol. 4, 652–654. [DOI] [PubMed] [Google Scholar]
- 40.Lenz, L. L., Mohammadi, S., Geissler, A. & Portnoy, D. A. (2003) Proc. Natl. Acad. Sci. USA 100, 12432–12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobayashi, K., Inohara N., Hernandez L. D., Galan J. E., Nunez G., Janeway C. A., Medzhitov R. & Flavell, R. A. (2002) Nature 416, 194–199. [DOI] [PubMed] [Google Scholar]
- 42.Chin, A. I., Dempsey, P. W., Bruhn, K., Miller, J. F., Xu, Y. & Cheng, G. (2002) Nature 416, 190–194. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.