Abstract
Stable gene transfer into target cell populations via integrating viral vectors is widely used in stem cell gene therapy (SCGT). Accurate vector copy number (VCN) estimation has become increasingly important. However, existing methods of estimation such as real-time quantitative PCR are more restricted in practicality, especially during clinical trials, given the limited availability of sample materials from patients. This study demonstrates the application of an emerging technology called droplet digital PCR (ddPCR) in estimating VCN states in the context of SCGT. Induced pluripotent stem cells (iPSCs) derived from a patient with X-linked chronic granulomatous disease were used as clonable target cells for transduction with alpharetroviral vectors harboring codon-optimized CYBB cDNA. Precise primer–probe design followed by multiplex analysis conferred assay specificity. Accurate estimation of per-cell VCN values was possible without reliance on a reference standard curve. Sensitivity was high and the dynamic range of detection was wide. Assay reliability was validated by observation of consistent, reproducible, and distinct VCN clustering patterns for clones of transduced iPSCs with varying numbers of transgene copies. Taken together, use of ddPCR appears to offer a practical and robust approach to VCN estimation with a wide range of clinical and research applications.
Keywords: : chronic granulomatous disease, droplet digital PCR, induced pluripotent stem cells, integrating vectors, vector copy number
Introduction
Integrating vectors effectively achieve long-term expression of a transgene in the treatment of monogenetic hematopoietic disorders.1 Gene insertion into stem/progenitor cells results in stable modification of all hematopoietic lineages. Vector biological potency correlates positively with per-cell provirus integrations, as does genotoxicity due to proto-oncogene activation.2 Precise estimation of average vector copy number (VCN) in targeted cell populations is important in defining therapeutic windows, with neither too few integrated vectors to affect transgene expression nor so many as to trigger malignancy. An early approach to estimating VCN, Southern blotting, permits quantitative determination of provirus insertions in a clone; however, this technique is somewhat time-consuming.3 More commonly, real-time quantitative PCR (qPCR) is used to determine VCN by fluorophore detection. This may be achieved with dyes such as SYBR green, which will intercalate into double-stranded amplified product. However, detection using intercalating dyes may lead to false positives occurring because the intercalation process is not specific. Where specificity is required in detecting particular target sequences, hydrolysis probes are preferably used. A signal is detected when a fluorescent reporter is cleaved during targeted-gene PCR extension.4 The threshold cycle (Ct) value for a particular transgene (the number of cycles required to reach an arbitrary value of fluorescence intensity during the exponential phase of amplification) is used to calculate VCN in reference to a standard curve or to the Ct value for an endogenous housekeeping gene.5 Reduced template purity or concentration, however, by reducing PCR amplification efficiency, compromises qPCR assay accuracy. Both Southern blotting and qPCR lack scalability, precluding high-throughput analysis when sample size is limited. Especially for the former, relatively large amounts of high-quality, purified genomic DNA (gDNA) are required. This is unpractical to achieve during follow-ups in gene therapy clinical trials, where it is difficult to obtain such large amounts of sample material from patients. For these reasons, a more sensitive, less laborious, and more efficient method of VCN estimation is required.
In droplet digital PCR (ddPCR), samples are partitioned into minute droplets, with the contents of each individual droplet discretely serving as a template for PCR amplification.6 Unlike qPCR, ddPCR uses an end-point approach, with samples defined as “positive” when a fluorescent signal exceeds a user-defined threshold. During partitioning, template gDNA is randomly distributed within volumetrically defined water-in-oil sample droplets. Fluorescence-labeled TaqMan hydrolysis primer probes are then used in a multiplex reaction, targeting a transgene of interest and an internal reference gene such as RPP30, which is present in two copies within a diploid genome. After PCR amplification of all individual droplets, a droplet reader detects fluorescence at wavelengths specific to the proprietary fluorophore FAM or VIC. The ratio between positive and negative droplets can be modeled mathematically by Poisson statistics, allowing calculation of the concentrations of targeted nucleic acid sequences within a known sample volume. Normalization against two-copy reference gene values yields VCN per cell.7 When primer probes are carefully designed, target amplification specificity of ddPCR and qPCR are similar. However, through Poisson statistics data modeling based on event frequency and probability, ddPCR in principle offers more precise VCN values. In theory, multiple partitioning of droplets (∼20,000) in ddPCR achieves high-resolution assay sensitivity, with scalability at a level not achievable through qPCR, enabling accurate yet reproducible detection of small fold PCR-amplicon concentration changes. Because absolute target quantification does not depend on Ct values normalized to a standard curve, ddPCR is much less reliant on amplification efficiency than qPCR. In addition, because all key steps are automated, the scope for manual error is less. Thus ddPCR is a substantial advance in several areas in absolute quantification of target DNA.
Viral transduction of induced pluripotent stem cells (iPSCs) is suited to demonstrate the practical use of ddPCR. Although lines such as human myeloid leukemic PLB-985 cells are simple to culture, they may become mitotically unstable and no longer retain a diploid genome. Central to any PCR-based method for VCN estimation including ddPCR is that all transduced target cells carry only two copies of an internal reference gene. Any additional copies caused by mitotic instability may compromise VCN estimation accuracy. For the purpose of assessing VCN distribution within a parental bulk-transduced population, iPSCs are considered to be more advantageous in terms of practicality over hematopoietic stem cells (HSCs), for example. This is because clonal populations of HSCs are typically obtained by colony pick-up from methylcellulose culture. However, HSCs are susceptible to spontaneous differentiation after prolonged in vitro culture. Differentiated cells are commonly granulocytes/monocytes. These cell types can migrate and cause overlap with originating cells from another colony. On the other hand, it is relatively simple to single-cell clone iPSCs and to further expand them under feeder-free culture conditions. Large amounts of highly purified gDNA can be extracted from expanded cells derived from only a single parental iPSC. Analysis of these cells would offer valuable insight into VCN distribution within bulk-transduced cell populations. It is expected that estimated VCN of individual clones would be similar to that of other clones derived from the same parental population but only if distribution profiles are determined with the anticipated accuracy of ddPCR.
In this study we attempted to demonstrate ease in practical application of ddPCR when estimating VCN in transduced cells. Precision was conferred by exact primer design. VCN was estimated by determining the ratio between the concentration of either codon-optimized (CO) CYBB (cytochrome b558 subunit β) or a puromycin resistance gene (PURO) and that of RPP30, a reference gene. Reliance on an external reference standard curve was not required. High sensitivity was apparent, as target gene detection was possible whether water or non-transgene-containing gDNA was used to dilute the template. Assay reliability was validated by observation of consistent, reproducible, and distinct VCN clustering patterns for clones of transduced iPSCs with varying numbers of transgene copies. Therefore, ddPCR may be considered as having potential value in clinical and research settings for VCN estimation.
Materials and Methods
Cells
PLB-985 (PLB) cells were maintained in RPMI 1640 with 10% fetal bovine serum and 1% penicillin–streptomycin–glutamate (all from Gibco/Thermo Fisher Scientific, Waltham, MA). X-linked chronic granulomatous disease (XCGD) PLB cells were generated from the immortalized PLB-985 myelomonocytic leukemic cell line by disruption of CYBB, encoding gp91phox via homologous recombination.8 M. Grez (Georg-Speyer-Haus, Frankfurt, Germany) kindly provided single-copy (SC) PLB cells constituting a clonal population in which each cell contains only one copy of the transgene-encoding CO gp91phox.9 In designations of all transduced cells used, “E” and “S” refer to the promoters EFS and SFFV, respectively, and “91,” “P,” and “N” refer to the transgenes gp91phox, PURO, and nerve growth factor receptor (NGFR), respectively. The number at the end, 1 or 10, refers to the multiplicity of infection used for transduction. E91P and S91P PLB cells are transduced XCGD PLB cell lines containing provirus insertions (Table 1). XCGD-iPSCs were generated and maintained as described.10
Table 1.
Vector copy number estimation in PLB cells
| Samplea | Provirus | Reaction type | Probe | Copies/μl | VCNb |
|---|---|---|---|---|---|
| SC91 | pAlpha.SIN.EFS.gp91s.oPRE | Singleplex | CYBB (CO) | 36.4 | 1.10 |
| PURO | 0.03 | 0.00 | |||
| RPP30 | 66.2 | NA | |||
| Multiplex | CYBB (CO) | 36.7 | 1.10 | ||
| RPP30 | 66.5 | NA | |||
| Multiplex | PURO | 0.04 | 0.00 | ||
| RPP30 | 61.1 | NA | |||
| E91P | pAlpha.SIN.EFS.gp91s.IRES.PURO.oPRE | Singleplex | CYBB (CO) | 23.2 | 1.12 |
| PURO | 24.3 | 1.17 | |||
| RPP30 | 41.6 | NA | |||
| Multiplex | CYBB (CO) | 23.9 | 1.24 | ||
| RPP30 | 38.7 | NA | |||
| Multiplex | PURO | 24.4 | 1.20 | ||
| RPP30 | 40.6 | NA | |||
| S91P | pAlpha.SIN.SFFV.gp91s.IRES.PURO.oPRE | Singleplex | CYBB (CO) | 16.7 | 1.12 |
| PURO | 18.4 | 1.23 | |||
| RPP30 | 29.8 | NA | |||
| Multiplex | CYBB (CO) | 16.3 | 1.25 | ||
| RPP30 | 26.0 | NA | |||
| Multiplex | PURO | 17.4 | 1.25 | ||
| RPP30 | 27.9 | NA |
CYBB (CO), codon-optimized CYBB (cytochrome b558 subunit β); NA, not applicable; PURO, puromycin; RPP30, ribonuclease P protein subunit p30; SFFV, spleen focus-forming virus; VCN, vector copy number.
SC91, the PLB clone having the indicated provirus as a single copy, which contains CYBB but no PURO sequence; E91P, the bulk PLB cell population transduced with the indicated viral vector (E stands for the EFS promoter whereas P indicates the presence of PURO); S91P, the bulk PLB cell population transduced with the indicated vector (S stands for SFFV whereas P stands for PURO).
VCN was calculated by the formula: CYBB or PURO copies/RPP30 copies × 2.
Alpharetroviral vectors and cell transduction
A. Schambach (Medizinische Hochschule Hannover, Hannover, Germany) kindly provided the alpharetroviral backbone vector containing CO CYBB cDNA.11 Construction of a series of alpharetroviral vectors (Supplementary Fig. S1; supplementary data are available online at http://online.liebertpub.com/hgtb) is described. The production of viral supernatant is described.12 In the case of XCGD-iPSCs, cells were first maintained under feeder-free conditions before virus transduction at the desired multiplicity of infection. To obtain clonal populations of transduced XCGD-iPSCs, dissociated cells were singly sorted by flow cytometry (FACSAria I sorter; BD Biosciences, San Jose, CA) into 96-well plates. Sorted cells were then expanded and maintained. All transduced XCGD-iPSCs and PLB cells were maintained under constant puromycin (Thermo Fisher Scientific) selection pressure to ensure retention of transgene-positive cells.
Sample predigestion with restriction enzymes
Approximately 400 ng of purified gDNA was digested with a 0.25-unit/μl concentration of NotI (New England BioLabs, Ipswich, MA) and BglII (Takara, Shiga, Japan) in a total reaction mixture of 20 μl at 37°C for 1 hr. No further purification was carried out, and 5 μl of the restriction mixture (∼100 ng) was used directly in subsequent ddPCR analysis. For simplicity, predigestion was carried out only to excise the vector fragment, but not the RPP30 reference sequence.
Estimation of target transgene concentration by ddPCR
A NucleoSpin tissue XS kit (Macherey-Nagel, Duren, Germany) was used according to the manufacturer's instructions to extract gDNA from a series of PLB cells and iPSCs. Supplementary Table S1 provides primer–probe sequences in detail. Probe #5 (Roche Diagnostics, Basel, Switzerland) from the Universal Probe Library, conjugated with FAM, was used to detect amplification of CO CYBB. The probe structures are described for PURO13 and RPP30.14 VCN was determined with the QX200 droplet digital PCR system (Bio-Rad, Hercules, CA) as per the manufacturer's instructions and as follows. A final reaction mixture (20 μl) contained the recommended 2 × supermix, primers (forward and reverse, 1000 nM final concentration), probe stock solution, and sample gDNA. The sample mixture was transferred to a DG8 cartridge. This was placed into the QX200 droplet generator. Sample droplets were transferred onto a 96-well PCR plate and sealed, using the recommended foil and sealer. Using a C1000 Touch thermal cycler (Bio-Rad), droplets were amplified to end point by heating to 95°C for 10 min followed by 40 cycles of 94°C for 30 sec and 53°C for 120 sec, with a final heating step of 98°C for 10 min. The reacted products were held at 4°C. The plate was placed into the QX200 droplet reader. Using the manufacturer's QuantaSoft software, the concentration of the target amplicon per unit volume of input for each sample was estimated for both CO CYBB/PURO and the RPP30 reference gene. Estimated VCN values were calculated by dividing twice the concentration of the target species by the concentration of the reference species.
VCN estimation by qPCR
The same primers and probes for detecting CO CYBB and RPP30 that were used in ddPCR were also used for VCN estimation in a duplex qPCR reaction (Supplementary Table S1). The cycling conditions for amplification were 95°C for 10 min followed by 40 cycles of 94°C for 30 sec and 53°C for 120 sec, with a final heating step of 98°C for 10 min. The reacted products were held at 4°C. Standard amplification curves were obtained by serial dilution of gDNA extracted from a transduced XCGD-iPSC clone with known VCN. Non-transgene-containing gDNA was used for dilution. The C1000 Touch thermal cycler was used for both amplification and fluorophore detection. Absolute VCN values was calculated by the ΔΔCt method with RPP30 as the calibrator gene according to the protocol described in a study aimed at optimizing duplex qPCR.15 A modified Excel (Office 2016; Microsoft, Redmond, WA) spreadsheet in this study was used for the calculation of gDNA standard curves and for the estimation of VCN (see the online supplement).15
Results
Precise primer design allows accurate detection of target transgene
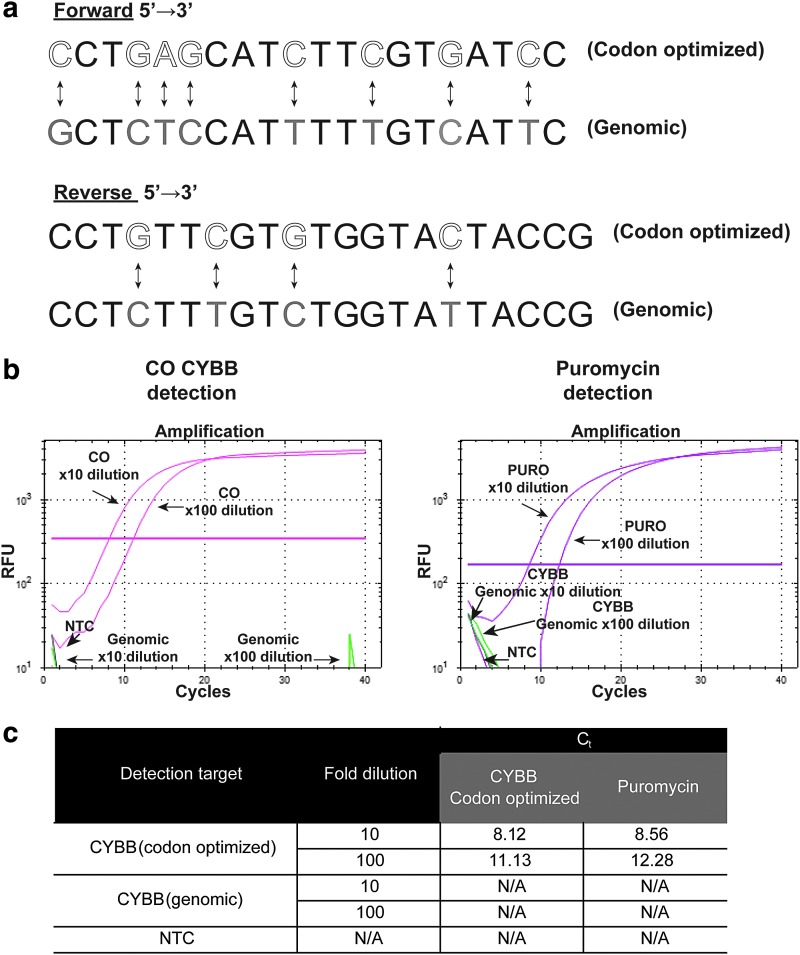
Like all PCR-based techniques, ddPCR can use target-specific primers sensitive to differences in only a few bases. In Fig. 1a, vertical two-headed arrows highlight specific differences between CO CYBB (white) and the corresponding genomic sequences (gray) encoding gp91phox (genomic). Two primer–probe pairs were designed to target either CO CYBB (an essential component of NADPH oxidase) or PURO (Supplementary Table S1). The forward primer for CO CYBB overlapped exon 1 to increase specificity; this avoids probe cross-reactivity, with binding to off-target sequences. To test specificity, the transfer plasmid pAlpha.SIN.EFS.gp91s.F2A.PURO.T2A.ΔLNGFR.oPRE (E91PN; Supplementary Fig. S1) was used as the template for PCR amplification. Two separate qPCRs were aimed at detection of CO CYBB or PURO. As a control for specificity, a plasmid DNA containing a non-codon-optimized CYBB cDNA, but no PURO sequence, was used as template for both reactions. Both CO CYBB and PURO were detectable at both 10- and 100-fold dilutions of template DNA (Fig. 1b). Changes in Ct (threshold cycle) values due to dilution did not affect the exponential phase of detection. The amplification efficiencies of CO CYBB and PURO appeared similar, as inferred from similar Ct values at the same template dilutions. No signal could be found with a control template, confirming that the approach to primer design conferred adequate specificity (Fig. 1c).
Figure 1.
Primer design and target specificity. (a) Bidirectional arrows indicate differences in primer design targeting either codon-optimized (CO) (white letters) or genomic (gray letters) CYBB sequences. (b) Amplification curves for the detection of CO CYBB and the puromycin resistance gene (PURO) (pink). As a control, primer probes targeting genomic CYBB (green) were included within the same reaction samples. The plasmid pAlpha.SIN.EFS.gp91s.F2A.PURO.T2A.ΔLNGFR.oPRE was diluted to 10 and 100 times. The horizontal pink line represents Ct (threshold cycle) values. NTC, no-template control; RFU, relative fluorescence units. (c) Tabular summary of CO CYBB and PURO detection alongside respective Ct values. N/A, not applicable (indicates no detection); NTC, no-template control.
VCN estimation in transduced PLB cells and XCGD-iPSCs
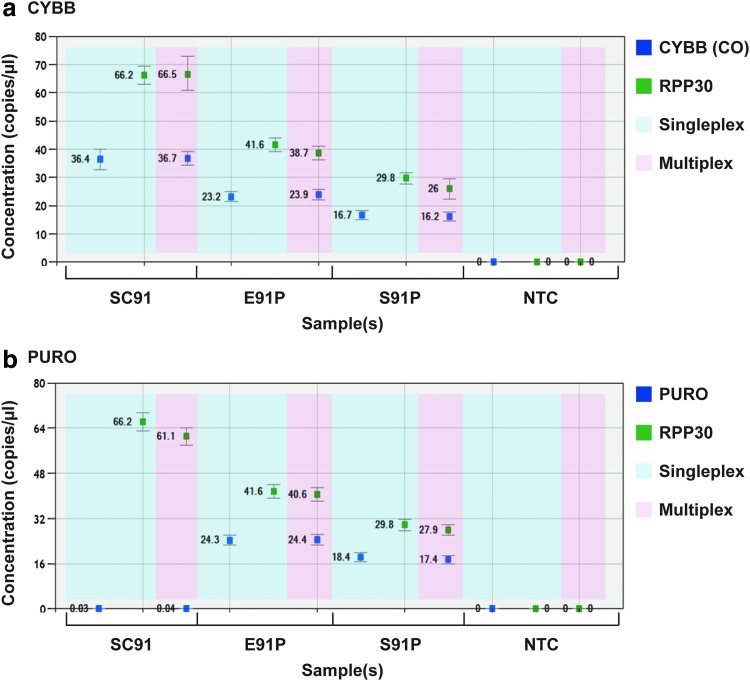
To estimate the number of provirus insertions in the genome of target cells, not only the concentration of the target must be measured but also that of a reference gene. In this study, the reference gene selected was RPP30 (RPP30, green; Fig. 2), normally present in two copies in a diploid genome. VCN can be determined by calculating the ratio of the concentrations of the target gene (CO CYBB or PURO, blue; Fig. 2) and the reference gene. To demonstrate and to test the efficiency of ddPCR, gDNA was extracted from PLB cells stably transduced with the provirus insertions pAlpha.SIN.EFS.gp91.IRES.PURO (E91P) and pAlpha.SIN.SFFV.gp91.IRES.PURO (S91P); numbers of provirus insertions were unknown. Transduced cells were maintained under puromycin selection pressure over several passages before gDNA was isolated to ensure that only cells with the transgene successfully integrated into the genome were selected. Cloned PLB cells with a known single-copy (SC91) provirus insertion served as a positive control (Table 1). Using ddPCR, the VCN value for SC91 PLB cells was approximately 1, using primer probes targeting CO CYBB, whether CO CYBB and RPP30 were amplified in singleplex or multiplex reactions (Fig. 2 and Table 1). The VCN values for the PLB samples E91P and S91P were approximately 1.2 whether the target gene was CO CYBB or PURO, and whether amplified in singleplex or multiplex reactions (Fig. 2 and Table 1). PLB cells, cancerous in origin, are genomically unstable, with a more-than-diploid genome.16 Genomically more stable iPSCs from a patient with XCGD were transduced, using slightly modified vector constructs, and the VCN was assessed. Detected VCN values ranged widely, from 1 to as high as 14 (Table 2). With gDNA from cells harboring both target sequences, VCN values when targeting PURO were comparable to those obtained when targeting CO CYBB, verifying the reliability of the assay system. In subsequent assays, only the primer probe targeting CO CYBB was used as a clinically relevant assay system (most clinical vectors contain no selection markers).
Figure 2.
VCN estimation in PLB cells. To estimate average VCN values, the concentrations (copies/μl) of either (a) CO CYBB (dark blue) or (b) PURO and of the RPP30 reference gene (green) were determined in each sample, using singleplex reactions (light blue, single primer probe) or multiplex reactions (pink, both primer probes). Samples of gDNA were extracted from three different types of PLB cells containing the provirus insertions SC91 (single-copy pAlpha.SIN.EFS.oPRE), E91P (pAlpha.SIN.EFS.IRES.PURO.oPRE), or S91P (pAlpha.SIN.SFFV.IRES.PURO.oPRE).
Table 2.
Vector copy number estimation in induced pluripotent stem cells
| Samplea | Provirus | MOI | Cell type | CYBB VCNb | PURO VCNb |
|---|---|---|---|---|---|
| EPN1 | pAlpha.SIN.EFS.PURO.T2A.NGFR.oPRE | 1 | XCGD-iPSC | 0.00 | 1.08 |
| E91PN1 | pAlpha.SIN.EFS.gp91s.F2A.PURO.T2A.NGFR.oPRE | 1 | XCGD-iPSC | 1.10 | 1.07 |
| SPN1 | pAlpha.SIN.SFFV.PURO.T2A.NGFR.oPRE | 1 | Healthy-iPSC | 0.00 | 7.81 |
| S91PN1 | pAlpha.SIN.SFFV.gp91s.F2A.PURO.T2A.NGFR.oPRE | 1 | Healthy-iPSC | 6.75 | 6.33 |
| E91PN10 | pAlpha.SIN.EFS.gp91s.F2A.PURO.T2A.NGFR.oPRE | 10 | XCGD-iPSC | 1.12 | 1.13 |
| S91P10 | pAlpha.SIN.SFFV.gp91s.IRES.PURO.oPRE | 10 | XCGD-iPSC | 14.15 | 13.75 |
CYBB, cytochrome b558 subunit β; gp91s, synthetic gp91 sequence; iPSC, induced pluripotent stem cell; MOI, multiplicity of infection; NGFR, nerve growth factor receptor; PURO, puromycin; VCN, vector copy number (copies/cell); XCGD, X-linked chronic granulomatous disease.
For sample names, each letter represents the following: E, EFS; P, PURO; N, NGFR; 91, gp91s.
VCN was calculated by the formula: CYBB or PURO copies/RPP30 copies × 2.
Cellular dilution permits sensitive detection of target transgene even at low concentrations of template within heterogeneous samples
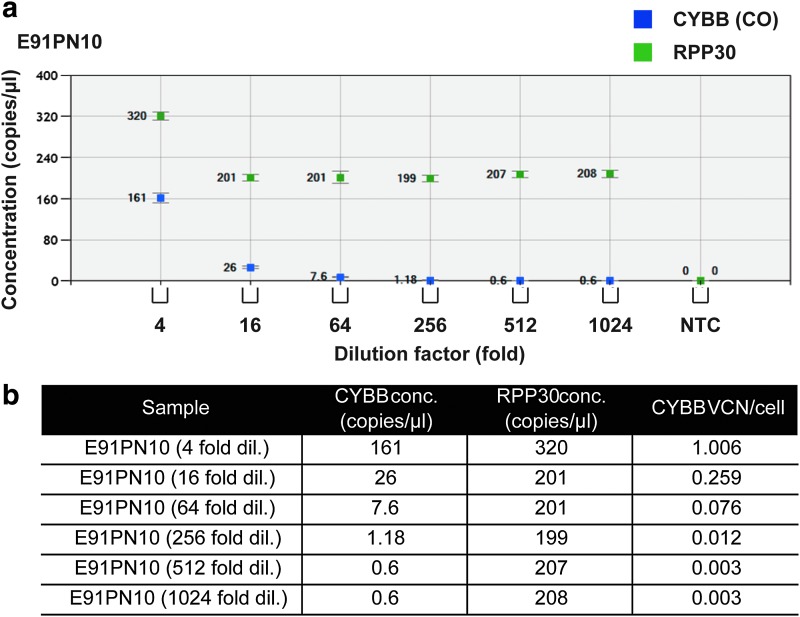
Detection of transgene-positive cells, present at low percentages among targeted populations, in clinical settings such as hematopoietic SCGT, resembles detection of a rare mutant allele in wild-type DNA.14 In unsorted material, concentrations of transgene-containing gDNA can be low. To model this scenario and to estimate the sensitivity limit of ddPCR, a sample of CO CYBB gDNA was diluted with non-CO CYBB-containing gDNA, thereby diluting CO CYBB while keeping the concentration of RPP30 constant (Fig. 3 and Supplementary Fig. S2). Using E91PN10 cells with a VCN of approximately 1, detection of CO CYBB was possible at dilutions of the original template up to 256-fold (approximately 0.4% of the total) (Fig. 3).
Figure 3.
Assay sensitivity after sample dilution with non-codon-optimized CYBB-containing gDNA. (a) To estimate the level of sensitivity of ddPCR, E91PN10 gDNA was diluted using gDNA extracted from transduced XCGD iPSCs that do not contain codon-optimized (CO) CYBB. Indicated are the concentrations of CO CYBB (dark blue) and RPP30 (green). NTC, no-template control. (b) Tabular summary of assay sensitivity. CO CYBB-non-containing gDNA was used to dilute E91PN10 gDNA samples, keeping the concentration of the reference RPP30 gene relatively constant while reducing the concentration of CO CYBB.
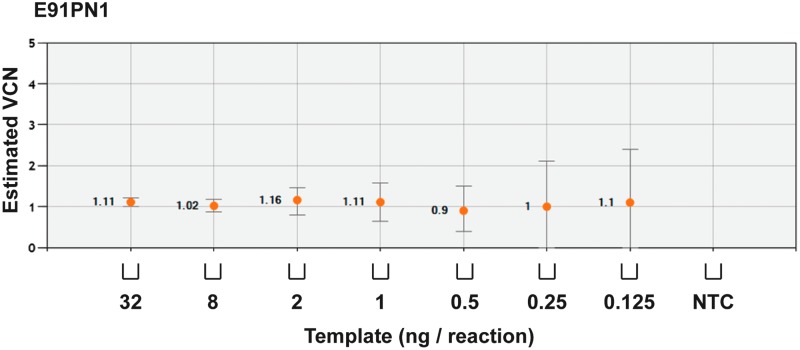
Minimum variability is observed in low VCN states
“Variability” refers to the range of estimated VCN values at a known concentration of template gDNA. Minimal variability is important in reliance on estimated VCN values. Furthermore, to determine the sensitivity and variability of ddPCR, gDNA from transduced iPSCs with a VCN of approximately 1.1 (E91PN1) was used. Starting from a fixed concentration of 32 ng, samples were diluted with water to as low as 0.125 ng per reaction, thereby reducing concentrations of both target gene and RPP30 reference gene (Supplementary Fig. S3). Variability in estimated VCN values was minimal with as little as 1 ng of template (cf. error bars; Fig. 4).
Figure 4.
Determination of assay variability. To determine assay variability with reducing amounts of template, samples were diluted with water to the stated mass of template gDNA. By this method the concentrations of both the target gene and RPP30 were reduced. gDNA was extracted from the E91PN1 line of transduced XCGD-iPSCs (estimated VCN value, 1.1). Error bars represent total error at a 95% confidence interval.
Optimum accuracy in VCN estimation is achieved by predigestion of samples with restriction enzymes
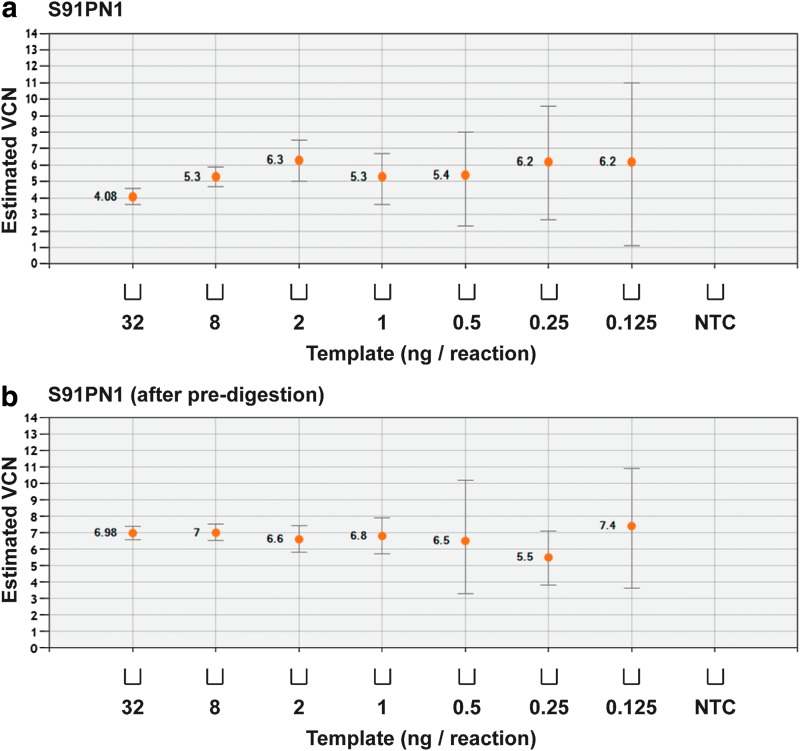
We next sought to determine variability at high VCN states, using gDNA from S91PN1 cells with a VCN of 6.75. Without predigestion of the template, in samples diluted with water as described previously, estimated VCN varied considerably at all template concentrations. This was true even with the use of substantial amounts of template gDNA, with average VCN values ranging from 4.08 (32 ng) to 6.3 (2 ng) (Fig. 5a). To reduce variability, undiluted S91PN1 gDNA was digested with NotI and BglII to excise fragments containing the target sequence recognized by the probe (Supplementary Fig. S4) for improving amplification efficiency. With ddPCR, predigestion also helps ensure that each partitioned droplet contains only one copy of the transgene-containing template. Predigestion considerably reduced estimated VCN variability among dilutions, with values closer to the prediluted value of 6.75, and restored sensitivity above the threshold of approximately 1 ng of template (Fig. 5b), as seen using E91PN1 gDNA.
Figure 5.
Sample predigestion with restriction enzymes reduces assay variability. The S91PN1 line of transduced XCGD-iPSCs was estimated to have an average VCN value of 6.75. Samples were diluted with water to the stated mass of template gDNA: (a) without predigestion or (b) with predigestion, using the restriction enzymes NotI and BglII. Error bars represent total error at a 95% confidence interval.
Application of ddPCR to evaluate VCN distribution in cloned iPSCs
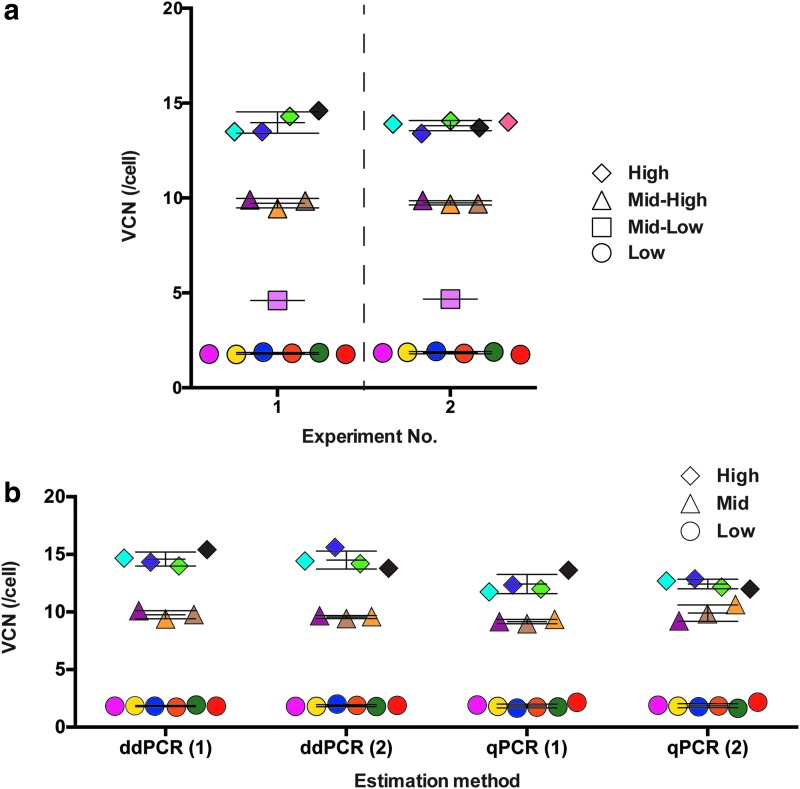
To assess VCN distribution within bulk-transduced iPSCs, single-cell cloning from three parental iPSC lines with low (E0.1, 1.93), mid-range (E10, 9.25), and high (S10, 14.15) average VCN values was conducted. A summary of VCN estimation of parental iPSC samples and their derivative clones is found in Supplementary Table S2. Fifteen clones were yielded from these three iPSC lines and were individually reassessed for VCN values (Supplementary Table S2). Shown side-by-side are the results from two independent experiments (Fig. 6a and Supplementary Table S2: ddPCR Exp #1 and Exp #2). When average values obtained from duplicate results were plotted for each clone, four distinct classes (low, mid-low, mid-high, and high VCN values) were observed. With the exception of one clone (mid-low), values in each category were tightly clustered, with little standard deviation from the mean value. These mean values closely reflected those of the parental populations in bulk culture. Of note is that profiles of VCN distribution were readily reproduced in independent experiments (Fig. 6a: Experiment No. 1, left; Experiment No. 2, right).
Figure 6.
Distribution of VCN in cloned iPSCs. Single-cell clones from three transduced iPSC lines were individually reassessed for VCN values. Each clone is represented by the same-shaped symbol and color. (a) On the basis of the estimated VCN value, clones were grouped into four arbitrary orders of magnitude (low, mid-low, mid-high, and high). Each symbol represents mean values of two independent experiments analyzing the same clone. (b) Direct comparison was made between ddPCR and qPCR by repeating VCN estimation of the same categories of clones (ddPCR Exp #3 and qPCR Exp #2; Supplementary Table S2). To simplify, mid-high becomes mid (mid-low not included). Each symbol represents a single measurement for each independent reaction by either method [ddPCR (1) and (2) or qPCR (1) and (2)]. Error bars (mean ± SD) represent the average VCN within each category and the distribution of each clone.
Because qPCR is currently the most common method for estimating VCN, a direct comparison with ddPCR is necessary to determine any differences in accuracy and reproducibility. Using selected cloned samples, we first confirmed that the same primer–probes could be used for qPCR analysis (results are shown in Supplementary Fig. S5 and Supplementary Table S2, qPCR Exp #1). Then, using the same 13 samples, a direct comparison was made between ddPCR and qPCR (Supplementary Table S2, ddPCR Exp #3 and qPCR Exp #2). Each sample, analyzed in duplicate, is represented by the same color [Fig. 6b: ddPCR (1) and (2); qPCR (1) and (2)]. By qPCR analysis, similar patterns of clustering are observed. Taking this similarity into consideration, we assumed uniform VCNs for clones within each category, and calculated mean values ± SD for comparison (Supplementary Tables S3 and S4). At high VCN states, there was a significant difference in the estimated mean values between ddPCR and qPCR (high vs. high, Supplementary Table S4; p < 0.0001), whereas at low and mid VCN states, there was no significant difference. However, greater standard deviation from the mean was observed when using qPCR, compared with ddPCR (Supplementary Table S3, Low VCN; Supplementary Fig. S5, Low, ddPCR-3 vs. qPCR-2). Of note, the same 13 clone samples were analyzed by ddPCR in a total of 6 independent reactions (duplicates in 3 independent experiments, each conducted by 3 different individuals), showing highly consistent VCN values (Supplementary Fig. S5 and Supplementary Table S2). These results suggest that at least for the assessment of mid to low VCN values, ddPCR is as accurate at reproducibly estimating VCN with less variability, compared with qPCR.
Discussion
In this study we demonstrate ddPCR as a user-friendly and clinically applicable approach to VCN estimation in an SCGT context for XCGD and address four critical parameters: specificity, sensitivity, accuracy, and practicality. Absolute quantification of concentrations of a target transgene and of the internal reference gene RPP30 made it possible to estimate VCNs in transduced cells. Specificity in target-gene amplification was conferred by precise primer design targeting codon-optimized CYBB cDNA, which is a feature unique to XCGD SCGT. Sensitivity was adequate as only 1 ng of gDNA template was required for accurate quantification of the target gene with minimum variability. Assay reliability was further verified by observing tight clustering of VCN values for cloned individual iPSCs derived each from a series of parental bulk-transduced populations. Although a similar clustering profile was obtained by qPCR analysis, for duplex reactions, it was necessary to use the more complicated ΔΔCt method for calculating the absolute VCN values. Yet despite applying a corrective measure by this approach, there appeared to be greater interexperimental variability by using qPCR in independent reactions (duplicate reactions for each clone; Supplementary Fig. S5 and Supplementary Table S3). On the other hand, VCN estimation by ddPCR was more straightforward, could be achieved without reliance on standard curves, and was independent of PCR amplification efficiency bias. In this study, the three independent ddPCR experiments were performed by different individuals, yet highly consistent values could be obtained. Thus ddPCR represents a more practical approach compared with previous PCR-based nucleic acid quantification techniques.
As with all PCR-based assays, any factor that can impair gene amplification efficiency will compromise accuracy in quantifying target nucleic acids.17 This applies to integrating vectors for which provirus insertion is semirandom. Depending on the site of integration, such as methylated genomic regions rich in CpG sequences,18 amplification may be either prohibited or severely compromised. Impurity in gDNA19 and the presence of inhibitors could also exert negative effects,20,21 most notably by raising the Ct value. However, the end-point approach to analysis that is inherent in ddPCR lessens the influence of these factors.22 Because VCN estimation by ddPCR is based on accurate distinction of droplet status as positive or negative for the target sequences, subtle changes in gene amplification efficiency do not affect the results.
In estimating VCN, multiplex reactions are preferred over separate monoplex reactions (target and reference genes) because the former reduce the accumulation of human manual errors and the overall required amount of template gDNA. However, the coexistence of two PCRs in one tube may affect the gene amplification efficiency of each other. Given the end-point nature of analysis, ddPCR may be considered less susceptible to this mutual interference than a comparable qPCR approach, where a subtle change in Ct may considerably affect VCN estimation. This was apparent in our own efforts at qPCR analysis where RPP30 acted as the calibrator gene. Similar to a previous report aimed at optimizing VCN estimation for duplex qPCR, it was necessary to use the more complicated ΔΔCt method for calculating the absolute VCN values.15 With ddPCR, by setting the user-defined thresholds for FAM (CO CYBB-derived) and VIC (RRP30-derived) fluorophore signals independently of one another, it is possible to clearly distinguish droplet status as positive or negative for the target sequences. In fact, multiplexing did not compromise estimated values by ddPCR in this study. The results closely resembled those obtained using monoplex reactions (Fig. 2 and Table 2).
Previously it had been reported that qPCR-based methods have shortcomings in sensitivity and precision when evaluating single-VCN or low-VCN states.23 Accuracy and reliability of qPCR diminish when discriminating less than a 2-fold difference in VCN values between samples, particularly at lower orders of magnitude. With ddPCR, the droplet partitioning gives the assay greater resolution than does qPCR and allows estimated VCN states to be resolved at 95% confidence intervals. Combined with the use of end-point analysis and Poisson statistics, ddPCR is more reproducibly accurate than qPCR in absolute quantification of target nucleic acids as less variability is observed at low VCN states (Fig. 6b). Some of these shortcomings of qPCR were apparent in a study using a qPCR- and standard curve-based method that yielded estimated values as much as 30 to 40% below true values.24 In the present study no underestimation was observed during initial ddPCR optimization using reference single-copy PLB cells. An estimated value of 1.1 for VCN was confirmed. Using transduced XCGD-iPSCs, the sensitivity threshold of the assay was estimated to be as little as 1 ng of total gDNA template (Figs. 4 and 5). With this amount of template, there was little variability in estimated VCN value compared with estimation using undiluted template. Therefore, ddPCR appears to be the preferred option where greater accuracy is desired.
One of the most promising areas for clinical use of ddPCR is hematopoietic SCGT. After the infusion of genetically modified autologous HSCs the percentage of gene marking in a peripheral blood target population can be as low as 1%. The most accurate method to quantify gene marking within transduced-cell progeny would involve a sorting procedure before gDNA extraction, but this approach may not always be practical. With ddPCR, 1% marking remains within the sensitivity limit of the technique. Thus ddPCR can be used with ease to simply track the presence of gene-marked cells in peripheral blood. However, actual VCN values for certain target gene-marked cell populations will be grossly underestimated because the template is diluted with non-transgene-containing gDNA. This may be overcome by applying a corrective factor if the percentages of transgene-containing cells are known.
In hematopoietic SCGT, early clinical trials relied on the use of integrating gammaretroviral vectors, where transgene expression was driven by viral promoters.25 Unfortunately, inherent risk proved unacceptably high, with mutagenic events detected for SCID-X1,26 Wiskott-Aldrich syndrome,27 and XCGD.28 With a shift toward safer lentiviral vectors designed to self-inactivate, similar adverse events have yet to be observed. As hematopoietic SCGT has become safer, researchers now strive to design protocols best suited for each target disease. For example, some diseases may require supraphysiologically high levels of transgene expression to be of measurable benefit to patients. This is the concept behind cell cross-correction therapy for metachromatic leukodystrophy, allowing neurons to benefit in vivo from glial cells that express a therapeutic gene originated from transduced hematopoietic stem/progenitor cells.29 It is likely that extremely high VCN values will be required to achieve such elevated expression levels. However, increasing gene transfer rates by raising the multiplicity of infection leads to an exponential increase in provirus insertions and an increased associated risk of insertional mutagenesis.30 Therefore having an accurate method of estimating average VCN values in transduced cells is thought to be essential for monitoring efficacy and safety in treated patients. Previous studies looking at copy number variants at specific loci of the genome found that qPCR-based methods lacked accuracy for copy number assessment at high integer values.31 This was thought to be attributed to a combination of measurement error and poor experiment reproducibility. In this study using ddPCR, initially it was found that at higher vector copy number states, there was significant variation in the estimated value with reducing concentration of template gDNA (Fig. 5a). The same sample was then predigested, using restriction enzymes, which helps to ensure that only a single transgene copy is present per partitioned droplet, a condition necessary to achieve optimum accuracy. Also, efficient predigestion of gDNA improves ddPCR assay accuracy by facilitating template access.32 Indeed, assay variability was reduced and greater consistency in estimated VCN values was observed with up to a minimum of approximately 1 ng of template (Fig. 5b). To further ascertain not only assay accuracy but also reproducibility of these results, we took advantage of an important characteristic of iPSCs and obtained single-cell clones from parental bulk-transduced populations with either low, mid, or high VCN (Fig. 6 and Supplementary Table S2). Tight clustering patterns representing consistent VCN values of cloned iPSCs were observed in separate experiments. This further validates the accuracy of ddPCR at estimating high VCN values and confirms experiment reproducibility at the same level of accuracy.
In summary, it may be concluded that ddPCR could be a more practical and user-friendly approach to estimating VCN values than is conventional qPCR. It is at least equivalent to if not better than qPCR in terms of accuracy. Specialized training is not necessary and the quantity of sample material required is minimal. Therefore, use of ddPCR technology may permit better evaluation of the therapeutic window for vector-mediated gene transfer and may thereby confer clinical benefits.
Supplementary Material
Acknowledgments
The authors thank Dr. Yumiko Ishii (University of Tokyo), who offered technical support for cell sorting by flow cytometry; all members in our group for their assistance; and Dr. A.S. Knisely for critical reading of the manuscript. This work was supported by funding from the Project for the Realization of Regenerative Medicine and Support for Core Institutes for iPS Cell Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT; to H.N.); and by a Grant-in-Aid for the Global COE Program from MEXT to the University of Tokyo. This work was also supported by the Program for Intractable Disease Research Utilizing Disease-Specific iPS Cells, Research Center Network for realization of Regenerative Medicine, from the Japan Science and Technology Agency (JST), and the Japan Agency for Medical Research and Development (AMED). H.-T.L. was supported by the Interchange Association of Japan, the Academic Research Grant for Graduate School of Frontier Sciences Doctor Course Students, the EGS research assistant program, IMSUT, and the Graduate Program in Gerontology, the University of Tokyo Global Leadership initiative for an Age-Friendly Society, University of Tokyo.
Author Disclosure
Y.Y. and S.I. are currently employed by Bio-Rad Laboratories.
References
- 1.Williams DA, Thrasher AJ. Concise review: lessons learned from clinical trials of gene therapy in monogenic immunodeficiency diseases. Stem Cells Transl Med 2014;3:636–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu C, Dunbar CE. Stem cell gene therapy: the risks of insertional mutagenesis and approaches to minimize genotoxicity. Front Med 2011;5:356–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cantsilieris S, Baird PN, White SJ. Molecular methods for genotyping complex copy number polymorphisms. Genomics 2013;101:86–93 [DOI] [PubMed] [Google Scholar]
- 4.VanGuilder HD, Vrana KE, Freeman WM. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques 2008;44:619–626 [DOI] [PubMed] [Google Scholar]
- 5.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 2008;3:1101–1108 [DOI] [PubMed] [Google Scholar]
- 6.Manoj P. Droplet digital PCR technology promises new applications and research areas. Mitochondrial DNA 2016;27:742–746 [DOI] [PubMed] [Google Scholar]
- 7.Pinheiro LB, Coleman VA, Hindson CM, et al. . Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem 2012;84:1003–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhen L, King AA, Xiao Y, et al. . Gene targeting of X chromosome-linked chronic granulomatous disease locus in a human myeloid leukemia cell line and rescue by expression of recombinant gp91phox. Proc Natl Acad Sci U S A 1993;90:9832–9836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaufmann KB, Brendel C, Suerth JD, et al. . Alpharetroviral vector-mediated gene therapy for X-CGD: functional correction and lack of aberrant splicing. Mol Ther 2013;21:648–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin HT, Masaki H, Yamaguchi T, et al. . An assessment of the effects of ectopic gp91phox expression in XCGD iPSC-derived neutrophils. Mol Ther Methods Clin Dev 2015;2:15046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suerth JD, Maetzig T, Galla M, et al. . Self-inactivating alpharetroviral vectors with a split-packaging design. J Virol 2010;84:6626–6635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suerth JD, Maetzig T, Brugman MH, et al. . Alpharetroviral self-inactivating vectors: long-term transgene expression in murine hematopoietic cells and low genotoxicity. Mol Ther 2012;20:1022–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakajima O, Akiyama H, Teshima R. Real-time polymerase chain reaction method for detecting contamination of beef by material from genetically engineered cattle. Biol Pharm Bull 2009;32:1313–1316 [DOI] [PubMed] [Google Scholar]
- 14.Hindson BJ, Ness KD, Masquelier DA, et al. . High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 2011;83:8604–8610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christodoulou I, Patsali P, Stephanou C, et al. . Measurement of lentiviral vector titre and copy number by cross-species duplex quantitative PCR. Gene Ther 2016;23:113–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bakhoum SF, Compton DA. Chromosomal instability and cancer: a complex relationship with therapeutic potential. J Clin Invest 2012;122:1138–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meijerink J, Mandigers C, van de Locht L, et al. . A novel method to compensate for different amplification efficiencies between patient DNA samples in quantitative real-time PCR. J Mol Diagn 2001;3:55–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson MD, Stirzaker C, Statham AL, et al. . Evaluation of affinity-based genome-wide DNA methylation data: effects of CpG density, amplification bias, and copy number variation. Genome Res 2010;20:1719–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bustin SA, Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech 2004;15:155–166 [PMC free article] [PubMed] [Google Scholar]
- 20.Dingle TC, Sedlak RH, Cook L, et al. . Tolerance of droplet-digital PCR vs real-time quantitative PCR to inhibitory substances. Clin Chem 2013;59:1670–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kontanis EJ, Reed FA. Evaluation of real-time PCR amplification efficiencies to detect PCR inhibitors. J Forensic Sci 2006;51:795–804 [DOI] [PubMed] [Google Scholar]
- 22.Hindson CM, Chevillet JR, Briggs HA, et al. . Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods 2013;10:1003–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scaramuzza S, Biasco L, Ripamonti A, et al. . Preclinical safety and efficacy of human CD34+ cells transduced with lentiviral vector for the treatment of Wiskott-Aldrich syndrome. Mol Ther 2013;21:175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charrier S, Ferrand M, Zerbato M, et al. . Quantification of lentiviral vector copy numbers in individual hematopoietic colony-forming cells shows vector dose-dependent effects on the frequency and level of transduction. Gene Ther 2011;18:479–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Booth C, Gaspar HB, Thrasher AJ. Gene therapy for primary immunodeficiency. Curr Opin Pediatr 2011;23:659–666 [DOI] [PubMed] [Google Scholar]
- 26.Hacein-Bey-Abina S, Garrigue A, Wang GP, et al. . Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest 2008;118:3132–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boztug K, Schmidt M, Schwarzer A, et al. . Stem-cell gene therapy for the Wiskott-Aldrich syndrome. N Engl J Med 2010;363:1918–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stein S, Ott MG, Schultze-Strasser S, et al. . Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat Med 2010;16:198–204 [DOI] [PubMed] [Google Scholar]
- 29.Biffi A, Montini E, Lorioli L, et al. . Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 2013;341:1233158. [DOI] [PubMed] [Google Scholar]
- 30.Kustikova OS, Wahlers A, Kuhlcke K, et al. . Dose finding with retroviral vectors: correlation of retroviral vector copy numbers in single cells with gene transfer efficiency in a cell population. Blood 2003;102:3934–3937 [DOI] [PubMed] [Google Scholar]
- 31.Khan FF, Carpenter D, Mitchell L, et al. . Accurate measurement of gene copy number for human alpha-defensin DEFA1A3. BMC Genomics 2013;14:719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernandez-Jimenez N, Castellanos-Rubio A, Plaza-Izurieta L, et al. . Accuracy in copy number calling by qPCR and PRT: a matter of DNA. PLoS One 2011;6:e28910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.