Abstract
Infections with the Gram-negative coccobacillus Acinetobacter baumannii are a major threat in hospital settings. The progressing emergence of multidrug-resistant clinical strains significantly reduces the treatment options for clinicians to fight A. baumannii infections. The current lack of robust methods to genetically manipulate drug-resistant A. baumannii isolates impedes research on resistance and virulence mechanisms in clinically relevant strains. In this study, we developed a highly efficient and versatile genome-editing platform enabling the markerless modification of the genome of A. baumannii clinical and laboratory strains, regardless of their resistance profiles. We applied this method for the deletion of AdeR, a transcription factor that regulates the expression of the AdeABC efflux pump in tigecycline-resistant A. baumannii, to evaluate its function as a putative drug target. Loss of adeR reduced the MIC90 of tigecycline from 25 μg/ml in the parental strains to 3.1 μg/ml in the ΔadeR mutants, indicating its importance in the drug resistance phenotype. However, 60% of the clinical isolates remained nonsusceptible to tigecycline after adeR deletion. Evolution of artificial tigecycline resistance in two strains followed by whole-genome sequencing revealed loss-of-function mutations in trm, suggesting its role in an alternative AdeABC-independent tigecycline resistance mechanism. This finding was strengthened by the confirmation of trm disruption in the majority of the tigecycline-resistant clinical isolates. This study highlights the development and application of a powerful genome-editing platform for A. baumannii enabling future research on drug resistance and virulence pathways in clinically relevant strains.
INTRODUCTION
One of the greatest global health problems results from the limited treatment options to fight bacterial infections caused by multidrug-resistant (MDR) organisms. The group of ESKAPE organisms that is comprised of Enterobacter spp., Staphylococcus aureus/epidermidis, Klebsiella pneumonia, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterococcus faecalis/faecium is considered to cause the vast majority of, often untreatable, nosocomial infections (1). Among these ESKAPE pathogens A. baumannii is most difficult to treat due to its multiple intrinsic and acquired resistance mechanisms that resulted in the development of MDR, extensively drug resistant (XDR), or even pan-drug-resistant (PDR) phenotypes (2–5).
Bacteria have evolved multiple ways to evade antibiotic-mediated cell death, such as (i) enzymatic modification/cleavage of the antibiotic (e.g., beta-lactams), (ii) modification/protection of the antibiotic target (e.g., fluoroquinolones), or (iii) reduction of the intracellular concentration by antibiotic efflux or reduced influx (e.g., tetracyclines) (6). The expression of such defense mechanisms may require an extensive metabolic investment, often leading to a reduced fitness of these resistant bacteria in the absence of the external selection pressure (7). To overcome these ecological drawbacks, bacteria have developed diverse transcriptional regulation mechanisms that ensure specific expression of the resistance genes only in the presence of poisoning antibiotics (8). The need for precise regulation of resistance gene cluster expression makes transcriptional regulators promising drug targets as part of a strategy to overcome antibiotic resistances. Small molecules that specifically interfere with the transcriptional regulator function may switch off the resistance mechanism. The combination of such inhibitors with a potent antibiotic as an adjuvant drug may provide a new tool to fight infections caused by MDR, XDR, or even PDR pathogens by restoring the efficacy of an approved antibiotic (9, 10).
In clinical settings the use of different and/or combinations of antibiotics to treat hazardous infections caused by the Gram-negative pathogen A. baumannii leads to a tremendous selection pressure toward the development of multidrug-resistant strains. Unfortunately, we currently have only limited understanding of the innate and acquired antibiotic resistance mechanisms of A. baumannii clinical isolates. This lack of knowledge is mainly based on the unavailability of robust methods that allow the genetic manipulation of MDR, XDR, and PDR patient-derived strains (11). In most studies investigating antibiotic resistance mechanisms in A. baumannii, laboratory strains were used, such as ATCC-19606 and ATCC-17978, which do not represent the problematic clonal lineages that are currently present in hospitals (12, 13).
In this study, we developed a novel method allowing the short-term construction of markerless gene knockouts in A. baumannii clinical isolates, regardless of their resistance profiles. We applied this powerful tool in 10 diverse A. baumannii strains that were resistant to multiple antibiotics to evaluate the transcriptional regulator AdeR as a potential drug target to rejuvenate the activity of tigecycline, a drug of last resort for treatment of XDR A. baumannii infections (14). We demonstrated that AdeR-regulated AdeABC-mediated drug efflux is not the exclusive tigecycline resistance mechanism in XDR A. baumannii clinical strains, disqualifying AdeR as a direct drug target.
MATERIALS AND METHODS
Bacterial strains, MICs, MLST, and oligonucleotides.
Ten XDR A. baumannii clinical isolates from the BioVersys proprietary strain collection were used in this study. As a control the tigecycline-susceptible A. baumannii ATCC-17978 strain from the American Type Culture Collection (ATCC) was used. Multilocus sequence type (MLST) analysis was performed according to the Pasteur scheme by using specific primers (source: http://pubmlst.org/abaumannii/) (12). MICs were determined by the microdilution method in cation-adjusted Mueller-Hinton broth (CA-MHB) according to CLSI guidelines (15). Bacteria were grown using Luria-Bertani (LB) broth or agar at 37°C unless otherwise stated. All oligonucleotides used in this study are listed in Table S1 in the supplemental material and were synthesized at Microsynth AG (Balgach, Switzerland).
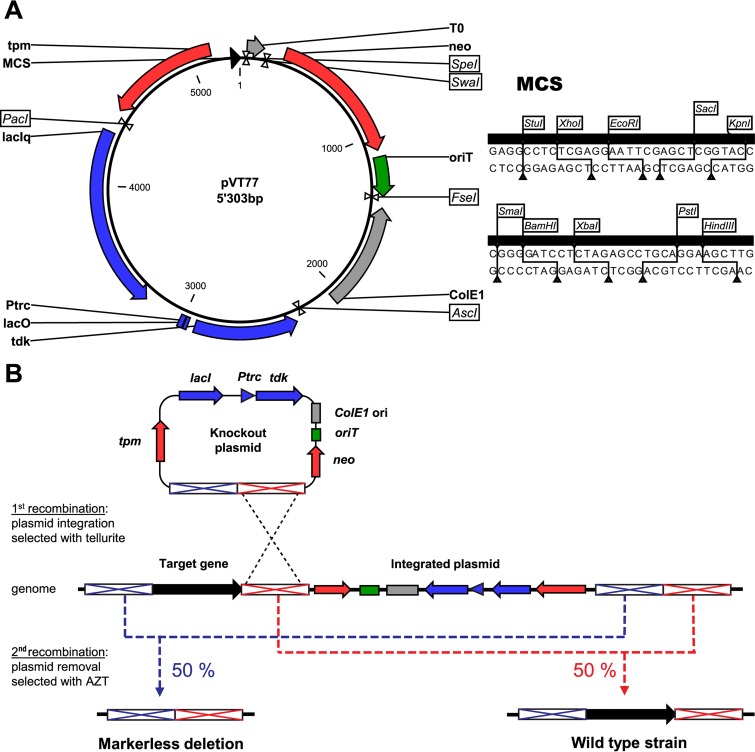
Cloning of the gene knockout vector backbone pVT77 suitable for genetic manipulation of drug-resistant A. baumannii strains.
The vector pSEVA238 (Standard European Vector Assembly platform [http://seva.cnb.csic.es/]) was used as the starting backbone for cloning of the pVT77 knockout plasmid (16). First, the pUC19 origin of replication (ColE1) was PCR amplified from the pUC19 plasmid using the primers oVT01/oVT02 and cloned into the AscI and FseI restriction sites of pSEVA238, creating plasmid pVT52. A synthetic cassette containing the thiopurine S-methyltransferase (tpm) gene of Acinetobacter baylyi (locus tag ACIAD2922) driven by the Burkholderia cenocepacia rpsL PCS12 promoter was cloned with EcoRI and PacI into pVT52 to allow antibiotic-independent selection for genomic plasmid integration by sodium tellurite (pVT59). For counterselection with 3′-azido-3′-deoxythymidine (AZT), the thymidine kinase (tdk) gene was PCR amplified from Escherichia coli DH5α genomic DNA using primers oVT113/oVT114 flanked with an upstream ribosome binding site (RBS) and cloned into pSEVA234, creating plasmid pVT66. On this plasmid tdk is placed under the repression of LacI. The lacI-tdk cassette was excised from pVT66 with PacI and AscI restriction enzymes and subsequently cloned into pVT59, resulting in plasmid pVT67. Further modifications were done to maximize the cloning possibilities in the multiple cloning site (MCS). The EcoRI restriction site present in the lacI-tdk cassette was removed by PCR mutagenesis using primers oVT141/oVT142. The present MCS was also replaced with a more complete MCS, including StuI, XhoI, EcoRI, SacI, KpnI, SmaI, BamHI, XbaI, PstI, and HindIII restriction sites, by cloning annealed oligonucleotides oVT149 and oVT150 into EcoRI and HindIII restriction sites of pVT67. The resulting plasmid was named pVT77, and it was used as a backbone for the cloning of flanking regions to knock out any specific targets in A. baumannii clinical isolates (Fig. 1A).
FIG 1.
Schematic representation of the knockout platform developed for targeted gene deletion in XDR A. baumannii. (A) Representation of the modular pVT77 cloning vector. Important features of the vector platform are shown: (i) a thiopurine S-methyltransferase (tpm) for antibiotic-free selection of clones after plasmid integration and a kanamycin resistance cassette (neo) for selection in E. coli (in red); (ii) a functional counterselection cassette (lacIq-Ptrc-lacO-tdk; shown in blue) for genomic plasmid removal in the presence of IPTG and AZT; (iii) an origin of transfer (oriT; in green) for conjugative plasmid transfer from E. coli to A. baumannii; (iv) ColE1 ori for efficient plasmid replication in E. coli (please note that this origin of replication is not active in A. baumannii, resulting in a suicide vector for this pathogen) and the transcriptional terminator T0 (in gray); and (v) a multiple cloning site (MCS; black) allowing the integration of the up- and downstream regions of the target gene. The plasmid map was generated with CLC Main Workbench, version 6.9.1. (B) Representation of the two-step allelic exchange method for targeted gene deletion using derivatives of the knockout plasmid pVT77. The knockout plasmid is site-specifically integrated next to a target gene by an initial homologous recombination event, and clones are selected with potassium tellurite. Afterwards, a second recombination event for the subsequent plasmid removal is produced by the induction of the counterselection cassette, which triggers cytotoxicity in the presence of AZT, leading to 50% knockout mutants or wild-type revertants.
Construction of adeR and trm gene deletions in A. baumannii isolates.
Scarless deletions of the adeR and trm genes were done in A. baumannii using a two-step allelic exchange method adapted from Amin and colleagues (Fig. 1B) (17). DNA fragments corresponding to 700-bp up- and downstream genomic regions of the genes to be deleted were amplified by PCR using primers oVT154/oVT106 and oVT107/oVT155 for adeR and oVT412/oVT413 and oVT414/oVT415 for trm, respectively. The up- and downstream fragments of each gene were ligated and introduced into pVT77 previously digested with XhoI/XbaI and EcoRI/XbaI for adeR and trm, respectively, using Gibson assembly. The resulting knockout plasmids were transformed in the E. coli conjugative strain MFDpir, which is auxotrophic for diaminopimelic acid (DAP) (18). The transfer of the plasmid in A. baumannii isolates was achieved by conjugation, as previously described (17). Briefly, 0.2-ml overnight cultures from the donor (MFDpir cells containing the knockout plasmid) and receiver (A. baumannii isolate) strains were mixed and washed twice with 1 ml of LB broth to remove residual antibiotics. The cells were resuspended in 50 μl of medium and transferred onto a 0.45-μm-pore-size nitrocellulose filter placed on LB agar containing 300 μM DAP. After overnight incubation at 37°C for conjugative plasmid transfer, the cells were scraped off the filter and resuspended in 0.4 ml of 0.85% (wt/vol) NaCl, and 0.1-ml aliquots were plated on LB agar plates containing 100 μg/ml sodium tellurite. After overnight selection at 37°C, clones were screened for genomic plasmid integration by PCR using primers oVT93/oVT08 and oVT423/oVT8 for adeR and trm, respectively. Clones containing up- and downstream plasmid integrations were used to inoculate 2 ml of fresh LB broth containing 1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) and cultured for 3 h at 37°C to express the heterologous thymidine kinase. Culture aliquots (0.1 ml) of a 1:10 dilution series (100 to 10−2) were plated on LB agar plates containing 200 μg/ml AZT and incubated overnight at 37°C for plasmid removal from the genome. Clones were screened for gene deletion and plasmid removal by PCR using primers oVT93/oVT94 and oVT423/oVT424 for adeR and trm, respectively. The genomic gene deletions were finally confirmed by DNA sequencing (Microsynth AG, Balgach, Switzerland).
Restoration of a functional trm gene by chromosomal single nucleotide knock-in in A. baumannii isolates.
For allelic replacement of the mutated trm, a 1.4-kb DNA fragment including the wild-type trm (lacking the A240 deletion) was PCR amplified from isolate BV186 using primers oVT416/oVT417. This fragment was cloned into pVT77 previously digested with KpnI/XbaI using Gibson assembly. The two-step allelic exchange method described above was used to restore wild-type trm, and the successful gene replacement was confirmed by sequencing using primers oVT312/oVT161.
qRT-PCR.
The expression of the efflux pump operon adeABC was evaluated by quantifying adeB expression using adeB-qRT-F/adeB-qRT-R primers. A. baumannii isolates were grown in LB broth at 37°C to mid-log phase (optical density at 600 nm [OD600] of 0.5), and total RNA was extracted using a PureLink RNA minikit (Ambion) according to the manufacturer's recommendations. Residual DNA contaminations were removed using a Turbo DNA-free kit (Ambion). Quantitative reverse transcription-PCR (qRT-PCR) was performed using a GoTaq 1-Step RT-qPCR System kit (Promega) on a StepOne Real-Time PCR LightCycler (Applied Biosystems). Extracted RNA (25 ng) was mixed with 10 μl of GoTaq MasterMix, 3 μl of 8 μM primers, 0.4 μl of GoScript RT mix, and 0.3 μl of carboxy-X-rhodamine (CXR) standard dye in a total volume of 20 μl. As a housekeeping gene, the RNA polymerase sigma factor D (rpoD) was quantified with primers rpoD-qRT-F/rpoD-qRT-R, and adeB expression was normalized to that of rpoD using the comparative ΔΔCT (where CT is threshold cycle) method. A detailed description of the DNA probes used for RNA detection is given in Table S1 in the supplemental material.
Genotyping of adeR, adeS, and trm in clinical isolates.
A genomic sequence including adeR and adeS was PCR amplified from all A. baumannii isolates using primers oVT05 and oVT134 (except for ATCC-17978, for which the oVT151 primer was used instead of oVT134). A genomic sequence including trm was PCR amplified from all A. baumannii isolates using primers oVT160 and oVT161. The PCR products were subsequently sent for sequencing (Microsynth AG, Balgach, Switzerland).
In vitro evolution of tigecycline resistance.
A. baumannii ATCC-17978 and its ΔadeR mutant (ATCC-17978ΔadeR) were subjected to serial passages to develop tigecycline resistance in vitro (see Fig. S1 in the supplemental material). In brief, LB broth liquid cultures were passaged every 48 h starting with a tigecycline concentration of 0.2 μg/ml and incubated at 37°C. For passaging, the cultures were diluted 1/100 and exposed to double the tigecycline concentration of the preceding culture until a concentration of 6.4 μg/ml tigecycline was reached. The artificially evolved tigecycline-resistant strains derived from strain A. baumannii ATCC-17978 and its ΔadeR mutant were designated ATCC-17978(TGC) and ATCC-17978ΔadeR(TGC), respectively.
Whole-genome sequencing.
Genomic DNA (gDNA) of ATCC-17978(TGC), ATCC-17978ΔadeR(TGC), and their parental strains was extracted using a GeneJET genomic DNA purification kit (Thermo Scientific) according to the manufacturer's recommendations. The four gDNA samples were multiplex sequenced using a Illumina MiSeq system (Illumina, Inc.) using a paired-end 250-bp protocol at the Quantitative Genomics Facility (Department of Biosystems Science and Engineering, ETH Zurich, Basel, Switzerland). Four to eight million high-quality reads per sample were generated and mapped against the reference genome of A. baumannii ATCC-17978 (GenBank accession number CP000521.1) using Bowtie2, achieving an average coverage above ×100 (19). Small nucleotide variations were spotted using SAMtools (20). Recombination events were screened with a series of in-house perl scripts based on (i) coverage fluctuations, (ii) the distribution of start/end positions of mapped reads, and (iii) reads that significantly mapped to at least two noncontiguous regions of the reference chromosome (i.e., regions including the newly formed recombination boundaries). In addition, de novo assembly of reads with poor mapping score values was performed with SOAPdenovo 2 and identified insertion sequences (IS) were aligned with the ISFinder insertion sequence database (https://www-is.biotoul.fr/) (21, 22).
Accession number(s).
The sequence of the pVT77 cloning vector has been deposited in the GenBank under accession number KX397287. Sequences of the adeR and adeS genes for the following isolates were deposited in GenBank under the indicated accession numbers: BV26 (KX819204), BV94 (KX819205), BV173 (KX819206), BV175 (KX819207), BV185 (KX819208), BV186 (KX819209), BV187 (KX819210), BV189 (KX819211), BV190 (KX819212), and BV191 (KX819213). Sequences of the trm gene were deposited in GenBank under the indicated accession numbers: BV26 (KX819214), BV94 (KX819215), BV173 (KX819216), BV175 (KX819217), BV185 (KX819218), BV186 (KX819219), BV187 (KX819220), BV189 (KX819221), BV190 (KX819222), and BV191 (KX819223). The reads of the whole-genome sequencing have been deposited in the GenBank Sequence Read Archive (SRA) as follows: ATCC-17978 (GenBank accession number SRR4176247), ATCC-17978ΔadeR (SRR4176248), ATCC-17978(TGC) (SRR4176249), and ATCC-17978ΔadeR(TGC) (SRR4176250).
RESULTS
Knockout technology for genetic manipulation of A. baumannii clinical isolates regardless of their antibiotic resistance profiles.
A. baumannii is among the ESKAPE pathogens most difficult to treat because it often combines a multitude of intrinsic and acquired drug resistance mechanisms (2). In order to investigate the function of putative drug targets in clinically problematic drug-resistant isolates of A. baumannii, we had to develop a novel technology platform enabling the genome modification of these MDR isolates because traditional methods using antibiotic selection cassettes cannot be applied in these pathogens. We screened a collection of clinically relevant A. baumannii isolates for their susceptibilities to various chemical agents and demonstrated that all strains were sensitive to sodium tellurite (MIC ≤ 8 μg/ml) (Table 1). Our results suggested that a tellurite resistance cassette might be a powerful tool to select for genetic manipulation of MDR, XDR, and PDR A. baumannii isolates independently of their antibiotic resistance profiles. A two-step homologous recombination approach was developed to target clinical isolates (Fig. 1B). Traditional antibiotic selection cassettes were replaced by the Acinetobacter baylyi-derived tellurite resistance marker gene tpm, encoding a thiopurine S-methyltransferase, to allow selection for chromosomal plasmid integration with sodium tellurite (23). In bacteria, expression of Tpm enzymatically detoxifies tellurite by a reduction and methylation of tellurite to produce volatile dimethyl telluride (24). The sacB gene, widely used as a counterselection cassette to initiate a second recombination event in different bacteria, was shown not to be capable of efficiently removing the integrated plasmid from the genome of A. baumannii and E. coli (17, 25). During our design process we also noticed that SacB expression is slightly toxic even in the absence of sucrose, leading to the selection of inactivating mutations in the sacB gene. As an alternative, we PCR amplified the E. coli-derived thymidine kinase gene (tdk) and cloned it under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter. The expression of thymidine kinase in the presence of the reverse transcriptase small-molecule inhibitor 3′-azido-3′-deoxythymidine (AZT) results in the incorporation of a chain-terminating thymidine analogue during DNA replication, which finally leads to cell death or removal of the tdk-expressing plasmid from the genome (26). This powerful setup selected for the second recombination event with an efficiency of >95% in A. baumannii clinical isolates. The application of our gene knockout platform enabled us to efficiently delete different target genes in A. baumannii strains of diverse origins that were antibiotic susceptible or that accumulated multiple antibiotic resistance mechanisms.
TABLE 1.
Characterization of the A. baumannii clinical isolate panel used in this study
| Strain designation | Strain isolation |
MLST | MIC (μg/ml)a |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Location | Date | GEN | MEM | CIP | TIM* | CTX | SXT* | SAM* | CST | TET | TEL | ||
| ATCC-17978 | France | 1951 | 77 | 2 | 0.5 | 1 | 16/2 | 16 | >8/152 | 4/2 | 0.5 | 2 | 4 |
| BV26 | Switzerland | 1979 | 1 | >128 | 1 | 4 | >256/2 | 16 | >8/152 | 64/32 | 0.5 | 128 | 4 |
| BV94 | USA | 2011 | 2 | >128 | 32 | 256 | >256/2 | >256 | >8/152 | 16/8 | 64 | 32 | 8 |
| BV173 | Greece | 2012 | 2 | >128 | >64 | 128 | >256/2 | >256 | >8/152 | 128/64 | 32 | >256 | 1 |
| BV175 | Turkey | 2012 | 2 | 128 | 32 | 256 | >256/2 | 256 | >8/152 | 32/16 | >256 | 256 | 0.5 |
| BV185 | Mexico | 2013 | 2 | >128 | >64 | 128 | >256/2 | >256 | >8/152 | 64/32 | >256 | 256 | 1 |
| BV186 | USA | 2013 | 2 | 16 | 64 | 256 | >256/2 | >256 | >8/152 | 32/16 | 16 | 8 | 4 |
| BV187 | USA | 2013 | 2 | 32 | 64 | 256 | >256/2 | >256 | >8/152 | 16/8 | 16 | 8 | 4 |
| BV189 | Spain | 2013 | 2 | 128 | 32 | 128 | >256/2 | 256 | >8/152 | 32/16 | 64 | 16 | 8 |
| BV190 | Greece | 2012 | 1 | >128 | 64 | 64 | >256/2 | >256 | >8/152 | 64/32 | 256 | 256 | 4 |
| BV191 | China | 2013 | 2 | >128 | >64 | 256 | >256/2 | >256 | >8/152 | 128/64 | >256 | >256 | 2 |
| ATCC-25922b | 1 | <0.06 | <0.25 | 8/2 | <0.25 | 0.125/2.34 | 4/2 | 1 | 2 | 4 | |||
MLST, multilocus sequence type; CIP, ciprofloxacin; CST, colistin; CTX, cefotaxime; GEN, gentamicin; MEM, meropenem; SAM, ampicillin-sulbactam; SXT, trimethoprim-sulfamethoxazole; TIM, ticarcillin-clavulanate; TEL, tellurite (nonantibiotic compound); TET, tetracycline. Classification of antibiotic resistance was done according to breakpoints published by the Clinical and Laboratory Standards Institute and is indicated as follows: susceptible, underlining; intermediate, italics; resistant, boldface (46). *, values for the components are separated by a slash.
Quality control.
Deletion of genomic adeR reveals an AdeABC-independent tigecycline resistance mechanism.
Tigecycline is considered to be a last-resort treatment regimen for infections with MDR/XDR A. baumannii (14). Resistance to tigecycline is mediated by the AdeABC efflux pump, which is controlled by the AdeRS two-component system (TCS) (27–31). This TCS was recently suggested as a putative drug target for small-molecule intervention to interfere with different antibiotic resistance pathways in A. baumannii (10). In order to evaluate the importance of the transcriptional regulator AdeR in mediating tigecycline resistance in clinical A. baumannii isolates, we customized our gene knockout platform (Fig. 1A) to produce genomic adeR deletions. We selected a diverse panel of 10 tigecycline-resistant clinical A. baumannii isolates using the following parameters: (i) their recent isolation from diverse geographic locations, (ii) their antibiotic resistance profile to first-, second-, and last-line drugs (carbapenems, colistin, and tigecycline, respectively), and (iii) their clonal lineage as determined by using MLST according to the Pasteur scheme (12). All strains were isolated between 2011 and 2013 (except for BV26, which was isolated in 1979) and classified as XDR according to the definition given by Magiorakos and colleagues (32). MLST analysis revealed that eight of the clinical isolates belonged to clonal cluster 2 (CC2), and two belonged to clonal cluster 1 (CC1), clusters that represent the most prominent worldwide epidemic clonal lineages (12, 13) (Table 1). Conjugation from E. coli was used to introduce the knockout plasmid into A. baumannii isolates. The plasmid conjugation/integration efficiency was measured as the number of transconjugants per receiver cell and was similar among different clinical isolates (1.8 × 10−7 ± 0.2 × 10−7 transconjugants/cell; n = 3) and one order of magnitude lower than for the reference strain ATCC-17978 (3.2 × 10−6 transconjugants/cell). We successfully created scarless adeR knockout (ΔadeR) mutants of these 10 XDR A. baumannii strains and also deleted adeR in the reference strain ATCC-17978. We characterized the tigecycline resistance profile of the ΔadeR mutants and their parental strains using the microdilution method. Deletion of adeR in 10 XDR A. baumannii strains reduced the MIC90 of tigecycline from 25 μg/ml to 3.1 μg/ml, demonstrating a significant shift of ΔadeR mutants toward tigecycline susceptibility (Table 2). Our data indicate that AdeR is important for A. baumannii to develop tigecycline resistance. However, in clinical practice the tigecycline susceptibility breakpoint for A. baumannii infections is ≤1 μg/ml (33). Our results demonstrated that 60% of the tested clinical isolates remained tigecycline nonsusceptible after adeR deletion and suggest that AdeR-regulated AdeABC-mediated tigecycline resistance is not the exclusive resistance mechanism rendering tigecycline inactive in these strains. We propose the existence of at least one AdeR-unrelated resistance mechanism that may be developed in A. baumannii clinical isolates to render tigecycline ineffective.
TABLE 2.
Tigecycline resistance profile and list of identified mutations in the clinical A. baumannii isolate panel and the corresponding ΔadeR strains
| Strain designation | Tigecycline MIC (μg/ml) |
AdeRS amino acid substitution(s) | Deletion in trm | |
|---|---|---|---|---|
| Wild type | ΔadeR strain | |||
| ATCC-17978 | 0.4 | 0.4 | Reference | |
| BV26 | 6.3 | 0.4 | AdeS E201K E237K | |
| BV94 | 25 | 3.1 | AdeS F170S | G1065 |
| BV173 | 6.3 | 0.8 | AdeR D21V D26N | A240 |
| BV175 | 3.1 | 0.8 | A240 | |
| BV185 | 6.3 | 3.1 | A240 | |
| BV186 | 3.1 | 3.1 | G1065 | |
| BV187 | 3.1 | 3.1 | G1065 | |
| BV189 | 6.3 | 3.1 | A240 | |
| BV190 | 6.3 | 0.8 | AdeS Q339K | T1050 |
| BV191 | 25 | 1.6 | AdeS G79S | A240 |
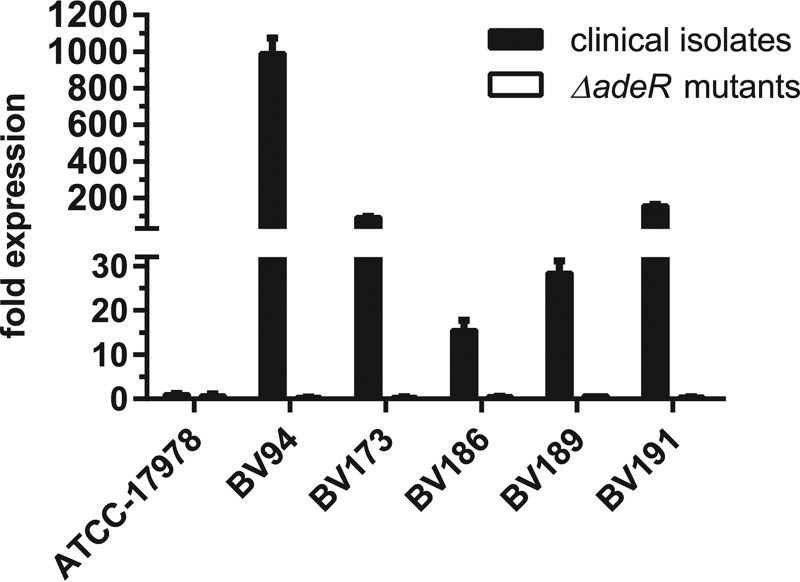
AdeR was previously shown to activate the expression of the RND (resistance-nodulation-cell division)-type AdeABC efflux pump and most likely controls the extrusion of tigecycline from the cells (31). However, the presence of an alternative activating mechanism for AdeABC mediated by the TCS BaeSR was recently suggested (34). In order to determine if overexpression of AdeABC was mediating tigecycline resistance despite the deletion of adeR, we quantified the expression level of adeB, which is the main component of the RND efflux pump. By using quantitative real-time PCR, we demonstrated general overexpression of adeB in all tested tigecycline-resistant clinical isolates (Fig. 2). The overexpression of the adeABC efflux pump correlated with elevated tigecycline MICs, as demonstrated by 990-fold and 160-fold overexpression levels of adeB in strains BV94 and BV191 with the highest tigecycline resistance levels, respectively. In contrast, all ΔadeR mutants lost the ability to express adeB, suggesting that AdeR is the main transcriptional activator of the AdeABC efflux pump. These data further indicate that an AdeR-unrelated tigecycline resistance mechanism in A. baumannii clinical isolates is not based on deregulated expression of the AdeABC efflux pump.
FIG 2.
Quantification of adeB expression levels in tigecycline-resistant A. baumannii clinical isolates and their ΔadeR mutants. The adeB expression levels of the individual strains were determined by quantitative real-time PCR and normalized to the adeB expression of the reference strain ATCC-17978.
To evaluate if the AdeABC efflux pump overexpression in tigecycline-resistant clinical isolates was triggered by mutations in the AdeRS TCS as previously described, we analyzed the genomic sequences of adeR and adeS (35). To distinguish between mutations involved in adeABC overexpression and those coming from clonal lineage evolution, we used as reference sequences the tigecycline-sensitive A. baumannii strains AYE and ACICU, which are representatives of clonal clusters CC1 and CC2, respectively (12). The only strain that contained mutations in the transcriptional regulator AdeR was BV173 (Table 2). One of the identified amino acid substitutions (D21V) in this strain was previously described to mediate adeABC overexpression (36). Mutations in the sensor kinase AdeS were much more abundant, and different amino acid substitutions were identified in four clinical isolates (Table 2). Interestingly, five tigecycline-resistant clinical isolates had wild-type AdeRS sequences without any amino acid substitutions. For these strains, we further analyzed the sequence of the intergenic regulatory region between adeRS and adeABC that encodes the AdeR DNA-binding site. However, we could not identify any mutations that could explain the overexpression of adeABC. These strains, with the exception of BV175, were not resensitized to tigecycline by the deletion of adeR (Table 2).
Taken together, these results indicate that the loss of adeR in XDR A. baumannii clinical isolates abolished the expression of adeB, leading to increased tigecycline sensitivity in 8 out of 10 ΔadeR mutants. However, 60% of the clinical isolates remained tigecycline nonsusceptible, with an MIC above the clinical susceptibility breakpoint of ≤1 μg/ml (33, 37). The requirement for AdeR to express the AdeABC efflux pump and the absence of mutation in four strains that were not resensitized to tigecycline indicate the presence of an alternative AdeR-AdeABC-independent tigecycline resistance pathway in these strains and disqualify AdeR as an adjuvant drug target.
Insights into alternative tigecycline resistance mechanisms.
To investigate putative AdeRS-AdeABC-unrelated tigecycline resistance mechanisms in A. baumannii, we developed artificial tigecycline-resistant strains by subjecting A. baumannii ATCC-17978 and its ΔadeR mutant (ATCC-17978ΔadeR) to liquid serial passage in the presence of increasing tigecycline concentrations (see Fig. S1 in the supplemental material). After 12 days (6 passages) we obtained wild-type and ΔadeR-derived strains able to grow at 6.4 μg/ml tigecycline and designated them ATCC-17978(TGC) and ATCC-17978ΔadeR(TGC), respectively. We analyzed the susceptibilities of these artificially evolved strains to various antibiotic classes. As expected, both strains were resistant to high tigecycline concentrations (see Table S2 in the supplemental material). In parallel with the tigecycline resistance, MICs of tetracycline increased to 32 μg/ml and 64 μg/ml, and MICs of ciprofloxacin increased to 2 μg/ml and 4 μg/ml for ATCC-17978(TGC) and ATCC-17978ΔadeR(TGC), respectively. In contrast, MICs to meropenem remained unchanged in both evolved strains. Both strains also showed a slight shift toward gentamicin susceptibility. To investigate if an alternative regulatory mechanism induced the adeABC expression, we analyzed the expression of adeB but could not detect an altered adeB expression pattern in the parental and evolved strains. This result was consistent with the presence of wild-type adeS and adeRS sequences in the evolved knockout and parental strains, respectively. In summary, our results suggest the presence of an AdeRS-unrelated tigecycline resistance mechanism leading to an increased resistance pattern to diverse antibiotics.
To unravel this new resistance mechanism, we sequenced the genome of the artificially evolved tigecycline-resistant strains ATCC-17978(TGC) and ATCC-17978ΔadeR(TGC) and compared the sequences with those of the parental strains. We identified the trm gene (A1S_2858) coding for a methyltransferase with unknown function as the only gene that was disrupted in both independently evolved strains (see Table S3 in the supplemental material). In ATCC-17978(TGC), we identified an insertion sequence (IS) of approximately 1 kb integrated into the trm gene at nucleotide position 747 (total trm length, 1,212 nucleotides [nt]). The IS shared 97% sequence identity with ISAba12, a transposon that is present in two copies in the genome of ATCC-17978 (22). In ATCC-17978ΔadeR(TGC), an adenine deletion at trm nucleotide 311 caused a frameshift leading to a premature stop codon. In conclusion, both strains expressed a truncated Trm variant that is most likely nonfunctional. We also identified several other mutated genes in the two independently evolved strains. However, these genes were mutated in one or the other strain and therefore cannot represent a general alternative tigecycline resistance mechanism (see Table S3). To evaluate the relevance of trm disruption in clinical strains, we sequenced the trm gene in our tigecycline-resistant A. baumannii strain panel. Except for BV26, all clinical isolates carried a nucleotide deletion in trm leading to an early stop codon and therefore a nonfunctional methyltransferase (Table 2). These data suggest that loss of Trm may potentially confer tigecycline resistance by using a pathway independent of the efflux pump AdeABC. To our surprise, three ΔadeR mutant strains (BV173, BV175, and BV190) contained a disrupted Trm variant but remained susceptible to tigecycline.
To characterize the role of trm disruption in tigecycline resistance we (i) deleted trm from the genome of the reference strain ATCC-17978, and (ii) we restored the functionality of chromosomal trm by reintroducing the deleted nucleotide A240 in clinical isolates. Loss of Trm in ATCC-17978 marginally increased tigecycline resistance whereas trm restoration in clinical isolates slightly decreased tigecycline resistance (see Table S4 in the supplemental material). Although these MIC shifts are considered in the standard deviation of the method, the consistency of the results among the strains suggests that loss of Trm plays a general role in tigecycline resistance in A. baumannii.
DISCUSSION
In contrast to the use of conventional antibiotics that target conserved essential mechanisms, the adjuvant approach aims to modulate nonessential mechanisms, which may vary significantly within the same species. Consequently, the identification of new drug targets for adjuvant therapy requires extensive validation in diverse clinical strains to ensure target relevance. Unfortunately, methods to manipulate these drug-resistant strains are lacking, rendering drug target validation difficult. Recently, a method based on the recombineering technology has been developed to produce gene deletion mutants in A. baumannii (11). This method requires the plasmid-encoded expression of a recombinase to enhance the efficiency of homologous recombination and electroporation to incorporate foreign DNA into the cells (11). Unfortunately, clone selection is performed with two different antibiotic resistance cassettes, and electroporation may be very inefficient in clinical isolates, which prevents an adaptation of this method for the manipulation of drug-resistant strains. Moreover, the recombinase expression vector has to be cured from the cells to allow the subsequent introduction of another vector that expresses the FLP recombinase, which once again has to be cured to finally produce markerless mutants (11). Overall, this recombineering-based method is a time-consuming procedure and is incompatible with multidrug-resistant isolates.
Amin and colleagues previously developed a two-step method to construct scarless knockout mutants in MDR A. baumannii by implementation of a tellurite resistance operon into their knockout plasmids that allowed targeting A. baumannii isolates regardless of their antibiotic resistance profiles (17). The applied operon used as selection marker consisted of three consecutive genes (kilA, telA, and telB) carried by a 3-kb DNA sequence. Its expression requires an enormous metabolic investment and confers resistance to 30 μg/ml tellurite (17). A sacB cassette was further used to select for the second recombination event and plasmid removal from the genome. Several sucrose passages were required for successful plasmid removal, indicating the inefficiency of this counterselection cassette (17). SacB is also prone to accumulate inactivation mutations due to its slight cytotoxicity even in the absence of sucrose, making the use of this counterselection marker disadvantageous in A. baumannii (25).
Here, we report the design of a highly efficient knockout platform for A. baumannii isolates, regardless of their antibiotic resistance profiles. The knockout platform takes advantage of an efficient thiopurine S-methyltransferase encoded by a single tpm gene of Acinetobacter baylyi (Fig. 1A). This resistance marker confers resistance to ≥100 μg/ml tellurite and requires a low fitness investment from the cells. We combined this selection marker with an inducible variant of an E. coli-derived thymidine kinase (tdk) to select for the second recombination event, allowing plasmid removal with an efficiency of >95% in A. baumannii isolates in a one-step counterselection passage on AZT. The developed knockout technology enables us to perform markerless gene deletions in A. baumannii clinical and laboratory strains in only 3 days, with the capability for repetitive application of the process to delete multiple genes from the same genome. The platform is suitable for a multitude of genome engineering techniques in drug-resistant bacteria. Besides producing targeted gene knockouts, we used this technology platform to do gene disruptions and targeted chromosomal insertions (knock-ins) of single nucleotides (point mutation) as well as longer DNA sequences to produce conditional knockout mutants in A. baumannii. Moreover, this platform has a modular architecture and allows the exchange of the individual genetic building blocks (e.g., origin of replication, selection marker, or counterselection cassette) by digestion/ligation reactions, enabling the potential for customization to target other pathogens (Fig. 1A). We successfully employed a modified version of the knockout platform to produce markerless gene deletions in MDR P. aeruginosa, highlighting the potential for expansion of the platform to other pathogenic species (data will be published elsewhere).
In a case study we demonstrated the efficient deletion of adeR from the genome of diverse XDR A. baumannii clinical isolates. We characterized these isolates with an antibiotic panel that was chosen to discriminate between non-MDR, MDR, and XDR strains and found that most of the strains were resistant to all tested antibiotics (Table 1) (32). Our data suggest that some of the strains may be even classified as PDR; however, further analyses are required to finalize this classification.
The analysis of AdeR as a potential adjuvant drug target confirmed the importance of AdeR in mediating tigecycline resistance in A. baumannii. Our data further suggest that there is at least one AdeR-unrelated mechanism that confers moderate resistance to tigecycline in A. baumannii clinical isolates (Table 2). The U.S. Food and Drug Administration (FDA) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) do not provide breakpoints for A. baumannii treatment with tigecycline. However, in clinical practice the susceptibility breakpoint of ≤1 μg/ml is commonly accepted (33, 37).
Because in the absence of AdeR the expression of adeB was abolished, this alternative resistance pathway is unrelated to AdeABC-mediated drug efflux (Fig. 2). It was recently shown that the TCS BaeSR may trigger the activation of adeABC expression in A. baumannii ATCC-17978 (34). Our results indicate that the BaeSR regulation of adeABC expression may occur through AdeR because the loss of adeR completely abolished adeABC expression under the tested conditions. This observation suggests a cross talk between both TCSs by either BaeS-mediated phosphorylation of AdeR or specific activation of adeR expression by BaeR. Further experiments are required to clarify a putative dynamic interaction between AdeRS and BaeSR.
Sequencing of adeRS in our strain panel revealed that five A. baumannii isolates contained mutations in AdeRS (Table 2). We found one strain with mutated AdeR, but four strains with alterations in the sequence of AdeS, suggesting that the sensor kinase is more prone to mutations, leading to an overexpression of AdeABC. With the exception of the D21V substitution mutation in AdeR of strain BV173, the identified mutations have not been previously described (27, 31, 35, 36, 38, 39). To our surprise, the tested XDR strain panel contained four tigecycline-resistant isolates with wild-type AdeRS that were not resensitized for tigecycline by deletion of adeR (Table 2). This finding highlights the presence of an alternative AdeABC-independent resistance mechanism mediating tigecycline protection in A. baumannii. The hypothesis of an alternative resistance pathway was further supported by the successful in vitro development of tigecycline resistance in ATCC-17978ΔadeR.
Whole-genome sequencing of artificially in vitro-evolved tigecycline-resistant strains identified the trm gene, coding for a methyltransferase, as the only gene that was disrupted in both independently evolved strains (see Table S3 in the supplemental material). Chen and colleagues recently developed artificial tigecycline resistance in A. baumannii ATCC-19606 and also found a mutated trm gene, suggesting an important role of this methyltransferase in conferring moderate tigecycline resistance (40). Methyltransferases are able to catalyze the methylation of ribosomal RNAs (rRNAs). The conformational modification of the rRNAs, which are the molecular targets of tigecycline, may result in an alteration of the antibiotic target interaction, suggesting that trm disruption could be a valid tigecycline resistance mechanism (41). We also sequenced the trm gene of our XDR strain panel and found in 9 out of 10 clinical isolates a disrupted trm gene (Table 2). Interestingly, the isolate that harbors a wild-type trm (BV26) was isolated in 1979, almost 25 years before tigecycline was approved by the FDA. Our finding suggests that in contrast to AdeABC efflux pump overexpression, which can be selected under pressure of different antibiotic classes, trm disruption may be a specific result of tigecycline selection pressure in clinical settings. We demonstrated that the disruption of trm plays only a moderate role in tigecycline resistance in clinical isolates, suggesting that loss of Trm may not be the only trigger for an alternative AdeABC-independent tigecycline resistance pathway.
Besides trm, we found other genes that were mutated in one of the two independently evolved strains (see Table S3 in the supplemental material) and that were previously linked to tigecycline resistance in A. baumannii, such as rpsJ (S10) or adeN (42–45). However, to our knowledge the identified mutations in the RNase E 23S rRNA pseudouridylate synthase and in the promoter of the MATE family efflux pump AbeM (see Table S3) were never linked to tigecycline resistance, and further studies are required to verify their impact on tigecycline resistance.
Since the development of our gene knockout platform enables efficient and quick genetic manipulation of A. baumannii clinical isolates regardless of their resistance profiles, this technology is a powerful tool to investigate the molecular nature of different pathways that allow A. baumannii to escape antibiotic-mediated killing. By using AdeR as an example, our study revealed the genetic variability of different tigecycline-resistant isolates, which may be a cause of the different selection pressures applied during patient treatment in clinical units, clearly demonstrating that the study of one model strain is not sufficient to obtain a global picture of species-related pathogenesis and antibacterial resistance.
Supplementary Material
ACKNOWLEDGMENT
We acknowledge funding for V.T. from the Marie Curie Initial Training Network ITN-2013-607694-Translocation.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01275-16.
REFERENCES
- 1.Pendleton JN, Gorman SP, Gilmore BF. 2013. Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther 11:297–308. doi: 10.1586/eri.13.12. [DOI] [PubMed] [Google Scholar]
- 2.Dijkshoorn L, Nemec A, Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol 5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 3.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Visca P, Seifert H, Towner KJ. 2011. Acinetobacter infection—an emerging threat to human health. IUBMB Life 63:1048–1054. doi: 10.1002/iub.534. [DOI] [PubMed] [Google Scholar]
- 5.Paterson DL, Harris PNA. 2015. Editorial commentary: the new Acinetobacter equation: hypervirulence plus antibiotic resistance equals big trouble. Clin Infect Dis 61:155–156. doi: 10.1093/cid/civ227. [DOI] [PubMed] [Google Scholar]
- 6.Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. 2015. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 7.Andersson DI, Hughes D. 2010. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol 8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 8.Depardieu F, Podglajen I, Leclercq R, Collatz E, Courvalin P. 2007. Modes and modulations of antibiotic resistance gene expression. Clin Microbiol Rev 20:79–114. doi: 10.1128/CMR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pieren M, Tigges M. 2012. Adjuvant strategies for potentiation of antibiotics to overcome antimicrobial resistance. Curr Opin Pharmacol 12:551–555. doi: 10.1016/j.coph.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 10.Worthington RJ, Blackledge MS, Melander C. 2013. Small-molecule inhibition of bacterial two-component systems to combat antibiotic resistance and virulence. Future Med Chem 5:1265–1284. doi: 10.4155/fmc.13.58. [DOI] [PubMed] [Google Scholar]
- 11.Tucker AT, Nowicki EM, Boll JM, Knauf GA, Burdis NC, Trent MS, Davies BW. 2014. Defining gene-phenotype relationships in Acinetobacter baumannii through one-step chromosomal gene inactivation. mBio 5:e01313-14. doi: 10.1128/mBio.01313-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. 2010. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. doi: 10.1371/journal.pone.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zarrilli R, Pournaras S, Giannouli M, Tsakris A. 2013. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents 41:11–19. doi: 10.1016/j.ijantimicag.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Peleg AY, Hooper DC. 2010. Hospital-acquired infections due to Gram-negative bacteria. N Engl J Med 362:1804–1813. doi: 10.1056/NEJMra0904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 9th ed CLSI document M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 16.Silva-Rocha R, Martínez-García E, Calles B, Chavarría M, Arce-Rodríguez A, de las Heras A, Páez-Espino AD, Durante-Rodríguez G, Kim J, Nikel PI, Platero R, de Lorenzo V. 2013. The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res 41:D666–D675. doi: 10.1093/nar/gks1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amin IM, Richmond GE, Sen P, Koh TH, Piddock LJ, Chua KL. 2013. A method for generating marker-less gene deletions in multidrug-resistant Acinetobacter baumannii. BMC Microbiol 13:158. doi: 10.1186/1471-2180-13-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrières L, Hémery G, Nham T, Guérout A-M, Mazel D, Beloin C, Ghigo J-M. 2010. Silent mischief: bacteriophage Mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery. J Bacteriol 192:6418–6427. doi: 10.1128/JB.00621-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu S-M, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam T-W, Wang J. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nowak J, Seifert H, Higgins P. 2013. The tellurite-resistance determinant Tpm of the Acinetobacter baylyi strain ADP1 as a useful non-antibiotic selection marker for genetic manipulation in Acinetobacter baumannii, abstr p1356 Abstr 23rd Eur Soc Clin Microbiol Infect Dis Conf, Berlin, Germany, 27 to 30 April 2013. [Google Scholar]
- 24.Prigent-Combaret C, Sanguin H, Champier L, Bertrand C, Monnez C, Colinon C, Blaha D, Ghigo J-M, Cournoyer B. 2012. The bacterial thiopurine methyltransferase tellurite resistance process is highly dependent upon aggregation properties and oxidative stress response. Environ Microbiol 14:2645–2660. doi: 10.1111/j.1462-2920.2012.02802.x. [DOI] [PubMed] [Google Scholar]
- 25.Wang H, Bian X, Xia L, Ding X, Müller R, Zhang Y, Fu J, Stewart AF. 2014. Improved seamless mutagenesis by recombineering using ccdB for counterselection. Nucleic Acids Res 42:e37. doi: 10.1093/nar/gkt1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holzer GW, Mayrhofer J, Gritschenberger W, Falkner FG. 2005. Dominant negative selection of vaccinia virus using a thymidine kinase/thymidylate kinase fusion gene and the prodrug azidothymidine. Virology 337:235–241. doi: 10.1016/j.virol.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 27.Hornsey M, Ellington MJ, Doumith M, Thomas CP, Gordon NC, Wareham DW, Quinn J, Lolans K, Livermore DM, Woodford N. 2010. AdeABC-mediated efflux and tigecycline MICs for epidemic clones of Acinetobacter baumannii. J Antimicrob Chemother 65:1589–1593. doi: 10.1093/jac/dkq218. [DOI] [PubMed] [Google Scholar]
- 28.Ruzin A, Keeney D, Bradford PA. 2007. AdeABC multidrug efflux pump is associated with decreased susceptibility to tigecycline in Acinetobacter calcoaceticus-Acinetobacter baumannii complex. J Antimicrob Chemother 59:1001–1004. doi: 10.1093/jac/dkm058. [DOI] [PubMed] [Google Scholar]
- 29.Sun J-R, Chan M-C, Chang T-Y, Wang W-Y, Chiueh T-S. 2010. Overexpression of the adeB gene in clinical isolates of tigecycline-nonsusceptible Acinetobacter baumannii without insertion mutations in adeRS. Antimicrob Agents Chemother 54:4934–4938. doi: 10.1128/AAC.00414-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peleg AY, Adams J, Paterson DL. 2007. Tigecycline efflux as a mechanism for nonsusceptibility in Acinetobacter baumannii. Antimicrob Agents Chemother 51:2065–2069. doi: 10.1128/AAC.01198-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchand I, Damier-Piolle L, Courvalin P, Lambert T. 2004. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob Agents Chemother 48:3298–3304. doi: 10.1128/AAC.48.9.3298-3304.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 33.Garnacho-Montero J, Dimopoulos G, Poulakou G, Akova M, Cisneros JM, De Waele J, Petrosillo N, Seifert H, Timsit JF, Vila J, Zahar J-R, Bassetti M. 2015. Task force on management and prevention of Acinetobacter baumannii infections in the ICU. Intensive Care Med 41:2057–2075. doi: 10.1007/s00134-015-4079-4. [DOI] [PubMed] [Google Scholar]
- 34.Lin M-F, Lin Y-Y, Yeh H-W, Lan C-Y. 2014. Role of the BaeSR two-component system in the regulation of Acinetobacter baumannii adeAB genes and its correlation with tigecycline susceptibility. BMC Microbiol 14:119. doi: 10.1186/1471-2180-14-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoon E-J, Courvalin P, Grillot-Courvalin C. 2013. RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii: major role for AdeABC overexpression and AdeRS mutations. Antimicrob Agents Chemother 57:2989–2995. doi: 10.1128/AAC.02556-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Higgins PG, Schneiders T, Hamprecht A, Seifert H. 2010. In vivo selection of a missense mutation in adeR and conversion of the novel blaOXA-164 gene into blaOXA-58 in carbapenem-resistant Acinetobacter baumannii isolates from a hospitalized patient. Antimicrob Agents Chemother 54:5021–5027. doi: 10.1128/AAC.00598-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brink AJ, Bizos D, Boffard KD, Feldman C, Grolman DC, Pretorius J, Richards GA, Senekal M, Steyn E, Welkovic N, Association of Surgeons of South Africa, Critical Care Society of Southern Africa, Federation of Infectious Diseases Societies of Southern Africa, South African Thoracic Society, Trauma Society of South Africa . 2010. Guideline: appropriate use of tigecycline. S Afr Med J 100:388–394. doi: 10.7196/SAMJ.4109. [DOI] [PubMed] [Google Scholar]
- 38.Rumbo C, Gato E, López M, Ruiz de Alegría C, Fernández-Cuenca F, Martínez-Martínez L, Vila J, Pachón J, Cisneros JM, Rodríguez-Baño J, Pascual A, Bou G, Tomás M, Spanish Group of Nosocomial Infections and Mechanisms of Action and Resistance to Antimicrobials (GEIH-GEMARA), Spanish Society of Clinical Microbiology and Infectious Diseases (SEIMC), Spanish Network for Research in Infectious Diseases (REIPI). 2013. Contribution of efflux pumps, porins, and β-lactamases to multidrug resistance in clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 57:5247–5257. doi: 10.1128/AAC.00730-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coyne S, Courvalin P, Périchon B. 2011. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother 55:947–953. doi: 10.1128/AAC.01388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Q, Li X, Zhou H, Jiang Y, Chen Y, Hua X, Yu Y. 2014. Decreased susceptibility to tigecycline in Acinetobacter baumannii mediated by a mutation in trm encoding SAM-dependent methyltransferase. J Antimicrob Chemother 69:72–76. doi: 10.1093/jac/dkt319. [DOI] [PubMed] [Google Scholar]
- 41.Morić I, Savić M, Ilić-Tomić T, Vojnović S, Bajkić S, Vasiljević B. 2010. rRNA methyltransferases and their role in resistance to antibiotics. J Med Biochem 29:165–174. [Google Scholar]
- 42.Beabout K, Hammerstrom TG, Perez AM, Magalhães B, de Prater FAG, Clements TP, Arias CA, Saxer G, Shamoo Y. 2015. The ribosomal S10 protein is a general target for decreased tigecycline susceptibility. Antimicrob Agents Chemother 59:5561–5566. doi: 10.1128/AAC.00547-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenfeld N, Bouchier C, Courvalin P, Périchon B. 2012. Expression of the resistance-nodulation-cell division pump AdeIJK in Acinetobacter baumannii is regulated by AdeN, a TetR-type regulator. Antimicrob Agents Chemother 56:2504–2510. doi: 10.1128/AAC.06422-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Damier-Piolle L, Magnet S, Brémont S, Lambert T, Courvalin P. 2008. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob Agents Chemother 52:557–562. doi: 10.1128/AAC.00732-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoon E-J, Chabane YN, Goussard S, Snesrud E, Courvalin P, Dé E, Grillot-Courvalin C. 2015. Contribution of resistance-nodulation-cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii. mBio 6:e00309-15. doi: 10.1128/mBio.00309-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; 24th informational supplement. CLSI document M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.