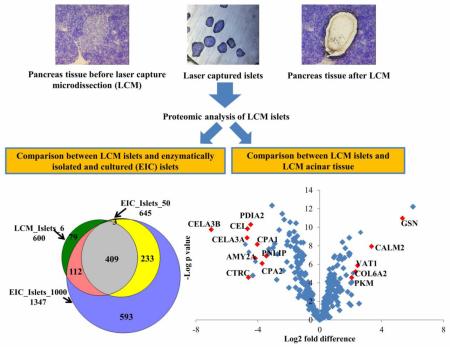
Abstract
The etiology of Type 1 Diabetes (T1D) remains elusive. Enzymatically isolated and cultured (EIC) islets cannot fully reflect the natural protein composition and disease process of in vivo islets, because of the stress from isolation procedures. In order to study islet protein composition in conditions close to the natural environment, we performed proteomic analysis of EIC islets, and laser capture microdissected (LCM) human islets and acinar tissue from fresh-frozen pancreas sections of three cadaveric donors. 1104 and 706 proteins were identified from 6 islets equivalents (IEQ) of LCM islets and acinar tissue, respectively. The proteomic profiles of LCM islets were reproducible within and among cadaveric donors. The endocrine hormones were only detected in LCM islets, whereas catalytic enzymes were significantly enriched in acinar tissue. Furthermore, high overlap (984 proteins) and similar function distribution were found between LCM and EIC islets proteomes, except that EIC islets had more acinar contaminants and stress-related signal transducer activity proteins. The comparison among LCM islets, LCM acinar tissue and EIC islets proteomes indicates that LCM combined with proteomic methods enables accurate and unbiased profiling of islet proteome from frozen pancreata. This paves the way for proteomic studies on human islets during the progression of T1D.
Keywords: Laser capture microdissection, Proteomics, Human pancreatic islets, Human pancreatic acinar tissue, Islet Proteome
Graphical abstract
1. Introduction
Pancreatic islets are composed of several types of endocrine (i.e., hormone-producing) cells, including insulin-secreting β-cells, glucagon-secreting α-cells, somatostatin-secreting δ-cells. Insulin-secreting β-cells are the major constituent of human islets, accounting for 60-87% of the total islet volume [1-3]. The autoimmune-mediated destruction of β-cells leads to the development of Type 1 Diabetes (T1D) [4, 5]. Among the various risk factors proposed for this disease, genetic determinants may contribute to 40-50% risk of T1D [6]. Human leukocyte antigen (HLA) regions were reported to be the strongest genetic determinants, as a strong linkage between T1D and certain variants of the highly polymorphic HLA class II immune recognition molecules has been established [7]. Viral infection was implicated as one of the environmental factors that may trigger autoimmunity and initiate the destruction of β-cells during the development of T1D [8]. Although there is consensus on the view that T1D results from a combination of genetic and environmental factors, the pathogenetic mechanisms are not fully understood. Therefore, investigation on the dysfunction and death of β-cells may contribute to better understanding the pathogenic mechanism of T1D.
Enzymatically isolated and cultured (EIC) islets [9-11] have been frequently used to investigate biochemical signaling pathways that could trigger β-cell changes and death in T1D. A wealth of information can be obtained from EIC islets, and the studies focusing on those from donors with T1D are providing insights of paramount importance to the field of T1D research[12]. However, such in vitro models have some limitations: they do not fully reflect what happens in vivo due to a lack of the natural environment where islets exist and due to the changes in cell physiology induced by isolation and culture. The procedure of enzymatic isolation of pancreatic islets causes major structural changes and induces up-regulation of stress-related genes in islets [13]. The cells’ phenotype, native functions, and responsiveness to stimuli can be altered during culture [14]. It is of note that mRNA levels of insulin and insulin promoter factor 1 decreased by 40% in EIC islets compared with islets in vivo [15], reflecting the inability to sustain mature beta cells in culture. Furthermore, EIC islets frequently contain a significant percentage of contaminating acinar cells and duct cells [16]. On the other hand, animal models exist, but they do not fully mimic human pancreas function, physiology, and disease due to inter-species differences [17-20]. On this note, significant differences between human and mouse islets were reported notably in the cytoarchitecture [1], immune responses, pathways controlling glucose-responsiveness, regenerative capacity, and response to therapy in diabetes [21]. In addition, obtaining fresh isolated human islet samples is challenging because of the limited material availability and because of the requirements in terms of expertise, technologies, and logistics. Networks such as the Integrated Islet Distribution Program (IIDP) <https://iidp.coh.org/>, the Alberta Islet Distribution Program (AIDP) in Canada, the Oxford Consortium for Islet Transplantation (OXCIT) in the UK, and the European Consortium for Islet Transplantation (ECIT) <http://ecit.dri-sanraffaele.org/en/index.html> provide human pancreatic EIC islets for basic research. It is noteworthy that islet isolation is frequently performed on pancreata considered not suitable for pancreas transplantation, and islet distribution for research is frequently performed when islet preparations fail to meet defined release criteria for transplantation (e.g. insufficient islet yield). Although biopsy specimens from the pancreata of living individuals can provide a wealth of information on T1D onset and recurrence [22], performing this procedure on living individuals with T1D is a challenging and risky procedure [23], that also raises important ethical concerns [24]. Alternatively, human pancreatic tissue can be collected from cadaveric individuals, from individuals at-risk for T1D, from diabetic and transplanted patients, and can be preserved frozen. Fresh-frozen pancreata can be obtained from tissue repositories such as nPOD [25, 26], it is frequently available or it can be easily prepared at most pathology facilities. The preparation requires minimal manipulation of the tissue and enables the preservation of the protein content along with the natural tissue architecture. For these reasons we decided to perform laser-capture microdissection (LCM) and proteomic analysis on islets from frozen pancreas sections.
LCM employs a high-energy laser source to separate the desired cells from the remaining tissue section, enabling isolation and downstream analysis [27]. This technique allowed the study of islet- and β-cell-specific genes [16, 28, 29] and proteins [30]. LCM enables the extraction of samples from an environment which is well conserved and close to the natural condition, to better investigate cell physiology [31], cell biology [32], cell transcriptome [13], and proteome [30]. Islets obtained with this method are expected to reflect closely the in vivo molecular composition and pathology of in situ islets. The exploration of the proteome signature of LCM islets with an unbiased method may provide information on the changes in protein composition occurring in dysfunctional islets, even with limited sample amounts, and may thus facilitate understanding of the pathogenesis of T1D.
In this study, we aimed at characterizing the proteome of a limited amount of LCM islets from frozen tissue blocks. In order to confirm the fidelity of the proteome of the LCM material and the accuracy of the microdissection, we compared the proteome of LCM islets from 3 cadaveric donors, each with 3 technique replicates, and we compared the protein profile of LCM islets with that of EIC islets and that of LCM pancreatic acinar tissue. We applied a mass spectrometry-based label-free proteomics approach with advanced instrumentation, nanoLC-Q Exactive HF Orbitrap mass spectrometry (QE-HF MS). With this study we aimed at establishing the feasibility of using LCM to isolate limited amounts of islets from frozen pancreatic tissue sections for proteomics analysis. This study opens the path to future endeavors aimed at investigating the pathophysiology of T1D.
2. Materials and Methods
2.1 Pancreatic tissue sourcing
Research pancreata from three non-diabetic deceased heart-beating donors were obtained from Organ Procurement Organizations, in accordance with federal guidelines for organ donation. Organs were processed at the cGMP facility of the Diabetes Research Institute, University of Miami, under Institutional Review Board-issued waiver that states that islet cells isolation from donors does not constitute human subject research. Organ recovery and transport met transplant-grade criteria. Tissue processing procedures were conducted by the Diabetes Research Institute’s (DRI) cGMP Cell Processing Facility personnel and Pathology core staff. Islet cell isolation was performed following the method reported in a previous study [33].
Briefly, the pancreatic ducts of Wirsung and Santorini were stapled at the pancreas/duodenum interface and the pancreas was separated from the duodenum and blunt-cut at the neck. A tissue block was resected in the region of the neck for immediate embedding in TissueTek OCT and freezing on dry ice. Pancreatic duct in the head and tail sections of the pancreas was cannulated with 18G angiocatheters, and 350 mL of 1.4 mg/mL of Liberase C/T GMP Grade (Roche Diagnostics GmbH, Cat # 05339880001) solution was introduced through the pancreatic duct. The enzymatic solution was injected while keeping the pancreas cold. Following organ distention, the tissue was cut into 7-9 pieces, and loaded in the 500 ml digestion chamber containing a stainless steel screen. Digestion was allowed to proceed at 37 °C until it was judged complete, according to the number of free islets and the amount of acinar tissues observed in the digestion samples collected every minute during the digestion process. Gradient centrifugation on a COBE cell processor 2991 enabled separation of islets from pancreatic acinar and ductal tissue. Following purification and washing, isolated islet cells were cultured overnight in Supplemented CMRL-1066 culture medium (Mediatech, Cat # 99-603-CV), at 22 °C, with 5% CO2. Preparation of EIC islets for proteomic analysis was started following 12 hour culture.
In addition, all work reported here was approved by the University of North Carolina at Greensboro.
2.2 Solvents and Chemicals
Common solvents were purchased from Sigma-Aldrich (St. Louis, MO). Tissue-Tek O.C.T. compound (Sakura 4583, Cat # 25608-930) and 100% ethanol (Decon Labs, Cat # 71006-012) were purchased from VWR. Toluidine blue O (Cat # 198161-25G) was purchased from Sigma-Aldrich. Anti-insulin antibody clone K36aC10 (Cat # I2018, 1:1000) was purchased from Sigma-Aldrich. PPS Silent Surfactant was purchased from Expedeon (San Diego, California, Cat # 21011). Histostain Plus Broadspectrum AEC kit was purchased from Invitrogen (Cat # 85-8943).
2.3 Preparation of frozen tissue sections
Tissue blocks were resected from the neck region of cadaveric pancreata from organ donors, embedded in Tissue-Tek O.C.T. compound (Sakura 4583, VWR Cat # 25608-930) and immediately frozen at −80 °C in dry ice. The blocks were cut with a Leica cryotome into sections of 10 μm thickness, in an environment controlled to −20 °C. The inner chamber and the stage of the cryotome were wiped with 100% ethanol (VWR, Decon Labs Cat # 71006-012) and the blade was changed after each sample in order to avoid contamination. Three pancreas sections were transferred onto each of 10 Leica PEN Membrane slides, i.e. polyethylene naphthalate (PEN) membrane slides (Leica Cat # 11505189) designed for laser capture microdissection. After the placement of three sections on PEN membrane slides, one extra reference section was prepared on a regular glass slide for insulin immunohistochemistry and mapping. Sections were maintained frozen and stored at −80 °C.
2.4 Immunohistochemical staining of reference sections
The reference sections, placed on regular glass slides, were fixed for 15min in 10% formalin, then washed 4 times in PBS (Gibco Life Technologies 10010-049) and stained with an immunohistochemical method to identify islets with β-cells. The immunohistochemical staining was performed with the Histostain Plus Broadspectrum AEC kit, following the manufacturer’s instruction, with the anti-insulin antibody clone K36aC10 (dilution 1:300) and counterstaining. The reference sections were scanned with a PathScan instrument to obtain maps of the entire sections and identify insulin-containing islets.
2.5 Staining and dehydration for LCM
Toluidine blue O staining of frozen pancreas sections enables good discrimination of islets and acinar tissue: islets appear lightly colored compared to the surrounding acinar tissue; moreover, endocrine cells have a characteristic ‘rugged’ aspect in phase contrast illumination. We mapped insulin-containing islets via conventional immunohistochemical staining of reference sections, and we used Toluidine Blue O staining to guide the laser-capture microdissection in ethanoldehydrated sections. One PEN membrane slide at a time was stained and dehydrated, in preparation for each LCM session. A 0.5% w/v Toluidine blue O staining solution was prepared in 70% ethanol. Fresh solution was prepared for each sample, using RNase free disposables.
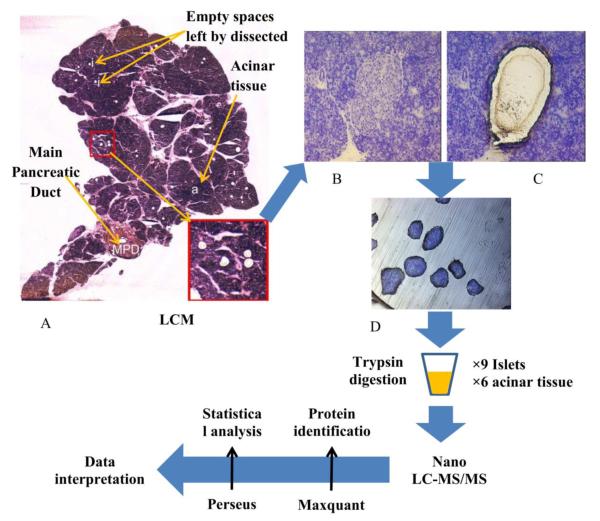
The staining and dehydration protocol was performed with 8 clean Coplin jars, prefilled with 50 mL of 70% (jar # 1-5), 90% (jar #6), and 100% (jar #7, 8) ethanol. The protocol was performed by dipping the slide for 30 seconds in each jar of the ethanol series, following the numerical order, to obtain dehydration. After jar 3, the slide was drained, placed horizontally, stained for 90 seconds with 200 uL of Toluidine blue O staining solution, then drained and transferred to jar 4, to continue dehydration. The stained and dehydrated slide was drained with a Kimwipe (Kimtech Science Kimberly-Clark, VWR cat #21905-026), transferred under a laminar flow hood for 4 minutes, then placed in a slide box containing the desiccant Drierite (WA Hammond, VWR Cat # 22890-902) wrapped in Kimwipes. The workflow of sample preparation is shown in Figure 1.
Figure 1.
Schematic representation of the experimental work flow: section of human pancreas after microdissection of islets (A); Human islet before (B) and after (C) Laser–capture microdissection; Laser-captured islets (D).
2.6 LCM of pancreatic islets and acinar tissue
LCM samples were collected with a Leica LMD system. Each LCM session lasted a total of 60 minutes, to avoid tissue rehydration and degradation. Pancreatic islets were identified in the tissue by visualizing in bright field and in phase-contrast with a lOx magnification. Islet borders can be clearly observed at 20x magnification. Scans of the insulin-stained reference sections were used to map the insulin-containing islets. We have optimized the setup for Laser capture microdissection of our samples. The configuration is shown in Table 1.
Table 1.
System configuration for laser capture microdissection with the Leica LMD instrument
| Parameters | 10x magnification | 20x magnification |
|---|---|---|
| Aperture | 10 | 13 |
| Intensity | 40 | 35 |
| Speed | 4 | 7 |
| Offset | 26 | 34 |
| Ap Diff | 8 | 8 |
| Option | Med | Med |
Samples of islets (LCM islets) and of acinar tissue (LCM acinar tissue) were microdissected from pancreatic tissue sections derived from three cadaveric donors (HP2146, HP2147, and HP2219). Donor characteristics are given in Table 2. Each sample was collected from 6-9 pancreatic sections. The total microdissected area was annotated and converted to islets equivalents (IEQ, see paragraph 2.7 and Table 2). The LCM tissues were collected into the cap of 500 μL sterile RNase/DNase/Protease free Eppendorf tubes and were resuspended with 50 μL of 50 mM NH4HCO3. The tubes were closed, frozen with dry ice, and subsequently maintained at −80 °C. LCM acinar tissue samples were collected and processed in the same way as LCM islets. An equivalent amount of LCM acinar tissue was obtained by laser-capturing the shape utilized for each islet on neighboring areas of acinar tissue, and collecting this tissue into separate tubes.
Table 2.
Details of cadaveric donors and tissue samples. Age, Sex and Body Mass Index (BMI) of the donors are reported. Total areas of pancreatic tissue collected during each LCM session are expressed in μm2; total volumes collected are expressed in Islet Equivalents, IEQ (1 IEQ = 1.77×106 μm3).
| Donor ID | HP2146 | HP2147 | HP2219 | HP2178 | HP2183 |
|---|---|---|---|---|---|
| Age | 23 | 41 | 43 | 35 | 25 |
| Sex | M | F | F | F | M |
| BMI | 22.0 | 33.2 | 31.0 | 34.8 | 30.1 |
|
| |||||
| EIC Islets Volume | 2000 IEQ | 2000 IEQ | |||
|
| |||||
|
LCM Islets1 Area
(Volume) |
1058810 μm2 (5.98 IEQ) |
1058048 μm2 (5.98 IEQ) |
825642 μm2 (4.66 IEQ) |
||
|
LCM Islets2 Area
(Volume) |
776607 μm2 (4.39 IEQ) |
1374966 μm2 (7.77 IEQ) |
825027 μm2 (4.66 IEQ) |
||
|
LCM Islets3 Area
(Volume) |
879671 μm2 (4.96 IEQ) |
1081700 μm2 (6.11 IEQ) |
825164 μm2 (4.66 IEQ) |
||
|
| |||||
|
LCM Acinar1 Area
(Volume) |
1058810 μm2 (5.98 IEQ) |
1058048 μm2 (5.98 IEQ) |
825642 μm2 (4.66 IEQ) |
||
|
LCM Acinar2 Area
(Volume) |
776607 μm2 (4.39 IEQ) |
1374966 μm2 (7.77 IEQ) |
825027 μm2 (4.66 IEQ) |
||
|
LCM Acinar3 Area
(Volume) |
879671 μm2 (4.96 IEQ) |
1081700 μm2 (6.11 IEQ) |
825164 μm2 (4.66 IEQ) |
||
2.7 Conversion of LCM areas to volumes and to IEQ
Following the consensus report of 1990 [34], the total volume of isolated islets can be expressed as number of islet equivalents (IEQ). An IEQ corresponds to the volume of a ‘standard’ islet, a sphere with a diameter d = 150 μm and a volume of VIEQ = 1.77×106 μm3. The area of laser captured tissue was annotated, and the volume was calculated by multiplying the area for the thickness of the tissue section (10 μm). The total volume of laser-captured islets was then divided by the standard volume of 1 IEQ to obtain the number of laser-captured IEQ. 1 IEQ contains approximately 1560 islet cells [3].
2.8 Enzymatically isolated and cultured islets preparation
Enzymatically isolated cultured islet samples (2000 IEQ /sample) were collected from cadaveric donors. The characteristics of the donors are reported in Table 2. After release from the cGMP organ processing facility, EIC islets samples (2000 IEQ /sample) were washed 3 times with PBS. After washing, the supernatant was removed and 50 μL of 50 mM NH4HCO3 were added to the pellet. These islets were resuspended and frozen immediately at −80°C. After thawing samples from two cadaveric donors at room temperature, the volume containing 2000 IEQ from each donor was divided into samples containing 50 (n = 2), 100 (n = 2), 200 (n = 1), 500 (n = 1), 1000 (n = 1) IEQ for further proteomic sample preparation. Each sample was analyzed in duplicate.
2.9 Preparation of samples for proteomic analysis
The sample preparation method was adapted from a method previously reported [10]. Briefly, 6 μL of 1% pps silent surfactant (PPS) was added to extract and solubilize hydrophobic proteins, followed by incubation with 1.5 μL of 50 mM DTT at 95 °C for 6 min. Samples were sonicated for 3 min before alkylation with 7.5 μL 50 mM iodoacetamide for 25 min at 45 °C in the dark. The samples were then digested using trypsin (trypsin : protein = 1:50) at 37 °C overnight. The PPS was hydrolyzed by adding 2 M HCl. The cleavage reaction proceeded for 2 h at room temperature. After that, the samples were spinned at 16,000 g for 12 min and the supernatant was separated for one-dimensional liquid chromatography-tandem mass spectrometry (1D-LCMS/MS) analysis.
2.10 LC-MS/MS analysis
The chromatographic separation of peptides was performed on a PepMap C18 analytical column (2 μm particle, 50 cm × 75 μm i.d., ThermoFisher Scientific). Mobile phases for separation (A: 0.1% FA in water; B: 0.1% FA in ACN) were delivered by an UltiMate 3000 UHPLC system (Thermo Scientific). For analysis, 5.0 μL of each peptide samples were loaded onto the column. A linear gradient of 40% B in 100 min was set to elute peptides. Eluted peptides were ionized by nano-electrospray and analyzed by a QE-HF MS (Thermo Scientific). MS/MS data was acquired in data dependent mode with one full MS scan (resolution 120,000 at m/z 200) followed by 10 MS/MS scans (resolution 15,000 at m/z 200). Other settings of the QE-HF MS used for analysis include: full MS AGC target of 3e6, MS/MS AGC target of 1e5, dynamic exclusion of 20 s, mass isolation window of 1.4, and normalized collision energy of 32.
2.11 Data Analysis
The acquired datasets were analyzed by using MaxQuant (Version 1.5.2.8, http://www.maxquant.org/) and the built-in Andromeda search engine with a UniProt human database (12/3/2014) containing 89,734 entries. The search parameters were as follows: variable modifications of protein N-terminal acetylation and methionine oxidation, and fixed modification of cysteine carbamidomethylation. The minimum peptide length was set to 7 amino acids and a maximum of 2 missed cleavages was allowed for the search. Trypsin was selected as the specific proteolytic enzyme. The global false discovery rate (FDR) cut off used for both peptides and proteins was 0.01 [35]. Label-free quantitation was performed in MaxQuant. To further improve the quantification accuracy, only the razor/unique peptides were used for quantitative calculations. The other parameters used were the default settings in MaxQuant software for processing MS/MS data.
2.12 Statistical Analysis
The resultant data matrix obtained from MaxQuant analysis was further analyzed by Perseus software (Version 1.5.1.6, http://141.61.102.17/perseus_doku/doku.php). Briefly, the proteins, including only identified by site, reverse hits, and potential contaminants (manually selected for contaminants with no protein names), were filtered out from the data matrix. The protein intensities were transformed to log2 scale before performing analysis using various built-in statistical functions of Perseus. A two sample t-test adapted from significance analysis of microarrays [36] was performed with Benjamini-Hochberg FDR cut off = 0.05, S0 = 0.02. S0 in the two sample t-test was applied to take into account both the p value and the difference between group means. The higher S0, the stricter of significant differences analysis is [36].
2.13 Gene Ontology (GO) Functional Annotation Analysis and Enrichment
Protein Analysis Through Evolutionary Relationships (PANTHER) classification system (http://www.pantherdb.org/) was used for GO functional annotation analysis [37] based on the UniProt accession IDs of the identified protein. PANTHER, version 10.0 released on May 15, 2015, includes 11,929 protein families with 83,190 functionally distinct protein subfamilies.
2.14 Protein-Protein interaction networks with the STRING database
The STRING database of known and predicted protein-protein interactions was used to map interaction networks [38]. The interactions include direct (physical) and indirect (functional) associations. We searched the database version 10.0 by accessing the website http://www.string-db.org/ and by making queries for Multiple proteins by identifier (gene names). Networks were designed to show a first shell of query proteins only and a second shell of no more than 5 interactors. All active interaction sources were selected. Minimum required interaction score was set at 0.400. Meaning of network edges was set to evidence.
3. Results
3.1 Overview of the proteome coverage of LCM islets, LCM acinar tissues, and EIC islets
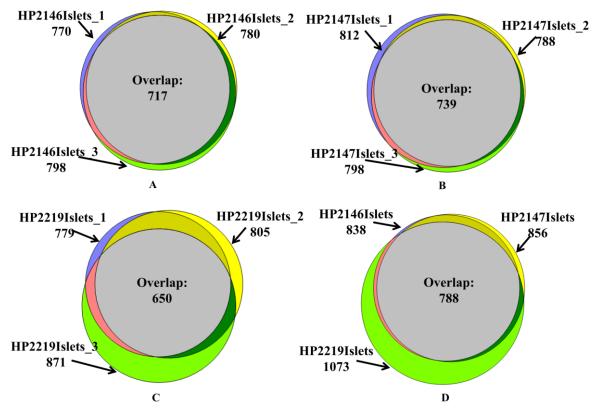
LCM islets samples (n = 9) collected from three cadaveric donors were used in this study. A schematic representation of the experimental workflow (processing of LCM samples, sample digestion, data collection, and data analysis) is indicated in Figure 1. After removing reverse hits and potential contaminants, a total of 1104 unique proteins were identified in 9 samples of LCM islets (Table S1). On average, 800 proteins were detected in LCM islet samples with 6 IEQ (Table S1, Table 2, and Figure 2). Figure 2A, 2B, and 2C show that the number of identified proteins in LCM islets has good reproducibility in the proteome coverage within cadaveric donor, reaching an average of 80% overlap. The proteome reproducibility among cadaveric donors is close to 71%, and is presented in Figure 2D. Of these 1104 proteins identified in LCM islets, 922 proteins were annotated as expressed in islets in the Human Protein Atlas (HPA). HPA is a database with curated annotation of transcriptomic and immunohistochemical (antibody staining) results [39]. Protein abundance in HPA was classified into four levels: high, medium, low abundance, and not detectable. Of those 922 islet proteins, HPA indicates that 169 are detected at high abundance, 347 at medium, and 170 at low abundance (Table S1). We compared the proteins detected in LCM islets with those expected from the transcriptome of the Beta Cell Gene Atlas (BCGA) [40], and 1065 out of 1104 detected were annotated in the pancreatic islet transcriptome. Within the BCGA, the transcriptomic level was categorized into enriched, moderate, low, and no expression [40]. Out of those 1065 LCM islets proteins, 997 derived from pancreatic islet transcripts with enriched expression, 12 with moderate, 16 with low and 32 with no expression (Table 3).
Figure 2.
Venn diagram representation of the proteins identified in LCM islets in 3 technical replicates for each of 3 cadaveric donors: HP2146 (A), HP2147 (B), HP2219 (C). The number of proteins identified in each replicate and in the intersection (Overlap) are indicated. Venn diagram analysis of the sets of all proteins identified in islets from the three cadaveric donor samples (D).
Table 3.
The number of proteins identified in LCM islet and in LCM acinar tissue against the Beta Cell Gene Atlas-BCGA (Transcriptome expression values for each gene were ranked on a scale of 0-100. Cutoffs for expression level are as follows: enriched (>75), moderate (50-75), low (25-50) and no expression (<25) [40])
| Expression level |
BCGA Duct |
BCGA Exocrine |
BCGA Pancreatic islets |
BCGA β- cell |
|
|---|---|---|---|---|---|
| LCM islets | Enriched | 288 | 965 | 997 | 556 |
| Moderate | 553 | 46 | 12 | 200 | |
| Low | 168 | 25 | 16 | 93 | |
| No expression | 88 | 61 | 66 | 248 | |
|
| |||||
| LCM acinar tissue |
Enriched | 214 | 644 | 655 | 397 |
| Moderate | 348 | 19 | 7 | 124 | |
| Low | 75 | 7 | 8 | 45 | |
| No expression | 67 | 34 | 34 | 138 | |
|
| |||||
| Proteins only identified in LCM islets |
Enriched | 93 | 394 | 423 | 187 |
| Moderate | 292 | 30 | 7 | 95 | |
| Low | 64 | 19 | 8 | 53 | |
| No expression | 26 | 32 | 37 | 140 | |
|
| |||||
| Proteins only identified in LCM acinar tissue |
Enriched | 19 | 74 | 76 | 28 |
| Moderate | 49 | 4 | 2 | 19 | |
| Low | 10 | 0 | 0 | 5 | |
| No expression | 5 | 5 | 5 | 31 | |
With the same sample preparation method, 706 proteins were identified in six LCM acinar tissues samples from the same three cadaveric donors as LCM islets. Figure S1 shows the reproducibility in the number of identified proteins within (80% on average) and among (70%) cadaveric donors. 88.9±1.5% of the proteins detected in LCM acinar tissue were also detected in LCM islets (Figure S2, see the overlaps). Overall, 623 proteins identified in LCM islets were in common with those identified in LCM acinar tissue. The known endocrine hormones insulin, somatostatin, and ghrelin-27 were only detected in LCM islets and not in LCM acinar tissue (Table S1). Glucagon (GCG) was only identified in two of the 6 LCM acinar tissues with a relative abundance 1000 fold lower than that of LCM islets (Table S1). Of the 706 proteins identified in LCM acinar tissue, 612 proteins were annotated as expressed in islets in the HPA database, 285 are detected at high abundance, 182 at medium, and 67 at low abundance (Table S1). The correlation of these proteins to BCGA is shown in Table 3.
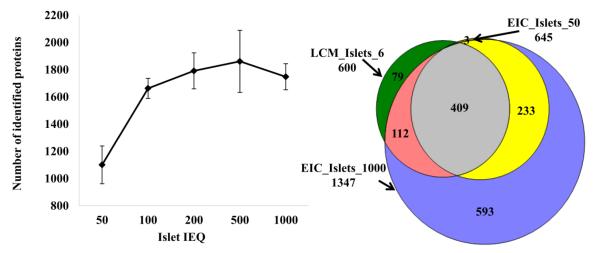
In comparison to the EIC islet proteome, 984 proteins (approximately 90%) identified in LCM islets were also detected in EIC islets (Table S2). The EIC islet proteome was conducted with different amounts of islets, ranging from 50 to 1000 IEQ (Table S3). The number of proteins identified increased with the increase of islets material from 50 to 500 IEQ, and slightly decreased when the islets volume used was 1000 IEQ (Figure 3A). The Venn diagram in Figure 3B shows a comparison of the proteins consistently detected in all replicates of LCM islets (overlap proteins from 9 samples) with approximately 6 IEQ (Table 2), 1000 IEQ of EIC islets (overlap proteins from replicates of 1000 IEQ each) and 50 IEQ of EIC islets (overlap proteins from replicates of 50 IEQ each). The number of proteins consistently detected on 6 IEQ of LCM islets (600) is similar to that of those consistently detected in 50 IEQ of EIC islets (645). Notably, out of 45 proteins reported by HPA to be highly expressed in pancreatic acinar tissue, 44 were detected in EIC islets and were not detected in LCM islets (Table S1).
Figure 3.
Number of proteins identified in EIC islets, in samples ranging from 50 to 1000 IEQ (A). Comparison of proteins identified in 6 IEQ of LCM islets (600 proteins were the overlap proteins from all LCM islets replicates with 6 IEQ) with proteins from 50 IEQ of EIC islets (645 proteins were overlap proteins of all EIC islets replicates with 50 IEQ) and from 1000 IEQ of EIC islets (1347 proteins were overlap proteins of all EIC islets replicates with 1000 IEQ) (B)
3.2 PANTHER Functional Classification
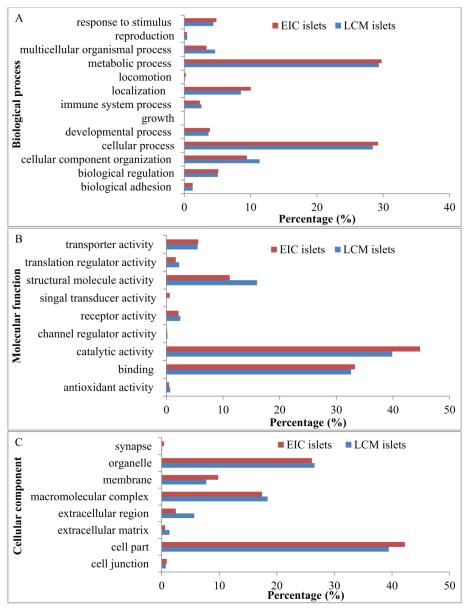
The 1104 UniProt accession IDs identified in LCM islets were analyzed by PANTHER functional classification [37]. Figure 4 shows the PANTHER classification of biological process, molecular function, and cellular component performed on the proteomes of LCM islets and EIC islets. 1803, 1018, and 956 annotation hits were found in the categories of biological process (A), molecular function (B), and cellular component (C) in LCM islets, and 4626, 3092, and 2402 were found in the categories of biological process (A), molecular function (B), and cellular component (C) in EIC islets. The higher number of annotation hits compared to the number of identified proteins is due to the fact that one protein can be involved in multiple biological processes. Metabolic process, catalytic activity, and cell part were the dominant categories in the biological process, molecular function, and cellular component annotations, respectively (Figure 4A, 4B, and 4C). The functional classification distribution of proteins from LCM islets was similar to that of proteins from EIC islets. However, signal transducer activity proteins were only detected in EIC islets and not in LCM islet, whereas extracellular region, extracellular matrix and structural molecule activity proteins were substantially more represented in LCM islets (Figure 4B). The details of signal transducer activity proteins identified in EIC islets but not in LCM islets are reported in Table 4 and their interaction network is shown in Figure S3A. The extracellular region, extracellular matrix, and structural molecular activity proteins identified in LCM islet but not in EIC islets are reported in Table 5, and their interaction network is shown in Figure S3B-S3D.
Figure 4.
PANTHER functional classification for proteins identified in LCM islets and EIC islets. Of the LCM islets proteins, 1803, 1018, and 956 annotation hits were found in the categories of biological process (A), molecular function (B), and cellular component (C), respectively; and for EIC islets proteins, 4626, 3092, and 2402 were found in the categories of biological process (A), molecular function (B), and cellular component (C), respectively.
Table 4.
Signal transducer activity proteins identified in EIC islets but absent in LCM islets (Bold proteins were quantified in more than 10 EIC samples)
| Uniprot ID | Protein name | Gene name |
|---|---|---|
| P29992 | Guanine nucleotide-binding protein subunit alpha-11 | GNA11 |
| Q14344 | Guanine nucleotide-binding protein subunit alpha-13 | GNA13 |
| P63096 | Guanine nucleotide-binding protein G(i) subunit alpha-1 | GNAI1 |
| P04899 | Guanine nucleotide-binding protein G(i) subunit alpha-2 | GNAI2 |
| P08754 | Guanine nucleotide-binding protein G(k) subunit alpha | GNAI3 |
| P09471 | Guanine nucleotide-binding protein G(o) subunit alpha | GNAO1 |
| P50148 | Guanine nucleotide-binding protein G(q) subunit alpha | GNAQ |
| P19086 | Guanine nucleotide-binding protein G(z) subunit alpha | GNAZ |
| P36507 | Dual specificity mitogen-activated protein kinase kinase 2 | MAP2K2 |
| P46734 | Dual specificity mitogen-activated protein kinase kinase 3 | MAP2K3 |
| P45985 | Dual specificity mitogen-activated protein kinase kinase 4 | MAP2K4 |
| O95819 | Mitogen-activated protein kinase kinase kinase kinase 4 | MAP4K4 |
| Q9H205 | Olfactory receptor 2AG1 | OR2AG1 |
| Q03405 | Urokinase plasminogen activator surface receptor | PLAUR |
| O60271 | C-Jun-amino-terminal kinase-interacting protein 4 | SPAG9 |
| Q9Y6E0 | Serine/threonine-protein kinase 24 | STK24 |
Table 5.
Extracellular region, extracellular matrix, and structural molecular activity proteins identified in LCM islets but absent in EIC islets
| Uniprot ID | Protein name | Gene name | |||
|---|---|---|---|---|---|
|
Extracellular
region |
P02671 | Fibrinogen alpha chain | FGA | ||
| P01871 | Ig mu chain C region | IGHM | |||
| P01857 | Ig gamma-1 chain C region | IGHG1 | |||
| P20774 | Mimecan | OGN | |||
| A0A0A0MS12 | Protein IGHV4-34 (Fragment) | IGHV4-34 | |||
| P01859 | Ig gamma-2 chain C region | IGHG2 | |||
| P25311 | Zinc-alpha-2-glycoprotein | AZGP1 | |||
| P02652 | Apolipoprotein A-II | APOA2 | |||
| P01876 | Ig alpha-1 chain C region | IGHA1 | |||
| P01008 | Antithrombin-III | SERPINC1 | |||
| P01019 | Angiotensinogen | AGT | |||
| P06727 | Apolipoprotein A-IV | APOA4 | |||
| P04196 | Histidine-rich glycoprotein | HRG | |||
| P51884 | Lumican | LUM | |||
| P0CG06 | Ig lambda-3 chain C regions | IGLC3 | |||
| O75830 | Serpin I2 | SERPINI2 | |||
| P02765 | Alpha-2-HS-glycoprotein | AHSG | |||
| P05155 | Plasma protease C1 inhibitor | SERPING1 | |||
| P02675 | Fibrinogen beta chain | FGB | |||
| P51888 | Prolargin | PRELP | |||
| P01860 | Ig gamma-3 chain C region | IGHG3 | |||
|
| |||||
|
Extracellular
matrix |
P20774 | Mimecan | OGN | ||
| P02462 | Collagen alpha-1(IV) chain | COL4A1 | |||
| P08572 | Collagen alpha-2(IV) chain | COL4A2 | |||
| P51884 | Lumican | LUM | |||
| P51888 | Prolargin | PRELP | |||
| P02790 | Hemopexin | HPX | |||
|
| |||||
|
Structure
molecular activity |
P09493 | Tropomyosin alpha-1 chain | TPM1 | ||
| P13646 | Keratin, type I cytoskeletal 13 | KRT13 | |||
| Q9H1K4 | Mitochondrial glutamate carrier 2 | SLC25A18 | |||
| P02538 | Keratin, type II cytoskeletal 6A | KRT6A | |||
| Q5T749 | Keratinocyte proline-rich protein | KPRP | |||
| P19013 | Keratin, type II cytoskeletal 4 | KRT4 | |||
| P13647 | Keratin, type II cytoskeletal 5 | KRT5 | |||
| P08729 | Keratin, type II cytoskeletal 7 | KRT7 | |||
| Q04695 | Keratin, type I cytoskeletal 17 | KRT17 | |||
| Q8N1N4 | Keratin, type II cytoskeletal 78 | KRT78 | |||
| P12035 | Keratin, type II cytoskeletal 3 | KRT3 | |||
| P05787 | Keratin, type II cytoskeletal 8 | KRT8 | |||
| P04259 | Keratin, type II cytoskeletal 6B | KRT6B | |||
| Q13835 | Plakophilin-1 | PKP1 | |||
| P35527 | Keratin, type I cytoskeletal 9 | KRT9 | |||
| A6NMB1 | Sialic acid-binding Ig-like lectin 16 | SIGLEC16 | |||
| P04264 | Keratin, type II cytoskeletal 1 | KRT1 | |||
| P13645 | Keratin, type I cytoskeletal 10 | KRT10 | |||
| Q86YZ3 | Hornerin | HRNR | |||
| P35908 | Keratin, type II cytoskeletal 2 epidermal |
KRT2 | |||
| P08779 | Keratin, type I cytoskeletal 16 | KRT16 | |||
| P02533 | Keratin, type I cytoskeletal 14 | KRT14 | |||
3.3 Quantitative results of protein profile in islets and acinar tissue
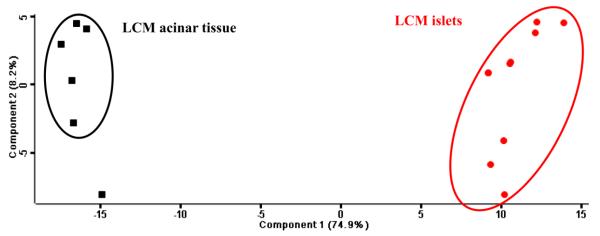
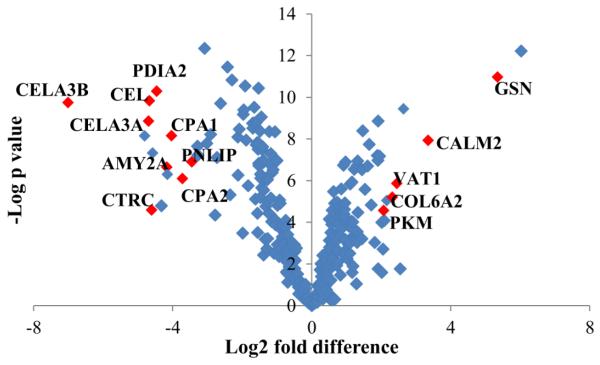
A label-free proteomics technique was applied in this study for the quantification of protein abundance, as shown in Table S1. In order to know whether LCM can accurately dissect the tissue sections as expected, we performed a comparison of the quantitative data associated to the proteomes of LCM islets and LCM acinar tissue. A principal component analysis (PCA) was implemented to identify the most differentially expressed proteins, and to visualize whether a clear separation exists based on the relative abundance of LCM islets and LCM acinar tissue proteins (Figure 5). The first principal component accounted for 74.9% of the variance in the scores plot. Two sample t-test (FDR = 0.05, S0 = 0.02) was conducted on the proteins quantified in all samples of LCM islets and of LCM acinar tissue, resulting in 219 significantly different proteins (Table S4). Figure 6 presents the fold change between LCM islets and LCM acinar tissue proteins, along with statistical significance, by using proteins quantified in all the samples. The abundance of gelsolin (GSN, 32 fold) and calmodulin-like protein 2 (CAML2, 6 fold), Synaptic vesicle membrane protein VAT-1 homolog (VAT1, 5 fold), Collagen alpha-2(VI) chain (COL6A2, 5 fold), Pyruvate kinase PKM (PKM, 4.5 fold) were significantly higher in islets than in acinar tissue (Figure 6). Catalytic enzymes such as pancreatic alpha-amylase (AMY2A), chymotrypsin-like elastase family member (CEL, CELA3A, CELA3B), chymotrypsin-C (CTRC), carboxypeptidase (CPA1, CPA2), protein disulfide-isomerase A2 (PDIA2), and pancreatic triacylglycerol lipase (PNLIP) were significantly higher (8-100 fold) in LCM acinar tissue compared to LCM islets (Figure 6). It is noteworthy that the endocrine hormones insulin, somatostatin, and ghrelin-27 were only detected in islets and not in acinar tissue - glucagon was detected in only a few acinar samples with extremely low abundance; therefore they were excluded in the t-test.
Figure 5.
Principal component analysis of LCM islet and LCM acinar tissue samples. Proteins quantified in both LCM islets and LCM acinar tissue were used in the analysis.
Figure 6.
Scatter plot for visualizing differences in protein abundance between LCM islets (right portion of the plot) and LCM acinar tissue (left portion of the plot). The plot shows - log p values versus the difference of mean (log2 scale, mean is calculated based on proteins quantified in three replicates) between protein quantifications in LCM islets and LCM acinar tissue. Proteins in red were few selected significantly different proteins between LCM islets and LCM acinar tissue (t-test with criterion of FDR = 0.05, S0 = 0.02).
4. Discussion
We applied liquid chromatography-tandem mass spectrometry (LC-MS/MS) to study the proteome of laser capture microdissected (LCM) islets and LCM acinar tissue from frozen sections of human pancreas, along with the proteome of enzymatically isolated and cultured (EIC) islets. The comparisons show that the LCM method enables accurate isolation of islets from frozen samples of pancreas and that the proteomic analysis of the LCM tissue with LCMS/MS yields reproducible protein expression profiles. The LCM method provides sufficient material for a good coverage of islets proteome with limited amounts of tissue, detecting an average of 800 proteins from approximately 6 IEQ per LC-MS/MS analysis (Table 2, Figure 2). The LCM method has been used to separate islets or beta cells for gene expression studies [16, 41]. This method has also been applied to isolate islets from formalin-fixed paraffin embedded slides for downstream protein content analyses [30]. Here we propose the use of LCM to isolate specific cell types from frozen sections for proteomics analysis, a method that avoids protein cross-linking issues in commonly preserved tissues [16, 41, 42]. This method could be applied to study the in situ islet proteome and to “take a snapshot” of the pathophysiological mechanism underlying islet and β-cell dysfunction in Type 1 Diabetes. In addition, the exocrine pancreas was reported to be involved in the pathogenesis of T1D [43, 44] and the method we propose enables proteomic analysis of purified pancreatic acinar tissue without enzymatic isolation. Previous studies on the cellular function of the pancreas were typically performed in vitro on two enzymatically isolated and separated functional units: the endocrine islets of Langerhans and the exocrine pancreatic acinar tissue [11, 45, 46]. Therefore, LCM appears as a promising technique for isolating these two units from their intact environment, in order to investigate the protein composition and pathophysiology of endocrine and exocrine cells in T1D. This may contribute to the understanding of the pathogenesis of T1D and provide potential biomarkers for the diagnosis of T1D at an early stage.
Label-free proteomics technique was used in this study to identify and quantify proteins of all the samples. The number of proteins identified in LCM islet (1104) and LCM acinar tissue (706) (Table S1) were relatively higher than those identified in human β-cells (462) and in human exocrine tissue (372) [47] when a similar 1D LC-MS/MS label-free proteomics platform was used. We identified 1861 proteins (average) in 500 EIC islets, which is lower than the number of proteins reported in the past for EIC islets (3365 proteins in [48] and 4450 proteins in [9]), but likely attributed to the multi-dimensional LC-MS/MS platform and the number of islets used in those studies. Strong cation exchange coupled with LC was used in both studies to broaden the proteomic coverage [9, 48], which is feasible when large numbers of islets are available (~4000 IEQ pooled from 5 donors [48] and ~13,000 IEQ pooled from 21 donors [9]). Despite the low number of islets and despite the fact that 1D-LC-MS/MS was used in this study, the identification of an average of 800 proteins in the LCM islets (Figure 2), with a volume corresponding to only ~6 IEQ without any fractionation indicates a great achievement of the LCM method in protein identification. In addition, the proteomes were reproducible (80%) in technical replicates of LCM islets (Figure 2) and of LCM acinar tissue (Figure S1), and proteome reproducibility was observed even among cadaveric donors (70%). The small variations in islets and acinar tissue proteomes could be related to threshold of detection, as well as to different location or different functional state of the samples collected. Human islets are known to have α-, β-, and δ-cells interspersed the islet core [49]. Many variables could influence protein composition of islet cells and of β-cells, including age of the donor, islet size, fractional content of different endocrine cells in an islet, anatomical location inside the pancreas, functional state, cell size, microenvironment, and timing of tissue harvesting. Differences in the gene expression of LCM β-cells and differences in islets size have been reported [16]. The high reproducibility in the LCM islets and acinar tissue proteome within and among subjects, and the high overlap with the proteomes reported in the HPA (Table S1) and in the BCGA (Table 2) databases suggest fidelity of the proteome obtained from LCM isolated islets.
Furthermore, the high proteome overlap of all the proteins identified in LCM islets with those of EIC islets (984 of 1104, 90%, Table S2) and the high proteome overlap of the common proteins of all replicates in LCM islets_6IEQ, with the common proteins of all replicates in EIC_1000 islets (521 of 600, 87%, Figure 3B) indicates that the LCM method can provide a protein profile comparable to that obtained with the EIC method. This can be observed in the similar distribution of proteins from LCM and EIC islets in the biological processes (Figure 4A), molecular functions (Figure 4B), and cellular components (Figure 4C). The similar distribution of dominant protein categories (Figure 4) further suggests that the LCM method doesn’t have a significant bias in identifying proteins compared to the EIC method. Conversely, the lower number of proteins identified in LCM islets compared to EIC islets can be attributed to the low number of LCM islets used in the study, which could result in protein amounts falling below the detection limits of the instrument. For instance, the proteins detected in EIC islets but absent in LCM islets occurred in relatively low abundance, and some proteins were only identified in one or two replicates (Proteins in yellow, Table S3).
It is noteworthy that 44 highly expressed acinar tissue proteins (acinar-specific, not expressed in islets, based on HPA) were detected in EIC islets and not in LCM islets. This suggests that EIC islets frequently contain a certain proportion of contaminating acinar cells [16]. Furthermore, a set of proteins were consistently identified in EIC islets and not in LCM islets: these include a subset of signal transducer activity proteins - reported in Table 4 and shown in Figure S3A. Among these we observed a series of proteins that can be activated by inflammatory cytokines and environmental stress signals: guanine nucleotide-binding protein (G proteins, GNAs) [50], mitogen-activated protein kinases (MAP Kinases) and serine/threonine-protein kinases[51, 52]. During EIC islet preparation, the exocrine pancreas is digested and its integrity is lost, resulting in the release and activation of a large number of digestive enzymes. These enzymes have a strong destructive effect on islet cells [53], which could induce stress on islet cells. The detection of these stress related proteins in EIC islets (Table 4) corresponds with up-regulation of stress genes in islets isolated with the EIC method [13]. We have also observed a set of proteins that were consistently identified in LCM islets and not in EIC islets. These include extracellular matrix components that may be lost in EIC islets as a result of the collagenase digestion, or as a result of the release of pancreatic digestive enzymes (Table 5, Figure S3B). Collagen 4A1 and 4A2, Lumican (LUM, involved in collagen fibril organization) and Prolargin (PRELP, involved in collagen binding) were not detected in EIC islets. The network of collagens could thus be significantly altered in EIC islets when compared to in situ (LCM) islets. This could result in structural changes inside cells. Supporting this view, a set of structure molecular activity proteins, that control structural integrity, cell organization and polarization[37], were detected in LCM islets and not detected in EIC islets: these include a large number of keratins (KRTs, see Table 5, Figure S3D). These findings could serve as an input for future studies aimed at exploring in great detail the differences between EIC and LCM islets. The absence of acinar tissue specific proteins in LCM islets, along with the absence of stress related proteins and the presence of intracellular structural and extracellular matrix components suggest that LCM on frozen section is a more conservative method, reflecting more closely the actual tissue composition and physiology, compared to the enzymatic isolation and culture of islets.
In addition, the clustering of LCM islets and LCM acinar tissue samples as shown in the principal component analysis (Figure 5), and the significant differences in protein abundance between LCM islets and LCM acinar tissue observed in two sample t-test (Figure 6) indicate that the LCM method reported is able to provide good separation of exocrine and endocrine pancreas. The detection of the well-known hormones, insulin, glucagon, somatostatin, and ghrelin-27 in LCM islets but not in LCM acinar tissue is in agreement with what have been reported in previous studies [54, 55]. The higher abundance of GSN, CALM2, VAT1, COL6A2, and PKM in LCM islets compared to LCM acinar tissue (Figure 6) is in line with the protein expression levels observed via antibody staining reported in HPA [39]. The higher abundance of the catalytic enzymes, AMY2A, CEL, CELA3A, CELA3B, CTRC, CPA1, CPA2, and PNLIP (Figure 6) in acinar tissue compared to islets is in agreement with data from HPA [39] and with previous studies on protein [56] and gene expression levels [13]. The high abundance of these digestive enzymes in acinar tissue (Figure 6) is associated with the main function of the exocrine pancreas, which is to secrete digestive enzymes into the intestine for digesting food to assist nutrient absorption [57]. Of these proteins found to be overexpressed in acinar tissue (Figure 6), CPA1, CELA3B, CEL, and CTRC are reported as pancreatic acinar tissue-specific in the HPA database. We detected these proteins also in islets but with relatively low abundance. Overall, this illustrates that the proteome of islets (and acinar tissue) collected via LCM from frozen sections closely reflects the protein composition of these tissues in their natural environment. Moreover, the fact that our method identified an average of 800 proteins from LCM islet samples of only 6 IEQ indicates that a very small amount of laser-capture microdissected material is sufficient to identify hundreds of proteins.
One factor that could have determined limited proteome coverage is the amount of material in input, i.e. the total volume of microdissected tissue. In this study we show that a volume corresponding to 6 IEQ is sufficient to obtain valuable information, but larger amounts of tissue are expected to provide a broader coverage of the proteome – as indicated by our studies with different amounts of EIC islets. In this study, we identified only a fraction of the proteins present in the LCM islets and EIC islets. Due to this fact, further improvements of the approach will be needed to uncover the actual differences between LCM islets and EIC islets. LCM instruments can be equipped with lasers that have higher power compared to the one we tested. These lasers deliver more energy over time, and thus enable a more efficient and more precise microdissection, boosting the yield. Novel functions in commercially available instruments enable the automation of the tissue area selection based on image recognition algorithms, a feature that largely accelerates tissue collection enabling higher output. Future endeavors will benefit from the advancement of the technologies for laser-capture microdissection and for proteomic analysis.
5. Conclusions
In sum, the LCM method combined with label-free based LC-MS/MS proteomics workflow described in this study allows for proteomics analysis of islets from frozen pancreata. The high similarity found between the proteomes of LCM islets from frozen sections and EIC islets demonstrates that LCM combined with proteomics technique is a reliable way to provide good and reproducible proteome coverage even with small amounts of material. This approach enables studies on the pathology of T1D islets using frozen and stored pancreatic samples. The different proteome profiles of LCM pancreatic islets, LCM acinar tissue, and EIC islets also strongly suggest the usefulness of the LCM method to isolate islets and acinar cells with an intact environment for studies on their molecular composition. Future applications of this LCM method may help us understand the pathogenesis of T1D and identify potential biomarkers for T1D diagnosis at an early stage. In addition, this study provides reference for the optimal islet number for proteomics analysis, which may be useful for planning future proteomic studies of islets isolated using the laser capture microdissection method or the enzymatic isolation method.
Supplementary Material
Figure S1. Venn diagram representation of the proteins identified in LCM acinar tissue in 2 technical replicates for each of 3 cadaveric donors: HP2146 (A), HP2147 (B), HP2219 (C). The number of proteins identified in each replicate and in the intersection (Overlap) are indicated. Venn diagram analysis of the sets of all proteins identified in acinar tissue from the three cadaveric donor samples (D).
Figure S2. Comparison in the identified proteins between LCM islets and LCM acinar tissue from three cadaveric donors: HP2146 (A), HP2147 (B), and HP2219 (C).
Figure S3 Protein-Protein interaction networks with the STRING database. Signal transducer activity proteins only identified in EIC islets (A); extracellular region proteins (B), extracellular matrix (C), and structural molecular activity proteins (D) only identified in LCM islets but not in EIC islets. Nodes represent proteins. Colored nodes represent query proteins; white nodes represent second shell of interactors. Edges represent protein-protein associations: known interactions (from curated databases and experimentally determined), predicted interactions (gene neighborhood, gene fusion, gene co-occurrence) and other (text mining, co-expression, protein homology).
Table S1. LCM Pancreatic islets and LCM acinar tissue proteome and their relative abundance (LFQ) on log2 scale
Table S2 Overlap proteins in LCM islets and EIC Islets (Bold proteins were quantified in more than 10 EIC samples)
Table S3 EIC islets proteome with input volumes ranging from 50 to 1000 IEQ and their relative abundance (LFQ) on log2 scale; proteins in yellow are few examples of proteins only quantified in few samples in EIC islets and absent in LCM islets.
Table S4 Significantly different protein between LCM islets and LCM acinar tissue with FDR=0.05, S0=0.02
Highlights.
The LCM method enables the isolation of islets cells with an intact environment.
LCM-based islet proteomics is a reliable way to provide good proteome coverage.
This study provides reference of the optimal islet equivalents for proteomic studies.
Significance.
The etiological agent triggering autoimmunity against beta cells in Type 1 diabetes (T1D) remains obscure. The in vitro models available (enzymatically isolated and cultured islets, EIC islets) do not accurately reflect what happens in vivo due to lack of the natural environment where islets exist and the preparation-induced changes in cell physiology. The importance of this study is that we investigated the feasibility of laser capture microdissection (LCM) for the isolation of intact islets from frozen cadaveric pancreatic tissue sections. We compared the protein profile of LCM islets (9 replicates from 3 cadaveric donors) with that of both LCM acinar tissues (6 replicates from the same 3 cadaveric donor as LCM islets) and EIC islets (at least 4 replicates for each sample with the same islets equivalents) by using proteomics techniques with advanced instrumentation, nanoLC-Q Exactive HF Orbitrap mass spectrometry (nano LC-MS/MS). The results demonstrate that the LCM method is reliable in isolating islets with an intact environment. LCM-based islet proteomics is a feasible approach to obtain good proteome coverage for assessing the pathology of T1D using cadaveric pancreatic samples, even from very small sample amounts. Future applications of this LCM-based proteomic method may help us understand the pathogenesis of T1D and identify potential biomarkers for T1D diagnosis at an early stage.
Acknowledgements
This work was supported by the National Institutes of Health (DK099174) and by the Diabetes Research Institute Foundation. The authors thank Dr. Chih-Wei Liu, Dr. Ricardo Pastori, Dr. Peter Buchwald and Dr. Elina Linetsky for their help in the preparation of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Cabrera O, Berman DM, Kenyon NS, Ricordi C, Berggren PO, Caicedo A. The unique cytoarchitecture of human pancreatic islets has implications for islet cell function. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(7):2334–2339. doi: 10.1073/pnas.0510790103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Brissova M, Fowler MJ, Nicholson WE, Chu A, Hirshberg B, Harlan DM, Powers AC. Assessment of human pancreatic islet architecture and composition by laser scanning confocal microscopy. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 2005;53(9):1087–97. doi: 10.1369/jhc.5C6684.2005. [DOI] [PubMed] [Google Scholar]
- [3].Pisania A, Weir GC, O'Neil JJ, Omer A, Tchipashvili V, Lei J, Colton CK, Bonner-Weir S. Quantitative analysis of cell composition and purity of human pancreatic islet preparations. Laboratory investigation; a journal of technical methods and pathology. 2010;90(11):1661–75. doi: 10.1038/labinvest.2010.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ziegler AG, Nepom GT. Prediction and pathogenesis in type 1 diabetes. Immunity. 2010;32(4):468–78. doi: 10.1016/j.immuni.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wyatt R, Williams AJ. Islet Autoantibody Analysis: Radioimmunoassays. Methods in molecular biology (Clifton, N.J.) 2015 doi: 10.1007/7651_2015_292. [DOI] [PubMed] [Google Scholar]
- [6].Xie Z, Chang C, Zhou Z. Molecular mechanisms in autoimmune type 1 diabetes: a critical review. Clinical reviews in allergy & immunology. 2014;47(2):174–92. doi: 10.1007/s12016-014-8422-2. [DOI] [PubMed] [Google Scholar]
- [7].Notkins AL, Lernmark A. Autoimmune type 1 diabetes: resolved and unresolved issues. The Journal of clinical investigation. 2001;108(9):1247–52. doi: 10.1172/JCI14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Colli ML, Nogueira TC, Allagnat F, Cunha DA, Gurzov EN, Cardozo AK, Roivainen M, Op de beeck A, Eizirik DL. Exposure to the Viral By-Product dsRNA or Coxsackievirus B5 Triggers Pancreatic Beta Cell Apoptosis via a Bim / Mcl-1 Imbalance. PLoS Pathog. 2011;7(9):e1002267. doi: 10.1371/journal.ppat.1002267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Schrimpe-Rutledge AC, Fontès G, Gritsenko MA, Norbeck Angela D., Anderson DJ, Waters M, Adkins JN, Smith RD, Poitout V, Metz TO. Discovery of Novel Glucose-Regulated Proteins in Isolated Human Pancreatic Islets Using LC–MS/MS-Based Proteomics. Journal of proteome research. 2012;11(7):3520–3532. doi: 10.1021/pr3002996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Waanders LF, Chwalek K, Monetti M, Kumar C, Lammert E, Mann M. Quantitative proteomic analysis of single pancreatic islets. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(45):18902–7. doi: 10.1073/pnas.0908351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Eizirik DL, Sammeth M, Bouckenooghe T, Bottu G, Sisino G, Igoillo-Esteve M, Ortis F, Santin I, Colli ML, Barthson J, Bouwens L, Hughes L, Gregory L, Lunter G, Marselli L, Marchetti P, McCarthy MI, Cnop M. The human pancreatic islet transcriptome: expression of candidate genes for type 1 diabetes and the impact of pro-inflammatory cytokines. PLoS genetics. 2012;8(3):e1002552. doi: 10.1371/journal.pgen.1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lanzoni G, Pokrywczynska M, Inverardi L. Team Science in Type 1 Diabetes: New Insights from the Network for Pancreatic Organ Donors with Diabetes (nPOD) CELLR4. 2016 In Press. [Google Scholar]
- [13].Marselli L, Thorne J, Ahn YB, Omer A, Sgroi DC, Libermann T, Otu HH, Sharma A, Bonner-Weir S, Weir GC. Gene expression of purified beta-cell tissue obtained from human pancreas with laser capture microdissection. The Journal of clinical endocrinology and metabolism. 2008;93(3):1046–53. doi: 10.1210/jc.2007-0931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kaur G, Dufour JM. Cell lines: Valuable tools or useless artifacts. Spermatogenesis. 2012;2(1):1–5. doi: 10.4161/spmg.19885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ahn YB, Xu G, Marselli L, Toschi E, Sharma A, Bonner-Weir S, Sgroi DC, Weir GC. Changes in gene expression in beta cells after islet isolation and transplantation using laser-capture microdissection. Diabetologia. 2007;50(2):334–42. doi: 10.1007/s00125-006-0536-5. [DOI] [PubMed] [Google Scholar]
- [16].Marselli L, Thorne J, Dahiya S, Sgroi DC, Sharma A, Bonner-Weir S, Marchetti P, Weir GC. Gene expression profiles of Beta-cell enriched tissue obtained by laser capture microdissection from subjects with type 2 diabetes. PloS one. 2010;5(7):e11499. doi: 10.1371/journal.pone.0011499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Haurogne K, Pavlovic M, Rogniaux H, Bach JM, Lieubeau B. Type 1 Diabetes Prone NOD Mice Have Diminished Cxcr1 mRNA Expression in Polymorphonuclear Neutrophils and CD4+ T Lymphocytes. PloS one. 2015;10(7):e0134365. doi: 10.1371/journal.pone.0134365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Vieira FS, Nanini HF, Takiya CM, Coutinho-Silva R. P2X7 receptor knockout prevents streptozotocin-induced type 1 diabetes in mice. Molecular and cellular endocrinology. 2016;419:148–57. doi: 10.1016/j.mce.2015.10.008. [DOI] [PubMed] [Google Scholar]
- [19].Yanagimachi T, Fujita Y, Takeda Y, Honjo J, Atageldiyeva KK, Takiyama Y, Abiko A, Makino Y, Kieffer TJ, Haneda M. Pancreatic glucose-dependent insulinotropic polypeptide (GIP) (1-30) expression is upregulated in diabetes and PEGylated GIP(1-30) can suppress the progression of low-dose-STZ-induced hyperglycaemia in mice. Diabetologia. 2015 doi: 10.1007/s00125-015-3842-y. [DOI] [PubMed] [Google Scholar]
- [20].Yoshida K, Kikutani H. Genetic and immunological basis of autoimmune diabetes in the NOD mouse. Reviews in immunogenetics. 2000;2(1):140–6. [PubMed] [Google Scholar]
- [21].King AJ. The use of animal models in diabetes research. British journal of pharmacology. 2012;166(3):877–94. doi: 10.1111/j.1476-5381.2012.01911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Vendrame F, Pileggi A, Laughlin E, Allende G, Martin-Pagola A, Molano RD, Diamantopoulos S, Standifer N, Geubtner K, Falk BA, Ichii H, Takahashi H, Snowhite I, Chen Z, Mendez A, Chen L, Sageshima J, Ruiz P, Ciancio G, Ricordi C, Reijonen H, Nepom GT, Burke GW, 3rd, Pugliese A. Recurrence of type 1 diabetes after simultaneous pancreas-kidney transplantation, despite immunosuppression, is associated with autoantibodies and pathogenic autoreactive CD4 T-cells. Diabetes. 2010;59(4):947–57. doi: 10.2337/db09-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Krogvold L, Edwin B, Buanes T, Ludvigsson J, Korsgren O, Hyoty H, Frisk G, Hanssen KF, Dahl-Jorgensen K. Pancreatic biopsy by minimal tail resection in live adult patients at the onset of type 1 diabetes: experiences from the DiViD study. Diabetologia. 2014;57(4):841–3. doi: 10.1007/s00125-013-3155-y. [DOI] [PubMed] [Google Scholar]
- [24].Atkinson MA. Pancreatic biopsies in type 1 diabetes: revisiting the myth of Pandora's box. Diabetologia. 2014;57(4):656–9. doi: 10.1007/s00125-013-3159-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Campbell-Thompson M, Wasserfall C, Kaddis J, Albanese-O'Neill A, Staeva T, Nierras C, Moraski J, Rowe P, Gianani R, Eisenbarth G, Crawford J, Schatz D, Pugliese A, Atkinson M. Network for Pancreatic Organ Donors with Diabetes (nPOD): developing a tissue biobank for type 1 diabetes. Diabetes/metabolism research and reviews. 2012;28(7):608–17. doi: 10.1002/dmrr.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Pugliese A, Yang M, Kusmarteva I, Heiple T, Vendrame F, Wasserfall C, Rowe P, Moraski JM, Ball S, Jebson L, Schatz DA, Gianani R, Burke GW, Nierras C, Staeva T, Kaddis JS, Campbell-Thompson M, Atkinson MA. The Juvenile Diabetes Research Foundation Network for Pancreatic Organ Donors with Diabetes (nPOD) Program: goals, operational model and emerging findings. Pediatric diabetes. 2014;15(1):1–9. doi: 10.1111/pedi.12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bonner RF, Emmert-Buck M, Cole K, Pohida T, Chuaqui R, Goldstein S, Liotta LA. Laser capture microdissection: molecular analysis of tissue. Science (New York, N.Y.) 1997;278(5342):1481–1483. doi: 10.1126/science.278.5342.1481. [DOI] [PubMed] [Google Scholar]
- [28].Ginsberg SD, Hemby SE, Mufson EJ, Martin LJ. Cell and Tissue Microdissection in Combination with Genomic and Proteomic Applications. In: Zaborszky L, Wouterlood FG, Lanciego JL, editors. Neuroanatomical Tract-Tracing 3: Molecules, Neurons, and Systems. Springer US; Boston, MA: 2006. pp. 109–141. [Google Scholar]
- [29].Altar CA, Vawter MP, Ginsberg SD. Target identification for CNS diseases by transcriptional profiling. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2009;34(1):18–54. doi: 10.1038/npp.2008.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nishida Y, Aida K, Kihara M, Kobayashi T. Antibody-validated proteins in inflamed islets of fulminant type 1 diabetes profiled by laser-capture microdissection followed by mass spectrometry. PloS one. 2014;9(10):e107664. doi: 10.1371/journal.pone.0107664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sturm D, Marselli L, Ehehalt F, Richter D, Distler M, Kersting S, Grutzmann R, Bokvist K, Froguel P, Liechti R, Jorns A, Meda P, Baretton GB, Saeger HD, Schulte AM, Marchetti P, Solimena M. Improved protocol for laser microdissection of human pancreatic islets from surgical specimens. Journal of visualized experiments. 2013;(71):50231. doi: 10.3791/50231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Marciniak A, Cohrs CM, Tsata V, Chouinard JA, Selck C, Stertmann J, Reichelt S, Rose T, Ehehalt F, Weitz J, Solimena M, Slak Rupnik M, Speier S. Using pancreas tissue slices for in situ studies of islet of Langerhans and acinar cell biology. Nature protocols. 2014;9(12):2809–22. doi: 10.1038/nprot.2014.195. [DOI] [PubMed] [Google Scholar]
- [33].Ricordi C, Lacy PE, Scharp DW. Automated islet isolation from human pancreas. Diabetes. 1989;38(Suppl 1):140–2. doi: 10.2337/diab.38.1.s140. [DOI] [PubMed] [Google Scholar]
- [34].Ricordi C, Gray DW, Hering BJ, Kaufman DB, Warnock GL, Kneteman NM, Lake SP, London NJ, Socci C, Alejandro R, et al. Islet isolation assessment in man and large animals. Acta diabetologica latina. 1990;27(3):185–95. doi: 10.1007/BF02581331. [DOI] [PubMed] [Google Scholar]
- [35].Michalski A, Cox J, Mann M. More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. Journal of proteome research. 2011;10(4):1785–93. doi: 10.1021/pr101060v. [DOI] [PubMed] [Google Scholar]
- [36].Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(9):5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mi H, Guo N, Kejariwal A. P.D. Thomas, PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic acids research. 2007;35(Database issue):D247–52. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic acids research. 2015;43(Database issue):D447–52. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F. Proteomics. Tissue-based map of the human proteome. Science (New York, N.Y.) 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- [40].Kutlu B, Burdick D, Baxter D, Rasschaert J, Flamez D, Eizirik DL, Welsh N, Goodman N, Hood L. Detailed transcriptome atlas of the pancreatic beta cell. BMC Med Genomics. 2009;2:3. doi: 10.1186/1755-8794-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Marselli L, Sgroi DC, Bonner-Weir S, Weir GC. Laser capture microdissection of human pancreatic beta-cells and RNA preparation for gene expression profiling. Methods in molecular biology (Clifton, N.J.) 2009;560:87–98. doi: 10.1007/978-1-59745-448-3_8. [DOI] [PubMed] [Google Scholar]
- [42].Shapiro JP, Biswas S, Merchant AS, Satoskar A, Taslim C, Lin S, Rovin BH, Sen CK, Roy S, Freitas MA. A quantitative proteomic workflow for characterization of frozen clinical biopsies: laser capture microdissection coupled with label-free mass spectrometry. J Proteomics. 2012;77:433–40. doi: 10.1016/j.jprot.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Liu CW, Atkinson MA, Zhang Q. Type 1 diabetes cadaveric human pancreata exhibit a unique exocrine tissue proteomic profile. Proteomics. 2016;16(9):1432–46. doi: 10.1002/pmic.201500333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Rodriguez-Calvo T, Ekwall O, Amirian N, Zapardiel-Gonzalo J, von Herrath MG. Increased immune cell infiltration of the exocrine pancreas: a possible contribution to the pathogenesis of type 1 diabetes. Diabetes. 2014;63(11):3880–90. doi: 10.2337/db14-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Choudhary A, Hu He K, Mertins P, Udeshi ND, Dancik V, Fomina-Yadlin D, Kubicek S, Clemons PA, Schreiber SL, Carr SA, Wagner BK. Quantitative-proteomic comparison of alpha and Beta cells to uncover novel targets for lineage reprogramming. PloS one. 2014;9(4):e95194. doi: 10.1371/journal.pone.0095194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Goke B. Islet cell function: alpha and beta cells--partners towards normoglycaemia. International journal of clinical practice. Supplement. 2008;(159):2–7. doi: 10.1111/j.1742-1241.2007.01686.x. [DOI] [PubMed] [Google Scholar]
- [47].Brackeva B, Kramer G, Vissers JPC, Martens GA. Quantitative proteomics of rat and human pancreatic beta cells. Data in brief. 2015;3:234–239. doi: 10.1016/j.dib.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Metz TO, Jacobs JM, Gritsenko MA, Fontes G, Qian WJ, Camp DG, 2nd, Poitout V, Smith RD. Characterization of the human pancreatic islet proteome by two-dimensional LC/MS/MS. Journal of proteome research. 2006;5(12):3345–54. doi: 10.1021/pr060322n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Weir GC, Marselli L, Marchetti P, Katsuta H, Jung MH, Bonner-Weir S. Towards better understanding of the contributions of overwork and glucotoxicity to the beta-cell inadequacy of type 2 diabetes. Diabetes, obesity & metabolism. 2009;11(Suppl 4):82–90. doi: 10.1111/j.1463-1326.2009.01113.x. [DOI] [PubMed] [Google Scholar]
- [50].Tokiwa G, Dikic I, Lev S, Schlessinger J. Activation of Pyk2 by stress signals and coupling with JNK signaling pathway. Science (New York, N.Y.) 1996;273(5276):792–4. doi: 10.1126/science.273.5276.792. [DOI] [PubMed] [Google Scholar]
- [51].Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiological reviews. 2001;81(2):807–69. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- [52].Cuenda A. Mitogen-activated protein kinase kinase 4 (MKK4) The international journal of biochemistry & cell biology. 2000;32(6):581–7. doi: 10.1016/s1357-2725(00)00003-0. [DOI] [PubMed] [Google Scholar]
- [53].Pang X, Xue W, Feng X, Tian X, Teng Y, Ding X, Pan X, Guo Q, He X. Experimental studies on islets isolation, purification and function in rats. International journal of clinical and experimental medicine. 2015;8(11):20932–8. [PMC free article] [PubMed] [Google Scholar]
- [54].Barreto SG, Carati CJ, Toouli J, Saccone GT. The islet-acinar axis of the pancreas: more than just insulin. American journal of physiology. Gastrointestinal and liver physiology. 2010;299(1):G10–22. doi: 10.1152/ajpgi.00077.2010. [DOI] [PubMed] [Google Scholar]
- [55].Andralojc KM, Mercalli A, Nowak KW, Albarello L, Calcagno R, Luzi L, Bonifacio E, Doglioni C, Piemonti L. Ghrelin-producing epsilon cells in the developing and adult human pancreas. Diabetologia. 2009;52(3):486–93. doi: 10.1007/s00125-008-1238-y. [DOI] [PubMed] [Google Scholar]
- [56].Gronborg M, Bunkenborg J, Kristiansen TZ, Jensen ON, Yeo CJ, Hruban RH, Maitra A, Goggins MG, Pandey A. Comprehensive proteomic analysis of human pancreatic juice. Journal of proteome research. 2004;3(5):1042–55. doi: 10.1021/pr0499085. [DOI] [PubMed] [Google Scholar]
- [57].Whitcomb DC, Lowe ME. Human pancreatic digestive enzymes. Digestive diseases and sciences. 2007;52(1):1–17. doi: 10.1007/s10620-006-9589-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Venn diagram representation of the proteins identified in LCM acinar tissue in 2 technical replicates for each of 3 cadaveric donors: HP2146 (A), HP2147 (B), HP2219 (C). The number of proteins identified in each replicate and in the intersection (Overlap) are indicated. Venn diagram analysis of the sets of all proteins identified in acinar tissue from the three cadaveric donor samples (D).
Figure S2. Comparison in the identified proteins between LCM islets and LCM acinar tissue from three cadaveric donors: HP2146 (A), HP2147 (B), and HP2219 (C).
Figure S3 Protein-Protein interaction networks with the STRING database. Signal transducer activity proteins only identified in EIC islets (A); extracellular region proteins (B), extracellular matrix (C), and structural molecular activity proteins (D) only identified in LCM islets but not in EIC islets. Nodes represent proteins. Colored nodes represent query proteins; white nodes represent second shell of interactors. Edges represent protein-protein associations: known interactions (from curated databases and experimentally determined), predicted interactions (gene neighborhood, gene fusion, gene co-occurrence) and other (text mining, co-expression, protein homology).
Table S1. LCM Pancreatic islets and LCM acinar tissue proteome and their relative abundance (LFQ) on log2 scale
Table S2 Overlap proteins in LCM islets and EIC Islets (Bold proteins were quantified in more than 10 EIC samples)
Table S3 EIC islets proteome with input volumes ranging from 50 to 1000 IEQ and their relative abundance (LFQ) on log2 scale; proteins in yellow are few examples of proteins only quantified in few samples in EIC islets and absent in LCM islets.
Table S4 Significantly different protein between LCM islets and LCM acinar tissue with FDR=0.05, S0=0.02