Abstract
Aspergillus fumigatus is capable of causing invasive aspergillosis or acute bronchopulmonary aspergillosis, and the current situation is alarming. There are no vaccine or allergen shots available for Aspergillus-induced allergies. Thus, a novel approach in designing of an effective vaccine or allergen shot candidate against A. fumigatus is needed. Using immunoinformatics approaches from the characterized A. fumigatus allergens, we have mapped epitopic regions to predict potential peptides that elicit both Aspergillus-specific T cells and B cell immune response. Experimentally derived immunodominant allergens were retrieved from www.allergen.org. A total of 23 allergenic proteins of A. fumigatus were retrieved. Out of 23 allergenic proteins, 13 of them showed high sequence similarity to both human and mouse counterparts and thus were eliminated from analysis due to possible cross-reactivity. Remaining allergens were subjected to T cell (major histocompatibility complex class I and II alleles) and B cell epitope prediction using immune epitope database analysis resource. Only five allergens have shown a common B and T cell epitopic region between human and mouse. They are Asp f1 {147–156 region (RVIYTYPNKV); Mitogillin}, Asp f2 {5–19 region (LRLAVLLPLAAPLVA); Hypothetical protein}, Asp f5 {305–322 region (LNNYRPSSSSLSFKY); Metalloprotease}, Asp f17 {98–106 region (AANAGGTVY); Hypothetical protein}, and Asp f34 {74–82 region (YIQDGSLYL); PhiA cell wall protein}. The epitopic region from these five allergenic proteins showed potential for development of single peptide- or multipeptide-based vaccine or allergen shots for experimental prioritization.
Keywords: : allergens, Aspergillus fumigatus, Asp f34, epitopes, vaccine, vaccine design
Introduction
Aspergillus species are the most common ubiquitous spore-bearing fungal pathogens. A. fumigatus is one of the leading causative agents of invasive aspergillosis and acute bronchopulmonary aspergillosis.1 A. fumigatus causes infection in the form of invasive aspergillosis in the allogeneic hematopoietic stem cell transplant, HIV patients and individuals having cancer. A. fumigatus causes allergy in asthmatic or cystic fibrosis patients.2,3 Allergy results from hypersensitive reaction to Aspergillus allergens in patients with atopic asthma or having cystic fibrosis disease.2 Diseases associated with A. fumigatus allergens are increasing compared with other fungal allergens and, furthermore, it adds problems to life-threatening infections in immunocompromised patients such as patients having cancer, HIV, and those who have undergone organ transplants.2,4 Globally, it has been estimated that of 193 million asthmatic patients, 4,837,000 have allergic bronchopulmonary aspergillosis (ABPA).5 Recent data suggested that the fungal-associated allergic reactions or infections are increasing worldwide.1 To control Aspergillus-associated problems, various studies have been conducted for the development of a vaccine candidate against aspergillosis that showed promising results in mouse models.6–8 However, the use of recombinant allergens (Asp f3 and Asp f2) or crude extract and homology to host protein showed certain limitations.6,7,9 Furthermore, the emergence of drug resistance isolate of A. fumigatus opens up new challenges for A. fumigatus-associated infections.10 Over the last few decades, the use of azole fungicides increased in agriculture that led to emergence of azole-resistant A. fumigatus strain.11 Other major hurdles in fungal vaccine designing are the pathogenesis process, evading of pathogen from the immune system, host genetic factors such as highly polymorphic nature of major histocompatibility complex (MHC) genes present in the population, and genetic variation in pathogen recognition receptors (PRRs).12,13 Polymorphisms in PRRs (TLR, Pentraxins, etc.) can modulate host response against the microbes and that needs to be addressed for better immune response against the vaccines.14,15 Till now, there is no vaccine or allergen shot therapy for Aspergillus-induced allergies.16 In a recent development, epitopic peptide-based approaches to map potential vaccine candidates have gained importance.17 Designing of vaccine against A. fumigatus possibly needs integration of the immunoinformatics or immunogenetic approach.12
Thus, to map the epitopic region from the reported allergens of A. fumigatus, we used different in silico approaches to predict potential human and mouse MHC class I and MHC class II T cell or B cell epitopic region from protein sequence of A. fumigatus's allergens. Mouse MHC class II and MHC class I T cell epitopes were predicted because common epitopes that recognize both human and mouse MHC T cell epitopes might be tested on model organism for their therapeutic potential and their results can be tested on human subjects.18 Another purpose for screening of epitopic peptides of antigens from A. fumigatus with no homologs in humans is that they recognize both MHC class I and MHC class T cells of human. Other than vaccine or allergy shot candidate, such peptides can be directly used ex vivo for the development of A. fumigatus-specific T cells (Asp-STs) for adoptive immunotherapy of invasive aspergillosis in the allogeneic hematopoietic stem cell transplant individuals having hematopoietic malignancies.4 With the advancement of technology or various omics approaches, they pave the way to discover novel therapeutic or drug targets for both communicable and noncommunicable diseases that have serious impact in both developed or developing countries.19 In this study, we used the reverse vaccinology approach that resulted in identification of potential peptides or allergen shot candidate against A. fumigatus-induced infections or allergies.
Materials and Methods
Retrieval of A. fumigatus allergens
A. fumigatus allergens known to date were retrieved from www.allergen.org, which provided the allergen data sets classified by WHO/IUIS/allergen nomenclature subcommittee, an international organization that is responsible for maintaining and developing a unique, unambiguous, and systematic nomenclature for allergenic proteins.
Protein sequence retrieval
The complete amino acid sequences of allergenic proteins were retrieved from www.allergen.org and National Center for Biotechnology Information database (NCBI) (www.ncbi.nlm.nih.gov). A total of 23 allergens of A. fumigatus were retrieved from NCBI database and further explored for vaccine or allergen shot candidates for A. fumigatus-induced infections.
Identification of protein sequence similarity with the host
Sequence similarity of the allergenic protein with host's protein sequences, for example, Homo sapiens (Taxid: 9606) and model organism Mus musculus (Taxid: 10090), was carried out using the basic local alignment search tool (BLASTp). The hit with an expectation value (E-value) less than 10−4 was excluded from the analysis and these protein sequences were assumed to have high sequence similarity with the host and model organism's proteome.18
Antigenicity prediction of allergens
Antigenicity of allergenic proteins was predicted by the use of VaxiJen v2.0 server, which provides the antigenic profile of bacterial, viral, parasitic, and fungal proteins. We choose the threshold value of 0.4 to increase the accurate antigenicity and to avoid false-positive results.19
Mapping of B cell epitope
Each allergen protein sequence was then subjected to B cell epitope prediction using immune epitope database analysis resource (IEDB-AR). It is a linear B cell epitope prediction software that uses a different method to predict the linear B cell epitope. In this software, we use the BepiPred method for the prediction of B cell epitope. BepiPred program uses a combination of hidden Markov and propensity scale methods to find out the linear B cell epitope in antigenic proteins.20,21
Mapping of T cell epitope
(1) T cell MHC class I epitope mapping
T cell MHC class I-restricted epitopes from the set of allergenic proteins were identified using IEDB-AR programs available at the IEDB-AR.21 This database contains data sets of experimentally characterized B cell and T cell epitopes for humans and other model organisms that are used for vaccine research (mouse and nonhuman primates). MHC class molecules bind with antigens and then these bound antigens or epitopes are recognized by T cells for further processing. Inhibitory concentration (IC50) values were calculated for peptide epitopes that bind to MHC alleles, and on the bases of IC value, T cell epitopes were classified as follows: low-affinity IC50 value <5000 nM, intermediate-affinity IC50 value <500 nM, and high-affinity IC50 value <50 nM. We considered only lower IC50 value epitopes because lower value indicates higher binding affinity of epitopes with host MHC alleles. We used all mouse MHC class I alleles (H-2-Db, H-2-Dd, H-2-Kb, H-2-Kd, H-2-Kk, and H-2-Ld)18 and eight human MHC class I alleles that cover about 85–90% of the world population (A*0101, A*0201, A*2402, A*0301, A*1101, B*0702, B*0801, and B*1501). The epitopes for T cell MHC class I alleles were identified by submitting the FASTA format of allergenic protein sequence to IEDB-AR. The artificial neural network (ANN) method was used to predict nine-mer sequence MHC class I epitopes.18
(II) Mapping of T cell MHC class II epitope
T cell MHC class II-restricted epitopes were identified using IEDB-AR.21 We used mouse MHC class II alleles and most common human MHC class II molecule DR alleles. The epitopes for T cell MHC class II alleles were identified by submitting the FASTA format of allergenic protein sequence to IEDB-AR. The 15-mer sequence epitope identification was performed using the consensus method.22 This method uses combination of stabilized matrix alignment and average relative binding matrix strategies to deduce MHC class II epitopes. This approach showed the best performance and is highly sensitive among other similar methods.18
Sequence identity mapping of epitopes with host proteome
The most common predicted B cell and T cell epitopic regions of allergenic proteins were further subjected for sequence similarity with protein sequences of human or mouse to eliminate any possible autoimmune response in the host. BLASTp program was used to predict the similarity.23
3D structure modeling and characterization of epitopes
Using 10 allergenic proteins, Asp f1, Asp f2, Asp f5, Asp f17, and Asp f34 allergenic proteins containing both T cell and B cell epitopes (in mouse and human) were subjected to 3D structure modeling for epitopic region characterization. The FASTA formats of these proteins were subjected to Phyre2 server to make the 3D structure of target allergenic protein.24 BLAST of protein sequences using Phyre2 server against the protein data bank (PDB) was performed and few best hits based on the structural alignment were used as template. Out of five allergens, the PDB template was predicted for only Asp f1 and Asp f5 allergenic proteins. For the best template, predicted PDB files were subjected to ModRefiner for refinement of structure.25 Energy minimization of these structures was carried out by YASARA force field minimization tool that improves overall quality of predicted protein structures.26 Furthermore, modeled structures were validated by RAMPAGE (http://mordred.bioc.cam.ac.uk/∼rapper/rampage.php), a program that has been extensively used for stereochemical characteristics of predicted structures of the protein. PyMOL program (www.pymol.org/) was used to illustrate the predicted structures of epitopes. The position of predicted epitopes was also visualized by PyMOL.
Result and Discussion
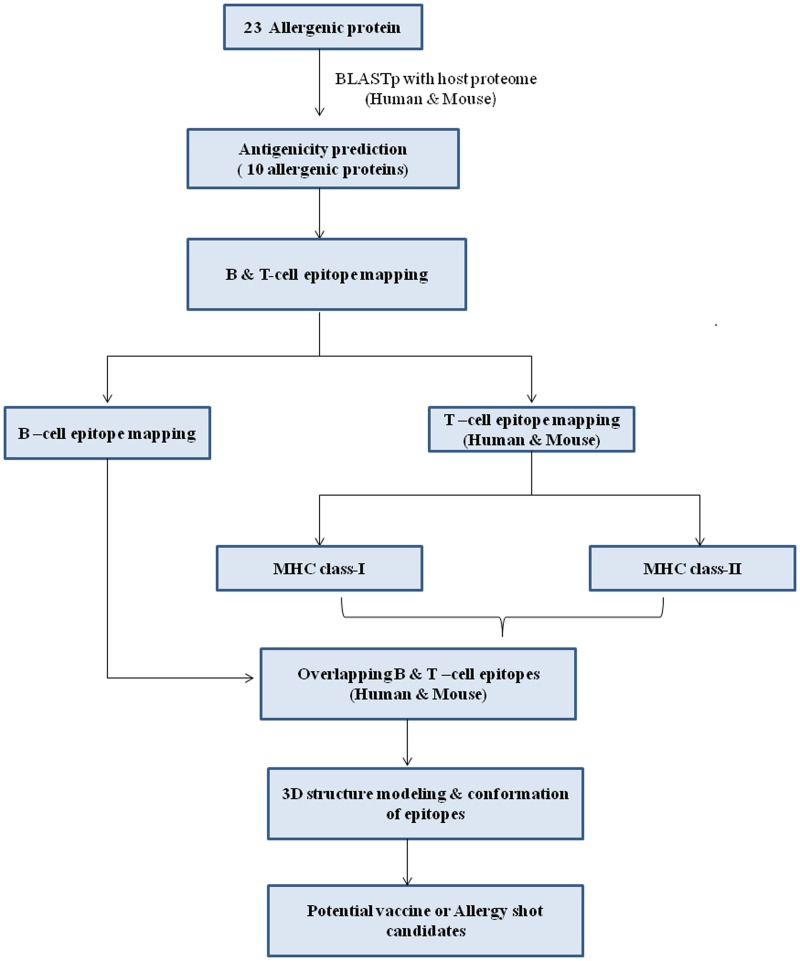
Allergic disorders such as asthma, atopic dermatitis, and allergic rhinitis caused by A. fumigatus have gained public attention. A. fumigatus not only causes ABPA but also is responsible for allergic Aspergillus sinusitis, hypersensitivity pneumonitis, and IgE-mediated asthma.27 Various strategies have been used to treat allergies such as allergen avoidance and elimination, subcutaneous injection of allergenic extract, and allergen shots.28 Immunotherapy involves the subcutaneous administration of gradually increasing quantities of allergens or allergen epitopic peptides until a dose has been reached that is effective enough to induce immunologic tolerance to these allergens. The goal of allergen-specific immunotherapy (SIT) is to subside the symptoms induced by allergens and further to reduce the recurrence of disease in the long term.29 In a recent report, it is observed that allergic incidence was caused by Alternaria alternata where whole crude antigens were used as SIT.30,31 So, attention has been focused on envisaging peptides that display both MHC class I and, especially, MHC class II T cell epitopes.32 A multitope vaccine or allergen shots having epitopes from several allergens may provide protection from A. fumigatus infections or allergies. In this direction, the reverse vaccinology approach has been employed to discover best epitopic peptides from A. fumigatus for experimental prioritization for vaccine or allergen shot candidates. The overall strategy used in this work is given in Figure 1.
FIG. 1.
Overall strategy used for prediction of vaccine or allergy shot candidates against Aspergillus-induced infections and allergy.
A total of 23 allergens of A. fumigatus were derived from allergen database and are presented in Table 1. These retrieved allergenic proteins of A. fumigatus were used to predict a vaccine or allergic shot candidate and have also been analyzed for ideal epitopic regions. Initially, these 23 allergenic proteins were subjected to homology search with host and mouse (model organism) proteome. A similar epitopic region, if selected for vaccine or allergy shots against A. fumigatus, may lead to devastating cross-reaction in host or it might lead to autoimmune diseases.33,34 Thus, it is important to screen the best allergenic protein that can be considered as potential vaccine or allergic shot candidate for experimental studies. Therefore, to obtain similarity between allergenic proteins and host or model organisms proteome, BLASTp was performed against mouse and human proteins. Of 23 allergenic proteins of A. fumigatus, 13 allergic proteins (Asp f3, Asp f6, Asp f8, Asp f10, Asp f11, Asp f12, Asp f13, Asp f18, Asp f22, Asp f23, Asp f27, Asp f28, and Asp f29) showed high sequence similarity with host and model organism. Thus, these allergenic proteins were eliminated from further analysis due to their role in potential cross-reactivity. Remaining 10 allergenic proteins (Asp f1, Asp f2, Asp f4, Asp f5, Asp f7, Asp f9, Asp f15, Asp f16, Asp f17, and Asp f 34) (Table 2) were considered for antigenicity analysis. All 10 allergenic proteins predicted to be most probable antigens by VaxiJen server having a threshold value >0.4. The antigenicity score of each of these allergens is given in Table 2. Furthermore, these allergens were subjected to map B and T cell epitopes.
Table 1.
Allergen Retrieved from www.allergen.org
| Aspergillus fumigatus | ||
|---|---|---|
| Allergen | GI number | Molecular weight (KDa) |
| Asp f1 | 166486 | 18 |
| Asp f2 | 1881574 | 37 |
| Asp f3 | 2769700 | 19 |
| Asp f4 | 3005839 | 30 |
| Asp f5 | 3776613 | 40 |
| Asp f6 | 1648970 | 26.5 |
| Asp f7 | 2879888 | 12 |
| Asp f8 | 6686524 | 11 |
| Asp f9 | 2879890 | 34 |
| Asp f10 | 963013 | 34 |
| Asp f11 | 5019414 | 24 |
| Asp f12 | 1930153 | 90 |
| Asp f13 | 2295 | 34 |
| Asp f15 | 3005841 | 16 |
| Asp f16 | 3643813 | 43 |
| Asp f17 | 2980819 | |
| Asp f18 | 2143220 | 34 |
| Asp f22 | 13925873 | 46 |
| Asp f23 | 21215170 | 44 |
| Asp f27 | 91680605 | 18 |
| Asp f28 | 91680607 | 13 |
| Asp f29 | 91680609 | 13 |
| Asp f34 | 133920236 | 20 |
Table 2.
Antigenicity of Allergen
| Antigen | GI number | Protein name | Antigenicity score (Threshold >0.4) |
|---|---|---|---|
| Asp f1 | 166486 | Mitogillin | 0.7540 |
| Asp f2 | 1881574 | Hypothetical protein | 0.8795 |
| Asp f4 | 3005839 | Hypothetical protein | 1.0311 |
| Asp f5 | 3776613 | Metalloprotease | 0.5683 |
| Asp f7 | 2879888 | Hypothetical protein | 0.8011 |
| Asp f9 | 2879890 | Hypothetical protein | 0.7615 |
| Asp f15 | 3005841 | Hypothetical protein | 0.8088 |
| Asp f16 | 3643813 | Hypothetical protein | 0.9120 |
| Asp f17 | 2980819 | IgE-binding protein | 0.9860 |
| Asp f34 | 133920236 | Cell wall protein PhiA | 0.5564 |
B and T cell epitope mapping
In silico tools become important for selecting good epitopic regions from immunodominant proteins that can save the screening time or expenses of synthetic peptides.13,19 It has been established that T and B lymphocytes act as antigenic determinants or epitopes of antigens instead of entire antigens. T cell recognizes epitopic peptides using T cell receptor that binds to either MHC I (CD8+ T cell) or MHC II (CD4+ T cells) class molecules or both present on antigen-presenting cells. Furthermore, T helper (CD4+ T cells) cells induce the B cells to activate humoral immune response.18 Ten antigenic allergenic proteins of A. fumigatus were subjected for mapping of linear B cell epitopes using the IEDB-AR BepiPred method. The identification of B cell epitopes is important for vaccine design, diagnosis, and antibody production.35,36 B cell epitopes are antigenic determinants that are recognized by the paratope region of membrane-bound antibodies or receptors on B-lymphocytes.18 All the identified B cell epitopes are listed in Table 3. Previously, it has been observed that allergen epitopes mainly comprised hydrophobic amino acids, and amino acids, Ser, Gly, Ala, and particularly Lys, play an important role in IgE antibody binding allergenic epitopic peptides.37,38 Our results showed very few lysine residues in predicted epitopic peptides from Asp f1, Asp f2, Asp f5, Asp f17, and Asp f34 allergens (Table 4).
Table 3.
Linear B Cell Epitopes for Allergen
| Serial No. | Allergen | GI number | Start | End | Epitope |
|---|---|---|---|---|---|
| 1 | Asp f1 | 166486 | 1 | 24 | MVAIKNLFLLAATAVSVLAAPSPL |
| 35 | 48 | QQLNPKTNKWEDKR | |||
| 104 | 118 | RPPKHSQNGMGKDDH | |||
| 132 | 142 | YKFDSKKPKED | |||
| 81 | 97 | GYDGNGKLIKGRTPIKF | |||
| 2 | Asp f2 | 1881574 | 20 | 37 | TLPTSPVPIAARATPHEP |
| 56 | 63 | CNATQRRQ | |||
| 97 | 105 | GNRPTMEAV | |||
| 124 | 133 | DNPDGNCALE | |||
| 136 | 146 | GGHWRGANATS | |||
| 169 | 179 | YTVAGSETNTF | |||
| 215 | 225 | SNGTESTHDSE | |||
| 242 | 304 | PGVGCAGESHGPDQGHDTGSASAPASTSTSSSSSGSGSGATTTPTDSPSATIDVPSNCHTHEG | |||
| 3 | Asp f4 | 3005839 | 21 | 44 | EWSGEAKTSDAPVSQATPVSNAVA |
| 46 | 97 | AAAASTPEPSSSHSDSSSSSGVSADWTNTPAEGEYCTDGFGGRTEPSGSGIF | |||
| 101 | 108 | NVGKPWGS | |||
| 111 | 120 | IEVSPENAKK | |||
| 128 | 135 | VGSDTDPW | |||
| 143 | 153 | IGPDGGLTGWY | |||
| 169 | 195 | YVAFDENSQGAWGAAKGDELPKDQFGG | |||
| 221 | 228 | IQAENAHH | |||
| 264 | 275 | VDGIGGKVVPGP | |||
| 4 | Asp f5 | 3776613 | 51 | 69 | TVIEAPSSFAPFKPQSYVE |
| 119 | 127 | NVGKDGKVF | |||
| 132 | 144 | SFYTGQIPSSAAL | |||
| 147 | 158 | RDFSDPVTALKG | |||
| 170 | 182 | DSASSESTEEKES | |||
| 255 | 274 | INDPTEGERTVIKDPWDSVA | |||
| 280 | 318 | ISDGSTNYTTSRGNNGIAQSNPSGGPSYLNNYRPSSSSL | |||
| 324 | 335 | YSVSSSPPSSYI | |||
| 360 | 376 | EKAGNFEYNTNGQGGLG | |||
| 385 | 405 | QDGSGTNNANFATPPDGQPGR | |||
| 471 | 510 | LKPGDKRSTDYTMGEWASNRAGGIRQYPYSTSLSTNPLTY | |||
| 541 | 559 | HGKNDAPKPTLRDGVPTDG | |||
| 5 | Asp f7 | 2879888 | 1 | 15 | SSGYSGPCSKGSPCV |
| 21 | 41 | YDTATSASAPSSCGLTNDGFS | |||
| 6 | Asp f9 | 2879890 | 31 | 58 | TWSKCNPLEKTCPPNKGLAASTYTADFT |
| 68 | 94 | VTAGKVPVGPQGAEFTVAKQGDAPTID | |||
| 110 | 116 | AAPGTGV | |||
| 196 | 207 | YNDAKGGTRFPQ | |||
| 217 | 231 | WAGGDPSNPKGTIEW | |||
| 233 | 243 | GGLTDYSAGPY | |||
| 252 | 270 | IENANPAESYTYSDNSGSW | |||
| 7 | Asp f15 | 3005841 | 18 | 32 | LAAPTPENEARDAIP |
| 34 | 55 | SVSYDPRYDNAGTSMNDVSCSN | |||
| 73 | 91 | FARIGGAPTIPGWNSPNCG | |||
| 109 | 117 | DAAPGGFN | |||
| 138 | 150 | ATYEEADPSHCAS | |||
| 8 | Asp f16 | 3643813 | 27 | 40 | PLAETCPPNKGLAA |
| 58 | 84 | VTAGKVPVGPQGAEFTVAKQGDAPTID | |||
| 127 | 160 | GDTTQVQTNYFGKGDTTTYDRGTYVPVATPQETF | |||
| 186 | 197 | YNDAKGGTRFPQ | |||
| 207 | 218 | GPAATPATPGHH | |||
| 271 | 337 | SSSSSVTSSTTSTASSASSTSSKTPSTSTLATSTKATPTPSGTSSGSNSSSSAEPTTTGGSGSSNTG | |||
| 351 | 378 | STGSSTSAGASATPELSQGAAGSIKGSV | |||
| 391 | 399 | CWHSKQNDD | |||
| 9 | Asp f17 | 2980819 | 3 | 11 | LVSREAPAV |
| 29 | 42 | SSYNGGDPSAVKSA | |||
| 51 | 65 | NSGVDTVKSGPALST | |||
| 98 | 106 | AANAGGTVY | |||
| 111 | 118 | AQYTAADS | |||
| 125 | 133 | AKVPESLSD | |||
| 10 | Asp f34 | 133920236 | 13 | 26 | AATASAAACQAPTN |
| 39 | 48 | AVQYQPFSAA | |||
| 58 | 71 | SQNASCDRPDEKSA | |||
| 75 | 92 | IQDGSLYLYAASATPQEI | |||
| 98 | 125 | GMGQGKIGYTTGAQPAPRNSERQGWAID | |||
| 154 | 165 | AGVANPAGNTDC | |||
| 173 | 182 | EDVTNPNSCV |
Table 4.
Selected High-Affinity Binding (IC50 < 50 nM) Nine-mer Mouse MHC Class I Epitopes
| Serial No. | Allergen | GI number | Start | End | Epitope |
|---|---|---|---|---|---|
| 1 | Asp f1 | 166486 | 2 | 10 | VAIKNLFLL |
| 148 | 156 | VIYTYPNKV | |||
| 87 | 95 | KLIKGRTPI | |||
| 2 | Asp f2 | 1881574 | 102 | 110 | MEAVGAYDV |
| 3 | Asp f4 | 3005839 | 8 | 16 | YATINGVLV |
| 162 | 170 | LEAGETKYV | |||
| 4 | Asp f5 | 3776613 | |||
| 5 | Asp f7 | 2879888 | 41 | 49 | SENVVALPV |
| 6 | Asp f9 | 2879890 | 244 | 252 | TMYVKSVRI |
| 167 | 175 | QETFHTYTI | |||
| 7 | Asp f15 | 3005841 | 25 | 33 | NEARDAIPV |
| 5 | 13 | TPISLISLF | |||
| 8 | Asp f16 | 3643813 | 157 | 165 | QETFHTYTI |
| 9 | Asp f17 | 2980819 | 6 | 14 | REAPAVGVI |
| 82 | 90 | VEGVIDDLI | |||
| 10 | Asp f34 | 133920236 | 67 | 75 | DEKSATFYI |
MHC, major histocompatibilty complex.
Furthermore, T cells and MHC-I and MHC-II class epitopes have been predicted by the ANN method.18 We considered a low IC50 value for epitope prediction. On the basis of IC50 value, epitopes were classified into three categories: high-affinity (IC50 < 50 nM), intermediate (IC50 < 500), and low-affinity (IC50<) binding epitopes. Two allergenic proteins, Asp f5 and Asp f7, did not contain any high-affinity binding MHC class I T cell epitopes for mouse and human, respectively. We use all mouse MHC class I alleles and eight human alleles (A*0101, A*0201, A*2402, A*0301, A*1101, B*0702, B*0801, and B*1501) that cover 90% of the world population39 (Tables 3–6). Furthermore, four allergenic proteins, Asp f1, Asp f2, Asp f4, and Asp f5, were predicted to have high-affinity binding mouse MHC class II-restricted epitopes, whereas all 10 allergenic proteins showed high-affinity human MHC class II-restricted T cell epitopes. The fifteen-mer MHC class II-restricted T cell epitopes are presented in Tables 6 and 7. Previously, Chaudhary et al. tested the therapeutic potential of Asp f1 allergen epitopes (INQQLNPKTNKWEDK, INQQLNPK, LNPKTNKWEDK) in sensitized BALB/c mice. They observed the increase in production of Th1 cytokines and suppression of lung eosinophilia by Asp f1 peptides. Thus, they establish the use of allergen peptides to control allergenic reactions in mice and open the way for human study.27 Our analysis also predicted the same B cell and T cell (MHC-II class) epitopic peptides that are used by Chaudhary et al. and suggested a strong correlation between in silico prediction and experimental evidences. We further analyze the epitopic data to screen common epitopic peptides for mouse and human so that they can be tested first on mouse model of A. fumigatus-induced allergy or infection model, and then the promising results from these studies can go for clinical trials for human use. Three allergenic proteins, Asp f1, Asp f2, and Asp f5, contained overlapping mouse and human MHC class I and II epitopes (Table 7), whereas only two allergic proteins, Asp f17 and Asp f34, contained overlapping human MHC class I and II epitopes (Table 8). It has been suggested that the cell wall proteins of A. fumigatus having no homology with humans, but showing homology with other fungal proteins, can be considered as ideal vaccine candidates against fungal pathogens.40 Recently, Tiwari et al. found the Asp fl 2 allergenic protein at germinating stage of Aspergillus flavus and showed no homology with human proteome.41 Previously, Gautam et al. have also reported Asp f2 and Asp f13 using the immunoproteomic approach and showed antibodies against these proteins in the serum samples of ABPA patients.42 Furthermore, Virginio et al. identified Asp f 12 and Asp f 22 from cell wall extracts of A. fumigatus's germinating conidia and also confirmed the presence of antibodies in patient serum samples against Asp f 12 and Asp f 22.43 Thus, the epitopic regions (predicted in our study) from these allergens may also be considered as promising vaccine candidates that potentially block the germinating conidia in the host. Furthermore, overlapping epitopes (MHC class I and II) were also recognized as B cell epitopes. So, these identified epitopes might be involved in both humoral and cell-mediated immunity (CD4+ and CD8+), which will be suitable for experimental studies in combination or alone in a mouse model of A. fumigatus-induced infection or for in vitro studies in human cell lines (Table 9). Previously, various studies showed the immunodominant role of allergens as vaccine or allergy shot candidates.7,44 Furthermore, allergen SIT or allergen shots balance the immune response, specially TH1 and TH2 immune response, and control the undesirable immune reactions.27,45
Table 5.
Selected High-Affinity Binding (IC50 < 50 nM) Nine-mer Human MHC Class I Epitopes
| Serial No. | Allergen | GI number | Start | End | Epitope |
|---|---|---|---|---|---|
| 1 | Asp f1 | 166486 | 118 | 126 | HYLLEFPTF |
| 9 | 17 | LLAATAVSV | |||
| 147 | 155 | RVIYTYPNK | |||
| 2 | Asp f2 | 1881574 | 9 | 17 | VLLPLAAPL |
| 181 | 189 | ASDLMHRLY | |||
| 198 | 206 | WVDHFADGY | |||
| 15 | 23 | APLVATLPT | |||
| 163 | 171 | SMCSQGYTV | |||
| 94 | 102 | KYFGNRPTM | |||
| 183 | 191 | DLMHRLYHV | |||
| 3 | Asp f4 | 3005839 | 244 | 252 | SIISHGLSK |
| 272 | 280 | VPGPTRLVV | |||
| 31 | 39 | APVSQATPV | |||
| 244 | 252 | SIISHGLSK | |||
| 91 | 99 | PSGSGIFYK | |||
| 4 | Asp f5 | 3776613 | 529 | 537 | MLYEVLWNL |
| 242 | 250 | YVAEADYQV | |||
| 312 | 320 | RPSSSSLSF | |||
| 76 | 84 | KMIAPDATF | |||
| 334 | 342 | YIDASIIQL | |||
| 19 | 27 | HPAHQSYGL | |||
| 495 | 503 | RQYPYSTSL | |||
| 125 | 133 | KVFSYGNSF | |||
| 4 | 12 | LLLAGALAL | |||
| 316 | 324 | SSLSFKYPY | |||
| 314 | 322 | SSSSLSFKY | |||
| 348 | 356 | IYHDLLYTL | |||
| 5 | Asp f7 | 2879888 | |||
| 6 | Asp f9 | 2879890 | 235 | 243 | LTDYSAGPY |
| 15 | 23 | YTAAALAAV | |||
| 47 | 55 | GLAASTYTA | |||
| 192 | 200 | RTLTYNDAK | |||
| 171 | 179 | HTYTIDWTK | |||
| 141 | 149 | QVQTNYFGK | |||
| 95 | 103 | TDFYFFFGK | |||
| 5 | 13 | ILRSADMYF | |||
| 7 | 15 | RSADMYFKY | |||
| 7 | Asp f15 | 3005841 | 96 | 104 | LQYEQNTIY |
| 8 | Asp f16 | 3643813 | 251 | 259 | HLLGQLWLL |
| 381 | 389 | ALWCSAPSL | |||
| 5 | 13 | YTAAALAAV | |||
| 285 | 293 | SSASSTSSK | |||
| 198 | 206 | TPMRLRLAA | |||
| 182 | 190 | RTLTYNDAK | |||
| 161 | 169 | HTYTIDWTK | |||
| 333 | 341 | SSNTGSWLR | |||
| 242 | 250 | RERQPRRVL | |||
| 131 | 139 | QVQTNYFGK | |||
| 245 | 253 | QPRRVLHLL | |||
| 85 | 93 | TDFYFFFGK | |||
| 19 | 206 | TPMRLRLAA | |||
| 285 | 293 | SSASSTSSK | |||
| 417 | 425 | FGIGVSPSF | |||
| 9 | Asp f17 | 2980819 | 84 | 92 | GVIDDLISK |
| 23 | 31 | ALASAVSSY | |||
| 130 | 138 | SLSDIAAQL | |||
| 118 | 126 | SLAKAISAK | |||
| 113 | 121 | YTAADSLAK | |||
| 98 | 106 | AANAGGTVY | |||
| 85 | 93 | VIDDLISKK | |||
| 118 | 126 | SLAKAISAK | |||
| 10 | Asp f34 | 133920236 | 74 | 82 | YIQDGSLYL |
| 175 | 183 | VTNPNSCVY | |||
| 175 | 183 | VTNPNSCVY | |||
| 45 | 53 | FSAAKSSIF | |||
| 65 | 73 | RPDEKSATF | |||
| 61 | 69 | ASCDRPDEK |
Table 6.
Selected High-Affinity Binding (IC50 < 50 nM) Fifteen-mer Mouse MHC Class II Epitopes
| Serial No. | Allergen | GI number | Start | End | Epitope |
|---|---|---|---|---|---|
| 1 | Asp f1 | 166486 | 9 | 23 | LLAATAVSVLAAPSP |
| 8 | 22 | FLLAATAVSVLAAPS | |||
| 2 | Asp f2 | 1881574 | 5 | 19 | LRLAVLLPLAAPLVA |
| 3 | Asp f4 | 3005839 | 39 | 53 | VSNAVAAAAAASTPE |
| 38 | 52 | PVSNAVAAAAAASTP | |||
| 4 | Asp f5 | 3776613 | 318 | 332 | LSFKYPYSVSSSPPS |
| 319 | 333 | SFKYPYSVSSSPPSS | |||
| 5 | Asp f17 | 2980819 | 93 | 108 | KDKFVAANAGGTVYED |
| 6 | Asp f34 | 133920236 | 75 | 89 | IQDGSLYLYAASATP |
Table 7.
Selected High-Affinity Binding (IC50 < 50 nM) Fifteen-mer Human MHC Class II Epitopes
| Serial No. | Allergen | GI number | Start | End | Epitope |
|---|---|---|---|---|---|
| 1 | Asp f1 | 166486 | 1 | 15 | MVAIKNLFLLAATAV |
| 39 | 53 | PKTNKWEDKRLLYSQ | |||
| 40 | 54 | KTNKWEDKRLLYSQA | |||
| 49 | 63 | LYSQAKAESNSHHAP | |||
| 75 | 89 | HWFTNGYDGNGKLIK | |||
| 2 | Asp f2 | 1881574 | 4 | 18 | LLRLAVLLPLAAPLV |
| 226 | 240 | AFEYFALEAYAFDIA | |||
| 15 | 29 | APLVATLPTSPVPIA | |||
| 204 | 218 | DGYDEVIALAKSNGT | |||
| 3 | Asp f4 | 3005839 | 5 | 20 | DTVYATINGVLVSWI |
| 37 | 51 | TPVSNAVAAAAAAST | |||
| 40 | 54 | GELCSIISHGLSKVI | |||
| 4 | Asp f5 | 3776613 | 1 | 15 | MRGLLLAGALALPAS |
| 179 | 193 | EKESYVFKGVSGTVS | |||
| 64 | 78 | PQSYVEVATQHVKMI | |||
| 576 | 590 | CNPNFVQARDAILDA | |||
| 505 | 519 | TNPLTYTSVNSLNAV | |||
| 308 | 322 | LNNYRPSSSSLSFKY | |||
| 305 | 319 | PSYLNNYRPSSSSLS | |||
| 5 | Asp f7 | 2879888 | 15 | 28 | VGQLTYYDTATSASA |
| 6 | Asp f9 | 2879890 | 9 | 23 | ADMYFKYTAAALAAV |
| 18 | 32 | AALAAVLPLCSAQTW | |||
| 238 | 252 | YSAGPYTMYVKSVRI | |||
| 274 | 288 | KFDGSVDISSSSSVT | |||
| 104 | 118 | AEVVMKAAPGTGVVS | |||
| 7 | Asp f15 | 3005841 | 68 | 82 | GSVPGFARIGGAPTI |
| 6 | 20 | PISLISLFVSSALAA | |||
| 1 | 15 | MKFTTPISLISLFVS | |||
| 8 | Asp f16 | 3643813 | 102 | 116 | GGTVYEDLKAQYTAA |
| 43 | 57 | SEKLVSTINSGVDTV | |||
| 100 | 114 | NAGGTVYEDLKAQYT | |||
| 114 | 128 | TAADSLAKAISAKVP | |||
| 15 | 29 | SDISAQTSALASAVS | |||
| 9 | Asp f17 | 2980819 | 1 | 15 | MYFKYTAAALAAVLP |
| 260 | 274 | AEHQVRRLRRYSSSS | |||
| 196 | 210 | PQTPMRLRLAAGPAA | |||
| 93 | 108 | AEVVMKAAPGTGVVS | |||
| 340 | 354 | LRLRLWLWLYSSTGS | |||
| 10 | Asp f34 | 133920236 | 1 | 15 | MQIKSFVLAASAAAT |
| 39 | 53 | AVQYQPFSAAKSSIF | |||
| 48 | 62 | AKSSIFAGLNSQNAS | |||
| 75 | 89 | IQDGSLYLYAASATP | |||
| 25 | 39 | TNKYFGIVAIHSGSA |
Table 8.
Common or Overlapping Epitopes of Allergens Recognizing MHC Class I and MHC Class II Alleles of Human and Mouse
| S. No. | Allergen | Mouse MHC class I | Mouse MHC class II | Human MHC class I | Human MHC class II |
|---|---|---|---|---|---|
| 1 | Asp f1 | 148–156 (VIYTYPNKV) | 147–155 (RVIYTYPNK) | ||
| 9–23 (LLAATAVSVLAAPSP) | 9–17 (LLAATAVSV) | 1–15 (MVAIKNLFLLAATAV) | |||
| 2 | Asp f2 | 5–19 (LRLAVLLPLAAPLVA) | 9–17 (VLLPLAAPL) | 4–18 (LLRLAVLLPLAAPLV) | |
| 3 | Asp f5 | 318–332 (LSFKYPYSVSSSPPS) | 316–324 (SSLSFKYPY) | 308–322 (LNNYRPSSSSLSFKY) | |
| 319–333 (SFKYPYSVSSSPPSS) | 314–322 (SSSSLSFKY) | 305–319 (PSYLNNYRPSSSSLS) | |||
| 4 | Asp f17 | 93–108 (DKFVAANAGGTVYED) | 98–106 (AANAGGTVY) | ||
| 5 | Asp f34 | 75–89 (IQDGSLYLYAASATP) | 74–82 (YIQDGSLYL) |
Table 9.
Potential Antigenic Allergen Proteins for Vaccine Candidate
| Serial No. | Allergen | GI Number | GenBank protein ID | Protein name | Immune response |
|---|---|---|---|---|---|
| 1 | Asp f1 | 166486 | AAB07779 | Mitogillin | Cellular and humoral |
| 2 | Asp f2 | 1881574 | AAC69357 | Hypothetical protein | Cellular and humoral |
| 3 | Asp f5 | 3776613 | CAA83015 | Metalloprotease | Cellular and humoral |
| 4 | Asp f17 | 2980819 | CAA12162 | IgE-binding protein | Cellular and humoral |
| 5 | Asp f34 | 133920236 | CAM54066 | cell wall protein PhiA | Cellular and humoral |
Modeling of tertiary structure
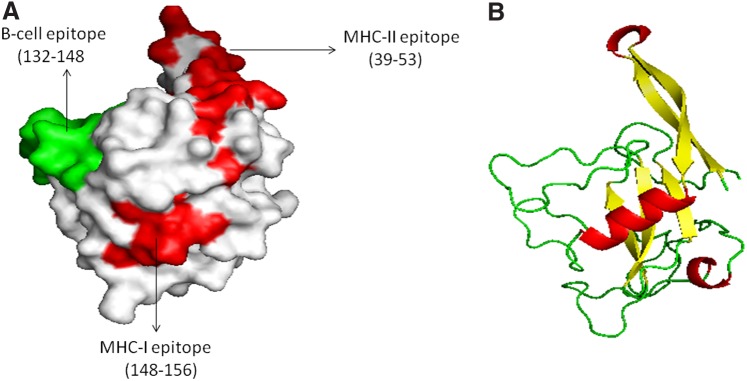
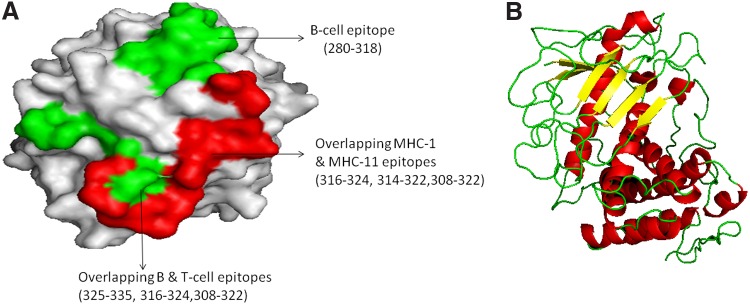
These five allergenic proteins that have overlapping MHC class I and MHC class II T cell epitopes were used to predict 3D modeled structure. Previously, Asp f1, Asp f2, Asp f3, and Asp f16 recombinant allergens have been tested as vaccine candidates.7,9,46 Of five promising allergens as vaccine or allergen shot candidates, Phyre2 server predicted 3D structure template for Asp f1 and Asp f5 only (Figs. 2 and 3). It identified multiple templates based on the best aligned sequence for some of the proteins. The best structural template was selected for Asp f1 and Asp f5 manually on the basis of best alignment length, a minimum number of gaps, and higher identity. For Asp f1 and Asp f5 structure models, unique template IDs (d1jbsa and c4k90A) were chosen. Asp f1 allergenic protein predicted to be a member of the ribonuclease family, whereas Asp f5 predicted to be an extracellular metalloproteinase. Furthermore, predicted model structures were submitted to energy minimization and structure refinement using ModRefiner and YASARA force field energy minimization server. After that modeled structures were validated by RAMPAGE. The Ramachandran plot predicted the structure stability of modeled structure. For Asp f1, 95.2% residues were found in the favored region, 4.8% in allowed region, and 0% in outlier region (Supplementary Fig. S1), and in case of Asp f5, 88.6% residues were in the favored region, 7.3% residues were in allowed region, and 4.1% residues were in outlier region (Supplementary Fig. S2). Furthermore, PyMOL was used to illustrate the spatial locations of residues in some epitopic peptides, which predicted to be located on the surface of the protein and presented at N-terminal of the protein. It is evident that T cell and B cell epitopes are exposed to the surface of the protein and therefore it supports that the predicted sequence may act as a potential vaccine peptide32 (Figs. 2 and 3). A similar method has been used for prediction of the 3D structure of proteins for vaccine candidate.19
FIG. 2.
Predicted 3D structure of Asp f1 and B cell and T cell epitopic regions. (A) The B and T cell epitopic region of Asp f1, red surface shows MHC-I T cell epitopic region, whereas green surface-exposed region shows overlapped T and B cell epitopes. (B) 3D structure of Asp f1. MHC, major histocompatibility complex.
FIG. 3.
Predicted 3D structure of Asp f5 and B cell and T cell epitopic regions. (A) The B and T cell epitopic region of Asp f5, red surface shows MHC-I and II T cell epitopic region, whereas green surface-exposed region shows overlapped T and B cell epitopes. (B) 3D structure of Asp f5.
Thus, the vaccination, alone and combination of selected peptides from these five allergenic proteins, can be used to combat Aspergillus-induced infection due to activation of both humoral and cell-mediated immune responses. On the other side, small T cell peptides (8–9 mer) (Table 10) can be used as allergen shot candidates because IgE antibody recognizes large epitopic peptides (B cell epitopes), thus these small peptides can activate T cell immune response and eliminate IgE activation.47
Table 10.
Potential Allergen Shot Peptides of Selected Allergenic Proteins
| Serial No. | Allergen | GI Number | T cell peptides |
|---|---|---|---|
| 1 | Asp f1 | 166486 | HYLLEFPTF |
| VIYTYPNKV | |||
| KLIKGRTPI | |||
| 2 | Asp f2 | 1881574 | MEAVGAYDV |
| 3 | Asp f17 | 2980819 | REAPAVGVI |
| VEGVIDDLI | |||
| 4 | Asp f34 | 133920236 | DEKSATFYI |
Conclusion
A total of five potential allergenic proteins (Asp f1, Asp f2, Asp f5, Asp f17, and Asp f34) from A. fumigatus as vaccine or allergy shot candidates were obtained. Epitopic peptides from these five proteins in combination or alone could be used to prioritize in experimental validation with human cell lines or in mouse model of A. fumigatus infection or allergic mouse models. Previously, Chaudhary et al. showed the therapeutic use of Asp f1 allergen epitopes (INQQLNPKTNKWEDK, INQQLNPK, LNPKTNKWEDK) in sensitized BALB/c mice. Chaudhary et al. observed increase in production of Th1 cytokines and suppression of lung eosinophilia by Asp f1 peptides. Thus, they established the use of allergen peptides to control allergenic reaction in mice. In addition, Gautam et al. identified Asp f2 using the immunoproteomic approach in ABPA patients, which correlates with our in silico results. Furthermore, we also analyzed the 3D structure of Asp f1 and Asp f5 allergenic proteins. Overall, resulting peptides from our analysis could be subjected to experimental prioritization to explore vaccine candidates or allergy immunotherapy against Aspergillus-mediated infections.
Supplementary Material
Abbreviations Used
- ABPA
allergic bronchopulmonary aspergillosis
- ANN
artificial neural network
- BLASTp
basic local alignment search tool
- IC50
inhibitory concentration
- IEDB-AR
immune epitope database analysis resource
- MHC
major histocompatibility complex
- NCBI
National Center for Biotechnology Information database
- PDB
protein data bank
- PRRs
pathogen recognition receptors
- SIT
specific immunotherapy
Acknowledgment
The authors are thankful to the Department of Biotechnology and Bioinformatics, Jaypee University of Information Technology, for providing research facilities and PhD fellowship.
Author Contributions
R.T. and J.S. conceived and designed the experiments. R.T. performed the experiments. R.T. and J.S. analyzed the data. J.S. contributed reagents/materials/analysis tools. R.T. and J.S. contributed in writing of the manuscript.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Thakur R, Anand R, Tiwari S, et al. Cytokines induce effector T-helper cells during invasive aspergillosis; what we have learned about T-helper cells? Front Microbiol. 2015;6:429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaudhary N, Marr KA. Impact of Aspergillus fumigatus in allergic airway diseases. Clin Transl Allergy. 2011;1:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shah A, Panjabi C. Allergic bronchopulmonary aspergillosis: a perplexing clinical entity. Allergy Asthma Immunol Res. 2016;8:282–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deo SS, Gottlieb DJ. Adoptive T-cell therapy for fungal infections in haematology patients. Clin Transl Immunol. 2015;4:e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denning DW, Pleuvry A, Cole DC. Global burden of allergic bronchopulmonary aspergillosis with asthma and its complication chronic pulmonary aspergillosis in adults. Med Mycol. 2013;51:361–370 [DOI] [PubMed] [Google Scholar]
- 6.Cenci E, Mencacci A, Bacci A, et al. T cell vaccination in mice with invasive pulmonary aspergillosis. J Immunol. 2000;165:381–388 [DOI] [PubMed] [Google Scholar]
- 7.Ito JI, Lyons JM, Hong TB, et al. Vaccinations with recombinant variants of Aspergillus fumigatus allergen Asp f 3 protect mice against invasive aspergillosis. Infect Immun. 2006;74:5075–5084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu M, Capilla J, Johansen ME, et al. Saccharomyces as a vaccine against systemic aspergillosis: “the friend of man” a friend again? J Med Microbiol. 2011;60:1423–1432 [DOI] [PubMed] [Google Scholar]
- 9.Diaz-Arevalo D, Bagramyan K, Hong TB, et al. CD4+ T cells mediate the protective effect of the recombinant Asp f3-based anti-aspergillosis vaccine. Infect Immun. 2011;79:2257–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verweij PE, Chowdhary A, Melchers WJ, et al. Azole resistance in aspergillus fumigatus: can we retain the clinical use of mold-active antifungal azoles? Clin Infect Dis. 2016;62:362–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chowdhary A, Kathuria S, Xu J, et al. Emergence of azole-resistant aspergillus fumigatus strains due to agricultural azole use creates an increasing threat to human health. PLoS Pathog. 2013;9:e1003633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iannitti RG, Carvalho A, Romani L. From memory to antifungal vaccine design. Trends Immunol. 2012;33:467–474 [DOI] [PubMed] [Google Scholar]
- 13.Patronov A, Doytchinova I. T-cell epitope vaccine design by immunoinformatics. Open Biol. 2013;3:120139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pellegrino P, Falvella FS, Cheli S, et al. The role of Toll-like receptor 4 polymorphisms in vaccine immune response. Pharmacogenomics J. 2016;16:96–101 [DOI] [PubMed] [Google Scholar]
- 15.Thakur R, Shankar J. In silico analysis revealed high-risk single nucleotide polymorphisms in human pentraxin-3 gene and their impact on innate immune response against microbial pathogens. Front Microbiol. 2016;7:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santos E, Levitz SM. Fungal vaccines and immunotherapeutics. Cold Spring Harb Perspect Med. 2014;4:a019711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell AW, McCluskey J, Rossjohn J. More than one reason to rethink the use of peptides in vaccine design. Nat Rev Drug Discov. 2007;6:404–414 [DOI] [PubMed] [Google Scholar]
- 18.Rana A, Rub A, Akhter Y. Proteome-wide B and T cell epitope repertoires in outer membrane proteins of Mycobacterium avium subsp. paratuberculosis have vaccine and diagnostic relevance: a holistic approach. J Mol Recognit. 2015;28:506–520 [DOI] [PubMed] [Google Scholar]
- 19.Vishnu US, Sankarasubramanian J, Gunasekaran P, et al. Novel vaccine candidates against brucella melitensis identified through reverse vaccinology approach. OMICS. 2015;19:722–729 [DOI] [PubMed] [Google Scholar]
- 20.Kim Y, Ponomarenko J, Zhu Z, et al. Immune epitope database analysis resource. Nucleic Acids Res. 2012;40:525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vita R, Overton JA, Greenbaum JA, et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015;43:405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang P, Sidney J, Dow C, et al. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol. 2008;4:e1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410 [DOI] [PubMed] [Google Scholar]
- 24.Kelley LA, Mezulis S, Yates CM, et al. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krieger E, Joo K, Lee J, et al. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins. 2009;77:114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu D, Zhang Y. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys J. 2011;101:2525–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaudhary N, Mahajan L, Madan T, et al. Prophylactic and therapeutic potential of Asp f1 epitopes in naive and sensitized BALB/c Mice. Immune Netw. 2009;9:179–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Behrmann J. The anti-vaccination movement and resistance to allergen-immunotherapy: a guide for clinical allergists. Allergy Asthma Clin Immunol. 2010;6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akdis CA, Akdis M. Mechanisms of allergen-specific immunotherapy and immune tolerance to allergens. World Allergy Organ J. 2015;8:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuna P, Kaczmarek J, Kupczyk M. Efficacy and safety of immunotherapy for allergies to Alternaria alternata in children. J Allergy Clin Immunol. 2011;127:502–508 [DOI] [PubMed] [Google Scholar]
- 31.Twaroch TE, Curin M, Valenta R, et al. Mold allergens in respiratory allergy: from structure to therapy. Allergy Asthma Immunol Res. 2015;7:205–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Yang H-W, Chen H, et al. In silico prediction of T and B Cell Epitopes of Der f 25 in Dermatophagoides farinae. Int J Genomics. 2014; DOI: 10.1155/2014/483905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shankar J, Nigam S, Saxena S, et al. Identification and assignment of function to the genes of Aspergillus fumigatus expressed at 37 degrees C. J Eukaryot Microbiol. 2004;51:428–432 [DOI] [PubMed] [Google Scholar]
- 34.Levitz SM. Aspergillus vaccines: hardly worth studying or worthy of hard study? Med Mycol. 2016; [Epub ahead of print]; DOI: 10.1093/mmy/myw081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bai L, Otsuki H, Sato H, et al. Identification and characterization of common B cell epitope in bovine leukemia virus via high-throughput peptide screening system in infected cattle. Retrovirology. 2015;12:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liljeroos L, Malito E, Ferlenghi I, et al. Structural and computational biology in the design of immunogenic vaccine antigens. J Immunol Res. 2015;2015:156241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolff N, Yannai S, Karin N, et al. Identification and characterization of linear B-cell epitopes of beta-globulin, a major allergen of sesame seeds. J Allergy Clin Immunol. 2004;114:1151–1158 [DOI] [PubMed] [Google Scholar]
- 38.Oezguen N, Zhou B, Negi SS, et al. Comprehensive 3D-modeling of allergenic proteins and amino acid composition of potential conformational IgE epitopes. Mol Immunol. 2008;45:3740–3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leblanc P, Moise L, Luza C, et al. VaxCelerate II: rapid development of a self-assembling vaccine for Lassa fever. Hum Vacci Immunother. 2014;10:3022–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Champer J, Ito J, Clemons K, et al. Proteomic analysis of pathogenic fungi reveals highly expressed conserved cell wall proteins. J Fungi. 2016;2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tiwari S, Thakur R, Goel G, et al. Nano LC-Q-TOF analysis of proteome revealed germination of Aspergillus flavus conidia is accompanied by MAPK signalling and cell wall modulation. Mycopathologia. 2016; [Epub ahead of print]; DOI: 10.1007/s11046-016-0056-x [DOI] [PubMed] [Google Scholar]
- 42.Gautam P, Sundaram CS, Madan T, et al. Identification of novel allergens of aspergillus fumigatus using immunoproteomics approach. Clin Exp Allergy. 2007;37:1239–1249 [DOI] [PubMed] [Google Scholar]
- 43.Virginio ED, Kubitschek-Barreira PH, Batista MV, et al. Immunoproteome of aspergillus fumigatus using sera of patients with invasive aspergillosis. Int J Mol Sci. 2014;15:14505–14530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jacquet A, Vanderschrick J-F, Vandenbranden M, et al. Vaccination with the recombinant allergen ProDer p 1 complexed with the cationic lipid DiC14-amidine prevents allergic responses to house dust mite. Mol Ther. 2005;11:960–968 [DOI] [PubMed] [Google Scholar]
- 45.Valenta R, Campana R, Focke-Tejkl M, et al. Vaccine development for allergen-specific immunotherapy based on recombinant allergens and synthetic allergen peptides: lessons from the past and novel mechanisms of action for the future. J Allergy Clin Immunol. 2016;137:351–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Banerjee B, Kurup VP, Greenberger PA, et al. Cloning and expression of aspergillus fumigatus allergen Asp f 16 mediating both humoral and cell-mediated immunity in allergic bronchopulmonary aspergillosis (ABPA). Clin Exp Allergy. 2001;31:761–770 [DOI] [PubMed] [Google Scholar]
- 47.Nilsson OB, Adedoyin J, Rhyner C, et al. In vitro evolution of allergy vaccine candidates, with maintained structure, but reduced B cell and T cell activation capacity. PLoS One. 2011;6:24558. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
Cite this article as: Thakur R, Shankar J (2016) In silico identification of potential peptides or allergen shot candidates against Aspergillus fumigatus, BioResearch Open Access 5:1, 330–341, DOI: 10.1089/biores.2016.0035.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.