Abstract
Insertion of biological nanopore into artificial membrane is of fundamental importance in nanotechnology. Many applications require control and knowledge of channel orientation. In this work, the insertion orientation of the bacteriophage SPP1 and phi29 DNA packaging motor into lipid membranes was investigated. Single molecule electrophysiological assays and Ni-NTA-nanogold binding assays revealed that both SPP1 and phi29 motor channels exhibited a one-way traffic property for TAT peptide translocation from N- to C-termini of the protein channels. SPP1 motor channels preferentially inserts into liposomes with their C-terminal wider region facing inward. Changing the hydrophobicity of the N- or C-termini of phi29 connector alters the insertion orientation, suggesting that the hydrophobicity and hydrophilicity of the termini of the protein channel governs the orientation of the insertion into lipid membrane. It is proposed that the specificity in motor channel orientation is a result of the hydrophilic/hydrophobic interaction at the air/water interface when the protein channels are incorporating into liposome membranes.
Keywords: Nanopore, Nanobiotechnology, Viral Assembly, single pore sensing, single pore DNA sequencing, viral DNA packaging motor
INTRODUCTION
Inserting protein nanochannels into liposomes or planar lipid membranes can mimic cell membrane channels. Membrane-embedded biological nanopores have a wide range of applications from disease-related biomarker detection [1; 2], nucleic acid characterization [3–10], protein and peptide sensing [11; 12], and so forth [13–17], by allowing target analytes to either pass though the channel or bind to specific probes on the pore. Due to asymmetry of channel structure and surface charge properties, those embedded channels display different behaviors as target molecules are transported across the lumen from different directions [18; 19]. Determining and eventually controlling the orientation of the protein channels in liposomes and planar membranes is thus significant for technological applications.
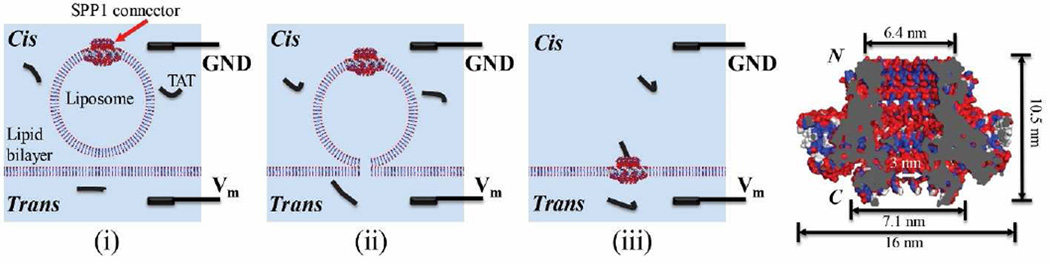
Bacteriophage SPP1 is a dsDNA bacterial virus that infects Bacillus subtilis. The central component of the DNA packaging motor is the head and tail connector that enables the viral genome to be packaged into procapsid during virus assembly [20–22]. The connector is composed of twelve copies of the protein subunit gp6 in virion [21] or thirteen copies in the recombinant complex assembled from the over-expressed gene product [23]. Based on the crystal structure [23; 24], the SPP1 connector has an overall diameter of 16 nm and a height of 10.5 nm. The narrowest constriction of the internal channel is ~3 nm (Fig. 1). We have previously reengineered the connector at the C-terminal end to introduce a 6×His-tag, purified the connector, and inserted it in planar lipid bilayer to serve as a membrane-embedded nanopore [25].
Figure 1.
Schematic showing fusion of C-His SPP1 connector-liposomes with planar lipid bilayer to generate membrane embedded SPP1 nanopores. Ground and reference electrodes are placed in cis- and trans- compartments of the chamber, respectively. TAT peptide is premixed in both compartments. N and C indicate the narrower N- and wider C-termini of the connector.
The well characterized bacterial virus phi29 motor channels were used as a control. The phi29 connector forms a dodecameric funnel shaped channel [26; 27] with an internal channel diameter of 6 nm at the wider end and 3.6 nm at the narrowest constriction. We have extensively investigated the membrane-embedded phi29 nanopore system [8; 28] for characterizing nucleic acids [8; 18; 29–34], and for sensing of proteins [2] and chemicals [35]. The phi29 connector exhibits a one-way traffic property for dsDNA translocation from N- to C-terminus [18]. Here we report that the channel of the DNA packaging motor of bacteriophage SPP1 has a highly oriented direction after self-assembling into DPhPC liposome without any special treatment, as determined by its one-way traffic property for TAT peptide translocation and Ni-NTA-nanogold binding assay [18].
MATERIALS AND METHODS
Materials
DPhPC (1,2-diphytanoyl-sn-glycerol-3-phosphocholine) phospholipid was purchased from Avanti Polar Lipids, Inc. Ni-NTA-nanogold was purchased from Nanoprobes LLC. TAT peptide (Cys-Tyr-Gly-Arg-Lys-Lys-Arg-Arg-Gln-Arg-Arg-Arg) was obtained from GenScript, Inc. and used without further purification. All other reagents were purchased from Sigma or Fisher.
Cloning and Purification of the SPP1 connector
The expression and purification of C-His SPP1 gp6 [25] and phi29 gp10 connector has been described previously [8; 28].
Self-assembling of SPP1 connector into liposome
C-his SPP1 (as well as C-His gp10 and N-His gp10) connector self-assembles into liposome during the dehydration-rehydration process as described previously [8; 28]. Briefly, chloroform was removed under vacuum from the DPhPC lipid suspension during dehydration step followed by rehydration in buffer containing 250 mM sucrose and purified connectors.
Insertion of the connector into planar lipid bilayer
The protocol for the incorporation of connectors into lipid bilayer have been described [8; 18; 28–30]. A planar DPhPC lipid bilayer was painted on a 200 µm aperture on a Teflon partition, isolating cis (1 M KCl; 5 mM HEPES, pH 7.9) and trans (0.15 M or 1 M KCl; 5 mM HEPES, pH 7.9) compartments containing conducting buffer. The liposomes were then added into the cis-compartment to fuse with planar lipid bilayer to insert the connector and form planar membrane-embedded nanopores (Fig. 1).
Electrophysiological measurements
Typically, a potential of −50 mV or +50 mV (all voltages refer to Vm) was applied across the connector by a pair of AgCl/Ag electrodes (Fig. 1) and a current passing through the connector was monitored. Driven by the bias voltage, premixed TAT peptide (positive charge with pI 12; MW: 1.6 kDa) was electrophoretically transported through the connector. While translocating, the peptide partially blocks the flow of ions through the connector, which was reflected by a transient current drop. The analog current signal was then amplified, filtered at 5 kHz, and digitized at 10 or 50 kHz by Axopatch 200B patch clamp and Axon 1440A (Axon Instruments, Inc.).
Ni-NTA-nanogold binding assay
Ni-NTA-nanogold binding assay was conducted as previously described [18]. Liposome was added into the cis-chamber with 1 M KCl buffer; Ni-NTA-nanogold (1.8 nm; 300 pM) was added into trans-chamber with 0.15 M KCl buffer to facilitate binding to His-tag. Ni-NTA-nanogold was added after formation of lipid-bilayer. We further ensured that the lipid-bilayer was intact during the course of the experiments to avoid Ni-NTA-nanogold diffusion into the cis-chamber.
Data analysis
The data were recorded with pClamp 10.2 software (Axon Instruments, Inc.). The blockades were calculated with a home-built Matlab program and statistically analyzed with OriginPro 8.1 (OriginLab Corporation). The capture rate was derived by exponentially fitting the time interval between two adjacent events as described previously [36].
RESULTS
Characterization of TAT peptide one-way traffic through C-His SPP1 channels
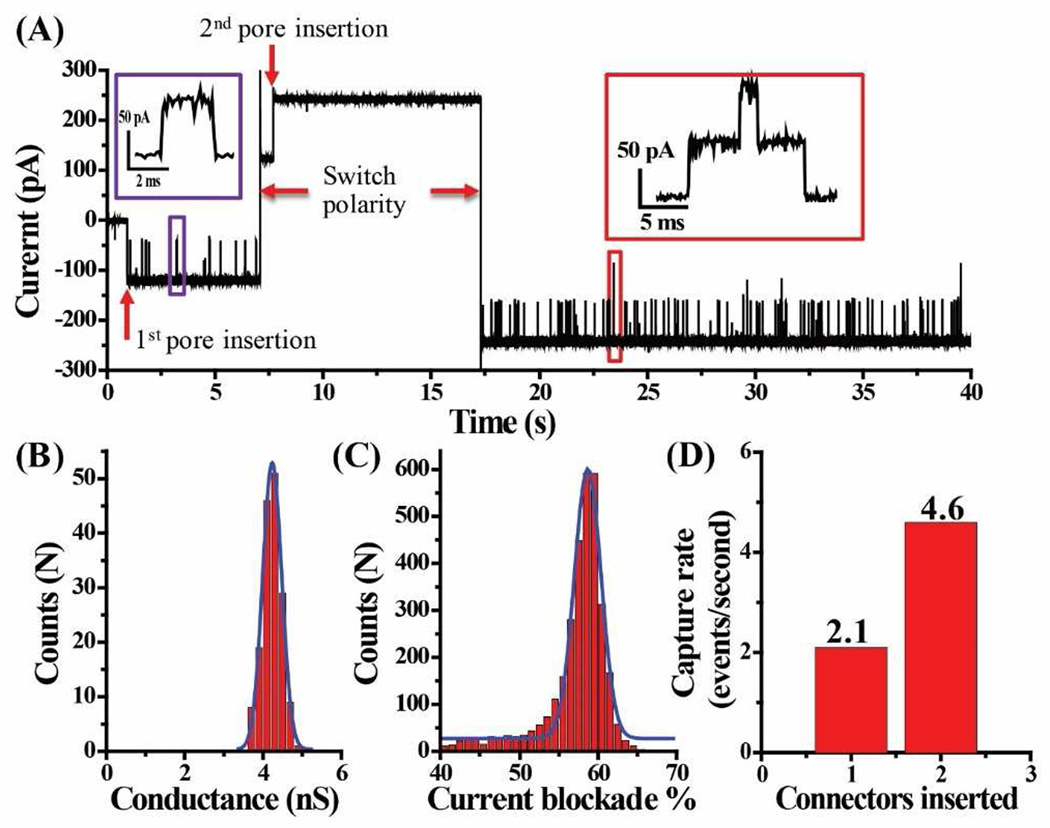
We characterized the electrophysiological properties of the connector after membrane insertion using single channel conduction assays. Fig. 2A represents a typical current trace with C-His SPP1 connector insertion and subsequent TAT translocation. The insertion of C-His SPP1 connector results in a current jump of ~120 pA or 2.4 nS under asymmetric (cis: 1 M KCl; trans: 0.15 M KCl) ionic strength. Under symmetric buffer conditions (1 M KCl buffer in both cis and trans chambers), the connector exhibits a conductance of 4.2±0.5 nS (Fig. 2B). In the absence of peptides, the current trace is quiescent and the connector does not show any gating (transient opening and closing of the channel) properties [29] under the experimental conditions used herein.
Figure 2.
Current trace and capture rate of C-His SPP1 connectors in planar lipid bilayer and subsequent TAT peptide translocation. (A) Representative current trace showing TAT (300 nM) peptide translocation after premixing in both compartments, translocation is only observed at −50 mV but not at +50mV, as demonstrated by polarity switching (cis: 1 M KCl; trans: 0.15 M KCl). Insert: A single peptide translocation (violet box) and two peptides translocation through two membrane-embedded connectors (red). (B) Histogram showing conductance profile of connectors from 163 events (1 M KCl in both cis- and trans-chamber). Blue line: Gaussian fit. (C) Histogram showing current blockade distribution of peptide translocation events (3776 events). Blue line: Gaussian fit. (D) Quantification of peptide capture rate in panel A upon insertion of connectors.
Under an applied voltage of −50 mV, TAT peptide is electrophoretically driven through the C-His SPP1 connector. Due to partial blocking of the channel, a transient drop in current is observed as each peptide translocates through the pore. Fig. 2A shows a typical current trace of TAT peptide translocation in presence of one and subsequently two C-His SPP1 connectors. The ~80 pA spikes (Fig. 2A, violet box insert) representing TAT peptide translocation events correspond to a current blockade of 58.7±3.4% (Fig. 2C). The current blockade % is calculated as [(I0 − Ib)/ I0] ×100, where I0 is the step size of one SPP1 channel insertion, and Ib is the current observed during TAT peptide translocation, normalized to one SPP1 channel. Such translocation events are only observed under −50 mV but not at +50 mV, as demonstrated by polarity switching, which indicates a one-way traffic property [18] for peptide translocation. A second channel inserted with the same orientation as evidenced by both the doubling of capture rate (Fig. 2D) and the distinctive current signature of two TAT peptide translocating through the two pores simultaneously (red insert box). The frequency of those multi-level translocation events increased when more nanopores were present in the lipid membrane (Suppl. Fig. 1).
Identifying the orientation of C-His SPP1 self-assembly into liposome
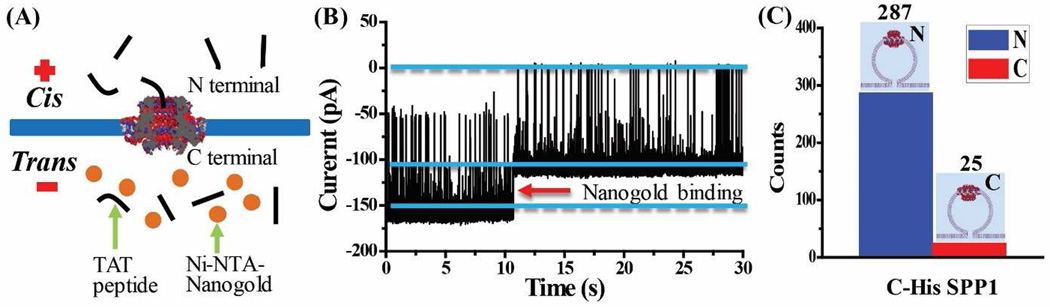
To identify the direction of TAT peptide translocation and the orientation of C-His SPP1 connector in lipid, Ni-NTA-nanogold was added into trans-chamber (Fig. 3A). If the C-terminal 6×His-tag of the connector is facing the trans-chamber, the electron donor groups on histidine imidazole rings will readily form coordinate bonds with the Ni-NTA complex on nanogold (KD of 390 nM) [37; 38].
Figure 3.
Ni-NTA-nanogold binding assay for determining the orientation of C-His SPP1 connector in the lipid membrane with 92% of the connectors with its N terminal outside the liposome. (A) Schematic. (B) Representative current trace showing TAT peptide translocation (300 nM) through a single connector and subsequent binding of Ni-NTA-nanogold (1.8 nm; 300 pM added into trans-chamber) to C-terminal His-tag of connector. (C) Quantification of the orientation adopted by connectors from 312 insertion events. Cis-chamber: 1 M KCl, 5 mM HEPES, pH 7.9; trans-chamber: 0.15 M KCl, 5 mM HEPES, pH 7.9. Applied voltage: −50 mV.
As shown in Fig. 3B, in the presence of a single SPP1 connector in the lipid membrane, positively charged TAT peptides translocate from the cis- to trans-chambers as demonstrated by transient current blockade events. Soon after, a permanent blockade event is observed due to the specific binding between the His-tag and Ni-NTA-nanogold, indicating that the connector is oriented with the C-terminal facing the trans-chamber (as illustrated in Fig. 3A) [18]. As a control, without adding Ni-NTA-nanogold, the current trace does not show any permanent blockade events over an extended period of time (30 mins) (Suppl. Fig. 2). The results demonstrate that C-His SPP1 connector exhibits a one-way traffic property for TAT peptide translocating from N- to C-termini of the connector, a direction of translocation same as the translocation of dsDNA through phi29 motor channel [18; 19].
To determine the orientation preference of the C-His SPP1 connector in the lipid membrane, we investigated a total of 312 connectors by TAT translocation assay. 287 C-His SPP1 connectors show TAT translocation under −50 mV with their N-terminus facing the cis-chamber, while only 25 are oriented with C-terminal facing the cis-chamber (Fig. 3C). Due to liposome fusion with the planar membrane, the C-His SPP1 terminal oriented outwards of the liposome faces the cis-chamber. Thus, the TAT translocation results demonstrate a highly oriented (~ 92%) self-assembly of C-His SPP1 connectors with the N termini outward of the liposomes.
Investigating the orientation of phi29 connector self-assembly into liposome
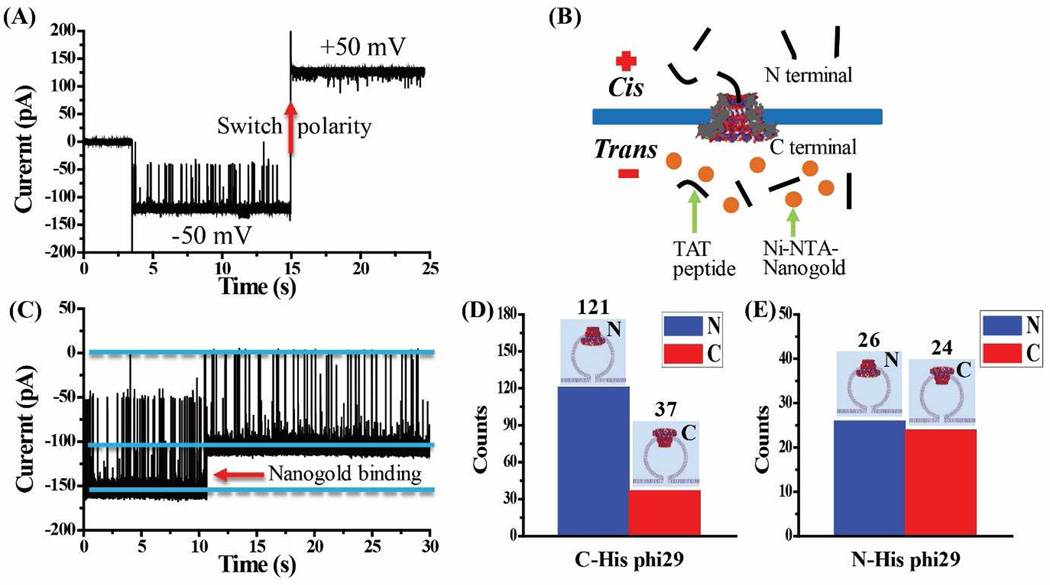
To investigate the mechanism for such orientation preference, the hydrophobicity and hydrophilicity of the N- and C-termini was under scrutiny with phi29 connector as control in this study. Accordingly, phi29 gp10 connector with 6×His-tag either at the C- or N-terminus, as developed previously [8; 39] was tested. Similar to C-His SPP1 connector, TAT peptide also shows a one-way traffic property in both C-His gp10 connector (Fig. 4A) and N-His gp10 connector (Suppl. Fig. 3A). Ni-NTA-nanogold binding assay (Fig. 4B-C) reveal that C-His gp10 connector translocates TAT peptide from N- to C-terminus, as observed in C-His SPP1 connector. Statistical analysis of 158 C-His gp10 connectors reveals that 77% (121 out of 158) of C-His gp10 connectors orient with N-terminus facing the cis-chamber (Fig. 4D) and outward in the liposomes. However, when six highly charged His-tag was attached to the N-termini instead of the C-termini of phi29 connector to enhance the hydrophilicity the N-termini, the ratio of the outward N-termini was reduced, and there is no obvious orientation preference for N-His gp10 connector insertion (Fig. 4E).
Figure 4.
C-His phi29 connectors as control for planar lipid bilayer insertion and subsequent TAT peptide translocation. (A) While TAT peptide is premixed in both compartments (300 nM), translocation is only observed at −50 mV but not at +50 mV, as demonstrated by polarity switching. (B) Schematic of the Ni- NTA-nanogold binding assay for determining the orientation of C-His phi29 connector in the lipid membrane. (C) Representative current trace showing TAT peptide translocation through a single C-His phi29 connector and subsequent binding of Ni-NTA-nanogold (1.8 nm; 300 pM added into trans-chamber) to C-terminal His-tag of C-His phi29 connector. Applied voltage: −50 mV. Quantification of the orientation adopted by (D) C-His phi29 connector from 158 insertion events and (E) N-His phi29 connector from 50 insertion events. Cis-chamber: 1 M KCl, 5 mM HEPES, pH 7.9; trans-chamber: 0.15 M KCl, 5 mM HEPES, pH 7.9.
DISCUSSION
C-His SPP1 connector had a highly preferred orientation when inserted in liposome with the N-terminal end facing outward of the liposomes during reconstitution and facing the cis-chamber after fusion into lipid bilayer. For comparison, C-His phi29 connectors display a weaker orientation preference, while N-His phi29 connectors has no obvious preference in the liposome.
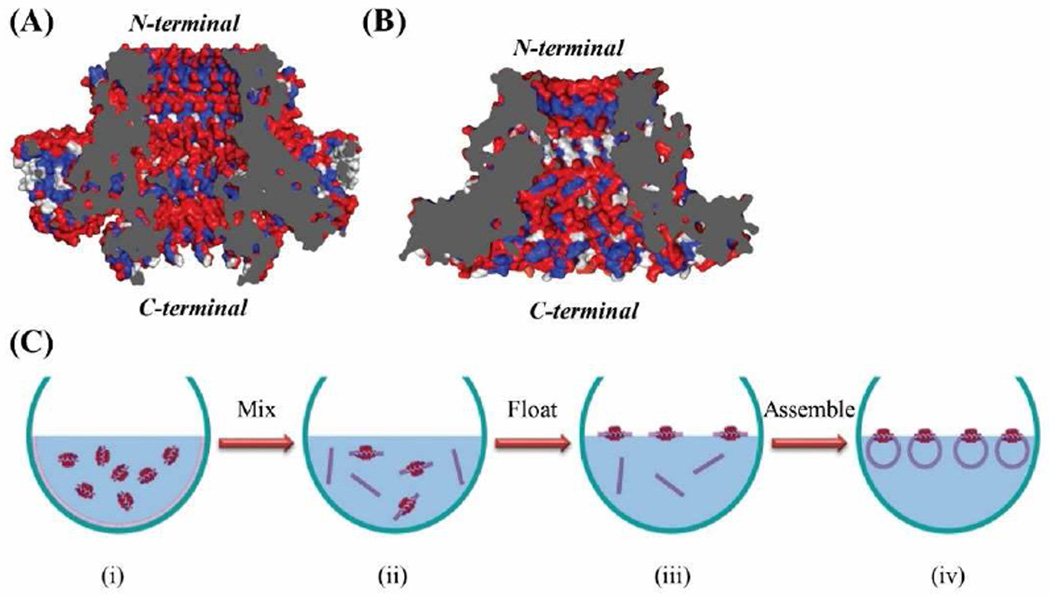
To investigate the mechanism for such orientation preference, the hydrophobicity and hydrophilicity of the N-and C termini of the SPP1 and phi29 was investigated. Liposome formation is governed mainly by hydrophilic and hydrophobic interactions and lowering the interfacial energy of the lipid-air-water system. Histidine is a hydrophilic amino acid and the presence of 6×His-tag can modulate the relative hydrophilicity of the terminal ends of the connector. Our results indicate that the more hydrophilic C-terminal of both C-His SPP1 and C-His phi29 prefer to face inwards in the liposome architecture. Analysis of the cross section of the crystal structures of SPP1 connector (PDB: 2JES) [23; 24] (Fig. 5A) and phi29 connector (PDB: 1FOU) [26; 27] (Fig. 5B) show the location of hydrophilic (red), hydrophobic (blue), and neutral (white) amino acids. For SPP1 connector, the C-terminal amino acids plus 72 hydrophilic histidine amino acids (6×His for each of the 12 subunits) results in significant hydrophilicity at the C-terminus, while the N-terminus is relatively more hydrophobic. For the native phi29 connector, the hydrophilic and hydrophobic groups at N- or C-termini are distributed more evenly, but the C-HIs phi29 connector with the addition of hexa-his-tag exhibits slightly higher hydrophilicity at the C-termini. However, when the hexa-his-tag was added into the N-termini instead of the C-termini, the ratio of the outward N-termini was reduced, and there is no obvious orientation preference. Considering that C-His phi29 has less orientation preference then C-His SPP1 and N-His phi29 has no obvious preference, it appears that whichever terminus is more hydrophilic will tend to face inwards in the liposome during assembly process.
Figure 5.
Cross-sectional structures of SPP1 (A) and phi29 (B) connectors. SPP1 gp6 connector (PDB: 2JES); Phi29 gp10 connector (PDB: 1FOU). Red: hydrophilic; blue: hydrophobic; and white: neutral amino acids. (C) Proposed mechanism of C-His SPP1 connector self-assembly into liposomes. The gray regions represent the cross-section area of the nanopore.
A possible mechanism of the highly oriented self-assembly of the C-His SPP1 connector into liposome is shown in Fig. 5C. When the dehydrated lipid film is mixed with C-His SPP1 connector and rehydration buffer, and vortexed, the lipids and connectors arrange themselves with the hydrophobic lipid tails aligned with hydrophobic sections of the connector, and the hydrophilic lipid head groups aligned with the hydrophilic sections of the connector. Multilamellar lipid fragments form and some of them have connectors embedded in them. The lipid fragments tend to float at the air/water interface to reduce surface tension [40]. In this step, the relatively more hydrophobic N-terminus will face up to contact with air phase and hydrophilic C-terminus will immerse in water phase, to reduce the interfacial energy of the system. During the extrusion process, small unilamellar vesicles form with the more hydrophilic connector terminal facing the interior of the liposomes and more hydrophobic terminal facing outward of the liposomes. Upon fusion of the liposomes with a planar lipid bilayer, the more hydrophobic N-terminal will face towards the cis-chamber, as observed in our studies.
Our findings will facilitate an easy way to obtain liposomes with highly oriented C-His SPP1 connectors embedded in the membrane. Due to the longitudinal asymmetry of the channel, the liposome could possibly be employed as bioreactor or carriers for drug delivery and drug release in a directional manner. By infusing the liposome into planar lipid bilayer, the C-His SPP1 connector can be a highly oriented nanopore for a wide range of single molecule sensing applications.
CONCLUSIONS
C-His connector of SPP1 DNA packaging motor has a highly preferred orientation when self-assembling into liposomes, as proved by single-molecule electrophoresis assay and Ni-NTA-nanogold binding assay. 92% of the C-His SPP1 connectors have their more hydrophilic C-termini facing inward of the liposomes. Changing the hydrophobicity of the N- or C-termini of phi29 connector alters the insertion orientation, suggesting that the hydrophobicity and hydrophilicity of the termini of the protein channel governs the orientation of the insertion into lipid membrane.
Supplementary Material
Acknowledgments
The research was supported by NIH grant R01 EB012135 to P.G. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. P.G's Sylvan G. Frank Endowed Chair position in Pharmaceutics and Drug Delivery is funded by the CM Chen Foundation. P.G. is a consultant of Oxford Nanopore Ltd and RNA Nanobio Ltd.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Wang Y, Zheng D, Tan Q, Wang MX, Gu LQ. Nanopore-based detection of circulating microRNAs in lung cancer patients. Nat Nanotechnol. 2011 Oct;6(10):668–674. doi: 10.1038/nnano.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang S, Haque F, Rychahou PG, Evers BM, Guo P. Engineered Nanopore of Phi29 DNA-Packaging Motor for Real-Time Detection of Single Colon Cancer Specific Antibody in Serum. ACS Nano. 2013 Oct 23;7:9814–9822. doi: 10.1021/nn404435v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, et al. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008 Oct;26(10):1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Venkatesan BM, Bashir R. Nanopore sensors for nucleic acid analysis. Nature Nanotechnology. 2011 Oct;6:615–624. doi: 10.1038/nnano.2011.129. [DOI] [PubMed] [Google Scholar]
- 5.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci U S A. 1996 Nov 26;93(24):13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manrao EA, Derrington IM, Laszlo AH, Langford KW, Hopper MK, Gillgren N, et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat Biotechnol. 2012 Apr;30(4):349–353. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laszlo Andrew H. Decoding long nanopore sequencing reads of natural DNA. Nature Biotechnol. 2014;32:829–833. doi: 10.1038/nbt.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wendell D, Jing P, Geng J, Subramaniam V, Lee TJ, Montemagno C, et al. Translocation of double-stranded DNA through membrane-adapted phi29 motor protein nanopores. Nat Nanotechnol. 2009 Nov;4:765–772. doi: 10.1038/nnano.2009.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franceschini Lorenzo. A nanopore machine promotes the vectorial transport of DNA across membranes. Nature Communications. 2013;4:1–8. doi: 10.1038/ncomms3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar Shiv. PEG-Labeled Nucleotides and Nanopore Detection for Single Molecule DNA Sequencing by Synthesis. Sci Rep. 2012;2:1–8. doi: 10.1038/srep00684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rotem D, Jayasinghe L, Salichou M, Bayley H. Protein detection by nanopores equipped with aptamers. J Am Chem Soc. 2012 Feb 8;134(5):2781–2787. doi: 10.1021/ja2105653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosen Christian B. Single-molecule site-specific detection of protein phosphorylation with a nanopore. Nature Biotechnol. 2014;32:179–181. doi: 10.1038/nbt.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howorka S, Siwy Z. Nanopore analytics: sensing of single molecules. Chem Soc Rev. 2009 Aug;38(8):2360–2384. doi: 10.1039/b813796j. [DOI] [PubMed] [Google Scholar]
- 14.Olasagasti F, Lieberman KR, Benner S, Cherf GM, Dahl JM, Deamer DW, et al. Replication of individual DNA molecules under electronic control using a protein nanopore. Nat Nanotechnol. 2010 Nov;5(11):798–806. doi: 10.1038/nnano.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reiner JE, Kasianowicz JJ, Nablo BJ, Robertson JW. Theory for polymer analysis using nanopore-based single-molecule mass spectrometry. Proc Natl Acad Sci U S A. 2010 Jul 6;107(27):12080–12085. doi: 10.1073/pnas.1002194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodrigues CG, Machado DC, Chevtchenko SF, Krasilnikov OV. Mechanism of KCl Enhancement in Detection of Nonionic Polymers by Nanopore Sensors. Biophysical Journal. 2008 Dec 1;95:5186–5192. doi: 10.1529/biophysj.108.140814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haque F, Li J, Wu H-C, Liang X-J, Guo P. Solid-state and biological nanopore for real-time sensing of single chemical and sequencing of DNA. Nano Today. 2013;8:56–74. doi: 10.1016/j.nantod.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jing P, Haque F, Shu D, Montemagno C, Guo P. One-Way Traffic of a Viral Motor Channel for Double-Stranded DNA Translocation. Nano Lett. 2010 Aug 19;10:3620–3627. doi: 10.1021/nl101939e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Z, Khisamutdinov E, Schwartz C, Guo P. Mechanism of one-way traffic of hexameric phi29 DNA packaging motor with four electropositive relaying layers facilitating antiparallel revolution. ACS Nano. 2013 May 28;7:4082–4092. doi: 10.1021/nn4002775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orlova EV, Gowen B, Droge A, Stiege A, Weise F, Lurz R, et al. Structure of a viral DNA gatekeeper at 10 A resolution by cryo-electron microscopy. EMBO J. 2003 Mar 17;22:1255–1262. doi: 10.1093/emboj/cdg123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lurz R, Orlova EV, Gunther D, Dube P, Droge A, Weise F, et al. Structural organisation of the head-to-tail interface of a bacterial virus 1. J Mol Biol. 2001 Jul 27;310(5):1027–1037. doi: 10.1006/jmbi.2001.4800. [DOI] [PubMed] [Google Scholar]
- 22.Camacho AG, Gual A, Lurz R, Tavares P, Alonso JC. Bacillus subtilis bacteriophage SPP1 DNA packaging motor requires terminase and portal proteins. J Biol Chem. 2003 Jun 27;278:23251–23259. doi: 10.1074/jbc.M301805200. [DOI] [PubMed] [Google Scholar]
- 23.Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, Turner J, et al. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007 Apr 4;26(7):1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lhuillier S, Gallopin M, Gilquin B, Brasiles S, Lancelot N, Letellier G, et al. Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. Proc Natl Acad Sci U S A. 2009 May 26;106(21):8507–8512. doi: 10.1073/pnas.0812407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S, Ji Z, Yan E, Haque F, Guo P. Three-step channel conformational changes common to DNA packaging motors of bacterial viruses T3, T4, SPP1, and Phi29. Virology. 2016 May 12; doi: 10.1016/j.virol.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simpson AA, Leiman PG, Tao Y, He Y, Badasso MO, Jardine PJ, et al. Structure determination of the head-tail connector of bacteriophage phi29. Acta Cryst. 2001;D57:1260–1269. doi: 10.1107/s0907444901010435. [DOI] [PubMed] [Google Scholar]
- 27.Guasch A, Pous J, Ibarra B, Gomis-Ruth FX, Valpuesta JM, Sousa N, et al. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage phi29 connector particle. J Mol Biol. 2002 Jan 25;315(4):663–676. doi: 10.1006/jmbi.2001.5278. [DOI] [PubMed] [Google Scholar]
- 28.Haque F, Geng J, Montemagno C, Guo P. Incorporation of Viral DNA Packaging Motor Channel in Lipid Bilayers for Real-Time, Single-Molecule Sensing of Chemicals and Double-Stranded DNA. Nat Protoc. 2013;8:373–392. doi: 10.1038/nprot.2013.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jing P, Haque F, Vonderheide A, Montemagno C, Guo P. Robust Properties of Membrane-Embedded Connector Channel of Bacterial Virus Phi29 DNA Packaging Motor. Mol Biosyst. 2010;6:1844–1852. doi: 10.1039/c003010d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geng J, Fang H, Haque F, Zhang L, Guo P. Three reversible and controllable discrete steps of channel gating of a viral DNA packaging motor. Biomaterials. 2011 Jul 31;32:8234–8242. doi: 10.1016/j.biomaterials.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang H, Jing P, Haque F, Guo P. Role of channel Lysines and "Push Through a One-way Valve" Mechanism of Viral DNA packaging Motor. Biophysical Journal. 2012 Jan 4;102:127–135. doi: 10.1016/j.bpj.2011.11.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geng J, Wang S, Fang H, Guo P. Channel size conversion of Phi29 DNA-packaging nanomotor for discrimination of single- and double-stranded nucleic acids. ACS Nano. 2013 Apr 23;7(4):3315–3323. doi: 10.1021/nn400020z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haque F, Wang S, Stites C, Chen L, Wang C, Guo P. Single pore translocation of folded, double-stranded, and tetra-stranded DNA through channel of bacteriophage Phi29 DNA packaging motor. Biomaterials. 2015 Mar 26;53:744–752. doi: 10.1016/j.biomaterials.2015.02.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De-Donatis G, Zhao Z, Wang S, Huang PL, Schwartz C, Tsodikov VO, et al. Finding of widespread viral and bacterial revolution dsDNA translocation motors distinct from rotation motors by channel chirality and size. Cell Biosci. 2014;4:30. doi: 10.1186/2045-3701-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haque F, Lunn J, Fang H, Smithrud D, Guo P. Real-Time Sensing and Discrimination of Single Chemicals Using the Channel of Phi29 DNA Packaging Nanomotor. ACS Nano. 2012 Mar 29;6:3251–3261. doi: 10.1021/nn3001615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Z, Hu Y, Shan X, Li W, Bai X, Wang P, et al. Revealing Three Stages of DNA-Cisplatin Reaction by a Solid-State Nanopore. Sci Rep. 2015;5:11868. doi: 10.1038/srep11868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahira I, Fuchida H, Tabata S, Shindo N, Uchinomiya S, Hamachi I, et al. Design of a binuclear Ni(II)-iminodiacetic acid (IDA) complex for selective recognition and covalent labeling of His-tag fused proteins. Bioorg Med Chem Lett. 2014 Jul 1;24(13):2855–2858. doi: 10.1016/j.bmcl.2014.04.096. [DOI] [PubMed] [Google Scholar]
- 38.Khan F, He M, Taussig MJ. Double-hexahistidine tag with high-affinity binding for protein immobilization, purification, and detection on ni-nitrilotriacetic acid surfaces. Anal Chem. 2006 May 1;78(9):3072–3079. doi: 10.1021/ac060184l. [DOI] [PubMed] [Google Scholar]
- 39.Cai Y, Xiao F, Guo P. The effect of N- or C-terminal alterations of the connector of bacteriophage phi29 DNA packaging motor on procapsid assembly, pRNA binding, and DNA packaging. Nanomedicine. 2008;4:8–18. doi: 10.1016/j.nano.2007.10.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yasmann Anthony, Sukharev Sergei. Properties of Diphytanoyl Phospholipids at the Air-Water Interface. Langmuir. 2015;31:350–357. doi: 10.1021/la503800g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.