Abstract
Chondrocytes are the sole cell type found in articular cartilage and are repeatedly subjected to mechanical loading in vivo. We hypothesized that physiological dynamic compression results in changes in energy metabolism to produce proteins for maintenance of the pericellular and extracellular matrices. The objective of this study was to develop an in-depth understanding for the short term (<30 min.) chondrocyte response to sub-injurious, physiological compression by analyzing metabolomic profiles for human chondrocytes harvested from femoral heads of osteoarthritic donors. Cell-seeded agarose constructs were randomly assigned to experimental groups, and dynamic compression was applied for 0, 15, or 30 minutes. Following dynamic compression, metabolites were extracted and detected by HPLC-MS. Untargeted analyses examined changes in global metabolomics profiles and targeted analysis examined the expression of specific metabolites related to central energy metabolism. We identified hundreds of metabolites that were regulated by applied compression, and we report the detection of 16 molecules not found in existing metabolite databases. We observed patient-specific mechanotransduction with aging dependence. Targeted studies found a transient increase in the ratio of NADP+ to NADPH and an initial decrease in the ratio of GDP to GTP, suggesting a flux of energy into the TCA cycle. By characterizing metabolomics profiles of primary chondrocytes in response to applied dynamic compression, this study provides insight into how OA chondrocytes respond to mechanical load. These results are consistent with increases in glycolytic energy utilization by mechanically induced signaling, and add substantial new data to a complex picture of how chondrocytes transduce mechanical loads.
Keywords: Mechanotransduction, metabolomics, osteoarthritis, systems biology, chondrocyte biology
Introduction
Osteoarthritis (OA) is the most common joint disorder, affecting more than 40 million individuals in the United States [1, 2]. OA is characterized by deterioration of the protective, low-friction, load-bearing cartilage that surrounds the joint. The highly specialized chondrocyte plays an important metabolic role in synthesizing, maintaining, and repairing the tissue and is the sole cell type in articular cartilage [3]. At these joint surfaces (e.g. the femoral head), the articular chondrocyte is subjected to repeated mechanical loading (e.g. standing, walking, running, etc.). Chondrocytes and other mammalian cells sense and respond to these mechanical stimuli through biochemical and biological outputs, but the intracellular pathways behind chondrocyte mechanotransduction remain unclear [4-6].
Mechanical stimulation has both anabolic and catabolic effects on articular chondrocytes [7, 8]. In chondrocytes, anabolic responses promote synthesis and production of extracellular matrix (ECM) and pericellular matrix (PCM) [7]. Catabolic responses involve secretion of proteases (e.g. MMP-13) which results in the breakdown of ECM molecules. Dynamic loading has been shown to promote these anabolic responses in chondrocytes; whereas, static loading has been shown to inhibit them [9, 10]. Dynamic compression has been shown to alter signal transduction including activation of GTPase signaling via the Rho-A and ROCK pathways, Erk-1 and -2, MAPK and SEK, and Smad2 [9, 11-13].
In addition to signal transduction, chondrocytes can alter their energy metabolism in response to mechanical loading. The enzyme AKT is important in regulating FoxO signaling for energy homeostasis [14], and cyclic loading has been shown to reduce phosphorylation of AKT in OA chondrocytes [15]; whereas, mechanical stimulation induces AKT phosphorylation in healthy cells [16]. These energy-related signaling responses may affect matrix synthesis because healthy chondrocytes show increases in sulfated glycosaminoglycans (sGAG) in response to mechanical loading in contrast to OA chondrocytes [17]. However, how loading might change chondrocyte metabolite levels, which can mark changes in biosynthetic activity, is unknown.
Previous research suggests that both inflammation and OA alter central energy metabolism, including the balance between glycolysis and oxidative phosphorylation [18]. One potential mechanism of energy-related mechanotransduction involves regulation of AMP-activated protein kinase which can prevent catabolism induced by mechanical injury [19]. Based on these and other data, we hypothesized that physiological dynamic compression increases chondrocyte glycolytic energy flux to promote the anabolic response to maintain the environment of the ECM and PCM. The objective of this study was to use both untargeted (global) and targeted metabolomics to identify candidate mediators of chondrocyte mechanotransduction in primary human OA chondrocytes. This study evaluates our hypothesis using primary human OA chondrocytes subjected to applied dynamic compression following encapsulation in physiologically stiff agarose.
Chondrocyte mechanotransduction happens on both short- and long- timescales. Because early responses have the potential to set the trajectory for longer-term behavior, in this study, we focus on short term (<30 min), loading-induced changes in small molecules (i.e. cellular metabolites smaller than ∼1000 Da). Building upon previous methodology to encapsulate chondrocytes in agarose similar in stiffness to the human PCM, we profiled the metabolomic responses of primary human OA chondrocytes in response to applied compression at physiological levels [20, 21].
This study identified changes in 1421 untargeted metabolites and 48 metabolites pertinent to central energy metabolites and protein production. Metabolites were identified via database searches (METLIN, and HMDB), and 16 of the 1421 metabolites were not found in database searches, potentially representing novel mediators of chondrocyte mechanotransduction. Future research may build on these results to test mechanical loading as an OA therapy.
Materials and Methods
Chondrocyte Culture and Encapsulation
Primary human chondrocytes were harvested from hip cartilage of five Grade IV OA patients undergoing joint replacement surgery (mean age: 63 years, age range: 54-80, mean weight: 80.4 kg, weight range: 56.9-99 kg). The cartilage was digested in Type IV collagenase (2 mg/mL for 12-14 hrs. at 37°C), and then cultured in DMEM with 10% fetal bovine serum and antibiotics (10,000 I.U./mL penicillin and 10000 μg/mL streptomycin) in 5% atmospheric CO2. Cells were encapsulated using previously optimized methods [22] at a concentration of ∼500,000 cells/gel (gel diameter = 7mm, gel height = 12.7mm).
Mechanical Stimulation
From each donor (N = 5), cell-seeded agarose gels were randomly assigned to a loading group (n = 5 biological replicates for each loading group). Loading groups consisted of unloaded controls (i.e. 0 minutes of loading), 15, or 30 minutes of dynamic, cyclic compression. A short loading timescale was used because initial early-time responses can set the trajectory for longer-term behavior. The loading protocol followed previously optimized methods [22] in which homogeneous deformations [20] were applied to the cell-seeded gels using a custom built bioreactor emulating physiological loading conditions: frequency of 1.1 Hz and average sinusoidal compressive strains of 5% with an amplitude of 1.9% based on initial gel height (Supplementary Figure 1). The loading frequency was selected based on the preferred stride rate in humans [23], and the applied strain profile was selected to be in the range of deformations measured in human patients using MRI [24].
Metabolite Extraction
Metabolite extraction was performed using identical methods from our previous study [22]. Gels were flash frozen, pulverized, and metabolites were extracted by adding 1 mL of a 70:30 Methanol:Acetone solution and vigorously vortexing the mixture every 5 minutes for 20 minutes. Samples were kept at -20°C overnight for further metabolite extraction. Proteins were removed by centrifugation, the supernatant extracted, and the solvent removed via centrifugation under a vacuum for 6.5 hours. The dried samples were then resuspended in 100 μL of mass spectrometry grade water and acetonitrile (50:50 v/v).
Untargeted and Targeted Metabolomic Profiling
Metabolomics is an experimental technique for characterizing large numbers of small molecules (<1000 Da) in biological samples [25]. Recent studies of joint tissues and fluids have used metabolomic analysis to examine OA phenotypes, identify candidate biomarkers, and explore the inflammatory responses [26-29]. In this study, metabolites were extracted following dynamic compression and analyzed by nano-liquid chromatography and mass spectrometry (Supplementary Methods, [22]). Untargeted metabolites were detected in positive mode on an Agilent 6538 Q-TOF spectrometer with a resolution of ∼20,000 and accuracy of ∼5 ppm. For the targeted approach, ∼50 metabolites known to be involved in central energy metabolism were analyzed using the Quantitative Analysis package within the Agilent MassHunter Workstation B.04.00 (Agilent Technologies) and a database of the calculated isotopic distributions (including H+ and Na+ adducts) of these targeted masses (Isotope Distribution Calculator, Agilent Technologies).
To assess the effects of physiological loading on chondrocyte biology, three experimental groups were used: unloaded control samples (UC) and samples that received either 15 (DL15) or 30 (DL30) minutes of dynamic compression. Principal components analysis (PCA) was utilized to assess metabolome-scale changes caused by mechanical loading. Pearson's correlation coefficients were used to estimate the flux of metabolite intensities over the timecourse of loading. To assess the differences in intensity distributions (m/z spectra plots for the various loading groups), two-sample Kolmogorov-Smirnov tests were implemented. Targeted metabolite profiles were analyzed by PCA, hierarchical agglomerative cluster analysis and correlation analyses. Additionally, the median ratios of NADP+:NADPH, NAD+:NADH, ATP:ADP, and GDP:GTP were calculated as a function of time to assess relative changes in energy metabolism.
Compound Identification and Enrichment Analysis
To putatively identify compounds, a batch search of all of the untargeted metabolite mass to charge (m/z) values was performed in METLIN and HMDB. Both databases contain over 80,000 identifiable metabolites [30, 31]. Search parameters included using a mass tolerance of 20 ppm, and positively charged molecules with potential +1H+ or +1Na+ adducts. Compounds with LipidMAPS identifications [32] were designated as human and not considered further if detected in a non-human species at the time of database search. Untargeted metabolites were then examined by unsupervised clustering to identify groups of co-regulated metabolites. Clusters of co-regulated metabolites were analyzed for pathway enrichment using IMPaLA [33].
Results
The objective of this study was to characterize the cellular response to applied compression for primary human OA chondrocytes by examining changes in metabolomic profiles. Chondrocytes were harvested from donor joint tissue, grown in tissue culture, embedded in agarose with stiffness similar to the human PCM, and dynamically loaded in tissue culture. Samples were then flash frozen, pulverized, and the metabolites were extracted and analyzed using HPLCMS. We analyzed untargeted metabolomics profiles to minimize bias from a priori selection of relevant pathways which inherently excludes potentially important data. To evaluate our hypothesis, we also analyzed metabolomics profiles of targeted metabolites related to energy metabolism and protein production.
These data demonstrate that applied dynamic compression alters both untargeted and targeted metabolomic responses of primary human OA chondrocytes in vitro. Using a systems biology approach, we find key differences between loaded and unloaded OA chondrocytes and identified 70 metabolites as potential mediators for chondrocyte mechanotransduction.
Untargeted Analysis
We detected 4955 metabolites present in at least one dataset (Table S1). We refined the 4955 metabolite list to 1421 metabolites by focusing on metabolites present in more than half of the samples for each loading group. To determine the effects of dynamic compression on chondrocyte metabolism, unloaded control samples were compared to samples subjected to either 15 or 30 minutes of dynamic compression. Statistical analyses found loading-associated differences that were both shared between human donors (n = 5) and specific to individual donors (examined with 5 replicates per donor). Enrichment analysis of detected metabolites identified galactose metabolism, chondroitin sulfate degradation, lipid and amino acid metabolism as chondrocyte pathways affected by mechanical loading (Table 1).
Table 1.
Pathways and metabolites altered by mechanical loading. Clusters refer to untargeted clusters identified in Figure 1A.
| Pathways | Cluster | Mechanosensitive Metabolites [KEGG ID] |
|---|---|---|
| Galactose metabolism | 1 | C06376;C00159;C00795;C00267;C01132;C00095;C00124;C00137 |
| 4 | C00243;C05400;C01235;C05402;C00089 | |
| Chondroitin sulfate degradation | 1 | C02336;C00267;C04053;C00532;C10906;C00379;C01132;C03878;C00221; C01904 |
| 3 | C00252;C01083;C00243;C02713;C08240;C08250;C05402;C00089 | |
| 4 | C00252;C01083;C00243;C08240;C08250;C05402;C00089 | |
| Lipid synthesis, transport, and metabolism | 1 | C00267;C01132;C00588;C01582 |
| 2 | C00588;C00157;C00246;C06429 | |
| 3 | C00350;C00157 | |
| 4 | C04230;C00157 | |
| Glycine, serine, and threonine metabolism | 3 | C00263;C05519;C06231;C00188 |
| 4 | C05519;C00263;C00188 |
Cluster 1: increase with loading. Cluster 2: increase after 30 min. loading. Cluster 3: decrease with loading. Cluster 4: Increase after 15 min. loading
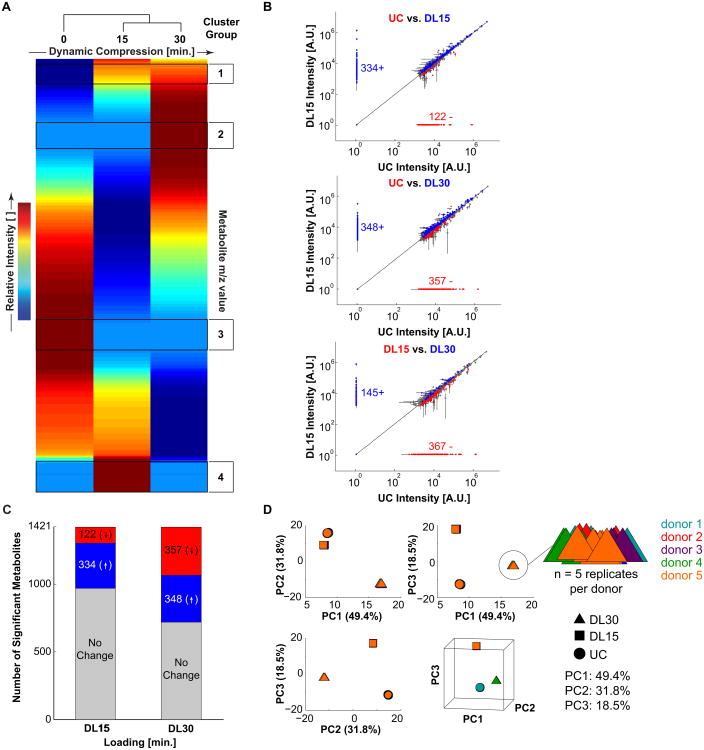
Unsupervised clustering identified four clusters of interest in the untargeted metabolomic profiles (Figure 1A). These clusters suggest that pathways related to calcium signaling, energy metabolism, redox regulation, amino acid and lipid metabolism are regulated by mechanical loading (Tables S2-S5). Comparisons between unloaded control (UC) and DL15 samples revealed changes in 456 metabolites (Figure 1B-C, Table S6). Of these 456 metabolites, 334 were increased (↑) with dynamic loading and 122 were decreased (↓) in response to loading (Figure 1B-C). Comparisons between UC and DL30 found changes in 705 metabolites, of which 348 increased and 357 decreased, in response to dynamic compression (Figure 1B-C, Table S7). Finally, we found differences between DL15 and DL30 groups in 512 metabolites. 145 metabolites increased and 367 decreased for 15 minutes of compression compared with 30 minutes of compression (Figure 1B-C, Table S8).
Figure 1. Applied compression resulted in distinct untargeted metabolomic profiles for primary OA chondrocytes.
(A) Patterns of metabolite expression displayed via heatmap following hierarchical clustering. Clusters of interest denoted on the right-hand side (see Table 1). for enrichment analysis).Cluster 1: increasing with loading. Cluster 2: on after 30 min. Cluster 3: off with loading. Cluster 4: on at 15 min. (B) Loading-induced up-down regulation plots for UC vs. DL15, UC vs. DL30, and DL15 vs. DL30, respectively. Up regulated metabolites represent molecules that accumulate with respect to mechanical loading, and down regulated metabolites are being depleted with respect to mechanical loading. (C) Of the 1421 metabolites in common to all groups, 334 were upregulated and 122 were downregulated after 15 minutes of compression. 348 were upregulated and 357 were downregulated after 30 minutes of compression. (D) Principal Components Analysis for the untargeted data to assess differences between sample groups. The first three principal components contained 99.7% of the overall variance. For each of the sample groups, UC (●), DL15 (■), and DL30 (▲), there are n = 5 donors each sampled with 5 replicates.
These differences in mechanically-induced metabolomic profiles are supported by both principal component analysis (PCA) and comparisons of metabolite distributions between experimental groups. The log-transformed and normalized metabolite intensity data showed distinct separation in the first 3 principal components between the experimental groups (UC, DL15, and DL30), with the first three principal components containing 99.7% of the overall variance (Figure 1D). Kolmogorov-Smirnov two-sample distribution tests revealed significant differences among distributions of the median metabolite expression levels for various loading groups (Supplementary Figure 2).
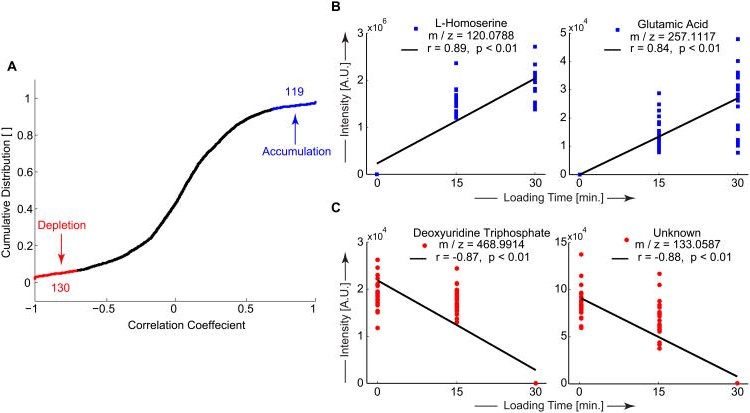
Correlation analysis examined the effects of dynamic compression (0, 15, or 30 minutes) on metabolite expression levels (Figure 2). There were 249 statistically significant metabolites that correlated with loading time. There were 119 positively correlated metabolites (i.e. accumulated) and 130 negatively correlated (i.e. depleted) with loading time (Figure 2A). The strongest positively and negatively correlated metabolites were defined as candidate mediators of chondrocyte mechanotransduction (Table S9).
Figure 2. Dynamic compression results in both accumulation and depletion of untargeted metabolites.
(A) The cumulative distribution of correlation coefficients of untargeted metabolites was used to identify metabolites that were either accumulating (blue) or being depleted (red). The strongest correlated metabolites were identified as candidate mediators of chondrocyte mechanotransduction. (B and C) Metabolite expression levels versus loading time used for correlation analysis. (B) Accumulation of selected candidate mediators of mechanotransduction which were database-matched as L-homoserine (left) and glutamic acid (right). (C) Depletion of selected candidate mediators of mechanotransduction. Deoxyuridine triphosphate as matched in the METLIN database (left) and a metabolite with m/z = 133.0587 which did not match any database molecules (unknown, right).
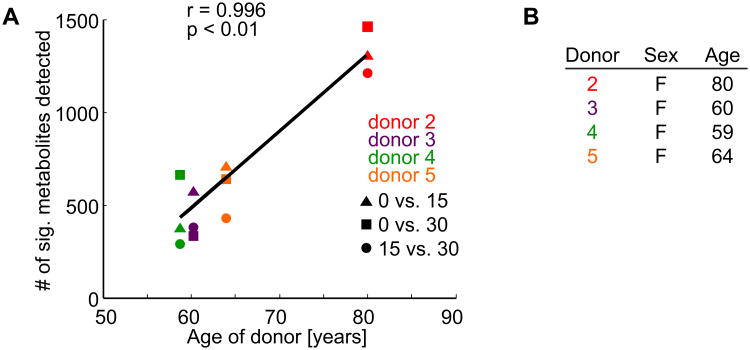
The number of significant mechanosensitive metabolites correlated with patient age (Figures 3-4, and Supplementary Figure 3). Different donors displayed heterogeneity in their responses to applied compression. For all donors, (1-5), both 15 and 30 minutes of dynamic compression resulted in hundreds of statistically significant changes in metabolite expression (Figure 4). These results indicate that both shared and patient-specific responses to applied compression are affected by aging.
Figure 3. Aging-related chondrocyte mechanotransduction in female OA chondrocytes.
We observed a significant correlation (r = 0.996, p < 0.01) when comparing the number of significant mechanosensitive metabolites with the age of the femoral head donor. (A) Age-correlated increases in the number of significant metabolites. (B) Characteristics of femoral head articular cartilage used in the correlation analysis. Note that Donor 1 was male and is included in Supplementary Figure 3.
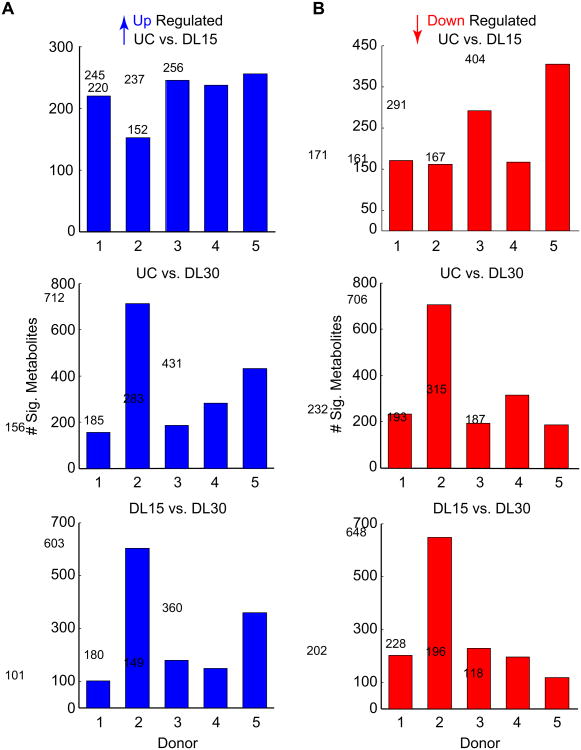
Figure 4. Patient-specific heterogeneity in chondrocyte mechanotransduction.
Upon compression, chondrocytes from each donor resulted in different numbers of significant mechanically up- (A) or down- (B) regulated metabolites. These differences may stem from variations in both phenotype of end-stage osteoarthritis and patient-specific differences in the cellular response to mechanical loading.
Targeted Analysis
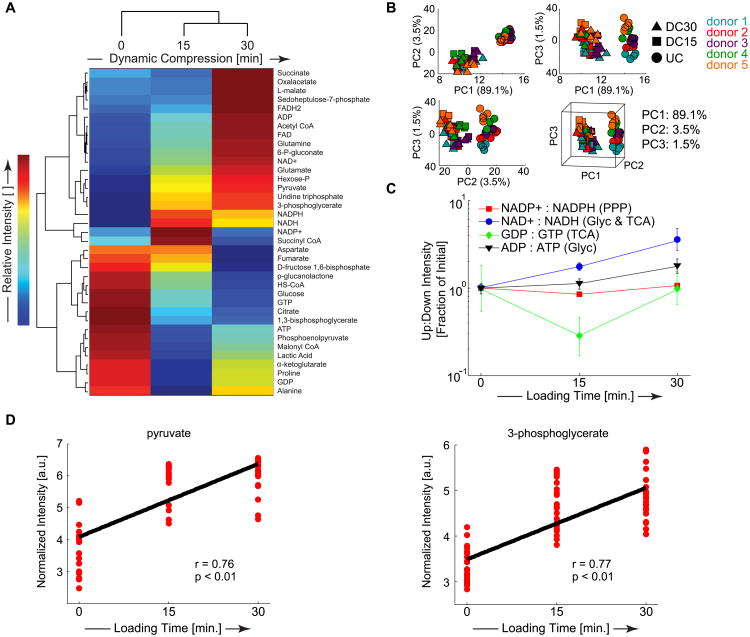
By focusing on metabolites common to central energy metabolism and protein production, we examined our hypothesis that physiological dynamic compression increases chondrocyte glycolytic energy flux to promote the anabolic response to maintain the environment of the ECM and PCM. In this analysis, central energy metabolism focused on glucose metabolism including the pentose phosphate pathway (PPP), glycolysis (Glyc), and the tricarboxylic acid (TCA) cycle (Table S10). Cluster analysis identified groups of metabolites with similar responses to applied dynamic compression (Figure 5A). PCA on targeted metabolites revealed that central energy metabolites are strongly regulated by applied compression (Figure 5B). Targeted metabolomics profiles from mechanically loaded groups clustered separately from unloaded controls, and the first three principal components contained 94.1% of the overall variance (1st component: 89.1%, 2nd component: 3.5%, and 3rd component: 1.5%).
Figure 5. Loading-induced changes in expression of targeted metabolites specific to central-energy-metabolism.
Metabolites associated with central energy metabolism and protein production (e.g. amino acids) were targeted for detailed analysis via detection of multiple ionized adducts and ions. (A) Hierarchical agglomerative cluster analysis reveals changes in metabolite intensities with respect to increased dynamic loading. (B) Principal Components Analysis for the targeted data was used for comparing differences/separations among sample groups. The first three principal components contained 94.1% of the overall variance. For each of the sample groups, UC (●), DL15 (■), and DL30 (▲), there were n = 5 donors sampled 5 times. (C) Ratios of upstream to downstream mediators of central energy metabolism. PPP = pentose phosphate pathway. Glyc = Glycolysis. TCA = Tricarboxylic Acid Cycle. (D) Significant accumulation of pyruvate and 3-phosphoglycerate in response to applied dynamic compression.
To estimate changes in energy flux through the PPP, Glyc, and the TCA cycle, the median ratios of upstream to downstream expression levels for specific targeted metabolites were analyzed (Figure 5C). The ratio of NADP+ to NADPH decreased after 15 minutes of loading prior to increasing after 30 minutes of loading, indicating an initial cessation in energy flow which then increases for the PPP. We observed continual increases in the ratio of ADP to ATP, indicating increased glycolytic energy flux consistent with our hypothesis. Finally, we found a substantial decrease in the ratio of GDP to GTP after 15 minutes that was restored after 30 minutes of loading, indicating an initial cessation of TCA-based energy flux that recovers with additional mechanical loading. Both pyruvate and 3-phosphoglycerate were positively correlated with dynamic loading time (Figure 5D, p < 0.01). Taken together, these results demonstrate that mechanical loading strongly regulates chondrocyte energy metabolism with upregulation of glycolytic energy flux and transient decreases in energy flow to both the PPP and the TCA cycle that are restored following 30 minutes of loading.
Discussion
Previous studies have found altered mechanotransduction between normal and OA chondrocytes. Kawakita et al. found a significant reduction of Akt phosphorylation in diseased (OA) chondrocytes in response to cyclic loading; whereas, Niehoff et al. found that cyclic loading induced Akt phosphorylation in healthy chondrocytes. Akt is serine/threonine kinase plays an important role in a variety of chondrocyte cellular mechanisms, including cell apoptosis [34], cell proliferation and growth [35], and proteoglycan synthesis [36]. Holledge et al. found that, in response to 0.33 Hz oscillatory fluid flow in monolayer, sulfated glycosaminoglycan production for normal chondrocytes was significantly higher than for OA chondrocytes. These studies emphasize the differences in cellular phenotypes between normal and OA chondrocytes in response to mechanical stimulation.
We applied dynamic compression to primary chondrocytes and observed both increases and decreases in hundreds of untargeted metabolites. These metabolomic changes represent an altered cellular phenotype resulting from exogenous, dynamic loading. The ability of metabolomic characterization to capture chondrocyte responses to compression is highlighted by our observation that, when comparing between 0, 15, and 30 minutes of loading, 99.7% of the variance is contained in the first three principal components (Figure 1D). The importance of compression in regulating chondrocyte behavior is further emphasized by numerous differences between unloaded and loaded samples (Figures 1-2, and Supplementary Figure 2). While this study describes the early (<30 minute) response to compression, one limitation is that the long-term (e.g. 24 hour) response remains to be determined. These significant differences demonstrate both the importance of mechanical loading to chondrocyte biology and the ability of metabolomics to describe large-scale changes in chondrocyte physiology.
The metabolome is defined as the set of small-molecules found within a biological sample including substrates, co-factors, and other cytosolic molecules that comprise the physiology of cellular biochemistry [37]. The metabolome can be viewed as a collection of state variables describing the cellular phenotype [25]. These measurements provide valuable insight into how chondrocytes sense and respond to applied loading. Future studies may build on this untargeted dataset by creating mathematical models (e.g. Hidden Markov Models) and by comparing differences in specific mechanically-induced metabolite expression between normal and OA cells to identify candidate pathways for therapeutic intervention. For example, if specific mechanosensitive pathways are found that enable anabolic processes in chondrocytes, mechanical loading (i.e. exercise) could be administered as an effective therapy for healing damaged cartilage. Many therapies available to OA patients (e.g. joint replacement) are extremely expensive, and inaccessible to certain patient populations. Extending these results to determine optimal daily loading of cartilage (e.g. exercise) as a treatment for OA would be extremely valuable to combat OA and associated comorbidities simultaneously.
In this study, we identify mechanosensitive pathways using enrichment analysis of clusters of co-regulated metabolites (Figure 1A, Table 1, Tables S2-S5). Dynamic compression notably decreased expression of several metabolites compared with unloaded controls (Figure 1A, Cluster 3), including valine, leucine, and isoleucine biosynthesis. Several other metabolites increase dynamic loading. These metabolites were enriched for ion-channel signaling and glutathione synthesis, which are consistent with prior chondrocyte data [38, 39] and support physiological compression as a mechanism to produce cartilage matrix. Riboflavin metabolism and glycogenoloysis also gradually increased with loading, suggesting that cellular energy utilization can be altered by the mechanical environment [40, 41].
Additionally, there was increased expression of metabolites specific to either 15 or 30 minutes of dynamic compression. Fifteen minutes of dynamic compression resulted in glycine, serine, and threonine metabolism (Cluster 4). Thirty minutes of dynamic compression resulted in upregulation of metabolites enriched for amino acid and lipid metabolism (Figure 1A, Cluster 2). Metabolites relating to phenylalanine, tyrosine, and tryptophan synthesis were upregulated, in addition to those for arginine and proline metabolism, consistent with biosynthetic effects of dynamic compression.
Certain pathways were represented in multiple clusters (Table 1). This result may represent differential regulation of distinct pathway components (e.g. upstream vs. downstream) or technical limitations. Technical limitations include the inability of mass spectrometry to distinguish between isomers (e.g. changes in phospholipid structure) or between identified masses within 20 ppm database search tolerances. Dynamic compression appeared to increase remodeling of chondroitin sulfate and increase glycine, serine, and threonine metabolism, emphasizing the importance of loading to matrix remodeling. Several metabolites related to lipid synthesis, transport, and metabolism were affected, suggesting cell membrane alterations in response to dynamic compression [42]. Finally, galactose metabolism was upregulated by loading, further suggesting the importance of energy metabolism in defining how chondrocytes respond to loading.
Because cartilage is a viscoelastic material, cyclical loading results in internal energy dissipation within the tissue [43, 44]. To date, it is unknown whether or not applied mechanical deformation can alter cellular energy utilization. We hypothesized that physiological dynamic compression will increase glycolytic energy flux to promote the anabolic response in chondrocytes to maintain the environment of the ECM and PCM. This project provides fundamental data for understanding potential mechanical-to-cellular energy conversion by quantifying relative levels of metabolites targeted to central energy metabolism (Figure 5).
Hierarchical clustering clearly demonstrated that dynamic compression regulates energy-related metabolites, and we found accumulation of key amino acid precursors, including pyruvate and 3-phosphoglycerate, which may indicate increased glycolytic flux (Figure 5C). These glycolytic changes were also marked by the consumption of fructose-1,6-bisphosphate and 1,3-bisphosphoglycerate which may serve to increase pyruvate flux for maximizing protein synthesis. Applying a stoichiometric matrix model for flux balance analysis will enable full evaluation of our hypothesis. By simultaneously analyzing the complete targeted dataset, we could infer whether observed consumption and accumulation indicate increased or decreased metabolic flux [45]. The significant changes in expression of these specific metabolites in response to dynamic loading may indicate energy utilization towards matrix protein production.
In this study, chondrocytes were harvested from hip replacement patients with Grade IV osteoarthritis. Future studies should compare these data from OA chondrocytes to the healthy chondrocyte response in an age- and gender-matched manner. In our analysis, we found a strong correlation between the number of mechanically sensitive metabolites and patient age (p < 0.01, r = 0.996). These data suggest a relationship between aging and mechanotransduction as measured by the chondrocyte metabolome in response to applied physiological compression. Future studies may identify differences in clinical patient populations by utilizing untargeted LC-MS-based metabolomics to identify biomarkers in osteoarthritis [46]. The candidate mediators identified in this study (Supplementary Tables) may prove useful as indicators of normal and OA mechanotransduction. To our knowledge, this is the first time that metabolomics has been used to assess the loading induced differences for primary human OA chondrocytes.
Conclusions
This study demonstrates the power of utilizing high-dimensional metabolomics as a tool for understanding chondrocyte mechanotransduction. In our targeted analysis, we discovered significant correlations with increased mechanical loading and central energy reorganization. Similarly, our untargeted analysis revealed hundreds of significant metabolites, including potential novel mediators for chondrocyte mechanotransduction via energy, lipid, and amino acid metabolism. Finally, we found a positive correlation between the number of mechanically-induced metabolites and patient age, suggesting that metabolomics can characterize aging-dependent changes in chondrocyte mechanotransduction. Metabolomics may yield important biomarker candidates for tracking progression of OA in clinical populations. Future work will expand on these data to elucidate the mechanosensitive differences between OA and normal human chondrocytes.
Supplementary Material
Acknowledgments
We acknowledge Drs. Ross Carlson, Brian Bothner, and Edward Dratz, MSU, for critical insight provided during discussions. We thank Timothy Hamerly for generating and sharing the standards data. Funding was provided by NIH P20GM10339405S1, NSF 1342420, Montana State University, and the Murdock Charitable Trust. The funding sources were not involved in the design or execution of this study.
Footnotes
Conflict of Interest: Dr. June owns stock in Beartooth Biotech LLC which has no relationship to this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cicuttini FM, Wluka AE, Urquhart D, Tanamas SK, Wang Y. Epidemiology Should Not Be Forgotten in Osteoarthritis Imaging. Osteoarthritis Cartilage. 2011;19(9):1165–6. doi: 10.1016/j.joca.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Woolf AD, Erwin J, March L. The Need to Address the Burden of Musculoskeletal Conditions. Best Practice & Research in Clinical Rheumatology. 2012;26(2) doi: 10.1016/j.berh.2012.03.005. pp. [DOI] [PubMed] [Google Scholar]
- 3.Sophia Fox AJ, Bedi A, Rodeo SA. The Basic Science of Articular Cartilage: Structure, Composition, and Function. Sports Health. 2009;1(6):461–8. doi: 10.1177/1941738109350438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haudenschild DR, D'lima DD, Lotz MK. Dynamic Compression of Chondrocytes Induces a Rho Kinase-Dependent Reorganization of the Actin Cytoskeleton. Biorheology. 2008;45(3-4) pp. [PubMed] [Google Scholar]
- 5.Grygorczyk R, Furuya K, Sokabe M. Imaging and Characterization of Stretch-Induced Atp Release from Alveolar A549 Cells. J Physiol. 2013;591(Pt 5):1195–215. doi: 10.1113/jphysiol.2012.244145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaalouk DE, Lammerding J. Mechanotransduction Gone Awry. Nat Rev Mol Cell Biol. 2009;10(1):63–73. doi: 10.1038/nrm2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruiz-Romero C, Carreira V, Rego I, Remeseiro S, López-Armada MJ, Blanco FJ. Proteomic Analysis of Human Osteoarthritic Chondrocytes Reveals Protein Changes in Stress and Glycolysis. Proteomics. 2008;8(3):495–507. doi: 10.1002/pmic.200700249. [DOI] [PubMed] [Google Scholar]
- 8.Farnsworth NL, Antunez LR, Bryant SJ. Dynamic Compressive Loading Differentially Regulates Chondrocyte Anabolic and Catabolic Activity with Age. Biotechnol Bioeng. 2013 doi: 10.1002/bit.24860. pp. [DOI] [PubMed] [Google Scholar]
- 9.Bougault C, Aubert-Foucher E, Paumier A, Perrier-Groult E, Huot L, Hot D, Duterque-Coquillaud M, Mallein-Gerin F. Dynamic Compression of Chondrocyte-Agarose Constructs Reveals New Candidate Mechanosensitive Genes. PloS one. 2012;7(5):e36964. doi: 10.1371/journal.pone.0036964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buschmann MD, Gluzband YA, Grodzinsky AJ, Hunziker EB. Mechanical Compression Modulates Matrix Biosynthesis in Chondrocyte/Agarose Culture. J Cell Sci. 1995;108(Pt 4):1497–508. doi: 10.1242/jcs.108.4.1497. [DOI] [PubMed] [Google Scholar]
- 11.Haudenschild DR, Chen J, Pang N, Lotz MK, D'lima DD. Rho Kinase-Dependent Activation of Sox9 in Chondrocytes. Arthritis Rheum. 2010;62(1) doi: 10.1002/art.25051. pp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Croos JN, Jang B, Dhaliwal SS, Grynpas MD, Pilliar RM, Kandel RA. Membrane Type-1 Matrix Metalloproteinase Is Induced Following Cyclic Compression of in Vitro Grown Bovine Chondrocytes. Osteoarthritis Cartilage. 2007;15(11):1301–10. doi: 10.1016/j.joca.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 13.Fitzgerald JB, Jin M, Chai DH, Siparsky P, Fanning P, Grodzinsky AJ. Shear- and Compression-Induced Chondrocyte Transcription Requires Mapk Activation in Cartilage Explants. Journal of Biological Chemistry. 2008;283(11) doi: 10.1074/jbc.M708670200. pp. [DOI] [PubMed] [Google Scholar]
- 14.Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villen J, Becker EB, Dibacco S, De La Iglesia N, Gygi S, Blackwell TK, Bonni A. A Conserved Mst-Foxo Signaling Pathway Mediates Oxidative-Stress Responses and Extends Life Span. Cell. 2006;125(5):987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 15.Kawakita K, Nishiyama T, Fujishiro T, Hayashi S, Kanzaki N, Hashimoto S, Takebe K, Iwasa K, Sakata S, Nishida K, Kuroda R, Kurosaka M. Akt Phosphorylation in Human Chondrocytes Is Regulated by P53r2 in Response to Mechanical Stress. Osteoarthritis Cartilage. 2012;20(12):1603–9. doi: 10.1016/j.joca.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 16.Niehoff A, Offermann M, Dargel J, Schmidt A, Bruggemann GP, Bloch W. Dynamic and Static Mechanical Compression Affects Akt Phosphorylation in Porcine Patellofemoral Joint Cartilage. J Orthop Res. 2008;26(5):616–23. doi: 10.1002/jor.20542. [DOI] [PubMed] [Google Scholar]
- 17.Holledge MM, Millward-Sadler SJ, Nuki G, Salter DM. Mechanical Regulation of Proteoglycan Synthesis in Normal and Osteoarthritic Human Articular Chondrocytes--Roles for Alpha5 and Alphavbeta5 Integrins. Biorheology. 2008;45(3-4):275–88. [PubMed] [Google Scholar]
- 18.Nishida T, Kubota S, Aoyama E, Takigawa M. Impaired Glycolytic Metabolism Causes Chondrocyte Hypertrophy-Like Changes Via Promotion of Phospho-Smad1/5/8 Translocation into Nucleus. Osteoarthritis Cartilage. 2013;21(5):700–9. doi: 10.1016/j.joca.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 19.Petursson F, Husa M, June R, Lotz M, Terkeltaub R, Liu-Bryan R. Linked Decreases in Liver Kinase B1 and Amp-Activated Protein Kinase Activity Modulate Matrix Catabolic Responses to Biomechanical Injury in Chondrocytes. Arthritis Res Ther. 2013;15(4):R77. doi: 10.1186/ar4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zignego DL, Jutila AA, Gelbke MK, Gannon DM, June RK. The Mechanical Microenvironment of High Concentration Agarose for Applying Deformation to Primary Chondrocytes. J Biomech. 2013 doi: 10.1016/j.jbiomech.2013.10.051. pp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jutila AA, Zignego DL, Schell WJ, June RK. Encapsulation of Chondrocytes in High-Stiffness Agarose Microenvironments for in Vitro Modeling of Osteoarthritis Mechanotransduction. Ann Biomed Eng. 2014 doi: 10.1007/s10439-014-1183-5. pp. [DOI] [PubMed] [Google Scholar]
- 22.Jutila AA, Zignego DL, Hwang BK, Hilmer JK, Hamerly T, Minor CA, Walk ST, June RK. Candidate Mediators of Chondrocyte Mechanotransduction Via Targeted and Untargeted Metabolomic Measurements. Arch Biochem Biophys. 2014;545:116–23. doi: 10.1016/j.abb.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Umberger BR, Martin PE. Mechanical Power and Efficiency of Level Walking with Different Stride Rates. J Exp Biol. 2007;210(Pt 18):3255–65. doi: 10.1242/jeb.000950. [DOI] [PubMed] [Google Scholar]
- 24.Sutter EG, Widmyer MR, Utturkar GM, Spritzer CE, Garrett WE, Jr, Defrate LE. In Vivo Measurement of Localized Tibiofemoral Cartilage Strains in Response to Dynamic Activity. Am J Sports Med. 2015;43(2):370–6. doi: 10.1177/0363546514559821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: The Apogee of the Omics Trilogy. Nat Rev Mol Cell Biol. 2012;13(4):263–9. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W, Likhodii S, Zhang Y, Aref-Eshghi E, Harper PE, Randell E, Green R, Martin G, Furey A, Sun G, Rahman P, Zhai G. Classification of Osteoarthritis Phenotypes by Metabolomics Analysis. BMJ Open. 2014;4(11):e006286. doi: 10.1136/bmjopen-2014-006286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mickiewicz B, Heard BJ, Chau JK, Chung M, Hart DA, Shrive NG, Frank CB, Vogel HJ. Metabolic Profiling of Synovial Fluid in a Unilateral Ovine Model of Anterior Cruciate Ligament Reconstruction of the Knee Suggests Biomarkers for Early Osteoarthritis. J Orthop Res. 2015;33(1):71–7. doi: 10.1002/jor.22743. [DOI] [PubMed] [Google Scholar]
- 28.Attur M, Dave M, Abramson SB, Amin A. Activation of Diverse Eicosanoid Pathways in Osteoarthritic Cartilage: A Lipidomic and Genomic Analysis. Bull NYU Hosp Jt Dis. 2012;70(2):99–108. [PubMed] [Google Scholar]
- 29.Mickiewicz B, Kelly JJ, Ludwig TE, Weljie AM, Wiley JP, Schmidt TA, Vogel HJ. Metabolic Analysis of Knee Synovial Fluid as a Potential Diagnostic Approach for Osteoarthritis. J Orthop Res. 2015 doi: 10.1002/jor.22949. pp. [DOI] [PubMed] [Google Scholar]
- 30.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelnikov I, Arndt D, Xia J, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R, Scalbert A. Hmdb 3.0--the Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41(Database issue):D801–7. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu ZJ, Schultz AW, Wang J, Johnson CH, Yannone SM, Patti GJ, Siuzdak G. Liquid Chromatography Quadrupole Time-of-Flight Mass Spectrometry Characterization of Metabolites Guided by the Metlin Database. Nat Protoc. 2013;8(3):451–60. doi: 10.1038/nprot.2013.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CR, Shimizu T, Spener F, Van Meer G, Wakelam MJ, Dennis EA. Update of the Lipid Maps Comprehensive Classification System for Lipids. J Lipid Res. 2009;(50 Suppl):S9–14. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cavill R, Kamburov A, Ellis JK, Athersuch TJ, Blagrove MS, Herwig R, Ebbels TM, Keun HC. Consensus-Phenotype Integration of Transcriptomic and Metabolomic Data Implies a Role for Metabolism in the Chemosensitivity of Tumour Cells. PLoS Comput Biol. 2011;7(3):e1001113. doi: 10.1371/journal.pcbi.1001113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh CD, Chun JS. Signaling Mechanisms Leading to the Regulation of Differentiation and Apoptosis of Articular Chondrocytes by Insulin-Like Growth Factor-1. J Biol Chem. 2003;278(38):36563–71. doi: 10.1074/jbc.M304857200. [DOI] [PubMed] [Google Scholar]
- 35.Priore R, Dailey L, Basilico C. Downregulation of Akt Activity Contributes to the Growth Arrest Induced by Fgf in Chondrocytes. J Cell Physiol. 2006;207(3):800–8. doi: 10.1002/jcp.20620. [DOI] [PubMed] [Google Scholar]
- 36.Starkman BG, Cravero JD, Delcarlo M, Loeser RF. Igf-I Stimulation of Proteoglycan Synthesis by Chondrocytes Requires Activation of the Pi 3-Kinase Pathway but Not Erk Mapk. Biochem J. 2005;389(Pt 3):723–9. doi: 10.1042/BJ20041636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliver SG, Winson MK, Kell DB, Baganz F. Systematic Functional Analysis of the Yeast Genome. Trends Biotechnol. 1998;16(9):373–8. doi: 10.1016/s0167-7799(98)01214-1. [DOI] [PubMed] [Google Scholar]
- 38.O'conor CJ, Leddy HA, Benefield HC, Liedtke WB, Guilak F. Trpv4-Mediated Mechanotransduction Regulates the Metabolic Response of Chondrocytes to Dynamic Loading. Proc Natl Acad Sci U S A. 2014;111(4):1316–21. doi: 10.1073/pnas.1319569111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knight MM, Roberts SR, Lee DA, Bader DL. Live Cell Imaging Using Confocal Microscopy Induces Intracellular Calcium Transients and Cell Death. Am J Physiol Cell Physiol. 2003;284(4):C1083–9. doi: 10.1152/ajpcell.00276.2002. [DOI] [PubMed] [Google Scholar]
- 40.Lakshmi AV. Riboflavin Metabolism--Relevance to Human Nutrition. Indian J Med Res. 1998;108:182–90. [PubMed] [Google Scholar]
- 41.Terkeltaub R, Yang B, Lotz M, Liu-Bryan R. Chondrocyte Amp-Activated Protein Kinase Activity Suppresses Matrix Degradation Responses to Proinflammatory Cytokines Interleukin-1beta and Tumor Necrosis Factor Alpha. Arthritis Rheum. 2011;63(7):1928–37. doi: 10.1002/art.30333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anishkin A, Loukin SH, Teng J, Kung C. Feeling the Hidden Mechanical Forces in Lipid Bilayer Is an Original Sense. Proc Natl Acad Sci U S A. 2014;111(22):7898–905. doi: 10.1073/pnas.1313364111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mow VC, Kuei SC, Lai WM, Armstrong CG. Biphasic Creep and Stress Relaxation of Articular Cartilage in Compression? Theory and Experiments. J Biomech Eng. 1980;102(1):73–84. doi: 10.1115/1.3138202. [DOI] [PubMed] [Google Scholar]
- 44.Grodzinsky AJ, Lipshitz H, Glimcher MJ. Electromechanical Properties of Articular Cartilage During Compression and Stress Relaxation. Nature. 1978;275(5679):448–50. doi: 10.1038/275448a0. [DOI] [PubMed] [Google Scholar]
- 45.Stephanopoulos GN, Aristidou AA, Nielsen J. Metabolic Engineering: Principles and Methodologies. Academic Press; 1998. [Google Scholar]
- 46.Adams SB, Jr, Setton LA, Kensicki E, Bolognesi MP, Toth AP, Nettles DL. Global Metabolic Profiling of Human Osteoarthritic Synovium. Osteoarthritis Cartilage. 2012;20(1):64–7. doi: 10.1016/j.joca.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.