SUMMARY
Many large viruses, including tailed dsDNA bacteriophages and herpesviruses, assemble their capsids via formation of precursors, called procapsids or proheads. The prohead has an internal core, made of scaffolding proteins, and an outer shell, formed by the major capsid protein. The prohead usually contains a protease which is activated during capsid maturation to destroy the inner core and liberate space for the genome. Here we report a 2.0 Å-resolution structure of the pentameric procapsid protease of bacteriophage T4, gene product (gp)21. The structure corresponds to the enzyme’s pre-active state in which its N-terminal region blocks the catalytic center, demonstrating that the activation mechanism involves self-cleavage of nine N-terminal residues. We describe similarities and differences between T4 gp21 and related herpesvirus proteases. We found that gp21 and the herpesvirus proteases have similarity with proteins forming the tubes of phage tails and bacterial type VI secretion systems, suggesting their common evolutionary origin.
Keywords: virus assembly, procapsid, prohead protease, herpesvirus, bacteriophage, self-cleavage, phage tail tube, bacterial type VI secretion system
Subject categories: Structural Biology, Virology, Enzymes, Protein Evolution
eTOC Paragraph
Fokine and Rossmann present the structure of the bacteriophage T4 capsid assembly protease, revealing that phage and herpesvirus proteases have similar folds but different activation mechanisms. The structure suggests that these proteases share common evolutionary origin with proteins forming the tubes of phage tails and bacterial type VI secretion systems.
INTRODUCTION
Bacterial viruses (bacteriophages or phages) are probably the most abundant biological entities on Earth with ~1031 virions in the biosphere (Hatfull and Hendrix, 2011; Hendrix, 2003). Most phages (96%) visualized by electron microscopy (Ackermann, 2007) have a tail and are classified into the Caudovirales order. Based on tail morphology the Caudovirales phages are divided into three families: Myoviridae, characterized by contractile tails; Siphoviridae, possessing long, non-contractile tails; and Podoviridae, characterized by short tails. Bacteriophage T4, a member of the Myoviridae family, has been used for many years as a model system to study virus assembly (Black and Rao, 2012; Leiman et al., 2010). The T4 virion consists of a 1200 Å-long and 860 Å-wide prolate head (Fokine et al., 2004) encapsidating a 172-kbp double-stranded DNA molecule, and a 1000 Å-long tail (Kostyuchenko et al., 2005; Leiman et al., 2004) containing a rigid inner tube surrounded by a contractile sheath. The tail terminates in a baseplate (Kostyuchenko et al., 2003; Taylor et al., 2016; Yap et al., 2016) to which are attached six long tail fibers. When the phage baseplate attaches to an E. coli cell the tail sheath contracts and drives the tail tube across the cell’s periplasmic space. The phage DNA genome then passes from the capsid into the cell through the tail tube.
The mature T4 capsid is a prolate icosahedron with icosahedral ends and an elongated midsection. The outer shell of the T4 head is formed by 930 copies of the major capsid protein, gene product 23 (gp23), which is organized into a hexagonal lattice characterized by the triangulation numbers Tend = 13 laevo for the icosahedral ends and Tmid = 20 for the midsection (Fokine et al., 2004). Eleven capsid vertices are occupied by pentamers of the special vertex protein, gp24 (Fokine et al., 2005), whereas the twelfth vertex is occupied by the dodecameric protein, gp20 (Sun et al., 2015), which is a portal for DNA packaging during the capsid assembly and exit during infection.
The T4 head and tail assemble via independent pathways and join together to form the mature virion. The head assembles via formation of a DNA-free precursor called a prohead or procapsid (Black et al., 1994). Prohead formation is initiated by the portal protein, gp20 (61 kDa), which nucleates the assembly of a scaffolding core. The main component of the core is the scaffolding protein, gp22 (30 kDa, with ~580 copy number). The other proteins constituting the core are gp67 (9 kDa, ~340 copies), gp68 (17 kDa, ~240 copies), gpalt (77 kDa, ~40 copies), IPI (10 kDa, ~360 copies), IPII (11 kDa, ~360 copies), and IPIII (22 kDa, ~370 copies) (Black et al., 1994; Leiman et al., 2003). In addition, the inner core contains the prohead maturation protease, gp21 (23 kDa, ~72 copies) which is initially inactive (Showe et al., 1976a, b). The major capsid protein, gp23 (56 kDa), forms an outer shell around the scaffolding core whereas the special vertex protein, gp24 (48 kDa), attaches to the prohead vertices (Fokine et al., 2006; Fokine et al., 2005). After completion of prohead assembly, the gp21 protease is converted to an active enzyme which cleaves most of the molecules of the core proteins gp22, gp67, and gp68 into small peptides. These peptides escape from the capsid, probably through holes in the major capsid protein capsomers, thus liberating space for the phage DNA genome. The protease also cuts off small N-terminal pieces (0.9 – 1.5 kDa) from IPI, IPII, IPIII, and gpalt (Black et al., 1994). In addition gp21 removes the 65-residue N-terminal region from gp23 and 10 N-terminal residues from gp24. The gp21 protease cleaves polypeptide chains after Leu/Ile-X-Glu motifs, where X can be any residue (Black et al., 1994). Although the protease was tentatively localized in the center of the scaffolding core (van Driel et al., 1980), studies of the T4 head-gene mutants suggested that activation of the protease is triggered by attachment of the gp24 vertex protein to the procapsid assembly (McNicol et al., 1977). In particular, a T4 mutant lacking gp24 produces proheads containing uncleaved core proteins (McNicol et al., 1977; Onorato et al., 1978). Addition of the gp24 protein to these proheads initiates cleavage of the core. As the isolated active gp21 enzyme was found to have a molecular weight of ~18.5 kDa, which is smaller than the weight of the full-length protein (23 kDa), it was suggested that the protease activates itself by self-cleavage (Keller and Bickle, 1986; Showe et al., 1976a).
After digestion of the core proteins, the gp21 protease finally cuts itself into small fragments which then leave the capsid. After destruction of the inner core, phage DNA is packaged into the head through the portal by the ATP-driven packaging motor, composed of gp17 (Sun et al., 2008). During DNA packaging the outer gp23/gp24 shell of the capsid expands, which results in an ~50% increase in capsid volume (Black et al., 1994; Leiman et al., 2003). The outer surface of the expanded capsid is then decorated by Hoc and Soc proteins (Fokine et al., 2011; Qin et al., 2010). After DNA packaging is complete the portal is sealed by gp13 and gp14 proteins which create the attachment site for an independently assembled phage tail (Akhter et al., 2007; Fokine et al., 2013).
In bacteriophage genomes, genes coding for the capsid assembly protease are usually located within the head morphogenesis modules. In these modules the protease gene (Cheng et al., 2004) is usually followed by the scaffolding protein gene which is, in turn, followed by the major capsid protein gene (e.g., genes 21, 22, and 23 of T4). In some phages, like λ and Mu, the protease and the scaffolding protein are coded by the same gene. The full-length product of this gene encodes a fusion protein consisting of the N-terminal protease domain and the C-terminal scaffolding region. The alternative predominant product of this gene, resulting from the use of an alternative start codon, is the scaffolding protein alone. Some phages, such as HK97 (Hendrix and Johnson, 2012; Veesler et al., 2014), do not have a separate scaffolding protein. In these cases the N-terminal part of the major capsid protein functions as the scaffold. Phage Gifsy-2 has an unusually large tripartite gene which encodes the protease at the 5′ end, the scaffolding protein in the middle, and the major capsid protein at the 3′ end (Effantin et al., 2010). Some phages, like φ29 and P22, do not encode a protease. Such phages have relatively small scaffolds which can dissociate without proteolytic cleavage and their scaffolding proteins can escape from the procapsid unprocessed during the genome packaging (Anderson and Reilly, 1993; Casjens and King, 1974; Morais et al., 2003). However, viruses with large capsids (like T4 and herpesviruses) usually need a protease to destroy the scaffolding core.
An unusual feature of the T4 gene 21 is the presence of a strong ribosome binding site which initiates translation of an alternative shorter version of the protein starting at Met45. Both the full-length gp21 and the short version were suggested to be important for phage development (Hintermann and Kuhn, 1992) although only the full-length gp21 is likely incorporated into the prohead.
Bioinformatic analysis of the phage protease sequences (Cheng et al., 2004; Liu and Mushegian, 2004) showed that the capsid assembly proteases of many tailed phages, including T4 and HK97, are related to the capsid assembly proteases (“assemblins”) of herpesviruses. Furthermore, most of the known prohead proteases from tailed dsDNA phages belong to one of two independent clans of serine proteases: herpesvirus assemblin-like or ClpP-like (Liu and Mushegian, 2004). Phylogenetic analysis of dsDNA bacteriophages suggested that the primordial common ancestor of the present-day phages might possess a herpesvirus assemblin-like protease and that, in some phages, this type of protease might be replaced by a ClpP-like protease during evolution (Liu and Mushegian, 2004).
Herpesviruses and tailed dsDNA phages have similar capsid assembly pathways and the major capsid proteins of these viruses have structural similarity (Baker et al., 2005; Brown and Newcomb, 2011; Cardone et al., 2012). As is the case for phages λ and Mu, the herpesvirus protease and the prohead scaffolding protein share one gene (Liu and Roizman, 1991; Zuhlsdorf et al., 2015). The full-length product of this herpesvirus gene is a fusion protein, UL26, which consists of the N-terminal protease domain and the C-terminal scaffolding domain. The alternative predominant product of the gene (produced from an alternative RNA transcript) is the scaffolding protein, UL26.5. In the procapsid the C-terminal part of the scaffolding protein interacts with the major capsid protein. During capsid maturation, the protease cleaves both the scaffolding protein and itself at the maturation (M) site located near the C-terminus. As a result, the scaffolding protein dissociates from the major capsid protein shell and later escapes from the procapsid during DNA packaging. In addition, the protease cleaves itself at the release (R) site resulting in release of the N-terminal protease domain from the C-terminal part containing the scaffolding protein.
Sequence alignment of the T4 gp21 protease with proteases of other tailed phages and herpesviruses (Liu and Mushegian, 2004; Thomas and Black, 2013) predicted Ser140 and His85 to be the gp21 residues involved in catalysis. To avoid possible self-cleavage, we produced a gp21 mutant which had alanines in positions 140 and 85. Here we present the structure of the mutant gp21 protein and describe the protease activation mechanism. The structure also explains the enzyme’s cleavage specificity. We describe similarities and differences between the T4 protease and the related herpesvirus proteases. We show that these proteases share a fold similarity with proteins forming the tubes of tailed phages and bacterial type VI secretion systems, suggesting a common evolutionary origin of all these proteins.
RESULTS AND DISCUSSION
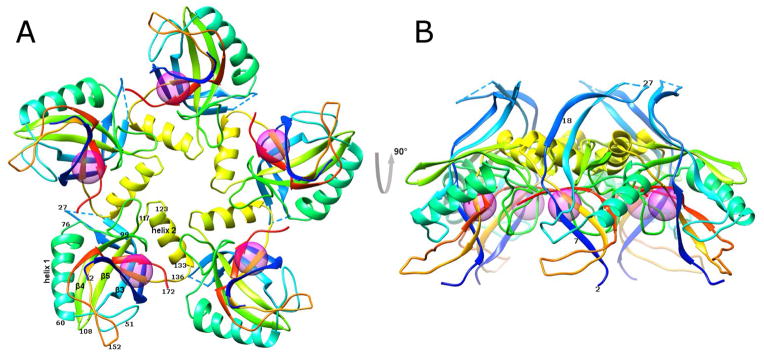
The structure of the bacteriophage T4 head maturation protease, gp21, has been determined using X-ray crystallography to 2.0 Å resolution. The crystals contained one pentamer of gp21 in the asymmetric unit. The gp21 pentamer has the shape of a starfish (Figure 1) with a diameter of ~95 Å and a height of ~50 Å. The core of each gp21 monomer consists of an orthogonal β-sandwich flanked by two α-helices.
Figure 1. Crystal structure of the T4 prohead protease pentamer.
The polypeptide chains, shown as ribbons, are rainbow-colored from blue at the N-terminus to red at the C-terminus. Positions of the catalytic centers are shown by semitransparent spheres.
(A) View along the five-fold axis of the pentamer.
(B) View perpendicular to the five-fold axis.
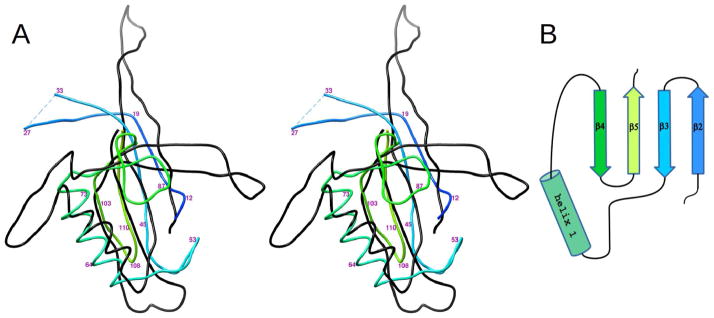
Each gp21 monomer interacts with two other monomers within the pentamer. The surface area buried for each gp21 monomer on formation of a pentamer is 1700 Å2. This large interface area shows (Janin, 1997; Janin and Rodier, 1995) that the monomers are bound tightly to their neighbors and, hence, that the pentamer is a stable entity. Most of the residues forming the interchain interfaces belong to the region 117–136 (Figure 2), which contains α-helix 2 (residues 123–133), and to the loop region 76–99, connecting α–helix 1 to β-strand 4. Helix 2 is inserted into a groove formed by an adjacent monomer and bound via electrostatic and hydrophobic interactions.
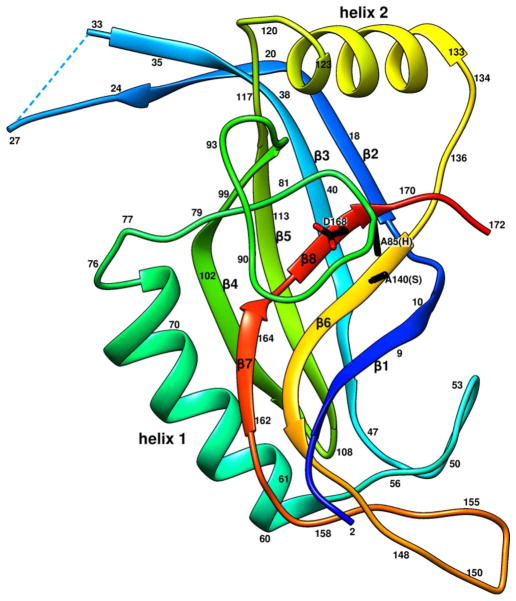
Figure 2. Ribbon diagram of the gp21 monomer rainbow-colored from blue at the N-terminus to red at the C-terminus.
Side chains of the residues 140 (Ser of the catalytic triad mutated to Ala), 85 (His of the triad mutated to Ala), and 168 (Asp of the catalytic triad) are shown in black.
Catalytic Center of gp21 and the Structural Basis of the Protease Cleavage Specificity
The five catalytic centers of the gp21 pentamer are located in the arms of the starfish (Figure 1). The catalytic triad of T4 gp21 is formed by Ser140, His85 and Asp168, consistent with earlier sequence analyses (Liu and Mushegian, 2004; Thomas and Black, 2013). Residues 140 and 85, which had been mutated to alanines (Figure 2), are located in β–strand 6, and in the loop connecting α-helix 1 to β–strand 4, respectively, whereas Asp168 is situated in β–strand 8. The loop region, containing His85 of the catalytic triad, is involved in interactions with other monomers within the gp21 pentamer. The pentameric interactions stabilize the conformation of this flexible region, suggesting that gp21 pentamerization is important for catalysis.
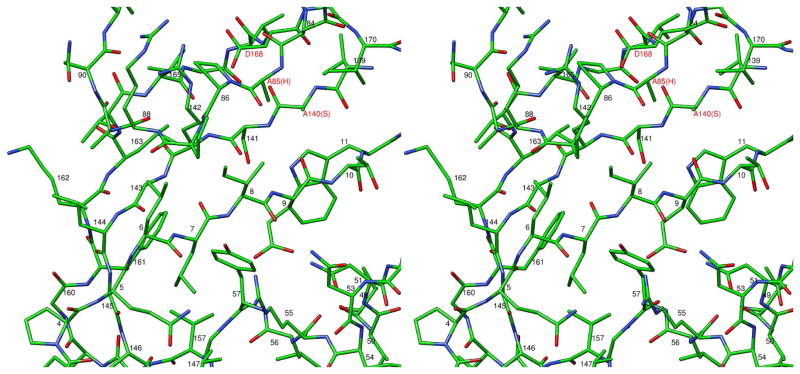
The T4 head maturation protease cleaves polypeptide chains after the specific motifs Ile/Leu-X-Glu, where X can be any residue (Black et al., 1994). In the gp21 crystal structure, the N-terminal region of gp21 containing the cleavage motif Leu7-Ile8-Glu9 is located in the active center and represents a substrate bound to the protease (Figure 3). This N-terminal region is a part of β-strand 1 and is involved in anti-parallel β-sheet interaction with the neighboring β-strand 6 (Figures 2 and 3). The side chain of Leu7 is located in the hydrophobic pocket (made by residues, Val157, Phe161, Tyr57, Gly145 and Leu147) explaining the requirement of Leu or Ile in this position of the cleavage motif. The side chain of Ile8 is exposed to the bulk solvent, explaining the absence of any specific side chain requirement for this position of the substrate. The side chain of Glu9 is located in the pocket created by residues Arg55, Asn51, and Tyr57, and make a salt bridge with Arg55, thus clarifying the requirement for the glutamate residue in this position of the cleavage motif.
Figure 3. Stereo view of the gp21 substrate binding site blocked by the N-terminal region of the polypeptide chain.
The peptide bond between Glu9 and Thr10 is located near the side chain of residue 140, which is the catalytic Ser mutated to Ala.
Activation of the Capsid Maturation Protease
The gp21 protease is initially produced in the host cell as an inactive enzyme, which is later activated in the assembled phage proheads. Our structure shows that the N-terminal region of gp21 blocks the catalytic center preventing binding of other substrates. Therefore, the N-terminal region of the protein should be cleaved off (after Glu9) and released from the active center during the enzyme’s activation process. In the gp21 crystal structure, which represents the protease in the pre-active state, the carbonyl carbon of Glu9 is located in proximity to the side chain of residue 140 which is the catalytic serine in native gp21. However, the orientation of the Glu9 carbonyl plane in the pre-active state is not favorable for catalysis. Thus interactions within the prohead, causing the protease activation, force the N-terminal region of gp21 to adopt a conformation favorable for the cleavage reaction.
Earlier biochemical studies of T4 mutants (McNicol et al., 1977) suggested that the gp21 protease is activated by attachment of the pentameric vertex protein, gp24, to the prohead assembly. These data, together with the pentameric nature of gp21, suggest that, in the inner core of the prohead, the gp21 pentamers are likely to be located near pentameric vertices. The number of gp21 monomers per T4 capsid is estimated from the SDS gel analysis (Black et al., 1994; Onorato et al., 1978) to be ~72, which is within a reasonable agreement with the assumption of there being one gp21 pentamer at each pentameric prohead vertex.
In the prohead the axes of the gp21 and gp24 pentamers are likely to be collinear, with gp21 being closer to the prohead center. Taking into account the curvature of the gp23/gp24 outer capsid shell, the convex top surface of the gp21 starfish pentamer (Figure 1B) is likely oriented towards the gp24 pentameric vertex, whereas the concave bottom surface of the gp21 starfish likely interacts with the inner core proteins. The gp21 structure contains a long extended region 2–27, which extends from top to bottom of the pentamer and includes the cleavable nine N-terminal residues of the protein (Figures 1B and 2). A part of this extended region (residues 18–24) protrudes from the top of the pentamer surface. Thus, this region probably serves as a sensor for gp24 attachment to the prohead. The attachment of gp24 may result in “squeezing” of this gp21 region, initiating the cleavage of the peptide bond between Glu9 and Thr10 of gp21 and the release of the cleaved N-terminal peptide.
Protease Release and Self-Degradation
After the cleaved N-terminal region is released, the active site becomes accessible for other cleavage substrates. The X-ray map does not show any interpretable density for the C-terminal part of gp21 after residue 172, suggesting that this region may be flexible. The flexibility of this region is consistent with the secondary structure prediction (using PsiPred, (Buchan et al., 2013)), which predicted a 25 residue-long coil region 173–197 followed by an α-helical region 198–212. In the T4 21amH29 mutant the gp21 protein lacks a small (> 4 kDa) C-terminal region and does not associate with the proheads (Onorato et al., 1978; Showe et al., 1976b). This observation, together with the fact that some phages and herpesviruses have scaffolding proteins fused to the C-terminus of the protease, suggests that the flexible C-terminal part of gp21 attaches to the scaffolding protein, gp22, of the assembling prohead core. In the fully assembled prohead the protease pentamers probably acquire fixed positions surrounded by other molecules.
The protease has two additional potential self-cleavage sites near its C-terminus: Ile185-Thr186-Glu187 and Leu204-Ala205-Glu206. Self-cleavage at these sites is probably necessary to detach the protease from the gp22 scaffold and to allow it to diffuse within the prohead and to cleave the prohead proteins. The last ordered residue in the gp21 model (172) is only ~10 Å apart from the catalytic center of the same gp21 monomer (Figure 2) and ~20 Å from the active site belonging to an adjacent gp21 chain within the pentamer. Thus, residues 185–187 and 204–206 belonging to the potential C-terminal cleavage sites can reach the active centers, assuming that the C-terminal part of gp21 is flexible and that the N-terminal regions 1–9 of gp21 have been cleaved off and released from the active centers. Study of the T4 gp24− mutant (Onorato et al., 1978; Showe et al., 1976b) suggested that the protease can slowly activate itself by self-cleavage within the proheads even in the absence of gp24 but cannot digest other prohead proteins. This observation suggests that self-cleavage is necessary, but not sufficient, to allow protease diffusion within the prohead. So, the binding of gp24 to the prohead not only initiates the protease self-cleavage, but also probably pushes the gp21 pentamer out from its fixed position. Thus the attachment of gp24 and self-cleavage at the C-terminal sites releases the protease pentamers and allows them to move freely and digest the core proteins. After cleavage of the inner core of the prohead, the gp21 protease also destroys itself into small cleavage products. The escape of all the degradation products from the prohead, probably through holes in immature gp23 capsomers, liberates space for genomic DNA entry.
In addition to the two potential cleavage motifs in the C-terminal region and the N-terminal cleavage site, the protease contains four potential Ile/Leu-X-Glu self-cleavage sites (Ile61-Leu62-Glu63; Ile70-Asn71-Glu72; Leu80-Gly81-Glu82; Ile99-Ile100-Glu101), located in the region between α-helix 1 and β-strand 4. As these cleavage sites are located in the ordered part of the structure and far from the active center of the enzyme, they should be available only to other gp21 pentamers. In this case at least one gp21 molecule/pentamer should remain in the capsid after maturation. The number of protease molecules in the mature head was estimated from analysis of SDS gels to be about four (Showe et al., 1976b). This is in reasonable agreement with one intact pentamer left per mature capsid.
Comparison of T4 gp21 to the Herpesvirus Proteases
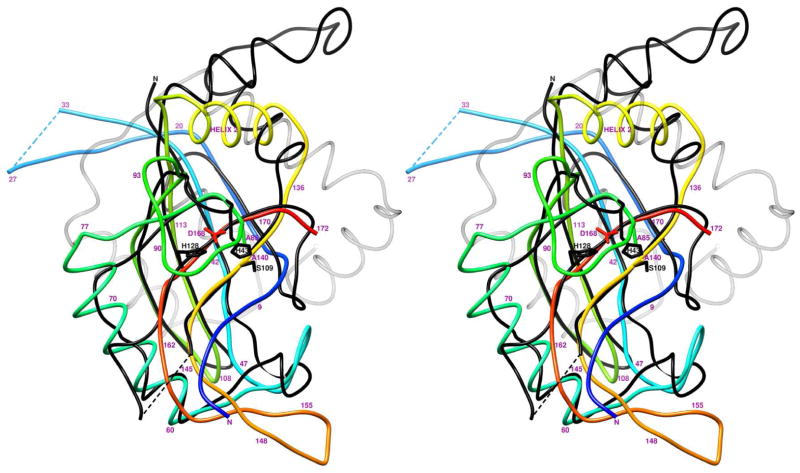
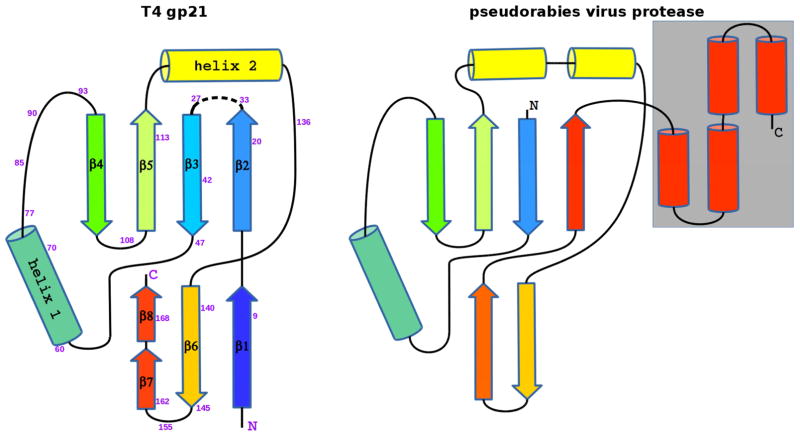
The structure of gp21 was found to be similar to herpesvirus proteases, consistent with earlier predictions based on sequence analyses (Cheng et al., 2004; Liu and Mushegian, 2004). Comparison of the gp21 structure with other known structures using the Dali algorithm (Holm et al., 2008) indicated that the closest structural relative was the capsid assembly protease from the pseudorabies virus ((Zuhlsdorf et al., 2015); PDB ID: 4v08), a member of the α-Herpesviridae subfamily. These two structures can be superimposed with RMSD of 2.2 Å using 91 equivalenced Cα atoms (Figures 4, 5), although the sequence identity between the equivalenced residues is only 7%. Most of the superimposable gp21 components are in the β-sandwich and helix 1. However, the β-strand 1 of gp21, which includes the cleavable N-terminal peptide, is not present in the herpesvirus proteases.
Figure 4. Stereo view of superposition of T4 gp21 with the pseudorabies virus protease.
The polypeptide chain of gp21 is rainbow-colored from blue at the N-terminus to red at the C- terminus. The polypeptide chain of the pseudorabies virus protease is shown in black. The C- terminal helical region of the pseudorabies virus protease, absent in gp21, is semitransparent. Residue numbers for the gp21 protein are shown in magenta.
Figure 5. Schematic diagrams showing topology of T4 gp21 (left) and the pseudorabies herpesvirus protease (right).
β–strands are represented by arrows, and α-helices are represented by cylinders. The C-terminal helical region of the pseudorabies virus protease, absent in gp21, is outlined by a grey rectangle.
Helix 2 is important for pentamerization of gp21. The corresponding region in the herpesvirus proteases is longer and contains two or three helices (Figure 4). This region of herpesvirus proteases is involved in protein dimerization (together with one of the C-terminal helices absent in the gp21 fold). Thus, this part of the structure is important for formation of protease oligomers in both virus families.
Unlike gp21, which has the classical catalytic triad Ser140/His85/Aps168, the herpesvirus proteases have unusual triads formed by a serine and two histidines, such as Ser109/His43/His128 of the pseudorabies virus protease. The superposition of the gp21 structure with the structure of pseudorabies virus protease (Figure 4) shows that the catalytic serines, as well as the histidines, acting as the second members of the triads, have similar positions in these proteins. However, the third members of the catalytic triads (Asp168 in gp21 and His128 in the pseudorabies virus protease) have different positions in these structures.
The capsid assembly proteases should be activated only in the assembled procapsids. The T4 gp21 protease is kept inactive by the N-terminal region, which blocks the active center. The herpesvirus proteases, lacking such an N-terminal region, use a different activation mechanism which presumably depends on the protein dimerization (Darke et al., 1996; Schmidt and Darke, 1997; Zuhlsdorf et al., 2015). Active dimers of herpesvirus proteases are presumably formed only in assembled procapsids.
Although the T4 gp21 and herpesvirus proteases have different amino acid sequences, oligomeric states, and activation mechanisms, these proteins have very similar folds suggesting their common evolutionary origin.
Common Evolutionary Origin of the T4 Procapsid Protease and the Tube Proteins of Phages and Bacterial Type VI Secretion Systems
The T4 head assembly protease, gp21, has structural similarity with proteins forming tubes in the long-tailed Siphoviridae and Myoviridae phages. Bacteriophage T4 tail tube protein gp19 (PDB ID: 5iv5 (Taylor et al., 2016)) can be superimposed onto the gp21 monomer with RMSD of 3.3 Å for 71 equivalenced Cα atoms (Figure 6), although the sequence identity between the structurally aligned residues is only 4%. This superposition places helix 1 of gp21 together with the β-sheet formed by strands 2, 3, 4, and 5, onto similar structural elements of gp19. In T4 virions the corresponding β-strands of gp19 form the inner surface of the tail tube.
Figure 6. Comparison of the T4 capsid assembly protease, gp21, with the T4 tail tube protein gp19.
(A) Stereo view of the superposition of gp21 with gp19 (PDB ID: 5iv5). Region 10 - 116 of gp21 is rainbow–colored from blue at the N-terminus to yellow-green at the C- terminus. Region 36 – 160 of gp19 is shown in black. Residue numbers for the gp21 chain are shown in magenta. (B) Schematic diagram showing topology of the conserved structural motif. See also Figure S1
The tail tube-like fold common to gp19 and gp21 has also been recognized in other bacteriophage proteins, including those located in the central part of phage baseplates and at the interface between capsids and tails (Cardarelli et al., 2010; Veesler and Cambillau, 2011). All these proteins assemble into rings associated with the tail tube. Together with the tail tube they form a continuous conduit for DNA ejection. These structural similarities, combined with similar functions, suggest that the tail tube-like proteins are likely to have originated from a common ancestor (Cardarelli et al., 2010; Veesler and Cambillau, 2011).
The phage tail tube-like fold has also been found in the tubes of the bacterial type VI secretion systems which shares structural similarity with the contractile tails of Myoviridae phages (Leiman et al., 2009; Mougous et al., 2006; Pell et al., 2009). The gp21 monomer can be superimposed on the tube protein Hcp1 from the type VI secretion system (PDB ID: 1y12) with RMSD of 4 Å between 74 equivalenced Cα atoms (Figure S1).
The structural data mentioned above show that the occurrence of the tail tube-like fold is widely dispersed. Thus, considering the enormous number of phages in the biosphere, it probably represents one of the most abundant protein folds on Earth. The structure of T4 gp21 shows that this fold is present not only in tubular structures but also in the proteins involved in the capsid assembly of phages and related herpesviruses. Therefore, this fold might originate in the procapsid or in the tail of a primordial virus.
EXPERIMENTAL PROCEDURES
Protein Production
The gene coding for the T4 gp21 mutant protein was chemically synthesized (GenScript, Inc., Piscataway, NJ), and cloned into the pET-28a(+) vector using the NdeI and XhoI restriction sites. In the mutant gene the codons of the catalytic Ser140 and His85 residues, were substituted by the Ala codons, CTG. To avoid the alternative translation start at Met45 (Hintermann and Kuhn, 1992), the alternative start codon was converted to the Leu codon, CTG, and the preceding Shine-Dalgarno sequence, GAAGGA, was altered by the silent base substitutions to GAGGGC. The vector was transformed into E. coli BL21(DE3) (Novagen) cells. The cells were grown to OD600 = 0.6 in 2YT media supplemented with kanamycin (50 μg/ml). The protein expression was induced with 1 mM IPTG for 4 hours at 37 °C. The cells were harvested by centrifugation and resuspended in buffer containing 20 mM Tris-HCl pH 8.0, 150 mM NaCl, and Pierce EDTA-free protease inhibitors. The cells were disrupted by sonication, and the insoluble cell debris was removed by centrifugation. The supernatant was applied to a HisTrap HP column (GE Healthcare). The protein was eluted from the column by 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 250 mM imidazole.
Crystallization and X-ray Data Collection
Crystals of gp21 were obtained by the hanging-drop vapor diffusion method at 20 °C using the crystallization reservoir solution containing 1.3 M ammonium sulfate, 0.1 M Hepes pH 7.5, and 0.1 M NaCl. Hanging drops, prepared by mixing 2 μl of the purified protein solution (15 mg/ml) and 2 μl of the reservoir solution, were equilibrated against 1 ml of the reservoir solution. Platinum derivatives were obtained by soaking crystals for 24 hours in the crystallization reservoir solution containing 1 mM K2PtCl4. Crystals were briefly dipped into the cryoprotectant solution containing 20 % glycerol, 1 M ammonium sulfate, 80 mM Hepes pH 7.5, 80 mM NaCl and flash-frozen in liquid nitrogen.
X-ray diffraction datasets were collected at GM/CA-CAT beamlines 23 ID-D and ID-B of the Advanced Photon Source (APS), Argonne National Laboratory. The resolution of the best native dataset was 1.94 Å, whereas the best platinum derivative crystal diffracted to 2.8 Å (Table 1).
Table 1.
Data collection and refinement statistics.
| X-ray dataset | Native | Pt derivative |
|---|---|---|
| Wavelength (Å) | 1.033 | 1.072 (peak) |
| Resolution range (Å) | 1.94 (1.98 – 1.94)* | 2.8 (2.85 – 2.8) |
| Space group | P21 | |
| Unit cell parameters | a=41.75 Å, b=124.04 Å, c=88.96 Å, β=89.96° | a=41.72 Å, b=123.58 Å, c=88.75 Å, β=90.21° |
| Completeness (%) | 93 (76) | 99.7 (99.7) |
| Redundancy | 3.6 (3.6) | 7.0 (7.1) |
| Mean I/sigma(I) | 11.9 (2.0) | 19.6 (5.0) |
| R-merge | 0.097 (0.54) | 0.096 (0.43) |
| Refinement using the native dataset | ||
| Number of reflections | 61875 | |
| Number of reflections used for R-free | 2355 | |
| Number of non-hydrogen atoms | 7033 | |
| Number of water molecules | 620 | |
| R-work | 0.172 | |
| R-free | 0.205 | |
| RMS bonds (Å) | 0.007 | |
| RMS angles (°) | 0.85 | |
| Ramachandran favored (%) | 98.8 | |
| Ramachandran outliers | 0 | |
| Rotamer outliers (%) | 0.15 | |
| Average B-factor (Å2) | 31.4 | |
Statistics for the highest-resolution shell are shown in parentheses.
Structure Determination, Model Building, and Refinement
The X-ray datasets were processed using HKL2000 (Otwinowski and Minor, 1997). The gp21 crystals belonged to the space group P21 with five molecules in the asymmetric unit. Determination of the heavy atom positions and phasing were performed using Phenix Autosol (Terwilliger et al., 2009). Five platinum sites were found in the asymmetric unit, which were related by a five-fold axis collinear with the crystallographic a axis. Phases were initially determined to 3.8 Å resolution by the single-wavelength anomalous diffraction (SAD) method and improved by density modification using non-crystallographic symmetry. The Resolve (Terwilliger, 2000) iterative phase extension procedure, employing NCS averaging, was used to determine the phases of the higher-resolution reflections. The atomic model was built using Phenix Autobuild (Terwilliger et al., 2008) and Coot (Emsley et al., 2010). The structure was refined using phenix.refine (Afonine et al., 2012). Final refinement statistics are summarized in Table 1. Structure superpositions were performed using Coot (Emsley et al., 2010; Krissinel and Henrick, 2004). Interface surface areas were calculated using PISA (Krissinel and Henrick, 2007). Figures were prepared using Chimera (Pettersen et al., 2004).
Supplementary Material
Highlights.
Phage T4 prohead protease, gp21, is activated by self-cleavage of its N-terminal part
Gp21 and herpesvirus proteases have similar folds but different activation mechanisms
Phage and herpesvirus proteases have a fold similarity to phage tail tube proteins
Acknowledgments
We thank Prof. Venigalla Rao (The Catholic University of America) for helpful discussions and critical reading of the manuscript. This work was supported by NIH grant AI081726 to M. G. R. and Venigalla Rao.
Footnotes
ACCESSION NUMBERS
The gp21 structure and associated structure factors have been deposited with the Protein Data Bank (PDB) under accession code 5JBL.
AUTHOR CONTRIBUTIONS
A. F. and M. G. R. designed research. A. F. performed research. A. F. and M. G. R. wrote the paper.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ackermann HW. 5500 Phages examined in the electron microscope. Arch Virol. 2007;152:227–243. doi: 10.1007/s00705-006-0849-1. [DOI] [PubMed] [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix. refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhter T, Zhao L, Kohda A, Mio K, Kanamaru S, Arisaka F. The neck of bacteriophage T4 is a ring-like structure formed by a hetero-oligomer of gp13 and gp14. Biochim Biophys Acta. 2007;1774:1036–1043. doi: 10.1016/j.bbapap.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Anderson D, Reilly B. Morphogenesis of bacteriophage phi29. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and Other Gram-Positive Bacteria: Biochemistry, Physiology, and Molecular Genetics. Washington, DC: American Society for Microbiology; 1993. pp. 859–867. [Google Scholar]
- Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol. 2005;79:14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black LW, Rao VB. Structure, assembly, and DNA packaging of the bacteriophage T4 head. Adv Virus Res. 2012;82:119–153. doi: 10.1016/B978-0-12-394621-8.00018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black LW, Showe MK, Steven AC. Morphogenesis of the T4 head. In: Karam JD, editor. Molecular Biology of Bacteriophage T4. Washington, D.C: American Society for Microbiology; 1994. pp. 218–258. [Google Scholar]
- Brown JC, Newcomb WW. Herpesvirus capsid assembly: insights from structural analysis. Curr Opin Virol. 2011;1:142–149. doi: 10.1016/j.coviro.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013;41:W349–357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardarelli L, Pell LG, Neudecker P, Pirani N, Liu A, Baker LA, Rubinstein JL, Maxwell KL, Davidson AR. Phages have adapted the same protein fold to fulfill multiple functions in virion assembly. Proc Natl Acad Sci U S A. 2010;107:14384–14389. doi: 10.1073/pnas.1005822107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone G, Heymann JB, Cheng N, Trus BL, Steven AC. Procapsid assembly, maturation, nuclear exit: dynamic steps in the production of infectious herpesvirions. Adv Exp Med Biol. 2012;726:423–439. doi: 10.1007/978-1-4614-0980-9_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, King J. P22 morphogenesis. I: Catalytic scaffolding protein in capsid assembly. J Supramol Struct. 1974;2:202–224. doi: 10.1002/jss.400020215. [DOI] [PubMed] [Google Scholar]
- Cheng H, Shen N, Pei J, Grishin NV. Double-stranded DNA bacteriophage prohead protease is homologous to herpesvirus protease. Protein Sci. 2004;13:2260–2269. doi: 10.1110/ps.04726004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darke PL, Cole JL, Waxman L, Hall DL, Sardana MK, Kuo LC. Active human cytomegalovirus protease is a dimer. J Biol Chem. 1996;271:7445–7449. doi: 10.1074/jbc.271.13.7445. [DOI] [PubMed] [Google Scholar]
- Effantin G, Figueroa-Bossi N, Schoehn G, Bossi L, Conway JF. The tripartite capsid gene of Salmonella phage Gifsy-2 yields a capsid assembly pathway engaging features from HK97 and lambda. Virology. 2010;402:355–365. doi: 10.1016/j.virol.2010.03.041. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokine A, Battisti AJ, Kostyuchenko VA, Black LW, Rossmann MG. Cryo-EM structure of a bacteriophage T4 gp24 bypass mutant: the evolution of pentameric vertex proteins in icosahedral viruses. J Struct Biol. 2006;154:255–259. doi: 10.1016/j.jsb.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Fokine A, Chipman PR, Leiman PG, Mesyanzhinov VV, Rao VB, Rossmann MG. Molecular architecture of the prolate head of bacteriophage T4. Proc Natl Acad Sci U S A. 2004;101:6003–6008. doi: 10.1073/pnas.0400444101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokine A, Islam MZ, Zhang Z, Bowman VD, Rao VB, Rossmann MG. Structure of the three N-terminal immunoglobulin domains of the highly immunogenic outer capsid protein from a T4-like bacteriophage. J Virol. 2011;85:8141–8148. doi: 10.1128/JVI.00847-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokine A, Leiman PG, Shneider MM, Ahvazi B, Boeshans KM, Steven AC, Black LW, Mesyanzhinov VV, Rossmann MG. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc Natl Acad Sci U S A. 2005;102:7163–7168. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokine A, Zhang Z, Kanamaru S, Bowman VD, Aksyuk AA, Arisaka F, Rao VB, Rossmann MG. The molecular architecture of the bacteriophage T4 neck. J Mol Biol. 2013;425:1731–1744. doi: 10.1016/j.jmb.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull GF, Hendrix RW. Bacteriophages and their genomes. Curr Opin Virol. 2011;1:298–303. doi: 10.1016/j.coviro.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix RW. Bacteriophage genomics. Curr Opin Microbiol. 2003;6:506–511. doi: 10.1016/j.mib.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Hendrix RW, Johnson JE. Bacteriophage HK97 capsid assembly and maturation. Adv Exp Med Biol. 2012;726:351–363. doi: 10.1007/978-1-4614-0980-9_15. [DOI] [PubMed] [Google Scholar]
- Hintermann E, Kuhn A. Bacteriophage T4 gene 21 encodes two proteins essential for phage maturation. Virology. 1992;189:474–482. doi: 10.1016/0042-6822(92)90571-6. [DOI] [PubMed] [Google Scholar]
- Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v. 3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janin J. Specific versus non-specific contacts in protein crystals. Nat Struct Biol. 1997;4:973–974. doi: 10.1038/nsb1297-973. [DOI] [PubMed] [Google Scholar]
- Janin J, Rodier F. Protein-protein interaction at crystal contacts. Proteins. 1995;23:580–587. doi: 10.1002/prot.340230413. [DOI] [PubMed] [Google Scholar]
- Keller B, Bickle TA. The nucleotide sequence of gene 21 of bacteriophage T4 coding for the prohead protease. Gene. 1986;49:245–251. doi: 10.1016/0378-1119(86)90285-4. [DOI] [PubMed] [Google Scholar]
- Kostyuchenko VA, Chipman PR, Leiman PG, Arisaka F, Mesyanzhinov VV, Rossmann MG. The tail structure of bacteriophage T4 and its mechanism of contraction. Nat Struct Mol Biol. 2005;12:810–813. doi: 10.1038/nsmb975. [DOI] [PubMed] [Google Scholar]
- Kostyuchenko VA, Leiman PG, Chipman PR, Kanamaru S, van Raaij MJ, Arisaka F, Mesyanzhinov VV, Rossmann MG. Three-dimensional structure of bacteriophage T4 baseplate. Nat Struct Biol. 2003;10:688–693. doi: 10.1038/nsb970. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Leiman PG, Arisaka F, van Raaij MJ, Kostyuchenko VA, Aksyuk AA, Kanamaru S, Rossmann MG. Morphogenesis of the T4 tail and tail fibers. Virol J. 2010;7:355. doi: 10.1186/1743-422X-7-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, Burley SK, Almo SC, Mekalanos JJ. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci U S A. 2009;106:4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell. 2004;118:419–429. doi: 10.1016/j.cell.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Leiman PG, Kanamaru S, Mesyanzhinov VV, Arisaka F, Rossmann MG. Structure and morphogenesis of bacteriophage T4. Cell Mol Life Sci. 2003;60:2356–2370. doi: 10.1007/s00018-003-3072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu FY, Roizman B. The herpes simplex virus 1 gene encoding a protease also contains within its coding domain the gene encoding the more abundant substrate. J Virol. 1991;65:5149–5156. doi: 10.1128/jvi.65.10.5149-5156.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Mushegian A. Displacements of prohead protease genes in the late operons of double-stranded-DNA bacteriophages. J Bacteriol. 2004;186:4369–4375. doi: 10.1128/JB.186.13.4369-4375.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNicol LA, Simon LE, Black LW. A mutation which bypasses the requirement for p24 in bacteriophage T4 capsid morphogenesis. J Mol Biol. 1977;116:261–283. doi: 10.1016/0022-2836(77)90216-9. [DOI] [PubMed] [Google Scholar]
- Morais MC, Kanamaru S, Badasso MO, Koti JS, Owen BA, McMurray CT, Anderson DL, Rossmann MG. Bacteriophage phi29 scaffolding protein gp7 before and after prohead assembly. Nat Struct Biol. 2003;10:572–576. doi: 10.1038/nsb939. [DOI] [PubMed] [Google Scholar]
- Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onorato L, Stirmer B, Showe MK. Isolation and characterization of bacteriophage T4 mutant preheads. J Virol. 1978;27:409–426. doi: 10.1128/jvi.27.2.409-426.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CWJ, Sweet RM, editors. Methods in Enzymology, Volume 276: Macromolecular Crystallography, part A. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Pell LG, Kanelis V, Donaldson LW, Howell PL, Davidson AR. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc Natl Acad Sci U S A. 2009;106:4160–4165. doi: 10.1073/pnas.0900044106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Qin L, Fokine A, O’Donnell E, Rao VB, Rossmann MG. Structure of the small outer capsid protein, Soc: a clamp for stabilizing capsids of T4-like phages. J Mol Biol. 2010;395:728–741. doi: 10.1016/j.jmb.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt U, Darke PL. Dimerization and activation of the herpes simplex virus type 1 protease. J Biol Chem. 1997;272:7732–7735. doi: 10.1074/jbc.272.12.7732. [DOI] [PubMed] [Google Scholar]
- Showe MK, Isobe E, Onorato L. Bacteriophage T4 prehead proteinase. I Purification and properties of a bacteriophage enzyme which cleaves the capsid precursor proteins. J Mol Biol. 1976a;107:35–54. doi: 10.1016/s0022-2836(76)80016-2. [DOI] [PubMed] [Google Scholar]
- Showe MK, Isobe E, Onorato L. Bacteriophage T4 prehead proteinase. II Its cleavage from the product of gene 21 and regulation in phage-infected cells. J Mol Biol. 1976b;107:55–69. doi: 10.1016/s0022-2836(76)80017-4. [DOI] [PubMed] [Google Scholar]
- Sun L, Zhang X, Gao S, Rao PA, Padilla-Sanchez V, Chen Z, Sun S, Xiang Y, Subramaniam S, Rao VB, et al. Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution. Nat Commun. 2015;6:7548. doi: 10.1038/ncomms8548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, Zhang Z, Hegde S, Fokine A, Rossmann MG, Rao VB. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Taylor NM, Prokhorov NS, Guerrero-Ferreira RC, Shneider MM, Browning C, Goldie KN, Stahlberg H, Leiman PG. Structure of the T4 baseplate and its function in triggering sheath contraction. Nature. 2016;533:346–352. doi: 10.1038/nature17971. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Adams PD, Read RJ, McCoy AJ, Moriarty NW, Grosse-Kunstleve RW, Afonine PV, Zwart PH, Hung LW. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr D Biol Crystallogr. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JA, Black LW. Mutational analysis of the Pseudomonas aeruginosa myovirus KZ morphogenetic protease gp175. J Virol. 2013;87:8713–8725. doi: 10.1128/JVI.01008-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Driel R, Traub F, Showe MK. Probable localization of the bacteriophage T4 prehead proteinase zymogen in the center of the prehead core. J Virol. 1980;36:220–223. doi: 10.1128/jvi.36.1.220-223.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veesler D, Cambillau C. A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol Mol Biol Rev. 2011;75:423–433. doi: 10.1128/MMBR.00014-11. first page of table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veesler D, Khayat R, Krishnamurthy S, Snijder J, Huang RK, Heck AJ, Anand GS, Johnson JE. Architecture of a dsDNA viral capsid in complex with its maturation protease. Structure. 2014;22:230–237. doi: 10.1016/j.str.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap ML, Klose T, Arisaka F, Speir JA, Veesler D, Fokine A, Rossmann MG. Role of bacteriophage T4 baseplate in regulating assembly and infection. Proc Natl Acad Sci U S A. 2016;113:2654–2659. doi: 10.1073/pnas.1601654113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuhlsdorf M, Werten S, Klupp BG, Palm GJ, Mettenleiter TC, Hinrichs W. Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin pUL26N, a Herpesvirus Serine Protease. PLoS Pathog. 2015;11:e1005045. doi: 10.1371/journal.ppat.1005045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.