SUMMARY
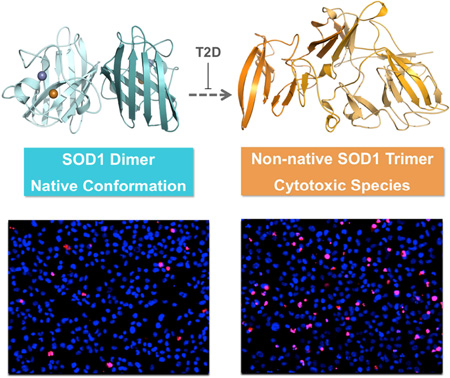
The majority of amyotrophic lateral sclerosis (ALS)-related mutations in the enzyme Cu, Zn superoxide dismutase (SOD1), as well as a post-translational modification, glutathionylation, destabilize the protein and lead to a misfolded oligomer that is toxic to motor neurons. The biophysical role of another physiological SOD1 modification, T2-phosphorylation, has remained a mystery. Here, we find that a phosphomimetic mutation, T2D, thermodynamically stabilizes SOD1 even in the context of a strongly SOD1-destabilizing mutation, A4V, one of the most prevalent and aggressive ALS-associated mutations in North America. This stabilization protects against formation of toxic SOD oligomers and positively impacts motor neuron survival in cellular assays. We solve the crystal structure of T2D-SOD1 and explain its stabilization effect using DMD simulations. These findings imply that T2-phosphorylation may be a plausible innate cellular protection response against SOD1-induced cytotoxicity, and stabilizing the SOD1 native conformation might offer us viable pharmaceutical strategies against currently incurable ALS.
Graphical Abstract
INTRODUCTION
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by gradual loss of motor neurons in the brain and spinal cord (Boillee et al., 2006; Cleveland and Rothstein, 2001; Redler and Dokholyan, 2012). Currently no effective therapeutics for ALS exist (Abel et al., 2012; Ilieva et al., 2009; Israelson et al., 2015). Genomic studies have associated approximately 12% of familial ALS cases and 1.5% of sporadic cases with mutations in the ubiquitous antioxidant enzyme Cu, Zn superoxide dismutase (SOD1) (Cirulli et al., 2015; Rosen et al., 1993). SOD1 aggregates have been discovered in the ventral horns of spinal cords from patients with familial or sporadic cases of ALS, indicating a contribution of non-genetic factors to disease progression (Shibata et al., 1996; Shibata et al., 1994). Recent studies have further implicated non-native SOD1 oligomers, instead of large aggregates, as the toxic species in cellular assays (Jonsson et al., 2004; Luchinat et al., 2014; Proctor et al., 2016; Zetterstrom et al., 2007). Indeed, several disease mutants (e.g. A4V and G93A) destabilize SOD1 and promote formation of a trimeric species (Proctor et al., 2016; Redler et al., 2014). Aberrant post-translational modifications such as glutathionylation, a result of oxidative stress (Barber et al., 2006; Townsend, 2007), also increase the rate of SOD1 oligomerization as well as the exposure of a toxicity-associated epitope (Bosco et al., 2010; Redler et al., 2014; Redler et al., 2011). Hence, understanding how somatic mutations and non-genetic factors affect the stability of the SOD1 dimer and non-native oligomers is vital to the development of novel therapeutic strategies. Identifying mechanisms to stabilize the native conformation of SOD1 and to eliminate potentially toxic species can shed light on new venues for drug discovery.
The stability of the SOD1 dimer is affected by ALS-related mutations and several post-translational modifications, which typically bias the SOD1 misfolding pathway toward dimmer dissociation and lead to larger populations of oligomers and aggregates (Broom et al., 2016; Khare et al., 2004; Redler et al., 2014). In familial and sporadic ALS cases, post-translational modifications induced by environmental factors play a significant role in disease progression (Barber and Shaw, 2010; Chattopadhyay et al., 2015; Pasinelli and Brown, 2006). Specifically, glutathionylation of C111, oxidative modification of W32, and phosphorylation of T2 or S58/T59 have been identified in human SOD1 (Coelho et al., 2014; Wilcox et al., 2009). While C111-glutathionylation and W32-oxidation have been studied thoroughly and linked to increased concentrations of non-native oligomers (Coelho et al., 2014; Redler et al., 2011), studies on phosphorylation have been limited due to a lack of knowledge of the corresponding kinases. A recent study demonstrated that S59-phosphorylated SOD1 relocates to the nucleus and functions as a transcription factor regulating oxidative resistance genes (Tsang et al., 2014). However, the effects of T2-phosphorylation, especially its impact on protein stability, have remained elusive.
Experimental characterization of phosphorylated SOD1 has been limited due to the inability to isolate pure T2-phosphorylated SOD1 or to artificially produce it via kinase reaction (Goscin and Fridovich, 1972). Phosphorylated SOD1 can be enriched through ion exchange chromatography, but the resulting sample is impure, containing a large proportion of unmodified and alternatively phosphorylated SOD1 (T2 or S58/T59) (Wilcox et al., 2009). Here we address this problem by producing the phosphomimetic mutant T2D-SOD1. We explore the properties of phosphomimetic mutant using a synergistic computational and experimental approach. Using molecular dynamics simulations, we demonstrate similarities between T2D-SOD1 and T2-phosphorylated SOD1 in terms of their energetic, thermodynamic, and structural characteristics, thus supporting the use of T2D as a phosphomimetic mutation. We solve the crystal structure of T2D-SOD1, and find that it closely matches the wild type (WT-SOD1) structure (Deng et al., 1993). In agreement with our computational predictions, the phosphomimetic T2D mutation stabilizes the SOD1 dimer without altering its native structure and preserves the same level of cellular viability when compared to WT-SOD1. In order to assess the role of phosphorylation in disease, we test the ability of the T2D mutation to mitigate the effects of an aggressive ALS mutation A4V. By expressing T2D-SOD1, A4V–SOD1 and T2D/A4V–SOD1 in hybridized motor neuron cells, we find that the T2D mutation rescues the cytotoxic effect of A4V–SOD1, demonstrating the benefit of stabilizing the SOD1 native conformation and offering a plausible pharmaceutical strategy to fight ALS.
RESULTS
Computational analysis suggests common characteristics shared by T2-phosphorylated SOD1 and T2D-SOD1
We perform all-atom DMD simulations to test whether the physical properties of the phosphomimetic mutant T2D-SOD1 reproduce those of T2-phosphorylated SOD1 (Pi-SOD1). We compare energetic, structural, and thermodynamic signatures of T2D-SOD1 and Pi-SOD1 (Figure S1). Specifically, we compare the potential energy distribution of each species, which are obtained through replica exchange simulations. The free energy histogram contains three distinct peaks representing three co-existing metastable states: a native low-energy dimeric structure, a partially unfolded dimer, and dissociated monomers with partial unfolding. T2D-SOD1 and Pi-SOD1 both have enriched populations of native dimers (peak 1) and decreased populations of dissociated monomers (peak 3) (Figure S1A, B), indicating reduced amounts of nonnative states in both species in comparison with the unmodified SOD1 (WT-SOD1). We also compare melting temperatures (Tm) corresponding to the unfolding of SOD1 monomers through specific heat analysis (Ding and Dokholyan, 2008). We find that the Tm of both T2D-SOD1 and Pi-SOD1 are lower than that of WT-SOD1 (Figure S1C, D). Furthermore, Pi-SOD1 exhibits a major peak with a shoulder in the specific heat plot against temperature (Figure S1C, D), in contrast to WT-SOD1 that shows a single peak in heat capacity. The single peak suggests the dimer dissociation and monomer unfolding are coupled processes for unmodified SOD1 and the shoulder peak represents a partial decoupling induced by phosphorylation (Proctor et al., 2011). We find that T2D-SOD1 reproduces this feature by preserving this decoupling. Together these results reveal similar monomer and dimer stabilities for T2D-SOD1 and Pi-SOD1.
The T2D-SOD1 dimer is structurally similar to the WT-SOD1 dimer
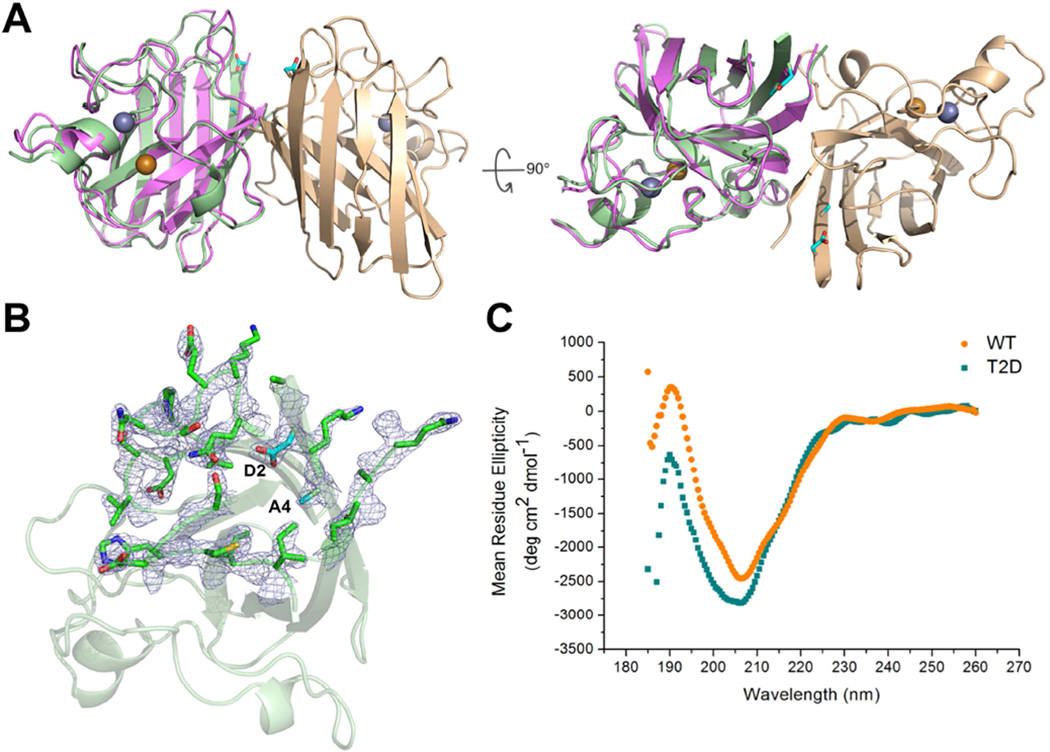
In order to study the structural effect of the phosphomimetic mutation T2D, we solve the crystal structure of T2D-SOD1 to resolution of 2.2 Å (PDB: 5K02, Table 1). Structure solution by molecular replacement with the model of WT-SOD1 (PDB: 1SPD) reveals a dimeric β-barrel occupying the center of an asymmetric unit (Figure 1). In the T2D-SOD1 crystals, the packing between the central dimers leads to a ternary complex in space group P1 (Figure S2). The structures of T2D-SOD1 and WT-SOD1 are similar to each other. By superimposing their backbone and side chains, we find that T2D-SOD1 deviates from WT-SOD1 with root mean square deviation of 0.93 Å (Figure 1A). This deviation largely originates from the flexible loops near the N terminus (Figure 1B). CD spectra also indicate that T2D-SOD1 and WT-SOD1 have similar secondary structure composition in solution (Figure 1C). To quantitate the rearrangement induced by the T2D mutation, we calculate the rotation angles between two monomers using previously described algorithms (Proctor et al., 2011). For T2D-SOD1 and WT-SOD1 these angles are 36.0° and 33.6°, respectively. Our DMD simulation predicts that both the T2D mutation and T2-phosphorylation induces an increase in the average monomer-monomer angle (Figure S3), suggesting that the T2D mutation may structurally mimic the steric effects of SOD1 phosphorylation at T2. Overall the crystal structure confirms that no large rearrangement of subunits or disruption of the dimer interface occurs in T2D-SOD1.
Table 1. Data collection and refinement statistics.
| Data Collection | |
|---|---|
| Space Group | P1 |
| Cell Dimensions | |
| a,b,c (Å) | 111.980, 111.961, 150.46 |
| α,β,γ (°) | 89.86, 89.85, 60.13 |
| Resolution (Å) | 40.00–2.10 (2.14–2.10) |
| Rmerge (%) | 6.1 |
| I / σI | 18.3 (1.6) |
| Completeness (%) | 80.41 |
| Redundancy | 2.5 (2.3) |
| Refinement Statistics | |
| Resolution (Å) | 40.00–1.99 |
| No. Reflections | 329360 |
| Rwork / Rfree (%) | 14.6/16.2 |
| No. atoms | |
| Protein Atoms | 27396 |
| Ligand/ion | 48 |
| Water | 686 |
| Overall B value (Å2) | 36.11 |
| Protein | 36.01 |
| Ligand/ion | 34.50 |
| Water | 40.09 |
| RMSD | |
| Bond Lengths (Å) | .006 |
| Bond Angles (°) | 1.084 |
| Ramachandran | |
| Most Favored (%) | 87.0 |
| Additionally Allowed (%) | 12.9 |
| Generously Allowed (%) | 0.1 |
| Disallowed (%) | 0.0 |
Figure 1. T2D-SOD1 and WT-SOD1 have similar structures in crystals and in solution state.
(A) Overall ribbon diagram of the T2D-SOD1 homodimer (PDB: 5K02) with Zn2+ (silver) and Cu2+ (golden) shown in spheres. The superposition of T2D- (light green and light brown) and WT-SOD1 (purple, PDB: 1SPD) in one subunit indicates the minor structural change in the loop regions. The side chains of D2 and A4 are indicated as cyan sticks. (B) Electron density map of D2 residue and surrounding residues (i. 22–29, 107–113) in one subunit. (C) CD spectra of T2D-SOD1 (cyan square) and WT-SOD1 (orange circle) indicate they both have β-sheet structure in solution. See also Figure S1, S2 and S3.
T2D mutation promotes the native dimer population in both WT- and A4V–SOD1
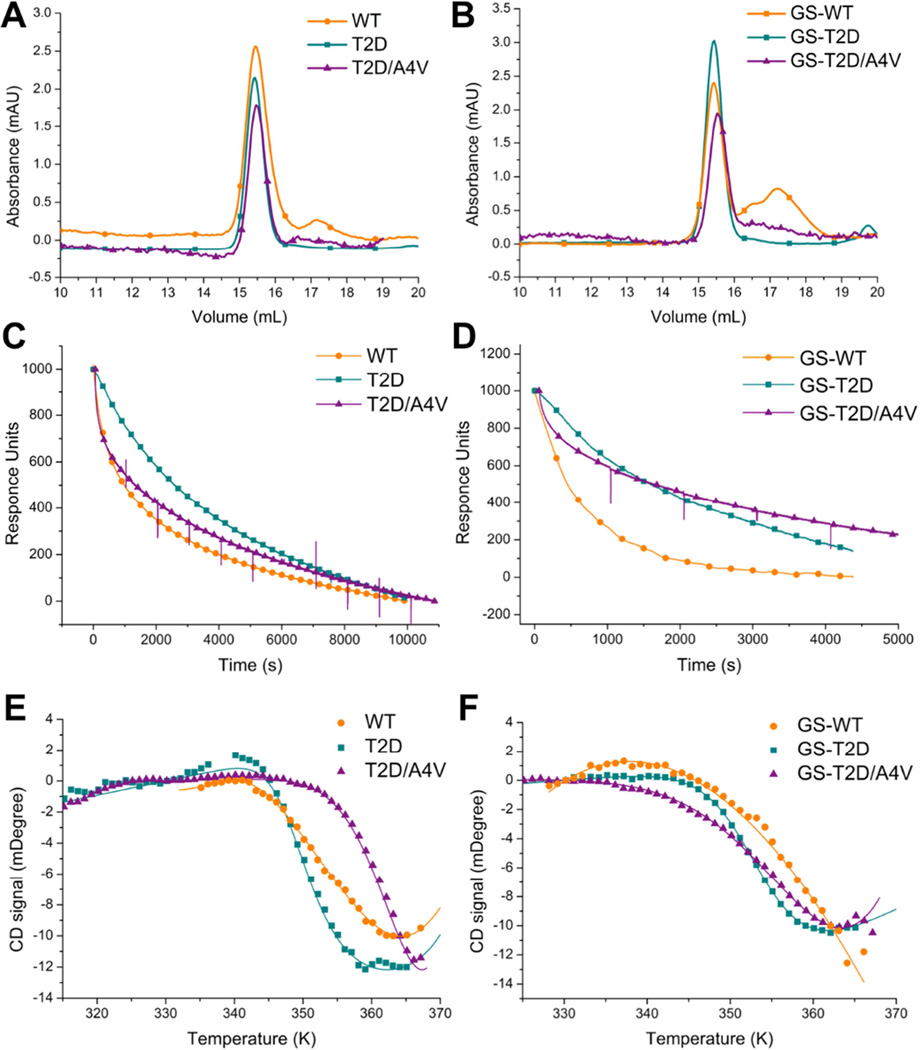
Many disease-linked SOD1 mutants undergo dimer dissociation with greater frequency than WT-SOD1, leading to an increase in the populations of monomeric species that form the available pool for further oligomerization (Broom et al., 2015; Khare and Dokholyan, 2006). In contrast, we found that T2D-SOD1 maintained a stable dimer population after incubation at 37 °C for up to seven days (Figure 2A). For comparison, WT-SOD1 forms a small population of monomeric proteins during the same time period (Figure 2A). Glutathionylation at C111 has been shown to promote SOD1 dimer dissociation in both wild type and mutant SOD1 (e.g. A4V–SOD1) by inducing steric clashing near the dimer interface (Redler et al., 2011; Proctor et al., 2011). The small difference in the monomer-dimer population distribution between T2D- and WT-SOD1 is exacerbated by glutathionylation. As seen previously, glutathionylated WT-SOD1 (GS-WT-SOD1) features an increased population of monomer as compared to unmodified WT-SOD1, yielding up to 50% of the entire protein population after an incubation period of seven days at 37°C (Figure 2B). However, GS-T2D-SOD1 and GS-T2D/A4V–SOD1 resist dimer dissociation, featuring significantly smaller proportions of monomer than GS-WT-SOD1. Furthermore, A4V–SOD1 has previously been shown to have a higher propensity to form both monomer and oligomers (Luchinat et al., 2014; Redler et al., 2014). However, no formation of higher oligomer is evident in any of the phosphomimetic samples (Figure 2 and Figure S4). These results suggest that the phosphomimetic T2D mutation prevents or decreases SOD1 dimer dissociation.
Figure 2. T2D stabilizes the native state of SOD1 through decreased dimer dissociation rates.
(A) Size exclusion chromatograms showing the populations of native dimer (15.5 ml) and monomer (17.4 ml) for unmodified T2D-, T2D/A4V- and WT-SOD1. Samples were taken after incubation at physiological conditions (30 µM SOD1, pH 7.4) and 37°C for 7 days. (B) Size exclusion chromatograms for glutathionylated T2D-, T2D/A4V- and WT-SOD1. Dissociation of immobilized dimers was monitored by surface plasmon resonance for T2D-, T2D/A4V- and WT-SOD1 (C) and glutathionylated species (D). Thermal denaturation curves of unmodified (E) and glutathionylated(F) SOD1 proteins indicate the Tm of T2D-SOD1 is lower than that of WT-SOD1 in the unfolding of SOD1 monomers. Fitting to a two-state model is represented as lines. Size exclusion chromatograms, dissociation profile and thermal denaturation curves of A4V–SOD1 and GS-A4V–SOD1 have been reported (Redler et al., 2011). See also Figure S4.
T2D mutation strengthens interactions at the SOD1 dimer Interface
In order to explore the mechanism of T2D dimer stabilization, we measure the dimer dissociation rates using SPR. In the case of glutathionylation-enriched SOD1 samples, Redler, et al showed that reassociation of previously dissociated monomers is inhibited (Redler et al., 2011). Together these studies determined the dissociation rate constants for WT-SOD1 and A4V–SOD1 as 1.72×10−4 s−1 and 2.60 ×10−3 s−1, respectively, indicating that A4V–SOD1 dissociates 15 times faster than does the WT-SOD1. We measure the dissociation rate constants for T2D-SOD1 as 1.01×10−4 s−1, approximately half that of WT-SOD1 (Table 2 and Figure 2C). For the glutathionylation-enriched samples, the rate constant of GS-T2D-SOD1 is reduced by a factor of 9 compared to GS-WT-SOD1, indicating GS-T2D-SOD1 dissociated significantly slower with an average half-time of 1.85 hr, in comparison to a half-time of 0.29 hr for GS-WT-SOD1 (Table 2 and Figure 2D). The dissociation rates of unmodified and glutathionylated T2D/A4V–SOD1 are similar to the respective values for T2D-SOD1 (16 times slower than A4V–SOD1), indicating an overall decrease in dimer dissociation consistent with our results from size exclusion chromatography. Because dimer dissociation and subsequent metal loss induce the structural distortions that promote protein aggregation, the decrease we find in dimer dissociation upon phosphomimetic T2D mutation suggests that phosphorylation at T2 may inhibit the formation of toxic SOD1 oligomers (Proctor et al., 2016).
Table 2. Biophysical characterization of T2D-, T2D/A4V- and WT-SOD1.
T2D, T2D/A4V and WT indicate unmodified species, GS-T2D, GS-T2D/A4V and GS-WT indicate C111-glutathionylation enriched species. Thermal melting temperatures (Tm) for the unfolding process of SOD1 monomers were measured by circular dichroism thermal denaturation. Dimer dissociation rate constants (koff) were measured by surface plasmon resonance. The standard deviation is indicated with ±.
| Tm (°C) | koff (s−1) | |
|---|---|---|
| T2D | 79.8 ± 1.5 | 1.01×10−4 ± 5.0×10−6 |
| T2D/A4V | 82.4 ± 0.7 | 1.56×10−4 ± 0.1×10−6 |
| WT | 81.5 ± 0.4 | 1.72×10−4 ± 0.2×10−6 |
| GS-T2D | 77.0 ± 0.5 | 1.50×10−4 ± 2.8×10−5 |
| GS-T2D/A4V | 77.0 ±0.4 | 1.61×10−4 ± 0.2×10−6 |
| GS-WT | 81.0 ± 1.8 | 9.49×10−4 ± 1.2×10−5 |
T2D mutation rescues cell viability in the context of an FALS mutation A4V
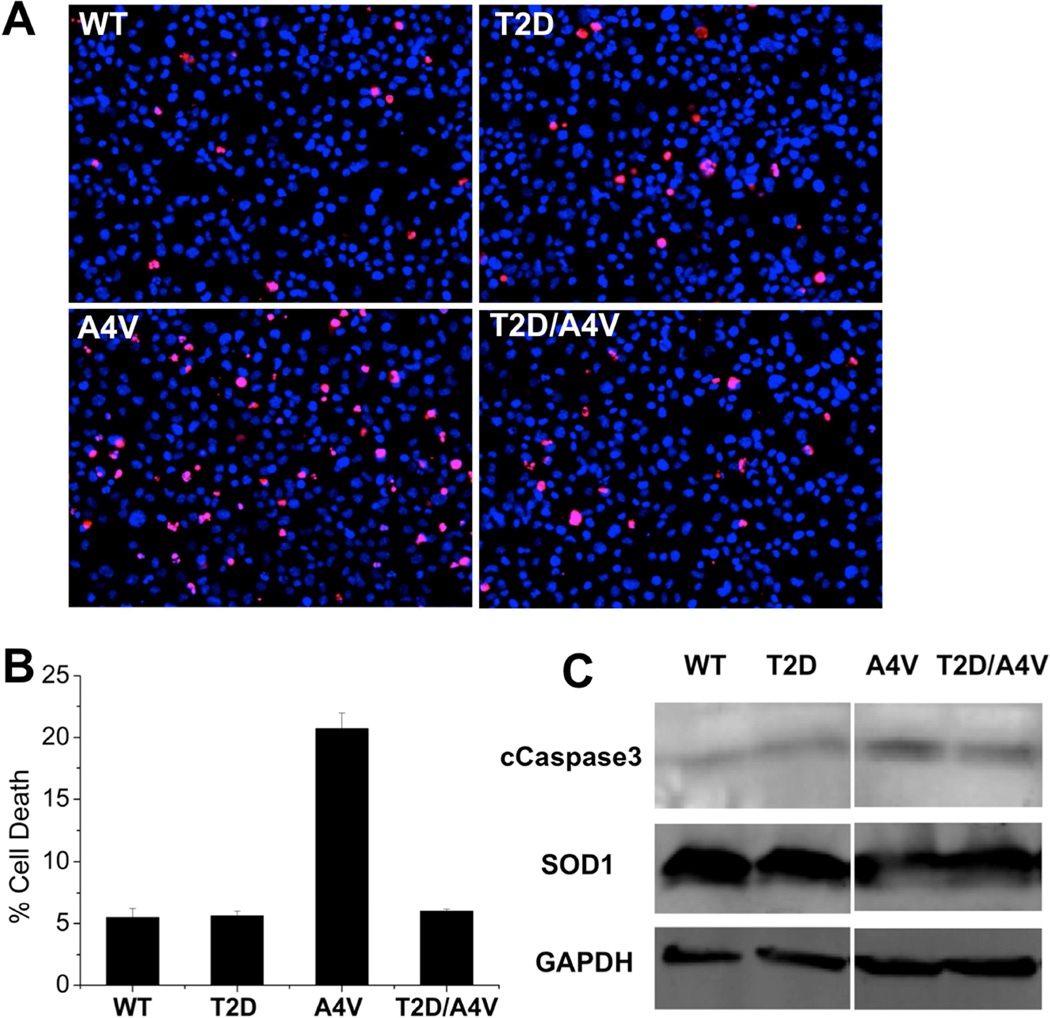
In order to gain an understanding of the physiological relevance of the phosphomimetic mutant SOD1 in cellular environment, we overexpress T2D-SOD1 and WT-SOD1 in immortalized mouse motor neuron-like cells (NSC-34) with similar expression levels for wild type and mutants (Figure 3) (Cashman et al., 1992). We measure the cell viability using propidium iodide to mark non-viable cells, in concert with Hoescht staining of viable cells (Figure 3A) (Latt and Stetten, 1976; Moore et al., 1998). Overexpression of T2D-SOD1 and WT-SOD1 yield cell death rates of 7% and 6%, respectively (Figure 3B), which are identical results within experimental error. For a disease-relevant comparison, we similarly overexpress an aggressive ALS mutant A4V–SOD1 and the double mutant T2D/A4V–SOD1 in NSC-34 cells. While A4V–SOD1 expression results in 20% cell death, T2D/A4V–SOD1 expression results in only 6% cell death, demonstrating the ability of the T2D mutation to rescue toxicity of one of the most aggressive ALS-associated SOD1 variants. Supporting this finding, we find that cells expressing SOD1 with the A4V mutation alone feature high expression of the apoptotic marker caspase3, in agreement with the known toxicity of the A4V–SOD1 mutant, while cells transfected with T2D/A4V–SOD1 express lower levels of caspase3, similar to the levels expressed by cells transfected with WT-SOD1 (Figure 3C). These results indicate that the T2D mutation ameliorates cytotoxicity in the context of a severe ALS related mutation.
Figure 3. T2D mutation reduces the cytotoxic effect of A4V in motor neuron-like cells.
(A) Transfection of NSC-34 cells with T2D-SOD1 and WT-SOD1 (negative control) led to similar cell death ratio. A4V–SOD1 transfection (positive control) decreased cell viability and T2D/A4V–SOD1 rescued cells from A4V–SOD1 mediated cytotoxicity. Applied three days post-transfection, red stain (propidium iodide) identifies dead cells, while blue stain (Hoechst) identifies all cell population. (B) Average cell death rates are measured as the percentage of red stained cells: WT 5.4%, T2D 5.6%, A4V 20.7% and T2D/A4V 6.0%. Each bar represents the standard error of the mean, n=3. (C) Levels of the apoptotic marker cCaspase 3 in NSC-34 cells demonstrated by western blot confirm the protection role of T2D mutation, as T2D/A4V leads to reduced expression of cCaspase 3. The normalized expression levels: WT 100%, T2D 89%, A4V 377% and T2D/A4V 135%.
Possible destabilizing effect of the T2D mutation on SOD1 trimers
Since cytotoxicity in motor neuron-like cells is found to correlate with the propensity of forming trimeric SOD1 (Proctor et al., 2016), we study the effect of T2D mutation on the stability of toxic SOD1 trimers. We use structural models of the trimer proposed by Proctor et al (Proctor et al., 2016) to computationally evaluate the change in free energy upon mutation (ΔΔGmut) using the Eris molecular design suite (Yin et al., 2007a, b). The T2D mutation destabilizes the SOD1 trimer structure (ΔΔGmut = 2.9 kcal/mol) (Figure S5), while the A4V mutation stabilizes the trimer (ΔΔGmut = −4.8 kcal/mol), in accordance with experimental observations (Redler et al., 2014). When combined in the same structure, the T2D mutation rescues the trimer-stabilizing effect of the A4V mutation (ΔΔGmut = 1.2 kcal/mol), suggesting that not only is the T2D mutation stabilizing to the native SOD1 dimer structure, but also inhibits formation of the toxic trimeric form by making the trimer structure less stable. Therefore, the phosphomimetic T2D mutation has a profound effect on SOD1 stability: by both stabilizing native dimer and destabilizing the toxic trimer states, this mutation strongly inhibits formation of toxic oligomers and promotes significant increase in cell viability.
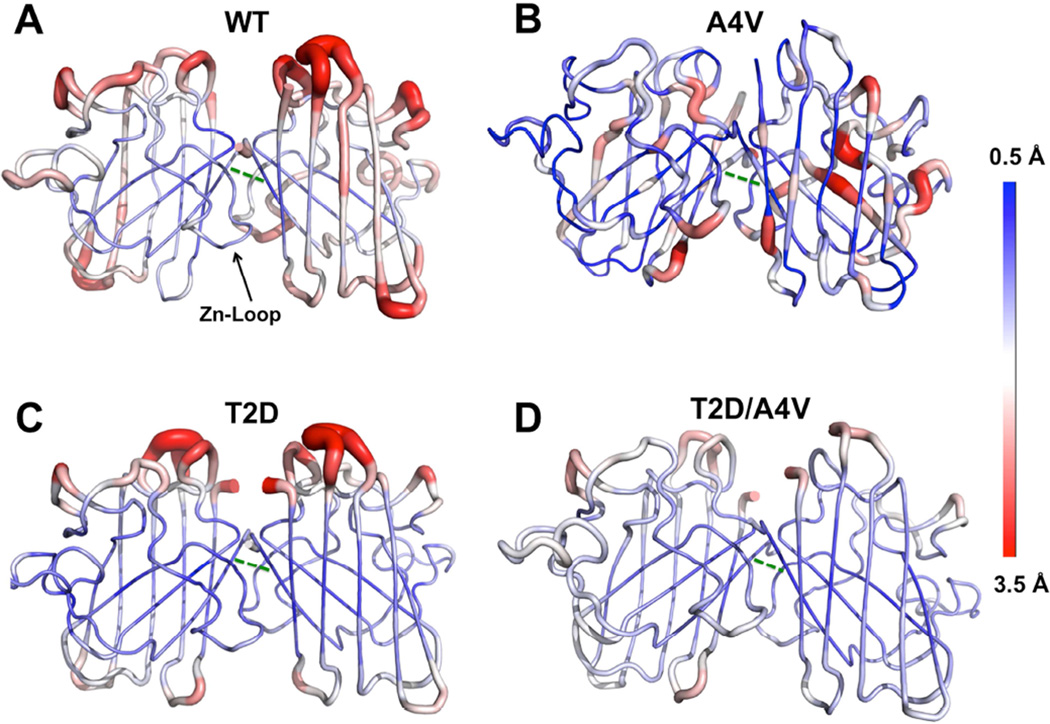
Previous studies indicate that ALS-associated mutations and aberrant post-translational modifications can disrupt the dynamic coupling between SOD1 monomers, which leads to disruption of stabilizing structural elements (the β-plug and zinc-binding loop) and global destabilization of the native state (Ding and Dokholyan, 2008; Khare and Dokholyan, 2006). To understand whether and how phosphomimetic mutation affect the dynamics of SOD1 dimer on a nanosecond time scale, we have performed short (50 ns) DMD simulations and observed different dynamic signatures of WT-SOD1, T2D-SOD1, T2D/A4V–SOD1, and A4V–SOD1 (Figure 4 and Figure S6). In contrast to WT, T2D, and T2D/A4V, the A4V mutation leads to higher fluctuation of the residues at dimer interface, as measured by root mean square fluctuation in the DMD trajectories (Figure 4). Specifically, V148-V148 and F50-I151 are key hydrophobic contacts between the SOD1 monomers. We observe high flexibility of Val148 and Ile 151 only in the A4V–SOD1 (Figure 4B, green dash line). We also find that T2/A4 and V148/151 have cross-correlated motions in the inter-residue covariance matrix (Figure S6), which indicates that the packing strain induced by the T2D or A4V mutation can affect these residues. The A4V mutation likely induces steric clash at the interface or weakens the V148-V148 and F50-I151 interactions, whereas in T2D-SOD1 and T2D/A4V–SOD1 these interactions are stable (Figure 4C, D). This observation is consistent with the high propensity of A4V–SOD1 to dissociate and form monomers in comparison with WT-SOD1, T2D-SOD1 and T2D/A4V–SOD1. Furthermore, we find strong correlations within the zinc-binding loop (residues 49–60) only in A4V–SOD1, whereas in WT-, T2D-, and T2D/A4V–SOD1 these correlated motions are suppressed (Figure S6). The zinc-binding loop has been suggested to allosterically regulate the conformation of the C4F6 epitope (Bosco et al., 2010), which is directly linked to neural-toxicity of SOD1 in a primary microglia activation assay (Furukawa et al., 2015; Rotunno et al., 2014). We propose that in A4V–SOD1 the cross-correlated motions promote the exposure of the disease-related epitope, and that the T2D/A4V double mutant prevents this effect. These effects may lead to the differential cytotoxicity observed in cells transfected with A4V–SOD1 and T2D/A4V–SOD1.
Figure 4. DMD simulation suggests the key interface interactions are disrupted in A4V–SOD1, in contrast to WT-, T2D- and T2D/A4V–SOD1.
The flexibility of the protein backbone is measured as root mean square fluctuation (RMSF, Red: 3.5 Å, Blue: 0.5 Å) and the results are an average of ten DMD trajectories: (A) WT-SOD1, (B) A4V–SOD1, (C) T2D-SOD1 and (D) T2D/A4V–SOD1. The hydrophobic contact between V148-V148 is indicated as green dash line. The zinc-binding loop (Zn-loop) which allosterically modulates the C4F6 epitope is indicated in the WT-SOD1 structure. See also Figure S5 and S6.
DISCUSSION
Recent studies suggest that non-native SOD1 oligomers are the cytotoxic species that promote disease progression and cause motor neuron degeneration in SOD1-mediated ALS (Jonsson et al., 2004; Proctor et al., 2016; Zetterstrom et al., 2007). The formation of SOD1 oligomers and aggregates is critically dependent on the dissociation of the native dimer into monomers, a process exacerbated by disease-linked mutations (Ding et al., 2012; Ivanova et al., 2014; Khare et al., 2004b). By reducing the population of SOD1 monomers, the formation of cytotoxic oligomers can be inhibited. Our findings demonstrate that the T2D mutation, which mimics phosphorylation at T2, increases the stability of the native conformation of SOD1 and reduces the cytotoxic effect of A4V–SOD1 in motor neuron-like cells. Furthermore, we suggest that T2-phosphorylation may be an intrinsic protective mechanism that stabilizes cytosolic SOD1. Our simulations and structural calculations for T2D-SOD1 and Pi-SOD1 not only demonstrate the stabilizing effect of the T2D mutation, but also establish the high degree of structural and energetic similarity between the phosphomimetic T2D-SOD1 construct and phosphorylated WT-SOD1. Quantitative analysis of the crystal structure of T2D reveals that T2D-SOD1 has a larger monomer-monomer angle in comparison to WT-SOD1, as predicted for the Pi-SOD1 (Figure S4). We also measure the thermal melting temperature (Tm) for T2D-SOD1 and find that T2D-SOD1 has a lower Tm than WT-SOD1 (T2D-SOD1: 79.8°C, WT-SOD1: 81.5°C) (Table 2 and Figure 2 E , F), which agrees with the shift of corresponding specific heat (Cv) peak to a lower temperature for Pi-SOD1 (Figure S1C, D). Based on these results, we posit that T2-phosphorylation of SOD1 has similar biophysical characteristics to T2D-SOD1 and stabilizes the native conformation of SOD1 dimer.
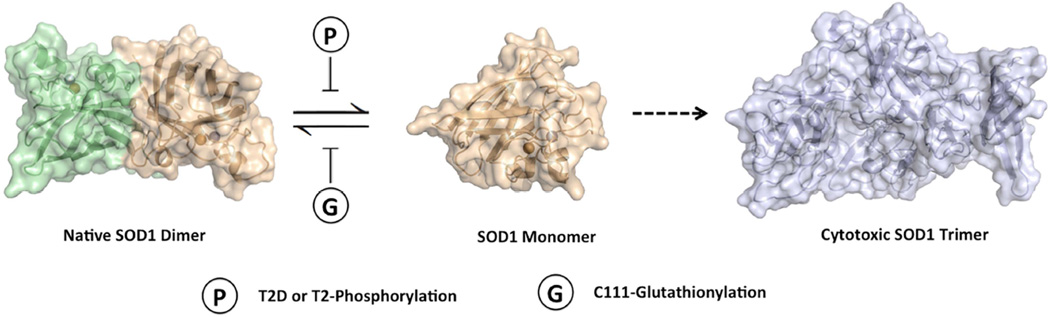
T2-phosphorylation and C111-glutathionylation have been identified as the dominant post-translational modifications to SOD1 in human erythrocytes (Wilcox et al., 2009). It is plausible that the phosphorylation is an intrinsic mechanism to offset the impact of oxidative stress or other deleterious effects on the SOD1 structure. The late onset of ALS symptoms (∼55 years old on average) suggests that such mechanisms may have evolved to protect the native SOD1 dimer (Ilieva et al., 2009; Redler and Dokholyan, 2012). Given the high cytosolic concentration (30–100 µM) of SOD1, a balancing mechanism that would offset environmental stressors, such as oxidative stress, may be important for the stability and function of SOD1 (Jaarsma et al., 2000). The protective role of phosphorylation has been discovered in other neurodegenerative diseases. In a rat model of Parkinson`s disease, researchers have applied phosphomimetic mutations and suggested phosphorylation at S87 inhibits the aggregation of human α-synuclein and protects against its toxicity (Oueslati et al., 2012). However, the roles of phosphorylation in neurodegenerative diseases are usually deleterious. Phosphorylation can promote the spreading of β-amyloid in Alzheimer pathogenesis (Rezaei-Ghaleh et al., 2016), reduce binding of Tau-protein to microtubules and correlate with severity of neuronal cytopathology (Augustinack et al., 2002; Lindwall and Cole, 1984). Together our findings and other studies reveal that while C111-glutathionylation promotes dimer dissociation by inhibiting the association of SOD1 monomers (decreased kon) (McAlary et al., 2013; Redler et al., 2011), T2-phosphorylation may inhibit dimer dissociation by stabilizing the dimer interface (decreased koff) (Figure 5).
Figure 5. A proposed mechanism of the regulation of dimer-monomer equilibrium in SOD1 by post-translational modifications.
T2-Phosphorylation and C-111 glutathionylation may regulate the dimer-monomer equilibrium and have opposing effects on the stability of native SOD1 dimers. The T2D mutation (or T2-Phosphorylation) inhibits the dimer dissociation (decreased koff), whereas C111-glutathionylation inhibits the association of monomers (decreased kon).
SOD1 represents a common factor in both familial and sporadic cases of ALS (Pasinelli and Brown, 2006; Renton et al., 2014). While most familial mutations destabilize SOD1, our work demonstrates the contrasting influence of a phosphomimetic mutation that stabilizes the native dimer and ameliorates toxicity induced by an aggressive disease mutation. Therefore, stimulating SOD1 phosphorylation in motor neurons or developing drugs to mimic the effects of T2D mutation may offer promising new directions for development of therapeutic strategies. The inhibition of small oligomer formation for disease-linked proteins represents a pharmaceutical approach broadly applicable to many neurodegenerative disorders (Haass and Selkoe, 2007; Kirkitadze et al., 2002). For SOD1, oligomerization and aggregation are proportional to the fraction of monomers (Khare et al., 2004). Researchers have developed various strategies to stabilize SOD1 dimer, including co-expression of chaperones and small molecule binding (Bruening et al., 1999; Limpert et al., 2013; Naik et al., 2012). T2D-SOD1 features a significantly reduce the population of monomers and oligomers, which underlies its beneficial effect to motor neuron-like cells. Hence, identification of T2D as a stabilizing mutation suggests new avenues for therapy development in currently incurable ALS.
EXPERIMENTAL PROCEDURES
Modeling of phosphorylated SOD1 and phosphomimetic mutants
All-atom discrete molecular dynamics (DMD) is a molecular dynamics algorithm that utilizes discrete step functions in place of continuous potentials to describe interaction energies between particles in a simulation (Ding et al., 2008; Proctor et al., 2011). The all-atom protein model features explicit modeling of all heavy atoms, polar hydrogens, and a reaction-based hydrogen bonding potential. We obtained the known structure of SOD1 from the Protein Data Bank (PDB: 1SPD, 1UXM) and performed computational mutagenesis using the Eris suite (Yin et al., 2007a, b). We made no structural adjustment to residues participating in metal-binding or disulfide bond interactions. Following mutation, we minimized the structure using an iterative relaxation and equilibration protocol (Proctor et al., 2011). We performed single-temperature and replica exchange DMD simulations of the optimized T2D-SOD1 structure. In the monomer species, we used 12 replicas for a simulation length of 100 ns at temperatures of 0.5 (∼252 K), 0.52 (∼262 K), 0.54 (∼272 K), 0.56 (∼282 K), 0.58 (∼292 K), 0.60 (∼302 K), 0.62 (∼312 K), 0.64 (∼322 K), 0.66 (∼332 K), 0.68 (∼343 K), 0.70 (∼353 K), and 0.72 (∼363 K) kcal mol−1. In the dimer species, we used 16 replicas for a simulation of 50 ns at temperatures of 0.48 (∼242 K), 0.495 (∼249 K), 0.51 (∼257 K), 0.525 (∼264 K), 0.54 (∼272 K), 0.555 (∼280 K), 0.57 (∼287 K), 0.585 (∼295 K), 0.60 (∼302 K), 0.615 (∼310 K), 0.63 (∼317 K), 0.645 (∼325 K), 0.65 (∼327 K), 0.67 (∼337 K), 0.69 (∼347 K), and 0.71 (∼357 K) kcal (mol kB)−1. While the temperatures used in MD simulations do not equate directly with physical temperatures, they can be used to evaluate relative changes in physical temperature. We combined replica trajectories using weighted histogram analysis method (WHAM) (Kumar et al., 1992). We utilized the MMTSB tool to calculate the constant volume heat capacity (Feig et al., 2004) and analyzed energetic states and molecular interactions as described previously (Proctor et al., 2011).
Cloning, expression, and purification of SOD1
We produced the SOD1 mutant vectors (T2D-SOD1, A4V–SOD1 and T2D/A4V–SOD1) with a Stratagene mutagenesis kit (Agilent Technologies, CA) following the provided protocol. Expression, purification, and separation of modified populations were carried out as described previously (Goscin and Fridovich, 1972; Wilcox et al., 2009).
Circular dichroism (CD) and thermal melting points analysis
We collected CD spectroscopy data using a Chirascan Plus instrument (Applied Photophysics, MA). We dialyzed protein samples against 10 mM phosphate buffer (pH 7.4) and diluted them to 0.2 mg/mL for analysis. CD spectra were measured from 260 nm to 185 nm at 20°C with 50,000 readings taken at each 0.5 nm increment. Melting curves were monitored from 20°C to 94°C at 205 nm. The apparent melting temperature was determined by fitting to a two-state model (Zhu et al., 2013).
Size exclusion chromatography
We prepared the samples (8.8 µM SOD1 in 20 mM Tris and 150 mM NaCl, pH 7.4) and incubated them at 37°C for 0 hr, 24 hr, or 7 days. After incubation the samples were analyzed using a Superdex 200 10/300 GL column (GE Healthcare, PA) described previously (Redler et al., 2011).
Surface plasmon resonance (SPR)
SOD1 samples were biotinylated on one subunit using the EZ-link NHS-LC-LC-biotin linker (Life Technologies, CA) (Khare et al., 2004). We then dialyzed the biotinylated SOD1 into PBS buffer (40 mM phosphate, 150 mM NaCl, pH 7.4) before loading onto a SA chip (Biorad, CA). Dimer dissociation was monitored for WT-SOD1, T2D-SOD1 and T2D/A4V–SOD1 samples using the Proteon XPR36 (Bio-Rad, CA) at 25 °C. The dissociation reaction was initiated by flowing PBS buffer with 0.005% Tween 20 over the SA chip. Each SPR curve is an average of six independent measurements Data was analyzed as describe previously (Redler et al., 2011).
Crystallization, data collection, and model building
Crystallization of T2D was accomplished using a hanging drop vapor diffusion method against a well buffer. T2D was crystallized in a buffer of 2 M (NH4)2SO4, 0.1 M Sodium Cacodylate, 20% MPD and 10 mM ZnCl2 (pH 6.5) after a period of approximately 5 months.
The T2D crystals were transferred into the well buffer plus 20% glycerol as cryoprotectant and then flash-frozen in liquid nitrogen. The X-ray diffraction data of T2D crystal were collected at 100K on Beamline X29 at Brookhaven National Laboratory. We processed the raw data with HKL2000 (Otwinowski and Minor, 1997). The structure of SOD T2D was solved by molecular replacement method (Navaza and Saludjian, 1997). We then performed the refinement with Program Refmac5 in CCP4 Suite (Murshudov et al., 2011). The resulted model was rebuilt by program O (Jones et al., 1991) and further refined by Chiron (Ramachandran et al., 2011).
Cell culture, transfection, and cell viability assay
We maintained the NSC-34 cells in Dulbecco’s Modified Eagle Medium growth medium containing 10% FBS (Cashman et al., 1992). We plated approximately 5000 NSC 34 cells in one well of 24-well format plate with differentiation medium (1% FBS+NEAA+10 µM Retinoid Acid). After two days of differentiation, we transfected cells with 2 µg of pCI neo WT SOD1 or pCI neo SOD1 mutants using Lipofectamine 3000 following the manufacturer’s instruction. We performed propidium iodide (for dead cells) and Hoechst (for all cells) staining (Fisher Scientific, PA) 3 days post transfection without fixation (Latt and Stetten, 1976). We fixed the cells 24 hours post-transfection for immunostaining. For Western blot, we used approximately 20,000 cells per well of 6-well format and collected the cells 2 days post-transfection.
Immunoblotting and immunostaining
We performed Western blotting as previously described (Tao et al., 2011). Briefly, cell lysates were denatured in SDS loading buffer, resolved by 18% or 4–20% SDS-PAGE and transferred to a PVDF membrane. Then we incubated the membranes with antibodies against cCaspase3 (Cell Signaling Technology, MA), SOD1 (Calbiochem, MA) and GAPDH, followed by secondary antibody incubation (Moore et al., 1998). The protein bands were visualized with an Odyssey infrared imaging system (LI-COR Biosciences, NE). The expression level of cCaspase 3 was normalized using ImageJ.
Supplementary Material
Highlights.
Stabilizing SOD1 increases the viability of motor neuron-like cells
Crystal structure of the stabilizing mutant T2D-SOD1 is reported
Biophysical characterizations of T2D-SOD1 reveal stabilizing role of T2-phosphorylation
In Brief.
SOD1 destabilization and misfolding are associated with the fatal neurodegenerative disease ALS. Fay and Zhu et al. report the discovery of a phosphomimetic mutation, T2D, which stabilizes SOD1 and rescues the cytotoxic effect of an ALS-associated mutant.
Acknowledgments
We thank Dr. Michael Caplow, Dr. Feng Ding and Dr. Marino Convertino for valuable discussions. We also thank Dr. Rachel Redler and Dr. Lanette Fee for outstanding support of the study. This work was supported by NIH Grant R01GM080742 (to N.V.D.). E.A.P. was supported by a Ruth L. Kirschstein National Research Service Award (F31AG039266) from the National Institute on Aging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The accession numbers for the structures reported in this paper are PDB: 5K02
AUTHOR CONTRIBUTIONS
J.M.F., C.Z., and N.V.D. designed research; J.M.F., C.Z., E.A.P., and Y.T. performed research; J.M.F., W.C., and H.K. obtained crystal structure; C.Z., E.A.P. and J.M.F. analyzed the data; C.Z., J.M.F., E.A.P. and N.V.D. wrote the paper.
REFERENCES
- Abel O, Powell JF, Andersen PM, Al-Chalabi A. ALSoD: A user-friendly online bioinformatics tool for amyotrophic lateral sclerosis genetics. Hum. Mutat. 2012;33:1345–1351. doi: 10.1002/humu.22157. [DOI] [PubMed] [Google Scholar]
- Augustinack JC, Schneider A, Mandelkow EM, Hyman BT. Specific tau phosphorylation sites correlate with severity of neuronal cytopathology in Alzheimer’s disease. Acta Neuropathol. 2002;103:26–35. doi: 10.1007/s004010100423. [DOI] [PubMed] [Google Scholar]
- Barber SC, Mead RJ, Shaw PJ. Oxidative stress in ALS: a mechanism of neurodegeneration and a therapeutic target. Biochim. Biophys. Acta. 2006;1762:1051–1067. doi: 10.1016/j.bbadis.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Barber SC, Shaw PJ. Oxidative stress in ALS: key role in motor neuron injury and therapeutic target. Free Radic. Biol. Med. 2010;48:629–641. doi: 10.1016/j.freeradbiomed.2009.11.018. [DOI] [PubMed] [Google Scholar]
- Boillee S, Vande Velde C, Cleveland DW. ALS: A disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Bosco DA, Morfini G, Karabacak NM, Song Y, Gros-Louis F, Pasinelli P, Goolsby H, Fontaine BA, Lemay N, McKenna-Yasek D, et al. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat. Neurosci. 2010;13:1396–1403. doi: 10.1038/nn.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broom HR, Rumfeldt JAO, Vassall KA, Meiering EM. Destabilization of the dimer interface is a common consequence of diverse ALS-associated mutations in metal free SOD1. Protein Sci. 2015;24:2081–2089. doi: 10.1002/pro.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broom HR, Vassall KA, Rumfeldt JA, Doyle CM, Tong MS, Bonner JM, Meiering EM. Combined Isothermal Titration and Differential Scanning Calorimetry Define Three-State Thermodynamics of fALS-Associated Mutant Apo SOD1 Dimers and an Increased Population of Folded Monomer. Biochemistry. 2016;55:519–533. doi: 10.1021/acs.biochem.5b01187. [DOI] [PubMed] [Google Scholar]
- Bruening W, Roy J, Giasson B, Figlewicz DA, Mushynski WE, Durham HD. Up-regulation of protein chaperones preserves viability of cells expressing toxic Cu/Zn-superoxide dismutase mutants associated with amyotrophic lateral sclerosis. J. Neurochem. 1999;72:693–699. doi: 10.1046/j.1471-4159.1999.0720693.x. [DOI] [PubMed] [Google Scholar]
- Cashman NR, Durham HD, Blusztajn JK, Oda K, Tabira T, Shaw IT, Dahrouge S, Antel JP. Neuroblastoma x spinal cord (NSC) hybrid cell lines resemble developing motor neurons. Dev. Dyn. 1992;194:209–221. doi: 10.1002/aja.1001940306. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay M, Nwadibia E, Strong CD, Gralla EB, Valentine JS, Whitelegge JP. The Disulfide Bond, but Not Zinc or Dimerization, Controls Initiation and Seeded Growth in Amyotrophic Lateral Sclerosis-linked Cu, Zn Superoxide Dismutase (SOD1) Fibrillation. J. Biol. Chem. 2015;290:30624–30636. doi: 10.1074/jbc.M115.666503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli ET, Lasseigne BN, Petrovski S, Sapp PC, Dion PA, Leblond CS, Couthouis J, Lu YF, Wang Q, Krueger BJ, et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–1441. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Rothstein JD. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat. Rev. Neurosci. 2001;2:806–819. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- Coelho FR, Iqbal A, Linares E, Silva DF, Lima FS, Cuccovia IM, Augusto O. Oxidation of the Tryptophan 32 Residue of Human Superoxide Dismutase 1 Caused by Its Bicarbonate-dependent Peroxidase Activity Triggers the Non-amyloid Aggregation of the Enzyme. J. Biol. Chem. 2014;289:30690–30701. doi: 10.1074/jbc.M114.586370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng HX, Hentati A, Tainer JA, Iqbal Z, Cayabyab A, Hung WY, Getzoff ED, Hu P, Herzfeldt B, Roos RP, et al. Amyotrophic lateral sclerosis and structural defects in Cu,Zn superoxide dismutase. Science. 1993;261:1047–1051. doi: 10.1126/science.8351519. [DOI] [PubMed] [Google Scholar]
- Ding F, Dokholyan NV. Dynamical roles of metal ions and the disulfide bond in Cu, Zn superoxide dismutase folding and aggregation. Proc. Natl. Acad. Sci. U S A. 2008;105:19696–19701. doi: 10.1073/pnas.0803266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Tsao D, Nie HF, Dokholyan NV. Ab initio folding of proteins with all-atom discrete molecular dynamics. Structure. 2008;16:1010–1018. doi: 10.1016/j.str.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Furukawa Y, Nukina N, Dokholyan NV. Local unfolding of Cu, Zn superoxide dismutase monomer determines the morphology of fibrillar aggregates. J. Mol. Biol. 2012;421:548–560. doi: 10.1016/j.jmb.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig M, Karanicolas J, Brooks CL. MMTSB Tool Set: enhanced sampling and multiscale modeling methods for applications in structural biology. J. Mol. Graph. Model. 2004;22:377–395. doi: 10.1016/j.jmgm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Furukawa Y, Anzai I, Akiyama S, Imai M, Cruz FJ, Saio T, Nagasawa K, Nomura T, Ishimori K. Conformational Disorder of the Most Immature Cu,Zn-Superoxide Dismutase Leading to Amyotrophic Lateral Sclerosis. J. Biol. Chem. 2015;291:4144–4155. doi: 10.1074/jbc.M115.683763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goscin SA, Fridovich I. The purification and properties of superoxide dismutase from Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1972;289:276–283. doi: 10.1016/0005-2744(72)90078-2. [DOI] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nat. Rev. Mol. Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- Ilieva H, Polymenidou M, Cleveland DW. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J. Cell Biol. 2009;187:761–772. doi: 10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelson A, Ditsworth D, Sun SY, Song SW, Liang JS, Hruska-Plochan M, McAlonis-Downes M, Abu-Hamad S, Zoltsman G, Shani T, et al. Macrophage Migration Inhibitory Factor as a Chaperone Inhibiting Accumulation of Misfolded SOD1. Neuron. 2015;86:218–232. doi: 10.1016/j.neuron.2015.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova MI, Sievers SA, Guenther EL, Johnson LM, Winkler DD, Galaleldeen A, Sawaya MR, Hart PJ, Eisenberg DS. Aggregation-triggering segments of SOD1 fibril formation support a common pathway for familial and sporadic ALS. Proc. Natl. Acad. Sci. U S A. 2014;111:197–201. doi: 10.1073/pnas.1320786110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaarsma D, Haasdijk ED, Grashorn JA, Hawkins R, van Duijn W, Verspaget HW, London J, Holstege JC. Human Cu/Zn superoxide dismutase (SOD1) overexpression in mice causes mitochondrial vacuolization, axonal degeneration, and premature motoneuron death and accelerates motoneuron disease in mice expressing a familial amyotrophic lateral sclerosis mutant SOD1. Neurobiol. Dis. 2000;7:623–643. doi: 10.1006/nbdi.2000.0299. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Jonsson PA, Ernhill K, Andersen PM, Bergemalm D, Brannstrom T, Gredal O, Nilsson P, Marklund SL. Minute quantities of misfolded mutant superoxide dismutase-1 cause amyotrophic lateral sclerosis. Brain. 2004;127:73–88. doi: 10.1093/brain/awh005. [DOI] [PubMed] [Google Scholar]
- Khare SD, Caplow M, Dokholyan NV. The rate and equilibrium constants for a multi-step reaction sequence for the aggregation of superoxide dismutase in ALS. Proc. Natl. Acad. Sci. U S A. 2004;101:15094–15099. doi: 10.1073/pnas.0406650101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare SD, Dokholyan NV. Common dynamical signatures of familial amyotrophic lateral sclerosis-associated structurally diverse Cu, Zn superoxide dismutase mutants. Proc. Natl. Acad. Sci. U S A. 2006;103:3147–3152. doi: 10.1073/pnas.0511266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkitadze MD, Bitan G, Teplow DB. Paradigm shifts in Alzheimer’s disease and other neurodegenerative disorders: the emerging role of oligomeric assemblies. J. Neurosci. Res. 2002;69:567–577. doi: 10.1002/jnr.10328. [DOI] [PubMed] [Google Scholar]
- Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The Weighted Histogram Analysis Method for Free-Energy Calculations on Biomolecules .1. The Method. J. Comput. Chem. 1992;13:1011–1021. [Google Scholar]
- Latt SA, Stetten G. Spectral studies on 33258 Hoechst and related bisbenzimidazole dyes useful for fluorescent detection of deoxyribonucleic acid synthesis. J. Histochem. Cytochem. 1976;24:24–33. doi: 10.1177/24.1.943439. [DOI] [PubMed] [Google Scholar]
- Limpert AS, Mattmann ME, Cosford ND. Recent progress in the discovery of small molecules for the treatment of amyotrophic lateral sclerosis (ALS) Beilstein J. Org. Chem. 2013;9:717–732. doi: 10.3762/bjoc.9.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindwall G, Cole RD. Phosphorylation affects the ability of tau protein to promote microtubule assembly. J. Biol. Chem. 1984;259:5301–5305. [PubMed] [Google Scholar]
- Luchinat E, Barbieri L, Rubino JT, Kozyreva T, Cantini F, Banci L. In-cell NMR reveals potential precursor of toxic species from SOD1 fALS mutants. Nat. Commun. 2014;5:5502. doi: 10.1038/ncomms6502. [DOI] [PubMed] [Google Scholar]
- McAlary L, Yerbury JJ, Aquilina JA. Glutathionylation potentiates benign superoxide dismutase 1 variants to the toxic forms associated with amyotrophic lateral sclerosis. Sci. Rep. 2013;3:3275. doi: 10.1038/srep03275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore A, Donahue CJ, Bauer KD, Mather JP. Simultaneous measurement of cell cycle and apoptotic cell death. Methods Cell Biol. 1998;57:265–278. doi: 10.1016/s0091-679x(08)61584-8. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Zhang N, Gao P, Fisher MT. On the design of broad based screening assays to identify potential pharmacological chaperones of protein misfolding diseases. Curr. Top. Med. Chem. 2012;12:2504–2522. doi: 10.2174/1568026611212220006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaza J, Saludjian P. AMoRe: An automated molecular replacement program package. Macromolecular Crystallography, Pt A. 1997;276:581–594. doi: 10.1016/S0076-6879(97)76079-8. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Oueslati A, Paleologou KE, Schneider BL, Aebischer P, Lashuel HA. Mimicking phosphorylation at serine 87 inhibits the aggregation of human alpha-synuclein and protects against its toxicity in a rat model of Parkinson’s disease. J. Neurosci. 2012;32:1536–1544. doi: 10.1523/JNEUROSCI.3784-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat. Rev. Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- Proctor EA, Ding F, Dokholyan NV. Structural and Thermodynamic Effects of Post-translational Modifications in Mutant and Wild Type Cu, Zn Superoxide Dismutase. J. Mol. Biol. 2011;408:555–567. doi: 10.1016/j.jmb.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor EA, Fee L, Tao Y, Redler RL, Fay JM, Zhang Y, Lv Z, Mercer IP, Deshmukh M, Lyubchenko YL, et al. Nonnative SOD1 trimer is toxic to motor neurons in a model of amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U S A. 2016;113:614–619. doi: 10.1073/pnas.1516725113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Kota P, Ding F, Dokholyan NV. Automated minimization of steric clashes in protein structures. Proteins. 2011;79:261–270. doi: 10.1002/prot.22879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redler RL, Dokholyan NV. The Complex Molecular Biology of Amyotrophic Lateral Sclerosis (ALS) Molecular Biology of Neurodegenerative Diseases. 2012;107:215–262. doi: 10.1016/B978-0-12-385883-2.00002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redler RL, Fee L, Fay JM, Caplow M, Dokholyan NV. Non-native soluble oligomers of Cu/Zn superoxide dismutase (SOD1) contain a conformational epitope linked to cytotoxicity in amyotrophic lateral sclerosis (ALS) Biochemistry. 2014;53:2423–2432. doi: 10.1021/bi500158w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redler RL, Wilcox KC, Proctor EA, Fee L, Caplow M, Dokholyan NV. Glutathionylation at Cys-111 induces dissociation of wild type and FALS mutant SOD1 dimers. Biochemistry. 2011;50:7057–7066. doi: 10.1021/bi200614y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Chio A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat. Neurosci. 2014;17:17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaei-Ghaleh N, Amininasab M, Kumar S, Walter J, Zweckstetter M. Phosphorylation modifies the molecular stability of beta-amyloid deposits. Nat. Commun. 2016;7:11359. doi: 10.1038/ncomms11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, Oregan JP, Deng HX, et al. Mutations in Cu/Zn Superoxide-Dismutase Gene Are Associated with Familial Amyotrophic-Lateral-Sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Rotunno MS, Auclair JR, Maniatis S, Shaffer SA, Agar J, Bosco DA. Identification of a Misfolded Region in Superoxide Dismutase 1 That Is Exposed in Amyotrophic Lateral Sclerosis. J. Biol. Chem. 2014;289:28527–28538. doi: 10.1074/jbc.M114.581801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata N, Asayama K, Hirano A, Kobayashi M. Immunohistochemical study on superoxide dismutases in spinal cords from autopsied patients with amyotrophic lateral sclerosis. Dev. Neurosci. 1996;18:492–498. doi: 10.1159/000111445. [DOI] [PubMed] [Google Scholar]
- Shibata N, Hirano A, Kobayashi M, Sasaki S, Kato T, Matsumoto S, Shiozawa Z, Komori T, Ikemoto A, Umahara T, et al. Cu/Zn Superoxide Dismutase-Like Immunoreactivity in Lewy Body-Like Inclusions of Sporadic Amyotrophic-Lateral-Sclerosis. Neurosci. Lett. 1994;179:149–152. doi: 10.1016/0304-3940(94)90956-3. [DOI] [PubMed] [Google Scholar]
- Tao Y, Neppl RL, Huang ZP, Chen J, Tang RH, Cao R, Zhang Y, Jin SW, Wang DZ. The histone methyltransferase Set7/9 promotes myoblast differentiation and myofibril assembly. J. Cell Biol. 2011;194:551–565. doi: 10.1083/jcb.201010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend DM. S-glutathionylation: indicator of cell stress and regulator of the unfolded protein response. Mol Interv. 2007;7:313–324. doi: 10.1124/mi.7.6.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang CK, Liu Y, Thomas J, Zhang YJ, Zheng XFS. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat. Commun. 2014;5:3446. doi: 10.1038/ncomms4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox KC, Zhou L, Jordon JK, Huang Y, Yu YB, Redler RL, Chen X, Caplow M, Dokholyan NV. Modifications of Superoxide Dismutase (SOD1) in Human Erythrocytes A POSSIBLE ROLE IN AMYOTROPHIC LATERAL SCLEROSIS. J. Biol. Chem. 2009;284:13940–13947. doi: 10.1074/jbc.M809687200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S, Ding F, Dokholyan NV. Eris: an automated estimator of protein stability. Nat Methods. 2007a;4:466–467. doi: 10.1038/nmeth0607-466. [DOI] [PubMed] [Google Scholar]
- Yin S, Ding F, Dokholyan NV. Modeling backbone flexibility improves protein stability estimation. Structure. 2007b;15:1567–1576. doi: 10.1016/j.str.2007.09.024. [DOI] [PubMed] [Google Scholar]
- Zetterstrom P, Stewart HG, Bergemalm D, Jonsson PA, Graffmo KS, Andersen PM, Brannstrom T, Oliveberg M, Marklund SL. Soluble misfolded subfractions of mutant superoxide dismutase-1s are enriched in spinal cords throughout life in murine ALS models. Proc. Natl. Acad. Sci. U S A. 2007;104:14157–14162. doi: 10.1073/pnas.0700477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Dai Z, Liang H, Zhang T, Gai F, Lai L. Slow and bimolecular folding of a de novo designed monomeric protein DS119. Biophys. J. 2013;105:2141–2148. doi: 10.1016/j.bpj.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.