Abstract
Purpose
Carfilzomib, while active in B-cell neoplasms displayed heterogeneous response in chronic lymphocytic leukemia (CLL) samples from patients and showed interpatient variability to carfilzomib-induced cell death. To understand this variability and predict patients that would respond to carfilzomib, we investigated the mechanism by which carfilzomib induces CLL cell death.
Experimental Design
Using CLL patient samples and cell lines, complementary knockdown and knockout cells, and carfilzomib-resistant cell lines, we evaluated changes in intracellular networks to identify molecules responsible for carfilzomib’s cytotoxic activity. Carfilzomib-treated cells were immunoblotted for molecules involved in ubiquitin, apoptotic, and endoplasmic reticulum (ER) stress response pathways and results correlated with carfilzomib cytotoxic activity. Co-immunoprecipitation and pull down assays were performed to identify complex interactions among MCL-1, Noxa, and Bak.
Results
Carfilzomib triggered ER stress and activation of both the intrinsic and extrinsic apoptotic pathways through alteration of the ubiquitin proteasome pathway. Consequently, the transcription factor CCAAT/enhancer-binding protein homology protein (CHOP) accumulated in response to carfilzomib, and CHOP depletion conferred protection against cytotoxicity. Carfilzomib also induced accumulation of MCL-1 and Noxa, whereby MCL-1 preferentially formed a complex with Noxa and consequently relieved MCL-1’s protective effect on sequestering Bak. Accordingly, depletion of Noxa or both Bak and Bax conferred protection against carfilzomib-induced cell death.
Conclusions
Collectively, carfilzomib induced ER stress culminating in activation of intrinsic and extrinsic caspase pathways, and we identified the CHOP protein level as a biomarker that could predict sensitivity to carfilzomib in CLL.
Keywords: Proteasome inhibitor, chronic lymphocytic leukemia, carfilzomib, apoptosis, Endoplasmic reticulum stress
INTRODUCTION
Chronic lymphocytic leukemia (CLL) (1) is an incurable malignancy that is characterized by the progressive accumulation of mature B cells in the peripheral blood, lymph nodes, bone marrow, spleen, and liver. CLL typically affects the elderly population, many of whom have preexisting conditions that restrict the use of the current standard treatment, which involves the combination of chemotherapeutic agents (fludarabine and cyclophosphamide) and an antibody (rituximab) (2, 3). Therefore, combination therapy that targets distinct biological pathways in CLL pathogenesis represents a significant step outside of the conventional chemotherapy-based approach. Accordingly, we recently identified carfilzomib, a Food and Drug Administration (FDA)-approved second-generation ubiquitin-proteasome pathway (UPP) inhibitor (4), as a potent pharmacologic agent that causes cytotoxicity in CLL cells before or after treatment with ibrutinib (5-9), an irreversible inhibitor of Bruton’s tyrosine kinase (10).
The UPP, which has been validated clinically as a therapeutic target (11), regulates many vital cellular processes including cell cycle progression, transcriptional regulation, signal transduction, apoptosis, immune response, and elimination of damaged proteins. Formation of a K48-linked polyubiquitin chain is the first step in the UPP that permits recognition of a protein targeted for degradation by the proteasome (12). The 26S proteasome is a proteolytic complex with caspases-like activity (beta 1 subunit), trypsin-like activity (beta 2 subunit), and chymotrypsin-like activity (beta 5 subunit) (13). Carfilzomib (also known as PR-171) is a tetrapeptide epoxyketone-based analog of the natural compound epoxomicin, which selectively and irreversibly inhibits the chymotrypsin-like activity of the proteasome (14-16).
Structurally, chemically, and mechanistically carfilzomib differs from bortezomib, the first proteasome inhibitor approved by the FDA, since bortezomib is a synthetic peptide borate that binds reversibly to the proteasome. Carfilzomib targets more selectively the chymotrypsin-like activity of the proteasome than bortezomib (i.e., carfilzomib has less selectivity to the caspaseand trypsin-like activity of the proteasome than bortezomib) in multiple myeloma (17) and earlier phase I clinical trials in multiple myeloma indicated that carfilzomib is a more robust inhibitor of the chymotrypsin-like activity than bortezomib (18, 19). Moreover, in multiple myeloma in vivo and in vitro studies indicated that carfilzomib can circumvent bortezomib resistance since some bortezomib resistant patients (20) and bortezomib resistant cell lines (21) remain responsive to carfilzomib therapeutic effects. In CLL, earlier preclinical studies indicated that in vitro treatment of CLL cells with bortezomib resulted in significant cell death by apoptosis (22, 23), however, in a phase II clinical trial conducted with bortezomib in fludarabine-refractory CLL patients, no patients achieved complete or partial responses. Though, some biological activity was observed (e.g. ≥50% decrease in absolute lymphocyte count, lymph nodes and spleen size was achieved in some of the patients) (24). Subsequently, it was shown that CLL refractory effect to bortezomib was due to its boronate moiety interaction with plasma components (25-27). Nevertheless, the biological activities observed with bortezomib in this high-risk factors and severe treatment-resistant group of CLL patients indicated that proteasome inhibitors with better efficacy and safety profiles in combination with agents with different toxicity profiles are warranted.
Carfilzomib cytotoxic activity in CLL has been described previously by our group (10) and by another (25); however, the molecular mechanism by which carfilzomib triggers cell death in CLL has not been elucidated. Furthermore, in our investigation, a bimodal distribution of cytotoxicity was observed in response to carfilzomib treatment: limited or substantial cell death (10). Therefore, in the present report, we first evaluated the cytotoxic effect of carfilzomib in 30 CLL patient samples at five different concentrations. We then used these CLL patient samples and additional B-cell lines to further examine the intracellular pathways implicated in carfilzomib-induced cell death. Finally, we investigated the molecular differences that could potentially be responsible for the heterogenic cytotoxic response to carfilzomib between patients. Thus, the aim of this study was to gain a better understanding of the molecular networks affected by carfilzomib, which could help identify CLL patients with a higher probability of responding to carfilzomib and carfilzomib-based combination therapies who could participate in further clinical studies. Our investigation identified the proapoptotic transcription factor CCAAT/enhancer-binding protein (C/EBP) homology protein (CHOP) as a biomarker that could predict sensitivity to carfilzomib in CLL.
MATERIALS AND METHODS
Reagents, cell lines, lentiviral vectors, and antibodies
The description, source, and location of all reagents, cell lines, lentiviral vectors, and antibodies that were used in this study are described in Supplementary Table S1.
Patient sample collection and characteristics
Peripheral blood samples were collected from CLL patients after written informed consent was obtained in accordance with the Declaration of Helsinki and under a protocol approved by the Institutional Review Board of The University of Texas MD Anderson Cancer Center. All patients and clinical characteristics are summarized in Table 1. Only 5 out of 30 patients received prior therapies (Patient 085: (1) Chlorambucil; (2) Fludarabine + Rituximab; (3) Rituximab; (4) Fludarabine + Rituximab; (5) Bendamustine + Rituximab; Patient 514: Ibrutinib; Patient 195: Fludarabine + Cyclophosphamide + Rituximab; Patient 575: (1) Fludarabine + Cyclophosphamide + Rituximab; (2) Ofatumumab + Revlimid; Patient 622: Ibrutinib).
Table 1.
Clinical Characteristics of 30 patients with CLL
| Pt | Sex | Age, y | Pr Rx | RAI Stage | WBC | IgVH gene | ZAP-70 IHC | B2M | ATM* | p53* |
|---|---|---|---|---|---|---|---|---|---|---|
| 514 | F | 69 | 1 | 0 | 35 | MUT | NEG | 1.8 | 0 | 0 |
| 302 | F | 59 | 0 | 0 | 27 | MUT | ND | 2.0 | 0 | 0 |
| 828 | M | 65 | 0 | 0 | 33 | MUT | NEG | 2.0 | 0 | 0 |
| 724 | M | 52 | 0 | 0 | 14 | MUT | POS | 1.7 | 0 | 0 |
| 295 | M | 71 | 0 | 0 | 104 | MUT | ND | 1.5 | 0 | 0 |
| 607 | F | 70 | 0 | 0 | 96 | MUT | NEG | 2.8 | 0 | 0 |
| 176 | M | 54 | 0 | 0 | 45 | MUT | NEG | 1.4 | 0 | 0 |
| 622 | M | 49 | 1 | 0 | 45 | MUT | POS | 1.7 | 0 | 0 |
| 043 | M | 52 | 0 | 1 | 75 | MUT | POS | 3.1 | 0 | 0 |
| 871 | M | 52 | 0 | 1 | 150 | MUT | ND | 3.4 | 0 | 0 |
| 916 | F | 60 | 0 | 1 | 62 | MUT | NEG | 2.4 | 0 | 0 |
| 417 | M | 41 | 0 | 2 | 158 | MUT | ND | 2.5 | 0 | 0 |
| 033 | M | 67 | 0 | 0 | 76 | UNMUT | POS | 2.7 | 0 | 0 |
| 708 | M | 54 | 0 | 1 | 44 | UNMUT | ND | 2.1 | ND | ND |
| 425 | M | 50 | 0 | 1 | 64 | UNMUT | POS | 2.4 | 0 | 29 |
| 454 | M | 57 | 0 | 1 | 89 | UNMUT | ND | 6.1 | 38 | 0 |
| 079 | F | 58 | 0 | 1 | 78 | UNMUT | ND | 2.1 | 81 | 0 |
| 315 | M | 59 | 0 | 3 | 103 | UNMUT | NEG | 6.5 | 0 | 91 |
| 195 | M | 50 | 1 | 4 | 43 | UNMUT | NEG | 2.2 | 0 | 0 |
| 820 | F | 58 | 0 | 1 | 72 | ND | POS | 2.0 | 0 | 0 |
| 089 | M | 66 | 0 | 1 | 45 | ND | NEG | 2.8 | 0 | 0 |
| 932 | F | 78 | 0 | 1 | 113 | ND | ND | 1.8 | ND | ND |
| 504 | F | 52 | 0 | 1 | 152 | ND | POS | 4.4 | 0 | 67 |
| 835 | F | 75 | 0 | 1 | 121 | ND | ND | 2.2 | 0 | 0 |
| 146 | M | 72 | 0 | 1 | 68 | ND | NEG | 4.9 | 34 | 0 |
| 575 | F | 76 | 2 | 1 | 107 | ND | ND | 3.9 | 0 | 0 |
| 002 | F | 67 | 0 | 1 | 37 | ND | POS | 1.6 | 19 | 0 |
| 166 | M | 63 | 0 | 2 | 75 | ND | POS | 2.5 | 0 | 0 |
| 967 | M | 77 | 0 | 4 | 63 | ND | NEG | 3.1 | 0 | 0 |
| 085 | F | 64 | 5 | 4 | 65 | ND | NEG | 1.6 | 0 | 0 |
ATM, Ataxia telangiectasia mutated; B2M, b-2-microglobulin level (mg/L); F, female; IgVH, immunoglobulin variable region heavy chain; IHC, immunohistochemistry; M, male; MUT, mutated; ND, not determined; NEG, negative; POS, positive; Pt, patient; Pr Rx, prior treatments; UNMUT, unmutated; WBC, white blood cell count (K/mL of blood); ZAP-70, Zeta-chain-associated protein kinase-70.
Percentage of positive cell with cytogenetic abnormality for the corresponding locus.
Cell culture, treatment, lentiviral infection
Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Hypaque density centrifugation (Atlanta Biologicals, Norcross, GA). PBMCs were cultured at 1 × 107/ml in RPMI supplemented with 10% human serum. The CLL cell lines (MEC1 and MEC2) and the mouse embryonic fibroblasts (MEFs) were cultured in RPMI 1640 supplemented with 20% fetal bovine serum and DMEM supplemented with 10% fetal bovine serum, respectively. Cells were left untreated or were treated with either the vehicle (DMSO) or the indicated concentrations of carfilzomib for the indicated times. HEK293 supernatants containing lentiviral particles were obtained and used to infect the MEC1 cell line as previously described (28). Carfilzomib-resistant MEC1 was generated by exposing the cells to gradually increasing nontoxic concentrations of carfilzomib (0.5 nM to 13 nM) for a period of 4 months. Selection was stopped for 10 days before experimental treatment.
Immunoblotting
Cell lysates preparation, immunoprecipitation assays, pull-down assays, and immunoblot analyses were performed as previously described (28).
Cell viability assay
Cells double positive for annexin V/PI were identified as previously described (29). Cell viability was also evaluated with use of the CellTiter-Glo Luminescent Cell Viability assay (Promega Corporation, Madison, WI) according to the manufacturer’s procedure.
Data analysis
Where shown, values are expressed as mean ± standard deviations. To evaluate changes between experimental groups, an unpaired two-tailed Student t test was used. P values of <0.05 were considered statistically significant.
RESULTS
Carfilzomib induces a heterogeneous cytotoxic response in CLL patient samples
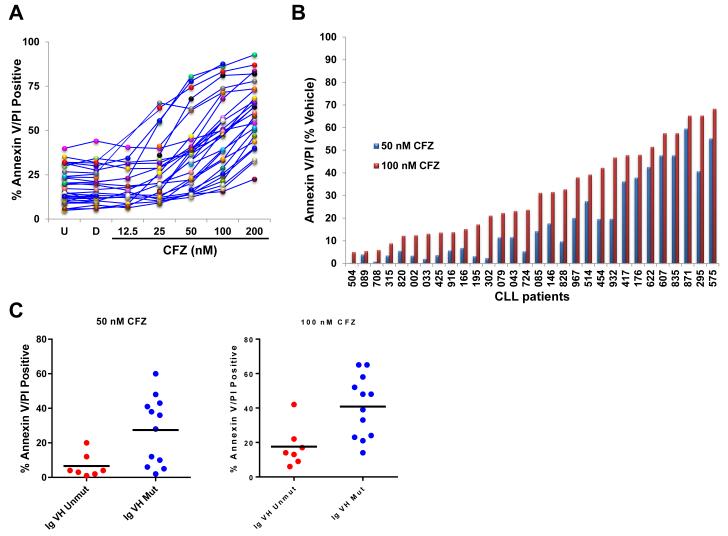
The cytotoxic effect of carfilzomib was evaluated in PBMCs isolated from 30 CLL patients (Table 1). Apoptosis assessed by annexin V/PI double positivity showed a concentration-dependent response effect after carfilzomib treatment for 16 h, with cytotoxic response variability between patient samples (Figure 1A). For instance, the cytotoxic median response was 11.6% (range: 0.3%–60%) and 27.5% (range: 5.1%–68.4%) with 50 and 100 nM carfilzomib (Figure 1B), respectively. Next, we investigated correlations between clinical and prognostic markers and the cytotoxic effect of carfilzomib. A strong association between IgVH unmutated status (unfavorable biological marker) and lower cytotoxic effect of carfilzomib was observed at both 50 nM (p=0.0157) and 100 nM (p=0.0070) (Figure 1C). Similar associations could not be made with other prognostic factors (data not shown).
Figure 1. Carfilzomib cytotoxic profile in 30 CLL patient samples.
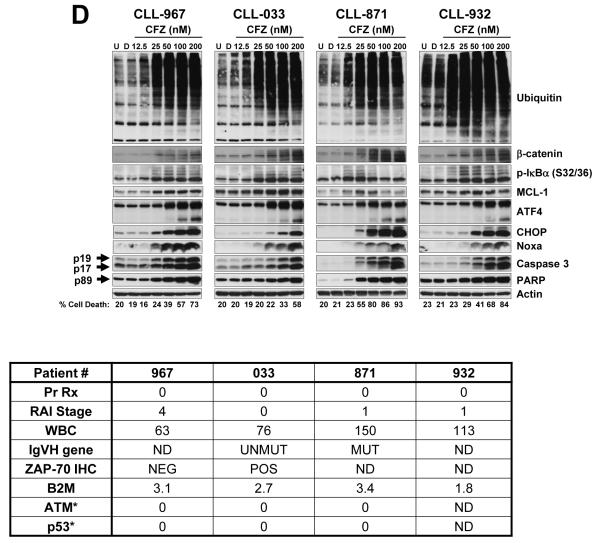
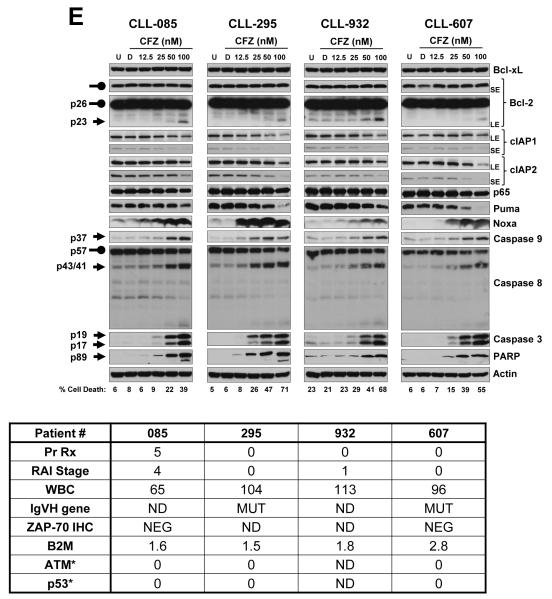
(A) Evaluation of carfilzomib apoptotic response in CLL. PBMCs of 30 patients with CLL were subjected to a concentration-dependent response of carfilzomib or were treated with vehicle (DMSO) or were left untreated for 16 h. Cell death was evaluated by annexin V/PI double positivity. U, untreated; D, DMSO; CFZ, carfilzomib. (B) Relative cytotoxic effect of carfilzomib at 50 nM and 100 nM. The percentage of cell death recorded in DMSO-treated cells was subtracted from the percentage of cell death recorded at 50 nM and 100 nM of carfilzomib, respectively. Results are shown with histogram graphs. (C) Correlation between IgVH unmutated or mutated status and cytotoxic response to carfilzomib in CLL patient samples. CLL patient samples with umutnated IgVH status (IgVH Unmut) (n=7; red dots) were less sensitive than CLL patient samples with mutated IgVH status (IgVH Mut) (n=12; blue dots) to the cytotoxic effect of carfilzomib at 50 nM (p=0.0157) and 100 nM (p=0.0070), respectively. Cytotoxic response was evaluated by annexin V/PI double positivity. Graph prism software was used to evaluate the mean (horizontal lines) and p values. (D and E) Intracellular molecular changes in CLL patient samples in response to carfilzomib treatment. PBMCs from 7 CLL patient samples, representing a wide range of cytotoxic profiles (median cytotoxicity at 100 nM: 63% (range 33%–86%), were isolated and either left untreated or treated with the indicated concentration of carfilzomib or with vehicle for 16 h. Cell lysates for each condition were processed for immunoblot analysis with the indicated antibodies. Cell death evaluated by annexin V/PI double positivity is indicated beneath the immunoblot panels. For each sample, patients’ characteristics are shown below the western blot. Arrow and rounded arrow indicate cleaved and full-length of the corresponding protein, respectively. U, untreated; D, DMSO; CFZ, carfilzomib; LE, long exposure; SE, short exposure; Pr Rx, prior treatments; WBC, white blood cell count (K/mL of blood); IgVH, immunoglobulin variable region heavy chain; ZAP-70, zeta-chain-associated protein kinase-70; IHC, immunohistochemistry; B2M, β-2-microglobulin level (mg/mL); ATM, Ataxia telangiectasia mutated; *, Percentage of positive cell with cytogenetic abnormality for the corresponding locus.
Expression of proteins affected by carfilzomib in CLL patient samples
Due to its inhibition of the proteasome, carfilzomib treatment resulted in a concentration-dependent accumulation of polyubiquitinated proteins and the stabilization of short-lived proteins (e.g., β-catenin and p-IκBα) (30, 31) (Figure 1D). We next evaluated carfilzomib treatment effect on levels of pro and antiapoptotic proteins in CLL samples (n=7) representing a wide range of cytotoxic profile. Bcl-xL, Bcl-2, cIAP1, cIAP2, p65, and Puma protein expression levels were not affected by carfilzomib treatment (Figure 1E). At higher concentrations of carfilzomib, however, some of these proteins showed decreased expression, likely due to their cleavage by caspases; for example, the appearance of a 23-kDa cleaved form of Bcl-2, previously shown to be generated by caspase activation and blocked by a pan caspase inhibitor (32), was observed (Figure 1E). In response to carfilzomib treatment, MCL-1 (Figure 1D) and Noxa (Figure 1D-E) protein levels increased in a concentration-dependent manner, with the exception of that in the CLL-871 sample that was highly sensitive to carfilzomib at higher concentrations, in which decrease of MCL-1 protein was likely due to its cleavage by caspases (33).
Carfilzomib induces an endoplasmic reticulum stress response in CLL patient samples
Carfilzomib treatment activated the endoplasmic reticulum (ER) stress pathway, as indicated by increased protein expression of two transcription factors, activator of transcription 4 (ATF4) and CHOP (34) (Figure 1D). Furthermore, both the intrinsic and extrinsic apoptosis pathways were activated by carfilzomib in a concentration-dependent manner as determined by the accumulation of the active forms of caspase 9 and caspase 8, respectively (Figure 1E). Activation of the initiator caspases resulted in activation of the downstream executioner caspase 3 and accumulation of cleaved PARP and cleaved Bcl-2 (Figure 1E-D).
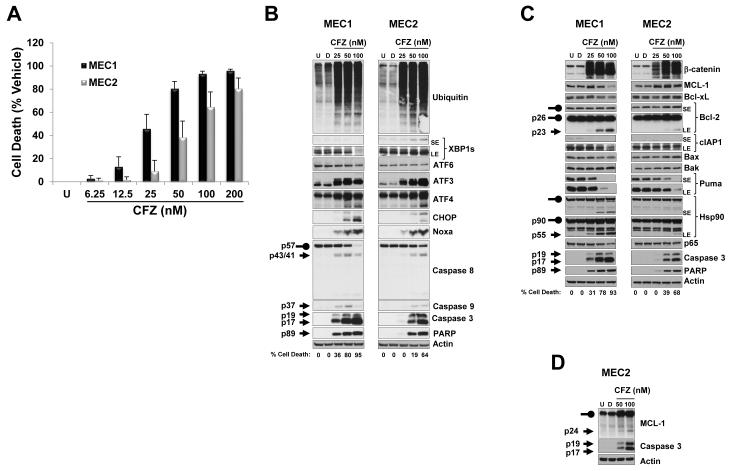
Carfilzomib induces a similar protein signature in MEC1 and MEC2 CLL cell lines
To dissect the molecular mechanism that triggers the cytotoxic response of carfilzomib, we chose to use the MEC1 and MEC2 CLL cell lines (35). Carfilzomib induced a similar cytotoxic and molecular response in these cell lines. Carfilzomib treatment for 24 h was cytotoxic to MEC cell lines in a concentration-dependent manner (Figure 2A). Molecular responses such as accumulation of polyubiquitinated proteins (Figure 2B) and stabilization of the short-lived protein β-catenin (Figure 2C) were comparable to those observed in primary CLL lymphocytes. Changes in pro and antiapoptotic protein expression indicated a similar response to those in the CLL patient samples (Figure 2B-C). The appearance of a 23-KDa, 55-kDa and 24-KDa cleaved form of Bcl-2, Hsp90 and MCL-1, respectively, previously shown to be generated by caspase activation and blocked by a pan caspase inhibitor (32, 33, 36), were observed (Figure 2C-D). Moreover, carfilzomib treatment did not affect the expression of the proapoptotic proteins Bax and Bak (Figure 2C).
Figure 2. Evaluation of the cytotoxic and molecular effects of carfilzomib in the MEC1 and MEC2 CLL cell lines.
(A) MEC1 and MEC2 were sensitive to carfilzomib-induced cell death. MEC1 and MEC2 were left untreated or were treated with vehicle or the indicated concentrations of carfilzomib for 24 h; cell survival was then evaluated by using the CellTiter-Glo Luminescent Cell Viability assay. Cytotoxic effect of carfilzomib is presented as percentage of cell death from vehicle (i.e., the percentage of cell death from DMSO treatment was subtracted from all samples). (B - D) MEC1 and MEC2 showed the same molecular signature as CLL patient samples in response to carfilzomib. Cell lysates from MEC1 and MEC2 untreated or treated with vehicle or with the indicated concentrations of carfilzomib for 24 h were processed for immunoblotting analysis with the indicated antibodies. Arrow and rounded arrow indicate cleaved and full-length of the corresponding protein, respectively. U, untreated; D, DMSO; CFZ, carfilzomib; LE, long exposure; SE, short exposure.
The unfolded protein response (UPR) is triggered when the ER function is disturbed (34) and consists of three signaling pathways controlled by transcription factors ATF4, ATF6, and X-box binding protein 1 (XBP1s) (34). ATF6 protein expression level did not change after carfilzomib treatment (Figure 2B), and XBP1s protein was not affected in MEC1, but showed a slight increase in MEC2 (Figure 2B). Similar to what we observed in CLL patient samples, carfilzomib treatment triggered a comparable ER stress response, as noted by the increased protein level of ATF4 and CHOP (Figure 2B). ATF3 protein, which is also regulated by the ATF4 UPR arm (37), was also increased by carfilzomib treatment (Figure 2B). These results suggest that ATF4 and CHOP are the prevalent UPR transcription factors induced after carfilzomib treatment in CLL cells. Finally, similar to findings in primary CLL lymphocytes, carfilzomib treatment resulted in activation of the intrinsic and extrinsic apoptotic pathways in both cell lines (Figure 2B-C).
Carfilzomib induces stable and polyubiquitinated forms of MCL-1 and Noxa
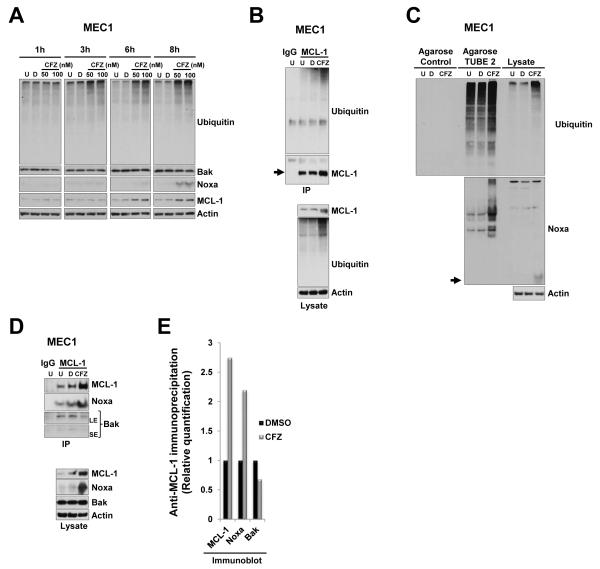
After 24 h of incubation, carfilzomib induced the accumulation of MCL-1 and Noxa protein in a concentration-dependent manner in both CLL patient samples and cell lines (Figure 1D and 2B-C). Treatment with carfilzomib (50 and 100 nM) showed that in the MEC1 line, MCL-1 and Noxa proteins visibly accumulated within 6 h and 8 h, respectively (Figure 3A), whereas the Bak protein level was not affected (Figure 3A). We next correlated accumulation of MCL-1 and Noxa proteins with their posttranslational modification by the ubiquitin-proteasome pathway. Carfilzomib treatment of MEC1 and MEC2 resulted in the accumulation of high-molecular-weight ubiquitinated forms of MCL-1 (Figure 3B and Supplementary Figure S1A), however, in a similar assay accumulation of ubiquitinated forms of Noxa was less obvious (Supplementary Figure S1B). However, a slower migrating band was present in the Noxa immunoblot, which likely represented an ubiquitinated form of Noxa (Supplementary Figure S1B, asterisk). To confirm that carfilzomib induces the accumulation of polyubiquitinated Noxa, we performed a pull-down assay with GST-TUBE 2 that specifically binds to polyubiquitinated chains (Figure 3C and Supplementary Figure S1C, upper panel); this assay revealed the presence of higher-molecular-weight forms recognized by anti-Noxa antibody, specifically in carfilzomib-treated cells (Figure 3C and Supplementary Figure S1C, lower panel). Together, these results show that carfilzomib treatment leads to the stabilization and polyubiquitination of both MCL-1 and Noxa.
Figure 3. Carfilzomib treatment resulted in accumulation of polyubiquitinated MCL-1 and Noxa.
(A) Carfilzomib treatment results in accumulation of MCL-1 and Noxa protein in a time dependent manner. MEC1 cells were incubated for the indicated time with vehicle or with the indicated concentrations of carfilzomib or left untreated. Cell lysates were subjected to immunoblotting analysis with the indicated antibodies. (B) Accumulated MCL-1 proteins were polyubiquitinated in response to carfilzomib. MEC1 cells were left untreated or were treated with vehicle or 25 nM carfilzomib for 24 h. Cell lysates were immunoprecipitated (IP) with MCL-1 antibodies. To show specificity, IgG antibodies were used in parallel in an immunoprecipitation assay with untreated cell lysate. Bound proteins were subjected to SDS-PAGE and then immunoblotted with ubiquitin and subsequently with MCL-1 after stripping of the membrane (upper panels). Cell lysates were also immunoblotted with the indicated antibodies (bottom panels). Arrow indicates full-length form of the corresponding protein at the expected molecular weight. (C) Accumulated Noxa proteins were polyubiquitinated in response to carfilzomib. Agarose beads or agarose TUBE 2 beads were mixed with cell lysates from MEC1 treated as indicated in (B). Bound proteins were subjected to SDS-PAGE and immunoblotted with ubiquitin (upper panel) or Noxa (lower panel). Arrow indicates full-length form of the corresponding protein at the expected molecular weight. Respective cell lysates were run in parallel on the same gel (right portion of the blot). (D) Accumulated MCL-1 and Noxa preferentially formed a complex after carfilzomib treatment. Cell lysates from MEC1 cells that were treated as indicated in (B) were subjected to immunoprecipitation (IP) with MCL-1 antibodies (upper panels). Bound proteins were subjected to SDS-PAGE and immunoblotted with Noxa and Bak antibodies and then with anti-MCL1 after stripping. To show specificity, IgG antibodies were used in parallel in an immunoprecipitation assay with untreated cell lysate. In parallel, respective cell lysates were also subjected to immunoblot analysis with the indicated antibodies (lower panels). U, untreated; D, DMSO; CFZ, carfilzomib; LE, long exposure; SE, short exposure. (E) MCL-1/Noxa complexes were prevalent from MCL-1/Bak complexes after carfilzomib treatment. Densitometry quantification of the immunoprecipitation shows in (D).
Carfilzomib induces preferential accumulation of an MCL-1/Noxa complex
MCL-1 exercises its antiapoptotic role by sequestering the proapoptotic Bak and does so by blocking its ability to oligomerize and form pores in the mitochondrial outer membrane (38). The BH3-only protein Noxa competes for MCL-1 binding, which results in the dissociation of the MCL-1/Bak complex, releasing the apoptotic protein Bak (39). Immunoprecipitation assays indicated that MCL-1 formed complexes with both Noxa and Bak in untreated or vehicle-treated cells (Figure 3D, U and D lanes). After carfilzomib treatment, the major anti-MCL-1 immunoprecipitation complex was between accumulated MCL-1 and Noxa proteins (Figure 3D anti-Noxa panel and 3E), and less Bak was found in the anti-MCL-1 immunoprecipitation complex (Figure 3D anti-Bak panel and 3E). These results indicated that carfilzomib favored the formation of a proapoptotic complex formed by MCL-1 and Noxa versus an antiapoptotic complex formed by MCL-1 and Bak.
BH3-only protein Noxa and proapoptotic protein Bak and Bax are major players in the carfilzomib cytotoxic response
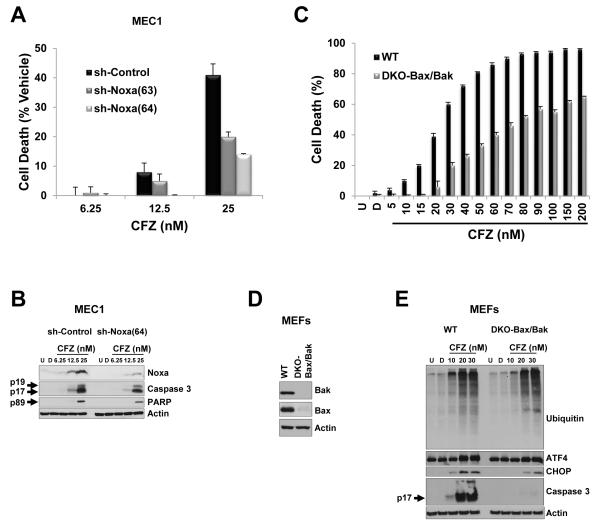
Our results indicated that carfilzomib treatment affects the dynamic between the pro-survival protein MCL-1 with the BH3-only protein Noxa and the proapoptotic executioner protein Bak. Therefore, we evaluated the role of Noxa and Bak in carfilzomib-induced cell death. We silenced Noxa protein by using a lentivirus expressing a shRNA-targeting mRNA of Noxa (sh-Noxa) in MEC1 cells. Among the five shRNAs tested, only sh-Noxa(63) and sh-Noxa(64) resulted in decreased Noxa protein at baseline (Supplementary Figure S2A), which correlated with a protective effect against the cytotoxicity induced by carfilzomib (Figure 4A and Supplementary Figure S2B). Furthermore, after a concentration-dependent response of carfilzomib, accumulation of Noxa protein was diminished in MEC1 expressing sh-Noxa(64) compared with MEC1 expressing sh-control (Figure 4B), which correlated with a delay in accumulation of the cleaved form of caspase 3 (Figure 4B) and protection against cell death (Figure 4A). Due to the fact that shRNA approaches do not lead to complete protein knockdown, we limited our incubation time to 24 h with 25 nM carfilzomib.
Figure 4. Depletion of Noxa, Bak, and Bak conferred a protective effect against carfilzomib-induced cell death.
(A and B) Knockdown of Noxa by an shRNA approach protected MEC1 against the cytotoxic effect of carfilzomib. MEC1 cells stably expressing the corresponding shRNA were incubated for 24 h with the indicated concentrations of carfilzomib. Cytotoxicity was evaluated by the CellTiter-Glo Luminescent Cell Viability assay, and cell death was represented as the percentage of cell death from vehicle (i.e., the percentage of cell death from DMSO treatment was subtracted from carfilzomib treatments) (A). Indicated cell lysates from (A) were subjected to immunoblot analysis with the indicated antibodies (B). (C–E) The cytotoxic effect of carfilzomib was attenuated in cells deficient for Bax and Bak genes. Double knockout (DKO) MEFs for Bak and Bax or wild-type (WT) MEFs were left untreated or were treated with vehicle or were subjected to a concentration-dependent response of carfilzomib for 24 h (C). The cytotoxic effect of carfilzomib was assessed as indicated in (A). Cell lysate from DKO-Bax/Bak and WT MEFs untreated or treated as indicated were subjected to immunoblot analysis with the indicated antibodies (D and E). Arrow indicates cleaved form of the corresponding protein. U, untreated; D, DMSO; CFZ, carfilzomib.
Bak and Bax are crucial executioners of the intrinsic apoptotic pathway because their activation and oligomerization result in mitochondrial outer membrane permeabilization. As a corollary, Bax and Bak double deficient cells are resistant to apoptotic stimuli that disrupt mitochondria integrity (40). To evaluate the importance of the carfilzomib-induced cytotoxic effect through the intrinsic apoptotic pathways, we used MEFs deficient in both Bax and Bak (DKO-Bax/Bak) (Figure 4D). Compared with wild-type (WT) MEFs, the inhibitory effect of carfilzomib on the proteasome or ER stress response were not affected in DKO-Bax/Bak MEFs, as shown by accumulation of polyubiquitinated proteins and an increase in ATF4 and CHOP proteins (Figure 4E). Notably, DKO-Bax/Bak MEFs, compared with WT MEFs, were less sensitive to carfilzomib-induced cell death in a concentration-dependent manner (Figure 4C), and consequently, activation of caspase 3 was significantly attenuated (Figure 4E). Taken together, these results indicated that carfilzomib induced an intrinsic apoptotic response that is dependent on the BH3 only protein Noxa and on the proapoptotic proteins Bak and Bax.
ER stress response plays a significant role in carfilzomib-induced cell death
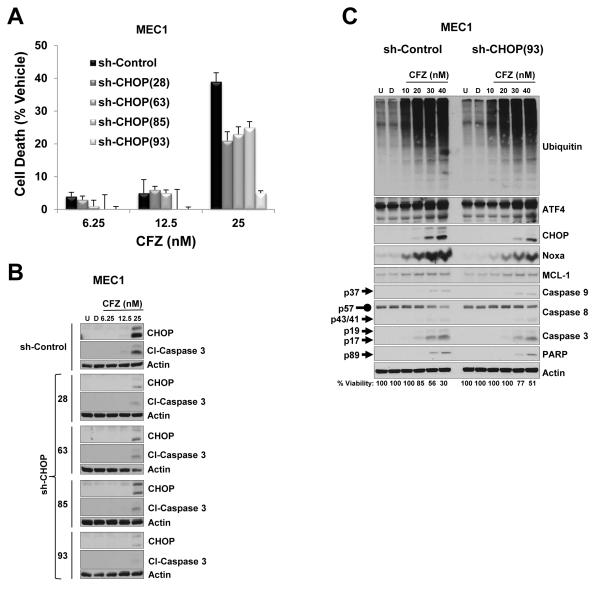
Disruption of ER function triggers a stress signaling network through the UPR in an attempt to restore protein homeostasis. However, an unresolved ER stress condition results in UPR-induced apoptosis. Besides activation of both extrinsic and intrinsic apoptotic pathways by carfilzomib treatment, we also observed increased expression of the transcription factor CHOP, a key mediator in ER stress–induced apoptosis (41, 42). To determine the role of CHOP in carfilzomib cytotoxic response, we silenced CHOP protein by using a lentivirus expressing a shRNA-targeting mRNA of CHOP (sh-CHOP) in MEC1 cells. Because CHOP protein is not present at baseline in MEC1 cells (Figure 2B), but its protein level increases after carfilzomib treatment (Figure 2B), we treated MEC1 cells that stably expressed different sh-CHOPs with varying concentrations of carfilzomib. Due to the fact that shRNA approaches do not lead to complete protein knockdown, we limited our incubation time to 24 h with 25 nM carfilzomib. Compared with MEC1 sh-control cells, a decrease of CHOP protein was clearly noted in cells from the four MEC1 cell lines expressing respective sh-CHOP; sh-CHOP(93) was the most effective (Figure 5B). Notably, downregulation of CHOP protein correlated with protection against the cytotoxic effect of carfilzomib (Figure 5A) and consequently, attenuation of activated caspase 3 was observed (Figure 5B). Knockdown of CHOP protein did not affect accumulation of polyubiquitinated proteins, ATF4, and MCL-1 after carfilzomib treatment (Figure 5C). Comparison of MEC1 sh-control and MEC1 sh-CHOP(93) cells treated with carfilzomib revealed a delay in protein accumulation of Noxa, cleaved form of caspase 3 and PARP, and cleavage of pro-caspase 8 in MEC1 sh-CHOP(93) cells (Figure 5C). In addition, the cytotoxic effect of carfilzomib was also attenuated in MEC1 sh-CHOP(93) (Figure 5C, bottom numbers). These results indicate that CHOP, a critical transcription factor in the unresolved ER stress response, plays a significant role in carfilzomib-induced cell death.
Figure 5. Cytotoxic effect of carfilzomib is attenuated by depletion of CHOP protein.
(A and B) Knockdown of CHOP by an shRNA approach protected MEC1 against the cytotoxic effect of carfilzomib. MEC1 cells expressing the corresponding shRNA were treated with vehicle or with the indicated concentrations of carfilzomib for 24 h. The CellTiter-Glo Luminescent Cell Viability assay was used to evaluate the cytotoxic effect of carfilzomib, and cell death was represented as the percentage of cell death from vehicle (i.e., the percentage of cell death from DMSO treatment was subtracted from carfilzomib treatments) (A). Cell lysates from (A) were analyzed by immunoblot assay with the indicated antibodies (B). (C) MEC1 expressing sh-control and sh-CHOP(93) were left untreated or treated with vehicle or the indicated concentrations of carfilzomib for 24 h. Cell lysates were subjected to immunoblot assay with the indicated antibodies. Cell viability, measured as indicated in (A), is indicated at the bottom of the immunoblot panels. Arrow and rounded arrow indicate cleaved and full-length of the corresponding protein, respectively. U, untreated; D, DMSO; CFZ, carfilzomib; Cl, cleaved.
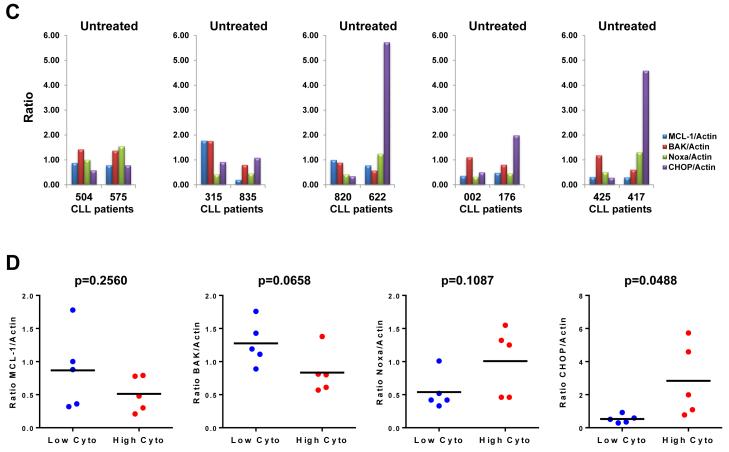
Higher levels of CHOP characterize CLL patient samples with high cytotoxic response to carfilzomib
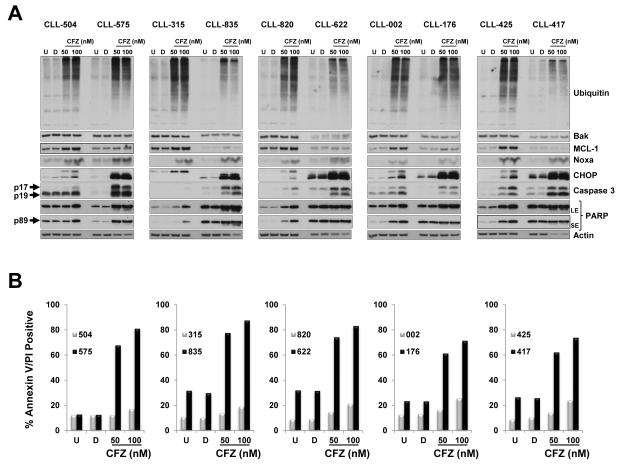
Cytotoxic profiling of carfilzomib in 30 CLL samples indicated high heterogeneity responses between patient samples (Figure 1A-B). Our preclinical investigations using shRNA knockdown approaches and genetically deficient cells indicated that the cytotoxic effect of carfilzomib is dependent on MCL-1 through the proapoptotic proteins Noxa and Bak and also involves a proapoptotic ER stress response through the transcription factor CHOP. Therefore, we decided to compare the expression level of the above proteins in five CLL patient samples with a low cytotoxicity profile (CLL-504, −315, −820, −002, −425) versus five CLL patient samples with a high cytotoxic profile (CLL-575, −835, −622, −176, −417) in response to 50 nM and 100 nM carfilzomib, respectively (Figure 6B). All patient samples responded to carfilzomib, as indicated by accumulation of polyubiquitinated proteins (Figure 6A). In all patient samples, carfilzomib treatment resulted in an apoptotic response, as indicated by accumulation of the cleaved form of caspase 3 and PARP (Figure 6A). After carfilzomib treatment, Noxa protein accumulation was noticeable with no clear differences between both sets of patient samples (Figure 6A) whereas the Bak protein level was not affected (Figure 6A). Noticeably, after carfilzomib treatment, the CHOP protein level was significantly higher in patient samples with high cytotoxic profiles than in those with low cytotoxic profiles (Figure 6A). After carfilzomib treatment, MCL-1 protein accumulation was noticeable in patients with low cytotoxic profiles but not in patients with high cytotoxic profiles, probably due to excessive cleavage of MCL-1 by caspases (Figure 6A). Furthermore, to determine whether there were differences in MCL-1, Bak, Noxa, and CHOP protein levels at baseline, and in MCL-1/Noxa and MCL-1/Bak ratios, we quantified the immunoblot band corresponding to these proteins in untreated samples for each patient (Figure 6C). Differences in MCL-1/Noxa (p=1.000) and MCL-1/Bak (p=0.9696) ratios were not significant between both sets of patients. Differences in protein expression at baseline between both sets of patients were significant only for CHOP (p=0.0488), with the set of patients with high cytotoxicity showing the highest level of CHOP protein (Figure 6D). These results identified the CHOP protein level as a biomarker to predict sensitivity to the cytotoxic effect of carfilzomib in CLL.
Figure 6. CHOP protein expression is greater in CLL patient samples with high cytotoxicity response to carfilzomib.
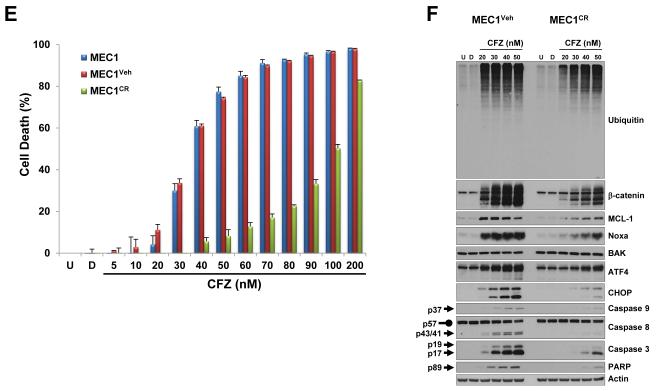
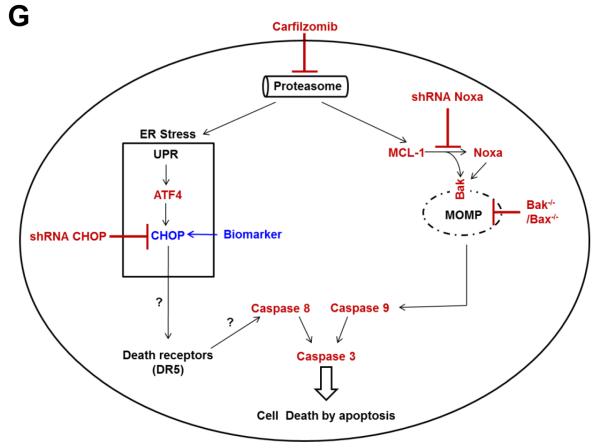
(A and B) The CHOP protein expression level was highly increased after carfilzomib treatment in CLL patient samples, representing a high cytotoxic profile. Extract from cells treated as indicated from 5 CLL patients samples with low cytotoxic response (median cytotoxicity with 100 nM carfilzomib: 21% (range: 17%–20%); grey bars) and 5 CLL patient samples with high cytotoxic response (median cytotoxicity with 100 nM carfilzomib: 81% (range: 72%–88%); black bars) (B) were analyzed by immunoblot assays with the indicated antibodies (A). The cytotoxic effect of carfilzomib for the indicated conditions was assessed by annexin V/PI double positivity (B). U, untreated; D, DMSO; CFZ, carfilzomib; LE, long exposure; SE, short exposure. (C and D) The CHOP protein expression level at baseline is higher in CLL patients with high cytotoxic profile in response to carfilzomib. Baseline (untreated) immunoblot bands for the indicated proteins for each patient sample were estimated by densitometry analysis (C). Graph prism software was used to evaluate the mean (horizontal line) and p values (D). (E and F) Carfilzomib-resistant MEC1 showed less sensitivity to carfilzomib treatment. MEC1 were passed for 4 months with no selection (MEC1) or with stepwise increasing concentrations of carfilzomib (MEC1CR) or the corresponding amount of vehicle (MEC1Veh). Selection was removed for 10 days and then the indicated cell lines were treated as indicated for 24 h. Cell viability was determined using the CellTiter-Glo Luminescent Cell Viability assay (E). Corresponding cell pellets were subjected to immunoblot assays (F). Arrow and rounded arrow indicate cleaved and full-length of the corresponding protein, respectively. (G) A wiring diagram describing the pathways involved in carfilzomib-induced cell death in CLL. Carfilzomib inhibitory effect on the proteasome induced a proapoptotic response involving Noxa, MCL-1, Bax and Bak to engage the intrinsic apoptotic pathways as well as an unresolved unfolded protein response (UPR) involving the transcription factors ATF4 and CHOP. CHOP, a key mediator in ER stress-induced apoptosis, may regulate DR5 expression to activate the extrinsic apoptotic pathway and was identified as a possible prognostic marker to predict cytotoxic efficacy of carfilzomib in CLL. U, untreated; D, DMSO; CFZ, carfilzomib; ER, reticulum endoplasmic; UPR, unfolded protein response; MOMP, mitochondrial outer membrane permeabilization.
Carfilzomib signature response is attenuated in carfilzomib-resistant MEC1
We next generated carfilzomib-resistant MEC1 to investigate the outcome on Noxa and CHOP protein expression after carfilzomib treatment. Carfilzomib-resistant MEC1 (MEC1CR) was less sensitive to carfilzomib cytotoxic effect than MEC1 subjected to no treatment (MEC1) or DMSO (MEC1Veh) (Figure 6E). As compared to MEC1Veh, higher concentrations of carfilzomib were required in MEC1CR to induce accumulation of polyubiquitinated protein, β-catenin, MCL-1, Noxa, ATF4, CHOP and activation of the intrinsic and extrinsic apoptotic pathways (Figure 6F). These results indicated that carfilzomib-induced accumulation of Noxa and CHOP proteins was altered in carfilzomib-resistant cells. Therefore, this model strongly indicates that less sensitivity to carfilzomib cytotoxicity is correlated with less Noxa protein accumulation and weaker ER stress responses.
Taken together, our results indicate that the extent of Noxa protein buildup and the strength of the ER stress response appear to be determinants for carfilzomib-mediated cytotoxicity response. Based on the results presented in this report, we propose a signaling model to summarize the different pathways involved in carfilzomib-induced cell death in CLL (Figure 6G).
DISCUSSION
CLL is the most common leukemia of the Western world and primarily affects the elderly population, who are often not healthy enough to undergo chemotherapy. Recently, we showed that carfilzomib, a selective and irreversible proteasome inhibitor, is a potent cytotoxic agent in CLL samples isolated from patients treated with ibrutinib, a well-tolerated, orally bioavailable Bruton’s tyrosine kinase inhibitor (10). While our present manuscript was under preparation a phase I trial conducted in a group of 19 patients with relapsed/refractory CLL showed modest activity of carfilzomib (43). In this study, the patient population was high risk with very unfavorable clinical and prognostic factors (i.e., 84% had a Rai stage between 3 and 4, 85% were IgVH unmutated, 47% had del(17p), 21% had del(11q), 68% were refractory to fludarabine and 70% had complex karyotype), which probably contributed to the poor response observed with carfilzomib. For instance, a recent report indicated that complex karyotype and fludarabine-refractory disease showed poor outcome for CLL patients treated with ibrutinib-based regimens (i.e. shorter event free survival and overall survival) (44). Nonetheless, this clinical trial opens up a question of what are the determinants of cytotoxicity for carfilzomib in CLL.
From the established prognostic markers in CLL (45-47), we report for the first time a correlation between IgVH unmutated status (unfavorable biologic marker) and lower sensitivity to carfilzomib’s cytotoxic effect in CLL patient samples. Correlation between ZAP-70 positivity (unfavorable biologic marker) and the carfilzomib cytotoxic effect was not significant at 50 nM (p=0.2028) or 100 nM (p=0.1417). A recent report showed that the cytotoxic effect of carfilzomib was irrespective of del (17p) (25), which is associated with loss of p53 and poor outcome with chemotherapy. However, our report could not illustrate a correlation between the cytotoxic effect of carfilzomib and del (11q), which is associated with loss of ATM and progressive disease, or del (17p) because only 13% and 10% of the patients, respectively, presented these unfavorable genetic abnormalities. Furthermore, the authors of the phase I trial with carfilzomib did not investigate the cytotoxic effect of carfilzomib ex vivo at baseline to assess any correlation between the carfilzomib-induced cytotoxic effects ex vivo versus in vivo (43). This is particularly relevant since here we used 30 CLL patient samples and showed high interpatient variability in response to the cytotoxic effect of carfilzomib. Notably, using carfilzomib-sensitive and carfilzomib-resistant CLL samples, we identified the protein level of CHOP as a biomarker that could predict sensitivity to carfilzomib in CLL. Nevertheless, this phase I trial showed that carfilzomib was well tolerated, with no concentration-limiting toxicity reported up to an amount of 56 mg/m2 and indicated that carfilzomib could be used in combination therapy with other agents. For instance, we previously demonstrated that carfilzomib caused potent cytotoxicity in CLL cells isolated from peripheral blood of patients undergoing ibrutinib therapy (10).
In 2012, carfilzomib was approved by the FDA for treatment of multiple myeloma (MM) (4, 48), and several preclinical studies have focused on its mode of action in this B-cell neoplasm. At the apex of carfilzomib-induced apoptotic sequelae is inhibition of chymotrypsin-like activity (49). However, investigation of downstream events of this action has suggested changes in several pathways leading to cell demise. For instance, activation of JNK (50), ER stress response (51), and oxidative stress (52) have been shown to be induced by carfilzomib preceding cell death in MM. Carfilzomib also improves MM-induced tumor burden on bone homeostasis by targeting osteoblasts and osteoclasts, possibly through β-catenin and IκBα protein stabilization, respectively (53-55). While our manuscript was under review, Narita et al showed a correlation between shorter progression-free survival and lower mRNA levels of ATF3 and ATF4, key players involved in ER stress response, at baseline and during therapy with bortezomib and dexamethasone in patients with refractory/relapsed multiple myeloma (56). However, similar studies need to be conducted to establish any correlation between ER stress response and clinical outcome during carfilzomib therapy.
Noxa protein accumulation (but not Noxa mRNA) after carfilzomib treatment in CLL patient samples has been reported previously (10, 25); however, the biological significance of this observation has not been investigated. In our study, carfilzomib treatment resulted in accumulation of both the proapoptotic BH3-only protein Noxa and the antiapoptotic protein MCL-1. Accumulated Noxa and MCL-1 were polyubiquitinated and preferentially formed a complex, causing a shift in the balance between the proapoptotic executioner Bak and MCL-1. Depletion of Noxa, Bak, and Bax proteins provided protection against carfilzomib’s cytotoxic effect. Notably, in contrast to this study and to our previous report (10), Gupta et al. (25) observed no changes in markers associated with activation of the ER stress response. Nonetheless, we showed that carfilzomib preferentially engaged the ATF4 branch of the UPR. The unresolved ER stress response resulted in the accumulation of the transcription factor CHOP, and its depletion had a protective effect against carfilzomib-induced cell death. Taken together, our results confirm the importance of the ER stress pathway and specifically the induction of proapoptotic proteins CHOP and Noxa in the cytotoxic activity of carfilzomib.
We were the first to report that carfilzomib treatment in CLL patient samples resulted in the activation of both the intrinsic and extrinsic apoptotic pathways (10), which we confirmed in the present study. Bcl-2 family members play an important role in the engagement of the intrinsic apoptotic pathways (57). Particularly, equilibrium between proapoptotic and antiapoptotic members is crucial to maintain the integrity of the outer membrane of the mitochondria and prevent the release of cytochrome c, a key component of the apoptosome complex responsible for the activation of the initiator caspase 9 (58). Our study indicated that carfilzomib treatment resulted in upregulation of a protein complex between the proapoptotic BH3-only protein Noxa and the antiapoptotic protein MCL-1, which probably contributed to the unleashing of Bak. Consequently, depletion of Noxa, or depletion of Bax and Bak, the two executioners of mitochondrial outer membrane permeabilization, resulted in a protective effect against the carfilzomib cytotoxic effect. In this study, we showed interplay between Noxa, MCL-1, and Bak; however, because we used cells deficient in both Bak and Bax, we could not determine whether Bak is the preferred apoptotic executioner. Furthermore, higher cytotoxic concentrations of carfilzomib were correlated with a decreased MCL-1 protein level, probably due to its cleavage by caspases that could also contribute to the release of Bak. Furthermore, while our manuscript was under submission, the group of Emily Cheng used biochemical assays as well as quadruple knockout cells deficient in Puma/Bim/Bid/Noxa to show that Noxa like Puma, Bim and tBid can directly activate Bak and Bax (59).
Carfilzomib treatment also resulted in activation of the extrinsic pathway that is activated by engagement of cell-surface death receptors by their respective ligands (60). Death receptor activation results in the formation of the death-inducing signaling complex (DISC), where the apical initiator pro-caspase 8 is recruited, activated, and triggers the apoptotic signal. Consequently, caspase 8 activation by carfilzomib indicates activation of death receptor signaling pathways. Since Death Receptor 5 (DR5) transcription has been shown to be regulated by CHOP (61, 62), we hypothesized that DR5 may be responsible for carfilzomib-induced caspase 8 activation. In addition, carfilzomib-induced cell surface expression of DR5 was recently reported in solid tumors (63). Moreover, implication of the extrinsic pathway is also suggested by the fact that deficiency in Bax and Bak proteins did not confer complete protection against carfilzomib-induced cell death.
In conclusion, by using primary CLL lymphocytes as well as cell lines, we showed that the cytotoxic effect of carfilzomib induced a proapoptotic response that involved the Bcl-2 family members Noxa, MCL-1, Bax, and Bak as well as an unresolved ER stress response that resulted in upregulation of the proapoptotic transcription factor CHOP. Accordingly, carfilzomib-induced cytotoxic, apoptotic and ER stress responses were significantly affected in carfilzomib resistant cells. Importantly, we identified the CHOP protein level as an important determinant in the cytotoxic efficacy of carfilzomib in CLL.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Carfilzomib, a second-generation proteasome inhibitor, is highly active in B-cell malignancies such as mantle cell lymphoma and multiple myeloma. In contrast, carfilzomib shows cytotoxicity heterogeneity in CLL. Because the mechanism of carfilzomib’s action has not been investigated in detail, the cytotoxicity heterogeneity cannot be explained. Here we show that carfilzomib has a wide range of cytotoxicity in CLL cells. Notably, we identified CHOP, a critical transcription factor in the unresolved endoplasmic reticulum (ER) stress response as a biomarker that could predict sensitivity to carfilzomib in CLL. At a molecular level, we showed that carfilzomib induced a proapoptotic response involving Noxa, MCL-1, Bax, and Bak and both the intrinsic and extrinsic caspase pathways as well as an unresolved ER stress response that resulted in upregulation of the proapoptotic transcription factor CHOP. Accordingly, carfilzomib-induced cytotoxic, apoptotic and ER stress responses were significantly affected in a carfilzomib resistant cell line.
ACKNOWLEDGMENTS
The authors thankfully acknowledge Dr. Bryant G. Darnay and Tamara K. Locke for critical comments and reading of the manuscript, Ben Hayes and Mark Nelson for coordinating CLL sample distribution and Xioyan Shao for maintenance of the CFZ resistant MEC1 lines. V.G., and W.G.W. are members of the CLL Research Consortium.
Financial Support: This work was supported in part by grant CLL P01 CA81534 from the National Cancer Institute, Department of Health and Human Services, and The University of Texas MD Anderson Moon Shot Program.
Footnotes
Conflict of Interest Disclosure: The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
B.L. designed and performed the experiments presented in this study, analyzed data, and wrote the manuscript. M.J.K. and W.G.W. provided clinical and patient-related information. V.G. obtained funding, participated in designing experiments, and reviewed the manuscript.
REFERENCES
- 1.Nabhan C, Rosen ST. Chronic lymphocytic leukemia: a clinical review. JAMA. 2014;312:2265–76. doi: 10.1001/jama.2014.14553. [DOI] [PubMed] [Google Scholar]
- 2.Hallek M. Chronic lymphocytic leukemia: 2015 Update on diagnosis, risk stratification, and treatment. Am J Hematol. 2015;90:446–60. doi: 10.1002/ajh.23979. [DOI] [PubMed] [Google Scholar]
- 3.Jain N, O'Brien S. Initial treatment of CLL: integrating biology and functional status. Blood. 2015;126:463–70. doi: 10.1182/blood-2015-04-585067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kortuem KM, Stewart AK. Carfilzomib. Blood. 2013;121:893–7. doi: 10.1182/blood-2012-10-459883. [DOI] [PubMed] [Google Scholar]
- 5.Pan Z, Scheerens H, Li SJ, Schultz BE, Sprengeler PA, Burrill LC, et al. Discovery of selective irreversible inhibitors for Bruton's tyrosine kinase. ChemMedChem. 2007;2:58–61. doi: 10.1002/cmdc.200600221. [DOI] [PubMed] [Google Scholar]
- 6.Honigberg LA, Smith AM, Sirisawad M, Verner E, Loury D, Chang B, et al. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc Natl Acad Sci U S A. 2010;107:13075–80. doi: 10.1073/pnas.1004594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrd JC, Furman RR, Coutre SE, Flinn IW, Burger JA, Blum KA, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369:32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woyach JA, Smucker K, Smith LL, Lozanski A, Zhong Y, Ruppert AS, et al. Prolonged lymphocytosis during ibrutinib therapy is associated with distinct molecular characteristics and does not indicate a suboptimal response to therapy. Blood. 2014;123:1810–7. doi: 10.1182/blood-2013-09-527853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maddocks KJ, Ruppert AS, Lozanski G, Heerema NA, Zhao W, Abruzzo L, et al. Etiology of Ibrutinib therapy discontinuation and outcomes in patients with chronic lymphocytic leukemia. JAMA Oncol. 2015;1:80–7. doi: 10.1001/jamaoncol.2014.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamothe B, Cervantes-Gomez F, Sivina M, Wierda WG, Keating MJ, Gandhi V. Proteasome inhibitor carfilzomib complements ibrutinib's action in chronic lymphocytic leukemia. Blood. 2015;125:407–10. doi: 10.1182/blood-2014-07-585364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crawford LJ, Irvine AE. Targeting the ubiquitin proteasome system in haematological malignancies. Blood Rev. 2013;27:297–304. doi: 10.1016/j.blre.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kish-Trier E, Hill CP. Structural biology of the proteasome. Annu Rev Biophys. 2013;42:29–49. doi: 10.1146/annurev-biophys-083012-130417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demo SD, Kirk CJ, Aujay MA, Buchholz TJ, Dajee M, Ho MN, et al. Antitumor activity of PR-171, a novel irreversible inhibitor of the proteasome. Cancer Res. 2007;67:6383–91. doi: 10.1158/0008-5472.CAN-06-4086. [DOI] [PubMed] [Google Scholar]
- 15.Harshbarger W, Miller C, Diedrich C, Sacchettini J. Crystal structure of the human 20S proteasome in complex with carfilzomib. Structure. 2015;23:418–24. doi: 10.1016/j.str.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 16.Arastu-Kapur S, Anderl JL, Kraus M, Parlati F, Shenk KD, Lee SJ, et al. Nonproteasomal targets of the proteasome inhibitors bortezomib and carfilzomib: a link to clinical adverse events. Clin Cancer Res. 2011;17:2734–43. doi: 10.1158/1078-0432.CCR-10-1950. [DOI] [PubMed] [Google Scholar]
- 17.Teicher BA, Tomaszewski JE. Proteasome inhibitors. Biochem Pharmacol. 2015;96:1–9. doi: 10.1016/j.bcp.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 18.Hamilton AL, Eder JP, Pavlick AC, Clark JW, Liebes L, Garcia-Carbonero R, et al. Proteasome inhibition with bortezomib (PS-341): a phase I study with pharmacodynamic end points using a day 1 and day 4 schedule in a 14-day cycle. J Clin Oncol. 2005;23:6107–16. doi: 10.1200/JCO.2005.01.136. [DOI] [PubMed] [Google Scholar]
- 19.O'Connor OA, Stewart AK, Vallone M, Molineaux CJ, Kunkel LA, Gerecitano JF, et al. A phase 1 dose escalation study of the safety and pharmacokinetics of the novel proteasome inhibitor carfilzomib (PR-171) in patients with hematologic malignancies. Clin Cancer Res. 2009;15:7085–91. doi: 10.1158/1078-0432.CCR-09-0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vij R, Siegel DS, Jagannath S, Jakubowiak AJ, Stewart AK, McDonagh K, et al. An open-label, single-arm, phase 2 study of single-agent carfilzomib in patients with relapsed and/or refractory multiple myeloma who have been previously treated with bortezomib. Br J Haematol. 2012;158:739–48. doi: 10.1111/j.1365-2141.2012.09232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn DJ, Hunsucker SA, Chen Q, Voorhees PM, Orlowski M, Orlowski RZ. Targeted inhibition of the immunoproteasome is a potent strategy against models of multiple myeloma that overcomes resistance to conventional drugs and nonspecific proteasome inhibitors. Blood. 2009;113:4667–76. doi: 10.1182/blood-2008-07-171637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelley TW, Alkan S, Srkalovic G, Hsi ED. Treatment of human chronic lymphocytic leukemia cells with the proteasome inhibitor bortezomib promotes apoptosis. Leuk Res. 2004;28:845–50. doi: 10.1016/j.leukres.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 23.Pahler JC, Ruiz S, Niemer I, Calvert LR, Andreeff M, Keating M, et al. Effects of the proteasome inhibitor, bortezomib, on apoptosis in isolated lymphocytes obtained from patients with chronic lymphocytic leukemia. Clin Cancer Res. 2003;9:4570–7. [PubMed] [Google Scholar]
- 24.Faderl S, Rai K, Gribben J, Byrd JC, Flinn IW, O'Brien S, et al. Phase II study of single-agent bortezomib for the treatment of patients with fludarabine-refractory B-cell chronic lymphocytic leukemia. Cancer. 2006;107:916–24. doi: 10.1002/cncr.22097. [DOI] [PubMed] [Google Scholar]
- 25.Gupta SV, Hertlein E, Lu Y, Sass EJ, Lapalombella R, Chen TL, et al. The proteasome inhibitor carfilzomib functions independently of p53 to induce cytotoxicity and an atypical NF-kappaB response in chronic lymphocytic leukemia cells. Clin Cancer Res. 2013;19:2406–19. doi: 10.1158/1078-0432.CCR-12-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia L, Liu FT. Why bortezomib cannot go with 'green'? Cancer Biol Med. 2013;10:206–13. doi: 10.7497/j.issn.2095-3941.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu FT, Agrawal SG, Movasaghi Z, Wyatt PB, Rehman IU, Gribben JG, et al. Dietary flavonoids inhibit the anticancer effects of the proteasome inhibitor bortezomib. Blood. 2008;112:3835–46. doi: 10.1182/blood-2008-04-150227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamothe B, Besse A, Campos AD, Webster WK, Wu H, Darnay BG. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of I kappa B kinase activation. J Biol Chem. 2007;282:4102–12. doi: 10.1074/jbc.M609503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel V, Chen LS, Wierda WG, Balakrishnan K, Gandhi V. Impact of bone marrow stromal cells on Bcl-2 family members in chronic lymphocytic leukemia. Leuk Lymphoma. 2014;55:899–910. doi: 10.3109/10428194.2013.819573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao C, Xiao G, Hu J. Regulation of Wnt/beta-catenin signaling by posttranslational modifications. Cell Biosci. 2014;4:13–32. doi: 10.1186/2045-3701-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays. 2000;22:442–51. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 32.Fadeel B, Hassan Z, Hellstrom-Lindberg E, Henter JI, Orrenius S, Zhivotovsky B. Cleavage of Bcl-2 is an early event in chemotherapy-induced apoptosis of human myeloid leukemia cells. Leukemia. 1999;13:719–28. doi: 10.1038/sj.leu.2401411. [DOI] [PubMed] [Google Scholar]
- 33.Herrant M, Jacquel A, Marchetti S, Belhacene N, Colosetti P, Luciano F, et al. Cleavage of Mcl-1 by caspases impaired its ability to counteract Bim-induced apoptosis. Oncogene. 2004;23:7863–73. doi: 10.1038/sj.onc.1208069. [DOI] [PubMed] [Google Scholar]
- 34.Szegezdi E, Logue SE, Gorman AM, Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 2006;7:880–5. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stacchini A, Aragno M, Vallario A, Alfarano A, Circosta P, Gottardi D, et al. MEC1 and MEC2: two new cell lines derived from B-chronic lymphocytic leukaemia in prolymphocytoid transformation. Leuk Res. 1999;23:127–36. doi: 10.1016/s0145-2126(98)00154-4. [DOI] [PubMed] [Google Scholar]
- 36.Park S, Park JA, Kim YE, Song S, Kwon HJ, Lee Y. Suberoylanilide hydroxamic acid induces ROS-mediated cleavage of HSP90 in leukemia cells. Cell Stress Chaperones. 2015;20:149–57. doi: 10.1007/s12192-014-0533-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang HY, Wek SA, McGrath BC, Lu D, Hai T, Harding HP, et al. Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol Cell Biol. 2004;24:1365–77. doi: 10.1128/MCB.24.3.1365-1377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gelinas C, White E. BH3-only proteins in control: specificity regulates MCL-1 and BAK-mediated apoptosis. Genes Dev. 2005;19:1263–8. doi: 10.1101/gad.1326205. [DOI] [PubMed] [Google Scholar]
- 39.Correia C, Lee SH, Meng XW, Vincelette ND, Knorr KL, Ding H, et al. Emerging understanding of Bcl-2 biology: Implications for neoplastic progression and treatment. Biochim Biophys Acta. 2015;1853:1658–71. doi: 10.1016/j.bbamcr.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–30. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot RT, Remotti H, et al. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–95. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol. 2011;13:184–90. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Awan FT, Flynn JM, Jones JA, Andritsos LA, Maddocks KJ, Sass EJ, et al. Phase I dose escalation trial of the novel proteasome inhibitor carfilzomib in patients with relapsed chronic lymphocytic leukemia and small lymphocytic lymphoma. Leuk Lymphoma. 2015:1–7. doi: 10.3109/10428194.2015.1014368. e-pub ahead of print 11 March 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson PA, O'Brien SM, Wierda WG, Ferrajoli A, Stingo F, Smith SC, et al. Complex karyotype is a stronger predictor than del(17p) for an inferior outcome in relapsed or refractory chronic lymphocytic leukemia patients treated with ibrutinib-based regimens. Cancer. 2015;121:3612–21. doi: 10.1002/cncr.29566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–6. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 46.Hallek M, Fischer K, Fingerle-Rowson G, Fink AM, Busch R, Mayer J, et al. Addition of rituximab to fludarabine and cyclophosphamide in patients with chronic lymphocytic leukaemia: a randomised, open-label, phase 3 trial. Lancet. 2010;376:1164–74. doi: 10.1016/S0140-6736(10)61381-5. [DOI] [PubMed] [Google Scholar]
- 47.Gonzalez D, Martinez P, Wade R, Hockley S, Oscier D, Matutes E, et al. Mutational status of the TP53 gene as a predictor of response and survival in patients with chronic lymphocytic leukemia: results from the LRF CLL4 trial. J Clin Oncol. 2011;29:2223–9. doi: 10.1200/JCO.2010.32.0838. [DOI] [PubMed] [Google Scholar]
- 48.Herndon TM, Deisseroth A, Kaminskas E, Kane RC, Koti KM, Rothmann MD, et al. U.S. Food and Drug Administration approval: carfilzomib for the treatment of multiple myeloma. Clin Cancer Res. 2013;19:4559–63. doi: 10.1158/1078-0432.CCR-13-0755. [DOI] [PubMed] [Google Scholar]
- 49.Parlati F, Lee SJ, Aujay M, Suzuki E, Levitsky K, Lorens JB, et al. Carfilzomib can induce tumor cell death through selective inhibition of the chymotrypsin-like activity of the proteasome. Blood. 2009;114:3439–47. doi: 10.1182/blood-2009-05-223677. [DOI] [PubMed] [Google Scholar]
- 50.Kuhn DJ, Chen Q, Voorhees PM, Strader JS, Shenk KD, Sun CM, et al. Potent activity of carfilzomib, a novel, irreversible inhibitor of the ubiquitin-proteasome pathway, against preclinical models of multiple myeloma. Blood. 2007;110:3281–90. doi: 10.1182/blood-2007-01-065888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kikuchi S, Suzuki R, Ohguchi H, Yoshida Y, Lu D, Cottini F, et al. Class IIa HDAC inhibition enhances ER stress-mediated cell death in multiple myeloma. Leukemia. 2015;29:1918–27. doi: 10.1038/leu.2015.83. [DOI] [PubMed] [Google Scholar]
- 52.Fink EE, Mannava S, Bagati A, Bianchi-Smiraglia A, Nair JR, Moparthy K, et al. Mitochondrial thioredoxin reductase regulates major cytotoxicity pathways of proteasome inhibitors in multiple myeloma cells. Leukemia. 2015 doi: 10.1038/leu.2015.190. e-pub ahead of print 24 July 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hurchla MA, Garcia-Gomez A, Hornick MC, Ocio EM, Li A, Blanco JF, et al. The epoxyketone-based proteasome inhibitors carfilzomib and orally bioavailable oprozomib have anti-resorptive and bone-anabolic activity in addition to anti-myeloma effects. Leukemia. 2013;27:430–40. doi: 10.1038/leu.2012.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu B, Chen Y, Usmani SZ, Ye S, Qiang W, Papanikolaou X, et al. Characterization of the molecular mechanism of the bone-anabolic activity of carfilzomib in multiple myeloma. PLoS One. 2013;8:e74191. doi: 10.1371/journal.pone.0074191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eda H, Santo L, Cirstea DD, Yee AJ, Scullen TA, Nemani N, et al. A novel Bruton's tyrosine kinase inhibitor CC-292 in combination with the proteasome inhibitor carfilzomib impacts the bone microenvironment in a multiple myeloma model with resultant antimyeloma activity. Leukemia. 2014;28:1892–901. doi: 10.1038/leu.2014.69. [DOI] [PubMed] [Google Scholar]
- 56.Narita T, Ri M, Masaki A, Mori F, Ito A, Kusumoto S, et al. Lower expression of activating transcription factors 3 and 4 correlates with shorter progression-free survival in multiple myeloma patients receiving bortezomib plus dexamethasone therapy. Blood Cancer J. 2015;5:e373. doi: 10.1038/bcj.2015.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 58.Wang C, Youle RJ. The role of mitochondria in apoptosis. Annu Rev Genet. 2009;43:95–118. doi: 10.1146/annurev-genet-102108-134850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen HC, Kanai M, Inoue-Yamauchi A, Tu HC, Huang Y, Ren D, et al. An interconnected hierarchical model of cell death regulation by the BCL-2 family. Nat Cell Biol. 2015;17:1270–81. doi: 10.1038/ncb3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dickens LS, Powley IR, Hughes MA, MacFarlane M. The 'complexities' of life and death: death receptor signalling platforms. Exp Cell Res. 2012;318:1269–77. doi: 10.1016/j.yexcr.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 61.Yamaguchi H, Wang HG. CHOP is involved in endoplasmic reticulum stress-induced apoptosis by enhancing DR5 expression in human carcinoma cells. J Biol Chem. 2004;279:45495–502. doi: 10.1074/jbc.M406933200. [DOI] [PubMed] [Google Scholar]
- 62.Lu M, Lawrence DA, Marsters S, Acosta-Alvear D, Kimmig P, Mendez AS, et al. Cell death. Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science. 2014;345:98–101. doi: 10.1126/science.1254312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Han B, Yao W, Oh YT, Tong JS, Li S, Deng J, et al. The novel proteasome inhibitor carfilzomib activates and enhances extrinsic apoptosis involving stabilization of death receptor 5. Oncotarget. 2015;6:17532–42. doi: 10.18632/oncotarget.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.