Abstract
Dystroglycanopathy is a major class of congenital muscular dystrophy caused by a deficiency of functional glycans on α-dystroglycan (αDG) with laminin-binding activity. Recent advances have led to identification of several causative gene products of dystroglycanopathy and characterization of their in vitro enzymatic activities. However, the in vivo functional roles remain equivocal for enzymes such as ISPD, FKTN, FKRP, and TMEM5 that are supposed to be involved in post-phosphoryl modifications linking the GalNAc-β3-GlcNAc-β4-Man-6-phosphate core and the outer laminin-binding glycans. Herein, by direct nano-LC-MS2/MS3 analysis of tryptic glycopeptides derived from a truncated recombinant αDG expressed in the wild-type and a panel of mutated cells deficient in one of these enzymes, we sought to define the full extent of variable modifications on this phosphorylated core O-glycan at the functional Thr317/Thr319 sites. We showed that the most abundant glycoforms carried a phosphorylated core at each of the two sites, with and without a single ribitol phosphate (RboP) extending from terminal HexNAc. At much lower signal intensity, a novel substituent tentatively assigned as glycerol phosphate (GroP) was additionally detected. As expected, tandem RboP extended with a GlcA-Xyl unit was only identified in wild type, whereas knocking out of either ISPD or FKTN prevented formation of RboP. In the absence of FKRP, glycoforms with single but not tandem RboP accumulated, consistent with the suggested role of this enzyme in transferring the second RboP. Intriguingly, the single GroP modification also required functional FKTN whereas absence of TMEM5 significantly hindered only the addition of RboP. Our findings thus revealed additional levels of complexity associated with the core structures, suggesting functional interplay among these enzymes through their interactions. The simplified analytical workflow developed here should facilitate rapid mapping across a wider range of cell types to gain better insights into its physiological relevance.
Dystroglycanopathy is a group of congenital muscular dystrophies that arise from glycosylation defects of α-dystroglycan (αDG).1 The hallmark and cause of these diseases, which include Fukuyama-type congenital muscular dystrophy, muscle-eye-brain diseases, Walker-Warburg syndrome, and congenital muscular dystrophy type 1D, are a lack of functional O-mannosyl glycans of αDG capable of binding laminin (1–3). To date, dozens of causative gene products of dystroglycanopathy have been identified, all of which have been demonstrated or are assumed to be involved in the synthesis of the laminin-binding glycans. A plethora of O-glycans including the normal O-GalNAc mucin types and all three core types of O-mannosyl glycans, namely, M1 (Gal-β4-GlcNAc-β2-Man), M2 [Gal-β4-GlcNAc-β2-(Gal-β4-GlcNAc-β6-)Man], and M3 (GalNAc-β3-GlcNAc-β4-Man) (4), have been identified on various sites of αDG. Remarkably, the functional laminin-binding glycans were found to be exclusively carried on the core M3 at specific Thr317/Thr319 sites (5). The formation of this innermost base structure involves at least three enzymes, namely, protein O-mannosyltransferase 1/2 heterodimer (POMT1/2), protein O-mannose β-1,4-N-acetylglucosaminyltransferase 2 (POMGNT2/AGO61), and β-1,3-N-acetylgalactosaminyltransferase 2 (B3GALNT2) (6–8). Notably, this trisaccharide core can be further phosphorylated at the 6-position of O-mannose by protein-O-mannose kinase (POMK/SGK196) (6). On the other hand, functional laminin binding is known to require a polymeric Xyl-GlcA repeat sequence, the elongation of which is catalyzed by LARGE (9). Because the Xyl-GlcA repeat is released from the core upon chemical hydrolysis of phosphoester linkages by hydrogen fluoride treatment, the laminin-binding glycan synthesized by LARGE was inferred to be extended from the phosphorylated core M3 via the phosphate added by POMK. However, the exact structural element bridging this missing link is lost by such chemical treatment and thus remains undescribed until very recently (10–13).
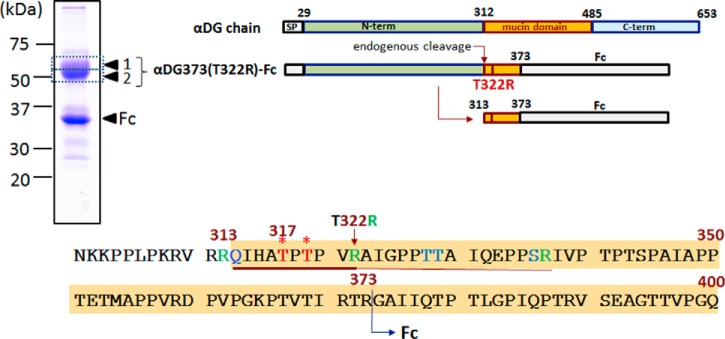
A major technical problem in identifying the elusive structural module linking the phosphorylated core M3 and the polymeric GlcA-Xyl units is the limiting mass spectrometry (MS) sensitivity in detecting a very large glycan or glycopeptide carrying multiple negative charges in the form of HexA and phosphate. Most of the analytical work to date has used recombinant αDG truncated at the C terminus to different extents but all containing at least the first 10 amino acid residues following the putative endogenous furin cleavage site R312 that releases the N-terminal domain (5, 14). For convenience in the purification, the truncated C terminus is also commonly fused to antibody Fc or other tags, which may or may not be further removed during sample preparation. Such a strategy typically yielded very large and heterogeneous tryptic glycopeptides that contain additional O-glycosylated serine/threonine downstream of the critical Thr317/Thr319 attachment sites (Fig. 1), which hampered high-quality MS/MS sequencing and unambiguous data interpretation (10). Alternatively, a tryptic site can be engineered by converting the first threonine after T319 to lysine (T322K), which would not only further truncate the target tryptic glycopeptide carrying Thr317/Thr319, but at the same time abolish the extra O-glycosylation. After further glycosidase treatment to remove mucin-type O-glycans, a recent study using such truncated αDG constructs expressed in and purified from mouse NIH 3T3 cells succeeded for the first time in detecting the further modified phosphorylated core by MS analysis in negative ion mode (11).
Fig. 1.
Recombinant αDG constructs and amino acid sequence of N301-Q400. The full-length αDG chain consists of an N-terminal domain (N-term) that is lost upon endogenous processing via furin-like cleavage (14), yielding a mature product starting at Q313. The inclusion of this domain in the recombinant αDG construct is, however, necessary as it is required for proper laminin-binding glycan formation at Thr317/Thr319 (5) (marked with asterisks). For the αDG373(T322R)-Fc product used in this study, an Fc domain is fused to R373, in place of the mucin-like and C-terminal domains. A tryptic cleavage site was introduced via T322R mutation, which allowed formation of a 10-amino acid tryptic peptide carrying only two threonine sites for O-glycosylation. The recombinant products, expressed and purified by protein G Sepharose from HEK293T cells, ran as two major bands on SDS-PAGE corresponding to ∼35 and 50–60 kDa. Only the upper band, excised, in-gel digested, and analyzed as bands 1 and 2 yielded similar tryptic peptides corresponding to the αDG mucin domain, whereas the lower band only gave peptides derived from the Fc domain. The entire amino acid sequence of αDG373(T322R)-Fc is shown in supplemental Fig. S1.
It was shown that a tandem ribitol phosphate unit (RboP-RboP) can be attached to the terminal GalNAc of phosphorylated core M3, which can then be extended further by repeating HexA-Pent units, consistent with the proposed model that is supported by in vitro enzymatic assays of the involved causative gene products including isoprenoid synthase domain containing (ISPD), fukutin (FKTN), and fukutin-related protein (FKRP) (11). It also indicated that ISPD is a cytidine diphosphate ribitol (CDP-Rbo) synthase and FKTN transfers a Rbo5P from CDP-Rbo to phosphorylated core M3, thereby providing an acceptor site for FKRP to form the RboP-RboP tandem repeat. The possibility has also been raised that TMEM5 serves as a xylosyltransferase using the RboP-RboP structure as an acceptor site to initiate the first step of Xyl-GlcA repeat sequence synthesis (10). However, this rather unique glycosylation structural motif has yet to be detected intact on αDG expressed in other cells, including the widely used HEK293T cells or indeed any human cell type, owing to the aforementioned technical difficulties. It is not known whether there is structural variation associated with core M3 modifications that contributes to the tissue-specific glycosylation status of αDG (4) and impacts on its functions as an extracellular matrix receptor in the brain, heart, skeletal muscle, and kidney, or correlates with its reduced expression in human breast and colon cancers in relation to tumor progression (15).
By adopting similar truncation and tryptic site engineering of recombinant αDG, we demonstrate a highly sensitive analytical workflow from in-gel digestion to direct nano-LC-MS2/MS3 analysis without additional chemo-enzymatic treatments, for unambiguous identification of the target glycopeptides carrying a further modified phosphorylated core M3. Diagnostic fragment ions were afforded by complementary modes of MS2 in positive ion mode, which can be programmed for target MS3 and/or used for rapid filtering of large spectral data sets to allow meaningful manual data interpretation in anticipation of novel substituents. We provide the requisite MS evidence for similar occurrence of tandem RboP-Xyl-GlcA substituent and its incompletely, extended forms on the phosphorylated core M3 in HEK293T cells. Moreover, we performed CRISPR/Cas9 genome editing to create a panel of mutated HCT116 colon cancer cells lacking ISPD, FKTN, FKRP, or TMEM5 and mapped the glycosylation variants of recombinant αDG expressed by these cells to address the in vivo functions of these causative gene products.
EXPERIMENTAL PROCEDURES
Antibodies
The following antibodies were used in this study: anti-Flag mAb (Sigma-Aldrich, St. Louis, MO); anti-myc (Wako, Osaka, Japan); anti-FKTN (Proteintech, Chicago, IL); IIH6 mAb (Millipore, Billerica, MA); anti-β-DG mAb (Novocastra, Newcastle, UK); horseradish peroxidase (HRP)-conjugated anti-human IgG-Fc goat Ab (Invitrogen, Carlsbad, CA); HRP-conjugated anti-mouse IgM mAb (Thermo Fisher Scientific, Waltham, MA); and HRP-conjugated anti-mouse IgG mAb (Invitrogen).
cDNA Construction
For the expression of Fc-fused αDG recombinant proteins, amino acid substitutions and deletion mutants of αDG (αDG373(T322R)) (Fig. 1) were made by standard PCR and genetic engineering techniques using the expression plasmids as constructed previously (7, 16). For Flag-tagged expression vectors of human FKTN, FKRP, TMEM5, and ISPD, the encoding sequence of individual open reading frames was amplified by PCR and cloned into pCMV14–3×FLAG vector (Sigma). For the human FKTN-myc expression vector, the FKTN coding sequence amplified by PCR was cloned into pSecTag2 (Invitrogen).
Generation of Knockout Cells Using CRISPR/Cas9 Genome Editing
The construction of vector (pKO) for TMEM5-KO cells was performed basically in accordance with previous reports (17, 18). The 1.5-kb fragment of the human TMEM5 gene used for the 5′ arm was amplified by PCR from HCT116 cell genomic DNA using the primers 5′-ggggtacCAAATTATGCAGTCATTTGC-3′ and 5′-ctagctagcGATAAGAAACGAGCAGAGCC-3′, and then subcloned between the KpnI and NheI sites to create the pKO 5′ arm (hTMEM5). The 1.5-kb fragment of the TMEM5 gene used for the 3′ arm was amplified similarly using the primers 5′-ataagaatgcggccGCCCTGTACTGCCTATTCTC-3′ and 5′-ataagaatgcggccgcGTTGACATATGCATTGCAATC-3′, and then subcloned into the NotI site of the pKO 5′ arm (hTMEM5) to create pKO-hTMEM5. The target DNA sequence of the guide RNA (gRNA) was as follows: GCCCGAAGAAGACGTGGTAGG, which was inserted into the pX330 vector (Addgene) to create pX330-guide-TMEM5. To generate the KO cells of TMEM5, pX330-guide-TMEM5 and pKO-TMEM5 were transiently transfected into HCT116 cells in a six-well plate by Lipofectamine 2000 (Invitrogen). At 96 h post-transfection, cells were incubated in puromycin (0.5 μg/ml). Approximately 40 colonies were obtained 20 days later. Homologous recombination in HCT116 cells was confirmed by genomic PCR and sequencing.
For the preparation of ISPD-, FKTN-, or FKRP-KO cells, the gRNAs flanking the target enhancer regions were designed by ToolGen (Seoul, South Korea) dRGEN synthesis services. The target DNA sequences of the gRNAs used in this study were as follows: for ISPD deletion, ATTGAAAATTGACCTGTGGCGGG; for FKTN deletion, GAGTAGAATCAATAAGAACGTGG; and for FKRP deletion, CATGCGGCTCACCCGCTGCCAGG. The gRNA expression plasmids (pRGEN-U6-sgRNA) were purchased from ToolGen. To generate the KO cells lacking ISPD, FKTN, or FKRP, individual gRNA plasmids and a plasmid expressing Cas9-GFP (Addgene Cambridge, MA,, catalog #44719) were transiently transfected into HCT116 cells in a six-well plate with a Cas9:gRNA molar ratio of 1:5 using Polyethylenimine Max (Polysciences, Inc., Warrington, PA), in accordance with a previous report (19). At 48 h post-transfection, cells with high green fluorescent protein (GFP) expression were identified using fluorescence-activated cell sorting with the Aria II cell sorter (BD Biosciences). Sorted cells were plated into individual wells of a 96-well plate and then re-plated as single cells in 6-cm dishes. The deletions in the clones were confirmed by sequencing.
Expression of Fc-fused αDG Recombinant Proteins
HEK293T as well as HCT116 and its mutated cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) in 5% CO2 at 37 °C. For the expression of αDG373(T322R)-Fc, cells were grown overnight and transfected using Polyethylenimine Max (Polysciences, Inc.), in accordance with a previous report (19). Protein expression was performed in DMEM supplemented with 10% FBS treated with a Protein G affinity column (GE Healthcare) to remove immunoglobulin G. The secreted Fc-fused αDG fragments in culture medium were purified using a Protein G affinity column.
Western Blot
Western blots were performed as described previously (16, 20).
Interaction Analyses by Pull-down Assay
Immunoprecipitation was performed in accordance with procedures described previously (17). At 24 h after transfection, cells were lysed in buffer A (50 mm Tris/Cl, pH 8.0, containing 1% Nonidet P-40, 150 mm NaCl, protease inhibitor mixture (Nacalai Tesque, Kyoto, Japan), 20 μm MG132 (Peptide Institute, Osaka, Japan), and 2 μm Z-VAD-fmk (Promega, Madison, WI)). Anti-Flag, anti-myc, and anti-FKTN antibodies and protein G-Sepharose beads (GE Healthcare) were used for the precipitation of FKRP-flag, TMEM5-flag, FKTN-myc and TMEM5-TAP. Beads were washed with high salt buffer (50 mm Tris/Cl, pH 8.0, containing 1% Nonidet P-40 and 150 mm NaCl) twice, washed with PBS, and then heated at 65 °C in Laemmli's sample buffer. Immunoprecipitates were subjected to SDS-PAGE and immunoblotting.
Identification of O-glycopeptides
The gel band containing αDG was excised and subjected to in-gel digestion by sequential steps of reduction with 10 mm dithiothreitol at 37 °C for 1 h, alkylation with 50 mm iodoacetamide in 25 mm ammonium bicarbonate buffer for 1 h in the dark at room temperature, destaining with 50% acetonitrile in 25 mm ammonium bicarbonate buffer, and then overnight digestion with sequencing-grade trypsin (Promega) at 37 °C. The digested products were sequentially extracted with distilled water, 1% formic acid, and 50% acetonitrile/1% formic acid, dried down, and then redissolved in 0.1% formic acid for further cleaned up by ZipTip C18 (Millipore) before analysis. The peptide mixtures were analyzed by nanospray LC-MS/MS on an Orbitrap Fusion Tribrid (Thermo Scientific) coupled to an UltiMate 3000 RSLCnano System (Dionex, Sunnyvale, CA). Peptide mixtures were loaded onto an Acclaim PepMap RSLC 25 cm × 75 μm i.d. column (Dionex) and separated at a flow rate of 500 nL/min using a gradient of 5% to 35% solvent B (100% acetonitrile with 0.1% formic acid) in 60 min. Solvent A was 0.1% formic acid in water. The parameters used for MS and MS/MS data acquisition under the HCD product ion trigger CID mode were: top speed mode with 3-s cycle time; FTMS: scan range (m/z) = 550–2000; resolution = 120 K; AGC target = 2 × 105; maximum injection time = 60 ms; monoisotopic precursor selection on; including charge state 2–6; dynamic exclusion after two times within 10 s and then exclusion for 40 s with 10-ppm tolerance. FTMSn (HCD): isolation mode = quadrupole; isolation window = 1.6; collision energy 28% with stepped collision energy 5%; resolution = 30 K; AGC target = 1 × 105; maximum injection time = 75 ms; HCD production ions m/z 204.0867 or 366.1396 (z = 1) within top 20 product ions were used to trigger CID; ITMSn (CID): isolation mode = quadrupole; isolation window = 1.6; collision energy = 30%; AGC target = 1 × 104; ion trap scan rate = rapid. HCD and CID MS2 data sets were filtered for candidate glycopeptide spectra based on the presence of MS2 ions corresponding to the expected tryptic peptide core containing the target O-glycosylation sites and/or peptide fragment ions, and then manually interpreted and assigned as described in the Results. Further rounds of searching through the acquired data set were based on thus identified diagnostic RboP/GroP-HexNAc+ oxonium ions. For MS3 analysis, HCD MS2 ions at m/z 358.0895 and 418.1105 were targeted for CID MS3 using the inclusion list feature. The parameters used for FTMS3 (CID) were: isolation mode = iontrap; MS isolation window = 1.6; MS2 isolation window = 3.0; scan range mode: auto normal; collision energy = 30%; detector type: orbitrap; resolution = 30 K; AGC target = 1 × 105; maximum injection time = 120 ms.
RESULTS AND DISCUSSION
The Identification of Additional Modifications on Phosphorylated O-glycans of αDG Expressed in HEK293T Cells
To facilitate direct detection and sequencing of the αDG glycopeptides carrying the target O-glycans, we have opted for an analytical strategy that would preserve as much of the native O-glycosylation and other modifications as possible without introducing any chemical cleavages or glycosidase digestions prior to the MS analysis. Our choice as minimum manipulation was truncation of αDG at R373 and fusion with Fc for secretion and ease of purification (Fig. 1 and supplemental Fig. S1). The resulting Fc-tagged and purified proteins appeared as two major bands on SDS-PAGE at positions corresponding to ∼35 kDa and just above 50 kDa (Fig. 1), neither of which was stained by IIH6, an anti-αDG antibody recognizing laminin-binding glycans, as would be expected from the apparent size. Following in-gel tryptic digestion and recovery of peptides/glycopeptides from the gel, we initially searched for the target glycopeptide Q313-R337, but were unsuccessful. Hence, we decided to introduce T322R mutation to shorten the resulting tryptic glycopeptide further and to confine the O-glycosylation to only Thr317 and Thr319, which have been implicated as sites carrying the functional laminin-binding glycans based on a mutagenesis study (5). Although the T322R substitution may affect the overall O-glycosylation occupancy and heterogeneity, previous study has shown that functional laminin-binding glycans could still be formed at Thr317 and Thr319 of the T322R mutant expressed in mouse NIH 3T3 cells (17). We then anticipated finding the resulting Q313-R322 and its companion A323-R337 tryptic glycopeptides by searching among the MS2 spectra for the presence of respective peptide core ions. However, we succeeded in identifying only the A323-R337 glycopeptides, all of which were O-glycosylated to various extents, including glycoforms that carried an additional phosphate, which could be assigned as phosphorylated core M3 (supplemental Fig. S2). No evidence could be found for the presence of any additional glycan modifications in this tryptic glycopeptide that contains T328, T329, and S336.
We suspected that the failure to detect the target Q313-R337 peptide core ion may have been caused by an unknown modification of the peptide core itself, particularly at the nascent N terminus generated by an endogenous cleavage. Our search strategy was modified accordingly and extended to use several of the expected peptide y ions, which led to the identification of a peptide core with a mass discrepancy of minus 17 u localized to the N terminus, likely because of pyroglutamylation of the terminal glutamine residue. In fact, a total of over 8000 MS2 spectra containing the modified peptide core ion at m/z 551.80382+ were filtered out using this strategy, which comprised 673 redundant spectra that could be collectively assigned to 65 unique precursor glycoforms (supplemental Table S1). From the compiled data, it was clear that there were glycoforms carrying only the core M1/M2 without an additional phosphate, some of which might correspond instead to mucin-type O-GalNAc cores. These nonphosphorylated core-type structures were difficult to distinguish unambiguously by current HCD/CID MS2 analyses alone and were not pursued further. We focused our efforts instead on glycoforms that contain at least a phosphorylated core M3 structure. Among the major structures satisfying this criterion were the glycoforms carrying two phosphorylated core M3 moieties, with and without additional modifications (supplemental Table S1, Fig. 2).
Fig. 2.
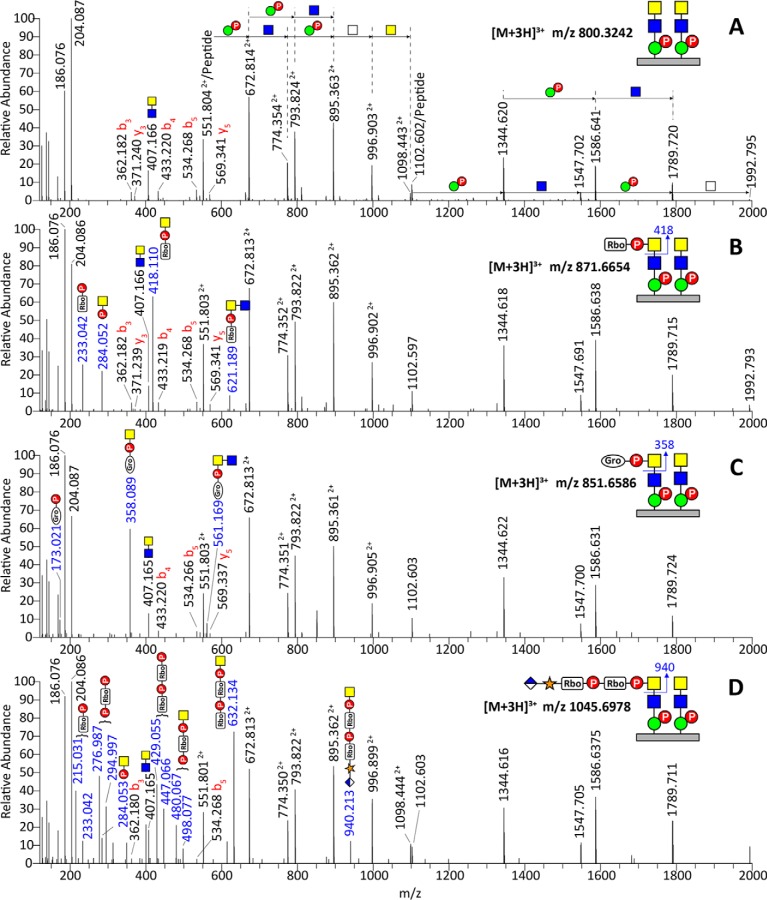
HCD MS2 spectra of tryptic αDG373(T322R)-Fc glycopeptides, 313pyrQIHATPTPVR322, carrying two phosphorylated core M3 moieties (A), with additional RboP (B), GroP (C), or HexA-Pent-RboP-RboP (D) substituents. Additional representative spectra for glycoforms carrying only one phosphorylated core M3, two phosphorylated core M3 moieties with two RboP, and two phosphorylated core M3 moieties with one RboP + one GroP are shown in supplemental Fig. S3. The pentitol phosphate substituent was assumed and annotated as ribitol phosphate based on recent reports (11), whereas the analogous three-carbon substituent was tentatively assigned as glycerol phosphate (GroP) based on accurate mass increment alone, without further analysis to define its stereochemistry. Similarly, the identities of GlcA, Xyl, and GalNAc versus GlcNAc and Man were inferred from the literature and are represented by symbols following the SNFG (Symbol Nomenclature for Glycans) system (26), the details of which can be found at NCBI [http://www.ncbi.nlm.nih.gov/books/NBK310273/]. Their linkages and anomericity were not further established in this work. In (D), the RboP, P-RboP, RboP-RboP, and P-RboP-HexNAc+ ions as annotated were each accompanied by further loss of a H2O moiety, giving an additional ion at 18 u lower.
All of the spectra of this series contain the same sets of ions corresponding to (1) HexNAc+ and HexNAc2+ at m/z 204.0866 and 407.1660, respectively; (2) singly and doubly charged peptide core at m/z 1102.6003 and 551.8038, respectively; (3) peptide b3, b4, b5, y3, and y5 ions confirming the core sequence; and (4) the peptide core extended by a string of phosphorylated Hex and HexNAc that can be mapped to two phosphorylated core M3 moieties attached at two separate sites (annotated in Fig. 2A). These common sets of ions indicate that the MS2 spectra filtered out for further manual assignment here all derived from the same glycopeptide core, differing only in additional modifications. Importantly, the prominent ion at m/z 418.110, representing a mass increment of 214.024 Da from the HexNAc+ oxonium ion at m/z 204.086, identifies the presence of a recently reported ribitol phosphate (RboP) substituent on HexNAc (Fig. 2B). This is further supported by ions at m/z 233.042 (RboP), 284.052 (phospho-HexNAc+), and 621.189 (RboP-HexNAc2+). More intriguingly, based on similar sets of ions, we identified another analogous substituent represented by a mass increment of 154.002 Da from HexNAc+, which can be tentatively assigned as glycerol phosphate (GroP) and supported by ions at m/z 173.021 (GroP), 358.089 (GroP-HexNAc+), and 561.169 (GroP-HexNAc2+) (Fig. 2C). By virtue of these diagnostic ions, the GroP-HexNAc+ and RboP-HexNAc+ substituents were found on several but not all glycoforms of αDG373(T322R)-Fc expressed in HEK293T cells (supplemental Table S1). Notably, a tandem RboP-RboP unit was only found in the context of a fully extended HexA-Pent-RboP-RboP-HexNAc substituent, identified by the corresponding oxonium ion at m/z 940.213, further supported by ions at m/z 632.134 (RboP-RboP-HexNAc+), 498.077 (P-RboP-HexNAc+), 447.066 (RboP-RboP), 294.997 (P-RboP), 284.053 (P-HexNAc+), and 233.042 (RboP) (Fig. 2D). This critical oxonium ion at m/z 940.2 was more prominent in the corresponding trap CID MS2 spectrum (Fig. 3D).
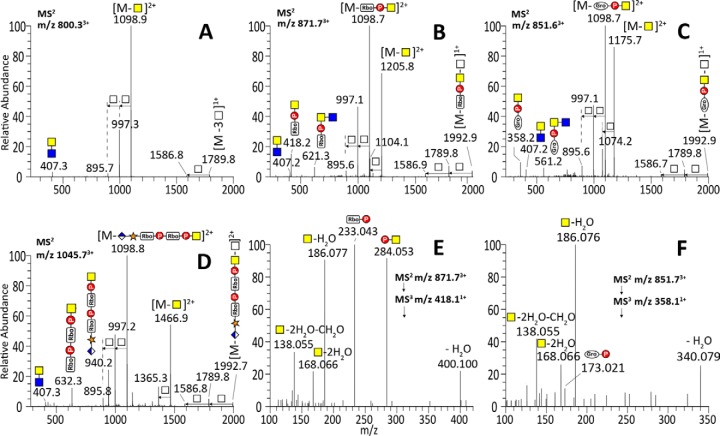
Fig. 3.
Ion trap CID MS2 spectra of tryptic αDG373(T322R)-Fc glycopeptides, 313pyrQIHATPTPVR322, carrying two phosphorylated core M3 moieties (A), with additional RboP (B), GroP (C), or HexA-Pent-RboP-RboP (D) substituents, along with the characteristic MS3 spectra of the RboP-HexNAc+ (E) and GroP-HexNAc+ (F) MS2 ions. Facile losses of terminal ±RboP/GroP-HexNAc moieties yielded the common peptide core ion retaining two phospho-Hex (m/z 1586.81+) and those with an additional one (m/z 895.72+, 1789.81+), two (m/z 997.32+, 1992.91+), or three HexNAc (m/z 1098.92+). The trap CID MS2 spectra presented here were acquired from the same 3+ precursors giving the HCD MS2 spectra shown in Fig. 2. The oxonium ions detected are similar to those afforded by HCD, but measured at lower accuracy, with the smaller ones being lost because of the low mass cut-off of trap CID MS2. The MS3 spectra contained secondary fragment ions at m/z 138.055, 168.066, and 186.076 typically associated with HexNAc+, but not m/z 204.087 itself. Instead, P-HexNAc+ at m/z 284.053 was detected.
The complementary trap CID MS2 spectra were in general characterized by prominent losses of terminal substituted and unsubstituted HexNAc, thereby producing the common doubly and singly charged peptide core ions retaining two phospho-Hex and 1–3 HexNAc (as annotated in Fig. 3A–3D). In particular, the common base peak ion was derived from the loss of terminal ±R-HexNAc, which corroborated the identification of terminal substituents (r = RboP, GroP, or HexA-Xyl-RboP-RboP) by the oxonium ions. Glycoforms carrying two phosphorylated core M3 moieties but only one terminal HexNAc of which was further substituted would thus afford the common doubly charged ion at m/z 1098.8 after losing ±R-HexNAc, accompanied by the one derived from losing an unsubstituted HexNAc. Such characteristics suggest that the R substituents are all carried on the terminal and not the penultimate HexNAc. We further subjected the prominent RboP-HexNAc+ and GroP-HexNAc+ ions at m/z 418.1 and 358.1, respectively, to MS3 (Fig. 3E and 3F), either by targeted MS2/MS3 or by product ion-dependent MS3 in additional LC-MS/MS runs. As expected, the RboP-HexNAc+ afforded MS3 ions at m/z 233.043 (RboP) and 284.053 (P-HexNAc+), which were also present in the MS2 spectrum (Fig. 2B and 2D). Likewise, GroP-HexNAc+ yielded MS3 ions at m/z 173.021 (GroP) and 284.053, albeit very weakly, consistent with the overall lower intensity of the precursor glycopeptide carrying this novel substituent. Although not an accurate quantitative profiling given the lack of knowledge on the response factors for each of the glycoforms carrying different modifications on 1–2 phosphorylated core M3, an extracted ion chromatogram overlay of select glycoforms indicates that glycoform with two phosphorylated core M3 moieties and that further substituted by a single RboP- were by far the more abundant compared with those substituted with GroP- or fully extended HexA-Xyl-RboP-RboP- (Fig. 4). Moreover, the glycoforms with both Thr317 and Thr319 occupied were also more prominent than those with only single-site occupancy.
Fig. 4.
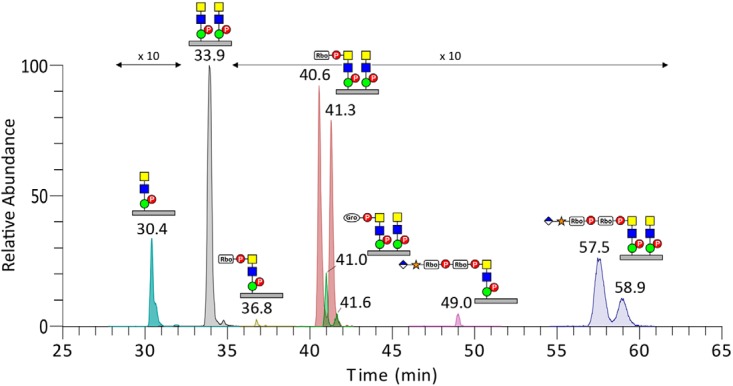
Overlays of the extracted ion chromatograms of select 313pyrQIHATPTPVR322 glycoforms carrying phosphorylated core M3 with and without additional substituents. The glycoforms with two phosphorylated core M3 moieties but carrying a single RboP, GroP, or GlcA-Xyl-RboP-RboP substituent were each clearly eluted as two major peaks, likely because of the substituent being carried on either of the two sites. For simplicity, not all of the glycoforms identified from the tryptic digest of αDG373(T322R)-Fc expressed in HEK293T cells (see supplemental Table S1) are plotted here.
Establishment of KO Cells of Individual Gene Products Causative of Dystroglycanopathy
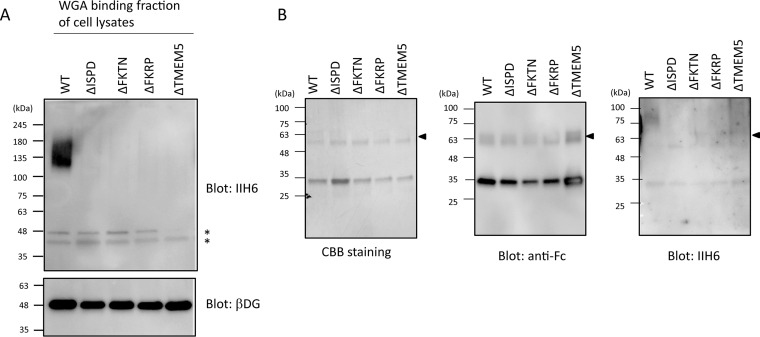
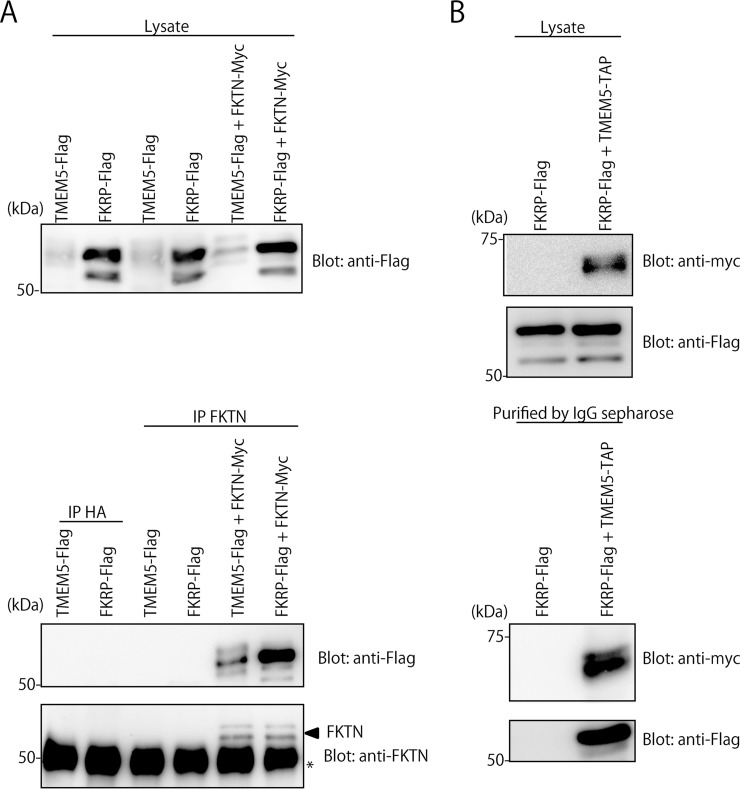
ISPD-, TMEM5-, FKTN-, or FKRP-deficient cells were established by using the CRISPR/Cas9 genome editing of HCT116 cells, a colon cancer cell line known to express the laminin-binding glycans on αDG (15). As expected, individual KO cells exhibited no laminin-binding glycans (Fig. 5A), whereas the target product expression rescued the defect (supplemental Fig. S4). It has been reported that ISPD is a cytosolic protein for the synthesis of CDP-Rbo (11, 13), whereas FKRP, FKTN, and TMEM5 are located at the Golgi apparatus (21–24). These three Golgi enzymes were not functionally compensating one another (supplemental Fig. S4). Because FKRP exists as a homodimer and in multimeric protein complexes (21), we conducted analyses of the interactions among these causative gene products by a pull-down assay (Fig. 6). FKRP-flag, TMEM5-flag, and/or FKTN-myc were transiently coexpressed in HEK293T cells. The immunoprecipitants with anti-FKTN antibody contained FKRP-flag and TMEM5-flag, individually (Fig. 6A). The lysate of HEK293 expressing FKRP-flag and TMEM5-TAP were pulled down with IgG-Sepharose. The pulled-down proteins were found to contain FKRP-flag as well as TMEM5-TAP (Fig. 6B). The results indicate that FKRP, FKTN, and TMEM5 form a hetero-oligomeric complex, suggesting that their cooperative interplay is indispensable for the formation of the laminin-binding glycans. To address the functional roles of these causative gene products in cells, glycopeptides derived from in-gel digestion of αDG373(T322R)-Fc expressed by the individual KO cells (Fig. 5B) were subjected to similar LC-MS/MS analyses.
Fig. 5.
Establishment of KO cells of the causative gene products and expression of Fc-fused αDG mutant. A, The cells with KO of ISPD, FKTN, FKRP, or TMEM5 displayed no detectable laminin-binding glycans. The lysates of individually mutated cells were subjected to immunoblot analysis using IIH6 and anti-β-DG antibodies. The bands indicated by asterisk were nonspecifically detected by the secondary anti-mouse IgM antibody. B, αDG373(T322R)-Fc, which was expressed by WT HCT116 and its mutated cells and then purified by protein G Sepharose. The purified αDG373(T322R)-Fc was subjected to SDS-PAGE followed by CBB staining and Western blotting with anti-Fc and -IIH6 antibodies. Arrow indicates bands corresponding to αDG373(T322R)-Fc.
Fig. 6.
Analyses of interactions among TMEM5, FKTN, and FKRP by pull-down assay. A, Co-immunoprecipitation of FKRP and FKTN as well TMEM5 and FKTN in HEK293T cells. Lysates of HEK293T cells transiently expressing FKRP-flag, TMEM5-flag, and/or FKTN-myc were immunoprecipitated (IP) with anti-HA (IP: HA) and anti-FKTN (IP: FKTN) antibodies, and subjected to SDS-PAGE and Western blotting (Blot) with anti-Flag or anti-FKTN antibodies. To examine the level of TMEM5-flag and FKRP-flag, the cell lysates were directly subjected to SDS-PAGE and Western blotting with anti-Flag or anti-FKTN antibodies. B, Co-immunoprecipitation of TMEM5 and FKRP in HEK293T cells. Lysates of HEK293T cells transiently expressing FKRP-flag and TMEM5-TAP were pulled down with IgG-Sepharose, and subjected to SDS-PAGE and Western blots (Blot) with anti-myc or anti-flag antibodies. To examine the levels of TMEM5-TAP and FKRP-flag, the cell lysates were directly subjected to SDS-PAGE and Western blots with anti-myc or anti-Flag antibodies. The tandem affinity purification (TAP) tag consists of two immunoglobulin (IgG)-binding domains of Staphylococcus aureus, protein A, tandem myc epitope and a calmodulin-binding peptide, separated by a cleavage site for the tobacco etch virus (TEV) protease.
The Additional Modifications on Phosphorylated O-glycans of αDG Expressed by HCT116 Cells Lacking Individual Causative Gene Products of Dystroglycanopathy
Taking advantage of the established fragmentation pattern, particularly the diagnostic oxonium ions, we were able to rapidly filter out relevant glycopeptide spectra for manual assignment and confirmed the similar presence of RboP/GroP modifications on phosphorylated core M3 attached to Thr317/Thr319 of αDG373(T322R)-Fc expressed in HCT116 cells (supplemental Table S1). However, unlike HEK293T cells, fully extended HexA-Pent-RboP-RboP-HexNAc could not be found among the αDG glycoforms identified for WT HCT116 cells or any of its derived mutants. This probably reflects the observation that HCT116 cells express a higher level of endogenous laminin-binding glycans relative to HEK293T cells and thus the HexA-Pent-primed core would be readily extended, albeit not fully on the truncated αDG373(T322R)-Fc constructs, rendering their detection difficult. Moreover, in HCT116 cells, the glycoforms with single RboP, two RboP, and one RboP + one GroP modifications were mostly detected in the FKRP mutant, or at much higher abundance in the FKRP mutant relative to the WT (Fig. 7), consistent with the lack of further extension and consequent accumulation because of impaired FKRP activity. It should also be pointed out that, in cases where two RboP could be found without HexA-Pent, they were not found as a tandem repeat unit but rather as a single RboP on each phosphorylated core M3 (supplemental Fig. S3B). Likewise, no supporting evidence could be found for a tandem RboP-GroP in either direction (supplemental Fig. S3C).
Fig. 7.

Graphical representation of the identified phosphorylated core M3-carrying glycoforms of 313pyrQIHATPTPVR322 derived from αDG373(T322R)-Fc expressed in HCT116 and its various KO mutants. Each of the detected glycopeptides was quantified based on XIC peak intensity, normalized to the protein abundance of αDG373(T322R)-Fc recovered from each mutant, and graphically represented here as “bands” with thickness corresponding to their calculated relative abundance (see supplemental Table S1 for details and actual data). The red bands indicate the glycoforms with compositions consistent with carrying 1–2 phosphorylated core M3, with and without additional glycosyl residues. Glycoforms of the same composition with additional RboP and GroP substituents are represented by green and blue bands, respectively. Only one glycoform with both GroP and RboP was detected and is represented here as a green band. The glycoforms with RboP substituents (green bands) are notably identified only in samples from the WT and the FKRP mutant. FKTN-KO apparently led to an inability to produce any glycoforms with either RboP or GroP substituent.
The absence of any RboP unit in ΔISPD and ΔFKTN mutants is expected because the former is required to synthesize the CDP-ribitol, whereas the latter is the transferase suggested to be responsible for the addition of RboP from CDP-ribitol onto phosphorylated core M3 (11–13). More puzzling is the near absence of this substituent also in the ΔTMEM5 mutant. TMEM5 was shown to be able to transfer Xyl from UDP-Xyl to CDP-ribitol and thus the true donor substrate of FKRP may be the presynthesized CDP-ribitol-Xyl instead of CDP-ribitol. It follows that both TMEM5 and FKTN may use and compete for the same CDP-ribitol pool. Proper in vivo transfer of RboP from CDP-ribitol to phosphorylated core M3 by FKTN would thus require its functional and coordinated interactions with TMEM5 facilitated by their physical association into an enzyme complex, as has been demonstrated (Fig. 6). Our data showing convergent glycosylation defects between ΔFKTN and ΔTMEM5 mutants are supportive of this model in which impairment of either would hinder the activity of the other. A different scenario may apply to GroP modification as we observed that this novel modification was prevented in ΔFKTN but not ΔTMEM5 mutant. This suggests that FKTN can equally transfer a GroP unit in place of RboP to the phosphorylated core M3 acceptor, independent of TMEM5.
Indeed, RboP and GroP modifications have been found in the structures of teichoic acids from Gram-positive bacteria cell walls (25). A series of enzymes are involved in the formation of RboP-RboP, GroP-GroP and GroP-RboP structures using CDP-ribitol and CDP-glycerol as donor substrates in the biosynthesis of teichoic acids. Although mammals do not synthesize teichoic acids, the evolutionary relationship between the biosynthesis pathways of teichoic acids and laminin-binding glycans is of interest. It is currently unclear whether GroP may substitute for RboP in linking the laminin-binding glycans to phosphorylated core M3 under unspecified physiological stages or in specific cell types, as we have not been able to detect such a functional structural unit, nor any tandem GroP or GroP-RboP modification.
CONCLUSION
In this study, using our nano-LC-MS/MS strategy that allows for the parallel acquisition of both HCD and trap CID MS2/MS3 data, we directly identified the previously reported HexA-Pent-RboP-RboP- and RboP- substituents, along with another unique modification tentatively assigned as GroP, on phosphorylated core M3 attached to the truncated αDG expressed in HEK293T cells. Moreover, many other glycoforms of Thr317/Thr319 carrying a combination of single RboP, GroP, or none of these modifications, on the two phosphorylated core M3 structures could be detected but, intriguingly, not tandem RboP modifications lacking further HexA-Xyl extension. The facile formation of ±X-RboP-HexNAc+ and ±X-GroP-HexNAc+ oxonium ions via HCD and their corroborative losses from precursors in trap CID can both be detected at high mass accuracy in the Orbitrap and be used as diagnostic ions to facilitate future rapid screening and identification of these unusual glycosyl modifications in a wide range of sample sources. The modifications identified in a panel of mutated HCT116 cells lacking ISPD, FKTN, FKRP, or TMEM5 not only confirmed the enzymatic activities of these causative gene products but also provided new insights into their functional roles during the formation of the laminin-binding glycans on αDG.
Supplementary Material
Acknowledgments
We thank the Laboratory for Animal Resources and Genetic Engineering, Center for Developmental Biology, Institute of Physical and Chemical Research, Kobe, for providing the DT-A-pA/loxP/PGK-NeopA/loxP vector. We also thank Kiyomi Senda (Nagoya City University) for her help in the preparation of the recombinant proteins. The mass spectrometry data were acquired at the Academia Sinica Common Mass Spectrometry Facilities for Proteomics and Protein Analysis located at the Institute of Biological Chemistry.
Footnotes
Author contributions: H.Y., K. Khoo, and K. Kato designed research; H.Y., C.K., T.O., and S.N. performed research; H.Y., C.K., T.O., S.N., and K. Khoo analyzed data; H.Y., K. Khoo, and K. Kato wrote the paper.
* This study was supported by Academia Sinica (to K.H.K.) and Grant-in-Aid for Scientific Research (C) (15K07935 to H.Y.) and Scientific Research on Innovative Areas (26110716 to H.Y. and 25102008 to K.K.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
 This article contains supplemental material.
This article contains supplemental material.
The authors declare no conflict of interest.
1 The abbreviations used are:
- αDG
- α-dystroglycan
- B3GALNT
- β-1,3-N-acetylgalactosaminyltransferase
- CID
- collision-induced dissociation
- core M1
- Gal-β4-GlcNAc-β2-Man
- core M2
- Gal-β4-GlcNAc-β2-(Gal-β4-GlcNAc-β6-)Man
- core M3
- GalNAc-β3-GlcNAc-β4-Man
- CDP-Rbo
- cytidine diphosphate ribitol
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- FKTN
- fukutin
- FKRP
- fukutin-related protein
- GlcA
- glucuronic acid
- Gro
- glycerol
- GroP
- glycerol phosphate
- HCD
- higher-energy collisional dissociation
- HRP
- horseradish peroxidase
- Hex
- hexose
- HexA
- hexuronic acid
- ISPD
- isoprenoid synthase domain containing
- KO
- knockout
- LC
- liquid chromatography
- HexNAc
- N-acetyl hexosamine
- POMT
- O-mannosyltransferase
- POMK
- O-mannose kinase
- Pent
- pentose
- PCR
- polymerase chain reaction
- POMGNT
- protein O-mannose β-1,4-N-acetylglucosaminyltransferase
- Rbo
- ribitol
- RboP
- ribitol phosphate
- MS/MS or MS2
- tandem mass spectrometry
- TAP
- tandem affinity purification
- TMEM5
- transmembrane protein 5
- Xyl
- xylose.
REFERENCES
- 1. Chiba A., Matsumura K., Yamada H., Inazu T., Shimizu T., Kusunoki S., Kanazawa I., Kobata A., and Endo T. (1997) Structures of sialylated O-linked oligosaccharides of bovine peripheral nerve a-dystroglycan. The role of a novel O-mannosyl-type oligosaccharide in the binding of a-dystroglycan with laminin. J. Biol. Chem. 272, 2156–2162 [DOI] [PubMed] [Google Scholar]
- 2. Holt K. H., Crosbie R. H., Venzke D. P., and Campbell K. P. (2000) Biosynthesis of dystroglycan: processing of a precursor propeptide. FEBS Lett. 468, 79–83 [DOI] [PubMed] [Google Scholar]
- 3. Wells L. (2013) The O-mannosylation pathway: glycosyltransferases and proteins implicated in congenital muscular dystrophy. J. Biol. Chem. 288, 6930–6935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yoshida-Moriguchi T., and Campbell K. P. (2015) Matriglycan: a novel polysaccharide that links dystroglycan to the basement membrane. Glycobiology 25, 702–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hara Y., Kanagawa M., Kunz S., Yoshida-Moriguchi T., Satz J. S., Kobayashi Y. M., Zhu Z., Burden S. J., Oldstone M. B., and Campbell K. P. (2011) Like-acetylglucosaminyltransferase (LARGE)-dependent modification of dystroglycan at Thr-317/319 is required for laminin binding and arenavirus infection. Proc. Natl. Acad. Sci. U.S.A. 108, 17426–17431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yoshida-Moriguchi T., Willer T., Anderson M. E., Venzke D., Whyte T., Muntoni F., Lee H., Nelson S. F., Yu L., and Campbell K. P. (2013) SGK196 Is a glycosylation-specific O-mannose kinase required for dystroglycan function. Science 341, 896–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yagi H., Nakagawa N., Saito T., Kiyonari H., Abe T., Toda T., Wu S. W., Khoo K. H., Oka S., and Kato K. (2013) AGO61-dependent GlcNAc modification primes the formation of functional glycans on a-dystroglycan. Sci. Rep. 3, 3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stevens E., Carss K. J., Cirak S., Foley A. R., Torelli S., Willer T., Tambunan D. E., Yau S., Brodd L., Sewry C. A., Feng L., Haliloglu G., Orhan D., Dobyns W. B., Enns G. M., Manning M., Krause A., Salih M. A., Walsh C. A., Hurles M., Campbell K. P., Manzini M. C., Stemple D., Lin Y. Y., and Muntoni F. (2013) Mutations in B3GALNT2 cause congenital muscular dystrophy and hypoglycosylation of a-dystroglycan. Am. J. Hum. Genet. 92, 354–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Inamori K., Yoshida-Moriguchi T., Hara Y., Anderson M. E., Yu L., and Campbell K. P. (2012) Dystroglycan function requires xylosyl- and glucuronyltransferase activities of LARGE. Science 335, 93–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Praissman J. L., Willer T., Sheikh M. O., Toi A., Chitayat D., Lin Y. Y., Lee H., Stalnaker S. H., Wang S., Prabhakar P. K., Nelson S. F., Stemple D. L., Moore S. A., Moremen K. W., Campbell K. P., and Wells L. (2016) The functional O-mannose glycan on a-dystroglycan contains a phospho-ribitol primed for matriglycan addition. Elife 5, e14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kanagawa M., Kobayashi K., Tajiri M., Manya H., Kuga A., Yamaguchi Y., Akasaka-Manya K., Furukawa J., Mizuno M., Kawakami H., Shinohara Y., Wada Y., Endo T., and Toda T. (2016) Identification of a post-translational modification with ribitol-phosphate and its defect in muscular dystrophy. Cell Rep. 14, 2209–2223 [DOI] [PubMed] [Google Scholar]
- 12. Gerin I., Ury B., Breloy I., Bouchet-Seraphin C., Bolsee J., Halbout M., Graff J., Vertommen D., Muccioli G. G., Seta N., Cuisset J. M., Dabaj I., Quijano-Roy S., Grahn A., Van Schaftingen E., and Bommer G. T. (2016) ISPD produces CDP-ribitol used by FKTN and FKRP to transfer ribitol phosphate onto a-dystroglycan. Nat. Commun. 7, 11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Riemersma M., Froese D. S., van Tol W., Engelke U. F., Kopec J., van Scherpenzeel M., Ashikov A., Krojer T., von Delft F., Tessari M., Buczkowska A., Swiezewska E., Jae L. T., Brummelkamp T. R., Manya H., Endo T., van Bokhoven H., Yue W. W., and Lefeber D. J. (2015) Human ISPD is a cytidyltransferase required for dystroglycan O-mannosylation. Chem. Biol. 22, 1643–1652 [DOI] [PubMed] [Google Scholar]
- 14. Kanagawa M., Saito F., Kunz S., Yoshida-Moriguchi T., Barresi R., Kobayashi Y. M., Muschler J., Dumanski J. P., Michele D. E., Oldstone M. B., and Campbell K. P. (2004) Molecular recognition by LARGE is essential for expression of functional dystroglycan. Cell 117, 953–964 [DOI] [PubMed] [Google Scholar]
- 15. Sgambato A., Migaldi M., Montanari M., Camerini A., Brancaccio A., Rossi G., Cangiano R., Losasso C., Capelli G., Trentini G. P., and Cittadini A. (2003) Dystroglycan expression is frequently reduced in human breast and colon cancers and is associated with tumor progression. Am. J. Pathol. 162, 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nakagawa N., Manya H., Toda T., Endo T., and Oka S. (2012) Human natural killer-1 sulfotransferase (HNK-1ST)-induced sulfate transfer regulates laminin-binding glycans on alpha-dystroglycan. J. Biol. Chem. 287, 30823–30832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ninagawa S., Okada T., Sumitomo Y., Horimoto S., Sugimoto T., Ishikawa T., Takeda S., Yamamoto T., Suzuki T., Kamiya Y., Kato K., and Mori K. (2015) Forcible destruction of severely misfolded mammalian glycoproteins by the non-glycoprotein ERAD pathway. J. Cell Biol. 211, 775–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fu Y., Sander J. D., Reyon D., Cascio V. M., and Joung J. K. (2014) Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32, 279–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Longo P. A., Kavran J. M., Kim M. S., and Leahy D. J. (2013) Transient mammalian cell transfection with polyethylenimine (PEI). Methods Enzymol. 529, 227–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yagi H., Yanagisawa M., Suzuki Y., Nakatani Y., Ariga T., Kato K., and Yu R. K. (2010) HNK-1 epitope-carrying tenascin-C spliced variant regulates the proliferation of mouse embryonic neural stem cells. J. Biol. Chem. 285, 37293–37301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alhamidi M., Kjeldsen Buvang E., Fagerheim T., Brox V., Lindal S., Van Ghelue M., and Nilssen O. (2011) Fukutin-related protein resides in the Golgi cisternae of skeletal muscle fibres and forms disulfide-linked homodimers via an N-terminal interaction. PLoS ONE 6, e22968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Matsumoto H., Noguchi S., Sugie K., Ogawa M., Murayama K., Hayashi Y. K., and Nishino I. (2004) Subcellular localization of fukutin and fukutin-related protein in muscle cells. J. Biochem. 135, 709–712 [DOI] [PubMed] [Google Scholar]
- 23. Esapa C. T., Benson M. A., Schroder J. E., Martin-Rendon E., Brockington M., Brown S. C., Muntoni F., Kroger S., and Blake D. J. (2002) Functional requirements for fukutin-related protein in the Golgi apparatus. Hum. Mol. Genet. 11, 3319–3331 [DOI] [PubMed] [Google Scholar]
- 24. Willer T., Inamori K., Venzke D., Harvey C., Morgensen G., Hara Y., Beltran Valero de Bernabe D., Yu L., Wright K. M., and Campbell K. P. (2014) The glucuronyltransferase B4GAT1 is required for initiation of LARGE-mediated a-dystroglycan functional glycosylation. Elife 3, e03941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qian Z., Yin Y., Zhang Y., Lu L., Li Y., and Jiang Y. (2006) Genomic characterization of ribitol teichoic acid synthesis in Staphylococcus aureus: genes, genomic organization and gene duplication. BMC Genomics 7, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Varki A., Cummings R. D., Aebi M., Packer N. H., Seeberger P. H., Esko J. D., Stanley P., Hart G., Darvill A., Kinoshita T., Prestegard J. J., Schnaar R. L., Freeze H. H., Marth J. D., Bertozzi C. R., Etzler M. E., Frank M., Vliegenthart J. F., Lutteke T., Perez S., Bolton E., Rudd P., Paulson J., Kanehisa M., Toukach P., Aoki-Kinoshita K. F., Dell A., Narimatsu H., York W., Taniguchi N., and Kornfeld S. (2015) Symbol nomenclature for graphical representations of glycans. Glycobiology 25, 1323–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.