Abstract
Ribosome-associated J protein-Hsp70 chaperones promote nascent polypeptide folding and normal translational fidelity. Though known to span the ribosome subunits, understanding of J protein Zuo1 function is limited. New structural and crosslinking data allow more precise positioning of Saccharomyces cerevisiae Zuo1 near the 60S polypeptide exit site, pointing to interactions with ribosomal protein eL31 and 25S rRNA helix 24. The junction between the 60S-interacting and subunit-spanning helices is a hinge, positioning Zuo1 on the 40S, yet accommodating subunit rotation. Interaction between C-terminus of Zuo1 and 40S occurs via 18S rRNA expansion segment 12 (ES12) of helix 44, which originates at the decoding site. Deletions in either ES12 or C-terminus of Zuo1 alter stop codon readthrough and −1 frameshifting. Our study offers insight into how this cotranslational chaperone system may monitor decoding site activity and nascent polypeptide transit, thereby coordinating protein translation and folding.
INTRODUCTION
Generating a functioning proteome is a challenge faced by all cells. Particularly demanding is the generation of properly folded nascent chains as they exit the ribosome, both because of the crowded environment, and because a protein domain cannot fold properly until its synthesis is completed1,2. To maintain overall cellular protein homeostasis, the rate at which polypeptides are synthesized must also be optimized relative to capacity for folding in the cytosol and productive translocation across membranes, as well as for translation fidelity3–6. The exit site of the ribosome is a hub for factors that promote protein quality control. In eukaryotes these include a complex Hsp70-based system that is implicated in both protein folding and translational fidelity7,8. Like all Hsp70 machines, the ribosome-based system includes a J protein (often called Zuotin, and more specifically Zuo1 and DnaJC2 in yeast and humans, respectively)9–11, which stimulates the ATPase activity of its partner Hsp70 (ribosome-associated Ssb in yeast)12. This J protein activity is universally required for an Hsp70 to functionally interact with its client proteins13, including nascent chains exiting the ribosome14.
Zuo1 forms a stable heterodimer (often called the ribosome associated complex, RAC) with an atypical Hsp70 (called Ssz1 and HspA14 in fungi and humans, respectively), tethering it to the ribosome15–18. Ssz1/HspA14 bind, but do not hydrolyze, ATP and thus cannot partner with a J protein and perform the classic client protein binding cycle12,19,20. These three proteins (Ssb, Zuo1 and Ssz1), which are often referred to as a chaperone triad, play a role together in folding of nascent polypeptides14. This chaperone triad has also been implicated in quality control pathways more closely tied to the process of protein synthesis per se. Translational readthrough of stop codons, a measure of translation fidelity, is increased in triad deletion mutants21. The ability of ribosomes to shift translational reading frame, particularly −1 frameshifting, is a capacity used to produce alternative proteins from the same mRNA transcript or to regulate premature termination22. −1 frameshifting is decreased in triad deletion mutants23.
It has been appreciated for some time that Zuo1 interacts with the 60S subunit close to the polypeptide exit site of the ribosome tunnel24,25. Recently, cryo-electron microscopic (cryo-EM) analysis revealed interaction with the 40S subunit as well26. Consistent with small angle X-ray crystallographic analysis25, Zuo1 was found to span approximately 190 Å across the subunits26 (Fig. 1a). Based on its position on the 40S subunit, the C-terminal 4-helix bundle (residues 348–433)25,27 of Zuo1 has been proposed to interact with expansion segment 12 (ES12), an extension of helix 44 (H44) of 18S rRNA26. A long internal alpha helix, called the middle domain (MD), is proposed to span the subunits. The remainder of RAC, that is Ssz1 and N-terminal segments of Zuo1, is in close proximity to the 60S subunit. The segment immediately adjacent to the MD, which is referred to as the “zuotin homology domain” (ZHD), is most relevant to this report. Zuo1’s ZHD has sequence identity to a second ribosome-associated J protein, Jjj1, which is involved in biogenesis of the 60S subunit. The ZHD is functionally important for association of both proteins with ribosomes28,29.
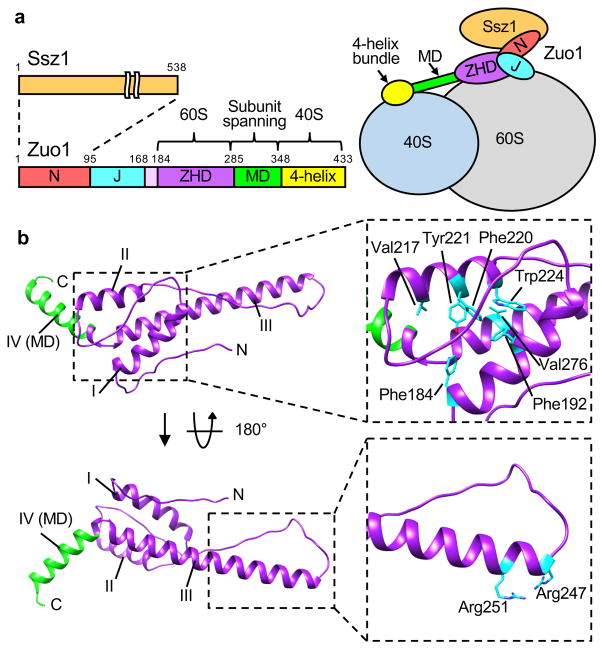
Figure 1.
Structure of zuotin homology domain (ZHD) of Zuo1. (a) Overall architecture of Zuo1 (left) and its interaction with the ribosome (right). N-terminus (N) of Zuo1, which interacts with the atypical Hsp70 Ssz1, is adjacent to the J domain (J). The ZHD spans residues 184–285. A predicted long alpha helix constituting the middle domain (MD, green) connects the ZHD and the C-terminal 4-helix bundle (yellow). Zuo1 interacts with the 60S and 40S subunits via the ZHD and 4-helix bundle, respectively. (b) Ribbon representation of crystal structure of residues 169–303 of Zuo1. ZHD (purple); MD (green). Helices I–IV indicated (I–IV). Hydrophobic amino acids constituting hydrophobic core of the helix bundle formed by helices I–III are highlighted by stick representation (top right). Arg247 and Arg251 at the N-terminal tip of helix III, which are important for Zuo1’s interaction with the ribosome, are highlighted by stick representation (bottom right).
Although our understanding of chaperone function on the ribosome has increased significantly in the past few years, major questions remain. Due to its inherent flexibility, structural information for the bulk of Zuo1 is lacking. Thus, how Zuo1 interacts with the 60S subunit has remained an enigma, and therefore progress towards understanding the relationship between the 60S interaction at the exit tunnel and the 40S interaction has been slow. Furthermore, the recent observation that Zuo1 binds the 40S subunit raises the question of whether this interaction is important for Zuo1’s role in translational fidelity26. This question is particularly intriguing as the proposed Zuo1 interaction site is the tip of 18S rRNA H44, which originates at the decoding site.
RESULTS
Structure of the ZHD of Zuo1
We pursued structural analysis of the region of Zuo1 that interacts with the 60S subunit by crystallizing the Zuo1 segment spanning residues 166–303. This segment extends through the ZHD and includes a segment of the MD domain (Fig. 1a). Initial attempts to solve the structure of the selenomethionine (SeMet) labeled wild type protein resulted in poorly diffracting crystals, and inadequate anomalous diffraction due to low methionine content. We therefore substituted codons Lys210 and Asn269 with methionine codons to enable additional SeMet incorporation. These sites were chosen for substitution based on the observation that methionine is present at these positions in some Zuo1 orthologs, including human DnaJC2. We determined the structure of the selenomethionine-substituted variant to a resolution of 1.85 Å using the single-wavelength anomalous diffraction (SAD) method (Fig. 1b; Table 1). Two molecules are present in the asymmetric unit (Supplementary Fig. 1a). Both contain four helices (Fig. 1b). Helices I–III form a three helix bundle, with helix IV positioned at a 120-degree angle relative to helix III. The major difference between the monomers of the asymmetric unit occurs in the long flexible loop between helices II and III (residues 230–247) (Supplementary Fig. 1a). Helix IV in the structure presented here is the N-terminal portion of the MD that spans the subunits. We note that throughout, we refer to residues 184–285 of the structure presented here as the ZHD. Previously, however, the segment encompassing residues 205–285, which does not include helix I, were referred to as the ZHD28,29. See Supplementary Fig. 1c for further explanation.
Table 1.
Data collection and refinement statistics
| Zuo1166–303 | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 60.33, 69.45, 95.08 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 47.67–1.85(1.92–1.85) |
| Rmerge | 0.1087(1.51) |
| Rmeas | 0.113 |
| CC1/2 | 0.999(0.636) |
| CC* | 1.000(0.882) |
| I / σI | 16.76(1.77) |
| Completeness (%) | 100(99) |
| Redundancy | 13.2(12.8) |
| Refinement | |
| Resolution (Å) | 1.85 |
| No. reflections | 35251 |
| Rwork / Rfree | 0.175/0.219 |
| No. atoms | |
| Protein | 2278 |
| Ligand/ion | 32 |
| Water | 96 |
| B-factors | |
| Protein | 38.5 |
| Ligand/ion | 52.9 |
| Water | 42.8 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.012 |
| Bond angles (°) | 1.23 |
Structure was determined from one crystal. Values in parentheses are for highest resolution shell
Helix I, helix II and the extreme C-terminal 11 residues of helix III (272–282) form the hydrophobic core of the ZHD three helix bundle. Residues Phe184, Phe192, Val217, Phe220, Tyr221, Trp224, and Val276 within this hydrophobic pocket are highly conserved among Zuo1 orthologs. The later 4 are highly conserved in Jjj1 and its orthologs as well (Fig. 1b). The 37 residue helix III is continuous and extended in both monomers of the asymmetric unit (Supplementary Fig. 1a). At its tip are two conserved arginines (Arg247, Arg251), which we previously reported to be important for ribosome association and function of both Zuo1 and Jjj128 (Fig. 1b). The 120-degree angle kink between helices III and IV is centered on proline 284, and is stabilized primarily by interactions involving the side chains of Asp283 and Arg285, which are invariant in both Zuo1 and Jjj1 orthologs (Supplementary Fig. 1b, d). Pro284 is not as highly conserved. Although most Zuo1 orthologs have a proline at this position, some insects and plants have lysine, glutamic acid or alanine. On the other hand, although several fungi have a proline at this position, Saccharomyces cerevisiae Jjj1 and most of its orthologs have a positively charged residue. Modeling of the ZHD region of Zuo1 and Jjj1 human orthologs using S. cerevisiae ZHD as a template generated similar structures of the ZHD and junction with helix IV (Supplementary Fig. 1c), consistent with the possibility that ZHD domains are structurally similar in both Jjj1 and Zuo1 orthologs.
The ZHD-60S subunit interaction
Using this new structural information as a guide, we performed in vivo site-specific cross-linking to probe the interaction of Zuo1 with two previously implicated surface exposed ribosomal proteins near the polypeptide exit site, eL22 and eL3124–26 (Fig. 2a). The nonnatural, photo-activable amino acid p-benzoyl-L-phenylalanine (Bpa) was incorporated in place of endogenous, surface exposed residues by enhanced nonsense suppression at 21 positions in eL22 and 12 positions in eL31 (Supplementary Fig. 2a). After exposure of cells to UV, strong crosslink products migrating at approximately 90 kDa were detected for three eL31 variants, those having Bpa at positions Arg79 and Glu81 in the surface exposed loop and at position Val7, in a nearby β-strand. Each reacted with Zuo1-specific antibodies (Fig. 2b). Weaker, Zuo1-reacting bands were detected when Bpa was incorporated at adjacent positions on the loop (Glu82, Glu83 or Asp84) (Supplementary Fig. 2b). These results give strong support to the idea that eL31 interacts with Zuo1. No eL22 crosslinks to Zuo1 were detected in our analysis of 21 Bpa variants.
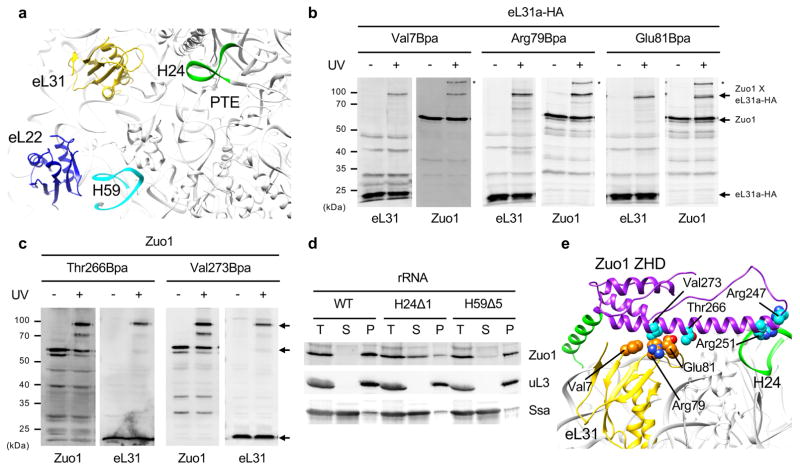
Figure 2.
Interaction of Zuo1 with the 60S subunit. (a) Overview of previously implicated contact sites for Zuo1 near the polypeptide tunnel exit (PTE) of 60S. (b & c) Site-specific cross-linking between eL31a and Zuo1. Cells expressing variants of HA-tagged eL31a or Zuo1 having Bpa incorporated at the indicated position, in b and c, respectively, were exposed to UV light (+), or as a control not exposed (−). Cross-linking was analyzed by immunoblotting using antibodies specific for Zuo1 or HA tag (eL31) after SDS-PAGE. Migration of molecular weight standards indicated in kDa (left). Zuo1 and HA reactive bands indicated with arrows. Two independent yeast strains were analyzed for each Bpa variant, with similar results. (d) Lysates from cells expressing wild type (WT) or mutant rRNAs having one or 5 base pairs of H24 (H24Δ1) or H59 (H59Δ5) deleted, respectively, were centrifuged through a sucrose cushion. Equivalent amounts of supernatant (S) and pellet (P) fractions, as well as total lysates (T), were analyzed by immunoblotting. Antibodies specific for Zuo1, and for uL3 and Ssa, as controls for ribosomes and a predominantly soluble protein, respectively, were used. One representative immunoblot (from three independent experiments, performed with different yeast cultures) is shown. (b,c,d) Uncropped gel images are shown in Supplementary Data Set 1. (e) Docking of Zuo1 ZHD to 60S. The atomic structures of Zuo1169–303 and ribosome (PDB 3J78) were fit to cryo-EM map of Zuo1-Ssz1 bound ribosome by rigid-body docking. Residues whose cross-linking are presented in b and c are shown in sphere representation (eL31 in orange and Zuo1 in cyan); Zuo1 residues Arg247 and Arg251 known to be important for ribosome association are also shown in sphere representation (cyan).
Bpa was also incorporated into Zuo1. We focused on the interval between residues 236 and 281, which encompasses helix III and a portion of the adjacent long loop, because it contains the most highly conserved surface exposed residues, including Arg247 and Arg251 known to be important for ribosome association28. Bpa was successfully incorporated at 26 positions (Supplementary Fig. 2c). No crosslinks were detected when Bpa was incorporated in the segment of Zuo1 surrounding Arg247 and Arg251. However, Thr266Bpa and Val273Bpa generated strong crosslinks to eL31, while 6 other variants having Bpa between positions 262 to 281 crosslinked weakly to eL31 (Fig. 2c; Supplementary Fig. 2c–e). Earlier reports indicated that deletion of the entire eL31 gene results in only slight destabilization of the Zuo1-ribosome interaction24. Therefore, we anticipated that residues interacting with eL31 would not substantially affect association of Zuo1 with the ribosome. Consistent with this view, a variant altered for three residues on the exposed surface of helix III, Asp262Ala, Thr266Ala and Val273Ala, co-migrated with ribosomes during centrifugation of cell lysates through sucrose cushions (Supplementary Fig. 2f).
Crosslinking of the C-terminus of helix III of Zuo1 to eL31 was key, as it allowed us to begin to position the ZHD on the 60S subunit. First, the face of the C-terminal segment of helix III having Thr266 and Val273 was manually docked to the surface-exposed loop and adjacent β-strand of eL31. The Arg247 and Arg251 residues at the N-terminal tip were pointed either towards H24 rRNA or towards H59 rRNA/eL22, as two reported cryo-EM analyses indicated these as sites of possible interaction of Zuo125,26. The interactions for both positions were then optimized by rigid body docking within the density map of Zuo1-Ssz1 bound 60S26 (Supplementary Fig. 3). Neither model fit perfectly within the 7.2 Å density dataset used in the docking. For example, relevant to the H24 model, density is lacking near the tip of the H24 rRNA helix. In the case of the H59/eL22 model, helix IV, that is the beginning of the MD, projected away from the 40S subunit and thus away from the density ascribed to the extended MD.
Therefore, we decided to experimentally test whether H24 or H59 is important for Zuo1’s interaction with the ribosome. We made use of a S. cerevisiae strain in which the ~150 chromosomal rDNA repeats are completely deleted and their essential function supported by rRNA transcribed from a plasmid carrying a single rDNA repeat30. The terminal base pair of H24 or the five most terminal base pairs of H59 were removed, resulting in strains rRNAH24Δ1 and rRNAH59Δ5 (Fig. 2d, Supplementary Fig. 2g). The vast majority of Zuo1 in extracts of rRNAH59Δ5 cells pelleted with ribosomes through a sucrose cushion, even though the deletion of 5 base pairs removed the surface exposed portion of the helix. On the other hand, most of Zuo1 was found in the supernatant fraction when extracts of rRNAH24Δ1 cells were analyzed, indicating that Zuo1’s interaction with the ribosome is destabilized upon alteration of H24.
Together, these genetic, crosslinking and modeling results are consistent with the idea that H24 interacts with Arg247 and Arg251 of ZHD’s helix III (Fig. 2e), and with the previously demonstrated importance of interactions with rRNA for association of Zuo1 with the ribosome9,24. As these arginines are conserved in Jjj1 (Arg221 and Arg225) and earlier cryo-EM studies pointed to interaction of Jjj1 with the exposed loop of eL3126,31 the conserved segment of Jjj1 may well interact with the 60S subunit in a similar manner. We also note that although we observed no crosslink between eL22 and Zuo1, two cryo-EM studies suggest an interaction between these two proteins. It is quite possible that a region of Zuo1 not tested (e.g. the J domain) interacts with eL22 and this interaction was not detected for technical reasons in our eL22 crosslinking analysis.
The Zuo1-40S subunit interaction
Recent cryo-EM analysis suggests that Zuo1 contacts the 40S subunit via interaction with ES12 of H4426. To verify this idea, we constructed a strain whose ribosomes lack the terminal 10 base pairs of ES12 (Fig. 3a; Supplementary Fig. 4a). The majority of Zuo1 remained in the supernatant after pelleting of ribosomes in extracts of rRNAES12Δ10 cells (Fig. 3a). The Zuo1 C-terminal 4-helix bundle and the C-terminal end of the MD is implicated in association with the 40S subunit26. Both this segment of the MD and helix 1 of the 4-helix bundle (348–363) contain positively charged residues, which are thus candidates for interaction with ES12. We altered 6 of these, Lys341, Lys342 and Lys344 in the MD and Lys348, Lys352 and Lys353 in helix 1, individually to alanines. Interaction of each variant with the ribosome was partially destabilized, as a portion did not co-sediment with ribosomes (Fig. 3b). We then constructed triple mutants, combining the alanine substitutions of the MD or helix 1. The vast majority of Zuo1Lys341/342/344Ala and Zuo1Lys348/352/353Ala did not co-sediment with ribosomes through a sucrose cushion (Fig. 3c, Supplementary Fig. 4b), indicating that most of the Zuo1-ribosome complexes were destabilized to the extent that they did not persist through the centrifugation procedure. Together, our results give strong support to the idea that ES12 of H44 of the 18S rRNA interacts with the C-terminal portion of the MD and helix I of Zuo1’s C-terminal 4-helix bundle.
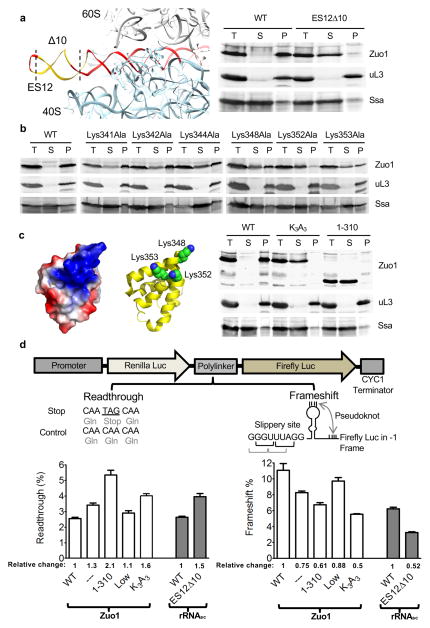
Figure 3.
Interaction of Zuo1 C-terminus with 40S ribosomal subunit and effect on translation fidelity. (a) rRNA ES12 (red). 10 base pairs deleted to yield ES12Δ10 (yellow) (left). Stability of Zuo1 interaction with ribosomes in cells expressing one copy of rDNA (right). Lysates were centrifuged through sucrose cushions. Equivalent amounts of total lysate (T), supernatant (S) and pellet (P) fractions subjected to electrophoresis and immunoblot analysis. Antibodies specific for Zuo1, and for uL3 and Ssa, as controls for ribosomes and a soluble protein, respectively. (b) Stability of ribosome interaction of indicated Zuo1 variants each having an alanine substituted for a single lysine. Analysis performed as in a. (c) Variants used for analysis of translational fidelity. Zuo1 C-terminal 4-helix bundle (residues 348–433; PDB 2LWX); Surface charge density showing distribution of basic (blue) and acidic (red) residues (far left). Three conserved lysine residues altered highlighted as spheres (middle). Δzuo1 cells expressing either Zuo1 (WT); Zuo1Lys348/352/353Ala (K3A3); Zuo11–310 (1–310) analyzed for ribosome association as in a (right). (a,b,c) One representative immunoblot (from three independent experiments, performed with different yeast cultures) is shown. Uncropped gel images are shown in Supplementary Data Set 1. (d) in vivo analyses of readthrough and −1 frameshifting. (top) Cartoon of dual-luciferase reporters. (bottom) % readthrough and −1 frameshifting compared to control reporters in which no stop codon between genes is present or genes are in frame, respectively. Relative change in readthrough and frameshifting are given; levels set at 1 in the case of Δzuo1 cells (open bars) expressing wild type Zuo1 and in the case of rRNASC, cells (i.e. cells carrying a single rDNA repeat; dark gray bars) expressing wild type rRNA. Strains as indicated in a and c with the addition of ZUO1 under MET3 promoter (Low) and no Zuo1 (---). Data shown are mean and s.e.m. from three different yeast cultures, each evaluated at least twice.
The base of H44 is in close proximity to the decoding center. Thus, binding of Zuo1 to ES12 raises the question as to the importance of the Zuo1-40S subunit interaction to Zuo1’s known effect on translational fidelity (i.e. readthrough and −1 frameshifting). To address this issue, we constructed a C-terminal deletion mutant, zuo11–310, which removes the segment of ZUO1 that encodes the 4-helix bundle and a portion of the MD helix, eliminating the possibility of interaction with ES12. As a control, we analyzed cells carrying wild type Zuo1 expressed under the control of the repressible promoter MET3 (called ZUO1Low); in the presence of high methionine Zuo1 is expressed at less than 5% of normal levels (Supplementary Fig. 4c). As previously reported32, such low levels of Zuo1 substantially suppressed the slow growth and cation-sensitive phenotypes of zuo1Δ cells, as do Zuo11–310 and Zuo1Lys348/352/353Ala (Supplementary Fig. 4d).
Translational readthrough and −1 frameshifting were assessed using a dual luciferase system in which reporter plasmids have a translational fusion between the sea pansy luciferase (RLuc) gene and the downstream firefly luciferase (Fluc) gene. As test sequences are located within the linker between the genes (Fig. 3d), Fluc serves as the reporter and Rluc as the internal control. To measure readthrough the test plasmid contained a TAG stop codon in the linker region. Wild type cells having this plasmid had 2.6% of the Fluc activity of cells harboring the control plasmid with a sense codon at that position (Fig. 3d). Readthrough in zuo1Lys348/352/353Ala and zuo11–310 cells was 160% and 210% of that in wild type cells, respectively. In ZUO1Low cells readthrough was only 110% of the wild type level. For assessing −1 frameshifting, the reporter plasmid, previously used to demonstrate a ~50% reduction in −1 frameshifting of a zuo1Δ strain23, which contains a −1 shift in reading frame and a viral sequence programmed for −1 frameshifting, was used (Fig. 3d). −1 frameshifting in zuo1Lys348/352/353Ala and zuo11–310 cells was 50% and 61% of that in ZUO1 cells, respectively, while that in ZUO1Low cells approached wild type levels at 88%. We also tested rRNAES12Δ10 cells in which the 40S Zuo1 binding site is deleted. Readthrough was increased (150% of wild type) and −1 frameshifting decreased (52% of wild type) in rRNAES12Δ10 (Fig. 3d). Together, these results are consistent with the idea that Zuo1’s interaction with the 40S subunit, more specifically with the tip of H44 (i.e. ES12) of 18S rRNA, is important for Zuo1 function in translational fidelity. In addition, consistent with a previous report21, the J domain, and thus functional interaction with Hsp70, is also required for Zuo1 function in translation fidelity, as alteration of the conserved HisProAsp motif of the J domain resulted in increased readthrough and decreased −1 frameshifting (Supplementary Fig. 4e).
Zuo1 spanning of the 40S and 60S subunits
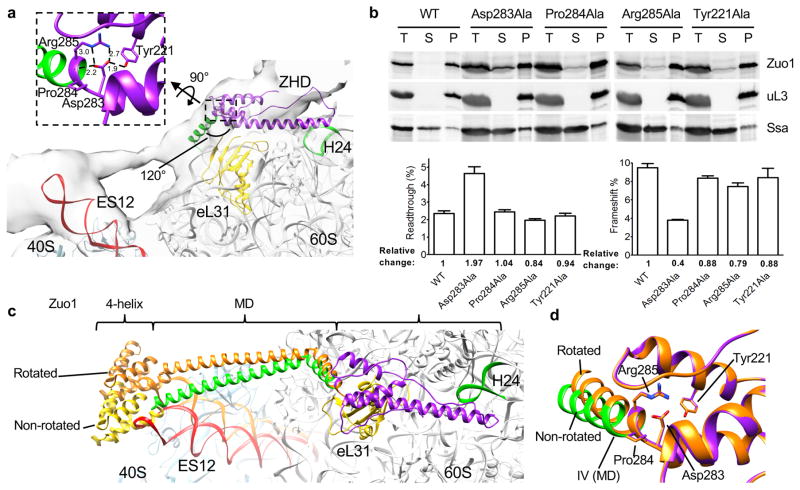
The 40S subunit rotates on the order of 11° relative to the 60S subunit during a round of peptide bond formation33. Zuo1 is able to bind to ribosomes that are in both the nonrotated and rotated states26. But, how Zuo1 accommodates such movement is not known. Recent results26 indicate that the rigidity of the MD helix, which links the ribosome interacting domains, is functionally important. Therefore, we more thoroughly analyzed the junction between the ZHD and the MD as a candidate for playing a role in orienting the C-terminal 4-helix bundle for interaction with the 40S subunit and for accommodating rotation of the subunits. The helix III-helix IV 120° angled junction is centered on Pro284 and stabilized by several interactions (Fig. 4a, Supplementary Fig. 1b): (1) the salt bridge between the side chains of Asp283 and Arg285, (2) a hydrogen bond between Asp283 and Tyr221 and (3) interaction of Asp283 with the amide backbone between Pro284 and Arg285. Residues Asp283, Arg285 and Tyr221 are all well resolved in the electron density maps with the side chain B-factor of 24.46 Å2, 20.67 Å2, and 22.91 Å2, respectively. We experimentally assessed the importance of this junction, altering Tyr221, Asp283, Pro284 and Arg285, each to alanine. Single alterations had minimal effects on cell growth even in the presence of cations (Supplementary Fig. 4f). The majority of all variants co-migrated with ribosomes through a sucrose cushion, but a portion of Zuo1Asp283Ala was present in the supernatant, suggesting an alteration in ribosome association (Fig. 4b). Cells expressing Zuo1Asp283Ala, but not other variants, were defective in translational readthrough and −1 frameshifting, having 176% higher readthrough and only 40% the level of frameshifting compared to wild type cells (Fig 4b). This in vivo effect of altering Asp283, the predominant interacting component of the junction, indicates that this junction is functionally important and is consistent with a role in orienting the interaction of Zuo1 with the 40S subunit.
Figure 4.
Positioning of Zuo1 C-terminal 4-helix bundle on 40S. (a) Junction between ZHD and MD. Atomic structure of ribosome (PDB 3J78) and Zuo1 169–303 fitted to cryo-EM density map of Zuo1-Ssz1 bound ribosome by rigid-body docking. Enlarged view of amino acids forming the 120° angled hinge between helix III of ZHD and N-terminus of MD shown. The salt bridge and hydrogen bonds are shown as dashed lines among Tyr221, Asp283, and Arg285. (b) Effects of alteration of residues at ZHD-MD junction. Stability of interaction of Zuo1 variants with ribosomes was analyzed by centrifugation of extracts through sucrose cushions; equivalent amounts of total lysate (T), supernatant (S) and pellet (P) fractions subjected to electrophoresis and immunoblot analysis using indicated antibodies (top). One representative immunoblot (from three independent experiments, performed with different yeast cultures) is shown. Uncropped gel images are shown in Supplementary Data Set 1. in vivo analyses of readthrough and −1 frameshifting; relative levels in cells expressing wild type Zuo1 set at 1 (bottom). Data shown are mean and s.e.m. from three different yeast cultures, each evaluated twice. (c) Model of Zuo1 binding to nonrotated and rotated states of ribosome. Zuo1 residues 173–433 (ZHD/MD/4-helix bundle) and a ribosome (PDB 3J78) were sequentially fitted to a cryo-EM map of Zuo1-Ssz1 bound nonrotated ribosome. Zuo1 bound to rotated state was modeled by moving Zuo1 MD and 4-helix bundle by 11 degrees relative to nonrotated state to mimic the ratchet-like motion of ribosome. (d) Accommodation of 11 degrees movement of ZHD-MD hinge. Minimum energy state of amino acids (orange) after 11 degrees rotation of MD superimposed on the junction shown in a that is in purple and green.
We next addressed the possible involvement of the ZHD-MD junction in accommodating subunit rotation. We modeled the entire ZHD-MD-4 helix bundle segment of Zuo1 (residues 169–433) using existing structural information. The entire segment was then positioned within the cryo-EM density of ribosome-bound Zuo1-Ssz1 in the nonrotated state26 (Fig. 4c), based on the positioning of the ZHD on the 60S subunit discussed in the previous section (Fig. 2e). The MD and 4 helix bundle segment of Zuo1 was then rotated 11 degrees relative to the ZHD. Upon this single change, the 4-helix bundle became positioned at the tip of ES12 in the rotated state, in a manner similar to that in the non-rotated state (Fig. 4c). To assess whether such movement could be accommodated by the junction without disrupting noncolavent interactions, the 11 degrees restraint was released for the Zuo1 ZHD-MD segment (169–303) through energy minimization. The distances between atoms forming the salt bridge and hydrogen bond at the junction changed minimally upon release, suggesting that the interactions could remain intact during subunit rotation (Fig. 4d). These results suggest a model in which a restrained flexibility of the ZHD-MD (i.e. helix III-helix IV) junction, which essentially forms a hinge, is important for accommodating subunit rotation, allowing Zuo1 interaction with both subunits during rounds of translation.
DISCUSSION
There is a growing appreciation of the importance of cellular strategies that ensure both the fidelity of the translation process itself and the productive folding of the protein synthesized34–36. Data, both presented here and previously published26, points to how Zuo1’s dual interaction with the 40S and the 60S ribosomal subunits may enable it to play a key role in such protein quality control (Fig. 5). It is tempting to speculate that Zuo1 serves a unique role as it is positioned to monitor and/or fine tune codon-anticodon interactions and thus peptide bond formation, as well as nascent polypeptide chain folding/transit within the tunnel.
Figure 5.
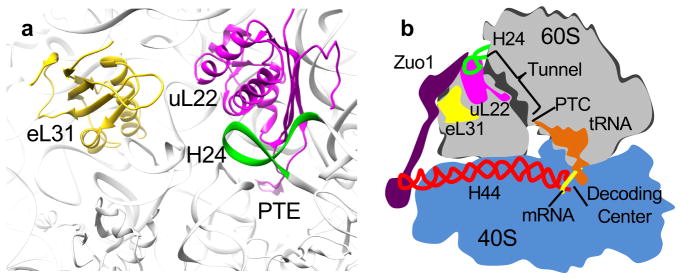
Model of the network of interaction of Zuo1 with the ribosome. (a) Proximity of H24, uL22 and ribosome tunnel. H24 (green) on the subunit surface directly interacts with uL22 (magenta), which extends into the interior of the subunit, forming a patch on the tunnel surface. (b) Schematic diagram of Zuo1’s interactions with the ribosome implying its function in coordinating protein translation and protein folding. Zuo1 monitors transit of nascent polypeptide chains in the tunnel via its interaction with H24 (green), which in turn interacts with uL22 (magenta). Zuo1 monitors or regulates peptide bond formation at the decoding center via interaction of its C-terminal 4-helix bundle with ES12 of H44 (red), which originates at the decoding center. Through these interactions Zuo1 functionally connects the two subunits of the ribosome.
A picture of Zuo1’s bipartite ribosome interaction is emerging. As suggested earlier26, and verified here, functional connection to the decoding center of the 40S subunit occurs via interaction of Zuo1’s C-terminal 4-helix bundle with ES12 of H44. Disruption of this interaction, either by altering rRNA or Zuo1 binding sites, increases translational readthrough of stop codons and decreases −1 frameshifting. Nucleotides near the base of H44 interact with codon-anticodons base pairs and release factors regulating translation rate and ensuring accuracy of translation elongation and termination37–39. Thus, through interaction with ES12, Zuo1 could monitor decoding or possibly affect the proofreading process itself. In regards to Zuo1 playing a signaling/monitoring role, it is also intriguing that helix III of the ZHD interacts with H24 of 25S rRNA on the surface of the 60S subunit (Fig 5). Interestingly, H24 also interacts with uL22 (previously called L17 in eukaryotes). uL22 extends from the subunit surface into the subunit interior forming a patch on the wall of the exit tunnel at its constriction point, which serves as a monitoring gate for translation arrest or pausing40–43. There is abundant evidence in both prokaryotes and eukaryotes that interaction of the nascent chain with the tunnel interior plays a signaling role44. In most cases this signaling is thought to affect the rate of peptide bond formation at the peptidyl transferase site. However, there is evidence that uL22 also signals to the ribosome surface near the exit site34,44,45.
The positioning of Zuo1 near the exit site and its contact with H44 suggests the possibility of regulatory functions for both interactions. In addition, structural and crosslinking data point to a possible line of communication between them, with helix III of the ZHD interacting with 60S subunit, and the adjacent MD helix extending to the 40S subunit. The intramolecular interactions at the junction between the ZHD and the MD (helices III and IV of the structure reported here) are predicted to be flexible enough to accommodate ribosome movement associated with a round of peptide bond formation, that is movement between the nonrotated and rotated states. In a purified system Zuo1 binding stabilized 80S ribosomes in the nonrotated state26, consistent with possible influence on the rate of peptide bond formation. Since translation rate has been linked to translation fidelity39,46, Zuo1 binding may affect fidelity in this way as well.
What role might Zuo1’s dual interaction with the ribosome play? One can imagine that it could either play a sensing role(s), signaling back to its partner Hsp70 Ssb, or act as a conduit for a signal from Ssb. For example, Zuo1 could serve as a sensor of activity at the decoding center and/or from within the tunnel, providing feedback to regulate Hsp70 activity. Such feedback might result in altering the timing of its J domain activity to stimulate Hsp70’s ATPase activity and thus Hsp70’s interaction with the nascent chain. Alternatively, Zuo1 could be the recipient of a signal from either Ssb or the atypical Hsp70 Ssz1. Upon interaction with a nascent chain exiting the tunnel, a signal to regulate peptide bond formation might be made through Zuo1’s contacts with the 40S and/or 60S subunits.
Regardless of exact mechanisms, such ideas are reminiscent of the well-established role of the multimeric RNA-protein complex, signal recognition particle (SRP)47. To our knowledge eukaryotic SRP is the only other factor known to span ribosomal subunits, connecting such distant sites. However, SRP action is both more specific and more direct than that of Zuo1. There is no evidence that Zuo1 directly binds nascent chains. SRP not only interacts directly with a signal sequence on the nascent chain upon its exit from the tunnel, it interacts directly with the decoding center to attenuate protein synthesis until the signal sequence contacts its receptor on the endoplasmic reticulum47,48. Zuo1 likely has a more general function, coupling protein folding to translation rate. A connection between translation rate and both translation fidelity and productive protein folding has been noted previously6,39. Thus, fine tuning of coupling of these events likely plays an important role in protein homeostasis.
It is also interesting to note that Zuo1 and its orthologs have diverse regulatory functions off the ribosome, consistent with evidence that its interaction with the ribosome is marginally stable and likely dynamic9,27. For example, S. cerevisiae Zuo1 activates Pdr1, a transcription factor implicated in quorum sensing27,49. Human DnaJC2 functions as a chromatin modulator, affecting development and DNA repair50,51. Thus, via both on- and off-ribosome functions, Zuo1/DnaJC2 may play a more global regulatory role than we presently appreciate, coordinating protein synthesis and folding with other aspects of gene expression and cell physiology.
ONLINE METHODS
Protein expression and purification
DNA encoding residues 166–303 of Zuo1 was amplified by PCR, and cloned into a modified pET-28a vector that contains N-terminal His6-tag, thioredoxin (TRX) tag and a cleavable tobacco etch virus (rTEV) protease site. Two mutations to substitute methionine for K210 and N269 were introduced into ZUO1166–303 via QuikChange PCR mutagenesis (Stratagene). Protein was expressed in Escherichia coli Rosetta 2 (DE3) pLysS using LB or autoinduction medium52 containing L-selenomethionine (SeMet). Cells were resuspended in lysis buffer (50 mM Tris pH 7.5, 250 mM NaCl, 10 mM imidazole, 5% glycerol) with cOmplete EDTA-free Protease Inhibitor Cocktail (Roche), and then lysed using a French press at 4°C. Lysates were clarified by centrifugation. Initial protein purification was done using gravity flow nickel column chromatography. Tags were removed by proteolysis with recombinant rTEV protease overnight at 4°C. Tagged protein, cleaved His6/TRX-tag, and His6-tagged rTEV were separated from Zuo1 fragment by gravity flow nickel chromatography. Protein was further purified by using S-75 size exclusion chromatography column (GE Healthcare) in 5 mM HEPES pH 7.5, 100 mM NaCl and 5 mM dithiothreitol. Protein was used immediately or stored at −80°C.
Crystallization, data collection, structure determination and refinement
Crystals were obtained at 4°C by the hanging drop vapor diffusion method. One microliter of SeMet labeled K210M N269M Zuo1166–303 (15 mg/ml) was mixed with one microliter of crystallization buffer (100 mM BisTris, pH 6.5, 30% PEG 3350, 30 mM MgCl2, 5 mM dithiothreitol). The reservoir volume was 0.5 ml.
Zuo1166–303 crystals were flash-frozen in liquid nitrogen after cryo-protecting the crystals in cryo-solution containing mother liquor and 5% ethylene glycol. X-ray diffraction data were collected at 100 K at the Advanced Photon Source beamline 23-ID-D at a wavelength of 0.95372 Å. Data were processed using XDS53. Phenix, AutoSol54 was used to identify the selenium atoms, and generate the electron density map and initial model. A final model was obtained after several rounds of model building using Coot55 and refining with Phenix.refine54. Ramachandran plot analysis was carried out using MolProbity56. 99.6% of the residues were in favored regions; 100% were in allowed regions.
Molecular modeling and docking
For modeling of ZHD homologs, sequences were first aligned by Clustal Omega57. Then, using the S. cerevisiae Zuo1 ZHD structure as a template, model structures were generated using Modeller58. All dockings were performed by initially positioning structures to the cryo-EM map of Zuo1/Ssz1 bound nonrotated ribosome and then optimizing by rigid-body docking using Chimera59. To model Zuo1 binding to the ribosome in the nonrotated state, ribosome (PDB 3J78), Zuo1 4-helix bundle (PDB 2LWX), Zuo1 MD and Zuo1 ZHD were sequentially fitted to the cryo-EM map of Zuo1/Ssz1 bound nonrotated ribosome and joined together. Since the structure of only a small portion of the MD has been experimentally determined, I-TASSER was used for modeling60. First, residues 284–365 were modeled using the α-helical keratin 14 (PDB 3TNU) as a template. The model was fit to the cryo-EM map, and then overlapping residues between the N- and C-termini portions of the model (284–300 and 348–365) and the experimental structures of Zuo1 ZHD and Zuo1 4-helix bundle were deleted for sequential docking. Zuo1 bound to the rotated state was modeled by rotating the modeled MD/4-helix bundle structure (285–433) by 11 degrees perpendicular to the plain of rotation of the tip of ES12 centered at the alpha carbon of R285, to mimic the ratchet-like motion. To examine the capability of the ZHD:MD hinge to accommodate such ratchet-like movement of ribosome, the minimum energy state of the amino acids forming the hinge (283–285 and 221) was generated, after rotation of 11 degrees, using Chimera.
Yeast strains and plasmids
Unless otherwise stated strains used were of the W303 genetic background. A list of yeast plasmids used in this study is shown in Supplementary Table 1. Mutagenesis was carried out by QuikChange PCR mutagenesis (Stratagene), using the standard protocol.
A previously described zuo1::HIS3 deletion9,61, was used for analysis of ZUO1 mutants. Strains were passaged thrice on rich medium containing 1 mM GuHCl to ensure the absence of prions, as the [PSI+] prion is known to affect stop codon readthrough. All plasmids used containing wild-type and mutant forms of ZUO1 were centromeric, based on the pRS plasmid series62,63. Reduced expression level of Zuo1 was achieved by growing cells expressing Zuo1 under the control of the MET3 promoter64 on selective minimal glucose media containing 400 μg/ml methionine.
For analysis of rDNA mutants, KAY48865, which has a deletion of the ~150 repeats of ribosomal DNA (RDN) encoding 5S, 5.8S, 18S and 25S rRNA, and harbors the plasmid pRDN-hyg1, was used. pNOY373, a 2μ plasmid containing the LEU2 gene and a RDN repeat, was used to construct variants of 18S and 25S rRNA. For ease of mutant creation, segments of RDN were subcloned, and then transferred back into pNOY373 after construction by QuikChange PCR mutagenesis (Stratagene). For construction of ES12 and H24 mutants, a Nde1-Mlu1 fragment was used; for the H59 mutant a Mlu1 and Xho1 fragment was used. KAY488 transformed with pNOY373 or derivatives were plated on 5-FOA to select against the original plasmid, which contains the URA3 gene. After selection, plasmids were rescued from KAY488 and sequenced to ensure the presence of the mutation prior to carrying out experiments.
Crosslinking
p-benzoyl-L-phenylalanine (Bpa) cross-linking was fundamentally carried out as previously described, using plasmid ptRNA-Bpa, which encodes a variant tRNA synthetase and tRNACUA for Bpa incorporation66,67. An amber stop codon (TAG) was introduced into the open reading frames of ZUO1, RPL31A or RPL22A on the plasmids pRS315-Zuo1, pRS416TEF-Rpl31A-HA or pRS316-Rpl22A-HA. Cells were grown in the presence of 2 mM Bpa at 30°C in 50 ml of minimal media overnight in the dark, with a starting OD600 of 0.06. Approximately 25 OD600 of cells treated with 100 μg/ml of cycloheximide were exposed to 365 nm UV (Stratalinker 1800 UV irradiator) for 1 hr at 4°C to activate Bpa, while the same amount of cells was kept at 4°C as a control. Cells were lysed via agitation for 5 min at 4°C in lysis buffer (300 mM sorbitol, 20 mM HEPES-KOH pH 7.4, 1 mM EGTA, 5 mM MgCl2, 10 mM KCl, 10% glycerol, 2 mM β-mercaptoethanol, RNasin Ribonuclease Inhibitor (Promega) at a dilution of 1:1000) with glass beads. 5 OD260 units of cell lysates were loaded onto a 2 ml sucrose cushion containing lysis buffer with 0.5 M sucrose instead of sorbitol. Ribosomes were pelleted by centrifugation for 3 hr at 50,000 rpm in a TLA 100.3 rotor (Beckman Coulter) at 4°C. The ribosome-containing pellet was suspended in SDS sample buffer (0.124M Tris-HCl buffer pH 6.8 containing 4% SDS, 10% glycerol, 0.02% bromophenol blue and 4.5% β-mercaptoethanol) and used for immunoblotting. HA tag-specific antibody (Covance, no longer available and Abcam, catalog number: ab9110) was used to detect HA-tagged eL31a and eL22a. A rabbit polyclonal antibody generated using a fusion between Zuo1166–284 and glutathionine-S-transferase (GST) was used to detect Zuo1 (see Supplementary Fig. 3e). To detect Ssz1, a rabbit polyclonal antibody to a GST fusion of the ATPase domain (residues 1–401) of Ssz168. Crosslinking was carried out with cells of the DS10 strain background69. For eL31a and eL22a cross-linking, rpl31aΔ::KanMX and rpl22a::TRP1 strains, respectively, were used. For Zuo1 cross-linking, a zuo1Δ rpl31aΔ strain carrying the pRS416TEF-Rpl31a-HA plasmid was constructed.
Ribosome sedimentation analysis
For preparation of yeast cell lysates, cells were grown at 30°C, to mid-log phase in selective minimal medium, 100 μg/ml of cycloheximide was added and cells immediately harvested by centrifugation at 4°C. Cells were washed and resuspended in ice-cold buffer A (20 mM Tris-HCl pH 7.5, 50 mM KCl and 5 mM MgCl2) plus 1.5 mM pepstatin, cOmplete Mini EDTA-free Protease Inhibitor Cocktail (Roche) and 20 units of Recombinant RNasin Ribonuclease Inhibitor (Promega). Cells were lysed using a Retsch MM400 mixer mill, after freezing in liquid nitrogen. After lysis, 0.2 ml of ice-cold buffer A was added and the cell lysate was clarified by centrifugation. To separate ribosomes from soluble proteins, 5 OD260 units of lysates were applied to the top of a 2 ml sucrose cushion (buffer A containing 0.5 M sucrose) and centrifuged for 3 hrs at 50,000 rpm at 4°C in a TLA100.3 rotor (Beckman Coulter). The supernatant was removed and precipitated with 86% acetone overnight at −20°C. The ribosome and precipitated protein pellets were resuspended in SDS sample buffer by shaking at 4°C. Equivalent amounts of total lysate, supernatant and ribosome fractions were analyzed by SDS-PAGE and immunoblot analysis. Antibodies used: Ssa, rabbit polyclonal using segment containing residues 239–589 fused to GST as an immunogen70; uL3, mouse monoclonal made to S. cerevisiae uL371 (kind gift of Jonathan Warner); Zuo1, rabbit polyclonal made using a segment containing residues 1–68 fused to GST as an immunogen9.
Comparison of protein expression levels using immunoblot analysis was done as described previously28. Briefly, equivalent numbers of cells were harvested by centrifugation, and lysed by incubation in 0.1 M NaOH for 5 min at room temperature. The lysate was centrifuged and the pellet was resuspended in 50 μl of SDS sample buffer and boiled for 5 min. Equal amounts of extracts were subjected to SDS-PAGE and immunoblot analysis.
Stop codon readthrough and −1 frameshifting analyses
Reporter plasmids for monitoring readthrough are based on pDB68872,73, which has a translational fusion of the Renilla and firefly luciferase genes. To quantitatively study readthrough, an amber stop codon (TAG) flanked by CAA codons was introduced in the linker region between Renilla and firefly luciferase genes, because CAA codons flanking a stop codon have been shown to increase readthrough74. As a control, another plasmid was generated that has a sense codon with CAA codons on either side. Plasmids used for assessing frameshift frequencies having the firefly luciferase gene in the 0 frame (control) or −1 frame relative to the Renilla luciferase gene were a gift of J. Dinman23.
Luciferase assays were performed in a 96-well format using BioTek Synergy™ 2 multi-mode microplate reader and the Dual-Luciferase Reporter 1000 Assay System (Promega) as described previously72. An equivalent number of cells were harvested and lysed using Passive Lysis Buffer according to the manufacturer’s protocol. 5 μl of each of the lysates was taken in Lumitrac 200 96-well microplate (Greiner Bio-One) and firefly and Renilla luciferase activities were measured by sequential injection of 25 μl each of Luciferase Assay Reagent II and Stop & Glo buffer as described by the manufacturer. For each yeast strain tested, three individual transformants were assayed. An extract was made from each and assayed in duplicate. Negative controls that contained only Passive Lysis Buffer were used to determine the background and deducted from all the experimental values. The firefly/Renilla activity ratio generated for each of the reporter plasmids was divided by that for the respective control plasmids to obtain readthrough and frameshift efficiencies, which are reported as mean ± standard error.
Phylogenetic tree analysis
Amino acid sequences of S. cerevisiae Zuo1 and Jjj1, A. thaliana ZRF1 and human DNAJC2p and DNAJC21p were used to identify the Zuo1 and Jjj1 orthologs used in this study by protein BLAST. The protein sequences were aligned using ClustalW conducted in MEGA775 and trees were constructed using the Maximum Likelihood (ML) method based on the JTT matrix-based model76.
Supplementary Material
Acknowledgments
We thank: A. Senes, J. Keck and S. Butcher for helpful discussions during the course of this work; D. Bedwell (U. Alabama), P. Farabaugh (U. Maryland Baltimore County) and J. Dinman (U. Maryland) for dual luciferase plasmids; K. Asano (Kansas State U.) for the rDNA deletion strain; J. Warner (Albert Einstein College of Medicine) for uL3 antibody. This work was supported by National Institutes of Health grants GM031107 and GM027870 (E.A.C.). C.A.B. was supported by NIH Grants GM094584, GM094622, and GM098248. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. GM/CA@APS has been funded in whole or in part with Federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006).
Footnotes
Accession code: Atomic coordinates and structure factors have been deposited to the Protein Data Bank with the accession code 5DJE.
AUTHOR CONTRIBUTIONS
K.L. performed in vivo crosslinking experiments, analyses of ribosome association, generation of mutants and other molecular biological experiments. R.S. performed in vivo misreading experiments, analyses of ribosome association, generation of mutants and other molecular biological experiments. O.K.S. performed crystallographic analysis, analyses of ribosome association and generation of mutants. C.B. performed crystallographic analysis. E.A.C. oversaw all aspects of the experiments and manuscript preparation. All authors participated in interpreting the data and writing the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–55. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 2.Gloge F, Becker AH, Kramer G, Bukau B. Co-translational mechanisms of protein maturation. Curr Opin Struct Biol. 2014;24:24–33. doi: 10.1016/j.sbi.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Wrobel L, et al. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature. 2015;524:485–8. doi: 10.1038/nature14951. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Chen XJ. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature. 2015;524:481–4. doi: 10.1038/nature14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korennykh A, Walter P. Structural basis of the unfolded protein response. Annu Rev Cell Dev Biol. 2012;28:251–77. doi: 10.1146/annurev-cellbio-101011-155826. [DOI] [PubMed] [Google Scholar]
- 6.Sherman MY, Qian SB. Less is more: improving proteostasis by translation slow down. Trends Biochem Sci. 2013;38:585–91. doi: 10.1016/j.tibs.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- 8.Preissler S, Deuerling E. Ribosome-associated chaperones as key players in proteostasis. Trends Biochem Sci. 2012;37:274–83. doi: 10.1016/j.tibs.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Yan W, et al. Zuotin, a ribosome-associated DnaJ molecular chaperone. EMBO J. 1998;17:4809–17. doi: 10.1093/emboj/17.16.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hundley HA, Walter W, Bairstow S, Craig EA. Human Mpp11 J protein: ribosome-tethered molecular chaperones are ubiquitous. Science. 2005;308:1032–4. doi: 10.1126/science.1109247. [DOI] [PubMed] [Google Scholar]
- 11.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–92. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang P, Gautschi M, Walter W, Rospert S, Craig EA. The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nat Struct Mol Biol. 2005;12:497–504. doi: 10.1038/nsmb942. [DOI] [PubMed] [Google Scholar]
- 13.Clerico EM, Tilitsky JM, Meng W, Gierasch LM. How hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J Mol Biol. 2015;427:1575–88. doi: 10.1016/j.jmb.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willmund F, et al. The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell. 2013;152:196–209. doi: 10.1016/j.cell.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gautschi M, et al. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc Natl Acad Sci U S A. 2001;98:3762–7. doi: 10.1073/pnas.071057198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gautschi M, Mun A, Ross S, Rospert S. A functional chaperone triad on the yeast ribosome. Proc Natl Acad Sci U S A. 2002;99:4209–14. doi: 10.1073/pnas.062048599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otto H, et al. The chaperones MPP11 and Hsp70L1 form the mammalian ribosome-associated complex. Proc Natl Acad Sci U S A. 2005;102:10064–9. doi: 10.1073/pnas.0504400102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiaux J, et al. Structural analysis of the ribosome-associated complex (RAC) reveals an unusual Hsp70/Hsp40 interaction. J Biol Chem. 2010;285:3227–34. doi: 10.1074/jbc.M109.075804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conz C, et al. Functional characterization of the atypical Hsp70 subunit of yeast ribosome-associated complex. J Biol Chem. 2007;282:33977–84. doi: 10.1074/jbc.M706737200. [DOI] [PubMed] [Google Scholar]
- 20.Jaiswal H, et al. The chaperone network connected to human ribosome-associated complex. Mol Cell Biol. 2011;31:1160–73. doi: 10.1128/MCB.00986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rakwalska M, Rospert S. The ribosome-bound chaperones RAC and Ssb1/2p are required for accurate translation in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:9186–97. doi: 10.1128/MCB.24.20.9186-9197.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caliskan N, Peske F, Rodnina MV. Changed in translation: mRNA recoding by −1 programmed ribosomal frameshifting. Trends Biochem Sci. 2015;40:265–74. doi: 10.1016/j.tibs.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muldoon-Jacobs KL, Dinman JD. Specific effects of ribosome-tethered molecular chaperones on programmed −1 ribosomal frameshifting. Eukaryot Cell. 2006;5:762–70. doi: 10.1128/EC.5.4.762-770.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peisker K, et al. Ribosome-associated complex binds to ribosomes in close proximity of Rpl31 at the exit of the polypeptide tunnel in yeast. Mol Biol Cell. 2008;19:5279–88. doi: 10.1091/mbc.E08-06-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leidig C, et al. Structural characterization of a eukaryotic chaperone--the ribosome-associated complex. Nat Struct Mol Biol. 2013;20:23–8. doi: 10.1038/nsmb.2447. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, et al. Structural basis for interaction of a cotranslational chaperone with the eukaryotic ribosome. Nat Struct Mol Biol. 2014;21:1042–6. doi: 10.1038/nsmb.2908. [DOI] [PubMed] [Google Scholar]
- 27.Ducett JK, et al. Unfolding of the C-terminal domain of the J-protein Zuo1 releases autoinhibition and activates Pdr1-dependent transcription. J Mol Biol. 2013;425:19–31. doi: 10.1016/j.jmb.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaschner LA, Sharma R, Shrestha OK, Meyer AE, Craig EA. A conserved domain important for association of eukaryotic J-protein co-chaperones Jjj1 and Zuo1 with the ribosome. Biochim Biophys Acta. 2015;1853:1035–45. doi: 10.1016/j.bbamcr.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Albanese V, Reissmann S, Frydman J. A ribosome-anchored chaperone network that facilitates eukaryotic ribosome biogenesis. J Cell Biol. 2010;189:69–81. doi: 10.1083/jcb.201001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wai HH, Vu L, Oakes M, Nomura M. Complete deletion of yeast chromosomal rDNA repeats and integration of a new rDNA repeat: use of rDNA deletion strains for functional analysis of rDNA promoter elements in vivo. Nucleic Acids Res. 2000;28:3524–34. doi: 10.1093/nar/28.18.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greber BJ, Boehringer D, Montellese C, Ban N. Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nat Struct Mol Biol. 2012;19:1228–33. doi: 10.1038/nsmb.2425. [DOI] [PubMed] [Google Scholar]
- 32.Hundley H, et al. The in vivo function of the ribosome-associated Hsp70, Ssz1, does not require its putative peptide-binding domain. Proc Natl Acad Sci U S A. 2002;99:4203–8. doi: 10.1073/pnas.062048399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svidritskiy E, Brilot AF, Koh CS, Grigorieff N, Korostelev AA. Structures of yeast 80S ribosome-tRNA complexes in the rotated and nonrotated conformations. Structure. 2014;22:1210–8. doi: 10.1016/j.str.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pechmann S, Willmund F, Frydman J. The ribosome as a hub for protein quality control. Mol Cell. 2013;49:411–21. doi: 10.1016/j.molcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hilal T, Spahn CM. Ribosome rescue and protein quality control in concert. Mol Cell. 2015;57:389–90. doi: 10.1016/j.molcel.2015.01.029. [DOI] [PubMed] [Google Scholar]
- 36.Shen PS, et al. Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science. 2015;347:75–8. doi: 10.1126/science.1259724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Youngman EM, Cochella L, Brunelle JL, He S, Green R. Two distinct conformations of the conserved RNA-rich decoding center of the small ribosomal subunit are recognized by tRNAs and release factors. Cold Spring Harb Symp Quant Biol. 2006;71:545–9. doi: 10.1101/sqb.2006.71.036. [DOI] [PubMed] [Google Scholar]
- 38.Carter AP, et al. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–8. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 39.Zaher HS, Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009;136:746–62. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–36. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 41.Lin PJ, Jongsma CG, Pool MR, Johnson AE. Polytopic membrane protein folding at L17 in the ribosome tunnel initiates cyclical changes at the translocon. J Cell Biol. 2011;195:55–70. doi: 10.1083/jcb.201103118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fulle S, Gohlke H. Statics of the ribosomal exit tunnel: implications for cotranslational peptide folding, elongation regulation, and antibiotics binding. J Mol Biol. 2009;387:502–17. doi: 10.1016/j.jmb.2009.01.037. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, Wolfle T, Rospert S. Interaction of nascent chains with the ribosomal tunnel proteins Rpl4, Rpl17, and Rpl39 of Saccharomyces cerevisiae. J Biol Chem. 2013;288:33697–707. doi: 10.1074/jbc.M113.508283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson DN, Beckmann R. The ribosomal tunnel as a functional environment for nascent polypeptide folding and translational stalling. Curr Opin Struct Biol. 2011;21:274–82. doi: 10.1016/j.sbi.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 45.Pool MR. A trans-membrane segment inside the ribosome exit tunnel triggers RAMP4 recruitment to the Sec61p translocase. J Cell Biol. 2009;185:889–902. doi: 10.1083/jcb.200807066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fluitt A, Pienaar E, Viljoen H. Ribosome kinetics and aa-tRNA competition determine rate and fidelity of peptide synthesis. Comput Biol Chem. 2007;31:335–46. doi: 10.1016/j.compbiolchem.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wild K, Halic M, Sinning I, Beckmann R. SRP meets the ribosome. Nat Struct Mol Biol. 2004;11:1049–53. doi: 10.1038/nsmb853. [DOI] [PubMed] [Google Scholar]
- 48.Elvekrog MM, Walter P. Dynamics of co-translational protein targeting. Curr Opin Chem Biol. 2015;29:79–86. doi: 10.1016/j.cbpa.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prunuske AJ, Waltner JK, Kuhn P, Gu B, Craig EA. Role for the molecular chaperones Zuo1 and Ssz1 in quorum sensing via activation of the transcription factor Pdr1. Proc Natl Acad Sci U S A. 2012;109:472–7. doi: 10.1073/pnas.1119184109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richly H, et al. Transcriptional activation of polycomb-repressed genes by ZRF1. Nature. 2010;468:1124–8. doi: 10.1038/nature09574. [DOI] [PubMed] [Google Scholar]
- 51.Gracheva E, et al. ZRF1 mediates remodeling of E3 ligases at DNA lesion sites during nucleotide excision repair. J Cell Biol. 2016;213:185–200. doi: 10.1083/jcb.201506099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blommel PG, Becker KJ, Duvnjak P, Fox BG. Enhanced bacterial protein expression during auto-induction obtained by alteration of lac repressor dosage and medium composition. Biotechnol Prog. 2007;23:585–98. doi: 10.1021/bp070011x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 56.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Webb B, Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr Protoc Bioinformatics. 2014;47:561–32. doi: 10.1002/0471250953.bi0506s47. [DOI] [PubMed] [Google Scholar]
- 59.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 60.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–38. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.James P, Pfund C, Craig EA. Functional specificity among Hsp70 molecular chaperones. Science. 1997;275:387–9. doi: 10.1126/science.275.5298.387. [DOI] [PubMed] [Google Scholar]
- 62.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–22. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 64.Mountain HA, Byström AS, Larsen JT, Korch C. Four major transcriptional responses in the methionine/threonine biosynthetic pathway of Saccharomyces cerevisiae. Yeast. 1991;7:781–803. doi: 10.1002/yea.320070804. [DOI] [PubMed] [Google Scholar]
- 65.Nemoto N, et al. Yeast 18 S rRNA is directly involved in the ribosomal response to stringent AUG selection during translation initiation. J Biol Chem. 2010;285:32200–12. doi: 10.1074/jbc.M110.146662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krishnamurthy M, et al. Caught in the act: covalent cross-linking captures activator-coactivator interactions in vivo. ACS Chem Biol. 2011;6:1321–6. doi: 10.1021/cb200308e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ting SY, Schilke BA, Hayashi M, Craig EA. Architecture of the TIM23 inner mitochondrial translocon and interactions with the matrix import motor. J Biol Chem. 2014;289:28689–96. doi: 10.1074/jbc.M114.588152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eisenman HC, Craig EA. Activation of pleiotropic drug resistance by the J-protein and Hsp70-related proteins, Zuo1 and Ssz1. Mol Microbiol. 2004;53:335–44. doi: 10.1111/j.1365-2958.2004.04134.x. [DOI] [PubMed] [Google Scholar]
- 69.Stone DE, Craig EA. Self-regulation of 70-kilodalton heat shock proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:1622–32. doi: 10.1128/mcb.10.4.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Halladay JT, Craig EA. A heat shock transcription factor with reduced activity suppresses a yeast HSP70 mutant. Mol Cell Biol. 1995;15:4890–7. doi: 10.1128/mcb.15.9.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vilardell J, Warner JR. Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol Cell Biol. 1997;17:1959–65. doi: 10.1128/mcb.17.4.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Salas-Marco J, Bedwell DM. Discrimination between defects in elongation fidelity and termination efficiency provides mechanistic insights into translational readthrough. J Mol Biol. 2005;348:801–15. doi: 10.1016/j.jmb.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 73.Kramer EB, Vallabhaneni H, Mayer LM, Farabaugh PJ. A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae. RNA. 2010;16:1797–808. doi: 10.1261/rna.2201210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Namy O, Hatin I, Rousset JP. Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep. 2001;2:787–93. doi: 10.1093/embo-reports/kve176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016 doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–82. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.