Abstract
Candida albicans is a major fungal pathogen causing lethal infections in immunocompromised patients. C. albicans forms antifungal tolerant biofilms contributing significantly to therapeutic failure. The recently established haploid C. albicans biofilm model provides a new toolbox to uncover the mechanism governing the higher antifungal tolerance of biofilms. Here, we comprehensively examined the proteomics and antifungal susceptibility of standard diploid (SC5314 and BWP17) and stable haploid (GZY792 and GZY803) strains of C. albicans biofilms. Subsequent downstream analyses identified alkyl hydroperoxide reductase 1 (AHP1) as a critical determinant of C. albicans biofilm's tolerance of amphotericin B. At 32 μg/ml of amphotericin B, GZY803 haploid biofilms showed 0.1% of persister population as compared with 1% of the diploid biofilms. AHP1 expression was found to be lower in GZY803 biofilms, and AHP1 overexpression in GZY803 restored the percentage of persister population. Consistently, deleting AHP1 in the diploid strain BWP17 caused a similar increase in amphotericin B susceptibility. AHP1 expression was also positively correlated with the antioxidant potential. Furthermore, C. albicans ira2Δ/Δ biofilms were susceptible to amphotericin B and had a diminished antioxidant capacity. Interestingly, AHP1 overexpression in the ira2Δ/Δ strain restored the antioxidant potential and enhanced the persister population against amphotericin B, and shutting down the AHP1 expression in ira2Δ/Δ biofilms reversed the effect. In conclusion, we provide evidence that the AHP1 gene critically determines the amphotericin B tolerance of C. albicans biofilms possibly by maintaining the persisters' antioxidant capacity. This finding will open up new avenues for developing therapies targeting the persister population of C. albicans biofilms. The mass spectrometry proteomics data are available via ProteomeXchange with identifier PXD004274.
Candida albicans is a major commensal fungal species in the human microbiota, which inhabits nearly 70% of healthy individuals (1). However, C. albicans transforms into an opportunistic pathogen in compromized host populations and accounts for ∼15% of healthcare infections with associated mortality as high as 40% (2–4). Ability to form surface-attached microbial communities or biofilm formation is a prominent virulence attribute of C. albicans (5, 6). Biofilms of C. albicans often develop on medical indwelling devices, producing dormant persister populations that largely contribute to the multidrug tolerance of biofilms (6, 7). Hence, biofilm growth of C. albicans is directly linked with therapeutic failure, which causes severe consequences to the host.
The higher antifungal resistance of C. albicans biofilms is well documented (6, 8). Although various hypotheses have been proposed to explain the higher resistance in the biofilm mode of growth, the mechanism is still poorly understood (9). In the present study, we took advantage of the recently discovered haploid strains of C. albicans (10) to uncover the mechanism behind the higher persistence of the C. albicans biofilms against amphotericin B, a drug known as “gold-standard” antifungal agent (11). We previously developed an efficient molecular method to generate mutants from haploid C. albicans (12) and used a novel haploid biofilm model of C. albicans to unravel a novel biofilm regulator (13).
In this study, we comprehensively conducted comparative ploidy proteomics and antifungal susceptibility tests of diploid and haploid C. albicans biofilms using two standard diploid strains, SC5314 and BWP17 and two stable haploid strains, GZY792 and GZY803. Subsequently, downstream analyses led us to identify AHP1 as a critical determinant of amphotericin B tolerance in the surviving cell population of C. albicans biofilms. Using targeted genetic approaches, we obtained new mechanistic insights about the role of AHP1 in determining the resistance of C. albicans biofilms against amphotericin B.
EXPERIMENT PROCEDURES
Fungal Strains and Culture Conditions
Two diploids, SC5314 and BWP17, and two stable haploids, GZY792 and GZY803, were used as standard C. albicans diploid and haploid strains. The constructions of yeast strains and plasmids used in this study are described in supplemental Table S1. Fungal cells were cultured at 30 °C in YPD (2% yeast extract, 1% peptone, and 2% glucose), or GMM (glucose minimal medium, 6.79 g/l yeast nitrogen base without amino acids, and 2% glucose) supplemented with appropriate amino acids (uridine 80 μg/ml, arginine 40 μg/ml, and histidine 40 μg/ml) if necessary. Solid culture plates were prepared by the addition of 2% agar.
Antifungal Agents
Amphotericin B, Caspofungin, Fluconazole, Ketoconazole, and Voriconazole (Sigma-Aldrich, St. Louis, MO) were used in this study.
Biofilm Formation
Biofilm development methodology was adapted from our well-established protocol (13, 14). For each strain, single colonies were inoculated in liquid GMM supplemented with required amino acids and incubated at 30 °C overnight. The cultures were then used to prepare cell suspension of MacFarland of 0.375 (equivalent to 107 cell/ml). An aliquot of 100 μl of the suspension was put in one well in a sterile flat bottom 96-well plate (Greiner bio-one). The plates were incubated at 37 °C for 1.5 h with the shaking speed of 80 rpm. After this adhesion phase, non-adhered cells were removed and 200 μl of fresh GMM were added to the adhered cells. The plates were incubated at 37 °C till 72 h; media were changed every 24 h to remove nonadhered cells.
Quantification of Biofilms by XTT and Colony Forming Unit Methods
Biofilms were quantified using 2, 3-bis (2-methoxy-4-nitro-5–16 sulfophenyl)-5-[(phenylamino) carbonyl]-2H-tetrazolium hydroxide (XTT) reduction assay and/or colony forming unit (CFU) counting method as previously described (13). For XTT assay, biofilms were incubated with XTT solution (4 μm of menadione and 0.2 mg/ml of XTT in PBS) at 37 °C and kept in dark for 20 min. Solutions were then transferred to a new 96-well plate and colorimetric changes were measured at 490 nm using a calibrated spectrophotometer (Multiskan™ GO, Thermo Scientific, Waltham, MA). For CFU assay, serial dilutions were performed on biofilm suspension and spread onto Sabouraud dextrose agar (SDA) plates to incubate at 30 °C for 48 h before counting the colonies.
Antifungal Susceptibility Testing for Planktonic C. albicans Cells
Determination of minimum inhibition concentration (MIC)1 was performed according to the Clinical and Laboratory Standards Institute (CLSI) guideline (15). Briefly, overnight C. albicans cultures at 30 °C in GMM supplemented with required amino acids were used to prepare a cell suspension of 0.38 McFarLand standard (equivalent to 107 cells/ml). The suspension was diluted with RPMI to yield an inoculum of ∼0.5 × 103 to 2 × 103 cells/ml. The MIC was performed in 96-well plate platform and each strain was exposed to the 2-fold diluted solutions of the mentioned antifungals. The plates were incubated at 30 or 37 °C for 48 h before MIC values were recorded following the guideline. In order to obtain comparable planktonic MIC to that of the biofilm mode, additional Antifungal susceptibility testing was performed with the final inoculum of 107 cells/ml as we have previously described (14).
Antifungal Susceptibility Testing for C. albicans Biofilm
Antifungal susceptibility testing for C. albicans biofilm was conducted via XTT method or CFU assay. After 72 h biofilm formation, media were removed and cells were incubated in different antifungal concentrations in RMPI. The plates were incubated at 37 °C for 24 h. XTT methods or CFU assays were performed as described above to estimate the biofilm susceptibility. For survival rate at 32 μg/ml amphotericin B, statistical analysis was performed using analysis of variance. Differences were considered significant when p values are less than 0.05.
Antioxidant Capacity Assay
Seventy-two-hour biofilm cells were collected in lysis buffer (50 mm Tris-HCl pH 7.4, 150 mm KCl, 1% Nonidet P-40). Samples were homogenized using glass beads (0.5 mm) and a high speed vortex (Vortex-genie, Scientific Industries, Bohemia, NY) following 7 cycles of 1 min on and 2 min off ice. Total antioxidant capacity assay kit (Abcam, Cambridge, MA) was used to estimate the total antioxidant potentials of the biofilms following manufacture protocol. Statistical analysis was performed using analysis of variance. Differences were considered significant when p values are less than 0.05.
RNA Extraction and Real-time PCR
RNA was extracted from 72 h biofilms cells using TRIzol following the manufacture's protocol (Invitrogen, Carlsbad, CA). RNA was used for reverse transcription (Promega, Singapore) and real-time PCR (KAPA biosystem) was performed to estimate the level of RNA of interests. Primer sequences for real-time PCR were adapted from previous publication (16, 17). Statistical analysis was performed using analysis of variance. Differences were considered significant when p values are less than 0.05.
Protein Extraction and Western Blot
Biofilm cells were harvested into 2-ml screw-cap microcentrifuge tubes, washed once by PBS, and resuspended into 9 m urea lysis buffer (20 mm HEPES [pH8.0], 9 m urea, 1 mm sodium orthovanadate, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate). After addition of acid-washed glass beads (Sigma-Aldrich), cells were broken by six rounds of 60-s beating at 5000 rpm in a MicroSmash MS-100 beater (TOMY Medico, Tokyo, Japan) with 1 min of cooling between rounds. The lysed cells were then centrifuged at 16,000 rpm for 15 min at RT. Supernatants were then collected and the protein concentrations were determined by Nanodrop 1000 (Thermo scientific). After mixed with 3× protein loading buffer (150 mm Tris-HCl, pH 6.8, 6% SDS, 30% glycerol, 3% β-mercaptoethanol, 37.5 mm EDTA, 0.06% bromphenol blue), normalized protein samples (100 μg) were separated by SDS-PAGE and subsequently transferred to a PVDF membrane (Bio-Rad Laboratories, Hercules, CA). For Western blot analysis, the membrane was first incubated with 5% BSA (dissolved in PBS containing 0.1% Tween-20, PBST) at room temperature for 1 h (or at 4 °C overnight). After a brief rinse with PBST, the membrane was incubated with PBST containing a 1:2500 diluted first antibody (mouse monoclonal Myc antibody, Nacalai Tesque, Tokyo, Japan) at RT for 1 h, followed by three rounds of 5-min wash with PBST. The membrane was then incubated with PBST containing a 1:5000 diluted secondary antibody (HRP-linked anti-mouse IgG from sheep; GE Healthcare UK). After three rounds of 5-min wash with PBST, the membrane was immersed in Pierce ECL WB substrate solution (Thermo Scientific) and exposed to X-film (Fujifilm). Next, the membrane was stripped with Restore Western blot Stripping Buffer (Thermo Scientific) as instructed by the manual and blocked with 5% BSA again. Subsequently, the membrane was reprobed with 1:2500 diluted first antibody PSTAIRE (Santa Cruz Biotechnology, Dallas, TX) and 1:2500 diluted secondary antibody (HRP-linked anti-rabbit IgG from goat; Santa Cruz Biotechnology) to detect the house keeping protein Cdc28.
Protein Extraction for iTRAQ Analysis
Seventy-two-hour biofilms of SC5314, BWP17, GZY792, and GZY803 were taken for protein extraction. Biofilm was washed once with PBS and cell pellets were solubilized in triethylammonium bicarbonate (TEAB)/urea/triton-X/SDS (TUTS) buffer. Samples were homogenized using glass bead (0.5 mm) in an Omni Bead rupter 24 (Omni International Inc., Kennesaw, GA) following operator protocol. Lysates were collected and protein concentration was determined using Bradford assay (Bio-Rad).
In-Solution Digestion and Samples Clean Up
For each sample, 100 μg of proteins were taken. Solution was diluted with 0.5 m TEAB to a SDS concentration of less than 0.05%; pH was adjusted to 8. Subsequently, samples were reduced with 5 mm TCEP at 65 °C for 60 min and alkylated with 10 mm MMTS for 15 min at room temperature. Following reduction and alkylation, 1 μg of trypsin per 20 μg of proteins were added and trypsinization was performed at 37 °C for 16 h. Digested peptides were stored at - 20 °C before LC separation and MS analysis. Samples were then acidified and subjected to strong cation-exchange chromatography using iTRAQ method development kit (SCIEX, Foster City, CA). Elutes were then desalted in Sep-Pak C18 cartridge (Waters, Milford, MA), dried, and then reconstituted in 25 μl of diluent (98% water, 2%ACN, and 0.05% formic acid) before sending for LC-MS/MS analysis.
LC-MS/MS Analysis
The detailed methodology for LC-MS/MS has been previously described by our group (18). In brief, the peptides were dissolved with 50 μl mobile phase A (2% acetonitrile, 0.1% formic acid). Water peptide separation was conducted using an EksigentnanoLC Ultra and ChiPLC-nanoflex (Eksigent, Dublin, CA) in TrapElute configuration. Samples were then loaded on a 200 μm × 0.5 mm column and eluted on an analytical 75 μm × 15 cm column (ChromXP C18-CL, 3 μm). An amount of 2 μl of the sample was separated by a gradient formed by mobile phase A (2% acetonitrile, 0.1% formic acid) and mobile phase B (98% acetonitrile, 0.1% formic acid) at 3 μl/min flow rate. The following gradient elution was used for peptide separation: 5 to 5% of mobile phase B in 1 min, 5 to 12% of mobile phase B in 15 min, 12 to 30% of mobile phase B in 104 min, 30 to 90% of mobile phase B in 2 min, 90 to 90% in 7min, 90 to 5% in 3 min, and finally held at 5% of mobile phase B for 13 min. The tandem MS analysis was performed using a 5600 TripleTOF system (SCIEX) under Information Dependent Mode. The mass range of 400–1800 m/z and accumulation times of 250 ms per spectrum were chosen for precursor ions selections. MS/MS analysis was performed on the 20 most abundant precursors (accumulation time: 100 ms) per cycle with 15 s dynamic exclusion. Recording of MS/MS was acquired under high sensitivity mode with rolling collision energy.
Protein Identification and Quantification
Peptide identification and quantification was carried out on the ProteinPilot 4.5 software Revision 1656 (AB SCIEX) using the Paragon database search algorithm (4.5.0.0.1654) and the integrated false discovery rate (FDR) analysis function. The data were searched against a protein sequence database downloaded from UniProtKB for C. albicans SC5314 on 25th May, 2015 (total 14,851 entries). The MS/MS spectra obtained were searched using the following user-defined search parameters: Sample Type: iTRAQ 8-plex (Peptide Labeled); Cysteine Alkylation: MMTS; Digestion: Trypsin; Instrument: TripleTOF5600; Special Factors: None; Species: None; ID Focus: Biological Modification; Database: Algae Transcriptome.fasta; Search Effort: Thorough; FDR Analysis: Yes; The MS/MS spectra were searched against a decoy database to estimate the false discovery rate (FDR) for peptide identification. The decoy database consisted of reversed protein sequences from the C. albicans database. The resulting data set was auto bias-corrected to remove any variations imparted because of the unequal mixing during the combination of different labeled samples. Different modification states of the same peptide sequences are considered distinct by the software. In this study, a strict unused score cut-off ≥1.3 was adopted as the qualification criterion, corresponding to a peptide confidence level of ≥95%. The identified proteins with ≥2 peptides corresponding to 95% confidence interval were selected and further filtered using population statistics (19). Population statistic was also applied to select the cut-off for up-regulated and down-regulated proteins. Protein expression profiling of haploid biofilms was also clustered using Cluster (version 3.0) and Treeview (version 1.1.6) software (20).
Gene Ontology Analysis
Identified proteins were subjected to gene ontology analysis using Cytoscape (v2.8.3) (21) with BINGO plugin (v2.44) (22). Figures generated were abstracted from Cytoscape as previously described (23).
Experimental Design and Statistical Rationale
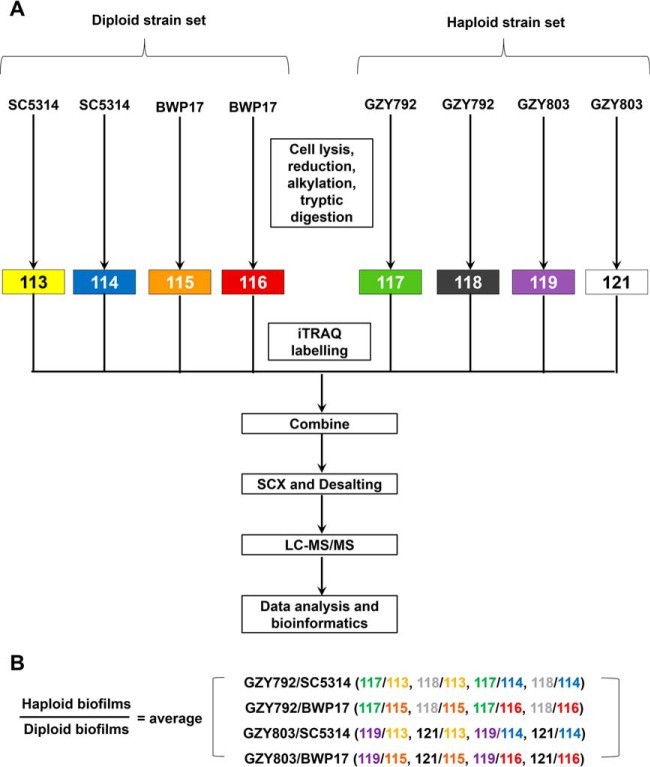
Seventy-two-hour C. albicans haploid and diploid biofilms were collected for iTRAQ-based mass spectrometry analysis. Two haploid (GZY792 and GZY803) and two diploid strains (SC5314 and BWP17) were used as haploid and diploid replicates, respectively. Two biofilm samples of each strain under identical environmental conditions were obtained as the technical replicates. In order to obtain a proteomic expression data set in C. albicans haploid biofilms, independent of strain variation, cross comparison was performed between biofilm proteomes of the two haploid strains and those of the two diploid strains (Fig. 1). In other words, proteins that were similarly expressed in all four strains were considered “common” for the biofilm proteome of C. albicans. The proteins that were differentially expressed between haploid and diploid biofilm proteomes were classified as “distinctly expressed.” For other biochemical, functional, and quantitative assays, such as transcriptional analysis, XTT and CFU quantitative methods, and total antioxidant capacity estimation, one representative haploid strain, GZY803, was examined and normalized to that of a diploid control strain, BWP17. Experiments were performed in at least three biological replicates and significance of the results was analyzed following analysis of variance. Differences were considered significant when p values are less than 0.05.
Fig. 1.
Experimental design for iTRAQ labeling showing biological replicates. A, Two replicates from 72-h biofilms of two control diploid strains (SC5314 and BWP17) and two haploid strains (GZY792 and GZY803) were labeled with iTRAQ labeling reagents 113, 114, 115, and 116, and 117, 118, 119, and 121, respectively. B, Protein expression in haploid biofilm compared with diploid was determined by taking average of all cross comparisons between each replicates of haploid biofilm versus diploid biofilm of each strain.
RESULTS
iTRAQ-based Quantitative Proteomics of Haploid and Diploid Biofilms of Candida albicans
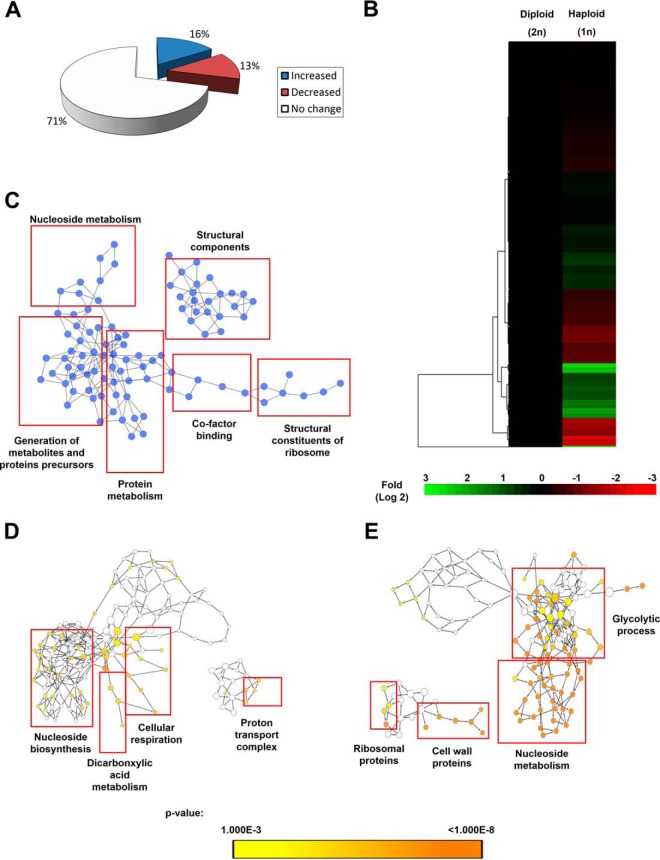
To understand the molecular mechanism operating in the biofilm mode of growth of C. albicans haploids, we performed an iTRAQ-based mass spectrometry (24) profiling of proteins of 72 h mature biofilms of the two haploids, GZY792 and GZY803 compared with that of the diploids, SC5314 and BWP17. The methodology that was used to analyze the changes in protein expression among different biofilm samples is illustrated in supplemental Fig. S1. Using a decoy database search strategy, FDRs for protein and peptides were estimated as 2.8% and 0.4%, respectively. From the search results, a cutoff of protein unused score ≥1.3 (corresponds to 95% confidence) was used as qualification criterion, which resulted in 511 proteins. No protein or peptide identified from the decoy database had ≥ 95% confidence based on our ProteinPilot search result. Among the proteins that satisfied the stringent cutoff of protein unused score ≥1.3, 374 proteins identified by more than one single peptide were selected for further study. Cross comparison was performed between biofilm proteomes of the two haploid and two diploid strains. Ratio of protein expression of haploid relative to diploid biofilms was estimated as average of each cross comparison (Fig. 1). Subsequently, deviation of ratios (i.e. 117/113) of individual proteins between various replicates was calculated as described previously (25). Geometric mean, standard deviation and percentage coefficient of variation (%CV) of 374 proteins were also determined. Next, to determine cutoff threshold, population statistical analyses (19) were applied which showed that %CV = 66% corresponded to 88% coverage of the data; therefore, the cut-off fold was fixed at 1.66-fold (66%) for increased, and 0.6-fold (1/1.66) for decreased proteins (supplemental Fig. S2). On the other hand, from the original 374 proteins, 39 proteins that showed large variation between haploid and diploid samples, indicated by % CV larger than 66% were filtered out. Thereafter, student-t statistical test was applied to the remaining 335 proteins. Expression was considered significant when p value was less than 0.05 between samples. Applying the cutoff threshold determined by population statistics (supplemental Fig. S2) to these significantly altered proteins resulted in 55 significantly increased proteins and 45 significantly decreased proteins (Fig. 2A). Hence, 235 remaining proteins were then classified as the common proteins between the haploid and diploid biofilms (Fig. 2A). Expression pattern of these proteins was also illustrated and organized by two-dimensional hierarchical clustering (Fig. 2B). Each protein was colored corresponding to its fold change. In addition, pathway analysis by Cytoscape (21) revealed that similarly expressed proteins were mapped into the pathways of nucleoside and protein metabolism, protein and metabolite precursor generation, co-factor binding, and cellular and ribosomal structures (Fig. 2C). On the other hand, mapping differently regulated proteins to their gene ontology revealed that nucleoside biosynthesis, dicarboxylic acid metabolism, cellular respiration, and ATP synthesis-coupled proton transport were significantly increased; and, nucleoside breakdown, glycolysis, cell wall and ribosomal proteins expression were suppressed in the haploid biofilm proteome as compared with the diploids (Fig. 2D, 2E, and supplemental Table S2). Full lists of similarly and distinctively expressed proteins between haploid and diploid biofilms are given in supplemental Table S3. In addition, to examine the reliability of our results, we confirmed expression of selected genes at the transcriptional level using real time PCR (supplemental Fig. S3).
Fig. 2.
Protein expression function classification of 72 h haploid versus diploid C. albicans biofilms. Two commonly used C. albicans diploids (SC5314 and BWP17) and two standard haploids (GZY792 and GZY803) were used in this study. Fungal cells cultured overnight at 30 °C in GMM (supplemented with required amino acids) were used to allow biofilm development at 37 °C. Proteins from 72-h biofilms were extracted and processed for iTRAQ mass spectrometry. A, Percentage of similarities and differences in proteomes of haploid compared with diploid biofilm were represented. B, Clustered display of haploid biofilms' protein expression profiles as normalized to diploids generated using Cluster and Tree view software. C, Biological processes, molecular functions, and cellular components that are similarly regulated in haploid and diploid biofilms. Biological processes, molecular functions, and cellular components that are significantly higher (D) and lower (E) in the haploid compared with the diploid biofilm.
Comparative Antifungal Susceptibility of C. albicans Haploid and Diploid Biofilms
Analysis of the proteomic data showed differential proteomics responses between haploid and diploid biofilms, providing information that may explain the phenotypic differences between the two types of C. albicans cells (26–28). To further examine the differential behavior, we performed antifungal susceptibility tests on C. albicans haploid and diploid strains in the planktonic and biofilm modes using standard micro-dilution assays (14). In the planktonic mode, C. albicans haploids showed a similar susceptibility as the diploids to all antifungals tested (Table I). In addition, the haploids in the biofilm mode exhibited higher resistance to the antifungals tested including caspofungin, fluconazole, ketoconazole, and voriconazole, similar to the biofilms of diploids (BWP17). Intriguingly, the haploid biofilm was more susceptible to amphotericin B than the diploid biofilm. This observation was further confirmed by counting colony forming units (CFU) (Fig. 3A). The minimum inhibitory concentration (MIC) of the haploid GZY803 biofilms against amphotericin B was 2 μg/ml whereas that of the diploid BWP17 biofilm was 16 μg/ml. At amphotericin B concentration of 2 μg/ml, less than 10% of GZY803 biofilm cells survived the treatment as compared with the untreated control. On the contrary, 16 μg/ml of amphotericin B was required to attain a similar level of effectiveness against the diploid biofilm (Fig. 3A). Furthermore, CFU analysis also revealed that the haploid biofilm was less tolerant to amphotericin B than the diploid biofilm. At the amphotericin B concentration of 32 μg/ml, GZY803 haploids showed a 10-fold reduction of the persister population i.e. 0.1% versus 1%, as compared with the diploids (Fig. 3B). Taken together, our data showed that the haploid biofilm is less tolerant to amphotericin B than the diploid biofilm.
Table I. Antifungal susceptibility of C. albicans in different modes of growth. Two commonly used C. albicans diploids (SC5314 and BWP17) and two standard haploids (GZY792 and GZY803) were used in this study. Fungal cells cultured overnight at 30 °C in GMM (supplemented with required amino acids) were used to prepare the required inoculum for the antifungal susceptibility test. Minimum inhibition concentration (MIC) was recorded by following the CLSI guideline, or optical density (planktonic), or XTT test (biofilm cells).
| MIC (μg/ml) | |||||
|---|---|---|---|---|---|
| Strains | 30 °C |
37 °C |
Biofilm 107/ml | ||
| CLSI 103/ml | Planktonic 107/ml | CLSI 103/ml | Planktonic 107/ml | ||
| Amphotericin B | |||||
| SC5314 | 0.25 | 0.5 | 0.25 | 0.5 | 16 |
| BWP17 | 0.25 | 0.5 | 0.25 | 0.5 | 16 |
| GZY792 | 0.25 | 0.5 | 0.25 | 0.5 | 2 |
| GZY803 | 0.25 | 0.5 | 0.25 | 0.5 | 2 |
| Caspofungin | |||||
| SC5314 | 0.5 | 2 | 0.5 | 2 | >32 |
| BWP17 | 0.5 | 2 | 0.5 | 2 | >32 |
| GZY792 | 0.5 | 2 | 0.5 | 2 | >32 |
| GZY803 | 0.5 | 2 | 0.5 | 2 | >32 |
| Fluconazole | |||||
| SC5314 | 0.5 | 2 | 0.25 | 2 | >32 |
| BWP17 | 0.5 | 2 | 0.25 | 2 | >32 |
| GZY792 | 0.25 | 2 | 0.25 | 2 | >32 |
| GZY803 | 0.25 | 2 | 0.25 | 2 | >32 |
| Ketoconazole | |||||
| SC5314 | 0.125 | 0.25 | 0.125 | 0.25 | >32 |
| BWP17 | 0.125 | 0.25 | 0.125 | 0.25 | >32 |
| GZY792 | 0.125 | 0.25 | 0.125 | 0.25 | >32 |
| GZY803 | 0.125 | 0.25 | 0.125 | 0.25 | >32 |
| Voriconazole | |||||
| SC5314 | 0.006 | 0.06 | 0.006 | 0.06 | >32 |
| BWP17 | 0.006 | 0.06 | 0.006 | 0.06 | >32 |
| GZY792 | 0.006 | 0.06 | 0.006 | 0.06 | >32 |
| GZY803 | 0.006 | 0.06 | 0.006 | 0.06 | >32 |
Fig. 3.
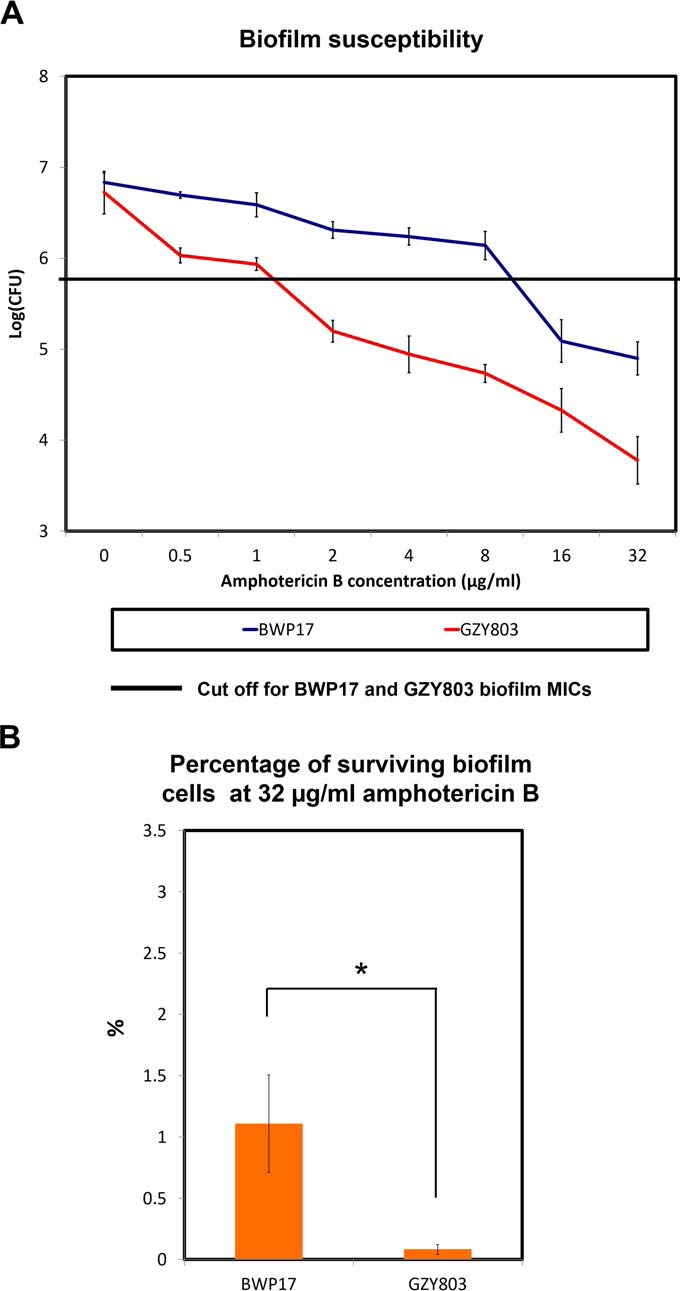
GZY803 mature biofilm showed increased susceptibility to amphotericin B as compared with that of BWP17. A, C. albicans diploid (BWP17) and haploid (GZY803) strains were cultured at 30 °C in GMM (supplemented with required amino acids). Overnight cultures were then used to inoculate and allow biofilm development at 37 °C. Seventy-two-hour biofilms were treated with different concentrations of amphotericin B in RPMI for 24 h and minimum inhibition concentrations were recorded using CFU assays. B, Percentage of surviving biofilm cells at 32 μg/ml was also estimated by comparing the CFU at 32 μg/ml to the fungal count of untreated biofilms. Mean of at least three replicates is shown, with error bars showing S.D. (*): p value <0.05.
Identification of AHP1 as a Critical Determinant of C. albicans Haploid Biofilm's Persistence Against Amphotericin B
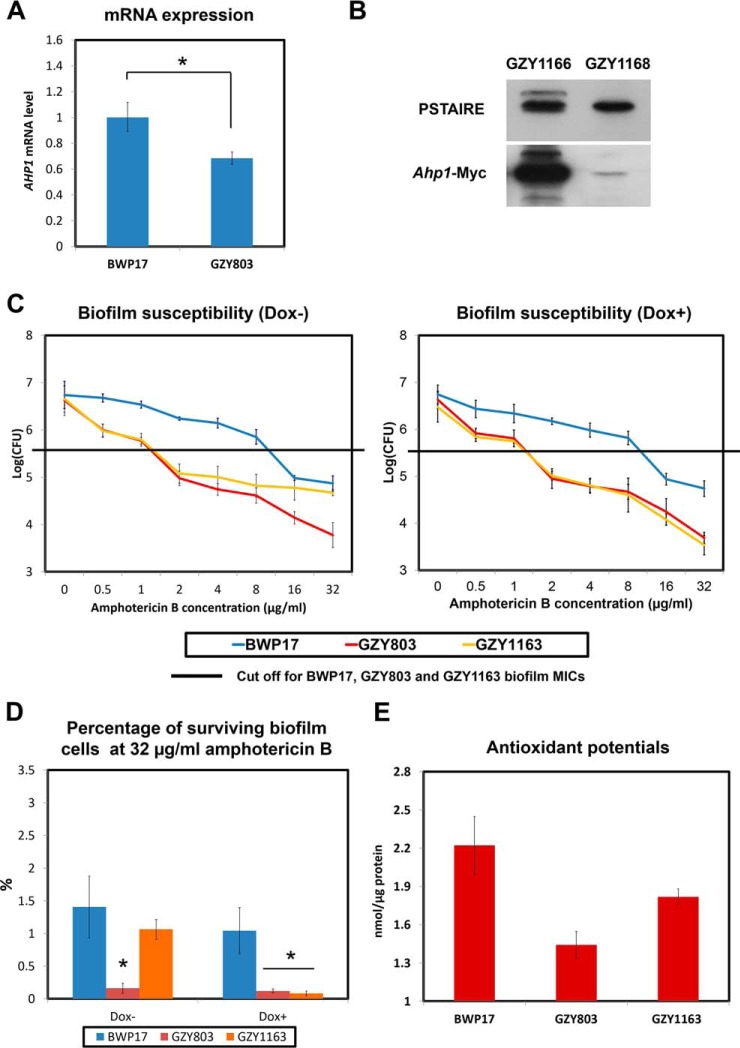
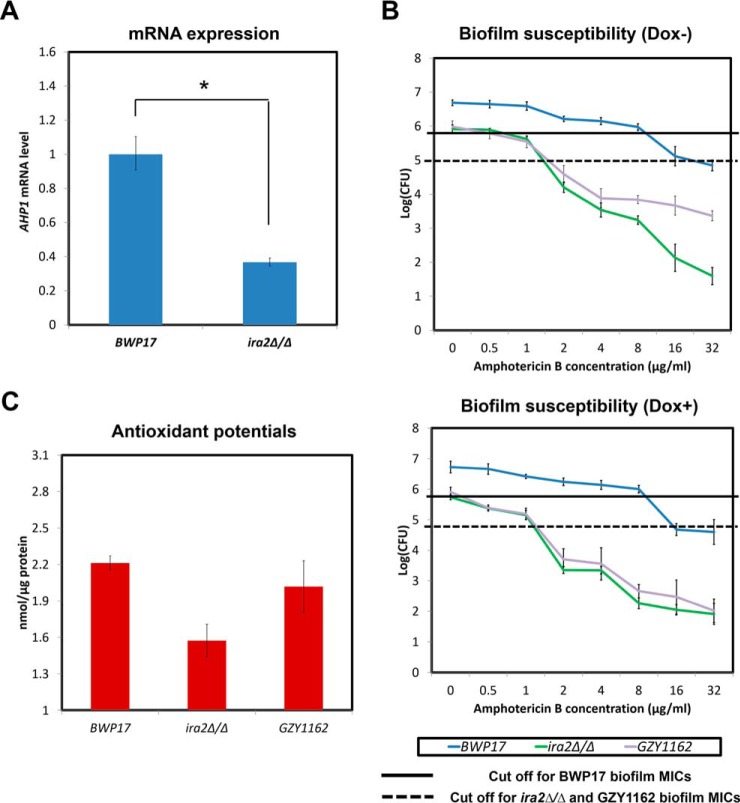
Amphotericin B is known to exert its antifungal effect via binding to ergosterol, causing pore formation and cell death via increased oxidative stress (29, 30). Previously, we have shown that higher antioxidant capacity is important for higher resistance in C. albicans biofilms (31). Therefore, we further analyzed the haploid and diploid biofilm proteomes of C. albicans with respect to antioxidant proteins in order to find an explanation for the differential susceptibility to amphotericin B. Proteomics analysis identified Ahp1, a cell wall peroxidase (32) that plays a vital role in oxidative stress response, whose expression was lower in the haploid biofilm proteome (supplemental Table S3). Subsequent Western blot and real-time PCR analyses corroborated that the haploid biofilm expressed a significantly less amount of AHP1 than the diploid biofilm at both mRNA and protein levels (Fig. 4A and 4B).
Fig. 4.
Reduction of AHP1 expression in GZY803 biofilms is linked with decreased antioxidant potentials and tolerance of amphotericin B. A, BWP17 and GZY803 cells were cultured in GMM supplemented with required amino acids. Overnight cultures were used to inoculate and allow biofilm development at 37 °C. RNA from 72-h biofilms was extracted and examined for AHP1 expression using real-time PCR. ACT1 and PMA1 were used as controls. B, GZY1166 (BWP17:AHP1-Myc) and GZY1168 (GZY803:AHP1-Myc) were cultured in GMM supplemented with required amino acids. Overnight cultures were used to inoculate and allow biofilm development at 37 °C. Proteins from 72-h biofilm cells were extracted and examined for AHP1 expression by Western blot. C, To examine amphotericin B resistance and tolerance relating to AHP1 expression in the haploid biofilm, 72-h biofilms of GZY1163 (GZY803+Tetoff-Myc-AHP1) were treated with different concentrations of amphotericin B (in the presence or absence of 20 μg/ml of Dox) in RPMI for 24 h and CFUs were recorded. D, Percentage of surviving biofilm cell population at 32 μg/ml amphotericin B was also estimated following CFU assay. Diploid strain (BWP17) and haploid parent strain (GZY803) strain were included as controls. E, To estimate total antioxidant capacity, proteins from 72-h biofilms of BWP17, GZY803 and GZY1163 were also used for the antioxidant test. For all graphs, the mean of at least three replicates is shown, with error bars showing S.D. (*): p value <0.05.
Antioxidant capacity is crucial for C. albicans to neutralize oxidative stress (33, 34). We hypothesized that the reduced AHP1 expression found in the haploid biofilm might have caused a significant drop of the antioxidant potential, which contributes to the higher susceptibility to amphotericin B. To test this hypothesis, we expressed AHP1 under the control of the TetOff promoter in the GZY803 background (GZY1163) and examined the susceptibility to amphotericin B. In GZY1163, AHP1 is overexpressed in the absence and shut down in the presence of doxycycline (20 μg/ml) (35). With or without AHP1 expression, the GZY1163 biofilm showed the same MIC (2 μg/ml) as the GZY803 biofilm (Fig. 4C). Interestingly, with AHP1 overexpression (Dox-), the GZY1163 biofilm displayed a higher survival rate at higher concentrations of amphotericin B than the GZY803 biofilm (Fig. 4C and 4D). Approximately 10-fold higher survival rates were recorded in the GZY1163 biofilm under Dox- conditions at amphotericin B concentrations of 32 μg/ml (Fig. 4D). Adding doxycycline to the GZY1163 cells abolished the increased tolerance (Fig. 4D). Increased AHP1 expression, therefore, seemed to augment the persister population and enhance C. albicans survival in the haploid biofilm to a level near the BWP17 diploid biofilm. Also, we noted that the GZY803 haploid biofilm showed reduced antioxidant capability than the diploid BWP17 biofilm. Overexpression of AHP1 in the GZY803 background significantly improved the antioxidant potential in the biofilm (Fig. 4E). In summary, AHP1 overexpression in GZY803 correlates well with the increased total antioxidant potential in biofilms and the tolerance against amphotericin B.
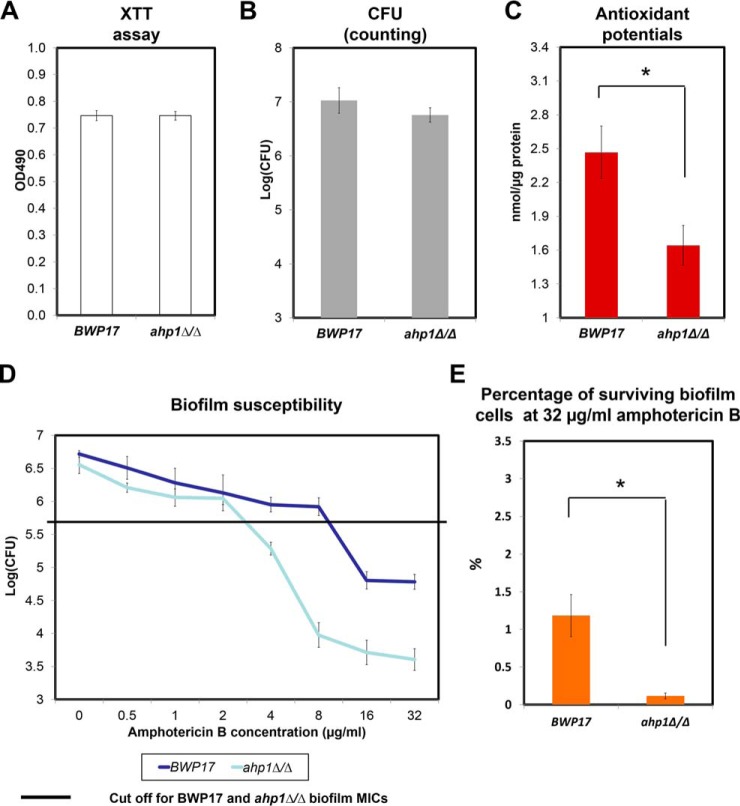
ahp1Δ/Δ C. albicans Biofilms Exhibited Reduced Persister Population and Increased Susceptibility to Amphotericin B
Preceding experiments revealed the critical contribution of AHP1 in the persister population of C. albicans haploid biofilms. To examine whether AHP1 plays the same role in diploid C. albicans, we generated the ahp1Δ/Δ mutant in the BWP17 background and assessed its biofilm formation and susceptibility to amphotericin B. Our XTT assays and CFU counts indicated that the ahp1Δ/Δ strain was capable of forming mature biofilm with a similar biomass as BWP17 (Fig. 5A and 5B). Despite comparable biomass, ahp1Δ/Δ biofilms displayed a lower antioxidant potential than BWP17 (Fig. 5C). Also, the CFU assays reflected a decrease in amphotericin B resistance in ahp1Δ/Δ biofilms (Fig. 5D). The MIC of ahp1Δ/Δ biofilms was reduced to 4 μg/ml, 4-fold lower than that of the BWP17 biofilms (16 μg/ml) (Fig. 5D). The persister population was also consistently reduced to 0.1% in the ahp1Δ/Δ biofilm as compared with 1% in that of BWP17 under 32 μg/ml amphotericin B treatment (Fig. 5E). These results corroborated findings in the haploids and confirmed Ahp1's regulatory role in biofilm persister formation and tolerance against amphotericin B.
Fig. 5.
C. albicans ahp1Δ/Δ biofilm exhibited reduced antioxidant potentials and diminished tolerance and resistance to amphotericin B. The overnight culture of ahp1Δ/Δ (GZY1210) C. albicans strain in GMM (supplemented with amino acids) was used inoculate and allow biofilm development at 37 °C. A, B, Quantification of biofilms formed by BWP17 and ahp1Δ/Δ after 72 h was conducted via XTT and CFU methods. C, Proteins from 72-h biofilms of BWP17 and ahp1Δ/Δ were also extracted and used to estimate the total antioxidant capacity. D, To examine amphotericin B resistance and tolerance, 72-h biofilm samples of BWP17 and ahp1Δ/Δ were treated with different concentrations of amphotericin B in RPMI for 24 h and CFUs were counted by plating the biofilm samples at each concentration on Sabouraud dextrose agar. E, Percentage of surviving biofilm cells at 32 μg/ml was also estimated by comparing the CFUs at 32 μg/ml to the fungal count of untreated biofilms. For all graphs, mean of at least three replicates is shown, with error bars showing S.D. (*): p value <0.05.
ira2Δ/Δ C. albicans Biofilms Displayed an Increased Susceptibility to Amphotericin B
Previously, we demonstrated that Ira2 regulates biofilm formation in C. albicans. IRA2 deletion was associated with defective biofilms characterized by reduced structural complexity (13). Next, we queried whether the ira2Δ/Δ biofilm is associated with reduced antifungal susceptibility like the ahp1Δ/Δ biofilm. Spot plate assays showed that ira2Δ/Δ biofilms exhibited increased susceptibility to amphotericin B but not to other antifungals (supplemental Fig. S4). The higher susceptibility of the ira2Δ/Δ biofilms to amphotericin B was confirmed by CFU assays (Fig. 6). The ira2Δ/Δ biofilms showed an MIC of 2 μg/ml whereas reintroducing IRA2 into the deletion strain restored the resistance (MIC = 8 μg/ml). Hence, the data demonstrated an association between IRA2 and amphotericin B susceptibility of C. albicans.
Fig. 6.
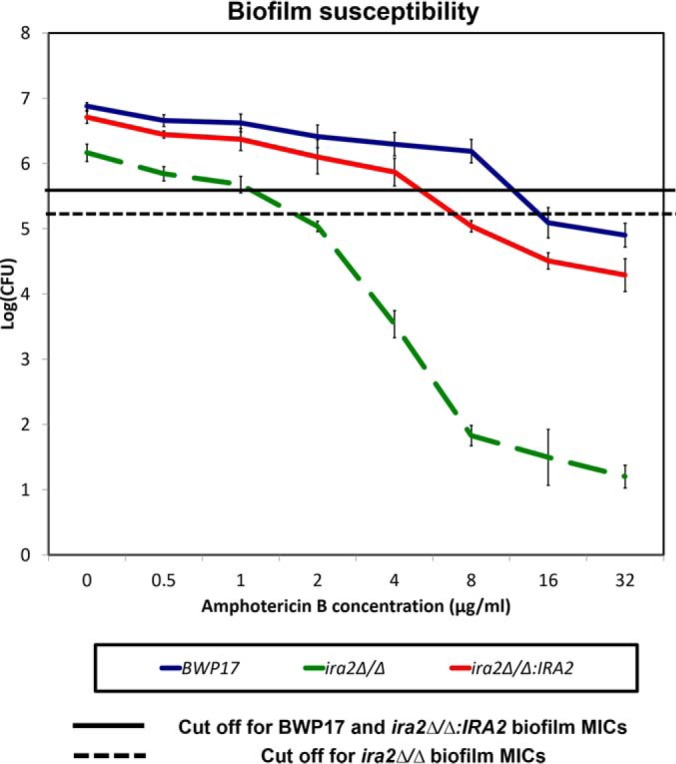
C. albicans ira2Δ/Δ biofilm showed increased susceptibility to amphotericin B. ira2Δ/Δ (GZY923) C. albicans strain was cultured in GMM supplemented with required amino acids. The overnight culture was used to inoculate and allow biofilm development at 37 °C. Seventy-two-hour biofilms were treated with different concentrations of amphotericin B in RPMI for 24 h and CFUs were recorded by plating the biofilm samples at each concentration on Sabouraud dextrose agar. Parent strain (BWP17) and ira2Δ/Δ rescued strain (ira2Δ/Δ:IRA2, GZY1022) were included as controls. For all graphs, mean of at least three replicates is shown, with error bars showing S.D.
As presented above, the first set of experiments derived from the comparative analysis of the proteomes of C. albicans haploid and diploid strains demonstrated that Ahp1 plays a pivotal role in the persister population of C. albicans biofilms against amphotericin B. The second set of experiments showed an association of Ira2 with amphotericin B susceptibility in C. albicans biofilms. We also found that IRA2 deletion results in lower expression of AHP1 at the transcriptional level (Fig. 7A). We next asked whether overexpression of AHP1 could protect the ira2Δ/Δ biofilm against amphotericin B and found that AHP1 overexpression in the ira2Δ/Δ strain (GZY1162) enhanced the tolerance of the ira2Δ/Δ biofilm against amphotericin B (Fig. 7B), leading to more than 10-fold increase in the survival rate under 16 μg/ml and 32 μg/ml amphotericin B treatments as compared with the ira2Δ/Δ strain (Fig. 7B); and shutting down the AHP1 overexpression with doxycycline treatment abolished the increment in the persister population and drug tolerance of the GZY1162 biofilm (Fig. 7B). In addition, the diminished antioxidant capacity in the ira2Δ/Δ strain was largely restored when AHP1 was overexpressed as compared with the BWP17 biofilm (Fig. 7C). Taken together, the comprehensive set of data firmly demonstrated a central role for Ahp1 in Ira2-mediated C. albicans biofilm's susceptibility to amphotericin B.
Fig. 7.
IRA2 regulation of AHP1 is linked with C. albicans biofilms' antioxidant potentials and tolerance of amphotericin B. A, ira2Δ/Δ (GZY923) C. albicans strain was cultured in GMM supplemented with required amino acids. The overnight culture was used to inoculate and allow biofilm development at 37 °C. RNA from 72-h biofilm cells was extracted and examined for AHP1 expression using real-time PCR. ACT1 and PMA1 were used as controls. B, To examine amphotericin B resistance and tolerance, 72-h biofilms of BWP17, ira2Δ/Δ and ira2Δ/Δ+TetOff-Myc-AHP1 (GZY1162) were treated with different concentrations of amphotericin B (in the presence or absence of 20 μg/ml of Dox) in RPMI for 24 h and CFUs were recorded by plating the biofilm samples at each concentration on Sabouraud dextrose agar. C, To estimate total antioxidant capacity, proteins from 72-h biofilms of BWP17, ira2Δ/Δ and ira2Δ/Δ+TetOff-Myc-AHP1 (GZY1162) were also used for the antioxidant test. For all graphs, mean of at least three replicates is shown, with error bars showing S.D. (*): p value <0.05.
DISCUSSION
The recent discovery of haploid strains of C. albicans and subsequent validation of a haploid biofilm model has offered a new toolbox necessary for genetic manipulation in C. albicans research (13). In this study, we performed a comparative proteomics analysis on haploid and diploid C. albicans biofilms and targeted gene expression study to uncover AHP1 as a pivotal antioxidant for the survival of persister cells against amphotericin B.
C. albicans haploid biofilm proteome shares a high similarity with that of the diploid biofilm with more than 70% of proteins expressed approximately at the same level, in particular that of structural proteins (Fig. 2). The high similarity explains our observation of haploid biofilms which comprised of comparable stacked yeast and hyphal cells encased in extracellular matrix as diploid biofilms (13). Expressions of proteins related to major biochemical pathways were considerably similar between haploid and diploid biofilms proteomes consistent with the comparable nature of the biofilms (Fig. 2).
On the other hand, some of the proteins were differentially expressed between the haploid and diploid biofilm proteomes. The haploid biofilms expressed a higher level of key molecules in ATP synthesis and respiration such as citrate synthase and ATP synthase subunits (Fig. 2 and supplemental Table S2). On the contrary, the diploid biofilms exhibited an increase in proteins from the glycolysis pathway (36) including hexokinase, phosphofructokinase, and pyruvate kinase as haploids. These implied that haploid biofilms might rely more on fermentation and less aerobic respiration and glycolysis to generate ATP as compared with diploid biofilms. The haploid biofilms also demonstrated increased expression of chorismate synthase and aspartate semialdehyde dehydrogenase, determinant proteins in dicarboxylic and aromatic amino acids metabolisms (37, 38). Also, compared with the diploids, the haploid biofilm proteome exhibited a reduction in the expression of ribosome structure proteins. Depletion in ribosome generation and reduced glycolysis are consistent with the reported delay in growth, development, and maturation in C. albicans haploid biofilms as compared with the diploids (13). Proteomic analysis of the haploid biofilms also revealed a diminished expression of molecules in cell wall composition (Fig. 2 and supplemental Table S2), which correlates with reduced in vivo virulence in mouse models of infection (10) and delayed ex vivo invasion in oral tissue models (13). Among the identified cell wall proteins, alkyl hydroperoxide reductase 1 (Ahp1) and coproporphyrinogen III oxidase have previously been shown to be associated with antifungal resistance (31, 39–41). Overexpression of the coproporphyrinogen III oxidase, an enzyme of heme biosynthesis pathway, is a survival strategy of Aspergillus fumigatus and Saccharomyces cerevisiae against antifungal treatment (40, 41). On the other hand, our previous study on C. albicans biofilm proteome found that Ahp1 is associated with the higher antioxidative capacity of the C. albicans biofilms (8). Therefore, in the present study, we aimed to comprehensively investigate the role of Ahp1 in antifungal susceptibility of C. albicans biofilms.
The haploid cells in biofilm mode exhibited high resistance to the many antifungals tested (caspofungin, fluconazole, ketoconazole, and voriconazole) as the diploid biofilm. Intriguingly, we found that the haploid biofilms, having an MIC value of 2 μg/ml, are more susceptible to amphotericin B as compared with the diploid biofilms with an MIC of 16 μg/ml (Fig. 3). Different mechanisms that have been explored to explain the higher drug resistance of C. albicans biofilms include the presence of the extracellular polymeric substance (EPS), higher antioxidative capacity, and persister populations (5, 9, 42). However, we have observed that the C. albicans haploids form comparable biofilms with the extracellular matrix similar to diploid biofilms (13). Therefore, EPS is unlikely the explanation of the observed difference in amphotericin B susceptibility between the haploid and diploid biofilms. Other studies in the literature corroborated that EPS may not be a contributory factor against amphotericin B in Candida biofilms (43, 44).
Another explanation for higher antifungal tolerance of the C. albicans biofilms is the subpopulation of cells called “persisters,” which are tolerant to repeated exposure to high concentration of antifungals (7). We observed that 0.1% of the cells in the haploid GZY803 biofilms survived at 32 μg/ml amphotericin B treatments whereas the diploid BWP17 biofilm had a 1% persister population (p value < 0.05). Hence, the haploid C. albicans possessed considerably fewer persister cells against amphotericin B than the diploids in biofilms (Fig. 3). To understand the differential behavior of the haploid and diploid biofilms, we examined the biofilm proteomes. The proteomics analysis demonstrated that the haploid biofilm has considerably less amount of Ahp1 (Fig. 4 and supplemental Table S3), a key protein of the antioxidant defense system, located in the cell wall (33). Ahp1 is a member of the peroxiredoxin family and plays a vital role in protecting the cells from reactive oxygen species as well as in stress adaptation (32). Ahp1 coordinates with other peroxidases such as Trx1 to protect the cells against oxidative stress (45). Hence, upon binding to its substrate, Ahp1 induces antioxidant response by activating the Cad1 transcriptional signaling pathway (46). Moreover, previous studies have shown that Ahp1 is an important antioxidant in the C. albicans biofilms (31, 47). These are particularly relevant in antifungals like amphotericin B that is known to generate reactive oxidative species (30, 48). Foregoing analysis provided clear evidence on the role of Ahp1 and antioxidant responses in the persister population of C. albicans biofilms against amphotericin B treatment. Our findings corroborate with previous works. Superoxide dismutases, important players in C. albicans antioxidant defense, were found to be associated with biofilm persistence to miconazole (16). Not only in fungi, hydrogen peroxide-inducible genes activator (OxyR), a sensor of hydrogen peroxide, was also shown as a determinant of bacterial persisters (49). Our study, therefore, has emphasized the crucial contribution of antioxidant protection in survival strategy against antifungal treatment.
We also found a link between IRA2 and AHP1 in amphotericin B resistance of C. albicans biofilms. Ira2 is a putative GTPase-regulating protein of Ras1, an activator of the cAMP/protein kinase A pathway (50). Ira2 is known to mediate fungal heat shock and stress response (51). Previously, we demonstrated that Ira2 regulates biofilm formation in C. albicans and a strain lacking IRA2 formed a thinner and less dense biofilm than wild-type strains (13). Here, we found that despite the structural defects, ira2Δ/Δ biofilms remain resistant to several antifungals but more susceptible to amphotericin B than the wild-type strain (Fig. 6 and supplemental Fig. S4). Therefore, we assumed that the persister population of ira2Δ/Δ biofilms lacked some elements required for survival against amphotericin B treatment. Our subsequent experiments uncovered hitherto a previously unknown link between AHP1 and IRA2 in C. albicans biofilms (Fig. 7). AHP1 expression in C. albicans persister population could be directly or indirectly regulated by Ira2. It has been shown that AHP1 is regulated by Yap1 and Skn7 (45). Interestingly, functions of both Yap1 and Skn7 are linked to GTPase molecules. In Saccharomyces cerevisiae, Skn7 is known to interact with Rho GTPases (52). On the other hand, ScYap1 translocation to the cytoplasm is required for the Ran GTPase activity (53). Ira2 is a regulator of Ras (51) which belongs to the superfamily of small GTPases including Rho and Ran (54). Therefore, it is possible that Ira2 is a direct or indirect upstream effector of Ran and Rho GTPases, which in turn regulates Skn7 and Yap1 and finally mediates AHP1 expression.
In conclusion, the present study, for the first time, unraveled the comparative ploidy proteome of haploid and diploid C. albicans biofilms. Subsequent studies on haploid mutagenesis provided new molecular insights to explain the higher resistance of C. albicans biofilms against amphotericin B, the gold standard antifungal agent. Taken together, we demonstrated that IRA2's role in biofilm regulation is related to that of AHP1, both increase the antioxidative capacities and survival rates of C. albicans cells in the biofilm under amphotericin B treatment. These findings will open up new avenues to develop targeted therapy to eradicate the persister population in C. albicans biofilms, which could benefit millions of patients suffering from biofilm-associated infections.
Supplementary Material
Acknowledgments
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (55) partner repository with the data set identifier PXD004274.
Footnotes
Author contributions: C. J. S., and Y. W. conceived, designed, provided general guidance, and co-wrote the manuscript. T. T. performed the biofilm experiments, protein extractions, proteomics analysis, and co-wrote the manuscript. G. Z. constructed most of the yeast strains, performed the western blot, and co-wrote the manuscript. L. Q., and L. T. K. conducted the mass spectrometry, gave advices to the proteomics analysis, and revised the manuscript. C. T. gave guidance and assisted in the biofilm experiments. F. Y. C. assisted strain construction and western blot analysis.
* This study was supported by NMRC-NIG (R-221-000-073-511), NUS-Start-up (R-221-000-064-133) to C.J.S. and funding from Agency for Science, Technology, and Research (A*STAR) to Y.W.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- MIC
- minimum inhibition concentration
- AHP1
- alkyl hydroperoxide reductase 1
- IRA2
- Inhibitory Regulator of the RAS-cAMP pathway
- iTRAQ
- Isobaric tags for relative and absolute quantitation.
REFERENCES
- 1. Mavor A. L., Thewes S., and Hube B. (2005) Systemic fungal infections caused by Candida species: epidemiology, infection process and virulence attributes. Curr. Drug Targets 6, 863–874 [DOI] [PubMed] [Google Scholar]
- 2. Gullo A. (2009) Invasive fungal infections: the challenge continues. Drugs 69, 65–73 [DOI] [PubMed] [Google Scholar]
- 3. Delaloye J., and Calandra T. (2014) Invasive candidiasis as a cause of sepsis in the critically ill patient. Virulence 5, 161–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wisplinghoff H., Bischoff T., Tallent S. M., Seifert H., Wenzel R. P., and Edmond M. B. (2004) Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39, 309–317 [DOI] [PubMed] [Google Scholar]
- 5. Seneviratne C. J., Jin L., and Samaranayake L. P. (2008) Biofilm lifestyle of Candida: a mini review. Oral Diseases 14, 582–590 [DOI] [PubMed] [Google Scholar]
- 6. Ramage G., Saville S. P., Thomas D. P., and Lopez-Ribot J. L. (2005) Candida biofilms: an update. Eukaryotic Cell 4, 633–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. LaFleur M. D., Kumamoto C. A., and Lewis K. (2006) Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrobial Agents Chemother. 50, 3839–3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chandra J., Kuhn D. M., Mukherjee P. K., Hoyer L. L., McCormick T., and Ghannoum M. A. (2001) Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J. Bacteriol. 183, 5385–5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mathe L., and Van Dijck P. (2013) Recent insights into Candida albicans biofilm resistance mechanisms. Curr. Genet. 59, 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hickman M. A., Zeng G., Forche A., Hirakawa M. P., Abbey D., Harrison B. D., Wang Y. M., Su C. H., Bennett R. J., Wang Y., and Berman J. (2013) The ‘obligate diploid’ Candida albicans forms mating-competent haploids. Nature 494, 55–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sarosi G. A. (1990) Amphotericin B. Still the ‘gold standard’ for antifungal therapy. Postgrad Med. 88, 151–152, 155- 161, 165–166 [DOI] [PubMed] [Google Scholar]
- 12. Zeng G., Wang Y. M., Chan F. Y., and Wang Y. (2014) One-step targeted gene deletion in Candida albicans haploids. Nat. Protoc. 9, 464–473 [DOI] [PubMed] [Google Scholar]
- 13. Seneviratne C. J., Zeng G., Truong T., Sze S., Wong W., Samaranayake L., Chan F. Y., Wang Y. M., Wang H., Gao J., and Wang Y. (2015) New “haploid biofilm model” unravels IRA2 as a novel regulator of Candida albicans biofilm formation. Sci. Reports 5, 12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Seneviratne C. J., Jin L. J., Samaranayake Y. H., and Samaranayake L. P. (2008) Cell density and cell aging as factors modulating antifungal resistance of Candida albicans biofilms. Antimicrobial Agents Chemother. 52, 3259–3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. National Committee for Clinical Laboratory Standards. (2002) Reference method for broth dilution antifungal susceptibility testing of yeasts—second edition: Approved standard M27-A2. Natl. Comm. Clin. Lab. Standards Wayne, PA [Google Scholar]
- 16. Bink A., Vandenbosch D., Coenye T., Nelis H., Cammue B. P., and Thevissen K. (2011) Superoxide dismutases are involved in Candida albicans biofilm persistence against miconazole. Antimicrobial Agents Chemother. 55, 4033–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vylkova S., Jang W. S., Li W., Nayyar N., and Edgerton M. (2007) Histatin 5 initiates osmotic stress response in Candida albicans via activation of the Hog1 mitogen-activated protein kinase pathway. Eukaryotic Cell 6, 1876–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ghosh D., Li Z., Tan X. F., Lim T. K., Mao Y., and Lin Q. (2013) iTRAQ based quantitative proteomics approach validated the role of calcyclin binding protein (CacyBP) in promoting colorectal cancer metastasis. Mol. Cell. Proteomics 12, 1865–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gan C. S., Chong P. K., Pham T. K., and Wright P. C. (2007) Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ). J. Proteome Res. 6, 821–827 [DOI] [PubMed] [Google Scholar]
- 20. Eisen M. B., Spellman P. T., Brown P. O., and Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maere S., Heymans K., and Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 23. Li P., Seneviratne C. J., Alpi E., Vizcaino J. A., and Jin L. (2015) Delicate Metabolic Control and Coordinated Stress Response Critically Determine Antifungal Tolerance of Candida albicans Biofilm Persisters. Antimicrobial Agents Chemother. 59, 6101–6112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wiese S., Reidegeld K. A., Meyer H. E., and Warscheid B. (2007) Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics 7, 340–350 [DOI] [PubMed] [Google Scholar]
- 25. Gao Y., Lim T. K., Lin Q., and Li S. F. (2016) Identification of cypermethrin induced protein changes in green algae by iTRAQ quantitative proteomics. J. Proteomics 139, 67–76 [DOI] [PubMed] [Google Scholar]
- 26. Lunt S. Y., and Vander Heiden M. G. (2011) Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 27, 441–464 [DOI] [PubMed] [Google Scholar]
- 27. Vander Heiden M. G., Cantley L. C., and Thompson C. B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chaffin W. L., Lopez-Ribot J. L., Casanova M., Gozalbo D., and Martinez J. P. (1998) Cell wall and secreted proteins of Candida albicans: identification, function, and expression. Microbiol. Mol. Biol. Rev. 62, 130–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mitri S., Xavier J. B., and Foster K. R. (2011) Social evolution in multispecies biofilms. Proc. Natl. Acad. Sci. U.S.A. 108, 10839–10846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mesa-Arango A. C., Trevijano-Contador N., Roman E., Sanchez-Fresneda R., Casas C., Herrero E., Arguelles J. C., Pla J., Cuenca-Estrella M., and Zaragoza O. (2014) The production of reactive oxygen species is a universal action mechanism of Amphotericin B against pathogenic yeasts and contributes to the fungicidal effect of this drug. Antimicrobial Agents Chemother. 58, 6627–6638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Seneviratne C. J., Wang Y., Jin L., Abiko Y., and Samaranayake L. P. (2008) Candida albicans biofilm formation is associated with increased anti-oxidative capacities. Proteomics 8, 2936–2947 [DOI] [PubMed] [Google Scholar]
- 32. Chauhan N., Inglis D., Roman E., Pla J., Li D., Calera J. A., and Calderone R. (2003) Candida albicans response regulator gene SSK1 regulates a subset of genes whose functions are associated with cell wall biosynthesis and adaptation to oxidative stress. Eukaryotic Cell 2, 1018–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chauhan N., Latge J. P., and Calderone R. (2006) Signalling and oxidant adaptation in Candida albicans and Aspergillus fumigatus. Nat. Rev. Microbiol. 4, 435–444 [DOI] [PubMed] [Google Scholar]
- 34. Dantas Ada S., Day A., Ikeh M., Kos I., Achan B., and Quinn J. (2015) Oxidative stress responses in the human fungal pathogen, Candida albicans. Biomolecules 5, 142–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Park Y. N., and Morschhauser J. (2005) Tetracycline-inducible gene expression and gene deletion in Candida albicans. Eukaryotic Cell 4, 1328–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wimhurst J. M., and Manchester K. L. (1973) Induction and suppression of the key enzymes of glycolysis and gluconeogenesis in isolated perfused rat liver in response to glucose, fructose and lactate. Biochem. J. 134, 143–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kitzing K., Auweter S., Amrhein N., and Macheroux P. (2004) Mechanism of chorismate synthase. Role of the two invariant histidine residues in the active site. J. Biol. Chem. 279, 9451–9461 [DOI] [PubMed] [Google Scholar]
- 38. Holland M. J., and Westhead E. W. (1973) Purification and characterization of aspartic -semialdehyde dehydrogenase from yeast and purification of an isozyme of glyceraldehyde-3-phosphate dehydrogenase. Biochemistry 12, 2264–2270 [DOI] [PubMed] [Google Scholar]
- 39. Gautam P., Upadhyay S. K., Hassan W., Madan T., Sirdeshmukh R., Sundaram C. S., Gade W. N., Basir S. F., Singh Y., and Sarma P. U. (2011) Transcriptomic and proteomic profile of Aspergillus fumigatus on exposure to artemisinin. Mycopathologia 172, 331–346 [DOI] [PubMed] [Google Scholar]
- 40. Gautam P., Shankar J., Madan T., Sirdeshmukh R., Sundaram C. S., Gade W. N., Basir S. F., and Sarma P. U. (2008) Proteomic and transcriptomic analysis of Aspergillus fumigatus on exposure to amphotericin B. Antimicrobial Agents Chemother. 52, 4220–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang L., Zhang Y., Zhou Y., An S., Zhou Y., and Cheng J. (2002) Response of gene expression in Saccharomyces cerevisiae to amphotericin B and nystatin measured by microarrays. J. Antimicrob. Chemother. 49, 905–915 [DOI] [PubMed] [Google Scholar]
- 42. Seneviratne C. J., Wang Y., Jin L., Wong S. S., Herath T. D., and Samaranayake L. P. (2012) Unraveling the resistance of microbial biofilms: has proteomics been helpful? Proteomics 12, 651–665 [DOI] [PubMed] [Google Scholar]
- 43. Baillie G. S., and Douglas L. J. (2000) Matrix polymers of Candida biofilms and their possible role in biofilm resistance to antifungal agents. J. Antimicrob. Chemother. 46, 397–403 [DOI] [PubMed] [Google Scholar]
- 44. Hawser S. P., and Douglas L. J. (1995) Resistance of Candida albicans biofilms to antifungal agents in vitro. Antimicrobial Agents Chemother. 39, 2128–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee J., Spector D., Godon C., Labarre J., and Toledano M. B. (1999) A new antioxidant with alkyl hydroperoxide defense properties in yeast. J. Biol. Chem. 274, 4537–4544 [DOI] [PubMed] [Google Scholar]
- 46. Iwai K., Naganuma A., and Kuge S. (2010) Peroxiredoxin Ahp1 acts as a receptor for alkylhydroperoxides to induce disulfide bond formation in the Cad1 transcription factor. J. Biol. Chem. 285, 10597–10604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sun X., Lu H., Jiang Y., and Cao Y. (2013) CaIPF19998 reduces drug susceptibility by enhancing the ability of biofilm formation and regulating redox homeostasis in Candida albicans. Curr. Microbiol. 67, 322–326 [DOI] [PubMed] [Google Scholar]
- 48. Phillips A. J., Sudbery I., and Ramsdale M. (2003) Apoptosis induced by environmental stresses and amphotericin B in Candida albicans. Proc. Natl. Acad. Sci. U.S.A. 100, 14327–14332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu N., He L., Cui P., Wang W., Yuan Y., Liu S., Xu T., Zhang S., Wu J., Zhang W., and Zhang Y. (2015) Ranking of persister genes in the same Escherichia coli genetic background demonstrates varying importance of individual persister genes in tolerance to different antibiotics. Front. Microbiol. 6, 1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tanaka K., Lin B. K., Wood D. R., and Tamanoi F. (1991) IRA2, an upstream negative regulator of RAS in yeast, is a RAS GTPase-activating protein. Proc. Natl. Acad. Sci. U.S.A. 88, 468–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tanaka K., Nakafuku M., Tamanoi F., Kaziro Y., Matsumoto K., and Toh-e A. (1990) IRA2, a second gene of Saccharomyces cerevisiae that encodes a protein with a domain homologous to mammalian ras GTPase-activating protein. Mol. Cell. Biol. 10, 4303–4313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Alberts A. S., Bouquin N., Johnston L. H., and Treisman R. (1998) Analysis of RhoA-binding proteins reveals an interaction domain conserved in heterotrimeric G protein beta subunits and the yeast response regulator protein Skn7. J. Biol. Chem. 273, 8616–8622 [DOI] [PubMed] [Google Scholar]
- 53. Yan C., Lee L. H., and Davis L. I. (1998) Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J. 17, 7416–7429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Goitre L., Trapani E., Trabalzini L., and Retta S. F. (2014) The Ras superfamily of small GTPases: the unlocked secrets. Methods Mol. Biol. 1120, 1–18 [DOI] [PubMed] [Google Scholar]
- 55. Vizcaino J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.