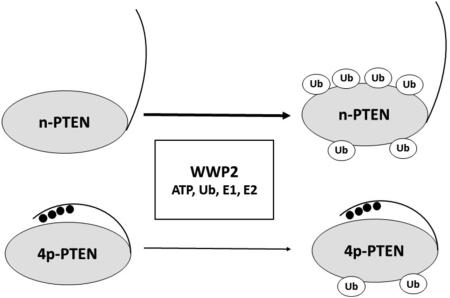
Abstract
PTEN is a lipid phosphatase that converts phosphatidylinositol 3,4,5-phosphate (PIP3) to phosphatidylinositol 4,5-phosphate (PIP2) and plays a critical role in the regulation of tumor growth. PTEN is subject to regulation by a variety of post-translational modifications, including phosphorylation on a C-terminal cluster of four Ser/Thr residues (380, 382, 383, and 385) and ubiquitylation by various E3 ligases, including NEDD4-1 and WWP2. It has previously been shown that C-terminal phosphorylation of PTEN can increase its cellular half-life. Using in vitro ubiquitin transfer assays, we show that WWP2 is more active than NEDD4-1 in ubiquitylating unphosphorylated PTEN. The mapping of ubiquitylation sites in PTEN by mass spectrometry showed that both NEDD4-1 and WWP2 can target a broad range of Lys residues in PTEN, although NEDD4-1 versus WWP2 showed a stronger preference for ubiquitylating PTEN's C2 domain. Whereas tetraphosphorylation of PTEN did not significantly affect its ubiquitylation by NEDD4-1, it inhibited PTEN ubiquitylation by WWP2. Single-turnover and pull-down experiments suggested that tetraphosphorylation of PTEN appears to weaken its interaction with WWP2. These studies reveal how the PTEN E3 ligases WWP2 and NEDD4-1 exhibit distinctive properties in Lys selectivity and sensitivity to PTEN phosphorylation. Our findings also provide a molecular mechanism for the connection between PTEN Ser/Thr phosphorylation and PTEN's cellular stability.
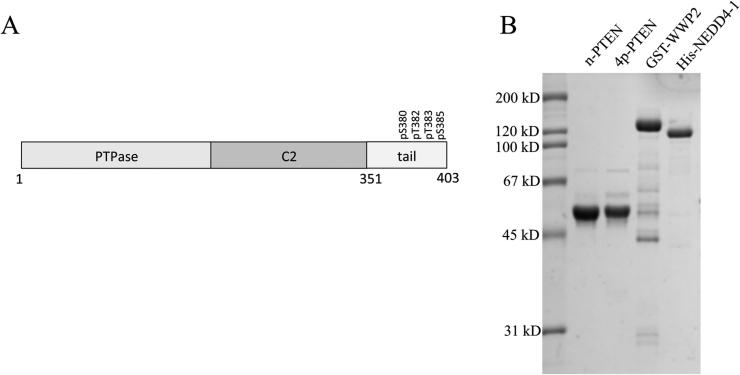
PTEN is a lipid phosphatase that hydrolyzes the 3-phosphate from phosphatidylinositol 3,4,5-phosphate (PIP3) to PIP2 and plays a major role in cell growth and motility.1–6 PTEN is a 403-residue protein composed of an N-terminal catalytic (PTPase, amino acids 15–185) domain followed by a lipid-binding C2 domain (amino acids 186–351) and terminating in an ~50-residue flexible regulatory domain (Figure 1).7 PTEN loss of function mutations occur commonly in a wide array of human cancers, allowing for the increase in PIP3 levels fueled by PI3 kinases that in turn allosterically activate Akt protein kinase and drive neoplastic growth.4–6,8,9 Beyond mutation, PTEN's activity is regulated by other mechanisms, including post-translational modification.10–14 Two types of post-translational modifications occurring on PTEN are phosphorylation and ubiquitylation.14,15 Of these, C-terminal phosphorylation on a cluster of Ser380, Thr382, Thr383, and Ser385 of PTEN, first identified in 2001, has attracted special attention as a key regulatory influence.10 Prior studies have suggested that this cluster of phosphorylations can drive an intramolecular interaction with the PTEN C2 domain, resulting in weakened membrane association and catalytic activity.16 Somewhat paradoxically, this phospho-cluster has also been reported to enhance the cellular stability of PTEN.10 However, the evidence of this effect of phosphorylation on PTEN stability has been indirect, relying on mutagenesis of the 380–385 cluster to Ala. Moreover, it is unknown if this PTEN instability is related to changes in its cellular localization, altered cellular signaling, or differential ubiquitylation.
Figure 1.
(A) Architectural layout of PTEN and (B) purified proteins (n-PTEN, 4p-PTEN, WWP2, and NEDD4-1) prepared in this study (sodium dodecyl sulfate–polyacrylamide gel electrophoresis stained with Coomassie blue).
Three PTEN ubiquityltransferase E3 ligases have been reported, NEDD4-1, WWP2, and XIAP.17–19 NEDD4-1 and WWP2 E3 ligases belong to the subset of ubiquityltransferases that have a catalytic HECT domain that operates with E1 and E2 ligases to catalyze the transfer of a ubiquityl group to Lys residues on target proteins.20,21 Unlike the more common RING domain E3 ligases, the HECT domain gets charged by ubiquitin on a nucleophilic Cys residue to form an intermediate E3-ubiquitin thioester.20,22 This E3 catalytic intermediate is in turn attacked by substrate Lys residues, affording ubiquitylated proteins.22 XIAP is a member of the larger family of RING domain E3 ligases that are Zn-binding proteins and serve as a template facilitating the transfer of ubiquitin from the E2 enzyme Cys thioester intermediate to a Lys on a target substrate.23,24
In this study, we analyze with purified proteins the NEDD4-1-, WWP2-, and XIAP-catalyzed ubiquitylation of PTEN in its unphosphorylated and phosphorylated states. We employed expressed protein ligation25 to generate semisynthetic forms of PTEN that were either tetraphosphorylated on the 380–385 Ser/Thr cluster (4p-PTEN) or unphosphorylated at these residues (n-PTEN). In our hands, XIAP did not catalyze PTEN ubiquitylation whereas both NEDD4-1 and WWP2 showed PTEN ubiquityltransferase activity. Compared with NEDD4-1, we found that WWP2 appears to more rapidly ubiquitylate n-PTEN. NEDD4-1 showed similar ubiquitylation kinetics toward both n-PTEN and 4p-PTEN. In contrast, WWP2 showed a marked reduction in its ability to ubiquitylate 4p-PTEN versus its ability to ubiquitylate n-PTEN. Mass spectrometric analysis suggests that the two E3 ligases possess overlapping but distinct ubiquitylation preferences for PTEN Lys specificity. Below, we describe these findings and discuss their relevance to PTEN regulation.
EXPERIMENTAL PROCEDURES
Reagents
The pGEX6p-2 WWP2 plasmid for bacterial expression of GST-WWP2 was a gift from W. Wei at Beth-Israel Deaconess Medical Center. pFastBac1-His-NEDD4-1 was described previously.17 Human recombinant XIAP protein was purchased from Sigma-Aldrich. Purified wild-type ubiquitin, human ubiquitin-activating enzyme UBE1, and human ubiquitin-conjugating enzyme UbcH5c were prepared as described previously.26 The lysine-free ubiquitin (K0-Ub) and human ubiquitin-conjugating enzyme UbcH7 were purchased from LifeSensors. The Colloidal Blue Staining Kit was purchased from Invitrogen. The anti-PTEN antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). Apyrase was from New England Biolabs. All other reagents of the highest quality were commercially purchased from either Sigma or Fisher.
Peptide Synthesis
Peptides were synthesized using a PS3 peptide synthesizer from Protein Technologies (Tuscon, AZ) using conventional FMOC peptide synthesis strategies. They were purified by reversed phase high-performance liquid chromatography on a C-18 column and the correct structures confirmed by matrix-assisted laser desorption ionization mass spectrometry.
Generation and Purification of Nonphosphorylated and Tetraphosphorylated Semisynthetic PTEN
We used expressed protein ligation25 to generate semisynthetic PTEN with a nonphosphorylated or tetraphosphorylated C-terminal tail as previously described by Bolduc et al.16 In brief, a C-terminally truncated PTEN (tPTEN, amino acids 1–378) fused to Mycobacterium xenopi GyrA intein and chitin-binding domain (tPTEN–intein–CBD) was expressed in High Five insect cells using a baculoviral expression system. The fusion protein was immobilized on the chitin resin and then incubated with 2 mM nonphosphorylated or tetraphosphorylated C-tail peptide in buffer containing 400 mM MESNA at room temperature for 48 h. The ligation was confirmed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) using Coomassie blue staining.16 Then the ligated PTEN was further purified by FPLC-anion exchange chromatography using a MonoQ column (GE Healthcare). Prior studies have established that 4p-PTEN and n-PTEN prepared in this way show the molecular weights predicted by mass spectrometry.16 The purified protein was concentrated to 5 mg/mL, flash-frozen, and kept at −80 °C.
Expression and Purification of GST-WWP2
BL-21 Codon Plus cells were transformed with the pGEX6p-2 WWP2 plasmid and cultured in LB medium at 37 °C to reach the optimal density (OD600 = 0.6) in a shaker incubator grown on a 1 L scale. GST-WWP2 expression was induced by 0.5 mM isopropyl β-d-1-thiogalactopyranoside at 16 °C for 20 h. The cells were resuspended in lysis buffer [25 mM Tris-HCl (pH 8.0), 250 mM NaCl, 1 mM phenylmethanesulfonyl fluoride (PMSF), and 1× Roche cocktail protease inhibitors] and lysed using a French press, and the lysate was loaded on glutathione agarose followed by washing with wash buffer [25 mM Tris-HCl (pH 8.0), 250 mM NaCl, and 0.1% Triton X-100]. The desired GST-WWP2 was eluted using 25 mM Tris-HCl (pH 8.0) and 250 mM NaCl containing 50 mM reduced glutathione (pH 8.0). Fractions containing GST-WWP2 were combined and dialyzed against a buffer consisting of 25 mM Tris-HCl (pH 7.5), 250 mM NaCl, 1 mM EDTA, 5 mM DTT, and 10% glycerol, and the protein was concentrated to 2–5 mg/mL and flash-frozen. The yield of GST-WWP2 was 0.5 mg/L of culture. GST-WWP2 was stored in aliquots at −80 °C after being flash-frozen.
Expression and Purification of His6-NEDD4-1
For NEDD4-1, pFascBac1-His-NEDD4-1 was used to generate Bacmid to infect Sf21 insect cells that produced the baculovirus for NEDD4-1 expression. The Sf9 insect cells were infected with baculovirus and cultured at 27 °C for 48 h. The cells were then harvested and resuspended in lysis buffer [25 mM HEPES (pH 7.5), 250 mM NaCl, 1 mM EDTA, 10% glycerol, 1 mM PMSF, and 1× Roche cocktail protease inhibitors]. The cells were lysed using a 40 mL homogenizer. The cell lysate from 1 L of culture was incubated with 2.5 mL of Ni-NTA resin for His-NEDD4-1 binding. After being washed with 200 mL of wash buffer [25 mM HEPES (pH 7.5), 250 mM NaCl, 1 mM EDTA, and 0.1% Triton X-100], the protein was eluted with a gradient of wash buffer now also containing 80–500 mM imidazole. The eluted protein was dialyzed overnight in 25 mM Tris-HCl (pH 8.0), 5 mM NaCl, and 5 mM DTT. Then His-NEDD4-1 was further purified by FPLC using an anion exchange MonoQ column with a gradient from 5 to 500 mM NaCl added to the dialysis buffer over 240 column volumes. Fractions corresponding to the desired protein were verified by SDS–PAGE, and highly purified fractions were combined. To the combined fractions was added 10% glycerol, and then the protein was concentrated to 5 mg/mL. The purified protein was flash-frozen and stored in aliquots at −80 °C. The yield of His-NEDD4-1 is around 5 mg/L of culture.
In Vitro Ubiquitylation Assay
The in vitro ubiquitylation reaction was conducted in microcentrifuge tubes in volumes of 20–50 μL containing 40 mM Tris-HCl (pH 7.5), 50 mM NaCl, 2 mM DTT or 0.5 mM TCEP, and 5 mM MgCl2, with 5 mM ATP, 100 μM ubiquitin, 50 nM E1 protein, 1 μM E2 protein, 1 μM E3 protein, and 1 or 10 μM PTEN. The reaction mixture without E1 protein was preincubated at 30 °C for 20 min, and then the reaction was initiated by adding E1. To quench the reaction, an aliquot of the reaction mixture was mixed with SDS loading dye at different time points and boiled for 5 min. The samples were then examined via SDS–PAGE and stained using a Colloidal Blue Staining Kit following the manufacturer's protocol. In addition, samples were also analyzed using Western blotting with an anti-PTEN antibody. The quenched reaction samples containing 50 ng of PTEN were loaded onto 10% SDS–PAGE gels. The protein was then transferred to nitrocellulose membranes using the iBlot Dry Blotting system (Thermo Fishier). The membranes were blocked with 5% BSA in TBST buffer for 30 min and then incubated with the anti-PTEN antibody (1:1000 dilution) at 4 °C overnight. Afterward, the membranes were washed with TBST and probed with the HRP-conjugated anti-mouse secondary antibody at a 1:10000 dilution. The bands were detected by chemiluminescence using an ECL Western blotting detection kit from GE Healthcare. All assays were repeated on at least two independent occasions with results and replicates being similar.
Single-Turnover WWP2 Ubiquitin Transfer Assay
These experiments were based on previously reported methods.27 Reaction mixtures containing 40 mM Tris-HCl (pH 7.5), 0.5 mM TCEP, 1 mM MgCl2, 1 mM ATP, 100 μM wild-type ubiquitin, 0.5 μM E1 protein, 5 μM E2 protein, and 2.5 μM WWP2 in a volume of 40 μL were incubated at 30 °C for 10 min. Then, 1 μM apyrase was added to the reaction mixtures, and they were incubated at 30 °C for 1 min, followed by addition of 1 μM PTEN. The final volume of the reaction mixtures was adjusted to 50 μL. Aliquots of the reaction mixture were quenched at different time points after the addition of PTEN. The samples were analyzed by Western blotting using an anti-PTEN antibody as described above.
GST-WWP2 Pull-Down Assay of PTEN Forms
Nonphosphorylated or tetraphosphorylated PTEN (1 μM) was mixed with or without 1 μM GST-WWP2 in 25 mM Tris (pH 7.5), 50 mM NaCl, 5 mM DTT, and 10 μL glutathione agarose in a total volume of 100 μL. The mixture was incubated at 4 °C with gentle agitation overnight. Afterward, the glutathione agarose was washed thrice with 1 mL of wash buffer [25 mM HEPES (pH 7.5), 250 mM NaCl, and 0.1% Triton X-100]. Twenty microliters of SDS loading dye was added to the resin, and the mixture was then boiled for 5 min. The samples were then analyzed by Western blotting using an anti-PTEN antibody as described above.
Data Analysis
To quantify the rate of the ubiquitylation reaction, the PTEN band was quantified by densitometric analysis using ImageJ software, and the rate of decrease of unmodified and phosphorylated PTEN (micromolar per minute) was calculated.
Enzymatic Digestion
Approximately 2.5 μg of protein from the PTEN ubiquitylation reactions was separated via SDS–PAGE and visualized with Coomassie blue staining. Protein bands of interest were excised, cut into 1 mm × 1 mm pieces, and dehydrated with methanol for 5 min. The gel pieces were then washed as follows: for 1 × 5 min with a 30% methanol/70% water mixture, 2 × 10 min with water, and 3 × 10 min with a 100 mM ammonium bicarbonate (NH4HCO3)/30% acetonitrile mixture. Gel pieces were dried in a SpeedVac. Protein disulfide bonds were reduced with 10 mM tris-(carboxyethyl)phosphine (TCEP) in 100 mM NH4HCO3 for 60 min at 56 °C, followed by alkylation with 55 mM 2-chloroacetamide in 100 mM NH4HCO3 for 45 min at room temperature in the dark. The gel pieces were washed with 100 mM NH4HCO3 for 15 min and dehydrated with acetonitrile followed by complete drying in a SpeedVac. Gel pieces were rehydrated in a trypsin solution (15 ng/μL trypsin in 50 mM NH4HCO3) on ice for 45 min. Excess trypsin solution was discarded, replaced with 50 mM NH4HCO3, and incubated overnight at 37 °C. Digestion buffer was collected and saved. Peptides were extracted once with 50 mM NH4HCO3, once with acetonitrile, and twice with 5% formic acid in 50% acetonitrile; each extraction was performed by incubation at 37 °C for 15 min with vortexing. All supernatants were combined and dried in a SpeedVac. The samples were reconstituted in 50 μL of AspN reaction buffer [50 mM Tris-HCl and 2.5 mM ZnSO4 (pH 8)], and 12.5 ng/μL AspN was added for overnight digestion at 37 °C. Digestion reactions were stopped by the addition of 50 μL of 0.2% trifluoroacetic acid, and the samples were desalted using C18 STAGE Tips.28 The samples were reconstituted in 0.2% formic acid in water prior to liquid chromatography–tandem mass spectrometry analysis, and one-third of each sample was injected.
Liquid Chromatography–Tandem Mass Spectrometry (LC–MS/MS)
Chromatographic separation was performed using a Dionex Ultimate 3000 RSLCnano system (Thermo Scientific) with a 75 μm × 15 cm Acclaim PepMap100 separating column (Thermo Scientific) protected by a 2 cm guard column (Thermo Scientific). The mobile phase flow rate was 300 nL/min and consisted of 0.1% formic acid in water (A) and 0.1% formic acid and 95% acetonitrile (B). MS analysis was performed using an LTQ Orbitrap Velos Pro mass spectrometer (Thermo Scientific). The spray voltage was set at 2.2 kV. Orbitrap spectra were collected from m/z 400 to 1800 at a resolution of 30000 followed by data-dependent HCD MS/MS (at a resolution of 7500, with a collision energy of 35% and an activation time of 0.1 ms) of the 10 most abundant ions using an isolation width of 2.0 Da. Charge-state screening was enabled to reject the generation of MS/MS spectra for unassigned and singly charged precursor ions. A dynamic exclusion time of 40 s was used to discriminate against previously selected ions. Data were searched using SEQUEST in Proteome Discoverer version 1.3 (Thermo Scientific) against a Homo sapiens PTEN UniProt database (UniProt accession number P60484). Database search parameters were as follows: enzyme, nonspecific (to allow for combined trypsin and AspN cleavage); precursor mass tolerance, 10 ppm; fragment ion tolerance, 0.03 Da; static modification, Cys carbamidomethylation; variable modifications, Met oxidation, Lys ubiquitylation and acetylation, and Ser/Thr/Tyr phosphorylation. The data were filtered using a 1% false discovery rate threshold and a maximum peptide rank of 1. All MS/MS spectra assigned to modified PTEN peptides were manually inspected.
RESULTS
Ubiquitylation of n-PTEN by NEDD4-1, WWP2, and XIAP
For these studies, we prepared 4p-PTEN and n-PTEN using expressed protein ligation as described previously.16 In this method, amino acids 1–378 of PTEN are prepared as a thioester via an intein using a baculovirus expression system, and this is chemoselectively ligated to N-Cys-containing synthetic peptides of amino acids 379–403 with or without phosphorylation at Ser380, Thr382, Thr383, and Ser385. These are wild-type in sequence except for the presence of a Y379C mutation necessitated by the ligation process that, as reported previously, does not alter the activity of PTEN.16 These semisynthetic PTEN proteins were shown to be >90% pure (see Figure 1). As shown previously,16 the unligated PTEN forms run just below the ligated forms on SDS–PAGE, so the high purity of the semisynthetic PTENs is achieved after anion exchange chromatography. NEDD4-1 protein was also prepared in a baculovirus expression system and contains an N-terminal His6 tag that was used in Ni resin affinity chromatography for purification (see Figure 1). WWP2 was prepared as a GST fusion protein using an Escherichia coli expression system and purified by glutathione affinity chromatography (see Figure 1). XIAP was obtained from commercial sources as a purified recombinant protein.
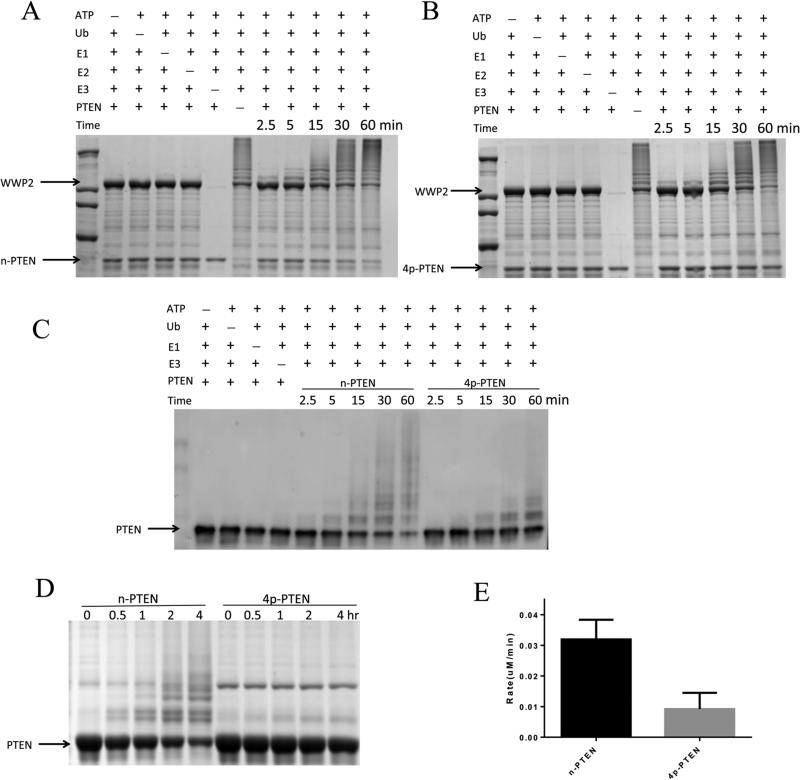
Ubiquitylation experiments with WWP2 were performed with 50 nM E1 ubiquitin activating enzyme, 1 μM UbcH5c (E2 ligase), 1 μM WWP2 (E3 ligase), 1 μM n-PTEN, 100 μM ubiquitin, and 5 mM MgATP. We monitored the ubiquitylation using a combination of both of Coomassie blue-stained SDS–PAGE as well as Western blots with the anti-PTEN Ab (Figure 2). The reaction mixture's multiband/streaking pattern readily visualized by both methods was attributed to various levels of ubiquitin and polyubiquitin chain attachment catalyzed by WWP2. There was a clear increase in the level of ubiquitylation with time over the course of the reaction, and the nonubiquitylated n-PTEN was largely depleted after 60 min under these conditions (see Figure 2). WWP2, E2, E1, and ATP were each necessary for the reaction to occur as their omission prevented detectable ubiquitylation. In the higher-molecular weight region of the Coomassie blue-stained SDS–PAGE gel, autoubiquitylation of WWP2 was also readily detected (Figure 2A,B). Using a higher concentration of n-PTEN (10 μM) relative to that of WWP2 (1 μM), perhaps better approximating steady-state conditions, the rate of depletion of n-PTEN was reduced, and it had a half-life of ~1 h (Figure 2D).
Figure 2.
In vitro ubiquitylation assay of n-PTEN and 4p-PTEN by WWP2. (A) In vitro ubiquitylation assay for n-PTEN. The reaction was conducted in the presence of 5 mM ATP, 100 μM wild-type ubiquitin, 50 nM E1, 1 μM E2 (UbcH5c), 1 μM WWP2, and 1 μM n-PTEN in assay buffer with 40 mM HEPES (pH 7.5) and 5 mM MgCl2. The reaction was quenched at 2.5, 5, 15, 30, and 60 min. Negative controls without ATP, ubiquitin, E1, E2, E3, or PTEN were assayed. The samples were resolved by SDS–PAGE, and the gel was stained with colloidal blue. (B) In vitro ubiquitylation assay for 4p-PTEN. The reaction was conducted for 4p-PTEN using the same conditions as in panel A. (C) n-PTEN and 4p-PTEN ubiquitylation analyzed by Western blotting using the anti-PTEN antibody. E2 was added to these experiments, but the minus E2 conditions are not shown in this blot. (D) In vitro ubiquitylation assay for n-PTEN and 4p-PTEN. The reaction conditions were the same as in panel A except that 10 μM PTEN was used instead of 1 μM PTEN. Ubiquitylation of PTEN at time points of 0, 0.5, 1, 2, and 4 h was analyzed by colloidal blue staining. (E) Quantification of the rate of PTEN ubiquitylation. The ubiquitylation assay performed in panel D was quantified by measuring the decrease in nonubiquitylated PTEN band intensities and calculating the decrease in nonubiquitylated PTEN protein levels as a function of time over replicates (n = 3) with the standard error shown.
NEDD4-1 (1 μM) was also tested as an E3 ligase with n-PTEN and showed an ability to ubiquitylate PTEN, although at a rate slower than that of WWP2 (Figure 3A,B). Given the complexity of the reactions and the wide range of product distributions, it is difficult to precisely quantify the relative rates, but monitoring depletion of the unmodified PTEN suggests an ~4-fold difference. In contrast to that with NEDD4-1 and WWP2, we were unable to detect n-PTEN ubiquitylation with XIAP (Figure 3). We are uncertain why XIAP, though undergoing autoubiquitylation in our hands, was inactive with PTEN in this assay. However, given the measurable rates with NEDD4-1 and WWP2, we elected not to pursue further experiments with XIAP.
Figure 3.
In vitro ubiquitylation assay of n-PTEN and 4p-PTEN by NEDD4-1 and XIAP. (A) The reaction for n-PTEN and 4p-PTEN ubiquitylation by NEDD4-1 was conducted with 5 mM ATP, 100 μM wild-type ubiquitin, 50 nM E1, 1 μM E2 (UbcH5c), 1 μM NEDD4-1, and 10 μM PTEN, for 0, 0.5, 1, 2, and 4 h. The samples were analyzed by colloidal blue staining. (B) n-PTEN and 4p-PTEN ubiquitylation (1 μM) by NEDD4-1 under the same reaction conditions was analyzed by Western blotting using the anti-PTEN antibody. (C) The ubiquitylation of n-PTEN and 4p-PTEN by XIAP was analyzed by colloidal blue staining. The reaction of the mixture containing 100 μM wild-type ubiquitin, 50 nM E1, 1 μM E2 (UbcH5c), 1 μM XIAP and 1 μM PTEN was quenched at 0, 1, 2, and 4 h and analyzed by colloidal blue staining. (D) Ubiquitylation of n-PTEN and 4p-PTEN under the same condition as in panel C was analyzed by Western blotting using the anti-PTEN antibody.
PTEN Phosphorylation Inhibits WWP2-Mediated Ubiquitylation
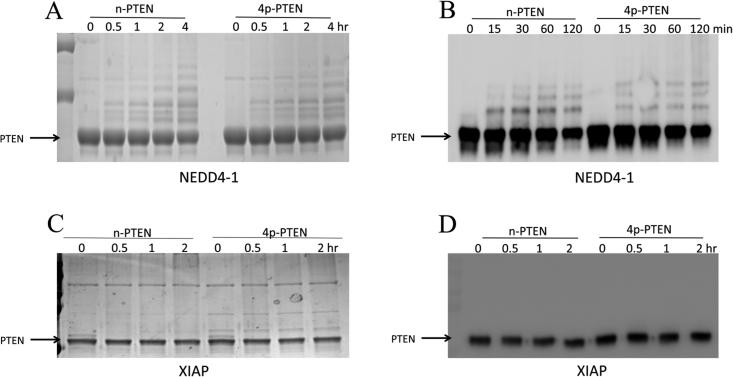
We then investigated the ubiquitylation of 4p-PTEN by WWP2 and NEDD4-1. Whereas the rate of ubiquitylation of 4p-PTEN by NEDD4-1 was similar to that of its reaction with the n-PTEN substrate (Figure 3), WWP2 showed a marked reduction in its level of ubiquitylation of 4p-PTEN (Figure 2). In principle, this reduction in the rate of WWP2 ubiquitylation of 4p-PTEN could be caused by an interference with steps up to and including E2's ubiquityl transthioesterification loading of E3. This was explored in two ways. First, we examined the WWP2/4p-PTEN reaction in the presence of an alternative E2 enzyme, UbCH7, in place of UbCH5c. We observed that UbCH7 could work in concert with WWP2 to ubiquitylate n-PTEN and 4p-PTEN, but this system showed a similar drop in efficiency with the 4p-PTEN substrate relative to that involving UbCH5c (Figure 4A). These results suggest that the differences in WWP2 ubiquitylation between 4p-PTEN and n-PTEN are not related to specific interactions between E2 UbCH5c and E3 WWP2. To examine this further, we performed a single-turnover experiment in which WWP2 was charged by E1 and E2 and then ATP is cleared from the reaction by apyrase-mediated degradation.27 The removal of ATP at this stage prevents recharging of the E3-ubiquityl thioester after it transfers its ubiquitin to PTEN. In this way, one can more specifically monitor the kinetics of the specific step involving the transfer of ubiquitin from WWP2 to PTEN. In this experiment, we showed that n-PTEN is still preferentially ubiquitylated over 4p-PTEN by WWP2 (Figure 4B). These results suggest that the specific enzymatic step that involves transfer of ubiquitin from WWP2 to PTEN is affected by PTEN phosphorylation.
Figure 4.
(A) In vitro ubiquitylation assay of PTEN by WWP2 using a different E2. The reaction conditions were the same as in Figure 2D except that E2 was UbcH7 instead of UbcH5c. The samples were analyzed by colloidal blue staining. (B) A single-turnover assay to measure ubiquitin transfer from WWP2 to n-PTEN and 4p-PTEN was conducted by charging WWP2 with ubiquitin and then adding PTEN after depletion of ATP using apyrase. The ubiquitylation of PTEN was analyzed by Western blotting with the anti-PTEN antibody.
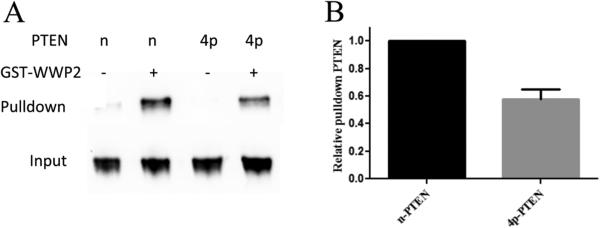
The observations described above suggest that the tetraphosphorylation of PTEN may directly disrupt WWP2-mediated ubiquitylation of the PTEN protein by weakening the interaction between the WWP2 and PTEN proteins. To assess this possibility, we performed a pull-down experiment in which GST-WWP2 was immobilized on glutathione resin, and these were incubated with either n-PTEN or 4p-PTEN proteins. After being washed, the glutathione resin was denatured with SDS and the PTEN was analyzed by Western blotting using anti-PTEN Ab, revealing that n-PTEN was more efficiently pulled down by WWP2 compared with that of 4p-PTEN (Figure 5). These findings suggest that tetraphosphorylation of PTEN does indeed reduce its affinity for WWP2, and this likely contributes to 4p-PTEN's weakened ubiquitylation by WWP2 compared with that of n-PTEN.
Figure 5.
GST pull-down assay of WWP2 and PTEN. (A) GST-WWP2 or GST (1 μM) and n-PTEN or 4p-PTEN (1 μM) were incubated with glutathione agarose resin overnight. The pull-down of PTEN by WWP2 was analyzed by Western blotting using the anti-PTEN antibody. (B) Quantification of 4p-PTEN relative to n-PTEN pull-down by WWP2, average over replicates (n = 3) with the standard error shown.
Site Specificity of WWP2- and NEDD4-1-Mediated PTEN Ubiquitylation
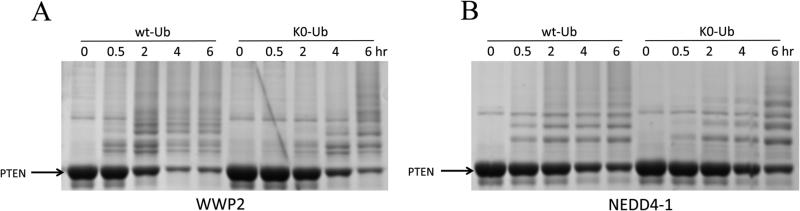
We next analyzed the Lys sites of ubiquitylation by WWP2 and NEDD4-1 using mass spectrometry. In these experiments, n-PTEN was exposed to the E3 ligases for 6 h using Lys-free ubiquitin. Although Lys-free ubiquitin compared with natural ubiquitin is less efficiently processed by WWP2 with a rate now similar to that of NEDD4-1 (Figure 6), its use significantly simplified the analysis because it prevented polyubiquitin chain formation. The reaction mixtures were subjected to SDS–PAGE and in-gel enzymatic digestion and analyzed using LC–MS/MS. To ensure the maximal sequence coverage of PTEN for these experiments, thereby optimizing the mass spectrometric detection of all the potential PTEN Lys ubiquitylation sites, sequential enzymatic digestion was conducted with trypsin followed by AspN. Of the 34 PTEN Lys residues, 29 were covered among the identified peptides from the NEDD4-1 ubiquitylation experiment, and 25 were covered among the identified peptides from the WWP2 ubiquitylation experiment. The total PTEN sequence coverage was 91.3 and 79.2% for the NEDD4-1 and WWP2 experiments, respectively (Figure 7A). As an indication of the comprehensive PTEN sequence coverage for the NEDD4-1 experiment, all the peptides that were not identified had an average length of four amino acids, which rendered them below the m/z detection range of the mass spectrometer used for this study. The PTEN peptides that were identified in the NEDD4-1 and WWP2 experiments are listed in Tables S1 and S2, respectively.
Figure 6.
In vitro ubiquitylation assay of n-PTEN with wild-type (wt-Ub) or lysine-free ubiquitin (K0-Ub). (A) The ubiquitylation reaction was conducted in 40 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 5 mM ATP, 50 nM E1, 1 μM E2, 1 μM WWP2, 10 μM n-PTEN, with 100 μM wt-Ub or K0-Ub. The reactions were quenched at 0, 0.5, 2, 4, and 6 h and the mixtures analyzed by colloidal blue staining, and the reaction with K0-Ub at 6 h was subjected to mass spectrometry analysis. (B) Same in vitro ubiquitylation conditions as in panel A using wt-Ub or K0-Ub with NEDD4-1.
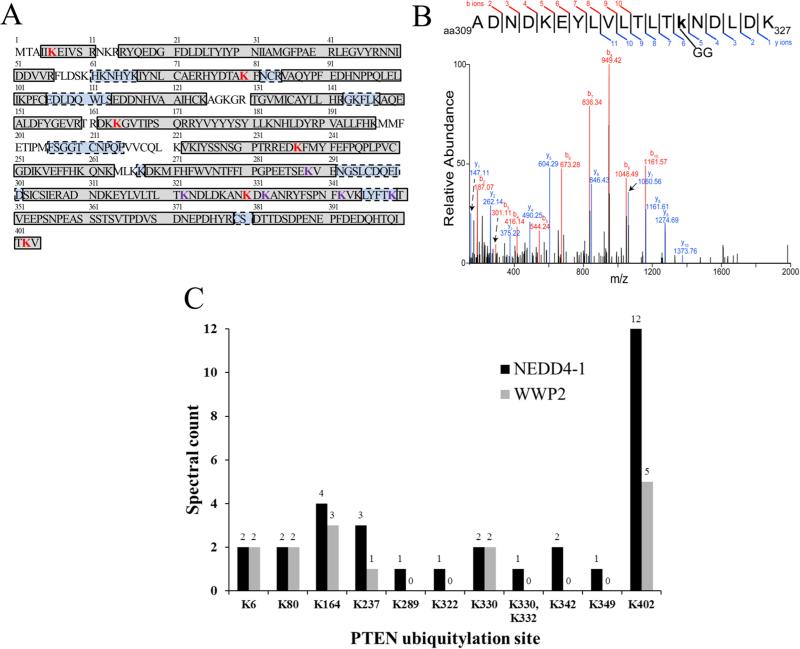
Figure 7.
Mass spectrometric identification of n-PTEN. (A) PTEN sequence coverage. Boxed, shaded regions indicate identified peptides. Gray-shaded boxes with solid outlines indicate regions identified in common between the NEDD4-1 and WWP2 experiments. Blue-shaded boxes with dashed outlines indicate regions that were identified only in the NEDD4-1 experiment. Bold, red letters are identified sites of Lys ubiquitylation. Bold, purple letters are sites of Lys ubiquitylation identified only in the NEDD4-1 experiment. (B) Representative MS/MS spectrum of the peptide containing Ub-Lys322. (C) Relative abundance of PTEN Lys ubiquitylation sites based on spectral counts.
A representative MS/MS spectrum of a ubiquitylated peptide (Ub-Lys322) is shown in Figure 7B. A total of 11 ubiquitylation sites were identified in the NEDD4-1 experiment compared to six in the WWP2 experiment (Figure 7A). The PTEN ubiquitylation sites that were shared by the two E3 ligases include Lys6, Lys80, Lys164, Lys237, Lys330, and Lys402. These Lys sites are relatively broadly distributed throughout the PTEN protein and its major domains, PTPase, C2, and C-terminal regulatory domains (see Figure 1A). It should be noted that of the five ubiquitylation sites that were identified exclusively in the NEDD4-1 experiment, only one of these Lys residues (Lys349) was in a region of PTEN that was not identified in the WWP2 experiment. With the exception of Lys349, each of the peptides containing the putative Lys ubiquitylation sites was also identified in their unmodified forms. Of the six ubiquitylation sites that were identified in common between the NEDD4-1 and WWP2 experiments, only Lys402 appears to be preferentially ubiquitylated by NEDD4-1 as indicated by the greater spectral counts of the peptides containing this ubiquitylated residue (Figure 7C). The five ubiquitin sites observed with only NEDD4-1 included Lys289, Lys322, Lys332, Lys342, and Lys349. Each of these sites occurs in the C2 domain. Interestingly, of the four known PTEN ubiquitin sites (Lys13, Lys66, Lys80, and Lys289) listed in Phosphosite plus (http://www.phosphosite.org/homeAction.action), only Lys289 overlaps with the 11 sites mapped here.
DISCUSSION
Because of PTEN's major role as a tumor suppressor PIP3 phosphatase, understanding how PTEN's cellular stability is controlled may offer new directions for therapeutic intervention. These studies have confirmed the ability of both WWP2 and NEDD4-1 to ubiquitylate PTEN using enzymatic assays with purified proteins, although WWP2 appeared to be more robust as a PTEN E3 ligase in our hands. We were unable to demonstrate that XIAP serves as a PTEN E3 ligase, but we cannot rule out the possibility that other factors in the cell may facilitate its potential to ubiquitylate PTEN.
We were especially interested in how phosphorylation of PTEN on a cluster of C-terminal Ser/Thr residues 380, 382, 383, and 385 might influence its potential for ubiquitylation, as it had been proposed that this phosphate cluster can increase PTEN's cellular half-life.10 Using tetraphosphorylated PTEN, we found that such phosphorylation antagonizes ubiquitylation by WWP2 but not NEDD4-1. Coupled with WWP2's enzymatic efficiency being greater than that of NEDD4-1 as a PTEN E3 ligase, these results suggest that WWP2 may be dominant in governing PTEN's cellular degradation when PTEN is nonphosphorylated. Perhaps NEDD4-1's more significant control of PTEN's stability occurs when PTEN exists in its phosphorylated state, although more in vivo experiments will be needed to probe this possibility.
The precise mechanism of how phosphorylation of PTEN inhibits its ubiquitylation by WWP2 is uncertain; however, it is improbable that the Ser/Thr phosphates are impeding Lys targeting through proximity-induced obstruction as most of the ubiquitylation sites are remote from the C-terminal tail and are spread throughout PTEN. The weakening of the WWP2–PTEN interaction observed by pull-down assays likely contributes to the reduced ubiquitylation efficiency. Such weakened interaction could plausibly result from the closed conformation of 4p-PTEN where surfaces of PTEN interaction would be less available to WWP2 binding.16 Perhaps NEDD4-1 binds to PTEN in a fashion different from the way in which WWP2 interacts with PTEN and is thus less affected by PTEN phosphorylation. Indeed, it has been reported that NEDD4-1 interacts with PTEN in a WW2 motif-independent manner.29 Further evidence of this appears to come from the enhanced targeting of the PTEN C2 domain Lys sites for ubiquitylation by NEDD4-1 versus WWP2.
It is also possible that phosphorylation of PTEN, by virtue of its weakening of PTEN membrane binding, may also protect PTEN from membrane-associated E3 ligases.30 The combination of cytosolic-nuclear localization of 4p-PTEN and 4p-PTEN's intrinsic resistance to WWP2-catalyzed ubiquitylation could synergize to stabilize PTEN in cells. It is not yet clear which of the large number of ubiquitylation sites identified here may be most physiologically relevant. It seems likely that the majority of them, if they occur in the cell, would help target PTEN toward the proteasome, but if not, they would likely interfere with PTEN's normal enzymatic function.
Prior studies have validated 4p-PTEN's reduced PIP3 phosphatase activity relative to that of n-PTEN.16 It is therefore somewhat paradoxical that tetraphosphorylation of PTEN, which inhibits its PIP3 phosphatase activity, also promotes a longer cellular half-life for the protein. We speculate that these apparently conflicting properties associated with phosphorylation of PTEN allow for a fine-tuning of signaling that maintains PIP3 levels in a precise range. This regulatory mechanism also can allow for strict kinetic control so that after PTEN is turned on by dephosphorylation and PIP3 levels drop, PTEN can be quickly eliminated by a proteasomally mediated pathway.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Dr. Wenyi Wei for the gift of the GST-WWP2 DNA plasmid.
Funding
We are grateful to the National Institutes of Health for financial support.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.6b00448.
n-PTEN peptides identified by LC–MS/MS from the NEDD4-1 ubiquitylation experiment (Table S1) (PDF) n-PTEN peptides identified by LC–MS/MS from the WWP2 ubiquitylation experiment (Table S2) (PDF)
REFERENCES
- 1.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, Parsons R. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 3.Myers MP, Pass I, Batty IH, Van der Kaay J, Stolarov JP, Hemmings BA, Wigler MH, Downes CP, Tonks NK. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun H, Lesche R, Li DM, Liliental J, Zhang H, Gao J, Gavrilova N, Mueller B, Liu X, Wu H. PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3,4,5,-trisphosphate and Akt/protein kinase B signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 1999;96:6199–6204. doi: 10.1073/pnas.96.11.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weng LP, Smith WM, Dahia PL, Ziebold U, Gil E, Lees JA, Eng C. PTEN suppresses breast cancer cell growth by phosphatase activity-dependent G1 arrest followed by cell death. Cancer research. 1999;59:5808–5814. [PubMed] [Google Scholar]
- 6.Vazquez F, Sellers WR. The PTEN tumor suppressor protein: an antagonist of phosphoinositide 3-kinase signaling. Biochim. Biophys. Acta, Rev. Cancer. 2000;1470:M21–35. doi: 10.1016/s0304-419x(99)00032-3. [DOI] [PubMed] [Google Scholar]
- 7.Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, Dixon JE, Pandolfi P, Pavletich NP. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 8.Perren A, Weng LP, Boag AH, Ziebold U, Thakore K, Dahia PL, Komminoth P, Lees JA, Mulligan LM, Mutter GL, Eng C. Immunohistochemical evidence of loss of PTEN expression in primary ductal adenocarcinomas of the breast. Am. J. Pathol. 1999;155:1253–1260. doi: 10.1016/S0002-9440(10)65227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao YJ, Ping XL, Zhang H, Chen FF, Lee PK, Ahsan H, Chen CJ, Lee PH, Peacocke M, Santella RM, Tsou HC. PTEN/MMAC1 mutations in hepatocellular carcinomas. Oncogene. 1999;18:3181–3185. doi: 10.1038/sj.onc.1202659. [DOI] [PubMed] [Google Scholar]
- 10.Vazquez F, Ramaswamy S, Nakamura N, Sellers WR. Phosphorylation of the PTEN tail regulates protein stability and function. Molecular and cellular biology. 2000;20:5010–5018. doi: 10.1128/mcb.20.14.5010-5018.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torres J, Pulido R. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. J. Biol. Chem. 2001;276:993–998. doi: 10.1074/jbc.M009134200. [DOI] [PubMed] [Google Scholar]
- 12.Al-Khouri AM, Ma Y, Togo SH, Williams S, Mustelin T. Cooperative phosphorylation of the tumor suppressor phosphatase and tensin homologue (PTEN) by casein kinases and glycogen synthase kinase 3beta. J. Biol. Chem. 2005;280:35195–35202. doi: 10.1074/jbc.M503045200. [DOI] [PubMed] [Google Scholar]
- 13.Odriozola L, Singh G, Hoang T, Chan AM. Regulation of PTEN activity by its carboxyl-terminal autoinhibitory domain. J. Biol. Chem. 2007;282:23306–23315. doi: 10.1074/jbc.M611240200. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Jiang X. Post-translational regulation of PTEN. Oncogene. 2008;27:5454–5463. doi: 10.1038/onc.2008.242. [DOI] [PubMed] [Google Scholar]
- 15.Trotman LC, Wang X, Alimonti A, Chen Z, Teruya-Feldstein J, Yang H, Pavletich NP, Carver BS, Cordon-Cardo C, Erdjument-Bromage H, Tempst P, Chi SG, Kim HJ, Misteli T, Jiang X, Pandolfi PP. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128:141–156. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolduc D, Rahdar M, Tu-Sekine B, Sivakumaren SC, Raben D, Amzel LM, Devreotes P, Gabelli SB, Cole P. Phosphorylation-mediated PTEN conformational closure and deactivation revealed with protein semisynthesis. eLife. 2013;2:e00691. doi: 10.7554/eLife.00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z, Wang J, Erdjument-Bromage H, Tempst P, Cordon-Cardo C, Pandolfi PP, Jiang X. NEDD4–1 is a protooncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Themsche C, Leblanc V, Parent S, Asselin E. X-linked inhibitor of apoptosis protein (XIAP) regulates PTEN ubiquitination, content, and compartmentalization. J. Biol. Chem. 2009;284:20462–20466. doi: 10.1074/jbc.C109.009522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maddika S, Kavela S, Rani N, Palicharla VR, Pokorny JL, Sarkaria JN, Chen J. WWP2 is an E3 ubiquitin ligase for PTEN. Nat. Cell Biol. 2011;13:728–733. doi: 10.1038/ncb2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. U. S. A. 1995;92:5249. doi: 10.1073/pnas.92.11.5249-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell. 2008;14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Berndsen CE, Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 2014;21:301–307. doi: 10.1038/nsmb.2780. [DOI] [PubMed] [Google Scholar]
- 23.Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitinconjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. U. S. A. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- 25.Muir TW, Sondhi D, Cole PA. Expressed protein ligation: a general method for protein engineering. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiener R, DiBello AT, Lombardi PM, Guzzo CM, Zhang X, Matunis MJ, Wolberger C. E2 ubiquitinconjugating enzymes regulate the deubiquitinating activity of OTUB1. Nat. Struct. Mol. Biol. 2013;20:1033–1039. doi: 10.1038/nsmb.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saha A, Lewis S, Kleiger G, Kuhlman B, Deshaies RJ. Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol. Cell. 2011;42:75–83. doi: 10.1016/j.molcel.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 29.Wang X, Shi Y, Wang J, Huang G, Jiang X. Crucial role of the C-terminus of PTEN in antagonizing NEDD4–1-mediated PTEN ubiquitination and degradation. Biochem. J. 2008;414:221–229. doi: 10.1042/BJ20080674. [DOI] [PubMed] [Google Scholar]
- 30.Rauch S, Martin-Serrano J. Multiple interactions between the ESCRT machinery and arrestin-related proteins: implications for PPXY-dependent budding. Journal of virology. 2011;85:3546–3556. doi: 10.1128/JVI.02045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.