Abstract
The small nuclear 7SK RNA regulates RNA polymerase II (RNA Pol II) transcription, by sequestering and inhibiting the positive transcription elongation factor b (P-TEFb). P-TEFb is stored in the 7SK ribonucleoprotein (RNP) that contains the three nuclear proteins Hexim1, LaRP7, and MePCE. P-TEFb interacts with the protein Hexim1 and the 7SK RNA. Once P-TEFb is released from the 7SK RNP, it activates transcription by phosphorylating the C-terminal domain of RNA Pol II. P-TEFb also plays a crucial role in the replication of the human immunodeficiency virus HIV-1, through its recruitment by the viral transactivator Tat. Previous work demonstrated that the protein Tat promotes the release of P-TEFb from the 7SK RNP through direct binding to the 7SK RNA. Hexim1 and Tat proteins both comprise conserved and similar arginine-rich motifs that were identified to bind the 7SK RNA at a repeated GAUC site located at the top of the 5′-terminal hairpin (HPI). Here, we report the solution structure of this region as determined by nuclear magnetic resonance, to identify HPI structural features recognized by Hexim1 and Tat. The HPI solution structure displays an elongated shape featuring four helical segments interrupted by one internal loop and three bulges with distinct folds. In particular, the repeated GAUC motif adopts a pre-organized geometry. Our results suggest that the binding of Hexim1 and Tat to the 7SK RNA could originate from a conformational selection of this motif, highlighting how RNA local structure could lead to an adaptive recognition of their partners.
Keywords: RNA, NMR, 7SK, Hexim, Tat, TAR
INTRODUCTION
The 7SK RNA is an abundant 331-nt small nuclear RNA (snRNA) transcribed by RNA polymerase III discovered in mammalian cells in the middle 1970s (Zieve et al. 1977; Gurney and Eliceiri 1980; Kruger and Benecke 1987; Murphy et al. 1987). 7SK regulates the transcriptional activity of RNA polymerase II (RNA Pol II) by sequestering the positive transcription elongation factor b (P-TEFb) into the 7SK ribonucleoprotein (RNP) (Gurney and Eliceiri 1980). This complex also contains the three nuclear proteins Hexim1, LaRP7, and MePCE (Fig. 1A; Nguyen et al. 2001; Yang et al. 2001; Michels et al. 2003; Egloff et al. 2006; Zhou and Yik 2006; Barrandon et al. 2007; Michels and Bensaude 2008; Diribarne and Bensaude 2009; Czudnochowski et al. 2010). The La-related protein LaRP7 and the methylphosphate capping enzyme (MePCE) act cooperatively to promote the 7SK RNP assembly (Jeronimo et al. 2007; He et al. 2008; Krueger et al. 2008; Markert et al. 2008; Xue et al. 2010). MePCE, which comprises a methyltransferase domain, adds one γ-monomethyl phosphate cap structure onto the 5′-end of the 7SK RNA while LaRP7 binds to its conserved 3′ UUU-OH region. P-TEFb is stored on the 7SK RNP by interacting in a reversible manner with Hexim1 and the 7SK RNA (Nguyen et al. 2001; Yang et al. 2001; Michels et al. 2003; Yik et al. 2005; Egloff et al. 2006). P-TEFb is a heterodimer comprising the cyclin T1/T2 and the cyclin-dependent kinase CDK9. Once P-TEFb is released from the 7SK RNP, CDK9 phosphorylates the C-terminal domain of RNA Pol II, the C-terminal region of the Spt5 subunit of the DRB sensitivity-inducing factor (DSIF), and the NELF-E subunit of the negative elongation factor (NELF) to restore the transcription (Marshall et al. 1996; Peterlin and Price 2006; Zhou and Yik 2006; Chapman et al. 2008; Egloff and Murphy 2008). The 7SK snRNA mediates the interaction of Hexim1 with P-TEFb, thereby inhibiting P-TEFb kinase activity. More recently, the PPM1G phosphatase has been shown to bind directly 7SK and Hexim1 once P-TEFb is released from the 7SK RNP (Gudipaty et al. 2015). In addition, P-TEFb plays a crucial role in regulating the human immunodeficiency virus HIV-1 replication. P-TEFb is recruited by the viral transactivator Tat that binds to cyclin T1 through its activation domain, while the RNA-binding domain of Tat associates with the trans-activating responsive RNA element (TAR) located at the 5′-untranslated region of the viral genome (Muesing et al. 1987; Marciniak et al. 1990; Mancebo et al. 1997; Isel and Karn 1999; Karn 1999; Berkhout and van Wamel 2000). TAR captures Tat and P-TEFb, resulting in the displacement of the 7SK RNP, which activates the phosphorylation of RNA Pol II (Barboric and Lenasi 2010; D'Orso and Frankel 2010). Interestingly, it was also demonstrated that Tat promotes the release of P-TEFb from the 7SK RNP (Barboric et al. 2007; Sedore et al. 2007; Muniz et al. 2010). Hexim1 and Tat proteins both comprise conserved and similar arginine-rich motifs (ARMs) that were found to bind the 7SK RNA (Weiss and Narayan 1998; Yik et al. 2004). Tat was shown to efficiently displace Hexim1 from the 7SK RNA, suggesting that the 7SK RNA contains a TAR RNA-like Tat-binding motif embedded with the Hexim1 recognition site (Muniz et al. 2010).
FIGURE 1.
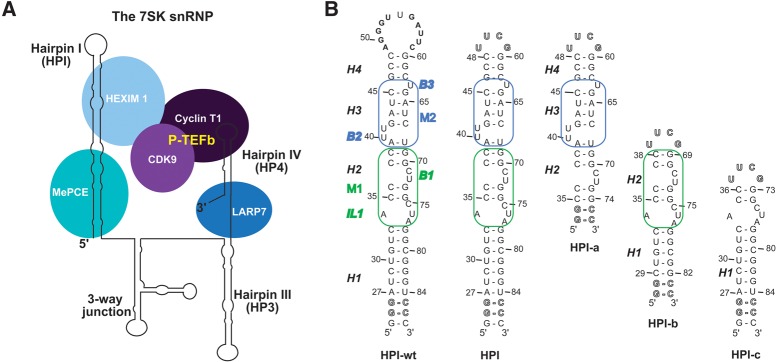
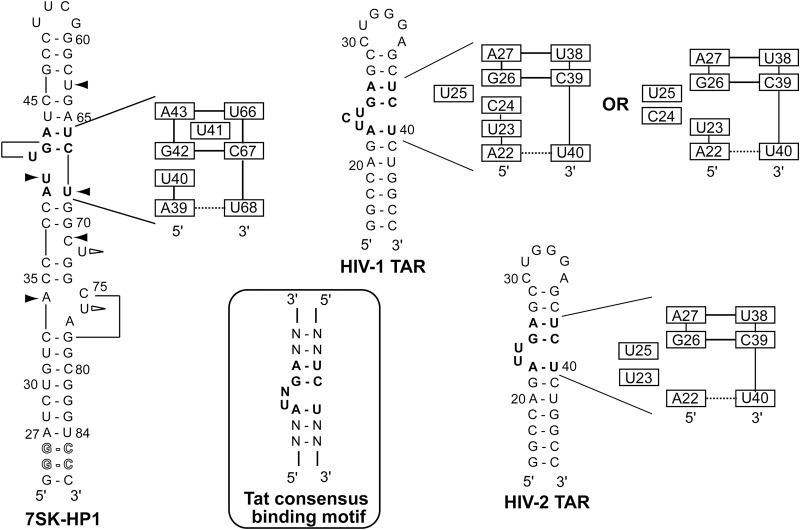
The 7SK RNA and its 5′-terminal hairpin HPI. (A) Schematic representation of the 7SK RNP complex. P-TEFb comprises the cyclin T1 and the cyclin-dependent kinase CDK9. (B) Sequences and secondary structures of (i) the 7SK 5′-terminal hairpin (HPI-wt), (ii) HPI, and (iii) the three subdomain constructs (HPI-a, HPI-b, and HPI-c) used for nuclear magnetic resonance (NMR) studies. Outlined nucleotides at 5′-end were changed to improve transcription efficiency. The 11-nt apical loop of HPI-wt was substituted by a thermo-stable UUCG tetraloop (HPI). The RNA oligonucleotides HPI-a, HPI-b, and HPI-c comprise nucleotides 35–74, 29–38/69–82, and 24–36/73–87, respectively. Both HPI-a and HPI-b contain two additional G-C base pairs. The internal loop IL1 comprises (34/75–77) nucleotides. The bulges B1, B2, and B3 comprise (71/72), (40/41), and (63) residues, respectively. The M1 motif (green) contains the helix H2, the internal loop IL1, and the bulge B1. The M2 motif (blue) comprises the helix H3 and bulges B2 and B3.
The sequence of the 7SK RNA is highly conserved in vertebrates (Wassarman and Steitz 1991; Egloff et al. 2006; Gruber et al. 2008a; Gursoy et al. 2000), and 7SK homologs exist in invertebrates and insects (Gruber et al. 2008b; Nguyen et al. 2012). Based on chemical and enzymatic probing experiments, the human 7SK RNA has been proposed to fold into a structure composed of four domains: highly conserved 5′- (HPI) and 3′- (HP4) hairpins, bordering one three-way junction and a middle hairpin (HP3) (Wassarman and Steitz 1991). More recently, an alternative secondary structure based on computational analysis has been proposed, with major differences in the middle part of the 7SK RNA while the upper part of HPI and HP4 structures were conserved (Marz et al. 2009). Several studies demonstrated that the 5′-terminal hairpin of 7SK includes the Hexim1 binding site, the (24–87) region being the minimal sequence required for interaction (Egloff et al. 2006; Belanger et al. 2009; Martinez-Zapien et al. 2015). The 3′-terminal hairpin was also identified as essential for P-TEFb regulation but not involved in Hexim1 binding (Muniz et al. 2013).
The recognition mechanism underlying the interaction between the 7SK RNA and Hexim1 or Tat is still an open question. A repeated (G42AUC45-G64AUC67) motif flanked by (U40U41) and U63 bulges is located in the top part of the (24–87) region in the Hairpin I (M2 region, Fig. 1B). This sequence, highly conserved in eukaryotes and other organisms (Wassarman and Steitz 1991; Gursoy et al. 2000; Gruber et al. 2008a,b), was previously shown to be essential for specific Hexim1 recognition (Lebars et al. 2010). The same region was shown to be involved in Tat binding (Muniz et al. 2010). Aiming at identifying structural features recognized by Hexim1 or Tat at the 5′-terminal hairpin of 7SK, we solved the solution structure of the (24–87) region using nuclear magnetic resonance (NMR). To overcome the challenging spectral crowding and transverse relaxation problems raised by the size of HPI (18.2 kDa), we used the “divide and conquer approach” (Lukavsky and Puglisi 2005). We divided the HPI hairpin into three smaller overlapping RNAs (Fig. 1B). The set of experimental restraints, including distances from NOE measurements, dihedral angles and orientational restraints from residual dipolar coupling constants, led to well-defined models of the solution structure of HPI. Its solution structure features four helical segments, interrupted by one internal loop (IL1) and three bulges (B1, B2, B3) with distinct structures. Our three-dimensional structure suggests that the differential binding of Hexim1 and Tat to the 7SK HPI could involve a conformational selection of a pre-organized motif. The solution structure of 7SK HPI provides structural basis to understand how RNA local structure could modulate the specificity of recognition.
RESULTS
Design of RNA oligonucleotides for NMR studies
The HPI RNA
In a previous study, the 5′-terminal hairpin (HPI) of 7SK RNA was shown to fold similarly whether isolated or in the context of the full-length RNA, and its secondary structure was determined (Lebars et al. 2010). The 11-nt apical loop of the 5′-terminal hairpin of the 7SK RNA (HPI-wt) was substituted with a thermo-stable UUCG tetraloop (HPI) to reduce spectral complexity arising from the superposition of high and low intensity signals (Fig. 1B; Varani et al. 1991; Molinaro and Tinoco 1995). The pattern of imino protons 1H and 15N resonances of HPI-wt and HPI are almost identical with the exception of the apical loop regions, indicating that the loop substitution does not alter the global fold of the RNA (Fig. 2A). In addition, carbon chemical shifts, which provide a sensitive probe of the chemical environment, indicate that the RNAs adopt the same conformation (Fig. 2B,C). Two distinct RNA samples were produced selectively: 13C/15N labeled at either G or A nucleotides. As shown in Figure 2B, the chemical shifts of C8H8 cross-peaks of G residues, are similar for HPI-wt and HPI RNAs, with the exception of G60 and the nucleotides located in the apical loops (Gloop). Chemical shifts of C2H2 cross-peaks of A residues are also similar (Fig. 2C). In particular, the chemical shifts of A34 and A77 are identical, indicating that HPI-wt and HPI adopt the same conformation in the bulged region. These observations show that the overall structure of the 7SK 5′-terminal hairpin is not altered upon the modification of the apical loop.
FIGURE 2.
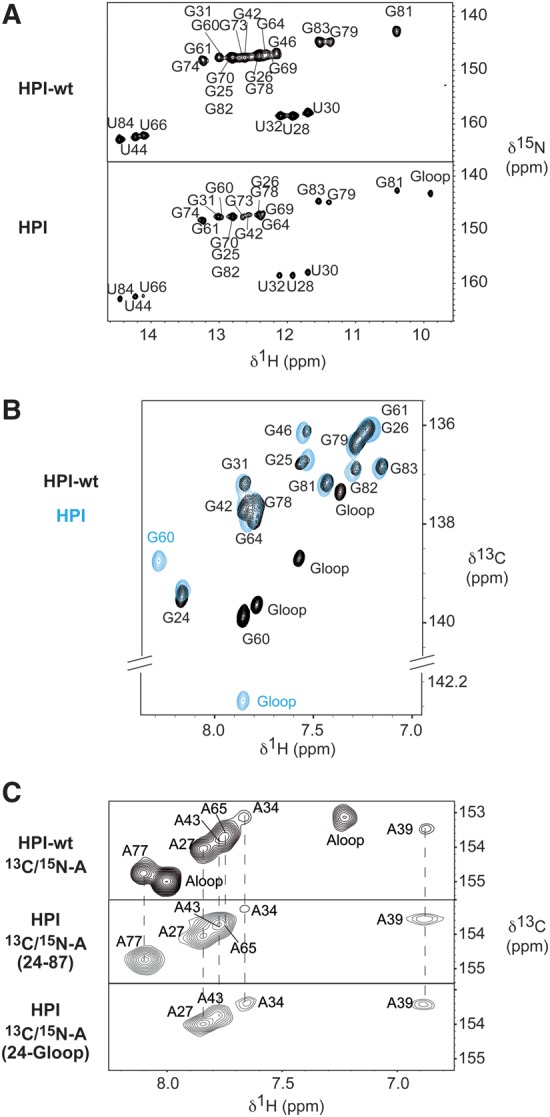
HPI-wt and HPI adopt the same conformation. (A) 1H–15N HSQC spectra showing imino protons region of HPI-wt (top) and HPI (bottom). All imino protons were assigned via sequential nuclear Overhauser effects (NOEs) observed in 2D-NOESY experiments. In the HPI construct, imino protons of the UUCG apical loop resonate at the expected chemical shifts typically observed, which ensures a proper folding of the RNA (Varani et al. 1991; Molinaro and Tinoco 1995). (B) 1H–13C HSQC spectra showing the aromatic H8-C8 of selectively labeled [13C/15N-G] HPI-wt (black) and HPI (blue), recorded at 30°C. (C) 1H–13C HSQC spectra showing the aromatic H2–C2 correlations of the selectively labeled [13C/15N-A] HPI-wt (top), the selectively labeled [13C/15N-A] HPI (middle), and the segmentally and selectively labeled [13C/15N-A(G24-G60)] HPI (bottom). HPI was selectively labeled at all A positions (middle), and segmentally and selectively labeled at A positions in the G24-G60 segment (bottom). Assignment of aromatic H2–C2 of HPI-wt had been previously reported (Lebars et al. 2010). Assignment of HPI was based on analysis of NOESY spectra recorded at 4°C, 10°C, 15°C, 20°C, 30°C in 90/10 H2O/D2O and at 20°C and 30°C in 99.9% D2O. Spectra were recorded in 50 mM sodium phosphate buffer at pH 6.2.
Divide and conquer approach: subdomain constructs of HPI (HPI-a, HPI-b, HPI-c)
Based on the chemical shift data described above, we designed three subdomain constructs of HPI: (i) HPI-a (nucleotides 35–74), (ii) HPI-b (nucleotides 29–38/69–82), and (iii) HPI-c (nucleotides 24–36/73–87) (Fig. 1B). The three fragments cover the entire 5′-terminal hairpin of HPI. Both HPI-a and HPI-b contained two additional G-C base pairs and a UUCG-tetraloop to stabilize their fold. The conservation of the subdomain construct structures was assessed by comparing their chemical shifts with those measured on HPI. The imino proton resonances of all three subdomains overlap with those of HPI for common regions (Supplemental Figs. S1, S2). Furthermore, the chemical shift values of C8H8 aromatic cross-peaks observed in 1H–13C HSQC spectra are very similar for common regions of HPI and the subdomain constructs (Supplemental Fig. S3). Notably, the chemical shifts of the C8H8 cross-peaks of A34 and A77, located at a stem–loop junction, are identical in HPI-b and HPI (Supplemental Fig. S3D). These observations suggest that the local RNA motifs adopt the same conformation in both HPI RNA and smaller constructs.
Resonance assignment of HPI and the three subdomains
The secondary structures of HPI-a, HPI-b, and HPI-c (Fig. 1B) were characterized by the analysis of 2D NOESY and 1H–15N HSQC experiments recorded in 90/10 H2O/D2O at various pHs and temperatures (see Materials and Methods section). All base-paired imino protons were assigned via sequential NOEs observed in 2D NOESY spectra at different mixing times (Supplemental Fig. S2). Watson-Crick 15N–1H–15N hydrogen bonds were directly observed in heteronuclear J(N,N)-HN COSY experiments performed in H2O (Supplemental Fig. S4A; Dingley and Grzesiek 1998; Hennig and Williamson 2000). The assignment of imino protons was possible for HPI except for U72 and U76, where no imino proton signal could be detected. In the bulges, U40 and U41 were partially protected from solvent exchange and could be assigned in NOESY spectra performed at 4°C with 50 msec and 75 msec mixing times (Supplemental Fig. S5A).
Complete assignments of nonexchangeable protons and their directly bound carbons and nitrogens were obtained for HPI-a, HPI-b, and HPI-c, using standard homonuclear and heteronuclear 2D and 3D correlation experiments (1H, 15N, 13C, 31P) (Supplemental Figs. S4, S5). Sequential H6/8-H1′ connectivities observed in a classical helix A were identified for all residues, except for U40, U41, G46, U63, U68, C71, U72, C75 to A77 and the apical loop. No sequential H6–H1′ connectivities were observed for U41, U68, U72, and U76. Weak sequential cross-peaks were observed in the case of U40, U63, and A77 while strong correlations were visible between G46 and C45, and C71 and G73 (Supplemental Fig. S5B).
To overcome strong overlaps of chemical shifts during HPI NOESY assignment, spectra were interpreted by superimposing the NOE patterns observed in spectra of the subdomain constructs. In addition, resonances were discriminated using 1H–13C HSQC experiments recorded on selectively 15N/13C-labeled HPI. Partial assignments (30%) of protons and their directly bound carbons and nitrogens were obtained.
Three-dimensional structure of HPI
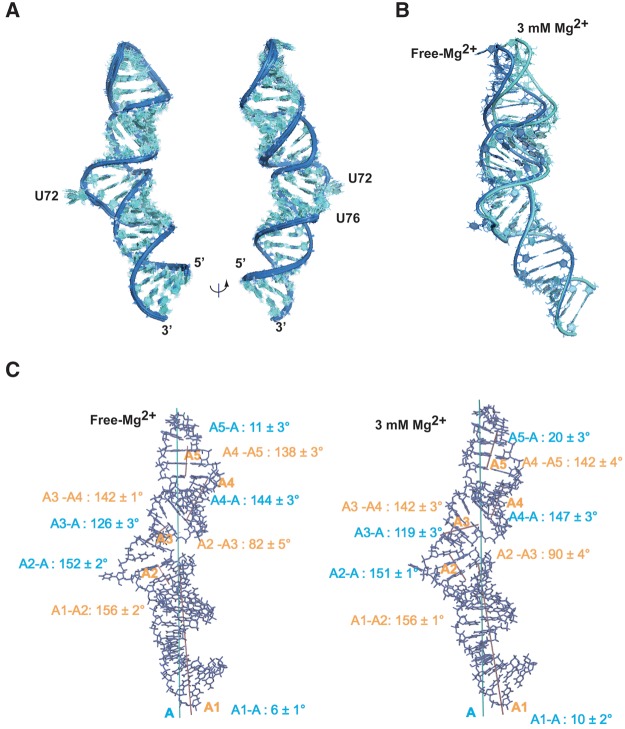
The three-dimensional structure of HPI was determined by combining local structural restraints obtained from the three subdomain constructs (HPI-a, HPI-b, and HPI-c) and global restraints from residual dipolar couplings (RDCs) measured on HPI RNA (Ottiger et al. 1998). Structures were calculated using 854 NOE distance restraints, 484 dihedral torsion restraints, and 51 residual dipolar coupling (RDC) constraints measured at 30°C (Supplemental Figs. S4, S6). The overall structure is well defined with a heavy atom root-mean-square deviation compared to the global average structure (RMSD) of 0.8 ± 0.1 Å (Table 1; Fig. 3A). HPI contains four helical segments comprising G24-C33/G78-C87 (H1), C35-C38/G69-G74 (H2), G42-C45/G64-C67 (H3), G46-C48/G60-C62 (H4), interrupted by one internal loop (IL1) and three bulges (B1, B2, and B3) with distinct structures (Fig. 1B). HPI RNA displays an elongated shape with a bend that takes place at motif M1 (Fig. 3C). The interhelix angle between (C35-36/G73-G74) and (C37-38/G69-G70) stems is 82 ± 5° (Fig. 3C, left panel).
TABLE 1.
NMR restraints and statistics for the 10 converged structures in the absence of magnesium and with 3 mM Mg2+
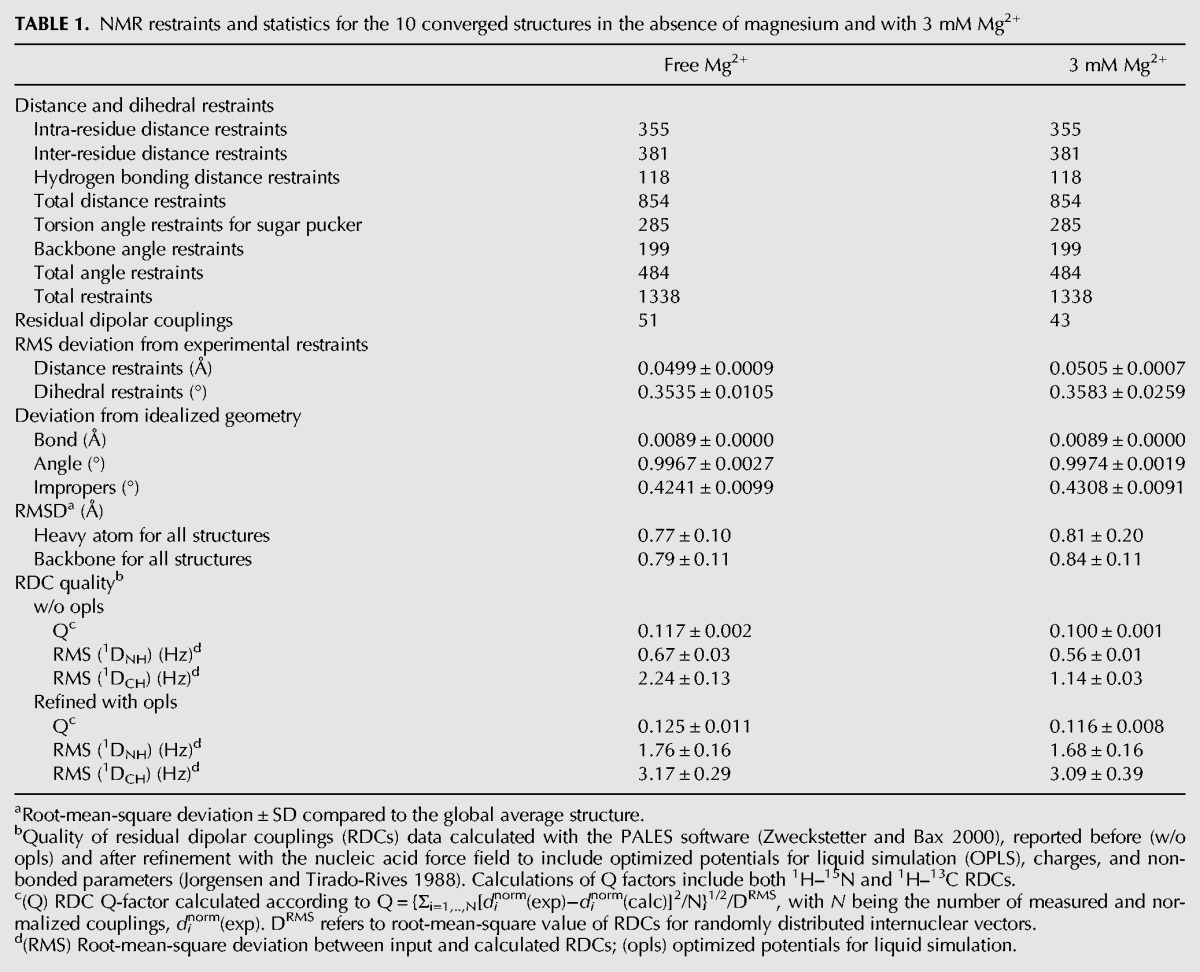
FIGURE 3.
Three-dimensional structure of HPI. (A) Superimposition of 10 converged NMR structures. Bases and sugars are shown in light blue and the backbone in dark blue. (B) Structures of HPI calculated using RDCs measured in the absence of Mg2+ (dark blue) and with 3 mM Mg2+ (light blue). The regions H1 are superimposed. (C) Inter-angle axis measurement for the free-Mg2+ structure (left) and 3 mM Mg2+ model (right). Overall axes (A) of RNAs are shown in blue. Five stems were defined using the 3DNA software (Colasanti et al. 2013). Helical axes for the five stems were generated using MOLMOL software and are indicated in pink (Koradi et al. 1996). Angles between each stem and the overall axis are indicated in blue. Angles between helices are indicated in orange.
The structure of the M1 motif is dominated by weak interactions
The M1 motif is formed by four Watson-Crick base pairs, C35:G74, C36:G73, C37:G70, and C38:G69, interrupted by one internal loop (IL1: 34/75-77 nucleotides) and one bulge (B1: 71–72 nucleotides) (Fig. 4A,B). These base pairs are characterized by weak interactions as indicated by enhanced line broadening of G69, G70, G73, and G74 imino protons upon increasing temperature (Supplemental Fig. S7). The destabilization of these base pairs in the M1 region may result from distortions within the flanking bulges.
FIGURE 4.
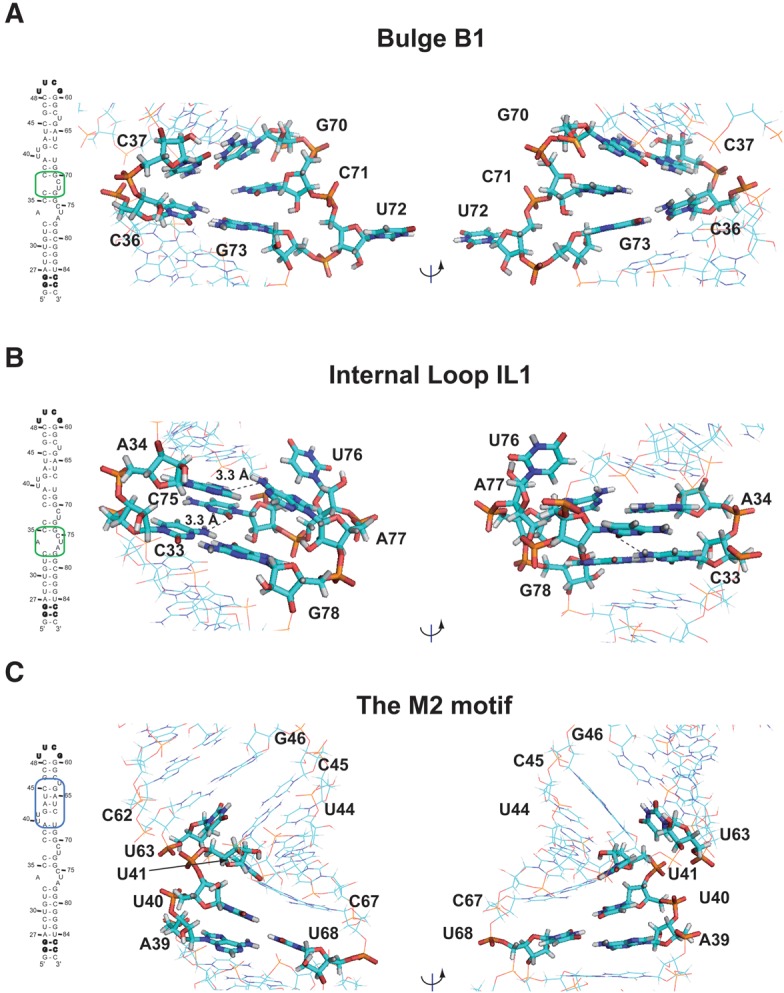
Bulges and internal loop. (A) View of the bulge B1 in the M1 Motif. (B) View of the internal loop IL1 in the M1 motif. (C) View of the M2 motif. Broken lines indicate possible hydrogen bonds.
In the bulge B1, C71 is stacked on residue G70, whereas U72 is swung outside the helix (Fig. 4A). This conformation is supported by the observation of sequential H6/8-H1′ connectivities between C71, G70, and G73. In the case of U72, only one inter-residue contact could be detected between G73-H8 and U72-H5. The H1′/H2′ cross-peak observable in COSY and TOCSY experiments indicated a C2′-endo conformation of C71 and U72 sugar puckers, whereas ε dihedral angles were constrained to gauche− conformations as determined from analysis of HP-COSY and HCP experiments.
In the internal loop (IL1), A34 is in a C3′-endo and anti conformations, stacked on residue C33 (Fig. 4B). Consistently, sequential H6/8-H1′ connectivities with C33 and C35 and NOEs involving A34-H2, C35-H1′, and G78-H1′ were observed. Several unambiguous NOE contacts place A77 inside, but devoid of stacking interaction. Weak H8-H1′ connectivity with G78 and NOEs involving A77-H2 and G78-H1′ were observed. A cross-strand NOE contact between A34-H2 and A77-H2 was also identified. As a result, the average distance between A34(N1) and A77(N6) in the ensemble of 10 structures is 3.31 ± 0.05 Å, which is compatible with a water-mediated hydrogen bond. In addition, as described above, NMR experiments (COSY-DQF, TOCSY, NOESY, and HCP) pointed out that C75 is in C2′-endo and syn conformations, with the ε torsion angle in the gauche− conformation. C75 is positioned deep into the major groove in the proximity of the C33:G78 base pair (Fig. 4B). Analysis of structures shows that the average distance between C75(O2) and C33(N4) is 3.3 ± 0.2 Å, which is compatible with the formation of a cWW-tHW (cis-Watson-Crick/Watson-Crick, trans-Hoogsteen/Watson-Crick) base triple involving C75:C33:G78 (Fig. 5A; Abu et al. 2012). Finally, U76 is swung outside the helix adopting C2′-endo and syn conformations with ε and β torsion angles in gauche− conformations (Fig. 4B).
FIGURE 5.
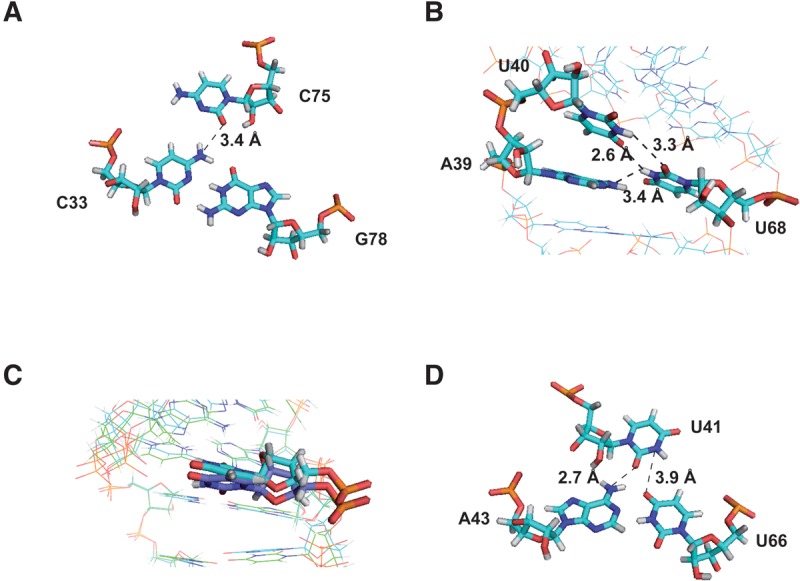
Hydrogen bond networks. (A) Base triple involving C33:G78:C75 in the free-Mg2+ structure. (B) Zoom on A39, U40, and U68 residues in the free-Mg2+ structure. (C) Comparison of the U41 residue from Mg2+-free (dark blue) and Mg2+-bound structures (light blue). (D) Base triple involving A43:U66:U41 in the 3 mM Mg2+ model. Distances consistent with the formation of hydrogen bonds are displayed. Broken lines indicate possible hydrogen bonds.
The M2 motif folds into a preformed platform
The M2 motif encompassing the repeated GAUC sequence contains a helical segment composed of four Watson-Crick base pairs (H3) and two bulges, B2 and B3, defining a preformed pocket (Fig. 4C).
In the bulge B3, U63 is unpaired and stacked on residue C62. This is supported by the observation of weak sequential H6/8-H1′ connectivities between U63, C62, and G64.
In the bulge B2, U40 is unpaired and stacked on residue A39. NOEs involving A39-H2 and U40-H1′, U40-H6 and A39-H1′, and U40-H3 and G42-H1 were observed (Supplemental Fig. S5B). Concerning U41, NOE contacts and dihedral angle constraints place this residue deep into the major groove in the proximity of A43-U66 and G42-C67 Watson-Crick base pairs. Indeed, the U41 imino proton H3 has NOE interactions with both U66-H3 and C67-H5 protons. In addition, the U41 sugar pucker adopts a C2′-endo conformation and the β and ε torsion angles gauche− conformations. The conformation of U41 suggests that it could form a base-triple with A43-U66 or G42-C67 (Fig. 4C). However, analysis of NOESY experiments at various pHs (from 6.5 to 5.5) and temperatures (from 4°C to 30°C) did not provide evidence for the formation of a U41-mediated hydrogen bond with either of these two base pairs. Finally, the A39:U68 closing base pair of the M2 motif deviates from classical Watson-Crick geometry. Indeed, the imino proton of U68 could not be detected unambiguously due to fast exchange with the solvent (Supplemental Fig S5A). Nevertheless, nonexchangeable proton NMR data place U68 inside, between C67 and G69 (Figs. 4C, 5B). Analysis of the ensemble of structures reveals that U68-H3 could be involved in two hydrogen bond networks (Fig. 5B). On one hand, the average distance between A39(N1) and U68(N3) in the ensemble of 10 structures is 4.1 ± 0.2 Å, which is compatible with the formation of a water-mediated hydrogen bond. On the other hand, the average distances between U40(H3) and U68(O2) as well as U40(O4) and U68(H3) in the ensemble of 10 structures are 3.3 ± 0.1 Å and 2.6 ± 0.2 Å respectively, which is compatible with a UU (2-carbonyl-N3, 4-carbonyl-N3) base pair. The U68 residue is in a weak stacking and could transiently be involved in these hydrogen bond networks. The destabilization of A39:U68 base pair probably originates from the distortion induced by the (U40/U41) dinucleotide bulge B2.
Effect of magnesium on HPI structure
Since Mg2+ ions are involved in RNA folding, stabilization of tertiary structure, and catalysis, we examined their impact on HPI structure.
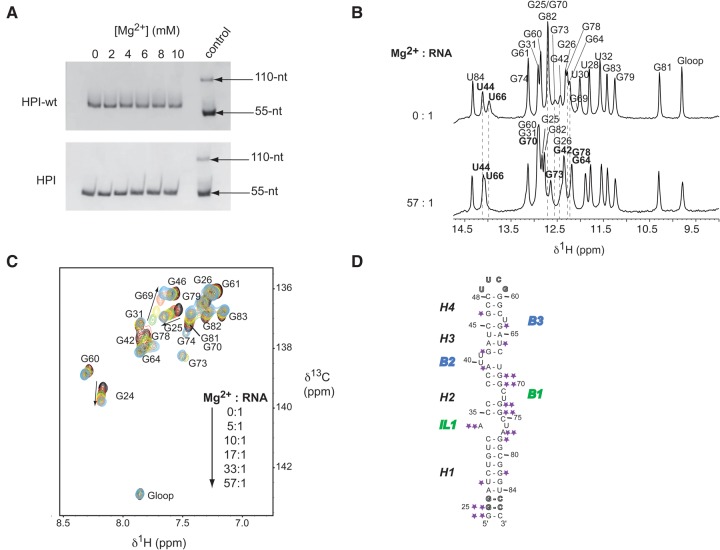
Native gel analysis was first performed to assess the global effect of magnesium on the conformation of both HPI and HPI-wt with respect to monomeric or dimeric forms. As illustrated in Figure 6A, both HPI and HPI-wt fold in a monomoneric form at all magnesium concentrations tested. Next, NMR titrations monitoring exchangeable and aromatic protons were performed to detect potential Mg2+ binding sites by chemical shift perturbation analysis (Fig. 6B,C; Supplemental Figs. S8, S9). In particular, the NH resonances of U30, G64, and G78 shift upfield, whereas G25, U66, G70, G82, and G73 shift downfield. Among these residues, G70 and G73, located in the M1 motif, exhibit the most drastic changes: G70-H1 undergoes a shift of 0.19 ppm whereas Mg2+ binding results in the narrowing of G73-H1 resonance (Fig. 6B; Supplemental Fig. S8). Interestingly, NOESY spectra analysis shows that base-pairing patterns remain unaltered at all magnesium concentrations tested. The Mg2+ induced effects were also analyzed by monitoring nonexchangeable protons in 1H–13C HSQC spectra (Fig. 6C; Supplemental Figs. S8C, S9). C8-H8 and C2-H2 cross-peaks undergo chemical shift changes, generally limited to <0.12 ppm for both 13C and 1H. Remarkably, G69, G70, G73, and G74, located in the M1 motif, showed up with increasing magnesium concentration. Altogether, these observations suggest that magnesium stabilizes the M1 motif and the stem–loop junctions of HPI, while the base-pairing and the global fold of the RNA are maintained.
FIGURE 6.
Effect of magnesium on HPI. (A) Native gel electrophoresis on 10% polyacrylamide. HPI-wt (top) and HPI (bottom) were loaded in the buffer used for NMR studies with increasing magnesium concentration. The last lane (control) contains a thio-modified RNA at its 5′-end, which allows the visualization of a monomer (55-nt: S-RNA) and a dimer (110-nt: RNA-S-S-RNA). (B) Titration of HPI by magnesium by monitoring exchangeable protons. The assignment of imino protons based on the analysis of NOESY experiments is indicated for the Mg2+-free RNA (top) and the 10 mM Mg2+-RNA corresponding to a ratio of 57:1 (bottom). Broken lines and bold assignments indicate imino protons that undergo chemical shifts changes. (C) Magnesium titration of HPI using nonexchangeable protons. Overlays of regions of 1H–13C HSQC spectra showing the aromatic H8-C8 of selectively labeled [13C/15N-G] HPI at 30°C, Mg2+-free (black) and upon successive additions of magnesium. Arrows indicate the cross-peaks that shifted upon addition of magnesium. (D) Summary of the effects observed in 1H and 13C experiments on HPI upon Mg2+ binding. Small effects are indicated by one star, while stronger effects are highlighted using two stars.
However, the additive effect of slight changes could lead to local and long-range helical distortions. To investigate this possibility, residual dipolar couplings (RDCs) were measured for HPI in the presence of 3 mM Mg2+ (Supplemental Fig. S6B,C). As chemical shifts and NOE patterns were not significantly affected upon magnesium addition, the same dihedral and distances restraints were used to generate a model of Mg2+-HPI. Structures were then calculated as previously described (Table 1). Analysis of converged structures shows that while the overall structure of HPI is maintained, Mg2+ binding induces a slight bending at the M1 motif where residues show the biggest changes in NMR data upon magnesium addition (Fig. 3B). As illustrated in Figure 3C, angles between (G24-C33/G78-C87), (C37-C38/G69-G70), and (G46-C48/G60-C62) stems and the principal axis of HPI undergo slight variations. The analysis of models obtained from RDC measured in the presence of 3 mM Mg2+ indicates little changes on the conformation, except for U41 in the B2 bulge that is shifted closer to the A43:U66 base pair upon magnesium addition (Fig. 5C). This is supported by the measurement of RDC constants in Mg2+-free and 3 mM Mg2+ samples, the values being, respectively, 13.4 Hz and 6.7 Hz for the U41(C5-H5). Analysis of Mg2+ model shows that the average distances between U41(O2) and A43(N6) and between U41(N3) and U66(O4) are 2.7 Å and 3.9 Å, respectively, which deviates from a cWW-tHW (cis-Watson-Crick/Watson-Crick, trans Hoogsteen/Watson-Crick) base triple (Fig. 5D).
Mapping of the interaction with HIV-1 Tat RNA-binding domain
In our previous work, we used a combined NMR and biochemical approach to identify the recognition elements in 7SK RNA for Hexim1 binding (Lebars et al. 2010). Our results demonstrated that the repeated GAUC motif (M2) was essential for Hexim1 recognition. Similarly, here we used NMR to investigate Tat binding to HPI-wt. Several studies showed that Tat contains a highly conserved arginine-rich motif (ARM) that binds to HIV-1 TAR RNA and 7SK HPI RNA (Weiss and Narayan 1998; Yik et al. 2004). Therefore, we used a 36 aa peptide encompassing the ARM motif (RKKRRQRRR) to map its interaction on HPI RNA (Fig. 7A).
FIGURE 7.
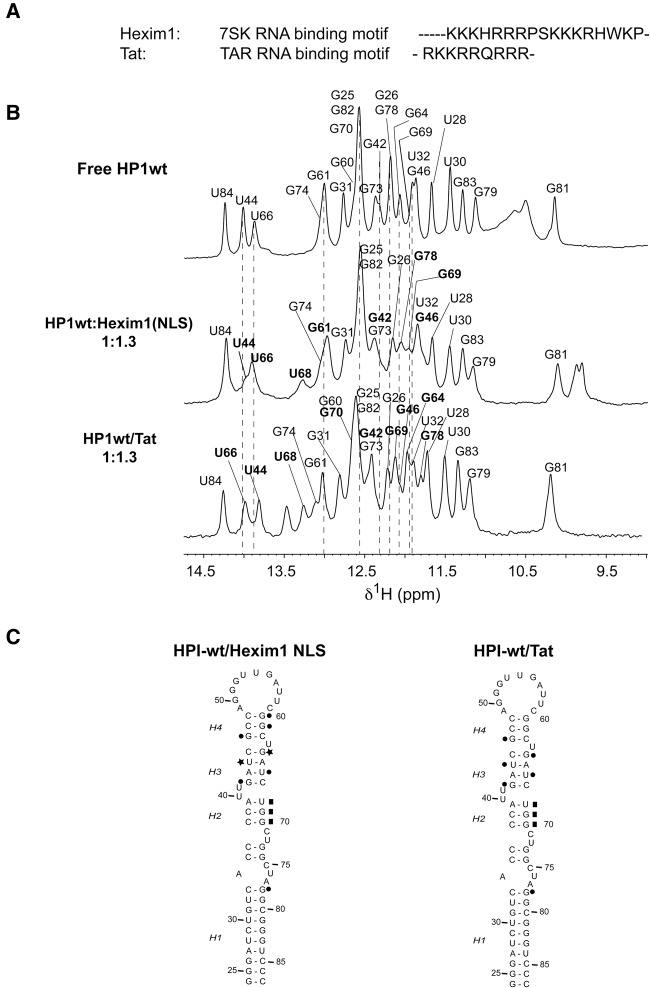
Interaction with Tat peptide and comparison with TAR RNA. (A) Comparison of Hexim1 and Tat RNA binding domains. (B) Titration of HPI RNA by Hexim1 NLS domain and Tat peptide. The imino protons region of 1D spectra recorded at 15°C in 50 mM sodium phosphate buffer in 90/10 H2O/D2O at pH 6.4. The assignment of imino proton based on analysis of NOESY experiments is indicated for the free HPI-wt (top), the HPI-wt:Hexim1(NLS) complex (middle), and the HPI-wt:Tat complex (bottom; Supplemental Fig. S10B). The assignment of HPI-wt:Hexim1(NLS) complex has already been reported in our previous work (Lebars et al. 2010). Broken lines and bold assignments indicate imino protons that undergo chemical shift changes. (C) Summary of the effects observed on HPI upon interaction. The results obtained with Hexim1 NLS have been previously reported (Lebars et al. 2010). The opening of base pairs is indicated with stars. The stabilization of base pairs is indicated with squares. Residues for which imino protons undergo chemical shifts changes upon addition of peptide are highlighted with circles.
The chemical shift perturbations of HPI-wt imino protons were monitored as a function of the peptide concentration and were used to identify the RNA helical regions in contact with Tat. Successive addition of Tat on HPI induced several changes in the imino protons region (Supplemental Fig. S10A). Distinct signals corresponding to the free and bound form of U44 and U66 imino protons were observed, consistent with a slow exchange between these two states and submicromolar dissociation constant (Supplemental Fig. S10A). Saturation of the RNA was observed at a Tat/RNA ratio of 1.3:1. Assignment of imino protons in HP1:Tat complex was based on analysis of NOESY experiments (Supplemental Fig. S10B). Tat peptide binding induced specific changes in imino protons that were mainly confined in the upper half of HPI-wt (Fig. 7B; Supplemental Fig. S10). First, imino protons of residues G42, U44, G46, G64, U66, G69, G70, and G78 undergo chemical shift variations upon the addition of Tat peptide, whereas the imino protons of the other part of the RNA molecule remain unchanged. Except for G78, no change is observed in the H1 stem. Secondly, two new resonances at 13.25 and 13.45 ppm show up, revealing lower solvent accessibility. Whereas the signal at 13.45 ppm could not be unambiguously assigned, the resonance at 13.25 ppm was assigned to the U68 imino proton that forms a Watson-Crick base pair with A39 residue upon Tat peptide interaction. It is worth noting that the binding of Tat peptide results in the narrowing of G69-H1 resonance, indicating that the Tat peptide binding to HPI stabilizes the H2 stem, similarly to Hexim1 NLS. In contrast to Hexim1 NLS, Tat binding does not induce an opening of Helix H3. Possible conformational changes of HPI RNA upon Tat peptide binding were further investigated using nonexchangeable protons (Supplemental Fig. S10C). 1H–13C HSQC spectrum recorded on a selectively 13C/15N G-labeled HPI shows the disappearance of G42, G46, and G64 cross-peaks, concomitant with the appearance of new cross-peaks. In conclusion, these experiments show that Tat peptide targets the same site as Hexim1 NLS with a different mechanism (Fig. 7C).
DISCUSSION
To understand the structural basis for HPI recognition by Hexim1 and Tat, we determined the solution structure of the 5′-terminal hairpin (HPI) of the 7SK snRNA.
The HPI RNA is highly structured
HPI is highly structured with four helical segments (H1 to H4), interrupted by one internal loop (IL1) and three partially structured bulges (B1, B2, B3). Two preformed “platforms” are located in M1 (IL1/H2/B1) and M2 (H3/B2/B3) motifs (Fig. 1B). The structural analysis of HPI reveals that the M1 motif adopts a fold defined by a consistent set of NOE and dihedral angles constraints. However, this structure lacks the stabilizing interaction provided by hydrogen bonding, indicating the possibility to adopt alternative local folds under the influence of external factors, such as the interaction with partners of the 7SK RNA. Indeed, the base-pairing interaction network is weak, as shown by the fast exchange with the solvent enhanced upon a mild increase in the temperature evidenced by NMR. Similar features were observed for the M2 motif that adopts a particular geometry structured with four Watson-Crick base pairs and the U41 residue positioned deep into the major groove. The plasticity of the M2 motif is evidenced by behaviors of U40 and U41 residues. Their imino protons are broad, which indicate that these residues are in weak stacking and interactions, thus able to adopt other conformations not detectable by NMR.
Our observations for M1 and M2 motifs are reminiscent of those reported for the 7SK HP4 RNA hairpin that comprises a dinucleotide CU bulge (Durney and D'Souza 2010). In HP4, the C residue was found to be positioned into the major groove in the proximity of a G:C base pair, although no direct hydrogen bond could be observed. The authors demonstrated that this geometry could constitute a preformed pocket for ligand binding.
The recognition of HPI could be driven by a conformational selection
Mutation studies within the M1 motif indicated that this region could constitute a secondary binding site for the Hexim1 protein (Lebars et al. 2010). The 359 aa protein Hexim1 comprises three functional domains: (i) an N-terminal unstructured part containing a proline-rich region (1–148), (ii) the ARM RNA-binding motif (149–179) overlapping with a nuclear localization signal (NLS), and (iii) a C-terminal region containing an acidic region and a coiled-coil domain involved in P-TEFb inhibition and binding, respectively (Weiss and Narayan 1998; Schulte et al. 2005). The ARM motif has been shown to bind the 5′-terminal hairpin (HPI) of the 7SK RNA (Weiss and Narayan 1998; Lebars et al. 2010). Previous work suggested that Hexim binds 7SK as a dimer (Blazek et al. 2005; Dulac et al. 2005; Li et al. 2005). The way Hexim interacts with the 7SK RNA is still an open question, and several possible binding mechanisms have been proposed (Martinez-Zapien et al. 2015). Among them, the repeated GAUC-GAUC bordered by U-bulges (motif M2) may provide a high-affinity primary binding site targeted by the ARM motif of Hexim, and the M1 region may provide a secondary weak binding site contacted by another part of the protein. The M2 motif is conserved in eukaryotes and several organisms (Gursoy et al. 2000; Gruber et al. 2008a,b; Marz et al. 2009; Nguyen et al. 2012). Interestingly, several studies demonstrated that the HIV-1 Tat also binds with high specificity and efficiency to the 5′-terminal hairpin of the 7SK snRNA (Muniz et al. 2010). The Tat binding site is embedded within the Hexim recognition motif. In addition, Tat was shown to efficiently displace Hexim on the 7SK RNA. In the present work, possible changes in HPI RNA upon Tat peptide binding were investigated using NMR by monitoring imino protons in helical regions. Our experiments revealed a close similarity between Tat and Hexim binding to 7SK HPI: the A39:U68 base pair that was not observable in the free RNA, closes upon binding and, G69:C38 and G70:C37 base pairs are stabilized. However, in contrast to Hexim1, the binding of Tat does not induce an opening of the helix H3 but rather a stabilization of the structure (Fig. 7C). Although Tat and Hexim1 contain a similar RNA-binding motif, our results imply that the two proteins interact with the same site through different mechanisms. Further experiments and modeling studies will be needed to establish accurate mechanisms of interaction.
Another intriguing question is how Tat discriminates TAR and 7SK-HPI RNAs? It has been proposed that Tat binds to the 7SK RNP to recruit the catalytically inactive form of P-TEFb. TAR captures Tat and P-TEFb from the 7SK RNP, leading to the displacement of the inhibitory complex 7SK RNP, which allows the phosphorylation of RNA Pol II (Barboric and Lenasi 2010; D'Orso and Frankel 2010). Comparison of the 7SK 5′-terminal hairpin with HIV-1 TAR and HIV-2 TAR revealed that HPI carries the conserved Tat-binding element (Fig. 8). Tat recognizes a consensus motif in TAR RNA, in which U23 and the adjacent base pairs G26:C39 and A22:U40 were identified as essential for Tat binding (Dingwall et al. 1989; Weeks et al. 1990; Sumner-Smith et al. 1991; Weeks and Crothers 1991). In the free HIV-1 TAR RNA, the bulged residues U23 and C24 were shown stacked within the helix, while U25 was swung out (Aboul-ela et al. 1996). Later, other research groups found different stacking arrangements at the trinucleotide bulge with U23 stacked on A22 and residues C24 and U25 in a predominant looped out conformation (Fig. 8; Long and Crothers 1999; Franck et al. 2009). In the case of HIV-2 TAR RNA, U23 has been shown to be involved in a weak or transient base stacking on A22, with U25 looped out (Farès et al. 2007). Additional studies based on combined molecular dynamics and measured RDCs demonstrated that U23 and U25 were looped out (Franck et al. 2009). Finally, all studies showed that the A22:U40 base pair was not stable.
FIGURE 8.
Comparison with TAR RNA from HIV-1 and HIV-2. Sequences and secondary structures of HPI, HIV-1 TAR, and HIV-2 TAR. The Tat consensus binding motif is encircled. Bold nucleotides indicate the Tat binding motif. For HPI: Black arrows indicate that the nucleotide is involved in a stacking interaction; white arrows indicate that the residue is looped outside the helix and lines indicate that U41 and C75 are positioned into the major groove. Schematic representations of the Tat and Hexim binding site are shown. Solid and dotted horizontal lines indicate stable and weak base-pairings, respectively. Representations of HIV-1 TAR and HIV-2 TAR in the free form are based on structures from previous works (Aboul-ela et al. 1996; Long and Crothers 1999; Franck et al. 2009).
The analysis of the solution structure of the 7SK HPI reveals that the repeated GAUC-GAUC motif (M2) adopts a geometry that slightly differs from those observed in the TAR RNA element (Fig. 8). The U40 residue in the 7SK HPI shows similarity with the U23 residue in HIV-1 TAR RNA. Both residues are stacked on the A:U adjacent base pair and the sequential walk observable in NMR experiments is interrupted at this position. Another common point relates to the U68 residue in HPI, the imino proton not being observable, as reported for the U40 residue from TAR RNA. This common feature present both in the 7SK HPI, HIV-1, and HIV-2 TAR RNA probably originates from the U-bulge. Finally, the position of U41 in the HPI 7SK RNA in the major groove of M2 differs from the looped out conformation of U25 found in HIV-1 and HIV-2 TAR RNA.
Conclusion
The three-dimensional solution structure of HPI from the 7SK RNA shows that it is highly structured. HPI contains four helical segments interrupted by one internal loop and three bulges with distinct structures. The stacked structure of the internal loop and bulges opposite helices induces minor distortions leading to a bending of the RNA that takes place at the M1 motif. HPI provides two pre-organized platforms for binding. In particular, the M2 motif exhibits high similarity with the TAR RNA element. How does Tat discriminate HPI and TAR? Analysis of the structures of HPI and TAR highlights subtle differences, such as the position of U41 in HPI compared to U25 in TAR. These results provide elements to understand how subtle structural changes of RNA local structure could lead to an adaptive recognition of their partners. Further investigations will be necessary to establish the detailed molecular and structural principles of the distinct binding of Hexim1 and Tat to the same site within the 7SK RNA.
MATERIALS AND METHODS
RNA samples preparation
Milligram quantities of RNAs were prepared either unlabeled, fully and selectively 13C/15N labeled by in vitro transcription with homemade T7 RNA polymerase from oligonucleotide templates containing a 2′-O-methyl modification at position 2 or, in the case of HPI-wt and HPI, from linearized plasmids (Milligan and Uhlenbeck 1989; Kao et al. 1999). Plasmids were obtained by cloning the HPI-wt or HPI sequence into pRZ or pHDV encoding for a hammerhead ribozyme in 3′ of the target RNA (Walker et al. 2003). The DNA templates were purchased from Sigma and Eurogentec. Transcription conditions were optimized according to previous protocols (Milligan and Uhlenbeck 1989; Wyatt et al. 1991). Labeled NTPs (nucleotide triphosphates) were purchased from Eurisotop and Sigma.
HPI was also segmentally 13C/15N labeled. The RNA segment (G24-Gloop) was prepared 13C/15N labeled. The RNA segment (G60-C87) was modified at its 5′-end by incorporating GMP nucleoside purchased from Sigma. After purification, the (G24-Gloop) RNA segment was ligated to the modified (G60-C87) fragment by T4 RNA ligase 2 using a complementary 43-nt DNA splint as described previously (Lebars et al. 2014). The T4 RNA ligase 2 was purchased from New England BioLabs. The 43-nt DNA splint (5′-GCCTAGCCAGCCAGATCAGCCCGAAGGCGATCAATGGGGTGAC-3′) was purchased from SIGMA.
All RNAs were purified on denaturing polyacrylamide gels (Wyatt et al. 1991). After electroelution and ethanol precipitation, RNAs were dialyzed for 24 h against the buffer used for NMR experiments. Samples were concentrated by lyophilization and resuspended in 90/10 H2O/D2O for experiments involving exchangeable protons and in 99.9% D2O for nonexchangeable proton experiments. Each sample was refolded by heating at 95°C (2 min) and snap-cooled at 4°C. The concentration of RNA samples was measured using a Nanodrop Spectrometer and calculated with molar extinction coefficients obtained from the Eurofins MWG Operon calculator (http://www.operon.com/tools/oligo-analysis-tool.aspx).
Native gel
The effect of magnesium on HPI-wt and HPI RNAs was characterized by electrophoresis under native conditions. The RNA samples and the control (at 10 μM) were heated at 95°C (2 min) and snap-cooled at 4°C in the NMR buffer (50 mM sodium phosphate at pH 6.2). After centrifugation, at 10,000 r.p.m., for 5 min at room temperature, increasing quantities of magnesium chloride (from 1 to 10 mM) were added. After incubation for 10 min at room temperature, the samples were examined by electrophoresis on a 10% polyacrylamide native gel. The gel was run in 0.5× TBE at 150 V, at 4°C. Bands were visualized with toluidine blue.
NMR spectroscopy
NMR experiments were recorded at 500, 600 MHz and 700 MHz on a DRX and Avance III Bruker spectrometers equipped with a z-gradient TBI probe (at 500 MHz), TCI (at 700 MHz) and TXI (at 600 MHz) z-gradient cryoprobes. NMR data were processed using TopSpin (Bruker) and analyzed with Sparky software packages (Goddard and Kneller 2004). NMR experiments were performed in 50 mM sodium phosphate buffer (pH 6.2). The concentration of unlabeled and labeled RNA samples ranged from 0.2 to 0.8 mM. Sample volumes were 280 µL in Shigemi NMR tubes and 150 µL in 3 mm NMR tubes.
1H, 13C, 15N, and 31P assignments were obtained using standard homonuclear and heteronuclear methods. NMR data were acquired at 4°C, 10°C, 15°C, 20°C, and 30°C for exchangeable protons and at 15°C, 20°C, and 30°C for nonexchangeable protons. Solvent suppression for samples in 90/10 H2O/D2O was achieved using combined “Jump and Return” and WATERGATE sequences (Plateau and Gueron 1982; Piotto et al. 1992; Sklenar et al. 1993). The residual HDO resonance in 99.9% D2O was suppressed using low power presaturation. Two-dimensional NOESY spectra in 90/10 H2O/D2O were acquired with mixing time of 50, 150 and 400 msec. Base-pairing was established via sequential nuclear Overhauser effects (nOes) observed in 2D NOESY spectra at different mixing times and directly observed from J(N,N)-HNN COSY experiments performed in 90/10 H2O/D2O at 4°C and 15°C (Dingley and Grzesiek 1998; Hennig and Williamson 2000). NOESY spectra with mixing times of 50, 150, and 400 msec in 99.9% D2O were acquired at 15°C, 20°C, and 30°C.
Heteronuclear NMR spectra were measured at either 20°C or 30°C in 99.9% D2O with the exception of 1H-15N HSQC experiments that were acquired at 4°C, 10°C, and 15°C in 90/10 H2O/D2O. 13C and/or 15N decoupling during acquisition was achieved using the GARP composite pulse sequence. Assignments of RNAs were achieved using 2D NOESY experiments at different mixing times, 2D TOCSY and 2D COSY-DQF. Assignments of nonexchangeable protons of labeled RNAs were completed using 13C filtered-NOESY, 1H–13C constant-time HSQC, 3D-HCCH-TOCSY, and HCN experiments (Santoro and King 1992; Zwahlen et al. 1997; Iwahara et al. 2001; Riek et al. 2001). 2D filtered/edited NOESY performed on selectively labeled samples with 13C/15N -G or -U or -A or -C were used to confirm NOE assignments. Dihedral restraints were obtained from HP-COSY and HCP experiments at 30°C (Sklenar et al. 1986; Marino et al. 1995).
NOESY experiments recorded in 90/10 H2O/D2O and in 99.9% D2O, at different mixing times and various temperatures were also performed in the presence of 3 mM Mg2+. Heteronuclear NMR spectra (1H-15N HSQC, 1H-13C HSQC) were also acquired with 3 mM Mg2+.
For residual dipolar coupling (RDC) experiments, partial alignment of 13C/15N HPI RNA was achieved by adding Pf1 filamentous bacteriophage supplied in 10 mM sodium phosphate buffer at pH 6.5 (ASLA Biotech), resulting in a splitting of the deuterium solvent line of 7 Hz. 1H–15N and 1H–13C RDCs were calculated as the difference between couplings measured for isotropic and partially aligned samples with ipap-HSQC and constant time HSQC experiments recorded at 30°C (Ottiger et al. 1998). RDCs could be measured for the Mg2+-free and 3 mM Mg2+ samples.
Distance and dihedral restraints for structure calculation
Distance restraints involving nonexchangeable protons were derived from cross-peak intensities in NOESY experiments using the H5/H6 cross-peaks of pyrimidines as internal standard. Interproton distances derived from NOE cross-peaks were classified into four distance bound ranges: strong (1.8–3.0 Å), medium (3.0–4.5 Å), weak (4.0–5.5 Å), and very weak (4.5–7.0 Å). Hydrogen bonding restraints were used for base pairs: from G24•C87 to C33•G78, C35•G74 to C36•G73, C37•G70 to C38•G69 and G42•C67 to C48•G60. Classical A-helix dihedral angle values were used for stem residues: α = −65 ± 30°, β = 178 ± 30° and ε = −155 ± 30°. For residues G24, A34, C35, C37, U40 to G42, G46, U1 to G60, U63, G64, U68, C71 to G73, C75 to G78, α was left unconstrained. β was constrained to the trans conformation (178 ± 30°) for most of residues, except U41 and U76, which were constrained to gauche− conformation (−60 ± 30°) as determined from the analysis of 3JP-H5′, 3JP-H5” and 3JP-C4′ from HP-COSY and HCP experiments. ε was constrained to the trans conformation (−155 ± 30°) for most of residues, with the exception of C36, U41, G46, U2, C3, C71, U72, C75, and U76, that were constrained to gauche− conformation (260 ± 30°) as determined from the analysis of 3JH3′-P, 3JC2′-P and 3JC4′-P derived from HP-COSY and HCP experiments. Torsion angles χ were derived from the observation of intra-residue H6/H8-H1′ cross-peak volumes. All bases were restrained to anti-conformation (−158 ± 30°), with the exception of U41, G46, U2, C3, G4, C71, U72, C75 to A77 which χ angles were loosely constrained. The ribose sugar pucker was determined from the analysis of H1′–H2′ coupling constants. Torsion angles ν0 to ν4 were derived from the analysis of COSY-DQF and TOCSY experiments. Nucleotides with no COSY and no TOCSY cross-peaks between H1′ and H2′ protons were restrained to the C3′-endo conformation (ν0 = 6 ± 15°, ν1 = −25 ± 15°, ν2 = 37 ± 15°, ν3 = −37 ± 15°, ν4 = 21 ± 15°). Nucleotides U2, C3, C71, U72, C75 to A77, with cross-peaks in the COSY spectrum, were classified as C2′-endo (ν0 = −21 ± 15°, ν1 = 35 ± 15°, ν2 = −35 ± 15°, ν3 = 24 ± 15°, ν4 = −2 ± 15°).
Structure calculations
Structures were calculated with CNS (crystallography and NMR system) torsion angle molecular dynamics (TAMD) protocol for nucleic acids using NOE and dihedral angle restraints (Brunger et al. 1998). One hundred structures were generated from a randomized extended strand. The first stage consisted of a high-temperature torsion angle dynamics for 30 psec at 20 000 K with a van der Waals scale factor of 0.1. During the second stage, the molecules were cooled for 40 psec of torsion angle dynamics with a van der Waals scale factor increased from 0.1 to 1. In the third stage, molecules were submitted to a second slow cooling for 35 psec in Cartesian space where the van der Waals scale factor increased from 1 to 4. Finally, 10 Powell cycles of energy minimization of 300 steps each were done.
In the second step, the structures were refined adding RDCs data to the structural restraints. Axial (Da) and rhombic (R) components of the aligned tensor were determined from structures calculated in the absence of RDCs with zero violation on NOE distance (0.2 Å) and dihedral (5°). The initial alignment tensor was first estimated from the best fit of observed RDCs to the calculated prefolded structures, using the PALES software (Zweckstetter and Bax 2000). An extensive grid search was then used to determine optimal Da and R values (Clore et al. 1998). Da and R were calculated at −20.2 Hz and 0.58, respectively, for the structures with no magnesium, and at −22.2 Hz and 0.58, respectively, for the structures with 3 mM magnesium. The prefolded structures were next submitted to a calculation including RDC data. The first stage consisted of a torsion angle dynamics for 10 psec at 300 K with a van der Waals scale factor of 0.1. During the second stage, the molecules were cooled for 9 psec of torsion angle dynamics with a van der Waals scale factor increased from 0.1 to 1. The SANI force constraint used for RDCs was gradually increased from 0.01 kcal mol−1 Hz−2 to 1.00 kcal mol−1 Hz−2. Finally, 10 Powell cycles of energy minimization of 800 steps each were done. These structures, with no violations and the best fit with RDCs data, were further refined using the nucleic acid force field to include optimized potentials for liquid simulation (opls), charges and nonbonded parameters (Jorgensen and Tirado-Rives 1988). Quality factors (Q) and root-mean-square deviations (RMSD) between input and calculated RDCs were calculated with the PALES software for the 10 final converged structures before and after refinement with the opls force field (Table 1; Zweckstetter and Bax 2000). Structures were visualized and analyzed with MOLMOL, Pymol and 3DNA software packages (Koradi et al. 1996; Delano 2005; Colasanti et al. 2013).
Coordinates
Atomic coordinates have been deposited in the Protein Data Bank (PDB ID code 5IEM) and the NMR data in the Biological Magnetic Resonance Bank (BMRB access code 30026).
HIV-1 Tat peptide synthesis, purification, and analysis
Tat peptide was synthesized on a Applied biosystem 433A peptide synthesizer using standard Fmoc chemistry and resins. The peptide was purified by HPLC using a preparative scale C18 column (Waters: PrepPak cartridge, 21 × 250 mm, 300 A, 5 µM) with an acetonitrile gradient in 0.1% trifluoroacetic acid. The molecular weight and purity of the peptide were confirmed by mass spectroscopy. The sequence of HIV-1 Tat peptide is FITKGLGISYGRKKRRQRRRPSQGGQTHQDPIPKQ. Tat peptide was dialyzed against the buffer used for NMR experiments. The concentration was determined on a Nanodrop Spectrometer using a molar extinction coefficient calculated with the ExPASy Proteomics (http://www.expasy.ch/tools/protparam.html).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Agence Nationale de la Recherche (grant RNASPIN ANR-2010-JCJC-1507-01 to I.L.); by the French Infrastructure for Integrated Structural Biology (FRISBI) (ANR-10-INSB-05-01); and by Instruct, as part of the European Strategy Forum on Research Infrastructures (ESFRI) and through national member agreements; CNRS, University of Strasbourg; and INSERM. Funding to pay publication charges was provided by Agence Nationale de la Recherche. The authors thank Claude Ling for technical assistance at the NMR spectrometer. We are also grateful to Katia Zanier and Robert Drillien for critical reading of the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.056523.116.
REFERENCES
- Aboul-ela F, Karn J, Varani G. 1996. Structure of HIV-1 TAR RNA in the absence of ligands reveals a novel conformation of the trinucleotide bulge. Nucleic Acids Res 24: 3974–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu A, Petrov A, Stombaugh J, Zirbel CL, Leontis NB. 2012. Comprehensive survey and geometric classification of base triples in RNA structures. Nucleic Acids Res 40: 1407–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboric M, Lenasi T. 2010. Kick-sTARting HIV-1 transcription elongation by 7SK snRNP deporTATion. Nat Struct Mol Biol 17: 928–930. [DOI] [PubMed] [Google Scholar]
- Barboric M, Yik JH, Czudnochowski N, Yang Z, Chen R, Contreras X, Geyer M, Matija Peterlin B, Zhou Q. 2007. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res 35: 2003–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrandon C, Bonnet F, Nguyen VT, Labas V, Bensaude O. 2007. The transcription-dependent dissociation of P-TEFb-HEXIM1–7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol Cell Biol 27: 6996–7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger F, Baigude H, Rana TM. 2009. U30 of 7SK RNA forms a specific photo-crosslink with Hexim1 in the context of both a minimal RNA-binding site and a fully reconstituted 7SK/Hexim1/P-TEFb ribonucleoprotein complex. J Mol Biol 386: 1094–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B, van Wamel JL. 2000. The leader of the HIV-1 RNA genome forms a compactly folded tertiary structure. RNA 6: 282–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazek D, Barboric M, Kohoutek J, Oven I, Peterlin BM. 2005. Oligomerization of HEXIM1 via 7SK snRNA and coiled-coil region direct the inhibition of P-TEFb. Nucleic Acids Res 33: 7000–7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, et al. 1998. Crystallography & NMR System (CNS), a new software suite for macromolecular structure determination. Acta Cryst D 54: 905–921. [DOI] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Hintermair C, Dirk E. 2008. Molecular evolution of the RNA polymerase II CTD. Trends Genet 24: 289–296. [DOI] [PubMed] [Google Scholar]
- Clore GM, Gronenborn AM, Tjandra N. 1998. Direct structure refinement against residual dipolar couplings in the presence of rhombicity of unknown magnitude. J Magn Reson 131: 159–162. [DOI] [PubMed] [Google Scholar]
- Colasanti A, Lu XJ, Olson W. 2013. Analyzing and building nucleic acid structures with 3DNA. J Vis Exp 74: e4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czudnochowski N, Vollmuth F, Baumann S, Vogel-Bachmayr K, Geyer M. 2010. Specificity of Hexim1 and Hexim2 complex formation with cyclin T1/T2, importin α and 7SK snRNA. J Mol Biol 395: 28–41. [DOI] [PubMed] [Google Scholar]
- DeLano WL. 2005. The PyMOL molecular graphics system. Delano Scientific LLC, San Francisco, CA. [Google Scholar]
- Dingley AJ, Grzesiek S. 1998. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide 2JNN couplings. J Am Chem Soc 120: 8293–8297. [Google Scholar]
- Dingwall C, Ernberg I, Gait MJ, Green SM, Heaphy S, Karn J, Lowe AD, Singh M, Skinner MA, Valerio R. 1989. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc Natl Acad Sci 86: 6925–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diribarne G, Bensaude O. 2009. 7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor. RNA Biol 6: 122–128. [DOI] [PubMed] [Google Scholar]
- D'Orso I, Frankel AD. 2010. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat Struct Mol Biol 17: 815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac C, Michels AA, Fraldi A, Bonnet F, Nguyen VT, Napolitano G, Lania L, Bensaude O. 2005. Transcription-dependent association of multiple positive transcription elongation factor units to a HEXIM multimer. J Biol Chem 280: 30619–30629. [DOI] [PubMed] [Google Scholar]
- Durney MA, D'Souza V. 2010. Preformed protein-binding motifs in 7SK snRNA: structural and thermodynamic comparisons with retroviral TAR. J Mol Biol 404: 555–567. [DOI] [PubMed] [Google Scholar]
- Egloff S, Murphy S. 2008. Cracking the RNA polymerase II CTD code. Trends Genet 24: 280–288. [DOI] [PubMed] [Google Scholar]
- Egloff S, Van Herreweghe E, Kiss T. 2006. Regulation of polymerase II transcription by 7SK snRNA: two distinct RNA elements direct P-TEFb and HEXIM1 binding. Mol Cell Biol 26: 630–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farès C, Amata I, Carlomagno T. 2007. 13C-detection in RNA bases: revealing structure-chemical shift relationships. J Am Chem Soc 129: 15814–15823. [DOI] [PubMed] [Google Scholar]
- Franck AT, Stelzer AC, Al-Hashimi HM, Andricioaei I. 2009. Constructing RNA dynamical ensembles by combining MD and motionally decoupled NMR RDCs: new insights into RNA dynamics and adaptive ligand recognition. Nucleic Acids Res 37: 3670–3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG. 2004. SPARKY3. University of California, San Francisco, CA. [Google Scholar]
- Gruber AR, Kilgus C, Mosig A, Hofacker IL, Hennig W, Stadler PF. 2008a. Arthropod 7SK RNA. Mol Biol Evol 25: 1923–1930. [DOI] [PubMed] [Google Scholar]
- Gruber AR, Koper-Emde D, Marz M, Tafer H, Bernhart S, Obernosterer G, Mosig A, Hofacker IL, Stadler PF, Benecke BJ. 2008b. Invertebrate 7SK snRNAs. J Mol Evol 66: 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipaty SA, McNamara RP, Morton EL, D'Orso I. 2015. PPM1G binds 7SK RNA and Hexim1 to block P-TEFB assembly into the 7SK snRNP and sustain transcription elongation. Mol Cell Biol 35: 3810–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney T Jr, Eliceiri GL. 1980. Intracellular distribution of low molecular weight RNA species in HeLa cells. J Cell Biol. 87: 398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gursoy HC, Koper D, Benecke BJ. 2000. The vertebrate 7SK RNA separates hagfish (Myxine glutinosa) and lamprey (Lampetra fluviatilis). J Mol Evol 50: 456–464. [DOI] [PubMed] [Google Scholar]
- He N, Jahchan NS, Hong E, Li Q, Bayfield MA, Maraia RJ, Luo K, Zhou Q. 2008. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol Cell 29: 588–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig M, Williamson JR. 2000. Detection N-H···N hydrogen bonding in RNA via scalar couplings in the absence of observable imino proton resonances. Nucleic Acids Res 28: 1585–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isel C, Karn J. 1999. Direct evidence that HIV-1 Tat stimulates RNA polymerase II carboxyl-terminal domain hyperphosphorylation during transcriptional elongation. J Mol Biol 290: 929–941. [DOI] [PubMed] [Google Scholar]
- Iwahara J, Wojciak JM, Clubb RT. 2001. Improved NMR spectra of a protein-DNA complex through rational mutagenesis and the application of a sensitivity optimized isotope-filtered NOESY experiment. J Biomol NMR 19: 231–241. [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Therien C, Bergeron D, Bourassa S, Greenblatt J, et al. 2007. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell 27: 262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen WL, Tirado-Rives J. 1988. The OPLS [Optimized Potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc 110: 1657–1666. [DOI] [PubMed] [Google Scholar]
- Kao C, Zheng M, Rudisser S. 1999. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA 5: 1268–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn J. 1999. Tackling Tat. J Mol Biol 293: 235–254. [DOI] [PubMed] [Google Scholar]
- Koradi R, Billeter M, Wuthrich K. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph 14: 51–55. [DOI] [PubMed] [Google Scholar]
- Krueger BJ, Jeronimo C, Roy BB, Bouchard A, Barrandon C, Byers SA, Searcey CE, Cooper JJ, Bensaude O, Cohen EA, et al. 2008. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res 36: 2219–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger W, Benecke BJ. 1987. Structural and functional analysis of a human 7SK RNA gene. J Mol Biol 195: 31–41. [DOI] [PubMed] [Google Scholar]
- Lebars I, Martinez-Zapien D, Durand A, Coutant J, Kieffer B, Dock-Bregeon AC. 2010. HEXIM1 targets a repeated GAUC motif in the riboregulator of transcription 7SK promotes base-pair rearrangements. Nucleic Acids Res 38: 7749–7763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebars I, Vileno B, Bourbigot S, Turek P, Wolff P, Kieffer B. 2014. A fully enzymatic method for site-directed spin labeling of long RNA. Nucleic Acids Res 42: e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Price JP, Byers S, Cheng D, Peng J, Price DH. 2005. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at Threonine 186. J Biol Chem 280: 28819–28826. [DOI] [PubMed] [Google Scholar]
- Long KS, Crothers DM. 1999. Characterization of the solution conformations of unbound and Tat peptide-bound forms of HIV-1 TAR RNA. Biochemistry 38: 10059–10069. [DOI] [PubMed] [Google Scholar]
- Lukavsky PJ, Puglisi JD. 2005. Structure determination of large biological RNAs. Methods Enzymol 394: 399–416. [DOI] [PubMed] [Google Scholar]
- Mancebo HS, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, et al. 1997. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev 11: 2633–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak RA, Garcia-Blanco MA, Sharp PA. 1990. Identification and characterization of a HeLa nuclear protein that specifically binds to the transactivation-response TAR element of human immunodeficiency virus. Proc Natl Acad Sci 87: 3624–3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino JP, Schwalbe H, Anklin C, Bermel W, Crothers DM, Griesinger C. 1995. Sequential correlation of anomeric ribose protons and intervening phosphorus in RNA oligonucleotides by a 1H, 13C, 31P triple resonance experiment : HCP-CCH-TOCSY. J Biomol NMR 5: 87–92. [DOI] [PubMed] [Google Scholar]
- Markert A, Grimm M, Martinez J, Wiesner J, Meyerhans A, Meyuhas O, Sickmann A, Fischer U. 2008. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep 9: 569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Peng J, Xie Z, Price DH. 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem 271: 27176–27183. [DOI] [PubMed] [Google Scholar]
- Martinez-Zapien D, Saliou JM, Han X, Atmanene C, Proux F, Cianferani S, Dock-Bregeon AC. 2015. Intermolecular recognition of the non-coding RNA 7SK and HEXIM protein in perspective. Biochimie 117: 63–71. [DOI] [PubMed] [Google Scholar]
- Marz M, Donath A, Verstraete N, Nguyen VT, Stadler PF, Bensaude O. 2009. Evolution of 7SK RNA and its protein partners in metazoa. Mol Biol Evol 26: 2821–2830. [DOI] [PubMed] [Google Scholar]
- Michels AA, Bensaude O. 2008. RNA-driven cyclin-dependent kinase regulation: when CDK9/cyclin T subunits of P-TEFb meet their ribonucleoprotein partners. Biotechnol J 3: 1022–1032. [DOI] [PubMed] [Google Scholar]
- Michels AA, Nguyen VT, Fraldi A, Labas V, Edwards M, Bonnet F, Lania L, Bensaude O. 2003. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol Cell Biol 23: 4859–4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan JF, Uhlenbeck OC. 1989. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol 180: 51–62. [DOI] [PubMed] [Google Scholar]
- Molinaro M, Tinoco I Jr. 1995. Use of ultra stable UNCG tetraloop hairpins to fold RNA structures: thermodynamic and spectroscopic applications. Nucleic Acids Res 23: 3056–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muesing MA, Smith DH, Capon DJ. 1987. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell 48: 691–701. [DOI] [PubMed] [Google Scholar]
- Muniz L, Egloff S, Ughy B, Jady BE, Kiss T. 2010. Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat. PLoS Pathog 6: e1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz L, Egloff S, Kiss T. 2013. RNA elements directing in vivo assembly of the 7SK/MePCE/Larp7 transcriptional regulatory snRNP. Nucleic Acids Res 41: 4686–4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S, Di Liegro C, Melli M. 1987. The in vitro transcription of the 7SK RNA gene by RNA polymerase III is dependent only on the presence of an upstream promoter. Cell 51: 81–87. [DOI] [PubMed] [Google Scholar]
- Nguyen VT, Kiss T, Michels AA, Bensaude O. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414: 322–325. [DOI] [PubMed] [Google Scholar]
- Nguyen D, Krueger BJ, Sedore SC, Brogie JE, Rogers JT, Rajendra TK, Saunders A, Matera AG, Lis JT, Uguen P, et al. 2012. The Drosophila 7SK snRNP and the essential role of dHEXIM in development. Nucleic Acids Res 40: 5283–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottiger M, Delaglio F, Bax A. 1998. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J Magn Reson 131: 373–378. [DOI] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol Cell 23: 297–305. [DOI] [PubMed] [Google Scholar]
- Piotto M, Saudek V, Sklenar V. 1992. Gradient-tailored excitation for single-quantum NMR-spectroscopy of aqueous-solutions. J Biomol NMR 2: 661–665. [DOI] [PubMed] [Google Scholar]
- Plateau P, Gueron M. 1982. Exchangeable proton NMR without base-line distortion, using new strong-pulse sequences. J Am Chem Soc 104: 7310–7311. [Google Scholar]
- Riek R, Pervushin K, Fernandez C, Kainosho M, Wüthrich K. 2001. [13C,13C]- and [13C,1H]-TROSY in a triple resonance experiment for ribose-base and intrabase correlations in nucleic acids. J Am Chem Soc 123: 658–664. [DOI] [PubMed] [Google Scholar]
- Santoro J, King GC. 1992. A constant-time 2D Overbodenhausen experiment for inverse correlation of isotopically enriched species. J Magn Reson 97: 202–207. [Google Scholar]
- Schulte A, Czudnochowski N, Barboric M, Schonichen A, Blazek D, Peterlin BM, Geyer M. 2005. Identification of a cyclin T-binding domain in Hexim1 and biochemical analysis of its binding competition with HIV-1 Tat. J Biol Chem 280: 24968–24977. [DOI] [PubMed] [Google Scholar]
- Sedore SC, Byers SA, Biglione S, Price JP, Maury WJ, Price DH. 2007. Manipulation of P-TEFb control machinery by HIV: recruitement of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res 35: 4347–4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklenar V, Miyashiro H, Zon G, Miles T, Bax A. 1986. Assignment of the 31P and 1H resonances in oligonucleotides by two-dimensional NMR spectroscopy. FEBS Lett 208: 94–98. [DOI] [PubMed] [Google Scholar]
- Sklenar V, Piotto M, Leppik R, Saudek V. 1993. Gradient-tailored water suppression for 1H-15N HSQC experiments optimized to retain full sensitivity. J Magn Reson A 102: 241–245. [Google Scholar]
- Sumner-Smith M, Roy S, Barnett R, Reid LS, Kuperman R, Delling U, Sonenberg N. 1991. Critical chemical features in trans-acting-responsive RNA are required for interaction with human immunodeficiency virus type 1 Tat protein. J Virol 65: 5196–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G, Cheong C, Tinoco I Jr. 1991. Structure of an unusually stable RNA hairpin. Biochemistry 30: 3280–3289. [DOI] [PubMed] [Google Scholar]
- Walker SC, Avis JM, Conn GL. 2003. General plasmids for producing RNA in vitro transcripts with homogeneous ends. Nucleic Acids Res 31: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman DA, Steitz JA. 1991. Structural analyses of the 7SK ribonucleoprotein (RNP), the most abundant human small RNP of unknown function. Mol Cell Biol 11: 3432–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks KM, Crothers DM. 1991. RNA recognition by Tat-derived peptides: interaction in the major groove? Cell 66: 577–588. [DOI] [PubMed] [Google Scholar]
- Weeks KM, Ampe C, Schultz SC, Steitz TA, Crothers DM. 1990. Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science 249: 1281–1285. [DOI] [PubMed] [Google Scholar]
- Weiss MA, Narayan N. 1998. RNA recognition by arginine-rich peptide motifs. Biopolymers 48: 167–180. [DOI] [PubMed] [Google Scholar]
- Wyatt JR, Chastain M, Puglisi JD. 1991. Synthesis and purification of large amounts of RNA oligonucleotides. Biotechniques 11: 764–769. [PubMed] [Google Scholar]
- Xue Y, Yang Z, Chen R, Zhou Q. 2010. A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP. Nucleic Acids Res 38: 360–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhu Q, Luo K, Zhou Q. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414: 317–322. [DOI] [PubMed] [Google Scholar]
- Yik JH, Chen R, Pezda AC, Samford CS, Zhou Q. 2004. A human immunodeficiency virus type-1 Tat-like arginine-rich RNA binding-domain is essential for HEXIM1 to inhibit RNA polymerase II transcription through 7SK snRNA-mediated inactivation of P-TEFb. Mol Cell Biol 24: 5094–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yik JH, Chen R, Pezda AC, Zhou Q. 2005. Compensatory contributions of HEXIM1 and HEXIM2 in maintaining the balance of active and inactive positive transcription elongation factor b complexes for control of trancription. J Biol Chem 280: 16368–16376. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Yik JH. 2006. The yin and yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol Mol Biol Rev 70: 646–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieve G, Benecke BJ, Penman S. 1977. Synthesis of two classes of small RNA species in vivo and in vitro. Biochemistry 16: 4520–4525. [DOI] [PubMed] [Google Scholar]
- Zwahlen C, Legault P, Vincent SJF, Greenblatt J, Konrat R, Lewis EK. 1997. Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: application to a bacteriophage λ N-peptide/boxB RNA complex. J Am Chem Soc 119: 6711–6721. [Google Scholar]
- Zweckstetter M, Bax Ad. 2000. Prediction of sterically induced alignment in a dilute liquid crystalline media phase: aid to protein structure determination by NMR. J Am Chem Soc 122: 3791–3792. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.